Abstract
The effects of different carbon sources on expression of the styrene catabolism genes in Pseudomonas fluorescens ST were analyzed by using a promoter probe vector, pPR9TT, which contains transcription terminators upstream and downstream of the β-galactosidase reporter system. Expression of the promoter of the stySR operon, which codes for the styrene two-component regulatory system, was found to be constitutive and not subject to catabolite repression. This was confirmed by the results of an analysis of the stySR transcript in P. fluorescens ST cells grown on different carbon sources. The promoter of the operon of the upper pathway, designated PstyA, was induced by styrene and repressed to different extents by organic acids or carbohydrates. In particular, cells grown on succinate or lactate in the presence of styrene started to exhibit β-galactosidase activity during the mid-exponential growth phase, before the preferred carbon sources were depleted, indicating that there is a threshold succinate and lactate concentration which allows induction of styrene catabolic genes. In contrast, cells grown on glucose, acetate, or glutamate and styrene exhibited a diauxic growth curve, and β-galactosidase activity was detected only after the end of the exponential growth phase. In each experiment the reliability of the reporter system constructed was verified by comparing the β-galactosidase activity and the activity of the styrene monooxygenase encoded by the first gene of the styrene catabolic operon.
Styrene is a chemical that is used extensively in the manufacturing of plastics and synthetic rubbers. This toxic compound is released into the environment mainly through factory wastewater, evaporation, and pyrolysis of polystyrene. Different routes for styrene catabolism in different microorganisms have been described (8, 9, 17, 21, 29, 31). Recently, strains belonging to the genus Pseudomonas have been studied more extensively both at the physiological level (21–23) and the molecular level (2, 17, 24, 30). In these strains the catabolic genes are organized in a cluster whose expression requires the presence of two genes, styS and styR, which are organized in an operon and code for a sensor kinase and a regulatory DNA binding protein, respectively. Two-component regulatory systems for genes involved in aromatic hydrocarbon degradation have been described previously only for toluene degradation in Pseudomonas putida F1 and Thauera sp. strain T1 (6, 15) and for degradation of biphenyls in Rhodococcus sp. strain M5 (14).
In our laboratory, Pseudomonas fluorescens ST, which is able to grow on styrene as a sole carbon source, has been characterized, and both the regulatory genes (styS and styR) and the upper pathway genes (styA, styB, styC, and styD), which code for conversion of styrene into phenylacetic acid, have been sequenced (2, 17, 18). At the moment, our interest is focused on characterization of the regulatory system and, in particular, on the effects of different carbon sources on styrene-induced expression of the regulatory and structural genes. Several examples of carbon catabolite repression of expression of catabolic pathways for aromatic and nonaromatic compounds have been described in Pseudomonas spp. (12, 20, 32). However, none of these studies dealt with catabolic operons regulated by a two-component regulatory system.
In this paper we describe the effects of growth on different carbon sources on expression of the styrene regulatory and degradative operons.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and chemicals.
The bacterial strains and plasmids used in this study are listed in Table 1. P. fluorescens ST and Escherichia coli cells were routinely grown at 30 and 37°C, respectively, in Luria-Bertani (LB) medium (19) or mineral salts medium (9) containing different carbon sources at the following concentrations: 0.2% succinate, 0.05% glucose, 0.1% lactate, and 0.1% acetate. In induction studies, styrene was added via the gas phase as previously described (17). When necessary, cultures were supplemented with ampicillin (100 μg/ml), tetracycline (15 μg/ml), or chloramphenicol (30 μg/ml). Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM), 5′-bromo-4′-chloro-3′-indolyl-β-d-galactopyranoside (X-Gal) (1 mM), and 2-nitrophenyl-β-d-galactopyranoside (1 mM) were added to the media when appropriate.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and/or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| P. fluorescens ST | Sty+ | 1 |
| E. coli DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA (Nalr) (lacIZYA-argF)U169 deoR [80dlac (lacZ)M15] | Bethesda Research Laboratories |
| E. coli S17.1 | recA pro thi hsdR RP4-2-Tc::Mu-Km::Tn7 Tra+ Tpr Strr | 27 |
| Plasmids | ||
| pPR9 | lacZ promoter probe vector RK2 replicon; Apr Cmr; 8.9 kb | Our laboratory |
| pBluescriptII KS+ | ColE1 replicon; Apr; 2.9 kb | Strategene |
| pPR9TT | pPR9 derivative in which a 410-bp BamHI-BglII fragment containing the rrnBT1T2 terminators was cloned into a BglII site of pPR9; Apr Cmr; 9.3 kb | Our laboratory |
| pTZ19R | ColE1 replicon; Apr; 2.9 kb | MBI Fermentas |
| pTE30 | pTZ19R derivative containing a chromosomal fragment of P. fluorescens ST carrying two truncated genes coding for paaK and styS; Apr; 6.0 kb | Our laboratory |
| pBSPs | pBluescriptII KS+ derivative in which a 409-bp PCR blunt-ended fragment containing PstySR was cloned into the HincII site of the vector; Apr; 3.3 kb | This study |
| pPR9Ps | pPR9 derivative in which the 411-bp fragment from pBSPs containing PstySR was cloned as a HindIII-PstI fragment into the same sites of the vector; Apr Cmr; 9.4 kb | This study |
| pPR9TTPs | pPR9TT derivative in which a 411-bp fragment from pBSPs containing PstySR was cloned as a HindIII-PstI fragment into the same sites of the vector; Apr Cmr; 9.7 kb | This study |
| pTPE30 | pTZ19R derivative containing a chromosomal fragment of P. fluorescens ST carrying the genes coding for styR, styA, and styB; Apr; 6.0 kb | 17 |
| pBSPa | pBluescriptII KS+ derivative in which a 492-bp PCR blunt-ended fragment containing PstyA was cloned into the HincII site of the vector; Apr; 3.3 kb | This study |
| pPR9TTPa | pPR9TT derivative in which a 560-bp BamHI-XhoI fragment from pBSPa containing PstyA was cloned into BglII-XhoI sites of the vector; Apr Cmr; 9.9 kb | This study |
Apr, ampicillin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; Sty, styrene metabolic phenotype; Strr, streptomycin resistance.
Conjugative mating.
Plasmids were transferred from E. coli S17.1 to P. fluorescens ST by mating on membranes, and the mixtures were incubated on nutrient-yeast extract agar at 30°C for 14 h. The mating mixtures were then plated onto selective media.
DNA manipulation.
Transformations of E. coli, restrictions, and ligations were carried out by using standard procedures (26). Plasmid DNA was prepared by the alkaline lysis protocol (26) or with a QIAGEN Midi isolation kit (Qiagen). DNA fragments were purified from agarose by using a Qiaquick gel extraction kit or a QIAEXII kit (Qiagen). DNA 5′ protruding ends and 3′ protruding ends were made blunt by using Klenow polymerase and T4 DNA polymerase, respectively. PCR amplification of the styrene monooxygenase promoter region (designated PstyA) from pTPE30 (17) was performed by using the following synthesized primers: 5′ GCTCTAGAATGTCAGATCTCTGGC 3′ and 5′ GGGGTACCTACGTAGTAGTAGTGG 3′ containing an XbaI site and a KpnI site (underlined nucleotides), respectively. PCR amplification of the regulatory gene promoter region (designated PstySR) from pTE30 (our laboratory) was performed by using the following synthesized primers: 5′ CAAGCTTGAATGCTTCATGTCGGC 3′ and 5′ GGAATTCCGATCCAGAATGATCCG 3′ containing an HindIII site and an EcoRI site (underlined nucleotides), respectively. PCR amplifications were performed by using standard procedures and, unless otherwise specified, Pfu polymerase from Stratagene.
All PCR fragments were controlled by sequencing them with an Applied Biosystems automated sequencer (model 373 Stretch) and a DyeDeoxy terminator cycle sequencing kit (Perkin-Elmer). Both commercially available and synthetic primers were used for sequencing reactions.
Northern blot analysis.
P. fluorescens ST cells were grown on glucose, succinate, and styrene to an optical density at 600 nm (OD600) of approximately 0.3. RNA was prepared and electrophoresis was performed essentially as described by Leoni et al. (16). RNAs were transferred onto nitrocellulose filters (Optitran BA-S 83; Schleicher & Schuell) as described by Sambrook et al. (26) and heat fixed. A 1.7-kb XhoI-BglII DNA fragment containing 1,100 nucleotides of styS and 600 nucleotides of styR was labeled with [α-32P]dATP (3.0 Ci/nmol; Amersham Corp.) by using a random priming labeling kit (Boehringer) and was purified with a Sephadex G-50 spin column. Filter hybridization and washing were performed by using standard procedures (26).
Induction conditions.
In induction assays, P. fluorescens ST cells harboring pPR9TTPa, pPR9Ps, or pPR9TTPs were pregrown overnight at 30°C in mineral salts medium supplemented with succinate, lactate, glucose, acetate, or styrene. The styrene-grown cells were transferred to styrene mineral medium, while the succinate-, lactate-, acetate-, and glucose-grown cells were inoculated into the corresponding mineral media with or without styrene. Cell growth was measured by monitoring the OD600.
SMO assays.
To quantify styrene monooxygenase (SMO) activity, production of indigo was assayed essentially as described by O'Connor et al. (23). Cells were harvested in the exponential and stationary phases by centrifugation, washed with 50 mM potassium phosphate buffer (pH 7.0), and resuspended in the same buffer to an OD600 of 3.0. One hundred to 600 μl of concentrated cells was added to 400 μl of 50 mM potassium phosphate buffer (pH 7) containing 0.25 mM indole in 1.5-ml polypropylene microcentrifuge tubes. The samples were incubated horizontally at 30°C with vigorous shaking for 30 min. The samples were then centrifuged at 14,000 rpm for 2 min, and the supernatants were carefully discarded. The cell pellets were resuspended in 1 ml of dimethylformamide and extracted by shaking for 15 min. The tubes were then centrifuged to remove the cell debris, and the OD600 of the supernatants were determined. The data presented below are the results obtained from at least three independent experiments with standard deviations ranging from 5 to 10%.
β-Galactosidase assay.
β-Galactosidase activity was measured as described by Miller (19) and was expressed in Miller units. The data presented below are to the results obtained from at least three independent experiments with a standard deviation of 10%.
RESULTS
Construction of PstySR-lacZ fusions.
In order to study the activity of the promoter of the styrene regulatory operon, designated PstySR, we used two new promoter probe vectors, pPR9 and pPR9TT, which were based on the RK2 replicon and contained lacZ as a reporter gene (Santos et al., unpublished data). pPR9 is a derivative of pJB653 (3) in which the Pm-xylS expression system has been replaced by the lacZ gene from PMC1871 (Pharmacia), which lacks transcription and translation signals. This plasmid contains the polylinker of pBluescriptII KS(+) and the transcriptional terminators of Ω-Km (3) located downstream of lacZ. Moreover, the Cmr marker was inserted into the unique HindIII site downstream of the trfA gene. pPR9TT is a derivative of pPR9 in which the strong ribosomal terminators rrnBT1T2 from pBTac1 are inserted upstream of the polylinker region. No β-galactosidase activity was detected with pPR9TT in the E. coli or Pseudomonas sp. strains tested, while low levels of such activity (5 to 20 Miller units) were observed with pPR9 depending on the host strain and the growth phase, indicating a possible weak read-through from the vector (data not shown). The putative promoter region, PstySR, was obtained by PCR as described above. This region is located upstream of the stySR genes, which encode a sensor histidine kinase and a response regulator (Fig. 1). The 409-bp PCR product included the stop codon of the upstream gene, paaK (18, 30), the intergenic region containing PstySR, and the first 46 codons of styS. We cloned PstySR in both pPR9 and pPR9TT, which generated pPR9Ps and pPR9TTPs, respectively (see below). To do this, PstySR first was cloned into the HincII site of pBluescriptII KS(+), which generated pBSPs, and then was transferred to pPR9 and pPR9TT as a 411-bp HindIII-PstI fragment at the same sites of the vectors, in frame with the lacZ gene.
FIG. 1.
Regulatory and catabolic operons of the styrene degradation system in P. fluorescens ST. styS, sensor; styR, regulator; styAB, SMO gene; styC, epoxystyrene isomerase gene; styD, phenylacetaldehyde dehydrogenase gene; paaK, phenylacetyl-coenzyme A ligase gene; IS1162, insertion sequence. PstySR, promoter of stySR; PstyA, promoter of styABCD; I1, putative styrene-sensing domain; HK1 and HK2, histidine kinase domains; R1, sensor receiver domain; I2, putative oxygen- and/or redox potential-sensing domain; R2, regulator receiver domain; B, DNA binding domain. The bent arrows indicate the promoter regions and the orientation of gene transcription.
Activity of the PstySR promoter under different growth conditions.
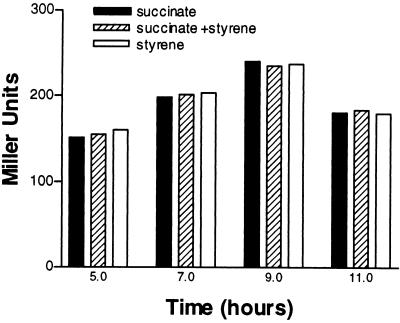
Previously, it was reported that a transcription termination-like sequence is present just downstream of the paaK stop codon (30). However, the effectiveness of this putative terminator has not been proven. In order to assess the functioning of such a putative terminator, we cloned PstySR in both pPR9 and pPR9TT. In order to study the activity of PstySR under different growth conditions in a homologous system, we transferred plasmids pPR9Ps and pPR9TTPs into strain ST, which contained the stySR and styABCD operons (Fig. 1) in its chromosome. The β-galactosidase activity of ST cells harboring pPR9Ps did not depend on the presence of styrene and was not influenced by additional carbon sources in any of the growth phases analyzed. The β-galactosidase activities of P. fluorescens ST(pPR9Ps) cells grown on succinate, succinate plus styrene, and styrene are shown in Fig. 2. The same results were obtained when cells were grown on glucose, on glutamate, or in LB medium (data not shown). To confirm that these results were not due to read-through from the vector because of the inefficiency of the putative terminator located upstream of PstySR, the same experiments were performed with pPR9TTPs. The results obtained with this vector were identical to the results obtained with pPR9Ps, indicating that the natural terminator is effective. However, since RK2-based vectors, such as pPR9 and pPR9TT, occur at levels of five to seven copies per chromosome (3), we examined the possibility that the presence of multiple copies of PstySR could result in apparent constitutive expression of PstySR. Therefore, we analyzed the transcripts of stySR genes in P. fluorescens ST grown on glucose, succinate, and styrene. The results obtained (Fig. 2) showed that a comparable amount of the stySR transcript (length, approximately 3.6 kb) was present under the growth conditions examined, which confirmed the data obtained with the β-galactosidase assay.
FIG. 2.
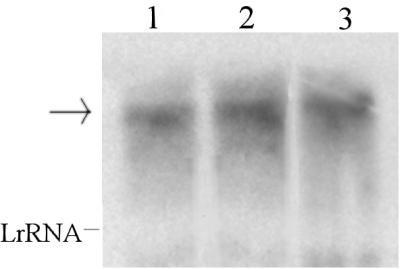
Activity of the PstySR promoter under induced and uninduced conditions. The graph shows the β-galactosidase activities of P. fluorescens ST(pPR9Ps) at different times during growth on 0.2% succinate, 0.2% succinate and styrene, and styrene. The gel shows stySR expression as determined by Northern blot analysis. P. fluorescens ST cells were grown on glucose (lane 1), succinate (lane 2), and styrene (lane 3) to an OD600 of 0.3. The experimental procedures used for RNA preparation and detection are described in the text. Each lane contained 20 μg of RNA. The probe was a 1.7-kb DNA fragment containing both styS and styR sequences. LrRNA, large rRNA. The arrow indicates the position of the StySR transcript.
Construction of PstyA-lacZ fusion.
The PstyA promoter (Fig. 1) is induced in the presence of styrene and is responsible for expression of the styrene catabolic operon (2, 17, 24, 30). Sequence analysis of the DNA region upstream of styA has shown that there is an inverted repeat that is located 75 bp upstream of the start codon and contains a sequence identical to the tod box sequence involved in toluene utilization in Pseudomonas putida F1 (15). It has been shown that this box is the DNA binding site of TodT, which belongs to a two-component regulatory system that is highly homologous to the StyS-StyR system. To study the activity of PstyA, a 492-bp PCR fragment that included 23 codons of the upstream styR gene, the intergenic region containing PstyA, and the first 81 codons of the styA gene was cloned into the HincII site of pBluescriptII KS(+), generating pBSPa. pPR9TTPa was constructed by cloning the 560-bp BamHI-XhoI fragment from pBSPa into the BglII-XhoI sites of pPR9TT in frame with the lacZ gene.
Effects of different carbon sources on PstyA activity.
pPR9TTPa was transferred by conjugation into P. fluorescens ST, and cells were grown on styrene mineral medium (Fig. 3A) and on mineral medium supplemented with different carbon sources in the presence or absence of styrene (Fig. 4A), as described above. Samples were harvested at different times during the exponential and stationary phases, and SMO and β-galactosidase activities were determined. SMO activity was determined by monitoring the conversion of indole to indoxyl, which spontaneously dimerizes to the blue dye indigo. Formation of indigo has been used extensively to select microorganisms that express dioxygenase or monooxygenase activities (2, 7). Previously, we demonstrated that E. coli expressing a DNA fragment containing styAB formed indigo from indole and styrene oxide from styrene, indicating that the two reactions are catalyzed by the same enzyme (2). In this way we could directly measure the activity of the styAB gene product together with cloned PstyA expression.
FIG. 3.
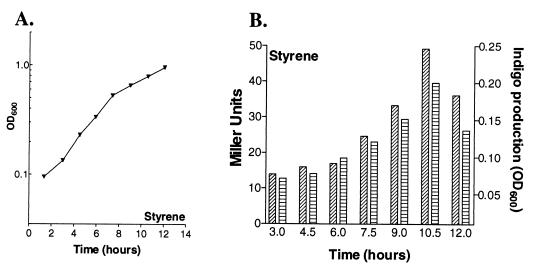
Activity of the PstyA promoter under induction conditions. (A) Growth curve for P. fluorescens ST(pPR9TTPa) when styrene was the sole carbon source. (B) β-Galactosidase activity (diagonally cross-hatched bars) and indigo production (horizontally cross-hatched bars) at different times.
FIG. 4.
Effects of different carbon sources on PstyA promoter activity. (A) P. fluorescens ST cells harboring pPR9TTPa were grown on different carbon sources in the presence (■) or in the absence (□) of styrene. (B) β-Galactosidase activity in the presence (solid bars) or in the absence (open bars) of styrene and indigo production in the presence (cross-hatched bars) or in the absence (stippled bars) of styrene at different times.
The results which we obtained showed that formation of indigo was induced only in the presence of styrene (Fig. 3B and 4B). When cells were grown on organic acids or carbohydrates, indigo was not formed and β-galactosidase activity was not detected (Fig. 4B). When styrene was used as the sole carbon source, formation of indigo and β-galactosidase activity were detected in the early exponential growth phase (Fig. 3B). However, cells grown on succinate or lactate and styrene started to accumulate indigo and to exhibit β-galactosidase activity during the mid-exponential growth phase (Fig. 4B). This suggests that cells started to grow by utilizing succinate or lactate and that the shift in substrate utilization from these organic acids to styrene occurred before the preferred carbon sources were depleted. The conclusion that during the early exponential growth phase these organic acids repressed PstyA induction was confirmed by the finding that the two enzymatic activities considered were easily detected in the early exponential phase of the growth when glycerol was the carbon source added (data not shown). The effects of succinate and lactate at concentrations ranging from 0.05 to 0.4% were also examined. A diauxic growth curve was not observed, indicating that there was not mutual exclusion by the two substrates and that there probably is a threshold succinate and lactate concentration which allows induction of the styrene catabolic genes.
It has been reported that in Pseudomonas lemoignei uptake of succinate depends on the pH (28), and the optimum pH range is 5.6 to 7.0. We performed experiments with P. fluorescens ST cells grown in pH 6.0 buffered mineral medium supplemented with succinate and styrene, and we found that the styrene catabolic operon was expressed only at the end of the exponential phase (data not shown). This higher level of repression could have been a result of a higher concentration of succinate inside the cells due to greater efficiency of its transport system. However, we were not able to obtain diauxic growth even at higher concentrations.
Cells grown on glucose or acetate and styrene started to accumulate indigo and to exhibit β-galactosidase activity only after the end of exponential growth phase (Fig. 4), indicating that these carbon sources do impose a high level of catabolite repression on expression of the styrene degradative operon. Furthermore, we examined the influence of the concentration of these carbon sources by using concentrations ranging from 0.05 to 0.4%, and we observed that an increase in concentration resulted in an increase in the time necessary for the shift to styrene utilization. This resulted in a prolonged second lag in diauxic growth (data not shown). Several other substrates were tested, and we found that arginine and glycerol did not affect PstyA induction, while glutamate and citrate strongly repressed PstyA induction (data not shown), as described above for glucose and acetate. Finally, the results of the assays performed in LB medium in the presence or in the absence of styrene showed that neither β-galactosidase nor SMO is expressed in this medium. A similar repressive effect of LB medium has been described previously for the majority of the aromatic or aliphatic catabolic operons that have been studied so far (12, 20, 32).
DISCUSSION
Our results show that PstyA expression is induced by styrene. In the presence of an additional carbon source, such as an organic acid or a carbohydrate, induction by styrene was affected, and the extent to which induction was affected depended on the carbon source and on its concentration. It is known that organic acids are usually the preferred carbon sources in Pseudomonas spp. cultures (5), but the mechanism of catabolite repression in these microorganisms is not understood yet. Our data confirm the results obtained for SMO activity in Pseudomonas putida CA-3 and support the hypothesis that also in this strain catabolite repression can occur at the transcriptional level (21).
Results obtained with pPR9Ps and in the transcript analysis showed that expression of the PstySR promoter is constitutive and does not depend on the type of carbon source. PstySR is the promoter of the operon coding for the two-component regulatory system, which includes a sensor (styS) and a regulator (styR), which are necessary for PstyA induction (24, 30). If there is no control at the translation level, StyS and StyR are constitutively present in a cell. This suggests that some steps in the signal transduction from styrene to PstyA activation are controlled by catabolite repression. The factor that is responsible for catabolite repression can affect the kinase activity of the sensor, can inhibit phosphorylation of the regulator or binding of the regulator to the promoter, or can directly bind to a specific sequence on the repressible promoter. However, analysis of different promoters of aromatic and aliphatic degradative operons did not reveal common sequences which could be the binding site for a common repressor. In Pseudomonas cultures, the presence of a solvent in the medium triggers a stress response which induces an overall readjustment of the cells through activation of defense mechanisms, including adaptation to the solvent (10, 11; for a review see reference 13). Many of these defense mechanisms are energy dependent so that growing cells in the presence of styrene leads to a requirement for more energy. This demand for extra energy is preferably met by using a readily utilizable carbon source rather than the solvent, whose utilization requires many steps to obtain an energy-yielding intermediate.
Finding a two-component regulatory system for degradation of aromatic compounds is not common. Such a system is usually associated with complex metabolic responses to environmental changes, such as nitrogen fixation, alginate production, nodulation, or virulence, or with a stress response (25, 33). It is possible that cells sense styrene as a stress factor or that styrene catabolism requires fine regulation linked to the redox status of the cell due to the toxicity of the catabolic intermediates styrene oxide and phenylacetaldehyde. We do not know if this kind of regulation is also associated with catabolite repression, but it is possible to look at this process as a response to a specific energetic state of the cells. Recently, the effect of IIANtr, a protein of the PTS-like transport system (4), on carbon catabolite repression of the ς54-dependent Pu promoter has been described (4). This protein seems to play a role in the relationship between some ς54-dependent promoters and nitrogen and carbon metabolism (4). This finding is consistent with the picture emerging from studies of catabolite repression of aromatic and aliphatic degradative operons in Pseudomonas spp. (12, 20, 32), which seems to indicate that the mechanism involved is a general mechanism related to cell metabolism, since a single carbon source has different repressive effects depending on the strain and the growth conditions.
ACKNOWLEDGMENTS
We thank Hermann Heipieper for useful discussions.
This work was supported by grant 9701252.49 from the Target Project on Biotechnology, Consiglio Nazionale delle Ricerche, Rome, Italy. P. M. Santos received a Ph.D. fellowship from the FCT, Portugal (grant PRAXIS XXI/BD/15899/98).
REFERENCES
- 1.Baggi G, Boga M M, Catelani D, Galli E, Treccani V. Styrene catabolism by a strain of Pseudomonas fluorescens. Syst Appl Microbiol. 1983;4:141–147. doi: 10.1016/S0723-2020(83)80042-3. [DOI] [PubMed] [Google Scholar]
- 2.Beltrametti F, Marconi A M, Bestetti G, Colombo C, Galli E, Ruzzi M, Zennaro E. Sequencing and functional analysis of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol. 1997;63:2232–2239. doi: 10.1128/aem.63.6.2232-2239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blatny J M, Brautaset T, Winther-Larsen H C, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol. 1997;63:370–379. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cases I, Pérez-Martín J, de Lorenzo V. The IIANtr (PtsN) protein of Pseudomonas putida mediates the C source inhibition of the ς54-dependent Pu promoter of the TOL plasmid. J Biol Chem. 1999;274:15562–15568. doi: 10.1074/jbc.274.22.15562. [DOI] [PubMed] [Google Scholar]
- 5.Collier D N, Hager P W, Phibbs P V., Jr Catabolite repression control in the pseudomonads. Res Microbiol. 1996;147:551–561. doi: 10.1016/0923-2508(96)84011-3. [DOI] [PubMed] [Google Scholar]
- 6.Coschigano P W, Young L Y. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl Environ Microbiol. 1997;63:652–660. doi: 10.1128/aem.63.2.652-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaton R W, Chapman P J. Formation of indigo and related compounds from indolecarboxylic acids by aromatic acid-degrading bacteria: chromogenic reactions for cloning genes encoding dioxygenases that act on aromatic acids. J Bacteriol. 1995;177:6983–6988. doi: 10.1128/jb.177.23.6983-6988.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmans S, Smits J P, Van der Werf M J, Volkering F, de Bont J A M. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl Environ Microbiol. 1989;55:2850–2855. doi: 10.1128/aem.55.11.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmans S, Van der Werf M J, de Bont J A M. Bacterial degradation of styrene involving a novel flavin adenine dinucleotide-dependent styrene monooxygenase. Appl Environ Microbiol. 1990;56:1347–1351. doi: 10.1128/aem.56.5.1347-1351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heipieper H J, de Bont J A M. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of the fatty acid composition of membranes. Appl Environ Microbiol. 1994;60:4440–4444. doi: 10.1128/aem.60.12.4440-4444.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heipieper H J, Meulenbeld G, van Oirschot Q, de Bont J A M. Effect of environmental factors on the trans/cis ratio of unsaturated fatty acids in Pseudomonas putida S12. Appl Environ Microbiol. 1996;62:2772–2777. doi: 10.1128/aem.62.8.2773-2777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holtel A, Marqués S, Mohler I, Jakubzik U, Timmis K. Carbon source-dependent inhibition of xyl operon expression of the Pseudomonas putida TOL plasmid. J Bacteriol. 1994;176:1173–1176. doi: 10.1128/jb.176.6.1773-1776.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isken S, de Bont J A M. Bacteria tolerant to organic solvents. Extremophiles. 1998;2:229–238. doi: 10.1007/s007920050065. [DOI] [PubMed] [Google Scholar]
- 14.Labbé D, Garmon J, Lau P C K. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J Bacteriol. 1997;179:2772–2776. doi: 10.1128/jb.179.8.2772-2776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau P C K, Wang Y, Patel A, Labbé D, Bergeron H, Brousseau R, Konisshi Y, Rawlings M. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leoni L, Ciervo A, Orsi N, Visca P. Iron-regulated transcription of the pvdA gene in Pseudomonas aeroginosa: effect of Fur and PvdS on promoter activity. J Bacteriol. 1996;178:2299–2313. doi: 10.1128/jb.178.8.2299-2313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marconi A M, Beltrametti F, Bestetti G, Solinas F, Ruzzi M, Galli E, Zennaro E. Cloning and characterization of styrene catabolism genes from Pseudomonas fluorescens ST. Appl Environ Microbiol. 1996;62:121–127. doi: 10.1128/aem.62.1.121-127.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marconi A M, Ruzzi M, Iurescia S, Zennaro E. Genetics of styrene in bacteria. Rec Res Dev Microbiol. 1998;2:95–105. [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Müller C, Petruschka L, Cuypers H, Burchhardt G, Herrmann H. Carbon catabolite repression of phenol degradation in Pseudomonas putida is mediated by the inhibition of the activator protein PhlR. J Bacteriol. 1996;178:2030–2036. doi: 10.1128/jb.178.7.2030-2036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connor K, Buckley C M, Hartmans S, Dobson A D W. Possible regulatory role for nonaromatic carbon source in styrene degradation by Pseudomonas putida CA-3. Appl Environ Microbiol. 1995;61:544–548. doi: 10.1128/aem.61.2.544-548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Connor K, Duetz W, Wind B, Dobson A D W. The effect of nutrient limitation on styrene metabolism in Pseudomonas putida CA-3. Appl Environ Microbiol. 1996;62:3594–3599. doi: 10.1128/aem.62.10.3594-3599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connor K E, Dobson A D W, Hartmans S. Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl Environ Microbiol. 1997;63:4287–4291. doi: 10.1128/aem.63.11.4287-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panke S, Witholt B, Schmid A, Wubbolts M. Towards a biocatalyst for (s)-styrene oxide production: characterization of styrene degradation pathway of Pseudomonas sp. strain VLB120. Appl Environ Microbiol. 1998;64:2032–2043. doi: 10.1128/aem.64.6.2032-2043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Manniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 28.Terpe K, Kerkhoff K, Pluta E, Jendrossek D. Relationship between succinate transport and production of extracellular poly(3-hydroxybutyrate) depolymerase in Pseudomonas lemoignei. Appl Environ Microbiol. 1999;65:1703–1709. doi: 10.1128/aem.65.4.1703-1709.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utkin I B, Yakimov M M, Matveeva L N, Kozlyak E I, Rogozhin I S, Solomon Z G, Bezborodov A M. Degradation of styrene and ethylbenzene by Pseudomonas species Y2. FEMS Microbiol Lett. 1991;77:237–242. [Google Scholar]
- 30.Velasco A, Alonso S, García J L, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warhurst A M, Clarke K F, Hill R A, Holt R A, Fewson C A. Metabolism of styrene by Rhodococcus rhodochrous NCIMB 13259. Appl Environ Microbiol. 1994;60:1137–1145. doi: 10.1128/aem.60.4.1137-1145.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuste L, Canosa I, Rojo F. Carbon-source-dependent expression of the PalkB promoter from the Pseudomonas oleovorans alkane degradation pathway. J Bacteriol. 1998;180:5218–5226. doi: 10.1128/jb.180.19.5218-5226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhulin I B, Taylor B L, Dixon R. PAS domain S-boxes in Archaea, bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]