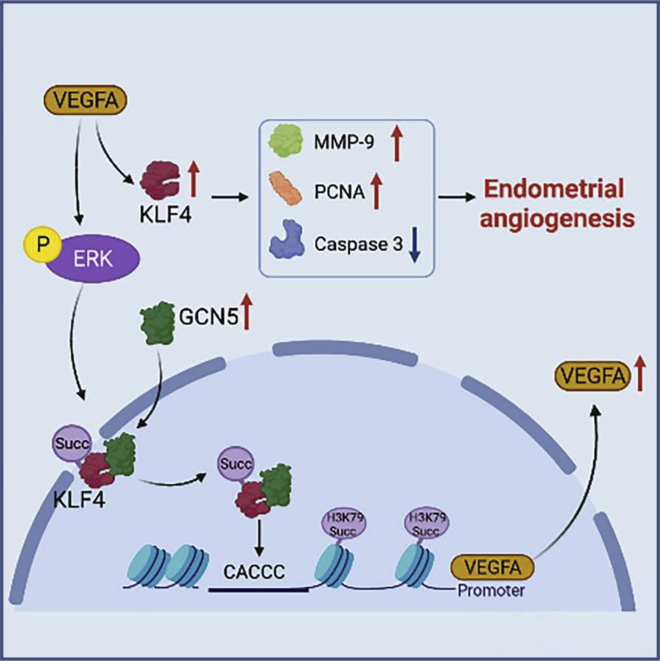
Summary
Endometrial angiogenesis is necessary for good endometrial receptivity. Krüppel-like factor 4 (KLF4) is a transcription factor that is essential for regulating angiogenesis. Here we found that vascular endothelial growth factor A (VEGFA) can form a positive feedback loop with KLF4 to promote the proliferation and migration of human endometrial microvascular endothelial cells (HEMECs) and inhibit cell apoptosis. General control non-derepressible 5 (GCN5) is also time-dependent on VEGFA and participates in the KLF4-VEGFA loop. In addition, we found that GCN5 is a succinyltransferase that modulates the succinylation of histones and nonhistones. GCN5 interacts with KLF4 and is recruited to the KLF4-binding site of the VEGFA promoter to succinylate H3K79, which initiates gene transcription epigenetically. For nonhistones, GCN5 succinylates KLF4 that is activated by ERK signaling in HEMECs treated with VEGFA to increase its transcription activity. These results demonstrate KLF4-VEGFA positive feedback loop is regulated by epigenetics, which contributes to endometrial angiogenesis.
Subject areas: Biological sciences, Molecular biology, Cell biology
Graphical abstract
Highlights
-
•
KLF4 mediates VEGFA-induced endometrial angiogenesis
-
•
VEGFA increases the interaction between KLF4 and GCN5
-
•
VEGFA promotes H3K79 succinylation by upregulating KLF4 and GCN5
-
•
VEGFA succinylates KLF4 and promotes interaction of KLF4 and GCN5 via ERK pathway
Biological sciences; Molecular biology; Cell biology;
Introduction
Recent decades have witnessed great progress in assisted reproductive technology (ART), which has become an integral element of care for many women struggling with infertility over the last 40 years (Niederberger et al., 2018). The main limiting factor in an effective ART therapy is implantation failure, which is responsible for over 72% of all failures (Achache and Revel, 2006). Human implantation is a complex process requiring interactions between a blastocyst and receptive endometrium. Endometrial receptivity refers to the ability to allow embryo positioning, adhesion, and implantation during a specific period (the implantation window, which is 4–6 days in the middle of the luteal phase) (Lessey and Young, 2019). Hence, maternal endometrial receptivity is the key to successful embryonic implantation.
During the implantation window, the endometrium undergoes a series of complex functional changes to ensure the ability to accept embryos, including the formation of pinocytoid process, the columnar growth of luminal epithelium, enlarged and highly crimped glands, endometrial stromal edema, and angiogenesis (Ashary et al., 2018). Endometrial angiogenesis, in which the vascular endothelial growth factor (VEGF) is critically involved, facilitates the invasion and nutrition of embryogenesis in the mid-luteal phase (Nayak and Brenner, 2002; Salmasi et al., 2021). VEGF family includes VEGFA, VEGFB, VEGFC, VEGFD, VEGFE, and placental growth factor (PlGF), and VEGFA appears to be the most significant mediators of angiogenesis in the growth of blood vessels in a variety of physiological and pathological conditions (Shibuya, 2006). The biological effects of VEGFA are mostly mediated by the tyrosine kinase receptor VEGFR-2 (Shibuya, 2006). VEGFA is essential for the permeability of endometrial blood vessels and the proliferation of blood vessels during implantation of fertilized eggs (Rabbani and Rogers, 2001). Controlled ovarian hyperstimulation (COH) is an important step in in vitro fertilization and embryo transfer (IVF-ET). However, animal and clinical studies show that uterine receptivity deteriorates during COH. COH reduces VEGFA during uterine receptivity (Nayak and Brenner, 2002). Thus, VEGFA and underlying molecular mechanisms in angiogenesis are necessary for receptive endometrium, which is important for embryo implantation.
Krüppel-like factor 4 (KLF4), which belongs to the KLF family, is a zinc finger transcription factor that binds to “CACCC” or “GT-box” sites through a DNA-binding domain consisting of three zinc fingers (Garrett-Sinha et al., 1996). Mounting evidence indicates that KLF4 is highly expressed in endothelial cells and vascular smooth muscle cells and acts as a critical regulator of vascular homeostasis through an epigenetic mechanism (Atkins and Jain, 2007). KLF4 could interact with histone acetyltransferases or deacetylases to increase or attenuate chromatin acetylation, respectively, which in turn affects transcription. For example, KLF4 recruits p300 to the KLF4-binding sites of TGF-β1-regulated target promoters, and the promoter chromatin-localized p300 acetylates histone H3, which initiates gene transcription (He et al., 2015). KLF4 undergoes posttranslational modification via phosphorylation (Li et al., 2010), acetylation (He et al., 2015), ubiquitination, and methylation (Hu et al., 2015), which fine-tunes the function of KLF4 by changing its protein stability. In addition, KLF4 can be used as a methylation reader that preferentially recognizes specific methylation sequences and induces chromatin remodeling and transcription activation by recruiting gene methylation cis-regulatory elements (Wan et al., 2017). Besides, although KLF4 interacts with a protein to change its function, it can recruit histone-modifying enzymes, which in turn change the adjunct histone modifications, affect chromatin remodeling, and regulate the expression of target genes. The function of KLF4 in angiogenesis appears to be highly cell type-specific and context-dependent. Several studies have described increased cell proliferation, migration, and tube formation as a result of KLF4 in human retinal microvascular endothelial cells and pulmonary artery endothelial cells (Liang et al., 2017; Wang et al., 2015), but contradictory findings show the anti-proliferative effects of KLF4 in cardiovascular and tumor diseases (Zeng et al., 2018; Zheng et al., 2013). However, the role of KLF4 in endometrial angiogenesis is not clear.
In the present study, we determined the role of KLF4 in human endometrial microvascular endometrial cells (HEMECs), which plays an important role in receptive endometrium. We provided the evidence that KLF4 regulates angiogenesis via epigenetic mechanism in HEMECs.
Results
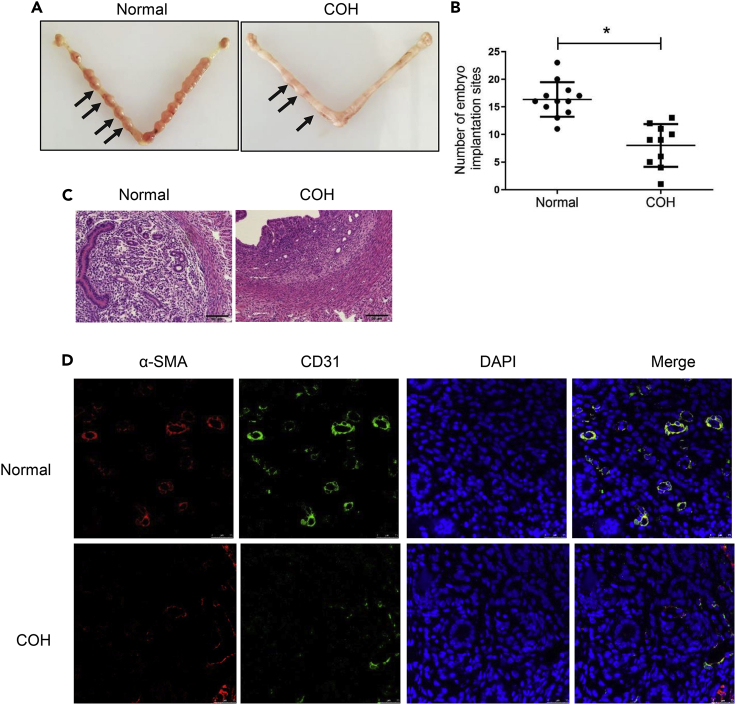
Effect of COH on endometrial receptivity in mice
Previous study has shown that the endometrium undergoes secretory changes prematurely in the postovulatory and early luteal phase of IVF cycles, resulting in a window of implantation appearing earlier and reduced endometrial receptivity (Bourgain, 2004). COH is one of the important steps of IVF-ET. As shown in Figure 1A, the implanted embryos in pregnant mice of the normal group were large and developed well, showing beaded arrangement after implantation, whereas the implanted embryos in mice of the COH group were small. The number of embryos implanted in the COH group was significantly reduced compared with the normal group (∗p < 0.05) (Figure 1B). HE staining showed that the endometrium of COH pregnant mice showed smaller glands, reduced cytosis, and sparse endometrium vessels compared with the normal group (Figure 1C). Mature vascular units are indicated by α-SMA. CD31 is commonly used to determine proliferation of endothelial cells located in blood vessels. Dual-immunofluorescence staining showed that compared with the normal group, the number of blood vessels marked by α-SMA and CD31 in the COH group was significantly reduced (Figure 1D), suggesting that COH decreased microvessel density, which may be related to the reduction of VEGFA expression, leading to inhibited endometrial receptivity.
Figure 1.
Effect of controlled hyperstimulation on endometrial receptivity in mice
(A) Representative images of the pregnant uterus of the normal and COH group.
(B) Comparison of the number of implanted embryos in normal and COH group (n = 10, each group). Data are represented as mean ± SD, ∗p < 0.05 vs. normal group, Student’s t-test.
(C) Representative hematoxylin-eosin (HE)-stained sections of each group of mice. Magnification, ×200, scale bar = 50 μm.
(D) The difference of α-SMA and CD31 expression in the uterus of mice in normal and COH groups were detected by immunofluorescence staining. The red staining represents α-SMA, the green staining represents CD31, and the blue staining represents the cell nucleus. Magnification, ×630, Scale bar = 25 μm.
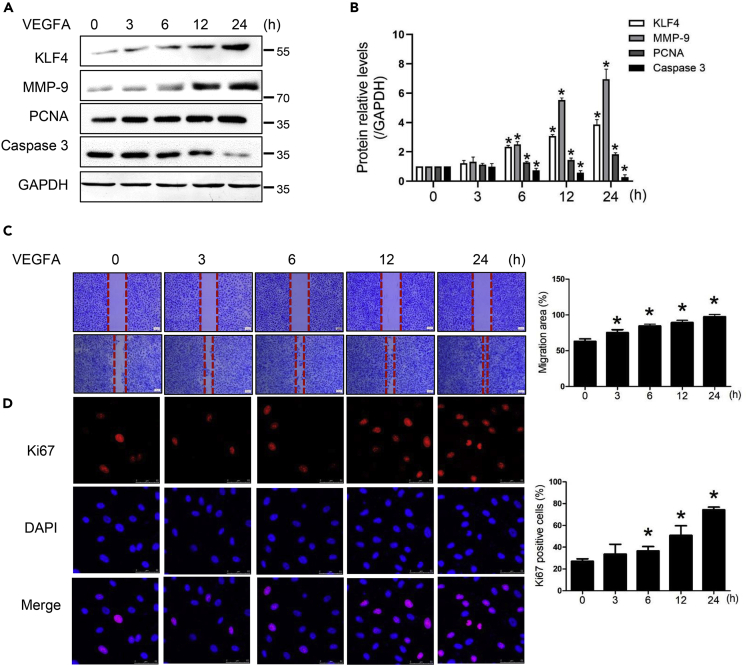
VEGFA promotes HEMEC angiogenesis
Numerous studies have reported that VEGFA promotes angiogenesis by promoting the expression of cell proliferation protein PCNA (Friedlander et al., 2015) and migration protein MMP-9 (Wen and Li, 2015), inhibiting the activity of apoptosis protein Caspase 3 (Wang et al., 2015) and regulating KLF4 (Wang et al., 2015). We incubated HEMECs with VEGFA (40 ng/mL) for multiple time points and performed Western blotting analysis. We found that the expression of MMP-9, PCNA, and KLF4 were increased in a time-dependent manner (∗p < 0.05). Conversely, as an effector of apoptosis, the expression of Caspase 3 was gradually decreased (Figures 2A and 2B). A scratch-wound assay indicated that HEMECs stimulated by VEGFA exhibited stronger migration ability and followed a time-dependent manner (Figure 2C). Ki67, which is a mitotic nuclear antigen, marks cells in the state of proliferation. The result of immunofluorescence staining showed that VEGFA stimulation increased the number of Ki67-positive cells, suggesting that VEGFA promoted the proliferation of HEMECs (Figure 2D).
Figure 2.
VEGFA promotes HEMEC angiogenesis
(A) HEMECs were stimulated with VEGFA (40 ng/mL) for different time points. Western blotting analysis was used to detect the protein expression of KLF4, MMP-9, PCNA, and Caspase 3.
(B) Densitometric scanning. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 h, one-way ANOVA.
(C) VEGFA promoted HEMEC migration by the scratch-wound assay. Statistical data of the results are shown on the right. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 h, one-way ANOVA.
(D) Immunofluorescence staining was performed for the expression of Ki67 in HEMECs. Red staining represents Ki67 and blue staining represents the nucleus. Scale bar = 50 μm. Statistical data of the results are shown on the right. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 h, one-way ANOVA.
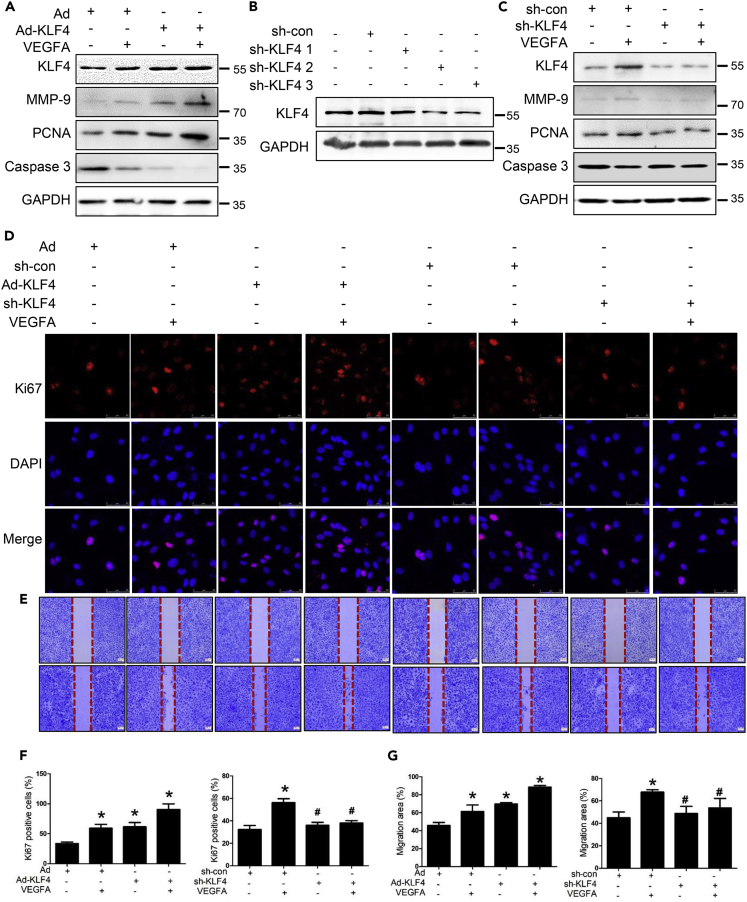
KLF4 mediates VEGFA-induced endometrial angiogenesis
KLF4 is closely related to angiogenesis and has various effects because of different cell backgrounds (Wang et al., 2015; Zheng et al., 2013). To validate the roles of KLF4 in VEGFA-induced endometrial angiogenesis, we performed Dual-immunofluorescence staining to detect α-SMA and KLF4 in endometrium and found KLF4 in endometrium was decreased in COH group compared with normal group. The expression regions of KLF4 and α-SMA were partially overlap, indicating that KLF4 may affect endometrial angiogenesis (Figure S1A). In vitro, HEMECs were co-treated with a KLF4 overexpression adenovirus (Ad-KLF4) or VEGFA. KLF4 overexpression further increased PCNA and MMP-9 by VEGFA and decreased Caspase 3 expression (Figures 3A and S1B). In contrast, when HEMECs were infected with three KLF4-specific shRNAs (sh-KLF4) adenoviruses to block endogenous KLF4 expression (sh-KLF4 2 and sh-KLF4 3 had high knockdown efficiency), VEGFA-induced expression of PCNA and MMP-9 and VEGFA-reduced Caspase 3 expression were abrogated (Figures 3B–3C and S1C–S1D). Moreover, cellular immunofluorescence assay indicated that KLF4 overexpression promoted Ki67 expression, and the number of Ki67-positive cells decreased after KLF4 knockdown (Figures 3D and 3F). Scratch-wound assay was performed and the co-treatment group with KLF4 overexpression and VEGFA showed stronger migration ability compared with the Ad group, whereas the co-treatment group with KLF4 knockdown and VEGFA showed weaker migration ability compared with the VEGFA group (Figures 3E and 3G). These results indicated that KLF4 is essential for the induction of endometrial angiogenesis by regulating proliferation, migration, and apoptosis responding to VEGFA signaling.
Figure 3.
KLF4 mediates VEGFA-induced endometrial angiogenesis
(A) After HEMECs were infected with Ad-KLF4 adenovirus 24 h and incubated with VEGFA another 12 h, the protein expression of KLF4, MMP-9, PCNA, Caspase 3 were detected by Western blotting.
(B) HEMECs were infected with three KLF4-specific knockdown adenoviruses (sh-KLF4 1, sh-KLF4 2, sh-KLF4 3). Western blotting analysis was performed for KLF4 expression.
(C) After HEMECs were infected with sh-KLF4 2 24 h and stimulated with VEGFA for 12 h, the protein expression of KLF4, MMP-9, PCNA, and Caspase 3 were detected by Western blotting.
(D) HEMECs were treated as (A) and (C). The expression of Ki67 was detected by immunofluorescence staining. Red staining represents Ki67 and blue staining represents the nucleus. Scale bar = 50 μm.
(E) HEMECs were treated as (A) and (C). The migration of HEMECs was detected by Scratch-wound assay.
(F) Statistical data of immunofluorescence staining results in (D). Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. Ad or sh-con, #p < 0.05 vs. VEGFA, one-way ANOVA.
(G) Statistical data of Scratch-wound assay results in (E), Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. Ad or sh-con, #p < 0.05 vs. VEGFA, one-way ANOVA.
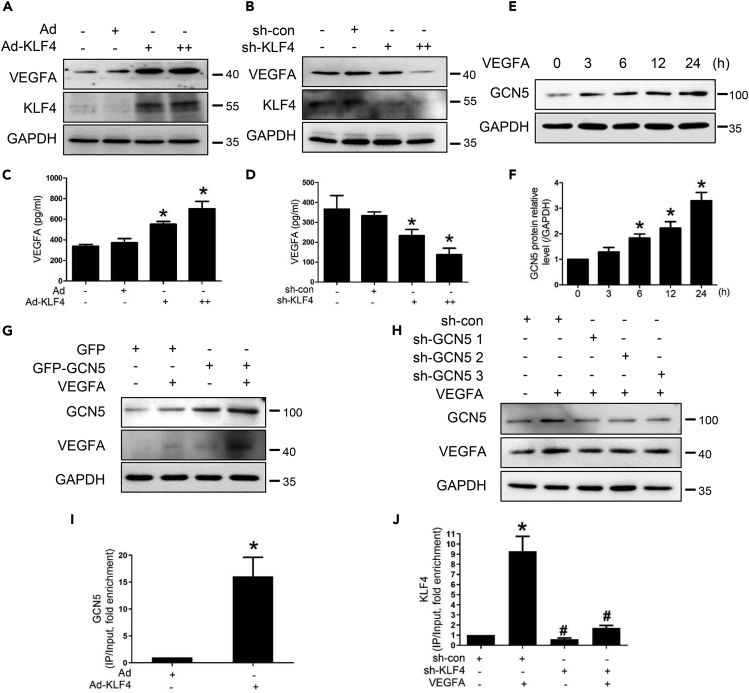
GCN5 participates in the feedback regulatory loop between KLF4 and VEGFA
Subsequently, we observed whether KLF4 could feedback-regulate the expression of VEGFA. Western blotting analysis was performed to detect the VEGFA protein expression. Firstly, we treated HEMECs and human umbilical vein endothelial cells (HUVECs) with Ad-KLF4 or not and found that the expression of VEGFA was increased in both HEMECs and HUVECs, suggesting that KLF4 can promote angiogenesis in HEMECs and HUVECs (Figure S2A). VEGFA protein level was increased in the KLF4 overexpression group but decreased in the KLF4 knockdown group (Figures 4A–4B and S2B–S2C). We also detected the VEGFA protein levels in the cell culture supernatant by ELISA and found VEGFA level in KLF4 overexpression group was higher than control group, knockdown of KLF4 reduced the level of VEGFA in HEMECs culture supernatant (Figures 4C and 4D), suggesting that KLF4 triggers VEGFA secretion. Recent studies have shown that GCN5 can upregulate VEGFA level by histone epigenetic modification, which accelerates bone angiogenesis to vascular remodeling (Jing et al., 2017). The expression of GCN5 gradually was increased in a time-dependent manner with VEGFA stimulation in HEMECs and peaked at 24 h, as detected by Western blotting (Figures 4E and 4F). Subsequently, we constructed the GCN5 overexpression plasmid (GFP-GCN5) and GCN5 knockdown plasmids (sh-GCN5). When HEMECs were transfected with GFP-GCN5, the expression level of VEGFA was increased (Figures 4G and S2D). When HEMECs were transfected with sh-GCN5, three GCN5 knockdown plasmids all exerted an inhibitory effect on VEGFA (Figures 4H and S2E). VEGFA promoter has three positions which contain “CACCC” sites that can bind to KLF4. To determine whether GCN5 is increased on the VEGFA gene promoter, which was activated by KLF4 in HEMECs, ChIP analysis was conducted. GCN5 was highly concentrated in the ∼0.6 kb region of the VEGFA promoter that contains “CACCC” sites in KLF4 overexpression-treated HEMECs compared with the cells infected with Ad (Figure 4I). The knockdown of KLF4 by sh-KLF4 reduced the inducing effect of VEGFA on the binding of KLF4, regardless of VEGFA treatment (Figure 4J).
Figure 4.
GCN5 participates in the feedback regulatory loop between KLF4 and VEGFA
(A–B) After HEMECs were infected with Ad-KLF4 or sh-KLF4 adenovirus for 36 h, VEGFA and KLF4 protein levels were detected by Western blotting.
(C–D) We treated HEMECs with Ad-KLF4 or sh-KLF4 adenovirus for 36 h and detected VEGFA levels in cell culture supernatant by ELISA. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. Ad or sh-con, #p < 0.05 vs. VEGFA; one-way ANOVA.
(E) HEMECs were stimulated with VEGFA for different time intervals, and GCN5 expression level was assessed by Western blotting.
(F) Densitometric analyses for Western blots in (E). Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 h, one-way ANOVA.
(G–H) HEMECs were transfected with expression plasmids for GCN5 or three GCN5-specific knockdown plasmids (sh-GCN5 1, sh-GCN5 2, and sh-GCN5 3) and treated with VEGFA. Western blotting analysis was performed for GCN5 and VEGFA expressions.
(I) ChIP analysis of GCN5 occupancy at the VEGFA promoter in HEMECs that were infected with Ad or Ad-KLF4. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. Ad, Student’s t-test.
(J) ChIP analysis of KLF4 occupancy at the VEGFA promoter in HEMECs that were infected with sh-con or sh-KLF4 for 24 h and then treated with VEGFA for an additional 12 h. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. sh-con, #p < 0.05 vs. VEGFA, one-way ANOVA.
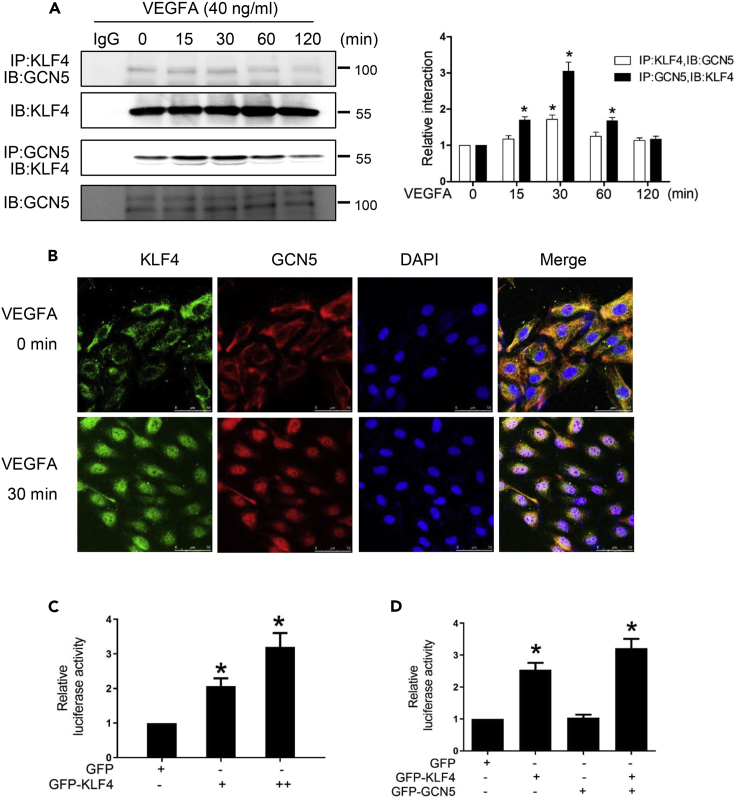
VEGFA increases the interaction between KLF4 and GCN5
After discovering that GCN5 participates in the feedback loop formed by KLF4 and VEGFA, we then explored the relationship between KLF4 and GCN5. Co-immunoprecipitation revealed that VEGFA increased the levels of GCN5 present in anti-KLF4 immunoprecipitates, peaking at 30 min and subsequently decreasing to control levels at 120 min. Similar results were obtained when cells were immunoprecipitated with GCN5, and the levels of KLF4 were assessed (Figure 5A). Dual-immunofluorescence staining revealed that KLF4 and GCN5 were almost all distributed in the cytoplasm before VEGFA treatment but translocated to the nuclear area in HEMECs treated with VEGFA for 30 min, and that KLF4 and GCN5 primarily co-localized during this period (Figure 5B), which coincide with the immunoprecipitation results (Figure 5A). To determine the functional significance of the association of KLF4 with GCN5, we constructed a VEGFA promoter-luciferase reporter construct, including the region spanning from −2,000 to +1 of the human VEGFA promoter, and performed a luciferase reporter assay using HEK293T cells. The results indicated that overexpression of KLF4 increased VEGFA promoter activity in a concentration-dependent manner (Figure 5C). GCN5 could not increase VEGFA promoter activity, whereas KLF4 and GCN5 overexpression resulted in a three-fold increase in activity compared with the GFP group. The VEGFA promoter activity increased by KLF4 and GCN5 overexpression was higher than activity increased by KLF4 overexpression, suggesting that KLF4 bound to “CACCC” sites and GCN5 synergistically induced the activation of the VEGFA promoter (Figure 5D).
Figure 5.
VEGFA increases the interaction of KLF4 with GCN5
(A) HEMECs were treated with VEGFA for the indicated times. The interaction between KLF4 and GCN5 was examined by reciprocal coimmunoprecipitation (CoIP). Densitometric analyses for CoIP results (right panel), Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 min, one-way ANOVA.
(B) Protein localization of KLF4 and GCN5 in HEMECs were detected by dual-immunofluorescence staining. The red staining represents GCN5, the green represents KLF4, and the blue staining represents the cell nucleus. Scale bar = 50 μm.
(C) HEK293T cells were transfected with the indicated VEGFA promoter luciferase reporters alone or in combination with the KLF4 expression vector. Cells were lysed, and the luciferase activity was measured using the dual luciferase reporter assay system. The data represent the relative VEGFA promoter activity normalized to renilla luciferase. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. GFP, one-way ANOVA.
(D) Luciferase reporter controlled by the VEGFA promoter was transfected into HEK293T cells with KLF4 or GCN5 expression plasmids. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. GFP, one-way ANOVA.
VEGFA promotes H3K79 succinylation by upregulating KLF4 and GCN5
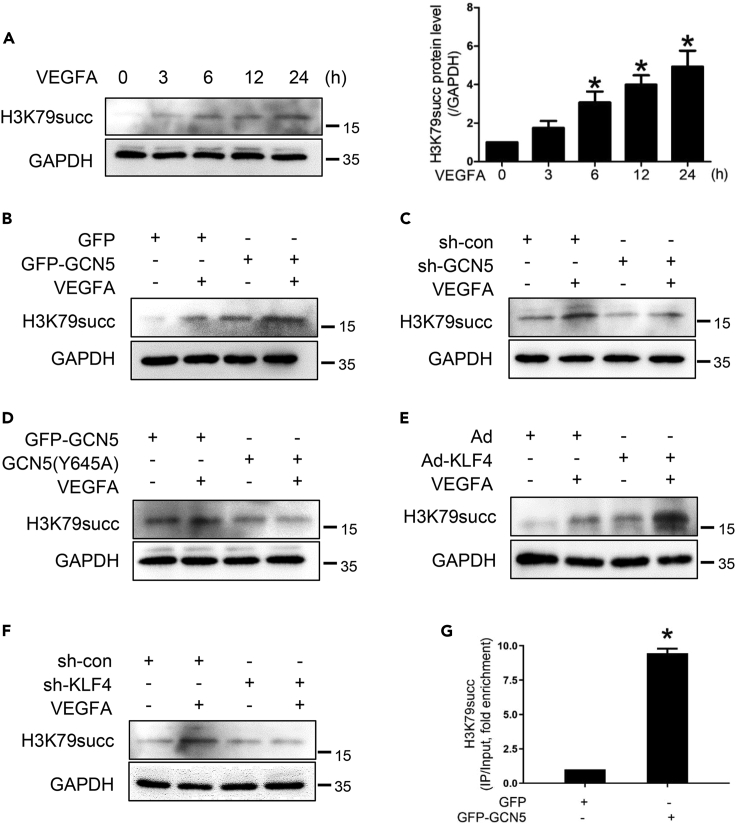
It has recently been reported that GCN5 can promote H3K79 succinylation (Tong et al., 2020). To determine whether the VEGFA-induced angiogenesis effect is related to H3K79 succinylation, Western blotting analysis was performed to assess the effect of VEGFA on H3K79 succinylation expression. Notably, H3K79 succinylation was significantly increased in the VEGFA-treated cells compared with the VEGFA-untreated group (Figure 6A). When the cells were transfected with GFP-GCN5, the increase in VEGFA-induced H3K79 succinylation was more obvious than the GFP group (Figures 6B and S3A). VEGFA-induced H3K79 succinylation was abrogated by sh-GCN5 (Figures 6C and S3B), suggesting that GCN5 is responsible for the succinylation of histone H3K79 in HEMECs. We reconstituted the expression of GFP-GCN5 or GFP-GCN5 Y645 mut (GCN5(Y645A)), which had impaired ability to bind to succinyl-CoA but not acetyl-CoA, leading to reduced H3K79 succinylation. These results showed that the Y645A site may be the key site of GCN5 regulation of H3K79 succinylation (Figures 6D and S3C). KLF4 can participate in the regulation of histone modification to affect gene expression such as methylate histone H3K79 (Wang et al., 2019). To examine whether KLF4 could regulate H3K79 succinylation, the cells were infected with Ad-KLF4 or sh-KLF4. KLF4 markedly increased VEGFA-induced H3K79 succinylation expression, and knockdown of KLF4 inhibited H3K79 succinylation (Figures 6E–6F and S3D–S3E). We performed ChIP assays to observe H3K79 succinylation occupancy on VEGFA promoter and found that the level of succinylated H3K79 at the VEGFA promoter was increased in GCN5-overexpressing HEMECs (Figure 6G). These results clearly suggest that VEGFA promotes the KLF4-mediated recruitment of GCN5 to the VEGFA promoter, subsequently leading to increased H3K79 succinylation of the VEGFA promoter.
Figure 6.
VEGFA promotes H3K79 succinylation by upregulating KLF4 and GCN5
(A) HEMECs were incubated with VEGFA for different time points. Western blotting was used to detect the succinylated H3K79. Densitometric scanning (right panel). Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 h, one-way ANOVA.
(B–C) HEMECs were transfected with expression plasmids for GFP-GCN5 or sh-GCN5 1 and treated with VEGFA. Western blotting analysis was performed for succinylated H3K79 expression.
(D) HEMECs were transfected with expression plasmids for GFP-GCN5 or its tyrosine 645-deficient mutant Y645A and treated with VEGFA. Western blotting analysis was performed for H3K79 succinylation.
(E–F) HEMECs were infected with Ad-KLF4 or sh-KLF4 adenovirus for 24 h and then incubated with VEGFA for 12 h. Succinylated H3K79 were assessed by Western blotting.
(G) ChIP analyses were used to detect succinylated H3K79 occupancy at the VEGFA promoter in HEMECs that were transfected with GFP-GCN5. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. GFP, Student’s t-test.
VEGFA-increased succinylation levels of KLF4 are regulated by GCN5
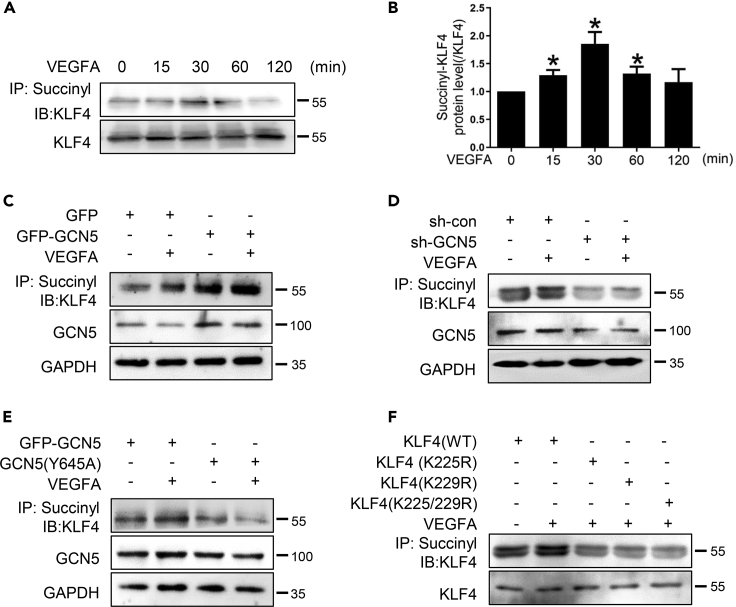
Because KLF4 interacts with GCN5, we sought to determine whether GCN5 succinylates KLF4. Firstly, we assessed the succinylation of KLF4. VEGFA-stimulated KLF4 succinylation began within 15 min, reached a maximum at 30 min, and returned to basal levels at 120 min. KLF4 expression did not change during the entire time course of the study (Figures 7A and 7B). Subsequently, we transfected GFP-GCN5, sh-GCN5, or GCN5(Y645A) into HEMECs. GCN5 overexpression further increased VEGFA-induced KLF4 succinylation (Figures 7C and S4A). GCN5 knockdown inhibited KLF4 succinylation, regardless of VEGFA stimulation (Figures 7D and S4B). In addition, GCN5 Y645 mut acting like its GCN5 depletion counterpart reduced the expression levels of KLF4 succinylation (Figures 7E and S4C). K225 and K229 are important lysine sites in the zinc structure of KLF4, which affect the activity of KLF4 (Yu et al., 2011b). To observe whether VEGFA succinylates these sites of KLF4, the cells were transfected with KLF4 mutants K225R, K229R, and K225/229R. Immunoprecipitation showed that KLF4 at Lys225, Lys229, or Lys225/229 has been succinylated (Figures 7F and S4D). Altogether, these findings suggested that GCN5 could succinylate these sites in KLF4.
Figure 7.
VEGFA induces KLF4 succinylation regulated by GCN5
(A) After HEMECs were stimulated with VEGFA for different time points, succinylated KLF4 was examined by immunoprecipitation.
(B) Quantitation of the succinylated KLF4 normalized to total KLF4. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 min, one-way ANOVA.
(C) After HEMECs were transfected with expression plasmids for GCN5 and stimulated with VEGFA for 30 min, succinylated KLF4 was examined by immunoprecipitation.
(D) HEMECs were transfected with expression plasmids for GFP-GCN5 or sh-GCN5 1 and stimulated with VEGFA for 30 min. Succinylated KLF4 was examined by immunoprecipitation.
(E) HEMECs were transfected with plasmids for GFP-GCN5 or GCN5(Y645A) and stimulated with VEGFA for 30 min. Succinylated KLF4 was detected by immunoprecipitation.
(F) HEMECs were transfected with expression plasmids for KLF4 or its acetylation-deficient mutants K225, K229R, and K225/229R, then treated with VEGFA. Succinylated KLF4 was examined by immunoprecipitation.
VEGFA succinylates KLF4 and promotes interaction of KLF4 and GCN5 via ERK signaling pathway
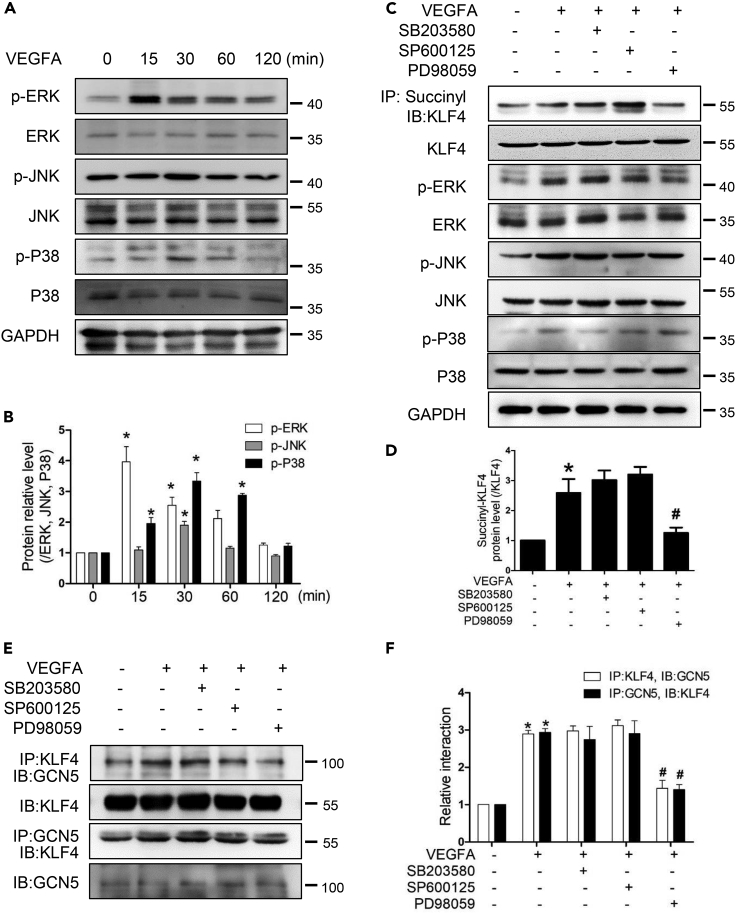
Based on the above experimental results, we then investigated which signaling pathway mediates the succinylation modification of KLF4 in response to VEGFA stimulation. VEGFA regulates multiple signaling pathways such as MAPK (Sun et al., 2018), so we studied the signaling pathway in HEMECs treated with VEGFA. p-ERK levels peaked at 15 min, p-JNK and p-P38 expression levels were the highest at 30 min, and returned to basal levels at 2 h (Figures 8A and 8B). HEMECs were incubated with the p38 inhibitor SB203580, the JNK inhibitor SP600125, or the ERK inhibitor PD98059 for 2 h before exposure to VEGFA. It was found that the administration of PD98059 efficiently blocked VEGFA-induced succinylated KLF4 levels (Figures 8C–8D and S4E). Interestingly, immunoprecipitation experiments showed that the binding of KLF4 and GCN5 was also decreased in the group given PD98059 compared with the VEGFA group (Figures 8E and 8F). These results indicate that the activation of ERK signaling pathway is essential for KLF4 succinylation and interaction of KLF4 with GCN5.
Figure 8.
VEGFA succinylates KLF4 and promotes interaction of KLF4 and GCN5 via ERK signaling pathway
(A) HEMECs were stimulated with VEGFA for different time points, Western blotting analysis was used to detect the protein levels of p-ERK, ERK, p-JNK, JNK, p-P38, and P38.
(B) Densitometric analyses for Western blots in (A). Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. 0 min, one-way ANOVA.
(C) HEMECs were pretreated with SB203580 (20 mM), SP600125 (5 mM), PD98059 (20 mM) for 2 h, followed by a 30 min incubation with VEGFA. Total proteins were used for the detection of succinylated KLF4 in anti-succinyllysine immunoprecipitates. Western blotting analysis was used to detect p-ERK, ERK, p-JNK, JNK, p-P38, and P38 expressions.
(D) Quantitation of the succinylated KLF4 normalized to total KLF4. Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. untreated group, #p < 0.05 vs. VEGFA, one-way ANOVA.
(E) HEMECs were treated as (B). Interactions of KLF4 with GCN5 in HEMECs were examined by CoIP.
(F) Densitometric analyses for CoIP in (D). Data are represented as mean ± SD, n = 3, ∗p < 0.05 vs. the untreated group, #p < 0.05 vs. VEGFA, one-way ANOVA.
Discussion
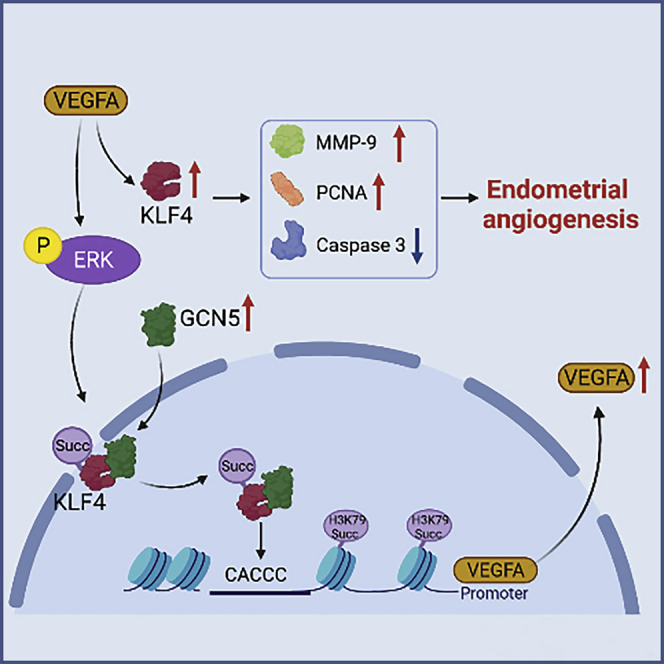
Low pregnancy rates caused by low endometrial receptivity have plagued ART. How to improve endometrial receptivity and increase pregnancy rate have become a challenge for many researchers. High blood vessel density is the material basis for ensuring good endometrial receptivity. In our study, we demonstrated that VEGFA and KLF4 formed a positive feedback loop to promote endometrial angiogenesis, which improves endometrial receptivity. VEGFA-promoted KLF4 expression upregulated HEMEC proliferation and migration and downregulated apoptosis. KLF4 could also feedback-regulate the expression of VEGFA in HEMECs. KLF4 interacted with GCN5, recruited it to the KLF4-binding site of VEGFA promoter, and succinylated H3K79, which initiated gene transcription (Figure 9). This study attempted to epigenetically elucidate the molecular mechanism of endometrial angiogenesis.
Figure 9.
Proposed the mechanism model of KLF4 and VEGFA positive feedback loop to promote endometrial angiogenesis
VEGFA-promoted KLF4 expression upregulated HEMEC proliferation and migration and downregulated apoptosis. KLF4 could also feedback-regulate the expression of VEGFA in HEMECs. KLF4 interacted with GCN5, recruited it to the KLF4-binding site of VEGFA promoter, and succinylated H3K79, which initiated gene transcription.
VEGFA plays an important role in regulating the growth of the endometrium, the development of the embryo, and the formation of ovarian blood vessels. During the implantation window, PR regulates its response factor VEGFA to regulate endometrial angiogenesis and decidualization to ensure successful embryo implantation (Bhurke et al., 2016). Kaya et al. identified several PR binding sites within a ±200 kb window of the transcription start site of the VEGFA gene (Kaya et al., 2015). The increase in VEGFA expression during the “implantation window” period helps to increase the receptivity of the endometrium and facilitates embryo implantation and formation of early villi vessels. When VEGFA expression is insufficient, angiogenesis at the implantation site decreases, which leads to poor early villi formation and decreased endometrial receptivity (Lash et al., 2012). Allegra et al.(Allegra et al. (2009) studied the pregnancy outcomes of infertile patients undergoing IVF-ET and found that the expression of VEGFA in the endometrial tissue “implantation window” pregnancy group was significantly higher than that of the nonpregnant group, and VEGFA can promote endothelial cell proliferation surrounding the implantation site and increase the vascular permeability of the endometrium. In vivo, the stimulation of exogenous ovarian hormones damages the endometrial homeostasis environment, resulting in the reduction of related angiogenic factors and low endometrial tolerance (Figure 1).
KLF4 is a transcription factor that exerts biological effects on the proliferation and differentiation of cells in a context-dependent manner. In the leukemia cells, Liu et al. found that KLF4 can act as a cooperative gene of RUNX1 to activate its target gene P57, inhibit the cell proliferation, and promote apoptosis (Liu et al., 2019). Conversely, KLF4 functions as an oncogene in human breast cancer (Yu et al., 2011a). Recent studies have found that KLF4 may be involved in the regulation of endometrial receptivity. In endometrial epithelial cells, estrogen can rapidly increase the expression of KLF4, which transactivates MCM2 and promotes DNA synthesis and cell proliferation. Progesterone disrupts the transcription complex consisting of KLF4, ER, and RNA polymerase II and favors the assembly of a PR-KLF15 complex at the Mcm2 promoter, leading to downregulation of the MCM2 proteins (Ray and Pollard, 2012). The regenerative capacity of the human endometrium requires a population of local stem cells. In a mouse menstruation model, uterine stromal SM22α+-derived CD34+KLF4+ stem cells are activated and augments ERα transcriptional activity and proliferative signaling, which make these cells then transdifferentiate to the endometrial epithelium (Yin et al., 2019). In our study, we confirmed that KLF4 is expressed in normal endometrium and decreased in endometrium of COH mice (Figure S1A). We first demonstrated the existence of a feedback loop between VEGFA and KLF4 in HEMECs. VEGFA promoted KLF4 expression, which upregulated HEMEC proliferation and migration and downregulated apoptosis (Figures 2 and 3). Meanwhile, KLF4 promoted the expression of VEGFA in HEMECs, triggered VEGFA secretion and subsequent blood vessel formation (Figures 4A–4D and S2B–S2C). VEGFA promoter contain share considerable binding sites for Sp1/Sp3, AP-1, Egr-1, STAT-3, HIF1α, KLF8, KLF4, YY1 at the proximal region of promoters from various species (Cheng et al., 2018; Sanchez-Navarro et al., 2021; Shima et al., 1996; Wang et al., 2015; Yang et al., 2019). In HEMECs, we found that KLF4 bound to ∼0.6 kb region of the VEGFA promoter which contain “CACCC” sites, suggesting that KLF4 can transcriptionally activate VEGFA (Figure 4J) and form a positive feedback loop with VEGFA to promote endometrial angiogenesis.
GCN5, also known as lysine acetyltransferase 2A (KAT2A), is a histone acetyltransferase (HAT) that binds to acetyl-CoA and transfers its acetyl group to histones. In addition, GCN5 functions as a histone succinyltransferase by directly transferring the succinyl group from succinyl-CoA to histone H3 lysine 79 (H3K79), which is important for the regulation of gene expression in tumor cells (Yuan et al., 2020). These features suggest that GCN5 regulates gene expression and chromosome structure mainly by modifying histones. In reproduction, GCN5-mediated histone acetylation promotes the expulsion of nucleosomes in sperm, and the lack of histone acetylation can lead to male reproductive disorders and negatively affect early embryo formation (Luense et al., 2019). GCN5 regulates the proangiogenic factor secretion of BMSCs in osteoporotic trabecular bone by enhancing acetylation of H3K9 on the promoter of the vegfa gene (Jing et al., 2017). In mouse embryos at various stages before implantation, including two-cell, four-cell, eight-cell, morula, and blastocyst stage embryos, the expression level of GCN5 in embryos at all stages, except four-cell, was significantly increased, suggesting that GCN5 is involved in early embryonic epigenetics and regulates gene expression(Liu et al., 2014; Peng et al., 2015). Reduced GCN5 expression may lead to early embryo damage, with about 25% of gCN5-deficient mice dying before birth, with embryos stunted and showing increased apoptosis (Phan et al., 2005). However, the role of GCN5 in angiogenesis, especially endometrial angiogenesis, has been poorly reported. In HEMECs, we found that VEGFA promoted GCN5 expression (Figures 4E and 4F), which is involved in the feedback loop between KLF4 and VEGFA. We cultured HEMECs with overexpressed (Figures 4G and S2D) or knockdowned GCN5 (Figures 4H and S2E) and found that GCN5 was potent in inducing VEGFA expression. However, the result of a luciferase reporter assay showed that GCN5 could not activate VEGFA promoter (Figure 5D). Moreover, ChIP assay revealed that overexpression of KLF4 increased the recruitment of GCN5 to the regions of VEGFA promoter, which contain “CACCC” sites (Figure 4I), suggesting that KLF4 recruits GCN5 by interacting with GCN5. KLF4 and CGN5 were translocated to the nuclear area of HEMECs after VEGFA treatment (Figure 5B) and formed a transcriptional activation complex (Figure 5A) on the VEGFA promoter. Although GCN5 cannot activate the VEGF-A promoter but can increase the activation of the VEGFA promoter by KLF4 (Figures 5C and 5D).
Histone modifications such as the frequently occurring lysine succinylation are central to the regulation of chromatin-based processes (Weinert et al., 2013). Succinylation of lysine participates in a variety of cellular regulatory processes, including DNA replication, transcription, and damage repair. In budding yeast, the K77 residue of histone H4 simulates succinylation, which accelerates nucleosome cleavage, enabling DNA to be fully exposed to binding proteins such as transcription factors (Jing et al., 2020). Histone H3K122 succinylation is enriched at promoters of active genes, can alter nucleosome stability, loosen chromatin, and regulate gene transcription levels (Zorro Shahidian et al., 2021). HAT1 is a histone succinyltransferase that enhances gene expression profiling by catalyzing histone H3K122 succinylation and facilitates glycolysis by catalyzing the succinylation of PGAM1 on K99 in cancer cells, suggesting that HAT1-mediated succinylation is essential to tumorigenesis (Yang et al., 2021). GCN5 is a histone succinyltransferase that is responsible for the succinylation of histone H3K79, with a maximum frequency around the transcription start sites of genes for the regulation of gene expression (Wang et al., 2017; Yuan et al., 2020). VEGFA can increase succinylation of histone H3K79 in HEMECs (Figure 6A), which may contribute to transcriptional activation. Overexpression or knockdown of GCN5 can promote or inhibit H3K79 succinylation (Figures 6B, 6C, 6G, and S3A–S3B). Expression of GCN5 mut (Y645A), which has impaired ability to bind to succinyl-CoA but not acetyl-CoA, leads to reduced H3K79 succinylation and gene expression (Tong et al., 2020; Wang et al., 2017), which is concordant with our results (Figures 6D and S3C). Furthermore, we found that KLF4 can regulate H3K79 succinylation level (Figures 6E–6F and S3D–S3E). These results indicate that KLF4-mediated recruitment of GCN5 may facilitate H3K79 succinylation on the promoter of VEGFA.
Lysine succinylation, an essential biological process that regulates cell activity, partially activates cell function. The highly succinylated lactate dehydrogenase (LDHA) at lysine K222 in gastric cancer cells can enhance cell viability and promote cell proliferation, invasion, and migration (Li et al., 2020). In the reproductive field, researchers have found that Lead (Pb)-induced low TCA circulating activity reduces lysine succinylation, thereby hindering the distribution of germ elongation in mice and germ cells in the sputum cells (Yang et al., 2020). VEGFA can promote KLF4 succinylation and peaks at 30 min, which is consistent with the results of interaction between KLF4 and GCN5 (Figures 7A and 7B). Similar to HAT1 succinylated nonhistone protein to facilitate glycolysis (Yang et al., 2021), GCN5 can succinylate KLF4 in HEMECs (Figures 7C–7E and S4A–S4C). Because succinylation sites and acetylation sites extensively overlap, we assessed the level of succinylation at known acetylation sites of KLF4 (K225 and K229) (He et al., 2015). The succinylation of KLF4 was decreased in HEMECs transfected with KLF4 mutants K225R, K229R, and K225/229R, demonstrating that KLF4 at Lys225 and Lys229 has been succinylated by GCN5 in cells treated with VEGFA (Figures 7F and S4D). Such findings provide strong evidence that KLF4 interacts with GCN5 to succinylate the Lys225 and Lys229sites of KLF4 and is recruited to the VEGFA promoter region and then succinylates H3K79 at the promoter, ultimately increasing VEGFA transcription levels.
Then, we detected MAPK signaling pathways, including ERK, JNK, and P38 signaling pathways (Figures 8A and 8B), and attempted to find which one affects KLF4 succinylation and KLF4-GCN5 interaction. VEGFA increased the level of p-ERK, p- JNK, and p-P38 in HEMECs (Figures 8A and 8B). The ERK inhibitor PD98059 reversed the level of succinylated KLF4 (Figures 8C and 8D). Similarly, PD98059 weakened the binding between KLF4 and GCN5 (Figures 8E and 8F). Therefore, we believe that the ERK signaling pathway is key to the succinylation of KLF4 in response to VEGFA and its binding with GCN5. Riverso et al. showed that RAS/RAF/MEK/ERK signaling positively modulates KLF4 expression through the transcription factor E2F1 to promote the development of melanoma (Riverso et al., 2017). miR-25 was found to target KLF4 and directly activate the ERK signaling pathway to promote cell migration and invasion (Ding et al., 2018). ERK is a downstream component of the phosphoric acid pathway that regulates various conserved cellular processes.
Development of the human endometrial vasculature is critical to the physiology of uterine growth, menstrual cycles, placentation, and support of the growing embryo. We have elucidated the mechanism underlying enhanced cell proliferation and migration of HEMECs, in which VEGFA and KLF4 form a positive feedback loop to promote endometrial angiogenesis.
Limitations of the study
Our current study elucidates the mechanism of GCN5 modulates KLF4-VEGFA positive feedback loop by epigenetics, which contributes to endometrial angiogenesis. However, we currently cannot exclude the transcriptional effect of KLF4 in other endometrial cells such as endometrial glandular epithelial cells, endometrial cavity epithelial cells, and endometrial stromal cells, which will facilitate understanding of human endometrial physiology and pathophysiology that important for implantation in ART. We are investigating the mechanism by which KLF4 improves endometrial receptivity in endometrial luminal epithelial cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| KLF4 | Abcam | Cat#ab215036 |
| VEGFA | Proteintech | Cat#19003-1-AP; RRID: AB_2212657 |
| MMP-9 | Abcam | Cat#ab38898; RRID: AB_776512 |
| PCNA | Proteintech | Cat#10205-2-AP; RRID: AB_2160330 |
| Ki67 | Abcam | Cat#ab16667; RRID: AB_302459 |
| α-SMA | Abcam | Cat#ab202509; RRID: AB_2868435 |
| CD31 | Servicebio | Cat#GB11063-2 |
| Caspase 3 | Abcam | Cat#ab32351; RRID: AB_725946 |
| GCN5 | PTM-biolab | Cat#PTM-5115 |
| Succinyl-H3K79 | PTM-biolab | Cat#PTM-412 |
| Succinyllysine | PTM-biolab | Cat#PTM-419 |
| IgG | Abcam | Cat#ab172730; AB_2687931 |
| ERK | Proteintech | Cat#16443-1-AP; RRID: AB_10603369 |
| phospho-ERK | Immunoway | Cat#YP0101 |
| JNK | Proteintech | Cat#24164-1-AP; RRID: AB_2879443 |
| phospho-JNK | Immunoway | Cat#YP0156; RRID: AB_2895031 |
| P38 | Proteintech | Cat#14064-1-AP; RRID: AB_2878007 |
| phospho-P38 | Immunoway | Cat#YP0338; RRID: AB_2814750 |
| GAPDH | Proteintech | Cat#60004-1-Ig; RRID: AB_2107436 |
| Chemicals, peptides, and recombinant proteins | ||
| ECM | Sciencell | Cat#1001 |
| DMEM | Gibco | Cat#8121409 |
| Fetal Bovine Serum | Sciencell | Cat#0025 |
| Endothelial cell growth supplement | Sciencell | Cat#1052 |
| Penicillin and streptomycin | Sciencell | Cat#0503 |
| Recombinant Human VEGF165 | Proteintech | Cat#HZ-1038 |
| GnRHa | GL Biochem | Cat#61703 |
| HMG | Livzon | Cat#150802 |
| hCG | Livzon | Cat#150802 |
| ERK inhibitor (PD98059) | MedChemExpress | Cat#HY-12028 |
| JNK inhibitor (SP600125) | MedChemExpress | Cat#HY-12041 |
| P38 inhibitor (SB203580) | MedChemExpress | Cat#HY-10256 |
| Lipofectamine 2000 | Invitrogen | Cat#11668 |
| RIPA lysis buffer | Best Bio | Cat#310011 |
| Crystal Violet Staining Solution | Beyotime | Cat#C0121 |
| Protein A-agarose | Santa cruz | Cat#sc-2001 |
| Critical commercial assays | ||
| ChIP assay Kit | Millipore | Cat#17-295 |
| Human VEGFA ELISA kit | Elabscience | Cat# E-EL-H0111c |
| Experimental models: Cell lines | ||
| HEMEC | Sciencell | Cat#7010 |
| HEK 293T | Procell | Cat#CL-0005 |
| Experimental models: Organisms/strains | ||
| Mice: Kunming | Charles River Laboratories | Strain Code: 202 |
| Oligonucleotides | ||
| Human VEGFA promoter | Sangon Biotech | 5′-GCTGTTTGGGAGGTCAGAAATAGG-3′; 5′-ACGCTGCTCGCTCCATTCAC-3′ |
| Recombinant DNA | ||
| Plasmid: VEGFA promoter | This paper | N/A |
| Plasmid: pEGFP-KLF4 | (Yu et al., 2011b) | N/A |
| Plasmid: KLF4(K225R) | (Yu et al., 2011b) | N/A |
| Plasmid: KLF4(K229R) | (Yu et al., 2011b) | N/A |
| Plasmid: KLF4 (K225/229R) | (Yu et al., 2011b) | N/A |
| Plasmid: GCN5 control vector | This paper | N/A |
| Plasmid: pEGFP-GCN5 | This paper | N/A |
| Plasmid: GCN5(Y645A) | (Tong et al., 2020) | N/A |
| Plasmid: sh-GCN5 control vector | This paper | N/A |
| Plasmid:sh-GCN5 1 | This paper | N/A |
| Plasmid:sh-GCN52 | This paper | N/A |
| Plasmid:sh-GCN53 | This paper | N/A |
| Adenovirus: sh-KLF4 control vector | This paper | N/A |
| Adenovirus: sh-KLF4 1 | This paper | N/A |
| Adenovirus: sh-KLF4 2 | This paper | N/A |
| Adenovirus: sh-KLF4 3 | This paper | N/A |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| GraphPad Prism Version 8.0 | GraphPad | https://www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ming He (heming@hebcm.edu.cn).
Materials availability
Plasmids generated in this study are available upon request from the lead contact, Ming He (heming@hebcm.edu.cn).
Experimental model and subject details
Establishment of COH model
Animal housing and procedures used in this study were approved by the local Animal Care and Use Committee of Hebei University of Chinese Medicine. A total of 40 Kunming female virgin mice (age range, 7-8 weeks old) were housed and maintained in a 12-h light/12-h dark cycle clean animal facility at Hebei University of Chinese Medicine. Mice were randomly divided into normal group and COH group. Cytology of vaginal shedding was performed for two estrous cycles. Mice in the COH group underwent treatment using the GnRHa long protocol for 11 days. Briefly, gonadotropin-releasing hormone agonist (GnRHa, alarelin acetate, China) was injected intraperitoneally at 40 IU/100g for 9 days. Human menopausal gonadotropin (HMG, menotropin injection, China) was administered by intraperitoneal injection at 400 IU/kg on 9th day after GnRHa injection, followed by human chorionic gonadotropin (hCG) injection at 100 IU/100g 48 h later. At 16:00 on the 11th day, mice in each group were caged according to the ratio of female to male = 2:1, and those with pubic embolus in the morning of the next day were recorded as the first day of pregnancy (D1). To assess the characteristics of the endometrium using a COH mouse model during implantation, the mice were subjected to anesthesia with ketamine (60 mg/kg of body weight) plus xylazine (5 mg/kg of body weight), which was injected intraperitoneally prior to implantation (D4). Embryo implantation numbers were counted at D8.
Cell culture and treatments
HEMECs, located in the endometrium, which involved in endometrial angiogenesis during the menstrual cycle with the rapid growth and shedding of the endometrium, are a useful model to elucidate the mechanisms of normal and pathological angiogenesis and develop treatments for female reproductive tract disorders. HEMECs (Sciencell, USA) were cultured in endothelial cell medium (ECM, Sciencell, USA) containing 5% fetal bovine serum (FBS), 1% endothelial cell growth supplement (ECGS), 1% penicillin and streptomycin, maintained in a humidified at mosphere with 5% CO2 at 37 °C. After the density of HEMECs reached 80%, the cells were incubated in serum-free medium for 24 h prior to carrying out cell experiments.
Human embryonic kidney 293T (HEK293T) cells (Procell, China) were cultured in high glucose DMEM containing 10% fetal bovine serum, 1% penicillin and 1% streptomycin and performed for Luciferase reporter assay.
Method details
Western blotting analysis
HEMECs lysates were prepared using ice-cold RIPA lysis buffer and lysed on the ice for 30min. The homogenates were centrifuged at 8,000 rpm for 10 min at 4 °C to obtain cells lysates. Protein extracted from HEMECs were electrophoresed on 10% SDS-PAGE gels and transferred onto PVDF membranes. Membranes were blocked with 5% dry milk in TTBS for 2 h at 37 °C and incubated overnight at 4 °C with the following primary antibodies: 1:1000 GAPDH, 1:500 PCNA, 1:500 Caspase 3, 1:500 MMP-9, 1:500 VEGFA, 1:500 KLF4, 1:500 GCN5, 1:500 succinyl-H3K79, 1:1000 p-ERK, 1:1000 ERK, 1:500 P38, 1:500 JNK, 1:500 p-P38, and 1:500 p-JNK. After incubation with the respective secondary antibody, antibody-antigen complexes were visualized using a Chemiluminescence Plus Western immunoblot analysis kit (Millipore, USA).
Scratch-wound assay
The HEMECs in the 6-well plate were scratched by the tip of a 200-μL pipette and treated with VEGFA, infected with KLF4 overexpression or knockdown adenovirus. After incubation for appropriate time, the cells were fixed with 4% paraformaldehyde and for 10 min and stained with crystal violet staining solution for 10 min. Phase-contrast microscopic images were captured indicated times after incubation. The experiments were performed in triplicate.
Morphology analysis
Uterine tissues were harvested and embedded in paraffin. 5-μm-thick sections were prepared for HE staining. Sections were successively dewaxed with xylene and hydrated with different concentrations of ethanol, followed by hematoxylin-eosin staining. Images were acquired using a microscope (BX53, Olympus, Japan).
Immunofluorescence staining
Uterine tissues of harvested and embedded in paraffin were deparaffinized with xylene and rehydrated. Sections were blocked using 10% goat serum and incubated overnight at 4 °C with anti-α-SMA, anti-CD31 or anti-KLF4 primary antibodies. After washing with PBS and hybridized with a fluorescein-labeled secondary antibody for 1 h at 37 °C, the sections were mounted with DAPI and visualized with a laser scanning confocal microscope.
HEMECs were fixed in 4% paraformaldehyde for 10 min at room temperature, washed with PBS, and then incubated with 10% goat serum blocking solution for 30 min in a humidified room at room temperature. The cells were incubated with anti-KLF4, anti-GCN5, anti-Ki67 primary antibodies at 4 °C overnight, wash with PBS, and then mix with appropriate fluorescein-labeled secondary antibody for 1 h. Finally, the cells were washed with PBS, fixed with DAPI, and observed with a laser scanning confocal microscope.
ELISA assay
HEMECs culture supernatant was collected by centrifugation for 10 min at 1500 rpm. VEGFA expression levels were measured using Human VEGFA ELISA kit according to the manufacturer’s instructions. Absorption at a wave length of 450 nm (A450) was determined using the microplate reader.
Co-immunoprecipitation (CoIP)
Co-immunoprecipitation was performed as described previously (Zhang et al., 2012). Briefly, the protein of HEMECs were immunoprecipitated with 2 μg of anti-KLF4, anti-GCN5, or anti-Succinyllysine antibodies for 2 h at 4 °C, followed by incubation with protein A-agarose overnight at 4 °C. Protein A-agarose-antigen-antibody complexes were washed with 5 times with IPH washing buffer (50 mM Tris-HCl, pH 8.0, 2 M NaCl, 5 mM EDTA, 0.5% NP-40, 0.1 mM PMSF) for 20 min at 4 °C. The pellets were suspended in 2×SDS loading buffer and resolved using SDS-PAGE, followed by Western blotting analysis with anti-GCN5, anti-KLF4 antibodies.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assays were performed with the ChIP assay Kit (Millipore, Germany) following the manufacturer’s instructions. Cells were cultured to 80-90% confluence, crosslinked with 1% paraformaldehyde for 10 min and then stoped the fixation with 1.25 M Glycine for 5 min. After lysis, DNA fragments in the cell lysates were sheared to size 200 to 1000 bp by the sonication system. The samples were diluted 10-fold and then precleared with protein A-agarose/salmon sperm DNA for 30 min at 4 °C. The DNA fragments were immunoprecipitated overnight at 4 °C with the anti-GCN5, anti-KLF4, anti-succinylated H3K79, or anti-IgG (as negative control) antibodies. Chromatin bound agarose beads were harvested and washed with Low Salt Wash Buffer, High Salt Wash Buffer, Lici Wash Buffer, TE Wash Buffer and Elution Buffer consecutively. After crosslinking was reversed in 0.5M EDTA, 1M Tris HCl (PH 6.5) and proteinase K Buffer for 1h at 65 °C, followed by DNA purification experiments. Purified DNA was used for qRT-PCR, the primer sequences are listed: Forward:5′-GCTGTTTGGGAGGTCAGAAATAGG-3′; Reverse:5′-ACGCTGCTCGCTCCATTCAC-3′ (VEGFA).
Plasmid constructs
The CDS regions of GCN5 and Y645 mutant (Y to A) GCN5 were synthesized and cloned into the plasmid of CMV-MCS-EGFP-SV40-Neomycin by GeneChem. This constructed plasmid was named GFP-GCN5 and GFP-GCN5(Y645A). GCN5 shRNA (GeneChem, China) was generated with CCTGGAGAAGTTCTTCTACTT (sh-GCN5 1), CACATCATCAAGAAGCAGAAA (sh-GCN5 2), CCACCTGAAGGAGTATCACAT (sh-GCN5 3). A control vector was generated with the control oligonucleotide TTCTCCGAACGTGTCACGT.
pEGFP-KLF4 and pAd-KLF4 were prepared as previously described (Yu et al., 2011b). Lysine-to-argininemutations of GFP-KLF4 at Lys225 (K225R), Lys229 (K229R), or Lys225/229 (K225/229R) have also been described (Yu et al., 2011b). KLF4 shRNA (GeneChem, China) was generated with TCGGAGAGAGACCGAGGAGTT (sh-KLF4 1), GCCAGAGGAGCCCAAGCCAAA (sh-KLF4 2), TCCCTTCAACCTGGCGGACAT (sh-KLF4 3). A control vector was generated with the control oligonucleotide TTCTCCGAACGTGTCACGT.
Luciferase reporter assay
HEK293T cells were seeded into each well of a 24-well plate and grown for 24 h prior to transfection with reporter plasmid, expression plasmids for KLF4, GCN5 and the control pTK-RL plasmid using Lipofectamine 2000 reagent. The cells were harvested and measured firefly luciferase activities as well as renilla luciferase activities were measured with the Dual-Luciferase Reporter System (Promega) according to the manufacturer’s protocol. Specific promoter activity was expressed as the relative ratio of firefly luciferase activity to renilla luciferase activity. All promoter constructs were evaluated in a minimum of three separate wells per experiment.
Quantification and statistical analysis
Data were expressed as bar graphs (mean ± SD) of at least three independent experiments. Statistical analyses were performed using Student’s t test or one-way ANOVA based on the number of groups compared. A value of p < 0.05 were considered statistically significant.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 82074247, No. 81603654), Natural Science Foundation of Hebei Province (No. 2021423031), S&T Program of Hebei (No. 21377771D, No.19277749D), “333 Talens Project” of Hebei Province (No. A202101064), Construction Program of New Research and Development Platform and Institution, Hebei Province Innovation Ability Promotion Plan (No. 20567624H), and Postgraduates Innovation Funding Program of Hebei Province (No. CXZZSS2021083).
Author contributions
Conceptualization, M.H.; Methodology, S.J.D. and L.J.F.; Investigation, C.C., Y.L.Z., Y.Z., R.B.N., and Y.M.Z.; Writing – Original Draft, C.C. and M.H.; Writing – Review & Editing, C.C. and M.H.; Funding Acquisition, M.H. and Y.Z.; Resources, Y.C.M; Supervision, Y.C.M. and L.J.F.
Declarations of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104509.
Supplemental information
Data and code availability
This paper does not report original code. All data needed to evaluate the conclusions in the paper are present in the paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Achache H., Revel A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum. Reprod. Update. 2006;12:731–746. doi: 10.1093/humupd/dml004. [DOI] [PubMed] [Google Scholar]
- Allegra A., Marino A., Coffaro F., Lama A., Rizza G., Scaglione P., Sammartano F., Santoro A., Volpes A. Is there a uniform basal endometrial gene expression profile during the implantation window in women who became pregnant in a subsequent ICSI cycle? Hum. Reprod. 2009;24:2549–2557. doi: 10.1093/humrep/dep222. [DOI] [PubMed] [Google Scholar]
- Ashary N., Tiwari A., Modi D. Embryo implantation: war in times of love. Endocrinology. 2018;159:1188–1198. doi: 10.1210/en.2017-03082. [DOI] [PubMed] [Google Scholar]
- Atkins G.B., Jain M.K. Role of Kruppel-like transcription factors in endothelial biology. Circ. Res. 2007;100:1686–1695. doi: 10.1161/01.RES.0000267856.00713.0a. [DOI] [PubMed] [Google Scholar]
- Bhurke A.S., Bagchi I.C., Bagchi M.K. Progesterone-regulated endometrial factors controlling implantation. Am. J. Reprod. Immunol. 2016;75:237–245. doi: 10.1111/aji.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgain C. Endometrial biopsy in the evaluation of endometrial receptivity. J. Gynecol. Obstet. Biol. Reprod. 2004;33:S13–S17. doi: 10.1016/s0368-2315(04)96397-1. [DOI] [PubMed] [Google Scholar]
- Cheng S., Zhang X., Xu Y., Dai X., Li J., Zhang T., Chen X. Krüppel-like factor 8 regulates VEGFA expression and angiogenesis in hepatocellular carcinoma. Sci. Rep. 2018;8:17415. doi: 10.1038/s41598-018-35786-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X., Zhong T., Jiang L., Huang J., Xia Y., Hu R. miR-25 enhances cell migration and invasion in non-small-cell lung cancer cells via ERK signaling pathway by inhibiting KLF4. Mol. Med. Rep. 2018;17:7005–7016. doi: 10.3892/mmr.2018.8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander L.T., Hussani H., Cullinan M.P., Seymour G.J., De Silva R.K., De Silva H., Cameron C., Rich A.M. VEGF and VEGFR2 in dentigerous cysts associated with impacted third molars. Pathology. 2015;47:446–451. doi: 10.1097/PAT.0000000000000272. [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha L.A., Eberspaecher H., Seldin M.F., de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J. Biol. Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- He M., Zheng B., Zhang Y., Zhang X.H., Wang C., Yang Z., Sun Y., Wu X.L., Wen J.K. KLF4 mediates the link between TGF-β1-induced gene transcription and H3 acetylation in vascular smooth muscle cells. Faseb. J. 2015;29:4059–4070. doi: 10.1096/fj.15-272658. [DOI] [PubMed] [Google Scholar]
- Hu D., Gur M., Zhou Z., Gamper A., Hung M.C., Fujita N., Lan L., Bahar I., Wan Y. Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis. Nat. Commun. 2015;6:8419. doi: 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H., Liao L., Su X., Shuai Y., Zhang X., Deng Z., Jin Y. Declining histone acetyltransferase GCN5 represses BMSC-mediated angiogenesis during osteoporosis. Faseb. J. 2017;31:4422–4433. doi: 10.1096/fj.201700118R. [DOI] [PubMed] [Google Scholar]
- Jing Y., Ding D., Tian G., Kwan K.C.J., Liu Z., Ishibashi T., Li X.D. Semisynthesis of site-specifically succinylated histone reveals that succinylation regulates nucleosome unwrapping rate and DNA accessibility. Nucleic Acids Res. 2020;48:9538–9549. doi: 10.1093/nar/gkaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H.S., Hantak A.M., Stubbs L.J., Taylor R.N., Bagchi I.C., Bagchi M.K. Roles of progesterone receptor A and B isoforms during human endometrial decidualization. Mol. Endocrinol. 2015;29:882–895. doi: 10.1210/me.2014-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash G.E., Innes B.A., Drury J.A., Robson S.C., Quenby S., Bulmer J.N. Localization of angiogenic growth factors and their receptors in the human endometrium throughout the menstrual cycle and in recurrent miscarriage. Hum. Reprod. 2012;27:183–195. doi: 10.1093/humrep/der376. [DOI] [PubMed] [Google Scholar]
- Lessey B.A., Young S.L. What exactly is endometrial receptivity? Fertil. Steril. 2019;111:611–617. doi: 10.1016/j.fertnstert.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Li H.X., Han M., Bernier M., Zheng B., Sun S.G., Su M., Zhang R., Fu J.R., Wen J.K. Krüppel-like factor 4 promotes differentiation by transforming growth factor-β receptor-mediated smad and p38 MAPK signaling in vascular smooth muscle cells. J. Biol. Chem. 2010;285:17846–17856. doi: 10.1074/jbc.M109.076992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang C., Zhao T., Su Z., Li M., Hu J., Wen J., Shen J., Wang C., Pan J., et al. Lysine-222 succinylation reduces lysosomal degradation of lactate dehydrogenase a and is increased in gastric cancer. J. Exp. Clin. Cancer Res. 2020;39:172. doi: 10.1186/s13046-020-01681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Yu H., Chen X., Shen T., Cui Z., Si G., Zhang J., Cheng Y., Jia S., Zhang X., et al. PDGF-BB/KLF4/VEGF signaling Axis in pulmonary artery endothelial cell angiogenesis. Cell. Physiol. Biochem. 2017;41:2333–2349. doi: 10.1159/000475652. [DOI] [PubMed] [Google Scholar]
- Liu S., Xing Y., Lu W., Li S., Tian Z., Xing H., Tang K., Xu Y., Rao Q., Wang M., Wang J. RUNX1 inhibits proliferation and induces apoptosis of t(8;21) leukemia cells via KLF4-mediated transactivation of P57. Haematologica. 2019;104:1597–1607. doi: 10.3324/haematol.2018.192773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhao D., Zheng Y., Wang L., Qian Y., Xu C., Huang H., Hwa Y.L., Jin F. Expression of histone acetyltransferase GCN5 and histone deacetylase 1 in the cultured mouse preimplantation embryos. Curr. Pharmaceut. Des. 2014;20:1772–1777. doi: 10.2174/13816128113199990521. [DOI] [PubMed] [Google Scholar]
- Luense L.J., Donahue G., Lin-Shiao E., Rangel R., Weller A.H., Bartolomei M.S., Berger S.L. Gcn5-Mediated histone acetylation governs nucleosome dynamics in spermiogenesis. Dev. Cell. 2019;51:745–758.e6. doi: 10.1016/j.devcel.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak N.R., Brenner R.M. Vascular proliferation and vascular endothelial growth factor expression in the rhesus macaque endometrium. J. Clin. Endocrinol. Metab. 2002;87:1845–1855. doi: 10.1210/jcem.87.4.8413. [DOI] [PubMed] [Google Scholar]
- Niederberger C., Pellicer A., Cohen J., Gardner D.K., Palermo G.D., O'Neill C.L., Chow S., Rosenwaks Z., Cobo A., Swain J.E., et al. Forty years of IVF. Fertil. Steril. 2018;110:185–324.e5. doi: 10.1016/j.fertnstert.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Peng M., Li Y., Huang H., Jin F. The expression of GCN5, HDAC1 and DNMT1 in parthenogenetically activated mouse embryos. J. Obstet. Gynaecol. 2015;35:131–135. doi: 10.3109/01443615.2014.942605. [DOI] [PubMed] [Google Scholar]
- Phan H.M., Xu A.W., Coco C., Srajer G., Wyszomierski S., Evrard Y.A., Eckner R., Dent S.Y. GCN5 and p300 share essential functions during early embryogenesis. Dev. Dynam. 2005;233:1337–1347. doi: 10.1002/dvdy.20445. [DOI] [PubMed] [Google Scholar]
- Rabbani M.L., Rogers P.A. Role of vascular endothelial growth factor in endometrial vascular events before implantation in rats. Reproduction. 2001;122:85–90. doi: 10.1530/rep.0.1220085. [DOI] [PubMed] [Google Scholar]
- Ray S., Pollard J.W. KLF15 negatively regulates estrogen-induced epithelial cell proliferation by inhibition of DNA replication licensing. Proc. Natl. Acad. Sci. U S A. 2012;109:E1334–E1343. doi: 10.1073/pnas.1118515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riverso M., Montagnani V., Stecca B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene. 2017;36:3322–3333. doi: 10.1038/onc.2016.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmasi S., Sharifi M., Rashidi B. Ovarian stimulation and exogenous progesterone affect the endometrial miR-16-5p, VEGF protein expression, and angiogenesis. Microvasc. Res. 2021;133:104074. doi: 10.1016/j.mvr.2020.104074. [DOI] [PubMed] [Google Scholar]
- Sanchez-Navarro A., Pérez-Villalva R., Murillo-de-Ozores A.R., Martínez-Rojas M.Á., Rodríguez-Aguilera J.R., González N., Castañeda-Bueno M., Gamba G., Recillas-Targa F., Bobadilla N.A. Vegfa promoter gene hypermethylation at HIF1α binding site is an early contributor to CKD progression after renal ischemia. Sci. Rep. 2021;11:8769. doi: 10.1038/s41598-021-88000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M. Differential roles of vascular endothelial growth factor receptor-1 and receptor-2 in angiogenesis. J. Biochem. Mol. Biol. 2006;39:469–478. doi: 10.5483/bmbrep.2006.39.5.469. [DOI] [PubMed] [Google Scholar]
- Shima D.T., Kuroki M., Deutsch U., Ng Y.S., Adamis A.P., D'Amore P.A. The mouse gene for vascular endothelial growth factor. J. Biol. Chem. 1996;271:3877–3883. doi: 10.1074/jbc.271.7.3877. [DOI] [PubMed] [Google Scholar]
- Sun J., Huang W., Yang S.F., Zhang X.P., Yu Q., Zhang Z.Q., Yao J., Li K.R., Jiang Q., Cao C. Gαi1 and Gαi3mediate VEGF-induced VEGFR2 endocytosis, signaling and angiogenesis. Theranostics. 2018;8:4695–4709. doi: 10.7150/thno.26203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Guo D., Yan D., Ma C., Shao F., Wang Y., Luo S., Lin L., Tao J., Jiang Y., et al. KAT2A succinyltransferase activity-mediated 14-3-3ζ upregulation promotes β-catenin stabilization-dependent glycolysis and proliferation of pancreatic carcinoma cells. Cancer Lett. 2020;469:1–10. doi: 10.1016/j.canlet.2019.09.015. [DOI] [PubMed] [Google Scholar]
- Wan J., Su Y., Song Q., Tung B., Oyinlade O., Liu S., Ying M., Ming G.L., Song H., Qian J., et al. Methylated cis-regulatory elements mediate KLF4-dependent gene transactivation and cell migration. Elife. 2017;6:e20068. doi: 10.7554/eLife.20068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Su Y., Huang C., Yin Y., Zhu J., Knupp A., Chu A., Tang Y. FOXH1 is regulated by NANOG and LIN28 for early-stage reprogramming. Sci. Rep. 2019;9:16443. doi: 10.1038/s41598-019-52861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Guo Y.R., Liu K., Yin Z., Liu R., Xia Y., Tan L., Yang P., Lee J.H., Li X.J., et al. KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature. 2017;552:273–277. doi: 10.1038/nature25003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang C., Gu Q., Sims M., Gu W., Pfeffer L.M., Yue J. KLF4 promotes angiogenesis by activating VEGF signaling in human retinal microvascular endothelial cells. PLoS One. 2015;10:e0130341. doi: 10.1371/journal.pone.0130341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert B.T., Schölz C., Scholz C., Wagner S.A., Iesmantavicius V., Su D., Daniel J.A., Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Wen Y.L., Li L. Correlation between matrix metalloproteinase-9 and vascular endothelial growth factor expression in lung adenocarcinoma. Genet. Mol. Res. 2015;14:19342–19348. doi: 10.4238/2015.December.29.44. [DOI] [PubMed] [Google Scholar]
- Yang G., Yuan Y., Yuan H., Wang J., Yun H., Geng Y., Zhao M., Li L., Weng Y., Liu Z., et al. Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Rep. 2021;22:e50967. doi: 10.15252/embr.202050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Liu X., Chen J., Wen Y., Liu H., Peng Z., Yeerken R., Wang L., Li X. Lead-mediated inhibition of lysine acetylation and succinylation causes reproductive injury of the mouse testis during development. Toxicol. Lett. 2020;318:30–43. doi: 10.1016/j.toxlet.2019.10.012. [DOI] [PubMed] [Google Scholar]
- Yang W., Li Z., Qin R., Wang X., An H., Wang Y., Zhu Y., Liu Y., Cai S., Chen S., et al. YY1 promotes endothelial cell-dependent tumor angiogenesis in hepatocellular carcinoma by transcriptionally activating VEGFA. Front. Oncol. 2019;9:1187. doi: 10.3389/fonc.2019.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M., Zhou H.J., Lin C., Long L., Yang X., Zhang H., Taylor H., Min W. CD34(+)KLF4(+) stromal stem cells contribute to endometrial regeneration and repair. Cell Rep. 2019;27:2709–2724.e3. doi: 10.1016/j.celrep.2019.04.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Li J., Chen H., Fu J., Ray S., Huang S., Zheng H., Ai W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Zheng B., Han M., Wen J.K. ATRA activates and PDGF-BB represses the SM22α promoter through KLF4 binding to, or dissociating from, its cis-DNA elements. Cardiovasc. Res. 2011;90:464–474. doi: 10.1093/cvr/cvr017. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Yuan H., Yang G., Yun H., Zhao M., Liu Z., Zhao L., Geng Y., Liu L., Wang J., et al. IFN-alpha confers epigenetic regulation of HBV cccDNA minichromosome by modulating GCN5-mediated succinylation of histone H3K79 to clear HBV cccDNA. Clin. Epigenet. 2020;12:135. doi: 10.1186/s13148-020-00928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Li Y., Pan Y., Lan X., Song F., Sun J., Zhou K., Liu X., Ren X., Wang F., et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Zheng B., Gu C., Fu J.R., Wen J.K. TGF-β1 downregulates AT1 receptor expression via PKC-δ-Mediated Sp1 dissociation from KLF4 and smad-mediated PPAR-γ association with KLF4. Arterioscler. Thromb. Vasc. Biol. 2012;32:1015–1023. doi: 10.1161/ATVBAHA.111.244962. [DOI] [PubMed] [Google Scholar]
- Zheng X., Li A., Zhao L., Zhou T., Shen Q., Cui Q., Qin X. Key role of microRNA-15a in the KLF4 suppressions of proliferation and angiogenesis in endothelial and vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 2013;437:625–631. doi: 10.1016/j.bbrc.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Zorro Shahidian L., Haas M., Le Gras S., Nitsch S., Mourão A., Geerlof A., Margueron R., Michaelis J., Daujat S., Schneider R. Succinylation of H3K122 destabilizes nucleosomes and enhances transcription. EMBO Rep. 2021;22:e51009. doi: 10.15252/embr.202051009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code. All data needed to evaluate the conclusions in the paper are present in the paper. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.