SUMMARY
Cold-induced thermogenesis in endotherms demands adaptive thermogenesis fueled by mitochondrial respiration and Ucp1-mediated uncoupling in multilocular brown adipocytes (BAs). However, dietary regulation of thermogenesis in BAs is not fully understood. Here we describe that the deficiency of Leucine-rich pentatricopeptide repeat containing protein (Lrpprc) in BAs reduces mtDNA-encoded ETC gene expression, causes ETC proteome imbalance, and abolishes the mitochondria-fueled thermogenesis. BA-specific Lrpprc knockout mice are cold-resistant in 4°C cold tolerance test in the presence of food, which is accompanied by the activation of transcription factor 4 (ATF4) and proteome turnover in BAs. ATF4 activation genetically by BA-specific ATF4 overexpression or physiologically by a low-protein diet feeding can improve cold tolerance in wild-type and Ucp1 knockout mice. Furthermore, ATF4 activation in BAs improves systemic metabolism in obesogenic environment regardless of Ucp1’s action. Therefore, our study reveals a diet-dependent but Ucp1-independent thermogenic mechanism in BAs that is relevant to systemic thermoregulation and energy homeostasis.
INTRODUCTION
The adaptive thermogenesis refers to the heat production in response to changes in ambient temperature and diet, which mainly occurs in brown adipose tissue (BAT) containing multilocular and Ucp1+ brown adipocytes (BAs) (Cannon and Nedergaard, 2004, 2011). The cold-induced adaptive thermogenesis, also called βAR-induced adaptive thermogenesis, requires mitochondrial respiration through electron transport chain (ETC) and uncoupling protein 1 (Ucp1)-mediated uncoupling in BAs (Cannon and Nedergaard, 2004; Lowell and Spiegelman, 2000). Brown fat depots in cold-stimulated humans can be recognized using 18F-fluoro-deoxyglucose positron emission tomography (18F-FDG PET) with computer-assisted tomography (CT). Brown fat 18F-FDG uptake activity gradually declines with aging and metabolic diseases (Betz and Enerback, 2015; Cypess et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009). Thus, increasing brown fat abundance and function to boost adaptive thermogenesis has been proposed as a therapeutic strategy to offset the positive energy balance and to improve metabolic health in humans (Bartelt and Heeren, 2014; Betz and Enerback, 2018).
We have recently described that BA-specific mitochondrial transcription factor A (Tfam) knockout mice (TfamBKO) exhibit a paradoxical trade-off between mitochondria-fueled βAR-induced adaptive thermogenesis in BAT and systemic metabolism in mice (Masand et al., 2018). In the current study, we further demonstrate that disrupting the synchronization of mtDNA- and nuclear-encoded ETC protein expression induces diet- and ATF4-dependent protein turnover in BAs, representing a Ucp1-independent thermogenic program in response to dietary changes. This ATF4 activation in BAs can contribute to organismal thermoregulation and promote metabolic health in obesogenic environment. Collectively, our observations suggest a new role of BAs in thermoregulation and energy metabolism.
RESULTS
Defective mtDNA ETC gene expression in brown adipocytes induces a diet-dependent thermogenic program
Leucine-rich pentatricopeptide repeat containing protein (LRPPRC) is a master regulator of mtDNA-encoded RNA maturation and stability (Ruzzenente et al., 2012; Siira et al., 2017; Spahr et al., 2016). Among different fat depots, Lrpprc expressed at a higher level in interscapular BAT (iBAT) than inguinal WAT (iWAT) and epididymal WAT (eWAT) (Figure S1A–B). We have generated the BA-specific Lrpprc knockout mice (Ucp1-Cre; Lrpprcf/f, LrpprcBKO). Q-PCR and western blot confirmed that Lrpprc was efficiently deleted in the BAT and slightly reduced in the iWAT of LrpprcBKO mice but not in other tissues (Figure S1A–B) at room temperature (RT). Thermogenic genes Ucp1 and Dio2 were reduced in these mice (Figure S1C). Ucp1 protein levels were not altered in the BAT of LrpprcBKO mice, neither did the sympathetic nerve-driven βAR signaling (using phosphor-PKA substrate immunoblot as the readout) (Figure S1D). Lrpprc deficiency-induced “whitening” of BAs at RT (Figure S1F). However, both wild-type and Lrpprc-deficient BAs exhibited unilocular morphology uniformly at thermoneutrality (30°C) (Figure S1F), where mitochondrial respiration is not needed for thermoregulation due to the absence of sympathetic inputs and βAR signaling in BAs.
Steady-state mRNA levels of most mtDNA-encoded genes were reduced in the BAT of LrpprcBKO mice at RT, without significant changes in nuclear-encoded ETC genes (Figure 1A) or mtDNA copy numbers (Figure S1E). This specific reduction of mtDNA-encoded ETC genes was also observed in the BAT of LrpprcBKO mice housed at 30°C, without changes in nuclear-encoded ETC genes (Figure 1B). Immunoblots from isolated BAT mitochondria further confirmed that mtDNA-encoded complex IV proteins, mt-Co1 and mt-Co2, were reduced in LrpprcBKO mice (Figure 1C–D). We performed mass spectrometry analysis of freshly isolated BAT mitochondria from control and LrpprcBKO mice at normal chow housed at both RT and 30°C (Supplementary Data 1, Figure S1G–I). Gene Ontology (GO) enrichment analysis showed that mitochondrial respiratory chain, NADH dehydrogenase (complex I) and cytochrome C oxidase (complex IV) activities were the most affected by Lrpprc deficiency at both ambient temperatures (Figure S1K). Indeed, all complex IV proteins were reduced accompanied by the increase of complex IV assembly factors (Figure S1J). To quantitate the BAT ETC proteome, we calculated the average log2 fold change (log2FC) values for all complex proteins identified and we found that complex IV was the most downregulated complex and complex V was the one upregulated complex (Figure 1E, S1J), suggesting that an ETC proteome imbalance is induced by Lrpprc deficiency in BAs. Clustering analysis showed that the proteomic changes induced by Tfam or Lrpprc deficiency were similar (Figure S1L) (Masand et al., 2018). Comparing the BAT mitochondrial proteome in TfamBKO and LrpprcBKO mice, we found that most of complex I and IV subunits, and some of complex III subunits, were downregulated in the BAT mitochondria of TfamBKO and LrpprcBKO mice at both temperatures (Figure S1M). On the other hand, proteins involved in mitochondrial protein import and proteases were selectively upregulated (Figure S1M). We conclude that Lrpprc-deficiency, like Tfam-deficiency, causes a specific reduction of mtDNA-encoded ETC gene and protein expression (Mootha et al., 2003), preferentially the activity of the complex IV (cytochrome C oxidase).
Figure 1. Lrpprc deficiency in the brown adipocytes triggers a dietary protein (or amino acid)-dependent thermogenic process.
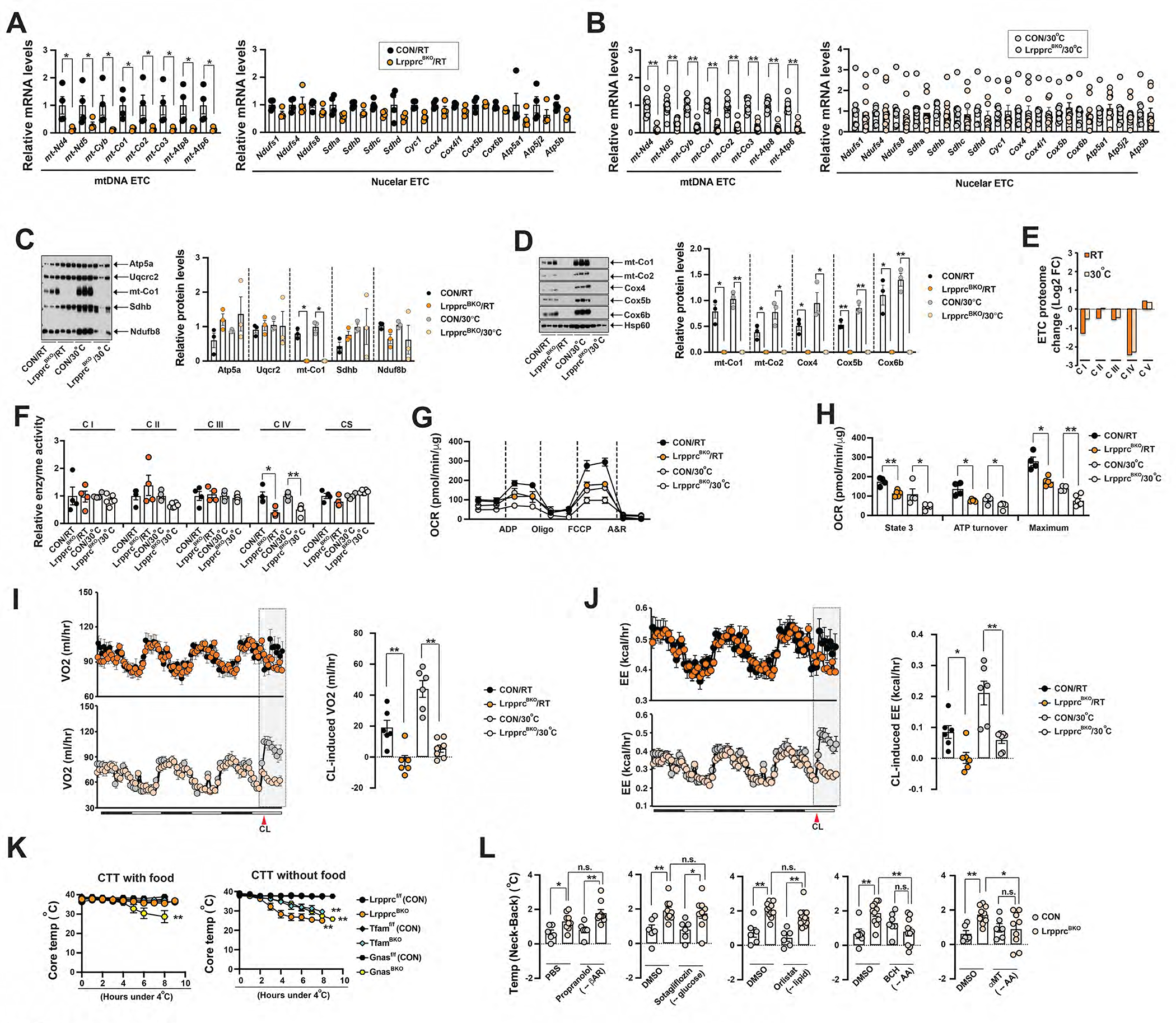
(A) Q-PCR analysis of mRNA levels of mtDNA-encoded and nuclear-encoded ETC gene in the BAT from ~8-week-old male CON and LrpprcBKO mice at RT. Sample size: CON/RT (n=4) and LrpprcBKO/RT (n=4). (B) Q-PCR analysis of mRNA levels of mtDNA-encoded and nuclear-encoded ETC gene in the BAT from ~10-week-old male CON and LrpprcBKO mice at 30°C. Sample size: CON/30°C (n=10) and LrpprcBKO/30°C (n=11). (C) Left: Mitochondrial cocktail immunoblot showing amounts of representative protein abundance of each ETC complex, Ndufb8 (complex I), Sdhb (complex II), Uqcrc2 (complex III), mt-Co1 (complex IV) and Atp5a (complex V), in isolated BAT mitochondria from ~8–12-week-old male CON and LrpprcBKO mice housed at RT and 30°C. Right: Relative protein abundance (to Hsp60) shown. Significance between genotypes indicated. (D) Left: Immunoblots of complex IV subunits (mt-Co1, mt-Co2, Cox4, Cox5b, Cox6b) and Hsp60 in isolated mitochondria from above mice. Right: Relative protein abundance (to Hsp60) shown. Significance between genotypes indicated. (E) Log2 fold change values of each ETC proteome from BAT of LrpprcBKO mice. (F) Relative in vitro enzyme activities of Complex I to IV and citrate synthase (CS) in BAT of ~8–12-week-old male CON and LrpprcBKO mice housed at RT and 30°C. Sample size: CON/RT (n=4), LrpprcBKO/RT (n=4), CON/30°C (n=4) and LrpprcBKO/30°C (n=6). (G) Seahorse experiments measuring oxygen consumption rates (OCRs) of isolated mitochondria from BAT of ~8–12-week-old male and female CON and LrpprcBKO mice housed at RT and 30°C, upon addition of ADP, Oligo, FCCP and A&R. (H) State 3, ATP turnover and maximum OCRs in Seahorse measurements. Values are normalized as per mitochondrial protein. Sample size: CON/RT (n=4), LrpprcBKO/RT (n=5), CON/30°C (n=4) and LrpprcBKO/30°C (n=6). (I) Left: Recordings of oxygen consumption (VO2) in ~8–12-week-old male CON and LrpprcBKO mice for three days at RT and 30°C. Red arrowhead: time of CL injection. Right: Average hourly CL-induced VO2 in the aforementioned mice. (J) Left: Recordings of energy expenditure (EE) in ~8–12-week-old male CON and LrpprcBKO mice for three days at RT and 30°C. Red arrowhead: time of CL injection. Right: Average hourly CL-induced EE in the aforementioned mice. Sample size: CON/RT (n=6), LrpprcBKO/RT (n=6), CON/30°C (n=6) and LrpprcBKO/30°C (n=7). (K) Left: 4°C Cold tolerance test (CTT) of ~8–12-week-old male and female LrpprcBKO, TfamBKO, GnasBKO and their relative control mice housed at RT with food. Note: data from GnasBKO and relative control was from our previous publication (Paulo et al., 2018b). Sample size: Lrpprcf/f (CON) (n=6), LrpprcBKO (n=9), Tfamf/f (CON) (n=4), TfamBKO (n=4), Gnasf/f (CON) (n=12), and GnasBKO (n=9). Right: CTT of ~8–12-week-old male and female LrpprcBKO, TfamBKO, GnasBKO and their relative control mice housed at RT without food. Sample size: Lrpprcf/f (CON) (n=7), LrpprcBKO (n=6), Tfamf/f (CON) (n=7), TfamBKO (n=9), Gnasf/f (CON) (n=4) and GnasBKO (n=5). (L) The Temp (Neck-Back), the difference of the surface temperatures between neck (overlaying BAT) and dorsal back (not overlay BAT) by infrared (IR) thermal imaging, of ~12-week-old male and female CON and LrpprcBKO mice 2 hours after injections of 10 mg kg−1 Propranolol or 10 mg kg−1 Sotagliflozin or 25 mg kg−1 Orlistat or 50mg kg−1 2-amino-2-norbornane-carboxylic acid (BCH) 100mg kg−1 α-methyl-dl-tryptophan (αMT) or their vehicles at thermoneutrality. Sample size: CON (n=6) and LrpprcBKO (n=11). Data were presented as average ± SEM. Unpaired t-test. n.s.: not significant, *: p<0.05 and **: p<0.01.
Since mitochondrial respiration charges βAR-induced adaptive thermogenesis in BAT (Cannon and Nedergaard, 2004), we first measured the mitochondrial respiration activity in vitro. Despite Lrpprc deficiency decreased the mtDNA-encoded gene expression by ~90%, complex IV enzymatic activity was only reduced by half in the LrpprcBKO mice housed at both temperatures (Figure 1F). Seahorse experiments also confirmed that isolated BAT mitochondria from the LrpprcBKO mice exhibited about 50% reduction of state 3, ATP turnover and maximum respiration rates (Figure 1G–H). We then used indirect calorimetry experiments to measure βAR-induced adaptive thermogenesis, as well as basal energy expenditure (EE, calculated from O2 consumption and CO2 production), respiratory exchange ratio (RER), average and accumulative food intake and physical activity of 8–10-week-old male control and LrpprcBKO mice at RT and 30°C. Body weight and fat mass of LrpprcBKO and control mice were not different at this age. At basal state, there were no differences in basal EE, RER, food intake (hourly average and cumulative) and physical activity (Figure 1I–J, S1N–T). However, b3 agonist CL 316 243 (CL) stimulation significantly induced EE (hereby heat production) in control mice, and this effect was absent in the LrpprcBKO mice (Figure 1I–J).
BAT thermogenesis is essential for organismal thermoregulation in rodents (Cannon and Nedergaard, 2004, 2011). Genetic ablation of BAT in mice can lead to increased cold sensitivity in 4°C cold tolerance test (CTT) (Lowell et al., 1993; Wang et al., 2017). Impaired mitochondrial biogenesis in BAs (in the BA-specific Gs alpha knockout mice (GnasBKO) (Paulo et al., 2018b)) also causes defective βAR-induced adaptive thermogenesis and increased cold sensitivity in CTT. Even though LrpprcBKO mice did not have respiration-capable mitochondria in BAs, they were cold-resistant during CTT when food was available during the cold challenge (Figure 1K). A similar diet-dependent cold resistance phenotype was also observed in the TfamBKO mice (Figure 1K). In contrast, the GnasBKO mice were cold sensitive regardless of the food availability during CTT (Figure 1K). Collectively, the LrpprcBKO mice are thermogenically competent in spite of the reduced activity of complex IV (the oxygen-consuming cytochrome C oxidase) in BAs.
WAT browning does not contribute to cold tolerance in LrpprcBKO mice
WAT browning, the expansion of Ucp1+ thermogenically active beige adipocytes in white adipose tissues, can be an alternative thermogenic mechanism. We investigated whether the WAT browning can account for the cold resistance phenotype in the LrpprcBKO mice under fed condition. Indeed, we observed elevated beige marker gene expression in the iWAT of LrpprcBKO mice at 8-week and 8-month of age at RT (Figure S2A). Immunoblot from iWAT of LrpprcBKO mice showed that Ucp1 protein levels were upregulated (Figure S2B). Histological examinations also confirmed the presence of the multilocular beige adipocytes in the iWAT at 8-week and 8-month of age (Figure S2C). Levels of phospho-PKA substrates in the iWAT were comparable, suggesting that there was no increase in sympathetic activity in the iWAT of LrpprcBKO mice (Figure S2B). The WAT browning phenotype in the LrpprcBKO and TfamBKO mice was absent under thermoneutral housing (Figure S2D–E) (Masand et al., 2018). Moreover, we did not observe significant changes in gene expression in creatine and calcium cycles that have been reported to be involved in Ucp1-independent thermogenesis specifically in beige fat (Figure S2F) (Ikeda et al., 2017; Kazak et al., 2015). White adipocytes in the epididymal white adipose tissue (eWAT) also process Ucp1-independent and creatine-dependent thermogenesis (Bertholet et al., 2017). We found that white adipocytes in the eWAT of LrpprcBKO mice were histologically indistinguishable from those in control mice at ~8-week of age (Figure S2G). And q-PCR analysis failed to reveal any difference in the expression of Ucp1 and genes involved in the creatine thermogenesis pathway (Figure S2H).
Previously we have characterized a Ucp1-iDTR system that enables us to genetically ablate Ucp1+-lineage beige adipocytes and address their physiological functions in vivo (Paulo et al., 2018a; Wang et al., 2017). After diphtheria toxin/DT-induced ablation of both Ucp1+-lineage brown and beige adipocytes initially, BAs in BAT will be fully regenerated within 3 weeks but beige adipocytes in the iWAT remains absent at RT (Paulo et al., 2018a). We have generated the following three groups of mice: LrpprcBKO;iDTR (Ucp1-Cre;Lrpprcf/f;ROSA-LSL-iDTR), LrpprcBKO (Ucp1-Cre;Lrpprcf/f) and CON (Lrpprcf/f). At RT, we injected DT in these three groups at 3-week of age (Figure S2I). We confirmed the absence of Ucp1 upregulation and multilocular beige adipocytes in the iWAT of ~8-week-old LrpprcBKO;iDTR mice, indicating a successful ablation of Lrpprc-deficient beige adipocytes in the iWAT (Figure S2J). Additionally, Lrpprc remained deleted and, consequently, mtDNA-encoded mt-Co1 was still reduced after a complete BAT regeneration (Figure S2K). However, these three groups of mice behaved similarly during CTT (Figure S2L), suggesting that expansion of Ucp1+ beige adipocytes in the LrpprcBKO mice did not functionally contribute to their cold resistance phenotype.
Dietary proteins drive BAT thermogenesis in the LrpprcBKO mice
The infrared (IR) thermal imaging can measure surface temperature on live mice, which has been used in brown adipocyte biology (Crane et al., 2014; Fischer et al., 2020). Compared to BAT Telemetry, confounding factors from skin and fur may affect IR imaging. Mice were not shaved to maintain consistent fur status and avoid the alterations of the natural thermoregulatory state. To determine the reliability of this IR approach, C57bl/6J mice housed at 30°C were subjected to 15-minute mild cold stimulation at RT. Surface temperatures were measured before and 15-minute after RT exposure by IR (Figure S2M–N). As expected, 15-minute RT stimulation increased the surface temperature at the neck (overlaying the BAT), but the surface temperature at the dorsal back (not overlaying BAT) and core temperature remained unchanged (Figure S2O–P). But BAT temperature measurement alone by IR (or BAT telemetry) does not accurately reflect BAT activation. We used the “Temp (Neck-Back)” value, the difference of surface temperature between the neck and the dorsal back, as a quantitative measure for BAT thermogenic activity. As expected, either RT stimulation or CL similarly increased the Temp (Neck-Back) under both fed and fasted conditions in wild-type C57bl/6J mice, and it was blocked by Propranolol (Figure S2Q–R). In contrast, nutrient status did not affect this RT-induced BAT activity, as short-term fasting (4 hours) didn’t affect RT’s effect (Figure S2Q).
However, LrpprcBKO mice, housed at thermoneutrality, showed an increased Temp (Neck-Back) on feeding condition, but not after fasting (Figure S2S–T). And this increase was not affected by RT stimulation or propranolol treatment (Figure 1L, S2S). Temp (Neck-Back) was not affected in the LrpprcBKO mice housed at RT (Figure S2U), which may be due to the fact the Ucp1-dependent thermogenesis is already active in the wild-type BAs housed at RT. The observations in IR and CTT experiments collectively suggest that a diet-dependent thermogenic process is present in the Lrpprc-deficient BAs.
Glucose and fatty acid from dietary food have been regarded as the preferred substrates for mitochondrial oxidation in BAs. BAT 18F-FDG uptake was reduced by half in the LrpprcBKO mice (Figure S2V). Fatty acids that fuel mitochondrial respiration in BAs either come from diet or lipolysis in WAT (Bartelt et al., 2011; Shin et al., 2017). Both CON and LrpprcBKO mice showed a similar rate of glycerol increase after CL injection, suggesting that systemic lipolytic activity was not affected in the LrpprcBKO mice (Figure S2W). We also examined the BAT lipolytic activity ex vivo. Upon lipolytic stimulation (mimicked by the addition of 10 mM Forskolin in the media), BAT from CON and LrpprcBKO mice housed at RT and 30°C exhibited a similar rate of glycerol release (Figure S2X), which was consistent with a similar expression level of key lipolytic proteins (Hsl and Atgl) (Figure S2Y).
We further used macronutrient absorption inhibitors to evaluate the dietary requirement of this diet-dependent BAT thermogenesis in the LrpprcBKO mice by IR at thermoneutrality. Modulating availability of fatty acid (by lipase inhibitor Orlistat (Ballinger and Peikin, 2002)) and glucose (dual SGLT inhibitor Sotagliflozin (Powell et al., 2014)) had no effect on Temp (Neck-Back) of the LrpprcBKO mice (Figure 1L, S2Z–AA). In contrast, 2-amino-2-norbornane-carboxylic acid (BCH), an uptake inhibitor of amino acid (AA) (Cha et al., 2018; Krokowski et al., 2013), reduced the Temp (Neck-Back) in the LrpprcBKO mice (Figure 1L). Another AA uptake inhibitor, α-methyl-dl-tryptophan (αMT, 100 mg kg−1), showed a similar effect (Figure 1L). Of note, all these macronutrient absorption inhibitors and propranolol have no effect on the Temp (Neck-Back) in control mice housed at thermoneutrality (Figure 1L), suggesting the presence of a cold-independent but dietary protein (or AA)-fueled thermogenic process specifically in the Lrpprc-deficient BAs.
ATF4 activation in brown adipocytes drives dietary protein (or AA)-fueled but Ucp1-independent thermoregulation
The postprandial metabolic response (also known as Luxuskonsumption, specific dynamic reaction, thermic effect of food, or diet-induced thermogenesis) has been observed for over a century (BORSOOK, 1936; Lusk, 1931). The LrpprcBKO mice may represent an experimental model system to dissect the molecular controls of BAT thermogenesis in response to dietary changes.
We first performed RNA-seq experiments from the BAT of male control and LrpprcBKO mice. Clustering analysis of differentially expressed genes (DEGs) suggested that thermoneutral housing affected BAT transcriptome more profoundly than Lrpprc deficiency (Figure S3A). Volcano plots of DEGs showed that there were approximately 6 times more up- and down-regulated DEGs in LrpprcBKO mice at 30°C (Figure S3B). Within downregulated DEGs, 132 genes were commonly observed at both RT and 30°C (Figure S3C). The oxidative phosphorylation was the most significantly enriched pathway in downregulated DEGs at both ambient temperatures (Figure S3D–E). On the other hand, pathways of immune cell activation and protein homeostasis were highly enriched among the upregulated 165 genes at both RT and 30°C (Figure S3F–G). Cis-regulatory sequence analysis using iRegulon predicted that activating transcription factor 4 (ATF4) and its downstream transcription factors ATF3 and DDIT3 were the top regulators of the upregulated DEGs (Figure 2A). For example, 39 (out of 165) genes were predicted to contain putative ATF4 response element “TTGCATCA” on their promoter regions. These genes regulated diverse cellular processes, such as response to ER stress, cellular amino acid metabolism, transport of small molecules, one-carbon metabolism, steroid metabolism and fibroblast proliferation (Figure 2B). Combined analysis of the transcriptomic and proteomic profiling of mitochondria-localized proteins further demonstrated that the non-synchronization of mtDNA- and nuclear-encoded ETC expression in the BAT of LrpprcBKO mice (Figure 2C). Thus, the aforementioned omics studies indicate that the ATF4 transcription network, also called the integrated stress response (ISR), is activated by Lrpprc deficiency in BAs.
Figure 2. ATF4 activation in brown adipocytes is necessary for thermogenic phenotypes in the LrpprcBKO mice.
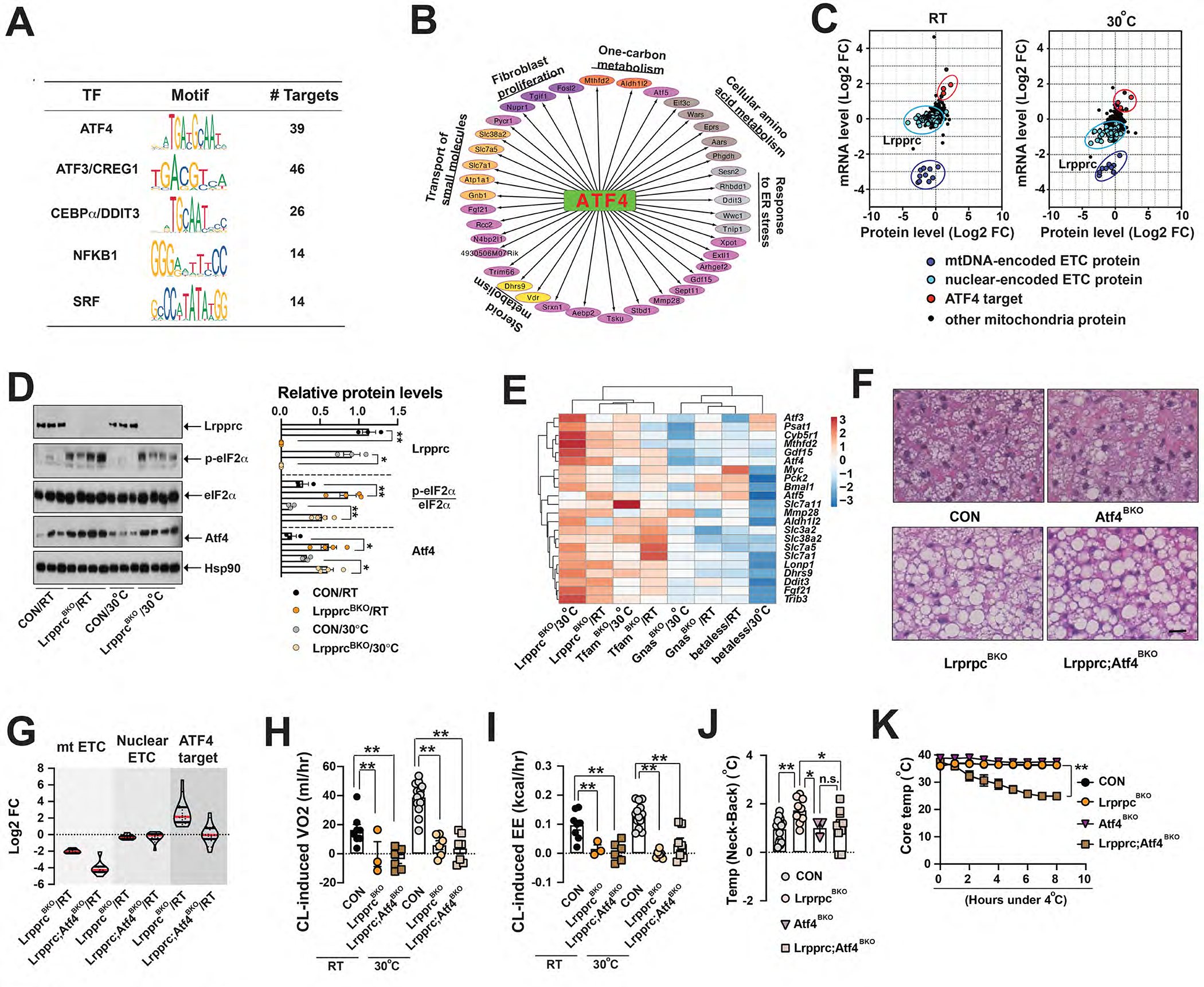
(A) List of enriched Transcript factors (TF) in commonly up-regulated DEGs in the BAT of CON and LrpprcBKO mice at RT and 30°C. Name, motif sequence and number of targets of each TF shown. (B) ATF4 signaling network. GO terms of ATF4 targets shown. (C) Scatter plots showing the correlation of log2 fold changes (Log2 FC)) of mRNA and protein levels of mitochondria-localized proteins from BAT of LrpprcBKO mice housed at RT and 30°C. mtDNA-encoded ETC, nuclear-encoded ETC, and ATF4 targets circled. (D) Left: Immunoblots showing amounts of Lrpprc, p-eIF2α, total eIF2α, Atf4 and Hsp90 in the BAT of ~8–12-week-old male CON and LrpprcBKO mice at normal chow at both RT and 30°C. Right: Relative protein abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between genotypes indicated. (E) Clustering analysis of log2 fold changes of known ATF4 target genes in the BAT of mouse models with defective mitochondrial function in BAs (LrpprcBKO, TfamBKO, GnasBKO and betaless mice, ~8–12-week-old male) at normal chow at both RT and 30°C. Sample size: n=5 for each genotype/condition. (F) Representative H&E staining of BAT from ~8–10-week-old male CON, LrpprcBKO, Atf4BKO, and Lrpprc;Atf4BKO mice housed at RT. Scale bar: 50 μm. (G) Violin plot of log2 fold change (FC) of 8 mtDNA-encoded ETC (mt ETC, listed in Fig.1A), 15 nuclear-encoded ETC (nuclear ETC, listed in Fig.1A) and 22 ATF4 targets (listed in Fig.2E) in the BAT of ~8–12-week old male LrpprcBKO and Lrpprc;Atf4BKO mice to their relative controls (Lrpprcf/f and Lrpprc;Atf4f/f) housed at RT. Red and black bars: median and quartiles. Hourly CL-induced VO2 (H) and EE (I) in ~10-week old male CON, LrpprcBKO and Lrpprc;Atf4BKO mice at RT and 30°C. Sample size: CON/RT(n=8), LrpprcBKO/RT (n=3), Lrpprc;Atf4BKO/RT(n=6), CON/30°C(n=13), LrpprcBKO/30°C (n=6) and Lrpprc;Atf4BKO/30°C (n=7). (J) The Temp (Neck-Back) of ~12-week-old male and female CON, LrpprcBKO, Atf4BKO, and Lrpprc;Atf4BKO mice at thermoneutrality. Sample size: CON (n=15), LrpprcBKO (n=9), Atf4BKO (n=3) and Lrpprc;Atf4BKO (n=10). (K) CTT of ~8–12-week old male and female CON, LrpprcBKO, Atf4BKO, and Lrpprc;Atf4BKO mice housed at RT. Sample size: CON (n=6), LrpprcBKO (n=9), Atf4BKO (n=4), and Lrpprc;Atf4BKO (n=8). Data were presented as average ± SEM. Unpaired t-test. n.s.: non-significant; *: p<0.05 and **: p<0.01.
The ISR is centrally controlled by the phosphorylation of eukaryotic translation initiation factor eIF2α. When phosphorylated, it specifically induces ATF4 translation and its target gene expression (Pakos-Zebrucka et al., 2016). Indeed, we observed p-eIF2α and Atf4 proteins were elevated, along with known ATF4 targets in the BAT of LrpprcBKO mice at RT and 30°C (Figure 2D). TfamBKO mice, but not betaless mice, showed similar induction of p-eIF2α in the BAT (Figure S3H–I). Additional clustering analysis using log2 fold change (Log2 FC) of these ATF4 targets in various mouse models with defective βAR-induced adaptive thermogenesis such as betaless, GnasBKO, LrpprcBKO, TfamBKO mice (Masand et al., 2018; Paulo et al., 2018b) demonstrated that ATF4 transcription network was specifically induced by Lrpprc (and Tfam) deficiency in BAs (Figure 2E).
To evaluate the functional relevance of ATF4 activation in BAs, we have generated BA-specific Atf4 knockout mice (Ucp1-Cre;Atf4f/f, Atf4BKO) and BA-specific Lrpprc and Atf4 double knockout mice (Ucp1-Cre;Lrpprcf/f;Atf4f/f, Lrpprc;Atf4BKO). Atf4 deficiency alone in BAs reduced ATF4 target gene expression but did not affect multilocular morphology of BAs at RT and mitochondrial ETC gene expression at RT and 30°C (Figure S3K–L). And indirect calorimetry experiments showed that Atf4 deficiency did not significantly affect basal and CL-induced EE, RER, food intake and physical activity (Figure S3M–T). Thus, the thermogenic phenotypes reported in the global Atf4 knockout mice might be contributed by Atf4 deletion in BAs and other cell types collectively (Liu et al., 2017; Wang et al., 2010; Wang et al., 2013). Lrpprc and Atf4 double deficient BAs were unilocular and exhibited reduced mtDNA ETC gene expression similar to those in the LrpprcBKO mice (Figure 2F–G, S3K). The Lrpprc;Atf4BKO mice also lacked CL-induced VO2 and EE due to the defect of mtDNA gene expression in BAs at RT and 30°C (Figure 2H–I), but without noticeable changes in basal EE, RER, food intake and physical activity (Figure S3U–Z). However, the upregulation of ATF4 target genes induced by Lrpprc deficiency was suppressed by additional Atf4 deficiency in BAs (Figure 2G). The Lrpprc;Atf4BKO mice were no longer cold-resistant during CTT, although Atf4 deficiency itself in BAs did not affect core body temperature during CTT (Figure 2K). A similar result was observed in IR, that the increased BAT activity in the LrpprcBKO mice at thermoneutrality was absent in the Lrpprc;Atf4BKO mice (Figure 2J). This epistasis analysis demonstrates that ATF4 activation in BAs is necessary for Lrpprc deficiency-induced thermoregulation.
Since elevated ATF4 expression is sufficient for transcriptional induction of ISR targets (Masuda et al., 2016; Shan et al., 2009), we then decided to ectopically overexpress ATF4 in BAs to examine its consequences on thermoregulation. We have crossed ROSA-LSL-ATF4 (Flag-tagged human ATF4 flanked by stop cassette in ROSA locus) with Ucp1-Cre mice, to generate BA-specific ATF4 overexpressing mice (ATF4BOX) and their controls (Figure 3A). Flag immunoblot confirmed the presence of Flag-ATF4 protein in the BAT of the ~8-week-old ATF4BOX mice, which resulted in a mild increase (~2–3-fold) in total ATF4 protein level (Figure 3B). The ATF4BOX mice may represent a useful model system to investigate the physiological regulation of this novel thermogenesis, if a similar extent of ATF4 overexpression in BAs can be achieved. ATF4 overexpression did not affect BA multilocular morphology, mitochondrial respiration, or Ucp1 and ETC (nuclear- and mtDNA-encoded) gene expression and protein levels in isolated mitochondria (Figure 3C–G). Similar to LrpprcBKO and TfamBKO mice, ATF4BOX mice exhibited marked upregulation of ATF4 target gene network in the BAT at both RT and 30°C (Figure 3H), further confirming the ISR activation by ATF4 overexpression in the BAs. Indirect calorimetry experiments showed that ATF4BOX mice exhibited similar βAR-induced adaptive thermogenesis, basal EE, RER and physical activity as control mice at both ambient temperatures (Figure S4A–E). ATF4BOX mice even exhibited a slight increase in cumulative food intake at thermoneutrality (Figure S4F–H). Therefore, BA-specific ATF4 overexpression does not affect mitochondrial ETC gene expression in BAT and βAR-induced adaptive thermogenesis in mice. When housed at RT, ATF4BOX mice were cold-resistant as the control mice during CTT from RT (Figure 3J), because mitochondria-fueled Ucp1-mediated thermogenesis was not affected by the ATF4 overexpression in BAs. Wild-type mice, acclimated to thermoneutrality, rapidly drop their core temperature upon CTT from 30°C due to diminished Ucp1-dependent thermogenesis. However, ATF4BOX mice exhibited enhanced cold resistance during CTT from 30°C (Figure 3J), further suggesting the presence of ATF4-induced thermogenesis in mice.
Figure 3. ATF4 activation in brown adipocytes induces Ucp1-independent thermogenesis in mice.
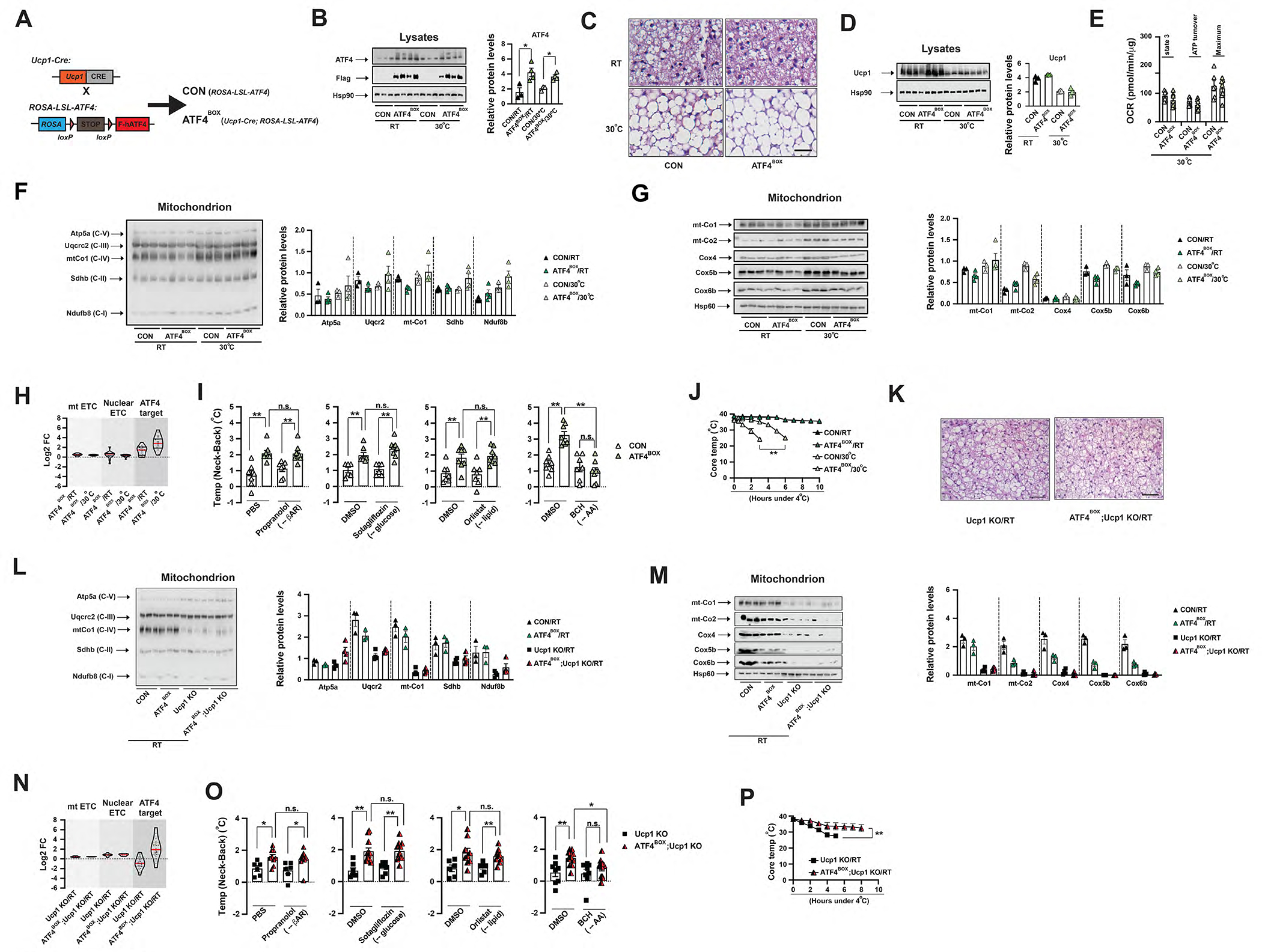
(A) Cross scheme to generate BA-specific ATF4 overexpression (ATF4BOX) and its control (CON). (B) Left: Immunoblots of total ATF4 (endogenous and exogenous), Flag-ATF4 (exogenous) and Hsp90 in the BAT from ~8-week-old male CON and ATF4BOX mice at RT and 30°C. Right: Relative abundance of total ATF4 protein (to Hsp90) amounts. Significance between genotypes indicated. (C) Representative H&E staining of BAT from ~8–10-week-old male CON, and ATF4BOX mice housed at RT and 30°C. Scale bar: 50 μm. (D) Left: Immunoblots of Ucp1 and Hsp90 form BAT lysates of ~8–10-week-old male CON and ATF4BOX mice at RT and 30°C. Right: Relative abundance of Ucp1 protein (to Hsp90) amounts. (E) State 3, ATP turnover and maximum OCRs in Seahorse experiments measuring oxygen consumption rates (OCRs) of isolated mitochondria from BAT of ~8–12-week-old male and female CON and ATF4BOX mice housed at 30°C, upon addition of ADP, Oligo, FCCP and A&R. Values are normalized as per mitochondrial protein. Sample size: CON (n=6), ATF4BOX (n=7). (F) Left: Mitochondrial cocktail immunoblot showing amounts of representative protein abundance of each ETC complex, Ndufb8 (complex I), Sdhb (complex II), Uqcrc2 (complex III), mt-Co1 (complex IV) and Atp5a (complex V), in isolated BAT mitochondria from ~8–10-week-old male CON and ATF4BOX mice housed at RT and 30°C. Right: Relative abundance (to Hsp60) amounts. (G) Left: Immunoblots of mt-Co1, mt-Co2, Cox4, Cox5b, Cox6b and Hsp60 in isolated BAT mitochondria from ~8–10-week-old male CON and ATF4BOX mice housed at RT and 30°C. Right: Relative abundance (to Hsp60) amounts. (H) Violin plot of log2 FC of mt ETC, nuclear ETC and ATF4 target in the BAT of ~8–12-week-old male ATF4BOX mice to their relative controls (ROSA-LSL-ATF4) housed at RT and 30°C. Red and black bars: median and quartiles. (I) The Temp (Neck-Back) of ~12-week-old male and female CON and ATF4BOX mice 2 hours after injections of Propranolol or Sotagliflozin or Orlistat or BCH or their vehicles at thermoneutrality. Sample size: CON (n=7) and ATF4BOX (n=8). (J) CTT of ~8–12-week-old male and female CON and ATF4BOX mice housed at RT and 30°C. Sample size: CON/RT (n=5), ATF4BOX/RT (n=5), CON/30°C (n=6) and ATF4BOX/30°C (n=7). (K) Representative H&E staining of BAT of ~8-week-old male Ucp1 KO and ATF4BOX;Ucp1 KO mice at RT. Scale bar: 100 μm. (L) Left: Mitochondrial cocktail immunoblot showing amounts of representative protein abundance of each ETC complex, Ndufb8 (complex I), Sdhb (complex II), Uqcrc2 (complex III), mt-Co1 (complex IV) and Atp5a (complex V), in isolated BAT mitochondria from ~8–10-week-old male CON, ATF4BOX, Ucp1 KO and ATF4BOX;Ucp1 KO mice at RT. Right: Relative abundance (to Hsp60) amounts. (M) Left: Immunoblots of mt-Co1, mt-Co2, Cox4, Cox5b, Cox6b and Hsp60 in isolated BAT mitochondria from ~8–10-week-old male CON, ATF4BOX, Ucp1 KO and ATF4BOX;Ucp1 KO mice at RT. Right: Relative abundance (to Hsp60) amounts. (N) Violin plot of log2 FC of mt ETC, nuclear ETC and ATF4 target in the BAT of ~8–12-week old male Ucp1 knockout (Ucp1 KO) and BA-specific ATF4 overexpression mice in Ucp1 KO background (ATF4BOX;Ucp1 KO) to their relative controls (ROSA-LSL-ATF4;Ucp1 KO or Ucp1-Cre;Ucp1 KO) housed at RT. Red and black bars: median and quartiles. (O) The Temp (Neck-Back) of ~12-week-old male and female Ucp1 KO and ATF4BOX;Ucp1 KO mice 30 minutes after injections of Propranolol or Sotagliflozin or Orlistat or BCH or their vehicles at thermoneutrality. Sample size: Ucp1 KO (n=6) and ATF4BOX;Ucp1 KO (n=9). (P) CTT of ~8–12-week old male and female Ucp1 KO and ATF4BOX;Ucp1 KO mice housed at RT. Sample size: Ucp1 KO/RT (n=14) and ATF4BOX;Ucp1 KO (n=7). Data were presented as average ± SEM. Unpaired t-test. n.s.: non-significant; *: p<0.05 and **: p<0.01.
To examine whether this ATF4 activation in BAs is competent to replace Ucp1-mediated thermogenesis in mice, we then generated BA-specific ATF4 overexpressing mice in the Ucp1 deficiency background (ATF4BOX;Ucp1 KO). At RT, Ucp1 deficiency led to reduced ETC protein expression per mitochondrion without significant changes of mRNA levels and exaggerated lipid filling in BAs as previously reported (Kazak et al., 2017). These phenotypes were not affected by ATF4 overexpression (Figure 3K–M). ATF4 overexpression in BAs did not affect EE, food intake and physical activity in the Ucp1 KO mice (Figure S4I–R). In contrast, ATF4 overexpression still induced ATF4 target gene expression in Ucp1 KO mice (Figure 3N), similar to its effect in wild-type mice (Figure 3H). Although Ucp1 KO mice rapidly lose their cold temperature upon CTT from RT, ATF4BOX;Ucp1 KO mice were cold-resistant (Figure 3P). Finally, IR also showed the dietary protein (or AA) drove the BAT thermogenesis, as BCH injection suppressed the increased Temp (Neck-Back) in the ATF4BOX mice (in either wild-type and Ucp1 KO background) (Figure 3I,O). Neither Orlistat (blocking fat absorption) nor Sotagliflozin (blocking glucose absorption) nor Propranolol (blocking βAR signaling) had any effect (Figure 3I,O). Therefore, we conclude that ATF4 activation in BAs can drive the AA-fueled but Ucp1-independent thermoregulation in mice.
Thermogenic capacity of cellular protein turnover in brown adipocytes
The presence of Ucp1-independent thermogenesis was first evident because the Ucp1 KO mice were cold-resistant in a hybrid background (Hofmann et al., 2001). Besides Ucp1-mediated uncoupling in mitochondria, heat can be generated through futile cycles where two metabolic pathways operate simultaneously in opposite directions (Qian and Beard, 2006; Tseng et al., 2010), such as calcium cycle, creatine cycle, triglyceride-fatty acid cycle, glycerol-3-phosphate cycle and glycolysis-gluconeogenesis cycle (Anunciado-Koza et al., 2008; Guan et al., 2002; Ikeda et al., 2017; Kazak et al., 2015; Shulman et al., 1985; Veliova et al., 2020). Protein turnover, the coupled protein synthesis and degradation could be a potential thermogenic mechanism especially in BAs, by wasting ATP to generate heat as the byproduct (Koeppen and Stanton, 2018; Qian and Beard, 2006; She et al., 2007). ATF4 transcriptional program regulates intracellular amino acid homeostasis, a conserved amino acid response (AAR) from yeast to mammals (Kilberg et al., 2009; Mirzaei et al., 2014; Soultoukis and Partridge, 2016). Particularly, amino acid transporters (AATs) that mediate the uptake of the circulating amino acids after digestion of dietary proteins were ATF4 target genes. Indeed, we observed that many AATs (Slc3a2, Slc7a1, Slc7a11 and Scl38a2) were transcriptionally upregulated by ATF4 activation in BAs (Figure 2E, S5C). We further confirmed that Slc3a2 mRNA and protein levels were also elevated in the BAT of the aforementioned mice (Figure S5A–F). We also observed that LrpprcBKO mice exhibited elevated Fluciclovine (18F leucine analog) uptake in the BAT in vivo (Figure 4A–B, S5G). In contrast, the Lrpprc;Atf4BKO mice did not show elevated Fluciclovine uptake in BAT (Figure S5H), suggesting that ATF4-dependent AA uptake is upregulated in the BAT of LrpprcBKO mice.
Figure 4. ATF4 activation induced cellular protein turnover in brown adipocytes.
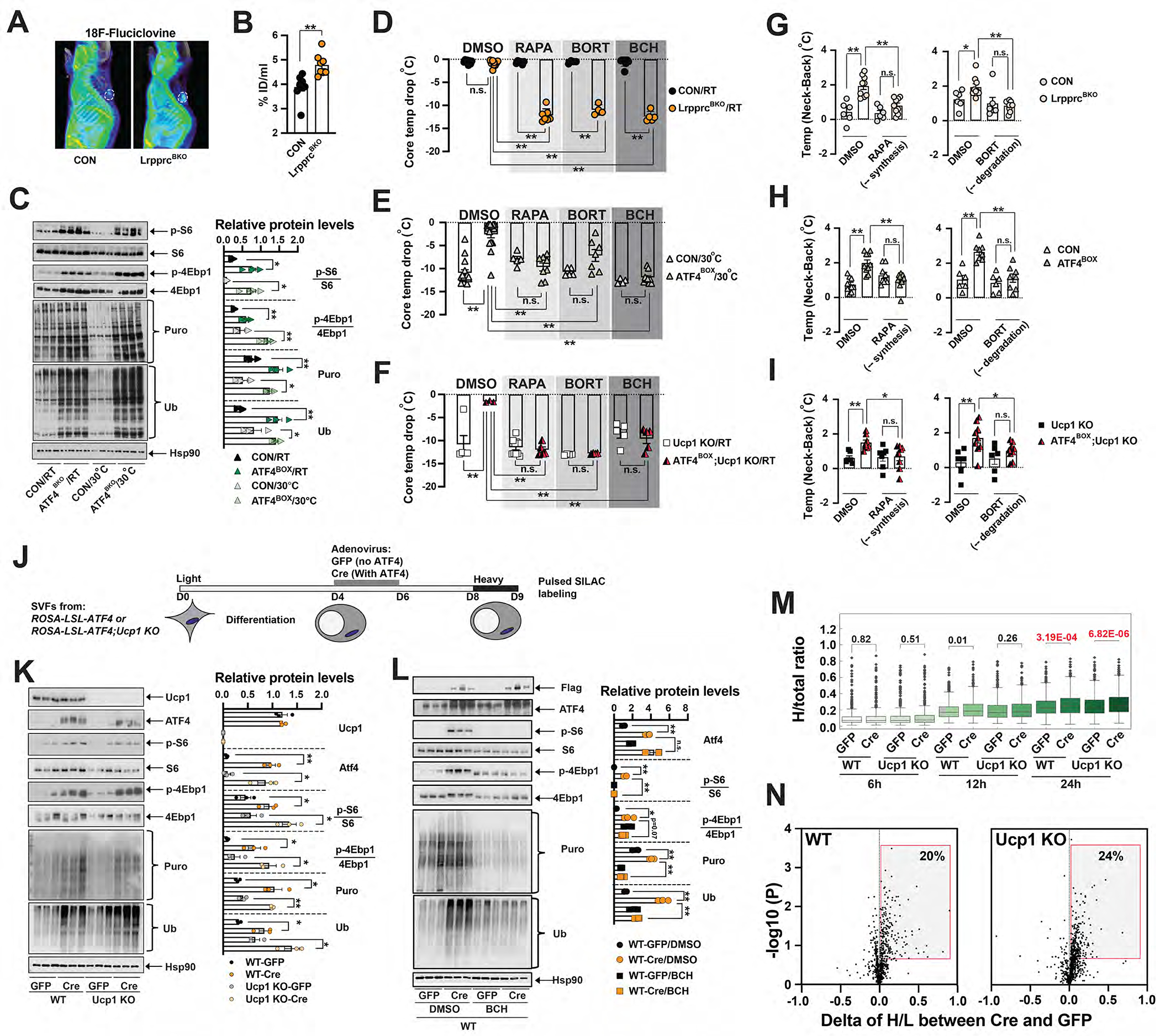
3D rendering (A) and average values of 18F-Fluciclovine uptake (B) in ~10–12-week-old male CON and LrpprcBKO mice. White cycle: BAT. Sample size: CON (n=9) and LrpprcBKO (n=8). (C) Left: Immunoblots of p-S6, total S6, p-4Ebp1, total 4Ebp1, puromycylated protein, ubiquitinated protein and Hsp90 in the BAT of ~10-week-old male CON and ATF4BOX mice at ad libitum feeding at RT and 30°C. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between genotypes indicated. (D) Core temperature drop of ~10-week-old male and female LrpprcBKO mice and their relative controls with pretreatment of DMSO, or 4mg kg−1 rapamycin (RAPA), or 0.625mg kg−1 bortezomib (BORT), or 50mg kg−1 BCH after 8 hours 4°C CTT from RT. Sample size: CON/RT/DMSO (n=9), LrpprcBKO/RT/DMSO (n=10), CON/RT/RAPA (n=6), LrpprcBKO/RT/RAPA (n=7), CON/RT/BORT (n=4), LrpprcBKO/RT/BORT (n=4), CON/RT/BCH (n=8) and LrpprcBKO/RT/BCH (n=5). (E) Core temperature drop of ~10-week-old male and female CON and ATF4BOX mice with pretreatment of DMSO or RAPA or BORT or BCH after 3 hours 4°C CTT from 30°C. Sample size: CON/30°C/DMSO (n=14), ATF4BOX/30°C/DMSO (n=14), CON/30°C/RAPA (n=5), ATF4BOX/30°C/RAPA (n=8), CON/30°C/BORT (n=5), ATF4BOX/30°C/BORT (n=7), CON/30°C/BCH (n=4) and ATF4BOX/30°C/BCH (n=9). (F) Core temperature drop of ~10-week-old male and female Ucp1 KO and ATF4BOX;Ucp1 KO mice with pretreatment of DMSO or RAPA or BORT or BCH after 3 hours 4°C CTT from RT. Sample size: Ucp1 KO/RT/DMSO (n=5), ATF4BOX;Ucp1 KO/RT/DMSO (n=3), Ucp1 KO/RT/RAPA (n=10), ATF4BOX;Ucp1 KO/RT/RAPA (n=5), Ucp1 KO/RT/BORT (n=4), ATF4BOX;Ucp1 KO/RT/BORT (n=4), Ucp1 KO/RT/BCH (n=5) and ATF4BOX;Ucp1 KO/RT/BCH (n=5). (G) The Temp (Neck-Back) of ~12-week-old male and female CON and LrpprcBKO mice 2 hours after injections of RAPA or BORT or DMSO at thermoneutrality. Sample size: CON (n=6) and LrpprcBKO (n=11). (H) The Temp (Neck-Back) of ~12-week-old male and female CON and ATF4BOX mice 2 hours after injections of RAPA or BORT or DMSO at thermoneutrality. Sample size: CON (n=7) and ATF4BOX (n=8). (I) The Temp (Neck-Back) of ~12-week-old male and female Ucp1 KO and ATF4BOX;Ucp1 KO mice 2 hours after injections of RAPA or BORT or DMSO at thermoneutrality. Sample size: Ucp1 KO (n=6) and ATF4BOX;Ucp1 KO (n=9). (J) Experimental setup of pSILAC in ATF4-expressing differentiated BAs. BAT SVF cells from ROSA-LSL-ATF4 or ROSA-LSL-ATF4;Ucp1 KO mice were cultured and differentiated in vitro. Four days after differentiation, GFP or Cre adenovirus were added at 100 MOI for two days. Cells were cultured in light medium from day 0 to 8, then transferred to heavy medium up to one day. Cells were collected at indicated time points for expression and mass spectrometry analyses. (K) Left: Immunoblots of Ucp1, Flag-ATF4, p-S6, total S6, p-4Ebp1, total 4Ebp1, puromycylated protein, ubiquitinated protein and Hsp90 in GFP or Cre-treated wild-type or Ucp1 KO BAs. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between genotypes indicated. (L) Left: Immunoblots of Flag-ATF4, total ATF4, p-S6, total S6, p-4Ebp1, total 4Ebp1, puromycylated protein, ubiquitinated protein and Hsp90 in GFP or Cre-treated wild-type BAs after 4-hour treatment of 10 mM BCH or DMSO. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between genotypes and treatment indicated. Data were presented as average ± SEM. Unpaired t-test. n.s.: non-significant; *: p<0.05 and **: p<0.01. (M) Box plot showing the fraction of heavy labeled over total protein in the GFP- or Cre-infected BAs in either wild-type (WT) or Ucp1 KO background at 6, 12, 24 hours post heavy medium switch. Welch’s 2-sided t-test performed. (N) Volcano plots showing the changes of H/L ratio between Cre- and GFP-infected BAs (X axis) and −log10 p value (Y axis) at 24-hour timepoint in WT and Ucp1 KO background. Grey box: proteins with higher turnover after ATF4-overexpression.
We then evaluated rates of protein synthesis in the BAT of LrpprcBKO mice at both RT and 30°C. First, ribosome protein S6 and mRNA translation repressor 4Ebp1 were highly phosphorylated in the BAT of LrpprcBKO mice (Figure S5I), reflecting a higher activity of mTORC1. Puromycin chasing experiments showed that global puromycylated proteins (an indicator of protein synthesis) were elevated in the BAT of LrpprcBKO mice (Figure S5I). Ubiquitinated proteins (an indicator of protein degradation) were simultaneously elevated in the BAT of LrpprcBKO mice, indicating the presence of an increased protein turnover (the coupled protein synthesis and degradation) (Figure S5I) (Harper and Bennett, 2016). In parallel, components of proteasome and autophagy degradation systems were transcriptionally upregulated in the BAT of the LrpprcBKO mice (Figure S5Q–R). For example, Lc3b expression levels were elevated without changes in the rate of processing/activation (Figure S5Q). This augmented protein turnover in BAT was dependent on ATF4 activity; it was absent in the Lrpprc;Atf4BKO mice (Figure S5J–K), but still present in the TfamBKO and ATF4BOX mice (with or without Ucp1) (Figure 4C, S5L–M). Our data suggest that cellular protein turnover is correlated with ATF4 activity, which may constitute a potential molecular mechanism underlying the ATF4-induced cold resistance in mice.
A single administration of 50mg kg−1 BCH (1 hour prior to CTT) suppressed S6 and 4Ebp1 hyperphosphorylation and simultaneous protein puromycylation and ubiquitination in the BAT of LrpprcBKO mice (Figure S5N). Consequently, the LrpprcBKO mice rapidly dropped their core temperature during CTT from RT to 4°C (Figure 4D). αMT also suppressed the cold resistance phenotype in the LrpprcBKO mice during CTT (Figure S5S). Similarly, we used rapamycin (RAPA) to inhibit protein synthesis and bortezomib (BORT) to inhibit protein degradation, and then examined their acute effects on cold tolerance. Pretreatment of 4mg kg−1 RAPA or 0.625mg kg−1 BORT also fully abolished the cold resistance phenotype in the LrpprcBKO mice (Figure 4D). The effects of BCH, RAPA and BORT on systemic thermoregulation were also observed in TfamBKO, ATF4BOX and ATF4BOX;Ucp1 KO mice (Figure 4E–F, S5T). Similarly, RAPA and BORT suppressed the BAT thermogenesis (determined by the Temp (Neck-Back) in IR) in the LrpprcBKO and ATF4BOX mice (in wild-type and Ucp1 KO background) (Figure 4G–I). These in vivo data suggest that the Ucp1-independent thermogenic mechanism in the aforementioned mouse models is fueled by ATF4-driven AA uptake and protein turnover. But these experiments with IP injections of inhibitors have two major caveats. First, these inhibitors may have systemic effects other than BAT, although IR studies have shown that inhibition of AA uptake (by BCH and αMT) and protein turnover (by RAPA and BORT) can suppress higher BAT temperature in the LrpprcBKO mice and ATF4BOX (in wild-type and Ucp1 KO background) mice (Figure 1L, 3I, O, 4D–I, S5S). These inhibitors had no effect on the CTT of control and GnasBKO mice (Figure 4D–F, S5T–U), suggesting that their systemic administration had minor effects on thermoregulation. In addition, RAPA only affected the cellular protein turnover in the BAT, not in the muscle (Figure S5O–P), indicating that the effect of RAPA in cold resistance phenotype in the ATF4BOX mice was due to the inhibition of mTORC1-dependent protein synthesis specifically in the BAT. Second, multiple downstream pathways are regulated by the AA-dependent mTORC1 activation, which can be suppressed by BCH and RAPA injection. Since multiple AATs of multiple classes are upregulated in the BAT in our mouse models (Figure 2E, S5C), their relative contributions to BAT thermogenesis need further investigations.
AA uptake from dietary protein has long been observed in rodents especially under cold exposure (López-Soriano and Alemany, 1987; López-Soriano et al., 1988). AAs can charge thermogenesis through multiple metabolic routes depending on the AA types (Owen et al., 2002). Branched-chain AAs can enter TCA cycle via succinyl-CoA or acetyl-CoA to fuel mitochondrial respiration (Neinast et al., 2019; Yoneshiro et al., 2019). AAs can also fuel the last step of glycolysis pathway to generate ATP when mitochondrial respiration is impaired. This metabolic flexibility (AA-driven energy production) is notably observed in cancer cells (DeBerardinis, 2011; Vincent et al., 2015; Wang and Dong, 2019). BAT tissue of the LrpprcBKO mice exhibited enhanced lactate production ex vivo (Figure S5V), indicating that a cytosolic ATP production might compensate for defective oxidative phosphorylation in the Lrpprc-deficient BAs. Interestingly, this increase in lactate production was blocked by the BCH treatment (Figure S5V), suggesting that AA can be substrates of energy production to fuel thermogenesis in Lrpprc-deficient BAs. This increased lactate production phenotype was not seen in the BAT of Lrpprc;Atf4BKO mice (Figure S5W), demonstrating the importance of ATF4-driven the AA uptake in this fuel switch. In contrast, ATF4 activation with functional mitochondria (in the ATF4BOX mice) did not significantly increase lactate production in the BAT (Figure S5X). Thus, LrpprcBKO and ATF4BOX mice may utilize different routes of ATP production to fuel the protein turnover in BAs.
Using BAT SVFs from ROSA-LSL-ATF4 and ROSA-LSL-ATF4;Ucp1 KO mice, we generated differentiated BAs overexpressing ATF4 in wild-type and Ucp1 KO background by infecting them with GFP or Cre adenovirus (Figure 4J). Q-PCR analysis showed that ATF4 expression induced ISR gene expression, but without noticeable differences in ETC gene expression (Figure S5Y), similar to the BAT with ATF4 overexpression in vivo (Figure 3H). In this in vitro system, the simultaneous increase of protein puromylation and ubiquitination by ATF4 activation was also evident (Figure 4K) independently of Ucp1, and it was blocked by the treatment of BCH (Figure 4L). Next, we performed a pulsed stable isotope labeling by amino acids in cell culture approach (pSILAC) to determine whether protein turnover is regulated by ATF4 in vitro (Figure 4J). Fully differentiated BAs were cultured in light (L) media until Day 8, then were switched to heavy (H) media for 6, 12 and 24 hours. The coverage of H-labeled peptides increased with labeling time, reaching ~1000 unique proteins (over 50% of detected proteins) at 24-hour (Supplementary Data 2). The H/L ratio of any given protein by mass spectrometry represents the relative abundance of newly synthesized peptides over preexisting peptides. At 24-hour, ATF4 overexpression increased the H/L ratio in both WT and Ucp1 deficient background (Figure 4M). We further analyzed the difference of H/L ratio caused by ATF4 overexpression; a positive H/L difference between Cre and GFP suggests an accelerated turnover. At the cutoff of (p<0.2), about one-fifth of detected proteins showed faster turnover in both WT and Ucp1 KO background (Figure 4N). More proteins showed increased protein turnover rates under a longer H labeling period; about 49% detected proteins exhibited accelerated turnover at 72 hours in WT BAs (Figure S5Z). Network analysis from the proteins with accelerated turnover at 24, 48 and 72 hours exhibited enrichment in protein homeostasis, such as folding, translation and transport (Figure S5AA).
Physiological regulation of ATF4 activation in brown adipocytes
Since 2–3-fold ATF4 overexpression in BAs is sufficient to drive this AA responsive and Ucp1-independent thermogenesis in mice, we reasoned that physiological stimuli that can activate ATF4 in BAs to a similar extent may promote BAT thermogenesis. Protein-restriction diet feeding induces ATF4 activity in the liver through GCN2-dependent and - independent mechanisms (Laeger et al., 2016; Maida et al., 2016), but whether it has a direct effect on BAT is still unknown.
We custom-made a low-protein diet (LP, 5%P/13%F/82%C, P: protein, F: fat, C: carbohydrate) and its isogenic control normal-protein diet (CP, 25%P/13%F/62%C) with the same calorie density (3.76 kcal g−1). Newly weaned wild-type (C57bl/6J) mice were housed at 30°C for ~4 weeks on regular-chow diet, then they were switched to either CP or LP diet for an additional 4 weeks (Figure 5A). 30°C housing is used here for two reasons: it can reduce cold-induced thermogenesis and it can better reflect human physiology, because humans most live at thermoneutrality (Ganeshan and Chawla, 2017; Maloney et al., 2014). Under LP feeding, BA morphology transformed from unilocularity to multilocularity even under 30°C (Figure 5B). Indirect calorimetry experiment showed that EE was elevated by LP diet independently of the difference of body weight (Figure S6A–B) (Tschop et al., 2012). RER, physical activity and food intake were not affected by LP feeding in indirect calorimetry measurement (Figure S6C–G), although other studies showed a similar low-protein diet either induced or had no effect on food intake over longer periods (Hill et al., 2017; Maida et al., 2016). Consistent with the notion that LP feeding can activate Ucp1-dependent thermogenesis in BAs due to augmented sympathetic inputs to BAT (Hill et al., 2017; Laeger et al., 2016; Laeger et al., 2014; Maida et al., 2016), LP feeding increased the expression of ETC mRNA and Ucp1 mRNA and protein in the BAT (Figure 5E, S6H–I) and LP-fed C57bl/6J mice exhibited enhanced BAT thermogenesis and cold resistance compared to CP-fed mice (Figure 5C–D). Moreover, Ucp1 KO mice housed at RT were cold sensitive in CTT (Enerback et al., 1997). Surprisingly, LP also increased the food-dependent BAT thermogenesis (by IR) and cold resistance (by CTT) of the Ucp1 KO mice housed at RT (Figure 5K–M, P), suggesting that low-protein diet feeding can promote cold tolerance through additional Ucp1-independent mechanisms.
Figure 5. A low-protein diet feeding activates ATF4 in brown adipocytes.
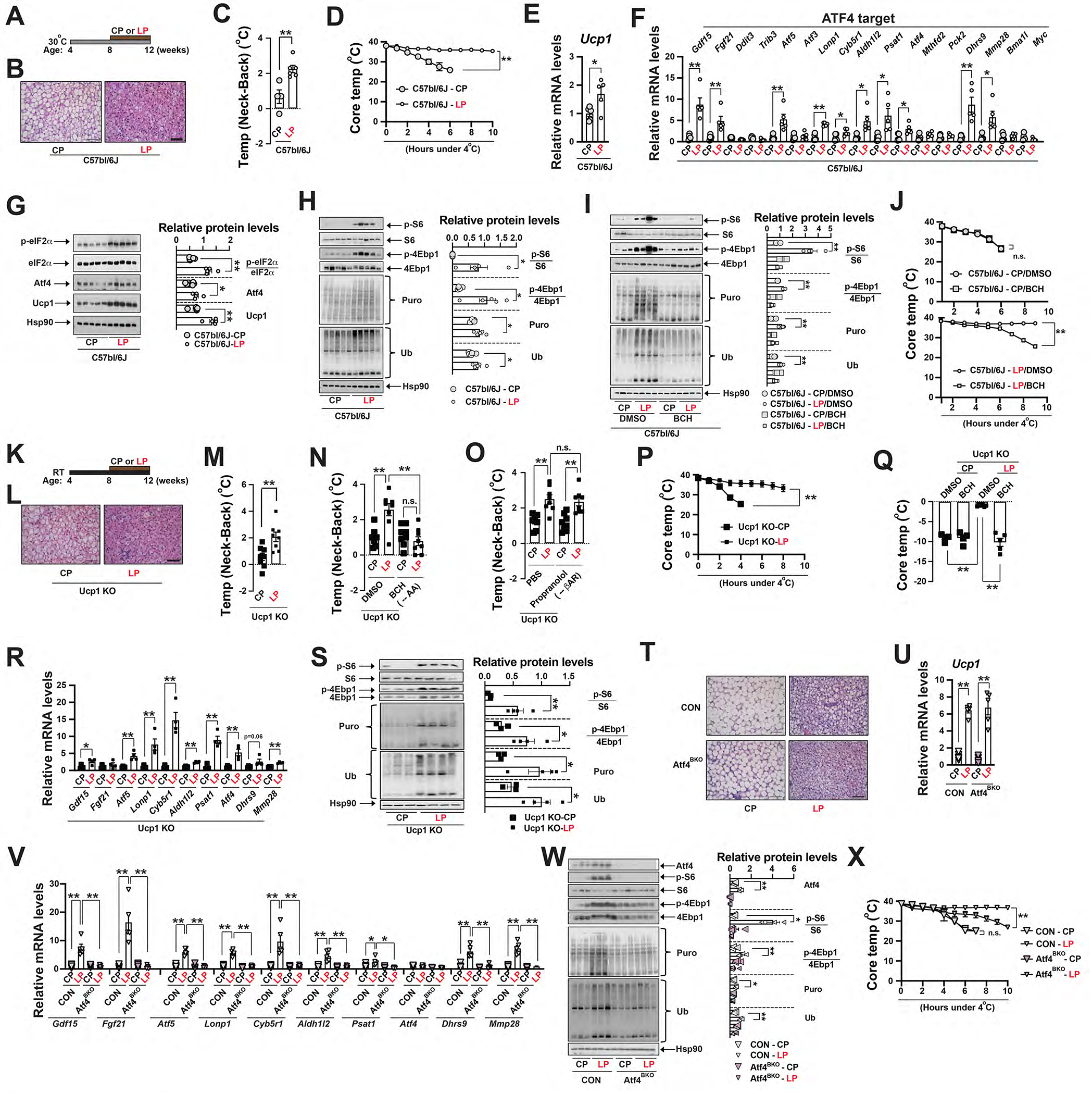
(A) Diagram of experimental design showing low-protein (LP) feeding of C57bl/6j mice at 30°C. (B) Representative H&E staining of BAT of ~12-week-old male C57bl/6J mice after 4-week CP or LP feeding at 30°C. Scale bar: 100 μm. (C) The Temp (Neck-Back) of ~12-week-old male C57bl/6J mice after 4-week CP or LP feeding at 30°C. Sample size: CP (n=4) and LP (n=6). (D) CTT of ~12-week-old male and female wild-type (C57bl/6J) mice between low-protein (LP) and control protein (CP) feeding. Sample size: C57bl/6J-CP (n=5) and C57bl/6J-LP (n=5). Q-PCR analysis of Ucp1 (E) and ATF4 target genes (F) in the BAT of ~12-week-old male C57bl/6J mice at ad libitum CP or LP feeding at 30°C. Sample size: CP (n=5) and LP (n=5). (G) Left: Immunoblots of p-eIF2α, total eIF2α, Atf4, Ucp1 and Hsp90 in the BAT of ~12-week-old male C57bl/6J mice after 4-week CP or LP feeding at 30°C. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between diets indicated. (H) Left: Immunoblots of p-S6, total S6, p-4Ebp1, total 4Ebp1, puromycylated protein, ubiquitinated protein and Hsp90 in the BAT of C57bl/6J mice at ad libitum CP or LP feeding at 30°C. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between diets indicated. (I) Left: Immunoblots of p-S6, total S6, p-4Ebp1, total 4Ebp1, puromycylated protein, ubiquitinated protein and Hsp90in the BAT of C57bl/6J mice at ad libitum feeding CP or LP feeding with DMSO or BCH injection at 30°C. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between diets indicated. (J) CTT of ~12-week-old male and female C57bl/6J mice after CP or LP feeding at 30°C. DMSO or BCH was injected 1 hour prior to CTT. Sample size: C57bl/6J-CP/DMSO (n=3), C57bl/6J-CP/BCH (n=4), C57bl/6J-LP/DMSO (n=7) and C57bl/6J-LP/BCH (n=9). (K) Diagram of experimental design showing LP feeding of Ucp1 KO mice at RT. (L) Representative H&E staining of BAT of ~12-week-old male Ucp1 KO mice after 4-week CP or LP feeding at RT. Scale bar: 100 μm. (M) The Temp (Neck-Back) of ~12-week-old male and female Ucp1 KO mice under 4-week CP or LP feeding. (N) The Temp (Neck-Back) of ~12-week-old male and female Ucp1 KO mice with or without BCH injection under 4-week CP or LP feeding. (O) The Temp (Neck-Back) of ~12-week-old male and female Ucp1 KO mice with or without propranolol injection under 4-week CP or LP feeding. Sample size: CP (n=8) and LP (n=8). (P) CTT of ~12-week-old male and female Ucp1 KO mice after CP or LP feeding at RT. Sample size: Ucp1 KO-CP (n=5) and Ucp1 KO-LP (n=7). (Q) Core temperature drop of ~12-week-old male and female Ucp1 KO mice fed with CP or LP after 4-hour CTT from RT. DMSO or BCH was injected 1 hour prior to CTT. Sample size: Ucp1 KO-CP/DMSO (n=4), Ucp1 KO-CP/BCH (n=4), Ucp1 KO-LP/DMSO (n=6) and Ucp1 KO-LP/BCH (n=5). (R) Q-PCR analysis of ATF4 target genes in the BAT of ~12-week-old male and female Ucp1 KO mice after 4-week CP or LP feeding at RT. Sample size: Ucp1 KO-CP (n=4) and Ucp1 KO-LP (n=4). (S) Left: Immunoblots of p-S6, total S6, p-4Ebp1, total 4Ebp1, puromycylated protein, ubiquitinated protein and Hsp90 in the BAT of ~12-week-old Ucp1 KO mice at ad libitum CP or LP feeding at RT. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between diets indicated. (T) Representative H&E staining of BAT of ~12-week-old male CON and Atf4BKO mice after 4-week CP or LP feeding at 30°C. Scale bar: 100 μm. Q-PCR analysis of Ucp1 (U) and ATF4 target genes (V) in the BAT of the aforementioned mice. Sample size: CON-CP (n=6), CON-LP (n=6), Atf4BKO-CP (n=5) and Atf4BKO-LP (n=5). (W) Left: Immunoblots of p-S6, total S6, p-4Ebp1, total 4Ebp1, puromycylated protein, ubiquitinated protein and Hsp90 in the BAT of CON and Atf4BKO mice at ad libitum CP or LP feeding at 30°C. Right: Relative abundance (to Hsp90) or phosphorylation (to total protein) shown. Significance between diets indicated. (X) CTT of ~12-week-old male and female CON and Atf4BKO mice after CP or LP feeding at 30°C. Sample size: CON-CP (n=5), CON-LP (n=12), Atf4BKO-CP (n=3) and Atf4BKO-LP (n=11). Data were presented as average ± SEM. Unpaired t-test. n.s.: non-significant; *: p<0.05 and **: p<0.01.
Given the connection between ATF4 activation and low-protein diet and the fact that pharmacological inhibition of AA uptake by BCH suppressed this ATF4 activation induced thermoregulation in vivo (Figure 4H–I), we then investigated whether ATF4 in BAs contributes to the LP-induced but Ucp1-independent thermogenesis. LP feeding activated upstream ATF4 activator (phosphorylated eIF2α), Atf4 protein accumulation, upregulation of many ATF4 target genes and increased protein turnover in the BAT of the wild-type mice at thermoneutrality (Figure 5F–H). In contrast, the LP-induced ATF4 transcriptional activation and protein turnover were not evident in skeletal muscles (Figure S6J–K). Acute blockade of AA uptake by BCH suppressed the LP-induced protein turnover and cold resistance phenotype in wild-type mice (Figure 5I–J). In Ucp1 KO mice, LP feeding still promoted the characteristic multilocular morphology of Ucp1-deficient BAs and induced ATF4 target gene expression and protein turnover in the BAT (Figure 5L,R–S). AA uptake inhibition by BCH abolished the LP-induced BAT thermogenesis and cold resistance phenotype in the Ucp1 KO mice (Figure 5N, Q), indicating that LP feeding can activate ATF4 in BAs independently of Ucp1. Notably, this LP-induced BAT thermogenesis in the Ucp1 KO mice was not suppressed by propranolol (Figure 5O), similar to the LrpprcBKO mice (Figure 1L).
Finally, LP-induced cold resistance was partially suppressed in BA-specific Atf4 knockout mice (Atf4BKO) mice, along with reductions of ATF4 target genes and protein turnover in BAT (Figure 5V–X). In contrast, LP-induced BA multilocular morphology, EE increase and upregulation of Ucp1 and ETC genes in the BAT were not affected by Atf4 deficiency (Figure 5T–U, S6L–T). Collectively, low-protein diet induces the ATF4-driven but Ucp1-independent thermogenesis, representing a new physiological axis of thermoregulation in mice.
Positive correlation between ATF4 activity in brown adipocytes and systemic metabolism
Next, we began to characterize the metabolic consequences of ATF4 activation in BAs in the aforementioned mouse models. Although male LrpprcBKO mice had similar body weight and adipose mass at ~8-week of age, they gained less adiposity under normal chow feeding at both ambient temperatures up to ~8-month of age (Figure S7A–C). Since the ATF4-dependent ISR remained elevated in the BAT of LrpprcBKO mice under high fat diet (HFD) (Figure S7D), we also determined their metabolic performance under HFD. Male LrpprcBKO mice gained less body weight under high fat diet (HFD) (Figure 6A). Fat percentage was increased in control mice by four-fold by 12-week HFD, but it barely increased in LrpprcBKO mice (Figure S6F). Fat depots like iWAT and eWAT showed progressively reduced weight under HFD and contained smaller adipocytes, and BAT only showed reduced weight after 12-week HFD (Figure 6B, S7E). The adipocyte size in eWAT was also reduced in LrpprcBKO mice after HFD (Figure S7G–H). We also measured other HFD-induced metabolic parameters in male LrpprcBKO mice. Systemic insulin sensitivity was improved in LrpprcBKO mice, and serum insulin levels were significantly reduced at 12-week HFD (Figure 6C–F). HFD-induced hypertriglyceridemia was completely inhibited in LrpprcBKO mice (Figure 6G). Similar results were also obtained at 30°C (Figure 6H). HFD also induced ectopic triglyceride accumulation in the liver in control mice, which was also absent in LrpprcBKO mice (Figure 6I–K). HFD-induced adipose inflammation was suppressed by Lrpprc deficiency in BAs. Q-PCR analysis showed macrophage markers (Cd68, F4/80 and Cd11c) and pro-inflammatory cytokine (Ccl2) were reduced in the eWAT of LrpprcBKO mice after 12-week HFD (Figure S7J) and serum leptin levels were also reduced (Figure S7I). Thus, Lrpprc deficiency in BAs in BAT leads to the protection against HFD-induced obesity, insulin resistance, hepatic steatosis, hypertriglyceridemia, and adipose inflammation, despite defective thermogenesis in BAT. The reduced adiposity and liver TG phenotypes were also observed in female LrpprcBKO mice (Figure S7K–N).
Figure 6. LrpprcBKO mice exhibit improved systemic metabolism due to ATF4 activation in brown adipocytes.
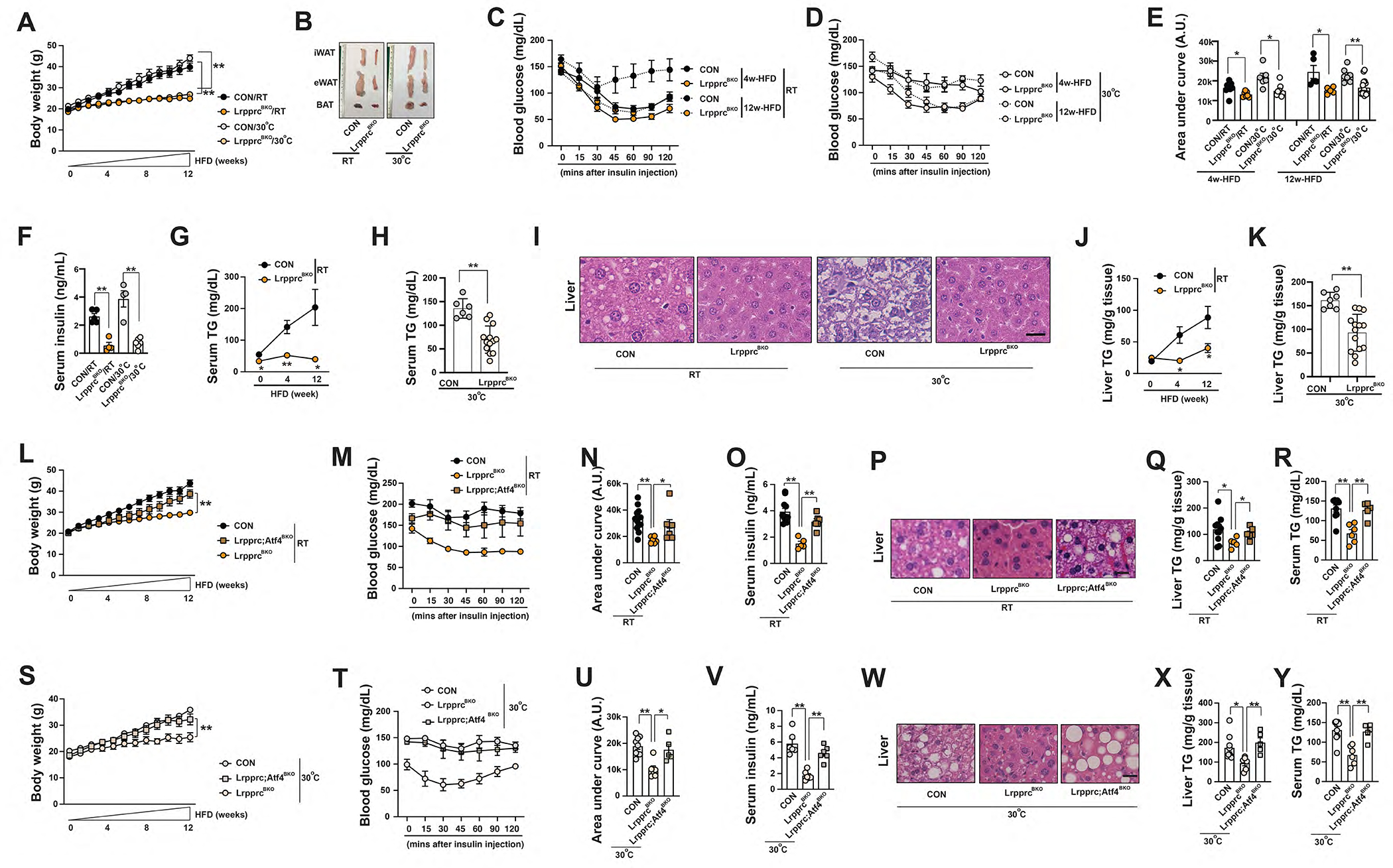
(A) Body weight of male CON and LrpprcBKO mice under 12-week HFD at RT and 30°C. Sample size: CON/RT (n=12), LrpprcBKO/RT (n=11), CON/30°C (n=7) and LrpprcBKO/30°C (n=15). (B) Representative images of dissected iWAT, eWAT and BAT from male CON and LrpprcBKO mice after 12-week HFD. Serum glucose levels during ITT in male CON and LrpprcBKO mice after 4-week and 12-week HFD at RT (C) and 30°C (D). Sample size: CON/4w-HFD/RT (n=8), LrpprcBKO/4w-HFD/RT (n=7), CON/12w-HFD/RT (n=5), LrpprcBKO/12w-HFD/RT (n=5), CON/4w-HFD/30°C (n=6), LrpprcBKO/4w-HFD/30°C (n=9), CON/12w-HFD/30°C (n=7) and LrpprcBKO/12w-HFD/30°C (n=15). (E) Area under the curve (AUC) values of glucose levels in ITTs showed. (F) Serum insulin levels in male CON and LrpprcBKO mice after 12-week HFD. Sample size: CON/RT (n=5), LrpprcBKO/RT (n=4), CON/30°C (n=4), LrpprcBKO/30°C (n=6). (G) Serum triglyceride contents of male CON and LrpprcBKO mice after HFD at RT. Sample size: Sample size: CON/NC/RT (n=6), LrpprcBKO/NC/RT (n=10), CON/4w-HFD/RT (n=8), LrpprcBKO/4w-HFD/RT (n=10), CON/12w-HFD/RT (n=5) and LrpprcBKO/12w-HFD/RT (n=5). (H) Serum triglyceride contents of male CON and LrpprcBKO mice after 12-week HFD at 30°C. Sample size: CON/12w-HFD/30°C (n=6) and LrpprcBKO/12w-HFD/30°C (n=13). (I) Representative H&E staining of liver from male CON and LrpprcBKO mice after 12-week HFD. Scale bar: 25 μm. (J) Liver triglyceride contents of male CON and LrpprcBKO mice after HFD at RT. Sample size: CON/NC/RT (n=6), LrpprcBKO/NC/RT (n=7), CON/4w-HFD/RT (n=7), LrpprcBKO/4w-HFD/RT (n=6), CON/12w-HFD/RT (n=5) and LrpprcBKO/12w-HFD/RT (n=5). (K) Liver triglyceride contents of male CON and LrpprcBKO mice after 12-week HFD at 30°C. Sample size: CON/12w-HFD/30°C (n=7) and LrpprcBKO/12w-HFD/30°C (n=14). (L) Body weight of male CON, LrpprcBKO and Lrpprc;Atf4BKO mice under 12-week HFD at RT. Sample size: CON/RT (n=13), LrpprcBKO/RT (n=6) and Lrpprc;Atf4BKO/RT (n=8). (M) Serum glucose levels during ITT in male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after 12-week HFD at RT. (N) Area under the curve (AUC) values of glucose levels in ITTs showed. (O) Serum insulin levels of male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after HFD at RT. (P) Representative H&E staining of liver from male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after 12-week HFD at RT. Scale bar: 25 μm. Liver triglyceride contents (Q) and serum triglyceride contents (R) of male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after HFD at RT. Sample size: CON (n=11), LrpprcBKO (n=5), and Lrpprc;Atf4BKO (n=6). (S) Body weight of male CON, LrpprcBKO and Lrpprc;Atf4BKO mice under 12-week HFD at 30°C. Sample size: CON/30°C (n=11), LrpprcBKO/30°C (n=7), and Lrpprc;Atf4BKO/30°C (n=6). (T) Serum glucose levels during ITT in male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after 12-week HFD at 30°C. (U) Area under the curve (AUC) values of glucose levels in ITTs showed. (V) Serum insulin levels of male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after HFD at 30°C. (W) Representative H&E staining of liver from male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after 12-week HFD at 30°C. Scale bar: 25 μm. Liver triglyceride contents (X) and serum triglyceride contents (Y) of male CON, LrpprcBKO and Lrpprc;Atf4BKO mice after HFD at 30°C. Sample size: CON (n=9), LrpprcBKO (n=6), and Lrpprc;Atf4BKO (n=5). Data were presented as average ± SEM. Unpaired t-test. *: p<0.05 and **: p<0.01.
The LrpprcBKO mice exhibit increased EE (at night), reduced RER and reduced food intake (at night) without any changes in physical activity under 4-week HFD at 30°C (Figure S7O,Q,S,U). Progression plot analyses suggest that the difference in body weight could also be one of the contributing factors for these changes in the LrpprcBKO mice (Figure S7P,R,T,V). The rapid divergence in their body weight gain under HFD complicates the interpretation of CLAMS analyses in HFD-fed mice. A recent report on the IMPC data (over 30,000 laboratory mice) also demonstrates a pronounced mass effect on energy expenditure, showing that bigger mice expend more energy under standard housing conditions (Corrigan et al., 2020).
Since Atf4 activation in BAs is required for protein turnover-fueled thermogenesis in mice, we then evaluated whether Atf4 activation also contributes to the metabolic fitness in the LrpprcBKO mice. We first examined the metabolic performance of male BA-specific Atf4 knockout mice (Atf4BKO). In fact, HFD-induced obesity, insulin resistance, hepatosteatosis and hyperlipidemia were not significantly altered by Atf4 deficiency alone (Figure S7W–AD). We then characterized the full spectrum of metabolic performance of Lrpprc;Atf4BKO mice. Compared to the LrpprcBKO mice, the Lrpprc;Atf4BKO mice were no longer protected against HFD-induced obesity, adipocyte hypertrophy, systemic insulin resistance, hepatosteatosis and hyperlipidemia at RT and 30°C (Figure 6L–Y, S7AE–AJ). Thus, Atf4 deletion reverses metabolic benefits without enhancing mitochondrial respiration and Ucp1-mediated thermogenesis in the LrpprcBKO mice.
Similar to the LrpprcBKO mice, the Lrpprc;Atf4BKO mice still exhibited elevated thermogenic gene expression and expansion of multilocular Ucp1+ beige adipocyte in the iWAT after 12-week HFD at RT (Figure S7AQ–AR), although they did not show the same metabolic benefits under HFD anymore. Thus, the abundance of multilocular Ucp1+ beige adipocytes in iWAT alone does not correlate with the improved systemic metabolism caused by ATF4 activation in BAs.
To determine whether ATF4 activation in wild-type BAs (without defective mitochondria) can induce systemic metabolic benefits, we performed HFD experiments similarly on ATF4BOX mice at both RT and 30°C. ATF4-ISR remained elevated in the BAT of ATF4BOX mice even after HFD (Figure S7AK), but persistent WAT browning was not observed (Figure S7AQ–AR). At RT and 30°C, both male and female ATF4BOX mice gained less body weight, which was contributed by reduced adiposity (Figure 7A–D, S7AN–AP). Again, other metabolic benefits, such as improved insulin sensitivity, reduced hepatosteatosis and hyperlipidemia and reduced adipose inflammation were all evident in ATF4BOX mice (Figure 7E–J, S7AL–AM). Importantly, these metabolic benefits caused by ATF4 overexpression in BAs were present even in the Ucp1 KO background (Figure 7O–S).
Figure 7. ATF4 activation in brown adipocytes is sufficient to improve systemic metabolism in wild-type and Ucp1 knockout mice.
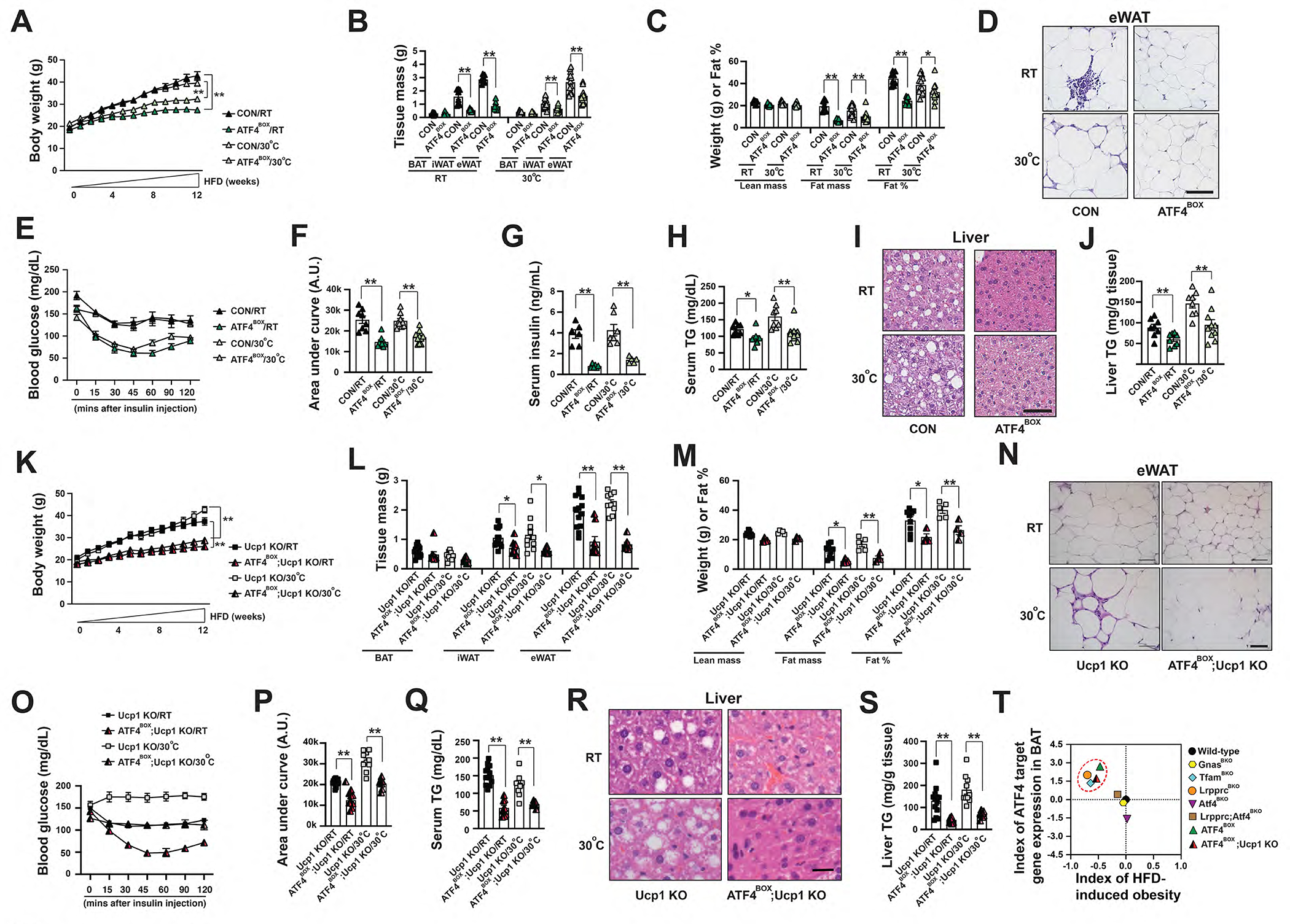
(A) Body weight of male CON and ATF4BOX mice after 12-week HFD at RT and 30°C. Sample size: CON/RT (n=8), ATF4BOX/RT (n=8), CON/30°C (n=17) and ATF4BOX/30°C (n=18). (B) Tissue mass of eWAT, iWAT, and BAT of male CON and ATF4BOX mice after 12-week HFD. Sample size: male CON/RT (n=8), ATF4BOX/RT (n=8), CON/30°C (n=13) and ATF4BOX/30°C (n=13). (C) Lean mass, fat mass, and fat percentage of male CON and LrpprcBKO mice after 12-week HFD. Sample size: CON/RT (n=8), ATF4BOX/RT (n=8), CON/30°C (n=13) and ATF4BOX/30°C (n=13). (D) Representative H&E staining of eWAT from male CON and ATF4BOX mice after 12-week HFD. Scale bar: 50 μm. (E) Serum glucose levels during ITT in male CON and ATF4BOX mice after 12-week HFD at RT and 30°C. (F) Area under the curve (AUC) values of glucose levels in ITTs showed. Sample size: CON/RT (n=8), ATF4BOX/RT (n=8), CON/30°C (n=9) and ATF4BOX/30°C (n=10). (G) Serum insulin levels in male CON and ATF4BOX mice after 12-week HFD. Sample size: CON/RT (n=6), ATF4BOX/RT (n=6), CON/30°C (n=7), ATF4BOX/30°C (n=5). (H) Serum triglyceride contents of male CON and ATF4BOX mice after 12-week HFD. Sample size: CON/RT (n=8), ATF4BOX/RT (n=8), CON/30°C (n=8) and ATF4BOX/30°C (n=10). (I) Representative H&E staining of liver from male CON and ATF4BOX mice after 12-week HFD. Scale bar: 50 μm. (J) Liver triglyceride contents of male CON and ATF4BOX mice after 12-week HFD. Sample size: CON/RT (n=8), ATF4BOX/RT (n=8), CON/30°C (n=8) and ATF4BOX/30°C (n=10). (K) Body weight of male Ucp1 KO and ATF4BOX;Ucp1 KO mice under 12-week HFD at RT and 30°C. Sample size: Ucp1 KO/RT (n=9), ATF4BOX;Ucp1 KO/RT (n=4), Ucp1 KO/30°C (n=5), and ATF4BOX;Ucp1 KO/30°C (n=4). (L) Tissue mass of eWAT, iWAT, and BAT of male Ucp1 KO and ATF4BOX;Ucp1 KO mice after 12-week HFD at RT and 30°C. (M) Lean mass, fat mass, and fat percentage of male Ucp1 KO and ATF4BOX;Ucp1 KO mice after 12-week HFD at RT and 30°C. Sample size: Ucp1 KO/RT (n=9), ATF4BOX;Ucp1 KO/RT (n=4), Ucp1 KO/30°C (n=5), and ATF4BOX;Ucp1 KO/30°C (n=4). (N) Representative H&E staining of eWAT from male CON and ATF4BOX mice after 12-week HFD. Scale bar: 50 μm. (O) Serum glucose levels during ITT in male Ucp1 KO and ATF4BOX;Ucp1 KO mice after 12-week HFD at RT and 30°C. (P) Area under the curve (AUC) values of glucose levels in ITTs showed. Sample size: Ucp1 KO/RT (n=9), ATF4BOX;Ucp1 KO/RT (n=4), Ucp1 KO/30°C (n=5), and ATF4BOX;Ucp1 KO/30°C (n=4). (Q) Serum triglyceride contents of male Ucp1 KO and ATF4BOX;Ucp1 KO mice after 12-week HFD at RT and 30°C. Sample size: Ucp1 KO/RT (n=9), ATF4BOX;Ucp1 KO/RT (n=4), Ucp1 KO/30°C (n=5), and ATF4BOX;Ucp1 KO/30°C (n=4). (R) Representative H&E staining of liver from male CON and ATF4BOX mice after 12-week HFD. Scale bar: 25 μm. (S) Liver triglyceride contents of male Ucp1 KO and ATF4BOX;Ucp1 KO mice after 12-week HFD at RT and 30°C. Sample size: Ucp1 KO/RT (n=9), ATF4BOX;Ucp1 KO/RT (n=4), Ucp1 KO/30°C (n=5), and ATF4BOX;Ucp1 KO/30°C (n=4). Data were presented as average ± SEM. Unpaired t-test. *: p<0.05 and **: p<0.01. (T) Scatter plots showing index of HFD-induced obesity (the Log2 fold-change values of body weight after HFD) as the function of ATF4 activation in BAs (the average Log2 fold-change of ATF4 target genes in BAT). Dashed red cycle: LrpprcBKO, TfamBKO, ATF4BOX and ATF4BOX;Ucp1 KO mice that exhibited enhanced cellular proteome turnover and reduced HFD-induced obesity.
We calculated the index of HFD-induced obesity (the Log2 fold-change values of body weight after HFD between mutants and controls) to quantitatively evaluate the effect of knockout or overexpression of a given gene on HFD-induced obesity in the individual experiment. For example, the indexes of HFD-induced obesity for LrpprcBKO and GnasBKO mice were −0.7 (a 38% decrease in body weight) and −0.046 (a non-significant 3% decrease) respectively, indicating that Lrpprc deficiency in BAs affected HFD-induced obesity much more profoundly than Gnas deficiency. The data from the above collection of BA-specific knockout or overexpression mouse models suggest that ATF4 activation in BAs is positively associated with a decreased susceptibility of HFD-induced obesity (Figure 7T).
DISCUSSION
Comparative studies have suggested that the appearance of thermogenically active BAs in endotherms provides an evolutionary advantage to survive in cold environment (Jastroch et al., 2018). Thermogenically active BAs in rodents and humans are defined by three criteria: multilocular in morphology, rich in mitochondria and positive for UCP1. Divergent from this “mitochondria-fueled and Ucp1-mediated thermogenesis”-centric viewpoint, this study demonstrates that BAs can efficiently generate heat via dietary amino acid uptake and protein turnover. Wild-type mice only process activated Ucp1-dependent thermogenesis. GnasBKO (or betaless or Ucp1 KO) mice at RT or wild-type mice housed at thermoneutrality lose the Ucp1-dependent thermogenesis, consequently showing defective thermoregulation in CTT. Even though TfamBKO and LrpprcBKO mice also have reduced mitochondrial respiration, they can still maintain their core temperature during CTT through augmented ATF4 activation in BAs only under fed condition. ATF4 overexpression in BAs even promoted cold resistance in wild-type mice at thermoneutrality and even Ucp1 KO mice at RT. And importantly, the unilocular BAs with low Ucp1, such as the ones present in mouse housed at thermoneutrality or maybe humans, can also be thermogenically competent due to this ATF4 activation in BAs.
Besides cold temperature, dietary changes also affect BAT thermogenesis through the diet-dependent thermogenesis process (Bonet et al., 2017; Kozak, 2010; Rothwell and Stock, 1979). In fact, this diet-dependent thermogenesis was also conspicuous in young pigs (without Ucp1 expression) (Dauncey and Ingram, 1979; Lucke and Hall, 1983) and humans with complete spinal cord injury (lacking sympatho-adrenal activity) (Aksnes et al., 1995). Noteworthily, this ATF4 activation in BAs can be gauged from low (under normal chow feeding) to high (under LP feeding) in wild-type mice, suggesting that it is a physiologically relevant process that can tailor energy expenditure to changes in macronutrient composition. More studies are needed to investigate this new thermoregulatory axis, which will further expand our knowledge regarding how dietary macronutrients and BAT thermogenesis are dynamically integrated in health and disease.
The “mitochondrial dysfunction→ATF4 activation” axis has been described in cells (Bao et al., 2016; Kuhl et al., 2017; Quiros et al., 2017), liver (Kang et al., 2021) and muscles (da Silveira et al., 2020; Forsström et al., 2019; Keipert et al., 2014; Khan et al., 2017; Kim et al., 2013). Although whether this axis regulates BAT thermogenesis is unknown, ATF4-dependent Fgf21 transcription has been observed in the BAT of Ucp1 knockout mice (Keipert et al., 2015), and the elevation of circulating Fgf21 levels contributes to the improved metabolic performance of the Ucp1 knockout mice at RT (Keipert et al., 2020). Fgf21 acts on its receptor in the central brain to regulate BAT thermogenesis via sympathetic nervous system and then βAR activation in BAs (Owen et al., 2014). Although Fgf21 transcription was significantly upregulated in the BAT of the LrpprcBKO mice, the circulating Fgf21 levels were comparable between CON and LrpprcBKO mice (Figure S3J). In addition, the LP diet feeding can also induce Fgf21 transcription in both BAT and liver and increase circulating Fgf21 levels by ~10-fold. But circulating Fgf21 levels were not affected in the Atf4BKO mice (Figure S6U), which might be due to the relatively smaller size of BAT (compared to the liver). It is unlikely that BAT-derived Fgf21 is the causal factor of this propranolol-insensitive thermogenesis induced by ATF4 activation in the BAs. A recent study showed that BA-specific Opa1 KO mice exhibited improved metabolism, which was ATF4-dependent but Fgf21-independent (Pereira et al., 2021). The metabolic contributions of BAT-derived Fgf21 require further investigations.
Promoting the thermogenic activity from BAs to increase total energy expenditure has been proposed as a therapeutic approach for obesity prevention and/or treatment. We have demonstrated that mice with elevated ATF4 actitivity in BAs (LrpprcBKO, TfamBKO, and ATF4BOX mice) exhibited a profound anti-obesity phenotype, regardless of their activity in Ucp1-dependent thermogenesis (Figure 7T). And removing the ATF4 (without affecting Ucp1-dependent thermogenesis) in LrpprcBKO mice reversed metabolic benefits, suggesting a positive correlation between the ATF4 activity in BAs and systemic metabolism (Figure 7T). Humans most live at thermoneutrality and don’t have abundant multilocular Ucp1+ BAs. Studies along this direction may unveil a translational perspective to link BAs to precision nutrition for the prevention or treatment of obesity and other metabolic dysfunctions in humans.
LIMITATION OF THE STUDY
Although brown adipocyte ATF4 activation improves thermoregulation in mice (e.g. the ATF4BOX mice) in the Ucp1-independent manner, the in vivo relevance and mechanistic details of this ATF4-driven heat production in brown adipocytes haven’t been established yet. Future studies will determine the proteome degradative mechanism and heat production in vitro cell autonomously in brown adipocytes. Low protein diet is a rather broad intervention that affects multiple tissues nutritionally and hormonally. The actual properties of low protein diet important of its capacity in promoting the propranolol-insensitive thermogenic program in BAT are unknown, which also warrants separate investigations.
STAR*METHODS
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Biao Wang (biao.wang@ucsf.edu).
Materials availability
Reagents in this study are available by request to lead contact, Biao Wang (biao.wang@ucsf.edu).
Data and code availability
The mass spectrometry data files (raw and search results) have been deposited to http://proteomecentral.proteomexchange.org, the ProteomeXchange Consortium, via the PRIDE partner repository with PXD008798. The raw RNA-seq data have been deposited to NCBI GEO (accession # GSE117985). No code is generated in this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse studies:
ROSA-LSL-ATF4 (JAX #029394), ROSA-LSL-iDTR (JAX #007900) and Ucp1 KO (JAX #003124) mice were purchased from JAX. Lrpprcf/f, Atf4f/f, and Ucp1-Cre (JAX #024670) mice were kindly provided by Drs. Nils-Göran Larsson, Christopher Adams, and Evan Rosen. TfamBKO and GnasBKO mice were characterized before (Masand et al., 2018; Paulo et al., 2018b). Mice were housed in a temperature-controlled environment at 22°C (RT) under a 12h light:dark cycle with free access to water and food (PicoLab® #5053). For thermoneutral experiments, ~4-week-old mice were placed in a 30°C rodent chamber (Power Scientific RIS52SD Rodent Incubator) for an additional 3–4 weeks to reach their thermoneutral zone. Low-protein diet (LP, 5%P/13%F/82%C, P: protein, F: fat, C: carbohydrate) and its isogenic control normal-protein diet (CP, 25%P/13%F/62%C) were custom-made from Research Diets. The calorie density for both diets was identical (3.76 kcal g−1). For LP or CP feeding, ~8-week-old mice housed at RT or 30°C were switched to either LP or CP diet for additional 4 weeks. For high-fat diet studies, ~8-week-old mice were transferred to a 60% fat diet (Research Diets, D12492) housed at RT or 30°C. GnasBKO mice were in129S6/SvEvTac Black Swiss background, and all other mice were in C57BL/6J background. All animal experiments were approved by the UCSF Institutional Animal Care and Use Committee in adherence to US National Institutes of Health guidelines and policies.
METHOD DETAILS
Indirect calorimetry measurements:
The Comprehensive Lab Animal Monitoring System (CLAMS) was used to measure metabolic parameters including oxygen consumption (VO2), carbon dioxide production (VCO2), food intake, water intake and physical activity for 3 days (after 2-day acclimation period). RER was calculated as the ratio of VCO2/VO2. Energy expenditure (EE) were calculated per mouse based on oxygen consumption using the formula EE=(3.815+1.232*RER)*VO2 (Butler and Kozak, 2010; Tschop et al., 2012). Hourly average and 3-day (from day 3 to day 5) cumulative food intakes were calculated separately. At 9–10am of the day 5, CL316 243 (CL, TOCRIS, #1499) at the dose of 1mg kg−1 was injected intraperitoneally. The CL-induced VO2 and EE were calculated as hourly average values of ~4–5 hours after CL injection. Data were exported from CLAX and analyzed by MATLAB. Investigators were blinded to the mouse genotypes for CLAMS, which was performed by the UCSF Diabetes and Endocrinology Research Center Metabolic Research Unit.
18F-fluorodeoxyglucose (FDG) uptake:
A dedicated small animal PET/CT (Inveon, Siemens Medical Solutions, Malvern, PA) was used for all imaging procedures at room temperature. For consistent data acquisition, all animals were fasted overnight, at least 12 hours, before administration of 18F-fluorodeoxyglucose (FDG). FDG (3.94±0.17 MBq, range: 3.67–4.17 MBq) was administered intravenously via tail vein under anesthesia (2–2.5% isoflurane). Uptake time of 55 min (±1 min) was strictly followed before the start of the scan. During the uptake time, the animals were awake and kept warm over a temperature-controlled heating pad at 37°C. There was no induced voiding during the uptake time. Ten-minute static PET data were acquired for all animals, followed by CT under isoflurane (2–2.5%) anesthesia. The total imaging time was under 20 minutes. Once the data for PET and CT were acquired, reconstructions were performed using vendor-provided software. An iterative reconstruction algorithm with CT-based attenuation correction was used for PET, and a Feldkamp reconstruction algorithm modified for conebeam was used for CT. The reconstructed volumes were 128×128×159 matrices with a voxel size of 0.776383 mm × 0.776383 mm × 0.796 mm for PET, and 512×512×700 matrices with an isotropic voxel size of 0.196797 mm × 0.196797 mm × 0.196797 mm for CT. The CT acquisition parameters were continuous 120 rotation steps over 220°, 80 kVp/500 μA tube voltage/current, and 175 ms exposure per step. Spherical VOIs (2 mm diameter) were drawn completely within brown adipose tissue, back of the cervical spine of each animal, and % injected dose per unit volume (%ID/ml) was calculated for analysis.
18F-Fluciclovine uptake:
After at least 6 hours of fasting, 18F-fluciclovine (approximately 3.663–4.181 MBq) in aqueous solution (~0.1 mL) was administered via tail vein, followed by saline flush. In order to capture 1-hour post-injection as the scan time, 10 min static positron emission tomography (PET) scan started at 55 minutes after the radiotracer injection. X-ray computed tomography followed PET scan with scan parameters: continuous 120 rotation steps over 220°, 80 kVp/500 μA tube voltage/current, and 175 ms exposure per step. Animals were kept anesthetized using isoflurane (2–2.5%) during the injection and the scan time. PET and CT reconstructions were performed using the manufacturer’s provided reconstruction algorithms. Reconstructed PET image pixel values were calibrated to be in the unit of activity concentration (Bq/ml) using a pre-calculated calibration ratio. All images were analyzed using an open-source image processing software, AMIDE (amide.sourceforge.org). Percent of injected dose per unit volume (%ID/ml) was calculated in AMIDE by entering the injected activity corrected for decay at the scan start time for each animal image. 3D volume rendering and cross-sectional (or multiplanar reconstruction, MPR) views were shown.
Temperature measurements:
~8–12-week-old male and female mouse was singly housed with free-access to food (unless specified in the text) and water during cold tolerance test (CTT). The core body temperatures prior to and during 4°C cold exposure (at one-hour interval) were measured using BAT-12 Microprobe Thermometer with probe RET-3 (Physitemp). 4mg kg−1 rapamycin (TCI America, #TCR0097), 0.625 mg kg−1 bortezomib (Selleck, #S1013), 50mg kg−1 BCH (Sigma, #A7902), 100mg kg−1 αMT (Sigma, #M8377) or DMSO (Sigma, #D8418) were injected intraperitoneally 1 hour prior to CTT. For infrared (IR) thermal imaging, mice were placed in a small box and pictures were taken by an E60 FLIR infrared camera (FLIR i3, FLIR Systems, Tokyo, Japan) positioned at 10 cm away from the mouse. Mice were then moved to cages at RT for 15 minutes (one mouse per cage), and the surface temperatures were recorded again. 10mg kg−1 propranolol (Sigma, #P0884) was injected 15 minutes prior to the transfer to RT. For IR imaging at basal condition (without RT stimulation), 4 mg kg−1 rapamycin (TCI America, #TCR0097), 0.625 mg kg−1 bortezomib (Selleck, #S1013), 50 mg kg−1 BCH (Sigma, #A7902), 100mg kg−1 αMT (Sigma, #M8377), Orlistat 25 mg kg−1 (Selleck, #S1729), Sotagliflozin 10 mg kg−1 (Selleck, #S8103) Propranolol 10 mg kg−1 (Sigma, #P0884) or DMSO (Sigma, #D8418) were injected intraperitoneally 2 hours prior to imaging. Mice were not shaved to maintain consistent fur status and avoid the alterations of the natural thermoregulatory state. FLIR quick report software was used to analyze thermal images. Briefly, two regions of interest (ROI) with same dimensions, one overlaying interscapular region (BAT) and another one at dorsal back (2 cm away from BAT in the image). The difference of surface temperature between the neck and the dorsal back (the “Temp (Neck-Back)”) was calculated to quantitate BAT thermogenic activity.
Metabolic measurements:
Body weight was monitored once a week. DEXA (or EchoMRI) was performed following manufacturer’s instructions. For insulin tolerance test (ITT), mice were fasted 4–6 hours before intraperitoneal administration of insulin (Humulin; 0.75U kg−1). Blood glucose was measured from tail vein at indicated time points with a glucometer (Contour, Bayer). Serum and liver TG contents were measured by Infinity Triglycerides Reagents (Thermo Scientific, #TR22421). Serum insulin, leptin and Fgf21 levels were measured by commercial ELISA kits (Alpco, #80-INSMSV-E01; Crystal Chem Inc, #90030; R&D, #MF2100).
Beige adipocyte ablation:
About 3-week-old male and female mice were administrated with diphtheria toxin (DT) at the dose of 10ng per mouse (Biological Laboratories Inc. #150) or PBS by intraperitoneal injection.
Histology:
Tissues were fixed in 10% formalin and processed and stained at AML Laboratories. Cell size was measured using Adiposoft software (Galarraga et al., 2012). Adipocyte size distribution was calculated using total adipocyte numbers counted in multiple images.
ETC Complex Activities:
Frozen BAT tissue from about 8-week-old male and female mice was homogenized in 250 μL homogenization buffer (120 mM KCl, 20mM HEPES, 1mM EGTA, pH 7.4) by sonication (5 second pulse × 5, 60% power) using a Microson XL2000 Ultrasonic Cell Disruptor (Misonix). Protein was quantitated using the Bradford assay and all samples were diluted to a final concentration of 1μg/μl of protein. The spectrophotometric kinetic assays were performed using a monochromator microplate reader (Tecan M200 Pro). Complex I activity (NADH:ubiquinone oxidoreductase) was determined by measuring oxidation of NADH at 340 nm (using ferricyanide as the electron acceptor) in a reaction mixture of 50 mM potassium phosphate (pH 7.5), 0.2 mM NADH, and 1.7 mM potassium ferricyanide. Complex II activity (Succinate Dehydrogenase) was determined by measuring the reduction of the artificial electron acceptor 2,6-dichlorophenol-indophenol (DCIP) at 600 nm in a reaction mixture of 50 mM potassium phosphate (pH 7.5), 20 mM succinate, 2 μM DCIP, 10 μM rotenone, and 1 mM potassium cyanide. Complex III activity (Ubiquinol:cytochrome c oxidoreductase) was determined by measuring the reduction of cytochrome c at 550 nm in a reaction mixture of 50 mM potassium phosphate (pH 7.5), 35 μM reduced decylubiquinone, 15 μM cytochrome c, 10 μM rotenone, and 1 mM potassium cyanide. Complex IV activity (Cytochrome c oxidase) was determined by measuring the oxidation of cytochrome c at 550 nm in a reaction mixture of 50 mM potassium phosphate (pH 7.0) and 100 μM reduced cytochrome c. Citrate synthase activity was determined by measuring the reduction of 5,5’-dithiobis (2-nitrobenzoic acid) (DTNB) at 412 nm which was coupled to the reduction of acetyl-CoA by citrate synthase in the presence of oxaloacetate. The reaction mixture consisted of 100 mM Tris-HCl (pH 8.0), 100 μM DTNB, 50 μM acetyl-CoA, and 425 μM oxaloacetate. All activities were calculated as nmoles/min/mg protein, normalized to citrate synthase (CS) activity and finally expressed as the percentage of wild-type activity.
Mitochondria Isolation:
Freshly dissected BAT tissue from ~8–12-week-old male and female mice was homogenized in a Dounce homogenizer with 5ml ice-cold mitochondria isolation buffer (210mM Mannitol, 70mM Sucrose, 1mM EGTA, 5mM HEPES pH7.5, 0.5% BSA). The homogenates were filtered through cheesecloth to remove residual particulates and intact mitochondria were isolated by differential centrifugation using a previously described protocol (Cannon and Lindberg, 1979). The mitochondrial pellet was resuspended in 25μL of isolation buffer and protein was quantitated using the Bradford assay (BioRad, #500–0006).
mtDNA Quantification:
The relative mtDNA content was measured using qPCR. The β2 microglobulin gene (B2m) was used as the nuclear gene (nDNA) normalizer for calculation of the mtDNA/nDNA ratio. A 322bp region of the mouse mtDNA was amplified using forward primer mtDNAF (CGACCTCGATGTTGGATCA) and the reverse primer mtDNAR (AGAGGATTTGAACCTCTGG). A fragment of the B2m gene was amplified using forward primer, B2mF (TCTCTGCTCCCCACCTCTAAGT), and reverse primer, B2mR (TGCTGTCTCGATGTTTGATGTATCT), giving an amplicon of 106 bp. The relative mtDNA content was calculated using the formula: mtDNA content = 1/2ΔCt, where ΔCt = CtmtDNA − CtB2m.
Mitochondrial Respiration Assay:
The XFe24 extracellular flux analyzer from Seahorse Biosciences was used to measure the rate of mitochondrial oxygen consumption, as previously described (Sadat et al., 2016). Mitochondria were isolated from 6–12-week-old male and female mice and immediately plated on the XFe24 cell culture microplate at a density of 2μg per well. The plate was centrifuged at 2000g for 20 mins at 4°C to enable the mitochondria to adhere to the plate. The XFe24 cartridge was equilibrated with the calibration solution overnight at 37°C. Mitochondrial Assay Solution (1X MAS) (70 mM sucrose, 220mM mannitol, 10 mM KH2PO4, 5 mM MgCl2, 2 mM HEPES, 1.0mM EGTA and 0.2% (w/v) fatty acid-free BSA) was freshly prepared and pH adjusted to 7.2. Unless otherwise stated, 10mM Pyruvate and 5mM Malate were used as substrates in the 1X MAS. 1X MAS was used to prepare cellular stress reagents, 5mM ADP, 2.5μM Oligomycin (Oligo), 4μM FCCP, 4μM Antimycin and 4μM Rotenone (A&R). All the reagents were loaded in the ports as suggested by Seahorse biosciences. Oxygen consumption rates (OCR) were measured for 2min with 1min of mixing. State 3 respiration was calculated as OCR after ADP minus OCR after A&R. ATP turnover was calculated as the OCR difference before and after Oligo. Maximum respiration was calculated as OCR after FCCP minus OCR after A&R. OCR was expressed as pMoles of oxygen/min/μg mitochondrial protein.
Mass spectrometry of BAT mitochondria:
Purified BAT mitochondria from 10–12-week-old male mice housed at RT or 30°C (n=3 for each genotype/condition) were resuspended in 8 M urea, 50 mM Tris, 5 mM CaCl2, 100 mM NaCl, and protease inhibitors. Mitochondria were lysed by probe sonication on ice, and proteins reduced by the addition of 5 mM DTT for 30 min at 37°C, followed cysteine alkylation by the addition of 15 mM iodoacetamide at RT for 45 min in the dark. The reaction was then quenched by the addition of 15 mM DTT for 15 minutes at RT. Proteins were first digested by the addition of endoproteinase LysC (Wako LC) at a 1:50 substrate:enzyme and incubated for 2h at RT. Next, samples were further digested by the addition of trypsin (Promega) at 1:100 substrate:enzyme, and incubated overnight at 37°C. Protein digests were then acidified by the addition of 0.5% triflororacetic acid, and samples desalted on C18 stage tips (Rainin). Peptides were resuspended in 4% formic acid and 3% acetonitrile, and approximately 1μg of digested mitochondria proteins was loaded onto a 75μm ID column packed with 25cm of Reprosil C18 1.9μm, 120Å particles. Peptides were eluted into an Orbitrap Fusion Tribrid (Thermo Fisher) mass spectrometer by gradient elution delivered by an Easy1200 nLC system (Thermo Fisher). The gradient was from 4.5% to 31% acetonitrile over 120 minutes. MS1 spectra were collected with oribitrap detection, while the 15 most abundant ions were fragmented by HCD and detected in the ion trap. All data were searched against the Mus musculus uniprot database (downloaded July 22, 2016). Peptide and protein identification searches were performed using the MaxQuant data analysis algorithm, and all peptide and protein identifications were filtered to a 1% false-discovery rate (Cox et al., 2014; Cox and Mann, 2008). Label free quantification analysis was performed using the MSstats R-package (Choi et al., 2014). Proteome changes of each ETC complex were calculated by averaging log2 values of fold change of all identified proteins within individual ETC complex (Masand et al., 2018).
Lactate production ex vivo:
50 mg of BAT tissue was isolated and placed in a 24 well plate with DMEM+ 10%FBS for 24 hours. Extracellular L-lactate concentrations were quantified in 96-well plate format using a commercial kit (Megazyme, #K-LATE) following the manufacturers’ protocol.
Lipolysis:
For lipolysis in vivo, mice were fasted for 6 hours and serum glycerol levels were measured before and after 1mg kg−1 CL316 243 (CL, TOCRIS, #1499) injection. Lipolysis ex vivo was performed as previously described (Paulo et al., 2018b). Briefly, tissue samples (~20 mg) of interscapular brown fat pads from overnight fasted mice were dissected and incubated at 37°C without shaking in 500 μl of KRB buffer (12 mM HEPES, 121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, and 0.33 mM CaCl2) containing 2% fatty acid free BSA and 0.1% glucose with or without 10 μM Forskolin (Sigma, F6886) at 37°C for 3 hours. The KRB buffer were collected and used free glycerol analysis using Free Glycerol Reagent (Sigma, F6428), respectively. The levels of free glycerol were normalized to the mass of the tissue sample.
Pulsed SILAC in differentiated brown adipocytes:
BAT SVF was isolated from ROSA-LSL-ATF4 or ROSA-LSL-ATF4;Ucp1 KO mice and digested in digestion buffer for 45 min: Ham’s F-10 1X Media (#10-070-CV, Corning), Collagenase 1.5 mg/ml (#C6885, Sigma) and Dispase II 1 mg/ml (#14549000, Roche). Cells were grown in DMEM media with 10% FBS until confluent. Adipocyte differentiation was induced by treating confluent cells for 48 hours in differentiation medium: DMEM 10% FBS, 20 nM insulin (#I9278, Sigma), 1 nM T3 (#T2877, Sigma), 0.5 mM IBMX (3-Isobutyl-1-methylxanthine, #I5879, Sigma), 0.5 μM Dexamethasone (#D8893, Sigma), 0.125 mM Indomethacin (#I7378, Sigma), 1 μM Rosiglitazone (#R2408, Sigma). For adenovirus infection at day 4, 50 MOI of Ad-GFP (#1060, Vector biolabs) or Ad-Cre (#1700, Vector biolabs) was incubated in differentiation media supplemented with 0.5 μg·mL−1 PLL (Poly-L-Lysine, #P2636, Sigma) for 5–10 min at room temperature. The virus-containing medium was then used to replace the culture media already on cells, and the cells were returned to the incubator. After a transduction period of 48 h, the viral medium was discarded and replaced with fresh differentiation media. All assays were conducted 4 days post-transduction to allow for optimal expression. Pulsed SILAC experiments were performed using the Thermo Scientific SILAC Protein Quantitation Kit (#A33969, Thermo Scientific). Briefly, cells were grown, differentiated and transfected in DMEM media containing light-leucine (12C6 L-lysine-2HCl). 4 days after transfection media was switch to heavy-leucine (13C6 L-lysine-2HCl) DMEM for the indicated times. Cell culture plates were washed 3 times with cold PBS and kept at −80°C. A lysis buffer consisting of 8 M urea, 100 mM ammonium bicarbonate pH 8.0, 150 mM NaCl, and Roche mini complete protease and phosphatase inhibitors, was added directly to the cell culture plate. Cells were then transferred to Eppendorf tube, and further lysed by probe sonication. Subsequent sample preparation was performed as described above for proteomic analysis of BAT mitochondria, with exception that only LysC was used for proteolytic digestion. Peptides were resuspended in 1% formic acid, and approximately 1μg of digested proteins was loaded onto a 75μm ID column packed with 15cm of Waters BEH C18 1.7μm, 130Å particles. Peptides were eluted into an Oribtrap Exploris 480 (Thermo Fisher) mass spectrometer by gradient elution delivered by an Easy1200 nLC system (Thermo Fisher). The gradient was from 4% to 24% acetonitrile over 110 minutes. MS1 spectra were collected with oribitrap detection, while the 20 most abundant ions were fragmented by HCD and detected in the orbitrap. All data were searched against the Mus musculus uniprot database (downloaded July 2, 2020). Peptide and protein identification searches were performed using the MaxQuant data analysis algorithm, and all peptide and protein identifications were filtered to a 1% false-discovery rate (Cox et al., 2014; Cox and Mann, 2008).
Immunoblots:
Puromycin (ThermoFisher, #A1113803) was injected intraperitoneally at the dose of 0.04 μmol g−1 30 minutes prior to tissue collection. For lysates, tissues were lysed in ice-cold lysis-buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 6 mM EGTA, 20 mM NaF, 1% Triton X-100, 1μM MG132 and protease inhibitors) using a TissueLyser II (Qiagen). After centrifugation at 13000 rpm for 15 min, supernatants were reserved for protein determinations and SDS-PAGE analysis. Mitochondria were lyzed in the above lysis buffer before immunoblotting. Antibodies used were: Ucp1 (Sigma, #U6382), FLAG (Sigma, #F1804), phosphor-PKA substrates (Cell Signaling Technology, #9624), Lrpprc (Santa Cruz Biotechnology, #SC-66844), Atf4 (ABclonal, #A0201), p-eIF2α (Cell Signaling Technology, #3398), eIF2α (Cell Signaling Technology, #5324), p-S6 (Cell Signaling Technology, #5364), S6 (Cell Signaling Technology, #2217X), p-4Ebp1 (Cell Signaling Technology, # 2855), 4Ebp1(Cell Signaling Technology, #9452), Hsp90 (Proteintech, #13171-1-AP), Slc3a2 (Proteintech, #15193-1-AP), total OXPHOS protein (Abcam, #ab110413), mt-Co2 (Proteintech, #55070-1-AP), Cox4 (Cell Signaling Technology, #4850), Cox5b (Bethyl, #A-305-523A), Cox6b (Abgent, # AP12336b), Hsp60 (Bethyl, #A302-846A), Puromycin (Kerafast, #EQ0001), Ubiquitin (Santa Cruz Biotechnology, #SC-8017), Lc3b (Cell Signaling Technology, #2775), Hsl (Cell Signaling Technology, #4107), Atgl (Cell Signaling Technology, #2138), and Actin (Cell Signaling Technology, #4967). Quantification of immunoblots were performed using NIH ImageJ software.
Q-PCR and RNA-seq:
Total RNA was extracted from tissues homogenized in TRIsure (Bioline, #BIO-38033) reagent and ISOLATE II RNA Mini kit (Bioline, #BIO-52073). Isolated RNA was reverse transcribed using iScript cDNA Synthesis Kit (Biorad, #170-8891), and the resulting cDNA was used for quantitative PCR on a CFX384 real-time PCR detection system (Bio-Rad). Relative mRNA expression level was determined using the 1/2ΔCt method with 36B4 as the internal reference control. Primer sequences were listed in Supplementary Table 4. RNA-seq was performed by Novogene Inc. Briefly, first strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Double-stranded cDNA was purified using AMPure XP beads. Remaining overhangs of the purified double-stranded cDNA were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150~200bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). The libraries were sequenced in Illumina for 20 million reads with pair-end 150 bp (PE150). Downstream analysis was performed using a combination of programs including STAR, HTseq and Cufflink. Alignments were parsed using Tophat program and differentially expressed genes (DEGs) were determined through DESeq2/edgeR. KEGG enrichment was implemented by ClusterProfiler. Cis-regulatory sequence analysis was performed using iRegulon plugin in Cytoscape.
Quantification and statistical analysis:
Data were presented as average ± SEM. Statistical significance was determined by unpaired t-test in GraphPad Prism 8. *: p<0.05 and **: p<0.01. Sample sizes for animal experiments were selected based on numbers typically used in our previous publications. No randomization of animals or predetermination of sample sizes by statistical methods were performed. No samples were measured repeatedly.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Lrpprc | Santa Cruz Biotechnology |
Cat#SC-66844 |
| Rabbit anti-phospho-PKA substrates | Cell Signaling | Cat#9624 |
| Rabbit anti-p-eIF2α | Cell Signaling | Cat#3398 |
| Rabbit anti-eIF2α | Cell Signaling | Cat#5324 |
| Rabbit anti-p-S6 | Cell Signaling | Cat#5364 |
| Rabbit anti-S6 | Cell Signaling | Cat#2217 |
| Rabbit anti-p-4Ebp1 | Cell Signaling | Cat#2855 |
| Rabbit anti-4Ebp1 | Cell Signaling | Cat#9452 |
| Rabbit anti-Atf4 | ABclonal | Cat#A0201 |
| Mouse anti-total OXPHOS rodent | Abcam | Cat#ab110413 |
| Rabbit anti-Hsp60 | Bethyl | Cat#A302-846A |
| Rabbit anti-mt-Co2 | Proteintech | Cat#55070-1-AP |
| Rabbit anti-Cox4 | Cell Signaling | Cat#4850 |
| Rabbit anti-Cox5b | Bethyl | Cat#A-305-523A-T |
| Rabbit anti-Cox6b | Abgent | Cat#AP20624a |
| Rabbit anti-Slc3a2 | Proteintech | Cat#15193-1-AP |
| Rabbit anti-Puromycin | Kerafast | Cat#EQ0001 |
| Mouse anti-Ubiquitin | Santa Cruz Biotechnology |
Cat#SC-8017 |
| Rabbit anti-Ucp1 | Sigma | Cat#U6382 |
| Mouse anti-Flag | Sigma | Cat#F1804 |
| Rabbit anti-Hsp90 | Santa Cruz Biotechnology |
Cat#SC-7949 |
| Rabbit anti-Lc3b | Cell Signaling | Cat#2775 |
| Rabbit anti-Hsl | Cell Signaling | Cat#4107 |
| Rabbit anti-Atgl | Cell Signaling | Cat#2138 |
| Rabbit anti-Actin | Cell Signaling | Cat#4967 |
| Bacterial and Virus Strains | ||
| GFP adenovirus | Vector biolabs | Cat#1060 |
| Cre adenovirus | Vector biolabs | Cat#1700 |
| Biological Samples | ||
| N/A | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| B-NADH | Sigma | Cat#N7410 |
| DCIP | Sigma | Cat#D1878 |
| DTNB | Sigma | Cat#D8130 |
| Acetyl Coenzyme A | Sigma | Cat#A2056 |
| Cytochrome C | Sigma | Cat#C3131 |
| Decylubiquinone | Sigma | Cat#D7911 |
| Potassium Borohydride | Sigma | Cat#P4129 |
| Potassium Cyanide | Sigma | Cat#20781 |
| Potassium Ferricyanide | Sigma | Cat#244023 |
| Sodium Hydrosulfite | Sigma | Cat#157953 |
| MG132 | Millipore | Cat#474791 |
| diphtheria toxin | Biological Laboratories Inc |
Cat#150 |
| Puromycin | ThermoFisher | Cat#A1113803 |
| Rapamycin | TCI America | Cat#TCR0097 |
| Bortezomib | Selleck | Cat#S1013 |
| Insulin solution human | Sigma | Cat#I9278-5ML |
| 2-amino-2-norbornane-carboxylic acid (BCH) |
Sigma | Cat#A7902 |
| α-methyl-dl-tryptophan (αMT) | Sigma | Cat#M8377 |
| Orlistat | Selleck | Cat#S1729 |
| Sotagliflozin | Selleck | Cat#M8377 |
| propranolol | Sigma | Cat#P0884 |
| CL316243 disodium salt | Tocris bioscience | Cat#1499 |
| Bio-Rad Protein Assay Solution | BioRad | Cat#500-0006 |
| TRIsure | Bioline | Cat#BIO-38033 |
| Collagenase | Sigma | Cat#C6885 |
| Dispase II | Roche | Cat##14549000 |
| Forskolin | Sigma | Cat#F6886 |
| 3,3′,5-Triiodo-L-thyronine (T3) | Sigma | Cat#T2877 |
| 3-Isobutyl-1-methylxanthine (IBMX) | Sigma | Cat#I5879 |
| Dexamethasone | Sigma | Cat#D8893 |
| Indomethacin | Sigma | Cat#I7378 |
| Rosiglitazone | Sigma | Cat#R2408 |
| Poly-L-Lysine | Sigma | Cat#P2636 |
| Critical Commercial Assays | ||
| Infinity Triglycerides Reagent | Thermo Scientific | Cat#TR22421 |
| Free Glycerol Reagent | Sigma | Cat#F6428 |
| QIAamp DNA Mini Kit | Qiagen | Cat#51304 |
| ISOLATE II RNA Mini Kit | Bioline | Cat#BIO-52073 |
| iScript cDNA Synthesis Kit | BioRad | Cat#170-8891 |
| Mouse insulin ultrasensitive ELISA kit | Alpco | Cat#80-INSMSV-E01 |
| Mouse leptin ELISA kit | Crystal Chem Inc | Cat#90030 |
| Mouse/Rat FGF-21 Quantikine ELISA Kit | R&D | Cat#MF2100 |
| L-Lactic Acid (L-Lactate) Assay Kit | Megazyme | Cat#K-LATE |
| SILAC Protein Quantitation Kit | Thermo Scientific | Cat##A33969 |
| Deposited Data | ||
| BAT mitochondrial mass spectrometry data | ProteomeXchange Consortium |
PXD008798 |
| BAT RNA-seq data | NCBI GEO | GSE117985 |
| Experimental Models: Cell Lines | ||
| N/A | ||
| Experimental Models: Organisms/Strains | ||
| ROSA-LSL-FlaghATF4 | JAX | Cat#029394 |
| ROSA-LSL-iDTR | JAX | Cat#007900 |
| Ucp1 KO | JAX | Cat#003124 |
| Lrpprcf/f | (Ruzzenente et al., 2012) | |
| Ucp1-Cre | JAX | Cat#024670 |
| Atf4f/f | (Ebert et al., 2012) | N/A |
| TfamBKO | (Masand et al., 2018) | N/A |
| Lkb1BKO | (Masand et al., 2018) | N/A |
| GnasBKO | (Paulo et al., 2018b) | N/A |
| betaless | (Bachman et al., 2002) | N/A |
| Oligonucleotides | ||
| Full sequences in Supplementary Table 4 | Elim Biopharm | N/A |
| Recombinant DNA | ||
| N/A | ||
| Software and Algorithms | ||
| ImageJ | NIH | |
| MaxQuant data analysis | (Cox et al., 2014) | |
| Adiposoft | (Galarraga et al., 2012) | |
| AMIDE | amide.sourceforge.org | |
| Cytoscape | cytoscape.org | |
| NetworkAnalyst | www.networkanalyst.ca/ | |
| FLIR QuickReport | www.flir.com | |
| Other | ||
| Low-protein diet (LP) | Research Diets | Supplementary Table 3 |
| Isogenic control protein diet (CP) | Research Diets | Supplementary Table 3 |
ACKNOWLEDGEMENTS
We thank Christophe Paillart and Vassily Kutyavin for the assistance with indirect calorimetry experiments. We thank Dr. Karl Kochanowski for his technical assistance with extracellular L-lactate quantification assays. This work is supported by National Institutes of Health grants DK105175 (B.W.), U54NS100717 (N.J.K.), P50GM082250 (N.J.K.), UCSF Diabetes Research Center P30DK063720 (B.W.), UCSF Nutrition Obesity Research Center P30DK098722 (B.W.), S10RR023051 (S.Y.). E.P. is supported by a fellowship grant from Hillblom foundation.
Footnotes
DECLARATION OF INTERESTS
Authors declare no competing financial interests.
References:
- Aksnes AK, Brundin T, Hjeltnes N, and Wahren J (1995). Metabolic, thermal and circulatory effects of intravenous infusion of amino acids in tetraplegic patients. Clin Physiol 15, 377–396. [DOI] [PubMed] [Google Scholar]
- Anunciado-Koza R, Ukropec J, Koza RA, and Kozak LP (2008). Inactivation of UCP1 and the glycerol phosphate cycle synergistically increases energy expenditure to resist diet-induced obesity. The Journal of biological chemistry 283, 27688–27697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman ES, Dhillon H, Zhang CY, Cinti S, Bianco AC, Kobilka BK, and Lowell BB (2002). betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science 297, 843–845. [DOI] [PubMed] [Google Scholar]
- Ballinger A, and Peikin SR (2002). Orlistat: its current status as an anti-obesity drug. European journal of pharmacology 440, 109–117. [DOI] [PubMed] [Google Scholar]
- Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. (2016). Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, et al. (2011). Brown adipose tissue activity controls triglyceride clearance. Nature medicine 17, 200–205. [DOI] [PubMed] [Google Scholar]
- Bartelt A, and Heeren J (2014). Adipose tissue browning and metabolic health. Nature reviews. Endocrinology 10, 24–36. [DOI] [PubMed] [Google Scholar]
- Bertholet AM, Kazak L, Chouchani ET, Bogaczynska MG, Paranjpe I, Wainwright GL, Betourne A, Kajimura S, Spiegelman BM, and Kirichok Y (2017). Mitochondrial Patch Clamp of Beige Adipocytes Reveals UCP1-Positive and UCP1-Negative Cells Both Exhibiting Futile Creatine Cycling. Cell metabolism 25, 811–822.e814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz MJ, and Enerback S (2015). Human Brown Adipose Tissue: What We Have Learned So Far. Diabetes 64, 2352–2360. [DOI] [PubMed] [Google Scholar]
- Betz MJ, and Enerback S (2018). Targeting thermogenesis in brown fat and muscle to treat obesity and metabolic disease. Nature reviews. Endocrinology 14, 77–87. [DOI] [PubMed] [Google Scholar]
- Bonet ML, Mercader J, and Palou A (2017). A nutritional perspective on UCP1-dependent thermogenesis. Biochimie 134, 99–117. [DOI] [PubMed] [Google Scholar]
- BORSOOK H (1936). THE SPECIFIC DYNAMIC ACTION OF PROTEIN AND AMINO ACIDS IN ANIMALS. Biological Reviews 11, 147–180. [Google Scholar]
- Butler AA, and Kozak LP (2010). A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes 59, 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon B, and Lindberg O (1979). Mitochondria from brown adipose tissue: isolation and properties. Methods in enzymology 55, 65–78. [DOI] [PubMed] [Google Scholar]
- Cannon B, and Nedergaard J (2004). Brown adipose tissue: function and physiological significance. Physiological reviews 84, 277–359. [DOI] [PubMed] [Google Scholar]
- Cannon B, and Nedergaard J (2011). Nonshivering thermogenesis and its adequate measurement in metabolic studies. The Journal of experimental biology 214, 242–253. [DOI] [PubMed] [Google Scholar]
- Cha YJ, Kim ES, and Koo JS (2018). Amino Acid Transporters and Glutamine Metabolism in Breast Cancer. International journal of molecular sciences 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B, and Vitek O (2014). MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics (Oxford, England) 30, 2524–2526. [DOI] [PubMed] [Google Scholar]
- Corrigan JK, Ramachandran D, He Y, Palmer CJ, Jurczak MJ, Chen R, Li B, Friedline RH, Kim JK, Ramsey JJ, et al. (2020). A big-data approach to understanding metabolic rate and response to obesity in laboratory mice. eLife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, and Mann M (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & cellular proteomics : MCP 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Crane JD, Mottillo EP, Farncombe TH, Morrison KM, and Steinberg GR (2014). A standardized infrared imaging technique that specifically detects UCP1-mediated thermogenesis in vivo. Molecular metabolism 3, 490–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, et al. (2009). Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine 360, 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silveira WA, Fazelinia H, Rosenthal SB, Laiakis EC, Kim MS, Meydan C, Kidane Y, Rathi KS, Smith SM, Stear B, et al. (2020). Comprehensive Multi-omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 183, 1185–1201.e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauncey MJ, and Ingram DL (1979). Effect of dietary composition and cold exposure on non-shivering thermogenesis in young pigs and its alteration by the beta-blocker propranolol. The British journal of nutrition 41, 361–370. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ (2011). Serine metabolism: some tumors take the road less traveled. Cell metabolism 14, 285–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert SM, Dyle MC, Kunkel SD, Bullard SA, Bongers KS, Fox DK, Dierdorff JM, Foster ED, and Adams CM (2012). Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. The Journal of biological chemistry 287, 27290–27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, and Kozak LP (1997). Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94. [DOI] [PubMed] [Google Scholar]
- Fischer AW, Cannon B, and Nedergaard J (2020). Leptin: Is It Thermogenic? Endocrine reviews 41, 232–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsström S, Jackson CB, Carroll CJ, Kuronen M, Pirinen E, Pradhan S, Marmyleva A, Auranen M, Kleine IM, Khan NA, et al. (2019). Fibroblast Growth Factor 21 Drives Dynamics of Local and Systemic Stress Responses in Mitochondrial Myopathy with mtDNA Deletions. Cell metabolism 30, 1040–1054.e1047. [DOI] [PubMed] [Google Scholar]
- Galarraga M, Campion J, Munoz-Barrutia A, Boque N, Moreno H, Martinez JA, Milagro F, and Ortiz-de-Solorzano C (2012). Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. Journal of lipid research 53, 2791–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan K, and Chawla A (2017). Warming the mouse to model human diseases. Nature reviews. Endocrinology 13, 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan HP, Li Y, Jensen MV, Newgard CB, Steppan CM, and Lazar MA (2002). A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nature medicine 8, 1122–1128. [DOI] [PubMed] [Google Scholar]
- Harper JW, and Bennett EJ (2016). Proteome complexity and the forces that drive proteome imbalance. Nature 537, 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, Laeger T, Albarado DC, McDougal DH, Berthoud HR, Munzberg H, and Morrison CD (2017). Low protein-induced increases in FGF21 drive UCP1-dependent metabolic but not thermoregulatory endpoints. Scientific reports 7, 8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann WE, Liu X, Bearden CM, Harper ME, and Kozak LP (2001). Effects of genetic background on thermoregulation and fatty acid-induced uncoupling of mitochondria in UCP1-deficient mice. The Journal of biological chemistry 276, 12460–12465. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Kang Q, Yoneshiro T, Camporez JP, Maki H, Homma M, Shinoda K, Chen Y, Lu X, Maretich P, et al. (2017). UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nature medicine 23, 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastroch M, Oelkrug R, and Keipert S (2018). Insights into brown adipose tissue evolution and function from non-model organisms. The Journal of experimental biology 221. [DOI] [PubMed] [Google Scholar]
- Kang SG, Choi MJ, Jung SB, Chung HK, Chang JY, Kim JT, Kang YE, Lee JH, Hong HJ, Jun SM, et al. (2021). Differential roles of GDF15 and FGF21 in systemic metabolic adaptation to the mitochondrial integrated stress response. iScience 24, 102181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Jedrychowski MP, Erickson BK, Shinoda K, Cohen P, Vetrivelan R, Lu GZ, Laznik-Bogoslavski D, Hasenfuss SC, et al. (2015). A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Stavrovskaya IG, Lu GZ, Jedrychowski MP, Egan DF, Kumari M, Kong X, Erickson BK, Szpyt J, et al. (2017). UCP1 deficiency causes brown fat respiratory chain depletion and sensitizes mitochondria to calcium overload-induced dysfunction. Proceedings of the National Academy of Sciences of the United States of America 114, 7981–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keipert S, Kutschke M, Lamp D, Brachthäuser L, Neff F, Meyer CW, Oelkrug R, Kharitonenkov A, and Jastroch M (2015). Genetic disruption of uncoupling protein 1 in mice renders brown adipose tissue a significant source of FGF21 secretion. Molecular metabolism 4, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keipert S, Lutter D, Schroeder BO, Brandt D, Stahlman M, Schwarzmayr T, Graf E, Fuchs H, de Angelis MH, Tschop MH, et al. (2020). Endogenous FGF21-signaling controls paradoxical obesity resistance of UCP1-deficient mice. Nature communications 11, 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keipert S, Ost M, Johann K, Imber F, Jastroch M, van Schothorst EM, Keijer J, and Klaus S (2014). Skeletal muscle mitochondrial uncoupling drives endocrine cross-talk through the induction of FGF21 as a myokine. American journal of physiology. Endocrinology and metabolism 306, E469–482. [DOI] [PubMed] [Google Scholar]
- Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivelä R, Pessia A, Velagapudi V, and Suomalainen A (2017). mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell metabolism 26, 419–428.e415. [DOI] [PubMed] [Google Scholar]
- Kilberg MS, Shan J, and Su N (2009). ATF4-dependent transcription mediates signaling of amino acid limitation. Trends in endocrinology and metabolism: TEM 20, 436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Jeong YT, Oh H, Kim SH, Cho JM, Kim YN, Kim SS, Kim DH, Hur KY, Kim HK, et al. (2013). Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nature medicine 19, 83–92. [DOI] [PubMed] [Google Scholar]
- Koeppen BM, and Stanton BA (2018). Berne & Levy physiology. (Philadelphia (PA): Elsevier; ). [Google Scholar]
- Kozak LP (2010). Brown fat and the myth of diet-induced thermogenesis. Cell metabolism 11, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokowski D, Han J, Saikia M, Majumder M, Yuan CL, Guan BJ, Bevilacqua E, Bussolati O, Broer S, Arvan P, et al. (2013). A self-defeating anabolic program leads to beta-cell apoptosis in endoplasmic reticulum stress-induced diabetes via regulation of amino acid flux. The Journal of biological chemistry 288, 17202–17213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl I, Miranda M, Atanassov I, Kuznetsova I, Hinze Y, Mourier A, Filipovska A, and Larsson NG (2017). Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Albarado DC, Burke SJ, Trosclair L, Hedgepeth JW, Berthoud HR, Gettys TW, Collier JJ, Munzberg H, and Morrison CD (2016). Metabolic Responses to Dietary Protein Restriction Require an Increase in FGF21 that Is Delayed by the Absence of GCN2. Cell reports 16, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Henagan TM, Albarado DC, Redman LM, Bray GA, Noland RC, Munzberg H, Hutson SM, Gettys TW, Schwartz MW, et al. (2014). FGF21 is an endocrine signal of protein restriction. The Journal of clinical investigation 124, 3913–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gu H, Gan L, Xu Y, Feng F, Saeed M, and Sun C (2017). Reducing Smad3/ATF4 was essential for Sirt1 inhibiting ER stress-induced apoptosis in mice brown adipose tissue. Oncotarget 8, 9267–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Soriano FJ, and Alemany M (1987). Effect of cold-temperature exposure and acclimation on amino acid pool changes and enzyme activities of rat brown adipose tissue. Biochimica et biophysica acta 925, 265–271. [DOI] [PubMed] [Google Scholar]
- López-Soriano FJ, Fernández-López JA, Mampel T, Villarroya F, Iglesias R, and Alemany M (1988). Amino acid and glucose uptake by rat brown adipose tissue. Effect of cold-exposure and acclimation. The Biochemical journal 252, 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, and Spiegelman BM (2000). Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660. [DOI] [PubMed] [Google Scholar]
- Lowell BB, V SS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, and Flier JS (1993). Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 366, 740–742. [DOI] [PubMed] [Google Scholar]
- Lucke J, and Hall G (1983). Thermogenesis in stress-susceptible pigs: a review. J R Soc Med 76, 514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk G (1931). The specific dynamic action. Journal of Nutrition 3, 519–530. [Google Scholar]
- Maida A, Zota A, Sjoberg KA, Schumacher J, Sijmonsma TP, Pfenninger A, Christensen MM, Gantert T, Fuhrmeister J, Rothermel U, et al. (2016). A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. The Journal of clinical investigation 126, 3263–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney SK, Fuller A, Mitchell D, Gordon C, and Overton JM (2014). Translating animal model research: does it matter that our rodents are cold? Physiology (Bethesda, Md.) 29, 413–420. [DOI] [PubMed] [Google Scholar]
- Masand R, Paulo E, Wu D, Wang Y, Swaney DL, Jimenez-Morales D, Krogan NJ, and Wang B (2018). Proteome Imbalance of Mitochondrial Electron Transport Chain in Brown Adipocytes Leads to Metabolic Benefits. Cell metabolism 27, 616–629.e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Miyazaki-Anzai S, Keenan AL, Shiozaki Y, Okamura K, Chick WS, Williams K, Zhao X, Rahman SM, Tintut Y, et al. (2016). Activating transcription factor-4 promotes mineralization in vascular smooth muscle cells. JCI insight 1, e88646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei H, Suarez JA, and Longo VD (2014). Protein and amino acid restriction, aging and disease: from yeast to humans. Trends in endocrinology and metabolism: TEM 25, 558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, Delmonte T, Villeneuve A, Sladek R, Xu F, et al. (2003). Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proceedings of the National Academy of Sciences of the United States of America 100, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neinast MD, Jang C, Hui S, Murashige DS, Chu Q, Morscher RJ, Li X, Zhan L, White E, Anthony TG, et al. (2019). Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell metabolism 29, 417–429.e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen BM, Ding X, Morgan DA, Coate KC, Bookout AL, Rahmouni K, Kliewer SA, and Mangelsdorf DJ (2014). FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell metabolism 20, 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, and Hanson RW (2002). The key role of anaplerosis and cataplerosis for citric acid cycle function. The Journal of biological chemistry 277, 30409–30412. [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, and Gorman AM (2016). The integrated stress response. EMBO reports 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo E, Wu D, Hecker P, Zhang Y, and Wang B (2018a). Adipocyte HDAC4 activation leads to beige adipocyte expansion and reduced adiposity. The Journal of endocrinology 239, 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulo E, Wu D, Wang Y, Zhang Y, Wu Y, Swaney DL, Soucheray M, Jimenez-Morales D, Chawla A, Krogan NJ, et al. (2018b). Sympathetic inputs regulate adaptive thermogenesis in brown adipose tissue through cAMP-Salt inducible kinase axis. Scientific reports 8, 11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RO, Marti A, Olvera AC, Tadinada SM, Bjorkman SH, Weatherford ET, Morgan DA, Westphal M, Patel PH, Kirby AK, et al. (2021). OPA1 deletion in brown adipose tissue improves thermoregulation and systemic metabolism via FGF21. eLife 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DR, DaCosta CM, Smith M, Doree D, Harris A, Buhring L, Heydorn W, Nouraldeen A, Xiong W, Yalamanchili P, et al. (2014). Effect of LX4211 on glucose homeostasis and body composition in preclinical models. The Journal of pharmacology and experimental therapeutics 350, 232–242. [DOI] [PubMed] [Google Scholar]
- Qian H, and Beard DA (2006). Metabolic futile cycles and their functions: a systems analysis of energy and control. Systems biology 153, 192–200. [DOI] [PubMed] [Google Scholar]
- Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, and Auwerx J (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of cell biology 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell NJ, and Stock MJ (1979). A role for brown adipose tissue in diet-induced thermogenesis. Nature 281, 31–35. [DOI] [PubMed] [Google Scholar]
- Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Camara Y, Milenkovic D, Zickermann V, Wibom R, Hultenby K, et al. (2012). LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. The EMBO journal 31, 443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadat R, Barca E, Masand R, Donti TR, Naini A, De Vivo DC, DiMauro S, Hanchard NA, and Graham BH (2016). Functional cellular analyses reveal energy metabolism defect and mitochondrial DNA depletion in a case of mitochondrial aconitase deficiency. Mol Genet Metab 118, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Ord D, Ord T, and Kilberg MS (2009). Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptional induction via CCAAT enhancer-binding protein-activating transcription factor response elements. The Journal of biological chemistry 284, 21241–21248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She P, Reid TM, Bronson SK, Vary TC, Hajnal A, Lynch CJ, and Hutson SM (2007). Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell metabolism 6, 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Ma Y, Chanturiya T, Cao Q, Wang Y, Kadegowda AKG, Jackson R, Rumore D, Xue B, Shi H, et al. (2017). Lipolysis in Brown Adipocytes Is Not Essential for Cold-Induced Thermogenesis in Mice. Cell metabolism 26, 764–777.e765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GI, Ladenson PW, Wolfe MH, Ridgway EC, and Wolfe RR (1985). Substrate cycling between gluconeogenesis and glycolysis in euthyroid, hypothyroid, and hyperthyroid man. The Journal of clinical investigation 76, 757–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siira SJ, Spahr H, Shearwood AJ, Ruzzenente B, Larsson NG, Rackham O, and Filipovska A (2017). LRPPRC-mediated folding of the mitochondrial transcriptome. Nature communications 8, 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultoukis GA, and Partridge L (2016). Dietary Protein, Metabolism, and Aging. Annual review of biochemistry 85, 5–34. [DOI] [PubMed] [Google Scholar]
- Spahr H, Rozanska A, Li X, Atanassov I, Lightowlers RN, Chrzanowska-Lightowlers ZM, Rackham O, and Larsson NG (2016). SLIRP stabilizes LRPPRC via an RRM-PPR protein interface. Nucleic acids research 44, 6868–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschop MH, Speakman JR, Arch JR, Auwerx J, Bruning JC, Chan L, Eckel RH, Farese RV Jr., Galgani JE, Hambly C, et al. (2012). A guide to analysis of mouse energy metabolism. Nature methods 9, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YH, Cypess AM, and Kahn CR (2010). Cellular bioenergetics as a target for obesity therapy. Nature reviews. Drug discovery 9, 465–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, and Teule GJ (2009). Cold-activated brown adipose tissue in healthy men. The New England journal of medicine 360, 1500–1508. [DOI] [PubMed] [Google Scholar]
- Veliova M, Ferreira CM, Benador IY, Jones AE, Mahdaviani K, Brownstein AJ, Desousa BR, Acín-Pérez R, Petcherski A, Assali EA, et al. (2020). Blocking mitochondrial pyruvate import in brown adipocytes induces energy wasting via lipid cycling. EMBO reports 21, e49634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent EE, Sergushichev A, Griss T, Gingras MC, Samborska B, Ntimbane T, Coelho PP, Blagih J, Raissi TC, Choinière L, et al. (2015). Mitochondrial Phosphoenolpyruvate Carboxykinase Regulates Metabolic Adaptation and Enables Glucose-Independent Tumor Growth. Molecular cell 60, 195–207. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, et al. (2009). Functional brown adipose tissue in healthy adults. The New England journal of medicine 360, 1518–1525. [DOI] [PubMed] [Google Scholar]
- Wang C, Huang Z, Du Y, Cheng Y, Chen S, and Guo F (2010). ATF4 regulates lipid metabolism and thermogenesis. Cell research 20, 174–184. [DOI] [PubMed] [Google Scholar]
- Wang C, Xia T, Du Y, Meng Q, Li H, Liu B, Chen S, and Guo F (2013). Effects of ATF4 on PGC1α expression in brown adipose tissue and metabolic responses to cold stress. Metabolism 62, 282–289. [DOI] [PubMed] [Google Scholar]
- Wang Y, Paulo E, Wu D, Wu Y, Huang W, Chawla A, and Wang B (2017). Adipocyte Liver Kinase b1 Suppresses Beige Adipocyte Renaissance Through Class IIa Histone Deacetylase 4. Diabetes 66, 2952–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, and Dong C (2019). Gluconeogenesis in Cancer: Function and Regulation of PEPCK, FBPase, and G6Pase. Trends Cancer 5, 30–45. [DOI] [PubMed] [Google Scholar]
- Yoneshiro T, Wang Q, Tajima K, Matsushita M, Maki H, Igarashi K, Dai Z, White PJ, McGarrah RW, Ilkayeva OR, et al. (2019). BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature 572, 614–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry data files (raw and search results) have been deposited to http://proteomecentral.proteomexchange.org, the ProteomeXchange Consortium, via the PRIDE partner repository with PXD008798. The raw RNA-seq data have been deposited to NCBI GEO (accession # GSE117985). No code is generated in this study.


