SUMMARY
The development of resistance to β-lactam antibiotics has made Staphylococcus aureus a clinical burden on a global scale. MRSA (methicillin-resistant S. aureus) is commonly known as a superbug. The ability of MRSA to proliferate in the presence of β-lactams is attributed to the acquisition of mecA, which encodes the alternative penicillin binding protein, PBP2A, which is insensitive to the antibiotics. Most MRSA isolates exhibit low-level β-lactam resistance, whereby additional genetic adjustments are required to develop high-level resistance. Although several genetic factors that potentiate or are required for high-level resistance have been identified, how these interact at the mechanistic level has remained elusive. Here, we discuss the development of resistance and assess the role of the associated components in tailoring physiology to accommodate incoming mecA.
KEYWORDS: Staphylococcus aureus, MRSA, MecA, antimicrobial resistance
INTRODUCTION
A dramatic increase in antimicrobial resistance (AMR) among human pathogens has been reported by the World Health Organization (1). This clinical challenge is compounded by the lack of a mechanistic understanding of the emergence of bacterial resistance as well as a decline in new antibiotic discoveries (2). Among human pathogens, Staphylococcus aureus is a prominent example of the spectre of AMR (3, 4).
Penicillin was first introduced to treat patients with S. aureus bacteremia in the early 1940s, but as early as 1942, the first penicillin-resistant strains of S. aureus were isolated (5). The development of penicillin resistance is mediated by an enzyme, penicillinase/β-lactamase, encoded by blaZ, which cleaves the β-lactam ring and destroys the action of the antibiotic (4, 6).
In response to the spread and emergence of penicillin resistance, a semisynthetic β-lactam named methicillin, impervious to β-lactamase, was developed and introduced into the clinic in 1959 (7, 8). One year later, methicillin-resistant S. aureus (MRSA) was recovered from a patient diagnosed with osteomyelitis and septic arthritis (9). Since then, antibiotic resistance in S. aureus has spread in epidemic waves, with MRSA initially being largely a nosocomial infection, leading to so-called hospital-associated MRSA (HA-MRSA) (10–13). More recently, community-associated MRSA (CA-MRSA) has emerged as a major clinical threat, creating a reservoir of MRSA in and out of health care settings (10, 11). CA-MRSA can be genetically distinguished from HA-MRSA, also having less of a portfolio of antibiotic resistance properties and often making the toxin Panton-Valentine leukocidin (PVL) (14). However, there are now many examples of how CA-MRSA has spread to health care settings, blurring the distinction between the two MRSA types (11). Additionally, MRSA can be harbored by livestock (livestock-associated MRSA [LA-MRSA]), where it can cause disease in those animals and also spread to humans via contact (15, 16).
MRSA is a clear clinical threat, whereby the outcomes of MRSA-mediated infections are worse than those of methicillin-sensitive S. aureus (MSSA) (17). In addition, MRSA means resistance to nearly all available β-lactams (18). Currently, MRSA-mediated infections account for significantly high morbidity and mortality rates (19, 20).
GENETIC FACTORS REQUIRED BY MRSA FOR β-LACTAM RESISTANCE
Penicillin binding proteins (PBPs) are essential for the final stages of bacterial cell wall peptidoglycan (PG) biosynthesis and are the targets of penicillin and other β-lactams (21, 22). Methicillin resistance is, most commonly, mediated by the acquisition of a novel penicillin-binding protein, 2A (PBP2A), which is able to take over the activities of endogenous enzymes (PBPs) due to its low affinity for β-lactam antibiotics (Table 1) (23). Using a penicillin-binding assay, several researchers identified the novel low-affinity PBP2A encoded by mecA in MRSA strains (21, 24, 25). It was confirmed that mec is found only on the chromosome of MRSA and not MSSA and is explicitly required for resistance (26). The mecA gene and regulatory elements are encoded on a mobile genetic element found in all MRSA strains called the staphylococcal cassette chromosome (SCCmec), which has been acquired from an exogenous, environmental source (27). The degree of expression of methicillin resistance is determined by mecI and mecR1 (28), regulatory elements encoded within the mec gene complex adjacent to mecA on the chromosomal SCCmec element (29). The regulatory genes mecR1 and mecI are structurally and functionally similar to the blaZ regulatory components blaR1 and blaI, which, in response to β-lactam antibiotics, induce the expression of mecA and blaZ, respectively (8, 30). It is important to note that some MRSA strains carry a modified version of the mec regulatory system that may lack mecI or include truncated versions of some genes (31, 32). The strains with the complete regulatory system tend to be slower in the induction of resistance when challenged with methicillin than those with an incomplete system (33).
TABLE 1.
Genetic factors required for MRSA β-lactam resistance
| Gene/operon | SAOUHSCa | Functional class | Function | Reference(s) |
|---|---|---|---|---|
| mecA | Extracellular PG synthesis | Encodes the heterologous penicillin-binding protein PBP2A with low affinity for β-lactam antibiotics | 158 | |
| mecB | Extracellular PG synthesis | Encodes a heterologous penicillin-binding protein PBP2B with low affinity for β-lactam antibiotics | 46 | |
| mecC | Extracellular PG synthesis | Encodes a heterologous penicillin-binding protein PBP2C with low affinity for β-lactam antibiotics | 41, 42 | |
| pbp4 | 00646 | Extracellular PG synthesis | PBP with transpeptidase activity | 47 |
S. aureus 8325 genome locus tag.
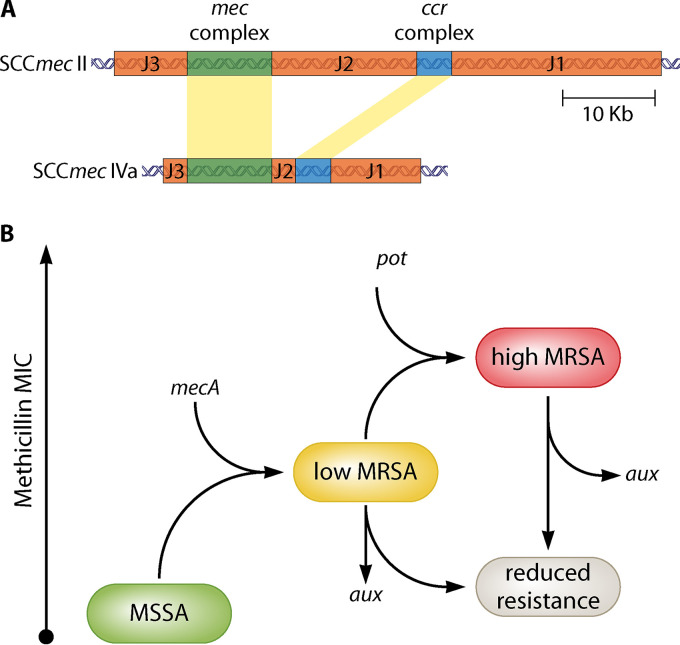
SCCmec elements contain three basic genetic components and share a structurally similar backbone that consists of (i) a mec gene complex carrying mecA and its regulators, surrounding open reading frames (ORFs), and insertion sequences, (ii) a cassette chromosome recombinase (ccr) gene complex containing ccrAB and/or ccrC, ensuring the mobility of SCCmec and surrounding ORFs, and (iii) the joining region (J region) (Fig. 1A) (34). Some SCCmec elements also carry housekeeping genes inside J regions as well as transposons (35). Based on the nature of the ccr and mec gene complexes, coupled with the location and DNA segments of the J regions, SCCmec elements are classified into types I to XIII and different subtypes (11, 36, 37). SCCmec types I, II, and III are large elements encoding resistance to several classes of antibiotics and have historically been associated with prevalent HA-MRSA clones (10). These include the archetypal strains COL (type I) and N315 (type II) (38). More recently, types IV and V have become more common, associated with CA-MRSA but also some widespread HA-MRSA strains, here again making it more difficult to designate isolates as HA/CA MRSA (39). Strain USA300 (type IV) has been used to generate an ordered transposon mutation library as a community-wide resource (40).
FIG 1.
Factors required and associated with β-lactam resistance by MRSA. (A) SCCmec element. The major features of SCCmec types II and IV are shown as examples of HA-MRSA and CA-MRSA, respectively (based on reference 11). The mecA gene is contextualized within SCCmec of different sizes, by the cassette chromosome recombinase (ccr) gene complex and the joining regions (J1 to J3) that can encode a variety of other functions, including housekeeping and transposons. (B) Roles of Aux and Pot factors. There are two steps required for an MRSA strain to develop high-level resistance to β-lactams. The first is acquisition of the mecA/SCCmec cassette that is required for resistance per se. This, however, provides only the ability to grow in the presence of low concentrations of β-lactams. High-level MRSA is supported by those factors associated with resistance. The second step, leading to high-level resistance, requires a mutation in a potentiator gene (pot). Auxiliary factors (aux) are required to support resistance.
Although different SCCmec elements share structural similarities and carry mecA, SCCmec type XI carries a mec homolog classified as mecC, originally designated mecALGA251 (Table 1) (41, 42). The mecC gene encodes PBP2C, which shares only 63% identity at the amino acid level with PBP2A encoded by mecA (41). However, strains carrying mecC exhibit methicillin resistance properties similar to phenotypes of strains with mecA in a temperature-sensitive manner (43). Also, mutagenesis experiments have confirmed that mecC mediates β-lactam resistance in different S. aureus strain backgrounds (42). The important difference between the PBPs encoded by mecC and mecA is that PBP2C has higher binding affinity for oxacillin than cefoxitin in comparison to PBP2A, offering an unusual antibiotic susceptibility profile for these two antibiotics and providing a useful tool to distinguish them (44). Also, in contrast to PBP2A, PBP2C does not require the transglycosylase activity of PBP2 to exhibit high-level oxacillin resistance, indicating that PBP2C might preferentially interact with monofunctional transglycosylase(s) (43). Very recently, mecC-derived MRSA has revealed exciting insights into the evolution of antibiotic resistance. Strains of LA-MRSA with mecC have been found to have been harbored by hedgehogs in the preantibiotic era and to have subsequently spread to livestock and humans (45). In the hedgehog reservoir, a dermatophyte fungus has been found to produce β-lactams that create a selective advantage for MRSA strains in the natural host, providing a nice example of the environmental development of antimicrobial resistance (45).
MRSA can also be attained via a plasmid-borne mecA homologue called mecB (Table 1) (46). Finally, in the laboratory setting, MRSA can be selected for without the need for mecA, or its homologues, via mutations associated with the gene encoding the endogenous PBP4 (Table 1) (47). This occurs via missense mutations surrounding the active site with unknown consequences and alterations to the promoter region resulting in overproduction of PBP4 (47).
Borderline oxacillin-resistant S. aureus (BORSA) is also clinically relevant (48). BORSA strains do not have mecA or mecC, demonstrate borderline levels of resistance to β-lactamase-resistant penicillins, and typically hyperproduce β-lactamase, sometimes associated with point mutations in native PBP-encoding genes (48).
GENETIC FACTORS ASSOCIATED WITH β-LACTAM RESISTANCE BY MRSA
Methicillin resistance requires PBP2A to take over all the transpeptidase functions of the endogenous PBPs that are inhibited by the β-lactams. This is an extraordinary feat for an enzyme that has evolved outside S. aureus, given the complex interactions that govern peptidoglycan (PG) homeostasis during growth and division (49). Once mecA is introduced into the genome of S. aureus, PBP2A therefore does not act in isolation from the endogenous systems required for cell growth and division, having a significant impact on cellular physiology (50). Those processes that are needed for cell wall homeostasis require a tight integration of many different factors (51), and thus it is no surprise that, despite being essential for methicillin resistance through its ability to function as an alternative transpeptidase, PBP2A is not the only factor associated with resistance to β-lactams (52). Indeed, early studies performed using transposon mutagenesis allowed the identification of a set of chromosomal genes whose function is required for mecA-associated methicillin resistance (51, 53, 54). As these genes were not the mediators of methicillin resistance per se, they were named auxiliary (aux) genes or factors (Fig. 1B). Here, the genes and proteins that have been published to act as auxiliary factors are highlighted in bold, listed in Table 2, and displayed in Fig. 2. Given the role of Aux factors in supporting the ability of PBP2A to facilitate PG synthesis, their study also sheds light on the interconnectedness of basic physiological processes that underpin growth and division. Many of the Aux group of components are those that provide precursors needed for correct synthesis of the cell wall, but this group also includes factors that are involved in a whole variety of cellular physiological processes, such as the GlnR repressor from nitrogen metabolism (55), the acyl carrier protein of fatty acid biosynthesis HmrB (56), the surface protein FmtB, and FmtC (MprF), which is required for lysinylation of phosphatidyl glycerol in the cell membrane (57, 58).
TABLE 2.
Auxiliary (Aux) factors: mutations reduce β-lactam resistance
| Gene/operon | SAOUHSC | Functional class | Function | Reference(s) |
|---|---|---|---|---|
| glmS | 02399 | Intracellular PG synthesis | Glucosamine-6-phosphate synthase | 78 |
| glmM (femD) | 02405 | Intracellular PG synthesis | Phosphoglucosamine mutase | 84 |
| murA | 01146 | Intracellular PG synthesis | Transferase; converts UDP-GlcNAc to UDP-GlcNAc-enoylpyruvate | 78 |
| murB | 00752 | Intracellular PG synthesis | Reductase; converts UDP-GlcNAc-enoylpyruvate to UDP-MurNAc | 78 |
| murC | 01856 | Intracellular PG synthesis | UDP-N-acetylmuramate-l-alanine ligase | 78 |
| murD | 01147 | Intracellular PG synthesis | UDP-N-acetylmuramoylalanine-d-glutamate ligase | 78 |
| murE | 00954 | Intracellular PG synthesis | Catalyses incorporation of lysine into the peptide stem | 78, 159 |
| murF | 02317 | Intracellular PG synthesis | UDP-N-acetylmuramoyl-tripeptide-d-alanyl-d-alanine ligase | 78 |
| femX (fmhB) | 02527 | Intracellular PG synthesis | Addition of the first glycine to the peptide stem | 78, 81 |
| femA | 01373 | Intracellular PG synthesis | Addition of the 2nd and 3rd glycine to the peptide stem | 82 |
| femB | 01374 | Intracellular PG synthesis | Addition of the 4th and 5th glycine to the peptide stem | 83 |
| murT | 02107 | Intracellular PG synthesis | Mur ligase homolog | 78, 160 |
| glyS | 01666 | Intracellular PG synthesis | Glycine tRNA synthetase | 78 |
| murJ | 01871 | Extracellular PG synthesis | Lipid II translocase | 161 |
| pbp1 (pbpA) | 01145 | Extracellular PG synthesis | PBP with transpeptidase activity | 78 |
| pbp2 | 01467 | Extracellular PG synthesis | PBP with transpeptidase and transglycosylase activity | 89 |
| pbp4 | 00646 | Extracellular PG synthesis | PBP with transpeptidase activity | 90 |
| ftsW | 01063 | Extracellular PG synthesis | Transglycosylase | 78 |
| tarO | 00762 | Cell wall synthesis | Forms 1st precursor in WTA synthesis | 162 |
| tarA | 00640 | Cell wall synthesis | Forms 2nd intermediate in WTA synthesis | 52 |
| tarB | 00643 | Cell wall synthesis | Forms 3rd intermediate in WTA synthesis | 52 |
| tarD | 00645 | Cell wall synthesis | Involved in WTA synthesis | 52 |
| tarL | 00227 | Cell wall synthesis | Polyribitol-phosphate extension of WTA | 78 |
| tarI | 00225 | Cell wall synthesis | WTA synthesis | 52 |
| tarS | 00228 | Cell wall synthesis | Glycosyltransferase | 92 |
| ltaS | 00728 | Cell wall synthesis | Lipoteichoic acid synthase | 54 |
| fmtB (mrp) | 02404 | Cell wall synthesis | Cell surface protein | 57 |
| ftsA | 01149 | Cell division | Divisome component | 78 |
| ftsZ | 01150 | Cell division | Divisome component | 78 |
| glnR | 01285 | Regulation and cell signaling | Glutamine synthetase repressor | 55 |
| fmtA | 00998 | Regulation and cell signaling | Membrane protein | 58 |
| fmtC (mprF) | 01359 | Regulation and cell signaling | Lysinylation of membrane phosphatidylglycerol | 58 |
| prsS | 00200 | Regulation and cell signaling | ECF sigma factor | 116 |
| sigB | 02298 | Regulation and cell signaling | Transcription factor | 112 |
| sarA | 00620 | Regulation and cell signaling | Accessory regulator A | 114 |
| pknB | 01187 | Regulation and cell signaling | Eukaryotic-like serine/threonine kinase | 54, 114 |
| vraSR | 02098/9 | Regulation and cell signaling | Two-component signal transduction sensor of cell wall stress | 99 |
| spsB | 00903 | Protein secretion | Signal peptidase I | 78 |
| prsA | 01972 | Protein folding and stabilization | Required for posttranslational maturation of PBP2A | 106 |
| htrA1 | 01838 | Protein folding and stabilization | Required for posttranslational maturation of PBP2A, acts in synergy with PrsA | 107 |
| hmrA | 02374 | Protein stability | Endopeptidase | 56, 117 |
| hmrB | 01201 | Metabolism | Homologue of acyl carrier protein | 56 |
| gatD | 02106 | Metabolism | Glutamine amidotransferase | 160 |
| sucC | 01216 | Posttranslational modification | β subunit of succinyl-coenzyme A synthetase | 97 |
| sucD | 01218 | Pos-translational modification | α subunit of succinyl-coenzyme A synthetase | 97 |
| cycA | 01803 | Transport | Putative amino acid permease gene | 74, 163 |
| auxA | 01025 | Hypothetical protein | Putative transmembrane transporter protein | 96 |
| auxB | 01050 | Hypothetical protein | Putative transmembrane protein | 96 |
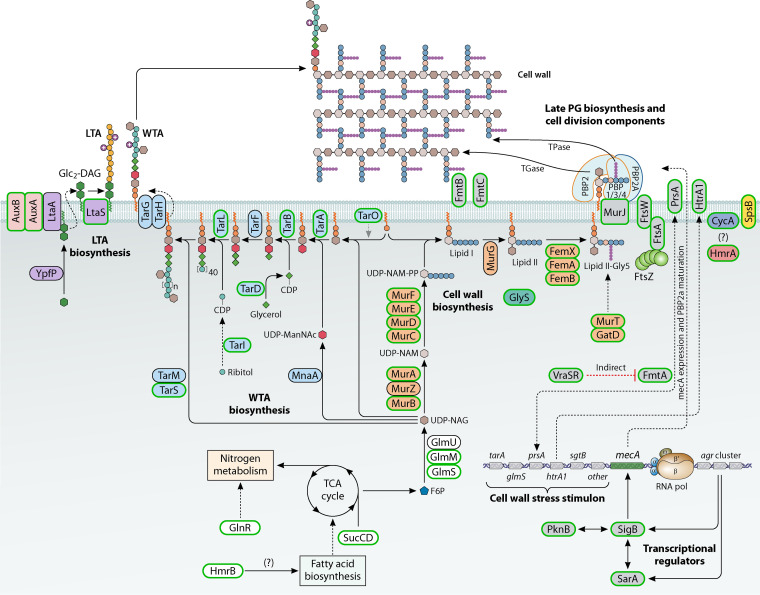
FIG 2.
Schematic model of the range of Aux factors involved in β-lactam resistance by MRSA. The Aux factors form a body of components largely involved in cell wall homeostasis and the response to stress. Auxiliary factors have a green outline, and those with a black outline have no demonstrated impact on methicillin resistance. Factors involved in the same biological pathway have a common shading color. TCA, tricarboxylic acid.
Interestingly, the levels of methicillin resistance of most MRSA isolates are not the same even if all Aux factors remain intact. One of the characteristics of MRSA isolates is their ability to express β-lactam resistance in so-called heterogeneous fashion (59). The majority of cells in a bacterial population are resistant only to low concentrations of methicillin (≤5 μg mL−1). However, a small minority of cells (10−4 to 10−3) exhibit high-level methicillin resistance (>50 μg mL−1). The heterogeneous culture, in turn, has the potential to develop homogeneous resistance, where all cells are uniformly resistant to high concentrations of methicillin (60). This conversion can be induced under laboratory conditions by exposure to β-lactams. Once strains have converted from heterogeneous to high-level homogeneous resistance, this remains even in the absence of antibiotic and is not connected with genetic mutations in mec or aux genes (61, 62). Constitutive expression of mecA does not lead to homogeneous resistance (32, 63). Most of the genes that have been identified to potentiate the conversion to high-level resistance are not directly involved in cell wall biosynthesis or linked to PBP2A. In many cases, they are responsible for regulation of cell physiology in the broader sense (50, 59, 64, 65). Genome sequencing of laboratory-derived mecA-containing mutants selected on high concentrations of methicillin (100 μg mL−1) has revealed that mutations in several genes can lead to high-level methicillin resistance (66). The genes in which mutations lead to increased β-lactam resistance we have called “potentiators” (pot), as opposed to auxiliary genes (aux), in which mutation leads to decreased resistance (Fig. 1B). The Pot factors are therefore both intriguing and perplexing as to how point mutations in genes not apparently linked to PBP2A function can tailor physiology and result in such a profound effect on resistance levels.
Given the wide range of SCCmec types (11) and clonal lineages (67, 68), it is likely that some of the aux and pot factors may be strain specific. High-level MRSA can be established in the laboratory using just mecA in an MSSA background (50, 69), suggesting that there are factors independent of SCCmec. Therefore, without systematic analysis across a broad range of backgrounds, the role of SCCmec cannot be verified. With that caveat, we have covered the portfolio of factors in order to begin to establish those common, underlying mechanisms that underpin the development and maintenance of resistance in MRSA.
Thus, high-level MRSA involves an underlying prerequisite for an exogenous PBP, set within the genetic and physiological context of an MSSA cell, where Aux and Pot factors provide a yin and yang of resistance capabilities.
Auxiliary (Aux) Factors: Mutations Reduce β-Lactam Resistance
Aux factors (Fig. 2; Table 2) support resistance by ensuring that PBP2A is able to carry out cell wall synthesis in the presence of antibiotics. Mutation of any of the Aux factors upsets the delicate framework that allows this to occur. It is currently unknown if PBP2A becomes part of the integrated cell growth and division machinery, which involves multiple protein-protein interactions (70, 71), or acts more independently. The presence of mecA in cells, in the absence of antibiotic stress, leads to a raft of transcriptional changes with consequential metabolic outputs (50). This suggests that having the active enzyme itself creates a stress on the cells, which have to alter their physiology to accommodate this. The Aux factors can therefore be categorized into three major groups: those are directly involved in cell wall homeostasis, those that control it (particularly in response to cell wall stress), and components involved in more general stress responses required to ensure cellular fitness in changing environments.
Cell wall homeostasis.
PG is a primary structural component of the bacterial wall that surrounds the cell, maintaining internal turgor and morphology (72). PG synthesis is the target for β-lactam antibiotics. Biochemically, PG is a single macromolecule made of glycan strands cross-linked via peptide side chains. These strands are made by polymerization of N-acetylglucosamine (GlcNAc) and N-acetyl-muramic acid (MurNAc) and interconnected by peptide side branches (73). Synthesis of the PG is a multistep process (Fig. 2) and occurs in three different locations in the cell. It starts in the cytoplasm, where in the first stage the starter unit of PG biosynthesis, fructose-6-phosphate, is converted into UPD-N-acetylglucosamine by a series of reactions catalyzed by GlmS, GlmM, and GlmU and then into UPD-N-acetylmuramate through the activities of MurA, MurZ, and MurB. In the next steps, performed by MurCDEF PG ligases, a pentapeptide side chain (l-Ala-d-Glu-l-Lys-d-Ala-d-Ala) on UDP-MurNAc is synthesized (73). Impaired transport of amino acids, particularly alanine, at this stage leads to increased susceptibility to β-lactams (74).
In the second stage, the UDP-MurNAc-pentapeptide together with a transport lipid bactoprenol form, in an MraY-catalyzed reaction, UDP-P-P-MurNAc-pentapeptide or lipid I. Then, MurG adds UDP-GlcNAc to lipid I, and thus, a final PG building block, UDP-P-P-(GlcNAc)-MurNAc-pentapeptide or lipid II, is synthesized (75). Subsequently, a family of peptidyltransferases (FemX, FemA, and FemB) complete a pentaglycine bridge on the pentapeptide chain, which results in lipid II-Gly5 (76). For this step, a constant supply of glycine is regulated by GlyS (77) and downregulation of glyS leads to immediate resensitization to β-lactams in MRSA (78). Additionally, the d-Glu in the side chain is amidated to d-glutamine by GatD and MurT enzymes (79). This is required for optimal polymerization of PG by PBPs. Next, the modified lipid II-Gly5 is exported to the outer leaflet of the plasma membrane by lipid translocase MurJ (80). Many of the genes involved in the intracellular stages of PG synthesis as well as genes that are crucial for divisome formation, ftsZ and ftsA, have been described as aux (Table 2) (54, 78, 81–85).
Translocation of the lipid II-Gly5 outside the cell serves as a signal for recruitment of PBP2 and PBP2A (in MRSA), and the third stage of PG biosynthesis begins. During this stage, linear chains of the lipid II-Gly5 are polymerized into mature PG by several penicillin-binding proteins and other enzymes (86). Each of these proteins plays an important role in this process: PBP1, a monofunctional transpeptidase, is required for initiation of PG biosynthesis and separation of the two daughter cells (87); PBP2, a bifunctional transpeptidase-transglycosylase, performs transglycosylation of disaccharide units and transpeptidation of the pentapeptide chains during synthesis of the main PG layer; whereas monofunctional transpeptidase PBP4, which is nonessential under laboratory conditions, performs additional polymerization of the PG layer (88). In the presence of β-lactam antibiotics, MRSA still requires the presence of the native PBP1 and PBP2 (78, 89). PBP2 remains essential, probably because of its transglycosylase activity, as the heterologous PBP2A can perform only transpeptidase cross-linking (89). In addition, as the catalytic activity of PBP2 is inhibited, the PG synthesized under such conditions has an unsatisfactory level of cross-linking that makes PBP4 an essential auxiliary factor for methicillin resistance (90).
The MurNAc residues of the PG can be decorated by a large glycopolymer known as wall teichoic acid (WTA). WTA plays a significant role in growth, morphology, and virulence of S. aureus, and the process of WTA biosynthesis has been elucidated (52). The first enzymatic reaction in WTA synthesis is performed by the MraY paralog TarO, which forms the initial WTA precursor (Fig. 2). Further extension and decoration of this precursor, its polymerization, glycosylation, export, and d-alanylation are catalyzed by different Tar and Dlt proteins, whereas its attachment to PG is mediated by the LCP family of enzymes (91). Screening of S. aureus mutants using an antisense-mRNA approach (78) has shown that depletion of Tar proteins has a severe negative effect on β-lactam resistance (Table 2). It has been speculated that in MRSA, WTA serves as an adjunct for PBP2A to perform PG cross-linking (92, 93).
The third important structural polymer of the bacterial cell wall is lipoteichoic acid (LTA). It consists of glycerol phosphate building blocks decorated with d-alanine residues. The biosynthesis of LTA has been studied and characterized (94). The key enzyme in this process is LtaS, LTA synthase, which polymerizes the LTA backbone chain at the outside of the cell membrane. Production of LTA is regulated at the posttranslational level by signal peptidase SpsB, and loss of it in the MRSA background leads to a decrease in β-lactam resistance, as does an ltaS mutant, which also leads to severe morphological defects and survival only under osmotically stabilizing conditions with high concentrations of sucrose (92). In fact, ΔltaS mutants tend to acquire compensatory mutations in other genes, such as gdpP, sgtB, mazE, clpX, or vraT (95). In a screen of an ordered library of transposon mutants, AuxA and AuxB have been identified as being required for both LTA stability and high-level β-lactam resistance (96).
An interesting interaction between metabolism, protein modification, and β-lactam resistance has recently been described (97). Mutations in sucC or sucD lead to reduced resistance, concomitant with an increase in the succinylation of multiple proteins. These include the major PG hydrolase Atl, which results in reduced enzyme activity. It is the modulation of cell wall homeostasis enzymes that is proposed to result in the perturbation of resistance (97).
The involvement of Aux factors from across the spectrum of different cell wall components highlights that optimal PG synthesis requires the wider context of overall cell wall homeostasis.
Cell wall stress stimulon (VraSR, TarA, GlmS, FmtA, PBP2, SgtB).
The sensor-regulator system VraSR is a sensor of cell wall integrity and activates the genes required for cell wall repair in case of inhibition of its synthesis or damage caused by cell wall-targeting antibiotics (98). This system consists of the histidine kinase VraS, which in response to cell wall damage signals undergoes autophosphorylation and then activates its cognate response regulator, VraR. When activated, VraR acts as a transcriptional regulator and modulates transcription of nearly 40 genes (99). The third component, VraT (YvqF), is a putative membrane protein. Its gene is cotranscribed with other components of the system, and VraT is required for correct induction of the VraSR-mediated cell damage response, but its role remains unclear (100). The VraSR-dependent regulon is known as the cell wall stress stimulon (CWSS) and consists of many genes required for cell wall synthesis and homeostasis and thus includes some auxiliary factors, namely, tarA (tagA), glmS, pbp2, sgtB, and the vraSR operon itself (99, 101). It has also been suggested that auxiliary factor FmtA is controlled by VraR, but in this case the modulation of expression is indirect (99, 101, 102). The exact nature of the signal(s) required for autophosphorylation of VraS and therefore the triggering of CWSS in the natural environment remains unknown. However, the VraSR-mediated response in S. aureus can be caused by loss of LCP proteins (103). The LCP proteins act as WTA ligases, and their loss results in defective cell separation, increased β-lactam susceptibility, changes in cell wall properties, and other defects (104, 105).
The VraSR regulon also contains prsA and htrA1, which are required for the correct maturation of PBP2A. The chaperone PrsA is needed for appropriate folding of PBP2A at physiological temperatures (106), whereas the serine protease HtrA1 ensures proteolysis of misfolded PBP2A molecules (107). The synergistic role of these proteins for PBP2A quality control is confirmed by the fact that single deletions of prsA or htrA1 in the model MRSA strain COL had only a mild effect on β-lactam resistance, whereas a double deletion led to a significant drop (107).
VraSR is not only limited to the regulation of genes involved in cell wall homeostasis but also, by downregulating the agr operon, can modulate quorum sensing and virulence of S. aureus (108). Natural products that effect VraSR have been found to resensitize MRSA to cell wall antibiotics, and thus this regulatory mechanism is a target for the development of new antimicrobial agents (109).
VraSR has likely evolved as a key regulator, monitoring the physiological status of the essential cell wall. To what it responds is currently unknown, and antibiotic resistance can be used as a tool to probe its important cellular functions.
Stress-associated sigma factors (SigB and PrsS).
Alternative sigma factors provide one of the many mechanisms for S. aureus to respond to its environment, both internal and external. SigB is an alternative sigma factor that is involved in a wide variety of cellular processes. This includes control of stress response (110) and biofilm formation (111). Early studies of S. aureus transposon mutants and deletion mutants (112, 113) have demonstrated that disruption or deletion of sigB leads to a decrease of methicillin resistance in strain COL. This could be due to reduced mecA expression in the sigB mutant (114). Another two proteins that interact with SigB, global regulator SarA and the PknB kinase, are important for methicillin resistance (114, 115), but the precise mechanism of this still has to be elucidated.
The extracytoplasmic function (ECF) sigma factor PrsS is another regulatory protein required to support methicillin resistance in USA300 (116). This could be due to decreased accumulation of PBP2A and another auxiliary factor, a zinc-dependent endopeptidase, HmrA (117).
Potentiator (Pot) Factors: Mutations Increase β-lactam Resistance
Pot factors (Fig. 3; Table 3) provide a window from which to view the impact of PBP2A on the cell and how its role can be most optimally accommodated. They provide something that PBP2A (and SCCmec) alone cannot do alone. Their effect is profound, leading to a multifold increase in resistance. However, their mode of action is largely a mystery. Do they act via PBP2A activity or independently? Unlike Aux, the Pot factors are a more eclectic group of components, whose effects span from pleiotropic regulation of gene expression through cell signaling pathways, protein stability, and cell wall homeostasis. The components that have been published as acting as potentiator factors are shown in parentheses in the headings of the sections below and in Table 3.
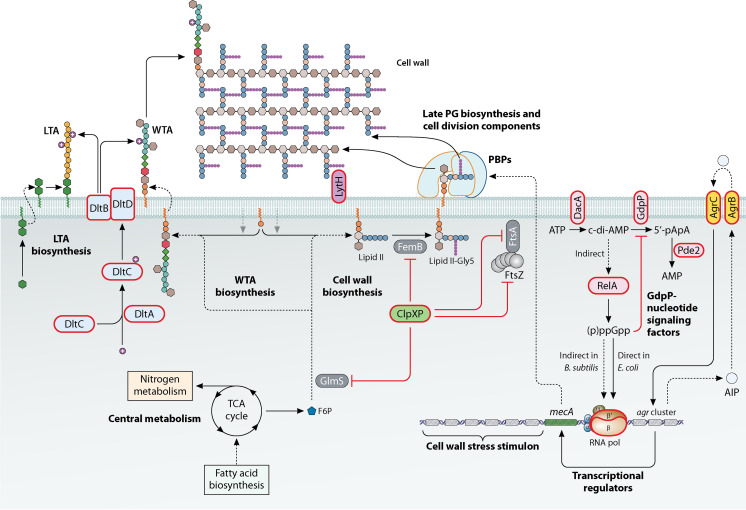
FIG 3.
Schematic model of the range of Pot factors involved in β-lactam resistance by MRSA. The Pot factors have a variety of largely pleiotropic functions in cellular physiology and cell wall homeostasis. Potentiators have a red outline; other components have a black outline.
TABLE 3.
Potentiator (Pot) factors: mutations increase β-lactam resistance
| Gene/operon | SAOUHSC | Functional class | Function | Reference(s) |
|---|---|---|---|---|
| clpXP | 01778/00790 | Protein stability | ATP-dependent Clp protease | 118 |
| gdpP | 00015 | Nucleotide signaling | Phosphodiesterase, hydrolyzes cyclic-di-AMP | 128 |
| pde2 | 01812 | Nucleotide signaling | Phosphodiesterase, hydrolyzes c-di-AMP and pApA to AMP | 129 |
| relA | 01742 | Nucleotide signaling | Bifunctional synthase and hydrolase of (p)ppGpp alarmone | 69 |
| rpoB/rpoC | 00524/5 | Genetic information processing | DNA-directed RNA polymerase β/β’ subunit | 50, 141 |
| agr | 02261–02265 | Quorum sensing | Global regulator of biofilm formation and toxin production | 149 |
| lytH | 01739 | Cell wall homeostasis | PG hydrolase | 155 |
| dlt operon | 00868–00872 | Cell wall homeostasis | Transfer of d-alanine onto teichoic acids | 152 |
Protein turnover and the ClpXP system (ClpXP).
The proteolytic activity of ClpXP is required in a wide range of cellular processes, including the modulation of β-lactam resistance (99, 118) and production of the major virulence factor Protein A (100, 119). Thus, inhibition of ClpXP proteolytic activity is an attractive target for developing new approaches for the treatment of infections (120). However, loss of ClpXP leads to conversion to homogeneous high-level β-lactam resistance (118), cell wall thickening, an increase in PG cross-linking, and reduced cell size.
The ATP-dependent ClpXP proteolytic complexes are widely distributed among different bacterial species and are responsible for targeted protein degradation during the bacterial life cycle (121). The complex is made of two distinct proteins, an ATPase called ClpX and a peptidase called ClpP. The function of ClpX is to recognize, unfold, and translocate proteins tagged for degradation in the proteolytic chamber of ClpP. Besides this, the ClpX subunit also can act independently as a molecular chaperone that facilitates correct folding of newly synthesized proteins (121). The ClpP subunit, as well, can act on its own, but because of the small size of its proteolytic chamber, ClpP is able to independently degrade only small peptides, or it may also interact with other ATPase subunits with different recognition specificities, such as ClpC or ClpA. Loss of ClpP has a more profound effect on β-lactam resistance than loss of ClpX, while ClpC has no role, suggesting that ClpP modulates β-lactam resistance through ClpX/C-independent mechanisms (118). Studies using an inactive version of ClpXP that traps protein substrates inside its proteolytic chamber (122) shows that some of the aux gene products, including femB, glmS, ftsZ, and ftsA, are direct targets for ClpXP. Thus, elimination of ClpXP proteolytic activity could stabilize auxiliary factors and lead to an increase in resistance. Comparison of transcriptomic data for an S. aureus clpX mutant (lacking both ClpX chaperone and ClpXP protease activities) and a mutant expressing a clpXI265E variant of the gene (only the chaperone activity is preserved) demonstrated the modulation of many auxiliary factors that are required for methicillin resistance (123). This includes components of PG biosynthesis (ftsL and pbp1) and members of the type VII secretion system. These findings suggest that changes in β-lactam tolerance might still be solely connected with the enzymatic properties of the ClpXP complex; however, in the case of clpX, advantages gained through abolishment of the ClpXP activity are simply counterbalanced by the negative impact that absence of the ClpX chaperone activity would have.
Deletion of clpX in S. aureus can lead to suppression of phenotypes observed as a result of loss of other factors involved in antibiotic resistance. For example, clinical isolates of a daptomycin-resistant S. aureus mutant (124) that contain rpoBA477D and exhibit high-level tolerance to oxacillin (>256 μg mL−1) during in vivo selection acquired a loss-of-function mutation in clpX. The clpX mutation in these strains partially compensated the negative fitness cost caused by rpoBA477D, but it did not affect the strain’s β-lactam or daptomycin tolerance (125). In another study, loss of ClpX activity rendered the auxiliary gene ltaS (LTA synthase), ordinarily essential under standard laboratory conditions, nonessential (126). The clpX mutation compensated for the septum placement defect presented by LTA-negative mutants of S. aureus.
Nucleotide-signaling pathways (GdpP, Pde2, RelA).
Nucleotide-signaling molecules are indispensable components as they regulate cellular pathways in all forms of life. The role of nucleotide-signaling molecules in methicillin resistance has been examined in a number of studies (50, 69, 127–129). The recent discovery of S. aureus c-di-AMP (128), a signaling nucleotide, has highlighted its importance in the homeostasis of cellular nucleotide concentrations during environmental changes (130). c-di-AMP is synthesized by diadenylyl cyclase (DacA) and hydrolyzed by c-di-AMP phosphodiesterase (GdpP) (127, 128). GdpP contains two N-terminal transmembrane domains, a PAS sensory domain, a GGDEF domain, a DHH domain, and a DHH/DHHA1 domain (128, 131). The C-terminal DHH and DHH/DHHA1 domains possess phosphodiesterase activity that hydrolyzes c-di-AMP to pApA (phosphadenylyl-adenosine) and then to AMP (130–132). Several studies have reported that the increased intracellular levels of c-di-AMP due to the disruption of gdpP lead to elevated resistance to cell wall-targeting antibiotics (50, 131, 133–136). Increased c-di-AMP levels have been shown to be associated with increased PG cross-linking, indicating the role of c-di-AMP in regulating cell wall characteristics and concomitant resistance to β-lactam antibiotics (128). Also, cells with depleted intracellular c-di-AMP levels are significantly more sensitive to oxacillin and larger in size (127, 137). This suggests a potential function for c-di-AMP in regulating components of the cell wall homeostasis machinery that control the strength of the cell wall. Another interesting observation is that the regulation of membrane potential also influences the level of resistance to β-lactam antibiotics in a gdpP mutant. Mutations in dacA or gdpP lead, respectively, to reduced or increased c-di-AMP, concomitant with reduced or increased membrane potential (137).
In addition to membrane-bound phosphodiesterase (PDE) gdpP, a cytoplasmic PDE encoded by pde2 which preferentially hydrolyzes pApA to AMP, ensuring tight control of cellular c-di-AMP levels, was recently characterized (129). Interestingly, pde2 mutation leads to increased resistance to oxacillin similar to that of gdpP mutation in strains of different backgrounds (50, 129). Collectively, the transition from low-level to high-level methicillin resistance is associated with c-di-AMP levels potentially by regulating cell wall synthesis and allowing cells to cope with membrane and cell wall damage upon exposure to β-lactam antibiotics.
Mwangi et al. (69) identified point mutations in the relA gene, encoding a (p)ppGpp synthetase which triggers the stringent response, to be a positive genetic determinant for methicillin resistance. The turnover of (p)ppGpp is tightly controlled by relA (134, 138); however, upon nutrient starvation, which causes the stringent response, cells accumulate high levels of (p)ppGpp in the bacterial cell which slow down the translation of gene products involved in macromolecular biosynthesis (139). Interestingly, Corrigan et al. (64) demonstrated that cells with high levels of (p)ppGpp inhibited the hydrolysis of intracellular c-di-AMP by GdpP, resulting in increased levels of cellular c-di-AMP. Moreover, high-levels of c-di-AMP were shown to activate the production of (p)ppGpp via an unknown mechanism, linking the two different nucleotide-signaling pathways (64). This, in combination with increased PG cross-linking associated with high-levels of intracellular c-di-AMP and upregulation of pbp4 in a gdpP mutant (64), suggests that these pathways prepare the cell to withstand antibiotic intervention, but mechanistic insights are elusive.
RNA polymerase (RpoB, RpoC).
Mutations in the genes rpoB and rpoC, which encode the two largest subunits of RNA polymerase, β and β′, respectively, are well known factors that can change levels of antibiotic resistance (140). In terms of methicillin resistance, however, the importance of rpoB/C mutations has quite often been overlooked, as these mutations are identified along with mutations in other potentiators, such as relA (66). Now that whole-genome sequencing is a routine procedure, the significance of rpo mutations in conversion from heterogeneous to homogeneous methicillin resistance is highlighted (50).
Those rpo mutants with an increased resistance to β-lactams have a pleiotropic phenotype (50, 125, 141). This includes a prolonged doubling time and increased cell wall thickness. In addition, S. aureus cells harboring rpoBA477D are smaller in size (125). In relation to changes in gene expression, β-lactam-associated mutations in the genes encoding RNA polymerase lead to increased expression of mecA (50), which also correlates with increased concentration of PBP2A (141). The increased resistance to methicillin cannot, however, be attributed to PBP2A levels, as overexpression of mecA from a multicopy plasmid did not result in high-level resistance (50). The introduction of mecA into S. aureus and subsequent acquisition of the rpo mutations leading to high-level resistance have been established (50). The introduction of mecA alone, in the MSSA SH1000 strain background, leads to increased expression of the genes involved in metabolism of pyruvate (ldh1, lctP, ald1, adhE, adh, pflA, and pflB) and nitrogen (nirR, nirB, nirD, nasF, narG, narH, narT, and narJ), as well as genes of oxygen-independent ribonucleotide reductase (nrdD and nrdG). This set of genes is usually associated with anaerobic growth. After the mecA-containing strains acquired rpo mutations, the expression level of these genes returned to the parental level. This suggests that respiration is impaired by mecA, and this is suppressed by an rpo mutation, as was verified by oxygen utilization experiments (50).
Biochemical characterization of RNA polymerase with mutated β or β′ subunits associated with high-level methicillin resistance (50) revealed changes in transcription initiation elongation and RNA polymerase, with a mutated β subunit having lower affinity for SigA. These changes could result in differential gene expression compared to that of the parental strain. It is important to point out an interaction loop that exists between stress response regulator Spx and two potentiators, RNA polymerase and the ClpXP proteolytic complex. Spx controls expression of its regulon by interacting directly with α subunit of RNA polymerase. Spx is essential, but its loss can be compensated by a mutation in rpoB, associated with rifampin resistance (142). The rpoBA477D mutation (125) led to an increased concentration of Spx. Finally, the level of Spx in the cell is regulated by ClpXP proteolysis (123). Thus, both potentiators, rpo and clpXP, increase resistance to β-lactams with a concomitant increase in Spx levels.
Quorum sensing and the accessory gene regulator system (Agr).
The accessory gene regulator cluster (agr) is a global regulatory system that governs quorum sensing and virulence in S. aureus. Inactivation of agr leads to a significant increase in the rate of conversion from heterogeneous to homogeneous resistance, whereas complementation of the agr mutant reverses this effect (143, 144).
The products of the agr cluster (AgrA to -D) produce and respond to the buildup of autoinducing peptide (AIP) in the environment as a quorum sensing system. The Agr system modulates the production of a range of other regulators, for example, SarA, SarR, SrrAB, SarX, CodY, and SigB, in a direct or indirect manner (145). Another gene in the agr cluster encodes the RNAIII transcript, which is a multifunctional RNA that can act as antisense controlling posttranscriptional regulation of many components (146). Transcriptomic studies (147) demonstrated that RNAIII modulates expression of major virulence factors and transcriptional regulators.
Health care-associated MRSA (HA-MRSA) strains have a correlation between high expression levels of mecA and a repressed or dysfunctional agr cluster, with concomitant reduced toxin production and lower virulence, whereas community-associated MRSA (CA-MRSA) strains usually have lower PBP2A but increased agr expression (136, 148). The rationale for this trade-off may relate to the changes in PG structure caused by PBP2A, thus reducing the ability of the Agr system to respond to increased AIP concentrations and to effectively autoactivate the agr cluster (148). Recent experiments with USA300, a CA-MRSA strain that has low-level mecA expression, showed that inactivation of the agr cluster led to changes in mecA expression (149), suggesting a cross-regulation between agr and mecA. Inactivation of the agr cluster also leads to decreased susceptibility to oxacillin and ampicillin. The agr mutation results in increased concentrations of long chain fatty acids in the cytoplasmic membrane, possibly contributing to increased membrane stability and thus affecting β-lactam resistance (149, 150).
Cell wall homeostasis (Dlt, LytH).
As described above, many components involved in PG biosynthesis are auxiliary factors required for high-level β-lactam resistance (Table 3). However, inactivation of genes within the dltABCDX operon (151) in MRSA strain KAN96 has the opposite effect (152). Products of this operon catalyze decoration of LTA and WTA by d-alanine residues and suggests that the resulting changes in cell wall structure may reduce its susceptibility to β-lactam antibiotics (152).
Cell wall homeostasis is governed by PG synthesis and hydrolysis to permit growth and division. The PG hydrolase LytH is an amidase that targets uncross-linked PG (153). In early studies, laboratory mutants as well as clinical isolates lacking LytH activity demonstrated increased methicillin resistance (154, 155), although this has been questioned (153).
CONCLUDING REMARKS
High-level β-lactam resistance requires the acquisition of a gene encoding an exogenous PBP, generally mecA. However, the presence of PBP2A brings with it several cellular challenges. This protein has to be able to take over the essential transpeptidase activity of endogenous PBPs in the presence of β-lactams but in doing so must not significantly interfere with PG synthesis in the absence of antibiotics. This creates a delicate balancing act for the cell, requiring the correct functioning of a number of components (Aux factors) to support resistance. These have roles in many processes that permit optimal PG synthesis and the flow of precursors. However, there is also an intriguing set of genes in which mutations promote the development of high-level resistance (Pot). Recent transcriptional analysis has provided a glimpse into how the potentiator rpo may facilitate the ability of PBP2A to function optimally, without disrupting important physiological processes (50). It is interesting to speculate as to whether the Pot factors all act independently or are part of a continuum that facilitates high-level resistance. Elucidating their individual and collective roles in MRSA and MSSA will reveal insights into fundamental physiology and how this underpins the development of clinically relevant antibiotic resistance.
The clinical importance of MRSA reveals that whatever molecular mechanisms are at play, they do not unduly affect the fitness of resistant strains that are able to proliferate and cause disease. This is important, as it reveals that harboring SCCmec and the pot mutation in rpo do not in themselves disadvantage MRSA in the hospital environment, as shown by the epidemic spread of strains around the world. The acquisition of mecA and rpo results in high-level MRSA with concomitant slower growth in vitro but no change in the ability of the strain to cause disease in a mouse sepsis model (50). However, this is nuanced by the SCCmec type, where CA-MRSA with the smaller type IV element is able to maintain growth rate and toxin production levels in vitro, compared to HA-MRSA with the larger type II, suggesting a role in the ability of strains to compete in the community setting (156). CA-MRSA has evolved several times, where an evolutionary compromise has been achieved between maintaining antibiotic resistance and enhanced pathogenic capability but without sacrificing overall fitness (157).
The complex set of genetic determinants that support β-lactam resistance sets up a number of questions to be addressed in order to explain how high-level resistance is maintained. These questions are not only interesting from an antibiotic resistance perspective—it must be remembered that resistance emerges from the basic mechanisms of bacterial physiology that underpin growth and division.
UNANSWERED QUESTIONS
What is the role of the SCCmec type and the chromosomal background in the complement of identified Aux and Pot factors? To date, studies have taken place over a large range of strains, which has made underlying principles difficult to elucidate.
What are the individual and collective roles of the Aux and Pot factors in resistance? While there are clear themes among the Aux factors, how they and the Pot factors work to support high-level resistance is largely obscure.
What is the effect of PBP2A on S. aureus in the absence of antibiotics? PBP2A is an exogenous enzyme that has evolved outside of S. aureus, and its presence stresses the cell through mechanisms unknown.
How is PBP2A able to substitute for the endogenous enzymes in PG synthesis in the presence of antibiotics? In the presence of antibiotics, PBP2A takes on the herculean task of all of the endogenous transpeptidases to allow growth and division.
Can an understanding of Aux and Pot factors be exploited to combat the scourge of MRSA? Approaches have already been developed to reduce high-level resistance, and it is these, and others, that may prove clinically important in the future.
What can we learn about the fundamental principles of growth and division from elucidation of MRSA resistance mechanisms? Being MRSA is not essential for the cell and provides a tractable experimental system in which to dissect those physiological processes required for life.
ACKNOWLEDGMENTS
This work was funded by the UKRI Strategic Priorities Fund (grant no. EP/T002778/1).
We declare there are no conflicts of interest.
REFERENCES
- 1.WHO. 2014. Antimicrobial resistance: global report on surveillance. https://apps.who.int/iris/handle/10665/112642.
- 2.da Cunha BR, Fonseca LP, Calado CRC. 2019. Antibiotic discovery: where have we come from, where do we go? Antibiotics (Basel) 8:45. 10.3390/antibiotics8020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabath LD, Wallace SJ. 1971. Factors influencing methicillin resistance in staphylococci. Ann N Y Acad Sci 182:258–266. 10.1111/j.1749-6632.1971.tb30662.x. [DOI] [PubMed] [Google Scholar]
- 4.Sabath LD. 1982. Mechanisms of resistance to beta-lactam antibiotics in strains of Staphylococcus aureus. Ann Intern Med 97:339–344. 10.7326/0003-4819-97-3-339. [DOI] [PubMed] [Google Scholar]
- 5.Rammelkamp CH, Maxon T. 1942. Resistance of Staphylococcus aureus to the action of penicillin. Exp Biol Med 51:386–389. 10.3181/00379727-51-13986. [DOI] [Google Scholar]
- 6.Bondi A, Dietz C. 1945. Penicillin resistant staphylococci. Am J Clin Pathol 60:55–58. 10.3181/00379727-60-15089. [DOI] [PubMed] [Google Scholar]
- 7.Jevons MP. 1961. Celbenin-resistant staphylococci. BMJ 1:124–125. 10.1136/bmj.1.5219.124-a. [DOI] [Google Scholar]
- 8.Lowy F. 2003. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest 111:1265–1273. 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jevons P, Coe AW, Parker MT. 1963. Methicillin resistance in staphylococci. Lancet 281:904–907. 10.1016/S0140-6736(63)91687-8. [DOI] [PubMed] [Google Scholar]
- 10.Chambers HF, DeLeo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakhundi S, Zhang K. 2018. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31:e00020-18. 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couto I, Melo-Cristino J, Fernandes ML, Garcia T, Serrano N, Salgado MJ, Torres-Pereira A, Sanches IS, De Lencastre H. 1995. Unusually large number of methicillin-resistant Staphylococcus aureus clones in a Portuguese hospital. J Clin Microbiol 33:2032–2035. 10.1128/jcm.33.8.2032-2035.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panlilio AL, Culver DH, Gaynes RP, Banerjee S, Henderson TS, Tolson JS, Martone WJ. 1992. Methicillin-resistant Staphylococcus aureus in U.S. hospitals, 1975–1991. Control Hosp Epidemiol 13:582–586. [DOI] [PubMed] [Google Scholar]
- 14.Uhlemann A-C, Otto M, Lowy FD, DeLeo FR. 2014. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect Genet Evol 21:563–574. 10.1016/j.meegid.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crespo-Piazuelo D, Lawlor PG. 2021. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir Vet J 74:21. 10.1186/s13620-021-00200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuny C, Wieler L, Witte W. 2015. Livestock-associated MRSA: the impact on humans. Antibiotics (Basel) 4:521–543. 10.3390/antibiotics4040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cosgrove S, Sakoulas G, Perencevich E, Schwaber M, Karchmer A, Carmeli Y. 2003. Comparison of mortality associated with methicillin resistant and methicillin susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis 36:53–59. 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 18.Stryjewski ME, Corey GR. 2014. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin Infect Dis 58(Suppl 1):S10–S19. 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- 19.Dantes R, Mu Y, Belflower R, Aragon D, Dumyati G, Harrison LH, Lessa FC, Lynfield R, Nadle J, Petit S, Ray SM, Schaffner W, Townes J, Fridkin S. 2013. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med 173:1970–1978. 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein E, Smith DL, Laxminarayan R. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis 13:1840–1846. 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown DF, Reynolds PE. 1980. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett 122:275–278. 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- 22.Hartman BJ, Tomasz A. 1986. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother 29:85–92. 10.1128/AAC.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim D, Strynadka NCJ. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol 9:870–876. 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- 24.Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol 158:513–516. 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Utsui Y, Yokota T. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother 28:397–403. 10.1128/AAC.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart GC, Rosenblum ED. 1980. Genetic behavior of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol 144:1200–1202. 10.1128/jb.144.3.1200-1202.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555. 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Archer GL, Bosilevac JM. 2001. Signaling antibiotic resistance in staphylococci. Science 291:1915–1916. 10.1126/science.1059671. [DOI] [PubMed] [Google Scholar]
- 29.Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus structural. Antimicrob Agents Chemother 45:1323–1336. 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregory PD, Lewis RA, Curnock SP, Dyke KG. 1997. Studies of the repressor (BlaI) of beta-lactamase synthesis in Staphylococcus aureus. Mol Microbiol 24:1025–1037. 10.1046/j.1365-2958.1997.4051770.x. [DOI] [PubMed] [Google Scholar]
- 31.Hurlimann-Dalel R, Ryffel C, Kayser F, Berger-Bächi B. 1992. Survey of the methicillin resistance-associated genes mecA, mecRl-mecI, and femA-femB in clinical isolates of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 36:2617–2621. 10.1128/AAC.36.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki E, Kuwahara-Arai K, Richardson JF, Hiramatsu K. 1993. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob Agents Chemother 37:1219–1226. 10.1128/AAC.37.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryffel C, Kayser FH, Berger-Bachi B. 1992. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother 36:25–31. 10.1128/AAC.36.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito T, Hiramatsu K, Oliveira DC, De Lencastre H, Zhang K, Westh H, O'Brien F, Giffard PM, Coleman D, Tenover FC, Boyle-Vavra S, Skov RL, Enright MC, Kreiswirth B, Kwan SK, Grundmann H, Laurent F, Sollid JE, Kearns AM, Goering R, John JF, Daum R, Soderquist B. 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turlej A, Hryniewicz W, Empel J. 2011. Staphylococcal cassette chromosome mec (SCCmec) classification and typing methods: an overview. Pol J Microbiol 60:95–103. 10.33073/pjm-2011-013. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Chen D, Peters BM, Li L, Li B, Xu Z, Shirliff ME. 2016. Staphylococcal chromosomal cassettes mec (SCCmec): a mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog 101:56–67. 10.1016/j.micpath.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 37.Urushibara N, Aung MS, Kawaguchiya M, Kobayashi N. 2019. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan. J Antimicrob Chemother 75:46–50. 10.1093/jac/dkz406. [DOI] [PubMed] [Google Scholar]
- 38.Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song J-H, Hiramatsu K. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother 50:1001–1012. 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bal AM, Coombs GW, Holden MTG, Lindsay JA, Nimmo GR, Tattevin P, Skov RL. 2016. Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J Glob Antimicrob Resist 6:95–101. 10.1016/j.jgar.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Álvarez L, Holden MTG, Lindsay H, Webb CR, Brown DFJ, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RLR, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis 11:595–603. 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ballhausen B, Kriegeskorte A, Schleimer N, Peters G, Becker K. 2014. The mecA homolog mecC confers resistance against β-lactams in Staphylococcus aureus irrespective of the genetic strain background. Antimicrob Agents Chemother 58:3791–3798. 10.1128/AAC.02731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim C, Milheiriço C, Gardete S, Holmes MA, Holden MTG, De Lencastre H, Tomasz A. 2012. Properties of a novel PBP2A protein homolog from Staphylococcus aureus strain LGA251 and its contribution to the β-lactam-resistant phenotype. J Biol Chem 287:36854–36863. 10.1074/jbc.M112.395962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cartwright EJP, Paterson GK, Raven KE, Harrison EM, Gouliouris T, Kearns A, Pichon B, Edwards G, Skov RL, Larsen AR, Holmes MA, Parkhill J, Peacock SJ, Török ME. 2013. Use of Vitek 2 antimicrobial susceptibility profile to identify mecC in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 51:2732–2734. 10.1128/JCM.00847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen J, Raisen CL, Ba X, Sadgrove NJ, Padilla-González GF, Simmonds MSJ, Loncaric I, Kerschner H, Apfalter P, Hartl R, Deplano A, Vandendriessche S, Černá Bolfíková B, Hulva P, Arendrup MC, Hare RK, Barnadas C, Stegger M, Sieber RN, Skov RL, Petersen A, Angen Ø, Rasmussen SL, Espinosa-Gongora C, Aarestrup FM, Lindholm LJ, Nykäsenoja SM, Laurent F, Becker K, Walther B, Kehrenberg C, Cuny C, Layer F, Werner G, Witte W, Stamm I, Moroni P, Jørgensen HJ, de Lencastre H, Cercenado E, García-Garrote F, Börjesson S, Hæggman S, Perreten V, Teale CJ, Waller AS, Pichon B, Curran MD, Ellington MJ, Welch JJ, et al. 2022. Emergence of methicillin resistance predates the clinical use of antibiotics. Nature 602:135–141. 10.1038/s41586-021-04265-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker K, van Alen S, Idelevich EA, Schleimer N, Seggewiß J, Mellmann A, Kaspar U, Peters G. 2018. Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg Infect Dis 24:242–248. 10.3201/eid2402.171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.da Costa T, de Oliveira C, Chambers H, Chatterjee S. 2018. PBP4: a new perspective on Staphylococcus aureus β-lactam resistance. Microorganisms 6:57–57. 10.3390/microorganisms6030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hryniewicz MM, Garbacz K. 2017. Borderline oxacillin-resistant Staphylococcus aureus (BORSA)—a more common problem than expected? J Med Microbiol 66:1367–1373. 10.1099/jmm.0.000585. [DOI] [PubMed] [Google Scholar]
- 49.Salamaga B, Kong L, Pasquina-Lemonche L, Lafage L, von und Zur Muhlen M, Gibson JF, Grybchuk D, Tooke AK, Panchal V, Culp EJ, Tatham E, O’Kane ME, Catley TE, Renshaw SA, Wright GD, Plevka P, Bullough PA, Han A, Hobbs JK, Foster SJ. 2021. Demonstration of the role of cell wall homeostasis in Staphylococcus aureus growth and the action of bactericidal antibiotics. Proc Natl Acad Sci USA 118:e2106022118. 10.1073/pnas.2106022118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panchal VV, Griffiths C, Mosaei H, Bilyk B, Sutton JAF, Carnell OT, Hornby DP, Green J, Hobbs JK, Kelley WL, Zenkin N, Foster SJ. 2020. Evolving MRSA: high-level β-lactam resistance in Staphylococcus aureus is associated with RNA polymerase alterations and fine tuning of gene expression. PLoS Pathog 16:e1008672. 10.1371/journal.ppat.1008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roemer T, Schneider T, Pinho MG. 2013. Auxiliary factors: a chink in the armor of MRSA resistance to β-lactam antibiotics. Curr Opin Microbiol 16:538–548. 10.1016/j.mib.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J, Su J, Pan J, Hailey J, McGuinness D, Tan CM, Flattery A, Walker S, Black T, Roemer T. 2013. Discovery of wall teichoic acid inhibitors as potential anti-MRSA β-lactam combination agents. Chem Biol 20:272–284. 10.1016/j.chembiol.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berger-Bächi B, Strässle a, Gustafson JE, Kayser FH. 1992. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 36:1367–1373. 10.1128/AAC.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Lencastre H, Wu SW, Pinho MG, Ludovice AM, Filipe S, Gardete S, Sobral R, Gill S, Chung M, Tomasz A. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb Drug Resist 5:163–175. 10.1089/mdr.1999.5.163. [DOI] [PubMed] [Google Scholar]
- 55.Gustafson J, Strassle A, Hachler H, Kayser FH, Berger-Bachi B. 1994. The femC locus of Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J Bacteriol 176:1460–1467. 10.1128/jb.176.5.1460-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kondo N, Kuwahara-Arai K, Kuroda-Murakami H, Tateda-Suzuki E, Hiramatsu K. 2001. Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob Agents Chemother 45:815–824. 10.1128/AAC.45.3.815-824.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Komatsuzawa H, Ohta K, Sugai M, Fujiwara T, Glanzmann P, Berger-Bächi B, Suginaka H. 2000. Tn551-mediated insertional inactivation of the fmtB gene encoding a cell wall-associated protein abolishes methicillin resistance in Staphylococcus aureus. J Antimicrob Chemother 45:421–431. 10.1093/jac/45.4.421. [DOI] [PubMed] [Google Scholar]
- 58.Komatsuzawa H, Ohta K, Fujiwara T, Choi GH, Labischinski H, Sugai M. 2001. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 203:49–54. 10.1111/j.1574-6968.2001.tb10819.x. [DOI] [PubMed] [Google Scholar]
- 59.Kim C, Mwangi M, Chung M, Milheiriço C, Milheirço C, de Lencastre H, Tomasz A. 2013. The mechanism of heterogeneous beta-lactam resistance in MRSA: key role of the stringent stress response. PLoS One 8:e82814. 10.1371/journal.pone.0082814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peacock SJ, Paterson GK. 2015. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem 84:577–601. 10.1146/annurev-biochem-060614-034516. [DOI] [PubMed] [Google Scholar]
- 61.Ryffel C, Strassle a, Kayser FH, Berger-Bachi B. 1994. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 38:724–728. 10.1128/AAC.38.4.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finan JE, Rosato AE, Dickinson TM, Ko D, Archer GL. 2002. Conversion of oxacillin-resistant staphylococci from heterotypic to homotypic resistance expression. Antimicrob Agents Chemother 46:24–30. 10.1128/AAC.46.1.24-30.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hiramatsu K. 1995. Molecular evolution of MRSA. Microbiol Immunol 39:531–543. 10.1111/j.1348-0421.1995.tb02239.x. [DOI] [PubMed] [Google Scholar]
- 64.Corrigan RM, Bowman L, Willis AR, Kaever V, Gründling A. 2015. Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J Biol Chem 290:5826–5839. 10.1074/jbc.M114.598300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pardos de la Gandara M, Borges V, Chung M, Milheirico C, Gomes J, de Lencastre H, Tomasz A. 2018. Genetic determinants of high-level oxacillin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 62:e00206-18. 10.1128/AAC.00206-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dordel J, Kim C, Chung M, Pardos de la Gándara M, Holden MTJ, Parkhill J, de Lencastre H, Bentley SD, Tomasz A. 2014. Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. mBio 5:e01000-13. 10.1128/mBio.01000-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lindsay JA. 2019. Staphylococci: evolving genomes. Microbiol Spectr 7:GPP3-0071-2019. 10.1128/microbiolspec.GPP3-0071-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitzgerald JR, Holden MTG. 2016. Genomics of natural populations of Staphylococcus aureus. Annu Rev Microbiol 70:459–478. 10.1146/annurev-micro-102215-095547. [DOI] [PubMed] [Google Scholar]
- 69.Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. 2013. Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb Drug Resist 19:153–159. 10.1089/mdr.2013.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bottomley AL, Liew ATF, Kusuma KD, Peterson E, Seidel L, Foster SJ, Harry EJ. 2017. Coordination of chromosome segregation and cell division in Staphylococcus aureus. Front Microbiol 8:1575. 10.3389/fmicb.2017.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Steele VR, Bottomley AL, Garcia-Lara J, Kasturiarachchi J, Foster SJ. 2011. Multiple essential roles for EzrA in cell division of Staphylococcus aureus. Mol Microbiol 80:542–555. 10.1111/j.1365-2958.2011.07591.x. [DOI] [PubMed] [Google Scholar]
- 72.Lund VA, Wacnik K, Turner RD, Cotterell BE, Walther CG, Fenn SJ, Grein F, Wollman AJ, Leake MC, Olivier N, Cadby A, Mesnage S, Jones S, Foster SJ. 2018. Molecular coordination of Staphylococcus aureus cell division. Elife 7:e32057. 10.7554/eLife.32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heijenoort JV. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R–36R. 10.1093/glycob/11.3.25R. [DOI] [PubMed] [Google Scholar]
- 74.Gallagher LA, Shears RK, Fingleton C, Alvarez L, Waters EM, Clarke J, Bricio-Moreno L, Campbell C, Yadav AK, Razvi F, O'Neill E, O'Neill AJ, Cava F, Fey PD, Kadioglu A, O'Gara JP. 2020. Impaired alanine transport or exposure to d-cycloserine increases the susceptibility of MRSA to β-lactam antibiotics. J Infect Dis 221:1006–1016. 10.1093/infdis/jiz542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Heijenoort J. 2007. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol Mol Biol Rev 71:620–635. 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rohrer S, Maki H, Berger-Bächi B. 2003. What makes resistance to methicillin heterogeneous? J Med Microbiol 52:605–607. 10.1099/jmm.0.05176-0. [DOI] [PubMed] [Google Scholar]
- 77.Apostolidi M, Saad NY, Drainas D, Pournaras S, Becker HD, Stathopoulos C. 2015. A glyS T-box riboswitch with species-specific structural features responding to both proteinogenic and nonproteinogenic tRNAGly isoacceptors. RNA 21:1790–1806. 10.1261/rna.052712.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee SH, Jarantow LW, Wang H, Sillaots S, Cheng H, Meredith TC, Thompson J, Roemer T. 2011. Antagonism of chemical genetic interaction networks resensitize MRSA to β-lactam antibiotics. Chem Biol 18:1379–1389. 10.1016/j.chembiol.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 79.Münch D, Roemer T, Lee SH, Engeser M, Sahl HG, Schneider T. 2012. Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog 8:e1002509. 10.1371/journal.ppat.1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Monteiro JM, Pereira AR, Reichmann NT, Saraiva BM, Fernandes PB, Veiga H, Tavares AC, Santos M, Ferreira MT, Macário V, VanNieuwenhze MS, Filipe SR, Pinho MG. 2018. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554:528–532. 10.1038/nature25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rohrer S, Ehlert K, Tschierske M, Labischinski H, Berger-Bächi B. 1999. The essential Staphylococcus aureus gene fmhB is involved in the first step of peptidoglycan pentaglycine interpeptide formation. Proc Natl Acad Sci USA 96:9351–9356. 10.1073/pnas.96.16.9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strandén AM, Ehlert K, Labischinski H, Berger-BÄChi B. 1997. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol 179:9–16. 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henze U, Sidow T, Wecke J, Labischinski H, Berger-Bachi B. 1993. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol 175:1612–1620. 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jolly L, Wu S, Heijenoort JV, De Lencastre H, Mengin-Lecreulx D, Tomasz A. 1997. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J Bacteriol 179:5321–5325. 10.1128/jb.179.17.5321-5325.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan CM, Therien AG, Lu J, Lee SH, Caron A, Gill CJ, Lebeau-Jacob C, Benton-Perdomo L, Monteiro JM, Pereira PM, Elsen NL, Wu J, Deschamps K, Petcu M, Wong S, Daigneault E, Kramer S, Liang L, Maxwell E, Claveau D, Vaillancourt J, Skorey K, Tam J, Wang H, Meredith TC, Sillaots S, Wang-Jarantow L, Ramtohul Y, Langlois E, Landry F, Reid JC, Parthasarathy G, Sharma S, Baryshnikova A, Lumb KJ, Pinho MG, Soisson SM, Roemer T. 2012. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci Transl Med 4:126ra35. 10.1126/scitranslmed.3003592. [DOI] [PubMed] [Google Scholar]
- 86.Fuda CCS, Fisher JF, Mobashery S. 2005. β-Lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell Mol Life Sci 62:2617–2633. 10.1007/s00018-005-5148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pereira SFF, Henriques AO, Pinho MG, De Lencastre H, Tomasz A. 2009. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol Microbiol 72:895–904. 10.1111/j.1365-2958.2009.06687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reed P, Atilano ML, Alves R, Hoiczyk E, Sher X, Reichmann NT, Pereira PM, Roemer T, Filipe SR, Pereira-Leal JB, Ligoxygakis P, Pinho MG. 2015. Staphylococcus aureus survives with a minimal peptidoglycan synthesis machine but sacrifices virulence and antibiotic resistance. PLoS Pathog 11:e1004891. 10.1371/journal.ppat.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pinho MG, Filipe SR, De Lencastre H, Tomasz a. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J Bacteriol 183:6525–6531. 10.1128/JB.183.22.6525-6531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother 52:3955–3966. 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Farha MA, Leung A, Sewell EW, D'Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED. 2013. Inhibition of WTA synthesis blocks the cooperative action of PBPs and sensitizes MRSA to β-lactams. ACS Chem Biol 8:226–233. 10.1021/cb300413m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S. 2012. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci USA 109:18909–18914. 10.1073/pnas.1209126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Foster TJ. 2019. Can β-Lactam antibiotics be resurrected to combat MRSA? Trends Microbiol 27:26–38. 10.1016/j.tim.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 94.Percy MG, Gründling A. 2014. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu Rev Microbiol 68:81–100. 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 95.Karinou E, Schuster CF, Pazos M, Vollmer W, Gründling A. 2019. Inactivation of the monofunctional peptidoglycan glycosyltransferase SgtB allows Staphylococcus aureus to survive in the absence of lipoteichoic acid. J Bacteriol 201:e00574-18. 10.1128/JB.00574-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mikkelsen K, Sirisarn W, Alharbi O, Alharbi M, Liu H, Nøhr-Meldgaard K, Mayer K, Vestergaard M, Gallagher LA, Derrick JP, McBain AJ, Biboy J, Vollmer W, O'Gara JP, Grunert T, Ingmer H, Xia G. 2021. The novel membrane-associated auxiliary factors AuxA and AuxB modulate β-lactam resistance in MRSA by stabilizing lipoteichoic acids. Int J Antimicrob Agents 57:106283. 10.1016/j.ijantimicag.2021.106283. [DOI] [PubMed] [Google Scholar]
- 97.Campbell C, Fingleton C, Zeden MS, Bueno E, Gallagher LA, Shinde D, Ahn J, Olson HM, Fillmore TL, Adkins JN, Razvi F, Bayles KW, Fey PD, Thomas VC, Cava F, Clair GC, O’Gara JP. 2021. Accumulation of succinyl coenzyme A perturbs the methicillin-resistant Staphylococcus aureus (MRSA) succinylome and is associated with increased susceptibility to beta-lactam antibiotics. mBio 12:e00530-21. 10.1128/mBio.00530-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gardete S, Wu SW, Gill S, Tomasz A. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob Agents Chemother 50:3424–3434. 10.1128/AAC.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 100.Boyle-Vavra S, Yin S, Jo DS, Montgomery CP, Daum RS. 2013. VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother 57:83–95. 10.1128/AAC.01651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sengupta M, Jain V, Wilkinson BJ, Jayaswal RK. 2012. Chromatin immunoprecipitation identifies genes under direct VraSR regulation in Staphylococcus aureus. Can J Microbiol 58:703–708. 10.1139/w2012-043. [DOI] [PubMed] [Google Scholar]
- 102.Zhao Y, Verma V, Belcheva A, Singh A, Fridman M, Golemi-Kotra D. 2012. Staphylococcus aureus methicillin-resistance factor fmtA is regulated by the global regulator SarA. PLoS One 7:e43998. 10.1371/journal.pone.0043998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chan YGY, Frankel MB, Dengler V, Schneewind O, Missiakas D. 2013. Staphylococcus aureus mutants lacking the LytR-CpsA-Psr family of enzymes release cell wall teichoic acids into the extracellular medium. J Bacteriol 195:4650–4659. 10.1128/JB.00544-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hübscher J, McCallum N, Sifri CD, Majcherczyk PA, Entenza JM, Heusser R, Berger-Bächi B, Stutzmann Meier P. 2009. MsrR contributes to cell surface characteristics and virulence in Staphylococcus aureus. FEMS Microbiol Lett 295:251–260. 10.1111/j.1574-6968.2009.01603.x. [DOI] [PubMed] [Google Scholar]
- 105.Dengler V, Meier PS, Heusser R, Kupferschmied P, Fazekas J, Friebe S, Staufer SB, Majcherczyk PA, Moreillon P, Berger-Bächi B, McCallum N. 2012. Deletion of hypothetical wall teichoic acid ligases in Staphylococcus aureus activates the cell wall stress response. FEMS Microbiol Lett 333:109–120. 10.1111/j.1574-6968.2012.02603.x. [DOI] [PubMed] [Google Scholar]
- 106.Jousselin A, Manzano C, Biette A, Reed P, Pinho MG, Rosato AE, Kelley WL, Renzoni A. 2015. The Staphylococcus aureus chaperone PrsA is a new auxiliary factor of oxacillin resistance affecting penicillin-binding protein 2A. Antimicrob Agents Chemother 60:1656–1666. 10.1128/AAC.02333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Roch M, Lelong E, Panasenko OO, Sierra R, Renzoni A, Kelley WL. 2019. Thermosensitive PBP2a requires extracellular folding factors PrsA and HtrA1 for Staphylococcus aureus MRSA β-lactam resistance. Commun Biol 2:417–417. 10.1038/s42003-019-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Taglialegna A, Varela MC, Rosato RR, Rosato AE. 2019. VraSR and virulence trait modulation during daptomycin resistance in methicillin-resistant Staphylococcus aureus infection. mSphere 4:e00557-18. 10.1128/mSphere.00557-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Renzoni A, Kelley WL, Rosato RR, Martinez MP, Roch M, Fatouraei M, Haeusser DP, Margolin W, Fenn S, Turner RD, Foster SJ, Rosato E. 2017. Molecular bases determining daptomycin resistance-mediated resensitization to β-lactam (seesaw effect) in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 61:e01634-16. 10.1128/AAC.01634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chan PF, Foster SJ, Ingham E, Clements MO. 1998. The Staphylococcus aureus alternative sigma factor σ(B) controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J Bacteriol 180:6082–6089. 10.1128/JB.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rachid S, Ohlsen K, Wallner U, Hacker J, Hecker M, Ziebuhr W. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J Bacteriol 182:6824–6826. 10.1128/JB.182.23.6824-6826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu S, De Lencastre H, Tomasz A. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol 178:6036–6042. 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Singh VK, Jayaswal RK, Wilkinson BJ. 2001. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiol Lett 199:79–84. 10.1016/S0378-1097(01)00163-X. [DOI] [PubMed] [Google Scholar]
- 114.Li L, Cheung A, Bayer AS, Chen L, Abdelhady W, Kreiswirth BN, Yeaman MR, Xiong YQ. 2016. The global regulon sarA regulates β-lactam antibiotic resistance in methicillin-resistant Staphylococcus aureus in vitro and in endovascular infections. J Infect Dis 214:1421–1429. 10.1093/infdis/jiw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tamber S, Schwartzman J, Cheung AL. 2010. Role of PknB kinase in antibiotic resistance and virulence in community-acquired methicillin-resistant Staphylococcus aureus strain USA300. Infect Immun 78:3637–3646. 10.1128/IAI.00296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Krute CN, Bell-Temin H, Miller HK, Rivera FE, Weiss A, Stevens SM, Shaw LN. 2015. The membrane protein PrsS mimics σs in protecting Staphylococcus aureus against cell wall-targeting antibiotics and DNA-damaging agents. Microbiology (Reading) 161:1136–1148. 10.1099/mic.0.000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Botelho TO, Guevara T, Marrero A, Arêde P, Fluxa VS, Reymond JL, Oliveira DC, Xavier Gomis-Rüth F. 2011. Structural and functional analyses reveal that Staphylococcus aureus antibiotic resistance factor HmrA is a zinc-dependent endopeptidase. J Biol Chem 286:25697–25709. 10.1074/jbc.M111.247437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bæk KT, Gründling A, Mogensen RG, Thøgersen L, Petersen A, Paulander W, Frees D. 2014. β-Lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob Agents Chemother 58:4593–4603. 10.1128/AAC.02802-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jelsbak L, Ingmer H, Valihrach L, Cohn MT, Christiansen MHG, Kallipolitis BH, Frees D. 2010. The chaperone ClpX stimulates expression of Staphylococcus aureus protein A by Rot dependent and independent pathways. PLoS One 5:e12752. 10.1371/journal.pone.0012752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gao P, Ho PL, Yan B, Sze KH, Davies J, Kao RYT. 2018. Suppression of Staphylococcus aureus virulence by a small-molecule compound. Proc Natl Acad Sci USA 115:8003–8008. 10.1073/pnas.1720520115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baker TA, Sauer RT. 2012. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta 1823:15–28. 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, Krieger S, Ohlsen K, Hecker M, Gerth U, Ingmer H, Frees D. 2013. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res 12:547–558. 10.1021/pr300394r. [DOI] [PubMed] [Google Scholar]
- 123.Stahlhut SG, Alqarzaee AA, Jensen C, Fisker NS, Pereira AR, Pinho MG, Thomas VC, Frees D. 2017. The ClpXP protease is dispensable for degradation of unfolded proteins in Staphylococcus aureus. Sci Rep 7:11739–11739. 10.1038/s41598-017-12122-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Peleg AY, Miyakis S, Ward DV, Earl AM, Rubio A, Cameron DR, Pillai S, Moellering RC, Eliopoulos GM. 2012. Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7:e28316. 10.1371/journal.pone.0028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bæk KT, Thøgersen L, Mogenssen RG, Mellergaard M, Thomsen LE, Petersen A, Skov S, Cameron DR, Peleg AY, Frees D. 2015. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother 59:6983–6991. 10.1128/AAC.01303-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bæk KT, Bowman L, Millership C, Søgaard D, Kaever V, Siljamäki P, Savijoki K, Dupont Søgaard M, Kaever V, Siljamäki P, Savijoki K, Varmanen P, Nyman TA, Gründling A, Frees D. 2016. The cell wall polymer lipoteichoic acid becomes nonessential in Staphylococcus aureus cells lacking the ClpX chaperone. mBio 7:e01228-16. 10.1128/mBio.01228-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dengler V, McCallum N, Kiefer P, Christen P, Patrignani A, Vorholt JA, Berger-Bächi B, Senn MM. 2013. Mutation in the C-di-AMP cyclase dacA affects fitness and resistance of methicillin resistant Staphylococcus aureus. PLoS One 8:e73512. 10.1371/journal.pone.0073512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. 2011. C-di-amp is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog 7:e1002217. 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bowman L, Zeden MS, Schuster CF, Kaever V, Gründling A. 2016. New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J Biol Chem 291:26970–26986. 10.1074/jbc.M116.747709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Corrigan RM, Gründling A. 2013. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol 11:513–524. 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 131.Griffiths JM, O'Neill AJ. 2012. Loss of function of the GdpP protein leads to joint β-lactam/glycopeptide tolerance in Staphylococcus aureus. Antimicrob Agents Chemother 56:579–581. 10.1128/AAC.05148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem 285:473–482. 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. 2010. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes. Antimicrob Agents Chemother 54:4900–4902. 10.1128/AAC.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Corrigan RM, Bellows LE, Wood A, Gründling A. 2016. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci USA 113:E1710–E1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Greninger AL, Chatterjee SS, Chan LC, Hamilton SM, Chambers HF, Chiu CY. 2016. Whole-genome sequencing of methicillin-resistant Staphylococcus aureus resistant to fifth-generation cephalosporins reveals potential non-mecA mechanisms of resistance. PLoS One 11:e0149541. 10.1371/journal.pone.0149541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pozzi C, Waters EM, Rudkin JK, Schaeffer CR, Lohan AJ, Tong P, Loftus BJ, Pier GB, Fey PD, Massey RC, O'Gara JP. 2012. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8:e1002626. 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zeden MS, Schuster CF, Bowman L, Zhong Q, Williams HD, Gründling A. 2018. Cyclic-di-adenosine monophosphate (c-di-AMP) is required for osmotic regulation in Staphylococcus aureus but dispensable for viability in anaerobic conditions. J Biol Chem 293:3180–3200. 10.1074/jbc.M117.818716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gentry D, Li T, Rosenberg M, McDevitt D. 2000. The rel gene is essential for in vitro growth of Staphylococcus aureus. J Bacteriol 182:4995–4997. 10.1128/JB.182.17.4995-4997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68:1128–1148. 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee YH, Nam KH, Helmann JD. 2013. A mutation of the RNA polymerase β′ subunit (rpoC) confers cephalosporin resistance in Bacillus subtilis. Antimicrob Agents Chemother 57:56–65. 10.1128/AAC.01449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Aiba Y, Katayama Y, Hishinuma T, Murakami-Kuroda H, Cui L, Hiramatsu K. 2013. Mutation of RNA polymerase β-subunit gene promotes heterogeneous-to-homogeneous conversion of β-lactam resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:4861–4871. 10.1128/AAC.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Villanueva M, Jousselin A, Baek KT, Prados J, Andrey DO, Renzoni A, Ingmer H, Frees D, Kelley WL. 2016. Rifampin resistance rpoB alleles or multicopy thioredoxin/thioredoxin reductase suppresses the lethality of disruption of the global stress regulator spx in Staphylococcus aureus. J Bacteriol 198:2719–2731. 10.1128/JB.00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cuirolo A, Plata K, Rosato AE. 2009. Development of homogeneous expression of resistance in methicillin-resistant Staphylococcus aureus clinical strains is functionally associated with a β-lactam-mediated SOS response. J Antimicrob Chemother 64:37–45. 10.1093/jac/dkp164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Plata KB, Rosato RR, Rosato AE. 2011. Fate of mutation rate depends on agr locus expression during oxacillin-mediated heterogeneous-homogeneous selection in methicillin-resistant Staphylococcus aureus clinical strains. Antimicrob Agents Chemother 55:3176–3186. 10.1128/AAC.01119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vuong C, Götz F, Otto M. 2000. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect Immun 68:1048–1053. 10.1128/IAI.68.3.1048-1053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bronesky D, Wu Z, Marzi S, Walter P, Geissmann T, Moreau K, Vandenesch F, Caldelari I, Romby P. 2016. Staphylococcus aureus RNAIII and its regulon link quorum sensing, stress responses, metabolic adaptation, and regulation of virulence gene expression. Annu Rev Microbiol 70:299–316. 10.1146/annurev-micro-102215-095708. [DOI] [PubMed] [Google Scholar]
- 147.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol 183:7341–7353. 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rudkin JK, Edwards AM, Bowden MG, Brown EL, Pozzi C, Waters EM, Chan WC, Williams P, O'Gara JP, Massey RC. 2012. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis 205:798–806. 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Song HS, Choi TR, Han YH, Park YL, Park JY, Yang SY, Bhatia SK, Gurav R, Kim YG, Kim JS, Joo HS, Yang YH. 2020. Increased resistance of a methicillin-resistant Staphylococcus aureus Δagr mutant with modified control in fatty acid metabolism. AMB Express 10:64. 10.1186/s13568-020-01000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rosado H, Turner RD, Foster SJ, Taylor PW. 2015. Impact of the β-lactam resistance modifier (−)-epicatechin gallate on the non-random distribution of phospholipids across the cytoplasmic membrane of Staphylococcus aureus. Int J Mol Sci 16:16710–16727. 10.3390/ijms160816710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in Gram-positive bacteria. Microbiol Mol Biol Rev 67:686–723. 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nakao A, Imai SI, Takano T. 2000. Transposon-mediated insertional mutagenesis of the d-alanyl-lipoteichoic acid (dlt) operon raises methicillin resistance in Staphylococcus aureus. Res Microbiol 151:823–829. 10.1016/S0923-2508(00)01148-7. [DOI] [PubMed] [Google Scholar]
- 153.Do T, Schaefer K, Santiago AG, Coe KA, Fernandes PB, Kahne D, Pinho MG, Walker S. 2020. Staphylococcus aureus cell growth and division are regulated by an amidase that trims peptides from uncrosslinked peptidoglycan. Nat Microbiol 5:291–303. 10.1038/s41564-019-0632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fujimura T, Murakami K. 2008. Staphylococcus aureus clinical isolate with high-level methicillin resistance with an lytH mutation caused by IS1182 insertion. Antimicrob Agents Chemother 52:643–647. 10.1128/AAC.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fujimura T, Murakami K. 1997. Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzymes. J Bacteriol 179:6294–6301. 10.1128/jb.179.20.6294-6301.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Collins J, Rudkin J, Recker M, Pozzi C, O'Gara JP, Massey RC. 2010. Offsetting virulence and antibiotic resistance costs by MRSA. ISME J 4:577–584. 10.1038/ismej.2009.151. [DOI] [PubMed] [Google Scholar]
- 157.Otto M. 2013. Community-associated MRSA: what makes them special? Int J Med Microbiol 303:324–330. 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Beck WD, Berger-Bachi B, Kayser FH. 1986. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol 165:373–378. 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ornelas-Soares A, De Lencastre H, De Jonge BLM, Tomasz A. 1994. Reduced methicillin resistance in a new Staphylococcus aureus transposon mutant that incorporates muramyl dipeptides into the cell wall peptidoglycan. J Biol Chem 269:27246–27250. 10.1016/S0021-9258(18)46975-X. [DOI] [PubMed] [Google Scholar]
- 160.Figueiredo TA, Sobral RG, Ludovice AM, de Almeida JMF, Bui NK, Vollmer W, de Lencastre H, Tomasz A. 2012. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog 8:e1002508. 10.1371/journal.ppat.1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Huber J, Donald RGK, Lee SH, Jarantow LW, Salvatore MJ, Meng X, Painter R, Onishi RH, Occi J, Dorso K, Young K, Park YW, Skwish S, Szymonifka MJ, Waddell TS, Miesel L, Phillips JW, Roemer T. 2009. Chemical genetic identification of peptidoglycan inhibitors potentiating carbapenem activity against methicillin-resistant Staphylococcus aureus. Chem Biol 16:837–848. 10.1016/j.chembiol.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 162.Maki H, Yamaguchi T, Murakami K. 1994. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J Bacteriol 176:4993–5000. 10.1128/jb.176.16.4993-5000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zeden MS, Burke Ó, Vallely M, Fingleton C, O'Gara JP. 2021. Exploring amino acid and peptide transporters as therapeutic targets to attenuate virulence and antibiotic resistance in Staphylococcus aureus. PLoS Pathog 17:e1009093. 10.1371/journal.ppat.1009093. [DOI] [PMC free article] [PubMed] [Google Scholar]