SUMMARY
As sessile organisms, plants are finely tuned to respond dynamically to developmental, circadian and environmental cues. Genome-wide studies investigating these types of cues have uncovered the intrinsically different ways they can impact gene expression over time. Recent advances in single-cell sequencing and time-based bioinformatic algorithms are now beginning to reveal the dynamics of these time-based responses within individual cells and plant tissues. Here, we review what these techniques have revealed about the spatiotemporal nature of gene regulation, paying particular attention to the three distinct ways in which plant tissues are time sensitive. (i) First, we discuss how studying plant cell identity can reveal developmental trajectories hidden in pseudotime. (ii) Next, we present evidence that indicates that plant cell types keep their own local time through tissue-specific regulation of the circadian clock. (iii) Finally, we review what determines the speed of environmental signaling responses, and how they can be contingent on developmental and circadian time. By these means, this review sheds light on how these different scales of time-based responses can act with tissue and cell-type specificity to elicit changes in whole plant systems.
Keywords: transcriptome dynamics, single-cell, development, circadian, environmental signaling, temporal regulatory networks
INTRODUCTION
Plants must regulate the timing of gene expression with exquisite precision to develop and thrive in a rapidly changing environment. Driven by the coordinated effort of hundreds of transcription factors (TFs) interacting with their genome-wide targets, gene expression programs reliably turn on and off to respond to such changes. To understand how such genome-wide regulatory mechanisms operate dynamically, the study of time has typically fallen into three broad paradigms. The first is through the context of plant development, where dynamic genome-wide expression events drive cell-type patterning. The second is the circadian clock, which introduces oscillations in genome-wide expression levels that align to a 24-h period. The third is the timing of genome-wide responses to environmental cues, which can vary depending on the plant’s developmental state or time of day. Importantly, because different plant cell types express unique sets of genes, the passage of time has different effects among and between different plant tissues.
Understanding how genome-wide responses play out over different timescales and in a cell-type-specific manner has been technically challenging. Historically, genome-wide investigations have been limited to a handful of time-points conducted using homogenized ‘bulk’ tissue samples. Such approaches can easily miss early and transient responses over time, as well as ones that occur only in specific cell types. Fortunately, advances in genomic technologies now allow scientists to better measure genome-wide responses with greater precision over time and in space. This has been achieved in part by the continuous drop in sequencing costs, the rise of single-cell transcriptomics, and the improvement of computational approaches that can learn the regulatory networks underlying these dynamic responses.
The purpose of this review is to describe the current state of the art in capturing gene regulatory dynamics over time, with particular attention on what these techniques have revealed about unique timing mechanisms at a cell-type level. To begin, we discuss how single-cell sequencing has led to characterization of plant cell identity at the transcriptional level, and how this has allowed developmental trajectories to be reconstructed in silico. We then cover evidence that suggests that the circadian clock works differently within different plant tissues. We also reflect on the rate at which environmental signaling events occur in real time, and how they can interact with developmental and circadian timing. Finally, by integrating genome-wide responses across time and space scales, we provide a forecast of what new questions of plant systems dynamics the community might address in the future (Box 1).
Box 1. Investigating the molecular dynamics underlying different timescales in plants.
Plants respond dynamically to developmental, circadian and environmental cues, and these responses vary over time and space.
Advances in single-cell sequencing have helped characterize plant cell identity at the transcriptional level, which has allowed developmental trajectories to be reconstructed in silico using pseudotime.
The circadian clock allows plant cells to maintain their own local time, which can vary among different cell and tissue types, and across developmental time.
Responses to environmental signals entail dynamic transcriptional cascades that can change rapidly and can be captured by applying machine learning algorithms to time-series data.
The timing of environmental signaling responses can be contingent on developmental and circadian time.
DEVELOPMENTAL PSEUDOTIME: PREDICTING THE PAST FROM PLANT CELL IDENTITY
Unlike mammals, plants grow and develop indeterminately. Furthermore, while mammalian organogenesis follows a strict timeline of cellular differentiation, plants are more flexible; their development can speed up or slow down depending on resource availability and environmental cues. For this reason, a central goal in plant genomics is to understand not only the molecular signaling networks that govern plant cell-type patterning, but also what controls their rate of development over time. However, mapping the different developmental trajectories that unfold at the genome-wide level over time is technically challenging. Fortunately, single-cell sequencing and pseudotime algorithms are enabling measurement of the genome-wide signaling events that underlie plant developmental trajectories. In this section, we first highlight what single-cell sequencing has revealed about the developmental differences between plant cell types. We then discuss how pseudotime is helping to capture the temporal nature of cell-type transitions as they occur over plant development.
Plant cell identity at the gene regulatory level
The ‘identity’ of a cell can be defined by its physiology, spatial position and developmental lineage (Morris, 2019; Scheres, 2001). Cell identity can also be described at the molecular level through the unique ways cell types regulate the expression of their genomes (Efroni, 2018; Pinheiro et al., 2021). Such signatures of cell identity can be detected by a variety of methods (Berkowitz et al., 2021; Birnbaum et al., 2003; Gifford et al., 2008; Knauer et al., 2019), with genome-wide transcriptional or chromatin profiling through single-cell sequencing becoming a dominant approach. While many different single-cell sequencing techniques exist (for review, see Birnbaum, 2018), the rise of single-cell investigations across eukaryotes in general, and in plants in particular, has been driven by the 10 × Genomics single-cell microfluidics-based platform (Birnbaum, 2018; McFaline-Figueroa et al., 2020). Currently, cell-type atlases that report plant cell identity exist for the Arabidopsis root (Denyer et al., 2019; Shahan et al., 2020; Shulse et al., 2019; Zhang et al., 2019), shoot (Kim et al., 2021; Lopez-Anido et al., 2021; Xu et al., 2021a) and seed (Picard et al., 2021), as well as other plant species (Kajala et al., 2021; Liu et al., 2021; Xu et al., 2021b; Figure 1a). Importantly, these cell-type atlases are generated through an unbiased clustering of single sequenced cells (or nuclei) using algorithms that exploit the variability between each cell’s transcriptome or epigenome (Satija et al., 2015). Given this unbiased nature, part of the promise of single-cell sequencing lies in its ability to detect rare or previously uncharacterized cell types, such as those found in the chalazal endosperm region in seed (Picard et al., 2021).
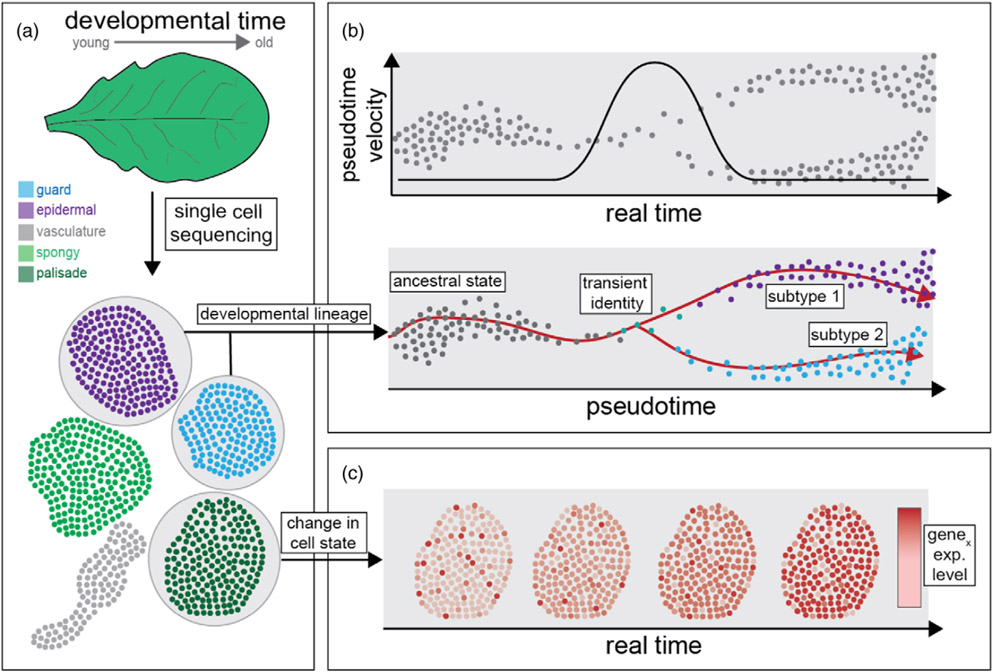
Figure 1.
Pseudotime versus real time. (a) Single-cell sequencing approaches can create plant cell atlases that reveal tissue-specific gene regulatory patterns that contribute to cell identity.
(b) Pseudotime algorithms can predict cell-type developmental lineages found within cell atlases. Importantly, pseudotime is not proportional to real time. For example, pseudotime’s velocity can increase at developmental junctions (Nelms and Walbot, 2019).
(c) Beyond pseudotime, cell-type-specific gene expression can also change in real time. Transcriptional changes in response to the environment over real time can change a cell’s ‘state’ without changing its identity (Morris, 2019).
However, the promise of single-cell sequencing in plant genomics lies not just in its ability to discover new cell types, but also in its ability to reveal the canonical expression patterns that make up cell identity. For some cell types, such transcriptional signatures differ dramatically. For example, even though they arise from the same developmental lineage, epidermal and guard cell transcriptomes are highly distinct (Lopez-Anido et al., 2021; Figure 1b). In contrast, for other cell types, such as palisade and spongy mesophyll, differences in cell identity at the transcriptome level have proven more subtle (Xie et al., 2020; Xu et al., 2021). Nevertheless, small differences in expression between cell types have helped reveal new cell functions. For example, small disparities in transcriptomes of abaxial and adaxial bundle sheath cells in maize have pointed to preferential phloem loading on the abaxial side (Bezrutczyk et al., 2021). Plant cell identity at the genome-wide level has also been revealed by assaying differences in the epigenome (Pinheiro et al., 2021). Performing single-nuclei ATAC-seq in the maize leaf found differences in chromatin accessibility between each cell type that contributed to cell identity (Marand et al., 2021). Nevertheless, epigenetic marks—such as regions of open chromatin or DNA methylation—appear to be less variable at the single-cell scale compared with gene expression (Birnbaum et al., 2003; Dorrity et al., 2021; Farmer et al., 2021; Kawakatsu et al., 2016). This is somewhat surprising, given that chromatin accessibility and DNA-methylation patterns show a high degree of cell-type specificity in mammals (Cusanovich et al., 2018; Gorkin et al., 2020; Luo et al., 2018). However, this discrepancy may lie in the technical limitations of current single-cell assays to plant tissues, and may be resolved through multi-omic methods that can report both the transcriptome and epigenome of the same cell (Zhu et al., 2019).
Plant cells not only express the core canonical gene regulatory programs that drive cell identity, but also express additional gene regulatory programs that are tailored to meet the demands of the environment. For example, guard cells in the leaf not only express the core set of genes that maintain cell identity, but can also express additional transcriptional programs, such as those that are specific for drought stress (Lopez-Anido et al., 2021; van Weringh et al., 2021). Such cell-type responses to external stimuli are considered changes in cell ‘state’ (Morris, 2019; Figure 1c). In systems with just one cell-type identity, changes in cell state can be clearly detected. For example, through single-cell sequencing of Chlamydomonas cell culture over time, the transcriptional changes driven by iron limitation and the circadian clock could be easily discerned (Ma et al., 2021). However, in multicellular systems, where many different cell identities exist, it is more difficult to dissociate cell identity from cell state. In these instances, changes in cell state can be detected through looking at variation in splicing (La Manno et al., 2018), tracking transcript accumulation over time (Erhard et al., 2019; Zhang et al., 2016), and with algorithms that try to distinguish between identity-driven and state-driven responses (Kotliar et al., 2019). Importantly, the distinction between cell identity and cell state will help parse out the different gene regulatory programs a cell type might be expressing at any one time, an important step in untangling the mix of developmental, circadian and environmental responses that each cell type can display.
Deciphering developmental trajectories hidden in pseudotime
Plant cell types arise from stem cell niches, and through a series of divisions arrive at their differentiated state. Because these divisions occur in a spatial progression, plant tissues consist of an ordered file of cells, with younger cells separated from their more mature counterparts (Figure 1a). Luckily for systems biologists, such an arrangement means that each plant organ holds a spatial map of developmental time, which can be interrogated using single-cell sequencing. Indeed, with the help of pseudotime algorithms, new insights are being made into how these developmental trajectories unfold over time. Briefly, these algorithms aim to draw the most parsimonious developmental lineages among cell types, beginning at the ancestral cell type and ending at the most differentiated cell types (Lederer and La Manno, 2020; Figure 1b). Many different pseudotime algorithms exist, where their predictive power depends on the underlying developmental trajectory; different flavors of algorithms are better suited to different types of trajectories (for review, see Saelens et al., 2019).
Pseudotime has proven useful in deciphering developmental timelines in both the root and shoot. For example, after single-cell sequencing captured the transcriptomes of cell types within the Arabidopsis root tip, pseudotime was able to predict the developmental trajectories that connect these cell types together and parse them into meristematic, elongation and maturation zones (Shahan et al., 2020). This approach confirmed the well-known mechanism that TFs SHORTROOT and SCARECROW play in early tissue patterning, as well as extended what is known about their mechanism of action by discovering that SHORTROOT can act to specify mature endodermal cell identity (Shahan et al., 2020). Indeed, along these lines pseudotime is helping revise the timing of well-known molecular signaling mechanisms that drive development. For example, in the Arabidopsis leaf a pseudotime model captured the developmental trajectory of guard cells within the leaf epidermis (Lopez-Anido et al., 2021). This model correctly predicted the key role SPEECHLESS plays in early patterning, but also found that SPEECHLESS was required much later in stomatal development to drive cell fate commitment (Lopez-Anido et al., 2021). Beyond transcriptomics, pseudotime can also be used to predict the developmental transitions that occur in the plant epigenome. This is useful because measuring how the chromatin landscape changes over pseudotime can predict which events precede gene expression (Ma et al., 2020a). Such an approach was taken to detect the changes in chromatin accessibility that occur between meristematic cells and phloem companion cells in maize (Marand et al., 2021).
Some developmental transitions in plants occur rapidly and thus can be difficult to detect in real time. Pseudotime has proven to be powerful in its ability to detect such transient events. For example, pseudotime was able to capture very early and transient transcriptional states that occur when lateral root primordia cells arise from xylem pole pericycle cells (Gala et al., 2021), or as mature conductive cells arise from the root stem cell niche (Roszak et al., 2021). In another example, when applied to tomato shoots, pseudotime was able to detect transitions in cell identity that lead to shoot-borne roots (Omary et al., 2020). In each of these studies, pseudotime led to the identification and functional validation of key candidate genes involved in these phenotypes. More broadly, pseudotime has also been able to indicate the rate or ‘velocity’ at which such developmental transitions occur (Figure 1b). The concept of pseudotime velocity was benchmarked by observing single-cell meiotic transcriptional events in maize, where an increased pseudotime velocity was associated with the transition of premeiotic cells to prophase I (Nelms and Walbot, 2019).
A key limitation of pseudotime is that it serves only as a proxy for real time, and often can predict trajectories that are far from the ground truth (Saelens et al., 2019). Combining pseudotime with real-time approaches can enhance the accuracy of pseudotime. This can be achieved through measuring developmental change over multiple time-points and then employing pseudotime to infer what happens during the time in between. For example, this time-based approach was taken in animals to study mouse gastrulation (Cao et al., 2019; Mittnenzweig et al., 2021). Here, pseudotime helped build a continuous model of fetal development from discrete time-points sampled over a 36-h period (Mittnenzweig et al., 2021). This approach revealed the genome-wide transcriptional signatures that drive cell-type development, finding that while some cell types were driven by a discrete set of factors, others were driven by a collection of factors that lacked clear expression boundaries (Mittnenzweig et al., 2021). Indeed, when applied to plants, combining real time with pseudotime approaches may help to parse out complex developmental transitions that may be difficult to predict from pseudotime alone. This method has already shown promise in ordering the expression changes that drive the transition to flowering in the tomato meristem (Meir et al., 2021).
CIRCADIAN TIME: ARE THERE LOCAL TIME ZONES WITHIN PLANT TISSUES?
Beyond developmental time, plants also have an internal molecular clock that keeps track of real time. The circadian clock—defined by a series of interlocked transcriptional feedback loops—ticks away within different plant cells and tissues. Through its action, the circadian clock creates oscillations in the expression of thousands of genes, all of which are entrained to the 24-h photoperiod (Harmer et al., 2000; Figure 2a). Such oscillations are crucial for ensuring that plant function is in rhythm with the environment. For example, the circadian clock sets the rate of starch degradation in the evening (Kim et al., 2017) and promotes carbon fixation during the daytime (Dodd et al., 2004). Given its enormous impact on gene regulation, understanding how the clock regulates its gene targets is a central challenge in systems biology. Complicating this task further, new research is indicating that different plant tissues regulate their own clocks in a cell-type-specific way. In this section, we first present evidence supporting how cell-type-specific circadian gene expression occurs in plants. We then assess what bioinformatic approaches can allow plant biologists to model such expression patterns over time with tissue specificity.
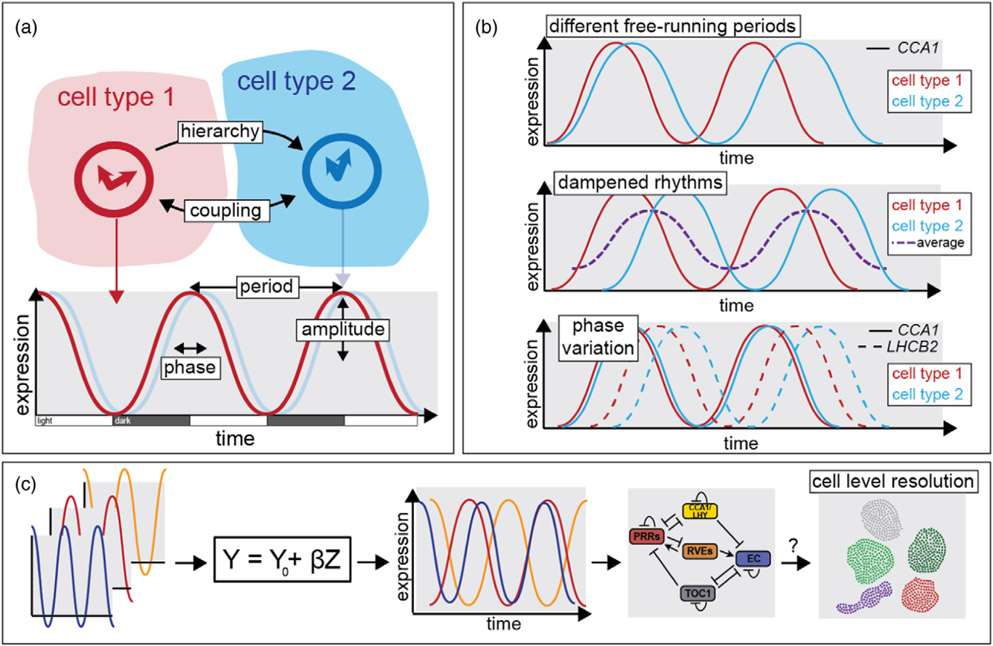
Figure 2.
Plant cell types run their own local circadian time.
(a) By introducing variation in period, phase and amplitude, the circadian clock aligns the expression levels of thousands of genes to the time of day. Each plant cell self-regulates its own clock gene regulatory mechanisms, which can lead to cell-type-specific outputs. Evidence suggests different cell types can synchronize their clock through coupling or hierarchical arrangements (Gould et al., 2018; Yakir et al., 2011).
(b) A difference in free-running periods between cell types (shown in red and blue), as seen under constant light conditions, gives evidence for cell-type-specific regulation of clock function (e.g. CCA1 expression; Gould et al., 2018; Yakir et al., 2011). Similarly, tissue-specific clock regulation can lead to different phasing of gene expression, which can appear as dampened rhythms at the whole-organ level (dashed purple line). Phase variation between cell types also exists (e.g. LHCB2; Endo et al., 2014).
(c) At the genome-wide level, linear models can compare circadian patterns (Greenham et al., 2020), leading to gene regulatory networks that can reveal the molecular mechanisms driving clock function. By employing single-cell sequencing, the next frontier is to understand how the circadian clock works with cell-type specificity at the genome-wide level.
Tissue-specific timekeeping
In animals, different organs and tissues can express their own distinct circadian rhythms. In mammals, such multiple tissue-specific oscillators are organized hierarchically, where a central clock located in the suprachiasmatic nucleus (SCN) of the brain provides temporal information that synchronizes peripheral oscillators (Mohawk et al., 2012; Tomioka and Matsumoto, 2010). Considerable evidence supports the model that plants also have multiple oscillators regulated tissue-specifically (Endo, 2016; Nohales, 2021; Sorkin and Nusinow, 2021). One of the first examples was shown in tobacco, where rhythms in cytosolic free calcium and LHCB (CAB) gene expression were shown to exhibit different free-running periods (Sai and Johnson, 1999; Figure 2b). Consistent with this, a subsequent study in Arabidopsis showed different temperature-responsiveness of epidermal and mesophyll clocks, as measured by CAT3 and LHCB1.1 (CAB2) expression (Michael et al., 2003). Similarly, rhythmic expression of the clock component CCA1 exhibited delayed phasing and longer periods in guard cells compared with adjacent mesophyll and epidermal cells, again supporting tissue specificity in oscillator function (Yakir et al., 2011). Crucially, the discovery of rhythms with distinct periods demonstrates the presence of multiple oscillators (Nohales, 2021). This is because while a single clock can drive rhythms with multiple phases, it cannot generate multiple periods, as the period is an intrinsic property of the oscillator. If distinct tissue-specific clocks exist, then these different clocks should be able to generate different periods and phases unique to each cell type (Figure 2b). To test this, tissue-specific promoters were used to overexpress the clock TF CCA1 specifically in the epidermis, mesophyll or the vasculature. Because CCA1 overexpression confers arrhythmicity, this approach allowed identification of which tissue-specific clock was necessary for individual output rhythms. This demonstrated that clock function in the leaf vasculature – but not in the epidermis or mesophyll—was necessary for photoperiodic expression of florigen (FT) and induction of flowering (Endo et al., 2014; Shimizu et al., 2015). In contrast, the epidermal clock was critical for the thermal regulation of cell elongation (Shimizu et al., 2015; Figure 2b).
Are tissue-specific circadian clocks in plants also organized hierarchically like those in animals? Early studies suggested otherwise and, instead, suggested that plant circadian clocks worked in a cell-autonomous manner. This was in part shown when each cotyledon in Arabidopsis was entrained by light–dark cycles to be 180° out of phase with each other, while the shoot apex was entrained to an intermediate phase. Remarkably, circadian rhythms monitored by luciferase activity driven by clock-regulated promoters showed that these three spatial domains retained distinct circadian phases when allowed to free-run for several days (Thain et al., 2000). This suggested that there was little or no cell-to-cell synchronization. However, several subsequent studies have shown weak, hierarchal coupling between different plant cell types (Fukuda et al., 2007; James et al., 2008; Wenden et al., 2012). For example, perturbing the vasculature clock changed the clock component TOC1 expression not only in the vasculature but also in the mesophyll. Perturbing the mesophyll clock did not alter the vasculature clock in the same way, suggesting that the vasculature clock is dominant over that of the mesophyll (Endo et al., 2014).
This same interplay between different clocks has been shown at the organ level. The clock in roots has been shown to be a simplified version of the shoot clock; the root clock preferentially expresses evening phase clock components, while morning phased components (e.g. CCA1 and LHY) are expressed at lower levels and display reduced target promoter binding (James et al., 2008; Lee and Seo, 2018). Perhaps for this reason the function of the root clock is modulated by signals arising from the shoot (Bordage et al., 2016; James et al., 2008; Takahashi et al., 2015). These signals include sucrose (Frank et al., 2018; James et al., 2008; Philippou et al., 2019) and light, with the latter at least in part resulting from light piping from shoot to root through the vasculature (Nimmo, 2018). Importantly, long-distance signaling from the shoot apical meristem to the root modulates rhythmicity in the root (Takahashi et al., 2015). Collectively, these findings suggest a hierarchical nature of plant clocks, with the shoot apical meristem playing a coordinating role reminiscent of the mammalian SCN (Takahashi et al., 2015).
Desynchronization of the clock between cells could explain why some plant tissues display dampened rhythms (Figure 2b). Specifically, in contrast to the robust rhythms displayed by the shoot apical meristem, root tip, cotyledons and hypocotyls of Arabidopsis (Fukuda et al., 2012; Gould et al., 2018; Takahashi et al., 2015), there are two distinct waves of circadian behavior in the root. The averaging of these two waves generates the dampened rhythms typically found when expression is averaged across the whole root (Gould et al., 2018). Similar evidence for desynchronization between plant tissues being the source of dampened rhythms was shown in a separate study looking at expression of CCA1 in single nuclei of leaf tissue over extended circadian conditions (i.e. under constant light; Yakir et al., 2011). One day after shifting into constant light, guard cells and non-guard (epidermal and mesophyll) cells showed very similar peak expression times, with 64% of guard cells and 81% of non-guard cells peaking 0–3 h after dawn. Following 7 days in constant light, guard cell nuclei peaked an average of 11 h later, while the non-guard cells exhibited only a slight shift of a few hours. Analysis of protoplasts isolated from these two tissue types confirmed the desynchronization of the single-cell clocks as the driving force behind the dampened rhythms (Yakir et al., 2011).
Due to its simple structure of small floating bodies and immobile leaf fronds, the duckweed Lemna gibba has proven an attractive model for studying the interaction between single-cell oscillators (Muranaka et al., 2015). Monitoring CCA1 oscillations using a luciferase reporter uncovered cell-to-cell variation in period resulting in desynchronized rhythms. While this was consistent with previous findings in Arabidopsis, experiments in L. gibba have also uncovered evidence of reduced variation in phase and period among neighboring cells, suggesting the coupling of some cells in close proximity. This spatial pattern of phase synchronicity correlated with photosynthetic activity rhythms, suggesting that this physiological output is driven by the circadian phase (Muranaka and Oyama, 2016). Indeed, local coupling effects have also been suggested by the robust rhythms in the shoot apical meristem and root tip, possibly due to relatively high cell density, which may facilitate coupling among cells (Fukuda et al., 2012; Gould et al., 2018; Takahashi et al., 2015). What are the signaling molecules driving this cell-density-dependent coupling? In the mammalian SCN oscillator, coupling is thought to be maintained through interneuronal signaling via neuropeptides (Pilorz et al., 2020). In plants, additional single-cell studies from intact tissue that incorporate peptide and metabolite level resolution are needed to elucidate possible intercellular signaling mechanisms.
Modeling circadian tissue-specific transcriptomics genome-wide
How can one measure the circadian clock’s impact on gene regulation at the genome-wide level? Capturing circadian oscillations transcriptome-wide over several days and integrating them into gene regulatory networks remains a significant challenge. Most circadian network studies in plants focus on the properties of the core oscillator and modeling the transcriptional network using mutant analysis as well as transcriptome and TF-binding assays (Nakamichi, 2020). Mathematical modeling of the plant circadian clock, primarily based on differential equation systems, has also been used to predict core oscillator function and rhythmic output (Figure 2c; Fogelmark and Troein, 2014; Locke et al., 2005; Pokhilko et al., 2010, 2012, 2013). However, few studies incorporate the regulatory connections between the core oscillator and the transcriptome (Bonnot and Nagel, 2021; Muller et al., 2020). A recent method for predicting transcriptional networks from time course data that incorporates expression level and rates of change to infer network connections offers an effective approach for building such circadian gene regulatory networks (Desai et al., 2017). Another challenge in modeling rhythmic expression patterns genome-wide is comparing between conditions or genotypes. One solution is to integrate time course transcriptome data into co-expression networks that capture representative transcript expression patterns over time and use the gene correlation scores in a linear model to indicate significant differences in pattern between samples or treatments (Greenham et al., 2020).
At present, the modeling approaches mentioned above have typically drawn on bulk RNA-seq transcriptomic data. Whole-organ sampling averages gene expression between tissues and thus is not an accurate representation of how the clock network functions throughout the plant, particularly given the evidence for tissue-specific regulation. Clearly single-cell sequencing approaches will be necessary to identify tissue-specific transcriptional circadian outputs, including clock genes and clock-regulated output pathways that are uniquely expressed among different tissues (Figure 2c). The challenge then becomes how to integrate these tissue-specific circadian networks to predict organ level responses.
Although single-cell circadian transcriptomic studies have yet to be done in plants, initial studies in non-plant systems are shedding light on the differences in the genome-wide regulation of the circadian clock across individual cells. In animals, laser capture microdissection (LCM) has revealed how asynchronous neurons within the SCN lead to precise outputs that synchronize the body clock (Park et al., 2016). Here, both community structure detection methods and multivariate analyses generated networks connecting different cell types together based on synaptic and paracrine signaling. The resulting transcriptional patterns revealed heterogeneity among neurons with subpopulation expression states. Integrating the transcriptional states of neuropeptide and receptor gene pairs with a correlation network predicted interactions between gene clusters, providing insight into the neuronal network structure (Park et al., 2016). These differences in clock output at the cell-type level were confirmed in a single-cell SCN atlas, which was created from a combination of single-cell sequencing, LCM and in situ hybridization approaches (Wen et al., 2020). This atlas revealed core clock gene expression holding similar phasing in non-neuronal cells, which was delayed compared with neuronal cells. Importantly, the SCN atlas was able to identify cell-type-specific TFs that were likely responsible for driving rhythms in each respective cell type (Wen et al., 2020). In the future, a similar approach will hopefully reveal the cell-type-specific TFs responsible for regulating unique clock functions across plant tissues.
ENVIRONMENTAL TIME: SYNCHRONIZING GENE EXPRESSION RESPONSES WITH THE SPEED OF EXTERNAL STIMULI
To adapt to changes in the environment, plants often begin by changing the way they regulate their genes. Indeed, a shift in an external cue, such as nutrient, water or light availability, can lead to dramatic and rapid changes in expression of hundreds to thousands of genes (Jing and Lin, 2020; Krouk et al., 2010a). Often, the tempo of such transcriptional responses is aligned to the timing of the environmental cue itself. For example, some cues, such as the introduction of light (Feng et al., 2017) or nutrients (Krouk et al., 2010b; Swift et al., 2019), elicit gene regulatory responses that unfold within minutes. Others – such as the changing of seasons—are met with equally slow responses, such as the histone modifications that alter over a period of months (Nishio et al., 2020). Importantly, the timing of environmental signaling responses can hinge on the plant’s developmental state, as well as the circadian clock (Schneider et al., 2019). In this section, we explore how the timing of environmental signaling is synchronized to environmental cues, and review how such timing can be contingent on developmental or circadian timescales. Additionally, we outline how environmentally responsive gene regulatory networks can be constructed using machine learning from RNA-seq datasets.
Aligning the timing of internal expression responses with dynamic changes in external stimuli
An environmental cue can lead a plant to rapidly change regulation of hundreds to thousands of genes. For example, upon sensing nitrogen—a key plant nutrient—Arabidopsis roots and shoots change the expression of thousands of genes within minutes (Brooks et al., 2019; Varala et al., 2018). How are such gene regulatory cascades initiated? In nitrogen signaling, early sensory mechanisms, such as the nitrate transceptor NRT1.1 and the master TF NLP7, play crucial roles in initiating such nitrate response transcriptional cascades (Alvarez et al., 2020; Ho et al., 2009; Marchive et al., 2013). These and other signaling factors act quickly; changes in genome-wide differential expression can be detected as early as 3 min after nitrate exposure (Krouk et al., 2010b). Included among early differentially expressed genes are TFs, which in turn activate downstream targets, thus increasing the number of genes within the transcriptional cascade (Alvarez et al., 2021; Bouguyon et al., 2015; Chang et al., 2013; Figure 3a). What determines the rate at which such transcriptional cascades proceed? In nitrogen signaling, it was found that the dose of nitrogen itself plays a key role in the time-based response. Specifically, the rate at which genes are expressed is determined by the dose of N sensed, where higher doses led to an increased rate of transcriptional output over time (Swift et al., 2020). Curiously, the relationship between the amount of N provided to the plant and the rate of transcriptional change over time could be modeled using the Michaelis–Menten equation, suggesting the dose–response relationship between the nutrient uptake and transcriptome output can in part mirror simple enzyme kinetics (Figure 3b). Importantly, modeling the dose–response relationship in this way identified TF TGA1 as a TF that regulates dose-dependent N signaling over time. Early evidence suggests that such TF-mediated responses are also at work in glucose (Singh et al., 2017) and sucrose signaling (Zhang et al., 2016) in plants.
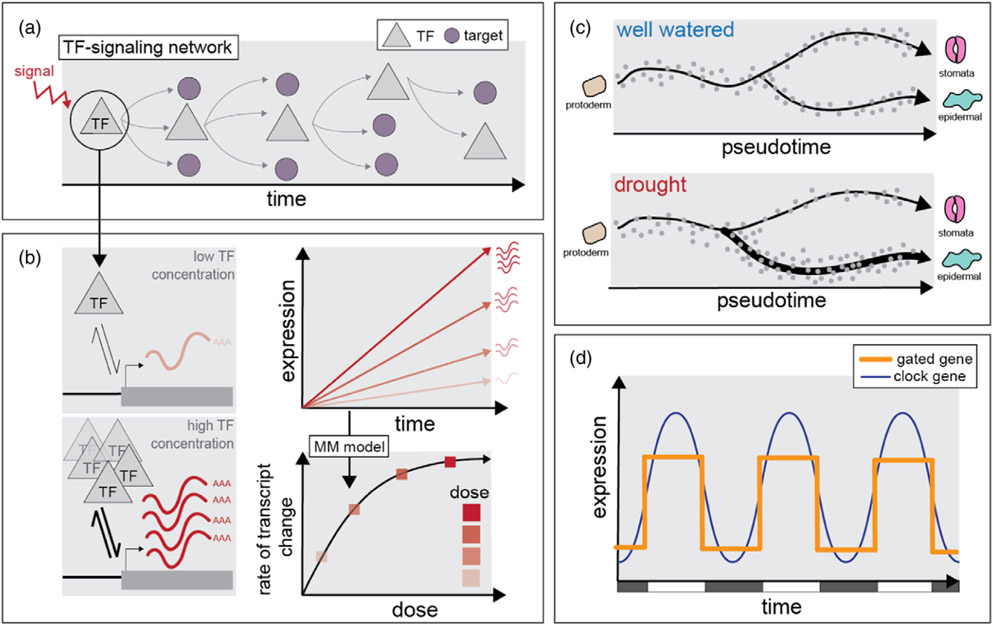
Figure 3.
The dynamic timing of plant responses to environmental signals.
(a) Transcription factor (TF) cascades are dynamic over time and can be triggered by an environmental cue.
(b) TF–target interactions underlie the kinetics of TF transcriptional cascades, where higher concentrations of a TF can lead to proportional changes in expression output. In this way, the rate of transcriptional change can be aligned to an environmental signal and explained by simple kinetic models (such as the Michaelis–Menten model; Swift et al., 2020).
(c) Environmental signaling can interface with developmental trajectories. For example, under drought conditions, developing leaves can produce fewer guard cells (Hamanishi et al., 2012).
(d) Transcriptional responses to the environment can also depend on the circadian clock. One key mechanism is ‘gating’, where the expression of an environmentally responsive gene (in orange) is restricted to a certain time of day by the circadian clock (in blue; Millar and Kay, 1996)
More broadly, the rate at which TFs bind and activate their gene targets is an important determinant in how rapid environmental signaling cascades progress. At the molecular level, TF–target binding is inherently stochastic; TFs are diffuse throughout the nucleus and only by chance find their target binding site. To improve the odds, genomes rely on a range of mechanisms to promote TF–target binding events, such as increasing TF concentration, or changing the epigenetic landscape (Figure 3b; for review, see Alvarez et al., 2021; Swift and Coruzzi, 2017). Importantly, these mechanisms enable plants to align the timing of the transcriptional output to the timing of the environmental cue. For example, in temperature sensing, through partnering with TFs the photoreceptor phytochrome B regulates target genes in a temperature-dependent manner; this is made possible by its temperature-dependent reversion from an active to inactive state (Jung et al., 2016; Legris et al., 2016). By such mechanisms, transcriptional activation of target genes over time can be proportional to the cue in question, ensuring that incremental changes in the environment over time are met with incremental changes in the transcriptome.
Environmental signaling happens in the context of developmental and circadian timescales
Crucially, the way environmental signaling responses unfold over time is often contingent on other dynamic processes. One key factor is the developmental stage of the plant. For example, a leaf’s developmental stage will inform the way it responds to drought stress. Unlike in fully developed leaves that have terminated cellular differentiation, developing leaves will respond differently; drought stress will push them into a developmental trajectory that ends cellular proliferation prematurely, resulting in comparably lower leaf area and lower levels of endoreduplication (Baerenfaller et al., 2012; Claeys and Inze, 2013). This physiological response is reflected at the transcriptional level, where leaves that are still developing have different drought-responsive expression profiles compared with developed leaves (Claeys and Inze, 2013). Indeed, within the developmental gradient of the growing leaf, cells within the expansion zone respond to drought markedly differently compared with those still undergoing proliferation (Skirycz et al., 2010; Verelst et al., 2013). Drought also impacts the proportion of cells entering different developmental lineages; to prevent water loss, the proportion of stomatal cells arising from the protodermal lineage is reduced (Figure 3c; Hamanishi et al., 2012). Such cell-type-specific responses to environmental stress will likely be further resolved using single-cell sequencing approaches, and some success has already been made for guard cell responses to drought (van Weringh et al., 2021), as well as in related stresses such as heat shock (Jean-Baptiste et al., 2019) and salinity (Dinneny et al., 2008) in the Arabidopsis root.
In a similar fashion, the circadian clock also plays an important role in determining the magnitude of the transcriptional response to an environmental perturbation. One such way is a process referred to as ‘gating’ (Figure 3d). For example, the ability of light to induce expression of LHCB (CAB) is a function of the time of day (Millar and Kay, 1996). This is, at least in part, because photoreceptor abundance varies with the circadian clock (Tóth et al., 2001), and clock components like TOC1, ELF3 and GI interact with light-signaling PIFs (Martin et al., 2018; Nieto et al., 2015; Nohales et al., 2019; Soy et al., 2016; Zhu et al., 2016). Similarly, extensive interplay exists between the circadian clock and hormone signaling, resulting in time-of-day-dependent sensitivity to various hormones, such as abscisic acid (ABA). This is mediated through ABA biosynthesis and signaling, both of which are regulated by several clock components including LHY and TOC1 (Adams et al., 2018; Legnaioli et al., 2009). In turn, ABA also influences circadian control through induction of TOC1 expression that is gated to the day, providing a mechanism for temporal drought responses (Singh and Mas, 2018). The role of the clock in ABA responses helps to explain why time of day informs transcriptional responses to drought stress in Brassica rapa (Greenham et al., 2017), Arabidopsis (Wilkins et al., 2010) and Populus (Wilkins et al., 2009), as well to related stresses such as cold (Fowler et al., 2005) and heat (Bonnot and Nagel, 2021; Grinevich et al., 2019). Indeed, for heat stress, an additional layer of post-transcriptional regulation exists – ribosomes were found to prioritize mRNAs in a circadian-dependent manner, including mRNAs encoding clock targeted TFs (Bonnot and Nagel, 2021). Hopefully, future studies will reveal which cell-specific clocks are sensing and gating these environmental cues, and how they are transmitted across tissues. At least in the case of Chlamydomonas, single-cell sequencing uncovered a later diurnal phase in iron-deficient cells (Ma et al., 2021) that is consistent with the long period observed in Arabidopsis plants under iron-limiting conditions (Chen et al., 2013; Hong et al., 2013; Salomé et al., 2013). This certainly argues for considering individual cell phase when determining the cell state, especially in multicellular organisms where phase response is likely dependent on cell identity.
Building environmentally responsive networks from time-series data
A central goal of systems biology is to map out the underlying gene regulatory networks that allow plants to respond dynamically to environmental cues (Alvarez et al., 2021). To detect these networks, one popular approach is to track how gene expression dynamics change over time using time-series RNA-seq. Assaying transcriptional change over time can provide two important clues about the structure of the underlying network (Alvarez et al., 2021). (i) First, because causality moves forward with time, fine-scale time-series experiments conducted over a period of minutes to hours can help provide an indication of network causality and hierarchy; upstream transcriptional regulators can be distinguished from downstream targets. Such information is useful, as early upstream TFs can play an outsized role on phenotype (Varala et al., 2018). (ii) Second, time-series data can catch genes whose expression turns on and off within a given time frame, and thus might be easily missed if assaying only at one time-point. For example, fine-scale time-series experiments, conducted in the order of minutes, can catch transient TF–target interactions, which are absent in steady-state data (Alvarez et al., 2014).
Unfortunately, the transcriptional cascades revealed by time-series data are typically too complex to allow one to easily reconstruct the underlying gene regulatory network. This is because often there are many TF–target interactions taking place at once, making it difficult to tease out which TFs regulate which genes. For this reason, scientists are turning to machine learning techniques to help predict the TF–target interactions within time-series data. Indeed, over the past decade many different algorithms have been applied in both plant and non-plant species to solve this question (Alvarez et al., 2021). The Inferelator is an example of a machine learning algorithm applied to model plant transcriptional responses over time (Bonneau et al., 2006; Wilkins et al., 2016). This algorithm works by assuming each gene’s expression pattern can be described as a combination of the expression of a set of unknown TFs. By these means, the Inferelator was able to predict the most likely TF–target interactions within the genome-wide expression response of rice to drought (Wilkins et al., 2016). Another example of a machine learning algorithm applied to model plant transcriptome dynamics is Dynamic Factor Graphs, which relies on linear regression to estimate the relative impact each expressed TF has on a gene’s expression (Cirrone et al., 2020). This algorithm was applied to predict the underlying transcriptional networks that govern nutrient signaling in Arabidopsis (Brooks et al., 2019; Varala et al., 2018), and has been used to forecast gene expression states at future, untested time-points (Cirrone et al., 2020). Importantly, including the rate at which genes change their expression over time (as described in the section above) has been shown to improve machine learning predictions of TF–target networks (Desai et al., 2017).
In response to the rise of single-cell methods, machine learning tools have been adapted to predict gene regulatory networks from single-cell transcriptome data. Such tools are built upon those designed to analyze bulk RNA-seq data, but hold additional parameters, such as the ability to handle sparse datasets (Pratapa et al., 2020). When applied to single-cell transcriptomic data, these methods can provide cell-type resolution of transcriptional networks (Tripathi and Wilkins, 2021), as well as give insight into the TF–target interactions that drive cell-type transitions (Matsumoto et al., 2017). An example of one method is Single-Cell Regulatory Network Inference and Clustering (SCENIC), which relies on the machine learning algorithm GENIE3. This algorithm uses random forest clustering to predict TF–target interactions within each clustered cell type. Importantly, it also draws upon cis-regulatory elements that sit upstream of regulated genes to assist in TF–target prediction (Aibar et al., 2017).
Indeed, to make predictions, most machine learning tools often need to be first provided a set of known TF–target interactions or information on the chromatin landscape (Alvarez et al., 2021). Such ‘priors’ can be computed in silico, for example by predicting which TFs bind cis-regulatory sites that sit upstream of expressed genes (Aibar et al., 2017). Priors can also be generated experimentally, typically by relying on techniques that can rapidly assay TF–target interactions. One such technique is DAP-seq, an ex vivo technique that can screen for genome-wide binding sites of a TF (O’Malley et al., 2016). Similarly, protoplast-based assays have been designed that can quickly report TF–target interactions that lead to changes in gene regulation (Bargmann et al., 2013; Brooks et al., 2019). At the single-cell level, multimodal assays that report both gene expression and regions of open chromatin mean priors can be generated from the same experimental sample (Zhu et al., 2020). Collectively, these techniques can provide machine learning algorithms with a set of ‘ground-truth’ interactions that help improve TF–target predictions.
THE FUTURE OF TIME
As described in the sections above, the way time is measured can reveal different types of gene regulation. For example, assaying transcriptional changes over real time can report how plant genomes respond to circadian or environmental cues. In contrast, pseudotime approaches are useful for detecting developmental trajectories and catching transient cell identities. However, developmental, environmental and circadian signaling do not occur in isolation to one another; they co-exist. Understanding how these timescales interact in plants is the next frontier. Plants display remarkable developmental plasticity in response to environmental change – they can grow taller to seek light or grow lateral roots to forage for nutrients. Such plasticity is in part driven by integrating developmental, environmental and circadian networks over time. Consequently, discovering the crosstalk among these cues can lead to discovering as yet unknown biological mechanisms. A fruitful means to detect such mechanisms will be to understand how these different timescales interact to regulate gene expression at the cell-type level.
Techniques are on the horizon to meet this challenge (Box 2). These include single-cell multi-omic methods that can report both the epigenome and transcriptome of different cell types (Zhu et al., 2019), CRISPR libraries that create cell-type-specific gene knockouts (Birnbaum, 2018), and single-cell proteomics (Alfaro et al., 2021). Spatial sequencing that can illustrate in situ differences in gene expression will likely prove an important technique in validating single-cell sequencing approaches (Alon et al., 2021; Cho et al., 2021; Takei et al., 2021). Protoplasting (Ortiz-Ramirez et al., 2018), LCM (Olsen and Krause, 2019) and nuclei isolation techniques (Thibivilliers et al., 2020) are constantly improving to ensure dynamic changes in plant gene regulation can be captured faithfully at the single-cell level. Dovetailing with these techniques is the growth in novel algorithms for analyzing single-cell transcriptomic data (Sagar and Grun, 2020), as well as community efforts that aim to benchmark gene regulatory markers of plant cell identity (Ma et al., 2020b; Plant Cell Atlas et al., 2021; Rhee et al., 2019). Importantly, the plant community works on a range of different plant models, many of which are important for agriculture. Thus, assaying for differences at the cell-type level between species is an important next step. Work in this direction has already indicated that while some tissues, such as the meristem, appear largely similar, others appear quite disparate (Kajala et al., 2021).
Box 2. Future directions for Plant Cell System Dynamics.
Plants display remarkable developmental plasticity in response to environmental change driven, at least in part, by integrating developmental, environmental and circadian networks over time, but the mechanisms by which such integration occurs remain largely unknown.
Advances in spatially resolved sequencing that illustrate in situ differences in gene expression are needed to validate single-cell sequencing approaches.
A continuing growth of novel algorithms will aid analysis of temporal single-cell transcriptomics datasets.
Community-wide efforts are beginning to benchmark gene regulatory markers of plant cell identity.
There is a pressing need for comparative work to determine the extent to which cell- and tissue-specific gene expression patterns vary among model species and species of agricultural and ecological importance.
In conclusion, plants are dynamic systems. No better example of this can be found than at the molecular level, where patterns of gene regulation are in constant movement. Here we have reviewed how these dynamics can unfold following developmental, circadian or environmental timescales, as well as how they can be localized in a cell- and tissue-specific manner. The challenge for the future will be to integrate across these different timescales, as well as cell types, to create a new field of Plant Cell System Dynamics, an area of research nucleated at the 2018 and 2022 Plant Molecular Biology Gordon Research Conferences, and which form the basis of this review.
FUNDING INFORMATION
Work in the area of Plant Cell System Dynamics was supported by NSF grants IOS-1547796 (to CRM), DBI-2042159 (to KG), and an NIH grant NIGMS R01-GM121753 (to GC). In addition, two conferences in the area of Plant Cell System Dynamics have been supported by NSF IOS-2005283 [for Plant Molecular Biology Gordon Research Conference: Spatial and temporal dynamics in Plant Biology, 2022 (to CRM)] and NSF IOS-1824578 [for Plant Molecular Biology Gordon Research Conference: Dynamic Plant Systems, 2018 (to GC)] and also by a Zegar Family Foundation Grant. JRE is an Investigator of the Howard Hughes Medical Institute. JS is an Open Philanthropy Awardee of the Life Sciences Research Foundation.
REFERENCES
- Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S et al. (2018) Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytologist, 220, 893–907. [DOI] [PubMed] [Google Scholar]
- Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G et al. (2017) SCENIC: single-cell regulatory network inference and clustering. Nature Methods, 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro JA, Bohlander P, Dai M, Filius M, Howard CJ, van Kooten XF et al. (2021) The emerging landscape of single-molecule protein sequencing technologies. Nature Methods, 18, 604–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon S, Goodwin DR, Sinha A, Wassie AT, Chen F, Daugharthy ER et al. (2021) Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems. Science, 371, eaax2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez JM, Brooks MD, Swift J & Coruzzi GM (2021) Time-based systems biology approaches to capture and model dynamic gene regulatory networks. Annual Review of Plant Biology, 72, 105–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez JM, Riveras E, Vidal EA, Gras DE, Contreras-Lopez O, Tamayo KP et al. (2014) Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal, 80, 1–13. [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Schinke AL, Brooks MD, Pasquino A, Leonelli L, Varala K et al. (2020) Transient genome-wide interactions of the master transcription factor NLP7 initiate a rapid nitrogen-response cascade. Nature Communications, 11, 1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Massonnet C, Walsh S, Baginsky S, Buhlmann P, Hennig L et al. (2012) Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Molecular Systems Biology, 8, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann BOR, Marshall-Colon A, Efroni I, Ruffel S, Birnbaum KD, Coruzzi GM et al. (2013) TARGET: a transient transformation system for genome-wide transcription factor target discovery. Molecular Plant, 6, 978–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz O, Xu Y, Liew LC, Wang Y, Zhu Y, Hurgobin B et al. (2021) RNA-seq analysis of laser microdissected Arabidopsis thaliana leaf epidermis, mesophyll and vasculature defines tissue-specific transcriptional responses to multiple stress treatments. The Plant Journal, 107, 938–955. [DOI] [PubMed] [Google Scholar]
- Bezrutczyk M, Zollner NR, Kruse CPS, Hartwig T, Lautwein T, Kohrer K et al. (2021) Evidence for phloem loading via the abaxial bundle sheath cells in maize leaves. The Plant Cell, 33, 531–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum KD (2018) Power in numbers: single-cell rna-seq strategies to dissect complex tissues. Annual Review of Genetics, 52, 203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW et al. (2003) A gene expression map of the Arabidopsis root. Science, 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Bonneau R, Reiss DJ, Shannon P, Facciotti M, Hood L, Baliga NS et al. (2006) The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biology, 7, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnot T & Nagel DH (2021) Time of day prioritizes the pool of translating mRNAs in response to heat stress. The Plant Cell, 33, 2164–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordage S, Sullivan S, Laird J, Millar AJ & Nimmo HG (2016) Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytologist, 212, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, Kubes M, Pervent M, Leran S et al. (2015) Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants, 1, 15015. [DOI] [PubMed] [Google Scholar]
- Brooks MD, Cirrone J, Pasquino AV, Alvarez JM, Swift J, Mittal S et al. (2019) Network Walking charts transcriptional dynamics of nitrogen signaling by integrating validated and predicted genome-wide interactions. Nature Communications, 10, 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ et al. (2019) The single-cell transcriptional landscape of mammalian organogenesis. Nature, 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KN, Zhong S, Weirauch MT, Hon G, Pelizzola M, Li H et al. (2013) Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis, eLIFE, 2, e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-Y, Wang Y, Shin L-J, Wu J-F, Shamugam V, Tsednee M et al. (2013) Iron is involved in maintenance of circadian period length in Arabidopsis. Plant Physiology, 161, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CS, Xi J, Si Y, Park SR, Hsu JE, Kim M et al. (2021) Microscopic examination of spatial transcriptome using Seq-Scope. Cell, 184 (3559–3572), e3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirrone J, Brooks MD, Bonneau R, Coruzzi GM & Shasha DE (2020) OutPredict: multiple datasets can improve prediction of expression and inference of causality. Scientific Reports, 10, 6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claeys H & Inze D (2013) The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiology, 162, 1768–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusanovich DA, Reddington JP, Garfield DA, Daza RM, Aghamirzaie D, Marco-Ferreres R et al. (2018) The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature, 555, 538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K & Timmermans MCP (2019) Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Developmental Cell, 48, 840–852. [DOI] [PubMed] [Google Scholar]
- Desai JS, Sartor RC, Lawas LM, Jagadish SVK & Doherty CJ (2017) Improving gene regulatory network inference by incorporating rates of transcriptional changes. Scientific Reports, 7, 17244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S et al. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science, 320, 942–945. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Parkinson K & Webb AAR (2004) Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytologist, 162, 63–70. [Google Scholar]
- Dorrity MW, Alexandre CM, Hamm MO, Vigil AL, Fields S, Queitsch C et al. (2021) The regulatory landscape of Arabidopsis thaliana roots at single-cell resolution. Nature Communications, 12, 3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I (2018) A conceptual framework for cell identity transitions in plants. Plant and Cell Physiology, 59, 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M (2016) Tissue-specific circadian clocks in plants. Current Opinion in Plant Biology, 29, 44–49. [DOI] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T & Kay SA (2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature, 515, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard F, Baptista MAP, Krammer T, Hennig T, Lange M, Arampatzi P et al. (2019) scSLAM-seq reveals core features of transcription dynamics in single cells. Nature, 571, 419–423. [DOI] [PubMed] [Google Scholar]
- Farmer A, Thibivilliers S, Ryu KH, Schiefelbein J & Libault M (2021) Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Molecular Plant, 14, 372–383. [DOI] [PubMed] [Google Scholar]
- Feng F, Mei H, Fan P, Li Y, Xu X, Wei H et al. (2017) Dynamic transcriptome and phytohormone profiling along the time of light exposure in the mesocotyl of rice seedling. Scientific Reports, 7, 11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogelmark K & Troein C (2014) Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Computational Biology, 10, e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SG, Cook D & Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiology, 137, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank A, Matiolli CC, Viana AJC, Hearn TJ, Kusakina J, Belbin FE et al. (2018) Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Current Biology, 28, 2597–2606.e2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H, Nakamichi N, Hisatsune M, Murase H & Mizuno T (2007) Synchronization of plant circadian oscillators with a phase delay effect of the vein network. Physical Review Letters, 99, 98102. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Ukai K & Oyama T (2012) Self-arrangement of cellular circadian rhythms through phase-resetting in plant roots. Physical Review E, 86, 41917. [DOI] [PubMed] [Google Scholar]
- Gala HP, Lanctot A, Jean-Baptiste K, Guiziou S, Chu JC, Zemke JE et al. (2021) A single-cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. The Plant Cell, 33, 2197–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM & Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences of the United States of America, 105, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorkin DU, Barozzi I, Zhao Y, Zhang Y, Huang H, Lee AY et al. (2020) An atlas of dynamic chromatin landscapes in mouse fetal development. Nature, 583, 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Domijan M, Greenwood M, Tokuda IT, Rees H, Kozma-Bognar L et al. (2018) Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. eLIFE, 7, e31700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, Guadagno CR, Gehan MA, Mockler TC, Weinig C, Ewers BE et al. (2017) Temporal network analysis identifies early physiological and transcriptomic indicators of mild drought in Brassica rapa. eLIFE, 6, e29655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenham K, Sartor RC, Zorich S, Lou P, Mockler TC & McClung CR (2020) Expansion of the circadian transcriptome in Brassica rapa and genome-wide diversification of paralog expression patterns, eLIFE, 9, e58993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinevich DO, Desai JS, Stroup KP, Duan JQ, Slabaugh E & Doherty CJ (2019) Novel transcriptional responses to heat revealed by turning up the heat at night. Plant Molecular Biology, 101, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanishi ET, Thomas BR & Campbell MM (2012) Drought induces alterations in the stomatal development program in Populus. Journal of Experimental Botany, 63, 4959–4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T et al. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science, 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC & Tsay YF (2009) CHL1 functions as a nitrate sensor in plants. Cell, 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hong S, Kim SA, Guerinot ML & McClung CR (2013) Reciprocal interaction of the circadian clock with the Fe homeostasis network in Arabidopsis thaliana. Plant Physiology, 161, 893–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI et al. (2008) The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science, 322, 1832–1835. [DOI] [PubMed] [Google Scholar]
- Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, Dorrity MW, Saunders L, Bubb KL et al. (2019) Dynamics of gene expression in single root cells of Arabidopsis thaliana. The Plant Cell, 31, 993–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y & Lin R (2020) Transcriptional regulatory network of the light signaling pathways. New Phytologist, 227, 683–697. [DOI] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science, 354, 886–889. [DOI] [PubMed] [Google Scholar]
- Kajala K, Gouran M, Shaar-Moshe L, Mason GA, Rodriguez-Medina J, Kawa D et al. (2021) Innovation, conservation, and repurposing of gene function in root cell type development. Cell, 184, 3333–3348. [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Stuart T, Valdes M, Breakfield N, Schmitz RJ, Nery JR et al. (2016) Unique cell-type-specific patterns of DNA methylation in the root meristem. Nature Plants, 2, 16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Kim HS, Choi SH, Jang JY, Jeong MJ & Lee SI (2017) The importance of the circadian clock in regulating plant metabolism. International Journal of Molecular Sciences, 18, 2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Symeonidi E, Pang TY, Denyer T, Weidauer D, Bezrutczyk M et al. (2021) Distinct identities of leaf phloem cells revealed by single cell transcriptomics. The Plant Cell, 33, 511–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S, Javelle M, Li L, Li X, Ma X, Wimalanathan K et al. (2019) A high-resolution gene expression atlas links dedicated meristem genes to key architectural traits. Genome Research, 29, 1962–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotliar D, Veres A, Nagy MA, Tabrizi S, Hodis E, Melton DA et al. (2019) Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-seq, eLIFE, 8, e43803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Crawford NM, Coruzzi GM & Tsay YF (2010a) Nitrate signaling: adaptation to fluctuating environments. Current Opinion in Plant Biology, 13, 266–273. [DOI] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE & Coruzzi GM (2010b) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biology, 11, R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V et al. (2018) RNA velocity of single cells. Nature, 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer AR & La Manno G (2020) The emergence and promise of single-cell temporal-omics approaches. Current Opinion in Biotechnology, 63, 70–78. [DOI] [PubMed] [Google Scholar]
- Lee HG & Seo PJ (2018) Dependence and independence of the root clock on the shoot clock in Arabidopsis. Genes Genomics, 40, 1063–1068. [DOI] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J & Mas P (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO Journal, 28, 3745–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A et al. (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science, 354, 897–900. [DOI] [PubMed] [Google Scholar]
- Liu Q, Liang Z, Feng D, Jiang S, Wang Y, Du Z et al. (2021) Transcriptional landscape of rice roots at the single-cell resolution. Molecular Plant, 14, 384–394. [DOI] [PubMed] [Google Scholar]
- Locke JCW, Southern MM, Kozma-Bognar L, Hibberd V, Brown PE, Turner MS et al. (2005) Extension of a genetic network model by iterative experimentation and mathematical analysis. Molecular Systems Biology, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Anido CB, Vaten A, Smoot NK, Sharma N, Guo V, Gong Y et al. (2021) Single-cell resolution of lineage trajectories in the Arabidopsis stomatal lineage and developing leaf. Developmental Cell, 56(1043–1055), e1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Hajkova P & Ecker JR (2018) Dynamic DNA methylation: in the right place at the right time. Science, 361, 1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Salome PA, Merchant SS & Pellegrini M (2021) Single-cell RNA sequencing of batch Chlamydomonas cultures reveals heterogeneity in their diurnal cycle phase. The Plant Cell, 33, 1042–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Zhang B, LaFave LM, Earl AS, Chiang Z, Hu Y et al. (2020a) Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell, 183, 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Denyer T & Timmermans MCP (2020b) PscB: a browser to explore plant single cell RNA-sequencing data sets. Plant Physiology, 183, 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marand AP, Chen Z, Gallavotti A & Schmitz RJ (2021) A cis-regulatory atlas in maize at single-cell resolution. Cell, 184(3041–3055), e3021. [DOI] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Brehaut V, Blondet E, Colot V et al. (2013) Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nature Communications, 4, 1713. [DOI] [PubMed] [Google Scholar]
- Martin G, Rovira A, Veciana N, Soy J, Toledo-Ortiz G, Gommers CMM et al. (2018) Circadian waves of transcriptional repression shape PIF-regulated photoperiod-responsive growth in Arabidopsis. Current Biology, 28(311–318), e315. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Kiryu H, Furusawa C, Ko MSH, Ko SBH, Gouda N et al. (2017) SCODE: an efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation. Bioinformatics, 33, 2314–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFaline-Figueroa JL, Trapnell C & Cuperus JT (2020) The promise of single-cell genomics in plants. Current Opinion in Plant Biology, 54, 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meir Z, Aviezer I, Chongloi GL, Ben-Kiki O, Bronstein R, Mukamel Z et al. (2021) Dissection of floral transition by single-meristem transcriptomes at high temporal resolution. Nature Plants, 7, 800–813. [DOI] [PubMed] [Google Scholar]
- Michael TP, Salomé PA & McClung CR (2003) Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proceedings of the National Academy of Sciences of the United States of America, 100, 6878–6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ & Kay SA (1996) Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 93, 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittnenzweig M, Mayshar Y, Cheng S, Ben-Yair R, Hadas R, Rais Y et al. (2021) A single-embryo, single-cell time-resolved model for mouse gastrulation. Cell, 184, 2825–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB & Takahashi JS (2012) Central and peripheral circadian clocks in mammals. Annual Review of Neuroscience, 35, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SA (2019) The evolving concept of cell identity in the single cell era. Development, 146, dev169748. [DOI] [PubMed] [Google Scholar]
- Müller LM, Mombaerts L, Pankin A, Davis SJ, Webb AAR, Goncalves J et al. (2020) Differential effects of day/night cues and the circadian clock on the barley transcriptome. Plant Physiology, 183, 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranaka T, Okada M, Yomo J, Kubota S & Oyama T (2015) Characterisation of circadian rhythms of various duckweeds. Plant Biology, 17, 66–74. [DOI] [PubMed] [Google Scholar]
- Muranaka T & Oyama T (2016) Heterogeneity of cellular circadian clocks in intact plants and its correction under light-dark cycles. Science Advances, 2, e1600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N (2020) The transcriptional network in the Arabidopsis circadian clock system. Genes, 11, 1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms B & Walbot V (2019) Defining the developmental program leading to meiosis in maize. Science, 364, 52–56. [DOI] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière J-M & Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Current Biology, 25, 187–193. [DOI] [PubMed] [Google Scholar]
- Nimmo HG (2018) Entrainment of Arabidopsis roots to the light:dark cycle by light piping. Plant, Cell and Environment, 41, 1742–1748. [DOI] [PubMed] [Google Scholar]
- Nishio H, Nagano AJ, Ito T, Suzuki Y & Kudoh H (2020) Seasonal plasticity and diel stability of H3K27me3 in natural fluctuating environments. Nature Plants, 6, 1091–1097. [DOI] [PubMed] [Google Scholar]
- Nohales MA (2021) Spatial organization and coordination of the plant circadian system. Genes, 12, e89030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales MA, Liu W, Duffy T, Nozue K, Sawa M, Pruneda-Paz JL et al. (2019) Multi-level modulation of light signaling by GIGANTEA regulates both the output and pace of the circadian clock. Developmental Cell, 49, 840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen S & Krause K (2019) A rapid preparation procedure for laser microdissection-mediated harvest of plant tissues for gene expression analysis. Plant Methods, 15, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley RC, Huang SS, Song L, Lewsey MG, Bartlett A, Nery JR et al. (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell, 165, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M, Gil-Yarom N, Yahav C, Steiner E & Efroni I (2020) A conserved superlocus regulates above- and belowground root initiation. bioRxiv. 10.1101/2020.11.11.377937 [DOI] [PubMed] [Google Scholar]
- Ortiz-Ramirez C, Arevalo ED, Xu X, Jackson DP & Birnbaum KD (2018) An efficient cell sorting protocol for maize protoplasts. Current Protocols in Plant Biology, 3, e20072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Zhu H, O’Sullivan S, Ogunnaike BA, Weaver DR, Schwaber JS et al. (2016) Single-cell transcriptional analysis reveals novel neuronal phenotypes and interaction networks involved in the central circadian clock. Frontiers in Neuroscience, 10, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippou K, Ronald J, Sánchez-Villarreal A, Davis AM & Davis SJ (2019) Physiological and genetic dissection of sucrose inputs to the Arabidopsis thaliana circadian system. Genes, 10, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard CL, Povilus RA, Williams BP & Gehring M (2021) Transcriptional and imprinting complexity in Arabidopsis seeds at single-nucleus resolution. Nature Plants, 7, 730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilorz V, Astiz M, Heinen KO, Rawashdeh O & Oster H (2020) The concept of coupling in the mammalian circadian clock network. Journal of Molecular Biology, 432, 3618–3638. [DOI] [PubMed] [Google Scholar]
- Pinheiro I, Torres-Padilla ME & Almouzni G (2021) Epigenomics in the single cell era, an important read out for genome function and cell identity. Epigenomics, 13, 981–984. [DOI] [PubMed] [Google Scholar]
- Plant Cell Atlas Consortium; Jha SG, Borowsky AT, Cole BJ, Fahlgren N, Farmer A, Huang SC et al. (2021), Vision, challenges and opportunities for a plant cell atlas. eLIFE, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ & Millar AJ (2012) The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Molecular Systems Biology, 8, 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW et al. (2010) Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Molecular Systems Biology, 6, 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Mas P & Millar AJ (2013) Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Systems Biology, 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratapa A, Jalihal AP, Law JN, Bharadwaj A & Murali TM (2020) Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nature Methods, 17, 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Birnbaum KD & Ehrhardt DW (2019) Towards building a plant cell atlas. Trends in Plant Science, 24, 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roszak P, Heo J-O, Blob B, Toyokura K, de Luis Balaguer MA Lau W et al. (2021) Analysis of phloem trajectory links tissue maturation to cell specialization. bioRxiv. 10.1101/2021.01.18.427084 [DOI] [Google Scholar]
- Saelens W, Cannoodt R, Todorov H & Saeys Y (2019) A comparison of single-cell trajectory inference methods. Nature Biotechnology, 37, 547–554. [DOI] [PubMed] [Google Scholar]
- Sagar & Grün D (2020) Deciphering cell fate decision by integrated single-cell sequencing analysis. Annual Review of Biomedical Data Science, 3, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai J & Johnson CH (1999) Different circadian oscillators control Ca(2+) fluxes and Lhcb gene expression. Proceedings of the National Academy of Sciences of the United States of America, 96, 11659–11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Oliva M, Weigel D & Krämer U (2013) Circadian clock adjustment to plant iron status depends on chloroplast and phytochrome function. EMBO Journal, 32, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF & Regev A (2015) Spatial reconstruction of single-cell gene expression data. Nature Biotechnology, 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B (2001) Plant cell identity. The role of position and lineage. Plant Physiology, 125, 112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Bolger A, Zeier J, Preiskowski S, Benes V, Trenkamp S et al. (2019) Fluctuating light interacts with time of day and leaf development stage to reprogram gene expression. Plant Physiology, 179, 1632–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahan R, Hsu CW, Nolan TM, Cole BJ, Taylor IW, Vlot AHC et al. (2020) A single cell Arabidopsis root atlas reveals developmental trajectories in wild type and cell identity mutants. bioRxiv. 10.1101/2020.06.29.178863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu H, Katayama K, Koto T, Torii K, Araki T & Endo M (2015) Decentralized circadian clocks process thermal and photoperiodic cues in specific tissues. Nature Plants, 1, 15163. [DOI] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Ciobanu D, Lin J, Yoshinaga Y, Gouran M et al. (2019) High-throughput single-cell transcriptome profiling of plant cell types. Cell Reports, 27(2241–2247), e2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Gupta A, Singh D, Khurana JP & Laxmi A (2017) Arabidopsis RSS1 mediates cross-talk between glucose and light signaling during hypocotyl elongation growth. Scientific Reports, 7, 16101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M & Mas P (2018) A functional connection between the circadian clock and hormonal timing in Arabidopsis. Genes, 9, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, De Bodt S, Obata T, De Clercq I, Claeys H, De Rycke R et al. (2010) Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant Physiology, 152, 226–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin ML & Nusinow DA (2021) Time will tell: intercellular communication in the plant clock. Trends in Plant Science, 26, 706–719. [DOI] [PubMed] [Google Scholar]
- Soy J, Leivar P, González-Schain N, Martín G, Diaz C, Sentandreu M et al. (2016) Molecular convergence of clock and photosensory pathways through PIF3–TOC1 interaction and co-occupancy of target promoters. Proceedings of the National Academy of Sciences of the United States of America, 113, 4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Adame M, Tranchina D, Henry A & Coruzzi GM (2019) Water impacts nutrient dose responses genome-wide to affect crop production. Nature Communications, 10, 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J, Alvarez JM, Araus V, Gutierrez RA & Coruzzi GM (2020) Nutrient dose-responsive transcriptome changes driven by Michaelis-Menten kinetics underlie plant growth rates. Proceedings of the National Academy of Sciences of the United States of America, 117, 12531–12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift J & Coruzzi GM (2017) A matter of time—how transient transcription factor interactions create dynamic gene regulatory networks. Biochim Biophys Acta-Gene Regulatory Mechanisms, 1860, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Hirata Y, Aihara K & Mas P (2015) A hierarchical multi-oscillator network orchestrates the Arabidopsis circadian system. Cell, 163, 148–159. [DOI] [PubMed] [Google Scholar]
- Takei Y, Yun J, Zheng S, Ollikainen N, Pierson N, White J et al. (2021) Integrated spatial genomics reveals global architecture of single nuclei. Nature, 590, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain SC, Hall A & Millar AJ (2000) Functional independence of multiple circadian clocks that regulate plant gene expression. Current Biology, 10, 951–956. [DOI] [PubMed] [Google Scholar]
- Thibivilliers S, Anderson D & Libault M (2020) Isolation of plant root nuclei for single cell RNA sequencing. Current Protocols in Plant Biology, 5, e20120. [DOI] [PubMed] [Google Scholar]
- Tomioka K & Matsumoto A (2010) A comparative view of insect circadian clock systems. Cellular and Molecular Life Sciences, 67, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R, Kevei É, Hall A, Millar AJ, Nagy F & Kozma-Bognár L (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiology, 127, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi RK & Wilkins O (2021) Single cell gene regulatory networks in plants: opportunities for enhancing climate change stress resilience. Plant, Cell and Environment, 44, 2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weringh A, Pasha A, Esteban E, Gamueda PJ & Provart NJ (2021) Generation of guard cell RNA-seq transcriptomes during progressive drought and recovery using an adapted INTACT protocol for Arabidopsis thaliana shoot tissue. bioRxiv. 10.1101/2021.04.15.439991 [DOI] [Google Scholar]
- Varala K, Marshall-Colon A, Cirrone J, Brook MD, Pasquino AV, Leran S et al. (2018) Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proceedings of the National Academy of Sciences of the United States of America, 115, 6494–6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verelst W, Bertolini E, De Bodt S, Vandepoele K, Demeulenaere M, Pe ME et al. (2013) Molecular and physiological analysis of growth-limiting drought stress in Brachypodium distachyon leaves. Molecular Plant, 6, 311–322. [DOI] [PubMed] [Google Scholar]
- Wen S, Ma D, Zhao M, Xie L, Wu Q, Gou L et al. (2020) Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nature Neuroscience, 23, 456–467. [DOI] [PubMed] [Google Scholar]
- Wenden B, Toner DLK, Hodge SK, Grima R & Millar AJ (2012) Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proceedings of the National Academy of Sciences of the United States of America, 109, 6757–6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Bräutigam K & Campbell MM (2010) Time of day shapes Arabidopsis drought transcriptomes. The Plant Journal, 63, 715–727. [DOI] [PubMed] [Google Scholar]
- Wilkins O, Hafemeister C, Plessis A, Holloway-Phillips MM, Pham GM, Nicotra AB et al. (2016) EGRINs (environmental gene regulatory influence networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. The Plant Cell, 28, 2365–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Waldron L, Nahal H, Provart NJ & Campbell MM (2009) Genotype and time of day shape the Populus drought response. The Plant Journal, 60, 703–715. [DOI] [PubMed] [Google Scholar]
- Xie Y, Jiang S, Li L, Yu X, Wang Y, Luo C et al. (2020) Single-cell RNA sequencing efficiently predicts transcription factor targets in plants. Frontiers in Plant Science, 11, 603302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Li X, Gao X, Jia Y, Li F & Zhang Z (2021) Decoding nucleosome positions with ATAC-seq data at single-cell level. bioRxiv. 10.1101/2021.02.07.430096 [DOI] [PubMed] [Google Scholar]
- Xu M, Du Q, Tian C, Wang Y & Jiao Y (2021a) Stochastic gene expression drives mesophyll protoplast regeneration. Science Advances, 7, eabg8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Crow M, Rice BR, Li F, Harris B, Liu L et al. (2021b) Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Developmental Cell, 56, 557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakir E, Hassidim M, Melamed-Book N, Hilman D, Kron I & Green RM (2011) Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. The Plant Journal, 68, 520–531. [DOI] [PubMed] [Google Scholar]
- Zhang TQ, Xu ZG, Shang GD & Wang JW (2019) A single-cell RNA sequencing profiles the developmental landscape of Arabidopsis root. Molecular Plant, 12, 648–660. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhu JY, Roh J, Marchive C, Kim SK, Meyer C et al. (2016) TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Current Biology, 26, 1854–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Preissl S & Ren B (2020) Single-cell multimodal omics: the power of many. Nature Methods, 17, 11–14. [DOI] [PubMed] [Google Scholar]
- Zhu C, Yu M, Huang H, Juric I, Abnousi A, Hu R et al. (2019) An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nature Structural & Molecular Biology, 26, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T & Wang ZY (2016) TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nature Communications, 7, 13692. [DOI] [PMC free article] [PubMed] [Google Scholar]