SUMMARY
Systemic immunity is stringently regulated by commensal intestinal microbes, including the pathobiont Candida albicans. This fungus utilizes various transcriptional and morphological programs for host adaptation, but how this heterogeneity affects immunogenicity remains uncertain. We show that UME6, a transcriptional regulator of filamentation, is essential for intestinal C. albicans-primed systemic Th17 immunity. UME6 deletion and constitutive overexpression strains are non-immunogenic during commensal colonization, whereas immunogenicity is restored by C. albicans undergoing oscillating UME6 expression linked with β-glucan and mannan production. In turn, intestinal reconstitution with these fungal cell wall components restores protective Th17 immunity to mice colonized with UME6-locked variants. These fungal cell wall ligands and commensal C. albicans stimulate Th17 immunity through multiple host pattern recognition receptors, including Toll-like receptor 2 (TLR2), TLR4, Dectin-1, and Dectin-2, which work synergistically for colonization-induced protection. Thus, dynamic gene expression fluctuations by C. albicans during symbiotic colonization are essential for priming host immunity against disseminated infection.
Graphical abstract
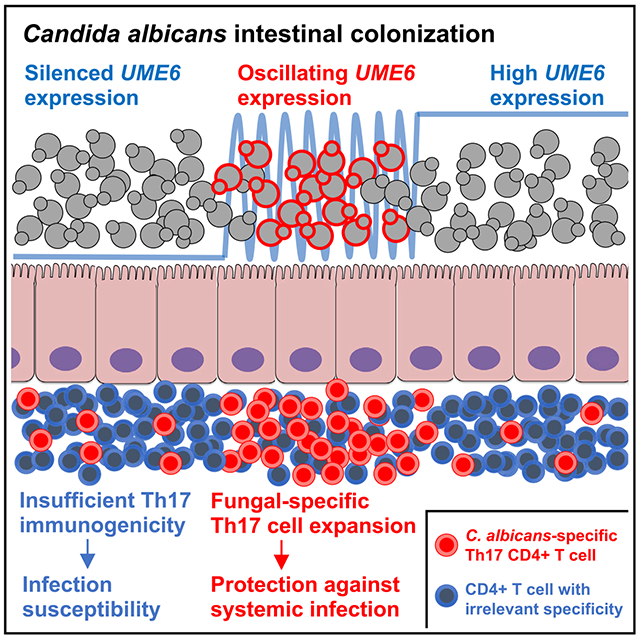
In brief
Intestinal colonization by commensal microbes, including the fungal pathobiont Candida albicans, primes protective immunological changes throughout the body. Shao et al. show that Th17 immunity primed by commensal C. albicans requires dynamic oscillations in expression of the transcriptional regulator UME6, which controls fungal morphology and virulence.
INTRODUCTION
Commensal microbes that colonize the intestine are increasingly recognized to stimulate systemic immunity and calibrate immune responses in non-mucosal tissues (Honda and Littman, 2016; Hooper et al., 2012; Iliev and Cadwell, 2021). Examples include susceptibility of mice to exogenous viral pathogens after antibiotic eradication of commensal bacteria (Abt et al., 2012; Ichinohe et al., 2011). Recent studies show that these protective effects of commensal microbiota extend to fungi (Aggor et al., 2019; Desai and Lionakis, 2019; Tso et al., 2018). The common pathobiont Candida albicans (Ca) is responsible for priming accumulation of circulating Th17 CD4+ T cells with fungal cross-reactivity in humans and is the primary target of natural anti-fungal antibodies in humans and mice (Bacher et al., 2019; Doron et al., 2021; Ost et al., 2021). Acquisition of commensal fungi in re-wilded mice or after Ca colonization are associated with increased circulating granulocytes and germinal center B cells (Lin et al., 2020). Our studies using Ca engineered to constitutively express model antigens show that gut colonization primes systemic expansion of fungus-specific Th17 CD4+ T cells (Shao et al., 2019). This is linked to accumulation of activated circulating neutrophils and protects against invasive infection by Ca and other pathogens susceptible to Th17 immunity, such as Staphylococcus aureus. Importantly, these antimicrobial benefits require tonic stimulation by commensal Ca because protective Th17 immunity is lost upon eradication of intestinal fungi (Shao et al., 2019). Thus, Ca commensalism is mutually beneficial to the host and microbe in that the hospitable intestinal niche promotes fungal propagation while protecting immune-competent hosts against infection by Ca or other pathogens susceptible to Th17 immunity (Bacher et al., 2019; Shao et al., 2019; Tso et al., 2018).
Symbiosis in this context also raises important questions regarding the features of commensal microbes responsible for priming protective Th17 immunity. These knowledge gaps are accentuated for fungi, given their polymorphic nature and dynamic response to environmental cues through transcriptional and morphological remodeling (Cottier and Hall, 2019; Kadosh and Mundodi, 2020; Mayer et al., 2013; Noble et al., 2017; Sudbery et al., 2004; Tyc et al., 2016). Ca responsiveness to environmental cues, including morphological changes and virulence phenotypes, are controlled by the transcriptional regulator Ume6. Ca ume6-null mutants are defective for filamentation under hypha-inducing growth conditions in vitro and cause reduced mortality after intravenous infection (Banerjee et al., 2008). Reciprocally, constitutive high-level UME6 expression drives Ca hypha formation independent of growth conditions and increased mortality of mice after intravenous inoculation (Banerjee et al., 2013; Carlisle et al., 2009). Co-colonization studies also show that ume6-deficient Ca have enhanced commensal fitness, whereas constitutive UME6 overexpression strains have reduced fitness compared with wild-type (WT) Ca (Witchley et al., 2019). Given the central role of UME6 in regulating Ca phenotypes, we reasoned that comparing the immunogenicity of a UME6-deficient strain with strains engineered for constitutive UME6 overexpression would unveil new insights into how Ca colonization primes systemic Th17 immunity.
RESULTS
UME6 expression oscillations during Ca colonization primes protective Th17 immunity
Ca was constructed in which both alleles of UME6 were placed under the control of a tetracycline-regulatable repressor (Carlisle et al., 2009; Witchley et al., 2019; Figures S1A and S1B). This allows UME6 transcription to be suppressed in the presence of doxycycline (DOX), resulting in an UME6 functionally deficient strain (UME6-off), whereas the absence of DOX leads to constitutive UME6 overexpression. This strain was also engineered to constitutively expresses the I-Ab restricted 2W1S55–68 peptide so that CD4+ T cells with this surrogate fungal specificity could be identified by I-Ab:2W1S tetramer staining (Igyarto et al., 2011; Moon et al., 2007). The expected differences in UME6 expression and associated morphological changes were verified for Ca-2W1S-tetO-UME6 compared with the parental WT Ca-2W1S strain (Figures S1C and S1D). Importantly, tetO regulation of UME6 did not affect expression of the recombinant protein containing the 2W1S peptide plus GFP (Figure S1E), allowing immunogenicity primed by Ca locked into UME6-off or overexpression states to be evaluated by tracking CD4+ T cells with I-Ab:2W1S specificity.
A defining feature of Ca symbiosis is protection against intravenous challenge in mice intestinally colonized with the fungus (Shao et al., 2019; Tso et al., 2018). To test how UME6 expression affects systemic immunity, we first compared the susceptibility of mice orally administered the Ca-2W1S-tetO-UME6 strain with continuous DOX supplementation (UME6-off) or without DOX supplementation (UME6-on) to a subsequent intravenous challenge with WT Ca (Figure 1A). Surprisingly, mice colonized with Ca locked into UME6-off or -on states remained highly susceptible because both groups showed ~1,000-fold higher kidney fungal burdens compared with mice colonized with WT Ca (Figure 1B). Loss of systemic immunity was not explained by reduced colonization because Ca was recovered at similar levels in the feces and in each intestinal segment of mice colonized with UME6-off, UME6-on, or WT cells (Figure S2A), consistent with prior studies showing that loss of UME6 does not decrease colonization levels in individually colonized mice (Witchley et al., 2019). Susceptibility also did not reflect the need for simultaneous colonization by UME6-off and -on cells because mice co-colonized with Ca-2W1S-tetO-UME6 (UME6-on) and a UME6-deficient strain remained susceptible to WT Ca infection (Figure S2B). These findings demonstrate an essential role of UME6 in commensal Ca-primed protective systemic immunity and suggest that dynamic regulation of UME6 is important because loss of UME6 or forced constitutive expression renders commensal Ca unable to prime normal systemic immune responses.
Figure 1. UME6 expression oscillations during Ca colonization primes protective Th17 immunity.
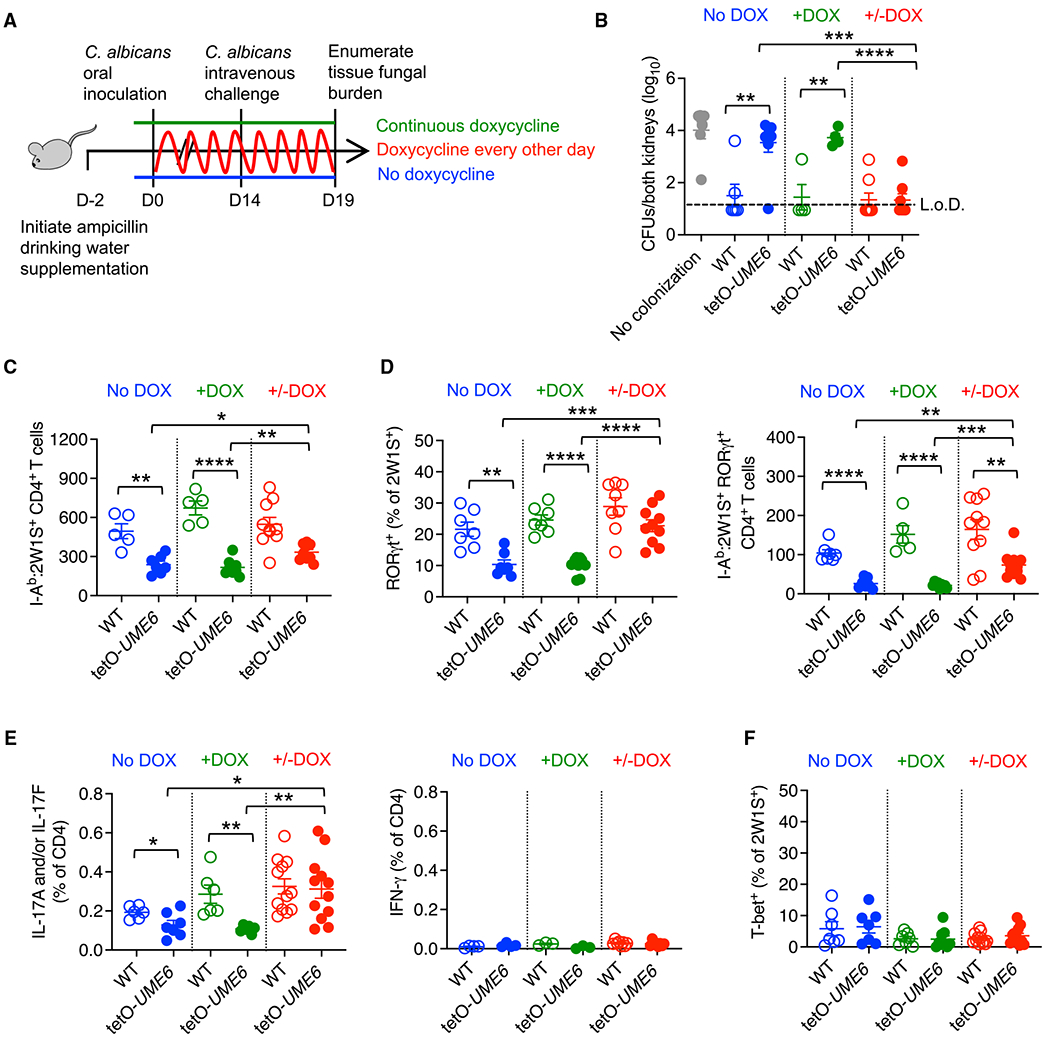
(A) Schematic of DOX drinking water supplementation to control UME6 expression by Ca in mice colonized by Ca-2W1S-tetO-UME6.
(B) Ca kidney CFUs 5 days after Ca-2W1S intravenous challenge for mice colonized with Ca-2W1S (WT) compared with Ca-2W1S-tetO-UME6 or no colonization controls maintained on ampicillin drinking water without (no DOX) or with (+DOX) continuous supplementation or oscillating DOX supplementation every other day.
(C) Number of I-Ab:2W1S CD4+ T cells in the spleen and pooled lymph nodes for mice described in (A) 14 days after Ca oral inoculation.
(D) Percentage of RORγt+ and number RORγt + I-Ab:2W1S CD4+ T cells from Ca-2W1S-tetO-UME6 (filled) or WT Ca-2W1S (open) colonized mice described in (C).
(E) Percentage of IL-17A- and/or IL-17F- or IFN-γ-producing CD4+ T cells in the spleen and lymph nodes after heat-killed WT Ca stimulation for mice described in (C).
(F) Percentage of Tbet+ cells among I-Ab:2W1S CD4+ T cells from Ca-2W1S-tetO-UME6 (filled) or WT Ca-2W1S (open) colonized mice described in (C).
Each data point represents the results from an individual mouse, representative of at least three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars, mean ± SEM. L.o.D., limit of detection.
Fluctuations in pH, nutrient, oxygen, and metabolite levels in the intestine, together with the ability of microbes to respond to these environmental perturbations (Cottier and Hall, 2019; Kadosh and Mundodi, 2020; Mayer et al., 2013; Noble et al., 2017; Sudbery et al., 2004; Tyc et al., 2016), led us to further investigate how dynamic shifts in Ca UME6 expression affect immunogenicity. The Ca-2W1S-tetO-UME6 strain was utilized and temporally forced into adopting UME6-off or -on phenotypes by the presence or absence, respectively, of DOX in the drinking water (Figure 1A). Remarkably, protection against Ca challenge was restored in Ca-2W1S-tetO-UME6-colonized mice when DOX supplementation occurred every other day compared with high susceptibility in mice colonized with Ca locked into UME6-on or -off states, whereas mice maintained on continuous DOX supplementation or without DOX remained susceptible (Figure 1B). Prior studies showed that virulence for yeast-locked Ca is restored by inducing a single transition to the hyphal morphology (Saville et al., 2003). However, we found that optimal priming of commensal-induced immunity required multiple oscillations between UME6-off and -on states because only partial protection was achieved when DOX supplementation was initiated a single time after colonization (Figure S2C). Protection was not simply due to oscillations in DOX administration because mice without fungal colonization remained susceptible, whereas mice colonized with WT Ca-2W1S (with no DOX-induced regulation of UME6 expression) showed the expected near-sterilizing reduction in kidney fungal burdens when DOX supplementation occurred every other day (Figure S2D).
The immunological basis for discordant systemic immunity primed by Ca was evaluated by comparing fungus-specific I-Ab:2W1S CD4+ T cells in the spleen and lymph nodes of colonized mice. Sharply reduced percentages and numbers of RORγt + I-Ab:2W1S CD4+ T cells were found in mice colonized with UME6-off- or -on-locked cells compared with WT Ca, which parallels their susceptibility to systemic infection (Figures 1B–1D and S3A). Comparatively, RORγt + I-Ab:2W1S CD4+ T cells were significantly increased in mice colonized with oscillating compared with UME6-off- or -on-locked cells, although the levels still remained reduced compared with mice colonized by WT Ca (Figures 1C, 1D, and S3A). Along with RORγt expression, interleukin (IL)-17A/F production by CD4+ T cells after stimulation with heat-killed Ca was increased in mice colonized with oscillating UME6 expression compared with UME6-off- or -on-locked cells and to levels comparable with mice colonized with WT Ca (Figures 1D, 1E, and S3B). Similar numbers of splenocyte and lymph node cells (~2 × 108 nucleated cells) and proportions of CD4+ T cells (~20%) between mice colonized with Ca in each context compared with no-colonization control mice and use of non-recombinant heat-killed Ca for stimulation indicate that the difference in the percentage of IL-17A/F-producing cells is representative of the fungus-specific Th17 response. Reduced Th17 priming in mice colonized with UME6-off or -on Ca was not associated with reciprocally increased Th1 cells because Tbet expression and interferon (IFN)-γ production remained at background levels (Figures 1E and 1F). A similarly reduced accumulation of fungus-specific Th17 CD4+ T cells was also found in the mesenteric lymph node in mice colonized by UME6-off- or -on-locked cells compared with WT Ca (Figure S3C), similar to prior studies showing that intestinal colonization does not prime enriched Candida-specific CD4+ T cell accumulation in the gut-draining lymph node compared with the spleen or other peripheral lymph nodes (Shao et al., 2019). Reduced colonization-induced Th17 priming by UME6-off- or -on-locked cells did not reflect immunological ignorance, given similarly increased CD44 expression, a mark of antigen experience (Moon et al., 2007), by I-Ab:2W1S CD4+ T cells in mice colonized with UME6-on cells, UME6-off cells, or cells with oscillating UME6 expression compared with WT Ca (Figure S3D). Thus, protective Th17 immunogenicity is primed by commensal Ca with dynamic shifts in UME6 expression.
Reconstitution with fungal cell wall moieties restores Th17 immunogenicity to UME6-locked Ca
Changes in expression and/or surface exposure of fungal cell wall moieties such as β-glucans and mannans can influence Ca immunogenicity (Chen et al., 2019; Cottier and Hall, 2019; Davis et al., 2014). To investigate whether these moieties contribute to commensal Ca immunogenicity, we examined whether addition of β-glucan and mannan restores Th17 systemic immunity primed by Ca locked into UME6-off or -on states (Figure 2A). The combination of UME6-off or -on cells effectively primed protective immunity against an intravenous challenge when administered with β-1,3-glucan and mannan (Figure 2B). Protection against systemic infection still required intestinal Ca colonization because fungal burdens in mice administered β-glucan plus mannan alone rebounded to levels comparable with mice colonized with UME6-off or -on cells (Figure 2B). These immunogenicity differences were not linked with intestinal colonization levels because β-glucan- and mannan-treated mice contained similar fecal colony-forming units (CFUs) regardless of being locked into UME6-off or -on states (Figure S4). Expansion of Th17 fungus-specific I-Ab:2W1S CD4+ T cells was also restored in mice administered β-1,3-glucan and mannan together with UME6-locked cells (Figure 2C). Thus, reconstitution with β-glucan plus mannan restores Th17 immunogenicity when provided in combination with Ca locked into UME6-off or -on states.
Figure 2. Intestinal reconstitution with fungal cell wall moieties restores Th17 immunogenicity to UME6-locked Ca.
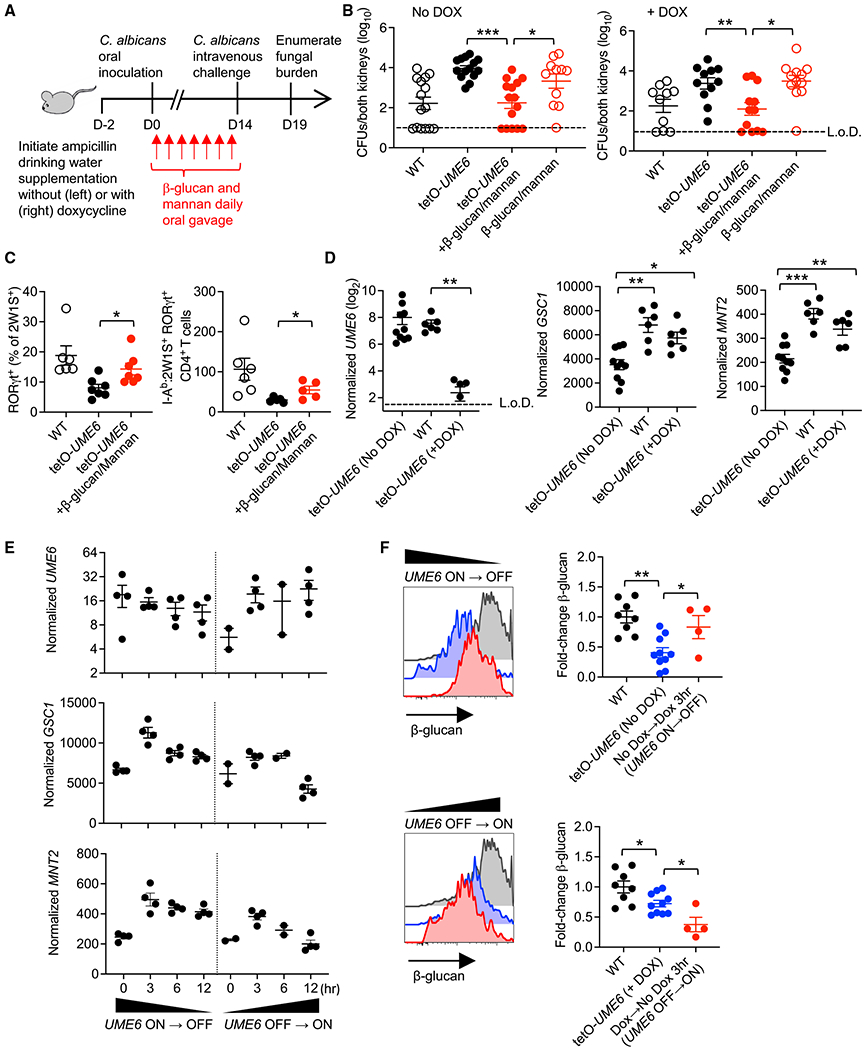
(A) Schematic of β-glucan plus mannan treatment of Ca-2W1S-tetO-UME6 colonized mice where DOX is used to control UME6 expression.
(B) Ca kidney CFUs 5 days after Ca-2W1S intravenous challenge for mice colonized with Ca-2W1S (WT), Ca-2W1S-tetO-UME6, or Ca-2W1S-tetO-UME6 and β-glucan plus mannan treatment with and without continuous DOX supplementation.
(C) Percentage of RORγt+ and number RORγt + I-Ab:2W1S CD4+ T cells from the spleen and lymph nodes of mice colonized with Ca-2W1S (WT), Ca-2W1S-tetOUME6, or Ca-2W1S-tetO-UME6 and β-glucan plus mannan treatment.
(D) UME6, GSC1, and MNT2 expression by Ca recovered from the feces of mice colonized with Ca-2W1S (WT), Ca-2W1S-tetO-UME6 without (UME6-on), or with (UME6-off) continuous DOX supplementation.
(E) UME6, GSC1, and MNT2 expression by Ca recovered from the feces of mice colonized with Ca-2W1S-tetO-UME6 at defined time points after discontinuation of DOX drinking water supplementation forcing the UME6-on → off transition and after initiation of DOX supplementation forcing the UME6-off → on transition.
(F) Relative anti-β-glucan staining intensity for Ca recovered from the feces of mice colonized with Ca-2W1S (WT) or Ca-2W1S-tetO-UME6 with continuous DOX deprivation (no DOX) and 3 h after initiating DOX drinking water supplementation forcing the UME6-on → off transition or with continuous DOX drinking water supplementation (+Dox) and 3 h after discontinuation forcing the UME6-off → on transition.
Each data point represents the results from an individual mouse, representative of at least two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. Error bars, mean ± SEM.
To investigate how UME6 controls expression of these cell wall moieties, expression of β-1,3 glucan and mannan biosynthesis genes was evaluated in the feces of mice colonized with WT Ca-2W1S or Ca-2W1S-tetO-UME6 under static UME6-off or -on conditions. As expected, UME6 expression by Ca-2W1S-tetO-UME6 was regulated by DOX, with high levels observed in the absence of DOX and much lower levels in the presence of DOX (Figure 2D). WT Ca-2W1S also showed relatively high UME6 expression, well above that observed for the tetO-UME6 strain with DOX supplementation (Figure 2D). Interestingly, expression of GSC1 and MNT2, required for β-1,3-glucan and mannan biosynthesis, respectively (Dutton et al., 2014; Mio et al., 1997; Munro et al., 2005), was repressed by UME6 because significantly reduced expression was observed in mice colonized with UME6-on compared with UME6-off cells (Figure 2D). UME6 repression of GSC1 is consistent with high GSC1 mRNA transcripts during Ca yeast-phase growth (Mio et al., 1997). However, GSC1 and MNT2 expression was also high in mice colonized with WT Ca cells despite relatively high UME6 expression (Figure 2D). A possible unifying explanation is that expression of these cell wall moieties involves fluctuating expression of UME6 during commensal colonization. This hypothesis was evaluated by comparing GSC1 and MNT2 expression by tetO-UME6 cells in the feces of colonized mice at defined time points after initiation (UME6-on → UME6-off) or discontinuation (UME6-off → UME6-on) of DOX supplementation. These experiments showed that oscillating UME6 drives amplified shifts in GSC1 and MNT2 expression, with most pronounced increased expression of both 3 h after initiating DOX supplementation (Figure 2E).
UME6 control of β-glucan exposure during colonization was further evaluated by staining Ca cells in the feces using an anti-β-glucan antibody. We found increased β-glucan exposure by WT Ca compared with cells locked into UME6-off or -on states (Figure 2F). β-Glucan exposure by UME6-on cells also increased 3 h after forced transition to the UME6-off state by DOX supplementation, which parallels increased GSC1 expression during the UME6-on → UME6-off transition (Figure 2E). Comparatively, β-glucan exposure by UME6-off cells was reduced after forced transition to the UME6-on state (Figure 2F). Thus, Ca β-glucan exposure is controlled by dynamic shifts in UME6 expression and in particular within the first 3 h during transition from UME6-on to UME6-off states.
A parallel approach was undertaken in which we examined Ca lacking MNT1/MNT2 genes, which encode proteins that add mannose residues during cell wall biosynthesis (Munro et al., 2005). An mnt1/mnt2-null Ca strain was used to investigate the role of mannan immunogenicity, given a lack of reagents for direct mannan staining. Despite colonization densities similar to WT Ca, mice colonized with mnt1/mnt2 cells remained highly susceptible to WT Ca intravenous challenge (Figures 3A and 3B). Systemic Th17 immunogenicity was further evaluated using mnt1/mnt2 cells constitutively expressing 2W1S55–68/GFP (Igyarto et al., 2011; Figure 3C). Colonization with mnt1/mnt2 cells resulted in decreased systemic accumulation of I-Ab:2W1S CD4+ T cells relative to the WT (Figure 3D) and their muted differentiation into Th17 effectors based on RORγt and IL-17A/F production (Figures 3E and 3F). As expected, anti-β-glucan staining was also sharply increased in mnt1/mnt2 cells, consistent with the mannan predominance in the outer cell wall masking β-glucan exposure (Figure 3G; Bain et al., 2014). Thus, mannan, in synergy with β-glucan, is required for priming systemic Th17 immunity by commensal Ca.
Figure 3. Ca MNT1/MNT2 is essential for colonization-induced Th17 immunogenicity.
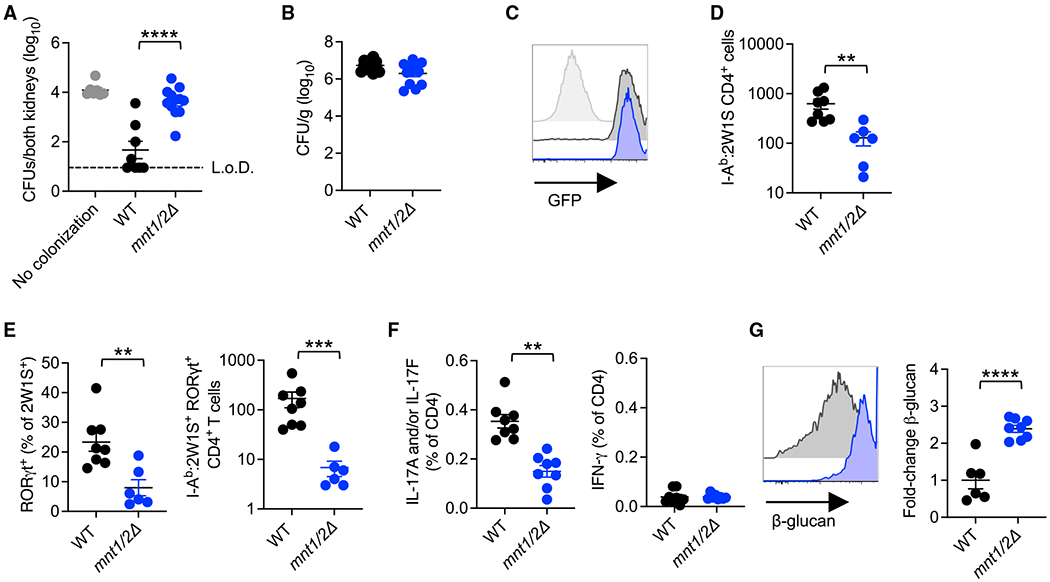
(A) Kidney CFUs 5 days after Ca-2W1S intravenous challenge for mice colonized with Ca-2W1S (WT) or mnt1/2Δ Ca in the preceding 14 days.
(B) Ca in the feces 14 days after oral inoculation with Ca-2W1S (WT) or mnt1/2Δ Ca for mice maintained on ampicillin drinking water.
(C) GFP fluorescence of Ca-2W1S (WT; black) or mnt1/2Δ Ca-2W1S (blue) compared with the parental non-recombinant mnt1/2Δ strain (gray shaded) cultured in Yeast Extract–Peptone–Dextrose (YPD) medium.
(D) Number of I-Ab:2W1S CD4+ T cells in the spleen and peripheral lymph nodes for mice described in (B) 14 days after Ca oral inoculation.
(E) Percentage of RORγt+ and number RORγt + I-Ab:2W1S CD4+ T cells from mice described in (B).
(F) Percentage of IL-17A- and/or IL-17F-, or IFN-γ-producingCD4+ T cells in the spleen and lymph nodes after heat-killed WT Ca stimulation for mice described in (B).
(G) Relative anti-β-glucan staining intensity for Ca recovered from the feces of mice described in (B).
Each data point represents the results from an individual mouse, representative of at least two independent experiments. **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars, mean ± SEM.
Multiple pattern recognition receptors work synergistically to optimize colonization-induced protection
We next investigated the pattern recognition receptors (PRRs) responsible for commensal Ca-primed Th17 immunogenicity. Because intestinal reconstitution with β-glucan plus mannan restores protective immunity with Ca locked into UME6-off or -on states, we first evaluated how these moieties stimulate immune cells. Neutralizing antibodies against defined PRRs were tested for their ability to block the macrophage nuclear factor κB (NF-κB) response to β-glucan or mannan stimulation (Rees and Lowy, 2018). We focused on Toll-like receptor 2 (TLR2), TLR4, Dectin-1, and Dectin-2 because fungal β-glucan and mannan have been shown to stimulate cells through these PRRs in other contexts (Ifrim et al., 2016; Jouault et al., 2003; Netea et al., 2006, 2008; Saijo et al., 2010; Saijo and Iwakura, 2011; Sato et al., 2006; Taylor et al., 2007). Interestingly, although neutralizing TLR2, TLR4, and Dectin-1 individually reduced activation in response to β-glucan stimulation, NF-κB expression levels still remained higher than no-stimulation controls, whereas β-glucan stimulation was suppressed to near background levels when TLR2, TLR4, Dectin-1, and Dectin-2 were neutralized simultaneously (Figure 4A). Likewise, TLR4 neutralization only partially reversed activation in response to mannan stimulation, whereas extinguishing NF-κB expression to background levels required concurrent blocking of TLR2, TLR4, Dectin-1, and Dectin-2 (Figure 4A).
Figure 4. Multiple PRRs work synergistically for optimizing colonization-induced protection.
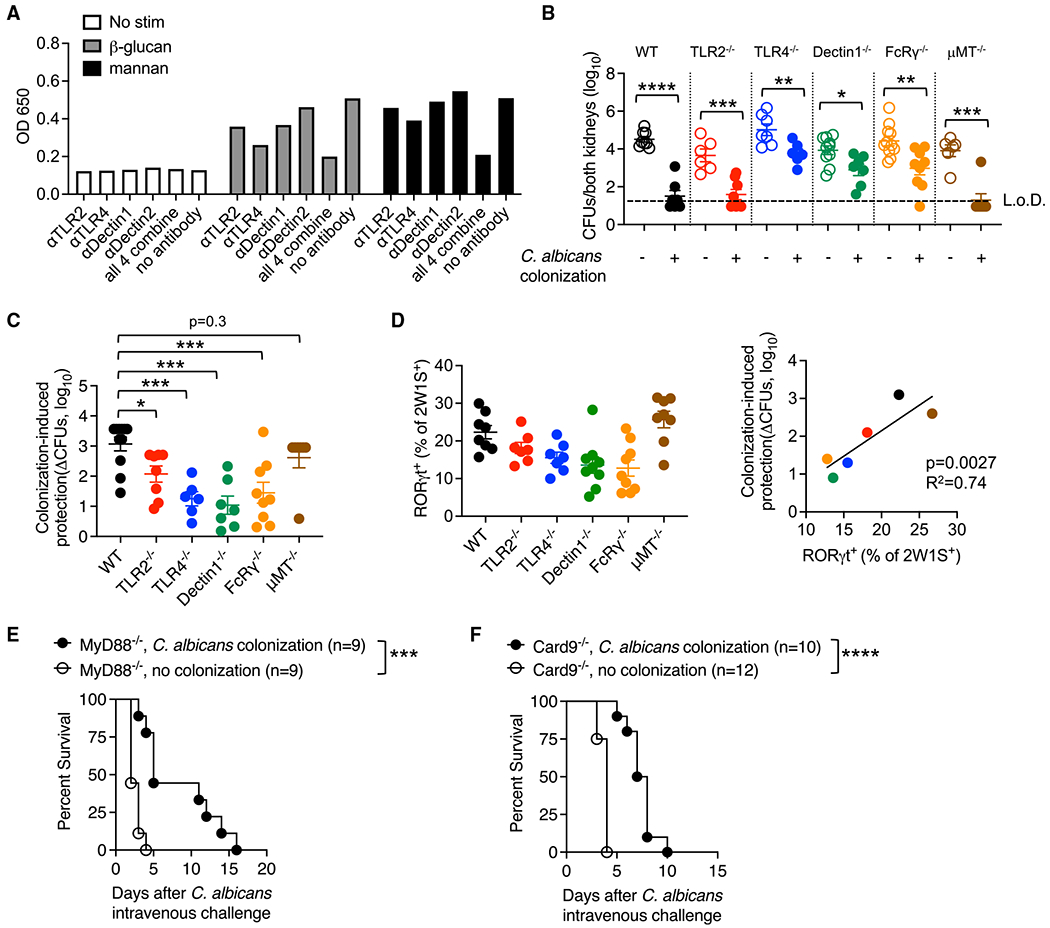
(A) NF-κB relative expression by RAW-blue cells stimulated with fungal β-glucan or mannan in the presence of neutralizing antibodies against each PRR.
(B) Ca kidney CFUs 5 days after Ca-2W1S intravenous infection occurring 14 days after Ca-2W1S oral inoculation (colonization) compared with no-colonization controls for each group of mice maintained on ampicillin drinking water.
(C) Fold reduction in Ca kidney CFUs for each group of mice described in (B).
(D) Percentage of RORγt + cells among I-Ab:2W1S CD4+ T cells 14 days after Ca-2W1S intestinal colonization and correlation with levels of colonization-induced protection (fold reduction in Ca kidney CFUs).
(E) Percentage of survival after Ca intravenous challenge in colonized compared with no-colonization control MyD88-deficient mice.
(F) Percentage of survival after Ca intravenous challenge in colonized compared with no-colonization control Card9-deficient mice.
Each data point represents the results from an individual mouse, representative of at least two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars, mean ± SEM.
To investigate the importance of these PRRs in sensing ligands expressed by WT Ca, colonization-induced protection against intravenous challenge was evaluated in mice with targeted defects in TLR2, TLR4, Dectin-1, or the FcRγ subunit of Dectin-2 (Sato et al., 2006). These experiments showed that each group of mice was partially protected when colonized by Ca compared with no-colonization controls (Figure 4B). The degree of colonization-induced protection for mice with defects in each PRR was reduced compared with WT control mice (Figure 4C). For example, the ~1,000-fold reduction in kidney burden associated with Ca colonization in WT mice was reduced to only ~10-fold in TLR4-or Dectin-1-deficient mice (Figure 4C). Analysis of I-Ab:2W1S CD4+ T cells showed direct correlations between Th17 differentiation and the degree of colonization-induced protection in each PRR-deficient strain compared with WT mice (Figure 4D).
Reduced colonization-induced protection in FcRγ-deficient mice does not reflect a protective role of antibodies because B cell-deficient (μMT−/−) mice lacking antibodies showed near-sterilizing immunity against Ca challenge primed by commensal colonization (Figures 4B–4D). Likewise, diminished levels of colonization-induced protection in TLR2-, TLR4-, Dectin-1-, or Dectin-2-deficient mice were also not explained by differences in colonization efficiency because fungal CFUs in the feces were similar between mice lacking each PRR compared with WT mice (Figure S5). Functional cooperativity between these PRRs is further supported by analysis of mice deficient in downstream signaling molecules. For example, enhanced survival of Ca-colonized MyD88-deficient mice, with defects in TLR2 and TLR4 responsiveness (Bellocchio et al., 2004), likely reflects stimulation via Dectin-1 or Dectin-2 (Figure 4E). In turn, enhanced survival of Ca-colonized Card9-deficient mice, with defects in Dectin-1 and Dectin-2 responsiveness (Saijo and Iwakura, 2011), may reflect TLR2- or TLR4-dependent signaling (Figure 4F). Thus, multiple PRRs play additive roles and work synergistically for optimal commensal Ca immunogenicity.
DISCUSSION
An increasingly recognized host benefit of Ca colonization is calibration of systemic Th17 immunity, which protects immune-competent individuals against invasive disseminated infection (Bacher et al., 2019; Shao et al., 2019; Tso et al., 2018). We previously showed that commensal Ca primes systemic expansion of fungus-specific Th17 CD4+ T cells, which, in turn, stimulates activation of circulating neutrophils and other phagocytic cells, which protect against intravenous Ca challenge (Shao et al., 2019). This symbiotic relationship is consistent with neutropenia, CD4+ T cell deficiency, or genetic defects in the Th17 axis representing primary risk factors for severe fungal infection (Boisson et al., 2013; Desai and Lionakis, 2018; Lionakis et al., 2014; Puel et al., 2011). Fungal virulence determinants responsible for invasive infection have been the focus of intense investigation, with morphological plasticity linked to fungal virulence (Kadosh and Mundodi, 2020; Saville et al., 2003). However, the fungal features responsible for symbiosis in the more ubiquitous context of commensal colonization are relatively undefined.
These knowledge gaps were examined using Ca strains engineered for regulated expression of UME6, a gene encoding a key transcriptional regulator responsible for filamentation and virulence (Banerjee et al., 2013; Carlisle et al., 2009). We show that commensal Ca-primed Th17 immunity is regulated via dynamic gene expression shifts during intestinal colonization. In particular, immunogenicity is restored by intestinal Ca oscillating between UME6-off and -on states, whereas cells locked into off/on states (or mixtures of these two states) did not induce protection. Thus, commensal Ca-primed systemic Th17 immunity is linked to dynamic transcriptional changes in fungal cells, highlighting exciting intricacies of this symbiotic relationship.
Although UME6 is considered a master regulator of the filamentation program (Banerjee et al., 2013; Carlisle et al., 2009), ume6 deletion mutants display similar proportions of yeast to hyphae throughout the intestine as WT Ca (Witchley et al., 2019), and only modest filamentation defects were observed during cutaneous infection (Wakade et al., 2021). Despite limited effects on morphology, UME6 has been linked to regulation of a large number of hypha-specific genes during intestinal colonization (Kadosh and Mundodi, 2020; Witchley et al., 2019). This includes a protease encoded by SAP6 and an adhesion protein encoded by HYR1, both of which suppress commensal fitness in the intestine (Witchley et al., 2019). Our results highlight that UME6 controls expression of GSC1 and MNT2 and likely other enzymes required for synthesis of immunogenic cell wall moieties. Dynamic shifts in UME6 expression are responsible for modulating the expression of GSC1 and MNT2, and these changes are linked to induction of systemic immunity. Masking the immune-stimulatory properties of β-glucan and mannans promotes immune evasion by Ca during invasive infection (Chen et al., 2019; Cottier and Hall, 2019; Davis et al., 2014). However, in the context of commensalism, when priming systemic Th17 immunity is beneficial through protection against disseminated infection, our data suggest that purposeful unmasking likely occurs.
Systemic immunity in mice reconstituted with β-glucan and mannan is not explained by trained immunity or activation of innate phagocytic cells (Quintin et al., 2012; Tso et al., 2018) because the presence of commensal Ca cells was also required. Although non-antigen-specific immune training by commensal Ca cannot be excluded, we propose that protection against disseminated infection is via systemic accumulation of fungus-specific Th17 cells, consistent with the necessity for CD4+ T cells (Shao et al., 2019). Although CD4+ T cells are essential, B cells and antibodies are non-essential because sterilizing immunity was observed in intestinally colonized B cell-deficient (μMT−/−) mice. These findings differ from recent studies showing that the susceptibility of μMT−/− mice to Ca infection can be rescued by donor B cells from germ-free mice monocolonized with Ca (Doron et al., 2021). These differences likely reflect our analysis of Ca immunogenicity in the presence of other commensal bacteria and in the physiological context of antibiotic-induced dysbiosis, which leads to enhanced fungal colonization.
Symbiotic priming of Th17 immunity by commensal fungi involved multiple PRRs, including TLR2, TLR4, Dectin-1, and Dectin-2, which work in combination for optimal colonization-induced protection. Redundancy between these receptors, which stimulate cells through unique signaling intermediates, highlights the importance of host sensing of Ca in the commensal state. Intricately coordinated expression of genes by Ca during commensalism further underscores benefits to Ca priming Th17 immunogenicity that protects against invasive infection and maintains the intestinal niche for colonization. The dynamic transcriptional regulation required for Ca-primed protective Th17 immunity further supports the altruistic explanation that sustaining homeostasis in mammalian hosts through protection against intestinal inflammation (Ost et al., 2021) and immunity against disseminated infection by fungi or other pathogens susceptible to Th17 immunity are dominant agenda items for Ca in the commensal state.
Limitations of the study
Colonization by Ca at the extremes of UME6 expression (constitutively on versus off), dual colonization with UME6-on and -off cells, and colonization with Ca forced to oscillate between extremes of UME6-on and -off states were compared with mice colonized with WT Ca and used to demonstrate the importance of dynamic shifts in UME6 expression for priming of Th17 immunogenicity. This indirect approach was utilized because tools to directly prove that UME6 is oscillating in individual cells during colonization are currently unavailable. For example, flow cytometry can be used to identify fungal cells, but reagents for intracellular UME6 staining in fungi are not available. We generated UME6 reporter strains to investigate expression using microscopy, but expression is hard to detect in the high-autofluorescence background of intestinal tissues. We focus on UME6, given its importance in morphological changes in response to environmental cues, and these results investigating a single transcription factor establish the foundation for evaluating dynamic expression of other fungal regulators in future studies. Th17 immunogenicity primed by commensal Ca was directly linked to shifts in expression of β-glucan plus mannan biosynthetic enzymes at the transcriptional level, and changes in β-glucan exposure was confirmed with anti-β-glucan antibodies. A role of mannan was verified using MNT1/MNT2-deficient strains for intestinal colonization. The lack of reagents for direct mannan staining and non-viability of GSC1-deficient Ca cells (Mio et al., 1997) necessitated use of these complementary approaches. More definitive analysis will require new tools for evaluating the expression of these immunogenic fungal cell wall moieties, ideally at the single-cell level, during intestinal colonization.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sing Sing Way (singsing.way@cchmc.org).
Materials availability
Ca strains and other reagent describe in this paper are available from the lead contact upon request with a completed Material Transfer Agreement.
Data and code availability
All data reported will be shared by the lead contact upon request. Raw sequence reads have been deposited to the National Library of Medicine (NCBI) and are publicly available through BioProject accession number PRJNA827179 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA827179) with identifiers for each individual sample provided in the Key resources table.
DESeq2 R code used to analyze RNA sequencing data is provided in Data S1.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-Cy7 anti-mouse CD4 (clone: GK1.5) | eBioscience | Cat# 25-0041-82; RRID:AB_469576 |
| PE-Cy5 anti-mouse CD8 (clone: 53–6.7) | eBioscience | Cat# 35-0081-82; RRID:AB_11217674 |
| PE-Cy5 anti-mouse CD11b (clone: M1/70) | Biolegend | Cat# 101210; RRID:AB_312793 |
| PE-Cy5 anti-mouse CD11c (clone: N418) | Biolegend | Cat# 15-0114-82; RRID:AB_468717 |
| PE-Cy5 anti-mouse F4/80 (clone: BM8) | eBioscience | Cat# 15-4801-82; RRID:AB_468798 |
| PE-Cy5 anti-mouse B220 (clone: RA3-6B2) | Biolegend | Cat# 15-0452-83; RRID:AB_468756 |
| Alexa Fluor 700 anti-mouse CD44 (clone: IM7) | Biolegend | Cat# 56-0441-82; RRID:AB_494011 |
| APC-eFluor780 anti-mouse CD45 (clone: 30F11) | invitrogen | Cat# 47-0451-82; RRID:AB_1548781 |
| FITC anti-mouse FOXP3 (clone: FJK-16S) | eBioscience | Cat# 11-5773-82; RRID:AB_465243 |
| AF647 anti-mouse RORγt (clone: Q31-378) | BD bioscience | Cat# 562682; RRID:AB_2687546 |
| PE-Cy7 anti-mouse IL17A (clone: TC 11-18H10.1) | Biolegend | Cat# 506922; RRID:AB_2125010 |
| PE anti-mouse IL17F (clone: 9D3.1C8) | Biolegend | Cat# 517008; RRID:AB_10690818 |
| Anti-mouse TLR2 (clone C9A12) | Invivogen | Cat#mabg-mtlr2; RRID:AB_11125339 |
| Anti-mouse TLR4/MD2 (clone MTS510) | Hycult Biotech | Cat#HM1029; RRID:AB_533218 |
| Anti-mouse Dectin-1 (clone R1-8g7) | Invivogen | Cat# mabg-mdect; RRID:AB_2753143 |
| Anti-mouse Dectin-2 (clone D2.11E4) | Thermo Fisher Scientific | Cat#MA1-82675; RRID:AB_930457 |
| Anti-(1-3)-beta-D-glucan | Biosupplies | Cat#400-2; RRID:AB_2747399 |
| Bacterial and virus strains | ||
| Candida albicans SC5314 | Kaplan Lab, University of Pittsburg (Igyarto et al., 2011) | N/A |
| Recombinant Candida albicans SC5314 | Kaplan Lab, University of Pittsburg (Igyarto et al., 2011) | CaURA3/CaURA3 pENO1-ENO1-GFP-2W1S-OVA323-339-OVA257-264-I-Eα50-66 |
| Recombinant Candida albicans SC5314-tetO-UME6 | This study | CaURA3/CaURA3 pENO1-ENO1-GFP-2W1S-OVA323-339-OVA257-264-I-Eα50-66 CaHygB-CaTAR-tetO-UME6/CaHygB-CaTAR-tetO-UME6 |
| SC5314-mnt1/2Δ | (Munro et al., 2005) | mnt1−/−;mnt2−/− |
| Recombinant Candida albicans SC5314-MNT1/2Δ | This study | N/A |
| Candida albicans ume6 deficient | (Carlisle and Kadosh, 2010) | ume6−/− |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin | Sigma-Aldrich | Catalog# A0166 |
| Gentamicin | Sigma-Aldrich | Catalog# G3632 |
| Metronidazole | Sigma-Aldrich | Catalog# M3761 |
| Neomycin | Sigma-Aldrich | Catalog# N6386 |
| Vancomycin | MP Biomedical | Catalog# 0219554005 |
| Doxycycline | Sigma-Aldrich | Catalog# D9891 |
| Mannan | Sigma-Aldrich | Catalog# M7504 |
| β-glucan | Millipore | Catalog# 346210 |
| QUANTI-Blue | Invivogen | Catalog# rep-qbs |
| Dehydrated Culture Media: Brain Heart Infusion | Thermo Fisher Scientific | Catalog# B11060 |
| Yeast extract | Boston Bio Product | Catalog# P-950 |
| Bactopeptone | BD Bioscience | Catalog# DF0118170 |
| Uridine | Alfa-Aesar | Catalog# A15227 |
| D-(+)-Glucose | Sigma-Aldrich | Catalog# G5767 |
| Agar | Bio Express | Catalog# J637 |
| DMEM | Gibco | Catalog# 10313-21 |
| Fetal bovine serum | GeneMate | Catalog# S-1200-500 |
| Glutamine (100X) | Gibco | Catalog# 25030-081 |
| HEPES 1M | Gibco | Catalog# 15630-080 |
| Penicillin-streptomycin solution (100X) | Gibco | Catalog# 15140-122 |
| BD Golgi Plug (Brefeldin A solution) | BD Bioscience | Catalog# 555029 |
| Foxp3/Transcription factor staining buffer set | eBioscience | Catalog# 00-5523-00 |
| Fixation/Permeabilization solution kit | BD Bioscience | Catalog# 554722 |
| APC Conjugation Kit-Lightning-Link | abcam | Catalog# ab201807 |
| Calcofluor White Stain | Biotium | Catalog# 29067 |
| Critical commercial assays | ||
| ZymoBIOMICS RNA Miniprep Kit | ZYMO RESEARCH | Catalog# R2001 |
| SuperScript™ II Reverse Transcriptase kit | Invitrogen | Catalog# 18064014 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Catalog# A25780 |
| SMARTer Stranded Total RNA-Seq Kit v3 | TaKaRa | Catalog# 634485 |
| Deposited data | ||
| WT C. albicans 1.1 | WT_1.1 | PRJNA827179 |
| WT C. albicans 1.2 | WT_1.2 | PRJNA827179 |
| WT C. albicans 3.1 | WT_3.1 | PRJNA827179 |
| WT C. albicans 3.2 | WT_3.2 | PRJNA827179 |
| WT C. albicans 3.3 | WT_3.3 | PRJNA827179 |
| WT C. albicans 3.4 | WT_3.4 | PRJNA827179 |
| UME6-ON C. albicans 2.1 | UME6ON_2.1 | PRJNA827179 |
| UME6-ON C. albicans 2.2 | UME6ON_2.2 | PRJNA827179 |
| UME6-ON C. albicans 2.3 | UME6ON_2.3 | PRJNA827179 |
| UME6-ON C. albicans 2.4 | UME6ON_2.4 | PRJNA827179 |
| UME6-ON C. albicans 2.5 | UME6ON_2.5 | PRJNA827179 |
| UME6-ON C. albicans 2.6 | UME6ON_2.6 | PRJNA827179 |
| UME6-ON C. albicans 3.1 | UME6ON_3.1 | PRJNA827179 |
| UME6-ON C. albicans 3.2 | UME6ON_3.2 | PRJNA827179 |
| UME6-ON C. albicans 3.3 | UME6ON_3.3 | PRJNA827179 |
| UME6-ON C. albicans 3.4 | UME6ON_3.4 | PRJNA827179 |
| UME6-OFF C. albicans 1.1 | UME6OFF_1.1 | PRJNA827179 |
| UME6-OFF C. albicans 1.2 | UME6OFF_1.2 | PRJNA827179 |
| UME6-OFF C. albicans 2.1 | UME6OFF_2.1 | PRJNA827179 |
| UME6-OFF C. albicans 2.2 | UME6OFF_2.2 | PRJNA827179 |
| UME6-OFF C. albicans 2.3 | UME6OFF_2.3 | PRJNA827179 |
| UME6-OFF C. albicans 2.4 | UME6OFF_2.4 | PRJNA827179 |
| UME6-ON to OFF (0 h) 1 | UME6ONtoOFF_0h_1 | PRJNA827179 |
| UME6-ON to OFF (0 h) 2 | UME6ONtoOFF_0h_2 | PRJNA827179 |
| UME6-ON to OFF (0 h) 3 | UME6ONtoOFF_0h_3 | PRJNA827179 |
| UME6-ON to OFF (0 h) 4 | UME6ONtoOFF_0h_4 | PRJNA827179 |
| UME6-ON to OFF (3 h) 1 | UME6ONtoOFF_3h_1 | PRJNA827179 |
| UME6-ON to OFF (3 h) 2 | UME6ONtoOFF_3h_2 | PRJNA827179 |
| UME6-ON to OFF (3 h) 3 | UME6ONtoOFF_3h_3 | PRJNA827179 |
| UME6-ON to OFF (3 h) 4 | UME6ONtoOFF_3h_4 | PRJNA827179 |
| UME6-ON to OFF (6 h) 1 | UME6ONtoOFF_6h_1 | PRJNA827179 |
| UME6-ON to OFF (6 h) 2 | UME6ONtoOFF_6h_2 | PRJNA827179 |
| UME6-ON to OFF (6 h) 3 | UME6ONtoOFF_6h_3 | PRJNA827179 |
| UME6-ON to OFF (6 h) 4 | UME6ONtoOFF_6h_4 | PRJNA827179 |
| UME6-ON to OFF (12 h) 1 | UME6ONtoOFF_12h_1 | PRJNA827179 |
| UME6-ON to OFF (12 h) 2 | UME6ONtoOFF_12h_2 | PRJNA827179 |
| UME6-ON to OFF (12 h) 3 | UME6ONtoOFF_12h_3 | PRJNA827179 |
| UME6-ON to OFF (12 h) 4 | UME6ONtoOFF_12h_4 | PRJNA827179 |
| UME6-OFF to ON (0 h) 1 | UME6OFFtoON_0h_1 | PRJNA827179 |
| UME6-OFF to ON (0 h) 2 | UME6OFFtoON_0h_2 | PRJNA827179 |
| UME6-OFF to ON (3 h) 1 | UME6OFFtoON_3h_1 | PRJNA827179 |
| UME6-OFF to ON (3 h) 2 | UME6OFFtoON_3h_2 | PRJNA827179 |
| UME6-OFF to ON (3 h) 3 | UME6OFFtoON_3h_3 | PRJNA827179 |
| UME6-OFF to ON (3 h) 4 | UME6OFFtoON_3h_4 | PRJNA827179 |
| UME6-OFF to ON (6 h) 1 | UME6OFFtoON_6h_1 | PRJNA827179 |
| UME6-OFF to ON (6 h) 2 | UME6OFFtoON_6h_2 | PRJNA827179 |
| UME6-OFF to ON (12 h) 1 | UME6OFFtoON_12h_1 | PRJNA827179 |
| UME6-OFF to ON (12 h) 2 | UME6OFFtoON_12h_2 | PRJNA827179 |
| UME6-OFF to ON (12 h) 3 | UME6OFFtoON_12h_3 | PRJNA827179 |
| UME6-OFF to ON (12 h) 4 | UME6OFFtoON_12h_4 | PRJNA827179 |
| Experimental models: Cell lines | ||
| Raw-Blue Macrophage Cells | Invivogen | Catalog# raw-sp RRID: CVCL_X594 |
| Experimental models: Organisms/strains | ||
| C57BL/6 | Charles River Laboratories | Strain Code: 027 |
| B6.129-Tlr2tm1Kir/J | The Jackson Laboratory | Stock No: 004650 |
| B6(Cg)-Tlr4tm1.2Karp/J | The Jackson Laboratory | Stock No: 029015 |
| B6.129-Card9tm1Xlin/J | The Jackson Laboratory | Stock No: 028652 |
| B6.129P2(SJL)-Myd88tm1.1Defr/J | The Jackson Laboratory | Stock No: 009088 |
| B6.129S6-Clec7atm1Gdb/J | The Jackson Laboratory | Stock No: 012337 |
| B6;129P2-Fcer1gtm1Rav/J | The Jackson Laboratory | Stock No: 002847 |
| B6.129S2-Ighmtm1Cgn/J | The Jackson Laboratory | Stock No: 002288 |
| Oligonucleotides | ||
| 5′junction, Primer1: GCACCAAACACCCGAAATTA; Primer2: CCATTTTGGCGTGAGGTAATCC | Thermo Fisher Scientific | N/A |
| 3′junction, Primer1: AGTGAAAGTCGAGTTTACCACTC; Primer2: GCTGCAGTTGCAGTTGTTGT | Thermo Fisher Scientific | N/A |
| Native ume6 upstream, Primer1: AAGCAAAGCAATAATGAGCCAA; Primer2: AGAATTTCCCGGGAGTTGTTTA | Thermo Fisher Scientific | N/A |
| Ume6qPCR; Primer1: GAACAATGGTGGTGGTAGTGG; Primer2: AATTCGACAAATCCAACATCC | Thermo Fisher Scientific | (Childers et al., 2014) |
| Act1qPCR; Primer1: TTGCTCCAGAAGAACATCCAG; Primer2: AGTAACACCATCACCAGAATCC | Thermo Fisher Scientific | (Childers et al., 2014) |
| 7707 Primer:GAGAATCGAAGAAGAA TTAGGTTCTGAAGCTATCTACGCTG GTAAAGATTTCCAAAAGGCTTCTCA ATTGGGTGGTGGTTCTAAAggTgAAg AATTATT |
Thermo Fisher Scientific | N/A |
| 7708 Primer:TTTAATTAGTTCATAT ATTCAAGATGTTCCTATAAAAGAA AAAAAAAGCACCAG CTTTTTTTTATTTAATCGTAAAACG ACGGCCAGTGAATTC |
Thermo Fisher Scientific | N/A |
| 5059 Primer: GCAAATGATTATACATGGGGATG | Thermo Fisher Scientific | N/A |
| 7722 Primer: ACAATCCAGTACATCAAACTCCAA | Thermo Fisher Scientific | N/A |
| 3925 Primer: GAACATAACCTTCTGGCATGGC | Thermo Fisher Scientific | N/A |
| 7763 Primer: GATGCTTGGGTCCACTTCT | Thermo Fisher Scientific | N/A |
| Recombinant DNA | ||
| pV1393 | (Vyas et al., 2015) | Resource of CaCAS9 |
| pYM70 | (Basso et al., 2010) | Resource of TEF2 promoter-CaHygB-ACT1 terminator |
| pLC1031 | (Kim etal., 2019) | Resource of CaHygB-CaTAR-tetO |
| Software and algorithms | ||
| Prism 9.3.1 | GraphPad | N/A |
| Flow Jo 9.9.6 | Treestar | N/A |
| Gene5 | BioTek | N/A |
| Subread | http://subread.sourceforge.net/ | N/A |
| DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | N/A |
| Other | ||
| 2W1S52-68:I-Ab Tetramer | Jenkins Lab, University of Minnesota (Moon et al., 2007) | N/A |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6 mice were purchased from Charles River laboratories. TLR2-deficient, TLR4-deficient, Card9-deficient, MyD88-deficient, Dectin-1-deficient, FcRγ-deficient, μMT-deficient mice each on the isogenic C57BL/6 background were purchased from The Jackson Laboratory. All mice were housed under specific pathogen-free conditions, and used between 8 and 12 weeks of age. All experiments were performed using sex- and age-matched controls under Cincinnati Children’s Hospital Research Foundation IACUC approved protocols.
Microbes
Recombinant Ca (Ca-2W1S) expressing the I-Ab:2W1S55-68 peptide plus GFP and the parental SC5314 strain were provided by Dr. Daniel Kaplan (Igyarto et al., 2011). Ca-2W1S-tetO-UME6 was generated in this study by replacing the endogenous UME6 upstream regulatory loci with a TEF2 promotor leading tetracycline repressible trans-activator and activator binding sequence as described (Basso et al., 2010; Carlisle et al., 2009; Kim et al., 2019; Vyas et al., 2015; Witchley et al., 2019). The plasmids used to generate Ca-2W1S-tetO-UME6 are listed in key resources table. Ca-ume6 deficient has been described (Carlisle and Kadosh, 2010). For oral inoculation, Ca-2W1S, Ca-2W1S-tetO-UME6, and Ca-ume6 deficient was cultured in Yeast Extract–Peptone–Dextrose (YPD) media with 20 μg/mL doxycycline at 30°C with shaking (200 rpm). To create the Ca mnt1/2 deletion strain expressing GFP-2W1S, a cassette was generated by PCR amplifying the GFP-2W1S:NatR fragment from pGM2278 (a gift from Dr. Judtih Berman, University of Tel Aviv) using oligos 7707/7708. These oligos incorporate approximately 80 bp of homology to target the cassette to the 3′ end of the ENO1 gene. The cassette was transformed into the mnt1/2 strain (NGY337; a gift from Dr. Neil Gow, University of Exeter) and transformants selected on YPD containing nourseothricin. Colonies that had bright nuclear fluorescence were PCR checked to confirm correct integration of the cassette at the 3′ end of ENO1. The 5′ junction of integration was checked by PCR with oligos 5059/7722 and the 3′ junction by PCR with 3925/7763.
METHOD DETAILS
Intestinal colonization and infection
For intestinal colonization, 80 μL of Ca-2W1S and Ca-2W1S-tetO-UME6 grown to stationary phase was orally administrated to mice. For co-colonization, the relative concentration of Ca-2W1S-tetO-UME6 and Ca-ume6 deficient in overnight culture was first determined by OD600, and combined at the indicated ratios for oral inoculation. To establish Ca intestinal colonization, the drinking water of mice was supplemented with ampicillin (1 mg/mL) beginning two days prior to oral Ca inoculation, and maintained throughout the experiment as described (Shao et al., 2019). The drinking water for some groups of mice were further supplemented with doxycycline (2 mg/mL). For infection, Ca-2W1S was cultured in Yeast Extract–Peptone–Dextrose (YPD) media at 30°C with shaking overnight and back-diluted to early log phase growth (OD600~0.1), then washed and diluted into sterile PBS, and injected via the lateral tail vein (5 × 104 CFUs) as described (Shao et al., 2019). For enumerating recoverable Ca pathogen burden, the kidneys, each intestinal tissue or fresh fecal pellets were weighed and homogenized into sterile PBS with 0.05% Triton X-. Serial dilutions of each tissue homogenate were plated into agar plates supplemented with antibiotics (ampicillin [2.5 μg/mL], gentamicin [2.5 μg/mL], neomycin [2.5 μg/mL], metronidazole [2.5 μg/mL], vancomycin [1.25 μg/mL], and doxycycline [20 μg/mL]), with enumeration after incubation at 37°C for 24 h as described (Shao et al., 2019). For intestinal reconstitution, purified fungal β-1,3-glucan and mannan were resuspended in sterile water (10 mg/mL each), and 100 μL orally administered to mice using a 20G curved feeding needle.
Cell staining, stimulation and flow cytometry
Fluorophore-conjugated antibodies used for cell analysis include anti-CD4 (clone GK1.5), anti-CD8a (clone 53–6.7), anti-CD11b (clone M1/70), anti-CD11c (clone N418), anti-F4/80 (clone BM8), anti-B220 (clone RA3-6B2), anti-CD44 (clone IM7), anti-CD45 (clone 30F11), anti-RORγt (clone Q31-378), anti-IL17A (clone TC 11-18H10.1), anti-IL17F (clone 9D3.1C8).
For detecting CD4+ T cells with I-Ab:2W1S55-68 surrogate Ca specificity, single cell suspension from spleen and peripheral lymph nodes of colonized mice were incubated with PE conjugated I-Ab:2W1S55-68 tetramer at room temperature for 60 min, and enrichment applied using anti-PE antibody conjugated beads as described (Moon et al., 2007). For cytokine production, 2 × 106 single cell suspension from spleen and peripheral lymph nodes was stimulated with 106 CFU heat-killed (65°C 30 min) Ca in 200 μL DMEM medium (supplemented with 10% fetal bovine serum, 1% L-glutamine, 10 mM HEPES, 1% penicillin-streptomycin) at 37°C with 5% CO2 for 24 h, with brefeldin A(GolgiPlug, BD Bioscience) supplementation in the final 6 h (Shao et al., 2019). Intracellular staining and intranuclear staining were performed after cell permeabilization using commercial kits (BD PharMingen and eBioscience) according to the manufacturers’ instructions. For detecting β-glucan on Ca cells, fresh fecal pellets from colonized mice were homogenized in sterile PBS and filtered through 70μm nylon mesh. The fecal homogenate was incubated with anti-β-glucan antibody conjugated with APC by lightning kit (abcam) in PBS with 10% FBSat 4°C for 1 h, followed by Calcofluor white (CFW; 5nM) staining with 10% FBS at 4°C for 5 min, with Ca further identified as GFP + CFW + cells. Samples were acquired using an BD FACS-Canto cytometer and analyzed using FlowJo software (Tree Star).
DNA/RNA isolation and PCR/qRT-PCR
DNA was prepared by resuspending Ca into 50 mM Tris buffer (pH 7.5) supplemented with 1 mM EDTA, 0.2% β-mercaptoethanol and 1000 units/mL of lyticase (Sigma), and incubation at 37°C for 30 min to disrupt fungal cells. Lysates were pelleted by centrifugation (15000 rpm for 8 min). DNA was precipitated from the supernatants using an equal volume of isopropanol (−20°C for 2 h), and then washed with 75% ethanol before resuspending with DNAase free water. RNA from broth culture cells or fresh fecal pellets of colonized mice was extracted using the ZymoBIOMOICS RNA Mini Kit following the manufacturer protocols. For quantitative PCR, 500 ng extracted RNA was reverse transcribed into cDNA by SuperScript™ II Reverse Transcriptase kit with random and oligo dT primers, and gene expression determined in qPCR (SYBR; Applied Biosystems) by 7500 Fast Real-Time PCR System (Applied Biosystems). For broader analysis of Ca gene expression, RNA was subjected to first strand cDNA synthesis and sequence library preparation using the SMARTer Stranded Total RNA-Seq Pico input Kit v3 (Takara Bio USA Inc) following the manufacturer’s recommendations. Briefly, approximately 10 ng RNA was added to first strand cDNA synthesis reaction, which was primed by random hexamers from the SMART Pico Oligos Mix. Second strand synthesis and addition of Illumina indexes was performed using 5 cycles of PCR amplification, followed by guide-RNA mediated enzymatic ribosomal RNA depletion and 15 cycles of final library amplification. Samples were pooled and subjected to sequencing on a NovaSeq 6000 sequencing machine (Illumina Corp.) to a mean depth of approximately 120 million 150 nucleotide paired-end reads per sample. Reads were subjected to quality control and trimming using the program Sickle then aligned to the Ca SC5314 genome (accession # NC_032089.1) using the program STAR using default settings, resulting in ~80 million read per sample after alignment Ca genome alignment. The number of reads aligned to each gene was extracted using the program feature Counts from the Subread software package (Liao et al., 2014). Count normalization and differential gene abundance was determined using DESeq2 (Love et al., 2014).
Raw cell culture
Raw-Blue cells (Invivogen) is derived from mouse macrophage RAW 264.7 cell line with chromosomal integrated a secreted embryonic alkaline phosphatase (SEAP) reporter construct inducible by NF-κB expression. 5 × 104 Raw-Blue cells were seeded with blocking antibodies against each pattern recognition receptor (used at 25 μg/mL each) 30 min prior to fungal β-1,3-glucan (10 μg/mL) or mannan (500 μg/mL) stimulation. Supernatants was collected 16 h after stimulation, and SEAP secretion reflective of NF-κB activation was detected by colorimetric assay using Quanti-Blue reagent (Invivogen).
C. albicans morphology analysis
Ca-2W1S and Ca-2W1S-tetO-UME6 were cultured at 37°C in YPD liquid media supplemented with 10% FBS with shaking (200 rpm) for 5 h. Thereafter, fungi were transferred to a clear Petri dish and morphology analyzed using a Cytation5 imaging reader (BioTek) and processed using Gen5 software.
QUANTIFICATIONS AND STATISTICAL ANALYSIS
The number of individual animals used per group is shown in each figure panel, or shown by individual data points or individual wells for cell stimulation assays. All statistical analysis was performed by Prism (Graphpad) software. Student’s t test was used to compared differences between two groups. One-way ANOVA with Tukey’s test for multiple comparisons was used to evaluate experiments containing more than two groups. The threshold for statistical significance for all experiments was set at p < 0.05.
Supplementary Material
Highlights.
C. albicans cells locked into UME6-on or -off states fail to prime immunogenicity
Forced oscillations of UME6 support C. albicans-induced Th17 immunogenicity
Intestinal fungal β-glucan and mannan stimulate systemic Th17 immunogenicity
Th17 immunogenicity requires signaling via multiple pattern recognition receptors
ACKNOWLEDGMENTS
This work was supported by NIH-NIAID through grants DP1AI131080 (to S.S.W.), R01AI141893 (to R.J.B.), R01AI081704 (to R.J.B.), and R01AI108992 (to S.M.N.). S.S.W. is supported by the HHMI Faculty Scholar’s program and Burroughs Wellcome Fund Investigator in the Pathogenesis Award. We thank Neil Gow for the gift of Ca strains and Judith Berman for the gift of plasmids and strains. We thank the staff of Research Animal Resources at Cincinnati Children’s Hospital led by Dr. Saigiridhar Tummala for the conscientious care of experimental mice, and the NIH Tetramer Core Facility (supported by NIH-NIAID contract 75N93020D00005) for providing MHC tetramer reagents. All flow cytometric data were acquired using equipment maintained by the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110837.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. (2012). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggor FEY, Way SS, and Gaffen SL (2019). Fungus among us: the frenemies within. Trends Immunol. 40, 469–471. 10.1016/j.it.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher P, Hohnstein T, Beerbaum E, Rocker M, Blango MG, Kaufmann S, Rohmel J, Eschenhagen P, Grehn C, Seidel K, et al. (2019). Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. Cell 176, 1340–1355.e15. 10.1016/j.cell.2019.01.041. [DOI] [PubMed] [Google Scholar]
- Bain JM, Louw J, Lewis LE, Okai B, Walls CA, Ballou ER, Walker LA, Reid D, Munro CA, Brown AJP, et al. (2014). Candida albicans hypha formation and mannan masking of beta-glucan inhibit macrophage phagosome maturation. mBio 5, e01874. 10.1128/mbio.01874-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Thompson DS, Lazzell A, Carlisle PL, Pierce C, Monteagudo C, Lopez-Ribot JL, and Kadosh D (2008). UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol. Biol. Cell 19, 1354–1365. 10.1091/mbc.e07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, Uppuluri P, Zhao XR, Carlisle PL, Vipulanandan G, Villar CC, Lopez-Ribot JL, and Kadosh D (2013). Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 12, 224–232. 10.1128/ec.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso LR Jr., Bartiss A, Mao Y, Gast CE, Coelho PSR, Snyder M, and Wong B (2010). Transformation of Candida albicans with a synthetic hygromycin B resistance gene. Yeast 27, 1039–1048. 10.1002/yea.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellocchio S, Montagnoli C, Bozza S, Gaziano R, Rossi G, Mambula SS, Vecchi A, Mantovani A, Levitz SM, and Romani L (2004). The contribution of the Toll-like/IL-1 receptor superfamily to innate and adaptive immunity to fungal pathogens in vivo. J. Immunol 172, 3059–3069. 10.4049/jimmunol.172.5.3059. [DOI] [PubMed] [Google Scholar]
- Boisson B, Wang C, Pedergnana V, Wu L, Cypowyj S, Rybojad M, Belkadi A, Picard C, Abel L, Fieschi C, et al. (2013). An ACT1 mutation selectively abolishes interleukin-17 responses in humans with chronic mucocutaneous candidiasis. Immunity 39, 676–686. 10.1016/j.immuni.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, and Kadosh D (2009). Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc. Natl. Acad. Sci. U S A 106, 599–604. 10.1073/pnas.0804061106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle PL, and Kadosh D (2010). Candida albicans Ume6, a filament-specific transcriptional regulator, directs hyphal growth via a pathway involving Hgc1 cyclin-related protein. Eukaryot. Cell 9, 1320–1328. 10.1128/ec.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Jackson JW, Tams RN, Davis SE, Sparer TE, and Reynolds TB (2019). Exposure of Candida albicans beta (1,3)-glucan is promoted by activation of the Cek1 pathway. PLoS Genet. 15, e1007892. 10.1371/journal.pgen.1007892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers DS, Mundodi V, Banerjee M, and Kadosh D (2014). A 5’ UTR-mediated translational efficiency mechanism inhibits the Candida albicans morphological transition. Mol. Microbiol 92, 570–585. 10.1111/mmi.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottier F, and Hall RA (2019). Face/off: the interchangeable side of Candida albicans. Front. Cell Infect. Microbiol 9, 471. 10.3389/fcimb.2019.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SE, Hopke A, Minkin SC Jr., Montedonico AE, Wheeler RT, and Reynolds TB (2014). Masking of β(1-3)-glucan in the cell wall of Candida albicans from detection by innate immune cells depends on phosphatidylserine. Infect. Immun 82, 4405–4413. 10.1128/iai.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai JV, and Lionakis MS (2018). The role of neutrophils in host defense against invasive fungal infections. Curr. Clin. Microbiol. Rep 5, 181–189. 10.1007/s40588-018-0098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai JV, and Lionakis MS (2019). Setting up home: fungal rules of commensalism in the mammalian gut. Cell Host Microbe 25, 347–349. 10.1016/j.chom.2019.02.012. [DOI] [PubMed] [Google Scholar]
- Doron I, Leonardi I, Li XV, Fiers WD, Semon A, Bialt-DeCelie M, Migaud M, Gao IH, Lin WY, Kusakabe T, et al. (2021). Human gut mycobiota tune immunity via CARD9-dependent induction of anti-fungal IgG antibodies. Cell 184, 1017–1031.e14. 10.1016/j.cell.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton LC, Nobbs AH, Jepson K, Jepson MA, Vickerman MM, Aqeel Alawfi S, Munro CA, Lamont RJ, and Jenkinson HF (2014). O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. mBio 5, e00911. 10.1128/mbio.00911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, and Littman DR (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, and Macpherson AJ (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1273. 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, and Iwasaki A (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U S A 108, 5354–5359. 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifrim DC, Quintin J, Courjol F, Verschueren I, van Krieken JH, Koentgen F, Fradin C, Gow NA, Joosten LA, van der Meer JW, et al. (2016). The role of dectin-2 for host defense against disseminated candidiasis. J. Interferon Cytokine Res 36, 267–276. 10.1089/jir.2015.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igyarto BZ, Haley K, Ortner D, Bobr A, Gerami-Nejad M, Edelson BT, Zurawski SM, Malissen B, Zurawski G, Berman J, and Kaplan DH (2011). Skin-resident murine dendritic cell subsets promote distinct and opposing antigen-specific T helper cell responses. Immunity 35, 260–272. 10.1016/j.immuni.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev ID, and Cadwell K (2021). Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology 160, 1050–1066. 10.1053/j.gastro.2020.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouault T, Ibata-Ombetta S, Takeuchi O, Trinel PA, Sacchetti P, Lefebvre P, Akira S, and Poulain D (2003). Candida albicans phospholipomannan is sensed through toll-like receptors. J. Infect. Dis 188, 165–172. 10.1086/375784. [DOI] [PubMed] [Google Scholar]
- Kadosh D, and Mundodi V (2020). A Re-evaluation of the relationship between morphology and pathogenicity in Candida species. J. Fungi (Basel) 6, 13. 10.3390/jof6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Iyer KR, Pardeshi L, Munoz JF, Robbins N, Cuomo CA, Wong KH, and Cowen LE (2019). Genetic analysis of Candida Auris implicates Hsp90 in morphogenesis and azole tolerance and Cdr1 in azole resistance. mBio 10. e02529–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin JD, Devlin JC, Yeung F, McCauley C, Leung JM, Chen YH, Cronkite A, Hansen C, Drake-Dunn C, Ruggles KV, et al. (2020). Rewilding Nod2 and Atg16l1 mutant mice uncovers genetic and environmental contributions to microbial responses and immune cell composition. Cell Host Microbe 27, 830–840.e4. 10.1016/j.chom.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Netea MG, and Holland SM (2014). Mendelian genetics of human susceptibility to fungal infection. Cold Spring Harb. Perspect. Med 4, a019638. 10.1101/cshperspect.a019638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer FL, Wilson D, and Hube B (2013). Candida albicans pathogenicity mechanisms. Virulence 4, 119–128. 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio T, Adachi-Shimizu M, Tachibana Y, Tabuchi H, Inoue SB, Yabe T, Yamada-Okabe T, Arisawa M, Watanabe T, and Yamada-Okabe H (1997). Cloning of the Candida albicans homolog of Saccharomyces cerevisiae GSC1/FKS1 and its involvement in beta-1,3-glucan synthesis. J. Bacteriol 179, 4096–4105. 10.1128/jb.179.13.4096-4105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, and Jenkins MK (2007). Naive CD4(+)T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213. 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro CA, Bates S, Buurman ET, Hughes HB, Maccallum DM, Bertram G, Atrih A, Ferguson MA, Bain JM, Brand A, et al. (2005). Mnt1p and Mnt2p of Candida albicans are partially redundant alpha-1,2-mannosyl-transferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem 280, 1051–1060. 10.1074/jbc.m411413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Brown GD, Kullberg BJ, and Gow NAR (2008). An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol 6, 67–78. 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, et al. (2006). Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Invest 116, 1642–1650. 10.1172/jci27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, Gianetti BA, and Witchley JN (2017). Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol 15, 96–108. 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost KS, O’Meara TR, Stephens WZ, Chiaro T, Zhou H, Penman J, Bell R, Catanzaro JR, Song D, Singh S, et al. (2021). Adaptive immunity induces mutualism between commensal eukaryotes. Nature 596, 114–118. 10.1038/s41586-021-03722-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M , Israel L, Chrabieh M, Audry M, et al. (2011). Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332, 65–68. 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J, Saeed S, Martens JHA, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg BJ, Wijmenga C, et al. (2012). Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe 12, 223–232. 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees PA, and Lowy RJ (2018). Measuring type I interferon using reporter gene assays based on readily available cell lines. J. Immunol. Methods 461, 63–72. 10.1016/j.jim.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. (2010). Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 32, 681–691. 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Saijo S, and Iwakura Y (2011). Dectin-1 and Dectin-2 in innate immunity against fungi. Int. Immunol 23, 467–472. 10.1093/intimm/dxr046. [DOI] [PubMed] [Google Scholar]
- Sato K, Yang XL, Yudate T, Chung JS, Wu J, Luby-Phelps K, Kimberly RP, Underhill D, Cruz PD Jr., and Ariizumi K (2006). Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J. Biol. Chem 281, 38854–38866. 10.1074/jbc.m606542200. [DOI] [PubMed] [Google Scholar]
- Saville SP, Lazzell AL, Monteagudo C, and Lopez-Ribot JL (2003). Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2, 1053–1060. 10.1128/ec.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao TY, Ang WG, Jiang TT, Huang FS, Andersen H, Kinder JM, Pham G, Burg AR, Ruff B, Gonzalez T, et al. (2019). Commensal Candida albicans positively calibrates systemic Th17 immunological responses. Cell Host Microbe 25, 404–417.e6. 10.1016/j.chom.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P, Gow N, and Berman J (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol. 12, 317–324. 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, and Brown GD (2007). Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol 8, 31–38. 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso GHW, Reales-Calderon JA, Tan ASM, Sem X, Le GTT, Tan TG, Lai GC, Srinivasan KG, Yurieva M, Liao W, et al. (2018). Experimental evolution of a fungal pathogen into a gut symbiont. Science 362, 589–595. 10.1126/science.aat0537. [DOI] [PubMed] [Google Scholar]
- Tyc KM, Herwald SE, Hogan JA, Pierce JV, Klipp E, and Kumamoto CA (2016). The game theory of Candida albicans colonization dynamics reveals host status-responsive gene expression. BMC Syst. Biol 10, 20. 10.1186/s12918-016-0268-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas VK, Barrasa MI, and Fink GR (2015). A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv 1, e1500248. 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakade RS, Huang M, Mitchell AP, Wellington M, and Krysan DJ (2021). Intravital imaging of Candida albicans identifies differential in vitro and in vivo filamentation phenotypes for transcription factor deletion mutants. mSphere 6, e0043621. 10.1128/msphere.00436-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP, and Noble SM (2019). Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host Microbe 25, 432–443.e6. 10.1016/j.chom.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported will be shared by the lead contact upon request. Raw sequence reads have been deposited to the National Library of Medicine (NCBI) and are publicly available through BioProject accession number PRJNA827179 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA827179) with identifiers for each individual sample provided in the Key resources table.
DESeq2 R code used to analyze RNA sequencing data is provided in Data S1.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE-Cy7 anti-mouse CD4 (clone: GK1.5) | eBioscience | Cat# 25-0041-82; RRID:AB_469576 |
| PE-Cy5 anti-mouse CD8 (clone: 53–6.7) | eBioscience | Cat# 35-0081-82; RRID:AB_11217674 |
| PE-Cy5 anti-mouse CD11b (clone: M1/70) | Biolegend | Cat# 101210; RRID:AB_312793 |
| PE-Cy5 anti-mouse CD11c (clone: N418) | Biolegend | Cat# 15-0114-82; RRID:AB_468717 |
| PE-Cy5 anti-mouse F4/80 (clone: BM8) | eBioscience | Cat# 15-4801-82; RRID:AB_468798 |
| PE-Cy5 anti-mouse B220 (clone: RA3-6B2) | Biolegend | Cat# 15-0452-83; RRID:AB_468756 |
| Alexa Fluor 700 anti-mouse CD44 (clone: IM7) | Biolegend | Cat# 56-0441-82; RRID:AB_494011 |
| APC-eFluor780 anti-mouse CD45 (clone: 30F11) | invitrogen | Cat# 47-0451-82; RRID:AB_1548781 |
| FITC anti-mouse FOXP3 (clone: FJK-16S) | eBioscience | Cat# 11-5773-82; RRID:AB_465243 |
| AF647 anti-mouse RORγt (clone: Q31-378) | BD bioscience | Cat# 562682; RRID:AB_2687546 |
| PE-Cy7 anti-mouse IL17A (clone: TC 11-18H10.1) | Biolegend | Cat# 506922; RRID:AB_2125010 |
| PE anti-mouse IL17F (clone: 9D3.1C8) | Biolegend | Cat# 517008; RRID:AB_10690818 |
| Anti-mouse TLR2 (clone C9A12) | Invivogen | Cat#mabg-mtlr2; RRID:AB_11125339 |
| Anti-mouse TLR4/MD2 (clone MTS510) | Hycult Biotech | Cat#HM1029; RRID:AB_533218 |
| Anti-mouse Dectin-1 (clone R1-8g7) | Invivogen | Cat# mabg-mdect; RRID:AB_2753143 |
| Anti-mouse Dectin-2 (clone D2.11E4) | Thermo Fisher Scientific | Cat#MA1-82675; RRID:AB_930457 |
| Anti-(1-3)-beta-D-glucan | Biosupplies | Cat#400-2; RRID:AB_2747399 |
| Bacterial and virus strains | ||
| Candida albicans SC5314 | Kaplan Lab, University of Pittsburg (Igyarto et al., 2011) | N/A |
| Recombinant Candida albicans SC5314 | Kaplan Lab, University of Pittsburg (Igyarto et al., 2011) | CaURA3/CaURA3 pENO1-ENO1-GFP-2W1S-OVA323-339-OVA257-264-I-Eα50-66 |
| Recombinant Candida albicans SC5314-tetO-UME6 | This study | CaURA3/CaURA3 pENO1-ENO1-GFP-2W1S-OVA323-339-OVA257-264-I-Eα50-66 CaHygB-CaTAR-tetO-UME6/CaHygB-CaTAR-tetO-UME6 |
| SC5314-mnt1/2Δ | (Munro et al., 2005) | mnt1−/−;mnt2−/− |
| Recombinant Candida albicans SC5314-MNT1/2Δ | This study | N/A |
| Candida albicans ume6 deficient | (Carlisle and Kadosh, 2010) | ume6−/− |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin | Sigma-Aldrich | Catalog# A0166 |
| Gentamicin | Sigma-Aldrich | Catalog# G3632 |
| Metronidazole | Sigma-Aldrich | Catalog# M3761 |
| Neomycin | Sigma-Aldrich | Catalog# N6386 |
| Vancomycin | MP Biomedical | Catalog# 0219554005 |
| Doxycycline | Sigma-Aldrich | Catalog# D9891 |
| Mannan | Sigma-Aldrich | Catalog# M7504 |
| β-glucan | Millipore | Catalog# 346210 |
| QUANTI-Blue | Invivogen | Catalog# rep-qbs |
| Dehydrated Culture Media: Brain Heart Infusion | Thermo Fisher Scientific | Catalog# B11060 |
| Yeast extract | Boston Bio Product | Catalog# P-950 |
| Bactopeptone | BD Bioscience | Catalog# DF0118170 |
| Uridine | Alfa-Aesar | Catalog# A15227 |
| D-(+)-Glucose | Sigma-Aldrich | Catalog# G5767 |
| Agar | Bio Express | Catalog# J637 |
| DMEM | Gibco | Catalog# 10313-21 |
| Fetal bovine serum | GeneMate | Catalog# S-1200-500 |
| Glutamine (100X) | Gibco | Catalog# 25030-081 |
| HEPES 1M | Gibco | Catalog# 15630-080 |
| Penicillin-streptomycin solution (100X) | Gibco | Catalog# 15140-122 |
| BD Golgi Plug (Brefeldin A solution) | BD Bioscience | Catalog# 555029 |
| Foxp3/Transcription factor staining buffer set | eBioscience | Catalog# 00-5523-00 |
| Fixation/Permeabilization solution kit | BD Bioscience | Catalog# 554722 |
| APC Conjugation Kit-Lightning-Link | abcam | Catalog# ab201807 |
| Calcofluor White Stain | Biotium | Catalog# 29067 |
| Critical commercial assays | ||
| ZymoBIOMICS RNA Miniprep Kit | ZYMO RESEARCH | Catalog# R2001 |
| SuperScript™ II Reverse Transcriptase kit | Invitrogen | Catalog# 18064014 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | Catalog# A25780 |
| SMARTer Stranded Total RNA-Seq Kit v3 | TaKaRa | Catalog# 634485 |
| Deposited data | ||
| WT C. albicans 1.1 | WT_1.1 | PRJNA827179 |
| WT C. albicans 1.2 | WT_1.2 | PRJNA827179 |
| WT C. albicans 3.1 | WT_3.1 | PRJNA827179 |
| WT C. albicans 3.2 | WT_3.2 | PRJNA827179 |
| WT C. albicans 3.3 | WT_3.3 | PRJNA827179 |
| WT C. albicans 3.4 | WT_3.4 | PRJNA827179 |
| UME6-ON C. albicans 2.1 | UME6ON_2.1 | PRJNA827179 |
| UME6-ON C. albicans 2.2 | UME6ON_2.2 | PRJNA827179 |
| UME6-ON C. albicans 2.3 | UME6ON_2.3 | PRJNA827179 |
| UME6-ON C. albicans 2.4 | UME6ON_2.4 | PRJNA827179 |
| UME6-ON C. albicans 2.5 | UME6ON_2.5 | PRJNA827179 |
| UME6-ON C. albicans 2.6 | UME6ON_2.6 | PRJNA827179 |
| UME6-ON C. albicans 3.1 | UME6ON_3.1 | PRJNA827179 |
| UME6-ON C. albicans 3.2 | UME6ON_3.2 | PRJNA827179 |
| UME6-ON C. albicans 3.3 | UME6ON_3.3 | PRJNA827179 |
| UME6-ON C. albicans 3.4 | UME6ON_3.4 | PRJNA827179 |
| UME6-OFF C. albicans 1.1 | UME6OFF_1.1 | PRJNA827179 |
| UME6-OFF C. albicans 1.2 | UME6OFF_1.2 | PRJNA827179 |
| UME6-OFF C. albicans 2.1 | UME6OFF_2.1 | PRJNA827179 |
| UME6-OFF C. albicans 2.2 | UME6OFF_2.2 | PRJNA827179 |
| UME6-OFF C. albicans 2.3 | UME6OFF_2.3 | PRJNA827179 |
| UME6-OFF C. albicans 2.4 | UME6OFF_2.4 | PRJNA827179 |
| UME6-ON to OFF (0 h) 1 | UME6ONtoOFF_0h_1 | PRJNA827179 |
| UME6-ON to OFF (0 h) 2 | UME6ONtoOFF_0h_2 | PRJNA827179 |
| UME6-ON to OFF (0 h) 3 | UME6ONtoOFF_0h_3 | PRJNA827179 |
| UME6-ON to OFF (0 h) 4 | UME6ONtoOFF_0h_4 | PRJNA827179 |
| UME6-ON to OFF (3 h) 1 | UME6ONtoOFF_3h_1 | PRJNA827179 |
| UME6-ON to OFF (3 h) 2 | UME6ONtoOFF_3h_2 | PRJNA827179 |
| UME6-ON to OFF (3 h) 3 | UME6ONtoOFF_3h_3 | PRJNA827179 |
| UME6-ON to OFF (3 h) 4 | UME6ONtoOFF_3h_4 | PRJNA827179 |
| UME6-ON to OFF (6 h) 1 | UME6ONtoOFF_6h_1 | PRJNA827179 |
| UME6-ON to OFF (6 h) 2 | UME6ONtoOFF_6h_2 | PRJNA827179 |
| UME6-ON to OFF (6 h) 3 | UME6ONtoOFF_6h_3 | PRJNA827179 |
| UME6-ON to OFF (6 h) 4 | UME6ONtoOFF_6h_4 | PRJNA827179 |
| UME6-ON to OFF (12 h) 1 | UME6ONtoOFF_12h_1 | PRJNA827179 |
| UME6-ON to OFF (12 h) 2 | UME6ONtoOFF_12h_2 | PRJNA827179 |
| UME6-ON to OFF (12 h) 3 | UME6ONtoOFF_12h_3 | PRJNA827179 |
| UME6-ON to OFF (12 h) 4 | UME6ONtoOFF_12h_4 | PRJNA827179 |
| UME6-OFF to ON (0 h) 1 | UME6OFFtoON_0h_1 | PRJNA827179 |
| UME6-OFF to ON (0 h) 2 | UME6OFFtoON_0h_2 | PRJNA827179 |
| UME6-OFF to ON (3 h) 1 | UME6OFFtoON_3h_1 | PRJNA827179 |
| UME6-OFF to ON (3 h) 2 | UME6OFFtoON_3h_2 | PRJNA827179 |
| UME6-OFF to ON (3 h) 3 | UME6OFFtoON_3h_3 | PRJNA827179 |
| UME6-OFF to ON (3 h) 4 | UME6OFFtoON_3h_4 | PRJNA827179 |
| UME6-OFF to ON (6 h) 1 | UME6OFFtoON_6h_1 | PRJNA827179 |
| UME6-OFF to ON (6 h) 2 | UME6OFFtoON_6h_2 | PRJNA827179 |
| UME6-OFF to ON (12 h) 1 | UME6OFFtoON_12h_1 | PRJNA827179 |
| UME6-OFF to ON (12 h) 2 | UME6OFFtoON_12h_2 | PRJNA827179 |
| UME6-OFF to ON (12 h) 3 | UME6OFFtoON_12h_3 | PRJNA827179 |
| UME6-OFF to ON (12 h) 4 | UME6OFFtoON_12h_4 | PRJNA827179 |
| Experimental models: Cell lines | ||
| Raw-Blue Macrophage Cells | Invivogen | Catalog# raw-sp RRID: CVCL_X594 |
| Experimental models: Organisms/strains | ||
| C57BL/6 | Charles River Laboratories | Strain Code: 027 |
| B6.129-Tlr2tm1Kir/J | The Jackson Laboratory | Stock No: 004650 |
| B6(Cg)-Tlr4tm1.2Karp/J | The Jackson Laboratory | Stock No: 029015 |
| B6.129-Card9tm1Xlin/J | The Jackson Laboratory | Stock No: 028652 |
| B6.129P2(SJL)-Myd88tm1.1Defr/J | The Jackson Laboratory | Stock No: 009088 |
| B6.129S6-Clec7atm1Gdb/J | The Jackson Laboratory | Stock No: 012337 |
| B6;129P2-Fcer1gtm1Rav/J | The Jackson Laboratory | Stock No: 002847 |
| B6.129S2-Ighmtm1Cgn/J | The Jackson Laboratory | Stock No: 002288 |
| Oligonucleotides | ||
| 5′junction, Primer1: GCACCAAACACCCGAAATTA; Primer2: CCATTTTGGCGTGAGGTAATCC | Thermo Fisher Scientific | N/A |
| 3′junction, Primer1: AGTGAAAGTCGAGTTTACCACTC; Primer2: GCTGCAGTTGCAGTTGTTGT | Thermo Fisher Scientific | N/A |
| Native ume6 upstream, Primer1: AAGCAAAGCAATAATGAGCCAA; Primer2: AGAATTTCCCGGGAGTTGTTTA | Thermo Fisher Scientific | N/A |
| Ume6qPCR; Primer1: GAACAATGGTGGTGGTAGTGG; Primer2: AATTCGACAAATCCAACATCC | Thermo Fisher Scientific | (Childers et al., 2014) |
| Act1qPCR; Primer1: TTGCTCCAGAAGAACATCCAG; Primer2: AGTAACACCATCACCAGAATCC | Thermo Fisher Scientific | (Childers et al., 2014) |
| 7707 Primer:GAGAATCGAAGAAGAA TTAGGTTCTGAAGCTATCTACGCTG GTAAAGATTTCCAAAAGGCTTCTCA ATTGGGTGGTGGTTCTAAAggTgAAg AATTATT |
Thermo Fisher Scientific | N/A |
| 7708 Primer:TTTAATTAGTTCATAT ATTCAAGATGTTCCTATAAAAGAA AAAAAAAGCACCAG CTTTTTTTTATTTAATCGTAAAACG ACGGCCAGTGAATTC |
Thermo Fisher Scientific | N/A |
| 5059 Primer: GCAAATGATTATACATGGGGATG | Thermo Fisher Scientific | N/A |
| 7722 Primer: ACAATCCAGTACATCAAACTCCAA | Thermo Fisher Scientific | N/A |
| 3925 Primer: GAACATAACCTTCTGGCATGGC | Thermo Fisher Scientific | N/A |
| 7763 Primer: GATGCTTGGGTCCACTTCT | Thermo Fisher Scientific | N/A |
| Recombinant DNA | ||
| pV1393 | (Vyas et al., 2015) | Resource of CaCAS9 |
| pYM70 | (Basso et al., 2010) | Resource of TEF2 promoter-CaHygB-ACT1 terminator |
| pLC1031 | (Kim etal., 2019) | Resource of CaHygB-CaTAR-tetO |
| Software and algorithms | ||
| Prism 9.3.1 | GraphPad | N/A |
| Flow Jo 9.9.6 | Treestar | N/A |
| Gene5 | BioTek | N/A |
| Subread | http://subread.sourceforge.net/ | N/A |
| DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | N/A |
| Other | ||
| 2W1S52-68:I-Ab Tetramer | Jenkins Lab, University of Minnesota (Moon et al., 2007) | N/A |


