Abstract
Secondary metabolism in fluorescent pseudomonads is globally regulated by gacS, which encodes a membrane-bound sensor kinase, and gacA, which encodes a transcriptional response regulator. Spontaneous mutation in either gene blocked biosynthesis of the antimicrobial compounds hydrogen cyanide, 2,4-diacetylphloroglucinol, pyoluteorin, and pyrrolnitrin by the model biocontrol strain Pseudomonas fluorescens CHA0. Spontaneous mutants also had altered abilities to utilize several carbon sources and to increase medium pH compared with the wild type, suggesting that gacS and gacA influence primary as well as secondary bacterial metabolism. Inoculant efficacy for biocontrol was significantly reduced by contamination with regulatory mutants which accumulated during inoculum production. Spontaneous mutants accumulated in all 192 separate liquid cultures examined, typically at a frequency of 1% or higher after 12 days. During scale-up in a simulated industrial fermentation process, mutants increased exponentially and accounted for 7, 23, and 61% of the total viable cells after transfer to 20-, 100-, and 500-ml preparations, respectively. GacS− and GacA− mutants had identical phenotypes and occurred at the same frequency, indicating that the selective pressures for the two mutants were similar. We developed a simple screening method for monitoring inoculant quality based on the distinctive appearance of mutant colonies (i.e., orange color, enlarged diameter, hyperfluorescence). Mutant competitiveness was favored in a nutrient-rich medium with a high electrolyte concentration (nutrient broth containing yeast extract). We were able to control mutant accumulation and to clean up contaminated cultures by using certain mineral amendments (i.e., zinc, copper, cobalt, manganese, and ammonium molybdate) or by diluting media 1/10. Spontaneous mutants and genetic constructs had the same response to culture conditions. Zinc and medium dilution were also effective for improving the genetic stability of other P. fluorescens biocontrol strains obtained from Ghana and Italy.
Certain plant-associated bacteria, particularly fluorescent Pseudomonas spp., have been exploited for suppression of crop diseases, and the importance of these bacteria in agriculture is expected to grow (5). Commercial development of biocontrol entails large-scale production of inoculants. Bacterial inoculants, regardless of their intended use (e.g., agricultural, pharmaceutical, food processing, manufacturing), are typically mass produced in industrial fermentors, and small batches are used to inoculate increasing fermentation volumes, a process referred to as scale-up (33). A stream-lined process (i.e., a cost-effective process) that results in a high yield and optimal efficacy is the primary objective in fermentations designed to recover viable cells. Culture media are prepared from an eclectic assortment of ingredients and are generally nutrient rich, which does not reflect the conditions in most natural bacterial environments. This is particularly evident for biocontrol agents originally isolated from the rhizosphere or phyllosphere, where nutrients are often limiting. Considering this and the scale-up process necessary to prepare large volumes, liquid fermentation of bacterial inoculants is disturbingly similar to repeated transferring and prolonged incubation in artificial growth media, laboratory practices long known to generate spontaneous mutations in microorganisms.
Genetic and molecular analysis has demonstrated that production of various antifungal compounds is a primary mechanism of biocontrol for many strains, accounting for as much as 90% of the disease-suppressing activity (36). As more biocontrol strains are analyzed, it is becoming apparent that biosynthesis of antifungal secondary metabolites in Pseudomonas spp. is commonly controlled by a two-component system comprising the sensor kinase GacS and the response regulator GacA. gacS is the new designation for a group of conserved genes in Pseudomonas spp. that encode functionally homologous cognate sensor kinases (e.g., apdA, lemA, pheN, and repA) (19). Genetically constructed GacS− and GacA− mutants are less inhibitory for fungal pathogens, presumably due to loss of antibiotics and hydrogen cyanide (HCN) (35). Recent evidence indicates that a gac mutation also negatively affects other regulatory elements, including autoinducers and sigma factors (1, 28, 40).
Despite obvious potential problems with instability in gacS and gacA or other genes important in biocontrol, little if any effort has been made to document, to understand, and to control these problems during inoculant production. Here we describe accumulation of a high number of spontaneous GacS− and GacA− mutants in liquid cultures of the Swiss biocontrol strain Pseudomonas fluorescens CHA0. The importance of gacA in biosynthesis of the antifungal metabolites 2,4-diacetylphloroglucinol (PHL), pyoluteorin (PLT), and HCN and the role of this gene in fungal inhibition and the biocontrol activity of strain CHA0 have previously been demonstrated by using gene replacement and transposon insertional mutation (21). Less is known about the function of gacS in this strain (3). Our objectives were to phenotypically characterize the spontaneous regulatory mutants and to determine their impact on the biocontrol efficacy of bacterial inoculants. We then tried to identify the selective pressures that favor mutant accumulation during inoculant production and to develop a cost-effective approach to minimize genetic instability in P. fluorescens biocontrol strains.
(A preliminary report of this work has been published previously [8].)
MATERIALS AND METHODS
Bacterial strains, mutant derivatives, and culture media.
The strains and plasmids used in this study are described in Table 1. Wild-type strain CHA0 was originally isolated in 1983 from a Swiss sandy loam soil that naturally suppressed tobacco black root rot (15). An archival sample from 1985, kept at −80°C, was used in this study. Strains CHA510, CHA89, and CHA96rif are genetically engineered regulatory mutant derivatives of CHA0. Spontaneous regulatory mutant derivatives CHAS9, CHAS17, and CHAS45 were isolated from stationary-phase nutrient broth cultures, and mutant CHASP1 was isolated from tobacco roots that had been inoculated with wild-type strain CHA0 and grown under gnotobiotic conditions for 6 weeks. Wild-type strains PGNL1, PGNR1, and PGNR4 were isolated from tobacco roots grown in a Ghana silt loam soil that suppressed tomato root diseases; and wild-type strains PINR2 and PINR3 were isolated from tomato roots grown in an Albenga, Italy, sandy loam soil that suppressed Fusarium wilt. Spontaneous mutants of these strains were isolated from orange sectors that appeared in colonies grown for 10 to 14 days on King's medium B (KB) agar (18). Plasmids used for genetic complementation were vectored by Escherichia coli. All bacteria were stored in dilute 0.08% nutrient broth (Difco, Detroit, Mich.) supplemented with 40% glycerol at −80°C. Fresh cultures were started from glycerol stocks for each experiment by plating portions onto KB agar.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| P. fluorescens strains | ||
| CHA0 | Ant+ HPT+ Flu+ DS+ Cmr, wild type | 15 |
| CHA89 | Ant− HPT− Flu++ DS− Kmr, gene replacement gacA mutant of CHA0 | 21 |
| CHA96rif | Ant− HPT− Flu++ DS− Kmr, Rifr derivative of CHA89 | 26 |
| CHA510 | Ant− HPT− Flu++ DS− Kmr, apdA::Tn5 mutant of CHA0 | Bull and Haasb |
| CHAS9 | Ant− HPT− Flu++ DS−, spontaneous gacA mutant of CHA0 | Bull and Haasb |
| CHAS45 | Ant− HPT− Flu++ DS−, spontaneous gacA mutant of CHA0 | This study |
| CHAS17 | Ant− HPT− Flu++ DS−, spontaneous apdA mutant of CHA0 | Bull and Haasb |
| CHASP1 | Ant− HPT− Flu++ DS−, spontaneous apdA mutant of CHA0 | This study |
| PGNL1 | Ant+ HPT+ Flu+ DS+, wild type | 16 |
| PGNL1S2 | Ant− HPT− Flu++, spontaneous gacA mutant of PGNL1 | This study |
| PGNR1 | Ant+ HPT+ Flu+ DS+, wild type | 16 |
| PGNR1S1 | Ant− HPT− Flu++, spontaneous gacA mutant of PGNR1 | This study |
| PGNR4 | Ant+ HPT+ Flu+ DS+, wild type | 16 |
| PGNR4S2 | Ant− HPT− Flu++, spontaneous gacA mutant of PGNR4 | This study |
| PINR2 | Ant+ HPT+ Flu+ DS+, wild type | 16 |
| PINR2S3 | Ant− HPT− Flu++, spontaneous gacA mutant of PINR2 | This study |
| PINR3 | Ant+ HPT+ Flu+ DS+, wild type | 16 |
| PINR3S1 | Ant− HPT− Flu++, spontaneous gacA mutant of PINR2 | This study |
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 (rK−mK+) supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15λ− | 31 |
| ED8767 | metB hsdS supE supF recA56 | 25 |
| HB101 | F−hsdS20 supE44 recA13 ara14 proA2 lacY1galK2 rpsL20 leu-6 thi-1 xyl-5 mtl-1 (str-20)Δ(mcrC-mrr) | 2 |
| Plasmids | ||
| pME3066 | IncP-1 replicon, Mob+ Tcr, contains a functional gacA (Y49) gene from CHA0 | 21 |
| pJEL5771 | Tcr, contains a functional apdA gene from P. fluorescens Pf-5 | 6 |
| pEMH97 | Tcr, contains a functional lemA gene from P. syringae pv. syringae B728a | 13 |
| pME497 | IncP-1 replicon, RepA(ts) Tra+ Kmr | 38 |
Ant, antibiotics (PHL and PLT); HPT, hydrogen cyanide, protease, TSO; DS, disease suppression; Flu, fluorescent siderophores (pyoverdine and pyochelin) (+, wild-type production level; ++, overproduction; −, reduced or no production); Apr, Cmr, Kmr, Tcr, and Rifr, resistance to ampicillin, chloramphenicol, kanamycin, tetracycline, and rifampin, respectively.
Bull and Haas, unpublished data.
Liquid cultures were grown in normal-strength nutrient broth containing yeast extract (NBY broth), which was prepared with 0.8% nutrient broth and 0.5% yeast extract (Difco) in twice-distilled H2O (pH 6.5). Single lots of nutrient broth and yeast extract were used throughout this study. Prepared NBY broth contained (per liter) 1,441.0 mg of total nitrogen, 604.0 mg of amino nitrogen, 600.1 mg of phosphate, 597.9 mg of potassium, 259.7 mg of sodium, 121.7 mg of chloride, 54.9 mg of sulfate, 22.9 mg of magnesium, 6.1 mg of calcium, 0.5 mg of zinc, and less than 0.1 mg each of cobalt, copper, iron, manganese, tin, and lead. Media conductivity, a measure of electrolyte concentration, was determined by using a conductivity meter (model LM20; Volmatic SARL, Mazé, Switzerland), and pH was determined with a digital meter (ABS, Zürich, Switzerland).
Mutant characterization.
A total of 578 spontaneous mutants with a distinct orange colony phenotype were isolated from 192 NBY broth cultures of wild-type strain CHA0 that were incubated for 12 days. The mutants were analyzed to determine their genetic similarity to the wild type by using a method based on a PCR performed with randomly amplified polymorphic DNA markers. Primer D7, obtained from a series of random oligonucleotides (Operon Technologies, Almeda, Calif.), provided consistent and distinct band patterns with polymorphic markers specific to strain CHA0 (16). Bacteria were grown in wells of microtiter plates containing 50 μl of dilute (0.1×) KB broth and were incubated for 24 h at 27°C with gentle agitation. The methods used for sample preparation, PCR amplification, and gel electrophoresis were methods that have been described previously (16).
All 578 mutants were tested at least twice to determine whether they produced HCN (16), extracellular proteases (30), and tryptophan side chain oxidase (TSO), an enzyme important in indoleacetic acid biosynthesis (27), by using standard methods. A random subsample consisting of 205 of the mutants were then screened to determine whether they exhibited genetic complementation with gacS and gacA clones. Mobilization of recombinant cosmids pJEL5771 and pME3066 from E. coli was accomplished by triparental mating with helper plasmid pME497 (38). Transconjugants were screened to determine whether they were restored for HCN, protease, and TSO production on milk agar (30). Genetically engineered derivatives CHA510 and CHA89 were routinely used as controls for complementation of gacS and gacA mutations, respectively.
Five mutants that were completely complemented with either gacS or gacA were further characterized to determine their reversion frequencies, cell lengths, carbon source utilization profiles, pH changes in NBY broth, antibiotic sensitivities, antibiotic and siderophore production profiles, in vitro fungal inhibition profiles and abilities to suppress cucumber damping-off. Reversion frequencies were estimated by determining the fractions of CFU obtained from 24-h NBY broth cultures of spontaneous mutants that were protease positive on milk agar. Cell length was determined after 24 h of growth in 20 ml of NBY broth by mounting cells on polycarbonate filters, staining them with CHA0-specific antisera and fluorescent antibodies, and measuring the lengths of 100 cells of each isolate with a Zeiss Axioskop epifluorescence microscope, as previously described (37). Carbon source utilization profiles were determined by using the Biolog GN and GP Microplate system according to the instructions of the manufacturer (Biolog Inc., Hayward, Calif.). Changes in pH were determined in NBY broth after 24 h of growth. Tolerance to synthetic PHL (200 to 1,000 μg/ml) and tolerance to PLT (50 to 500 μg/ml) were determined in NBY broth as described by Keel et al. (17). High-performance liquid chromatography was used to quantify production of pyochelin, salicylic acid, PLT, and pyrrolnitrin in NBY broth after 48 h of incubation; and production of PHL was quantified in NBY broth amended with 1% glucose as previously described (7). The abilities of mutants to inhibit Pythium ultimum growth were determined on KB agar with and without 100 μM FeCl3 by spotting 5-μl portions of overnight NBY broth cultures at opposite sides of plates 5 mm from the edge. After 24 h of incubation at 27°C, fluorescence around the bacterial colonies was observed with a UV lamp. Then the plates were inoculated with P. ultimum by inverting a 4-mm-diameter agar plug from a 3-day-old culture in the center. In each case the distance between the edges of the bacterial and fungal colonies (inhibition zone) was measured after 36 h.
Suppression of cucumber damping-off caused by P. ultimum was evaluated with Eschikon sandy loam soil (26). The soil was sieved (2.0-mm mesh), infested with 0.5% crushed millet seed colonized by P. ultimum (particle diameter, <1.0 mm), and incubated for 24 h at 20°C before it was distributed into plastic pots (diameter, 7.5 cm; depth, 5.5 cm). Bacteria were grown for 24 h in NBY broth and collected by centrifugation. Suspensions containing approximately 1011 CFU/ml were prepared with 0.5% medium-viscosity sodium carboxymethylcellulose (Fluka, Buchs, Switzerland). Pregerminated (2 days, 24°C, 0.85% water agar) surface-disinfected seeds of cucumber (Cucumis sativus ‘Chinesische Schlange’) were submerged in bacterial suspensions for 5 min and planted 0.5 cm deep in the infested soil; 10 to 15 seeds were planted in each pot. Plants were grown in a climate chamber at 22°C with 70% relative humidity by using a 16-h photoperiod. The percentages of seedlings that emerged and were standing were determined after 10 days.
Influence of mutant contamination on inoculant efficacy.
Suspensions of wild-type strain CHA0, gacS mutant strain CHAS17, and gacA mutant strain CHAS33 cells were prepared from NBY broth cultures as described above. Suspensions were combined to obtain mutant concentrations ranging from 0 to 100%. Pregerminated cucumber seeds were soaked in the suspensions and planted in Pythium-infested soil. The percentages of seedlings that emerged and were standing were determined after 10 days. The treatments consisted of three replicate pots containing 15 seeds each, and the experiment was repeated once. Nonbacterized seeds served as a disease control that was not included in the analysis.
Four assays to determine the influence of mineral amendments on mutant accumulation.
Unless otherwise indicated, bacteria were grown in 20-ml portions of NBY broth in 100-ml Erlenmeyer flasks and were incubated for 48 h at 27°C with shaking at 140 rpm in the dark. Filter-sterilized mineral solutions were added to autoclaved media to obtain a concentration of 1.0 mM [B(OH3), CaCl2 · 2H2O, FeSO4 · 7H2O, LiCl, MgSO4 · 7H2O, Mo7(NH4)6O24 · 4H2O, MnCl2 · 4H2O, NaCl], 0.7 mM (CuSO4, ZnSO4 · 7H2O), or 0.1 mM (CoCl2 · 6H2O). Cultures were inoculated with 10-μl portions of overnight precultures that were diluted 1/10 so that the concentrations were approximately 103 to 104 CFU/ml. Wild-type precultures contained no detectable mutants (<1 × 10−4 CFU/ml). Mixtures of the wild type and mutants were prepared by combining precultures and then were used to inoculate cultures. Sampling was done by plating appropriate serial dilutions onto KB agar amended with chloramphenicol (30 μg/ml) (KBcm agar), a natural antibiotic resistance marker for strain CHA0 (38). Other P. fluorescens strains were plated onto nonamended KB agar. Colonies were enumerated, and the percentage of orange mutants relative to nonpigmented wild-type colonies was determined after 5 days.
In the first experiment, the effects of media on accumulation of mutants from a wild-type culture were determined. CHA0 was grown for 12 days in cultures containing NBY broth, NBY broth supplemented with 0.7 mM CuSO4, dilute (0.1×) NBY broth, and dilute NBY broth adjusted to a conductivity of 4.0 mS with 30 mM NaCl (this was the approximate conductivity of 1× NBY broth cultures after 48 h of bacterial growth). The cultures were incubated for 12 days, and serial dilutions were plated onto KBcm agar. The total number of CFU and the percentage of orange mutants were determined by using 500 to 3,000 colonies per treatment. As a second measure of mutant accumulation, 94 random colonies subjected to each treatment were tested to determine whether they produced HCN, proteases, and TSO. Each treatment was replicated 10 times, and two samples were used for each replicate; the experiment was conducted four times.
The second experiment was designed to mimic industrial fermentation processes, in which typically there is stepwise scale-up of batch size (20, 32). Samples (10 μl) taken from the 12-day NBY broth cultures described above, which contained moderate levels of mutants (approximately 1.3%), were used to seed 20-ml fresh portions of NBY broth, dilute NBY broth, or NBY broth containing CuSO4. After 48 h of shaking at approximately 110 rpm, the total number of CFU and the percentage of mutants were determined in each case, and 100-μl portions of the cultures were used to inoculate 100-ml portions of fresh media in 500-ml Erlenmeyer flasks. The resulting cultures were in turn used to inoculate 500-ml portions in 1-liter flasks. The treatments each consisted of three to six replicates, each of which was started by using an independent seed culture, and the experiment was conducted three times.
In the third experiment we examined the influence of a wider range of minerals on the accumulation of orange mutants from cultures containing initially low but detectable levels of mutants (approximately 0.3%). Bacteria were grown in 20-ml portions of NBY broth, dilute (0.1×) NBY broth, dilute NBY broth containing NaCl, and NBY broth containing 1 of 11 minerals. After 48 h, the total number of CFU and the percentage of mutants were determined in each case. Each treatment consisted of four replicates, and the experiment was conducted four times.
In the fourth experiment we examined the influence of minerals on competition between coinoculated wild-type strain CHA0 and gacS mutants (CHAS17 and CHASP1) or gacA mutants (CHAS9 and CHAS45). Test cultures were inoculated with a mixture containing 80% wild-type strain CHA0 and 20% mutant. After 48 h, the total number of CFU and the percentage of mutants were determined in each case. The experiment was arranged as a 5 × 14 factorial in a split-plot design with a main plot for the wild type-mutant combination and a subplot for culture medium. Because of the large number of treatments, the experiment consisted of eight replicates studied over time. An extension of this fourth experiment was designed to determine the relationship between zinc concentration and mutant accumulation. In this experiment we used spontaneous mutants (CHAS17 and CHAS45) and compared them with genetically engineered mutants (CHA510 and CHA96rif). Mixtures containing 90% wild-type strain CHA0 and 10% mutant were used to inoculate NBY broth amended with a range of ZnSO4 · 7H2O concentrations (0 to 1.1 mM). The percentage of mutants was determined after 48 h by plating onto KBcm agar. The percentage of CHA96rif was also determined by plating onto KBcm agar containing 100 μg of rifampin per ml. The experiment consisted of six replicates studied over time.
Effect of zinc and medium dilution on mutant accumulation in other biocontrol strains.
For each strain, mixtures containing 99% wild-type strain CHA0 and 1% gacA mutant were used to inoculate NBY broth, dilute (0.1×) NBY broth, and NBY broth containing 0.7 mM ZnSO4 · 7H2O. Mutants of each strain were HCN and protease negative and had an orange colony phenotype identical to that of CHA0 mutants, which was used to determine the percentage of mutants after 48 h. Treatments were arranged as a 5 × 3 factorial with a main plot for strain and a subplot for medium. Treatments consisted of three replicates, and the experiment was conducted twice.
Influence of minerals and medium dilution on growth of wild-type strain CHA0 and mutants.
Wild-type strain CHA0 and spontaneous mutants were grown individually in NBY broth, dilute NBY broth, dilute NBY broth containing NaCl, and NBY broth containing minerals. After 48 h, the numbers of CFU per milliliter were determined by plating samples onto KBcm agar. Treatments were arranged as a 6 × 14 factorial in a split-plot design with a main plot for bacterial strain and a subplot for medium treatment. The experiment consisted of six replicates studied over time. The growth rates of CHA0, CHAS17, and CHAS33 were determined by measuring the optical densities at 600 nm from zero time to 48 h in 150-ml portions of NBY broth, dilute (0.1×) NBY broth, dilute NBY broth containing 30 mM NaCl, and NBY broth containing 0.7 mM CuSO4 or 0.7 mM ZnSO4 · 7H2O. Treatments consisted of two replicates.
Data analysis.
Bacterial CFU data were transformed by using the logarithmic base 10, and percentage data were transformed by using the arcsine of square roots prior to analysis of variance. Unless indicated otherwise, treatments were arranged in a randomized complete block design, and experiments were repeated two to four times. Data from repeated trials were pooled after we confirmed in a preliminary analysis that the trial-main effects interaction was not significant and/or that variances between trials were homogeneous as determined by an F test or Bartlett's test. For most experiments, main effects and interactions were further analyzed for significance with the SAS general linear models procedure (Statistical Analysis Systems Institute, Cary, N.C.), with the mean comparisons performed by using Fisher's protected least-significant-difference (P = 0.05) (LSD0.05) test. SAS regression procedures were used to determine relationships between mutant content and inoculum efficacy and between zinc concentration and mutant accumulation.
RESULTS
Mutant phenotypic, genotypic, and biochemical characterization.
Spontaneous mutants appeared at a high frequency (approximately 1%) in stationary-phase cultures of CHA0. Mutants were easily distinguished from the wild type in dilution-plated samples based on the unusual appearance of colonies (i.e., orange color; flattened; expanded; often transluscent; surrounded by a more intense, diffusible, yellow, fluorescent pigment, whose intensity increased over a period of 5 days). The correlation between the orange colony color and the loss of HCN, protease, and TSO production was approximately 98%. Orange mutants were indistinguishable from the wild type by PCR-randomly amplified polymorphic DNA analysis. A total of 49.7% of 205 orange mutants were restored to a wild-type phenotype with a gacS clone, and 48.2% were restored with a gacA clone. Of the remaining 2.1% pleiotropic mutants not restored with either of the single-gene clones, none required both clones for complementation. Generally, GacS− and GacA− mutants behaved similarly in all tests throughout this study, and spontaneous mutants were indistinguishable from genetically engineered derivatives.
Spontaneous mutants exhibited no signs of reversion to HCN-, protease-, or TSO-positive status (<10−5 revertants per ml) after three 48-h subcultures in NBY broth. Compared to the wild type, GacS− and GacA− exhibited mutants clearly reduced and delayed production of HCN, protease, and TSO and produced no detectable antibiotics. As in previous studies, mutants were found to be simply negative for these characteristics (Table 2). However, leaky metabolite production was occasionally observed with both spontaneous and genetically engineered mutants, particularly when incubation periods were long (e.g., >48 h instead of 24 h for HCN determination). Mutants produced significantly more pyochelin and salicylic acid, had significantly larger cells, and raised the pH of NBY broth (normally pH 6.5) significantly more than the wild type (Table 2). A total of 128 carbon sources were tested, and differences were observed in the ability of spontaneous mutants to utilize alaninamide, d-malic acid, and mono-methyl succinate (increased), as well as dl-α-glycerol phosphate, glycyl-l-glutamic acid, and glycyl-l-aspartic acid (decreased), compared to the wild type. No differences were observed in tolerance to PHL and PLT compared to the wild type.
TABLE 2.
Phenotypic characterization of P. fluorescens CHA0 spontaneous regulatory mutantsa
| Characteristic | Wild type | gacA mutant | gacS mutant |
|---|---|---|---|
| Colony morphology on KB agar and NBY agar | Circular, smooth, convex, opaque, beige | Generally flat, translucent, orange, 10 to 50% greater diam | Generally flat, translucent, orange, 10 to 50% greater diam |
| Cell length (μm) | 3.3 (0.5) | 5.8 (1.1) | 5.3 (0.8) |
| Pyochelin concn (ng/108 CFU) | 19.2 (4.6) | 125.5 (19.1) | 143.9 (11.3) |
| Salicylic acid concn (ng/108 CFU) | 0.6 (0.3) | 4.2 (1.1) | 7.1 (2.9) |
| Extracellular protease | +b | − | − |
| TSO | + | − | − |
| Hydrogen cyanide | + | − | − |
| PHL concn (ng/108 CFU) | 61.8 (2.5) | <0.07 | <0.07 |
| Pyrrolnitrin concn (ng/108 CFU) | 2.5 (0.9) | <0.1 | <0.1 |
| PLT concn (ng/108 CFU) | 12.5 (1.5) | <0.07 | <0.07 |
| Nutrient broth pH after 48 h | 7.77 (0.02) | 8.04 (0.03) | 8.09 (0.04) |
| Inhibition zone for P. ultimum growth (mm) on: | |||
| KB agar | 1.03 (0.12) | 0.97 (0.09) | 0.93 (0.03) |
| KB agar containing 100 μM FeCl3 | 1.43 (0.09) | 0.03 (0.03) | 0.07 (0.07) |
| % Cucumber seedlings standing after 7 days in P. ultimum-infested soil | 82.5 (3.8) | 35.7 (5.4) | 31.1 (4.2) |
The values are data for wild-type strain CHA0 and 5 to 10 mutants complemented with gacS and gacA clones. The values in parentheses are standard errors. Each treatment was replicated two to five times.
+, strong positive reaction; −, strong negative reaction, although some leakiness was observed in mutants.
Spontaneous mutation compromised the biocontrol efficacy of inoculants.
Wild-type strain CHA0 inhibited the growth of Pythium spp. on KB agar with or without supplemental iron (Table 2). Added iron actually improved the inhibitory activity of CHA0, probably by stimulating antibiotic biosynthesis. In contrast, GacS− and GacA− mutants lost the ability to inhibit Pythium growth on KB agar amended with iron, which repressed production of diffusible fluorescent pigments typical of pyoverdine siderophores. In the absence of added iron, mutants overproduced fluorescent pigment, and their inhibitory activity was identical to that of the wild type.
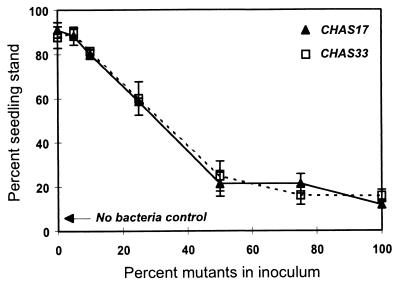
When cucumber seeds were treated with single strains, spontaneous mutants were significantly less effective than the wild type for controlling Pythium damping-off, which confirmed the results of previous studies in which genetically constructed mutants were used (Table 2). More importantly though, when CHA0 inoculants were contaminated with mutants at different ratios, the level of protection for cucumber decreased significantly as the level of contamination increased (P = 0.0001; r2 = 0.82) (Fig. 1). Inoculant efficacy was significantly reduced by as little as 10% mutant contamination and was essentially lost when the level of contamination was more than 50%. Contamination with either GacS− or GacA− mutants had the same detrimental impact.
FIG. 1.
Influence of mutant contamination on biocontrol efficacy of CHA0 inoculants. Cucumber seeds were treated with suspensions of wild-type CHA0 inoculum contaminated by adding a gacS (CHAS17) or gacA (CHAS33) mutant at a range of concentrations from 0 to 100%. Seeds were grown in soil infested with P. ultimum. The percentages of seedlings that emerged and were standing after 10 days are shown. The values are means based on six replicates; the error bars indicate standard errors.
Certain mineral amendments and medium dilution improved the genetic stability of CHA0 cultures.
Four approaches were used to evaluate the influence of culture conditions on the accumulation of regulatory mutants in NBY broth. First, we determined the effects of copper amendment, medium dilution, and electrolyte concentration on the appearance of mutants in wild-type cultures that contained no detectable mutants at the start of the experiment. All treatments significantly reduced the accumulation of orange mutants compared to normal-strength (1×) NBY broth, and dilute (0.1×) NBY broth provided the best control of mutant accumulation (Table 3). The validity of using orange colony color to identify mutants was supported by the fact that nearly identical results were obtained when randomly sampled colonies from each treatment were tested to determine whether they produced HCN, protease, and TSO. In NBY broth, approximately 1% of the colonies were negative for these metabolites. In comparison, no negative colonies were observed in dilute NBY broth, and only 0.2 and 0.3% of the colonies were negative in copper-amended NBY broth and dilute NBY broth containing NaCl, respectively. Total bacterial growth was less for all treatments than total bacterial growth in normal NBY broth (Table 3).
TABLE 3.
Mutant accumulation in wild-type strain CHA0 cultures after 12 days of incubation in NBY broth containing copper or dilute NBY brotha
| Medium | Total bacterial concn (log10 CFU/ml) | % of orange mutants |
|---|---|---|
| NBY broth | 9.16 | 1.19 |
| NBY broth containing 0.7 mM CuSO4 | 9.05 | 0.52 |
| Dilute (0.1×) NBY broth | 8.99 | 0.02 |
| Dilute (0.1×) NBY broth containing 30 mM NaCl | 8.92 | 0.25 |
Broth media were inoculated with overnight CHA0 cultures that contained no detectable mutants. The total numbers of viable bacteria and the percentages of orange-colony mutants were determined after 12 days. The values are means based on 40 replicate cultures from four trials (no significant treatment-trial interaction). Each main effect was significant (P ≤ 0.0001). The means were compared by using Fisher's protected LSD test. The Fisher's LSD0.05 values for total bacterial concentration and percentage of orange mutants were 0.04 and 0.45, respectively.
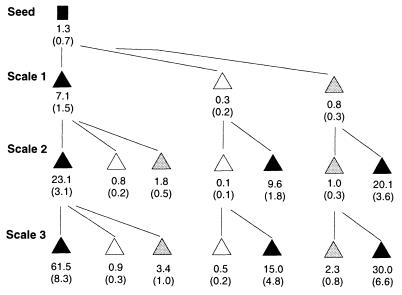
Second, we examined the problems that might be expected in large-scale fermentations, in which typically small batches are used to inoculate increasingly larger volumes of media. When a medium with a selective pressure for mutants (i.e., NBY broth) was used for scale-up from 20 to 100 to 500 ml, an exponential increase in the number of mutants was observed (Fig. 2). In contrast, when a medium that favored the wild type over mutants was used (i.e., dilute NBY broth or copper-amended NBY broth), mutant accumulation was arrested at all of the stages of scale-up. Not only did switching from NBY broth to dilute medium or copper-amended medium at any stage stop further mutant accumulation, but it essentially restored the culture to predominantly wild type cells (Fig. 2). Transferring clean cultures (i.e., dilute or copper-amended cultures) to full-strength NBY broth, even for just one cycle, had the opposite effect of polluting them with mutants.
FIG. 2.
Mutant accumulation from contaminated seed cultures through three stages of scale-up from 20 to 100 to 500 ml (Scale 1 to 3). The inoculation and sampling methods used are described in Materials and Methods. The lines indicate origins of inocula. Cultures were grown for 48 h in NBY broth (solid triangles), dilute (0.1×) NBY broth (open triangles), and NBY broth containing 0.7 mM CuSO4 (grey triangles). The values below the symbols are the average percentages (standard errors) of mutants based on 14 replicate broth preparations.
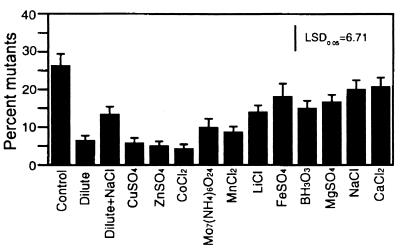
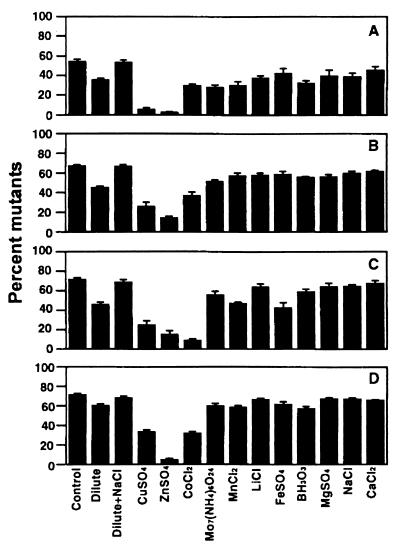
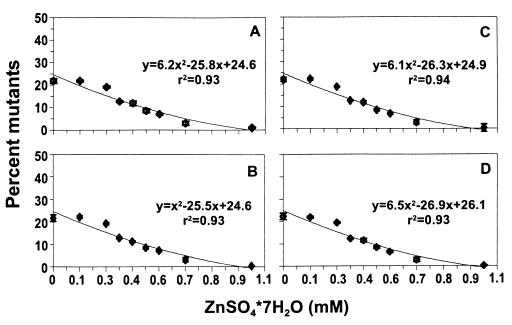
Building on the promising results obtained with copper, we next examined a larger range of minerals. When cultures contaminated with approximately 0.3% orange mutants were used to inoculate dilute medium and NBY broth amended with 1 of 11 minerals, we observed dramatic reductions in mutant accumulation ranging from 25% in the NBY broth control to approximately 5% in dilute (0.1×) NBY broth and NBY broth amended with copper, zinc, or cobalt (P = 0.0001) (Fig. 3). Ammonium molybdate and manganese reduced mutant accumulation to approximately 10%. Lithium, iron, boron, and magnesium slightly reduced mutant accumulation, and sodium and calcium had no effect compared to the NBY broth control. The beneficial effect of diluting NBY broth was slightly but significantly diminished by raising the electrolyte concentration with NaCl (Fig. 3). A similar effect of NaCl when it was added to dilute NBY broth was observed in another set of experiments in which cultures were inoculated with 10% GacS− and GacA− mutants (P = 0.0001) (Fig. 4). Zinc, copper, and cobalt were consistently the most effective treatments, and the reduction in mutant accumulation obtained when NBY broth was diluted was always lost when NaCl was added. There was a significant inverse relationship between mutant accumulation and zinc sulfate concentration (P = 0.0001; r2 = 0.88) (Fig. 5). Mutant accumulation was cut in half at concentrations of 0.5 mM and almost completely controlled at concentrations of ≥1.0 mM, regardless of whether mutants had defects in gacS or gacA (Fig. 5A and C) or were spontaneous or genetically engineered (Fig. 5B and D). For CHA96rif, plating onto rifampin-amended KBcm agar and using orange colony color as a marker for determining mutant accumulation gave nearly identical results, which further validated our mutant detection method (data not shown). The total number of bacterial CFU after 48 h was approximately log 9.4 per ml in nonamended NBY broth and was essentially unchanged by zinc sulfate concentrations of <0.8 mM. Increasing toxicity was observed at concentrations of 1.0 and 1.5 mM, and the average growth reductions were 0.2 and 0.9 log units, respectively (data not shown).
FIG. 3.
Effects of minerals on competition between CHA0 and spontaneous orange mutants. NBY broth (control), dilute (0.1×) NBY broth, dilute NBY broth containing NaCl, or NBY broth containing minerals was inoculated with a low but detectable level of orange mutants (approximately 0.3%). After 48 h, the percentage of mutants was determined. The bars indicate means based on 16 cultures; error bars indicate standard errors. Fisher's protected LSD0.05 value was 6.71%.
FIG. 4.
Effects of minerals on competition between CHA0 and gacA mutant CHAS9 (A) or CHAS45 (B) or gacS mutant CHASP1 (C) or CHAS17 (D). Broth preparations (see the legend to Fig. 3) were inoculated with a bacterial mixture containing 90% wild-type strain CHA0 and 10% mutant. After 48 h, the percentages of mutants were determined. The bars indicate means based on eight cultures; the error bars indicate standard errors. Fisher's protected LSD0.05 values were 10.4% (A), 8.2% (B), 11.3% (C), and 6.8% (D).
FIG. 5.
Relationship between zinc concentration and mutant accumulation. NBY broth preparations amended with a range of zinc concentrations were inoculated with a mixture containing 90% wild-type strain CHA0 and 10% spontaneous mutant CHAS17 (A) or CHAS45 (C) or genetically engineered mutant CHA510 (B) or CHA96rif (D). After 48 h, the percentages of mutants were determined. The values are means based on six cultures; the error bars indicate standard errors. The regression lines approximate 6x2 − 26x + 25 (P ≤ 0.0001; r2 ≥ 0.93).
Reduction of spontaneous mutation in other biocontrol strains.
Spontaneous mutants defective for HCN, protease, and the antibiotics PHL and PLT were readily recovered from five wild-type biocontrol pseudomonads isolated from tobacco roots grown in soil from Ghana (PGNR1, PGNR4, PGNL1) and Italy (PINR2, PINR3). Orange translucent sectors composed of regulatory mutants appeared in colonies grown for an extended period (about 10 days) on KB agar. Over time, mutants eventually overgrew the wild type. These orange mutants were phenotypically identical to those observed with strain CHA0 and were complemented with a gacA or gacS clone. When a wild type was combined with a corresponding GacA− mutants at a concentration of 10%, mutant accumulation was reduced by zinc amendment and medium dilution (P = 0.0289) compared to mutant accumulation with full-strength NBY broth (Table 4). This was true for all five strains. However, a significant strain-medium interaction (P = 0.0113) indicated that some strains responded better than others to zinc amendment and medium dilution. In zinc-amended NBY broth, the reductions in mutant accumulation compared to nonamended NBY broth ranged from 4.6-fold for PGNR4 to 17.2-fold for PINR2. In diluted NBY broth, the reductions ranged from 4.3-fold for PINR3 to 19.8-fold for PGNL1. It was also evident that some strains (e.g., PINR3) were more susceptible to mutant accumulation than others regardless of medium, which indicated that genetic stability varied among biocontrol isolates (Table 4).
TABLE 4.
Influence of zinc amendment and medium dilution on accumulation of spontaneous gacA regulatory mutants of biocontrol strains from Ghana and Italya
| Origin | Strain | % of mutantsb
|
Fisher's LSD0.05 | ||
|---|---|---|---|---|---|
| NBY broth | NBY broth containing zinc | Dilute NBY broth | |||
| Ghana | PGNL1 | 31.7 | 2.3 | 1.6 | 8.0 |
| PGNR1 | 16.8 | 2.6 | 2.7 | 8.6 | |
| PGNR4 | 22.3 | 4.8 | 2.3 | 6.8 | |
| Albenga, Italy | PINR2 | 29.2 | 1.7 | 3.0 | 3.7 |
| PINR3 | 40.4 | 4.0 | 9.3 | 14.2 | |
NBY broth, NBY broth containing 0.7 mM ZnSO4 · 7H4O (zinc), and dilute (0.1×) NBY broth were inoculated with 99% wild-type strain and 1% gacA mutant. Percentages of mutants were determined after 48 h of growth as described in Materials and Methods.
The strain-medium interaction was significant (P = 0.0113), and data were analyzed based on response to individual main effects. The values are means based on six replicates. The differences within a row or column as determined by Fisher's protected LSD test are significant at P ≤ 0.0289. The Fisher's LSD0.05 values for NBY broth, NBY broth containing zinc, and dilute NBY broth were 14.7, 1.4, and 2.2, respectively.
Mineral and medium dilution effects on mutant accumulation were independent of any effect on growth of individual bacteria.
A significant mineral-strain interaction and significant main effects (P = 0.0001) indicated that medium treatments had differential effects on the yield of culturable bacteria after 48 h of growth. Generally, for any given medium there were no consistent differences between the wild type and mutants (Table 5) and no apparent relationship between medium effects on growth and mutant accumulation (Fig. 4). For example, in zinc-amended NBY broth growth of one GacA− mutant, CHAS9, was slightly reduced compared with growth in nonamended NBY broth, but growth of another GacA− mutant, CHAS45, was not affected (Table 5). In copper-amended NBY broth, the reverse was true, and growth of CHAS45 but not growth of CHAS9 was reduced. Zinc and copper did not affect growth of either of the GacS− mutants or the wild type. However, both zinc and copper reduced accumulation of all mutants in competition experiments (Fig. 4). Furthermore, cobalt, which also reduced mutant accumulation (Fig. 4), did not affect growth of any of the mutants but reduced growth of the wild type (Table 5). Growth of all strains was reduced in dilute NBY broth and dilute NBY broth containing NaCl, but there were generally no differences among strains (Table 5).
TABLE 5.
Effects of liquid media on bacterial growtha
| Medium | Conductivity (mS) | Log CFU/mlb
|
Fisher's LSD0.05 | ||||
|---|---|---|---|---|---|---|---|
| CHA0 | CHAS17 | CHASP1 | CHAS9 | CHAS45 | |||
| NBY broth | 2.4 | 9.67 | 9.48 | 9.57 | 9.39 | 9.55 | 0.15 |
| NBY broth containing CuSO4 | 2.4 | 9.66 | 9.52 | 9.54 | 9.28 | 9.00 | 0.16 |
| NBY broth containing ZnSO4 | 2.3 | 9.61 | 9.58 | 9.69 | 9.19 | 9.60 | 0.20 |
| NBY broth containing CoCl2 | 2.4 | 9.37 | 9.46 | 9.36 | 9.38 | 9.45 | NS |
| NBY broth containing Mo7(NH4)6O24 | 2.9 | 9.59 | 9.55 | 9.67 | 9.44 | 9.66 | 0.11 |
| NBY broth containing MnCl2 | 2.3 | 9.68 | 9.49 | 9.63 | 9.40 | 9.57 | 0.13 |
| NBY broth containing LiCl | 2.4 | 9.61 | 9.53 | 9.61 | 9.39 | 9.51 | 0.16 |
| NBY broth containing FeSO4 | 2.3 | 9.54 | 9.50 | 9.59 | 9.38 | 9.57 | 0.12 |
| NBY broth containing BH3O3 | 2.2 | 9.65 | 9.58 | 9.63 | 9.44 | 9.58 | 0.13 |
| NBY broth containing MgSO4 | 2.4 | 9.67 | 9.53 | 9.68 | 9.51 | 9.48 | 0.14 |
| NBY broth containing NaCl | 2.3 | 9.67 | 9.48 | 9.59 | 9.35 | 9.57 | 0.18 |
| NBY broth containing CaCl2 | 2.4 | 9.67 | 9.56 | 9.65 | 9.51 | 9.53 | 0.11 |
| Dilute (0.1×) NBY broth | 0.2 | 8.86 | 8.86 | 8.94 | 8.82 | 8.89 | 0.07 |
| Dilute (0.1×) NBY broth containing 30 mM NaCl | 4.0 | 8.93 | 8.89 | 8.98 | 8.85 | 9.01 | 0.08 |
Wild-type strain CHA0, gacS mutants CHAS17 and CHASP1, and gacA mutants CHAS9 and CHAS45 were inoculated individually into 20 ml of NBY broth, NBY broth amended with a mineral, or dilute NBY broth. Sodium chloride was added to dilute NBY broth to increase the medium conductivity. Total viable cell counts (log CFU) were determined after 48 h.
The values are means based on six replicates. The medium-strain interaction was significant (P ≤ 0.0001), and data were analyzed on the basis of the response to medium (P ≤ 0.0001 within a column) and strain (P ≤ 0.0429 within a row, except where NS indicates that the differences were not significant as determined by analysis of variance). The means were compared by using Fisher's protected LSD test. The LSD0.05 values for strains CHA0, CHAS17, CHASP1, CHAS9, and CHAS45 were 0.10, 0.14, 0.13, 0.16, and 0.14, respectively.
DISCUSSION
Our results document for the first time that maintaining genetic stability during liquid fermentation is critical for production of high-quality biocontrol inoculants (32, 39). The gacS and gacA global regulatory genes of several P. fluorescens strains were very unstable in certain media. Even though all strains were susceptible, the degrees of instability of strains varied. The ability of an inoculant to protect cucumber from disease was significantly reduced by as little as 10% mutants and was essentially abolished when the level of contamination reached 50%. Such levels occur because mutants multiply exponentially when processes are scaled up to increasingly larger fermentor volumes. Our method for identifying gac mutants by dilution plating and visual assessment of distinct colony phenotypes (e.g., darker pigmentation, sprawling, hyperfluorescence) offers an inexpensive and simple option for routinely monitoring the quality of inoculant fermentations. Mutants of our biocontrol strains had the same orange-brown color, compared to beige for the wild type, but in other strains the color to watch for may be different (4, 11).
Mutants lost the ability to produce all of the key antifungal metabolites (e.g., PHL, PLT, hydrogen cyanide). Increased siderophore production, which compensated for the loss of these compounds in in vitro inhibition assays, did not compensate in the rhizosphere, where sufficient iron may have been available (24). There was a significant inverse relationship between inoculant efficacy and mutant contamination which reflected a dose-response relationship like that defined in various biocontrol systems. A threshold population density of bacteria is required for significant disease suppression, and relatively small decreases in the size of a population can dramatically reduce the level of protection (14). We extended this idea by specifying that a threshold population of biocontrol-active cells is needed for effective disease suppression. Thus, mutant contamination compromises inoculant performance by diluting the dose of biocontrol-active cells in the final product.
Having identified gacS-gacA instability as a problem, we set out to understand why mutants accumulate during inoculum production and to develop an approach to manage this problem. The results of scale-up experiments, in which small amounts of mutants were repeatedly transferred into fresh media, indicated that mutant accumulation resulted primarily from greater mutant competitiveness rather than increased frequency of mutational events. Further evidence that competition is the mechanism behind mutant accumulation comes from the fact that when mutants and wild-type strain CHA0 were grown in various media individually, the numbers of CFU at the end of fermentation were similar. However, we cannot eliminate the possibility that minerals and medium dilution have different effects on growth rates. Other workers have recently sequenced gacA alleles from several mutants, which revealed a surprising diversity of mutational events (point, deletion, and frame-shift mutations involving as few as 3 bp) (C. T. Bull and D. Haas, unpublished data). This indicates that selection is for the Gac− phenotype rather than for a particular mutational hot spot. The main selective factor for gac mutants appears to have been the nutrient content of the media because accumulation was far greater in full-strength NBY broth than in 0.1× NBY broth. However, because the beneficial effect of medium dilution was largely negated by raising the medium conductivity to the same level as the conductivity of NBY broth, the real culprit is more likely higher osmotic potential and not excess nutrient availability per se.
It is still possible that a gac mutation may be a starvation response akin to the GASP phenotype described for Escherichia coli and other bacteria, in which subpopulations which have a growth advantage during the stationary phase are selected (42). Even if our media retained an excess of most nutrients, certain key factors (possibly minerals) were gradually depleted during culture growth, which may have resulted in a shift towards mutants. This would explain why gac mutants typically appeared in older cultures. Compensating for such a deficiency may be the mechanism by which mineral amendments repressed mutants during the stationary phase, but this does not explain how minerals worked when they were added to fresh media. Mutation did not appear to increase the sensitivity to toxic metals like zinc, because growth characteristics were similar for mutants and wild-type strain CHA0 when they were inoculated individually. Overproduction of metal-chelating siderophores may have provided a competitive advantage to mutants as trace minerals became scarce, an advantage that was lost when excess amounts of the metals were supplied. In mycopathogenic Pseudomonas tolassii, gacS mutants are thought to appear because the gene is downregulated under starvation conditions, something like gac atrophy (11). Interestingly, most of the minerals that we found which repressed mutants have previously been shown to stimulate biosynthesis of antifungal metabolites regulated by gacS-gacA (7). Another unusual, but possible, explanation is that a compound in the media triggered mutation, in the same way that the plant phenolic compound acetosyringone selects for nonpathogenic variants of the pathogenic bacterium Agrobacterium tumefaciens (10) or benzyl alcohol selects for Tol− variants in the bioremediation bacterium Pseudomonas putida (22).
Factors that influence the fitness of gac mutants could have long-term implications after inoculant application. Although in nature we have only rarely seen gac mutants like our tobacco root isolate CHASP1, such mutants appear to be at least as competitive as the wild-type strain in certain environments. Natsch et al. (26) found that a gacA insertion mutant of CHA0 was slightly less competitive in bulk soil, equally competitive in the rhizosphere, and more competitive on the rhizoplane and in the root interior compared to the wild type. Similar results were observed with a gacS insertion mutant of the phytopathogenic organism P. syringae pv. syringae which exhibited reduced colonization on bean leaves in the field but was equally competitive on germinating bean seeds (12). Understanding the conditions that modulate mutation and/or mutant accumulation may enable us to prevent gac mutants that contaminate inoculants from displacing biocontrol-active cells after application to plants. On the other hand, identifying environmental signals that are conducive to gac mutants may inspire new disease control strategies that can be used against pathogenic bacteria which also have very unstable gac genes and which are rendered generally avirulent by mutation (9, 11, 23, 29, 34, 41).
ACKNOWLEDGMENTS
We thank C. Bull, D. Haas, J. Loper, and P. Schmidli-Sacherer for providing strains and plasmids and D. Weller (USDA-ARS, Pullman, Wash.) and E. Frossard (ETH Eschikon, Switzerland) for providing comments on the manuscript.
This work was supported in part by the Swiss National Science Foundation (grant 3100-50522.97) and by the European Union IMPACT Framework (project PL 920053).
REFERENCES
- 1.Blumer C, Heeb S, Pessi G, Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Sci USA. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 3.Carruthers F, Haas D. Signal transduction and the two-component regulatory system encoded by the lemA/gacA genes in Pseudomonas fluorescens CHA0: implications for biocontrol. IOBC WPRS Bull. 1997;21(9):151. [Google Scholar]
- 4.Chancey S T, Wood D W, Pierson L S., III Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl Environ Microbiol. 1999;65:2294–2299. doi: 10.1128/aem.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook R J. Making greater use of introduced microorganisms for biological control of plant pathogens. Annu Rev Phytopathol. 1993;31:53–80. doi: 10.1146/annurev.py.31.090193.000413. [DOI] [PubMed] [Google Scholar]
- 6.Corbell N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duffy B K, Défago G. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl Environ Microbiol. 1999;65:2429–2438. doi: 10.1128/aem.65.6.2429-2438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duffy B K, Défago G. Influence of cultural conditions on spontaneous mutations in Pseudomonas fluorescens CHA0. Phytopathology. 1995;85:1146. [Google Scholar]
- 9.Eriksson A R B, Andersson R A, Pirhonen M, Palva E T. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol Plant-Microbe Interact. 1998;11:743–752. doi: 10.1094/MPMI.1998.11.8.743. [DOI] [PubMed] [Google Scholar]
- 10.Fortin C, Nester E W, Dion P. Growth inhibition and loss of virulence in cultures of Agrobacterium tumefaciens treated with acetosyringone. J Bacteriol. 1992;174:5676–5685. doi: 10.1128/jb.174.17.5676-5685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grewal S I S, Han B, Johnstone K. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus bisporus. J Bacteriol. 1995;177:4658–4668. doi: 10.1128/jb.177.16.4658-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirano S S, Ostertag E M, Savage S A, Baker L S, Willis D K, Upper C D. Contribution of the regulatory gene lemA to field fitness of Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1997;63:4304–4312. doi: 10.1128/aem.63.11.4304-4312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson K B. Dose-response relationships and inundative biological control. Phytopathology. 1994;84:780–784. [Google Scholar]
- 15.Keel C, Défago G. Interactions between beneficial soil bacteria and root pathogens: mechanisms and ecological impact. In: Gange A C, Brown V K, editors. Multitrophic interactions in terrestrial systems. London, United Kingdom: Blackwell Scientific Publishers; 1996. pp. 27–46. [Google Scholar]
- 16.Keel C, Weller D M, Natsch A, Défago G, Cook R J, Thomashow L S. Conservation of the 2,4-diacetylphloroglucinol biosynthetic locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62:552–563. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keel C, Schnider U, Maurhofer M, Voisard C, Laville J, Burger U, Wirthner P, Haas D, Defago G. Suppression of root diseases by Pseudomonas fluorescens CHA0: importance of the bacterial secondary metabolite 2,4-diacetylphloroglucinol. Mol Plant-Microbe Interact. 1992;5:4–13. [Google Scholar]
- 18.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 19.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 20.Lam K S, Gustavson D R, Veitch J M, Forenza S, Ross J, Miller D, Roach J, Lebherz III W B, Poole K. Large scale production and semi-purification of kedarcidin in a 1000-L fermentor. J Ind Microbiol. 1994;13:356–360. doi: 10.1007/BF01577219. [DOI] [PubMed] [Google Scholar]
- 21.Laville J, Voisard C, Keel C, Maurhofer M, Défago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leddy M B, Phipps D W, Ridgway H F. Catabolite-mediated mutations in alternate toluene degradative pathways in Pseudomonas putida. J Bacteriol. 1995;177:4713–4720. doi: 10.1128/jb.177.16.4713-4720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao C H, McCallus D E, Wells J M, Tzean S S, Kang G Y. The repB gene required for production of extracellular enzymes and fluorescent siderophores in Pseudomonas viridiflava is an analog of the gacA gene of Pseudomonas syringae. Can J Microbiol. 1996;42:177–182. doi: 10.1139/m96-026. [DOI] [PubMed] [Google Scholar]
- 24.Loper J E, Henkels M D. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl Environ Microbiol. 1997;63:99–105. doi: 10.1128/aem.63.1.99-105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray N E, Brammer W J, Murray K. Lamboid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977;150:53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- 26.Natsch A, Keel C, Pfirter H A, Haas D, Défago G. Contribution of the global regulator gene gacA to persistence and dissemination of Pseudomonas fluorescens biocontrol strain CHA0 introduced into soil microcosms. Appl Environ Microbiol. 1994;60:2553–2560. doi: 10.1128/aem.60.7.2553-2560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oberhänsli T, Défago G, Haas D. Indole-3-acetic acid (IAA) synthesis in the biocontrol strain CHA0 of Pseudomonas fluorescens: role of tryptophan side chain oxidase. J Gen Microbiol. 1991;137:2273–2279. doi: 10.1099/00221287-137-10-2273. [DOI] [PubMed] [Google Scholar]
- 28.Pierson L S, III, Pierson E A. Phenazine antibiotic production in Pseudomonas aureofaciens: role in rhizosphere ecology and pathogen suppression. FEMS Microbiol Lett. 1996;136:101–108. [Google Scholar]
- 29.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacherer P, Défago G, Haas D. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHA0. FEMS Microbiol Lett. 1994;16:155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Schroth M N, Loper J E, Hildebrand D C. Bacteria as biocontrol agents of plant disease. In: Klug M J, Reddy C A, editors. Current perspectives in microbial ecology. Washington, D.C.: American Society for Microbiology; 1984. pp. 362–369. [Google Scholar]
- 33.Smith R S. Production and quality control of inoculants. In: Elkan G H, editor. Symbiotic nitrogen fixation technology. New York, N.Y: Marcel Dekker; 1987. pp. 391–411. [Google Scholar]
- 34.Tan M W, Rahme L G, Sternberg J A, Tompkins R G, Ausubel F M. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc Natl Acad Sci USA. 1999;96:2408–2413. doi: 10.1073/pnas.96.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomashow L S, Mavrodi D V. The genetics and regulation of antibiotic production by PGPR. In: Ogoshi A, Kobayashi K, Homma Y, Kodama F, Kondo N, Akino S, editors. Plant growth-promoting rhizobacteria: present status and future prospects. OECD Workshop. Sapporo, Japan: Hokkaido University Press; 1997. pp. 108–115. [Google Scholar]
- 36.Thomashow L S, Weller D M. Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites. In: Stacey G, Keen N T, editors. Plant-microbe interactions. Vol. 1. New York, N.Y: Chapman and Hall; 1996. pp. 187–235. [Google Scholar]
- 37.Troxler J, Zala M, Moënne-Loccoz Y, Keel C, Défago G. Predominance of nonculturable cells of the biocontrol strain Pseudomonas fluorescens CHA0 in the surface horizon of large outdoor lysimeters. Appl Environ Microbiol. 1997;63:3776–3782. doi: 10.1128/aem.63.10.3776-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voisard C, Rella M, Haas D. Conjugative transfer of plasmid RP1 to soil isolates of Pseudomonas fluorescens is facillitated by certain large RP1 deletions. FEMS Microbiol Lett. 1988;55:9–14. [Google Scholar]
- 39.Weller D M. Biological control of soilborne plant pathogens in the rhizosphere with bacteria. Annu Rev Phytopathol. 1988;26:379–407. [Google Scholar]
- 40.Whistler C A, Corbell N A, Sarniguet A, Ream W, Loper J E. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor ςs and the stress response in Pseudomonas fluorescens Pf-5. J Bacteriol. 1998;180:6635–6641. doi: 10.1128/jb.180.24.6635-6641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong S M, Carroll P A, Rahme L G, Ausubel F M, Calderwood S B. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect Immun. 1998;66:5854–5861. doi: 10.1128/iai.66.12.5854-5861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zambrano M M, Kolter R. GASPing for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]