Abstract
Neuroblastoma (NB) is the most common extracranial pediatric solid cancer originating from undifferentiated neural crest cells. NB cells express EZH2 and GLI1 genes that are known to maintain the undifferentiated phenotype of cancer stem cells (CSC) in NB. Recent studies suggest that tumor-derived extracellular vesicles (EVs) can regulate the transformation of surrounding cells into CSC by transferring tumor-specific molecules they contain. However, the horizontal transfer of EVs molecules in NB remains largely unknown. We report the analysis of NB-derived EVs in bioengineered models of NB that are based on a collagen 1/hyaluronic acid scaffold designed to mimic the native tumor niche. Using these models, we observed an enrichment of GLI1 and EZH2 mRNAs in NB-derived EVs. As a consequence of the uptake of NB-derived EVs, the host cells increased the expression levels of GLI1 and EZH2. These results suggest the alteration of the expression profile of stromal cells through an EV-based mechanism, and point the GLI1 and EZH2 mRNAs in the EV cargo as diagnostic biomarkers in NB.
Keywords: Neuroblastoma, Extracellular vesicles, EZH2, GLI1, 3D tumor models
Introduction
Neuroblastoma (NB) that originates from neural crest stem cells is the most common extracranial solid tumor of the childhood1,2. NB belongs to a family of poorly differentiated tumors known as small-blue-round-cell tumors (SBRCT)3, and is characterized by the presence of cancer stem cells (CSC)4–6. Experimental evidence demonstrates that CSC are involved in drug resistance, aggressive tumor behavior and relapse in neuroblastoma6–8. CSC display unique features as they show the ability to retain stem cell properties (self-renewing and expression of stemness genes) and differentiation into multiple cell lineages7,8 CSC populations exist in dynamic equilibrium between the bidirectional conversion of CSC and differentiated non-CSCs that constitute the bulk of the tumor7,8.
Conventional treatments are highly effective in eliminating differentiated non-CSCs cells, but fail to target CSCs. Thus, disrupting the dynamic equilibrium between CSC and non-CSC by induction of differentiation is a current strategy to convert resistant cells into more sensitive to therapeutic intervention7,8. For example, 13-cis-retinoic acid is a wide used differentiating agent reported to reduce stemness characteristics in NB 6. Another well-known strategy consists in depleting stemness-related factors involved in CSC maintenance8. Among them, the epigenetic regulator EZH2 is of special interest within the CSC context9. EZH2 is essential for stem cell renewal and maintenance, and highly expressed in NB-CSC 6,9–12. Many studies have specifically addressed the impact of depleting EZH2 in a number of cancers e.g. gliobastoma, medulloblastoma, breast, ovarian, prostate, leukemia, colorectal, pancreatic, among others9,13–19. All of these reports conclude that inhibiting EZH2, either by RNA interference or pharmacological compounds such as DZnep, results in a reduction of CSC properties and invasion.
The Hedgehog (HH) signaling cascade is a conserved developmental pathway, whose physiological roles in humans consist of organogenesis, stem cell maintenance, and tissue repairing 20,21. To date, numerous reports have shown that the nuclear mediator of the HH pathway, named GLI1, has a critical function in maintaining the CSC undifferentiated phenotype in NB 22–24, and other solid tumors (e.g lung adenocarcinoma, anaplastic thyroid cancer, glioma, myeloma, breast, colon, gastric, pancreatic and prostate cancers) 21,25–29. As reported for EZH2, suppression of GLI1, by RNA silencing or drug-mediated using for example GANT61 or cyclopamine, inhibits stemness features and triggers CSC differentiation.
Investigation of the dedifferentiation process of non-CSCs into CSC is also of paramount relevance for the development and improvement of anti-cancer therapies30. In the last decade, many studies have described dedifferentiation of non-CSCs from different cancer types by overexpression of stemness-related molecules 30–33. Together with exogenous overexpression of reprogramming factors (OCT3/4, SOX2 and KLF4)30, ectopic expression of EZH2 is most widely used to promote dedifferentiation of non-CSC into CSC. In vitro induction of EZH2 expression has been shown to lead the CSC phenotype and CSC maintenance in laryngeal squamous cell carcinoma, breast, colorectal and gastric cancers, among others34–37. Although activated Hh signaling and increased levels of GLI1 has been identified in CSC, the effect of artificial-induced overexpression of GLI1 is poorly investigated 29. Heiden et.al. demonstrated that GLI1 plays an important role in thyroid CSC maintenance by using thyroid cancer cells stably transfected with a human GLI1 cDNA encoding26. Similar results were reported for gastric CSC by Yu et.al. In addition, it was demonstrated that exogenous GLI1 expression decreased the sensitivity to cisplatin38.
Recent studies reveal that bidirectional interconversion of CSC and non-CSC is, in part, regulated by surrounding cells or stroma cells, such as carcinoma associated fibroblasts (CAFs) or mesenchymal stem cells (MSCs) 39,40. Pro-stemness stromal cells can secrete paracrine signals, mainly cytokines, to enhance cancer stemness. In addition, stromal cells can also alter their expression profile39,40. In the case of neuroblastoma, Pelizzo et. al. reported that MSC isolated from the NB stroma exhibited traits of NB cancer cells, such as high expression levels of CXCR4 41. In this regard, recent evidence that extracellular vesicles (EVs) mediate the interactions between CSC, non-CSC and stromal cells in the tumor microenvironment is providing important insights into physiological mechanisms of cell dedifferentiation and maintenance 42,43.
EVs are small membrane vesicles secreted by cells into the extracellular space. Some EVs are shed from the surface of the cell, but other small EVs such as exosomes (30–150 nm) come from an endocytic origin and are released through exocytosis 44. EVs contain cell-specific molecules such as proteins, mRNA, microRNA, DNA and metabolites, which can be taken up by cells to modify their phenotypes and behaviors44. EVs are found to be involved in cell-cell communication and cell and metabolic reprogramming of their target cells42,43,45,46.
Since the presence of CSC is critical for tumor maintenance, resistance and metastasis, several studies focused on the effort to identify molecular cargos in EVs related to CSC, as well as on the interconversion between CSC and non-CSC and the possible alteration of the expression profile of stromal cells. It has been reported that glioblastoma-derived EVs induce the expression of CSC markers (CD133 and Nestin) in stromal MSC47, gastric cancer EVs induce differentiation of MSC into CAFs48, and melanoma EVs can cause oncogenic reprogramming of MSC in vitro49. Interestingly, Gyukity-Sebestyén et al. showed that internalization of melanoma EVs by MSC resulted in higher expression of melanoma-specific markers MITF and MLANA, and the induction of malignant transformation of the recipient MSC49.
Currently, monolayer cultures represent the major source for isolation of EVs. However, they are not a desirable source of EVs for identifying new cargo molecules. This is because culture dishes do not provide the microenvironment and signaling present in native tumors, which affect many cancer cell processes including EVs properties. We previously reported the first analysis of EVs in a 3D tumor model that was bioengineered using Ewing’s sarcoma tumor cells in porous scaffolds50. The 3-dimensionality, composition and stiffness of the cell environment were all involved in regulating the size of EVs released by the cells, and importantly, the content of the EVs50. These findings demonstrated the notion that EVs should be obtained from cells cultured within a native-like environment, rather than in simple cell monolayers. We therefore developed a tissue-engineered model of neuroblastoma, designed to mimic the native tumor microenvironment for studies of EVs.
Although there is a growing interest in the role of EVs as regulators of disease progression, little is known about EVs-mediated communication in the NB niche. In fact, no previous research has identified the molecular cargo in EVs related to NB-CSC markers. Here, we analyze for the first time the presence of two key molecules that maintain NB-CSC properties - EZH2 and GLI1, in NB-derived EVs. We also investigated the possibility of EVs-mediated horizontal transfer of RNA from NB cells to stromal MSC in the NB microenvironment.
Materials and Methods
Neuroblastoma samples from patients.
Fully de-identified samples of Neuroblastoma tumors were obtained from the Columbia University Tissue Bank, on an IRB-approved protocol. Frozen tissue samples from four different patients were cut into sets of contiguous sections for histological and immunohistochemical studies.
Scaffold preparation.
Porous scaffolds were produced from collagen I – hyaluronate (Col1-HA) solutions by freeze-drying. A 1% (wt/v) solution was prepared from low molecular weight (10–20 kDa) Sodium Hyaluronate (HA, Lifecore, US) in distilled water using a previously described method 50. Lyophilized scaffolds were cross-linked with a water-soluble carbodiimide. The resulting scaffolds were incubated in 95% ethanol solution containing 33 mM EDC (Sigma–Aldrich Co. Ltd., UK) and 6 mM NHS (Sigma–Aldrich Co. Ltd., UK) for 4 h at 25°C. After crosslinking, the scaffolds were washed thoroughly in distilled water (5 min × 5 times), frozen and lyophilized.
Cell culture
Neuroblastoma cell lines
SK-N-BE(2) (from the American Type Culture Collection, ATCC) was cultured according to the manufacturer’s specifications, in ATCC-formulated Eagle’s Minimum Essential Medium (EMEM) supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin. LAI-5s and SK-N-LP cell lines were kindly shared by Dr. Mora at Sant Joan de Deu Hospital, and cultured in RPMI medium supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin.
Cells were cultured at 37°C and 5% CO2 in a humidified incubator. Each scaffold was seeded with 2 ×106 neuroblastomacells, as previously reported50. Cell-seeded scaffolds were transferred to non-treated 12-multiwell plates (Nunc) and cultured in 2 mL of medium at 37°C / 5% CO2. Cell numbers were determined by Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) according to the manufacturer’s instructions.
Human Mesenchymal Stem Cells (hMSC, obtained from Lonza) were cultured in DMEM supplemented with 10% (v/v) Hyclone FBS, 1% penicillin/streptomycin and 1 ng/mL of basic fibroblast growth factor, bFGF).
Scanning Electron Microscopy (SEM).
For SEM analysis of bioengineered tumors, samples were washed twice in PBS and fixed in 4% paraformaldehyde in PBS (Santa Cruz, US) for 1 hour. Fixed specimens underwent a graded dehydration in a series of ethanol solutions (70, 85, 95, 100%, 5 min each) and hexamethyldisilazane (HMDS, Sigma) (15 min). Samples were dried overnight in a fume hood, sputter-coated with gold and palladium, and imaged using SEM (Hitachi S-4700).
Immunohistochemistry (IHC).
Frozen sections of the native NB tumors were fixed in pre-cooled acetone (−20°C) for 10 min, washed with PBS and treated with 0.3% H2O2 solution in PBS at room temperature for 10 min to block endogenous peroxidase activity. The samples were blocked for 1 h in 5% normal horse serum (Vectastain Elite ABC HRP Kit R.T.U.; Vector laboratories, PK-7200). The samples were then incubated with the following primary antibodies diluted in antibody diluent (Dako, S3022), in a humid chamber overnight at 4°C: Collagen 1 (dilution 1:500; Abcam, ab34710), EZH2 (dilution 1:50; Millipore # 07–689) and GLI1 (GLI1-rabbit dilution 1:50, Cell Signaling, #2553). The next day, the sections were washed (3 times, 5 min each) with PBST and incubated with secondary antibodies (Vectastain Elite ABC HRP Kit R.T.U. from Vector laboratories, PK-7200), following manufacturer instructions, and developed using Impact DAB (Vector Laboratories, SK4105). Negative controls were prepared by omitting the primary antibody step. Slides were counterstained with Hematoxylin QS (Vector Labs).
For the hyaluronan acid binding protein (HABP) staining, the sections were blocked using 1% BSA in HBSS at room temperature for 30 min, and incubated with a biotinylated HABP antibody (dilution 1:100; Millipore #385911). A Streptavidin Alexa Fluor 488 conjugate (dilution 1:500, Molecular Probes) was used as a secondary antibody.
Live-Dead assay.
Bioengineered tumors were incubated in EMEM medium containing 2μM Calcein and 4μM of ethidium homodimer-1 for 30 min at 37°C, 5% CO2, as indicated by the manufacturer’s protocol (LIVE/DEAD® Viability/Cytotoxicity Kit, Molecular Probes). Samples were imaged with a fluorescence microscope (Olympus IX81 light microscope, Center Valley PA).
Extracellular vesicles isolation.
Cells cultured in monolayers and scaffolds were washed with PBS twice and cultured in EMEM or RPMI supplemented with 10% (v/v) EVs-depleted FBS (SBI) and 1% penicillin/streptomycin for 12h. The supernatants were collected, and extracellular vesicles were isolated from culture media using the total EVs isolation kit (Invitrogen), according to the manufacturer’s protocol. EVs were resuspended in 50 ml PBS and particles size distribution and concentration were measured by nanoparticle tracking analysis (NanoSight NS500).
Transmission Electron Microscopy (TEM).
For negative staining, 3 μl of each sample was applied onto glow-discharged, carbon-coated, electron microscopy grids. After the sample was adsorbed and excess was blotted, grids were deposited successively on top of three different ~50-μl drops of MilliQ water and blotted. Then, grids were laid on top of two distinct ~50-μl drops of 1% uranyl acetate, stained for 1 min and air dried. Grids were visualized on a Tecnai 12 transmission electron microscope (Thermo Fisher Scientific) with a lanthanum hexaboride cathode operated at 120 keV. Images were collected at nominal magnification x21900 with a 4k×4k TemCam-F416 CMOS camera (TVIPS).
Quantitative Real-Time PCR (qRT-PCR).
Total RNA from cells was obtained using Trizol (Life Technologies) and the total RNA from extracellular vesicles was isolated using the Total EVs RNA & Protein Isolation Kit (Thermo Fisher Scientific), following the manufacturer’s instructions. RNA preparations were treated with “Ready-to-go you-prime first-strand beads” (GE Healthcare) to obtain cDNA. Quantitative real-time PCR was performed using DNA Master SYBR Green I mix (Applied Biosystems). mRNA expression levels in cells were quantified applying the ΔCt method, ΔCt = (Ct of the gene of interest - Ct of Actin). mRNA loading in EVs were quantified applying the ΔCt method but EZH2 and GLI1 levels were normalized using U6Sn levels as internal control. EZH2 and GLI1 primers were obtained from the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/).
Datasets for genomics analysis.
R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl.) was used to study EZH2 and GLI1 mRNA levels in Neuroblastoma tumors as previously described 50. The R2 platform is an online genomics analysis tool that allows the user to analyze publicly available microarray datasets. Briefly, we selected EZH2 or GLI1 as genes of interest to generate a MegaSampler using the following dataset that were profiled on Affymetrix U133 (u133p2) arrays and normalized using MAS5.0.
Cell line Neuroblastoma. Versteeg. Source: GEO ID: gse28019; R2 internal identifier: ps_avgpres_gse28019geo24_u133p2–24 standard human Neuroblastoma Cell lines were profiled without applying any transfections in order to measure the expression profiles.
Tumor Neuroblastoma public. Versteeg. Source: GEO ID: GSE16476; R2 internal identifier: ps_avgpres_nbadam88_u133p2–88 human Neuroblastoma samples were analyzed.
Normal Bone marrow Mesenchymal stem cells. Yamaguchi. Source: GEO ID: gse7637; R2 internal identifier: ps_avgpres_gse7637geo30_u133p2. 30 samples of human mesenchymal stem cells were cultured and subjected to RNA extraction and hybridization on microarrays.
RNA quality
RNA quality and size distribution of the cells and cell-secreted EVs were determined by electropherograms from the Bioanalyzer 2100 using the RNA Pico Chip kit (Agilent Technologies).
Western blot.
Cells were lysed in RIPA buffer containing protease inhibitors (Sigma-Aldrich, P8340) and extracellular vesicles extracts were obtained using the Total EVs RNA & Protein Isolation Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Cell preparations were centrifugated at 12,000 g for 10 min and supernatants containing soluble proteins were collected for analysis. 20μg of cells or extracts of the extracellular vesicles were loaded on 4–12% gradient Bis-Tris gels (BioRad), transferred to a nitrocellulose membrane and incubated with antibodies Calnexin (1:500; Santa Cruz, sc-11397, CD81 (1:500; Santa Cruz, sc-7637), EZH2 (dilution 1:100; Millipore # 07–689) and GAPDH (1:5000; Invitrogen 437000) at 4°C, overnight. For detection, membranes were incubated with a secondary antibody anti-rabbit or anti-mouse conjugated with Alexa Fluor 680 dye (1:5000; Thermo Fisher Scientific) at room temperature for one hour and imaged on Licor Odyssey scanner.
EVs-mediated transfer of RNA.
SK-N-BE(2), LAI-5s and SK-N-LP cells were cultured on Col1-HA scaffolds for 7 days in EMEM or RPMI supplemented with 10% (v/v) FBS and 1% penicillin/streptomycin. For EVs isolation, cells were cultured with 10% EVs-depleted FBS (SBI) and 1% penicillin/streptomycin for 12h. Supernatants were harvested, and extracellular vesicles were isolated. Protein concentration was measured by Bradford assay. 10μg samples of extracellular vesicle protein were labeled with SYTO RNA Select green fluorescent (Invitrogen) during 30 min at 37°C/5% CO2 at a final dye concentration of 10 μM. EVs Spin Columns (MW 3000) were used to remove unincorporated dye from EVs labeling. The same volume of PBS without extracellular vesicles was also treated with SYTO RNA and EVs spin columns to serve as a control. 5,000 hMSC per well were seeded into a 8-well chamber slide a day before the EVs-mediated transferring assay. 10 μg of labeled extracellular vesicles in PBS, or the same volume of PBS (control) were incubated with hMSC passage 3 during 2h at 37°C/5% CO2. Cells were fixed for 20 min in 4% PFA solution in PBS, and mounted with Vectashield-DAPI.
hMSC were incubated with 10 μg of EVs protein for 12h or 48h for analyses of mRNA expression by qRT-PCR
Results
Tissue-engineered model of neuroblastoma.
Neuroblastoma is a pediatric tumor characterized by the presence of small round blue cells (Figure 1A) and extracellular matrix enriched in collagen 1 (Col1) and hyaluronic acid (HA) proteins (Figure 1B). In order to build a tissue-engineered model of neuroblastoma (TE-NB), we first fabricated a 3-dimensional porous scaffold. To mimic neuroblastoma extracellular matrix composition, the biomaterial was made of natural preparations of Col1 and HA, and was stabilized by the freeze-drying technique and carbodiimide crosslinking chemistry with 1-Ethyl-3-(3-Dimethylaminopropyl) Carbodiimide (EDC) and N-Hydroxysuccinimide (NHS) (Figure 1C–E).
Figure 1. Biomaterial scaffold that mimics the extracellular matrix of neuroblastoma.
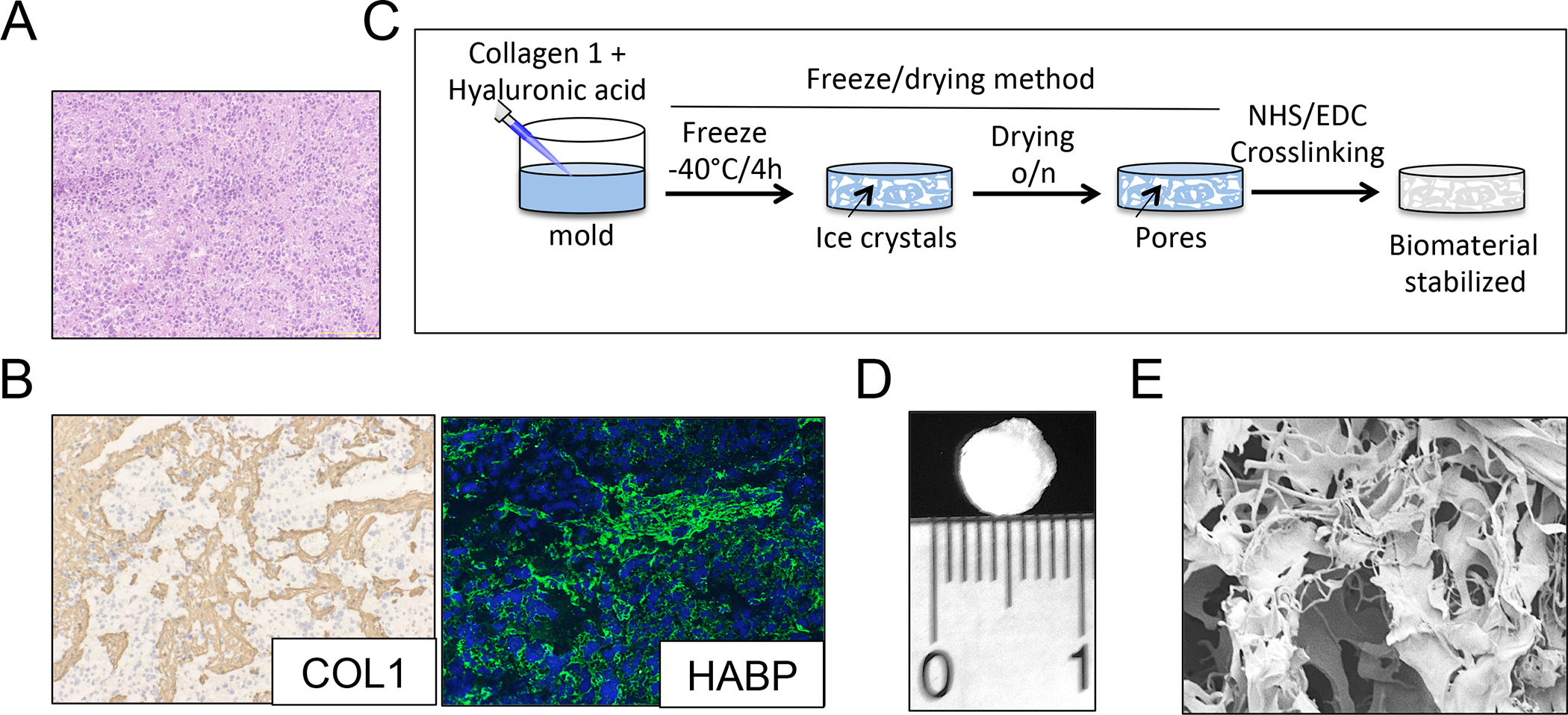
(A) Representative image of a neuroblastoma tumor showing the presence of small round blue cells by Hematoxylin and Eosin staining. (B) Characterization of the extracellular matrix composition for neuroblastoma tumors. (Left): Immunohistochemical staining for Collagen 1 (COL1); Counterstaining with hematoxylin QS (blue). (Right): Immunofluorescence image of hyaluronan acid binding protein (green); cell nuclei were stained by Hoechst 33342. Representative images are shown (n=3 per condition). (C) Preparation of Collagen1-Hyaluronic acid (Col1-HA) scaffolds. (D) Representative image of a Collagen1-Hyaluronic acid (Col1-HA) biomaterial fabricated by freeze-drying. (E) Scanning Electron Microscopy (SEM) image of a Col1-HA scaffold.
We previously demonstrated that the degradation rate of Col1-HA low molecular weight (LMW) scaffolds is much slower than that of Col1-HA high molecular weight (HMW) scaffolds due to a higher density of chemical cross-linking 50. We also showed that Col1-HA LMW scaffolds are more suitable for supporting attachment and proliferation of cancer cells for long-term studies50. Therefore, we used Col1-HA LMW scaffolds as the biomimetic environment for neuroblastoma cells.
Neuroblastoma cells in vitro are classified by three distinct cellular phenotypic variants. The first one is the neuroblastic phenotype (termed N) that grows as small aggregates of small cells. A second cell type is substrate-adherent (termed S). It displays large and flattened cells that show contact inhibition. Finally, the third one is termed I and exhibits an intermediate morphology to those of N and S51,52. We first cultured neuroblastoma N-type cells (SK-N-BE(2)) on Col1-HA LMW scaffolds to fabricate N-type TE-NB models. TE-NB resembled native tumor morphology with small round blue cells that formed typical aggregates that grow over time (Figure 2A, B), and integrate within the extracellular matrix (Figure 2B). As expected, cancer cells cultured in this 3D setting proliferated slower than in monolayers, mimicking the proliferation rate of tumor cells in patients 50,53 (Figure 2C). Live/Dead analysis demonstrated full viability of the TE-NB after 7 days of culture (Figure 2D).
Figure 2. Characterization of the Tissue-engineered model of N-type Neuroblastoma (TE-NB).
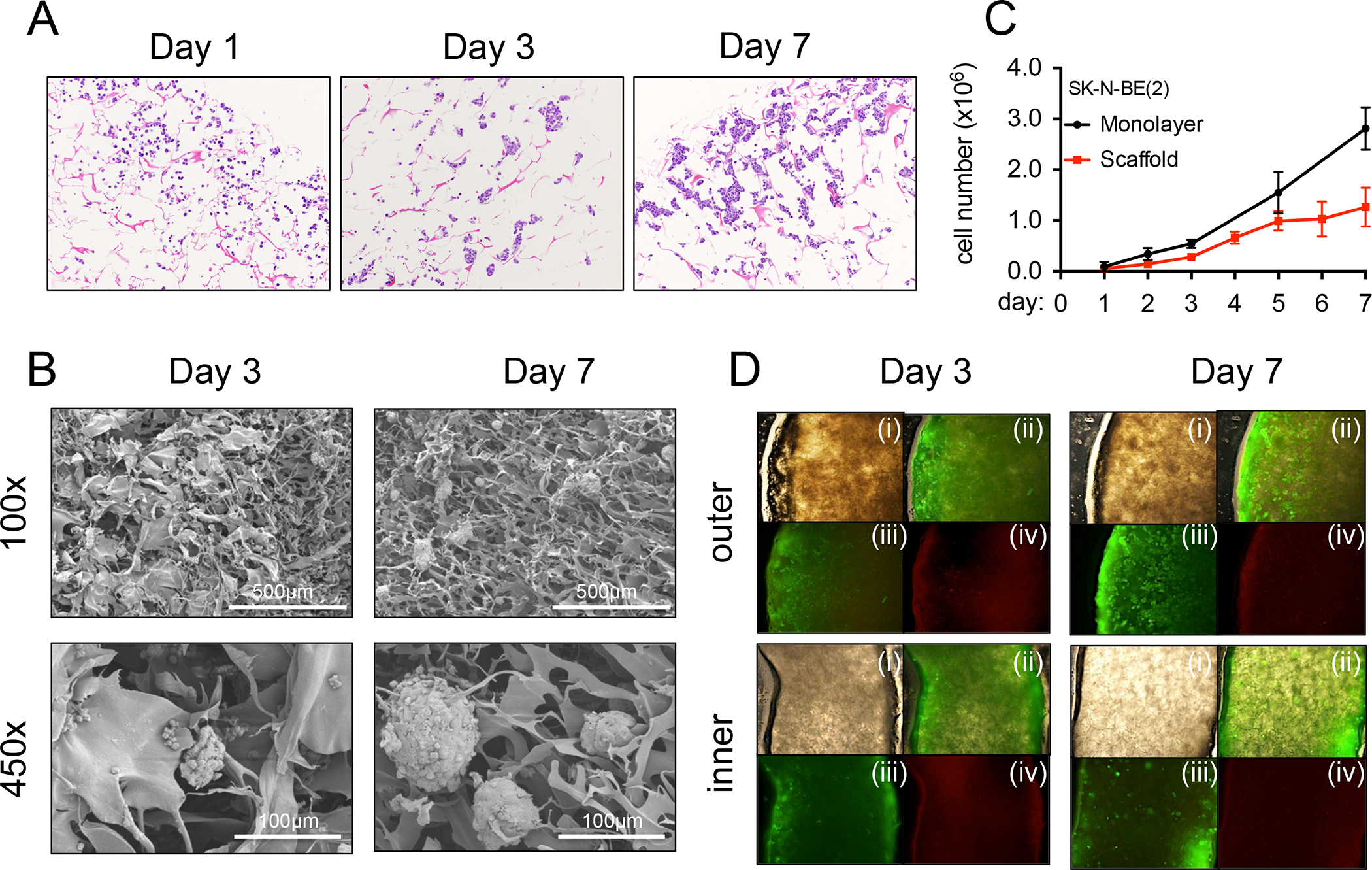
(A) Hematoxylin and Eosin staining images showing SK-N-BE(2) cell aggregates growing over-time, at day 1,3 and 7 after cell seeding. (B) Formation of cell aggregates and cell-matrix integration in TE-NB models at day 3 and day 7 by Scanning Electron Microscopy (SEM). (C) Cell proliferation for SK-N-BE(2) cells cultured in monolayers and in scaffolds. (D) Live/dead staining images of the inner and outer sides of TE-NB models at day 3 and 7 (n=4). (i) brightfield, (ii) merge, (iii) Calcein staining (green-live cells), (iv) ethidium homodimer-1 staining (red-dead cells).
Purity of extracellular vesicle preparations.
Once the 3D tumor model was established, we isolated EVs released from the N-type NB cells cultured in monolayers and in bioengineered tumors, and evaluated the purity of their preparations. We first analyzed the concentration and size distribution of EVs isolated from monolayers (ML) and TE-NB at day 7 by Nanoparticle Tracking Analysis (NTA) (Figure 3A,B). The concentration of particles was similar for both conditions. Samples were heterogeneous regarding size, but mostly between the average sizes described for small extracellular vesicles (sEVs) (Figure 3A). Modal size confirmed the majority presence of 100 nm particles (Figure 3B). Next, we assessed particle morphology by TEM (Figure 3C). TEM analyses confirmed our findings from NTA; we observed heterogeneous preparations of sEVs. We detected particles with the typical lipid bilayer-enclosed nanoparticles spherical structure (Figure 3C).
Figure 3. Evaluation of the purity of N-type extracellular vesicles preparations.
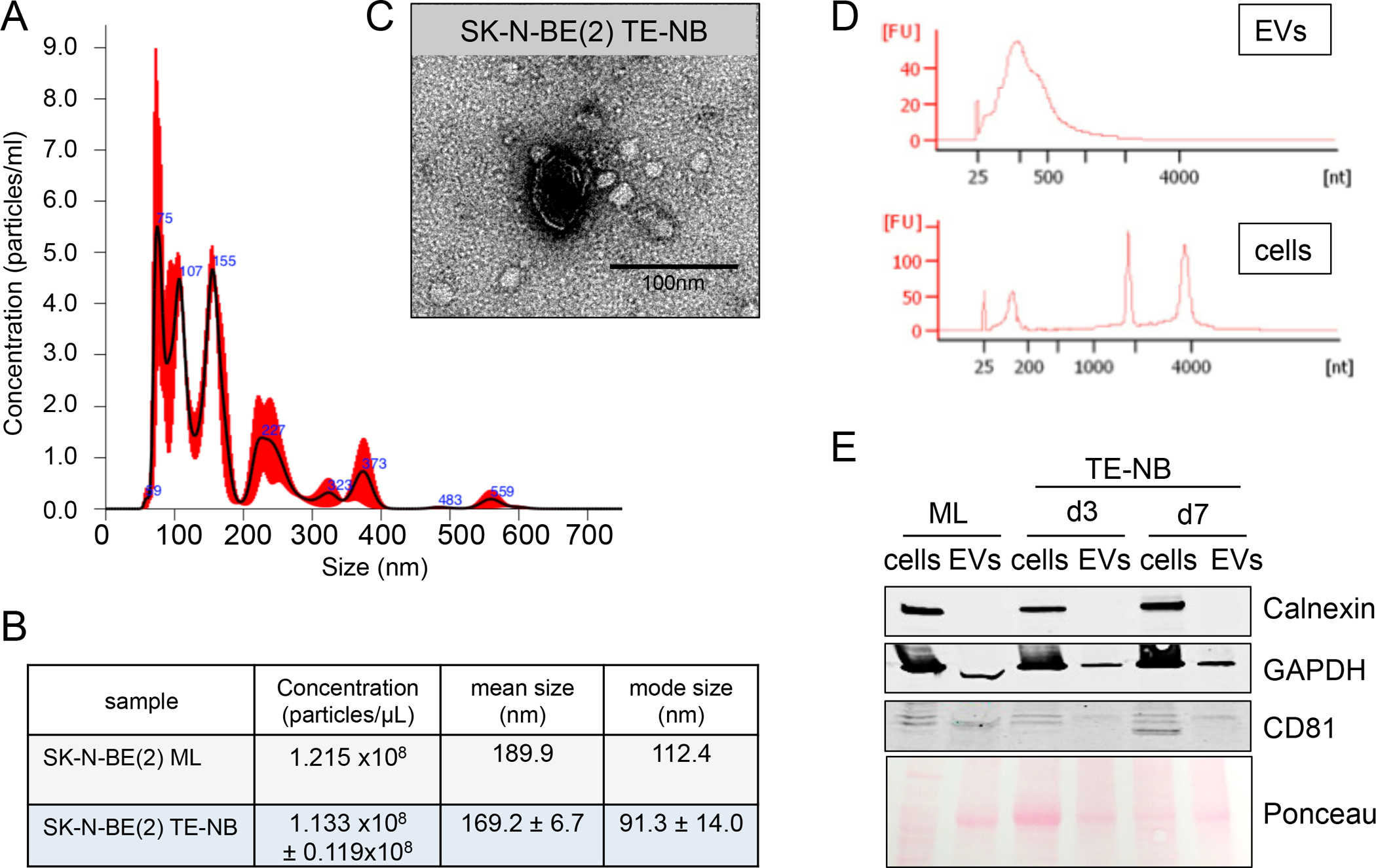
(A) Characterization of particle size distribution of EVs by Nanoparticle tracking analysis (NTA). Representative size distribution of EVs isolated from N-type TE-NB models at day 7. (B) EVs concentration, mean size and mode size of EVs released by N-type NB cells (SK-N-BE(2)) cultured in monolayer (ML) or in the TE-NB at day 7. Values were directly obtained from the NTA analyses. (C) Evaluation of EVs morphology by transmission electron microscopy (TEM). Representative image of N-type TE-NB-derived EVs showing an isolated spherical EV. (D) Electropherograms of total RNA isolated from cells and extracellular vesicles (EVs) from the TE-NB model at day 7. FU, fluorescent units; nt, nucleotides (RNA size). (E) Western blot of the indicated proteins in whole extracts of cells and extracellular vesicles (EV) from monolayer cultures (ML) at day 7 and TE-NB models at day 3 (d3) and day 7 (d7).
In addition, we performed the EVs isolation protocol using supernatants from scaffolds cultured in culture medium without cells at day 7, to discard EVs contamination from the biomaterial in our preparations. NTA analyses (Supp. Figure 1A,B) and TEM images (Supp. Figure 1C) demonstrated that the biomaterial did not release any detectable EV. Thus, we confirmed that EVs isolated from the TE-NB were produced by NB cells.
For determining purity of the EVs cargo we analyzed RNA profiles for the cells and EVs released from the TE-NB at day 7. The RNA profiles of the cells and EVs are completely different, and are used for determining the quality of the EV isolation. Specifically, the RNA profile from EVs lacked the peaks for ribosomal RNA subunits 18S and 28S, and showed enrichment in small RNAs (Fig. 3D). On the contrary, the RNA profile from cells showed a clear peak at ~1900 nt, that belongs to the 18S rRNA, and a second one at ~3800 nt for the 28S rRNA. These results confirmed that there was no cellular contamination of EVs preparations. To further evaluate the purity of the EV preparations, we checked the protein levels for the exosomal marker CD81 (not detectable in cells) and cellular marker calnexin (not detectable in EVs). Analyses were done for the protein extracts of NB cells and EVs obtained from cells cultured in monolayers and in the TE-NB, at day 3 and day 7 (Fig. 3E), to confirm the presence of CD81 and absence of calnexin in EV preparations, and the absence of CD81 and presence of calnexin in cell preparations. Both sets of analyses (RNA profiles and presence of specific proteins) thus confirmed that the isolated products are pure EVs.
Studies of EVs cargo
GLI1 and EZH2 are important mediators for maintaining the stemness phenotype of neuroblastoma. We hypothesized that NB-derived EVs could carry GLI1 and/or EZH2 mRNAs, and could thereby transfer them to surrounding stromal cells to modify their behavior, as reported for glioblastoma and melanoma47,49.
To test this hypothesis, we first confirmed the presence of neuroblastoma cells positive for EZH2 and GLI1 in native tumors from patients (Supp. Figure 2A). We then compared EZH2 and GLI1 gene expression profiles of normal human mesenchymal stem cells (hMSC) (by an array from Yamaguchi), neuroblastoma tumors and neuroblastoma cell lines cultured in monolayer (by arrays from Versteeg), using R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl). We confirmed that EZH2 and GLI1 genes were highly expressed in NB tumors and cell lines, and low expressed in hMSC. Interestingly, EZH2 and GLI1 levels were higher in cells cultured in monolayers than in tumors (Supp. Figure 2B).
For further validation of the N-type TE-NB, we compared EZH2 and GLI1 mRNA levels from NB cells cultured in monolayer (ML) and in the TE-NB model at day 3 and 7 (Figure 4A,B). Although we did not find significant differences in EZH2 mRNA expression levels between monolayers and TE-NB models (Figure 4A), EZH2 protein levels were lower in the TE-NB model at day 7 compared to day 3 and to NB cells cultured in monolayer (Supp Figure 3). These data suggest that 7 days are not enough for cells to regulate the transcriptional mechanisms to decrease EZH2 mRNA levels. But interestingly, translational or post-translational mechanisms seems to be modified after 7 days of culture in 3D to downregulate EZH2 protein levels in the cell. On the contrary, GLI1 mRNA expression levels were lower in the TE-NB than in monolayer, which better mimics the levels obtained from arrays for native tumors (Figure 4B).
Figure 4. Evaluation of EZH2 and GLI1 mRNAs in N-type NB cells and Extracellular vesicles.
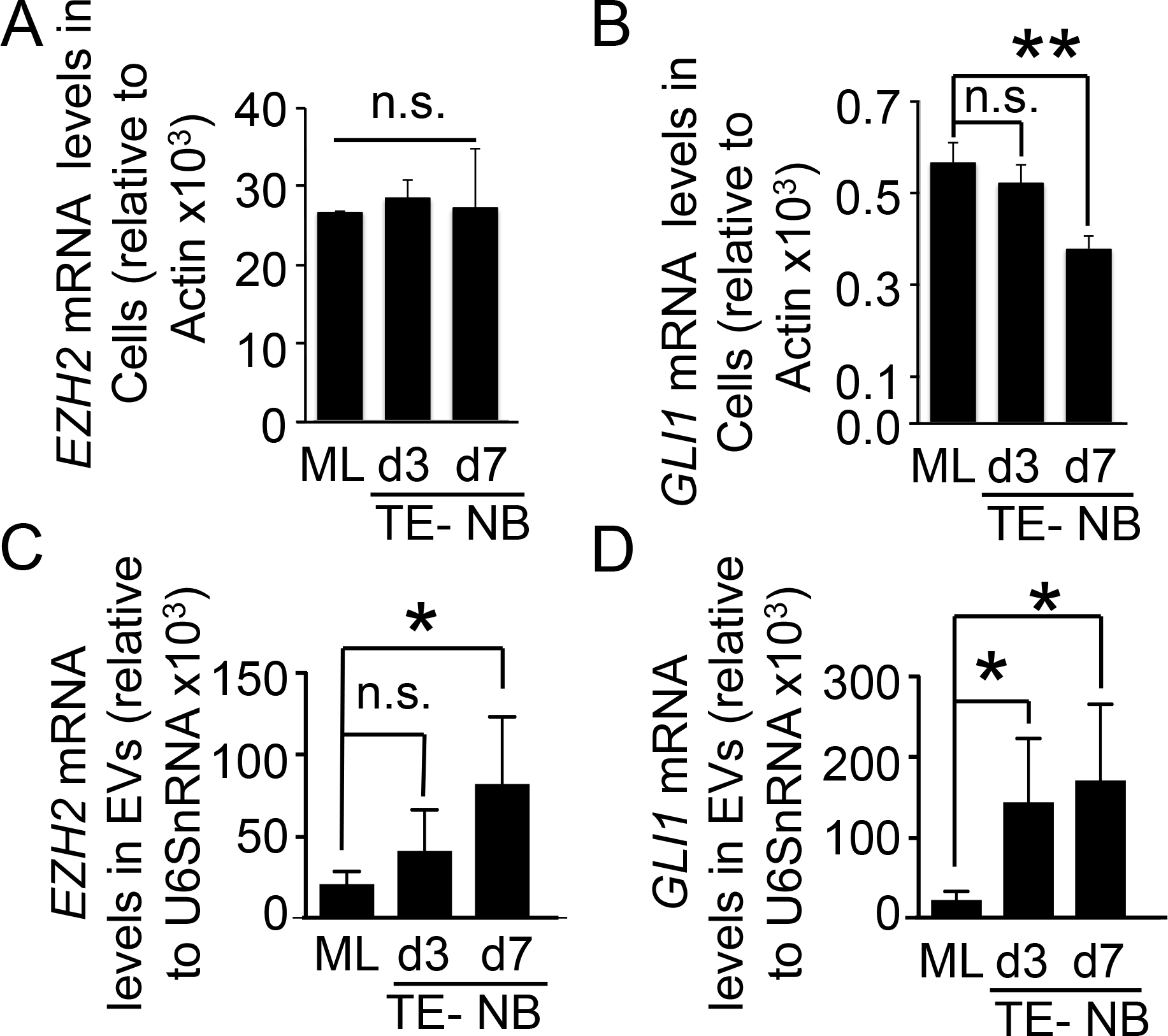
(A) qRT-PCR analysis of EZH2 in the N-type neuroblastoma cell line (SK-N-BE(2)) cultured in monolayer (ML) and in the tissue-engineered model (TE-NB) for 3 and 7 days. In all cases, relative endogenous expression of EZH2 was normalized to actin expression. Two-tailed Studentś t-test was used to determine statistical significance. ns, not significant. (B) qRT-PCR of GLI1 expression in the neuroblastoma cell line SK-N-BE(2) cultured during 3 and 7 days in Col1-HA scaffold (TE-NB) compared to monolayer (ML). Relative endogenous expression of GLI1 was normalized to actin and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test.**p < 0.01; ns, not significant. (C) EZH2 mRNA cargo levels in extracellular vesicles from SK-N-BE(2) cells cultured in monolayer (ML), and in Col1-HA scaffold for 3 and 7 days (TE-NB). EZH2 mRNA amount was normalized to U6SnRNA and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test. *p < 0.05; ns, not significant. (D) GLI1 mRNA detection in extracellular vesicles released from SK-N-BE(2) cultured in monolayer and scaffold (TE-NB) at days 3 and 7 by qRT-PCR. GLI1 mRNA amount was normalized to U6SnRNA and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test. *p < 0.05; ns, not significant.
We then isolated NB-derived EVs from N-type NB cells cultured in monolayers and TE-NB models, and detected high levels of both EZH2 and GLI1 mRNA in EVs from TE-NB models at day 7(Figure 4C, D). Interestingly, enrichment of EZH2 and GLI1 mRNA in EVs was higher in the TE-NB than in monolayer. EZH2 mRNA levels were significantly higher in EVs from the TE-NB than cells cultured in monolayer at day 7 (Figure 4C). Enrichment of GLI mRNA began to become significantly higher in TE-NB-derived EVs at day 3 (Figure 4D). Together these results suggest that different mechanisms may control the sorting of molecules into EVs between cells cultured in monolayer and cells in 3D, and that the cellular context may affect the specific sorting of mRNA and loading of EVs.
Next, to figure it out if the loading of EZH2 and GLI1 mRNAs is N-type specific or a general phenomenon in neuroblastoma, we built two different TE-NB using S-type and I-type cells.
LAI-5s was chosen as the S-type cell line to be assessed. LAI-5s is MYCN amplified and p53 mutated, as described for the N-type cell line SK-N-BE(2). S-type cells cultured into the Col1-HA scaffold did not show cell death at d3 and d7 of culturing (Supp.Fig 4A), and grew forming tissue layers within the biomaterial (Supp.Fig 4B,C) instead of aggregates as observed for SK-N-BE(2). Thus, we confirmed that the biomaterial is also suitable for culturing S-type cells and developing a living S-type TE-NB model. Next, we checked EZH2 and GLI1mRNA expression levels by the cells in 3D compared to monolayer (Supp.Figure 4D). LAI-5s cells in monolayer expressed significantly higher levels of both EZH2 and GLI1 genes than the TE-NB model, as observed in patients’ tumors. Again, this result supports the idea that the biomaterial induces the cells to recapitulate the expression profile observed in native tumors.
After validating the S-type TE-NB model, we isolated and characterized EVs released by LAI 5-S cells in monolayer and 3D by NTA analyses (Figure 5A,B). As observed for SK-N-BE(2) cells, we distinguished an heterogeneous population of sEVs at day 7 (Figure 5A). The modal size for LAI5s sEVs in monolayer and the TE-NB was around 100nm (Figure 5B). The morphology of sEVs was spherical, as expected (Figure 5C). Analyses of enrichment of EZH2 and GLI1mRNAs by qRT-PCR demonstrated that both molecules are loaded at higher levels into TE-NB-derived EVs than in monolayer (Figure 5D). This data confirmed again that the microenvironment plays a critical role in modifying the regulatory mechanisms of sorting of molecules in EVs. We concluded that the loading of EZH2 and GLI1 mRNAs also occurs in S-type cells when cultured within the Col1-HA biomaterial.
Figure 5. Characterization of S-type Neuroblastoma-derived EVs.
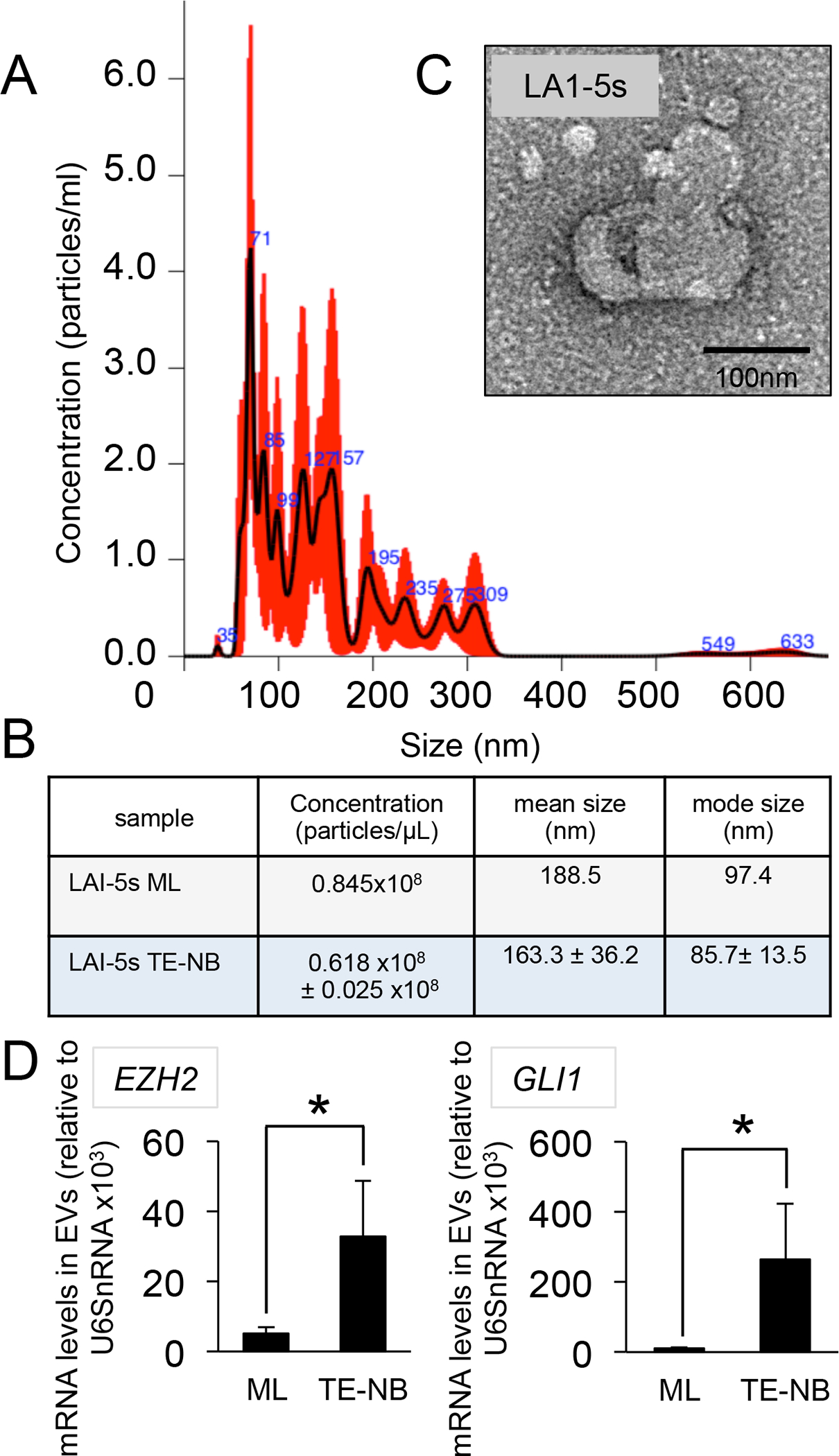
(A) Representative Particle size distribution of EVs isolated from S-type tissue-engineered neuroblastoma (TE-NB) models determined by NTA (n=3). (B) Analyses of concentration, mean size and mode size of EVs isolated from LAI-5s cells cultured in monolayer (ML) and scaffold (TE-NB) at day 7 (n=3). Values were obtained by NTA. (C) Morphology of EV visualized by TEM. (D) EZH2 and GLI1 mRNA enrichment in S-type TE-NB-derived EVs compared to EVs released by cells cultured in monolayer (ML). mRNA amount was normalized to U6SnRNA and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test. *p < 0.05
We developed a third TE-NB model using the SK-N-LP cell line for further investigating if I-type cells-derived EVs are also enriched in EZH2 and GLI1 mRNAs. SK-N-LP cells display I-type phenotype, p53 wild type, and MYCN amplified as the other two cell lines studied. The SK-N-LP TE-NB model is also a living tissue as determined by live/dead analysis (Supp. Figure 5A), and cells can form cell sheets within the material at day 7 (Supp. Figure 5B,C), as observed for the S-type TE-NB model (Supp.Fig 4B,C). Although EZH2 mRNA levels significantly increased at day 3 with respect to the monolayer, the TE-NB model expressed lower levels of both EZH2 and GLI1 mRNAs at day 7, compared to monolayer (Supp. Figure 5D). Thus, we confirmed one more time, that the TE-NB better mimics the situation in patients independently of the NB cell type studied.
As we performed for the N-type and S-type TE-NB models, we assessed the concentration, size and morphology of EVs released by I-type cells (Figure 6A, B, C). The morphology was the “cup-shaped” appearance commonly seen among sEVs, also observed for N- and S-type EVs. However, the modal size for both monolayer and TE-NB was higher than 100nm. Importantly, we confirmed that I-type EVs released from the TE-NB were enriched in EZH2 and GLI1 mRNAs (Figure 6D).
Figure 6. Characterization of I-type Neuroblastoma-derived EVs.
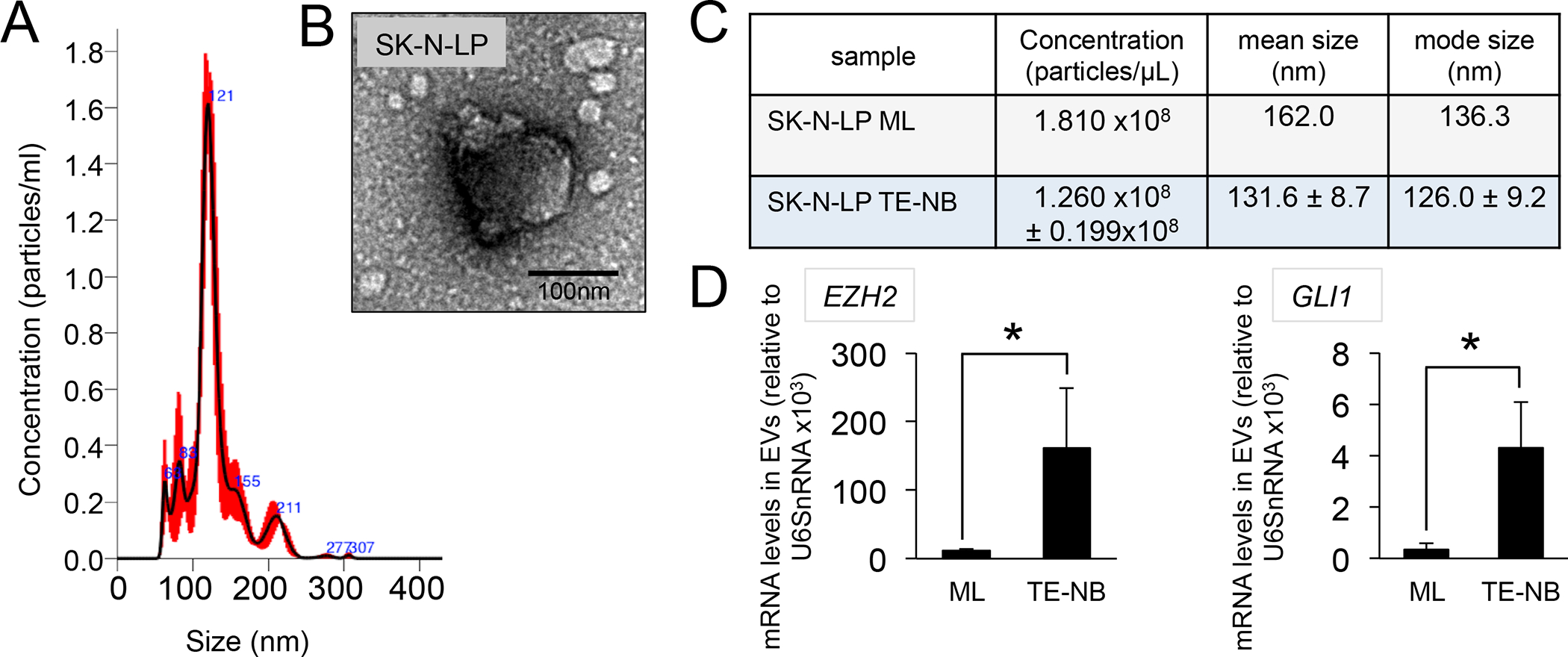
(A) Representative size distribution profiles of EVs released by SK-N-LP cells cultured in Col1-HA scaffolds (tissue engineered neuroblastoma model -TE-NB) for 7 days determined by NTA. (B) Analysis of morphology of EVs isolated from TE-NB models by TEM. (C) Concentration (particles/μL), mean size (nm) and modal size (nm) of EVs isolated from SK-N-LP cells cultured in monolayer cells (SK-N-LP ML) or tissue-engineered neuroblastoma models (SK-N-LP TE-NB). (C) EZH2 and GLI1 mRNA levels in SK-N-LP TE-NB-derived EVs compared to monolayer (ML). mRNA amount was normalized to U6SnRNA and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test. *p < 0.05
Studies of transferring of mRNA-loaded EVs.
After identifying GLI1 and EZH2 mRNAs in the cargo of N, S and I-type NB-derived EVs, we investigated if TE-NB-derived EVs could be transferred to stromal MSC, as described for gliobastoma and melanoma. To this end, we labeled N, S and I-type TE-NB-derived EVs isolated at day 7 with SYTO RNASelect (reagent that selectively stains RNA in green). hMSCs showed green RNA label already after 2 hours of incubation with SYTO RNASelect-TE-NB derived EVs, confirming the uptake of EVs by MSC (Figure 7A). MSC-derived EVs were used as experimental control. Labeled MSC-derived EVs were also uptaken by the MSC (Supp. Figure 6A).
Figure 7. EV-mediated transfer of EZH2 and GLI1 mRNAs to human mesenchymal stem cells.
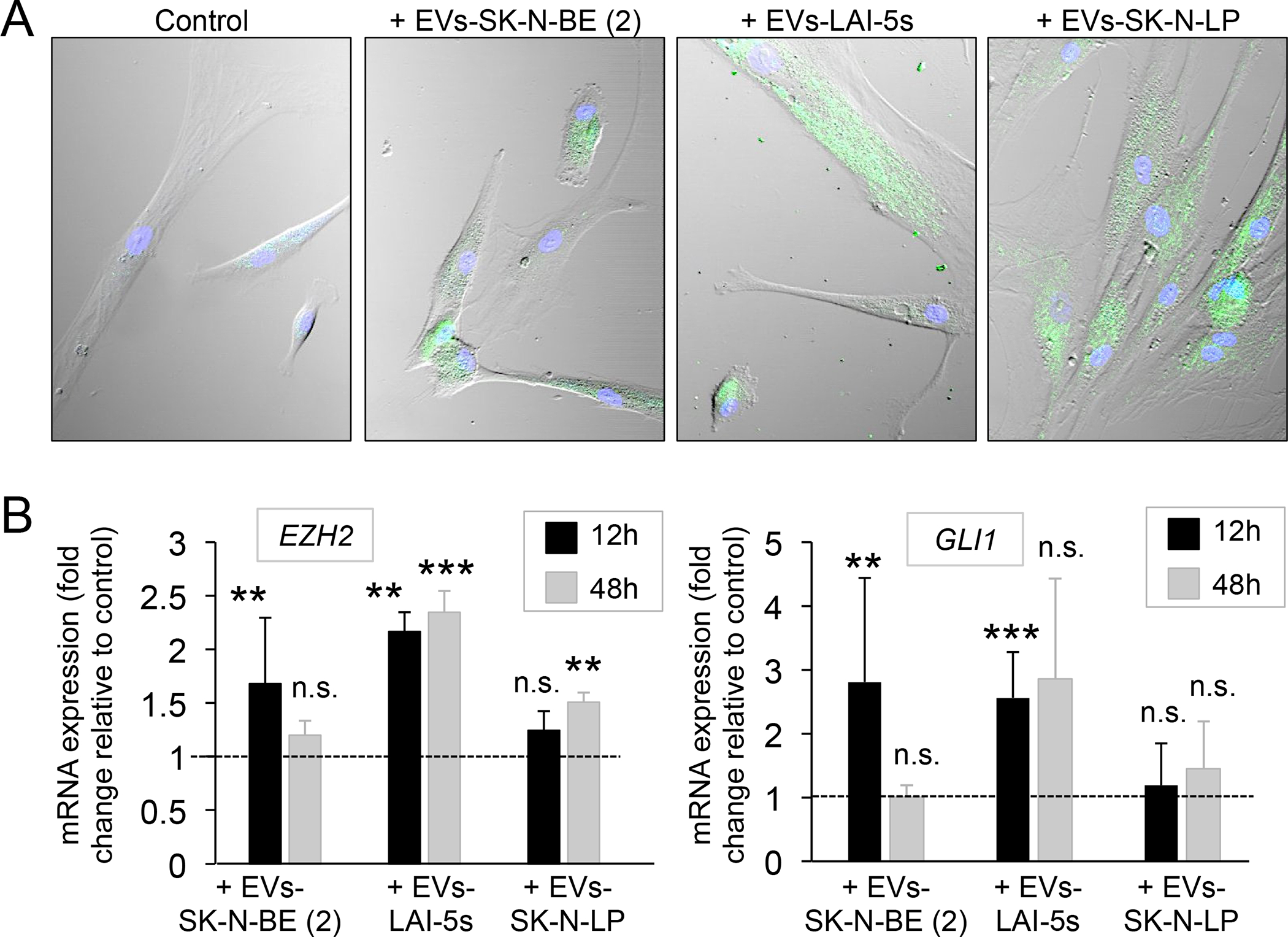
(A) Confocal images of human mesenchymal stem cells (hMSC) after 2h of incubation with PBS (control), and neuroblastoma-derived extracellular vesicles isolated from SK-N-BE(2), LAI-5s and SK-N-LP cells cultured during 7 days on Col1-HA scaffold. Extracellular vesicles were labeled with SYTO RNA Select green fluorescent. Representative images of 3 independent assays. (B) EZH2 mRNA levels and GLI1 mRNA levels in hMSC treated with extracellular vesicles from the indicated cell lines cultured in Col1-HA scaffold during 7 days in comparison to hMSC treated with PBS (control), after 12 and 48 hours of incubation. Statistical significance was determined by the two-tailed Student’s t test. *p < 0.05; **p < 0.01; ***p < 0.001; ns, not significant.
We found that EZH2 and GLI1 mRNA molecules are cargo of EVs in N, S and I-type TE-NB models. Also, we observed that RNA molecules loaded into EVs were transferred to hMSC for the three types of TE-NB constructs. Now, we wondered if specifically both EZH2 and GLI1 could be transferred from EVs to the host cells. For investigating this possibility, we incubated hMSC with 10 μg of EVs protein from each TE-NB (N, S and I-type TE-NB) for 12 and 48 hours.
hMSC incubated with EVs from SK-N-BE (2) TE-NB exhibited significantly high levels of EZH2 and GLI1 compared to untreated MSCs (control) at 12 hours, but expression decreased for both genes at 48 hours (Figure 7B). Similar behavior was observed for hMSC incubated with EVs from LAI-5s TE-NB. But in this case, EZH2 levels remained high at 48 hours (Figure 7B). Therefore, we can confirm that both mRNAs were transferred from the EVs to the hMSC. This is explained by the initial increase in mRNA expression observed at 12 hours of incubation followed by a decrease to reach the levels of the untreated control (basal levels) at 48 hours. The fact that EZH2 mRNA levels remained high at 48 hours for LAI-5s EVs could make us think about the possibility of not only mRNA transferring but also a gene regulation mechanism. Surprisingly, EZH2 mRNA levels were significantly higher in respect to the control at 48 hours of incubation with SK-N-LP EVs (Figure 7B). This suggests a possible mechanism of gene regulation, as well. However, neither EZH2 nor GLI1 levels were altered in hMSC at 24 hours of incubation with SK-N-LP EVs.
hMSC express EZH2 and GLI1 but at lower levels than NB cells and tumors (Supp. Figure 2). As an experimental control, we wondered if EVs from hMSC could also induce overexpression of both markers upon EVs treatment. We found that hMSC EVs did not induce overexpression of any of both mRNAs in the host MSC compared to the control (Supp. Figure 6B).
Collectively, these data confirm that mRNA can be transferred from NB-EVs to human mesenchymal stromal cells, and open the possibility of a mechanism of gene regulation EVs-mediated to induce over-expression of both the EZH2 and GLI1 in the host cells.
Discussion
We investigated whether EVs released by NB cells carry EZH2 and GLI1 mRNAs, two key molecules involved in NB-CSC maintenance. Based on current reports, we hypothesized that the cargo of NB-derived EVs could be transferred to stromal MSC to induce over-expression of these two NB stemness markers. To test this hypothesis, we analyzed NB-EVs in three different types of 3D tissue-engineered models of neuroblastoma (TE-NB) formed by cultivation of NB cells in porous scaffolds designed to mimic the native tumor niche.
We and others have demonstrated that bioengineered 3D tumors are powerful tools for cancer research54,55. Bioengineered tumor models bridge the gap between 2D cultures (used for drug discovery and screening) and animal models (used for efficacy and safety assessment before proceeding to clinical trials) in a predictive, inexpensive, and low time-consuming fashion. 2D models have largely failed as model systems in preclinical studies 56,57. In the last decade, it has been a significant effort to develop human tumor models. However, the development of 3D tumor models for the study of EVs has not been as explored to a great extent. In 2016, we published the first study of EVs using a bioengineered 3D Ewing’s sarcoma (EWS) model50. After that time, several other studies reported on the effects of 3-dimensionality and tumor composition on EVs biogenesis, cargo and surface markers58–61. A number of recent reports confirmed that EVs secreted by cells cultured in monolayers show different profiles of signaling molecules (RNA, DNA, proteins) and surface lipids and proteins, when compared to the EVs secreted by 3D models and found in vivo 58–61.
Given the importance of the cell context for the sorting of molecules to EVs and for the use of EVs as biomarkers, it is anticipated that a 3D model is more suitable to study these processes than 2D cell monolayers. Therefore, we decided to bioengineered 3D models of the three different types of NB cells described to date. We first analyzed the extracellular matrix composition of primary NB from adrenal glands of infants, and observed that its was enriched in collagen 1 and hyaluronic acid proteins. We used these two proteins to fabricate a biomaterial scaffold mimicking the composition of native NB and cultured NB cells in this scaffold for isolation of NB-derived EVs.
GLI1 mRNA was lower expressed in monolayer than in 3D at day 7 for the three TE-NB models developed. While, EZH2 mRNA levels were significantly lower in monolayer at day 7 for the S and I-type TE-NB models. These results suggest that the scaffold may be altering the expression profiles, consistent with recent reports on 3D tumor models of cervical58 and gastric cancer 61, and our previous results for the bioengineered Ewing’s sarcoma model50. Although culturing cells in 3D for one week was enough for mimicking EZH2 mRNAs levels in Ewing’s sarcoma, and S and I-type TE-NB models, one-week culture duration was not sufficient to show effects for N-type neuroblastoma. We did not observe differences in EZH2 mRNA expression between monolayer cells and cells cultured in the TE-NB model. However, EZH2 protein levels decayed at this time point suggesting an effect of the 3D context on the translational, post-translational or post-transcriptional cell machinery.
We do not know why exactly the artificial extracellular matrix was capable of readjust GLI1 and EZH2 expression to more physiological levels in NB cells. GLI1 is a transcription factor that mediates the highly conserved evolutionary Hedgehog signal transduction pathway. Transcriptional activation of GLI1 is auto-regulated by GLI1 and also by GLI2, EWSR1-FLI1, c-MYC, through KRAS, TGFβ, WNT pathways, and epigenetically regulated by H3K27ac and H3K4me362. EZH2 is the enzymatic subunit of the PRC2 complex, which mediates mono-, di-, and tri-methylation of histone H3 lysine 27 to regulate epigenetically gene transcription of downstream target genes 9. Research of the EZH2 has been much more focused on regulation of EZH2 targets genes than on deciphering how EZH2 expression is controlled. In the last decade, numerous studies have demonstrated that EZH2 expression is controlled by a plethora of miRNAs, non-coding RNAs and various oncogenic transcription factors63. EZH2 activity is also controlled by post-translational modifications 64. However, the precise regulatory mechanisms underlying EZH2 function remain largely unclear. It is possible that over longer times of culture the cells within the biomaterial scaffold could modify the transcriptional expression of EZH2. We previously observed that time is a crucial factor for cell adaptation to the new environment and to start to express physiological gene levels65.
Despite the fact that EZH2 mRNA intracellular levels did not change in N-type 3D cultures compared to 2D culture after 7 days of culture, mRNA enrichment in EVs was altered. Levels of EZH2 mRNA were higher in TE-NB-derived EVs than in monolayer-derived EVs for the three 3D models, as previously observed for Ewing’s sarcoma50. Enrichment of GLI1 mRNA was also observed in EVs isolated from all the TE-NB models developed. These data suggest the utility of EZH2 mRNA and GLI1 mRNA found in EVs as diagnostic biomarkers for neuroblastoma. Also, these data highlight the existence of regulatory sorting mechanisms in 3D that differ from the 2D systems.
Despite the growing interest in EVs functions, the mechanism of how specific mRNA sequences are selectively loaded into EVs remains poorly understood. A number of studies in 2D cell cultures involve RNA-binding proteins (RBPs) in EVs cargo sorting66. RBPs can bind to specific sequences present in the 3′-UTR of the exosomal mRNAs in such a way that each mRNA is paired with specific RBPs regulatory molecules66,67. Different reports also support the fact that ESCRT complexes (related to membrane-involving processes) participate in mRNA sorting to EVs68. Thus, many different studies demonstrate that EVs are loaded with specific mRNAs. But more research and predictive models are necessary to elucidate the precise mechanisms behind EVs molecule sorting. Our data suggest that 3D models are unique tools for studying these processes.
Importantly, Koppers-Lalic et al. demonstrate that RNA post-transcriptional modifications also have a role to specific sorting to EVs69. Also, Batagov et al. demonstrated that RNAs carrying specific sequences for exosomal secretion had a significant difference in half-life time distribution compared to intracellular RNAs70. Hence, they found that the mean half-life time for intracellular RNAs was 1.8-times longer than for exosomal RNAs in B-cells, while for fibroblasts was 1.3-times longer (but statistically significant) 70. These findings could suggest that although we found the same EZH2 mRNA intracellular levels between monolayer and the N-type TE-NB, the mRNA from the N-type TE-NB could be higher modified with specific exosomal sequences, and therefore would remain less time inside the cell. This would explain less EZH2 protein in total cell extracts and enrichment of EZH2 mRNA in EVs at day 7. The same process would also explain the results for the observed enrichment of EZH2 mRNA in EVs in the other two models, and GLI1 enrichment in EVs in all the 3D models developed, although further research will be crucial for revealing the mechanisms behind exosomal mRNA regulation.
Reciprocal communication between CSC, non-CSC and stromal cells is inherent to tumor development39,40. Microenvironment supports CSC maintenance, initiation and nutrition of solid tumors, cell proliferation, and distant colony formation, with CSC and non-CSC influencing the surrounding cells towards showing distinct characteristics of the tumor cells 39,40,71. We investigated if NB cells could influence the surrounding stromal cells to show overexpression of EZH2 and/or GLI1. To this end, we co-cultured TE-NB-derived EVs loaded with EZH2 mRNA and GLI1 mRNA and stromal mesenchymal stem cells. N and S-type TE-NB-derived EVs were taken up by the stromal cells, which in turn increased the mRNA levels of both regulators of the NB-CSC phenotype at 12 hours. However, this not was the case for the I-type EVs. Only EZH2 levels were higher after 48 hours of incubation, which suggests the possibility of a mechanism of gene regulation by EVs. This phenomenon was also observed for S-type EVs; EZH2 levels remained high at 48 hours after EVs treatment. Together we can confirm that there is a horizontal transfer of both EZH2 and GLI mRNAs from N and S-type NB-derived EVs to the host. However, how I-type EVs can induce EZH2 expression in hMSC after 48 hours is unknown, as well as, how EZH2 abnormal levels persist in hMSC after 48 hours of initial incubation with S-type NB-derived EVs.
Besides the enrichment of mRNAs in EVs, it is crucial to investigate the presence of both EZH2 and GLI proteins as EVs cargo, and protein transferring to the stromal cells.
One of the advantages of our TE-NB models is that they need a small number of cells and reagents. They are cheap, small, and compatible with high-throughput screenings. However, this is also a limitation for molecular biology techniques. Because of the small number of cells that a TE-NB uses, we also obtained a small amount of EVs per construct. We can easily detect mRNAs and small RNAs in EVs because the sensitivity of the qRT-PCR technique is higher than the Western blot; and because the qRT-PCR requires less amount of initial biological material than the Western blot. GAPDH and CD81 proteins were easily detected by Western blot because they are abundant as cargo. However, proteins that are present at low levels are very difficult to detect. We could not find any of both EZH2 and GLI1 proteins as cargo of TE-NB EVs (data not shown). This could be for the reason that none of both proteins are present in TE-NB EVs or for the mentioned technical limitation of the models Detection of proteins present at low levels in EVs could be achievable by increasing the size of the TE-NB models that would increase the amount of the biological material to work with. In that case, it would necessary to couple the TE-NB model to a perfusion bioreactor to avoid a necrotic core of cells generated by the low diffusion of nutrients and oxygen. The perfusable model would be also useful for transferring assays and later analysis of protein expression in the host cells. That model would produce a higher yield of EVs, and more than 10μg of EVs could be assayed. In addition, it is important to say that transferring experiments were performed with a single dose of EVs and cell harvesting at different time points of incubation, using hMSC in monolayer. Although the monolayer model and experimental design gave us fruitful insights, this does not represent the situation in the human body. Cells from the human tissues are in 3D, and continuously receiving tumor-derived EVs. Therefore, to perform relevant functional assays for an understanding of the biological process it would be necessary to develop complex model systems to mimic the continuous bombardment of EVs to which the cells of the body are subjected.
Reprogramming of non-CSC and stromal cells towards CSC-derived EVs has been described, but is still poorly understood 42,43. Studies in melanoma demonstrated that transferring of cargo of EVs to MSCs is involved in malignant transformation 49. Our data support that EVs are transferred to MSCs, and open the door to analyze the involvement of neuroblastoma EVs in the possible alteration of the expression profile of stromal MSCs. However, the important question is if MSC stromal cells could transition into NB-CSC after the acquisition of CSC features, or if they would gain a new status of reactive/supportive stroma for CSC maintenance, as previously reported 48. Studies of 3D tumor models and the EVs they secrete will help identify and characterize the mechanisms underlying phenotypic changes in cancer-associated stroma, as well as the riles of EVs as mediators of CSC signaling. However, much more work is necessary to deeply understanding of all of these processes.
Supplementary Material
Supplementary Figure 1. Evaluation of the presence of EVs in the biomaterial without cells. (A) Representative size distribution profile of isolated particles from Col1/HA biomaterial incubated in culture medium during 7 days compared to PBS. Supernatants were treated with Total Exosome Isolation Reagent and the final product was resuspended in PBS for NTA analysis (B) Concentration of particles isolated from the biomaterial compared to the control (PBS). Data obtained from the NTA analyses. (C) TEM image of the final product isolated from the biomaterial supernatants.
Supplementary Figure 2. (A) EZH2 and GLI1 expression in neuroblastoma tumors from three different patients (NB-1, NB-2 and NB-3) by immunohistochemistry. Counterstaining was performed with hematoxylin QS (blue).(B) EZH2 and GLI1 mRNA average expression in human mesenchymal stem cells, neuroblastoma tumors and neuroblastoma cell lines on public available expression array data by BoxBlot presentation using a comparative study of the amc onco-genomics software tool (www.amc.com).
Supplementary Figure 3. Western blot analysis for EZH2 protein in N-type TE-NB models at day 3 and day 7 and in N-type neuroblastoma cells cultured in monolayer. GAPDH was used as loading control. SK-N-BE(2) was the N-type cell line assayed.
Supplementary Figure 4. Characterization of the Tissue-engineered model of S-type Neuroblastoma (TE-NB) (A) Live/dead staining images of the S-type neuroblastoma cell line LAI-5s cultured within Col1/HA scaffold for 3 and 7 days. (n=3). Hoechst dye (blue) stains DNA, Calcein stains green-live cells, and ethidium homodimer-1 stains red-dead cells. (B) Hematoxylin and Eosin staining images showing LAI-5s cells growing within the biomaterial over-time, at day 3 and 7 after cell seeding. (C) Scanning Electron Microscopy (SEM) representative images showing cell-matrix integration in TE-NB models at day 3 and formation of tumoral tissue and day 7. (D) EZH2 and GLI1 mRNA intracellular levels in the S-type TE-NB models generated with LAI-5s cells at day 3 and 7 compared to monolayer (ML) conditions. Relative endogenous expression of GLI1 was normalized to actin and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test.**p < 0.01; .***p < 0.001; ns, not significant.
Supplementary Figure 5. Characterization of the Tissue-engineered model of I-type Neuroblastoma (TE-NB) generated with SK-N-LP cells (A) Live/dead staining images of the I-type neuroblastoma cell line SK-N-LP cultured within Col1/HA scaffold for 3 and 7 days. (n=3). Hoechst dye (blue) stains DNA, Calcein stains green-live cells, and ethidium homodimer-1 stains red-dead cells. (B) Hematoxylin and Eosin staining images of I-type TE-NB at day 3 and 7 after cell seeding. (C) (left) Representative image showing SK-N-LP cells attached to the scaffold and forming patches of tissue day 3 by SEM; (right) SEM image of SK-N-LP cells forming compact tumoral tissue in the TE-NB and day 7. (D) Analysis of EZH2 and GLI1 expression by qRT-PCR; mRNA expression of the indicated genes in I-type TE-NB models is represented as the fold change in expression of the target gene relative to the monolayer condition (ML). Relative endogenous expression of both genes was normalized to actin and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test. **p < 0.01; .***p < 0.001.
Supplementary Figure 6. Analysis of transferring of human mesenchymal stem cells-derived EV. (A) Confocal images of human mesenchymal stem cells (hMSC) treated with PBS (control) or hMSC-derived extracellular vesicles (EVs-hMSC) for 2 hours. EVs were labeled with SYTO RNA Select green fluorescent before incubation with hMSC. (B) EZH2 and GLI1 mRNA levels in hMSC incubated with EVs released by hMSC (EVs-hMSC) in comparison to the PBS control, after 12 hours of incubation. Statistical significance was determined by the two-tailed Student’s t test. ns, not significant.
ACKNOWLEDGMENTS
The authors gratefully acknowledge The Spanish Association Against Cancer (AECC) and NIH funding of this work (EB27062, EB 25765, CA 249799). The IBEC Group has support from the Commission for Universities and Research of the Department of Innovation, Universities, and Enterprise of the Generalitat de Catalunya (2017 SGR 1079). The authors acknowledge the support of CERCA Programme/Generalitat de Catalunya. We thank the Molecular Pathology Tumor Bank for providing deidentified tumor samples, Histology Facility at Columbia University for histological services, University of Barcelona Science and Technology Centers (CCiTUB) for SEM technical assistance, Dr. Tao Su at Columbia University Molecular Biology service for Agilent Bioanalyzer analyses, and Mr. Almici at IBEC for the assistance given with the confocal microscope.
All authors have read the journal’s policy on conflicts of interest. All authors have read the journal’s authorship agreement.
Footnotes
COMPETING INTERESTS
The authors declare that they have no competing interests
AVAILABILITY OF DATA AND MATERIALS
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Matthay KK et al. Neuroblastoma. Nat Rev Dis Primers 2, 16078, doi: 10.1038/nrdp.2016.78 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Cheung NK & Dyer MA Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nature reviews. Cancer 13, 397–411, doi: 10.1038/nrc3526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajwanshi A, Srinivas R & Upasana G Malignant small round cell tumors. J Cytol 26, 1–10, doi: 10.4103/0970-9371.54861 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Schaft DW et al. Tumor cell plasticity in Ewing sarcoma, an alternative circulatory system stimulated by hypoxia. Cancer Res 65, 11520–11528, doi: 10.1158/0008-5472.CAN-05-2468 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti L, Abou-Antoun T, Vukmanovic S & Sandler AD Reversible adaptive plasticity: a mechanism for neuroblastoma cell heterogeneity and chemo-resistance. Front Oncol 2, 82, doi: 10.3389/fonc.2012.00082 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veschi V, Verona F & Thiele CJ Cancer Stem Cells and Neuroblastoma: Characteristics and Therapeutic Targeting Options. Front Endocrinol (Lausanne) 10, 782, doi: 10.3389/fendo.2019.00782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayob AZ & Ramasamy TS Cancer stem cells as key drivers of tumour progression. J Biomed Sci 25, 20, doi: 10.1186/s12929-018-0426-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saygin C, Matei D, Majeti R, Reizes O & Lathia JD Targeting Cancer Stemness in the Clinic: From Hype to Hope. Cell Stem Cell 24, 25–40, doi: 10.1016/j.stem.2018.11.017 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Wen Y, Cai J, Hou Y, Huang Z & Wang Z Role of EZH2 in cancer stem cells: from biological insight to a therapeutic target. Oncotarget 8, 37974–37990, doi: 10.18632/oncotarget.16467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamijo T Role of stemness-related molecules in neuroblastoma. Pediatr Res 71, 511–515, doi: 10.1038/pr.2011.54 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Li Z et al. EZH2 regulates neuroblastoma cell differentiation via NTRK1 promoter epigenetic modifications. Oncogene 37, 2714–2727, doi: 10.1038/s41388-018-0133-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C et al. EZH2 Mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res 72, 315–324, doi: 10.1158/0008-5472.CAN-11-0961 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suva ML et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res 69, 9211–9218, doi: 10.1158/0008-5472.CAN-09-1622 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Liu H et al. MELK and EZH2 Cooperate to Regulate Medulloblastoma Cancer Stem-like Cell Proliferation and Differentiation. Mol Cancer Res 15, 1275–1286, doi: 10.1158/1541-7786.MCR-17-0105 (2017). [DOI] [PubMed] [Google Scholar]
- 15.van Vlerken LE et al. EZH2 is required for breast and pancreatic cancer stem cell maintenance and can be used as a functional cancer stem cell reporter. Stem Cells Transl Med 2, 43–52, doi: 10.5966/sctm.2012-0036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong X & Nephew KP Ovarian Cancer Stem Cells: Role in Metastasis and Opportunity for Therapeutic Targeting. Cancers (Basel) 11, doi: 10.3390/cancers11070934 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorodetska I, Lukiyanchuk V, Peitzsch C, Kozeretska I & Dubrovska A BRCA1 and EZH2 cooperate in regulation of prostate cancer stem cell phenotype. Int J Cancer 145, 2974–2985, doi: 10.1002/ijc.32323 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Lund K, Adams PD & Copland M EZH2 in normal and malignant hematopoiesis. Leukemia 28, 44–49, doi: 10.1038/leu.2013.288 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Singh AK et al. Salinomycin inhibits epigenetic modulator EZH2 to enhance death receptors in colon cancer stem cells. Epigenetics, 1–18, doi: 10.1080/15592294.2020.1789270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peer E, Tesanovic S & Aberger F Next-Generation Hedgehog/GLI Pathway Inhibitors for Cancer Therapy. Cancers (Basel) 11, doi: 10.3390/cancers11040538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Didiasova M, Schaefer L & Wygrecka M Targeting GLI Transcription Factors in Cancer. Molecules 23, doi: 10.3390/molecules23051003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Zapico ME GLI1 finds a new role in cancer stem cell biology. EMBO Mol Med 5, 483–485, doi: 10.1002/emmm.201302505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahi MH et al. Expression and epigenetic modulation of sonic hedgehog-GLI1 pathway genes in neuroblastoma cell lines and tumors. Tumour Biol 32, 113–127, doi: 10.1007/s13277-010-0105-x (2011). [DOI] [PubMed] [Google Scholar]
- 24.Xu L et al. Sonic Hedgehog pathway is essential for neuroblastoma cell proliferation and tumor growth. Mol Cell Biochem 364, 235–241, doi: 10.1007/s11010-011-1222-6 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Po A et al. Noncanonical GLI1 signaling promotes stemness features and in vivo growth in lung adenocarcinoma. Oncogene 36, 4641–4652, doi: 10.1038/onc.2017.91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heiden KB et al. The sonic hedgehog signaling pathway maintains the cancer stem cell self-renewal of anaplastic thyroid cancer by inducing snail expression. J Clin Endocrinol Metab 99, E2178–2187, doi: 10.1210/jc.2014-1844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Regan JL et al. Non-Canonical Hedgehog Signaling Is a Positive Regulator of the WNT Pathway and Is Required for the Survival of Colon Cancer Stem Cells. Cell Rep 21, 2813–2828, doi: 10.1016/j.celrep.2017.11.025 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Cochrane CR, Szczepny A, Watkins DN & Cain JE Hedgehog Signaling in the Maintenance of Cancer Stem Cells. Cancers (Basel) 7, 1554–1585, doi: 10.3390/cancers7030851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mastrangelo E & Milani M Role and inhibition of GLI1 protein in cancer. Lung Cancer (Auckl) 9, 35–43, doi: 10.2147/LCTT.S124483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedmann-Morvinski D & Verma IM Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep 15, 244–253, doi: 10.1002/embr.201338254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litviakov N et al. Amplifications of stemness genes and the capacity of breast tumors for metastasis. Oncotarget 11, 1988–2001, doi: 10.18632/oncotarget.27608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song WS et al. Sox2, a stemness gene, regulates tumor-initiating and drug-resistant properties in CD133-positive glioblastoma stem cells. J Chin Med Assoc 79, 538–545, doi: 10.1016/j.jcma.2016.03.010 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Kumar SM et al. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene 31, 4898–4911, doi: 10.1038/onc.2011.656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J et al. EZH2 is overexpressed in laryngeal squamous cell carcinoma and enhances the stem-like properties of AMC-HN-8 cells. Oncol Lett 12, 837–846, doi: 10.3892/ol.2016.4704 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CJ et al. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell 19, 86–100, doi: 10.1016/j.ccr.2010.10.035 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JF et al. EZH2 promotes colorectal cancer stem-like cell expansion by activating p21cip1-Wnt/beta-catenin signaling. Oncotarget 7, 41540–41558, doi: 10.18632/oncotarget.9236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan L et al. The polycomb group protein EZH2 induces epithelial-mesenchymal transition and pluripotent phenotype of gastric cancer cells by binding to PTEN promoter. J Hematol Oncol 11, 9, doi: 10.1186/s13045-0170547-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu B et al. GLI1-mediated regulation of side population is responsible for drug resistance in gastric cancer. Oncotarget 8, 27412–27427, doi: 10.18632/oncotarget.16174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan TS, Shaked Y & Tsai KK Targeting the Interplay Between Cancer Fibroblasts, Mesenchymal Stem Cells, and Cancer Stem Cells in Desmoplastic Cancers. Front Oncol 9, 688, doi: 10.3389/fonc.2019.00688 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plaks V, Kong N & Werb Z The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 16, 225–238, doi: 10.1016/j.stem.2015.02.015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelizzo G et al. Microenvironment in neuroblastoma: isolation and characterization of tumor-derived mesenchymal stromal cells. BMC Cancer 18, 1176, doi: 10.1186/s12885-018-5082-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun Z, Wang L, Dong L & Wang X Emerging role of exosome signalling in maintaining cancer stem cell dynamic equilibrium. J Cell Mol Med, doi: 10.1111/jcmm.13676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J, Liao K & Zhou W Exosomes Regulate the Transformation of Cancer Cells in Cancer Stem Cell Homeostasis. Stem Cells Int 2018, 4837370, doi: 10.1155/2018/4837370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathieu M, Martin-Jaular L, Lavieu G & Théry C Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21, 9–17, doi: 10.1038/s41556-018-0250-9 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Jung YJ et al. Cell reprogramming using extracellular vesicles from differentiating stem cells into white/beige adipocytes. Science Advances 6, eaay6721, doi: 10.1126/sciadv.aay6721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang E et al. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduction and Targeted Therapy 5, doi: 10.1038/s41392-020-00359-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma Z et al. Exosomes from glioma cells induce a tumor-like phenotype in mesenchymal stem cells by activating glycolysis. Stem Cell Res Ther 10, 60, doi: 10.1186/s13287-019-1149-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Gu J et al. Gastric cancer exosomes trigger differentiation of umbilical cord derived mesenchymal stem cells to carcinoma-associated fibroblasts through TGF-beta/Smad pathway. PLoS One 7, e52465, doi: 10.1371/journal.pone.0052465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gyukity-Sebestyen E et al. Melanoma-Derived Exosomes Induce PD-1 Overexpression and Tumor Progression via Mesenchymal Stem Cell Oncogenic Reprogramming. Front Immunol 10, 2459, doi: 10.3389/fimmu.2019.02459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villasante A et al. Recapitulating the Size and Cargo of Tumor Exosomes in a Tissue-Engineered Model. Theranostics 6, 1119–1130, doi: 10.7150/thno.13944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walton JD et al. Characteristics of Stem Cells from Human Neuroblastoma Cell Lines and in Tumors. Neoplasia 6, 838–845, doi: 10.1593/neo.04310 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campos Cogo S et al. An overview of neuroblastoma cell lineage phenotypes and in vitro models. Exp Biol Med (Maywood) 245, 1637–1647, doi: 10.1177/1535370220949237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fischbach C et al. Engineering tumors with 3D scaffolds. Nat Methods 4, 855–860, doi: 10.1038/nmeth1085 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Villasante A & Vunjak-Novakovic G Tissue-engineered models of human tumors for cancer research. Expert Opin Drug Discov 10, 257–268, doi: 10.1517/17460441.2015.1009442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hapach LA, Mosier JA, Wang W & Reinhart-King CA Engineered models to parse apart the metastatic cascade. NPJ Precis Oncol 3, 20, doi: 10.1038/s41698-019-0092-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langhans SA Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front Pharmacol 9, 6, doi: 10.3389/fphar.2018.00006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brancato V, Oliveira JM, Correlo VM, Reis RL & Kundu SC Could 3D models of cancer enhance drug screening? Biomaterials 232, 119744, doi: 10.1016/j.biomaterials.2019.119744 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Thippabhotla S, Zhong C & He M 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci Rep 9, 13012, doi: 10.1038/s41598-019-49671-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haraszti RA et al. Exosomes Produced from 3D Cultures of MSCs by Tangential Flow Filtration Show Higher Yield and Improved Activity. Mol Ther 26, 2838–2847, doi: 10.1016/j.ymthe.2018.09.015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carfi Pavia F et al. A 3Dscaffold of PLLA induces the morphological differentiation and migration of primary astrocytes and promotes the production of extracellular vesicles. Mol Med Rep 20, 1288–1296, doi: 10.3892/mmr.2019.10351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rocha S et al. 3D Cellular Architecture Affects MicroRNA and Protein Cargo of Extracellular Vesicles. Adv Sci (Weinh) 6, 1800948, doi: 10.1002/advs.201800948 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor R et al. Regulation of GLI1 by cis DNA elements and epigenetic marks. DNA Repair (Amst) 79, 10–21, doi: 10.1016/j.dnarep.2019.04.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamaguchi H & Hung MC Regulation and Role of EZH2 in Cancer. Cancer Res Treat 46, 209–222, doi: 10.4143/crt.2014.46.3.209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu H et al. Regulation and role of post-translational modifications of enhancer of zeste homologue 2 in cancer development. Am J Cancer Res 6, 2737–2754 (2016). [PMC free article] [PubMed] [Google Scholar]
- 65.Villasante A, Marturano-Kruik A & Vunjak-Novakovic G Bioengineered human tumor within a bone niche. Biomaterials 35, 5785–5794, doi: 10.1016/j.biomaterials.2014.03.081 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Liegro CM, Schiera G & Di Liegro I Extracellular Vesicle-Associated RNA as a Carrier of Epigenetic Information. Genes (Basel) 8, doi: 10.3390/genes8100240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szostak N et al. Sorting signal targeting mRNA into hepatic extracellular vesicles. RNA Biol 11, 836–844, doi: 10.4161/rna.29305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Juan T & Furthauer M Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol 74, 66–77, doi: 10.1016/j.semcdb.2017.08.022 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Koppers-Lalic D et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8, 1649–1658, doi: 10.1016/j.celrep.2014.08.027 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Batagov AO, Kuznetsov VA & Kurochkin IV Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics 12 Suppl 3, S18, doi: 10.1186/1471-2164-12-S3-S18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wortzel I, Dror S, Kenific CM & Lyden D Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell 49, 347–360, doi: 10.1016/j.devcel.2019.04.011 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Evaluation of the presence of EVs in the biomaterial without cells. (A) Representative size distribution profile of isolated particles from Col1/HA biomaterial incubated in culture medium during 7 days compared to PBS. Supernatants were treated with Total Exosome Isolation Reagent and the final product was resuspended in PBS for NTA analysis (B) Concentration of particles isolated from the biomaterial compared to the control (PBS). Data obtained from the NTA analyses. (C) TEM image of the final product isolated from the biomaterial supernatants.
Supplementary Figure 2. (A) EZH2 and GLI1 expression in neuroblastoma tumors from three different patients (NB-1, NB-2 and NB-3) by immunohistochemistry. Counterstaining was performed with hematoxylin QS (blue).(B) EZH2 and GLI1 mRNA average expression in human mesenchymal stem cells, neuroblastoma tumors and neuroblastoma cell lines on public available expression array data by BoxBlot presentation using a comparative study of the amc onco-genomics software tool (www.amc.com).
Supplementary Figure 3. Western blot analysis for EZH2 protein in N-type TE-NB models at day 3 and day 7 and in N-type neuroblastoma cells cultured in monolayer. GAPDH was used as loading control. SK-N-BE(2) was the N-type cell line assayed.
Supplementary Figure 4. Characterization of the Tissue-engineered model of S-type Neuroblastoma (TE-NB) (A) Live/dead staining images of the S-type neuroblastoma cell line LAI-5s cultured within Col1/HA scaffold for 3 and 7 days. (n=3). Hoechst dye (blue) stains DNA, Calcein stains green-live cells, and ethidium homodimer-1 stains red-dead cells. (B) Hematoxylin and Eosin staining images showing LAI-5s cells growing within the biomaterial over-time, at day 3 and 7 after cell seeding. (C) Scanning Electron Microscopy (SEM) representative images showing cell-matrix integration in TE-NB models at day 3 and formation of tumoral tissue and day 7. (D) EZH2 and GLI1 mRNA intracellular levels in the S-type TE-NB models generated with LAI-5s cells at day 3 and 7 compared to monolayer (ML) conditions. Relative endogenous expression of GLI1 was normalized to actin and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test.**p < 0.01; .***p < 0.001; ns, not significant.
Supplementary Figure 5. Characterization of the Tissue-engineered model of I-type Neuroblastoma (TE-NB) generated with SK-N-LP cells (A) Live/dead staining images of the I-type neuroblastoma cell line SK-N-LP cultured within Col1/HA scaffold for 3 and 7 days. (n=3). Hoechst dye (blue) stains DNA, Calcein stains green-live cells, and ethidium homodimer-1 stains red-dead cells. (B) Hematoxylin and Eosin staining images of I-type TE-NB at day 3 and 7 after cell seeding. (C) (left) Representative image showing SK-N-LP cells attached to the scaffold and forming patches of tissue day 3 by SEM; (right) SEM image of SK-N-LP cells forming compact tumoral tissue in the TE-NB and day 7. (D) Analysis of EZH2 and GLI1 expression by qRT-PCR; mRNA expression of the indicated genes in I-type TE-NB models is represented as the fold change in expression of the target gene relative to the monolayer condition (ML). Relative endogenous expression of both genes was normalized to actin and error bars represent standard deviations of relative expression. Statistical significance was determined by the two-tailed Student’s t test. **p < 0.01; .***p < 0.001.
Supplementary Figure 6. Analysis of transferring of human mesenchymal stem cells-derived EV. (A) Confocal images of human mesenchymal stem cells (hMSC) treated with PBS (control) or hMSC-derived extracellular vesicles (EVs-hMSC) for 2 hours. EVs were labeled with SYTO RNA Select green fluorescent before incubation with hMSC. (B) EZH2 and GLI1 mRNA levels in hMSC incubated with EVs released by hMSC (EVs-hMSC) in comparison to the PBS control, after 12 hours of incubation. Statistical significance was determined by the two-tailed Student’s t test. ns, not significant.
Data Availability Statement
The datasets during and/or analyzed during the current study are available from the corresponding author on reasonable request.


