Abstract
The Mediator complex, which in humans is 1.4 MDa in size and includes 26 subunits, controls many aspects of RNA polymerase II (Pol II) function. Apart from its size, a defining feature of Mediator is its intrinsic disorder and conformational flexibility, which contributes to its ability to undergo phase separation and to interact with a myriad of regulatory factors. In this Review, we discuss Mediator structure and function, with emphasis on recent cryogenic electron microscopy data of the 4.0-MDa transcription preinitiation complex. We further discuss how Mediator and sequence-specific DNA-binding transcription factors enable enhancer-dependent regulation of Pol II function at distal gene promoters, through the formation of molecular condensates (or transcription hubs) and chromatin loops. Mediator regulation of Pol II reinitiation is also discussed, in the context of transcription bursting. We propose a working model for Mediator function that combines experimental results and theoretical considerations related to enhancer–promoter interactions, which reconciles contradictory data regarding whether enhancer–promoter communication is direct or indirect. We conclude with a discussion of Mediator’s potential as a therapeutic target and of future research directions.
Subject terms: Electron microscopy, Transcription
The Mediator complex is an important regulator of RNA polymerase II. This Review discusses recent structural insights into Mediator function and proposes a model that reconciles contradictory data on whether enhancer–promoter communication during transcription is direct or indirect.
Introduction
The RNA polymerase II (Pol II) complex initiates transcription as part of a preinitiation complex (PIC), which is about 4 MDa in size in humans1. The PIC contains the general transcription factors (TFs) TF IIA (TFIIA), TFIIB, TFIID, TFIIE, TFIIF and TFIIH, Pol II and Mediator (Fig. 1). Because Pol II transcribes all protein-coding genes and many non-coding RNAs, including enhancer RNAs (eRNAs), it initiates transcription at tens of thousands of sites in a typical human cell. Pol II is recruited to correct transcription start sites through the coordinated action of the PIC and sequence-specific DNA-binding TFs such as p53. Whereas some Pol II transcripts are short (for example, eRNAs or small nuclear RNAs), others can extend to 100 kb or more in human cells. Numerous factors, distinct from those in the PIC, interact with elongating Pol II to control its function2.
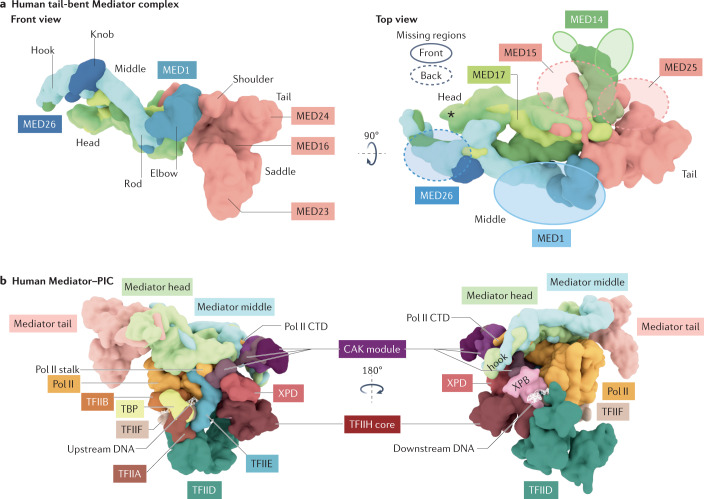
Fig. 1. Structures of human Mediator and Mediator–PIC.
a | Structural features of Mediator in the tail-bent conformation18. In this conformation (compared with the tail-extended conformation), MED23 and MED24 separate in combination with a shift in the MED16 propeller domain. The scaffold subunits MED17 and MED14 run through the head and middle modules and contact the tail. For the ‘top view’ shown on the right, structural disorder among Mediator subunits is indicated as semi-transparent ovals. The size of the oval correlates with the size of the disordered region unresolved by cryogenic electron microscopy18. The location of MED6 is denoted with an asterisk. The middle module includes MED1, MED4, MED7, MED9, MED10, MED19, MED21, MED26 and MED31; the head module includes MED6, MED8, MED11, MED14, MED17, MED18, MED20, MED22, MED27, MED28 and MED30; the tail module includes MED15, MED16, MED23, MED24, MED25 and MED29. See also Supplementary Movie 1. b | Surface renderings of two views of the Mediator–preinitiation complex (PIC) structure18. The Mediator complex is shown in the tail-bent conformation, suggesting it contains MED16 isoform 1 (ref.18). Mediator sits atop the PIC, with the cyclin-dependent kinase (CDK)-activating kinase (CAK) module of transcription factor IIH (TFIIH) sandwiched between the Mediator hook and MED6. The CAK module is tethered to core TFIIH through its flexible MAT1 subunit. Consistent with its role in promoting phosphorylation of the RNA polymerase II (Pol II) caboxy-terminal domain (CTD), Mediator positions the CTD near the CAK, which contains CDK7. The TFIID subunit TATA box-binding protein (TBP) binds DNA upstream of the transcription start site. The TAF2 subunit of TFIID contacts the TFIIH subunits p52 and p8 (not shown) to help position the DNA translocase XPB, thereby promoting its interaction with downstream DNA, the Pol II jaw (not shown) and the Mediator hook domain. Finally, the Pol II stalk is an interaction hub, making contact with several Mediator subunits and with TFIIE, TFIIH and TFIIB in the PIC. See also Supplementary Movie 2.
The Mediator complex exists as two compositionally and functionally distinct entities, depending upon whether it is bound to a Mediator kinase module (MKM), to form a larger cyclin-dependent kinase (CDK)–Mediator complex. The basic functions of Mediator or CDK–Mediator are to activate or block Pol II transcription initiation, and to regulate Pol II elongation. Throughout this Review, we discuss many fundamental aspects of Pol II transcription regulation — including initiation, pausing, chromatin architecture, enhancer function, molecular condensates and transcription bursting — focusing on the mechanistic roles of Mediator. We also propose a model for Mediator function at enhancers that reconciles evidence for their direct or indirect regulation of Pol II activity at promoters. We conclude with a discussion of key topics for future research. For additional information about Pol II-mediated transcription that is not focused on Mediator, we direct readers to other reviews on this topic1,3–5. We note there is extensive literature on the Mediator complex in plants, a topic we do not discuss here but that is covered in other reviews6–8.
Mediator composition and structure
Although Mediator is conserved from yeast to humans, its subunit composition and sequences and its overall structure have evolved considerably (Box 1). Yeast (Saccharomyces cerevisiae) Mediator includes 21 subunits and is approximately 0.8 MDa in size, whereas human Mediator includes 26 subunits and is 1.4 MDa in size (Fig. 1a and Supplementary Movie 1). The yeast Mediator complex has a modular organization with subcomplexes called the ‘head’, ‘middle’ and ‘tail’, which can each be biochemically reconstituted. The reconstituted yeast modules helped to establish the first set of high-resolution structural data for the Mediator complex9–13. Through genetic knockout or depletion experiments, individual modules have been shown to function independently in yeast14–16; the head and middle modules associate directly with Pol II, whereas the tail does not. This modular architecture is roughly conserved in mammalian Mediator complexes.
Mouse and human Mediator complexes are highly conserved, each containing 26 subunits that are extensively interconnected17–20 (Fig. 1a and Supplementary Movie 1), suggesting greater functional coordination between modules compared with yeast. In both yeast and mammalian Mediator, the MED14 and MED17 subunits span nearly the entire length of the complex and contact regions in the head, middle and tail modules (Box 1). MED14 and MED17 are also part of an α-helical bundle formed by nine subunits that serves as a major structural scaffold17–19 (Supplementary Movie 1). In humans, this α-helical bundle is twice as large as in yeast, supporting a larger tail module that may integrate more diverse regulatory inputs and may direct conformational changes21. In both yeast and human Mediator, the tail module is more flexible compared with the head and middle modules22, and can adopt distinct conformational states18.
The four-subunit MKM reversibly associates with Mediator (Box 2). Unlike the other Mediator modules (head, middle and tail), the kinase module exists as a stable entity in cells and likely functions independently of Mediator. For example, the MKM can be isolated as a stable entity from human cells23, and in yeast, the kinase module is selectively degraded following nutrient stress24 and can be recruited to genomic loci separately from Mediator14,25,26. The kinase module subunits CDK8, cyclin C, MED12 and MED13 are conserved from yeast to humans, although the sequences and subunit sizes have diverged. A structure for the yeast kinase module has been determined27, but high-resolution data for the human complex are not yet available. The human kinase module is about 600 kDa in size, and when bound to Mediator, it markedly alters Mediator function by preventing its association with Pol II28–30; furthermore, MKM binding is mutually exclusive with inclusion of the metazoan-specific subunit MED26 in Mediator31–33, resulting in a 29-subunit CDK–Mediator complex.
Box 1 Similarities and differences between the human and yeast Mediator complexes.
Research in yeast has driven our understanding of Mediator structure and function. Many basic Mediator functions, such as binding the RNA polymerase II (Pol II) carboxy-terminal domain, conformational flexibility and transcription factor (TF)-dependent activation of Pol II, are conserved from yeast to humans. However, the regulation of Pol II-mediated transcription is more complex in mammals, and mechanistic findings from yeast are not always relevant in human cells. Recent data suggest that perhaps 80% of yeast genes do not require TF-dependent regulation240, in apparent contrast to mammalian cells. Some additional distinctions between human and yeast Mediator are summarized below.
Different subunit composition and structure: whereas yeast and human Mediator share a similar core architecture (for example, similar organization of orthologous subunits), the structures of yeast Mediator and human Mediator are different; the figure shows human Mediator in the tail-extended conformation, which best resembles the Saccharomyces cerevisiae conformation. Structured regions of MED14 and MED17 are depicted as ribbons in each structure. The structural distinctions result primarily from the additional subunits in human Mediator (MED23, MED24, MED25, MED26, MED28 and MED30) and expanded sequence in MED1 and MED14, which collectively account for about 600 kDa of additional molecular mass.
Diversification of subunit functions: alternative splicing can alter protein structure and function and is widespread in humans but rare in yeast. Human Mediator subunits include 363 exons (261 among the 26 Mediator subunits; 102 in the four kinase module subunits), whereas S. cerevisiae Mediator includes 27 exons (54 exons in Schizosaccharomyces pombe). Moreover, vertebrates have paralogues of Mediator kinase module subunits CDK8, MED12 and MED13 (Box 2).
An expanded set of TF–Mediator interactions: in accordance with the diversity of cell types in humans, the number of TFs has expanded greatly from yeast to humans, from approximately 300 to more than 1,600 (ref.81). The metazoan-specific Mediator subunits are commonly bound by TFs (Table 2), suggesting that these subunits evolved in part to accommodate an expanding set of TFs. In yeast, the tail subunit Med15 is a major target of TF binding43, whereas TFs target many subunits in human Mediator (Table 2), and complexes lacking the tail can still respond to TFs241.
Regulation through enhancer–promoter interactions: Pol II transcription in humans is controlled by arrays of cell type-specific enhancers, which interact with promoters across large stretches of genomic DNA. Such long-range regulation is absent in yeast151. In humans, an expanded Pol II carboxy-terminal domain (52 repeats of the general consensus YSPTSPS, compared with 26 in S. cerevisiae) and Mediator interactions with cohesin31,171, which is dependent on MED30, a metazoan-specific subunit242, may contribute to enhancer–promoter interactions167.
Postinitiation transcription control: Pol II promoter-proximal pausing is largely absent in S. cerevisiae, which lacks negative elongation factor (NELF) and some subunits of the super elongation complex. Human Mediator regulates Pol II pausing and elongation in a variety of ways, including through its metazoan-specific MED26 subunit77 or through the Mediator kinase module (Box 2).
Box 2 The MKM.
The Mediator kinase module (MKM) contains four subunits in yeast or human cells: cyclin-dependent kinase 8 (CDK8), cyclin C, MED12 and MED13. Paralogues of CDK8, MED12 and MED13 emerged in vertebrates (CDK19, MED12L and MED13L, respectively); the biological roles of these paralogues remain poorly understood, but they appear to be functionally distinct. The subunit organization of the MKM is approximated in the figure (CDK8, cyclin C, MED12 and MED13). Through its association with Mediator (forming the CDK–Mediator complex), the kinase module delivers the only known enzymatic activity to the complex. Transcription factors (TFs) are a major class of CDK8 phosphorylation targets in human and yeast cells, although a more diverse set of substrates have been identified in humans (see the figure). The MKM phosphorylates itself (not shown) and the Mediator complex (see the figure inset; sites labelled ‘P’ approximate phosphorylation locations in MED14 and MED26). In general, the MKM appears to be especially important for driving changes in gene expression, such as during a stress response or during development70,71,74,243,244. Because changes in gene expression are controlled by TFs, these CDK8-dependent or CDK19-dependent changes likely reflect their regulation of TF function (see the figure).
The kinase module controls Mediator function by preventing its association with RNA polymerase II (Pol II), as observed in both yeast and human cells29,30. Consequently, the kinase module may help to shut down transcription from an active promoter and/or it may serve to mark a genetic locus for future activation. In support of the latter hypothesis, several laboratories have concluded that CDK–Mediator can mark genes for future activation213,245, which presumably occurs upon dissociation of the kinase module from Mediator.
Major biological roles of the human MKM include the control of enhancer function and the regulation of transcription elongation (see the figure). Many laboratories have demonstrated that disruption of MED12 function or inhibition of Mediator kinase activity prevents normal enhancer function and disrupts activation of gene expression programmes70,74,113,114,246. Human MED12 may also contribute to enhancer function through binding enhancer RNA112 and/or through regulation of CDK8 or CDK19 function23,247. The MKM also controls Pol II promoter-proximal pausing and elongation in human cells, most likely through its association with the super elongation complex (SEC)72. It remains unclear whether the kinase module mediates these functions as CDK–Mediator or as an independent entity. Structural data for the human preinitiation complex reveal that the kinase module and the CDK-activating kinase module of TF IIH (TFIIH) bind overlapping sites on Mediator (the hook region; dashed oval in the figure inset), suggesting that TFIIH will dissociate from the preinitiation complex upon MKM binding. The putative location of the kinase module in promoter-bound CDK–Mediator would position it downstream of the transcription start site (TSS), which could enable its phosphorylation of, and interaction with, pausing and elongation factors such as AF4/FMR2 family member 4 (AFF4), negative elongation factor (NELF) and positive transcription elongation factor b (P-TEFb). Inset shows top view of Mediator in tail-extended conformation.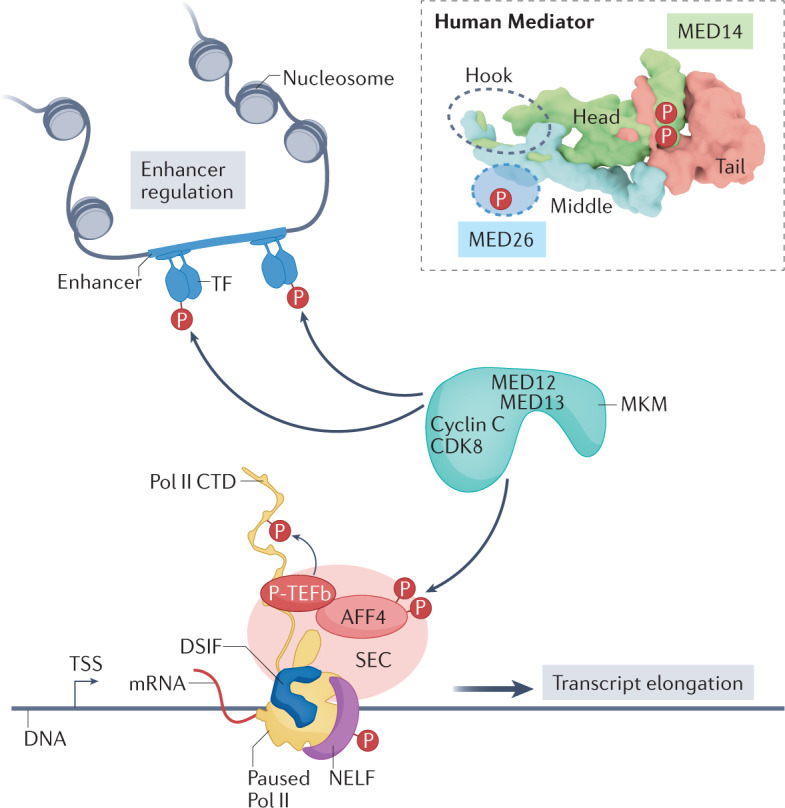
CTD, carboxy-terminal domain; DSIF, 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor.
Disorder and conformational allostery
Disorder and/or conformational flexibility is a common characteristic of proteins and protein complexes that regulate transcription34. Mediator is a quintessential example of disorder and flexibility, having subunits that contain more intrinsically disordered regions (IDRs) than other protein complexes of comparable size35 (Fig. 1a and Table 1). Large sets of disordered sequences contribute to Mediator’s ability to undergo phase separation into molecular condensates36, an aspect of Mediator function that we discuss later.
Table 1.
Disordered regions in Mediator subunits
| Subunita | Disordered residuesb | Propensity score |
|---|---|---|
| MED1 | 518–1581c | 4.0 |
| MED4 | 192–270 | 2.0 |
| MED6 | 192–246 | −0.6 |
| MED7 | 175–-233 | 1.0 |
| MED8 | 192–268 | 1.0 |
| MED9 | 1–65 | 2.0 |
| MED10 | None | NA |
| MED11 | None | NA |
| MED14 | 1–50, 965–1454 | 4.2 |
| MED15 | 1–615c | 5.4 |
| MED16 | None | 0.6 |
| MED17 | None | 0.9 |
| MED18 | None | −1.5 |
| MED19 | 1–62, 159–244 | 2.8 |
| MED20 | None | 0.1 |
| MED21 | None | −1.6 |
| MED22 | 139–200 | 0.7 |
| MED23 | 1334–1368 | 1.0 |
| MED24 | None | 0.3 |
| MED25 | 199–747 | 2.7 |
| MED26 | 1–480c | 2.8 |
| MED27 | None | −0.8 |
| MED28 | 1–42, 146–178 | 1.3 |
| MED29 | 1–55 | 0.4 |
| MED30 | 1–29 | −0.8 |
| MED31 | None | NA |
| Kinase module | ||
| MED12 | 1–75, 308–356, 619–728, 1731–2177 | 3.9 |
| MED12L | 1–58, 633–717, 1750–2189 | 2.3 |
| MED13 | 429–591, 674–887, 1472–1630 | 3.7 |
| MED13L | 310–1109, 1529–1680 | 4.6 |
| CDK8 | 1–49, 340–464 | 2.4 |
| CDK19 | 1–50, 321–502 | 3.4 |
| Cyclin C | 245–283 | −1.2 |
CDK, cyclin-dependent kinase. aSubunits with unresolved regions in the structure provided in ref.18 are listed, with disordered residues indicated. The propensity score is also shown, which estimates the propensity for intrinsic disorder219. Some Mediator subunits (designated ‘NA’) contain too few amino acids to be evaluated by the propensity score metric. Structurally unresolved regions from subunits highlighted in bold are shown in Fig. 1a. bPredicted disordered residues are given for the kinase module. cRegions that were built by polyalanine chains18.
Structural disorder and conformation dynamics are hallmarks of information processing in biology, and Pol II transcription requires integration of a large number of regulatory inputs37. As the primary intermediate between TFs and the PIC (see later), Mediator serves as a hub through which diverse and combinatorial inputs can be simultaneously coordinated to yield context-specific outcomes; Mediator’s role as a ‘signal integrator’ is likely enabled by its intrinsic disorder and conformational allostery38. In support of this concept, Mediator adopts distinct conformations and interacts with different regulatory factors in distinct biological contexts. For example, Mediator undergoes structural shifts upon association with Pol II39,40 or upon assembly into the PIC18. TF binding can also trigger conformational shifts in Mediator33, and proteomics experiments suggest that some factors preferentially bind Mediator conformations induced by TF binding, including the cohesin, CREB-binding protein (CBP; also known as KAT3A)–p300 (also known as KAT3B) and SAGA complexes31. Mediator structural shifts also allow stable, high-affinity Pol II carboxy-terminal domain (CTD) binding18,22 (Supplementary Movie 1) and may activate Pol II within the PIC41.
Although yeast and human Mediator are divergent in amino acid sequence, disordered regions and conformational flexibility are conserved in yeast Mediator complexes35, which similarly show evidence of structural changes upon binding TFs or Pol II13,40. Moreover, a recent structure of Mediator from the fungus Chaetomium thermophilum showed evidence of structural shifts and conformational coupling between head, middle and tail modules even in the absence of other bound factors42.
A recent study reported human Mediator structures with the tail module in two different conformations (Supplementary Movie 1), called ‘extended’ and ‘bent’18. The structural change appeared to result from incorporation of distinct MED16 isoforms: a MED16 isoform lacking only 36 residues at the carboxyl terminus (isoform 1) caused a β-propeller domain to shift about 55 Å, thereby bending the tail and shifting the MED23 subunit about 25 Å away from MED24 (at the tail) towards the head and shoulder region (Supplementary Movie 1). This ‘tail-bent’ conformation establishes new MED23 contacts with MED14 and MED15. Furthermore, large portions of MED16 and MED25 become disordered during the transition from the tail-extended conformation to the tail-bent conformation18, which raises the question of whether other Mediator structured domains become disordered (or vice versa) in different contexts (for example, binding to a TF).
Given that IDRs commonly represent protein–protein interfaces, Mediator IDRs may function, at least in part, as sites of interaction with other regulatory proteins43. This possibility is consistent with the diverse array of cofactors that interact with Mediator44. Different sets of IDRs may be exposed when Mediator is bound to different factors, and this may contribute to its conformational allostery and may to help regulate the timing of Mediator’s interactions with other proteins. However, because IDRs cannot be resolved to high resolution, it is difficult to determine whether Mediator IDRs undergo structural reorganization (for example, upon TF binding or during PIC assembly). Indeed, removal of the MED1 IDR increased structural homogeneity of Mediator for cryogenic electron microscopy (cryo-EM) analysis18.
Mediator controls PIC function
Recent cryo-EM structures of the human PIC complement earlier structural data from yeast45,46. Here we focus on Mediator and the human (rather than the yeast) PIC, because the recent cryo-EM data include the entire TFIID complex (Fig. 1b and Supplementary Movie 2); by contrast, structural data for yeast PICs thus far have contained TATA box-binding protein (TBP), but not the other TFIID subunits. TFIID binds promoter DNA at three different sites: the TATA box sequence upstream of the transcription start site, the initiator element at the transcription start site and the downstream promoter element. These interactions are mediated, respectively, by the TFIID subunits TBP, TAF1 and TAF1 with TAF2 (refs47,48). Most human promoters contain at least one of these sequences, but few contain all three49. Nevertheless, TFIID binds promoters with different combinations of sequence elements in a similar but not identical fashion47.
Structural data for the human PIC reveal that Mediator and TFIID work together to organize the PIC on promoter DNA (Fig. 1b and Supplementary Movie 2), in agreement with prior biochemical and cellular data50–54. Mediator and TFIID cooperate to position TFIIH within the PIC, through multiple contacts18. TAF2 contacts the p8 and p52 subunits of TFIIH to help position XPB for interaction with the Pol II jaw (RPB5 subunit), whereas Mediator contacts XPB through its hook domain (Fig. 1b and Supplementary Movie 2). Thus, Mediator and TFIID sandwich XPB and position its ATP-dependent translocase for promoter opening18, a first step in Pol II transcription initiation (see later). Mediator also positions the CDK-activating kinase (CAK) module of TFIIH, which includes CDK7, cyclin H and MAT1, within the PIC, through contacts with MED6 and the hook domain (Fig. 1a and Supplementary Movie 2). This Mediator–CAK interaction properly orients the kinase CDK7 for Pol II CTD phosphorylation17,18. Furthermore, CDK7 was observed to adopt an active conformation (compared with autoinhibited) upon binding Mediator in the PIC17. These data provide a structural basis for prior observations that Mediator activates the kinase activity of TFIIH41,55,56.
TFIID and Mediator also recruit57 and orient Pol II within the PIC. This sets up the entire PIC structure, because the remaining PIC factors assemble around Pol II: TFIIA and TFIIB bind opposite sides of the TFIID subunit TBP, whereas TFIIE and TFIIF bind directly to Pol II (Fig. 1b and Supplementary Movie 2). TFIIB also directly binds Pol II; therefore, TFIID affects the TFIIB–Pol II interaction through TBP. Mediator interacts extensively with Pol II19, making contact with the Pol II stalk (RPB4 and RPB7 subunits), the Pol II dock domain (within RPB1), the Pol II RPB3–RPB11 dimer, RPB8 and the Pol II CTD17,18.
Biochemical and proteomics data indicate that Mediator and TFIID directly interact through the TFIID subunit TAF7 and the amino terminus of MED26 (ref.58). Whereas the MED26 amino terminus is structured on its own59, it is not resolved in current PIC structures17,18, suggesting it is not stably bound. Notably, biochemical and cell-based experiments suggest that Mediator converts TFIID into an active structural state54. In agreement with these experiments, the structure of TFIID shifts upon Mediator binding to the PIC18,47.
The PIC is structurally dynamic
Although the PIC structures show a single conformational state (Fig. 1b and Supplementary Movie 2), the complex is highly dynamic. TFIID, TFIIH and Mediator account for most of the protein density, and each of these factors contains disordered regions and can adopt different structures33,60,61. Furthermore, Pol II is an enzyme that generates a substantial pulling force to move along DNA62, including proofreading and translocation that require coordinated movements involving the bridge helix and trigger loop with each newly incorporated RNA base63. Cryo-EM analysis of the complete human PIC (that is, containing TFIID and Mediator) revealed distinct structures, in which XPB, TBP and promoter DNA were not correctly positioned for transcription initiation18. To orient the PIC for transcription initiation, conformational changes involving TFIID, TFIIH and TFIIE were observed; moreover, rearrangement of the Mediator hook and knob, combined with a 20° rotation of Mediator around the Pol II stalk, was required (Supplementary Movie 2). In addition, biochemical and structural data suggest that a flexible MED7–MED21 ‘hinge’ promotes stable Mediator–Pol II association39. These results underscore the structural dynamics associated with PIC function.
Structural changes occur not only from spontaneous, thermodynamically permissible fluctuations (for example, from the conformational rearrangement of the PIC to an initiation-competent form) but also from energy-dependent enzymatic processes. The XPB subunit of TFIIH is an ATP-dependent translocase that activates Pol II transcription initiation by opening the DNA duplex at the transcription start site, through the generation of torsional strain64. XPB translocation not only opens the DNA duplex but also shifts the position of the Pol II stalk; this shift disrupts both the contact of MAT1 with the stalk and a structural interface between the TFIIH subunits XPB and XPD65. Because the stalk is an interaction hub within the PIC (Fig. 1b), we speculate that this XPB-induced structural shift may trigger additional conformational changes among its associated factors (for example, Mediator, TFIIE and TFIIH) during transcription initiation. Likewise, XPB is a PIC interaction hub, and its ATPase-dependent translocation may initiate conformational rearrangements in TFIID and/or Mediator that could facilitate Pol II promoter escape.
Mediator undergoes a structural shift upon binding the Pol II CTD18, which orients the CTD for phosphorylation by the TFIIH-associated kinase CDK717,18 (Fig. 1b and Supplementary Movie 2). During transcription initiation, CDK7 phosphorylates the CTD, and it has been proposed that Mediator may sequentially bind and release the CTD through a structural gating mechanism to allow multiple CTD repeats to access the CDK7 active site17,18. Because Mediator does not bind phosphorylated CTD repeats66, this mechanism would help to release Pol II from contacts with the PIC; that is, it would facilitate promoter escape, which must occur for the next phase of transcription: promoter-proximal pausing.
Pol II pausing and transcript elongation
Following promoter escape, Pol II typically pauses after transcribing 20–80 nucleotides67,68. Pol II promoter-proximal pausing is regulated by a host of factors, including 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole sensitivity-inducing factor (DSIF) and negative elongation factor (NELF), which bind Pol II surfaces that in the PIC are occupied by TFIIB, TFIIE and TFIIF69. Pol II pausing serves numerous biological functions and coincides with 5′ capping of the nascent pre-mRNA. The ‘release’ of pausing is regulated in part by the kinase CDK9, which is a component of the super elongation complex (SEC).
The CDK–Mediator complex has a role in controlling Pol II pausing, at least in certain biological contexts. Inhibition of Mediator kinase CDK8 inhibits Pol II pause release (increases pausing) during interferon-γ activation70; likewise, CDK8 depletion increases pausing at hypoxia-inducible genes71. Biochemical and proteomics data revealed a physical association of SEC subunits with CDK–Mediator, but not with Mediator lacking the CDK module31,72 (but see below). The mechanism by which CDK–Mediator may cooperate with the SEC at Pol II promoter-proximal pause sites remains unclear; however, Mediator kinase activity may contribute to pause release, as CDK8 or CDK19 phosphorylate factors that regulate Pol II pausing, including NELF and SEC subunits73.
The MKM also influences Pol II-mediated transcript elongation. Depletion of CDK8 or inhibition of its kinase activity correlates with slower Pol II elongation rates at gene bodies71,72,74. Moreover, MED12 depletion causes elongation defects and reduced CDK9 occupancy at gene bodies75, underscoring a functional link between the MKM and the SEC. Perhaps related to this functional coordination, acute (2-h degron-mediated) depletion of MED14 in human cells caused a global reduction in Pol II transcription76; however, many genes maintained expression through compensatory activation of CDK9, which promotes pause release.
Finally, the Mediator subunit MED26 has been linked to control of Pol II elongation through interactions with SEC subunits77. This is notable because MED26 is mutually exclusive with CDK module–Mediator association31–33, suggesting the existence of distinct, co-dependent or redundant functions for SEC regulation through MED26 or through the MKM.
Mediator–TF reciprocal regulation
Mediator subunits lack any discernible DNA-binding domains, and there is no available evidence that Mediator can bind DNA in a sequence-specific manner. In this section, we discuss how Mediator is recruited to specific genomic locations.
TFs recruit Mediator to genomic loci
Sequence-specific DNA-binding TFs appear to be the main mechanism by which Mediator is recruited to the genome78, although transcription hubs or molecular condensates79 may also contribute (see later). In general, TFs activate transcription by recruiting Mediator and chromatin modifying factors such as CBP or chromatin remodelling complexes such as SWI/SNF to specific genomic locations. TF-dependent recruitment of these factors helps to establish conditions favourable for transcription, through histone acetylation (for example, by CBP or p300) and nucleosome remodelling (for example, by SWI/SNF) around transcription start sites80. Although the human genome encodes more than 1,600 TFs81, only a fraction of these TFs have well-characterized molecular mechanisms, and many interact with different Mediator subunits (Table 2), suggesting that subunit-specific TF–Mediator interactions are required to activate TF-specific gene sets. In agreement with this hypothesis, removal of individual Mediator subunits has been shown to block gene activation by TFs that bind the missing subunit. For example, in mouse embryonic fibroblasts, knockout of Med1 blocks activation by nuclear receptors82, and knockout of Med23 blocks activation by ELK1 (refs83,84).
Table 2.
A representative set of mammalian Mediator subunits and transcription factors that bind them
| Subunit | Transcription factors | Refs |
|---|---|---|
| MED1 | TRα, TRβ, RARα, RXRα, PPARγ, VDR, PPARα, ER, AR, GR, HNF4, PGC1α, POU2AF1 | 95,220–226 |
| MED14 | HNF4, PPARγ | 224,227 |
| MED15 | SMAD2–SMAD4, SMAD3–SMAD4, SREBP1A | 90,228 |
| MED17 | VP16, p53 | 229 |
| MED19 | REST | 230 |
| MED23 | RUNX2, E1A, ELK1 | 83,231,232 |
| MED24 | TR | 221 |
| MED25 | ETS factors, ATF6α | 233,234 |
| CDK8a | MYC | 235 |
| MED12a | β-Catenin, REST, GLI3 | 236–238 |
The published evidence of transcription factor–Mediator interactions is too extensive to catalogue here; this is a curated list. AR, androgen receptor; ATF6α, activating transcription factor 6α; CDK8, cyclin-dependent kinase 8; E1A, adenovirus early region 1A; ER, oestrogen receptor; ETS, E26 transformation-specific; GLI3, GLI family zinc-finger 3; GR, glucocorticoid receptor; HNF4, hepatocyte nuclear factor 4; PGC1α, proliferator-activated receptor-γ coactivator 1α; POU2AF1, POU class 2 homeobox-associating factor 1; PPAR, peroxisome proliferator-activated receptor; RARα, retinoic acid receptor-α; REST, RE1 silencing transcription factor; RUNX2, RUNX family transcription factor 2; RXRα, retinoid X receptor-α; SREBP1A, sterol regulatory element-binding protein 1A; TR, thyroid hormone receptor; VDR, vitamin D receptor. aSubunit of the Mediator kinase module.
Few structural details of TF–Mediator interactions are available, owing to the disorder and dynamics of TF activation domains. Available evidence shows that TF activation domains retain their disorder even upon Mediator binding, adopting a so-called fuzzy interface85. By contrast, the TF-binding surface of Mediator is structured, at least in reported cases. We emphasize that disordered or fuzzy interfaces are compatible with high affinity and selectivity. For example, an interaction between histone H1 and its chaperone yields binding affinities in the picomolar range at physiological ionic strength, yet maintains structural disorder at the interface86. A key feature of the interaction is a large, multivalent interaction surface that involves many dispersed amino acids. Such multivalent interfaces are observed among TF–Mediator interactions as well, and binding affinities are typically measured in the 10–100-nM range87–90.
TFs control Mediator function
TFs control Mediator function in part by recruiting Mediator to specific genomic sequences, such as enhancers (Fig. 2), to facilitate interactions with Pol II and other PIC factors. However, TFs also appear to activate Mediator function through additional mechanisms. Biochemical experiments have shown that TFs stabilize Mediator-containing PICs at transcription start sites, providing a longer time frame for Pol II to successfully initiate (or reinitiate) transcription91–94. In agreement with these findings, transcriptional output is increased in a Mediator-dependent and TF-dependent manner in reconstituted transcription systems56,95,96. TF–Mediator binding coincides with conformational changes in the Mediator complex (Fig. 2a), which can be observed even at low resolution13,33, and TF–Mediator binding correlates with changes in Pol II activity41,97. Consequently, TF-directed structural shifts in Mediator may contribute to Mediator-dependent activation of Pol II transcription. Such a regulatory mechanism could help to ensure that Mediator is activated at the appropriate ‘time and place’ in the nucleus: when bound by a TF on genomic DNA.
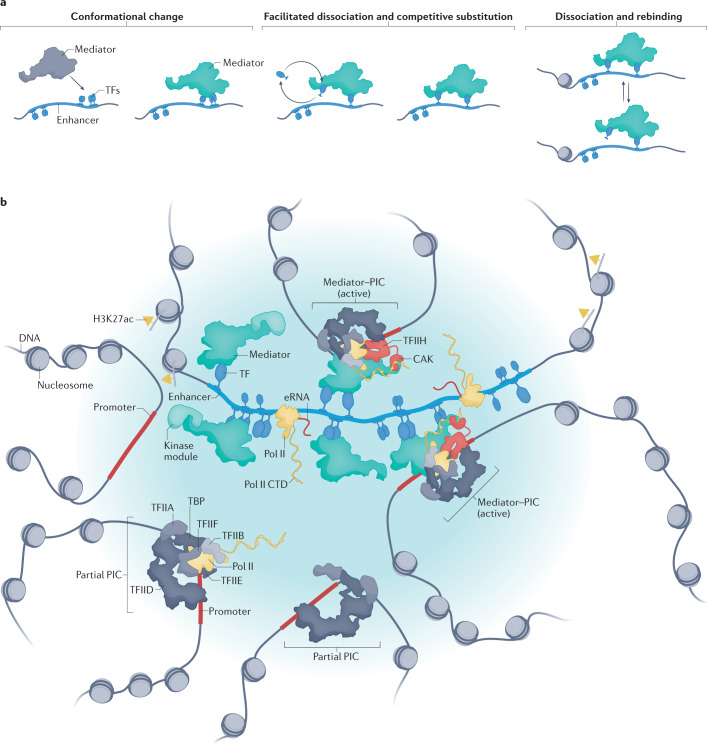
Fig. 2. Models for Mediator function at enhancers.
a | Potential mechanisms of reciprocal Mediator–transcription factor (TF) regulation: TFs can induce conformational changes in Mediator upon binding (left), which may influence Mediator function. Different molecules of a given TF could exchange with each other on DNA over time, especially if their local concentration is high100, but this may not alter Mediator occupancy if it is bound by multiple TFs (middle). Also, intrinsically disordered activation domains of TFs may displace each other on Mediator, in a process called ‘competitive substitution’101, which is favoured at high local concentrations (middle). Finally, TFs may transiently dissociate from DNA, but if Mediator-bound they would remain positioned to rebind DNA (right). b | Mediator function at a super-enhancer, which is shown at the centre, densely bound by TFs, Mediator complexes and other preinitiation complex (PIC) factors (not shown). This clustering of factors favours a high local concentration of intrinsically disordered regions (not shown), which are present in TFs, Mediator and other PIC components. Mutual weak attraction forces among intrinsically disordered regions will enforce a high local concentration of these factors (blue shading). Multiple Mediator complexes can bind the super-enhancer and can be oriented in all directions, to enable a single super-enhancer to activate multiple promoters at once if they are in spatial proximity. Enhancer RNAs (eRNAs), which are transcribed bidirectionally from the enhancers, may contribute to gene activation through the formation of additional multivalent weak interactions239 or through binding TFs or other regulatory factors. Over time, many promoters may colocalize with a super-enhancer, but activation will not occur unless a PIC can be fully assembled for activation. CAK, cyclin-dependent kinase-activating kinase module of transcription factor IIH; CTD, carboxy-terminal domain; H3K27ac, acetylated histone H3 Lys27; Pol II, RNA polymerase II; TBP, TATA box-binding protein; TFIIA, transcription factor IIA; TFIIB, transcription factor IIB; TFIID, transcription factor IID; TFIIE, transcription factor IIE; TFIIF, transcription factor IIF, TFIIH, transcription factor IIH.
TF-dependent control of Mediator recruitment, structure and function highlights the biological importance of the TF–Mediator interface. Evidence of TF-induced conformational shifts suggests that TF binding to a Mediator subunit will change its structure. However, few data are available, and the results are mixed: NMR data for the yeast Med15 interaction with the TF Gal4 or Gcn4 show no evidence of Med15 structural rearrangement upon binding85; by contrast, NMR data for human MED25 bound to several different E26 transformation-specific (ETS) family TFs98 or NMR data of the human vitamin D receptor–retinoid X receptor heterodimer bound to MED1 (ref.99) show evidence of structural changes at the Mediator subunit interface upon binding the TF activation domain. Notably, in each of these cases (MED25 or MED1), amino acids peripheral to the main TF–Mediator interface influenced the structural shift, further highlighting the extended and multivalent nature of TF–Mediator interactions. To date, no high-resolution crystallography or cryo-EM data have been obtained for any TF–Mediator interface, presumably because such interactions are too dynamic, despite their high affinity.
Mediator controls TF function
Evidence that Mediator reciprocally controls TF function has emerged in recent years. For example, structural and biochemical experiments revealed that a domain within MED25 binds and sequesters ETS TF sequences that are autoinhibitory for DNA binding. Thus, ETS TF occupancy on DNA was enhanced in the presence of MED25 (ref.87). Separately, other regions of MED25 participated in multivalent binding of the ETS TF activation domains. This distinct TF–Mediator interaction (that is, MED25 binding of TF sequences inhibitory to DNA binding) may also help to control ETS TF function by increasing its residence time on DNA.
Mediator could influence TF residence time on genomic DNA in other ways. At transcriptionally active loci, a high local concentration of unbound TFs would compete with DNA-bound TFs and reduce their residence time, through a mechanism called ‘facilitated dissociation’100. If there is high TF occupancy at enhancer regions, multiple TFs may bind Mediator simultaneously, which would increase Mediator residence time, because Mediator would remain tethered to DNA even upon dissociation of one of its bound TFs (Fig. 2a). Identical TFs, through their disordered activation domains, may also displace each other on Mediator, through competitive substitution101 (Fig. 2a). Finally, the TF could be retained near the enhancer following its dissociation from DNA if the TF activation domain remained bound to Mediator. The TF–Mediator interaction would facilitate TF rebinding to DNA (Fig. 2a, right). In this way, Mediator and TFs may cooperate to maintain enhancer occupancy and activity over physiologically relevant time frames.
Another means by which Mediator controls TF function is through the Mediator-associated kinases CDK8 and CDK19 (Box 2). This was first observed in yeast, which expresses a CDK8 orthologue but lacks a CDK19 orthologue. Numerous studies demonstrated that TF phosphorylation by yeast Cdk8 could either activate or inhibit TF function, depending on the TF and the cellular context102,103. In human cells, CDK8 and CDK19 have an expanded set of substrates compared with yeast73, but TFs are common targets. On the basis of existing data, CDK8 and CDK19 can positively or negatively regulate TF activity in mammalian cells. For example, sterol regulatory element-binding protein 1 (SREBP1) activity is repressed104 whereas STAT1, NOTCH and SMAD TFs are activated by Mediator kinases70,105,106. Whether CDK8 or CDK19 phosphorylates TFs as a separate entity (that is, the MKM) and/or as part of the CDK–Mediator complex remains an open question.
Mediator function at enhancers
Enhancers are metazoan-specific DNA regulatory elements that are dispersed throughout the genome. It is estimated that the human genome contains approximately 150,000 enhancers107, but ‘only’ about 10,000–50,000 of them are active in any given cell type108. Active enhancers are characterized by bound TFs, acetylated histone H3 Lys27 and bidirectional transcription. Importantly, enhancers are mobile in 3D space109, and therefore have the potential to interact with multiple promoters over time.
The bidirectional transcription at active enhancers generates eRNAs, which are unstable and typically 1 kb or shorter in size110. The biological functions of eRNAs remain poorly understood; however, Mediator is capable of binding eRNAs, perhaps through MED1 (ref.111) and/or the kinase module subunit MED12 (ref.112). Binding of eRNA to MED12 was linked to changes in Mediator kinase function and contributed to the activation of nearby protein-coding genes112. Consistent with these results, other laboratories have linked MED12 to enhancer function113,114, although potential links to eRNAs were not examined. Enhancer activation during induction of oestrogen receptor-α required Mediator at all stages, starting with rapid initiation of eRNA transcription, followed by target gene activation and stable recruitment of co-activators such as p300 (ref.115).
The MKM also appears to affect enhancer function in cell type-specific and context-specific ways. For example, Mediator kinase activity appears to restrain TF function at super-enhancers — clusters of enhancers that collectively span several kilobases or more in mammalian genomes — in acute myeloid leukaemia cells116 and in embryonic stem cells117; by contrast, CDK8 activity is required for maximal activation of enhancer-bound JAK–STAT pathway TFs during the interferon response in mouse embryonic fibroblasts or human cells70. This discrepancy in functional outcomes likely results from CDK8-dependent or CDK19-dependent phosphorylation of enhancer-bound TFs, which will differ between cell types and biological contexts.
Enhancer sequences are characterized by clusters of TF-binding sites, and, consequently, active enhancers will be bound by an array of TFs (Fig. 2b). Because typical mammalian enhancers encompass several hundred base pairs, and because TFs recruit Mediator to enhancers through high-affinity binding interactions, it is plausible that multiple Mediator complexes14 could simultaneously occupy an active enhancer (Fig. 2b). In agreement with this possibility, chromatin immunoprecipitation followed by sequencing (ChIP–seq) experiments provide evidence of high Mediator occupancy at enhancers118,119.
Transcription hubs or condensates?
Cell imaging experiments have revealed the existence of Pol II clusters in mammalian cells120,121 that correspond with sites of active transcription. Mediator appears to stabilize these clusters, as they are lost upon its depletion76. Combined with measurements of 3D genome topology, factor occupancy and gene expression, a model has emerged in which active enhancers and promoters are juxtaposed in cell nuclei, whereas transcriptionally silent regions are sequestered elsewhere122. Clustering of enhancers and promoters through spatial proximity will result in high local concentrations of bound factors such as chromatin remodellers, TFs, Mediator and Pol II (Fig. 2b). Why did this phenomenon evolve? What are the biological consequences of this clustering?
IDRs are common among eukaryotic proteins and are over-represented in transcription regulators123. A common feature among TFs is clusters of IDRs within their activation domains34. These IDR-containing activation domains directly bind Mediator and also allow TFs to undergo phase separation at physiologically relevant concentrations36,124. Because TF-binding sites are clustered at enhancers and promoters, TF binding will similarly cluster the multivalent IDRs on each TF. This clustering may promote liquid–liquid phase separation125 and the formation of molecular condensates. Interestingly, the TF Krüppel-like factor 4 can form localized condensates on DNA at concentrations that are an order of magnitude lower than required for phase separation in bulk water (that is, in the absence of a consensus DNA-binding surface)126. These ‘surface condensates’ were observed in vitro but are too small for reliable identification in live cells, at least with current techniques. It remains challenging to experimentally verify condensate formation at transcriptionally active loci in cells, and precisely how or whether molecular condensates contribute to Pol II transcription remains controversial, in part because high local concentrations of TFs, Pol II, Mediator and other factors can be achieved in the absence of phase separation127–129.
Live-cell imaging experiments in S. cerevisiae show that Mediator, TFIID and Pol II control PIC assembly in real time as other PIC factors rapidly converge at sites co-occupied by these three factors57. In addition, compared with smaller PIC factors such as TFIIB and TFIIE, Mediator and TFIID show limited diffusion in yeast nuclei, suggesting they are constrained to scan specific genomic regions57. These observations are consistent with the hypothesis that molecular condensates (or hubs that do not undergo phase separation but retain high local concentrations of TFs and PIC factors) help to direct PIC assembly in cell nuclei. Mediator, TFIID and Pol II each possesses IDRs and are predicted to undergo phase separation under physiologically relevant conditions79, which has been confirmed experimentally for Mediator and Pol II120,121,130,131. Moreover, Mediator and Pol II condensates will merge and incorporate TFs within the same condensate36,124,132.
These and other results133 suggest that condensates or hubs regulate gene expression through compartmentalization, which helps to maintain a high local concentration of TFs, Pol II, Mediator and other factors, thereby increasing factor residency time on genomic DNA and facilitating PIC–promoter assembly. Intrinsic disorder alone has also been shown to increase rates of binding interactions134, independently of condensate formation. We emphasize that well-characterized high-affinity protein–protein interactions such as TF–Mediator interactions and protein–DNA interactions such as TATA box binding by TBP remain essential for Pol II regulation regardless of whether condensate formation occurs.
Interestingly, among the cohort of transcription regulation proteins, Mediator is an outlier in terms of the number and length of IDRs within its subunits35 (Table 1), suggesting that Mediator evolution was driven in part by optimizing its phase separation properties135.
Super-enhancers
A functionally relevant distinction among enhancers is their size; large enhancer regions, ranging between 5 and 50 kb, are often designated as ‘super-enhancers’136. Because of their size, super-enhancers will bind more TFs compared with typical enhancers; more bound TFs will recruit more chromatin remodellers, Mediator, Pol II and other PIC factors119,137–139. This concentration of factors at super-enhancers may promote liquid–liquid phase separation, which, in turn, could influence chromatin architecture and help to retain active promoters and enhancers in spatial proximity122,140. Indeed, the surface tension of molecular condensates establishes a force that can help to pull genomic loci together141. In this way, super-enhancer condensates (or hubs) may complement other factors such as cohesin, TFs and/or Mediator to maintain enhancers and promoters in spatial proximity.
Super-enhancers appear to be especially important for driving expression of lineage-specific genes142, which are among the most highly transcribed genes in any given cell type. This high level of expression suggests a requirement for stable clustering of Pol II, TFs and Mediator, which could be established with the formation of molecular condensates5,140. Mediator occupancy is exceptionally high at super-enhancers (Fig. 2b); estimates of Mediator occupancy based on ChIP–seq data are a common means to identify super-enhancers in human cells119. The ability of Mediator to promote phase separation while interacting with TFs, Pol II and other factors is consistent with its central role in super-enhancer function119. Also consistent with this model, rapid depletion of Mediator (through a MED14 degron) was shown to disproportionately affect the expression of lineage-specific genes in human cells76.
Transcription bursting
Bursting involves multiple transcription initiation events from the same promoter, in a short time frame (for example, up to 10 min)143–145. In mammalian cells, bursting appears to be a general phenomenon, generating numerous transcripts before turning off146, with prolonged dormancy periods147. Following a pioneering round of transcription, additional Pol II complexes may assemble at the promoter to reinitiate transcription at the transcription start site (Fig. 3). On the basis of live-cell imaging experiments, burst initiation (that is, activation of transcription at a promoter) correlates with enhancer–promoter proximity148, TF binding149 and recruitment of Mediator and/or TFIIH to the promoter150. These events are not mutually exclusive, and others have proposed that chromatin looping through enhancer–promoter interactions may allow complete PIC assembly by delivering Mediator to gene promoters151. Consistent with this model, forced enhancer–promoter looping increased the frequency of transcription bursts from the β-globin gene promoter in mammalian cells152. A Mediator requirement for transcription bursting was suggested by MED11 depletion experiments in human cells145, but the precise mechanisms by which Mediator may control bursting remain unclear.
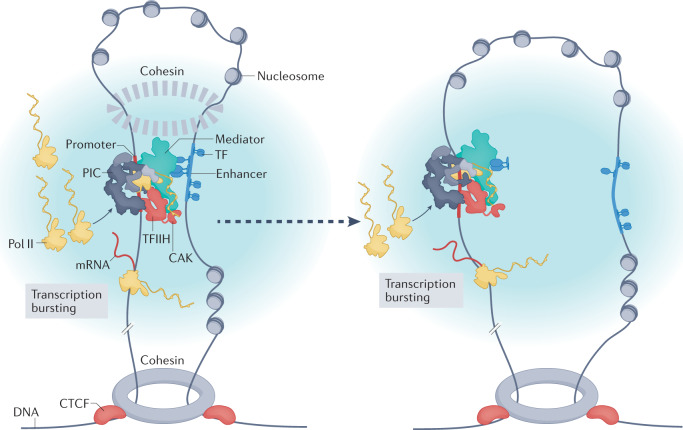
Fig. 3. A working model for Mediator function.
An enhancer–promoter interaction (loop) is shown on the left, within a larger topologically associating domain formed by CTCF and cohesin. Mediator is bound to one or more transcription factors (TFs) that occupy the enhancer, and the preinitiation complex (PIC) at the promoter is fully assembled and active. Such local architecture of enhancer–promoter chromatin looping could be further stabilized by Mediator-associated cohesin167, but this association would be transient (dashed circle) relative to topologically associating domain boundaries (solid circle). Following a brief, direct enhancer–promoter interaction, the enhancer detaches from the promoter (for example, through dissociation of TFs from enhancer DNA); however, if one or more TFs remain bound to Mediator, the complex could remain in an active conformational state. This state could allow continued transcription reinitiation (bursting) from the PIC scaffold complex, provided RNA polymerase II (Pol II) and other PIC factors continue to associate for reinitiation (right). Ultimately, reinitiation may stop (not shown), because of TF–Mediator dissociation, binding of the kinase module to Mediator (which would block Mediator–Pol II interaction) or PIC disassembly. The light blue shading represents a hub or condensate that establishes a high local concentration of PIC components that promotes transcription initiation and bursting. TFIIH, transcription factor IIH.
A basic requirement for bursting is reinitiation, which may be facilitated by a PIC scaffold complex that remains at the promoter after the initial round of Pol II transcription. Biochemical experiments have shown that reinitiation occurs more rapidly compared with a pioneering round of transcription153, in agreement with the PIC scaffold model. ChIP–seq data provide general support for scaffold PICs, because bound PIC factors are commonly observed at loci that are not undergoing active transcription50 (note that such partial PIC assemblies may also facilitate rapid induction of transcription from a dormant state). Direct evidence for the existence of a reinitiation scaffold comes from biochemical experiments with yeast nuclear extracts in which PICs were assembled on promoter DNA and bound factors were probed by western blotting before and after addition of nucleoside triphosphates94. On the basis of these results, the scaffold PIC retained TFIIA, TFIID, TFIIE, TFIIH and Mediator — that is, Pol II, TFIIF and TFIIB were missing. Furthermore, the PIC scaffold was stabilized by a TF (GAL4–VP16 in this case), and TF binding resulted in a higher rate of transcription reinitiation94.
TFIID also contributes to transcription bursting and reinitiation154. Mutations in TFIID-binding sites on promoter DNA decrease burst size or frequency, suggesting that transcription initiation or reinitiation is affected145,155,156. TFIID is structurally dynamic60 and undergoes reorganization during PIC assembly157 and following a pioneering round of transcription158. Before transcription initiation, TFIID binds DNA downstream of the promoter through its ‘lobe C’ subunits TAF1, TAF2 and TAF7 (refs47,48). For Pol II to transcribe this region, the contact between TFIID and the downstream DNA must be released. Biochemical data suggest that TFIID does not re-engage downstream DNA following an initial round of transcription158, which probably favours reinitiation from a scaffold PIC. Whereas cooperativity between Mediator and TFIID is widely reported50–54, it is unclear whether Mediator contributes to TFIID function during reinitiation and transcription bursting.
Although studies are limited due to the technical challenges associated with live-cell imaging of transcription, reinitiation was suggested to occur every 4 s during a bursting event145. This fast time frame suggests that barriers to a pioneering round of transcription, including de novo PIC assembly, are removed for reinitiation. We hypothesize that promoter DNA may remain open (single-stranded) at the transcription start site, to bypass the requirement for XPB to rebind downstream DNA and hydrolyse ATP to melt the template (again) for reinitiation. Evidence for the maintenance of an open promoter at highly transcribed genes has been reported159,160. Interestingly, evaluation of Mediator’s role in bursting kinetics by comparing wild type and MED11-depleted HeLa cells showed that the reinitiation rate was slowed from 4 s to approximately 8 s in MED11-depleted cells145. Moreover, the burst duration was reduced from 90 s in wild type cells to 68 s in MED11-depleted cells, resulting in reduced transcriptional output145. These results suggest Mediator is required for rapid transcription reinitiation and that a scaffold PIC remains poised for reinitiation.
Looping: direct or indirect contacts?
In mammalian cells, genomic regions of 1 Mb or more in size are organized into topologically associating domains (TADs), which are formed and maintained by CTCF and cohesin161. Within TADs, formation of chromatin loops of 10–100 kb in size may juxtapose regulatory regions such as enhancers and promoters (Fig. 3). However, because enhancer–promoter loops within TADs are more dynamic and transient compared with TAD boundaries162, they have been challenging to detect. This challenge is compounded by cell-to-cell variability in genome architecture: enhancer–promoter interactions are not uniform across a population of cells163. Experiments that assess 3D genome architecture across a population of cells have shown little correlation between enhancer–promoter interactions and gene expression patterns164. These data could reflect biological variability and the limitations of existing assays, or they could indicate that enhancers affect promoter activity at a distance (that is, indirectly).
Perhaps the best evidence for direct interactions between enhancers and promoters derives from innovative experiments that showed that enforcement of enhancer–promoter loops increases transcriptional output in mammalian cells165,166. Moreover, a technique called ‘Micro-Capture-C’ provided evidence of direct enhancer–promoter contacts within TADs that appeared to be maintained in part through cohesin, CTCF and TFs167. Prior chromatin conformation analyses similarly inferred the existence of direct enhancer–promoter contacts during active transcription, resulting in looping of the intervening chromatin168,169, and many laboratories have provided evidence of Mediator-dependent enhancer–promoter looping170–173. Furthermore, data from yeast suggest Mediator physically connects TF-bound upstream activating sequences with adjacent gene promoters25,26,174,175. Collectively, these results suggest that Pol II is activated through the formation of direct enhancer–promoter interactions, and that enhancers may help to deliver TF-bound Mediator complexes to gene promoters, as a final step in PIC assembly.
By contrast, several groundbreaking reports have argued against the existence of direct enhancer–promoter contacts, and suggest that Mediator does not directly tether promoters and enhancers, but instead implicate enhancer function at a distance21,176. Other studies have shown that enhancer–promoter proximity appears to be maintained by active transcription177. A model21 consistent with these findings proposes that TFs bound to enhancers that are close in space to promoters can diffuse and bind Mediator at promoter-bound PICs; TF–Mediator binding then activates Pol II through induction of conformational changes.
Technological improvements in live-cell imaging have allowed observation of Pol II transcription and estimation of enhancer–promoter distances in real time, representing an exciting advance. Measured enhancer–promoter distances have ranged between 100 and 350 nm at actively transcribed loci120,148,178–181. These measurements support a model in which enhancer–promoter proximity, not direct contact, is the predominant means by which Pol II activity is regulated in mammalian cells.
The seemingly contradictory results summarized above raise the question of whether enhancer–dependent activation of PICs at promoters is direct or indirect. Addressing this question is extremely challenging given the transient and dynamic nature of chromatin loops, their cell-to-cell variability and the requirement of averaging results of chromosome conformation experiments across cell populations162. Current estimates of enhancer–promoter distances are at the practical resolution limits of fluorescence microscopy. Even at the technical resolution limit (50 nm or greater) and in the absence of measurement error, it would be impossible to resolve enhancer–promoter separation182 given the size of Mediator–PIC (approximately 40 nm). Many live-cell imaging experiments also rely on fluorophores tethered to 5′ ends of nascent RNA transcripts, which may diffuse away from Pol II, and this complicates interpretation of data. Moreover, imaging transcription in live cells requires averaging fluorescent signals over time frames of seconds to minutes, which may prevent detection of transient or infrequent enhancer–promoter interactions183.
Despite these limitations (all experimental methods have limitations), live-cell imaging data184 and chromosome conformation methods108 have transformed our understanding of mammalian enhancers and genome organization. On the basis of available data and the limitations outlined above, we propose a working model (see the next section) in which enhancer function requires direct contact with PICs at promoters; however, this ‘direct’ model retains key aspects of other models that invoke action at a distance21.
A working model of Mediator function
A model for TF-dependent and Mediator-dependent activation of Pol II transcription is summarized in Fig. 3, which attempts to reconcile a diverse and sometimes contradictory set of experimental data, and draws on concepts proposed by others. The enhancer (or super-enhancer) shown in Fig. 3 is mobile109 and densely occupied by TFs, Mediator, Pol II and other factors, such as CBP or p300 (ref.108). At any point in time, the enhancer may contact nearby promoters, and the probability of interaction will increase with prolonged proximity, which could be favoured by tethering elements185–187. Enhancer–promoter contacts could also be favoured by Mediator interactions with the extended, disordered Pol II CTD22. In agreement with this concept, defects in enhancer-dependent gene activation occur upon Pol II CTD truncation188, which may partially reflect a reduced probability of Pol II–Mediator interaction. Note that the model in Fig. 2b allows a single enhancer to activate multiple promoters at once, an observation verified in Drosophila melanogaster and mammalian cells143,189. Simultaneous enhancer interactions with several promoters could be feasible if enhancer–promoter contacts occur through multiple distinct enhancer-bound Mediator complexes.
To reconcile this model with observations that active enhancer–promoter pairs are maintained in spatial proximity with little evidence for stable, direct interaction, we combine the well-established structural plasticity of Mediator with a hysteresis model proposed recently190. The hysteresis model postulates that promoters can remain active after a transient enhancer interaction; that is, the enhancer does not need to maintain direct contact with the promoter to continue to influence its function, because the brief enhancer–promoter interaction stably alters the structure and function of the promoter-bound PIC. How could this occur? We propose that TFs (one or more) could remain bound to Mediator after the enhancer region moves away from the PIC (Fig. 3). This would require dissociation of the TF DNA-binding domain from the enhancer, but TF dwell times on DNA are unexpectedly short in cells, likely due to binding competition with other TFs191. TFs can alter Mediator structure33, and TF–Mediator binding correlates with activation of Pol II within the PIC41,97 and with increased rates of transcription reinitiation94. Thus, as long as at least one TF remains bound to Mediator, and as long as Pol II and other PIC factors remain clustered around the promoter at high concentration, repeated rounds of initiation (bursting) could occur in the absence of direct enhancer–promoter contacts. This ‘Mediator-centric’ model is also consistent with an independent theoretical explanation for prolonged promoter activity in the absence of continual, direct enhancer–promoter interactions192.
We emphasize that despite the Mediator-centric aspects of the model shown in Fig. 3, the key to transcription activation depends on promoter-bound PIC factors. A stabler PIC (or PIC scaffold) would increase the probability of successful transcription initiation and reinitiation. This is consistent with data that show a correlation between promoter elements that bind TFIID, such as the TATA box and initiator, and the burst size (that is, the number of Pol II complexes that initiate transcription) and duration155,156. Finally, we note that maintenance of a high local concentration of TFs and PIC factors through condensates or hubs79 would favour prolonged Pol II activity.
Mediator as a therapeutic target
Mediator has been implicated in myriad diseases, such as developmental disorders, cardiovascular diseases and cancer193, and viral pathogenesis, including HIV and severe acute respiratory syndrome coronavirus 2 pathogenesis194,195. This broad spectrum of diseases reflects the general requirement of Mediator for Pol II transcription.
Enzymes are common targets for molecular therapeutics, and an array of CDK8 and CDK19 inhibitors have been discovered. Existing inhibitors target the ATP-binding site, which is virtually identical in CDK8 and CDK19, and as a result are unable to selectively target either kinase. Nevertheless, several Mediator kinase inhibitors are in clinical trials, which may ultimately yield treatments for cancers or developmental disorders linked to CDK8 or CDK19 function196,197.
Recent insights into the importance of phase separation in Pol II transcription, and the discovery that Mediator itself undergoes phase separation, suggest new therapeutic strategies to influence Mediator function. Altered expression of Mediator subunits is commonly observed in cancer (for example, increased MED1 expression)198,199, and we speculate that in some cases this change may disrupt Mediator-dependent condensate formation and contribute to disease. In support of this concept, elevated expression of MED1 altered the properties of transcriptional condensates in cells and disrupted oestrogen receptor-dependent gene activation200. Recent data suggest that beneficial therapeutic outcomes can be achieved through modulation of condensate properties201, and new targeting strategies are being developed202.
TFs are high-impact therapeutic targets because they establish cell type-specific gene expression programmes and direct transcriptional responses that are relevant to all physiological processes. Many oncoproteins and tumour suppressors are TFs, and many developmental diseases are caused by mutations in TFs that disrupt normal TF function or expression203. TFs activate Pol II through their interactions with Mediator, suggesting that disruption of TF–Mediator binding could effectively block TF function (Fig. 4).
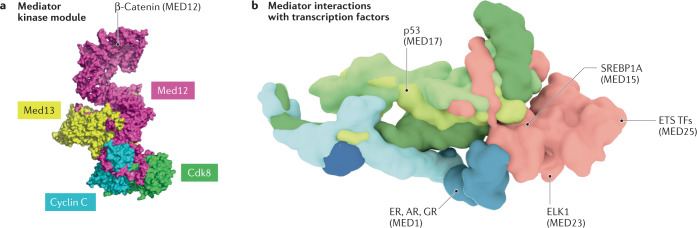
Fig. 4. Mediator as a therapeutic target.
a | Structure of the Mediator kinase module of Saccharomyces cerevisiae27, in which a MED12 interaction with β-catenin in human cells is indicated238. b | The human Mediator structure is shown, with a subset of identified transcription factor (TF)-binding sites highlighted. A potential strategy to manipulate TF function is to block specific TF–Mediator interactions, or to degrade a Mediator subunit targeted by a specific TF. Because different TFs bind Mediator at different sites, this strategy may allow gene activation by other TFs to occur normally. AR, androgen receptor; CDK8, cyclin-dependent kinase 8; ER, oestrogen receptor; ETS, E26 transformation-specific; GR, glucocorticoid receptor; SREBP1A, sterol regulatory element-binding protein 1A.
In support of this concept, TF-specific transcriptional responses are blocked in cells lacking specific Mediator subunits204,205. For example, knockout of Med1, which is bound by nuclear receptors, blocked nuclear receptor-dependent gene activation in mouse embryonic fibroblasts82; similarly, a MED23 knockout prevented activation of ELK1 target genes83,206. Importantly, different TFs target different Mediator subunits (Table 2), suggesting that blocking one TF–Mediator interface will not adversely affect activation by other TFs. Indeed, transcriptional responses directed by other signal-specific TFs occurred normally in cells lacking individual Mediator subunits83; thus, Mediator subunit loss of function provides a means to selectively regulate entire gene expression programmes. Validation of this concept has emerged in recent years using various strategies to target specific TF–Mediator interfaces to block TF function207–209. Whereas these proof-of-concept examples used drug-like compounds or stapled peptides to target a TF-binding site on Mediator, proteolysis-targeting chimaeras210 may also be effective, provided subunit-specific targeting could be established211.
Concluding remarks
The many ways in which Mediator affects Pol II-mediated transcription create many experimental questions, ranging from the structure of a TF–Mediator interface to the role of Mediator in chromosome organization. Because Mediator is a large complex that affects Pol II transcription genome-wide, it is a challenging factor to study. Biochemical experiments, including structural analysis and in vitro transcription, can best assess mechanistic questions, but they require isolation of the complex to near homogeneity. Common cell-based approaches such as Mediator subunit depletion or knockout trigger a cascade of events due to Mediator’s general role in Pol II transcription. Furthermore, compensatory mechanisms involving other transcription co-activators76 may be triggered upon loss of specific Mediator subunits. Such indirect effects create challenges in data interpretation. Fortunately, innovative methods such as rapid subunit depletion using degrons continue to improve our understanding of Mediator, in part by helping to distinguish direct versus indirect effects in cells. Here we highlight some areas that are poised for new discoveries in the coming years.
Enhancer function: the mechanisms by which Mediator functions at enhancers (for example, direct, indirect or both direct and indirect) remain unclear, due in part to the experimental challenges. Related to this question, whether Mediator binds eRNAs specifically or promiscuously, such as in the case of Polycomb repressive complex 2 (ref.212), remains to be determined.
Tethering elements: several research groups have identified DNA elements that lack enhancer activity but promote enhancer–promoter interactions through a ‘tethering’ function185–187. It remains unclear whether Mediator might affect the function of these tethering elements, but data from mouse embryonic stem cells suggest tethering elements influence Mediator recruitment to nearby enhancers186, and we speculate that such elements may act in part through recruitment of the CDK–Mediator complex213.
Molecular condensates: the realization that liquid–liquid phase separation contributes to Pol II transcription regulation has been revolutionary. Mediator appears to have a central role in the regulation of transcription condensates, but it remains unclear whether Mediator condensates possess biophysical properties that directly affect Mediator functions, or how its condensate properties could be altered, for example through post-translational modifications, conformation switching or alternative splicing of subunits.
Transcription bursting: although Mediator is known to regulate bursting145, the molecular mechanisms by which Mediator might promote rapid reinitiation at scaffold PIC assemblies is not understood.
Structure and function: despite major advances in our understanding of Mediator structure, many interesting questions remain due to the varied roles of Mediator in Pol II transcription and its conformational flexibility. For instance, structural and biophysical experiments may reveal how Mediator acts to thread the Pol II CTD through the CDK7 active site during transcription initiation, and additional Mediator conformational changes may be resolved to high resolution using cryo-EM.
Compensatory responses and Mediator subcomplexes: is Mediator required for all Pol II transcription? Rapid, 2-h depletion of MED14, which disrupts Mediator structure, showed that Pol II can continue to transcribe some genes despite a 90% reduction in MED14 levels; this condition revealed a compensatory mechanism involving positive transcription elongation factor b (P-TEFb)76. Although MED14 loss over longer time frames (60 h) revealed expected global reductions in Pol II transcription21, these results collectively suggest that Pol II transcription can occur in the absence of Mediator, at least temporarily. Mediator subcomplexes may contribute to P-TEFb compensation, but it remains to be determined whether Mediator complexes with altered subunit composition (for example, missing MED14 and other subunits) could help to control gene expression in mammalian cells. Similarly, there is great potential for variation through alternative splicing of Mediator subunits, but few isoform-specific functional roles have been reported214.
Regulatory functions beyond enhancers and promoters: studies in model organisms and in mammalian cells have implicated Mediator, at least peripherally, in the control of co-transcriptional splicing215, termination216,217 and mRNA export218.
Supplementary information
Acknowledgements
Due to space and citation limitations, the authors regret that they were unable to cite all relevant articles or to discuss all aspects of Mediator function in biology. The Taatjes laboratory is funded in part by the NIH (R35 GM139550 to D.J.T.) and the NSF (MCB-1818147 to D.J.T.); the Iwasa laboratory is funded in part by the NSF (MCB-190330 to J.I.).
Glossary
- Mediator complex
In humans, a 26-subunit complex that lacks the Mediator kinase module.
- Mediator kinase module
(MKM). A four-subunit complex that includes cyclin-dependent kinase 8 (CDK8), cyclin C, MED12 and MED13; vertebrates express also the paralogues CDK19, MED12L and MED13L.
- Cyclin-dependent kinase (CDK)–Mediator complex
Mediator bound to the Mediator kinase module (MKM), which may contain CDK8 or CDK19; because MED26 is mutually exclusive with MKM, CDK–Mediator consists of 29 subunits.
- Carboxy-terminal domain
(CTD). The disordered carboxyl terminus of the RNA polymerase II (Pol II) subunit RPB1, composed of heptad repeats of the general sequence YSPTSPS, which are differentially phosphorylated during Pol II transcription initiation, pausing, elongation and termination.
- Pol II jaw
RNA polymerase II domain composed of subunits RBP1 and RBP5 that contacts DNA downstream of the transcription start site.
- Hook domain
A region within the Mediator middle module, at the opposite end of the tail, which is formed by MED10, MED19 and the amino-terminal portion of MED14.
- Stalk
Composed of the RNA polymerase II subunits RPB4 and RPB7, the stalk serves as an interaction hub within the preinitiation complex.
- Bridge helix
An α-helix that spans the RNA polymerase II (Pol II) active site and undergoes structural changes in coordination with the trigger loop during nucleotide incorporation and Pol II translocation.
- Trigger loop
A domain near the RNA polymerase II active site that transitions between an open state and a closed state with each nucleoside triphosphate added to the nascent RNA; helps to detect base pair mismatches.
- TF activation domains
Regions of transcription factors (TFs) that interact with other proteins, such as chromatin remodellers or Mediator; activation domains are typically disordered with low-complexity sequences and may phase separate at physiological concentrations.
- Hysteresis
In the context of Mediator and transcription, hysteresis could involve a structural isomerization to achieve a more active state, triggered by transcription factor–Mediator binding and/or Mediator–preinitiation complex association. These interaction-induced structural changes may persist, rendering Mediator activity dependent on prior protein–protein interactions.
- Proteolysis-targeting chimaeras
Bivalent small molecules that bind a protein of interest and target it to an E3 ubiquitin ligase, thereby promoting its ubiquitylation and degradation.
Author contributions
D.J.T. and W.F.R. wrote the article; W.F.R., S.N. and J.I. helped research data for the article.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Gang Wang, Yanhui Xu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
D.J.T. is a member of the scientific advisory board of Dewpoint Therapeutics. All the other authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41580-022-00498-3.
References
- 1.Schier AC, Taatjes DJ. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 2020;34:465–488. doi: 10.1101/gad.335679.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen FX, Smith ER, Shilatifard A. Born to run: control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2018;19:464–478. doi: 10.1038/s41580-018-0010-5. [DOI] [PubMed] [Google Scholar]
- 3.Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. doi: 10.1038/s41586-019-1517-4. [DOI] [PubMed] [Google Scholar]
- 4.Furlong EEM, Levine M. Developmental enhancers and chromosome topology. Science. 2018;361:1341–1345. doi: 10.1126/science.aau0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buendia-Monreal M, Gillmor CS. Mediator: a key regulator of plant development. Dev. Biol. 2016;419:7–18. doi: 10.1016/j.ydbio.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Dolan WL, Chapple C. Conservation and divergence of mediator structure and function: insights from plants. Plant Cell Physiol. 2017;58:4–21. doi: 10.1093/pcp/pcw176. [DOI] [PubMed] [Google Scholar]
- 8.Malik N, Agarwal P, Tyagi A. Emerging functions of multi-protein complex mediator with special emphasis on plants. Crit. Rev. Biochem. Mol. Biol. 2017;52:475–502. doi: 10.1080/10409238.2017.1325830. [DOI] [PubMed] [Google Scholar]
- 9.Imasaki T, et al. Architecture of the mediator head module. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lariviere L, et al. Structure of the mediator head module. Nature. 2012;492:448–451. doi: 10.1038/nature11670. [DOI] [PubMed] [Google Scholar]
- 11.Nozawa K, Schneider TR, Cramer P. Core mediator structure at 3.4 a extends model of transcription initiation complex. Nature. 2017;545:248–251. doi: 10.1038/nature22328. [DOI] [PubMed] [Google Scholar]
- 12.Robinson PJ, et al. Molecular architecture of the yeast mediator complex. eLife. 2015;4:e08719. doi: 10.7554/eLife.08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai KL, et al. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell. 2014;157:1430–1444. doi: 10.1016/j.cell.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anandhakumar J, Moustafa YW, Chowdhary S, Kainth AS, Gross DS. Evidence for multiple mediator complexes in yeast independently recruited by activated heat shock factor. Mol. Cell Biol. 2016;36:1943–1960. doi: 10.1128/MCB.00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knoll ER, Zhu ZI, Sarkar D, Landsman D, Morse RH. Role of the pre-initiation complex in mediator recruitment and dynamics. eLife. 2018;7:e39633. doi: 10.7554/eLife.39633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleh MM, Jeronimo C, Robert F, Zentner GE. Connection of core and tail mediator modules restrains transcription from TFIID-dependent promoters. PLoS Genet. 2021;17:e1009529. doi: 10.1371/journal.pgen.1009529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdella R, et al. Structure of the human mediator-bound transcription preinitiation complex. Science. 2021;372:52–56. doi: 10.1126/science.abg3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, et al. Structures of the human mediator and mediator-bound preinitiation complex. Science. 2021;372:eabg0635. doi: 10.1126/science.abg0635. [DOI] [PubMed] [Google Scholar]
- 19.Rengachari S, Schilbach S, Aibara S, Dienemann C, Cramer P. Structure of human mediator-RNA polymerase II pre-initiation complex. Nature. 2021;594:129–133. doi: 10.1038/s41586-021-03555-7. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, et al. Structure of mammalian mediator complex reveals tail module architecture and interaction with a conserved core. Nat. Commun. 2021;12:1355. doi: 10.1038/s41467-021-21601-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Khattabi L, et al. A pliable mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell. 2019;178:1145–1158.e20. doi: 10.1016/j.cell.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson PJ, et al. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell. 2016;166:1411–1422. doi: 10.1016/j.cell.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/S0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 25.Jeronimo C, et al. Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol. Cell. 2016;64:455–466. doi: 10.1016/j.molcel.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrenko N, Jin Y, Wong KH, Struhl K. Mediator undergoes a compositional change during transcriptional activation. Mol. Cell. 2016;64:443–454. doi: 10.1016/j.molcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li YC, et al. Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing mediator kinase module. Sci. Adv. 2021;7:eabd4484. doi: 10.1126/sciadv.abd4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elmlund H, et al. The cyclin-dependent kinase 8 module sterically blocks mediator interactions with RNA polymerase II. Proc. Natl Acad. Sci. USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator co-activator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai KL, et al. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat. Struct. Mol. Biol. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebmeier CC, Taatjes DJ. Activator-mediator binding regulates mediator-cofactor interactions. Proc. Natl Acad. Sci. USA. 2010;107:11283–11288. doi: 10.1073/pnas.0914215107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paoletti AC, et al. Quantitative proteomic analysis of distinct mammalian mediator complexes using normalized spectral abundance factors. Proc. Natl Acad. Sci. USA. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 34.Soto LF, et al. Compendium of human transcription factor effector domains. Mol. Cell. 2022;82:514–526. doi: 10.1016/j.molcel.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toth-Petroczy A, et al. Malleable machines in transcription regulation: the mediator complex. PLoS Comput. Biol. 2008;4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boija A, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–1855.e16. doi: 10.1016/j.cell.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biddle JW, Martinez-Corral R, Wong F, Gunawardena J. Allosteric conformational ensembles have unlimited capacity for integrating information. eLife. 2021;10:e65498. doi: 10.7554/eLife.65498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berlow RB, Dyson HJ, Wright PE. Expanding the paradigm: intrinsically disordered proteins and allosteric regulation. J. Mol. Biol. 2018;430:2309–2320. doi: 10.1016/j.jmb.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato S, et al. Role for the MED21-MED7 hinge in assembly of the mediator-RNA polymerase II holoenzyme. J. Biol. Chem. 2016;291:26886–26898. doi: 10.1074/jbc.M116.756098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai KL, et al. Mediator structure and rearrangements required for holoenzyme formation. Nature. 2017;544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer KD, Lin S, Bernecky C, Gao Y, Taatjes DJ. p53 activates transcription by directing structural shifts in mediator. Nat. Struct. Mol. Biol. 2010;17:753–760. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, et al. Mediator structure and conformation change. Mol. Cell. 2021;81:1781–1788.e4. doi: 10.1016/j.molcel.2021.01.022. [DOI] [PubMed] [Google Scholar]
- 43.Sanborn AL, et al. Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to mediator. eLife. 2021;10:e68068. doi: 10.7554/eLife.68068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huttlin EL, et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell. 2021;184:3022–3040.e28. doi: 10.1016/j.cell.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plaschka C, et al. Architecture of the RNA polymerase II-mediator core initiation complex. Nature. 2015;518:376–380. doi: 10.1038/nature14229. [DOI] [PubMed] [Google Scholar]
- 46.Schilbach S, et al. Structures of transcription pre-initiation complex with TFIIH and mediator. Nature. 2017;551:204–209. doi: 10.1038/nature24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, et al. Structural insights into preinitiation complex assembly on core promoters. Science. 2021;372:eaba8490. doi: 10.1126/science.aba8490. [DOI] [PubMed] [Google Scholar]
- 48.Patel AB, et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science. 2018;362:eaau8872. doi: 10.1126/science.aau8872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vo Ngoc L, Wang YL, Kassavetis GA, Kadonaga JT. The punctilious RNA polymerase II core promoter. Genes Dev. 2017;31:1289–1301. doi: 10.1101/gad.303149.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grunberg S, Henikoff S, Hahn S, Zentner GE. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J. 2016;35:2435–2446. doi: 10.15252/embj.201695020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guermah M, Malik S, Roeder RG. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-kappaB and Sp1. Mol. Cell. Biol. 1998;18:3234–3244. doi: 10.1128/MCB.18.6.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson KM, Wang J, Smallwood A, Arayata C, Carey M. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 2002;16:1852–1863. doi: 10.1101/gad.995702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu C, Fuller MT. Recruitment of mediator complex by cell type and stage-specific factors required for tissue-specific TAF dependent gene activation in an adult stem cell lineage. PLoS Genet. 2015;11:e1005701. doi: 10.1371/journal.pgen.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marr MT, Isogai Y, Wright KJ, Tjian R. Coactivator cross-talk specifies transcriptional output. Genes Dev. 2006;20:1458–1469. doi: 10.1101/gad.1418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang YW, et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl Acad. Sci. USA. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 57.Nguyen VQ, et al. Spatiotemporal coordination of transcription preinitiation complex assembly in live cells. Mol. Cell. 2021;81:3560–3575.e6. doi: 10.1016/j.molcel.2021.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takahashi H, et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat. Commun. 2015;6:5941. doi: 10.1038/ncomms6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lens Z, et al. Solution structure of the N-terminal domain of mediator subunit MED26 and molecular characterization of its interaction with EAF1 and TAF7. J. Mol. Biol. 2017;429:3043–3055. doi: 10.1016/j.jmb.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 60.Cianfrocco MA, et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell. 2013;152:120–131. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greber BJ, Toso DB, Fang J, Nogales E. The complete structure of the human TFIIH core complex. eLife. 2019;8:e44771. doi: 10.7554/eLife.44771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 63.Cheung AC, Cramer P. A movie of RNA polymerase II transcription. Cell. 2012;149:1431–1437. doi: 10.1016/j.cell.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Fishburn J, Tomko E, Galburt E, Hahn S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl Acad. Sci. USA. 2015;112:3961–3966. doi: 10.1073/pnas.1417709112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aibara S, Schilbach S, Cramer P. Structures of mammalian RNA polymerase II pre-initiation complexes. Nature. 2021;594:124–128. doi: 10.1038/s41586-021-03554-8. [DOI] [PubMed] [Google Scholar]
- 66.Sogaard TM, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 2007;282:14113–14120. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- 67.Core L, Adelman K. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 2019;33:960–982. doi: 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vos SM, Farnung L, Urlaub H, Cramer P. Structure of paused transcription complex Pol II-DSIF-NELF. Nature. 2018;560:601–606. doi: 10.1038/s41586-018-0442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steinparzer I, et al. Transcriptional responses to IFN-gamma require mediator kinase-dependent pause release and mechanistically distinct CDK8 and CDK19 functions. Mol. Cell. 2019;76:485–499. doi: 10.1016/j.molcel.2019.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galbraith MD, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–1339. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poss ZC, et al. Identification of mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep. 2016;15:436–450. doi: 10.1016/j.celrep.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen M, et al. CDK8/19 mediator kinases potentiate induction of transcription by NFkappaB. Proc. Natl Acad. Sci. USA. 2017;114:10208–10213. doi: 10.1073/pnas.1710467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bhagwat AS, et al. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 2016;15:519–530. doi: 10.1016/j.celrep.2016.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jaeger MG, et al. Selective mediator dependence of cell-type-specifying transcription. Nat. Genet. 2020;52:719–727. doi: 10.1038/s41588-020-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takahashi H, et al. Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alerasool N, Leng H, Lin ZY, Gingras AC, Taipale M. Identification and functional characterization of transcriptional activators in human cells. Mol. Cell. 2022;82:677–695.e7. doi: 10.1016/j.molcel.2021.12.008. [DOI] [PubMed] [Google Scholar]
- 79.Palacio M, Taatjes DJ. Merging established mechanisms with new insights: condensates, hubs, and the regulation of RNA polymerase II transcription. J. Mol. Biol. 2022;434:167216. doi: 10.1016/j.jmb.2021.167216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Markert J, Luger K. Nucleosomes meet their remodeler match. Trends Biochem. Sci. 2021;46:41–50. doi: 10.1016/j.tibs.2020.08.010. [DOI] [PubMed] [Google Scholar]
- 81.Lambert SA, et al. The human transcription factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 82.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell. 2000;5:683–693. doi: 10.1016/S1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 83.Stevens JL, et al. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 84.Wang W, et al. Mediator MED23 links insulin signaling to the adipogenesis transcription cascade. Dev. Cell. 2009;16:764–771. doi: 10.1016/j.devcel.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tuttle LM, et al. Mediator subunit Med15 dictates the conserved “fuzzy” binding mechanism of yeast transcription activators Gal4 and Gcn4. Nat. Commun. 2021;12:2220. doi: 10.1038/s41467-021-22441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borgia A, et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555:61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Currie SL, et al. ETV4 and AP1 transcription factors form multivalent interactions with three sites on the MED25 activator-interacting domain. J. Mol. Biol. 2017;429:2975–2995. doi: 10.1016/j.jmb.2017.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milbradt AG, et al. Structure of the VP16 transactivator target in the mediator. Nat. Struct. Mol. Biol. 2011;18:410–415. doi: 10.1038/nsmb.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vojnic E, et al. Structure and VP16 binding of the mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol. 2011;18:404–409. doi: 10.1038/nsmb.1997. [DOI] [PubMed] [Google Scholar]
- 90.Yang F, et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 91.Cantin GT, Stevens JL, Berk AJ. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl Acad. Sci. USA. 2003;100:12003–12008. doi: 10.1073/pnas.2035253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu SY, Zhou T, Chiang CM. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol. Cell Biol. 2003;23:6229–6242. doi: 10.1128/MCB.23.17.6229-6242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 95.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Naar AM, et al. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 97.Wang G, et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 98.Henley MJ, et al. Unexpected specificity within dynamic transcriptional protein-protein complexes. Proc. Natl Acad. Sci. USA. 2020;117:27346–27353. doi: 10.1073/pnas.2013244117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Belorusova AY, et al. Molecular determinants of MED1 interaction with the DNA bound VDR-RXR heterodimer. Nucleic Acids Res. 2020;48:11199–11213. doi: 10.1093/nar/gkaa775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Erbas A, Marko JF. How do DNA-bound proteins leave their binding sites? The role of facilitated dissociation. Curr. Opin. Chem. Biol. 2019;53:118–124. doi: 10.1016/j.cbpa.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sottini A, et al. Polyelectrolyte interactions enable rapid association and dissociation in high-affinity disordered protein complexes. Nat. Commun. 2020;11:5736. doi: 10.1038/s41467-020-18859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chi Y, et al. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hirst M, Kobor MS, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell. 1999;3:673–678. doi: 10.1016/S1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 104.Zhao X, et al. Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest. 2012;122:2417–2427. doi: 10.1172/JCI61462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-b pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fryer CJ, White JB, Jones KA. Mastermind recruits CycC:Cdk8 to phosphorylate the notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 107.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015;16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat. Rev. Genet. 2019;20:437–455. doi: 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- 109.Gu B, et al. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science. 2018;359:1050–1055. doi: 10.1126/science.aao3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cardiello JF, Sanchez GJ, Allen MA, Dowell RD. Lessons from eRNAs: understanding transcriptional regulation through the lens of nascent RNAs. Transcription. 2020;11:3–18. doi: 10.1080/21541264.2019.1704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hsieh CL, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc. Natl Acad. Sci. USA. 2014;111:7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lai F, et al. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aranda-Orgilles B, et al. MED12 regulates HSC-specific enhancers independently of mediator kinase activity to control hematopoiesis. Cell Stem Cell. 2016;19:784–799. doi: 10.1016/j.stem.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moyo MB, Parker JB, Chakravarti D. Altered chromatin landscape and enhancer engagement underlie transcriptional dysregulation in MED12 mutant uterine leiomyomas. Nat. Commun. 2020;11:1019. doi: 10.1038/s41467-020-14701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Murakami S, Nagari A, Kraus WL. Dynamic assembly and activation of estrogen receptor alpha enhancers through coregulator switching. Genes Dev. 2017;31:1535–1548. doi: 10.1101/gad.302182.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pelish HE, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lynch CJ, et al. Global hyperactivation of enhancers stabilizes human and mouse naive pluripotency through inhibition of CDK8/19 Mediator kinases. Nat. Cell Biol. 2020;22:1223–1238. doi: 10.1038/s41556-020-0573-1. [DOI] [PubMed] [Google Scholar]
- 118.Quevedo M, et al. Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat. Commun. 2019;10:2669. doi: 10.1038/s41467-019-10502-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cho WK, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sabari BR, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eear3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bhat P, Honson D, Guttman M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 2021;22:653–670. doi: 10.1038/s41580-021-00387-1. [DOI] [PubMed] [Google Scholar]
- 123.Liu J, et al. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zamudio AV, et al. Mediator condensates localize signaling factors to key cell identity genes. Mol. Cell. 2019;76:753–766.e6. doi: 10.1016/j.molcel.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shrinivas K, et al. Enhancer features that drive formation of transcriptional condensates. Mol. Cell. 2019;75:549–561.e7. doi: 10.1016/j.molcel.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Morin JA, et al. Sequence-dependent surface condensation of a pioneer transcription factor on DNA. Nat. Phys. 2022;18:271–276. doi: 10.1038/s41567-021-01462-2. [DOI] [Google Scholar]
- 127.Chong S, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chong S, et al. Tuning levels of low-complexity domain interactions to modulate endogenous oncogenic transcription. Mol. Cell. 2022;82:2084–2097.e5. doi: 10.1016/j.molcel.2022.04.007. [DOI] [PubMed] [Google Scholar]
- 129.Trojanowski J, et al. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell. 2022;82:1878–1893.e10. doi: 10.1016/j.molcel.2022.04.017. [DOI] [PubMed] [Google Scholar]
- 130.Boehning M, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018;25:833–840. doi: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- 131.Kwon I, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guo YE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Brodsky S, et al. Intrinsically disordered regions direct transcription factor in vivo binding specificity. Mol. Cell. 2020;79:459–471.e4. doi: 10.1016/j.molcel.2020.05.032. [DOI] [PubMed] [Google Scholar]
- 134.Shammas SL, Travis AJ, Clarke J. Remarkably fast coupled folding and binding of the intrinsically disordered transactivation domain of cMyb to CBP KIX. J. Phys. Chem. B. 2013;117:13346–13356. doi: 10.1021/jp404267e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nagulapalli M, Maji S, Dwivedi N, Dahiya P, Thakur JK. Evolution of disorder in mediator complex and its functional relevance. Nucleic Acids Res. 2016;44:1591–1612. doi: 10.1093/nar/gkv1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Blobel GA, Higgs DR, Mitchell JA, Notani D, Young RA. Testing the super-enhancer concept. Nat. Rev. Genet. 2021;22:749–755. doi: 10.1038/s41576-021-00398-w. [DOI] [PubMed] [Google Scholar]
- 137.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sabari BR, Dall’Agnese A, Young RA. Biomolecular condensates in the nucleus. Trends Biochem. Sci. 2020;45:961–977. doi: 10.1016/j.tibs.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shin Y, et al. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175:1481–1491.e13. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fukaya T, Lim B, Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stavreva DA, et al. Transcriptional bursting and co-bursting regulation by steroid hormone release pattern and transcription factor mobility. Mol. Cell. 2019;75:1161–1177.e11. doi: 10.1016/j.molcel.2019.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tantale K, et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 2016;7:12248. doi: 10.1038/ncomms12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wan Y, et al. Dynamic imaging of nascent RNA reveals general principles of transcription dynamics and stochastic splice site selection. Cell. 2021;184:2878–2895.e20. doi: 10.1016/j.cell.2021.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rodriguez J, et al. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2019;176:213–226.e18. doi: 10.1016/j.cell.2018.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen H, et al. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 2018;50:1296–1303. doi: 10.1038/s41588-018-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Donovan BT, et al. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 2019;38:e100809. doi: 10.15252/embj.2018100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bartman CR, et al. Transcriptional burst initiation and polymerase pause release are key control points of transcriptional regulation. Mol. Cell. 2019;73:519–532.e4. doi: 10.1016/j.molcel.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: transcription enters a new era. Cell. 2014;157:13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Bartman CR, Hsu SC, Hsiung CC, Raj A, Blobel GA. Enhancer regulation of transcriptional bursting parameters revealed by forced chromatin looping. Mol. Cell. 2016;62:237–247. doi: 10.1016/j.molcel.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hawley DK, Roeder RG. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem. 1987;262:3452–3461. doi: 10.1016/S0021-9258(18)61372-9. [DOI] [PubMed] [Google Scholar]
- 154.Joo YJ, et al. Downstream promoter interactions of TFIID TAFs facilitate transcription reinitiation. Genes Dev. 2017;31:2162–2174. doi: 10.1101/gad.306324.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Larsson AJM, et al. Genomic encoding of transcriptional burst kinetics. Nature. 2019;565:251–254. doi: 10.1038/s41586-018-0836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pimmett VL, et al. Quantitative imaging of transcription in living Drosophila embryos reveals the impact of core promoter motifs on promoter state dynamics. Nat. Commun. 2021;12:4504. doi: 10.1038/s41467-021-24461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yakovchuk P, Gilman B, Goodrich JA, Kugel JF. RNA polymerase II and TAFs undergo a slow isomerization after the polymerase is recruited to promoter-bound TFIID. J. Mol. Biol. 2010;397:57–68. doi: 10.1016/j.jmb.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 158.Zhang Z, et al. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. eLife. 2015;4:e07777. doi: 10.7554/eLife.07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li J, et al. Kinetic competition between elongation rate and binding of NELF controls promoter-proximal pausing. Mol. Cell. 2013;50:711–722. doi: 10.1016/j.molcel.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pavelitz T, Bailey AD, Elco CP, Weiner AM. Human U2 snRNA genes exhibit a persistently open transcriptional state and promoter disassembly at metaphase. Mol. Cell Biol. 2008;28:3573–3588. doi: 10.1128/MCB.00087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci. Adv. 2019;5:eaaw1668. doi: 10.1126/sciadv.aaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Finn EH, Misteli T. Molecular basis and biological function of variability in spatial genome organization. Science. 2019;365:eaaw9498. doi: 10.1126/science.aaw9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Misteli T. The self-organizing genome: principles of genome architecture and function. Cell. 2020;183:28–45. doi: 10.1016/j.cell.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ghavi-Helm Y, et al. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 2019;51:1272–1282. doi: 10.1038/s41588-019-0462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Deng W, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deng W, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hua P, et al. Defining genome architecture at base-pair resolution. Nature. 2021;595:125–129. doi: 10.1038/s41586-021-03639-4. [DOI] [PubMed] [Google Scholar]
- 168.Fullwood MJ, et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kagey M, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Siersbaek R, et al. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol. Cell. 2017;66:420–435.e5. doi: 10.1016/j.molcel.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 174.Jeronimo C, Robert F. Kin28 regulates the transient association of mediator with core promoters. Nat. Struct. Mol. Biol. 2014;21:449–455. doi: 10.1038/nsmb.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wong KH, Jin Y, Struhl K. TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell. 2014;54:601–612. doi: 10.1016/j.molcel.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sun F, et al. The Pol II preinitiation complex (PIC) influences mediator binding but not promoter-enhancer looping. Genes Dev. 2021;35:1175–1189. doi: 10.1101/gad.348471.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Alexander JM, et al. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife. 2019;8:e41769. doi: 10.7554/eLife.41769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Heist T, Fukaya T, Levine M. Large distances separate coregulated genes in living Drosophila embryos. Proc. Natl Acad. Sci. USA. 2019;116:15062–15067. doi: 10.1073/pnas.1908962116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li J, et al. Single-molecule nanoscopy elucidates RNA polymerase II transcription at single genes in live cells. Cell. 2019;178:491–506.e28. doi: 10.1016/j.cell.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li J, et al. Single-gene imaging links genome topology, promoter-enhancer communication and transcription control. Nat. Struct. Mol. Biol. 2020;27:1032–1040. doi: 10.1038/s41594-020-0493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Valli J, et al. Seeing beyond the limit: a guide to choosing the right super-resolution microscopy technique. J. Biol. Chem. 2021;297:100791. doi: 10.1016/j.jbc.2021.100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Patange S, Ball DA, Karpova T, Larson DR. Towards a ‘spot on’ understanding of transcription in the nucleus. J. Mol. Biol. 2021;433:167016. doi: 10.1016/j.jmb.2021.167016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rodriguez J, Larson DR. Transcription in living cells: molecular mechanisms of bursting. Annu. Rev. Biochem. 2020;89:189–212. doi: 10.1146/annurev-biochem-011520-105250. [DOI] [PubMed] [Google Scholar]
- 185.Batut PJ, et al. Genome organization controls transcriptional dynamics during development. Science. 2022;375:566–570. doi: 10.1126/science.abi7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Pachano T, et al. Orphan CpG islands amplify poised enhancer regulatory activity and determine target gene responsiveness. Nat. Genet. 2021;53:1036–1049. doi: 10.1038/s41588-021-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Levo M, et al. Transcriptional coupling of distant regulatory genes in living embryos. Nature. 2022;605:754–760. doi: 10.1038/s41586-022-04680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sawicka A, et al. Transcription activation depends on the length of the RNA polymerase II C-terminal domain. EMBO J. 2021;40:e107015. doi: 10.15252/embj.2020107015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Allahyar A, et al. Enhancer hubs and loop collisions identified from single-allele topologies. Nat. Genet. 2018;50:1151–1160. doi: 10.1038/s41588-018-0161-5. [DOI] [PubMed] [Google Scholar]
- 190.Xiao JY, Hafner A, Boettiger AN. How subtle changes in 3D structure can create large changes in transcription. eLife. 2021;10:e64320. doi: 10.7554/eLife.64320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Paramanathan T, Reeves D, Friedman LJ, Kondev J, Gelles J. A general mechanism for competitor-induced dissociation of molecular complexes. Nat. Commun. 2014;5:5207. doi: 10.1038/ncomms6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Grah R, Zoller B, Tkacik G. Nonequilibrium models of optimal enhancer function. Proc. Natl Acad. Sci. USA. 2020;117:31614–31622. doi: 10.1073/pnas.2006731117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Compe E, Egly JM. The long road to understanding RNAPII transcription initiation and related syndromes. Annu. Rev. Biochem. 2021;90:193–219. doi: 10.1146/annurev-biochem-090220-112253. [DOI] [PubMed] [Google Scholar]
- 194.Ruiz A, et al. Characterization of the influence of mediator complex in HIV-1 transcription. J. Biol. Chem. 2014;289:27665–27676. doi: 10.1074/jbc.M114.570341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schneider WM, et al. Genome-scale identification of SARS-CoV-2 and Pan-coronavirus host factor networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Calpena E, et al. De novo missense substitutions in the gene encoding CDK8, a regulator of the mediator complex, cause a syndromic developmental disorder. Am. J. Hum. Genet. 2019;104:709–720. doi: 10.1016/j.ajhg.2019.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Roninson IB, et al. Identifying cancers impacted by CDK8/19. Cells. 2019;8:821. doi: 10.3390/cells8080821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bragelmann J, et al. Pan-cancer analysis of the mediator complex transcriptome identifies CDK19 and CDK8 as therapeutic targets in advanced prostate cancer. Clin. Cancer Res. 2017;23:1829–1840. doi: 10.1158/1078-0432.CCR-16-0094. [DOI] [PubMed] [Google Scholar]
- 199.Syring I, et al. Comprehensive analysis of the transcriptional profile of the mediator complex across human cancer types. Oncotarget. 2016;7:23043–23055. doi: 10.18632/oncotarget.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Klein IA, et al. Partitioning of cancer therapeutics in nuclear condensates. Science. 2020;368:1386–1392. doi: 10.1126/science.aaz4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Risso-Ballester J, et al. A condensate-hardening drug blocks RSV replication in vivo. Nature. 2021;595:596–599. doi: 10.1038/s41586-021-03703-z. [DOI] [PubMed] [Google Scholar]
- 202.Sanders DW, et al. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020;181:306–324.e28. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ito M, Okano HJ, Darnell RB, Roeder RG. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002;21:3464–3475. doi: 10.1093/emboj/cdf348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lee YL, et al. Mediator subunit MED1 is required for E2A-PBX1-mediated oncogenic transcription and leukemic cell growth. Proc. Natl Acad. Sci. USA. 2021;118:e1922864118. doi: 10.1073/pnas.1922864118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yang X, et al. Selective requirement for Mediator MED23 in Ras-active lung cancer. Proc. Natl Acad. Sci. USA. 2012;109:E2813–E2822. doi: 10.1073/pnas.1204311109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Allen BL, et al. Suppression of p53 response by targeting p53-Mediator binding with a stapled peptide. Cell Rep. 2022;39:110630. doi: 10.1016/j.celrep.2022.110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Garlick JM, et al. Norstictic acid is a selective allosteric transcriptional regulator. J. Am. Chem. Soc. 2021;143:9297–9302. doi: 10.1021/jacs.1c03258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nishikawa JL, et al. Inhibiting fungal multidrug resistance by disrupting an activator-mediator interaction. Nature. 2016;530:485–489. doi: 10.1038/nature16963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Burslem GM, Crews CM. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell. 2020;181:102–114. doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schneider M, et al. The PROTACtable genome. Nat. Rev. Drug Discov. 2021;20:789–797. doi: 10.1038/s41573-021-00245-x. [DOI] [PubMed] [Google Scholar]
- 212.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Dimitrova E, et al. FBXL19 recruits CDK-mediator to CpG islands of developmental genes priming them for activation during lineage commitment. eLife. 2018;7:e37084. doi: 10.7554/eLife.37084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Immarigeon C, et al. Drosophila mediator subunit Med1 is required for GATA-dependent developmental processes: divergent binding interfaces for conserved coactivator functions. Mol. Cell Biol. 2019;39:e00477-18. doi: 10.1128/MCB.00477-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huang Y, et al. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell. 2012;45:459–469. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mukundan B, Ansari A. Srb5/Med18-mediated termination of transcription is dependent on gene looping. J. Biol. Chem. 2013;288:11384–11394. doi: 10.1074/jbc.M112.446773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Takahashi H, et al. The role of mediator and little elongation complex in transcription termination. Nat. Commun. 2020;11:1063. doi: 10.1038/s41467-020-14849-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Schneider M, et al. The nuclear pore-associated TREX-2 complex employs mediator to regulate gene expression. Cell. 2015;162:1016–1028. doi: 10.1016/j.cell.2015.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Vernon RM, et al. Pi-pi contacts are an overlooked protein feature relevant to phase separation. eLife. 2018;7:e31486. doi: 10.7554/eLife.31486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 221.Yuan C, Ito M, Fondell JD, Fu Z, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl Acad. Sci. USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- 223.Hittelman AB, Burakov D, Iniguez-Lluhi JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1 associated proteins. EMBO J. 1999;18:5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Malik S, Wallberg AE, Kang YK, Roeder RG. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell Biol. 2002;22:5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell. 2003;12:1137–1149. doi: 10.1016/S1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 226.Chu CS, et al. Unique immune cell coactivators specify locus control region function and cell stage. Mol. Cell. 2020;80:845–861.e10. doi: 10.1016/j.molcel.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Grontved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol. Cell Biol. 2010;30:2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kato Y, Habas R, Katsuyama Y, Naar A, He X. A component of the ARC/mediator complex required for TGF beta/nodal signalling. Nature. 2002;418:641–646. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- 229.Ito M, et al. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell. 1999;3:361–370. doi: 10.1016/S1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 230.Ding N, et al. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J. Biol. Chem. 2009;284:2648–2656. doi: 10.1074/jbc.M806514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Boyer TG, Martin MED, Lees E, Riccardi RP, Berk AJ. Mammalian Srb/mediator complex is targeted by adenovirus E1a protein. Nature. 1999;399:276–279. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- 232.Liu Z, et al. Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat. Commun. 2016;7:11149. doi: 10.1038/ncomms11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Verger A, et al. The mediator complex subunit MED25 is targeted by the N-terminal transactivation domain of the PEA3 group members. Nucleic Acids Res. 2013;41:4847–4859. doi: 10.1093/nar/gkt199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sela D, et al. Role for human mediator subunit MED25 in recruitment of mediator to promoters by endoplasmic reticulum stress-responsive transcription factor ATF6alpha. J. Biol. Chem. 2013;288:26179–26187. doi: 10.1074/jbc.M113.496968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 236.Ding N, et al. Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol. Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zhou H, Kim S, Ishii S, Boyer TG. Mediator modulates Gli3-dependent Sonic hedgehog signaling. Mol. Cell Biol. 2006;26:8667–8682. doi: 10.1128/MCB.00443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/beta-catenin signaling. J. Biol. Chem. 2006;281:14066–14075. doi: 10.1074/jbc.M602696200. [DOI] [PubMed] [Google Scholar]
- 239.Henninger JE, et al. RNA-mediated feedback control of transcriptional condensates. Cell. 2021;184:207–225.e24. doi: 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rossi MJ, et al. A high-resolution protein architecture of the budding yeast genome. Nature. 2021;592:309–314. doi: 10.1038/s41586-021-03314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Cevher MA, et al. Reconstitution of active human core mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol. 2014;21:1028–1034. doi: 10.1038/nsmb.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Pherson M, Misulovin Z, Gause M, Dorsett D. Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila. Genome Res. 2019;29:602–612. doi: 10.1101/gr.243832.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development. 2010;137:2723–2731. doi: 10.1242/dev.053660. [DOI] [PubMed] [Google Scholar]
- 244.Westerling T, Kuuluvainen E, Makela TP. Cdk8 is essential for preimplantation mouse development. Mol. Cell Biol. 2007;27:6177–6182. doi: 10.1128/MCB.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.D’Urso A, et al. Set1/COMPASS and mediator are repurposed to promote epigenetic transcriptional memory. eLife. 2016;5:e16691. doi: 10.7554/eLife.16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sooraj D, et al. MED12 and BRD4 cooperate to sustain cancer growth upon loss of mediator kinase. Mol. Cell. 2022;82:123–139.e7. doi: 10.1016/j.molcel.2021.11.015. [DOI] [PubMed] [Google Scholar]
- 247.Park MJ, et al. Oncogenic exon 2 mutations in mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J. Biol. Chem. 2018;293:4870–4882. doi: 10.1074/jbc.RA118.001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.