Abstract
Pseudomonas aeruginosa dominates the complex polymicrobial cystic fibrosis (CF) airway and is a leading cause of death in persons with CF. Oral streptococcal colonization has been associated with stable CF lung function. However, no studies have demonstrated how Streptococcus salivarius, the most abundant streptococcal species found in individuals with stable CF lung disease, potentially improves lung function or becomes incorporated into the CF airway biofilm. By utilizing a two-species biofilm model to probe interactions between S. salivarius and P. aeruginosa, we discovered that the P. aeruginosa exopolysaccharide Psl promoted S. salivarius biofilm formation. Further, we identified a S. salivarius maltose-binding protein (MalE) that is required for promotion of biofilm formation both in vitro and in a Drosophila melanogaster co-infection model. Finally, we demonstrate that promotion of dual biofilm formation with S. salivarius is common among environmental and clinical P. aeruginosa isolates. Overall, our data supports a model in which S. salivarius uses a sugar-binding protein to interact with P. aeruginosa exopolysaccharide, which may be a strategy by which S. salivarius establishes itself within the CF airway microbial community.
Subject terms: Bacteriology, Biofilms, Microbiome, Clinical microbiology
Introduction
Cystic fibrosis (CF) is the most common lethal genetic disorder in Caucasian populations [1]. Individuals with this disorder accumulate thick mucus in the lungs, and the inability to clear this mucus from the airways facilitates the colonization of microbes [1]. The bacterial species Pseudomonas aeruginosa is the leading cause of death for individuals with CF [2]. The prevalence of P. aeruginosa colonization in the CF population increases with age, with up to 60% of CF adults being colonized in their lifetime [3]. P. aeruginosa uses various strategies to cause persistent infections in the lung, including evasion of the host immune system, conversion to a mucoid phenotype, and biofilm formation [2]. These adaptation mechanisms, particularly the ability to form recalcitrant biofilms, render P. aeruginosa difficult to treat with antibiotics and permit the development of lifelong chronic infections that lead to rapid lung deterioration and mortality [4].
Biofilms are defined as a community of microbes that are attached to a surface and embedded in a protective extracellular matrix [5]. P. aeruginosa produces multiple exopolysaccharides that comprise its biofilm matrix. Non-mucoid strains of P. aeruginosa, which typically colonize the CF lung initially, produce the exopolysaccharides Pel and Psl [6]. Over the course of infection, non-mucoid strains will accumulate mutations and convert to a mucoid phenotype by switching to production of the exopolysaccharide alginate [7]. Pel, Psl, and alginate play an important role in antimicrobial resistance by preventing penetration of antibiotics into the P. aeruginosa biofilm [8, 9]. Additionally, Psl is important for P. aeruginosa integration into polymicrobial biofilms [10].
Only in recent years have researchers began to study how cross-species interactions in biofilms influence the composition of polymicrobial communities [10]. The importance of studying P. aeruginosa interactions with other microbes in the CF airway polymicrobial community is becoming increasingly recognized [11, 12]. For instance, P. aeruginosa has been found to synthesize glutamate from precursor molecules secreted by Rothia mucilaginosa, another common microbe found in the CF lung [13]. Additionally, Psl produced by P. aeruginosa has been shown to interact with the staphylococcal protein A of Staphylococcus aureus to increase P. aeruginosa resistance to antibiotics [14]. Lastly, colonization of Stenotrophomonas maltophilia, an emerging CF pathogen, is promoted in murine lungs through integration into P. aeruginosa biofilms [15]. Relevant to our study, oral streptococci are increasingly recognized as core residents of the CF lung microbiota. Historically, these bacterial taxa have been thought to reside solely in the oral cavity and any detection of these microbes outside of this environment, particularly in the CF lung, was thought to be transient or attributed to oral contamination during sample collection. Multiple independent microbiome studies using sputum and bronchoalveolar lavage fluid have confirmed the presence of mitis and salivarius group oral streptococci in the CF airway [16–19]. Oral commensal streptococci have been shown to be associated with lung stability and increased microbial diversity in CF individuals. Streptococcus salivarius is the most prevalent streptococcal species found in the lungs of individuals with stable CF lung disease [16]. We previously reported that the second most abundant oral commensal found in the CF airway, Streptococcus parasanguinis, adheres to the mucoid P. aeruginosa exopolysaccharide alginate, resulting in the promotion of S. parasanguinis biofilm formation [20]. Therefore, it is important to understand how these commensals are incorporated into the CF polymicrobial community and potentially impact lung function in the CF population.
S. salivarius has been shown to commonly colonize the upper respiratory tract of infant children [21, 22]. Because many individuals with CF become colonized with P. aeruginosa during adulthood, it is likely that S. salivarius is present in the lungs during the early stages of infection with non-mucoid P. aeruginosa [3]. In an effort to better understand how S. salivarius incorporates into biofilms with the major CF pathogen P. aeruginosa, we utilized two species in vitro and in vivo biofilm models to identify mechanisms that facilitate S. salivarius colonization. Here, we report that S. salivarius exploits the non-mucoid P. aeruginosa exopolysaccharide Psl to promote streptococcal biofilm formation. This enhanced biofilm phenotype was consistent when P. aeruginosa environmental and clinical isolates were grown with S. salivarius. Moreover, we found that the presence of a streptococcal maltose-binding surface protein, MalE, potentially facilitates the interaction between S. salivarius and P. aeruginosa Psl. Finally, we show that P. aeruginosa promotes S. salivarius colonization in a Drosophila melanogaster in vivo model of co-infection. Taken together, our study highlights a unique mechanism by which S. salivarius utilizes P. aeruginosa extracellular components to influence the CF airway microbial community by initiating and sustaining streptococcal colonization within the CF lung.
Results
Non-mucoid Pseudomonas aeruginosa promotes Streptococcus salivarius biofilm formation
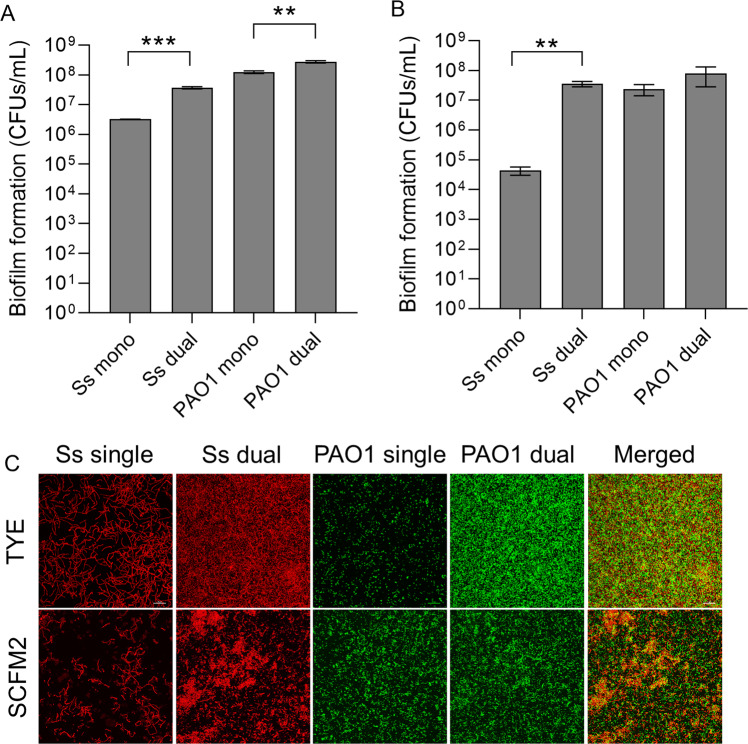
To characterize interactions between the oral commensal S. salivarius and the CF lung pathogen P. aeruginosa, we co-cultured S. salivarius with an acute wound isolate (PAO1) and chronic CF isolate (FRD1). S. salivarius and the acute isolate PAO1 formed significantly more biofilm biomass when co-cultured compared to single species controls (Fig. 1). When we co-cultured S. salivarius with the mucoid isolate FRD1, biofilm formation did not increase compared to the single species controls (Fig. 1). To examine the relative species contribution in dual species biofilms with S. salivarius and PAO1, both planktonic and biofilm colony forming units (CFUs) were enumerated (Fig. 2A). When co-cultured in TSBYE, both S. salivarius and P. aeruginosa biofilm CFUs increased significantly compared to single species controls. In contrast, no change in planktonic cell number was observed for either S. salivarius or P. aeruginosa in the presence of the other species (Fig S1A). We also co-cultured S. salivarius and P. aeruginosa in a synthetic cystic fibrosis sputum medium known as SCFM2, which mimics the nutrient profile found in the sputum of persons with CF [23]. When cultured in SCFM2, we observed an even greater increase in S. salivarius biofilm CFUs in the presence of P. aeruginosa (Fig. 2B). However, P. aeruginosa planktonic and biofilm cell number did not increase in the presence of S. salivarius in SCFM2, suggesting that this interspecies interaction exclusively enhances S. salivarius biofilm growth in synthetic CF sputum (Fig. 2 (Fig S1B)), which better recapitulates the nutritional environment in the CF airway [23]. We confirmed these observations via confocal laser microscopy and observed an increase in S. salivarius and P. aeruginosa biofilm formation in TSBYE when co-cultured, and an increase only in S. salivarius biofilm formation in SCFM2 (Fig. 2C). Additionally, propidium iodide staining, which stains dead cells or nucleic acids [24, 25] was prominent in our dual biofilm TSBYE group (Fig S2). We did not observe a reduction in planktonic cells for either species in the dual biofilm group, suggesting that this staining is likely due to extracellular DNA, an important component of the biofilm matrix [24, 26]. Enhanced S. salivarius biofilm formation in the presence of P. aeruginosa strain PAO1, but not FRD1 suggests that PAO1 possesses a distinct feature that promotes S. salivarius biofilm formation.
Fig. 1. S. salivarius and P. aeruginosa produce an enhanced biofilm in a dual species model.
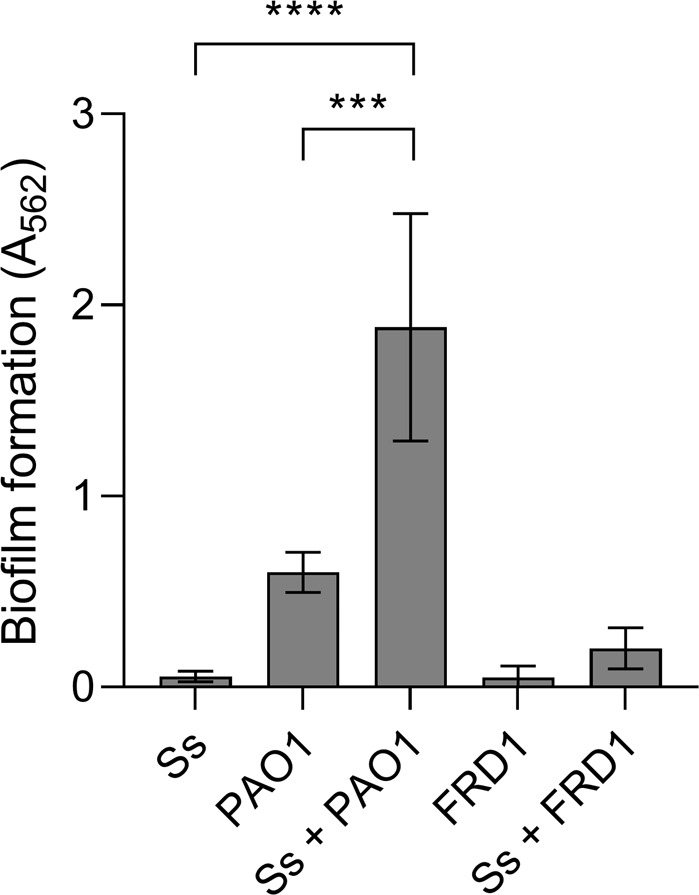
S. salivarius (Ss) was co-cultured with either of two P. aeruginosa strains (PAO1 and FRD1) in TSBYE medium in a 96-well plate for 16 h at 37 °C (n = 3 biological replicates, 6 technical). Biofilm biomass was then measured using crystal violet staining. One-way ANOVA with Šίdák’s multiple comparisons test. Error bars indicate standard deviation. ***p < 0.001, ****p < 0.0001.
Fig. 2. Non-mucoid P. aeruginosa strain PAO1 promotes S. salivarius biofilm formation.
A Quantification of Ss and PAO1 biofilm CFUs was performed in a 6-h, 6-well mono- and dual-species biofilm model. Samples were cultured in TSBYE medium and B synthetic CF sputum (SCFM2), serially diluted and plated on THB agar. Data represent three biological replicates performed in triplicate. Student’s t test. Error bars indicate mean ± SD. C Confocal microscopy was performed on Ss and PAO1-GFP single and dual species biofilms in both TSBYE and SCFM2. Ss was stained with hexidium iodide. **p < 0.01, ***p < 0.001.
P. aeruginosa exopolysaccharide Psl enhances S. salivarius biofilm formation
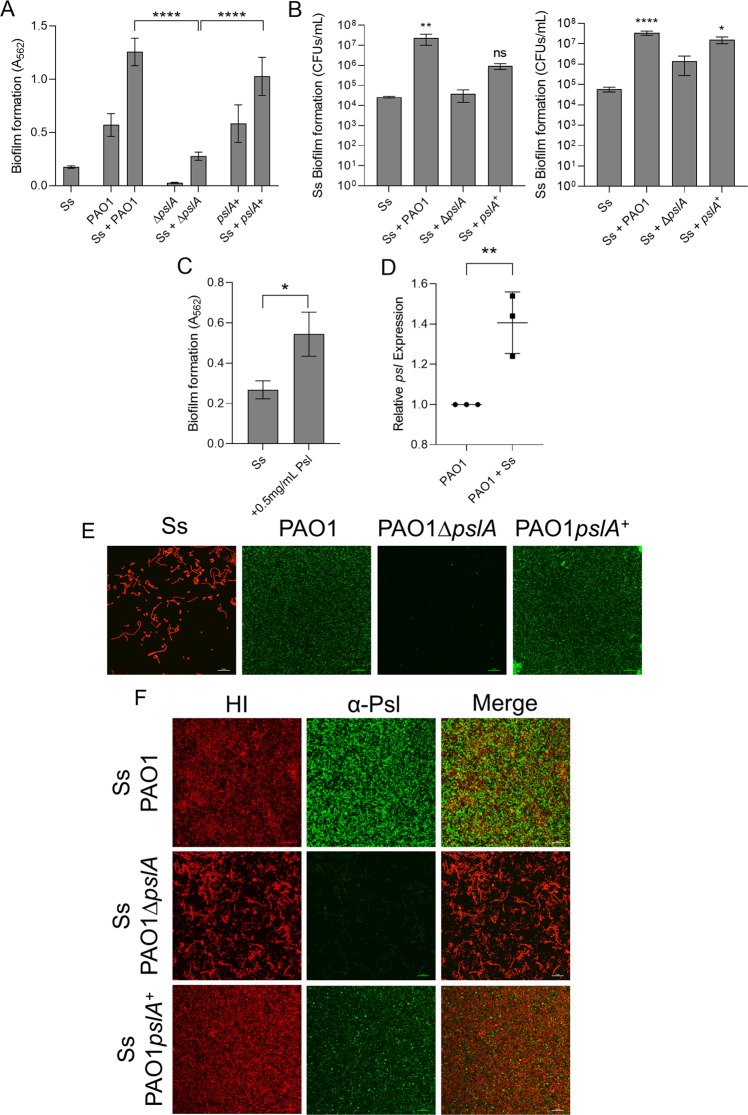
One major difference between the two strains is the exopolysaccharides produced within their respective biofilm matrices. Initial CF infecting P. aeruginosa strains that are acquired from the environment resemble PAO1 and produce Pel and Psl, whereas CF adapted strains like FRD1 overproduce alginate, largely due to accumulation of mutations in the mucA gene [6, 7]. To investigate whether production of the exopolysaccharide Psl, the most prominent exopolysaccharide made by PAO1, promotes S. salivarius biofilm formation, we co-cultured S. salivarius biofilms with a PAO1 mutant deficient in Psl (PAO1ΔpslA), and PAO1ΔpslA complemented with a wild-type copy of pslA (PAO1pslA+) [27]. When S. salivarius was co-cultured with PAO1ΔpslA, we observed significantly less dual biofilm formation compared to wild-type PAO1 co-cultures with S. salivarius. Co-culturing of S. salivarius with PAO1pslA+ restored the dual species biofilm to levels similar to the wild-type PAO1 dual biofilm (Fig. 3A). When quantifying S. salivarius biofilm CFUs in both TSBYE and SCFM2, we found that co-culture of S. salivarius with PAO1ΔpslA resulted in a significantly decreased S. salivarius biofilm cell number compared to S. salivarius co-cultured with wild-type PAO1 (Fig. 3B). When co-cultured with PAO1pslA+, S. salivarius biofilm was restored to levels similar to wild-type PAO1 in SCFM2 and partially restored in TSBYE, further confirming that Psl enhances S. salivarius biofilm formation. We quantified and compared planktonic CFUs of PAO1ΔpslA and PAO1pslA+ to wildtype PAO1 planktonic CFUs to confirm the lack of S. salivarius biofilm formation in the presence of PAO1ΔpslA was not caused by growth defects and lower cell density in the mutant strain co-culture (Figs. S3, S4). Additionally, we saw no growth advantage over time when we performed 16-h growth curves of wild-type PAO1 cultured with and without S. salivarius, further confirming that S. salivarius biofilm promotion is not due to increased growth of P. aeruginosa (Fig. S5). To further confirm that P. aeruginosa Psl does indeed contribute to enhanced biofilm formation by S. salivarius, we co-cultured S. salivarius with two P. aeruginosa isolates known to produce little to no Psl and measured changes in biofilm biomass [28, 29]. P. aeruginosa exopolysaccharide biosynthetic genes compete for the same precursor sugars, therefore, induction of one polysaccharide decreases the production of alternative exopolysaccharides [30]. Hence, overproduction of alginate in the mucA deficient strain of PAO1 results in a reduction in Psl production [29, 30]. Significantly less dual biofilm formation was observed with the PAO1ΔmucA-alginate overproducing strain in comparison to wild-type PAO1 (Fig. S6A). We observed a similar result when co-culturing S. salivarius with the P. aeruginosa strain PA14, which lacks essential Psl biosynthetic genes and solely produces the exopolysaccharide Pel (Fig. S6B) [31]. Additionally, purified Psl significantly increased the single species S. salivarius biofilm, further confirming the role of Psl in promoting S. salivarius (Fig. 3C). Lastly, pslA, a gene required for Psl production, was shown to be significantly upregulated in the presence of S. salivarius (Fig. 3D).
Fig. 3. P. aeruginosa exopolysaccharide Psl promotes S. salivarius biofilm formation.
Ss was co-cultured with P. aeruginosa PAO1 strains (A) PAO1∆pslA and PAO1 pslA+ in TSBYE medium with 1% sucrose in a 96-well plate for 16 h at 37 °C with 5% CO2 (n = 3 biological replicates, 3 technical). Biofilm biomass was then measured using crystal violet staining. One-way ANOVA with Dunnett’s multiple comparisons test. B Quantification of Ss biofilm-forming cells after co-culturing with PAO1, PAO1∆pslA, and PAO1 pslA+ in TSBYE (left) and SCFM2 [50] in a 6-h, 6-well model at 37 °C with 5% CO2 (n = 3 biological replicates, each with 3 technical replicates). One-way ANOVA with Šίdák’s multiple comparisons test. C 0.5 mg/mL purified Psl was added to Ss single cultures in TSBYE with 1% sucrose in a 96-well 16-h biofilm. Crystal violet staining was used to quantify biofilm biomass. D qPCR quantification of P. aeruginosa pslA expression compared to 16S rRNA control. Student’s t test. Fluorescence microscopy images at 60× magnification of 16-h single (E) and dual species (F) biofilms of Ss and PAO1, PAO1∆pslA, and PAO1pslA+ cultured in TSBYE supplemented with 1% sucrose. Ss was stained with hexidium iodide, and Psl was stained with a FITC-conjugated α-Psl monoclonal antibody. Scale bar: 20 μm. *p < 0.05, **p < 0.01, ****p < 0.0001.
To understand the spatial relationship between S. salivarius and Psl within a dual biofilm, we performed confocal laser scanning microscopy on single and dual biofilms of S. salivarius and P. aeruginosa strains PAO1, PAO1ΔpslA, and PAO1pslA+ stained with a FITC-conjugated α-Psl antibody to further characterize the role of Psl in promotion of S. salivarius biofilm formation (Fig. 3E, F). Consistent with our biofilm CFU quantifications, we observed a significant increase in S. salivarius biofilm in the presence of wild-type PAO1 as well as PAO1pslA+. When co-cultured with PAO1ΔpslA, no increase in S. salivarius biofilm formation was observed. Both S. salivarius and Psl were dispersed throughout the wild-type PAO1 and PAO1pslA+ dual biofilms. Additionally, changes in S. salivarius biofilm architecture were observed in the presence of Psl (Fig. 3F). Overall, our findings show that Psl not only promotes S. salivarius biofilm development, but also modifies S. salivarius biofilm structure in dual species biofilms with P. aeruginosa.
To determine whether S. salivarius utilizes Psl only as a biofilm matrix scaffold or also metabolizes Psl, we tested whether S. salivarius could utilize purified Psl as a carbon source by monitoring planktonic growth in full-strength and 1:1 diluted THB media that was supplemented with Psl or glucose (Fig. S7). S. salivarius grew similarly in the presence of Psl compared to the no-sugar control in full strength THB. Conversely, S. salivarius growth was enhanced in the presence of purified Psl in diluted THB media compared to the no-sugar control. This finding suggests that S. salivarius metabolizes Psl under conditions in which preferred carbon sources are limited. Our results demonstrate that Psl promotes S. salivarius biofilm formation, as well as S. salivarius planktonic growth via metabolism of Psl under specific nutrient-limited conditions.
S. salivarius maltose-binding protein MalE plays a role in promotion of S. salivarius biofilm formation both in vitro and in vivo
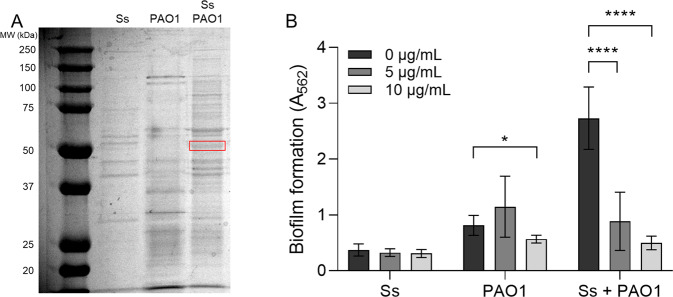
To identify candidate S. salivarius proteins that could be involved in P. aeruginosa-dependent biofilm promotion, we examined the protein profile of whole-cell lysates of S. salivarius and P. aeruginosa single and dual cultures (Fig. 4A). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis revealed the overexpression of one ~50 kDa protein that was identified as the S. salivarius maltose-binding protein MalE. MalE was overexpressed in dual cultures with P. aeruginosa, but not in single S. salivarius cultures. To examine whether MalE is involved in P. aeruginosa-dependent promotion of S. salivarius biofilm formation, we added anti-MalE antibodies to single and dual cultures of S. salivarius and P. aeruginosa. We found that anti-MalE antibodies significantly inhibited dual biofilm formation in a dose-dependent manner, while growth of single species biofilms was unaffected (Fig. 4B). These findings suggest that MalE is involved in promotion of S. salivarius biofilm formation in the presence of P. aeruginosa. To determine whether P. aeruginosa promotes S. salivarius colonization in an in vivo model of co-infection, Drosophila melanogaster were co-infected with subcultures of S. salivarius and P. aeruginosa, and bacterial CFUs were enumerated after 24 h. Colonization was performed with and without 10 µg/mL of α-MalE antibodies to test whether colonization of S. salivarius is MalE-dependent. S. salivarius colonization significantly increased in the presence of P. aeruginosa, while no change in P. aeruginosa colonization between groups was observed (Fig. 5A). Consistent with our in vitro data, the addition of 10 µg/mL α-MalE antibodies caused a significant decrease in S. salivarius colonization in the dual infection group but had no effect on colonization during single species infections (Fig. 5A). These findings suggest that MalE is required for increased colonization by S. salivarius in the presence of P. aeruginosa.
Fig. 4. S. salivarius maltose-binding protein MalE promotes dual biofilm formation.
A Ss and PAO1 were cultured individually and dually in TSBYE medium at 37 °C while shaking until OD600 = 1.8 was reached. Cells were resuspended in Tris-Buffered Saline (TBS) and lysed using the Bead Blaster 24 (Benchmark). Supernatant was run on SDS page gel, and overproduced bands in the dual sample were sent to the UAB Mass Spectrometry core for identification. B Ss and PAO1 were cultured in TSBYE with 1% sucrose in a 96-well 16-h biofilm model in the presence of 0 µg/mL, 5 µg/mL or 10 µg/mL α-MalE mAbs. Biofilms were stained with crystal violet to measure biofilm biomass (n = 3 biological, 3 technical). Error bars indicate mean ± SD. Two-way ANOVA with Tukey’s multiple comparisons test for post-hoc analysis. *p < 0.05, ****p < 0.0001.
Fig. 5. Promotion of S. salivarius colonization of Drosophila is MalE and Psl dependent.
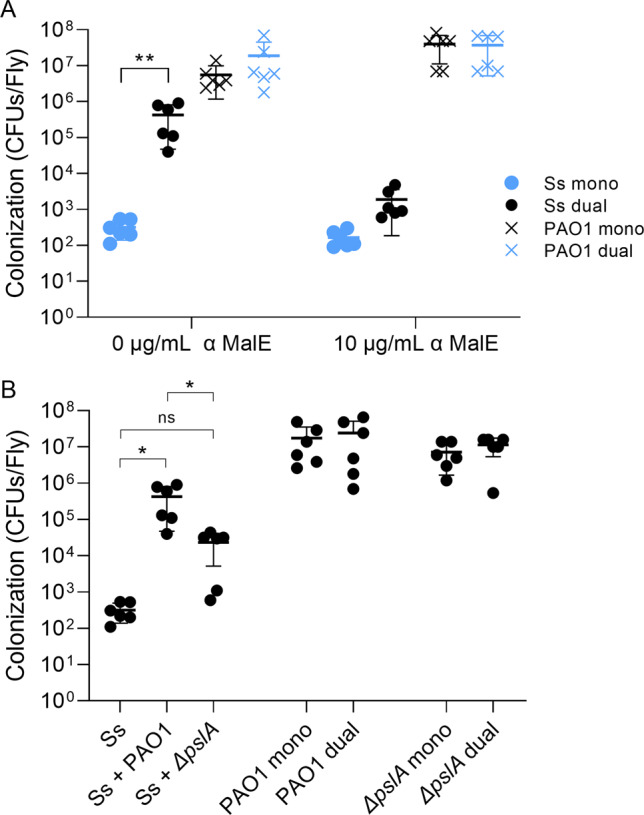
A After antibiotic treatment, Drosophila were infected with subcultures of Ss, PAO1, or both species with or without 10 µg/mL α-MalE mAbs. After 24 h, bacterial colony-forming units were enumerated (n = 6 biological replicates, 10 flies per replicate). Two-way ANOVA with Šίdák’s multiple comparisons test. B Quantification of bacterial CFUs per fly after 24-hour colonization with Ss, PAO1, and PAO1ΔpslA. Error bars indicate mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05, **p < 0.01.
To examine whether the promotion of S. salivarius colonization is also Psl-dependent in an in vivo model of infection, we infected flies with subcultures of PAO1 and PAO1ΔpslA with and without S. salivarius, then quantified CFUs per fly after 24 h. The presence of wild-type PAO1 significantly promoted S. salivarius colonization, while the presence of PAO1ΔpslA marginally, but not significantly, promoted S. salivarius colonization (Fig. 5B). Additionally, S. salivarius did not promote P. aeruginosa colonization. Our results demonstrate that in vivo colonization of S. salivarius is both MalE-dependent and Psl-dependent.
Environmental and clinical non-mucoid isolates of P. aeruginosa enhance dual biofilms with S. salivarius
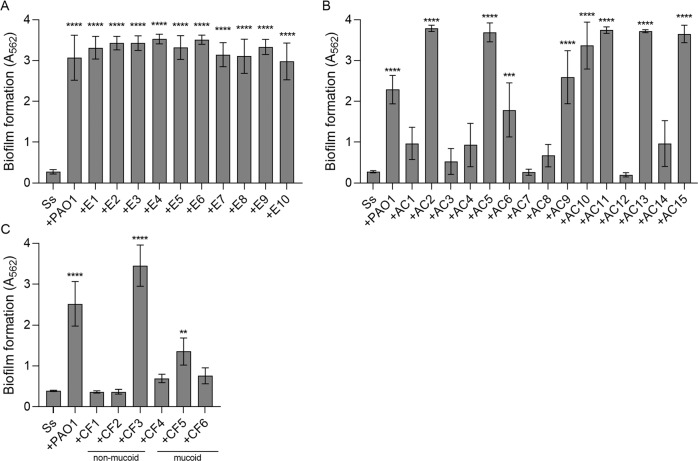
To test whether our enhanced dual biofilm phenotype was consistent with non-lab adapted strains of P. aeruginosa, we isolated ten non-mucoid environmental P. aeruginosa isolates from local water sources in Birmingham, Alabama. Biofilm formation significantly increased to levels similar to that of PAO1 when all ten isolates were co-cultured with S. salivarius (Fig. 6A). S. salivarius was also co-cultured with fifteen non-CF clinical P. aeruginosa isolates. Out of fifteen isolates, nine produced an enhanced dual biofilm in combination with S. salivarius (Fig. 6B). Lastly, three non-mucoid and three mucoid CF clinical P. aeruginosa isolates were co-cultured with S. salivarius (Fig. 6C). One out of the three non-mucoid CF isolates promoted biofilm formation to a greater degree than PAO1, while one out of the three mucoid isolates significantly promoted dual biofilm formation to a lesser extent than PAO1. Overall, these findings indicate that the ability to promote S. salivarius biofilm formation is common among both environmental and clinical isolates of P. aeruginosa.
Fig. 6. Environmental and clinical isolates of P. aeruginosa promote dual biofilm formation.
Ss was co-cultured with A environmental, B non-CF acute, or C CF non-mucoid and mucoid isolates of P. aeruginosa in TSBYE with 1% sucrose in a 96-well 16-h biofilm model. Biofilms were stained with crystal violet to measure biofilm biomass (n = 3 biological, 3 technical). Error bars indicate mean ± SD. One-way ANOVA with Dunnett’s multiple comparisons test (A + B). One-way ANOVA with Holm-Šίdák’s multiple comparisons test (C). **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
Research in recent years has emphasized that a large percentage of bacteria present within any given environment exists in biofilms, rather than a free-floating, planktonic state. Additionally, bacterial species usually exist within polymicrobial communities rather than mono-species infections [32]. However, few studies have examined the role of polysaccharides in the development of mixed-species biofilms. In this paper, we demonstrate a mechanism by which a commensal bacterium utilizes a polysaccharide from a pathogenic species to promote its own biofilm formation. Our results are significant because this mechanism could potentially change the microbial composition of the CF lung by promoting commensal streptococci colonization.
The importance of studying bacterial infections in the context of the CF lung polymicrobial community is becoming increasingly recognized. A seminal study that reported oral commensal streptococci were associated with improved lung function identified S. salivarius as the most abundant oral streptococcus found in the lungs of clinically stable individuals with CF [16]. However, no studies have examined how S. salivarius colonizes the CF lung polymicrobial environment and impacts lung function. In this study, we demonstrate S. salivarius may incorporate itself into polymicrobial biofilms by using a streptococcal sugar-binding protein, MalE, to interact with the P. aeruginosa exopolysaccharide Psl in both in vitro and in vivo fly models of co-infection.
Our findings are significant because interactions between S. salivarius and P. aeruginosa may have an impact on P. aeruginosa virulence and pathogenesis. Although P. aeruginosa viability was not reduced in our in vitro and in vivo models of co-infection, S. salivarius may potentially interfere with P. aeruginosa pathogenesis and impact CF lung function by disrupting Psl availability to P. aeruginosa. Psl is a polysaccharide known for its role in resistance against antibiotic treatment and persistence of lung infections [9, 33, 34]. Our findings demonstrate that S. salivarius not only uses Psl as a biofilm scaffold, but can also metabolize Psl. This sequestering of Psl by S. salivarius could interfere with the ability of P. aeruginosa to use Psl as a mechanism for persistence.
We have previously shown that oral commensal streptococci can use cell surface adhesins to bind to the P. aeruginosa exopolysaccharide alginate to promote its own biofilm formation [20]. MalE has been shown to be anchored to the cell surface of Gram-positive bacteria, therefore allowing it to interact with extracellular molecules [35]. MalE in Group A Streptococcus has a wide range of sugar substrates, which has been implicated in helping Streptococcus species adapt to different host environments and support colonization [36]. Additionally, the maltose-binding protein of Streptococcus mutans along with many other bacterial species has been shown to also bind and transport sucrose in addition to maltose [37]. The Psl structure contains multiple sugars, including galactose, mannose, rhamnose, and glucose [38], which S. salivarius MalE may be utilizing to promote biofilm formation. Our previous findings and current results suggest MalE as a possible candidate protein that facilitates oral commensal biofilm promotion by binding to exopolysaccharides produced by P. aeruginosa.
Numerous studies have demonstrated the potential health benefits of S. salivarius. In combination with Streptococcus oralis, S. salivarius has been shown to inhibit biofilm formation by six pathogens that commonly infect the upper respiratory tract, including Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus pyogenes, Streptococcus pneumoniae, Propionibacterium acnes, and Moraxella catarrhalis [39]. Persons with CF infected with P. aeruginosa experience respiratory exacerbation episodes characterized by a large inflammatory response associated with increased pro-inflammatory cytokines IL-1, IL-6, IL-8, and TNF-α [40]. S. salivarius downregulates the innate immune response to P. aeruginosa in infected human epithelial cells [41] and inhibits both the pro-inflammatory NF- κB pathway in vitro and inflammation in an in vivo colitis mouse model [42]. Additionally, S. salivarius has been shown to downregulate IL-8 production induced by P. aeruginosa in human bronchial epithelial cells [41]. These observations warrant further studying of interactions between S. salivarius and P. aeruginosa to understand how S. salivarius affects the inflammatory host response to P. aeruginosa and how, in turn, this affects CF lung tissue damage.
Because S. salivarius colonizes the upper respiratory tract early in life and P. aeruginosa colonizes individuals with CF more commonly as they age, we would expect S. salivarius to be present in the CF lung during the early stages of infection when non-mucoid P. aeruginosa is acquired from the environment. All ten of our non-mucoid environmental P. aeruginosa isolates were able to produce an enhanced dual biofilm when co-cultured with S. salivarius.. The ability of S. salivarius to create an enhanced biofilm with environmental strains suggests that S. salivarius could be utilizing P. aeruginosa Psl from these non-mucoid strains to colonize the lungs during early P. aeruginosa infection. Further studies are required to understand how S. salivarius colonization impacts early P. aeruginosa infection. While only one of the three non-mucoid CF isolates produced an enhanced dual biofilm, the two other isolates displayed colony morphology differing from that of high Psl-producing strains such as PAO1, suggesting that their biofilm matrix is not primarily comprised of Psl. When S. salivarius was co-cultured with mucoid CF isolates of P. aeruginosa, one isolate produced a significantly enhanced dual biofilm. These results are consistent with previous literature that demonstrates Psl contributes to biofilm formation in some mucoid CF P. aeruginosa isolates [43].
In summary, we report a novel mechanism by which S. salivarius and P. aeruginosa interact within a biofilm and in our in vivo model. Furthermore, we have previously shown that the oral commensal S. parasanguinis interacts with the P. aeruginosa exopolysaccharide alginate to promote streptococcal biofilm formation [20]. Collectively, these studies illustrate potential mechanisms by which oral commensal streptococci interact with P. aeruginosa to alter the composition of the CF airway microbial community. In conclusion, our data suggest a model in which oral streptococci exploit P. aeruginosa exopolysaccharides, resulting in enhanced commensal biofilm development. The novel interactions between S. salivarius and P. aeruginosa revealed in this study could have implications for CF airway microbial community development and warrant further study.
Materials and methods
Bacterial strains, culture conditions, and reagents
Strains S. salivarius K12, P. aeruginosa PAO1, P. aeruginosa PAO1∆pslA, P. aeruginosa PAO1pslA+, P. aeruginosa PA14, P. aeruginosa FRD1, and P. aeruginosa environmental, acute clinical, and CF isolates were used in this study (Table 1). S. salivarius was grown on Todd-Hewitt Broth (THB) agar (Becton Dickinson) and cultured statically at 37 °C in 5% CO2 in THB. P. aeruginosa was grown on Pseudomonas Isolation Agar (PIA; Becton Dickinson) and cultured in Luria broth (LB; Fisher) and incubated while shaking (250 rpm) at 37 °C. PAO1pslA+ and PAO1-GFP were selected for on PIA with 100 μg/mL carbenicillin (Sigma-Aldrich) and were cultured in LB with 100 μg/mL carbenicillin. DH10b (E. coli) was cultured while shaking in LB at 37 °C. After transformation, DH10b was cultured in SOC medium (Fisher) while shaking at 37 °C for an hour.
Table 1.
Bacterial strains and plasmids.
| Strain | Characteristics | Reference/source |
|---|---|---|
| K12 (S. salivarius) | Wildtype | [51] |
| FRD1 (P. aeruginosa) | CF isolate, mucoid | [52] |
| PAO1 (P. aeruginosa) | Wound isolate, non-mucoid | [53] |
| PAO1ΔpslA (P. aeruginosa) | In-frame deletion of pslA | [27] |
| PAO1 pslA+ (P. aeruginosa) | Complemented PAO1ΔpslA with pslA gene | This study |
| PAO1ΔmucA (P. aeruginosa) | Deletion of mucA (PDO300) | [28] |
| PA14 (P. aeruginosa) | Wildtype | [29] |
| PAO1-GFP | PAO1 with carbR GFP plasmid | This study |
| Environmental isolates (P. aeruginosa) | E1-E10 non-mucoid, water isolate | This study |
| AC1 (P. aeruginosa) | Non-mucoid, urine isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC2 (P. aeruginosa) | Non-mucoid, wound isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC3 (P. aeruginosa) | Non-mucoid, urine isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC4 (P. aeruginosa) | Non-mucoid, bronchial wash isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC5 (P. aeruginosa) | Non-mucoid, urine isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC6 (P. aeruginosa) | Non-mucoid, blood isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC7 (P. aeruginosa) | Non-mucoid, bronchoalveolar lavage isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC8 (P. aeruginosa) | Non-mucoid, urine isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC9 (P. aeruginosa) | Non-mucoid |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC10 (P. aeruginosa) | Non-mucoid, tracheal aspiration isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC11 (P. aeruginosa) | Non-mucoid, urine isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC12 (P. aeruginosa) | Non-mucoid, urine isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC13 (P. aeruginosa) | Non-mucoid, urine isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC14 (P. aeruginosa) | Non-mucoid, nasal isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| AC15 (P. aeruginosa) | Non-mucoid, maxillary sinus isolate |
Dr. Bill Benjamin UAB Clinical Microbiology Lab |
| CF clinical isolates (P. aeruginosa) | CF1-CF3 non-mucoid, CF4-CF6 mucoid |
Dr. Susan Birket UAB CF Center |
| DH10b (E. coli) | Host strain for cloning | Thermo Fisher |
| pBKSNS1 | pBluescript K(+) ligated to pslA gene | This study |
Biofilm formation assays
Overnight cultures of S. salivarius and P. aeruginosa were sub-cultured in THB and LB, respectively, and grown to exponential phase (OD600 0.5–0.8). Sub-cultures were then inoculated into Tryptic Soy Broth (MP Biomedicals) with 0.5% yeast extract (Fisher) (TSBYE) containing 1% sucrose at a dilution of 1:1000 for S. salivarius and 1:100 for P. aeruginosa. The two strains were inoculated either separately for the single species biofilm or together for the dual species biofilm assays. 200 μL of each sample was added to a 96-well plate (Nunc) in triplicate and incubated statically at 37 °C in 5% CO2 for 16 h. The biofilms were then stained with 0.1% crystal violet and dissolved in 30% acetic acid [44]. Absorbance was measured at 562 nm to quantify biofilm biomass using the Synergy HTX Multi-Mode Microplate Reader (BioTek).
Quantification of P. aeruginosa and S. salivarius in co-cultures
Cultures were grown in either TSBYE with 1% sucrose, or synthetic cystic fibrosis sputum (SCFM2). SCFM2 was made as previously described [23]. To quantify colony forming units of each species, serial dilutions in TSBYE of planktonic samples from a 6-h six-well biofilm assay were plated on THB agar square plates (Fisher) using the track dilution method [45]. Remaining planktonic cells were aspirated off, and adherent biofilm cells were then washed two times with phosphate-buffered saline (PBS), scraped and resuspended in 3 mL of TSBYE. The resulting suspension was serially diluted and plated.
Construction of the PAO1 pslA+ complemented strain
The pslA gene was cloned by PCR amplifying ~500 bp upstream and downstream of the coding region from the wildtype PAO1 strain using primer sequences described (Table 2). The PCR product was cloned into the EcoRI and BamHI sites of the pBluescript K(+) shuttle vector (Addgene). The resulting plasmid, referred to as pBKSNS1, was converted to a mobilizable plasmid by incorporation of a moriT into the HindIII site [46] and transformed into competent E. coli strain DH10b using a standard transformation method [47]. The pBKSNS1 plasmid was introduced into PAO1ΔpslA through triparental mating [46]. Plasmid conjugation events were selected for using PIA with 100 μg/mL carbenicillin.
Table 2.
Primer sequences.
| Gene | Forward or Reverse | Sequence (5′−3′) | Amplicon size (bp) |
|---|---|---|---|
| pslA | Forward | GGATTGGCGGCGTCAGATTT | 2207 |
| Reverse | TCGATATAGCCGAAGCCGGT | ||
| pslA (qPCR) | Forward | CATGCACCTGGCCGAATA | 109 |
| Reverse | CGGCAGCGAGTTGTAGTT | ||
| 16S rRNA | Forward | GCTGGACTATCGCCGCTG | 150 |
| 16S rRNA | Reverse | ATCTCGTAACCGGTGAAGGTG |
Psl purification
Purification was performed on PAO1pslA+ cultures grown overnight at 37 °C in six-well plates in TSBYE supplemented with 1% sucrose and 100 µg/mL carbenicillin. Cultures were pooled together, diluted 1:1 with 0.9% NaCl, and agitated with 0.01 M EDTA by centrifuging at 200 rpm for 30 min at 4 °C to detach cell-associated Psl. Cultures were then centrifuged at 10,000 g for 15 min at 4 °C to remove bacterial cells. The resulting supernatant was filtered with a 0.22 μm vacuum filter to remove excess cell debris. Exopolysaccharide was then precipitated with 1:1 volume of cold 100% ethanol for 1 h at −80 °C, and resulting precipitate was centrifuged at 15,000 g for 15 min at 4 °C. The pellet was resuspended in PBS containing 1 mM CaCl2 and 10 mM MgCl2 and was subsequently treated with DNase I (100 µg/mL), RNase A (100 µg/mL), and Proteinase K (100 µg/mL) for 2 h at 37 °C, then lyophilized [48].
pslA quantification
Biofilms were cultured in TSBYE supplemented with 1% sucrose in six-well plates for 6 h in 5% CO2. Biofilms were washed with PBS, and adherent bacteria was resuspended. RNA was isolated using the Direct-zol RNA Mini Prep kit (Zymo Research). cDNA conversion was performed with the iScript cDNA Synthesis kit (Bio-Rad), on the CFX96 Real-Time PCR System (Bio-Rad). P. aeruginosa 16S rRNA was used to quantify total RNA present in samples. Primers specific to P. aeruginosa pslA and 16S rRNA are listed in Table 2. The delta-delta CT method was used to calculate fold change of gene expression.
SDS-PAGE and mass spectrometry protein analysis
S. salivarius and P. aeruginosa were cultured planktonically in TSBYE medium at 37 °C in single and dual cultures until OD600 ~1.8 was reached. Cultures were spun down, resuspended in tris-buffered saline, and lysed using the Bead Blaster 24 (Benchmark Scientific). Cell debris was centrifuged, and the resulting supernatant was mixed with 6× Laemmli buffer and ran on an SDS Page gel. Overexpressed bands were excised from the gels and digested with trypsin. The digested peptide fragments were analyzed for protein identification by LC-MS/MS as described previously [49] and sent to the UAB Mass Spectrometry Core for identification. We repeated the SDS-PAGE experiment three times, the only reproducible change was this ~50 kDa band. We also excised the bands from three biologic replicates and analyzed by mass spectrometry.
Immunofluorescence and confocal laser scanning microscopy
Bacterial strains were grown in either TSBYE supplemented with 1% sucrose or in SCFM2 in a sterile eight-well treated μSlide (Ibidi). Samples were incubated at 37 °C under 5% CO2 for 16 h. Biofilm samples were gently washed twice with PBS to eliminate planktonic bacteria and then incubated in PBS with 1% bovine serum albumin (BSA) for 10 min. Samples were then stained for 10 min with hexidium iodide (Thermo Fisher) at a 1:1000 dilution and a FITC-conjugated α-Psl antibody (Creative Biolabs) at a 1:100 dilution in PBS with 1% BSA. The samples were washed with PBS once more and then analyzed for fluorescence using the Nikon A1R Confocal Laser Scanning Microscope (Nikon Instruments Inc.) at the University of Alabama at Birmingham High Resolution Imaging Facility.
Drosophila melanogaster colonization assay
Drosophila melanogaster flies were maintained on Jazz-mix Drosophila food (Fisher). Three to seven-day-old flies were treated with antibiotics for 3 days (50 µg/mL vancomycin, 50 µg/mL erythromycin, and 50 µg/mL ampicillin). Flies were separated into vials in groups of ten and subsequently starved for 3 h prior to infection. S. salivarius and P. aeruginosa cultures were grown to A600 2.0. For single species infection groups, 1.5 mL of the respective culture was centrifuged for 6 min at 6000 g and resuspended in 100 µl of 5% sucrose. For dual infection groups, 0.75 mL of each culture were combined, centrifuged, and resuspended in 5% sucrose. Resuspended cultures were then pipetted onto sterile 21 mm filter disks (Whatman) and placed into plastic vials containing 5 mL of 5% sucrose agar. After 24 h of infection, flies were briefly washed in 70% ethanol to remove outside contaminants and then washed with sterile PBS. Flies were crushed and resuspended in 500 µl PBS. The resulting homogenate was then serially diluted and plated on THB agar plates using the track dilution method to quantify bacterial colony-forming units inside flies.
Statistical analysis
All graphs represent sample means ± SD. The Shapiro-Wilk normality test was used to determine distribution of datasets. Statistical analysis of normally distributed data was performed using either Student’s t test or one-way ANOVA with Tukey’s multiple comparisons test. For experiments with two or more factors (for example, absorbance and treatment), a two-way ANOVA was used with either Tukey’s or Šίdák’s multiple comparisons test. Tests were performed using Graphpad Prism version 9 for Windows, La Jolla California USA, www.graphpad.com. Data were considered statistically significant if p < 0.05.
Supplementary information
Acknowledgements
This work was supported by grants awarded to JAS from the National Institute of Dental and Craniofacial Research (R00-DE025913), the National Institute of General Medical Sciences (R35GM142748), the UAB Microbiome Center, and start-up funds from the UAB Department of Microbiology. SNS was supported by the Alabama Louis Stokes for Minority for Participation fellowship funded by the National Science Foundation (1806130), the National Heart, Lung, and Blood Institute T32 UAB pre-doctoral training program in lung diseases (T32HL134640-03), and is now supported by the National Heart, Lung, and Blood Institute NRSA Fellowship (F31HL162487-01). The authors would like to thank Shawn Williams at the UAB High Resolution Imaging Facility for his assistance with the Nikon A1 Confocal microscope and imaging analysis. We thank Landon Wilson at the UAB Targeted Metabolomics and Proteomics Laboratory for his assistance in performing Mass Spectrometry and subsequent data analysis. We thank Michael Crowley at the UAB Heflin Center for Genomic Sciences and William Van Der Pol and Elliot Lefkowitz at the UAB Center for Clinical and Translational Science for their help with RNA sequencing analysis. This publication was made possible by the UAB Center for Clinical and Translational Science Grant Number UL1TR001417 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH). We also thank Dr. Bill Benjamin at the UAB Clinical Microbiology Laboratory for the acute clinical P. aeruginosa isolates and Dr. Susan Birket at the UAB Cystic Fibrosis Center for the CF clinical P. aeruginosa isolates. Lastly, we thank Natalie Lindgren at UAB for providing the water samples from which we isolated our environmental P. aeruginosa strains.
Author contributions
SNS and JAS designed the study. SNS, JAS, and JJB performed experiments and conducted subsequent data analysis. SNS and JAS wrote the manuscript, and all authors reviewed and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01221-y.
References
- 1.Davidson DJ, Porteous DJ. The genetics of cystic fibrosis lung disease. Thorax. 1998;53:389–97. doi: 10.1136/thx.53.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray TS, Egan M, Kazmierczak BI. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr Opin Pediatr. 2007;19:83–8. doi: 10.1097/MOP.0b013e3280123a5d. [DOI] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis Foundation Patient Registry. Bethesda, MD: Cystic Fibrosis Foundation 2020.
- 4.Sousa AM, Pereira MO. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens. 2014;3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–90. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin M, Nivens D, Weadge J, Howell P. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2. [DOI] [PMC free article] [PubMed]
- 7.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, et al. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goltermann L, Tolker-Nielsen T. Importance of the exopolysaccharide matrix in antimicrobial tolerance of Pseudomonas aeruginosa aggregates. Antimicrob Agents Chemother. 2017;61:e02696–16. doi: 10.1128/AAC.02696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billings N, Ramirez Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, et al. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLOS Pathog. 2013;9:e1003526. doi: 10.1371/journal.ppat.1003526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Periasamy S, Nair HAS, Lee KWK, Ong J, Goh JQJ, Kjelleberg S, et al. Pseudomonas aeruginosa PAO1 exopolysaccharides are important for mixed species biofilm community development and stress tolerance. Front Microbiol. 2015;6:851. doi: 10.3389/fmicb.2015.00851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibley CD, Parkins MD, Rabin HR, Duan K, Norgaard JC, Surette MG. A polymicrobial perspective of pulmonary infections exposes an enigmatic pathogen in cystic fibrosis patients. Proc Natl Acad Sci USA. 2008;105:15070–5. doi: 10.1073/pnas.0804326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surette MG. The cystic fibrosis lung microbiome. Ann Am Thorac Soc. 2014;11:S61–5. doi: 10.1513/AnnalsATS.201306-159MG. [DOI] [PubMed] [Google Scholar]
- 13.Gao B, Gallagher T, Zhang Y, Elbadawi-Sidhu M, Lai Z, Fiehn O, et al. Tracking polymicrobial metabolism in cystic fibrosis airways: Pseudomonas aeruginosa metabolism and physiology are influenced by Rothia mucilaginosa-derived metabolites. mSphere. 2018;3:e00151–18. doi: 10.1128/mSphere.00151-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armbruster CR, Wolter DJ, Mishra M, Hayden HS, Radey MC, Merrihew G, et al. Staphylococcus aureus protein A mediates interspecies interactions at the cell surface of Pseudomonas aeruginosa. mBio. 2016;7. [DOI] [PMC free article] [PubMed]
- 15.McDaniel MS, Schoeb T, Swords WE. Cooperativity between Stenotrophomonas maltophilia and Pseudomonas aeruginosa during polymicrobial airway infections. Infect Immun. 2020;88. [DOI] [PMC free article] [PubMed]
- 16.Filkins LM, Hampton TH, Gifford AH, Gross MJ, Hogan DA, Sogin ML, et al. Prevalence of streptococci and increased polymicrobial diversity associated with cystic fibrosis patient stability. J Bacteriol. 2012;194:4709–17. doi: 10.1128/JB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Almeida OGG, Capizzani CPdC, Tonani L, Grizante Barião PH, da Cunha AF, De Martinis ECP, et al. The lung microbiome of three young Brazilian patients with cystic fibrosis colonized by fungi. Front Cell Infect Microbiol. 2020;10. [DOI] [PMC free article] [PubMed]
- 18.Maeda Y, Elborn JS, Parkins MD, Reihill J, Goldsmith CE, Coulter WA, et al. Population structure and characterization of viridans group streptococci (VGS) including Streptococcus pneumoniae isolated from adult patients with cystic fibrosis (CF) J Cyst Fibros. 2011;10:133–9. doi: 10.1016/j.jcf.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Hogan DA, Willger SD, Dolben EL, Hampton TH, Stanton BA, Morrison HG, et al. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLOS ONE. 2016;11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scoffield JA, Duan D, Zhu F, Wu H. A commensal streptococcus hijacks a Pseudomonas aeruginosa exopolysaccharide to promote biofilm formation. PLOS Pathog. 2017;13:e1006300. doi: 10.1371/journal.ppat.1006300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kononen E, Jousimies-Somer H, Bryk A, Kilp T, Kilian M. Establishment of streptococci in the upper respiratory tract: longitudinal changes in the mouth and nasopharynx up to 2 years of age. J Med Microbiol. 2002;51:723–30. doi: 10.1099/0022-1317-51-9-723. [DOI] [PubMed] [Google Scholar]
- 22.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018;24:133–45.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci USA. 2015;112:4110–5. doi: 10.1073/pnas.1419677112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulos L, Prévost M, Barbeau B, Coallier J, Desjardins R. LIVE/DEAD BacLight: application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water. J Microbiol Methods. 1999;37:77–86. doi: 10.1016/S0167-7012(99)00048-2. [DOI] [PubMed] [Google Scholar]
- 25.Okshevsky M, Meyer RL. Evaluation of fluorescent stains for visualizing extracellular DNA in biofilms. J Microbiol Methods. 2014;105:102–4. doi: 10.1016/j.mimet.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol. 2015;41:341–52. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 27.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–38. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathee K, Ciofu O, Sternberg C, Lindum PW, Campbell JIA, Jensen P, et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–57. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 29.Rahme Laurence G, Stevens Emily J, Wolfort Sean F, Shao J, Tompkins Ronald G, Ausubel, et al. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 30.Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, et al. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ Microbiol. 2012;14:1995–2005. doi: 10.1111/j.1462-2920.2012.02753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–75. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabrilska RA, Rumbaugh KP. Biofilm models of polymicrobial infection. Future Microbiol. 2015;10:1997–2015. doi: 10.2217/fmb.15.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami K, Ono T, Viducic D, Somiya Y, Kariyama R, Hori K, et al. Role of psl genes in antibiotic tolerance of adherent Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017;61:e02587–16. doi: 10.1128/AAC.02587-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris AJ, Jackson L, Cw Yau Y, Reichhardt C, Beaudoin T, Uwumarenogie S, et al. The role of Psl in the failure to eradicate Pseudomonas aeruginosa biofilms in children with cystic fibrosis. NPJ Biofilms Microbiomes. 2021;7:63. doi: 10.1038/s41522-021-00234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sutcliffe IC, Harrington DJ. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology. 2002;148:2065–77. doi: 10.1099/00221287-148-7-2065. [DOI] [PubMed] [Google Scholar]
- 36.Shelburne SA, 3rd, Fang H, Okorafor N, Sumby P, Sitkiewicz I, Keith D, et al. MalE of group A Streptococcus participates in the rapid transport of maltotriose and longer maltodextrins. J Bacteriol. 2007;189:2610–7. doi: 10.1128/JB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilic AO, Honeyman AL, Tao L. Overlapping substrate specificity for sucrose and maltose of two binding protein-dependent sugar uptake systems in Streptococcus mutans. FEMS Microbiol Lett. 2007;266:218–23. doi: 10.1111/j.1574-6968.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 38.Ma L, Lu H, Sprinkle A, Parsek Matthew R, Wozniak, Daniel J. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J Bacteriol. 2007;189:8353–6. doi: 10.1128/JB.00620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bidossi A, De Grandi R, Toscano M, Bottagisio M, De Vecchi E, Gelardi M, et al. Probiotics Streptococcus salivarius 24SMB and Streptococcus oralis 89a interfere with biofilm formation of pathogens of the upper respiratory tract. BMC Infect Dis. 2018;18:653. doi: 10.1186/s12879-018-3576-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, Ghnaim H, et al. Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit. 1995;152:2111–8. doi: 10.1164/ajrccm.152.6.8520783. [DOI] [PubMed] [Google Scholar]
- 41.Cosseau C, Devine DA, Dullaghan E, Gardy JL, Chikatamarla A, Gellatly S, et al. The commensal Streptococcus salivarius K12 downregulates the innate immune responses of human epithelial cells and promotes host-microbe homeostasis. Infect Immun. 2008;76:4163–75. doi: 10.1128/IAI.00188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaci G, Goudercourt D, Dennin V, Pot B, Doré J, Ehrlich SD, et al. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol. 2014;80:928–34. doi: 10.1128/AEM.03133-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma L, Wang S, Wang D, Parsek MR, Wozniak DJ. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2012;65:377–80. doi: 10.1111/j.1574-695X.2012.00934.x. [DOI] [PubMed] [Google Scholar]
- 44.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jett BD, Hatter KL, Huycke MM, Gilmore MS. Simplified agar plate method for quantifying viable bacteria. BioTechniques. 1997;23:648–50. doi: 10.2144/97234bm22. [DOI] [PubMed] [Google Scholar]
- 46.Suh SJ, Silo-Suh LA, Ohman DE. Development of tools for the genetic manipulation of Pseudomonas aeruginosa. J Microbiol Methods. 2004;58:203–12. doi: 10.1016/j.mimet.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 47.Seidman CE, Struhl K, Sheen J, Jessen T. Introduction of plasmid DNA into cells. Curr Protoc Mol Biol. 1997;37:1.8.1-.8.10. [DOI] [PubMed]
- 48.Pedersen SS, Espersen F, Høiby N, Shand GH. Purification, characterization, and immunological cross-reactivity of alginates produced by mucoid Pseudomonas aeruginosa from patients with cystic fibrosis. J Clin Microbiol. 1989;27:691–9. doi: 10.1128/jcm.27.4.691-699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bu S, Li Y, Zhou M, Azadin P, Zeng M, Fives-Taylor P, et al. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 2008;190:1256–66. doi: 10.1128/JB.01078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan BK, Sistrom M, Wertz JE, Kortright KE, Narayan D, Turner PE. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep. 2016;6:26717. doi: 10.1038/srep26717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tagg JR. Prevention of streptococcal pharyngitis by anti-Streptococcus pyogenes bacteriocin-like inhibitory substances (BLIS) produced by Streptococcus salivarius. Indian J Med Res. 2004;119:13–6. [PubMed] [Google Scholar]
- 52.Ohman DE, Chakrabarty AM. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–8. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holloway BW, Krishnapillai V, Morgan AF. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.