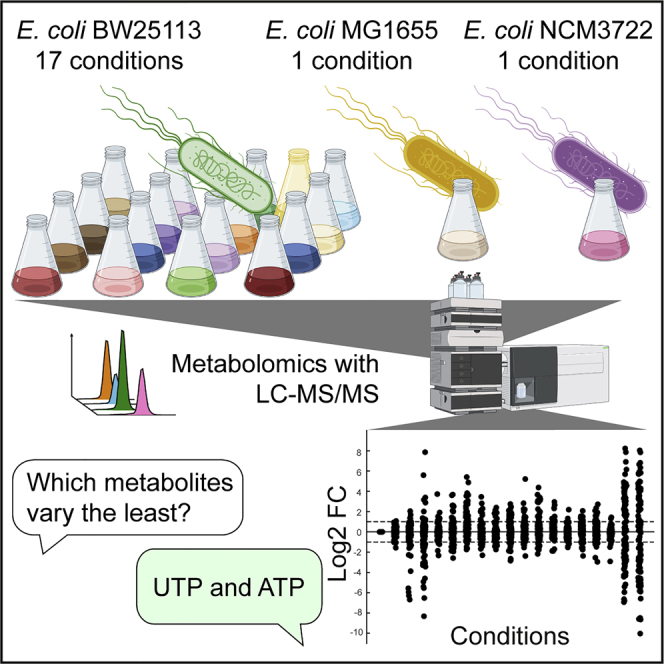
Summary
Metabolite concentrations vary across conditions and such metabolome changes are relevant for metabolic and gene regulation. Here, we used LC-MS/MS to explore metabolite concentration changes in Escherichia coli. We measured 101 primary metabolites in 19 experimental conditions that include various nutrients and stresses. Many metabolites showed little variation across conditions and only few metabolites correlated with the growth rate. The least varying metabolites were nucleotides (e.g. UTP had 10% variation) and amino acids (e.g. methionine had 13% variation). These results show that E. coli maintains protein and RNA building blocks within narrow concentration ranges, thus indicating that many feedback mechanisms in biosynthetic pathways contribute to end-product homeostasis.
Subject areas: Microbiology, Microbial physiology, Omics, Metabolomics
Graphical abstract
Highlights
-
•
101 E coli metabolites were measured in 19 conditions
-
•
Biosynthetic end-products vary little between conditions
-
•
Few metabolites correlate with the growth rate
-
•
Metabolome data identify active regulatory metabolites
Microbiology; Microbial physiology; Omics; Metabolomics
Introduction
Cells must maintain intracellular metabolites at physiological concentrations, despite changes of metabolic flux (Gerosa et al., 2015; Park et al., 2016; Schafer et al., 2004). High metabolite levels can inhibit enzymes that generate them (Alam et al., 2017), whereas low metabolite levels lead to insufficient active site occupancies of enzymes that consume them (Bennett et al., 2009). Thus, metabolite levels can only vary within certain ranges and theoretical analyses suggested that metabolite concentration homeostasis is a primary objective of metabolic regulation (Hofmeyr and Cornish-Bowden, 1991; Thomas and Fell, 1996). A number of studies measured microbial metabolites in different experimental conditions (Bennett et al., 2009; Brauer et al., 2006; Chubukov et al., 2013; Gerosa et al., 2015; Kochanowski et al., 2017; Park et al., 2016), but have not provided a systematic analysis of metabolite concentration homeostasis.
Here, we measured 101 E coli metabolites in the same 19 experimental conditions that were recently used to measure the E. coli proteome (Schmidt et al., 2016). We found that many metabolites vary little across conditions, which is in contrast to the high condition-dependency of the proteome. The least varying metabolites were UTP and ATP, with less than 15% variation between all conditions that supported growth. Similarly, most amino acids remained within narrow concentration ranges, and methionine was the least varying amino acid. The high constancy of nucleotide and amino acid end-products may facilitate their accurate supply and demand, as suggested by metabolic control theory (Hofmeyr and Cornish-Bowden, 2000). In conclusion, the condition-dependent E. coli metabolome revealed that nucleotides and amino acids are the least varying class of metabolites, thus supporting theories (Hofmeyr and Cornish-Bowden, 2000; Schafer et al., 2004), which suggest that end-product homeostasis is an important objective of metabolic regulation in biosynthetic pathways.
Results
The condition-dependent E. coli metabolome
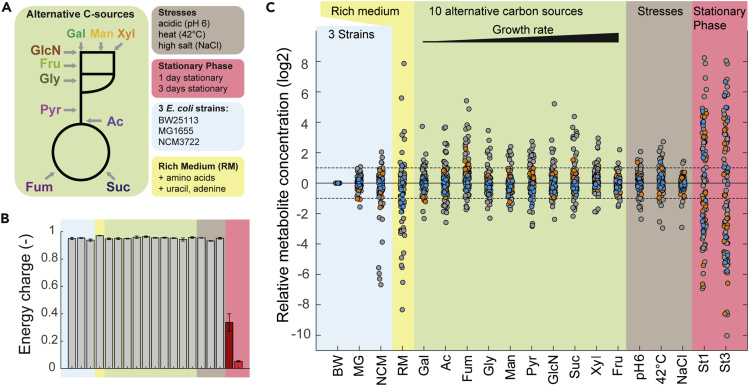
To assess the variability of intracellular metabolites, we measured 101 E coli metabolites in 19 experimental conditions (Figure 1A and Table S1). For 17 conditions, we used the E. coli strain BW25113, which was grown in: 1) minimal glucose medium, 2–11) minimal medium with 10 alternative carbon sources, 12–14) minimal glucose medium under three stresses (low pH of 6, high salt concentration, and high temperature of 42°C), 15–16) stationary phase (1 day and 3 days), and 17) synthetic-rich medium (minimal medium supplemented with glycerol, 20 amino acids, adenine, and uracil). In the remaining two conditions, we cultured the E. coli strains NCM3722 and MG1655 in minimal glucose medium. The proteome was previously measured in the same conditions (Schmidt et al., 2016), and we observed almost the same biomass-specific growth rates, which varied between 0.2 and 1.0 per h (Figure S1). Metabolomics samples were collected with a filter method (Bennett et al., 2008; Gerosa et al., 2015; Kochanowski et al., 2017), and metabolite extracts were analyzed with liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Guder et al., 2017). Measured metabolite concentrations were reproducible between three biological replicates, with a median error of 14% across the 101 metabolites (Figure S2).
Figure 1.
The condition-dependent E. coli metabolome
(A) The 19 conditions used in this study were adapted from the proteome study (Schmidt et al., 2016): three glucose-fed E. coli strains (BW25113, NCM3722, MG1655), 10 alternative carbon sources, three stresses (low pH, high salt, and high temperature), and stationary phase (1 day and 3 days). See also Table S1.
(B) Adenylate energy charge in the 19 conditions ([ATP]+ 0.5.[ADP])/([ATP]+[ADP]+[AMP]). Bars are means from n = 3 replicates and error bars show the SD.
(C) Relative concentration of 101 intracellular E. coli metabolites. Concentrations are shown as log2 fold changes normalized to the glucose condition with E. coli BW25113. Dots show the mean of n = 3 replicates. Amino acids are shown in orange and nucleotides in blue. See also Table S2. Note that if a particular metabolite is supplemented to the cultivation medium, it is not possible to differentiate between its intracellular and extracellular concentration. Therefore, amino acids and adenine are excluded from the rich medium condition. Succinate is excluded from the succinate condition. See Figure S1–S3.
We determined absolute concentrations of ATP, ADP, and AMP using authentic standards and an internal 13C-labeled standard (Bennett et al., 2008; Guder et al., 2017). This allowed us to calculate the energy charge in the 19 conditions as: ([ATP]+0.5.[ADP])/([ATP]+[ADP]+[AMP]). The energy charge was high and remained almost constant in all conditions that supported growth (Figure 1B), thus indicating that cells maintain energy homeostasis despite the different quality of nutrients in the medium. Only stationary cells had a low energy charge, which is consistent with previous reports (Brauer et al., 2006). The energy charge reflects the energetic status of the cell (Atkinson, 1968), and it has been described before that growing bacteria maintain the energy charge within a narrow range (De la Fuente et al., 2014) and that it gradually decreases in non-growing bacteria (Bergkessel et al., 2016). Moreover, the high energy charge of growing cells shows that our sampling protocol was fast and efficient, because ineffective sampling and quenching would immediately affect ATP levels, which have a turn-over time of a few seconds (Heijnen, 2010).
Stationary phase and rich medium cause widespread metabolome changes
Next, we sought to systematically analyze metabolite concentration changes across conditions. In most conditions, the majority of metabolites varied less than 2-fold, when normalized to the glucose condition with the BW25113 strain (Figure 1C and Table S2). However, the stationary phase conditions and the rich medium condition led to higher metabolite concentration changes (Figure 1C). Cells in stationary phase degrade proteins and RNA (Bergkessel et al., 2016), and accordingly, we observed increases of metabolites in degradation pathways of amino acids and nucleotides (Figure S3). Biosynthetic intermediates, in contrast, decreased in stationary phase cells, presumably because biosynthetic pathways are less active in non-growing cells (Figure S3). Biosynthetic intermediates were also low in E. coli cells that grew in the rich medium condition, which contains external amino acids and the nucleobases uracil and adenine (Figure S3).
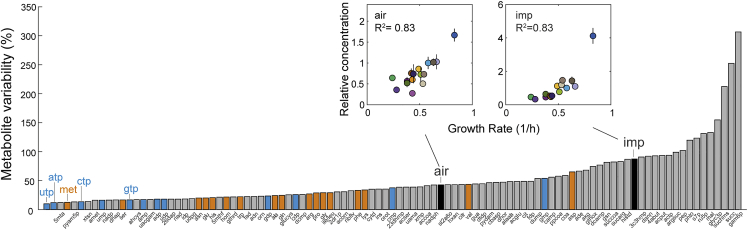
Nucleotide triphosphates are the least varying class of metabolites
Next, we analyzed metabolome variability based on the relative SD (RSD) of metabolite levels across 16 of the 19 conditions (Figures 2, S4, and S5). The RSD was calculated from the SD of the concentration of a particular metabolite in the 16 conditions. The SD was then normalized to the mean concentration of the metabolite in the 16 conditions, to obtain the RSD across conditions. For this calculation of the RSD, we used the mean metabolite concentration of the three replicates. Thus, the RSD indicates to what extent a metabolite varies across the 16 conditions. We excluded the two stationary phase conditions from this analysis, due to the higher metabolome changes in non-growing bacteria (Figure 1C). The rich medium was also excluded because the medium contains high amounts of extracellular amino acids, which confounds measurements of intracellular amino acids (residuals of the cultivation medium cling to cells and the filter).
Figure 2.
Variability of metabolites across conditions
The variability of 101 measured metabolites is shown as relative SD(RSD) across 16 conditions (stationary phase and rich medium were not considered). The RSD is the SD across conditions normalized to the mean. Amino acids are shown in orange and nucleotides in blue. Inserts show the relationship of the growth rate with concentrations of IMP and 5-Amino-1-(5-phospho-D-ribosyl)imidazole (air). Colors indicate the experimental condition. R is the Pearson correlation coefficient of growth rates and metabolite levels. See Figure S4–S6.
First, we confirmed that metabolome variability across conditions does not correlate with the error between replicates (Figure S6). This means that a high relative SD across conditions is not due to a higher SD between experimental replicates. Additionally, we measured response curves with authentic standards to confirm that metabolite concentrations in our samples fall into the linear range of our LC-MS/MS method (Figures S7–S9). The response curves were linear over several orders of magnitude, which are consistent with our previous report of the LC-MS/MS method (Guder et al., 2017). The high linearity is important to ensure that a 1% change in the LC-MS/MS signal (the 12C/13C ratio) corresponds to a 1% change in metabolite concentration.
Out of 101 measured metabolites, 42 metabolites varied less than 30% across conditions (Figure 2). These stable metabolites included 13 of 17 measured amino acids, and 10 of 12 nucleotides. The least varying metabolites were UTP and ATP (10% and 12% variability), suggesting that end-product homeostasis is especially important in the case of nucleotide biosynthesis. According to metabolic control analysis, a high stability of biosynthetic end-products can facilitate their accurate supply and demand (Hofmeyr and Cornish-Bowden, 2000), and thus the stable pool of nucleotide triphosphates could ensure proper control over RNA synthesis rates. Indeed, all four nucleotide classes (adenosine-phosphates, guanosine-phosphates, cytidine-phosphates, and uridine-phosphates) varied less in their triphosphate form than the respective di- and monophosphates (Figure 2). The most variable nucleotide was GMP, which varied 54% across conditions, mainly because GMP increased 2.6-fold in the 42°C condition.
Intermediates in nucleotide metabolism varied more than the end-products. Inosine-monophosphate (IMP), for instance, is a central intermediate in purine nucleotide biosynthesis, and IMP was among the metabolites with a high variability (87%). Among all measured metabolites, IMP had the highest correlation with the growth rate (insert in Figure 2). Another intermediate in purine nucleotide biosynthesis, 5-Amino-1-(5-phospho-D-ribosyl)imidazole (AIR), had the second highest growth rate correlation among all 101 metabolites. Such “growth-rate effects” are typically attributed to the interdependency of gene expression and growth (Gerosa et al., 2013; Scott et al., 2010), and the condition-dependent proteome study (Schmidt et al., 2016) found that many proteins correlated with the growth rate. Here, we found that except IMP and AIR only few metabolites correlated with growth rate.
Adenine and uracil salvage inactivate de novo biosynthesis of nucleotides
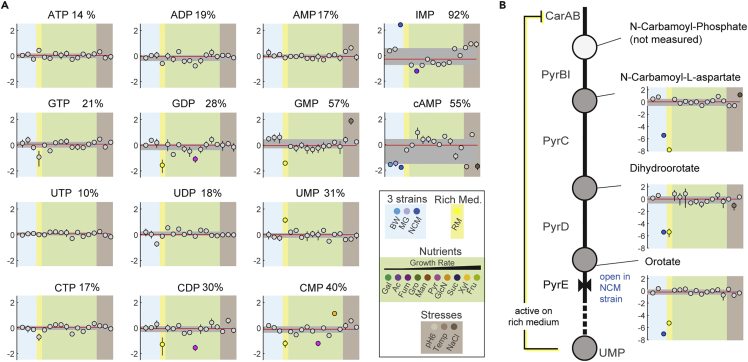
The rich medium contained the nucleobases adenine and uracil, which are converted into nucleotides via salvage pathways. Despite this, the concentration of UTP and ATP in rich medium was similar to the other 16 conditions (Figure 3A), showing that E. coli maintains homeostasis of UTP and ATP even if they are synthesized via completely different pathways (de novo biosynthesis of nucleotides vs. nucleotide salvage).
Figure 3.
Variability of nucleotides and feedback regulation of the pyrimidine pathway
(A) Relative concentration of nucleotides in 19 conditions. Metabolite concentrations are shown as log2 fold changes that are normalized to the glucose condition with the BW25113 strain. Dots show the mean of n = 3 replicates, lines show the SD. Log2 fold changes > 1 or < −1 are indicated by the color of the respective condition. Numbers next to the metabolite show the relative SD(RSD) across 17 conditions (stationary phase conditions were not considered). Note that RSD values differ from Figure 2 because the rich medium is considered here.
(B) Same as (A) for intermediates in pyrimidine nucleotide biosynthesis. The metabolic map shows the pyrimidine pathway and allosteric feedback inhibition of carbamoyl phosphate synthetase (CarAB) by UMP. The metabolic bottleneck at orotate phosphoribosyltransferase (PyrE) is present in all conditions except the NCM3722 strain, due to a frameshift mutation in MG1655-derived E. coli (Jensen, 1993).
See Figure S7.
Next, we examined metabolic regulation mechanisms that could be responsible for the activation of salvage pathways in the rich medium condition. Supplementation of adenine in the rich medium condition led to high levels of intracellular hypoxanthine in E. coli (Figure S3), which is an intermediate in the adenine salvage pathway. Hypoxanthine is an allosteric activator of the transcription factor PurR that controls expression of genes in purine and pyrimidine nucleotide metabolism (Cho et al., 2011). Thus, the high hypoxanthine levels may activate PurR, which in turn represses expression of genes in de novo biosynthesis of purine and pyrimidine nucleotides. The proteome data (Schmidt et al., 2016) confirm that this regulation is active in the rich medium condition and that it led to lower abundances of most enzymes in those pathways. Apart from enzyme-level regulation via the hypoxanthine-PurR circuit, allosteric regulation of enzyme activity may also switch from de novo nucleotide biosynthesis to nucleotide salvage. Several nucleotides feedback inhibits enzymes in their own biosynthesis pathways. For example, UTP inhibits aspartate transcarbamoylase, and UMP together with CMP inhibit carbamoyl phosphate synthetase (Reaves et al., 2013) (Figure 3B). Because UTP varied little, it is unlikely that the UTP feedback was active in the rich medium condition. However, UMP levels increased more than 2-fold on rich medium and presumably feedback inhibited carbamoyl phosphate synthase (Figure 3B).
To confirm that this regulation inactivates de novo nucleotide biosynthesis during growth on rich medium, we took a closer look at intermediates in the pathways (Figure 2B). Two of three measured intermediates in the purine pathway decreased on rich medium (IMP and adenylosuccinate, Figure S3), and all three measured pyrimidine intermediates decreased (carbamoyl-aspartate, orotate, and dihydroorotate, Figure 3B). Thus, the low concentration of nucleotide intermediates on rich medium supports the hypothesis that de novo nucleotide biosynthesis is inactive in this condition, and that cells salvage externally supplied uracil and adenine.
We noticed that all three pyrimidine intermediates were less abundant in the E. coli strain NCM3722 than in the other two strains under same conditions (Figure 3B). The higher concentration of pyrimidine intermediates in E. coli BW25113 and MG1655 is probably due to the known frameshift mutation in MG1655-derived E. coli strains (Jensen, 1993) that causes low expression of pyrE-encoded orotate phosphoribosyltransferase (PyrE). The proteome data confirm this pyrimidine bottleneck at PyrE, because BW25113 and MG1655 have less than 50 copies of the enzyme, compared to 568 PyrE copies in NCM3722 (Schmidt et al., 2016). Despite this strong perturbation of PyrE levels in MG1655 and BW25113, the concentration of UTP and CTP was similar in all three E. coli strains, which further underlines the important role of nucleotide homeostasis.
Methionine is the least varying amino acid
Next, we inspected variability of the 18 measured amino acids. The least varying amino acid was methionine and the most variable amino acid was aspartate (Figure 2). The high stability of methionine may reflect its role in translation initiation and that methionine biosynthesis limits overall protein synthesis in E. coli (Li et al., 2014). The high variability of aspartate is probably due to an entry-point effect of the alternative carbon sources (Figure S10): aspartate is only one step away from the TCA cycle metabolite oxaloacetate, and aspartate increased when E. coli grew on the TCA cycle metabolites fumarate and succinate. Similarly, many TCA-derived amino acids (lysine, asparagine, glutamine, aspartate, and arginine) had the highest concentration in fumarate-fed cells (Figure S10). Fumarate had a stronger entry-point effect than succinate, presumably because fumarate has a direct effect on aspartate: high fumarate concentrations can drive the amination of fumarate to aspartate (via aspartate ammonia-lyase). Xylose enters metabolism via the pentose phosphate pathway (PPP) and it led to high levels of PPP-derived phenylalanine.
Apart from these entry point effects of the alternative carbon sources, amino acids were among the least varying metabolites (Figure 2). This confirms previous theories, which suggest that some metabolic regulation mechanisms are geared to maintain homeostasis of biosynthetic end-products (Hofmeyr and Cornish-Bowden, 2000; Schafer et al., 2004). Because growth rates varied up to 3-fold between conditions (Figure S1), we inspected the mechanisms that could adapt amino acid biosynthesis flux to different demands for protein synthesis. E. coli can modulate biosynthetic fluxes in amino acid pathways by changes of enzyme activity or enzyme abundance. The proteome data indicated that changes of enzyme abundance were probably the main driver of flux changes, because almost all enzyme levels in amino acid pathways correlated linearly with the growth rate (Schmidt et al., 2016). These changes in enzyme abundance are either due to negative feedback of amino acids via transcription factors (Cho et al., 2012) or transcriptional attenuation (Yanofsky, 1981), or due to global growth-rate-dependent regulation (Scott et al., 2010).
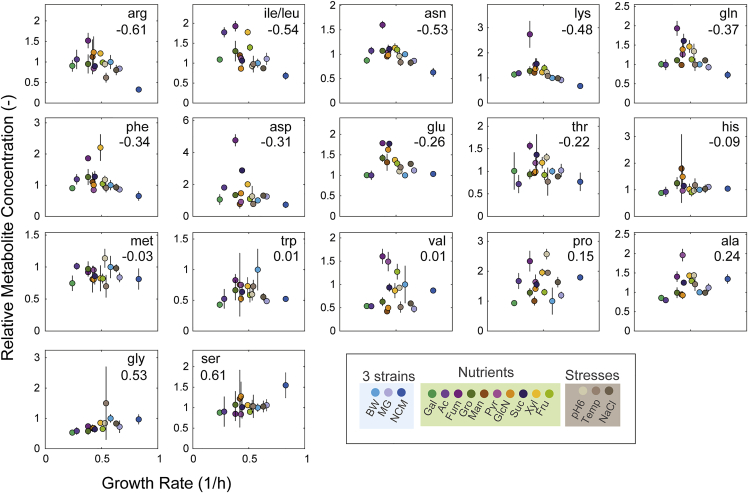
If negative feedback of amino acids adapts enzyme abundance to growth rates, we expected a negative relationship between amino acid levels and growth rates, because lower amino acid levels would de-repress transcription. To test this, we correlated all 18 measured amino acids with the growth rate (Figure 4). While most amino acids did not correlate with growth, arginine and the pool of leucine/isoleucine showed a modest negative correlation with growth rates (Figure 4). Arginine and leucine are allosteric activators of the transcription factors ArgR and Lrp, which control almost all amino acid metabolisms due to a hierarchical structure of the transcription regulation network in E. coli (Cho et al., 2012). Thus, decreases of arginine and leucine at higher growth rates may drive increases of enzyme levels via transcriptional regulation.
Figure 4.
Relationship between amino acids and growth rates
The relative concentration of amino acids is plotted against the growth rate in the respective condition. Metabolite concentrations are shown as fold changes that are normalized to the glucose condition with the BW25113 strain. Dots are the mean of n = 3 replicates and lines are the SD. Numbers in the plots are the Pearson correlation coefficient (R).
Discussion
In this study, we measured 101 E coli metabolites in 19 experimental conditions, and several metabolites, especially biosynthetic end-products, showed remarkably little variation across conditions. An exception were stationary phase conditions (1 day and 3 days), which led to higher metabolome changes including biosynthetic end-products (e.g. phenylalanine increased >10-fold). We assume that end-product homeostasis is less relevant in stationary phase cells, because protein and RNA synthesis are low in non-growing cells compared to growing cells (Bergkessel et al., 2016). Instead, our data indicated that stationary phase cells degrade protein and RNA, because they had high levels of metabolites that are involved in degradation of amino acids and nucleotides (Figure S3).
Biosynthetic end-products were the least varying class of metabolites in all conditions that supported growth. Especially, UTP, ATP, and methionine were remarkably stable and varied less than 15% between conditions. Our observation that UTP is the least varying metabolite matches the previous discovery of a dual regulatory strategy in the pyrimidine pathway of E. coli, where classical feedback inhibition and directed overflow work together to ensure end-product homeostasis (Reaves et al., 2013). Similar dual regulatory strategies exist in amino acid pathways (Sander et al., 2019), and a hypothesis is that the resulting homeostasis of biosynthetic end-products is important to seamlessly link metabolism with protein and RNA synthesis. This hypothesis was also described within the framework of metabolic control theory (Hofmeyr and Cornish-Bowden, 2000), which suggested that “linking” metabolites at the interface of macromolecular synthesis and metabolism should remain stable to coordinate their demand and supply.
However, not all amino acids remained within narrow concentration ranges. For example, arginine and (iso-)leucine varied more than 2-fold across conditions and their concentration showed a modest growth dependency (Figure 4). The growth dependency indicates that arginine and leucine adapt amino acid fluxes, since both amino acids have the potential to control almost all amino acid biosynthesis pathways via interactions with the transcription factors ArgR and Lrp (Cho et al., 2012). Thus, amino acids levels may have different regulatory roles: i) stable amino acids like methionine can coordinate supply and demand for protein synthesis, and ii) arginine and leucine can drive changes of biosynthetic fluxes. However, it is important to underline that arginine and leucine show only a modest growth dependency, and probably other factors contribute to changes in biosynthetic flux, e.g. growth dependencies of gene expression (Scott et al., 2010). In general, we were surprised that very few metabolites showed growth dependencies, which is in contrast to the proteome that had pervasive growth-rate effects (Schmidt et al., 2016). The metabolite with highest growth-rate dependency was IMP (Figure 2), and future studies could explore feedback between IMP and gene expression by mapping IMP-binding proteins with proteomic-approaches (Piazza et al., 2018).
Moreover, the metabolome data indicated regulatory mechanisms in pyrimidine nucleotide biosynthesis that might be active in the rich medium condition. The data suggested that in rich medium, E. coli synthesizes pyrimidine nucleotides by uracil salvage instead of de novo synthesis, and that de novo synthesis is inactivated by two regulatory metabolites: high hypoxanthine levels (Figure S3) downregulated enzyme abundance (via PurR), and high UMP levels (Figure 3A) feedback inhibited carbamoyl phosphate synthase.
In conclusion, by measuring the E. coli metabolome in different experimental conditions, we found that biosynthetic end-products are the least varying class of metabolites. Especially, nucleotide triphosphates and methionine changed less than 15% between conditions, despite more than 3-fold difference in growth rates. These results suggest that many feedbacks in biosynthetic pathways of amino acids (Cho et al., 2012; Sander et al., 2019) and nucleotides (Reaves et al., 2013) are designed to maintain end-product homeostasis and explains why deviations from end-product homeostasis are sensed by other cellular processes, e.g. via nucleotides like ppGpp (Büke et al., 2022).
Limitations of the study
A limitation of this study is that we did not quantify the full metabolome, which includes hundreds of primary and secondary metabolites. Although untargeted metabolome methods capture a much larger fraction of the metabolome, they are not as quantitative as the isotope ratio LC-MS/MS method used in this study. From our experience, the 13-C internal standard is essential to obtain a sufficiently large dynamic range, as well as a linear relationship between LC-MS/MS signal and metabolite concentration. Another limitation is that we measured only one bacterial species (E. coli) and the jury is out if other bacteria and other organisms maintain their biosynthetic metabolome also within a narrow concentration range.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli BW2511330 | Baba et al., 2006 | N/A |
| Escherichia coli MG1655 | DSMZ | 18039 |
| Escherichia coli NCM372231 | Soupene et al., 2003 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Acetonitrile | Honeywell Riedel-de Haen | Cat#14261-2L |
| Methanol | VWR | Cat#83638.320 |
| Uniformly 13C-labeled glucose | Cambridge Isotope Laboratories | N/A |
| Deposited data | ||
| Metabolome data at MetaboLights | This study | MTBLS4249 |
| Software and algorithms | ||
| Matlab R2018b for analysis of experimental data | Mathworks.com | N/A |
| Other | ||
| Filter membrane | Merck Millipore | HVLP02500 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hannes Link (hannes.link@uni-tuebingen.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cultivation media and strains
E. coli strains BW25113 (Baba et al., 2006), MG1655 (DSMZ No. 18039) and NCM3722 (Soupene et al., 2003) were used in this study. The cultivation conditions were the same as described by Schmidt and coworkers (Schmidt et al., 2016). The M9 minimal medium consisted of: 200 mL of 5 × base salt solution (211 mM Na2HPO4, 110 mM KH2PO4, 42.8 mM NaCl, 56.7 mM (NH4)2SO4), 10 mL of trace elements (0.63 mM ZnSO4, 0.7 mM CuCl2, 0.71 mM MnSO4, 0.76 mM CoCl2), 1 mL 0.1 M CaCl2 solution, 1 mL 1 M MgSO4 solution, 2 mL of 500 × thiamine solution (1.4 mM) and 0.6 mL 0.1 M FeCl3 solution. The resulting solution was filled up to 1 L with water and was filter sterilized. 40 x stock solutions of individual carbon sources were prepared in H2O and pH was set to 7 with 1 M NaOH or HCl. Final concentrations of individual carbon sources were: sodium acetate 3.5 g/L, disodium fumarate 2.8 g/L, galactose 2.3 g/L, glucosamine 2.1 g/L, glycerol 2.2 g/L, sodium pyruvate 3.3 g/L, disodium succinate hexahydrate 5.7 g/L, glucose, fructose, mannose and xylose 5 g/L. For the salt stress condition, NaCl was added to M9 to a concentration of 50 mM. For the pH stress condition, the pH was set to 6.0 by titrating the M9 medium with HCl. For the rich medium, amino acids were dissolved in M9 to the following concentrations: alanine 1.0 mg/L, adenine 10.2 mg/L, arginine 51.1 mg/L, asparagine 1.6 mg/L, aspartic acid 81.8 mg/L, cysteine 1.2 mg/L, glutamate 15.2 mg/L, glutamine 13.9 mg/L, glycine 0.4 mg/L, histidine 20.5 mg/L, isoleucine 51.1 mg/L, leucine 102.3 mg/L, lysine 51.1 mg/L, methionine 20.5 mg/L, phenylalanine 51.1 mg/L, proline 5.2 mg/L, serine 9.2 mg/L, threonine 102.3 mg/L, tryptophan 51.1 mg/L, tyrosine 51.1 mg/L, valine 143.2 mg/L and uracil 20.5 mg/L. Glycerol was added to the rich medium to a final concentration of 2.2 g/L. All chemicals were from Sigma-Aldrich.
Cultivation conditions
Glycerol-stocks of E. coli strains were kept at -80°C and streaked out on LB plates (Luria Miller, Carl Roth), and incubated overnight at 37°C. Single colonies were picked and inoculated into a M9 preculture (7.5 mL in 100-ml flasks), which was incubated for 6 – 10 h, to be subsequently diluted into a second preculture. The second preculture was used to inoculate the main culture at a starting OD of 0.05 in 35 mL medium in 500-mL non-baffled wide-neck shake flasks, covered by a 38-mm silicone sponge closure (BellCo glass). Cultivations were performed at 37°C, 200 rpm and 5-cm shaking diameter (Infors HT Minitron), except for the heat-stressed cells which were grown at 42°C. Growth was monitored by measuring the optical density at 600 nm (OD600). Specific growth rates were calculated by linear regression of ln(OD600) against the time during the exponential growth phase (Table S1).
Method details
Metabolite measurements
Samples for metabolomics were collected at an OD600 of ca. 0.5, and 2 ml of the culture was vacuum-filtered through a filter membrane (HVLP02500, Merck Millipore). Subsequently, the filter was immersed into 1 mL of acetonitrile:methanol:H2O (40:40:20) at -20°C. Extraction was performed overnight at -20°C. Cell extracts were then centrifuged for 20 min at –9°C, 13 000 rpm (HeraerusTM Pico 17TM ThermoScientific).
The supernatant of metabolite extracts was mixed with 13C internal standard in equal proportion and stored at -80°C until analysis by LC-MS/MS. To prepare 13C internal standard, E. coli MG1655 was grown in a 1-L Bioreactor with 500 mL of M9 minimal medium containing 4 g L−1 uniformly 13C-labeled glucose (Cambridge Isotope Laboratories) to an OD of 1. The bioreactor was a BioFlo115 bioprocess system (Eppendorf, Hamburg, Germany), equipped with a pH-sensor (Mettler Toledo, Colombus, OH) and a DO-sensor (Mettler Toledo, Colombus, OH). The bioreactor was aerated using air passed through 4 M potassium hydroxide in order to avoid incorporation of 12C by CO2. Aliquots of 20 mL were vacuum-filtered using 0.45 μm pore size filters (HVLP02500, Merck Millipore) and immediately transferred into 5 mL of 40:40:20 (v-%) acetonitrile/methanol/water kept at −20°C. Half of the culture was treated with a mixture of ampicillin, chloramphenicol, rifampicin, and trimethoprim for 15 min before sampling to enforces increases of metabolites. Final extracts from glucose-grown cells and inhibitor treated cells were mixed 1:1 and absence of 12C peaks in this 13C standard was confirmed by LC-MS. Aliquots of the 13C standard were stored at −80°C.
Samples were analyzed by LC-MS/MS, with an Agilent 6495 triple quadrupole mass spectrometer (Agilent Technologies) as described previously (Guder et al., 2017). An Agilent 1290 Infinity II UHPLC system (Agilent Technologies) was used for liquid chromatography using two columns: i) an Acquity UPLC BEH Amide (Waters) for acidic conditions and ii) a iHILIC-Fusion(P) (HILICON AB) for basic conditions. The column oven was at 30°C. LC solvents were: solvent A: water with ammonium formate (10 mM) and formic acid (0.1% v/v) for acidic conditions, and water with ammonium carbonate (10 mM) and ammonium hydroxide (0.2%) for basic conditions; solvents B: acetonitrile with formic acid (0.1% v/v) for acidic, and acetonitrile for basic conditions. The LC gradient was: 0 min 90% B, 1.3 min 40% B, 1.5 min 40% B, 1.7 min 90% B, 2 min 90% B. The flow rate was 0.4 mL/min. The injection volume was 3 μL. Settings of the ESI source were: 200°C source gas, 14 L/min drying gas and 24 psi nebulizer pressure. The sheath gas temperature was at 300°C and flow at 11 L/min. The electrospray nozzle was set to 500 V and capillary voltage to 2500 V.
Alpha-ketoglutarate was derivatized prior to LC-MS/MS analysis (Zimmermann et al., 2014), using 50 μM phenylhydrazine (Sigma-Aldrich). The mixture was incubated for 1 h at -20°C in acetonitrile:methanol:H2O (40:40:20), and analyzed with LC-MS/MS on the Acquity UPLC BEH Amide column.
Quantification and statistical analysis
Quantification and statistical analysis were done with MATLAB. The statistical details of each experiment can be found in the respective figure caption. For metabolomics data, n = 3 is the number of replicates, which are n = 3 samples from independent shake flask cultures. Quantification of intracellular metabolite concentrations was based on the ratio of 12C and 13C peak heights as described previously14. 12C/13C ratios of peak heights were normalized to the OD-specific cell volume at the time point of sampling. It was previously shown that the OD-specific cell volume is constant (Volkmer and Heinemann, 2011), and we used an OD-specific cell volume of 0.74 μL/mL/OD for all conditions. The mean values of the three replicates of the OD-normalized 12C/13C ratios were used to calculate the RSD. First, the standard deviation (SD) between the different conditions was calculated. Then the SD was normalized to the mean concentration of the metabolite in all conditions, to obtain the RSD across conditions.
Acknowledgments
This work has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 715650, ERC Starting Grant MapMe). H.L. and J.R. acknowledge funding from the Cluster of Excellence EXC 2124 from the Deutsche Forschungsgemeinschaft.
Author contributions
D.R. performed experiments, analyzed data, and co-wrote the manuscript. S.D. analyzed data and co-wrote the manuscript. M.L. analyzed data. J.R. analyzed data. H.L. analyzed data and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104503.
Supplemental information
See also Figure 1A.
Concentrations are given as fold-changes normalized to the glucose condition with E. coli BW25113. The data is given as mean and standard deviation of n = 3 replicates. See also Figure 1C.
Data and code availability
The metabolome data are available at MetaboLights under the accession code MTBLS4249. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alam M.T., Olin-Sandoval V., Stincone A., Keller M.A., Zelezniak A., Luisi B.F., Ralser M. The self-inhibitory nature of metabolic networks and its alleviation through compartmentalization. Nat. Commun. 2017;8:16018. doi: 10.1038/ncomms16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson D.E. Energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 1968;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B.D., Yuan J., Kimball E.H., Rabinowitz J.D. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergkessel M., Basta D.W., Newman D.K. The physiology of growth arrest: uniting molecular and environmental microbiology. Nat. Rev. Microbiol. 2016;14:549–562. doi: 10.1038/nrmicro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M.J., Yuan J., Bennett B.D., Lu W., Kimball E., Botstein D., Rabinowitz J.D. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. U S A. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büke F., Grilli J., Cosentino Lagomarsino M., Bokinsky G., Tans S.J. ppGpp is a bacterial cell size regulator. Curr. Biol. 2022;32:870–877.e5. doi: 10.1016/j.cub.2021.12.033. [DOI] [PubMed] [Google Scholar]
- Cho B.-K., Federowicz S., Park Y.-S., Zengler K., Palsson B.Ø. Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat. Chem. Biol. 2012;8:65–71. doi: 10.1038/nchembio.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho B.-K., Federowicz S.A., Embree M., Park Y.-S., Kim D., Palsson B.Ø. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2011;39:6456–6464. doi: 10.1093/nar/gkr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubukov V., Uhr M., Le Chat L., Kleijn R.J., Jules M., Link H., Aymerich S., Stelling J., Sauer U. Transcriptional regulation is insufficient to explain substrate-induced flux changes in Bacillus subtilis. Mol. Syst. Biol. 2013;9:709. doi: 10.1038/msb.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Fuente I.M., Cortés J.M., Valero E., Desroches M., Rodrigues S., Malaina I., Martínez L. On the dynamics of the adenylate energy system: homeorhesis vs homeostasis. PLoS One. 2014;9:e108676. doi: 10.1371/journal.pone.0108676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerosa L., Haverkorn van Rijsewijk B.R.B., Christodoulou D., Kochanowski K., Schmidt T.S.B., Noor E., Sauer U. Pseudo-transition analysis identifies the key regulators of dynamic metabolic adaptations from steady-state data. Cell Syst. 2015;1:270–282. doi: 10.1016/j.cels.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Gerosa L., Kochanowski K., Heinemann M., Sauer U. Dissecting specific and global transcriptional regulation of bacterial gene expression. Mol. Syst. Biol. 2013;9:658. doi: 10.1038/msb.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guder J.C., Schramm T., Sander T., Link H. Time-optimized isotope ratio LC–MS/MS for high-throughput quantification of primary metabolites. Anal. Chem. 2017;89:1624–1631. doi: 10.1021/acs.analchem.6b03731. [DOI] [PubMed] [Google Scholar]
- Heijnen J.J. In: Biosystems Engineering II: Linking Cellular Networks and Bioprocesses Advances in Biochemical Engineering/Biotechnology. Wittmann C., Krull R., editors. Springer; 2010. Impact of thermodynamic principles in systems biology; pp. 139–162. [DOI] [PubMed] [Google Scholar]
- Hofmeyr J.H.S., Cornish-Bowden A. Quantitative assessment of regulation in metabolic systems. Eur. J. Biochem. 1991;200:223–236. doi: 10.1111/j.1432-1033.1991.tb21071.x. [DOI] [PubMed] [Google Scholar]
- Hofmeyr J.-H.S., Cornish-Bowden A. Regulating the cellular economy of supply and demand. FEBS Lett. 2000;476:47–51. doi: 10.1016/s0014-5793(00)01668-9. [DOI] [PubMed] [Google Scholar]
- Jensen K.F. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanowski K., Gerosa L., Brunner S.F., Christodoulou D., Nikolaev Y.V., Sauer U. Few regulatory metabolites coordinate expression of central metabolic genes in Escherichia coli. Mol. Syst. Biol. 2017;13:903. doi: 10.15252/msb.20167402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.-W., Burkhardt D., Gross C., Weissman J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.O., Rubin S.A., Xu Y.-F., Amador-Noguez D., Fan J., Shlomi T., Rabinowitz J.D. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 2016;12:482–489. doi: 10.1038/nchembio.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I., Kochanowski K., Cappelletti V., Fuhrer T., Noor E., Sauer U., Picotti P. A map of protein-metabolite interactions reveals principles of chemical communication. Cell. 2018;172:358–372.e23. doi: 10.1016/j.cell.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Reaves M.L., Young B.D., Hosios A.M., Xu Y.-F., Rabinowitz J.D. Pyrimidine homeostasis is accomplished by directed overflow metabolism. Nature. 2013;500:237–241. doi: 10.1038/nature12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander T., Farke N., Diehl C., Kuntz M., Glatter T., Link H. Allosteric feedback inhibition enables robust amino acid biosynthesis in E. coli by enforcing enzyme overabundance. Cell Syst. 2019;8:66–75.e8. doi: 10.1016/j.cels.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer J.R.A., Fell D.A., Rothman D., Shulman R.G. Protein phosphorylation can regulate metabolite concentrations rather than control flux: the example of glycogen synthase. Proc. Natl. Acad. Sci. U S A. 2004;101:1485–1490. doi: 10.1073/pnas.0307299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Kochanowski K., Vedelaar S., Ahrné E., Volkmer B., Callipo L., Knoops K., Bauer M., Aebersold R., Heinemann M. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M., Gunderson C.W., Mateescu E.M., Zhang Z., Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- Soupene E., van Heeswijk W.C., Plumbridge J., Stewart V., Bertenthal D., Lee H., Prasad G., Paliy O., Charernnoppakul P., Kustu S. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 2003;185:5611–5626. doi: 10.1128/jb.185.18.5611-5626.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S., Fell D.A. Design of metabolic control for large flux changes. J. Theor. Biol. 1996;182:285–298. doi: 10.1006/jtbi.1996.0166. [DOI] [PubMed] [Google Scholar]
- Volkmer B., Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS One. 2011;6:e23126. doi: 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zimmermann M., Sauer U., Zamboni N. Quantification and mass isotopomer profiling of α-keto acids in central carbon metabolism. Anal. Chem. 2014;86:3232–3237. doi: 10.1021/ac500472c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See also Figure 1A.
Concentrations are given as fold-changes normalized to the glucose condition with E. coli BW25113. The data is given as mean and standard deviation of n = 3 replicates. See also Figure 1C.
Data Availability Statement
The metabolome data are available at MetaboLights under the accession code MTBLS4249. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.