Summary
The insulin receptor (INSR) is an evolutionarily conserved signaling protein that regulates development and cellular metabolism. INSR signaling promotes neurogenesis in Drosophila; however, a specific role for the INSR in maintaining adult neural stem cells (NSCs) in mammals has not been investigated. We show that conditionally deleting the Insr gene in adult mouse NSCs reduces subventricular zone NSCs by ∼70% accompanied by a corresponding increase in progenitors. Insr deletion also produced hyposmia caused by aberrant olfactory bulb neurogenesis. Interestingly, hippocampal neurogenesis and hippocampal-dependent behaviors were unperturbed. Highly aggressive proneural and mesenchymal glioblastomas had high INSR/insulin-like growth factor (IGF) pathway gene expression, and isolated glioma stem cells had an aberrantly high ratio of INSR:IGF type 1 receptor. Moreover, INSR knockdown inhibited GBM tumorsphere growth. Altogether, these data demonstrate that the INSR is essential for a subset of normal NSCs, as well as for brain tumor stem cell self-renewal.
Keywords: cancer stem cell, insulin-like growth factors, IGF-II, subventricular zone, subgranular zone, stem cell niche, mouse, glioblastomas, insulin receptor
Highlights
-
•
Insulin receptor (INSR) is essential for adult SVZ neural stem cell self-renewal
-
•
INSR deletion causes hyposmia with increased olfactory bulb neurogenesis
-
•
Hippocampal stem cells (and associated behaviors) do not require INSR
-
•
Glioblastomas overexpress INSR pathway components required for tumorsphere growth
Levison, Wood, and colleagues (Chidambaram et al.) show that conditionally deleting the insulin receptor (INSR) in adult murine neural stem cells (NSCs) depletes NSCs in the subventricular zone and correspondingly increases progenitors, resulting in hyposmia caused by aberrant olfactory bulb neurogenesis. In contrast, hippocampal neurogenesis is unperturbed. They also show that the INSR is necessary for human glioblastoma tumorsphere self-renewal.
Introduction
Insulin or insulin-like signaling is a highly conserved signaling system that regulates development, as well as key cellular functions, including glucose and lipid metabolism. Whereas Drosophila insulin-like peptides signal through a single InR, mammals possess several insulin-related signaling receptors that include the well-known metabolic insulin receptor B (INSRB) isoform, as well as the insulin receptor A (INSRA) splice variant of the INSR (lacking exon 11 containing 12 amino acids), and the insulin-like growth factor type 1 receptor (IGF1R). Unlike INSRB, the INSRA and IGF1R regulate cell proliferation and regulate stem cell self-renewal and progenitor amplification in various cell populations (Andres et al., 2013; Massimino et al., 2021; Wamaitha et al., 2020; Wang et al., 2015). The family of ligands for these receptors includes insulin, insulin-like growth factor (IGF)-I, and IGF-II with varying affinities for the different receptors (Belfiore et al., 2017; Boucher et al., 2014). At physiological levels, insulin binds with high affinity only to the two INSR isoforms. Conversely, IGF-I binds with high affinity only to the IGF1R. IGF-II, however, is unique in that it binds with high affinity to both IGF1R and INSRA (Frasca et al., 1999). Prior investigations demonstrated that the IGF-II/INSRA signaling loop is (1) essential for embryonic development (Efstratiadis, 1998; Louvi et al., 1997), (2) elevated in certain types of tumor (Belfiore, 2007; Frasca et al., 1999), and (3) necessary in vitro for self-renewal of normal neural stem cells (NSCs) (Ziegler et al., 2012, 2014, 2015) and thyroid tumor stem cells (Malaguarnera et al., 2012).
Our recent studies demonstrated that IGF-II is an essential niche factor for several populations of adult stem cells, including the NSCs of the subventricular zone (SVZ) and the subgranular zone (SGZ) (Ziegler et al., 2019). In the adult central nervous system (CNS), IGF-II is predominantly produced by the choroid plexus, although it also is expressed at lower levels by hippocampal progenitors and brain endothelial cells (Bracko et al., 2012; Ferron et al., 2015; Logan et al., 1994). However, there is considerable controversy over which receptor mediates IGF-II actions in the adult CNS. Two reports concluded that IGF-II induces neural stem/progenitor cell (NSP) proliferation through the IGF1R either in primary neurosphere cultures (Ferron et al., 2015; Lehtinen et al., 2011) or in transit-amplifying progenitors (Ferron et al., 2015; Lehtinen et al., 2011). Another report concluded that IGF-II promotes hippocampal learning and memory consolidation indirectly through its binding to the cation-independent mannose-6-phosphate (M6P) receptor (M6P/IGF2R), generally considered a scavenger receptor for IGF-II and M6P-tagged proteins, but with no inherent signaling capability (Chen et al., 2011). Although our in vitro studies support the hypothesis that IGF-II promotes NSC self-renewal through the INSRA (Ziegler et al., 2012, 2014), no studies have directly tested the function of the INSR on NSC self-renewal in vivo. In the studies reported here, we used both in vitro and in vivo genetic approaches to delete the Insr in NSCs to test its function in NSC self-renewal, as well as to evaluate the downstream consequences on neurogenesis and behavior.
The INSR also is important in tumor biology. INSR overexpression in thyroid tumors stimulates the self-renewal of cancer stem cells (Malaguarnera et al., 2012; Vella et al., 2001). INSRA expression in particular is associated with a more aggressive, undifferentiated tumor phenotype in several types of solid tumor (Frasca et al., 1999; Scalia et al., 2020; Vella et al., 2002). Glioblastoma multiforme (GBM) is the most common type of malignant brain tumor in adults, accounting for 78% of all CNS tumors. Among the GBMs, the proneural and mesenchymal subtypes are the hardest to treat with high incidences of relapse. A prevailing view is that these relapses are due to the presence of self-renewing cancer stem cells that are resistant to chemotherapy and radiation therapies (Chandran et al., 2015). The INSR is commonly expressed in surgical specimens from GBM patients and contributes to GBM cell survival and growth through Akt-dependent signaling (Gong et al., 2016). Moreover, an ATP-competitive IGF1R/INSR inhibitor has shown promising results in decreasing cell viability and migration both in vitro and in vivo in temozolomide-resistant gliomas (Zhou, 2015). Based on our previous studies showing that IGF-II signaling through the INSR promotes NSC self-renewal in vitro and on data from other investigators showing that an IGF-II/INSR signaling loop enriches for cancer stem cells in other tumors (Sciacca et al., 1999; Vella et al., 2002), we tested the hypothesis that INSR and IGF-related genes are expressed at high levels in the most aggressive GBM subtypes and regulate growth of GBM cancer stem cells.
Results
Insr deletion decreases SVZ-derived NSCs and increases progenitor lineages
To test the function of the INSR in NSPs in vitro, we initially cultured NSPs from Insrfl/fl mice and used adenovirus (Ad)-Cre-GFP to delete the Insr or Ad-GFP or no virus as controls (see Figures S1A–S1D). A measurement of transfection efficiency at 24 h post-infection revealed no significant differences between the Ad-Cre-GFP- and control Ad-GFP-infected populations (see Figure S1E). Insr deletion in NSPs resulted in a 74% decrease in secondary neurosphere number (p = 0.0001; see Figure S1F) and a 78% decrease in neurosphere size between NSPs infected with Ad-Cre-GFP compared with control Ad-GFP virus (p < 0.0001; see Figure S1G).
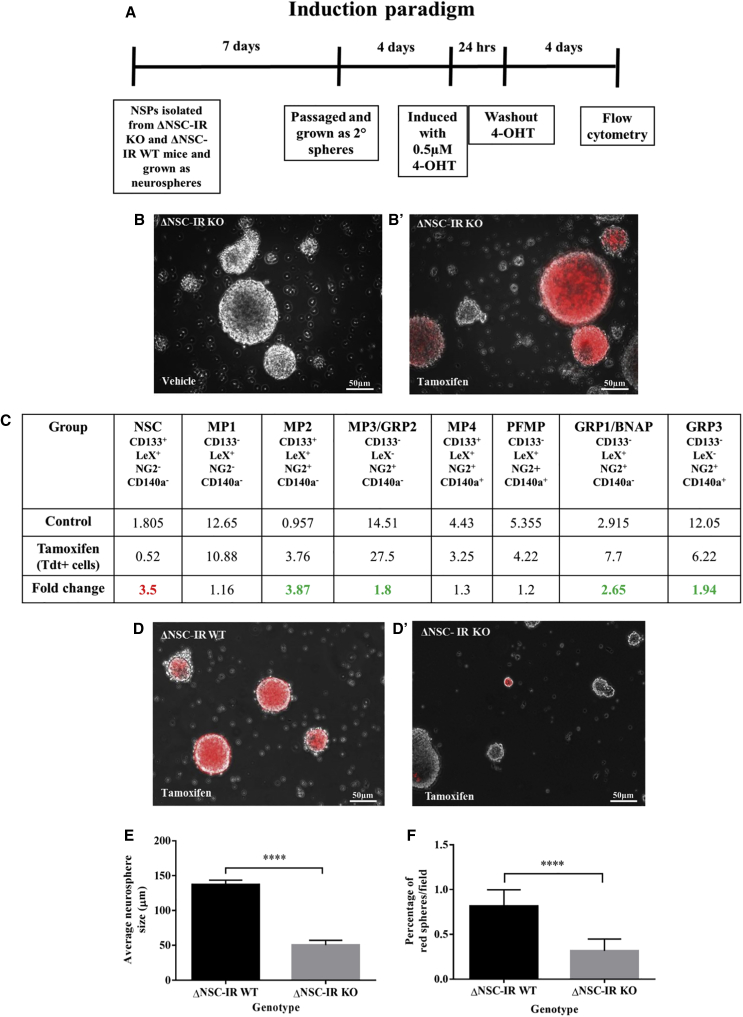
To determine whether deleting the Insr reduced the frequency of NSCs, we established an Insr knockout mouse line where the Insr is deleted specifically in adult NSCs through the nestin-CreERT2 promoter after tamoxifen administration. These mice also carry flox-stop tdTomato (tdT) reporter alleles: NestinCreERT2+/−tdT+/+Insr fl/fl (ΔNSC-IRKO) and NestinCreERT2+/−tdT+/+Insr WT (ΔNSC-IR wild-type [WT]; controls). SVZ cells from P4 ΔNSC-IRKO pups were grown in vitro as neurospheres and treated with 4-OH tamoxifen (0.5 μM) or vehicle for 24 h to induce Cre recombination. The treated cells were propagated for an additional 5 days and then dissociated for flow cytometric analysis as outlined in Figure 1A. Vehicle-treated cells were tdT negative, whereas tamoxifen-treated cells were strongly tdT positive (Figures 1B and B′). Using a flow cytometry panel established previously in our lab (Buono et al., 2012; Velloso et al., 2022), we parsed cells within the neurosphere into NSCs and seven different intermediate progenitors (Figure 1C). This analysis revealed that the ΔNSC-IRKO spheres had 3.5-fold fewer NSCs compared with the ΔNSC-IR WT when only the tdT+ cells were evaluated. Correspondingly, the ΔNSC-IRKO spheres had increased progenitor populations: Multipotential Progenitor-2 (MP2) (3.8-fold), MP3 (1.8-fold), Glial Restricted Progenitor-1 (GRP1) (2.6-fold), and GRP3 (1.9-fold) (Figure 1C). These changes were not due to altered expression levels of the cell surface markers because levels of CD133, LeX, NG2, and CD140a were not different between INSR WT cells, vehicle-treated cells (ΔNSC-IRKO mice treated with corn oil; tdT−), ΔNSC-IR WT (tdT+ and tdT− cells), and ΔNSC-IRKO tdT− cells (see Figure S2). A subset of the cells used for the flow cytometric analysis was re-plated to analyze sphere-forming ability. Consistent with an essential role for INSR in NSC self-renewal, fewer tdT+ spheres formed from the ΔNSC-IRKO cells compared with ΔNSC-IR WT cells (Figures 1D–1F).
Figure 1.
Insr deletion in NSCs in vitro decreases NSCs and alters lineage
(A) Induction paradigm of experiment. Cells from ΔNSC-IR WT and ΔNSC-IRKO mice were treated with 0.5 μM 4-OH tamoxifen or vehicle for 24 h.
(B and B′) Neurospheres from vehicle- and tamoxifen-treated cells 5 days after induction.
(C) ΔNSC-IR KO mice have fewer NSCs and a greater proportion of intermediate progenitors within the neurospheres than WT mice (red numbers indicate decreases in fold change, green numbers indicate increases in fold change) (n = 2 independent experiments).
(D and D′) Representative images of tertiary spheres after plating to measure sphere-forming ability.
(E) Average sizes of tertiary tdTomato red+ neurospheres.
(F) Percentage of tertiary tdTomato red+ neurospheres per field were reduced in ΔNSC-IRKO mice (unpaired t test, p < 0.0001; n = 3 independent experiments).
BNAP, bipotential neuron-astrocyte progenitor; GRP, glial restricted progenitor; MP, multipotent progenitor; NSC, neural stem cell; PFMP, platelet-derived growth factor-fibroblast growth factor 2 (PDGF-FGF2)-responsive MP.
Insr deletion decreases forebrain SVZ NSCs in vivo and compromises olfaction
To determine whether the INSR is essential for NSC self-renewal in vivo, we first analyzed tdT reporter expression in brains of Cre+/− and Cre-negative ΔNSC-IR mice given tamoxifen at 4–5 weeks of age. Two days after tamoxifen administration (see Figure S3A), tdT was strongly expressed by nestin+ cells of the SVZ, SGZ, and in the α-tanycytes of the hypothalamus in mice that were Cre+ (see Figures S3C, S3C′, S3F, and S3F′), whereas tdT+ cells were not evident in tamoxifen-administered Cre-mice (see Figures S3B and S3B′).
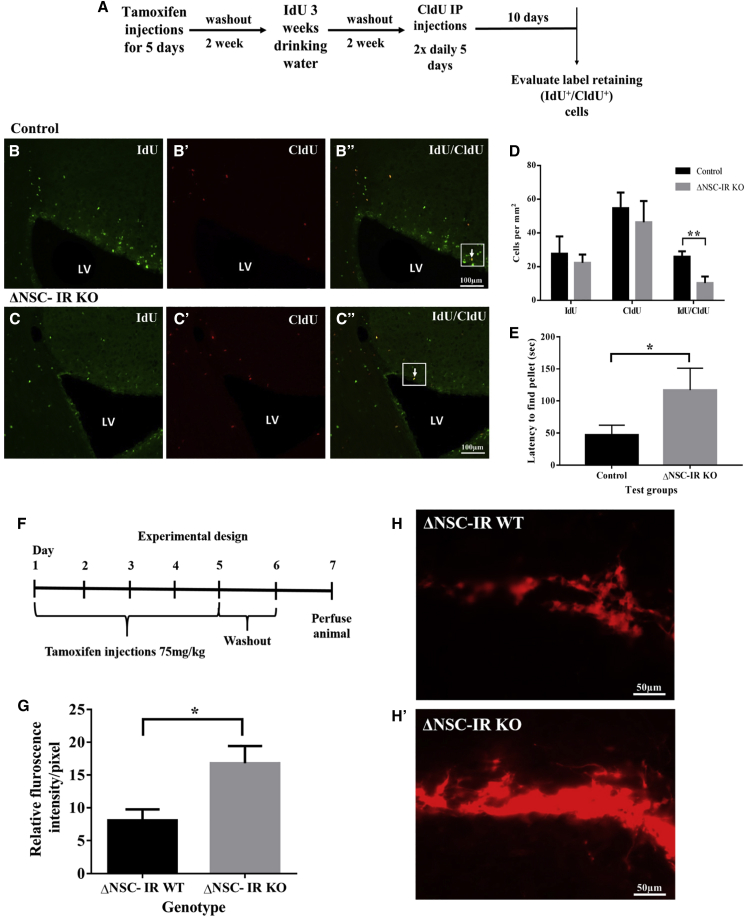
To analyze NSC populations in vivo, we administered tamoxifen or vehicle at 4–5 weeks of age to establish ΔNSC-IRKO and ΔNSC-IR WT mice, respectively. To determine whether there were fewer label-retaining cells as a measure of NSC self-renewal in the ΔNSC-IRKO neurogenic regions, we performed a double-thymidine analog labeling protocol where iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) were administered at equimolar concentrations on different schedules over the course of 5 weeks using a modification of the original protocol from Vega and Peterson (2005; Llorens-Martin and Trejo, 2011) as validated in our prior studies (Ziegler et al., 2019) (Figure 2A). Using stereology methods, we counted the number of IdU and CldU single- and double-positive cells in the SVZ in ΔNSC-IRKO and ΔNSC-IR WT mice. This analysis revealed a 66% reduction in the number of IdU+/CldU+ double-positive cells, representing the label-retaining, slowly cycling NSCs in the SVZ (cell numbers: ΔNSC-IR WT versus ΔNSC-IRKO, 26.5 ± 2.2 versus 8.8 ± 2.3; unpaired t test, p = 0.03) (Figures 2B–2D). These results confirm the in vitro findings that the INSR regulates NSC number in the SVZ.
Figure 2.
ΔNSC-IRKO mice have decreased numbers of SVZ NSCs, increased numbers of tdTomato+ SVZ progenitors, and impaired olfaction
(A) Schematic of experimental timeline depicting timing of tamoxifen and IdU and CldU administration.
(B–B″) Representative images of IdU and CldU labeling in control SVZ.
(C–C″) Representative images of IdU and CldU labeling in ΔNSC-IRKO SVZ. White arrows represent IdU+/CldU+ double-positive cells (scale bar represents 100 μm).
(D) Numbers of IdU and CldU single- and double-positive cells that were located within 80 μm of the lateral ventricle were reduced by 50% in the ΔNSC-IRKO SVZ (mean ± SEM, unpaired t test, ∗p = 0.03; n = 4 control, n = 5 IRKO).
(E) ΔNSC-IRKO mice required a significantly greater amount of time to find a buried food pellet compared with WT mice (mean ± SEM, Mann-Whitney test, ∗p = 0.01; n = 10 control, n = 12 IRKO).
(F) Schematic representation of Insr deletion paradigm.
(G) Relative SVZ fluorescence intensity in ΔNSC-IRKO and ΔNSC-IR WT mice (mean ± SEM, ∗p < 0.05, unpaired t test; n = 3 per group).
(H and H′) Representative images of tdTomato expression in ΔNSC-IRKO and ΔNSC-IR WT mice.
Because neuroblasts produced within the SVZ perpetually regenerate interneurons in the olfactory bulbs (OBs), we next tested the mice for deficits in olfaction using the buried food behavioral assay (Yang and Crawley, 2009). Consistent with aberrant neurogenesis in the OB, the ΔNSC-IRKO mice took twice as long to find a hidden food pellet compared with the ΔNSC-IR WT mice (ΔNSC-IR WT versus ΔNSC-IRKO: 19.25 ± 3.1 versus 55.8 ± 19.2 s; Mann-Whitney test, p = 0.01), indicating deficits in olfactory sensitivity (Figure 2E).
Reduction in NSC number with loss of Insr is not due to cell death
The results above support an essential role for the INSR in NSC number, but this could be because of INSR promoting survival and/or self-renewal of the NSCs. To assess whether the reduction in SVZ NSC number in the ΔNSC-IRKO was due to cell death, we induced ΔNSC-IR KO and ΔNSC-IR WT mice with tamoxifen for 5 days and sacrificed them on day 6 (Figure 2F). Cell death in the ΔNSC-IRKO SVZ would be expected to result in a reduction in the numbers of tdT+ cells in this short-term paradigm. However, we observed no reduction in tdT+ cells in the ΔNSC-IRKO SVZ. Rather, the density of tdT+ cells increased in the ΔNSC-IRKO compared with the ΔNSC-IR WT mice (ΔNSC-IR WT versus ΔNSC-IRKO: 8.1 ± 1.7 versus 17 ± 2.6 fluorescence intensities/pixel; unpaired t test, p < 0.05) (Figures 2G–2H′, p = 0.018). These results further support the conclusion that loss of the INSR causes the NSCs to shift their mode of division to produce more progenitors because of reduced self-renewal.
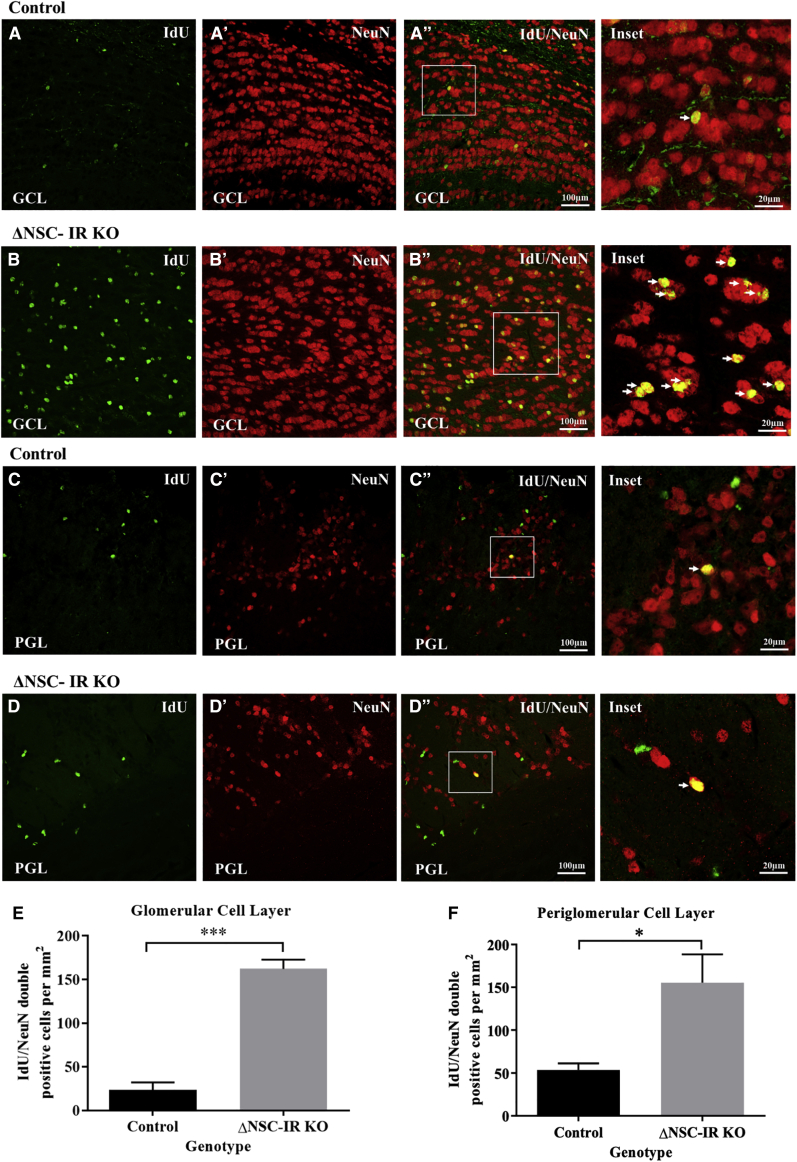
Insr deletion in NSCs increases production of OB granule cell and periglomerular neurons
As described above, Insr deletion compromised NSC self-renewal with a corresponding increase in several progenitor cell populations and a net increase in tdT+ cells within the SVZ. Consistent with the increase in tdT+ SVZ cells, there was an increase in the numbers of IdU+/NeuN+ double-positive cells in the granule cell layer (ΔNSC-IR WT versus ΔNSC-IRKO: 23.3 ± 8.7 versus 162.2 ± 10.4 cells; unpaired t test, p < 0.001) (Figures 3A–3B″ and 3E) and in the periglomerular layer (ΔNSC-IR WT versus ΔNSC-IRKO: 53.6 ± 7.6 versus 155.5 ± 33 cells; unpaired t test, p < 0.05) (Figures 3C–D″ and 3F) of the OB in the ΔNSC-IRKO versus ΔNSC-WT mice when analyzed 9 weeks after Insr deletion. Taken together, these results support an essential function for the INSR in the SVZ in promoting NSC self-renewal and regulating the production of certain progenitor populations.
Figure 3.
ΔNSC-IRKO mice have increased neurogenesis in the olfactory bulb
(A–A″) Granule cell layer of control mice stained for IdU+(green)/NeuN+ (red) cells in the olfactory bulb 9 weeks after tamoxifen administration.
(B–B″) ΔNSC-IRKO mice stained for IdU+/NeuN+ (arrows) in the granule cell layer at 9 weeks after tamoxifen administration.
(C–C″) Periglomerular layer of control mice stained for IdU+(green)/NeuN+ (red) cells of the olfactory bulb 9 weeks after tamoxifen administration (mean ± SEM, p < 0.001).
(D–D″) ΔNSC-IRKO mice showing increased IdU+/NeuN+ (arrows) in the periglomerular layer 9 weeks after tamoxifen administration.
(E) Quantification of IdU+/NeuN+ cells in the granule cell layer of control and ΔNSC-IRKO mice (mean ± SEM, p < 0.001).
(F) Quantification of IdU+/NeuN+ cells in the periglomerular layer of control and ΔNSC-IRKO mice (mean ± SEM, p < 0.05).
Scale bars represent 100 μm (A–D) or 20 μm (insets). Images are representative of three animals per genotype.
Hippocampal neurogenesis and hippocampal-dependent behaviors are unchanged in ΔNSC-IRKO mice
An important question that arises from the studies on the INSR in the SVZ NSCs is whether this receptor functions similarly in other NSC populations. To establish whether the INSR is required to maintain the NSCs of the hippocampal SGZ, we performed stereology to count IdU single-labeled, CldU single-labeled, and IdU+/CldU+ double-labeled cells within the SGZ of the same mice analyzed for changes in the SVZ. In contrast with the SVZ, similar numbers of labeled cells were observed in both ΔNSC-IRKO and ΔNSC-WT mice (see Figures S4A–S4C). To assess hippocampal spatial learning, we evaluated the mice using the Morris water maze test. Neither the acquisition rate (latency to find platform and path efficiency), nor the total time spent in the quadrant that contained the platform during the probe test was different between the ΔNSC-IRKO and ΔNSC-IR WT mice (ΔNSC-IR WT versus ΔNSC-IRKO: 24.5 ± 1.8 versus 27 ± 4.1 s) (see Figures S4D and S4F). ΔNSC-IRKO mice were further evaluated for performance in the elevated plus maze to determine whether function of the ventral hippocampus was altered with deletion of the Insr in the NSCs. There was no difference in the amount of time that ΔNSC-IRKO and ΔNSC-IR WT mice spent in the open arms of the maze (ΔNSC-IR WT versus ΔNSC-IRKO: 60.4 ± 10.1 versus 46.7 ± 12.7 s) (see Figure S4E).
In addition to the SVZ and SGZ NSCs, label-retaining α-tanycytes have been reported along the walls of the third ventricle that produce new neurons and glial cells in the paraventricular, supraoptic, and arcuate nuclei of the hypothalamus (Robins et al., 2013). Within the median eminence of the hypothalamus there is another set of progenitors that can generate neurons involved in metabolism (Lee et al., 2012). Therefore, we analyzed the relative numbers of IdU, CldU, and double-labeled cells in the hypothalamus and median eminence of the ΔNSC-IRKO versus ΔNSC-IR WT mice. IdU+ and CldU+ cells were much sparser in the hypothalamus than in the SVZ and the SGZ, and stereological counts of cells in the lateral wall of the third ventricle failed to reveal any change in the number of α-tanycytes between the ΔNSC-IRKO and ΔNSC-IR WT mice (see Figures S5A–S5C). Because the α-tanycytes can give rise to the neurons in the arcuate nucleus, which controls food intake, we measured the body weight over the 5-day course of tamoxifen injection. Although there was a trend toward decreased body weight in ΔNSC-IRKO versus control mice, the difference did not reach statistical significance (see Figure S5D). In the median eminence, both the β2-tanycytes and rapidly proliferating CldU+ transit-amplifying cells showed a trend toward a decrease in number (see Figures S6A–S6C). Taken together, these data indicate that the INSR has a specific role in SVZ NSC self-renewal but is not essential for maintaining hippocampal NSCs or MPs of the hypothalamus.
In silico data analysis reveals elevated expression of insulin/IGF genes and splice enzymes in proneural and mesenchymal subtypes of GBM
A number of studies have implicated overexpression of INSR/IGF-related genes in cancer (Chettouh et al., 2013; Kim et al., 2012; Vella et al., 2001; Vigneri et al., 2015; Wang et al., 2013). Therefore, we queried expression data for genes in the INSR/IGF growth factor system from the The Cancer Genome Atlas (TCGA) database for proneural (p) (n = 31) and mesenchymal (m) (n = 55) GBM subtypes. The proneural and mesenchymal GBM subtypes had higher expression of the INSR (proneural [5.6-fold], mesenchymal [4.9-fold]), IGF1R (proneural [10.4-fold], mesenchymal [11.3-fold]), and IGF2 (proneural [59-fold], mesenchymal [60-fold]) when compared with normal astrocytes (Table 1). Chettouh et al. (2013) showed that in hepatocellular carcinoma the change in ratio of INSRA to INSRB was due to alterations in splice enzymes. Therefore, we queried the expression of the enzymes known to be involved in splicing the mRNA for the INSR. In agreement with the earlier paper, we observed increased expression of several splice enzymes involved in INSR alternative splicing (CUCGBP1: proneural [4.9-fold], mesenchymal [4.5-fold]; HNRNPHI: proneural [2.9-fold], mesenchymal [2.3-fold]; HNRNPA2B1: proneural [2.5-fold], mesenchymal [2.1-fold]; SFRS1/AS2: proneural [2.8-fold], mesenchymal [2.4-fold]) (Table 1).
Table 1.
Insulin/IGF growth factor system genes and INSR mRNA splicing factor genes are induced in glioblastomas
| No. | Gene name | Fold change in expression: proneural | Fold change in expression: mesenchymal |
|---|---|---|---|
| 1 | INSR | 5.6 | 4.9 |
| 2 | IGF1R | 10.4 | 11.3 |
| 3 | IGF2 | 59 | 60 |
| 4 | (CUCGBP1) CELF1 | 4.9 | 4.5 |
| 5 | HNRNPH1 | 2.9 | 2.3 |
| 6 | HNRNPA2B1 | 2.5 | 2.1 |
| 7 | SFRS1 | 2.8 | 2.4 |
For rows 1–3, TCGA data mining for genes involved in the insulin/IGF growth factor system reveals higher expression of INSR, IGF1R, and IGF2 in proneural and mesenchymal subtypes versus normal astrocytes (proneural: n = 31, mesenchymal: n = 55). For rows 4–7, TCGA data mining for enzymes involved in INSR mRNA splicing in proneural and mesenchymal subtypes (proneural: n = 31, mesenchymal: n = 55). Data represent the mean of all values.
Proneural and mesenchymal subtype GBM stem cells increase INSRA/IGF1R ratio
Our prior studies demonstrated that: (1) IGF-II promotes stemness in adult NSCs (Ziegler et al., 2012, 2019), (2) IGF-II mediates SVZ NSC self-renewal through INSR in vitro (Ziegler et al., 2014), and (3) the Insra is highly expressed in the medial SVZ (Ziegler et al., 2012). Thus, we performed studies to establish whether gene expression changes seen in entire tumors reflected changes occurring within the GBM stem cells. Multiple studies in different cancer types have shown a switch in the ratio of INSRA/INSRB and INSRA/IGF1R that allow cancer cells to shift from a metabolic to a more proliferative phenotype (Andres et al., 2013; Chettouh et al., 2013; Garofalo et al., 2013; Lodhia et al., 2015; Ulanet et al., 2010). We generated tumorspheres from human gliomas and performed an RNA transcriptome analysis. Again, the ratio of the INSR to the IGF1R was 3.8-fold higher in GBM spheres compared with normal human NSCs (Table 2). Interestingly, INSRB (which contains exon 11) was not detected in either normal neurospheres or tumorspheres, consistent with our prior findings in the mouse SVZ in vivo (Ziegler et al., 2012, 2014).
Table 2.
INSRA/IGF1R ratio increases in GBM stem cells
| Gene | hNSC1 | hNSC2 | GSC1 | GSC2 | GSC3 | GSC4 | GSC5 | GSC6 |
|---|---|---|---|---|---|---|---|---|
| INSR | 11.4395 | 2.97384 | 6.75019 | 3.62512 | 7.17077 | 3.15267 | 3.06491 | 4.17927 |
| IGF1R | 57.0076 | 24.9964 | 6.06387 | 10.2188 | 12.2469 | 5.49724 | 3.69863 | 21.8827 |
| INSR/IGF1R | 0.20 | 0.12 | 1.11 | 0.35 | 0.59 | 0.57 | 0.83 | 0.19 |
| Average (Avg.) of INSR/IGF-1R ratios | 0.16 | 0.61 | ||||||
| Fold change | 1 | 3.8a | ||||||
INSR and IGF1R mRNA levels were quantified from GBM neurospheres from deep sequencing using Roche 454/GS FLX Sequencing Technology. Exon 11 (present only in INSRB) was undetectable in the human NSC (hNSC) populations glioma stem cells (GSC) and hNSC. Thus, the INSR in NSCs corresponds to INSRA (glioma stem cells: n = 6, normal human neural stem cells: n = 2). Data represent mean values of RNA-seq reads.
Increased expression.
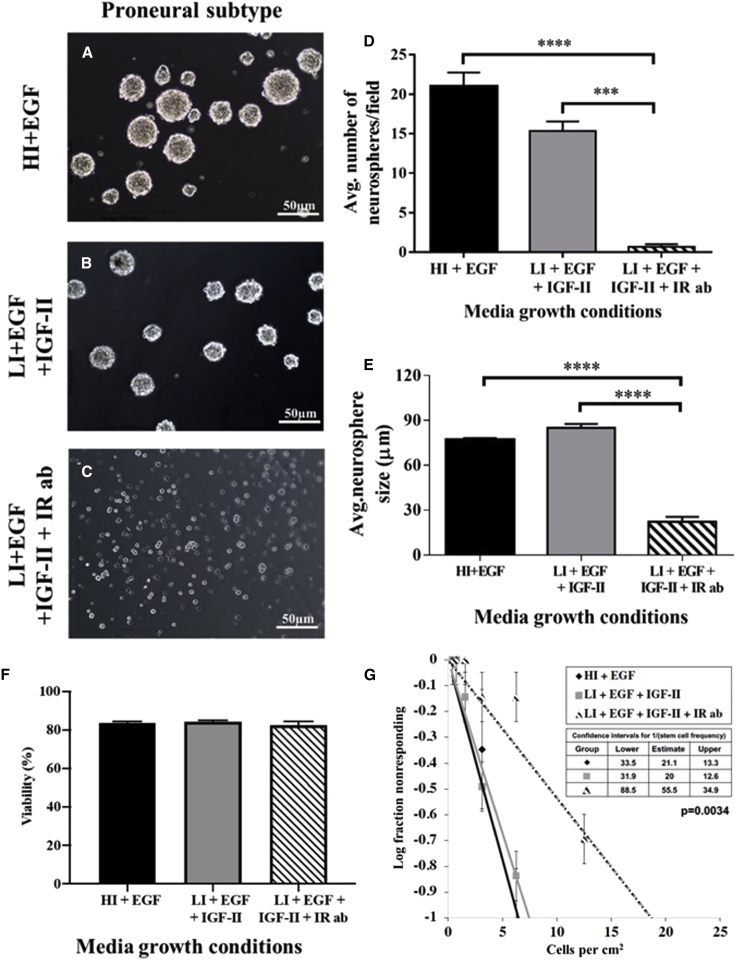
Blocking the INSR alters the number and size of proneural and mesenchymal GBM tumorspheres
Sphere-forming assays have been widely used to evaluate the self-renewal properties of stem cells, including cancer stem cells, with repeated passage used as an index of self-renewal (Molofsky et al., 2003; Raitano et al., 2013; Remboutsika et al., 2011; Zelentsova-Levytskyi et al., 2017). Our lab previously showed that lowering the concentration of insulin to physiological levels (low insulin [LI], 4.4 nM), to avoid cross-stimulating the IGF1R, and then adding IGF-II (224 ng/mL) increased numbers of murine SVZ neurospheres formed on subsequent passages, consistent with enhanced stemness via IGF-II actions. Although IGF-II can stimulate both the INSRA and IGF1R, we further demonstrated that IGF-II was acting through the INSR and not the IGF1R to promote NSC self-renewal in these studies. Therefore, we asked whether similarly lowering the concentration of insulin and adding IGF-II to the medium used to propagate human tumorspheres would enhance GBM tumorsphere growth. Contrary to our findings with normal neurospheres grown in vitro, there was no difference in the number or size of proneural GBM tumorspheres formed when the cells were grown in LI medium supplemented with IGF-II compared with control conditions containing high insulin (HI) (4.4 μM) (Figures 4A–4D). This result was likely due to the high endogenous expression of IGF-II in the GBMs. However, addition of an INSR blocking antibody (Ab) resulted in a 35-fold reduction in the number of proneural tumorspheres formed in epidermal growth factor (EGF)-supplemented media (HI + EGF versus LI + IGF-II + INSR Ab: 21 ± 1.7 versus 0.6 ± 0.3; one-way ANOVA, p < 0.0001) and an 18.5-fold reduction in the number of tumorspheres in EGF + IGF-II-supplemented media (LI + EGF + IGF-II versus LI + EGF + IGF-II + INSR Ab: 11.1 ± 1.2 versus 0.6 ± 0.3; one-way ANOVA, p < 0.001) (Figures 4A–4D). Similarly, blocking the INSR reduced the size of the tumorspheres by 3.5-fold in EGF-supplemented media (HI + EGF versus LI + IGF-II + IR Ab: 77 ± 1.1 versus 22 ± 3.17; one-way ANOVA, p < 0.0001) and 3.8-fold in EGF + IGF-II-supplemented media (LI + EGF + IGF-II versus LI + EGF + IGF-II + IR Ab: 85 ± 2.64 versus 22 ± 3.17; one-way ANOVA, p < 0.001) (Figure 4E). To investigate whether reduced number and tumorsphere size were due to increased cell death, we evaluated cell viability in tumorspheres grown for 7 days and found no difference between the three different defined media conditions (Figure 4F). To evaluate tumorsphere formation at the clonal level, we performed an Extreme Limiting Dilution Assay (Nguyen et al., 2018). At 21 days in vitro (DIV), proneural GBM tumorspheres in both conditions where INSR was activated (HI + EGF and LI + EGF + IGF-II) showed an ∼2.5× higher capacity for tumorsphere formation than cells where INSR was blocked (LI + EGF + IGF-II + INSR Ab) (chi-square test [Chisq] = 11.4; degrees of freedom [df] = 2; p = 0.0034) (Figure 4G).
Figure 4.
A function blocking antibody (Ab) to the INSR decreases self-renewal and proliferation of proneural GBM tumorspheres
(A) Representative images of proneural GBM tumorspheres grown in defined culture medium.
(B) Representative images of proneural GBM tumorspheres grown in defined medium with physiological concentrations of insulin but supplemented with IGF-II (LI + EGF + IGF-II).
(C) Representative images of proneural GBM tumorspheres grown in LI + EGF + IGF-II with a function blocking INSR Ab (IR Ab).
(D) Average number of tumorspheres/field ±SEM after 7 days of growth under different conditions (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(E) Average sizes of tumorspheres/field ±SEM after 7 days of growth under different conditions (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(F) Cell viability assay in proneural tumorspheres grown for 7 days in defined growth media (HI + EGF, LI + EGF + IGF-II, and LI + EGF + IGF-II + IR Ab) (p = not significant [n.s]).
(G) Extreme Limiting Dilution Assay (ELDA) performed on proneural tumorspheres grown for 21 days in vitro in the defined media conditions (HI + EGF, LI + EGF + IGF-II, and LI + EGF + IGF-II + IR Ab).
Data are representative of three independent experiments. Scale bars represent 50 μm. HI, high insulin; IR, INSR; LI, low insulin.
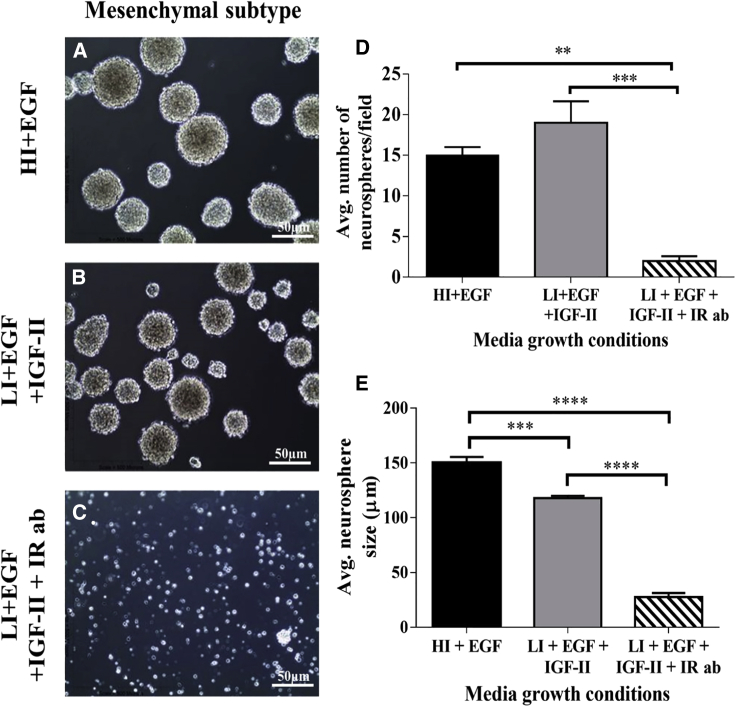
In mesenchymal tumorspheres supplemented with the INSR blocking Ab, we observed a 7.5-fold reduction in tumorsphere number compared with media with EGF (HI + EGF versus LI + IGF-II + INSR Ab: 15 ± 1 versus 2 ± 0.5; one-way ANOVA, p < 0.01) and a 9.5-fold reduction in tumorsphere number compared with media supplemented with EGF + IGF-II (LI + EGF + IGF-II versus LI + EGF + IGF-II + INSR Ab: 19 ± 2.6 versus 2 ± 0.5; one-way ANOVA, p < 0.001) formed on passage (Figures 5A–5D). As with the proneural GBM tumorspheres, blocking the INSR reduced sphere size in mesenchymal tumorspheres by 5.3-fold in EGF-supplemented media (HI + EGF versus LI + EGF + IGF-II + IR Ab: 150 ± 5.5 versus 28 ± 3.7; one-way ANOVA, p < 0.0001) and by 4.2-fold in EGF + IGF-II-supplemented media (LI + EGF + IGF-II versus LI + EGF + IGF-II + INSR Ab: 118 ± 2.03 versus 28 ± 3.7; one-way ANOVA, p < 0.0001) (Figure 5E). These data support the conclusion that the INSR maintains the stemness of proneural and mesenchymal GBM cancer stem cells.
Figure 5.
A function blocking Ab to the INSR decreases self-renewal and proliferation of mesenchymal GBM tumorspheres
(A) Representative images of mesenchymal GBM tumorspheres in medium with superphysiological levels of insulin (HI + EGF).
(B) Representative images of mesenchymal GBM tumorspheres grown with physiological levels of insulin + IGF-II (LI + EGF + IGF-II).
(C) Representative images of mesenchymal GBM tumorspheres grown in LI + EGF + IGF-II with a function blocking IR Ab.
(D) Average number of tumorspheres/field ±SEM after 7 days of growth under different conditions (∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
(E) Average size of tumorspheres/field ±SEM after 7 days of growth under different conditions (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Data are representative of three independent experiments. Scale bars represent 50 μm.
In breast and colorectal cancers, IGF1R promotes the proliferation of the progenitors within the tumor (Farabaugh et al., 2015; Rota et al., 2014). As shown in Table 2, there is an increase in the ratio of INSRA to IGF1R in the GBM spheres compared with normal human NSCs. To establish whether the INSR promotes growth of the tumorspheres, we grew proneural GBM tumorspheres in medium containing the INSR blocking Ab. When the sphere sizes were measured as an index of growth, inhibiting the INSR reduced sphere sizes by 3.5-fold in EGF-supplemented media (HI + EGF versus LI + IGF-II + INSR Ab: 77 ± 1.1 versus 22 ± 3.17; one-way ANOVA, p < 0.0001) and by 3.8-fold in EGF + IGF-II media supplemented with the INSR blocking Ab for the proneural subtype (LI + EGF + IGF-II versus LI + EGF + IGF-II + INSR Ab: 85 ± 2.64 versus 22 ± 3.17; one-way ANOVA, p < 0.001) (Figures 4A–4C and 4E). Similarly, inhibiting the INSR in the mesenchymal subtype reduced tumorsphere growth by 5.3-fold in EGF-supplemented media (HI + EGF versus LI + EGF + IGF-II + INSR Ab: 150 ± 5.5 versus 28 ± 3.7; one-way ANOVA, p < 0.0001) and by 4.2-fold in EGF + IGF-II media supplemented with the INSR blocking Ab (LI + EGF + IGF-II versus LI + EGF + IGF-II + INSR Ab: 118 ± 2.03 versus 28 ± 3.7; one-way ANOVA, p < 0.0001) (Figures 5A–5C and 5E). The addition of a non-specific IgG had no effect on tumorsphere size or number (see Figure S7).
Discussion
Although the INSRA isoform of the INSR is known to regulate NSCs in vitro and is elevated in several tumor types, INSR function in regulating NSC homeostasis in vivo has not been previously investigated. The findings reported here demonstrate that the INSR is necessary to maintain a subset of normal adult NSCs. Deleting the Insr in SVZ-derived NSPs in vitro decreased the numbers of NSCs with a corresponding increase in a subset of multipotential and bipotential neural progenitors. Consistent with the reduction in NSCs, Insr deletion reduced the numbers of secondary neurospheres after passage. Deleting the Insr in NSCs in vivo similarly reduced the number of NSCs located in the medial SVZ with a corresponding expansion in neural progenitors accompanied by olfactory deficits. Interestingly, our findings revealed that the INSR uniquely regulates NSCs in the SVZ because NSC numbers in the SGZ of the hippocampus and along the lateral wall of the third ventricle (α-tanycytes) were unaffected by Insr deletion. Altogether our results demonstrate that the INSR is an essential receptor for self-renewal of a subset of NSCs, and that its loss results in their depletion with functional consequences.
INSRA is necessary to exert effects of IGF-II on adult stem cell self-renewal
Our prior data strongly support the conclusion that it is the INSRA isoform, and not the metabolic INSRB isoform, that is responsible for SVZ NSC self-renewal. Laser microdissection studies revealed that the Insra is more highly expressed than the Igf1r within the medial aspect of the SVZ, whereas the Igf1r is more highly expressed than Insra in the lateral SVZ, which is the progenitor cell domain of the SVZ (Ziegler et al., 2012). By contrast, the Insrb was undetectable in the SVZ. The Insra also is the predominant isoform expressed in neurospheres and, as cells become lineage restricted, the levels of Insra decrease (Ziegler et al., 2012). There are numerous lines of evidence supporting the conclusion that IGF-II is the endogenous ligand that signals through the INSRA to maintain the NSCs in the adult SVZ. Our prior studies showed that IGF-II promotes stemness of SVZ-derived NSCs in vitro through the INSRA. Moreover, the alterations resulting from our in vivo deletion of the Insr in SVZ NSCs largely phenocopy the effect of Igf2 deletion from the adult SVZ (Ziegler et al., 2019). Igf2 deletion decreased NSCs in the SVZ, expanded the transit-amplifying progenitors, and caused olfaction deficits. These data support the conclusion that an IGF-II/INSRA signaling loop regulates NSC homeostasis (Ziegler et al., 2012, 2014, 2015, 2019). Although insulin also binds to the INSRA with high affinity, the biological effects of insulin versus IGF-II on INSR intracellular signaling have not been studied in NSCs. Studies using murine fibroblasts engineered to express only human INSRA or INSRB have established that the gene expression profiles for the two ligands differ (Pandini et al., 2003). Other studies have shown that activating INSRA stimulates cell proliferation and promotes survival, whereas the INSRB regulates metabolism (Belfiore et al., 2009; Belfiore and Malaguarnera, 2011; Sacco et al., 2009; Sciacca et al., 2003). Data elucidating the mechanism for IGF-II binding to INSRA (Alvino et al., 2011; Andersen et al., 2017; Rajapaksha et al., 2012) point to a role for IGF-II via INSRA that is distinct from the actions of insulin (Rajapaksha and Forbes, 2015). Using the neurosphere assay to analyze NSC self-renewal across passage, we demonstrated that IGF-II, but not insulin or IGF-I, maintained SVZ NSCs and also expanded their numbers, suggesting that IGF-II promotes symmetrical versus asymmetrical divisions (Ziegler et al., 2012).
IGF-II acts through both the IGF1R and the INSR to support adult NSC self-renewal
It is becoming increasingly clear that there is not a single NSC, but rather that multiple NSC subtypes exist in the adult CNS. The lack of a phenotype in the SGZ NSCs with Insr deletion provides yet another example of NSC heterogeneity. Our prior data taken together with the data presented here show that IGF-II acts through the INSRA to maintain SVZ NSCs, whereas the INSR is dispensable for SGZ NSC self-renewal. There are a number of studies that show that IGF-II is important for hippocampal neurogenesis (Ferron et al., 2015; Ouchi et al., 2013; Tao et al., 2015). Our recent characterization of adult neurogenesis on adult deletion of Igf2 showed that the number of NSCs in the SGZ declined with loss of Igf2. Moreover, deleting Igf2 in adulthood also caused deficits in spatial learning and increased anxiety (Ziegler et al., 2019). Bracko et al. (2012) suggested that SGZ NSC self-renewal is regulated by IGF-II via IGF1R and AKT-dependent signaling. Consistent with their findings, deleting the Insr in NSCs had no effect on label-retaining cells in the SGZ or on spatial learning and memory ability or anxiety. It should be noted that the original genetic experiments on Igf2 demonstrated that this growth factor promotes fetal growth through both the IGF1R and INSR (Baker et al., 1993; Liu et al., 1993; Louvi et al., 1997). Altogether these data support the conclusion that IGF-II acts through the INSR in the SVZ NSCs, while it signals through the IGF1R in the SGZ NSCs.
There are multiple types of adult stem cell in the SVZ
Accumulating evidence shows that there are multiple subtypes of NSC within the adult SVZ. Here we show that deleting the Insr within the NSPs in vitro decreased the NSC population by ∼70%. Similarly, Insr deletion from the NSCs in vivo resulted in a ∼70% reduction in label-retaining cells in the SVZ. Interestingly, deleting Igf2 in the adult mouse produced a ∼50% decrease in the percentage of NSCs (Ziegler et al., 2019). Altogether these observations indicate that IGF-II signaling via the INSR is necessary to maintain a subset of NSCs within the adult SVZ. Other studies have described subsets of NSCs within the SVZ. Azim et al. (2014) showed that glutamatergic neuronal progenitors and oligodendrocyte precursors are derived from the dorsal SVZ under the control of Wnt/β-catenin signaling, while GABAergic neural precursors are derived from the lateral/ventral SVZ and are independent of Wnt/β-catenin signaling (Azim et al., 2014). Merkle et al. (2007) demonstrated that spatially segregated NSPs give rise to different OB neurons. Using Ad-Cre-GFP virus to label SVZ radial glia dorsally and ventrally, the authors showed that the NSCs in the dorsal region produced tyrosine hydroxylase-positive periglomerular cells, while the ventrally located labeled NSCs gave rise to calbindin-positive periglomerular neurons. These data support the conclusion that there is significant functional heterogeneity among the SVZ NSCs.
Heterogeneity in INSR expression also has been documented in NSPs across species. Pollen et al. (2019) showed that INSR was higher at the protein level and mRNA level in human radial glia compared with primate radial glial cells. This finding complemented an earlier study showing strong immunoreactivity for the downstream kinase mammalian target of rapamycin (mTOR) and its substrate the ribosomal protein S6 in outer SVZ radial glia cells (Nowakowski et al., 2017). Together these findings and ours suggest that INSR may be promoting self-renewal of human outer SVZ cells, which is required for the dramatic expansion of the upper neocortical layers that is characteristic of the human brain.
INSRA signaling in SVZ NSCs is not necessary for cell survival
The INSR is important for the survival of cells throughout the body; therefore, the loss of the INSR in the NSCs might have resulted in their premature demise. However, instead of NSC loss, Insr deletion led to a loss of stemness in the neurosphere assay with an increase in the more rapidly dividing progenitors. These findings were corroborated by flow cytometric experiments that revealed a decrease in NSC number with a compensatory increase in the other intermediate progenitors, indicating that the decreased NSC number was not due to cell death. Similarly, in vivo deletion of the Insr further demonstrated that SVZ NSCs were reduced with a corresponding increase in OB neurogenesis.
The IGF-II/INSRA signaling loop regulates stem phenotypes in tumor CSCs
Accumulating evidence over the last few decades has shown that the INSR is abnormally expressed in many malignancies, such as breast, colon, lung, and thyroid (Chettouh et al., 2013; Kim et al., 2012; Vella et al., 2001; Vigneri et al., 2015; Wang et al., 2013). In particular, the INSRA is often more highly expressed than INSRB in these cancers. A number of studies have reported an IGF-II/INSRA autocrine loop that drives tumor progression (Sciacca et al., 1999; Vella et al., 2002). Unlike the binding of insulin, IGF-II binding to INSRA does not cause internalization of the receptor, leading to prolonged mitogenic activation allowing cancer cell proliferation (Morcavallo et al., 2012). Other studies have shown that a switch in the ratio of Insr isoforms leads to the malignant transformation of cancer cells. Chettouh et al. (2013) showed that in hepatocellular carcinoma, a switch in the ratio of Insra/Insrb is caused by alterations in the splice enzymes involved in Insr mRNA splicing. Our data mining results reveal that there is a similar overexpression of the enzymes involved in Insr mRNA splicing in proneural and mesenchymal GBM, leading to higher expression of the mitogenic Insra. This would contribute to an IGF-II/INSRA autocrine signaling loop to promote the self-renewal of GBM stem cells. In addition to documented changes in levels of these components of the IGF-II system in cancer, there also are studies that have implicated the IGF-II mRNA binding proteins. These RNA binding proteins increase the stability of IGF2 mRNA, resulting in production of higher levels of IGF-II protein. Interestingly, IGF-II binding protein-2 is overexpressed in GBMs, and increased expression of IGF-II binding protein-2 correlates with a poor prognosis for proneural GBMs (Cao et al., 2018).
Our findings here, taken together with our previously published data, support the conclusion that the INSRA is essential for maintenance of a subset of NSCs, and that the aggressive subtypes of GBM, i.e., proneural and mesenchymal, use an IGF-II/INSRA signaling loop to enhance self-renewal of tumor stem cells. These findings raise the possibility that IGF-II and INSRA should be considered as targets for new therapeutics either to enhance normal NSC function or to inhibit stem cell populations in GBMs.
Experimental procedures
Neurosphere propagation and quantification
Neurospheres were generated by enzymatically dissociating the periventricular region of Insrfl/fl mouse pups (post-natal days [P] 4–5) (see supplemental methods). Ad carrying the Cre recombinase gene with a GFP reporter (catalog [Cat] #1700; Vector Biolabs) or Ad carrying only GFP reporter (Cat #1060) (control) were infected at an MOI of 1,000 during passage into secondary spheres. For in vitro induction of recombination, NestinCreERT2−/+tdT+/+Insrfl/fl-generated secondary spheres were allowed to grow for 4 DIV, and then 0.5 μM 4-hydroxy tamoxifen (Sigma-Aldrich) or vehicle (ethanol) was added to the media for 24 h. Prior to counting the spheres, the plates were shaken to ensure uniform distribution of the spheres in the well. The number of neurospheres in six random fields under 4× magnification was determined for each well (Alagappan et al., 2009). Within each field the total number of red spheres (ΔNSC-IRKO) was counted, and the data were represented as the percentage of red spheres per field. ImageJ software was used to measure the size of the spheres and represented as average size of neurospheres between red ΔNSC-IRKO and WT red spheres (ΔNSC-IR WT). The results were obtained from three independent experiments.
Animals
All experiments were performed in accordance with protocols approved by the institutional animal care and use committee of Rutgers-New Jersey Medical School and in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80–23) revised in 1996. Tamoxifen (T5648; Sigma) was dissolved in a corn oil/ethanol (9:1) mixture and given intraperitoneally (i.p.) to mice 4–5 weeks of age at 75 mg/kg for 5 days. Mouse strains included Insrfl/fl mice, where exon 4 of the Insr is flanked by loxP sites (B6.129S4(FVB)-Insrtm1Khn/J) and a ΔNSC-IRKO mouse line that was established by crossing nestin-CreERT2 mice, where Cre expression is driven by the second intron of the nestin promoter (https://www.jax.org/strain/016261), with Insrfl/fl mice previously mated with a tdT reporter mouse line (B6;129S6-Gt(ROSA)26Sortm9(CAG-tdT)Hze/J; Jax mouse stock #007905). From these crosses, we generated NesCreERT2+/−tdT+/+Insr fl/fl mice. Both male and female heterozygous Cre mice were used for the experiments. ΔNSC-IR WT mice were the same genotype but given vehicle instead of tamoxifen. Progeny from breedings consisted of homozygous tdT and homozygous floxed Insr littermates that were either heterozygous Cre or Cre negative (WT). Cre-negative mice given tamoxifen were used in tdt reporter expression experiments as indicated.
Behavioral tests
The buried food test for mouse olfaction and Morris water maze (hidden platform) were conducted as for our previous study (Ziegler et al., 2019). Elevated plus maze to test anxiety was performed according to Walf and Frye (2007) (see supplemental methods).
Statistical analyses
All statistical analyses were performed using GraphPad Prism software. Unpaired t test was used for analysis of two groups. One-way ANOVA with Tukey’s post hoc was used for more than two groups. In all cases, p < 0.05 was considered to be statistically significant.
Subtyping GBM samples and in silico data mining
GBM tumorspheres were obtained from Dr. Nikos Tapinos (Brown University). The subtype was confirmed using a combination of pathology reports and a PCR mutation panel from SA Biosciences (qBiomarker Somatic Mutation PCR Array: Human Brain Cancers). The characterization, tumor-initiating ability, and transcriptomic profile of these glioma stem cells have been published previously (Zepecki et al., 2019, 2021).
RNAseqV2 data matrix was obtained from the level 3 data of all batches available for public access in the TCGA database. For each TCGA barcode, the RSEM normalized genes file was downloaded. These files were consolidated onto one Excel spreadsheet. Using the supplemental information in Brennan et al. (2013), the files from each TCGA barcode were classified to the subtype of GBM that they belonged to according to proneural and mesenchymal GBM subtypes. Genes measured were normalized to RNA-seq data of normal human astrocytes expression data from a Stanford database (http://web.stanford.edu/group/barres_lab/brain_rnaseq.html).
Expression analysis of RNA-seq data
RNA was obtained from stem cell cultures isolated from two GBM tumors (code-named GB2, WCR8) and from one normal hNSC culture. Next-generation RNA-seq was performed in the Epigenomics Core Facility of the Albert Einstein College of Medicine (NY, USA) using an Illumina HiSeq2500 machine (see supplemental methods).
Tumorsphere propagation and quantification
GBM stem cells from the two subtypes of GBM, namely, GB2, proneural, and WCR8, mesenchymal, were propagated; collected after 5–7 days; dissociated; and plated at 1 × 105 cells/mL in the same media with HI (4.4 μM) or LI (4.4 nM) plus/minus human recombinant IGF-II (224 ng/mL) and IgG isotype Ab (Santa Cruz) or with IGF-II and INSR blocking Ab. Tumorspheres were quantified as previously described. Viability was assessed by trypan blue dye exclusion method using ViCell automated cell counter. Clonal tumorsphere potential was evaluated using the Extreme Limiting Dilution Assay (Nguyen et al., 2018). Cells were grown at 50, 25, 12.5, 6.25, 3.12, and 1.56 cells/cm2 for 21 days under the culture conditions described above. Inequality in frequency between multiple groups was analyzed by likelihood ratio tests using the asymptotic chi-square approximation to the log ratio (Hu and Smyth, 2009).
Author contributions
Conceptualization, S.W.L., T.L.W., and N.T.; methodology, S.C., F.J.V., A.F., S.W.L., T.L.W., and N.T.; investigation, S.C., D.R., K.D., Y.C., K.M.S., E.F., and A.F.; formal analysis, K.D.; writing – original draft, S.C. and S.W.L.; writing – review & editing, S.C., S.W.L., T.L.W., and N.T.; funding acquisition, S.W.L., T.L.W., A.F., and N.T.; supervision, S.W.L., T.L.W., A.F., and N.T.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
This work was supported by grant R21 NS076874 (awarded to S.W.L. and T.L.W.); National Cancer Institute (NCI) grant R21CA235415 and a Warren Alpert Foundation grant (to N.T.); and NIH grants GM136357 and AI141816 (to A.F.).
Published: May 5, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2022.04.007.
Supplemental information
Data and code availability
Details for the use of code supporting RNA sequencing (RNA-seq) data analysis of the current study are available from Dr. Nikos Tapinos on request. The RNA-seq data discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus database and are accessible through GEO Series accession number GEO: GSE11422.
References
- Alagappan D., Lazzarino D.A., Felling R.J., Balan M., Kotenko S.V., Levison S.W. Brain injury expands the numbers of neural stem cells and progenitors in the SVZ by enhancing their responsiveness to EGF. ASN NEURO. 2009;1:e00009. doi: 10.1042/an20090002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvino C.L., Ong S.C., McNeil K.A., Delaine C., Booker G.W., Wallace J.C., Forbes B.E. Understanding the mechanism of insulin and insulin-like growth factor (IGF) receptor activation by IGF-II. PLoS One. 2011;6:e27488. doi: 10.1371/journal.pone.0027488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M., Norgaard-Pedersen D., Brandt J., Pettersson I., Slaaby R. IGF1 and IGF2 specificities to the two insulin receptor isoforms are determined by insulin receptor amino acid 718. PLoS One. 2017;12:e0178885. doi: 10.1371/journal.pone.0178885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres S.F., Simmons J.G., Mah A.T., Santoro M.A., Van Landeghem L., Lund P.K. Insulin receptor isoform switching in intestinal stem cells, progenitors, differentiated lineages and tumors: evidence that IR-B limits proliferation. J. Cell Sci. 2013;126:5645–5656. doi: 10.1242/jcs.132985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim K., Fischer B., Hurtado-Chong A., Draganova K., Cantu C., Zemke M., Sommer L., Butt A., Raineteau O. Persistent wnt/β-catenin signaling determines dorsalization of the postnatal subventricular zone and neural stem cell specification into oligodendrocytes and glutamatergic neurons. Stem Cells. 2014;32:1301–1312. doi: 10.1002/stem.1639. [DOI] [PubMed] [Google Scholar]
- Baker J., Liu J.P., Robertson E.J., Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75:73–82. doi: 10.1016/s0092-8674(05)80085-6. [DOI] [PubMed] [Google Scholar]
- Belfiore A. The role of insulin receptor isoforms and hybrid insulin/IGF-I receptors in human cancer. Curr. Pharm. Des. 2007;13:671–686. doi: 10.2174/138161207780249173. [DOI] [PubMed] [Google Scholar]
- Belfiore A., Frasca F., Pandini G., Sciacca L., Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 2009;30:586–623. doi: 10.1210/er.2008-0047. [DOI] [PubMed] [Google Scholar]
- Belfiore A., Malaguarnera R. Insulin receptor and cancer. Endocr. Relat. Cancer. 2011;18:R125–R147. doi: 10.1530/erc-11-0074. [DOI] [PubMed] [Google Scholar]
- Belfiore A., Malaguarnera R., Vella V., Lawrence M.C., Sciacca L., Frasca F., Morrione A., Vigneri R. Insulin receptor isoforms in physiology and disease: an updated view. Endocr. Rev. 2017;38:379–431. doi: 10.1210/er.2017-00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher J., Kleinridders A., Kahn C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb. Perspect. Biol. 2014;6:a009191. doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracko O., Singer T., Aigner S., Knobloch M., Winner B., Ray J., Clemenson G.D., Jr., Suh H., Couillard-Despres S., Aigner L., et al. Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J. Neurosci. 2012;32:3376–3387. doi: 10.1523/jneurosci.4248-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buono K.D., Vadlamuri D., Gan Q., Levison S.W. Leukemia inhibitory factor is essential for subventricular zone neural stem cell and progenitor homeostasis as revealed by a novel flow cytometric analysis. Dev. Neurosci. 2012;34:449–462. doi: 10.1159/000345155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Mu Q., Huang H. The roles of insulin-like growth factor 2 mRNA-binding protein 2 in cancer and cancer stem cells. Stem Cells Int. 2018;2018:1–15. doi: 10.1155/2018/4217259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran U.R., Luthra S., Santana-Santos L., Mao P., Kim S.H., Minata M., Li J., Benos P.V., DeWang M., Hu B., et al. Gene expression profiling distinguishes proneural glioma stem cells from mesenchymal glioma stem cells. Genom. Data. 2015;5:333–336. doi: 10.1016/j.gdata.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.Y., Stern S.A., Garcia-Osta A., Saunier-Rebori B., Pollonini G., Bambah-Mukku D., Blitzer R.D., Alberini C.M. A critical role for IGF-II in memory consolidation and enhancement. Nature. 2011;469:491–497. doi: 10.1038/nature09667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettouh H., Fartoux L., Aoudjehane L., Wendum D., Claperon A., Chretien Y., Rey C., Scatton O., Soubrane O., Conti F., et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013;73:3974–3986. doi: 10.1158/0008-5472.can-12-3824. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A. Genetics of mouse growth. Int. J. Dev. Biol. 1998;42:955–976. [PubMed] [Google Scholar]
- Farabaugh S.M., Boone D.N., Lee A.V. Role of IGF1R in breast cancer subtypes, stemness, and lineage differentiation. Front. Endocrinol. 2015;6:59. doi: 10.3389/fendo.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron S.R., Radford E.J., Domingo-Muelas A., Kleine I., Ramme A., Gray D., Sandovici I., Constancia M., Ward A., Menheniott T.R., Ferguson-Smith A.C. Differential genomic imprinting regulates paracrine and autocrine roles of IGF2 in mouse adult neurogenesis. Nat. Commun. 2015;6:8265. doi: 10.1038/ncomms9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca F., Pandini G., Scalia P., Sciacca L., Mineo R., Costantino A., Goldfine I.D., Belfiore A., Vigneri R. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol. Cell. Biol. 1999;19:3278–3288. doi: 10.1128/mcb.19.5.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo C., Capristo M., Manara M.C., Mancarella C., Landuzzi L., Belfiore A., Lollini P.L., Picci P., Scotlandi K. Metformin as an adjuvant drug against pediatric sarcomas: hypoxia limits therapeutic effects of the drug. PLoS One. 2013;8:e83832. doi: 10.1371/journal.pone.0083832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Ma Y., Sinyuk M., Loganathan S., Thompson R.C., Sarkaria J.N., Chen W., Lathia J.D., Mobley B.C., Clark S.W., Wang J. Insulin-mediated signaling promotes proliferation and survival of glioblastoma through Akt activation. Neuro Oncol. 2016;18:48–57. doi: 10.1093/neuonc/nov096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Kim E.S., Liu D., Lee J.J., Solis L., Behrens C., Lippman S.M., Hong W.K., Wistuba I.I., Lee H.Y. Prognostic impact of insulin receptor expression on survival of patients with nonsmall cell lung cancer. Cancer. 2012;118:2454–2465. doi: 10.1002/cncr.26492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.A., Bedont J.L., Pak T., Wang H., Song J., Miranda-Angulo A., Takiar V., Charubhumi V., Balordi F., Takebayashi H., et al. Tanycytes of the hypothalamic median eminence form a diet-responsive neurogenic niche. Nat. Neurosci. 2012;15:700–702. doi: 10.1038/nn.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen M.K., Zappaterra M.W., Chen X., Yang Y.J., Hill A.D., Lun M., Maynard T., Gonzalez D., Kim S., Ye P., et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.P., Baker J., Perkins A.S., Robertson E.J., Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75:59–72. doi: 10.1016/s0092-8674(05)80084-4. [DOI] [PubMed] [Google Scholar]
- Llorens-Martin M., Trejo J.L. Multiple birthdating analyses in adult neurogenesis: a line-up of the usual suspects. Front. Neurosci. 2011;5:76. doi: 10.3389/fnins.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhia K.A., Tienchaiananda P., Haluska P. Understanding the key to targeting the IGF Axis in cancer: a biomarker assessment. Front. Oncol. 2015;5:142. doi: 10.3389/fonc.2015.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan A., Gonzalez A.M., Hill D.J., Berry M., Gregson N.A., Baird A. Coordinated pattern of expression and localization of insulin-like growth factor-II (IGF-II) and IGF-binding protein-2 in the adult rat brain. Endocrinology. 1994;135:2255–2264. doi: 10.1210/endo.135.5.7525264. [DOI] [PubMed] [Google Scholar]
- Louvi A., Accili D., Efstratiadis A. Growth-promoting interaction of IGF-II with the insulin receptor during mouse embryonic development. Dev. Biol. 1997;189:33–48. doi: 10.1006/dbio.1997.8666. [DOI] [PubMed] [Google Scholar]
- Malaguarnera R., Morcavallo A., Giuliano S., Belfiore A. Thyroid cancer development and progression: emerging role of cancer stem cells. Minerva Endocrinol. 2012;37:103–115. [PubMed] [Google Scholar]
- Massimino M., Sciacca L., Parrinello N.L., Scalisi N.M., Belfiore A., Vigneri R., Vigneri P. Insulin receptor isoforms differently regulate cell proliferation and apoptosis in the ligand-occupied and unoccupied state. Int. J. Mol. Sci. 2021;22:8729. doi: 10.3390/ijms22168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle F.T., Mirzadeh Z., Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcavallo A., Genua M., Palummo A., Kletvikova E., Jiracek J., Brzozowski A.M., Iozzo R.V., Belfiore A., Morrione A. Insulin and insulin-like growth factor II differentially regulate endocytic sorting and stability of insulin receptor isoform A. J. Biol. Chem. 2012;287:11422–11436. doi: 10.1074/jbc.m111.252478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.P., Daniel P.M., Filiz G., Mantamadiotis T. Investigating neural stem cell and glioma stem cell self-renewal potential using extreme limiting dilution analysis (ELDA) Bio Protoc. 2018;8:e2991. doi: 10.21769/bioprotoc.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski T.J., Bhaduri A., Pollen A.A., Alvarado B., Mostajo-Radji M.A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S.J., Velmeshev D., et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science. 2017;358:1318–1323. doi: 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi Y., Banno Y., Shimizu Y., Ando S., Hasegawa H., Adachi K., Iwamoto T. Reduced adult hippocampal neurogenesis and working memory deficits in the Dgcr8-deficient mouse model of 22q11.2 deletion-associated schizophrenia can be rescued by IGF2. J. Neurosci. 2013;33:9408–9419. doi: 10.1523/jneurosci.2700-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandini G., Medico E., Conte E., Sciacca L., Vigneri R., Belfiore A. Differential gene expression induced by insulin and insulin-like growth factor-II through the insulin receptor isoform A. J. Biol. Chem. 2003;278:42178–42189. doi: 10.1074/jbc.m304980200. [DOI] [PubMed] [Google Scholar]
- Pollen A.A., Bhaduri A., Andrews M.G., Nowakowski T.J., Meyerson O.S., Mostajo-Radji M.A., Di Lullo E., Alvarado B., Bedolli M., Dougherty M.L., et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell. 2019;176:743–756.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitano S., Verfaillie C.M., Petryk A. Self-renewal of neural stem cells: implications for future therapies. Front. Physiol. 2013;4:49. doi: 10.3389/fphys.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapaksha H., Alvino C., McCarthy P., Forbes B.E. The insulin-like growth factor mutation database (IGFmdb) Growth Horm. IGF Res. 2012;22:158–166. doi: 10.1016/j.ghir.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Rajapaksha H., Forbes B.E. Ligand-binding affinity at the insulin receptor isoform-A and subsequent IR-A tyrosine phosphorylation kinetics are important determinants of mitogenic biological outcomes. Front. Endocrinol. 2015;6:107. doi: 10.3389/fendo.2015.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remboutsika E., Elkouris M., Iulianella A., Andoniadou C.L., Poulou M., Mitsiadis T.A., Trainor P.A., Lovell-Badge R. Flexibility of neural stem cells. Front. Physiol. 2011;2:16. doi: 10.3389/fphys.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins S.C., Stewart I., McNay D.E., Taylor V., Giachino C., Goetz M., Ninkovic J., Briancon N., Maratos-Flier E., Flier J.S., et al. α-Tanycytes of the adult hypothalamic third ventricle include distinct populations of FGF-responsive neural progenitors. Nat. Commun. 2013;4:2049. doi: 10.1038/ncomms3049. [DOI] [PubMed] [Google Scholar]
- Rota L.M., Albanito L., Shin M.E., Goyeneche C.L., Shushanov S., Gallagher E.J., LeRoith D., Lazzarino D.A., Wood T.L. IGF1R inhibition in mammary epithelia promotes canonical Wnt signaling and Wnt1-driven tumors. Cancer Res. 2014;74:5668–5679. doi: 10.1158/0008-5472.can-14-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco A., Morcavallo A., Pandini G., Vigneri R., Belfiore A. Differential signaling activation by insulin and insulin-like growth factors I and II upon binding to insulin receptor isoform A. Endocrinology. 2009;150:3594–3602. doi: 10.1210/en.2009-0377. [DOI] [PubMed] [Google Scholar]
- Scalia P., Giordano A., Williams S.J. The IGF-II-insulin receptor isoform-A autocrine signal in cancer: actionable perspectives. Cancers (Basel) 2020;12:366. doi: 10.3390/cancers12020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacca L., Costantino A., Pandini G., Mineo R., Frasca F., Scalia P., Sbraccia P., Goldfine I.D., Vigneri R., Belfiore A. Insulin receptor activation by IGF-II in breast cancers: evidence for a new autocrine/paracrine mechanism. Oncogene. 1999;18:2471–2479. doi: 10.1038/sj.onc.1202600. [DOI] [PubMed] [Google Scholar]
- Sciacca L., Prisco M., Wu A., Belfiore A., Vigneri R., Baserga R. Signaling differences from the A and B isoforms of the insulin receptor (IR) in 32D cells in the presence or absence of IR substrate-1. Endocrinology. 2003;144:2650–2658. doi: 10.1210/en.2002-0136. [DOI] [PubMed] [Google Scholar]
- Tao X., Jin G., Zou L., Li H., Qin J. IGF2 regulates neuronal differentiation of hippocampal radial glial cells in vitro. J. Cytol. Histol. 2015;6:1. [Google Scholar]
- Ulanet D.B., Ludwig D.L., Kahn C.R., Hanahan D. Insulin receptor functionally enhances multistage tumor progression and conveys intrinsic resistance to IGF-1R targeted therapy. Proc. Natl. Acad. Sci. U S A. 2010;107:10791–10798. doi: 10.1073/pnas.0914076107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega C.J., Peterson D.A. Stem cell proliferative history in tissue revealed by temporal halogenated thymidine analog discrimination. Nat. Methods. 2005;2:167–169. doi: 10.1038/nmeth741. [DOI] [PubMed] [Google Scholar]
- Vella V., Pandini G., Sciacca L., Mineo R., Vigneri R., Pezzino V., Belfiore A. A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. J. Clin. Endocrinol. Metab. 2002;87:245–254. doi: 10.1210/jcem.87.1.8142. [DOI] [PubMed] [Google Scholar]
- Vella V., Sciacca L., Pandini G., Mineo R., Squatrito S., Vigneri R., Belfiore A. The IGF system in thyroid cancer: new concepts. Mol. Pathol. 2001;54:121–124. doi: 10.1136/mp.54.3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velloso F.J., Kumari E., Buono K.D., Frondelli M.J., Levison S.W. Analyzing mouse neural stem cell and progenitor cell proliferation using EdU incorporation and multicolor flow cytometry. STAR Protoc. 2022;3:101065. doi: 10.1016/j.xpro.2021.101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneri P.G., Tirro E., Pennisi M.S., Massimino M., Stella S., Romano C., Manzella L. The insulin/IGF system in colorectal cancer development and resistance to therapy. Front. Oncol. 2015;5:230. doi: 10.3389/fonc.2015.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walf A.A., Frye C.A. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat. Protoc. 2007;2:322–328. doi: 10.1038/nprot.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamaitha S.E., Grybel K.J., Alanis-Lobato G., Gerri C., Ogushi S., McCarthy A., Mahadevaiah S.K., Healy L., Lea R.A., Molina-Arcas M., et al. IGF1-mediated human embryonic stem cell self-renewal recapitulates the embryonic niche. Nat. Commun. 2020;11:764. doi: 10.1038/s41467-020-14629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Cai S., Teng Z., Zhao X., Chen X., Bai X. Insulin therapy contributes to the increased risk of colorectal cancer in diabetes patients: a meta-analysis. Diagn. Pathol. 2013;8:180. doi: 10.1186/1746-1596-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang X., Wu Y., Han C. IGF-1R signaling is essential for the proliferation of cultured mouse spermatogonial stem cells by promoting the G2/M progression of the cell cycle. Stem Cells Dev. 2015;24:471–483. doi: 10.1089/scd.2014.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Crawley J.N. Simple behavioral assessment of mouse olfaction. Curr. Protoc. Neurosci. 2009 doi: 10.1002/0471142301.ns0824s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelentsova-Levytskyi K., Talmi Z., Abboud-Jarrous G., Capucha T., Sapir T., Burstyn-Cohen T. Protein S negatively regulates neural stem cell self-renewal through bmi-1 signaling. Front. Mol. Neurosci. 2017;10:124. doi: 10.3389/fnmol.2017.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepecki J.P., Karambizi D., Fajardo J.E., Snyder K.M., Guetta-Terrier C., Tang O.Y., Chen J.S., Sarkar A., Fiser A., Toms S.A., Tapinos N. miRNA-mediated loss of m6A increases nascent translation in glioblastoma. PLoS Genet. 2021;17:e1009086. doi: 10.1371/journal.pgen.1009086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepecki J.P., Snyder K.M., Moreno M.M., Fajardo E., Fiser A., Ness J., Sarkar A., Toms S.A., Tapinos N. Regulation of human glioma cell migration, tumor growth, and stemness gene expression using a Lck targeted inhibitor. Oncogene. 2019;38:1734–1750. doi: 10.1038/s41388-018-0546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. BMS-536924, an ATP-competitive IGF-1R/IR inhibitor, decreases viability and migration of temozolomide-resistant glioma cells in vitro and suppresses tumor growth in vivo. Onco Targets Ther. 2015;8:689–697. doi: 10.2147/ott.s80047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A.N., Schneider J.S., Qin M., Tyler W.A., Pintar J.E., Fraidenraich D., Wood T.L., Levison S.W. IGF-II promotes stemness of neural restricted precursors. Stem Cells. 2012;30:1265–1276. doi: 10.1002/stem.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A.N., Chidambaram S., Forbes B.E., Wood T.L., Levison S.W. Insulin-like growth factor-II (IGF-II) and IGF-II analogs with enhanced insulin receptor-a binding affinity promote neural stem cell expansion. J. Biol. Chem. 2014;289:4626–4633. doi: 10.1074/jbc.m113.537597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A.N., Levison S.W., Wood T.L. Insulin and IGF receptor signalling in neural-stem-cell homeostasis. Nat. Rev. Endocrinol. 2015;11:161–170. doi: 10.1038/nrendo.2014.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A.N., Feng Q., Chidambaram S., Testai J.M., Kumari E., Rothbard D.E., Constancia M., Sandovici I., Cominski T., Pang K., et al. Insulin-like growth factor II: an essential adult stem cell niche constituent in brain and intestine. Stem Cell Rep. 2019;12:816–830. doi: 10.1016/j.stemcr.2019.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Details for the use of code supporting RNA sequencing (RNA-seq) data analysis of the current study are available from Dr. Nikos Tapinos on request. The RNA-seq data discussed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus database and are accessible through GEO Series accession number GEO: GSE11422.