Abstract
Patients with BRAFV600E-mutant melanoma are effectively treated with the BRAF-inhibiting drug, vemurafenib, but soon develop drug resistance, limiting vemurafenib’s therapeutic efficacy. Constitutive activation of STAT3 in cancer cells and immune cells in the tumor microenvironment (TME) is a crucial contributor to the development of drug resistance and immune evasion in most cancers. Here, we investigated the antitumor efficacy and TME remodeling by APTSTAT3-9R, a cell-permeable STAT3 inhibitory peptide, as a strategy to treat vemurafenib-resistant melanoma. We found that vemurafenib-resistant melanoma remodels into immunosuppressive TME by increasing the expression of specific chemokines to facilitate the infiltration of immunosuppressive immune cells, such as myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs). Intratumoral treatment of APTSTAT3-9R led to a reduction in the population of MDSCs and TAMs, while increasing infiltration of cytotoxic T lymphocytes in the TME. Moreover, combination therapy with APTSTAT3-9R and an anti-PD-1 antibody enhanced significant suppression of tumor growth by decreasing infiltration of these immunosuppressive immune cells while increasing the infiltration and cytotoxicity of CD8+ T cells. These findings suggest that combined blockade of STAT3 and PD-1 signaling pathways may be an effective treatment option for overcoming poor therapeutic outcomes associated with drug-resistant BRAF-mutant melanoma.
Keywords: immunotherapy, melanoma, STAT3 inhibitors, tumor microenvironment, vemurafenib
Graphical abstract

A new treatment option is proposed for overcoming poor therapeutic outcomes associated with vemurafenib-resistant melanoma. Combined blockade of STAT3 signaling and PD-1 signaling pathways will be an effective treatment regimen for other drug-resistant tumors.
Introduction
Although most patients with BRAFV600E-mutant melanoma showing constitutive activation of the BRAF/MEK/ERK-signaling pathway experience a remarkable initial response to the BRAF inhibitor (BRAFi), vemurafenib,1,2 long-term therapeutic outcomes are compromised by acquired drug resistance.3 BRAFi resistance is known to be acquired through either de novo generation of secondary mutations in cancer cells or activation of other compensatory downstream effector pathways.4,5 A recent comprehensive secretome analysis of BRAFi-resistant tumors revealed that drug resistance results in the development of an immunosuppressive tumor microenvironment (TME) that limits the therapeutic efficacy of immune checkpoint inhibitor (ICI)-based immunotherapy.6 However, investigations into how acquired BRAFi resistance shapes the TME and how the resulting immunosuppressive TME can be mitigated have been lacking. Thus, there is an urgent need for a new treatment strategy capable of ameliorating the immunosuppressive TME of BRAFi-resistant tumors.
It was recently reported that constitutive activation of the signal transducer and activator transcription 3 (STAT3) pathway in cancer cells and immune cells in the TME, such as myeloid-derived suppressor cells (MDSCs) and M2-type tumor-associated macrophages (TAMs), contributes to tumor immune evasion or immune tolerance.7, 8, 9 Thus, targeting the STAT3 signaling pathways in both cancer cells and tumor-infiltrating immune cells may be a feasible strategy for ameliorating BRAFi resistance in melanoma through reshaping the immunosuppressive TME.10,11 We recently reported a high-affinity peptide binder for STAT3, designated APTSTAT3, that inhibits phosphorylation of STAT3.12, 13, 14, 15, 16 Upon modification with a cell-penetrating 9-arginine motif (APTSTAT3-9R), the peptide was readily internalized into the cytoplasm of cancer cells where it inhibited STAT3 downstream signaling pathways, thereby exerting high antitumor efficacy in vivo.13 However, the antitumor efficacy of APTSTAT3-9R against BRAFi-resistant melanoma and its effects on the TME have not been studied.
In this study, we investigated whether inhibition of STAT3 by APTSTAT3-9R not only induces cancer cell apoptosis but also remodels the immunosuppressive TME of BRAFi-resistant melanoma so as to potentiate antitumor efficacy in the drug-resistant tumor. We also examined the effects of combining APTSTAT3-9R with anti-PD-1 antibody (αPD-1) immunotherapy on TME remodeling and antitumor efficacy. We first analyzed changes in populations of MDSCs, TAMs, and CD8+ T cells in the TME of BRAFV600E-mutant melanoma with acquired resistance to vemurafenib. We further analyzed the chemokines secreted by BRAFi-resistant melanoma that are responsible for trafficking of these immunosuppressive cells to the TME. Finally, we examined the effects of APTSTAT3-9R-mediated inhibition of STAT3 on reshaping of the TME and antitumor efficacy, alone or in combination with ICIs, in BRAFi-resistant melanoma.
Results
Vemurafenib-resistant melanoma exhibits enhanced tumor formation and potentiates an immunosuppressive TME
To establish vemurafenib-resistant melanoma cells, we chronically exposed parental BRAFV600E-mutant melanoma cells, designated YUMM1.7P, to vemurafenib for 8–10 weeks, starting at a low (condition 1) or medium (condition 2) drug concentration and gradually increasing it (Figure S1A). Cell-viability assays revealed that this treatment successfully generated vemurafenib-resistant cells, designated YUMM1.7R, which exhibited half-maximal inhibitory concentration (IC50) values of 10.37 μM (condition 1) and 9.48 μM (condition 2)—values much higher than the IC50 value of vemurafenib for YUMM1.7P cells (3.12 μM) (Figure S1B). Because extracellular signal-regulated kinase (ERK) signaling is known to be constitutively activated in BRAFV600E-mutant melanoma,17 we assessed the effect of vemurafenib treatment on phosphorylation of ERK in both YUMM1.7P and YUMM1.7R cells. While treatment with vemurafenib led to a considerable reduction in ERK phosphorylation in YUMM1.7P cells, this reduction was dramatically suppressed in one of the generated YUMM1.7R cell lines (condition 1) (Figure S1C), which was chosen as a BRAFi-resistant cell line for subsequent experiments.
Using these YUMM1.7R cells, we first examined tumor-forming ability and resistance to vemurafenib in vivo upon transplantation into syngeneic immunocompetent C57BL/6 mice. YUMM1.7R-derived tumors grew more rapidly than those derived from YUMM1.7P cells; moreover, in contrast to YUMM1.7P cells, which were highly sensitive to inhibition of tumor growth by vemurafenib, BRAFi-resistant YUMM1.7R cell tumor growth was unaffected by vemurafenib treatment (Figure 1A), indicating successful establishment of BRAFi-resistant melanoma tumors. Next, we analyzed the TME of these vemurafenib-sensitive and -resistant melanoma tumors, with and without vemurafenib treatment. Previous studies have shown that treatment of vemurafenib-sensitive melanoma tumors with vemurafenib enhances melanoma antigen expression, promotes T cell cytotoxicity, and induces a more favorable TME.18,19 Accordingly, we assessed changes in populations of MDSCs, TAMs, and CD8+ T cells, which are key mediators of tumor immunity in the TME. The gating strategy for tumor-infiltrating immune cells is shown in Figure S2. The frequency of CD11b+Gr1+ MDSCs and CD11b+F4/80+Gr1− TAMs was dramatically increased in YUMM1.7R-derived tumors, which also showed a significant reduction in the frequency of tumor-infiltrating CD8+ T cells compared with vemurafenib-sensitive YUMM1.7P-derived tumors (Figures 1B–1D), indicating formation of an augmented immunosuppressive TME in the vemurafenib-resistant melanoma. As expected, vemurafenib treatment led to a significant decrease in the frequency of MDSCs and TAMs but increased the frequency of CD8+ T cells in YUMM1.7P-derived tumors. Although treatment of YUMM1.7R-derived tumors with vemurafenib also led to a reduction in the frequency of MDSCs and TAMs, unlike the case of vemurafenib-sensitive melanoma tumors, it failed to increase the frequency of CD8+ T cells. No significant change was observed in the frequency of CD3+CD4+ T cells by vemurafenib treatments in both YUMM1.7P- and YUMM1.7R-derived tumors (Figure S3). These results suggest that vemurafenib-resistant melanoma tumors develop an immunosuppressive TME by recruiting tumor-promoting MDSCs and TAMs while limiting CD8+ T cell infiltration into the tumor.
Figure 1.
Vemurafenib-resistant melanoma cells exhibit enhanced tumor formation and a potentiated immunosuppressive TME
(A) YUMM1.7P or YUMM1.7R cells (3 × 105 cells each) were subcutaneously injected into C57BL/6 mice. Vehicle control (veh.) or vemurafenib (VEM) (20 mg/kg) was injected i.p. daily into tumor-bearing mice. Arrows indicate the injection day. Tumor size is expressed as means ± SEM (n = 6 mice/group; ∗∗∗∗p < 0.0001 versus YUMM1.7P + veh; two-way ANOVA followed by Tukey’s post hoc test). (B–D) Left: representative contour plots showing the frequency of each individual immune cell population (MDSCs, TAMs, and CD8+ T cells) among live/dead−CD45+ cells in both control and treated groups. Single-cell suspensions of immune cells were isolated from tumors and analyzed by flow cytometry. Cell populations were identified as (B) MDSCs (CD45+CD11b+Gr1+), (C) TAMs (CD45+Gr1−CD11b+F4/80+), and (D) CD8+ T cells (CD45+CD3+CD8+). Right: quantification of results. Data are presented as means ± SEM (n = 6 mice/group; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; unpaired two-tailed Student’s t test); ns, not significant. (E) Total RNA was isolated from YUMM1.7P or YUMM1.7R cells for real-time qRT-PCR. mRNA levels of the chemokines CCL2, CXCL1, CXCL2, and CXCL5 were determined. Representative results from one of three independent experiments are shown. Data are presented as means ± SEM (n = 3; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001; unpaired two-tailed Student’s t test); ns, not significant.
Chemokines are known to play a crucial role in recruiting immune cells to the TME of a tumor;20 in particular, it has been shown that both CCL2 and CXCL1 are key chemokines involved in the recruitment of immunosuppressive cells such as MDSCs.21 Accordingly, we analyzed and compared expression levels of these chemokines in both YUMM1.7P and YUMM1.7R cells by real-time qRT-PCR. We found that expression levels of CCL2 and CXCL1 were remarkably higher in YUMM1.7R cells compared with YUMM1.7P cells; in contrast, CXCL2 and CXCL5 chemokines were not significantly different between the two cancer cell lines (Figure 1E). Collectively, these findings indicate that vemurafenib-resistant BRAFV600E-mutant melanoma cells exhibit enhanced tumor growth and form a potentiated immunosuppressive TME by excessively secreting CCL2 and CXCL1 chemokines compared with drug-sensitive parental melanoma cells.
APTSTAT3-9R suppresses the proliferation of vemurafenib-resistant melanoma cells and resensitizes them to vemurafenib
Since activation of STAT3 signaling pathways contributes to drug resistance of cancer cells and tumor immune evasion or immune tolerance,7, 8, 9 we examined the effect of the cell-permeable STAT3 binding peptide, APTSTAT3-9R, which can inhibit STAT3 phosphorylation, on vemurafenib-resistant YUMM1.7R cells. The key features in structure and sequence of APTSTAT3-9R are illustrated in Figure 2A. A control peptide (APTSCR-9R) that forms a structure similar to that of APTSTAT3-9R but possesses a scrambled sequence in the target-binding region was used for comparison throughout experiments. Whereas interleukin-6 (IL-6) treatment led to upregulation of phosphorylated STAT3 (P-STAT) in both YUMM1.7P and YUMM1.7R cells, this effect was markedly suppressed in both melanoma cell lines by APTSTAT3-9R treatment (10 μM); as expected, the control APTSCR-9R (10 μM) failed to inhibit STAT3 phosphorylation in either cell line (Figure 2B). This result indicates that APTSTAT3-9R is able to specifically inhibit STAT3 activation. Furthermore, inhibition of STAT3 phosphorylation by APTSTAT3-9R in both YUMM1.7P and YUMM1.7R cells led to a reduction in cell viability, whereas APTSCR-9R treatment caused no appreciable decrease in cell viability Figure 2C). Notably, the effect of APTSTAT3-9R was concentration dependent, becoming apparent beyond a concentration of 15 μM (Figure S4), results that are well correlated with those of cell-viability assays. Because it has been suggested that inhibition of STAT3 signaling pathways may overcome the acquired resistance of melanoma to vemurafenib,22 we examined whether APTSTAT3-9R treatment could resensitize YUMM1.7R cells to vemurafenib. To this end, we pretreated YUMM1.7R cells for 2 h with APTSCR-9R or APTSTAT3-9R at a concentration of 10 μM, which has little effect on cell viability but still inhibits STAT3 activation, and subsequently treated these cells with vemurafenib (5 μM) or vehicle. Treatment with either APTSTAT3-9R or vemurafenib alone did not reduce the viability of YUMM1.7R cells, whereas sequential treatment with APTSTAT3-9R and vemurafenib at the indicated concentrations considerably decreased the viability of vemurafenib-resistant melanoma cells (Figure 2D), suggesting resensitization of YUMM1.7R cells to vemurafenib.
Figure 2.
Effects of APTSTAT3-9R treatments on STAT3 phosphorylation, sensitivity to vemurafenib, and antitumor efficacy in melanomas
(A) Structure and characteristics of the cell-permeable STAT3-inhibiting peptide, APTSTAT3-9R. (B) Western blotting results for P-STAT3 (Tyr705), STAT3, and GAPDH in YUMM1.7P and YUMM1.7R cells pretreated with 10 μM APTSCR-9R or APTSTAT3-9R for 2 h and subsequently treated with IL-6 (20 ng/mL). (C) Viability of YUMM1.7P and YUMM1.7R cells after treatment with increasing concentrations of either APTSTAT3-9R or APTSCR-9R for 48 h. Representative results from one of three independent experiments are shown. Data are presented as means ± SEM (n = 5; ∗∗∗∗p < 0.0001, ∗∗p < 0.01; one-way ANOVA followed by Sidak’s post hoc test). (D) Viability of YUMM1.7R cells pretreated with 10 μM APTSTAT3-9R or APTSCR-9R for 2 h and subsequently treated with vemurafenib (VEM; 5 μM) for 48 h. Representative results from one of three independent experiments are shown. Data are presented as means ± SEM (n = 5; ∗∗∗∗p < 0.0001; one-way ANOVA followed by Sidak’s post hoc test). (E) Schematic depiction of the experimental design. C57BL/6 mice were injected s.c. with YUMM1.7R cells (3 × 105 cells). The mice were randomly allocated to each treatment group (n = 5 mice/group). (F) Growth curves of YUMM1.7R-derived melanoma tumors upon treatment with each modality. Tumor size is expressed as mean ± SEM (n = 5 mice/group; ∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05 versus untreated; two-way ANOVA followed by Tukey’s post hoc test). (G) Kaplan-Meier curves showing survival of each treatment condition (∗∗∗p < 0.0002, APTSTAT3-9R + VEM; Mantel-Cox log rank test). (H) Changes in body weight of each group were measured.
Next, we evaluated antitumor efficacy of APTSTAT3-9R and its combination with vemurafenib in YUMM1.7R-derived tumors. When YUMM1.7R-derived tumors reached ∼100 mm3, APTSTAT3-9R was intratumorally (i.t.) or intravenously (i.v.) injected at a dose of 10 mg/kg every other day for a total of four injections, whereas vemurafenib was administered intraperitoneally (i.p.) daily at a dose of 20 mg/kg for a total of seven injections (Figure 2E). As expected, vemurafenib (i.p.) or APTSCR-9R (i.t.) treatment was unable to inhibit growth of vemurafenib-resistant melanoma, whereas APTSTAT3-9R (i.t.) treatment led to inhibition of tumor growth by ∼35% relative to untreated control on day 15 (Figure 2F). Furthermore, combination of APTSTAT3-9R (i.t.) with vemurafenib (i.p.) was more effective than APTSTAT3-9R (i.t.) monotherapy in inhibiting tumor growth and increasing survival (Figure 2G), suggesting the possibility of mitigating the drug resistance by inhibition of STAT3 signaling pathways. Unlike the case of intratumorally injected APTSTAT3-9R (i.t.), APTSTAT3-9R (i.v.) was ineffective in inhibiting tumor growth, presumably due to limited tumor targeting efficiency of the small-sized peptide upon systemic delivery. There was no significant difference in the body-weight change of mice treated with each modality (Figure 2H). Collectively, these results demonstrate that APTSTAT3-9R is capable of inhibiting STAT3 signaling in vemurafenib-resistant YUMM1.7R cells and tumors, causing cell death and mitigating the acquired drug resistance of melanoma to vemurafenib.
APTSTAT3-9R remodels the TME of vemurafenib-resistant melanoma
As we confirmed that vemurafenib-resistant YUMM1.7R-derived tumors form an immunosuppressive TME and that APTSTAT3-9R (i.t.) inhibits tumor growth of the drug-resistant melanoma, we next investigated the effects of APTSTAT3-9R treatment on TME remodeling. After YUMM1.7R-derived tumors reached ∼100 mm3, APTSTAT3-9R or APTSCR-9R was injected i.t. at a dose of 10 mg/kg every other day for a total of four injections, whereas vemurafenib was administered i.p. daily at a dose of 20 mg/kg for a total of seven injections (Figure 3A). As expected, neither vemurafenib nor APTSCR-9R alone was effective in inhibiting the growth of drug-resistant melanoma tumors, whereas APTSTAT3-9R-treated mice showed considerable reductions in tumor growth (∼64%) relative to untreated controls (Figure 3B). We next analyzed the TME of tumors treated with each modality using flow cytometry. As expected, the frequencies of CD11b+Gr1+ MDSCs, CD11b+F4/80+Gr1- TAMs, and CD3+CD8+ T cells in the TME were unaltered by treatment with APTSCR-9R; notably, however, APTSTAT3-9R-treated tumors showed a significant reduction in both MDSCs and TAMs (Figures 3C and 3D). Moreover, APTSTAT3-9R treatment increased the CD8+/CD4+ T cell ratio by increasing the frequency of CD3+CD8+ T cells without affecting the frequency of CD3+CD4+ T cells in YUMM1.7R-derived tumors (Figure S5), suggesting mitigation of the immunosuppressive TME in the vemurafenib-resistant melanoma (Figure 3E). Interestingly, although vemurafenib treatment alone also led to a reduction in the frequencies of MDSCs and TAMs within YUMM1.7R-derived tumors, it was unable to effectively suppress tumor growth. We speculate that this limited antitumor efficacy of vemurafenib is attributable to the lack of an appreciable increase in the frequency of CD8+ T cells in the TME.
Figure 3.
APTSTAT3-9R inhibits growth of vemurafenib-resistant melanoma tumors and suppresses recruitment of immunosuppressive cells into the TME
(A) Schematic depiction of the experimental design. YUMM1.7R-derived tumors were formed by subcutaneous injection of 3 × 105 cells into C57BL/6 mice. On day 6, each treatment modality was initiated. Both APTSTAT3-9R and APTSCR-9R (10 mg/kg) were injected i.t. every other day, and vemurafenib (20 mg/kg) was injected i.p. daily as indicated. Tumors (n = 5 mice/group) were harvested on day 14 for flow-cytometry analysis. (B) Growth curves of YUMM1.7R-derived melanoma tumors upon treatment with each modality. Tumor size is expressed as mean ± SEM (n = 5 mice/group; ∗∗∗∗p < 0.0001 versus APTSTAT3-9R; two-way ANOVA followed by Tukey’s post hoc test). (C–E) Left: representative contour plots showing the frequency of (C) MDSCs, (D) TAMs, and (E) CD3+CD8+ T cell immune cell populations among live/dead−CD45+ cells in both control and treated groups. Right: quantification of results. Data are presented as means ± SEM (n = 5 mice/group; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; unpaired two-tailed Student’s t test); ns, not significant.
To further investigate the possible heterogeneity among the broadly defined immune cell subsets, we applied single-cell RNA sequencing (scRNA-seq) for dissociated cells of freshly resected tumors from untreated and APTSTAT3-9R-treated mice. We profiled two samples from APTSTAT3-9R-treated tumor (11,770 cells) and untreated control tumor (13,706 cells), respectively, at an average depth of ∼30,000 reads per cell. After quality control, we performed clustering and cell-type identification analysis using characteristic canonical cell markers (Figures S6A and S6B). As expected, there was considerable reconstitution of the infiltrated immune cells in APTSTAT3-9R-treated tumor compared with untreated tumor. The most prominent changes detected in APTSTAT3-9R-treated tumor was a notable decrease in immunosuppressive immune cells such as MDSCs and TAMs but a considerable increase in T cells, including CD4+ T cells, CD8+ T cells, and cytotoxic T cells (Figures S6C and S6D). Interestingly, there was an appreciable change in the composition ratio of M1- and M2-macrophages, showing an increase in M1-macrophages but a decrease in M2-macrophages (Figure S6C). Moreover, we recognized that cytotoxic CD8+ T cells in T cell cluster were significantly enriched in the APTSTAT3-9R-treated tumor compared with that in untreated tumor (Figure S6D). This scRNA-seq result is consistent with that obtained by flow-cytometry analysis (Figures 3C–3E) and suggests that APTSTAT3-9R treatment mitigated immunosuppressive TME of vemurafenib-resistant melanoma.
Because we found that the chemokines CCL2 and CXCL1, which recruit immunosuppressive MDSCs and TAMs to the TME, were upregulated in YUMM1.7R-derived tumors (Figure 1E), we examined the effect of APTSTAT3-9R on expression of these two chemokines in YUMM1.7R cells. APTSTAT3-9R treatment caused a significant, concentration-dependent decrease in the expression of these chemokines in YUMM1.7R cells (Figure S7), suggesting that this peptide led to a reduction in the recruitment of immunosuppressive MDSCs and TAMs to the TME by decreasing CCL2 and CXCL1 secretion. Taken together, these findings indicate that APTSTAT3-9R mitigates the immunosuppressive TME of vemurafenib-resistant melanoma by reducing chemokine production, thereby suppressing tumor growth.
Combining APTSTAT3-9R treatment with an anti-PD-1 antibody enhances antitumor efficacy against vemurafenib-resistant melanoma
Because APTSTAT3-9R treatment decreases the frequency of immunosuppressive MDSCs and TAMs but increases the frequency of CD8+ T cells in vemurafenib-resistant melanoma, we reasoned that combining APTSTAT3-9R with immunotherapy using an anti-PD-1 antibody (αPD-1) would improve antitumor efficacy. To test this, we administered APTSTAT3-9R (i.t.), αPD-1 (i.p.), or the combination thereof every other day for a total of four injections after YUMM1.7R-derived tumors reached a size of ∼100 mm3 (day 6) (Figure 4A). Although each monotherapy significantly inhibited tumor growth compared with untreated controls, combination therapy led to greater inhibition of tumor growth than either monotherapy Figure 4B). At the end of each treatment, we analyzed the TME using flow cytometry. Whereas αPD-1 alone treatment did not affect recruitment of immunosuppressive MDSCs or TAMs, APTSTAT3-9R and APTSTAT3-9R + αPD-1 treatments led to substantial—and similar—reductions in the frequencies of those immune cells in YUMM1.7R-derived tumors compared with untreated controls (Figures 4C and 4D), suggesting that APTSTAT3-9R, and not αPD-1, may be responsible for the reduced recruitment of MDSCs and TAMs into the TME. However, although treatment with APTSTAT3-9R or αPD-1 alone also led to a significant increase in the frequency of tumor-infiltrating CD8+ T cells (Figure 4E) as well as interferon-γ (IFN-γ)-producing functional CD8+ T cells (Figure 4F) compared with untreated controls, the combination of APTSTAT3-9R with αPD-1 further increased these cell populations to levels much greater than those produce by either monotherapy, potentially contributing to the greater antitumor efficacy.
Figure 4.
Effects of combined treatment with APTSTAT3-9R and anti-PD-1 antibody on tumor growth and TME remodeling
(A) Schematic depiction of the experimental design. YUMM1.7R-derived tumors were formed by s.c. injection of 3 × 105 cells into C57BL/6 mice; each treatment modality was initiated on day 6. APTSTAT3-9R (10 mg/kg) was injected i.t. every other day and αPD-1 (200 μg per mouse) was injected i.p. daily as indicated. Tumors (n = 5 mice/group) were harvested on day 14 for flow-cytometry analysis. (B) Growth curves of YUMM1.7R-derived melanoma tumors upon treatment with each modality. Tumor size is expressed as mean ± SEM (n = 5 mice/group; ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05 versus αPD-1 + APTSTAT3-9R; two-way ANOVA followed by Tukey’s post hoc test). (C–E) Quantitative data showing the frequency of tumor-infiltrating (C) MDSCs, (D) TAMs, and (E) CD8+ T cells in each group, expressed as a percentage. Data are presented as means ± SEM (n = 5 mice/group; ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; one-way ANOVA followed by Sidak’s post hoc test); ns, not significant. (F) Left: representative contour plots of IFN-γ-producing tumor-infiltrating CD8+ T cells produced by gating on live/dead−CD45+ cells. Right: quantitative result showing the proportion of IFN-γ-producing CD8+ T cells in each group, expressed as a percentage. Data are presented as means ± SEM (n = 5 mice/group; ∗∗∗∗p < 0.0001, ∗∗p < 0.01; one-way ANOVA followed by Sidak’s post hoc test); ns, not statistically significantly changed.
We next examined the effects of combination therapy on tumor growth and survival through another set of independent experiments (Figure S8A). Although αPD-1 (i.p.) or APTSTAT3-9R (i.t.) monotherapy could effectively inhibit tumor growth by day 13 compared with untreated control (Figures S8B and S8C), only slightly improved survival benefit was observed (Figure S8D). However, the combination of APTSTAT3-9R (i.t.) with αPD-1 resulted in much greater tumor growth inhibition and much longer overall survival than the other groups without showing appreciable body-weight changes (Figure S8E), indicating effectiveness of the optimal combination therapy. Taken together, these results indicate that mitigating the immunosuppressive TME through combined blockade of STAT3 and PD-1 signaling pathways may be an effective strategy for treating vemurafenib-resistant melanoma.
APTSTAT3-9R exerts its therapeutic efficacy against cancer cells and immune cells in the TME
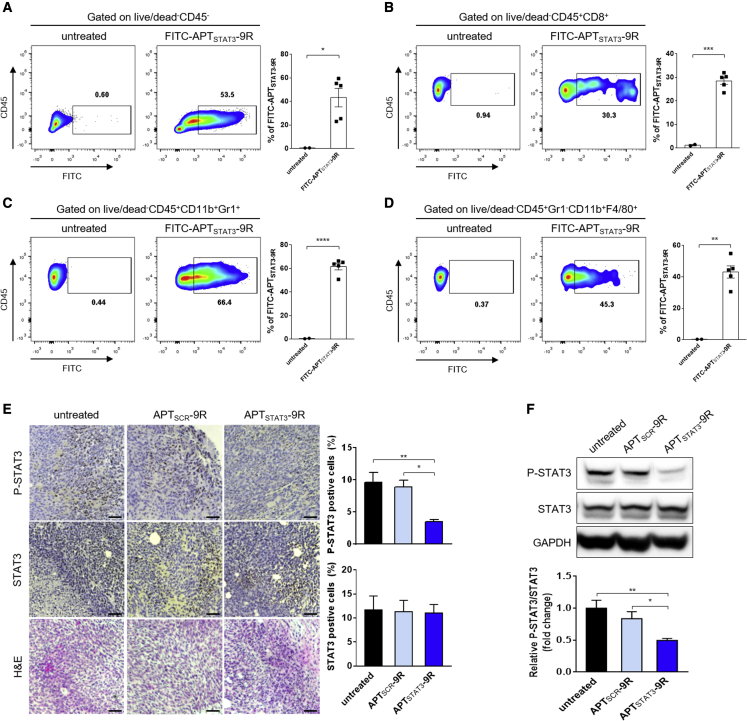
Next, to determine which cells take up APTSTAT3-9R, we injected (i.t.) fluorescein isothiocyanate (FITC)-labeled APTSTAT3-9R (FITC-APTsSTAT3-9R) into YUMM1.7R-derived tumors after tumors reached ∼200–300 mm3, and isolated cancer cells and immune cells in the TME 4 h after injection for flow-cytometry analysis. The gating strategy for isolating CD45-negative cells, CD8+ T cells, MDSCs, and TAMs from the TME is shown in Figure S9. Flow-cytometry analysis revealed considerable uptake of FITC-APTSTAT3-9R by MDSCs, TAMs, CD8+ T cells, and CD45-negative cells (i.e., cancer cells and fibroblasts) (Figures 5A–5D), suggesting that APTSTAT3-9R inhibits STAT3 signaling pathways in these cells and thereby attenuates the immunosuppressive function of MDSCs, TAMs, and cancer cells while promoting cytotoxic T cell activity. Furthermore, histopathological analyses of tumors treated with APTSTAT3-9R (i.t.) showed that the percentage of P-STAT3-positive cells was significantly reduced compared with untreated control and APTSCR-9R-treated tumors (Figure 5E). However, there was little difference in the percentage of STAT3-positive cells in the treated tumors, suggesting that APTSTAT3-9R treatment did not affect production of total STAT3 but could inhibit STAT3 phosphorylation. Western blot analyses of tumor lysates further confirmed a substantial reduction in STAT3 activation (P-STAT3) by APTSTAT3-9R treatment (i.t.) (Figure 5F), indicating inhibition of STAT3 activation in whole tumor tissues of vemurafenib-resistant melanoma. Collectively, these results indicate that, upon i.t. injection, APTSTAT3-9R is taken up by both immune cells and cancer cells in the TME and inhibits STAT3 activation in these cells, thereby contributing to antitumor efficacy.
Figure 5.
APTSTAT3-9R is effectively taken up by cells within the TME and inhibits STAT3 signaling in vemurafenib-resistant melanoma tumors
(A–D) Flow-cytometry analysis of TME cells positively stained with FITC-APTSTAT3-9R. When YUMM1.7R-derived tumors reached ∼300 mm3, mice were injected i.t. with FITC-APTSTAT3-9R (10 mg/kg), and tumors were harvested after 4 h for flow-cytometry analysis. Representative contour plots of (A) FITC-positive tumor/cancer-associated fibroblasts, obtained by gating on live/dead−CD45−; (B) MDSCs, obtained by gating on live/dead−CD45+CD11b+Gr1+; (C) TAMs, obtained by gating on live/dead−CD45+Gr1−CD11b+F4/80+; and (D) CD8+ T cells, obtained by gating on live/dead−CD45+CD8+. Data are presented as means ± SEM (n = 2 mice for the untreated group and n = 5 mice for FITC-APTSTAT3-9R-treated groups; ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; unpaired two-tailed Student’s t test). (E) Representative images showing immunohistochemical detection of P-STAT3 (Tyr705) and STAT3, and H&E staining in tissues of YUMM1.7R-derived tumors treated with each modality. When YUMM1.7R-derived tumors reached ∼100 mm3 (day 6), mice were injected i.t. with APTSCR-9R or APTSTAT3-9R (10 mg/kg) every other day for a total of four injections. Tumor sections were obtained on day 14. Scale bar, 100 μm. The bar graph shows quantification of P-STAT3 (Tyr705) and STAT3-positive cells. Data are presented as means ± SEM (n = 4 mice/group; ∗∗p < 0.01, ∗p < 0.05 versus untreated; one-way ANOVA followed by Sidak’s post hoc test). (F) Expression of P-STAT3 (Tyr705), STAT3, and GAPDH protein in YUMM1.7R-derived tumor tissues was determined by western blot analysis of lysates of tumors treated with either APTSCR-9R or APTSTAT3-9R as indicated in (A). Data are presented as means ± SEM (n = 4 mice/group; ∗∗p < 0.01, ∗p < 0.05 versus untreated; one-way ANOVA followed by Sidak’s post hoc test).
Discussion
Development of drug resistance to targeted therapy is a serious problem commonly encountered by cancer patients.23,24 This has also proven to be the case for vemurafenib-resistant melanoma. Although attempts have been made to combine BRAF inhibitors such as vemurafenib with other targeting agents (e.g., MEK inhibitors) to counter resistance,25 such approaches have not yet been successful.26 It was recently reported that activation of STAT3 signaling pathways is associated with the development of drug resistance in most cancers.27,28 In the present study, we confirmed that treatment with a cell-permeable STAT3-inhibiting peptide (APTSTAT3-9R) could resensitize vemurafenib-resistant YUMM1.7R melanoma cells to vemurafenib, thereby resulting in a reduction in cell viability (Figure 2). It has also been shown that STAT3 activation in tumor cells induces upregulation of multiple cytokines and chemokines that can recruit immunosuppressive MDSCs and TAMs to the TME.29 Indeed, we found that vemurafenib-resistant YUMM1.7R cells form tumors much more rapidly than vemurafenib-sensitive YUMM1.7P cells, and the TME of the resulting YUMM1.7R-derived tumors also showed greater infiltration of MDSCs and TAMs but lower CD8+ T cell infiltration (Figure 1). Furthermore, we demonstrated that vemurafenib-resistant YUMM1.7R cells substantially upregulated two chemokines, CCL2 and CXCL1, compared with vemurafenib-sensitive YUMM1.7P melanoma (Figure 1E). These findings reveal for the first time that STAT3 activation-mediated secretion of CCL2 and CXCL1 by vemurafenib-resistant melanoma cells may contribute to the recruitment of MDSCs and TAMs and formation of the immunosuppressive TME that is responsible for drug resistance and poor therapeutic response to ICI-based immunotherapy.30
Along the same lines, it has been shown that STAT3 activation in MDSCs induces expansion of these cells, compromises the function of effector T cells, and promotes the development of TAMs, collectively potentiating immunosuppressive tumorigenesis.31 Likewise, STAT3 activation in macrophages can enhance TAM reprogramming from antitumor M1 to tumor-promoting M2-like polarization32 and impair antigen-specific T cell responses in the TME.33 In contrast, ablating STAT3 in CD8+ T cells enables expression of CXCR3 (CXCL10 receptor), resulting in increased infiltration of CD8+ T cells into the TME.34 Using an adoptive transfer therapeutic strategy in mice, researchers have also shown that transfer of STAT3-depleted CD8+ T cells enables efficient tumor infiltration and robust CD8+ T cell proliferation, resulting in increased tumor antigen-specific T cell activity and tumor growth inhibition.35 In this context, inhibition of STAT3 signaling pathways in immune cells of the TME may not only suppress tumor-promoting functions of MDSCs and TAMs but also alleviate the inhibitory effect of those cells on tumor-infiltrating CD8+ T cells, thus promoting an antitumor immune response. Collectively, these observations indicate that STAT3 activation is a key signal associated with drug resistance in cancer cells and an immunosuppressive TME. Thus, we hypothesized that combining ICI therapy with a STAT3 inhibitor is a rational and attractive strategy for treatment of vemurafenib-resistant melanoma tumors. Indeed, we demonstrated that, upon i.t. injection, APTSTAT3-9R mitigated the immunosuppressive TME of vemurafenib-resistant melanoma tumors by reducing production of the two chemokines, CCL2 and CXCL1, and reducing tumor infiltration of MDSCs and TAMs, thereby inhibiting tumor growth (Figure 3). We further confirmed that APTSTAT3-9R treatment significantly increased tumor-infiltrating cytotoxic CD8+ T cells in the TME. For these reasons, we combined APTSTAT3-9R with αPD-1 therapy, which resulted in considerably improved antitumor efficacy in BRAFi-resistant melanoma tumors through combined blockade of STAT3 and PD-1 signaling (Figure 4), providing proof of concept of our hypothesis. Furthermore, since it has been reported that blockade of STAT3 signaling in immune cells within the TME can promote conversion of “cold tumors” to “hot tumors” by regulating immunosuppressive molecules,36,37 we anticipate that the combination regimen between APTSTAT3-9R and αPD-1 demonstrated in this study may be applied to patients with drug-resistant tumors or cold tumors.
We found that i.t. injected APTSTAT3-9R was considerably taken up by MDSCs, TAMs, and CD8+ T cells in the TME, as well as by cancer cells (Figures 5A–5D). Although we did not measure inhibition of STAT3 signaling in each cell type isolated from the tumor, we did confirm inhibition of STAT3 activation in whole tumor tissue of vemurafenib-resistant tumors by immunohistochemistry (Figure 5E) and in tumor lysates by western blotting (Figure 5F). Considering that STAT3 activation promotes immunosuppressive function in MDSCs, TAMs, and cancer cells, but impairs CD8+ T cell function,37 it is most likely that STAT3 inhibition by APTSTAT3-9R in these cells collectively leads to the observed potent antitumor efficacy in BRAFi-resistant melanoma through remodeling of the immunosuppressive TME as well as by directly killing cancer cells. Because APTSTAT3-9R used in this study was administered via i.t. injection, which is not a preferable treatment route in the clinic, we are currently developing an appropriate nanoparticle-based formulation that may enable delivery of this peptide therapeutic to the tumor site via i.v. injection. On the other hand, it seems reasonable to consider the use of previously developed small-molecule STAT3 inhibitors for combination therapy. However, this raises concerns of potential toxicity from systemic exposure to such inhibitors, given the vital importance of STAT3 signaling pathways in most tissues. This highlights the need to develop a drug-delivery system for STAT3 inhibitors. In light of this, i.t. injection of APTSTAT3-9R may be a feasible approach to minimize systemic toxicity while achieving potent therapeutic efficacy.
In conclusion, we demonstrated that inhibition of STAT3 signaling using a cell-permeable STAT3-inhibiting peptide (APTSTAT3-9R) in cancer cells and immune cells of the TME mitigated the immunosuppressive TME and resulted in effective inhibition of vemurafenib-resistant melanoma tumor growth. Furthermore, we showed that combined blockade of STAT3 signaling and PD-1 signaling pathways resulted in greater antitumor efficacy than APTSTAT3-9R monotherapy, suggesting this combination therapy as a new treatment option for overcoming poor therapeutic outcomes associated with drug-resistant BRAF-mutant melanoma.
Materials and methods
Mice
C57BL/6 mice were obtained from Samtako Bio Korea (Osan, South Korea) and housed under pathogen-free conditions. Animal care and experimental procedures were performed with approval of the Animal Care and Use Committees of the Korea Advanced Institute of Science and Technology (KAIST) (accreditation number: KA2021-028).
Cell line and culture
YUMM1.7 cells, a mouse melanoma cell line, were purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM/F12 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution at 37°C in a humidified 5% CO2 incubator. Vemurafenib-resistant YUMM1.7 cells (YUMM1.7R) were maintained under conditions of continuous exposure to 5 μM vemurafenib.
Antibodies and materials
Two peptides, APTSTAT3-9R and APTSCR-9R, were custom synthesized by AnyGen (Gwangju, South Korea). Antibodies against the following proteins were used for western blotting: P-STAT3 (clone D3A7, catalog no. 9145; Cell Signaling Technology, Danvers, MA, USA); STAT3 (clone 124H6, catalog no. 9139; Cell Signaling); P-p44/42 MAPK (ERK1/2) (clone D13.14.4E, catalog no. 4370; Cell Signaling); p44/42 MAPK (ERK1/2) (clone 137F5, catalog no. 4695; Cell Signaling); and GAPDH (catalog no. MA5-15738; Invitrogen, Carlsbad, CA, USA). The antibody use for immune checkpoint blockade therapy was InVivoPlus anti-PD-1 (clone RMP1-14, catalog no. BP0146; BioXcell, Lebanon, NH, USA). Vemurafenib was purchased from LC Laboratories (Woburn, MA, USA).
Syngeneic mouse melanoma model
C57BL/6 mice (6–8 weeks old) were subcutaneously injected with YUMM1.7P (3 × 105) or vemurafenib-resistant YUMM1.7R (3 × 105) melanoma cells, and tumor growth was monitored. Mice with a tumor size >1,500 mm3, which was considered lethal, were euthanized according to protocols approved by the Animal Care and Use Committees of the Korea Advanced Institute of Science and Technology. At the indicated time points, all mice were euthanized and tumors were harvested for flow-cytometry analysis of immune cells.
Flow cytometry and intracellular cytokine staining
Fluorescence-conjugated antibodies against CD8, CD11b, CD45, F4/80, and IFN-γ were purchased from eBioscience (Carlsbad, CA, USA). The fluorescence-conjugated antibody against Gr1 was purchased from BD Bioscience (San Jose, CA, USA). Dead cells were excluded by staining with the fixable viability dye, eFluor 506 (65-0866-14; eBioscience), followed by staining for cell-surface markers. Cells were immunostained by incubating with the following antibodies for 20 min at 4°C in the dark: PerCP-Cyanine 5.5-conjugated anti-CD45 (45-0451-82), BV605-conjugated anti-CD3 (564009; BD Bioscience), FITC-conjugated anti-CD4 (11-0041-85), phycoerythrin (PE)-conjugated anti-CD8 (12-0081-83), allophycocyanin-conjugated anti-IFN-γ (17-7311-82), FITC-conjugated anti-CD11b (11-0112-82), PE-Cyanine7-conjugated anti-CD11b (25-0112-82), BV605-conjugated anti-Gr1 (563299), PE-conjugated anti-F4/80 (12-4801-82), and eFluor 450-conjugated anti-F4/80 (48-4801-82). Following single-cell and live-cell selection, the following immune cell subsets were characterized using the indicated set of markers: CD8+ T cells (CD45+/CD3+/CD8+), MDSCs (CD45+/CD11b+/Gr1+), and TAMs (CD45+/Gr1-/CD11b+/F4/80+).
For measurement of cytokine production in tumor-infiltrated CD8+ T cells by flow cytometry, tumor tissues were first cut into small pieces, then enzymatically and mechanically dissociated into single-cell suspensions using a mouse tumor dissociation kit (Miltenyi Biotec, Auburn CA, USA) and gentleMACS Dissociator (Miltenyi Biotec). Cell suspensions were filtered through a cell strainer (70 μm) and washed with Dulbecco’s PBS. Red blood cells (RBCs) were removed by incubation with 2 mL of RBC lysis buffer (420301; BioLegend, San Diego, CA, USA) for 2 min at room temperature with gentle shaking. Isolated cells were stimulated with soluble anti-CD3e antibody (553057; BD Bioscience) and soluble anti-CD28 antibody (553294; BD Bioscience) for 5 h at 37°C in a humidified 5% CO2 atmosphere. After washing, cells were stained with the fixable viability dye eFluor 506, followed by immunostaining for cell-surface markers using fluorescently labeled anti-CD8 and anti-CD45 antibodies. Following surface staining, samples were fixed and permeabilized using an Intracellular Fixation & Permeabilization buffer set (88-8824-00; eBioscience). Cells were then stained with fluorescently labeled anti-IFN-γ antibodies for measurement of cytokine expression in CD8+ T cells. Flow cytometry was performed using an LSR Fortessa system (BD Biosciences) after compensation with UltraComp Compensation beads (01-2222-41; Invitrogen). Data were analyzed using FlowJo software (TreeStar, Ashland, OR, USA).
Single-cell RNA sequencing
The 3′ digital gene expression of 500–10,000 individual cells per sample was profiled using the Chromium Single Cell 3′ protocol. Single cells, reagents, and a single Gel Bead containing barcoded oligonucleotides are encapsulated into nanoliter-scale GEMs (Gel Bead in emulsion) using the Next GEM Technology. Libraries were prepared using the Chromium controller according to the 10x Chromium Next GEM Single Cell 3′ v3.1 protocol (CG000315). In brief, the cell suspensions were diluted in nuclease-free water to achieve a targeted cell count of 10,000. Cell suspension was mixed with master mix and loaded with Single Cell 3′ v3.1 Gel Beads and Partitioning Oil into a Chromium Next GEM chip G. RNA transcripts from single cells were uniquely barcoded and reverse transcribed within droplets. cDNA molecules were pooled, and the cDNA pool was then put through an end repair process, the addition of a single “A” base, and ligation of the adapters. The products were then purified and enriched with PCR to create the final cDNA library. The purified libraries were quantified using qPCR according to the KAPA qPCR Quantification Protocol Guide (Roche Sequencing, Pleasanton, CA, USA) and qualified using the Agilent Technologies 4200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). The libraries were then sequenced using the HiSeq platform (Illumina, San Diego, CA, USA) according to the read length in the user guide. After quality control filtering to remove cells with low gene detection (<500 genes) and high mitochondrial gene content (>8%), cell clustering and cell-type identification analysis were carried out using Seurat, which is well known as an automatic annotation method for scRNA-seq data.
Immunohistochemistry
Tumor samples were immediately washed with 1× PBS, fixed by incubating with 4% paraformaldehyde for 12 h, dehydrated, processed into paraffin blocks, and sectioned (4 μm thickness) onto HistoBond glass slides (Marienfeld Superior, Lauda-Königshofen, Germany) using a CM 1850 microtome (Leica, Wetzlar, Germany). Prepared tissue samples were deparaffinized using histological grade xylene (Sigma-Aldrich, St. Louis, MO, USA) and rehydrated for hematoxylin and eosin (H&E) staining. For immunohistochemical analyses, 4-μm sections were deparaffinized and immunostained using antibodies specific for P-STAT3 or STAT3. Excess dye was removed by washing stained samples three times with distilled water, followed by serial dehydration with two changes of 95% ethanol and 100% ethanol. Stained tissues were cleared with two changes of xylene (3 min each) and mounted with Permount mounting medium. The histology of mounted tumor samples was visualized using an inverted microscope (Eclipse Ti2; Nikon, Tokyo, Japan). 3,3′-Diaminobenzidine staining was quantified using the Color Deconvolution II plugin in ImageJ software.
RNA isolation and real-time PCR
Total RNA was isolated using an RNA purification kit (NANOHELIX, Daejeon, South Korea), and the indicated mouse target genes were analyzed by qRT-PCR using the SYBR reagent (Bio-Rad, Hercules, CA, USA) and the following primer pairs: CCL2, 5′-GTT GGC TCA GCC AGA TGC A-3′ (forward) and 5′-AGC CTA CTC ATT GGG ATC TTG-3′ (reverse); CXCL1, 5′-CCG AAG TCA TAG CCA CAC TCA A-3′ (forward) and 5′-GCA GTC TGT CTT TCT CCG TTA-3′ (reverse); CXCL2, 5′-GAA GTC ATA GCC ACT CTC AAG G-3′ (forward) and 5′-CCT CCT TTC CAG GTC AGT TAG C-3′ (reverse); CXCL5, 5′-GGT CCA CAG TGC CCT ACG-3′ (forward) and 5′-GCG AGT GCA TTC CGC TTA-3′ (reverse). Sequence-specific amplification was analyzed on a CFX96 Real-Time PCR Detection System (Bio-Rad). Data were normalized to TATA-binding protein expression, and relative expression levels were calculated using the 2−ΔΔCt method.
Statistical analysis
Data are presented as means ± standard errors of the mean (SEM), and were analyzed with GraphPad Prism Software v6.0 for Windows (GraphPad Software, San Diego, CA, USA). Comparisons between two different groups were performed using unpaired two-tailed Student’s t test; comparisons among more than two groups were performed using two-way analysis of variance (ANOVA) followed by Tukey’s post hoc test or one-way ANOVA followed by Sidak’s post hoc test. p values of less than 0.05 were considered statistically significant.
Data and materials availability
All data are available in the main text or supplemental information.
Acknowledgments
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF2018R1A3B1052661) funded by the Ministry of Science and ICT.
Author contributions
S.J. and T.W.K. conceived the project. T.W.K. performed most of the experiments. Y.K. and W.J. assisted with the in vivo experiments. H.K. performed certain in vitro experiments and analyzed the data. M.K. performed transcriptomic analyses. T.W.K. and S.J. analyzed most of the data and wrote the manuscript. All authors approved the final version of the manuscript.
Declaration of interests
S.J. is a co-founder of BiliX and a non-executive director of Ubiquoss Holdings. None of these activities is related to the paper.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.06.001.
Supplemental information
References
- 1.Wan P.T., Garnett M.J., Roe S.M., Lee S., Niculescu-Duvaz D., Good V.M., Jones C.M., Marshall C.J., Springer C.J., Barford D., et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty K.T., Puzanov I., Kim K.B., Ribas A., McArthur G.A., Sosman J.A., O'Dwyer P.J., Lee R.J., Grippo J.F., Nolop K., Chapman P.B. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villanueva J., Vultur A., Lee J.T., Somasundaram R., Fukunaga-Kalabis M., Cipolla A.K., Wubbenhorst B., Xu X., Gimotty P.A., Kee D., et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazarian R., Shi H., Wang Q., Kong X., Koya R.C., Lee H., Chen Z., Lee M.K., Attar N., Sazegar H., et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obenauf A.C., Zou Y., Ji A.L., Vanharanta S., Shu W., Shi H., Kong X., Bosenberg M.C., Wiesner T., Rosen N., et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–372. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu G., Bowman T., Huang M., Shivers S., Reintgen D., Daud A., Chang A., Kraker A., Jove R., Yu H. Roles of activated Src and Stat3 signaling in melanoma tumor cell growth. Oncogene. 2002;21:7001–7010. doi: 10.1038/sj.onc.1205859. [DOI] [PubMed] [Google Scholar]
- 8.Kortylewski M., Kujawski M., Wang T., Wei S., Zhang S., Pilon-Thomas S., Niu G., Kay H., Mule J., Kerr W.G., et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 9.Jones L.M., Broz M.L., Ranger J.J., Ozcelik J., Ahn R., Zuo D., Ursini-Siegel J., Hallett M.T., Krummel M., Muller W.J. STAT3 establishes an immunosuppressive microenvironment during the early stages of breast carcinogenesis to promote tumor growth and metastasis. Cancer Res. 2016;76:1416–1428. doi: 10.1158/0008-5472.CAN-15-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hossain D.M., Pal S.K., Moreira D., Duttagupta P., Zhang Q., Won H., Jones J., D'Apuzzo M., Forman S., Kortylewski M. TLR9-Targeted STAT3 silencing abrogates immunosuppressive activity of myeloid-derived suppressor cells from prostate cancer patients. Clin. Cancer Res. 2015;21:3771–3782. doi: 10.1158/1078-0432.CCR-14-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proia T.A., Singh M., Woessner R., Carnevalli L., Bommakanti G., Magiera L., Srinivasan S., Grosskurth S., Collins M., Womack C., et al. STAT3 antisense oligonucleotide remodels the suppressive tumor microenvironment to enhance immune activation in combination with anti-PD-L1. Clin. Cancer Res. 2020;26:6335–6349. doi: 10.1158/1078-0432.CCR-20-1066. [DOI] [PubMed] [Google Scholar]
- 12.Kim S., Kim D., Jung H.H., Lee I.H., Kim J.I., Suh J.Y., Jon S. Bio-inspired design and potential biomedical applications of a novel class of high-affinity peptides. Angew. Chem. Int. Ed. Engl. 2012;51:1890–1894. doi: 10.1002/anie.201107894. [DOI] [PubMed] [Google Scholar]
- 13.Kim D., Lee I.H., Kim S., Choi M., Kim H., Ahn S., Saw P.E., Jeon H., Lee Y., Jon S. A specific STAT3-binding peptide exerts antiproliferative effects and antitumor activity by inhibiting STAT3 phosphorylation and signaling. Cancer Res. 2014;74:2144–2151. doi: 10.1158/0008-5472.CAN-13-2187. [DOI] [PubMed] [Google Scholar]
- 14.Kim J.Y., Ahn J., Kim J., Choi M., Jeon H., Choe K., Lee D.Y., Kim P., Jon S. Nanoparticle-assisted transcutaneous delivery of a signal transducer and activator of transcription 3-inhibiting peptide ameliorates psoriasis-like skin inflammation. ACS Nano. 2018;12:6904–6916. doi: 10.1021/acsnano.8b02330. [DOI] [PubMed] [Google Scholar]
- 15.Keum H., Kim J., Yoo D., Kim T.W., Seo C., Kim D., Jon S. Biomimetic lipid Nanocomplexes incorporating STAT3-inhibiting peptides effectively infiltrate the lung barrier and ameliorate pulmonary fibrosis. J. Control Release. 2021;332:160–170. doi: 10.1016/j.jconrel.2021.02.022. [DOI] [PubMed] [Google Scholar]
- 16.Saw P.E., Xu X., Kim S., Jon S. Biomedical applications of a novel class of high-affinity peptides. Acc. Chem. Res. 2021;54:3576–3592. doi: 10.1021/acs.accounts.1c00239. [DOI] [PubMed] [Google Scholar]
- 17.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 18.Sumimoto H., Imabayashi F., Iwata T., Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J. Exp. Med. 2006;203:1651–1656. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frederick D.T., Piris A., Cogdill A.P., Cooper Z.A., Lezcano C., Ferrone C.R., Mitra D., Boni A., Newton L.P., Liu C., et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin. Cancer Res. 2013;19:1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagarsheth N., Wicha M.S., Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017;17:559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar V., Patel S., Tcyganov E., Gabrilovich D.I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F., Cao J., Wu J., Sullivan K., Shen J., Ryu B., Xu Z., Wei W., Cui R. Stat3-targeted therapies overcome the acquired resistance to vemurafenib in melanomas. J. Invest. Dermatol. 2013;133:2041–2049. doi: 10.1038/jid.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 24.Vasan N., Baselga J., Hyman D.M. A view on drug resistance in cancer. Nature. 2019;575:299–309. doi: 10.1038/s41586-019-1730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagle N., Van Allen E.M., Treacy D.J., Frederick D.T., Cooper Z.A., Taylor-Weiner A., Rosenberg M., Goetz E.M., Sullivan R.J., Farlow D.N., et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moriceau G., Hugo W., Hong A., Shi H., Kong X., Yu C.C., Koya R.C., Samatar A.A., Khanlou N., Braun J., et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell. 2015;27:240–256. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H.J., Zhuang G., Cao Y., Du P., Kim H.J., Settleman J. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 28.Zhao C., Li H., Lin H.J., Yang S., Lin J., Liang G. Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol. Sci. 2016;37:47–61. doi: 10.1016/j.tips.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat. Rev. Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petitprez F., Meylan M., de Reynies A., Sautes-Fridman C., Fridman W.H. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front Immunol. 2020;11:784. doi: 10.3389/fimmu.2020.00784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabrilovich D.I., Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Farrell A.M., Liu Y., Moore K.W., Mui A.L. IL-10 inhibits macrophage activation and proliferation by distinct signaling mechanisms: evidence for Stat3-dependent and -independent pathways. EMBO J. 1998;17:1006–1018. doi: 10.1093/emboj/17.4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng F., Wang H.W., Cuenca A., Huang M., Ghansah T., Brayer J., Kerr W.G., Takeda K., Akira S., Schoenberger S.P., et al. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425–436. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 34.Yue C., Shen S., Deng J., Priceman S.J., Li W., Huang A., Yu H. STAT3 in CD8+ T cells inhibits their tumor accumulation by downregulating CXCR3/CXCL10 Axis. Cancer Immunol. Res. 2015;3:864–870. doi: 10.1158/2326-6066.CIR-15-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kujawski M., Zhang C., Herrmann A., Reckamp K., Scuto A., Jensen M., Deng J., Forman S., Figlin R., Yu H. Targeting STAT3 in adoptively transferred T cells promotes their in vivo expansion and antitumor effects. Cancer Res. 2010;70:9599–9610. doi: 10.1158/0008-5472.CAN-10-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu H., Kortylewski M., Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 37.Huynh J., Chand A., Gough D., Ernst M. Therapeutically exploiting STAT3 activity in cancer - using tissue repair as a road map. Nat. Rev. Cancer. 2019;19:82–96. doi: 10.1038/s41568-018-0090-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.