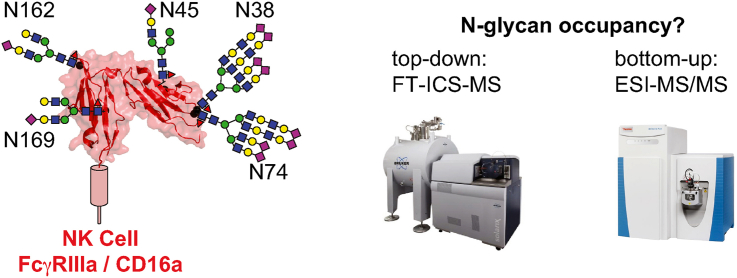
Abstract
The antibody-binding Fc γ receptors (FcγRs) trigger life-saving immune responses and many therapeutic monoclonal antibodies require FcγR engagement for full effect. One proven strategy to improve the efficacy of antibody therapies is to increase receptor binding affinity, in particular binding to FcγRIIIa/CD16a. Currently, affinities are measured using recombinantly-expressed soluble extracellular FcγR domains and CD16a-mediated antibody-dependent immune responses are characterized using cultured cells. It is notable that CD16a is highly processed with multiple N-glycosylation sites, and preventing individual N-glycan modifications affects affinity. Furthermore, multiple groups have demonstrated that CD16a N-glycan composition is variable and composition impacts antibody binding affinity. The level of N-glycosylation at each site is not known though computational prediction indicates low to moderate potential at each site based on primary sequence (40–70%). Here we quantify occupancy of the extracellular domains using complementary mass spectrometry-based methods. All five sites of the tighter-binding CD16a V158 allotype showed 65–100% N-glycan occupancy in proteomics-based experiments. These observations were confirmed using intact protein mass spectrometry that demonstrated the predominant species corresponded to CD16a V158 with five N-glycans, with a smaller contribution from CD16a with four N-glycans. Occupancy was likewise high for the membrane-bound receptor at all detected N-glycosylation sites using CD16a purified from cultured human natural killer cells. Occupancy of the N162 site, critical for antibody binding, appeared independent of N169 occupancy based on analysis of the T171A mutant protein. The weaker-binding CD16a F158 allotype showed higher occupancy of >93% at each site.
Keywords: Glycobiology, PNGase-F, Antibody-binding receptor, Mass spectrometry
Graphical abstract
Highlights
-
•
The soluble extracellular FcγRIIIa CD16a (V158) domains are highly N-glycosylated.
-
•
The F158 allotype is likewise highly glycosylated.
-
•
Here we present a method for estimating error from N-glycan occupancy measurements.
1. Introduction
The interaction of an antibody-coated target with leukocyte-expressed Fc γ receptors (FcγRs) has the potential to stimulate multiple types of protective immune responses (de Taeye et al., 2019). Several reports indicate that the sensitivity and the strength of FcγR-mediated responses may be increased to improve the treatment of multiple diseases including cancers, infection and autoimmunity (Bibeau et al., 2009; Zhang et al., 2007; Cartron et al., 2002; Weng and Levy, 2003; Musolino et al., 2008). For example, improving the FcγR-binding affinity of select antibodies that require FcγR interactions for full therapeutic effect improved efficacy, including the remarkable doubling of life expectancy in one obinutuzumab trial (Cheson et al., 2018; Pott et al., 2020; Niederfellner et al., 2011; Salles et al., 2012; Mossner et al., 2010; Herter et al., 2013). These observations support the effort to develop antibodies with improved FcγR-binding affinity, largely by engineering either the Fc N-glycan or the Fc polypeptide (Gunn et al., 2021; Liu et al., 2020). Among the five cognate activating FcγRs, FcγRIIIa/CD16a, is the primary target for engineered antibodies because it is primarily responsible for eliciting antibody-dependent cell-mediated cytotoxicity (ADCC) in natural killer cells (Wu et al., 2019).
CD16a is heavily processed and contains five N-glycosylation sites. Prior studies showed a high degree of glycan heterogeneity at different N-glycosylation sites using recombinant CD16a or CD16a purified from primary human leukocytes (Zeck et al., 2011; Patel et al., 2019, 2020, 2021; Roberts et al., 2020). Furthermore, CD16a with minimally-processed N-glycans increases IgG1 Fc binding affinity (Hayes et al., 2017; Patel et al., 2018; Subedi and Barb, 2018). Among the five N-glycosylation sites, both N162 and N45 impact antibody binding affinity. Removing the N162-glycan increased affinity towards the most common fucosylated IgG1 Fc glycoform, but eliminates the increased affinity observed with minimally processed CD16a N-glycans (Subedi and Barb, 2018; Drescher et al., 2003). The role of the N45-glycan is under debate, with different groups showing it either promotes or inhibits antibody binding (Subedi and Barb, 2018; Shibata-Koyama et al., 2009). The three remaining sites may contribute to yet undefined properties of CD16a on the cell surface considering the high degree of processing at N38 and N74 20. However, though the prior glycoproteomics approaches identified the presence of N-glycans at each of the five sites, these data do not distinguish the relative levels of N-glycosylation at each site; it is possible that each site is modified at a different frequency and impacts function. For example, low occupancy of the N162 glycan on the cell surface might promote binding to fucosylated antibodies, and different occupancies between the naturally occurring CD16a variants including V158 and F158 would be expected to differentially impact function.
Accurate estimates of N-glycan occupancy are available from dedicated experiments. Because carbohydrates reduce the ionization of peptides, comparing the signal intensities of glycosylated peptides to aglycosylated peptides in glycoproteomics experiments without an internal standard for each species potentially leads to errors (Riley et al., 2020). Occupancy may be estimated using PNGaseF, an enzyme that cleaves the bond between the Cγ atom and sidechain N of Asn, releasing the N-glycan and generating an Asp residue where an N-glycan was formerly attached (Plummer et al., 1984). This reaction increases the mass of the peptide by 0.984 Da, and the glycosylation percentage is determined by calculating the individual contribution of Asn and Asp containing peptides to the signal intensity in an MS spectrum (Carr and Roberts, 1986). There are a few notable caveats to this approach. First, substitution of an Asp residue changes the retention time for some peptides; this situation may be addressed by carefully selecting the window of retention times included for the calculation. Second, unmodified Asn residues may spontaneously deamidate, forming an Asp residue without N-glycosylation or PNGaseF activity. This outcome is reduced by preparing samples for analysis immediately prior to MS analysis, or by storing samples in a lyophilized state prior to analysis. Third, it is also possible that confounding peaks may obscure the isotopologue patterns and thus hinder accurate deconvolution of the Asn and Asp containing peptides. This scenario may be addressed by using specialized hardware capable of high-resolution detection, or by incorporating 18O into the Asp residue in place of 16O, and observing a mass increase of 2.988 Da relative to an unmodified Asn residue (Gonzalez et al., 1992). However, 18O incorporation can be complicated by [18O]-water concentrations below 100% that requires accounting for both 16O and 18O incorporation into Asp and acid-catalyzed exchange of 18O atoms at other sites.
Intact protein mass spectrometry may also provide a route to estimating N-glycan occupancy (Chen et al., 2021; Rogals et al., 2021). Measurements with multiply glycosylated proteins, like CD16a, may be particularly sensitive to N-glycan heterogeneity. Expressing proteins with HEK293 cells that lack a key N-glycan processing enzyme, Gnt1/MGAT1, reduces heterogeneity (Reeves et al., 2002). These cells express proteins with predominantly Man5 N-glycans at each N-glycosylation site and are expected to glycosylate sites with the same or similar efficiency as the wild-type cells because the oligosaccharyltransferase and MGAT1 occupy separate locations in the ER and medial Golgi, respectively (Burke et al., 1992).
We investigated CD16a using multiple mass spectrometry-based approaches to estimate the N-glycan occupancy at each individual site. We compared different single amino acid variants including the naturally occurring high affinity V158 allotype and the low affinity F158 allotype that is more common in the human population to identify any potential differences in N-glycan occupancy (Bruhns et al., 2009). We also analyzed the V158/T171A variant which disrupts the sequence for N169 glycosylation because the proximity of the site at only five residues from the C-terminus indicates N-glycan occupancy may be low and potentially impact N162 glycosylation (Bano-Polo et al., 2011). Lastly, we compared these occupancy estimates to CD16a purified from natural killer cells.
1.1. Experimental methods
Materials – All materials were purchased from Millipore Sigma unless otherwise noted.
Protein expression and purification – The soluble extracellular antibody-binding domain of CD16a V158 corresponding to residue R1-G174 was prepared as previously described using HEK293F and HEK293S cells (Subedi et al., 2014; Subedi and Barb, 2015). TEV digestion followed by size exclusion chromatography and EndoF1 digestion were performed as previously described, except that a Superdex75 column was used (Subedi et al., 2017). Plasmids encoding the soluble CD16a F158, and T171A variants were prepared using fusion PCR (Ho et al., 1989). YTS-CD16a cells were cultured and CD16a purified as described (Patel et al., 2021).
PNGaseF digestion using [16O]water – Sample (20 μg) in 50 μL of 50 mM ammonium carbonate, 10% MeOH, pH 8, was boiled for 5 min. Then, 1 μL of 0.25 M dithiothreitol (DTT) was added to reduce the proteins' disulfide bonds. The samples were then incubated at 37 °C for 1 h. Iodoacetamide (1.4 μL of 0.5 M) was then added followed by incubation at RT for 30 min in the dark. The reaction was quenched with an additional 1 μL of 0.25 M DTT and an additional 15 min incubation at RT in the dark. The endoproteases GluC (1 μL of 1 mg/mL; Promega) and chymotrypsin (1.5 μL of 1 mg/mL) were added to the samples, which were then incubated overnight at 37 °C. Some samples were boiled for 5 min following proteolysis. Peptides were isolated using a C18 resin according to the manufacturer's instructions. Fractions eluted from the C18 resin in 80:20:1 acetonitrile:water:formic acid and were frozen and then lyophilized. The lyophilized product was resuspended in 2 μL 50 mM sodium phosphate, pH 7.5 and 17 μL of water. PNGase F (1 μL; New England Biolabs) was added to each sample and then incubated overnight at 37 °C. Peptides were lyophilized, resuspended with water, and purified with a C18 step prior to MS. Some samples, as noted, were boiled for 5 min prior to the C18 purification. Efficiency of the PNGase F reaction was monitored with an intact (non-proteolyzed) CD16a V158 sample using an SDS-PAGE and compared to a sample prepared without PNGase F addition.
PNGaseF digestion using [18O]water – Peptides from samples (20 μg) were prepared as described above, except without C18 purification. Lyophilized peptides were resuspended with 19 μL of 97% 1H2–18O. Simultaneously, glycerol-free PNGase F (New England Biolabs) was lyophilized, resuspended in an equal volume of 1H2–18O, lyophilized again, and finally resuspended in an equal volume of 1H2–18O. [18O-exchanged]-PNGase F (1 μL) was added to 19 μL of lyophilized peptides and then incubated overnight at 37 °C. Peptides were lyophilized, resuspended with water, and purified with a C18 step prior to MS.
Peptide mass spectrometry – Lyophilized peptides were resuspended in 100 μL 0.1% HFBA (Heptafluorobutyric acid, Fluka, 77249) and cleaned-up using C18 pipette tips (Bond Elut OMIX C18, Agilent, A7003100). Briefly, tips were wetted with 2 × 100 μL 50% ACN (acetonitrile, Optima™ LC/MS Grade, Fisher Chemical, A955-4), equilibrated with 2 × 100 μL 1% HFBA and sample was loaded by pipetting back and forth ∼ 20 times. Tips were then washed with 2 × 100 μL 0.1% HFBA and eluted sequentially in 100 μL 50% acetonitrile (ACN, Optima™ LC/MS Grade, Fisher Chemical, A955-4), 0.1% formic acid (FA, LC-MS Grade, Pierce, 28905) followed by 100 μL 75% ACN, 0.1% FA. Eluted peptides were vacuum dried, reconstituted in 100 μL 5% ACN, 0.1% TFA (Trifluoroacetic acid, LC-MS grade, Pierce, 28904), and analyzed by nanoLC-MS/MS.
Samples (1–10 μL) of the reconstituted (glyco)peptides were loaded onto an Acclaim PepMap C18 trap column (300 μm, 100 Å) in 2% ACN, 0.05% TFA at 5 μL/min, eluted onto an Acclaim PepMap RSLC C18 column and separated on an Acclaim PepMap RSLC C18 column (75 μm × 150 mm, 2 μm, 100 Å) with the following gradient: 1.5%–6% solvent B (solvent A: 0.1% FA; solvent B: 90% ACN, 0.08% FA) over 10 min, 6%–17% solvent B over 35 min, 17%–31% solvent B over 25 min, 31%–62% solvent B over 10 min, and finally 62%–90% solvent B over 10 min, with a total run time of 95 min at a flow rate of 300 nL/min in an Ultimate 3000 RSLCnano UHPLC system, and eluted into the ion source of an Orbitrap QE + mass spectrometer (Thermo Fisher Scientific). The spray voltage was set to 1.9 kV and the temperature of the heated capillary was set to 280 °C. Full MS scans were acquired from m/z 266 to 2500 at 70,000 resolution. MS (Bibeau et al., 2009) scans following higher energy collision-induced dissociation (HCD, stepped 17, 27, 37) were collected for the Top20 most intense ions, with a first fixed mass of 125 m/z at 17,500 resolution. Data was acquired using Xcalibur Software (Version 2.January 8, 2806; Thermo) in positive ion mode. Ion m/z values and intensity was exported and analyzed using Xcalibur Qual Browser Software (Version 4.0.27.19; Thermo). Peptides were identified using Byonic (ProteinMetrics). RAW files for each proteomics sample are deposited in the MASSIVE server (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under the accession number MSV000088916 (massive.ucsd.edu).
The Asn v. Asp content of peptides containing N-glycosylation sites were determined by extracting the MS1 spectra containing both peptides using an R script (Supplemental Methods). A second R script (Supplemental Methods) fitted the coefficients for the amount of Asn and Asp peptides to the intensity data for each peak. The expected isotope distributions were calculated for each CD16a glycosylation site using ProteinProspector MS-Isotope (Chalkley et al., 2008). The percentage of N-glycosylation at each site was then determined by dividing the intensity of the Asp-containing peptide by the sum of the intensities for the Asp and Asn containing peptides.
Intact protein mass spectrometry – Protein samples were buffer exchanged using 10 kDa MWCO centrifugal spin filters into 50 mM ammonium acetate (SigmaUltra >98%) in HPLC grade water, and concentrated to a final protein concentration of 1–7 μM. Mass spectra were collected using a SolariX XR 12 T Fourier transform ion cyclotron resonance mass spectrometer (Bruker Scientific). Samples were introduced by direct infusion into an electrospray ionization source at a rate of 120 μL/min, using a capillary inlet voltage of 4200–4500 V. Mass spectra were collected in positive mode in the range of m/z 1000–3000. The transient length was 2.7962 s and 1 M points were collected. Mass spectra were analyzed using Data Analysis (Bruker Scientific), and protein monoisotopic masses were determined using the SNAP™ algorithm.
2. Results
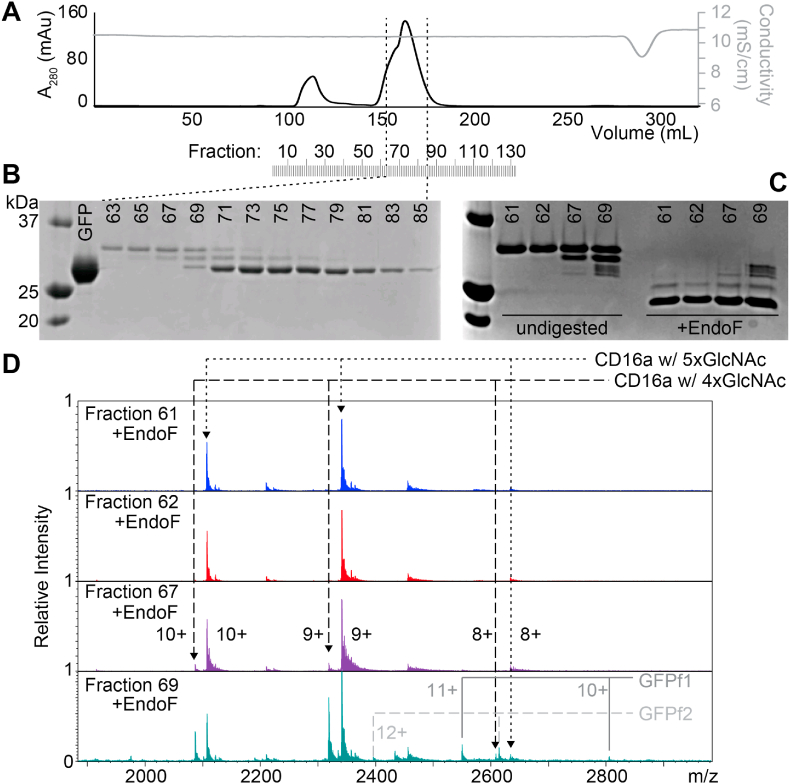
Intact CD16a mass spectrometry – Proteolytic removal of the GFP fused to the CD16a N-terminus revealed CD16a heterogeneity not evident in the full-length fused proteins (Fig. 1A). SDS-PAGE analysis revealed two distinct bands, of similar intensities, both separated from GFP (Fig. 1B). Selection of fractions 61, 62, 67 and 69 included an increasing amount of the smaller CD16a band and GFP (Fig. 1C). Incubation with endoglycosidase F1 (EndoF) efficiently trimmed the CD16a N-glycans to a single GlcNAc residue, collapsing the distinct CD16a bands into a single band with greater mobility (Fig. 1C). By comparison, GFP lacks an N-glycan and showed no change in mobility.
Fig. 1.
Analysis of intact CD16a V158. A. Gel filtration of CD16a using a Superdex 75 column following TEV digestion. B. SDS-PAGE analysis of the central peak. C. SDS-PAGE analysis of select CD16a fractions before and after treatment with EndoF. D. FT-ICR MS analysis of EndoF-treated CD16a fractions. The observed masses are reported in Table 2. GFP form 1 includes residues KIEW through the TEV cleavage site, and GFP form 2 starts with DIFEA and ends with the TEV cleavage site. “XX+” indicates charge state.
Analyses of both the EndoF-treated and untreated Superdex75 fractions by FT-ICR MS revealed high molecular weight species with multiple charge states. The two highest intensity peaks observed in fraction 61 following EndoF treatment represented a single species with three charge states (Fig. 1D). The observed mass calculated from the major species observed in fraction 61, following deconvolution, proved highly comparable to the calculated mass of CD16a with five GlcNAc residues with a mass error of 0.618 ppm (Table 1). Fraction 61 prior to EndoF treatment corresponded to CD16a with five Man5 N-glycans with a mass error of 0.176 ppm.
Table 1.
Mass accuracy of intact CD16a measurements using FT-ICR MS.
| Species | Observed [M+H]+ (Da) | Observed neutral mass (Da) | Calculated mass (Da) | mass error (ppm) |
|---|---|---|---|---|
| CD16a w/5xMan5 | 26127.0390 | 26126.0317 | 26126.0271 | 0.176 |
| CD16a w/4xMan5 | 24910.6084 | 24909.6011 | 24909.6043 | 0.128 |
| CD16a w/5xGlcNAc | 21060.3039 | 21059.2967 | 21059.3097 | 0.618 |
| CD16a w/4xGlcNAc | 20857.2508 | 20856.2435 | 20856.2303 | 0.632 |
| GFP form1 | 28032.9850 | 28031.9772 | 28032.0056 | 1.013 |
| GFP form2 | 28736.3098 | 28735.3020 | 28735.3233 | 0.741 |
Fraction 69 contained two major species, each with three charge states (Fig. 1D). The most abundant species matched that observed in fraction 61. The observed mass calculated from the second most intense species observed in fraction 69, following deconvolution, proved highly comparable to the calculated mass of CD16a with four GlcNAc residues with a mass error of 0.632 ppm (Table 1). This same species from the same fraction, prior to EndoF treatment, revealed a lower mass error value of 0.128 ppm and corresponded to CD16a with four Man5 N-glycans. In total, these results indicate that the two major CD16a species contained five or four N-glycans.
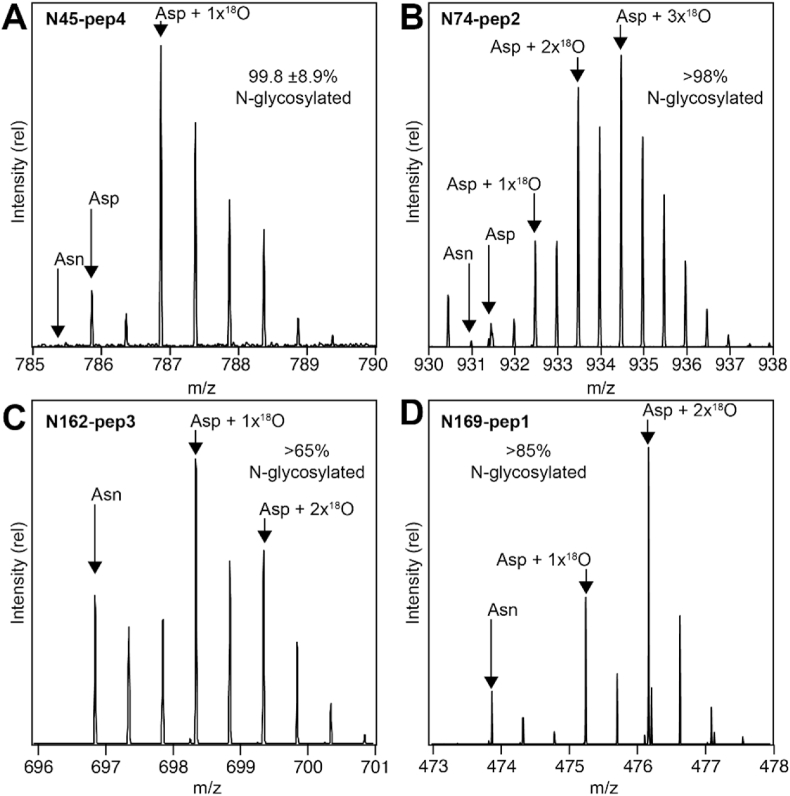
Site-specific N-glycan occupancy – We next determined N-glycan occupancy at each site using a proteomics-based approach. This approach does not require N-glycan homogeneity because N-glycans are removed during the analysis, providing an opportunity to characterize CD16a N-glycan occupancy with highly heterogeneous N-glycans (Patel et al., 2021).
We identified peptides covering each of the five N-glycosylation sites (Supplemental MSExcel worksheet). MS2 spectra for each peptide confirmed the assignments, with exception of the smallest N45 FHNE peptide (N45-2) and the intermediate N38 peptide (N38-2) found in only two samples (Figure S1). We previously characterized these peptides using CD16a from monocytes (Roberts et al., 2020).
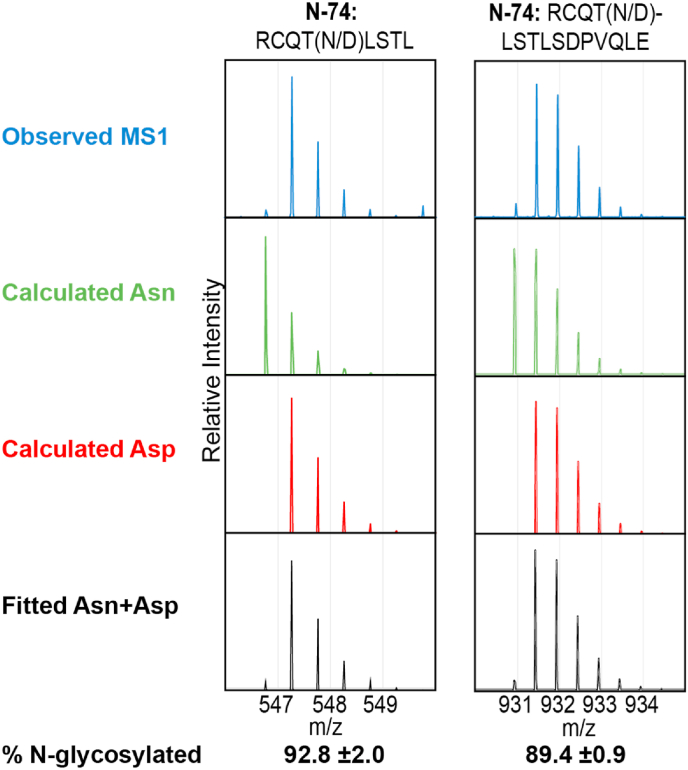
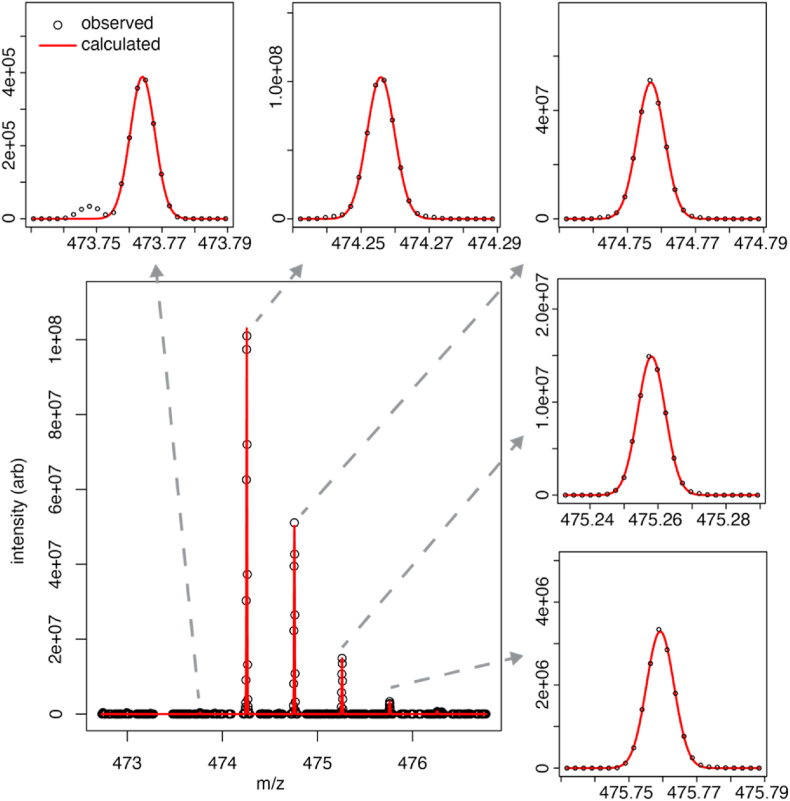
The multiplet pattern corresponding to each peptide showed evidence for both Asn and Asp residues. An example of MS1 spectra for two N74-containing peptides from CD16a V158 is shown in Fig. 2. The lowest m/z peak of the multiplet corresponds to the monoisotopic Asn-containing peptide. However, both the monoisotopic Asp-containing mass and the Asn-containing peptide with one heavy isotope contribute to the second peak, and the relative contributions must be deconvoluted for an accurate estimate of occupancy. We first quantified the intensity of each MS1 peak in the multiplet by fitting a Gaussian line shape function (Fig. 3). Fitting these intensity data with the calculated multiplet intensities of both the Asp- and Asn-containing peptides deconvoluted the relative contribution from each peptide to the observed spectrum, and allowed an estimate of the experimental error by using all peaks from the multiplet. Finally, N-glycan occupancy was determined by dividing the abundance of the Asp-peptide by the total of the Asn- and Asp-peptides. The error in this estimate is largely due to differences between the calculated and observed multiplet pattern for each species.
Fig. 2.
N-glycan occupancy for the N74 site of CD16a V158 as observed with two peptides in [16O]-water. Recorded spectra corresponding to each peptide are shown in the top row. The calculated isotope distributions for Asn- and Asp-containing peptides are shown in the second and third rows, respectively. The final row shows the fitted Asn + Asp data to determine the percentage of peptides that showed N-glycosylation.
Fig. 3.
A Gaussian line shape fitted to the individual isotopologue peaks to determine N-glycan occupancy. This MS1 spectrum shows the N169-containing peptide from the CD16a V158 allotype, following PNGaseF-catalyzed N-glycan removal in [16O]-water. The major peak in the upper left panel contains only an unglycosylated Asn at position 169. The major peak in the upper middle panel contains contributions for both Asn and Asp (formed by PNGaseF digestion of an N-glycosylated peptide) at this position, with the Asp-containing peptide predominant in this example.
CD16a V158 peptides containing the N45, N74 and N169 glycosylation sites showed minimal contribution to the observed intensities from masses calculated with Asn residues. Peptides with Asp residues at these sites dominated with a contribution of 88.0–92.8% of the intensities observed in MS1 spectra (Table 2). Asp residues proved more abundant at these sites for the F158 variant (98.0–99.6%). The V158/T171A variant likewise revealed high Asp content for the N45 and N74 sites, but lacks the N169 site due to the T171A mutation that destroys the consensus N-glycosylation sequence. These data indicate high levels of N-glycosylation at N45, N74 and N169 (Table 2).
Table 2.
N-glycosylation site occupancy of three CD16a variants (n.a. - not applicable, *- lower bound due to peak overlap, ** - no N peak observed, *** - lower bound due to supplemental 18O incorporation).
|
16O-water |
18O-water |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CD16a variant: |
V158 |
F158 |
T171A |
YTS |
V158 |
F158 |
T171A |
YTS |
|
| Native Peptide | site - peptide # | % | % | % | % | % | % | % | % |
| DNSTQWF | N38-1 | 91 ± 14 | |||||||
| SPEDNSTQW | N38-2 | ≥92.5 * | |||||||
| KCQGAYSPEDNSTQW | N38-4 | ≥60 * | 80 ± 22 | 100 ** | |||||
| STQWFHNE | N45-1 | 98 ± 11 | 95.8 ± 1.7 | >98.9 *** | |||||
| FHNE | N45-2 | 81 ± 28 | |||||||
| FHNESLISSQASSY | N45-4 | 92 ± 14 | 99.8 ± 8.9 | 96 ± 12 | |||||
| RCQTNLSTL | N74-1 | 92.8 ± 2.0 | 94.6 ± 2.5 | 97.3 ± 9.3 | >96.3 *** | 100 *** | >98.9 *** | 100 ± 9 | |
| RCQTNLSTLSDPVQLE | N74-2 | 89.4 ± 0.9 | 98.8 ± 2.0 | 98.4 ± 3.2 | >98.0 *** | >99.8 *** | |||
| VGSKNVSSE | N162-1 | NA | 96 ± 18 | ||||||
| GLVGSKNVSSE | N162-2 | 79.6 ± 2.2 | NA | 99.6 ± 2.8 | >67.2 *** | >99.8 *** | |||
| CRGLVGSKNVSSE | N162-3 | 70 ± 15 | NA | >64.9 *** | |||||
| (F158) FGSKNVSSE (only) | N162-4 | NA | 98.9 ± 4.8 | NA | NA | NA | >99 *** | NA | |
| TVNITITQG | N169-1 | 88.0 ± 1.1 | 99.6 ± 1.6 | NA | 100 ± 22 | >84.9 *** | 100 *** | NA | 93 ± 10 |
| TVNIAITQG (T171A only) | N169-2 | NA | NA | 0.0 ± 1.2 | NA | NA | NA | 0.0 ± 2.2 | |
The CD16a V158 N162 site appeared 69.5–79.6% occupied, in contrast to F158 and V158/T171A with 89.9–95.9% occupancy. The N38 site revealed the weakest intensities, and in some cases with overlapping peaks that limited complete analysis. Despite these limitations, we obtained occupancy estimates between 60% and 92.5% for the three CD16a variants.
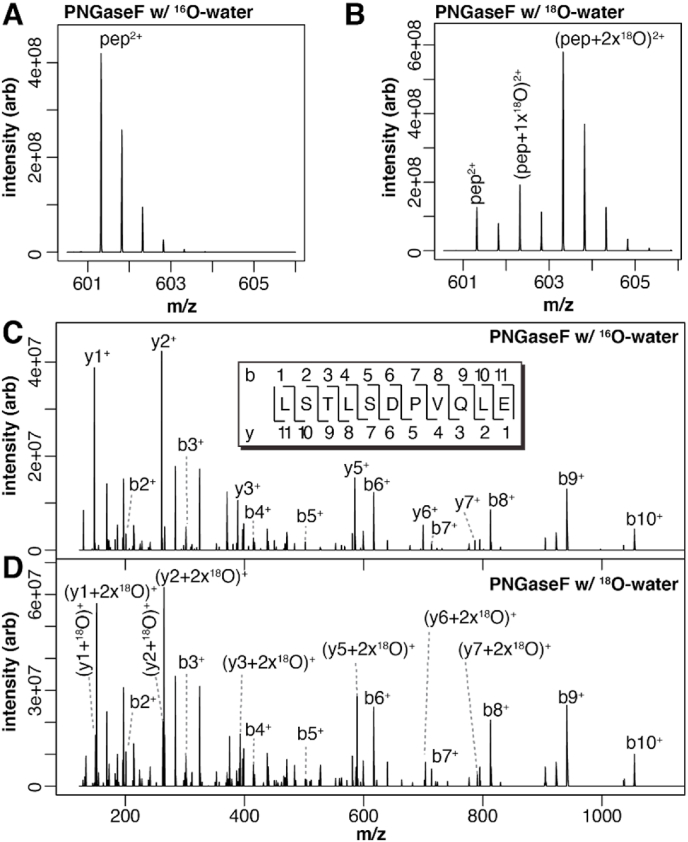
Site occupancy estimates using18O-water – We performed the PNGaseF reaction in [18O]-water to provide greater separation between the Asp and Asn containing peptides. One factor complicating the PNGaseF reaction is that reagents or impure [18O]-water can introduce [16O]-water, requiring careful separation of peptide peaks containing Asn, [16O]-Asp or [18O]-Asp (Gonzalez et al., 1992). This may be reduced by introducing [18O]-water following lyophilization of the substrate peptides. To our surprise, PNGaseF retained activity following two lyophilization steps, allowing near complete exchange to [18O]-water and reducing the contribution of [16O]-water in the reaction (Figure S2).
Peptides containing the N38, N45, N162 and N169 glycosylation sites revealed substantial [18O] incorporation. MS2 spectra of these peptides indicated that the majority of the Asp residues in the D-X-S/T sequon contained at least one [18O] atom (Fig. 4). These results were consistent with measurements performed using [16O]-water (Table 2).
Fig. 4.
N-glycosylation of CD16a V158 determined by treating with PNGaseF in [18O]-water. The monoisotopic masses for the Asn- and Asp-containing peptides are indicated with black arrows. N-glycan occupancy at each site as estimated from these spectra is indicated in the upper portion of each panel.
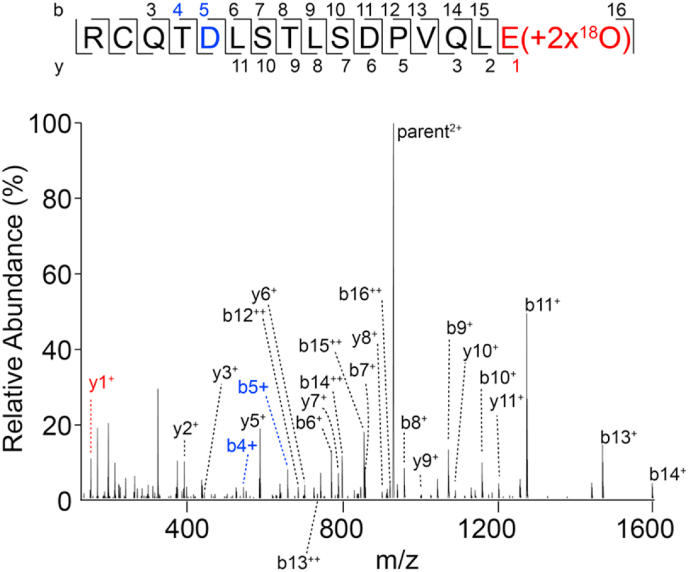
Supplemental18O incorporation – A detailed examination of mass spectra generated from reactions in [18O]-water revealed additional [18O] labels incorporated into peptides containing the N74, N162 and N169 glycosylation sites (Fig. 4). An MS2 spectrum of the longer N74 peptide (N74-2) identified two additional 18O atoms as part of the C-terminal Glu residue (Fig. 5). Other groups have noted that auxiliary labeling reactions resulting from the presence of residual protease activity (Shajahan et al., 2017; Alley et al., 2013). We examined the effect of residual GluC activity on 18O incorporation. We did not find evidence of residual GluC activity in our spectra. For example, the N74 peptides would be degraded upon PNGase digestion because GluC cleaves after Glu or Asp residues. In order to examine the potential that GluC activity contributed to the extra [18O] incorporation, we preserved GluC activity by eliminating a heat treatment step that occurs following proteolysis but prior to reversed-phase peptide purification followed by PNGaseF digestion. In this case, the N74 peptides were efficiently cleaved, generating an L75 peptide with additional 18O atoms in the C-terminal Glu residue (Fig. 6). Heat treatment eliminated GluC proteolytic activity and thus the auxiliary labeling is likely not catalyzed by the GluC protease. It is possible a buffer component or the peptides themselves catalyzed exchange of the O atoms in the peptide.
Fig. 5.
MS2 spectrum of the most abundant peptide species from CD16a V158 containing the N74 glycosylation site showing incorporation of two 18O atoms to the C-terminal glutamate residue (shown in red) and the presence of an Asp residue within the N-glycosylation sequon (shown in blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 6.
The L75 peptide is only expected to result from GluC-digestion following PNGaseF-digestion of a glycosylated N74-containing peptide. MS1 spectra of the L75 peptide identified in spectra collected using CD16a V158 following PNGaseF digestion in either 16O (A) or 18O (B) water. The positions of monoisotopic peaks for each species are indicated. (C–D) MS2 spectra of the major species from panels A and B, respectively, indicate the ion composition as well as demonstrate 18O incorporation in the C-terminal Glu residue.
CD16a N-glycan occupancy on a NK cell – The preceding experiments utilized soluble CD16a, truncated to remove the 7 kDa C-terminal transmembrane and intracellular portions. It is possible membrane localization during translation and protein folding affects N-glycosylation. We purified CD16a V158 from YTS-CD16a cells, a natural killer cell capable of ADCC and natural cytotoxicity to estimate the N-glycan occupancy following PNGaseF digestion. We identified peptides corresponding to each N-glycosylation site except N162. The peptide signal intensity observed using CD16a purified from YTS cells was lower than that observed with recombinantly-expressed proteins, and thus the N162 peptide was not observed but is likely still present. The N45, N74 and N169 sites all showed high levels of N-glycosylation, and the N38 site revealed a weak signal with only a peak for the Asp-containing peptide (Table 2). The resulting occupancy estimates are comparable to those obtained for the soluble CD16a V158 form.
3. Discussion
These data provide a thorough characterization of CD16a N-glycan occupancy. Among the five sites, occupancy at N162 and N38 appear lower, particularly for the recombinant V158 allotype. It is somewhat surprising that the CD16a N169 and N162 sites are predominantly modified. N162 and N169 are located close to the C-terminus of the soluble construct (G174). This high occupancy likely results from efficient STT3B modification because STT3A is expected to modify the C-terminal N-glycosylation sites with very low efficiency (Shrimal et al., 2013). The high levels of N162 occupancy in the V158/T171A variant indicate that glycosylation at N162 is independent from N169 glycosylation (Table 2). One previous observation with the highly homologous receptor, CD16b, found high N-glycan occupancy for the N45, N162 and N169 sites, with N74 occupied from 58 to 80% based on a comparison of the intensities for the glycosylated and unmodified peptides (Wojcik et al., 2020). CD16a and CD16b share 97.7% amino acid identity for the extracellular domains but two different N-glycosylation sites.
It is notable that the soluble CD16a V158 protein is somewhat less glycosylated at N162 and N169 compared to F158 (Table 2). Though the F158 allotype binds with reduced affinity relative to V158 34, these minor glycan occupancy differences are unlikely to account for differences in affinity. The factor(s) contributing to the affinity difference remain undefined.
With respect to future efforts measuring N-glycan occupancy with [18O] incorporation, the preservation of PNGaseF activity following two lyophilization steps allows a significant reduction of the [16O]-water content when isotope labeling is desired (Figure S2). The spectra for the V158 allotype collected with samples digested in [18O]-water reveal a small amount of peptide corresponding to Asp without [18O] incorporation, which is expected following incorporation of an [16O] atom during the PNGaseF reaction. This is not uniform and is most notable with the MS1 spectrum of the N169-containing peptide that shows low levels of the [16O]-Asp peptide (Fig. 4D). It is interesting that the N45–4 and N74-2 peptides showed an increased [16O] content that could be explained by deamidation of an unoccupied Asn prior to the PNGaseF reaction.
Finally, there is currently a growing interest in CD16a as a therapeutic, as a target for antibodies to recruit and/or activate NK cells, or for engineering to increase effector response efficiency (Zhao et al., 2020; Bogen et al., 2021; Zhu et al., 2020). A complete description of N-glycan occupancy, like that described here, is essential to identify variables that potentially impact function. Furthermore, the presence of an N-glycan dramatically alters how that portion of the protein surface is perceived by the environment and thus represents an important intrinsic feature of any glycosylated protein.
Funding sources
Funding to A.W.B. by the National Institutes of Health under Awards No. U01 AI148114 (NIAID). I. J. A. is grateful for funding of the 12 T FTICR mass spectrometer by the National Institutes of Health, award S10 OD025118. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CRediT authorship contribution statement
Elizabeth A. Lampros: Writing – original draft, performed experiments, wrote the first draft. Paul G. Kremer: Conceptualization, Writing – original draft, performed experiments, wrote the first draft. Jesús S. Aguilar Díaz de León: performed experiments. Elijah T. Roberts: performed experiments. Maria Carolina Rodriguez Benavente: performed experiments. I. Jonathan Amster: Supervision, Funding acquisition, Conceptualization. Adam W. Barb: Conceptualization, Writing – original draft, wrote the first draft, Funding acquisition, Supervision, All authors approve and final draft of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank Dr. Elisabet Gas Pascual (UGA) for operating the Q Exactive Plus mass spectrometer.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crimmu.2022.05.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Alley W.R., Jr., Mann B.F., Novotny M.V. High-sensitivity analytical approaches for the structural characterization of glycoproteins. Chem. Rev. 2013;113(4):2668–2732. doi: 10.1021/cr3003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano-Polo M., Baldin F., Tamborero S., Marti-Renom M.A., Mingarro I. N-glycosylation efficiency is determined by the distance to the C-terminus and the amino acid preceding an Asn-Ser-Thr sequon. Protein Sci. 2011;20(1):179–186. doi: 10.1002/pro.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibeau F., Lopez-Crapez E., Di Fiore F., Thezenas S., Ychou M., Blanchard F., Lamy A., Penault-Llorca F., Frebourg T., Michel P., Sabourin J.C., Boissiere-Michot F. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J. Clin. Oncol. 2009;27(7):1122–1129. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- Bogen J.P., Carrara S.C., Fiebig D., Grzeschik J., Hock B., Kolmar H. Design of a trispecific checkpoint inhibitor and natural killer cell engager based on a 2 + 1 common light chain antibody architecture. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.669496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhns P., Iannascoli B., England P., Mancardi D.A., Fernandez N., Jorieux S., Daeron M. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113(16):3716–3725. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- Burke J., Pettitt J.M., Schachter H., Sarkar M., Gleeson P.A. The transmembrane and flanking sequences of beta 1,2-N-acetylglucosaminyltransferase I specify medial-Golgi localization. J. Biol. Chem. 1992;267(34):24433–24440. [PubMed] [Google Scholar]
- Carr S.A., Roberts G.D. Carbohydrate mapping by mass spectrometry: a novel method for identifying attachment sites of Asn-linked sugars in glycoproteins. Anal. Biochem. 1986;157(2):396–406. doi: 10.1016/0003-2697(86)90643-3. [DOI] [PubMed] [Google Scholar]
- Cartron G., Dacheux L., Salles G., Solal-Celigny P., Bardos P., Colombat P., Watier H. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood. 2002;99(3):754–758. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- Chalkley R.J., Baker P.R., Medzihradszky K.F., Lynn A.J., Burlingame A.L. In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell. Proteomics. 2008;7(12):2386–2398. doi: 10.1074/mcp.M800021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Wu D., Robinson C.V., Struwe W.B. Native mass spectrometry meets glycomics: resolving structural detail and occupancy of glycans on intact glycoproteins. Anal. Chem. 2021;93(30):10435–10443. doi: 10.1021/acs.analchem.1c01460. [DOI] [PubMed] [Google Scholar]
- Cheson B.D., Chua N., Mayer J., Dueck G., Trneny M., Bouabdallah K., Fowler N., Delwail V., Press O., Salles G., Gribben J.G., Lennard A., Lugtenburg P.J., Fingerle-Rowson G., Mattiello F., Knapp A., Sehn L.H. Overall survival benefit in patients with rituximab-refractory indolent non-hodgkin lymphoma who received obinutuzumab plus bendamustine induction and obinutuzumab maintenance in the GADOLIN study. J. Clin. Oncol. 2018;36(22):2259–2266. doi: 10.1200/JCO.2017.76.3656. [DOI] [PubMed] [Google Scholar]
- de Taeye S.W., Rispens T., Vidarsson G. The ligands for human IgG and their effector functions. Antibodies (Basel) 2019;8(2) doi: 10.3390/antib8020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher B., Witte T., Schmidt R.E. Glycosylation of FcgammaRIII in N163 as mechanism of regulating receptor affinity. Immunology. 2003;110(3):335–340. doi: 10.1046/j.1365-2567.2003.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez J., Takao T., Hori H., Besada V., Rodriguez R., Padron G., Shimonishi Y. A method for determination of N-glycosylation sites in glycoproteins by collision-induced dissociation analysis in fast atom bombardment mass spectrometry: identification of the positions of carbohydrate-linked asparagine in recombinant alpha-amylase by treatment with peptide-N-glycosidase F in 18O-labeled water. Anal. Biochem. 1992;205(1):151–158. doi: 10.1016/0003-2697(92)90592-u. [DOI] [PubMed] [Google Scholar]
- Gunn B.M., Lu R., Slein M.D., Ilinykh P.A., Huang K., Atyeo C., Schendel S.L., Kim J., Cain C., Roy V., Suscovich T.J., Takada A., Halfmann P.J., Kawaoka Y., Pauthner M.G., Momoh M., Goba A., Kanneh L., Andersen K.G., Schieffelin J.S., Grant D., Garry R.F., Saphire E.O., Bukreyev A., Alter G. A Fc engineering approach to define functional humoral correlates of immunity against Ebola virus. Immunity. 2021;54(4):815–828 e5. doi: 10.1016/j.immuni.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J.M., Frostell A., Karlsson R., Muller S., Martin S.M., Pauers M., Reuss F., Cosgrave E.F., Anneren C., Davey G.P., Rudd P.M. Identification of Fc gamma receptor glycoforms that produce differential binding kinetics for rituximab. Mol. Cell. Proteomics. 2017;16(10):1770–1788. doi: 10.1074/mcp.M117.066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herter S., Herting F., Mundigl O., Waldhauer I., Weinzierl T., Fauti T., Muth G., Ziegler-Landesberger D., Van Puijenbroek E., Lang S., Duong M.N., Reslan L., Gerdes C.A., Friess T., Baer U., Burtscher H., Weidner M., Dumontet C., Umana P., Niederfellner G., Bacac M., Klein C. Preclinical activity of the type II CD20 antibody GA101 (obinutuzumab) compared with rituximab and ofatumumab in vitro and in xenograft models. Mol. Cancer Therapeut. 2013;12(10):2031–2042. doi: 10.1158/1535-7163.MCT-12-1182. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Liu R., Oldham R.J., Teal E., Beers S.A., Cragg M.S. Fc-engineering for modulated effector functions-improving antibodies for cancer treatment. Antibodies (Basel) 2020;9(4) doi: 10.3390/antib9040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossner E., Brunker P., Moser S., Puntener U., Schmidt C., Herter S., Grau R., Gerdes C., Nopora A., van Puijenbroek E., Ferrara C., Sondermann P., Jager C., Strein P., Fertig G., Friess T., Schull C., Bauer S., Dal Porto J., Del Nagro C., Dabbagh K., Dyer M.J., Poppema S., Klein C., Umana P. Increasing the efficacy of CD20 antibody therapy through the engineering of a new type II anti-CD20 antibody with enhanced direct and immune effector cell-mediated B-cell cytotoxicity. Blood. 2010;115(22):4393–4402. doi: 10.1182/blood-2009-06-225979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musolino A., Naldi N., Bortesi B., Pezzuolo D., Capelletti M., Missale G., Laccabue D., Zerbini A., Camisa R., Bisagni G., Neri T.M., Ardizzoni A. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J. Clin. Oncol. 2008;26(11):1789–1796. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- Niederfellner G., Lammens A., Mundigl O., Georges G.J., Schaefer W., Schwaiger M., Franke A., Wiechmann K., Jenewein S., Slootstra J.W., Timmerman P., Brannstrom A., Lindstrom F., Mossner E., Umana P., Hopfner K.P., Klein C. Epitope characterization and crystal structure of GA101 provide insights into the molecular basis for type I/II distinction of CD20 antibodies. Blood. 2011;118(2):358–367. doi: 10.1182/blood-2010-09-305847. [DOI] [PubMed] [Google Scholar]
- Patel K.R., Roberts J.T., Subedi G.P., Barb A.W. Restricted processing of CD16a/Fc gamma receptor IIIa N-glycans from primary human NK cells impacts structure and function. J. Biol. Chem. 2018;293(10):3477–3489. doi: 10.1074/jbc.RA117.001207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.R., Nott J.D., Barb A.W. Primary human natural killer cells retain proinflammatory IgG1 at the cell surface and express CD16a glycoforms with donor-dependent variability. Mol. Cell. Proteomics. 2019;18(11):2178–2190. doi: 10.1074/mcp.RA119.001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.R., Roberts J.T., Barb A.W. Allotype-specific processing of the CD16a N45-glycan from primary human natural killer cells and monocytes. Glycobiology. 2020;30(7):427–432. doi: 10.1093/glycob/cwaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.R., Rodriguez Benavente M.C., Lorenz W.W., Mace E.M., Barb A.W. Fc gamma receptor IIIa/CD16a processing correlates with the expression of glycan-related genes in human natural killer cells. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.015516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer T.H., Jr., Elder J.H., Alexander S., Phelan A.W., Tarentino A.L. Demonstration of peptide:N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. J. Biol. Chem. 1984;259(17):10700–10704. [PubMed] [Google Scholar]
- Pott C., Sehn L.H., Belada D., Gribben J., Hoster E., Kahl B., Kehden B., Nicolas-Virelizier E., Spielewoy N., Fingerle-Rowson G., Harbron C., Mundt K., Wassner-Fritsch E., Cheson B.D. MRD response in relapsed/refractory FL after obinutuzumab plus bendamustine or bendamustine alone in the GADOLIN trial. Leukemia. 2020;34(2):522–532. doi: 10.1038/s41375-019-0559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P.J., Callewaert N., Contreras R., Khorana H.G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. U. S. A. 2002;99(21):13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley N.M., Bertozzi C.R., Pitteri S.J. A pragmatic guide to enrichment strategies for mass spectrometry-based glycoproteomics. Mol. Cell. Proteomics. 2020;20 doi: 10.1074/mcp.R120.002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.T., Patel K.R., Barb A.W. Site-specific N-glycan analysis of antibody-binding Fc gamma receptors from primary human monocytes. Mol. Cell. Proteomics. 2020;19(2):362–374. doi: 10.1074/mcp.RA119.001733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogals M.J., Yang J.Y., Williams R.V., Moremen K.W., Amster I.J., Prestegard J.H. Sparse isotope labeling for nuclear magnetic resonance (NMR) of glycoproteins using 13C-glucose. Glycobiology. 2021;31(4):425–435. doi: 10.1093/glycob/cwaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles G., Morschhauser F., Lamy T., Milpied N., Thieblemont C., Tilly H., Bieska G., Asikanius E., Carlile D., Birkett J., Pisa P., Cartron G. Phase 1 study results of the type II glycoengineered humanized anti-CD20 monoclonal antibody obinutuzumab (GA101) in B-cell lymphoma patients. Blood. 2012;119(22):5126–5132. doi: 10.1182/blood-2012-01-404368. [DOI] [PubMed] [Google Scholar]
- Shajahan A., Heiss C., Ishihara M., Azadi P. Glycomic and glycoproteomic analysis of glycoproteins-a tutorial. Anal. Bioanal. Chem. 2017;409(19):4483–4505. doi: 10.1007/s00216-017-0406-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata-Koyama M., Iida S., Okazaki A., Mori K., Kitajima-Miyama K., Saitou S., Kakita S., Kanda Y., Shitara K., Kato K., Satoh M. The N-linked oligosaccharide at Fc gamma RIIIa Asn-45: an inhibitory element for high Fc gamma RIIIa binding affinity to IgG glycoforms lacking core fucosylation. Glycobiology. 2009;19(2):126–134. doi: 10.1093/glycob/cwn110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrimal S., Trueman S.F., Gilmore R. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. J. Cell Biol. 2013;201(1):81–95. doi: 10.1083/jcb.201301031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi G.P., Barb A.W. The structural role of antibody N-glycosylation in receptor interactions. Structure. 2015;23(9):1573–1583. doi: 10.1016/j.str.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi G.P., Barb A.W. CD16a with oligomannose-type N-glycans is the only "low-affinity" Fc gamma receptor that binds the IgG crystallizable fragment with high affinity in vitro. J. Biol. Chem. 2018;293(43):16842–16850. doi: 10.1074/jbc.RA118.004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi G.P., Hanson Q.M., Barb A.W. Restricted motion of the conserved immunoglobulin G1 N-glycan is essential for efficient FcgammaRIIIa binding. Structure. 2014;22(10):1478–1488. doi: 10.1016/j.str.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subedi G.P., Falconer D.J., Barb A.W. Carbohydrate-polypeptide contacts in the antibody receptor CD16A identified through solution NMR spectroscopy. Biochemistry. 2017;56(25):3174–3177. doi: 10.1021/acs.biochem.7b00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng W.K., Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J. Clin. Oncol. 2003;21(21):3940–3947. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- Wojcik I., Senard T., de Graaf E.L., Janssen G.M.C., de Ru A.H., Mohammed Y., van Veelen P.A., Vidarsson G., Wuhrer M., Falck D. Site-specific glycosylation mapping of Fc gamma receptor IIIb from neutrophils of individual healthy donors. Anal. Chem. 2020;92(19):13172–13181. doi: 10.1021/acs.analchem.0c02342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Mishra H.K., Walcheck B. Role of ADAM17 as a regulatory checkpoint of CD16A in NK cells and as a potential target for cancer immunotherapy. J. Leukoc. Biol. 2019;105(6):1297–1303. doi: 10.1002/JLB.2MR1218-501R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeck A., Pohlentz G., Schlothauer T., Peter-Katalinic J., Regula J.T. Cell type-specific and site directed N-glycosylation pattern of FcgammaRIIIa. J. Proteome Res. 2011;10(7):3031–3039. doi: 10.1021/pr1012653. [DOI] [PubMed] [Google Scholar]
- Zhang W., Gordon M., Schultheis A.M., Yang D.Y., Nagashima F., Azuma M., Chang H.M., Borucka E., Lurje G., Sherrod A.E., Iqbal S., Groshen S., Lenz H.J. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J. Clin. Oncol. 2007;25(24):3712–3718. doi: 10.1200/JCO.2006.08.8021. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Li Y., Wu X., Li L., Liu J., Wang Y., Liu Y., Li Q., Wang Z. Identification of anti-CD16a single domain antibodies and their application in bispecific antibodies. Cancer Biol. Ther. 2020;21(1):72–80. doi: 10.1080/15384047.2019.1665953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Blum R.H., Bjordahl R., Gaidarova S., Rogers P., Lee T.T., Abujarour R., Bonello G.B., Wu J., Tsai P.F., Miller J.S., Walcheck B., Valamehr B., Kaufman D.S. Pluripotent stem cell-derived NK cells with high-affinity noncleavable CD16a mediate improved antitumor activity. Blood. 2020;135(6):399–410. doi: 10.1182/blood.2019000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.