Abstract
For hazard identification and classification and labeling purposes, animal testing guidelines are required by law to evaluate developmental toxicity for new and existing chemical products. However, guideline developmental toxicity studies are costly, time-consuming, and require many laboratory animals. Computational modeling has emerged as a promising, animal-sparing, and cost-effective method for evaluating the developmental toxicity potential of chemicals, such as endocrine disruptors, without the use of animals. We aimed to develop a predictive and explainable computational model for developmental toxicants. To this end, a comprehensive dataset of 1,244 chemicals with developmental toxicity classifications was curated from public repositories and literature sources. Data from 2,140 toxicological high throughput screening (HTS) assays were extracted from PubChem and the ToxCast program for this dataset and combined with information about 834 chemical fragments to group assays based on their chemical-mechanistic relationships. This effort revealed two assay clusters containing 83 and 76 assays, respectively, with high positive predictive rates for developmental toxicants identified with animal testing guidelines (PPV = 72.4% and 77.3% during cross-validation). These two assay clusters can be used as developmental toxicity models and were applied to predict new chemicals for external validation. This study provides a new strategy for constructing alternative chemical developmental toxicity evaluations that can be replicated for other toxicity modeling studies.
Keywords: Big data, Chemical fragments, Developmental toxicity, High-throughput screening data, Read-across
Introduction
Traditional chemical toxicity evaluation methods rely on animal testing guidelines for hazard identification. These animal studies are required by law and are considered the gold standard for safety assessment testing. However, these studies are expensive, time-consuming, and require highly specialized study designs to detect developmental toxicants1,2. Furthermore, these methods raise ethical concerns because of the many laboratory animals required. The European Registration, Evaluation, Authorization, and Restriction of Chemicals (REACH) regulations have extensive developmental and reproductive toxicity (DART) testing requirements for industrial and consumer chemicals3,4. DART testing represented 90% of animal use and 70% of chemical toxicity testing costs associated with completing phase one of REACH3, with individual testing protocols sometimes requiring up to 3,200 animals per chemical4. One of the most well-known regulatory testing requirements for this purpose is the prenatal developmental toxicity study, described as the Organization for Economic Co-operation and Development (OECD) Test No. 4145 and the United States Environmental Protection Agency (US EPA) Office of Prevention, Pesticides, and Toxic Substances (OPPTS) test guideline 870.37006. Briefly, this protocol requires the administration of a test chemical to 4 groups of animals (e.g., at least 20 pregnant rats or rabbits with litters per group) at three different concentrations from the time of implantation of the embryo in utero and continuing throughout pregnancy until one dayterm before the expected day of delivery. A comprehensive array of developmental endpoints is defined based on the testing results, fetal body weight, embryo-fetal survival, fetal morphology (external, visceral, skeletal), and endocrine endpoints, to determine No Observed Adverse Effect Levels (NOAELs) or Low Observed Adverse Effect Levels (LOAELs) values for maternal and developmental toxicity effects.
The complexity and cost of the associated animal testing guidelines for prenatal developmental toxicity have produced a critical need to develop alternative approaches based on non-animal models, such as computational models. However, most current computational toxicology models for developmental toxicity predictions were developed using Quantitative Structure-Activity Relationship (QSAR) modeling and other structure-based approaches. These approaches only incorporated structural and physicochemical information of known toxicants7–13. Unfortunately, for complex in vivo endpoints such as developmental toxicity, relying on structural and physicochemical information alone for modeling and evaluations can be error-prone14,15. For example, compounds with similar structures may exhibit dissimilar toxicities, a phenomenon called activity cliffs, and cause incorrect predictions for new compounds16,17. Furthermore, international guidance regarding the chemical risk assessments requires that the toxicity predictions of new compounds need to have identified toxicity mechanisms18–20.
Over the past twenty years, advances in high-throughput screening (HTS) protocols and combinatorial chemistry revolutionized the environmental and health science data landscape21–23. Initiatives such as the US EPA’s Toxicity Forecaster (ToxCast) program, which screened approximately 1,800 chemicals in over 700 HTS assays, generated large amounts of biological data for mechanistic toxicity evaluations24,25. At the same time, a collaboration among the US EPA, National Center for Advancing Translational Sciences (NCATS), and the National Toxicology Program (NTP) launched a parallel initiative called Toxicity in the 21st Century (Tox21). The goal of Tox21 was to generate more detailed toxicity data, such as concentration-responses, for a larger chemical library using quantitative HTS protocols. The collaborative Tox21 initiative is ongoing, now including the Food and Drug Administration (FDA) and working toward a goal of testing about 10,000 chemicals in about 70 HTS assays26–29. Public databases such as PubChem host much of this data, making it available for modeling studies30,31. Integrating biological data into computational toxicity evaluations has shown great promise in addressing the backlog of registered chemicals that have not undergone complete safety assessments32–40. However, these models only incorporated manually selected biological data covering a well-known and narrow scope of possible mechanisms relevant to developmental toxicity38–41. Automatic data mining methods that can extract relevant public biological data offer a new strategy to shed light on unknown toxicity mechanisms that are not incorporated into the existing models23.
This study aimed to address the above limitations of the existing computational approaches for evaluating developmental toxicity using a data-driven read-across approach by combining chemical information and biological data (Figure 1). To this end, a large dataset for developmental toxicity was collected from public databases and literature sources. An automated data mining web tool was then used to extract biological data from PubChem for all of the chemicals in this dataset42. Additional biological data generated as part of the ToxCast initiative for these chemicals were collected from the US EPA’s CompTox Chemistry Dashboard43. Then, the collected PubChem and ToxCast assays were clustered based on their biological mechanisms using a recently developed method that incorporates the structural features of the chemicals in the developmental toxicity dataset33. This process revealed chemical in vitro-in vivo relationships used to reveal suites of assays representing developmental toxicity mechanisms. The assays with extra chemical information can be used as models to screen new compounds to prioritize potential toxicants.
Figure 1:
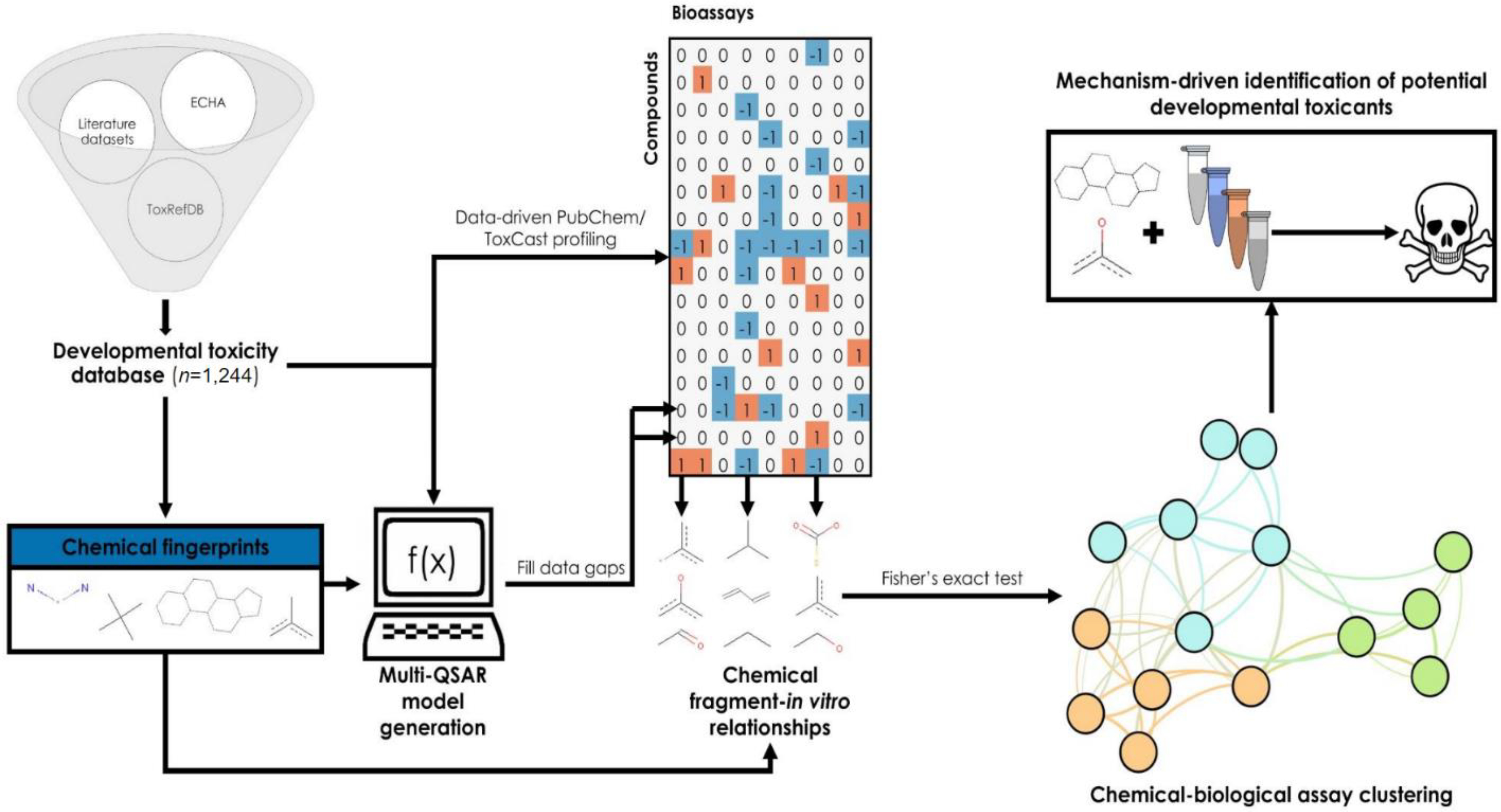
Overview of the workflow used in this study. The data-driven workflow employed in this study consists of four main stages: generation of chemical fingerprints and retrieval of in vitro bioassay data for each compound in the training set, implementation of QSAR models to fill data gaps, clustering PubChem and ToxCast assays based on correlations between chemical fragments and bioactivity, and identification of potential developmental toxicants by mechanism-driven read-across. Orange cells in the bioprofile with a value of 1 represent active results, gray cells with a value of 0 represent inconclusive/missing results, and blue cells with a value of −1 represent inactive results.
Methods
In Vivo Prenatal Developmental Toxicity Database
Chemicals with prenatal developmental toxicity data were collected from public database resources44–46 and individual datasets in the literature7. The definition of developmental toxicant was limited to adverse effects selective to the developing embryo or fetus after pregnant animal exposure. Therefore, non-toxicants in this study were limited to chemicals showing no adverse effects in the developing embryo or fetus in prenatal testing or only showing non-selective embryo-fetal effects (i.e., secondary effects from toxicity in the pregnant test animal).
The European Chemical Hazards Agency (ECHA) and Toxicity Reference Database (ToxRefDB)44,45 datasets consisted of 529 and 156 chemicals, respectively, with experimental data generated by test protocols similar to OECD 4145/OPPTS 870.37006. These datasets contained NOAEL values associated with prenatal developmental toxicity testing of chemicals administered to preclinical mammalian species (e.g., dogs, monkeys, rabbits, and rodents) orally or by inhalation. First, results marked as unreliable or unacceptable by ECHA or the EPA were removed from the datasets. Then, the NOAEL values were converted to developmentally toxic and nontoxic classifications. Chemicals with oral NOAEL values below the OECD 414/OPPTS 870.3700 limit dose of 1,000 mg/kg/day and inhalation NOAEL values below the OPPTS 798.4350 limit dose of 5,100 mg/m3 were classified as developmentally toxic. Because this study focuses on identifying selective prenatal developmental toxicants, chemicals were classified as developmentally toxic only if they had lower developmental NOAEL values than pregnant test animal NOAEL values derived from the same prenatal study. In some cases, chemicals had study results available in more than one species. Where the presence of developmental effects varied across species for a chemical, the chemical was conservatively considered toxic if it exhibited selective prenatal developmental toxicity in at least one species.
The Proctor & Gamble7 dataset consisted of 637 compounds with primarily mammalian data. Chemicals were classified into five categories: D (developmental toxicity in the absence of maternal toxicity), D(MT) (developmental toxicity only in the presence of maternal toxicity), DTer (teratogenicity in the absence of maternal toxicity), DTer(MT) (teratogenicity in the presence of maternal toxicity), and No Evidence (no developmental toxicity observed)7. These categories were derived using similar criteria to those used to classify the ECHA and ToxRefDB chemicals (i.e., chemicals classified as D(MT) and DTer(MT) showed developmental or teratogenic NOAEL below the pregnant test animal NOAEL). Therefore, chemicals with the D and DTer classifications were considered developmentally toxic, and those with all other categories were considered nontoxic, consistent with this study’s selective developmental toxicity focus.
Chemical structure information was collected for the chemicals from each dataset using PubChem’s identifier exchange service47 using their CAS number, European Community (EC) number, common name, or Distributed Structure-Searchable Toxicity (DSSTox) identifiers as input48. The CASE Ultra v1.8.0.0 DataKurator tool curated the individual datasets by standardizing their chemical structures. Salts were treated as their corresponding organic acids. Positively charged nitrogen atoms were neutralized by subtracting hydrogens, and all negatively charged atoms were neutralized by adding hydrogens. After these curation steps, the datasets were merged. In some cases, chemicals with duplicate curated structures showed conflicting designations across the component datasets, and the most conservative designation (i.e., developmentally toxic) was kept. The fully compiled in-house prenatal developmental toxicity database was used as the training set in this study and contained 1,244 unique chemicals, of which 660 were active, and 584 were inactive (Supplementary Table 1).
In Vitro Data Collection and Bioassay Clustering
A bioprofile was generated for the chemicals in the developmental toxicity database using public in vitro bioassay hit calls. PubChem bioassay hit calls were automatically collected for these chemicals using our in-house Chemical In Vitro-In Vivo Profiling tool (http://ciipro.rutgers.edu/)37,42. ToxCast hit calls were retrieved from the EPA ToxCast/Tox21 summary files for invitroDBv3.243,49. Bioassays with at least five active responses among the training set chemicals were retained for the final bioprofile to reduce the chances of overfitting due to limited data33.
These assays were then clustered into mechanistically related groups using statistically significant relationships among active results and the presence of specific chemical fragments using a recently published workflow33. First, Saagar fingerprints were generated for all training set chemicals50. The Saagar fingerprints consist of binary vectors denoting the presence or absence of approximately 1,000 chemical fragments for chemical toxicity studies50. Next, each substructure was compared pairwise with each bioassay using Fisher’s exact test to determine the statistical significance of the fragment’s presence in a chemical with its activity in the bioassay. The Fisher’s exact test’s outcome is a p-value, which indicates the probability of a chemical fragment-in vitro bioassay response relationship existing by random chance. The relationship between the existence of a chemical fragment in an assay dataset and these chemicals’ active responses was considered statistically significant if its corresponding p-value was less than 0.05 (i.e., a random-chance probability of less than 5%). Compounds showing active results in mechanistically related bioassays are likely to have the same key chemical fragments. Therefore, the bioassays can be clustered based on their statistically significant relationships to chemical fragments.
Biological Read-Across to Determine Developmental Toxicity Potential
Each mechanistically related cluster of bioassays was used to perform read-across to assess the chemical developmental toxicity using a biosimilarity search (Figure 1). Biosimilarity refers to the similarity between patterns of results in a battery of bioassays, such that chemicals showing similar patterns have higher biosimilarity42,51. A frequent challenge associated with using HTS screening data for toxicity evaluations is the bias toward inactive results, which is confounded by the relatively lower importance of inactive responses than active responses22,23. Therefore, a biosimilarity search assigns a lower weight to inactive responses to minimize the impact of this bias (Equation 1)42. In this equation, Aa and Ba represent the active responses in two bioassays within the same cluster, and Ai and Bi represent their inactive responses. In addition, the parameter w, which equals the ratio of active to inactive responses present among the bioassays in each cluster, lessens the impact of the bias usually present toward inactive responses.
| (1) |
However, missing data are common when profiling target compounds against public data22,23. Therefore, a high biosimilarity value does not always ensure a confident prediction. For example, the value may have been generated by comparing two chemicals with only one shared active bioassay response among many inactive and missing results. For this reason, an additional confidence parameter was calculated for each read-across prediction, representing the richness of assay data used to make the prediction and, therefore, indicates the prediction reliability (Equation 2).
| (2) |
Five-fold cross-validation was performed for each bioassay cluster to assess its ability to predict developmental toxicity potentials for new compounds. First, the target chemicals (i.e., the compounds in the developmental toxicity modeling set) tested in at least one bioassay within a cluster were randomly divided into five equally sized groups. Then, in each of five iterations, each chemical in one group was compared to the compounds of the other four groups, and its developmental toxicity was predicted by its most biosimilar neighbor. This procedure optimized the minimum biosimilarity between two compounds and associated confidence for each cluster using an exhaustive grid-search algorithm implemented in scikit-learn v0.24.152. Briefly, read-across predictions were made using each combination of parameters to identify the best-performing conditions, which were retained and then used to predict external compounds. When multiple bioassays within the same cluster contained at least ten mutual responses across the represented chemicals and had a Pearson correlation coefficient greater than 0.9, one was selected at random and retained for read-across predictions. The others were removed to reduce inflation of biosimilarity values by bioassays that may be represented more than once in the dataset (e.g., a bioassay deposited into PubChem twice with slightly different interpretations).
The predictions resulting from the cross-validation procedure were statistically evaluated using various universal parameters, including specificity (Equation 3), sensitivity (Equation 4), Correct Classification Ratio (CCR, Equation 5), and positive predictive value (PPV, Equation 6). The number of true positives (TP) represents correctly predicted prenatal developmental toxicants in each equation. The number of false positives (FP) represents nontoxic chemicals incorrectly predicted as prenatal developmental toxicants. The number of true negatives (TN) represents correctly predicted nontoxic chemicals. The number of false negatives (FN) represents prenatal developmental toxicants incorrectly predicted as nontoxic. Because toxic predictions are more meaningful than nontoxic predictions, only the PPV was used during each cluster’s read-across parameter optimization.
| (3) |
| (4) |
| (5) |
| (6) |
Quantitative Structure-Activity Relationship Modeling to Fill Data Gaps
Read-across predictions are only reliable if their associated confidence value is above a minimum threshold. Therefore, chemicals with substantial amounts of missing bioassay data cannot be predicted without filling these gaps. This data gap filling was accomplished using QSAR modeling, similar to previous studies33,41. First, all the chemicals tested in a given bioassay were retrieved along with their structural information and activities. This information was collected by querying the PUG-REST web service47 for PubChem bioassays or invitroDBv3.243,49 for ToxCast bioassays. After removing inconclusive results, each bioassay’s dataset was balanced by randomly removing inactive chemicals until reaching an equal number of active and inactive chemicals. For large bioassay datasets, the number of chemicals used for QSAR model development was limited to 10,000 to save computational time.
Four machine-learning algorithms were then used to develop the QSAR models for each assay in the target clusters: Bernoulli naïve Bayes (BNB), k-nearest neighbors (kNN), random forest (RF), and support vector machines (SVM). Each algorithm was implemented using the Python machine-learning library scikit-learn v0.19.0 (http://scikit-learn.org/)52 using the RDKit v2019.09.1.0 (http://rdkit.org/) implementation of Functional Connectivity FingerPrints (FCFPs) using a bond radius of 353. Finally, the models’ predictive performances were evaluated using the CCR from a five-fold cross-validation procedure (Equation 6).
Each of the four algorithms’ hyperparameters was previously described in detail54, along with the procedure used to optimize them during model training53,54. BNB models “naively” assume that the presence or absence of each FCFP descriptor is independent of all others to calculate the probability that a chemical will be active in a particular bioassay by applying Bayes’ Theorem55,56. kNN models predict a new chemical’s activity by a majority vote of its k most similar training set chemicals57. RF models build decision trees based on randomly selected FCFP descriptors and average their outputs to predict new chemicals’ activities58. Finally, SVM models optimize a set of thresholds for each descriptor used in model development that best distinguishes between active and inactive training chemicals59.
The predictions generated by each of the four algorithms for a single chemical were averaged to form a consensus prediction. Consensus predictions are robust because they leverage the strengths of various algorithms and have shown advantages for predicting new compounds in previous studies53,60–63. A chemical similarity-based applicability domain was also implemented by only reporting predictions for chemicals with a minimum of 40% similarity using FCFPs across their three nearest neighbors among the QSAR model training set for a particular assay. However, the consensus predictions showed poor predictive performance despite these precautions in rare cases (i.e., CCR < 0.6). In these cases, missing data were not populated with QSAR predictions.
Results
Prenatal Developmental Toxicity Database Overview
The prenatal developmental toxicity database used in this study consisted of data generated by regulatory animal test guidelines (i.e., OECD 414/OPPTS 870.3700) or guideline-like protocols compiled from the literature7 and public databases44. Because the sources had different data types (e.g., study-derived NOAELs versus data sorted into categories), original data were harmonized into two groups for model development. In the final database, chemicals labeled as toxic were selective embryo-fetal toxicants associated with adverse effects on developing embryos and fetuses. Chemicals labeled as nontoxic showed no adverse developmental effects or only showed adverse effects in the presence of pregnant animal toxicity. Among the three data sources, 68 chemicals existed more than once. Twenty-six (38.2%) of these chemicals had conflicting toxicity classifications across the data sources. In these circumstances, the most conservative classification was retained, such that any chemical with at least one result indicating toxicity was considered toxic in the database. The final database contained 1,244 unique chemicals, of which 660 (53.1%) were toxic, and 584 (46.9%) were nontoxic (Supplementary Table 1). Principal Component Analysis (PCA) was implemented using 206 descriptors available within the Molecular Operating Environment (MOE) software v2018.01 to visualize the chemical space covered by the final database. The top three principal components explain 54.5% of the total variance of the final database and show a sufficiently large and diverse chemical space, aside from several outliers from both classes (Supplementary Figure 1).
Profiling and Assay Clustering
The public in vitro bioassay data for the target chemicals in the training set were extracted from PubChem30,31 and the EPA ToxCast/Tox21 summary files for invitroDBv3.243,49. The resulting in vitro bioprofile used for clustering contained 2,140 bioassays, of which 1,590 (74.3%) were from PubChem and 550 (25.7%) were from invitroDBv3.2 (Figure 2). In total, this bioprofile contained 41,570 active results (1.8%), 325,900 inactive results (13.9%), and 1,967,270 inconclusive results or untested chemical-bioassay pairs (84.3%) for 1,091 training chemicals (Figure 2). This profiling result was then used to cluster mechanistically related bioassays based on the presence of specific chemical fragments. To this end, the presence or absence of 834 Saagar fragments was identified for each chemical.
Figure 2:
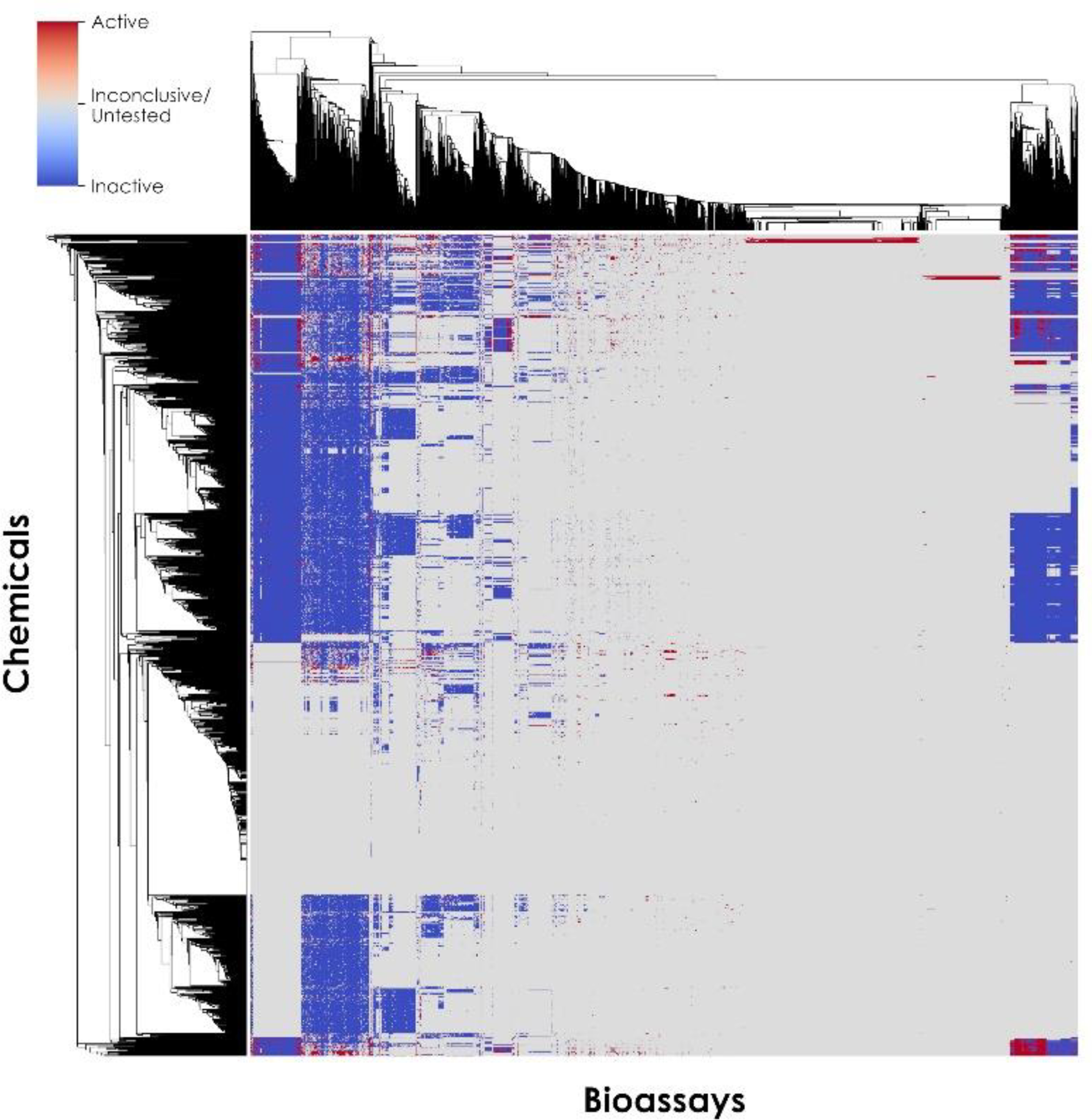
Bioprofile of 1,091 in vivo developmental toxicity database chemicals (y-axis) used for training across 2,140 PubChem and ToxCast bioassays (x-axis). Active results are shown as red squares, inactive results are shown as blue squares, and inconclusive results and untested chemical-bioassay pairs are shown as gray squares.
Among the 2,140 bioassays and 834 fragments, 61,928 statistically significant chemical-in vitro relationships were identified (p < 0.05) (Supplementary Figure 2). Next, mechanistically related bioassays were clustered by calculating the Jaccard distance between all pairs of bioassays using their profiles of existing significant chemical fragments. Bioassay pairs with a Jaccard similarity of at least 25% were used for clustering. After applying this minimum similarity threshold, 1,091 bioassays (51.0%) were considered to have “unique” mechanisms (i.e., < 25% similarity to all other individual bioassays) for the training set compounds and therefore excluded from further modeling. Therefore, 1,049 bioassays with presumed mechanistic connections to other bioassays remained available for the clustering analysis.
A network graph was generated to show the bioassay clusters (Figure 3). Each node represented one of the remaining 1,049 bioassays. Each edge connected two bioassays and had a length inversely representing the Jaccard similarity between the two bioassays (i.e., shorter edges indicate higher similarity). This network graph was then clustered using the Louvain modularity algorithm64 with resolution 0.35 and visualized using the Force Atlas algorithm65 with default parameters, as implemented in Gephi v0.9.2 (https://gephi.org/) (Figure 3). This process resulted in 68 clusters (Supplementary Table 2). Among the resulting clusters, 37 contained mixtures of bioassays from the PubChem and ToxCast databases. In Figure 3, each bioassay is represented by colored circles, and edges represent potential mechanistic relationships (i.e., bioassay pairs with > 25% similarity).
Figure 3:
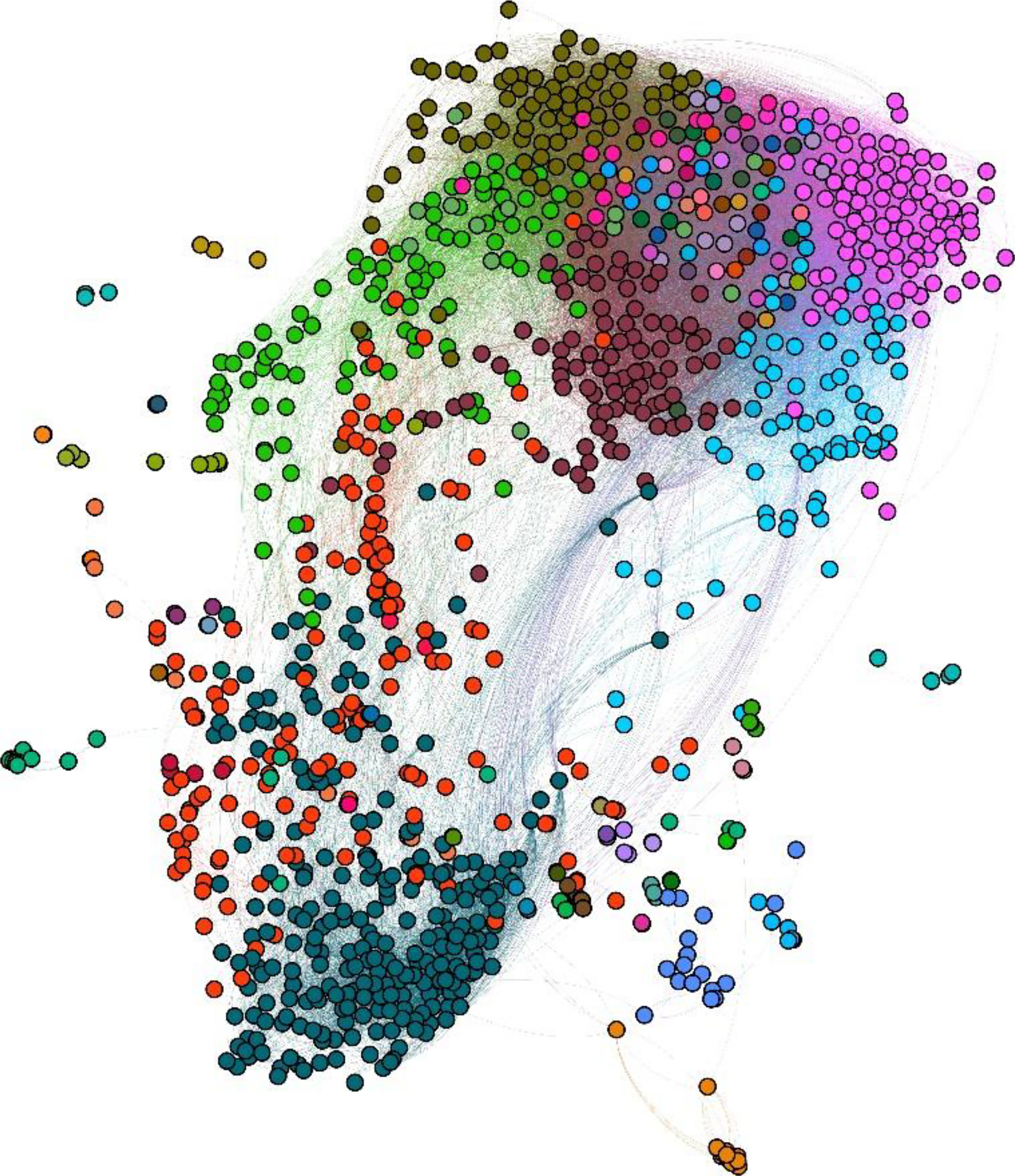
Cluster map of in vitro PubChem and Toxicity Forecaster (ToxCast) assays based on correlations between chemical fragments and assay activities. The Louvain modularity algorithm identified 68 clusters, each represented by a different color, as outlined in Supplementary Table 2. Each colored circle represents one bioassay such that circles of the same color belong to the same cluster. Each edge inversely correlates to the similarity between the connected assays, with longer edges representing lower similarity.
Read-Across Model Selection
Each of the 68 clusters depicted in Figure 3 was evaluated for their ability to predict prenatal developmental toxicity. First, 47 clusters containing less than five bioassays were removed to ensure the predictions could be biologically interpretable (Supplementary Table 2). Next, the remaining 21 clusters were evaluated to predict prenatal developmental toxicity using biological read-across studies. Each read-across study was evaluated using a five-fold cross-validation procedure.66 The minimum biosimilarity and confidence required to make read-across predictions were optimized during cross-validation. Then, the optimized PPV for each cluster was recorded, resulting in PPV values ranging from 51.4% to 100.0% (Equation 7, Figure 4A). Among the 21 clusters, 9 showed high predictive performance for prenatal developmental toxicity (PPV > 70%). Then, four of these nine clusters with confidence values equal to one were removed to reduce the effect of missing data on the resulting model reliability (Figure 4B). The read-across cross-validation results showed that three of the remaining five clusters had high sensitivity (Equation 4, ranging from 83.3% to 100.0%), low specificity (Equation 5, 0% for all three clusters), and low CCR (Equation 6, ranging from 41.7% to 50.0%) (Supplementary Table 3). These results indicated that the chemicals that had enough results in these clusters’ included in vitro assays for performing read-across were disproportionally in vivo developmental toxicants, limiting the model’s ability to optimize read-across parameters to predict nontoxic results properly. After excluding these three clusters, two potentially viable bioassay cluster models remained candidates to predict prenatal developmental toxicity.
Figure 4:
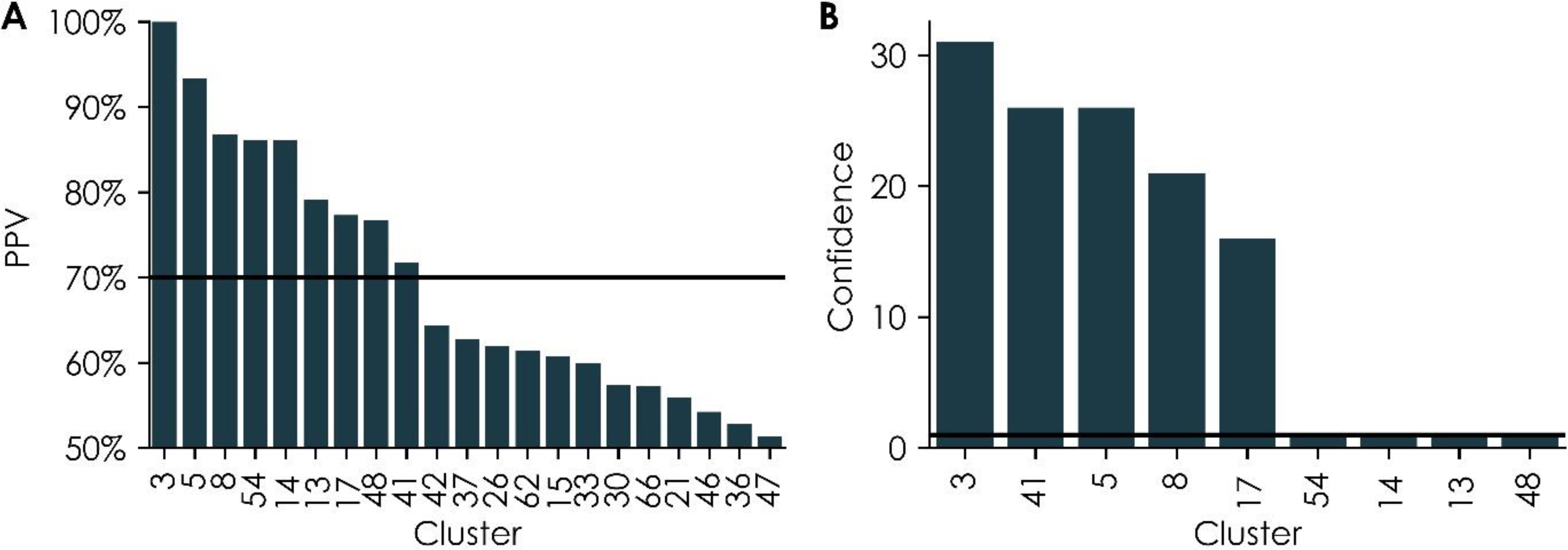
Cross-validation (A) positive predictive values (PPVs) for individual clusters containing at least five bioassays and (B) optimized confidence values for individual clusters with acceptable PPVs, as shown in (A). In both panels, the numbers along the x-axis represent cluster numbers as identified within the cluster map in Figure 3. The solid lines represent the thresholds used for cluster selection (PPV = 70% and confidence = 1).
The first model was Cluster 17, which had a higher predictive performance during cross-validation (PPV = 77.3%). This cluster contained 83 bioassays, of which 82 were collected from the PubChem database, and one was collected from the ToxCast database (Supplementary Table 2). The included bioassays collected from PubChem were deposited by Tox21 (48 bioassays, 55.4%), the National Center for Advancing Translational Sciences (NCATS) (29 bioassays, 39.8%), the Broad Institute (1 bioassay, 1.2%), New Mexico Molecular Libraries Screening Center (1 bioassay, 1.2%), Southern Research Institute (1 bioassay, 1.2%), the Scripps Research Institute Molecular Screening Center (1 bioassay, 1.2%), and the Vanderbilt High-Throughput Screening Facility (1 bioassay, 1.2%). Of these bioassays, 45 (54.2%) measured cell viability or cytotoxicity as counter screens for functional bioassays or drug repurposing screens (i.e., to treat various cancers67,68 or inhibit the growth of infectious bacteria69 or parasites70). Four additional bioassays were drug screens to identify chemicals that blocked cell entry of viruses, including those causing hemorrhagic fevers71 (i.e., Ebola, Lassa virus, and Marburg virus) and respiratory syndromes72 (i.e., the Middle East and severe acute respiratory syndrome-related coronaviruses). In addition, one bioassay identified chemicals that could inhibit antifungal efflux pumps73. The remaining 30 bioassays were functional and associated with protein targets, such as nuclear receptors, cytochrome P450 enzymes, G-protein coupled receptors, and transcription factors (Table 1).
Table 1.
Cluster 17 bioassay targets
| Target | PubChem AIDs | ToxCast AEIDs | References suggesting relevance to developmental toxicity |
|---|---|---|---|
|
| |||
| Muscarinic acetylcholine receptor M1 (CHRM1) | 628, 943, 944, 588852 | - | 96 |
| Cytochrome P450 2D6 (CYP2D6) | 891, 1851, 1645840 | 90,91,97 | |
| Caspase 3, apoptosis-related cysteine peptidase (CASP3) | 1346980, 1347034 | - | 98,99 |
| Cytochrome P450 3A4 (CYP3A4) | 1851, 1645841 | - | 90,91 |
| Thyroid-stimulating hormone receptor (TSHR) | 1259385, 1259395 | - | 100 |
| Atrial natriuretic peptide receptor 3 precursor (NPR2) | 1347050 | - | 84 |
| Chromobox protein homolog 1 (CBX1) | 488953 | - | 101 |
| Cytochrome P450 (CYP) 1A2, 2C9, 2C19 | 1851 | - | 90,91 |
| Cytochrome P450 19A1 (CYP19A1, aromatase) | 743083 | - | 92 |
| D(1A) dopamine receptor (DRD1) | 488983 | - | 97,102 |
| D(2) dopamine receptor isoform long (DRD2) | 485344 | - | 102,103 |
| Estrogen receptor beta (ERβ) | 1259378 | - | 97,104 |
| FAD-linked sulfhydryl oxidase ALR (GFER) | 485317 | - | 105 |
| Firefly luciferase | 1224835 | - | - |
| Heat shock transcription factor 1 (HSF1) | 743228 | - | 106 |
| Jun proto-oncogene (JUN) | 1159528 | - | 97,107–109 |
| Nuclear receptor subfamily 1, group I, member 2 (NR1I2) | 1346977 | - | 97,110 |
| One cut homeobox 1 (ONECUT1) | - | 83 | 87,88 |
| Peroxisome proliferator-activated receptor delta (PPARD) | 743215 | - | 111 |
| Potassium voltage-gated channel subfamily H member 2 isoform d (KCNH2) | 588834 | - | 112,113 |
| SMAD family member 1 (SMAD1) | 1347032 | - | 80 |
| Tyrosyl-DNA phosphodiesterase 1 (TDP1) | 686979 | - | - |
Cluster 41, which contained 76 bioassays, also emerged as a potential model for prenatal developmental toxicity predictions, showing a PPV of 71.7% during cross-validation. These 76 bioassays were collected from both the ToxCast (55 bioassays, 72.4%) and PubChem (21 bioassays, 27.6%) databases (Supplementary Table 2). Unlike Cluster 17, only a low percentage of these bioassays measured cell viability or cytotoxicity (9 bioassays, 11.8%). Additional bioassays from the ToxCast and Tox21 programs measured mitochondrial membrane potential disruptions74,75 (three bioassays, 3.9%), microtubule stability74 (two bioassays, 2.6%), and cytochrome P450 metabolism (eight bioassays, 10.5%). One further bioassay was an in vivo zebrafish assay developed as an alternative to mammalian tests to identify chemicals that induce embryonic death and structural anomalies76. The remaining 52 bioassays in Cluster 41 were functional, associated with protein targets, such as transcription factors, metalloproteinases, transmembrane receptors, and chemokines (Table 2).
Table 2.
Cluster 41 bioassay targets
| Target | PubChem AIDs | ToxCast AEIDs | References suggesting relevance to developmental toxicity |
|---|---|---|---|
|
| |||
| Androgen receptor (AR) | 588516, 743042, 1259243 | 1856 | 114,115 |
| Nuclear receptor subfamily 1, group I, member 2 (NR1I2) | 720659, 1346982, 1347033 | 103 | 97,110 |
| Aryl hydrocarbon receptor (AHR) | 743085, 743122 | 63 | 114,116,117 |
| Nuclear receptor subfamily 1, group I, member 3 (NR1I3) | 1224839, 1224892 | 101 | 118,119 |
| Estrogen receptor alpha (ERα) | 743080, 1259244 | - | 97,104 |
| Major histocompatibility complex, class II, DR alpha (HLA-DRA) | - | 147, 185 | 97,110 |
| Progesterone receptor (PGR) | 1346795, 1347031 | - | 104 |
| Acetylcholinesterase | 1347395 | - | 120 |
| cAMP responsive element binding protein 3 (CREB3) | - | 69 | 97,117 |
| CCAAT/enhancer binding protein (C/EBP), beta (CEBPB) | - | 67 | 117 |
| CD69 molecule | - | 303 | 97 |
| Chemokine (C-C motif) ligand 2 (CCL2) | - | 173 | 97,110,117 |
| Chemokine (C-X-C motif) ligand 10 (CXCL10) | - | 241 | 117 |
| Chemokine (C-X-C motif) ligand 8 (CXCL8) | - | 307 | 110 |
| Colony stimulating factor 1 (macrophage) (CSF1) | - | 243 | 121 |
| Cytochrome P450 19A1 (CYP19A1, aromatase) | 743139 | - | 92 |
| Estrogen-related nuclear receptor alpha (ERRα) | 1259401 | - | 122 |
| FBJ murine osteosarcoma viral oncogene homolog (FOS) | - | 64 | 123 |
| Forkhead box O3 (FOXO3) | - | 1426 | - |
| Matrix metallopeptidase 1 (interstitial collagenase) (MMP1) | - | 248 | - |
| Nuclear factor erythroid 2-related factor 2 isoform 1 (NFE2L2) | 651741 | 97 | 97,117,124 |
| Nuclear factor I/A (NFIA) | - | 95 | 125 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (NFKB1) | - | 94 | - |
| Nuclear respiratory factor 1 (NRF1) | - | 1460 | 124 |
| Peroxisome proliferator-activated receptor delta (PPARD) | - | 102 | 126 |
| Peroxisome proliferator-activated receptor gamma (PPARG) | - | 134 | 127 |
| POU class 2 homeobox 1 (POU2F1) | - | 98 | 97 |
| Prostaglandin E receptor 2 (subtype EP2) (PTGER2) | - | 289 | 110 |
| RAR-related orphan receptor B (RORB) | - | 104 | 97 |
| Retinoic acid receptor, alpha (RARA) | - | 136 | 110 |
| Retinoic acid receptor, beta (RARB) | - | 71 | 110 |
| Selectin E (SELE) | - | 305 | 128 |
| SMAD family member 1 (SMAD1) | - | 66 | 80 |
| Sterol regulatory element binding transcription factor 1 (SREBF1) | - | 107 | - |
| Thrombomodulin (THBD) | - | 162 | 129 |
| Upstream transcription factor 1 (USF1) | - | 72 | - |
| Vitamin D (1,25-dihydroxyvitamin D3) receptor (VDR) | - | 113 | 97 |
| X-box binding protein 1 (XBP1) | - | 114 | 130 |
External Validation by Predicting Test Chemicals
External validation with new chemicals is necessary to prove the utility of the selected models. To this end, Clusters 17 and 41 were used to predict the toxicity of chemicals outside of the training set77. Of the full in vivo developmental toxicity database compiled for this study, only 1,090 chemicals had bioassay data for the model training procedure. Therefore, the remaining 154 chemicals were used as an external validation set, with bioassay results populated by QSAR model predictions, consistent with previous computational toxicology studies32,33,41,53. Each QSAR model was first evaluated for its predictive performance before filling data gaps using their CCR (Equation 6). The QSAR models used to fill data gaps for these 154 chemicals had an average CCR of 68.4% (Supplementary Figure 3). Although this reliance on QSAR predictions adds uncertainty to the resulting predictions, this procedure mimics the model’s future use to fast screen new and untested chemicals early in the discovery and development process.
When using the cross-validation optimized confidence value for predictions (Equation 3), the Cluster 17 (confidence = 16) and 41 (confidence = 26) models showed high predictive performance for developmentally toxic chemicals in the external validation set (PPV = 100%) (Figure 5). Both clusters 17 and 41 were identified as potentially viable models for predicting developmental toxicity due partly to their high optimized confidence values compared to other clusters, which minimized the effects of missing data on the cluster selection process. Understandably, this low tolerance for missing data combined with the stringent applicability domain used for QSAR predictions proved to limit predictions of new and untested chemicals, resulting in low coverage of the overall dataset (4.5% and 3.9% for Clusters 17 and 41, respectively) and low predictive performance for nontoxic chemicals (Figure 5). This issue can, in some cases, be resolved by testing new chemicals in the relevant bioassays rather than relying on QSAR predictions. However, especially early in the discovery and development process, this solution is not always viable (e.g., when chemicals are not yet synthesized). In this circumstance, a potential solution is to increase the tolerance for missing data by lowering the confidence value required to report a prediction. High confidence values can limit the number of nontoxic predictions made since nontoxic chemicals often have a high proportion of inactive responses in the relevant bioassays.
Figure 5.
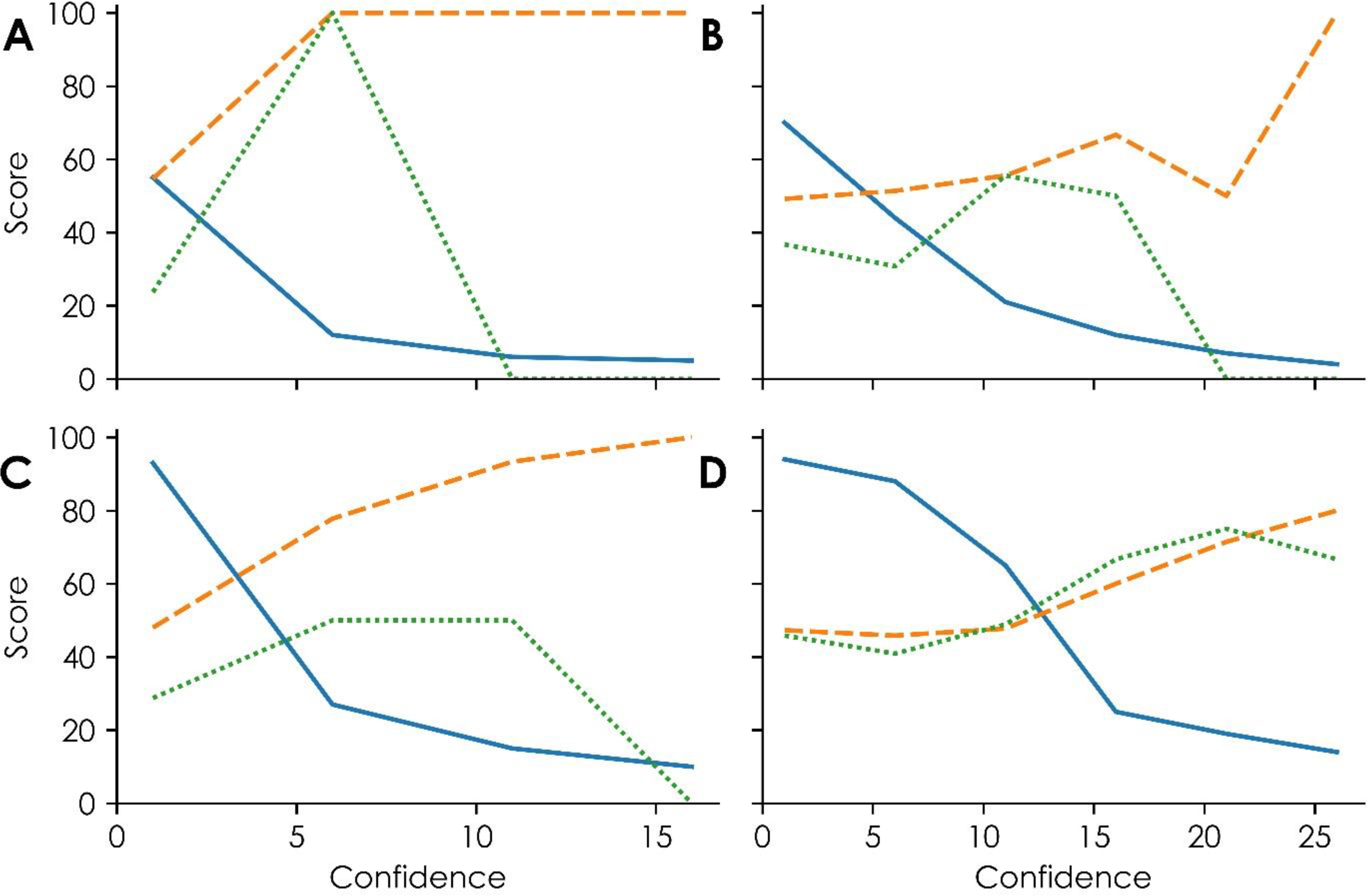
Clusters 17 and 41 models’ predictive performances, as shown by external validations. A and B show the performance of clusters 17 and 41, respectively, using activities from QSAR for chemicals whose three nearest neighbors in an assay showed at least 40% similarity. Panels C and D show the performance of clusters 17 and 41, respectively, using activities from QSAR for chemicals whose three nearest tested neighbors in an assay showed at least 30% similarity. The solid blue line in each graph represents the percent coverage of the external validation set containing 154 chemicals. The dashed orange lines represent the positive predictive values, or the fraction of predicted active chemicals predicted correctly, at varying confidence levels. The green dotted curves represent the specificity, or the fraction of correctly predicted in vivo inactive chemicals, at varying confidence levels.
For this reason, lowering the confidence value required to report read-across predictions increased both the coverage of the new chemicals and the balance of correctly predicted toxic and nontoxic chemicals. For example, using a confidence value of 6 instead of 16 for Cluster 17 model predictions showed high predictive performance for toxic and nontoxic chemicals (PPV = 100%, specificity = 100%) (Figure 5A). Similarly, using a confidence value of 16 instead of 26 for Cluster 41 model predictions improves the balance of predictive performance between toxicants and non-toxicants by retaining acceptable predictive performance for toxic chemicals (PPV = 66.7%) and increasing the cluster’s predictive performance for nontoxic chemicals (specificity = 50%) (Figure 5B). Further, using these adjusted confidence values increases the coverage more than two-fold for each cluster.
By lowering the similarity threshold required to report individual QSAR assay results to fill missing data before read-across, the coverage of predicting new chemicals further increased. Figures 5C and 5D show the results of varying confidence values with a lower similarity threshold (i.e., 30% instead of 40% similarity to three nearest tested neighbors required to report an assay result for a specific chemical). For example, using a confidence value of 11 for Cluster 17 predictions yields a high predictive performance for toxic chemicals (PPV = 93%), improved predictive performance for nontoxic chemicals (specificity = 50%), and improved coverage of 14.9%. Similarly, a confidence value of 21 for Cluster 41 predictions yields acceptable predictive performances for both toxic and nontoxic chemicals (PPV = 71.4%, specificity = 75.0%) and improved coverage of 19.5%.
Discussion
Implementing computational approaches can reduce the need for time-consuming, cost-inefficient, and often ethically undesirable animal tests and is particularly attractive for endpoints such as prenatal developmental toxicity where highly specialized animal study designs are necessary. Although previous computational approaches to evaluating new chemicals for prenatal developmental toxicity have shown promise, they have been limited in biological interpretability by only incorporating chemical structural information7–13,78,79 or focusing on limited and specific well-understood biological mechanisms38–41. Here, we presented a prenatal developmental toxicity database of over 1,200 chemicals spanning various use categories that underwent mammalian prenatal tests similar to the OECD 414/OPPTS 870.3700 protocols. Then, chemical structural information and biological data were integrated into a workflow that allowed for increased biological interpretability of the resulting predictions (Figure 1). As a result, clusters of bioassays were formed based on chemical-in vitro relationships, and two bioassay clusters, 17 and 41, were identified as models for predicting prenatal developmental toxicity.
These clusters contained several bioassays that align with plausible mechanisms of prenatal developmental toxicity (Tables 1 and 2). For example, the transforming growth factor beta (TGFβ)/SMAD pathway is well-known for its importance in embryonic development80. Bioassays identifying agonists and antagonists of this pathway included in Clusters 41 and 17, respectively, were, therefore, previously identified as relevant to predicting prenatal developmental toxicity [ToxCast Assay Endpoint Identifier (AEID) 66 and PubChem Assay Identifier (AID) 1347032]81. Similarly, endocrine disruption by drugs or environmental chemicals is a well-established mechanism of developmental toxicity82. Bioassays measuring endocrine disruption by binding to hormone receptors (e.g., nuclear estrogen, androgen, progesterone, and thyroid-stimulating hormone receptors) were present in both clusters.
Besides these well-known developmental toxicity mechanisms, some less established but potentially relevant targets were captured by this study. For example, one Cluster 17 bioassay measured natriuretic polypeptide receptor B (NPR-B) antagonism (PubChem AID 1347050)83 (Table 1). In previous studies, loss-of-function mutations in human NPR-B’s precursor gene were associated with impaired skeletal growth, suggesting that disrupting this receptor’s function may interfere with skeletal development84. An additional Cluster 17 bioassay was associated with hepatocyte nuclear factor 6 (HNF6)’s precursor gene ONECUT1 [ToxCast Assay Endpoint Identifier (AEID) 83]85,86. HNF6 was identified in previous studies as a regulator of liver87 and pancreatic88 development. Finally, in Cluster 41, several bioassays associated with various chemokines associated with immune response and inflammation were represented, along with matrix metallopeptidase 1 (MMP1) (Table 2). These targets were recently included in a proposed embryonic vascular disruption adverse outcome pathway (AOP) with endocrine disruption targets and TGFβ89.
Interestingly, both clusters also contained assays measuring the activity of cytochrome P450 enzymes. Although the contributions of these enzymes to developmental toxicity are not fully understood, previous studies showed that cytochrome P450 enzyme expressions varied by developmental stages and modulated fetal exposures to toxicants such as carcinogens, drugs, and alcohol90,91. Further, aromatase (cytochrome P450 19A1), a target of assays present in both clusters, plays a key role in hormone regulation by converting androgens to estrogens92.
This study highlighted the benefits and opportunities of using the publicly available biological data for computational toxicity predictions to resolve common issues in classic modeling studies (e.g., QSAR) for complex toxicity endpoints. Both well-established and putative prenatal developmental toxicity mechanisms were incorporated in the developed models, including endocrine disruption and embryonic blood vessel development. By relying on relevant biological data, this workflow provides users with an increased capacity to mechanistically interpret predictions, as required by international regulatory guidelines such as those by the OECD18,93. Further, using biological data combined with chemical structural information reduced the frequency of activity cliffs in previous studies by incorporating information about chemicals’ interactions with biomolecules into the prediction process14,34,51.
The predictions for external validation set chemicals, especially developmentally toxic ones, could be explained by looking at their nearest biological neighbors in the training set (Figure 6). For example, one developmentally toxic external validation set chemical was 2,2’,4,4’,5-pentachlorodiphenyl ether (CAS 60123–64-0). Chlorodiphenyl ethers are generated as byproducts during the manufacture of chlorinated phenols such as fungicides and wood preservatives94. Based on the Cluster 17 bioassays, this chemical was correctly predicted as toxic based on its most biologically similar nearest neighbor (confidence = 16), as identified by comparing their bioassay responses using the biosimilarity metric (Equation 2). This neighbor was tributyltetradecylphosphonium chloride (CAS 81741–28-8), a developmentally toxic training chemical used as a biocide in hydraulic fracking (Figure 6A)95. The high biosimilarity between these two chemicals was influenced mostly by their shared active responses in 11 viability/cytotoxicity assays and five functional assays. The functional assays represented four targets: the jun proto-oncogene, thyroid-stimulating hormone receptor, pregnane X receptor (NR1I2), and Small Mothers Against Decapentaplegic (SMAD) family member 1 (Table 1). Similarly, one example among Cluster 41’s toxic external validation predictions was 2,4-difluoro-1-(4-nitrophenoxy)benzene (CAS 28280–37-7). In the training set, based on the Cluster 41 assays, this chemical’s biological nearest neighbor was prochloraz (CAS 67747–09-5), a developmentally toxic fungicide (Figure 6B). These two chemicals shared active responses in 40 assays. Of these assays, five were viability/cytotoxicity assays. Two additional assays measured disruption of the mitochondrial membrane potential in vitro74,75 and embryonic death and structural anomalies in vivo using zebrafish [ToxCast assay endpoint identifier (AEID) 1507]76. The remainder were functional assays representing protein targets, including endocrine targets, cytochrome P450 enzymes, the aryl hydrocarbon receptor, and chemokines (Table 2).
Figure 6.
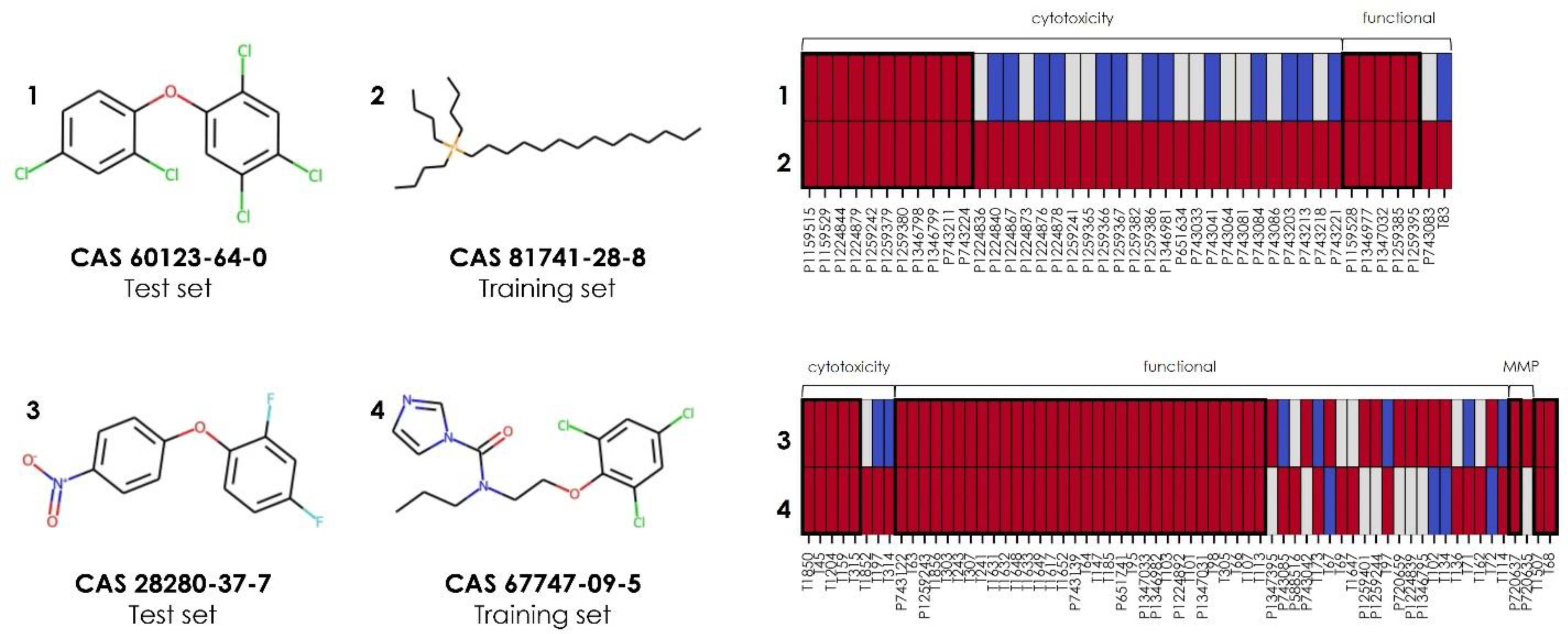
Sample toxic external validation predictions from the cluster models. Predictions are shown for 2,2’,4,4’,5-pentachlorodiphenyl ether (CAS 60123–64-0) based on its biological nearest neighbor within the training set chemicals tested in Cluster 17 bioassays, tributyltetradecylphosphonium chloride (CAS 81741–28-8), and for 2,4-difluoro-1-(4-nitrophenoxy)benzene (CAS 28280–37-7) based on its biological nearest neighbor within the training set chemicals tested in Cluster 41 bioassays, prochloraz (CAS 67747–09-5). A subset of the complete profile is shown in both panels, which includes the most influential bioassays on the resulting toxic prediction (i.e., those containing at least one active response between the validation chemical and its biological nearest neighbor). The columns in each profile represent assay identifiers from PubChem (prefixed with “P”) and ToxCast (prefixed with “T”), as listed in Tables 1 and 2. Active results are shown as red rectangles, inactive results are shown as blue rectangles, and untested chemical-bioassay pairs are shown as gray rectangles. Thick bold boxes surround shared active responses.
The computational workflow described here automatically identified bioassay data relevant to prenatal developmental toxicity from public databases and created a new strategy for predicting untested chemicals early in the discovery and development procedure or existing chemicals with limited safety data. Although the read-across models developed were insufficient to encompass all possible prenatal developmental toxicity mechanisms, this study highlighted the benefits and opportunities of using the publicly available biological data for computational toxicity predictions. Both well-established and putative prenatal developmental toxicity mechanisms were incorporated in the developed models, including endocrine disruption and embryonic blood vessel development. In addition, the adaptability built into the workflow paves the way for the easy incorporation of new bioassays as they are submitted to public database resources in future studies and applied to other complex toxicity endpoints.
Supplementary Material
Synopsis:
A computational approach can quickly and cost-efficiently identify potential prenatal developmental toxicants based on biological data to reduce, refine, and eventually replace animal testing.
Acknowledgment:
This project was partially supported by the National Institute of Environmental Health Sciences (Grant numbers R01ES031080, R15ES023148, and R35ES031709) and a Lubrizol research grant for Rutgers University.
Funding Sources
HLC, DPR, SS, and HZ were partially supported by National Institute of Environmental Health Sciences (NIEHS) grants (R01ES031080, R15ES023148, and R35ES031709) and a Lubrizol research grant for Rutgers University.
Footnotes
The authors declare that they have no competing interests.
Supporting Information
Chemical space of the prenatal developmental toxicity database (n=1,244) based on the top three principal components calculated from 206 two-dimensional Molecular Operating Environment (MOE) software v2018.01 descriptors (54.5% variance explained) (Figure S1). Chemical-in vitro relationship profile across 1,224 PubChem and ToxCast bioassays (y-axis) having a statistically significant relationship with at least one Saagar fragment (x-axis) (Figure S2). Five-fold cross-validation scores of Quantitative Structure-Activity Relationship (QSAR) models used to impute missing bioassay data (Figure S3).
Prenatal developmental toxicity database (Table S1). Assays included in clustering (Table S2). Read-across statistics (Table S3).
REFERENCES
- (1).Meigs L; Smirnova L; Rovida C; Leist M; Hartung T Animal Testing and Its Alternatives – the Most Important Omics Is Economics. ALTEX 2018, 35 (3), 275–305. 10.14573/altex.1807041. [DOI] [PubMed] [Google Scholar]
- (2).Hartung T Toxicology for the Twenty-First Century. Nature 2009, 460 (7252), 208–212. 10.1038/460208a. [DOI] [PubMed] [Google Scholar]
- (3).Rovida C; Hartung T Re-Evaluation of Animal Numbers and Costs for in Vivo Tests to Accomplish REACH Legislation Requirements for Chemicals - A Report by the Transatlantic Think Tank for Toxicology (T4). ALTEX 2009, 26 (3), 187–208. 10.14573/altex.2009.3.187. [DOI] [PubMed] [Google Scholar]
- (4).Corvi R; Spielmann H; Hartung T Alternative Approaches for Carcinogenicity and Reproductive Toxicity. In History of Toxicology and Environmental Health, The History of Alternative Test Methods in Toxicology; Balls M, Combes R, Worth A, Eds.; Academic Press, 2019; pp 209–217. 10.1016/B978-0-12-813697-3.00024-X. [DOI] [Google Scholar]
- (5).Organisation for Economic Co-operation and Development. OECD 414: Prenatal Developmental Toxicity Study. In OECD Guidelines for the Testing of Chemicals, Section 4; OECD Publishing: Paris, France, 2018. 10.1787/9789264070820-en. [DOI] [Google Scholar]
- (6).United States Environmental Protection Agency. Health Effects Test Guidelines OPPTS 870.3700 Prenatal Developmental Toxicity Study. In OPPTS Harmonized Test Guidelines, Series 870; 1998. https://doi.org/EPA-HQ-OPPT-2009-0156-0017. [Google Scholar]
- (7).Wu S; Fisher J; Naciff J; Laufersweiler M; Lester C; Daston G; Blackburn K Framework for Identifying Chemicals with Structural Features Associated with the Potential to Act as Developmental or Reproductive Toxicants. Chem. Res. Toxicol. 2013, 26 (12), 1840–1861. 10.1021/tx400226u. [DOI] [PubMed] [Google Scholar]
- (8).Hewitt M; Ellison CM; Enoch SJ; Madden JC; Cronin MTD Integrating (Q)SAR Models, Expert Systems and Read-across Approaches for the Prediction of Developmental Toxicity. Reprod. Toxicol. 2010, 30 (1), 147–160. 10.1016/j.reprotox.2009.12.003. [DOI] [PubMed] [Google Scholar]
- (9).Matthews EJ; Kruhlak NL; Daniel Benz R; Ivanov J; Klopman G; Contrera JF A Comprehensive Model for Reproductive and Developmental Toxicity Hazard Identification: II. Construction of QSAR Models to Predict Activities of Untested Chemicals. Regul. Toxicol. Pharmacol. 2007, 47 (2), 136–155. 10.1016/j.yrtph.2006.10.001. [DOI] [PubMed] [Google Scholar]
- (10).Cassano A; Manganaro A; Martin T; Young D; Piclin N; Pintore M; Bigoni D; Benfenati E CAESAR Models for Developmental Toxicity. Chem. Cent. J. 2010, 4 (Suppl 1), S4. 10.1186/1752-153X-4-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Arena VC; Sussman NB; Mazumdar S; Yu S; Macina OT The Utility of Structure-Activity Relationship (SAR) Models for Prediction and Covariate Selection in Developmental Toxicity: Comparative Analysis of Logistic Regression and Decision Tree Models. SAR QSAR Environ. Res. 2004, 15 (1), 1–18. 10.1080/1062936032000169633. [DOI] [PubMed] [Google Scholar]
- (12).Basant N; Gupta S; Singh KP In Silico Prediction of the Developmental Toxicity of Diverse Organic Chemicals in Rodents for Regulatory Purposes. Toxicol. Res. (Camb). 2016, 5 (3), 773–787. 10.1039/c5tx00493d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Zhang H; Mao J; Qi HZ; Ding L In Silico Prediction of Drug-Induced Developmental Toxicity by Using Machine Learning Approaches. Mol. Divers. 2020, 24 (4), 1281–1290. 10.1007/s11030-019-09991-y. [DOI] [PubMed] [Google Scholar]
- (14).Zhu H; Bouhifd M; Donley E; Egnash L; Kleinstreuer N; Kroese ED; Liu Z; Luechtefeld T; Palmer J; Pamies D; Shen J; Strauss V; Wu S; Hartung T Supporting Read-across Using Biological Data. ALTEX 2016, 33 (2), 167–182. 10.14573/altex.1601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Cherkasov A; Muratov EN; Fourches D; Varnek A; Baskin II; Cronin M; Dearden J; Gramatica P; Martin YC; Todeschini R; Consonni V; Kuz’Min VE; Cramer R; Benigni R; Yang C; Rathman J; Terfloth L; Gasteiger J; Richard A; Tropsha A QSAR Modeling: Where Have You Been? Where Are You Going To? J. Med. Chem. 2014, 57 (12), 4977–5010. 10.1021/jm4004285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Stouch TR; Kenyon JR; Johnson SR; Chen XQ; Doweyko A; Li Y In Silico ADME/Tox: Why Models Fail. J. Comput. Aided. Mol. Des. 2003, 17 (2–4), 83–92. 10.1023/A:1025358319677. [DOI] [PubMed] [Google Scholar]
- (17).Maggiora GM On Outliers and Activity Cliffs - Why QSAR Often Disappoints. J. Chem. Inf. Model. 2006, 46 (4), 1535. 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- (18).Organisation for Economic Co-operation and Development. Guidance Document on the Validation of (Quantitative)Structure-Activity Relationship [(Q)SAR] Models. OECD Environ. Heal. Saf. Publ. Ser. Test. Assess. 2007, 69, 1–154. 10.1787/9789264085442-en. [DOI] [Google Scholar]
- (19).Gramatica P Principles of QSAR Models Validation: Internal and External. QSAR Comb. Sci. 2007, 26 (5), 694–701. 10.1002/qsar.200610151. [DOI] [Google Scholar]
- (20).Benigni R; Netzeva TI; Benfenati E; Bossa C; Franke R; Helma C; Hulzebos E; Marchant C; Richard A; Woo YT; Yang C The Expanding Role of Predictive Toxicology: An Update on the (Q)SAR Models for Mutagens and Carcinogens. J. Environ. Sci. Heal. - Part C Environ. Carcinog. Ecotoxicol. Rev. 2007, 25 (1), 53–97. 10.1080/10590500701201828. [DOI] [PubMed] [Google Scholar]
- (21).Zhu H; Zhang J; Kim MT; Boison A; Sedykh A; Moran K Big Data in Chemical Toxicity Research: The Use of High-Throughput Screening Assays to Identify Potential Toxicants. Chem. Res. Toxicol. 2014, 27 (10), 1643–1651. 10.1021/tx500145h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Zhu H Big Data and Artificial Intelligence Modeling for Drug Discovery. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 1–17. 10.1146/annurev-pharmtox-010919-023324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ciallella HL; Zhu H Advancing Computational Toxicology in the Big Data Era by Artificial Intelligence: Data-Driven and Mechanism-Driven Modeling for Chemical Toxicity. Chem. Res. Toxicol. 2019, 32 (4), 536–547. 10.1021/acs.chemrestox.8b00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Dix DJ; Houck KA; Martin MT; Richard AM; Setzer RW; Kavlock RJ The ToxCast Program for Prioritizing Toxicity Testing of Environmental Chemicals. Toxicol. Sci. 2007, 95 (1), 5–12. 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- (25).Judson RS; Houck KA; Kavlock RJ; Knudsen TB; Martin MT; Mortensen HM; Reif DM; Rotroff DM; Shah I; Richard AM; Dix DJ In Vitro Screening of Environmental Chemicals for Targeted Testing Prioritization: The ToxCast Project. Environ. Health Perspect. 2010, 118 (4), 485–492. 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Shukla SJ; Huang R; Austin CP; Xia M The Future of Toxicity Testing: A Focus on in Vitro Methods Using a Quantitative High-Throughput Screening Platform. Drug Discov. Today 2010, 15 (23–24), 997–1007. 10.1016/j.drudis.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Thomas RS; Paules RS; Simeonov A; Fitzpatrick SC; Crofton KM; Casey WM; Mendrick DL The US Federal Tox21 Program: A Strategic and Operational Plan for Continued Leadership. ALTEX 2018, 35 (2), 163–168. 10.14573/altex.1803011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hsu C-W; Huang R; Attene-Ramos MS; Austin CP; Simeonov A; Xia M Advances in High-Throughput Screening Technology for Toxicology; 2017; Vol. 20. 10.1504/IJRAM.2017.082562. [DOI] [Google Scholar]
- (29).Collins FS; Gray GM; Bucher JR Transforming Environmental Health Protection. Science (80-. ). 2008, 319 (5865), 906–907. 10.1126/science.1154619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wang Y; Xiao J; Suzek TO; Zhang J; Wang J; Bryant SH PubChem: A Public Information System for Analyzing Bioactivities of Small Molecules. Nucleic Acids Res. 2009, 37, W623–W633. 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang Y; Bolton E; Dracheva S; Karapetyan K; Shoemaker BA; Suzek TO; Wang J; Xiao J; Zhang J; Bryant SH An Overview of the PubChem BioAssay Resource. Nucleic Acids Res. 2010, 38, D255–D266. 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhao L; Russo DP; Wang W; Aleksunes LM; Zhu H Mechanism-Driven Read-Across of Chemical Hepatotoxicants Based on Chemical Structures and Biological Data. Toxicol. Sci. 2020, 174 (2), 178–188. 10.1093/toxsci/kfaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Russo DP; Strickland J; Karmaus AL; Wang W; Shende S; Hartung T; Aleksunes LM; Zhu H Nonanimal Models for Acute Toxicity Evaluations: Applying Data-Driven Profiling and Read-Across. Environ. Health Perspect. 2019, 127 (4), 1–14. 10.1289/EHP3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Guo Y; Zhao L; Zhang X; Zhu H Using a Hybrid Read-across Method to Evaluate Chemical Toxicity Based on Chemical Structure and Biological Data. Ecotoxicol. Environ. Saf. 2019, 178, 178–187. 10.1016/j.ecoenv.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhu H; Rusyn I; Richard A; Tropsha A Use of Cell Viability Assay Data Improves the Prediction Accuracy of Conventional Quantitative Structure–Activity Relationship Models of Animal Carcinogenicity. Environ. Health Perspect. 2008, 116 (4), 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Kim MT; Huang R; Sedykh A; Wang W; Xia M; Zhu H Mechanism Profiling of Hepatotoxicity Caused by Oxidative Stress Using Antioxidant Response Element Reporter Gene Assay Models and Big Data. Environ. Health Perspect. 2016, 124 (5), 634–641. 10.1289/ehp.1509763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zhang J; Hsieh JH; Zhu H Profiling Animal Toxicants by Automatically Mining Public Bioassay Data: A Big Data Approach for Computational Toxicology. PLoS One 2014, 9 (6). 10.1371/journal.pone.0099863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Browne P; Judson RS; Casey WM; Kleinstreuer NC; Thomas RS Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ. Sci. Technol. 2015, 49 (14), 8804–8814. 10.1021/acs.est.5b02641. [DOI] [PubMed] [Google Scholar]
- (39).Judson RS; Magpantay FM; Chickarmane V; Haskell C; Tania N; Taylor J; Xia M; Huang R; Rotroff DM; Filer DL; Houck KA; Martin MT; Sipes N; Richard AM; Mansouri K; Woodrow Setzer R; Knudsen TB; Crofton KM; Thomas RS Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 in Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci. 2015, 148 (1), 137–154. 10.1093/toxsci/kfv168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kleinstreuer NC; Ceger P; Watt ED; Martin M; Houck K; Browne P; Thomas RS; Casey WM; Dix DJ; Allen D; Sakamuru S; Xia M; Huang R; Judson R Development and Validation of a Computational Model for Androgen Receptor Activity. Chem. Res. Toxicol. 2017, 30 (4), 946–964. 10.1021/acs.chemrestox.6b00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Ciallella HL; Russo DP; Aleksunes LM; Grimm FA; Zhu H Revealing Adverse Outcome Pathways from Public High-Throughput Screening Data to Evaluate New Toxicants by a Knowledge-Based Deep Neural Network Approach. Environ. Sci. Technol. 2021, 55, 10875–10887. 10.1021/acs.est.1c02656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Russo DP; Kim MT; Wang W; Pinolini D; Shende S; Strickland J; Hartung T; Zhu H CIIPro: A New Read-across Portal to Fill Data Gaps Using Public Large-Scale Chemical and Biological Data. Bioinformatics 2017, 33 (3), 464–466. 10.1093/bioinformatics/btw640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Williams AJ; Grulke CM; Edwards J; McEachran AD; Mansouri K; Baker NC; Patlewicz G; Shah I; Wambaugh JF; Judson RS; Richard AM The CompTox Chemistry Dashboard: A Community Data Resource for Environmental Chemistry. J. Cheminform. 2017, 9 (1), 1–27. 10.1186/s13321-017-0247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Knudsen TB; Martin MT; Kavlock RJ; Judson RS; Dix DJ; Singh AV Profiling the Activity of Environmental Chemicals in Prenatal Developmental Toxicity Studies Using the U.S. EPA’s ToxRefDB. Reprod. Toxicol. 2009, 28 (2), 209–219. 10.1016/j.reprotox.2009.03.016. [DOI] [PubMed] [Google Scholar]
- (45).Watford S; Ly Pham L; Wignall J; Shin R; Martin MT; Friedman KP ToxRefDB Version 2.0: Improved Utility for Predictive and Retrospective Toxicology Analyses. Reprod. Toxicol. 2019, 89 (May), 145–158. 10.1016/j.reprotox.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).European Chemicals Agency. Registered substances https://echa.europa.eu/information-on-chemicals/registered-substances.
- (47).Kim S; Thiessen PA; Bolton EE; Bryant SH PUG-SOAP and PUG-REST: Web Services for Programmatic Access to Chemical Information in PubChem. Nucleic Acids Res. 2015, 43 (W1), W605–W611. 10.1093/nar/gkv396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Richard AM; Williams CLR Distributed Structure-Searchable Toxicity (DSSTox) Public Database Network: A Proposal. Mutat. Res. - Fundam. Mol. Mech. Mutagen. 2002, 499 (1), 27–52. 10.1016/S0027-5107(01)00289-5. [DOI] [PubMed] [Google Scholar]
- (49).Judson R; Richard A; Dix D; Houck K; Elloumi F; Martin M; Cathey T; Transue TR; Spencer R; Wolf M ACToR - Aggregated Computational Toxicology Resource. Toxicol. Appl. Pharmacol. 2008, 233 (1), 7–13. 10.1016/j.taap.2007.12.037. [DOI] [PubMed] [Google Scholar]
- (50).Sedykh AY; Shah RR; Kleinstreuer NC; Auerbach SS; Gombar VK Saagar-A New, Extensible Set of Molecular Substructures for QSAR/QSPR and Read-Across Predictions. Chem. Res. Toxicol. 2021, 34 (2), 634–640. 10.1021/acs.chemrestox.0c00464. [DOI] [PubMed] [Google Scholar]
- (51).Ribay K; Kim MT; Wang W; Pinolini D; Zhu H Predictive Modeling of Estrogen Receptor Binding Agents Using Advanced Cheminformatics Tools and Massive Public Data. Front. Environ. Sci. 2016, 4 (March), 1–9. 10.3389/fenvs.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Pedregosa F; Varoquaux G; Gramfort A; Michel V; Thirion B; Grisel O; Blondel M; Prettenhofer P; Weiss R; Dubourg V; Vanderplas J; Passos A; Cournapeau D; Brucher M; Perrot M; Duchesnay É Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. 10.1007/s13398-014-0173-7.2. [DOI] [Google Scholar]
- (53).Ciallella HL; Russo DP; Aleksunes LM; Grimm FA; Zhu H Predictive Modeling of Estrogen Receptor Agonism, Antagonism, and Binding Activities Using Machine- and Deep-Learning Approaches. Lab. Investig. 2020, 101 (4), 490–502. 10.1038/s41374-020-00477-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Korotcov A; Tkachenko V; Russo DP; Ekins S Comparison of Deep Learning with Multiple Machine Learning Methods and Metrics Using Diverse Drug Discovery Data Sets. Mol. Pharm. 2017, 14 (12), 4462–4475. 10.1021/acs.molpharmaceut.7b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Manning CD; Raghavan P; Schuetze H The Bernoulli Model. In Introduction to Information Retrieval; Cambridge University Press, 2009; pp 234–265. [Google Scholar]
- (56).McCallum A; Nigam K A Comparison of Event Models for Naive Bayes Text Classification. EACL ‘03 Proc. tenth Conf. Eur. chapter Assoc. Comput. Linguist. 2003, 1, 307–314. 10.3115/1067807.1067848. [DOI] [Google Scholar]
- (57).Cover TM; Hart PE Nearest Neighbor Pattern Classification. IEEE Trans. Inf. Theory 1967, 13 (1), 21–27. 10.1109/TIT.1967.1053964. [DOI] [Google Scholar]
- (58).Breiman L Random Forests. Mach. Learn. 2001, 45 (1), 5–32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- (59).Vapnik VN Methods of Pattern Recognition. In The Nature of Statistical Learning Theory; Springer Science & Business Media: New York, 2000; pp 123–170. [Google Scholar]
- (60).Mansouri K; Karmaus AL; Fitzpatrick J; Patlewicz G; Pradeep P; Alberga D; Alepee N; Allen TEH; Allen D; Alves VM; Andrade CH; Auernhammer TR; Ballabio D; Bell S; Benfenati E; Bhattacharya S; Bastos JV; Boyd S; Brown JB; Capuzzi SJ; Chushak Y; Ciallella H; Clark AM; Consonni V; Daga PR; Ekins S; Farag S; Fedorov M; Fourches D; Gadaleta D; Gao F; Hartung T; Hirn M; Karpov P; Korotcov A; Lavado GJ; Lawless M; Li X; Luechtefeld T; Lunghini F; Mangiatordi GF; Marcou G; Marsh D; Martin T; Mauri A; Muratov EN; Myatt GJ; Nguyen D; Nicolotti O; Note R; Pande P; Parks AK; Peryea T; Polash AH; Rallo R; Roncaglioni A; Rowlands C; Ruiz P; Russo DP; Sayed A; Sayre R; Sheils T; Siegel C; Tetko IV; Thomas D; Tkachenko V; Todeschini R; Toma C; Tripodi I CATMoS : Collaborative Acute Toxicity Modeling Suite. Environ. Health Perspect. 2021, 129 (April), 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Mansouri K; Kleinstreuer N; Abdelaziz AM; Alberga D; Alves VM; Andersson PL; Andrade CH; Bai F; Balabin I; Ballabio D; Benfenati E; Bhhatarai B; Boyer S; Chen J; Consonni V; Farag S; Fourches D; García-Sosa AT; Gramatica P; Grisoni F; Grulke CM; Hong H; Horvath D; Hu X; Huang R; Jeliazkova N; Li J; Li X; Liu H; Manganelli S; Mangiatordi GF; Maran U; Marcou G; Martin T; Muratov E; Nguyen DT; Nicolotti O; Nikolov NG; Norinder U; Papa E; Petitjean M; Piir G; Pogodin P; Poroikov V; Qiao X; Richard AM; Roncaglioni A; Ruiz P; Rupakheti C; Sakkiah S; Sangion A; Schramm KW; Selvaraj C; Shah I; Sild S; Sun L; Taboureau O; Tang Y; Tetko IV; Todeschini R; Tong W; Trisciuzzi D; Tropsha A; Van Den Driessche G; Varnek A; Wang Z; Wedebye EB; Williams AJ; Xie H; Zakharov AV; Zheng Z; Judson RS CoMPARA: Collaborative Modeling Project for Androgen Receptor Activity. Environ. Health Perspect. 2020, 128 (2), 1–17. 10.1289/EHP5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Mansouri K; Abdelaziz A; Rybacka A; Roncaglioni A; Tropsha A; Varnek A; Zakharov A; Worth A; Richard AM; Grulke CM; Trisciuzzi D; Fourches D; Horvath D; Benfenati E; Muratov E; Wedebye EB; Grisoni F; Mangiatordi GF; Incisivo GM; Hong H; Ng HW; Tetko IV; Balabin I; Kancherla J; Shen J; Burton J; Nicklaus M; Cassotti M; Nikolov NG; Nicolotti O; Andersson PL; Zang Q; Politi R; Beger RD; Todeschini R; Huang R; Farag S; Rosenberg SA; Slavov S; Hu X; Judson. CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ. Health Perspect. 2016, 124 (7), 1023–1033. 10.1289/ehp.1510267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Solimeo R; Zhang J; Kim M; Sedykh A; Zhu H Predicting Chemical Ocular Toxicity Using a Combinatorial QSAR Approach. Chem. Res. Toxicol. 2012, 25 (12), 2763–2769. 10.1021/tx300393v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Blondel VD; Guillaume JL; Lambiotte R; Lefebvre E Fast Unfolding of Communities in Large Networks. J. Stat. Mech. Theory Exp. 2008, 2008 (10). 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- (65).Jacomy M; Venturini T; Heymann S; Bastian M ForceAtlas2, a Continuous Graph Layout Algorithm for Handy Network Visualization Designed for the Gephi Software. PLoS One 2014, 9 (6), 1–12. 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Tropsha A; Gramatica P; Gombar VK The Importance of Being Earnest: Validation Is the Absolute Essential for Successful Application and Interpretation of QSPR Models. QSAR Comb. Sci. 2003, 22 (1), 69–77. 10.1002/qsar.200390007. [DOI] [Google Scholar]
- (67).Xia M; Huang R; Sakamuru S; Alcorta D; Cho MH; Lee DH; Park DM; Kelley MJ; Sommer J; Austin CP Identification of Repurposed Small Molecule Drugs for Chordoma Therapy. Cancer Biol. Ther. 2013, 14 (7), 638–647. 10.4161/cbt.24596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Shen M; Asawa R; Zhang YQ; Cunningham E; Sun H; Tropsha A; Janzen WP; Muratov EN; Capuzzi SJ; Farag S; Jadhav A; Blatt J; Simeonov A; Martinez NJ Quantitative High-Throughput Phenotypic Screening of Pediatric Cancer Cell Lines Identifies Multiple Opportunities for Drug Repurposing. Oncotarget 2018, 9 (4), 4758–4772. 10.18632/oncotarget.23462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Ananthan S; Faaleolea ER; Goldman RC; Hobrath JV; Kwong CD; Laughon BE; Maddry JA; Mehta A; Rasmussen L; Reynolds RC; Secrist JA; Shindo N; Showe DN; Sosa MI; Suling WJ; White EL High-Throughput Screening for Inhibitors of Mycobacterium Tuberculosis H37Rv. Tuberculosis 2009, 89 (5), 334–353. 10.1016/j.tube.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Yuan J; Cheng KCC; Johnson RL; Huang R; Pattaradilokrat S; Liu A; Guha R; Fidock DA; Inglese J; Wellems TE; Austin CP; Su XZ Chemical Genomic Profiling for Antimalarial Therapies, Response Signatures, and Molecular Targets. Science (80-. ). 2011, 333 (6043), 724–729. 10.1126/science.1205216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Kouznetsova J; Sun W; Martínez-Romero C; Tawa G; Shinn P; Chen CZ; Schimmer A; Sanderson P; McKew JC; Zheng W; García-Sastre A Identification of 53 Compounds That Block Ebola Virus-like Particle Entry via a Repurposing Screen of Approved Drugs. Emerg. Microbes Infect. 2014, 3 (1), 1–7. 10.1038/emi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Millet JK; Tang T; Nathan L; Jaimes JA; Hsu HL; Daniel S; Whittaker GR Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J. Vis. Exp. 2019, 2019 (145), 1–9. 10.3791/59010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Lamping E; Cannon RD Use of a Yeast-Based Membrane Protein Expression Technology to Overexpress Drug Resistance Efflux Pumps. Methods Mol. Biol. 2010, 666, 219–250. 10.1007/978-1-60761-820-1_15. [DOI] [PubMed] [Google Scholar]
- (74).Giuliano KA; Gough AH; Lansing Taylor D; Vernetti LA; Johnston PA Early Safety Assessment Using Cellular Systems Biology Yields Insights into Mechanisms of Action. J. Biomol. Screen. 2010, 15 (7), 783–797. 10.1177/1087057110376413. [DOI] [PubMed] [Google Scholar]
- (75).Sakamuru S; Li X; Attene-Ramos MS; Huang R; Lu J; Shou L; Shen M; Tice RR; Austin CP; Xia M Application of a Homogenous Membrane Potential Assay to Assess Mitochondrial Function. Physiol. Genomics 2012, 44 (9), 495–503. 10.1152/physiolgenomics.00161.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Padilla S; Corum D; Padnos B; Hunter DL; Beam A; Houck KA; Sipes N; Kleinstreuer N; Knudsen T; Dix DJ; Reif DM; Carolina N Zebrafish Developmental Screening of the ToxCast TM Phase I Chemical Library. Reprod. Toxicol. 2012, 33 (2), 174–187. 10.1016/j.reprotox.2011.10.018. [DOI] [PubMed] [Google Scholar]
- (77).Tropsha A Best Practices for QSAR Model Development, Validation, and Exploitation. Mol. Inform. 2010, 29 (6–7), 476–488. 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- (78).Laufersweiler MC; Gadagbui B; Baskerville-Abraham IM; Maier A; Willis A; Scialli AR; Carr GJ; Felter SP; Blackburn K; Daston G Correlation of Chemical Structure with Reproductive and Developmental Toxicity as It Relates to the Use of the Threshold of Toxicological Concern. Regul. Toxicol. Pharmacol. 2012, 62 (1), 160–182. 10.1016/j.yrtph.2011.09.004. [DOI] [PubMed] [Google Scholar]
- (79).Kroes R; Renwick AG; Cheeseman M; Kleiner J; Mangelsdorf I; Piersma A; Schilter B; Schlatter J; Van Schothorst F; Vos JG; Würtzen G Structure-Based Thresholds of Toxicological Concern (TTC): Guidance for Application to Substances Present at Low Levels in the Diet. Food Chem. Toxicol. 2004, 42 (1), 65–83. 10.1016/j.fct.2003.08.006. [DOI] [PubMed] [Google Scholar]
- (80).Wu MY; Hill CS TGF-β Superfamily Signaling in Embryonic Development and Homeostasis. Dev. Cell 2009, 16 (3), 329–343. 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- (81).Wei Z; Sakamuru S; Zhang L; Zhao J; Huang R; Kleinstreuer NC; Chen Y; Shu Y; Knudsen TB; Xia M Identification and Profiling of Environmental Chemicals That Inhibit the TGFβ/SMAD Signaling Pathway. Chem. Res. Toxicol. 2019, 32 (12), 2433–2444. 10.1021/acs.chemrestox.9b00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).van Gelder MMHJ; van Rooij IALM; Miller RK; Zielhuis GA; de Jong-van den Berg, L. T. W.; Roeleveld, N. Teratogenic Mechanisms of Medical Drugs. Hum. Reprod. Update 2010, 16 (4), 378–394. 10.1093/humupd/dmp052. [DOI] [PubMed] [Google Scholar]
- (83).Solinski HJ; Dranchak P; Oliphant E; Gu X; Earnest TW; Braisted J; Inglese J; Hoon MA Inhibition of Natriuretic Peptide Receptor 1 Reduces Itch in Mice. Sci. Transl. Med. 2019, 11 (500), 1–15. 10.1126/scitranslmed.aav5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Bartels CF; Bükülmez H; Padayatti P; Rhee DK; Van Ravenswaaij-Arts C; Pauli RM; Mundlos S; Chitayat D; Shih LY; Al-Gazali LI; Kant S; Cole T; Morton J; Cormier-Daire V; Faivre L; Lees M; Kirk J; Mortier GR; Leroy J; Zabel B; Kim CA; Crow Y; Braverman NE; Van Den Akker F; Warman ML Mutations in the Transmembrane Natriuretic Peptide Receptor NPR-B Impair Skeletal Growth and Cause Acromesomelic Dysplasia, Type Maroteaux. Am. J. Hum. Genet. 2004, 75 (1), 27–34. 10.1086/422013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Martin MT; Dix DJ; Judson RS; Kavlock RJ; Reif DM; Richard AM; Rotroff DM; Romanov S; Medvedev A; Poltoratskaya N; Gambarian M; Moeser M; Makarov SS; Houck KA Impact of Environmental Chemicals on Key Transcription Regulators and Correlation to Toxicity End Points within EPA’s ToxCast Program. Chem. Res. Toxicol. 2010, 23 (3), 578–590. 10.1021/tx900325g. [DOI] [PubMed] [Google Scholar]
- (86).Romanov S; Medvedev A; Gambarian M; Poltoratskaya N; Moeser M; Medvedeva L; Gambarian M; Diatchenko L; Makarov S Homogeneous Reporter System Enables Quantitative Functional Assessment of Multiple Transcription Factors. Nat. Methods 2008, 5 (3), 253–260. 10.1038/nmeth.1186. [DOI] [PubMed] [Google Scholar]
- (87).Odom DT; Dowell RD; Jacobsen ES; Nekludova L; Rolfe PA; Danford TW; Gifford DK; Fraenkel E; Bell GI; Young RA Core Transcriptional Regulatory Circuitry in Human Hepatocytes. Mol. Syst. Biol. 2006, 2, 1–5. 10.1038/msb4100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Benitez CM; Goodyer WR; Kim SK Deconstructing Pancreas Developmental Biology. Cold Spring Harb. Perspect. Biol. 2012, 4 (6), 1–17. 10.1101/cshperspect.a012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Kleinstreuer N; Dix D; Rountree M; Baker N; Sipes N; Reif D; Spencer R; Knudsen T A Computational Model Predicting Disruption of Blood Vessel Development. 2013, 9 (4). 10.1371/journal.pcbi.1002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Oesterheld JR A Review of Developmental Aspects of Cytochrome P450. J. Child Adolesc. Psychopharmacol. 1998, 8 (3), 161–174. 10.1089/cap.1998.8.161. [DOI] [PubMed] [Google Scholar]
- (91).Miller MS; Juchau MR; Guengerich FP; Nebert DW; Raucy JL Drug Metabolic Enzymes in Developmental Toxicology. Fundam. Appl. Toxicol. 1996, 34 (2), 165–175. 10.1006/faat.1996.0187. [DOI] [PubMed] [Google Scholar]
- (92).Simpson ER; Clyne C; Rubin G; Wah CB; Robertson K; Britt K; Speed C; Jones M Aromatase - A Brief Overview. Annu. Rev. Physiol. 2002, 64, 93–127. 10.1146/annurev.physiol.64.081601.142703. [DOI] [PubMed] [Google Scholar]
- (93).Organization for Economic Co-operation and Development. OECD Principles for the Validation, for Regulatory Purposes, of (Quantitative) Structure-Activity Relationship Models; 2004. [Google Scholar]
- (94).Ahlborg UG; Thunberg TM; Spencer HC Chlorinated Phenols: Occurrence, Toxicity, Metabolism, and Environmental Impact. Crit. Rev. Toxicol. 1980, 7 (1), 1–35. 10.3109/10408448009017934. [DOI] [PubMed] [Google Scholar]
- (95).Stringfellow WT; Domen JK; Camarillo MK; Sandelin WL; Borglin S Physical, Chemical, and Biological Characteristics of Compounds Used in Hydraulic Fracturing. J. Hazard. Mater. 2014, 275, 37–54. 10.1016/j.jhazmat.2014.04.040. [DOI] [PubMed] [Google Scholar]
- (96).Jett DA; Guignet M; Supasai S; Lein PJ Developmental Toxicity within the Central Cholinergic Nervous System, Second Edition.; Elsevier Inc., 2018. 10.1016/B978-0-12-809405-1.00016-X. [DOI] [Google Scholar]
- (97).Leung MCK; Phuong J; Baker NC; Sipes NS; Klinefelter GR; Martin MT; McLaurin KW; Woodrow Setzer R; Darney SP; Judson RS; Knudsen TB Systems Toxicology of Male Reproductive Development: Profiling 774 Chemicals for Molecular Targets and Adverse Outcomes. Environ. Health Perspect. 2016, 124 (7), 1050–1061. 10.1289/ehp.1510385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Yamashita M Apoptosis in Zebrafish Development. Comp. Biochem. Physiol. - B Biochem. Mol. Biol. 2003, 136 (4), 731–742. 10.1016/j.cbpc.2003.08.013. [DOI] [PubMed] [Google Scholar]
- (99).Wang C; Wang T; Lian BW; Lai S; Li S; Li YM; Tan WJ; Wang B; Mei W Developmental Toxicity of Cryptotanshinone on the Early-Life Stage of Zebrafish Development. Hum. Exp. Toxicol. 2021, 40 (12_suppl), S278–S289. 10.1177/09603271211009954. [DOI] [PubMed] [Google Scholar]
- (100).Noyes PD; Friedman KP; Browne P; Haselman JT; Gilbert ME; Hornung MW; Barone S; Crofton KM; Laws SC; Stoker TE; Simmons SO; Tietge JE; Degitz SJ Evaluating Chemicals for Thyroid Disruption: Opportunities and Challenges with in Vitro Testing and Adverse Outcome Pathway Approaches. Environ. Health Perspect. 2019, 127 (9). 10.1289/EHP5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Singh AV; Knudsen KB; Knudsen TB Computational Systems Analysis of Developmental Toxicity: Design, Development and Implementation of a Birth Defects Systems Manager (BDSM). Reprod. Toxicol. 2005, 19 (3 SPEC. ISS.), 421–439. 10.1016/j.reprotox.2004.11.008. [DOI] [PubMed] [Google Scholar]
- (102).Waye A; Trudeau VL Neuroendocrine Disruption: More than Hormones Are Upset. J. Toxicol. Environ. Heal. - Part B Crit. Rev. 2011, 14 (5–7), 270–291. 10.1080/10937404.2011.578273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Souza BR; Romano-Silva MA; Tropepe V Dopamine D2 Receptor Activity Modulates Akt Signaling and Alters GABaergic Neuron Development and Motor Behavior in Zebrafish Larvae. J. Neurosci. 2011, 31 (14), 5512–5525. 10.1523/JNEUROSCI.5548-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Iguchi T; Watanabe H; Katsu Y; Mizutani T; Miyagawa S; Suzuki A; Kohno S; Sone K; Kato H Developmental Toxicity of Estrogenic Chemicals and Other Species. Congenit Anom 2002, 42 (2), 94–105. 10.1111/j.1741-4520.2002.tb00858.x. [DOI] [PubMed] [Google Scholar]
- (105).Li Y; Farooq M; Sheng D; Chandramouli C; Lan T; Mahajan NK; Kini RM; Hong Y; Lisowsky T; Ge R Augmenter of Liver Regeneration (Alr) Promotes Liver Outgrowth during Zebrafish Hepatogenesis. PLoS One 2012, 7 (1), e30835. 10.1371/journal.pone.0030835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Christians E; Davis AA; Thomas SD; Benjamin IJ Maternal Effect of Hsf1 on Reproductive Success. Nature 2000, 407 (6805), 693–694. 10.1038/35037669. [DOI] [PubMed] [Google Scholar]
- (107).Johnson RS; Van Lingen B; Papaioannou VE; Spiegelman BM A Null Mutation at the C-Jun Locus Causes Embryonic Lethality and Retarded Cell Growth in Culture. Genes Dev. 1993, 7 (7 B), 1309–1317. 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- (108).Hilberg F; Aguzzi A; Howells N; Wagner EF C-Jun Is Essential for Normal Mouse Development and Hepatogenesis. Nature 1993, 365, 179–181. 10.1038/366368a0. [DOI] [PubMed] [Google Scholar]
- (109).Jochum W; Passegué E; Wagner EF AP-1 in Mouse Development and Tumorigenesis. Oncogene 2001, 20 (19 REV. ISS. 2), 2401–2412. 10.1038/sj.onc.1204389. [DOI] [PubMed] [Google Scholar]
- (110).Sipes NS; Martin MT; Reif DM; Kleinstreuer NC; Judson RS; Singh A V; Chandler KJ; Dix DJ; Kavlock RJ; Knudsen TB Predictive Models of Prenatal Developmental Toxicity from ToxCast High-Throughput Screening Data. Toxicol. Sci. 2011, 124 (1), 109–127. 10.1093/toxsci/kfr220. [DOI] [PubMed] [Google Scholar]
- (111).Barak Y; Sadovsky Y; Shalom-Barak T PPAR Signaling in Placental Development and Function. PPAR Res. 2008, 2008. 10.1155/2008/142082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Azarbayjani F; Danielsson BR Embryonic Arrhythmia by Inhibition of HERG Channels: A Common Hypoxia-Related Teratogenic Mechanism for Antiepileptic Drugs? Epilepsia 2002, 43 (5), 457–468. 10.1046/j.1528-1157.2002.28999.x. [DOI] [PubMed] [Google Scholar]
- (113).Danielsson BR; Sköld AC; Johansson A; Dillner B; Blomgren B Teratogenicity by the HERG Potassium Channel Blocking Drug Almokalant: Use of Hypoxia Marker Gives Evidence for a Hypoxia-Related Mechanism Mediated via Embryonic Arrhythmia. Toxicol. Appl. Pharmacol. 2003, 193 (2), 168–176. 10.1016/j.taap.2003.07.002. [DOI] [PubMed] [Google Scholar]
- (114).Knapen D; Vergauwen L; Villeneuve DL; Ankley GT The Potential of AOP Networks for Reproductive and Developmental Toxicity Assay Development. Reprod. Toxicol. 2015, 56, 52–55. 10.1016/j.reprotox.2015.04.003. [DOI] [PubMed] [Google Scholar]
- (115).Kelce WR; Monosson E; Gamcsik MP; Laws SC; Gray LE Environmental Hormone Disruptors: Evidence That Vinclozolin Developmental Toxicity Is Mediated by Antiandrogenic Metabolites. Toxicology and Applied Pharmacology. 1994, pp 276–285. 10.1006/taap.1994.1117. [DOI] [PubMed] [Google Scholar]
- (116).Hahn ME Aryl Hydrocarbon Receptors: Diversity and Evolution. Chem. Biol. Interact. 2002, 141 (1–2), 131–160. 10.1016/s0009-2797(02)00070-4. [DOI] [PubMed] [Google Scholar]
- (117).Knudsen TB; Kleinstreuer NC Disruption of Embryonic Vascular Development in Predictive Toxicology. Birth Defects Res. Part C - Embryo Today Rev. 2011, 93 (4), 312–323. 10.1002/bdrc.20223. [DOI] [PubMed] [Google Scholar]
- (118).Chen F; Zamule SM; Coslo DM; Chen T; Omiecinski CJ The Human Constitutive Androstane Receptor Promotes the Differentiation and Maturation of Hepatic-like Cells. Dev. Biol. 2013, 384 (2), 155–165. 10.1016/j.ydbio.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (119).Wyde ME; Kirwan SE; Zhang F; Laughter A; Hoffman HB; Bartolucci-Page E; Gaido KW; Yan B; You L Di-n-Butyl Phthalate Activates Constitutive Androstane Receptor and Pregnane X Receptor and Enhances the Expression of Steroid-Metabolizing Enzymes in the Liver of Rat Fetuses. Toxicol. Sci. 2005, 86 (2), 281–290. 10.1093/toxsci/kfi204. [DOI] [PubMed] [Google Scholar]
- (120).Farag AT; El Okazy AM; El-Aswed AF Developmental Toxicity Study of Chlorpyrifos in Rats. Reprod. Toxicol. 2003, 17, 203–208. 10.1006/rtph.1998.1261. [DOI] [PubMed] [Google Scholar]
- (121).Chitu V; Stanley ER Regulation of Embryonic and Postnatal Development by the CSF-1 Receptor, 1st ed.; Elsevier Inc., 2017; Vol. 123. 10.1016/bs.ctdb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Sakamoto T; Matsuura TR; Wan S; Ryba DM; Kim J; Won KJ; Lai L; Petucci C; Petrenko N; Musunuru K; Vega RB; Kelly DP A Critical Role for Estrogen-Related Receptor Signaling in Cardiac Maturation. Circ. Res. 2020, 126 (12), 1685–1702. 10.1161/CIRCRESAHA.119.316100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Wagner EF Functions of AP1 (Fos/Jun) in Bone Development. Ann. Rheum. Dis. 2002, 61 (SUPPL. 2), 40–42. 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Leung L; Kwong M; Hou S; Lee C; Chan JY Deficiency of the Nrf1 and Nrf2 Transcription Factors Results in Early Embryonic Lethality and Severe Oxidative Stress. J. Biol. Chem. 2003, 278 (48), 48021–48029. 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- (125).Das Neves L; Duchala CS; Godinho F; Haxhiu MA; Colmenares C; Macklin WB; Campbell CE; Butz KG; Gronostajski RM Disruption of the Murine Nuclear Factor I-A Gene (Nfia) Results in Perinatal Lethality, Hydrocephalus, and Agenesis of the Corpus Callosum. Proc. Natl. Acad. Sci. U. S. A. 1999, 96 (21), 11946–11951. 10.1073/pnas.96.21.11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (126).Nishimura K; Nakano N; Chowdhury VS; Kaneto M; Torii M; Hattori M aki; Yamauchi, N.; Kawai, M. Effect of PPARβ/δ Agonist on the Placentation and Embryo-Fetal Development in Rats. Birth Defects Res. Part B - Dev. Reprod. Toxicol. 2013, 98 (2), 164–169. 10.1002/bdrb.21052. [DOI] [PubMed] [Google Scholar]
- (127).Fang H; Fang W; Cao H; Luo S; Cong J; Liu S; Pan F; Jia X Di-(2-Ethylhexyl)-Phthalate Induces Apoptosis via the PPARγ/PTEN/AKT Pathway in Differentiated Human Embryonic Stem Cells. Food Chem. Toxicol. 2019, 131 (November 2018), 110552. 10.1016/j.fct.2019.05.060. [DOI] [PubMed] [Google Scholar]
- (128).Vazão H; Rosa S; Barata T; Costa R; Pitrez PR; Honório I; De Vries MR; Papatsenko D; Benedito R; Saris D; Khademhosseini A; Quax PHA; Pereira CF; Mercader N; Fernandes H; Ferreira L High-Throughput Identification of Small Molecules That Affect Human Embryonic Vascular Development. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (15), E3022–E3031. 10.1073/pnas.1617451114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Tal T; Kilty C; Smith A; LaLone C; Kennedy B; Tennant A; McCollum CW; Bondesson M; Knudsen T; Padilla S; Kleinstreuer N Screening for Angiogenic Inhibitors in Zebrafish to Evaluate a Predictive Model for Developmental Vascular Toxicity. Reprod. Toxicol. 2017, 70, 70–81. 10.1016/j.reprotox.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (130).Kupsco A; Schlenk D Oxidative Stress, Unfolded Protein Response, and Apoptosis in Developmental Toxicity; Elsevier, 2015; Vol. 317. 10.1016/bs.ircmb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


