Abstract
Cells are equipped with numerous sensors that recognize nucleic acids, which probably evolved for defence against viruses. Once triggered, these sensors stimulate the production of type I interferons and other cytokines that activate immune cells and promote an antiviral state. The evolutionary conserved enzyme cyclic GMP–AMP synthase (cGAS) is one of the most recently identified DNA sensors. Upon ligand engagement, cGAS dimerizes and synthesizes the dinucleotide second messenger 2′,3′-cyclic GMP–AMP (cGAMP), which binds to the endoplasmic reticulum protein stimulator of interferon genes (STING) with high affinity, thereby unleashing an inflammatory response. cGAS-binding DNA is not restricted by sequence and must only be >45 nucleotides in length; therefore, cGAS can also be stimulated by self genomic or mitochondrial DNA. This broad specificity probably explains why the cGAS–STING pathway has been implicated in a number of autoinflammatory, autoimmune and neurodegenerative diseases; this pathway might also be activated during acute and chronic kidney injury. Therapeutic manipulation of the cGAS–STING pathway, using synthetic cyclic dinucleotides or inhibitors of cGAMP metabolism, promises to enhance immune responses in cancer or viral infections. By contrast, inhibitors of cGAS or STING might be useful in diseases in which this pro-inflammatory pathway is chronically activated.
Subject terms: Innate immunity, Acute kidney injury, Chronic kidney disease, Autoimmunity
The cyclic GMP–AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway not only is involved in host defence against infection but can lead to immune dysregulation. Here, the authors examine the biology and biochemistry of cGAS–STING and discuss its role in disease and potential approaches to therapeutic targeting.
Key points
Cyclic GMP–AMP synthase (cGAS) is an evolutionarily conserved cytosolic nucleic acid sensor that synthesizes the cyclic dinucleotide second messenger 2′,3′-cyclic GMP–AMP (cGAMP), which engages stimulator of interferon genes (STING) to trigger the production of inflammatory cytokines, including type I interferons.
Extracellular cGAMP is hydrolysed by transmembrane and soluble ectonucleotide pyrophosphatase/phosphodiesterase 1, and enters neighbouring cells via cGAMP importers. Transfer to adjoining and distant cells enables cGAMP to act as an immunotransmitter and modulate antiviral responses, antitumour immunity and tumour metastasis.
Activation of cGAS–STING by genomic or mitochondrial self DNA has been implicated in numerous autoinflammatory, autoimmune and neurodegenerative diseases, cell senescence and ageing, as well as in acute and chronic kidney injury.
The microbiome can stimulate or inhibit cGAS–STING via cGAS sensing of commensal DNA, microorganism-induced endogenous retroviruses, or self DNA released owing to infection-induced cell damage. These interactions can affect gut and skin homeostasis.
Modulating the proteins involved in the activation of the cGAS–STING pathway and its regulators provides therapeutic opportunities to attenuate or enhance cGAS–STING-driven inflammatory responses.
Introduction
Throughout evolution, from prokaryotes to mammals, host defence mechanisms have co-evolved to limit the deleterious effects of invading organisms. One such defence mechanism comprises three main components — cyclic GMP–AMP synthase (cGAS), a cyclic dinucleotide (CDN) and stimulator of interferon genes (STING)1. The structure and function of each of these three components have evolved in different ways in diverse organisms but the essential function of the cGAS–STING pathway is to detect and limit the propagation of foreign DNA. In mammals, the most prominent functions of this pathway in response to microbial nucleic acids is to generate inflammatory and antiviral proteins, and is dominated by the production of type I interferon (IFN-I)2. However, DNA binding to cGAS is not sequence specific; therefore, in addition to responding to pathogen-derived DNA, this pathway can generate an inflammatory response to self nucleic acids. Chronic activation of cGAS–STING by self DNA underlies rare autoinflammatory diseases such as Aicardi–Goutières syndrome and might also contribute to other disease processes related to autoimmunity, neurodegeneration, ageing and cancer3 (Fig. 1 and Table 1). For example, activation of cGAS–STING, predominantly by mitochondrial DNA (mtDNA), has been implicated in both acute and chronic kidney disease (CKD)4–7. However, deep knowledge of the biochemistry of the cGAS–STING cassette has also enabled investigators to harness this pathway to augment immune responses against tumours and reverse cancer-associated immune suppression (reviewed in refs8,9).
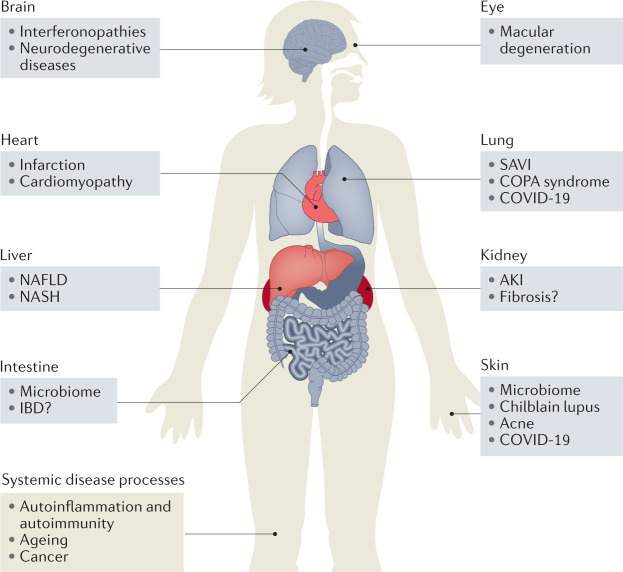
Fig. 1. Involvement of cGAS and/or STING in disease.
The cyclic GMP–AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway has been implicated in systemic disease processes, including autoimmunity, autoinflammation, ageing and cancer, as well as organ-specific diseases that target the eyes, brain, lungs, heart, kidneys, skin and intestines. cGAS–STING is also involved in the response to many viral infections, which can be systemic or local. AKI, acute kidney injury; COPA, coatomer protein subunit-α; IBD, inflammatory bowel disease; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; SAVI, STING-associated vasculopathy with onset in infancy.
Table 1.
Systemic diseases associated with cGAS–STING activation in humans or mice
| Disease names | Mouse models | Human diseases |
|---|---|---|
| AGS | Activation of cGAS–STING by loss of function mutations of DNases or nucleic acid metabolizing enzymes43,65,86,87 | Mutations in TREX1, RNASEH and other nucleic acid processing enzymes lead to the activation of cGAS and the production of IFNβ83,84 |
| SAVI | Gain-of-function mutations in Sting1 lead to SAVI-like diseases90–92 | Gain-of-function mutations in STING1 result in a systemic inflammatory disease with vasculitis and pulmonary fibrosis88,89 |
| SLE | cGAS–STING is activated in the lupus-prone TREX1 D18N mutant95 and Fcgr2b-knockout mice96 |
TREX1 heterozygous mutations associated with familial chilblain lupus100 cGAMP expression in PBMCs of 15% of patients with SLE and in microvesicles from SLE blood cells102,103 Mitochondria-rich red blood cells activate IFN-I through cGAS–STING in lupus monocytes108 |
| Ageing and senescence |
Activation of cGAS–STING-triggered immune senescence123,126,127 cGAS drives non-canonical inflammasome activation in age-related macular degeneration180 |
Activation of cGAS–STING triggered immune senescence123,126,127 cGAS drives non-canonical inflammasome activation in age-related macular degeneration180 |
AGS, Aicardi–Goutières syndrome; cGAS, cyclic GMP–AMP synthase; IFN-I, type I interferon; PBMCs, peripheral blood mononuclear cells; SAVI, STING-associated vasculopathy with onset in infancy; SLE, systemic lupus erythematosus; STING, stimulator of interferon genes.
In this Review, we first introduce the cytoplasmic cGAS–STING pathway and contrast cGAS with endosomal sensors from the Toll-like receptor (TLR) family. Next, we discuss the cell biology and biochemistry of cGAS–STING and highlight the specific role of the CDN 2′,3′-cyclic GMP–AMP (cGAMP) in spreading the inflammatory response to neighbouring cells. We then focus on the regulatory mechanisms that prevent activation by self DNA under steady-state conditions, and why and how cGAS–STING becomes activated in disease states. Finally, we consider therapeutic approaches designed to either attenuate cGAS–STING activation in inflammatory diseases or to stimulate inflammation in immune-suppressed states such as cancer.
Immune sensing and inflammation
In the field of medicine, inflammation is generally considered a pathological process, yet inflammation is a necessary process for survival. Inflammatory responses initiated by innate immune cells are not only triggered by viruses, bacteria and fungi as an essential response to eliminating infection, but inflammation is also necessary to repair tissue damage. TLRs are the best known innate sensors responsible for host immune defence. These receptors were first discovered in the fruit fly, Drosophila, and homologs were subsequently identified in all multicellular organisms (reviewed in ref.10). Although discovered more recently in mammals than TLRs2,11, ancient homologs of cGAS and STING have been detected in bacteria and in unicellular choanoflagellates, where they might have similar functions to those of the vertebrate proteins12–14. For example, in bacteria, cGAS activates a phospholipase to destroy the bacterial cell membrane and cause cell death. This mechanism probably functions as a bacterial defence against virus (that is, bacteriophage) reproduction14.
Although host sensors have evolved to detect and respond to microbial proteins, carbohydrates and lipids, these ligands can be modified by the microorganism to escape detection. By contrast, nucleic acids cannot be readily modified by the pathogens without affecting function. Unsurprisingly, host cells are equipped with multiple sensors for nucleic acid detection. In mammals, cytoplasmic nucleic acid sensors have a key role in the detection of pathogens that have breached membrane barriers15. Binding of double-stranded DNA to cGAS triggers a protein conformational change that results in the formation of cGAS dimers, activation of its catalytic site and synthesis of cGAMP (reviewed in ref.16; Fig. 2). cGAMP then functions as an endogenous second messenger and binds to STING, which resides in the endoplasmic reticulum (ER). After activation by cGAMP, STING is transferred from the ER to the Golgi via the ER–Golgi intermediate compartment17. Here, STING oligomerizes and acts as a platform to recruit and phosphorylate the serine/threonine-protein kinase TBK1. This kinase, in turn, phosphorylates the transcription factor interferon regulatory factor 3 (IRF3), which dimerizes and translocates to the nucleus to stimulate the expression of IFNβ. In addition to the canonical pathway of cGAS-mediated activation of STING, other DNA sensors (for example, the inflammasome receptor AIM2 and IFNγ-inducible protein 16, (IFI-16)), as well as RNA sensors (for example, retinoic acid-inducible gene-I, (RIG-I), and interferon-induced helicase C domain-containing protein 1 (MDA5)) signal through or cooperate with STING to drive IFN-I production. For example, etoposide-induced nuclear DNA damage in keratinocytes involves TNF receptor-associated factor 6 (TRAF6)-mediated assembly of K63-linked ubiquitin chains on STING and downstream activation of NF-κB, rather than IRF3 (ref.18). These non-canonical pathways of STING activation are independent of cGAS.
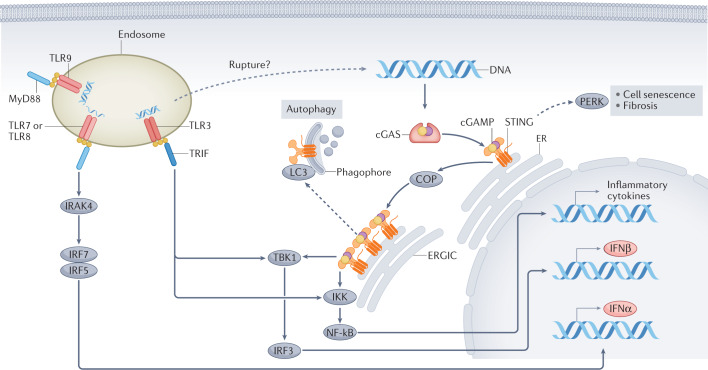
Fig. 2. Signalling pathways downstream of cGAS and of endosomal TLRs.
Binding to double-stranded DNA induces dimerization of cyclic GMP–AMP synthase (cGAS), which, in turn, leads to the synthesis of the second messenger 2′,3′-cyclic GMP–AMP (cGAMP). cGAMP binds with high affinity to stimulator of interferon genes (STING) leading to STING oligomerization and exit from the endoplasmic reticulum (ER); coatomer protein (COP) complexes facilitate the transport of STING. In the ER–Golgi intermediate compartment (ERGIC), STING recruits serine/threonine-protein kinase TBK1. The transcription factor interferon regulatory factor 3 (IRF3), which is a key TBK1 substrate, enters the nucleus following phosphorylation and promotes the transcription of IFNβ. Oligomerization of STING also enables activation of the inhibitor of nuclear factor-κB (NF-κB) kinase (IKK) complex, which promotes activation of the NF-κB transcription factor. NF-κB translocates to the nucleus where it promotes transcription of inflammatory cytokines tumour necrosis factor (TNF), IL-6 and IL-1β. By contrast, endosomal Toll-like receptors (TLRs) use the MyD88 or TRIF to generate platforms for downstream signal transduction. The DNA sensor TLR9 and the single-stranded RNA sensors, TLRs 7 and 8, activate IRF5 and IRF7 via Myd88 and IL-1 receptor-associated kinase 4 (IRAK4), which results in the synthesis of IFNα. The double-stranded RNA sensor TLR3, engages the TRIF platform, which leads to the activation of TBK1 and IKK, leading to IFNβ transcription. In non-classical pathway activation (dashed arrows), activated STING molecules on the ERGIC can bind to microtubule-associated protein 1 light chain 3 (MAP1LC3; also known as LC3) on the phagophore, which leads to STING degradation through autophagy. Moreover, STING activation in the ER might lead to PKR-like ER kinase (PERK) activation and subsequent cell senescence or fibrosis.
The IFN-I family comprises a large number of related proteins19 — 13 IFNα proteins, 1 IFNβ and several less-studied members, including IFNε, IFNκ, IFNτ and IFNω, all of which act on the IFN-I receptor (IFNAR). IFNβ can be produced by almost any cell type following stimulation by nucleic acids. Release of this cytokine serves to prime or amplify IFN-I production by other cells, including plasmacytoid dendritic cells, which are the main producers of IFNα20. IFN-I exerts its effector functions through the activation of >200 interferon-stimulated genes (ISGs), which in turn regulate translational control, and viral RNA stability and editing21. Of high relevance to autoimmunity, IFN-I is highly immunostimulatory and can induce the activation and maturation of dendritic cells, upregulation of MHC and co-stimulatory molecule expression, activation of natural killer (NK) cells, T cells and B cells, and suppression of regulatory T cells20. STING also forms a scaffold for activation of IKKε, leading to the nuclear translocation of NF-κB and the production of other inflammatory cytokines such as tumour necrosis factor (TNF), IL-6 and IL-1β, which have very broad activating potential both within and outside of the immune system (Fig. 2).
cGAS–STING and endosomal TLRs (E-TLRs; comprising TLRs 3, 7, 8 and 9) signal through overlapping downstream pathways to activate IFN-I and IFN-III, respectively, as well as inflammatory cytokines (Fig. 2). Only ligands that enter endosomes through phagocytosis or endocytosis can engage E-TLRs and their common downstream pathways require one adapter, either MyD88 or TRIF10. By contrast, the cGAS–STING pathway has several unique features: its DNA-sensing proteins are located in the cytoplasm, cGAS serves both as a DNA receptor and as a nucleotidyl transferase enzyme, and cGAS communicates with its effector platform STING at a distance by generating a CDN messenger (Fig. 2). Of note, although the E-TLR and cGAS–STING pathways are generally regarded as distinct, under certain conditions, membrane rupture might allow the escape of endosomal or lysosomal contents into the cytoplasm where they might be recognized by the cGAS–STING pathway22–24 (Fig. 2). The proteins RIG-I and MDA5 use the adaptor protein mitochondrial antiviral-signalling protein (MAVS) and act as cytoplasmic RNA sensors that can also stimulate the production of IFN-I25; however, they are not considered further here.
cGAS–STING biochemistry and cell biology
To understand how cGAS–STING functions in response to activating stimuli, it is useful to consider the subcellular localization of these proteins and their relationship to different organelles as well as the regulation of cGAMP as a mediator of cGAS activation.
Regulation and transport of cGAS
Subtle biochemical features unique to microbial DNA or RNA might influence the avidity of binding to intracellular sensors26 but both E-TLRs and cGAS can be activated by host and microbial nucleic acids. Consequently, self versus non-self discrimination is not particularly relevant to the regulation of these receptors. Rather, several strategies, such as rapid phagocytosis and efficient processing of host apoptotic cells, absence of nucleic acid sensors on most cell surface membranes and an abundance of extracellular and intracellular nucleases, prevent these pathways from promoting the development of autoimmune and autoinflammatory diseases (reviewed in ref.27).
When cGAS was initially characterized and identified as a cytoplasmic DNA sensor, overexpression of Flag-tagged cGAS indicated that the protein localized to the cytoplasm; a minor fraction could be detected in the nuclear or perinuclear regions2. However, subsequent studies showed that, under steady-state conditions, cGAS was predominantly detected in the nucleus, where it was tightly sequestered to and inhibited by chromatin in a high-avidity salt-resistant interaction28. This nuclear tethering prevents cGAS dimerization and activation, maintaining its resting state. Cryo-EM structures of the human cGAS–nucleosome core particle complex revealed that cGAS anchors to the nucleosome acidic patch on the histone H2A–H2B dimer through two conserved arginines. In this configuration, the three cGAS–DNA binding sites that are required for cGAS activation become inaccessible and cGAS dimerization is inhibited29–34. In addition, in the nucleus, the barrier to autointegration factor (BAF) protein competes with DNA for binding to cGAS35 and, in the cytoplasm, low levels of DNA fail to trigger cGAS activation owing to a threshold requirement for ligand-induced phase separation36. Notably, during mitosis, the N terminus of cGAS is hyperphosphorylated by kinases, including Aurora kinase B, thereby preventing its interaction with nuclear chromatin37. Other post-translational modifications of cGAS and their functional consequences are discussed elsewhere38,39. Of note, following DNA damage or under conditions of DNA instability, genomic DNA might form micronuclei that mis-segregate, rupture and interact with cGAS during mitosis40. Damaged DNA might also generate chromatin bridges through chromosome breakage–fusion–bridge cycles that are associated with weakened nuclear membranes and DNA release41. These findings suggest that cGAS might participate in the immunosurveillance of mitotic errors and/or the generation of micronuclei. Despite its nuclear location, cGAS activation and downstream signal transduction occur in the cytoplasm. Although the signals required to release cGAS from nuclear tethering remain unclear, a functional nuclear export signal (NES; 169LEKLKL174) has been identified on cGAS, which allows it to traffic from the nucleus to the cytoplasm42. cGAS can encounter genomic DNA in the cytoplasm under several circumstances (Fig. 3).
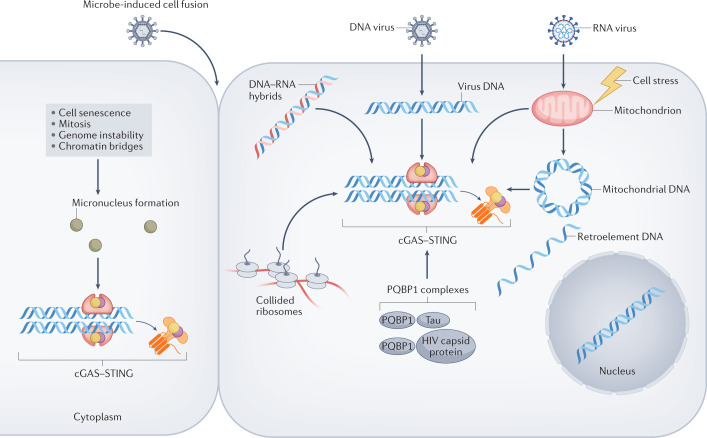
Fig. 3. Mechanisms of activation of the cGAS–STING pathway.
In the cytoplasm, cyclic GMP–AMP synthase (cGAS) can be activated by microbial or host DNA. The best-studied sources of cGAS-activating nucleic acids are DNA viruses and host mitochondria. However, several other DNA sources and cGAS-activating stimuli, such as endogenous retroelement DNA, micronuclei generated in senescent cells or owing to genomic instability, and the formation of chromatin bridges, have been reported40,41,44,195. Microbe-induced cell fusion also enables the release of chromatin into the cytosol and RNA viruses can promote cGAS activation through ribosomal collision79,81. Finally, certain DNA-binding proteins such as polyglutamine-binding protein 1 (PQBP1) might co-activate cGAS in combination with the HIV capsid protein77 or Tau138.
Nucleases such as DNase1, DNase1L3 and RNase1 provide an additional layer of protection against chronic responses to nucleic acids released from cells into the extracellular environment. For example, DNase1L3 contains a C-terminal tail that interacts with and degrades chromatin on microparticles43. Nucleases are also abundant within the cell; some are present in the cytoplasm, such as DNase III (also known as TREX1), and others are compartmentalized within endosomes (for example, DNase II). TREX1 is an abundant 3′–5′ exonuclease with a preference for single-stranded DNA and is thought to degrade DNA arising from retroelements and from mitochondria44. Furthermore, DNA–RNA hybrids that are produced by an infection with retroviruses or that occur in the life cycle of endogenous retroelements are degraded by RNaseH2 enzymes45. The action of these enzymes might be attenuated during inflammation because digestion is less efficient if the DNA is oxidized (that is, contains 8-OH-deoxyguanine), which can occur following the generation of reactive oxygen intermediates46.
cGAMP metabolism and transport
cGAMP is a small charged CDN that mediates cGAS activation and must also be regulated to avoid chronic activation of STING. CDNs were initially discovered as bacterial second messengers that have crucial roles in bacterial virulence, motility, metabolism and survival. Various bacteria produce CDNs, including cGMP–GMP, cAMP–AMP and 3′,3′-cGAMP47. These CDNs comprise two nucleotide monophosphates interlinked by phosphodiester bonds to form a cyclic structure. Unlike bacterial CDNs, which have two 3′–5′ bonds, mammalian cells produce a 2′,3′-cGAMP, which has 2′–5′ as well as 3′–5′ phosphodiester bonds. Although bacterial CDNs can also activate STING, 2′,3′-cGAMP binds to STING with a much higher affinity48, leading to cellular production of IFN-I and inflammatory cytokines (Fig. 2). Interestingly, Drosophila cells contain a STING orthologue that responds to exogenously administered 2′,3′-cGAMP by activating the transcription factor Relish, which is an orthologue of NF-κB49,50. The discovery of two cGAS-like receptors (cGLRs) in Drosophila helps to explain cGLR–STING antiviral immunity in the absence of IFN. cGLR1 is activated by double-stranded RNA to produce 3′,2′-cGAMP, whereas cGLR2 produces a combination of 2′,3′-cGAMP and 3′,2′-cGAMP, which shows the remarkable diversity of responses triggered by the cGAS–cGAMP–STING triad49,50.
The mechanisms that prevent accumulation of cGAMP and continuous stimulation of STING are incompletely defined. Whereas intracellular phosphodiesterases that hydrolyse cGAMP have not yet been identified51,52, evidence suggests that cells continuously export cGAMP into the extracellular space, where it might either be imported into neighbouring cells or hydrolysed by the multifunctional single-pass transmembrane protein ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1)51,52 (Fig. 4). The catalytic domain of ENPP1 faces the extracellular space enabling this enzyme to hydrolyse the extracellular pool of cGAMP into AMP and GMP52. ENPP1 is expressed on the surface of many cells, including skin cells, plasma cells and tumour cells, and can also be cleaved to form a highly bioactive serum protein53,54.
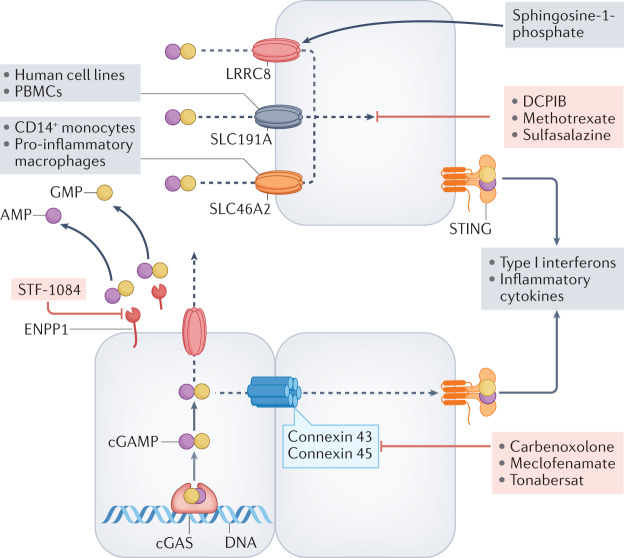
Fig. 4. Transport and metabolism of cGAMP.
Following activation, cyclic GMP–AMP synthase (cGAS) produces 2′,3′-cyclic GMP–AMP (cGAMP), which binds to stimulator of interferon genes (STING) within cells. The fate of intracellular cGAMP depends on its rate of synthesis, the cell type in which it is produced and the biological context. Excess cGAMP is transported out of cells and is hydrolysed by membrane-bound or soluble ectonucleotide pyrophosphatase phosphodiesterase 1 (ENPP1) to yield AMP and GMP in the extracellular space51,52. When production and export of cGAMP exceeds the catabolic capacity of ENPP1, cGAMP is imported into adjacent or distal cells through specific transporters, some of which can mediate bidirectional transport (LRRC8). The best-studied transporter in vascular endothelial cells is the volume-regulated anion (VRAC) channel LRRC8A:C heteromer, whereas the solute carrier SLC19A1 is functional in human cell lines and peripheral blood mononuclear cells, and SLC46A2 is active in CD14+ monocytes and pro-inflammatory macrophages60,62–64. In tumours, cGAMP can be transported between adjoining cells through gap junctions (for example, connexins 43 and 45)55,57,59, which can be blocked by the compounds shown. Several chemical compounds can block ENPP1 and cGAMP import as indicated. Sphingosine 1-phosphate can activate LRRC8 to potentiate cGAMP transport64.
cGAMP transport is highly relevant in inflammation, tumour immunity and autoimmunity. The intercellular transport of cGAMP was first recognized in virus-infected cells, where cGAMP trafficked between cells through gap junctions (that is, intercellular channels) formed by clusters of connexin proteins55. Connexins 43 and 45 were required for cGAMP transfer to adjoining cells but other connexins in this family of 20 proteins might also be involved. Of note, cGAMP transport via connexin 43 from virus-infected endothelial cells to macrophages amplified the antiviral response, which suggested that this mechanism promotes antiviral immunity56. Connexin-mediated transfer of cGAMP has also been reported to modulate inflammatory responses in cancer57, alcohol-induced hepatocyte injury58 and in inflamed joints56. Studies of tumour models and immunotherapy revealed that extracellular cGAMP acts as a soluble immune mediator in the tumour microenvironment (TME) to propagate the production of inflammatory cytokines, including IFN-I, and promote immune cell activation52. By acting as an immunotransmitter, cGAMP can modulate local antitumour immunity and affect tumour metastasis57,59.
In addition to transmission through channels that link two adjoining cells, cGAMP can enter the cytoplasm of surrounding cells via fusion of extracellular vesicles or through specialized cell-surface importers that can handle its negative charge (Fig. 4). Initial genome-wide CRISPR genetic screens identified two cGAMP importers — SLC19A1 and LRRC8. These large channel complexes enable cGAMP-mediated communication in non-adjacent or migrating cells. SLC19A1 is a carrier that transports folates and a variety of organic phosphates, such as thiamine derivatives and nucleotides, including cGAMP60,61. The requirement for SLC19A1 was demonstrated in primary human peripheral mononuclear cells and in lineage-specific cell lines (for example, in haematological but not epithelial or endothelial cell lines)60,62. LRRC8A is a crucial component of the ubiquitous volume-regulated anion channel (VRAC) and enables intercellular communication by transporting Cl− and osmolytes across the plasma membrane63. Unlike SLC19A1, which was dispensable for cGAMP transport in endothelial cells, the LRRC8A:C heteromer was required for cGAMP import into both immortalized human microvascular endothelial cells and primary human endothelial cells64. Depending on the concentration gradient of cGAMP, LRRC8A can act bi-directionally and export cGAMP out of endothelial cells that overexpress cGAS63. Although LRRC8A is ubiquitously expressed across cell types, other components of the heterodimeric VRAC complex (such as LRRC8C or LRRC8E) differentially contribute to cGAMP transport in a cell- and species-specific manner; LRRC8D inhibits cGAMP transport63,64. The discovery of LRRC8 as a cGAMP transporter enabled in vivo studies that revealed that cGAMP transport is required for clearance of herpes simplex virus 1 (HSV1)63. In 2021, SLC46A2 was identified as a third cGAMP importer that is specifically expressed in human CD14+ monocytes and in monocyte-derived macrophages62.
Further studies are required to elucidate the roles of different cGAMP transporters in disease states associated with cGAS–STING activation. Although cGAMP export and import by adjacent cells may be advantageous to protect against viral infection, persistent cGAMP spreading and STING activation might lead to excessive IFN-I production and pathogenic autoinflammatory or autoimmune responses in genetically predisposed individuals. The findings that cGAMP functions as an immunotransmitter and can be imported into naïve cells also have important therapeutic implications. Of note, although cGAS–STING activation can be assessed by quantifying cGAMP, detection of this CDN is technically difficult because the gold standard method is mass spectrometry and concentrations in tissues are low65 (nanomolar range); the increasing availability of cGAMP ELISAs might facilitate the detection of cGAS activation in the future.
Autophagy and cell death
Autophagy is a primordial function of cGAS–STING observed in invertebrates that do not synthesize IFN-I, such as the sea anemone12. Nonetheless, studies in this organism suggest that cGAS activation stimulates antiviral and antibacterial responses through ISG homologs. In mammalian cells, STING orchestrates several functions within the cell66. Activated STING molecules on the ER–Golgi intermediate compartment bind to microtubule-associated protein 1 light chain 3 (MAP1LC3; also known as LC3) on the autophagy membrane (that is, the phagophore), which leads to degradation of STING and termination of the activation signal (Fig. 2). This degradation process also helps to remove cytoplasmic DNA of host or microbial origin through enzymatic destruction in the autolysosome67. In clinical studies, cGAS–STING-induced autophagy was observed after radiotherapy or infection with Mycobacterium tuberculosis68,69; the bacterial DNA–STING interaction stimulated resistance to infection by eliciting a T helper 17 cell immune response70.
In certain cell types, intense STING activation drives apoptosis. In mice, this pro-apoptotic programme was driven by activation of mitochondrial B cell lymphoma 2-homology domain 3 only (BH3-only) proteins and was observed in T cells but not in macrophages nor dendritic cells71. The BH3-only protein PUMA also caused an increase in necroptosis, which required activation of STING following the release of mtDNA72. Certain viruses, such as murine γ-herpesvirus 68, induce necroptosis in a TNF- and STING-dependent manner73.
cGAS–STING in disease
The cGAS-STING pathway can be activated by a number of microbes that infect humans. In addition, substantial evidence implicates the cGAS–STING pathway in mouse models of inflammatory and neurodegenerative diseases, suggesting that this pathway is activated in several human diseases (Fig. 1; Tables 1 and 2).
Table 2.
Organ-specific diseases associated with cGAS–STING activation in humans or mice
| Disease names and/or affected tissues | Mouse models | Human diseases |
|---|---|---|
| ALS | Loss or blockade of STING rescued ALS phenotype In Prp-TDP-43Tg/+ mice133 | cGAMP detected in iPSC-derived motor neurons from patients with TARDBP mutations; increased cGAMP concentrations in spinal cord extracts from patients with ALS133 |
| PD | Lack of PINK1 or parkin led to oxidation of mtDNA and cGAS–STING activation135 | PINK1 or PRKN mutations associate with early-onset PD134 |
| AD | NAD+ supplementation reduced neuroinflammation by suppressing cGAS–STING137 | NA |
| Kidney disease | Cisplatin-induced tubular injury was attenuated by STING deficiency and inhibition5. Blocking STING trafficking from the ER improved glomerular pathology scores122 | cGAS and STING expression correlated with kidney fibrosis in patients with CKD7. cGAS–STING pathway implicated in diabetic kidney disease116 |
| Intestinal inflammation and microbiome |
STING deficiency or antagonism exacerbates gut inflammation142,144,146 STING contributes to gut microbial homeostasis143 Constitutive STING activation leads to dysbiosis and intestinal inflammation145 |
Increased STING levels in the colon of patients with ulcerative colitis145 |
| Familial chilblain lupus, UV skin responses, skin microbiome and acne |
Skin colonization with S. epidermidis led to cGAS–STING activation in keratinocytes140 UV light induces a IFN-I signature dependent on cGAS–STING106,107 |
Cutibacterium acnes associated with acne vulgaris stimulates IFN-I production in macrophages via cGAS–STING149 |
| Myocardial infarction, heart failure | cGAS- or STING-deficient mice are protected against myocardial infarction181,182. cGAS has an essential pathogenic role in pressure overload-induced heart failure182 | NA |
| Viral hepatitis | HBV evades immune sensing by inhibiting cGAS–STING expression and signalling183 | Decreased STING expression in peripheral blood of patients with HBV infection184 |
| NAFLD and NASH | Reduced liver injury in mice with impaired STING and high fat diet-induced NAFLD185. Macrophage activation via STING stimulates hepatocyte fat deposition and activates hepatic stellate cells186, possibly through sensing of mtDNA187 | Increased STING levels in livers of patients with NAFLD186,188 |
| ALD |
Ethanol-triggered ER stress causes activation of cGAS–STING58, STING-mediated apoptosis of hepatocytes189, and liver injury and fibrosis190 cGAMP transport through hepatic gap junctions amplifies tissue injury58 |
Increased cGAS–STING activation in the liver of patients with ALD58 |
| Obesity |
In DsbA-L KO, cGAS–STING activation suppresses thermogenesis and stimulates metabolic disorder117,191 High-fat diet causes metabolic stress and STING activation via mtDNA; STING deficiency partially prevented inflammation of adipose tissue, insulin resistance and obesity192 |
NA |
| Arthritis |
Sting1 loss protects Dnase2−/−Ifnar−/− mice from arthritis193 cGAS activation by self DNA causes arthritis in Dnase2−/−Ifnar1−/− mice65 TNF induces release of mtDNA, which activates cGAS; Cgas KO mice are protected from arthritis in the K/BxN arthritis model194 |
NA |
AD, Alzheimer disease; ALD, alcoholic liver disease; ALS, amyotrophic lateral sclerosis; cGAMP, 2′,3′-cyclic GMP–AMP; cGAS, cyclic GMP–AMP synthase; CKD, chronic kidney disease; ER, endoplasmic reticulum; HBV, hepatitis B virus; IFN-I, type I interferon; iPSCs, induced pluripotent stem cells; KO, knockout; mtDNA, mitochondrial DNA; NA, not available; NAFLD, non-alcoholic fatty liver disease; NASH, non-alcoholic steatohepatitis; PD, Parkinson disease; STING, stimulator of interferon genes; TNF, tumour necrosis factor; UV, ultraviolet. cGAS involvement in microbial infections is discussed in the main text.
Viruses and other infections
cGAS can be activated by a considerable number of DNA-containing viruses (for example, vaccinia, herpes simplex, hepatitis B and cytomegalovirus) given its broad DNA ligand specificity74. Surprisingly, RNA viruses such as HIV and SARS-CoV-2 can also activate cGAS, although this activation occurs through mechanisms distinct from those involving DNA viruses. For example, during HIV infection, a Y-form DNA intermediate with an overhanging stretch of guanines activates cGAS75, possibly through a protein intermediate, polyglutamine-binding protein 1 (PQBP1)76 (Fig. 3). In addition, the DNA-binding HIV capsid protein is recognized by the nuclear non-POU domain-containing octamer-binding protein (NONO), which results in cGAS activation77; cGAS was engaged in the nucleus but the location of the downstream signal transduction remains to be determined. By contrast, the positive-sense RNA virus SARS-CoV-2 activated cGAS by inducing mitochondrial damage78, micronuclei generation and cell fusion, which led to nuclear export of chromatin DNA into the cytoplasm, which can also be induced by bacteria with a type VI secretion system 5 (ref.79) (Fig. 3). Accordingly, in COVID-19, cGAS–STING activation has been implicated in lung inflammation and in skin manifestations of the disease through the induction of endothelial cell death and IFN-I production78. Single-stranded RNA viruses such as dengue virus can also activate cGAS by causing mitochondrial damage and release of mtDNA80. Notably, the translational stress induced by DNA or RNA viruses in infected cells frequently results in slowing of ribosomal movement (stalling) and can lead to collisions. Collided ribosomes also bind to cGAS and reduce its threshold of activation by DNA 15-fold, so viruses that induce ribosome collisions act as cGAS co-activators79,81 (Fig. 3). These studies indicate that, although DNA is the prototypic cGAS ligand, several factors can influence the threshold for cGAS activation, including infection with an RNA virus. Intracellular bacteria such as M. tuberculosis, Listeria and Chlamydia, as well as protozoan parasites such as Plasmodium, activate cGAS with variable effects on the host immune response (reviewed in ref.82).
Autoinflammatory and autoimmune diseases
The disease-causing potential of nucleic acid ligands, cGAS–STING and its downstream pathways is underscored by monogenic interferonopathies, which are disorders associated with the aberrant production of IFN-I. Over more than a decade, several studies have revealed that interferonopathies are caused by a variety of mutations in genes that are involved in nucleic acid metabolism. For example, mutations in TREX1, RNASEH2B and SAMHD1 lead to activation of cGAS and production of IFNβ83,84. Many patients with interferonopathies produce autoantibodies85, which supports the hypothesis that elevated levels of IFN-I contribute to lupus-like features owing to the aforementioned adjuvant effects of IFN-I on the immune system. Moreover, observations from studies of loss-of-function mutations in genes encoding DNases or certain nucleic acid-metabolizing enzymes substantiate the requirement for chronic activation of cGAS–STING to maintain the high IFN state typical of interferonopathies and of some systemic autoimmune diseases43,65,86,87. STING1 gain-of-function mutations cause STING-associated vasculopathy with onset in infancy (SAVI), which is a systemic inflammatory disease characterized by vasculitis and pulmonary fibrosis88,89. In mouse models, SAVI-like disease is largely, but not entirely, IFN-I dependent, and innate activation of monocytes and neutrophils has a key pathogenic role90–92. Coatomer protein subunit-α (COPA) syndrome, which is a disease with overlapping lung, joint and kidney involvement similar to SAVI, is also caused by chronic stimulation of STING owing to retention of STING at the Golgi89,93,94. Moreover, cGAS–STING activation has been demonstrated in certain lupus-prone mouse strains, including the TREX1 D18N mutant95 and Fcgr2b-knockout mice96.
Chronically elevated levels of IFN-I are observed in the blood of patients with many systemic autoimmune disorders, including systemic lupus erythematosus (SLE), Sjögren syndrome, scleroderma and dermatomyositis97. Although TLRs almost certainly have a role in perpetuating the IFN response following entry of immune complexes containing nucleic acid antigens into plasmacytoid dendritic cells98, emerging evidence indicates that the cGAS–STING pathway is also engaged, at least in some patients; 1–2% of patients with SLE have TREX1 mutations99,100, which, in mice, result in IFN-I production exclusively through the cGAS–STING pathway87,101. Moreover, in SLE, cGAMP could be detected in the peripheral blood cells of 15% of patients102,103 and in microvesicles released from blood cells102,103. Oxidized mtDNA released in the context of neutrophil extracellular traps formation in SLE was also linked to cGAS activation in humans104,105. Of note, ultraviolet light, which is known to precipitate cutaneous lupus and exacerbations of systemic lupus, induces a IFN-I signature that, in mice, was cGAS–STING dependent106,107. Mitochondria-rich red blood cells can also activate IFN-I through cGAS–STING in lupus monocytes108. In dermatomyositis, detection of IFNβ and phosphorylated IRF3 in skin biopsy samples suggested cGAS–STING activation, but this hypothesis has not been tested109.
In addition to the key role of IFN-I in host defence and tumour regulation, clear examples exist of IFN-I-independent STING activities that contribute to an inflammatory immune response. For example, several groups showed that mice with the STING S365A mutation, in which IRF3 cannot be recruited and activated to transcribe IFNβ, are still able to respond to a virus infection110–112. DNase II-deficient mice developed autoinflammatory arthritis and retained the ability to restrict HSV-1 infection and control tumour immune evasion. Although Dnase2−/− STING-S365A mice had reduced ability to control viral infection and tumour growth, they still exhibited severe polyarthritis.
Kidney disease
IFN-I might cause collapsing glomerulopathy, proliferative nephritis and vasculitis in cases of SAVI and coatomer protein subunit-α syndrome113. The cGAS–STING pathway might also contribute to acute kidney injury (AKI) and CKD characterized by kidney fibrosis114. In mice, the nephrotoxic agent cisplatin stimulated the expression of cGAS and STING in the damaged tubules, which mimicked high STING levels observed in the kidneys of patients with AKI6. Cisplatin-induced tubular injury, including apoptosis and inflammation, as well as kidney failure, were attenuated in STING-deficient mice and by pharmacological inhibition of STING with C-176 or H151 (ref.5). Researchers hypothesized that mtDNA leaked into the cytoplasm through the pores of damaged mitochondria and activated the cGAS–STING pathway in these AKI models. Mitochondrial damage is a well-recognized trigger of AKI (reviewed in ref.4) and targeting STING might therefore help to prevent AKI in patients receiving platinum-based chemotherapy who have increased plasma levels of mtDNA5. The cGAS–STING pathway was similarly activated in the ischaemia–reperfusion injury model of AKI6.
Given that mitochondrial damage leads to tubular cell injury, the mitochondrial transcription factor A (TFAM), which is downregulated in patients with CKD, was deleted in kidney tubule cells specifically to create a mouse model of CKD7. Akin to the AKI models, absence or inhibition of STING attenuated tubular structural damage, and reduced inflammatory cytokine levels and tubular apoptosis. Moreover, in 433 kidney tissue samples from patients with CKD, CGAS and STING1 expression correlated positively with the degree of kidney fibrosis7. Although these studies did not interrogate the downstream signalling cascade, cGAS–STING activation was shown to mediate kidney fibrosis through a non-canonical pathway (that is, independently of the cGAS–STING–TBK1–IRF3 pathway)115. In the unilateral ureteral obstruction model of kidney fibrosis, upon binding to cGAMP, STING was shown to interact directly with PKR-like ER kinase (PERK), which triggered phosphorylation of eukaryotic initiation factor 2α (eIF2α) and led to increased collagen expression and kidney fibrosis by modulating damage-initiated mRNA translation115. The cGAS–STING pathway has also been implicated in diabetic kidney disease116, where mitochondrial stress and release of mtDNA have important roles in disease pathogenesis117. cGAS–STING stimulation with mtDNA might also participate in metabolic diseases of the liver and adipose tissue (Table 2), which, in humans, are associated with CKD118, and can lead to tubular injury and kidney fibrosis in murine models of obesity119.
In contrast to AKI and the aforementioned CKD models, the role of cGAS–STING in lupus nephritis has been controversial. Initial studies in lupus mouse models indicated that STING suppresses systemic autoimmunity, including kidney disease120. Accordingly, loss of Cgas on the lupus-prone MRL/Fas.lpr background resulted in increased proteinuria and cellular infiltration in the kidney121. By contrast, a pro-inflammatory effect of STING activation was reported to drive lupus pathology in FcgR2b−/− mice96,122. Administration of the ISD017 peptide, which blocks STING trafficking from the ER, improved glomerular pathology scores and reduced IgG deposition in the kidney, which suggested that STING-targeted therapeutics might be useful in lupus nephritis122. The opposing findings in these studies suggest that the roles of cGAS and STING in autoimmune kidney diseases are complex and likely model dependent. Moreover, phenotypes might differ between models in which the cGAS–STING pathway is defective from birth, in which case compensatory mechanisms might be triggered, and those in which the pathway is inhibited pharmacologically in animals with established disease.
Immune senescence and ageing
Cells are constantly exposed to stress. When stress-induced damage is irreparable, cells undergo apoptosis but milder forms of injury can drive aged or stressed cells to exit the cell cycle and become senescent123. Senescent cells are characterized by elevated expression of the cell cycle inhibitor p16Ink4a and by major changes in transcriptional profiles, metabolism and chromatin organization124. These cells have a distinctive secretory phenotype known as the senescence-associated secretory phenotype, which is defined by changes in the expression of metalloproteinases, chemokines and cytokines (especially TNF, IL-6 and IL-1β)125. The senescence-associated secretory phenotype is thought to contribute to many chronic diseases associated with ageing, including atherosclerosis, osteoarthritis and possibly neurodegeneration. In the last 5 years, several studies have shown that immune senescence is often triggered by cGAS–STING activation following the release of chromatin fragments into the cytoplasm, possibly owing to defects in nuclear lamins123,126,127. Activating DNA ligands are likely to originate from multiple sources, including mitochondria and aberrant cytoplasmic chromatin fragments that emerge following oxidative stress, exposure to certain drugs and irradiation.
Neurodegenerative diseases
Accumulation of aggregated misfolded proteins in neurons and the surrounding glia is a pathological hallmark of many neurodegenerative diseases, including Alzheimer disease (AD), Parkinson disease (PD) and amyotrophic lateral sclerosis (ALS). Each of these diseases has a variable inflammatory component, which might depend on genetic susceptibility factors and stage of disease. The presence of aggregated or misfolded proteins triggers a stress response leading to cell death or activation of inflammatory cytokines, including IFN-I, which has long been known to exert neurocytotoxic effects. These effects were demonstrated clearly in transgenic mice that overexpress IFNα128, in studies of neurotropic viruses129 and in studies of interferonopathies. Moreover, selected mouse models of AD, PD and ALS are associated with a high IFN-I signature. In patients with ALS, several reports demonstrate that inflammatory cytokine levels, including IFN-I levels, as well as the frequency of autoimmune disorders, are high130–132. Moreover, cGAMP was detected in iPSC-derived motor neurons from patients with ALS associated with TARDBP mutations and cGAMP concentrations were elevated in spinal cord extracts from patients with spontaneous ALS133. Mutations that prevent the normal expression of the mitochondrial proteins PINK1 or parkin, are linked to early-onset PD in humans134. In mice that lack these mitochondrial-protective proteins, cell stress induces mitochondrial damage and mtDNA oxidation, which, in turn, activates the cGAS–STING pathway135. In patients with AD, ISG-expressing microglia surrounded amyloid plaques and IFN-I expression correlated with disease severity136. Of note, NAD+ supplementation reduced neuroinflammation in a mouse model of AD at least partly by suppressing cGAS–STING137. In mice, PQBP1 binds to aggregated tau proteins and activates cGAS–STING in microglia, resulting in neuroinflammation in the brain138. These findings from mouse models might have important implications for PQBP1-associated neurodegenerative diseases such as Huntington’s disease and spinocerebellar ataxia type 1 in humans.
Relationship with the microbiome
Commensal bacteria can alter IFN-I production via cGAS–STING signalling at multiple epithelial sites139–141. Conversely, STING expression levels affect commensal bacteria in the intestine, skin, lung and the TME.
Intestine
The IFN-I response triggered by cGAS sensing of DNA derived from commensal bacteria influences intraepithelial lymphocyte numbers, the distribution of different types of innate lymphoid cells and the function of regulatory T cells142. The effects of STING deficiency and overactivation in the intestine are controversial. Loss of Sting in mice led to altered development and function of regulatory T cells and was accompanied by dysbiosis, rendering the mice more prone to gut inflammation and enteric infection than controls142. Similarly, another study reported that Sting−/− mice were more susceptible than controls to inflammatory colitis owing to loss of IL-10 that was indirectly stimulated by STING143. By contrast, STING deficiency was reported to prevent the induction of colitis, which was exacerbated by a STING agonist144. Others observed that constitutive activation of STING (mouse model of SAVI) led to intestinal dysbiosis and spontaneous colitis; the altered microbiome propagated intestinal inflammation through STING ubiquitination and stabilization in myeloid cells145. The same study found elevated STING levels in the colon of patients with ulcerative colitis145. These findings demonstrate a reciprocal relationship between STING and the microbiome in shaping intestinal health and response to disease. Whether differences between the observations in mouse models can be explained by different approaches to genetic disruption of Sting1 (Sting−/− versus Sting1gt/gt), environmental factors or other experimental approaches, remains to be determined.
Additional studies revealed that microbial insults resulting from pathogenic bacteria triggered the release of mtDNA from dying intestinal endothelial cells, which resulted in STING activation and intestinal barrier dysfunction146. Furthermore, STING antagonists prevented sepsis-induced intestinal injury and protected mice from lipopolysaccharide-induced endotoxaemia146,147. Therefore, commensal and pathogenic bacteria can both lead to STING-mediated intestinal inflammation, although potentially through different mechanisms.
Lung
In addition to the intestinal inflammation discussed above, SAVI mice (carrying a Sting1 N153S gain-of-function mutation) develop lung disease141. Depleting gut commensals with oral antibiotics eliminated lung disease141 and prevented intestinal inflammation145. In this model, STING-mediated lung inflammation was attenuated in the presence of Bacteroides thetaiotaomicron, suggesting that specific commensal species might have inhibitory effects on the cGAS–STING pathway. However, in the intestine, bacterial products (for example, c-GMP–GMP), rather than a specific bacterial species, promoted STING-mediated inflammation145. These seminal studies introduced the potential therapeutic role of commensals and their products in regulating the cGAS–STING pathway and IFN-I production.
Skin
Skin colonization with Staphylococcus epidermidis led to cGAS–STING activation in keratinocytes, namely the sensing of microbe-induced endogenous retroviruses140. This cGAS-mediated IFN-I response was required for the induction of homeostatic T cell responses to this skin microorganism. Microbe-induced IFN-I responses have also been reported to promote tissue repair in the skin148, although the role of the cGAS–STING pathway was not investigated. By contrast, this pathway might contribute to inflammation in certain skin diseases. For example, the Gram-positive bacterium Cutibacterium acnes, which is associated with the inflammatory skin disease acne vulgaris, stimulates IFN-I production in macrophages via cGAS–STING149. Whether the cGAS–STING pathway links dysbiosis and high IFN-I in autoimmune diseases with skin involvement such as lupus, psoriasis, dermatomyositis and scleroderma warrants further investigation.
Tumour microenvironment
Therapeutic approaches to modulate the cGAS–STING pathway have been of particular interest in tumour therapy as IFN-I signalling can enhance antitumour immunity. A 2021 study150 showed that CDNs from commensal bacteria in the TME activate STING in monocytes, thereby potentiating the IFN-I response and anti-tumorigenic properties of NK cells and DCs. Faecal transplants from patients with melanoma who responded to the checkpoint inhibitor anti-PD-1 induced an intra-tumoural IFN-I signature in non-responders150, which supports the therapeutic use of commensal bacteria to modulate cGAS–STING function. Indeed, in murine models of melanoma and colon cancer, oral administration of Lactobacillus rhamnosus GG improved tumour responses to anti-PD-1 therapy via cGAS–STING-mediated IFNβ production in DCs151. This treatment altered the composition of the gut microbiome, and increased DC infiltration in the tumours and activation of cytotoxic CD8+ T cells in the TME. Of note, commensal bacteria (Lachnospiraceae species) suppressed radiation therapy-induced IFN-I production in murine colon and metastatic melanoma cancers by secreting butyrate, which inhibited the activation of TBK1 and IRF3 downstream of cGAS–STING152 (Fig. 5). Modulation of gut microbiota by vancomycin restored the antitumour effects of radiation therapy, which further supports the notion that altering the gut microbiome might have a therapeutic effect in cancer152.
Fig. 5. Targeting the cGAS–STING pathway.
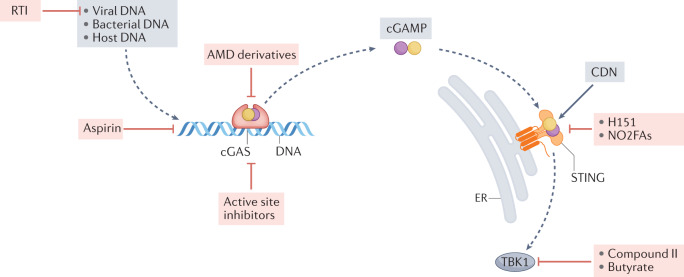
The cyclic GMP–AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway can be inhibited at many stages. Reverse transcriptase inhibitors (RTI) block the synthesis of cGAS-activating complementary DNA from endogenous retroelements. Aminoquinoline-type antimalarial drugs (AMDs) block DNA binding to cGAS166, whereas aspirin acetylates the cGAS residues Lys414, Lys384 and Lys394, which leads to its inactivation167. Active site inhibitors block the synthesis of the second messenger 2′,3′-cyclic GMP–AMP (cGAMP). For STING inhibition, both naturally occurring nitro-fatty acids (NO2-FAs) and the nitrofuran derivative, H-151, block palmitoylation of crucial cysteine residues on STING, preventing oligomerization of the protein171. A number of different TBK1 inhibitors, mostly belonging to the aminopyrimidine class, such as compound II, have been generated and used to treat autoimmune disorders in mice. In contrast to attenuation of the cGAS–STING pathway, STING can be activated by synthetic cyclic dinucleotides (CDNs), which might be useful in enhancing antitumour immunity.
Modulation of cGAS–STING in disease
As discussed above, cGAS, cGAMP, STING, as well as downstream kinases, are implicated in many diseases and are therefore therapeutic targets. Importantly, compounds that enhance or inhibit cGAS–STING have theoretical risks associated with them, and whether unwanted effects will be observed in humans treated with cGAS–STING-modifying compounds remains to be determined. The risk–benefit ratio of cGAS–STING modulators will also need to be compared with other approaches that target upstream or downstream molecules. These alternative targets for therapy would include oxidized mtDNA104 (upstream) or IFN receptor (IFNAR) blocking antibodies153 (downstream) of cGAS–STING. Here, we discuss the use of small molecules to activate or inhibit cGAS–STING in preclinical studies.
Pathway activation
In diseases such as cancer and in certain viral infections, enhancing the IFN-I response might have beneficial adjuvant effects on immune cells, especially CD8 cytotoxic T cells and antigen-presenting cells154.
Direct STING activation
Administration of IFNα has been used as adjunctive therapy in patients with certain types of cancer or chronic viral infections such as hepatitis C infection154. 5,6-Dimethylxanthenone-4-acetic acid (also known as vadimezan or ASA-404), is a potent selective agonist of the STING pathway and exerted significant antitumour and antiviral effects in mice owing to its stimulation of IFN-I and other cytokines155,156. However, 5,6-dimethylxanthenone-4-acetic acid does not activate human STING. Many different CDNs with agonist activity on human STING have been developed and are being evaluated as primary or adjunctive therapy in human cancers (reviewed in refs8,9). Unsurprisingly, CDNs have also been used to enhance inflammatory responses and reduce SARS-CoV-2 infection of human bronchial epithelial cells in vitro and mouse cells in vivo157. Of note, STING activation can induce apoptosis in T cells; therefore, careful evaluation of the differential effects on immune cell populations is necessary when assessing the therapeutic potential of STING agonists.
Targeting ENPP1 or cGAMP transport
Tumour cells upregulate ENPP1 on their surface, potentially as an immune evasion mechanism59. High ENPP1 expression has been associated with worse prognosis and response to therapy in different murine models of breast and colorectal cancer, probably owing to low extracellular cGAMP levels coupled with high levels of adenosine, which is a breakdown product of cGAMP hydrolysis and suppresses immune activation59,158. Given that secreted cGAMP activates and recruits both innate and adaptive immune cells that can reduce tumour growth and spreading59,159–161, extending the lifespan of extracellular cGAMP by targeting ENPP1 (Fig. 4) is of therapeutic interest in cancer. This approach is supported by evidence that inhibiting ENPP1 enhances the efficacy of current standard-of-care cancer therapies. ENPP1 inhibition with the phosphonate analogue STF-1084 not only enhanced IFNβ production and improved the response to radiation therapy in breast and pancreatic cancer54 but also potentiated the effects of immune checkpoint blockade on antitumour immunity59,159. Although an ENPP1 inhibitor was not effective when used alone in human cancers, clinical trials are evaluating its potential as a combination therapy.
Other cGAMP targeting approaches might involve the use of compounds that block gap junctions, such as carbenoxolone55,56, meclofenamate56 and tonabersat57. Pharmacological inhibition of cGAMP transport through gap junctions with these drugs controlled viral dissemination55, infection-induced toxic inflammation56 and tumour growth in a model of established brain metastasis57. Another therapeutic approach involves the targeting of cGAMP transporters. Two FDA-approved drugs, methotrexate and sulfasalazine, blocked cGAMP import through SLC19A1 (refs60,61) and new chemical tools to modulate LRRC8 channels on vascular endothelial cells have been developed64: moreover, sphingosine 1-phosphate (S1P) potentiates cGAMP import by endothelial cells, whereas the small molecule VRAC inhibitor DCPIB blocks LRRC8A-dependent uptake of extracellular cGAMP (Fig. 5).
Of note, selective delivery to the tumour site while avoiding the unwanted effects of systemic IFN-I induction, including the risk of autoimmunity, remains a considerable challenge for the implementation of CDN therapy. However, the cell type-specific expression of cGAMP importers and differential responsiveness to extracellular cGAMP provide opportunities for targeted therapy. For example, sulfasalazine inhibited cGAMP transport in macrophages and T cell-specific loss of LRRC8A:C channels in mice boosted T cell-driven immune responses162.
Pathway inhibition
In diseases where chronic IFN-I production is thought to contribute to autoinflammatory and autoimmune pathogenesis, attenuation of IFN-I activity might improve outcomes. Many small compounds inhibit cGAS, STING or downstream kinases such as TBK1 (Fig. 5). When targeting cGAS, small molecules that bind to and inhibit the nucleotidyl transferase active site of cGAS with nanomolar affinity have, so far, only been useful in vitro163–165. By contrast, the antimalarial aminoquinoline compound X6 inhibited DNA binding to cGAS in vitro and in Trex1-knockout mice166. Treatment with X6 attenuated the production of cGAMP and IFN-I, and reduced inflammation in the mouse heart166. Notably, sodium salicylate (also known as aspirin) inhibited cGAS activation by acetylating cGAS residues (specifically, Lys414, Lys384 and Lys394) that mediate DNA–cGAS interactions167. cGAS inhibition was also demonstrated in the Trex1-knockout mouse model but only at high concentrations of aspirin (equivalent to 3.5 g in a 70-kg adult, which would result in a blood concentration of ~1.5 mM), thus precluding its therapeutic use168.
Palmitoylation of Cys88 and Cys91 is necessary for the multimerization of STING, which, in turn, enables the binding of TBK1 (ref.169) (Fig. 2). Both naturally occurring nitro-fatty acids (NO2-FAs) and the nitrofuran derivative H-151 interact with one or more of these cysteines and attenuate STING-mediated inflammatory pathways170,171 (Fig. 5). The inhibitory effect of H-151 was demonstrated in Trex1−/− mice and in mouse models of ALS. The small molecule SN-011, which interacts with the cGAMP binding pocket was also an efficient STING inhibitor and reduced disease severity in Trex1−/− mice172. Reverse transcriptase inhibitors that prevent the accumulation of ligands such as retroelements or DNA–RNA hybrids have had some therapeutic success in murine lupus173, but whether they can prevent cGAS–STING activation remains to be determined.
Importantly, long-term inhibition of the cGAS–STING pathway could potentially increase susceptibility to infection and cancer. To date, spontaneous infections or increased cancer incidence in mice deficient for cGAS or STING has not been reported. However, specific pathogen-free environment and differences in mouse and human predisposition to cancers should be kept in mind.
The majority of compounds that target TBK1, which acts downstream of STING, TLR3, TLR-4 and TRIF pathways, have low nanomolar affinity (for example, aminopyrimidines) and also target other serine/threonine kinases174. More selective inhibitors have been subsequently developed and they exert inhibitory effects in Trex1−/− mice175 and mice with arthritis176,177.
Hydroxychloroquine (HCQ) is widely used to treat SLE and other autoimmune diseases. In vitro and animal studies suggest that this drug can attenuate both E-TLR and cGAS pathways166,178. In patients with SLE, HCQ is effective in the treatment of skin and joint disease but does not reduce life-threatening complications such as kidney disease179. Whether this limitation is due to inadequate dosing (HCQ at doses >400 mg/day might be associated with retinal and heart complications) or the presence of additional mechanisms of tissue injury is unclear. Nonetheless, more potent inhibitors of E-TLR and cGAS-STING are needed and are already in preclinical development. Given that cGAS activation seems to only occur in a subset of patients with SLE, such compounds might require a precision medicine approach guided by potential biomarkers such as cGAMP102.
Conclusions
Despite only being discovered in 2013, progress in delineating the mode of action of the DNA sensor cGAS, its crucial partnership with STING and its roles in host defence and inflammation have been remarkably rapid. At present, substantial evidence implicates the cGAS–STING pathway in mouse models of disease and accumulating data suggest that this pathway is also active in several human diseases. Over the next few years, improvements in cGAMP detection methods and in the evaluation of response to therapies designed to enhance or attenuate the cGAS–STING pathway will clarify its role in human diseases. Whether cGAS, cGAMP or STING will be easiest to target and have the fewest side-effects remains to be determined. Given that all of these molecules are upstream of IFN-I, targeting the cGAS–STING pathway is likely to be less disabling to host defences than blocking IFN-I, for example, through the common IFNAR.
Key questions remain to be addressed, for example, with regard to the cellular localization of cGAS and its regulation. The nature of the DNA ligands that trigger cGAS in autoinflammatory, autoimmune and degenerative disease states must also be elucidated. Finally, although mtDNA and retroelement DNA are frequently implicated in activation of the cGAS–STING pathway, precisely how and why these types of DNA activate cGAS remains to be clarified. Understanding these mechanisms might allow the pathway to be therapeutically targeted upstream of cGAS.
Acknowledgements
We thank Tomas Mustelin, Christian Lood and Grant Hughes for helpful discussions. This work was supported by the following grants: 1R21AR079661-01, DOD LR200061 and Hitchcock Foundation Scholar Award (to SSG) and R21AR077842 and the Alliance for Lupus Research (to K.B.E.).
Glossary
- Micronuclei
Small membrane-bound compartments that contain genomic DNA and form during mitosis in certain pathological states.
- Retroelements
Genetic elements that can be incorporated into a DNA sequence after reverse transcription of an RNA molecule.
- Necroptosis
A form of regulated cell death involving the activation of receptor-interacting protein kinases-1 (RIPK1) and RIPK3, and cell rupture (that is, necrosis).
Author contributions
All authors researched data for the article, made substantial contributions to discussions of the content and wrote, reviewed or edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Nephrology thanks A. Davidson, L. Li, J.T. Wilson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
K.B.E. and J.A. are co-founders of Amdax Therapeutics, LLC. S.S.-G. declares no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ablasser A, Chen ZJ. cGAS in action: expanding roles in immunity and inflammation. Science. 2019 doi: 10.1126/science.aat8657. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Agborbesong E, Li X. The role of mitochondria in acute kidney injury and chronic kidney disease and its therapeutic potential. Int. J. Mol. Sci. 2021;22:11253. doi: 10.3390/ijms222011253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong W, et al. The novel STING antagonist H151 ameliorates cisplatin-induced acute kidney injury and mitochondrial dysfunction. Am. J. Physiol. Renal Physiol. 2021;320:F608–F616. doi: 10.1152/ajprenal.00554.2020. [DOI] [PubMed] [Google Scholar]
- 6.Maekawa H, et al. Mitochondrial damage causes inflammation via cGAS-STING signaling in acute kidney injury. Cell Rep. 2019;29:1261–1273.e6. doi: 10.1016/j.celrep.2019.09.050. [DOI] [PubMed] [Google Scholar]
- 7.Chung KW, et al. Mitochondrial damage and activation of the STING pathway lead to renal inflammation and fibrosis. Cell Metab. 2019;30:784–799.e5. doi: 10.1016/j.cmet.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding C, Song Z, Shen A, Chen T, Zhang A. Small molecules targeting the innate immune cGAS–STING–TBK1 signaling pathway. Acta Pharm. Sin. B. 2020;10:2272–2298. doi: 10.1016/j.apsb.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garland KM, Sheehy TL, Wilson JT. Chemical and biomolecular strategies for STING pathway activation in cancer immunotherapy. Chem. Rev. 2022;122:5977–6039. doi: 10.1021/acs.chemrev.1c00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margolis SR, Wilson SC, Vance RE. Evolutionary origins of cGAS-STING signaling. Trends Immunol. 2017 doi: 10.1016/j.it.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Morehouse BR, et al. STING cyclic dinucleotide sensing originated in bacteria. Nature. 2020;586:429–433. doi: 10.1038/s41586-020-2719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen D, et al. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature. 2019;574:691–695. doi: 10.1038/s41586-019-1605-5. [DOI] [PubMed] [Google Scholar]
- 15.Elkon KB. Review: cell death, nucleic acids, and immunity: inflammation beyond the grave. Arthritis Rheumatol. 2018;70:805–816. doi: 10.1002/art.40452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato K, Omura H, Ishitani R, Nureki O. Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu. Rev. Biochem. 2017;86:541–566. doi: 10.1146/annurev-biochem-061516-044813. [DOI] [PubMed] [Google Scholar]
- 17.Dobbs N, et al. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe. 2015;18:157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunphy G, et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell. 2018;71:745–760.e5. doi: 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borden EC, et al. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug. Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronnblom L, Elkon KB. Cytokines as therapeutic targets in SLE. Nat. Rev. Rheumatol. 2010;6:339–347. doi: 10.1038/nrrheum.2010.64. [DOI] [PubMed] [Google Scholar]
- 21.Schoggins JW, et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monteith AJ, et al. Defects in lysosomal maturation facilitate the activation of innate sensors in systemic lupus erythematosus. Proc. Natl Acad. Sci. USA. 2016;113:E2142–E2151. doi: 10.1073/pnas.1513943113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon RE, Nemeth JF, Singh S, Lingham RB, Grewal IS. Harnessing SLE autoantibodies for intracellular delivery of biologic therapeutics. Trends Biotechnol. 2021;39:298–310. doi: 10.1016/j.tibtech.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Liu B, Gao C. Regulation of MAVS activation through post-translational modifications. Curr. Opin. Immunol. 2018;50:75–81. doi: 10.1016/j.coi.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Schlee M, Hartmann G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016;16:566–580. doi: 10.1038/nri.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elkon KB. Cell death, nucleic acids and immunity: inflammation beyond the grave. Arthritis Rheumatol. 2018;70:805–816. doi: 10.1002/art.40452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volkman HE, Cambier S, Gray EE, Stetson DB. Tight nuclear tethering of cGAS is essential for preventing autoreactivity. eLife. 2019 doi: 10.7554/eLife.47491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao B, et al. The molecular basis of tight nuclear tethering and inactivation of cGAS. Nature. 2020;587:673–677. doi: 10.1038/s41586-020-2749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathare GR, et al. Structural mechanism of cGAS inhibition by the nucleosome. Nature. 2020;587:668–672. doi: 10.1038/s41586-020-2750-6. [DOI] [PubMed] [Google Scholar]
- 31.Boyer JA, et al. Structural basis of nucleosome-dependent cGAS inhibition. Science. 2020;370:450–454. doi: 10.1126/science.abd0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kujirai T, et al. Structural basis for the inhibition of cGAS by nucleosomes. Science. 2020;370:455–458. doi: 10.1126/science.abd0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michalski S, et al. Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature. 2020;587:678–682. doi: 10.1038/s41586-020-2748-0. [DOI] [PubMed] [Google Scholar]
- 34.Ablasser A. DNA sensor in standby mode during mitosis. Science. 2021;371:1204–1205. doi: 10.1126/science.abg7422. [DOI] [PubMed] [Google Scholar]
- 35.Guey B, et al. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science. 2020;369:823–828. doi: 10.1126/science.aaw6421. [DOI] [PubMed] [Google Scholar]
- 36.Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li T, et al. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science. 2021 doi: 10.1126/science.abc5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y, Li S. Role of post-translational modifications of cGAS in innate immunity. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms21217842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong L, et al. Phosphorylation of cGAS by CDK1 impairs self-DNA sensing in mitosis. Cell Discov. 2020;6:26. doi: 10.1038/s41421-020-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackenzie KJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flynn PJ, Koch PD, Mitchison TJ. Chromatin bridges, not micronuclei, activate cGAS after drug-induced mitotic errors in human cells. Proc. Natl Acad. Sci. USA. 2021 doi: 10.1073/pnas.2103585118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun H, et al. A nuclear export signal is required for cGAS to sense cytosolic DNA. Cell Rep. 2021;34:108586. doi: 10.1016/j.celrep.2020.108586. [DOI] [PubMed] [Google Scholar]
- 43.Sisirak V, et al. Digestion of chromatin in apoptotic cell microparticles prevents autoimmunity. Cell. 2016;166:88–101. doi: 10.1016/j.cell.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nat. Immunol. 2014;15:415–422. doi: 10.1038/ni.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee-Kirsch MA, Wolf C, Gunther C. Aicardi-Goutieres syndrome: a model disease for systemic autoimmunity. Clin. Exp. Immunol. 2014;175:17–24. doi: 10.1111/cei.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gehrke N, et al. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 2013;39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Danilchanka O, Mekalanos JJ. Cyclic dinucleotides and the innate immune response. Cell. 2013;154:962–970. doi: 10.1016/j.cell.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holleufer A, et al. Two cGAS-like receptors induce antiviral immunity in Drosophila. Nature. 2021;597:114–118. doi: 10.1038/s41586-021-03800-z. [DOI] [PubMed] [Google Scholar]
- 50.Slavik KM, et al. cGAS-like receptors sense RNA and control 3′2′-cGAMP signalling in Drosophila. Nature. 2021;597:109–113. doi: 10.1038/s41586-021-03743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carozza JA, et al. Extracellular cGAMP is a cancer cell-produced immunotransmitter involved in radiation-induced anti-cancer immunity. Nat. Cancer. 2020;1:184–196. doi: 10.1038/s43018-020-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li L, et al. Hydrolysis of 2′3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 2014;10:1043–1048. doi: 10.1038/nchembio.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belli SI, van Driel IR, Goding JW. Identification and characterization of a soluble form of the plasma cell membrane glycoprotein PC-1 (5′-nucleotide phosphodiesterase) Eur. J. Biochem. 1993;217:421–428. doi: 10.1111/j.1432-1033.1993.tb18261.x. [DOI] [PubMed] [Google Scholar]
- 54.Carozza JA, et al. Extracellular cGAMP is a cancer cell-produced immunotransmitter involved in radiation-induced anti-cancer immunity. Nat. Cancer. 2020;1:184–196. doi: 10.1038/s43018-020-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ablasser A, et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pépin G, et al. Connexin-dependent transfer of cGAMP to phagocytes modulates antiviral responses. mBio. 2020;11:e03187-19. doi: 10.1128/mBio.03187-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Q, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. 2016;533:493–498. doi: 10.1038/nature18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luther J, et al. Hepatic gap junctions amplify alcohol liver injury by propagating cGAS-mediated IRF3 activation. Proc. Natl Acad. Sci. USA. 2020;117:11667. doi: 10.1073/pnas.1911870117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, et al. Metastasis and immune evasion from extracellular cGAMP hydrolysis. Cancer Discov. 2021;11:1212–1227. doi: 10.1158/2159-8290.CD-20-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luteijn RD, et al. SLC19A1 transports immunoreactive cyclic dinucleotides. Nature. 2019;573:434–438. doi: 10.1038/s41586-019-1553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ritchie C, Cordova AF, Hess GT, Bassik MC, Li L. SLC19A1 is an importer of the immunotransmitter cGAMP. Mol. Cell. 2019;75:372–381.e5. doi: 10.1016/j.molcel.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cordova AF, Ritchie C, Böhnert V, Li L. Human SLC46A2 is the dominant cGAMP importer in extracellular cGAMP-sensing macrophages and monocytes. ACS Cent. Sci. 2021;7:1073–1088. doi: 10.1021/acscentsci.1c00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou C, et al. Transfer of cGAMP into bystander cells via LRRC8 volume-regulated anion channels augments STING-mediated interferon responses and anti-viral immunity. Immunity. 2020;52:767–781.e6. doi: 10.1016/j.immuni.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 64.Lahey LJ, et al. LRRC8A:C/E heteromeric channels are ubiquitous transporters of cGAMP. Mol. Cell. 2020;80:578–591.e5. doi: 10.1016/j.molcel.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao D, et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl Acad. Sci. USA. 2015;112:E5699–E5705. doi: 10.1073/pnas.1516465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu D, et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 2019;26:1735–1749. doi: 10.1038/s41418-018-0251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liang Q, et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamazaki T, et al. Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nat. Immunol. 2020;21:1160–1171. doi: 10.1038/s41590-020-0751-0. [DOI] [PubMed] [Google Scholar]
- 69.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Dis E, et al. STING-activating adjuvants elicit a Th17 immune response and protect against mycobacterium tuberculosis infection. Cell Rep. 2018;23:1435–1447. doi: 10.1016/j.celrep.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gulen MF, et al. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 2017;8:427. doi: 10.1038/s41467-017-00573-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen D, et al. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc. Natl Acad. Sci. USA. 2018;115:3930–3935. doi: 10.1073/pnas.1717190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schock SN, et al. Induction of necroptotic cell death by viral activation of the RIG-I or STING pathway. Cell Death Differ. 2017;24:615–625. doi: 10.1038/cdd.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma Z, Damania B. The cGAS-STING defense pathway and its counteraction by viruses. Cell Host Microbe. 2016;19:150–158. doi: 10.1016/j.chom.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herzner AM, et al. Sequence-specific activation of the DNA sensor cGAS by Y-form DNA structures as found in primary HIV-1 cDNA. Nat. Immunol. 2015;16:1025–1033. doi: 10.1038/ni.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoh SM, et al. PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell. 2015;161:1293–1305. doi: 10.1016/j.cell.2015.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lahaye X, et al. NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell. 2018;175:488–501.e22. doi: 10.1016/j.cell.2018.08.062. [DOI] [PubMed] [Google Scholar]
- 78.Domizio JD, et al. The cGAS-STING pathway drives type I IFN immunopathology in COVID-19. Nature. 2022;603:145–151. doi: 10.1038/s41586-022-04421-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou Z, et al. Sensing of cytoplasmic chromatin by cGAS activates innate immune response in SARS-CoV-2 infection. Signal. Transduct. Target. Ther. 2021;6:382. doi: 10.1038/s41392-021-00800-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun B, et al. Dengue virus activates cGAS through the release of mitochondrial DNA. Sci. Rep. 2017;7:3594. doi: 10.1038/s41598-017-03932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan L, et al. Translation stress and collided ribosomes are co-activators of cGAS. Mol. Cell. 2021;81:2808–2822 e2810. doi: 10.1016/j.molcel.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hopfner KP, Hornung V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020;21:501–521. doi: 10.1038/s41580-020-0244-x. [DOI] [PubMed] [Google Scholar]
- 83.Crow YJ. Type I interferonopathies: Mendelian type I interferon up-regulation. Curr. Opin. Immunol. 2015;32:7–12. doi: 10.1016/j.coi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 84.Rice GI, et al. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat. Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cuadrado E, et al. Aicardi-Goutieres syndrome harbours abundant systemic and brain-reactive autoantibodies. Ann. Rheum. Dis. 2014 doi: 10.1136/annrheumdis-2014-205396. [DOI] [PubMed] [Google Scholar]
- 86.Grieves JL, et al. Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease. Proc. Natl Acad. Sci. USA. 2015;112:5117–5122. doi: 10.1073/pnas.1423804112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gray EE, Treuting PM, Woodward JJ, Stetson DB. Cutting edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi-Goutieres syndrome. J. Immunol. 2015;195:1939–1943. doi: 10.4049/jimmunol.1500969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Y, et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fremond ML, et al. Overview of STING-associated vasculopathy with onset in infancy (SAVI) among 21 patients. J. Allergy Clin. Immunol. Pract. 2021;9:803–818.e11. doi: 10.1016/j.jaip.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 90.Warner JD, et al. STING-associated vasculopathy develops independently of IRF3 in mice. J. Exp. Med. 2017;214:3279–3292. doi: 10.1084/jem.20171351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Siedel H, Roers A, Rosen-Wolff A, Luksch H. Type I interferon-independent T cell impairment in a Tmem173 N153S/WT mouse model of STING associated vasculopathy with onset in infancy (SAVI) Clin. Immunol. 2020;216:108466. doi: 10.1016/j.clim.2020.108466. [DOI] [PubMed] [Google Scholar]
- 92.Bouis D, et al. Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J. Allergy Clin. Immunol. 2019;143:712–725.e5. doi: 10.1016/j.jaci.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 93.Fremond ML, Crow YJ. STING-mediated lung inflammation and beyond. J. Clin. Immunol. 2021;41:501–514. doi: 10.1007/s10875-021-00974-z. [DOI] [PubMed] [Google Scholar]
- 94.Lepelley A, et al. Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J. Exp. Med. 2020 doi: 10.1084/jem.20200600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simpson SR, et al. T cells produce IFN-α in the TREX1 D18N model of lupus-like autoimmunity. J. Immunol. 2020;204:348–359. doi: 10.4049/jimmunol.1900220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thim-Uam A, et al. STING mediates lupus via the activation of conventional dendritic cell maturation and plasmacytoid dendritic cell differentiation. iScience. 2020;23:101530. doi: 10.1016/j.isci.2020.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barrat FJ, Crow MK, Ivashkiv LB. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 2019;20:1574–1583. doi: 10.1038/s41590-019-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hagberg N, et al. IFN-α production by plasmacytoid dendritic cells stimulated with RNA-containing immune complexes is promoted by NK cells via MIP-1β and LFA-1. J. Immunol. 2011;186:5085–5094. doi: 10.4049/jimmunol.1003349. [DOI] [PubMed] [Google Scholar]
- 99.Lee-Kirsch MA, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 100.Namjou B, et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 2011;12:270–279. doi: 10.1038/gene.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen, Z. J. cGAS deficiency protects TREX1 deficient mice from disease and early mortality (Lupus Research Institute, 2014).
- 102.An J, et al. cGAMP and cGAS are expressed in a subset of patients with systemic lupus erythematosus and associate with disease activity. Arthritis Rheum. 2017;69:800–807. doi: 10.1002/art.40002. [DOI] [Google Scholar]
- 103.Kato Y, et al. Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type I IFN production in systemic lupus erythematosus. Ann. Rheum. Dis. 2018 doi: 10.1136/annrheumdis-2018-212988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lood C, et al. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016 doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Caielli S, et al. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sontheimer C, Liggitt D, Elkon KB. Ultraviolet B irradiation causes stimulator of interferon genes-dependent production of protective type I interferon in mouse skin by recruited inflammatory monocytes. Arthritis Rheumatol. 2017;69:826–836. doi: 10.1002/art.39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Skopelja-Gardner S, et al. The early local and systemic Type I interferon responses to ultraviolet B light exposure are cGAS dependent. Sci. Rep. 2020;10:7908. doi: 10.1038/s41598-020-64865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caielli S, et al. Erythroid mitochondrial retention triggers myeloid-dependent type I interferon in human SLE. Cell. 2021;184:4464–4479 e4419. doi: 10.1016/j.cell.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Patel J, Maddukuri S, Li Y, Bax C, Werth VP. Highly multiplexed mass cytometry identifies the immunophenotype in the skin of dermatomyositis. J. Invest. Dermatol. 2021;141:2151–2160. doi: 10.1016/j.jid.2021.02.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu J, Dobbs N, Yang K, Yan N. Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity. 2020;53:115–126.e5. doi: 10.1016/j.immuni.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamashiro LH, et al. Interferon-independent STING signaling promotes resistance to HSV-1 in vivo. Nat. Commun. 2020;11:3382. doi: 10.1038/s41467-020-17156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li T, et al. TBK1 recruitment to STING mediates autoinflammatory arthritis caused by defective DNA clearance. J. Exp. Med. 2022 doi: 10.1084/jem.20211539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lodi L, et al. Type I interferon-related kidney disorders. Kidney Int. 2022 doi: 10.1016/j.kint.2022.02.031. [DOI] [PubMed] [Google Scholar]
- 114.Bai J, Liu F. cGAS–STING signaling and function in metabolism and kidney diseases. J. Mol. Cell Biol. 2021;13:728–738. doi: 10.1093/jmcb/mjab066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang D, et al. A non-canonical cGAS–STING–PERK pathway facilitates the translational program critical for senescence and organ fibrosis. Nat. Cell Biol. 2022 doi: 10.1038/s41556-022-00894-z. [DOI] [PubMed] [Google Scholar]
- 116.Khedr S, Dissanayake LV, Palygin O, Staruschenko A. Potential role of cGAS-STING pathway in the induction of diabetic kidney disease. FASEB J. 2020;34:1–1. [Google Scholar]
- 117.Bai J, et al. DsbA-L prevents obesity-induced inflammation and insulin resistance by suppressing the mtDNA release-activated cGAS-cGAMP-STING pathway. Proc. Natl Acad. Sci. USA. 2017;114:12196. doi: 10.1073/pnas.1708744114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang T-Y, et al. Association of metabolic dysfunction-associated fatty liver disease with kidney disease. Nat. Rev. Nephrol. 2022;18:259–268. doi: 10.1038/s41581-021-00519-y. [DOI] [PubMed] [Google Scholar]
- 119.Yang M, et al. DsbA-L ameliorates high glucose induced tubular damage through maintaining MAM integrity. EBioMedicine. 2019;43:607–619. doi: 10.1016/j.ebiom.2019.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sharma S, et al. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc. Natl Acad. Sci. USA. 2015;112:E710–E717. doi: 10.1073/pnas.1420217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Motwani M, et al. cGAS-STING pathway does not promote autoimmunity in murine models of SLE. Front. Immunol. 2021;12:689. doi: 10.3389/fimmu.2021.605930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Prabakaran T, et al. A STING antagonist modulating the interaction with STIM1 blocks ER-to-Golgi trafficking and inhibits lupus pathology. EBioMedicine. 2021;66:103314. doi: 10.1016/j.ebiom.2021.103314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang H, Wang H, Ren J, Chen Q, Chen ZJ. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA. 2017;114:E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baker DJ, et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Faget DV, Ren Q, Stewart SA. Unmasking senescence: context-dependent effects of SASP in cancer. Nat. Rev. Cancer. 2019;19:439–453. doi: 10.1038/s41568-019-0156-2. [DOI] [PubMed] [Google Scholar]
- 126.Dou Z, et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature. 2017;550:402–406. doi: 10.1038/nature24050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gluck S, et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017;19:1061–1070. doi: 10.1038/ncb3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Campbell IL, et al. Structural and functional neuropathology in transgenic mice with CNS expression of IFN-α. Brain Res. 1999;835:46–61. doi: 10.1016/S0006-8993(99)01328-1. [DOI] [PubMed] [Google Scholar]
- 129.Bhaskar M, Mukherjee S, Basu A. Involvement of RIG-I pathway in neurotropic virus-induced acute flaccid paralysis and subsequent spinal motor neuron death. mBio. 2021;12:e0271221. doi: 10.1128/mBio.02712-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miller ZA, et al. Increased prevalence of autoimmune disease within C9 and FTD/MND cohorts: completing the picture. Neurol. Neuroimmunol. Neuroinflamm. 2016;3:e301. doi: 10.1212/NXI.0000000000000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Miller ZA, et al. TDP-43 frontotemporal lobar degeneration and autoimmune disease. J. Neurol. Neurosurg. Psychiatry. 2013;84:956–962. doi: 10.1136/jnnp-2012-304644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Turner MR, Goldacre R, Ramagopalan S, Talbot K, Goldacre MJ. Autoimmune disease preceding amyotrophic lateral sclerosis: an epidemiologic study. Neurology. 2013;81:1222–1225. doi: 10.1212/WNL.0b013e3182a6cc13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yu CH, et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell. 2020;183:636–649.e18. doi: 10.1016/j.cell.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Newman LE, Shadel GS. Pink1/Parkin link inflammation, mitochondrial stress, and neurodegeneration. J. Cell Biol. 2018;217:3327–3329. doi: 10.1083/jcb.201808118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sliter DA, et al. Parkin and PINK1 mitigate STING-induced inflammation. Nature. 2018;561:258–262. doi: 10.1038/s41586-018-0448-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 136.Roy ER, et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J. Clin. Investig. 2020;130:1912–1930. doi: 10.1172/JCI133737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hou Y, et al. NAD+ supplementation reduces neuroinflammation and cell senescence in a transgenic mouse model of Alzheimer’s disease via cGAS-STING. Proc. Natl Acad. Sci. USA. 2021 doi: 10.1073/pnas.2011226118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jin M, et al. Tau activates microglia via the PQBP1-cGAS-STING pathway to promote brain inflammation. Nat. Commun. 2021;12:6565. doi: 10.1038/s41467-021-26851-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schaupp L, et al. Microbiota-induced type I interferons instruct a poised basal state of dendritic cells. Cell. 2020;181:1080–1096.e19. doi: 10.1016/j.cell.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 140.Lima-Junior DS, et al. Endogenous retroviruses promote homeostatic and inflammatory responses to the microbiota. Cell. 2021;184:3794–3811.e19. doi: 10.1016/j.cell.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Platt DJ, et al. Transferrable protection by gut microbes against STING-associated lung disease. Cell Rep. 2021;35:109113. doi: 10.1016/j.celrep.2021.109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Canesso MCC, et al. The cytosolic sensor STING is required for intestinal homeostasis and control of inflammation. Mucosal Immunol. 2018;11:820–834. doi: 10.1038/mi.2017.88. [DOI] [PubMed] [Google Scholar]
- 143.Ahn J, Son S, Oliveira SC, Barber GN. STING-dependent signaling underlies IL-10 controlled inflammatory colitis. Cell Rep. 2017;21:3873–3884. doi: 10.1016/j.celrep.2017.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Martin GR, Blomquist CM, Henare KL, Jirik FR. Stimulator of interferon genes (STING) activation exacerbates experimental colitis in mice. Sci. Rep. 2019;9:14281–14281. doi: 10.1038/s41598-019-50656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shmuel-Galia L, et al. Dysbiosis exacerbates colitis by promoting ubiquitination and accumulation of the innate immune adaptor STING in myeloid cells. Immunity. 2021;54:1137–1153.e1138. doi: 10.1016/j.immuni.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hu Q, et al. STING-mediated intestinal barrier dysfunction contributes to lethal sepsis. EBioMedicine. 2019;41:497–508. doi: 10.1016/j.ebiom.2019.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zeng L, et al. ALK is a therapeutic target for lethal sepsis. Sci. Transl. Med. 2017 doi: 10.1126/scitranslmed.aan5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Di Domizio J, et al. The commensal skin microbiota triggers type I IFN-dependent innate repair responses in injured skin. Nat. Immunol. 2020;21:1034–1045. doi: 10.1038/s41590-020-0721-6. [DOI] [PubMed] [Google Scholar]
- 149.Fischer K, et al. Cutibacterium acnes infection induces type I interferon synthesis through the cGAS-STING pathway. Front. Immunol. 2020;11:571334–571334. doi: 10.3389/fimmu.2020.571334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lam KC, et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184:5338–5356.e21. doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Si W, et al. Lactobacillus rhamnosus GG induces cGAS/STING-dependent type I interferon and improves response to immune checkpoint blockade. Gut. 2021 doi: 10.1136/gutjnl-2020-323426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang K, et al. Suppression of local type I interferon by gut microbiota-derived butyrate impairs antitumor effects of ionizing radiation. J. Exp. Med. 2021;218:e20201915. doi: 10.1084/jem.20201915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Morand EF, et al. Trial of anifrolumab in active systemic lupus erythematosus. N. Engl. J. Med. 2020;382:211–221. doi: 10.1056/NEJMoa1912196. [DOI] [PubMed] [Google Scholar]
- 154.Antonelli G, Scagnolari C, Moschella F, Proietti E. Twenty-five years of type I interferon-based treatment: a critical analysis of its therapeutic use. Cytokine Growth Factor Rev. 2015;26:121–131. doi: 10.1016/j.cytogfr.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weiss JM, et al. The STING agonist DMXAA triggers a cooperation between T lymphocytes and myeloid cells that leads to tumor regression. Oncoimmunology. 2017;6:e1346765. doi: 10.1080/2162402X.2017.1346765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Downey CM, Aghaei M, Schwendener RA, Jirik FR. DMXAA causes tumor site-specific vascular disruption in murine non-small cell lung cancer, and like the endogenous non-canonical cyclic dinucleotide STING agonist, 2′3′-cGAMP, induces M2 macrophage repolarization. PLoS One. 2014;9:e99988. doi: 10.1371/journal.pone.0099988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li M, et al. Pharmacological activation of STING blocks SARS-CoV-2 infection. Sci. Immunol. 2021 doi: 10.1126/sciimmunol.abi9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Takahashi R-u, et al. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nat. Commun. 2015;6:7318. doi: 10.1038/ncomms8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schadt L, et al. Cancer-cell-intrinsic cGAS expression mediates tumor immunogenicity. Cell Rep. 2019;29:1236–1248.e7. doi: 10.1016/j.celrep.2019.09.065. [DOI] [PubMed] [Google Scholar]
- 160.Marcus A, et al. Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity. 2018;49:754–763.e4. doi: 10.1016/j.immuni.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Carozza JA, et al. Structure-aided development of small-molecule inhibitors of ENPP1, the extracellular phosphodiesterase of the immunotransmitter cGAMP. Cell Chem. Biol. 2020;27:1347–1358.e5. doi: 10.1016/j.chembiol.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Concepcion AR, et al. The volume-regulated anion channel LRRC8C suppresses T cell function by regulating cyclic dinucleotide transport and STING-p53 signaling. Nat. Immunol. 2022;23:287–302. doi: 10.1038/s41590-021-01105-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hall J, et al. Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS One. 2017;12:e0184843. doi: 10.1371/journal.pone.0184843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lama L, et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 2019;10:2261. doi: 10.1038/s41467-019-08620-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Vincent J, et al. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat. Commun. 2017;8:750. doi: 10.1038/s41467-017-00833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.An J, et al. Inhibition of cyclic GMP-AMP synthase using a novel antimalarial drug derivative in Trex1-deficient mice. Arthritis Rheumatol. 2018;70:1807–1819. doi: 10.1002/art.40559. [DOI] [PubMed] [Google Scholar]
- 167.Dai J, et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell. 2019;176:1447–1460.e14. doi: 10.1016/j.cell.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Elkon KB. Aspirin meets cGAS. Nat. Rev. Rheumatol. 2019;15:254–255. doi: 10.1038/s41584-019-0205-y. [DOI] [PubMed] [Google Scholar]
- 169.Mukai K, et al. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 2016;7:11932. doi: 10.1038/ncomms11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hansen AL, et al. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl Acad. Sci. USA. 2018;115:E7768–E7775. doi: 10.1073/pnas.1806239115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Haag SM, et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 172.Hong Z, et al. STING inhibitors target the cyclic dinucleotide binding pocket. Proc. Natl Acad. Sci. USA. 2021 doi: 10.1073/pnas.2105465118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Beck-Engeser G, Eilat D, Wabl M. An autoimmune disease prevented by anti-retroviral drugs. Retrovirology. 2011;8:91. doi: 10.1186/1742-4690-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hasan M, Yan N. Therapeutic potential of targeting TBK1 in autoimmune diseases and interferonopathies. Pharmacol. Res. 2016;111:336–342. doi: 10.1016/j.phrs.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hasan M, et al. Cutting edge: inhibiting TBK1 by compound II ameliorates autoimmune disease in mice. J. Immunol. 2015;195:4573–4577. doi: 10.4049/jimmunol.1500162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hammaker D, Boyle DL, Firestein GS. Synoviocyte innate immune responses: TANK-binding kinase-1 as a potential therapeutic target in rheumatoid arthritis. Rheumatology. 2012;51:610–618. doi: 10.1093/rheumatology/ker154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Louis C, et al. Therapeutic effects of a TANK-binding kinase 1 inhibitor in germinal center-driven collagen-induced arthritis. Arthritis Rheumatol. 2019;71:50–62. doi: 10.1002/art.40670. [DOI] [PubMed] [Google Scholar]
- 178.An J, Minie M, Sasaki S, Woodward JJ, Elkon KB. Anti-malarial drugs as immune modulators: new mechanisms for old drugs. Annu. Rev. Med. 2016;68:317–330. doi: 10.1146/annurev-med-043015-123453. [DOI] [PubMed] [Google Scholar]
- 179.Wallace DJ, Gudsoorkar VS, Weisman MH, Venuturupalli SR. New insights into mechanisms of therapeutic effects of antimalarial agents in SLE. Nat. Rev. Rheumatol. 2012;8:522–533. doi: 10.1038/nrrheum.2012.106. [DOI] [PubMed] [Google Scholar]
- 180.Kerur N, et al. cGAS drives noncanonical-inflammasome activation in age-related macular degeneration. Nat. Med. 2018;24:50–61. doi: 10.1038/nm.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.King KR, et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat. Med. 2017;23:1481–1487. doi: 10.1038/nm.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cao DJ, et al. Cytosolic DNA sensing promotes macrophage transformation and governs myocardial ischemic injury. Circulation. 2018;137:2613–2634. doi: 10.1161/CIRCULATIONAHA.117.031046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Verrier ER, et al. Hepatitis B virus evasion from cyclic guanosine monophosphate-adenosine monophosphate synthase sensing in human hepatocytes. Hepatology. 2018;68:1695–1709. doi: 10.1002/hep.30054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Karimi-Googheri M, Daneshvar H, Khaleghinia M, Bidaki R, Kazemi Arababadi M. Decreased expressions of STING but not IRF3 molecules in chronic HBV infected patients. Arch. Iran. Med. 2015;18:351–354. [PubMed] [Google Scholar]
- 185.Qiao JT, et al. Activation of the STING-IRF3 pathway promotes hepatocyte inflammation, apoptosis and induces metabolic disorders in nonalcoholic fatty liver disease. Metabolism. 2018;81:13–24. doi: 10.1016/j.metabol.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 186.Luo X, et al. Expression of STING is increased in liver tissues from patients with NAFLD and promotes macrophage-mediated hepatic inflammation and fibrosis in mice. Gastroenterology. 2018;155:1971–1984.e4. doi: 10.1053/j.gastro.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yu Y, et al. STING-mediated inflammation in Kupffer cells contributes to progression of nonalcoholic steatohepatitis. J. Clin. Invest. 2019;129:546–555. doi: 10.1172/JCI121842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Wang X, et al. STING expression in monocyte-derived macrophages is associated with the progression of liver inflammation and fibrosis in patients with nonalcoholic fatty liver disease. Lab. Invest. 2020;100:542–552. doi: 10.1038/s41374-019-0342-6. [DOI] [PubMed] [Google Scholar]
- 189.Petrasek J, et al. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl Acad. Sci. USA. 2013;110:16544–16549. doi: 10.1073/pnas.1308331110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Iracheta-Vellve A, et al. Endoplasmic reticulum stress-induced hepatocellular death pathways mediate liver injury and fibrosis via stimulator of interferon genes. J. Biol. Chem. 2016;291:26794–26805. doi: 10.1074/jbc.M116.736991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bai J, et al. Mitochondrial stress-activated cGAS-STING pathway inhibits thermogenic program and contributes to overnutrition-induced obesity in mice. Commun. Biol. 2020;3:257. doi: 10.1038/s42003-020-0986-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Mao Y, et al. STING-IRF3 triggers endothelial inflammation in response to free fatty acid-induced mitochondrial damage in diet-induced obesity. Arterioscler. Thromb. Vasc. Biol. 2017;37:920–929. doi: 10.1161/ATVBAHA.117.309017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Baum R, et al. Cutting edge: AIM2 and endosomal TLRs differentially regulate arthritis and autoantibody production in DNase II-deficient mice. J. Immunol. 2015;194:873–877. doi: 10.4049/jimmunol.1402573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Willemsen J, et al. TNF leads to mtDNA release and cGAS/STING-dependent interferon responses that support inflammatory arthritis. Cell Rep. 2021;37:109977. doi: 10.1016/j.celrep.2021.109977. [DOI] [PubMed] [Google Scholar]
- 195.Ku JWK, et al. Bacterial-induced cell fusion is a danger signal triggering cGAS-STING pathway via micronuclei formation. Proc. Natl Acad. Sci. USA. 2020;117:15923–15934. doi: 10.1073/pnas.2006908117. [DOI] [PMC free article] [PubMed] [Google Scholar]