Abstract
Eukaryotic initiation factor 2B, eIF2B is a guanine nucleotide exchange, factor with a central role in coordinating the initiation of translation. During stress and disease, the activity of eIF2B is inhibited via the phosphorylation of its substrate eIF2 (p-eIF2α). A number of different kinases respond to various stresses leading to the phosphorylation of the alpha subunit of eIF2, and collectively this regulation is known as the integrated stress response, ISR. This targeting of eIF2B allows the cell to regulate protein synthesis and reprogramme gene expression to restore homeostasis. Advances within structural biology have furthered our understanding of how eIF2B interacts with eIF2 in both the productive GEF active form and the non-productive eIF2α phosphorylated form. Here, current knowledge of the role of eIF2B in the ISR is discussed within the context of normal and disease states focusing particularly on diseases such as vanishing white matter disease (VWMD) and permanent neonatal diabetes mellitus (PNDM), which are directly linked to mutations in eIF2B. The role of eIF2B in synaptic plasticity and memory formation is also discussed. In addition, the cellular localisation of eIF2B is reviewed and considered along with the role of additional in vivo eIF2B binding factors and protein modifications that may play a role in modulating eIF2B activity during health and disease.
Keywords: cellular targeting, eukaryotic gene expression, Protein synthesis, stress response, Translational Control
Introduction
Eukaryotic cells are constantly exposed to a multitude of stress conditions, and the capacity to respond and adapt to this ever-changing environment is a driving factor behind our evolutionary success. The fundamental response to stress requires a rapid alteration of gene expression, to promote cell survival. However, if the stress is sustained or the stress cannot be overcome, this alteration of gene expression can induce cell death. Although transcriptional responses are crucial for controlling changes in gene expression, regulation at the translational level often allows for a faster response which permits immediate adaptation.
Translation can be divided into three main stages: initiation, elongation and termination. While regulation occurs at all stages of translation, the initiation stage is generally regarded as the rate limiting step and thus critical for ensuring efficient protein synthesis. This regulation is tightly controlled via the activity of the eukaryotic initiation factor 2B (eIF2B), which coordinates the ability of cells to maintain cellular proteostasis during both health and disease. This targeting of eIF2B is controlled via the coordinated activation of a series of signalling cascades collectively known as the integrated stress response, ISR. Dysregulation of the ISR pathway has been implicated in the pathological mechanisms of a broad spectrum of clinical conditions. This article focuses on the role of eIF2B in a number of these conditions, for a wider review the authors direct the readers to the excellent review by Costa-Mattioli & Walter [1]. The central role of eIF2B in synaptic plasticity and cognitive function has been elucidated by many research groups [2–4]. The development of the eIF2B GEF activity enhancers ISRIB and 2BAct [5,6] has shown the therapeutic benefit of the modulation of eIF2B activity in a range of cognitive disorders, as discussed below. Loss-of-function mutations in the EIF2B1-5 genes are associated with the rare, frequently fatal neurological disorder, leukoencephalopathy with vanishing white matter (VWMD) [7]. In this review, we discuss how VWMD mutations within eIF2B lead to phenotypic changes in specific cell types. We also review studies of cellular localisation of eIF2B subunits, an area which may further functional characterisation of pathogenic VWMD missense mutations. Finally, we discuss the role of eIF2B in the pathogenic mechanism of permanent neonatal diabetes mellitus (PNDM), a heterogeneous group of conditions diagnosed before the age of 6 months.
eIF2 and eIF2B in translation
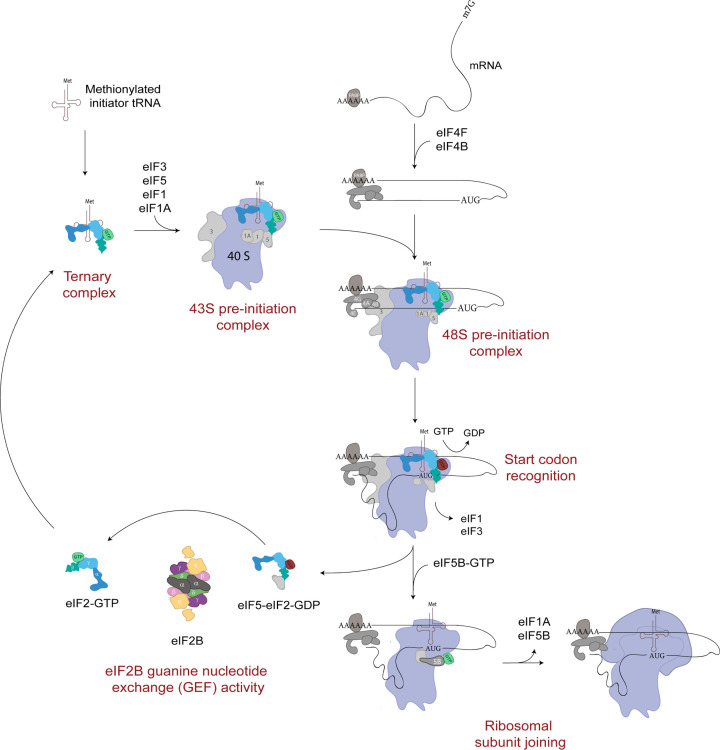
Central to the regulation of translation initiation is the availability of sufficient levels of Met-tRNAi, which is delivered to the 43S preinitiation complex (PIC) through interaction with the GTP-binding protein eukaryotic initiation factor, eIF2 to form the eIF2-GTP.Met-tRNAi ternary complex, (TC) (Figure 1). A detailed review of translation initiation is out of the scope of this review and the authors direct the readers to the following review articles [8,9]. In addition to the TC, the 43S PIC contains the small ribosomal subunit (40S) bound by the initiation factors eIF1, eIF1A and eIF3 (Figure 1) [9]. The 43S PIC scans the mRNA until a start codon is detected forming the 48S PIC. At this timeeIF2-GTP is hydrolysed via the activity of the GTP-activating protein, eIF5 and the inactive eIF2-GDP, which has a much lower affinity for Met-tRNAi is released still associated with eIF5 (Figure 1) [9,10]. Key to the continued success of translation initiation is the recycling of the released eIF2-GDP to active eIF2-GTP and the recruitment of Met-tRNAi to form new TC. This recycling role is carried out by the guanine nucleotide exchange factor, eIF2B (Figure 1). As eIF2-GDP is released from the 48S PIC as an eIF2-GDP/eIF5 complex this enables eIF5 to function as a GDP dissociation inhibitor (GDI) inhibiting any spontaneous exchange activity [11]. In yeast, eIF2B has also been shown to act as a GDI displacement factor (GDF) to release eIF2-GDP from eIF5 [12,13]. The exact mechanism for how new active TC is formed is still unclear however recent evidence suggests that a complex interplay between eIF2B, eIF5 and Met-tRNAi can influence the stability of TC (Figure 1) [14].
Figure 1. Translation initiation pathway.
A ternary complex formed of eIF2-GTP and a methionylated initiator tRNA is recruited to the 40S ribosomal subunit by various eIFs to form a 43 S PIC. Facilitated by other eIFs, the 43 S PIC is loaded onto a target mRNA molecule and scans the mRNA sequence for a start codon. Upon recognition of a start codon, eIF2-GTP is hydrolysed and released in complex with eIF5. eIF5B accommodates the binding of the 60S ribosomal subunit to the 40S subunit forming the elongation ready 80S ribosome. eIF2- GDP-eIF5 is recycled to eIF2-GTP by eIF2B.
While the eIF2-GTP is critical for recruiting Met-tRNAi to the start codon, alternative factors, such as eIF2A have been identified which can also promote this recruitment albeit when eIF2 levels are low and can result in targeted translation of specific mRNAs. A detailed analysis of the role of eIF2A and other alternative factors is outside the scope of this review, but the authors direct the readers to an detailed review on the topic [15].
eIF2B exhibits a greater level of complexity within its quaternary structure. It is composed of five nonidentical subunits, encoded by the genes EIF2B1–5 and termed eIF2Bα through to ε, respectively (Figure 2Ai). The γ and ε subunits catalyse the guanine nucleotide exchange activity of eIF2B, whereas the α, β and δ subunits are required to regulate this activity in response to various cellular signals [16–19]. While eIF2Bɛ on its own can carry out exchange activity (the C-terminal domain of eIF2Bɛ facilitates binding of eIF2 while the HEAT domain can catalyse eIF2 nucleotide exchange), the rate of this exchange is greatly enhanced through joining of the other eIF2B subunits [20].
The eIF2B regulatory subunits are responsible for modulating levels of eIF2B activity, dependent on the cellular environment. In response to conditions of cellular stress, specific eIF2 kinases (discussed in more detail later) phosphorylate the α subunit of eIF2 (p-eIF2α), converting eIF2 from a substrate to a competitive inhibitor of eIF2B GEF activity [21,22]. eIF2Bα in particular is required to facilitate this inhibition however detailed mutational analysis of eIF2Bβ and eIF2Bδ highlights that these subunits also contribute [17,23–29]. Precise expression levels of the eIF2B subunits within the cell is crucial for tight control of eIF2B activity [30]. Wortham et al. demonstrated that stable expression of eIF2Bε requires stoichiometric expression of eIF2Bγ and that stable expression of eIF2Bδ requires stoichiometric expression of eIF2Bβ. In the absence of co-stoichiometric expression eIF2Bε and δ subunits are ubiquitinated, targeting them for ubiquitin-proteasome degradation.
Prior to 2014, eIF2B was believed to be a pentameric complex comprises one copy of each of its subunits; however, mass spectrometry analysis has revealed that eIF2B exists as a decameric complex, comprises two copies of each of its subunits [31,32]. The first crystal structure of eIF2B was solved from Schizosaccharomyces pombe and revealed a central core composed of two copies of each of the regulatory subunits (α, β and δ), flanked at opposite sides by a heterodimer of the catalytic subunits (γ and ε) (Figure 2Ai) [33]. It is believed that these decameric eIF2B complexes form through the stabilisation of two eIF2B(βδγε) tetramers with one eIF2Bα homodimer [32–34].
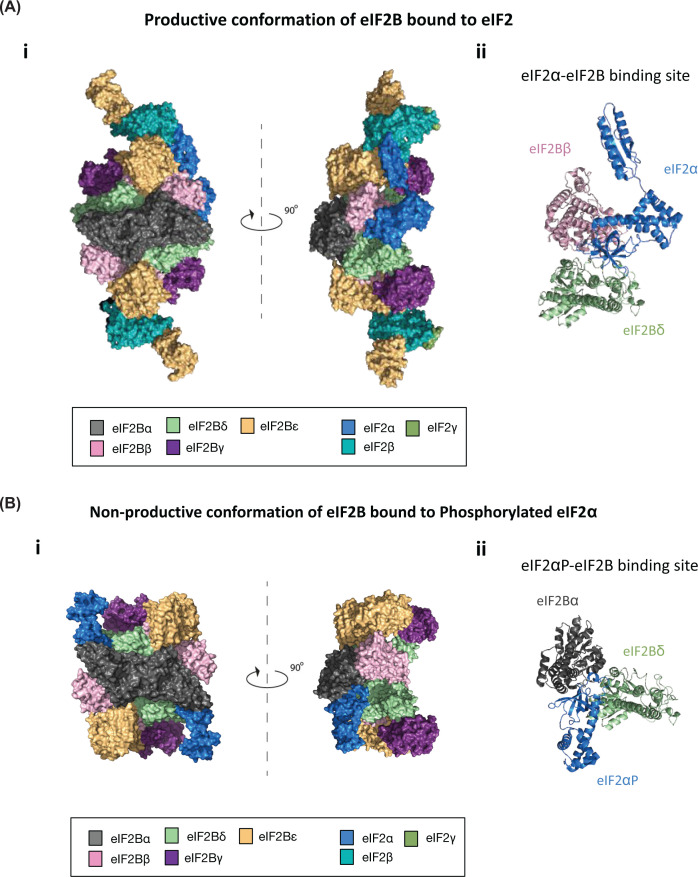
A number of cryo EM structures for mammalian and yeast eIF2B have now been solved together with the substrate eIF2 and the phosphorylated form of eIF2α [35–41]. These structures have provided detailed observations of structural rearrangements that take place through the binding of p-eIF2α to eIF2B (Figure 2A,B). In its productive or apostate conformation, eIF2γ (which contains the GTP binding site) binds primarily to eIF2Bε, and the α subunit of eIF2 makes contact with the regulatory subunits eIF2Bβ and δ (Figure 2Aii) [35–40]. However, when eIF2α is phosphorylated, structural changes are observed which are critical for the inhibition of eIF2B activity. The N-terminal domain of p-eIF2α now makes contact between the eIF2Bα and eIF2Bδ subunits which induces a conformational change and inhibits the interaction of eIF2γ with the eIF2Bε subunit (Figure 2Bii). This inhibitory structural rearrangement has been termed the non-productive conformation (Figure 2B).
Figure 2. CryoEM structures resolved for eIF2B bound to non-phosphorylated or phosphorylated eIF2.
(A) Orthogonal views of (i) decameric eIF2B in its productive conformation bound to a non-phosphorylated eIF2 heterotrimer shown in surface representation, (ii) with a cartoon representation of the eIF2α-eIF2B binding site (PDB 6O81 [27]). (B) Orthogonal views of (i) decameric eIF2B in its non-productive conformation bound to two phosphorylated eIF2α subunits shown in surface representation, (ii) with a cartoon representation of the phosphorylated eIF2α-eIF2B binding site (PDB 6O9Z [27]).
Detailed reviews of these structures have been previously published and the authors direct the readers to these [42,43].
eIF2B and the integrated stress response
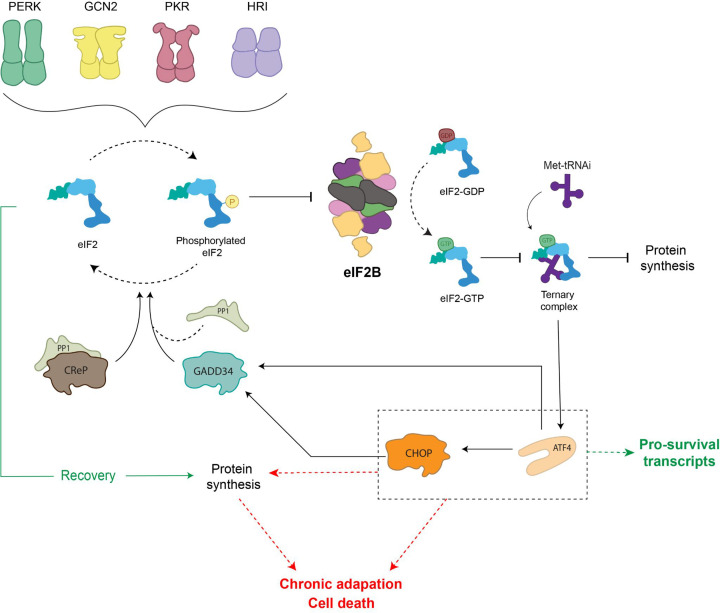
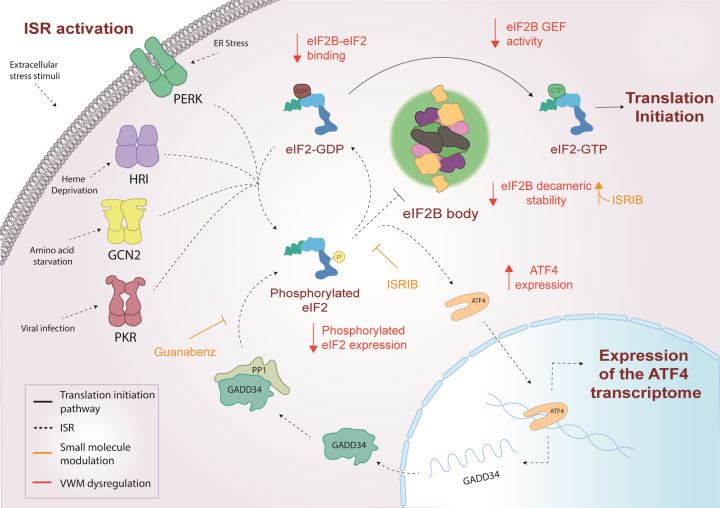
As mentioned above, the eIF2Bα, β and δ subunits are required to regulate eIF2B activity in response to various cellular signals via recognising and interacting with p-eIF2α, thereby causing structural rearrangements of eIF2B which inhibit its GEF activity. This phosphorylation event is carried out by four eIF2α kinases which when activated via specific stresses, phosphorylate the same single serine residue (S51) of eIF2α (Figure 3) [44]. Collectively, the function of these stress-sensing kinases and the subsequent effect on eIF2 and eIF2B activity is known as the integrated stress response, ISR [45,46]. These kinases are termed: GCN2 (general amino acid control nonderepressible 2), PKR (protein kinase RNA-like), PERK (protein kinase RNA-like endoplasmic reticulum kinase) and HRI (heme-regulated inhibitor) (Figure 3). Recently, it has been suggested that there may be a fifth kinase (MARK2) which can respond to proteotoxic stress and phosphorylate eIF2α [47]. While the eIF2α kinases share extensive homology in their catalytic domains (a dimerisation interface, crucial for kinase activation and catalytic function), each kinase has an unique regulatory domain which allows for activation of the ISR by a range of cellular stresses [48–52]. GCN2 is activated in response to amino acid deprivation in order to reduce the cellular demand for amino acids and is the only kinase conserved from yeast to mammalian cells [53]. PKR was thought to be activated primarily by double-stranded RNA (dsRNA) during viral infection; however, it can also be activated in response to oxidative stress, serum deprivation and more recently has been associated with the activation of proinflammatory signalling in response to pathogens [54–55]. Interestingly, while dsRNA can activate PKR directly, the activation of PKR by other stresses relies on its interaction with the protein activator of PKR (PACT) 1 [56]. PERK is principally activated in response to endoplasmic reticulum (ER) stress, commonly caused by the accumulation of unfolded proteins in the ER. PERK activation alleviates this stress by decreasing the level of proteins localised to the ER [57,58]. HRI while originally thought to protect erythroid cell against toxic globin aggregates, it can also be activated in non-erythroid cells in response to arsenite induced oxidative stress and more recently has been shown to play an important role in protecting cells from proteotoxicity [59–61].
Figure 3. Activation of the ISR pathway.
In response to various cellular stress stimuli eIF2α kinase molecules are activated through dimerisation. eIF2α kinases phosphorylate the α subunit of eIF2. In its phosphorylated form, eIF2 is a competitive inhibitor of eIF2B activity preventing replenishment of eIF2-GTP within the cell. This leads to inhibition of global protein synthesis while the translation of specific stress response mRNAs, including ATF4, is up-regulated. During episodes of acute ISR, ISR effectors are able to restore homeostasis and ATF4-mediated activation of CHOP induces the transcription of GADD34 to promote dephosphorylation of eIF2α. In cases where ISR effectors are unable to restore homeostasis, the cell transitions into a chronically activated ISR. Protein synthesis is restored via an eIF2B independent mechanism and ATF4-mediated activation of CHOP promotes proapoptotic gene expression.
The phosphorylation of eIF2α caused by these kinases converts eIF2 into a competitive inhibitor of eIF2B activity [62,63]. This results in a depleted level of eIF2-GTP within the cell, thereby decreasing levels of ternary complex which leads to a global translational attenuation of protein synthesis (Figure 3). In a recent review, the molar concentrations of a number of translation initiation factors from both yeast and HeLa cells were collated [9]. Here, it was shown that cells have approximately 3- to 5-fold less eIF2B complexes than eIF2; this highlights the significance of regulating eIF2B activity as even a minimal level of p-eIF2α can decrease eIF2B activity.
Paradoxically, during this time the translation of a number of stress responsive proteins is upregulated. The translation of these mRNAs is most commonly regulated by the presence of upstream open reading frames (uORFs) in their 5' UTR [64]. uORFs are generally inhibitory for the translation of a mRNA transcript under normal cellular conditions; however, during episodes of cellular stress, they can promote the translation. Key to the ISR regulation is the mRNA for the transcription factor, Activating Transcription Factor 4 (ATF4). ATF4 mRNA is ubiquitously expressed; however, under normal cellular conditions protein levels are low. ATF4 mRNA contains two uORFs [64]. Under normal cellular conditions, the first uORF which encodes a short polypeptide (three amino acids) is translated. This up-stream uORF acts as a positive element allowing ribosomes to scan and initiate translation at the next uORF. The second uORF sequence (59 amino acids) overlaps with the coding sequence of ATF4 in an out-of-frame manner, and, therefore, the translation of uORF2 inhibits the translation of ATF4 [64]. However during stress when eIF2B activity in inhibited and TC levels are low the rate of re-initiation at the second uORF decreases thereby increasing re-initiation of the ATF4 ORF. Increased ATF4 expression can activate prosurvival mechanisms within the cell through a number of different pathways. This mechanism is highly conserved and in yeast the transcription factor Gcn4p has 4 uORFs which regulate its expression upon ISR activation [65]. More detailed reviews of uORFs and their role in translational control are available [66].
If ISR signalling leads to the restoration of cellular homeostasis, ATF4-mediated activation of the transcription factor C/EBP homologous protein (CHOP) can contribute to the restoration of global translation. CHOP induces the transcription of growth arrest and DNA damage inducible protein (GADD34), an eIF2α phosphatase regulatory subunit which binds to PP1c to allow the regulated dephosphorylation of eIF2α (Figure 3) [67]. In cases of severe cellular stress where the pro-survival mechanisms induced by the ISR are unable to restore homeostasis, the ISR promotes programmed cell death signalling (Figure 3). As the ISR provides a central network for maintaining cellular homeostasis, dysregulation of ISR signalling has numerous pathological consequences and has been linked to conditions such as: cancer, diabetes, cardiovascular disease and neurodegeneration [1]. Thus, pharmacological modulation of the ISR has become a key area for therapeutic research. A detailed appreciation of these targets and therapeutics has been reviewed elsewhere [46,68], so only a few will be highlighted and discussed in this review.
Of particular interest is the small molecule known as Integrated Stress Response InhiBitor (ISRIB) [69]. ISRIB is a direct activator of eIF2B and was originally identified following a screen of compounds that could reduce the ISR [71]. Recent analysis of ISRIB binding to eIF2B and its competitive relationship with p-eIF2α has provided insight into the mode of action of ISRIB [37,38]. ISRIB binding has been shown to favour stabilisation of the productive form of eIF2B thereby making eIF2B relatively resistant to the inhibitory effects of p-eIF2α [37,38]. ISRIB does not affect the level of p-eIF2α but rather through interactions with eIF2Bβ and δ subunits; ISRIB promotes stabilisation of two tetramers to form an eIF2B(βδγɛ)2 octamer that favours eIF2Bα homodimer binding thus promoting decameric formation and enhancing eIF2B GEF activity [71,34–37]. Interestingly, ISRIB treatment has been shown to enhance cognition in aged mice while also reversing cognitive deterioration associated with neurodegeneration and traumatic brain injury [72–77]. A number of derivatives of ISRIB have been produced which show similar effects of cognitive enhancement and are currently in phase 1 clinical trials [77,79].
eIF2B in acute and chronic activation of the integrated stress response
It has been commonly reported that an acute and transient activation of the ISR prompts a global reduction of translation and induction of genes involved in supporting cellular recovery to regain homeostasis. In contrast, transition to a chronically activated ISR is widely reported as adaptive to prolonged stress, ultimately pro-apoptotic when cells are unable to overcome sustained stress with pathological consequences [80–82]. One of the best studied mechanisms of ISR-induced cell death involves ATF4-mediated activation of and interaction with CHOP [83–86]. CHOP has been shown to induce apoptosis via a number of mechanisms including the repression of anti-apoptotic proteins [87] and up-regulation of death receptors [88]. Hence, ATF4 and CHOP have intricate roles in the ISR and function to produce tailored responses, both pro- and anti-survival dependent on the cellular stress stimuli (Figure 3).
The role of eIF2B during the acute phase of the ISR has been defined and reviewed elsewhere [46]. eIF2B activity is inhibited to prevent ternary complex formation and allow ATF4-induced reprogramming, followed by restoration of eIF2B activity and protein synthesis hence hallmarking the termination of the acute ISR. However, its specific role during chronic stress is still unclear. Interestingly, as a cell transitions into a chronically activated ISR, it has been suggested that the restoration of protein synthesis does not require the recovery of eIF2B activity (Figure 3) [89]. Guan et al. suggest that a sustained reduction in eIF2B activity during chronic ER stress results in transcriptional and translational remodelling which is dependent on the activation of PERK and mediated by eIF3 [89]. In addition, pharmacological activation of eIF2B in chronic ER-stressed human fibroblast cells synergistically reduced viability by triggering caspase-3/7-mediated apoptosis [70], suggesting that the cellular response to eIF2B activation is dependent on the level and duration of ER stress.
Cellular adaptation to stress may correlate with levels of p-eIF2α and cell type-specific demand of global protein synthesis. Secretory cells, such as pancreatic β-cells, require basal p-eIF2α to prevent oxidative damage and rapid unburden of ER load [90–92]. Increased levels of p-eIF2α, hence sustained eIF2B inhibition, sensitises β-cells to CHOP-mediated apoptosis [93]. In contrast, hypo-phosphorylation of eIF2α renders β-cells with uncontrolled translation and deregulated antioxidant levels, highlighting the narrow threshold of eIF2α modulation and eIF2B inhibition [94–96]. Therefore, the cellular control of eIF2B activity in the transition from acute to chronic stress is critical and allows them to adapt and survive the shift from a protective short-lived ISR to adaptation to chronic stress.
eIF2B bodies and cellular localisation
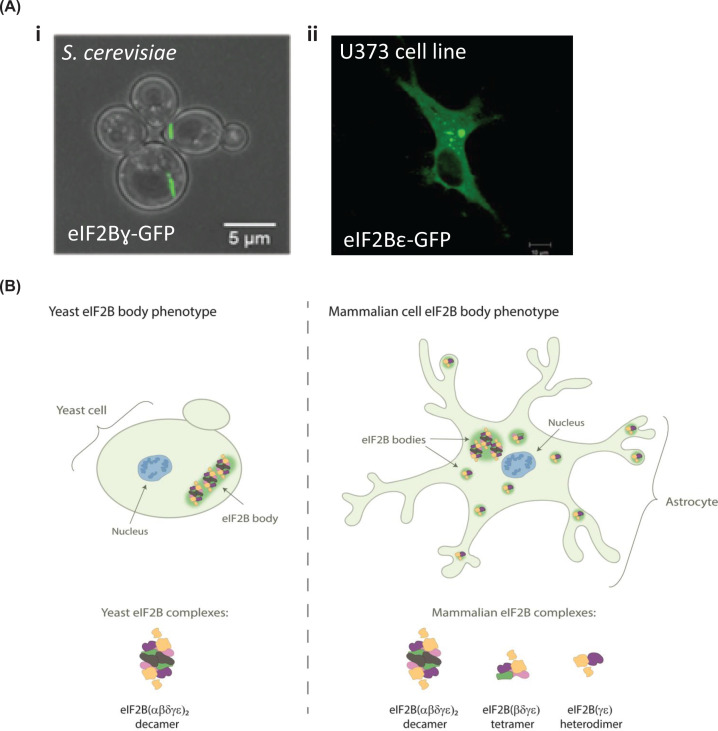
The cellular localisation of eIF2B has gained interest over the past two decades. In 2005, the Ashe group first identified cytoplasmic accumulations of eIF2B subunits in Saccharomyces cerevisiae (S. cerevisiae) and termed them eIF2B bodies [97]. Phenotypically each yeast was found to harbour one eIF2B body that formed a large filament shaped structure (Figure 4). These eIF2B bodies were distinct from accumulations of mRNA and other translation initiation factors, with the exception of the three subunits of eIF2. Given eIF2B’s role as a GEF for eIF2, eIF2B bodies were hypothesised to be sites of eIF2B GEF activity, required for translation initiation. Indeed, eIF2 was shown to dynamically localise to the eIF2B body, in a manner that correlated to eIF2B GEF activity within the cell [97]. A later study revealed that S. cerevisiae eIF2B bodies are motile within the cytosol and that inhibition of this motility is associated with the inhibition of translation [98]. More recently, deletion of eIF2Bα in yeast has been shown to disperse eIF2B bodies, suggesting that the decameric conformation of eIF2B is required for eIF2B body stability [99]. Interestingly, an eIF2BαV184D point mutation that does not destabilise the decameric conformation of eIF2B [100,33] and disrupts eIF2B body integrity in yeast [99]. In eIF2BαV184D mutant yeast, eIF2B localises to multiple small punctate foci, which have been termed microfoci. These microfoci were also observed in the presence of eIF2Bα gcd− (gcn2 depressible) mutations, associating catalytic impairment and eIF2B body stability. Through combining SEM imaging and a subtomogram-averaging approach, Marini et al. have obtained a 3D model of eIF2B bodies in yeast upon energy depletion [101]. Comparing the volume of the eIF2B decamer, the authors hypothesise that eIF2B bodies are repeats of decameric units, stacked through contacts between eIF2Bε subunits of adjacent decamers. Further investigation into the stability and structural arrangement of eIF2B bodies may provide a greater insight into their function.
Figure 4. eIF2B bodies in yeast and mammalian cells.
(A) Confocal images of eIF2B bodies in (i) S. cerevisiae strain yMK880 (eIF2Bγ-GFP) (ii) a human U373 astrocytoma cell overexpressing eIF2Bε-GFP. (B) Schematic representation of decameric eIF2B(αβδγε)2 and eIF2B(βδγε) tetrameric and eIF2B(γε) heterodimeric subcomplexes that have been identified in yeast and mammalian cells localising to eIF2B bodies.
The physiological relevance of eIF2B body formation within yeast has been debated. A number of studies have demonstrated that eIF2B bodies form more readily under conditions of glucose starvation and cytoplasmic acidification [102–105]. These are conditions that promote translational inhibition and thus, as eIF2B bodies appear to be sites of GEF activity, an increased formation under these conditions appears counterintuitive. Norris et al. hypothesised that as different yeast strains respond differently to environmental stresses, eIF2B body formation may also vary between yeast strains [99]. In support of this it was found that, the percentage of cells with eIF2B bodies in two different strains of S. cerevisiae varied significantly under steady state growth. The percentage of cells with eIF2B bodies increased within both strains following amino acid starvation, but rather unexpectedly an increase was not observed during glucose starvation. Additionally, it may be interesting to speculate whether differences may also be explained by variations in molar concentrations of the eIF2 and eIF2B protein complexes across the different yeast strains which could influence the formation of eIF2B bodies [9,106]. Further investigation into strain specific stress responses and concentrations of eIF2 and eIF2B subunits may provide a greater insight into the driving force behind eIF2B body formation.
Although the mechanism that promotes eIF2B body formation in yeast remains unclear, in mammalian cells eIF2B bodies have been shown to form under normal culture conditions. Hodgson et al. showed endogenously expressed eIF2B and eIF2 subunits localise to discrete cytoplasmic foci [107]. Fluorescently tagged proteins were used to analyse the dynamics between eIF2B subunits and eIF2α within these foci. eIF2B was found to be a stable component, whereas eIF2 is mobile; shuttling through the body in a manner that correlates with cellular eIF2B GEF activity. Interestingly, the composition and function of these eIF2B bodies appears to be regulated by the ISR. Unlike in yeast where a single eIF2B body exists, mammalian cells contain a number of eIF2B bodies that vary in size and eIF2B subunit-makeup (Figure 4). A high degree of co-localisation for all five eIF2B subunits was observed in larger eIF2B bodies. As expected, upon acute ISR activation, the association of p-eIF2α with these bodies was increased and the movement of eIF2 decreased, suggesting impaired GEF activity. Regulatory eIF2B subunits were rarely observed to co-localise with smaller eIF2B bodies (made of predominantly γε subunits), and thus it was hypothesised that ISR induction would not impact on the GEF activity of these bodies (Figure 4). Intriguingly, the movement of eIF2 through these smaller bodies was found to increase upon ISR stimulation, suggesting that eIF2B GEF activity is increased. This was accompanied by an increased degree of co-localisation with eIF2Bδ, suggesting that novel eIF2B(δγε) subcomplexes potential form following ISR activation. These data suggest that pools of eIF2B subcomplexes exist in mammalian cells and may have a role in mediating cellular stress responses. Subcomplexes of eIF2B have previously been shown in mammalian cells through native mass spectrometry (MS) [32]. eIF2B(βδγε) tetramers, eIF2B(γε) heterodimers and eIF2B(α2) homodimers were identified but not eIF2B(δγε) subcomplexes. Previous studies have shown eIF2Bβ and δ subunits can heterodimerise through C-terminal domain interactions [108,109]. Furthermore, eIF2Bβ is required for stable expression of eIF2Bδ [30] and thus the observed co-localisation of eIF2Bδ, but not eIF2Bβ with small eIF2B bodies in stressed cells is rather intriguing. Although Wortham et al. did not identify eIF2B(δγε) subcomplexes by native MS [32], application of high collision energy to disrupt the eIF2B(βδγε) tetramer led to the dissociation of eIF2Bβ but not eIF2Bδ from the complex, suggesting that eIF2Bδ can interact with eIF2Bγε in the absence of eIF2Bβ. Native MS analysis of eIF2B complexes during ISR stimulation could provide further insight into the existence of an eIF2B(δγε) subcomplex.
eIF2B post-translational modifications and interacting molecules
Post-translational modifications (PTMs) have been shown to influence eIF2B activity. Phosphorylation of eIF2Bε is crucial for eIF2B function [110]. Glycogen synthase kinase-3 (GSK3) catalyses phosphorylation of eIF2Bε at Ser535 and limits eIF2B activity [111]. Inhibition of GSK3 activity by insulin leads to Ser535 dephosphorylation and eIF2B activation. Two other phosphorylation sites at the C-terminus of eIF2Bε facilitate eIF2 binding [112]. Similarly, another study highlighted increased phosphorylation sites in yeast catalytic eIF2Bγ- and ε-, where the latter subunit showed phosphorylation site clustering located in the highly unstructured flexible loop connecting the ε/γ dimer [113]. eIF2 relies on PTM-mediated adaptable conformational changes according to its functional role [113]. Like eIF2, the subunits of eIF2B could use PTMs to switch between productive and non-productive conformations towards eIF2 binding, however the structural impact and the functional role of these PTMs in eIF2B activity remains largely unknown. Novel acetylation sites have also been identified in eIF2B. Acetylation has been broadly linked to the regulation of phosphorylation susceptibility of large protein complexes [114]. Interestingly, eIF2Bε remains largely absent of acetylation sites, whereas the eIF2B regulatory subunits reveal a remarkably conserved N-terminal acetylation site [113]. Such sites may have a role in stabilising complex formation as N-terminal acetylation prevents protein degradation [115]. On the contrary, N-terminally acetylated proteins can also be directed towards degradation [116]. Wortham et al. (2016) reported stoichiometric expression of eIF2B subunits is mediated by ubiquitin-mediated degradation [30]. A further understanding of the significance of these PTMs sites could uncover new regulation layers of eIF2B function.
In addition to PTMs, molecular interactions with eIF2B are critical for its function. eIF2α phosphorylation status defines the affinity fate of eIF2 to eIF2B [117] while eIF2-GDP is complexed with eIF5, to regulate GEF activity [11]. More recently, it has been shown that the binding of GTP to eIF2Bγ not only serves to provide a pool of available GTP but also enhances GEF activity [118]. Natural sugar metabolites bind to eIF2Bα2 dimers and promote eIF2B decameric formation [119]. In yeast, mutations in these sugar phosphate binding sites of eIF2Bα have been shown to disperse eIF2B bodies [100]. Viral proteins counteract translational shutdown by binding to host eIF2B and induce its productive state independently of p-eIF2α [120,121]. While some additional protein factors have been shown to bind to eIF2B [122,123], the contribution of other binding partners to eIF2B activity has not received much attention. Analysis of published data of protein–protein interactors of each eIF2B subunit curated by BioGRID (release 4.4.201) [124] is outlined in Table 1. The list of identified genes was annotated with respective slim GO terms through GOnet (update January 2020) [125]. This analysis has shown that there are many potential interactors of eIF2B, and these seem to share similar gene functions which go far beyond the scope of translation (Table 1). Future investigation of these interactors could provide new avenues of investigation for novel roles of eIF2B.
Table 1. Biological processes of eIF2B protein–protein interactors.
| Go term ID | GO term annotation | Number of genes involved per eIF2B subunit | ||||
|---|---|---|---|---|---|---|
| eIF2Bα | eIF2Bβ | eIF2Bγ | eIF2Bδ | eIF2Bε | ||
| GO:0007165 | Signal transduction | 40 | 37 | 39 | 48 | 46 |
| GO:0006810 | Transport | 34 | 38 | 31 | 30 | 43 |
| GO:0048856 | Anatomical structure development | 40 | 42 | 37 | 43 | 42 |
| GO:0002376 | Immune system process | 28 | 28 | 25 | 27 | 34 |
| GO:0006950 | Response to stress | 35 | 26 | 36 | 28 | 33 |
| GO:0030154 | Cell differentiation | 35 | 30 | 24 | 33 | 30 |
| GO:0034641 | Cellular nitrogen compound metabolic process | 33 | 41 | 32 | 28 | 27 |
| GO:0009058 | Biosynthetic process | 23 | 33 | 24 | 20 | 27 |
| GO:0022607 | Cellular component assembly | 14 | 22 | 17 | 16 | 25 |
| GO:0042592 | Homeostatic process | 16 | 20 | 17 | 18 | 24 |
| GO:0006464 | Cellular protein modification process | 18 | 22 | 17 | 19 | 23 |
| GO:0040011 | Locomotion | 12 | 14 | 10 | 14 | 21 |
| GO:0015031 | Protein transport | 20 | 18 | 12 | 13 | 19 |
| GO:0016192 | Vesicle-mediated transport | 13 | 16 | 11 | 11 | 19 |
| GO:0065003 | Protein-containing complex assembly | 11 | 16 | 13 | 11 | 18 |
| GO:0009056 | Catabolic process | 17 | 17 | 10 | 11 | 16 |
| GO:0048870 | Cell motility | 9 | 10 | 8 | 11 | 16 |
| GO:0007155 | Cell adhesion | 7 | 11 | 8 | 12 | 15 |
| GO:0007267 | Cell–cell signalling | 11 | 11 | 11 | 12 | 15 |
| GO:0008219 | Cell death | 9 | 9 | 9 | 9 | 15 |
| GO:0044281 | Small molecule metabolic process | 6 | 12 | 12 | 8 | 14 |
| GO:0055085 | Transmembrane transport | 8 | 11 | 9 | 9 | 13 |
| GO:0044403 | Symbiotic process | 16 | 12 | 11 | 10 | 12 |
| GO:0061024 | Membrane organisation | 11 | 13 | 11 | 9 | 12 |
| GO:0006412 | Translation | 13 | 14 | 9 | 7 | 11 |
| GO:0006629 | Lipid metabolic process | 6 | 8 | 6 | 5 | 11 |
| GO:0000003 | Reproduction | 15 | 13 | 12 | 10 | 10 |
| GO:0007049 | Cell cycle | 8 | 7 | 6 | 4 | 10 |
| GO:0008283 | Cell population proliferation | 4 | 7 | 7 | 6 | 9 |
| GO:0050877 | Nervous system process | 5 | 6 | 7 | 6 | 9 |
| GO:0007010 | Cytoskeleton organisation | 2 | 2 | 4 | 4 | 9 |
| GO:0051276 | Chromosome organisation | 6 | 11 | 10 | 9 | 8 |
| GO:0051301 | Cell division | 4 | 2 | 3 | 2 | 8 |
| GO:0000902 | Cell morphogenesis | 7 | 10 | 4 | 7 | 7 |
| GO:0006259 | DNA metabolic process | 4 | 8 | 7 | 6 | 7 |
| GO:0000278 | Mitotic cell cycle | 4 | 2 | 4 | 2 | 6 |
| GO:0007568 | Aging | 4 | 4 | 3 | 4 | 5 |
| GO:0006914 | Autophagy | 5 | 5 | 4 | 3 | 5 |
| GO:0003013 | Circulatory system process | 6 | 4 | 5 | 5 | 4 |
| GO:0034655 | Nucleobase-containing compound catabolic process | 5 | 7 | 2 | 5 | 4 |
| GO:0022618 | Ribonucleoprotein complex assembly | 3 | 6 | 4 | 2 | 4 |
| GO:0007034 | Vacuolar transport | 3 | 1 | 2 | 1 | 4 |
| GO:0007005 | Mitochondrion organisation | 3 | 2 | 4 | 0 | 4 |
| GO:0009790 | Embryo development | 5 | 7 | 7 | 8 | 3 |
| GO:0006605 | Protein targeting | 7 | 7 | 3 | 4 | 3 |
| GO:0006790 | Sulfur compound metabolic process | 3 | 3 | 1 | 2 | 3 |
| GO:0021700 | Developmental maturation | 2 | 2 | 0 | 2 | 3 |
| GO:0007059 | Chromosome segregation | 1 | 1 | 2 | 1 | 3 |
| GO:0042254 | Ribosome biogenesis | 5 | 4 | 2 | 8 | 2 |
| GO:0006397 | mRNA processing | 6 | 9 | 6 | 5 | 2 |
| GO:0048646 | Anatomical structure formation involved in morphogenesis | 1 | 3 | 2 | 5 | 2 |
| GO:0006457 | Protein folding | 1 | 2 | 1 | 2 | 2 |
| GO:0034330 | Cell junction organisation | 1 | 2 | 2 | 2 | 2 |
| GO:0040007 | Growth | 2 | 1 | 0 | 2 | 2 |
| GO:0006091 | Generation of precursor metabolites and energy | 1 | 2 | 2 | 1 | 2 |
| GO:0006913 | Nucleocytoplasmic transport | 2 | 2 | 2 | 1 | 2 |
| GO:0030705 | Cytoskeleton-dependent intracellular transport | 1 | 3 | 2 | 1 | 2 |
| GO:0140014 | Mitotic nuclear division | 0 | 0 | 1 | 0 | 2 |
| GO:0051186 | Cofactor metabolic process | 2 | 2 | 2 | 2 | 1 |
| GO:0005975 | Carbohydrate metabolic process | 0 | 0 | 0 | 1 | 1 |
| GO:0007009 | Plasma membrane organisation | 1 | 2 | 0 | 1 | 1 |
| GO:0006091 | Extracellular matrix organisation | 0 | 1 | 0 | 0 | 1 |
| GO:0019748 | Secondary metabolic process | 1 | 0 | 0 | 0 | 1 |
| GO:0043473 | Pigmentation | 0 | 1 | 0 | 0 | 1 |
| GO:0051604 | Protein maturation | 1 | 0 | 0 | 1 | 0 |
| GO:0006399 | tRNA metabolic process | 1 | 3 | 0 | 0 | 0 |
| GO:0006520 | Cellular amino acid metabolic process | 4 | 2 | 2 | 0 | 0 |
Identification of genes involved in protein–protein interactions was performed and curated by BioGRID (release 4.4.201). Gene ontology classification was performed using GOnet (DICE database, build January 2020). The whole human repository annotation was used as reference set.
eIF2B in synaptic plasticity and cognitive decline
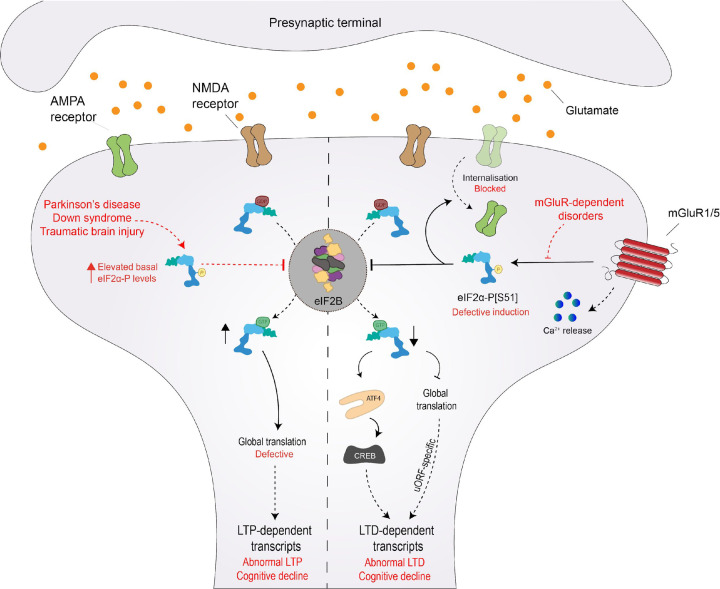
eIF2B and the regulation of protein synthesis plays a key role in synaptic plasticity and cognitive function [2–4]. Synaptic plasticity can be defined by the activity of synaptic connections, which ultimately coordinates the basis of learning and memory storage. High activity strengthens synapses, prompting long-term potentiation (LTP), while low activity weakens it, resulting in long-term depression (LTD) [126]. Interestingly, the phosphorylation status of eIF2α and therefore eIF2B activity, can dictate the fate of a given synapse, either facilitating LTP or LTD (Figure 5). Synapses undergoing local reductions of p-eIF2α are predicted to be potentiated. Upon eIF2α phosphorylation, mRNA translation of ATF4 suppresses CREB, a major transcription factor of plasticity-relevant proteins (Figure 5) [127,128]. In support of this, mutant eIF2α heterozygous mice (eIF2α+/S51A) displayed improved LTP and long-term memory consolidation [2]. In contrast, LTD relies on increased levels of p-eIF2α. Prisco et al. (2014) elegantly reported that uORF-driven translation remodels expression of cell surface receptors at synapses required for mGluR-LTD (Figure 5) [129]. It is the current view that this modulation of the eIF2α phosphorylation status can be adjusted to support a given learning task. LTP-dependent paradigms, such as contextual fear conditioning, shifts synapses to repress p-eIF2α, while LTD learning programs, such as object-in-place learning, demands the regulated translation of transcripts containing uORFs [129,130].
Figure 5. eIF2α phosphorylation and eIF2B modulation are key events in LTP- and LTD-dependent translation.
eIF2α phosphorylation status is a major switch of synaptic plasticity, dictating whether a given synapse undergoes long-term potentiation (LTP) or long-term depression (LTD). Suppression of eIF2α phosphorylation induces LTP by favouring new protein synthesis of ‘LTP proteins’. In contrast, increased levels of eIF2α phosphorylation drives the expression of LTD-specific proteins. Impairment of these synaptic changes triggers cognitive decline (highlighted in red), inherently associated with neurodegenerative disorders that bidirectionally affect levels of eIF2α phosphorylation.
As the hub of adaptability to learning and long-term memory storage, eIF2α phosphorylation, and thus modulation of eIF2B activity, has been studied in detail for cognition improvement. ISRIB, the eIF2B GEF activity enhancer, has been shown to attenuate p-eIF2α-dependent translational control without changes to eIF2α phosphorylation status per se or evidence of toxicity [74,131,132]. Indeed, eIF2B activation strengthens synaptic plasticity and memory consolidation in healthy rodents [70]. ISRIB also proved beneficial to counteract abnormally elevated levels of p-eIF2α and LTP-impairment in models of Parkinson’s [133], Down syndrome [74,134], traumatic brain injury [77], alcohol addiction [135] and drug abuse [136]. However, less is known of the impact of eIF2B activity modulation on LTD synapses which require p-eIF2α. Conflicting studies in Alzheimer’s disease (AD) models have shed some light on the involvement of eIF2B activation in LTD. Amyloid-β oligomer (AβO) accumulation is an age-related pathological hallmark of AD triggering ATF4-dependent neuronal cell death, resulting in progressive cognitive decline [137]. ISRIB ameliorated AβO-induced cognitive deficiency in rodents, is potentially linked to loss of eIF2B content observed in post-mortem AD brains [138]. Surprisingly, two other studies failed to recapitulate the beneficial cognitive effects of eIF2B activation in AD mice models [139,140]. Although the authors suggest different administration regimens and absence of ISR markers as causative reasoning for these unexpected results, AβO accumulation has been previously reported to selectively elevate LTD [141], favouring the p-eIF2α-dependent axis of synaptic plasticity. In support of this, eIF2B activation prevented proper object-placing learning of healthy rodents, which requires p-eIF2α-dependent translation [129]. Accordingly, augmenting p-eIF2α corrected deficient LTD in dystonia mice models [142]. Therefore, tailoring eIF2B function depending on the level of dependence of p-eIF2α could offer new avenues of therapeutic interventions.
Potential roles of eIF2B PTM modulation in cognition also warrants further investigation. A recent report has shown a novel role for eIF2B modulation during axonal wiring [143]. Rapid protein synthesis in growing axons overloads the ER, alleviated by eIF2α phosphorylation which paradoxically prevents key bursts of global translation. Guidance-cue Sema3A signalling overcomes this constraint by transiently supressing GSK-3β-mediated phosphorylation of eIF2Bε (Ser535), enhancing eIF2B activity to rescue global translation over specific time courses [143]. Additionally, lithium treatment in Down syndrome rodent models has been shown to inhibit GSK-3β activity [144], and thereby increase eIF2B activity, and improve synaptic strength [145].
Recent studies suggest that normal cognition relies on eIF2α-dependent translation within specific neuronal subtypes. Learning tasks in mice models reduced p-eIF2α in specific subsets of excitatory and inhibitory neurons [146]. Similarly, selective manipulation of PERK-eIF2α signalling in dopaminergic neurons resulted in multiple cognitive failures [147]. Additionally, conflicting reports highlight the need to address the involvement of other cell types in cognitive decline. Growth factor BDNF has been shown to up-regulate ATF4 mRNA translation independently of p-eIF2α in hippocampal neurons [148], whilst others report enhanced eIF2B activity upon BDNF treatment in similar cultured models [149]. It would be worthwhile to investigate how eIF2B activation within different cell types such as microglia, astrocytes and oligodendrocytes are involved in cognitive decline.
Mutations in eIF2B linked to vanishing white matter disease (VWMD)
The importance of eIF2B function within the cell is highlighted by the fact that loss of function mutations in any of the five subunits of eIF2B lead to the fatal neurological disorder, leukoencephalopathy with vanishing white matter (VWMD), also known as childhood ataxia with central nervous system hypomyelination (CACH) [7]. Although eIF2B is a global regulator of translation, glial cells appear selectively vulnerable, with astrocytes playing a central role in VWMD pathophysiology [150,151]. Patients suffer chronic degradation of the cerebral white matter associated with increased numbers of oligodendrocyte precursor cells and immature astrocytes [7]. In classical cases, disease onset occurs in childhood and is symptomatically characterised by cerebellar ataxia, spasticity, mild mental decline. Less commonly, patients may also present with ovarioleukodystrophy, loss of vision and epilepsy, and in very rare cases multiorgan defects have been observed [7]. However, phenotypically, symptoms and disease progression vary dramatically, likely due to the genetic complexity of VWMD. Inheritance is autosomal recessive, and mutations are most commonly missense, existing in homozygous or heterozygous states [152], and close to 200 mutations have been described in the Human Genome Database (www.hgmd.cf.ac.uk). Currently, no treatment is available for this disease although promising therapeutics have emerged, including ISRIB, 2BAct and Guanabenz, the latter of which is currently being used as part of the first VWMD clinical trials [76,153,6,154].
While the genetic link between eIF2B and VWMD is clear, the precise contributing role of mutated eIF2B in the progression of VWMD remains elusive. Biochemical analyses have investigated the functional effects that VWMD mutations have on eIF2B. Some mutations destabilise interactions between eIF2B subunits affecting complex formation, whereas other mutations can affect the GEF activity of eIF2B or the modulation of this activity by p-eIF2α [155–160] (Figure 6). The recent discovery of ISRIB is a promising avenue in the treatment of VWMD caused by mutations that destabilise the decameric conformation of eIF2B [75,34,35] (Figure 6). In vitro analyses of VWMD mutations associated with decreased eIF2B decameric stability, have demonstrated that ISRIB can rescue the stability of the eIF2B decamer and subsequently the GEF activity (Figure 6) [75].
Figure 6. Integrated stress response (ISR) dysregulation and modulation by small molecules in VWMD.
In response to various cellular stress stimuli, eIF2α kinases (PERK, HRI, GCN2, PKR) are selectively activated and phosphorylate eIF2α, resulting in the inhibition of eIF2B GEF activity and suppression of global translation. The translation of ISR effector mRNAs such as ATF4 is up-regulated promoting stress response gene expression. If stress is overcome GADD34 promotes dephosphorylation of eIF2α restoring homeostatic translation. VWM mutations can decrease eIF2B GEF activity, complex formation or eIF2B body formation, resulting in ISR dysregulation including increased ATF4 expression and decreased eIF2α phosphorylation. ISRIB and Guanabenz modulate ISR signalling through increasing eIF2B complex formation and decreasing dephosphorylation of eIF2α respectively, highlighting therapeutic potential for the treatment of VWM.
However, VWMD mutations have been identified that have no impact on the biochemically characterised in vitro functions of eIF2B but cause some of the most severe forms of disease [160,161], suggesting that eIF2B has functions in vivo that are not accounted for in vitro. Recently, two studies have investigated the therapeutic potential of ISRIB, and its derivative 2BAct, in VWMD mouse models, homozygous for eIF2BεR191H (R195H in humans) (2B5ho mice) or homozygous for eIF2BδR484W (R483W in humans) and heterozygous for eIF2BεR191H (2B4ho2B5he mice) [76,6]. In vitro studies have shown both eIF2BεR195H and eIF2BR483W mutations are associated with decreased decameric stability and eIF2B GEF activity, which can be rescued by ISRIB [161,75]. Clinical signs of VWMD were normalised in 2BAct treated 2B5ho mice and ISRIB treated 2B4ho2B5he mice; although clinical symptoms and pathology were significantly improved in ISRIB treated 2B5ho mice, mild signs of neurological deterioration were still observed [76,6]. Therefore, these data suggest that other in vivo factors could influence ISRIB’s effect. In order for ISRIB to act as an eIF2B activator, a pool of unassembled eIF2B tetramers must be available [1]. Hodgson et al. identified the presence of eIF2B bodies with varying degrees of eIF2B subunits in mammalian cells [107]. Additionally, it has been shown that VWMD mutations can impact upon the integrity and functionality of eIF2B bodies in yeast model systems [99,104], highlighting a possible involvement of localised pools of eIF2B in VWMD pathology. Deletion of eIF2Bα causes dispersal of eIF2B bodies. It could therefore be hypothesised that ISRIB could rescue eIF2B body formation in the presence of these VWMD mutations that disrupt body formation. However, mutational defects which impact upon catalytic function disrupt eIF2B body integrity, and it is currently unclear whether ISRIB’s ability to enhance eIF2B catalytic function would be sufficient to rescue localisation in the case of these mutants. Coupling in vitro analysis with in-cell assays could provide further insight into the influence of ISRIB on specific VWMD mutations.
VWMD mutations have also been found to dysregulate the ISR (Figure 6), and whilst the impact of VWMD mutations on eIF2B structure and function are diverse, dysregulation of the ISR appears to be a common consequence. Increased expression of the ATF4 driven transcriptome has been observed in a VWMD mouse model, homozygous for eIF2BεR191H (R195H in humans), and in VWMD patient brain samples [76,6]. Characteristic of a chronic ISR response, increased levels of p-eIF2α were not observed, likely explained by an increased GADD34 expression. Authors suggest that decreased eIF2B GEF activity caused by VWMD mutations is the driving force behind ISR induction. The ATF4 transcriptome is unable to restore homeostatic levels of eIF2B GEF activity thus inducing chronic stress. These findings have highlighted Guanabenz as a promising therapeutic for VWMD. Guanabenz is a potent agonist of the α2-adrenergic receptor and indirectly regulates eIF2B activity, by targeting the activity of the eIF2α phosphatase, GADD34, prolonging eIF2α phosphorylation during cellular stress, thereby decreasing the burden of global translation allowing more energy for homeostatic reprogramming [162] (Figure 6). However, guanabenz has also been shown to impact protein trafficking in the absence of any changes to eIF2α phosphorylation [163]. While the exact mode of action of guanabenz with respect to VWMD is unclear, administration of Guanabenz to VWMD mice (homozygous for eIF2BεR191H) has been shown to improve clinical symptoms of VWMD [153] and is now undergoing clinical trials [154].
Interestingly, in both VWMD mouse and patient brain, dysregulation of the ISR was observed in astrocytes and myelinating oligodendrocytes [76,6]. In vitro studies have highlighted that in the presence of VWMD mutations, specific cell-types appear more vulnerable to episodes of acute stress. These cells exhibit a heightened stress response, characterised by a hyper-induction of ATF4 and hyper-suppression of translation [75,164–167]. Chronic expression of ATF4 has also been observed in mouse models of mitochondrial dysfunction, and mutations in mitochondrial proteins have been associated with leukoencephalopathies [168–175]. Interestingly, transcriptome and proteome datasets generated from embryonic fibroblasts and whole brains of VWMD eIF2BεR132H mice (R136H in humans) have highlighted an impaired mitochondrial phenotype [176]. Although eIF2B does not regulate translation of the mitochondrial genome directly, the mitochondrial protein synthesis machinery itself is cytoplasmically translated and thus under the control of eIF2B. Fibroblast, oligodendrocyte precursor and astrocyte cells from the eIF2BεR132H mouse model suffer impairment of oxidative phosphorylation and ATP production [177]. While an adaptive increase in mitochondrial abundance restores this decreased oxidative phosphorylation in fibroblasts, increased mitochondrial abundance cannot rescue this phenotype in astrocytes or oligodendrocyte precursor cells [177,178]. VWMD highlights the dependence of glial cells on the precise accuracy of eIF2B mediated translational control.
eIF2B mutations and their link to neonatal diabetes
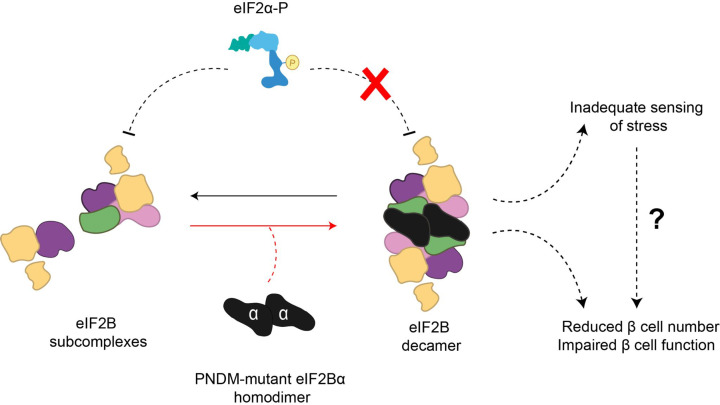
Recently, an increased incidence of heterozygous de novo missense variants in the EIF2B1 gene, which encodes eIF2Bα has been identified in patients with permanent neonatal diabetes mellitus (PNDM), a disorder resulting in early onset diabetes, typically diagnosed within the first 6 months of life [179]. In silico protein analysis revealed that the eIF2Bα missense variations identified in PNDM patients were present in either the binding surface occupied by p-eIF2α or altered residues involved in the interaction with eIF2 (Figure 7) [175]. Mutations in eIF2Bα have been previously characterised in the yeast orthologue of eIF2Bα, Gcn3p, and were categorised as either general control non-derepressible (gcn− mutations), which affects the regulatory activity of eIF2B, or as general control de-repressible (gcd− mutations), which affects the catalytic activity of eIF2B [27]. Interestingly two of the mutations identified in PNDM involve residue 44 on eIF2Bα. Mutations in this residue have previously been analysed in yeast and classified as gcn− with reduced sensitivity to p-eIF2α [17]. This suggests that the PNDM eIF2Bα mutations may hinder or impede the binding between eIF2Bα and p-eIF2α, thus leading to an inadequate sensing and response to cellular stress (Figure 7).
Figure 7. eIF2Bα PNDM mutations may lead to inadequate stress sensing and ISR in pancreatic β cells.
eIF2Bα missense mutations that lead to PNDM are present in residues which interact with eIF2 or the binding surface of p-eIF2α. Additionally, yeast studies have found that specific PNDM mutations can also disrupt eIF2B bodies and impact eIF2B decameric formation. This suggests that p-eIF2α binding to eIF2B is impeded, indicating a hindrance in stress-sensing which may lead to pancreatic β cell dysfunction or reduced number.
Additionally, in yeast, mutations in eIF2BαE44 also compromise the formation of eIF2B bodies [99], either by the decrease of eIF2Bα levels, leading to a decameric instability or by destabilising eIF2B subunit binding. It is known that for the recognition and binding of p-eIF2α, complete eIF2B decamer complex configuration is essential [36,37,180]. As such, the faulty sensing of stress in PNDM mutations may be also linked to the incorrect formation of eIF2B bodies. Furthermore, the assembly of these bodies may play an important role in the regulatory function of this complex.
When juxtaposed with eIF2Bα mutations associated with VWMD, the sites of PNDM eIF2Bα mutations differ greatly. The VWMD mutations are predominantly located in the C-terminal of eIF2Bα and appear to disrupt the formation of the eIF2B decamer, whereas PNDM mutations occur within the N-terminal region and appear to disrupt activation of the ISR [37,179]. However, interestingly in a VWMD patient that exhibited diabetic ketoacidosis, the homozygous missense variant was present in the N-terminal (eIF2BαL49R) [181], which would suggest a similar eIF2Bα defect observed in PNDM mutations and could explain the diabetic presentation.
While PNDM associated with heterozygous eIF2Bα variants do not exhibit severe neurological features, two reported cases displayed mild learning disability or attention deficit disorder [181], highlighting the link between cognitive abilities and eIF2B, which was discussed previously.
The most common monogenic PNDM subtype is Wolcott-Rallison syndrome (WRS; OMIM 226980) and is caused by recessive missense and truncation mutations in the EIF2AK3 gene, which encodes the eIF2 alpha kinase, PERK [182–185]. It is thought that since these mutations inhibit activation of the ISR the cells are unable to appropriately respond to stress and this ultimately leads to β-cell death, an analogous consequence of the eIF2Bα loss of function mutations. Interestingly, while β-cell death was not observed in knockout animal models of HRI [186], PKR [187] and GCN2 [188], PERK−/− mutants displayed degenerated pancreatic β-cells coupled with deficient function, leading to the development of diabetes mellitus [189].
High levels of glucose have been shown to lead to an increase of eIF2B activity and an increase in insulin protein synthesis in pancreatic β-cells [190]. Additionally, acute to moderate intensity resistance exercise has been shown to increase rates of protein synthesis and eIF2B activity in non-diabetic and moderately diabetic animal models. However, in severe insulin deficiency models this increase in eIF2B activity is inhibited and consequently, decreased levels of protein production are maintained [191,192]. GSK3 inhibition has been linked to insulin-stimulated liver glycogen synthesis [193], leading to eIF2Bε Ser535 dephosphorylation and eIF2B activation. This mechanism would account for the lack of eIF2B activation in severe hypoinsulinemia. Taken together, there is a clear direct association between eIF2B and pancreatic β-cell function and physiology.
Given the essential roles that PERK activation and eIF2B regulation play in β-cells, therapeutic targets that may lead to an abatement of stress, i.e., key factors in these stress pathways or rescue of function of those same factors, are potential and exciting approaches to restore β-cell health in diabetes. Therefore, targeting eIF2B could be a novel and effective avenue for therapy in β-cell disorders, such as PNDM or even diabetes mellitus [194].
ISRIB stabilises the productive conformation of decameric eIF2B, thus concurrently promoting eIF2B GEF activity and attenuating ISR effects [71,34,35]. However, the impact of the PNDM-associated eIF2Bα variants in the formation and localisation of eIF2B bodies, and the resulting impact on the GEF activity in mammalian cells is still unknown. Future studies concerning these areas may define the role of eIF2B in β-cells and in PNDM, and possibly open new therapeutic opportunities, such as ISRIB.
Conclusions and future directions
eIF2B is recognised as a critical control point in the regulation of translation. The inhibition of eIF2B GEF activity by p-eIF2α serves as the core event of the ISR, coordinating cellular responses to a vast range of cellular stresses. Dysregulation of eIF2B GEF activity and the ISR is now well established in a number of diseases and small molecules that target eIF2B have been shown to alleviate this dysfunction in many disease situations. Recent structural developments have provided extensive knowledge of the mechanisms that control eIF2B GEF activity and its inhibition by p-eIF2α, enhancing our understanding of how eIF2B mutations impact the ISR and the mechanism through which small molecules, such as ISRIB, can modulate eIF2B activity.
An important consideration for future research is the impact of cell-type specificity on eIF2B GEF activity and ISR regulation. It is clear that different cell types are selectively vulnerable to certain conditions of cellular stress and that mutations in specific ISR genes can cause detrimental effects in certain cell types. Uncovering the mechanism by which specific cells can activate the ISR will advance our molecular understanding of the ISR in disease. In-cell analysis of eIF2B has demonstrated the presence of eIF2B bodies; accumulations of functional eIF2B at specific foci within the cell cytoplasm. The eIF2B subunit composition of eIF2B bodies is suggested to influence GEF activity and is regulated by the ISR and the small molecule ISRIB. Characterisation of eIF2B bodies in different cell types could highlight cell-type specific dependencies for threshold levels of eIF2B GEF activity and regulation during the ISR as well as identifying new eIF2B protein–protein interactors that may influence function and body formation. The association of specific binding factors to eIF2B and the role of post translational modifications on eIF2B subunits may also provide critical information regarding cell specific phenotypes and response mechanisms.
Abbreviations
- ATF4
activating transcription factor 4
- CHOP
C/EBP homologous protein
- eIF2
eukaryotic initiation factor 2
- eIF2B
eukaryotic initiation factor 2B
- ER
endoplasmic reticulum
- GADD34
growth arrest and DNA damage inducible protein
- GCN2
general amino acid control nonderepressible 2
- GEF
guanine nucleotide exchange factor
- HRI
heme-regulated inhibitor
- ISR
integrated stress response
- ISRIB
integrated stress response inhibitor
- LTD
long-term depression
- LTP
long-term potentiation
- NTD
N-terminal domain
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- PKR
protein kinase R
- PNDM
permanent neonatal diabetes mellitus
- PTM
post-translational modification
- uORF
upstream open reading frame
- VWMD
vanishing white matter disease
Author Contribution
All authors contributed to the conception and writing of the text.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Costa-Mattioli M. and Walter P. (2020) The integrated stress response: from mechanism to disease. Science 368, eaat5314 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa-Mattioli M., Gobert D., Harding H., Herdy B., Azzi M., Bruno M.et al. (2005) Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 436, 1166–1173 10.1038/nature03897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton M.A. and Schuman E.M. (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58 10.1016/j.cell.2006.09.014 [DOI] [PubMed] [Google Scholar]
- 4.Huber K.M., Kayser M.S. and Bear M.F. (2000) Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science 288, 1254–1257 10.1126/science.288.5469.1254 [DOI] [PubMed] [Google Scholar]
- 5.Sidrauski C., McGeachy A.M., Ingolia N.T. and Walter P. (2015) The small molecule ISRIB reverses the effects of eIF2alpha phosphorylation on translation and stress granule assembly. eLife 4, 1–16 10.7554/eLife.05033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong Y.L., LeBon L., Basso A.M., Kohlhaas K.L., Nikkel A.L., Robb H.M.et al. (2019) eIF2B activator prevents neurological defects caused by a chronic integrated stress response. Elife 8, e42940 10.7554/eLife.42940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton E.M.C., van der Lei H.D.W., Vermeulen G., Gerver J.A.M., Lourenço C.M., Naidu S.et al. (2018) Natural History of Vanishing White Matter. Ann. Neurol. 84, 274–288 10.1002/ana.25287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershey J.W.B., Sonenberg N. and Mathews M.B. (2019) Principles of translational control. Cold Spring Harb. Perspect. Biol. 11, a032607 10.1101/cshperspect.a032607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrick W.C. and Pavitt G.D. (2018) Protein synthesis initiation in eukaryotic cells. (2018). Cold Spring Harb. Perspect. Biol. 10, a033092 10.1101/cshperspect.a033092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapp L.D. and Lorsch J.R. (2004) GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J. Mol. Biol. 335, 923–936 10.1016/j.jmb.2003.11.025 [DOI] [PubMed] [Google Scholar]
- 11.Jennings M. and Pavitt G. (2010) eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 465, 378–381 10.1038/nature09003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jennings M.D., Zhou Y., Mohammad-Qureshi S.S., Bennett D. and Pavitt G.D. (2013) eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev. 27, 2696–2707 10.1101/gad.231514.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh C.R., Lee B., Udagawa T., Mohammad-Qureshi S.S., Yamamoto Y., Pavitt G.D.et al. (2006) An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 25, 4537–4546 10.1038/sj.emboj.7601339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jennings M.D., Kershaw C.J., Adomavicius T. and Pavitt G.D. (2017) Fail-safe control of translation initiation by dissociation of eIF2α phosphorylated ternary complexes. Elife 6, e24542 10.7554/eLife.24542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komar A.A. and Merrick W.C. (2020) A Retrospective on eIF2A-and Not the Alpha Subunit of eIF2. Int. J. Mol. Sci. 21, 2054 10.3390/ijms21062054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimball S.R., Fabian J.R., Pavitt G.D., Hinnebusch A.G. and Jefferson L.S. (1998) Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eIF2B. J. Biol. Chem. 273, 12841–12845 10.1074/jbc.273.21.12841 [DOI] [PubMed] [Google Scholar]
- 17.Pavitt G.D., Yang W. and Hinnebusch A.G. (1997) Homologous segments in three 193 subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 17, 1298–1313 10.1128/MCB.17.3.1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavitt G.D., Ramaiah K.V., Kimball S.R. and Hinnebusch A.G. (1998) eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 12, 514–526 10.1101/gad.12.4.514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams D.D., Price N.T., Loughlin A.J. and Proud C.G. (2001) Characterization of the mammalian initiation factor eIF2B complex as a GDP dissociation stimulator protein. J. Biol. Chem. 276, 24697–24703 10.1074/jbc.M011788200 [DOI] [PubMed] [Google Scholar]
- 20.Gomez E. and Pavitt G.D. (2000) Identification of domains and residues within the epsilon subunit of eukaryotic translation initiation factor 2B (eIF2Bepsilon) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol. Cell. Biol. 20, 3965–3976 10.1128/MCB.20.11.3965-3976.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dever T.E., Yang W., Aström S., Byström A.S. and Hinnebusch A.G. (1995) Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol. Cell. Biol. 15, 6351–6363 10.1128/MCB.15.11.6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowlands A.G., Panniers R. and Henshaw E.C. (1988) The Catalytic Mechanism of Guanine Nucleotide Exchange Factor Action and Competitive Inhibition by Phosphorylated Eukaryotic Initiation Factor 2. J. Biol. Chem. 263, 5526–5533 10.1016/S0021-9258(18)60596-4 [DOI] [PubMed] [Google Scholar]
- 23.Dev K., Qiu H., Dong J., Zhang F., Barthlme D. and Hinnebusch A.G. (2010) The beta/Gcd7 subunit of eukaryotic translation initiation factor 2B (eIF2B), a guanine nucleotide exchange factor, is crucial for binding eIF2 in vivo. Mol. Cell. Biol. 30, 5218–5233 10.1128/MCB.00265-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dever T.E., Chent J.J., Barbert G.N., Cigan A.M., Feng L., Donahue T.F.et al. (1993) Mammalian eukaryotic initiation factor 2a kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast (phosphorylation/initiation factors/double-stranded RNA-dependent eIF-2a kinase/p68 kinase/heme-regulate. Biochemistry 90, 4616–4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elsby R., Heiber J.F., Reid P., Kimball S.R., Pavitt G.D. and Barber G.N. (2011) The Alpha Subunit of Eukaryotic Initiation Factor 2B (eIF2B) Is Required for eIF2- Mediated Translational Suppression of Vesicular Stomatitis Virus. J. Virol. 85, 9716–9725 10.1128/JVI.05146-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabian J.R., Kimball S.R., Heinzinger N.K. and Jefferson L.S. (1997) Subunit assembly and guanine nucleotide exchange activity of eukaryotic initiation factor 2B expressed in Sf9 cells. J. Biol. Chem. 272, 12359–12365 10.1074/jbc.272.19.12359 [DOI] [PubMed] [Google Scholar]
- 27.Hannig E.M., Williams N.P., Wek R.C. and Hinnebusch A.G. (1990) The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics 126, 549–562 10.1093/genetics/126.3.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimball S.R., Fabian J.R., Pavitt G.D., Hinnebusch A.G. and Jefferson L.S. (1998) Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eIF2B. J. Biol. Chem. 273, 12841–12845 10.1074/jbc.273.21.12841 [DOI] [PubMed] [Google Scholar]
- 29.Krishnamoorthy T., Pavitt G.D., Zhang F., Dever T.E. and Hinnebusch A.G. (2001) Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 21, 5018–5030 10.1128/MCB.21.15.5018-5030.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wortham N.C., Stewart J.D., Harris S., Coldwell M.J. and Proud C.G. (2016) Stoichiometry of the eIF2B complex is maintained by mutual stabilization of subunits. Biochem. J. 473, 571–580 10.1042/BJ20150828 [DOI] [PubMed] [Google Scholar]
- 31.Gordiyenko Y., Schmidt C., Jennings M.D., Matak-Vinkovic D., Pavitt G.D. and Robinson C.V. (2014) eIF2B is a decameric guanine nucleotide exchange factor with a γ2ε2 tetrameric core. Nat. Commun. 5, 3902 10.1038/ncomms4902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wortham N.C., Martinez M., Gordiyenko Y., Robinson C.V. and Proud C.G. (2014) Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J. 28, 2225–2237 10.1096/fj.13-243329 [DOI] [PubMed] [Google Scholar]
- 33.Kashiwagi K., Takahashi M., Nishimoto M., Hiyama T.B., Higo T., Umehara T.et al. (2016) Crystal structure of eukaryotic translation initiation factor 2B. Nature 531, 122–125 10.1038/nature16991 [DOI] [PubMed] [Google Scholar]
- 34.Tsai J.C., Miller-Vedam L.E., Anand A.A., Jaishankar P., Nguyen H.C., Renslo A.R.et al. (2018) Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science 359, eaaq0939 10.1126/science.aaq0939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zyryanova A.F., Weis F., Faille A., Alard A.A., Crespillo-Casado A., Sekine Y.et al. (2018) Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science 359, 1533–1536 10.1126/science.aar5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zyryanova A.F., Kashiwagi K., Rato C., Harding H.P., Crespillo-Casado A., Perera L.A.et al. (2021) ISRIB Blunts the Integrated Stress Response by Allosterically Antagonising the Inhibitory Effect of Phosphorylated eIF2 on eIF2B. Mol. Cell 81, 88.e6–103.e6 10.1016/j.molcel.2020.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoof M., Boone M., Wang L., Lawrence R., Frost A. and Walter P. (2021) eIF2B conformation and assembly state regulate the integrated stress response. Elife 10, e65703 10.7554/eLife.65703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenner L.R., Anand A.A., Nguyen H.C., Myasnikov A.G., Klose C.J., McGeever L.A.et al. (2019) eIF2B-catalyzed nucleotide exchange and phosphoregulation by the integrated stress response. Science 364, 491–495 10.1126/science.aaw2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adomavicius T., Guaita M., Zhou Y., Jennings M.D., Latif Z., Roseman A.M.et al. (2019) The structural basis of translational control by eIF2 phosphorylation. Nat Commun. 10, 2136 10.1038/s41467-019-10167-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordiyenko Y., Llácer J.L. and Ramakrishnan V. (2019) Structural basis for the inhibition of translation through eIF2α phosphorylation. Nat. Commun. 10, 2640 10.1038/s41467-019-10606-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kashiwagi K., Yokoyama T., Nishimoto M., Takahashi M., Sakamoto A., Yonemochi M.et al. (2019) Structural basis for eIF2B inhibition in integrated stress response. Science 364, 495–499 10.1126/science.aaw4104 [DOI] [PubMed] [Google Scholar]
- 42.Marintchev A. and Ito T. (2020) eIF2B and the integrated stress response: a structural and mechanistic view. Biochemistry 59, 1299–1308 10.1021/acs.biochem.0c00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anand A.A. and Walter P. (2020) Structural insights into ISRIB, a memory-enhancing inhibitor of the integrated stress response. FEBS J. 287, 239–245 10.1111/febs.15073 [DOI] [PubMed] [Google Scholar]
- 44.Donnelly N., Gorman A.M., Gupta S. and Samali A. (2013) The eIF2α kinases: their structures and functions. Cell. Mol. Life Sci. 70, 3493–3511 10.1007/s00018-012-1252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M.et al. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 11, 619–633 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- 46.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A. and Gorman A.M. (2016) The integrated stress response. EMBO Rep. 17, 1374–1395 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y.N., Kavianpour S., Zhang T., Zhang X., Nguyen D., Thombre R.et al. (2021) MARK2 phosphorylates eIF2α in response to proteotoxic stress. PLoS Biol. 19, e3001096 10.1371/journal.pbio.3001096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berlanga J.J., Herrero S. and de Haro C. (1998) Characterization of the heminsensitive eukaryotic initiation factor 2alpha kinase from mouse nonerythroid cells. J. Biol. Chem. 273, 32340–32346 10.1074/jbc.273.48.32340 [DOI] [PubMed] [Google Scholar]
- 49.Chen J.J., Throop M.S., Gehrke L., Kuo I., Pal J.K., Brodsky M.et al. (1991) Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc. Natl Acad. Sci. 88, 7729–7733 10.1073/pnas.88.17.7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harding H.P., Zhang Y. and Ron D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- 51.Meurs E., Chong K., Galabru J., Thomas N.S., Kerr I.M., Williams B.R.et al. (1990) Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62, 379–390 10.1016/0092-8674(90)90374-N [DOI] [PubMed] [Google Scholar]
- 52.Shi Y., Vattem K.M., Sood R., An J., Liang J., Stramm L.et al. (1998) Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18, 7499–7509 10.1128/MCB.18.12.7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vazquez de Aldana C.R., Wek R.C., Segundo P.S., Truesdell A.G. and Hinnebusch A.G. (1994) Multicopy tRNA genes functionally suppress mutations in yeast eIF-2 alpha kinase GCN2: evidence for separate pathways coupling GCN4 expression to unchanged tRNA. Mol. Cell. Biol. 14, 7920–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clemens M.J. and Elia A. (1997) The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interf. Cytokine Res. 17, 503–524 10.1089/jir.1997.17.503 [DOI] [PubMed] [Google Scholar]
- 55.Lemaire P.A., Anderson E., Lary J. and Cole J.L. (2008) Mechanism of PKR activation by dsRNA. J. Mol. Biol. 381, 351–360 10.1016/j.jmb.2008.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chukwurah E., Farabaugh K.T., Guan B.J., Ramakrishnan P. and Hatzoglou M. (2021) A tale of two proteins: PACT and PKR and their roles in inflammation. FEBS J. 22, 6365–6391 10.1111/febs.15691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harding H.P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M.et al. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 10.1016/S1097-2765(00)00108-8 [DOI] [PubMed] [Google Scholar]
- 58.Patil C. and Walter P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 10.1016/S0955-0674(00)00219-2 [DOI] [PubMed] [Google Scholar]
- 59.McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A.et al. (2005) Heme-regulated inhibitor kinase mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280, 16925–16933 10.1074/jbc.M412882200 [DOI] [PubMed] [Google Scholar]
- 60.Han A.P., Yu C., Lu L., Fujiwara Y., Browne C., Chin G.et al. (2001) Heme-regulated eIF2alpha kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20, 6909–6918 10.1093/emboj/20.23.6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mukherjee T., Ramaglia V., Abdel-Nour M., Bianchi A.A., Tsalikis J., Chau H.N.et al. (2021) The eIF2α kinase HRI triggers the autophagic clearance of cytosolic protein aggregates. J. Biol. Chem. 296, 100050 10.1074/jbc.RA120.014415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dever T.E., Yang W., Aström S., Byström A.S. and Hinnebusch A.G. (1995) Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol. Cell. Biol. 15, 6351–6363 10.1128/MCB.15.11.6351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rowlands A.G., Panniers R. and Henshaw E.C. (1988) The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 263, 5526–5533 10.1016/S0021-9258(18)60596-4 [DOI] [PubMed] [Google Scholar]
- 64.Vattem K.M. and Wek R.C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. 101, 11269–11274 10.1073/pnas.0400541101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hinnebusch A.G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 10.1146/annurev.micro.59.031805.133833 [DOI] [PubMed] [Google Scholar]
- 66.Young S.K. and Wek R.C. (2016) Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J. Biol. Chem. 291, 16927–16935 10.1074/jbc.R116.733899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brush M.H., Weiser D.C. and Shenolikar S. (2003) Growth arrest and DNA damage inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 23, 1292–1303 10.1128/MCB.23.4.1292-1303.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marciniak S.J., Chambers J.E. and Ron D. (2021) Pharmacological targeting of endoplasmic reticulum stress in disease. Nat. Rev. Drug Discov. 2, 115–140 10.1038/s41573-021-00320-3 [DOI] [PubMed] [Google Scholar]
- 69.Anand A.A. and Walter P. (2020) Structural insights into ISRIB, a memory-enhancing inhibitor of the integrated stress response. FEBS J. 287, 239–245 10.1111/febs.15073 [DOI] [PubMed] [Google Scholar]
- 70.Sidrauski C., Acosta-Alvear D., Khoutorsky A., Vedantham P., Hearn B.R., Li H.et al. (2013) Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife 2, e00498 10.7554/eLife.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sekine Y., Zyryanova A., Crespillo-Casado A., Fischer P.M., Harding H.P. and Ron D. (2015) Stress responses. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science 348, 1027–1030 10.1126/science.aaa6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krukowski K., Nolan A., Frias E.S., Boone M., Ureta G., Grue K.et al. (2020) Small molecule cognitive enhancer reverses age-related memory decline in mice. Elife 29, e62048 10.7554/eLife.62048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Halliday M., Radford H., Sekine Y., Moreno J., Verity N., le Quesne J.et al. (2015) Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 6, e1672 10.1038/cddis.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu P.J., Khatiwada S., Cui Y., Reineke L.C., Dooling S.W., Kim J.J.et al. (2019) Activation of the ISR mediates the behavioral and neurophysiological abnormalities in Down syndrome. Science 366, 843–849 10.1126/science.aaw5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong Y.L., LeBon L., Edalji R., Lim H.B., Sun C. and Sidrauski C. (2018) The small molecule ISRIB rescues the stability and activity of Vanishing White Matter Disease eIF2B mutant complexes. Elife 7, e32733 10.7554/eLife.32733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abbink T.E.M., Wisse L.E., Jaku E., Thiecke M.J., Voltolini-González D., Fritsen H.et al. (2019) Vanishing white matter: deregulated integrated stress response as therapy target. Ann. Clin. Transl. Neurol. 6, 1407–1422 10.1002/acn3.50826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chou A., Krukowski K., Jopson T., Zhu P.J., Costa-Mattioli M., Walter P.et al. (2017) Inhibition of the integrated stress response reverses cognitive deficits after traumatic brain injury. Proc. Natl Acad. Sci. 114, E6420–E6426 10.1073/pnas.1707661114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. ALZFORUM - Networking for a cure: DNL343, October 2021 https://www.alzforum.org/therapeutics/dnl343.
- 79. Calico and AbbVie Share Update on Early-Stage Clinical Programs, February 2021 https://www.calicolabs.com/press/calico-and-abbvie-share-update-on-early-stage-clinical-programs.
- 80.Bond S., Lopez-Lloreda C., Gannon P.J., Akay-Espinoza C. and Jordan-Sciutto K.L. (2020) The integrated stress response and phosphorylated eukaryotic initiation factor 2α in neurodegeneration. J. Neuropathol. Exp. Neurol. 79, 123–143 10.1093/jnen/nlz129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rutkowski D.T., Arnold S.M., Miller C.N., Wu J., Li J., Gunnison K.M.et al. (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 11, e374 10.1371/journal.pbio.0040374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ghaddar N., Wang S., Woodvine B., Krishnamoorthy J., van Hoef V., Darini C.et al. (2021) The integrated stress response is tumorigenic and constitutes a therapeutic liability in KRAS-driven lung cancer. Nat. Commun. 12, 4651 10.1038/s41467-021-24661-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han J., Back S.H., Hur J., Lin Y.H., Gildersleeve R., Shan J.et al. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 10.1038/ncb2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marciniak S.J., Yun C.Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R.et al. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 10.1101/gad.1250704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teske B.F., Fusakio M.E., Zhou D., Shan J., McClintick J.N., Kilberg M.S.et al. (2013) CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell. 24, 2477–2490 10.1091/mbc.e13-01-0067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaspar S., Oertlin C., Szczepanowska K., Kukat A., Senft K., Lucas C.et al. (2021) Adaptation to mitochondrial stress requires CHOP-directed tuning of ISR. Sci Adv. 7, eabf0971 10.1126/sciadv.abf0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCullough K.D., Martindale J.L., Klotz L.-O., Aw T.-Y. and Holbrook N.J. (2001) Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol. Cell. Biol. 21, 1249–1259 10.1128/MCB.21.4.1249-1259.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yamaguchi H. and Wang H.-G. (2004) CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J. Biol. Chem. 279, 45495–45502 10.1074/jbc.M406933200 [DOI] [PubMed] [Google Scholar]
- 89.Guan B.J., van Hoef V., Jobava R., Elroy-Stein O., Valasek L.S., Cargnello M.et al. (2017) A unique ISR program determines cellular responses to chronic stress. Mol. Cell 68, 885.e6–900.e6 10.1016/j.molcel.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wek R.C. and Anthony T.G. (2009) Beta testing the antioxidant function of eIF2alpha phosphorylation in diabetes prevention. Cell Metab. 10, 1–2 10.1016/j.cmet.2009.06.005 [DOI] [PubMed] [Google Scholar]
- 91.Lewerenz J. and Maher P. (2009) Basal levels of eIF2alpha phosphorylation determine cellular antioxidant status by regulating ATF4 and xCT expression. J. Biol. Chem. 284, 1106–1115 10.1074/jbc.M807325200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Back S.H., Scheuner D., Han J., Song B., Ribick M., Wang J.et al. (2009) Translation attenuation through eIF2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 10, 13–26 10.1016/j.cmet.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R.T., Remotti H.et al. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 10.1101/gad.12.7.982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H.et al. (2001) Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol. Cell 6, 1153–1163 10.1016/S1097-2765(01)00264-7 [DOI] [PubMed] [Google Scholar]
- 95.Choi W.G., Han J., Kim J.H., Kim M.J., Park J.W., Song B.et al. (2017) eIF2α phosphorylation is required to prevent hepatocyte death and liver fibrosis in mice challenged with a high fructose diet. Nutr. Metab. 14, 48 10.1186/s12986-017-0202-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rajesh K., Papadakis A.I., Kazimierczak U., Peidis P., Wang S., Ferbeyre G.et al. (2013) eIF2α phosphorylation bypasses premature senescence caused by oxidative stress and pro-oxidant antitumor therapies. Aging 12, 884–901 10.18632/aging.100620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Campbell S.G., Hoyle N.P. and Ashe M.P. (2005) Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translation control. J. Cell Biol. 170, 925–934 10.1083/jcb.200503162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taylor E.J., Campbell S.G., Griffiths C.D., Reid P.J., Slaven J.W., Harrison R.J.et al. (2010) Fusel alcohols regulate translation initiation by inhibiting eIF2B to reduce ternary complex in a mechanism that may involve altering the integrity and dynamics of the eIF2B body. Mol. Biol. Cell 21, 2202–2216 10.1091/mbc.e09-11-0962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Norris K., Hodgson R.E., Dornelles T., Allen K.E., Abell B.M., Ashe M.P.et al. (2021) Mutational analysis of the alpha subunit of eIF2B provides insights into the role of eIF2B bodies in translational control and VWM disease. J. Biol. Chem. 296, 100207 10.1074/jbc.RA120.014956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wortham N.C. and Proud C.G. (2015) Biochemical effects of mutations in the gene encoding the alpha subunit of eukaryotic initiation factor (eIF) 2B associated with Vanishing White Matter disease. BMC Med. Genet. 16, 64 10.1186/s12881-015-0204-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marini G., Nüske E., Leng W., Alberti S. and Pigino G. (2020) Reorganization of budding yeast cytoplasm upon energy depletion. Mol. Biol. Cell 31, 1232–1245 10.1091/mbc.E20-02-0125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Petrovska I., Nüske E., Munder M.C., Kulasegaran G., Malinovska L., Kroschwald S.et al. (2014) Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. Elife 3, e02409 10.7554/eLife.02409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Munder M.C., Midtvedt D., Franzmann T., Nüske E., Otto O., Herbig M.et al. (2016) A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife 5, e09347 10.7554/eLife.09347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moon S.L. and Parker R. (2018) Analysis of eIF2B bodies and their relationships with stress granules and P-bodies. Sci. Rep. 8, 12264 10.1038/s41598-018-30805-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nüske E., Marini G., Richter D., Leng W., Bogdanova A., Franzmann T.M.et al. (2020) Filament formation by the translation factor eIF2B regulates protein synthesis in starved cells. Biol. Open. 9, bio046391 10.1242/bio.046391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.von der Haar T. and McCarthy J.E. (2002) Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2, 531–544 10.1046/j.1365-2958.2002.03172.x [DOI] [PubMed] [Google Scholar]
- 107.Hodgson R.E., Varanda B.A., Ashe M.P., Allen K.E. and Campbell S.G. (2019) Cellular eIF2B subunit localization: implications for the integrated stress response and its control by small molecule drugs. Mol. Biol. Cell 30, 942–958 10.1091/mbc.E18-08-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bogorad A.M., Xia B., Sandor D.G., Mamonov A.B., Cafarella T.R., Jehle S.et al. (2014) Insights into the architecture of the eIF2Bα/β/δ regulatory subcomplex. Biochemistry 53, 3432–3445 10.1021/bi500346u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kuhle B., Eulig N.K. and Ficner R. (2015) Architecture of the eIF2B regulatory subcomplex and its implications for the regulation of guanine nucleotide exchange on eIF2. Nucleic Acids Res. 43, 9994–10014 10.1093/nar/gkv930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X. and Proud C.G. (2008) A novel mechanism for the control of translation initiation by amino acids, mediated by phosphorylation of eukaryotic initiation factor 2B. Mol. Cell. Biol. 28, 1429–1442 10.1128/MCB.01512-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Welsh G.I., Miller C.M., Loughlin A.J., Price N.T. and Proud C.G. (1998) Regulation of eukaryotic initiation factor eIF2B: glycogen synthase kinase-3 phosphorylates a conserved serine which undergoes dephosphorylation in response to insulin. FEBS Lett. 421, 125–130 10.1016/S0014-5793(97)01548-2 [DOI] [PubMed] [Google Scholar]
- 112.Wang X., Paulin F.E., Campbell L.E., Gomez E., O'Brien K., Morrice N.et al. (2001) Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo. EMBO J. 20, 4349–4359 10.1093/emboj/20.16.4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Beilsten-Edmands V., Gordiyenko Y., Kung J.C., Mohammed S., Schmidt C. and Robinson C.V. (2015) eIF2 interactions with initiator tRNA and eIF2B are regulated by post-translational modifications and conformational dynamics. Cell Discov. 1, 15020 10.1038/celldisc.2015.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C.et al. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- 115.Arnesen T. (2011) Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 9, e1001074 10.1371/journal.pbio.1001074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nguyen K.T., Lee C.S., Mun S.H., Truong N.T., Park S.K. and Hwang C.S. (2019) N-terminal acetylation and the N-end rule pathway control degradation of the lipid droplet protein PLIN2. J. Biol. Chem. 294, 379–388 10.1074/jbc.RA118.005556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sudhakar A., Ramachandran A., Ghosh S., Hasnain S.E., Kaufman R.J. and Ramaiah K.V. (2000) Phosphorylation of serine 51 in initiation factor 2 alpha (eIF2 alpha) promotes complex formation between eIF2 alpha(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry 39, 12929–12938 10.1021/bi0008682 [DOI] [PubMed] [Google Scholar]
- 118.Kershaw C.J., Jennings M.D., Cortopassi F., Guaita M., Al-Ghafli H. and Pavitt G.D. (2021) GTP binding to translation factor eIF2B stimulates its guanine nucleotide exchange activity. iScience 24, 103454 10.1016/j.isci.2021.103454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hao Q., Heo J.M., Nocek B.P., Hicks K.G., Stoll V.S., Remarcik C.et al. (2021) Sugar phosphate activation of the stress sensor eIF2B. Nat. Commun. 12, 3440 10.1038/s41467-021-23836-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kashiwagi K., Shichino Y., Osaki T., Sakamoto A., Nishimoto M., Takahashi M.et al. (2021) eIF2B-capturing viral protein NSs suppresses the integrated stress response. Nat. Commun. 12, 7102 10.1038/s41467-021-27337-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rabouw H.H., Visser L.J., Passchier T.C., Langereis M.A., Liu F., Giansanti P.et al. (2020) Inhibition of the integrated stress response by viral proteins that block p-eIF2-eIF2B association. Nat. Microbiol. 11, 1361–1373 10.1038/s41564-020-0759-0 [DOI] [PubMed] [Google Scholar]
- 122.Browne C.M., Samir P., Fites J.S., Villarreal S.A. and Link A.J. (2013) The yeast eukaryotic translation initiation factor 2B translation initiation complex interacts with the fatty acid synthesis enzyme YBR159W and endoplasmic reticulum membranes. Mol. Cell. Biol. 33, 1041–1056 10.1128/MCB.00811-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Klein U., Ramirez M.T., Kobilka B.K. and von Zastrow M. (1997) A novel interaction between adrenergic receptors and the alpha-subunit of eukaryotic initiation factor 2B. J. Biol. Chem. 272, 19099–19102 10.1074/jbc.272.31.19099 [DOI] [PubMed] [Google Scholar]
- 124.Oughtred R., Rust J., Chang C., Breitkreutz B.J., Stark C., Willems A.et al. (2021) The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 30, 187–200 10.1002/pro.3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pomaznoy M., Ha B. and Peters B. (2018) GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinformatics 19, 470 10.1186/s12859-018-2533-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Neves G., Cooke S.F. and Bliss T.V. (2008) Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat. Rev. Neurosci. 9, 65–75 10.1038/nrn2303 [DOI] [PubMed] [Google Scholar]
- 127.Kida S.A. (2012) Functional role for CREB as a positive regulator of memory formation and LTP. Exp. Neurobiol. 21, 136–140 10.5607/en.2012.21.4.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang Z., Belforte J.E., Lu Y., Yabe Y., Pickel J., Smith C.B.et al. (2010) eIF2alpha Phosphorylation-dependent translation in CA1 pyramidal cells impairs hippocampal memory consolidation without affecting general translation. J. Neurosci. 30, 2582–2594 10.1523/JNEUROSCI.3971-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Di Prisco G.V., Huang W., Buffington S.A., Hsu C.C., Bonnen P.E., Placzek A.N.et al. (2014) Translational control of mGluR-dependent long-term depression and object-place learning by eIF2α. Nat. Neurosci. 17, 1073–1082 10.1038/nn.3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Costa-Mattioli M., Gobert D., Stern E., Gamache K., Colina R., Cuello C.et al. (2007) eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129, 195–206 10.1016/j.cell.2007.01.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guthrie L.N., Abiraman K., Plyler E.S., Sprenkle N.T., Gibson S.A., McFarland B.C.et al. (2016) Attenuation of PKR-like ER Kinase (PERK) signaling selectively controls endoplasmic reticulum stress-induced inflammation without compromising immunological responses. J. Biol. Chem. 291, 15830–15840 10.1074/jbc.M116.738021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rabouw H.H., Langereis M.A., Anand A.A., Visser L.J., de Groot R.J., Walter P.et al. (2019) Small molecule ISRIB suppresses the integrated stress response within a defined window of activation. Proc. Natl Acad. Sci. 116, 2097–2102 10.1073/pnas.1815767116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Costa C., Sgobio C., Siliquini S., Tozzi A., Tantucci M., Ghiglieri V.et al. (2012) Mechanisms underlying the impairment of hippocampal long-term potentiation and memory in experimental Parkinson's disease. Brain 135, 1884–1899 10.1093/brain/aws101 [DOI] [PubMed] [Google Scholar]
- 134.Lanzillotta C. and Di Domenico F. (2021) Stress responses in down syndrome neurodegeneration: state of the art and therapeutic molecules. Biomolecules 11, 266 10.3390/biom11020266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Izumi Y. and Zorumski C.F. (2020) Inhibitors of cellular stress overcome acute effects of ethanol on hippocampal plasticity and learning. Neurobiol. Dis. 141, 104875 10.1016/j.nbd.2020.104875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Placzek A.N., Prisco G.V., Khatiwada S., Sgritta M., Huang W., Krnjević K.et al. (2016) eIF2α-mediated translational control regulates the persistence of cocaine-induced LTP in midbrain dopamine neurons. Elife 5, e17517 10.7554/eLife.17517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Oliveira M.M. and Klann E. (2021) eIF2-dependent translation initiation: memory consolidation and disruption in Alzheimer’s disease. Semin. Cell Dev. Biol. 125, 101–109S1084-9521(21)00196-8, 10.1016/j.semcdb.2021.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oliveira M.M., Lourenco M.V., Longo F., Kasica N.P., Yang W., Ureta G.et al. (2021) Correction of eIF2-dependent defects in brain protein synthesis, synaptic plasticity, and memory in mouse models of Alzheimer’s disease. Sci. Signal. 14, eabc5429 10.1126/scisignal.abc5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johnson E.C. and Kang J. (2016) A small molecule targeting protein translation does not rescue spatial learning and memory deficits in the hAPP-J20 mouse model of Alzheimer’s disease. Peer J. 4, e2565 10.7717/peerj.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Briggs D.I., Defensor E., Memar Ardestani P., Yi B., Halpain M.et al. (2017) Role of endoplasmic reticulum stress in learning and memory impairment and Alzheimer’s disease-like neuropathology in the PS19 and APPSwe mouse models of tauopathy and amyloidosis. eNeuro 4, ENEURO.0025-17.2017 10.1523/ENEURO.0025-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shankar G.M., Li S., Mehta T.H., Garcia-Munoz A., Shepardson N.E., Smith I.et al. (2008) Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat. Med. 14, 837–842 10.1038/nm1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rittiner J.E., Caffall Z.F., Hernández-Martinez R., Sanderson S.M., Pearson J.L., Tsukayama K.K.et al. (2016) Functional genomic analyses of mendelian and sporadic disease identify impaired eIF2α signaling as a generalizable mechanism for dystonia. Neuron 92, 1238–1251 10.1016/j.neuron.2016.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cagnetta R., Wong H.H., Frese C.K., Mallucci G.R., Krijgsveld J. and Holt C.E. (2019) Noncanonical modulation of the eIF2 pathway controls an increase in local translation during neural wiring. Mol. Cell 73, 474.e5–489.e5 10.1016/j.molcel.2018.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bertsch S., Lang C.H. and Vary T.C. (2011) Inhibition of glycogen synthase kinase 3[beta] activity with lithium in vitro attenuates sepsis-induced changes in muscle protein turnover. Shock 35, 266–274 10.1097/SHK.0b013e3181fd068c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Contestabile A., Greco B., Ghezzi D., Tucci V., Benfenati F. and Gasparini L. (2013) Lithium rescues synaptic plasticity and memory in Down syndrome mice. J. Clin. Invest. 123, 348–361 10.1172/JCI64650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sharma V., Sood R., Khlaifia A., Eslamizade M.J., Hung T.Y., Lou D.et al. (2020) eIF2α controls memory consolidation via excitatory and somatostatin neurons. Nature 586, 412–416 10.1038/s41586-020-2805-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Longo F., Mancini M., Ibraheem P.L., Aryal S., Mesini C., Patel J.C.et al. (2021) Cell-type-specific disruption of PERK-eIF2α signaling in dopaminergic neurons alters motor and cognitive function. Mol. Psychiatry 11, 6427–6450 10.1038/s41380-021-01099-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu J., Amar F., Corona C., So R.W.L., Andrews S.J., Nagy P.L.et al. (2018) Brain-derived neurotrophic factor elevates activating transcription factor 4 (ATF4) in neurons and promotes ATF4-dependent induction of Sesn2. Front. Mol. Neurosci. 11, 62 10.3389/fnmol.2018.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Takei N., Kawamura M., Hara K., Yonezawa K. and Nawa H. (2001) Brain-derived neurotrophic factor enhances neuronal translation by activating multiple initiation processes: comparison with the effects of insulin. J. Biol. Chem. 276, 42818–42825 10.1074/jbc.M103237200 [DOI] [PubMed] [Google Scholar]
- 150.Dooves S., Bugiani M., Postma N.L., Polder E., Land N., Horan S.T.et al. (2016) Astrocytes are central in the pathomechanisms of vanishing white matter. J. Clin. Invest. 126, 1512–1524 10.1172/JCI83908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Herrero M., Daw M., Atzmon A. and Elroy-Stein O. (2021) The energy status of astrocytes is the Achilles' Heel of eIF2B-Leukodystrophy. Cells 10, 1858 10.3390/cells10081858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pavitt G.D. and Proud C.G. (2009) Protein synthesis and its control in neuronal cells with a focus on vanishing white matter disease. Biochem. Soc. Trans. 37, 1298–1310 10.1042/BST0371298 [DOI] [PubMed] [Google Scholar]
- 153.Dooves S., Bugiani M., Wisse L.E., Abbink T.E.M., van der Knaap M.S. and Heine V.M. (2018) Bergmann glia translocation: a new disease marker for vanishing white matter identifies therapeutic effects of Guanabenz treatment. Neuropathol. Appl. Neurobiol. 44, 391–403 10.1111/nan.12411 [DOI] [PubMed] [Google Scholar]
- 154.European Union Clinical Trials Register (2021) A Study to Explore the Safety, Tolerability, Pharmacokinetic Profile, and Potential Efficacy of Guanabenz in Patients With Early Childhood Onset Vanishing White Matter (VWM) retrieved from. https://www.clinicaltrialsregister.eu/ctr-search/trial/2017-001438-25/NL [Google Scholar]
- 155.de Almeida R.A., Fogli A., Gaillard M., Scheper G.C., Boesflug-Tanguy O. and Pavitt G.D. (2013) A Yeast Purification System for Human Translation Initiation Factors eIF2 and eIF2Bε and Their Use in the Diagnosis of CACH/VWM Disease. PloS ONE 8, e53958 10.1371/journal.pone.0053958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fogli A. and Boespflug-Tanguy O. (2006) The large spectrum of eIF2B-related diseases. Biochem. Soc. Trans. 34, 22 10.1042/BST0340022 [DOI] [PubMed] [Google Scholar]
- 157.Li W., Wang X., Van Der Knaap M.S. and Proud C.G. (2004) Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell. Biol. 24, 3295-188 3306 10.1128/MCB.24.8.3295-3306.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Richardson J.P., Mohammad S.S. and Pavitt G.D. (2004) Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol. Cell. Biol. 24, 2352–2363 10.1128/MCB.24.6.2352-2363.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Scheper G.C., Proud C.G. and van der Knaap M.S. (2006) Defective translation initiation causes vanishing of cerebral white matter. Trends Mol. Med. 12, 159–166 10.1016/j.molmed.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 160.Wortham N.C. and Proud C.G. (2015) eIF2B: recent structural and functional insights into a key regulator of translation. Biochem. Soc. Trans. 43, 1234–1240 10.1042/BST20150164 [DOI] [PubMed] [Google Scholar]
- 161.Liu R., van der Lei H.D.W., Wang X., Wortham N.C., Tang H., van Berkel C.G.M.et al. (2011) Severity of vanishing white matter disease does not correlate with deficits in eIF2B activity or the integrity of eIF2B complexes. Hum. Mutat. 32, 103–1045 10.1002/humu.21535 [DOI] [PubMed] [Google Scholar]
- 162.Tsaytler P., Harding H.P., Ron D. and Bertolotti A. (2011) Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332, 91–94 10.1126/science.1201396 [DOI] [PubMed] [Google Scholar]
- 163.Krokowski D., Guan B.J., Wu J., Zheng Y., Pattabiraman P.P., Jobava R.et al. (2017) GADD34 function in protein trafficking promotes adaptation to hyperosmotic stress in human corneal cells. Cell Rep. 21, 2895–2910 10.1016/j.celrep.2017.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kantor L., Harding H.P., Ron D., Schiffmann R., Kaneski C.R., Kimball S.R.et al. (2005) Heightened stress response in primary fibroblasts expressing mutant eIF2B genes from CACH/VWM leukodystrophy patients. Hum. Genet. 118, 99–106 10.1007/s00439-005-0024-x [DOI] [PubMed] [Google Scholar]
- 165.Kantor L., Pinchasi D., Mintz M., Hathout Y., Vanderver A. and Elroy-Stein O. (2008) A point mutation in translation initiation factor 2B leads to a continuous hyper stress state in oligodendroglial-derived cells. PloS ONE 3, e3783 10.1371/journal.pone.0003783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Horzinski L., Kantor L., Huyghe A., Schiffmann R., Elroy-Stein O., Boespflug-Tanguy O.et al. (2010) Evaluation of the endoplasmic reticulum-stress response in eIF2B-mutated lymphocytes and lymphoblasts from CACH/VWM patients. BMC Neurol. 10, 94 10.1186/1471-2377-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Moon S.L. and Parker R. (2018) EIF2B2 mutations in vanishing white matter disease hypersuppress translation and delay recovery during the integrated stress response. RNA 24, 841–852 10.1261/rna.066563.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Carvalho K.S. (2013) Mitochondrial dysfunction in demyelinating diseases. Semin. Pediatr. Neurol. 20, 194–201 10.1016/j.spen.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 169.Dogan S.A., Pujol C., Maiti P., Kukat A., Wang S., Hermans S.et al. (2014) Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 19, 458–469 10.1016/j.cmet.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 170.Huang M.L., Sivagurunathan S., Ting S., Jansson P.J., Austin C.J., Kelly M.et al. (2013) Molecular and functional alterations in a mouse cardiac model of friedreich ataxia: activation of the integrated stress response, eIF2a phosphorylation, and the induction of downstream targets. Am. J. Clin. Pathol. 183, 745–757 10.1016/j.ajpath.2013.05.032 [DOI] [PubMed] [Google Scholar]
- 171.Mendes M.I., Gutierrez Salazar M., Guerrero K., Thiffault I., Salomons G.S., Gauquelin L.et al. (2018) Bi-allelic mutations in EPRS, encoding the glutamyl-prolylaminoacyl-tRNA synthetase, cause a hypomyelinating leukodystrophy. Am. J. Hum. Genet. 102, 676–684 10.1016/j.ajhg.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Moisoi N., Klupsch K., Fedele V., East P., Sharma S., Renton A.et al. (2009) Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 16, 449–464 10.1038/cdd.2008.166 [DOI] [PubMed] [Google Scholar]
- 173.Pereira R.O., Tadinada S.M., Zasadny F.M., Oliveira K.J., Pires K.M.P., Olvera A.et al. (2017) OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. EMBO J. 36, 2126–2145 10.15252/embj.201696179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Quirós P.M., Prado M.A., Zamboni N., D'Amico D., Williams R.W., Finley D.et al. (2017) Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 10.1083/jcb.201702058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Taylor R.W., Pyle A., Griffin H., Blakely E.L., Duff J., He L.et al. (2014) Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA 312, 68–77 10.1001/jama.2014.7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Gat-Viks I., Geiger T., Barbi M., Raini G. and Elroy-Stein O. (2015) Proteomics-level analysis of myelin formation and regeneration in a mouse model for Vanishing White Matter disease. J. Neurochem. 134, 513–526 10.1111/jnc.13142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Raini G., Sharet R., Herrero M., Atzmon A., Shenoy A., Geiger T.et al. (2017) Mutant eIF2B leads to impaired mitochondrial oxidative phosphorylation in vanishing white matter disease. J. Neurochem. 141, 694–707 10.1111/jnc.14024 [DOI] [PubMed] [Google Scholar]
- 178.Herrero M., Mandelboum S. and Elroy-Stein O. (2019) eIF2B mutations cause mitochondrial malfunction in oligodendrocytes. Neuromolecular Med. 21, 303–313 10.1007/s12017-019-08551-9 [DOI] [PubMed] [Google Scholar]
- 179.De Franco E., Caswell R., Johnson M.B., Wakeling M.N., Zung A., Dũng V.C.et al. (2020) De novo mutations in EIF2B1 affecting eIF2 Signaling cause neonatal/early-onset diabetes and transient hepatic dysfunction. Diabetes 69, 477–483 10.2337/db19-1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bogorad A.M., Lin K.Y. and Marintchev A. (2017) Novel mechanisms of eIF2B action and regulation by eIF2α phosphorylation. Nucleic Acids Res. 45, 11962–11979 10.1093/nar/gkx845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Alamri H., Al Mutairi F., Alothman J., Alothaim A., Alfadhel M. and Alfares A. (2016) Diabetic ketoacidosis in vanishing white matter. Clin. Case Rep. 4, 717–720 10.1002/ccr3.597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ozbek M.N., Senée V., Aydemir S., Kotan L.D., Mungan N.O., Yuksel B.et al. (2010) Wolcott-Rallison syndrome due to the same mutation (W522X) in EIF2AK3 in two unrelated families and review of the literature. Pediatr. Diabetes 11, 279–285 10.1111/j.1399-5448.2009.00591.x [DOI] [PubMed] [Google Scholar]
- 183.Fatani T.H. (2019) EIF2AK3 novel mutation in a child with early-onset diabetes mellitus, a case report. BMC Pediatrics 19, 85 10.1186/s12887-019-1432-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Julier C. and Nicolino M. (2010) Wolcott-Rallison syndrome. Orphanet. J. Rare Dis. 5, 29 10.1186/1750-1172-5-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Delépine M., Nicolino M., Barrett T., Golamaully M., Lathrop G.M. and Julier C. (2000) EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat. Genet. 25, 406–409 10.1038/78085 [DOI] [PubMed] [Google Scholar]
- 186.Han A., Yu C., Lu L., Fujiwara Y., Browne C., Chin G.et al. (2001) Heme-regulated eIF2α kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 20, 6909–6918 10.1093/emboj/20.23.6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang Y.L., Reis L.F., Pavlovic J., Aguzzi A., Schäfer R., Kumar A.et al. (1995) Deficient signaling in mice devoid of double‐stranded RNA‐dependent protein kinase. EMBO J. 14, 6095–6106 10.1002/j.1460-2075.1995.tb00300.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang P., McGrath B.C., Reinert J., Olsen D.S., Lei L., Gill S.et al. (2002) The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22, 6681–6688 10.1128/MCB.22.19.6681-6688.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Harding H.P., Zeng H., Zhang Y., Jungries R., Chung P., Plesken H.et al. (2001) Diabetes mellitus and exocrine pancreatic dysfunction in Perk−/− Mice reveals a role for translational control in secretory cell survival. Mol. Cell 7, 1153–1163 10.1016/S1097-2765(01)00264-7 [DOI] [PubMed] [Google Scholar]
- 190.Gilligan M., Welsh G.I., Flynn A., Bujalska I., Diggle T.A., Denton R.M.et al. (1996) Glucose Stimulates the activity of the guanine nucleotide-exchange factor eIF-2B in Isolated rat islets of langerhans. J. Biol. Chem. 271, 2121–2125 10.1074/jbc.271.4.2121 [DOI] [PubMed] [Google Scholar]
- 191.Farrell P.A., Fedele M.J., Vary T.C., Kimball S.R., Lang C.H. and Jefferson L.S. (1999) Regulation of protein synthesis after acute resistance exercise in diabetic rats. Am. J. Physiol. 276, 721–727 10.1152/ajpendo.1999.276.4.E721 [DOI] [PubMed] [Google Scholar]
- 192.Kostyak J.C., Kimball S.R., Jefferson L.S. and Farrell P.A. (2001) Severe diabetes inhibits resistance exercise-induced increase in eukaryotic initiation factor 2B activity. J. App. Physiol. 91, 79–84 10.1152/jappl.2001.91.1.79 [DOI] [PubMed] [Google Scholar]
- 193.McManus E.J., Sakamoto K., Armit L.J., Ronaldson L., Shpiro N., Marquez R.et al. (2005) Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. EMBO J. 24, 1571–1583 10.1038/sj.emboj.7600633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cnop M., Toivonen S., Igoillo-Esteve M. and Salpea P. (2017) Endoplasmic reticulum stress and eIF2α phosphorylation: The Achilles heel of pancreatic β cells. Mol. Metab. 6, 1024–1039 10.1016/j.molmet.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]