Abstract
The members of the radical S-adenosylmethionine (SAM) enzyme superfamily are responsible for catalyzing a diverse set of reactions in a multitude of biosynthetic pathways. Many members of this superfamily accomplish their transformations using the catalytic power of a 5′-deoxyadenosyl radical (5′-dAdo•), but there are also enzymes within this superfamily that bind auxiliary cofactors and extend the catalytic repertoire of SAM. In particular, the cobalamin (Cbl)-dependent class synergistically uses Cbl to facilitate challenging methylation and radical rearrangement reactions. Despite identification of this class by Sofia et al. 20 years ago, the low sequence identity between members has led to difficulty in predicting function of uncharacterized members, pinpointing catalytic residues, and elucidating reaction mechanisms. Here, we capitalize on the three recent structures of Cbl-dependent radical SAM enzymes that use common cofactors to facilitate ring contraction as well as radical-based and non-radical-based methylation reactions. With these three structures as a framework, we describe how the Cbl-dependent radical SAM enzymes repurpose the traditional SAM- and Cbl-binding motifs to form an active site where both Cbl and SAM can participate in catalysis. In addition, we describe how, in some cases, the classic SAM- and Cbl-binding motifs support the diverse functionality of this enzyme class, and finally, we define new motifs that are characteristic of Cbl-dependent radical SAM enzymes.
Keywords: metalloenzyme, cobalamin, S-adenosylmethionine, radical SAM, vitamin B12
Introduction
The radical S-adenosylmethionine (SAM) enzymes, so-named by Sofia et al., are characterized by a conserved series of Cys residues (Cys-X3-Cys-X2-Cys) that are involved in coordinating a [4Fe–4S] cluster.1 Most radical SAM enzymes reductively cleave SAM to form a 5′-deoxyadenosyl radical (5′-dAdo•) and methionine (Figure 1A). This reactive radical species can be used to initiate catalysis in a wide range of biosynthetic pathways, including those of vitamins, cofactors, and antibiotics.2 Current estimates suggest that, despite the identification of only 645 radical SAM enzyme sequences in the pioneering paper by Sofia et al., hundreds of thousands of sequences are part of the radical SAM enzyme superfamily today (radicalsam.org). These enzymes vary widely in their reactivity and, in many cases, invoke auxiliary cofactors to expand their catalytic repertoire.3 For example, the glycyl radical activase pyruvate–formate lyase activating enzyme (PFL-AE) employs a cation for catalysis,4 and lipoyl synthase (LipA),5 biotin synthase (BioB),6 and enzymes belonging to the “SPASM/TWITCH” class of the radical SAM enzyme superfamily employ auxiliary Fe–S clusters.7,8 Most notably, one of the largest classes of radical SAM enzymes, also originally highlighted in the Sofia et al. paper, are typified by a domain that has sequence similarity to proteins including methionine synthase (MetH), glutamate mutase, and methyl–malonyl CoA mutase that bind different flavors of Nature’s most beautiful cofactor, vitamin B12, or cobalamin (Cbl).1 The presence or absence of different axial ligands to the Co-containing macrocyclic cofactor differentiates the types of Cbl that are found in Nature, which include adenosyl (Ado)-Cbl, methyl (Me)-Cbl, and open Cbl, which lacks an upper axial ligand.9−11
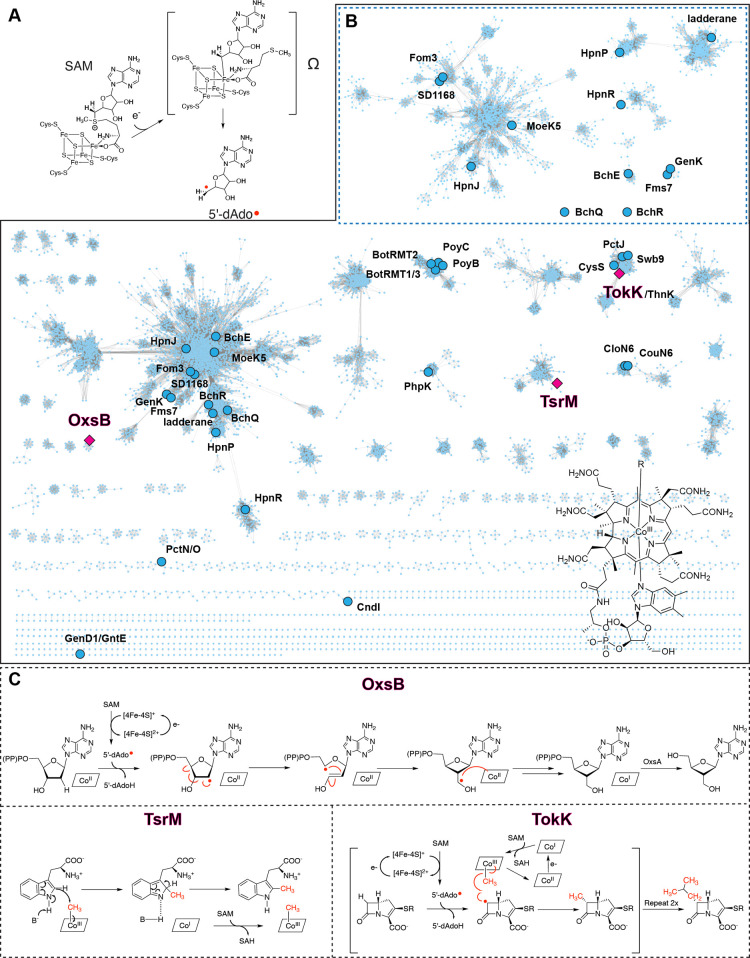
Figure 1.
Cbl-dependent radical SAM enzymes are widespread in Nature and showcase diverse reactivity. (A) [4Fe–4S] cluster-mediated reductive cleavage of SAM results in formation of a spectroscopically characterized organometallic intermediate and methionine (Ω).66 (B) Sequence similarity network (SSN) for the Cbl-dependent radical SAM enzymes is shown with an alignment score of 60.13−15 Nodes highlighted as pink squares represent the structurally characterized enzymes OxsB,18 TsrM,20 and TokK.21 The highlighted nodes (blue circles) are enzymes that have been biochemically investigated. The top right inset is a sub-SSN of the largest cluster from the main SSN that is analyzed with a more stringent alignment score of 80. The bottom right shows the complex structure of Cbl. (C) Reactions catalyzed by the structurally characterized Cbl-dependent radical SAM enzymes include a radical rearrangement to form OXT-A (OxsB), a non-radical based methylation of tryptophan (TsrM) that uses SAM and generates methylated Trp and S-adenosylhomocysteine (SAH), and a series of radical-dependent methylation reactions in asparenomycin A biosynthesis (TokK). SSNs were visualized with cytoscape.67
Remarkably, today this class of Cbl-dependent radical SAM enzymes, which started as a founding list of 25 protein sequences that included enzymes involved in the biosynthesis of oxetanocin A (OxsB), fosfomycin (Fom3), fortimicin (Fms7), bacteriochlorophyll (BchE), and l-phosphinothricylalanylalanine (BcpD/PhpK),1 has grown into a superfamily of its own. The large number of annotated Cbl-dependent radical SAM enzyme sequences are now best visualized and interpreted using a sequence similarity network (SSN), which emphasizes the widespread use of these enzymes in Nature (Figure 1B).12−15 Mapping of the biochemically and structurally characterized enzymes onto such an SSN also highlights the vast unexplored sequence space of this enzyme class (Figure 1B).12−15 The lack of details regarding how members of the Cbl-dependent enzyme class function is largely due to challenges associated with insolubility of these enzymes, a shortage of information regarding their physiological substrates, and lack of knowledge regarding what constitutes the active form of an enzyme.16,17
Consistent with what is observed across the radical SAM enzyme superfamily, the functions of the Cbl-dependent enzymes are diverse (Figure 1B). The majority of annotated Cbl-dependent radical SAM enzymes are proposed to use radical SAM chemistry and MeCbl to methylate inert substrate centers. Other members showcase more divergent functionality: OxsB is involved in facilitating a complex radical rearrangement reaction,18,19 and TsrM does not use radical chemistry at all20 (Figure 1C). Recent advances including new purification strategies17 and three structures of Cbl-dependent radical SAM enzymes that are implicated in radical rearrangement,18 non-radical-based methylation,20 and radical-based methylation21 have laid the groundwork for understanding how these enzymes catalyze their challenging reactions (Figure 1C). Here, we review the three recently determined structures of Cbl-dependent radical SAM enzymes.18,20,21 Through comparison of these structures with other structurally characterized Cbl-binding and radical SAM proteins, we detail the architectural features that support the use of both Cbl and SAM in catalysis.18,20,21 Further, we highlight how the traditional radical SAM- and Cbl-binding motifs have been repurposed to support the diverse chemistry of the Cbl-dependent radical SAM enzymes.
Extensive Reaction Scope of Cbl-Dependent Radical SAM Enzymes
As described above, most annotated Cbl-dependent radical SAM enzymes appear to be radical-based methylases, but the first structure determined of any Cbl-dependent radical SAM enzyme was that of the non-methylase OxsB.18 This protein is part of the pathway that produces the antiviral compound oxetenocin-A (OXT-A). OxsB is involved in contracting the five-membered deoxyribose ring of 2′-deoxyadenosine-5′-monophosphate (dAMP), 2′-deoxyadenosine-5′-diphosphate (dADP), and 2′-deoxyadenosine-5′-triphosphate (dATP) to form the four-membered oxetane ring of OXT-A (Figure 1C).18 The OxsB-mediated ring contraction requires that one molecule of SAM is reductively cleaved. The resulting 5′-dAdo• abstracts a hydrogen atom from the 2′ position of the substrate to form 5′-dAdoH and a ribose-derived radical species (Figure 1C).18 Subsequent rearrangement of the radical intermediate results in formation of the oxetane ring. Although the need for Cbl in this transformation is not immediately obvious, it is known that the presence of hydroxo (HO)-Cbl is required for the reaction to occur.18 Current proposals suggest that Cbl is involved in facilitating the ring closure step via formation of a Co substrate adduct and/or that Cbl is used as an electron acceptor for the product radical.10,18 Of note, OXT-A production also relies on OxsB working in concert with the HD domain phosphohydrolase enzyme OxsA and a promiscuous cellular dehydrogenase (Figure 1C).18,19,22
The second structure determined in this family was of a homologue of the Streptomyces enzyme TsrM. Like OxsB, TsrM from Kitasatospora setae does not perform a radical-based methylation. Instead, TsrM catalyzes the methylation of l-tryptophan in the biosynthetic pathway of thiostrepton using polar chemistry.20,23 More specifically, a proposed base-mediated removal of a proton from the C2 position of l-tryptophan forms a carbanionic intermediate that displaces a methyl cation from MeCbl (Figure 1C).20 One molecule of SAM is subsequently required to re-form MeCbl from Co(I)–Cbl at the end of the catalytic cycle (Figure 1C).20,23 The role of the radical SAM [4Fe–4S] cluster in TsrM is currently unknown but is proposed to be important for reducing adventitiously formed Co(II)–Cbl back to the Co(I)–Cbl state for methylation and subsequent catalytic cycling.20
The third Cbl-dependent radical SAM enzyme to be structurally characterized is TokK.21,24 This methylase is implicated in catalyzing serial methylation reactions to form the C6 isopropyl group of a carbapenem scaffold in the biosynthesis of asparenomycin A (Figure 1C).21 TokK is believed to perform radical-based methylation reactions, the type of chemistry expected for the majority of Cbl-dependent radical SAM enzymes. In the proposed TokK mechanism, one molecule of SAM is reductively cleaved to form 5′-dAdo•.21,25−27 This species is used to generate a substrate radical that is subsequently quenched via transfer of a methyl radical from MeCbl.21,25−27 A second molecule of SAM and an electron are then used to regenerate MeCbl from Co(II)–Cbl (Figure 1C).21,25−27 Subsequent passes through this reaction sequence would account for the iterative methylation reactions needed to complete formation of the isopropyl group.21
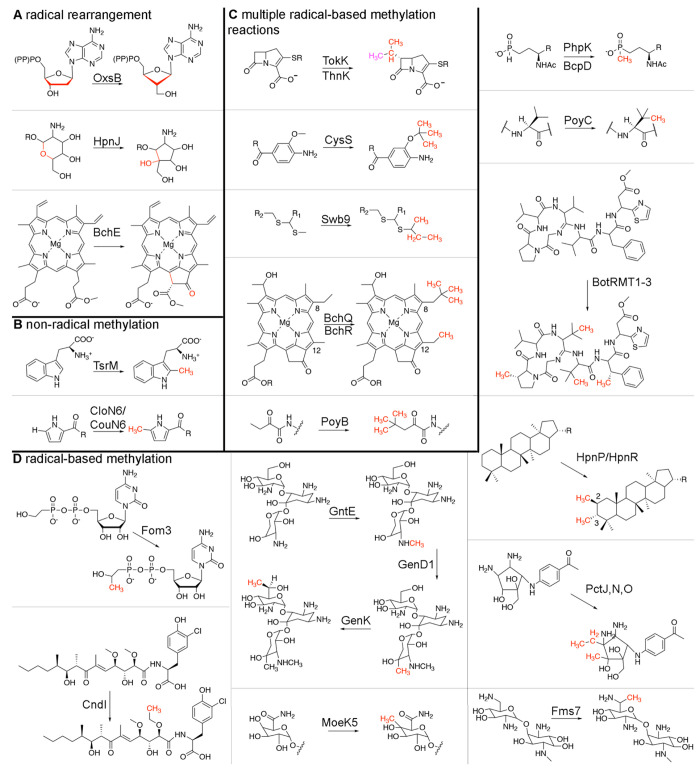
In addition to these three reactions, Cbl-dependent radical SAM enzymes have been linked to many other pathways. Similar to OxsB, both HpnJ and BchE do not catalyze methyl transfer reactions (Figure 2A). HpnJ from Burkholderia cenocepacia is proposed to mediate the contraction of a six-membered oxane ring into a five-membered ring in hopanoid biosynthesis.28 BchE catalyzes both oxidation of the C-131 carbon of Mg-protoporphyrin-IX monomethylester and a subsequent cyclization reaction to form the isocyclic ring of bacteriochlorophyll.29,30 In addition, the Cbl-dependent radical SAM enzyme, Kuste2803, has been identified as a potential catalyst for formation of the cyclobutane rings of ladderane lipids.31 HpnJ, BchE, and the ladderane biosynthetic enzyme cluster together in the main cluster of the SSN away from OxsB (Figure 1B). This distinct clustering of these enzymes (Figure 1B) likely reflects differences in substrates and also in enzyme mechanistic features. Indeed, BchE catalyzes an oxygen insertion in addition to the radical rearrangement reaction,29,30 and HpnJ must contract a six-membered ring of a large bacteriohopanetetrol glucosamine substrate to form the five-membered ring of bacteriohopanetetrol cyclitol ether.28 Regarding the need for Cbl in these reactions, it is known that OxsB requires HOCbl for turnover18 and that defects in Cbl biosynthesis disrupt bacteriochlorophyll formation in two facultative anaerobes that use BchE.29,30 However, the role and type of Cbl for these non-methylase reactions is understudied. Similarly, TsrM is the only member of this enzyme family that is currently known to function without radical chemistry, but CloN6 and CouN6 also methylate sp2-hybridized carbon centers23,32 and thus may catalyze their methylations using a TsrM-like mechanism (Figure 2B).23 Despite the identification of CloN6 and CouN6 in 2003,32 they have not been mechanistically or structurally investigated, leaving many open questions regarding whether this non-radical-based mode of catalysis will be used by additional members of this enzyme class.
Figure 2.
Cbl-dependent radical SAM enzymes catalyze a diverse set of reactions that are important to pathways that produce environmentally and medically relevant compounds. (A) Like OxsB, several Cbl-dependent radical SAM enzymes including HpnJ and BchE function as non-methylases. (B) Like TsrM, CloN6 and CouN6 have been proposed to use their cofactors to catalyze a non-radical-based methylation reaction.23 (C) Analogous to TokK, ThnK,33 CysS,34 Swb9,36 BchQ,37 and PoyB38 have been suggested to catalyze multiple radical-based methylation reactions. In the top panel, two methyl groups are added by ThnK (one to the β-lactam ring and then one to the resultant methyl group, which is shown in pink). TokK, on the other hand, is implicated in forming the appended isopropyl moiety. (D) A large majority of Cbl-dependent radical SAM enzymes are implicated in catalyzing single radical-based methylation reactions.
Akin to the reaction catalyzed by TokK, enzymes that include ThnK,33 CysS,34 PctJ,35 Swb9,36 BchQ,37 and PoyB38 appear to catalyze multiple methylation reactions on a substrate (Figure 2C). ThnK methylates its substrate’s β-lactam ring and then methylates the initially added methyl group,33 and CysS catalyzes iterative methylation reactions on a methyl group to form ethyl-, isopropyl-, sec-butyl-, and tert-butyl-containing cystobactamids.34 These enzymes, along with PctJ and Swb9, cluster together in the SSN (Figure 1B). The physiological substrate of Swb9 is unknown, but it has been proposed to iteratively methylate a quinomcyin precursor to generate ethyl, isopropyl, and sec-butyl moieties.36 PctJ is thought to be involved in a late-stage methylation reaction in pactamycin biosynthesis.35 Outside of this cluster, only two additional proteins, BchQ37 and PoyB,38 have been implicated in catalyzing serial methylation reactions (Figure 1B). BchQ adds one, two, or three methyl groups to the C8-ethyl position of a bacteriochlorophyll c substrate,37 and PoyB has been suggested to form the N-terminal tert-butyl group of polytheonamide A38 (Figure 2C).
Finally, there are also Cbl-dependent enzymes that, like TokK, are implicated in radical-based methylation reactions but only append one methyl group to a substrate (Figure 2D). This category of methylases includes Fom3, CndI, GenK/GntE/GenD1, Fms7, MoeK5, BchR, and HpnP/HpnR, which catalyze methylation of carbon centers in the biosynthesis and tailoring of fosfomycin,39−41 chondrochloren B,42 gentamicin C1,43−45 fortimicin A, moenomycin A,46 bacteriochlorophyll c,37 and hopanoids,47 respectively. Each of these proteins except GenD1, GntE, and CndI cluster together in the SSN (Figures 1B and 2D). A more stringent threshold applied to this cluster reveals colocalization of Fom3, MoeK5, and HpnJ as well as GenK and Fms7 (Figures 1B and 2D). SD1168 is also found in this main cluster and is proposed to catalyze either a C- or P-methylation, but its physiological function and substrate remain unknown.48 Related reactions are also catalyzed by a set of Cbl-dependent radical SAM enzymes found in a distinct cluster that are each involved in amino acid side chain methylation in polytheonamide A (PoyC)38 and bottromycin (BotRMT1, BotRMT2, and BotRMT3) biosynthesis49 (Figure 2D). Likewise, two additional pactamycin biosynthetic genes, PctN and PctO, which have been proposed to catalyze methylation reactions on the cyclopentane ring of a pactamycin precursor,35 are also implicated in catalyzing a single radical-based methylation but cluster in a distinct region of the SSN (Figure 1B). Last, BcpD and PhpK are known as P-methylases.50−52 Thus far, the PhpK homologue from Kitasatospora phosalacinea has been shown to require an electron donor and MeCbl for activity.50 However, the physiological substrate of this enzyme is still in debate17 (Figure 2D).
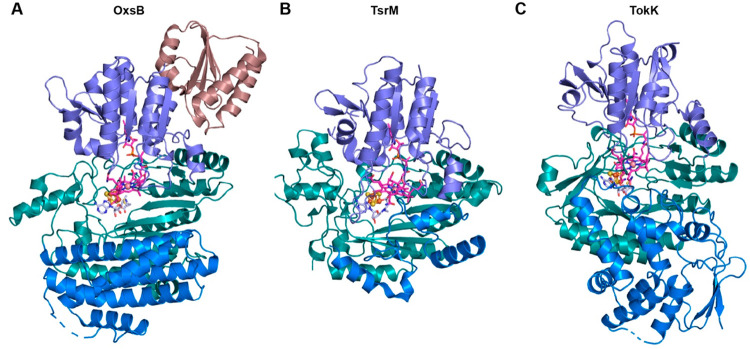
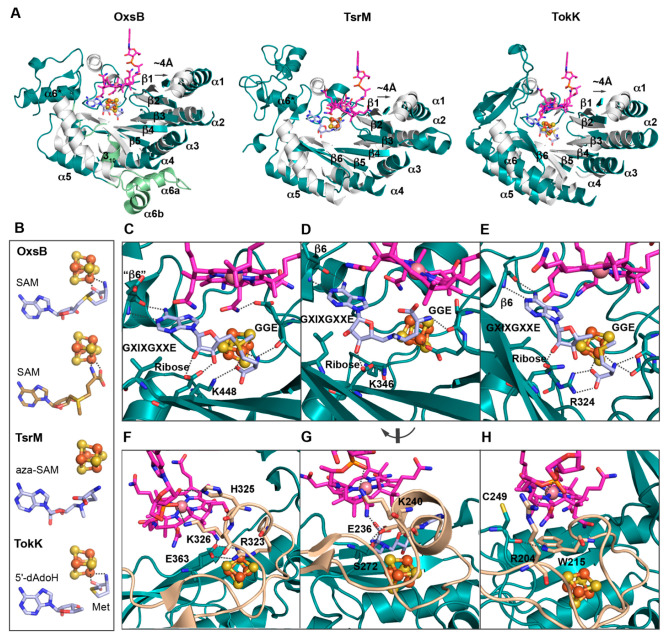
Thus, with just three structures to represent this broad range of divergent enzymes, it is clear that we still have a lot to learn about the structure–function relationships in this enzyme class. However, thus far, the three structures of OxsB, TsrM, and TokK have revealed that these proteins use a modular architecture. They are differentiated by architectural features including an N-terminal domain of unknown function and C-terminal helical bundle in OxsB, as well by the distinct C-terminal domains of TsrM and TokK that are involved in substrate binding (Figure 3). In general, the structures of OxsB, TsrM, and TokK are related by the use of traditional Cbl-binding and radical SAM domains that show subtle variations. As described below, these small changes appear to support different uses of SAM and Cbl and different chemical outcomes.
Figure 3.
The Cbl-dependent radical SAM enzymes OxsB, TsrM, and TokK showcase similar architectures that capitalize on the traditional Cbl-binding and radical SAM domains.18,20,21 (A) OxsB is composed of four domains: an N-terminal domain of unknown function (deep mauve), a Cbl-binding domain (purple), a radical SAM domain (dark teal), and a helix bundle domain (blue). (B) Structure of a homologue of the Streptomyces enzyme TsrM has a domain arrangement similar to that of OxsB but lacks the N-terminal domain and has an abbreviated C-terminal helical bundle domain (blue). (C) TokK also has a domain arrangement similar to that of OxsB but again lacks the N-terminal domain and showcases a new architecture in the C-terminal domain (blue). In all panels, Cbl is shown as pink sticks, SAM (aza-SAM in TsrM and SAM cleavage products in TokK) is shown in light blue sticks, and the radical SAM [4Fe–4S] cluster is shown in orange and brown spheres.
Recycled Cbl-Binding Motif Supports Divergent Reactivity
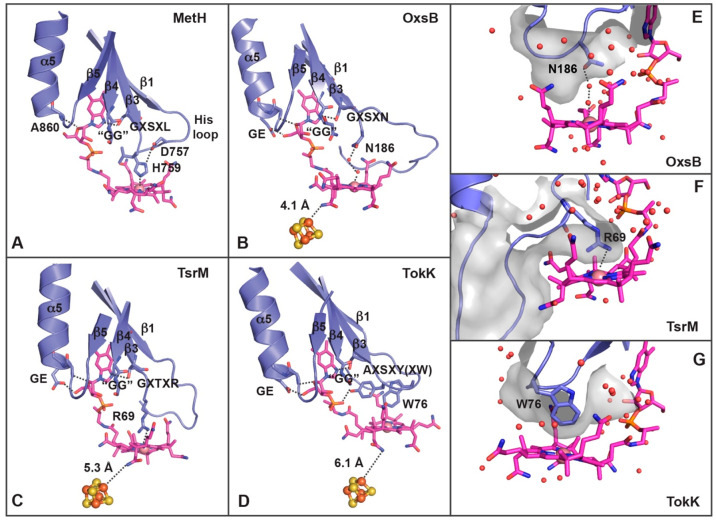
Cbl contains a corrin ring that is decorated with acetamide, methyl, and propionamide groups, as well as a dimethylbenzimidazole (DMB) tail. This DMB tail can serve as a lower axial ligand to Co, an orientation referred to as base-on, or with the DMB tail displaced from Co, which is known as base-off.9,10,18,53 The base-on mode of binding is found in proteins that use variable architectures, whereas most Cbl-dependent enzymes that bind Cbl in the base-off mode have been shown to use a Rossmann fold that consists of a core set of five parallel β-strands that are surrounded by helices (Figure 4A).9,10,18,53 This latter base-off binding mode allows the protein to tune the reactivity of Cbl using the protein environment rather than the DMB tail.53 For example, the lower axial ligand to the Cbl of MetH is a conserved His residue from an Asp757-X-His759-X-X-Gly762 motif54 that is found on a six-residue loop (His loop in Figure 4A).53 This basic His ligand in the His-on state is proposed to stabilize the Me–Co(III) state of MetH and also be able to rearrange to His-off to favor a square-planar Co(I) state.53,55 Additional residues downstream of this motif in the residue sequences Gly802-X-Ser804-X-Leu806 and Gly833-Gly834 are involved in hydrogen bonding with N3 of DMB (Ser804), stacking against the axial His ligand (Leu806), and forming a pocket that accommodates the DMB (Gly833-Gly834 residues) (Figure 4A).53 This base-off, His-on mode is also found in several other Cbl-dependent enzymes.56,57 However, the three structurally defined Cbl-dependent radical SAM enzymes described here bind Cbl in a base-off, His-off mode (Figure 4B–D).18,20,21
Figure 4.
Cbl binds to the Cbl-dependent radical SAM enzymes OxsB, TsrM, and TokK in a base-off, His-off mode. (A) In MetH, the closest ligand to Co is a His residue found on the loop (“His loop”) that connects β1 to α1 in the Cbl-binding domain.53 The residues from the MetH Cbl-binding domain that interact with Cbl are highlighted and shown as sticks, including the lower axial ligand (His759), the GG motif, and the downstream Cbl-binding motif (Gly-X-Ser-X-Leu). (B) The closest residue to Co of Cbl in OxsB is found on the loop that connects β3 to α3 (Asn186).18 Residues from the OxsB Cbl-binding domain that are involved in Cbl-binding are highlighted, including those that make up the GG motif, the downstream Cbl-binding motif, and a GE sequence of residues found on α5.18 The extra length of the “His loop” seems to correlate with the positioning of the corrin ring close to the radical SAM cluster (orange and brown spheres).18 (C) The closest residue to Co of Cbl in TsrM is found on the loop that connects β3 to α3 (Arg69).20 Other residues of interest that interact with Cbl come from the GG motif and the GE sequence found on α5. (D) Like OxsB and TsrM, the closest residue to Co of Cbl in TokK is found in the sequence Ala-X-Ser-X-Tyr-X-Trp.21 Here, Tyr maps to the position of Asn in OxsB and Arg in TsrM, but Trp is the closest protein residue to Co. (E) The lower axial face of Cbl in OxsB is found in a solvent-filled pocket.18 (F) The presence of a large Arg residue on the long loop that connects β3 to α3 in TsrM does not permit solvent to get near the Co ion of Cbl.20 (G) Similar to TsrM, the lower axial face of Cbl in TokK is also solvent-free.21 Panels E–G are each shown with a surface rendering, and water molecules are shown as red spheres.
A comparison of the Cbl-binding domains of OxsB, TsrM, and TokK to MetH reveals that the equivalent loops to MetH’s “His loop” are longer and are notably missing histidine residues. OxsB, TsrM, and TokK appear to modulate the reactivity of the Cbl using distinct residues of motifs that are downstream of MetH’s His motif. In OxsB, the loop connecting β1 to α1 is eight residues longer than that in MetH.18,53 The length of this so-called “His loop” shields the Cbl-binding site and seems to spatially dictate that Cbl sits approximately 4 Å away from the [4Fe–4S] cluster (Figure 4B). The closest protein side chain to Co of Cbl is Asn186, which is located on the loop that connects β3 to α3 in a sequence of residues (Gly182-X-Ser184-X-Asn186) that corresponds to the downstream Cbl-binding motif, for which the sequence in MetH is Gly802-X-Ser804-X-Leu806.18,53 Ser residues of this motif hydrogen bond to the DMB of Cbl. Whereas Leu806 in MetH stacks against the His ligand to the Co ion, Asn186 of OxsB is part of a hydrogen-bonding network that anchors a water molecule in place to serve as the lower axial ligand of Cbl (Figure 4B).53 A similar repurposing of this downstream Cbl-binding motif is also noted in the structures of TsrM and TokK. In TsrM and TokK, the “His loop” is 11 amino acids long, and the closest ligands to Cbl arise from the loop that connects α3 and β3 (Figure 4C,D). In TsrM, the closest residue to Co of Cbl is Arg69 (Gly65-X-Thr67-X-Arg69)20 of the downstream Cbl-binding motif, and the corrin ring is located 5.3 Å away from the [4Fe–4S] cluster (Figure 4C). In TokK, the closest residue is Trp76 (Ala-70-X-Ser72-X-Tyr74-X-Trp76)21 of the downstream Cbl-binding motif, and Cbl is approximately 6 Å away from the [4Fe–4S] cluster (Figure 4D). These large bulky ligands in TsrM and TokK, unlike Asn186 in OxsB, create an environment that occludes solvent from nearing the Co center of Cbl (Figure 4E–G).18,20,21
The lower Cbl environment in TsrM is reminiscent of that observed in the RNA modification enzyme QueG, where water coordination at the lower axial Co position is also prevented by a noncoordinating Arg side chain.58 However, the reactions catalyzed are very different: QueG catalyzes an epoxide reduction in queuosine biosynthesis and TsrM is proposed to facilitate a non-radical-based methylation. In both cases, the lack of a lower ligand is expected to facilitate a transition into the preferred four-coordinate geometry for formation of a Co(I)–Cbl species. This ligand environment in QueG is proposed to allow for formation of Co(I)–Cbl and a subsequent Cbl-tRNA intermediate.58 In TsrM, this environment instead appears to be a design strategy used to promote transfer of a methyl cation to substrate, permitting cycling between Me–Co(III)–Cbl and Co(I)–Cbl states.59 This proposal was recently described by Booker and co-workers who posit that Arg69 weakens the Co–C bond and favors nucleophilic displacement of the methyl group by the Trp substrate.20,23 In TokK, Trp76 should also disfavor formation of a hexacoordinate Cbl species. However, unlike TsrM, where substitution of the Arg residue results in substantial activity loss, there is more flexibility in the identity of the residue in this position in TokK.21 This flexibility was recently demonstrated through mutagenesis studies that showed that substitution of Trp76 in TokK with Phe, Ala, or His had only a minor effect on activity.21 This flexibility was not without limit, however. A Trp76Lys TokK variant, which contains a longer positively charged residue than His and more closely resembles TsrM, showed activity lower than that of any other tested variants.21 Although some substitutions at the lower axial position of the Cbl may be tolerated, collectively these results highlight the importance of local environment in the tuning of Cbl chemistry to afford different reactions.
Finally, OxsB, TsrM, and TokK all contain a conserved set of protein residues that accommodate and interact with the DMB tail of Cbl. As observed in MetH,53 each of these proteins contains one or multiple Gly residues on β4 of the Cbl-binding domain that form a pocket to house the DMB and ribose of Cbl (Figure 4A–D). This motif for base-off Cbl binding is known as the Gly-Gly motif. In all three structures, a Gly-Glu residue sequence at the start of α5 in the Cbl-binding domain is also present. These residues engage in two hydrogen bonds with the tail of Cbl (Figure 4B–D). This interaction (“GE”), along with the longer “His loop” and the use of the downstream Cbl-binding motif, appears to support the different reactivity of these enzymes and the close juxtaposition of Cbl with the [4Fe–4S] cluster and SAM found in the adjacent radical SAM domain (Figure 4B–D).18
Re-engineered Barrel Architecture Allows for the Juxtaposition of SAM and Cbl
Partial (β/α)6 and full (β/α)8 triose phosphate isomerase (TIM) barrel folds are common to both radical SAM and Cbl-dependent proteins.9,10 These enzymes use barrels to bind their cofactors and substrates and perform their reactions in an isolated environment. By sequestering reactive intermediates within the protein architecture, these folds mitigate the risk of propagating radical chemistry into the cell.10 In Cbl-dependent radical SAM enzymes, as first visualized in the structure of OxsB,18 the barrel fold is re-engineered via outward movement of each of the β-strands in the barrel not only to bind the radical SAM machinery but also to accommodate the corrin ring of Cbl (Figure 5A).10,18 The [4Fe–4S] cluster is ligated directly to Cys residues found in a Cys-X3-Cys-X2-Cys motif that follows the first β-strand of the TIM barrel.60,61 Together with SAM, the [4Fe–4S] cluster occupies a traditional C-terminal position in the barrel (Figure 5). Cbl, from the Cbl-binding domain, also sits within the interior of the barrel but is rotated 90° from its traditional position (Figure 5A).18 The preferential placement of the radical SAM machinery in this position and the conservation among the Cbl-dependent radical SAM enzymes discussed here suggest that these enzymes are radical SAM enzymes that have been repurposed to also bind Cbl.10,18 Importantly, this arrangement creates an active site within the barrel architecture where a substrate can bind and react with both SAM and Cbl (Figures 3–5).18,20,21
Figure 5.
Traditional SAM-binding motifs interact with Cbl. (A) Despite housing the corrin ring of Cbl (pink sticks) in the radical SAM domain (dark cyan), OxsB, TsrM, and TokK18,20,21 retain the traditional partial (β/α)6 TIM barrel fold albeit with some deviations relative to the prototypic radical SAM enzyme PFL-AE (PDB: 3CB8,62 gray). (B) The structure of OxsB has two orientations of SAM bound, one that appears competent for radical chemistry (top, light blue) and one that does not (bottom, brown). The aza-SAM in TsrM binds in a remarkably different orientation that does not coordinate the [4Fe–4S] cluster (light blue). TokK was crystallized with SAM cleavage products (light blue). (C) OxsB has all characteristic radical SAM-binding motifs, including the GGE (Glu363), ribose (Glu436), GXIXGXXE (Ile474), and the β6 motif (Glu545).18 In OxsB, the GGE motif interacts with SAM, Cbl, and R323 (not shown). Following a short α4a helix that connects β4 and α4, Lys448 interacts with SAM.18 (D) TsrM also contains the traditional SAM-binding motifs, including the GGE motif (Asp-271-Ser272-Glu273), ribose (Asp335), GXIXGXXE (Leu379), and the β6 motif (Arg412). Lys346, which follows α4a, also interacts with SAM. The GGE motif in TsrM serves as a ligand to the radical SAM cluster in lieu of SAM.20 (E) Radical SAM motifs can also be identified in TokK, including the GGE motif (Ala251-Asn252), ribose (Gln312), GXIXGXXE (Ile351), and the β6 motif (Gly384). Arg234, which follows α4a, coordinates to SAM. (F) In OxsB, residues from the Cys loop (wheat) coordinate the [4Fe–4S] cluster, interact with the GGE motif (Arg323), and interact with Cbl (His325-Lys326).18 (G) In TsrM, residues in the Cys loop bind the [4Fe–4S] cluster and interact with SAM (Glu236), Cbl (Glu 236 and Lys240), and the GGE motif (Glu236). (H) Finally, Cys loop residues in TokK interact with Cbl (Arg204) and stack between cofactors (Trp215).
Similar to how the structure of OxsB uses architectural features found in the Cbl-dependent protein MetH, the radical SAM domain of OxsB resembles that of the prototypical radical SAM enzyme PFL-AE.18 Differences between OxsB and PFL-AE structures include the presence of three extra helices following α5 in OxsB, a long loop in place of β6, and a CX4CX2C cluster-binding motif rather than the typical CX3CX2C motif (Figure 5A).18 Subsequently determined structures of TsrM20 and TokK21 show broadly similar domain architectures and cofactor placement to OxsB. Interestingly, TsrM and TokK do not share the differences noted above between OxsB and PFL-AE (Figure 5A–C).
Repurposed Classical Radical SAM-Binding Motifs Give Rise to New Function
In the majority of radical SAM enzymes, three irons of the [4Fe–4S] cluster are coordinated by cysteines of the Cys-X3-Cys-X2-Cys motif, and the fourth iron is coordinated by the amino and carboxylate moieties of SAM. SAM is further positioned in the active site by residues of the partial TIM barrel that can include a polar residue from the α4a helix in addition to a series of classic SAM-binding motifs: GGE, ribose, GXIXGXXE, and the β6 motif.60,61 Below, we consider a subset of these motifs that appear to be the most altered when OxsB, TsrM, and TokK are compared to non-Cbl-dependent radical SAM enzymes. Although structural data on SAM binding is limited to OxsB, we can also utilize a TsrM structure that has the SAM analogue, S-5′-azamethionine-5′-deoxyadenosine (aza-SAM) bound, and a TokK structure that is complexed with SAM cleavage products, Met and 5′-dAdoH (Figure 5A,B) in our analyses.
The most interesting variation in a motif is found in the so-called GGE motif. The GGE motif, for which the residue identity varies, traditionally provides a backbone hydrogen bond to the methionyl amino group of SAM (Figure 5C–E). However, in OxsB, the GGE motif (Ala361-Asp362-Glu363) also helps to secure structural elements around Cbl. Glu363 hydrogen bonds to a Cbl acetamide18 and forms a salt bridge to Arg323, a residue that is downstream of the CX4CX2C motif (Figure 5C,F). In TsrM, the side chain of Glu273 of the GGE motif (Asp271-Ser272-Glu273) ligates the unique iron of the [4Fe–4S] cluster instead of SAM ligating the cluster (Figure 5D), representing a substantial departure from other radical SAM enzymes. In TokK, the GGE motif (Asp250-Ala251-Asn252) appears more traditional in function if not in sequence, serving only to hydrogen bond with the methionyl moiety of SAM and to orient SAM for reductive cleavage (Figure 5E). Notably, SAM binds in two orientations in OxsB (Figure 5B): SAM binds to the [4Fe–4S] cluster in the canonical orientation as well as in a second orientation that loses the GGE interaction and does not appear competent to generate 5′-dAdo• due to an increased distance between the S atom of SAM and the [4Fe–4S] cluster (Figure 5B).18 This second orientation instead seems poised to methylate Cbl and could represent an evolutionary relic of the superfamily, simply retained as an orientation that could be sampled to prevent formation of 5′-dAdo• in the absence of a substrate.18 Intriguingly, recent work has shown that OxsB can convert both SAM into S-adenosylhomocysteine (SAH) and dAMP into a methylated derivative.19 These results suggest that the latter SAM binding mode may be competent to form MeCbl and/or a methylated product.19 However, the mechanism by which this methylated compound is formed, the structure of this compound, and the relevance of this compound (if any) to OXT-A biosynthesis await further discovery.19
The Cys loops of OxsB, TsrM, and TokK are also deserving of comment as they have modifications that allow residues of these loops to interact with the Cbl cofactors. In OxsB, two residues from the Cys loop, His325 and Lys326, are positioned to interact with Cbl (Figure 5F).18 Likewise, in TsrM and TokK, Cys loop residues Lys240 and Arg204 interact with the Cbl DMB tail (Figure 5G,H). In addition, each protein has a residue in the Cys loop, located two residues downstream of the Cys motif (Cys-X3–4-Cys-X2-Cys-XX) that appears to help communication between SAM and Cbl. In OxsB, this residue is Arg323, which as mentioned above interacts with Glu363 from the GGE motif (Figure 5F). In TsrM, this residue corresponds to Glu236, which like OxsB’s GGE motif residue Glu363, serves to hold SAM and Cbl near each other by interacting with both the amino group of SAM and an acetamide side chain of Cbl (Figure 5G). Finally, in TokK, residue Trp215 is found at the analogous position and is sandwiched between the [4Fe–4S] cluster and Cbl (Figure 5H). Based on the placement of this residue and experiments that show mutation of Trp215 to Phe, Ala, or Tyr markedly affects product formation,21 it is tempting to consider that this Trp residue could serve as a conduit for electron transfer from the [4Fe–4S] cluster to Co(II)–Cbl. This electron transfer step would generate the needed Co(I) species for remethylation of the cofactor.
A final structural commonality between OxsB, TsrM, and TokK is the presence of an interaction between a polar residue from the α4a helix of the radical SAM domain and SAM. This same interaction is observed in the radical SAM enzymes BioB,6 BtrN,63 and QueE.64 In OxsB and TokK, residues Lys448 and Arg324, respectively, interact with the carboxylate of SAM (Figure 5C,E).18,21 In TsrM, Lys346 from this protein region instead interacts with the ribosyl portion of SAM via ribose motif residue Asp33520 (Figure 5D).
Collectively, these observations suggest that the more divergent chemical roles played by members of the Cbl-dependent radical SAM enzymes are supported, at least in part, by repurposing the canonical radical SAM architecture. In particular, the use of the Glu of the GGE to secure Cbl close to the radical SAM machinery in OxsB, potentially aiding in electron transfer between cofactors (Figure 1C), is an appealing adaptation.18 In TsrM, the use of the Glu of the GGE motif to coordinate the unique iron of the cluster is a noteworthy modification that likely blocks SAM from binding the [4Fe–4S] cluster, explaining the lack of radical chemistry performed by TsrM (Figure 1C).20
C-Terminal Cap Domain is Involved in Substrate-Binding Conformational Change
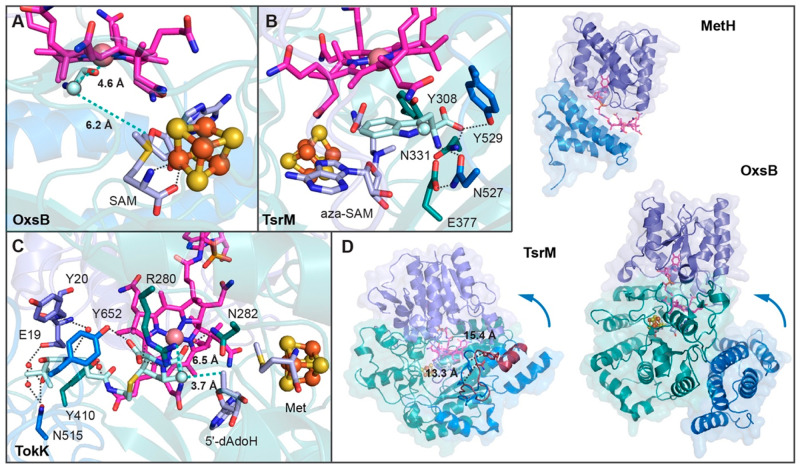
As described above, the arrangement of Cbl-binding and radical SAM domains in OxsB, TsrM, and TokK effectively creates an active site where a substrate can bind and be positioned to react with both SAM and Cbl (Figure 6). However, for OxsB, there is not a substrate-bound structure available.18 The inability to obtain a substrate-bound structure has been suggested to be due to the “open conformation” assumed by OxsB. In this conformation, the C-terminal helical bundle domain is far from Cbl, and the active site is filled with solvent.18 As this orientation would not be conducive to radical chemistry, it has been hypothesized that a conformational change that closes off the active site would correctly position the needed residues for substrate binding (Figure 6D).18 Nevertheless, mapping of the substrate position from PFL-AE onto the active site of OxsB reveals a location where substrate could bind and be poised equidistant between the 5′-carbon of SAM and Co of Cbl (Figure 6A).18 Consistent with the need for a conformational change to mediate the correct contacts with substrate, the structure of substrate-bound TsrM revealed that substrate binding is accompanied by a sizable 16 Å movement of a C-terminal domain loop to interact with substrate and close off the active site (Figure 6D).20 One interesting caveat of this structure, however, is that the substrate does not appear bound in a catalytically competent orientation.20 This unproductive orientation is proposed to be due to the presence of aza-SAM and HOCbl in the active site rather than the native SAM and MeCbl cofactors. Even so, this structure coupled with docking studies establishes a location for the active site that is similar to the one proposed for OxsB, where a substrate could bind to react with both cofactors (Figure 6B).20 For TokK, there is a substrate-bound structure that shows the carbapenam in a catalytically competent orientation (Figure 6C).21 The substrate is making contacts with residues from both the radical SAM domain and C-terminal domain. This structure provides an explanation for the observed nonprocessive nature of the TokK catalyzed reaction in that a new molecule of SAM cannot replace SAM cleavage products Met and 5′-dAdoH to afford the second methylation while substrate is bound.21
Figure 6.
Substrate binding in Cbl-dependent radical SAM enzymes requires residues in the radical SAM and C-terminal domains. (A) Modeling of the PFL-AE peptide substrate (cyan) into the active site of OxsB establishes a position where substrate could bind to react with SAM and Cbl. (B) Both the C-terminal domain (blue) and radical SAM domain (dark cyan) comprise the substrate binding pocket in TsrM as mapped out by a structure with substrate bound in a nonproductive orientation (cyan). (C) Like TsrM, the substrate of TokK (cyan) is tethered in place by interactions with residues from the C-terminal (blue) and radical SAM (dark cyan) domains. (D) MetH has a Cbl-binding domain and a helix bundle domain.53 The helix bundle domain acts as a cap that protects MeCbl from being photolyzed and/or demethylated by an adventitious biomolecule.68 Like MetH, OxsB has a Cbl-binding and a helix bundle domain (the N-terminal domain is omitted in this panel for clarity). However, in the current structure of OxsB, the helix bundle domain is displaced from Cbl, and the active site is open. Although some movement of the bundle domain was observed in crystallo,18 it has been proposed that additional movement is likely to occur that will facilitate substrate binding and catalysis. A C-terminal movement of TsrM residues Asn257 and Tyr259 found on a loop toward the substrate is observed through comparison of the structures determined in the presence (blue) and absence (dark red) of substrate.20
Collectively, conformational changes would seem to be required for substrate binding and/or product departure in all three of the structurally characterized Cbl-dependent radical SAM proteins.18,20,21 The region that moves in OxsB and TsrM appears to be the helical C-terminal domain (Figure 6D). Interestingly, the use of a helical domain to alter the accessibility of the upper face of Cbl is out of the MetH playbook (Figure 6D).53 In MetH, a helical bundle domain protects the Cbl in the resting state68 and must move to allow for catalysis. Although domain arrangements and exact domain topologies are not strictly conserved, these Cbl-dependent enzymes do appear to employ variations on a molecular theme to afford sequestration and thus protection of their Cbl cofactors during turnover.
Outlook
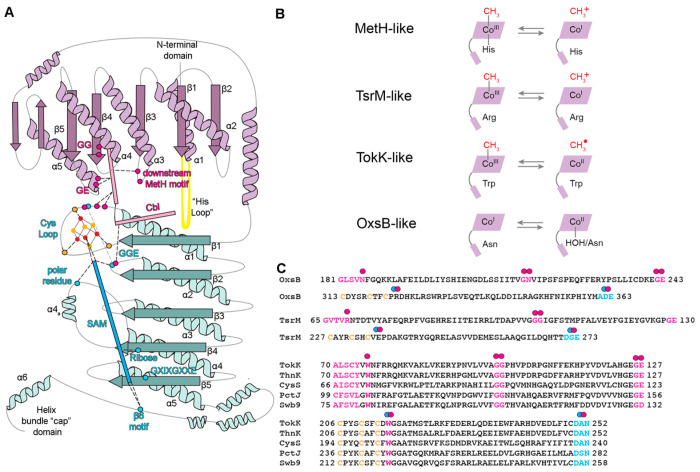
Twenty years ago, Sofia et al.1 used bioinformatics analysis to put forth the proposal that a subset of radical SAM enzymes would also be Cbl-dependent. This proposal was based on the presence of sequence motifs that were similar to the His-on/base-off Cbl-binding sequence motifs of MetH (Asp-X-His-X-X-Gly, Gly-X-Ser-X-Leu, Gly-Gly). With the recent structure determinations of OxsB, TsrM, and TokK, we now have experimental evidence that these residues identified as having a binding Cbl function do, in fact, bind Cbl and that the Cbl is base-off. Interestingly, the His loop using sequence Asp-X-His-X-X-Gly, which was so important to Sofia et al.’s prediction, turned out to be a bit of a red herring. Although it is quite possible that some family members will employ a His-on Cbl, we suspect that in the majority of cases, the “His loop” will play a role in orienting the Cbl cofactor close to the radical SAM cluster rather than in providing a histidine to ligate the Cbl. We further anticipate that “Gly-X-Ser-Leu” or the “downstream Cbl-binding motif” that provides Asn186/H2O as a ligand to the Co ion in OxsB, provides Arg69 in TsrM, and provides Trp76 in TokK will be a hotspot for substitutions that customize the chemistry of these enzymes (Figure 7A,B). It has long been known that strength of the Me–Co bond and the propensity for homolytic versus heterolytic cleavage can be modulated by the identity of the lower ligand, making substitutions in the “downstream Cbl-binding motif” potentially chemically determinative. It is incredible that the three structures determined thus far all appear to be in different subclasses (Figure 7B), raising the question of how many additional substitutions we should expect. The Gly-Gly motif predicted by Sofia et al. to be a base-off Cbl motif was correct. Outside the scope of the Sofia et al. paper, the structure of OxsB revealed another motif for Cbl-binding: a Gly-Glu motif found in α5 of the Cbl-binding domain and appears to aid in the relative positioning of the radical SAM and Cbl cofactors (Figure 7A,C). Finally, additional hotspots for chemistry-customizing substitutions can be found in the radical SAM domain, including the Cys loop and the GGE motif (Figure 7A,C).
Figure 7.
Hotspots in Cbl-dependent radical SAM enzymes for binding cofactors, tethering Cbl and SAM close together, and influencing reactivity. (A) Topology diagram of OxsB that shows the Cbl-binding (purple) and radical SAM domains (teal) that are also found in the other structurally characterized enzymes TsrM and TokK. The C-terminal “cap” domain is likely involved in conformational changes related to substrate binding.20,21 Residues involved in coordinating or creating space for Cbl are highlighted as pink spheres, residues involved in interacting with SAM are shown as blue spheres, and the Cys residues involved in coordinating the [4Fe–4S] cluster are shown as orange spheres. The location of the Cbl domain “His loop” is shown in yellow. As described in the text, the downstream Cbl-binding motif residues interact with Cbl and maintain the environment of the lower axial face. The “His loop” and the GE residues help spatially position Cbl. The radical SAM GGE motif serves additional purposes in OxsB and TsrM20 but is more traditional in TokK. Residues from the Cys loop bind the cluster, interact with Cbl, and facilitate communication between the cofactors. (B) Different environments of Cbl facilitate formation of the needed oxidation states for catalysis. There are currently four definable classes including MetH. (C) Downstream Cbl-binding, GG, and GE motifs are labeled in OxsB and TsrM from left to right in pink (top OxsB/TsrM sequences). In addition, the Cys loop (Cys-X3–4-Cys-X2-Cys-XX) and GGE radical SAM motifs are labeled in blue (bottom OxsB/TsrM sequences). A sequence alignment of TokK, ThnK, CysS, Swb9, and PctJ reveals conservation of the downstream Cbl-binding, GG, GE, Cys loop, and GGE motifs. All pink and blue dots correspond to the topology diagram shown in panel A. A double dot is used for motifs that facilitate communication between cofactors.
Specific predictions based on sequence can also be made using the newly available structures.18,20,21 As described by Booker and co-workers,21 ThnK and CysS have sequence motifs in common with TokK (Figure 7C) that suggest that a Trp residue will block ligation to the Co ion of Cbl in those enzymes as Trp76 does in TokK (Figure 4D). We can also predict that ThnK, CysS, PctJ, and Swb9 will use the “GGE motif” as traditionally employed by radical SAM enzymes, assisting in SAM ligation to the cluster. This prediction is based on the conservation of the TokK sequence, Asp250-Ala251-Asn252 (Figure 7C), which occupies the location of the “GGE” motif in the TokK structure. We additionally predict conservation of TokK’s Trp215 in ThnK, CysS, PctJ, and Swb9. Trp215 is one residue downstream of the Cys loop motif (Cys-X3-Cys-X2-Cys-X-Trp), and this Trp stacks between the radical SAM cluster and Cbl cofactor (Figure 7C). Swb9, like TokK, ThnK, and CysS, is implicated in catalyzing multiple methylation reactions.36 Currently, the reaction catalyzed by PctJ is unknown, but based on residue conservation described here and PctJ’s location in the SSN (Figure 1B), we suspect that PctJ will catalyze multiple methylation reactions.
Additional sequence alignments of OxsB and TsrM with enzymes that we categorize as nonradical methylases or non-methylases suffer from low sequence similarity, making it difficult to pinpoint residues in our predicted hotspots. Thus, despite the invaluable contributions that the structures of OxsB, TsrM, and TokK have made to our understanding of how Cbl-dependent radical SAM enzymes function, it is clear that we still have a lot to learn. We suspect, in fact, that we have only scratched the surface in our understanding of this enzyme family. The uncharted regions of the SSN (Figure 1) and the recent identification of family member Mmp10, which binds Cbl in its C-terminal domain (rather than the N-terminal domain),65 are indicative that additional studies are needed. Future mechanistic and structural characterization of Cbl-dependent radical SAM enzymes will be essential and will likely lead to new insights into the repurposing of traditional motifs to give rise to new functions.
Acknowledgments
This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Award Number DE-SC0021240 (J.B.R.), and the National Institutes of Health [R35 GM126982 (C.L.D.)]. C.L.D. is a HHMI Investigator, and J.B.R. is a Searle Scholar.
The authors declare no competing financial interest.
References
- Sofia H. J.; Chen G.; Hetzler B. G.; Reyes-Spindola J. F.; Miller N. E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001, 29 (5), 1097–1106. 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick J. B.; Duffus B. R.; Duschene K. S.; Shepard E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114 (8), 4229–4317. 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiva E.; Brown S.; Almonacid D. E.; Barber A. E. 2nd; Custer A. F.; Hicks M. A.; Huang C. C.; Lauck F.; Mashiyama S. T.; Meng E. C.; Mischel D.; Morris J. H.; Ojha S.; Schnoes A. M.; Stryke D.; Yunes J. M.; Ferrin T. E.; Holliday G. L.; Babbitt P. C. The Structure Function Linkage Database. Nucleic Acids Res. 2014, 42, D521–D530. 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisler K. A.; Hutcheson R. U.; Horitani M.; Duschene K. S.; Crain A. V.; Byer A. S.; Shepard E. M.; Rasmussen A.; Yang J.; Broderick W. E.; Vey J. L.; Drennan C. L.; Hoffman B. M.; Broderick J. B. Monovalent Cation Activation of the Radical SAM Enzyme Pyruvate Formate-Lyase Activating Enzyme. J. Am. Chem. Soc. 2017, 139 (34), 11803–11813. 10.1021/jacs.7b04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M. I.; Lanz N. D.; Goldman P. J.; Lee K. H.; Booker S. J.; Drennan C. L. Crystallographic snapshots of sulfur insertion by lipoyl synthase. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (34), 9446–9450. 10.1073/pnas.1602486113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovitch F.; Nicolet Y.; Wan J. T.; Jarrett J. T.; Drennan C. L. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 2004, 303 (5654), 76–79. 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D. H.; Basu M. K. Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 2011, 193 (11), 2745–2755. 10.1128/JB.00040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft D. H. Bioinformatic evidence for a widely distributed, ribosomally produced electron carrier precursor, its maturation proteins, and its nicotinoprotein redox partners. BMC Genomics 2011, 12, 21. 10.1186/1471-2164-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D. P.; Croft A. K.; Drennan C. L. Radical use of Rossmann and TIM barrel architectures for controlling coenzyme B12 chemistry. Annu. Rev. Biophys 2012, 41, 403–427. 10.1146/annurev-biophys-050511-102225. [DOI] [PubMed] [Google Scholar]
- Bridwell-Rabb J.; Grell T. A.J.; Drennan C. L. A Rich Man, Poor Man Story of S-Adenosylmethionine and Cobalamin Revisited. Annu. Rev. Biochem. 2018, 87, 555–584. 10.1146/annurev-biochem-062917-012500. [DOI] [PubMed] [Google Scholar]
- Bridwell-Rabb J.; Drennan C. L. Vitamin B12 in the spotlight again. Curr. Opin. Chem. Biol. 2017, 37, 63–70. 10.1016/j.cbpa.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson H. J.; Morris J. H.; Ferrin T. E.; Babbitt P. C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 2009, 4 (2), e4345. 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt J. A.; Bouvier J. T.; Davidson D. B.; Imker H. J.; Sadkhin B.; Slater D. R.; Whalen K. L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854 (8), 1019–1037. 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R.; Oberg N.; Gerlt J. A. The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019, 58 (41), 4169–4182. 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallot R.; Oberg N. O.; Gerlt J. A. ’Democratized’ genomic enzymology web tools for functional assignment. Curr. Opin Chem. Biol. 2018, 47, 77–85. 10.1016/j.cbpa.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk A. J.; Wang R. X.; Booker S. J. TsrM as a Model for Purifying and Characterizing Cobalamin-Dependent Radical S-Adenosylmethionine Methylases. Methods Enzymol 2017, 595, 303–329. 10.1016/bs.mie.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz N. D.; Blaszczyk A. J.; McCarthy E. L.; Wang B.; Wang R. X.; Jones B. S.; Booker S. J. Enhanced Solubilization of Class B Radical S-Adenosylmethionine Methylases by Improved Cobalamin Uptake in Escherichia coli. Biochemistry 2018, 57 (9), 1475–1490. 10.1021/acs.biochem.7b01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridwell-Rabb J.; Zhong A.; Sun H. G.; Drennan C. L.; Liu H.-w. A B12-dependent radical SAM enzyme involved in oxetanocin A biosynthesis. Nature 2017, 544 (7650), 322–326. 10.1038/nature21689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong A.; Lee Y. H.; Liu Y. N.; Liu H. W. Biosynthesis of Oxetanocin-A Includes a B12-Dependent Radical SAM Enzyme That Can Catalyze both Oxidative Ring Contraction and the Demethylation of SAM. Biochemistry 2021, 60 (7), 537–546. 10.1021/acs.biochem.0c00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox H. L.; Chen P. Y.; Blaszczyk A. J.; Mukherjee A.; Grove T. L.; Schwalm E. L.; Wang B.; Drennan C. L.; Booker S. J. Structural basis for non-radical catalysis by TsrM, a radical SAM methylase. Nat. Chem. Biol. 2021, 17 (4), 485–491. 10.1038/s41589-020-00717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox H. L.; Sinner E. K.; Townsend C. A.; Boal A. K.; Booker S. J.. Structural basis for carbapenam C-alkylation by TokK, a B12-dependent radical SAM enzyme. bioRxiv 2021, https://www.biorxiv.org/content/10.1101/2021.07.10.451909v1 (accessed 2022-01-19). [Google Scholar]
- Bridwell-Rabb J.; Kang G.; Zhong A.; Liu H. W.; Drennan C. L. An HD domain phosphohydrolase active site tailored for oxetanocin-A biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (48), 13750–13755. 10.1073/pnas.1613610113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaszczyk A. J.; Wang B.; Silakov A.; Ho J. V.; Booker S. J. Efficient methylation of C2 in l-tryptophan by the cobalamin-dependent radical S-adenosylmethionine methylase TsrM requires an unmodified N1 amine. J. Biol. Chem. 2017, 292 (37), 15456–15467. 10.1074/jbc.M117.778548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner E. K.; Lichstrahl M. S.; Li R.; Marous D. R.; Townsend C. A. Methylations in complex carbapenem biosynthesis are catalyzed by a single cobalamin-dependent radical S-adenosylmethionine enzyme. Chem. Commun. (Camb) 2019, 55 (99), 14934–14937. 10.1039/C9CC07197K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Donk W. A. Rings, radicals, and regeneration: The early years of a bioorganic laboratory. J. Org. Chem. 2006, 71 (26), 9561–9571. 10.1021/jo0614240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodyer R. D.; Li G.; Zhao H.; van der Donk W. A. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem. Commun. (Camb) 2007, 4, 359–361. 10.1039/B614678C. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; van der Donk W. A.; Liu W. Radical-mediated enzymatic methylation: a tale of two SAMS. Acc. Chem. Res. 2012, 45 (4), 555–564. 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerk C. L.; Welander P. V.; Hamad M. A.; Bain K. L.; Bernards M. A.; Summons R. E.; Valvano M. A. Elucidation of the Burkholderia cenocepacia hopanoid biosynthesis pathway uncovers functions for conserved proteins in hopanoid-producing bacteria. Environ. Microbiol 2015, 17 (3), 735–750. 10.1111/1462-2920.12509. [DOI] [PubMed] [Google Scholar]
- Gough S. P.; Petersen B. O.; Duus J. O. Anaerobic chlorophyll isocyclic ring formation in Rhodobacter capsulatus requires a cobalamin cofactor. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (12), 6908–6913. 10.1073/pnas.97.12.6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchane S.; Steunou A. S.; Picaud M.; Astier C. Aerobic and anaerobic Mg-protoporphyrin monomethyl ester cyclases in purple bacteria: a strategy adopted to bypass the repressive oxygen control system. J. Biol. Chem. 2004, 279 (8), 6385–6394. 10.1074/jbc.M309851200. [DOI] [PubMed] [Google Scholar]
- Rattray J. E.; Strous M.; Op den Camp H. J.; Schouten S.; Jetten M. S.; Damste J. S. A comparative genomics study of genetic products potentially encoding ladderane lipid biosynthesis. Biol. Direct 2009, 4, 8. 10.1186/1745-6150-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich L.; Heide L.; Li S. M. CloN6, a novel methyltransferase catalysing the methylation of the pyrrole-2-carboxyl moiety of clorobiocin. Chembiochem 2003, 4 (8), 768–773. 10.1002/cbic.200300609. [DOI] [PubMed] [Google Scholar]
- Marous D. R.; Lloyd E. P.; Buller A. R.; Moshos K. A.; Grove T. L.; Blaszczyk A. J.; Booker S. J.; Townsend C. A. Consecutive radical S-adenosylmethionine methylations form the ethyl side chain in thienamycin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (33), 10354–10358. 10.1073/pnas.1508615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Schnell B.; Baumann S.; Muller R.; Begley T. P. Biosynthesis of Branched Alkoxy Groups: Iterative Methyl Group Alkylation by a Cobalamin-Dependent Radical SAM Enzyme. J. Am. Chem. Soc. 2017, 139 (5), 1742–1745. 10.1021/jacs.6b10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo F.; Zhang J.; Sato S.; Hirayama A.; Eguchi T. Functional Characterization of 3-Aminobenzoic Acid Adenylation Enzyme PctU and UDP-N-Acetyl-d-Glucosamine: 3-Aminobenzoyl-ACP Glycosyltransferase PctL in Pactamycin Biosynthesis. Chembiochem 2019, 20 (19), 2458–2462. 10.1002/cbic.201900239. [DOI] [PubMed] [Google Scholar]
- Watanabe K.; Hotta K.; Nakaya M.; Praseuth A. P.; Wang C. C.; Inada D.; Takahashi K.; Fukushi E.; Oguri H.; Oikawa H. Escherichia coli allows efficient modular incorporation of newly isolated quinomycin biosynthetic enzyme into echinomycin biosynthetic pathway for rational design and synthesis of potent antibiotic unnatural natural product. J. Am. Chem. Soc. 2009, 131 (26), 9347–9353. 10.1021/ja902261a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew A. G. M.; Frigaard N.-U.; Bryant D. A. Bacteriochlorophyllide c C-8(2) and C-12(1) methyltransferases are essential for adaptation to low light in Chlorobaculum tepidum. J. Bacteriol. 2007, 189 (17), 6176–6184. 10.1128/JB.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent A.; Guillot A.; Benjdia A.; Chartier G.; Leprince J.; Berteau O. The B12-Radical SAM Enzyme PoyC Catalyzes Valine Cbeta-Methylation during Polytheonamide Biosynthesis. J. Am. Chem. Soc. 2016, 138 (48), 15515–15518. 10.1021/jacs.6b06697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S.; Kudo F.; Kim S.-Y.; Kuzuyama T.; Eguchi T. Methylcobalamin-dependent radical SAM C-methyltransferase Fom3 recognizes cytidylyl-2-hydroxyethylphosphonate and catalyzes the nonstereoselective C-methylation in fosfomycin biosynthesis. Biochemistry 2017, 56 (28), 3519–3522. 10.1021/acs.biochem.7b00472. [DOI] [PubMed] [Google Scholar]
- Allen K. D.; Wang S. C. Initial characterization of Fom3 from Streptomyces wedmorensis: The methyltransferase in fosfomycin biosynthesis. Arch. Biochem. Biophys. 2014, 543, 67–73. 10.1016/j.abb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin M. I.; van der Donk W. A. Stereospecific Radical-Mediated B12-Dependent Methyl Transfer by the Fosfomycin Biosynthesis Enzyme Fom3. Biochemistry 2018, 57 (33), 4967–4971. 10.1021/acs.biochem.8b00616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachid S.; Scharfe M.; Blocker H.; Weissman K. J.; Muller R. Unusual chemistry in the biosynthesis of the antibiotic chondrochlorens. Chem. Biol. 2009, 16 (1), 70–81. 10.1016/j.chembiol.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; McCarty R. M.; Ogasawara Y.; Liu Y. N.; Mansoorabadi S. O.; LeVieux J.; Liu H. W. GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme. J. Am. Chem. Soc. 2013, 135 (22), 8093–8096. 10.1021/ja312641f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Huang F.; Moison E.; Guo J.; Jian X.; Duan X.; Deng Z.; Leadlay P. F.; Sun Y. Delineating the biosynthesis of gentamicin x2, the common precursor of the gentamicin C antibiotic complex. Chem. Biol. 2015, 22 (2), 251–261. 10.1016/j.chembiol.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J.; Liu Y.-n.; McCarty R. M.; Liu H.-w. Reaction Catalyzed by GenK, a Cobalamin-Dependent Radical S-Adenosyl-l-methionine Methyltransferase in the Biosynthetic Pathway of Gentamicin, Proceeds with Retention of Configuration. J. Am. Chem. Soc. 2017, 139 (45), 16084–16087. 10.1021/jacs.7b09890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruszczycky M. W.; Ogasawara Y.; Liu H. W. Radical SAM enzymes in the biosynthesis of sugar-containing natural products. Biochim. Biophys. Acta 2012, 1824 (11), 1231–1244. 10.1016/j.bbapap.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welander P. V.; Summons R. E. Discovery, taxonomic distribution, and phenotypic characterization of a gene required for 3-methylhopanoid production. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (32), 12905–12910. 10.1073/pnas.1208255109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen K. D.; Wang S. C. Spectroscopic characterization and mechanistic investigation of P-methyl transfer by a radical SAM enzyme from the marine bacterium Shewanella denitrificans OS217. Biochim. Biophys. Acta 2014, 1844 (12), 2135–2144. 10.1016/j.bbapap.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L.; Rachid S.; Stadler M.; Wenzel S. C.; Muller R. Synthetic biotechnology to study and engineer ribosomal bottromycin biosynthesis. Chem. Biol. 2012, 19 (10), 1278–1287. 10.1016/j.chembiol.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Werner W. J.; Allen K. D.; Hu K.; Helms G. L.; Chen B. S.; Wang S. C. In vitro phosphinate methylation by PhpK from Kitasatospora phosalacinea. Biochemistry 2011, 50 (42), 8986–8988. 10.1021/bi201220r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigiri K.; Hidaka T.; Imai S.; Murakam T.; Seto H. Studies on the biosynthesis of bialaphos (SF-1293) 12. C-P bond formation mechanism of bialaphos: discovery of a P-methylation enzyme. J. Antibiot (Tokyo) 1992, 45 (5), 781–787. 10.7164/antibiotics.45.781. [DOI] [PubMed] [Google Scholar]
- Seto H.; Kuzuyama T.; Seto H.; Kuzuyama T. Bioactive natural products with carbon-phosphorus bonds and their biosynthesis. Nat. Prod Rep 1999, 16 (5), 589–596. 10.1039/a809398i. [DOI] [PubMed] [Google Scholar]
- Drennan C. L.; Huang S.; Drummond J. T.; Matthews R. G.; Ludwig M. L. How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase. Science 1994, 266 (5191), 1669–1674. 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- Marsh E. N. G.; Holloway D. E. Cloning and Sequencing of Glutamate Mutase Component-S from Clostridium-Tetanomorphum - Homologies with Other Cobalamin-Dependent Enzymes. Febs Letters 1992, 310 (2), 167–170. 10.1016/0014-5793(92)81321-C. [DOI] [PubMed] [Google Scholar]
- Jarrett J. T.; Amaratunga M.; Drennan C. L.; Scholten J. D.; Sands R. H.; Ludwig M. L.; Matthews R. G. Mutations in the B12-binding region of methionine synthase: how the protein controls methylcobalamin reactivity. Biochemistry 1996, 35 (7), 2464–2675. 10.1021/bi952389m. [DOI] [PubMed] [Google Scholar]
- Berkovitch F.; Behshad E.; Tang K. H.; Enns E. A.; Frey P. A.; Drennan C. L. A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (45), 15870–15875. 10.1073/pnas.0407074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancia F.; Keep N. H.; Nakagawa A.; Leadlay P. F.; McSweeney S.; Rasmussen B.; Bosecke P.; Diat O.; Evans P. R. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution. Structure 1996, 4 (3), 339–350. 10.1016/S0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- Dowling D. P.; Miles Z. D.; Kohrer C.; Maiocco S. J.; Elliott S. J.; Bandarian V.; Drennan C. L. Molecular basis of cobalamin-dependent RNA modification. Nucleic Acids Res. 2016, 44 (20), 9965–9976. 10.1093/nar/gkw806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexa D.; Saveant J. M. The Electrochemistry of Vitamin-B12. Acc. Chem. Res. 1983, 16 (7), 235–243. 10.1021/ar00091a001. [DOI] [Google Scholar]
- Dowling D. P.; Vey J. L.; Croft A. K.; Drennan C. L. Structural diversity in the AdoMet radical enzyme superfamily. Biochim. Biophys. Acta 2012, 1824 (11), 1178–1195. 10.1016/j.bbapap.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey J. L.; Drennan C. L. Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 2011, 111 (4), 2487–2506. 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vey J. L.; Yang J.; Li M.; Broderick W. E.; Broderick J. B.; Drennan C. L. Structural basis for glycyl radical formation by pyruvate formate-lyase activating enzyme. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (42), 16137–16141. 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman P. J.; Grove T. L.; Booker S. J.; Drennan C. L. X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (40), 15949–15954. 10.1073/pnas.1312228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling D. P.; Bruender N. A.; Young A. P.; McCarty R. M.; Bandarian V.; Drennan C. L. Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nat. Chem. Biol. 2014, 10 (2), 106–112. 10.1038/nchembio.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radle M. I.; Miller D. V.; Laremore T. N.; Booker S. J. Methanogenesis marker protein 10 (Mmp10) from Methanosarcina acetivorans is a radical S-adenosylmethionine methylase that unexpectedly requires cobalamin. J. Biol. Chem. 2019, 294 (31), 11712–11725. 10.1074/jbc.RA119.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horitani M.; Shisler K.; Broderick W. E.; Hutcheson R. U.; Duschene K. S.; Marts A. R.; Hoffman B. M.; Broderick J. B. Radical SAM catalysis via an organometallic intermediate with an Fe-[5′-C]-deoxyadenosyl bond. Science 2016, 352 (6287), 822–825. 10.1126/science.aaf5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13 (11), 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett J. T.; Drennan C. L.; Amaratunga M.; Scholten J. D.; Ludwig M. L.; Matthews R. G. A protein radical cage slows photolysis of methylcobalamin in methionine synthase from Escherichia coli. Bioorg. Med. Chem. 1996, 4 (8), 1237–1246. 10.1016/0968-0896(96)00119-8. [DOI] [PubMed] [Google Scholar]