Summary
Telomere length has been implicated in the organismal response to stress, but the underlying mechanisms are unknown.
Here we examine the impact of telomere length changes on the responses to three contrasting abiotic environments in Arabidopsis, and measure 32 fitness, developmental, physiological and leaf-level anatomical traits.
We report that telomere length in wild-type and short-telomere mutants is resistant to abiotic stress, while the elongated telomeres in ku70 mutants are more plastic. We detected significant pleiotropic effects of telomere length on flowering time and key leaf physiological and anatomical traits. Furthermore, our data reveal a significant genotype by environment (G × E) interaction for reproductive fitness, with the benefits and costs to performance depending on the growth conditions.
These results imply that life-history trade-offs between flowering time and reproductive fitness are impacted by telomere length variation. We postulate that telomere length in plants is subject to natural selection imposed by different environments.
Keywords: Arabidopsis thaliana, environment, flowering time, plants, seeds, stress, telomere dynamics, tert
Introduction
Telomeres are evolutionarily conserved protein–DNA complexes at the physical ends of linear eukaryotic chromosomes. Telomeric DNA consists of multiple copies of short G-rich repeats: TTAGGG in vertebrates and TTTAGGG in most plants. In the absence of telomerase, the end replication problem causes telomeres to shorten with each somatic cell division, eventually leading to cellular senescence or death (Bonnell et al., 2021). Thus, telomere length is viewed as an accurate cellular marker of biological age, and its improper maintenance has been linked to cancer and human aging-associated diseases (Maciejowski & de Lange, 2017).
Telomere length is highly dynamic, but the opposing actions of telomerase and the end replication problem ultimately establish a species-specific telomere length set point, which may reflect a fine compromise between potential positive gains in organismal survival and associated negative energy costs (Olsson et al., 2018). For instance, telomerase inhibition in human somatic cells is a known defense mechanism against tumorigenesis, but it eventually leads to cell senescence and death (Wright & Shay, 2005). By contrast, mice do not regulate telomerase in somatic tissues, and develop cancer more often (Blasco, 2005). These findings suggest that telomere length is under stabilizing selection, with intermediate lengths favored. Average telomere length also differs considerably between genotypes of the same species (Hemann & Greider, 2000; Shakirov & Shippen, 2004; Gatbonton et al., 2006; Cook et al., 2016). Hence, telomere length is a quantitative trait that is controlled by many genes (Ding et al., 2004; Levy et al., 2010; Codd et al., 2013; Abdulkina et al., 2019) and is possibly influenced by the environment (Harari et al., 2013; Romano et al., 2013). Intra-species variation in telomere length is consistent with ongoing evolution, but the degree to which this evolution is neutral or adaptive is unknown.
Telomere ecology, the study of telomere dynamics in an ecological context, is an emerging field that seeks to the elucidate evolution of fitness trade-offs as they relate to telomere length (Monaghan & Haussmann, 2006; Bize et al., 2009; Horn et al., 2010). Some of the initial work was conducted in birds. Such studies indicated that the 3-yr survival rate of Tachycineta bicolor birds was higher in individuals with longer telomeres compared to those with shorter telomeres (Haussmann et al., 2005). In Sterna hirundo, telomere length serves as a biomarker of reproductive success (Bauch et al., 2014), while in Acrocephalus arundinaceus malaria infection correlates with a long-term degradation of telomere length, shortened lifespan and decreased reproductive success (Asghar et al., 2015). Despite these intriguing observations, little is known about the interplay between telomere length control and organismal fitness under environmental change.
Studies manipulating environmental conditions simultaneously with telomere length could provide new insight into the adaptive role of telomeres. The model plant Arabidopsis provides a valuable system for examining the influence of environmental conditions on telomere biology, organismal fitness and plant physiology. Several Arabidopsis mutants with well-defined positive or negative changes in telomere length have been described. For example, in plants deficient in the Ku heterodimer (Ku70/Ku80) blunt-end telomeres are deprotected, allowing telomerase unfettered access (Kazda et al., 2012). As a consequence, telomeres expand several-fold (Riha et al., 2002; Gallego et al., 2003). By contrast, plants lacking functional TERT, the catalytic subunit of telomerase, undergo progressive shortening at a rate of up to 500 bp per plant generation, ultimately triggering massive genome instability and developmental arrest (Riha et al., 2001). Mutations in two other genes cause telomeres to reach a shorter than in wild-type, but stable set point length. One is NOP2A, a putative ribosomal RNA maturation and ribosome assembly factor, which was recently identified through QTL mapping as a positive regulator of telomere length (Abdulkina et al., 2019). A second gene, TAD3, encodes a tRNA adenosine deaminase and promotes telomerase-independent telomere length maintenance (Bose et al., 2020).
We previously reported that natural variation in telomere length set point in rice, maize and Arabidopsis genotypes affects flowering time, a key aspect of life history strategy, and showed that accessions with longer telomeres flower earlier (Choi et al., 2021). Here we explore the interplay between the genetic and environmental factors driving telomere biology in Arabidopsis by evaluating telomere length plasticity and pleiotropy in five genotypes grown under varying environmental conditions. We provide evidence that genotypes with distinct telomere lengths exhibit differential phenotypic responses under changing abiotic conditions, implying that mutations affecting telomere length homeostasis also impact other plant traits and, thus, show pleiotropic effects. Our experiments also reveal interesting genotype by environment (G × E) interactions for telomere length and several major plant phenotypes, suggesting that natural telomere length variation in plants could be under environment-driven selection pressures.
Materials and Methods
Plant genotypes
Seeds for Arabidopsis thaliana (L.) Heynh. wild-type accession Col-0 (CS6673) were obtained from the Arabidopsis Biological Resource Center (ABRC). Arabidopsis mutant lines tad3-2 (SALK_121147), tert-1 (5th generation), oli2-2/nop2a-2 (SALK_129648; 3rd generation) and ku70-2 (SALK_123114, 2nd generation) have been described previously (Fitzgerald et al., 1999; Kannan et al., 2008; Fujikura et al., 2009; Bose et al., 2020). All mutant lines are in the Col-0 genetic background (Supporting Information Table S1), and all seeds were bulked at the same time.
Plant growth and experimental conditions
Seeds were cold-stratified in 1.5 ml microcentrifuge tubes at 4°C for 3 d, and subsequently sown on ½ Murashige & Skoog (½MS) medium and grown in a culture room at 22°C under a 16 h : 8 h, light : dark photoperiod. On experimental day zero (Fig. S1), 7-d old seedlings were transplanted into soil (3 : 1 ratio of BioFungicide (Pro-Mix; Hummert International, Earth City, MO, USA) with Field and Fairway Calcined Clay (Profile; Hummert International)) in 9 cm2 plastic pots (two plants of the same genotype per pot) and grown in a plant growth chamber (CO2-BP; Conviron, Pembina, ND, USA). Six biological replicates (individual pots with plants) of all genotypes were used for each experimental condition. Pots with seedlings were labeled with randomly assigned numbers, and their positions in trays were randomized at the beginning of the experiment. Pot positions in trays were rerandomized weekly to minimize spatial effects within growth chambers. We manipulated both water availability and temperature for the following three treatment conditions: control (well-watered/normal temperature; wet|22°C), moderately dry (water limited/normal temperature; mod|22°C), and high temperature (well-watered/high temperature; wet|30°C). Conditions were as follows: wet|22°C (95% soil water content (SWC), 22°C); mod|22°C (35% SWC, 22°C); wet|30°C (95% SWC, 30°C).
Mod|22°C and wet|22°C treatments
Before the start of the experiment, each pot was filled with dry soil and weighed to obtain 0% SWC values, then left in standing water overnight to soak to 100% SWC and weighed again. Based on the known weight values for 0% and 100% SWC, 95% (wet|22°C treatment) and 35% (mod|22°C treatment) SWC values were calculated, as described previously (Campitelli et al., 2016). After transferring plants to soil, pots were weighed daily to monitor evaporation and SWC. All pots with plants in wet|22°C and mod|22°C conditions were allowed to reach 95% and 35% SWC values, respectively, by evaporation, after which individual pots were watered daily as needed to maintain target weight throughout the rest of the experiment. All plants in dry conditions reached the target 35% SWC in 23–28 d after they were moved from plates to soil. To independently confirm SWC values, we measured soil moisture twice throughout the experiment using a time-domain reflectometry (TDR) soil moisture probe (Campbell Scientific Hydrosense, Logan, UT, USA).
Wet|30°C treatment
It is known that growth at 30°C does not kill Arabidopsis plants but substantially stresses them, often leading to seed abortion or poor seed set (Chen et al., 2006). The high temperature wet|30°C treatment started at day 17 after seedlings were transferred to soil and was conducted in a dedicated Conviron CO2-BP growth chamber set at 30°C but with otherwise identical settings as used in the wet|22°C experiment. To make sure that plants were stressed specifically by high temperature and not by substantial water loss due to increased transpiration at 30°C, SWC was kept at > 80% by bottom-watering the plants daily. Under these conditions, many plants grown at 30°C flowered and subsequently senesced earlier than their counterparts grown at 22°C; thus, the experiment was terminated 1 wk earlier for this treatment.
Sample collection
Initially, each pot contained two plants of the same genotype. Leaves from the first plant were used throughout the experiment to obtain data for telomere length dynamics and to analyze phenological, physiological, leaf anatomical and morphological parameters before and after treatment started (0–15 d after reaching target SWC values, depending on the trait). The second plant was kept until the end of the experiment (when plants showed clear characteristics of physiological death, such as leaf yellowing and death, flowering cessation, seed maturation in siliques). These plants were then used to evaluate other raw traits, as described below and in Methods S1.
Telomere length measurements
To initially evaluate bulk telomere length in each genotype, terminal restriction fragment (TRF) assays were performed with genomic DNA digested with the Tru1I (Fermentas, Hanover, MD, USA) restriction enzyme. A 32P 5′-end-labeled (T3AG3)4 oligonucleotide was used as a probe (Fitzgerald et al., 1999). Radioactive signals were scanned with a Pharos FX Plus Molecular Imager (Bio-Rad), and the data were analyzed using the Quantity One v.4.6.5 software package (Bio-Rad). Mean telomere length (mean TRF) was calculated using the TeloTool program (Göhring et al., 2014). To evaluate telomere length values for leaf DNA samples before and after each experimental treatment, we performed the polymerase chain reaction (PCR)-based primer extension telomere repeat amplification (PETRA) assay, which allows precise telomere length measurement on individual chromosome arms in Arabidopsis (Heacock et al., 2004). Telomere length was measured on three chromosome arms (5R, 1L and 2R) that are representative, relatively easy to measure, and commonly used in Arabidopsis telomere research (Shakirov & Shippen, 2004; Surovtseva et al., 2007). The mean values from TeloTool were used to evaluate the effects of abiotic environments on telomere dynamics.
To explore telomere dynamics, we measured and averaged telomere lengths from all three chromosome arms the day before (T0) and 11 d after (Tf) the onset of treatments, and estimated the change in telomere length for each arm separately, as well as the average change across arms. To control for the possibility that initial telomere length may influence the degree of loss of telomeric DNA, we also explored the proportional change in telomere length by dividing the average change by initial telomere length. In total, we evaluated 13 different telomere traits (Table S2).
Phenotypes measured
To explore the relationship between telomere length variation and other plant traits, we measured an additional 18 raw phenotypes, and used nine of these to further calculate 10 composite phenotypes, so that a total of 19 phenotypes (18 − 9 + 10) were included in the analyses. These phenotypes covered a range of plant features: five phenological, two morphological, two fitness, six leaf anatomical, and four physiological traits (Table S2).
Missing data and imputation
Given the large number of traits measured in this experiment (13 telomere and 19 phenotypic traits with up to six biological replicates), missing data became problematic for the multivariate analyses (see ‘Principal component analysis, multivariate analyses of variance and analyses of variance’, later); these methods drop individuals from the analysis if there is a single missing trait value, decreasing overall power. In our experiment, phenological, morphological and fitness phenotypes had little to zero missing data (1–1.8%). Telomere length on individual chromosome arms is notoriously difficult to measure, but we were able to obtain data for 3–6 biological replicates for all conditions except for 2R, for which we had lower numbers but still at least 2–3 biological replicates. Missing data for physiological traits varied from 13% to 42%.
To account for missing data, we used the ‘impute missing data’ function in Jmp genomics (v.9.1) which utilizes the mean and covariance matrix, estimated using the least squares method, to impute missing values before analyses. We note that missing values were randomly and evenly spread across all genotypes in all environments, and that an initial comparison of running the analyses on raw (Dataset S1) and imputed (Dataset S2) data sets revealed that they were very similar, and that our statistical findings using the imputed data set were more conservative, in that 10 fewer significant results were discovered. Specifically, our analyses of variance (ANOVAs) yielded 50 instances of significance when carried out on our raw data (Dataset S3) vs 40 instances of significance when carried out on imputed data (Table 3). Thus, we chose to use the imputed data set for all analyses performed here. Dataset S4 provides boxplots displaying all 32 traits considered in our analyses (13 telomere traits and 19 plant traits) as well as imputed values overlaying them.
Table 3.
Analysis of variance (ANOVA) results for Arabidopsis plant traits.
| Phenology | |||||
|---|---|---|---|---|---|
|
| |||||
| Effect | Flowering day | Primary bolts | Secondary bolts | Height | Bolt thickness |
| Genotype | 13.62 (4)*** | 4.51 (4)** | 10.37 (4)*** | 3.41 (4)* | 4.83 (4)** |
| Treatment | 5.70 (2)** | 74.11 (2)*** | 244.17 (2)*** | 47.32 (2)*** | 1.48 (2) |
| Genotype × Treatment | 0.82 (8) | 1.81 (8) | 6.09 (8)*** | 1.31 (8) | 1.03 (8) |
| Physiology | ||||
|---|---|---|---|---|
|
| ||||
| Effect | Relative water content | Osmotic potential | Total Chl | Proline |
| Genotype | 3.74 (4)** | 7.36 (4)*** | 0.64 (4) | 2.94 (4)* |
| Treatment | 7.72 (2)*** | 13.66 (2)*** | 39.15 (2)*** | 16.90 (2)*** |
| Genotype × Treatment | 1.19 (8) | 2.17 (8)* | 0.58 (8) | 3.59 (8)** |
| Leaf anatomy | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Effect | Stomatal area | Stomatal density | Stomatal area per leaf area | Epidermal cell area | Stomatal index | Stomatal to epidermal area ratio |
| Genotype | 0.11 (4) | 0.56 (4) | 1.11 (4) | 1.10 (4) | 2.34 (4) | 1.05 (4) |
| Treatment | 1.68 (2) | 9.03 (2)*** | 8.39 (2)*** | 5.28 (2)** | 6.76 (2)** | 4.12 (2)* |
| Genotype × Treatment | 1.32 (8) | 2.43 (8)* | 1.53 (8) | 2.79 (8)** | 1.87 (8) | 1.42 (8) |
| Fitness | ||
|---|---|---|
|
| ||
| Effect | Total seeds | Biomass |
| Genotype | 2.23 (4) | 0.64 (4) |
| Treatment | 16.44 (2)*** | 2.11 (2) |
| Genotype × Treatment | 3.82 (8)*** | 1.24 (8) |
| Morphology | ||
|---|---|---|
|
| ||
| Effect | Leaf count | Leaf thickness |
| Genotype | 0.57 (4) | 0.90 (4) |
| Treatment | 0.40 (2) | 3.15 (2)* |
| Genotype × Treatment | 1.05 (8) | 0.71 (8) |
Model: trait = genotype + treatment + (genotype × treatment). Genotypes: Col-0; ku70; nop2a; tad3-2; tert. Numbers represent F-values with degrees of freedom in parentheses.
, P < 0.05;
, P < 0.01;
, P < 0.0001.
Principal component analysis, multivariate analyses of variance and analyses of variance
Principal component analysis (PCA) was performed to acquire a multidimensional overview of phenotypic trait variations and integration using the proc princomp function in Sas (SAS Institute, Cary, NC, USA). To further explore global patterns of trait variation while controlling for the likely correlation between traits within a trait grouping (identified in Table S2), we constructed a series of eight multivariate analyses of variance and analyses (MANOVAs) in Sas v.9.4 using the MANOVA option of the proc glm command. MANOVA models took the following form:
In this model, (‘yi + yj + …’, phenotypes within a trait grouping; ‘Treatment’, three environments; ‘Genotype’, five genotypes included in this study; ‘|’, indicates that all terms were considered both separately and as interaction combinations).
We then evaluated how all phenotypes individually responded to the environmental treatments by statistically comparing least-squared means (‘lsmeans’) using general linear mixed model ANOVAs (proc mixed) in Sas v.9.4 (Littell et al., 2006):
In this model, ‘yi’ represents the focal phenotype, and all other terms are the same as in the MANOVA model. To compare lsmeans we used pairwise contrasts (‘pdiff’ statement in Sas) and employed Tukey’s honestly significant difference (Tukey’s HSD; ‘adjust=tukey’ in Sas) test to control for multiple comparisons.
Within-genotype telomere length variation was typically unequal between genotypes, resulting in the violation of the assumption of equal variances. We note that this was driven by longer telomere genotypes (particularly ku) exhibiting relatively high telomere length variation, which commonly occurs due to hyperactive telomerase activity in this genotype (Riha & Shippen, 2003). To account for this, we ran this model, allowing for heterogeneity of variances among genotypes using the ‘repeated’ statement in proc mixed; for all telomere length traits this heterogeneous variance model resulted in a better fit than the null homogeneous variance model, based on null model likelihood ratio tests that compare the −2 restricted log likelihood ratios using a χ2-test.
Response strength and difference from the wild-type
To assess how any deviation away from the wild-type telomere length impacted the phenotypes, we determined the absolute value of the average difference of phenotypes for all genotypes relative to the Col-0 wild-type. We applied the following transformation to all individuals: (1) We first calculated the mean trait value for Col-0 for each phenotype in each of the three treatments to generate a matrix of wild-type phenotypes; (2) next, we estimated the difference from the wild-type by subtracting the observed phenotypic value for each individual from the mean value for Col-0 in the appropriate treatment; (3) we then estimated the absolute value by transforming all of the differences from wild-type estimates to positive values; (4) finally, we performed linear mixed model ANOVAs (as stated in ‘principal component analysis, multivariate analyses of variance and analyses of variance’, above) on these transformed values to obtain a least-squared mean difference from the wild-type for each genotype in each treatment. Note that because we performed an absolute value transformation (step 3) before estimating least-squared means, it is possible for Col-0 to have a nonzero estimate. We further explored differences from the wild-type by estimating both the fold difference and standard deviation from wild-type by dividing the absolute difference by the mean and standard deviation for Col-0, respectively, separately for the three treatments. These additional models confirmed the patterns for the absolute difference from wild-type (Dataset S5).
A unique feature of these transformations that compare the magnitude of trait differences of the mutant genotypes from the wild-type (Col-0), is that they remove the directionality of any differences (by taking the absolute value), therefore enabling a more direct analysis of the correlation between telomere length changes and plant traits. In this sense, trait values for a genotype that is furthest from the wild-type suggest that the telomere length characteristic of a specific genotype and the focal traits are more strongly related.
Results
Experimental setup
To explore the relationship between telomere length and life history strategies, we experimentally manipulated telomere length in Arabidopsis genotypes and measured the impact of these changes on fitness, development, physiology and leaf-level anatomy under varying environmental conditions. We analyzed five Arabidopsis wild-type and mutant genotypes with different telomere lengths: short (< 2 kb; tert (generation 5, G5) and nop2a mutants), medium (2–3 kb; wild-type Col-0 accession and tad3-2 mutant) and long (> 3 kb; ku70 mutant) (Fig. 1; Table S1). Telomeres in G5 tert mutants are not critically shortened (Riha et al., 2001), but since changes in plant physiology or gene expression can occur before telomere failure (Riha et al., 2001; Amiard et al., 2014), we selected two genotypes each for short and medium telomere length categories to detect both genotype-specific and telomere length-specific patterns in plant traits. Ku mutants represent the only stable, long-telomere mutants described for the Arabidopsis Col-0 background that do not have major genome abnormalities (Riha et al., 2002; Gallego et al., 2003). Unlike the situation in the tert and ku70 mutants, telomere length changes in nop2a and especially tad3 mutants are relatively minor and likely caused by indirect effects (Abdulkina et al., 2019; Bose et al., 2020), and these genotypes were used as internal controls in the experiment. All selected single-copy homozygous T-DNA mutant lines with no background contamination were previously generated in the same reference Col-0 background and are well-characterized (Fitzgerald et al., 1999; Kannan et al., 2008; Fujikura et al., 2009; Bose et al., 2020).
Fig. 1.
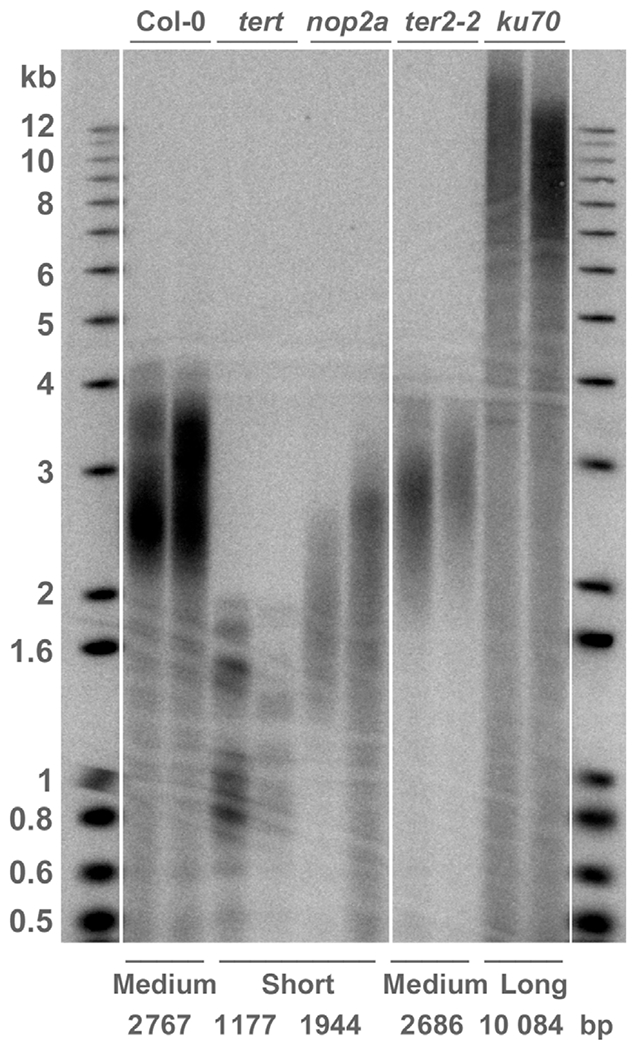
Telomere length in Arabidopsis genotypes used in the study. A representative terminal restriction fragment Southern blot is shown. Each lane represents genomic DNA from one individual plant of the corresponding genotype. All Arabidopsis genotypes were analyzed individually and also grouped as medium (Col-0, tad3-2), short (tert, nop2a) and long (ku70) telomere genotypes. The mean telomere length for each analyzed genotype is indicated at the bottom (in base pairs). DNA molecular weight markers and their corresponding sizes in kb are shown.
We manipulated soil moisture content (wet and moderately dry) and ambient temperature (22°C and 30°C) in an experimental design consisting of three contrasting abiotic environments: wet|22°C, mod|22°C and wet|30°C. We selected soil moisture levels and ambient temperatures that represent increasing levels of environmental pressure (from control wet|22°C to moderate dry mod|22°C to high temperature wet|30°C) expected to generate detectable and differential changes in fitness and physiological responses (Chen et al., 2006; Campitelli et al., 2016) without irreversibly impairing the plants (Fig. S1).
Global patterns suggest strong genotypic, treatment and genotype by environment effects on many, but not all, trait groups
We measured 24 raw traits in all three environments. After removing eight raw traits that were used to generate an additional 16 composite traits, a total of 32 traits covering six major phenotype groups, defined as telomere length, phenology, morphology, fitness, leaf anatomy, and physiology, were used for our statistical analyses (Table S2). We performed a PCA on all traits to identify unique patterns and structures of individual genotypes and treatments in multivariate space. PC1 clearly partitioned the treatment effects separating the harsher wet|30°C environment from the other two treatments, and partially separated ku70 and tert (genotypes with the longest and shortest telomere lengths, respectively) from the other three genotypes (Fig. S2). Overall, the first two PC axes explain 42.5% of phenotypic variation, demonstrating that telomere length genotypes and the applied treatments are strong drivers of observed variation.
To explore the global responses of traits within each group, we constructed a series of eight separate MANOVAs (Scheiner, 2001), each targeting a specific group of traits (Table S2). Our results indicate strong genotypic effects across all trait groups except morphological features and final change in average telomere length, very strong treatment effects (plasticity) across all trait groups, except, notably, telomere length, and evidence for G × E interactions among many trait groups (Table 1). To disentangle observed global patterns, we further analyzed individual traits within these groupings by exploring how telomere length variation in the Arabidopsis mutants pleiotropically impacts various trait sets.
Table 1.
Multivariate analysis of variance (MANOVA) results for Arabidopsis trait groups.
| Trait group | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Effect | Morphology | Phenology | Leaf anatomy | Physiology | Fitness | Initial telomere length (T0) | Final telomere length (Tf) | Change in telomere length (AvgTf – AvgT0) |
| Genotype | 1.31 (4) | 23.19 (5)*** | 4.15 (6)** | 9.02 (4)*** | 5.46 (4)** | 19.02 (4)*** | 23.32 (4)*** | 2.47 (4) |
| Treatment | 3.78 (2)* | 105.67 (5)*** | 5.22 (6)** | 27.21 (4)*** | 69.45 (2)*** | 2.63 (3) | 2.54 (3) | 0.31 (3) |
| Genotype × Treatment | 1.10 (8) | 7.63 (8)*** | 4.22 (8)** | 4.01 (8)** | 9.08 (8)*** | 1.82 (8) | 2.37 (8)* | 1.63 (8) |
Model: traitA + traitB etc = genotype + treatment + (genotype × treatment). Genotypes: Col-0, ku70, nop2a; tad3-2; tert. Numbers represent F-values with degrees of freedom in parentheses.
, P < 0.05;
, P < 0.01;
, P < 0.0001.
Telomere length plasticity in response to environment is only observed in the long-telomere ku70 mutants
Telomere lengths for three individual Arabidopsis chromosome arms (1L, 2R and 5L) were assayed before experimental treatments were initiated (T0) and after target treatments were achieved (Tf) using the PETRA assay (Heacock et al., 2004). Similar to the STELA approach used for human cells (Baird et al., 2003), this PCR-based assay measures telomere length on individual chromosome arms and produces distinct bands (Fig. S3), as opposed to the broad smears observed in TRF assays. As expected, significant differences in telomere length were observed between genotypes before (T0; F = 19.02, P < 0.0001) and after (Tf; F = 23.32, P < 0.0001) treatments (Table 1). We estimated the change in telomere length over time for individual chromosome arms (Tf – T0), and in aggregate across all three arms as an average (AvgTf – AvgT0) and proportional ((AvgTf – AvgT0)/AvgT0) change. Telomere length tended to slightly shorten over time, irrespective of treatment or genotype when considered in aggregate (AvgTf – AvgT0; Fig. 2a), despite some stochastic variation for individual chromosome arms (Fig. S4). Hence, regardless of the environment, telomere length in somatic tissues (leaves) generally shortens over the plant lifespan.
Fig. 2.
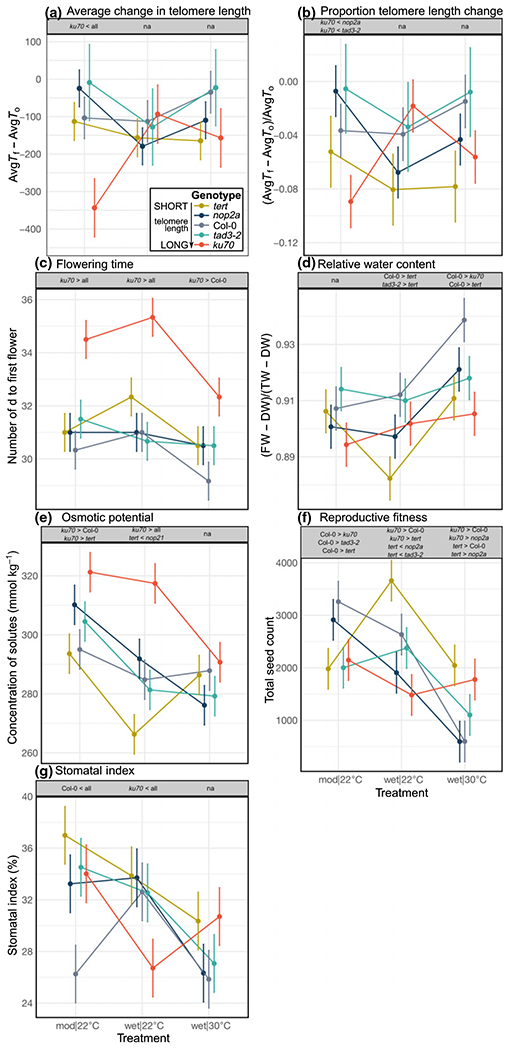
Changes in telomere length, and developmental, physiological, fitness, and anatomical leaf parameters in Arabidopsis genotypes exposed to contrasting abiotic environments. The following parameters were analyzed for all genotypes: (a) Telomere length changes over time for the average of three chromosome arms (AvgTf – AvgT0); (b) Telomere length changes over time as a proportion of initial telomere length (AvgTf – AvgT0)/AvgT0; (c) Flowering day was analyzed as a developmental parameter; relative water content RWC (fresh weight – dry weight)/(turgid weight – dry weight); (d) and osmotic potential OP; (e) were analyzed as physiological leaf parameters; (f) Total seed count was selected as a reproductive fitness proxy; (g) stomatal index was analyzed as an anatomical leaf parameter. Points show least-squared means ± SE. Relative telomere length for all genotypes (from the shortest, tert, to the longest, ku70) is indicated in the in-panel legend. The gray box above each panel lists significant pairwise differences between genotypes within that treatment following a Tukey–Kramer adjustment for multiple comparisons. >, genotype has a significantly larger trait value; <, genotype has a significantly smaller trait value; all, genotype is significantly different from all other genotypes; DW, dry weight; FW, fresh weight; na, no significant pairwise differences detected in that treatment; TW, turgid weight.
We next analyzed all genotypes for telomere length plasticity (changes in telomere length in response to the environment) over time (Tf – T0) in different treatments. We also explored interactions between genotypes and treatment (G × E) using MANOVAs that consider all tested chromosome arms simultaneously (Table 1). Unsurprisingly, we found no evidence for plasticity before experimental treatments (T0; no significant treatment and genotype × treatment effects). Interestingly, while we did not observe a direct treatment effect on telomere length after treatments were completed (Tf), we did detect significant G × E for Tf (F = 2.37, P < 0.05) (Table 1). This effect appears to be driven almost exclusively by ku70 mutants experiencing a relatively large telomere length decrease of c. 340 (± 70) bp in the mod|22°C environment (Fig. 2a), which is supported by a proportionally larger drop in telomere size (Fig. 2b), suggesting that dryer soils may accelerate telomere shortening for genotypes with initially longer telomeres. However, this G × E pattern of length plasticity for aggregate telomeres in ku70 mutants disappears when considering all genotypes and chromosome arms separately using ANOVAs (Table 2), and results in nonsignificant treatment (F = 0.31) and G × E (F = 1.63) effects when assessing the average change in telomere length over time (AvgTf – AvgT0) using MANOVAs (Table 1). Overall, our data provide some evidence for telomere length plasticity in the ku70 genotype with overly long telomeres, but not in genotypes with medium or short telomeres.
Table 2.
Analysis of variance (ANOVA) results for Arabidopsis telomere length traits.
| Telomeres | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||
| Effect | T0_1L | Tf_1L | Tf_T0_1L | T0_2R | Tf_2R | Tf_T0_2R | T0_5R | Tt_5R | Tf_T0_5R | AvgT0 | AvgTf | AvgTf – AvgT0 | Prop. telo. change |
| Genotype | 13.36 (4)*** | 15.39 (4)*** | 1.05 (4) | 10.63 (4)*** | 9.20 (4)*** | 0.97 (4) | 10.92 (4)*** | 11.48 (4)*** | 0.40 (4) | 17.68 (4)*** | 18.40 (4)*** | 1.56 (4) | 1.83 (4) |
| Treatment | 1.99 (2) | 2.33 (2) | 0.04 (2) | 3.75 (2)* | 3.60 (2)* | 0.06 (2) | 0.10 (2) | 0.07 (2) | 0.41 (2) | 2.98 (2) | 2.93 (2) | 0.34 (2) | 0.23 (2) |
| Genotype × Treatment | 1.54 (8) | 1.25 (8) | 0.60 (8) | 1.08 (8) | 1.14 (8) | 0.54 (8) | 1.30 (8) | 1.51 (8) | 1.13 (8) | 1.24 (8) | 0.85 (8) | 1.39 (8) | 1.62 (8) |
1L, left arm of chromosome 1; 2R, right arm of chromosome 2; 5R, right arm of chromosome 5; AvgTf, average telomere length at the end of the experiment; AvgTf – AvgT0, average change in telomere length over time; AvgT0, average telomere length at the start of the experiment; Prop. telo. change, proportion of change in telomere length after treatment relative to initial telomere length; Tf, telomere length at the end of the experiment; Tf_T0, change in telomere length over time; T0, telomere length at the start of the experiment.
Model: trait = genotype + treatment + (genotype × treatment). Genotypes: Col-0; ku70; nop2a; tad3-2; tert. Numbers represent F-values with degrees of freedom in parentheses.
, P < 0.05;
, P < 0.01;
, P < 0.0001.
Telomere length pleiotropically affects plant development, phenology, leaf physiology and anatomy
To more fully characterize the response of Arabidopsis genotypes with different telomere lengths to changing abiotic environments, we assessed several phenological traits. We observed indications of genotypic variation in flowering day (F = 13.62, P < 0.0001), primary (F = 4.51, P < 0.01) and secondary inflorescence bolts (F = 10.37, P < 0.0001), height (F = 3.41, P < 0.05) and bolt thickness (F = 4.83, P < 0.01) (Table 3). Flowering time phenotypes were particularly striking, with the long-telomere ku70 mutants being the most divergent from all other genotypes. On average, ku70 mutants began flowering 3–4 d later than wild-type plants across treatments (Fig. 2c). Additionally, flowering was delayed in all other mutant genotypes by 1–2 d in the wet|30°C environment, compared to the Col-0 wild-type. These data complement our recent findings on the impact of natural telomere length variation on flowering time in three different plant species (Choi et al., 2021), though the direction of flowering time changes in the T-DNA mutants is reversed.
Environmental conditions associated with drought and high heat stress are known to induce substantial changes in plant physiology, including shifts in leaf osmotic potential (OP), proline concentration, Chl and relative water content (RWC) (Chen et al., 2006; Verslues et al., 2014). Multivariate analyses of variance detected strong genotypic (F = 9.02, P < 0.0001), treatment (F = 27.21, P < 0.0001) and G × E (F = 4.01, P < 0.01) responses across these physiology traits (Table 1). As expected for plants grown in more stressful environments, the decrease in Chl content was significant in mod|22°C and wet|30°C conditions for all genotypes, as compared to the control wet|22°C environment (Table 3; Fig. S5A). Similarly, proline concentrations increased in all genotypes in response to mod|22°C and wet|30°C environments (Table 3; Fig. S5B).
Unexpectedly, plants showed a genotype-specific response for the two leaf-level water balance traits, RWC and OP. Under the control wet|22°C conditions, RWC was reduced in mutant lines with either short (tert, nop2a) or long (ku70) telomeres, but not in Col-0 or tad3-2 genotypes with a medium telomere length set point (Fig. 2d). This decrease in RWC is largely driven by tert mutants, which have the shortest telomeres among the genotypes tested. A more diverse response was observed for OP values. In the control wet|22°C environment, tert plants showed the lowest OP values and ku70 mutants displayed the highest (Fig. 2e).
Finally, we looked at several anatomical leaf traits. Analysis of treatment effect indicated that the environment had a substantial impact on five out of six anatomical leaf traits (Table 3), confirmed by a MANOVA (F = 5.22, P < 0.01) (Table 1). For two of these traits we detected significant G × E interactions (stomatal density, epidermal cell area) (Table 3), which were confirmed by a MANOVA (F = 4.22, P < 0.01). These G × E effects appear to result from nearly all genotypes exhibiting radically different phenotypes in the control wet|22°C environments compared to mod|22°C and wet|30°C treatments. Taken together, these data indicate that changes in telomere length homeostasis correlate with significant pleiotropic effects (sensu lato, the number of phenotypes impacted by a single gene knockout) in key plant development, phenology and leaf-level anatomy and physiology traits.
Telomere length impacts reproductive but not vegetative fitness parameters
We next tested whether telomere length changes can impact aboveground plant biomass and estimated total seed production, which we defined as parameters of vegetative and reproductive fitness, respectively. Multivariate analysis of variance results revealed a significant effect of genotype (F = 5.46, P < 0.01) and treatment (F = 69.45, P < 0.0001) on overall fitness, and a significant G × E interaction (F = 9.08, P < 0.0001) (Table 1). Interestingly, an examination of vegetative fitness (biomass) and reproductive fitness (total seeds) separately revealed differential contributions of these two traits to overall fitness. While biomass showed some variation across genotypes and treatments, an ANOVA did not detect a significant biomass difference between genotypes across all three growth conditions (F = 0.64, P = 0.79) (Table 3). These findings suggest that telomere length, or the genes governing telomere length homeostasis, do not significantly affect overall vegetative fitness.
By contrast, analysis of the reproductive fitness parameter, total seeds, revealed a markedly different profile. Under normal growth conditions (wet|22°C), ku70 and especially tert mutants appeared as clear outliers on the opposite sides of the seed quantity spectrum (Fig. 2f), with tert (shortest telomeres) producing the most seeds and ku70 mutants (longest telomeres) producing the fewest. Additionally, seed numbers for most genotypes plummeted in the wet|30°C environment, while ku70 produced slightly more seeds relative to wet|22°C (Fig. 2f), resulting in a rank change and a significant G × E interaction for reproductive fitness (F = 3.82, P < 0.0001) (Tables 1, 3). Collectively, these findings indicate that plant genotypes with telomeres on the opposite ends of the length spectrum show significant variation in reproductive fitness.
As an additional measure of reproductive success, we explored seed quality by comparing germination efficiency of seeds produced by all genotypes grown in wet|22°C and mod|22°C. Because most seeds produced in wet|30°C did not separate well from siliques, seeds from this treatment were omitted from the analysis. Overall, > 79% of all seeds germinated irrespective of the genotype or treatment they originated from (Fig. S6), and we did not detect significant genotypic (F = 0.99, P = 0.44), treatment (F = 0, P = 0.99) or G × E (F = 0.49, P = 0.75) effects. These results confirm that total seed count is an accurate and consistent proxy for reproductive fitness, and that telomere length mutants tested here do not have major defects in seed quality.
Evidence of genotype by environment interactions leading to life history trade-offs
We discovered several instances of G × E interactions, implying that telomere length may differentially impact plant phenotypes depending on the environment (Table 1, 3). Of particular note is a G × E interaction for reproductive fitness (Table 3), driven by clear rank changes in seed set for genotypes with the most extreme telomere sizes, tert and ku70 (Fig. 2f). Overall, these G × E interactions indicate that telomere length variation can exact environment-dependent fitness costs. Thus, the natural genetic variation for telomere length previously observed in plants (Choi et al., 2021) may be maintained through varying selection pressures imposed by different environmental conditions.
Scanning across all measured phenotypes revealed interesting patterns that may partially account for the observed fitness G × E interactions. For example, in line with higher reproductive fitness, ku70 and tert individuals had a higher stomatal index (SI) than all other genotypes in the wet|30°C environment (Fig. 2g), indicating that these genotypes develop a higher ratio of stomatal to epidermal cell density on their leaf surfaces. Given that stomata regulate transpiration and, by extension, leaf cooling (Nilson & Assmann, 2007; Daszkowska-Golec & Szarejko, 2013), higher SI in ku70 and tert may enable plants to maintain slightly cooler leaves, conferring a physiological advantage at higher temperatures. In support of SI as a driver of fitness differences, Col-0 individuals exhibited lower SI in the cooler and dryer mod|22°C treatment (Fig. 2g), where it is presumably beneficial to conserve water, potentially explaining why the wild-type outperformed most other genotypes in this environment (Fig. 2f). In addition, in the wet|30°C environment ku70 individuals developed fewer epidermal cells (Fig. S5C), further contributing to a high SI, while tert individuals developed smaller epidermal cells overall (Fig. S5D), suggesting that other leaf anatomical features may also be contribute to an elevated fitness in this environment. Further experiments that explore transpiration, gas exchange and leaf temperature regulation may provide insight into how cell size and structure influence the physiological and fitness trade-offs of these genotypes in varying environments.
Phenotypic response strength is proportional to telomere length divergence from wild-type
To further address correlations between telomere length and measured traits, phenotypic ‘response strength’ was defined as the absolute (positive or negative) divergence from the Col-0 wild-type trait values and was plotted for all genotypes, arranged from the shortest (tert) to the longest (ku70) average telomere length (Figs 3, S7). For several traits, we detected a U-shape profile, with the shortest and the longest telomere genotypes having the most variation from the Col-0 norm. For reproductive fitness (seeds), this effect was most apparent and was observed in all three environmental growth conditions (Fig. 3a). For flowering time, relative water content RWC and OP (Fig. 3b–d), and for several other plant phenological traits (Fig. S7), the U-shape response was also observed in one or more conditions. Overall, the ‘divergence from the wild-type’ analysis demonstrated a strong correlation between the extent of telomere length change and the degree of measured phenotypic response, with trait variation generally greater in genotypes that differ the most from the Col-0 wild-type norm (tert and ku70). Furthermore, these phenomena span multiple biological levels, from plant morphology and physiology to development to fitness, and can be detected in one, two or all three environments. We conclude that genotypes with the highest degree of deviation from the Col-0 telomere length set point in either direction show the largest effect on plant biology traits.
Fig. 3.
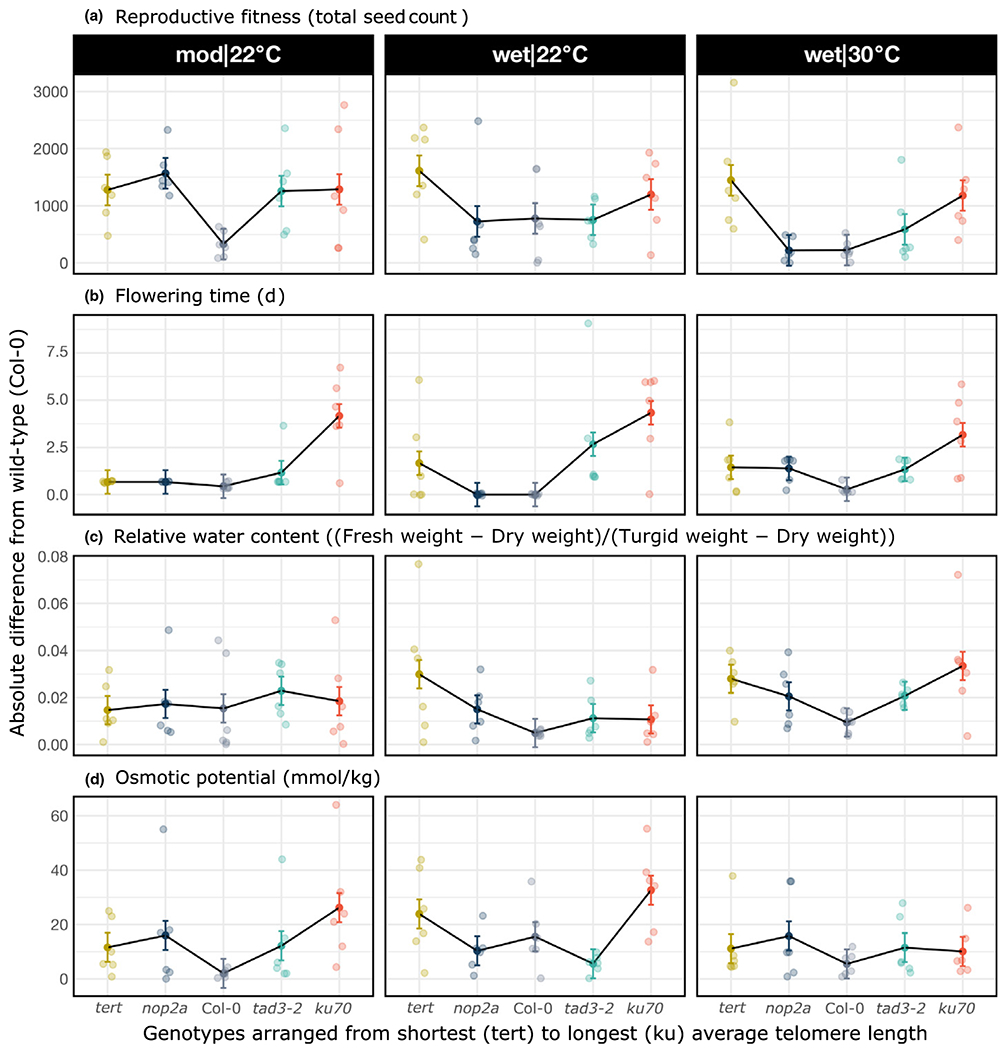
Analysis of response strength in Arabidopsis plant traits. Response strength is defined as the absolute (i.e. positive or negative) variation from the Col-0 wild-type trait values. Response strength was analyzed for reproductive fitness (a), flowering time (b), relative water content (c), and osmotic potential (d). Trait values of the Col-0 genotype were subtracted from the corresponding values for all other genotypes, transformed to positive values, and plotted, with genotypes from left to right arranged from the shortest (tert) to the longest (ku70) telomere length. Points show least-squared means ± SE.
Discussion
In humans, the subject of telomere length dynamics and plasticity in response to environmental or lifestyle factors has received substantial attention due to compelling connections to aging and disease (Victorelli & Passos, 2017). Consistent with these findings, our data show that telomere length in plant somatic tissues (leaves) decreases slightly over time in all environmental conditions tested. Interestingly, most cells in mature leaves are noncycling, and hence telomere attrition in leaves is not replication dependent and is, possibly, stochastic in nature. In marked contrast to several animal studies (Haussmann & Marchetto, 2010; Horn et al., 2010), we found that Arabidopsis genotypes with short and medium telomere length displayed little evidence for telomere length plasticity in response to environmental conditions. However, the long-telomere ku70 mutant emerged as an outlier, exhibiting some evidence of additional telomere shortening in response to the dry mod|22°C environment. Abnormally long telomeres in Arabidopsis ku70 mutants are inherently unstable and prone to trimming through a telomere rapid deletion-like mechanism (Watson & Shippen, 2007). Notably, abrupt telomere shortening in Arabidopsis has also been reported for mutants of a nucleosome remodeler, DDM1, and a chromosome end protection factor, TEN1, in response to environmental and genome-wide stressors (Lee et al., 2016; Xie & Shippen, 2018). These observations suggest that the plastic telomere length response observed in ku70 mutants under stress may be more prominent in mutant plants with major defects in telomere homeostasis.
In humans and animals, short telomere length is indicative of poor biological state, higher disease risk, poor survival and low reproductive success (Haussmann et al., 2005; Pauliny et al., 2006; Kappei & Londono-Vallejo, 2008; Bize et al., 2009). Similarly, we observe major changes in plant physiology and fitness associated with short telomeres. Under the control wet|22°C conditions, short telomere tert mutants showed significant changes in leaf physiology parameters, such as reduced leaf water content and OP, and yet surprisingly also displayed higher reproductive fitness. Since the tert mutants used in our study do not yet show major developmental or reproductive defects (Riha et al., 2001), our findings imply that telomere length status is consistent with pleiotropic effects on many physiological and fitness parameters.
Less is known about the fitness effects of abnormally long telomeres. Elongated telomeres in humans are rare and are typically associated with cancer (Stanley & Armanios, 2015; Gong et al., 2020; Schmutz et al., 2020). Studies in yeast revealed no fitness impacts of longer telomeres (Harari & Kupiec, 2018). Similarly, longer telomeres in wild Caenorhabditis elegans isolates do not correlate with alterations in offspring production or longevity (Cook et al., 2016). While we observed no substantial vegetative fitness (biomass) effects in long-telomere Arabidopsis ku70 mutants, we did detect significant and complex reproductive fitness associations for seed counts in these plants. ku70 mutants set fewer seeds under the control wet|22°C growth conditions than wild-type plants or mutants with shorter or medium telomeres, but unlike most other genotypes they remained relatively fertile in the most extreme wet|30°C environment. These fitness effects were accompanied by significant changes in plant physiology (higher OP), development (later flowering) and other traits in all growth conditions. Thus, our analysis of ku70 mutants for the first time uncovered interesting correlations between very long telomeres and significant changes in fitness and physiology, connecting improper telomere elongation with both positive and negative changes in plant biology, depending on the environment.
Our analyses detected widespread pleiotropy and provided clear evidence that mutations affecting telomere length genes also impact many other phenotypes. Signatures of functional pleiotropy for telomere length genes have been described in different eukaryotes (Gatbonton et al., 2006; Liu et al., 2010; Abdulkina et al., 2019), and thus appear to be evolutionarily conserved. The TERT gene is a classic example. This gene is essential for telomere elongation, but it is also implicated in mitochondrial biology (Sharma et al., 2012). Similarly, Ku functions in both DNA damage repair and telomere protection (Boulton & Jackson, 1996; Kazda et al., 2012), while NOP2A impacts both cell size and telomere length (Fujikura et al., 2009; Abdulkina et al., 2019).
Despite evidence of multiple pleiotropic actions of telomere-related genes, it is unknown whether such outcomes occur directly or indirectly, and whether they function in linear or parallel fashions (Paaby & Rockman, 2013; Solovieff et al., 2013). Several mechanisms can be envisioned. For example, telomere length impact on flowering time, now detected in three plant species (this study; Choi et al., 2021) could occur in a linear sequential fashion (gene mutations > telomere length > flowering time) or in parallel modes (i.e. gene mutations > telomere length and, independently, gene mutations > flowering time). Both mechanisms are supported by the available literature. Human NOP2 and TERT have nontelomere functions in cell cycle control through direct transcriptional activation of G1 phase cyclin D1 expression (Hong et al., 2016). Similarly, the direct pleiotropic effects of mutations in Dyskerin and NOP10, which function in telomere biology and ribosomal RNA maturation, could account for the remarkable similarities in human telomere disorders and diseases of ribosome biogenesis (Orsolic et al., 2016). Conversely, it is unlikely that hundreds of yeast genes previously discovered in genetic telomere length screens all have a direct role in telomere metabolism (Askree et al., 2004; Gatbonton et al., 2006; Liu et al., 2010). Hence, the question of causation needs to be carefully considered. While telomere length could causally drive differences in performance, an alternative indirect mechanism through pleiotropy is also plausible. A better understanding of the molecular mechanisms of telomere length control is needed to disentangle the many different aspects of pleiotropy observed in our study.
Our previous analysis of telomere length variation in 653 natural Arabidopsis accessions indicated that genotypes with longer telomeres flower earlier (Choi et al., 2021). These experiments provided preliminary evidence that telomere length may be an adaptive trait in plants and suggested an intriguing link between life history strategies and chromosome structure. Curiously, the data we present here are consistent with the opposite outcome – mutants with longer telomeres flower later. Several scenarios can be envisioned to reconcile these differences. First, the observed contrast may simply reflect differences in the experimental approaches and the choice of Arabidopsis genotypes: T-DNA mutant plants experimentally subjected to different growth regimes (this study) vs a purely computational study with natural Arabidopsis accessions (Choi et al., 2021). Second, the flowering time data for the Choi et al. (2021) study was collected for lower temperatures (10° and 16°C), while T-DNA mutants in this study were grown at higher temperatures (22°C and 30°C) and, additionally, in contrasting soil moisture regimes. Thus, both growth temperature and water availability could have largely contributed to the differences in flowering phenotypes. One approach to reconcile these differences might be to analyze a panel of flowering time Arabidopsis mutants for variation in telomere length set point, essentially reversing this study. These tests may contribute to experimental reconciliation of our data, as they will utilize T-DNA mutants (as in this study), but with mutations in major genes known to affect flowering time in natural Arabidopsis accessions (like the ones used by Choi et al. (2021)). Nevertheless, our new findings confirm the major role of telomere length homeostasis in plant life history strategies, including the developmental decision to initiate flowering.
Our data are also consistent with a recent ‘pace-of-life’ theory addressing the evolution of life history strategies as they relate to telomere length (Giraudeau et al., 2019). According to this hypothesis, shorter telomeres would correspond to a faster pace-of-life strategy with less investment in self-maintenance as a means of conserving more energy for reproduction. Inversely, longer telomeres could be indicative of increased investment in soma maintenance and late reproduction. Our findings support this model. Under optimal wet|22°C conditions, the medium and short telomere genotypes flowered faster than the long-telomere ku70 mutants, while producing significantly more seeds (especially the shortest-telomere tert mutant). By contrast, long-telomere ku70 mutants favor a long-term strategy with a reduced investment in reproduction (less seeds under control wet|22°C conditions), but an increased allocation of resources toward self-maintenance processes (late flowering).
Genotype by environment interactions are hallmarks of species adaptations to specific environmental conditions and have been proposed to play a role in telomere biology (Dugdale & Richardson, 2018). We found significant telomere length G × E effects on many plant traits, including reproductive fitness. We postulate that the reproductive fitness G × E, and the relatively higher performance of ku70 and tert in wet|30°C, at least partially reflects changes in leaf SI and other anatomical traits. Our data further suggest that the benefits of a particular telomere length in one environment, which occur as a result of its impact on specific phenotypes (e.g. SI), may come at a cost to performance in alternative environments, as a result of its impact on other phenotypes. For example, the ku70 mutants in the more stressful environment (i.e. wet|30°C) produce more seeds than other genotypes in our study, suggesting that in this environment longer telomeres provide selective advantages. However, early flowering is often favored by natural selection (Franks et al., 2007; Austen et al., 2017), which may explain why late-flowering ku70 plants exhibit lower fitness in the cooler mod|22°C and wet|22°C conditions.
Overall, our experimental findings provide intriguing evidence for the existence of life history trade-offs associated with a differential impact of telomere length variation (or the genetic variants underlying telomere length) on leaf anatomy, physiology and reproductive fitness under different environmental conditions. We identify heavy competitive costs between one such trade-off pair, flowering time and the number of seeds, providing the first evidence that the maintenance of overly long telomeres can indeed be advantageous or unfavorable, depending on the environment, and can substantially impact life history strategies.
Supplementary Material
Fig. S1 Soil water content of pots with different Arabidopsis genotypes grown under control (wet|22°C), moderately dry (mod|22°C) and high-temperature (wet|30°C) environments throughout the experiment.
Fig. S2 Principal component analysis of measured Arabidopsis traits.
Fig. S3 Representative primer extension telomere repeat amplification assays using primers specific to individual Arabidopsis chromosome arms.
Fig. S4 Average telomere length changes across all three tested chromosome arms in Arabidopsis genotypes grown in three environmental conditions.
Fig. S5 Analysis of additional physiological and anatomical leaf parameters in Arabidopsis genotypes grown in three abiotic environments.
Fig. S6 Germination efficiency of seeds produced by different Arabidopsis genotypes grown in wet|22°C and mod|22°C environments.
Fig. S7 Analysis of response strength in additional plant traits.
Methods S1 Additional methods information.
Table S1 Arabidopsis genotypes used in the study.
Table S2 List of raw and composite traits used in the analyses.
Dataset S1 Raw dataset for all plant traits.
Dataset S2 Imputed dataset for all plant traits.
Dataset S3 Univariate analyses of variance (ANOVAs) of raw trait data.
Dataset S5 Output of different methods of exploring the differences from wild-type (Col-0) plants.?
Dataset S4 Boxplots for imputed values for all 32 analyzed plant traits.
Acknowledgements
We thank Marisa Fajardo and Galina Aglyamova for help with plant growth, and other members of our labs for insightful discussions. This work was supported in part by the National Institutes of Health (R01 GM065383 to DES; R01 GM127402 to EVS), National Science Foundation (IOS-0922457 to TEJ) and the Kazan Federal University Strategic Academic Leadership Program. Russian Science Foundation project 21-14-00147 supported OLI2/NOP2A genotype data collection and analysis by LRA. The authors declare no competing financial interests.
Footnotes
Supporting Information
Additional Supporting Information may be found online in the Supporting Information section at the end of the article.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data availability
The data that support the findings of this study are available in the Supporting Information documents that accompany this article.
References
- Abdulkina LR, Kobayashi C, Lovell JT, Chastukhina IB, Aklilu BB, Agabekian IA, Suescún AV, Valeeva LR, Nyamsuren C, Aglyamova GV et al. 2019. Components of the ribosome biogenesis pathway underlie establishment of telomere length set point in Arabidopsis. Nature Communications 10: 5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiard S, Da Ines O, Gallego ME, White CI. 2014. Responses to telomere erosion in plants. PLoS ONE 9: e86220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar M, Hasselquist D, Hansson D, Zehtindjiev P, Westerdahl H, Bensch S. 2015. Chronic infection. Hidden costs of infection: chronic malaria accelerates telomere degradation and senescence in wild birds. Science 347: 436–438. [DOI] [PubMed] [Google Scholar]
- Askree SH, Yehuda T, Smolikov S, Gurevich R, Hawk J, Coker C, Krauskopf A, Kupiec M, McEachern MJ. 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proceedings of the National Academy of Sciences, USA 101: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austen EJ, Rowe L, Stinchcombe JR, Forrest JRK. 2017. Explaining the apparent paradox of persistent selection for early flowering. New Phytologist 215: 929–934. [DOI] [PubMed] [Google Scholar]
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D. 2003. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nature Genetics 33: 203–207. [DOI] [PubMed] [Google Scholar]
- Bauch C, Becker PH, Verhulst S. 2014. Within the genome, long telomeres are more informative than short telomeres with respect to fitness components in a long-lived seabird. Molecular Ecology 23: 300–310. [DOI] [PubMed] [Google Scholar]
- Bize P, Criscuolo F, Metcalfe NB, Nasir L, Monaghan P. 2009. Telomere dynamics rather than age predict life expectancy in the wild. Proceedings of the Royal Society of London. Series B: BiologicalSciences 276: 1679–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA. 2005. Mice with bad ends: mouse models for the study of telomeres and telomerase in cancer and aging. EMBO Journal 24: 1095–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnell E, Pasquier E, Wellinger RJ. 2021. Telomere replication: solving multiple end replication problems. Frontiers in Cell and Developmental Biology 9: 668171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S, Suescún AV, Song J, Castillo-Gonzalez C, Aklilu BB, Branham E, Lynch R, Shippen DE. 2020. tRNA ADENOSINE DEAMINASE 3 is required for telomere maintenance in Arabidopsis thaliana. Plant Cell Reports 39: 1669–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Research 24: 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campitelli BE, Des Marais DL, Juenger TE. 2016. Ecological interactions and the fitness effect of water-use efficiency: competition and drought alter the impact of natural MPK12 alleles in Arabidopsis. Ecology Letters 19: 424–434. [DOI] [PubMed] [Google Scholar]
- Chen J, Burke JJ, Velten J, Xin Z. 2006. FtsH11 protease plays a critical role in Arabidopsis thermotolerance. The Plant Journal 48: 73–84. [DOI] [PubMed] [Google Scholar]
- Choi JY, Abdulkina LR, Yin J, Chastukhina IB, Lovell JT, Agabekian IA, Young PG, Razzaque S, Shippen DE, Juenger TE et al. 2021. Natural variation in plant telomere length is associated with flowering time. Plant Cell 33: 1118–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd V, Nelson CP, Albrecht E, Mangino M, Deelen J, Buxton JL, Hottenga JJ, Fischer K, Esko T, Surakka I et al. 2013. Identification of seven loci affecting mean telomere length and their association with disease. Nature Genetics 45: 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Zdraljevic S, Tanny RE, Seo B, Riccardi DD, Noble LM, Rockman MV, Alkema MJ, Braendle C, Kammenga JE et al. 2016. The genetic basis of natural variation in Caenorhabditis elegans telomere length. Genetics 204: 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszkowska-Golec A, Szarejko I. 2013. Open or close the gate – stomata action under the control of phytohormones in drought stress conditions. Frontiers in Plant Science 4: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E et al. 2004. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117: 873–886. [DOI] [PubMed] [Google Scholar]
- Dugdale HL, Richardson DS. 2018. Heritability of telomere variation: it is all about the environment! Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences 373: 20160450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. 1999. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proceedings of the National Academy of Sciences, USA 196: 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE. 2007. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proceedings of the National Academy of Sciences, USA 104: 1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H. 2009. Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. The Plant Journal 59: 499–508. [DOI] [PubMed] [Google Scholar]
- Gallego ME, Jalut N, White CI. 2003. Telomerase dependence of telomere lengthening in Ku80 mutant Arabidopsis. Plant Cell 15: 782–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genetics 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudeau M, Angelier F, Sepp T. 2019. Do telomeres influence pace-of-life-strategies in response to environmental conditions over a lifetime and between generations? BioEssays 41: e1800162. [DOI] [PubMed] [Google Scholar]
- Göhring J, Fulcher N, Jacak J, Riha K. 2014. A new tool for telomere length measurement from terminal restriction fragment analysis with improved probe intensity correction. Nucleic Acids Research 42: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Stock AJ, Liu Y. 2020. The enigma of excessively long telomeres in cancer: lessons learned from rare human POT1 variants. Current Opinion in Genetics & Development 60: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari Y, Kupiec M. 2018. Do long telomeres affect cellular fitness? Current Genetics 64: 173–176. [DOI] [PubMed] [Google Scholar]
- Harari Y, Romano GH, Ungar L, Kupiec M. 2013. Nature vs nurture: interplay between the genetic control of telomere length and environmental factors. Cell Cycle 12: 3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussmann MF, Marchetto NM. 2010. Telomeres: linking stress and survival, ecology and evolution. Current Zoology 56: 714–727. [Google Scholar]
- Haussmann MF, Winkler DW, Vleck CM. 2005. Longer telomeres associated with higher survival in birds. Biology Letters 1: 212–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. 2004. Molecular analysis of telomere fusions in Arabidopsis: multiple pathways for chromosome end-joining. EMBO Journal 23: 2304–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Greider CW. 2000. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Research 28: 4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Lee JH, Chung IK. 2016. Telomerase activates transcription of cyclin D1 gene through an interaction with NOL1. Journal of Cell Science 129: 1566–1579. [DOI] [PubMed] [Google Scholar]
- Horn T, Robertson BC, Gemmell NJ. 2010. The use of telomere length in ecology and evolutionary biology. Heredity 105: 497–506. [DOI] [PubMed] [Google Scholar]
- Kannan K, Nelson AD, Shippen DE. 2008. Dyskerin is a component of the Arabidopsis telomerase RNP required for telomere maintenance. Molecular and Cellular Biology 28: 2332–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappei D, Londono-Vallejo JA. 2008. Telomere length inheritance and aging. Mechanisms of Ageing and Development 129: 17–26. [DOI] [PubMed] [Google Scholar]
- Kazda A, Zellinger B, Rössler M, Derboven E, Kusenda B, Riha K. 2012. Chromosome end protection by blunt-ended telomeres. Genes & Development 26: 1703–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Xie X, Yang K, Zhang J, Lee SY, Shippen DE. 2016. Dynamic interactions of Arabidopsis TEN1: stabilizing telomeres in response to heat stress. Plant Cell 28: 2212–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Neuhausen SL, Hunt SC, Kimura M, Hwang S-J, Chen W, Bis JC, Fitzpatrick AL, Smith E, Johnson AD et al. 2010. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proceedings of the National Academy of Sciences, USA 107: 9293–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell R, Milliken G, Stroup W, Wolfinger R, Schabenberger O. 2006. SAS for mixed models, vol. 53. Cary, NC, USA: SAS Institute. [Google Scholar]
- Liu N-N, Han TX, Du L-L, Zhou J-Q. 2010. A genome-wide screen for Schizosaccharomyces pombe deletion mutants that affect telomere length. Cell Research 20: 963–965. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, de Lange T. 2017. Telomeres in cancer: tumour suppression and genome instability. Nature Reviews Molecular Cell Biology 18: 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan P, Haussmann MF. 2006. Do telomere dynamics link lifestyle and lifespan? Trends in Ecology & Evolution 21: 47–53. [DOI] [PubMed] [Google Scholar]
- Nilson SE, Assmann SM. 2007. The control of transpiration. Insights from Arabidopsis. Plant Physiology 143: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Wapstra E, Friesen CR. 2018. Evolutionary ecology of telomeres: a review. Annals of the New York Academy of Sciences 1422: 5–28. [DOI] [PubMed] [Google Scholar]
- Orsolic I, Jurada D, Pullen N, Oren M, Eliopoulos AG, Volarevic S. 2016. The relationship between the nucleolus and cancer: current evidence and emerging paradigms. Seminars in Cancer Biology 37–38: 36–50. [DOI] [PubMed] [Google Scholar]
- Paaby AB, Rockman MV. 2013. The many faces of pleiotropy. Trends in Genetics 29: 66–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauliny A, Wagner RH, Augustin J, Szép T, Blomqvist D. 2006. Age-independent telomere length predicts fitness in two bird species. Molecular Ecology 15: 1681–1687. [DOI] [PubMed] [Google Scholar]
- Riha K, McKnight TD, Griffing LR, Shippen DE. 2001. Living with genome instability: plant responses to telomere dysfunction. Science 291: 1797–1800. [DOI] [PubMed] [Google Scholar]
- Riha K, Shippen DE. 2003. Ku is required for telomeric C-rich strand maintenance but not for end-to-end chromosome fusions in Arabidopsis. Proceedings of the National Academy of Sciences, USA 100: 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riha K, Watson JM, Parkey J, Shippen DE. 2002. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO Journal 21: 2819–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano GH, Harari Y, Yehuda T, Podhorzer A, Rubinstein L, Shamir R, Gottlieb A, Silberberg Y, Pe’er D, Ruppin E et al. 2013. Environmental stresses disrupt telomere length homeostasis. PLoS Genetics 9: e1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner SM. 2001. MANOVA: multiple response variables and multispecies interactions. In: Scheiner SM, Gurevitch J, eds. Design and analysis of ecological experiments, 2nd edn. NewYork, NY, USA: Oxford University Press, 99–115. [Google Scholar]
- Schmutz I, Mensenkamp AR, Takai KK, Haadsma M, Spruijt L, de Voer RM, Choo SS, Lorbeer FK, van Grinsven EJ, Hockemeyer D et al. 2020. TINF2 is a haploinsufficient tumor suppressor that limits telomere length. eLife 9: e61235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakirov EV, Shippen DE. 2004. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16: 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma NK, Reyes A, Green P, Caron MJ, Bonini MG, Gordon DM, Holt IJ, Santos JH. 2012. Human telomerase acts as a hTR-independent reverse transcriptase in mitochondria. Nucleic Acids Research 40: 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. 2013. Pleiotropy in complex traits: challenges and strategies. Nature Reviews Genetics 14: 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SE, Armanios M. 2015. The short and long telomere syndromes: paired paradigms for molecular medicine. Current Opinion in Genetics & Development 33: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. 2007. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO Journal 26: 3653–3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Badiger BG, Kesari R, Kumar MN. 2014. Drought tolerance mechanisms and their molecular basis. In: Jenks MA, Hasegawa PM, eds. Plant abiotic stress, 2nd edn. New York, NY, USA: John Wiley & Sons, 15–46. [Google Scholar]
- Victorelli S, Passos JF. 2017. Telomeres and cell senescence – size matters not. EBioMedicine 21: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JM, Shippen DE. 2007. Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Molecular and Cellular Biology 27: 1706–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE, Shay JW. 2005. Telomere biology in aging and cancer. Journal of the American Geriatrics Society 53: S292–S294. [DOI] [PubMed] [Google Scholar]
- Xie X, Shippen DE. 2018. DDM1 guards against telomere truncation in Arabidopsis. Plant Cell Reports 37: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Soil water content of pots with different Arabidopsis genotypes grown under control (wet|22°C), moderately dry (mod|22°C) and high-temperature (wet|30°C) environments throughout the experiment.
Fig. S2 Principal component analysis of measured Arabidopsis traits.
Fig. S3 Representative primer extension telomere repeat amplification assays using primers specific to individual Arabidopsis chromosome arms.
Fig. S4 Average telomere length changes across all three tested chromosome arms in Arabidopsis genotypes grown in three environmental conditions.
Fig. S5 Analysis of additional physiological and anatomical leaf parameters in Arabidopsis genotypes grown in three abiotic environments.
Fig. S6 Germination efficiency of seeds produced by different Arabidopsis genotypes grown in wet|22°C and mod|22°C environments.
Fig. S7 Analysis of response strength in additional plant traits.
Methods S1 Additional methods information.
Table S1 Arabidopsis genotypes used in the study.
Table S2 List of raw and composite traits used in the analyses.
Dataset S1 Raw dataset for all plant traits.
Dataset S2 Imputed dataset for all plant traits.
Dataset S3 Univariate analyses of variance (ANOVAs) of raw trait data.
Dataset S5 Output of different methods of exploring the differences from wild-type (Col-0) plants.?
Dataset S4 Boxplots for imputed values for all 32 analyzed plant traits.
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information documents that accompany this article.


