Summary
Intestinal organoids are physiologically relevant tools used for cellular models. However, the suitability of organoids to examine biological functions over existing established cell lines lacks sufficient evidence. Cytochrome P450 3A4 (CYP3A4) induction by pregnane X receptor ligands, glucose uptake via sodium/glucose cotransporter 1, and microsomal triglyceride transfer protein-dependent ApoB-48 secretion, which are critical for human intestinal metabolism, were observed in organoid-derived two-dimensional cells but little in Caco-2 cells. CYP3A4 induction evaluation involved a simplified method of establishing organoids that constitutively expressed a reporter gene. Compound screening identified several anticancer drugs with selective activities toward Caco-2 cells, highlighting their characteristics as cancer cells. Another compound screening revealed a decline in N-(4-hydroxyphenyl)retinamide cytotoxicity upon rifampicin treatment in organoid-derived cells, under CYP3A4-induced conditions. This study shows that organoid-derived intestinal epithelial cells (IECs) possess similar physiological properties as intestinal epithelium and can serve as tools for enhancing the prediction of biological activity in humans.
Subject areas: Human metabolism, Tissue engineering, Cell biology, Stem cells research
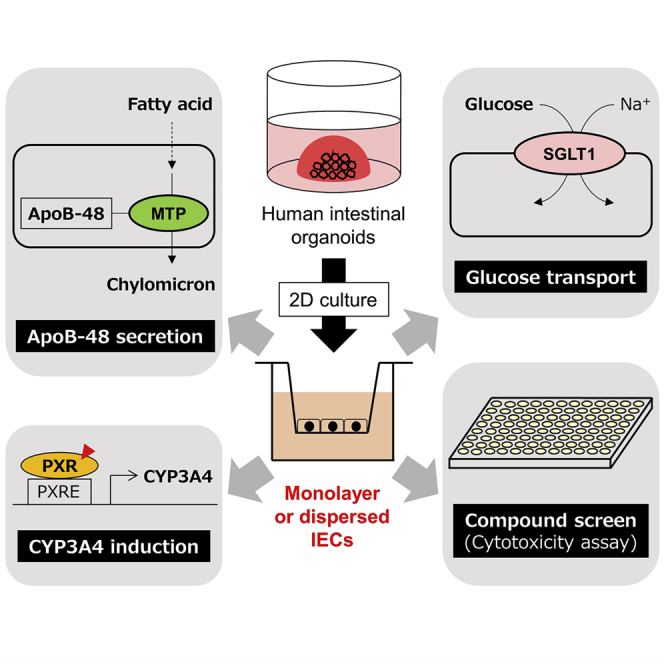
Graphical abstract
Highlights
-
•
Comparison of mRNA expression between organoid-derived intestinal epithelial cells (IECs) and Caco-2 cells
-
•
Evaluation of CYP3A4, SGLT1, and MTP protein function in organoid-derived IECs
-
•
Identification of anti-cancer drugs as selective cytotoxicity against Caco-2 cells
-
•
Reduction of N-(4-hydroxyphenyl)retinamide (4-HPR) cytotoxicity by rifampicin in organoid-derived IECs
Human metabolism; Tissue engineering; Cell biology; Stem cells research
Introduction
The small intestine is a vital organ that is responsible for many biological functions, including acting as a physical barrier against various pathogens, digestion, and absorption of dietary nutrients, and chylomicron synthesis for lipid transport. The molecular mechanism of the small intestinal functions has been thoroughly investigated both in vivo and in vitro using animal models, such as mice, and established cell lines, such as Caco-2 cells. For instance, knockout mice experiments have demonstrated the critical roles of numerous intestinal genes. However, their functions in humans are often considered speculative. Several differences in species in terms of gene expression and metabolism have been illustrated until now. Caco-2 cells, which have been established from human colon adenocarcinoma, have been widely used for decades that mimic human intestinal epithelium in vitro (Rousset, 1986). Because they can spontaneously differentiate into absorptive epithelial-like cells with a brush border layer on being cultured for more than 2 weeks after confluence, they are widely used as a model that can mimic intestinal absorption and permeability, especially in pharmacology (Meunier et al., 1995).
Nevertheless, Caco-2 cells are not reliable in terms of reciprocating the physiological function of intestinal epithelial cells (IECs) because several differences exist between Caco-2 cells and in vivo tissue. For instance, glucose and galactose are mainly absorbed in enterocytes via sodium/glucose cotransporter 1 (SGLT1) encoded by SLC5A1; however, this gene is expressed at low levels in Caco-2 cells (Kwon et al., 2021; Sun et al., 2002; Turner et al., 1996), although the gene expression is likely to be influenced by the nutrient environment, and low-glucose-consuming Caco-2 cells are reported to express SGLT1 (Mahraoui et al., 1994). Furthermore, following the absorption and resynthesis of emulsified dietary free fatty acids and 2-monoacylglycerol into triglycerides in enterocytes, they are assembled with apolipoprotein (Apo) B-48 by microsomal triglyceride transfer protein (MTP), encoded by MTTP, and secreted into the bloodstream by the lymphatic system. However, this pathway is seemingly inactive in Caco-2 cells; therefore, the chylomicron secretion capacity is restricted (Levy et al., 1995). Moreover, cytochrome P450 (CYP) 3A4 is involved in drug metabolism and is the most abundant CYP enzyme in the small intestine; however, its induction by chemicals is not observed most likely because one of the nuclear receptors called pregnane X receptor (PXR), which is encoded by NR1I2 and regulates cytochrome P450 3A4 (CYP3A4) expression, is either scarcely expressed in Caco-2 cells or is not expressed at all (Küblbeck et al., 2016; Prueksaritanont et al., 1996). Therefore, alternative biological tools are essential to understand the detailed and precise mechanism of IEC function in human biology.
Drug-induced intestinal toxicity can cause diarrhea, nausea, and bleeding, resulting in the discontinuation of the production of the drug or the impairment of quality of life (Rodrigues et al., 2019). Therefore, an accurate prediction and evaluation of the intestinal toxicity of bioactive molecules, particularly in preclinical stages, is essentially important. Direct injury to mucosal integrity, which is frequently accompanied by IEC death, is one of the major causes of intestinal toxicity. For instance, irinotecan (or its metabolite SN-38) and 5-fluorouracil can cause mucosal damage and diarrhea (Gupta et al., 1994). Nevertheless, no suitable in vitro model is available at present to predict intestinal toxicity in humans in a better manner.
Recently, organoids have been established as in vitro organ models that are composed of tissue-specific stem cells and differentiated cells in three-dimensional (3D) culture. They are self-organizing cell aggregations that are highly physiological owing to their potential to give rise to functional organs being transplanted into the animal model (Dutta et al., 2017; Rossi et al., 2018). Intestinal organoids are a promising alternative intestinal epithelial model that can replace Caco-2 cells. Wnt3a, R-spondin1, and Noggin are known as key factors essential for maintaining the stemness of intestinal stem cells that reside in the organoids (Sato et al., 2011). Application of intestinal organoids in nutrient and food science research, such as nutrient transport, metabolism, and function, is expanding (Cai et al., 2018; Huang et al., 2020; Zietek et al., 2020); however, evidence show that intestinal organoids possess better metabolic functions that are similar to in vivo IECs is insufficient. Our previous study demonstrated ways to reduce the cost and labor for organoid culture (Takahashi et al., 2018). Monolayer IECs were also developed simply by disrupting organoids and seeding them onto collagen I-coated plates (Takahashi et al., 2017). IECs generated by this method possess fundamental characteristics that are similar to in vivo IECs, although their biological functions have not been thoroughly investigated. Therefore, this study examined the physiology of IECs derived from human intestinal organoids by assessing the expression and function of genes that play an essential role in the small intestine but are expressed to a limited extent in Caco-2 cells. We assumed that monolayer or dispersed IECs are superior to organoids compared with Caco-2 cells in terms of their physiological characteristics because organoids are 3D structures and complexities of different cultural environments which may affect both the expression and function of genes. We showed that SLC5A1, MTTP, and NR1I2 expression was remarkably higher in organoid-derived monolayer IECs than in Caco-2 cells. Moreover, the products of these three genes were functional. Interestingly, we were able to establish organoids that stably express luciferase reporter gene under human CYP3A4 promoter and determine CYP induction in response to PXR activation in organoid-derived IECs. Furthermore, compound screening was conducted, and several compounds that induce selective cytotoxicity against IECs or Caco-2 cells were identified. Compounds with selective cytotoxicity against Caco-2 cells include histone deacetylase (HDAC) inhibitors that cause apoptosis in cancer cells, indicating that Caco-2 cells originated from colon cancer cells. Moreover, cytotoxicity of N-(4-hydroxyphenyl)retinamide (4-HPR) in IECs was found to decrease by cotreatment with rifampicin, a CYP3A4 inducer, suggesting that 4-HPR was enzymatically metabolized into inactive forms. Based on these results, it can be inferred that organoid-derived IECs exhibit better physiology that is closer to in vivo IECs compared with Caco-2 cells. Therefore, utilizing the IECs can enhance our understanding of human intestinal biology through molecular and biological approaches and thus prevent and mitigate intestinal diseases using detailed mechanisms of IEC function based on advanced physiological systems.
Results
Unlike Caco-2 cells, organoid-derived intestinal epithelial cells appropriately express intestinal genes
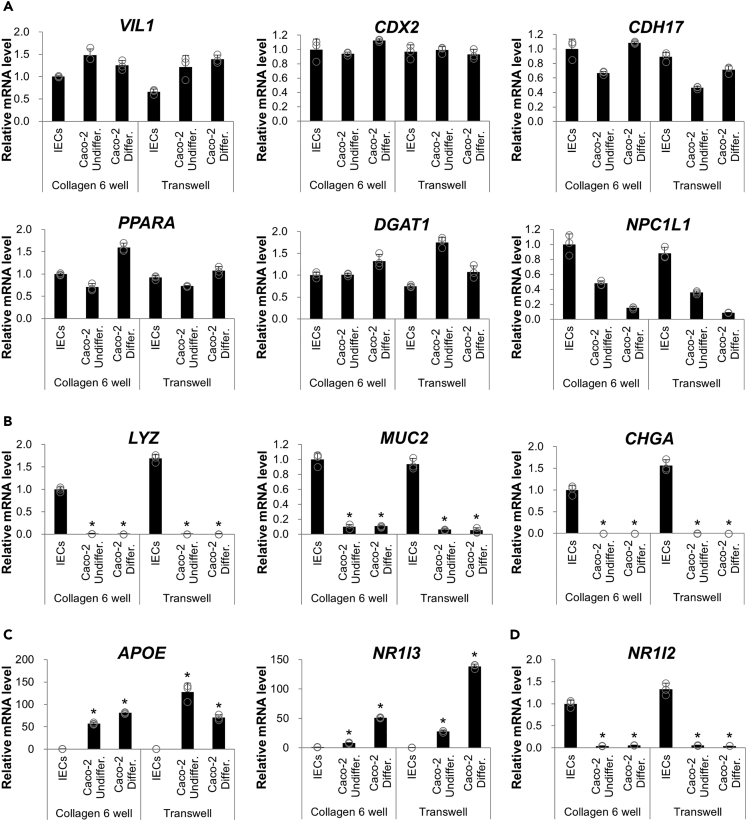
We previously efficiently differentiated human iPS cells into intestinal organoids with ileum-like properties (Takahashi et al., 2018). Through extensive gene expression analysis, it was observed that metabolically essential genes substantially expressed in the ileum, including APOA1, APOA4, and NPC1L1, were confirmed to be expressed in both human iPS cell (TkDN4-M)-derived organoids (iPSOs) and primary ileum organoids (PIOs; Figure S1). This indicates their suitability for use as a small intestinal model. Therefore, intestinal gene expression of IECs from iPSOs and undifferentiated and differentiated Caco-2 cells was compared. To avoid the effect of the culture environment between two-dimensional (2D) and 3D conditions, IECs derived from intestinal organoids in our previous studies (Sato et al., 2019; Takahashi et al., 2017) were adopted rather than the organoids themselves. Recently, we demonstrated that culture conditions from 3D to 2D have no considerable impact on the fundamental properties of IECs in terms of intestinal gene expression and ligand responsiveness to ligand-activated transcription factors (Takahashi et al., 2021). Moreover, culturing on a transparent membrane confers cellular polarity and epithelial electrical resistance to both cells, which may affect cellular characteristics. Cells grown on collagen-coated six-well plates and Transwells, generally used for standard cultures, were harvested. It was confirmed that both organoid-derived IECs and Caco-2 cells express PPARA, DGAT1, and NPC1L1, the critical genes involved in fatty acid and cholesterol homeostasis in the small intestine and the intestinal epithelial marker genes, VIL1, CDX2, and CDH17 (Figure 1A). In contrast, Caco-2 cells exhibited considerably low levels of LYZ, a Paneth cell gene, and MUC2, a goblet cell marker gene. This observation was consistent with previous findings in that organoid-derived IECs contained Paneth and goblet cells, as observed in the parental organoids (Takahashi et al., 2017) (Figure 1B). In contrast, the expression of APOE, a gene encoding Apo E, and NR1I3, a gene encoding the constitutive androstane receptor belonging to the xenobiotic-activated receptor family, was much higher in Caco-2 cells than in IECs (Figure 1C). Most importantly, both genes are reportedly expressed at low levels in the small intestine but at aberrantly high levels in Caco-2 cells (Bruck et al., 2017; Reisher et al., 1993). Unlike NR1I3, NR1I2, which encodes another xenobiotic-activated receptor, PXR, is highly expressed in the small intestine and scarcely expressed in Caco-2 cells (Bruck et al., 2017); it is also highly expressed in organoid-derived IECs (Figure 1D). Of note, a significant difference in mRNA levels between cell culture vessels (six-well plates and Transwells) was not observed. Furthermore, we performed an RNA sequence to compare the overall characteristics among primary human IECs, organoid-derived IECs, and differentiated Caco-2 cells. Clustering analysis revealed more similarity between primary human IECs and organoid-derived IECs than between primary human IECs and differentiated Caco-2 cells (Figure S2A). From the analysis, we were able to identify many genes highly or exclusively expressed in primary human IECs and organoid-derived IECs but not in Caco-2 cells (Figure S2B), including NR1I2. Gene expression analysis suggested that organoid-derived monolayer IECs exhibit physiological characteristics that are similar to in vivo IECs, which are not necessarily reciprocated in Caco-2 cells.
Figure 1.
Comparison of gene expression between monolayer IECs from hiPSOs and Caco-2 cells
(A–D) hiPSO-derived dispersed IECs or Caco-2 cells were seeded on collagen I-coated six-well plates or Transwells and cultured as monolayers as described in “STAR Methods.” After cells were harvested, the mRNA levels of each gene were determined by qRT-PCR and normalized to 18S rRNA levels. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was assessed by one-way analysis of variance with the Bonferroni test. ∗p < 0.05 (versus each group of IECs). (A) Intestinal genes are expressed in both monolayer IECs and Caco-2 cells. (B) Intestinal epithelium marker genes are exclusively expressed in monolayer IECs. (C) Genes are unphysiologically highly expressed in Caco-2 cells. (D) Identification of NR1I2 as a highly expressed gene in monolayer IECs.
See also Figure S2.
CYP3A4 induction by PXR can be evaluated in organoid-derived intestinal epithelial cells
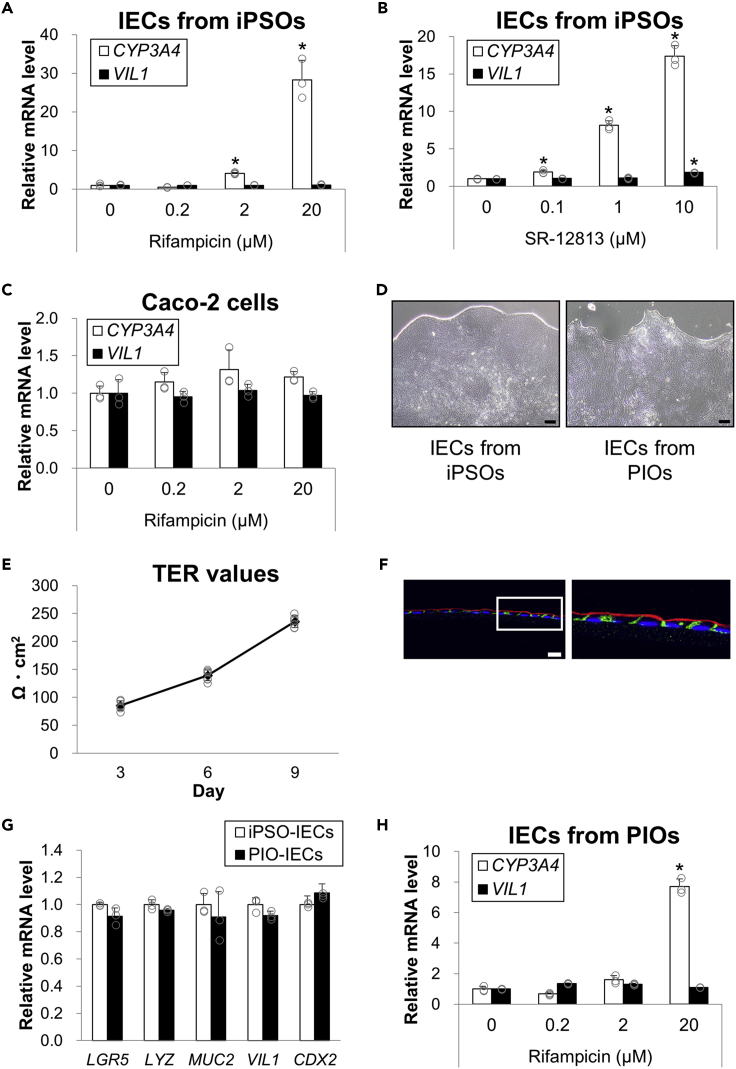
Because organoid-derived IECs express NR1I2, they would enable the evaluation of PXR ligand activities by monitoring CYP3A4 mRNA levels (Fahmi et al., 2010). Cells grown on collagen-coated six-well plates were treated with rifampicin, a well-known ligand for human PXR but not rodent PXR, and the expression of CYP3A4 was induced in a dose-dependent manner up to more than 20-fold (Figure 2A). The induction was similarly observed when collagen-coated Transwells were used (Figure S3), indicating that the culture equipment does not affect this phenomenon. Furthermore, SR-12813, a synthetic PXR agonist (Watkins et al., 2001), induced CYP3A4 mRNA levels in a dose-dependent manner (Figure 2B). Consistent with a previous study (Yamaura et al., 2016), rifampicin failed to cause CYP3A4 induction in Caco-2 cells (Figure 2C). To avoid the possibility that CYP3A4 induction is only observed in IECs derived from iPSOs, monolayer IECs were also established from PIOs. IECs from PIOs were observed to be morphologically similar to iPSO-derived IECs by phase-contrast microscopy (Figure 2D), and transepithelial electrical resistance (TEER) values increased in a culture time-dependent manner (Figure 2E) as well as IECs from iPSOs (Takahashi et al., 2017), which indicates tight junction formation. Their characteristics as IECs were further confirmed by immunofluorescent staining of frozen sections seeded on Transwells with anti-Villin1 and anti-E-cadherin antibodies, thereby demonstrating expression of the apical brush border protein, Villin1 on the apical side and the adherens junction marker protein, E-cadherin between cells (Figure 2F). Expression levels of several key marker genes between iPSO- and PIO-derived IECs under the basal culture condition were indistinguishable (Figure 2G), which also supports the similarity of IECs developed from iPSOs and PIOs. It was found that rifampicin induced CYP3A4 mRNA levels in IECs derived from PIOs (Figure 2H). These data indicate that endogenous PXR in monolayer IECs, generated from intestinal organoids, is functional, and IECs enable the evaluation of CYP3A4 induction mediated by PXR activation.
Figure 2.
CYP3A4 induction by PXR ligands in monolayer organoid-derived IECs
(A and B) hiPSO-derived monolayer IECs in collagen I-coated 12-well plates were treated with 0, 0.2, 2, and 20 μM rifampicin (A) or 0, 0.1, 1, 10 μM SR-12813 (B) for 48 h. Cells were harvested, and CYP3A4 and VIL1 mRNA levels were determined by qRT-PCR and normalized to 18S rRNA levels. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by one-way analysis of variance with the Bonferroni test. ∗p < 0.05 (versus no treatment group).
(C) Differentiated Caco-2 cells cultured in collagen I-coated 12-well plates were treated with 0, 0.2, 2, and 20 μM rifampicin for 48 h. After cells were harvested, the mRNA levels of each gene were determined by qRT-PCR and normalized to 18S rRNA levels. Assays were performed in n = 3 independent biological replicates (mean ± S.D.).
(D) Representative bright-field images of hiPSO- or PIO-derived IECs observed under a phase-constant microscope. Scale bar, 200 μm.
(E) TER values of PIO-derived IECs cultured in collagen-coated Transwells were measured on the indicated days. Assays were performed in n = 4 independent biological replicates (mean ± S.D.).
(F) Immunohistochemistry analysis of frozen sections of PIO-derived monolayer IECs stained with 4′,6-Diamidino-2-phenylindole (DAPI) (blue), anti-E-cadherin (green), and anti-Villin1 (red) antibodies. Scale bar, 20 μm. Right, magnified image of the boxed area.
(G) hiPSO- or PIO-derived monolayer IECs cultured in collagen I-coated 6-well plates were harvested, and the mRNA levels of each gene were determined by qRT-PCR and normalized to 18S rRNA levels. Assays were performed in n = 3 biological replicates (mean ± S.D.).
(H) PIO-derived monolayer IECs or cultured in collagen I-coated 12-well plates were treated with 0, 0.2, 2, and 20 μM rifampicin for 48 h. After cells were harvested, the mRNA levels of each gene were determined by qRT-PCR and normalized to 18S rRNA levels. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by one-way analysis of variance with the Bonferroni test. ∗p < 0.05 (versus no treatment group).
See also Figure S3.
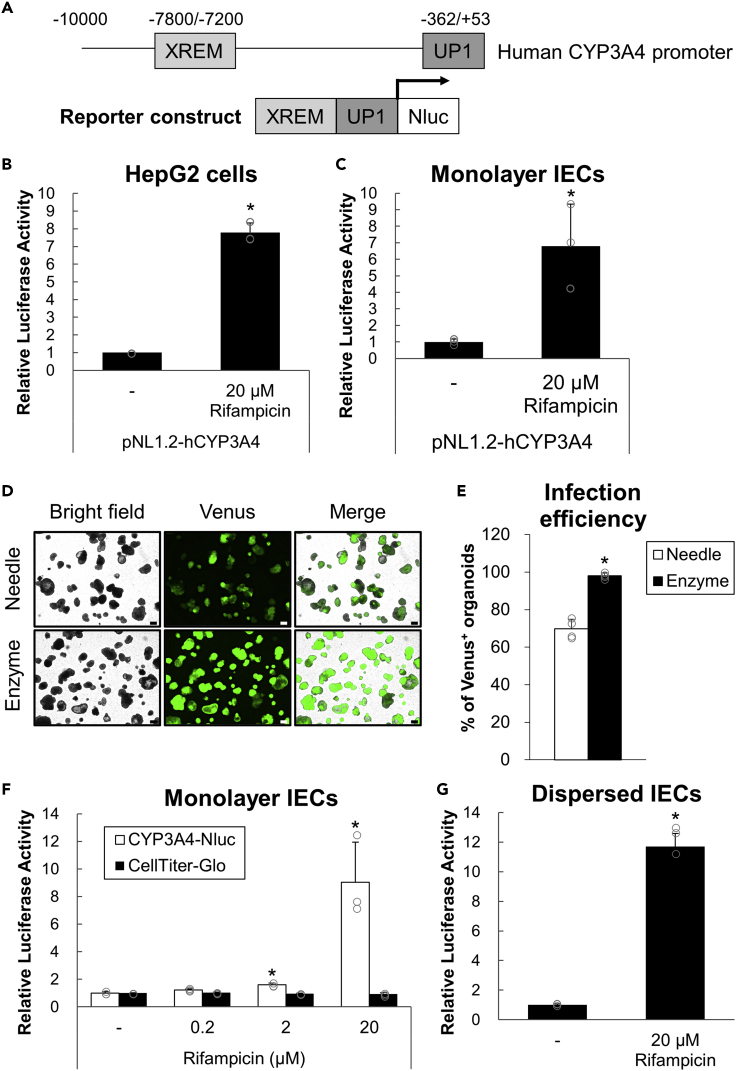
Although CYP induction can be routinely estimated by determining mRNA expression or protein activities, this process is expensive and laborious, and its throughput is usually low. Therefore, a luciferase reporter gene assay system was implemented to further explore the method that can readily evaluate CYP3A4 induction. A reporter plasmid construct containing a human CYP3A4 promoter that combines proximal (UP1) and distal (XREM) regions with each identified functional PXR-responsive element (Figure 3A) was created based on a previous study (Coumoul et al., 2002). The reporter plasmid was transiently transfected into HepG2 cells, a hepatoma-oriented cell line expressing endogenous PXR (Al-Dosari and Parvez, 2018), and luciferase activities were induced upon rifampicin treatment (Figure 3B).
Figure 3.
Evaluation of CYP3A4 by reporter gene assay in organoid-derived IECs
(A) Part of a reporter gene construct (pNL1.2-hCYP3A4) with human PXR promoter combined with two regions (XREM and UP1) containing functional PXR response elements.
(B and C) HepG2 cells (B) or hiPSO-derived monolayer IECs (C) were transiently transfected with pNL1.2-hCYP3A4 with pSV40-β-Gal. After 24 h, cells were treated with 0 or 20 μM rifampicin for 24 h. Luciferase activities were measured and normalized to β-galactosidase activities. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by Student’s t test. ∗p < 0.05. (D, E) After being disrupted by physical breaking with a 26-gauge needle or dispersed by enzymatic digestion with TrypLE Express solution followed by vigorous pipetting, hiPSO-derived intestinal organoids were seeded on collagen I-coated plates. Cells were cultured for four days (physical breaking, “Needle”) or one day (enzymatic digestion, “Enzyme”), infected with undiluted CSII-EF-MCS-IRES2-Venus lentiviruses, and embedded in Matrigel to regenerate organoids.
(D) bright-field or fluorescent images of organoids after 9 days of infection were taken. Scale bar, 200 μm.
(E) The proportions of Venus-positive organoids per microscopic bright field after 9 days of infection were calculated. Assays were performed in n = 4 independent images (mean ± S.D.). Statistical significance was determined by Student’s t test. ∗p < 0.05 (versus Needle).
(F and G) Monolayer (F) or dispersed (G) IECs developed from hiPSOs stably expressing human CYP3A4-Nluc were treated with 0-20 μM rifampicin for 24 h, and luciferase activities were determined. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by one-way analysis of variance with the Bonferroni test (F) or by Student’s t test (G). ∗p < 0.05 (versus no treatment group).
See also Figure S4.
Gene transduction into organoids with high efficiency is often considered challenging because they form 3D cell clumps. A previous study developed a method to efficiently express exogenous genes by lipofection or lentiviral infection via transient 2D culture of IECs (Takahashi et al., 2018). Therefore, transient gene transduction was performed by lipofection in IECs derived from iPSOs, and rifampicin treatment was found to induce luciferase activities in cells (Figure 3C). To further develop the assay that enables a more convenient evaluation of CYP3A4 induction, organoids that stably express a luciferase reporter gene under human CYP3A4 promoter with PXR-responsive elements were established by lentiviral infection. Because the organoid disruption procedure was switched from physical breaking (previous study) to enzymatic digestion (this study) before culturing to obtain IECs, the timing of infection with lentiviruses for Venus expression was optimized. As a result, the enzymatic digestion method was able to achieve almost 100% infection efficiency with both undiluted and 4-fold diluted viruses, whereas the physical breaking method achieved 70% and 40% infection efficiency with undiluted and 4-fold diluted viruses, respectively (Figures 3D, 3E, and S4). The result indicates that a higher infection efficiency was successfully achieved compared with the previous method, prompting us to consider that luciferase activity induction can be determined without cloning organoids after infection. As expected, rifampicin treatment induces luciferase activities in organoid-derived IECs that stably express the luciferase gene under the CYP3A4 promoter (Figure 3F). These results indicate that intestinal CYP3A4 induction by PXR activation could be determined by luciferase reporter gene assay in monolayer IECs by expressing the reporter gene both transiently and constitutively. Unlike organoids with 3D complexities and variations, monolayer IECs are homogeneous and would be suitable for screening studies. However, it takes a long time to obtain monolayer IECs from organoids, resulting in low throughput. Therefore, dispersed single IECs obtained by trypsin-treated organoids were used instead, followed by vigorous pipetting. The cells also responded to rifampicin, and the induction levels of luciferase activity were nearly the same when monolayer IECs were used (Figure 3G). Thus, CYP3A4 induction can be determined by luciferase assay in IECs of both monolayer and dispersed states, which would help perform high-throughput or large-scale studies.
SLC5A1 and MTTP are expressed and functional in organoid-derived intestinal epithelial cells
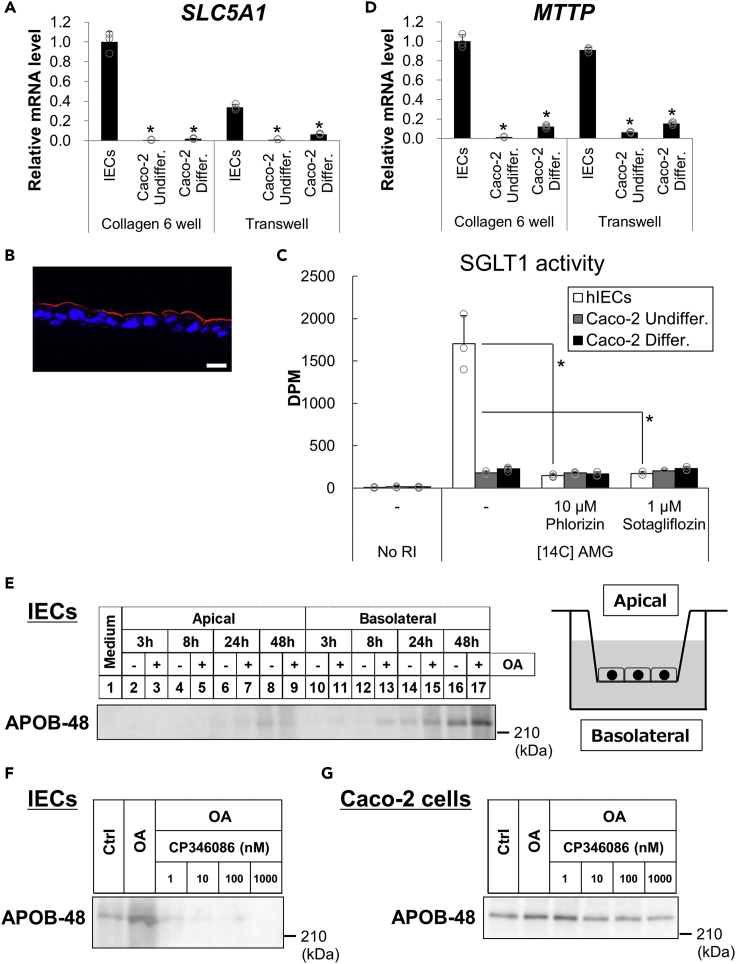
We next focused on another gene whose expression is reported to be low in Caco-2 cells, namely SLC5A1 (Sun et al., 2002; Turner et al., 1996; Kwon et al., 2021), which encodes SGLT1. SLC5A1 was found to be expressed in significantly higher levels in organoid-derived monolayer IECs compared with Caco-2 cells (Figure 4A). Immunofluorescent staining revealed that SGLT1 was expressed in the apical side of the IECs (Figure 4B). Rifampicin treatment failed to induce SLC5A1 (Figure S5), which indicates that PXR is not involved in the transcriptional regulation of SLC5A1. SGLT1 is exclusively expressed in the small intestine and is critically involved in sodium-dependent glucose and galactose absorption. To assess whether SGLT1 in IECs is functional as a transporter, [14C]-labeled α-methyl-D-glucopyranoside (AMG) uptake was quantified to evaluate SGLT1 transporter activity (Takasu et al., 2019). At first, we generated Caco-2 cells that overexpress Flag-tagged SGLT1 by transient transfection and found that [14C]-AMG uptake was enhanced by exogenous SGLT1 (Figure S6), which shows that the evaluation method works properly. We also found that [14C]-AMG uptake was much more prominent in IECs than in Caco-2 cells regardless of the differentiation state (Figure 4C). Moreover, phlorizin and sotagliflozin, which are SGLT1/2 dual inhibitors, effectively inhibited the uptake in IECs but not in Caco-2 cells. Given the low SGLT2 expression in the small intestine, these findings indicate that SGLT1 was responsible for the uptake in IECs.
Figure 4.
Determination of expression and activity of SGLT1 and MTP in monolayer IECs
(A and D) Monolayer IECs from hiPSOs and undifferentiated and differentiated Caco-2 cells were cultured in either collagen I-coated six-well plates or Transwells, as in Figure 1. mRNA levels of each gene were determined by qRT-PCR and normalized to 18S rRNA levels. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by one-way analysis of variance with the Bonferroni test. ∗p < 0.05 (versus each group of IECs).
(B) Immunohistochemistry analysis of frozen sections of hiPSO-derived IECs stained with DAPI (blue) and an anti-SGLT1 (red) antibody. Scale bar, 20 μm.
(C) Monolayer IECs from hiPSOs and undifferentiated and differentiated Caco-2 cells were treated with 10 μM phlorizin or 1 μM sotagliflozin for 2 h before treatment with 100 μM AMG and 1 μM [14C]-AMG for 1 h. A scintillation counter determined radioactivity. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by one-way analysis of variance with the Bonferroni test. ∗p < 0.05 (versus no inhibitor treatment group). No asterisk indicates no significance (p > 0.05).
(E) Monolayer IECs from hiPSOs were treated with or without 500 μM OA for 3, 8, 24, and 48 h in a basal medium with 20% FBS. Supernatants were collected from the apical or basolateral side. The same volume of the supernatant was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blot analysis was performed using an anti-ApoB antibody. Right, an illustration of Transwell insert.
(F and G) Monolayer IECs (F) or differentiated Caco-2 cells (G) were treated with or without 500 μM OA for 24 h in a basal medium with 20% FBS. Cells were after that treated with 0, 1, 10, 100, and 1000 nM CP346086 in the presence of 500 μM OA After 24 or 48 h, supernatants from the basolateral side were collected. The same volume of the supernatant was subjected to SDS-PAGE, and Western blot analysis was performed using an anti-ApoB antibody.
See also Figures S5 and S6.
Furthermore, this study noted the impaired function of Caco-2 cells as limited lipid export capacity, especially in terms of chylomicron secretion (Levy et al., 1995). Gene expression involved in chylomicron synthesis and secretion was explored. MTTP expression, encoding MTP involved in lipids and ApoB-48 assembly, which is an important event in chylomicron synthesis, was much higher in IECs than in Caco-2 cells even in the differentiated stage (Figure 4D). Consistent with SLC5A1, MTTP expression in IECs was not increased with rifampicin treatment (Figure S5), suggesting no involvement of PXR in its transcriptional regulation. Because chylomicron is generated from the resynthesized triacylglycerol in the endoplasmic reticulum and secreted into the basolateral side of lymph, Transwells that provide polarity to cells were used to examine MTP-mediated lipoprotein secretion. Apical and basolateral compartments of Transwell correspond to intestinal lumen and bloodstream sides, respectively. A time-dependent analysis revealed that ApoB-48, a marker of chylomicron particles, was secreted preferentially into the basolateral side of monolayer IECs (Figure 4E). Based on the results obtained, whether MTP mediates ApoB-48 secretion was verified. Both IECs generated from human iPSOs (hiPSOs) and differentiated Caco-2 cells were incubated with oleic acid (OA) with or without MTP inhibitor (CP346086). The secreted amounts of ApoB-48 were considerably suppressed in a dose-dependent manner by CP346086 treatment in the basolateral side of IECs (Figure 4F) but not in Caco-2 cells (Figure 4G), which implies that MTP has an essential role in chylomicron secretion in organoid-derived IECs. From these findings, organoid-derived IECs exhibit more physiological features than Caco-2 cells from the perspective of intestinal nutrient metabolism.
High-throughput screening identified Caco-2-selective compounds with anticancer drugs
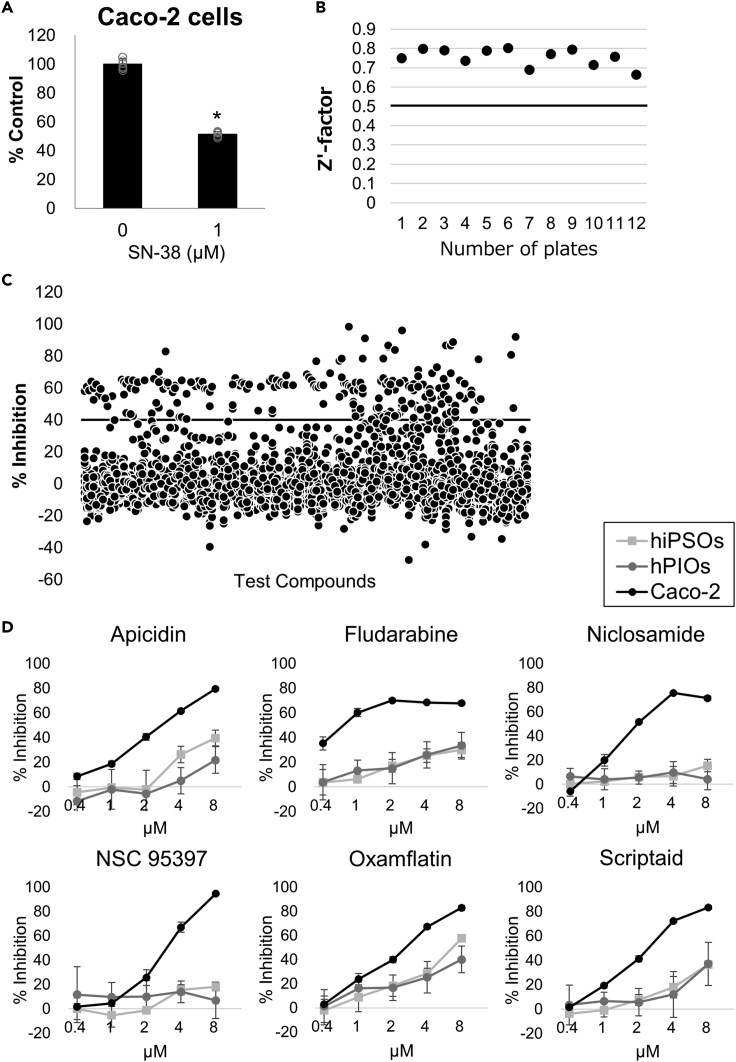
Because Caco-2 cells are derived from colon cancer cells (Ghadimi et al., 2000), their response to drugs, especially sensitivity to cytotoxicity, might differ from that of normal IECs. Therefore, to validate the hypothesis, compound screening was performed using approximately 3500 chemical libraries composed of drugs and pharmacologically active compounds to compare sensitivities between Caco-2 cells and organoid-derived IECs. Dispersed cells of each cell type were used rather than monolayer cells to improve assay throughput and reduce measurement variation. In addition, as a positive control compound that causes cytotoxicity against IECs, SN-38, which is a topoisomerase I inhibitor and an active metabolite of irinotecan, was chosen because it is known to cause intestinal toxicity and diarrhea as side effects (Gupta et al., 1994). The cytotoxic effect of SN-38 was confirmed, and 1 μM SN-38 was employed to secure the assay quality of each plate (Figure 5A).
Figure 5.
Identification of compounds by compound screening with selective cytotoxicity against Caco-2 cells
(A) Cytotoxic activity of 1 μM SN-38 against Caco-2 cells. Assays were performed in n = 6 independent biological replicates (mean ± S.D.). Statistical significance was determined by Student’s t test. ∗p < 0.05.
(B) Z′-factor values of the overall primary screening using Caco-2 cells.
(C) Scatterplots of primary screening data at a single concentration of 2 μM in Caco-2 cells and selection of hit compounds exhibiting >40% cytotoxic inhibition activity determined by CellTiter-Glo reagents.
(D) Dose-dependent analysis of compounds with prominently selective cytotoxicity against Caco-2 cells using dispersed IECs from hiPSOs, PIOs, and Caco-2 cells. Assays were performed in n = 4 independent biological replicates (mean ± S.D.).
The first screening of chemical library compounds using Caco-2 cells was performed at a single concentration of 2 μM of each compound in 0.2% dimethyl sulfoxide. The average Z′-factor of the screening exceeded 0.6, indicating the robust assay performance, which was adequate to filter compounds (Figure 5B). As initial hits, 150 compounds with an inhibitory activity of >40% were chosen (Figure 5C). Subsequently, counter screening of hiPSO-derived IECs was performed with 45 selected compounds having an inhibitory activity of <40%. Dose-dependent analysis of these compounds was subsequently performed using Caco-2 cells and IECs derived from hiPSOs and PIOs. As a result, compounds with dose-dependent cytotoxicity against Caco-2 cells and superior selectivity against IECs were successfully identified. The compounds include apicidin, fludarabine, niclosamide, NSC 95397, oxamflatin, and scriptaid (Figure 5D), all of which are known as anticancer drugs with antiproliferative activities (Monneret, 2005; Frank et al., 1999; Ren et al., 2010; Vogt et al., 2008).
Cytotoxic activity of 4-HPR is attenuated by rifampicin treatment in organoid-derived intestinal epithelial cells
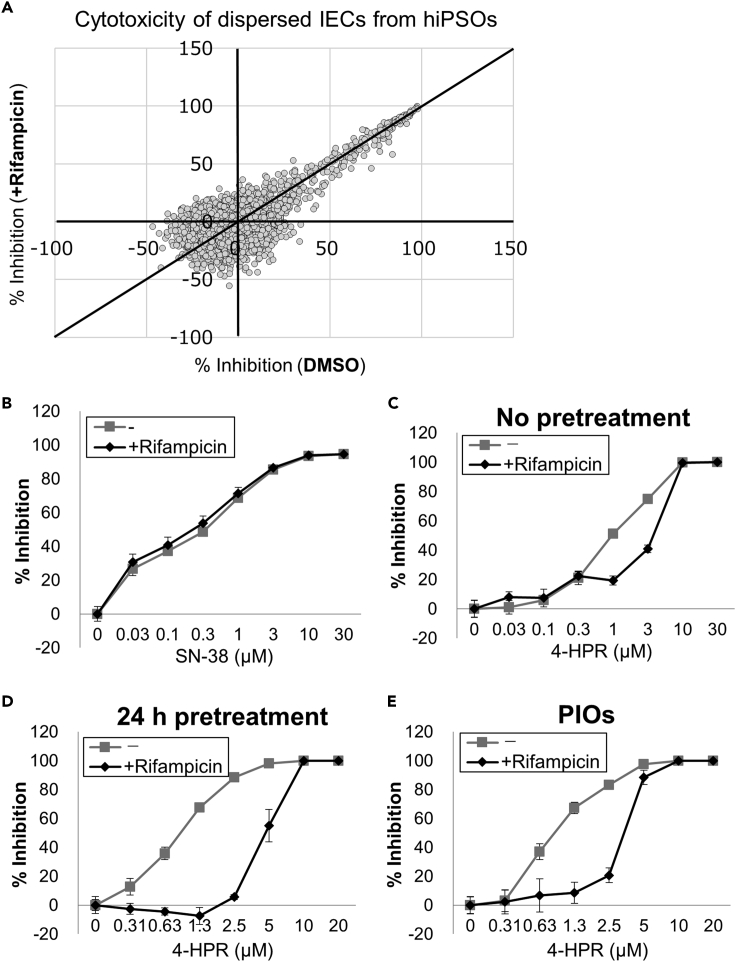
This study demonstrated that rifampicin treatment induces CYP3A4 expression in organoid-derived IECs (Figure 2), prompting speculation that the metabolism of certain drugs can be enhanced with rifampicin, thereby altering their overall pharmacological activities in cells. To elucidate this hypothesis, another compound screening was conducted using the same chemical library as in Figure 5, and the cytotoxicity of compounds against organoid-derived IECs was compared in the presence or absence of 20 μM rifampicin (Figure 6A). As a result, most compounds had little impact on the cytotoxic activity by simultaneous addition of rifampicin, including a positive control compound, SN-38 (Figure 6B). Interestingly, we discovered that 4-HPR exhibited cytotoxicity against IECs, and its activity was decreased by rifampicin treatment (Figure 6C). Notably, this decrease in its cytotoxicity was amplified by a 24-h pretreatment of rifampicin before the addition of 4-HPR (Figure 6D), with 50% inhibitory concentration (IC50) values of 0.87 μM (−rifampicin) and 4.8 μM (+rifampicin) (Table S1). These findings strongly support that rifampicin induces the conversion of 4-HPR into its inactive forms. In addition, the decrease in 4-HPR cytotoxicity was confirmed by cotreatment with rifampicin in IECs derived from PIOs (Figure 6E). From these results, we concluded that organoid-derived IECs have a great potential to be applied to the prediction of drug metabolism and interaction in the small intestine.
Figure 6.
Decreased cytotoxic activity of 4-HPR by rifampicin treatment in organoid-derived IECs
(A) Scatterplots of primary screening data of cytotoxicity determined by CellTiter-Glo reagents at a single concentration of 2 μM with or without 20 μM rifampicin in dispersed hiPSO-derived IECs.
(B and C) Dose-dependent cytotoxic activity of SN-38 (B) or 4-HPR (C) with or without 20 μM rifampicin in dispersed hiPSO-derived IECs. Assays were performed in n = 6 independent biological replicates (mean ± S.D.).
(D and E) Dose-dependent cytotoxic activity of 4-HPR in dispersed hiPSO-derived (D) or PIO-derived (E) IECs after treatment with or without 20 μM rifampicin for 24 h. Assays were performed in n = 6 independent biological replicates (mean ± S.D.).
See also Table S1.
Discussion
Although established from colon adenocarcinoma, Caco-2 cells have been widely used as modeled human small intestinal epithelium to investigate its function for decades. However, they have also been reported to exhibit properties that are similar to colon IECs and cancer cells owing to their origins. In addition to chromosome aneuploidy and mutations (Ghadimi et al., 2000), Caco-2 cells possess a limited capacity to induce CYP3A4 (Küblbeck et al., 2016) and export lipids (Levy et al., 1995). Alternatively, we previously established monolayer IECs from intestinal organoids that exhibited better physiological characteristics than Caco-2 cells, including Paneth and goblet cells and valid TEER values, which were unphysiologically high in Caco-2 cells (Takahashi et al., 2017). We hypothesized that the organoid-derived IECs would overcome the drawbacks of Caco-2 cells in terms of physiological intestinal function and metabolism. Using monolayer or dispersed IECs rather than organoids is a reasonable strategy to compare their cellular properties to Caco-2 cells because both cells are cultured in 2D conditions using the same culture equipment. This study verified IEC phenotypes from the perspective of intestinal biology. We focused on essential genes associated with human intestinal metabolism, including NR1I2, SLC5A1, and MTTP, which are expressed at low levels in Caco-2 cells, and found that these genes were functional in IECs; therefore, we believe that they possess physiologically relevant properties of small intestines that are similar to the in vivo tissue. Notably, PXR ligand specificity can differ among species (Xie et al., 2000); for instance, rifampicin and pregnenolone 16α-carbonitrile selectively activate human and rodent PXR, respectively. Therefore, data extrapolation from mice to humans should be avoided, and a system to precisely evaluate PXR activity in the human intestine is required. This study showed evidence that ligand activity of human PXR can be assessed in organoid-derived IECs. Besides mRNA levels, CYP3A4 induction by PXR activation can be determined by luciferase reporter gene assay in both monolayer or dispersed IECs. To the best of our knowledge, this is the first example of a biochemical assay that uses organoids that stably express a reporter gene, which may further accelerate the use of organoids in molecular and cellular biology, in terms of their extensive application such as for the purpose of screening. It should be emphasized that dispersed cells can be easily prepared from organoids, and monolayer IECs can be maintained for >1 week (Takahashi et al., 2017), whose flexible handling and cell homogeneities would contribute to expanding their applications. Moreover, unlike the dispersed cells, the monolayer IECs have the polarity of luminal and basolateral sides, and little chance to exhibit anoikis.
Nevertheless, why SLC5A1, MTTP, and NR1I2 expression is high in organoid-derived IECs and low in Caco-2 cells remains elusive. Although it is reported that MTP expression can be regulated by PXR (Meng et al., 2019), SLC5A1 and MTTP expression in IECs was not induced upon rifampicin treatment. Therefore, there is higher chance that important transcriptional regulators other than NR1I2 are expressed at low levels in Caco-2 cells but at high levels in organoids and organoid-derived IECs, which play critical roles in gene regulation. The expression of SLC5A1 and MTTP is reported to be regulated by HNF1 and HNF4 (Koepsell, 2020; Hussain et al., 2011). However, these transcription factors would not be candidates responsible for the gene expression regulation in the IECs, as Caco-2 cells highly express these transcription factors (Hu and Perlmutter, 1999). There is a possibility that such transcriptional regulators might be identified through the analysis of RNA sequence in Figure S2. In any case, based on this study, the molecular mechanism regarding the law of various gene expressions would be gradually clarified in the future for a better insight into the physiological function of the human intestinal epithelium.
Several compounds from the chemical library that selectively induce cellular toxicity against Caco-2 cells were identified. These compounds include drugs that cause cytotoxicity, specifically against cancer cells and not normal cells, such as HDAC inhibitors, including apicidin, oxamflatin, and scriptaid, which display the properties of Caco-2 cells as cancer-oriented cells. Therefore, Caco-2 cells would not be appropriate for screening compounds with potential intestinal toxicity in normal tissue. In contrast, whether organoid-derived IECs can be a potential biological tool to predict the intestinal toxicity accompanied by IEC death requires both basic and clinical investigation in the future. Another screening study showed that 4-HPR, which exerts cytotoxicity against IECs, was effectively inhibited by rifampicin treatment. 4-HPR was reported to be a substrate of CYP3A4 (Cooper et al., 2011); however, its inactive form metabolized by CYP3A4 has not been identified yet. Despite this problem, although it is reported that 4-HPR exhibits cytotoxicity against breast cancer cells (Simeone et al., 2002), whether intestinal toxicity in vivo can be caused by 4-HPR is worth further examination. It should be noted that these types of experiments using organoid-derived IECs would enable us to assess the change in pharmacological activities within the small intestine, mediated by endogenously induced CYP3A4. Therefore, the IECs have more potential to be used as a noble and essential tool to thoroughly investigate human physiological intestinal metabolism. Any future studies conducted to deduce a comparison between Caco-2 cells and organoid-derived IECs would help better understand the physiological and unphysiological aspects of Caco-2 cells, in addition to the elucidation of function and metabolism-associated signals conserved in vivo in intestinal tissue.
Recently, Kwon et al. established and physiologically characterized the monolayer human intestinal epithelial models derived from iPS cells or organoids (Kwon et al., 2021). The authors demonstrated the expression and activities of intestinal transporters and drug-metabolizing enzymes, such as CYP3A4, in the IECs, suggesting their usefulness for the better prediction of drug absorption and bioavailability in humans. Despite the differences in the establishment, culture methods, and medium ingredients, we believe that both IECs similarly exhibit fundamental properties that resemble the intestinal epithelium in vivo. In this study, we focused not only on drug metabolism (CYP3A4) but also on nutrient metabolism and drug responsiveness; therefore, together with the evidence provided by Kwon et al., these characterizations would further strengthen the usefulness of organoid-derived IECs in various research areas.
Withdrawal of Wnt signal activators such as Wnt3a has been reported to enhance differentiation toward enterocyte lineage. In the previous studies, intestinal organoids cultured with a differentiation medium without Wnt3a and SB202190 were used to examine enterocyte function or responsiveness, including ion transport (Foulke-Abel et al., 2016), norovirus replication (Ettayebi et al., 2016), and nutrient transport and metabolism (Zietek et al., 2020). In this study, we also used the differentiation medium to examine CYP3A4 induction and SGLT1 activity in the monolayer IECs. Given that switching from the human organoid culture medium to differentiation medium would yield more terminally differentiated IECs, the use of IECs cultured with a differentiation medium may enable us to evaluate the physiological function of mature enterocytes in a variety of assays in a more precise way.
In summary, IECs generated from human intestinal organoids exhibit pivotal physiological phenotypes in terms of intestinal metabolism, preserved in in vivo tissue but not in Caco-2 cells. Their use would aid in a better understanding of human intestinal physiology, including nutrient absorption and metabolism. Furthermore, responsiveness to bioactive compounds can be different in IECs and Caco-2 cells. Several compounds that selectively showed cytotoxicity against Caco-2 cells are anticancer drugs; therefore, Caco-2 cells would not be suitable for predicting intestinal toxicity in normal intestinal tissue. Instead, the use of IECs might open new avenues of understanding human intestinal biology, which has not been achieved yet using in vitro cell lines and in vivo animals. In addition to this, as the dog can represent closer gastrointestinal physiology to humans than mice (Ambrosini et al., 2020; Chandra et al., 2019; Mochel et al., 2017), the IECs have the potential to be applied not only to the substitution of preclinical animal studies but also to the development of veterinary medicine in the future.
Limitations of the study
Organoid-derived IECs exhibit characteristics that are more physiological than Caco-2 cells; however, to what extent and how IECs are similar to in vivo human IECs would need further investigation. Examination with organoids from more different donors would contribute to expanding the application of methods presented in this study. Besides mRNA expression, protein expression, localization, or function other than PXR, SGLT1, and MTP should be comprehensively analyzed, including intestinal marker genes. Studies on actual metabolism in IECs would be desired, especially in the research field of food/nutrient science and pharmacology. Organoid-derived IECs lack immune cells and bacterial flora, unlike in vivo IECs, which enable simplified analysis but may affect cellular characteristics. Their applications in basic and clinical sciences would help in understanding their usefulness in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-Villin1 | Abcam | Cat#ab3304; RRID: AB_303688 |
| Rabbit Anti-E-cadherin | Cell Signaling Technology | Cat#3195; RRID: AB_2291471 |
| Rabbit Anti-SGLT1 | Merck | Cat#07-1417 |
| Goat Anti-Apolipoprotein B-100 | Abcam | Cat#ab7616; RRID: AB_305987 |
| Cy3-conjugated donkey anti-mouse IgG | Jackson ImmunoResearch | Cat#711-166-150 |
| Cy3-conjugated donkey anti-rabbit IgG | Jackson ImmunoResearch | Cat#711-166-152 |
| Alexa Fluor 488-conjugated donkey anti-rabbit IgG | Jackson ImmunoResearch | Cat#711-546-152 |
| HRP-conjugated donkey anti-goat IgG | Jackson ImmunoResearch | Cat#705-035-147 |
| Bacterial and virus strains | ||
| One Shot™ Stbl3™ Chemically Competent E. coli | Thermo Fisher Scientific | C737303 |
| Mix & Go! Competent Cells-TG1 | Zymo Research | T3017 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM, high glucose | FUJIFILM Wako | 044-29765 |
| E-MEM with L-Glutamine and Phenol Red | FUJIFILM Wako | 051-07615 |
| Advanced DMEM/F12 | Thermo Fisher Scientific | 12634-028 |
| MEM Non-Essential Amino Acids Solution (100X) | Thermo Fisher Scientific | 11140050 |
| Fetal bovine serum | Thermo Fisher Scientific | 10270-106 |
| GlutaMAX Supplement (100X) | Thermo Fisher Scientific | 35050061 |
| HEPES (1 M) | Thermo Fisher Scientific | 15630080 |
| 30% Bovine serum albumin | FUJIFILM Wako | 015-23871 |
| Matrigel | Corning | 354234 |
| Y-27632 | FUJIFILM Wako | 257-00511 |
| SB202190 | Sigma-Aldrich | S7067 |
| CultureSure A83-01 | FUJIFILM Wako | 039-24111 |
| Gentamicin Sulfate Solution (50 mg/mL) | Nacalai Tesque | 11980-14 |
| Hexadimethrine bromide (Polybrene) | Sigma-Aldrich | 107689 |
| Rifampicin | FUJIFILM Wako | 189-01001 |
| Oleic acid | Nacalai Tesque | 25630-51 |
| CP-346086 dihydrate | Sigma-Aldrich | PZ0103 |
| α-methyl-D-glucopyranoside (AMG) | Tokyo Chemical Industry | M0228 |
| [14C] AMG | PerkinElmer | NEC659 |
| N-(4-hydroxyphenyl)retinamide (4-HPR) | Tokyo Chemical Industry | H1464 |
| Wnt3a, R-spondin1, Noggin CM | Takahashi et al., 2018 | N/A |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | L300015 |
| Lipofectamine Stem Transfection Reagent | Thermo Fisher Scientific | STEM00015 |
| TrypLE Express | Thermo Fisher Scientific | 12604013 |
| Cell Recovery Solution | BD Biosciences | 354253 |
| Recombinant mouse EGF | PeproTech | 315-09 |
| Recombinant human HGF | R&D systems | 294-HG-005 |
| Tissue-Tek Optimal Cutting Temperature compound | Sakura Finetechnical | 4583 |
| Fluoromount | Diagnostic BioSystems | K024 |
| Prestained XL-Ladder Broad | Intégrale | SP-2120 |
| 30% (w/v)-Acrylamide-bis (29:1) | Nacalai Tesque | 06141-35 |
| DAPI Fluorescence Stain, 1000X | Cell Biolabs | 112002 |
| Immobilon-P | Merck | IPVH00010 |
| Blocking One | Nacalai Tesque | 03953-95 |
| Critical commercial assays | ||
| SmGM-2 Bullet Kit | Lonza | CC-3182 |
| ISOGEN | NIPPON GENE | 319-90211 |
| RNeasy Mini Kit | QIAGEN | 74104 |
| RNase free DNase set | QIAGEN | 79254 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | 4368814 |
| FASTStart Universal SYBR Green Master (ROX) | Roche | 4913914001 |
| TaqMan Gene Expression Master Mix | Thermo Fisher Scientific | 4369016 |
| QIAGEN Plasmid Midi Kit | QIAGEN | 12145 |
| BD Cytofix/Cytoperm Fixation/Permeabilization Solution Kit | BD Biosciences | 554714 |
| ECL Western Blotting Detection Reagents | Cytiva | RPN2106 |
| Immobilon Western Chemiluminescent HRP Substrate | Merck | WBKLS0500 |
| Nano-Glo Luciferase Assay System | Promega | N1130 |
| HS RNA Kit | Agilent | DNF-472-0500 |
| MGIEasy RNA Directional Library Prep Set | MGI Tech | 1000006386 |
| Dynabeads mRNA Purification Kit | Thermo Fisher Scientific | 61006 |
| dsDNA HS Assay Kit | Thermo Fisher Scientific | Q32851 |
| DNBSEQ-G400RS High-throughput Sequencing Kit | MGI Tech | 1000016941 |
| CellTiter-Glo 3D Cell Viability Assay | Promega | G9682 |
| Deposited data | ||
| RNA-seq (primary-hIECs, Organoid-IECs, Differ-Caco-2) | This paper | DRA accession number: DRA013757 |
| Experimental models: Cell lines | ||
| Mouse: L cells | ATCC | CRL-2648 |
| Human: Caco-2 cells | ATCC | HTB-37 |
| InEpC – Human Intestinal Epithelial Cells | Lonza | CC-2931 |
| TkDN4-M | Stem Cell Bank (The University of Tokyo) | N/A |
| Human iPS cell-derived intestinal organoids | Takahashi et al. (2018) | N/A |
| Oligonucleotides | ||
| See Table S2 for information on qPCR probes and primers. | ||
| Recombinant DNA | ||
| p3xFLAG-CMV-10 | Sigma-Aldrich | E7658 |
| p3xFLAG-CMV-10-hSGLT1 | This paper | N/A |
| pNL1.2 | Promega | N1011 |
| pNL1.2-hCYP3A4 | This paper | N/A |
| pSV40-β-gal | Promega | E1081 |
| pCAG-HIVgp | RIKEN BioResource Research Center | N/A |
| pCMV-VSV-G-RSV-Rev | RIKEN BioResource Research Center | N/A |
| CS-UbC-hSox2-IRES2-Venus | RIKEN BioResource Research Center | N/A |
| CS-hCYP3A4-Nluc | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Cutadapt (v1.9.1) | Martin (2011) | https://github.com/marcelm/cutadapt |
| Sickle (v1.33) | Joshi and Fass, 2011 | https://github.com/najoshi/sickle |
| Hisat2 (v2.2.1) | Kim et al. (2019) | http://daehwankimlab.github.io/hisat2/ |
| Samtools (v1.11) | Li et al. (2009) | http://www.htslib.org/ |
| featureCounts (v2.0.0) | Liao et al. (2014) | https://www.rdocumentation.org/packages/Rsubread/versions/1.22.2/topics/featureCounts |
| R | R Core Team 2020 | https://www.r-project.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Ryuichiro Sato (roysato@g.ecc.u-tokyo.ac.jp).
Materials availability
Plasmids generated in this study are available from lead contact upon reasonable request. However, lentiviral plasmids and TkDN4-M are only available to academia with a material transfer agreement with the RIKEN BioResource Research Center (Ibaraki, Japan) and the University of Tokyo (Tokyo), respectively.
Experimental model and subject details
Cell culture
Caco-2 cells were cultured in Eagle’s minimum essential medium (MEM) with 10% fetal bovine serum (FBS), 1× MEM non-essential amino acid solution, 100 units/mL penicillin, and 100 μg/mL streptomycin. For differentiation, Caco-2 cells were monolayer cultured in type I collagen-coated 6-well plates, 12-well plates, or Transwells (all from Corning) for more than two weeks after they reached confluency. When cultured in Transwell, induction of stable TEER values was confirmed using Millicell ERS-2 (Merck Millipore) before use in assays. The cells were cultured in a 5% CO2 incubator at 37°C.
L cells stably expressing human R-spondin1 and human Noggin with (L-WRN) or without (L-RN) mouse Wnt3a were established by lentiviral infection as described previously (Takahashi et al., 2018). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Each conditioned medium was prepared from supernatants seeded at 1.4 × 106 cells/35 mm dish for 72 h. The cells were cultured in a 5% CO2 incubator at 37°C.
Primary human fetal small intestinal epithelial cells were purchased from Lonza (CC-2931). The cells were cultured in a 5% CO2 incubator at 33°C using SmGM-2 Bullet Kit (CC-3182) following the manufacture’s instructions.
Differentiation of human-induced pluripotent stem (iPS) cells (TkDN4-M) into intestinal organoids were conducted as described previously (Takahashi et al., 2018). According to the previous procedure, human intestinal organoid culture and passage were conducted with small modifications (Sato et al., 2019). Organoids embedded in Matrigel were washed with phosphate-buffered saline (−) and treated with TrypLE Express solution (Thermo Fisher Scientific) for 5 min at 37°C in a water bath. Recovered organoids were disrupted by vigorous pipetting 30 times and collected by centrifugation at 440 × g for 3 min. After removing supernatants, organoids were washed with 10 mL basal medium [Advanced DMEM/F-12 supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7.3), 2 mM GlutaMAX I, 100 units/mL penicillin, and 100 μg/mL streptomycin]. After centrifugation at 440 × g for 3 min, organoids were resuspended in Matrigel with 20% human organoid culture medium [Advanced DMEM/F-12 with 25% WRN CM, 10 mM HEPES (pH 7.3), 1% bovine serum albumin (BSA), 2 mM GlutaMAX I, 50 ng/mL mouse epidermal growth factor (mEGF), 50 ng/mL human hepatocyte growth factor, 10 μM Y-27632, 10 μM SB202190, 500 nM A83-01, 100 μg/mL gentamicin, 100 units/mL penicillin, and 100 μg/mL streptomycin] on ice. Suspension aliquots were added to wells of Nunc Multidish four-well plates (Thermo Fisher Scientific), leaving the border of each well untouched, and solidified in a 5% CO2 incubator at 37°C for 15 min. Human organoid culture medium (500 μL) was added to each well. The entire medium was changed every 3 days. Organoid passage was carried out every 6 or 7 days. The passaging ratio was from 1:8 to 1:16. The cells were cultured in a 5% CO2 incubator at 37°C. The experiments using primary human organoids complied with the Declaration of Helsinki and were approved by the human ethical committee of The University of Tokyo and Osaka University. All tissues were sampled with informed consent.
Monolayer culture of organoid-derived cells
After harvesting from Matrigel, digestion, and disruption by pipetting 30 times in TrypLE Express solution, organoids were collected by centrifugation at 440 × g for 3 min. Alternatively, after treated with cell recovery solution (BD Biosciences) for 30 min on ice, organoids were harvested by centrifugation at 440 × g for 3 min and disrupted in 1 mL of basal medium by passing through a 26-gauge needle 10 times, followed by centrifugation at 440 × g for 3 min. After washing with basal medium and harvest by centrifugation, cells were resuspended with human organoid culture medium, filtered through a 40 μm nylon mesh (Corning), and seeded on type I collagen-coated 6-well plates, 12-well plates, or Transwells. The medium was replaced every 2–3 days. Cell confluence was determined by microscope observation (6- or 12-well plates) or induction of TEER values (Transwell). To determine CYP3A4 induction upon ligand treatment and SGLT1 activity, the medium was replaced with differentiation medium [Advanced DMEM/F-12 with 12.5% RN CM, 10 mM HEPES (pH 7.3), 1% BSA, 2 mM GlutaMAX I, 50 ng/mL mEGF, 10 μM Y-27632, 500 nM A83-01, 100 μg/mL gentamicin, 100 units/mL penicillin, and 100 μg/mL streptomycin] and cultured in a 5% CO2 incubator at 37°C. The cells were used for assays when they reached confluency.
Method details
Plasmid construction
An expression plasmid for human SGLT1 was constructed by inserting a PCR fragment encoding human SGLT1 into p3xFLAG-CMV-10 (Sigma-Aldrich). A fragment of human CYP3A4 promoter with two PXR-responsive elements (Coumoul et al., 2002) was amplified by PCR and inserted into pNL1.2 plasmid (Promega) to generate pNL1.2-hCYP3A4. Lentiviral expression plasmid of CS-hCYP3A4-Nluc was constructed by replacing UbC-hSox2-IRES2-Venus of CS-UbC-hSox2-IRES2-Venus with the PCR fragment of NanoLuc gene under human CYP3A4 promoter. All lentiviral plasmids were amplified in Stbl3 E.coli (ThermoFisher Scientific), and other plasmids were in TG1 E.coli (Zymo Research).
Lentiviral production and infection
HEK293T cells seeded at 4 × 106 per dish (100 mm dish) were transfected with a lentiviral expression plasmid (10 μg/dish) with a packaging (pCAG-HIVgp; 4 μg/dish) and VSV-G- and Rev-expressing plasmid (pCMV-VSV-G-RSV-Rev; 4 μg/dish) using Lipofectamine 3000 Reagent (Thermo Fisher Scientific; 40 μL/dish) and P3000 Reagent (Thermo Fisher Scientific; 35 μL/dish) according to the supplier’s protocols. After 6 h, the culture medium was replaced by a fresh medium with 10 μM forskolin. After 48 h later, the medium containing lentiviruses was collected and filtered. Organoid-derived monolayer IECs seeded in six-well plates were infected with the medium containing 10 μg/mL polybrene using a centrifugation method (2500 rpm for 90 min). After centrifugation, cells were washed twice and refed with a fresh culture medium. Cells were harvested with TrypLE Express solution and embedded in Matrigel to regenerate organoids.
Reporter gene assay
HepG2 cells seeded at 1 × 105 per well (12-well plates) were transiently transfected with a reporter gene plasmid, pNL1.2-hCYP3A4 (0.25 μg/well), together with pSV40-β-Galactosidase (0.75 μg/well) using Lipofectamine 3000 Reagent (1.5 μL/well) and P3000 Reagent (2 μL/well). Alternatively, organoid-derived dispersed IECs on collagen I-coated 12-well plates were transiently transfected with pNL1.2-hCYP3A4 (0.25 μg/well), with pSV40-β-Gal (0.75 μg/well), using Lipofectamine Stem Transfection Reagent (Thermo Fisher Scientific; 2 μL/well). After 24 h transfection, cells were treated with 20 μM rifampicin and incubated for another 24 h. Luciferase activities were determined by the Nano-Glo Luciferase Assay System (Promega) according to the manufacturer’s protocol and normalized to β-galactosidase activities determined by absorbance at 405 nm, which was a result of conversion from o-nitrophenyl-β-D-galactopyranoside to o-nitrophenyl.
Quantitative reverse transcription–polymerase chain reaction (qRT–PCR)
Total cellular RNA was extracted by Isogen (Nippon Gene) or RNeasy Mini Kit (Qiagen). Reverse transcription was performed using High-Capacity cDNA Reverse Transcription Kits (Thermo Fisher Scientific). mRNA levels were measured by fluorescence real-time PCR on a StepOnePlus (Thermo Fisher Scientific) using PrimeTime qPCR Assays (Integrated DNA Technologies). 18S rRNA levels were used as an internal control to normalize the mRNA levels of each gene.
RNA sequencing
RNA sequencing was performed on our behalf by Bioengineering Lab (Kanagawa, Japan). Total RNA was extracted by RNeasy Mini Kit (Qiagen) with an additional step of on-column DNase I digestion. The RNA sample quality was checked with a 5200 Fragment Analyzer System (Agilent Technologies, Santa Clara, CA, USA) and Agilent HS RNA Kit (Agilent Technologies). Libraries for the sequencing were constructed using a MGIEasy RNA Directional Library Prep Set (MGI Tech, China), together with Dynabeads mRNA Purification Kit (Thermo Fisher Scientific) for removing the ribosomal RNA, according to the manufacturer’s instructions. After the measurement of concentrations with a Qubit 3.0 Fluorometer (Thermo Fisher Scientific) and dsDNA HS Assay Kit (Thermo Fisher Scientific), the prepared library was circulated using a MGIEasy Circularization Kit (MGI Tech). DNA Nanoballs (DNBs) were subsequently prepared with a DNBSEQ-G400RS High-throughput Sequencing Kit (MGI Tech). The sequence of these DNBs was analyzed on the DNBSEQ-G400 with 100-bp pair-end reads. The raw sequencing data were deposited in the DDBJ Sequence Read Archive (Kodama et al., 2012) (DRA accession number: DRA013757).
Processing and analysis of RNA sequencing data
Adaptor sequences and low-quality reads were removed using the Cutadapt (version 1.9.1) and Sickle software (version 1.33) (Quality score < 20, read length < 40), respectively. Hisat2 (version 2.2.1) was used to map the processed reads against the human genome (GRCh38.p13). SAM files were converted to BAM files, and then sorted and indexed using Samtools (version 1.11). The number of reads aligned to the genome was counted using featureCounts (version 2.0.0), followed by normalization of raw counts to the total gene length and the number of reads by transcripts per million (TPM). Clustering analysis was conducted by determination of Spearman's rank correlation coefficient-based distance from TPM values using the R function hclust. A heat map of genes was prepared with TPM values using Gene Cluster 3.0.
Western blot analysis
After independent collection of culture supernatant from both apical (100 μL) and basolateral (600 μL) sides of Transwells, the apical medium was diluted 6-fold with basal medium. After the addition of 6 × Laemmli sample buffer [1 mM Tris‒HCl (pH 6.8), 30% glycerol, 10% sodium dodecyl sulfate, 600 mM dithiothreitol, and 0.03% bromophenol blue] followed by boiling for 5 min, equal volumes of the samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride membrane. The membrane was blocked with Blocking One (Nacalai Tesque, Japan) for 1 h at room temperature (R.T.), incubated overnight at 4°C with a primary antibody against ApoB (1:200), and incubated with a secondary antibody for 1 h at R.T. Chemiluminescent signals were determined by Fusion Solo S (Vilber).
Immunofluorescence staining
Immunofluorescence staining of frozen sections of IEC was performed according to a previous protocol (Takahashi et al., 2017). Briefly, the Transwell membrane was fixed with 10% formalin, washed three times in 70% ethanol, and cut out by a surgical blade. The membrane was subsequently embedded in Tissue-Tek Optimal Cutting Temperature compound (Sakura Finetechnical) and frozen with liquid nitrogen. After blocking with 5% normal donkey serum, cryostat sections were incubated with mouse anti-Villin1 (1:50) and rabbit anti-E-cadherin (1:200) antibodies, followed by Cy3-conjugated donkey anti-mouse IgG (1:100), Alexa Fluor 488-conjugated donkey anti-rabbit IgG (1:100), and 4′,6-diamidino-2-phenylindole (DAPI; 1:1000). Alternatively, after blocking with Fixation and Permeabilization Solution (BD Biosciences), cryostat sections were incubated with rabbit anti-SGLT1 (1:25), followed by Cy3-conjugated donkey anti-rabbit IgG (1:100) and DAPI (1:1000). Fluorescence signals were visualized using a Leica TCS SP2 confocal laser scanning microscope.
Measurement of SGLT activity
Caco-2 cells seeded at 1 × 105 per well (12-well plates) were transiently transfected with human SGLT1 expression plasmid (1.3 μg/well) using Lipofectamine 3000 Reagent (3 μL/well) and P3000 Reagent (2.5 μL/well). SGLT activity assay was conducted after 72 h of transfection. Organoid-derived IECs were cultured with differentiation medium for two days before determining SGLT1 activity. Cells cultured in collagen I-coated 12-well plates were replaced with choline assay buffer [140 mM choline chloride, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 5 mM Tris-HCl (pH 7.4)] and incubated at 37°C for 20 min. The buffer was subsequently replaced with sodium assay buffer [140 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 5 mM Tris-HCl (pH 7.4)] with or without 10 μM phlorizin or 1 μM sotagliflozin. After incubation at 37°C for 2 h, cells were treated with 100 μM nonlabeled α-methyl-D-glucopyranoside (AMG) with or without 1 μM [14C]-AMG at 37°C for another 1 h. Cells were washed twice with cold choline assay buffer with 10 mM AMG and solubilized with 150 μL lysis buffer [2 mM dithiothreitol, 2 mM trans-1,2-diaminocyclohexane-N, N, N′, N′-tetraacetic acid, 10% glycerol, 1% Triton X-100, and 25 mM Tris-phosphate (pH 7.8)]. Cell lysates (20 μL) were mixed with 3 mL Ultima Gold (Perkin-Elmer). Radioactivity was determined by Tri-Carb 4810TR (Perkin-Elmer).
Determination of cell viability
After harvesting from Matrigel and disruption by pipetting 30 times in TrypLE Express solution, organoids were collected by centrifugation at 440 × g for 3 min. After washing with basal medium and harvest by centrifugation, cells were resuspended with human organoid culture medium and filtered through a 40 μm nylon mesh, resulting in the dispersed state at single-cell levels. Cells were seeded at 5.0 × 103 per well (96-well plate) or 1.2 × 103 per well (384-well plate) with or without compounds and incubated for 48 h in a 5% CO2 incubator at 37°C. Next, CellTiter-Glo® 3D reagent (Promega) was added to each well and incubated for 10 min at R.T.; luminescence signal was measured by TriStar2 (Berthold) or PHERAstar (BMG Labtech) according to the manufacturer’s protocols.
Data analysis
Assay validation was assessed using the Z′-factor calculated using Equation (1) as follows:
| Z′ = 1 − [(3S.D.ctrl + 3S.D.blank)/(Meanctrl − Meanblank)], | Equation (1) |
where ctrl is the 100% viable cell signals, blank is the no cell signals, and S.D. is the standard deviation. Concentration-response data were fitted to Equation (2) with fixed maximum inhibition values of 100% using GraphPad Prism 9 software (GraphPad Software) as follows:
| % inhibition = 100/(1 + 10(Log IC50 − Log[comp]) × Hill)) | Equation (2) |
Quantification and statistical analysis
Quantification was represented as the mean ±S.D. Data were analyzed using Student’s t test for two groups and one-way analysis of variance with the post hoc Bonferroni test for more than three groups. Differences were considered significant at p < 0.05, indicated by asterisks. All quantifications included at least n = 3 independent biological replicates.
Acknowledgments
We thank Dr. Atsushi Miyawaki (Brain Science Institute, RIKEN, Japan) for providing Venus-containing plasmids, Dr. Makoto Otsu for delivering human iPS cells, Ms. Akane Oku for technical support, and Dr. Riyo Imamura for offering helpful discussion. This study was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan under Grant-in-Aid for Scientific Research C (30835377 to Y.T.); the Agency for Medical Research and Development (AMED-CREST; 16gm0910008h0001 to R.S.); and the Plat-form Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research) from AMED (grant no. JP20a.m.0101086; support no. 2472).
Author contributions
Y.T. has designed the study, performed most of the experiments, and wrote the article. M.N. and Y.I. performed the experiments. S.S. and H.Kiyono provided human intestinal organoids, engaged in discussion with Y.T., and provided critical advice. H.Kojima and T.O. provided a chemical library, supported compound screening, and gave vital suggestions. M.S., Y.Y., and R.S. supported data analysis. Y.T. and R.S. supervised the project and secured the funding. All authors approved the final version of the article.
Declaration of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104542.
Contributor Information
Yu Takahashi, Email: ayutaka@g.ecc.u-tokyo.ac.jp.
Ryuichiro Sato, Email: roysato@g.ecc.u-tokyo.ac.jp.
Supplemental information
Data and code availability
RNA sequencing data have been deposited at DDBJ and are publicly available as of the date of publication. The accession number is in the key resources table. This study does not generate original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Al-Dosari M.S., Parvez M.K. Novel plant inducers of PXR-dependent cytochrome P450 3A4 expression in HepG2 cells. Saudi Pharmaceut. J. 2018;26:1069–1072. doi: 10.1016/j.jsps.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini Y.M., Park Y., Jergens A.E., Shin W., Min S., Atherly T., Borcherding D.C., Jang J., Allenspach K., Mochel J.P., Kim H.J. Recapitulation of the accessible interface of biopsy-derived canine intestinal organoids to study epithelial-luminal interactions. PLoS One. 2020;15:e0231423. doi: 10.1371/journal.pone.0231423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brück S., Strohmeier J., Busch D., Drozdzik M., Oswald S. Caco-2 cells - expression, regulation and function of drug transporters compared with human jejunal tissue. Biopharm. Drug Dispos. 2017;38:115–126. doi: 10.1002/bdd.2025. [DOI] [PubMed] [Google Scholar]
- Cai T., Qi Y., Jergens A., Wannemuehler M., Barrett T.A., Wang Q. Effects of six common dietary nutrients on murine intestinal organoid growth. PLoS One. 2018;13:e0191517. doi: 10.1371/journal.pone.0191517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra L., Borcherding D.C., Kingsbury D., Atherly T., Ambrosini Y.M., Bourgois-Mochel A., Yuan W., Kimber M., Qi Y., Wang Q., et al. Derivation of adult canine intestinal organoids for translational research in gastroenterology. BMC Biol. 2019;17:33. doi: 10.1186/s12915-019-0652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J.P., Hwang K., Singh H., Wang D., Reynolds C.P., Curley Jr R.W., Williams S.C., Maurer B.J., Kang M.H. Fenretinide metabolism in humans and mice: utilizing pharmacological modulation of its metabolic pathway to increase systemic exposure. Br. J. Pharmacol. 2011;163:1263–1275. doi: 10.1111/j.1476-5381.2011.01310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumoul X., Diry M., Barouki R. PXR-dependent induction of human CYP3A4 gene expression by organochlorine pesticides. Biochem. Pharmacol. 2002;64:1513–1519. doi: 10.1016/s0006-2952(02)01298-4. [DOI] [PubMed] [Google Scholar]
- Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Ettayebi K., Crawford S.E., Murakami K., Broughman J.R., Karandikar U., Tenge V.R., Neill F.H., Blutt S.E., Zeng X.L., Qu L., et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmi O.A., Kish M., Boldt S., Obach R.S. Cytochrome P450 3A4 mRNA is a more reliable marker than CYP3A4 activity for detecting pregnane X receptor-activated induction of drug-metabolizing enzymes. Drug Metab. Dispos. 2010;38:1605–1611. doi: 10.1124/dmd.110.033126. [DOI] [PubMed] [Google Scholar]
- Foulke-Abel J., In J., Donowitz M.J., Yin J., Zachos N.C., Kovbasnjuk O., Estes M.K., de Jonge H., Donowitz M. Human enteroids as a model of upper small intestinal ion transport physiology and pathophysiology. Gastroenterology. 2016;150:638–649.e8. doi: 10.1053/j.gastro.2015.11.047.e638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.A., Mahajan S., Ritz J. Fludarabine-induced immunosuppression is associated with inhibition of STAT1 signaling. Nat. Med. 1999;5:444–447. doi: 10.1038/7445. [DOI] [PubMed] [Google Scholar]
- Ghadimi B.M., Sackett D.L., Difilippantonio M.J., Schröck E., Neumann T., Jauho A., Auer G., Ried T. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–190. doi: 10.1002/(sici)1098-2264(200002)27:2<183::aid-gcc10>3.0.co;2-p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta E., Lestingi T.M., Mick R., Ramirez J., Vokes E.E., Ratain M.J. Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea. Cancer Res. 1994;54:3723–3725. [PubMed] [Google Scholar]
- Hu C., Perlmutter D.H. Regulation of α1-antitrypsin gene expression in human intestinal epithelial cell line Caco-2 by HNF-1α and HNF-4. Am. J. Physiol. 1999;276:G1181–G1194. doi: 10.1152/ajpgi.1999.276.5.g1181. [DOI] [PubMed] [Google Scholar]
- Huang W.K., Xie C., Young R.L., Zhao J.B., Ebendorff-Heidepriem H., Jones K.L., Rayner C.K., Wu T.Z. Development of innovative tools for investigation of nutrient-gut interaction. World J. Gastroenterol. 2020;26:3562–3576. doi: 10.3748/wjg.v26.i25.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M.M., Nijstad N., Franceschini L. Regulation of microsomal triglyceride transfer protein. Clin. Lipidol. 2011;6:293–303. doi: 10.2217/clp.11.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Shumway M., Leinonen R. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsell H. Glucose transporters in the small intestine in health and disease. Pflugers Arch. Eur. J. Physiol. 2020;472:1207–1248. doi: 10.1007/s00424-020-02439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küblbeck J., Hakkarainen J.J., Petsalo A., Vellonen K.S., Tolonen A., Reponen P., Forsberg M.M., Honkakoski P. Genetically modified Caco-2 cells with improved cytochrome P450 metabolic capacity. J. Pharm. Sci. 2016;105:941–949. doi: 10.1016/s0022-3549(15)00187-2. [DOI] [PubMed] [Google Scholar]
- Kwon O., Jung K.B., Lee K.R., Son Y.S., Lee H., Kim J.J., Kim K., Lee S., Song Y.K., Jung J., et al. The development of a functional human small intestinal epithelium model for drug absorption. Sci. Adv. 2021;7:eabh1586. doi: 10.1126/sciadv.abh1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy E., Mehran M., Seidman E. Caco-2 cells as a model for intestinal lipoprotein synthesis and secretion. FASEB J. 1995;9:626–635. doi: 10.1096/fasebj.9.8.7768354. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Mahraoui L., Rodolosse A., Barbat A., Dussaulx E., Zweibaum A., Rousset M., Brot-Laroche E., Rodolosse A., BArbAt A., ZweibAum A. Presence and differential expression of SGLT1, GLUT1, GLUT2, GLUT3 and GLUT5 hexose-transporter mRNAs in Caco-2 cell clones in relation to cell growth and glucose consumption. Biochem. J. 1994;298:629–633. doi: 10.1042/bj2980629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Meng Z., Gwag T., Sui Y., Park S.H., Zhou X., Zhou C. The atypical antipsychotic quetiapine induces hyperlipidemia by activating intestinal PXR signaling. JCI Insight. 2019;4:125657. doi: 10.1172/jci.insight.125657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier V., Bourrié M., Berger Y., Fabre G. The human intestinal epithelial cell line Caco-2; pharmacological and pharmacokinetic applications. Cell Biol. Toxicol. 1995;11:187–194. doi: 10.1007/bf00756522. [DOI] [PubMed] [Google Scholar]
- Mochel J.P., Jergens A.E., Kingsbury D., Kim H.J., Martín M.G., Allenspach K. Intestinal stem cells to advance drug development, precision, and regenerative medicine: a paradigm Shift in translational research. AAPS J. 2017;20:17. doi: 10.1208/s12248-017-0178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monneret C. Histone deacetylase inhibitors. Eur. J. Med. Chem. 2005;40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Prueksaritanont T., Gorham L.M., Hochman J.H., Tran L.O., Vyas K.P. Comparative studies of drug-metabolizing enzymes in dog, monkey, and human small intestines, and in Caco-2 cells. Drug Metab. Dispos. 1996;24:634–642. [PubMed] [Google Scholar]
- Reisher S.R., Hughes T.E., Ordovas J.M., Schaefer E.J., Feinstein S.I. Increased expression of apolipoprotein genes accompanies differentiation in the intestinal cell line Caco-2. Proc. Natl. Acad. Sci. USA. 1993;90:5757–5761. doi: 10.1073/pnas.90.12.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Duan L., He Q., Zhang Z., Zhou Y., Wu D., Pan J., Pei D., Ding K. Identification of niclosamide as a new small-Molecule inhibitor of the STAT3 signaling pathway. ACS Med. Chem. Lett. 2010;1:454–459. doi: 10.1021/ml100146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D., Souza T., Jennen D.G.J., Lemmens L., Kleinjans J.C.S., de Kok T.M. Drug-induced gene expression profile changes in relation to intestinal toxicity: state-of-the-art and new approaches. Cancer Treat. Rev. 2019;77:57–66. doi: 10.1016/j.ctrv.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Rossi G., Manfrin A., Lutolf M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018;19:671–687. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- Rousset M. The human colon carcinoma cell lines HT-29 and Caco-2: two in vitro models for the study of intestinal differentiation. Biochimie. 1986;68:1035–1040. doi: 10.1016/s0300-9084(86)80177-8. [DOI] [PubMed] [Google Scholar]
- Sato S., Hisaie K., Kurokawa S., Suzuki A., Sakon N., Uchida Y., Yuki Y., Kiyono H. Human norovirus propagation in human induced pluripotent stem cell-derived intestinal epithelial cells. Cell. Mol. Gastroenterol. Hepatol. 2019;7:686–688.e5. doi: 10.1016/j.jcmgh.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Simeone A.M., Ekmekcioglu S., Broemeling L.D., Grimm E.A., Tari A.M. A novel mechanism by which N-(4-hydroxyphenyl)retinamide inhibits breast cancer cell growth: the production of nitric oxide. Mol. Cancer Therapeut. 2002;1:1009–1017. [PubMed] [Google Scholar]
- Sun D., Lennernas H., Welage L.S., Barnett J.L., Landowski C.P., Foster D., Fleisher D., Lee K.D., Amidon G.L. Comparison of human duodenum and Caco-2 gene expression profiles for 12, 000 gene sequences tags and correlation with permeability of 26 drugs. Pharm. Res. 2002;19:1400–1416. doi: 10.1023/a:1020483911355. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Inoue Y., Kuze K., Sato S., Shimizu M., Kiyono H., Yamauchi Y., Sato R. Comparison of gene expression and activation of transcription factors in organoid-derived monolayer intestinal epithelial cells and organoids. Biosci. Biotechnol. Biochem. 2021;85:2137–2144. doi: 10.1093/bbb/zbab136. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Sato S., Kurashima Y., Lai C.Y., Otsu M., Hayashi M., Yamaguchi T., Kiyono H. Reciprocal inflammatory signaling between intestinal epithelial cells and adipocytes in the absence of immune cells. EBioMedicine. 2017;23:34–45. doi: 10.1016/j.ebiom.2017.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Sato S., Kurashima Y., Yamamoto T., Kurokawa S., Yuki Y., Takemura N., Uematsu S., Lai C.Y., Otsu M., et al. A refined culture system for human induced pluripotent stem cell-derived intestinal epithelial organoids. Stem Cell Rep. 2018;10:314–328. doi: 10.1016/j.stemcr.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu T., Yokono M., Tahara A., Takakura S. In vitro pharmacological profile of ipragliflozin, a sodium glucose Co-transporter 2 inhibitor. Biol. Pharm. Bull. 2019;42:507–511. doi: 10.1248/bpb.b18-00728. [DOI] [PubMed] [Google Scholar]
- Turner J.R., Lencer W.I., Carlson S., Madara J.L. Carboxy-terminal vesicular stomatitis virus G protein-tagged intestinal Na+-dependent glucose cotransporter (SGLT1): maintenance of surface expression and global transport function with selective perturbation of transport kinetics and polarized expression. J. Biol. Chem. 1996;271:7738–7744. doi: 10.1074/jbc.271.13.7738. [DOI] [PubMed] [Google Scholar]
- Vogt A., McDonald P.R., Tamewitz A., Sikorski R.P., Wipf P., Skoko J.J., 3rd, Lazo J.S. A cell-active inhibitor of mitogen-activated protein kinase phosphatases restores paclitaxel-induced apoptosis in dexamethasone-protected cancer cells. Mol. Cancer Therapeut. 2008;7:330–340. doi: 10.1158/1535-7163.mct-07-2165. [DOI] [PubMed] [Google Scholar]
- Watkins R.E., Wisely G.B., Moore L.B., Collins J.L., Lambert M.H., Williams S.P., Willson T.M., Kliewer S.A., Redinbo M.R. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001;292:2329–2333. doi: 10.1126/science.1060762. [DOI] [PubMed] [Google Scholar]
- Xie W., Barwick J.L., Downes M., Blumberg B., Simon C.M., Nelson M.C., Neuschwander-Tetri B.A., Brunt E.M., Guzelian P.S., Evans R.M. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- Yamaura Y., Chapron B.D., Wang Z., Himmelfarb J., Thummel K.E. Functional comparison of human colonic carcinoma cell lines and primary small intestinal epithelial cells for investigations of intestinal drug permeability and first-pass metabolism. Drug Metab. Dispos. 2016;44:329–335. doi: 10.1124/dmd.115.068429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietek T., Giesbertz P., Ewers M., Reichart F., Weinmüller M., Urbauer E., Haller D., Demir I.E., Ceyhan G.O., Kessler H., Rath E. Organoids to study intestinal nutrient transport, drug uptake and metabolism - update to the human model and expansion of applications. Front. Bioeng. Biotechnol. 2020;8:577656. doi: 10.3389/fbioe.2020.577656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited at DDBJ and are publicly available as of the date of publication. The accession number is in the key resources table. This study does not generate original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.