Abstract
Life-threatening ‘breakthrough’ cases of critical COVID-19 are attributed to poor or waning antibody response to the SARS-CoV-2 vaccine in individuals already at risk. Pre-existing autoantibodies (auto-Abs) neutralizing type I IFNs underlie at least 15% of critical COVID-19 pneumonia cases in unvaccinated individuals; however, their contribution to hypoxemic breakthrough cases in vaccinated people remains unknown. Here, we studied a cohort of 48 individuals (age 20-86 years) who received 2 doses of an mRNA vaccine and developed a breakthrough infection with hypoxemic COVID-19 pneumonia 2 weeks to 4 months later. Antibody levels to the vaccine, neutralization of the virus, and auto-Abs to type I IFNs were measured in the plasma. Forty-two individuals had no known deficiency of B cell immunity and a normal antibody response to the vaccine. Among them, ten (24%) had auto-Abs neutralizing type I IFNs (aged 43-86 years). Eight of these ten patients had auto-Abs neutralizing both IFN-α2 and IFN-ω, while two neutralized IFN-ω only. No patient neutralized IFN-β. Seven neutralized 10 ng/mL of type I IFNs, and three 100 pg/mL only. Seven patients neutralized SARS-CoV-2 D614G and the Delta variant (B.1.617.2) efficiently, while one patient neutralized Delta slightly less efficiently. Two of the three patients neutralizing only 100 pg/mL of type I IFNs neutralized both D61G and Delta less efficiently. Despite two mRNA vaccine inoculations and the presence of circulating antibodies capable of neutralizing SARS-CoV-2, auto-Abs neutralizing type I IFNs may underlie a significant proportion of hypoxemic COVID-19 pneumonia cases, highlighting the importance of this particularly vulnerable population.
Type I IFN auto-Abs are found in 20% of hypoxemic, mRNA vaccinated COVID-19 patients despite SARS-CoV-2 neutralizing antibodies.
INTRODUCTION
Since the start of the coronavirus disease 19 (COVID-19) pandemic ( 1 ), caused by severe respiratory syndrome coronavirus 2 (SARS-CoV-2), at least 6 million people have died from COVID-19 ( 2 ). Although the majority of infected individuals recover, it remains important to identify factors that put patients at greater risk for severe disease. Age is the major epidemiological risk factor of death from pneumonia, the risk doubling every five years of age from childhood onward ( 3 – 5 ). Patients with inborn errors (IE) of immunity affecting the production of, or response to type I IFNs, or both, are prone to critical COVID-19 pneumonia ( 6 – 8 ). These findings established the crucial role of type I IFNs in fending off SARS-CoV-2 ( 9 ). Moreover, auto-Abs neutralizing high concentrations (10 ng/mL in plasma diluted 1/10) of IFN-α2 and/or IFN-ω were found in at least 10% of individuals with critical COVID-19 ( 10 ), an observation replicated in various regions of the world ( 11 – 21 ). Patients with autoimmune polyendocrine syndrome type I (APS-1) harbor these neutralizing auto-Abs from early childhood and are at high risk of life-threatening COVID-19 ( 20 , 21 ). Moreover, at least 13.6% of unvaccinated patients with critical COVID-19 had auto-Abs neutralizing lower, more physiological concentrations (100 pg/mL in plasma diluted 1/10) of IFN-α2 and/or IFN-ω, while auto-Abs neutralizing IFN-β were found in another 1% of patients ( 22 ). In more than 34,000 uninfected individuals aged 18 to 100 years, the prevalence of auto-Abs neutralizing 10 ng/mL (or 100 pg/mL) of IFN-α2 or IFN-ω increased significantly with age, with 0.17% (1.1%) of individuals positive for these auto-Abs under 70 years old, and more than 1.4% (4.4%) positive over 70 years old, consistent with the higher risk of life-threatening COVID-19 in the elderly population ( 22 ). These auto-Abs thus precede infection and are strong determinants of critical disease, only second to age among common risk factors ( 23 ). The odds ratios (ORs) of critical disease are the highest in individuals with auto-Abs neutralizing 10 ng/mL of both IFN-α2 and IFN-ω (OR = 67; p-value = 7.8x10−13) ( 22 , 23 ).
RNA vaccines are highly effective at protecting against severe COVID-19 pneumonia ( 24 , 25 ). Despite their efficacy, ‘breakthrough’ cases, i.e., individuals diagnosed with SARS-CoV-2 infection despite being vaccinated with 2 doses, have been reported worldwide ( 26 , 27 ). Most breakthrough cases are asymptomatic or mild ( 26 ), but in rare cases they are severe, critical, or even fatal ( 28 , 29 ). It is thought that these severe or critical cases can result from a pathologically deficient (including inherited and acquired deficiencies of adaptive immunity) or a physiologically waning antibody response to the vaccine (especially in aging individuals). Incomplete protection from viral genotypes with vaccine-resilient mutations (such as Delta or Omicron), can also result in insufficient viral neutralization in vivo, in individuals otherwise at risk of hypoxemic pneumonia (for example, due to their age, sex, co-morbidity, rare or common genetic variant, or auto-Abs to type I IFNs) ( 30 ). In other words, breakthrough critical cases are thought to be due to a poor antibody response to the vaccine in at-risk individuals ( 31 ). Yet, the human genetic and immunological determinants of critical ‘breakthrough’ cases remain unclear, especially in patients with normal antibody response to the vaccine. Moreover, the biological and clinical efficacy of RNA vaccines in patients with known genetic or immunological determinants of critical COVID-19 pneumonia, i.e., in patients with IE of, or auto-Abs to type I IFNs, is not clear. With the COVID Human Genetic Effort (CHGE, www.covidhge.com), we recruited and tested patients with breakthrough COVID-19 and hypoxemic pneumonia. We tested the double hypothesis that some of these breakthrough cases of severe or critical COVID-19 pneumonia may have a normal antibody response to the vaccine and may also harbor auto-Abs to type I IFNs.
RESULTS
Fourty-two of 48 patients have normal antibody response to the vaccine
Forty-eight patients who suffered from hypoxemic COVID-19 pneumonia (severe or critical), despite having received 2 doses of mRNA vaccine, at least 2 weeks and up to 16 weeks (mean: 8 weeks) before infection were recruited from 6 countries (France, Greece, North Macedonia, Turkey, Ukraine, and United States of America). All COVID Human Genetic Effort (CHGE) patients whose samples were available were recruited; they had not been previously infected with SARS-CoV-2, as attested by the clinical information collected and/or a negative serology at the time of vaccination or performed at the onset of disease. These patients were aged 20 to 86 years (mean 53 years old) and included 34 men and 14 women. Five of them had a known deficiency of B cell immunity (immunosuppressive therapy in 3 individuals, and HIV infection in 1, and lymphoma with CAR-T cell treatment in one). We tested the 48 patients for their antibody response to SARS-CoV-2 mRNA vaccines. We found one of the 43 patients did not have a known B cell deficiency, but had an insufficient antibody response to the vaccine (defined as within 3 standard deviations from the mean of unvaccinated controls) (Arrow, Fig. 1A, S1A). The other patients had levels of antibody response to the vaccine similar to those of vaccinated controls (t-test, Supplementary Table 1). Of note, 3 of the 5 patients with a known B cell deficiency had a normal antibody response (above 3 standard deviations) (Fig. 1A). Overall, 42 patients had both no B cell deficiency and a normal antibody response to the vaccine, thus were further investigated.
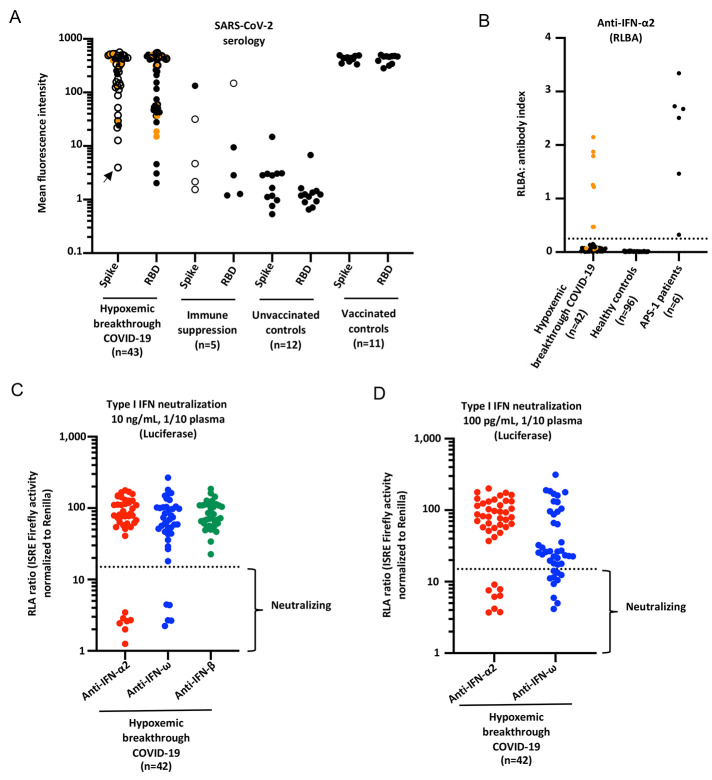
Fig. 1.
Neutralizing auto-antibodies (Abs) against IFN-α2 and IFN-ω in patients with hypoxemic breakthrough COVID-19 despite a normal serological response to SARS-CoV-2 mRNA vaccine. (A) SARS-CoV-2 serology against spike(S)-protein and receptor binding domain (RBD) in hypoxemic breakthrough COVID-19 (N=43), patients with immune suppression (n=5), unvaccinated controls (N=12), and vaccinated and uninfected healthy controls (n=11). Mean fluorescence intensity is shown. The orange dots correspond to the 10 individuals with auto-Abs neutralizing type I IFNs. Empty circles represent either Spike or RBD serology, to outline the highest value for one patient. The arrow represents the patient without B cell deficiency but with an insufficient Ab response to the virus. (B) Radioligand binding assay (RLBA) results for auto-Abs against IFN-α2 in patients with hypoxemic breakthrough COVID-19 pneumonia without immune suppression or low Ab response to the vaccine (N=42), uninfected controls (N=96), and uninfected APS-1 patients (N=6). (C) Neutralization of 10 ng/mL IFN-α2, IFN-ω or IFN-β in the presence of plasma 1/10 from patients with hypoxemic breakthrough COVID-19 pneumonia with a good Ab response to the vaccine (N=42). Relative luciferase activity is shown (ISRE dual luciferase activity, with normalization against Renilla luciferase activity) after stimulation with 10 ng/mL IFN-α2 or IFN-ω in the presence of plasma 1/10. RLA: relative luciferase activity. (D) Neutralization of 100 pg/mL IFN-α2 or IFN-ω in the presence of plasma 1/10 from patients with hypoxemic breakthrough COVID-19 pneumonia with a good Ab response to the vaccine (N=42).
Auto-Abs against type I IFNs in 10 of 42 patients with normal Ab response to the vaccine
We next tested all the samples from the 42 patients without known B cell deficiency and with a normal Ab response to the mRNA vaccine for IgG auto-Ab to type I IFN levels using a radioligand binding assay (RLBA). Seven of 42 patients tested had elevated titers of anti-IFN-α2 auto-Abs in RLBA (Fig. 1B). We then tested all these samples for their neutralization activity against IFN-α2, IFN-, and IFN-β at 10 ng/mL, 100 pg/mL, and 10 ng/mL respectively. We identified ten (24%) patients with IgG auto-Abs neutralizing IFN-α2 and/or IFN-ω, as did the APS-1 positive controls, while the healthy controls did not (Fig. 1C, D). Patients with neutralizing auto-Abs have lower luciferase induction (below threshold in dotted lines). All these patients had normal anti-SARS-CoV-2 Spike antibody response to the vaccine (Fig. S1D, E). In contrast, auto-Abs to type I IFN were not found in any of the 6 patients previously excluded because of a known B cell immunodeficiency (n=5) or an insufficient antibody response to the vaccine (n=1) (Fig. S1B, C). Of note, 8 of these 10 individuals (80%) had circulating auto-Abs neutralizing both IFN-α2 and IFN-ω, while two neutralized IFN-ω only (20%), and none neutralized IFN-β (Fig. 1C-D and Table 2). In addition, plasma from 7 patients (diluted 1/10) neutralized a high concentration (10 ng/mL) of type I IFNs (70%), while 3 neutralized only the lower, more physiological, dose (100 pg/mL) of type I IFNs (including the 2 neutralizing IFN-ω only) (30%) (Fig. 1C, D and Table 2). Overall, auto-Abs neutralizing IFN-α2 and/or IFN-ω were found at the onset of disease in 10 of 42 patients (24%) with breakthrough COVID-19 who suffered from hypoxemic pneumonia, despite having a normal antibody response to an mRNA vaccine.
Table 2. Auto-Abs neutralized in the 10 patients.
1: neutralizing. 0: non-neutralizing.
| Patient | anti-IFN-α2 auto-Abs (10 ng/mL) | anti-IFN-β auto-Abs (10 ng/mL) | anti-IFN-ω, auto-Abs (10 ng/mL) | anti-IFN-α2 auto-Abs (100 pg/mL) | anti-IFN-ω, auto-Abs (100 pg/mL) |
| P1 | 1 | 0 | 1 | 1 | 1 |
| P2 | 1 | 0 | 0 | 1 | 1 |
| P3 | 1 | 0 | 0 | 1 | 1 |
| P4 | 0 | 0 | 0 | 0 | 1 |
| P5 | 1 | 0 | 1 | 1 | 1 |
| P6 | 0 | 0 | 0 | 1 | 1 |
| P7 | 0 | 0 | 0 | 0 | 1 |
| P8 | 1 | 0 | 1 | 1 | 1 |
| P9 | 1 | 0 | 1 | 1 | 1 |
| P10 | 1 | 0 | 1 | 1 | 1 |
Demographic, clinical, and virological features of the 10 patients with auto-Abs to type I IFNs
The patients with hypoxemic breakthrough COVID-19 pneumonia and auto-Abs neutralizing type I IFNs included three women and seven men. They were aged 43 to 86 years old (mean: 75 years old) (Table 1). All were of European ancestry, except one Cambodian, and they originated from France (n=3), Greece (n=5), and the USA (n=2). None of these individuals reported having previously suffered from other severe viral infections. All 10 patients were hospitalized during COVID-19 for oxygen supplementation, including 5 hospitalized in an intensive care unit (ICU) who received mechanical ventilation, and one who received nasal oxygen high flow therapy but was recused of ICU because of age (P8). All of them survived. All presented with bilateral COVID-19 pneumonia and had a positive SARS-CoV-2 RT-PCR in the respiratory tract. The SARS-CoV-2 variants involved were unknown but most likely to be Delta variant, given the epidemiology at the location and time of sampling (i.e., before October 2021 for all samples tested). They had been vaccinated 2 to 16 weeks prior to the diagnosis of COVID-19. Of note, one individual (P2) had at least two auto-immune conditions (myasthenia gravis and Hashimoto’s thyroiditis), while another (P10) had APS-1. Myasthenia gravis and APS-1 are associated with auto-Abs to type I IFNs, which had however not been measured prior to COVID-19 in these two individuals. Finally, one individual (P1) belonged to a large family, whose members had all been fully vaccinated, and many were infected at the same time as he did ( 32 ). He was nevertheless the only one to suffer from critical disease, and also the only one to harbor neutralizing auto-Abs to type I IFNs. None of the 10 patients died of COVID-19, while more than 20% of unvaccinated individuals who died of COVID-19 harbored neutralizing auto-Abs ( 22 ) and 5-10% of unvaccinated patients with these auto-Abs died of COVID-19 ( 23 ), suggesting that although insufficient to prevent hypoxemic pneumonia, vaccination may have protected these patients from a fatal outcome. Overall, auto-Abs to type I IFNs can underlie hypoxemic breakthrough COVID-19 infection in previously healthy individuals who developed normal antibody responses after SARS-CoV-2 mRNA vaccination.
Table 1. Clinical and demographic information of the 10 patients with hypoxemic breakthrough COVID-19 infection and auto-Abs neutralizing type I IFNs.
HTN: hypertension, AF: atrial fibrillation. APS-1: auto-immune polyendocrine syndrome type 1.
| Patient | Origin | Residence | Sex | Age | Comorbidities | Vaccine source | Doses number | Time of disease post vaccination (weeks) | ICU | Classification | Outcome |
| P1 | American | USA | M | 80 | Diabetes, asthma | Pfizer | 2 | 2 | Yes | Critical | Alive |
| P2 | Greek | Greece | F | 82 | HTN, myasthenia gravis, hashimoto, dyslipidemia | Pfizer | 2 | 4 | Yes | Critical | Alive |
| P3 | Greek | Greece | M | 73 | HTN, diabetes, dyslipidemia, glaucome | Pfizer | 2 | 2 | Yes | Critical | Alive |
| P4 | Greek | Greece | M | 86 | HTN, diabetes, dyslipidemai, AF, benign prostate hyperplasia, parkinson | Pfizer | 2 | 12 | Yes | Critical | Alive |
| P5 | Greek | Greece | M | 73 | Diabetes, coronary heart disease | Pfizer | 2 | 3 | No | Severe | Alive |
| P6 | Greek | Greece | F | 77 | HTN, diabetes, dyslipidemia | Pfizer | 2 | 16 | No | Severe | Alive |
| P7 | Cambodian | France | M | 71 | HTN | Pfizer | 2 | 15 | Yes | Critical | Alive |
| P8 | French | France | F | 86 | NA | Pfizer | 2 | 6 | No | Critical | Alive |
| P9 | American | USA | M | 80 | NA | Pfizer | 2 | 2 | No | Critical | Alive |
| P10 | French | France | M | 43 | APS-1 | Pfizer | 2 | 2 | No | Severe | Alive |
Antibodies neutralizing SARS-CoV-2 in all 10 patients
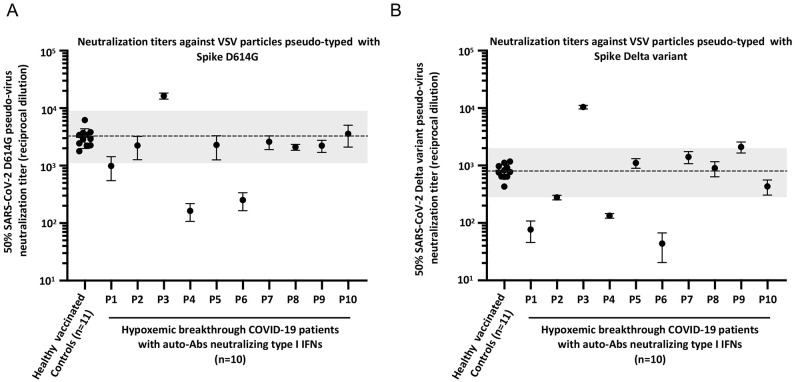
To further test the hypothesis that the hypoxemic breakthrough cases were driven by the auto-Abs neutralizing type I IFNs and not by an insufficient antibody response to the vaccine, we assessed the neutralizing activity in all 10 patients’ plasma against SARS-CoV-2. Although we did not collect blood samples prior to COVID-19 diagnosis, we collected them in the first 3 days of hospitalization. As we did not determine with which viral strain the patients had been infected, we performed the neutralization assay with pseudoviruses representing both the previously globally dominant D614G strain and the Delta variant (B.1.617.2), which was dominant when and where the patients were infected. We compared the patients’ results with the neutralization titers of healthy vaccinated donors 2-8 weeks after the 2nd dose of the mRNA vaccine. All 10 individuals tested had a neutralization capacity, when compared with the healthy vaccinated controls, although it was slightly reduced for 2 individuals (P4 and P6) for the D614G strain and for 3 individuals (P1, P4 and P6) for the Delta variant (Fig. 2A, B, S1D, E). Although P1 neutralized 10 ng/mL of type I IFNs, P4 and P6 only neutralized low concentrations of type I IFNs. Specifically, P4 neutralized both IFN-α2 and IFN-ω but only at 100 pg/mL, while P6 neutralized only IFN-ω at 100 pg/mL. This observation suggests that in patients whose auto-Abs neutralized only low concentrations of type I IFNs, sub-optimal antibody response to the vaccine may have also contributed to hypoxemic pneumonia. Overall, this suggested that hypoxemic COVID-19 pneumonia can occur in individuals with a normal antibody reponse to two doses of mRNA vaccine (42 of 48 patients tested). Moreover, in about 20% of the beakthrough cases (10 of 42 cases), hypoxemic pneumonia was probably due to auto-Abs neutralizing IFN-α2 and/or IFN-ω (and typically at high concentration of both IFNs). Finally, 70% of the latter cases (7 of 10 cases), plasma neutralization of two viral strains was normal, while one had a lower neutralization against the delta strain, and the remaining 2 had a subnormal neutralization of both viral strains (D614G, and Delta).
Fig. 2.
Neutralization titers against SARS-CoV-2 in the patients with auto-Abs against type I IFNs. Neutralization titers against SARS-CoV-2 for healthy vaccinated donors 2-8 weeks after the second dose of mRNA vaccine (n=11), and patients with hypoxemic breakthrough COVID-19 pneumonia and auto-Abs to type I IFNs (n=10). The dashed line shows the geometric mean of healthy donor titers, the box shows interquartile range, and the shaded region is the full range. (A) Neutralization assay performed with pseudoviruses representing the D614G strain, and (B) the Delta variant (B.1.617.2).
DISCUSSION
The pathogenesis of life-threatening COVID-19 pneumonia involves two steps, with a deficiency of respiratory type I IFN immunity in the first days of infection resulting in viral spread, which triggers excessive systemic and pulmonary inflammation ( 30 , 33 , 34 ). The vaccination of billions of individuals has efficiently reduced the number of critical cases. Nevertheless, breakthrough hypoxemic COVID-19 pneumonia can occur in previously healthy individuals who are vaccinated against SARS-CoV-2, which is assumed to be due to a poor antibody response to the vaccine ( 31 ). Our findings suggest that most breakthrough hypoxemic cases (42 of 48 tested) did not have a known B cell deficiency and also had a normal antibody response to the vaccine, although no samples were available before SARS-CoV-2 infection. Moreover, we showed that about 20% (10 of 42) of these breakthrough cases with normal antibody response to the vaccine also carried auto-Abs neutralizing IFN-α2 and/or IFN-ω (10 ng/mL for 7 patients and 100 pg/mL for 3 patients). In addition, the plasma of 7 of the 10 patients with auto-Abs to type I IFNs efficiently neutralized SARS-CoV-2 in vitro, while one had a lower neutralization against the delta strain, and plasma from the remaining 2 neutralized the two viral strains tested sub-optimally. Both patients had auto-Abs neutralizing only 100 pg/mL of type I IFNs. Plasma (diluted 1/10) from seven of the 10 individuals with these auto-Abs neutralized a high concentration (10 ng/mL) of both IFN-α2 and IFN-ω, consistent with unvaccinated individuals carrying such auto-Abs being at the greatest risk of critical COVID-19 among individuals carrying any combinations of auto-Abs to type I IFNs ( 22 , 23 , 30 ). The proportion of individuals with hypoxemic COVID-19 due to neutralizing both IFN-α2 and IFN-ω at the high dose (10 ng/mL) is even higher in the breakthrough cohort reported here (7 of 42, 16%) than in the previously described unvaccinated cohort (175 of 3,136, 7.1%) (P = 0.015) ( 22 ). Two of the 3 patients neutralizing only 100 pg/mL of type I IFNs, also had a slightly diminished neutralization capacity against SARS-CoV-2, suggesting in these individuals a combination of 2 factors: the presence of auto-Abs to low concentration of type I IFNs, and a suboptimal antibody response to the vaccine.
Nevertheless, as we were not able to identify and study auto-Ab positive individuals who were vaccinated and efficiently protected against severe infection, we cannot estimate the percentage of breakthrough cases with hypoxemic pneumonia in individuals with auto-Abs neutralizing type I IFNs infected with SARS-CoV-2. Until 70 years old, the proportion of individuals from the general population sampled prior to the pandemic that carry auto-Abs against both IFN-α2 and IFN-ω is 0.02% and 0.03% for the neutralization of 10 ng/mL and 100 pg/mL, respectively, while it reaches 0.6% and 1.6% over 70 years old. As mRNA vaccines have high efficacy to prevent critical pneumonia, it is probable that most patients with auto-Abs against type I IFNs benefit from vaccination, although the protection might not be sufficient in individuals neutralizing high concentrations of multiple type I IFNs. It is also not unreasonable to speculate that, despite an infection with a vaccine-covered viral variant and a normal antibody response to the vaccine, a small proportion of the patients with such auto-Abs might not be fully protected by the vaccine, especially if infected with a high viral inoculum. By inference from previous studies, the auto-Abs of the 8 patients neutralizing IFN-α2 also probably neutralizes the 13 types of IFN-α ( 10 , 20 , 22 , 35 , 36 ). These findings suggest that a potent post-vaccine humoral immunity can be insufficient to fight SARS-CoV-2 infection, especially in patients with auto-Abs neutralizing both IFN-α2 and IFN-ω, and even more so at high concentration.
Our results here suggest it may be beneficial to test for auto-Abs to type I IFN in vaccinated patients diagnosed with breakthrough COVID-19 pneumonia of varying severity. Testing uninfected people, including vaccinated individuals, may also be considered, especially in those over 70 years old given the high prevalence of auto-Abs to type I IFNs in this population (>4%) and their lower global type I IFN immunity ( 30 , 36 ). One of the 10 patients suffered from APS-1 and thus most likely harbored these auto-Abs since early childhood ( 20 , 21 , 37 ), while another patient had myasthenia gravis, which is also commonly associated with these auto-Abs ( 38 ). Testing patients with conditions known to be associated with these auto-Abs may benefit these patients. All individuals with auto-Abs to IFNs might benefit not only from vaccine boosters but perhaps from recurrent vaccinations. Prospective studies assessing vaccine-induced immunity before infection in patients with auto-Abs to type I IFNs would be informative, for example in the setting of vaccine trials. Systematic screening at hospital admission for auto-Abs to type I IFNs would also be of help for the management of vaccinated or unvaccinated individuals with hypoxemic pneumonia. Indeed, monoclonal antibodies (mAbs) neutralizing the virus could also be administered promptly ( 39 ), as shown for an IRF9-deficient patient ( 40 ), especially in patients with the highest titers of auto-Abs to type I IFNs. Anti-viral compounds, such as remdesivir ( 41 , 42 ) or molnupiravir ( 43 ), may also benefit these patients if administered early in the course of infection. Conversely, in ambulatory patients with these auto-Abs, early recombinant IFN-β therapy may also be considered, to prevent the development of hypoxemic pneumonia ( 44 ). In sum, our findings indicate that auto-Abs to type I IFNs is a susceptibility factor for a severe clinical course of COVID-19 even in vaccinated subjects with a breakthrough infection.
MATERIALS AND METHODS
Study Design
We enrolled 48 patients with proven hypoxemic COVID-19 pneumonia, 12 unvaccinated controls, and 11 vaccinated controls from 6 countries in this study. We collected plasma or serum samples for all these individuals to test for the presence of IgG Abs against SARS-CoV-2 and auto-Abs to type I IFNs by immuno-assay. All individuals were recruited according to protocols approved by local Institutional Review Boards (IRBs).
COVID-19 classification
The severity of COVID-19 was assessed for each patient as follows ( 6 , 10 ): “critical COVID-19 pneumonia” was defined as pneumonia developing in patients with critical disease, whether pulmonary, with high-flow oxygen, mechanical ventilation (continuous positive airway pressure, bilevel positive airway pressure, intubation), septic shock, or with damage to any other organ requiring admission to the intensive care unit. “Severe COVID-19” was defined as pneumonia developing in patients requiring low-flow oxygen (<6L/min). The controls were individuals infected with SARS-CoV-2 (as demonstrated by a positive PCR and/or serological test and/or displaying typical symptoms, such as anosmia/ageusia after exposure to a confirmed COVID-19 case) who remained asymptomatic or developed mild, self-healing, ambulatory disease with no evidence of pneumonia.
Statistics
For comparison of groups in Fig. 1a, a two-sided t test was performed using a Python library (SciPy) for both Spike and RBD. Briefly, all groups were compared to the unvaccinated control group (n=12). In addition, the group of auto-Ab positive breakthrough cases were compared to the group of auto-Ab negative breakthrough cases.
Detection of anti-cytokine auto-Abs by a high throughput automated ELISA (Gyros)
Cytokines, recombinant human (rh)IFN-α2 (Milteny Biotec, ref. number 130-108-984) or rhIFN-ω (Merck, ref. number SRP3061), were first biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific, cat. number A39257), according to the manufacturer’s instructions, with a biotin-to-protein molar ratio of 1:12. The detection reagent contained a secondary antibody Alexa Fluor 647 goat anti-human IgG (Thermo Fisher Scientific, ref. number A21445) diluted in Rexip F (Gyros Protein Technologies, ref. number P0004825; 1/500 dilution of the 2 mg/mL stock to yield a final concentration of 4 μg/mL). Buffer PBS-T 0.01% and Gyros Wash buffer (Gyros Protein Technologies, ref. number P0020087) were prepared according to the manufacturer’s instructions. Plasma or serum samples were then diluted 1/100 in PBS-T 0.01% and tested with the Bioaffy 1000 CD (Gyros Protein Technologies, ref. number P0004253), and the Gyrolab X-Pand (Gyros Protein Technologies, ref. number P0020520). Cleaning cycles were performed in 20% ethanol.
RLBA for anti-IFN-α2 auto-Ab detection
A DNA plasmid containing full-length cDNA sequence with a Flag-Myc tag (OriGene, #RC221091) was verified by Sanger sequencing and used as template in T7-promoter–based in vitro transcription/translation reactions (Promega, #L1170) using [S35]-methionine (PerkinElmer, #NEG709A). IFN-α2 protein was column-purified using NAP-5 columns (GE Healthcare, #17-0853-01); incubated with 2.5 μl of serum, 2.5 μl of plasma, or 1 μl of anti-myc–positive control antibody (Cell Signaling Technology, #2272); and immunoprecipitated with Sephadex protein A/G beads (4:1 ratio; Sigma-Aldrich, #GE17-5280-02 and #GE17-0618-05) in 96-well polyvinylidene difluoride filtration plates (Corning, #EK-680860). The radioactive counts [counts per minute (cpm)] of immunoprecipitated protein were quantified using a 96-well MicroBeta TriLux liquid scintillation plate reader (PerkinElmer). Antibody index for each sample was calculated as follows: (sample cpm value – mean blank cpm value)/(positive control antibody cpm value – mean blank cpm value). For the COVID-19 patient and CCP cohorts, a positive signal was defined as greater than 6 standards deviations above the mean of pre–COVID-19 blood bank non-inflammatory controls.
Functional evaluation of anti-cytokine auto-Abs by luciferase reporter assays
The blocking activity of anti-IFN-α2 and anti-IFN-ω auto-Abs was determined with a reporter luciferase activity. Briefly, HEK293T cells were transfected with a plasmid containing the Firefly luciferase gene under the control of the human ISRE promoter in the pGL4.45 backbone, and a plasmid constitutively expressing Renilla luciferase for normalization (pRL-SV40). Cells were transfected in the presence of the X-tremeGene9 transfection reagent (Sigma-Aldrich, ref. number 6365779001) for 24 hours. Cells in Dulbecco’s modified Eagle medium (DMEM, Thermo Fisher Scientific) supplemented with 2% fetal calf serum (FCS) and 10% healthy control or patient serum/plasma (after inactivation at 56°C, for 20 min) were either left unstimulated or were stimulated with IFN-α2 (Milteny Biotech, ref. number 130-108-984), IFN-ω (Merck, ref. number SRP3061), at 10 ng/mL or 100 pg/mL, or IFN-β (Milteny Biotech, ref. number: 130-107-888) at 10 ng/mL, for 16 hours at 37°C. Each sample was tested once for each cytokine and dose. Finally, cells were lysed for 20 min at room temperature and luciferase levels were measured with the Dual-Luciferase® Reporter 1000 assay system (Promega, ref. number E1980), according to the manufacturer’s protocol. Luminescence intensity was measured with a VICTOR-X Multilabel Plate Reader (PerkinElmer Life Sciences, USA). Firefly luciferase activity values were normalized against Renilla luciferase activity values. These values were then normalized against the median induction level for non-neutralizing samples, and expressed as a percentage. Samples were considered neutralizing if luciferase induction, normalized against Renilla luciferase activity, was below 15% of the median values for controls tested the same day.
SARS-CoV-2 serological studies
Serum collection
Control serum was collected under informed consent from healthy recipients of BNT162b2 vaccine (vaccines based on the Wuhan spike protein -S protein- sequence), which were confirmed to have no prior SARS-CoV-2 infection by anti-SARS-CoV-2 nucleocapsid (N protein) IgG assay ( 45 ). All serum samples were heat inactivated at 56°C for 30 min prior to neutralization experiments.
Luminex Assay
Luminex immunoassays for SARS-CoV-2 serology studies were performed as previously described using proteins from the Wuhan strain of the virus ( 46 ). Briefly, whole N protein, trimeric Spike ectodomain (residues 1-1213) and receptor binding domain (residues 328-533, all generously provided by Dr. John Pak, Chan Zuckerberg Biohub) were each conjugated to a unique spectrally encoded bead using manufacturer instructions (Luminex Antibody Coupling Kit, #40-50016) with 5 μg of protein per 1 million beads. All beads were blocked overnight before use in PBST supplemented with 0.1% BSA and pooled on day of use. 2000-2500 beads per ID were pooled per replicate. Patient serum or plasma was incubated with beads at a final dilution of 1:250 for 1 hour, washed twice in PBST, stained with an anti-IgG (human) pre-conjugated to phycoerythrin (Thermo Scientific, #12-4998-82) for 30 min at 1:2000, then washed thrice in PBST. Primary incubations were done in PBST supplemented with 2% nonfat milk and secondary incubations were done in PBST. Beads were processed in duplicate in 96 well format and analyzed on a Luminex LX 200 cytometer. Median Fluorescence Intensity from each set of beads within each bead ID were retrieved directly from the LX200 after normalizing to the intra-assay negative controls (Bovine Serum Albumin (BSA) conjugated beads).
Pseudovirus production
SARS-CoV-2 pseudoviruses were generated using a previously described recombinant vesicular stomatitis virus expressing GFP in place of the VSV glycoprotein (rVSV∆G-GFP) ( 47 ). The SARS-CoV-2 spike gene bearing the D614G mutation or the set of mutations in the B.1.617.2/Delta variant (T19R, T95I, G142D, ∆157-158, L452R, T478K, P681R, D614G, D950N) were cloned in a CMV-driven expression vector and used to produce SARS-CoV-2 spike reporter pseudoviruses. Pseudoviruses were titered on Huh7.5.1 cells overexpressing ACE2 and Transmembrane protease, serine 2 (TMPRSS2) (gift of Andreas Puschnik) using GFP expression to measure the concentration of focus forming units (ffu).
Pseudovirus neutralization experiments
Huh7.5.1-ACE2-TMPRSS2 cells were seeded in 96-well plates at a density of 7000 cells/well one day prior to pseudovirus inoculation. Cells were verified to be free of mycoplasma contamination with the MycoAlert Mycoplasma detection kit (Lonza). Serum samples were diluted into complete culture media (DMEM with 10% FBS, 10mM HEPES, 1x Pen-Strep-Glutamine) using the LabCyte Echo 525 liquid handler and 1500 ffu of SARS-CoV-2 pseudovirus was added to each well to reach final dilutions ranging from 1:20-1:10240, including no-serum and no-pseudovirus controls. Serum/pseudovirus mixtures were incubated at 37°C for 1h before being added directly to cells. Cells inoculated with serum/pseudovirus mixtures were incubated at 37°C and 5% CO2 for 24h, resuspended using 10x TrypLE Select (Gibco), and cell fluorescence was measured with the BD Celesta flow cytometer. All neutralization assays were repeated for a total of three independent experiments with each experiment containing two technical replicates for each condition. Flow cytometry data was analyzed with FlowJo to determine the percentage of cells transduced with pseudovirus (GFP-positive). Percent neutralization for each serum dilution was calculated by normalizing GFP-positive cell percentage to no-serum control wells. Fifty percent neutralization titers (NT50) were calculated from ten-point response curves generated in GraphPad Prism 7 using four-parameter logistic regression.
Acknowledgments
We thank the patients and their families for placing their trust in us. We warmly thank the members of both branches of the Laboratory of Human Genetics of Infectious Diseases. We warmly thank Y. Nemirovskaya, M. Woollett, D. Liu, S. Boucherit, C. Rivalain, M. Chrabieh and L. Lorenzo for administrative assistance. We warmly thank Helen C. Su for fruitful discussions. We warmly thank Yuna Muyshondt and Suzanne Sandmeyer for their assistance.
Funding: The Laboratory of Human Genetics of Infectious Diseases is supported by the Howard Hughes Medical Institute, the Rockefeller University, the St. Giles Foundation, the National Institutes of Health (NIH) (R01AI088364 and R01AI163029), the National Center for Advancing Translational Sciences (NCATS), NIH Clinical and Translational Science Award (CTSA) program (UL1 TR001866), a Fast Grant from Emergent Ventures, Mercatus Center at George Mason University, the Fisher Center for Alzheimer’s Research Foundation, the Meyer Foundation, the JPB Foundation, the program “Investissement d’Avenir” launched by the French Government and implemented by the Agence Nationale de la Recherche (ANR) with references ANR-21-RHUS-08 and ANR-10-IAHU-01, the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (FRM) (EQU201903007798),, the ANRS-COV05, ANR GENVIR (ANR-20-CE93-003),ANR AABIFNCOV (ANR-20-CO11-0001) and ANR GenMISC (ANR-21-COVR-0039) projects, the European Union’s Horizon 2020 research and innovation program under grant agreement No 824110 (EASI-genomics), the Square Foundation, Grandir - Fonds de solidarité pour l’enfance, the Fondation du Souffle, the SCOR Corporate Foundation for Science, The French Ministry of Higher Education, Research, and Innovation (MESRI-COVID-19), Institut National de la Santé et de la Recherche Médicale (INSERM), REACTing-INSERM and the University of Paris. CRG, RPD and CF were funded by Instituto de Salud Carlos III (COV20_01333, COV20_01334), the Spanish Ministry of Science and Innovation (RTC-2017-6471-1; AEI/FEDER, UE), the Fundación Canaria Instituto de Investigación Sanitaria de Canarias (FIISC19/43), Grupo DISA (OA18/017), Fundación MAPFRE Guanarteme (OA21/131), and Cabildo Insular de Tenerife (CGIEU0000219140 and “Apuestas científicas del ITER para colaborar en la lucha contra la COVID-19”).EA is supported by the Hellenic Foundation for Research and Innovation (INTERFLU, no. 1574) and the European Commission’s Horizon 2020 research and innovation program (CURE, grant no. 767015 and IMMUNAID, grant no. 779295). LR and LFP were supported by Singapore National Medical Research Council COVID-19 Research Fund (COVID19RF-001; COVID19RF-007; COVID19RF-0008; COVID19RF-060) and A*STAR COVID-19 Research funding (H/20/04/g1/006).The UCSF COMET Consortium is supported by the University of California, San Francisco Clinical and Translational Science Institute and the National Institutes of Health (U19AI077439). AA, GRB and XS are funded by the 202115-31 project, funded by La Marató de TV3. L.D.N. was supported by the intramural Research Program of the NIAID, NIH. J.L.D. is supported by the Chan Zuckerberg Biohub. The authors wish to acknowledge the support of the Chao Family Comprehensive Cancer Center Experimental Tissue Shared Resource, supported by the National Cancer Institute of the National Institutes of Health under award number P30CA062203. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. PB was supported by the French Foundation for Medical Research (FRM, EA20170638020). PB and TLV were supported by the MD-PhD program of the Imagine Institute (with the support of the Fondation Bettencourt-Schueller).
Author contributions: P.B. and J.-L.C. wrote the manuscript, with the assistance of S.V., Q.Z., S.-Y.Z., E.J., M.S.A and J.L.D. who edited the manuscript, tables and figures. P.B, S.V, J.L., L.M., A.G., T.L.V., J.R., Q.P., L.B., J.M., A.P., E.J., C.J.Y., A.C., L.T., performed the experiments, generated data or analyzed it. P.B., S.D., L.R., D.C.V., E.C., L.F.R., C.M., V.E., C.Z., E.C., A.W., A.M., G.W., R.B., A.F., S.S., I.T., P.G., Y.Z., S.D., D.A., E.P.A., L.R., L.R., D.C.V., L.F.R., M.G., N.H., D.B., Y.T.L., R.H., N.S.S.A., J.D., S.B., Y.S., O.B., C.M., M.N., B.N., K.S., O.A., A.A., E.J., G.R.B., C.T., M.L., P.L.T., T.B., K.D., G.G., G.R.G., L.F.P.N, L.R., A.P., A.B., F.R., T.O., S.K., J.M.P., L.D.N., J.T., X.S.M., S.T.A., L.M.A., and E.A. treated and recruited the patients. Y.A.J., S.A., S.B., S.A.C., S.C., Z.C., G.K.F., R.G., A.J., K.N.K., T.L., D.L., A.L., C.L., R.L., M.M., V.N., R.P., L.P., P.P., A.A.R., A.R., N.R., B.S., C.S., A.S., K.T., L.T.A., A.W., A.W., M.W. recruited patients, and collected and managed samples and data. C.S.C., D.J.E., C.M.H., M.F.K., C.R.L., P.G.W. founded and led the COMET Consortium. Q.Z., S.-Y.Z., E.J., M.S.A, J.-L.C and J.L.D. supervised the study.
Competing interests: J.-L.C. reports being an inventor on patent application PCT/US2021/042741, filed 22 July 2021, submitted by The Rockefeller University, which covers the diagnosis of, susceptibility to, and treatment of viral disease and viral vaccines, including COVID-19 and vaccine-associated diseases. C.S.C., D.J.E., C.M.H., M.F.K., C.R.L., P.G.W. report funding from Genentech. C.S.C. reports funding from The National Heart, Lung, and Blood Institute (NHLBI), The U.S. Food and Drug Administration (FDA), U.S. Department of Defense (DOD), and Quantum Leap Healthcare Collaborative, and serves on consulting and advisory boards for Vasomune, Gen1e Life Sciences, Janssen, and Cellenkos. C.M.H. reports funding from NHLBI and consulting fees for clinical trial design with Spring Discovery. The other authors have no conflicts of interest to report.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.
Appendix: List of COVID HGE Consortium authors
Laurent Abel1 Cristian Achille2 Alessandro Aiuti3 Saleh Al-Muhsen4 Fahd Al-Mulla5 Mark S. Anderson6 Evangelos Andreakos7 Micol Angelini8 Andrés A. Arias9 Gokhan Aytekin10 Fausto Baldanti11 Hagit Baris Feldman12 Alexandre Belot13 Federica Bergami14 Catherine M. Biggs15 Dusan Bogunovic16 Alexandre Bolze17 Anastasiia Bondarenko18 Ahmed A. Bousfiha19 Petter Brodin20 Yenan Bryceson21 Carlos D. Bustamante22 Manish J. Butte23 Giorgio Casari24 John Christodoulou25 Antonio Condino-Neto26 Stefan N. Constantinescu27 Francesca Conti28 Megan A. Cooper29 Clifton L. Dalgard30 Murkesh Desai31 Beth A. Drolet32 Jamila El Baghdadi33 Recai Ergun34 Dilek Ergun35 Sara Espinosa-Padilla36 Jacques Fellay37 Carlos Flores38 José Luis Franco39 Antoine Froidure40 Stefano Ghirardello41 Peter K. Gregersen42 Bodo Grimbacher43 Filomeen Haerynck44 David Hagin45 Rabih Halwani46 Lennart Hammarström47 James R. Heath48 Sarah E. Henrickson49 Elena W.Y. Hsieh50 Eystein Husebye51 Kohsuke Imai52 Yuval Itan53 Erich D. Jarvis54 Fikret Kanat55 Timokratis Karamitros56 Kai Kisand57 Vasyl Kopcha58 Mykhaylo Korda59 Cheng-Lung Ku60 Yu-Lung Lau61 Yun Ling62 Carrie L. Lucas63 Tom Maniatis64 Davood Mansouri65 László Maródi66 Isabelle Meyts67 Joshua D. Milner68 Kristina Mironska69 Trine H. Mogensen70 Francesco Mojoli71 Francisco Morandeira72 Tomohiro Morio73 Lisa F.P. Ng74 Luigi D. Notarangelo75 Antonio Novelli76 Giuseppe Novelli77 Cliona O'Farrelly78 Satoshi Okada79 Keisuke Okamoto80 Tayfun Ozcelik81 Michele Pagani82 Qiang Pan-Hammarström83 Jean W. Pape84 Rebeca Perez de Diego85 David S. Perlin86 Graziano Pesole87 Andrea Pession88 Antonio Piralla89 Anna M. Planas90 Carolina Prando91 Aurora Pujol92 Lluis Quintana-Murci93 Sathishkumar Ramaswamy94 Laurent Renia95 Igor Resnick96 Raúl Rigo-Bonnin97 Carlos Rodríguez-Gallego98 Vanessa Sancho-Shimizu99 Anna Sediva100 Mikko R.J. Seppänen101 Mohammed Shahrooei102 Anna Shcherbina103 Ondrej Slaby104 Andrew L. Snow105 Pere Soler-Palacín106 András N. Spaan107 Ivan Tancevski108 Stuart G. Tangye109 Ahmad Abou Tayoun110 Baykal Tulek111 Stuart E. Turvey112 K M Furkan Uddin113 Mohammed Uddin114 Bénédicte Clément115
1Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; University of Paris, Imagine Institute, Paris, France. 2Neonatal Intensive Care Unit, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 3San Raffaele Telethon Institute for Gene Therapy, IRCCS Ospedale San Raffaele, and Vita Salute San Raffaele University, Milan, Italy. 4Immunology Research Lab, Department of Pediatrics, College of Medicine, King Saud University, Riyadh, Saudi Arabia. 5Dasman Diabetes Institute, Department of Genetics and Bioinformatics, Dasman, Kuwait. 6Diabetes Center, University of California San Francisco, San Francisco, CA, USA. 7Laboratory of Immunobiology, Center for Clinical, Experimental Surgery and Translational Research, Biomedical Research Foundation of the Academy of Athens, Athens, Greece. 8Neonatal Intensive Care Unit, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 9St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA; Primary Immunodeficiencies Group, Department of Microbiology and Parasitology, School of Medicine, University of Antioquia, Medellín, Colombia; School of Microbiology, University of Antioquia UdeA, Medellín, Colombia. 10Department of Immunology and Allergy, Konya City Hospital 11Department of Clinical, Surgical, Diagnostic and Pediatric Sciences, University of Pavia, Pavia, Italy 12The Genetics Institute, Tel Aviv Sourasky Medical Center and Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel. 13Pediatric Nephrology, Rheumatology, Dermatology, HFME, Hospices Civils de Lyon, National Referee Centre RAISE, and INSERM U1111, Université de Lyon, Lyon, France. 14Molecular Virology Unit, Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 15Department of Pediatrics, BC Children's and St. Paul's Hospitals, University of British Columbia, Vancouver, BC, Canada. 16Icahn School of Medicine at Mount Sinai, New York, NY, USA. 17Helix, San Mateo, CA, USA. 18Shupyk National Healthcare University of Ukraine, Kyiv, Ukraine. 19Department of Pediatric Infectious Diseases and Clinical Immunology, CHU Ibn Rushd and LICIA, Laboratoire d'Immunologie Clinique, Inflammation et Allergie, Faculty of Medicine and Pharmacy, Hassan II University, Casablanca, Morocco. 20SciLifeLab, Department Of Women’s and Children’s Health, Karolinska Institutet, Stockholm, Sweden. 21Department of Medicine, Center for Hematology and Regenerative Medicine, Karolinska Institutet, Stockholm, Sweden. 22Stanford University, Stanford, CA, USA. 23Division of Immunology, Allergy, and Rheumatology, Department of Pediatrics and the Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, CA, USA. 24Clinical Genomics, IRCCS San Raffaele Scientific Institute and Vita-Salute San Raffaele University, Milan, Italy. 25Murdoch Children's Research Institute and Department of Pediatrics, University of Melbourne, Melbourne, VIC, Australia. 26Department of Immunology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil. 27de Duve Institute and Ludwig Cancer Research, Brussels, Belgium. 28Pediatric Unit, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy 29Washington University School of Medicine, St. Louis, MO, USA. 30Department of Anatomy, Physiology & Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA. 31Bai Jerbai Wadia Hospital for Children, Mumbai, India. 32School of Medicine and Public Health, University of Wisconsin, Madison, WI, USA. 33Genetics Unit, Military Hospital Mohamed V, Rabat, Morocco. 34Selcuk University, Department of Pulmonology, Konya, Turkey 35Selcuk University, Department of Pulmonology, Konya, Turkey 36Instituto Nacional de Pediatria (National Institute of Pediatrics), Mexico City, Mexico. 37School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; Precision Medicine Unit, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland. 38Research Unit, Hospital Universitario Nuestra Señora de Candelaria, Santa Cruz de Tenerife; CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Madrid; Genomics Division, Instituto Tecnológico y de Energías Renovables (ITER), Santa Cruz de Tenerife, Spain. 39Group of Primary Immunodeficiencies, University of Antioquia UDEA, Medellin, Colombia. 40Pulmonology Department, Cliniques Universitaires Saint-Luc ; Institut de Recherche Expérimentale et Clinique (IREC), Université Catholique de Louvain, Brussels, Belgium. 41Neonatal Intensive Care Unit, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 42Feinstein Institute for Medical Research, Northwell Health USA, Manhasset, NY, USA. 43Center for Chronic Immunodeficiency & Institute for Immunodeficiency, Medical Center, Faculty of Medicine, University of Freiburg, Freiburg, Germany. 44Department of Pediatric Immunology and Pulmonology, Centre for Primary Immunodeficiency Ghent (CPIG), PID Research Laboratory, Jeffrey Model Diagnosis and Research Centre, Ghent University Hospital, Ghent, Belgium. 45The Genetics Institute Tel Aviv Sourasky Medical Center, Tel Aviv, Israel. 46Sharjah Institute of Medical Research, College of Medicine, University of Sharjah, Sharjah, United Arab Emirates. 47Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden. 48Institute for Systems Biology, Seattle, WA, USA. 49Department of Pediatrics, Division of Allergy Immunology, Children’s Hospital of Philadelphia, Philadelphia, PA, USA; Department of Microbiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA. 50Departments of Pediatrics, Immunology and Microbiology, University of Colorado, School of Medicine, Aurora, CO, USA. 51Department of Medicine, Haukeland University Hospital, Bergen, Norway. 52Department of Community Pediatrics, Perinatal and Maternal Medicine, Tokyo Medical and Dental University (TMDU), Tokyo, Japan. 53Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA; Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA. 54Laboratory of Neurogenetics of Language and Howard Hughes Medical Institute, The Rockefeller University, New York, NY, USA. 55Selcuk University, Department of Pulmonology, Konya, Turkey 56Bioinformatics and Applied Genomics Unit, Hellenic Pasteur Institute, Athens, Greece. 57Molecular Pathology, Department of Biomedicine, Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu Estonia. 58Horbachevsky Ternopil National Medical University 59Horbachevsky Ternopil National Medical University 60Chang Gung University, Taoyuan County, Taiwan. 61Department of Pediatrics & Adolescent Medicine, The University of Hong Kong, Hong Kong, China. 62Shanghai Public Health Clinical Center, Fudan University, Shanghai, China. 63Department of Immunobiology, Yale University School of Medicine, New Haven, CT, USA. 64Zukerman Mind Brain Behavior Institute, Columbia University, New York, NY, USA. 65Department of Clinical Immunology and Infectious Diseases, National Research Institute of Tuberculosis and Lung Diseases, The Clinical Tuberculosis and Epidemiology Research Center, National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Masih Daneshvari Hospital, Shahid Beheshti, University of Medical Sciences, Tehran, Iran. 66Primary Immunodeficiency Clinical Unit and Laboratory, Department of Dermatology, Venereology and Dermatooncology, Semmelweis University, Budapest, Hungary. 67Department of Pediatrics, University Hospitals Leuven; KU Leuven, Department of Microbiology, Immunology and Transplantation; Laboratory for Inborn Errors of Immunity, KU Leuven, Leuven, Belgium. 68Department of Pediatrics, Columbia University Irving Medical Center, New York, NY, USA. 69University Clinic for Children's Diseases, Department of Pediatric Immunology, Medical Faculty, University “ St.Cyril and Methodij” Skopje, North Macedonia. 70Department of Biomedicine, Aarhus University, Aarhus, Denmark. 71Anesthesia and Intensive Care, Rianimazione I, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 72Department of Internal Medicine, Hospital Universitari de Bellvitge, IDIBELL, Barcelona, Spain. 73Tokyo Medical & Dental University Hospital, Tokyo, Japan. 74A*STAR Infectious Disease Labs, Agency for Science, Technology and Research, Singapore; Lee Kong Chian School of Medicine, Nanyang Technology University, Singapore. 75National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA. 76Laboratory of Medical Genetics, IRCCS Bambino Gesù Children’s Hospital, Rome, Italy. 77Department of Biomedicine and Prevention, Tor Vergata University of Rome, Rome, Italy. 78Comparative Immunology Group, School of Biochemistry and Immunology, Trinity Biomedical Sciences Institute, Trinity College Dublin, Ireland. 79Department of Pediatrics, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan. 80Tokyo Medical and Dental University, Tokyo, Japan. 81Department of Molecular Biology and Genetics, Bilkent University, Bilkent - Ankara, Turkey. 82Anesthesia and Intensive Care, Rianimazione I, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 83Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden. 84Haitian Study Group for Kaposi's Sarcoma and Opportunistic Infections (GHESKIO), Port-au-Prince, Haiti. 85Institute of Biomedical Research of IdiPAZ, University Hospital “La Paz”, Madrid, Spain. 86Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, NJ, USA. 87Department of Biosciences, Biotechnology and Biopharmaceutics, University of Bari A. Moro, Bari, Italy. 88Pediatric Unit, IRCCS Azienda Ospedaliero-Universitaria di Bologna, Bologna, Italy 89Molecular Virology Unit, Microbiology and Virology Department, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy 90IIBB-CSIC, IDIBAPS, Barcelona, Spain. 91Faculdades Pequeno Príncipe, Instituto de Pesquisa Pelé Pequeno Príncipe, Curitiba, Brazil. 92Neurometabolic Diseases Laboratory, Bellvitge Biomedical Research Institute (IDIBELL), L'Hospitalet de Llobregat, Barcelona, Spain; Catalan Institution of Research and Advanced Studies (ICREA), Barcelona, Spain; Center for Biomedical Research on Rare Diseases (CIBERER), ISCIII, Barcelona, Spain. 93Human Evolutionary Genetics Unit, CNRS U2000, Institut Pasteur, Paris, France; Human Genomics and Evolution, Collège de France, Paris, France. 94Al Jalila Children's Hospital, Dubai, UAE. 95A*STAR Infectious Disease Labs, Agency for Science, Technology and Research, Singapore; Lee Kong Chian School of Medicine, Nanyang Technology University, Singapore. 96University Hospital St. Marina, Varna, Bulgaria. 97Department of Clinical Laboratory, Hospital Universitari de Bellvitge, IDIBELL, Barcelona, Spain. 98Department of Immunology, University Hospital of Gran Canaria Dr. Negrín, Canarian Health System, Las Palmas de Gran Canaria; Department of Clinical Sciences, University Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain. 99Department of Pediatric Infectious Diseases and Virology, Imperial College London, London, UK; Centre for Pediatrics and Child Health, Faculty of Medicine, Imperial College London, London, UK. 100Department of Immunology, Second Faculty of Medicine Charles University, V Uvalu, University Hospital in Motol, Prague, Czech Republic. 101Adult Immunodeficiency Unit, Infectious Diseases, Inflammation Center, University of Helsinki and Helsinki University Hospital, Helsinki, Finland; Rare Diseases Center and Pediatric Research Center, Children's Hospital, University of Helsinki and Helsinki University Hospital, Helsinki, Finland. 102Specialized Immunology Laboratory of Dr. Shahrooei, Ahvaz, Iran; Department of Microbiology and Immunology, Clinical and Diagnostic Immunology, KU Leuven, Leuven, Belgium. 103Department of Immunology, Dmitry Rogachev National Medical Research Center of Pediatric Hematology, Oncology and Immunology, Moscow, Russia. 104Central European Institute of Technology & Department of Biology, Faculty of Medicine, Masaryk University, Brno, Czech Republic. 105Department of Pharmacology & Molecular Therapeutics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA. 106Pediatric Infectious Diseases and Immunodeficiencies Unit, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Catalonia, Spain. 107St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA; Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, Netherlands. 108Department of Internal Medicine II, Medical University of Innsbruck, Innsbruck, Austria. 109Garvan Institute of Medical Research, Darlinghurst, NSW, Australia; St Vincent’s Clinical School, Faculty of Medicine, UNSW Sydney, NSW, Australia. 110Al Jalila Children's Hospital, Dubai, UAE. 111Selcuk University, Department of Pulmonology, Konya, Turkey 112BC Children's Hospital, The University of British Columbia, Vancouver, Canada. 113Centre for Precision Therapeutics, Genetics & Genomic Medicine Centre, NeuroGen Children's Healthcare and Lecturer, Holy Family Red Crescent Medical College Dhaka, Bangladesh. 114 Mohammed Bin Rashid University of Medicine and Health Sciences, Dubai, UAE; GenomeArc Inc., Toronto, ON, Canada. 115 Service des Urgences, Groupement Hospitalier Nord, Hospices Civils de Lyon, Lyon, France.
Appendix: List of COMET Consortium authors
Yumiko Abe-Jones, MS1, Saurabh Asthana, PhD2,3,4, Sharvari Bhide, BA5, Carolyn S. Calfee, M 6,7, Sidney A. Carrillo, MPH6, Suzanna Chak, BA6, Zachary Collins, BA2,3,4, David J. Erle, MD3,4,7,8, Gabriela K. Fragiadakis, PhD3,4,9, Rajani Ghale, MS6, Carolyn M. Hendrickson, MD5,8, Alejandra Jauregui, BA6, Kirsten N. Kangelaris, MD1, Matthew F. Krummel, PhD2,3,4, Charles R. Langelier, MD, PhD10,11, Tasha Lea, MS2, Deanna Lee, BA5,8, Aleksandra Leligdowicz, MD, PhD12, Carolyn Leroux, BS6, Raphael Lota, BA13, Michael Matthay, MD7, Viet Nguyen, BA5,8, Ravi Patel, PhD3,4, Logan Pierce, MD1, Priya Prasad, PhD1, Arjun Arkal Rao, PhD2,3,4, Ahmad Rashid, BS13, Nicklaus Rodriguez, BA13, Bushra Samad, MS2,3,4, Cole Shaw, MSEE 2 , 3 , 4 , Austin Sigman, BS6, Kevin Tang, MS13, Luz Torres Altamirano, BA13, Alyssa Ward, PhD9, Andrew Willmore, BS6, Michael Wilson, MD14, Prescott G. Woodruff, MD6,7
1. Division of Hospital Medicine, University of California, San Francisco, CA, USA.
2. Department of Pathology, University of California, San Francisco, CA, USA.
3. UCSF CoLabs, University of California, San Francisco, CA, USA.
4. Bakar ImmunoX Initiative, University of California, San Francisco, CA, USA.
5. Division of Pulmonary and Critical Care Medicine, Department of Medicine, Zuckerberg San Francisco General Hospital and Trauma Center, University of California, San Francisco, CA, USA.
6. Division of Pulmonary and Critical Care Medicine, Department of Medicine, University of California San Francisco, San Francisco, California, USA.
7. Cardiovascular Research Institute, University of California, San Francisco, San Francisco, CA, USA.
8. Lung Biology Center, University of California, San Francisco, San Francisco, CA, USA
9. Division of Rheumatology, Department of Medicine, University of California, San Francisco, CA, USA.
10. Division of Infectious Diseases, University of California, San Francisco, CA, USA.
11. Chan Zuckerberg Biohub, University of California, San Francisco, CA, USA.
12. Division of Critical Care Medicine, Robarts Research Institute, University of Western Ontario, London, ON, Canada.
13. Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco, CA, USA.
14. Weill Institute for Neurosciences, Department of Neurology, University of California, San Francisco, CA, USA.
Appendix: List of French COVID study group authors
Laurent ABEL1, Clotilde ALLAVENA2, Claire ANDREJAK3, François ANGOULVANT4, Cecile AZOULAY5, Delphine BACHELET6, Marie BARTOLI7, Romain BASMACI8, Sylvie BEHILILL9, Marine BELUZE10, Nicolas BENECH11, Dehbia BENKERROU12, Krishna BHAVSAR6, Laurent BITKER11, Lila BOUADMA6, Maude BOUSCAMBERT13, Pauline CARAUX PAZ14, Minerva CERVANTES-GONZALEZ6, Anissa CHAIR6, Catherine CHIROUZE15, Alexandra COELHO16, Hugues CORDEL17, Camille COUFFIGNAL6, Sandrine COUFFIN-CADIERGUES18, Eric d’ORTENZIO7, Etienne DE MONTMOLLIN6, Alexa DEBARD19, Marie-Pierre DEBRAY6, Dominique DEPLANQUE20, Diane DESCAMPS6, Mathilde DESVALLÉE21, Alpha DIALLO7, Jean-Luc DIEHL22, Alphonsine DIOUF16, Céline DORIVAL12, François DUBOS23, Xavier DUVAL6, Philippine ELOY6, Vincent ENOUF9, Olivier EPAULARD24, Hélène ESPEROU18, Marina ESPOSITO-FARESE6, Manuel ETIENNE25, Denis GAROT26, Nathalie GAULT6, Alexandre GAYMARD13, Jade GHOSN6, Tristan GIGANTE27, Morgane GILG27, François GOEHRINGER28, Jérémie GUEDJ29, Alexandre HOCTIN16, Isabelle HOFFMANN6, Ikram HOUAS18, Jean-Sébastien HULOT22, Salma JAAFOURA18, Ouifiya KAFIF6, Florentia KAGUELIDOU30, Sabrina KALI6, Younes KERROUMI31, Antoine KHALIL6, Coralie KHAN21, Antoine KIMMOUN28, Fabrice LAINE32, Cédric LAOUÉNAN6, Samira LARIBI6, Minh LE6, Cyril LE BRIS33, Sylvie LE GAC 6, Quentin LE HINGRAT6, Soizic LE MESTRE7, Hervé LE NAGARD29, Adrien LEMAIGNEN26, Véronique LEMEE25, François-Xavier LESCURE6, Sophie LETROU6, Yves LEVY34, Bruno LINA13, Guillaume LINGAS29, Jean Christophe LUCET6, Moïse MACHADO35, Denis MALVY36, Marina MAMBERT16, Aldric MANUEL37, France MENTRÉ6, Amina MEZIANE12, Hugo MOUQUET9, Jimmy Mullaert6, Nadège NEANT29, Duc NGUYEN36, Marion NORET38, Aurélie PAPADOPOULOS18, Christelle PAUL7, Nathan PEIFFER-SMADJA6, Vincent PEIGNE39, Ventzislava PETROV-SANCHEZ7, Gilles PEYTAVIN6, Huong PHAM6, Olivier PICONE8, Valentine PIQUARD6, Julien POISSY23, Oriane PUÉCHAL40, Manuel ROSA-CALATRAVA13, Bénédicte ROSSIGNOL27, Patrick ROSSIGNOL28, Carine ROY6, Marion SCHNEIDER6, Richa SU6, Coralie TARDIVON6, Marie-Capucine TELLIER6, François TÉOULÉ12, Olivier TERRIER13, Jean-François TIMSIT6, Christelle TUAL41, Sarah TUBIANA6, Sylvie VAN DER WERF9, Noémie VANEL42, Aurélie VEISLINGER41, Benoit VISSEAUX6, 29, Aurélie WIEDEMANN34, Yazdan YAZDANPANAH6
1Inserm UMR 1163, Paris, France. 2CHU Nantes, France. 3CHU Amiens, France. 4Hôpital Necker, Paris, France. 5Hopitâl Cochin, Paris, France. 6Hôpital Bichat, Paris, France. 7ANRS, Paris, France. 8Hôpital Louis Mourier, Colombes, France. 9Pasteur Institute, Paris, France. 10F-CRIN Partners Platform, Paris, France. 11CHU Lyon, France. 12Inserm UMR 1136, Paris, France. 13Inserm UMR 1111, Lyon, France. 14CH Villeneuve Saint Georges, France. 15CHRU Jean Minjoz, Besançon, France. 16Inserm UMR 1018, Paris, France. 17Hôpital Avicenne, Bobigny, France. 18Inserm sponsor, Paris, France. 19CHU Toulouse, France. 20CIC 1403 Inserm - CHU Lille, France. 21Inserm UMR 1219, Bordeaux, France. 22Hôpital Européen Georges Pompidou, Paris, France. 23CHU Lille, France. 24CHU Grenoble, France. 25CHU Rouen, France. 26CHU Tours, France. 27F-CRIN INI-CRCT, Nancy, France. 28CHU Nancy, France. 29Inserm UMR 1137, Paris, France. 30Hôpital Robert Debré, Paris, France. 31GH Diaconesses, Paris, France. 32CHU Rennes, France. 33CH Beziers, France. 34Vaccine Research Insitute (VRI), Inserm UMR 955, Créteil, France. 35Grand Hôpital de l’Est Francilien, Marne-la-Vallée, France. 36CHU Bordeaux, France. 37CH Annecy, France. 38RENARCI, Annecy, France. 39CH Métropole Savoie, Cambery, France. 40REACTing, Paris, France. 41Inserm CIC-1414, Rennes, France. 42Hôpital la Timone, Marseille, France.
Contributor Information
Collaborators: Laurent Abel, Cristian Achille, Alessandro Aiuti, Saleh Al-Muhsen, Fahd Al-Mulla, Mark S. Anderson, Evangelos Andreakos, Micol Angelini, Andrés A. Arias, Gokhan Aytekin, Fausto Baldanti, Hagit Baris Feldman, Alexandre Belot, Federica Bergami, Catherine M. Biggs, Dusan Bogunovic, Alexandre Bolze, Anastasiia Bondarenko, Ahmed A. Bousfiha, Petter Brodin, Yenan Bryceson, Carlos D. Bustamante, Manish J. Butte, Giorgio Casari, John Christodoulou, Antonio Condino-Neto, Stefan N. Constantinescu, Francesca Conti, Megan A. Cooper, Clifton L. Dalgard, Murkesh Desai, Beth A. Drolet, Jamila El Baghdadi, Recai Ergun, Dilek Ergun, Sara Espinosa-Padilla, Jacques Fellay, Carlos Flores, José Luis Franco, Antoine Froidure, Stefano Ghirardello, Peter K. Gregersen, Bodo Grimbacher, Filomeen Haerynck, David Hagin, Rabih Halwani, Lennart Hammarström, James R. Heath, Sarah E. Henrickson, Elena W.Y. Hsieh, Eystein Husebye, Kohsuke Imai, Yuval Itan, Erich D. Jarvis, Fikret Kanat, Timokratis Karamitros, Kai Kisand, Vasyl Kopcha, Mykhaylo Korda, Cheng-Lung Ku, Yu-Lung Lau, Yun Ling, Carrie L. Lucas, Tom Maniatis, Davood Mansouri, László Maródi, Isabelle Meyts, Joshua D. Milner, Kristina Mironska, Trine H. Mogensen, Francesco Mojoli, Francisco Morandeira, Tomohiro Morio, Lisa F.P. Ng, Luigi D. Notarangelo, Antonio Novelli, Giuseppe Novelli, Cliona O'Farrelly, Satoshi Okada, Keisuke Okamoto, Tayfun Ozcelik, Michele Pagani, Qiang Pan-Hammarström, Jean W. Pape, Rebeca Perez de Diego, David S. Perlin, Graziano Pesole, Andrea Pession, Antonio Piralla, Anna M. Planas, Carolina Prando, Aurora Pujol, Lluis Quintana-Murci, Sathishkumar Ramaswamy, Laurent Renia, Igor Resnick, Raúl Rigo-Bonnin, Carlos Rodríguez-Gallego, Vanessa Sancho-Shimizu, Anna Sediva, Mikko R.J. Seppänen, Mohammed Shahrooei, Anna Shcherbina, Ondrej Slaby, Andrew L. Snow, Pere Soler-Palacín, András N. Spaan, Ivan Tancevski, Stuart G. Tangye, Ahmad Abou Tayoun, Baykal Tulek, Stuart E. Turvey, K M Furkan Uddin, Mohammed Uddin, Bénédicte Clément, Yumiko Abe-Jones, Saurabh Asthana, Sharvari Bhide, Carolyn S. Calfee, Sidney A. Carrillo, Suzanna Chak, Zachary Collins, David J. Erle, Gabriela K. Fragiadakis, Rajani Ghale, Carolyn M. Hendrickson, Alejandra Jauregui, Kirsten N. Kangelaris, Matthew F. Krummel, Charles R. Langelier, Tasha Lea, Deanna Lee, Aleksandra Leligdowicz, Carolyn Leroux, Raphael Lota, Michael Matthay, Viet Nguyen, Ravi Patel, Logan Pierce, Priya Prasad, Arjun Arkal Rao, Ahmad Rashid, Nicklaus Rodriguez, Bushra Samad, Cole Shaw, Austin Sigman, Kevin Tang, Luz Torres Altamirano, Alyssa Ward, Andrew Willmore, Michael Wilson, Prescott G. Woodruff, Laurent Abel, Clotilde Allavena, Claire Andrejak, François Angoulvant, Cecile Azoulay, Delphine Bachelet, Marie Bartoli, Romain Basmaci, Sylvie Behilill, Marine Beluze, Nicolas Benech, Dehbia Benkerrou, Krishna Bhavsar, Laurent Bitker, Lila Bouadma, Maude Bouscambert, Pauline Caraux Paz, Minerva Cervantes-Gonzalez, Anissa Chair, Catherine Chirouze, Alexandra Coelho, Hugues Cordel, Camille Couffignal, Sandrine Couffin-Cadiergues, Eric d’Ortenzio, Etienne De Montmollin, Alexa Debard, Marie-Pierre Debray, Dominique Deplanque, Diane Descamps, Mathilde Desvallée, Alpha Diallo, Jean-Luc Diehl, Alphonsine Diouf, Céline Dorival, François Dubos, Xavier Duval, Philippine Eloy, Vincent Enouf, Olivier Epaulard, Hélène Esperou, Marina Esposito-Farese, Manuel Etienne, Denis Garot, Nathalie Gault, Alexandre Gaymard, Jade Ghosn, Tristan Gigante, Morgane Gilg, François Goehringer, Jérémie Guedj, Alexandre Hoctin, Isabelle Hoffmann, Ikram Houas, Jean-Sébastien Hulot, Salma Jaafoura, Ouifiya Kafif, Florentia Kaguelidou, Sabrina Kali, Younes Kerroumi, Antoine Khalil, Coralie Khan, Antoine Kimmoun, Fabrice Laine, Cédric Laouénan, Samira Laribi, Minh Le, Cyril Le Bris, Sylvie Le Gac, Quentin Le Hingrat, Soizic Le Mestre, Hervé Le Nagard, Adrien Lemaignen, Véronique Lemee, François-Xavier Lescure, Sophie Letrou, Yves Levy, Bruno Lina, Guillaume Lingas, Jean Christophe Lucet, Moïse Machado, Denis Malvy, Marina Mambert, Aldric Manuel, France Mentré, Amina Meziane, Hugo Mouquet, Jimmy Mullaert, Nadège Neant, Duc Nguyen, Marion Noret, Aurélie Papadopoulos, Christelle Paul, Nathan Peiffer-Smadja, Vincent Peigne, Ventzislava Petrov-Sanchez, Gilles Peytavin, Huong Pham, Olivier Picone, Valentine Piquard, Julien Poissy, Oriane Puéchal, Manuel Rosa-Calatrava, Bénédicte Rossignol, Patrick Rossignol, Carine Roy, Marion Schneider, Richa Su, Coralie Tardivon, Marie-Capucine Tellier, François Téoulé, Olivier Terrier, Jean-François Timsit, Christelle Tual, Sarah Tubiana, Sylvie Van Der Werf, Noémie Vanel, Aurélie Veislinger, Benoit Visseaux, Aurélie Wiedemann, and Yazdan Yazdanpanah
Supplementary Materials
This PDF file includes:
Materials and Methods
Fig. S1
Table S1
Other Supplementary Material for this manuscript includes the following:
Data file S1
Reference and notes
- 1. Zhou P., Yang X. L., Wang X. G., Hu B., Zhang L., Zhang W., Si H. R., Zhu Y., Li B., Huang C. L., Chen H. D., Chen J., Luo Y., Guo H., Jiang R. D., Liu M. Q., Chen Y., Shen X. R., Wang X., Zheng X. S., Zhao K., Chen Q. J., Deng F., Liu L. L., Yan B., Zhan F. X., Wang Y. Y., Xiao G. F., Shi Z. L., A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worldometer. (2021), vol. 2021.
- 3. Levin A. T., Hanage W. P., Owusu-Boaitey N., Cochran K. B., Walsh S. P., Meyerowitz-Katz G., Assessing the age specificity of infection fatality rates for COVID-19: Systematic review, meta-analysis, and public policy implications. Eur. J. Epidemiol. 35, 1123–1138 (2020). 10.1007/s10654-020-00698-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O’Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D. A. T., Azman A. S., Paireau J., Fontanet A., Cauchemez S., Salje H., Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 590, 140–145 (2021). 10.1038/s41586-020-2918-0 [DOI] [PubMed] [Google Scholar]
- 5. Bogunovic D., Merad M., Children and SARS-CoV-2. Cell Host Microbe 29, 1040–1042 (2021). 10.1016/j.chom.2021.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I. K. D., Hodeib S., Korol C., Rosain J., Bilguvar K., Ye J., Bolze A., Bigio B., Yang R., Arias A. A., Zhou Q., Zhang Y., Onodi F., Korniotis S., Karpf L., Philippot Q., Chbihi M., Bonnet-Madin L., Dorgham K., Smith N., Schneider W. M., Razooky B. S., Hoffmann H. H., Michailidis E., Moens L., Han J. E., Lorenzo L., Bizien L., Meade P., Neehus A. L., Ugurbil A. C., Corneau A., Kerner G., Zhang P., Rapaport F., Seeleuthner Y., Manry J., Masson C., Schmitt Y., Schlüter A., Le Voyer T., Khan T., Li J., Fellay J., Roussel L., Shahrooei M., Alosaimi M. F., Mansouri D., Al-Saud H., Al-Mulla F., Almourfi F., Al-Muhsen S. Z., Alsohime F., Al Turki S., Hasanato R., van de Beek D., Biondi A., Bettini L. R., D’Angio’ M., Bonfanti P., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Oler A. J., Tompkins M. F., Alba C., Vandernoot I., Goffard J. C., Smits G., Migeotte I., Haerynck F., Soler-Palacin P., Martin-Nalda A., Colobran R., Morange P. E., Keles S., Çölkesen F., Ozcelik T., Yasar K. K., Senoglu S., Karabela S. N., Rodríguez-Gallego C., Novelli G., Hraiech S., Tandjaoui-Lambiotte Y., Duval X., Laouénan C., Snow A. L., Dalgard C. L., Milner J. D., Vinh D. C., Mogensen T. H., Marr N., Spaan A. N., Boisson B., Boisson-Dupuis S., Bustamante J., Puel A., Ciancanelli M. J., Meyts I., Maniatis T., Soumelis V., Amara A., Nussenzweig M., García-Sastre A., Krammer F., Pujol A., Duffy D., Lifton R. P., Zhang S. Y., Gorochov G., Béziat V., Jouanguy E., Sancho-Shimizu V., Rice C. M., Abel L., Notarangelo L. D., Cobat A., Su H. C., Casanova J. L.; COVID-STORM Clinicians; COVID Clinicians; Imagine COVID Group; French COVID Cohort Study Group; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; NIAID-USUHS/TAGC COVID Immunity Group , Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370, eabd4570 (2020). 10.1126/science.abd4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang S. Y., Zhang Q., Casanova J. L., Su H. C.; COVID Team , Severe COVID-19 in the young and healthy: Monogenic inborn errors of immunity? Nat. Rev. Immunol. 20, 455–456 (2020). 10.1038/s41577-020-0373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khanmohammadi S., Rezaei N., Khazaei M., Shirkani A., A Case of Autosomal Recessive Interferon Alpha/Beta Receptor Alpha Chain (IFNAR1) Deficiency with Severe COVID-19. J. Clin. Immunol. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Q., Bastard P., Cobat A., Casanova J. L.; COVID Human Genetic Effort , Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 603, 587–598 (2022). 10.1038/s41586-022-04447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bastard P., Rosen L. B., Zhang Q., Michailidis E., Hoffmann H. H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., Manry J., Shaw E., Haljasmägi L., Peterson P., Lorenzo L., Bizien L., Trouillet-Assant S., Dobbs K., de Jesus A. A., Belot A., Kallaste A., Catherinot E., Tandjaoui-Lambiotte Y., Le Pen J., Kerner G., Bigio B., Seeleuthner Y., Yang R., Bolze A., Spaan A. N., Delmonte O. M., Abers M. S., Aiuti A., Casari G., Lampasona V., Piemonti L., Ciceri F., Bilguvar K., Lifton R. P., Vasse M., Smadja D. M., Migaud M., Hadjadj J., Terrier B., Duffy D., Quintana-Murci L., van de Beek D., Roussel L., Vinh D. C., Tangye S. G., Haerynck F., Dalmau D., Martinez-Picado J., Brodin P., Nussenzweig M. C., Boisson-Dupuis S., Rodríguez-Gallego C., Vogt G., Mogensen T. H., Oler A. J., Gu J., Burbelo P. D., Cohen J. I., Biondi A., Bettini L. R., D’Angio M., Bonfanti P., Rossignol P., Mayaux J., Rieux-Laucat F., Husebye E. S., Fusco F., Ursini M. V., Imberti L., Sottini A., Paghera S., Quiros-Roldan E., Rossi C., Castagnoli R., Montagna D., Licari A., Marseglia G. L., Duval X., Ghosn J., Tsang J. S., Goldbach-Mansky R., Kisand K., Lionakis M. S., Puel A., Zhang S. Y., Holland S. M., Gorochov G., Jouanguy E., Rice C. M., Cobat A., Notarangelo L. D., Abel L., Su H. C., Casanova J. L.; HGID Lab; NIAID-USUHS Immune Response to COVID Group; COVID Clinicians; COVID-STORM Clinicians; Imagine COVID Group; French COVID Cohort Study Group; Milieu Intérieur Consortium; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort , Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370, eabd4585 (2020). 10.1126/science.abd4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koning R., Bastard P., Casanova J. L., Brouwer M. C., van de Beek D.; with the Amsterdam U.M.C. COVID-19 Biobank Investigators , Autoantibodies against type I interferons are associated with multi-organ failure in COVID-19 patients. Intensive Care Med. 47, 704–706 (2021). 10.1007/s00134-021-06392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Troya J., Bastard P., Planas-Serra L., Ryan P., Ruiz M., de Carranza M., Torres J., Martínez A., Abel L., Casanova J. L., Pujol A., Neutralizing Autoantibodies to Type I IFNs in >10% of Patients with Severe COVID-19 Pneumonia Hospitalized in Madrid, Spain. J. Clin. Immunol. 41, 914–922 (2021). 10.1007/s10875-021-01036-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vazquez S. E., Bastard P., Kelly K., Gervais A., Norris P. J., Dumont L. J., Casanova J. L., Anderson M. S., DeRisi J. L., Neutralizing Autoantibodies to Type I Interferons in COVID-19 Convalescent Donor Plasma. J. Clin. Immunol. 41, 1169–1171 (2021). 10.1007/s10875-021-01060-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goncalves D., Mezidi M., Bastard P., Perret M., Saker K., Fabien N., Pescarmona R., Lombard R., Walzer T., Casanova J.-L., Belot A., Richard J.-C., Trouillet-Assant S., Antibodies against type-I Interferon: detection and association with severe clinical outcome in COVID-19 patients. medRxiv, (2021). 10.1101/2021.04.02.21253262 [DOI] [PMC free article] [PubMed]
- 15. Wang E. Y., Mao T., Klein J., Dai Y., Huck J. D., Jaycox J. R., Liu F., Zhou T., Israelow B., Wong P., Coppi A., Lucas C., Silva J., Oh J. E., Song E., Perotti E. S., Zheng N. S., Fischer S., Campbell M., Fournier J. B., Wyllie A. L., Vogels C. B. F., Ott I. M., Kalinich C. C., Petrone M. E., Watkins A. E., Dela Cruz C., Farhadian S. F., Schulz W. L., Ma S., Grubaugh N. D., Ko A. I., Iwasaki A., Ring A. M.; Yale IMPACT Team , Diverse functional autoantibodies in patients with COVID-19. Nature 595, 283–288 (2021). 10.1038/s41586-021-03631-y [DOI] [PubMed] [Google Scholar]
- 16. van der Wijst M. G. P., Vazquez S. E., Hartoularos G. C., Bastard P., Grant T., Bueno R., Lee D. S., Greenland J. R., Sun Y., Perez R., Ogorodnikov A., Ward A., Mann S. A., Lynch K. L., Yun C., Havlir D. V., Chamie G., Marquez C., Greenhouse B., Lionakis M. S., Norris P. J., Dumont L. J., Kelly K., Zhang P., Zhang Q., Gervais A., Le Voyer T., Whatley A., Si Y., Byrne A., Combes A. J., Rao A. A., Song Y. S., Fragiadakis G. K., Kangelaris K., Calfee C. S., Erle D. J., Hendrickson C., Krummel M. F., Woodruff P. G., Langelier C. R., Casanova J. L., Derisi J. L., Anderson M. S., Ye C. J.; UCSF COMET consortium , Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci. Transl. Med. 13, eabh2624 (2021). 10.1126/scitranslmed.abh2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Acosta-Ampudia Y., Monsalve D. M., Rojas M., Rodríguez Y., Gallo J. E., Salazar-Uribe J. C., Santander M. J., Cala M. P., Zapata W., Zapata M. I., Manrique R., Pardo-Oviedo J. M., Camacho B., Ramírez-Santana C., Anaya J. M.; CP-COVID-19 group , COVID-19 convalescent plasma composition and immunological effects in severe patients. J. Autoimmun. 118, 102598 (2021). 10.1016/j.jaut.2021.102598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Solanich X., Rigo-Bonnin R., Gumucio V. D., Bastard P., Rosain J., Philippot Q., Perez-Fernandez X. L., Fuset-Cabanes M. P., Gordillo-Benitez M. A., Suarez-Cuartin G., Boza-Hernandez E., Riera-Mestre A., Parra-Martínez A., Colobran R., Antolí A., Navarro S., Rocamora-Blanch G., Framil M., Calatayud L., Corbella X., Casanova J. L., Morandeira F., Sabater-Riera J., Pre-existing Autoantibodies Neutralizing High Concentrations of Type I Interferons in Almost 10% of COVID-19 Patients Admitted to Intensive Care in Barcelona. J. Clin. Immunol. 41, 1733–1744 (2021). 10.1007/s10875-021-01136-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. Chauvineau-Grenier, P. Bastard, A. Servajean, A. Gervais, J. Rosain, E. Jouanguy, A. Cobat, J. L. Casanova, B. Rossi, Autoantibodies neutralizing type I interferons in 20% of COVID-19 deaths in a French hospital. Res Sq, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bastard P., Orlova E., Sozaeva L., Lévy R., James A., Schmitt M. M., Ochoa S., Kareva M., Rodina Y., Gervais A., Le Voyer T., Rosain J., Philippot Q., Neehus A. L., Shaw E., Migaud M., Bizien L., Ekwall O., Berg S., Beccuti G., Ghizzoni L., Thiriez G., Pavot A., Goujard C., Frémond M. L., Carter E., Rothenbuhler A., Linglart A., Mignot B., Comte A., Cheikh N., Hermine O., Breivik L., Husebye E. S., Humbert S., Rohrlich P., Coaquette A., Vuoto F., Faure K., Mahlaoui N., Kotnik P., Battelino T., Trebušak Podkrajšek K., Kisand K., Ferré E. M. N., DiMaggio T., Rosen L. B., Burbelo P. D., McIntyre M., Kann N. Y., Shcherbina A., Pavlova M., Kolodkina A., Holland S. M., Zhang S. Y., Crow Y. J., Notarangelo L. D., Su H. C., Abel L., Anderson M. S., Jouanguy E., Neven B., Puel A., Casanova J. L., Lionakis M. S., Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J. Exp. Med. 218, e20210554 (2021). 10.1084/jem.20210554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meisel C., Akbil B., Meyer T., Lankes E., Corman V. M., Staudacher O., Unterwalder N., Kölsch U., Drosten C., Mall M. A., Kallinich T., Schnabel D., Goffinet C., von Bernuth H., Mild COVID-19 despite autoantibodies against type I IFNs in autoimmune polyendocrine syndrome type 1. J. Clin. Invest. 131, e150867 (2021). 10.1172/JCI150867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H. H., Eto S., Garcia-Prat M., Bizien L., Parra-Martínez A., Yang R., Haljasmägi L., Migaud M., Särekannu K., Maslovskaja J., de Prost N., Tandjaoui-Lambiotte Y., Luyt C. E., Amador-Borrero B., Gaudet A., Poissy J., Morel P., Richard P., Cognasse F., Troya J., Trouillet-Assant S., Belot A., Saker K., Garçon P., Rivière J. G., Lagier J. C., Gentile S., Rosen L. B., Shaw E., Morio T., Tanaka J., Dalmau D., Tharaux P. L., Sene D., Stepanian A., Megarbane B., Triantafyllia V., Fekkar A., Heath J. R., Franco J. L., Anaya J. M., Solé-Violán J., Imberti L., Biondi A., Bonfanti P., Castagnoli R., Delmonte O. M., Zhang Y., Snow A. L., Holland S. M., Biggs C., Moncada-Vélez M., Arias A. A., Lorenzo L., Boucherit S., Coulibaly B., Anglicheau D., Planas A. M., Haerynck F., Duvlis S., Nussbaum R. L., Ozcelik T., Keles S., Bousfiha A. A., El Bakkouri J., Ramirez-Santana C., Paul S., Pan-Hammarström Q., Hammarström L., Dupont A., Kurolap A., Metz C. N., Aiuti A., Casari G., Lampasona V., Ciceri F., Barreiros L. A., Dominguez-Garrido E., Vidigal M., Zatz M., van de Beek D., Sahanic S., Tancevski I., Stepanovskyy Y., Boyarchuk O., Nukui Y., Tsumura M., Vidaur L., Tangye S. G., Burrel S., Duffy D., Quintana-Murci L., Klocperk A., Kann N. Y., Shcherbina A., Lau Y. L., Leung D., Coulongeat M., Marlet J., Koning R., Reyes L. F., Chauvineau-Grenier A., Venet F., Monneret G., Nussenzweig M. C., Arrestier R., Boudhabhay I., Baris-Feldman H., Hagin D., Wauters J., Meyts I., Dyer A. H., Kennelly S. P., Bourke N. M., Halwani R., Sharif-Askari N. S., Dorgham K., Sallette J., Sedkaoui S. M., AlKhater S., Rigo-Bonnin R., Morandeira F., Roussel L., Vinh D. C., Ostrowski S. R., Condino-Neto A., Prando C., Bonradenko A., Spaan A. N., Gilardin L., Fellay J., Lyonnet S., Bilguvar K., Lifton R. P., Mane S., Anderson M. S., Boisson B., Béziat V., Zhang S. Y., Vandreakos E., Hermine O., Pujol A., Peterson P., Mogensen T. H., Rowen L., Mond J., Debette S., de Lamballerie X., Duval X., Mentré F., Zins M., Soler-Palacin P., Colobran R., Gorochov G., Solanich X., Susen S., Martinez-Picado J., Raoult D., Vasse M., Gregersen P. K., Piemonti L., Rodríguez-Gallego C., Notarangelo L. D., Su H. C., Kisand K., Okada S., Puel A., Jouanguy E., Rice C. M., Tiberghien P., Zhang Q., Cobat A., Abel L., Casanova J. L.; HGID Lab; COVID Clinicians; COVID-STORM Clinicians; NIAID Immune Response to COVID Group; NH-COVAIR Study Group; Danish CHGE; Danish Blood Donor Study; St. James’s Hospital; SARS CoV2 Interest group; French COVID Cohort Study Group; Imagine COVID-Group; Milieu Intérieur Consortium; CoV-Contact Cohort; Amsterdam UMC Covid-19; Biobank Investigators; COVID Human Genetic Effort; CONSTANCES cohort; 3C-Dijon Study; Cerba Health-Care; Etablissement du Sang study group , Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol. 6, eabl4340 (2021). 10.1126/sciimmunol.abl4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.J. Manry, P. Bastard, A. Gervais, T. L. Voyer, J. Rosain, Q. Philippot, E. Michailidis, H. H. Hoffmann, S. Eto, M. Garcia-Prat, L. Bizien, A. Parra-Martinez, R. Yang, L. Haljasmagi, M. Migaud, K. Sarekannu, J. Maslovskaja, N. de Prost, Y. Tandjaoui-Lambiotte, C. E. Luyt, B. Amador-Borrero, A. Gaudet, J. Poissy, P. Morel, P. Richard, F. Cognasse, J. Troya, S. Trouillet-Assant, A. Belot, K. Saker, P. Garcon, J. G. Riviere, J. C. Lagier, S. Gentile, L. Rosen, E. Shaw, T. Morio, J. Tanaka, D. Dalmau, P. L. Tharaux, D. Sene, A. Stepanian, B. Megarbane, V. Triantafyllia, A. Fekkar, J. Heath, J. Franco, J. M. Anaya, J. Sole-Violan, L. Imberti, A. Biondi, P. Bonfanti, R. Castagnoli, O. Delmonte, Y. Zhang, A. Snow, S. Holland, C. Biggs, M. Moncada-Velez, A. Arias, L. Lorenzo, S. Boucherit, D. Anglicheau, A. Planas, F. Haerynck, S. Duvlis, R. Nussbaum, T. Ozcelik, S. Keles, A. Bousfiha, J. E. Bakkouri, C. Ramirez-Santana, S. Paul, Q. Pan-Hammarstrom, L. Hammarstrom, A. Dupont, A. Kurolap, C. Metz, A. Aiuti, G. Casari, V. Lampasona, F. Ciceri, L. Barreiros, E. Dominguez-Garrido, M. Vidigal, M. Zatz, D. van de Beek, S. Sahanic, I. Tancevski, Y. Stepanovskyy, O. Boyarchuk, Y. Nukui, M. Tsumura, L. Vidaur, S. Tangye, S. Burrel, D. Duffy, L. Quintana-Murci, A. Klocperk, N. Kann, A. Shcherbina, Y. L. Lau, D. Leung, M. Coulongeat, J. Marlet, R. Koning, L. Reyes, A. Chauvineau-Grenier, F. Venet, G. Monneret, M. Nussenzweig, R. Arrestier, I. Boudhabhay, H. Baris-Feldman, D. Hagin, J. Wauters, I. Meyts, A. Dyer, S. Kennelly, N. Bourke, R. Halwani, F. Sharif-Askari, K. Dorgham, J. Sallette, S. Mehlal-Sedkaoui, S. AlKhater, R. Rigo-Bonnin, F. Morandeira, L. Roussel, D. Vinh, C. Erikstrup, A. Condino-Neto, C. Prando, A. Bondarenko, A. Spaan, L. Gilardin, J. Fellay, S. Lyonnet, K. Bilguvar, R. Lifton, S. Mane, M. Anderson, B. Boisson, V. Beziat, S. Y. Zhang, E. Andreakos, O. Hermine, A. Pujol, P. Peterson, T. H. Mogensen, L. Rowen, J. Mond, S. Debette, X. deLamballerie, C. Burdet, L. Bouadma, M. Zins, P. Soler-Palacin, R. Colobran, G. Gorochov, X. Solanich, S. Susen, J. Martinez-Picado, D. Raoult, M. Vasse, P. Gregersen, C. Rodriguez-Gallego, L. Piemonti, L. Notarangelo, H. Su, K. Kisand, S. Okada, A. Puel, E. Jouanguy, C. Rice, P. Tiberghien, Q. Zhang, J. L. Casanova, L. Abel, A. Cobat, The risk of COVID-19 death is much greater and age-dependent with type I IFN autoantibodies. Res Sq, (2022). [Google Scholar]
- 24. Polack F. P., Thomas S. J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Moreira E. D., Zerbini C., Bailey R., Swanson K. A., Roychoudhury S., Koury K., Li P., Kalina W. V., Cooper D., Frenck R. W. Jr., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Jansen K. U., Gruber W. C.; C4591001 Clinical Trial Group , Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). 10.1056/NEJMoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas S. J., Moreira E. D. Jr., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J. L., Pérez Marc G., Polack F. P., Zerbini C., Bailey R., Swanson K. A., Xu X., Roychoudhury S., Koury K., Bouguermouh S., Kalina W. V., Cooper D., Frenck R. W. Jr., Hammitt L. L., Türeci Ö., Nell H., Schaefer A., Ünal S., Yang Q., Liberator P., Tresnan D. B., Mather S., Dormitzer P. R., Şahin U., Gruber W. C., Jansen K. U.; C4591001 Clinical Trial Group , Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine through 6 Months. N. Engl. J. Med. 385, 1761–1773 (2021). 10.1056/NEJMoa2110345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E. G., Rubin C., Indenbaum V., Tal I., Zavitan M., Zuckerman N., Bar-Chaim A., Kreiss Y., Regev-Yochay G., Covid-19 Breakthrough Infections in Vaccinated Health Care Workers. N. Engl. J. Med. 385, 1474–1484 (2021). 10.1056/NEJMoa2109072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuhlmann C., Mayer C. K., Claassen M., Maponga T., Burgers W. A., Keeton R., Riou C., Sutherland A. D., Suliman T., Shaw M. L., Preiser W., Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 399, 625–626 (2022). 10.1016/S0140-6736(22)00090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tenforde M. W., Self W. H., Adams K., Gaglani M., Ginde A. A., McNeal T., Ghamande S., Douin D. J., Talbot H. K., Casey J. D., Mohr N. M., Zepeski A., Shapiro N. I., Gibbs K. W., Files D. C., Hager D. N., Shehu A., Prekker M. E., Erickson H. L., Exline M. C., Gong M. N., Mohamed A., Henning D. J., Steingrub J. S., Peltan I. D., Brown S. M., Martin E. T., Monto A. S., Khan A., Hough C. L., Busse L. W., Ten Lohuis C. C., Duggal A., Wilson J. G., Gordon A. J., Qadir N., Chang S. Y., Mallow C., Rivas C., Babcock H. M., Kwon J. H., Halasa N., Chappell J. D., Lauring A. S., Grijalva C. G., Rice T. W., Jones I. D., Stubblefield W. B., Baughman A., Womack K. N., Rhoads J. P., Lindsell C. J., Hart K. W., Zhu Y., Olson S. M., Kobayashi M., Verani J. R., Patel M. M.; Influenza and Other Viruses in the Acutely Ill (IVY) Network , Association Between mRNA Vaccination and COVID-19 Hospitalization and Disease Severity. JAMA 326, 2043–2054 (2021). 10.1001/jama.2021.19499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leshem E., Nelson K., Lopman B. A., Severe breakthrough COVID-19 infections in Scotland-implications for immunisation programmes. Lancet Respir. Med. 9, 1354–1356 (2021). 10.1016/S2213-2600(21)00413-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Q., Bastard P., Cobat A., Casanova J. L.; COVID Human Genetic Effort , Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 603, 587–598 (2022). 10.1038/s41586-022-04447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lipsitch M., Krammer F., Regev-Yochay G., Lustig Y., Balicer R. D., SARS-CoV-2 breakthrough infections in vaccinated individuals: Measurement, causes and impact. Nat. Rev. Immunol. 22, 57–65 (2022). 10.1038/s41577-021-00662-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J., Laurie M. T., Rubio L., Vazquez S. E., Sunshine S., Mitchell A. M., Hapte-Selassie M., Mann S. A., Pilarowski G., Black D., Marquez C., Rojas S., Lionakis M. S., Petersen M., Whitman J. D., Jain V., Anderson M., Havlir D., DeRisi J., SARS-CoV-2 transmission dynamics and immune responses in a household of vaccinated persons. Clin. Infect. Dis. ciac029 (2022). 10.1093/cid/ciac029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carvalho T., Krammer F., Iwasaki A., The first 12 months of COVID-19: A timeline of immunological insights. Nat. Rev. Immunol. 21, 245–256 (2021). 10.1038/s41577-021-00522-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stertz S., Hale B. G., Interferon system deficiencies exacerbating severe pandemic virus infections. Trends Microbiol. 29, 973–982 (2021). 10.1016/j.tim.2021.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bastard P., Michailidis E., Hoffmann H. H., Chbihi M., Le Voyer T., Rosain J., Philippot Q., Seeleuthner Y., Gervais A., Materna M., de Oliveira P. M. N., Maia M. L. S., Dinis Ano Bom A. P., Azamor T., Araújo da Conceição D., Goudouris E., Homma A., Slesak G., Schäfer J., Pulendran B., Miller J. D., Huits R., Yang R., Rosen L. B., Bizien L., Lorenzo L., Chrabieh M., Erazo L. V., Rozenberg F., Jeljeli M. M., Béziat V., Holland S. M., Cobat A., Notarangelo L. D., Su H. C., Ahmed R., Puel A., Zhang S. Y., Abel L., Seligman S. J., Zhang Q., MacDonald M. R., Jouanguy E., Rice C. M., Casanova J. L., Auto-antibodies to type I IFNs can underlie adverse reactions to yellow fever live attenuated vaccine. J. Exp. Med. 218, e20202486 (2021). 10.1084/jem.20202486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manry J.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Michailidis E., Hoffmann H. H., Eto S., Garcia-Prat M., Bizien L., Parra-Martínez A., Yang R., Haljasmägi L., Migaud M., Särekannu K., Maslovskaja J., de Prost N., Tandjaoui-Lambiotte Y., Luyt C. E., Amador-Borrero B., Gaudet A., Poissy J., Morel P., Richard P., Cognasse F., Troya J., Trouillet-Assant S., Belot A., Saker K., Garçon P., Rivière J. G., Lagier J. C., Gentile S., Rosen L. B., Shaw E., Morio T., Tanaka J., Dalmau D., Tharaux P. L., Sene D., Stepanian A., Mégarbane B., Triantafyllia V., Fekkar A., Heath J. R., Franco J. L., Anaya J. M., Solé-Violán J., Imberti L., Biondi A., Bonfanti P., Castagnoli R., Delmonte O. M., Zhang Y., Snow A. L., Holland S. M., Biggs C. M., Moncada-Vélez M., Arias A. A., Lorenzo L., Boucherit S., Anglicheau D., Planas A. M., Haerynck F., Duvlis S., Ozcelik T., Keles S., Bousfiha A. A., El Bakkouri J., Ramirez-Santana C., Paul S., Pan-Hammarström Q., Hammarström L., Dupont A., Kurolap A., Metz C. N., Aiuti A., Casari G., Lampasona V., Ciceri F., Barreiros L. A., Dominguez-Garrido E., Vidigal M., Zatz M., van de Beek D., Sahanic S., Tancevski I., Stepanovskyy Y., Boyarchuk O., Nukui Y., Tsumura M., Vidaur L., Tangye S. G., Burrel S., Duffy D., Quintana-Murci L., Klocperk A., Kann N. Y., Shcherbina A., Lau Y. L., Leung D., Coulongeat M., Marlet J., Koning R., Reyes L. F., Chauvineau-Grenier A., Venet F., Monneret G., Nussenzweig M. C., Arrestier R., Boudhabhay I., Baris-Feldman H., Hagin D., Wauters J., Meyts I., Dyer A. H., Kennelly S. P., Bourke N. M., Halwani R., Sharif-Askari F. S., Dorgham K., Sallette J., Sedkaoui S. M., AlKhater S., Rigo-Bonnin R., Morandeira F., Roussel L., Vinh D. C., Erikstrup C., Condino-Neto A., Prando C., Bondarenko A., Spaan A. N., Gilardin L., Fellay J., Lyonnet S., Bilguvar K., Lifton R. P., Mane S., Anderson M. S., Boisson B., Béziat V., Zhang S. Y., Andreakos E., Hermine O., Pujol A., Peterson P., Mogensen T. H., Rowen L., Mond J., Debette S., de Lamballerie X., Burdet C., Bouadma L., Zins M., Soler-Palacin P., Colobran R., Gorochov G., Solanich X., Susen S., Martinez-Picado J., Raoult D., Vasse M., Gregersen P. K., Piemonti L., Rodríguez-Gallego C., Notarangelo L. D., Su H. C., Kisand K., Okada S., Puel A., Jouanguy E., Rice C. M., Tiberghien P., Zhang Q., Casanova J. L., Abel L., Cobat A.; HGID Lab; COVID Clinicians; COVID-STORM Clinicians; NIAID Immune Response to COVID Group; NH-COVAIR Study Group; Danish CHGE; Danish Blood Donor Study; St. James’s Hospital, SARS CoV2 Interest Group; French COVID Cohort Study Group; Imagine COVID-Group; Milieu Intérieur Consortium; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank Investigators; COVID Human Genetic Effort; CP-COVID-19 Group; CONSTANCES cohort; 3C-Dijon Study; Cerba Health-Care; Etablissement Français du Sang Study group , The risk of COVID-19 death is much greater and age dependent with type I IFN autoantibodies. Proc. Natl. Acad. Sci. U.S.A. 119, e2200413119 (2022). 10.1073/pnas.2200413119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meager A., Visvalingam K., Peterson P., Möll K., Murumägi A., Krohn K., Eskelin P., Perheentupa J., Husebye E., Kadota Y., Willcox N., Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLOS Med. 3, e289 (2006). 10.1371/journal.pmed.0030289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meager A., Wadhwa M., Dilger P., Bird C., Thorpe R., Newsom-Davis J., Willcox N., Anti-cytokine autoantibodies in autoimmunity: Preponderance of neutralizing autoantibodies against interferon-alpha, interferon-omega and interleukin-12 in patients with thymoma and/or myasthenia gravis. Clin. Exp. Immunol. 132, 128–136 (2003). 10.1046/j.1365-2249.2003.02113.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gupta A., Gonzalez-Rojas Y., Juarez E., Crespo Casal M., Moya J., Falci D. R., Sarkis E., Solis J., Zheng H., Scott N., Cathcart A. L., Hebner C. M., Sager J., Mogalian E., Tipple C., Peppercorn A., Alexander E., Pang P. S., Free A., Brinson C., Aldinger M., Shapiro A. E.; COMET-ICE Investigators , Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing Antibody Sotrovimab. N. Engl. J. Med. 385, 1941–1950 (2021). 10.1056/NEJMoa2107934 [DOI] [PubMed] [Google Scholar]
- 40. Lévy R., Zhang P., Bastard P., Dorgham K., Melki I., Hadchouel A., Hartoularos G. C., Neven B., Castelle M., Roy C., Toin T., Berteloot L., Bizien L., Abid H., Burgard M., Houhou-Fidouh N., Rozenberg F., Jouanguy E., Ye C. J., Gorochov G., Zhang Q., Casanova J. L., Monoclonal antibody-mediated neutralization of SARS-CoV-2 in an IRF9-deficient child. Proc. Natl. Acad. Sci. U.S.A. 118, e2114390118 (2021). 10.1073/pnas.2114390118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Beigel J. H., Tomashek K. M., Dodd L. E., Mehta A. K., Zingman B. S., Kalil A. C., Hohmann E., Chu H. Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R. W., Dierberg K., Tapson V., Hsieh L., Patterson T. F., Paredes R., Sweeney D. A., Short W. R., Touloumi G., Lye D. C., Ohmagari N., Oh M. D., Ruiz-Palacios G. M., Benfield T., Fätkenheuer G., Kortepeter M. G., Atmar R. L., Creech C. B., Lundgren J., Babiker A. G., Pett S., Neaton J. D., Burgess T. H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H. C.; ACTT-1 Study Group Members , Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 383, 1813–1826 (2020). 10.1056/NEJMoa2007764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gottlieb R. L., Vaca C. E., Paredes R., Mera J., Webb B. J., Perez G., Oguchi G., Ryan P., Nielsen B. U., Brown M., Hidalgo A., Sachdeva Y., Mittal S., Osiyemi O., Skarbinski J., Juneja K., Hyland R. H., Osinusi A., Chen S., Camus G., Abdelghany M., Davies S., Behenna-Renton N., Duff F., Marty F. M., Katz M. J., Ginde A. A., Brown S. M., Schiffer J. T., Hill J. A., Early Remdesivir to Prevent Progression to Severe Covid-19 in Outpatients. N. Engl. J. Med. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jayk Bernal A., Gomes da Silva M. M., Musungaie D. B., Kovalchuk E., Gonzalez A., Delos Reyes V., Martin-Quiros A., Caraco Y., Williams-Diaz A., Brown M. L., Du J., Pedley A., Assaid C., Strizki J., Grobler J. A., Shamsuddin H. H., Tipping R., Wan H., Paschke A., Butterton J. R., Johnson M. G., De Anda C., Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N. Engl. J. Med. (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Monk P. D., Marsden R. J., Tear V. J., Brookes J., Batten T. N., Mankowski M., Gabbay F. J., Davies D. E., Holgate S. T., Ho L. P., Clark T., Djukanovic R., Wilkinson T. M. A.; Inhaled Interferon Beta COVID-19 Study Group , Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 9, 196–206 (2021). 10.1016/S2213-2600(20)30511-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Elledge S. K., Zhou X. X., Byrnes J. R., Martinko A. J., Lui I., Pance K., Lim S. A., Glasgow J. E., Glasgow A. A., Turcios K., Iyer N. S., Torres L., Peluso M. J., Henrich T. J., Wang T. T., Tato C. M., Leung K. K., Greenhouse B., Wells J. A., Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 39, 928–935 (2021). 10.1038/s41587-021-00878-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zamecnik C. R., Rajan J. V., Yamauchi K. A., Mann S. A., Loudermilk R. P., Sowa G. M., Zorn K. C., Alvarenga B. D., Gaebler C., Caskey M., Stone M., Norris P. J., Gu W., Chiu C. Y., Ng D., Byrnes J. R., Zhou X. X., Wells J. A., Robbiani D. F., Nussenzweig M. C., DeRisi J. L., Wilson M. R., ReScan, a Multiplex Diagnostic Pipeline, Pans Human Sera for SARS-CoV-2 Antigens. Cell Rep. Med. 1, 100123 (2020). 10.1016/j.xcrm.2020.100123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hoffmann M., Kleine-Weber H., Pöhlmann S., A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 Is Essential for Infection of Human Lung Cells. Mol. Cell 78, 779–784.e5 (2020). 10.1016/j.molcel.2020.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Fig. S1
Table S1
Data file S1