LETTER
Carbapenem-resistant Acinetobacter spp. are commonly reported worldwide, which becomes a serious public health issue (1). The genetic plasticity of Acinetobacter allowed this bacterium to rapidly acquire and accumulate carbapenemase genes, such as blaOXA-23 and blaNDM-1, making it a problematic nosocomial persister as well as an important reservoir of resistance genes (2). Plasmid-mediated horizontal gene transfer is relevant to the dissemination of carbapenemase genes (2, 3). To date, 58 plasmid groups (GR1 to GR58) have been proposed in Acinetobacter based on the nucleotide identity of the rep genes (4, 5). Here, we report a multidrug-resistant (MDR) plasmid carrying blaOXA-58 and blaNDM-1, which represents a new plasmid type, GR59.
Strain SCLZS30 was recovered from the sewage outlet of Luzhou People’s Hospital, Sichuan Province, China, in August 2019. It was resistant to meropenem, cefotaxime, cefoxitin, tetracycline, and gentamicin, intermediate to ciprofloxacin and sulfamethoxazole/trimethoprim, and susceptible to tigecycline and colistin. The whole-genome sequence of SCLZS30 was determined using the PacBio RS II and Illumina HiSeq 2000 platforms. The genome of SCLZS30 consists of a 2,916,803-bp circular chromosome (41.1% GC content) and two circular plasmids, pNDM_SCLZS30 (47,845 bp) and p1_SCLZS30 (64,918 bp). An additional 23,275-bp contig could not be circularized but has two putative plasmid replication genes. Average nucleotide identity (ANI) analysis showed that SCLZS30 belongs to Acinetobacter towneri, as it has 95.27% identity (77.31% query coverage) to the A. towneri reference strain, GX3 (CP071766) (6). SCLZS30 contains 13 antibiotic resistance genes (ARGs), conferring resistance to aminoglycosides, β-lactams, macrolides, tetracyclines, sulfonamides, and amphenicols. Among them, aadB-blaOXA-392 (blaOXA-1 family)-catB3-qacEΔ1-sul1 and aadA2b are located on the chromosome, and aacC2d, aphA6, blaNDM-1, blaOXA-58, mph(E), msr(E), and tet(39) are carried by pNDM_SCLZS30.
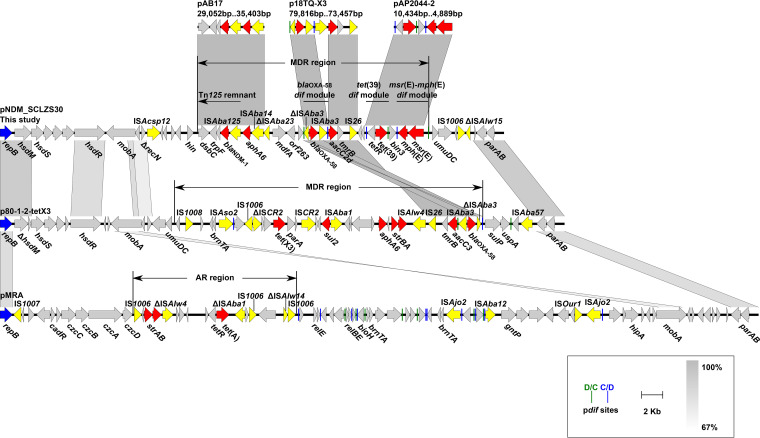
pNDM_SCLZS30 has a replication gene, repB, encoding a replicase protein of the Rep_3 family (Pfam:01051). As the repB gene has <74% nucleotide identity to the rep genes of all 58 existing Acinetobacter plasmid groups (4, 5), pNDM_SCLZS30 defines a novel group designated GR59. The backbone of pNDM_SCLZS30, with an average GC content of 36.59%, has only 88.6% identity (45% coverage) with its closest match, p80-1-2-tetX3 (CP041297, Acinetobacter indicus, animal origin, China). Determinants for conjugation were not identified on pNDM_SCLZS30, while a mobA gene for plasmid mobilization is present (Fig. 1). Transconjugants of Escherichia coli J53/EC600 were not obtained for pNDM_SCLZS30 after repeated attempts.
FIG 1.
Genetic characteristics of pNDM_SCLZS30. The MDR region of pNDM_SCLZS30 is compared with segments from pAB17 (MT002974, Acinetobacter baumannii, patient origin, Brazil), p18TQ-X3 (CP045132, Acinetobacter indicus, animal origin, China), and pAP2044-2 (CP087718, Acinetobacter pittii, patient origin, China). The complete nucleotide sequence of pNDM_SCLZS30 is compared with GR59 plasmids p80-1-2-tetX3 (CP041297, A. indicus, animal origin, China) and pMRA (CP079749, Acinetobacter johnsonii, environmental origin, China). Genes are indicated by arrows. repB, resistance genes, and mobile genetic elements are highlighted in blue, red and yellow, respectively. Short vertical lines represent pdif sites, D/C orientation in green and C/D in blue. Regions of >67% nucleotide sequence identity are indicated by gray shading. MDR, multidrug-resistant; AR, antibiotic resistance.
All seven resistance genes in pNDM_SCLZS30 are clustered in a 21,469-bp MDR region, which has a higher GC content (43.46%) than the backbone, suggesting different origins. Also, the MDR region is mosaic with areas of high and low GC contents, indicating that it might be shaped by multiple genetic events. The blaNDM-1 gene is found in a 4,149-bp Tn125 remnant (56.35% GC), with the genetic structure dsbC-trpF-ble-blaNDM-1-ISAba125, downstream of the ISAba14-aphA6 segment. As previously found in many Acinetobacter plasmids, blaOXA-58 and its flanking inverted ISAba3, one of which is truncated, are harbored in a 2,313-bp dif module (35.15% GC) (7), which is located within an mdfA-orf263-orf159-pdif-ΔISAba3-blaOXA-58-ISAba3-pdif-aacC2d-tmrB-IS26 region, revealing a novel genetic context. In addition, a tet(39) dif module (39.15% GC content) and an msr(E)-mph(E) dif module (36.67% GC content) (8), separated by a resolvase gene, are successively located 3.7-kb downstream of the blaOXA-58 dif module. The three dif modules were previously commonly found in Acinetobacter (8, 9), but their coexistence on a single plasmid is rare. BLASTn analysis showed that the three dif modules are also simultaneously present on pDETAB2 (CP047975) and pNDM_SCLZS86 (CP090865), but the genetic context of each dif module is different (10, 11). These results suggest that site-specific recombination via the pdif sites may account for the acquisition of ARGs and the rearrangement of multidrug resistance regions (8–10).
Three complete GR59 plasmids, p80-1-2-tetX3 (CP041297), pMRA (CP079749), and pXMC5X702-195k (CP084301), which contain a repB gene >79.64% identical (>98% coverage) to that of pNDM_SCLZS30 (4), were identified in GenBank. A 3.8-kb region, covering repB and parAB genes, is conserved in these GR59 plasmids (see Fig. S1 in the supplemental material). Six tandem 22-bp repeats were found 535 to 562 bp upstream of repB, likely iterons (Fig. S2). A potential origin of plasmid replication, characterized by an array of AT-rich repeat sequences, was identified close to these tandem repeats in each GR59 plasmid (Fig. S2). In addition to pNDM_SCLZS30, p80-1-2-tetX3 and pMRA also contain multiple clinically important ARGs (Fig. 1), and p80-1-2-tetX3 harbors similar blaOXA-58 and aacC modules as pNDM_SCLZS30, suggesting this plasmid group as a potential vehicle in mediating the dissemination of resistance determinants.
In summary, this study identifies a novel plasmid type, designated GR59, from an environmental A. towneri isolate, which carries both blaNDM-1 and blaOXA-58. This plasmid group may serve as an important platform to allow evolution and rearrangement of ARGs to adapt to different antimicrobial selection pressures. More studies are needed to monitor the prevalence of GR59 plasmids and their role in the spread of ARGs in Acinetobacter.
Data availability.
Complete sequences of the chromosome, pcontig1, pNDM_SCLZS30, and p1_SCLZS30 of SCLZS30 were deposited in GenBank under accession numbers CP090382 to CP090385.
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (31900125), the Scientific and Technological Project in Sichuan Province (2022JDRC0144), and the Joint Funds of the Luzhou and Southwest Medical University Natural Science Foundation (2019LZXNYDJ47). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiaoyi Dai, Email: daixiaoyi@swmu.edu.cn.
Luhua Zhang, Email: zhluhua@swmu.edu.cn.
REFERENCES
- 1.Hamidian M, Nigro SJ. 2019. Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom 5:e000306. 10.1099/mgen.0.000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramirez MS, Bonomo RA, Tolmasky ME. 2020. Carbapenemases: transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules 10:720. 10.3390/biom10050720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salto IP, Torres Tejerizo G, Wibberg D, Puhler A, Schluter A, Pistorio M. 2018. Comparative genomic analysis of Acinetobacter spp. plasmids originating from clinical settings and environmental habitats. Sci Rep 8:7783. 10.1038/s41598-018-26180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salgado-Camargo AD, Castro-Jaimes S, Gutierrez-Rios RM, Lozano LF, Altamirano-Pacheco L, Silva-Sanchez J, Perez-Oseguera A, Volkow P, Castillo-Ramirez S, Cevallos MA. 2020. Structure and evolution of Acinetobacter baumannii plasmids. Front Microbiol 11:1283. 10.3389/fmicb.2020.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro-Jaimes S, Guerrero G, Bello-Lopez E, Cevallos MA. 2022. Replication initiator proteins of Acinetobacter baumannii plasmids: an update note. Plasmid 119–120:102616. 10.1016/j.plasmid.2021.102616. [DOI] [PubMed] [Google Scholar]
- 6.Ma J, Wang J, Feng J, Liu Y, Yang B, Li R, Bai L, He T, Wang X, Yang Z. 2020. Characterization of three porcine Acinetobacter towneri strains co-harboring tet(X3) and blaOXA-58. Front Cell Infect Microbiol 10:586507. 10.3389/fcimb.2020.586507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Wang Y, Wu H, Wang ZY, Shen PC, Tian YQ, Sun F, Pan ZM, Jiao X. 2020. Coexistence of blaOXA-58 and tet(X) on a novel plasmid in Acinetobacter sp. from pig in Shanghai, China. Front Microbiol 11:578020. 10.3389/fmicb.2020.578020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell GA, Hall RM. 2017. The tet39 determinant and the msrE-mphE genes in Acinetobacter plasmids are each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob Agents Chemother 61:e00780-17. 10.1128/AAC.00780-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cameranesi MM, Moran-Barrio J, Limansky AS, Repizo GD, Viale AM. 2018. Site-specific recombination at XerC/D sites mediates the formation and resolution of plasmid co-integrates carrying a blaOXA-58- and TnaphA6-resistance module in Acinetobacter baumannii. Front Microbiol 9:66. 10.3389/fmicb.2018.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Moran RA, Chen Y, Doughty EL, Hua X, Jiang Y, Xu Q, Zhang L, Blair JMA, McNally A, van Schaik W, Yu Y. 2021. Transferable Acinetobacter baumannii plasmid pDETAB2 encodes OXA-58 and NDM-1 and represents a new class of antibiotic resistance plasmids. J Antimicrob Chemother 76:1130–1134. 10.1093/jac/dkab005. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Qiu Y, Fang C, Tang M, Dai X, Zhang L. 2022. Characterisation of a novel GR31 plasmid co-harbouring blaNDM-1 and blaOXA-58 in an Acinetobacter sp. isolate. J Glob Antimicrob Resist 29:212–214. 10.1016/j.jgar.2022.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download aac.00206-22-s0001.pdf, PDF file, 0.3 MB (348.8KB, pdf)
Data Availability Statement
Complete sequences of the chromosome, pcontig1, pNDM_SCLZS30, and p1_SCLZS30 of SCLZS30 were deposited in GenBank under accession numbers CP090382 to CP090385.