SUMMARY
ArcAB, also known as the Arc system, is a member of the two-component system family of bacterial transcriptional regulators and is composed of sensor kinase ArcB and response regulator ArcA. In this review, we describe the structure and function of these proteins and assess the state of the literature regarding ArcAB as a sensor of oxygen consumption. The bacterial quinone pool is the primary modulator of ArcAB activity, but questions remain for how this regulation occurs. This review highlights the role of quinones and their oxidation state in activating and deactivating ArcB and compares competing models of the regulatory mechanism. The cellular processes linked to ArcAB regulation of central metabolic pathways and potential interactions of the Arc system with other regulatory systems are also reviewed. Recent evidence for the function of ArcAB under aerobic conditions is challenging the long-standing characterization of this system as strictly an anaerobic global regulator, and the support for additional ArcAB functionality in this context is explored. Lastly, ArcAB-controlled cellular processes with relevance to infection are assessed.
KEYWORDS: facultative anaerobes, global regulatory networks, metabolic regulation, metabolism, two-component regulatory systems
INTRODUCTION
The ability to regulate and optimize metabolism based on environmental conditions is a hallmark of life. The metabolic capacity of bacteria is extremely diverse and for facultative anaerobes can include mechanisms for aerobic respiration, anaerobic respiration, and fermentation to generate energy in the form of ATP (1). Accordingly, many genes encoding metabolic functions are transcriptionally regulated by multiple systems to optimize metabolism depending on the substrates and electron acceptors available (2).
The average bacterial species encodes 30 two-component regulatory systems, but extreme examples have been described with more than 200 systems encoded in a single genome (3, 4). A canonical two-component system consists of a sensor kinase and a response regulator. Upon recognizing a specific stimulus through a sensor domain or by interaction with an adaptor molecule, the sensor kinase phosphorylates and thus activates the response regulator. Once activated, the response regulator can bind DNA, RNA, or other proteins, or act as an enzyme itself depending on the system (5). Hundreds of two-component regulatory systems have been characterized and regulate a multitude of processes, including pathogenesis, stress responses, and symbiotic interactions (6, 7). Despite substantial advancements in understanding bacterial two-component regulatory systems, there is still much to learn about specific systems and the processes they regulate.
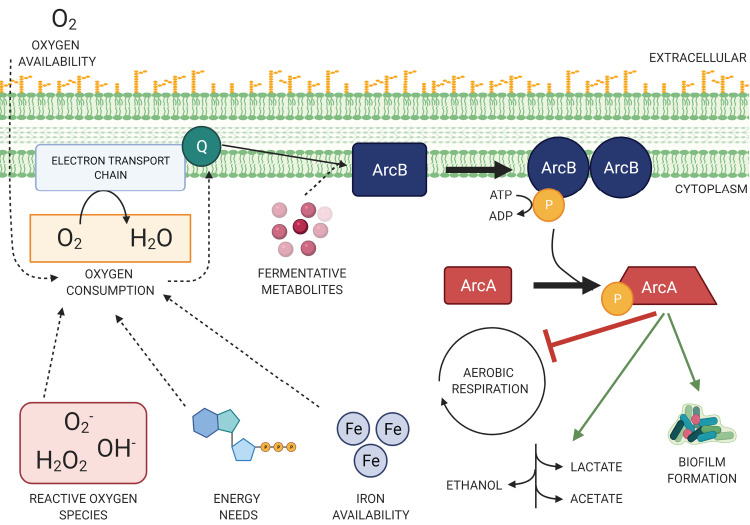
The anoxic redox control (or aerobic respiration control) (Arc) two-component regulatory system senses the modulation of oxygen availability for use as an electron receptor (8). The Arc system is found in facultatively anaerobic bacteria that can switch from utilizing aerobic respiration to fermentation or anaerobic respiration when oxygen is not being consumed. During fermentation and anaerobic respiration, bacteria continue to utilize glycolysis, but intermediate metabolites are shuttled to different pathways depending on the availability of alternative electron acceptors. The Arc system is involved in mediating the switch to fermentation and was touted early for its potential for global control of gene expression (9, 10). The sensor kinase ArcB is typically described as sensing microaerobic and anaerobic conditions, and after autophosphorylation, ArcB transphosphorylates the response regulator ArcA (11–13). Phosphorylated ArcA promotes fermentation as a primary energy-generating pathway by mainly repressing pathways associated with aerobic respiration. The active form of ArcA, often denoted as P-ArcA, is a cytosolic transcription factor and is projected to regulate over 1,100 genes directly or indirectly in Escherichia coli (Table 1) (14). Together, these proteins are referred to as the ArcAB or ArcBA system (Fig. 1). In an analysis of 698 bacterial species, >110 orthologs of ArcA and >130 orthologs of ArcB have been identified (15). Genomes have also been identified in which only ArcA or ArcB are encoded, but the significance of this finding remains unclear (15).
TABLE 1.
Studies defining the ArcA regulon
| Study | Species | Medium/media | Oxygenation | Method(s) | Results |
|---|---|---|---|---|---|
| Iyer 2021 (38) | Escherichia coli | M9 minimal medium + glucose | Anaerobic | RNA sequencing | 119 genes downregulated, 61 genes upregulated |
| Federowicz 2014 (39) | Escherichia coli | M9 minimal medium + glucose + ammonium chloride | Fermentative and nitrate respiratory conditions | ChIP-chip | Fermentative: 47/21operons repressed/activated; nitrate respiration: 67/47 operons repressed/activated |
| Park 2013 (36) | Escherichia coli | MOPS minimal medium + glucose | Anaerobic | ChIP-chip, ChIP-seq, DNase I footprinting assay | Directly repress 74 operons, directly activate 11 operons, 229 operons differentially expressed, 176 chromosomal binding regions |
| Morales 2013 (199) | Salmonella Typhimurium | LB | Aerobic | Microarrays | Aerobic conditions: 220/122 genes upregulated/downregulated; H2O2: 117/175 genes upregulated/downregulated |
| Yun 2012 (229) | Mannheimia succiniciproducens | TSB | 10% CO2 | Microarrays | 82 genes upregulated, 79 genes downregulated |
| Evans 2011 (95) | Salmonella Typhimurium | MOPS-buffered LB + xylose | Anaerobic | Microarrays | 147 genes upregulated, 245 genes downregulated |
| Gao 2010 (230) | Shewanella oneidensis | In silico | NA | Regulatory Sequence Analysis Tools (RSAT) | 214 genes with ArcA-binding motif |
| Wong 2007 (200) | Haemophilus influenzae | Brain heart infusion + NAD + hemin | Anaerobic | Microarrays | 23/1,697 genes differentially regulated |
| Salmon 2005 (14) | Escherichia coli | MOPS medium + glucose | Aerobic and anaerobic | Microarrays | 1,139 genes predicted to be directly/indirectly regulated |
| Liu 2004 (90) | Escherichia coli | MOPS-buffered LB + xylose | Anaerobic | Microarrays, footprinting | ~85 operons in ArcA regulon |
FIG 1.
Working model of ArcAB in detecting oxygen consumption for redox maintenance. The two-component regulatory system ArcAB responds to changes in oxygen consumption within the bacterial cell. Oxygen consumption can be influenced by a multitude of factors, including oxygen availability and the energetic needs of the cell. As activity at the electron transport chain decreases, quinones (Q) interact with sensor kinase ArcB, causing a conformational change and homodimerization. Now active, ArcB autophosphorylates and transphosphorylates response regulator ArcA via a phosphoryl relay. In turn, phosphorylated ArcA multimerizes and serves as a global transcription factor suppressing aerobic metabolic pathways and promoting fermentation among other processes.
ArcAB has been the subject of extensive research over the past 35 years. Most of this work was conducted in E. coli and has uncovered the structure and function of the Arc system (16). Since ArcB is often referred to as a sensor of anaerobic and microaerobic conditions, the system is implied to be primarily relevant to bacteria when encountering low oxygen conditions. An increasing number of recent studies, however, have reported phenotypes for ArcAB mutants under higher-oxygen conditions, establishing that ArcAB is responsive to more than oxygen availability alone (17, 18). In strains in which the genes encoding terminal oxidases are removed, ArcA is activated in aerobic conditions due to the inability of the cell to utilize oxygen as a terminal electron acceptor (19, 20). Accordingly, ArcB has been more accurately described as sensing the oxygen consumption rate or the degree to which bacteria acquire and utilize oxygen, rather than simply dissolved oxygen concentrations (21). Oxygen consumption may decrease even when abundant oxygen is available, so this distinction is important when considering the context of ArcA regulation. ArcAB has been described as not only a sensor of oxygen consumption but also as a general redox sensor (22). Metabolic activity and redox regulation are intimately linked processes, making these various descriptions of ArcAB compatible (23). Many studies included in this review report functions of ArcAB in microaerobic and anaerobic conditions, but these findings may apply to other instances where oxygen consumption is also affected. ArcAB functions in conjunction with multiple other transcriptional regulators of metabolism, especially the fumarate and nitrate reductase regulator (FNR) (24, 25). Since they are often studied together, the relationship between ArcAB and FNR will specifically be reviewed here.
An exciting area of current research aims to determine how ArcAB is activated and deactivated. Some groups report that the system is controlled by the cellular pool of quinones, while others suggest different modulators, including fermentation products (26–30). These competing hypotheses indicate that ArcAB is at the intersection of multiple signaling pathways and underscores again that this system may be active in multiple conditions, including aerobiosis. Because of its extensive regulatory capabilities, ArcAB systems have been linked to a wide array of cellular processes beyond central metabolism in recent years, including bacterial conjugation, acid tolerance, biofilm formation, and even bioluminescence (31–35). ArcAB may govern the elegant coordination of these processes with the appropriate metabolic pathways to provide the energy and resources cells require to perform them (36–39). The significance of ArcAB regulation is perhaps especially evident during infection, an instance where pathogens must balance metabolic needs with survival in the host environment. Over the last 35 years since the discovery of ArcAB in E. coli, much progress has been made in understanding this complex regulatory system in multiple bacterial species. Here, we explore the seminal discoveries made along the way and highlight active and future areas of ArcAB research.
ORGANIZATION OF THE Arc SYSTEM
Expression of arcB and arcA
E. coli cells have approximately 1,088 to 8,000 molecules of ArcA and 85 to 226 copies of ArcB in each cell, depending on media and experimental techniques (28, 40–42). The two genes encoding these proteins are not found together in the same operon in contrast with many other two-component systems (7, 43, 44). Their expression and regulation must therefore be considered independently. The arcB gene encoding the sensor kinase was initially thought to be expressed constitutively and not influenced by respiration (45). It is intuitive that a basal level of arcB expression is needed to ensure that at least some amount of sensor is present and responsive to stimulus (7). More recently, however, arcB transcription was shown to be increased at lower oxygen concentrations (3.3-fold higher at 1% O2 versus 10% O2 in the headspace of cultures), and ArcA itself may be a regulator of arcB transcription (46). Expression of arcB is also known to be affected after treatment with antibiotics including gentamicin and polymyxin B, suggesting that cell envelope stress influences ArcAB activity (41, 47). The small regulatory RNA ArcZ can destabilize arcB mRNA in aerobic conditions, which demonstrates posttranscriptional control of arcB is also possible and provides a potential mechanism by which ArcAB activity is limited (48).
The arcA gene is under the control of FNR, as well as ArcA itself, and is upregulated about 4-fold once E. coli encounters increasingly anaerobic levels (46, 49, 50). FNR and ArcAB respond to oxygen availability and consumption, respectively (21, 51, 52). Regulation of arcA by FNR thus demonstrates a coordinated response to related stimuli. Interestingly, a study simultaneously analyzing the regulons of ArcA, FNR, and IHF (integration host factor) found that arcA expression was upregulated in the ihf mutant but not the fnr mutant, a contradiction suggesting that additional levels of regulation are likely present (38). Expression of arcA is also upregulated in acidic relative to neutral microaerobic conditions, which is further indicative of ArcAB’s role as responding to redox changes (53). The degree to which changes in ArcA abundance affect activity remains an open question since contrary evidence has shown that ArcA protein levels do not change between aerobic and anaerobic conditions (28, 54). Groisman describes in a review of mechanisms of two-component system regulation the potential impact of length of stimulus exposure on response regulators’ autoregulatory abilities (7). It is unknown whether ArcA autoregulation is also temporally dependent, but this type of mechanism could explain the difference here. Although regulation of the expression of ArcA may influence overall activity, the primary determinant of ArcA regulatory activity is likely its phosphorylation state.
Structure of ArcB
ArcB’s structure is atypically configured compared to canonical sensor kinases (Fig. 2) (11). It has two transmembrane helices that function solely as an anchor into the membrane (55, 56). The intramembrane domains are connected by a conspicuously short periplasmic domain of only 16 amino acids (57). Many sensor kinases that are directly involved in interacting with external stimuli have more complex periplasmic domains (58). The periplasmic bridge of ArcB’s two intramembrane domains has been shown not to be involved in signaling, providing evidence that ArcB requires interactions with adaptor molecules to ultimately sense stimuli (55). Canonical sensor kinases have a single domain with an invariant histidine residue for autophosphorylation whereas ArcB is a tripartite sensor kinase, meaning it has two additional domains (59–63). These domains have an additional histidine residue and an aspartate residue to participate in the phosphorylation process. Early studies examining the structure and phosphorylation of ArcB were indeed confounded by three domains participating in the enzymatic reaction (29, 45, 64, 65). The catalytic region of the protein was subsequently identified to be composed of a primary transmitter domain, a central receiver domain, and a secondary transmitter (or phosphotransfer) domain (66). A single residue in each of these domains was later determined to play a role in phosphorylation and were identified as His292, Asp576, and His717, respectively (12, 29). The membrane anchor domain of ArcB is connected to these three catalytic domains by a linker slightly less than 200 amino acids in length. Within the linker are a leucine zipper, which is necessary for ArcB homodimer formation, and a Per-Arnt-Sim (PAS) domain, which is important for signal transduction (67–70). Within the PAS domain are two redox-sensitive cysteine residues (Cys180 and Cys241), which are in proximity with the cysteine residues of a second ArcB protein (57, 67). The oxidation state of these residues determines whether ArcB functions as a kinase or phosphatase (57). Two intermolecular disulfide bridges form between adjacent ArcB proteins when the cysteines are oxidized, preventing kinase activity. In contrast, the ArcB proteins can homodimerize to facilitate kinase function when the cysteine residues in the PAS domain are reduced and disulfide bridges are absent. Interestingly, the PAS domain is not present in the ArcB of Haemophilus influenzae, suggesting that activation and deactivation of this homolog is accomplished via a different mechanism than E. coli (71).
FIG 2.
Schematic of ArcA and ArcB domains. Sensor kinase ArcB is composed of two transmembrane domains connected by a short periplasmic domain. This anchor to the membrane is connected to the catalytic region of the protein by a linker. Within this linker are domains involved in interaction with regulators of ArcB and homodimerization. Once active, the atypical tripartite kinase region of ArcB can transphosphorylate ArcA. ArcA is composed of a receiver domain that once phosphorylated causes a conformational change in the helix-turn-helix domain. This change allows the second domain to bind DNA elements for ArcA-mediated transcriptional regulation.
Structure of ArcA
ArcA is configured in a manner characteristic of the OmpR/PhoB superfamily of two-component system response regulators (reviewed by Nguyen et al. [72]) (Fig. 2). At the N terminus of ArcA is a receiver domain (also referred to as a regulatory domain) where the protein is phosphorylated and dephosphorylated by ArcB at an aspartate residue (Asp56) (9, 29, 64). The receiver domain is connected to the C-terminal output or effector domain consisting of a helix-turn-helix (HTH) for DNA binding (72, 73). Phosphorylation of the ArcA receiver domain results in a conformational change that allows the effector domain to bind DNA targets (74). Connecting the receiver and effector domains is a short linker of unknown function (72). The output domain can further be described as a winged HTH domain as with other OmpR-like regulators (75–77). The wings refer to additional small β sheets that can impact the DNA binding properties of the response regulator. The HTH motif subtype of the OmpR superfamily is further identifiable by a characteristic string of four additional β strands, forming an antiparallel β sheet, in front of the prototypical winged HTH. Variation in residues contained within these β strands is proposed to be instrumental in determining which DNA sequences the domain can bind (72, 78). Techniques have thus been developed to predict sequence binding by these transcription factors based on residue differences (72, 78).
ArcB-to-ArcA Phosphorelay
The passage of phosphoryl groups from ArcB to ArcA, known as a phosphorelay, results in the activation of ArcA and has been well characterized (12, 13, 61, 64). Under reducing conditions and subsequent homodimerization, ArcB autophosphorylates via an intramolecular reaction with ATP as the phospho-donor (79). In a study comparing 25 histidine kinases, a population of ArcB was able to autophosphorylate most quickly (44). Relative to the other kinases, ArcB had a considerably lower level of saturation, referring to the percentage of the ArcB population that ultimately is phosphorylated. Yamamoto et al. suggested rapid autophosphorylation coupled with a low level of saturation implies ArcB is also quickly dephosphorylated. Following autophosphorylation, ArcB transphosphorylates ArcA via a ArcB1His292 → ArcB1Asp576 → ArcB2His717 → ArcAAsp54 phosphorelay, ending in P-ArcA activation (13, 80). Early findings suggested His292 of ArcB could directly phosphorylate Asp54 of ArcA, bypassing the other catalytic domains, but no evidence was found later to support this transfer (12, 13). The molecular mechanism of this phosphorelay, including the uncovering of intramolecular versus intermolecular interactions of the homodimers, was described by Teran-Melo et al. (80). Under conditions that activate ArcB kinase activity, the phosphoryl group is initially passed from the primary transmitter domain (His292) to the central receiver domain (Asp576) of the same ArcB molecule. Then, the phosphoryl group is transferred to the phosphotransfer domain of another proximal ArcB molecule (His717), which further illustrates the importance of homodimerization. From here, the phosphoryl group is passed to the receiver domain of ArcA (Asp56). In some cases the phosphotransfer to the receiver domain of ArcA has been described as a dynamic process requiring catalytic contributions from ArcA (81). ArcA can in fact be autophosphorylated in the presence carbamoyl phosphate and acetyl phosphate, but this connection has not been found to be physiologically relevant (82–84).
DNA Binding by ArcA
Initial studies of ArcA-DNA complexes at the promoters of ArcA-controlled genes predicted that ArcA binds to multiple sites or that ArcA multimerizes following phosphorylation (82, 85). The ability of ArcA to multimerize has since been shown to be dependent on both the receiver domain and DNA binding domain and may be a requirement before proper DNA binding (73, 86). The ArcA dimer ultimately forms at the α4-β5-α5 faces within the DNA-binding domain with α referring to alpha helixes and β referring to beta strands (72, 73). Multiple studies have provided additional evidence that ArcA forms dimers, but it is possible that higher orders of oligomerization also exist (72, 86). Only phosphorylated forms of ArcA are thought to form higher order oligomers (73, 87). Other evidence suggests that nonphosphorylated ArcA can form dimers and bind regulatory regions, though this has been attributed to nonspecific binding by some groups (86, 87). Regulation is dependent on phosphorylated ArcA binding to multiple repeats in the promoter of target genes, but it has not yet been fully determined to what extent dimers of P-ArcA interact with one another at these direct repeat binding sites (54). P-ArcA has been shown to bind the –35 and –10 elements of promoters, at the transcription start site itself, and even at loci almost 500 nucleotides upstream, depending on the target gene being regulated (36, 39). The consensus ArcA binding box, the DNA motif to which activated ArcA binds, has been extensively studied (36, 39, 74, 88–95). The prototypical ArcA consensus binding motif is approximately 15 bp in length and is highly conserved. The most common consensus sequence 5′–3′ GTTAATTAAATGTTA has been identified in multiple species including E. coli, Salmonella enterica, and Shewanella oneidensis (91–93, 95). Within this sequence are two direct repeats (GTTA). Notably, the oscillation of the motif (11 bp) aligns well with the length of helical turns of DNA (10.5 bp) (39, 78). Park et al. noted that binding sequences longer than 15 nucleotides have been identified with more advanced techniques and further described an extended 18-bp consensus sequence in E. coli (36). Analysis from this study showed that sites bound by ArcA contain up to five direct repeats with various lengths of space between each repeat. The amount of phosphorylated ArcA at each repeat can be variable and appears to be related to the number of repeats present (36). Bidirectional transcriptional regulation by ArcA has also been reported and can result in dual activation/repression or inverse regulation of opposing operons sharing a promoter region (39).
Dephosphorylation of ArcAB
The ability of cells to deactivate regulatory systems is as important as the mechanisms required to turn them on. During signal decay, ArcB kinase activity ceases, and ArcB in turn acts as a phosphatase to directly dephosphorylate ArcA (29). P-ArcA is dephosphorylated via an ArcAAsp56 → ArcB1His717 → ArcB1Asp576 → Pi intermolecular phosphorelay in which the domains of only a single ArcB protein are involved (96, 97). The phosphoryl group is transferred from the ArcA receiver domain (Asp56) back to the phosphotransfer domain (His717) of ArcB during this process. From here, the signal is passed to the central receiver domain (Asp576) of the same ArcB molecule before finally being released as inorganic phosphate. Following cessation of the stimulus, ArcB likely dephosphorylates due to the instability of the phosphoryl group bound at the aspartate residue in the central receiver domain (Asp576) (12, 97). Evidence has been described that ArcB can also be dephosphorylated by SixA during this process (98–100). SixA, a phosphohistidine phosphatase, is one of the first proteins described to have this enzymatic ability. However, a recent study challenges the connection of SixA to the ArcAB system, demonstrating that deletion of sixA did not impact a P-ArcA reporter assay (101). Pending further studies, the role of SixA in the dephosphorylation of ArcB remains open but highlights the potential for modulation of ArcAB by additional regulatory components. If SixA can dephosphorylate ArcB, more research will be needed to determine whether SixA activity is condition dependent and if it works in tandem with spontaneous release of the phosphoryl group. It is unlikely that ArcB-independent spontaneous hydrolysis of the phosphoryl group from ArcA contributes significantly to the decay pathway as the half-life of phosphorylated ArcA is long (30 min to longer than 1 h) (44, 86, 96).
REGULATION BY QUINONE POOLS
The nature of the signal for the Arc system has been a consistent focus of the field since the initial identification and characterization of the protein components themselves. arcA and arcB were identified in an E. coli genetic screen performed under anaerobic conditions for the purpose of identifying mutants that upregulate sdh, an operon that is normally repressed in the absence of oxygen (9, 10). The genes subsequently determined to comprise the ArcA regulon suggested that ArcAB was involved in regulating metabolism during anaerobiosis (11). arcA was also determined to be the same gene previously identified as dye, so named because loss of dye/arcA resulted in increased sensitivity to toluidine blue (9, 102). Toluidine blue induces photosensitizer-mediated oxidative stress and further served as an indication that ArcAB is linked to redox control or metabolism (9, 10, 103, 104). The short length of the periplasmic domain of ArcB and lack of potential redox reaction sites in this region suggested that the sensor kinase does not directly interact with a molecular signal, such as oxygen (57). Molecular oxygen was subsequently discounted as the signal because ArcA activity was shown to also decrease when alternative electron acceptors are present (9). Fermentation products were found to enhance ArcB kinase activity and inhibit phosphatase activity in anaerobic conditions, which supported ArcAB as a two-component system for anaerobiosis (28, 29, 105, 106). In this model, fermentative metabolites such as d-lactate, acetate, and pyruvate are the dominating influence on ArcB activity and thus serve as the signal for the system. Initial studies reported inhibition of ArcB phosphatase by such metabolites as an important mechanism by which ArcB transphosphorylates ArcA following autophosphorylation in anaerobic conditions (29). d-Lactate was also found to amplify ArcB kinase activity but ultimately was not alone sufficient for activation, suggesting other layers of regulation (106). Along these lines, Georgellis et al. highlighted that the genuine signal needs to be able to suppress ArcB kinase under oxidizing conditions (30). While fermentative metabolites may enhance ArcB kinase activity, the absence of these metabolites under most oxidizing conditions likely disqualifies them from being the primary contributors to regulation.
Identification of Quinone Involvement
The observed derepression of an ArcA-regulated promoter during anaerobic respiration indicated that a reduced component of the electron transport chain such as quinol may be the stimulus for ArcB (107). Quinones are comprised of a hydrophobic isoprenoid tail attached to a polar head group with a characteristic cyclic dione moiety (108). Quinones were considered good candidates for modulating ArcB since they are intricately linked to the redox state of the cell and located at the membrane. Indeed, the connection between quinones and ArcAB was firmly established in the early 2000s following a key set of experiments (30, 57). Radiolabeled ATP was used to track ArcB autophosphorylation, which was shown to be inhibited in the presence of soluble analogs of ubiquinone and menaquinone (30). When the quinones were reduced in the presence of hydrosulfite, they no longer inhibited ArcB kinase activity. Conclusively, oxidized forms of quinones were shown to repress ArcB autophosphorylation. These results ushered in a new focus of ArcAB research in which different quinone species were tested for their capability of interacting with ArcB.
Overview of Quinone Structure and Role
A combination of ubiquinone (UQ), menaquinone (MK), and demethylmenaquinone (DMK) are typically found in Gram-negative facultative anaerobes (109). UQ is classified as a benzoquinone and MK and DMK are classified as naphthoquinones based on their variable head groups (110, 111). UQ is the dominant quinone during aerobiosis, while MK and DMK are the major quinones during fermentation and anaerobic respiration. Each type interacts with different enzymes to modulate aerobic and anaerobic respiratory pathways (112, 113). The structure and function of quinones aligns well with the role of these molecules as excellent candidate transmitters of the cellular redox state to ArcB. The small size of quinones in general and overall hydrophobic nature allow for movement within the cellular membrane; the hydrophilic ringed head group can then interact with proteins imbedded within the membrane (108). As part of the electron transport chain (ETC), quinones accept both electrons and protons but only donate electrons. Quinones receive electrons from dehydrogenases specific to substrates such as NADH and succinate (114). The electrons are then shuttled to various reductases depending on the quinone and the electron acceptor. For example, UQ passes electrons to cytochrome bo3 which reduces oxygen to water and MK can pass electrons to fumarate via fumarate reductase (114–116). The quinone pool is an important component in the ETC, which supports the notion that quinones rapidly reflect the redox conditions within the cell. In turn, quinones can interact with ArcB, resulting in a response to changes in metabolic activity. Notably, ArcA downregulates expression of the nuo and sdh operons, which encode the dehydrogenases at the beginning of complexes I and II, respectively (36, 38). As complexes I and II feed directly into the quinone pool, ArcA regulation of these complexes serves as example of a feedback loop linking metabolic activity with maintenance of the ETC, a concept that warrants further interrogation.
When oxygen is present, the quinone pool is oxidized as electrons are shuttled through to terminal oxidoreductases (oxidases), ultimately reducing oxygen to water (114). Oxidized quinones function as a negative signal and prevent ArcB kinase activity under aerobic conditions (30). The two aforementioned redox-sensitive cysteines in the linker region of ArcB are the site of quinone-mediated regulation (57). The redox state of the quinone pool becomes reduced when electron acceptors are absent and cannot complete the ETC. Under anaerobic conditions, reduced quinones (quinols) reduce the cysteines of the PAS domain, resulting in the breakage of disulfide bonds between ArcB homodimers. When oxygen is not being utilized as an electron acceptor, ArcB’s kinase is “on,” resulting in autophosphorylation and subsequent transphosphorylation of ArcA. In contrast, oxidized quinones turn “off” the ArcAB system as electrons are transferred from the cysteine residues to the quinones. The pool of oxidized quinones is theoretically maintained under conditions where electrons are continuously transferred to oxygen (30). The maintenance of the disulfide bonds between the ArcB cysteines silences kinase activity (57). The position of the cysteines in relation to the inner membrane of the cell ensures proximity to quinones and further supports the working model of the adaptor molecule of ArcB activation being kept in or close to the inner membrane (30, 117). Quinones are also hypothesized to be reduced under conditions in which an abundance of carbon sources results in flooding of the ETC with electrons, which may be an avenue by which ArcAB becomes activated in aerobic conditions (87). Overall, the proposed regulation of ArcAB by the quinone pool couples ETC activity with transcriptional regulation and provides a mechanism by which ArcAB receives and transmits respiratory activity by an organism.
Modeling Quinone-Based Regulation of ArcAB
Although a compelling link has been established between the redox state of quinones and the activity of ArcB, there are discrepancies regarding which subsets in the quinone pool regulate the sensor kinase and when they do so (26, 27, 118). Quinones are difficult to isolate in the laboratory due to their hydrophobic nature, and in instances where they can be isolated from bacterial cells, maintaining the native oxidation state ex vivo has proven to be challenging (27, 119, 120). Characterizing the overall quinone profile of cells cultured in different conditions is therefore not trivial. Many studies rely on the analysis of mutants lacking specific species of quinones (26, 27, 118). These studies utilizing quinone mutants are complicated by the fact that different species of quinones share precursor molecules and biosynthesis pathway components. DMK for example is a direct precursor for MK, making it difficult to separate the roles of these quinone molecules (118). Furthermore, UbiE, the enzyme catalyzing the conversion of DMK to MK, is also required in the UQ synthesis pathway (27, 121). When studying mutant strains lacking specific quinones, the possibility of redundancy and compensation should be considered. E. coli mutants lacking ubiquinone have been shown to have higher levels of menaquinone and demethylmenaquinone for example (113). Functional substitutions by these quinones could impact how the different species interact with ArcB in mutant strains. There is also the concern that mutation of synthesis genes (ex. ubiE) to engineer strains lacking specific quinones may result in the accumulation of quinone intermediates (27). The ability of these intermediates to interact with ArcB is unknown. Despite these experimental considerations, two prevailing models regarding regulation of the Arc system by the quinone pool have been developed.
Model A
Model A (Fig. 3) is based on comparing reduction potentials (E°red) of the quinone species with ArcB (26). The E°red values for UQ, MK, and DMK are approximately +110, –80, and +36 mV, respectively (116). Based on its cysteine residues, the midpoint potential of ArcB was calculated to be –41 mV (26). MK is the only quinone of the three species to have a more negative reduction potential than ArcB. When menaquinol (reduced state) and ArcB interact, electrons flow from menaquinol to ArcB, reducing the ArcB cysteine residues and activating kinase activity. MK is the dominant species in anaerobic conditions, so the redox coupling of MK and ArcB would provide a mechanism for how ArcB can be activated in the absence of oxygen consumption. Conversely, when ubiquinone (oxidized state) and ArcB are in proximity, electrons flow from ArcB to ubiquinone since the E°red of ubiquinone is more positive (26). The result of such an interaction would be an oxidation of ArcB and silencing of kinase activity. UQ is the most abundant quinone under aerobic conditions, and it is expected that the UQ pools leads to repression of ArcAB function as the cell utilizes oxygen (112). This model therefore relies on the abundance of reduced MK relative to oxidized UQ for ArcAB to be active in anaerobic conditions. DMK can also be reduced by ArcB based on reduction potentials and thereby inactivate ArcB kinase activity. At first glance, this interaction appears counterintuitive since DMK is associated with anaerobic conditions in which ArcB should be active. In cells transitioning into anaerobic conditions in minimal media, the DMK pool increases more rapidly compared to the MK pool (120). One may hypothesize that DMK serves as a direct counter to MK and that this may be important in fine tuning ArcB activity in microaerobic conditions where ArcB could be partially active and UQ is not as abundant (26). Such a relationship is consistent with the notion that the overall enzymatic activity of ArcB as a kinase or phosphatase functions on a continuum. The activity of the ArcAB system can therefore not be simplified to function as an on/off switch, especially in intermediate conditions. The shift from UQ to MK in the quinone pool during anaerobiosis and the subsequent effect on ArcAB activity was shown previously (19). The same change in relative abundance of quinone species and activation of the Arc system was also seen in aerobic conditions in E. coli strains lacking terminal oxidases, providing more evidence that it is the utilization rather than the presence of oxygen that impacts the quinone profile of the cell and its interaction with ArcB (19). In support of this model is a study that measured ArcA activity as a readout of quinone modulation using an expression reporter system and included promoters for which ArcA acts as either an activator (cydA) or a repressor (sdh) (26). ArcA phosphorylation levels are not measured directly in these experiments, so there is a possibility of other regulators interfering with the reporter expression. An ArcA-P specific promoter system edited to remove binding sites of coregulator FNR was not used in this case but may be useful in addressing these concerns (119).
FIG 3.
Quinone regulation of ArcAB. Activity of the sensor kinase ArcB is controlled by redox sensitive cysteine residues within its linker domain. Quinones are a major determinant of the oxidation state of the cysteine residues. In reducing conditions (e.g., anaerobic culture), quinones reduces these residues, allowing for ArcB homodimerization. ArcB then functions as a kinase, and a phosphorelay ends with phosphorylation of the phosphotransfer domain of the partner ArcB molecule. In oxidizing conditions (e.g., aerobic culture), quinones oxidize the cysteines, which form disulfide bonds with the corresponding cysteines of a neighboring ArcB molecule. This formation silences ArcB kinase activity and phosphorylation does not occur. Ubiquinone, menaquinone, and demethylmenaquinone can activate or inactivate ArcB kinase activity depending on their own oxidation state and abundance. See the text for further details. Key: C-S-H, reduced cysteine residue; C-S-S-C, disulfide bond; L, linker domain; H1, primary transmitter domain; D, central receiver domain; H2, phosphotransfer domain. This model has been adapted from Alvarez et al. (26) and Bekker et al. (119) with permission.
Model B
The next model (Fig. 3) operates under the hypothesis that any quinone species can reduce or be reduced by ArcB (27). In this model, the redox reaction is dependent on the overall redox potential of the quinone and ArcB complex rather than just the midpoint potential of the individual cysteines of the ArcB linker region (27). The first study reporting that ArcB autophosphorylation is blocked by quinones demonstrated this relationship with both ubiquinone-0 and menadione (30). Multiple quinone species simultaneously contributing to the redox state of ArcB could help explain findings that there is not a linear correlation between oxygen availability and ArcA activity (21, 22, 119, 122). In utilizing single quinone mutants, van Beilen and Hellingwerf demonstrated that ArcA can still be phosphorylated and dephosphorylated in response to aerobic transitions, referred to as the “ArcB activation/deactivation cycle” (27, 118). A strength of the latest study is the use of a nitrogen gas sparging system to capture quinone composition of E. coli cells during the transition from aerobic to anaerobic conditions (27, 118, 119). The timing of ArcA phosphorylation here clearly correlates with the changes in the quinone profile of wild-type E. coli. While the study demonstrates that three mutant strains, each employing a different quinone, can still phosphorylate ArcA during changes in oxygen availability, the rate at which the different oxidized quinone species activate ArcB has not yet been determined. This model is therefore contingent on the overall amount of the quinone pool with the possibility that individual quinone species may differ in magnitude of effect on ArcB. Since the study relied on phosphorylation status of ArcA as a readout of system activity, additional investigation of target gene transcription activity would provide additional support for this model. All the quinone mutant strains in this study showed stunted growth during the sparging experiment, so the overall physiology of the mutant strains may also need further analysis.
Consolidation of ArcAB Regulation Data
Although the above two models share some commonalities with regard to quinone function in ArcB activation, variations in growth phenotypes of quinone mutants and specific roles of quinone species currently limit consolidation of these studies. Importantly, mutation of arcA and arcB did not impact the quinone profile of cells in either aerobic or anaerobic conditions, ruling out the possibility of direct regulation of quinone production by ArcAB (123). Mutants lacking UQ have been shown by others to have normal levels of ArcA phosphorylation in aerobic conditions (113). This finding is in opposition to both models which agree that UQ is necessary to inhibit ArcB kinase function in aerobic conditions and thus prevent ArcA phosphorylation (26, 27). The same group also reported nearly normal fermentative growth in anaerobic conditions in the mutant strains lacking UQ despite lower ArcA phosphorylation levels (113). The authors note these discrepancies and speculate that the differences may be due to effects of short-term versus long-term changes to aerobiosis. Disrupting quinone production affected ArcA phosphorylation only in anaerobic conditions in this study and was contingent on the presence of MK or MK and DMK. The requirement of MK for ArcA phosphorylation in anaerobic conditions may lend support to either model. Model A relies on MK as being the sole quinone capable of reducing ArcB, and MK is the dominant anaerobic quinone in facultative anaerobes. Model B infers that MK must be the quinone reducing ArcB as it is the most abundant quinone in anaerobic conditions in which ArcB is activated. Furthermore, the follow-up study by Nitzschke and Betterbrock demonstrates the importance of all three quinone species for proper growth in aerobic and anaerobic conditions and highlights that the single quinone mutants likely do not reflect quinone activity of wild-type strains (113).
Modulation of ArcA-P activity through ArcB phosphatase activity has not been fully explored but should not be discounted because ArcA can be phosphorylated even in relatively aerobic conditions (28). In this scenario, ArcB would constitutively phosphorylate ArcA, and the changes in ArcB phosphatase activity would ultimately control ArcA. This case is not likely to be the main mechanism of regulation given the extended evidence that ArcB kinase activity is indeed inhibited by quinones. The phosphatase function of ArcB should still be studied in the context of quinone-based regulation to more fully encompass the physiological continuum of ArcB function. The discrepancies between various models of ArcAB regulation may also be due to the use of different strains of E. coli, culture conditions, or the presence or absence of additional regulators. Multiple questions certainly remain regarding regulation of ArcAB. Are quinones the main regulator of ArcB function in species other than E. coli? Which quinones modulate ArcAB activity in periods of transition during anaerobiosis such as from fermentation to anaerobic respiration? Which other factors impact the redox state of quinones and ultimately their interaction with ArcB?
CONTROL OF CENTRAL METABOLISM
ArcA meets the definition of a global regulator set forth by Martínez-Antonio and Collado-Vides based on the large number of operons under its control and its ability to regulate diverse metabolic pathways (124). Global regulation of metabolism by ArcA is thought to be achievable because of the flexibility of its DNA-binding architecture (54). Up to 150 different operons across various networks are under the direct control of ArcA in E. coli (36, 90). ArcA coregulates these networks not only in conjunction with specific transcription factors but also with other global regulators (36, 124). Indeed, ArcA has been identified as one of five major global regulators of anaerobic fermentation in E. coli. (reviewed by Kargeti and Venkatesh [125]). Further complicating this global regulatory network is the regulation of other transcription factors by ArcA. Park et al. determined that 17 transcription factors are under direct regulatory control of ArcA in strict anaerobic conditions (36). Analysis of ArcA’s influence of vital cellular processes includes energy production and redox balance, demonstrating the importance of this global regulator. Catabolism of carbon sources to generate usable energy is highly dependent on the availability of electron acceptors and thus is presented as three possibilities: dependency on oxygen (aerobic respiration), dependency on alternative acceptors for the ETC (anaerobic respiration), or fermentation. It is important to consider, however, that the switch between aerobiosis and anaerobiosis works on a continuum and regulatory systems allow for fine tuning of the response to these changes (126). The activity of the Arc system has an inverse linear correlation to increasing aerobiosis, conferring maximal ArcA regulation under total anaerobic conditions (28). This description is somewhat at odds with earlier reports that found ArcA activity was most relevant in microaerobic conditions (22, 122). Once again, the observed differences may be due to variable laboratory methods that impact oxygen consumption and testing metrics. A review of the literature regarding ArcAB contributions to growth phenotypes reveals a wide range of results for arcA and arcB mutants across multiple species and in various conditions. For example, one study demonstrated growth defects with E. coli arcB deletion mutants in aerobic conditions in contrast to another that showed arcB deletion mutants of E. coli did not exhibit growth defects in these conditions (127, 128). Despite some lab-to-lab discrepancies in defining ArcAB regulon function, much progress has been made toward the study of metabolism controlled by the Arc system.
ArcA Regulon
Because of ArcA’s extensive regulatory network, studies exploring global gene expression have been important in pinpointing the control of metabolism by this transcription factor (Table 1). Many of these studies are performed in aerobic or anaerobic conditions, but ArcA’s role is perhaps best showcased during periods of transition when the availability of electron acceptors varies (39). The presence of alternative electron acceptors such as nitrate have indeed been shown to heavily influence activity of ArcA and the ArcA regulon independent of oxygen availability, providing yet further evidence that ArcAB is more closely linked to the redox state of the cell than oxygen availability alone (39, 129). Key themes of ArcA’s regulatory function have emerged and will be covered in depth here. The most notable function of ArcA is repression of oxidative metabolic pathways. Closer inspection of the ArcA regulon reveals a role for ArcA in promoting fermentative pathways, the stress response, contributing to acquisition and utilization of the key elements nitrogen and iron, and metabolic overflow. Despite reportedly high homology of ArcA proteins and conservation of its binding consensus sequence, the ArcA regulon can be highly variable between bacterial species. For example, only six genes are shared between the E. coli and S. oneidensis ArcA regulons (93). It is therefore pertinent to reiterate that the metabolic themes described here apply largely to work done in the model E. coli system and that the regulons of other species are currently less well defined.
Repression of Carbon Oxidation Pathways
ArcA represses genes of multiple pathways needed for the oxidation of carbon sources. In studies in which glucose is the sole carbon source for cells cultured anaerobically, ArcA directly downregulates catabolism of fatty acids, aromatic compounds, amino acids, and polyamines, among other compounds (36, 130). These pathways can all feed into the tricarboxylic acid (TCA) cycle, which is noteworthy as ArcA represses each gene corresponding to an active enzyme in the oxidative TCA cycle (39). Repression by ArcA thus targets pathways needed for the catabolism of carbon sources downstream of glycolysis. Notably, ArcA regulation has been tied to maintenance of the NADH/NAD+ ratio, an electron carrier and critical cofactor that connects pathways of central metabolism (22, 131). Furthermore, this ratio has been linked to metabolic flexibility and the redox state of the cell – concepts that have already been described in the context of ArcAB regulation (23). After exposure to increasingly aerobic conditions, arcA mutants had a significantly higher NADH/NAD+ ratio in comparison to wild-type cells (22). This effect of a skewed NADH/NAD+ ratio on metabolism in arcA mutants is evident by elevated levels of acetate production, which can be prevented by overexpression of NADH hydrogenase (131). Intriguingly, there is evidence of diminished activity in the TCA cycle in arcA mutants even though ArcA is a prominent repressor of key genes in this pathway (38). While there are inherent difficulties in measuring metabolic flux alongside transcriptional regulation, these results nevertheless serve as foundational evidence for two important points. First, global regulators of metabolism, including ArcA, cooperate to optimize metabolism, and impairment of only one such regulator can have dramatic consequences. Indeed, deletion of arcA results in increased flux through the TCA cycle in anaerobic conditions rather than fermentation, wasting metabolic substrates and energy (39, 132). Second, metabolism is regulated not only at the transcriptional level but also at the enzymatic level (i.e., by posttranslational modification and allosteric control). Predicting metabolic activity based on gene regulation alone is therefore difficult, especially when regulation by multiple transcriptional factors is considered. Application of different approaches, including transcriptomics and metabolomics, must be employed in multidisciplinary studies to understand the role of ArcA and other global regulators at the cellular level.
Promotion of Fermentation
In addition to repression of pathways necessary for oxidation of non-glycolytic carbon sources, P-ArcA promotes expression of genes involved in mixed acid fermentation, the primary metabolic pathway of E. coli cells that are unable to respire (133). Glycolysis is the main energy-producing pathway of fermentation, and certain enzymes in the glycolytic pathway are in fact upregulated by ArcA (39). Some groups have manipulated (or suggested the manipulation of) the ArcAB system in industry and biotechnology for over-producing a variety of fermentative compounds such as derivatives of acetyl coenzyme A (acetyl-CoA) (19, 134–140). In a strain of E. coli engineered to overproduce ArcA, fermentative pathways were induced based on an increased secretion of acetate (135). As expected, arcA and arcB mutant strains of E. coli produce less acetate relative to wild-type strains (132, 134). Deletion of arcA had been shown earlier to negatively affect expression of ackA, the gene that encodes the second enzyme in the acetyl-CoA to acetate pathway. Regulatory control of the genes in this pathway are more directly under the control of FNR. ArcA has, however, been well documented to strongly repress production of acetyl-CoA synthetase, which ultimately would prevent the conversion of acetate back to acetyl-CoA (36, 38). A decrease in acetate production in arcA and arcB mutants is thus likely a combinatorial effect of an increase in carbon flux through nonglycolytic pathways and decrease in expression of acetate-related genes.
Dysregulation of fermentation is also evident in E. coli strains lacking arcA or arcB through increased secretion of succinate and lactate relative to the isogenic wild-type strains (132, 134, 139). E. coli strains lacking ubiquinone also produce more lactate relative to the wild-type strain during changes in oxygen availability, which supports the notion that disruption of the ArcA activation pathway impacts fermentation based on the close link previously described between ArcB and quinones (27). The change in fermentative products for mutant strains indicates a skew toward reduction reactions regenerating NAD+ (e.g., pyruvate to lactate) over oxidation reactions producing ATP via substrate-level phosphorylation (acetyl-CoA to acetate) (134). These changes in metabolic flux of arcA and arcB mutants underscore the link between redox state and metabolic activity, as well as the difficulty in predicting metabolic activity of global regulator mutants (38, 134, 141). One might expect all fermentative products to decrease when ArcAB is absent if this regulatory system promotes fermentation. The overall change in profile of mixed acids produced during fermentation seen in arcA and arcB, however, is more telling when considering the number of metabolic pathways affected. Again, global metabolic changes rather than just individual metabolites must be accounted for when analyzing ArcAB mutants.
Although ArcA regulates anaerobic processes such as fermentation, there are exceptions to the classical description of ArcA as functional under anoxic conditions. Early studies focused on nitrate respiration found that ArcA-controlled genes encoding succinate and lactate dehydrogenases were less repressed in the presence of nitrate under low-oxygen conditions (142, 143). In an experiment where trimethylamine‐N‐oxide was added to anaerobically cultured E. coli as a terminal electron acceptor, cells shifted from fermentation to anaerobic respiration, and ArcA activity was momentarily repressed (144). The reductive TCA cycle shares some enzymes with the oxidative TCA cycle, which is why repression of ArcA activity in the context of anaerobic respiration is necessary. A study by Federowicz et al. also highlights genes in pathways downregulated by ArcA are de-repressed while transitioning from fermentation to nitrate respiration (39). These results further underscore the complexity of metabolic regulatory circuits and demonstrates the ability of cells to prioritize metabolic pathways based on energetic output, among other factors.
Nitrogen Metabolism
Catabolic processes controlled by ArcA inevitably affect the flux of glycolytic and TCA intermediates into anabolic pathways needed for the synthesis of the four major macromolecules (amino acids, nucleotides, lipids, and carbohydrates). ArcAB is likewise associated with nitrogen metabolism, an essential nutrient critical to anabolism. The cell must balance activity of anabolic pathways with overlapping pathways utilized for energy production via chemiosmosis (39). Major disruptions in nitrogen homeostasis have been observed in arcA mutants, affecting the balance between energy production pathways and protein synthesis. An arcA deletion mutant of E. coli was shown to have a decreased intake rate of ammonia, an important source of nitrogen, relative to an isogenic wild-type strain under strict anaerobic conditions with glucose as the sole carbon source (38). Nitrogen metabolism is of course critical to amino acid synthesis, and ArcA is an important regulator of amino acid fate. For instance, multiple studies have corroborated that ArcA upregulates arginine transporter gene transcription and downregulates the arginine degradation pathway (36, 38). Unexpectedly however, arginine was among the amino acids that accumulated in an arcA E. coli mutant strain. Loss of ArcA was subsequently associated with major metabolic dysregulation impacting the biosynthesis and utilization of amino acids which could not be predicted by examination of the ArcA regulon alone (38). Further, the rate of metabolic flux relative to translation was calculated to be higher in this arcA deletion mutant, indicating an imbalance of cellular activities critical to homeostasis and growth (38). When the proteome of wild-type cells was compared to that of arcA mutant cells in fermentative conditions, a larger portion of the proteome deemed “unused” or “unnecessary” was found in the mutant cells. The authors classified metabolic proteins as unnecessary if they had potential to be beneficial during adaptation to environmental conditions but were otherwise burdensome during fermentation. The wild-type cells were deemed to be more metabolically efficient as a result. Therefore, while cells can survive without an intact ArcAB system, they are at a clear disadvantage regarding balancing metabolic and translational efficiencies. In effect, ArcA regulation contributes to optimized cellular activity based on availability and utilization of metabolic precursors for energy production and biomass.
RESPONSE TO GROWTH LIMITATION
Stress Response
The ability of a cell to switch from generating energy for growth to conserving energy and making repairs under stress relies on careful metabolic regulation. Control of the bacterial stress response is indeed linked to ArcA-mediated regulation of central carbon metabolism such as the TCA cycle (145). The general stress response involves a metabolic shift during less favorable conditions, including nutrient deprivation and the entry into stationary phase (146). σS (or RpoS) is a sigma factor that coordinates the stress response in E. coli and influences the regulation of a wide range of genes that promote survival (147). Because multiple factors can lead to cellular stress, the complexity in connecting σS to simultaneous responses cannot be overstated (146, 148). With this in mind, σS activity has been shown to be influenced by central metabolic pathways of the cell, which in turn are regulated by σS. For example, mutation of pyruvate dehydrogenase, which provides acetyl-CoA for the TCA cycle, leads to an increase in σS levels (148). The relationship between metabolic enzymes and σS levels suggests an interesting connection to ArcA, an important repressor of the TCA cycle. ArcA exerts partial control of σS through the small activating RNA ArcZ, which binds rpoS transcripts (48). When ArcA is active, such as in anaerobic conditions, ArcZ is repressed and translation of RpoS is low. ArcZ has been proposed to support translation of rpoS while also repressing the Arc system. ArcAB was connected to the coordination of synthesis and proteolysis of RpoS with RssB, which facilitates degradation of RpoS by the protease ClpXP (87). During exponential phase, rpoS is directly repressed by ArcA and kept at basal levels (48, 87, 149). During rapid cell growth, quinones are expected to be reduced following flooding of the ETC with electrons, corresponding to ArcB kinase activity and phosphorylated ArcA. As cells enter stationary phase, phosphorylated ArcA level are thought to diminish and σS levels in turn increase (87). Comparable to conditions in which high levels of oxygen result in oxidized quinones, nutrient-poor conditions might also result in oxidized quinones as fewer electrons pass through the ETC (87). Oxidized quinones would lower ArcB activity and subsequently lead to de-repression of σS under the current mechanistic model. Although ArcA represses rpoS, deletion of arcA results in increased sensitivity to dehydration in E. coli (149). This phenotype is striking as σS is thought to be an important regulator active during dehydration tolerance and raises the question of how does ArcA coordinate with σS to resist the effects of dehydration. Expanding studies to include interrogation of ArcA alongside σS during various stressors will not only further untangle the connection of these two vast regulators but also provide more insight into cellular stress overall.
Starvation
Starvation is one potential trigger of the stress response, as noted above. Cells experience starvation when key nutrients such as amino acids become limited and cellular processes must be modified as a result. In culture, bacteria often begin to experience starvation when transitioning from exponential to stationary phase growth. ArcAB has been identified as an important metabolic regulator during stationary phase in which cells must balance a lack of available nutrients with a potentially harmful accumulation of metabolic by-products (150). Nyström et al. demonstrated that gene expression during anaerobiosis and aerobic stationary phase was similar. Despite the availability of oxygen and usable carbon sources, cells shutdown the TCA cycle via ArcA during stationary phase. The authors concluded that during starvation in stationary phase, cells lessen respiratory activity to prevent oxidative damage and to conserve endogenous resources. ArcAB plays a protective role in this case by repressing aerobic pathways, ultimately aiding cells in the aging process (150). These results may, however, be dependent on carbon source availability since the finding that ArcAB is important for survival during stationary phase was not apparent when arcA mutant cells were cultured in rich Luria-Bertani (LB) medium instead of minimal medium (151, 152). Intriguingly, TCA cycle mutants did live longer than wild-type counterparts in a rich medium during stationary phase, which may be due to lower reactive oxygen species (ROS) levels (152). Because active ArcA can downregulate the genes of the TCA cycle, one might expect that loss of ArcA resulting in increased TCA cycle activity would lower levels of survival. The authors speculate that in LB medium, starving cells must continue to rely on the TCA cycle to generate energy, so ArcA function would be counterintuitive. This same study also revealed that mutants lacking lipA, which encodes lipoyl synthase involved in lipoic acid biosynthesis, outlive wild-type cells in rich media but that this increased life span was in fact ArcA dependent (152). lipA mutants are unable to convert pyruvate to acetyl-CoA to enter the TCA cycle. The result is that pyruvate is converted to acetate, which can occur during fermentation and overflow metabolism, a concept described later. Therefore, ArcA repression of the TCA cycle would indeed be advantageous in conserving energy and resources to promote survival in the lipA mutant.
Iron Limitation
Iron is a critical nutrient which cells must acquire from the extracellular environment, and ArcAB is implicated in iron homeostasis in anaerobic conditions (153). Iron homeostasis and metabolism are closely connected as enzymes found in the TCA cycle and the ETC rely on the availability of iron for proper regulation and function (154, 155). Predictably, cells are likely to switch from using aerobic respiratory enzymes when iron is scarce and not available as an enzymatic cofactor (156, 157). Under conditions of iron starvation, cells then rely on alternative metabolic pathways, including fermentation. Accordingly, an arcB mutant of Actinobacillus actinomycetemcomitans grew poorly under iron-limiting conditions aerobically relative to an arcB complemented strain (158). This is a clear example in which sensing of environmental conditions through ArcAB leads to metabolic changes to optimize growth. When considering the regulatory response to iron, the regulons of Fur, the master regulator of iron metabolism, and ArcA have in fact been found to overlap (38, 159). ArcA specifically downregulates genes encoding enzymes that contain iron-sulfur clusters, including succinate dehydrogenase and NADH:ubiquinone oxidoreductase (160, 161). Of note, all of these genes are also controlled by at least one other regulator such as FNR, further demonstrating the ability of ArcA to interconnect multiple regulatory networks (160). ArcA is also well established as a regulator of genes encoding cytochromes that utilize heme-bound iron. ArcA can upregulate cydAB and downregulate cyoABCDE, which both encode terminal oxidases of the ETC (36, 162). Interestingly, less ArcA protein is found in iron-limited conditions (163), though, as stated earlier, the regulatory outcomes of a transcription factors cannot be determined by protein abundance alone. Nevertheless, the relationship of ArcAB activity to iron availability and utilization is evidently intricate, and future research should focus on further integrating ArcA-controlled metabolism into iron homeostasis.
Overflow Metabolism
Optimal metabolism is not solely based on the availability of substrates; therefore, the role of ArcA in repression of the TCA cycle needs to be considered from other perspectives. For example, E. coli can perform fermentation during periods of rapid growth in the presence of oxygen to quickly produce ATP (164, 165). It is hypothesized that the growing cell membrane may be unable to accommodate room for respiratory enzymes, so the cell must utilize other metabolic processes (164). Not surprisingly, ArcAB has been implicated in this process, known as “overflow metabolism,” as it controls multiple operons involved in these pathways (18, 131, 160). The term “overflow” refers to the increased production and release of metabolites such as acetate as the cell oxidizes substrates that could have otherwise been further catabolized through aerobic respiration. Because of the extensive regulatory network of ArcA, investigators have also noted that the connection between this system and an overflow of acetate could be an indirect relationship (131). Increased activity of the TCA cycle in arcA mutants may incidentally result in decreased acetate production because acetyl-CoA, a potential precursor of acetate, is continuing to be used in the oxidative TCA cycle. At the cellular level, overflow metabolism is perhaps the most striking example in which ArcA is active in the presence of oxygen and an abundance of carbon sources.
INTERACTIONS WITH OTHER REGULATORY SYSTEMS
Fumarate Nitrate Reductase
Multiple transcription factors regulate metabolism during anaerobiosis, but fumarate nitrate reductase (FNR) is the one most closely associated with ArcAB in E. coli. Together, ArcA and FNR control more than 80% of metabolic flux during anaerobiosis (39). Unlike ArcA, FNR uses iron sulfur clusters to directly senses molecular oxygen, which passes freely through the cellular membrane (25, 166, 167). Upon encountering oxygen-limited conditions, iron-sulfur clusters become reduced and FNR is activated. There is no concrete evidence that FNR can also be affected by other oxidizing agents that reduce the iron-sulfur cluster, implying a specificity for oxygen (166). As its name implies, FNR promotes the transition from aerobic respiration to anaerobic respiration with alternative terminal electron acceptors such as fumarate and nitrate. When fumarate and nitrate are available as electron acceptors during anaerobic respiration, FNR upregulates reductases encoded by the frdABCD and nirBD genes to properly utilize them as part of its role in controlling metabolism (39, 168). The apparent overlap between the ArcAB and FNR regulons was noted early on after it was reported that the expression of genes related to anaerobiosis was dependent on both systems (24, 49, 162, 169). For example, the binding of FNR at the promoter of cydAB, a component of the ETC, is contingent upon the presence of ArcA also binding in some cases (162). One group initially reported that 303 genes are regulated by both ArcAB and FNR, demonstrating extensive overlap of their regulons (14). FNR and ArcA dually contribute to repression of the tricarboxylic acid pathway (38). Both regulators repress the operons encoding α-ketoglutarate dehydrogenase, succinyl coenzyme A synthetase, and succinate dehydrogenase, enzymes critical to the oxidative tricarboxylic acid pathway (38). Like ArcAB, FNR also promotes fermentation by upregulating genes involved in non-oxidative pyruvate catabolism (pflB) for example (168). Multiple hypotheses have been proposed to differentiate the seemingly redundant roles of these regulators (14). It is speculated that ArcAB is more critical in microaerobic conditions, while FNR becomes the major regulator as cells encounter strict anaerobic environments (22, 122, 130, 170). This notion is complicated by studies that show ArcA becomes increasingly phosphorylated as conditions become more anaerobic. Whether this directly correlates with increasing ArcA activity or whether maximal ArcA function is reached before complete anaerobic conditions could inform the functionality of ArcA versus FNR. Curiously, mutations of arcA and arcB are epistatic over mutation of fnr under nitrate respiratory conditions (142). Another theory for explaining how the two systems work in tandem is FNR mediating a fast initial response to low oxygen levels with ArcAB becoming active only after sustained exposure (171).
Despite the partial overlap between the ArcA and FNR regulons, the transcription factors also serve distinctive functions. At the cellular level, ArcA’s major role is repression of catabolism while FNR activates chemiosmotic and anabolic pathways (39). In anaerobic conditions with glucose as the sole carbon source, there are only seven reported operons directly regulated by both ArcA and FNR in E. coli (36). There are additionally instances of ArcA and FNR having opposing activities on coregulated genes (130, 160, 171). This is especially evident in the regulation of cytochromes of the ETC. ArcA is involved in the upregulation and downregulation of cydAB and cyoABCDE, respectively; however, FNR downregulates both of these operons (20, 172). This multilayer regulation of cytochromes has again been attributed to the varying activity of both regulators under microaerobic and anaerobic conditions. Under microaerobic conditions, ArcA-mediated transcription of cydAB ensures the presence of cytochrome bd-I oxidase, which has a higher affinity for oxygen than cytochrome bo3 oxidase from cyoABCDE and therefore is useful for scavenging (162). When levels become anaerobic, FNR is then active to downregulate both cytochrome complexes as oxygen can no longer be utilized. In summary, ArcA and FNR activity is dependent on available substrates and electron acceptors, and they coordinate metabolism in complex networks with other regulators (160). The functionality of these intertwined regulators further illustrates the ability of bacterial cells to integrate multiple signals to optimize metabolic activity.
Phage Shock Protein System
The simplest two-component regulatory systems have one sensor and one response regulator. Cross-talk occurs when the sensor kinase of one system phosphorylates the response regulator of another system. Conventionally, coordinated cross talk between bacterial two-component systems, also referred to as cross-regulation, is thought to be kept to a minimum to promote specificity during a response to a stimulus (173). An investigation into cross talk of ArcAB with the two-component systems UhpBA, NtrBC, and PhoRB provided no evidence for physiologically relevant cross talk between these tested systems (174). Evidence has been presented suggesting ArcB can phosphorylate the previously mentioned orphan response regulator RssB, but the significance of this interaction has not been fully elucidated (44, 87). Regulatory networks involving multiple kinases have been identified and may be useful in integrating multiple stimuli (175). Along these lines, studies have suggested that ArcB interacts with targets in addition to ArcA (87, 176). For instance, ArcB reportedly cooperates with the phage shock protein (Psp) system in E. coli cells (177, 178). While initially identified in the response to damage by bacteriophage, the Psp response is now associated with various agents that result in compromised membrane integrity (reviewed by Joly et al. [179]). Homologs of the Psp system have been found in many organisms (15). In Gram-negative bacteria, where it was first identified, it is thought that the Psp system maintains the proton motive force when the inner membrane is damaged. It has been proposed that a not yet fully characterized direct interaction between ArcB and PspB, a membrane-bound component of the Psp system, is important for this function (178). The ArcB and PspB interaction was shown to be conditional on microaerobiosis. It was also proposed in the same study that the ArcAB regulon amplifies expression of the psp system. Although the initial stimulus of the Psp system has yet to be definitively identified, the contribution of ArcB to the activation of the Psp system in microaerobic conditions suggests that the redox state of the cell is an important aspect of Psp activation. Maintaining integrity of the cell envelope is critical for redox homeostasis, so the contribution of redox status to Psp activity is logical. The connection between a system that ultimately senses changes in redox conditions (ArcAB) to a system that senses membrane damage (Psp) would be largely based on the notion that the proton motive force generated at the membrane needs to be maintained following envelope stress. In agreement with this concept, the Arc system has been referred to as a “protometer,” describing how activation of ArcAB coincides with changes in the electrochemical gradient at the inner membrane (180).
Cell Envelope Maintenance
The role of ArcAB in response to perturbations at the cell envelope has also been demonstrated in Shewanella oneidensis in connection with the sigma factor σE (encoded by rpoE) (181). σE becomes active when envelope stress results in accumulation of misfolded outer membrane proteins and lipopolysaccharides in the periplasm and modulates envelope biogenesis as a result. Under stable conditions, σE is sequestered by the anti-sigma factor RseA. In arcA mutants of S. oneidensis, rseA gene expression is considerably elevated. Through a yet undefined mechanism, ArcAB and RseA are accordingly theorized to cooperate in mediating σE activity. The relationship between ArcAB and σE has been further supported by evidence that proper functioning of the lipopolysaccharide transport system relies on both systems (182). rpoE is upregulated in ΔarcA E. coli mutants relative to the wild-type strain, indicating the connection extends to other species (38). Interestingly, these studies connect ArcA function specifically to the outer membrane. As ArcAB has now been linked to inner and outer membrane maintenance (albeit in different organisms), general envelope integrity preservation by this system may be necessary for optimal redox homeostasis. It is noteworthy that in clinical strains of Klebsiella pneumoniae, arcB is upregulated following exposure to polymyxin B, a cationic peptide used as a model for antimicrobial peptides of the immune system (47). This study highlights that metabolic rewiring was an important aspect of response to this antibiotic. Specifically, genes encoding respiratory enzymes repressed by ArcA are downregulated following polymyxin B exposure. As the cellular response to polymyxin B includes a metabolic shift to fermentation, it is not surprising that ArcAB was implicated as a mediator in the process (47). Polymyxins are well recognized for perforating the outer membrane of bacterial cells, but the precise mechanism of how this leads to cell death is unclear. From inducing ROS damage to stiffening the membrane, a growing body of evidence implicates perturbations at the inner membrane by polymyxins (183–185). We can speculate that these changes can influence or impede ETC and specifically quinone activity at the membrane, which would in turn impact activation of the ArcB kinase. Damage by polymyxins has indeed been shown to impact enzyme activity at the respiratory chain (186). The Arc system has also been implicated in the response to multiple classes of antibiotics, including aminoglycosides and the cephalosporin cefixime that inhibits cell wall synthesis (41, 187, 188). This is notable since ArcAB has also been connected to cell envelope maintenance in S. oneidensis (181, 189, 190). These findings further display the complex networks dedicated to responding to stressors. Whether ArcAB controls functions directly in structural maintenance of the cell envelope, or whether redox homeostasis is so tightly dependent on envelope integrity that ArcAB metabolic regulation is inextricably tied to perturbations, remains to be determined.
RESPONSE TO REACTIVE OXYGEN SPECIES
Aerobic metabolism efficiently provides ATP to cells, but this benefit must be balanced with the cost of redox stress produced when using these pathways (191, 192). Cells have evolved sophisticated mechanisms for responding to these stressors and ultimately maintaining redox balance within the cell (193, 194). With the redox state of the cell closely linked to metabolism, involvement of ArcAB activity in maintaining redox balance within the cell across oxygen conditions are expected (137, 138). As noted above, ArcAB has been linked to the ratio of NADH/NAD+ in the cell and by proxy also ATP/ADP (129). Redox homeostasis by ArcAB may also involve maintenance of the extracellular microenvironment, such as release of reactive oxygen species (ROS), in addition to ROS levels inside the cell (195). In conditions in which the cell is not using oxidative phosphorylation, ArcA regulation promotes glutathione export as well as extracellular superoxide production (128). As already described, ArcAB has been implicated in “overflow metabolism” in which low-yield metabolic pathways replace higher-yield pathways. This diversion also helps maintain redox balance within the cell (196). The role of ArcA in responding to various forms of redox stress not only highlights the broad array of cellular processes impacted by ArcAB but also serves as an indication for other potential mechanisms of ArcA activation.
Mechanisms of ROS Resistance
The cell must respond to ROS, which are oxygen and its derivatives including peroxides, superoxide, and hydroxyl radicals, as they can damage bacterial DNA, proteins, and lipids (197). Accordingly, bacteria have evolved ways to scavenge and reduce ROS into less harmful products (193). ArcAB has been characterized by multiple studies to play a role in such ROS resistance. Indeed, in E. coli deletion of arcA or arcB resulted in increased susceptibility to hydrogen peroxide in aerobic conditions (198). In Salmonella enterica serovar Typhimurium, the ArcA regulon overlapped very little in comparison to the regulon of untreated aerobically grown cells following hydrogen peroxide treatment (199). Accordingly, the function of ArcA in the context of ROS may be difficult to predict utilizing established regulons defined in anaerobic conditions (36, 38, 39). The role of ArcAB in resisting hydrogen peroxide was shown to be independent from the ability to detoxify ROS since arcA and arcB E. coli mutants were shown to neutralize peroxide to wild-type levels (198). Similarly, the gene encoding superoxide dismutase in Haemophilus influenzae is not under the control of arcA despite arcA mutants being more susceptible to hydrogen peroxide (200). Other studies have since explored alternative mechanisms of ArcAB-mediated hydrogen peroxide resistance in Salmonella enterica, S. oneidensis, and H. influenzae (Fig. 4) (190, 200, 201). S. Typhimurium arcA mutants are more sensitive to the effects of hydrogen peroxide because ArcA downregulates a porin that enhances entry of this ROS (201). In H. influenzae, ArcA controlled expression of dps contributes to resistance via Dps-dependent protection against hydrogen peroxide-mediated DNA damage (200). Since ROS are also primary anti-bacterial effectors of innate immune cells, it is noteworthy that ArcAB has been described as promoting survival of S. Typhimurium in macrophages and neutrophils (202). Killing by the host cells in this study was not shown to be explicitly ROS-mediated, but ROS levels were not different in immune cells infected with wild-type or arcA/arcB mutant cells. Direct links between metabolic regulation by ArcA and oxidative stress have already been suggested based on findings that resistance to hydrogen peroxide is restored via amino acid supplementation in E. coli (198). It has been demonstrated in S. Typhimurium that arcA mutants have a skewed NADH/NAD+ ratio, which may result in a higher proportion of ferrous versus ferric iron and thus more hydroxyl radical formation via Fenton chemistry (199, 203). Because of the pleiotropic effects of mutating arcA or arcB, it is unlikely that a single mechanism for ROS resistance exists. Strikingly, ArcA has been implicated in the regulation of soxS, a gene encoding an important mediator of the ROS response (38, 204). Considering the multiple studies indicating that ArcAB does not play a role in the neutralization of ROS such as hydrogen peroxide, the biological significance of ArcAB-controlled soxS expression remains unclear. Ubiquinone has also been identified an important antioxidant within the cell membrane preventing peroxidation of lipids by hydrogen peroxide (205). Based on the earlier description of quinones as the main signal for ArcAB activation, this finding may serve as a direction for future research investigating how ArcAB can respond to oxidative stress through interaction with other molecules.
FIG 4.
ArcA-mediated responses to hydrogen peroxide. Following reactive oxygen stress from exposure to hydrogen peroxide (H2O2), cells utilize ArcA to respond by downregulating porins of H2O2 (S. enterica), producing proteins such as Dps to protect DNA from oxidative damage (H. influenzae), and promote maintenance of the outer membrane following ROS damage (S. oneidensis).
Activation of ArcAB during ROS Response
ArcAB actively regulates the response to reactive oxygen derivatives yet is canonically known to be active in conditions in which oxygen consumption drops. This apparent contradiction could be a starting point for novel research exploring an unknown role for ArcAB in aerobic growth and may uncover other regulators of the system beyond the quinone pool. The mechanisms for a relationship between ArcA-mediated control of metabolism and the ROS response may already be evident. The metabolic pathways ArcA controls are large sources of internal ROS production and thus may need to be repressed when the cell faces an exogenous ROS threat (206, 207). In this scenario, ArcA would need to be active under aerobic conditions, which could rely on one of three mechanisms: (i) binding/regulatory activity by unphosphorylated ArcA; (ii) an alternative kinase of ArcA; or (iii) kinase activity of ArcB under oxidizing conditions. No direct evidence has been presented for points 2 or 3, but noteworthy evidence has been reported for point 1. There are existing examples of response regulators not needing phosphorylation to function (208). Unphosphorylated ArcA has indeed been shown to bind the same DNA fragments as phosphorylated ArcA in experiments examining plasmid recombination (209). Subsequent DNase footprinting, however, showed that the binding pattern by unphosphorylated ArcA was different (36, 86, 87). In addition, apparent activity of ArcA under aerobic conditions may be attributed to small amounts of phosphorylated ArcA (54). Most recently, it has been found that S. enterica ArcA becomes partially active in the presence of ROS in a phosphorylation-independent manner (210). Zhou and colleagues demonstrated that ROS exposure leads to disulfide bond formation in ArcA, allowing for ArcA dimers to form without activation from ArcB. The oxidized cysteines forming these bonds are reportedly highly conserved, suggesting this new mechanism may be conserved in other species as well. Barring the existence other ArcB activators, ROS damage would ultimately have to impact quinone oxidation or abundance for ArcB to become active. One could theorize damage at the ETC affects the oxidation state of the quinone pool and ultimately its connection to ArcB. Future studies could also examine whether high levels of ROS result in the activation or release of other ArcB regulators. With the capability of ROS to cause cell-wide damage and the capacity of ArcA to affect global change at the cellular level, the role of ArcA in maintaining metabolic homeostasis during ROS response in aerobic conditions cannot be overlooked.
CONTRIBUTION TO PATHOGENESIS
Cellular replication is one component of bacterial fitness during infection. Furthermore, it is intuitive that transcriptional regulation is critical for optimal fitness and pathogenesis in the host. Dynamic bacterial interactions with the host include responses to changes in nutrient availability, oxygenation, and immune defenses, all of which are heavily dependent on the site and stage of infection (211, 212). This may be especially true during the initial stages of infection and during dissemination from an initial site, in which the transition between environments can be dramatic. Examples of two-component regulatory systems in pathogenic bacteria are so ubiquitous that the regulatory pathways have now become attractive targets of antimicrobial therapies (see reviews by Tiwari et al. [213], Rajput et al. [214], and Hirakawa et al. [215]). Likewise, the function of ArcAB in controlling metabolic functions in response to oxygen availability or consumption has prompted several groups to investigate the impact of the ArcAB system on infection processes and outcomes. Indeed, we have determined that arcB mutants of the opportunistic pathogens Serratia marcescens and Citrobacter freundii exhibit a significant loss of fitness as assessed by Tn-seq of bacteria recovered from an experimental bloodstream infection model (216, 217). S. marcescens arcA was also identified as a fitness gene in the same study. Similarly, a loss of fitness was also associated with a Klebsiella pneumoniae arcB transposon insertion mutant in a murine lung infection model (218). Growing evidence from the literature supports the notion that ArcAB control of gene expression is required for optimal bacterial fitness in the host environment. The diversity of organisms and infection models described in the sections that follow suggests that the importance of ArcAB is widespread among bacterial pathogens. Understanding the function of the ArcAB system during infection will aid in constructing models of pathogenesis and may provide viable targets for future therapeutic development.
Bacterial Survival during Infection
A role for ArcAB in directly and indirectly supporting growth during infection has been identified in several important pathogens. Disruption of arcA in S. Typhimurium significantly reduces the recovery of bacteria from mice following oral inoculation or during systemic infection following intraperitoneal injection (202, 219). Mutation of S. Typhimurium arcA furthermore limits intracellular survival in epithelial cells, macrophages, and neutrophils (202). So important is ArcAB to the pathogenesis of this species that mutating arcA alongside fnr and fliC has been proposed in developing an attenuated live S. Typhimurium vaccine (220). A second example of ArcA contributions to gastrointestinal infections has been observed using Vibrio cholerae in which arcA mutants exhibit reduced virulence and colonization in an infant mouse cholera model (221, 222). ArcA function was also shown to increase pathogenesis in a porcine pneumonia model using Actinobacillus pleuropneumoniae, with arcA mutant bacteria exhibiting lower clinical scores and reduced bacterial recovery compared to wild-type A. pleuropneumoniae (223). Finally, multiple bacterial species have been shown to require ArcAB function for optimal fitness during bloodstream infections, as previously noted (216, 217). The mammalian bloodstream presents an interesting paradox of oxygen availability in that the blood is an oxygen-rich environment, yet it is likely that little oxygen is available to extracellular pathogens in circulation as 98% of oxygen here is tightly bound to host hemoglobin within erythrocytes (224). Similarly, little is known about bacterial oxygen availability in the spleen and liver, which often harbor high concentrations of bacteria in murine models of bloodstream infections (225). H. influenzae, a natural inhabitant of the human nasopharynx, is also capable of causing invasive infections. H. influenzae arcA mutants required a higher lethal dose than wild-type bacteria following intraperitoneal injection of mice and decreased bloodstream recovery of an independent arcA mutant strain has also been observed (200, 226). All the bloodstream pathogens highlighted above can cause both localized and systemic infections and can be found as nonpathogenic colonizers of the human microbiota, further highlighting the need for pathogen adaptation during infection. For bacterial species that additionally occupy niches outside the mammalian host, such as S. marcescens and C. freundii, the transition to colonization from environmental reservoirs undoubtedly represents a shift in the metabolic requirements for fitness. The evidence for ArcAB function during pathogenic bacterial interactions solidifies the importance of this regulatory system and highlights the need for further investigation of the specific ArcAB-controlled metabolic pathways that contribute to each type of infection and in different infection niches.
Virulence
While metabolic coordination is likely to be a critical function of ArcAB transcriptional control during infection, additional ArcAB-regulated functions have also been implicated in pathogenesis. The S. Typhimurium gene loiA encodes a positive regulator of Salmonella pathogenicity island 1 (SPI-1) genes that are required for pathogenesis in a murine infection model, and a loss of Salmonella cellular invasion and reduced mouse virulence was observed when loiA was disrupted (219). Interestingly, expression of loiA is controlled by oxygen availability in an arcA- and arcB-dependent manner (219). Virulence genes encoded in SPI-1 were shown to be dependent on ArcA at the transcriptional and protein level in a subsequent S. Typhimurium study analyzing the proteome of this serovar in anaerobic conditions (227). These examples of regulation of SPI-1 effector genes by ArcAB in Salmonella are a clear demonstration of the diversity of ArcAB regulatory targets and expands the mechanisms by which this global regulator modulates infection processes. A second example of ArcAB-mediated control of virulence genes has been observed in V. cholerae. Mutation of arcA in V. cholerae results in a loss of cholera toxin production, the major virulence factor of this species and was further correlated to a loss of ctxAB (cholera toxin) and tcpA (toxin-coregulated pilus) gene expression in aerobic conditions (221). It was further suggested that the dysregulation of V. cholerae virulence genes was due to ArcA-dependent modulation of ToxT, a known transcriptional activator of ctxAB and tcpA expression (221). Finally, arcA mutants have been demonstrated to increase the sensitivity of H. influenzae to the bactericidal activity of human serum (94, 226). In nontypeable H. influenzae clinical isolate NT127, this loss of serum resistance was attributed to ArcA-dependent transcriptional modulation of the lipooligosaccharide glycosyltransferase gene lic2B, which participates in the lipooligosaccharide (LOS) biosynthesis pathway of this strain (94). ArcA and FNR have also been shown to participate in lipid A modifications in Salmonella Enteritidis based on oxygen availability (228).
CONCLUDING REMARKS
The two-component regulatory system ArcAB has been the subject of foundational research for more than 3 decades. The uniqueness of the structure of its sensor kinase ArcB and the complex binding architecture of the response regulator ArcA are some of the attributes that have made this system a fascinating one for study. Future work is still needed to address the discrepancies between models of how quinones regulate ArcAB under differing oxygenation conditions (see Text Box 1). Further development of techniques allowing for individual study of all three quinones within the same cell in situ will greatly aid in this matter. ArcA is a powerful repressor of central metabolic pathways, and this function is critical in mediating global metabolic changes not only during anaerobiosis but also in situations where the use of specific pathways is no longer efficient or even detrimental. From this perspective, ArcAB can incorporate feedback from the cell, including electron acceptor availability, enzymatic capabilities, and redox homeostasis, into a harmonized response. We have yet to fully characterize the operons under ArcA’s control and how this control may extend to other conditions in which oxygen consumption is impacted. This is especially true in species beyond E. coli and S. enterica. These discoveries will be important in uncovering all the cellular functions, including ROS response and cell envelope maintenance, that are becoming increasingly associated with ArcAB regulation. While the concept of this two-component system is strikingly straightforward, ArcAB and its vast regulon represent a sophisticated coordination of cellular processes. The numerous examples of ArcAB contributions to bacterial infections further suggests this global regulatory system plays an important role in responding to the host environment and additional work is needed to elucidate the specific nutrient and oxygenation requirements of each species to better understand the mechanisms of ArcAB function during infection.
BOX 1: KEY AREAS FOR FUTURE RESEARCH
Further characterization of ArcA oligomerization at target DNA binding sites and regulatory control by partially active ArcA
Clarification of control of ArcB phosphatase activity and effects of sustained stimulus exposure on ArcA activity
Continued profiling of quinone-based regulation of ArcB with respect to each quinone species
Integration of ArcA regulons from more culture conditions into global regulatory networks with a focus on coregulators such as FNR and σS
Elucidation of the significance of ArcA-controlled metabolic pathways during pathogenesis
ACKNOWLEDGMENTS
We thank Dimitris Georgellis and Klaas Hellingwerf for their permission to adapt figures from their aforementioned manuscripts for this review. All figures were created with Biorender.com.
This study was supported by Predoctoral Fellowship award 828854 to A.N.B. from the American Heart Association and Public Health Service awards AI148767 to M.T.A. and AI134731 to H.L.T.M. and M.A.B. from the National Institutes of Health. The authors have no competing interests to declare.
REFERENCES
- 1.Ward B. 2015. Bacterial energy metabolism, p 201–233. In Tang Y-W, Sussman M, Liu D, Poxton I, Schwartzman J (ed), Molecular medical microbiology, 2nd ed. Academic Press, Boston, MA. [Google Scholar]
- 2.Shimizu K. 2013. Metabolic regulation of a bacterial cell system with emphasis on Escherichia coli metabolism. ISRN Biochem 2013:645983. 10.1155/2013/645983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi X, Wegener-Feldbrügge S, Huntley S, Hamann N, Hedderich R, Søgaard-Andersen L. 2008. Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. J Bacteriol 190:613–624. 10.1128/JB.01502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaller GE, Shiu S-H, Armitage JP. 2011. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 21:R320–R330. 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 5.Gao R, Mack TR, Stock AM. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 32:225–234. 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groisman EA. 2016. Feedback control of two-component regulatory systems. Annu Rev Microbiol 70:103–124. 10.1146/annurev-micro-102215-095331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malpica R, Sandoval GRP, Rodríguez C, Franco B, Georgellis D. 2006. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal 8:781–795. 10.1089/ars.2006.8.781. [DOI] [PubMed] [Google Scholar]
- 9.Iuchi S, Lin EC. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA 85:1888–1892. 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iuchi S, Cameron DC, Lin EC. 1989. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol 171:868–873. 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iuchi S, Matsuda Z, Fujiwara T, Lin EC. 1990. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol 4:715–727. 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 12.Georgellis D, Lynch AS, Lin EC. 1997. In vitro phosphorylation study of the arc two-component signal transduction system of Escherichia coli. J Bacteriol 179:5429–5435. 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon O, Georgellis D, Lin EC. 2000. Phosphorelay as the sole physiological route of signal transmission by the Arc two-component system of Escherichia coli. J Bacteriol 182:3858–3862. 10.1128/JB.182.13.3858-3862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salmon KA, Hung S, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. 2005. Global gene expression profiling in Escherichia coli K12 effects of oxygen availability and ArcA. J Biol Chem 280:15084–15096. 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 15.Huvet M, Toni T, Sheng X, Thorne T, Jovanovic G, Engl C, Buck M, Pinney JW, Stumpf MPH. 2011. The evolution of the phage shock protein response system: interplay between protein function, genomic organization, and system function. Mol Biol Evol 28:1141–1155. 10.1093/molbev/msq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez AF, Georgellis D. 2010. In vitro and in vivo analysis of the ArcB/A redox signaling pathway. Methods Enzymol 471:205–228. 10.1016/S0076-6879(10)71012-0. [DOI] [PubMed] [Google Scholar]
- 17.Cabezas CE, Laulié AM, Briones AC, Pardo-Esté C, Lorca DE, Cofré AA, Morales EH, Mora AY, Krüger GI, Bueno SM, Hidalgo AA, Saavedra CP. 2021. Activation of regulator ArcA in the presence of hypochlorite in Salmonella enterica serovar Typhimurium. Biochimie 180:178–185. 10.1016/j.biochi.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Peebo K, Valgepea K, Nahku R, Riis G, Õun M, Adamberg K, Vilu R. 2014. Coordinated activation of PTA-ACS and TCA cycles strongly reduces overflow metabolism of acetate in Escherichia coli. Appl Microbiol Biotechnol 98:5131–5143. 10.1007/s00253-014-5613-y. [DOI] [PubMed] [Google Scholar]
- 19.Portnoy VA, Scott DA, Lewis NE, Tarasova Y, Osterman AL, Palsson BØ. 2010. Deletion of genes encoding cytochrome oxidases and quinol monooxygenase blocks the aerobic-anaerobic shift in Escherichia coli K-12 MG1655. Appl Environ Microbiol 76:6529–6540. 10.1128/AEM.01178-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iuchi S, Chepuri V, Fu HA, Gennis RB, Lin EC. 1990. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J Bacteriol 172:6020–6025. 10.1128/jb.172.10.6020-6025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nochino N, Toya Y, Shimizu H. 2020. Transcription factor ArcA is a flux sensor for the oxygen consumption rate in Escherichia coli. Biotechnol J 15:1900353. 10.1002/biot.201900353. [DOI] [PubMed] [Google Scholar]
- 22.Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J Bacteriol 185:204–209. 10.1128/JB.185.1.204-209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Graef MR, Alexeeva S, Snoep JL, Teixeira de Mattos MJ. 1999. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J Bacteriol 181:2351–2357. 10.1128/JB.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gunsalus RP, Park S-J. 1994. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol 145:437–450. 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 25.Green J, Paget MS. 2004. Bacterial redox sensors. Nat Rev Microbiol 2:954–966. 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez AF, Rodriguez C, Georgellis D. 2013. Ubiquinone and menaquinone electron carriers represent the yin and yang in the redox regulation of the ArcB sensor kinase. J Bacteriol 195:3054–3061. 10.1128/JB.00406-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Beilen JWA, Hellingwerf KJ. 2016. All three endogenous quinone species of Escherichia coli are involved in controlling the activity of the aerobic/anaerobic response regulator ArcA. Front Microbiol 7:1339. 10.3389/fmicb.2016.01339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolfe MD, Beek AT, Graham AI, Trotter EW, Asif HMS, Sanguinetti G, de Mattos JT, Poole RK, Green J. 2011. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem 286:10147–10154. 10.1074/jbc.M110.211144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iuchi S. 1993. Phosphorylation/dephosphorylation of the receiver module at the conserved aspartate residue controls transphosphorylation activity of histidine kinase in sensor protein ArcB of Escherichia coli. J Biol Chem 268:23972–23980. 10.1016/S0021-9258(20)80480-3. [DOI] [PubMed] [Google Scholar]
- 30.Georgellis D, Kwon O, Lin EC. 2001. Quinones as the redox signal for the Arc two-component system of bacteria. Science 292:2314–2316. 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- 31.Serna A, Espinosa E, Camacho EM, Casadesús J. 2010. Regulation of bacterial conjugation in microaerobiosis by host-encoded functions ArcAB and SdhABCD. Genetics 184:947–958. 10.1534/genetics.109.109918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng C, Chen J, Shan Y, Fang C, Liu Y, Xia Y, Song H, Fang W. 2013. Listeria monocytogenes ArcA contributes to acid tolerance. J Med Microbiol 62:813–821. 10.1099/jmm.0.055145-0. [DOI] [PubMed] [Google Scholar]
- 33.Longo P, Ota-Tsuzuki C, Nunes A, Fernandes B, Mintz K, Fives-Taylor P, Mayer M. 2009. Aggregatibacter actinomycetemcomitans arcB influences hydrophobic properties, biofilm formation and adhesion to hydroxyapatite. Braz J Microbiol 40:550–562. 10.1590/S1517-838220090003000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol Microbiol 65:538–553. 10.1111/j.1365-2958.2007.05809.x. [DOI] [PubMed] [Google Scholar]
- 35.Lu J, Peng Y, Wan S, Frost LS, Raivio T, Glover JNM. 2019. Cooperative function of TraJ and ArcA in regulating the F plasmid tra operon. J Bacteriol 201:e00448-18. 10.1128/JB.00448-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park DM, Akhtar MS, Ansari AZ, Landick R, Kiley PJ. 2013. The bacterial response regulator ArcA uses a diverse binding site architecture to regulate carbon oxidation globally. PLoS Genet 9:e1003839. 10.1371/journal.pgen.1003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun H, Song Y, Chen F, Zhou C, Liu P, Fan Y, Zheng Y, Wan X, Feng L. 2020. An ArcA-modulated small RNA in pathogenic Escherichia coli K1. Front Microbiol 11:574833. 10.3389/fmicb.2020.574833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iyer MS, Pal A, Srinivasan S, Somvanshi PR, Venkatesh KV. 2021. Global transcriptional regulators fine-tune the translational and metabolic efficiency for optimal growth of Escherichia coli. mSystems 6:e00001-21. 10.1128/mSystems.00001-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Federowicz S, Kim D, Ebrahim A, Lerman J, Nagarajan H, Cho B, Zengler K, Palsson B. 2014. Determining the control circuitry of redox metabolism at the genome-scale. PLoS Genet 10:e1004264. 10.1371/journal.pgen.1004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G-W, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ćudić E, Surmann K, Panasia G, Hammer E, Hunke S. 2017. The role of the two-component systems Cpx and Arc in protein alterations upon gentamicin treatment in Escherichia coli. BMC Microbiol 17:197. 10.1186/s12866-017-1100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gunsalus RP. 1992. Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J Bacteriol 174:7069–7074. 10.1128/jb.174.22.7069-7074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capra EJ, Laub MT. 2012. Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem 280:1448–1456. 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 45.Iuchi S, Lin EC. 1992. Mutational analysis of signal transduction by ArcB, a membrane sensor protein responsible for anaerobic repression of operons involved in the central aerobic pathways in Escherichia coli. J Bacteriol 174:3972–3980. 10.1128/jb.174.12.3972-3980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shalel Levanon S, San K-Y, Bennett GN. 2005. Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol Bioeng 92:147–159. 10.1002/bit.20583. [DOI] [PubMed] [Google Scholar]
- 47.Ramos PIP, Custódio MGF, del Rosario Quispe Saji G, Cardoso T, da Silva GL, Braun G, Martins WMBS, Girardello R, de Vasconcelos ATR, Fernández E, Gales AC, Nicolás MF. 2016. The polymyxin B-induced transcriptomic response of a clinical, multidrug-resistant Klebsiella pneumoniae involves multiple regulatory elements and intracellular targets. BMC Genomics 17:737. 10.1186/s12864-016-3070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandin P, Gottesman S. 2010. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 29:3094–3107. 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Compan I, Touati D. 1994. Anaerobic activation of arcA transcription in Escherichia coli: roles of Fnr and ArcA. Mol Microbiol 11:955–964. 10.1111/j.1365-2958.1994.tb00374.x. [DOI] [PubMed] [Google Scholar]
- 50.Ireland WT, Beeler SM, Flores-Bautista E, McCarty NS, Röschinger T, Belliveau NM, Sweredoski MJ, Moradian A, Kinney JB, Phillips R. 2020. Deciphering the regulatory genome of Escherichia coli, one hundred promoters at a time. eLife 9:e55308. 10.7554/eLife.55308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jordan PA, Thomson AJ, Ralph ET, Guest JR, Green J. 1997. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett 416:349–352. 10.1016/s0014-5793(97)01219-2. [DOI] [PubMed] [Google Scholar]
- 52.Crack J, Green J, Thomson AJ. 2004. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR). J Biol Chem 279:9278–9286. 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- 53.Marzan LW, Hasan CMM, Shimizu K. 2013. Effect of acidic condition on the metabolic regulation of Escherichia coli and its phoB mutant. Arch Microbiol 195:161–171. 10.1007/s00203-012-0861-7. [DOI] [PubMed] [Google Scholar]
- 54.Park DM, Kiley PJ. 2014. The influence of repressor DNA binding site architecture on transcriptional control. mBio 5:e01684-14. 10.1128/mBio.01684-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwon O, Georgellis D, Lynch AS, Boyd D, Lin ECC. 2000. The ArcB sensor kinase of Escherichia coli: genetic exploration of the transmembrane region. J Bacteriol 182:2960–2966. 10.1128/JB.182.10.2960-2966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maslennikov I, Klammt C, Hwang E, Kefala G, Okamura M, Esquivies L, Mörs K, Glaubitz C, Kwiatkowski W, Jeon YH, Choe S. 2010. Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis. Proc Natl Acad Sci USA 107:10902–10907. 10.1073/pnas.1001656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA 101:13318–13323. 10.1073/pnas.0403064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev 70:910–938. 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinoshita-Kikuta E, Kinoshita E, Eguchi Y, Koike T. 2016. Validation of cis and trans modes in multistep phosphotransfer signaling of bacterial tripartite sensor kinases by using Phos-Tag SDS-PAGE. PLoS One 11:e0148294. 10.1371/journal.pone.0148294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato M, Mizuno T, Shimizu T, Hakoshima T. 1997. Insights into multistep phosphorelay from the crystal structure ofl the C-terminal HPt domain of ArcB. Cell 88:717–723. 10.1016/s0092-8674(00)81914-5. [DOI] [PubMed] [Google Scholar]
- 61.Tsuzuki M, Ishige K, Mizuno T. 1995. Phosphotransfer circuitry of the putative multi-signal transducer, ArcB, of Escherichia coli: in vitro studies with mutants. Mol Microbiol 18:953–962. 10.1111/j.1365-2958.1995.18050953.x. [DOI] [PubMed] [Google Scholar]
- 62.Matsushika A, Mizuno T. 1998. A dual-signaling mechanism mediated by the ArcB hybrid sensor kinase containing the histidine-containing phosphotransfer domain in Escherichia coli. J Bacteriol 180:3973–3977. 10.1128/JB.180.15.3973-3977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsushika A, Mizuno T. 1998. The structure and function of the histidine-containing phosphotransfer (HPt) signaling domain of the Escherichia coli ArcB sensor. J Biochem 124:440–445. 10.1093/oxfordjournals.jbchem.a022132. [DOI] [PubMed] [Google Scholar]
- 64.Iuchi S, Lin EC. 1992. Purification and phosphorylation of the Arc regulatory components of Escherichia coli. J Bacteriol 174:5617–5623. 10.1128/jb.174.17.5617-5623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iuchi S, Weiner L. 1996. Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments. J Biochem 120:1055–1063. 10.1093/oxfordjournals.jbchem.a021519. [DOI] [PubMed] [Google Scholar]
- 66.Ishige K, Nagasawa S, Tokishita S, Mizuno T. 1994. A novel device of bacterial signal transducers. EMBO J 13:5195–5202. 10.1002/j.1460-2075.1994.tb06850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nuñez Oreza LA, Alvarez AF, Arias-Olguín II, Torres Larios A, Georgellis D. 2012. The ArcB leucine zipper domain is required for proper ArcB signaling. PLoS One 7:e38187. 10.1371/journal.pone.0038187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. 10.1128/MMBR.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsushika A, Mizuno T. 2000. Characterization of three putative sub-domains in the signal-input domain of the ArcB hybrid sensor in Escherichia coli. J Biochem 127:855–860. 10.1093/oxfordjournals.jbchem.a022679. [DOI] [PubMed] [Google Scholar]
- 70.Zhulin IB, Taylor BL, Dixon R. 1997. PAS domain S-boxes in archaea, bacteria and sensors for oxygen and redox. Trends Biochem Sci 22:331–333. 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 71.Georgellis D, Kwon O, Lin ECC, Wong SM, Akerley BJ. 2001. Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J Bacteriol 183:7206–7212. 10.1128/JB.183.24.7206-7212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen M-P, Yoon J-M, Cho M-H, Lee S-W. 2015. Prokaryotic 2-component systems and the OmpR/PhoB superfamily. Can J Microbiol 61:799–810. 10.1139/cjm-2015-0345. [DOI] [PubMed] [Google Scholar]
- 73.Toro-Roman A, Mack TR, Stock AM. 2005. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the α4-β5-α5 Face. J Mol Biol 349:11–26. 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lynch AS, Lin EC. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol 178:6238–6249. 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martínez-Hackert E, Stock AM. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol 269:301–312. 10.1006/jmbi.1997.1065. [DOI] [PubMed] [Google Scholar]
- 76.Kenney LJ. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol 5:135–141. 10.1016/s1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- 77.Galperin MY. 2010. Diversity of structure and function of response regulator output domains. Curr Opin Microbiol 13:150–159. 10.1016/j.mib.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joyce AP, Havranek JJ. 2018. Deciphering the protein-DNA code of bacterial winged helix-turn-helix transcription factors. Quant Biol 6:68–84. 10.1007/s40484-018-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peña-Sandoval GR, Georgellis D. 2010. The ArcB sensor kinase of Escherichia coli autophosphorylates by an intramolecular reaction. J Bacteriol 192:1735–1739. 10.1128/JB.01401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Teran-Melo JL, Peña-Sandoval GR, Silva-Jimenez H, Rodriguez C, Alvarez AF, Georgellis D. 2018. Routes of phosphoryl group transfer during signal transmission and signal decay in the dimeric sensor histidine kinase ArcB. J Biol Chem 293:13214–13223. 10.1074/jbc.RA118.003910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26:369–376. 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 82.Drapal N, Sawers G. 1995. Purification of ArcA and analysis of is specific interaction with the pfl promoter-regulatory region. Mol Microbiol 16:597–607. 10.1111/j.1365-2958.1995.tb02422.x. [DOI] [PubMed] [Google Scholar]
- 83.Strohmaier H, Noiges R, Kotschan S, Sawers G, Högenauer G, Zechner EL, Koraimann G. 1998. Signal transduction and bacterial conjugation: characterization of the role of ArcA in regulating conjugative transfer of the resistance plasmid R1. J Mol Biol 277:309–316. 10.1006/jmbi.1997.1598. [DOI] [PubMed] [Google Scholar]
- 84.Liu X, Peña Sandoval GR, Wanner BL, Jung WS, Georgellis D, Kwon O. 2009. Evidence against the physiological role of acetyl phosphate in the phosphorylation of the ArcA response regulator in Escherichia coli. J Microbiol 47:657–662. 10.1007/s12275-009-0087-9. [DOI] [PubMed] [Google Scholar]
- 85.Shen J, Gunsalus RP. 1997. Role of multiple ArcA recognition sites in anaerobic regulation of succinate dehydrogenase (sdhCDAB) gene expression in Escherichia coli. Mol Microbiol 26:223–236. 10.1046/j.1365-2958.1997.5561923.x. [DOI] [PubMed] [Google Scholar]
- 86.Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J Biol Chem 276:40873–40879. 10.1074/jbc.M104855200. [DOI] [PubMed] [Google Scholar]
- 87.Mika F, Hengge R. 2005. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli. Genes Dev 19:2770–2781. 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pellicer MT, Lynch AS, De Wulf P, Boyd D, Aguilar J, Lin EC. 1999. A mutational study of the ArcA-P binding sequences in the aldA promoter of Escherichia coli. Mol Gen Genet 261:170–176. 10.1007/s004380050954. [DOI] [PubMed] [Google Scholar]
- 89.McGuire AM, De Wulf P, Church GM, Lin EC. 1999. A weight matrix for binding recognition by the redox-response regulator ArcA-P of Escherichia coli. Mol Microbiol 32:219–221. 10.1046/j.1365-2958.1999.01347.x. [DOI] [PubMed] [Google Scholar]
- 90.Liu X, De Wulf P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem 279:12588–12597. 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 91.Favorov AV, Gelfand MS, Gerasimova AV, Ravcheev DA, Mironov AA, Makeev VJ. 2005. A Gibbs sampler for identification of symmetrically structured, spaced DNA motifs with improved estimation of the signal length. Bioinformatics 21:2240–2245. 10.1093/bioinformatics/bti336. [DOI] [PubMed] [Google Scholar]
- 92.Wang X, Gao H, Shen Y, Weinstock GM, Zhou J, Palzkill T. 2008. A high-throughput percentage-of-binding strategy to measure binding energies in DNA–protein interactions: application to genome-scale site discovery. Nucleic Acids Res 36:4863–4871. 10.1093/nar/gkn477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gao H, Wang X, Yang ZK, Palzkill T, Zhou J. 2008. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9:42. 10.1186/1471-2164-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong SMS, St Michael F, Cox A, Ram S, Akerley BJ. 2011. ArcA-regulated glycosyltransferase Lic2B promotes complement evasion and pathogenesis of nontypeable Haemophilus influenzae. Infect Immun 79:1971–1983. 10.1128/IAI.01269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, Hassan HM. 2011. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol 11:58. 10.1186/1471-2180-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Georgellis D, Kwon O, De Wulf P, Lin EC. 1998. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem 273:32864–32869. 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 97.Peña-Sandoval GR, Kwon O, Georgellis D. 2005. Requirement of the receiver and phosphotransfer domains of ArcB for efficient dephosphorylation of phosphorylated ArcA in vivo. J Bacteriol 187:3267–3272. 10.1128/JB.187.9.3267-3272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matsubara M, Mizuno T. 2000. The SixA phospho‐histidine phosphatase modulates the ArcB phosphorelay signal transduction in Escherichia coli. FEBS Lett 470:118–124. 10.1016/S0014-5793(00)01303-X. [DOI] [PubMed] [Google Scholar]
- 99.Ogino T, Matsubara M, Kato N, Nakamura Y, Mizuno T. 1998. An Escherichia coli protein that exhibits phosphohistidine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol Microbiol 27:573–585. 10.1046/j.1365-2958.1998.00703.x. [DOI] [PubMed] [Google Scholar]
- 100.Hakoshima T, Ichihara H. 2007. Structure of SixA, a histidine protein phosphatase of the ArcB histidine-containing phosphotransfer domain in Escherichia coli. Methods Enzymol 422:288–304. 10.1016/S0076-6879(06)22014-7. [DOI] [PubMed] [Google Scholar]
- 101.Schulte JE, Goulian M. 2018. The phosphohistidine phosphatase SixA targets a phosphotransferase system. mBio 9:e01666-18. 10.1128/mBio.01666-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roeder W, Somerville RL. 1979. Cloning the trpR gene. Mol Gen Genet 176:361–368. 10.1007/BF00333098. [DOI] [PubMed] [Google Scholar]
- 103.Kashef N, Hamblin MR. 2017. Can microbial cells develop resistance to oxidative stress in antimicrobial photodynamic inactivation? Drug Resist Updat 31:31–42. 10.1016/j.drup.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Alvarez AF, Malpica R, Contreras M, Escamilla E, Georgellis D. 2010. Cytochrome d but not cytochrome o rescues the toluidine blue growth sensitivity of arc mutants of Escherichia coli. J Bacteriol 192:391–399. 10.1128/JB.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Georgellis D, Kwon O, Lin EC. 1999. Amplification of signaling activity of the Arc two-component system of Escherichia coli by anaerobic metabolites: an in vitro study with different protein modules. J Biol Chem 274:35950–35954. 10.1074/jbc.274.50.35950. [DOI] [PubMed] [Google Scholar]
- 106.Rodriguez C, Kwon O, Georgellis D. 2004. Effect of d-lactate on the physiological activity of the ArcB sensor kinase in Escherichia coli. J Bacteriol 186:2085–2090. 10.1128/JB.186.7.2085-2090.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin ECC, Iuchi S. 1991. Regulation of gene expression in fermentative and respiratory systems in Escherichia coli and related bacteria. Annu Rev Genet 25:361–387. 10.1146/annurev.ge.25.120191.002045. [DOI] [PubMed] [Google Scholar]
- 108.Nowicka B, Kruk J. 2010. Occurrence, biosynthesis, and function of isoprenoid quinones. Biochim Biophys Acta Bioenerg 1797:1587–1605. 10.1016/j.bbabio.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 109.Collins MD, Jones D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol Rev 45:316–354. 10.1128/mr.45.2.316-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meganathan R, Kwon O. 2009. Biosynthesis of menaquinone (vitamin K2) and ubiquinone (coenzyme Q). EcoSal Plus 3:ecosalplus.3.6.3.3. 10.1128/ecosalplus.3.6.3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.IUPAC-IYB Commission on Biochemical Nomenclature. 1965. Tentative rules on nomenclature of quinones with isoprenoid side-chains. Biochim Biophys Acta 107:5–10. 10.1016/0304-4165(65)90382-X. [DOI] [PubMed] [Google Scholar]
- 112.Unden G, Bongaerts J. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta Bioenerg 1320:217–234. 10.1016/S0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 113.Nitzschke A, Bettenbrock K. 2018. All three quinone species play distinct roles in ensuring optimal growth under aerobic and fermentative conditions in E. coli K12. PLoS One 13:e0194699. 10.1371/journal.pone.0194699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Borisov VB, Verkhovsky MI. 2015. Oxygen as acceptor. EcoSal Plus 6:ecosalplus.ESP-0012-2015. 10.1128/ecosalplus.ESP-0012-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Søballe B, Poole RK. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817–1830. 10.1099/13500872-145-8-1817. [DOI] [PubMed] [Google Scholar]
- 116.Sharma P, Teixeira de Mattos MJ, Hellingwerf KJ, Bekker M. 2012. On the function of the various quinone species in Escherichia coli. FEBS J 279:3364–3373. 10.1111/j.1742-4658.2012.08608.x. [DOI] [PubMed] [Google Scholar]
- 117.Weiss SA, Jeuken LJC. 2009. Electrodes modified with lipid membranes to study quinone oxidoreductases. Biochem Soc Trans 37:707–712. 10.1042/BST0370707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sharma P, Stagge S, Bekker M, Bettenbrock K, Hellingwerf KJ. 2013. Kinase activity of ArcB from Escherichia coli is subject to regulation by both ubiquinone and demethylmenaquinone. PLoS One 8:e75412. 10.1371/journal.pone.0075412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bekker M, Alexeeva S, Laan W, Sawers G, Teixeira de Mattos J, Hellingwerf KJ. 2010. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J Bacteriol 192:746–754. 10.1128/JB.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bekker M, Kramer G, Hartog AF, Wagner MJ, de Koster CG, Hellingwerf KJ, Teixeira de Mattos MJ. 2007. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153:1974–1980. 10.1099/mic.0.2007/006098-0. [DOI] [PubMed] [Google Scholar]
- 121.Aussel L, Pierrel F, Loiseau L, Lombard M, Fontecave M, Barras F. 2014. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta 1837:1004–1011. 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 122.Alexeeva S, de Kort B, Sawers G, Hellingwerf KJ, Teixeira de Mattos MJ. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J Bacteriol 182:4934–4940. 10.1128/JB.182.17.4934-4940.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shestopalov AI, Bogachev AV, Murtazina RA, Viryasov MB, Skulachev VP. 1997. Aeration-dependent changes in composition of the quinone pool in Escherichia coli. FEBS Lett 404:272–274. 10.1016/s0014-5793(97)00143-9. [DOI] [PubMed] [Google Scholar]
- 124.Martínez-Antonio A, Collado-Vides J. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol 6:482–489. 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 125.Kargeti M, Venkatesh KV. 2017. The effect of global transcriptional regulators on the anaerobic fermentative metabolism of Escherichia coli. Mol Biosyst 13:1388–1398. 10.1039/c6mb00721j. [DOI] [PubMed] [Google Scholar]
- 126.Kothamachu VB, Feliu E, Wiuf C, Cardelli L, Soyer OS. 2013. Phosphorelays provide tunable signal processing capabilities for the cell. PLoS Comput Biol 9:e1003322. 10.1371/journal.pcbi.1003322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Perrenoud A, Sauer U. 2005. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J Bacteriol 187:3171–3179. 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Smirnova GV, Tyulenev AV, Muzyka NG, Oktyabrsky ON. 2020. Study of the relationship between extracellular superoxide and glutathione production in batch cultures of Escherichia coli. Res Microbiol 171:301–310. 10.1016/j.resmic.2020.07.004. [DOI] [PubMed] [Google Scholar]
- 129.Toya Y, Nakahigashi K, Tomita M, Shimizu K. 2012. Metabolic regulation analysis of wild-type and arcA mutant Escherichia coli under nitrate conditions using different levels of omics data. Mol Biosyst 8:2593–2604. 10.1039/c2mb25069a. [DOI] [PubMed] [Google Scholar]
- 130.Shalel-Levanon S, San K-Y, Bennett GN. 2005. Effect of oxygen, and ArcA and FNR regulators on the expression of genes related to the electron transfer chain and the TCA cycle in Escherichia coli. Metab Eng 7:364–374. 10.1016/j.ymben.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 131.Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. 2006. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol 72:3653–3661. 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Waegeman H, Beauprez J, Moens H, Maertens J, De Mey M, Foulquié-Moreno MR, Heijnen JJ, Charlier D, Soetaert W. 2011. Effect of iclR and arcA knockouts on biomass formation and metabolic fluxes in Escherichia coli K12 and its implications on understanding the metabolism of Escherichia coli BL21(DE3). BMC Microbiol 11:70. 10.1186/1471-2180-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Förster AH, Gescher J. 2014. Metabolic engineering of Escherichia coli for production of mixed-acid fermentation end products. Front Bioeng Biotechnol 2:16. 10.3389/fbioe.2014.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bidart GN, Ruiz JA, de Almeida A, Méndez BS, Nikel PI. 2012. Manipulation of the anoxic metabolism in Escherichia coli by ArcB deletion variants in the ArcBA two-component system. Appl Environ Microbiol 78:8784–8794. 10.1128/AEM.02558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Basan M, Hui S, Williamson JR. 2017. ArcA overexpression induces fermentation and results in enhanced growth rates of E. coli. Sci Rep 7:11866. 10.1038/s41598-017-12144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu M, Yao L, Xian M, Ding Y, Liu H, Zhao G. 2016. Deletion of arcA increased the production of acetyl-CoA-derived chemicals in recombinant Escherichia coli. Biotechnol Lett 38:97–101. 10.1007/s10529-015-1953-7. [DOI] [PubMed] [Google Scholar]
- 137.Ruiz JA, de Almeida A, Godoy MS, Mezzina MP, Bidart GN, Méndez BS, Pettinari MJ, Nikel PI. 2012. Escherichia coli redox mutants as microbial cell factories for the synthesis of reduced biochemicals. Comput Struct Biotechnol J 3:e201210019. 10.5936/csbj.201210019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Matsuoka Y, Kurata H. 2017. Modeling and simulation of the redox regulation of the metabolism in Escherichia coli at different oxygen concentrations. Biotechnol Biofuels 10:183. 10.1186/s13068-017-0867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Egoburo DE, Diaz Peña R, Alvarez DS, Godoy MS, Mezzina MP, Pettinari MJ. 2018. Microbial cell factories à la carte: elimination of global regulators Cra and ArcA generates metabolic backgrounds suitable for the synthesis of bioproducts in Escherichia coli. Appl Environ Microbiol 84:e01337-18. 10.1128/AEM.01337-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pettinari MJ, Nikel PI, Ruiz JA, Méndez BS. 2008. ArcA redox mutants as a source of reduced bioproducts. J Mol Microbiol Biotechnol 15:41–47. 10.1159/000111991. [DOI] [PubMed] [Google Scholar]
- 141.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. 2004. Integrating high-throughput and computational data elucidates bacterial networks. Nature 429:92–96. 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 142.Iuchi S, Aristarkhov A, Dong JM, Taylor JS, Lin EC. 1994. Effects of nitrate respiration on expression of the Arc-controlled operons encoding succinate dehydrogenase and flavin-linked L-lactate dehydrogenase. J Bacteriol 176:1695–1701. 10.1128/jb.176.6.1695-1701.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Park S-J, Tseng C-P, Gunsalus RP. 1995. Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coli in response to carbon supply and anaerobiosis: role of ArcA and Fnr. Mol Microbiol 15:473–482. 10.1111/j.1365-2958.1995.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 144.Denby KJ, Rolfe MD, Crick E, Sanguinetti G, Poole RK, Green J. 2015. Adaptation of anaerobic cultures of Escherichia coli K-12 in response to environmental trimethylamine-N-oxide. Environ Microbiol 17:2477–2491. 10.1111/1462-2920.12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yoshida H, Shimada T, Ishihama A. 2018. Coordinated hibernation of transcriptional and translational apparatus during growth transition of Escherichia coli to stationary phase. mSystems 3:e00057-18. 10.1128/mSystems.00057-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gottesman S. 2019. Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J Biol Chem 294:11685–11700. 10.1074/jbc.REV119.005593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Battesti A, Majdalani N, Gottesman S. 2015. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci USA 112:5159–5164. 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen AI, Goulian M. 2018. A network of regulators promotes dehydration tolerance in Escherichia coli. Environ Microbiol 20:1283–1295. 10.1111/1462-2920.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nyström T, Larsson C, Gustafsson L. 1996. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J 15:3219–3228. 10.1002/j.1460-2075.1996.tb00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gonidakis S, Finkel SE, Longo VD. 2010. E. coli hypoxia-inducible factor ArcA mediates lifespan extension in a lipoic acid synthase mutant by suppressing acetyl-CoA synthetase. Biol Chem 391:1139–1147. 10.1515/bc.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gonidakis S, Finkel SE, Longo VD. 2010. Genome-wide screen identifies Escherichia coli TCA cycle-related mutants with extended chronological lifespan dependent on acetate metabolism and the hypoxia-inducible transcription factor ArcA. Aging Cell 9:868–881. 10.1111/j.1474-9726.2010.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hidese R, Mihara H, Kurihara T, Esaki N. 2014. Global identification of genes affecting iron-sulfur cluster biogenesis and iron homeostasis. J Bacteriol 196:1238–1249. 10.1128/JB.01160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. 2013. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta 1827:455–469. 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 155.Cotter PA, Darie S, Gunsalus RP. 1992. The effect of iron limitation on expression of the aerobic and anaerobic electron transport pathway genes in Escherichia coli. FEMS Microbiol Lett 100:227–232. 10.1111/j.1574-6968.1992.tb14045.x. [DOI] [PubMed] [Google Scholar]
- 156.Chareyre S, Barras F, Mandin P. 2019. A small RNA controls bacterial sensitivity to gentamicin during iron starvation. PLoS Genet 15:e1008078. 10.1371/journal.pgen.1008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Folsom JP, Parker AE, Carlson RP. 2014. Physiological and proteomic analysis of Escherichia coli iron-limited chemostat growth. J Bacteriol 196:2748–2761. 10.1128/JB.01606-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fong KP, Gao L, Demuth DR. 2003. luxS and arcB control aerobic growth of Actinobacillus actinomycetemcomitans under iron limitation. Infect Immun 71:298–308. 10.1128/IAI.71.1.298-308.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tardat B, Touati D. 1993. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol Microbiol 9:53–63. 10.1111/j.1365-2958.1993.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 160.Shimizu K. 2016. Metabolic regulation and coordination of the metabolism in bacteria in response to a variety of growth conditions. Adv Biochem Eng Biotechnol 155:1–54. 10.1007/10_2015_320. [DOI] [PubMed] [Google Scholar]
- 161.Liu J, Chakraborty S, Hosseinzadeh P, Yu Y, Tian S, Petrik I, Bhagi A, Lu Y. 2014. Metalloproteins containing cytochrome, iron–sulfur, or copper redox centers. Chem Rev 114:4366–4469. 10.1021/cr400479b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cotter PA, Melville SB, Albrecht JA, Gunsalus RP. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol 25:605–615. 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 163.Zupok A, Gorka M, Siemiatkowska B, Skirycz A, Leimkühler S. 2019. Iron-dependent regulation of molybdenum cofactor biosynthesis genes in Escherichia coli. J Bacteriol 201:e00382-19. 10.1128/JB.00382-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Szenk M, Dill KA, de Graff AMR. 2017. Why do fast-growing bacteria enter overflow metabolism? Testing the membrane real estate hypothesis. Cell Syst 5:95–104. 10.1016/j.cels.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 165.Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, Hwa T. 2015. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528:99–104. 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kiley PJ, Beinert H. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev 22:341–352. 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 167.Crack JC, Green J, Thomson AJ, Le Brun NE. 2014. Iron–sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Acc Chem Res 47:3196–3205. 10.1021/ar5002507. [DOI] [PubMed] [Google Scholar]
- 168.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187:1135–1160. 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sawers G, Suppmann B. 1992. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J Bacteriol 174:3474–3478. 10.1128/jb.174.11.3474-3478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Levanon SS, San K-Y, Bennett GN. 2005. Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol Bioeng 89:556–564. 10.1002/bit.20381. [DOI] [PubMed] [Google Scholar]
- 171.Partridge JD, Sanguinetti G, Dibden DP, Roberts RE, Poole RK, Green J. 2007. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J Biol Chem 282:11230–11237. 10.1074/jbc.M700728200. [DOI] [PubMed] [Google Scholar]
- 172.Cotter PA, Gunsalus RP. 1992. Contribution of the fnr and arcA gene products in coordinate regulation of cytochrome o and d oxidase (cyoABCDE and cydAB) genes in Escherichia coli. FEMS Microbiol Lett 91:31–36. 10.1111/j.1574-6968.1992.tb05179.x. [DOI] [PubMed] [Google Scholar]
- 173.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu Rev Genet 41:121–145. 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 174.Verhamme DT, Arents JC, Postma PW, Crielaard W, Hellingwerf KJ. 2002. Investigation of in vivo cross-talk between key two-component systems of Escherichia coli. Microbiology (Reading) 148:69–78. 10.1099/00221287-148-1-69. [DOI] [PubMed] [Google Scholar]
- 175.Francis VI, Porter SL. 2019. Multikinase networks: two-component signaling networks integrating multiple stimuli. Annu Rev Microbiol 73:199–223. 10.1146/annurev-micro-020518-115846. [DOI] [PubMed] [Google Scholar]
- 176.Matsubara M, Kitaoka S, Takeda S, Mizuno T. 2000. Tuning of the porin expression under anaerobic growth conditions by His-to-Asp cross-phosphorelay through both the EnvZ-osmosensor and ArcB-anaerosensor in Escherichia coli. Genes Cells 5:555–569. 10.1046/j.1365-2443.2000.00347.x. [DOI] [PubMed] [Google Scholar]
- 177.Jovanovic G, Lloyd LJ, Stumpf MPH, Mayhew AJ, Buck M. 2006. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem 281:21147–21161. 10.1074/jbc.M602323200. [DOI] [PubMed] [Google Scholar]
- 178.Jovanovic G, Engl C, Buck M. 2009. Physical, functional and conditional interactions between ArcAB and phage shock proteins upon secretin-induced stress in Escherichia coli. Mol Microbiol 74:16–28. 10.1111/j.1365-2958.2009.06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MPH, Buck M. 2010. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev 34:797–827. 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 180.Bogachev AV, Murtazina RA, Shestopalov AI, Skulachev VP. 1995. Induction of the Escherichia coli cytochrome d by low δμ H+ and by sodium ions. Eur J Biochem 232:304–308. 10.1111/j.1432-1033.1995.tb20812.x. [DOI] [PubMed] [Google Scholar]
- 181.Liang H, Zhang Y, Wang S, Gao H. 2020. Mutual interplay between ArcA and σE orchestrates envelope stress response in Shewanella oneidensis. Environ Microbiol 23:652–668. 10.1111/1462-2920.15060. [DOI] [PubMed] [Google Scholar]
- 182.Xie P, Liang H, Wang J, Huang Y, Gao H. 2021. Lipopolysaccharide transport system links physiological roles of σE and ArcA in the cell envelope biogenesis in Shewanella oneidensis. Microbiol Spectr 9:e0069021. 10.1128/Spectrum.00690-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ayoub Moubareck C. 2020. Polymyxins and bacterial membranes: a review of antibacterial activity and mechanisms of resistance. Membranes 10:181. 10.3390/membranes10080181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Fu L, Wan M, Zhang S, Gao L, Fang W. 2020. Polymyxin B loosens lipopolysaccharide bilayer but stiffens phospholipid bilayer. Biophys J 118:138–150. 10.1016/j.bpj.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sabnis A, Hagart KLH, Klöckner A, Becce M, Evans LE, Furniss RCD, Mavridou DAI, Murphy R, Stevens MM, Davies JC, Larrouy-Maumus GJ, Clarke TB, Edwards AM. 2021. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife 10:e65836. 10.7554/eLife.65836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Deris ZZ, Akter J, Sivanesan S, Roberts KD, Thompson PE, Nation RL, Li J, Velkov T. 2014. A secondary mode of action of polymyxins against Gram-negative bacteria involves the inhibition of NADH-quinone oxidoreductase activity. J Antibiot (Tokyo) 67:147–151. 10.1038/ja.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. 2008. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated oxidative stress and cell death. Cell 135:679–690. 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Horinouchi T, Maeda T, Kotani H, Furusawa C. 2020. Suppression of antibiotic resistance evolution by single-gene deletion. Sci Rep 10:4178. 10.1038/s41598-020-60663-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yuan J, Wei B, Lipton MS, Gao H. 2012. Impact of ArcA loss in Shewanella oneidensis revealed by comparative proteomics under aerobic and anaerobic conditions. Proteomics 12:1957–1969. 10.1002/pmic.201100651. [DOI] [PubMed] [Google Scholar]
- 190.Wan F, Mao Y, Dong Y, Ju L, Wu G, Gao H. 2015. Impaired cell envelope resulting from arcA mutation largely accounts for enhanced sensitivity to hydrogen peroxide in Shewanella oneidensis. Sci Rep 5:10228. 10.1038/srep10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Imlay JA. 2019. Where in the world do bacteria experience oxidative stress? Environ Microbiol 21:521–530. 10.1111/1462-2920.14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Knoefler D, Leichert LIO, Thamsen M, Cremers CM, Reichmann D, Gray MJ, Wholey W-Y, Jakob U. 2014. About the dangers, costs and benefits of living an aerobic lifestyle. Biochem Soc Trans 42:917–921. 10.1042/BST20140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Imlay JA. 2015. Transcription factors that defend bacteria against reactive oxygen species. Annu Rev Microbiol 69:93–108. 10.1146/annurev-micro-091014-104322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Imlay JA, Sethu R, Rohaun SK. 2019. Evolutionary adaptations that enable enzymes to tolerate oxidative stress. Free Radic Biol Med 140:4–13. 10.1016/j.freeradbiomed.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Diaz JM, Hansel CM, Voelker BM, Mendes CM, Andeer PF, Zhang T. 2013. Widespread production of extracellular superoxide by heterotrophic bacteria. Science 340:1223–1226. 10.1126/science.1237331. [DOI] [PubMed] [Google Scholar]
- 196.van Hoek MJ, Merks RMH. 2012. Redox balance is key to explaining full vs. partial switching to low-yield metabolism. BMC Syst Biol 6:22. 10.1186/1752-0509-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ezraty B, Gennaris A, Barras F, Collet J-F. 2017. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 15:385–396. 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 198.Loui C, Chang AC, Lu S. 2009. Role of the ArcAB two-component system in the resistance of Escherichia coli to reactive oxygen stress. BMC Microbiol 9:183. 10.1186/1471-2180-9-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Morales EH, Collao B, Desai PT, Calderón IL, Gil F, Luraschi R, Porwollik S, McClelland M, Saavedra CP. 2013. Probing the ArcA regulon under aerobic/ROS conditions in Salmonella enterica serovar Typhimurium. BMC Genom 14:626. 10.1186/1471-2164-14-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wong SMS, Alugupalli KR, Ram S, Akerley BJ. 2007. The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol 64:1375–1390. 10.1111/j.1365-2958.2007.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Calderón IL, Morales E, Caro NJ, Chahúan CA, Collao B, Gil F, Villarreal JM, Ipinza F, Mora GC, Saavedra CP. 2011. Response regulator ArcA of Salmonella enterica serovar Typhimurium downregulates expression of OmpD, a porin facilitating uptake of hydrogen peroxide. Res Microbiol 162:214–222. 10.1016/j.resmic.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 202.Pardo-Esté C, Hidalgo AA, Aguirre C, Briones AC, Cabezas CE, Castro-Severyn J, Fuentes JA, Opazo CM, Riedel CA, Otero C, Pacheco R, Valvano MA, Saavedra CP. 2018. The ArcAB two-component regulatory system promotes resistance to reactive oxygen species and systemic infection by Salmonella Typhimurium. PLoS One 13:e0203497. 10.1371/journal.pone.0203497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Winterbourn C. 1995. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett 82–83:969–974. 10.1016/0378-4274(95)03532-X. [DOI] [PubMed] [Google Scholar]
- 204.Li Z, Demple B. 1994. SoxS, an activator of superoxide stress genes in Escherichia coli: purification and interaction with DNA. J Biol Chem 269:18371–18377. 10.1016/S0021-9258(17)32317-7. [DOI] [PubMed] [Google Scholar]
- 205.Søballe B, Poole RK. 2000. Ubiquinone limits oxidative stress in Escherichia coli. Microbiology 146:787–796. 10.1099/00221287-146-4-787. [DOI] [PubMed] [Google Scholar]
- 206.Messner KR, Imlay JA. 2002. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem 277:42563–42571. 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- 207.González-Flecha B, Demple B. 1995. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem 270:13681–13687. 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 208.Desai SK, Kenney LJ. 2017. To ~P or not to ~P? Non-canonical activation by two-component response regulators. Mol Microbiol 103:203–213. 10.1111/mmi.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Colloms SD, Alén C, Sherratt DJ. 1998. The ArcA/ArcB two-component regulatory system of Escherichia coli is essential for Xer site-specific recombination at psi. Mol Microbiol 28:521–530. 10.1046/j.1365-2958.1998.00812.x. [DOI] [PubMed] [Google Scholar]
- 210.Zhou Y, Pu Q, Chen J, Hao G, Gao R, Ali A, Hsiao A, Stock AM, Goulian M, Zhu J. 2021. Thiol-based functional mimicry of phosphorylation of the two-component system response regulator ArcA promotes pathogenesis in enteric pathogens. Cell Rep 37:110147. 10.1016/j.celrep.2021.110147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Passalacqua KD, Charbonneau M-E, O’Riordan MXD. 2016. Bacterial metabolism shapes the host-pathogen interface. Microbiol Spectr 4. 10.1128/microbiolspec.VMBF-0027-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Reniere ML. 2018. Reduce, induce, thrive: bacterial redox sensing during pathogenesis. J Bacteriol 200:e00128-18. 10.1128/JB.00128-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tiwari S, Jamal SB, Hassan SS, Carvalho PVSD, Almeida S, Barh D, Ghosh P, Silva A, Castro TLP, Azevedo V. 2017. Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Front Microbiol 8:1878. 10.3389/fmicb.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rajput A, Seif Y, Choudhary KS, Dalldorf C, Poudel S, Monk JM, Palsson BO. 2021. Pangenome analytics reveal two-component systems as conserved targets in ESKAPEE pathogens. mSystems 6:e00981-20. 10.1128/mSystems.00981-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hirakawa H, Kurushima J, Hashimoto Y, Tomita H. 2020. Progress overview of bacterial two-component regulatory systems as potential targets for antimicrobial chemotherapy. Antibiotics (Basel) 9:635. 10.3390/antibiotics9100635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Anderson MT, Mitchell LA, Zhao L, Mobley HLT. 2017. Capsule production and glucose metabolism dictate fitness during Serratia marcescens bacteremia. mBio 8:e00740-17. 10.1128/mBio.00740-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Anderson MT, Mitchell LA, Zhao L, Mobley HLT. 2018. Citrobacter freundii fitness during bloodstream infection. Sci Rep 8:11792. 10.1038/s41598-018-30196-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W, Cavalcoli JD, Mobley HLT. 2015. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio 6:e00775-15. 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jiang L, Feng L, Yang B, Zhang W, Wang P, Jiang X, Wang L. 2017. Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella Typhimurium invasion. PLoS Pathog 13:e1006429. 10.1371/journal.ppat.1006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhao X, Zeng X, Dai Q, Hou Y, Zhu D, Wang M, Jia R, Chen S, Liu M, Yang Q, Wu Y, Zhang S, Huang J, Ou X, Mao S, Gao Q, Zhang L, Liu Y, Yu Y, Cheng A. 2021. Immunogenicity and protection efficacy of a Salmonella enterica serovar Typhimurium fnr, arcA and fliC mutant. Vaccine 39:588–595. 10.1016/j.vaccine.2020.12.002. [DOI] [PubMed] [Google Scholar]
- 221.Sengupta N, Paul K, Chowdhury R. 2003. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect Immun 71:5583–5589. 10.1128/IAI.71.10.5583-5589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Xi D, Yang S, Liu Q, Li Y, Li Y, Yan J, Wang X, Ning K, Cao B. 2020. The response regulator ArcA enhances biofilm formation in the vpsT manner under the anaerobic condition in Vibrio cholerae. Microb Pathog 144:104197. 10.1016/j.micpath.2020.104197. [DOI] [PubMed] [Google Scholar]
- 223.Buettner FFR, Maas A, Gerlach G-F. 2008. An Actinobacillus pleuropneumoniae arcA deletion mutant is attenuated and deficient in biofilm formation. Vet Microbiol 127:106–115. 10.1016/j.vetmic.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 224.Pittman RN. 2011. Oxygen transport. Morgan & Claypool Life Sciences, San Rafael, CA. [PubMed] [Google Scholar]
- 225.Smith SN, Hagan EC, Lane MC, Mobley HLT. 2010. Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. mBio 1:e00262-10. 10.1128/mBio.00262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.De Souza-Hart JA, Blackstock W, Di Modugno V, Holland IB, Kok M. 2003. Two-component systems in Haemophilus influenzae: a regulatory role for ArcA in serum resistance. Infect Immun 71:163–172. 10.1128/IAI.71.1.163-172.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang Z, Sun J, Xia T, Liu Y, Fu J, Lo YK, Chang C, Yan A, Liu X. 2018. Proteomic delineation of the ArcA regulon in Salmonella Typhimurium during anaerobiosis. Mol Cell Proteomics 17:1937–1947. 10.1074/mcp.RA117.000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Fernández PA, Velásquez F, Garcias-Papayani H, Amaya FA, Ortega J, Gómez S, Santiviago CA, Álvarez SA. 2018. Fnr and ArcA regulate lipid A hydroxylation in Salmonella Enteritidis by controlling lpxO expression in response to oxygen availability. Front Microbiol 9:1220. 10.3389/fmicb.2018.01220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yun S, Shin JM, Kim O-C, Jung YR, Oh D-B, Lee SY, Kwon O. 2012. Probing the ArcA regulon in the rumen bacterium Mannheimia succiniciproducens by genome-wide expression profiling. J Microbiol 50:665–672. 10.1007/s12275-012-2007-7. [DOI] [PubMed] [Google Scholar]
- 230.Gao H, Wang X, Yang ZK, Chen J, Liang Y, Chen H, Palzkill T, Zhou J. 2010. Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis. PLoS One 5:e15295. 10.1371/journal.pone.0015295. [DOI] [PMC free article] [PubMed] [Google Scholar]