Key Points
Transactivation assays are appropriate for functional characterization of the majority of RUNX1 missense variants observed in RUNX1-FPD.
Implementation of transactivation assays for RUNX1 variants with unknown function accelerates their translation into clinical care.
Abstract
Familial platelet disorder with associated myeloid malignancies (RUNX1-familial platelet disorder [RUNX1-FPD]) is caused by heterozygous pathogenic germline variants of RUNX1. In the present study, we evaluate the applicability of transactivation assays to investigate RUNX1 variants in different regions of the protein. We studied 11 variants to independently validate transactivation assays supporting variant classification following the ClinGen Myeloid Malignancies Variant Curation Expert Panel guidelines. Variant classification is key for the translation of genetic findings. We showed that new assays need to be developed to assess C-terminal RUNX1 variants. Two variants of uncertain significance (VUS) were reclassified to likely pathogenic. Additionally, our analyses supported the (likely) pathogenic classification of 2 other variants. We demonstrated functionality of 4 VUS, but reclassification to (likely) benign was challenging and suggested the need for reevaluating current classification guidelines. Finally, clinical utility of our assays was illustrated in the context of 7 families. Our data confirmed RUNX1-FPD suspicion in 3 families with RUNX1-FPD-specific family history, whereas for 3 variants identified in RUNX1-FPD-nonspecific families, no functional defect was detected. Applying functional assays to support RUNX1 variant classification can be essential for adequate care of index patients and their relatives at risk. It facilitates translation of genetic data into personalized medicine.
Introduction
Heterozygous pathogenic germline variants of RUNX1 cause familial platelet disorder with associated myeloid malignancies (RUNX1-familial platelet disorder [RUNX1-FPD], FPDMM, FPD/AML).1,2 Patients typically show mild to moderate thrombocytopenia or even platelet counts in the low-normal range as well as qualitative platelet defects.2-7 Affected individuals face an ∼40% risk of developing hematologic malignancies.8 Germline variants of RUNX1 are classified following American College of Medical Genetics/Association for Molecular Pathology guidelines9 specified by recommendations of the ClinGen Myeloid Malignancies Variant Curation Expert Panel (MM-VCEP).8 In general, classification is based on criteria such as population, computational, segregation, and functional data. RUNX1 missense variants must frequently be classified as variants of uncertain significance (VUS) not allowing clinical conclusions because only variants considered as (likely) pathogenic confirm diagnoses. As demonstrated,10 functional analyses of VUS can contribute to their reclassification to (likely) pathogenic. Independent of patients’ phenotype and family history, transactivation assays (TAs) address RUNX1’s major function and are, therefore, suitable for first-line screening.10 In the present study, we evaluate applicability of TAs10 for different regions of RUNX1 and applied them to 11 RUNX1 variants for independent validation of their clinical utility.
Methods
We analyzed the ability of variants to activate transcription of RUNX1 target genes CSF1R, ETV1, and MYL9 by applying luciferase reporter assays in nephrogenic HEK293T and hematopoietic HEL cells.10 As controls, wild-type (WT) RUNX1b, the pathogenic variant Arg139Gln, and the benign variant Leu29Ser were included. The ability to activate transcription was scored into categories according to the MM-VCEP recommendations: (1) <20% (PS3 moderate); (2) 20% to 80%; (3) >80% to 115% (BS3 supporting); and (4) >115% (PS3 supporting) of WT activity.8 Since there are no specific instructions on combining results of multiple TAs, we followed general recommendations9,11 and concluded the applicability of the functional BS3/PS3 criterion (for details, please refer to Figure 1C). If available in the literature, we included data from secondary assays to further adapt the strength of BS3/PS3 criteria (supplemental Table 3; Figure 1C). This led to a final functional class independent of patients’ phenotype and family history. When both BS3 and PS3 or neither could be applied, variants were classified as variants of uncertain function. Finally, variants were classified following MM-VCEP recommendations including all available data.8 Variant nomenclature refers to transcript variant 2 (NM_001001890.3) encoding for RUNX1b (for RUNX1c, please refer to supplemental Table 1).
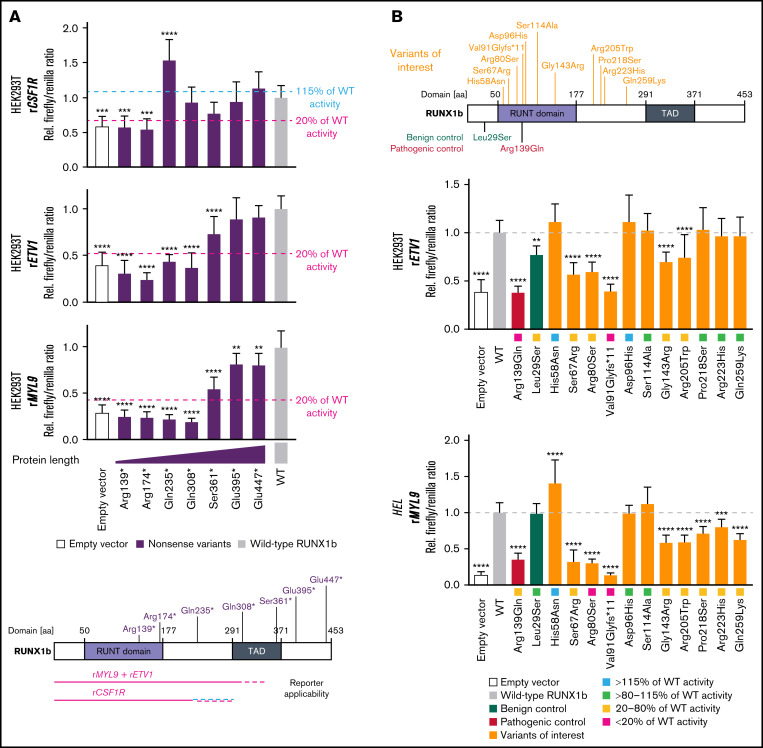
Figure 1.
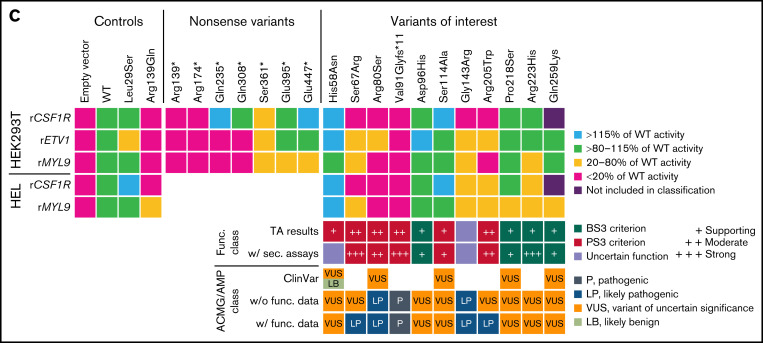
Validation of transactivation assays and characterization of RUNX1 variants of interest to support variant classification. Variant nomenclature refers to RUNX1 transcript variant 2 (NM_001001890.3) encoding for isoform RUNX1b. (A) Schematic overview of the investigated set of RUNX1b nonsense variants. Protein domains refer to Lam and Zhang.21 The bar graphs displays the firefly/renilla ratios relative to wild-type (WT) RUNX1b of applied reporter constructs rCSF1R, rETV1, and rMYL9 analyzing the set of RUNX1b nonsense variants in HEK293T cells (mean + standard deviation [SD]; 3 biological with 3 technical replicates; 1-way ANOVA in comparison with WT; Dunnett post hoc test; *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001). The threshold 115% and 20% of WT activity is highlighted regarding the MM-VCEP recommendations defining these thresholds for the application of BS3 and PS3, respectively.8 Differing results of variants for different reporters might be explained by effects of different coactivators/corepressors required. (B) Schematic overview of the analyzed set of RUNX1b variants of interest and used controls. Representative bar graphs displaying firefly/renilla ratios relative to WT RUNX1b of reporter constructs rETV1 in HEK293T cells and rMYL9 in HEL cells analyzing the set of RUNX1b variants of interest. Besides the potential impact of different coactivators/corepressors, differing results for individual reporters might be explained by effects of the cellular context (mean + SD; HEK293T: 3 biological with 6 technical replicates; HEL: 2 biological with 5 technical replicates; 1-way ANOVA in comparison with WT; Dunnett post hoc test; *P ≤ .05; **P ≤ .01; ***P ≤ .001; ****P ≤ .0001). (C) At the top is a heat map summarizing analyses of RUNX1 variants scored relative to WT RUNX1b activity. Purple results were not included in the classification because of the detection limit of rCSF1R. Below the heat map, resulting PS3/BS3 criterion based on TA results is shown as functional class and concluded as follows: (1) PS3 moderate, ≥2 TA results with <20% of WT activity without conflicting results (ie, ≥80% of WT activity); (2) PS3 supporting, ≥2 TA results >115% of WT; and (3) BS3 supporting, ≥2 TA results with >80% to 115% of WT without conflicting results (ie, ≥1 with <20% or ≥2 with >115% of WT activity). Subsequently, we included already published results from secondary assays (eg, dimerization and DNA binding, for detailed references see supplemental Table 3) to adapt the strength of the criterion according to the MM-VCEP recommendations.8 Our data provide evidence for the applicability of PS3 supporting for His58Asn, but other studies demonstrate functionality of this variant.22 No functional class was assigned (ie, variants of uncertain function) according to Brnich et al.11 At the bottom, available ClinVar classifications (not following MM-VCEP guidelines, except for the likely benign classification of His58Asn), classification based on American College of Medical Genetics/Association for Molecular Pathology (ACMG/AMP) and MM-VCEP guidelines without (w/o), and with (w/) applied functional criterion is shown. ANOVA, analysis of variance; aa, amino acid; TAD, transactivation domain.
Results and discussion
First, we aimed to validate if TAs are suitable to detect pathogenic RUNX1 variants affecting different regions of the protein. We chose a set of 7 RUNX1 (likely) pathogenic variants leading to premature termination codons. We studied them in TAs using CSF1R, ETV1, and MYL9 reporters (Figure 1A). Using ETV1 and MYL9 reporters, premature termination codon variants up to codon 308 displayed less than 20% of WT activity. The CSF1R reporter was suitable to detect pathogenic variants up to codon 235 because for Gln235*, enhanced transactivation of the CSF1R reporter was seen. In conclusion, present TAs can be applied to variants N-terminal of codon 308 (rETV1, rMYL9) or codon 235 (rCSF1R) covering most of the missense variants in RUNX1-FPD.3,12 Of note, of 31 reported missense germline RUNX1 variants classified as VUS in the RUNX1 Database, 6 are located C-terminal of codon 308.12
In the present study, we focused on 11 RUNX1 variants affecting codons before residue 308 (Figure 1B). All variants have been identified as heterozygous germline variants in patients with thrombocytopenia, myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), or as secondary finding. We observed transactivation defects for Ser67Arg, Arg80Ser, Val91Glyfs*11, Arg205Trp, and partly for Gly143Arg (Figure 1B-C). Following present classification standards,8,9 we reclassified Ser67Arg and Arg205Trp from VUS to likely pathogenic. Our data functionally confirmed the (likely) pathogenic classification of Arg80Ser and Val91Glyfs*11. Additionally, TAs demonstrated functionality of Asp96His, Pro218Ser, Arg223His, and Gln259Lys, but the lack of further benign criteria does not allow reclassification to (likely) benign.
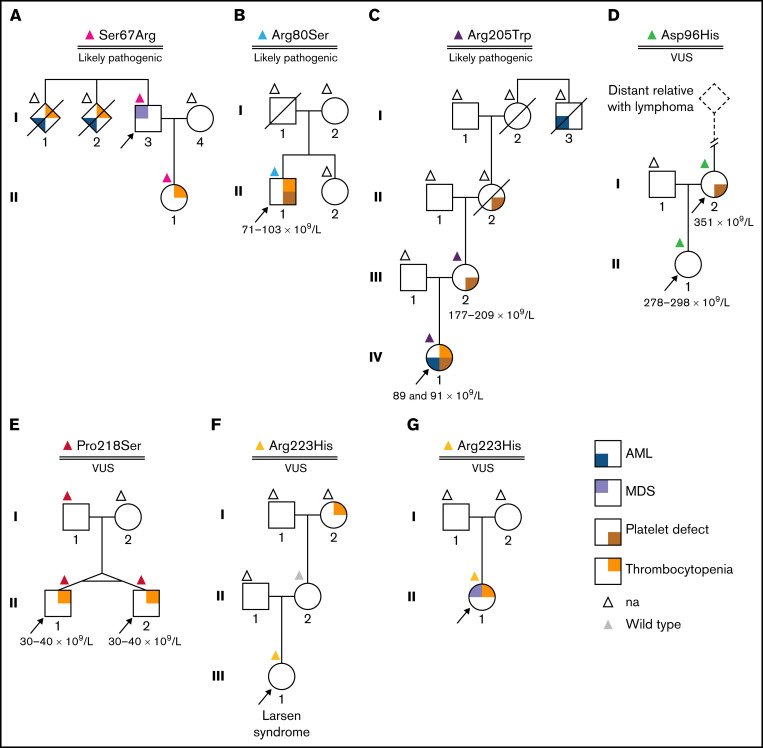
For 6 of the 11 protein variants included in our study, clinical data were available to assess the clinical utility of our functional investigations (Figure 2). Families A-C were characterized by RUNX1-FPD-typical phenotypes including AML, MDS, thrombocytopenia, and platelet defects. Our functional analyses of Ser67Arg, Arg80Ser, and Arg205Trp,13 identified in these families, revealed functional defects and led to or confirmed their likely pathogenic classification. Molecularly confirmed RUNX1-FPD allowed targeted management of index patients, predictive genetic testing, and, subsequently, surveillance of carriers.14 Surveillance permits early detection of MDS and, thus, may improve prognosis because stem cell transplantation can be timely performed.15 In family D, 2 carriers of the variant Asp96His were identified, 1 of them presenting with platelet defects. Owen et al16 have reported a classical RUNX1-FPD family also carrying this variant. It was recently reclassified to VUS following expert guidelines8 and present TAs displayed functionality. In pedigree E, Pro218Ser was identified in twins presenting with thrombocytopenia17,18 and, later, also in their father having normal platelet counts and function. This raised suspicion regarding the causality of the variant. In line with this, no experimental evidence for a functional impairment was observed. The variant Arg223His was reported to us in 2 independent families. In family F, it was identified by whole exome sequencing as a secondary finding in a girl with a complex phenotype. It was additionally reported in the index patient in family G having thrombocytopenia and MDS, but no RUNX1-FPD-specific family history. No functional impairment was observed by our TAs because all reporters showed at least 70% of WT activity. In conclusion, our functional data and the resulting variant classification confirmed RUNX1-FPD suspicion in families A-C, which was consistent with patients’ phenotypes and family histories. In families D-F, no FPD-suspicious family history was reported; from our perspective, this is a rare occasion in known RUNX1-FPD families. In line with this, we demonstrated functionality of the reported variants. Implementation of easy-to-apply TAs for missense VUS N-terminal of codon 308 is recommended and allows functional classification independent of patients’ phenotype. This improves translation of genetic data into personalized clinical care by supporting RUNX1-FPD diagnoses or denying functional impact of variants investigated.
Figure 2.
Pedigrees of investigated patients. Panels A-G display pedigrees of investigated patients. Color-coded triangles above individual symbols refer to their genotypes (NM_001001890.3). Numbers below individuals indicate platelet counts. Arrows indicate index patients. (A) Ser67Arg. Familial leukemia was suspected in a man with MDS because 2 siblings had thrombocytopenia and died of AML. Germline RUNX1:c.201C>G p.(Ser67Arg) was identified in the index, and his daughter had mild thrombocytopenia. At 49 years of age, clonal hematopoiesis was observed in the peripheral blood of the daughter (ie, DNMT3A, NM_022552.4:c.216del p.[Leu723Serfs*56], variant allele frequency [VAF] 32.5%; TP53, NM_000546.4:c.722C>A p.[Ser241Tyr], VAF 12.4%). RUNX1:c.199A>C p.(Ser67Arg) had been previously reported in the RUNX1 Database and was included in our studies. Functional data obtained were applied to classify the patient’s variant. (B) Arg80Ser. The variant was identified in a 19-year-old patient with quantitative and qualitative (ie, storage pool disease) platelet defects, psoriasis, and allergic rhinitis. No hematologic malignancy or additional relatives with platelet defects were reported. (C) Arg205Trp. The index patient had storage pool disease, thrombocytopenia, and developed AML at 26 years of age when RUNX1:c.263C>A p.(Ala88Asn) was identified as an additionally acquired somatic RUNX1 variant. Arg205Trp was inherited from III-2 having qualitative (ie, storage pool disease) but no quantitative platelet defects. Storage pool disease was also known in the maternal grandmother (II-2) whose maternal uncle (I-3) died of AML. (D) Asp96His. This variant was identified in a mother and her daughter reported minor bleeding issues while having normal platelet counts. Functional platelet tests displayed qualitative defects only in I-2. No MDS and/or AML was reported in the family. Whole-exome sequencing in II-1 did not reveal alternative candidate variants, so RUNX1-FPD was diagnosed, but following the MM-VCEP criteria publication, Asp96His was reclassified to VUS. (E) Pro218Ser. The variant was identified in monozygotic twins with thrombocytopenia. RUNX1-FPD was diagnosed.17 However, the variant was also identified in the twins’ father, who had no quantitative and no qualitative platelet defects. This raised suspicion regarding pathogenicity of the variant. To date, even after an extended thrombocytopenia panel, the genetic origin of the thrombocytopenia remains unknown. (F) Arg223His. The variant was reported as secondary finding by whole-exome sequencing in a 5-year-old girl who, following preterm birth, had developed bronchopulmonary dysplasia with subsequent chronic lung disease, hearing loss, and skeletal dysplasia with infantile scoliosis. Larsen syndrome was diagnosed. No thrombocytopenia or abnormal whole blood cell counts were seen. Functional platelet tests had not been performed. Idiopathic thrombocytopenia was reported for the maternal grandmother; however, her mother did not carry the variant. (G) Arg223His. In this family, the same variant as in family F was identified in a 12-year-old patient with refractory cytopenia in childhood. She was tested for hereditary bone marrow failure while being scheduled for hematopoietic stem cell transplantation (HSCT). na, no genetic testing.
In summary, we validated the clinical utility of our previously reported TAs10 by studying additional RUNX1 variants and discussing them in their clinical context. Limitations of TAs regarding variants C-terminal of the codon 308 have to be considered, but, in the context of RUNX1-FPD, this applies only to a minority of observed RUNX1 missense variants.3,12 Implementation of adequately performed TAs in the MM-VCEP classification scheme can be key to reclassify VUS observed in RUNX1, which is essential for patient care because only (likely) pathogenic variants molecularly confirm diagnoses. Following the present guidelines, the majority of VUS without functional impairments cannot be reclassified to (likely) benign, even if secondary assays demonstrate functionality.10 This limitation illustrates the need to consider a careful reevaluation of the present MM-VCEP guidelines. TAs support translation of genetic data into personalized clinical care, regarding both index patients and relatives at risk and, if applicable, should be integrated as first-line screening when VUS are identified. Future attempts must focus on the establishment of functional assays covering the C-terminal part of the protein and consider high-throughput assays that allow fast and accurate classification of RUNX1 variants. Because RUNX1-FPD may be underdiagnosed among patients with AML12,13,19 and germline testing of all patients with hematologic malignancies might be considered in the future,20 variant classification will become even more important to support clinical decision-making.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank the RUNX1 research program and ERN GENTURIS for referral of clinicians regarding functional investigation of RUNX1 variants and are most grateful for getting access to the Amaxa Nucleofector IIb of the research group of the Institute of Clinical Chemistry at Hannover Medical School.
The project was funded by the European Hematology Association John Goldman Clinical Research Grant 2017 (T.R.) and the Research Network MyPred (01GM1911B) funded by the German Federal Ministry of Education and Research (M.D., A.F., T.I., and T.R.).
Authorship
Contribution: T.R. designed the study; M.D. and T.R. designed and established experiments, which were performed, analyzed, and visualized by M.D. and T.R.; M.D., A.F., and T.R. performed the variant classification; Gene variants and clinical data were provided by A.B., J.C., H.H.-V., I.H., M.N., A.V., A.A., A.D., M.P.T.E., M.H.G.P.R., T.H.A.T., and N.D.; and M.D. and T.R. wrote the manuscript critically revised by all coauthors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tim Ripperger, Department of Human Genetics, Hannover Medical School (MHH), OE 6300, Carl-Neuberg-Str. 1, 30625 Hannover, Germany; e-mail: ripperger.tim@mh-hannover.de.
References
- 1.Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23(2):166-175. [DOI] [PubMed] [Google Scholar]
- 2.Schlegelberger B, Heller PG. RUNX1 deficiency (familial platelet disorder with predisposition to myeloid leukemia, FPDMM). Semin Hematol. 2017;54(2):75-80. [DOI] [PubMed] [Google Scholar]
- 3.Brown AL, Arts P, Carmichael CL, et al. RUNX1-mutated families show phenotype heterogeneity and a somatic mutation profile unique to germline predisposed AML. Blood Adv. 2020;4(6):1131-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown AL, Hahn CN, Scott HS. Secondary leukemia in patients with germline transcription factor mutations (RUNX1, GATA2, CEBPA). Blood. 2020;136(1):24-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glembotsky AC, Bluteau D, Espasandin YR, et al. Mechanisms underlying platelet function defect in a pedigree with familial platelet disorder with a predisposition to acute myelogenous leukemia: potential role for candidate RUNX1 targets. J Thromb Haemost. 2014;12(5):761-772. [DOI] [PubMed] [Google Scholar]
- 6.Rao AK, Poncz M. Defective acid hydrolase secretion in RUNX1 haplodeficiency: evidence for a global platelet secretory defect. Haemophilia. 2017;23(5):784-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latger-Cannard V, Philippe C, Bouquet A, et al. Haematological spectrum and genotype-phenotype correlations in nine unrelated families with RUNX1 mutations from the French network on inherited platelet disorders. Orphanet J Rare Dis. 2016;11(1):49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo X, Feurstein S, Mohan S, et al. ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants. Blood Adv. 2019;3(20):2962-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decker M, Lammens T, Ferster A, et al. Functional classification of RUNX1 variants in familial platelet disorder with associated myeloid malignancies. Leukemia. 2021;35(11):3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brnich SE, Abou Tayoun AN, Couch FJ, et al. ; Clinical Genome Resource Sequence Variant Interpretation Working Group . Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homan CC, King-Smith SL, Lawrence DM, et al. The RUNX1 database (RUNX1db): establishment of an expert curated RUNX1 registry and genomics database as a public resource for familial platelet disorder with myeloid malignancy. Haematologica. 2021;106(11):3004-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst MPT, Kavelaars FG, Löwenberg B, Valk PJM, Raaijmakers MHGP. RUNX1 germline variants in RUNX1-mutant AML: how frequent? Blood. 2021;137(10):1428-1431. [DOI] [PubMed] [Google Scholar]
- 14.Deuitch N, Broadbridge E, Cunningham L, Liu P. RUNX1 familial platelet disorder with associated myeloid malignancies. In: Adam MP, Ardinger HH, Pagon RA, et al., eds. GeneReviews(®). Vol. updated 2021 May 6. Seattle: University of Washington; 2021. [PubMed] [Google Scholar]
- 15.Desai AV, Perpich M, Godley LA. Clinical assessment and diagnosis of germline predisposition to hematopoietic malignancies: the University of Chicago experience. Front Pediatr. 2017;5:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owen CJ, Toze CL, Koochin A, et al. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood. 2008;112(12):4639-4645. [DOI] [PubMed] [Google Scholar]
- 17.Antony-Debré I, Duployez N, Bucci M, et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia. 2016;30(4):999-1002. [DOI] [PubMed] [Google Scholar]
- 18.Bluteau O, Sebert M, Leblanc T, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018; 131(7):717-732. [DOI] [PubMed] [Google Scholar]
- 19.Simon L, Spinella JF, Yao CY, et al. High frequency of germline RUNX1 mutations in patients with RUNX1-mutated AML. Blood. 2020;135(21):1882-1886. [DOI] [PubMed] [Google Scholar]
- 20.Feurstein S, Drazer M, Godley LA. Germline predisposition to hematopoietic malignancies. Hum Mol Genet. 2021;30(20):R225-R235. [DOI] [PubMed] [Google Scholar]
- 21.Lam K, Zhang DE. RUNX1 and RUNX1-ETO: roles in hematopoiesis and leukemogenesis. Front Biosci. 2012;17(1):1120-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koh CP, Wang CQ, Ng CE, et al. RUNX1 meets MLL: epigenetic regulation of hematopoiesis by two leukemia genes. Leukemia. 2013;27(9):1793-1802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.