SUMMARY
The past decade has revolutionized our understanding of regulatory noncoding RNAs (ncRNAs). Among the most recently identified ncRNAs are downstream-of-gene (DoG)-containing transcripts that are produced by widespread transcriptional readthrough. The discovery of DoGs has set the stage for future studies to address many unanswered questions regarding the mechanisms that promote readthrough transcription, RNA processing, and the cellular functions of the unique transcripts. In this review, we summarize current findings regarding the biogenesis, function, and mechanisms regulating this exciting new class of RNA molecules.
INTRODUCTION
Fifty-two years ago, our understanding of transcriptional regulation was transformed by the discovery of three nuclear RNA polymerases (Roeder, 2019). Since this monumental breakthrough, numerous studies have advanced our understanding of the highly regulated steps of the RNA polymerase II (RNAPII) transcription cycle, including initiation, elongation, and termination. The initiation step involves core promoter recognition and assembly of the preinitiation complex (PIC) that controls DNA opening and the start of RNAPII-mediated transcription (Buratowski, 1994; Conaway and Conaway, 1993; Jiang et al., 1996; Nikolov and Burley, 1997; Orphanides et al., 1996; Petrenko and Struhl, 2021; Roeder, 1996; Schier and Taatjes, 2020). Following initiation, RNAPII undergoes promoter-proximal pausing, a process that is regulated by several factors that stabilize paused RNAPII and control its release into productive elongation (Chen et al., 2018; Core and Adelman, 2019; Jonkers et al., 2014; Laitem et al., 2015; Mayer et al., 2015; Rahl et al., 2010; Wissink et al., 2019). Finally, upon reaching the 3′ end of genes, elongating RNAPII transitions into transcription termination, a regulatory step that involves RNA cleavage, polyadenylation, and RNAPII release (Core et al., 2008; Davidson et al., 2014; Fong et al., 2015; Hsin and Manley, 2012; Mandel et al., 2006; Meinhart and Cramer, 2004; Miki et al., 2017; Proudfoot, 2011, 2016; Tian and Manley, 2013; Zaborowska et al., 2016; Zhang et al., 2015). Numerous reviews have detailed the intricate regulatory steps of the transcription cycle (Core and Adelman, 2019; Dollinger and Gilmour, 2021; Porrua and Libri, 2015; Proudfoot, 2016; Schier and Taatjes, 2020). In addition to its role in the production of protein-coding mRNAs, RNAPII is also responsible for the production of noncoding RNAs (ncRNAs) that execute a diverse array of cellular functions (Cech and Steitz, 2014).
DoG RNAs REPRESENT A UNIQUE CLASS OF RNAPII-TRANSCRIBED ncRNAs
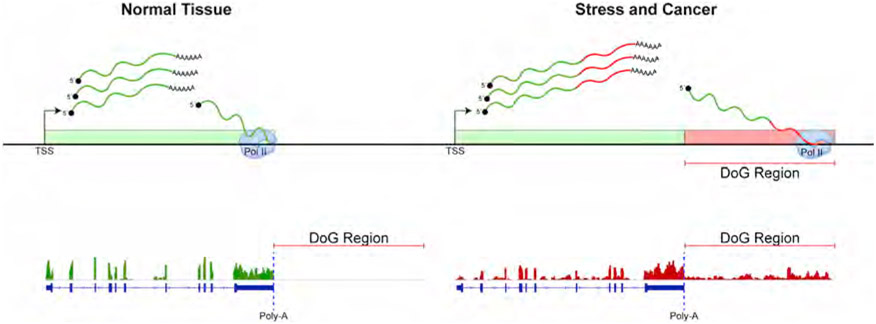
Downstream-of-gene (DoG)-containing RNAs are a unique class of ncRNA that possesses three defining features (Figure 1) (Rosa-Mercado and Steitz, 2022; Vilborg and Steitz, 2017). First, DoG RNAs initiate at the promoter of a protein-coding host gene. Second, DoG transcripts extend as long continuous transcripts for at least 5 kb beyond the 3′ terminal polyadenylation signals (PASs) of their host gene. Third, the DoG RNAs that have been studied to date are retained in the nucleus and are likely to remain associated with chromatin (Vilborg et al., 2015; Vilborg et al., 2017), although it is unknown if this is a universal feature. These properties set DoGs apart from many other ncRNAs, which do not originate from protein-coding genes and may have well-defined termination sites. Moreover, the production of DoG RNAs is triggered by various cellular stress responses and involves a disruption of RNAPII transcription termination and RNA polyadenylation (Grosso et al., 2015; Heinz et al., 2018; Nemeroff et al., 1998; Rosa-Mercado et al., 2021; Rutkowski et al., 2015; Shalgi et al., 2014; Vilborg et al., 2015; Vilborg and Steitz, 2017).
Figure 1. Molecular features that define readthrough transcripts (DoGs).
Schematic diagram to depict DoG biogenesis that occurs in response to various stress stimuli and in cancer versus normal tissue (Cardiello et al., 2018; Cugusi et al., 2022; Heinz et al., 2018; Nemeroff et al., 1998; Rosa-Mercado et al., 2021; Rutkowski et al., 2015; Shalgi et al., 2014; Vilborg et al., 2015; Vilborg et al., 2017; Vilborg and Steitz, 2017). DoGs are unidirectional, continuous transcripts that initiate from the promoter of the DoG-producing gene and extend at least >5 kb beyond the mRNA 3′ end processing polyadenylation (Poly-A) signals (Vilborg et al., 2015).
Pervasive transcription readthrough gives rise to the long DoG transcripts that are believed to account for 15%–30% of intergenic transcription (Rosa-Mercado and Steitz, 2022; Vilborg and Steitz, 2017). Unlike long intergenic ncRNAs (lincRNAs) that are autonomously transcribed, DoGs are transcripts generated from the upstream DoG-producing gene, which is capable of producing both spliced mRNA and RNA that is continuous with the long noncoding DoG transcript (Figure 1). This is best supported by a previous study (Vilborg et al., 2015), which revealed that DoG-producing genes contain only one transcription start site (TSS), as determined by CapSeq (Illingworth et al., 2010). Moreover, antisense oligonucleotide (ASO)-mediated knockdown of mRNA, synthesized from the DoG-producing host gene, results in a corresponding change in DoG levels, which is consistent with DoG production being dependent on continued transcription from the host gene promoter (Vilborg et al., 2015). Also, whereas lincRNAs are expressed at levels that are positively correlated with the expression levels of nearby genes (Andersson et al., 2014; Cheng et al., 2015; Kaikkonen et al., 2013; Lai et al., 2015; Rahnamoun et al., 2018; Sanyal et al., 2012), DoGs are produced regardless of the transcriptional levels of their host gene (Rosa-Mercado et al., 2021). For example, DoGs were found to be produced from DoG-producing genes that are activated, repressed, or not changing in response to hyperosmotic stress (Rosa-Mercado et al., 2021). These findings suggest that DoG versus DoG host gene regulatory mechanisms are uncoupled.
ESTABLISHED MECHANISMS REGULATING DoG RNA EXPRESSION
Although a number of cellular processes and factors have been linked to DoG biogenesis, the full breadth of mechanisms underlying transcriptional readthrough and various DoG properties remains to be fully elucidated. Strong evidence linking cellular stress responses to the expression of DoGs suggests a rapid mechanism underlying the switch producing spliced and polyadenylated protein-coding transcripts to long ncRNAs. DoGs are induced in response to various stress stimuli including osmotic stress (Rosa-Mercado et al., 2021; Vilborg et al., 2015), heat shock (Cardiello et al., 2018; Cugusi et al., 2022; Shalgi et al., 2014; Vilborg et al., 2017), influenza A virus (IAV) (Heinz et al., 2018; Nemeroff et al., 1998), and herpes simplex virus (HSV) infection (Rutkowski et al., 2015). Precisely how each of these diverse stress signals triggers DoG RNA expression is not fully understood; however, several involve alterations in RNAPII termination and RNA processing factors.
DoG expression is driven by RNAPII termination defects
Termination of gene transcription remains the least well-understood regulatory step of the transcription cycle (Proudfoot, 2016). Yet, defects in termination are becoming more readily identified and quantified by high-throughput nascent RNA sequencing methods that reveal “readthrough transcription” that maps downstream of the canonical polyadenylation sites to the 3′ gene boundary of protein-coding genes. The majority of studies to date suggest that DoG expression is tightly linked to termination defects.
Transcription termination is linked to mRNA 3′ end processing by polyadenylation and cleavage factors (Bauer et al., 2018; Nemeroff et al., 1998; Wang et al., 2020). Specifically, the PAS within the nascent transcript is recognized by the cleavage and PAS specificity factor (CPSF) that contains CPSF73, an endonuclease that cleaves the nascent RNA (Chan et al., 2014). Consistent with DoG production resulting from reduced transcription termination is the finding that the loss of CPSF73 results in partial induction of DoGs (Vilborg et al., 2015). The depletion of additional polyadenylation/termination factors, including 5′–3′ exoribonuclease, Xrn2, has also been shown to result in an enrichment of DoG production (Eaton et al., 2018). In addition, DoG-producing genes exhibit a reduced frequency of strong PASs compared with non-DoG-producing genes, which leads to less efficient termination in these regions and the ability of RNAPII to remain engaged in processive elongation past the normal termination sites (Rosa-Mercado et al., 2021). Similarly, nuclear poly(A)-binding protein, Nab2, which is a regulator of co-transcriptional splicing, has also been implicated in 3′ end cleavage of nascent RNA (Alpert et al., 2020). Consistent with this role, the loss of Nab2 was found to coincide with aberrant cleavage and termination, which in turn resulted in DoG production (Alpert et al., 2020).
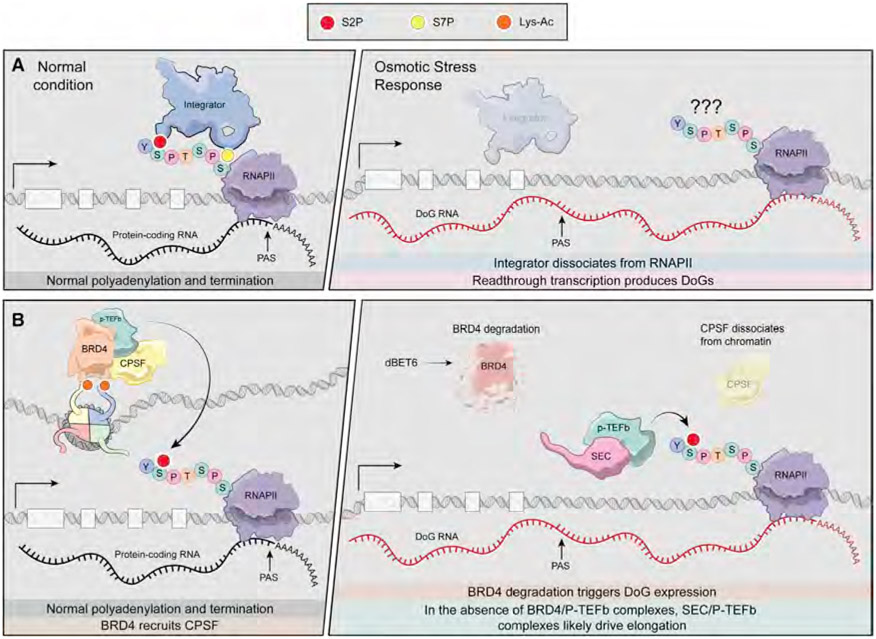
In addition to canonical cleavage and polyadenylation factors, the Integrator complex is involved in regulating DoG expression in the context of cellular stress (Figure 2). The Integrator complex is a large macromolecular assembly that possesses a ribonuclease catalytic activity that is involved in an array of RNA-based processes including RNA 3′ end processing, RNAPII pause-release, and termination (Kirstein et al., 2021). Integrator directly interacts with RNAPII by binding to phosphorylation sites in its C-terminal domain (CTD) (Baillat et al., 2005; Egloff et al., 2010). Profiling of the RNAPII protein interactome under the condition of osmotic shock revealed that this cellular stress triggers the dissociation of the Integrator complex from RNAPII (Rosa-Mercado et al., 2021). Importantly, several groups have demonstrated that disrupting the function of Integrator subunit INTS11 is sufficient to induce DoG expression (Dasilva et al., 2021; Rosa-Mercado et al., 2021). Thus, Integrator function appears to be a key player in the suppression of DoG expression under normal conditions, and its function is perturbed during osmotic stress, leading to DoG RNA production.
Figure 2. Mechanisms driving DoG expression.
(A) Integrator is involved in the suppression of DoG expression. Under normal conditions, the Integrator complex associates with RNAPII CTD through interaction with the Serine 2 and Serine 7 double phosphorylation (S2P/S7P) mark (Egloff et al., 2010). During hyperosmotic stress, Integrator dissociates from RNAPII, leading to defective termination and the production of termination readthrough DoG RNAs (Rosa-Mercado et al., 2021). The phosphorylation status of the RNAPII CTD at DoG-producing genes during osmotic stress has not been determined; however, alteration in CTD phosphorylation could contribute to this process. PAS, polyadenylation site.
(B) Degradation of BRD4 triggers DoG expression. P-TEFb can be recruited to target genes through interaction with bromodomain-containing protein 4 (BRD4), which is a reader of acetylated nucleosomes (Peterlin and Price, 2006). P-TEFb promotes transcriptional elongation by phosphorylation of the Serine 2 residue of the RNAPII CTD (Peterlin and Price, 2006; Yang et al., 2005). In addition to binding to P-TEFb, BRD4 interacts with cleavage and polyadenylation specificity factor (CPSF) (Arnold et al., 2021). When BRD4 is degraded by the small molecule dBET6, this results in dissociation of CPSF from chromatin and causes an accumulation of DoG RNAs (Arnold et al., 2021). In the absence of BRD4-P-TEFb complexes, it is likely that super elongation complex (SEC)-P-TEFb is responsible for driving transcription elongation of DoGs through RNAPII S2 phosphorylation (Luo et al., 2012).
DoG expression and human disease
In addition to activation of DoGs by cellular stress responses, DoGs have also been linked to human disease. Correlative evidence in clear cell renal cell carcinoma (ccRCC) suggests that inactivation of the histone H3 lysine 36 (H3K36) methyltransferase, SETD2, may promote DoG production (Grosso et al., 2015). SETD2 is highly mutated in renal cancer and its loss is associated with a dramatic decrease of the H3K36me3 histone modification (Cancer Genome Atlas Research, 2013; Dalgliesh et al., 2010; Duns et al., 2010; Grosso et al., 2015; Varela et al., 2011). H3K36me3 is typically associated with the bodies of actively transcribed genes, and several studies have demonstrated its involvement in RNA processing through the recruitment of RNA processing factors (Fahey and Davis, 2017; Kim et al., 2011; Kolasinska-Zwierz et al., 2009). Intriguingly, in ccRCC SETD2, loss is correlated with the production of DoG RNAs (Grosso et al., 2015). Recent studies in Drosophila have also linked the H3K36me3 modification to RNA polyadenylation, suggesting a potential mechanism for SETD2’s involvement in DoG expression (Meers et al., 2017). However, it will be important to examine DoG expression in isogenic SETD2 loss-of-function experiments to establish a direct link between SETD2, H3K36me3, and DoGs.
It has recently been demonstrated that treatment of cancer cells with the bromodomain-containing protein 4 (BRD4) degrader dBET6 can trigger the expression of DoGs (Arnold et al., 2021) (Figure 2). In recent years, inhibitors and degraders of BRD4 have emerged as a promising cancer therapy (Filippakopoulos et al., 2010; Shi and Vakoc, 2014; Winter et al., 2017). BRD4 is recruited to chromatin through an interaction between its bromodomains and acetylated histones (Filippakopoulos et al., 2010). BRD4 functions as an RNAPII elongation factor (EF) in complex with the RNAPII CTD kinase P-TEFb, and its inhibition results in a variety of transcriptional defects (Jang et al., 2005; Shi and Vakoc, 2014; Yang et al., 2005). BRD4 degradation through the proteolysis-targeting chimeric (PROTAC) molecule dBET6, or by genetic tagging of BRD4 with an inducible degron tag (dTag), triggers widespread transcriptional termination defects resulting in DoG expression (Arnold et al., 2021). Moreover, these effects are dependent on BRD4 protein degradation, as opposed to BRD4 inhibition, as treatment of cells with the BRD4 bromodomain inhibitor JQ1 does not result in DoG expression, implying that these effects require almost complete loss of BRD4 function. Mechanistically, BRD4 interacts with RNA 3′ end processing factors such as CPSF as well as RNAPII EFs including PAF, DRB sensitivity inducing factor (DSIF), and NELF, and upon BRD4 degradation, these proteins exhibit severely impaired binding to chromatin (Arnold et al., 2021). This study revealed an unexpected link between an RNAPII EF and the control of termination. It will be important for future studies to examine if DoGs play a functional role in the cellular response to BRD4 degradation.
MECHANISMS INVOLVED IN RNA BIOGENESIS WITH POTENTIAL LINKS TO DoG EXPRESSION
Termination defects are clearly central to the production of DoG RNAs. However, there are a number of other transcriptional regulatory pathways that may be relevant to DoGs and are worthy of further examination. It is widely accepted that mRNA processing through splicing, polyadenylation, and capping occurs co-transcriptionally and is tightly linked to RNAPII elongation (Bentley, 2014; Neugebauer, 2002; Proudfoot et al., 2002). As these processes are molecularly coupled, this raises the possibility that alterations in termination, elongation, or splicing could potentially result in DoG expression. Consistent with this possibility, in both yeast and mammalian cells, unspliced mRNAs are typically not cleaved at their 3′ ends and often present as readthrough transcripts (Alpert et al., 2020; Dye and Proudfoot, 1999; Herzel et al., 2018; Reimer et al., 2021). Future studies will be required to define the splicing status of DoGs, which currently remains unclear.
RNAPII CTD phosphorylation recruits Integrator
Alterations of RNAPII CTD phosphorylation may be involved in DoG production with respect to Integrator recruitment. Post-translational modifications (PTMs) of the RNAPII CTD coordinate RNAPII activity by facilitating its association with a number of different factors, including transcription elongation and termination factors (Hintermair et al., 2012; Hsin and Manley, 2012; Mayer et al., 2012; Zaborowska et al., 2016). The RNAPII CTD also recruits RNA processing factors that do not necessarily coordinate RNAPII transcriptional activity (Hsin and Manley, 2012; Zaborowska et al., 2016). The RNAPII CTD consists of 52 heptad repeats of the consensus sequence (YSPTSPS) and is phosphorylated at Y1, S2, T4, S5, and S7 (Hirose and Manley, 2000; Proudfoot et al., 2002; Shatkin and Manley, 2000). Phosphorylated S2/S5 residues serve as hallmarks for active transcription of protein-coding genes (Corden, 1990; Eick and Geyer, 2013; West and Corden, 1995). Specifically, S5P levels are enriched at the 5′ ends of coding regions, and as RNAPII elongates, S5P levels decrease while S2P levels increase (Corden, 1990; Eick and Geyer, 2013; West and Corden, 1995). RNAPII CTD Y1 phosphorylation is predominantly localized in the antisense orientation at promoter regions and is significantly enriched at active enhancers (Hsin et al., 2014). CTD S7 phosphorylation is associated with RNAPII-transcribed small nuclear RNAs (snRNAs) and is required for their expression (Egloff and Murphy, 2008; Egloff et al., 2007).
The Integrator complex was initially characterized through its ability to interact with the RNAPII CTD (Baillat et al., 2005). Considering that osmotic stress impairs the interaction between RNAPII and Integrator (Rosa-Mercado et al., 2021), it is likely that RNAPII CTD PTMs may play a role in this process. Integrator binds most efficiently to a S2P/S7P double modification, which is implemented by a combination of P-TEFb (S2P) and DNA-PK (S7P) kinase activity (Egloff et al., 2010). Considering that BRD4 degradation disrupts the function of a subset of P-TEFb-containing complexes, it will be important to investigate whether DoG expression is linked to defective Integrator-RNAPII interactions similarly to osmotic stress. Recent studies have also implicated RNAPII CTD Y1 phosphorylation as being involved in Integrator recruitment. Mutation of Y1 to phenylalanine (Y1F) in 39 of the 52 heptad repeats results in widespread transcription termination readthrough leading to DoG production (Shah et al., 2018). The Y1F RNAPII also displays impaired interactions with the Integrator complex, although a direct interaction between Integrator and Y1P has not been demonstrated (Shah et al., 2018). Another caveat of this work is that the Y1F mutant may not fully mimic the non-phosphorylated tyrosine residue and also will disrupt any function that the unmodified tyrosine residue plays. Therefore, a more definitive answer to the role of Y1 phosphorylation will require the inhibition of the Y1 kinase.
RNAPII elongation factors in DoG expression
DoG expression requires both a failure to terminate normally as well as continued RNAPII elongation far beyond the normal gene 3′ end. This implicates RNAPII EFs as potential players in DoG biogenesis. Indeed, a molecular shift that favors continued transcriptional elongation over termination would be expected to promote DoG-producing expression. A large number of EFs are dedicated to the control of various aspects of RNAPII elongation (Chen et al., 2018; Zhou et al., 2012). Whereas many EFs, such as the super elongation complex (SEC) and P-TEFb, function as positive EFs that predominantly stimulate the elongation process, others exhibit more dynamic roles in regulating elongation (Chen et al., 2018; Sims et al., 2004; Zhou et al., 2012). For instance, DSIF, composed of the SPT4 and SPT5 subunits, inhibits elongation until it is phosphorylated by the CDK9 kinase, which converts it to a positive EF (Chen et al., 2018; Zhou et al., 2012). In addition, more recent studies have revealed an additional role for SPT5 in preventing proteasomal degradation of the RNAPII large subunit, Rpb1 (Aoi et al., 2021; Hu et al., 2021). Similarly, the RNAPII-associated factors complex (PAFc) associates with elongating polymerase and was presumed to function largely as a positive EF, but it was recently demonstrated that depletion of PAFc results in pause-release from a subset of genes (Chen et al., 2015; Chen et al., 2017). Thus, although a large number of factors have been implicated in regulating RNAPII elongation through in vitro assays, the complex function and dynamic regulation of EFs in a cellular context is only beginning to become understood.
P-TEFb, in addition to its crucial role in promoting RNAPII elongation through phosphorylation of S2 of the RNAPII CTD, also phosphorylates a number of EFs, including SPT5 and NELFE (Egloff, 2021), to promote transcriptional elongation. P-TEFb also exists in several distinct complexes (Peterlin and Price, 2006). For example, if P-TEFb is bound to the 7SK snRNA and HEXIM1/2 RNA-binding proteins, then it is maintained in an inactive pool that is available for release and incorporation into other complexes (Peterlin and Price, 2006). P-TEFb can also complex with both SEC and BRD4 (Jonkers and Lis, 2015), although these complexes likely serve distinct functions, with SEC being recruited to rapidly induced stimulus-responsive genes and BRD4 targeting to chromatin through its histone lysine acetylation reader bromodomains (Jang et al., 2005; Yang et al., 2005). Since BRD4 and SEC represent the two major activating P-TEFb-containing complexes, the observation that degradation of BRD4 results in activation of DoG expression strongly suggests that SEC is responsible for driving elongation of DoG RNAs.
THE FUNCTIONAL ROLES OF READTHROUGH TRANSCRIPTION AND DoGs
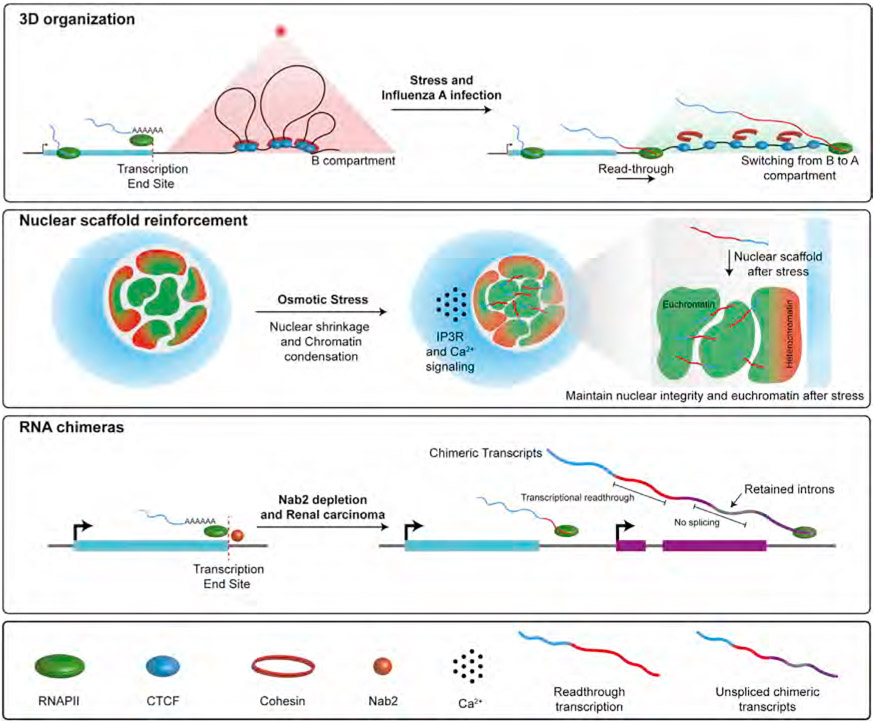
Despite DoGs becoming increasingly identified as a hallmark of termination defects (Heinz et al., 2018; Nemeroff et al., 1998; Rosa-Mercado et al., 2021; Rutkowski et al., 2015; Shalgi et al., 2014; Vilborg et al., 2015), it remains to be resolved whether readthrough transcription, DoGs themselves, or both exhibit consequences on cellular function. To date, there are few studies demonstrating that readthrough transcription can exhibit severe consequences on gene expression (Greger and Proudfoot, 1998; Shearwin et al., 2005). One such example is when the resulting readthrough transcripts run into the promoter of a nearby gene or ncRNA and restrict its activity through transcriptional interference or lead to the production of RNA chimeras (Greger and Proudfoot, 1998; Shearwin et al., 2005). Chimeric transcripts spanning multiple genes with retained introns have been shown to result when there is a failure in splicing due to a disruption in Nab2, which is required to ensure proper 3′ end cleavage (Alpert et al., 2020; Herzel et al., 2018) (Figure 3). In addition to transcriptional interference, the act of readthrough transcription has been shown to contribute to host genome 3D organization following IAV infection (Heinz et al., 2018). Specifically, an investigation of chromatin changes following IAV infection revealed that non-structural (NS1) protein of IAV induces readthrough transcription of highly active genes that results in the displacement of cohesin from chromatin, elimination of chromatin loops, and decompacted chromatin in the readthrough regions (Heinz et al., 2018) (Figure 3). These NS1-dependent changes in chromatin may contribute to NS1-dependent IAV virulence. Also consistent with a role for NS1 in contributing to transcription termination defects is the finding that NS1 inhibits the CPSF complex and disrupts mRNA cleavage and polyadenylation (Noah et al., 2003). Further studies are required to determine the relative contributions of NS1-dependent regulation of mRNA processing, pervasive readthrough transcription, and chromatin structure, and its ability to subvert host antiviral responses and increase virulence.
Figure 3. The roles of readthrough transcription and DoGs.
Schematic representation of the impact that readthrough transcription and DoGs have on the production of RNA chimeras, 3D chromatin organization, and nuclear scaffold reinforcement. In support of readthrough transcription leading to chimeric transcripts are several studies revealing aberrant coupling of splicing and 3′ end cleavage that leads to readthrough transcription and nascent transcripts with retained introns (Herzel et al., 2018). It has also been demonstrated that readthrough transcription leading to RNA chimeras originate following the depletion of nuclear export and splicing regulator, Nab2 (Alpert et al., 2020). RNAPII elongation has also been linked to chromatin structure in mammalian cells. Specifically, this was demonstrated by a study revealing influenza A virus (IAV)/NS1-dependent inhibition of transcription termination in which RNAPII elongates past termination sites leading to a loss of chromatin loops and local chromatin decompaction (Heinz et al., 2018). Emerging evidence also suggests that the DoGs themselves may function by reinforcing the nuclear scaffold during stress responses. This was supported by a study revealing that preventing DoG induction after osmotic stress, which leads to nuclear shrinkage as water is forced from the cell (Finan and Guilak, 2010), results in a more severe level of nuclear shrinkage and chromatin collapse (Vilborg et al., 2015). Together, these recent findings reveal the importance of further dissecting the relationship between readthrough transcription and DoGs to understand key elements of their functional mechanisms.
The identification that readthrough transcription through DoG-producing regions is a regulated process suggests that DoGs are functional (Alpert et al., 2020; Arnold et al., 2021; Cardiello et al., 2018; Dasilva et al., 2021; Grosso et al., 2015; Hennig et al., 2018; Melnick et al., 2019; Rosa-Mercado et al., 2021; Sharma et al., 2014). To our knowledge, however, there are no studies to date that have directly manipulated the DoG itself to discern its direct (causal) functions. Thus, it largely remains a mystery as to whether DoGs are functional molecules in the cell. DoGs have been shown to be found in the chromatin-bound fraction where they appear to colocalize with their upstream DoG-producing mRNA (Vilborg et al., 2015). This finding, together with the large size of DoGs, suggests that they may be involved in nuclear scaffold reinforcement. Consistent with this possibility is the finding that disrupting DoG induction by IP3R inhibition aggravates osmotic-stress-associated chromatin collapse (Vilborg et al., 2015) (Figure 3). Additional investigation is needed to determine whether DoGs affect gene expression in addition to maintaining nuclear integrity and stabilizing genomic regions that support DoG production.
FUTURE PERSPECTIVES
Readthrough transcription is an emerging phenotype, and DoG discovery opens the door for an exciting time defined by cataloging additional DoGs, identifying various DoG properties, and determining the particular genes that are subject to transcription readthrough in various cellular conditions and organisms of interest. In addition, it will be important for the field to provide key insights into unrecognized mechanisms underlying transcriptional readthrough and determine whether DoGs exhibit functions that impact cellular processes.
The process of cataloging additional DoGs across tissue types, developmental stages, and disease states, and distinguishing them from other transcripts, will be an important but also challenging effort because of the read-in transcription of DoGs into neighboring genes (Cardiello et al., 2018; Dasilva et al., 2021; Hennig et al., 2018; Rosa-Mercado et al., 2021; Roth et al., 2020; Rutkowski et al., 2015). Proper assignment of DoGs in different conditions and cell types requires the ability to identify DoGs that do not overlap with neighboring regions that are transcriptionally active on either DNA strand. As demonstrated in Table 1, we highlight a few of the packages that have been developed to streamline the process of DoG identification and quantification (Melnick et al., 2019; Roth et al., 2020; Wiesel et al., 2018). The recent development of long-read sequencing technologies will allow for the sequencing of full-length DoG transcripts, resolving issues such as the splicing status of DoGs and determining whether these transcripts are enriched or depleted for RNA modifications (Logsdon et al., 2020; Lorenz et al., 2020; Yue et al., 2015). There is also an imminent need to develop tools to experimentally manipulate and examine the direct functions of the plethora of ncRNAs that include DoGs. With an important consideration of any of these methods comes the challenge of discerning the functions of specific DoGs, while ensuring to discount any potential bleed through of another neighboring transcript. In Table 2, we highlight some of the commonly used practices in the field for studying RNA function that could be applied to probing the potential functional significance of DoGs (Abudayyeh et al., 2017; Arun et al., 2016; Bensaude, 2011; Daneshvar et al., 2020; Lai et al., 2020; Lee and Mendell, 2020; Leppek and Stoecklin, 2014; McHugh and Guttman, 2018; Rahnamoun et al., 2018; Vilborg et al., 2015). Taken together, this information will enable researchers to explore the readthrough phenomenon and advance our understanding of the widespread transcriptional readthrough that is arising in response to various stress conditions in mammalian cells.
Table 1.
Tools for identifying readthrough transcription in a variety of systems and contexts
| Tool | Tool Description | References |
|---|---|---|
| DoGFinder | Python based software package that identifies DoGs and quantifies their expression levels from any RNA-seq dataset. DoGFinder identifies DoGs by requiring a minimal coverage over a minimal initial length of continuous read density downstream of the 3’ end of every gene locus. | Wiesel et al., 2018 |
| Automatic Readthrough Transcription Detection (ARTDeco) | Python based software that provides the framework for quantification and characterization of readthrough transcription from NGS data. ARTDeco quantifies the degree of readthrough transcription using read-in and readthrough levels and the detection of DoG transcripts. ARTDeco varies from other programs by considering the discovered transcripts that extend into annotated genes to avoid the potential of arbitrary truncation. |
Roth et al., 2020 |
| Dogcatcher | Python based software that identifies DoG locations, genes that overlap with DoGs on the same or opposite strand, and an optional pipeline to provide differential expression of DoGs. | Melnick et al., 2019 |
| Long-read RNA-sequencing platforms | Entire nascent transcripts are sequenced from the 5’ end (defined by the transcription start site [TSS]) to the 3’ end (defined as the position of RNA Pol II at the time of isolation). | Logsdon et al., 2020 |
Table 2.
Tools for exploring functional significance of DoGs
| Tool | Tool description | References |
|---|---|---|
| Inhibiting eukaryotic transcription using widely used compounds | Actinomycin D targets DNA and inhibits RNAPll elongation. α-amanitin targets RNAPll to inhibit RNA synthesis, and CDK9 inhibitors such as DRB and flavopiridol inhibit RNAPll elongation and rRNA processing. | Yue et al., 2015 |
| RNAi-mediated knockdown or incorporation of antisense oligonucleotides (ASOs) | Loss-of-function studies have been instrumental in discerning ncRNA functions. ASOs revealed that knocking down mRNA of the host DoG-producing genes results in reduced levels of the corresponding DoGs. Thus, ASOs could be particularly useful with the targeting of antisense oligos to facilitate disruptions in termination at different regions of the DoG-producing host gene since ASOs are known to result in premature transcription termination. |
Vilborg et al., 2015
Rahnamoun et al., 2018 Bensaude, 2011 Arun et al., 2016 Lai et al., 2020 |
| Cas13 Rnase | The Cas9 system has been deployed in mammalian cells to target the degradation of specific RNA molecules, including lncRNAs. The potential advantages over oligonucleotide-based DoG depletion systems is in terms of limiting off-target effects and may be applicable to a wide-array of biological systems because it can be genetically encoded. Moreover, catalytically dead Cas13 is able to bind to its target RNAs without degrading them and thus would provide a useful tool for visualizing DoG transcripts and could potentially be used to disrupt DoG interactions with RNA binding proteins. | Abudayyeh et al., 2017 |
| RNA antisense purification (RAP) and streptavidin aptamers for purification of ribonucleoprotein complexes | Proteome-wide screens for identifying the interactome of DoGs could be employed. This approach would take advantage of established methodologies that have been coupled with mass spectrometry (MS) to identify proteins that interact with lncRNAs in cells, including RNA antisense purification (RAP and streptavidin aptamers for purification of ribonucleoprotein complexes. |
Daneshvar et al., 2020 Leppek and Stoecklin, 2014 McHugh and Guttman, 2018 |
ACKNOWLEDGMENTS
We thank Pedro Antonio Avila Lopez and Brianna Monroe for contributing to figure preparation for this review article. We apologize for any oversights or omissions. Research in the Lauberth lab is supported by a grant from the NIH/National Institute of General Medical Sciences (R35 GM128900) to S.M.L.; research in the Shilatifard laboratory is supported by NIH (R35CA197569 and U54CA231638) to A.S.; and research in the Shiekhattar laboratory is supported by funding from the University of Miami Miller School of Medicine, Sylvester Comprehensive Cancer Center and grants R01 GM078455, R01 GM105754, P30 CA240139, and DP1 CA228041 from the National Institute of Health to R.S.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert T, Straube K, Oesterreich FC, Herzel L, and Neugebauer KM (2020). Widespread transcriptional readthrough caused by Nab2 depletion leads to chimeric transcripts with retained introns. Cell Rep. 33, 108496. 10.1016/j.celrep.2020.108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Takahashi YH, Shah AP, Iwanaszko M, Rendleman EJ, Khan NH, Cho BK, Goo YA, Ganesan S, Kelleher NL, and Shilatifard A (2021). SPT5 stabilization of promoter-proximal RNA polymerase II. Mol. Cell 81, 4413.e5. 4424.e5. 10.1016/j.molcel.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M, Bressin A, Jasnovidova O, Meierhofer D, and Mayer A (2021). A BRD4-mediated elongation control point primes transcribing RNA polymerase II for 3′-processing and termination. Mol. Cell 81, 3589.e13. 3603.e13. 10.1016/j.molcel.2021.06.026. [DOI] [PubMed] [Google Scholar]
- Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, et al. (2016). Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 30, 34–51. 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, and Shiekhattar R (2005). Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276. 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bauer DLV, Tellier M, Martínez-Alonso M, Nojima T, Proudfoot NJ, Murphy S, and Fodor E (2018). Influenza virus mounts a two-pronged attack on host RNA polymerase II transcription. Cell Rep. 23, 2119.e3. 2129.e3. 10.1016/j.celrep.2018.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O (2011). Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity? Transcription 2, 103–108. 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2014). Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet 15, 163–175. 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S (1994). The basics of basal transcription by RNA polymerase II. Cell 77, 1–3. 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2013). Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49. 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiello JF, Goodrich JA, and Kugel JF (2018). Heat shock causes a reversible increase in RNA polymerase II occupancy downstream of mRNA genes, consistent with a global loss in transcriptional termination. Mol. Cell. Biol 38. e00181–1. 10.1128/MCB.00181-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, and Steitz JA (2014). The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94. 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, Weng L, Moresco JJ, Yates JR 3rd, Ule J, Manley JL, and Shi Y (2014). CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3′ processing. Genes Dev. 28, 2370–2380. 10.1101/gad.250993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Smith ER, and Shilatifard A (2018). Born to run: control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol 19, 464–478. 10.1038/s41580-018-0010-5. [DOI] [PubMed] [Google Scholar]
- Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, Shiekhattar R, and Shilatifard A (2015). PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell 162, 1003–1015. 10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FX, Xie P, Collings CK, Cao K, Aoi Y, Marshall SA, Rendleman EJ, Ugarenko M, Ozark PA, Zhang A, et al. (2017). PAF1 regulation of promoter-proximal pause release via enhancer activation. Science 357, 1294–1298. 10.1126/science.aan3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JH, Pan DZ, Tsai ZT, and Tsai HK (2015). Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Sci. Rep 5, 12648. 10.1038/srep12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, and Conaway JW (1993). General initiation factors for RNA polymerase II. Annu. Rev. Biochem 62, 161–190. 10.1146/annurev.bi.62.070193.001113. [DOI] [PubMed] [Google Scholar]
- Corden JL (1990). Tails of RNA polymerase II. Trends Biochem. Sci 15, 383–387. 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Core L, and Adelman K (2019). Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982. 10.1101/gad.325142.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, and Lis JT (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848. 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugusi SM, Mitter R, Kelly GP, Walker J, Han Z, Pisano P, Wierer M, Stewart A, and Svejstrup JQ (2022). Heat shock induces premature transcript termination and reconfigures the human transcriptome. Mol. Cell 82, 1–16. 10.1016/j.molcel.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. (2010). Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363. 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K, Ardehali MB, Klein IA, Hsieh FK, Kratkiewicz AJ, Mahpour A, Cancelliere SOL, Zhou C, Cook BM, Li W, et al. (2020). lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat. Cell Biol 22, 1211–1222. 10.1038/s41556-020-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasilva LF, Blumenthal E, Beckedorff F, Cingaram PR, Gomes dos Santos H, Edupuganti RR, Zhang A, Dokaneheifard S, Aoi Y, Yue J, et al. (2021). Integrator enforces the fidelity of transcriptional termination at protein-coding genes. Sci. Adv 7, eabe3393. 10.1126/sciadv.abe3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, Muniz L, and West S (2014). 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 28, 342–356. 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollinger R, and Gilmour DS (2021). Regulation of promoter proximal pausing of RNA polymerase II in metazoans. J. Mol. Biol 433, 166897. 10.1016/j.jmb.2021.166897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM, and Kok K (2010). Histone methyltransferase gene SETD2 is a novel tumorsuppressorgene in clear cell renal cell carcinoma. CancerRes. 70, 4287–4291. 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- Dye MJ, and Proudfoot NJ (1999). Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3, 371–378. 10.1016/s1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- Eaton JD, Davidson L, Bauer DLV, Natsume T, Kanemaki MT, and West S (2018). Xrn2 accelerates termination by RNA polymerase II, which is underpinned by CPSF73 activity. Genes Dev. 32, 127–139. 10.1101/gad.308528.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S (2021). CDK9 keeps RNA polymerase II on track. Cell. Mol. Life Sci 78, 5543–5567. 10.1007/s00018-021-03878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, and Murphy S (2008). Cracking the RNA polymerase II CTD code. Trends Genet. 24, 280–288. 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, and Murphy S (2007). Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318, 1777–1779. 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, and Murphy S (2010). The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem 285, 20564–20569. 10.1074/jbc.M110.132530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D, and Geyer M (2013). The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev 113, 8456–8490. 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- Fahey CC, and Davis IJ (2017). SETting the stage for cancer development: SETD2 and the consequences of lost methylation. Cold Spring Harb. Perspect. Med 7, a026468. 10.1101/cshperspect.a026468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. (2010). Selective inhibition of BET bromodomains. Nature 468, 1067–1073. 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan JD, and Guilak F (2010). The effects of osmotic stress on the structure and function of the cell nucleus. J. Cell. Biochem 109, 460–467. 10.1002/jcb.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, and Bentley DL (2015). Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol. Cell 60, 256–267. 10.1016/j.molcel.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, and Proudfoot NJ (1998). Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 17, 4771–4779. 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vítor AC, Desterro JM, Carmo-Fonseca M, and de Almeida SF (2015). Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife 4, e09214. 10.7554/eLife.09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. (2018).Transcription elongation can affect genome 3D structure. Cell 174, 1522.e22. 1536.e22. 10.1016/j.cell.2018.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig T, Michalski M, Rutkowski AJ, Djakovic L, Whisnant AW, Friedl MS, Jha BA, Baptista MAP, L’Hernault A, Erhard F, et al. (2018). HSV-1-induced disruption of transcription termination resembles a cellular stress response but selectively increases chromatin accessibility downstream of genes. PLoS Pathog. 14, e1006954. 10.1371/journal.ppat.1006954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel L, Straube K, and Neugebauer KM (2018). Long-read sequencing of nascent RNA reveals coupling among RNA processing events. Genome Res. 28, 1008–1019. 10.1101/gr.232025.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, et al. (2012). Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 31, 2784–2797. 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, and Manley JL (2000). RNA polymerase II and the integration of nuclear events. Genes Dev. 14, 1415–1429. [PubMed] [Google Scholar]
- Hsin JP, Li W, Hoque M, Tian B, and Manley JL (2014). RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. eLife 3, e02112. 10.7554/eLife.02112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin JP, and Manley JL (2012). The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26, 2119–2137. 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Peng L, Xu C, Wang Z, Song A, and Chen FX (2021). SPT5 stabilizes RNA polymerase II, orchestrates transcription cycles, and maintains the enhancer landscape. Mol. Cell 81, 4425.e6. 4439.e6. 10.1016/j.molcel.2021.08.029. [DOI] [PubMed] [Google Scholar]
- Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, and Bird AP (2010). Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 6, e1001134. 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, and Ozato K (2005). The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534. 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yan M, and Gralla JD (1996). A three-step pathway of transcription initiation leading to promoter clearance at an activation RNA polymerase II promoter. Mol. Cell. Biol 16, 1614–1621. 10.1128/MCB.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, and Lis JT (2014). Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3, e02407. 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, and Lis JT (2015). Getting up to speed with transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol 16, 167–177. 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, and Glass CK (2013). Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325. 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, and Bentley DL (2011). Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc. Natl. Acad. Sci. USA 108, 13564–13569. 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein N, Gomes dos Santos H, Blumenthal E, and Shiekhattar R (2021). The integrator complex at the crossroad of coding and noncoding RNA. Curr. Opin. Cell Biol 70, 37–43. 10.1016/j.ceb.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, and Ahringer J (2009). Differential chromatin marking of introns and expressed exons by H3K36me3. Nat. Genet 41, 376–381. 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Damle SS, Ling KK, and Rigo F (2020). Directed RNase H cleavage of nascent transcripts causes transcription termination. Mol. Cell 77, 1032.e4. 1043.e4. 10.1016/j.molcel.2019.12.029. [DOI] [PubMed] [Google Scholar]
- Lai F, Gardini A, Zhang A, and Shiekhattar R (2015). Integrator mediates the biogenesis of enhancer RNAs. Nature 525, 399–403. 10.1038/nature14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitem C, Zaborowska J, Isa NF, Kufs J, Dienstbier M, and Murphy S (2015). CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat. Struct. Mol. Biol 22, 396–403. 10.1038/nsmb.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, and Mendell JT (2020). Antisense-mediated transcript knockdown triggers premature transcription termination. Mol. Cell 77, 1044.e3. 1054.e3. 10.1016/j.molcel.2019.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, and Stoecklin G (2014). An optimized streptavidin-binding RNA aptamer for purification of ribonucleoprotein complexes identifies novel ARE-binding proteins. Nucleic Acids Res. 42, e13. 10.1093/nar/gkt956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon GA, Vollger MR, and Eichler EE (2020). Long-read human genome sequencing and its applications. Nat. Rev. Genet 21, 597–614. 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz DA, Sathe S, Einstein JM, and Yeo GW (2020). Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26, 19–28. 10.1261/rna.072785.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Lin C, and Shilatifard A (2012). The super elongation complex (SEC) family in transcriptional control. Nat. Rev. Mol. Cell Biol 13, 543–547. 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, and Tong L (2006). Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444, 953–956. 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, and Churchman LS (2015). Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554. 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, and Cramer P (2012). CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336, 1723–1725. 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- McHugh CA, and Guttman M (2018). RAP-MS: a method to identify proteins that interact directly with a specific RNA molecule in cells. Methods Mol. Biol 1649, 473–488. 10.1007/978-1-4939-7213-5_31. [DOI] [PubMed] [Google Scholar]
- Meers MP, Henriques T, Lavender CA, McKay DJ, Strahl BD, Duronio RJ, Adelman K, and Matera AG (2017). Histone gene replacement reveals a post-transcriptional role for H3K36 in maintaining metazoan transcriptome fidelity. eLife 6, e23249. 10.7554/eLife.23249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A, and Cramer P (2004). Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature 430, 223–226. 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Melnick M, Gonzales P, Cabral J, Allen MA, Dowell RD, and Link CD (2019). Heat shock in C. elegans induces downstream of gene transcription and accumulation of double-stranded RNA. PLoS One 14, e0206715. 10.1371/journal.pone.0206715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki TS, Carl SH, and Großhans H (2017). Two distinct transcription termination modes dictated by promoters. Genes Dev. 31, 1870–1879. 10.1101/gad.301093.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, and Krug RM (1998). Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1, 991–1000. 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM (2002). On the importance of being co-transcriptional. J. Cell Sci 115, 3865–3871. 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- Nikolov DB, and Burley SK (1997). RNA polymerase II transcription initiation: a structural view. Proc. Natl. Acad. Sci. USA 94, 15–22. 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah DL, Twu KY, and Krug RM (2003). Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307, 386–395. 10.1016/s0042-6822(02)00127-7. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, and Reinberg D (1996). The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657–2683. 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Peterlin BM, and Price DH (2006). Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305. 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Petrenko N, and Struhl K (2021). Comparison of transcriptional initiation by RNA polymerase II across eukaryotic species. eLife 10, e67964. 10.7554/eLife.67964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, and Libri D (2015).Transcription termination and the control of the transcriptome: why, where and how to stop. Nat. Rev. Mol. Cell Biol 16, 190–202. 10.1038/nrm3943. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ (2011). Ending the message: poly(A) signals then and now. Genes Dev. 25, 1770–1782. 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2016). Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352, aad9926. 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, and Dye MJ (2002). Integrating mRNA processing with transcription. Cell 108, 501–512. 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, and Young RA (2010). c-Myc regulates transcriptional pause release. Cell 141, 432–445. 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahnamoun H, Lee J, Sun Z, Lu H, Ramsey KM, Komives EA, and Lauberth SM (2018). RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol 25, 687–697. 10.1038/s41594-018-0102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer KA, Mimoso CA, Adelman K, and Neugebauer KM (2021). Co-transcriptional splicing regulates 3′ end cleavage during mammalian erythropoiesis. Mol. Cell 81, 998–1012.e7. 10.1016/j.molcel.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG (1996). The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci 21, 327–335. [PubMed] [Google Scholar]
- Roeder RG (2019). 50+ years of eukaryotic transcription: an expanding universe of factors and mechanisms. Nat. Struct. Mol. Biol 26, 783–791. 10.1038/s41594-019-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Mercado NA, and Steitz JA (2022). Who let the DoGs out? – biogenesis of stress-induced readthrough transcripts. Trends Biochem. Sci 47, 206–217. 10.1016/j.tibs.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Mercado NA, Zimmer JT, Apostolidi M, Rinehart J, Simon MD, and Steitz JA (2021). Hyperosmotic stress alters the RNA polymerase II interactome and induces readthrough transcription despite widespread transcriptional repression. Mol. Cell 81, 502.e4. 513.e4. 10.1016/j.molcel.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth SJ, Heinz S, and Benner C (2020). ARTDeco: automatic readthrough transcription detection. BMC Bioinformatics 21, 214. 10.1186/s12859-020-03551-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski AJ, Erhard F, L’Hernault A, Bonfert T, Schilhabel M, Crump C, Rosenstiel P, Efstathiou S, Zimmer R, Friedel CC, and Dölken A (2015). Widespread disruption of host transcription termination in HSV-1 infection. Nat. Commun 6, 7126. 10.1038/ncomms8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, and Dekker J (2012). The long-range interaction landscape of gene promoters. Nature 489, 109–113. 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AC, and Taatjes DJ (2020). Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34, 465–488. 10.1101/gad.335679.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Maqbool MA, Yahia Y, El Aabidine AZ, Esnault C, Forné I, Decker TM, Martin D, Schüller R, Krebs S, et al. (2018). Tyrosine-1 of RNA polymerase II CTD controls global termination of gene transcription in mammals. Mol. Cell 69, 48.e6. 61.e6. 10.1016/j.molcel.2017.12.009. [DOI] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Lindquist S, and Burge CB (2014). Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 7, 1362–1370. 10.1016/j.celrep.2014.04.044. [DOI] [PubMed] [Google Scholar]
- Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, and Lou H (2014). Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc. Natl. Acad. Sci. USA 111, E4920–E4928. 10.1073/pnas.1408964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ, and Manley JL (2000). The ends of the affair: capping and polyadenylation. Nat. Struct. Biol 7, 838–842. 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, and Egan JB (2005). Transcriptional interference–a crash course. Trends Genet. 21, 339–345. 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, and Vakoc CR (2014). The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol. Cell 54, 728–736. 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ 3rd, Belotserkovskaya R, and Reinberg D (2004). Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18, 2437–2468. 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Tian B, and Manley JL (2013). Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci 38, 312–320. 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. (2011). Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542. 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, and Steitz JA (2015). Widespread inducible transcription downstream of human genes. Mol. Cell 59, 449–461. 10.1016/j.molcel.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Sabath N, Wiesel Y, Nathans J, Levy-Adam F, Yario TA, Steitz JA, and Shalgi R (2017). Comparative analysis reveals genomic features of stress-induced transcriptional readthrough. Proc. Natl. Acad. Sci. USA 114, E8362–E8371. 10.1073/pnas.1711120114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, and Steitz JA (2017). Readthrough transcription: how are DoGs made and what do they do? RnA Biol. 14, 632–636. 10.1080/15476286.2016.1149680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hennig T, Whisnant AW, Erhard F, Prusty BK, Friedel CC, Forouzmand E, Hu W, Erber L, Chen Y, et al. (2020). Herpes simplex virus blocks host transcription termination via the bimodal activities of ICP27. Nat. Commun 11, 293. 10.1038/s41467-019-14109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, and Corden JL (1995). Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140, 1223–1233. 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel Y, Sabath N, and Shalgi R (2018). DoGFinder: a software for the discovery and quantification of readthrough transcripts from RNA-seq. BMC Genomics 19, 597. 10.1186/s12864-018-4983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, et al. (2017). BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol. Cell 67, 5.e19. 18.e19. 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink EM, Vihervaara A, Tippens ND, and Lis JT (2019). Nascent RNA analyses: tracking transcription and its regulation. Nat. Rev. Genet 20, 705–723. 10.1038/s41576-019-0159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JHN, Chen R, He N, Jang MK, Ozato K, and Zhou Q (2005). Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545. 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Yue Y, Liu J, and He C (2015). RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355. 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska J, Egloff S, and Murphy S (2016).The Pol II CTD: new twists in the tail. Nat. Struct. Mol. Biol 23, 771–777. 10.1038/nsmb.3285. [DOI] [PubMed] [Google Scholar]
- Zhang H, Rigo F, and Martinson HG (2015). Poly(A) signal-dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the poly(A) site. Mol. Cell 59, 437–448. 10.1016/j.molcel.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li T, and Price DH (2012). RNA polymerase II elongation control. Annu. Rev. Biochem 81, 119–143. 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]