Abstract
Immunotherapies have drastically improved clinical outcomes in a wide range of malignancies. Nevertheless, patient responses remain highly variable, and reliable biomarkers that predict responses accurately are not yet fully understood. Compelling evidence from preclinical studies and observational data from clinical cohorts have shown that commensal microorganisms that reside in the human gastrointestinal tract, collectively termed the ‘microbiome’, can actively modify responses to chemotherapeutic agents and immunotherapies by influencing host immunosurveillance. Notably, microbial correlates are largely context specific, and response signatures may vary by patient population, geographic location and type of anticancer treatment. Therefore, the incongruence of beneficial microbiome signatures across studies, along with an emerging understanding of the mechanisms underlying the interactions between the microbiome, metabolome and host immune system, highlight a critical need for additional comprehensive and standardized multi-omics studies. Future research should consider key host factors, such as diet and use of medication, in both preclinical animal models and large-scale, multicenter clinical trials. In addition, there is a strong rationale to evaluate the microbiome as a tumor-extrinsic biomarker of clinical outcomes and to test the therapeutic potential of derived microbial products (e.g. defined microbial consortia), with the eventual goal of improving the efficacy of existing anticancer treatments. This review discusses the importance of the microbiome from the perspective of cancer immunotherapies, and outlines future steps that may contribute to wide-ranging clinical and translational benefits that may improve the health and quality of life of patients with cancer.
Key words: gut microbiome, immunotherapy, oncology, therapeutic response, reverse translation, next-generation microbial therapeutics
Highlights
-
•
The gut microbiome impacts the outcomes of cancer treatment by influencing host immunosurveillance.
-
•
Modulation of microbiota represents a novel therapeutic strategy to improve responses.
-
•
Incongruent beneficial bacterial signatures complicate the design of modulators.
-
•
Reverse translation processes can be used to characterize candidate bacteria.
-
•
Rationally designed microbial consortia catalyze transition to a healthy ecology.
Introduction
The development of novel immunotherapies, including immune checkpoint inhibitor (ICI) agents which target regulatory pathways in T cells to enhance antitumor immune responses, has led to remarkable improvements in clinical outcomes for patients with cancer. Nevertheless, patient responses to these treatments remain variable, with some patients responding exceptionally well to therapy and others exhibiting no response.1, 2, 3, 4, 5, 6 Overall, there is an imminent need for robust and accurate predictive markers. While compelling, existing tumor intrinsic and extrinsic biomarkers that predict clinical outcomes have proven to be inconsistent within and across tumor types, and have therefore not entered routine clinical practice.7, 8, 9
There is substantial interest in using the gastrointestinal (gut) microbiota as a biomarker and/or target with the potential to modulate patient responses to cancer immunotherapy.7,10, 11, 12 The microbiome is a complex collection of microorganisms comprising bacteria, viruses, fungi, protozoa and their collective genomes.10 Microbiota have a beneficial impact on human health through their interactions with host cells and tissues by providing accessibility to nutrients, maintaining the integrity of the mucosal barrier, contributing to the development of the immune system, and maintaining metabolic and inflammatory homeostasis.7 Additionally, bacterial metabolites can enhance barrier function to prevent translocation events,13 and reduce colonization and infection by pathobionts that can influence mortality in patients with cancer.14 Continual advances in next-generation analytic tools have expedited our understanding of this vast ecology by facilitating the rapid characterization of trillions of microorganisms and their comparisons across clinical phenotypes.15, 16, 17
The overall effect of gut-derived microbiota on cancer treatment stems from the complex interplay among the microbiota, the tumor and the immune system.11 This active and ongoing cross-talk with the host has been reported to influence downstream immunological changes in preclinical disease models.10, 11, 12, 13, 14, 15, 16, 17, 18 Clinical studies have further demonstrated that various bacterial taxa correlate with response to immunotherapy in patients with cancer. However, since results are inconsistent between studies, the impact of biological versus methodological differences is unclear.19,20
Nevertheless, the promise of these initial findings warrants additional studies to better understand the relevance of the gut microbiome, its impact on cancer immunotherapies and its potential to predict clinical response. Several key questions remain unanswered, including the optimal composition of the gut microbiome for successful cancer treatment, the most important metrics that should be used to determine the potential clinical benefits of treatment, and how well the human gut microbiome is recapitulated in preclinical models. Technical and logistical challenges include the need to standardize sample handling procedures (e.g. collection, storage and processing), discovery techniques (e.g. sequencing), data processing and analysis methods including the reference databases on which they depend, and choice of animal models to perform translational experiments.7,11 This review discusses the rationale for combining ICIs and other targeted immunotherapies with microbiome therapeutics, and provides an overview of current research and approaches to study design for the use of such combinatorial regimens.
Gut microbiome signatures correlate with the efficacy of anticancer immunotherapies
The advent of ICIs has led to fundamental changes in our approach to cancer therapy.21 At the same time, a growing body of evidence has indicated the existence of a direct link between favorable bacterial signatures in the gut and enhanced antitumor immunity. These developments have sparked tremendous interest in exploring the utility of the gut microbiome, both as a biomarker to reliably identify patient subsets most likely to benefit from therapy and as an adjunct therapy to improve clinical outcomes.22, 23, 24, 25, 26
The precise mechanisms by which the gut microbiota can affect clinical responses are yet to be completely elucidated, although a number of theories have been advanced to explain the multifactorial role of gut commensals.27, 28, 29 The reported immunomodulatory role of the gut microbiota ranges from suppressive to stimulatory effects, and mechanisms of suppression include the modulation of T-regulatory and myeloid-derived suppressive cell function.22,30 A stimulatory role is represented by priming adaptive immune responses through the interaction of toll-like receptors on antigen-presenting cells and microbial components including pathogen-associated molecular patterns or microbial metabolites at the gut mucosal interface. Additional stimulatory effects can be brought about by induction of inflammatory signaling pathways through microbiota-induced cytokine production by lymphocytes, and systemic dissemination of microbial products or metabolites that can skew the inflammatory milieu.31
Critical initial insights have been drawn from preclinical mouse models in which the effectiveness of conventional chemotherapeutic agents (e.g. cyclophosphamide) and platinum-based agents (e.g. oxaliplatin) was reported to be dependent on the gut microbiota.32 These actions were mediated by a therapy-induced increase in gut permeability, resulting in translocation of beneficial bacteria, such as Lactobacillus johnsonii and Enterococcus hirae, into the mesenteric lymph nodes and priming of robust Th17 and memory Th1 responses,33 or by generation of reactive oxygen species by tumor-associated myeloid cells triggered by MyD88-dependent microbial sensing in the gut.34 Bacteria may also be detrimental to chemotherapeutic efficacy, as observed in colorectal cancer through activation of the autophagy pathway by Fusobacterium nucleatum,35 the breakdown of gemcitabine to its inactive form by intratumoral bacteria in pancreatic cancer,36 or reactivation of irinotecan to its toxic by-product by microbial beta-glucuronidases.37
Given their strong influence on the host immune system, it is not surprising that gut commensal bacteria can also modulate responses to ICIs (i.e. anti-PD-1, anti-PD-L1 and anti-CTLA-4 antibody therapy).7,10,20,31 A study in germ-free mice reported a lack of efficacy with anti-CTLA-4 antibody treatment in the absence of microbiota, and subsequent augmentation of its action upon oral feeding with Bacteroides fragilis in combination with either Bacteroides thetaiotaomicron or Burkholderia cepacia, or upon fecal microbiota transplant (FMT; the transfer of stool from a healthy screened donor to a recipient) using a Bacteroides-rich post-anti-CTLA-4 stool sample from a patient.38 Similarly, contrasting efficacies in mice treated with anti-PD-1 and reared at different sites were associated with selective enrichment of the genus Bifidobacterium, which was closely linked to maturation of dendritic cells and increased tumor-specific CD8+ T-cell activity. Strikingly, these beneficial effects could be transferred by co-housing the animals, thereby facilitating the transfer of bacteria, or by oral supplementation with Bifidobacterium species.39
Results from recent studies in multiple patient cohorts have lent further credence to the reported associations between the gut microbiota and the efficacy of immunotherapeutic agents.22, 23, 24, 25, 26 Broadly, a more diverse microbiota and differential enrichment of specific bacterial taxa have been associated with improved responses. Characterization of the microbiome in a unique cohort of patients with late-stage melanoma beginning treatment with anti-PD-1 therapy revealed high alpha diversity of the gut microbiome to be positively associated with treatment response, and identified two distinct patient clusters on the basis of bacterial abundances. One of these clusters was composed exclusively of patients who responded beneficially to treatment (responders) and was characterized by a bacterial signature (Type-I signature) rich in Ruminococcaceae, a member family of the Firmicutes, which play a dominant role in gut homeostasis. Furthermore, the presence of favorable bacteria in the gut was correlated with increased density of an antitumor cytolytic immune infiltrate at the tumor site and in the peripheral circulation.22 Similar observations have been made in unrelated cohorts in which members of the Ruminococcaceae family, such as Faecalibacterium prausnitzii, were consistently found to be associated with beneficial treatment outcomes.25,26 Complementary results were also noted in a separate study of patients with late-stage melanoma, wherein differences in baseline microbiota were reported between responders and non-responders. In this case, the gut microbiota of responders was found to be enriched in Bifidobacterium longum, Collinsella aerofaciens and Enterococcus faecium.23 In an independent cohort of patients with epithelial cancer (e.g. lung, bladder, renal) undergoing treatment with anti-PD-1 agents, concurrent treatment with broad-spectrum systemic antibiotics was found to hamper the efficacy of ICIs. Responders were characterized by enrichment of Akkermansia muciniphila; subsequent animal studies suggest that this species is sufficient to restore anti-PD-1 activity in germ-free mice mediated by an increase in the ratio of CD4+ T cells to CD4+ FoxP3+ regulatory T cells.24
Taken together, these data suggest that certain bacterial populations are strongly associated with treatment response. The studies showed conclusively that FMT from responder and non-responder patients into germ-free mice, followed by tumor engraftment and blockade of the PD-1 axis, recapitulated the clinical donor phenotype and produced better responses in mice receiving responder FMT than in mice receiving non-responder FMT.22, 23, 24 Such findings have spurred collaborative efforts across academic, biotechnological and pharmaceutical entities to further delineate and independently validate favorable microbiome signatures (taxonomic and functional) of response, with the eventual goal of developing microbiome-derived therapeutics that can be combined with ICI agents.
Despite these advancements, it is also important to consider that there is modest overlap in bacterial taxa that have been associated with improved ICI treatment responses across cohorts. This variability may result from several inherent differences across studies, such as geographic, dietary and lifestyle characteristics of the patient populations, as well as a lack of standardized sample preparation and analysis methods, as noted previously. These discrepancies highlight the need for continued validation, prospective investigation and mechanism-based microbiome studies to further delineate the underlying targets, including their relationships with other factors that contribute to treatment response.11,40 More importantly, these taxonomic differences might be moot in light of the increasing appreciation of functional redundancy, whereby groups of phylogenetically distinct bacteria may perform similar metabolic functions to alter the host immune system.
Strategies to modulate the microbiome
A growing body of preclinical and observational studies7,12,20,22,23,25,26,31,33,36,38 suggest that microbiome modulation may be a potential therapeutic strategy to improve the proportion of patients who exhibit clinical benefit from anti-PD1 therapy. A number of investigational strategies are currently being explored such as dietary interventions, prebiotics, probiotics and, most recently, FMT. Donors are generally selected based on a favorable microbial profile or preferred clinical phenotype. As the entire microbial ecosystem is transplanted, usually after antibiotic pretreatment, this strategy has the benefit of achieving robust engraftment with less competition from the recipient's preexisting microbiome. FMT's are being studied as an investigational treatment for a variety of diseases, including recurrent Clostridium difficile infection (CDI), which is characterized by low microbial diversity.41, 42, 43, 44 A proof-of-concept has been established for the use of FMT for CDI, although first-dose efficacy rates range from 52% to 96% depending on dose, route of administration, patient selection factors and the quality of the trial design.45,46 Clinical resolution is associated with increased microbial diversity and increased concentrations of secondary bile acids, providing evidence of the critical role of microbiome restoration in preventing recurrent infection.47, 48, 49 Although there is general agreement of the importance of screening stool donors for infectious diseases and underlying medical conditions,50 no mandatory guidelines exist, leading to inconsistency in donor stool screening and processing. Whole stool transplants carry the risk of transmission of emerging pathogens, which limits the viability of FMT as a therapeutic option, particularly with repeated dosing. A recent safety report by the US Food and Drug Administration (FDA) documented transmission of drug-resistant bacteria from donor material to two immunocompromised hosts, which resulted in hospitalizations from bacteremia and one death.51 A national FMT registry, supported by the American Gastroenterology Association, has been initiated as the long-term risk of whole stool transplants is unknown.52,53 A more selective approach is clearly needed to define the microbial components required to improve gastrointestinal health while mitigating patient risk.
Commercially available probiotics have been studied in preclinical models and are currently undergoing testing in clinical trials, including in patients with cancer (NCT03072641, NCT03358511), but have thus far yielded mixed results.31 Most probiotics are marketed as dietary supplements and are not subjected to a rigorous review process by the US FDA.54 Moreover, the composition of these supplements is inadequately studied and regulated, and their purported health benefits do not have the backing of robust scientific evidence derived from placebo-controlled clinical studies.
Enzymatic and chemical food digestion is one of the primary functions of gut bacteria, as the host depends on gut commensal bacteria to metabolize several key nutrients.55 It follows that diet and the composition of the gut microbiota, along with their transcriptional profiles and metabolites, are also closely linked.56 Changes in dietary intake, especially fiber, have been reported to cause notable changes in the abundance of bacteria that can influence immunologic and metabolic changes in the host.57, 58, 59, 60, 61, 62 Furthermore, prebiotics, such as inulin and fructans,63 and postbiotics, such as butyrate,64 can also shape the microbial niche by allowing preferential growth of certain bacteria. Dietary intervention studies are attractive as a means to study these interactions due to their favorable safety profile, low cost and ease of implementation, and are currently being evaluated in patients with cancer (NCT02843425, NCT02079662). More recently, targeted modulation with bacteriophages has also gained attention, as data have demonstrated that this approach can yield results equal to those of antibiotics in targeting specific bacteria while sparing beneficial commensals,64 and can lead to significant improvement in the efficacy of FMT.65
Intervention studies and the design of next-generation microbial therapeutics
Clinical intervention studies, such as randomized controlled trials comparing the standard-of-care regimen with the addition of tailored microbial consortia specifically intended to alter the microbiome, are a powerful alternative to observational cohort-based studies. In principle, interventional studies can be substantially smaller than observational studies because they are more easily controlled through randomization of a predefined patient group. These include prospective, placebo-controlled designs and provide the opportunity to sample patients' microbiota at multiple time points to generate causal links with clinical outcomes through the use of metagenomics, metabolomics and related omics technologies. This also enables evaluation of the clinical consequences of compositional and functional changes in the microbiome. Importantly, microbes that have been shown to correlate with improved outcomes in these studies may themselves be immediate candidates for drug development through reverse translation. Several groups have endeavored to characterize the change in microbial phenotype associated with a particular disease state, and design a single- or multistrain bacterial therapy to modulate the patient's microbiome (Figure 1). These individual strains or consortia of commensal microbes are thus designed to catalyze the transition to a predisease healthy ecology state within the gut, resulting in functional changes that can ameliorate human illness. Clinical success has been achieved with this strategy in patients infected with C. difficile.66,67 However, the use of tailored microbial consortia in ICI-treated patients is complicated by the lack of a universal signature associated with a beneficial treatment response, underlining the importance of continued validation in independent cohorts.68
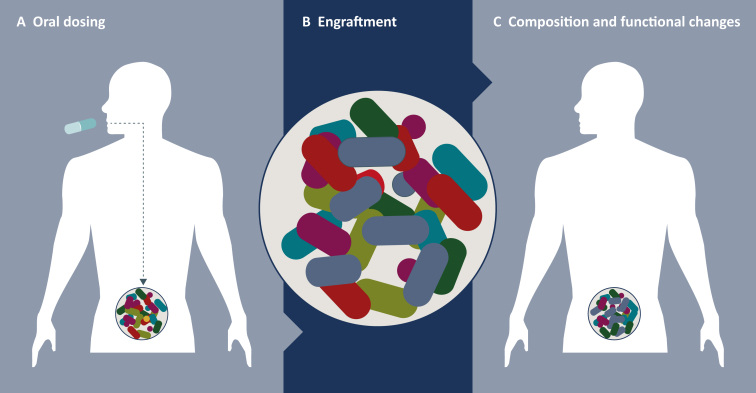
Figure 1.
Restructuring the microbiome.
Designer microbial consortia consisting of rationally chosen therapeutic interventions that can alter the microbiome composition to catalyze a change from a disease-associated state to a healthy state. (A) Engraftment can be influenced by dose titer and frequency, antibiotic preconditioning, identity and diversity of preexisting bacteria, and patient's lifestyle factors. (B) Engraftment is established via the germination of spores, rehydration of lyophilized bacteria, and subsequent growth and expansion in the host's microbiome. (C) Impacts of competition and cooperation between bacterial species on the composition and function of the host's microbiome lead to changes in local and systemic host gene expression.
The design of multistrain bacterial therapies is generally based on the application of comparative genomics and systems biology technologies coupled with isolation and characterization of difficult-to-culture strains (Figure 2). This type of reverse translational process starts with target identification by creating, curating and analyzing data on the human microbiome from observational and interventional human datasets, followed by the interrogation of phenotypic differences. Despite the large amount of normal person-to-person variation that exists within the human microbiome, the underlying functions of these ecologies have been found to be highly similar across a wide range of individuals for a given disease or health state.69,70 Knowledge of the underlying functional networks among different health states can provide practical guidance to inform the selection of combinations of commensal strains to create a drug candidate.
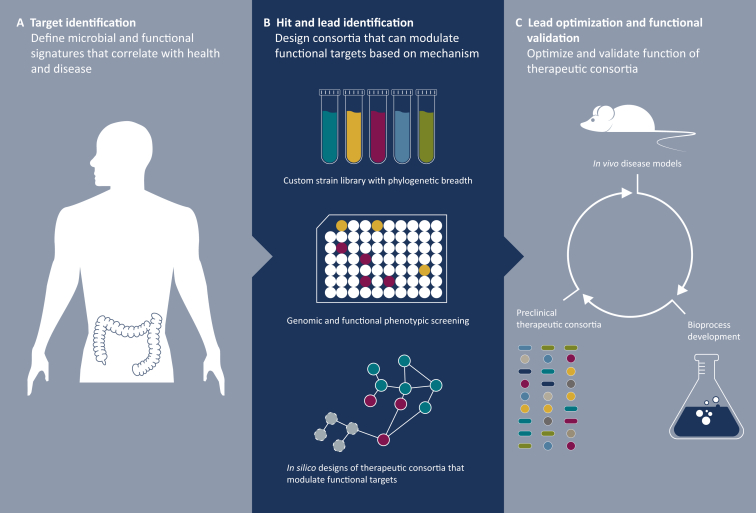
Figure 2.
Reverse translation in microbiome studies using tailored microbial consortia.
(A) The process begins with the deep characterization of interventional and observational datasets, using proprietary computational algorithms and systems biology analytics to deconvolute microbial signatures and targets of disease. (B) This is followed by the design of tailored microbial consortia intended specifically to modulate functional targets based on hypothesized mechanisms. (C) Finally, therapeutic consortia are optimized using in vivo disease models, advanced strain libraries and data integration platforms.
In the discovery phase, advanced computational approaches are used to analyze processed data from one of several assays, including next-generation sequencing approaches (e.g. 16S rRNA sequencing, whole metagenomic sequencing and whole metatranscriptomic sequencing), metabolomics and culturomics. Further downstream assessment is usually centered around differential enrichment of bacteria taxa and/or their functional groupings such as gene ontologies, protein families, metabolic pathways or metabolites to identify and characterize the underlying organismal networks of disparate microbial communities by phenotype. There is also a growing appreciation of the importance of multi-omics integration methodologies to comprehensively understand the biological interactions underlying these phenomena. Following this, lead identification is accomplished by designing a bacterial consortia that can modulate the target biology. Custom strain libraries representing a wide diversity of human commensals spanning large phylogenetic and functional breadth can be used to cover the search space of potential therapeutic solutions. These libraries are screened extensively to derive phenotype data on a per-strain basis. Their functional repertoire is then used as input for the in silico design process where different mixes of therapeutic consortia are rationally designed to have specific pharmacological properties to modulate, balance and regulate the targets of disease relevance identified in early discovery. Functional readouts can be numerous and are not limited to clinical outcome alone. These may include characterization of immune cell populations in the tumor microenviroment and systemic circulation, and quantification of fecal metabolites and other inflammatory markers such as calprotectin in the stool.
After designed consortia are manufactured to cover the disease-modifying targets adequately, they enter the final phase of the reverse translation process: lead optimization and functional validation. Relevant in vivo and ex vivo disease models and screens are utilized to test and optimize lead microbial consortia and fine-tune strain composition. Compositions are optimized to contain strains that are highly suitable for manufacturing and meet key drug formulation criteria. Proprietary data integration platforms are leveraged to develop and mature these therapeutic bacterial compositions to optimize both pharmacological and manufacturing properties of the consortia. The pharmacological properties are then confirmed using in vivo and ex vivo models relevant to the disease targets.
Phylogenetic and metagenomic assessments reveal significant differences in the identity and composition of intestinal microbiota in inbred mice and humans. To translate mechanisms and outcomes in mouse studies to humans, two types of models have been utilized to assess the effects of human-FMT,single bacterium strains and combinations of commensal strains. The gold standard is the use of germ-free mice colonized with human-derived bacterial compositions. A more accessible and less expensive alternative uses conventional inbred mice administered a broad-spectrum antibiotic regimen to deplete the endogenous gut microbiota before engraftment of human-derived material.71,72 Either mouse model can be used to assess the ability of FMT or commensal strains to reverse dysbiosis and other functional activities, including enhancement of immunotherapeutic activity, in syngeneic tumor models. The most efficacious candidates identified through the various preclinical assessments are further advanced for testing in the clinic.
Another therapeutic approach is to use bacterial spores purified from healthy donors. Approximately 50–60% of the bacteria resident in the human gut form spores.73 Spore formers represent a diverse set of taxa that modulate the immune system via production of short-chain fatty acids,30,74, 75, 76 metabolism of bile acids and breakdown of tryptophan.77 Furthermore, stable dormant spores, unlike vegetative bacteria, have the unique survival ability to tolerate the harsh acidic conditions of the gut; the low pH acts as a germinant resulting in an outgrowth of viable bacterial cells that can populate the lower gastrointestinal tract.78 These properties are ideal from the standpoint of pharmaceutical development, as spores can be formulated for long-term stability and convenient oral delivery.
For these reasons, bacteria that have evolved the ability to sporulate in the human gut are strongly represented in therapeutic interventions. Recently, a phase 1b study of SER-287, a consortium of bacterial spores, was completed in 58 enrolled patients with mild-to-moderate ulcerative colitis who were randomized to one of four treatment arms (Henn et al., in preparation). The hypothesis was that SER-287, a novel microbiome therapeutic, would reduce colonic inflammation by modulating metabolites critical to gut homeostasis through engraftment of donor species. In this trial, daily dosing of SER-287 (preceded by antibiotic preconditioning to facilitate engraftment) achieved significantly higher rates of clinical remission than placebo, with a favorable safety profile. Efficacy was associated with changes in pharmacokinetics and pharmacodynamics as assessed by dose-species engraftment (i.e. detection of organisms after treatment which were absent before treatment) and microbe-derived metabolites in stool over time.
This summary of SER-287 treatment results in ulcerative colitis is relevant to the burgeoning field of the microbiome in the context of cancer immunotherapy. Most importantly, it demonstrates that manipulation of the microbiome can modulate the immune system in humans, and can have a beneficial impact on disease outcome. In addition, these data demonstrate that conventional concepts of drug development, using pharmacokinetic and pharmacodynamic measures, can be adapted to therapeutic microbiome interventions, and that substantive pharmacological effects can be achieved in humans by altering the microbiome. Moreover, data from donor-derived commensals such as SER-287 may facilitate the discovery of the underlying drivers of clinical response. This information from donor-derived bacterial ecologies can, in turn, be used to develop defined microbial compositions that can be fermented in the laboratory.
Although microbiome intervention studies are highly promising, they remain in their infancy in terms of ICI treatment in cancer, and represent an imminent unmet need that is the focus of collaborations between and within academia and industry. Several factors need careful consideration in the design of such studies, including donor selection for sourcing of therapeutic microbiome material, identification of patient subsets that are most likely to benefit from therapy, and the timing and frequency of dosing, among others.
At the time of writing, at least seven clinical trials across five different organizations are underway to treat patients with various solid tumors using therapeutic microbiome material in combination with conventional ICI therapy (Table 1). Each microbial product is being evaluated with ICI agents for safety, engraftment of candidate donor-derived bacteria, efficacy in potentiating anticancer therapies, and ability to modulate immune tone. The Parker Institute for Cancer Immunotherapy, in collaboration with Seres Therapeutics and the MD Anderson Cancer Center, launched a phase 1b trial (NCT03817125) to treat patients with metastatic melanoma with nivolumab (anti-PD-1 antibody) in conjunction with a donor-derived bacterial consortia rich in Ruminococcaceae. Evelo Biosciences is conducting a phase 2 trial (NCT03595683) in which a cohort of patients with advanced melanoma are treated with Bifidobacterium species and pembrolizumab (anti-PD-1 antibody), in addition to a phase 1 trial for solid tumors (NCT03775850). Similarly, 4D Pharma is using Enterococcus gallinarum with pembrolizumab to treat both solid tumors (NCT03934827) and anti-PD-1-treated, relapsed solid tumors (NCT03637803). Also focusing on solid tumors is Nubiyota, a company which has a microbiome therapeutics platform that is undergoing a phase 1 trial (NCT03686202) with a consortium of bacteria along with approved anti-PD-1/PD-L1 antibody therapies. Finally, Vedanta Biosciences is investigating the use of their 11-strain consortium with nivolumab (anti-PD-1 antibody) on patients with melanoma, gastric/gastroesophageal junction adenocarcinoma, and colorectal cancer (NCT04208958).
Table 1.
Clinical trials currently testing the activity and safety of microbiome therapeutic agents alone or in combination with an immune checkpoint inhibitor in patients with cancer.
| Organization | Product | Indication | Study | Dose | Cohorts |
|---|---|---|---|---|---|
| Parker Institute for Cancer Immunotherapy |
SER-401 (donor derived, enriched in Ruminococcaceae) | Metastatic melanoma | Phase 1b (NCT03817125) | Initial daily loading dose (one capsule) for 7 days, followed by maintenance dose (one capsule) + nivolumab for 8 weeks | N = 30 |
| 4D Pharma |
MRx0518 (Enterococcus gallinarum) |
Solid tumors, PD-1 relapsed Solid tumors |
Phase 1/2 (NCT03637803) Phase 1/2 (NCT03934827a) |
1 capsule BID + pembrolizumab 1 capsule BID for 2–4 weeks |
N = 132 Open label Part A: open label (N = 20) Part B: 4:1 versus placebo (N = 100) |
| Evelo Biosciences |
EDP1503 (Bifidobacterium spp.) |
Solid tumors, PD-1 relapsed Advanced melanoma |
Phase 1/2 (NCT03775850) Phase 2 (NCT03595683b) |
Two capsules BID (3 × 1011 CFU) + pembrolizumab Two capsules BID (3 × 1011 CFU) + pembrolizumab |
N = 120 Open-label cohorts based on indication N = 70 Two open-label cohorts based on PD-1 response |
| Nubiyota | MET-4 (defined consortium) | Solid tumors | Phase 1 (NCT03686202) | Initial daily loading dose of 5 g (10 capsules) for 2 days, followed by maintenance doses of 1.5 g (three capsules) + checkpoint inhibitor |
N = 65 Three groups |
| Vedanta Biosciences | VE800 (11-strain defined consortium) | Advanced metastatic cancer | Phase 1/2 (NCT04208958c) | Daily dosing + nivolumab every 4 weeks | N = 111 |
BID, twice daily; CFU, colony-forming units.
Sponsored by Imperial College London.
Sponsored by the University of Chicago.
In collaboration with Bristol Myers Squibb.
These trials are geared to generate important early data on product formulation, dosage, scheduling and engraftment, as well as optimal clinical endpoints, including safety and efficacy. Together, the data from these studies can improve our understanding of the microbiome in the context of cancer therapy, and inform the design of future intervention studies using therapeutic microbiome material.
Conclusions and future directions
We are at a critical juncture in the field of microbiome research and cancer immunotherapy. It is likely that the existing data in the field represent only the beginning of the discoveries and opportunities that will lead to the improvement of clinical outcomes. Emerging data continue to suggest that commensal microbiota have the potential for significant therapeutic value to patients undergoing cancer treatment. Future studies designed to generate comprehensive data through the use of sophisticated multiomics approaches, the functional characterization of strains and a combination of in vivo and ex vivo disease models will undoubtedly help to advance the field and further define the diagnostic and therapeutic potential of the human microbiome in patients with cancer.
Pharmaceutical companies, which are testing an array of immunotherapeutic agents in clinical trials, can provide invaluable opportunities to thoroughly characterize the metagenome, metatranscriptome, metaproteome and metabolome of patient samples with next-generation sequencing tools. The industry is also uniquely poised to integrate these complex datasets by using novel machine learning and multiomics approaches, and can therefore serve as an ideal partner for companies with expertise in formulating safe and efficacious microbial modulators. The intent of conducting such translationally rich clinical trials is to extend beyond known findings by identifying novel bacterial signatures while providing an in-depth characterization of mechanisms of action. Equally important is the establishment of standardized end-to-end protocols that include consistent sampling methods (e.g. timing, frequency, collection, storage, processing) and bioinformatics analyses across all clinical studies. This enables the testing of hypotheses generated by the characterization of human samples in preclinical models in both germ-free and conventional mice, thereby enabling the development of interventional strategies that, in turn, can be reverse translated into novel therapeutic modalities in clinical trials.
An integral component of these clinical studies will be a comprehensive biomarker assessment that can be used across different tissues and materials, including longitudinal fecal, blood and tumor sampling, to determine the effects of such interventions on host metabolism and immune response. These trials will also provide a basis for testing microbial modulators as therapeutic adjuncts to ICI agents. By elucidating the complex bidirectional relationship between the microbiome and the host in the context of treatment response, this research provides the hope that we will be able to intervene effectively at multiple levels of this relationship to improve outcomes for patients with cancer.
Acknowledgments
Funding
None declared.
Disclosure
The authors have declared no conflicts of interest.
References
- 1.Eroglu Z., Zaretsky J.M., Hu-Lieskovan S., et al. High response rate to PD-1 blockade in desmoplastic melanomas. Nature. 2018;553(7688):347–350. doi: 10.1038/nature25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schvartsman G., Peng S.A., Bis G., et al. Response rates to single-agent chemotherapy after exposure to immune checkpoint inhibitors in advanced non-small cell lung cancer. Lung Cancer. 2017;112:90–95. doi: 10.1016/j.lungcan.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 3.Yip S.M., Wells C., Moreira R., et al. Checkpoint inhibitors in patients with metastatic renal cell carcinoma: results from the International Metastatic Renal Cell Carcinoma Database Consortium. Cancer. 2018;124(18):3677–3683. doi: 10.1002/cncr.31595. [DOI] [PubMed] [Google Scholar]
- 4.Ferris R.L., Blumenschein G., Jr., Fayette J., et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garon E.B., Rizvi N.A., Hui R., et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 6.Bellmunt J., de Wit R., Vaughn D.J., et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376(11):1015–1026. doi: 10.1056/NEJMoa1613683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McQuade J.L., Daniel C.R., Helmink B.A., Wargo J.A. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20(2):e77–e91. doi: 10.1016/S1470-2045(18)30952-5. [DOI] [PubMed] [Google Scholar]
- 8.Chan T.A., Yarchoan M., Jaffee E., et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garassino M., Rodriguez-Abreu D., Gadgeel S., et al. September 7–10, 2019. Evaluation of TMB in KEYNOTE-189: Pembrolizumab Plus Chemotherapy vs Placebo Plus Chemotherapy for Nonsquamous NSCLC. Presentation OA04.06. Presented at 2019 World Conference on Lung Cancer. Barcelona. [Google Scholar]
- 10.Cogdill A.P., Gaudreau P.O., Arora R., Gopalakrishnan V., Wargo J.A. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol. 2018;39(11):900–920. doi: 10.1016/j.it.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Elinav E., Garrett W.S., Trinchieri G., Wargo J. The cancer microbiome. Nat Rev Cancer. 2019;19(7):371–376. doi: 10.1038/s41568-019-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Routy B., Gopalakrishnan V., Daillere R., Zitvogel L., Wargo J.A., Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 13.Singh R., Chandrashekharappa S., Bodduluri S.R., et al. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10(1):89. doi: 10.1038/s41467-018-07859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z., Quan G., Jiang X., et al. Effects of metabolites derived from gut microbiota and hosts on pathogens. Front Cell Infect Microbiol. 2018;8:314. doi: 10.3389/fcimb.2018.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mallick H., Ma S., Franzosa E.A., Vatanen T., Morgan X.C., Huttenhower C. Experimental design and quantitative analysis of microbial community multiomics. Genome Biol. 2017;18(1):228. doi: 10.1186/s13059-017-1359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIver L.J., Abu-Ali G., Franzosa E.A., et al. bioBakery: a meta'omic analysis environment. Bioinformatics. 2018;34(7):1235–1237. doi: 10.1093/bioinformatics/btx754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franzosa E.A., Hsu T., Sirota-Madi A., et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13(6):360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo E., Taddei A., Ringressi M.N., Ricci F., Amedei A. The interplay between the microbiome and the adaptive immune response in cancer development. Therap Adv Gastroenterol. 2016;9(4):594–605. doi: 10.1177/1756283X16635082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sfanos K.S., Markowski M.C., Peiffer L.B., et al. Compositional differences in gastrointestinal microbiota in prostate cancer patients treated with androgen axis-targeted therapies. Prostate Cancer Prostatic Dis. 2018;21(4):539–548. doi: 10.1038/s41391-018-0061-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopalakrishnan V., Helmink B.A., Spencer C.N., Reuben A., Wargo J.A. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell. 2018;33(4):570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 22.Gopalakrishnan V., Spencer C.N., Nezi L., et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matson V., Fessler J., Bao R., et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Routy B., Le Chatelier E., Derosa L., et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 25.Chaput N., Lepage P., Coutzac C., et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 26.Frankel A.E., Coughlin L.A., Kim J., et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19(10):848–855. doi: 10.1016/j.neo.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viaud S., Daillere R., Boneca I.G., et al. Gut microbiome and anticancer immune response: really hot sh∗t! Cell Death Differ. 2015;22(2):199–214. doi: 10.1038/cdd.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shui L., Yang X., Li J., Yi C., Sun Q., Zhu H. Gut microbiome as a potential factor for modulating resistance to cancer immunotherapy. Front Immunol. 2020;10:2989. doi: 10.3389/fimmu.2019.02989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elinav E., Garrett W.S., Trinchieri G., Wargo J. The cancer microbiome. Nat Rev Cancer. 2019;19(7):371–376. doi: 10.1038/s41568-019-0155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith P.M., Howitt M.R., Panikov N., et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helmink B.A., Khan M.A.W., Hermann A., Gopalakrishnan V., Wargo J.A. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 32.Panebianco C., Andriulli A., Pazienza V. Pharmacomicrobiomics: exploiting the drug–microbiota interactions in anticancer therapies. Microbiome. 2018;6(1):92. doi: 10.1186/s40168-018-0483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viaud S., Saccheri F., Mignot G., et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iida N., Dzutsev A., Stewart C.A., et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu T., Guo F., Yu Y., et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geller L.T., Barzily-Rokni M., Danino T., et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357(6356):1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guthrie L., Gupta S., Daily J., Kelly L. Human microbiome signatures of differential colorectal cancer drug metabolism. NPJ Biofilms Microbiomes. 2017;3(1):27. doi: 10.1038/s41522-017-0034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vetizou M., Pitt J.M., Daillere R., et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivan A., Corrales L., Hubert N., et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knights D., Parfrey L.W., Zaneveld J., Lozupone C., Knight R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe. 2011;10(4):292–296. doi: 10.1016/j.chom.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly C.R., Kunde S.S., Khoruts A. Guidance on preparing an investigational new drug application for fecal microbiota transplantation studies. Clin Gastroenterol Hepatol. 2014;12(2):283–288. doi: 10.1016/j.cgh.2013.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly C.R., Kahn S., Kashyap P., et al. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology. 2015;149(1):223–237. doi: 10.1053/j.gastro.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore T., Rodriguez A., Bakken J.S. Fecal microbiota transplantation: a practical update for the infectious disease specialist. Clin Infect Dis. 2014;58(4):541–545. doi: 10.1093/cid/cit950. [DOI] [PubMed] [Google Scholar]
- 44.Mintz M., Khair S., Grewal S., et al. Longitudinal microbiome analysis of single donor fecal microbiota transplantation in patients with recurrent Clostridium difficile infection and/or ulcerative colitis. PLoS One. 2018;13(1):e0190997. doi: 10.1371/journal.pone.0190997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tariq R., Pardi D.S., Bartlett M.G., Khanna S. Low cure rates in controlled trials of fecal microbiota transplantation for recurrent Clostridium difficile infection: a systematic review and meta-analysis. Clin Infect Dis. 2019;68:1351–1358. doi: 10.1093/cid/ciy721. [DOI] [PubMed] [Google Scholar]
- 46.Bafeta A., Yavchitz A., Riveros C., Batista R., Ravaud P. Methods and reporting studies assessing fecal microbiota transplantation: a systematic review. Ann Intern Med. 2017;167(1):34–39. doi: 10.7326/M16-2810. [DOI] [PubMed] [Google Scholar]
- 47.van Nood E., Vrieze A., Nieuwdorp M., et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 48.Weingarden A.R., Chen C., Bobr A., et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am J Physiol Gastrointest Liver Physiol. 2014;306(4):G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Z.D., Jenq R.R., Ajami N.J., et al. Safety and preliminary efficacy of orally administered lyophilized fecal microbiota product compared with frozen product given by enema for recurrent Clostridium difficile infection: a randomized clinical trial. PLoS One. 2018;13(11):e0205064. doi: 10.1371/journal.pone.0205064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cammarota G., Ianiro G., Tilg H., et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66(4):569–580. doi: 10.1136/gutjnl-2016-313017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grady D. Fecal transplant is linked to a patient's death, the F.D.A. warns. The New York Times. June 13, 2019 [Google Scholar]
- 52.Kelly C.R., Kim A.M., Laine L., Wu G.D. The AGA's fecal microbiota transplantation national registry: an important step toward understanding risks and benefits of microbiota therapeutics. Gastroenterology. 2017;152(4):681–684. doi: 10.1053/j.gastro.2017.01.028. [DOI] [PubMed] [Google Scholar]
- 53.DeFilipp Z., Bloom P.P., Torres Soto M., et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N Engl J Med. 2019;381(21):2043–2050. doi: 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- 54.Venugopalan V., Shriner K.A., Wong-Beringer A. Regulatory oversight and safety of probiotic use. Emerg Infect Dis. 2010;16(11):1661–1665. doi: 10.3201/eid1611.100574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 56.Carmody R.N., Gerber G.K., Luevano J.M., Jr., et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17(1):72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benus R.F., van der Werf T.S., Welling G.W., et al. Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr. 2010;104(5):693–700. doi: 10.1017/S0007114510001030. [DOI] [PubMed] [Google Scholar]
- 58.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 59.Wu G.D., Chen J., Hoffmann C., et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.David L.A., Maurice C.F., Carmody R.N., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu G.D., Bushmanc F.D., Lewis J.D. Diet, the human gut microbiota, and IBD. Anaerobe. 2013;24:117–120. doi: 10.1016/j.anaerobe.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 62.Schulz M.D., Atay C., Heringer J., et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature. 2014;514(7523):508–512. doi: 10.1038/nature13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taper H.S., Roberfroid M.B. Possible adjuvant cancer therapy by two prebiotics – inulin or oligofructose. In Vivo. 2005;19(1):201–204. [PubMed] [Google Scholar]
- 64.O'Keefe S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13(12):691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cieplak T., Soffer N., Sulakvelidze A., Nielsen D.S. A bacteriophage cocktail targeting Escherichia coli reduces E. coli in simulated gut conditions, while preserving a non-targeted representative commensal normal microbiota. Gut Microbes. 2018;9(5):391–399. doi: 10.1080/19490976.2018.1447291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Petrof E.O., Gloor G.B., Vanner S.J., et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome. 2013;1(1):3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khanna S., Pardi D.S., Kelly C.R., et al. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J Infect Dis. 2016;214(2):173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- 68.McShane L.M., Cavenagh M.M., Lively T.G., et al. Criteria for the use of omics-based predictors in clinical trials. Nature. 2013;502(7471):317–320. doi: 10.1038/nature12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moya A., Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24(5):402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 70.Tian L., Wu A.-K., Friedman J., Waldor M.K., Weiss S.T., Liu Y.-Y. Deciphering functional redundancy in the human microbiome. bioRxiv. 2017:176313. doi: 10.1038/s41467-020-19940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhatt A.P., Redinbo M.R., Bultman S.J. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. 2017;67(4):326–344. doi: 10.3322/caac.21398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kennedy E.A., King K.Y., Baldridge M.T. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Browne H.P., Forster S.C., Anonye B.O., et al. Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533(7604):543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goverse G., Molenaar R., Macia L., et al. Diet-derived short-chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J Immunol. 2017;198(5):2172–2181. doi: 10.4049/jimmunol.1600165. [DOI] [PubMed] [Google Scholar]
- 75.Asarat M., Apostolopoulos V., Vasiljevic T., Donkor O. Short-chain fatty acids regulate cytokines and Th17/Treg cells in human peripheral blood mononuclear cells in vitro. Immunol Invest. 2016;45(3):205–222. doi: 10.3109/08820139.2015.1122613. [DOI] [PubMed] [Google Scholar]
- 76.Park J.H., Kotani T., Konno T., et al. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS One. 2016;11(5):e0156334. doi: 10.1371/journal.pone.0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yano J.M., Yu K., Donaldson G.P., et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Browne H.P., Neville B.A., Forster S.C., Lawley T.D. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol. 2017;15(9):531–543. doi: 10.1038/nrmicro.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]