ABSTRACT
Bacterial symbionts often provide critical functions for their hosts. For example, wood-boring bivalves called shipworms rely on cellulolytic endosymbionts for wood digestion. However, how the relationship between shipworms and their bacterial symbionts is formed and maintained remains unknown. Quorum sensing (QS) often plays an important role in regulating symbiotic relationships. We identified and characterized a QS system found in Teredinibacter sp. strain 2052S, a gill isolate of the wood-boring shipworm Bactronophorus cf. thoracites. We determined that 2052S produces the signal N-decanoyl-l-homoserine lactone (C10-HSL) and that this signal controls the activation of a biosynthetic gene cluster colocated in the symbiont genome that is conserved among all symbiotic Teredinibacter isolates. We subsequently identified extracellular metabolites associated with the QS regulon, including ones linked to the conserved biosynthetic gene cluster, using mass spectrometry-based molecular networking. Our results demonstrate that QS plays an important role in regulating secondary metabolism in this shipworm symbiont. This information provides a step toward deciphering the molecular details of the relationship between these symbionts and their hosts. Furthermore, because shipworm symbionts harbor vast yet underexplored biosynthetic potential, understanding how their secondary metabolism is regulated may aid future drug discovery efforts using these organisms.
IMPORTANCE Bacteria play important roles as symbionts in animals ranging from invertebrates to humans. Despite this recognized importance, much is still unknown about the molecular details of how these relationships are formed and maintained. One of the proposed roles of shipworm symbionts is the production of bioactive secondary metabolites due to the immense biosynthetic potential found in shipworm symbiont genomes. Here, we report that a shipworm symbiont uses quorum sensing to coordinate activation of its extracellular secondary metabolism, including the transcriptional activation of a biosynthetic gene cluster that is conserved among many shipworm symbionts. This work is a first step toward linking quorum sensing, secondary metabolism, and symbiosis in wood-boring shipworms.
KEYWORDS: quorum sensing, metabolomics, shipworm, symbiosis
INTRODUCTION
Wood-boring shipworms are bivalves that harbor intracellular gammaproteobacteria in their gills that express cellulases for wood digestion (1, 2). However, many details of the molecular mechanisms that govern the selection and maintenance of symbiotic bacteria by shipworms remain unknown. An analysis of published endosymbiont genomes and shipworm-associated metagenomes has indicated that these gill endosymbionts are also capable of producing a plethora of secondary metabolites comparable to well-known producers, such as Streptomyces spp. (3). Several predicted biosynthetic gene clusters (BGCs) are conserved among shipworm symbionts (3), which could indicate that the products of these clusters play a role in the symbiotic relationship. For example, the boronated antibiotic tartrolon, isolated from the shipworm symbiont Teredinibacter turnerae T7901, is hypothesized to participate in the inhibition of competing parasites in the shipworm gills and/or cecum (4).
Bacterial symbionts often use quorum sensing (QS) to coordinate group behavior, which is thought to help differentiate between a low-density, free-living state and high-density, host-associated state (5). In many proteobacteria, QS is mediated by acyl-homoserine lactone (acyl-HSL) signals produced by LuxI-family synthases (6). In this type of QS system, genes are regulated by members of the LuxR family of transcription factors which bind and respond to acyl-HSLs (6). The first QS system was characterized in the invertebrate symbiont Aliivibrio fischeri, which uses 3-oxo-hexanoyl-l-homoserine lactone (3-oxo-C6-HSL) to regulate bioluminescence in the light organ of its host squid, Euprymna scolopes (7, 8). Characterization of QS systems in shipworm symbionts therefore has the potential to provide insight into the details of their relationship with their host.
QS often regulates the production of extracellular factors, including secondary metabolites and enzymes, such as proteases (6, 9–11). A common example is the plant-associated pathogen Erwinia carotovorum, which is known to produce the antibiotic carbapenem in response to QS (9). In many cases, QS systems regulate adjacent genes in bacterial genomes, and a recent genome mining effort discovered that BGCs neighboring luxR homologs are widespread in proteobacteria (12). Interestingly, only a small percentage of QS-linked BGCs identified in this study were found in free-living and invertebrate-associated bacteria, while plant- and human-associated bacteria made up the majority (12).
One BGC of interest that is found in all cellulolytic shipworm symbionts isolated to date is a predicted hybrid trans-acyltransferase polyketide synthase-nonribosomal peptide synthetase (trans-AT PKS-NRPS) gene cluster termed GCF_3 (3). The product of GCF_3 has not been isolated or characterized. Teredinibacter sp. strain PMS-2052S.S.stab0a.01 (referred to here as 2052S) is a cellulolytic bacterial strain isolated from the gills of a specimen of the shipworm Bactronophorus cf. thoracites collected in Butuan, Agusan del Norte, Philippines. In the genome of 2052S, the GCF_3 BGC is adjacent to a predicted QS system. Determining how this BGC is regulated in a symbiont may enable the identification and characterization of its product.
In this work, we characterized the QS system used by the shipworm endosymbiont 2052S. We identified the acyl-HSL signal and linked it with its cognate synthase and receptor. We then determined that this QS system regulates the neighboring GCF_3 BGC and used untargeted metabolomics and molecular networking to identify metabolites associated with the QS regulon, including potential products of the GCF_3 BGC. To our knowledge, this is the first characterization of a shipworm endosymbiont QS system, which extends our understanding of the molecular details of this symbiosis.
RESULTS AND DISCUSSION
A conserved biosynthetic gene cluster in cellulolytic shipworm symbionts is adjacent to quorum sensing genes in strain 2052S.
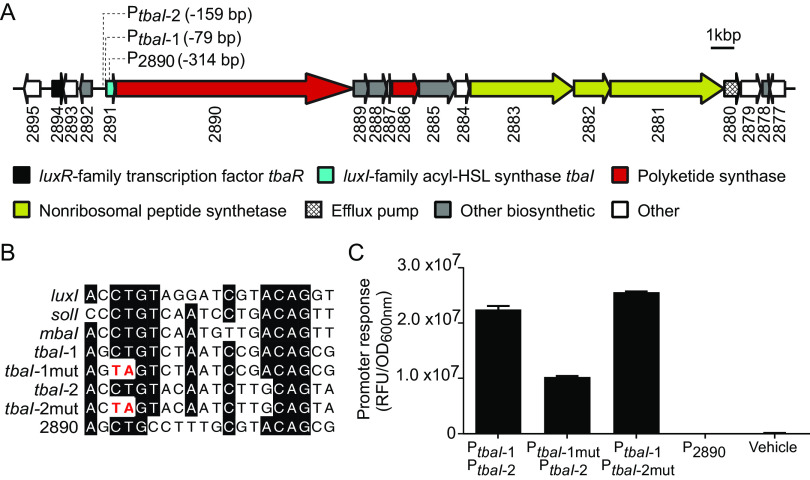
The cellulolytic strain 2052S was isolated from the gills of a specimen of the wood-boring shipworm Bactronophorus cf. thoracites (see Table S1 in the supplemental material for strain isolation information) (3). It is likely an intracellular symbiont like other Teredinibacter species (1); however, more studies will be needed to determine this classification definitively. In the genome of 2052S, the conserved BGC GCF_3 is adjacent to a luxR-family transcription factor gene (K256DRAFT_2894, tbaR) and an acyl-HSL synthase gene (K256DRAFT_2891, tbaI) (Fig. 1A). We therefore hypothesized that GCF_3 may be regulated by QS in this strain, as is true with other QS-linked BGCs in proteobacteria (10, 11). In other shipworm symbionts, GCF_3 is not adjacent to QS genes (see Fig. S1 in the supplemental material), indicating that QS may have been lost in those isolates or gained in 2052S.
FIG 1.
A quorum sensing system in the Teredinibacter sp. strain 2052S genome is adjacent to a conserved biosynthetic gene cluster. (A) Quorum sensing genes (tbaI and tbaR) neighbor a predicted hybrid trans-AT-PKS-NRPS biosynthetic gene cluster (3). Identified putative TbaR-binding sites are represented by dotted lines from their position in the cluster and list the number of base pairs they are located upstream of the start codon of the indicated gene. Numbers correspond to locus tags (K256DRAFT_XXXX) in the Joint Genome Institute Integrated Microbial Genomes (IMG) system (25). Genes are colored according to predicted function in antiSMASH 6.0 (26). (B) Comparison of known LuxR-type binding sites in the promoter sequences of Aliivibrio fischeri luxI, Ralstonia solanacearum solI, and Methylobacter tundripaludum mbaI with the putative TbaR-binding sites upstream of the acyl-HSL synthase gene tbaI and predicted PKS gene K256DRAFT_2890. (C) Response of E. coli reporter strains containing gfp fused to different promoter regions with putative TbaR-binding sites shown in B to 100 nM C10-HSL or ethyl acetate (vehicle). Data are the mean ± standard deviation of three technical replicates and are representative of two independent experiments. RFU, relative fluorescence units; OD, optical density.
We also found QS genes adjacent to BGCs in the genomes of other shipworm symbionts (see Fig. S2 in the supplemental material). However, these BGCs had low similarity to GCF_3, suggesting that QS may regulate the production of other secondary metabolites in these strains. Notably, Teredinibacter turnerae T7901, the most well-characterized shipworm symbiont, does not harbor a luxI-family synthase gene or complete luxR-family transcription factor gene. We have thus far only identified predicted QS systems in isolates from wood-boring shipworms that are not dominated by T. turnerae (3).
2052S produces and responds to the quorum sensing signal N-decanoyl-l-homoserine lactone.
We characterized the 2052S QS system by first determining if this strain can produce and respond to a QS signal under laboratory conditions. We identified two potential LuxR-family binding sites upstream of the tbaI acyl-HSL synthase gene (Fig. 1A and B), which is often positively autoregulated by its cognate LuxR-family transcription factor upon signal binding (6). We then constructed a two-plasmid reporter system (PtbaI-gfp) in Escherichia coli in which one plasmid expresses tbaR under its native promoter and the other plasmid contains the tbaI promoter, which includes the putative LuxR-family binding sites, fused to gfp. Adding an organic extract of the supernatant from a 2052S culture to the PtbaI-gfp reporter strain resulted in a significant increase in GFP fluorescence compared with a solvent control (see Fig. S3 in the supplemental material). This finding confirms that 2052S produces a QS signal and that the LuxR-family homolog TbaR binds this signal and activates tbaI expression in a positive feedback loop.
We then determined which of the two putative binding sites is primarily used by TbaR by constructing two separate reporter strains containing a CT-to-TA mutation in the conserved region of each site (Fig. 1B). We found that GFP fluorescence was unaffected when the mutation was introduced in PtbaI-2, suggesting that PtbaI-1 is the primary TbaR binding site (Fig. 1C). However, mutating PtbaI-1 did not completely abolish gfp activation, suggesting that TbaR can also bind PtbaI-2.
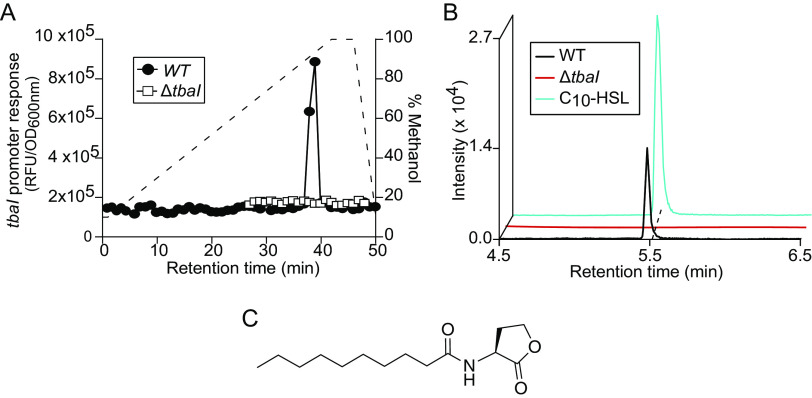
In order to isolate and characterize the acyl-HSL signal produced by 2052S, we separated organic supernatant extract by high-performance liquid chromatography (HPLC) and tested each fraction using the PtbaI-gfp reporter strain. This procedure resulted in one peak of GFP fluorescence in two adjacent fractions, which was not present in the supernatant from an unmarked, in-frame ΔtbaI mutant we constructed using sucrose counterselection (Fig. 2A). We detected a feature with an m/z of 256 in the pooled active fractions using liquid chromatography-mass spectrometry (LC-MS), which is consistent with the protonated mass of N-decanoyl-l-homoserine lactone (C10-HSL).
FIG 2.
Teredinibacter sp. strain 2052S produces the quorum sensing signal C10-HSL. (A) PtbaI-gfp activity of HPLC-fractionated culture supernatant extracts from 2052S and the ΔtbaI mutant. The dashed line shows the methanol gradient. (B) Extracted ion chromatogram of supernatant extracts of 2052S and the ΔtbaI mutant compared with a commercial C10-HSL signal for m/z 279.1812, corresponding to the sodiated adduct of C10-HSL. Mass tolerance, <5 ppm. (C) Structure of C10-HSL. RFU, relative fluorescence units; OD, optical density.
We confirmed that 2052S produces C10-HSL by using high-resolution LC-MS/MS to compare organic supernatant extracts from the 2052S and ΔtbaI strains with a commercial standard of C10-HSL (Fig. 2B). The retention time and fragmentation pattern of the signal produced by 2052S were indistinguishable from the commercial standard (Fig. 2B; see Table S2 in the supplemental material). Furthermore, the PtbaI-gfp reporter strain was responsive to the commercial standard (see Fig. S4 in the supplemental material), and we used this reporter assay to determine that 2052S produces approximately 250 ± 22 nM signal during early stationary phase. Together, these results demonstrate that the bacterial endosymbiont 2052S produces and responds to the QS signal C10-HSL.
Transcription of the conserved biosynthetic gene cluster GCF_3 is regulated by quorum sensing in 2052S.
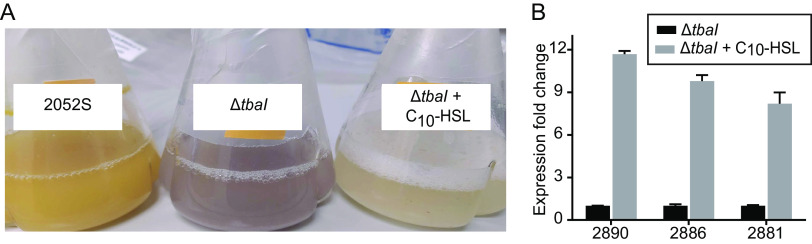
We next sought to determine what 2052S regulates using QS. The ΔtbaI mutant is still capable of using cellulose as its primary carbon source, indicating that cellulase production is not regulated by QS in 2052S (all experiments used cellulose as the carbon source, see also Fig. S5 in the supplemental material). We noticed a significant change in the pigmentation of the ΔtbaI culture, which we could partially complement by adding exogenous C10-HSL (Fig. 3A). Researchers have also observed a change in pigmentation in Prosthecomicrobium hirschii upon disruption of its QS system (13).
FIG 3.
The conserved BGC is transcriptionally activated by the addition of C10-HSL to the ΔtbaI mutant. (A) Pigmentation phenotype of 2052S (left) and ΔtbaI mutant (middle) cultures, and partial reversion to wild-type phenotype upon addition of QS signal to the ΔtbaI culture (right). (B) RT-qPCR results showing relative expression of GCF_3 genes K256DRAFT_2890, 2886, and 2881 upon addition of C10-HSL to the ΔtbaI strain normalized to ΔtbaI expression in the absence of signal. The 16S rRNA gene was used as a reference gene. Data are the mean ± standard deviation of three technical replicates and are representative of two independent experiments.
The observed change in pigmentation indicated that QS may play a role in regulating secondary metabolite production in 2052S. We used reverse transcription quantitative PCR (RT-qPCR) to determine if the conserved GCF_3 BGC adjacent to the QS genes in the 2052S genome is regulated by QS. We quantified the transcription of core genes in this cluster (K256DRAFT_2890, K256DRAFT_2886, and K256DRAFT_2881) in 2052S, the ΔtbaI mutant, and the ΔtbaI mutant chemically complemented with C10-HSL. All three core biosynthetic genes were found to be expressed in a QS signal-dependent manner in a late log-phase culture (Fig. 3B).
The first gene in the GCF_3 cluster is K256DRAFT_2890, which encodes a predicted multidomain, trans-AT-PKS. We identified a putative TbaR-binding site upstream of K256DRAFT_2890 within the tbaI gene (Fig. 1A and B). However, when we created a reporter strain containing this upstream region, it did not drive gfp expression in response to C10-HSL (Fig. 1C). This finding suggests that both tbaI and K256DRAFT_2890 are transcribed together in the same operon, which we confirmed by RT-PCR (see Fig. S6 in the supplemental material). Together, these results demonstrate that 2052S uses QS to coordinate the activation of the conserved GCF_3 BGC.
QS regulates the majority of the extracellular metabolome of 2052S.
Because GCF_3 contains predicted efflux pumps (Fig. 1A), we sought to identify changes in the extracellular metabolome of 2052S that are controlled by QS in order to determine which secondary metabolites are linked to this cluster. We first constructed an unmarked, in-frame deletion mutant of the 1.3-kb polyketide synthase gene K256DRAFT_2886 (Δ2886) and also complemented this mutant using a plasmid containing K256DRAFT_2886 driven by the 2052S rpoD promoter (pAWP275). We observed the same pigmentation in the Δ2886 mutant as that in the wild-type 2052S, which suggests that GCF_3 is not responsible for the change in pigmentation observed in the ΔtbaI mutant. We then used untargeted metabolomics to compare crude organic supernatant extracts from stationary-phase cultures of 2052S, the ΔtbaI mutant, the ΔtbaI mutant supplemented with C10-HSL, the Δ2886 mutant, and the Δ2886 mutant complemented with pAWP275.
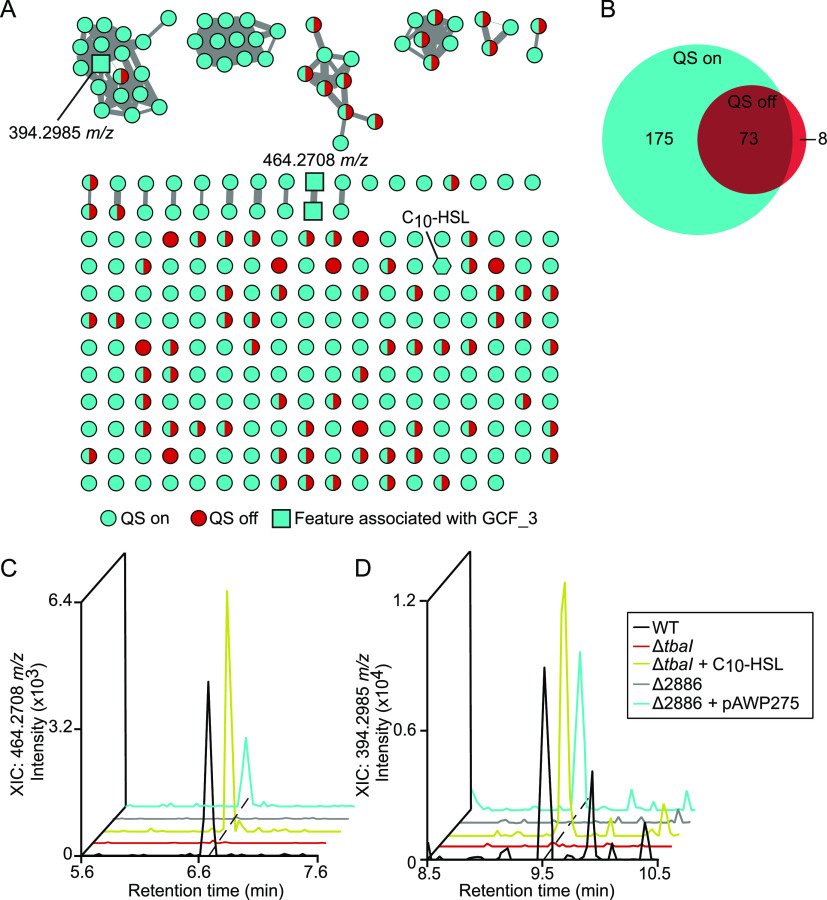
We analyzed the data using tandem MS (MS/MS)-based molecular networking within the Global Natural Products Social molecular networking (GNPS) platform, which clusters metabolites based on MS/MS fragmentation patterns (14). This analysis produced a primary data set of 442 features from raw LC-MS/MS scans. We further refined this data set by removing features found in the uninoculated growth medium as well as those not found in both independent replicates, resulting in a final data set of 256 features (Fig. 4A). The majority (175/256, 68%) of features were detectable only in the supernatant of QS-active strains (Fig. 4B). This result indicates that QS plays an important role in the regulation of secondary metabolism in this shipworm endosymbiont. Notably, none of the 2052S extracellular secondary metabolites had matches to compounds in the GNPS spectral library. This finding highlights the potentially unique biosynthetic potential of shipworm endosymbionts.
FIG 4.
Quorum sensing regulates the majority of extracellular metabolites produced by 2052S. (A) Molecular network of untargeted metabolomics data of supernatant extracts from cultures of 2052S, ΔtbaI, and ΔtbaI supplemented with C10-HSL. Features found in samples where QS is on (2052S and ΔtbaI + C10-HSL) are shown as cyan nodes; features found in the ΔtbaI culture, where QS is off, are shown as red nodes; and features found in both samples where QS is on and off are shown as split cyan and red nodes. Features identified as associated with GCF_3 (present in 2052S, ΔtbaI + C10-HSL, and Δ2886 + pAWP275, but absent in ΔtbaI and Δ2886) are shown as square nodes, and C10-HSL is shown as a hexagonal node. Edge width is scaled with cosine similarity score. (B) Venn diagram of features associated with the QS regulon. (C and D) Extracted ion chromatograms of m/z 464.2708 (C) and m/z 394.2985 (D) in culture supernatant extracts. Mass tolerance, <5 ppm.
To identify the putative product of the GCF_3 BGC in 2052S, we focused on extracellular metabolites present in cultures of 2052S, the ΔtbaI mutant supplemented with C10-HSL, and the Δ2886 mutant complemented with pAWP275, but absent in the ΔtbaI and Δ2886 mutants. We identified two putative metabolites matching this pattern that may be products of this gene cluster (Fig. 4C and D), including a feature with a precursor ion mass of 394.2985 m/z found in the largest cluster in the network (Fig. 4A). This cluster was found almost exclusively in QS-active samples. These metabolites will require further investigation to determine their structure and function.
We have identified and characterized a QS system in Teredinibacter sp. strain 2052S, a symbiont of the wood-boring shipworm B. cf. thoracites. We determined that 2052S produces and responds to the signal C10-HSL and that this signal regulates the activation of a BGC that is conserved among all wood-boring shipworm symbiont isolates, termed GCF_3. It is possible that secondary metabolites produced by shipworm endosymbionts play a role in establishing and maintaining the relationship between these bacteria and their host. The discovery of a symbiont that regulates its extracellular secondary metabolism using QS is consistent with this hypothesis, as QS is often thought to enable bacterial symbionts to differentiate between planktonic and host-associated states (5, 9). More studies will be needed to understand the role of these metabolites, as well as QS, in this symbiotic relationship.
MATERIALS AND METHODS
Plasmid construction.
All plasmids were constructed using Gibson assembly (15), with the exception of the reporter plasmids pAWP239, pAWP381, pAWP479, and pAWP480, which were constructed by inserting upstream gene sequences into the promoter probe plasmid pPROBE-GFP[LVA] (16) at the EcoRI and SacI restriction sites. The upstream sequences in pAWP479 and pAWP480, which contain CT-to-TA mutations, were ordered as gBlocks from Integrated DNA Technologies. Plasmids and primers used in this study are listed in Table 1 and 2, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Puri lab strain/plasmid collection no. | Description | Source or reference |
|---|---|---|---|
| Strain | |||
| E. coli TOP10 | EAWP2 | F– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| E. coli S17-1 λpir | EAWP3 | Donor strain. Tpr Smr recA thi pro hsd(r-m+) RP4-2-Tc::Mu::Km Tn7 λpir | 18 |
| Teredinibacter sp. 2052S.S.stab0a.01 | AWP283 | Shipworm symbiont isolated from the gills of Bactronophorus cf. thoracites. | 3, Table S1 |
| Teredinibacter sp. 2052S.S.stab0a.01 ΔtbaI | AWP284 | Shipworm symbiont isolated from the gills of Bactronophorus cf. thoracites. ΔtbaI | This study |
| Teredinibacter sp. 2052S.S.stab0a.01 Δ2886 | AWP285 | Shipworm symbiont isolated from the gills of Bactronophorus cf. thoracites. ΔK256DRAFT_2886 | This study |
| Teredinibacter sp. 2052S.S.stab0a.01 Δ2886 + pAWP275 | AWP286 | Shipworm symbiont isolated from the gills of Bactronophorus cf. thoracites. ΔK256DRAFT_2886 + pAWP275, KanR | This study |
| E. coli reporter strain PtbaI-gfp | EAWP128 | Acyl-HSL reporter strain; E. coli TOP10 with pAWP239 and pAWP240, Kanr Cmr | This study |
| E. coli reporter strain P2886-gfp | EAWP201 | Acyl-HSL reporter strain; E. coli TOP10 with pAWP381 and pAWP240, Kanr Cmr | This study |
| E. coli reporter PtbaI-1mut-gfp | EAWP202 | Acyl-HSL reporter strain; E. coli TOP10 with pAWP479 and pAWP240, Kanr Cmr | This study |
| E. coli reporter strain PtbaI-2mut-gfp | EAWP203 | Acyl-HSL reporter strain; E. coli TOP10 with pAWP480 and pAWP240, Kanr Cmr | This study |
| Plasmids | |||
| pPROBE-gfp[LVA] | Promoter probe vector | 16 | |
| pACYC184 | Replicating vector containing the p15A origin of replication | 27 | |
| pCM433kanT | Sucrose counterselection vector for constructing in-frame, unmarked deletion mutants | 28 | |
| pBBR1MCS-2 | Broad host range replicating vector | 29 | |
| pAWP239 | pPROBE-gfp[LVA] containing the tbaI promoter (−400 bp to +21 bp of the translational start site of K256DRAFT_2891) fused to gfp | This study | |
| pAWP240 | pACYC184 expressing tbaR K256DRAFT_2894) under its native promotor (400 bp upstream sequence) | This study | |
| pAWP381 | pPROBE-gfp[LVA] containing the P2886 promoter (−400 bp to +21 bp of the translational start site of K256DRAFT_2886) fused to gfp | This study | |
| pAWP378 | pCM433kanT containing flanks to create in-frame, unmarked deletion of tbaI (K256DRAFT_2891) | This study | |
| pAWP387 | pCM433kanT containing flanks to create in-frame, unmarked deletion of K256DRAFT_2886 | This study | |
| pAWP275 | pBBR1MCS-2 containing K256DRAFT_2886 driven by the 2052S rpoD promoter (300 bp upstream sequence of K256DRAFT_2811) | This study | |
| pAWP479 | pPROBE-gfp[LVA] containing the PtbaI-1mut promoter fused to gfp, pAWP239 with mutation of CT to TA in tbaI promoter in putative TbaR binding site PtbaI-1 (AGCTGTCTAATCCGACAGCG to AGTAGTCTAATCCGACAGC) (Fig. 1B) | This study | |
| pAWP480 | pPROBE-gfp[LVA] containing the PtbaI-2mut promoter fused to gfp, pAWP239 with mutation of CT to TA in tbaI promoter in putative TbaR binding site PtbaI-2 (ACCTGTACAATCTTGCAGTA to ACTAGTACAATCTTGCAGTA) (Fig. 1B) | This study |
TABLE 2.
Cloning and diagnostic primers used in this study
| Primer | Sequence (5′–3′)a | Description |
|---|---|---|
| oAWP186_433KTV1_fwd | ATGTGCAGGTTGTCGGTGTC | For amplifying the pCM433kanT backbone in two pieces. |
| oAWP160_433KTV1_rev | ATAAAGGTGAATCCCATAGGGCAGGA GCTATAATCTCGAGTCCCGTCAAG | |
| oAWP159_433KTV2_fwd | TAGCTCCTGCCCTATGGGAT | |
| oAWP187_433KTV2_rev | TGGTAACTGTCAGACCAAGTTTACTC | |
| oAWP937_239I_SacI_fwd | GGTGGTGAGCTCAATGATTGTGCCGAAATTATTG | For amplifying the tbaI upstream region to insert into pPROBE-gfp[LVA] promoter probe vector. Also used on gBlocks of PtbaI-1mut and PtbaI-2mut. |
| oAWP938_239I_EcoRI_rev | GGTGGTGAATTCAGTAATTACGATTGTGTTCA | |
| oAWP939_240I_fwd | CCTAATGCAGGAGTCGCATAATGGGGGTAATATTTAACACGC | For amplifying tbaR and 400 bp upstream to insert into pACYC184. |
| oAWP940_240I_rev | CGTTGACTCTCAGTCATAGTTCAGGGCGTCGGCTTAATTAA | |
| oAWP1558 | ATATGAGTAAACTTGGTCTGACAGTTACCAGACAGGCTGTTGATCTTTCCA | For amplifying flanks to create the ΔtbaI in-frame, unmarked deletion. |
| oAWP1559 | GGTGTGGATTGCGAGCATATCCTTTTCGCCCCCGTT | |
| oAWP1560 | GGCGAAAAGGATATGCTCGCAATCCACACCCACTGA | |
| oAWP1561 | CGTGCATCACGACACCGACAACCTGCACATCGAGCGGCTTCAGCAGCAAAA | |
| oAWP1426_010U_fwd | TCCTGTTGAAAGTAACATAGTGAGAGTCCTTCTTGCT | For amplifying flanks to create the Δ2886 in-frame, unmarked deletion. |
| oAWP1432_010U_rev | ATATGAGTAAACTTGGTCTGACAGTTACCACCGAGGAAAATACTGCCCATT | |
| oAWP1428_010D_fwd | AGGACTCTCACTATGTTACTTTCAACAGGACAATGACC | |
| oAWP1429_010D_rev | CGTGCATCACGACACCGACAACCTGCACATTCGAGCATGTAGACCTCATGG | |
| oAWP1418_009I_rev | GCGGGGATCTCATGCTGGAGTTCTTCGCCCTCATTGTCCTGTTGAAAGTAAA | For amplifying K256DRAFT_2886 to fuse downstream of 2052S rpoD promoter and insert into pBBR1MCS-2. |
| oAWP1419_009I_fwd | TCCGCAGGACTTTTAATGTCTCAAGTATTTATGTTTCC | |
| oAWP1420_PrpoD_009I_fwd | CAGTCACGACGTTGTAAAACGACGGCCAGTAGGAAGCCAGCAAGCGGGAA | For amplifying 2052S rpoD promoter to fuse with the K256DRAFT_2886 gene. |
| oAWP1421_PrpoD_009I_rev | AAATACTTGAGACATTAAAAGTCCTGCGGAGATTGGAG | |
| oAWP1402_004I_SacI_fwd | GGTGGTGAGCTCGAACTTCTTCAGGGTGAAGAAGCTC | For amplifying K256DRAFT_2886 upstream region to insert into pPROBE-gfp[LVA] promoter probe vector. |
| oAWP1403_004I_EcoRI_rev | GGTGGTGAATTCGCCAATAATTGCAATATCCATGTG | |
| oAWP1488_rev | GCGCCGTCACTATCGATCTTGT | For determining if tbaI and K256DRAFT_2890 are transcribed in the same operon. |
| oAWP1489_fwd | TTCGCCAGTCCCAACGACTTGC |
Homology regions used for Gibson assembly and restriction enzymes sites are underlined.
Strain growth.
Strains used in this study are listed in Table 1. Escherichia coli strains were grown in lysogeny broth (LB) at 37°C. Teredinibacter sp. strain PMS-2052S.S.stab0a.01 (2052S) was isolated from Bactronophorus cf. thoracites (PMS-1959H) collected from Butuan, Agusan del Norte, Philippines (Table S1), and was grown on shipworm basal medium (SBM) (30°C and 200 rpm) as described previously by Distel et al. (17). SBM contains NaCl (19.8 g/L), NH4Cl (267.5 mg/L), MgCl2·6H2O (8.95 g/L), Na2SO4 (3.31 g/L), CaCl2·2H2O (1.25 g/L), NaHCO3 (0.162 g/L), Na2CO3 (10 mg/L), KCl (0.552 mg/L), KBr (81 mg/L), H3BO3 (21.5 mg/L), SrCl2·6H2O (19.8 mg/L), KH2PO4 (3.82 mg/L), NaF (2.48 mg/L), Na2MoO4·2H2O (2.5 mg/L), MnCl2·4H2O (1.8 mg/L), ZnSO4. 7H2O (0.22 mg/L), CuSO4·5H2O (0.079 mg/L), Co(NO3)2·6H2O (0.049 mg/L), Fe-EDTA complex (4.15 mg/L), and HEPES (4.76 g/L) adjusted to pH 8.0. The carbon source used was Sigmacell cellulose type 101 (0.2 g/L). We determined a growth curve for 2052S grown on cellulose by spot plating 10 μL of the culture periodically and calculating the CFU per mL for 36 h (see Fig. S7 in the supplemental material).
Genetic manipulation.
Genetic manipulation of all strains derived from 2052S was performed at 30°C. Verified plasmids were conjugated into 2052S using the E. coli donor strain S17-1 (18) using the following method. 500 μL of exponentially growing cultures of donor and recipient strains were pelleted and washed with sterile ultrapure water. The two pellets were then combined in a total volume of 50 μL and spotted onto an SBM plate containing 10% (vol/vol) nutrient broth and subsequently incubated for 2 days. Successful conjugants were selected on SBM plates containing kanamycin (50 μg mL−1). To construct the unmarked deletion mutants ΔtbaI and Δ2886, kanamycin-resistant integrants (single crossovers) were restreaked and grown in SBM broth with no kanamycin before being spread onto an SBM plate containing 5% (vol/vol) sucrose for counterselection. The resulting colonies were screened for double crossovers by kanamycin sensitivity and colony PCR before the final mutants were verified by PCR and Sanger sequencing.
Identification of LuxR-type binding site.
Nucleotide sequences were identified as putative LuxR-type binding sites if they satisfied the following criteria: (i) located within 400 bp upstream of the translational start site, (ii) matched the general NNCTG-N10-CAGNN pattern with one mismatch or less, and (iii) contained eight or more base pairs with dyad symmetry (10, 19).
Acyl-HSL reporter assay.
The reporter assay was performed as described in reference 20. Briefly, cultures were centrifuged at 16,000 × g and the supernatant from a culture of 2052S was extracted twice with an equal volume of ethyl acetate containing 0.01% (vol/vol) acetic acid. The organic phase was subsequently dried under a nitrogen stream. The dried extract was then resuspended in acidified ethyl acetate, aliquoted into 1.5-mL tubes, and redried before a stationary-phase E. coli reporter strain diluted to an optical density (OD) of 0.1 was added to the tube. After 4 h of incubation (37°C and 200 rpm), GFP fluorescence (485-nm excitation, 510-nm emission) and absorbance at 600 nm were measured in a 96-well black, clear-bottomed plate by using a plate reader (SpectraMax i3x).
Isolation and characterization of the QS signal.
The acyl-HSL signal produced by 2052S was extracted from the supernatant of a 50-mL culture grown to early stationary phase as described above. The supernatant extract was resuspended in methanol, and 20% (i.e., from 10 mL of culture) was separated by HPLC using a Waters SunFire C18 column (4.6 by 100 mm, 5 μm) at 1.0 mL/min using a linear gradient of 10% to 100% methanol in water over 50 min. A 5-μL aliquot of each 1.0-mL fraction was analyzed using the E. coli reporter strain described above. The pooled adjacent fractions that showed GFP activity were then analyzed using LC-MS. Confirmation of the identified signal was performed using a C10-HSL standard purchased from Cayman Chemical.
RNA preparation.
Exponentially growing cultures of 2052S were diluted to an optical density at 600 nm (OD600) of 0.01 and were grown until log phase prior to RNA extraction. For the ΔtbaI + C10-HSL signal, a stock of 2 mM C10-HSL in dimethyl sulfoxide (DMSO) was added to a final concentration of 2 μM every 12 h. Subsequently, cultures were chilled on ice and then centrifuged at 4,700 rpm for 15 min at 4°C. Pellets were then stored at −80°C until further processing. Cell pellets were lysed by bead beating with 0.1-mm zirconia-silica beads in 1 mL TRIzol (ThermoFisher). A total of 200 μL of chloroform was then added, and the mixture was separated by centrifugation using phasemaker tubes (ThermoFisher). Subsequently 1.5 volumes of 100% ethanol was added to the aqueous phase of the extract, which was then used for DNase I treatment (Invitrogen) and cleanup using an Invitrogen RNA PureLink minikit according to the manufacturer’s instructions. The resulting purified RNA was checked for DNA contamination by Nanodrop and PCR using the degenerate 16S primers 27F and 1492R.
RT-qPCR.
cDNA was prepared for RT-qPCR by reverse transcribing 1 microgram of the extracted RNA using iScript reverse transcription Supermix (Bio-Rad). qPCR was performed using iTaq-universal SYBR green Supermix (Bio-Rad) containing 400 nM primers, and cDNA was normalized across all samples in a total volume of 10 μL. qPCRs were performed on a Bio-Rad CFX Opus 96 thermal cycler, and threshold cycle (CT) values were calculated using Bio-Rad CFX Maestro software. All primers and their corresponding gene targets are listed in Table 3.
TABLE 3.
Reverse transcription quantitative PCR primers used in this study
| Primer | Sequence (5′–3′) | Target |
|---|---|---|
| oAWP1064_qPCR_16s_fwd | AAGCAACGCGAAGAACCTTA | 16s rRNA reference gene |
| oAWP1065_qPCR_16s_rev | CACCGGCAGTCTCCTTAGAG | |
| oAWP1564_qPCR_ctg2827_fwd | AAATACCTGCTCGCGTCCGCT | 2052S trans-AT PKS gene, K256DRAFT_2890 |
| oAWP1565_qPCR_ctg2827_rev | TCGCTTTATGGACGCCTGCG | |
| oAWP1068_qPCR_ctg2823_fwd | GTAGCACTCGGGGTGATTGT | 2052S PKS gene, K256DRAFT_2886 |
| oAWP1069_qPCR_ctg2823_rev | ACAGCCTTGGGGAATATGTG | |
| oAWP1566_qPCR_ctg2819_fwd | GTCACCTGCAATTCCGGTGTG | 2052S NRPS gene, K256DRAFT_2881 |
| oAWP1567_qPCR_ctg2819_rev | ATGCCGGCGCAATTTGTGGTG |
High-resolution LC-MS/MS for acyl-HSL identification.
Mass spectrometry data were collected using a Waters Acquity I-class ultra-high pressure liquid chromatograph coupled to a Waters Xevo G2-S quadrupole time-of-flight mass spectrometer. An Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (2.1 by 50 mm) was used for separation of samples. Solvent A included water + 0.1 % (vol/vol) formic acid and solvent B included acetonitrile + 0.1% (vol/vol) formic acid. The sample was eluted from the column using a 10-min linear solvent gradient as follows: 0 to 0.1 min, 1% B; 0.1 to 10 min, 1 to 100% B. The solvent flow rate was 0.45 mL min−1. Mass spectra were collected in positive ion mode, with following parameters: 3 kV capillary voltage, 25 V sampling cone voltage, 150°C source temperature, 500°C desolvation temperature, and nitrogen desolvation at 800 L/h. The fragmentation spectra were collected using the same parameters with a 10- to 25-eV collision energy ramp. The lockspray solution was 200 pg/μL leucine enkephalin. The lockspray flow rate was 6 μL/min. Sodium formate was used to calibrate the mass spectrometer.
Untargeted high-resolution (HR) LC-MS/MS for molecular networking.
HR-MS/MS data were obtained from a 100% methanol elution of HP-20 Diaion resin (Sigma) that had been incubated with culture supernatant for at least 4 h collected from a late log phase (48 h) culture of 2052S, ΔtbaI, Δ2886, Δ2886 + pAWP275, or ΔtbaI + C10-HSL. Samples were passed through an C18 solid-phase extraction cartridge prior to being dissolved in 50% ACN/H2O at a final concentration of 1.0 mg/mL. For data collection, the suggested settings for Waters mass spectrometer in reference 19 were used. An Acquity UPLC BEH C18 column (2.1 by 50 mm) was used for the separation of samples. Solvent A included water + 0.1 % (vol/vol) formic acid and solvent B included acetonitrile + 0.1% (vol/vol) formic acid. A flow rate of 0.6 mL/min was used with the following gradient: 0 to 12 min, 1 to 100% B; 12 to 13 min, 100% B; and a column reconditioning phase until 15 min. The following parameters were used: 2.5 kV capillary voltage, 20 V sampling cone voltage, 120°C source temperature, 350°C desolvation temperature, and desolvation gas flow at 800 L/hr. MS1 acquisition range was set to m/z 100 to 1,500 with a scan time of 0.1 s in data-dependent acquisition mode. The top 5 most abundant MS1 ions were selected in each scan, and up to five MS2 scans in collision-induced dissociation (CID) mode was acquired with a 0.1-s scan time in positive mode. The MS survey was set to switch to MS2 acquisition when total ion chromatogram (TIC) rises above an intensity of 5.0 × 103, and MS2 acquisition switches back to MS survey after 0.25 s has elapsed. The collision energy gradient was set to gradient parameters as follows: 20 to 40 V for 100 Da to 60 to 80 V for 1,500 Da.
Molecular networking.
A molecular network was created using the online workflow (https://ccms-ucsd.github.io/GNPSDocumentation/) on the GNPS website (http://gnps.ucsd.edu). The data were filtered by removing all MS/MS fragment ions within ± 17 Da of the precursor m/z. MS/MS spectra were window filtered by choosing only the top 6 fragment ions in the ± 50 Da window throughout the spectrum. The precursor ion mass tolerance was set to 0.02 Da and the MS/MS fragment ion tolerance to 0.02 Da. A network was then created where edges were filtered to have a cosine score above 0.7 and more than 6 matched peaks. Furthermore, edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. Finally, the maximum size of a molecular family was set to 100, and the lowest scoring edges were removed from molecular families until the molecular family size was below this threshold. The spectra in the network were then searched against GNPS spectral libraries (21, 22). The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a cosine score above 0.7 and at least 6 matched peaks. DEREPLICATOR was used to annotate MS/MS spectra (23). The molecular networks were visualized using Cytoscape software (24).
Data availability.
The mass spectrometry data were deposited in the public repository MassIVE (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=f823719f2c5f4fca903bfe49f6964f45). The molecular networking job can be publicly accessed online at: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=d4ce2cc7d4db423eb00a4d2876993b50.
ACKNOWLEDGMENTS
We thank D. Petras (University of Tübingen) for reading the manuscript and providing helpful advice on the molecular networking analysis. We thank H. Naka (University of Utah) for initial help with T. turnerae genetics.
This work was supported by National Institutes of Health grant R00 GM118762 (to A.W.P.) and National Institutes of Health Fogarty International Center Philippine Mollusk Symbiont-International Cooperative Biodiversity Group (PMS-ICBG) grant U19TW008163 (to G.P.C. and M.G.H.). This work was also supported by funding from the Undergraduate Research Opportunities Program at the University of Utah (to E.G.M.).
J.M.D.R. and A.W.P. designed experiments. J.M.D.R. and E.G.M. performed experiments. M.A.A. isolated 2052S. G.P.C., M.G.H., and A.W.P. oversaw and supported the research. J.M.D.R. and A.W.P. wrote the manuscript. All authors have read and approved of the final version of the manuscript.
We declare no conflict of interest.
The work was completed under supervision of the Department of Agriculture-Bureau of Fisheries and Aquatic Resources, Philippines (DA-BFAR), in compliance with all required legal instruments and regulatory issuances covering the conduct of the research.
Footnotes
Supplemental material is available online only.
Contributor Information
Aaron W. Puri, Email: a.puri@utah.edu.
Laura Villanueva, Royal Netherlands Institute for Sea Research.
REFERENCES
- 1.Distel DL, Beaudoin DJ, Morrill W. 2002. Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl Environ Microbiol 68:6292–6299. 10.1128/AEM.68.12.6292-6299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterbury JB, Calloway CB, Turner RD. 1983. A cellulolytic nitrogen-fixing bacterium cultured from the gland of deshayes in shipworms (Bivalvia: Teredinidae). Science 221:1401–1403. 10.1126/science.221.4618.1401. [DOI] [PubMed] [Google Scholar]
- 3.Altamia MA, Lin Z, Trindade-Silva AE, Uy ID, Shipway JR, Wilke DV, Concepcion GP, Distel DL, Schmidt EW, Haygood MG. 2020. Secondary metabolism in the gill microbiota of shipworms (Teredinidae) as revealed by comparison of metagenomes and nearly complete symbiont genomes. mSystems 5:e00261-20. 10.1128/mSystems.00261-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Connor RM, V FJN, Abenoja J, Bowden G, Reis P, Beaushaw J, Relat RMB, Driskell I, Gimenez F, Riggs MW, Schaefer DA, Schmidt EW, Lin Z, Distel DL, Clardy J, Ramadhar TR, Allred DR, Fritz HM, Rathod P, Chery L, White J. 2020. A symbiotic bacterium of shipworms produces a compound with broad spectrum anti-apicomplexan activity. PLoS Pathog 16:e1008600. 10.1371/journal.ppat.1008600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster M, Sexton JD, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol 67:43–63. 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 7.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444–2449. 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 8.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–4586. 10.1128/JB.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGowan S, Sebaihia M, Jones S, Yu B, Bainton N, Chan PF, Bycroft B, Stewart GSAB, Williams P, Salmond GPCY. 1995. Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology (Reading) 141:1268–1268. 10.1099/13500872-141-5-1268. [DOI] [PubMed] [Google Scholar]
- 10.Puri AW, Schaefer AL, Fu Y, Beck DAC, Greenberg EP, Lidstrom ME. 2016. Quorum sensing in a methane-oxidizing bacterium. J Bacteriol 199:e00773-16. 10.1128/JB.00773-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seyedsayamdost MR, Chandler JR, Blodgett JAV, Lima PS, Duerkop BA, Oinuma K-I, Greenberg EP, Clardy J. 2010. Quorum-sensing-regulated bactobolin production by Burkholderia thailandensis E264. Org Lett 12:716–719. 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brotherton CA, Medema MH, Greenberg EP. 2018. luxR Homolog-linked biosynthetic gene clusters in Proteobacteria. mSystems 3:e00208-17. 10.1128/mSystems.00208-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao L, Schaefer AL, Coutinho BG, Brown PJB, Greenberg EP. 2018. An aryl-homoserine lactone quorum-sensing signal produced by a dimorphic prosthecate bacterium. Proc Natl Acad Sci USA 115:7587–7592. 10.1073/pnas.1808351115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aron AT, Gentry EC, McPhail KL, Nothias L-F, Nothias-Esposito M, Bouslimani A, Petras D, Gauglitz JM, Sikora N, Vargas F, van der Hooft JJJ, Ernst M, Kang KB, Aceves CM, Caraballo-Rodríguez AM, Koester I, Weldon KC, Bertrand S, Roullier C, Sun K, Tehan RM, Boya P CA, Christian MH, Gutiérrez M, Ulloa AM, Tejeda Mora JA, Mojica-Flores R, Lakey-Beitia J, Vásquez-Chaves V, Zhang Y, Calderón AI, Tayler N, Keyzers RA, Tugizimana F, Ndlovu N, Aksenov AA, Jarmusch AK, Schmid R, Truman AW, Bandeira N, Wang M, Dorrestein PC. 2020. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat Protoc 15:1954–1991. 10.1038/s41596-020-0317-5. [DOI] [PubMed] [Google Scholar]
- 15.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 16.Miller WG, Leveau JH, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact 13:1243–1250. 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 17.Distel DL, Morrill W, MacLaren-Toussaint N, Franks D, Waterbury J. 2002. Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int J Syst Evol Microbiol 52:2261–2269. 10.1099/00207713-52-6-2261. [DOI] [PubMed] [Google Scholar]
- 18.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat Biotechnol 1:784–791. 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 19.Antunes LCM, Ferreira RBR, Lostroh CP, Greenberg EP. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J Bacteriol 190:4392–4397. 10.1128/JB.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antunes LCM, Schaefer AL, Ferreira RBR, Qin N, Stevens AM, Ruby EG, Greenberg EP. 2007. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol 189:8387–8391. 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu W-T, Crüsemann M, Boudreau PD, Esquenazi E, Sandoval-Calderón M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu C-C, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw C-C, Yang Y-L, Humpf H-U, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, et al. 2016. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 34:828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. 2010. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45:703–714. 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 23.Mohimani H, Gurevich A, Shlemov A, Mikheenko A, Korobeynikov A, Cao L, Shcherbin E, Nothias L-F, Dorrestein PC, Pevzner PA. 2018. Dereplication of microbial metabolites through database search of mass spectra. Nat Commun 9:4035. 10.1038/s41467-018-06082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen I-MA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, Roux S, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2021. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49:D751–D763. 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T. 2021. antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 49:W29–W35. 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol 134:1141–1156. 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puri AW, Owen S, Chu F, Chavkin T, Beck DAC, Kalyuzhnaya MG, Lidstrom ME. 2014. Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense. Appl Environ Microbiol 81:1775–1781. 10.1128/AEM.03795-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovach ME, Elzer PH, Steven Hill D, Robertson GT, Farris MA, Roop RM, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S7 and Tables S1 and S2. Download aem.00270-22-s0001.pdf, PDF file, 1.0 MB (1MB, pdf)
Data Availability Statement
The mass spectrometry data were deposited in the public repository MassIVE (https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=f823719f2c5f4fca903bfe49f6964f45). The molecular networking job can be publicly accessed online at: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=d4ce2cc7d4db423eb00a4d2876993b50.