Summary
The origin, composition, distribution, and function of cells in the human intervertebral disc (IVD) have not been fully understood. Here, cell atlases of both human neonatal and adult IVDs have been generated and further assessed by gene ontology pathway enrichment, pseudo-time trajectory, histology, and immunofluorescence. Comparison of cell atlases revealed the presence of two subpopulations of notochordal cells (NCs) and their associated markers in both the neonatal and adult IVDs. Developmental trajectories predicted 7 different cell states that describe the developmental process from neonatal to adult cells in IVD and analyzed the NC’s role in the IVD development. A high heterogeneity and gradual transition of annulus fibrosus cells (AFCs) in the neonatal IVD was detected and their potential relevance in IVD development assessed. Collectively, comparing single-cell atlases between neonatal and adult IVDs delineates the landscape of IVD cell biology and may help discover novel therapeutic targets for IVD degeneration.
Subject areas: complex system biology, developmental biology, transcriptomics
Graphical abstract

Highlights
-
•
Compared scRNA-seq between human neonatal and adult IVD
-
•
Identified two notochordal cell populations in adults and their novel markers
-
•
Notochordal cells preserved their identity and functions into adulthood
-
•
Unveiled heterogeneity of nucleus pulposus and annulus fibrosus cells in human IVD
Complex system biology; Developmental biology; Transcriptomics
Introduction
At least 30% of total adult population is suffering from lower back pain, which originates in intervertebral disc (IVD) degeneration (Macfarlane et al., 1999; Andersson, 1999; Frank et al., 1996; de Schepper et al., 2010) To date, most treatments of IVD degeneration are limited to invasive surgical interventions, such as disc replacement and spinal fusion, or pain management that is not addressing the underlying cause of IVD degeneration (Knezevic et al., 2017).
The IVD is a shock-absorbing structure that connects two adjacent vertebrae and enables spine movement (Vergroesen et al., 2015; Buckley et al., 2018). The human IVD cell composition is highly heterogeneous; nucleus pulposus (NP) is the inner core and the annulus fibrosus (AF) is the outer region that is confined by two endplates sandwiching the disc (Sun et al., 2020). It is widely accepted that the embryonic notochord develops into the NP and that notochordal cells (NCs) disappear within the first decade of life (Séguin et al., 2018). However, the NC remnants or dormant NC can be found in the adult human IVD (McCann and Séguin, 2016; Wang et al., 2008). Additional study showed that the NCs or NC-like NP cells (NPCs) in humans persist throughout life (Risbud and Shapiro, 2011). The eventual fate of human NCs is not conclusive to date, but understanding its development has considerable potential to benefit cell therapies of IVD degeneration. Animals that keep the NCs in adulthood like cats and pigs do not exhibit disc degeneration (Hunter et al., 2003; Sheyn et al., 2019). NCs hold great potential to rejuvenate a degenerated human IVD and this potential has already been demonstrated in a mini pig model (Sheyn et al., 2019).
Recent advances in single-cell RNA sequencing (scRNA-seq) allow for creation of cell atlases that unveil rare subpopulations, delineate cellular heterogeneity, identify new markers, and predict developmental trajectories (Mathys et al., 2019; Stark et al., 2019; Kelly et al., 2020; Setty et al., 2019; Ho et al., 2021). Unraveling the single-cell atlas and transcriptomes of the human IVD at different ages will largely extend our understanding of its cell biology and development. To date, its heterogeneity, particularly in the early development, has not been sufficiently shown because of the limitation of traditional bulk-sequencing (Fernandes et al., 2020). Previously, multiple bulk proteomics studies delineated the general cell heterogeneity of human IVD (Rodrigues-Pinto et al., 2018; Tam et al., 2020b) and made comparisons at different developmental stages (Tam et al., 2020b). Recent studies of single-cell atlases in nonhuman vertebrates resolved the cellular heterogeneity in bovine caudal IVD (Panebianco et al., 2021) and found stem cells in rat IVD (Wang et al., 2021). Another study provided single-cell analysis of human adult IVD that identified the NPC and AFC populations (Fernandes et al., 2020). A recent study identified chondrocytes as dominating cell population in human IVD and found small NC subpopulations (Gan et al., 2021). Yet, no study has investigated cell atlases from neonatal human IVD and thus elaborated the role of NCs in development and hemostasis at a single-cell resolution.
Here, we analyze a human IVD single-cell atlas with direct head-to-head comparison between neonatal and adult human subjects to reveal the identities of all IVD populations and subpopulations of both developmental stages, discover rare cell populations and novel markers thereof, and predict the developmental trajectories from neonatal to adult IVD. We isolated cells from neonatal and adult IVDs postmortem, and analyzed them with scRNA-seq. An in-depth gene ontology term enrichment and pathway analysis was performed and developmental trajectories from neonatal to adult IVD were predicted. We further validated our findings with histology and immunofluorescence.
Results
Human neonatal and adult IVD cells are comprised of 15 distinct subpopulations
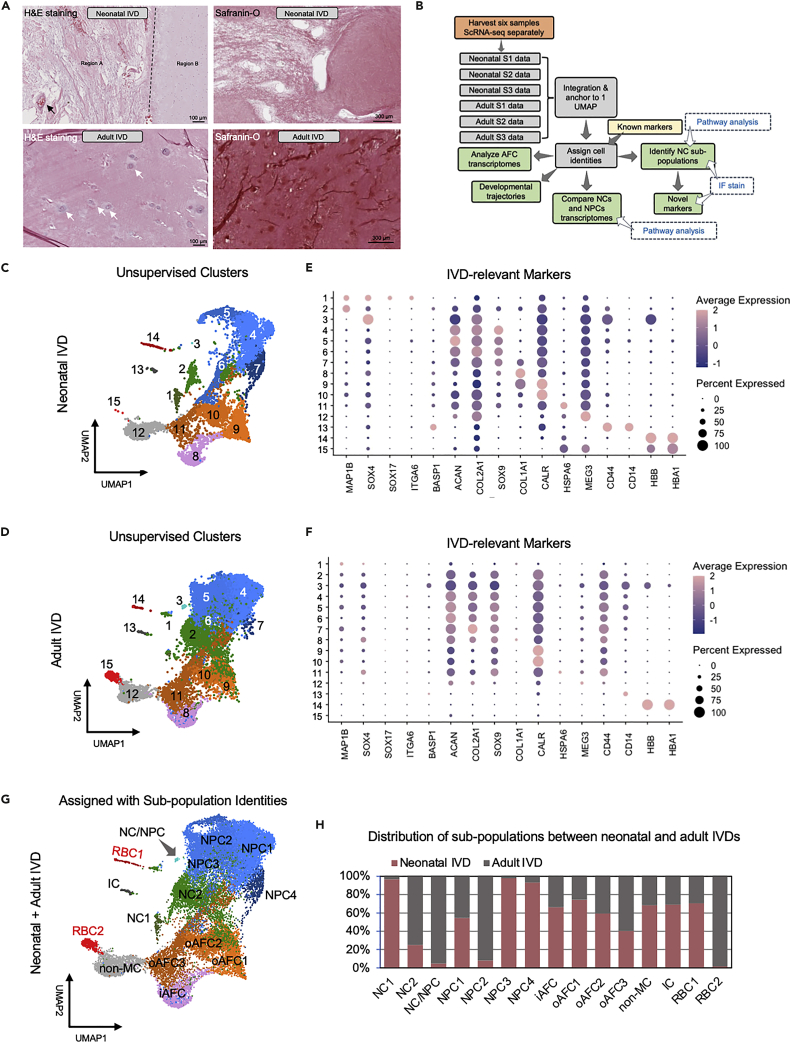
Histological analysis of neonatal IVD tissue demonstrated two distinct tissue regions. Region A contained aggregated cells and loose extracellular matrix (ECM) structures and region B contained sparsely distributed cells and dense ECM tissue (Figure 1A). Vessels and red blood cells were clearly identified in region A (Figure 1A). In contrast, the adult IVD did not exhibit different regions. The uniform and homogeneous tissue contained sparsely distributed cells whose cytoplasm was greatly larger and cell density lower than in neonatal IVD (Figure 1A).
Figure 1.
Sorting of Cells from Human Neonatal and Adult IVD of Distinct Tissue Morphologies into 15 Unsupervised Clusters Based on Their Expression of IVD Markers
(A) H&E and Safranin-O stains of neonatal and adult IVDs. The black dashed line shows the boundary separating two distinct regions. Black arrow points to a blood vessel. The white arrows point to NPCs.
(B) Illustrative scheme shows the workflow and outcome measures in this study.
(C and D) UMAP shows 15 unsupervised cell clusters that were identified in both neonatal (C) and adult (D) samples.
(E and F) Dot plots showing the expression level of classical IVD markers in each cluster of neonatal (E) and adult (F) IVD. Dot size indicates percent of cells expressing the genes and dot color intensity the average expression level (normalized to −2 to 2).
(G) Combined UMAP for all cells from neonatal and adult IVDs with clusters assigned to subpopulations based on their marker expressions in (C and D).
(H) Cell distribution between two ages (neonatal and adult) for each subpopulation. Numbers were normalized to total cell counts for each age.
Following workflow as shown in Figure 1B, the dimensional reduction based on the unsupervised UMAP method sorted all cells into 15 clusters that could be classified as 15 cell subpopulations residing in both neonatal (Figure 1C) and adult IVDs (Figure 1D). The expression levels of classical IVD markers in each cluster were shown as dot plot for neonatal IVD (Figure 1C) and adult IVD (Figure 1D). The selection of classical IVD-relevant markers includes NC markers MAP1B (Rodrigues-Pinto et al., 2018), SOX4 (Bhattaram et al., 2010), SOX17 (D'Amour et al., 2005; Sheyn et al., 2019), ITGA6 and BASP1 (Sheyn et al., 2019), NPC markers ACAN (Tam et al., 2020a; Fernandes et al., 2020; Risbud et al., 2015), COL2A1 (Tam et al., 2020a; Fernandes et al., 2020; Risbud et al., 2015) and SOX9 (Sheyn et al., 2019), AFC markers COL1A1 (van den Akker et al., 2017; Tam et al., 2020b), CALR (Tam et al., 2020b) and HSPA6 (Takao and Iwaki, 2002), as well as other previously shown markers like MEG3 for non-mesenchymal cells (Chen et al., 2017), CD44 (Schumann et al., 2015) and CD14 (Ziegler-Heitbrock and Ulevitch, 1993) for monocytes, and HBB (Talamo et al., 2003) and HBA1 (Pandey and Rizvi, 2011) for red blood cells (RBC, Figures 1E and 1F). Based on these markers we annotated each cluster (Figure 1G). The distribution of cells from neonatal vs. adult IVD in each annotated subpopulation, normalized by total cells of their respective age, shows that neonatal IVD disproportionally contributed to NC1, NPC3-4, (inner) iAFC, and (outer) oAFC1 clusters (Figure 1H). Adult IVD disproportionally contributed to the NC2, NC/NPC, NPC2, and RBC2 clusters. The contribution to rest of the subpopulations was more or less evenly distributed.
Human neonatal and adult intervertebral disc contain two distinct notochordal cell subpopulations
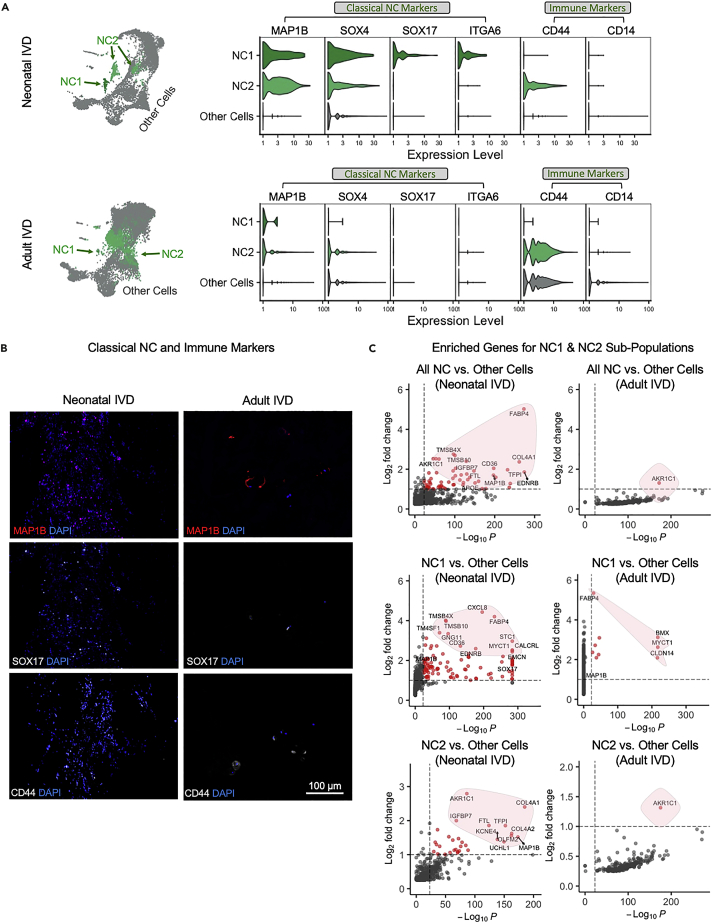
Cluster 1 and 2 in the neonatal IVD were identified as NC subpopulations (Figures 1C–1H) based on their expression of NC markers, MAP1B (Rodrigues-Pinto et al., 2018) and SOX4 (Bhattaram et al., 2010), and have been assigned to NC1 and NC2 (Figure 2A). NC1 and NC2 were found to be distinct from each other: NC1 expressed SOX17 and ITGA6, whereas NC2 expressed CD44 (Figure 2A). The NC subpopulations were also found in adult IVD (Figure 2A). NC2 showed overwhelmingly higher cell counts than NC1 in adults (Figures 1H and 2A). Immunostaining of the NC markers, MAP1B and SOX17, confirmed their expression in both neonatal and adult IVDs (Figure 2B) on the protein level. These proteins showed a higher overall expression in neonatal than in adult IVD. We also observed the NC2 expressed immune-relevant marker CD44 (Figure 2A) and confirmed the presence of CD44 proteins in both ages (Figure 2B).
Figure 2.
Identification of NC Subpopulations and Their Pathway Network Analysis
(A) The two NC subpopulations in UMAP and their expression of classical markers in both neonatal and adult IVD; two MAP1B+/SOX4+ NC subpopulations in the UMAP are colored green, and all other cells are colored gray; violin plots show NC expression levels of classical markers of NCs and immune cells for human neonatal and adult IVD. Other cells refer to the expression level for all cells in all clusters other than NC1 and NC2.
(B) The immunostaining of classical NC and immune markers in neonatal and adult IVD are shown. Scale bar = 100 μm; original magnification = 20x.
(C) Volcano plots showing the enriched genes for the entire populations of NC, subpopulations of NC1 or NC2, in neonatal or adult IVD. The y axis in the volcano plots shows the -log10p where the p value was about an enriched gene expression level in a specific subpopulation in a specific age (neonatal or adult) against all other cells the same age. The x-axis shows the log2FC (FC - fold change) of the expression level. The cut-off threshold was set to FC > 2 and p < 1×10−24. The enriched genes meeting the cut-off threshold are colored with red. The genes not meeting the cut-off threshold are colored with gray.
Pathway networks were further created based on the list of all enriched genes for NC1 and NC2 (Figures S1A and S1B), that is, 860 genes for NC1 and 583 genes for NC2. Our results show NC1 preferentially demonstrated the activities of cancer-related pathway STAT1 and the deactivation of Brachyury (TBXT), Microphthalmia-associated Transcription Factor (MITF), and SWI/SNF-related, Matrix-Associated, Actin-Dependent Regulator of Chromatin (SMARCD3), which is further associated with the deactivation of connective tissue cell pathways and the collagen-relevant Glycoprotein VI (GP6) signaling pathway, and the deactivation of connective tissue activities (Figure S2B). MITF was previously shown to be associated with osteoclast activities (Lu et al., 2010) and GP6 was shown to induce collagen deposition (Stegner et al., 2014). NC2 preferentially demonstrated stem cell pluripotency factor FGF2 (Figure S2A), which led to the organization of cytoplasm and organization of cytoskeleton.
AKR1C1, APOE, and FABP4 are novel markers for notochordal cells in human intervertebral disc
We selected several NC markers from top enriched genes that are shown in Figure 2C. Specifically, FABP4 is a strong marker for NC1 in both neonatal and adult IVD, AKR1C1 is the NC2 marker for neonatal IVD, and APOE is a relatively weak marker but can be identified in the entire NC population (Figure 2C). Interestingly, AKR1C1 is the only marker common to the entire NC population in adult IVD (Figure 2C). We compared the expression levels of FABP4, AKR1C1, and APOE in neonatal and adult IVDs due to their possible age-dependent specificity (Figure 3A). FABP4 is only specific to neonatal NC1. AKR1C1 lost certain specificity in adult IVD (Figure 3A) but is still specific to the entire NC population and NC2 (Figure 2C). The immunostaining of novel NC markers, AKR1C, FABP4, and APOE, confirmed their expression in both neonatal and adult IVDs. All markers show overlap with the cell nucleus besides AKR1C that appears in the cytoplasm.
Figure 3.
Discovery of Novel NC Markers, FABP4, AKR1C1, APOE, and their pathway/network analysis
(A) Comparison of the expression level of novel top markers in human neonatal versus adult IVD.
(B) Immunostaining of novel NC markers in neonatal and adult IVD are shown. The scale bar = 100 μm and original magnification = 20x. Qiagen IPA software was used to plot the (C) VEGF-regulated network involving FABP4 marker in NC1 subpopulation in neonatal IVD and P38-MAPK-regulated network involving FABP4 marker in NC1 subpopulation in adult IVD.
(D) TGFB1-regulated network involving AKR1C1 marker in NC2 subpopulation in neonatal IVD.
(E) Top biological functions activated by AKR1C1/AKR1C2 and ranked by the activation score in NC1 subpopulation in neonatal IVD.
We further investigated the role of the new markers in the regulatory networks and biofunctions. We found that FABP4 plays a critical role in the VEGF-regulated network in neonatal IVD, in which the upregulation of VEGF leads to the activation of NC markers FABP4 and ITGA6, inflammatory factor CXCL8, and downregulate ACAN (Figure 3C). In adult IVD, however, FABP4 is activated by the P38 MAPK regulator (Figure 3C). AKR1C1 and AKR1C2 play a critical role in the TGFB1 cascade in the neonatal NC2 subpopulation (Figure 3D). This network seems to be highly relevant to NCs, as it also leads to the activation of classical NC markers MAP1B and immune-relevant CD44 (Figure 3D). The activation of AKR1C1 is associated with multiple biological functions as shown in Figure 3E. Specifically, the upregulation of AKR1C1/AKR1C2 activates multiple tumor and cancer-related functions (Figure 3E). Lastly, we noticed the novel APOE marker was in the IL1 cascade, which also regulates AKR1C1/AKR1C2. (Figure S1C). We also report the potential role of these markers in regulatory networks and their association with important biological functions in Figure S3. Specifically, FABP4 expressed by NC1 in both neonatal and adult may be a downstream effect of VEGFA factor and KDR phosphatase. In neonatal NC1 only, APOE is upregulated by APP, a gene that inhibits the development of cytoplasm and formation of cytoskeleton biofunctions. AKR1C1 is upregulated by NPM1 in the NC2.
Nucleus pulposus cell populations contain four similar subpopulations with different biological functions in neonatal versus adult intervertebral disc
NPCs expressing their classical markers, ACAN (Tam et al., 2020a; Fernandes et al., 2020; Risbud et al., 2015), COL2A1 (Tam et al., 2020a; Fernandes et al., 2020; Risbud et al., 2015), and SOX9 (Sheyn et al., 2019) were detected in both neonatal IVD (Figure 4A) and adult IVD (Figure 4B). The entire NPC population contained four different subpopulations identified by their specific markers: NPC1 identified by OGN, NPC2 identified by MMP13, NPC3 identified by COL9A1, and NPC4 identified by DPT. The clusters 4–7 (Figures 1C and 1D) thus have been designated as NPC1-4, respectively. Cluster 3 (Figures 1C and 1D) partly expresses NC markers (SOX4+ but no MAP1B) and partly expresses NPC markers (COL2A1+ but no ACAN and SOX9), and thus it has been designated as NC/NPC. It may represent a transition state of cells from NC to NPC or a reservoir of NP progenitors. The expression of the NPC markers is shown in violin plots (Figures 4A and 4B). However, four NPC subpopulations from the same age are barely distinguishable (Figures 4A and 4B). The subpopulation markers are not very specific as the gene expression distributed on the UMAP did not always match the clustering on the UMAP (Figures 4A and 4B). The four NPC subpopulations may have similar transcriptomic profiles. The subtle difference among the four NPC subpopulations may be shown by their different enriched functions (see supplemental materials Figure S4). The AFC marker levels were used as controls (Figures 4A and 4B), and their expression was found very low among all NPCs in both the neonatal and adult IVD, which confirmed the NPC clusters are not overlapping with AFCs. Only NPC4 in neonatal IVD expressed AFC marker COL1A1 (Figure 4A).
Figure 4.
Identification of 4 similar NPC subpopulations and comparison of their NPC gene expression, pathways, and biofunctions with NC populations in neonatal and adult IVD
(A and B) Violin plots showing differences in the expression levels of NPC markers in four NPC subpopulations (NPC1-4) identified among the ACAN+/COL2A1+/SOX9 + NPC populations in UMAP (colored in blue) in both neonatal (A) and adult IVD (B). AFC markers were shown for comparison. Expression levels of each NPC subtype marker were projected via UMAP.
(C) Immunostaining of extracellular matrix (ECM) proteins in neonatal and adult IVD is shown. Scale bar = 100 μm; Original magnification = 20x.
(D) Heatmap shows biological functions enriched from all DEGs of NCs or NPCs in neonatal or adult IVD. Orange indicates activation, deactivation, and gray data that were not detected or did not pass the filtering.
(E) Circos plot showing the overlap between four different enriched gene lists: neonatal NCs, adult NCs, neonatal NPCs, and adult NPCs. Each purple line links the same gene expressed in both enriched gene lists. Red and orange show the shared and unique genes.
(F) Descriptive summary terms on the biofunctions enriched from each list.
NPCs are critical to the hemostasis and deposition of ECM (Frapin et al., 2019). We observed that the age greatly affected the critical ECM proteins (Figure 4C). Figure 4C shows the abundant presence of collagen type 1 (COL1A1) in the extracellular space and aggrecan in the cytoplasm in the neonatal IVD. The signal of intracellular COL1A1 appears to be weak in neonatal IVD (Figure 4C). Figure 4C shows the opposite trend for adult IVD: the staining of COL1A1 in the extracellular space of neonatal IVD is negligible, but the staining in the cytoplasm is strong; the staining of aggrecan in the extracellular space is strong, but weak in the cytoplasm (Figure 4C).
Notochordal cell and nucleus pulposus cell populations exhibit different gene ontology enrichments, pathways, and regulators
NC and NPC populations were compared in both neonatal and adult IVDs (Figures 4D–4F). In this analysis, NC1-2 were combined as NCs, and NPC1-4 were combined as NPCs (Figure 4D). NC populations were preferentially enriched to cancer and tumor activities, cell metabolism, cellular homeostasis, angiogenesis, and cell adhesion and spreading, whereas NPC populations were preferentially enriched to cytoskeletal tissue formation, the development of cytoplasm, wounding healing, and ECM development (Figure 4D). We also showed top-scored key regulatory networks in neonatal NCs and adult NPCs in Figure S1D. Several inflammatory and tumorigenic factors and pathways (IL1B, VEGF, and HGF) were upregulated in NC (Figure S1D). In NPCs, upregulation of the osteogenic factor BMP2 (Cai et al., 2021), osteoclast regulator MITF (Lu et al., 2010), DNA-binding transcription factor-related BCL6, NPC and skeletal development marker SOX9 (Sheyn et al., 2019; Bi et al., 1999; Zhou et al., 2006) were detected (Figure S1D). This trend seems to be age-independent except for BCL6, which was not detected in the adult IVD (Figure S1D). Different cell types (NC vs. NPC) demonstrated hugely distinct transcriptomes, but the differences between the ages (neonatal vs. adult) was much smaller, as summarized in the Circos plot (Figure 4E), where the overlap of expressed genes mostly occurred among the same cell types. The summary of enriched functional terms is presented in Figure 4F.
Annulus fibrosus cells in neonatal IVD are highly heterogeneous, and their expression of collagen-relevant genes is location-dependent
COL1A1 is the most common ECM protein secreted by AFCs (van den Akker et al., 2017; Tam et al., 2020b). CALR is a marker for the outer AF (Tam et al., 2020b) and heat shock proteins (HSPs, e.g., HSPA6, HSPA1A, etc.) are markers for the most outer layer of the AF (Takao and Iwaki, 2002). Thus, the COL1A1, CALR, and HSPA6 were used to identify AFC populations (Figures 5A and 5B). Cluster 8 (Figures 1C and 1D) has been classified as the inner AFC subpopulation (iAFC) because of its expression of COL1A1 (Figures 5A and 5B). The clusters 9–11 (Figures 1C and 1D) have been assigned to the outer AFC subpopulation (oAFC1-3) because of their expression of CALR (cluster 9, 10) and HSPA6 (cluster 11, Figures 5A and 5B). We detected a high degree of heterogeneity among neonatal AFC populations (Figure 5C). We identified LGALS1, HES1, HERPUD1, and DNAJB1 as AFC subpopulation markers for iAFC and oAFC1-3. Moreover, several types of collagens were also detected in neonatal IVD, specifically COL1A1, COL3A1, COL5A1, COL5A2, COL6A3, and COL12A1 (Figure 5D). Strikingly, their expression showed a decreasing trend assuming the location of subtypes follows the order of iAFC, oAFC1, oAFC2, and oAFC3 from inner core to the outer region (Figure 5D). A similar trend was observed in the enriched biofunctions and key pathway analyses (Figure 5E) across the neonatal iAFC and oAFC populations. As part of the pathways listed in Figure 5E, we show the mechanistic scheme of Unfolded Protein Response (UPR) signaling pathway and the relevant gene expression in oAFC2 in Figure S2. The adult IVD did not exhibit a similar trend of collagen-relevant gene expression (Figure S5). The illustration in Figure 5F shows the postulated structure transition from neonatal to adult IVD based on these spatially dependent gene expression patterns.
Figure 5.
Heterogeneity of Annulus Fibrosus Cells in Human Neonatal and Adult IVDs
(A and B) Expression levels of classical AFC markers (COL1A1, CALR, and HSPA6), classical NPC markers (ACAN, COL2A1, and SOX9), and classical NC markers (MAP1B) are shown for comparison, for both (A) neonatal IVD and (B) adult IVD.
(C) The AFC populations showed strong heterogeneity in neonatal IVD by having 4 distinct subtypes and their heterogenous markers. We demonstrated the expression distribution projected on UMAP and their quantitative expression levels.
(D) Among the 5 heterogeneous subpopulations detected in (C), the neonatal AFCs exhibited a decreasing trend of expression levels of ECM-relevant, collagen-producing genes from inner core NPCs to the outer region AFCs following the order of iAFC, oAFC1-3.
(E) The biological function and key canonical pathway enrichment results followed the same gradient.
(F) Predicted scheme for spatial distribution of the 4 AFC populations, along with inner NPC core and notochord, in neonatal IVD, as compared with the classical IVD structure in adult humans.
Resolved single-cell atlas for human neonatal and adult IVD with clusters assigned to subpopulations
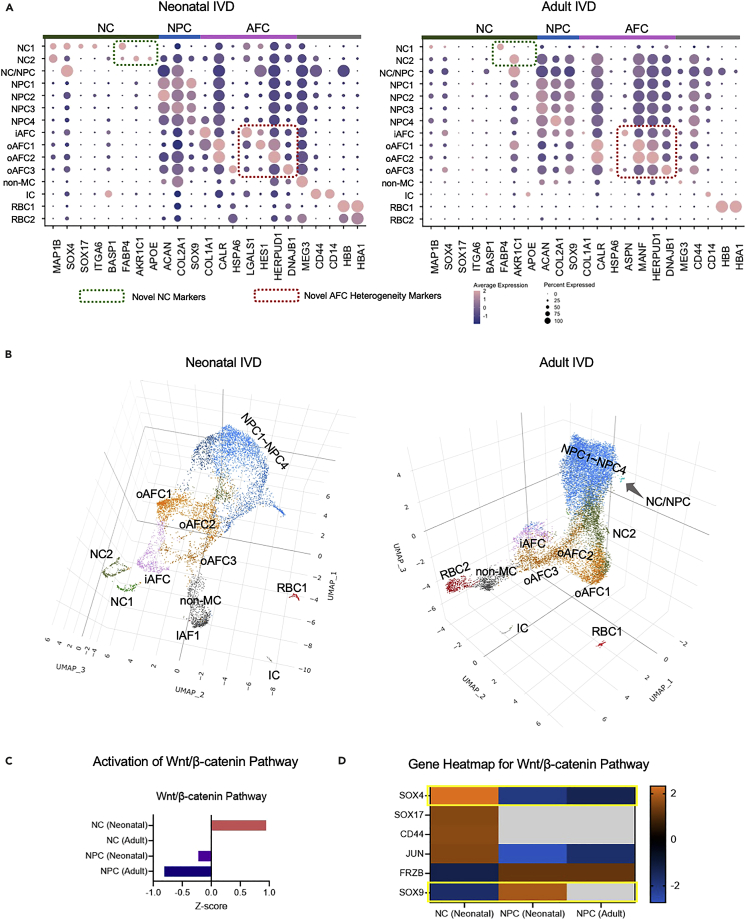
We have assigned cluster 12 into non-mesenchymal cells (non-MC) because of their MEG3 expression, a marker for non-mesenchymal, tumor-suppressing cells (Chen et al., 2017). Cluster 13 is a satellite cell population on the UMAP (Figures 1C and 1D). It shared the CD44 expression as NC2 but lacked all other NC markers (Figures 1C and 1D). Thus, we have assigned cluster 13 to immune-like cells (IC) because of its expression of immune cell marker CD14 (Ziegler-Heitbrock and Ulevitch, 1993) and CD44 (Schumann et al., 2015) (Figure 6A). Cluster 14 and 15 have been assigned to RBC2 and RBC1 because of its expression of hemoglobin markers HBB (Talamo et al., 2003) and HBA1 (Pandey and Rizvi, 2011) (Figure 6A). In addition to classical markers that were used to assign cell identities, Figure 6A also includes novel markers we found for NC, AFC and NPC populations. A heatmap of the expression levels of top enriched genes for all subpopulations shows the high specificity of our enriched genes and the high cellular heterogeneity in IVD structures (Figure S6). Collectively, the single-cell atlas of human IVD heterogeneously comprised of NC, NPC, AFC, IC, non-MC, and RBC populations, which can be more precisely classified into 15 different subtypes: NC1-2, NC/NPC, NPC1-4, iAFC, oAFC1-3, non-MC, IC, and RBC1-2 (Figure 6A).
Figure 6.
Complete Single-cell Atlases of Human Neonatal and Adult IVD
(A) Complete dot plots of marker expression of all subpopulations in the neonatal and adult IVD, including classical markers and novel markers found in the present study. The percent expressed is shown by the size of the dot, and the average expression level (normalized to −2 to 2) is shown by the color scale.
(B) Complete single-cell atlases visualized with 3D UMAP method, for both neonatal and adult IVD. The gene markers in the green rectangle are the novel markers found in the present study.
(C) Activation Z-score rated by Qiagen IPA software showing the canonical Wnt/β-catenin pathway has been differentially regulated in NCs and NPCs in neonatal or adult IVD.
(D) Wnt/β-catenin pathway key molecules’ expression levels are shown in the heatmap. The orange indicates the activation, the blue indicates the deactivation, and the gray indicates the data either was not detected or did not pass the filtering.
The fully resolved single-cell atlases were plotted in three-dimensional (3D) UMPAs (Figure 6B) for a more accurate and intuitive visualization. In the 3D atlas, NC1, NC2, IC, RBC, and non-MC visually appear to be away from the main continents. The NPC subpopulations (NPC1-4) tended to cluster together with vague boundaries whereas AFC subpopulations (iAFC, oAFC1-3) are distant from each other with clear boundaries.
One of the key pathways involved in the IVD is the Wnt/β-catenin pathway (Kondo et al., 2011). We found that the Wnt/β-catenin pathway to be activated in neonatal NC but downregulated in neonatal and adult NPCs (Figure 6C). We further expanded the key genes in the Wnt/β-catenin pathway and showed the expression of SOX4 regulator (also the NC marker) was upregulated in neonatal NCs compared to neonatal and adult NPCs, whereas another regulator gene — SOX9 — was downregulated in neonatal NCs compared to neonatal NPCs (Figure 6D).
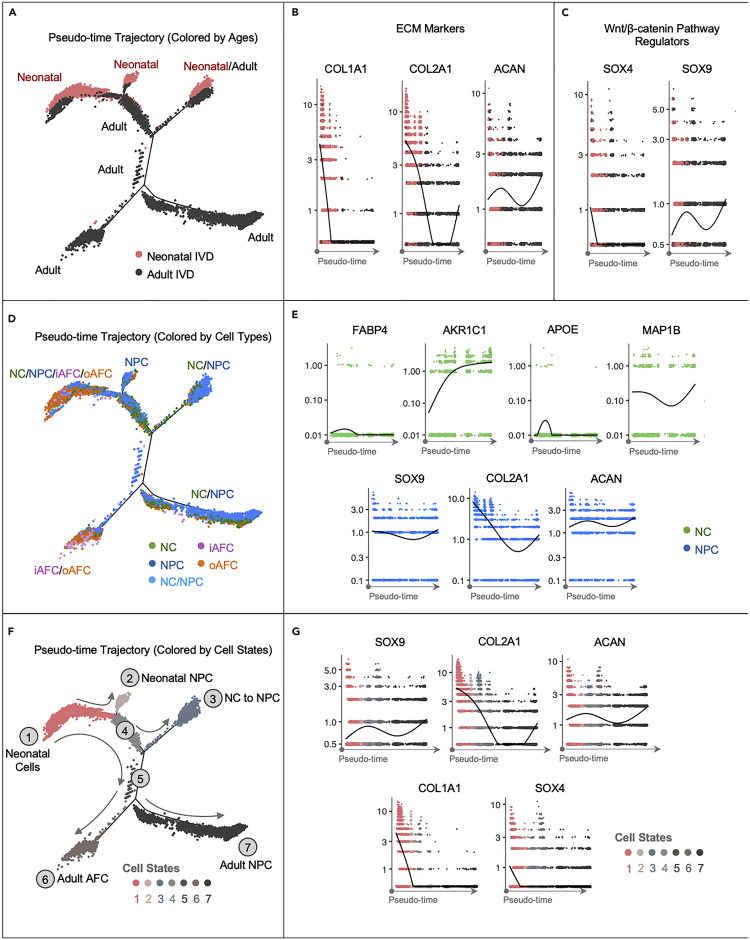
The pseudo-time trajectory predicted five developmental states from neonatal cells into adult nucleus pulposus cells with some notochordal cells preserved throughout adulthood
The trajectory, color coded by age, shows that neonatal cells occupy two branches and share one branch with the adult cells (Figure 7A). Adult cells occupy the other two branches (Figure 7A). ECM gene expression has changed over pseudo-time. COL1A1 expression decreased over pseudo-time, whereas COL2A1 decreased in the early stage but increased in the later stage. ACAN expression followed a swinging increase-decrease-increase pattern (Figure 7B). SOX4, a positive regulator of the Wnt/β-catenin canonical pathway (Sinner et al., 2007), decreased over pseudo-time, but SOX9 — a negative regulator (Sinha et al., 2021) — increased (Figure 7C), which is consistent with Figure 6D.
Figure 7.
Pseudo-time Trajectories Predicted IVD Cellular and Molecular Transition During Development
(A) Cell trajectories predicted based on 500 top genes enriched in each subpopulation and color coded based on different ages.
(B and C) The pseudo-time transition of (B) ECM markers and (C) Wnt/β-catenin canonical pathway regulators.
(D) Cell trajectories color coded by different cell types, including NC, NPC, iAFC, and oAFC, and (E) the change of classical and novel NC markers in NCs over pseudo-time, and change of classical NPC markers in NPCs over pseudo-time.
(F) Cell trajectories color coded by different cell states. Solid gray line shows the predicted developmental direction, and the dashed gray line shows the possible but inconclusively predicted developmental direction.
(G) Pseudo-time transitions of molecules of interest are displayed and colored with cell states.
NC populations are mainly positioned in the proximity to NPCs. NCs were present along the trajectory from the very early neonatal stage toward the later stage and eventually aggregate around NC/NPC in the adult branch (Figure 7D). The two adult IVD-prevalent branches show very distinct cell compositions, one is mainly composed of NCs and NPCs, the other one is mainly composed of AFCs (Figure 7D). The expression of novel and classical NC markers in the NC populations changed over pseudo-time, as shown in Figure 7E. The expression of NPC markers in NPC populations also changed over pseudo-time (Figure 7E). Specifically, COL2A1 decreased most dramatically over pseudo-time but bounced back in the end, whereas SOX9 and ACAN maintained relatively stable levels in NPC populations.
We found 7 cell states along the trajectory (Figure 7F). The annotation of each state depends on the trajectories of age (Figure 7A) and cell types (Figure 7D). State 1 has been annotated as neonatal cells because of its mixed cell composition originated in neonatal IVD. State 2 has been annotated as neonatal NPC, as it is still in the neonatal stage but mostly consists of NPCs. State 3 has been annotated as NC to NPC, because it comprises both neonatal and adult cells and consists of both NCs and NPCs. State 4 and State 5 are two intermediate stages that contain NPCs, NCs, and AFCs. State 6 has been annotated as adult AFC, because it is dominated by adult cells, mainly of iAFCs and oAFCs. State 7 has been annotated as adult NPC, because it is dominated by adult NPCs with a small quantity of NCs and AFCs. The direction of trajectory is determined by the direction of pseudo-time. The change of expression levels of key molecules during the developmental process are also shown over pseudo-time in Figure 7G, including SOX4, COL2A1, ACAN, COL1A1, and SOX4. Collectively, state 1 might indicate an early developmental stage whereas state 2 and 3 may indicate the process of NCs developing into NPCs which may occur during the neonatal stage. State 6 might indicate that AFCs are present in both ages during the development, but they may develop from neonatal AFCs into adult AFCs. State 7 may indicate the development of large quantities of NPCs in adults that are likely originating from neonatal NCs. It also indicates the continuing presence of NCs in adult IVDs.
Discussion
This study provides the first direct comparison of cell atlases at single-cell level between neonatal and adult human IVDs. With the help of scRNA-seq technology, we discovered FABP4, AKR1C1, and APOE as novel markers for human NCs. Our data also supported the previous assumption of the presence of NCs in the adult. It also models the developmental process from neonatal IVD, composed of various cell types, into adult IVD, composed mainly of NPCs and AFCs. Lastly, AFCs in neonatal IVD were found to be highly heterogeneous with a strong gradient profile that most probably correlated with spatial distribution.
This study identifies FABP4, AKR1C1, and APOE as highly specific markers for NC populations in the human neonatal and adult IVD (Figures 3A and 3B). Although our network analysis demonstrated both AKR1C1 and FABP4 to be associated with tumorigenicity and inflammation (Figures 3C–3E), these genes have also been associated with cell metabolism and may be therefore linked to increased cell activity and biosynthesis in early development. For example, AKR1C1 is an enzyme that catalyzes NADPH-dependent reductions (Zeng et al., 2017) and is responsible for hormone secretion (Marín et al., 2009). FABP4 is expressed in embryonic cartilage and bone tissues (Urs et al., 2006). In addition, AKR1C1 and FABP4 also mark the inflammatory response in early-stage development, as shown by VEGF-regulated FABP4 in neonatal NC1 subpopulation that also activated CXCL8 (Figure 3C). Similarly, AKR1C1 and APOE are involved in another IL1-activated inflammation network (Figure S1C). In previous research, the IL1 cascade was already considered an inflammatory and catabolic marker for adult and degenerative human IVD (Johnson et al., 2015). To our knowledge, its occurrence in neonatal IVD has not been previously reported. Noteworthy, FABP4 appears to be a robust NC1 marker in both neonatal and adult IVD. Although AKR1C1 can be used as NC2 marker, it is less specific in adult NC2 (Figure 3A). This may indicate a reduced NC cell metabolism in the adult IVD compared to neonatal.
Our results show that APOE is highly specific to NCs in neonatal IVD (Figure 2C), in contrast to a previous study in mouse IVD that demonstrated APOE to be expressed in both AFCs and NPCs (Zhang et al., 2013). Differences in findings may be because of the different species investigated.
Isolating and identifying rare NC populations in adult IVD is a challenging task because of their small quantity. To our knowledge, only one recent single-cell study by Gan et al. reported a NC population in adult IVD. However, they used TBXT to identify NCs, whereas we used MAP1B (Gan et al., 2021). Other studies, including proteomics (Tam et al., 2020b) and scRNA-seq (Fernandes et al., 2020) of human adult IVD, reported the presence of NPCs and AFCs but did not report NC populations. The differences in findings might be a result of different samples analyzed or analysis techniques (Tam et al., 2020b; Fernandes et al., 2020). Partly in line with our study, Richardson et al. demonstrated the presence of a subpopulation with NC-like phenotype but NPC-like morphology in adult IVD, using qPCR and immunohistochemical analysis (Richardson et al., 2017). In contrast to our findings (Figure 2A), they detected an expression of CD24 and TBXT. In a later study, Rodrigues-Pinto et al. from the same group reported the pre-selected CD24+ cells contained NCs expressing MAP1B (Rodrigues-Pinto et al., 2018), in fetal IVD. To our knowledge, our study is the only report demonstrating two distinct MAP1B + NC subpopulations in adult IVD. Differences in results between Richardson et al., Rodrigues-Pinto et al. and our study might be because of several reasons: The work by Richardson et al. focused on the CD24+ subpopulation as NC-like phenotype and observed their NP-like morphology (Richardson et al., 2017), whereas in our study, we did not find CD24+ cells as a distinct cluster. Therefore, their CD24+ subpopulations may be a different, more heterogeneous population across our clusters. Rodrigues-Pinto et al. sorted and specifically analyzed CD24+ cells (Rodrigues-Pinto et al., 2018), whereas our study did not include any preselection process and instead unbiasedly analyzed all cells. Lastly, our scRNA-seq technique identified two NC populations, which we divided into NC1 and NC2 (Figure 2A), which were not detected in previous studies (Fernandes et al., 2020; Gan et al., 2021).
Historically, NPCs are considered to be derived from NC lineage (Risbud et al., 2010). We confirmed the close lineage relations between NCs and NPCs and further unveiled the transition process with developmental trajectories (Figure 7F). To the best of our knowledge, no previous study predicted the developmental trajectories crossing ages (early vs. late development) and cell types (NC versus NPC). We found neonatal NC and adult NPC populations very distinct. At the molecular level, neonatal NCs have some tumor-like properties and are involved in tissue growth (Figures 4D–4F and S1D showing upregulation of IL1B, VEGF, and HGT), but adult NPCs appear to be more mature, with closer association to fully differentiated cells and developed cytoskeletal tissues (Figures 4D–4F and S1D showing upregulation of BMP2 and SOX9). BMP2 and SOX9 expression was shown in some adult subpopulations (Gan et al., 2021). At the cellular level, the adult NPCs have large cytoplasm, as opposed to the small neonatal NPCs (Figures 1A and 1B). This is in line with the enriched function of development of cytoplasm for adult NPCs (Figure 4D). The large cytoplasm of NPCs (Figure 1B) contradicts a previous report stating that NCs are larger than NPCs (Rodrigues-Pinto et al., 2018). At the tissue level, we found the matrix also changed from a fibrous, less-developed appearance to a more mature cartilage-like, (Figures 1A and 1B), as well as the ECM gene expression transformed from a collagens-rich (COL1A1 and COL2A1) to aggrecan-rich (ACAN) in the later stage (Figure 7B). It is known that COL2A1 ensures the removal of notochord during the development (Aszódi et al., 1998). Thus, the decrease of COL2A1 (Figures 7B and 7G) may mark the successful removal of notochord and completion of NC to NPC transition.
Though most NCs differentiated into adult NPCs, some NCs kept their identity throughout the entire trajectory (Figure 7D). The NC quantity is significantly decreased in adults (NCs constituted 7.5% of total cell counts in neonatal IVD and 17.5% in adult IVD, data not shown). Previous studies were not conclusive on their presence (Séguin et al., 2018; Fernandes et al., 2020; McCann and Séguin, 2016; Wang et al., 2008; Risbud and Shapiro, 2011). It should be noted that NC1 almost disappeared in adult IVD but NC2 percentage significantly elevated (Figure 1H). This indicates that NCs became more involved in ECM interactions and tissue developments but maintained robust pluripotency during the development. (Figures S1A and S1B). Our study shows that the NCs kept expressing NC markers, MAP1B and AKR1C1, in later pseudo-time and even elevated in the very late stage (Figure 7E). Therefore, the NCs may have preserved their capability to differentiate into NPCs during their continuing presence of NCs throughout adulthood.
The high heterogeneity of AFCs found in neonatal IVD is in line with recent studies that also described spatially defined AFC populations divided into inner and outer regions (Shi et al., 2015; Tam et al., 2020a). Our study presented a very detailed classification of AFCs into one subpopulation to inner AF regions (iAFC), and three distinct subpopulations in the outer AF regions (oAFC1-3), which has not been reported in previous relevant studies (Shi et al., 2015; Tam et al., 2020a).
A striking trend of decreasing expression levels of collagen-associated genes (Figure 5D) appeared when the order of the five AFC subpopulations from neonatal IVD was re-arranged from the inner region to the outer region to iAFC, oAFC1, oAFC2, and oAFC3 (Figure 5D). Interestingly, another proteomics study by Tam et al. (2020b) showed ECM proteins in 16-year to 17-year-olds increased from the inner NP to the outer AF region of the IVD, which seems to be contradictory to our findings of the decreasing expression of collagen-producing genes (Figure 5D). The protein content and RNA level may not necessarily be aligned. In line with Tam et al., we found GP6 signaling pathways relevant to cell-ECM interaction show gradients of expression from inner AF to outer AF (Figure 5E). The oAFC and iAFC have distinct gene expression profiles possibly due to their different spatially defined roles. AFCs are in a position that connects endplate and inner NP core, as suggested in Figure 5F. The promoted Unfolded Protein Response (UPR) we detected in the outer AFC region (Figure 5E) may be a sign of external stressors (Hetz et al., 2020; Wang et al., 2010). The UPR also orchestrates the enforcement of adaptive mechanisms to maintain an optimal rate of protein production and rapidly reacts to diverse stimuli, including extracellular responses to hormones, growth factors and small ligands that bind cell-surface receptors (Hetz et al., 2020). UPR is also a major contributor of the CALR marker for oAFC subpopulations (Figure S2). CALR was previously found to be expressed in the oAF as well (Tam et al., 2020b) and it is a key gene in the UPR pathway (Figure S2). In addition to UPR signaling, the platelet-derived growth factor (PDGF) signaling also follows the increasing trend from inner to outer AF regions (Figure 5F). PDGF expression has been shown to promote the proliferation of AFCs and NPCs (Pratsinis and Kletsas, 2007), suggesting a potential increased cell proliferation in the outer region. In short, the UPR and PDGF signaling may indicate an increased cell proliferation in response to external stimuli and stress applied to the outer AFCs. This theory was additionally supported by our detection of the expressed heat shock protein, HSPA6, in the outermost area of oAFC3 subpopulation. Heat shock proteins (HSPs, Figure 5A) were previously found to be strongly expressed in the outermost area of IVD and endplate cartilage in response to mechanical stress (Tam et al., 2020a; Takao and Iwaki, 2002). Collectively, the gradient trend of AFCs in neonatal IVD might be associated with certain biological functions, for instance, the elevated stress-induced cell proliferation and remodeling process among oAFCs. However, this needs to be confirmed by spatial single-cell analysis in the future.
In conclusion, we identified FABP4, AKR1C1 and APOE as novel markers for human NC populations. We demonstrated the first direct comparison of human neonatal and adult IVD at a single-cell level, finding two distinct NC subpopulations in both neonatal and adult IVD. A pseudo-time trajectory analysis predicted the developmental process from neonatal IVD, composed of various cell types, into adult IVD, composed of mainly NPCs and AFCs. We also found that NCs preserve their cell identity and some of their markers and function into adulthood. Lastly, we demonstrated a high heterogeneity of AFC populations in neonatal IVD and showed their gradual transitions of cell types, gene expression, function, signaling pathways, as well as their potential relevance in IVD development, which appears to be linked to a spatial gradient. Better understanding of these processes may help to identify the underlying factors contributing to age-associated diseases of the IVD and to develop tailored therapeutics that may delay or reverse these processes.
Limitations of the study
The present work is not without limitations: our scRNA-seq lacks spatial resolution. The recently emerging single-cell spatial sequencing technology may be required in the next study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Polyclonal Anti-HSPA6 | Abcam | ab212044 |
| Rabbit Monoclonal Anti-Apolipoprotein E [EP1374Y] | Abcam | ab52607 |
| Goat Polyclonal Anti-Collagen I | Biorad | 131001 |
| Mouse Monoclonal Anti-CD44 (Clone Bu52) | Biorad | MCA2504T |
| Mouse Monoclonal Anti-SOX17 | Invitrogen | MA5-24885 |
| Rabbit Polyclonal Anti-MAP1B | Novus Biologicals | NBP3-04801-20ul |
| Mouse Monoclonal Aldo-keto Reductase 1C1/AKR1C | R&D Systems | MAB6529-SP |
| Goat Polyclonal Anti-FABP4/A-FABP | R&D Systems | AF3150-SP |
| Goat Polyclonal Anti-Aggrecan G1-IGD-G2 | R&D Systems | AF1220-SP |
| Cy™2 AffiniPure Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch | 705-225-147 |
| Cy™3 AffiniPure F(ab')₂ Fragment Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | 711-166-152 |
| Alexa Fluor® 647 AffiniPure F(ab')₂ Fragment Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | 715-606-150 |
| Biological samples | ||
| Human neonatal IVD | NDRI | ND18842 |
| Human adult IVD | Cedars-Sinai Medical Center | Patient Cadaver |
| Chemicals, peptides, and recombinant proteins | ||
| Chromium Single-cell 3' Reagent Kits | 10x Genomics | PN-1000121 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) | Invitrogen | D1306 |
| Deposited data | ||
| Single-cell RNA-seq data for all samples used in this study | GSE189916 | Gene Expression Omnibus |
| Original code | https://github.com/jason199112345/scRAN-seq-neonatal-adult-spine-paper | GitHub |
| Software and algorithms | ||
| ImageJ (v2.1.0/1.53c) | (Schneider et al., 2012) | https://imagej.nih.gov/ij/download.html |
| Fiji | (Schindelin et al., 2012) | https://imagej.net/software/fiji/downloads |
| R (v4.0.3) | R Development Core Team | https://www.R-project.org |
| RStudio (v1.4.1103) | RStudio Team (2020). RStudio: Integrated Development for R. RStudio, PBC | https://www.rstudio.com/products/rstudio/ |
| Cell Ranger (v3.0.0) | 10x Genomics | |
| Loupe Cell Browser (v3.0.0) | 10x Genomics | |
| Seurat (v4.0.1) | (Hao et al., 2021; Stuart et al., 2019; Butler et al., 2018; Satija et al., 2015) | https://satijalab.org/seurat/ |
| Monocle (v2.18.0) | (Qiu et al., 2017b; Trapnell et al., 2014; Qiu et al., 2017a) | http://cole-trapnell-lab.github.io/monocle-release/ |
| Qiagen Ingenuity Pathway Analysis (v01-20-04) | v01-20-04 | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dmitriy Sheyn (dmitriy.sheyn@csmc.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
The Cedars-Sinai Medical Center Institutional Review Board approved this study (IRB number Pro00052234). Human neonatal and adult IVDs have both been investigated with scRNA-seq to reveal their cell atlas of respective age and to determine the difference between them. The basic information of the samples, human subjects, and the data sequencing are shown in the following Table S1. Specifically, the neonatal samples were collected from 4 different spine levels (L1-L5) from a 6-h postnatal male provided by the National Disease Research Interchange (NDRI, ND18842), 3 levels were used for the scRNA-seq analysis (3 neonatal samples sent for scRNA-seq, n = 1) and one for histology. The adult samples were collected from lumbar discs of 2 cadaveric spines and 1 clinical sample of removed and otherwise discarded disc. IVD tissues were harvested from three neonatal spinal disc levels and adult cadavers or clinical samples of patients with no back pain history (over 65 years old) were collected and analyzed via scRNA-seq (3 adult samples sent for scRNA-seq, n = 3). Additional lumbar levels were processed for histological analysis.
Method details
Single-cell isolation and sequencing
The IVDs were dissected and washed with Phosphate Buffered Saline (PBS). The tissue was manually minced to ∼1 mm pieces in a sterile environment and enzymatically digested. Tissue was digested for 1 h at 37°C in 2 mg/mL of Pronase Protease (Millipore, Temecula, CA) in growth media containing 1mM L-glutamine, 1% antibiotic-antimycotic solution (Gibco, Carlsbad, CA) and 10% fetal bovine serum (FBS, Gemini Bioproducts, West Sacramento, CA) in Dulbecco’s modified eagle media-F12 media (DMEM/F12, GIBCO, Carlsbad, CA). This was followed by ∼18 h digestion at 37°C in 0.25 mg/mL of Collagenase Type 1S (Sigma Aldrich) in growth media. The resulting sample was pushed through a 70 μm cell strainer and cells were isolated via centrifugation at 1,200 rpm for ten minutes, The pellet was resuspended in phosphate buffered saline at a concentration of 1,500 cells/μL and 100uL of sample was processed for single-cell RNA sequencing using the Chromium Single-cell 3' Reagent Kits from 10X Genomics.
Chromium Single-cell 3' v2 libraries with ∼3,000 cells were prepared on a Chromium Controller with chips and reagents from Single-cell Gene Expression v2 kits following the manufacturer’s protocols (10x Genomics). The libraries were then sequenced using paired-end sequencing (28 bp Read 1 libraries, and 91 bp Read 2) with a single sample index (8 bp) on an Illumina NovaSeq. Samples were sequenced to a depth of > 70,000 raw reads per cell, with raw sequencing data analyzed and visualized with pre-release versions of Cell Ranger 3.0.0 and Loupe Cell Browser 3.0.0. Raw sequencing data was demultiplexed and converted to fastq format by using bcl2fastq v2.20 (Illumina, San Diego, CA). Fragment analysis of indexed libraries was performed on the Agilent 4200 TapeStation (Agilent Technologies, Santa Clara, CA). The sequencing information is shown in Table S1, including number of cells sequenced, mean reads per cell, median genes per cell, and median UMI counts per cells. The barcode rank plot showing filtered UMI counts distributed over barcodes is shown in Figure S7.
Integration, pre-processing, and dimensional reduction of the single-cell dataset of neonatal and adult IVD
Seurat package (v4.0.1) in R (v4.0.3) has been used to process the data of six samples. We firstly used CreateSeuratObject function to transform the loaded data into a Seurat object for each sample. Next, the four Seurat objects were combined into one matrix. NormalizeData and FindVariableFeatures were used. Afterwards, FindIntegrationAnchors and IntegrateData were used to integrate and anchor the 6 samples’ data together. RunPCA function was used to for principal component analysis with PC = 30. DimPlot function was then used with “UMAP” reduction to obtain the 2D Uniform Mani-fold Approximation and Projection (UMAP) plot. FindNeighbors and FindClusters functions further classified all cells in 6 samples into 15 clusters at resolution of 0.5 (unsupervised clustering). The UMAP was then split by their sample type (neonatal or adult IVD) for comparison.
Detection of enriched genes and novel markers for notochordal cells
The expression levels of NC relevant genes in neonatal or adult IVD were separately visualized using violin plot in log scale with split view. In the volcano plots, we set the cutoff threshold of FC to be > 2 and the cutoff threshold of adjusted p value to be <1×10−24. Any genes falling out of this range were not considered as enriched genes or markers. Genes falling into this range were labeled with red color in the volcano plot. Next, top enriched genes from the red colored area were manually selected as novel in silico markers for specific cluster from neonatal IVD or adult IVD. Their expression levels were presented quantitatively in violin plots.
Visualization of single-cell atlases with 3D UMAP plots
The cell coordination of the top three dimensions (PC1, PC2, and PC3) were extracted from the Seurat object integrating neonatal IVD and adult IVD. FetchData function was used to create the data frame for the 3D UMAP plot. The plot_ly function in Plotly package was used to obtain the final 3D UMAP plot. The color for each type was plot as the same as in 2D UMAP.
Gene ontology term enrichment and pathway analysis
Gene ontology (GO) term enrichment and pathway analysis was performed using Qiagen Ingenuity Pathway Analysis (IPA, version 65367011). A list of enriched genes for a specific cell sub-population (as compared against all other cells) was firstly generated in R studio using FindMarkers function. The specific cell sub-populations include NC1-2, iAFC, oAFC1-3 respectively. In other attempts, we did multiple further analyses: we created enriched gene list for NC by combining NC1 and NC2 cell sub-populations followed by FindMarkers function against all other cells; we created enriched gene list for NPC by combining NPC1-4 cell sub-populations followed by FindMarkers function against all other cells. All the above enriched gene lists were generated for both neonatal IVD and adult IVD. The adjusted p value and fold change for genes with p < 0.0001 were loaded into the IPA software. We included the database for human tissue and primary cells but excluded those for cell lines, as our samples were harvested from human samples only. The analyses covered canonical pathways, upstream analysis, diseases and biological functions, and regulatory networks. The heatmaps for enriched biological functions, enriched pathways, and key genes in the pathways were replotted using GraphPad Prism (v9.2.0). We demonstrated the most relevant results in figures.
The comparison of gene overlap in NCs and NPCs in both neonatal and adult IVD was performed at Metascape.org using Circos plot. The enriched genes with p < 0.0001 and FC > 1 were selected for Circos plot analysis.
Histology and immunofluorescence
IVD samples from the sample were fixed in 10% phosphate buffered formalin for three days, decalcified by incubation in 0.5M EDTA (pH 7.4, Sigma Aldrich) for three weeks, and embedded in paraffin. Five-micron-thick sections were cut from the paraffin blocks. Hematoxylin and eosin (H&E) staining was performed to evaluate the morphological features. Images were captured using a Carl Zeiss Axio Imager Z1 fluorescent microscope (Carl Zeiss, Germany) equipped with ApoTome and AxioCam HRc cameras. Images were analyzed using QuPath software (v0.2.3).
For immunofluorescence, sections were deparaffinized, rehydrated in DPBS, treated with antigen retrieval solution at room temperature for 20 min (Dako #S3020, Agilent Technologies). Slides were washed with PBS, blocked with 3% normal donkey serum (Jackson ImmunoResearch Laboratories Inc., West Grove, PA) in 0.3% Triton-X (Sigma Aldrich, St. Louis, MO) and again washed with PBS. Slides were stained with primary antibodies (anti-human HSPA6, CD44, SOX17, AKR1C1, FABP4, APOE). The primary antibodies were applied to the slides, after which the slides were incubated at 4°C overnight and washed using PBS; the slides were then incubated with secondary antibodies for 1 h at room temperature. Finally, the slides were stained with 4',6-diamidino-2-phenylindole dihydrochloride (DAPI, 0.28 μg/mL) for 5 min in the dark. Subsequently, sections were washed three times in dPBS and mounted with Prolong Gold with DAPI (Life Technologies).
Pseudo-time analysis of developmental trajectories
The pseudo-time developmental trajectory analysis was performed using Monocle package (v2.18.0). The original integrated Seurat object was subset into a new Seurat object that contains the following cell populations: NC (as the combination of NC1-2), NPC (as the combination of NPC1-4), iAFC, and oAFC (as the combination of oAFC1-3). Low-quality cells were filtered by the following standards: 1) Cells whose total mRNAs >106 were filtered; 2) Cells whose total mRNAs were out of a range of mean ±2 × standard deviations were filtered. The gene marker was also filtered out when less than 10 cells expressed this marker. The data matrix was then log-transformed and standardized to the same scale. We chose up to 500 top globally enriched genes for each sub-populations to determine cell progress. The dimensions of the dataset were reduced using DDRTree method. The trajectories were plotted using plot_cell_trajectory function. The cells in trajectories were colored with either their ages, cell types, or pseudo-time states.
Quantification and statistical analysis
The quantification and statistical analysis in single cell RNA-seq was performed using Seurat package in R. The n = 1 for neonatal human and n = 3 for adult human. The n number is specific for the number of human subjects. Different samples collected from the same patient is considered as the one n. Z-score was calculated with Qiagen IPA software.
Acknowledgments
This study was supported by NIH/NIAMS, Grant/Award Number: K01AR071512. The authors wish to acknowledge the Cedars-Sinai Genomics Core for next generation services and the Cedars-Sinai Biobank for human adult cadaver samples. The authors thank National Disease Research Interchange (NDRI) for human neonatal samples and Mrs. Julia Sheyn for the fluorescent images acquisition.
Author contributions
Conceptualization, W.J., D.S., J.G., and R.H.; Methodology, W.J., R.H., D.S., P.M., and A.W.; Software, W.J., R.H., and A.W.; Validation, K.S and G.K.; Formal Analysis, W.J., R.H., and D.S.; Investigation, W.J., D.S., and R.H.; Resources, R.H., D.S., and W.J.; Data Curation, W.J. and R.H.; Writing – Original Draft, W.J.; Writing – Review & Editing, J.G., D.S., W.J., R.H., and G.K.; Visualization, W.J., D.S., and R.H.; Supervision, D.S., and R.H.; Funding Acquisition, D.S.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104504.
Supplemental information
Data and code availability
-
•
The raw sequencing data and original data are accessible in Gene Expression Omnibus: GSE189916.
-
•
The original code is accessible at GitHub: https://github.com/jason199112345/scRAN-seq-neonatal-adult-spine-paper.
References
- Andersson G.B. Epidemiological features of chronic low-back pain. Lancet. 1999;354:581–585. doi: 10.1016/S0140-6736(99)01312-4. [DOI] [PubMed] [Google Scholar]
- Aszódi A., Chan D., Hunziker E., Bateman J.F., Fässler R. Collagen II is essential for the removal of the notochord and the formation of intervertebral discs. JCB (J. Cell Biol.) 1998;143:1399–1412. doi: 10.1083/jcb.143.5.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattaram P., Penzo-Méndez A., Sock E., Colmenares C., Kaneko K.J., Vassilev A., DePamphilis M.L., Wegner M., Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Deng J.M., Zhang Z., Behringer R.R., de Crombrugghe B. Sox9 is required for cartilage formation. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- Buckley C.T., Hoyland J.A., Fujii K., Pandit A., Iatridis J.C., Grad S. Critical aspects and challenges for intervertebral disc repair and regeneration—harnessing advances in tissue engineering. JOR Spine. 2018;1:e1029. doi: 10.1002/jsp2.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Zou J., Wang W., Yang A. BMP2 induces hMSC osteogenesis and matrix remodeling. Mol. Med. Rep. 2021;23:125. doi: 10.3892/mmr.2020.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.K., Yu X.H., Yang W., Wang C., He W.S., Yan Y.G., Zhang J., Wang W.J. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50:e12313. doi: 10.1111/cpr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour K.A., Agulnick A.D., Eliazer S., Kelly O.G., Kroon E., Baetge E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- de Schepper E.I.T., Damen J., van Meurs J.B.J., Ginai A.Z., Popham M., Hofman A., Koes B.W., Bierma-Zeinstra S.M. The association between lumbar disc degeneration and low back pain: the influence of age, gender, and individual radiographic features. Spine. 2010;35:531–536. doi: 10.1097/BRS.0b013e3181aa5b33. [DOI] [PubMed] [Google Scholar]
- Fernandes L.M., Khan N.M., Trochez C.M., Duan M., Diaz-Hernandez M.E., Presciutti S.M., Gibson G., Drissi H. Single-cell RNA-seq identifies unique transcriptional landscapes of human nucleus pulposus and annulus fibrosus cells. Sci. Rep. 2020;10:15263. doi: 10.1038/s41598-020-72261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J.W., Kerr M.S., Brooker A.S., DeMaio S.E., Maetzel A., Shannon H.S., Sullivan T.J., Norman R.W., Wells R.P. Disability resulting from occupational low back pain. Part I: what do we know about primary prevention? A review of the scientific evidence on prevention before disability begins. Spine. 1996;21:2908–2917. doi: 10.1097/00007632-199612150-00024. [DOI] [PubMed] [Google Scholar]
- Frapin L., Clouet J., Delplace V., Fusellier M., Guicheux J., Le Visage C. Lessons learned from intervertebral disc pathophysiology to guide rational design of sequential delivery systems for therapeutic biological factors. Adv. Drug Deliv. Rev. 2019;149–150:49–71. doi: 10.1016/j.addr.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Gan Y., He J., Zhu J., Xu Z., Wang Z., Yan J., Hu O., Bai Z., Chen L., Xie Y., et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus progenitors in human intervertebral discs. Bone Res. 2021;9:37. doi: 10.1038/s41413-021-00163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., III, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho R., Workman M.J., Mathkar P., Wu K., Kim K.J., O’Rourke J.G., Kellogg M., Montel V., Banuelos M.G., Arogundade O.A., et al. Cross-comparison of human iPSC motor neuron models of familial and sporadic ALS reveals early and convergent transcriptomic disease signatures. Cell Syst. 2021;12:159–175.e9. doi: 10.1016/j.cels.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Matyas J., Duncan N. The notochordal cell in the nucleus pulposus: a review in the context of tissue engineering. Tissue Eng. 2003;9:667–677. doi: 10.1089/107632703768247368. [DOI] [PubMed] [Google Scholar]
- Johnson Z.I., Schoepflin Z.R., Choi H., Shapiro I.M., Risbud M.V. Disc in flames: roles of TNF-α and IL-1β in intervertebral disc degeneration. Eur. Cell. Mater. 2015;30:104–117. doi: 10.22203/ecm.v030a08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly N.H., Huynh N.P.T., Guilak F. Single cell RNA-sequencing reveals cellular heterogeneity and trajectories of lineage specification during murine embryonic limb development. Matrix Biol. 2020;89:1–10. doi: 10.1016/j.matbio.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic N.N., Mandalia S., Raasch J., Knezevic I., Candido K.D. Treatment of chronic low back pain–new approaches on the horizon. J. Pain Res. 2017;10:1111–1123. doi: 10.2147/JPR.S132769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N., Yuasa T., Shimono K., Tung W., Okabe T., Yasuhara R., Pacifici M., Zhang Y., Iwamoto M., Enomoto-Iwamoto M. Intervertebral disc development is regulated by Wnt/β-catenin signaling. Spine. 2011;36:E513–E518. doi: 10.1097/BRS.0b013e3181f52cb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.-Y., Li M., Lin Y.-L. Mitf induction by RANKL is critical for osteoclastogenesis. Mol. Biol. Cell. 2010;21:1763–1771. doi: 10.1091/mbc.e09-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane G.J., Thomas E., Croft P.R., Papageorgiou A.C., Jayson M.I.V., Silman A.J. Predictors of early improvement in low back pain amongst consulters to general practice: the influence of pre-morbid and episode-related factors. Pain. 1999;80:113–119. doi: 10.1016/S0304-3959(98)00209-7. [DOI] [PubMed] [Google Scholar]
- Marín Y.E., Seiberg M., Lin C.B. Aldo-keto reductase 1C subfamily genes in skin are UV-inducible: possible role in keratinocytes survival. Exp. Dermatol. 2009;18:611–618. doi: 10.1111/j.1600-0625.2008.00839.x. [DOI] [PubMed] [Google Scholar]
- Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., Menon M., He L., Abdurrob F., Jiang X., et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann M.R., Séguin C.A. Notochord cells in intervertebral disc development and degeneration. J. Dev. Biol. 2016;4:3. doi: 10.3390/jdb4010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey K.B., Rizvi S.I. Biomarkers of oxidative stress in red blood cells. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2011;155:131–136. doi: 10.5507/bp.2011.027. [DOI] [PubMed] [Google Scholar]
- Panebianco C.J., Dave A., Charytonowicz D., Sebra R., Iatridis J.C. Single-cell RNA-sequencing atlas of bovine caudal intervertebral discs: discovery of heterogeneous cell populations with distinct roles in homeostasis. FASEB J. 2021;35:e21919. doi: 10.1096/fj.202101149R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratsinis H., Kletsas D. PDGF, bFGF and IGF-I stimulate the proliferation of intervertebral disc cells in vitro via the activation of the ERK and Akt signaling pathways. Eur. Spine J. 2007;16:1858–1866. doi: 10.1007/s00586-007-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H., Trapnell C. Reversed graph embedding resolves complex single-cell developmental trajectories. bioRxiv. 2017 doi: 10.1101/110668. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.M., Ludwinski F.E., Gnanalingham K.K., Atkinson R.A., Freemont A.J., Hoyland J.A. Notochordal and nucleus pulposus marker expression is maintained by sub-populations of adult human nucleus pulposus cells through aging and degeneration. Sci. Rep. 2017;7:1501. doi: 10.1038/s41598-017-01567-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud M.V., Shapiro I.M. Notochordal cells in the adult intervertebral disc: new perspective on an old question. Crit. Rev. Eukaryot. Gene Expr. 2011;21:29–41. doi: 10.1615/critreveukargeneexpr.v21.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud M.V., Schaer T.P., Shapiro I.M. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev. Dynam. 2010;239:2141–2148. doi: 10.1002/dvdy.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risbud M.V., Schoepflin Z.R., Mwale F., Kandel R.A., Grad S., Iatridis J.C., Sakai D., Hoyland J.A. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J. Orthop. Res. 2015;33:283–293. doi: 10.1002/jor.22789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Pinto R., Ward L., Humphreys M., Zeef L.A.H., Berry A., Hanley K.P., Hanley N., Richardson S.M., Hoyland J.A. Human notochordal cell transcriptome unveils potential regulators of cell function in the developing intervertebral disc. Sci. Rep. 2018;8:12866. doi: 10.1038/s41598-018-31172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J., Stanko K., Schliesser U., Appelt C., Sawitzki B. Differences in CD44 surface expression levels and function discriminates IL-17 and IFN-γ producing helper T cells. PLoS One. 2015;10:e0132479. doi: 10.1371/journal.pone.0132479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séguin C.A., Chan D., Dahia C.L., Gazit Z. Latest advances in intervertebral disc development and progenitor cells. JOR Spine. 2018;1:e1030. doi: 10.1002/jsp2.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M., Kiseliovas V., Levine J., Gayoso A., Mazutis L., Pe’er D. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheyn D., Ben-David S., Tawackoli W., Zhou Z., Salehi K., Bez M., De Mel S., Chan V., Roth J., Avalos P., et al. Human iPSCs can be differentiated into notochordal cells that reduce intervertebral disc degeneration in a porcine model. Theranostics. 2019;9:7506–7524. doi: 10.7150/thno.34898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Wang F., Hong X., Wang Y.T., Bao J.P., Cai F., Wu X.T. The presence of stem cells in potential stem cell niches of the intervertebral disc region: an in vitro study on rats. Eur. Spine J. 2015;24:2411–2424. doi: 10.1007/s00586-015-4168-7. [DOI] [PubMed] [Google Scholar]
- Sinha A., Fan V.B., Ramakrishnan A.-B., Engelhardt N., Kennell J., Cadigan K.M. Repression of Wnt/β-catenin signaling by SOX9 and Mastermind-like transcriptional coactivator 2. Sci. Adv. 2021;7:eabe0849. doi: 10.1126/sciadv.abe0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner D., Kordich J.J., Spence J.R., Opoka R., Rankin S., Lin S.-C.J., Jonatan D., Zorn A.M., Wells J.M. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol. Cell Biol. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R., Grzelak M., Hadfield J. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- Stegner D., Haining E.J., Nieswandt B. Targeting Glycoprotein VI and the immunoreceptor tyrosine-based activation motif signaling pathway. Arterioscler. Thromb. Vasc. Biol. 2014;34:1615–1620. doi: 10.1161/atvbaha.114.303408. [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., III, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Liu B., Luo Z.-J. The immune privilege of the intervertebral disc: implications for intervertebral disc degeneration treatment. Int. J. Med. Sci. 2020;17:685–692. doi: 10.7150/ijms.42238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao T., Iwaki T. A comparative study of localization of heat shock protein 27 and heat shock protein 72 in the developmental and degenerative intervertebral discs. Spine. 2002;27:361–367. doi: 10.1097/00007632-200202150-00007. [DOI] [PubMed] [Google Scholar]
- Talamo F., D'Ambrosio C., Arena S., Del Vecchio P., Ledda L., Zehender G., Ferrara L., Scaloni A. Proteins from bovine tissues and biological fluids: defining a reference electrophoresis map for liver, kidney, muscle, plasma and red blood cells. Proteomics. 2003;3:440–460. doi: 10.1002/pmic.200390059. [DOI] [PubMed] [Google Scholar]
- Tam V., Chen P., Yee A., Solis N., Klein T., Kudelko M., Sharma R., Chan W.C., Overall C.M., Haglund L., et al. DIPPER: a spatiotemporal proteomics atlas of human intervertebral discs for exploring ageing and degeneration dynamics. bioRxiv. 2020 doi: 10.7554/elife.64940. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V., Chen P., Yee A., Solis N., Klein T., Kudelko M., Sharma R., Chan W.C.W., Overall C.M., Haglund L., et al. DIPPER, a spatiotemporal proteomics atlas of human intervertebral discs for exploring ageing and degeneration dynamics. Elife. 2020;9:e64940. doi: 10.7554/eLife.64940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs S., Harrington A., Liaw L., Small D. Selective expression of an aP2/Fatty Acid Binding Protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res. 2006;15:647–653. doi: 10.1007/s11248-006-9000-z. [DOI] [PubMed] [Google Scholar]
- van den Akker G.G.H., Koenders M.I., van de Loo F.A.J., van Lent P.L.E.M., Blaney Davidson E., van der Kraan P.M. Transcriptional profiling distinguishes inner and outer annulus fibrosus from nucleus pulposus in the bovine intervertebral disc. Eur. Spine J. 2017;26:2053–2062. doi: 10.1007/s00586-017-5150-3. [DOI] [PubMed] [Google Scholar]
- Vergroesen P.P.A., Kingma I., Emanuel K.S., Hoogendoorn R.J.W., Welting T.J., van Royen B.J., van Dieën J., Smit T.H. Mechanics and biology in intervertebral disc degeneration: a vicious circle. Osteoarthritis Cartilage. 2015;23:1057–1070. doi: 10.1016/j.joca.2015.03.028. [DOI] [PubMed] [Google Scholar]
- Wang J., Huang Y., Huang L., Shi K., Wang J., Zhu C., Li L., Zhang L., Feng G., Liu L., Song Y. Novel biomarkers of intervertebral disc cells and evidence of stem cells in the intervertebral disc. Osteoarthritis Cartilage. 2021;29:389–401. doi: 10.1016/j.joca.2020.12.005. [DOI] [PubMed] [Google Scholar]
- Wang W.L., Abramson J.H., Ganguly A., Rosenberg A.E. The surgical pathology of notochordal remnants in adult intervertebral disks: a report of 3 cases. Am. J. Surg. Pathol. 2008;32:1123–1129. doi: 10.1097/PAS.0b013e3181757954. [DOI] [PubMed] [Google Scholar]
- Wang Z., Butler P., Ly D., Spiotto M., Koong A., Yang G. Activation of the unfolded protein response in wound healing. J. Surg. Res. 2010;158:209. doi: 10.1016/j.jss.2009.11.111. [DOI] [Google Scholar]
- Zeng C.-M., Chang L.-L., Ying M.-D., Cao J., He Q.-J., Zhu H., Yang B. Aldo-Keto reductase AKR1C1-AKR1C4: functions, regulation, and intervention for anti-cancer therapy. Front. Pharmacol. 2017;8:119. doi: 10.3389/fphar.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Jin L., Reames D.L., Shen F.H., Shimer A.L., Li X. Intervertebral disc degeneration and ectopic bone formation in apolipoprotein E knockout mice. J. Orthop. Res. 2013;31:210–217. doi: 10.1002/jor.22216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Zheng Q., Engin F., Munivez E., Chen Y., Sebald E., Krakow D., Lee B. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc. Natl. Acad. Sci. U S A. 2006;103:19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock H.W., Ulevitch R.J. CD14: cell surface receptor and differentiation marker. Immunol. Today. 1993;14:121–125. doi: 10.1016/0167-5699(93)90212-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw sequencing data and original data are accessible in Gene Expression Omnibus: GSE189916.
-
•
The original code is accessible at GitHub: https://github.com/jason199112345/scRAN-seq-neonatal-adult-spine-paper.