ABSTRACT
Bacillus subtilis exhibits swarming motility, a flagellar-mediated form of surface motility. Here, we use transposon mutagenesis and sequencing (TnSeq) to perform a high-throughput screen for candidate genes required for swarming. The TnSeq approach identified all of the known genes required for flagellar biosynthesis and nearly all of the previously reported regulators that promote swarming. Moreover, we identified an additional 36 genes that improve swarming and validated them individually. Among these, two mutants with severe defects were recovered, including fliT, required for flagellar biosynthesis, and a gene of unknown function, yolB, whose defect could not be attributed to a lack of flagella. In addition to discovering additional genes required for B. subtilis swarming, our work validates TnSeq as a powerful approach for comprehensively identifying genes important for nonessential processes such as colony expansion on plates.
IMPORTANCE In TnSeq, transposons are randomly inserted throughout the chromosome at a population level, but insertions that disrupt genes of essential function cause strains that carry them to fall out of the population and appear underrepresented at the sequence level. Here, we apply TnSeq to the nonessential phenotype of motility in B. subtilis and spatially select for cells proficient in swarming. We find that insertions in nearly all genes previously identified as required for swarming are underrepresented in TnSeq analysis, and we identify 36 additional genes that enhance swarming. We demonstrate that TnSeq is a powerful tool for the genetic analysis of motility and likely other nonlethal screens for which enrichment is strong.
KEYWORDS: Bacillus, swarming, TnSeq, YolB, flagella, motility
INTRODUCTION
The ancestral strain of Bacillus subtilis NCIB3610 is capable of swarming motility, an active and social form of flagellar-mediated movement that occurs atop a solid surface (1, 2). Swarming over surfaces is thought to be different from swimming in liquid despite the fact that B. subtilis uses the same flagellar system for both behaviors. For example, when swimming cells are inoculated onto a soft agar surface, a lag period of immobility precedes the initiation of swarming, suggesting that a physiological change is necessary for the behavior (1, 3). One contributor to the lag is production of an extracellular surfactant that reduces surface tension and creates a thin layer of fluid within which to move (4, 5). Another factor that contributes to the lag is the time it takes to increase flagellar biosynthesis and increase the flagellar density (3, 6, 7). To determine the genetic requirements for swarming, a low-throughput transposon-based genetic screen was performed in which approximately 12,000 mutant colonies were picked, individually inoculated on miniature swarm agar plates, and manually screened for the inability to completely colonize the surface after overnight incubation (8). The forward genetic screen revealed a wide variety of “fla” mutants defective in flagellar biosynthesis as well as a mutant class called “swr” that abolished swarming motility but not swimming.
Study of swarming defective mutants has informed our understanding of how swarming and swimming differ. As one example, some mutants defective in the synthesis or activation of the extracellular lipopeptide surfactin abolished or impaired swarming, but these mutants could be rescued by providing purified surfactant exogenously (8–12). Other mutants exhibited cell-autonomous defects related to the flagellum. As examples, cells mutated for the cytoplasmic proteins SwrA and EF-P decreased transcription and translation of flagellar genes, respectively, while cells mutated for the membrane protein SwrB decreased the frequency of flagellar assembly (3, 13–15). In addition, cells mutated for the putative efflux protein SwrC were impaired for flagellar function, apparently due to membrane accumulation of endogenously synthesized surfactin (8, 16). The original transposon screen was not saturating, however, as other mutants have been discovered to have swarming defects like the two-component response regulator DegU and the polynucleotide phosphorylase PnpA that directly and indirectly activate flagellar gene expression, respectively (17–20). Thus, many of the genes that cell-autonomously promote swarming increase flagellar function or number.
B. subtilis synthesizes approximately 15 to 30 flagella per cell organized in a nonrandom pattern along the cell body (7, 13). The biosynthesis of each flagellum involves highly conserved proteins that are assembled sequentially from the inside out (21, 22). First, a basal body containing a type III secretion system is assembled in the membrane with a gear-like rotor docked on the cytoplasmic surface. Next, the type III secretion system exports structural components for the axle-like rod and universal joint-like hook. Once the rod-hook is polymerized to a particular length, the secretion system changes specificity and exports components of the helical propeller-like filament. Finally, proton motive force-consuming stator channels associate with the flagellar rotor and create torque. The B. subtilis genome encodes many but not all of the accepted flagellar structural proteins (22). Seemingly missing from the genome are homologs for the rod-cap protein that catalyzes rod polymerization and bushing proteins that surround the rod to stabilize flagellar rotation in the context of the envelope. Finally, while B. subtilis genes encode many hydrolase enzymes that remodel peptidoglycan, there is currently no known hydrolase required to create space in the wall for rod assembly (23, 24). Thus, in B. subtilis, the “missing parts” are either unnecessary or await discovery.
Here, we sought to identify additional genes required for flagellar assembly and/or swarming motility in B. subtilis. Previous work used transposon mutagenesis with a Tn10-based transposon somewhat prone to hot spot insertion, and manual screening of individual mutants was both cumbersome and limited by the diminishing return of the large set of genes already known to abolish motility when mutated (8). Here, we revisit the forward genetic approach but use a mariner-based transposon for mutagenesis and screen with systems-level high-throughput transposon sequencing (TnSeq) (25–29). As proof-of-concept, almost all genes previously reported to be required for flagellar biosynthesis and swarming motility were underrepresented in the TnSeq data set. Moreover, individual testing of predicted candidates revealed 36 newly identified mutants that exhibited swarming defects. All told, we show that TnSeq is a powerful approach to the genetic analysis of swarming motility. Moreover, the failure to reveal additional genes required for flagella structure in B. subtilis makes it increasingly likely that the missing parts do not exist, are not required for flagella biosynthesis in this organism, or will be missed through single-gene mutant analysis due to redundancy in the B. subtilis genome.
RESULTS
TnSeq analysis of swarming motility.
Transposon mutagenesis and high-throughput sequencing (TnSeq) requires a large library of mutants with diverse insertion sites. We chose a mariner-based transposon for library generation because it has been found to insert randomly at TA sequences, a pattern that is enriched in the low G+C content bacterium B. subtilis (29–31). Based on preliminary mutagenesis using standard conditions and plating, approximately 7% of CFU were transposants, which survived on spectinomycin plates at 42°C and were subsequently sequenced. Using this information, we generated a control mutant library containing approximately 1 million CFU from which chromosomal DNA was purified, processed, and sequenced using an Illumina platform. Separately, this control library was inoculated to the center of swarm agar plates (Fig. 1A). When the swarm radius reached 25 mm, the outer edge (greater than 15 mm off the center) was harvested by swabbing, and cells from 10 plates were combined to form a motility-enriched pool (Fig. 1A). Five different motility-enriched pools were generated from which chromosomal DNA was purified and sequenced.
FIG 1.
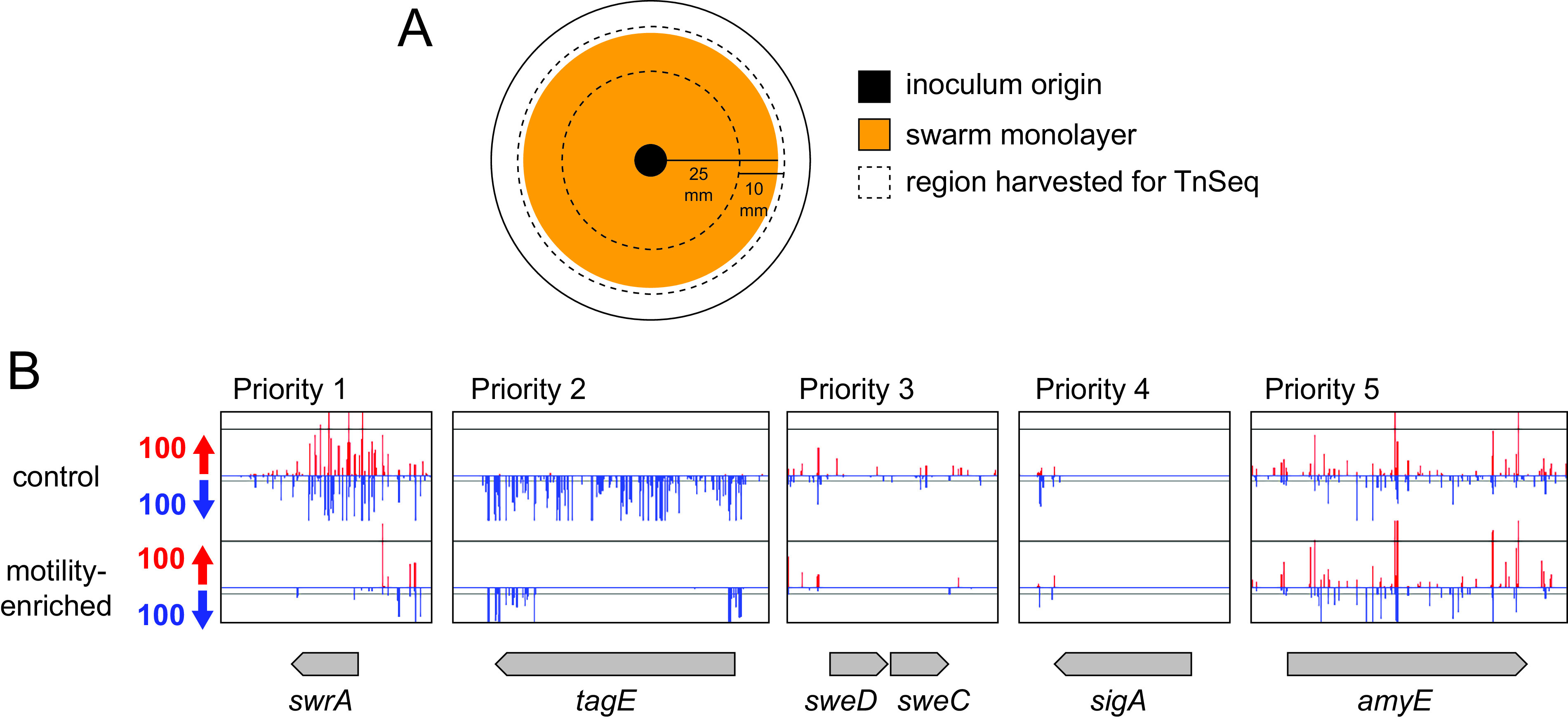
TnSeq priority classification of candidate genes defective in swarming motility. (A) Top view diagram of a petri plate used in the TnSeq selection for swarming motility. Outer circle represents perimeter of a petri plate. Black circle indicates the origin where the transposon mutagenized pool was inoculated. Orange circle indicates the area of the swarm with ~25 mm radius. The region between the dotted lines indicates the region from each plate that was harvested for pooling the cells prior to DNA isolation and transposon sequencing. (B) Sample genes with transposon insertion density graph generated using Artemis software. Transposon insertions on the top strand of the genome are colored red, while insertions on the bottom strand are colored blue. y axis indicates the number of sequencing reads at that insertion position. Gene length and orientation indicated as a gray arrow. Top data set is from the control library, and bottom data set is from one of the motility-enriched pools. Priority 1 represented genes that passed mathematical criteria for differential insertion, and the difference was readily visible on manual inspection. Priority 2 represented those genes that failed at least one of the mathematical criteria but were rescued by a clear insertion differential by manual inspection. Priority 3 genes were those that passed mathematical criteria but did not appear to have a dramatic insertion differential by manual inspection. Priority 4 genes were those genes that passed mathematical criteria but were discarded for having few, if any, transposon insertions. Priority 5 genes were those genes that passed neither mathematical nor manual inspection criteria and were discarded. Scale bar is 1 kb.
An average of 14 million reads were obtained for both the control and motility-enriched samples, ~97% of which were mapped to the B. subtilis genome. The complexity of a library was determined by comparing the number of possible TA sequences (mariner-insertion sites) in the chromosome relative to the number of TA sites that actually experienced transposon insertion in the pool. The B. subtilis wild-type genome has 218,028 possible TA sites, and the control library had at least two sequencing reads in 53% of the TA sites and at least 10 reads in 40% of the TA sites. Thus, the control library sampled insertions that both were diverse and exhibited high redundancy. Next, the insertions sequenced from the motility-enriched pools were compared to the control data set. We hypothesized that candidate genes required for swarming motility would be those genes that experienced a reduction in transposon insertion density specifically after selection for swarming proficiency.
To identify candidate genes worthy of further consideration, the transposon insertion density per open reading frame was compared by a combination of both mathematical and subjective criteria. Genes were mathematically included as potential candidates if they contained at least 10 potential TA sites within the coding region (i.e., were not so small that TA sites were inherently rare) and they experienced a statistically significant (P value of <0.05) 10-fold reduction in representation after swarming selection for at least 3 of the 5 motility-enriched transposon mutant pools. Genes were also subjectively curated by manual scanning of the graphical representation of transposon peak insertion density using Artemis software (32). Manual scanning could either increase confidence in the mathematical inclusion of a particular gene or add additional candidates that appeared to have a visually conspicuous reduction in insertion density after motility enrichment. Last, manual curation also excluded some mathematically supported candidates if the number of sequencing reads in that particular gene appeared to be low such that loss of those insertions during swarming could be either stochastic or due to a compounding growth defect. After the combined mathematical and manual screening of the data, genes were assigned a priority score from 1 to 5, with 1 being the highest priority and 5 being the lowest (Fig. 1B; see Fig. S1 in the supplemental material).
Genes were given a priority score of 1 if they passed all mathematical criteria and were also supported by manual screening with an insertion differential that was noticeable by eye. Genes were given a priority score of 2 if they failed one or more of the mathematical criteria but were rescued by manual screening because there was a visually dramatic reduction in insertion sites after motility enrichment. Genes were given a priority score of 3 if they passed the mathematical criteria but manual screening indicated that the insertion density differential did not appear to be dramatic, often because the insertion density in the control condition was low. Genes were given a priority score of 4 if they passed the mathematical criteria but were excluded by manual analysis for having too few insertions under both control and motility-selected conditions. Finally, genes were given a priority score of 5 if they failed to meet both the mathematical criteria and failed to be rescued by manual screening. Only those genes with priority scores of 1 to 3 were considered worthy of further analysis and individual phenotypic testing for swarming motility.
To determine whether the TnSeq screen for swarming motility-defective mutants was functioning as expected, we analyzed the list of candidates for genes known to be required for flagellar assembly and function (Table 1). Many of the genes required for the early stages of flagellar assembly, including the flagellar basal body, rod, and hook, are co-transcribed in the 32-gene fla/che operon, and the insertion density in this region was reduced in the motility-enriched population (Fig. 2A) (33–35). The genes dedicated to late-stage flagellar assembly, including the flagellar filament, are organized in multiple cistrons in a region called the “second flagellar cluster” located 180° from the fla/che operon on the circular chromosome, and flagellar genes in this region also experienced a reduction in insertion density (Fig. 2B). Two other loci are required for flagellar assembly and/or function: the flhO-flhP operon encoding the distal rod proteins FlhO and FlhP and the motA-motB operon encoding the force generating MotA-MotB stator units, and these operons also experienced a reduced insertion density (36–39). We conclude that the TnSeq screen for swarming-essential genes recovered every gene previously identified as being required for flagellar structure and function.
TABLE 1.
Genes predicted by TnSeq that were previously shown to be required for swarming motilitya
| Gene | PSb | Categoryc | Annotationd | Reference |
|---|---|---|---|---|
| cheA | 1 | Motility | Chemotaxis, run generator | 1 |
| cheC | 1 | Motility | fla/che operon, polar | 8 |
| cheD | 1 | Motility | fla/che operon, polar | 8 |
| cheW | 1 | Motility | fla/che operon, polar | 8 |
| cheY | 1 | Motility | Chemotaxis, run generator | 1 |
| cwlO | 1 | Envelope | PG hydrolase | 24 |
| degU | 1 | Regulation | Flagellar regulation | 18 |
| flgB | 1 | Motility | Flagellar structure | 82 |
| flgC | 1 | Motility | Flagellar structure | 82 |
| flgD | 1 | Motility | Flagellar structure | 36 |
| flgE | 1 | Motility | Flagellar structure | 36 |
| flgK | 1 | Motility | Flagellar structure | 38 |
| flgL | 1 | Motility | Flagellar structure | 38 |
| flgM | 1 | Motility | Flagellar regulation, polar | 82 |
| flgN | 1 | Motility | Flagellar structure | 83 |
| flhA | 1 | Motility | Flagellar assembly | 82 |
| flhB | 1 | Motility | Flagellar assembly | 82 |
| flhF | 1 | Motility | Flagellar regulation, polar | 13 |
| flhG | 1 | Motility | Flagellar regulation, polar | 13 |
| flhO | 1 | Motility | Flagellar structure | 36 |
| flhP | 1 | Motility | Flagellar structure | 36 |
| fliD | 1 | Motility | Flagellar structure | 84 |
| fliE | 1 | Motility | Flagellar structure | 82 |
| fliF | 1 | Motility | Flagellar structure | 82 |
| fliG | 1 | Motility | Flagellar structure | 82 |
| fliH | 1 | Motility | Flagellar assembly | 82 |
| fliI | 1 | Motility | Flagellar assembly | 82 |
| fliJ | 1 | Motility | Flagellar assembly | 82 |
| fliK | 1 | Motility | Flagellar regulation | 36 |
| fliL | 1 | Motility | Flagellar function, polar | 82 |
| fliM | 1 | Motility | Flagellar structure | 82 |
| fliO | 1 | Motility | Flagellar assembly | 82 |
| fliP | 1 | Motility | Flagellar assembly | 82 |
| fliQ | 1 | Motility | Flagellar assembly | 82 |
| fliR | 1 | Motility | Flagellar assembly | 82 |
| fliS | 1 | Motility | Flagellar assembly | 84 |
| fliY | 1 | Motility | Flagellar structure | 82 |
| gsaB | 1 | Translation | EF-P modification | 45 |
| hag | 1 | Motility | Flagellar structure | 1 |
| motA | 1 | Motility | Flagellar function | 37 |
| motB | 1 | Motility | Flagellar function | 37 |
| pdeH | 1 | Regulation | Flagellar function | 85 |
| pnpA | 1 | Regulation | Flagellar regulation | 19 |
| sigD | 1 | Motility | Flagellar regulation | 86 |
| swrA | 1 | Motility | Flagellar regulation | 8 |
| swrB | 1 | Motility | Flagellar assembly | 8 |
| swrC | 1 | Motility | Surfactin resistance | 8 |
| swrD | 1 | Motility | Flagellar function | 61 |
| ylxF | 1 | Motility | fla/che operon, polar | 82 |
| ymfI | 1 | Translation | EF-P activation | 44 |
| ynbB | 1 | Translation | EF-P modification | 45 |
| cheB | 2 | Motility | fla/che operon, polar | 8 |
| cwlQ | 2 | Envelope | PG hydrolase | 24 |
| minJ | 2 | Division | FtsZ localization factor | 87 |
| yabR | 2 | Unknown | Unknown | 8 |
| clpC | 3 | Regulation | AAA+ protease, unknown | 7 |
| clpX | 3 | Regulation | AAA+ protease, unknown | 7 |
| efp | 3 | Translation | Elongation factor P | 8 |
| rho | 3 | Transcription | Transcriptional terminator | 88 |
Table first sorted by priority score (PS) and second alphabetically by gene name.
Each gene was assigned a priority score (PS) based on data analysis. A score of 3 indicates that the gene passed the criteria of a ratio of 0.1 or lower and a statistical P value of <0.05 and could not be eliminated by manual evaluation of the data. A score of 2 indicates that the gene did not pass the ratio cutoff but was rescued because it appeared to have an insertion differential by visual scanning of the data set. A score of 1 indicates that the gene passed the quantitative criteria and also showed strong insertion differential by visual scanning of the data set.
Category is a general category of function assigned to the presumed or known function.
Annotation is a brief specific description of the known or predicted function of the gene product. “Polar” indicates that deletion of the gene does not impair swarming but transposon insertions within it are known to have polar effects on motility-essential genes downstream.
FIG 2.

TnSeq identified flagellar structural genes necessary for swarming motility. Transposon insertion densities within (A) the fla/che operon and (B) the second flagellar cluster graphed by Artemis software. Transposon insertions on the top strand of the genome are colored red, while insertions on the bottom strand are colored blue. y axis indicates the number of reads at that Tn insertion position. Gene length and orientation are indicated as arrows. Colored arrows indicate flagellar structural genes, and the colors match the relative location in the flagellum as indicated in panel C. (C) Cartoon diagram of the Gram-positive flagellum with parts colorcoded to the match the genes that make the corresponding structure in panels A and B.
Our TnSeq screen also identified a number of regulatory genes that have previously been reported to specifically confer defects in swarming motility (Table 1). For example, reduced insertion density was observed in the genes encoding proteins necessary for increased flagellar number during swarming, including the response regulator DegU, the master flagellar activator SwrA, and the activator of type III secretion SwrB (14, 40–42). Genes encoding other proteins known to be strictly required for swarming, such as the protease subunit ClpX, the polynucleotide phosphorylase PnpA, and the putative surfactin efflux protein SwrC, also experienced reduced insertion (7, 8, 19, 20). Finally, insertions within the gene encoding EF-P that enhances translation of a limiting flagellar structural protein and genes encoding YmfI and GsaB involved in EF-P activation were also underrepresented or absent (15, 43–45). We conclude that the TnSeq screen was successful at predicting regulators previously demonstrated to be required for swarming.
Some genes required for swarming motility were not predicted by the TnSeq analysis. For instance, the long srfAA-srfAD operon required for synthesis of the swarming surfactant, surfactin, the Com quorum sensing system that activates srfAA-srfAD gene expression, and the sfp gene required for enzymatic activation of the surfactin synthase complex all did not appear to be required (10, 46, 47). Their absence from the candidate list is likely explained because each gene contributes to the production of surfactin, and surfactin defects can be extracellularly complemented for swarming motility (1, 48). Presumably, the other members of the mixed mutant pool provided surfactin sufficient for the rare deficient mutants. One other gene known to be required for swarming, which encodes the biofilm matrix and flagellar clutch repressor SinR, was not predicted to be required by the TnSeq analysis, but this was likely because sinR experienced a low overall insertion density in the control library (49, 50). The failure to obtain insertions in the sinR gene may have been stochastic or because sinR mutants derepress biofilm matrix genes and excessive cell clumping may have impaired DNA recovery. All told, the TnSeq screen provided proof-of-principle results by predicting 59 out of 60 genes previously reported to be required for swarming in a cell-autonomous manner.
TnSeq identifies new genes required for swarming motility.
In addition to identifying the known motility genes, TnSeq analysis predicted 78 new candidate genes (Table S1), with priority scores from 1 to 3 (Fig. 1B). To validate these candidates, we made disruptions of those genes and quantitatively assayed the mutant strains for swarming motility. While many had behavior indistinguishable from that of the wild type (Table S1), 36 of the mutants exhibited a defect in swarming motility (Table 2). Twelve of the mutants were phenotypically classified as “slow” because the defect in swarming motility appeared to be due to a reduced rate of colony expansion (Fig. 3, cyan). Fifteen of the mutants were phenotypically classified as “extended lag” because the rate of swarm expansion appeared to be comparable to that of wild type but the period of immotility prior to swarming initiation was prolonged (Fig. 3, purple). Seven of the mutants were phenotypically classified as “terracing” because they exhibited a periodic cessation and reinitiation of swarming motility that gave rise to a terraced appearance of the colony after overnight incubation (Fig. 3, orange; Fig. S2). Finally, two of the mutants were phenotypically classified as “severe” because they failed to expand substantially from the inoculation origin during the period of measurement and even after overnight incubation (Fig. 3, red). We conclude that the TnSeq approach was successful at identifying new genes involved in swarming motility.
TABLE 2.
Candidate genes which when mutated conferred a defect in swarming motilitya
| Gene | PSb | Categoryc | Annotationd | Phenotype | Strain |
|---|---|---|---|---|---|
| dacA | 1 | Envelope | Penicillin binding protein 5 | Slow | DK654 |
| glnA | 3 | Metabolism | Glutamine synthetase | Slow | DK8619 |
| ksgA | 2 | Translation | rRNA methyltransferase | Slow | DS1342 |
| mecA | 3 | Regulation | Proteolytic adaptor | Slow | DS5334 |
| prmC | 1 | Translation | Release factor modification | Slow | DK8460 |
| whiA | 1 | Division | DNA binding protein | Slow | DK7713 |
| yaaR | 2 | Unknown | Unknown | Slow | DK9253 |
| ybaF | 2 | Envelope | Putative transporter | Slow | DK8614 |
| ylbM | 1 | Translation | tRNA modification protein | Slow | DK9257 |
| yopQ | 1 | Unknown | Unknown | Slow | DK8572 |
| ytlQ | 1 | Unknown | Unknown | Slow | DK4956 |
| yvcJ | 1 | Unknown | Unknown | Slow | DK8626 |
| greA | 3 | Transcription | Transcription elongation factor | Extended lag | DK9259 |
| lpdV | 1 | Envelope | Oxoisovalerate dehydrogenase | Extended lag | DK9234 |
| pgcA | 1 | Envelope | Phosphoglucomutase | Extended lag | DS2802 |
| rnhC | 2 | Transcription | RNase HIII | Extended lag | DK7763 |
| rnr | 1 | Regulation | RNase R | Extended lag | DK8421 |
| rsiX | 1 | Regulation | Anti-SigX anti-sigma factor | Extended lag | DK9233 |
| secDF | 1 | Secretion | Preprotein translocase | Extended lag | DS1138 |
| sweC | 3 | Envelope | Regulator of CwlO | Extended lag | DK8056 |
| sweD | 3 | Envelope | Regulator of CwlO | Extended lag | DK8058 |
| yaaT | 2 | Regulation | Regulator of RNase Y/Spo0A | Extended lag | DS6674 |
| yeaC | 2 | Unknown | Unknown | Extended lag | DK8525 |
| yeaD | 2 | Unknown | Unknown | Extended lag | DK4825 |
| yebA | 2 | Unknown | Unknown | Extended lag | DK8417 |
| ylbF | 1 | Regulator | Regulator of RNaseY/Spo0A | Extended lag | DS678 |
| ymcA | 1 | Regulator | Regulator of RNaseY/Spo0A | Extended lag | DS677 |
| yvpB | 1 | Unknown | Unknown | Terracing | DK7305 |
| ftsE | 1 | Envelope | Activator of CwlO | Terracing | DS1240 |
| ftsX | 3 | Envelope | Activator of CwlO | Terracing | DS1242 |
| gtaB | 1 | Envelope | G1P-uridylyltransferase | Terracing | DK7855 |
| walH | 1 | Envelope | Inhibitor of WalK | Terracing | DK8424 |
| yjbH | 2 | Regulation | Proteolytic adaptor-Spx | Terracing | DS8875 |
| yjcN | 1 | Unknown | Unknown | Terracing | DK8570 |
| fliT | 2 | Motility | Filament cap chaperone | Severe | DK9265 |
| yolB | 1 | Unknown | Unknown | Severe | DK8419 |
Table sorted by phenotype first and alphabetically by gene name second.
Each gene was assigned a priority score (PS) based on data analysis. A score of 3 indicates that the gene passed the criteria of a ratio of 0.1 or lower and a statistical P value of <0.05 and could not be eliminated by manual evaluation of the data. A score of 2 indicates that the gene did not pass the ratio cutoff but was rescued because it appeared to have a strong insertion differential by visual scanning of the data set. A score of 1 indicates that the gene passed the quantitative criteria and also showed strong insertion differential by visual scanning of the data set.
Category is a general category of function assigned to the presumed or known function.
Annotation is a brief specific description of the known or predicted function of the gene product.
FIG 3.
Newly identified genes which when mutated exhibit defects in swarming motility. (A) Quantitative swarm expansion assays of wild type (open circles) and mutant (closed circles) as indicated by the gene name in the upper left-hand corner of the graph. Some wild type data have been duplicated, as the same wild type data were used as a control for mutants that were analyzed for swarming concurrently. Each data point is the average of three replicates. Swarm graphs are grouped according to their respective class of defect: cyan, slow swarm expansion; purple, extended lag period; orange, terracing mutants; red, severe swarm defect. (B) Quantitative swarm expansion assays of the indicated mutants that had been backcrossed and maintained under high Mg2+ conditions and swarmed on standard conditions of 0.7% agar LB (black circles) and 0.7% agar TY (gray circles). The swarming behavior of wild type on 0.7% agar TY is indicated by white circles, and the data are repeated as a standard comparator for the simultaneously conducted experiments in panel B. Each data point is the average of three replicates. Coloring of class mutants is the same as that in panel A, save that a gray letter is included to show that swarming is indistinguishable from that of the wild type on TY. The following strains were used in panel B: wild type (DK1042), dltD (DK9658), ggaB (DK9659), tagE (DK9660), pgcA (DK9661), ugtP (DK9663), and gtaB (DK9664).
While successful in some regards, TnSeq also gave rise to a high frequency of false-positive candidates. We noted that insertions in genes related to the Gram-positive cell envelope component teichoic acid appeared to be strongly discriminated against after motility selection in TnSeq but some mutants swarmed like the wild type when tested individually (e.g., tagE, Fig. 1; Table S1). Teichoic acid synthesis mutants grow poorly with cell shape defects when cultivated under low magnesium (Mg2+) conditions (51–55), and thus propagation on LB may have inadvertently selected for suppressors that simultaneously improved growth and swarming motility. Six mutants defective in genes related to teichoic acid and cell envelope synthesis were chosen for further analysis, two of which had swarming phenotypes (gtaB and pgcA) and four of which did not (dltD, ggaB, tagE, and ugtP). Chromosomal DNA from each strain was purified, transformed, and selected on media containing high Mg2+. Finally, each resulting mutant was maintained on high Mg2+ and assayed for swarming on soft agar separately made with our standard condition LB and high Mg2+ TY medium. Two mutants presented new swarming defects on TY swarm agar (ggaB and tagE), supporting the idea that genetic suppression may have produced a false-negative result. Conversely, two mutants (gtaB and pgcA) reduced or abolished the swarming defect on TY agar, suggesting that genetic suppression may have exacerbated the original swarming defect. None of the backcrossed mutants exhibited a severe nonswarming phenotype. We conclude that mutants defective in envelope-related genes should be treated with care during growth and should be genetically complemented before further exploring their role in motility.
We noted that some of the mutants with swarming defects grew slowly in liquid LB medium, and we wondered how frequently growth defects were correlated with impaired swarming. Each mutant with a swarming defect was grown in a microtiter plate reader in triplicate, and the maximum growth rate during exponential growth was calculated. Relatively few mutants exhibited a dramatic decrease in growth rate relative to that of the wild type (Fig. 4). Reduction in growth rate, however, was more common in those mutants that exhibited a slow swarming phenotype. We infer that a reduction in growth rate is potentially one way to cause a defect in swarming motility, but whether the swarming defect is directly due to the growth rate change or some other consequence of the mutation is unknown. Interestingly, mutants with reduced growth rate did not appear to increase the lag time prior to swarming (e.g., glnA), suggesting that growth rate and lag period were unrelated (Fig. 4). Finally, we note that the mutants that conferred a severe defect in swarming motility, fliT and yolB, exhibited growth rates comparable to those of the wild type, and we infer that the swarming defect was due to some other factor.
FIG 4.
Growth rates of mutants with swarming defects. Cells were grown in a 96-well microtiter plate, and rates are the average of three replicate experiments. Error bars represent the standard deviation. Dashed lines are extensions of the standard deviation of the wild type for quick comparison.
One way in which a mutation can result in a severe defect in swarming motility is if the mutant is defective in flagellar biosynthesis. To detect flagellar biosynthesis, a version of the flagellar filament protein flagellin that can be fluorescently labeled with a maleimide derivative (hagT209C) (50) was introduced into both the fliT and yolB mutant backgrounds. Whereas the wild type was proficient for flagella biosynthesis, the fliT mutant was defective (Fig. 5). The fliT gene encodes a homolog of FliT, a bifunctional protein in Salmonella that serves primarily as the secretion chaperone for the flagellar cap protein FliD (56–60). The fliT flagellar filament defect observed in B. subtilis is consistent with a FliD secretion defect, and FliT is likely the last flagellar protein homolog that could have been predicted to have a motility defect by a reverse genetic approach. In contrast to fliT, the yolB mutant was proficient in flagellar biosynthesis (Fig. 5). The yolB gene encodes the protein of unknown function YolB, and the reason a yolB mutant is defective in swarming is unknown. As flagella are required for swarming motility, we infer that all of the other swarming mutants with partial defects in behavior are flagella proficient. Thus, the TnSeq screen identified new genes involved in swarming motility but seemingly no additional B. subtilis-specific genes required for flagellar assembly.
FIG 5.
Cells mutated for yolB are proficient for flagellar biosynthesis. Fluorescent micrographs of wild type (WT; DS1916), fliT (DK9348), and yolB (DK8486) mutants expressing the flagellar filament allele HagT209C and stained for membrane with FM 4 to 64 (false colored red) and flagella with Alexa fluor maleimide (false colored green).
DISCUSSION
The transposon mutagenesis with high-throughput sequencing (TnSeq) protocol described here was highly successful in identifying genes required for swarming motility. By selecting the subpopulation of the transposon mutant pool that was swarming proficient, we excluded individuals with transposon insertions in swarming-essential genes, and such genes experienced a reduction in insertion density after sequencing. Ultimately, our mathematical and manual screening of the insertion data was able to predict 59 of 60 genes previously reported to be required for swarming. Moreover, the data indicated that another 78 genes experienced a similar reduction in insertion density after motility enrichment, and these became candidate genes for individual validation. Manual testing of individual mutants found that roughly half of the candidates exhibited a swarming defect when tested in isolation, proving that the approach could identify new genes involved in the phenotype. Moreover, we found that many of the mutants that reduced swarming were associated with either envelope maintenance or efficient translation, perhaps consistent with the need for high numbers of proteinaceous transenvelope flagella. Finally, mutation of one gene, yolB, resulted in a severe defect in swarming, consistent with being a new member of the swr genetic class.
The swr genetic class is defined as those genes which, when mutated, abolish swarming motility but not swimming (8). Moreover, the swarming defect of a swr mutant is cell-autonomous and cannot be complemented extracellularly, thus excluding genes producing shared secreted products like surfactants and quorum signals. Preliminarily, yolB meets the criteria of a swr gene, as mutation results in a severe swarming defect both in mixed mutant pools and in isolation, while seemingly not impairing the production of flagella or impairing swimming, as motility was observed by wet mount observation. The yolB gene encodes YolB, an 118-amino-acid protein of unknown function that has no domains conserved with other proteins in the database. How YolB might promote swarming motility is unclear. Perhaps it is needed to increase flagellar number on surfaces like SwrA or to increase the power to individual flagellar motors like SwrD (13, 61). Whatever the case, YolB is encoded within the B. subtilis chromosomal prophage Spβ, and its activity may represent a form of phenotypic conversion by a horizontally transferred element.
Another class of mutants that causes a severe defect in swarming motility is the fla class, whose mutation abolishes flagellar biosynthesis (1). Only one new gene, fliT, met the criteria for a fla class mutant. While fliT could no doubt have been predicted by sequence analysis, we include it here, as, to the best of our knowledge, it has not been shown to have a motility defect in B. subtilis per se. We note, however, that the fliT mutant flagellar filament defect is entirely consistent with the biochemically supported role of FliT in B. subtilis as a secretion chaperone for the extracellular filament cap FliD (59). The FliT homolog in Salmonella enterica is multifunctional, serving both as a FliD secretion chaperone and as an adaptor for the regulatory proteolysis of the master activator of flagellar transcription (58, 60). Whether B. subtilis FliT is also multifunctional remains to be determined, but we note that the proteolytic adaptor function (targeting the master activator SwrA) is performed by SmiA, the protein encoded immediately downstream of FliT (7, 23, 62). Whether B. subtilis FliT plays multiple roles in B. subtilis flagellar regulation, including the proteolytic modulation of the master activator of flagellar transcription SwrA, is unknown.
In addition to the mass confirmation of genes involved in swarming and flagellar biosynthesis, the large TnSeq data set indicates that the original manual screen used to identify important genes was rather effective, as few additional mutants with severe defects were discovered (8). In addition, the system-wide analysis and large data set allow interpretation of genes that were not identified as candidates. For example, the putative missing parts in flagellar biosynthesis, specifically the rod polymerization cap protein and peptidoglycan bushings, remain undiscovered. Either the B. subtilis genome encodes multiple proteins that are both sequence-divergent and functionally redundant, or the rod-cap and bushing proteins are unnecessary and should perhaps be considered modifications associated with a Gram-negative envelope rather than essential structural components. We note that no indication of a bushing has been seen associated with flagella purified from B. subtilis (63–66) and that the outer membrane bushings and rod cap are absent from a number of bacteria, including those outside the Gram-positive phylogeny (66–70). While we acknowledge that it remains a possibility that these structural proteins were missed for technical (e.g., low insertion density in control) or biological reasons (e.g., essentiality) inherent to the screen, we must also acknowledge that there is no evidence a priori that such proteins either exist or are necessary in B. subtilis.
Despite the fact that data from B. subtilis originated the idea that flagellar assembly requires a dedicated peptidoglycan hydrolase, to date no peptidoglycan hydrolase has been identified to support the hypothesis in B. subtilis (13, 22, 23, 71, 72). Here, TnSeq analysis also failed to reveal a candidate for a peptidoglycan hydrolase supporting flagellar assembly. TnSeq and other genetic approaches may have failed to identify a target due to functional redundancy, but simultaneous deletion of the five seemingly most relevant candidates failed to abolish flagella (24). Instead of flagellar assembly directing peptidoglycan remodeling, perhaps peptidoglycan structure dictates the location of flagellar insertion. B. subtilis inserts flagella in a nonrandom peritrichous arrangement, and TnSeq indicated that the vegetative rod elongation hydrolase CwlO, its regulator WalH, and its modulators FtsEX and SweCD were required for optimal motility (13, 73–75). The diameter of the flagellar rod exceeds the average pore size of peptidoglycan (64, 66, 76, 77), but holes of sufficient dimension, perhaps mediated in part by CwlO, may be preexistent, and perhaps flagellar assembly is guided to these locations. Even so, mutations of WalH, FtsEX, SweCD, or CwlO impaired but did not abolish motility and thus likely did not abolish flagella (24). How the B. subtilis flagellar rod transits the thick peptidoglycan layer of the Gram-positive envelope remains unknown.
MATERIALS AND METHODS
Strains and growth conditions.
B. subtilis strains were grown in lysogeny broth (LB; 10 g tryptone, 5 g yeast extract, 5 g NaCl per L) or on LB plates fortified with 1.5% Bacto agar at 37°C. The high Mg2+ medium TY was made by adding 10 mM MgSO4 (final concentration) and 1 mM MnSO4 (final concentration) to LB. When appropriate, antibiotics were included at the following concentrations: 10 μg/mL tetracycline, 100 μg/mL spectinomycin, 5 μg/mL chloramphenicol, 5 μg/mL kanamycin, and 1 μg/mL erythromycin plus 25 μg/mL lincomycin (mls). Isopropyl β-d-thiogalactopyranoside (IPTG; Sigma) was added to the medium at the indicated concentration when appropriate.
Swarm expansion assay.
Cells were grown to mid-log phase at 37°C in LB and resuspended to an optical density at 600 nm (OD600) of 10 in distilled water containing 0.5% India ink (Higgins). Freshly prepared LB fortified with 0.7% Bacto agar (25 mL/plate) was dried for 10 min in a laminar flow hood, centrally inoculated with 10 μL of the cell suspension, dried for another 10 min, and incubated at 37°C. The India ink demarks the origin of the colony, and the swarm radius was measured relative to the origin. For consistency, an axis was drawn on the back of the plate and swarm radii measurements were taken along this transect.
Plate reader growth curves.
Colonies were inoculated into 3 mL LB broth, grown at 37°C in a roller drum until cultures were turbid, and back diluted to an OD600 of 0.001. Next, 1 mL of each of the cultures was loaded in technical triplicate into a Greiner Bio-One Cellstar 24-well cell culture plate with no lid. All remaining wells in the culture plate were filled with 1 mL of LB broth, both to serve as a sterile control and to minimize water evaporation. The plate was loaded onto a BioTek Synergy H1 microplate reader, and growth curves were measured using BioTek microplate reader and imager Gen5 3.11 software via the following protocol. Procedure was set as a “kinetic run” for 15 h with absorbance readings of 600 A every 20 min. The plate was set to shake in a continuous double-orbital at 37°C. At the end of the kinetic run, the temperature was set to turn off and the plate was held at room temperature. OD600 readings were exported to an Excel spreadsheet, growth rate was calculated over a series of 1-h windows, and the maximum growth rate was calculated.
Microscopy.
Fluorescence microscopy was performed with a Nikon 80i microscope with a phase contrast objective Nikon Plan Apo 100× and an Excite 120 metal halide lamp. FM4-64 (Molecular Probes) was visualized with a C-FL HYQ Texas Red filter cube (excitation filter 532 to 587 nm, barrier filter >590 nm). Alexa Fluor 488 C5 maleimide was visualized using a C-FL HYQ FITC filter cube (FITC; excitation filter 460 to 500 nm, barrier filter 515 to 550 nm). Images were captured with a Photometrics Coolsnap HQ2 camera in black and white, false colored, and superimposed using Metamorph image software.
For fluorescence microscopy of flagella, 0.5 mL of broth culture was harvested at an OD600 of ~1 and washed once in 1.0 mL of pH 8.0 phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4). The suspension was pelleted, resuspended in 50 μL of PBS containing 5 μg/mL Alexa Fluor 488 C5 maleimide (Molecular Probes), and incubated for 5 min at room temperature (50). One milliliter of PBS was added, and cells were pelleted and resuspended in 50 μL of 5 μg/mL FM4-64. Three microliters of suspension were placed on a microscope slide and immobilized with a poly-l-lysine-treated coverslip.
Strain construction.
Mutants were assembled from three primary approaches. First, we used transposon insertions in the gene of interest that were preexistent in our strain database from past screens. Second, we obtained insertion-deletion alleles in which the kanamycin resistance gene replaced the gene of interest from the BGSC (Bacillus Genetic Stock Center, Columbus, OH) originally generated as part of a high-throughput mutagenesis approach and used SPP1-mediated generalized transduction to transfer the insertion-deletion mutation into the swarming proficient wild type (78, 79). Third, we generated our own insertion-deletion alleles with a kanamycin resistance cassette replacement by long-flanking homology with adjacent DNA sequence and Gibson isothermal assembly (62). All strains used in the manuscript are listed in Table S2, and all primers are listed in Table S3.
For long-flanking homology replacements by isothermal assembly, the regions upstream and downstream of each indicated gene were PCR amplified using the two pairs of primers indicated in the parentheses. Next, DNA containing a kanamycin resistance gene was amplified from plasmid pDG780 (80) using universal primers 3250/3251. The three DNA fragments were combined at equimolar amounts to a total volume of 5 μL and added to a 15-μL aliquot of prepared master mix (see below). The reaction mixture was incubated for 60 min at 50°C. The completed reaction was then PCR amplified using the outside primers to amplify the assembled product. The amplified product was transformed into competent cells of DK1042. Insertions were verified by PCR amplification of mutant chromosomal DNA using the outside primers used for construction. The following primer pairs were used to build the following mutants: cshA (6861/6862, 6863/6864), cshB (6865/6866, 6867/6866), dltB (7175/7176, 7177/7178), gtaB (5421/5420, 5419/5418), rnhC (7238/7239, 7240/7241), rnmV (7179/7180, 7181/7182), whiA (7146/7147, 7148/7149), ykaA (6876/6877, 6878/6879), ypmA (7167/7168, 7169/7170), ypmB (7142/7143, 7144/7145), ytlQ (5769/5770, 5771/5772), and yvpB (6853/6854, 6855/6856).
To prepare the isothermal assembly reactions, a 5× isothermal assembly reaction buffer (500 mM Tris-HCl [pH 7.5], 50 mM MgCl2, 50 mM dithiothreitol [DTT; Bio-Rad], 31.25 mM PEG-8000 [Fisher Scientific], 5.02 mM NAD [Sigma-Aldrich], and 1 mM each dNTP [New England BioLabs]) was aliquoted and stored at –80°C. An assembly master mixture was made by combining prepared 5× isothermal assembly reaction buffer (131 mM Tris-HCl, 13.1 mM MgCl2, 13.1 mM DTT, 8.21 mM PEG-8000, 1.32 mM NAD, and 0.26 mM each dNTP) with Phusion DNA polymerase (New England BioLabs; 0.033 units/μL), T5 exonuclease diluted 1:5 with 5× reaction buffer (New England BioLabs; 0.01 units/μL), Taq DNA ligase (New England BioLabs; 5,328 units/μL), and additional dNTPs (267 μM). The master mix was aliquoted as 15 μL and stored at –80°C.
SPP1 phage transduction.
To 0.1 mL of dense culture grown in TY broth (LB supplemented after autoclaving with 10 mM MgSO4 and 100 μM MnSO4), serial dilutions of SPP1 phage stock were added and statically incubated for 15 min at 37°C. To each mixture, 3 mL TYSA (molten TY supplemented with 0.5% agar) was added, poured atop fresh TY plates, and incubated at 37°C overnight. Top agar from the plate containing near confluent plaques was harvested by scraping into a 50-mL conical tube, vortexed, and centrifuged at 5,000 × g for 10 min. The supernatant was treated with 25 μg/mL DNase final concentration before being passed through a 0.45-μm syringe filter and stored at 4°C. Recipient cells were grown to stationary phase in 2 mL TY broth at 37°C. One milliliter of cells was mixed with 5 μL of SPP1 donor phage stock. Nine milliliters of TY broth was added to the mixture and allowed to stand at 37°C for 30 min. The transduction mixture was then centrifuged at 5,000 × g for 10 min, the supernatant was discarded, and the pellet was resuspended in the remaining volume. One hundred microliters of cell suspension was then plated on TY fortified with 1.5% agar, the appropriate antibiotic, and 10 mM sodium citrate.
TnSeq.
We performed TnSeq using the mariner transposon delivery plasmid pWX642 (31). This plasmid contains (i) a spectinomycin resistance gene for selection, (ii) an MmeI site inside one of the inverted repeats to facilitate DNA library prep and determine the orientation of Tn insertion, (iii) the HiMar transposase under constitutive expression to allow Tn hopping, and (iv) a temperature-sensitive replication origin, which allows plasmid loss at high temperature. After plasmid loss, the spec resistant colonies contain a permanent chromosomal Tn insertion that is no longer mobile due to the loss of transposase.
To generate the strain for mutagenesis, DK1042 was transformed with 1 μL of pWX642 (31), plated on LB agar containing mls, and incubated at 30°C for 48 h. This step allows for the uptake of the Tn delivery plasmid. To avoid “jackpot,” four individual colonies were inoculated into one tube containing 4 mL LB spec100 broth and rolled at 22°C for 20 h, and this growth procedure was repeated with 4 parallel tubes. The 4 cultures were pooled, OD600 was measured, the cultures were diluted to an OD600 of 1 using LB, and 500 μL of culture was mixed with glycerol to a final concentration of 14%. Cultures were stored in cryotubes and frozen using liquid nitrogen. To determine the number of transposants per mL, the thawed library was serially diluted, separately plated on LB and LB spec100 plates, and incubated overnight at 42°C. Because pWX642 has a temperature-sensitive replication origin, only the Tn insertions on the genome will allow the cells to grow on LB spec100 plates. The transposition efficiency, determined by dividing the number of spectinomycin-resistant colonies by the total number of colonies, was approximately 7%.
To generate the mutant libraries, the mutagenized pools were diluted appropriately, plated on 20 large (150 by 15 mm) LB spec100 agar plates, and incubated at 42°C overnight such that each plate had ~50,000 colonies. Colonies were harvested with a rubber scraper, the slurry was diluted to an OD600 of 5, and genomic DNA was purified using the DNeasy blood and tissue DNA purification kit (Qiagen). In parallel, 10 μL of glycerol stock was spotted in the center of 10 0.7% LB agar plates and was incubated at 37°C until the swarm edge reached about 25 mm from the origin. The 10 mm edge of the swarm was harvested with a cotton swab into LB medium, and chromosomal DNA was purified as described above from the resulting slurry. The procedure was repeated in parallel to produce five motility-enriched libraries.
Next, 200 ng of genomic DNA from each library was digested with MmeI, and each library was separately ligated in barcode-tagged adapters with T4 DNA ligase incubated overnight at 16°C. Reactions were PCR amplified, the products were purified, and ~20 ng of each was sequenced on an Illumina NextSeq 500 machine in the Center for Genomics and Bioinformatics (CGB) at Indiana University with a setup of 82-nucleotide (nt) single-end reads, but only the first 35 nt were needed for analysis. We found that for the genome size of B. subtilis (~4,200 kb), 4 million reads were sufficient to get a complex library and reliable statistical analysis. For our samples, an average of 14 million reads were used for each sample, ~97% of which were mapped to the B. subtilis genome.
The sequencing reads obtained from the sequencer were first trimmed and demultiplexed using CLC genomics workbench (Qiagen). To allow comparison between samples, each sample was normalized by the total number of sequencing reads. The trimmed and normalized reads were mapped to the B. subtilis 3610 genome (NCBI accession CP020102) (81) using Bowtie. Tn insertions in a certain gene were treated equally with no discrimination of direction or position of insertions. Between the control and the motility-enriched libraries, the distribution of sequencing reads at the TA sites within each gene was compared using Mann Whitney U test. Genes in which reads were statistically underrepresented (P value of <0.05) in the swarming library compared to the control library were identified. Visual inspection of the transposon insertion profiles was performed with the Sanger Artemis Genome Browser and Annotation tool (32). The accession numbers for the TnSeq samples used in this study are listed in Table S4. Raw data spreadsheet and triage annotation are provided in Table S5.
Data availability.
TnSeq raw data were deposited to the NCBI Sequence Read Archive (accession no. PRJNA827353).
ACKNOWLEDGMENTS
We thank Ankur Dalia and Doug Rusch for the initial TnSeq experiment and analysis, Xheni Karaboja for assisting with TnSeq sample preparation, Zhongqing Ren for helping with TnSeq analysis, and the Center for Genomics and Bioinformatics at Indiana University for Illumina sequencing. This work was funded by NIH grants R01 GM141242 (X.W.) and R35 GM131783 (D.B.K.).
Footnotes
Supplemental material is available online only.
Contributor Information
Daniel B. Kearns, Email: dbkearns@indiana.edu.
Elizabeth Anne Shank, University of Massachusetts Medical School.
REFERENCES
- 1.Kearns DB, Losick R. 2003. Swarming motility in undomesticated Bacillus subtilis. Mol Microbiol 49:581–590. 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 2.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev 19:3083–3094. 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Be'er A, Harshey RM. 2011. Collective motion of surfactant-producing bacteria imparts superdiffusivity to the upper surface. Biophys J 101:1017–1024. 10.1016/j.bpj.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y, Berg HC. 2012. Water reservoir maintained by cell growth fuels the spreading of a bacterial swarm. Proc Natl Acad Sci USA 109:4128–4133. 10.1073/pnas.1118238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senesi S, Ghelardi E, Celandroni F, Salvetti S, Parisio E, Galizzi A. 2004. Surface-associated flagellum formation and swarming differentiation in Bacillus subtilis are controlled by the ifm locus. J Bacteriol 186:1158–1164. 10.1128/JB.186.4.1158-1164.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee S, Bree AC, Liu J, Patrick JE, Chien P, Kearns DB. 2015. Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci USA 112:250–255. 10.1073/pnas.1417419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearns DB, Chu F, Rudner R, Losick R. 2004. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol Microbiol 52:357–369. 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- 9.Arima K, Kakinuma A, Tamura G. 1968. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem Biophys Res Commun 31:488–494. 10.1016/0006-291x(68)90503-2. [DOI] [PubMed] [Google Scholar]
- 10.Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol 8:821–831. 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 11.Magnuson R, Solomon J, Grossman AD. 1994. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell 77:207–216. 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 12.Solomon JM, Magnuson R, Srivastava A, Grossman AD. 1995. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev 9:547–558. 10.1101/gad.9.5.547. [DOI] [PubMed] [Google Scholar]
- 13.Guttenplan SB, Shaw S, Kearns DB. 2013. The cell biology of peritrichous flagella in Bacillus subtilis. Mol Microbiol 87:211–219. 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips AM, Calvo RA, Kearns DB. 2015. Functional activation of the flagellar type III secretion apparatus. PLoS Genet 11:e1005443. 10.1371/journal.pgen.1005443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hummels KR, Kearns DB. 2019. Suppressor mutations in ribosomal proteins and FliY restore Bacillus subtilis swarming motility in the absence of EF-P PLoS Genet 15:e1008179. 10.1371/journal.pgen.1008179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuge K, Ohata Y, Shoda M. 2001. Gene yerP, involved in surfactin self-resistance in Bacillus subtilis. Antimicrob Agents Chemother 45:3566–3573. 10.1128/AAC.45.12.3566-3573.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol 66:395–409. 10.1111/j.1365-2958.2007.05923.x. [DOI] [PubMed] [Google Scholar]
- 18.Verhamme DT, Kiley TB, Stanley-Wall NR. 2007. DegU co-ordinates multicellular behavior exhibited by Bacillus subtilis. Mol Microbiol 65:554–568. 10.1111/j.1365-2958.2007.05810.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu B, Deikus G, Bree A, Durand S, Kearns DB, Bechhofer DH. 2014. Global analysis of mRNA decay intermediates in Bacillus subtilis wild-type and polynucleotide phosphorylase-deletion strains. Mol Microbiol 94:41–55. 10.1111/mmi.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Kearns DB, Bechhofer DH. 2016. Expression of multiple Bacillus subtilis genes is controlled by decay of slrA mRNA from Rho-dependent 3’ ends. Nucleic Acids Res 44:3364–3372. 10.1093/nar/gkw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chevance FF, Hughes KT. 2008. Coordinating the assembly of a bacterial nanomachine. Nat Rev Microbiol 6:455–465. 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee S, Kearns DB. 2014. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet 48:319–340. 10.1146/annurev-genet-120213-092406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen R, Guttenplan SB, Blair KM, Kearns DB. 2009. Role of the σD-dependent autolysins in Bacillus subtilis population heterogeneity. J Bacteriol 191:5775–5784. 10.1128/JB.00521-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez S, Dunn CM, Kearns DB. 2021. CwlQ is required for swarming motility but not flagellar assembly in Bacillus subtilis. J Bacteriol 203:e00029-21. 10.1128/JB.00029-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodman AL, Wu M, Gordon JI. 2011. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Protoc 6:1969–1980. 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson CM, Grossman AD. 2014. Identification of host genes that affect acquisition of an integrative and conjugative element in Bacillus subtilis. Mol Microbiol 93:1284–1301. 10.1111/mmi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meeske AJ, Rodrigues CDA, Brady J, Lim HC, Bernhardt TG, Rudner DZ. 2016. High-throughput genetic screens identify a large and diverse collection of new sporulation genes in Bacillus subtilis. PLoS Biol 14:e1002341. 10.1371/journal.pbio.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dempwolff F, Sanchez S, Kearns DB. 2020. TnFLX: a third-generation mariner-based transposon system for Bacillus subtilis. Appl Environ Microbiol 86:e02893-19. 10.1128/AEM.02893-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Breton Y, Mohapatra NP, Haldenwang WG. 2006. In vivo random mutagenesis of Bacillus subtilis by use of TnYLB-1, a mariner-based transposon. Appl Environ Microbiol 72:327–333. 10.1128/AEM.72.1.327-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobihal GS, Flores-Kim J, Roney IJ, Wang X, Rudner DZ. 2022. The WalR-WalK signaling pathway modulates the activities of both CwlO and LytE through control of the peptidoglycan deacetylase PdaC in Bacillus subtilis. J Bacteriol 204:e0053321. 10.1128/JB.00533-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28:464–469. 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuberi AR, Ying C, Weinreich MR, Ordal GW. 1990. Transcriptional organization of a cloned chemotaxis locus of Bacillus subtilis. J Bacteriol 172:1870–1876. 10.1128/jb.172.4.1870-1876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albertini AM, Caramori T, Crabb WD, Scoffone F, Galizzi A. 1991. The flaA locus of Bacillus subtilis is part of a large operon coding for flagellar structures, motility functions, and an ATPase-like polypeptide. J Bacteriol 173:3573–3579. 10.1128/jb.173.11.3573-3579.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Márquez-Magaña LM, Chamberlin MJ. 1994. Characterization of the sigD transcription unit of Bacillus subtilis. J Bacteriol 176:2427–2434. 10.1128/jb.176.8.2427-2434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Courtney CR, Cozy LM, Kearns DB. 2012. Molecular characterization of the flagellar hook in Bacillus subtilis. J Bacteriol 194:4619–4629. 10.1128/JB.00444-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. 2013. A mechanical signal transmitted by the flagellum controls signaling in Bacillus subtilis. Mol Microbiol 90:6–21. 10.1111/mmi.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan JM, Guttenplan SB, Kearns DB. 2014. Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis. J Bacteriol 196:740–753. 10.1128/JB.01217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrage AM, Vanderpool E, Kearns DB. 2018. Assembly order of flagellar rod subunits in Bacillus subtilis. J Bacteriol 200:e00425-18. 10.1128/JB.00425-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukahara K, Ogura M. 2008. Promoter selectivity of the Bacillus subtilis response regulator DegU, a positive regulator of the fla/che operon and sacB. BMC Microbiol 8:8. 10.1186/1471-2180-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogura M, Tsukahara K. 2012. SwrA regulates assembly of Bacillus subtilis DegU via its interaction with N-terminal domain of DegU. J Biochem 151:643–655. 10.1093/jb/mvs036. [DOI] [PubMed] [Google Scholar]
- 42.Mordini S, Osera C, Marini S, Scavone F, Bellazzi R, Galizzi A, Calvio C. 2013. The role of SwrA, DegU, and PD3 in fla/che expression in B. subitlis. PLoS One 8:e85065. 10.1371/journal.pone.0085065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajkovic A, Hummels KR, Witzky A, Erickson S, Gafken PR, Whitelegge JP, Faull KF, Kearns DB, Ibba M. 2016. Translational control of swarming proficiency in Bacillus subtilis by 5-amino-pentanolylated elongation factor P J Biol Chem 291:10976–10985. 10.1074/jbc.M115.712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hummels KR, Witzky A, Rajkovic A, Tollerson R, II, Jones LA, Ibba M, Kearns DB. 2017. Carbonyl reduction by YmfI in Bacillus subtilis prevents accumulation of an inhibitory EF-P modification state. Mol Microbiol 106:236–251. 10.1111/mmi.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witzky A, Hummels KR, Tollerson R, II, Rajkovic A, Jones LA, Kearns DB, Ibba M. 2018. EF-P posttranslational modification has variable impact on polyproline translation in Bacillus subtilis. mBio 9:e00306-18. 10.1128/mBio.00306-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakano MM, Xia L, Zuber P. 1991. Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J Bacteriol 173:5487–5493. 10.1128/jb.173.17.5487-5493.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano MM, Corbell N, Besson J, Zuber P. 1992. Isolation and characterization of sfp: a gene that functions in the production of the lipopeptide biosurfactant, surfactin, in Bacillus subtilis. Mol Gen Genet 232:313–321. 10.1007/BF00280011. [DOI] [PubMed] [Google Scholar]
- 48.Julkowska D, Obuchowski M, Hollan IB, Séror SJ. 2005. Comparative analysis of the development of swarming communities of Bacillus subtilis 168 and a natural wild type: critical effects of surfactin and the composition of the medium. J Bacteriol 187:65–76. 10.1128/JB.187.1.65-76.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. 2005. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol 55:739–749. 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 50.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 51.Rogers HJ, Thurman PF, Buxton RS. 1976. Magnesium and anion requirements of rodB mutants of Bacillus subtilis. J Bacteriol 125:556–564. 10.1128/jb.125.2.556-564.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Formstone A, Errington J. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol 55:1646–1657. 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 53.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. 2006. Wall teichoic acid polymers are dispensible for cell viability in Bacillus subtilis. J Bacteriol 188:8313–8316. 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wecke J, Madela K, Fischer W. 1997. The absence of D-alanine from lipoteichoic acid and wall teichoic acid alters surface charge, enhances autolysis and increases susceptibility to methicillin in Bacillus subtilis. Microbiology (Reading) 143:2953–2960. 10.1099/00221287-143-9-2953. [DOI] [PubMed] [Google Scholar]
- 55.Schirner K, Marles-Wright J, Lewis RJ, Errington J. 2009. Distinct and essential morphogenetic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J 28:830–842. 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fraser GM, Bennett JCQ, Hughes C. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol 32:569–580. 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- 57.Bennett JCQ, Thomas J, Fraser GM, Hughes C. 2001. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol Microbiol 39:781–791. 10.1046/j.1365-2958.2001.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamamoto S, Kutsukake K. 2006. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol 188:6703–6708. 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bange G, Kümmerer N, Engel C, Bozkurt G, Wild K, Sinning I. 2010. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci USA 107:11295–11300. 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato Y, Takaya A, Mouslim C, Hughes KT, Yamamoto T. 2014. FliT selectively enhances proteolysis of FlhC subunit in FlhD4C2 complex by an ATP-dependent protease, ClpXP. J Biol Chem 289:33001–33011. 10.1074/jbc.M114.593749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall AN, Subramanian S, Oshiro RT, Canzoneri AK, Kearns DB. 2018. SwrD (YlzI) promotes swarming in Bacillus subtilis by increasing power to flagellar motors. J Bacteriol 200:e00529-17. 10.1128/JB.00529-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 63.Abram D, Vatter AE, Koffler H. 1966. Attachment and structural features of flagella of certain bacilli. J Bacteriol 91:2045–2068. 10.1128/jb.91.5.2045-2068.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DePamphilis ML, Adler J. 1971. Fine structure and isolation of the hook-basal body complex of flagella from Escherichia coli and Bacillus subtilis. J Bacteriol 105:384–395. 10.1128/jb.105.1.384-395.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dimmit K, Simon M. 1971. Purification and thermal stability of intact Bacillus subtilis flagella. J Bacteriol 105:369–375. 10.1128/jb.105.1.369-375.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kubori T, Okumura M, Kobayashi N, Nakamura D, Iwakura M, Aizawa S-I. 1997. Purification and characterization of the flagellar hook-basal body complex of Bacillus subtilis. Mol Microbiol 24:399–410. 10.1046/j.1365-2958.1997.3341714.x. [DOI] [PubMed] [Google Scholar]
- 67.Pallen MJ, Penn CW, Chaudhuri RR. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol 13:143–149. 10.1016/j.tim.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 68.Pallen MJ, Matzke NJ. 2006. From The Origin of Species to the origin of bacterial flagella. Nat Rev Microbiol 4:784–790. 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- 69.Liu R, Ochman H. 2007. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci USA 104:7116–7121. 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao X, Zhang K, Boquoi T, Hu B, Motaleb MA, Miller KA, James ME, Charon NW, Manson MD, Norris SJ, Li C, Liu J. 2013. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgorferi. Proc Natl Acad Sci USA 110:14390–14395. 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fein D. 1979. Possible involvement of bacterial autolytic enzymes in flagellar morphogenesis. J Bacteriol 137:933–946. 10.1128/jb.137.2.933-946.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dijkstra AJ, Keck W. 1996. Peptidoglycan as a barrier to transenvelope transport. J Bacteriol 178:5555–5562. 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meisner J, Montero Llopis P, Sham L-T, Garner E, Bernhardt TG, Rudner DZ. 2013. FtsEX is required for CwlO peptidoglycan hydrolase activity during cell elongation in Bacillus subtilis. Mol Microbiol 89:1069–1083. 10.1111/mmi.12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brunet YR, Wang X, Rudner DZ. 2019. SweC and SweD are essential co-factors of the FtsEX-CwlO cell wall hydrolase complex in Bacillus subtilis. PLoS Genet 15:e1008296. 10.1371/journal.pgen.1008296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dobihal GS, Brunet YR, Flores-Kim J, Rudner DZ. 2019. Homeostatic control of cell wall hydrolysis by the WalRK two-component signaling pathway in Bacillus subtilis. Elife 8:e52088. 10.7554/eLife.52088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Demchick P, Koch AL. 1996. The permeability of the wall fabric of Escherichia coli and Bacillus subtilis. J Bacteriol 178:768–773. 10.1128/jb.178.3.768-773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pasquina-Lemonche L, Burns J, Turner RD, Kumar S, Tank R, Mullin N, Wilson JS, Chakrabarti B, Bullough PA, Foster SJ, Hobbs JK. 2020. The architecture of the Gram-positive bacterial cell wall. Nature 582:294–297. 10.1038/s41586-020-2236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koo B-M, Kritikos G, Farelli JD, Todor H, Tong K, Kimsey H, Wapinski I, Galardini M, Cabal A, Peters JM, Hachmann A-B, Rudner DZ, Allen KN, Typas A, Gross CA. 2017. Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst 4:291–305. 10.1016/j.cels.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14:1343–1348. 10.1128/JVI.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336. 10.1016/0378-1119(95)00652-4. [DOI] [PubMed] [Google Scholar]
- 81.Nye TM, Schroeder JW, Kearns DB, Simmons LA. 2017. Complete genome sequence of undomesticated Bacillus subtilis strain NCIB 3610. Genome Announc 5:e00364-17. 10.1128/genomeA.00364-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calvo RA, Kearns DB. 2015. FlgM is secreted by the flagellar export apparatus in Bacillus subtilis. J Bacteriol 197:81–91. 10.1128/JB.02324-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cairns LS, Marlow VL, Kiley TB, Birchall C, Ostrowski A, Aldridge PD, Stanley-Wall NR. 2014. FlgN is required for flagellum-based motility by Bacillus subtilis. J Bacteriol 196:2216–2226. 10.1128/JB.01599-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mukherjee S, Babitzke P, Kearns DB. 2013. FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis. J Bacteriol 195:297–306. 10.1128/JB.01654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y, Chai Y, Guo JH, Losick R. 2012. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J Bacteriol 194:5080–5090. 10.1128/JB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Márquez LM, Helmann JD, Ferrari E, Parker HM, Ordal GW, Chamberlin MJ. 1990. Studies of sigma D-dependent functions in Bacillus subtilis. J Bacteriol 172:3435–3443. 10.1128/jb.172.6.3435-3443.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patrick JE, Kearns DB. 2008. MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol Microbiol 70:1166–1179. 10.1111/j.1365-2958.2008.06469.x. [DOI] [PubMed] [Google Scholar]
- 88.Bidnenko V, Nicolas P, Grylak-Mielnicka A, Delumeau O, Auger S, Aucouturier A, Guerin C, Repoila F, Bardowski J, Aymerich S, Bidnenko E. 2017. Termination factor Rho: from the control of pervasive transcription to cell fate determination in Bacillus subtilis. PLoS Genet 13:e1006909. 10.1371/journal.pgen.1006909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2; Tables S1 to S4. Download jb.00089-22-s0001.pdf, PDF file, 0.5 MB (467.5KB, pdf)
Table S5. Download jb.00089-22-s0002.xlsx, XLSX file, 1.0 MB (1,001.6KB, xlsx)
Data Availability Statement
TnSeq raw data were deposited to the NCBI Sequence Read Archive (accession no. PRJNA827353).





