Abstract
Background:
Most available 4D-MRI techniques are limited by insufficient image quality and long acquisition times or require specially designed sequences or hardware that are not available in the clinic. These limitations have greatly hindered the clinical implementation of 4D-MRI.
Purpose:
This study aims to develop a fast ultra-quality (UQ) 4D-MRI reconstruction method using a commercially available 4D-MRI sequence and dual-supervised deformation estimation model (DDEM).
Methods:
Thirty-nine patients receiving radiotherapy for liver tumors were included. Each patient was scanned using a TWIST-VIBE MRI sequence to acquire 4D-MR images. They also received 3D T1-/T2-weighted MRI scans as prior images and UQ 4D-MRI at any instant was considered a deformation of them. A DDEM was developed to obtain a 4D deformable vector field (DVF) from 4D-MRI data, and the prior images were deformed using this 4D-DVF to generate UQ 4D-MR images. The registration accuracies of the DDEM, VoxelMorph (normalized cross-correlation (NCC) supervised), VoxelMorph (end-to-end point error (EPE) supervised), and the parametric total variation (pTV) algorithm were compared. Tumor motion on UQ 4D-MRI was evaluated quantitatively using region-of-interest (ROI) tracking errors, while image quality was evaluated using the contrast-to-noise ratio (CNR), lung–liver edge sharpness, and perceptual blur metric (PBM).
Results:
The registration accuracy of the DDEM was significantly better than those of VoxelMorph (NCC supervised), VoxelMorph (EPE supervised) and the pTV algorithm (all, p < 0.001), with an inference time of 69.3 ± 5.9 ms. UQ 4D-MRI yielded ROI tracking errors of 0.79 ± 0.65, 0.50 ± 0.55, and 0.51 ± 0.58 mm in the superior–inferior, anterior–posterior, and mid–lateral directions, respectively. From the original 4D-MRI to UQ 4D-MRI, the CNR increased from 7.25 ± 4.89 to 18.86 ± 15.81; the lung–liver edge full-width-at-half-maximum decreased from 8.22 ± 3.17 to 3.65 ± 1.66 mm in the in-plane direction and from 8.79 ± 2.78 to 5.04 ± 1.67 mm in the cross-plane direction, and the PBM decreased from 0.68 ± 0.07 to 0.38 ± 0.01.
Conclusion:
This novel DDEM method successfully generated UQ 4D-MR images based on a commercial 4D-MRI sequence. It shows great promise for improving liver tumor motion management during radiation therapy.
Keywords: Motion Management, 4D-MRI, Deformable Image Registration, Deep Learning
1. Introduction
Primary liver cancer and the metastasis of other cancers to the liver are leading causes of cancer-related death worldwide. Liver cancer accounted for 4.7% (905,677) of new cases and 8.3% (830,130) of deaths from all cancers in 20201. Radiation therapy (RT) is a major treatment modality for cancer. Historically, however, the role of RT in liver cancer has been limited to mostly palliative applications due to the potential for radiation-induced liver disease (RILD)2. The recent introduction of stereotactic body radiation therapy (SBRT) for liver cancer treatment3, however, has led to local tumor control rates of up to 86% at 3 years4, indicating its effectiveness. Despite these exciting improvements, RT is associated with Grade ≥ 3 toxicity event rates as high as 38%5, especially for patients with large tumors and poor liver function6. Use of an imaging modality with good soft tissue contrast during planning can help to manage tumor motion and improve the accuracy of beam delivery to the target, thus sparing normal tissues and reducing treatment toxicity.
Currently, four-dimensional computed tomography (4D-CT) is the standard imaging technique used for respiratory motion management in RT, especially when treating tumors in the chest region7. Unlike conventional CT, 4D-CT can show the motion trajectories of tumors during respiration; additionally, the internal target volumes (ITVs) defined on 4D-CT are significantly smaller than those on conventional CT8, 9, while maintaining the dose coverage10, 11. However, management of liver tumor motion using 4D-CT is difficult due to the poor soft-tissue contrast of CT. Also, the radiation dose received by patients in 4D-CT can be up to one order of magnitude higher than that in conventional 3D CT since multiple scans at the same couch position are needed for respiratory phase sorting12. To address these difficulties, various 4D-MRI techniques have been proposed13 and can be categorized as either retrospective or prospective 4D-MRI.
Most 4D-MRI techniques are retrospective, which means that images are continuously acquired over the whole region of interest (ROI) and retrospectively sorted into respiratory phases. For example, Yang et al. evaluated the correlation between diaphragm and liver tumor motion14, 15 and performed T2/T1-weighted 4D-MRI at a speed of 10–15 s per slice using a body area-based sorting method15. Liu et al.16 developed a k-space re-ordering method that uses breathing signals as the surrogate and verified its feasibility through studies of digital phantoms and healthy volunteers. Keijnemans et al.17 sorted images based on the liver dome position and achieved a total image acquisition time of 5 minutes. Other types of contrasts, including T2-weighted (T2w) imaging18, diffusion-weighted imaging (DWI)19, and quantitative magnetic resonance fingerprinting (MRF)20, can also be obtained via similar approaches. Despite these successes, retrospective 4D-MRI still faces challenges. First, sorting algorithms are sensitive to patients’ irregular breathing patterns, resulting in image artifacts, such as stitching artifacts. Second, the spatiotemporal resolution is compromised, especially with respect to large slice thicknesses (>5 mm) obtained with 2D acquisition-based methods. Third, long acquisition times (typically 10–15 minutes) are typically needed to satisfy the data sufficiency requirement in each phase bin.
Prospective 4D-MRI is achieved by using fast 3D acquisition or respiratory-gated 2D acquisition. For example, Yuan et al. 21 used a 3D spoiled gradient-echo sequence to image the abdomen for motion monitoring, with a temporal resolution of 0.615 s and voxel size of 2.7 mm × 2.7 mm × 4 mm. Due to the emphasis on imaging speed, prospective methods mainly use gradient-echo sequences and provide T1w images and are thus susceptible to field inhomogeneity. As the image quality and contrast are also compromised by the highly undersampled data, these images cannot be used for treatment planning. Using respiratory surrogates, Li et al. 22 proposed a prospective T2w 4D-MRI method involving 2D acquisition. Although the authors overcame the issue of data incompetency in sorting, scanning was time-consuming (~10 minutes), and the effects of irregular respiration on image quality were not resolved.
Despite recent advances in 4D-MRI development, current techniques still lack sufficient and consistent image quality for use in clinical applications due to at least one of the following deficiencies: inconsistent tumor contrast, image artifacts caused by breathing variations, and inadequate spatiotemporal resolution. In addition, some advanced 4D-MRI techniques require specially designed sequences or hardware and advanced image reconstruction methods that are not available in the clinic. These limitations have greatly hindered the clinical implementation of 4D-MRI.
This study aims to overcome the aforementioned deficiencies of existing 4D-MRI techniques by developing a ultra-quality (UQ) 4D-MRI method that uses a novel dual-supervised deformation estimation model (DDEM) based on a commercially available low-quality (LQ) 4D-MRI sequence. Specifically, respiratory motion patterns in LQ 4D-MRI were estimated using the DDEM and then applied to pre-aligned 3D-MRI data to reconstruct UQ 4D-MR images. We successfully generated T1w and T2w UQ 4D-MR images that exhibited significantly improved image quality and accurate tumor motion trajectory when compared with the original LQ 4D-MR images. Therefore, this new technique shows promise for wide clinical application.
2. Materials and Methods
2.1. Patient Data
The data used in this study were obtained from 39 patients undergoing RT for liver tumors. The study protocol was approved by the institutional review board. Among the 39 patients, 29 were male and 8 were female. 22 patients were diagnosed with primary liver cancer and 15 patients were diagnosed with liver metastases, and their age at diagnosis was 54.9 ± 10.9 years old.
Each patient underwent 4D-MRI using the TWIST volumetric interpolated breath-hold examination (TWIST-VIBE) MRI sequence23, which utilized view sharing technique with 20% central region and 20% sampling density in the peripheral region; hereafter, this is referred to as “original 4D-MRI.” This commercially available sequence was initially designed for fast volumetric imaging rather than 4D imaging. In this study, we decreased the acquisition time to 0.69s and continuously acquired 72 3D frames to achieve a 4D-MRI scan. Of the 39 patients, 27 also underwent regular T1w (free-breathing) and T2w (breath-holding) 3D MRI scans. Details of the imaging parameters are listed in Table 1. All the MR scans were done by a 3T scanner (Skyra, Siemens, Erlangen, Germany). All of the image intensities were normalized to 0-1 as pre-processing. For DDEM training, the original 4D-MR images were sorted into 10 phase bins based on the body area15 and paired up with a phase distance of 1, 2, 3, and 4, respectively. As a results, every patient contributes 40 image pairs. Patients with a low breathing amplitude or whose data contained severe artifacts were excluded from training. Cross-validation was conducted to fully utilize the data. Consequently, 25 patients were divided into 5 groups, each containing 5 patients. In each time, 4 groups with 800 image pairs were used for training and the remaining one with 200 image pairs was used for evaluation. Image binning was not needed for UQ 4D-MRI reconstruction, as deformations were directly estimated on time-resolved original 4D-MRI frames.
Table 1:
Imaging parameters of the enrolled patients.
| Original 4D-MRI | T1w 3D MRI | T2w 3D MRI | |
|---|---|---|---|
| No. of Patients | 39 | 27 | 27 |
| Contrast-enhanced | No | Yes | No |
| Acquisition mode | Free breathing | Free breathing | Gated |
| Sequence | TWIST-VIBE | Star-VIBE | TSE |
| Fat suppression | N/A | SPAIR | SPAIR |
| Turbo factor | N/A | N/A | 43 |
| Flip angle (°) | 5 | 9 | 72 |
| Echo trains per slice | N/A | N/A | 6 |
| Parallel imaging (factor) | CAIPIRINHA (4) | N/A | GRAPPA |
| Acceleration factor (PE) | 2 | N/A | 3 |
| Partial Fourier | 6/8 | 7/8 | N/A |
| TR (ms) | 3.44 | 2.83 | 1090.0 |
| TE (ms) | 1.23/2.45 | 1.48 | 84.0 |
| Bandwidth (Hz/pixel) | 1420 | 820 | 781 |
| Matrix size | 160 × 128 × 64 | 320 × 320 × 72 | 256 × 256 × 40 |
| Voxel size (mm) | 2.7 × 2.7 × 2.7 | 1.2 × 1.2 × 3.0 | 1.5 × 1.5 × 5.0 |
| Acquisition time | 0.69 s per 3D frame | 173 s | 60 s |
| No. of frames | 72 | N/A | N/A |
3D: Three-dimensional; 4D: Four-dimensional; CAIPIRINHA: Controlled aliasing in parallel imaging results in higher acceleration; GRAPPA: Generalized autocalibrating partial parallel acquisition; MRI: Magnetic resonance imaging; SPAIR: Spectral attenuated inversion recovery; T1w: T1-weighted; T2w: T2-weighted; TE: Echo time; TR: Repetition time; TSE: Turbo spin echo; TWIST: time-resolved imaging with interleaved stochastic trajectories; VIBE: volumetric interpolated breath-hold examination.
2.2. UQ 4D-MRI Reconstruction
The retrospective 4D-MRI method provides good spatial resolution and image contrast, while the prospective 4D-MRI method provides fine temporal resolution with few irregular breathing-induced artifacts. To combine the advantages of both methods, a prior image-based UQ 4D-MRI method was proposed. An overview of the method is shown in Figure 1. 3D MR images were deformed to each corresponding frame of the original 4D-MRI to reconstruct UQ 4D-MRI. Two aspects of this approach are challenging: potential differences in the respiratory phases of the 3D MR images used as prior images and those of the 4D-MRI frames, and the use of deep learning for image registration with several different types of image contrast24–26. Therefore, an automatic frame selection strategy was used instead of registering 3D MR images to every original 4D-MRI frame. Cross-correlation was computed between all 4D-MRI frames and the corresponding 3D MRI (T1w or T2w). The 4D-MRI frame of the highest cross-correlation value was considered at the closest respiratory phase of the corresponding 3D MRI, which was then registered to the 3D MRI using Elastix (Available at https://github.com/raacampbell/matlab_elastix) to account for residual mismatches. The registration is hierarchical and includes rigid, affine, and non-rigid B-spline stages to address coarse-to-fine deformations. Normalized correlation coefficient is the cost function because of the multi-parametric MRI registration. More details can be found at Mengler et al.’s work27. The alignment was manually verified before UQ 4D-MRI reconstruction. After these pre-processing procedures, the aligned frames were considered representations of the 3D MR images and registered to all other original 4D-MRI frames using DDEM to obtain the deformations along with the respiratory cycles (i.e., a 4D-DVF), which were then applied to the 3D MR images to reconstruct UQ 4D-MR images at corresponding frames.
Figure 1:
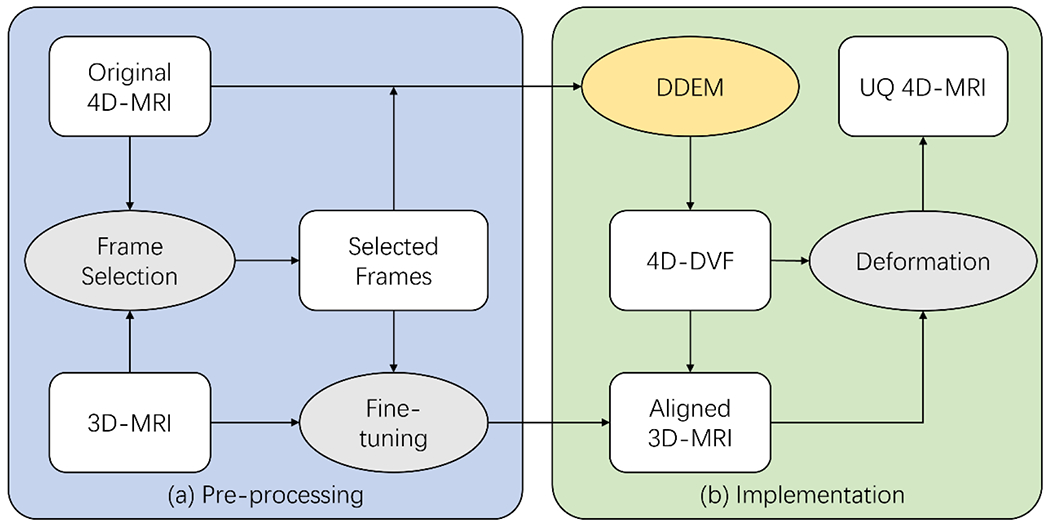
The overall study design of the proposed UQ 4D-MRI technique. (a) The original 4D-MRI frames whose respiratory phases were closest to that of the 3D-MR images were selected, and Elastix was used to fine-tune the residual mismatching. (b) A 4D-DVF between the selected frames and other frames was obtained via DDEM and applied to the aligned 3D-MR image to reconstruct the UQ 4D-MR images using corresponding frames. 4D: four-dimensional; DDEM: dual-supervised deformation estimation model; MRI: magnetic resonance imaging; DVF: displacement vector field UQ: ultra-quality.
2.3. DDEM
The original 4D-MR images were subject to noise and artifacts due to acquisition limitations, and thus, unsupervised training was challenging. Dual supervision was used to provide more accurate DVFs for UQ 4D-MRI reconstruction28. From each original 4D-MR image pair, a reference DVF was calculated using the parametric total variation (pTV) algorithm with the local correlation coefficient as the cost function29. This method was designed explicitly for respiratory motion registration and has achieved the best performance to date on the benchmark DIR-Lab30, 31. A difference map between the image pair was also fed to the DDEM to improve registration accuracy28, 32 Consequently, each training sample consisted of a moving phase, a fixed phase, a difference map, and a reference DVF. During training, the 4D-MR images and reference DVFs were cropped to fit the patients’ body shapes and resized to 128 × 128 × 64 voxels to save GPU memory. In addition to the end-to-end point error (EPE) between the reference deformation DVFr and the predicted deformation DVFp, the negative normalized correlation coefficient (NCC) between the warped volume Vw and fixed volume Vf was also included in the loss function L to compensate for potentially imperfect reference DVFs:
| (1.1) |
| (1.2) |
| (1.3) |
where p represents a voxel in space denotes the local mean intensity around ; and the window size W = 9. α and β are weighting factors and varied dynamically during training. A larger α during the initial training can encourage convergence, and a larger β during fine-tuning at a later stage can further optimize the registration. We empirically set α = 1 and β = 1 during the first 150 epochs, and α = 1 and β = 5 during the last 50 epochs.
The network architecture of DDEM was based on VoxelMorph33, as shown in Figure 2. It receives two phases of the original 4D-MRI as moving and fixed volumes and a difference map between them. The first half of the DDEM is an encoder consisting of 4 sequential convolutional blocks used to extract image features. The first three blocks have a 3 × 3 × 3 convolutional layer, with a stride of 2, followed by a leaky rectified linear unit (ReLU) activation layer, while the stride in the last one is 1. The numbers of convolutional kernels in these blocks are 16, 32, 32, and 32, respectively. Using the decoder, the extracted image features were then processed to generate a DVF between the moving and fixed images. The decoder uses similar convolutional blocks with 32 kernels each, and 2 × 2 × 2 up-sampling layers are applied after each ReLU activation to expand the feature volume. The two parts are linked by concatenation connections, which convey local information captured by the encoder to the decoder to better utilize the features. The last convolutional layer contains 3 convolutional kernels, and the resulting three output channels represent motion in the x-, y-, and z-directions. The deformation was then applied to the moving volume to generate a warped volume Vw as another output. Another two models with the identical network architecture but supervised by NCC and EPE were trained as comparison baselines, respectively. All models were trained by an ADAM optimizer for 200 epochs at an initial learning rate of 10−4, which was halved every 50 epochs. Training was conducted using with Pytorch 1.7.0 on an Nvidia RTX 3090 GPU and an Intel I7-9700K CPU.
Figure 2:
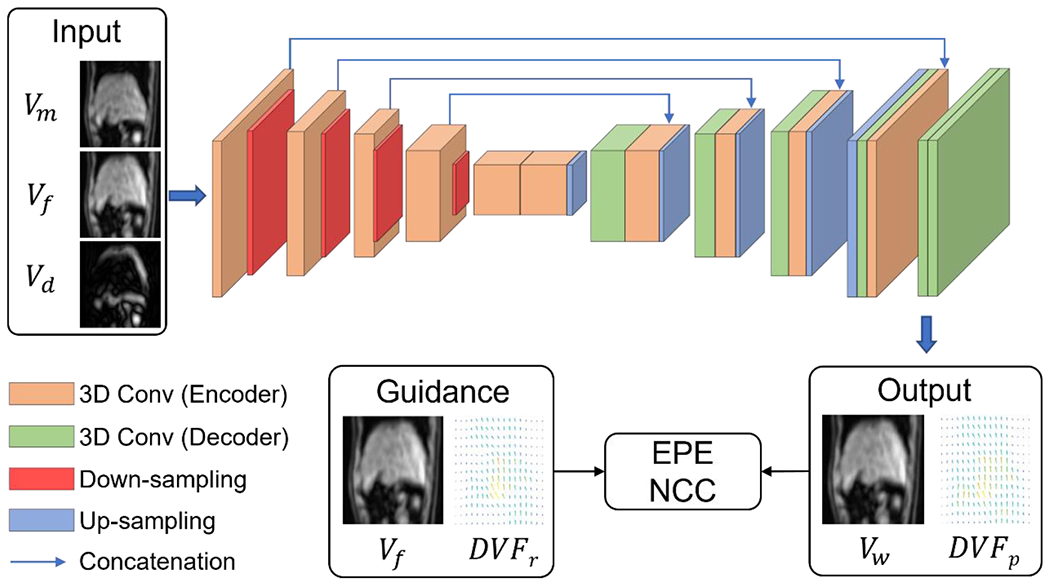
Architecture of the proposed DDEM. The moving volume (Vm), fixed volume (Vf), and map of differences map between these volumes (Vd) were input to the network. The output consisted of a predicted deformation (DVFp) and warped volume (Vw). The training was dually supervised by the reference deformation (DVFr) and Vf. 3D: three-dimensional; Conv: convolution; EPE: end-to-end point error; DVF: displacement vector field; NCC: normalized cross correlation.
2.4. Evaluation
The registration performances of DDEM, VoxelMorph, and pTV algorithm on the original 4D-MRI data were evaluated using intensity-based metrics, including the structural similarity index (SSIM), peak signal-to-noise ratio (PSNR), mean squared error (MSE), and NCC. Each image was restored to its original shape prior to the metric calculations. T-tests were performed on the metrics to evaluate the significance with a threshold of 0.05. The inference times of the methods were also recorded for comparison.
4D imaging is used primarily for motion management, and therefore, motion accuracy is an essential component of UQ 4D-MR images. A region matching-based method for ROI tracking was developed in MATLAB. 3D ROI bounding boxes were defined on the first frames of 4D images and used to screen subsequent volumetric frames to identify the locations with maximum image similarity in each frame. The displacements between the positions of the initial bounding box and the positions in subsequent frames were calculated as the ROI motion trajectory. 4D images were interpolated before measurement to ensure a motion measurement resolution of 0.33 mm. ROI motion amplitude was recorded in the superior–inferior (SI), anterior–posterior (AP), and mid–lateral (ML) directions, and the relative motion error was calculated as the difference in ROI motion amplitude between the original and the UQ 4D-MR images.
The contrast-to-noise ratio (CNR) was used to quantify tumor contrast enhancement in the UQ 4D MR images. Precisely, the mean voxel intensities of the tumor and surrounding normal liver tissue were calculated, and the difference between these values was divided by the standard deviation sampled from the normal liver tissue.
A no-reference perceptual blur metric (PBM) proposed by Crete et al.34 was used to evaluate the overall image quality of the original and UQ 4D MR images. Briefly, PBM compares variations in the intensities of neighboring voxels between the original image and an image to which a low-pass filter has been applied to cause blurring. Less variation in intensity represents a more blurred original image, resulting in a higher PBM value.
The lung–liver edge full-width at half-maximum (FWHM) was also used to quantify the sharpness of the organ boundaries35. Linear profiles were taken at the liver–lung interfaces on MR images in the cross-plane and in-plane directions, and curves were fitted along with them:
| (2) |
where p is the voxel position; f is the voxel intensity at p; a0, a1, and a2 are fitted constants; and s represents the sharpness of the curve. Thresholds of 5% and 95% of the maximum were applied to the curves to define an edge, and the distance between the two thresholds was considered the FWHM of the edge.
Because the CNR and edge FWHM calculations involved manually selected ROIs, each measurement was repeated three times, and the average value was reported. Some patients had already undergone surgery before RT, and their MR images did not have solid tumors. In such cases, another organ, such as the gallbladder, was chosen as a substitute ROI in the motion trajectory measurement, and the CNR was not calculated.
3. Results
3.1. Performance of the DDEM
The results of a comparison of the registration performance of the DDEM, VoxelMorph, and reference pTV algorithm are shown in Table 2. Both the DDEM and VoxelMorph models had a faster inference time than the pTV algorithm. In terms of image similarity, the DDEM had significantly better NCC, MSE, PSNR, and SSIM than VoxelMorph (NCC supervised), VoxelMorph (EPE supervised), and pTV algorithm (p< 0.001 for all metrics). These results suggest that dual supervision can help the model to converge and surpass the reference labels.
Table 2:
Registration results of all of the methods applied to the test set.
| MSE (10−3) | NCC | PSNR | SSIM | Time | |
|---|---|---|---|---|---|
| w/o Registration | 1.60±1.10 | 0.772±0.053 | 28.95±3.21 | 0.912±0.050 | N/A |
| pTV algorithm | 0.95±0.53 | 0.825±0.037 | 31.02±2.88 | 0.960±0.015 | 28.4±3.4 s |
| VoxelMorph (NCC supervised) | 0.56±0.32 | 0.856±0.030 | 33.28±2.81 | 0.963±0.016 | 69.1±5.9 ms |
| VoxelMorph (EPE supervised) | 1.00±0.54 | 0.817±0.038 | 30.78±2.79 | 0.955±0.018 | 69.9±5.3 ms |
| DDEM | 0.43±0.28 | 0.908±0.022 | 34.72±3.16 | 0.978±0.009 | 69.3±5.9 ms |
DDEM: dual-supervised deformation estimation model; EPE: end-to-end point error; MSE: Mean squared error; NCC: Normalized cross-correlation; PSNR: Peak signal-to-noise ratio; pTV: parametric total variation; SSIM: Structure similarity index measure.
3.2. UQ 4D-MRI
The original 4D-MR and reconstructed UQ T1w and T2w 4D-MR images of an example patient in the axial and sagittal views are shown in Figure 3(a) and (b), respectively. Six consecutive frames are presented, and the arrows indicate the tumor position in the first frame. As shown, the original 4D-MR images have insufficient image contrast, such that the tumor is almost invisible, with a tumor CNR of only 1.9. Motion artifacts can also be observed, resulting in inhomogeneous patterns in the liver and blurred lung–liver boundaries. The PBM of those images was 0.700, and the FWHM of the lung–liver edge was 8.92 and 12.64 mm in the cross-plane and in-plane directions, respectively. In comparison, the reconstructed UQ T1w 4D-MR images exhibit better tumor contrast, with a higher CNR of 7.7, and less severe motion artifacts. In the reconstructed UQ T1w images, the PBM decreased to 0.426, and the FWHM of the lung–liver edge decreased to 11.86 and 7.38 mm in the cross-plane and in-plane directions, respectively. Additional comparison reveals that the tumor visibility was further improved in the UQ T2w 4D-MR images, with the highest CNR of 14.4, a considerable increase in lung–liver edge sharpness, and a reduction in motion artifacts. The PBM decreased to 0.397, and the FWHM of the lung–liver edge decreased to 5.12 and 3.21 mm in the cross-plane and in-plane directions, respectively.
Figure 3:
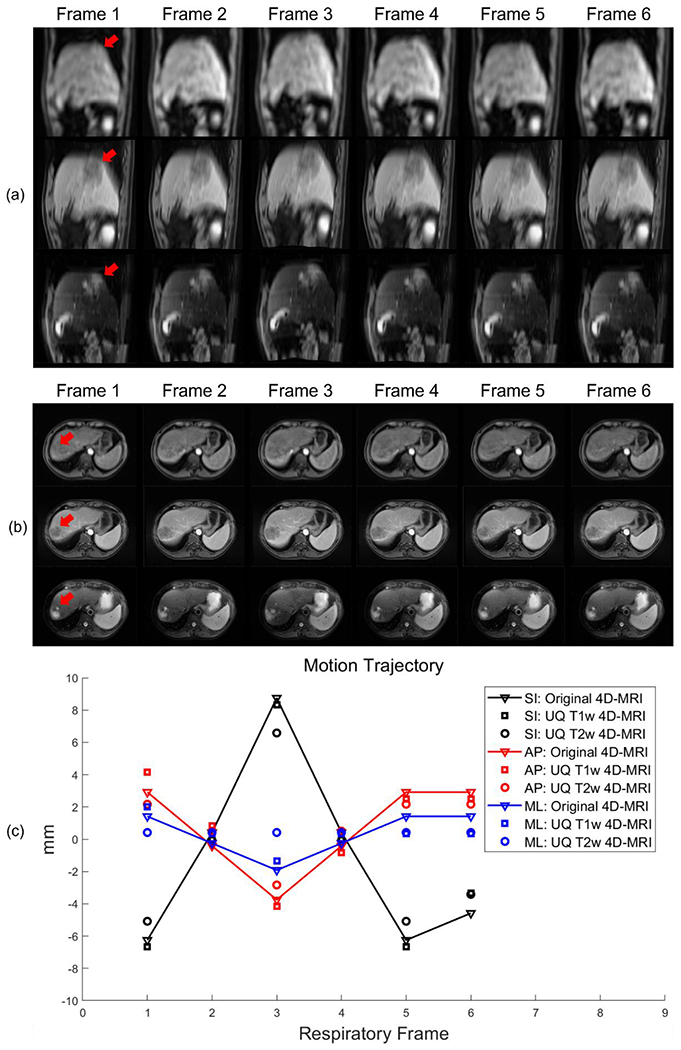
Reconstruction of an image from a sample patient using 6 consecutive frames. (a, b) Original 4D-MR (top), UQ T1w 4D-MR (mid), and UQ T2w 4D-MR images (bottom) of the patient in sagittal and axial views, respectively. Arrows indicate the tumor in the first frame. The images represent 3D volumes; only 2D slices are shown here for demonstration. (c) The tumor motion trajectory of this patient in the SI, AP, and ML directions. UQ 4D-MR images show good matching of tumor motion with the original 4D-MR data. 4D-MRI: four-dimensional magnetic resonance imaging; AP: anterior-posterior; ML: mid-lateral; SI: superior-inferior; T1w: T1-weighted; T2w: T2-weighted; UQ: ultra-quality.
In addition to image quality, motion was accurately reflected in the generated UQ 4D-MR images. Figure 3(c) shows the motion amplitude of the tumor in 6 consecutive frames. As expected in respiration, the motion amplitude was largest in the SI direction. In the UQ T1w and T2w 4D-MR images, the tumor motion matched well with that in the original 4D-MR images, and most of the relative motion errors were within 2 mm. The respective mean motion errors in all 72 frames in the AP, ML, and SI directions were 0.70, 0.76, and 0.62 mm for the UQ T1w 4D-MR images and 0.87, 0.90, and 0.82 mm for the UQ T2w 4D-MR images.
3.3. Statistical Results
UQ T1w and T2w 4D-MRI were obtained on 27 patients who had both 4D-MRI and 3D MRI scans, and ROI motion amplitude was measured on all of them. The results of the statistical analysis of the mean motion error are shown as box plots in Figure 4(a, b). The relative ROI motion errors of UQ T1w 4D-MRI in the SI, AP, and ML directions were 0.80 ± 0.36, 0.50 ± 0.36, and 0.50 ± 0.39 mm, respectively. The relative ROI motion errors of UQ T2w 4D-MRI in the SI, AP, and ML directions were 0.78 ± 0.42, 0.51 ± 0.36, and 0.52 ± 0.43 mm, respectively. For both contrasts, relative ROI motion errors were 0.79 ± 0.65, 0.50 ± 0.55, and 0.51 ± 0.58 mm in the SI, AP, and ML directions. The relative ROI motion errors were < 1 mm in all directions. The images of 19 of these 27 patients contained visible solid tumors, and CNR was measured only in these images. The results of statistical analysis of the tumor CNR in different MR images are shown as box plots in Figure 4(c). The CNR increased from 7.25 ± 4.89 in the original 4D-MR images to 7.86 ± 5.37 and 18.86 ± 15.81 in the UQ T1w and T2w 4D-MR images, respectively. Plots of the lung–liver edge FWHM in the in-plane and cross-plane directions are displayed in Figure 4(d) and Figure 4(e), respectively. The lung–liver edge FWHM in the in-plane direction was reduced from 8.22 ± 3.17 mm in the original 4D-MR images to 4.51 ± 1.98 mm and 3.65 ± 1.66 mm in the UQ T1w and T2w 4D-MR images, respectively. In the cross-plane direction, the lung–liver edge FWHM was reduced from 8.79 ± 2.78 mm in the original 4D-MR images to 7.27 ± 2.71 mm and 5.04 ± 1.67 mm in the UQ T1w and T2w 4D-MR images, respectively. The PBM results of different 4D-MR images are shown in Figure 4(f). The PBM in the original 4D-MRI was 0.68 ± 0.07, and this value decreased to 0.38 ± 0.01 and 0.40 ± 0.01 in the UQ T1w and T2w 4D-MR images, respectively.
Figure 4:
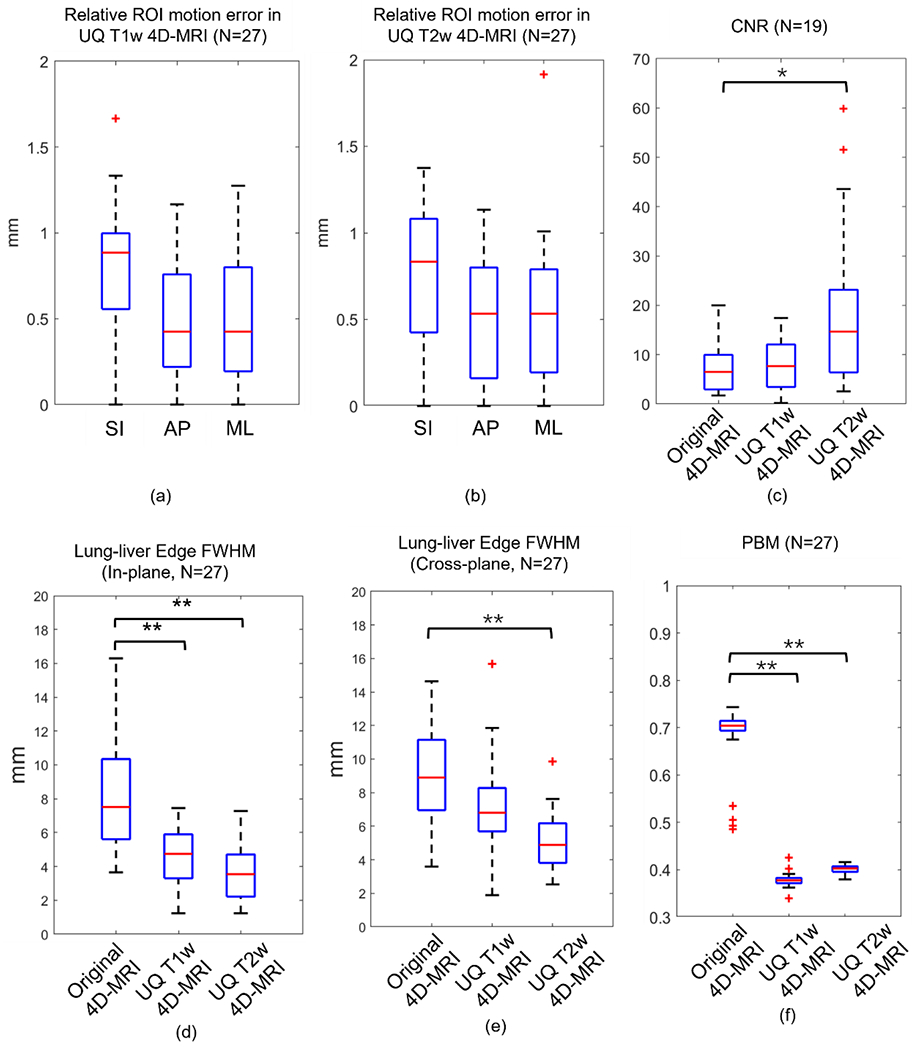
Statistical analysis of the patient data. (a, b) Relative ROI motion error of the UQ T1w and T2w 4D-MR images in the SI, AP, and ML directions, respectively. (c) CNR of the original 4D-MR and reconstructed UQ T1w and T2w 4D-MR images. (d, e) Lung–liver edge FWHM of the original 4D-MR and reconstructed UQ T1w and T2w 4D-MR images in the in-plane and cross-plane directions, respectively. (f) PBM of the original 4D-MR and reconstructed UQ T1w and T2w 4D-MR images. *: P-value < 0.05; **: P-value < 0.001; 4D-MRI: four-dimensional magnetic resonance imaging; AP: anterior-posterior; CNR: contrast-to-noise ratio; FWHM: full width at half maximum; ML: mid-lateral; PMB: perceptual blur metric; ROI: region of interest; SI: superior-inferior; T1w: T1-weighted; T2w: T2-weighted; UQ: ultra-quality.
4. Discussion
In this paper, we report the initial results of UQ 4D-MRI using a DDEM based on a commercially available 4D-MRI sequence. The registration performance of DDEM outperformed those of VoxelMorph (NCC supervised), VoxelMorph (EPE supervised), and the pTV algorithm, and the reconstructed UQ 4D-MR images showed significantly improved image quality compared with that of commercial 4D-MRI and yielded accurate tumor motion measurements with a fast processing time.
The use of dual supervision is believed to explain the superior performance of DDEM over VoxelMorph (NCC supervised), VoxelMorph (EPE supervised), and the pTV algorithm. Medical image registration has been a hot research topic for decades, and numerous methods have been proposed. Despite huge successes, low-quality image registration remains a big challenge, and the accuracy of both iterative and deep learning methods worsens as the down-sampling factor increases36. Guidance from reference DVFs can encourage the convergence and improve the performance of DDEM., even if the original 4D-MR images are noisy and blurred. On the other hand, guidance from image similarity can further fine-tune the registration and help surpass the reference DVFs. Fan et al.28 demonstrated a similar improvement in registration accuracy with dual supervision, although their model was patch-based and evaluated using high-quality brain MR images. We extended our method to predict the deformation of the whole ROI and 4D-MR images with respiratory motion. Preparing reference DVFs can be time-consuming and may limit the application of dual supervision24–26. However, the reference DVFs were only needed during model training and dual supervision did not introduce any additional latency in inference, as shown in Table 2.
The reconstructed UQ 4D-MR images generated via DDEM exhibited significant improvements in image quality, including tumor CNR and image texture, compared with the original 4D-MR images. In particular, the lung–liver edges were sharper, and blurring artifacts were considerably decreased. The tumor motion trajectories derived from UQ 4D-MR images were comparable to those derived from the original 4D-MR images, with a mean error < 1 mm in all directions, suggesting that our method accurately reflects tumor motion. Also, the motion trajectories show no significant difference between UQ T1w and T2w 4D-MRI, which is expected since they were derived from the same DVT. However, potential errors might exist in pre-processing, such as inaccurate alignment in frame selection. The consistent tumor motion trajectories in both UQ 4D-MR images can be a quality assurance of the entire workflow. The image quality of reconstructed UQ 4D-MRI is inherited from 3D MRI, and so are the artifacts. For example, the T1w 3D MRI was acquired under free-breathing and volume-averaging artifacts present, especially at the lung-liver interface. The T2w 3D MRI was acquired under breath-holding with multiple concatenations (typically 4~5), resulting in the bright or dark slabs. The reconstructed UQ 4D-MRI also shows these artifacts. However, these artifacts do not affect the accuracy of DDEM and UQ 4D-MRI reconstruction, since registration was only conducted between the original 4D-MRI frames.
We have shown that our UQ 4D-MRI method has major advantages over current retrospective 4D-MRI methods, which rely on sorting algorithms and are thus vulnerable to irregular breathing and can cause stitching artifacts or missing slices. Furthermore, the spatial resolution of existing methods is often compromised due to the time efficiency and data sufficiency requirements of retrospective 4D-MRI, and the acquisition times are relatively long. In contrast, our method uses 3D MR images as prior knowledge. The resulting UQ 4D-MR images inherit quality from these prior images and thus can achieve a comparable level of image quality, which is indicated by smooth and complete organ surfaces and fine voxel sizes. In addition, apart from image acquisition, our method generates UQ 4D-MR images at a speed of 60 ms per volumetric frame, making it highly applicable in the clinic.G
Our UQ 4D-MRI method also has advantages over current prospective 4D-MRI methods, which often produce images with compromised quality, have limited image contrast choices, and are sensitive to field inhomogeneity. As noted above, the quality of images generated using our method is comparable to that of the prior 3D MR images, which can be acquired via sequences robust to field inhomogeneity, such as fast spin echo. Our method also provides versatile image contrast choices. Although only T1w and T2w 4D-MR images were reconstructed in this study, our method can be further extended to multiple types of image contrast if corresponding 3D MR images are provided.
Our method can also be used to obtain accurate 3D tumor motion trajectories, whereas other studies investigating prior image-based time-resolved 4D-MRI have only evaluated tumor motion trajectories on 2D images or projections. For example, Harris et al. modeled a patient’s basic deformation patterns using principal component analysis (PCA) and calculated the coefficients for those patterns based on a single 2-dimensional (2D) cine MRI37, multiple 2D cine MRI38, or on-board kV projections39. The deformation was then applied to a prior MR image to generate real-time volumetric cine MRI for lung tumor patients. Pham et al. 40 developed a similar method but used a neural network to predict pattern coefficients. In that study, the high contrast between the tumor and normal tissue in the lungs made it relatively easy to capture and model tumor motion. In contrast, we focused on liver tumors when developing our method, which are more challenging due to the poor contrast. Additionally, both of their ground truth motion trajectories were obtained from one or multiple 2D images and thus may not reflect accurate 3D tumor motion, resulting in misleading 3D image deformation. In contrast, our method directly tracks tumor motion on 3D images and yields more reliable results. Finally, the deformation reported in earlier studies involved a combination of basic patterns, and small motions may have been omitted during motion modeling. Our method uses DIR for deformation estimation, which theoretically can capture any motion.
Our UQ method can be used to facilitate the application of 4D-MRI for clinical purposes, including target volume delineation and on-board treatment guidance. Zhang et al. showed that a fusion of multisource MRI could significantly improve the tumor CNR compared with a single type of contrast41, leading to improved inter-observer consistency during gross tumor volume delineation42. Due to monetary and temporal limitations, it may be impractical to require patients to undergo multiple 4D-MRI scans. Our method enables patients to undergo multiple 3D MRI scans and obtain UQ 4D-MR images with various contrasts. Current MRI-based on-board treatment guidance relies on 2D cine MRI with orthogonal views, which can only provide information about tumor motion on a couple of projections43. In contrast, our method enables real-time 4D-MRI if the temporal resolution of the original 4D-MRI is sufficient, thus providing more accurate information for treatment monitoring and tracking.
One limitation of our study is the absence of functional images. DWI and dynamic contrast-enhanced (DCE) images provide additional information beyond anatomical images, which can potentially facilitate the localization of liver tumors44 and metastases45. However, this study focused only on T1w and T2w images to demonstrate the feasibility of our method. In future research, we will enroll additional patients with functional MR image data, including DWI and DCE images, to further verify the generalizability of our method. Another limitation is the temporal resolution of UQ 4D-MRI. Currently, the acquisition time of conventional 4D-MRI is 0.69 s per frame to achieve good image quality, which yields around six frames per typical respiratory cycle (~4 s). Some studies have demonstrated the feasibility of DVF estimation using highly undersampled images36. In the future, we will further reduce the acquisition time of the original commercial 4D-MRI scans to improve the temporal resolution. Finally, it is noted that the proposed method may benefit from more validation and testing before its clinical implementation. We did not introduce any specially designed types of processing to the dataset, and we expect that our method will be robust to other patient cohorts and commercial MRI sequences. In our future studies, we plan to use digital human phantoms, such as the 4D Digital Extended Cardiac-Torso (XCAT) Phantom46, 47, to comprehensively and thoroughly evaluate the robustness and accuracy of our proposed method. We will also enroll patients at other hospitals that use different scanning protocols to further evaluate the generalizability of our method.
5. Conclusion
This study developed a novel DDEM for fast UQ 4D-MRI reconstruction based on a commercially available 4D-MRI sequence. Compared with the original 4D-MR images, the reconstructed UQ 4D-MR images provided versatile image contrast, significantly improved image quality, and accurate tumor motion trajectories within a short processing time. This method overcomes the limitations of current retrospective and prospective 4D-MRI methods and shows great promise as a means of expanding the clinical implementation of 4D-MRI for liver cancer motion management during RT.
Acknowledgements
We would like to thank the support from the 2022 Exchange Program for Teachers and Students of Higher Education Institutions in Hong Kong, Macao and MainlandChina. Dr. Jing Cai received research grants from General Research Fund (GRF 15102118, GRF 15102219), the University Grants Committee, Health and Medical Research Fund (HMRF 06173276), the Food and Health Bureau, Hong Kong Special Administrative Regions, and National Institutes of Health (NIH R01 CA226899), The United States of America.
Conflict of Interest Statement for All Authors
Dr. Jing Cai received research grants from General Research Fund (GRF 15102118, GRF 15102219), the University Grants Committee, Health and Medical Research Fund (HMRF 06173276), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Regions, and National Institutes of Health (NIH R01 CA226899), Department of Health & Human Services, The United States of America. Other authors have nothing to disclose.
Funding Statement
This research was partly supported by research grants of General Research Fund (GRF 15102118, GRF 15102219), the University Grants Committee, Health and Medical Research Fund (HMRF 06173276), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Regions, and National Institutes of Health (NIH R01 CA226899), Department of Health & Human Services, The United States of America.
Data Availability Statement for this Work
Research data are not available at this time.
Reference:
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Chen CP. Role of radiotherapy in the treatment of hepatocellular carcinoma. J Clin Transl Hepatol. 2019;7(2):183–190. doi: 10.14218/JCTH.2018.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerum S, Heinz C, Belka C, et al. Stereotactic body radiation therapy (SBRT) in patients with hepatocellular carcinoma and oligometastatic liver disease. Radiation Oncology. 2018/05/29 2018; 13( 1): 100. doi: 10.1186/s13014-018-1048-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rim CH, Kim HJ, Seong J. Clinical feasibility and efficacy of stereotactic body radiotherapy for hepatocellular carcinoma: a systematic review and meta-analysis of observational studies. Radiother Oncol. 2019/02/01/ 2019;131:135–144. doi: 10.1016/j.radonc.2018.12.005 [DOI] [PubMed] [Google Scholar]
- 5.Murray LJ, Dawson LA. Advances in stereotactic body radiation therapy for hepatocellular carcinoma. Semin Radiat Oncol. 2017/07/01/ 2017;27(3):247–255. doi: 10.1016/j.semradonc.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 6.Scorsetti M, Comito T, Cozzi L, et al. The challenge of inoperable hepatocellular carcinoma (HCC): results of a single-institutional experience on stereotactic body radiation therapy (SBRT). J Cancer Res Clin Oncol. 2015/07/01 2015; 141(7): 1301–1309. doi: 10.1007/s00432-015-1929-y [DOI] [PubMed] [Google Scholar]
- 7.Keall PJ, Mageras GS, Balter JM, et al. The management of respiratory motion in radiation oncology report of AAPM Task Group 76. Med Phys. Oct 2006;33(10):3874–3900. doi: 10.1118/1.2349696 [DOI] [PubMed] [Google Scholar]
- 8.Hof H, Rhein B, Haering P, Kopp-Schneider A, Debus J, Herfarth K. 4D-CT-based target volume definition in stereotactic radiotherapy of lung tumours: comparison with a conventional technique using individual margins. Radiother Oncol. 2009;93(3):419–423. doi: 10.1016/j.radonc.2009.08.040 [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Hayes S, Paskalev K, et al. Dosimetric comparison of stereotactic body radiotherapy using 4D CT and multiphase CT images for treatment planning of lung cancer: evaluation of the impact on daily dose coverage. Radiother Oncol. 2009;91(3):314–324. doi: 10.1016/j.radonc.2008.11.018 [DOI] [PubMed] [Google Scholar]
- 10.Zhao L, Sandison GA, Farr JB, Hsi WC, Li XA. Dosimetric impact of intrafraction motion for compensator-based proton therapy of lung cancer. Phys Med Biol. Jun 21 2008;53(12):3343. doi: 10.1088/0031-9155/53/12/019 [DOI] [PubMed] [Google Scholar]
- 11.Underberg RW, Lagerwaard FJ, Cuijpers JP, Slotman BJ, De Koste JRVS, Senan S. Four-dimensional CT scans for treatment planning in stereotactic radiotherapy for stage I lung cancer. Int J Radiat Oncol Biol Phys. Nov 15 2004;60(4):1283–1290. doi: 10.1016/j.ijrobp.2004.07.665 [DOI] [PubMed] [Google Scholar]
- 12.Li T, Schreibmann E, Thorndyke B, et al. Radiation dose reduction in four-dimensional computed tomography. Med Phys. 2005;32(12):3650–3660. doi : 10.1118/1.2122567 [DOI] [PubMed] [Google Scholar]
- 13.Stemkens B, Paulson ES, Tijssen RH. Nuts and bolts of 4D-MRI for radiotherapy. Phys Med Biol. 2018;63(21):21TR01. doi: 10.1088/1361-6560/aae56d [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Cai J, Wang H, et al. Is diaphragm motion a good surrogate for liver tumor motion? Int J Radiat Oncol Biol Phys. Nov 15 2014;90(4):952–8. doi: 10.1016/j.ijrobp.2014.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J, Cai J, Wang H, et al. Four-dimensional magnetic resonance imaging using axial body area as respiratory surrogate: initial patient results. Int J Radiat Oncol Biol Phys. 2014;88(4):907–912. doi: 10.1016/j.ijrobp.2013.11.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Yin F-F, Chen N-k, Chu M-L, Cai J. Four dimensional magnetic resonance imaging with retrospective k-space reordering: A feasibility study. 2015;42(2):534–541. doi: 10.1118/1.4905044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keijnemans K, Borman PTS, van Lier A, Verhoeff JJC, Raaymakers BW, Fast MF. Simultaneous multi-slice accelerated 4D-MRI for radiotherapy guidance. Phys Med Biol. Apr 28 2021;66(9)doi: 10.1088/1361-6560/abf591 [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Yin FF, Czito BG, Bashir MR, Cai J. T2-weighted four dimensional magnetic resonance imaging with result-driven phase sorting. Med Phys. Aug 2015;42(8):4460–71. doi: 10.1118/1.4923168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Zhong X, Czito BG, et al. Four-dimensional diffusion-weighted MR imaging (4D-DWI): a feasibility study. Med Phys. 2017;44(2):397–406. doi: 10.1002/mp.12037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Cui D, Hui ES, Cai J. Time-resolved magnetic resonance fingerprinting for radiotherapy motion management. Med Phys. Dec 2020;47(12):6286–6293. doi: 10.1002/mp.14513 [DOI] [PubMed] [Google Scholar]
- 21.Yuan J, Wong OL, Zhou Y, Chueng KY, Yu SK. A fast volumetric 4D-MRI with sub-second frame rate for abdominal motion monitoring and characterization in MRI-guided radiotherapy. Quant Imaging Med Surg. 2019;9(7):1303. doi: 10.21037/qims.2019.06.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, Wei J, Olek D, et al. Direct comparison of respiration-correlated four-dimensional magnetic resonance imaging reconstructed using concurrent internal navigator and external bellows. International Journal of Radiation Oncology*Biology*Physics. 2017/03/01/ 2017;97(3):596–605. doi: 10.1016/j.ijrobp.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Block KT, Chandarana H, Fatterpekar G, et al. Improving the robustness of clinical T1-weighted MRI using radial VIBE. Magnetom Flash. 2013;5:6–11. [Google Scholar]
- 24.Fu Y, Lei Y, Wang T, Curran WJ, Liu T, Yang X. Deep learning in medical image registration: a review. Phys Med Biol. Oct 22 2020;65(20):20TR01. doi: 10.1088/1361-6560/ab843e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haskins G, Kruger U, Yan R Deep learning in medical image registration: a survey. Machine Vision and Applications. 2020;31(1):8. doi: 10.1007/s00138-020-01060-x [DOI] [Google Scholar]
- 26.Xiao H, Teng X, Liu C, et al. A review of deep learning-based three-dimensional medical image registration methods. Quant Imaging Med Surg. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mengler L, Khmelinskii A, Diedenhofen M, et al. Brain maturation of the adolescent rat cortex and striatum: changes in volume and myelination. Neuroimage. 2014/01/01/ 2014;84:35–44. doi: 10.1016/j.neuroimage.2013.08.034 [DOI] [PubMed] [Google Scholar]
- 28.Fan JF, Cao XH, Yap EA, Shen DG. BIRNet: Brain image registration using dual-supervised fully convolutional networks. Med Image Anal. May 2019;54:193–206. doi: 10.1016/j.media.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vishnevskiy V, Gass T, Szekely G, Tanner C, Goksel O. Isotropic total variation regularization of displacements in parametric image registration. IEEE Tram Med Imaging. Feb 2016;36(2):385–395. doi: 10.1109/TMI.2016.2610583 [DOI] [PubMed] [Google Scholar]
- 30.Castillo R, Castillo E, Guerra R, et al. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol. 2009;54(7):1849. doi: 10.1088/0031-9155/54/7/001 [DOI] [PubMed] [Google Scholar]
- 31.Castillo R, Castillo E, Fuentes D, et al. A reference dataset for deformable image registration spatial accuracy evaluation using the COPDgene study archive. Phys Med Biol. May 7 2013;58(9):2861–77. doi: 10.1088/0031-9155/58/9/2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan J, Cao X, Yap P-T, Shen D. BIRNet: Brain image registration using dual-supervised fully convolutional networks. Med Image Anal. 2019;54:193–206. doi: 10.1016/j.media.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, Dalca AV. VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging. Feb 4 2019;doi: 10.1109/TMI.2019.2897538 [DOI] [PubMed] [Google Scholar]
- 34.Crete F, Dolmiere T, Ladret P, Nicolas M. The blur effect: perception and estimation with a new no-reference perceptual blur metric. International Society for Optics and Photonics; 2007:64920I. [Google Scholar]
- 35.Ahmad R, Ding Y, Simonetti OP. Edge sharpness assessment by parametric modeling: application to magnetic resonance imaging. Concepts Magn Reson Part A Bridg Educ Res. May 1 2015;44(3):138–149. doi: 10.1002/cmr.a.21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terpstra ML, Maspero M, D’Agata F, et al. Deep learning-based image reconstruction and motion estimation from undersampled radial k-space for real-time MRI-guided radiotherapy. Phys Med Biol. 2020;65(15):155015. doi: 10.1088/1361-6560/ab9358 [DOI] [PubMed] [Google Scholar]
- 37.Harris W, Ren L, Cai J, Zhang Y, Chang Z, Yin F-F. A technique for generating volumetric cine-magnetic resonance imaging. International Journal of Radiation Oncology*Biology*Physics. 2016/06/01/ 2016;95(2):844–853. doi: 10.1016/j.ijrobp.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris W, Yin F-F, Cai J, Ren L. Volumetric cine magnetic resonance imaging (VC-MRI) using motion modeling, free-form deformation and multi-slice undersampled 2D cine MRI reconstructed with spatio-temporal low-rank decomposition. Ouant Imaging Med Surg. 2020;10(2):432–450. doi: 10.21037/qims.2019.12.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris W, Wang C, Yin F-F, Cai J, Ren L. A novel method to generate on-board 4D MRI using prior 4D MRI and on-board kV projections from a conventional LINAC for target localization in liver SBRT. Med Phys. 2018;45(7):3238–3245. doi: 10.1002/mp.12998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pham J, Harris W, Sun W, Yang Z, Yin FF, Ren L. Predicting real-time 3D deformation field maps (DFM) based on volumetric cine MRI (VC-MRI) and artificial neural networks for on-board 4D target tracking: a feasibility study. Phys Med Biol. Aug 21 2019;64(16): 165016. doi: 10.1088/1361-6560/ab359a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Yin F-F, Moore B, Han S, Cai J. A multisource adaptive magnetic resonance image fusion technique for versatile contrast magnetic resonance imaging. Original Article. Cancer Translational Medicine. May 1, 2018 2018;4(3):65–69. doi: 10.4103/ctm.ctm_21_18 [DOI] [Google Scholar]
- 42.Zhang J, Srivastava S, Wang C, et al. Clinical evaluation of 4D MRI in the delineation of gross and internal tumor volumes in comparison with 4DCT. J Appl Clin Med Phys. 2019;20(9):51–60. doi: 10.1002/acm2.12699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Yin F-F. 4D-MRI in radiotherapy. IntechOpen; 2019. [Google Scholar]
- 44.Kim J-E, Kim SH, Lee SJ, Rhim H. Hypervascular hepatocellular carcinoma 1 cm or smaller in patients with chronic liver disease: characterization with gadoxetic acid–enhanced MRI that includes diffusion-weighted imaging. American Journal of Roentgenology. 2011/06/01 2011;196(6):W758–W765. doi: 10.2214/AJR.10.4394 [DOI] [PubMed] [Google Scholar]
- 45.Colagrande S, Castellani A, Nardi C, Lorini C, Calistri L, Filippone A. The role of diffusion-weighted imaging in the detection of hepatic metastases from colorectal cancer: A comparison with unenhanced and Gd-EOB-DTPA enhanced MRI. Ear J Radiol. 2016/05/01/ 2016;85(5):1027–1034. doi: 10.1016/j.ejrad.2016.02.011 [DOI] [PubMed] [Google Scholar]
- 46.Segars WP, Tsui BMW, Jing C, Fang-Fang Y, Fung GSK, Samei E. Application of the 4-D XC AT phantoms in biomedical imaging and beyond. IEEE Trans Med Imaging. 2018;37(3):680–692. doi: 10.1109/TMI.2017.2738448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panta RK, Segars P, Yin F-F, Cai J. Establishing a framework to implement 4D XCAT phantom for 4D radiotherapy research. J Cancer Res Ther. Oct-Dec 2012;8(4):565. doi: 10.4103/0973-1482.106539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not available at this time.


