Abstract
Recombinant phages are generated when Lactococcus lactis subsp. lactis harboring plasmids encoding the abortive type (Abi) of phage resistance mechanisms is infected with small isometric phages belonging to the P335 species. These phage variants are likely to be an important source of virulent new phages that appear in dairy fermentations. They are distinguished from their progenitors by resistance to Abi defenses and by altered genome organization, including regions of L. lactis chromosomal DNA. The objective of this study was to characterize four recombinant variants that arose from infection of L. lactis NCK203 (Abi+) with phage φ31. HindIII restriction maps of the variants (φ31.1, φ31.2, φ31.7, and φ31.8) were generated, and these maps revealed the regions containing recombinant DNA. The recombinant region of phage φ31.1, the variant that occurred most frequently, was sequenced and revealed 7.8 kb of new DNA compared with the parent phage, φ31. This region contained numerous instances of homology with various lactococcal temperate phages, as well as homologues of the lambda recombination protein BET and Escherichia coli Holliday junction resolvase Rus, factors which may contribute to efficient recombination processes. A sequence analysis and phenotypic tests revealed a new origin of replication in the φ31.1 DNA, which replaced the φ31 origin. Three separate HindIII fragments, accounting for most of the recombinant region of φ31.1, were separately cloned into gram-positive suicide vector pTRK333 and transformed into NCK203. Chromosomal insertions of each plasmid prevented the appearance of different combinations of recombinant phages. The chromosomal insertions did not affect an inducible prophage present in NCK203. Our results demonstrated that recombinant phages can acquire DNA cassettes from different regions of the chromosome in order to overcome Abi defenses. Disruption of these regions by insertion can alter the types and diversity of new phages that appear during phage-host interactions.
Bacteriophages continue to be a significant economic problem for the dairy industry (9, 30). Starter culture strains which contain naturally occurring phage defense mechanisms can control the problem if they are used in conjunction with sanitation measures. Bacteriophages, nevertheless, have the capacity to evolve and rapidly overcome host defense mechanisms (9, 13, 22, 29). The logical routes of phage evolution include point mutations, deletions, and acquisition of new DNA. Coinfecting phages and the genomic contents of the host cell, which can include functional or defective prophages, are potential sources of new DNA. Identification of the genetic routes by which phages adapt and evolve against industrial starter cultures will be an important part of controlling the appearance of new phages in fermentation environments.
Lactococcal phages are classified into 12 species based on morphology and DNA homologies (20). The c2 (prolate-headed) species and the 936 and P335 (small isometric-headed) species are the most important taxa, since these are the major organisms that disrupt dairy fermentations worldwide. While the 936 species is composed of only lytic phages, P335 species exhibit high levels of DNA homology between temperate and lytic members (17, 20). P335 phages have been appearing in cheese plants with increasing frequency in recent years and are now considered members of an important new phage species (1, 17, 21, 28).
Workers in our laboratory have previously characterized a number of phages belonging to the P335 species which are virulent for Lactococcus lactis NCK203. In response to the inhibitory pressure of an abortive type of defense (AbiC) (12), phage ul36 can acquire chromosomal sequences from the host in a recombinational process that generates a related but new virulent phage, ul37 (8, 29). Phage ul37 is not inhibited by AbiC and differs in its base plate and tail morphology. The nature and extent of the sequences and their number or location in the Lactococcus chromosome have not been determined. Knockout insertions directed into the bacterial chromosome in the regions acquired by ul37 eliminated the AbiC-induced metamorphosis of ul36 to ul37.
Recombinant derivatives of φ31, a P335 phage distinct from ul36, have also appeared after infection of NCK203 harboring either AbiA or Per31 defense mechanisms on high-copy-number replicons (8, 31). Both AbiA and Per31 are abortive infection defense mechanisms that interfere with the replication of the phage genome in the host. While the mechanism by which abiA interferes with DNA replication remains to be elucidated, per31 cloned in trans on a plasmid appears to titrate phage replication factors to a false origin after infection (14, 26, 31). Collectively, these observations suggest that the recombinational events that lead to new virulent phages are not limited to a single phage-host Abi combination and may be characteristic of the evolutionary routes exploited by P335 phages to overcome abortive defense mechanisms. The objectives of the present study were to characterize the number and variety of recombinant phages that can arise from NCK203 harboring per31 after challenge with phage φ31, to identify the chromosomal sequences which contribute to the events, and to determine if disruption of selected chromosomal loci alters the types and ratios of recombinant phages that emerge.
MATERIALS AND METHODS
Bacterial strains, plasmids, and bacteriophages.
Table 1 lists the strains, plasmids, and bacteriophages used in this work. L. lactis subsp. lactis strains were grown at 30°C in M17 medium (Difco Laboratories, Detroit, Mich.) supplemented with 0.5% glucose (M17G). Erythromycin and chloramphenicol were added as needed at concentrations of 1.5 and 7.5 μg/ml, respectively. Escherichia coli strains were grown in Luria-Bertani medium (36) or brain heart infusion medium (Difco Laboratories) supplemented with 200 μg of erythromycin per ml or 50 μg of ampicillin per ml as needed. Bacterial stock cultures were stored at −20°C in the appropriate medium supplemented with 10% (vol/vol) glycerol. Phages were propagated by using L. lactis subsp. lactis NCK203 or one of its derivatives, and phage titers were determined by standard double-layer agar plate methods (40). Individual plaques were propagated by transferring them into 3.5 to 5 ml of M17G containing 100 mM CaCl2 and inoculating the preparations with 35 to 50 μl of an overnight culture of L. lactis. The tubes were incubated at 30°C and, after cell lysis, centrifuged to pellet the cellular debris. The phage lysates were then filtered through a 0.45-μm-pore-size syringe filter (Nalgene Co., Rochester, N.Y.).
TABLE 1.
Bacterial strains, plasmids, and phages
| Strain, plasmid, or phage | Relevant characteristics(s)a | Source or reference(s) |
|---|---|---|
| L. lactis subsp. lactis strains | ||
| NCK203 | Propagating host for phage φ31, the φ31 variants, ul36, and ul37 | 15, 37 |
| NCK203-A | Insertion mutant of NCK203 produced by single crossover insertion with pTRK333-A | This study |
| NCK203-B | Insertion mutant of NCK203 produced by single crossover insertion with pTRK333-B | This study |
| NCK203-D | Insertion mutant of NCK203 produced by single cross-over insertion with pTRK333-D | This study |
| E. coli MC1061 | Transformation host | 16 |
| Plasmids | ||
| pTRK333 | E. coli-gram-positive suicide shuttle vector, Apr Cmr Tcr, 5.8 kb | 29 |
| pTRK333-A | pTRK333 + 3.4-kb fragment A of φ31.1, Cmr, 9.2 kb | This study |
| pTRK333-B | pTRK333 + 1.8-kb fragment B of φ31.1, Cmr, 7.5 kb | This study |
| pTRK333-D | pTRK333 + 1.8-kb fragment D of φ31.1, Cmr, 7.5 kb | This study |
| pTRK361 | Per31+, Emr, 11.4 kb | 31 |
| pTRK637 | pTRKH2 + 2.0-kb fragment B of φ31.1, Emr, 8.7 kb | This study |
| pTRK638 | pTRKH2 + 1.5-kb subfragment of fragment B of φ31.1, Emr, 8.2 kb | This study |
| pTRK639 | pTRKH2 + 0.5-kb subfragment of fragment B of φ31.1, Emr, 7.2 kb | This study |
| pTRK640 | pTRKH2 + 0.4 kb subfragment of fragment B of φ31.1, Emr, 7.1 kb | This study |
| pSA3 | E. coli-gram-positive shuttle vector, Cmr Tcr Emr, 10.2 kb | 5 |
| Bacteriophages | ||
| φ31 | SI, P335 species, 31.9 kb | 1 |
| φ31.1 | Recombinant variant of φ31, 30.1 kb | 31 |
| φ31.2 | Recombinant variant of φ31, 29.9 kb | 31 |
| φ31.7 | Recombinant variant of φ31, 29.1 kb | This study |
| φ31.8 | Recombinant variant of φ31, 30.5 kb | This study |
| ul36 | SI, P335 species, 28.8 kb | 27 |
| ul37 | Recombinant variant of ul36, 31.1 kb | 29 |
Abbreviations: Apr, ampicillin resistance; Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Tcr, tetracycline resistance; SI, small isometric headed.
DNA isolation.
E. coli plasmid DNA was isolated by using standard alkaline lysis procedures (36). Plasmids were isolated from lactococcal cells as described by O'Sullivan and Klaenhammer (32), except that ethidium bromide was not used. Genomic DNA was isolated from Lactococcus strains as follows. An overnight culture in M17G (1.5 to 4.0 ml) was centrifuged, and the pellet was resuspended in 500 μl of 50 mM Tris-HCl buffer (pH 8.0). Approximately 1 mg of powdered lysozyme was added, and the preparation was incubated for 15 to 20 min at 37°C. The cell suspension was then extracted twice with 450 μl of phenol and 50 μl of chloroform-isoamyl alcohol (23:1, vol/vol); this was followed by two extractions with 500 μl of chloroform-isoamyl alcohol. The nucleic acids were precipitated with 0.1 volume of 3 M sodium acetate and 2 volumes of ethanol and pelleted with a microcentrifuge. The final pellet was washed with 70% ethanol and resuspended in 15 to 30 μl of TE buffer (pH 7.6) containing RNase.
Phage DNA was prepared as follows. Four to five milliliters of phage lysate was incubated for 1 h at 37°C after 3 μl of a solution containing 3 mg of DNase per ml and 3 mg of RNase per ml was added. Polyethylene glycol 8000 and NaCl were added to final concentrations of 10% and 0.5 M, respectively. After gentle mixing the preparations were incubated overnight at 4°C. The phage was pelleted by centrifugation at 4,000 × g and air dried. The phage pellets were resuspended in 500 μl of 50 mM Tris (pH 8.0) for DNA extraction. The procedure for lactococcal genomic DNA extraction described above was used, except that the lysozyme step was omitted.
DNA manipulations.
Restriction endonuclease digestion and ligation were performed as described by Sambrook et al. (36). For gene cloning, DNA fragments were isolated from agarose gel slices by using a GeneClean II kit (Bio 101, La Jolla, Calif.) according to the manufacturer's instructions. Vector fragments were dephosphorylated with shrimp alkaline phosphatase (Amersham Pharmacia Biotech, Piscataway, N.J.). Electroporation of both L. lactis and E. coli cells was carried out as described by Dower et al. (11) with a Gene Pulser apparatus (Bio-Rad, Richmond, Calif.) set at 25 μF, 2.0 kV, and 200 Ω; 0.2-cm cuvettes were used.
Southern transfer of DNA from electrophoresis gels was accomplished by using Magnacharge nylon transfer membranes (MSI, Westboro, Mass.) and the instructions of the manufacturer for alkaline transfer. Hybridizations in which 32P-labeled probes were used were carried out in a hybridization oven (Robbins, Sunnyvale, Calif.) by following the manufacturer's protocol at 65°C for 4 to 18 h; 7% sodium dodecyl sulfate (SDS)–0.25 M NaH2PO4 hybridization buffer (pH 7.4) was used. The membranes were washed at room temperature with two wash buffers, 2× SSC–0.1% SDS and 0.1× SSC–0.1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
DNA sequencing was accomplished by using Tn1000 insertions as described by Strathmann et al. (39) and instructions kindly provided by Gold Biotechnology, Inc. (St. Louis, Mo.). Each of the five HindIII fragments of the recombinant region of φ31.1 was separately cloned into the vector pMOB. Isolates with Tn1000 insertions were sequenced from both ends of the inserted transposon with the following primers: G186 (ATATAAACAACGAATTATCTCC) and G187 (GTATTATAATCAATAAGTTATACC). Double-stranded DNA for sequencing was isolated by using standard miniprep procedures or a PERFECTprep plasmid DNA kit (5 Prime-3 Prime, Inc., Boulder, Colo.). Sequencing was accomplished by using T7 Sequenase 2.0 DNA sequencing kits or a Thermo Sequenase cycle sequencing kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. After assembly of the contigs with DNASIS for Windows (Hitachi Software, San Bruno, Calif.), oligo primers were designed by using Primer Designer software (Scientific and Educational Software, Durham, N.C.) to sequence through gaps and to link the separate HindIII fragments. The sequence was analyzed with Clone Manager software (Scientific and Educational Software). Sequence homology searches were carried out by using the BLAST algorithms of the National Center for Biotechnology Information.
Mitomycin C induction.
L. lactis NCK203 and its derivatives were grown in M17G at 30°C to an optical density at 600 nm of 0.2. Mitomycin C (Sigma Chemical Co., St. Louis, Mo.) was added to a final concentration of 10 μg/ml. The optical density at 600 nm was monitored for 4 h, and a cell sample was removed after 100 min. The cells were pelleted and the DNA was extracted by the method described above for lactococcal genomic DNA.
PCR.
Phage DNA was isolated from φ31.1 and genomic DNAs were isolated from NCK203 and its derivatives as described above, and these DNAs were used as PCR templates. PCR products were generated by using Taq DNA polymerase obtained from Boehringer Mannheim (Indianapolis, Ind.) by following the manufacturer's protocols. For one experiment (see Fig. 6), the following primer sets were used: set A, consisting of ATAGGGCCTCAAACGAGCTTATCAAATTATCA and TCTACTGCTCAGGATTAGTG; set B, consisting of GTTGCAGAATATCCGGCCAC and TTGACTTCTTCGCCATCTGC; set D, consisting of TCACATTCTGGACATTCTAA and AATTACGGAATCTTGAGCGCTT; and the amp set, consisting of GCAGCAGATTACGCGCAGAA and TTAGACGTCAGGTGGCACTT. Reactions were performed by using an annealing temperature of 52°C and an extension temperature of 68°C for 3.5 min. To subclone regions of φ31.1 (see Fig. 4), the following primer sets were used: for pTRK637, pTRK638, and pTRK639, left primer GCAAGAGCATTATCTCAACCGGAAGTAG and right primers TCCATAACCGTCACATCTTGCTTTCT, CTGATAGCCCGATTTAATTC, and CCGTAAGAATTGGCCATAGTATATATTT, respectively; and for pTRK640, ATACTATGGCCAATTCTTACGGAAGTAT and TAATCTCTTCGTCTGTCGTTCCAGATTT. Reactions were performed by using an annealing temperature of 50°C and an extension temperature of 68°C for 2 min.
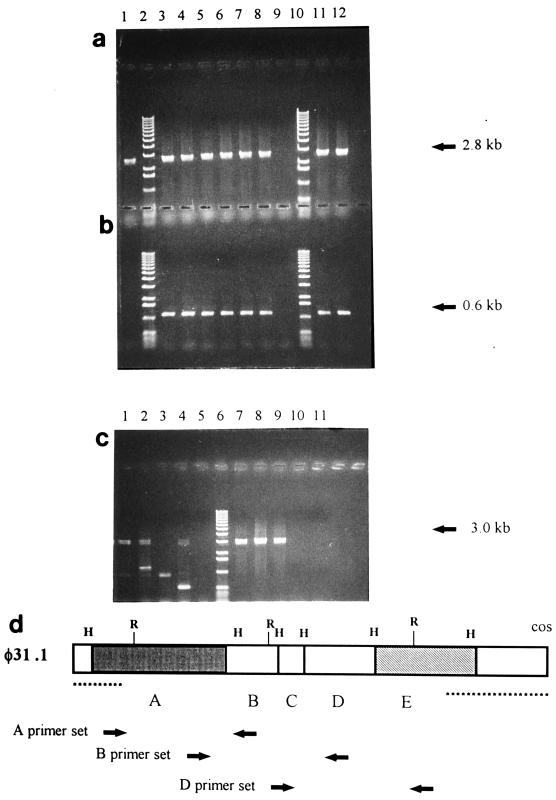
FIG. 6.
PCR products, showing that the pTRK333A chromosomal insertion did not occur in the region contributing to φ31.1. (a and b) Template DNA from NCK203 (lane 1), six single-colony isolates of NCK203-A (lanes 3 through 8), NCK203-B (lane 11), or NCK203-D (lane 12) and markers (lanes 2 and 10). Lane 9 contained a PCR control without template DNA. The primers used were set A (a) (expected product size, 2.8 kb) and the vector set (b) (expected product size, 0.6 kb). (c) Template DNA from NCK203 (lanes 1 and 7), NCK203-A (lanes 2 and 8), NCK203-B (lanes 3 and 9), or NCK203-D (lanes 4 and 10) and markers (lane 6). Lanes 5 and 11 contained a PCR control without template DNA. The primers used were set B (lanes 1 through 5) (expected product size, 3.0 kb) and set D (lanes 7 through 11) (expected product size, 3.1 kb). The smaller products obtained with primer set B were artifacts of one of the primers, which annealed to the vector, pTRK333. (d) Map of the recombinant region of φ31.1, similar to the map in Fig. 2b, showing the locations of the primer sets designed and used for the PCR experiment.
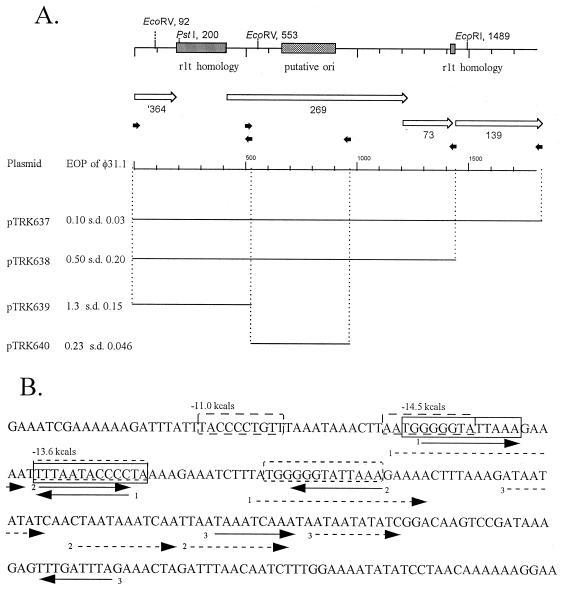
FIG. 4.
Localization of the putative origin region of phage φ31.1. (A) Schematic drawing of the region, showing predicted ORFs and regions of phage rlt homology. The small arrows indicate the positions of oligo primers used in PCR experiments to create subclones. The bars indicate the fragments cloned into pTRKH2. The EOPs are averages based on three or four replicates. (B) Sequence of the 240-bp shaded ori region. The solid arrows indicate indirect repeats, the dashed arrows indicate direct repeats, and the boxes indicate the stem regions of predicted stem-loops regions. s.d., standard deviation.
RESULTS
Isolation of recombinant variants of φ31.
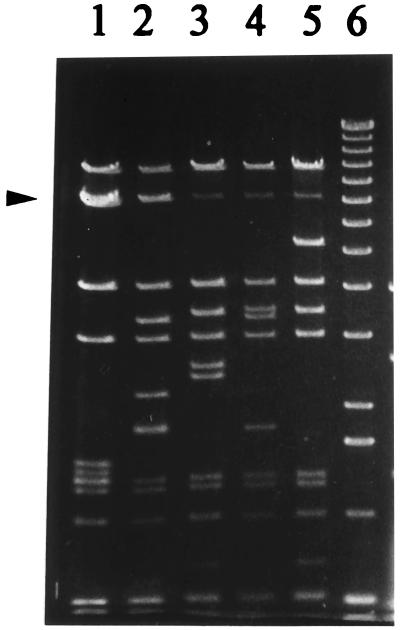
L. lactis NCK203(pTRK361), which encoded per31 on high-copy-number replicon pTRKH2, was challenged in multiple experiments with phage φ31 (Table 1). Consistent with previous reports (8, 31), the average efficiency of plaquing (EOP) of φ31 on this host was 7 × 10−7, and the plaques which formed at this very low frequency appeared to be normal with no reduction in size despite the extremely efficient Abi defense mechanism provided by Per31. Phage were purified from isolated plaques, propagated on NCK203, and then titrated on NCK203(pTRK361). All of these phage were completely resistant to the Per31 phenotype (Perr) and exhibited an EOP of 1.0 on this host. Electron micrographs of φ31 and four of the mutant phages revealed no discernable morphological differences among the phages (data not shown). DNAs were isolated from Per31r phages and were digested with HindIII and EcoRI. Although many bands were identical to bands in the φ31 pattern, four characteristic fragmentation patterns were observed for the new phages; these phages were designated φ31.1, φ31.2, φ31.7, and φ31.8, and the HindIII fragments are shown in Fig. 1. The most frequently found type of Per31r phage occurred in 30 of the 48 plaques examined (Table 2); this phage was similar, as determined by restriction analysis, to φ31.1, which was initially described by O'Sullivan et al. (31). HindIII and EcoRI fingerprints characteristic of phages φ31.2, φ31.7, and φ31.8 occurred in 9, 5, and 3 of the 48 plaques examined, respectively. A Per31r variant that produced the same restriction pattern as φ31 was found in 1 of the 48 plaques examined (Table 2).
FIG. 1.
HindIII restriction digestion of φ31 (lane 1), φ31.1 (lane 2), φ31.2 (lane 3), φ31.7 (lane 4), and 31.8 (lane 5). Lane 6 contained a 1-kb marker (Gibco-BRL, Gaithersburg, Md.). The φ31 7-kb band (doublet) which is replaced in the recombinant phages is indicated by an arrowhead.
TABLE 2.
Frequency of phage types occurring after plaquing φ31 on Per31+ NCK203 and its Per31+ insertion derivatives, NCK203-A, NCK203-B, and NCK203-C
| Per31r strain harboring pTRK361 | Frequencies of recombinant phage typesa
|
Total no. of plaques examined | ||||
|---|---|---|---|---|---|---|
| φ31b | φ31.1 | φ31.2 | φ31.7 | φ31.8 | ||
| NCK203 | 1c | 30 | 9 | 5 | 3 | 48 |
| NCK203-A | 8 | 8 | 0 | 4 | 0 | 20 |
| NCK203-B | 4 | 0 | 13 | 0 | 7 | 24 |
| NCK203-D | 1 | 0 | 7 | 4 | 3 | 15 |
Each occurrence of a phage type was based on the HindIII or EcoRI band pattern of DNA from an isolated single plaque. The numbers are total numbers based on results obtained by using two or three separately propagated phage lysates.
The EOP for φ31 on all Per31+ strains was 10−6 to 10−7.
Per31r variant of φ31 which had the same restriction fragment pattern as φ31.
Mapping of the φ31-Per31r variants.
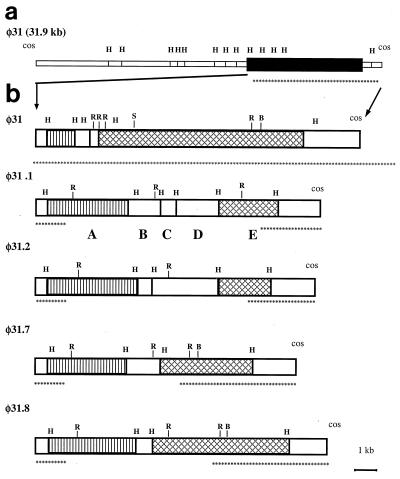
When the common HindIII fragments of the DNAs from phages φ31, φ31.1, φ31.2, φ31.7, and φ31.8 were compared with the φ31 map (1), the results indicated that ca. 8- to 9-kb region (between HindIII sites) of φ31 DNA had been replaced with new DNA in all of the variants (Fig. 1). Maps of all four phage variants were prepared by hybridizing partial HindIII digests with the labeled cos fragments of φ31; to do this, we used an approach described by Rackwitz et al. (35) for mapping DNA cloned in lambda phage vectors. The following two hybridization probes were used: a 32P-labeled 1.6-kb HindIII fragment containing the right cohesive end and a labeled 0.5-kb PvuII fragment containing the left cohesive end of φ31. The maps showed that all four variant phages and φ31 were identical except for the ca. 8- to 9-kb region where the suspected exchange occurred in the φ31 genome (Fig. 2a). In this region the number and sizes of HindIII fragments varied considerably among the recombinant phages. However, cross-hybridization experiments (data not shown) revealed that the leftmost recombinant fragments (Fig. 2b) of all of the phages were homologous, at least in part, to each other and to a smaller 1.5-kb fragment of φ31. The rightmost recombinant fragments of the phages (Fig. 2b) exhibited homology to each other and to a 7.4-kb HindIII fragment of φ31. These homologous right-end fragments of new DNA were subcloned and analyzed. Mapping of the BamHI, SalI, and EcoRI sites in these fragments indicated that phages φ31.1 and φ31.2 had acquired more DNA during the DNA exchange (approximately 9 and 8 kb, respectively) than φ31.7 and φ31.8 had acquired (approximately 5.5 and 6.5 kb, respectively).
FIG. 2.
(a) HindIII map of the complete φ31 genome. The solid box represents the region replaced in the recombinant variants, which is expanded in panel b. (b) HindIII maps of φ31 and variants φ31.1, φ31.2, φ31.7, and φ31.8, showing the recombinant regions. Similar boxes indicate HindIII fragments that exhibit DNA homology, as determined by Southern blotting. HindIII fragments found in recombinant φ31.1 are labeled A through E. The sites marked H, R, B, and S are restriction sites for HindIII, EcoRI, BamHI, and SalI, respectively. The asterisks identify areas identical to areas in φ31, as determined by restriction mapping.
The maps also revealed that the four phages could be grouped into two pairs. Phages φ31.1 and φ31.7 were identical at the left end of the recombinant region, and φ31.1 had acquired more new DNA than φ31.7 had acquired. Likewise, φ31.8 was identical to φ31.2 in the left-end region, and φ31.2 had acquired more new DNA than φ31.8 had acquired.
Sequencing of the region of φ31.1 that was acquired from L. lactis.
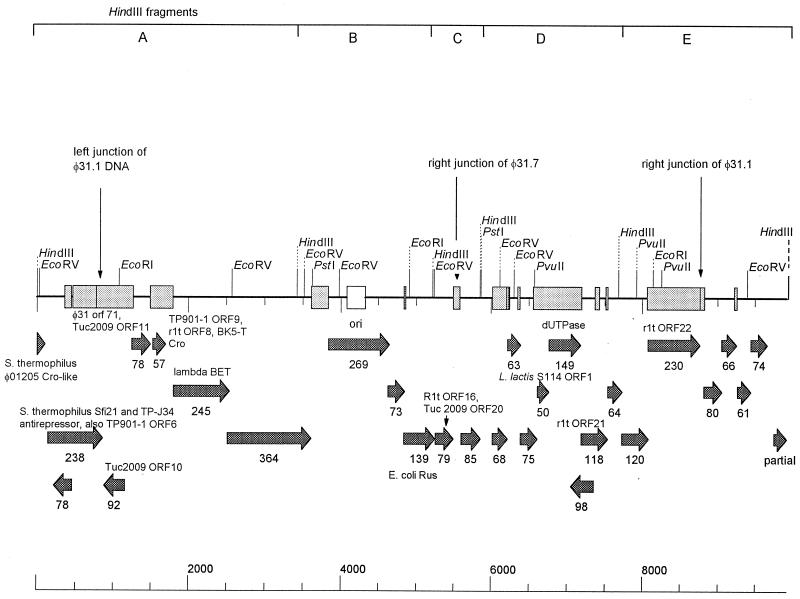
The DNA of each of the five HindIII fragments in the region (Fig. 2b, fragments A through E) was separately subcloned and sequenced. The accession number of the sequence is AF208055, and the characteristics of the sequence are shown in Fig. 3. The junctions between φ31 DNA and the region acquired from NCK203 were determined by comparing the sequence with the φ31 DNA sequence determined previously (10; S. A. Walker, personal communication) (GenBank accession no. U51128 and AF022773). Additional sequencing and PCR performed with φ31.7 confirmed that the left portion of the φ31.7 sequence was the same as the left portion of the φ31.1 sequence, but the exchanged region was smaller and the right junction for recombination with φ31 occurred upstream of the φ31.1 junction position (data not shown). The 7.8-kb portion of φ31.1 DNA-encoded sequences exhibited numerous DNA sequence level homologies (levels of identity, 91 to 99%) with lactococcal temperate phage sequences of phages rlt (41), BK5-T (3), TP901-1 (24), and Tuc2009 (26), as well as with chromosomal DNA of L. lactis subsp. cremoris S114 sequenced in conjunction with the abiN gene (34). Many of these sequences overlapped, and many were short sequences that occurred at the beginning of predicted coding regions. Some longer regions which exhibited DNA level homology encoded complete genes; these regions included open reading frame 92 (ORF92) (hypothetical protein of TP901-1, 97% identity, 98% positive), ORF57 (rlt ORF8, 89% identity, 95% positive), ORF68 (coding region of L. lactis subsp. cremoris S114, 97% identity, 97% positive), ORF149 (dUTPase of rlt, 97% identity, 99% positive), and ORF230 (rlt ORF22, 96% identity, 97% positive). Overall, the recombinant region contained 18 predicted ORFs. Three of these exhibited amino acid level homology with lactococcal and Streptococcus thermophilus predicted proteins (ORF238, ORF79, and ORF118) (Fig. 3). Two ORFs exhibited weak homology with proteins involved in DNA recombination and repair. ORF245 was homologous to the lambda recombination protein BET (25% identity, 40% positive). This protein plays a role in general recombination and in the late, rolling-circle mode of lambda DNA replication (38); it is a single-stranded DNA binding protein that can promote renaturation of DNA. ORF139 of φ31.1 was homologous to the E. coli Rus protein (25) (29% identity, 46% positive). The Rus protein is an endonuclease that can resolve Holliday junction intermediates and correct defects in genetic recombination and DNA repair.
FIG. 3.
Genetic map of the recombinant region of φ31.1. The stipplied boxes indicate regions of DNA level homology with temperate lactococcal phages. The arrows indicate ORFs predicted from the φ31.1 sequence, and the numbers of amino acids are indicated below the arrows. Amino acid level homologies to other phage sequences are also indicated. The vertical arrows indicate junctions of the recombinant regions.
Localization of the φ31.1 putative origin of replication.
Since the recombinant phages were resistant to Per31, they were expected to have acquired new origins of replication (ori). Examination of the DNA sequence of φ31.1 revealed two potential origin regions (Fig. 4A) encoded in HindIII fragment B of φ31.1 (Fig. 2b). First, between φ31.1 ORF364 and ORF269 there was a 222-bp region homologous to phage rlt from its origin region (41). Second, in ORF269 there was a 240-bp region with the increased secondary structure that is typical of origins (21a) (Fig. 4B). This region, designated putative ori, contained direct repeats that were 19, 11, and 10 bp long, two inverted repeats that were 11 bp long, one inverted repeat that was 9 bp long, and three predicted hairpin loops with energies of −14.5, −13.6, and −11 kcal (as determined with Clone Manager 5) (Fig. 4B). While φ31.1 ORF269 and ORF73 in this region did not exhibit significant homology, ORF139 was homologous to the E. coli Rus protein.
HindIII fragment B of φ31.1 was subcloned into pTRKH2 to investigate which regions, if any, elicited a Per+ phenotype against φ31.1. Oligonucleotide primers were used to amplify the regions shown in Fig. 4A by PCR for cloning into the high-copy-number vector pTRKH2. The EOPs of φ31 and the various recombinant phages were determined for NCK203 harboring the various plasmids shown in Fig. 4A. The presence of complete fragment B, slightly extended to include complete ORF139 (pTRK637), decreased the φ31.1 EOP 10-fold, and the sizes of the plaques formed were reduced, which is characteristic of a Per+ phenotype. A smaller fragment, which lacked ORF139 (pTRK638), decreased the EOP to 0.5 and reduced the plaque size. The ori region alone (pTRK640) decreased the EOP to 0.2 and reduced the plaque size. Each of the subcloned fragments had similar effects on φ31.7 and on φ31.1. These results indicate that a Per+ phenotype can be attributed to the ori region cloned in pTRK640. The region that exhibited homology to rlt (cloned in pTRK639) had no effect on the EOP of φ31.1 but did reduce the plaque size slightly. This plasmid also reduced the plaque sizes of φ31 and φ31.2 without reducing the EOPs (data not shown). In fact, the EOPs of φ31 and φ31.2 were not affected by any of the cloned fragments.
Single crossover insertions of φ31.1 fragments into the L. lactis NCK203 chromosome.
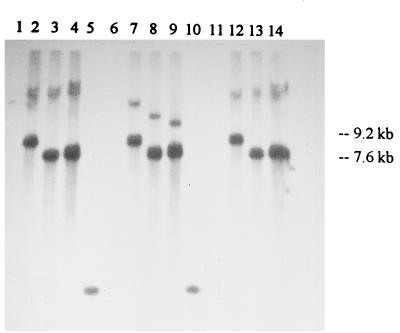
The gram-positive suicide plasmid pTRK333 (29), which is composed of pBR322 with the chloramphenicol resistance gene from pGK12 (23), was used to direct insertions into regions of the NCK203 chromosome implicated in the recombination events. HindIII fragments A, B, and D from φ31.1 (Fig. 2b) were individually cloned into pTRK333 to create pTRK333-A, pTRK333-B, and pTRK333-D, respectively. The identities of the constructs were verified by restriction analysis, and then the constructs were electroporated into L. lactis NCK203. The genomic DNA from each class of Cmr transformants and NCK203 was recovered and separately digested with BamHI, PvuI, and SalI, and Southern hybridization was performed with pTRK333 DNA (Fig. 5). Each of the enzymes used cut pTRK333 once and did not cut within any of the subcloned phage fragments. In each case, hybridizing bands were not detected in NCK203 (Fig. 5, lanes 1, 6, and 11). Strong bands corresponding to the sizes of the inserted plasmids (pTRK333-A, 9.2 kb; pTRK333-B and pTRK333-D, 7.6 kb) were detected in all of the transformants, which indicated that integration and tandem amplification of the plasmids had occurred. Higher-molecular-weight and more weakly hybridizing bands, representing junction fragments, were also detected. The insertional derivatives of NCK203 were designated NCK203-A (with insertion of pTRK333-A), NCK203-B (with insertion of pTRK333-B), and NCK203-D (with insertion of pTRK333-D).
FIG. 5.
Southern hybridization showing insertion of the pTRK333-A, pTRK333-B, and pTRK333-C suicide plasmids into the chromosome of NCK203. Genomic DNA from NCK203 (lanes 1, 6, and 11), NCK203-A (lanes 2, 7, and 12), through NCK203-B (lanes 3, 8, and 13), or NCK203-D (lanes 4, 9, and 14) was digested with BamHI (lanes 1 through 4), PvuI (lanes 6 through 9), or SalI (lanes 11 through 14). Lanes 5 and 10 contained markers (one low-molecular-weight band in the marker lanes did hybridize with the probe). The Southern blot was probed with 32P-labeled pTRK333 DNA.
A PCR-based strategy was used to determine whether each pTRK333 construct was inserted into its corresponding region of the chromosome (Fig. 6). We designed primers that flanked fragments A, B, and D (Fig. 6) (see above). Genomic DNAs from NCK203-B and NCK203-D and from single-colony isolates of NCK203-A were used as templates with primer sets A, B, and D. Vector primers flanking the ampicillin resistance gene in pTRK333 were included as a positive control. PCR products that were 0.6 kb long were obtained with the vector primers for the six NCK203-A isolates, NCK203-B, and NCK203-D but not for the NCK203 control (Fig. 6b), which confirmed that the vector was integrated in the NCK203 derivatives.
In the reactions in which the primers flanking fragment A were used, a 2.8-kb amplicon was generated for all six NCK203-A isolates, as well as the controls, NCK203, NC203-B, and NCK203-D (Fig. 6a). The pTRK333-A insertion did not occur in the region flanked by the primers in primer set A in any of the single-colony isolates of NCK203-A. When fragment A of φ31.1 was labeled and used as a probe with BclI and PvuII digests of NCK203 chromosomal DNA (these enzymes did not digest fragment A), two separate hybridizing bands were detected (data not shown). Therefore, there were at least two separate loci of the NCK203 genome which were homologous to fragment A and could potentially have been involved in the recombinations that resulted in the φ31 variants.
In contrast, neither the NCK203-B templates (primer set B) (Fig. 5c, lane 3) nor the NCK203-D templates (primer set D) (Fig. 5c, lane 10) generated amplicons of the expected size. In the case of primer sets A, B, and D the presence of a product indicated the presence of the wild type without an insertion in the region flanked by the primers. Absence of a product indicated that the insertion occurred in the region flanked by the primers, since the size of the fragment plus the tandemly amplified insert (Fig. 5) would exceed the size of possible PCR products of the reaction. These data confirmed hybridization results which showed that the B and D insertions occurred in the regions flanked by the primers.
NCK203 integrants produced different classes of recombinant phages.
We performed experiments to evaluate whether the integration events altered the frequency of appearance and the types of recombinant phages generated. Plasmid pTRK361 (Per31+) was electroporated into each of the three NCK203 insertional derivatives. Acquisition of the plasmid and retention of the chromosomal insertions were confirmed. The clones, which were designated NCK203-A(pTRK361), NCK203-B(pTRK361), and NCK203-D(pTRK361), all limited the EOP of phage φ31 to 10−6.
We thought that it was possible that standard φ31 lysates prepared with L. lactis NCK203 could contain one or more of the recombinant phages. Therefore, the phage lysates used for these experiments were specially prepared with the NCK203 insertional derivatives NCK203-A, NCK203-B, and NCK203-D. For example, phage φ31 lysates prepared with NCK203-A were used to challenge NCK203-A(pTRK361). Two or three independently propagated phage lysates were used to test each NCK203 derivative. Plaques appearing on lawns of NCK203(pTRK361), NCK203-A(pTRK361), NCK203-B(pTRK361), and NCK203-D(pTRK361) were picked and propagated with NCK203, NCK203-A, NCK203-B, and NCK203-D, respectively. Phage DNA was isolated and restricted with EcoRI, and the fragmentation patterns were examined in order to classify the phages by comparing the results with known types. The results are shown in Table 2.
Phage φ31.1 was not recovered from strain NCK203-B(pTRK361) or NCK203-D(pTRK361) but was recovered from NCK203-A(pTRK361). On the other hand, NCK203-A(pTRK361) failed to produce either φ31.2 or φ31.8. Phage φ31.7 was recovered from NCK203-D(pTRK361) but not from NCK203-B(pTRK361). Thus, the insertion in NCK203-A eliminated the appearance of one pair, φ31.2 and φ31.8, whereas the insertion in NCK203-B eliminated the other pair, φ31.1 and φ31.7. While the NCK203-B and NCK203-D insertions eliminated φ31.1, the NCK203-A insertion did not, providing additional evidence that the NCK203-A insertion did not occur in the region of NCK203 DNA that contributed to the formation of φ31.1. Collectively, the results indicate that the disruptions resulting from using φ31.1 fragments A and B could eliminate the appearance of all four recombinant phages.
Mitomycin C induction.
NCK203 harbors an inducible prophage (29). This prophage was not implicated in the recombination event that led to the appearance of phage ul37 (29). After mitomycin C was added to NCK203 and the insertion derivatives NCK203-A and NCK203-B, the optical densities of all three cultures decreased over 3 h, and prophage DNAs with identical restriction fragment patterns were detected in all three of the induced cultures (data not shown). Therefore, the chromosomal insertions in NCK203-A and NCK203-B did not disrupt the inducible prophage residing in NCK203.
DISCUSSION
Newly evolving phages which are not sensitive to the phage defense mechanisms of lactococcal starter cultures pose an economic threat to the cheese industry (1, 28, 29). In this paper we provide the first detailed description of virulent recombinant lactococcal phages which acquire chromosomal DNA from Lactococcus sp. as a mechanism to adapt to pressures imposed by abortive defense systems. In this study the DNA sequence of the exchanged region of φ31.1 was elucidated, and an origin of replication was identified. We found that a least two distinct chromosomal loci gave rise to recombinant phages. Finally, chromosomal insertions into two regions eliminated the appearance of all four types of recombinant phage.
A number of observations in this study indicated that the φ31 recombinant variants arose from two different loci on the NCK203 chromosome. First, an analysis of the restriction maps of the four phages indicated that they could be grouped into two pairs. The phages in each pair shared similar new DNA fragments, but the lengths of the DNA fragments acquired from NCK203 differed. Second, two separate chromosomal disruptions of NCK203 (when φ31.1 fragment A or B was used) were required to prevent the appearance of the four phages. Third, hybridization of NCK203 chromosomal DNA with HindIII fragment A of φ31.1 revealed two distinct areas of homology. Thus, we propose that there are at least two areas of the NCK203 chromosome which can contribute DNA to the formation of new, virulent recombinant phages. The consistent appearance of the four types of recombinants which we obtained strongly suggests that discrete cassettes of DNA must be acquired during the recombination events that generate viable lytic phages, and the sizes of these cassettes can vary considerably.
Because of the homology which phages ul36 and φ31 exhibit with the NCK203 chromosome, a form of homologous recombination is a likely route by which ul37 and the φ31 variants acquired chromosomal DNA. The inducible prophage of NCK203 does not appear to be involved. However, it is entirely possible that the source of chromosomal DNA acquired by the incoming phages was a defective or noninducible temperate phage or prophage remnants. The sequence data for φ31.1 strongly support this theory. First, the recombinant regions of φ31.1 and φ31.7 contain numerous regions that exhibit DNA homology and identity with lactococcal temperate phages and with φ31. Second, the presence of two proteins homologous to lambda and E. coli proteins involved in recombination suggests that a recombination-competent but defective prophage in NCK203 was the source of φ31.1 and φ31.7 recombinant DNA.
Selection pressure provided by Per31 allowed us to recover the recombinant phages. It has been proposed that Per31 is the origin of replication of φ31, which, when cloned on a plasmid, retards replication of superinfecting phage (14, 31). Acquisition of a different origin of replication by φ31 could potentially circumvent this phage defense. Indeed, the 7.4-kb HindIII DNA fragment encoding the φ31 origin (34) is partially replaced in all of the recombinant Per31r variants. However, none of the new regions (fragments A, B, and D) of φ31.1 were found to replicate independently in NCK203 when they were cloned into pTRK333, which lacks a gram-positive origin of replication, nor were they amplified by superinfection with φ31.1 after they were cloned into pSA3 (unpublished data). However, fragment B subclones expressed a Per phenotype when they were cloned into pTRKH2. The putative origin has been localized by sequence analysis and Per experiments to a region with increased secondary structure encoded in a predicted ORF whose function is unknown. A nearby small region of homology with rlt slightly reduces the plaque size of φ31.1 without affecting the EOP.
Increasing reliance on starter strains that have been engineered to be highly phage resistant places tremendous evolutionary pressure on phages to overcome the resistance mechanisms. The source of new lytic phages in lactococcal fermentations, whether prophage are a contributing factor or not, has long been a subject of speculation. It has been reported that most lactococci used in defined starters in cheese making are lysogenic (6, 18; for a review see reference 22). No direct connection, however, has been established between temperate and lytic phages. Recently, the P335 phage species, which contains both temperate and lytic phages, has been recognized (20). This species is appearing with increasing frequency in cheese plants (1, 21, 28, 29) and is the only group in which a direct link between temperate and lytic phages can be inferred due to shared DNA homology. From the results of this study, it is clear that P335 phage remnants scattered about the Lactococcus genome can contribute to the evolution and adaptation of new phage strains.
The sequence information available for lactococcal phage genomes and searches for sequence similarities have revealed numerous sequences shared by lytic and temperate phages. The sequence homologies strongly suggest that lytic and lysogenic phages are related and may have evolved through cassette exchanges (4, 7, 10, 19, 33). Phage φ31 and three lactococcal temperate phages, φLC3 (2), BK5-T (3), and rlt (41), have been linked by establishing sequence identity (10, 33, 42). An 888-bp fragment of φ31 DNA which was cloned in a promoter-probe vector was sequenced and shown to contain a middle phage promoter region (33). The last 176 bp of this fragment was almost identical (level of identity, 96.5%) to a region of phage φLC3 described previously. In addition, approximately 2.5 kb of the φ31 sequence, including the middle promoter and flanking regions, exhibit 94 to 95% homology with the temperate phage rlt sequence (42). The homologous sequences have been mapped close to the cos sites of these phages. In an investigation of the mechanism of AbiA, a 1.7-kb section of φ31 DNA was sequenced, which revealed three ORFs (10). Two of the ORFs exhibited significant amino acid homology with two ORFs of the temperate phage BK5-T DNA (levels of homology, 98 and 62 to 66%). The level of DNA homology in the region of the first homologous ORF was 84%. The sequence homologies between φ31 and the two temperate phages provide evidence of a close evolutionary relationship. Furthermore, they provide direct evidence of a genetic exchange between prophages and a commercial virulent phage in lactococci and suggest that the recent evolution of the P335 virulent phages probably occurred via cassette exchanges with lactococcal prophages and phage remnants located around the genome.
As more lactococcal phage sequences become available and the genetic routes by which the phages evolve are elucidated, the relationships between temperate and lytic phages are likely to be determined. Basic research into the evolution and adaptive genetics of lactococcal phages, which is rewarding in its own right, should also lead to significant improvements in dairy starter cultures. One application will most certainly be the use of molecular techniques to eliminate the genetic routes and sequences by which new phages that are virulent for Lactococcus species evolve.
ACKNOWLEDGMENTS
This work was supported by Rhodia, Inc., Madison, Wis., by the USDA NRICGP under project 97-35503-4368, and by the North Carolina Agricultural Research Service (project NC06369).
We thank Mary Allison Beauchamp for clerical assistance and Dan O'Sullivan, Gordana Djordjevic, Gwen Allison, Martin Kullen, Shirley Walker, Soren Madsen, and Michael Callanan for helpful discussions and critical reviews of the manuscript.
Footnotes
Paper FSR98-2 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Alatossava T, Klaenhammer T R. Molecular characterization of three small isometric-headed bacteriophages which vary in their sensitivity to the lactococcal phage resistance plasmid pTR2030. Appl Environ Microbiol. 1991;57:1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birkeland N-K, Lonneborg A-M. The cos region of lactococcal bacteriophage φLC3. DNA Sequence J. 1993;4:211–214. doi: 10.3109/10425179309015634. [DOI] [PubMed] [Google Scholar]
- 3.Boyce J D, Davidson B E, Hillier A J. Sequence analysis of the Lactococcus lactis temperate bacteriophage BK5-T and demonstration that the phage DNA has cohesive ends. Appl Environ Microbiol. 1995;61:4089–4098. doi: 10.1128/aem.61.11.4089-4098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandry P S, Moore S C, Boyce J D, Davidson B E, Hillier A J. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol. 1997;26:49–64. doi: 10.1046/j.1365-2958.1997.5491926.x. [DOI] [PubMed] [Google Scholar]
- 5.Dao M L, Ferretti J J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985;49:115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson B E, Powell I B, Hillier A J. Temperate bacteriophages and lysogeny in lactic acid bacteria. FEMS Microbiol Rev. 1990;87:79–90. doi: 10.1111/j.1574-6968.1990.tb04880.x. [DOI] [PubMed] [Google Scholar]
- 7.Desiere F, Lucchini S, Bruttin A, Zwahlen M-C, Brussow H. A highly conserved DNA replication module from Streptococcus thermophilus phages is similar in sequence and topology to a module from Lactococcus lactis phages. Virology. 1997;234:372–382. doi: 10.1006/viro.1997.8643. [DOI] [PubMed] [Google Scholar]
- 8.Dinsmore P K, Klaenhammer T R. Phenotypic consequences of altering the copy number of abiA, a gene responsible for aborting bacteriophage infections in Lactococcus lactis. Appl Environ Microbiol. 1994;60:1129–1136. doi: 10.1128/aem.60.4.1129-1136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinsmore P K, Klaenhammer T R. Bacteriophage resistance in Lactococcus. Mol Biotechnol. 1995;4:297–314. doi: 10.1007/BF02779022. [DOI] [PubMed] [Google Scholar]
- 10.Dinsmore P K, Klaenhammer T R. Molecular characterization of a genomic region in a Lactococcus bacteriophage that is involved in its sensitivity to the phage defense mechanism AbiA. J Bacteriol. 1997;179:2949–2957. doi: 10.1128/jb.179.9.2949-2957.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durmaz E, Higgins D L, Klaenhammer T R. Molecular characterization of a second abortive phage resistance gene present in Lactococcus lactis ME2. J Bacteriol. 1992;174:7463–7469. doi: 10.1128/jb.174.22.7463-7469.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill C. Bacteriophage and bacteriophage resistance in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:87–108. [Google Scholar]
- 14.Hill C, Massey I J, Klaenhammer T R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991;57:283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill C, Pierce K, Klaenhammer T R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction/modification (R/M) and abortive infection (Hsp) Appl Environ Microbiol. 1989;55:2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huynh T V, Young R A, Davis R W. Construction and screening cDNA libraries in λgt10 and λgt11. In: Glover D M, editor. DNA cloning. I. Oxford, United Kingdom: IRL Press Ltd.; 1985. pp. 49–78. [Google Scholar]
- 17.Jarvis A W. DNA-DNA homology between lactic streptococci and their temperate and lytic phages. Appl Environ Microbiol. 1984;47:1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis A W. Bacteriophages of lactic acid bacteria. J Dairy Sci. 1989;72:3406–3428. [Google Scholar]
- 19.Jarvis A W. Relationships by DNA homology between lactococcal phages 7–9, P335, and New Zealand industrial lactococcal phages. Int Dairy J. 1995;5:355–366. [Google Scholar]
- 20.Jarvis A W, Fitzgerald G F, Mata M, Mercenier A, Neve H, Powell I A, Ronda C, Saxelin M, Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32:2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- 21.Josephsen J, Andersen N, Behrndt H, Brandsborg E, Christiansen G, Hansen M B, Hansen S, Nielsen E W, Vogensen F K. An ecological study of lytic bacteriophages of Lactococcus lactis subsp. cremoris isolated in a cheese plant over a five year period. Int Dairy J. 1994;4:123–140. [Google Scholar]
- 21a.Keppel F, Fayet O, Georgopoulos C. Strategies of bacteriophage DNA replication. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Press; 1988. pp. 145–264. [Google Scholar]
- 22.Klaenhammer T R, Fitzgerald G F. Bacteriophages and bacteriophage resistance. In: Gasson M J, de Vos W, editors. Applied genetics of lactic acid bacteria. Glasgow, United Kingdom: Blackie and Son, Ltd.; 1994. pp. 106–168. [Google Scholar]
- 23.Kok J, van der Vossen J M B M, Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen P L, Hammer K. Temporal transcription of the lactococcal temperate phage TP901-1 and DNA sequence of the early promoter region. Microbiology. 1998;144:2203–2215. doi: 10.1099/00221287-144-8-2203. [DOI] [PubMed] [Google Scholar]
- 25.Mahdi A A, Sharples G J, Mandal T N, Lloyd R G. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J Mol Biol. 1996;257:561–573. doi: 10.1006/jmbi.1996.0185. [DOI] [PubMed] [Google Scholar]
- 26.McGrath S, Seegers J F M L, Fitzgerald G F, van Sinderen D. Molecular characterization of a phage-encoded resistance system in Lactococcus lactis. Appl Environ Microbiol. 1999;65:1891–1899. doi: 10.1128/aem.65.5.1891-1899.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moineau S, Fortier J, Ackermann H W, Pandian S. Characterization of lactococcal bacteriophages from Quebec cheese plants. Can J Microbiol. 1992;38:875–882. [Google Scholar]
- 28.Moineau S, Pandian S, Klaenhammer T R. Restriction/modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the industry. Appl Environ Microbiol. 1993;59:197–202. doi: 10.1128/aem.59.1.197-202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moineau S, Pandian S, Klaenhammer T R. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl Environ Microbiol. 1994;60:1832–1841. doi: 10.1128/aem.60.6.1832-1841.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neve H. Bacteriophage. In: Cogan T M, Accolas J-P, editors. Dairy starter cultures. New York, N.Y: VCH Publishers, Inc.; 1996. pp. 157–189. [Google Scholar]
- 31.O'Sullivan D J, Hill C, Klaenhammer T R. Effect of increasing the copy number of bacteriophage origins of replication in trans on incoming-phage proliferation. Appl Environ Microbiol. 1993;59:2449–2456. doi: 10.1128/aem.59.8.2449-2456.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Sullivan D J, Klaenhammer T R. Rapid miniprep isolation of high-quality plasmid DNA from Lactococcus and Lactobacillus spp. Appl Environ Microbiol. 1993;59:2730–2733. doi: 10.1128/aem.59.8.2730-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Sullivan D J, Walker S A, West S G, Klaenhammer T R. Development of an expression strategy using a lytic phage to trigger explosive plasmid amplification and gene expression. Bio/Technology. 1996;14:82–87. doi: 10.1038/nbt0196-82. [DOI] [PubMed] [Google Scholar]
- 34.Prevots F, Tolou S, Delpech B, Kaghad M, Daloyau M. Nucleotide sequence and analysis of the new chromosomal abortive infection gene abiN of Lactococcus lactis subsp. cremoris S114. FEMS Microbiol Lett. 1998;159:331–336. doi: 10.1111/j.1574-6968.1998.tb12879.x. [DOI] [PubMed] [Google Scholar]
- 35.Rackwitz H-R, Zehetner G, Frischauf A-M, Lehrach H. Rapid restriction mapping of DNA cloned in lambda phage vectors. Gene. 1984;30:195–200. doi: 10.1016/0378-1119(84)90120-3. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sanders M E, Leonhard P J, Sing W D, Klaenhammer T R. Conjugal strategy for construction of fast acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986;52:1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanger F, Coulson A R, Hong G F, Hill D F, Petersen G B. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 39.Strathmann M, Hamilton B A, Mayeda C A, Simon M I, Meyerowitz E M, Palazzolo M J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci USA. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terazghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters M H J, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage rlt. Mol Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 42.Walker S A, Dombroski C S, Klaenhammer T R. Common elements regulating gene expression in temperate and lytic bacteriophages of Lactococcus species. Appl Environ Microbial. 1998;64:1147–1152. doi: 10.1128/aem.64.3.1147-1152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]