Summary
Conventional dendritic cells (cDCs), cDC1 and cDC2, act both to initiate immunity and maintain self-tolerance. The tryptophan metabolic enzyme indoleamine 2,3-dioxygenase 1 (IDO1) is used by cDCs in maintaining tolerance, but its role in different subsets remains unclear. At homeostasis, only mature CCR7+ cDC1 expressed IDO1 that was dependent on IRF8. Lipopolysaccharide treatment induced maturation and IDO1-dependent tolerogenic activity in isolated immature cDC1, but not isolated cDC2. However, both human and mouse cDC2 could induce IDO1 and acquire tolerogenic function when co-cultured with mature cDC1 through the action of cDC1-derived l-kynurenine. Accordingly, cDC1-specific inactivation of IDO1 in vivo exacerbated disease in experimental autoimmune encephalomyelitis. This study identifies a previously unrecognized metabolic communication in which IDO1-expressing cDC1 cells extend their immunoregulatory capacity to the cDC2 subset through their production of tryptophan metabolite l-kynurenine. This metabolic axis represents a potential therapeutic target in treating autoimmune demyelinating diseases.
Keywords: immunotolerance, dendritic cells, tryptophan metabolism, IDO1, metabolites, kynurenine, IL-6, AhR, RelB, neuroinflammation
Graphical abstract
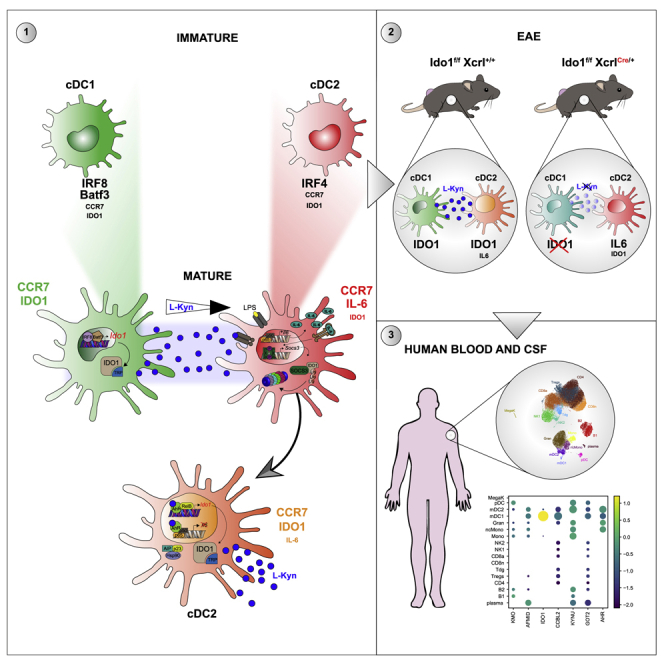
Highlights
-
•
The tolerogenic IDO1 pathway is expressed in mature cDC1 but not in cDC2
-
•
Mature IDO1+ cDC1 are regulatory in vitro and in vivo
-
•
IDO1 competent cDC1 induce regulatory cDC2 via Trp metabolism
-
•
l-kynurenine recruits AhR competent cDC2 into a tolerogenic pool
Activation of the tryptophan metabolic enzyme indoleamine 2,3-dioxygenase 1 (IDO1) induces DC tolerance, but how this pathway is used by selected cDC subsets is currently unclear. Gargaro et al. show that activation of the IDO1 pathway, which is expressed in mature cDC1 but not in cDC2, induces regulatory cDC2 via AhR-mediated metabolic communication.
Introduction
Dendritic cells (DCs) play a vital role in innate and adaptive host immunity, efficiently priming naive lymphocytes (Bošnjak et al., 2022; Steinman and Hemmi, 2006). However, DCs are also important in maintaining immune homeostasis and self-tolerance (Pulendran et al., 2000; Robertson et al., 2021; Steinman et al., 2003). The type of DC as well as the extent of maturation or activation can profoundly affect their capability to induce immunity or tolerance (Merad et al., 2013). Distinct types of DC subsets have different functions in local immunosurveillance, migration, and antigen presentation (Durai and Murphy, 2016). Under steady-state conditions, DCs consist of three major types, based on developmental origin, surface markers, and functions. In particular, plasmacytoid DCs (pDCs) produce high amounts of type-1 interferon (IFN) (Blasius et al., 2006), whereas conventional or classical DCs (cDCs) comprise two major subsets, conventional type-1 DCs (cDC1s) and conventional type-2 DCs (cDC2s). cDC1 (Naik et al., 2007) are specialized for cross-presentation and CD8+ T-cell priming (den Haan et al., 2000) and express CD8α and CD24 in mouse spleens or CD103 in the periphery (Askew and Harding, 2008). Development of the cDC1 lineage requires the transcription factors Irf8 (Tamura et al., 2005) and Id2 (Hacker et al., 2003; Spits et al., 2000). cDC2s express CD172 or CD11b in lymphoid organs and are characterized by expression of the transcription factor Irf4 (Suzuki et al., 2004). cDC subset’s ability to induce immune activation or tolerance may be influenced by environmental conditions (Pulendran, 2015).
One mechanism by which DCs regulate tolerance involves the enzyme indoleamine 2,3-dioxygenase 1 (IDO1). IDO1 is a tryptophan (Trp)-metabolizing enzyme, which exerts potent immunoregulatory effects when expressed in DCs (Grohmann et al., 2003; Sharma et al., 2021). IDO1 is the rate-limiting enzyme in the Trp metabolic pathway, which produces a series of immunoregulatory molecules, known as kynurenines (Mellor and Munn, 2004; Puccetti and Grohmann, 2007; Terness et al., 2002). Both Trp depletion and kynurenine production are involved in the conversion of naive CD4+ T cells into Foxp3+ regulatory T (Treg) cells (Fallarino et al., 2006). In addition, the main IDO1 Trp metabolite, l-kynurenine (l-kyn), exerts tolerogenic effects by activating the aryl hydrocarbon receptor (AhR) (Mezrich et al., 2010; Opitz et al., 2011), a ligand-activated transcription factor and immune sensor in several immune cells, including DCs (Di Meglio et al., 2014). AhR activation by the prototype ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) or by l-kyn induces expression of IDO1 in human and mouse DCs, resulting in an increase of Treg cells (Manni et al., 2020).
DCs induce high amounts of IDO1 mRNA in response to stimulation by cytokines such as type-I and type-II IFNs, transforming growth factor β (TGF-β) (Pallotta et al., 2011), and by Toll-like receptor (TLR) ligands, including lipopolysaccharide (LPS) (Bessede et al., 2014). IDO1 in DCs is also controlled by post-transcriptional mechanisms. IL-6 impairs DC tolerogenic activity by inducing proteasome-dependent IDO1 degradation through a suppressor of cytokine signaling 3 (SOCS3)-dependent mechanism (Orabona et al., 2008). Because different cDC subsets use distinct transcriptional programs (Murphy et al., 2016), they may conceivably regulate IDO1 expression in a differential fashion in response to disparate stimuli. Crosstalk among distinct DC subsets has been proposed (den Haan et al., 2000; Yoneyama et al., 2005). However, its impact in regulating DC tolerogenic functions has not been explored. Because AhR and IDO1 are critical regulators of tolerance, their regulation in different DC subsets is of potential significance.
Although sparce data suggest that Trp metabolism might be preferentially expressed in specific DC subsets (Ardouin et al., 2016), the differential regulation of the IDO1 pathway in selected DC subsets and their impact in conditioning immunogenic or tolerogenic functions in other DC subsets has not been explored. In this study, using physiologically relevant in vitro and in vivo genetic approaches, we investigated the mechanisms by which various DC subsets acquire the IDO1 tolerogenic pathway and respond to immune-active Trp metabolites. Our study highlights principles underlying selective control of inducible IDO1 in DC subsets and identifies a Trp metabolite for cDC2 immune education.
Results
LPS induces IDO1 in purified cDC1, but not in cDC2 nor pDCs
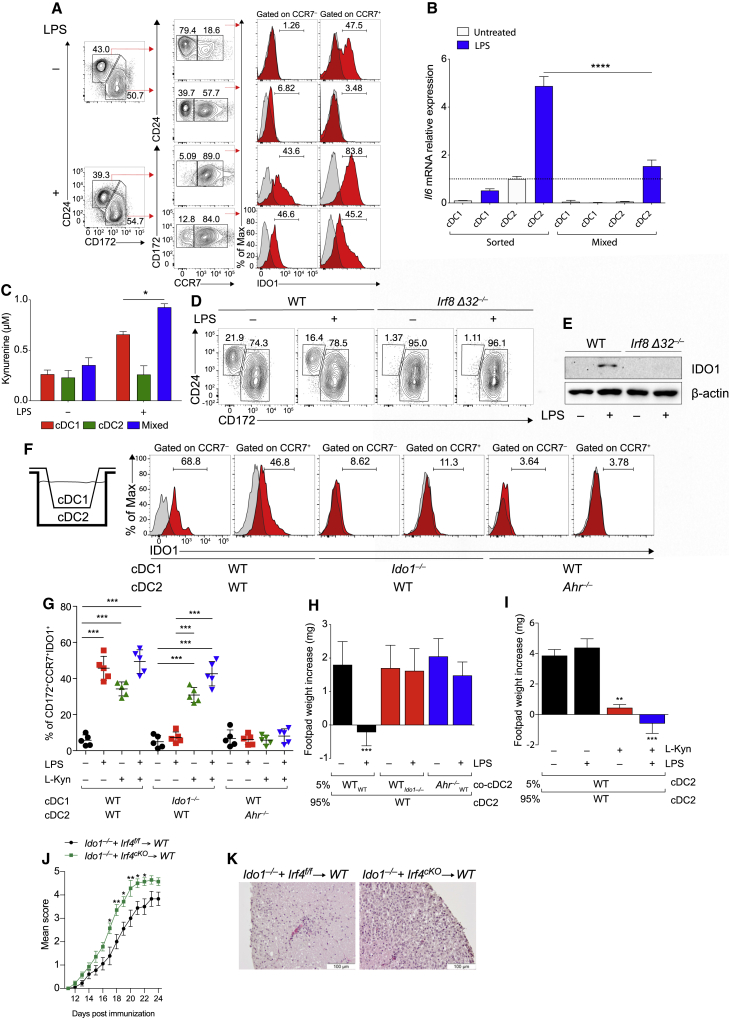
Both human and murine DCs express IDO1 in response to LPS stimulation (Nguyen et al., 2010; Von Bubnoff et al., 2011). Using an antibody specifically reactive to murine IDO1, but not IDO2 or TDO2 (Figure S1A), we observed a low baseline expression of IDO1 protein in untreated cDCs, from Flt3L-treated bone marrow (BMDC) cultures (Figure S1B), which increased at 48 h of LPS stimulation (Figure S1C). We developed an intracellular staining (IS) assay to detect IDO1 expression (Figure S1D). We found that both cDC1 and cDC2 were able to upregulate IDO1 upon LPS treatment (Figure 1A).
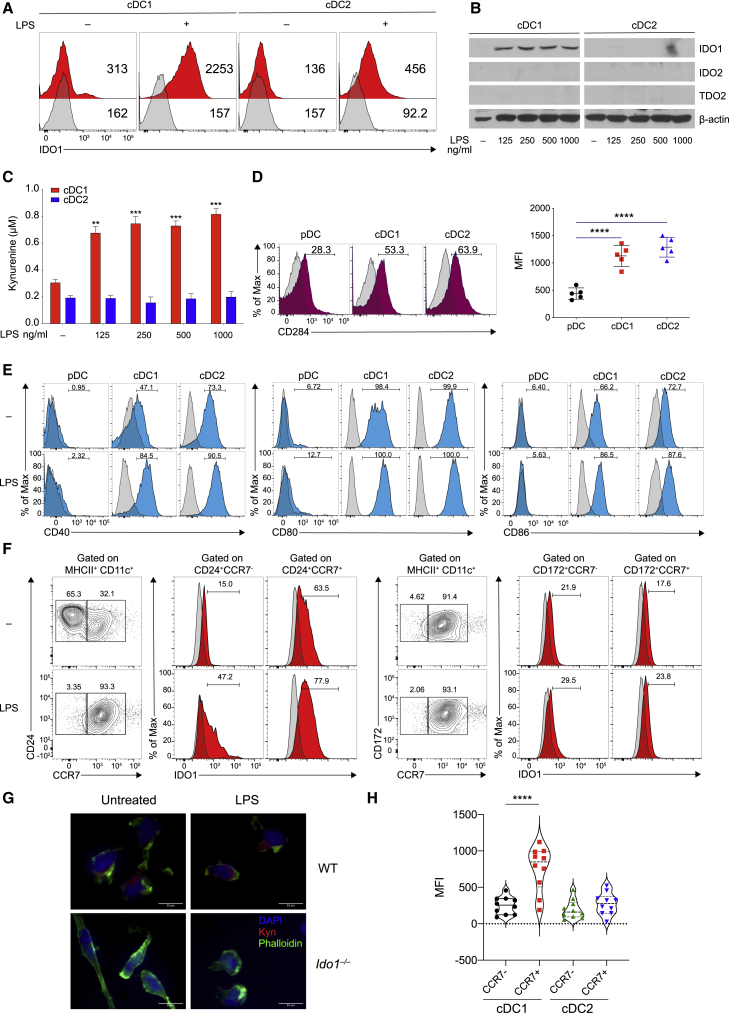
Figure 1.
IDO1 is selectively induced in cDC1 cells following LPS stimulation
(A) IDO1 expression was analyzed by IS in BMDCs (n = 5).
(B) Immunoblot analysis was carried out for IDO1, IDO2, TDO2, and β-actin expression (n = 3).
(C) Supernatants from cells prepared as in (B) were analyzed for l-kynurenine content by HLPC.
(D and E) Flow plot (left) and quantification (right) of CD284 (D), and flow plot of CD40, CD80, and CD86 (E) on DC subsets (n = 3).
(F) BM-derived cDC1 and cDC2 were treated as in (B) and IDO1 expression evaluated in CCR7− and CCR7+ populations treated as in (B), pre-gated on cDC1 and cDC2 (n = 3).
(G) Immunofluorescence analysis of l-kynurenine expression in sorted CCR7+ cDC1 of different genotypes treated as in (B) (n = 3).
(H) IDO1 expression (MFI) in thymic CCR7− and CCR7+ dendritic cell subsets, gated on CD11c+MHCII+XCRI+CD117− and CD11c+MCHII+CD172+CD117− (n = 3).
Data are shown as mean ± SD. ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, one-way (D and H) or two-way (C) ANOVA followed by Bonferroni multiple comparison test. Isotype control as gray histogram. Please also see Figure S1.
To study IDO1 expression in the respective cDC subsets, we sort-purified, cDC1 and cDC2 subsets from BMDC cultures (Figure S1E). LPS treatment induced IDO1 protein expression only in cDC1, but not in cDC2 (Figure 1B). IDO2 or TDO2 remained undetectable (Figure 1B). Microarray analysis indicated that LPS induced Ido1 in cDC1, but not in cDC2 and pDCs (Figure S1F), and l-kyn was detected only in supernatants of cDC1, but not of cDC2 (Figure 1C).
We found that cDC1 and cDC2 expressed similar amounts of TLR 4 (CD284) protein (Deng et al., 2016; Uehori et al., 2005), whereas pDCs expressed a low amount (Figure 1D). LPS treatment could cause an increase in CD80, CD86, and CD40 mRNA (Figure S1G) and protein (Figure 1E) in cDC1 and cDC2, but not in pDCs, although, in untreated cells, CCR7 was more expressed in sorted cDC2 than in cDC1 (Figure 1F). In BMDC cultures, we found that IDO1 was selectively expressed and induced in CCR7+ cDC1 cultures, both at the steady state and after treatment with LPS. However, IDO1 was not expressed in either untreated or activated cDC2 cells (Figure 1F). Accordingly, l-kyn was detected in WT CCR7+ cDC1 but not in the Ido1-deficient (Ido1−/−) counterpart (Figure 1G). Similar to LPS, treatment of sorted cDC1 cells with other TLR ligands led to IDO1 expression in CCR7+ cDC1 (Figure S1H), but not in CCR7+ cDC2. The expression of IDO1 in mature cDC1 in vivo confirmed Ido1 transcript expression in mature CCR7+ cDC1 but not in the CCR7− counterpart (Figure S1I) and IDO1 protein expression in thymic CCR7+ cDC1 but not in CCR7+ cDC2 (Figure 1H). Overall, we found that both cDC1 and cDC2 were able to express IDO1 in mixed cDCs from BMDCs; in contrast, in isolated cDC populations, IDO1 was expressed or induced only in mature cDC1, but not in mature cDC2.
IRF8 and Batf3 are required for IDO1 expression in CCR7+ cDC1
The transcription factors Batf3 and Irf8 are both required for the development of cDC1 (Durai et al., 2019a; Grajales-Reyes et al., 2015). Chromatin immunoprecipitation sequencing (ChIP-seq) for H3K27ac binding showed accessible chromatin in the Ido1 promoter in cDC1, but not cDC2 and pDCs (Figure 2A). ChIP-seq for BATF3 and IRF8 revealed coincident binding of BATF3 and IRF8 near the Ido1 transcriptional start site in cDC1, in a region containing two AP-1-IRF composite elements (AICEs) at −126 bp (AICE1) and +495 bp (AICE2) (Figures S2A and 2A). Using a retroviral reporter vector (RV) in BMDCs (Figure S2B), we found that the region containing the AICE1 site at −126 bp conferred higher reporter activity in cDC1 as than cDC2 cells, whereas the AICE2 site at +495 bp had no activity (Figures 2B–2D). ChIP-PCR for IRF8 binding confirmed specific enrichment in the −126-bp region in CCR7+ cDC1, but not CCR7− cDC1 (Figure 2E). We expressed single-guide RNAs (sgRNAs) (Cong et al., 2013) directed at AICE1 site (Figure S2C) in BMDCs from Cas9 genetically targeted mice (Platt et al., 2014) (Figure S2D). IDO1 expression was significantly reduced by sgRNAs directed to the −126 bp AICE1 in mature CCR7+ cDC1 (Figure 2F). To test whether Batf3 was also required, we used two methods for restoring cDC1 in Batf3-deficient (Batf3−/−) mice. First, cDC1 were regenerated by IL-12 treatment of BMDCs from Batf3−/− mice, as reported (Tussiwand et al., 2012). Although both WT and Batf3−/− cDC1 expressed similar activation markers (Figure S2E), we found that unlike WT cDC1, Batf3−/− CCR7+ cDC1 did not express IDO1 (Figure 2G). Second, CCR7+ cDC1 from Batf3−/− Irf8VENUS mice, which contain a transgene carrying three copies of a phage artificial chromosome (PAC) containing a 130 kb Irf8 genomic region (Grajales-Reyes et al., 2015; Schönheit et al., 2013; Theisen et al., 2019), also did not express IDO1 protein (Figure 2H). Overall, these results suggest that IDO1 expression in cDC1 may require cooperation between BATF3 and IRF8, as well as a maturation signal.
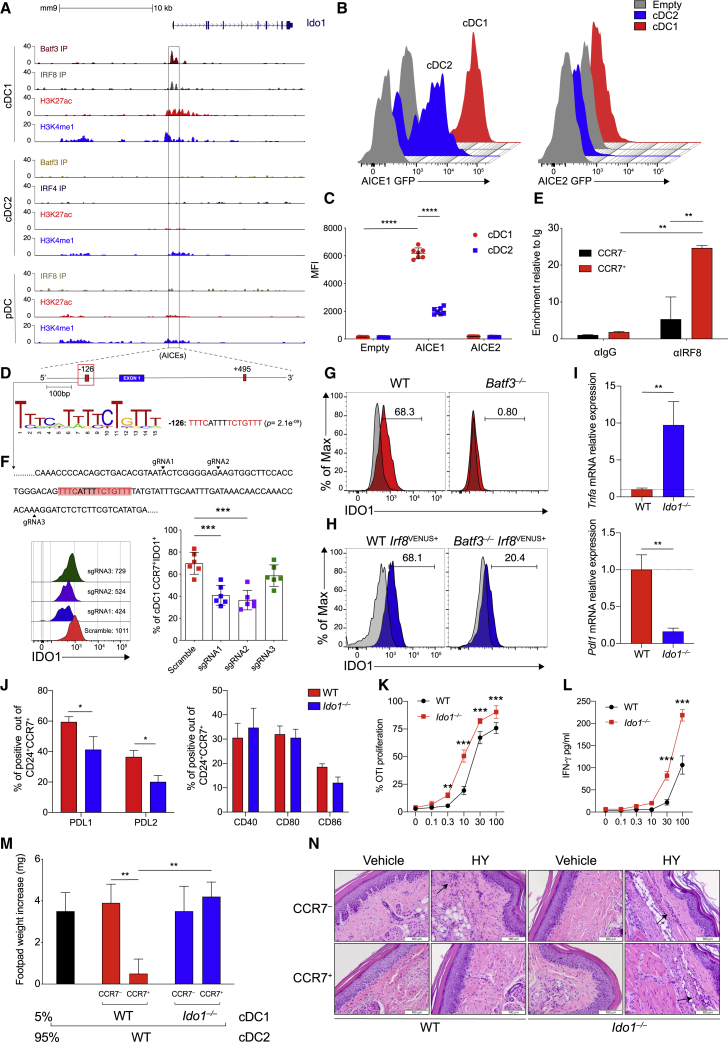
Figure 2.
IRF8 imprints constitutive IDO1 expression in cDC1
(A) ChIP-seq tracks display open chromatin areas and bindings of IRF8, BATF3, IRF4, H3K27ac, and H3K4me1 around Ido1 locus. Boxed areas at −126 bp or +495 bp from IDO1 TSS indicate regions assessed for enhancer activity.
(B and C) Flow cytometric analysis showing GFP-reporter activities (B) and quantification (C) in cDC1s and cDC2s expressing IDO1 −126 bp and + 495-bp enhancers (n = 7).
(D) FIMO analysis depicting p values of the two predicted AICEs (red boxes) in mouse Ido1 chr8: 25,694,453–25,713,138 (−126 bp from Ido1 TSS).
(E) IRF8 enrichment at the AICEs sequences of Ido1 promoter in sorted CCR7+ and CCR7− cDC1 (n = 2).
(F) IDO1 expression (MFI) in CCR7+ cDC1s differentiated from Rosa26Cas9−GFP/+ CD117hi BM progenitors expressing scramble RNA or sgRNA(s) (black arrowheads) targeting Ido1 −126 bp AICE1, as depicted in the single-color histograms (n = 3).
(G and H) IDO1 expression by flow in WT and Batf3−/− (H) and WT Irf8VENUS+ and Batf3−/−Irf8VENUS+ cDC1 (n = 3).
(I) Tnf and Pdl1 mRNA expression in sorted CCR7+ cDC1 of indicated phenotypes (n = 4).
(J) PDL1, PDL2, CD40, CD80, and CD86 expressions by flow in CCR7+ cDC1 (n = 3).
(K) Sorted WT or Ido1−/− cDC1 assayed for presentation to OT-I T cells in response to soluble OVA protein (n = 2).
(L) IFN-γ production in supernatants from (K) (n =2).
(M) Analysis of DTH is presented as change in footpad weight. n = 5 mice/group for (n = 2).
(N) H&E staining of mice footpad from mice in (M).
Data are shown as means ± SD. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, two-way ANOVA followed by Bonferroni multiple comparison test (C, E, F, J, K, and L) or Tukey’s multiple comparison test (M) and unpaired t test (I). Isotype control is shown as gray histogram. Please also see Figure S2.
CCR7+IDO1+ cDC1s are characterized by regulatory function
We evaluated the regulatory function of IDO1 in CCR7+ cDC1. Increased Tnf and reduced Cd274 transcripts were found in CCR7+ cDC1 isolated from Ido1−/− mice compared with CCR7+ cDC1 isolated from WT mice (Figure 2I). Accordingly, a lower frequency of PDL1- and PDL2-positive cells was found in the CCR7+ Ido1−/− cDC1 than in the CCR7+ Ido1+/+ cDC1 counterpart, whereas CD80 and CD86 were not increased (Figure 2J). OT-I transgenic CD8+ T cells proliferated to a greater extent (Figure 2K) and produced higher amounts of IFN-γ (Figure 2L) when stimulated with Ido1−/− cDC1 than Ido1+/+ cDC1. In addition, stimulation of Ido1−/− CCR7+ cDC1 with LPS resulted in higher IL-12 p40 production compared with the WT counterpart (Figure S2F). Furthermore, using an in vivo model of delayed-type hypersensitivity (DTH) response (Pallotta et al., 2011), we found that WT CCR7+ cDC1 reduced reactivity to HY antigen, an effect abrogated when Ido1−/− CCR7+ cDC1s were used as the minority fraction (Figure 2M). Detectable DTH response was associated with the production of IFN-γ (Figure S2G) and an immune infiltrate in the HY-challenged footpads (Figure 2N).
cDCs control inflammation in experimental autoimmune encephalomyelitis (EAE) (Paterka et al., 2017; Yogev et al., 2012). In order to elucidate the role of cDC1 in controlling autoimmune central nervous system (CNS) inflammation, Irf8 Δ32−/− mice, lacking cDC1 (Durai et al., 2019a) and WT counterparts were subjected to MOG35–55-induced EAE. Compared with WT mice, lack of cDC1 (Figure S3A) worsened EAE (Figure 3A), increased antigen-elicited production of interleukin-17 (IL-17) and granulocyte-macrophage colony-stimulating factor (GM-CSF), and reduced TGF-β (Figure 3B), both by CNS-draining lymph nodes (LNs) and in plasma (Figure S3B). Consistent with the more severe of EAE symptoms, Irf8 Δ32−/− mice displayed a greater CNS immune cell infiltration (Figure 3C) and reduced T regulatory (Treg) cell frequency (Figure 3D). In stark contrast, IFN regulatory factor 4 conditional deleted mice (Irf4f/f ItgaxCre+), characterized by a deficit of the cDC2 lineage (Bajaña and Turner, 2016), were protected from EAE (Figure S3C). These data suggest a major role of the cDC2 subset in sustaining the CNS inflammation.
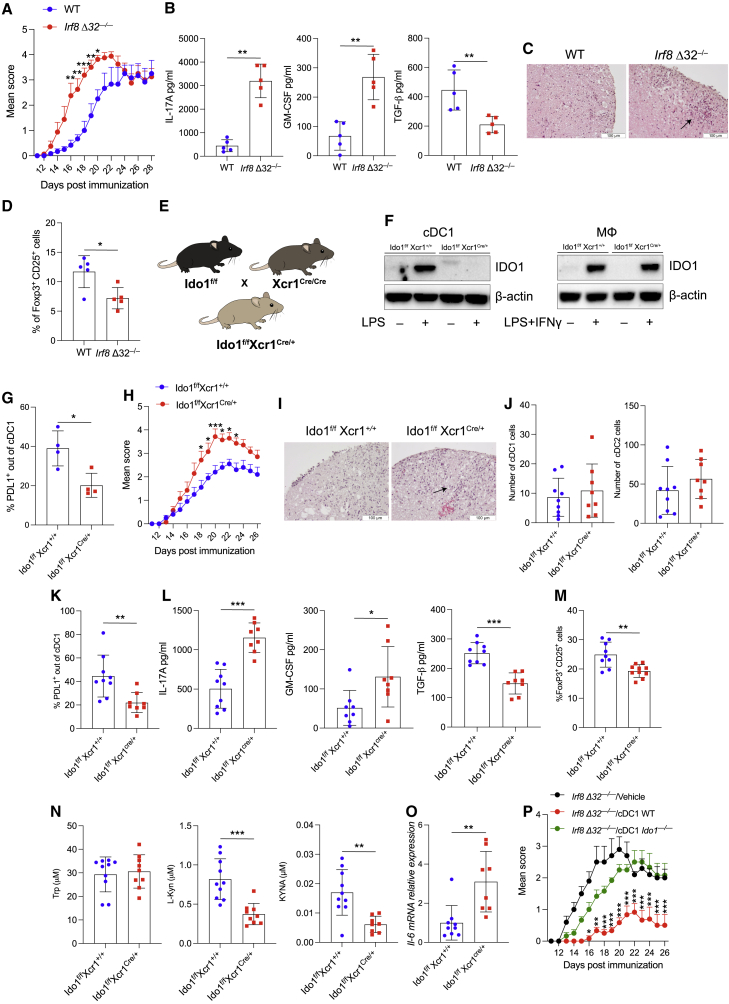
Figure 3.
Selective IDO1 gene deletion in cDC1 worsens EAE
(A) EAE score in WT and Irf8 Δ32−/− mice. Five mice per group, n = 3.
(B) IL-17, GM-CFS, and TGF-β in supernatant of cervical LNs from EAE mice in (A) at day 28 and restimulated in vitro with MOG 20 μg/mL for 5 days.
(C) H&E staining of spinal cord sections from EAE mice in (A).
(D) Flow cytometric analysis of FoxP3+CD25+ cells in cervical LNs of mice immunized as in (A).
(E) Schematic representation of Ido1f/fXcr1Cre/+ strain generation.
(F) IDO1 immunoblot. Left: cDC1 untreated or treated with LPS 250 ng/mL for 48 h. Right: macrophages (MΦ) untreated or treated with LPS 250 ng/mL and IFN-γ 15 ng/mL for 48 h β-actin was used as loading control (n = 3).
(G) Flow cytometric analysis of PDL1+ in thymic cDC1.
(H) EAE score in Ido1f/f Xcr1+/+ and Ido1f/fXcr1cre/+ mice. 9 mice per group, n = 2.
(I) H&E staining of spinal cord sections from mice treated in (H).
(J) Quantification of cDC1 and cDC2 in spinal cords from EAE mice in (H) at day 26.
(K) PDL1+ cDC1 frequency out cDC1 by flow cytometry in spinal cords of EAE mice in (H).
(L) Cytokine analysis in supernatants from cervical LNs of EAE mice in (H) at day 26 stimulated as in (B).
(M) Flow cytometric analysis of FoxP3+CD25+ cells in cervical LNs of mice immunized as in (H).
(N) Tryptophan metabolites in plasma from EAE mice in (H) at day 26.
(O) Il6 mRNA expression in cervical LNs from EAE mice in (H).
(P) EAE score in Irf8 Δ32−/− mice transferred with cDC1 genotypes at days 4 and 7 post immunization. 5 mice per group, n = 3.
Data are mean ± SD. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, two-way ANOVA followed by Bonferroni multiple comparison test (A, H, and P) or unpaired t test (B, D, G, and J–O). Each dot represents an individual value. Please also see Figure S3.
To dissect the contribution of IDO1 in the cDC1-dependent effect, mice with a floxed Ido1 gene (Bishnupuri et al., 2019) were crossed to Xcr1-Cre mice (Ferris et al., 2020; Wohn et al., 2020) (Figure 3E). The resulting mouse line (Ido1f/fXcr1Cre/+) demonstrated effective deletion of IDO1 protein expression in cDC1, but not in macrophages (Figure 3F). cDC1 isolated from these mice exhibited lower PDL-1 expression compared with WT (Ido1f/fXcr1+/+) (Figure 3G). When EAE was induced in Ido1f/fXcr1Cre/+ mice, disease severity (Figure 3H) and CNS immune infiltrate (Figure 3I) were increased compared with littermate controls, although no major differences in the number of cDCs was revealed in CNS (Figure 3J). In addition, PDL-1 expression was reduced in cDC1 from Ido1f/fXcr1Cre/+ mice compared with WT controls (Figure 3K), whereas IL-17 and GM-CSF were increased (Figures 3L and S3D) and TGF-β (Figures 3L and S3D) and Treg cell frequency (Figure 3M) were reduced in Ido1f/fXcr1Cre/+ mice, compared with WT. Among Trp metabolites, l-kyn and kynurenic acid were reduced in plasma of Ido1f/fXcr1Cre/+ mice compared with WT littermates (Figures 3N and S3E). Increased Il-6 transcripts were found in ex-vivo CNS-draining LNs at the peak of the disease in Ido1f/fXcr1Cre/+ mice compared with WT controls (Figure 3O), and cDC2 expressed low Ido1 and increased Il6 transcripts compared with cDC2 from Ido1f/fXcr1+/+ (Figure S3F). Last, to functionally validate the immunoregulatory phenotype of IDO1-competent mature cDC1, we transferred either IDO1 WT cDC1 or IDO1-deficient cDC1 pulsed with MOG peptide into mice with ongoing disease 4 and 7 days after immunization. Transfer of IDO1+ cDC1 protected against disease, whereas transfer of Ido1−/− cDC1 did not alter disease progression (Figure 3P). These data suggest that selective disruption of IDO1 function in cDC1 cells is sufficient to increase EAE severity as IDO1+ cDC1 are characterized by an intrinsic immunoregulatory phenotype linked to suppression of proinflammatory responses and generation of tolerogenic activity in EAE.
IL-6 production by cDC2 represses IDO1 protein expression in response to LPS
IL-6 is induced in DCs in response to inflammatory stimuli including LPS (Heink et al., 2017). cDC2 expressed higher amounts of Il6 transcripts compared with cDC1 after LPS treatment (Figure 4A), and IL-6 protein was produced more rapidly and in a greater amount by cDC2 relative to cDC1 (Figures 4B and 4C). Although LPS induced IDO1 protein expression only in cDC1, but neither in cDC2 nor pDCs (Figures 1B and S1C), Ido1 transcripts could be induced in cDC2, suggesting that cDC2 have the potential to produce the IDO1 protein (Figure S4A). Notably, LPS-activated cDC2 produced IDO1 protein only after IL-6 neutralization combined with IL-6 receptor (IL-6R) blockade, (Figure 4D), whereas IDO1 expression was not affected in cDC1 (Figure 4D). Ido1 transcripts were not affected by IL-6 neutralization in cDC2 (Figure S4B), consistent with the notion of IL-6 regulation of IDO1 protein stability. Hence, SOCS3 expression was low and unaffected by LPS treatment in cDC1, whereas unstimulated cDC2 showed constitutive SOCS3 expression, which only marginally increased by LPS treatment (Figure 4E). Accordingly, IDO1 protein accumulated in LPS-treated cDC2 cells upon proteasome inhibition (Figure 4F) or Socs3 silencing (Figure S4C). Notably, IL-6 neutralization in LPS-conditioned cDC2, co-cultured with CD4+ T cells derived from OT-II Foxp3 YFP mice, restored basal Foxp3 expression, an effect lost using IDO1-deficient cDC2 (Figure S4D). These results suggest that although Ido1 mRNA could be detected in LPS-treated cDC2 cells, failure to accumulate IDO1 protein is due to IL-6-driven proteasomal degradation.
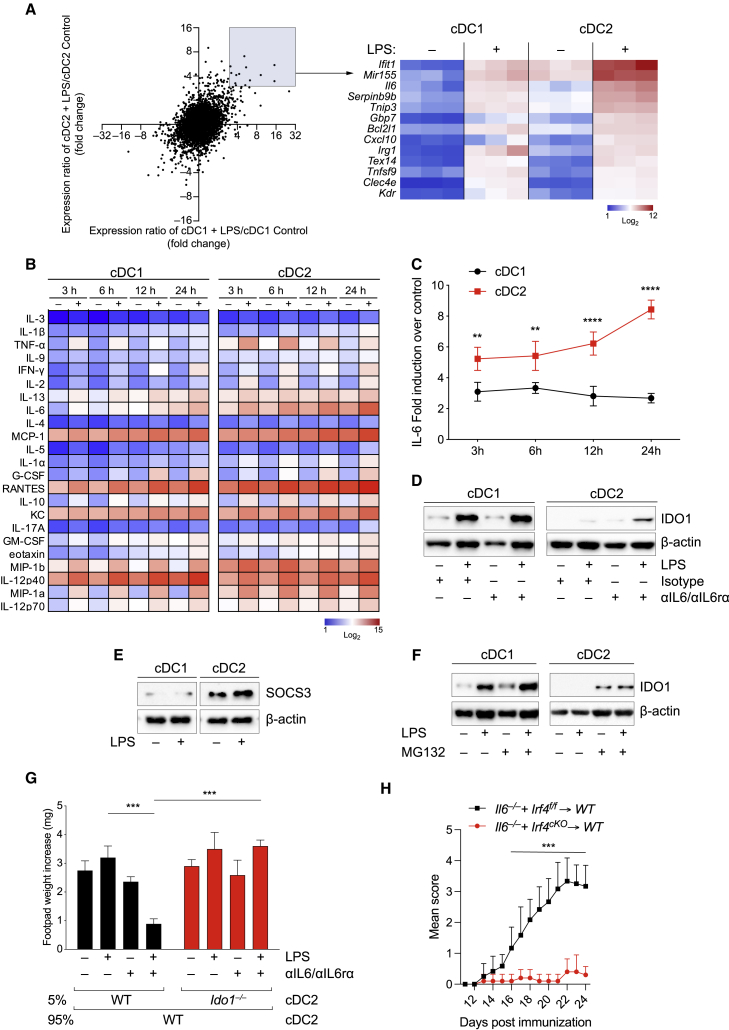
Figure 4.
IL-6 production by cDC1 and cDC2 regulates differential IDO1 expression
(A) Gene expression microarray analysis of cDC1 and cDC2 untreated or treated with LPS (250 ng/mL) for 24 h. Bright blue (lowest) to bright red (highest).
(B) Heatmap of cytokines in supernatants from cDC1 and cDC2 cultures treated as (A) (n = 2).
(C) IL-6-fold induction in LPS (250 ng/mL) stimulated cDC1 and cDC2 over the untreated control (n = 3).
(D) IDO1 immunoblot in cDC1 and cDC2 treated as depicted for 48 h (n = 3).
(E) SOCS3 immunoblot in cDC1 and cDC2 treated as depicted for 24 h (n = 3).
(F) IDO1 immunoblot in cDC1 and cDC2 treated as depicted for 48 h (n = 4).
(G) Analysis of skin reactivity is presented as change in footpad weight. n = 6 mice per group (n = 2).
(H) EAE score of mixed bone marrow chimeras with bone marrow of indicated genotypes. 6 mice per group, n = 2. Data are shown as means ± SD. ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, two-way ANOVA followed by Bonferroni multiple comparison test (C and H) or Tukey’s multiple comparison test (G). β-Actin used as loading control (D–F). Please also see Figure S4.
Next, we tested the potential regulatory functions of cDC2 under conditions that restored IDO1 function, using the DTH assay to the HY peptide. DTH (Figure 4G) and T helper type-1 responses (Figures S4E and S4F) were significantly diminished when HY-bearing cDC2, used as minority fraction, were treated with LPS in combination with neutralization of IL-6 and IL-6R blockade an effect abrogated when Ido1−/− cDC2 were used (Figure 4G).
It is known that IL-6 from T cells, B cells, and DCs promotes the clinical signs of EAE (Heink et al., 2017) and the transfer of Il6−/− BM results in protection against EAE. Furthermore, on testing the effect of selective Il6 deletion in cDC2 obtained by the use of mixed BM chimeras—e.g., Irf4 conditional gene deletion and Il6−/− BM (Kopf et al., 1994)—resulted in less severe EAE scores, compared with WT counterparts (Figure 4H). These data suggested that neutralization of IL-6 and receptor blockade act to confer tolerogenic functions on inflammatory cDC2 cells potentially by a mechanism requiring IDO1.
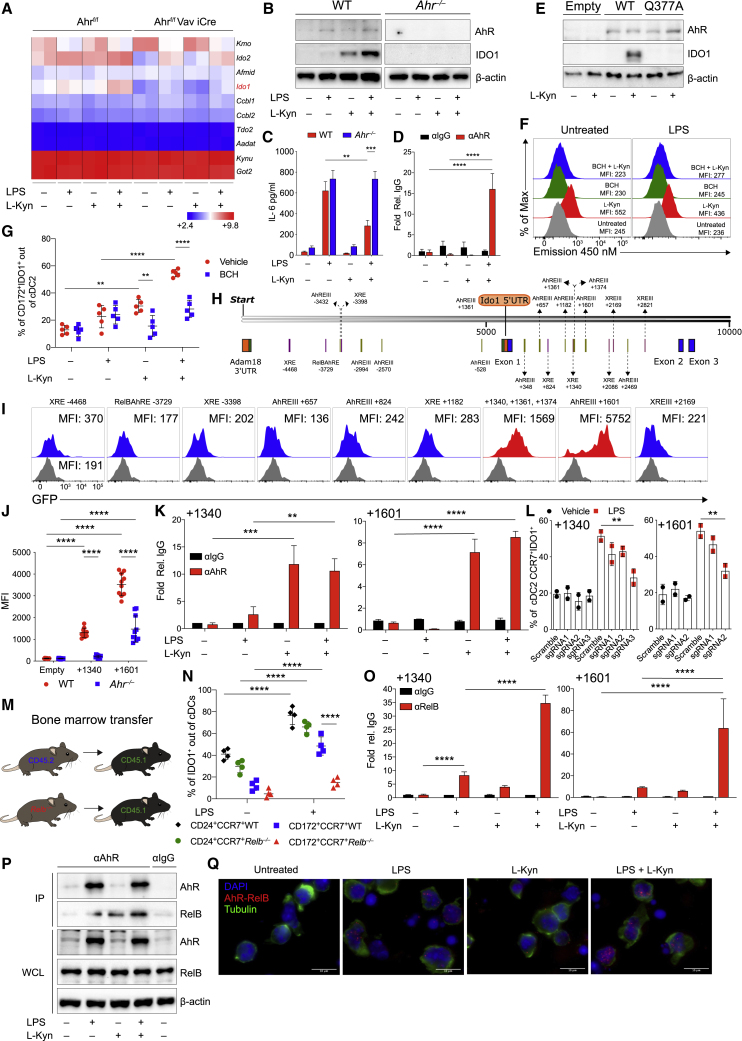
cDC2 acquires conditional expression of IDO1 dependent on l-kynurenine production by cDC1
Because we found a different pattern of IDO1 expression in DCs in isolation or in co-cultures (Figure 1), we tested any possible crosstalk between cDC subsets in conditioning IDO1 expression. Approximately 50% CCR7+ cDC1 expressed IDO1 in untreated unfractionated DC cultures, and 80% CCR7+ cDC1 expressed IDO1 upon LPS treatment (Figures 5A and S5A). However, unlike isolated cDC2 cells—which did not express IDO1 even after LPS stimulation (Figure 1B)—approximately 50% CCR7+cDC2 from unfractionated cultures expressed IDO1 following LPS treatment (Figures 5A, S5A, and S5B). cDC2 isolated from these mixed cDC cultures produced less Il6 than when cultured as segregated cells (Figure 5B). In accordance, l-kyn was increased in mixed cDC cultures activated with LPS, compared with LPS-treated isolated cDC1 counterparts (Figure 5C), indicating a higher amount of total IDO1.
Figure 5.
cDC2s cultured with cDC1s under LPS stimulation exhibit conditional and AhR-dependent IDO1 induction
(A) IDO1 expression analyzed by IS in BMDCs (n = 5).
(B) Il6 mRNA expression in sorted cDC1 and cDC2 treated in isolation or purified mixed LPS-treated or LPS-untreated BMDCs (n =3).
(C) l-kynurenine content in supernatants from B (n = 3).
(D) Flow cytometric analysis of cDCs differentiated from WT and Irf8 Δ32−/− treated as depicted for 48 h.
(E) IDO1 and β-actin immunoblot in BMDCs as in (D) (n = 3).
(F) IDO1 expression by IS in cDC2 harvested from cDC1 and cDC2 co-cultures in trans-well plates with or without LPS for 48 h (n = 3).
(G) CCR7+IDO1+cDC2 cell frequency in cultures established as in (F) treated as depicted (n = 3).
(H) Skin reactivity in mice treated with cDC2 either alone or in combination with 5% of WT, Ido1−/− or Ahr−/− cDC2 (co-cDC2) conditioned with cDC1 as in (F). 6 mice per group, n = 2.
(I) Skin reactivity in mice transferred with cDC2 combinations treated as depicted. 6 mice per group, n = 2.
(J) EAE score of mixed bone marrow chimeras with bone marrow of indicated genotypes. 8 mice per group, n = 2.
(K) H&E staining of spinal cord sections from mice treated in (J). Data are shown as means ± SD. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, one-way (B) or two-way (G and J) ANOVA followed by Bonferroni multiple comparison test. Two-way ANOVA followed by Tukey’s multiple comparison test (C, H, and I). Isotype control as gray histogram. Please also see Figure S5.
To understand whether cDC1 are required for IDO1 induction in mixed BMDCs, we differentiated conventional DCs from WT and Irf8 Δ32−/−. In accordance with previous results (Durai et al., 2019b), cDCs derived from Irf8 Δ32−/− mice did not differentiate cDC1 in response to LPS (Figure 5D), and the loss of cDC1 in BMDC cultures impaired IDO1 overall expression in response to LPS (Figure 5E).
To test whether IDO1 activity in cDC1 was required for cDC2 to acquire IDO1 expression in mixed cultures, we treated cultures with or without the IDO1 inhibitor, 1-l-MT, in the absence or presence of LPS. 1-l-MT treatment blocked IDO1 induction in cDC2, decreased IDO1 expression in cDC1 (Figure S5C), and suppressed l-kyn production by cDC mixed cultures overall (Figure S5D), in agreement with the notion of the Trp metabolic pathway acting so as to facilitate IDO1 expression by cDC2.
Then, we used the trans-well system to examine if direct cell contacts between cDC1 and cDC2 were required for induction of IDO1 in cDC2 in unfractionated cultures. Here, following LPS treatment, IDO1 was induced in cDC2 even when Ido1+/+ cDC1s were separated from cDC2s by a trans-well membrane (Figure 5F), and they expressed less Il6 and Socs3 transcripts compared with those cultured with Ido1−/− cDC1 (Figures S5E and 5F) excluding direct cell contact as a requirement. Induction of IDO1 and suppression of Il6 and Socs3 in cDC2 required cDC1 to express IDO1 and cDC2 to express AhR in LPS-treated trans-well cultures (Figure 5F), although IDO1 was still induced in cDC1 (Figure S5G). These results suggest that soluble l-kyn produced by cDC1 permeating trans-well membranes may mediate induction of IDO1 in cDC2 cells. In fact, externally added l-kyn during LPS stimulation induced IDO1 expression in WT cDC2, but not Ahr−/− cDC2 or when cDC2 were cultured with Ido1−/− cDC1 (Figure 5G). To investigate whether similar crosstalk between subsets of cDCs also occurs in natural DCs, we analyzed IDO1 expression in cDCs in mesenteric LNs. Similar to BMDCs, IDO1 was mainly expressed in mature cDC1 (Figure S5H), whereas its expression was impaired in cDC2 from Ido1f/fXcr1Cre/+ mice (Figure S5I). Moreover, LPS maturation of splenic cDCs promoted IDO1 expression and catalytic function in cDC2 (Figure S5J), requiring IDO1 enzymic function in cDC1 (Figure S5K).
Finally, we employed DTH experiment to test whether IDO1-expressing cDC2 cells derived from trans-well cultures exerted in vivo regulatory capacity. We found that immunostimulatory activity of cDC2 was greatly reduced by the inclusion of a minor fraction of cDC2 cells, derived from co-cultures with untreated or LPS-treated WT cDC1 (Figure 5H). However, this reduction was lost using a minority fraction of WT cDC2 cultured with Ido1−/− cDC1 or of Ahr−/− cDC2 (Figure 5H). A DTH reduction was also observed by l-kyn treatment of the cDC2 minority fraction, with or without LPS (Figure 5I). Consistently, selective Ido1 deletion in cDC2 obtained by a mixed BM chimeras with Irf4 conditional genetic deletion and Ido1−/− BM, resulted in a more severe EAE clinical scores (Figure 5J) and increased CNS infiltrates (Figure 5K) compared with WT BM. Overall, these data suggest that IDO1+ cDC1 are required to imprint IDO1 expression in cDC2, contributing to the acquisition of a regulatory function in this latter cell subset. The observation that Irf4-deficient mixed BM chimeras were as susceptible to EAE as those lacking Ido1 in cDC1 suggests that IDO1+ cDCs are critical regulators of pathogenic response in this experimental model.
Isolated cDC2 treated with l-kyn acquires IDO1 expression in the absence of cDC1
Because l-kyn restored IDO1 expression in cDC2 cultured with Ido1−/−cDC1 (Figure 5G), we asked whether l-kyn could directly induce IDO1 expression in cDC2 even in the absence of cDC1. First, in cDC2, exogenous l-kyn treatment of WT cDC2, but not Ahr−/− cDC2, increased both IDO1 mRNA expression (Figures 6A and S6A) and protein as well (Figure 6B), with or without LPS, whereas the Trp catabolic enzymes were not induced (Figure 6A). Furthermore, treatment with l-kyn suppressed IL-6 production in LPS-stimulated WT cDC2, but not in Ahr−/− cDC2 cells derived from BM cultures (Figure 6C). Chip-PCR analysis confirmed AhR binding on previously described p65 responsive elements (Kimura et al., 2009) in cDC2 in response to LPS + l-kyn treatment (Figure 6D). Consistently, Socs3 mRNA expression was higher both in untreated and LPS-treated Ahr−/− cDC2 as well as in those cultured in combination with l-kyn compared with WT cDC2 (Figure S6B). In addition, cDC2 lacking AhR or IDO1 secreted high amounts of IL-12, IL-6, RANTES, and to a lesser extent, IFN-γ (Figure S6C). Next, we generated cDC2 cells from Ahr−/− BM and reconstituted AhR expression using either WT or Q377A AhR-expressing RV, the latter unable to be activated by l-kyn (Nuti et al., 2014). Consistent with previous data, l-kyn induced IDO1 mRNA and protein in cDC2 reconstituted with WT AhR, but not in those with the Q377A AhR mutant (Figures 6E and S6D).
Figure 6.
AhR cooperates with RelB to induce IDO1 in isolated cDC2 treated with l-kynurenine
(A) Heatmap of tryptophan metabolic enzymes gene expression in cDC2 from AhRf/f mice versus Ahrf/fVav1 iCre mice treated as depicted. Bright blue (lowest) to bright red (highest).
(B) IDO1 immunoblot in cDC2 treated as shown for 48 h (n = 4).
(C) IL-6 analysis in supernatants of cDC2 treated as in (A) for 48 h (n = 4).
(D) AhR enrichment at the κB sequences of Il6 promoter by ChIP in sorted cDC2 untreated or treated overnight with LPS and conditioned for 2 h with l-kynurenine (n = 2).
(E) IDO1 immunoblot in purified cDC2 were transfected and treated as depicted for 48 h (n = 4).
(F) l-kynurenine uptake in purified cDC2. MFI is shown (n = 4).
(G) IDO1 expression by IS in cDC2 treated as (A) in the presence or absence of BCH for 48 h (n = 5).
(H) Predicted canonical AhR binding sites in the murine Ido1 promoter.
(I) MFI of GFP expression in pre-gated as Thy1.1+ cDC2 transduced with RV vector containing regions as described in (H). Gray histograms show empty reporter (n = 4).
(J) Quantification of the +1,340 bp and +1,601 bp Ido1 enhancer activity in WT and Ahr−/− cDC2s using retroviral reporters as in (I) (n = 3).
(K) ChIP-PCR analysis of AhR binding on +1,340 bp and +1,601 bp Ido1 enhancer elements in cDC2 treated as in (C) (n = 3).
(L) IDO1 expression by IS in gated CCR7+cDC2 derived from Rosa26Cas9−GFP/+ c-Kithi progenitors infected with RV expressing sgRNAs targeting +1,340 bp and +1,601 bp Ido1 enhancer elements treated as shown for 48 h (n = 2).
(M) Schematic representation of BM chimera model.
(N) IDO1 expression by IS in cDC1 and cDC2 from BMDC derived as in (M) and cultured as shown (n = 3).
(O) ChIP-PCR analysis of RelB binding on +1,340 bp and +1,601 bp Ido1 enhancer elements in cDC2 treated as in (C) (n = 3).
(P) AhR and RelB Immunoblot in purified cDC2 treated as in (K) where AhR was immunoprecipitated (n = 3).
(Q) AhR and RelB interaction in purified cDC2 treated as indicated by PLA. Red spots show a single AhR/RelB interaction. Scale bars, 10 μm (n = 3).
Data are shown as means ± SD. ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, one-way (L) or two-way (C, D, G, J, K, N, and O) ANOVA followed by Bonferroni multiple comparison test. Dots represent a biological replicate (G and N). β-actin used as loading control (B, E, and P). Data are normalized to IgG control (D, K, and O). Please also see Figure S6.
l-kyn can activate AhR only in cells that express the large neutral amino acid transporter (LAT) solute carrier family 7 member 5 SLC7A5 (LAT1) (Sinclair et al., 2018). A slight upregulation of SLC7A5 carrier was found upon treatment of the cells with LPS or l-kyn that did not require Ahr expression (Figures S6E and S6F). 2-Amino-2-norbornanecarboxylic acid (BCH) is an inhibitor of all large amino acid transporters (Gomes and Soares-da-Silva, 1999). By a flow cytometry-based assay, we confirmed that BCH inhibited l-kyn uptake by cDC2 (Figures 6F and S6G) and impaired l-kyn-mediated IDO1 induction in cDC2 (Figure 6G), suggesting that BCH sensitive transporters are required for IDO1 induction by l-kyn in cDC2.
AhR cooperates with RelB to induce IDO1 in cDC2
To gain insights into this kyn and AhR-mediated Ido1 induction in cDC2, we searched for AhR-responsive elements in the Ido1 promoter. We explored AhR canonical binding sites (Guyot et al., 2013) in Ido1 promoter using Snap gene. We found that the Ido1 5′ upstream region, but also Ido1 internal noncoding region, contains canonical xenobiotic responsive elements (XREs) and recognizable AhR target sequences (AhREs) (Bacsi and Hankinson, 1996) (Figure 6H). Notably, by reporter analysis, constructs containing the region encompassing the three AhR-responsive elements (+ 1,340 bp, +1,361 bp, and +1,374 bp) as well as that containing the AhRE at +1,601 were each sufficient to confer specific reporter activity in cDC2 cells (Figure 6I) in AhR-dependent manner (Figure 6J). Hence, treatment of cDC2 with l-kyn, either alone or in combination with LPS increased the binding of AhR to both regions approximately 5-fold over binding in untreated cDC2 as detected by ChIP-PCR (Figure 6K). Notably, editing of those regions in cDC2 by RVs (Figure S2C) carrying specific sgRNAs (Figure S6H) caused a significant reduction of Ido1 expression in mature CCR7+ cDC2s (Figure 6L).
Previous co-immunoprecipitation studies have reported that RelB can physically interact with AhR in BM-derived DCs (Vogel et al., 2013). Thus, to test for a requirement for Relb in Ido1 expression by cDC2 cells, we generated BM chimeras (Figure 6M) in which WT or Relb−/− BM was transferred into WT recipients, which allows for normal development of Relb−/− DCs (Figure S6I) (Briseño et al., 2017). Notably, IDO1 was reduced in Relb−/− CCR7+ cDC2 chimera-derived upon LPS treatment compared with WT CCR7+ cDC2, conversely Relb−/− cDC1 cells expressed similar IDO1 protein (Figure 6N). These data suggest that in cDC2 treated with LPS, IDO1 induction is RelB dependent, although it is dispensable in cDC1. Accordingly, in ChIP-PCR assays, cDC2 treated with LPS and l-kyn, increased the binding of RelB to AhR-responsive sites located at +1,340 bp and +1,601 bp in the Ido1 gene (Figure 6O).
Specific AhR-RelB interaction was confirmed by AhR immunoprecipitation (IP) in cDC2 treated with LPS and l-kyn (Figure 6P), the Duolink proximity ligation assay (DPLA), providing real time in situ interaction between AhR and RelB (Figure 6Q) and AhR and RelB nuclear translocation in cDC2 (Figure S6J). These results were also confirmed in Ahr−/− MEF treated with l-kyn reconstituted with WT but not in those with the Q377A AhR mutant (Figure S6K). Accordingly, l-kyn treatment induced AhR and RelB nuclear translocation in Ahr+/+ but not Ahr−/− MEF either reconstituted with the Q377A AhR mutant form (Figure S6L) or the WT AhR but pre-treated with BCH before treatment with l-kyn (Figure S6M). Finally, we tested whether LPS-conditioned cDC2 treated with l-kyn acquired in vitro regulatory phenotype. OT-II transgenic CD4+ T cells proliferate less when stimulated with LPS-conditioned cDC2 treated with l-kyn, compared with those treated with LPS alone (Figure S6N). Such effect was lost when cDC2s lacking AhR were used (Figure S6N). Similarly, l-kyn treatment of LPS-conditioned cDC2, restored Foxp3 expression in CD4+ T cells in cDC2-OT-II Foxp3 YFP cell co-cultures, an effect abolished using AhR-deficient cDC2 (Figure S6O). Altogether, these data show that l-kyn promotes AhR and RelB interaction, leading to IDO1 expression and acquisition of immune regulatory functions in LPS-treated cDC2.
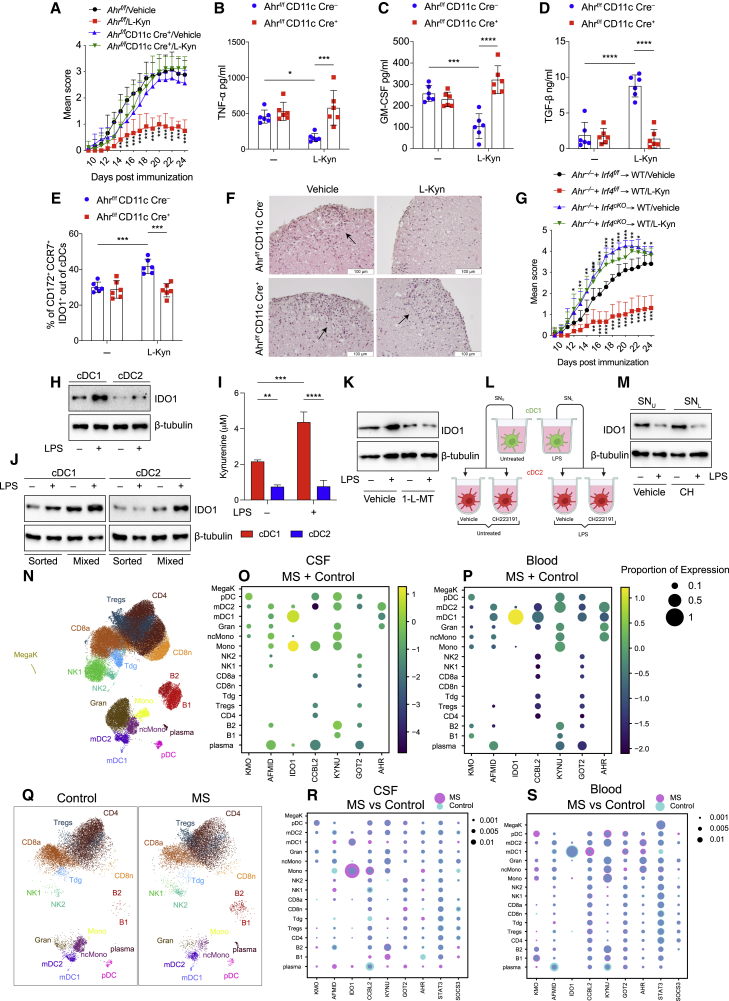
l-kynurenine administration in vivo suppresses EAE by inducing immunoregulatory cDCs
EAE development is limited by the metabolism of dietary Trp, generating AhR agonists (Gutiérrez-Vázquez and Quintana, 2018; Rothhammer et al., 2016), and AhR expression in cDCs is responsible for controlling CNS inflammation (Duarte et al., 2013).
Based on our findings, we tested the ability of l-kyn supplementation to induce immunoregulatory properties in EAE and assessed the critical role of cDCs in this effect. Using Ahrf/f Itgax Cre− and Ahrf/f Itgax Cre+ mice, the latter lacking AhR selectively in CD11c+ cells, we found that oral l-kyn administration reduced EAE scores and CNS inflammation in Ahrf/f Itgax Cre−, but not in Ahrf/f Itgax Cre+ mice (Figure 7A). Similar results were obtained in a more cDC-specific Zbtb46 Cre+ crossed with Ahrf/f (Figure S7A), suggesting that AhR expressed by cDCs was crucial in EAE amelioration. Multiplex cytokine analysis confirmed that l-kyn treatment reduced proinflammatory cytokines, including TNF-α (Figure 7B) and GM-CSF (Figure 7C), and increased the anti-inflammatory cytokine TGF-β (Figure 7D) in the plasma in Ahrf/f Itgax Cre− but not Ahrf/f Itgax Cre+ mice. In addition, draining LNs from l-kyn-treated Ahrf/f Itgax Cre− mice showed reduced amounts of IL-17 and IFN-γ, compared with untreated or l-kyn Ahrf/f Itgax Cre+-treated mice (Figure S7B). Consistently, in vivo l-kyn treatment increased IDO1 protein in cDC2, but not in cDC1 (Figure S7C), from Ahrf/f Itgax Cre− cervical LNs but not from Ahrf/f Itgax Cre+ (Figure 7E). Notably, Ahrf/f Itgax Cre+ mice had more immune infiltrates in the spinal cords following l-kyn treatment, compared with Ahrf/f Itgax Cre− mice (Figure 7F). Similarly, selective AhR deletion in cDC2 by a mixed BM chimeras impaired the in vivo l-kyn protective effect in the EAE model (Figure 7G). Collectively, these findings suggested that Ahr deletion renders cDCs, and mostly pathogenic cDC2, unresponsive to the anti-inflammatory effects of l-kyn. Moreover, these data imply a dominant paracrine effect (in trans) of the AhR-sufficient cDC2s when stimulated with l-kyn, although half of the cDC2s in the Irf4f/f control are still Ahr−/− and therefore presumably hyper-inflammatory.
Figure 7.
l-kynurenine induces immune regulatory functions in EAE
(A) EAE score in Ahrf/fItgax Cre− or Ahrf/fItgax Cre+ mice treated with l-kyn or vehicle, from 11 to 22 days post immunization. 6 mice per group, n = 3.
(B–D) TNF-α (B), GM-CSF (C), and TGFβ (D) in plasma from EAE mice in (A).
(E) IDO1+CCR7+ cDC2 of cDCs by IS in cervical LNs from EAE mice treated as in (A).
(F) H&E staining of spinal cord sections from mice treated in (A). Scale bar, 100 μM.
(G) EAE score of mixed bone marrow chimeras with bone marrow of indicated genotypes. 9 mice per group, n = 2.
(H) IDO1 immunoblot in cDC1 and cDC2, sorted from human PBMCs, were treated as depicted for 36 h (n = 3).
(I) l-kynurenine content in supernatants from cells prepared as in (H) (n = 3).
(J) IDO1 immunoblot in purified human cDC1 and cDC2 cultured alone (sorted) or together (mixed), in trans-well plates, with or without LPS for 36 h (n = 3).
(K) IDO1 immunoblot in human cDC1 and cDC2 co-cultured in trans-well plates and treated as depicted for 36 h (n = 3).
(L) Schematic representation of human cDC2 treatment with cDC1 conditioned media. cDC1 untreated (SNU), LPS treated (SNL) for 36 h, CH2223191 (CH) cDC2 pre-treated for 2 h and incubated with cDC1 conditioned media for 36 h.
(M) IDO1 immunoblot in cDC2 treated as in (L) (n = 3).
(N) UMAP plot representing 17 color-coded cell clusters identified in merged single-cell transcriptomes of blood (42,969) and CSF (22,357) cells from control (n = 4) and multiple sclerosis (MS; n = 4) patients.
(O and P) Dot plot depicting selected genes of tryptophan metabolic pathway in cell clusters of CFS (O) and blood (P).
(Q) Comparative UMAP plots depicting only CSF cells from control (12,705 cells, left plot) and MS (9,652 cells, right plot) donors.
(R and S) Dot plot depicting selected genes differentially expressed in at least one cluster of MS cells compared with controls in CSF (R) or blood (S). Purple indicates higher and turquoise indicates lower expression in MS, respectively.
Data are shown as means ± SD. ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ∗∗∗∗ p < 0.0001, two-way ANOVA followed by Bonferroni multiple comparison test (A–E, G, and I). Dots represent a biological replicate (B–E). β-tubulin used as loading control (H, J, K, and M). Dot size encodes percentage of cells expressing the gene (O, P, R, and S). Please also see Figure S7.
Human IDO1+ cDC1 promotes IDO1 expression in human cDC2
It has been recently reported that cDC1 express IDO1, and TLR ligation further increases IDO1 expression in cDC1s, whereas induces only modest expression in cDC2s (Sittig et al., 2021). To exploit the pattern of IDO1 expression in human cDCs, cDC1 and cDC2 were isolated from human PBMCs from healthy donors, and IDO1 expression was measured after LPS treatment. Similar to the mouse model, LPS treatment of purified human cDCs induced IDO1 protein expression (Figure 7H) and l-kyn production (Figure 7I). Similarly, cDC2 co-cultured with cDC1 in trans-well acquired IDO1 expression by a mechanism requiring IDO1 activity in cDC1 (e.g., it was abolished by 1-l-MT) and AhR expression in cDC2 (e.g., it was prevented by AhR inhibitor CH223191 in cDC2) (Figures 7J–7M). These results suggest that human IDO1+ cDC1 educates human cDC2 to acquire the IDO1 tolerogenic pathway via a mechanism that requires AhR in cDC2.
A recent study used single-cell transcriptomics to identify a specific location-associated cellular composition and transcriptomic profile of cerebrospinal fluid (CSF) and blood leukocytes in relapsing remitting (RR) multiple sclerosis (MS) patients and controls, finding that cDC1 and cDC2 clusters had a higher proportion in CSF than that in blood (Schafflick and Xu, 2020). We interrogated the same single-cell RNAseq datasets to investigate the expression of Trp metabolic enzymes, including Ido1 and AhR in the different cell clusters (Figure 7N). In agreement with what we found in mouse cells (Figure 1), we confirmed that Ido1 was expressed in cDC1, whereas AhR was more broadly expressed in the myeloid compartment, both in blood and CSF (Figures 7O and 7P). In addition, cDC1 cells in CSF did not express other kynurenine enzymes, whereas cDC1 from blood did express other kynurenine metabolic enzymes, suggesting a location-specific presence of Trp metabolism in CSF located cDC1. Similarly, only in CSF, a distinct type of monocytes was found expressing Ido1 but not in blood (Figures 7O and 7P).
We next aimed to identify compartment-specific gene expression signatures on a per cluster level in MS versus control using the same model (Figures 7Q and S7D). We focused our attention on genes encoding Trp metabolic enzymes, AhR, SOCS3, and STAT3. We found that a greater proportion of those genes was differentially expressed in blood than in CSF (Figures 7R and 7S). We identified a slight increase of cDC1 cells expressing IDO1 only in control blood compared with MS blood, whereas in CSF, IDO1 was highly expressed in one type of monocyte cluster (Figure 7R). Moreover, CSF MS cDC1 expressed the kynurenine enzymes, such as KYNU, suggesting that l-kyn might be faster catabolized by these cells and not used to activate the immunoregulatory factor AhR. Overall, in this analysis focused on genes related to Trp metabolisms and metabolite sensing, we identified a compartment-specific leukocyte transcriptome and composition including an enrichment of cDC1 in the CSF. These findings emphasized the differential immune microenvironment of the CSF and blood related to those pathways in MS.
Discussion
The main finding of this study is that cDCs use a pathway of metabolic communication to maintain self-tolerance, in which the cDC1 subset controls the ability of cDC2 subset to become tolerogenic. Previous studies reported that Ido1 mRNA was expressed by a small subset of XCR1+CCR7+cDC1 (Ardouin et al., 2016; Breton et al., 2016; Lindenbergh and Stoorvogel, 2018) and that homeostatic cDC maturation was associated with acquired regulatory function (Ardouin et al., 2016), including promotion of tolerance in T cells and conversion to Treg cells (Nguyen et al., 2010). However, these studies did not functionally link these effects to IDO1 expression in this subset. Here, using cellular and molecular tools, we show that cDC1 maturation is accompanied by the acquisition of an IDO1-dependent regulatory function.
In agreement with recent studies into the molecular basis of cDC1-selective IRF8-mediated gene expression (Kim et al., 2020), we also show that the initial IDO1 expression by cDC1 is based on the usage of specific AICE regions in the Ido1 enhancer. Specifically, of two AICEs identified in the Ido1 promoter, one is used preferentially by cDC1, but not by cDC2 or pDCs and conditioning IDO1 expression in cDC1. Moreover, LPS treatment increases IDO1 in cDC1, whereas it increases IL-6 and SOCS3 expressions in cDC2.Thus, we clarify that the previously recognized action of IL-6 to promote IDO1 degradation (Orabona et al., 2008) appears to be manifested preferentially in cDC2 and not in cDC1. This finely tuned balance of IDO1 production and degradation suggests that under some inflammatory conditions, such as those mimicked by the presence of LPS or in autoimmunity, cDC2 could be resistant to acquiring a regulatory function, unless they receive effective communication from cDC1, via activation of Trp metabolism. This feature has a broader significance since it might apply to human cDC2 that share a similar cytokine profile secretion, including the production of IL-6 with murine cDC2 (Schlitzer et al., 2013). Moreover, our results indicate that the functional phenotype of a specific DC subset can be profoundly modulated by other neighboring DCs.
To date, very little is known about molecular mechanisms underlying crosstalk between different cDCs, resulting in the transfer of tolerogenic functions from a cDC subset to another and on the role of the metabolism of a specific aminoacidic in these processes. DC subset communication with other immune cells may involve several mechanisms, including cell-to-cell contact (Gao et al., 2017; Pasqual et al., 2018), extracellular vesicles (Kowal and Tkach, 2019; Torralba et al., 2018), and release of specific molecules by DCs, including cytokines, chemokines, and lipids (Minarrieta et al., 2017). Here, we show that an efficient system to bias the function of cDCs toward a regulatory and tolerogenic function is the release of Trp metabolites, l-kyn, through the action of IDO1 enzyme active in cDC1. l-kyn can act in both autocrine as well as paracrine fashion, influencing the activity of inflammatory cDC2 that express l-kyn specific carriers and receptor. Communication between cDC1 and cDC2, involving the Trp catabolite l-kyn, acts as rapid and efficient metabolic system to spread tolerogenic activity from cDC1 to the cDC2 subset. Specifically, to respond to l-kyn produced by cDC1 or provided exogenously, cDC2s need to express both AhR acting as l-kyn receptor and RelB as an AhR coactivator. Such condition often occurs upon cDC contact with LPS, leading to AhR transcriptional activity culminating in binding to AhR-RelB responsive elements in the Ido1 gene (Vogel et al., 2013). Dysregulation of these coordinated responses may lead to excessive immune activation and immunopathology, as that found in selected autoinflammatory conditions or in autoimmunity. Accordingly, Ido1−/− mice exhibit more severe clinical scores in EAE, compared with WT mice (Yan et al., 2010). Furthermore, modulation of the kynurenine pathway by inhibition of kynurenine monoxigenase (KMO) with the drug Ro 61-8048 resulted in increase in Foxp3 cell numbers and marked amelioration of EAE disease (Sundaram et al., 2020). Accordingly, specific Trp metabolites are frequently dysregulated in both EAE and MS patients (Gaetani et al., 2020; Takenaka et al., 2019). Our data suggest that this may be mainly due to decreased IDO1 expression and activity, particularly in cDC1, leading to decreased kynurenine production and thus affecting the regulatory functions of other inflammatory cDCs.
The fact that cDC1-less mice developed more severe EAE suggests that the regulatory function of IDO1 resides primarily in cDCs and the absence of IDO1 in other cell types, such as the cells in the gut, could be compensated by microbiota production of immune-active Trp metabolites (Zelante et al., 2013).
Higher amounts of IL-6 have been observed in plasma of MS patients compared with controls (Frei et al., 1991). Our results suggest that increased IL-6 production might be suppressive of IDO1 in cDC2, thus sustaining autoimmune Th17 and Th1 cell autoreactivity. In agreement, we found that cDC2 reduced IL-6 production and acquired IDO1 activity when in contact with IDO1 competent cDC1 in vitro or in mice treated with the Trp metabolite l-kyn in vivo. Our data also suggest that Ido1 mRNA and protein could become potential markers for assessing the presence of specific regulatory functions in cDCs of MS patients. Indeed, we found expression of IDO1 in mature human cDC1 and only minimal in cDC2 both at the steady state and after treatment with LPS. Re-analysis of single-cell mRNA expression of CSF cell clusters of MS patients revealed that Ido1 mRNA expression was found only in cDC1 but not in cDC2, suggesting a potential defect of cDC cross-regulation in MS patients. Accordingly, in patients with early MS, specific alterations were detected in DCs expressing the CD141hiIRF8hiCXCR3+CD68− markers (Böttcher et al., 2019). Altogether, our results highlight that applying effective means to activate antigen-specific tolerogenic responses, potentially in cDCs, may lead to the establishment of long-term tolerance by promoting active myelin reconstitution in EAE.
Limitations of the study
At present, little is known about whether cDC communication may be dysfunctional in specific autoimmune diseases, including MS, and future work will be needed to address this issue and to determine whether other DCs, such as pDCs, which also express AhR and RelB, or monocyte-derived DCs might be targeted by regulatory cDC1 expressing IDO1 or other immune regulatory pathways. Finally, whether additional inflammatory signals or other TLR ligands, besides LPS, may be able to activate similar cDC communication remains a subject for future investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BV786 rat anti-mouse CD117 (clone: 2B8) | BD Bioscience | Cat#: 564012; RRID: AB_2732005 |
| PeCF594 rat anti-mouse CD135 (clone: A2F10.1) | BD Bioscience | Cat#: 562537; RRID: AB_2737639 |
| 500 rat anti-mouse I-A/I-E (clone: M5/114.15.2) | BD Bioscience | Cat#: 562366; RRID: AB_11153488 |
| BV421 mouse anti-mouse CCR9 (clone: CW-1.2) | BD Bioscience | Cat#: 565412; RRID: AB_2739223 |
| AF700 rat anti-mouse Ly6C (clone: AL-21) | BD Bioscience | Cat#: 561237; RRID: AB_10612017 |
| AF488 rat anti-mouse CD127 (clone: SB/199) | BD Bioscience | Cat#: 561533; RRID: AB_10892634 |
| BV421 mouse anti-mouse LAP (clone: TW7-16B4) | BD Bioscience | Cat#: 565638; RRID: AB_2739315 |
| BV786 rat anti-mouse B220 (clone RA3-6B2) | BD Bioscience | Cat#: 563894; RRID: AB_2738472 |
| BV605 rat anti-mouse CD24 (clone: M1/69) | BD Bioscience | Cat#: 563060; RRID: AB_2737981 |
| APC rat anti-mouse CD172 (clone: P84) | BD Bioscience | Cat#: 46-1721-82; RRID: AB_10804639 |
| APC Cy7 or BV786 rat anti-mouse CD44 (clone: IM7) | BD Bioscience | Cat#: 560568; RRID: AB_1727481 Cat#: 563736, RRID: AB_2738395 |
| FITC rat anti-mouse CD86 (clone: GL-1) | BD Bioscience | Cat#: 553691; RRID: AB_394993 |
| BV650 rat anti-mouse CD197 (CCR7) (clone 4B12) | BD Bioscience | Cat#: 564356; RRID: AB_2738766 |
| BV421 rat anti-human Cleac9A (clone: 34A/Cleac9A) | BD Bioscience | Cat#: 564266, RRID: AB_2738716 |
| PerCP ef710 or biotin rat anti-mouse CD172 (clone: P84) | eBioscience | Cat#: 46-1721-82; RRID: AB_10804639 Cat#: 13-1721-82; RRID: AB_1963572 |
| APC rat anti-mouse CD317 (clone: eBio927) | eBioscience | Cat#: 17-3172-82; RRID: AB_10596356 |
| PerCP-eFluor710 rat anti-mouse SiglecH (clone: eBio-440c) | eBioscience | Cat#: 46-0333-82; RRID: AB_1834443 |
| PE mouse anti-mouse TLR4/MD2 complex (clone: MTS510) | eBioscience | Cat#: 2-9041-80, RRID: AB_466236) |
| Biotin rat anti-mouse CD105 (clone: MJ/18) | eBioscience | Cat#: 13-1051-82; RRID: AB_466556 |
| APC-eFluor 780 armenian hamster anti-mouse/human CD11c (clone: N418) | eBioscience | Cat#: 47-0114-82; RRID: AB_1548652 |
| PE rat anti-mouse CD40 (clone: 1C10) | eBioscience | Cat#: 12-0401-81; RRID: AB_465648 |
| Biotin rat anti-mouse CD117 (clone: 2B8) | BioLegend | Cat#: 105804; RRID: AB_313212 |
| Biotin rat anti-mouse Ly6G (clone: AL-21) | BioLegend | Cat#: 127604; RRID: AB_1186108 |
| Biotin rat anti-mouse/human B220 (clone: RA3-6B2) | BioLegend | Cat#: 103204; RRID: AB_312989 |
| Biotin rat anti-mouse Ter119 (clone: TER-119) | BioLegend | Cat#: 116204; RRID: AB_313705 |
| Biotin rat anti-mouse CD3e (clone: 17A2) | BioLegend | Cat#: 100244; RRID: AB_2563947 |
| APC armenian hamster anti-mouse CD3e (clone: 145-2C11) | BioLegend | Cat#: 100312, RRID: AB_312677 |
| Biotin rat anti-mouse CD19 (clone: 6D5) | BioLegend | Cat#: 115504; RRID: AB_313639 |
| Biotin, BV711 or Pacific Blue mouse anti-mouse CD90/CD90.1 (Thy1.1) (clone: OX-7) | BioLegend | Cat#:202510; RRID: AB_2201417 Cat#: 202539; RRID: AB_2562645 Cat#: 202522; RRID: AB_1595477 |
| Biotin rat anti-mouse CD4 (clone: GK1.5) | BioLegend | Cat#: 100404; RRID: AB_312689 |
| Biotin rat anti-mouse CD8a (clone: 53-6-7) | BioLegend | Cat#: 100704; RRID: AB_312743 |
| FITC rat anti-mouse I-A/I-E (clone: M5/114.15.2) | BioLegend | Cat#: 107605; RRID: AB_313321 |
| PE or biotin rat anti-mouse CD197 (CCR7) (clone 4B12) | BioLegend | Cat#: 120106; RRID: AB_389358 Cat# 120104; RRID: AB_389232 |
| PeCy7 rat anti-mouse CD24 (clone: M1/69) | BioLegend | Cat#: 101822; RRID: AB_756048 |
| PE or BV421 mouse anti-mouse/rat XCR1 (clone: ZET) | BioLegend | Cat#: 148212; RRID: AB_2564367 Cat#: 148216; RRID: AB_2565230 |
| Biotin or PE rat anti-mouse TCR Vα2 (clone: B20.1) | BioLegend | Cat#: 127803; RRID: AB_1134185 Cat#: 127808; RRID: AB_1134183 |
| Biotin or FITC mouse anti-human CD3 (clone: UCHT1) | BioLegend | Cat#: 302403; RRDI: AB_314058 Cat#: 3024055; RRDI: AB_2562046 |
| Biotin or BV711 mouse anti-human CD14 (clone: M5E2) | BioLegend | Cat#: 301826; RRDI: AB_2291250 Cat#: 301837; RRDI: AB_2562909 |
| Biotin or FITC mouse anti-human CD66b (clone: G10F5) | BioLegend | Cat#: 305120; RRDI: AB_2566608 Cat#: 302103; RRDI: AB_314496 |
| Biotin or FITC mouse anti-human CD19 (clone: HIB19) | BioLegend | Cat#: 302204; RRDI: AB_314233 Cat#: 302205; RRDI: AB_314235 |
| Biotin or FITC mouse anti-human CD20 (clone: 2H7) | BioLegend | Cat#: 302349; RRDI: AB_2565523 Cat#: 302303; RRDI: AB_314252 |
| Biotin or FITC mouse anti-human CD335 (clone: 9E2) | BioLegend | Cat#: 331906; RRDI: AB_1027671 Cat#: 331921; RRDI: AB_2561964 |
| APC Cy7 mouse anti-human CD16 (clone: 3G8) | BioLegend | Cat#: 302017; RRDI: AB_314218 |
| PeCy7 mouse anti-human CD1c (clone: L161) | BioLegend | Cat#: 331515; RRDI: AB_1953227 |
| BV510 mouse anti-human CD11c (clone: 3.9) | BioLegend | Cat#: 301634, RRID: AB_2563795 |
| PerCP/Cy5.5 mouse anti-human CD123 (clone: 6H6) | BioLegend | Cat#: 306016; RRDI: AB_2264693 |
| BV785 mouse anti-human HLA-DR (clone: L243) | BioLegend | Cat#: 307642, RRID: AB_2563461 |
| BV605 mouse anti-human CD303 (Clone: 201A) | BioLegend | Cat#: 354224, RRID: AB_2572149 |
| PE armenian hamster CD80 (clone: 16-10A1) | BioLegend | Cat#: 104707; RRID: AB_313128 |
| PE rat anti-mouse B220 (clone: clone RA3-6B2) | BioLegend | Cat#: 103208; RRID: AB_312993 |
| PE/Dazzle 594 rat anti-mouse CD223 (clone: C9B7W) | BioLegend | Cat#: 125224, RRID: AB_2572082 |
| APC Cy7 rat anti-mouse CD25 (clone: PC61) | BioLegend | Cat#: 102026; RRID: AB_830745 |
| PE goat anti-mouse IgG (minimal x-reactivity) (clone: Poly4053) | BioLegend | Cat#: 405307; RRID: AB_315010 |
| Rat IgG2a,k Isotype Ctrl (clone: RTK2758) | BioLegend | Cat#: 400502; RRID: AB_326523 |
| Mouse IgG1k Isotype Ctrl (clone: MG1-45) | BioLegend | Cat#: 401402; RRID: AB_2801451 |
| Biotin or APC human anti-human CD141(clone: REA674) | Milteny Biotech | Cat#: 130-114-186, RRID: AB_2751231 Cat#: 130-114-184, RRID: AB_2751230 |
| Biotin human anti-human CD1c (clone: REA694) | Milteny Biotech | Cat#: 130-110-593, RRID: AB_2656043 |
| Monoclonal Anti-alpha-Tubulin-FITC antibody produced in mouse | Sigma Aldrich | Cat#: F2168; RRID: AB_476967 |
| Mouse anti-mouse IDO1 (clone: 8G11) | Merck | Cat#: MABS485 |
| Mouse anti l-kynurenine (clone 3D4-F29) | ImmuSmol | Cat#: IS0003 |
| Polyclonal sheep anti-mouse/human AhR | R&DSYSTEM | Cat#: AF6697; RRID: AB_10891869 |
| Normal Sheep IgG Isotype Ctrl | R&DSYSTEM | Cat#: 5-001-A; RRID: AB_10141430 |
| Mouse anti-mouse RelB (clone: A-9) | Santa Cruz Biotechnology | Cat#: sc-166416; RRID: AB_2179178 |
| Mouse anti-mouse RelB (clone: D-4) | Santa Cruz Biotechnology | Cat#: sc-48366; RRID: AB_628212 |
| Mouse anti-mouse ICSKB x (clone: E-9) | Santa Cruz Biotechnology | Cat#: sc-365042 X; RRID: AB_10850401 |
| Polyclonal rabbit anti-mouse SOCS3 | Cell Signaling | Cat#: 2923; RRID: AB_2255132 |
| Rabbit monoclonal anti-mouse RelB (clone C1E4) | Cell Signaling | Cat#: 4922; RRID: AB_2179173 |
| Mouse monoclonal anti-beta-Actin (clone: AC-40) | Sigma Aldrich | Cat#: A3853; RRID: AB_262137 |
| Mouse monoclonal anti-beta-Tubulin (clone: AA2) | Sigma Aldrich | Cat#: T8328; RRID: AB_1844090 |
| Lamin B polyclonal antibody | Thermo Fisher Scientific | Cat#: PA519468; RRID: AB_10985414 |
| Mouse monoclonal anti-mouse AhR (clone: RPT1) | Thermo Fisher Scientific | Cat#: MA1-514; RRID: AB_2273723 |
| Polyclonal rabbit anti-mouse IDO2 | Dept. Experimental Medicin, Phamacology Section, Univeristy of Perugia | N/A |
| Polyclonal rabbit anti-mouse TDO2 | Dept. Experimental Medicin, Phamacology Section, University of Perugia | N/A |
| Mouse monoclonal anti-huma IDO1 (clone 10.1) | Sigma Aldrich | Cat#:05-840; RRID: AB_310044 |
| Rabbit anti-Sheep IgG (H+L) HPR conjugate | Thermo Fisher Scientific | Cat#: 31480; RRID: AB_228457 |
| Goat anti-Mouse IgG (H+L) HPR conjugate | Thermo Fisher Scientific | Cat#: 31430; RRID: AB_228307 |
| Rabbit anti-Rabbit IgG (H+L) HPR conjugate | Thermo Fisher Scientific | Cat#: 31460; RRID: AB_228341 |
| Bacterial and virus strains | ||
| One Shot™ TOP10 Chemically Competent E.Coli | Thermo Fisher Scientific | Cat#: C404006 |
| Subcloning Efficiency DH5a competent cells | Invitrogen™ | Cat#: 18265017 |
| Chemicals, peptides, and recombinant proteins | ||
| TransIT-LTI | Mirus Bio | Cat#: MIR2300 |
| PMA (phorbol 12-myristate 13-acetate) | Sigma-Aldrich | Cat#: P1585 |
| Ionomycin | Sigma-Aldrich | Cat#: I9657 |
| Brefeldin A | eBioscience | Cat#: 00-4506-51 |
| Paraformaldehyde | Electron Microscopy Science | Cat#: 15714 |
| Poly-L-lysin solution 0.1% | Sigma-Aldrich | Cat#: P8920 |
| Saponin form quillaja bark | Sigma-Aldrich | Cat#: S7900 |
| Lipopolysaccharide from Escherichia coli (055: B5) | Sigma-Aldrich | Cat#: L2880 |
| Albumin from chicken egg white (OVA) | Sigma-Aldrich | Cat#: A5503 |
| l-kynurenine | Sigma-Aldrich | Cat#: K8625 |
| Soluble Toxoplasma gondii antigen (STAg) | Tussiwand et al., 2012 | N/A |
| 2-Amino-2-norbomanecarboxylic acid | Sigma-Aldrich | Cat#: A7902 |
| Bovine serum albumin Fraction V (Immunoglobulin and Protease Free) (BSA) | Rockland | Cat#: BSA-50 |
| Blotting-Grade Blocker (nonfat dry milk) | BioRad | Cat#: 170-6404 |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat#: 1000-4D |
| Proteinase K | Sigma-Aldrich | Cat#: P6556 |
| Deoxyribonuclease I from bovine pancreas | Sigma-Aldrich | Cat#: D4527 |
| Collagenase B from Clostridium histolyticum | Roche | Cat#: 11088815001 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat#:11697498001 |
| Ficoll-Paque™ Plus | GE Healthcare | Cat#:17-1400-02 |
| Sodium Orthovanadate | Sigma-Aldrich | Cat#: 450243 |
| Sodium Fluoride | Sigma-Aldrich | Cat#: 215309 |
| Aprotin from bovine lung | Sigma-Aldrich | Cat#: A1153 |
| Leupeptin hydrocloride | Sigma-Aldrich | Cat#: L9783 |
| Pepstatin A | Sigma-Aldrich | Cat#: P5318 |
| Phenylmethanesulfonyl fluoride (PMFS) | Sigma-Aldrich | Cat#: P7625 |
| Histopaque 1119 | Sigma-Aldrich | Cat#: 1119 |
| Hexadimethrine bromide (Polybrene) | Sigma-Aldrich | Cat#: H9268 |
| Anti-IL-6 25 μg/ml (clone 20 F3) | Division of Biological Chemestry, Biocente, Innsbruck Medical University, Austria Louis Boon |
N/A |
| LEAFTM Purified anti-mouse/rat CD126 (IL-6Rα chain, clone D7715A7) | BioLegend | Cat#: 115815; RRID: AB_2810349 |
| Incomplete Freund’s Adjuvant | BD Bioscience | Cat#: 263910 |
| MOG35–55 peptide (MEVGWYRSPFSRVVHLYRNGK) | Cambridge Research Biochemicals | Cat#: crb1000205n |
| Mycobacterium tuberculosis TB H37 Ra | BD Bioscience | Cat#: 231141 |
| Pertussis toxin from B. pertussis | List Biological Laboratories | Cat#: 180 |
| H-2Db-restricted HY peptide (WMHHNMDLI) | BioFab Research | N/A |
| Iscove’s Modified Dulbecco’s Media | GIBCO™ | Cat#: 12440-053 |
| Opti-MEM Reduced Serum Medium | GIBCO™ | Cat#: 31985-070 |
| Hanks’ Balanced Salt Solution (HBSS) | GIBCO™ | Cat#: 14170-112 |
| Fetal Bovine Serum (Characterized) | GIBCO™ | Cat#: #10270-106 |
| Sodium pyruvate | GIBCO™ | Cat#: 11360-070 |
| L-Glutamine | GIBCO™ | Cat#: 25030-024 |
| Pen Strep (Penicillin Streptomycin) | GIBCO™ | Cat#: 15140-122 |
| 2-Mercaptoethanol | GIBCO™ | Cat#: 31350-010 |
| MEM Non-essential Amino Acid Solution (100X) | GIBCO™ | Cat#: 11140-035 |
| Trypsin-EDTA (0.05%), phenol red | GIBCO™ | Cat#: 25300-062 |
| MG-132 Ready Made solution | Sigma-Aldrich | Cat#: M7449 |
| Recombinant murine IL-12 | Peprotech | Cat#: 210-12 |
| L-1-methyltryptophan | Sigma-Aldrich | Cat#: 44739 |
| HindIII | New England BioLab | Cat#: R0104S |
| BamHI | New England BioLab | Cat#: R0136S |
| BbsI | New England BioLab | Cat#: R0539S |
| MagniSort™ streptavidin negative selection beads | Invitrogen™ | Cat#: MSNB-6002 |
| MojoSort™ Streptavidin Nanobeads | Biolegend | Cat#: 480016 |
| Qdot™605 Streptavidin conjugate | Invitrogen™ | Cat#: Q10101MP |
| APC or PeCy7 Streptavidin | BioLegend | Cat#: 405207, Cat#: 405206 |
| Anti-Biotin MicroBeads UltraPure | Milteny Biotech | Cat#: 130-105-637; RRID: AB_2811216 |
| CD4 (L3T4) microbeads mouse | Milteny Biotech | Cat#: 130-117-043 |
| CD8 (Ly-2) microbeads mouse | Milteny Biotech | Cat#: 130-117-044 |
| Clean-Blot™ IP Detection Reagent (HRP) | BioRad | Cat#: 21230; RRID: AB_2864363 |
| Clarity Western ECL Substrate | BioRad | Cat#: 1705061 |
| Clarity Max Western ECL Substrate | BioRad | Cat#: 1705062 |
| RLT buffer | Qiagen | Cat: #79216 |
| Duolink™ In Situ Mounting Medium with DAPI | Sigma-Aldrich | Cat#: DUO82040 |
| Alexa Fluor™ 488 Phalloidin | Invitrogen™ | Cat#: 12379 |
| ProLong™ Gold Antifade Mountant with DAPI | Invitrogen™ | Cat#: P36931 |
| Critical commercial assays | ||
| QIAamp DNA Micro Kit | Qiagen | Cat#: 56304 |
| IL6 Mouse Uncoated ELISA Kit | Thermo Fisher Scientific | Cat#: 88-7064-88; RRID: AB_2574990 |
| IFN gamma Mouse Uncoated ELISA Kit | Thermo Fisher Scientific | Cat#: 88-7314-88; RRID: AB_2575070 |
| Mouse TGF-beta 1 DuoSet ELISA | R&DSYSTEM | Cat#: DY679-05; RRID: AB_2797393 |
| Bio-Plex Pro Mouse Cytokines Grp I Panel 23-plex | BioRad | Cat#: M60009RDPD; RRID: AB_2857368 |
| CellTrace™ CFSE Cell Proliferation Kit | Thermo Fisher Scientific | Cat#: C34554 |
| Quantitect Reverse transcription kit | Qiagen | Cat#: 205313 |
| RNeasy Mini Kit | Qiagen | Cat#: 74104 |
| iTaq Universal SYBR Green Supermix | BioRad | Cat#:1725124 |
| Phusion High-Fidelity DNA Polymerase | New England Biolab | Cat#: M0530S |
| Bradford protein assay | BioRad | Cat#: 500-0006 |
| Duolink™ In Situ Detection Reagent Red | Sigma-Aldrich | Cat#: DUO92008 |
| Deposited data | ||
| Microarrays of BM-derived cDC1, cDC2, and pDC | This study | GSE203449 and GSE203450 |
| Microarrays for mature cDC1 | Ardouin et al., 2016 | GSE71171 |
| ChIP-seq datasets for IRF4, IRF8, BATF3, H3K4me1, H3K27ac | Grajales-Reyes et al., 2015 | GSE66899 |
| scRNA-seq datasets for MS and control patients | Schafflick and Xu, 2020 | https://github.com/chenlingantelope/MSscRNAseq2019.git |
| Experimental models: Cell lines | ||
| Mouse: Ahr−/− MEFs | Manni et al., 2020 | N/A |
| Mouse: MEF MSCV-Ahr-IRES-hCD4 | This paper | N/A |
| Mouse: MEF MSCV-Ahr Q377A-IRES-hCD4 | This paper | N/A |
| Mouse: Raw 264.7 | ATCC | Cat#: ATCC TIB-71; RRID: CVCL_0493 |
| Mouse: Raw 264.7 MSCV-Ido1-IRES-GFP | This paper | N/A |
| Mouse: Raw 264.7 MSCV-Ido2-IRES-GFP | This paper | N/A |
| Mouse: Raw 264.7 MSCV-Tdo2-IRES-GFP | This paper | N/A |
| Human: Platinum-E retroviral packaging cell line | Morita et al., 2000 | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Charles River Laboratories | Crl: 027; RRID: IMSR_CRL:027 |
| Mouse: B6.129-Ido1tm1Alm/J (Ido1−/−) | Jackson Laboratory | JAX: 005867; RRID: IMSR_JAX:005867 |
| Mouse: B6.129- Ahrtm1Bra/J mice (Ahr−/−) | MRC National Institute for Medical Research, London, UK, B. Stockinger | N/A |
| Mouse: Ahrtm3.1Bra/J (Ahrf/f) | Jackson Laboratory | JAX: 006203; RRID: IMSR_JAX:006203 |
| Mouse: B6.Cg-Tg (Itgax-cre) 1-1Reiz/J (CD11c-Cre) | Jackson Laboratory | JAX:008068; RRID: IMSR_JAX:008068 |
| Mouse: Ahrf/f CD11c Cre | This paper | N/A |
| Mouse: Ahrf/f Vav iCre | This paper | N/A |
| Mouse: B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J (Vav-iCre) | Jackson Laboratory | JAX:008610; RRID: IMSR_JAX:008610 |
| Mouse: B6J.129(Cg)-Gt(ROSA) 26Sortm1.1(CAG-cas9∗,-EGFP)Fezh/J (Rosa26Cas9-GFP /Cas9-GFP) | Jackson Laboratory | JAX: 026179; RRID: IMSR_JAX:026179 |
| Mouse: B6.129S(C)-Batf3tm1Kmm/J (Batf3−/−) | Jackson Laboratory | JAX: 013755; RRID: IMSR_JAX:013755 |
| Mouse: Irf8VENUS | Grajales-Reyes et al., 2015 | N/A |
| Mouse: Batf3−/−Irf8VENUS | Grajales-Reyes et al., 2015 | N/A |
| Mouse: Relb−/− | Briseño et al., 2017, Briseño et al., 2017 | N/A |
| Mouse: B6.SJLPtprcaPepcb/BoyCrl (CD45.1) | Charles River Laboratories | Crl: 494; RRID: IMSR_CRL:494 |
| Mouse: C57BL/6 Tg(TcraTcrb)1100Mjb/Crl (OT-I) | Charles River Laboratories | Crl: 642; RRID: IMSR_CRL:642 |
| Mouse: C57BL/6-Tg(TcraTcrb)425Cbn/Crl (OT-II) | Charles River Laboratories | Crl: 643; RRID: IMSR_CRL:643 |
| Mouse: B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J (Foxp3YFP-cre) | Jackson Laboratory | JAX: 016959; RRID :IMSR_JAX:016959 |
| Mouse: OT-II Foxp3 YFP | This paper | N/A |
| Mouse: Irf4f/fCD11c Cre | This paper | N/A |
| Mouse: B6.129S2-Il6tm1Kopf/J (Il6−/−) | Jackson Laboratory | JAX: 002650; RRID: IMSR_JAX:002650 |
| Mouse: Ido1f/fXcr1Cre/+ | This paper | N/A |
| Mouse: C57BL/6J-Irf8em1Kmm/J (Irf8 Δ32−/−) | Jackson Laboratory | JAX: 032744; RRID: IMSR_JAX:032744 |
| Mouse: Ahrf/f Zbtb46 Cre | This paper | N/A |
| Oligonucleotides | ||
| CpG 1826 PTO (T∗C∗C∗A∗T∗G∗A∗C∗G∗T∗T∗C∗C∗T∗G∗A∗C∗G∗T∗T) | BioFab Research | N/A |
| Polyinosinic:polycytidylic acid Poly I:C | BioFab Research | N/A |
| Duolink™ In Situ Probemarker PLUS | Sigma-Aldrich | Cat#: DUO92009 |
| Duolink™ In Situ Probemarker MINUS | Sigma-Aldrich | Cat#: DUO92010 |
| Sequence of primer used for real time PCR (5’>3’) | N/A | |
| Ido1 (FW): CGATGTTCGAAAGGTGCTGC | Sigma-Aldrich | N/A |
| Ido1 (RV): GCAGGAGAAGCTGCGATTTC | Sigma-Aldrich | N/A |
| Ido2 (FW): TCAGACTTCCTCACTTAATCG | Sigma-Aldrich | N/A |
| Ido2 (RV): GCTGCTCACGGTAACTCT | Sigma-Aldrich | N/A |
| Tdo2 (FW): GTGAAGGACGACTGTCATACCG | Sigma-Aldrich | N/A |
| Tdo2 (RV): GCTGAAAGGGACCTGGAAT | Sigma-Aldrich | N/A |
| Il-6 (FW): CCGGAGAGGAGACTTCACAG | Sigma-Aldrich | N/A |
| Il-6 (RV): TCCACGATTTCCCAGAGAAC | Sigma-Aldrich | N/A |
| Socs-3 (FW): CAGCCTGCGCCTCAAGACCTT | Sigma-Aldrich | N/A |
| Socs-3 (RV): GCACCAGCTTGAGTACACAGTCG | Sigma-Aldrich | N/A |
| RelB (FW): GCCAGAGAGTCCGCGCC | Sigma-Aldrich | N/A |
| RelB (RV): CAATTCATCTGTGGTCCTGGAGA | Sigma-Aldrich | N/A |
| AhR (FW): TCTGTTCTTAGGCTCAGCGTC | Sigma-Aldrich | N/A |
| AhR (RV): GCGCCTGTAACAAGAACTCTC | Sigma-Aldrich | N/A |
| Slc7a5 (FW): CAGCTCCCTGAGTATGAAAGC | Sigma-Aldrich | N/A |
| Slc7a5 (RV): CCATTCCAGTAGACACCCCTTC | Sigma-Aldrich | N/A |
| Tnf-α (FW): TCTACTGAACTTCGGGGTGA | Sigma-Aldrich | N/A |
| Tnf-α (RV): CACTTGGTGGTTTGCTACGA | Sigma-Aldrich | N/A |
| Pdl1(FW): ACACTCTGAAGGAGGCTT | Sigma-Aldrich | N/A |
| Pdl1 (RV): ACGCCACATTTCTCCACATCT | Sigma-Aldrich | N/A |
| Actin (FW): GGC TCC TAG CAC CAT GAA GA | Sigma-Aldrich | N/A |
| Actin (RV): AGC TCA GTA ACA GTC CGC C | Sigma-Aldrich | N/A |
| Sequence of primer used for ChIP PCR (5’>3’) | Sigma-Aldrich | N/A |
| AICE-1 (FW): GCCATCAGAGCCAACAGTTG | Sigma-Aldrich | N/A |
| AICE-1 (RV): TTGGCCACTGAAGGAATGTC | Sigma-Aldrich | N/A |
| XRE +1340 (FW): CCTTGCTACAAGTGTCCTGG | Sigma-Aldrich | N/A |
| XRE +1340 (RV): GTCCGGGGTCATCACAACAT | Sigma-Aldrich | N/A |
| AhRE III +1601 (FW): TCTGTGTTTTCCGATGGTCTT | Sigma-Aldrich | N/A |
| AhRE III +1601 (RV): AGGCAATGTCCCCAGATGAA | Sigma-Aldrich | N/A |
| IL-6 (FW): AGCACACTTTCCCCTTCCTA | Sigma-Aldrich | N/A |
| IL-6 (Rev): TGAGCTACAGACATCCCCAG | Sigma-Aldrich | N/A |
| Sequences of primers used for cloning IDO1 enhancer elements (5’>3’) | Sigma-Aldrich | N/A |
| AICE 1 -126 bp (FW): GGAAAAGCTTGCCATCAGAGCCAACAG TTG |
Sigma-Aldrich | N/A |
| AICE 1 -126 bp (RV): GACCGGATCCTTGGCCACTGAAGGAAT GTC. |
Sigma-Aldrich | N/A |
| AICE 2 +495 bp (FW): GCTCAAGCTTGGGAAGCCAGTTTGTCA CAA |
Sigma-Aldrich | N/A |
| AICE 2 +495 bp (RV): TCCAGGATCCGCACAAAAGCACAATCAA GGT |
Sigma-Aldrich | N/A |
| XRE -4468 bp (FW): TGGAAAGCTTACTAGTGCCAGTGTAACCAG | Sigma-Aldrich | N/A |
| XRE -4468 bp (RV): ACAAGGATCCCAAACACAACCCCTGACCTG | Sigma-Aldrich | N/A |
| XRE -3398 bp (FW): GGCTAAGCTTCACAGGGCAATTAGGGAGC | Sigma-Aldrich | N/A |
| XRE -3398 bp (RV): GAGGGGATCCAGGAGGGTAAAACAGTTGG AAAGG |
Sigma-Aldrich | N/A |
| XRE +824 bp (FW): TAGCAAGCTTGCTGAGAACGCACTGAACAA | Sigma-Aldrich | N/A |
| XRE +824 bp (RV): ACCCGGATCCGAGACTAGGTGAGCATGGGG | Sigma-Aldrich | N/A |
| XRE +1340 bp (FW): AGCCAAGCTTCCTTGCTACAAGTGTCCTGG | Sigma-Aldrich | N/A |
| XRE +1340 bp (RV): ATTAGGATCCGTCCGGGGTCATCACAACAT | Sigma-Aldrich | N/A |
| RelAhRE -3729 bp (FW): GTCAAAGCTTGCTACATAGAGAGTGC AAGGC |
Sigma-Aldrich | N/A |
| RelAhRE -3729 bp (RV): AGGGGGATCCTCAAGCAAGTGAAGTTT TAGACC |
Sigma-Aldrich | N/A |
| XREIII +2169 bp (FW): CGGCAAGCTTTCCTGCAGCACCCTAGG | Sigma-Aldrich | N/A |
| XREIII +2169 bp (RV): ATATGGATCCGAATGGTCACCTTGTCACGT | Sigma-Aldrich | N/A |
| AhREIII +257 bp (FW): GGTAAAGCTTACCTTGATTGTGCTTTTGTGC | Sigma-Aldrich | N/A |
| AhREIII +257 bp (RV): TGCAGGATCCGACTGTCTGGCCTTGAATT TGA |
Sigma-Aldrich | N/A |
| AhREIII +1182 bp (FW): TCTAAAGCTTGTGGGCTAGAGACTGGG ATC |
Sigma-Aldrich | N/A |
| AhREIII +1182 bp (RV): CTGCGGATCCACTGGATTGGATGGGAG CTG |
Sigma-Aldrich | N/A |
| AhREIII +1601 bp (FW): AATGAAGCTTTCTGTGTTTTCCGATGGT CTT |
Sigma-Aldrich | N/A |
| AhREIII +1601 bp (RV): ATGTGGATCCAGGCAATGTCCCCAGAT GAA |
Sigma-Aldrich | N/A |
| Sequences of sgRNA for targeting IDO1 enhancer (+146 bp AICE-1) (5’>3’) | Sigma-Aldrich | N/A |
| sgRNA 1 (FW): CACCGAGTATTACGTGTCAGCTGTG | Sigma-Aldrich | N/A |
| sgRNA 1 (RV): AAACCACAGCTGACACGTAATACTA | Sigma-Aldrich | N/A |
| sgRNA 2 (FW): CACCGCTTCTCCCCGAGTATTACGT | Sigma-Aldrich | N/A |
| sgRNA 2 (RV): AAACACGTAATACTCGGGGAGAAGC | Sigma-Aldrich | N/A |
| sgRNA 3 (FW): CACCGTGACGAAGAGAGATCCTTTG | Sigma-Aldrich | N/A |
| sgRNA 3 (RV): AAACCAAAGGATCTCTCTTCGTCAC | Sigma-Aldrich | N/A |
| Sequences of sgRNA for targeting IDO1 enhancer (+1340 bp XRE) (5’>3’) | Sigma-Aldrich | N/A |
| sgRNA 1 (FW): CACCGGCATCTTTGGGTGCCCTGGC | Sigma-Aldrich | N/A |
| sgRNA 1 (RV): AAACGCCAGGGCACCCAAAGATGCC | Sigma-Aldrich | N/A |
| sgRNA 2 (FW): CACCGAATGTGGAACTGACCACCTC | Sigma-Aldrich | N/A |
| sgRNA 2 (RV): AAACGAGGTGGTCAGTTCCACATTC | Sigma-Aldrich | N/A |
| sgRNA 3 (FW): CACCGGGCGTGCCTGGTTTTGAGGT | Sigma-Aldrich | N/A |
| sgRNA 3 (RV): AAACACCTCAAAACCAGGCACGCCC | Sigma-Aldrich | N/A |
| Sequences of sgRNA for targeting IDO1 enhancer (+1601 bp AhREIII) (5’>3’) | Sigma-Aldrich | N/A |
| sgRNA 1 (FW): CACCGTCTAACAGGAAGCAGCCCTA | Sigma-Aldrich | N/A |
| sgRNA 1 (RV): AAACTAGGGCTGCTTCCTGTTAGAC | Sigma-Aldrich | N/A |
| sgRNA 2 (FW): CACCGTAGGGGTGGGACATTAGGAC | Sigma-Aldrich | N/A |
| sgRNA 2 (RV): AAACGTCCTAATGTCCCACCCCTAC | Sigma-Aldrich | N/A |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad | https://www.graphpad.com |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| FlowJo v10 | Tree Star | https://www.flowjo.com/solutions/flowjo |
| FACSDiva | BD Biosciences | https://www.bdbiosciences.com/en-eu/products/software/instrument-software/bd-facsdiva-software#Overview |
| Primer3 | RRID:SCR_003139 | http://bioinfo.ut.ee/primer3-0.4.0/ |
| BioRender | BioRender | https://biorender.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Francesca Fallarino (francesca.fallarino@unipg.it).
Materials availability
All animal strains used in this study are available from The Jackson Laboratory or Charles River Breeding Laboratories. No new animal strains were generated for this study.
Data and code availability
The accession number for the microarrays reported in this paper are available on the GEO database with the following accession number: GSE203449 and GSE203450.
Following datasets were downloaded and reanalyzed: microarrays for mature cDC1 (GSE71171) (Ardouin et al., 2016), ChIP-seq datasets for IRF4, IRF8, BATF3, H3K4me1, H3K27ac (GSE66899) (Grajales-Reyes et al., 2015), scRNA-seq datasets for MS and control patients (https://github.com/chenlingantelope/MSscRNAseq2019.git.) (Schafflick and Xu, 2020).
Experimental model and subject details
Mice
Wild type C57BL/6 mice were obtained from The Jackson Laboratory, Charles River Breeding Laboratories, or bred in our facility. Ido1−/− mice (B6.129-Ido1tm1Alm/J, JAX: 005867), Ahrf/f mice (Ahrtm3.1Bra/J, JAX: 006203), Itgax-Cre mice (B6.Cg-Tg (Itgax-cre) 1-1Reiz/J, JAX: 008068), Vav1-iCre mice (B6.Cg-Commd10Tg(Vav1-icre)A2Kio/J, JAX: 008610), Zbtb46-Cre mice (B6.Cg-Zbtb46tm3.1(cre)Mnz/J, JAX: 028538), Rosa26Cas9-GFP /Cas9-GFP mice (B6J.129(Cg)-Gt(ROSA) 26Sortm1.1(CAG-cas9∗,-EGFP)Fezh/J, JAX: 026179), OT-I mice (C57BL/6-Tg(TcraTcrb)1100Mjb/Crl, Crl: 642), OT-II mice (C57BL/6- Tg(TcraTcrb)425Cbn/Crl, Crl: 643), Batf3−/− mice (B6.129S(C)-Batf3tm1Kmm/J, JAX: 013755), Irf8VENUS, Batf3−/−Irf8VENUS (Grajales-Reyes et al., 2015), Irf4f/f mice (B6.129S1-Irf4tm1Rdf/J, JAX: 009380), Il6−/− (B6.129S2-Il6tm1Kopf/J, JAX: 002650), Irf8 Δ32−/− (C57BL/6J-Irf8em1Kmm/J, JAX: 032744) and Relb−/−(Briseño et al., 2017, Briseño et al., 2017) were all maintained on the C57BL/6 background. OT-II Foxp3 YFP mice were generated by crossing Foxp3YFP-cre (B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J, JAX: 016959) with OT-II mice. B6.129- Ahrtm1Bra/J mice (Ahr−/−) were kindly supplied by B. Stockinger (MRC National Institute for Medical Research, London, UK). Ahrf/f mice were backcrossed with Itgax-Cre, Vav1-iCre or Zbtb46-Cre mice. IRF4f/f mice were backcrossed with Itgax-Cre mice. Heterozygous Ido1tm1a(EUCOMM)Wtsi were purchased from Wellcome Trust Sanger Institute. The tm1a 201D-first allele contains a lacZ trapping cassette flanked by FRT sites and a human βactin promoter driven neo cassette flanked by loxP sites inserted in the intron sequence of the Ido1 gene disrupting the gene function. Tm1A mice were later bred with FLPeR mice (The Jackson Laboratory, Bar harbor, ME; strains 012930) to generate tm1c conditional allele with a restored gene function. Tm1c allele containing mice were bred with mice expressing Cre recombinase under control of Xcr1 cDC1 specific promoter (Bishnupuri et al., 2019). For bone marrow chimeras CD45.1 mice (B6.SJLPtprcaPepcb/BoyCrl, Crl: 494) were purchased from Charles River Laboratories. All mice were maintained in a specific pathogen-free animal facility following institutional guidelines and with protocols approved by the Animal Studies Committee at Washington University in St. Louis in compliance with National (Italian Parliament DL 26/2014) and Perugia University Animal Care and Use Committee guidelines. Most of the experiments were performed with mice 8–12 weeks of age, using sex-matched littermates.
Construction of retroviral vectors (RVs)
PCR products containing Ahr or Ahr Q377A cDNA were cloned in frame with MSCV-IRES-hCD4 (MSCV-Ahr-IRES-hCD4 or MSCV-Ahr Q377A-IRES-hCD4) and Ido1, Ido2 and Tdo2 cDNA were cloned in frame with MSCV-IRES-GFP (MSCV-Ido1-IRES-GFP, MSCV-Ido2-IRES-GFP and MSCV-Tdo2-IRES-GFP). Construction of the retroviral reporter vector (Thy1.1 pA GFP CMVp_min PmeI MSCV) for assessing transcriptional activities of enhancer elements for Ido1 are described in our previous studies (Durai et al., 2019a; Grajales-Reyes et al., 2015; Tussiwand et al., 2012). The enhancer regions of mouse Ido1 promoter containing: AICEs sequence at –126 bp and +495 bp; XREs sequence at –4468 bp, –3398 bp, +824 bp and +2169 bp; RelBAhRE sequence at –3729 bp; AhREIIIs sequence at +657 bp, +1182 bp and +1601 bp; XRE-AhREIII sequence encompassing +1340 bp–+1374 bp, were amplified from genomic DNA using Phusion High-Fidelity DNA Polymerase (#M0530S, New England Biolab). Purified PCR products were digested with HindIII and BamHI and cloned into the HindIII and BamHI digested retroviral reporter vector Thy1.1 pA GFP CMVp RV (Durai et al., 2019a). Sequences of oligonucleotides used for cloning the enhancer elements (200-350 bp) and the mutants are listed in the key resources table.
Cell lines
Ahr−/− MEFs were prepared as described (Manni et al., 2020). MSCV-Ahr-IRES-hCD4 or MSCV-Ahr Q377A-IRES-hCD4 retroviral vectors were transfected into Platinum-E cells with TransIT-LTI (Mirus Bio, #MIR2300) and viral supernatants were collected two days later. 5×104 cells were plated in 96 well plates and were infected one day after plating, with virus conditioned media by centrifugation with 2 μg/ml polybrene (#H9268, Sigma-Aldrich) at 2.250 r.p.m for 60 min. Infected MEF were sorted as hCD4+ Thy1.1+ cells and used for experiments. Raw 264.7 (TIB-71) were purchased from ATCC. MSCV-Ido1-IRES-GFP, MSCV-Ido2-IRES-GFP and MSCV-Tdo2-IRES-GFP were transfected into Platinum-E cells with TransIT-LTI and viral supernatants were collected two days later. 5×104 Raw 264.7 cells were transduced with the supernatant containing retroviruses by centrifugation with 2 μg/ml polybrene at 2.250 r.p.m for 60 min. Infected Raw 264.7 were sorted as GFP+ and used for experiments.
Platinum-E retroviral packaging cell line (Plat-E)
Plat-E cells (Morita et al., 2000) were cultured in complete IMDM. When reaching 70%–80% confluence, the cells were split by trypsinization.
Bacterial strains
Subcloning Efficiency DH5α competent cells and One Shot™ TOP10 were purchased from Invitrogen.
Method details
Cell isolation and culture
In vitro bone marrow-derived dendritic cells
Bone marrow cells were isolated from C57BL/6 or Ahr−/−, Relb−/− and Ido1−/− mice as previously described (Briseño et al., 2017, Briseño et al., 2017). BM was harvested from femur, tibia and pelvis using mortar and pestle in 1x PBS supplemented with 0.5% BSA and 2 mM EDTA (MACS buffer), passed through a 70 μm cell strainer and centrifuged at 1400 r.p.m for 5 min. Red blood cells were lysed with ACK lysis buffer (Ammonium Chloride 0.15 M, Potassium Carbonate 10 mM) and debris were removed by a gradient centrifugation using Histopaque1119 (#11191, Sigma-Aldrich) prior to culture. Cells were resuspended at 2×106 cells/ml in Iscove’s Modified Dulbecco’s Media (IMDM, #12440053, Thermo Fisher) supplemented with 0.1 mM Non-essential Amminoacids (#11140-035 Thermo Fisher), 1 mM Sodium Pyruvate (#11360-070, Thermo Fisher), 5 mM glutamine (#25030-024, Thermo Fisher), 50 μM 2-Mercaptoethanol (#31350-010, Thermo Fisher), 100 U/ml penicillin, 100 g/ml streptomycin (#15140-122, Thermo Fisher) and 10% FBS (#10270-106, Thermo Fisher) (complete IMDM) containing 5% murine Flt3-L and were seed 5 ml/well in 6-plate tissue culture plates at 37°C for 8-10 days. For all culture experiments, loosely adherent and suspension cells were harvested by gentle pipetting at the indicated time point. pDCs, cDC1 and cDC2 were sorted into complete IMDM by FACSAria Fusion as B220+Bst2+ (pDCs), B220–CD11c+MHCII+CD24+CD172α– (cDC1) and B220–CD11c+MHCII+CD24–CD172α+ (cDC2). Sort purity of > 95% was confirmed by post-sort analysis before cells were used for further experiments. For microarray analysis, some of the sorted cDC1, cDC2 and pDCs were stimulated with 250 ng/ml of Escherichia coli LPS (055:B5) o/n or some cDC2 were primed with LPS and then stimulated with 50 μM of l-kyn for 12h. RNA was purified from the stimulated or unstimulated cDCs using a NucleoSpin RNA XS kit and then subjected to microarray analysis. Transfection of cDC2 with Socs3-specific siRNA was done as described (Orabona et al., 2008). Gene-specific siRNAs were predesigned by Ambion, which also supplied negative control siRNA. For retroviral reporter assays and in vitro CRISPR/Cas9 deletion, BM cells from WT and R26Cas9 mice were depleted of CD3, CD19, B220, CD105, Ly6G, Ter119-expressing cells by staining with the corresponding biotinylated antibodies followed by depletion with MagniSort Streptavidin Negative Selection Bead (#MSNB-6002, Thermo Fisher). Lin–CD117high were cultured in complete IMDM + 5% Flt3-L overnight followed by infection.
Isolation of OT-I, OT-II and OT-II Foxp3 YFP cells from spleen
Splenocytes were isolated as previously (Grajales-Reyes et al., 2015). Briefly, excised spleens were incubated in complete IMDM supplemented with 250 mg/mL collagenase B (#11088815001, Roche) and 30 U/mL DNaseI (#D4527, Sigma-Aldrich) at 37° C for 30 min, and red blood cells were lysed with ACK. OT-I cells were sorted from the spleen as B220–CD11c–CD4–CD8+Vα2+. OT-II cells were sorted from the spleen as B220–CD11c–CD8–CD4+CD44–Vα2+. Cells were labelled with 1 μM CFSE (#C34554, Thermo Fisher Scientific) proliferation dye. OT-II Foxp3 YFP cells were sorted from the spleen as B220–CD11c–CD8–CD4+CD44–CD25–Vα2+.
Human dendritic cells isolation
Peripheral blood (buffy coat) was obtained from healthy blood donor drawn at blood Transfusion Center (Azienda Ospedaliera di Perugia) in accordance with the written approval of the Director of the Blood Transfusion Service. Peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation using Ficoll-Paque medium (#17-1400-02, GE Healthcare) from buffy coat diluted 1:4 with PBS 1X. Dendritic cell was enriched from PBMCs by immunomagnetic depletion of monocytes and macrophages (CD14+ CD16+), B cells (CD19+), T cell (CD3+), granulocytes (CD66b+), neutrophils (CD16+) and NK cells (CD335+). Cells were resuspended in RPMI 1640 (#11875093, Thermo Fisher) supplemented with 0.1 mM Non-essential Amminoacids (#11140-035 Thermo Fisher), 1 mM Sodium Pyruvate (#11360-070, Thermo Fisher), 5 mM glutamine (#25030-024, Thermo Fisher), 50 μM 2-Mercaptoethanol (#31350-010, Thermo Fisher), 100 U/ml penicillin, 100 g/ml streptomycin (#15140-122, Thermo Fisher) and 10% FBS (#10270-106, Thermo Fisher) (complete RPMI). cDC1 and cDC2 were sorted into complete RPMI by FACSAria Fusion as CD141+ Cleac9+ HLA-DR+CD11c–CD3–CD14–CD16–CD19–CD335–CD66b–(cDC1) and CD1c+ HLA-DR+CD11c+CD3–CD14–CD16–CD19–CD335–CD66b– (cDC2). Sort purity of > 95% was confirmed by post-sort analysis before cells were used for further experiments.
Transfection and transduction
Plat-E cells were plated in 6- or 12-well cell culture plates at a density of 0.4×106 cells/mL and incubated overnight. Retroviral plasmid DNAs mixed with TransIT-LT1 in Opti-MEM™ (#31985-070, GIBCO™) reduced serum medium were transfected into the PlatE cells and incubated overnight. The culture media was changed, and the supernatant containing retroviruses was collected 24 hours later. For retroviral transduction, lineage– BM progenitors (CD117hi cells) were used. BM progenitors were sort-purified and cultured in complete IMDM supplemented with appropriate cytokines overnight. After removing the culture media, the cells were transduced with the supernatant containing retroviruses in the presence of 2 μg/ml polybrene by spinoculation at 2.250 r.p.m for 1 h at 32°C. The culture media was changed 24 hours later, and the cells were further cultured. Details for the cell culture are described in the cell isolation and culture section.
CRISPR-Cas9 deletion of enhancer elements
Oligonucleotides containing guide RNA sequences with BbsI compatible overhangs were annealed and ligated into single guide RNA (sgRNA) vector Thy1.1-hU6-gRNA-BbsI stuffer-MSCV (Theisen et al., 2018). Oligonucleotide sequences for generating sgRNA are listed in the key resources table. Plasmids containing either scramble RNA or guide RNA were transfected into Plat-E packaging cells and retroviruses were collected 48 hours later as described in the ‘Transfection and transduction’ section. Lineage–CD117hi BM progenitors’ sort-purified from Rosa26Cas9-GFP/+ mice were transduced with the retroviruses and then cultured with 5% Flt3L conditioned medium for 7 days.
In silico IDO1 promoter analysis
The M. musculus (NCBI37/mm9) Ido1 genomic sequences were obtained using UCSC Genome Browser database. The DNA sequences 5306 bp upstream and 12854 bp downstream to Ido1 start codon were analyzed. XRE, AhREIII and RelBAhRE transcription factor binding sites were identified using Snapgene software. XRE, AhREIII and RelBAhRE DNA binding sites were defined as those possessing respectively consensus binding sequence, 5’-GCGTG-3’ (XRE), 5’-TCGTG-3’(AhREIII) and 5’-GGGTG-3’ (RelBAhRE) (Guyot et al., 2013).
cDC1 and cDC2 experimental treatments
At least 1×106 cDC1 and cDC2 were cultured in complete IMDM (mouse) or complete RPMI (human) and stimulated for 24, 36 or 48 hours. Compounds used are: LPS 055:B5 1-0.125 μg/ml (#L2880-10MG, Sigma Aldrich, 100 ng/ml stock in 1 X PBS), l-kynurenine (#K86251G, Sigma Aldrich, 25 mM stock in HCl 0.5 M), CpG 1826 PTO 10 μg/ ml (BioFab Research, 1 mg/ml in 1x PBS), polyinosinic:polycytidylic acid 25 μg/μl (Poly I:C, BioFab Research, 1 mg/ml stock), soluble tachyzoite antigen (STAg) 1 μg/μl (1 mg/ml stock) (Tussiwand et al., 2012), anti-IL-6 25 μg/ml (clone 20 F3, kindly provided by Louis Boon), LEAF™ Purified anti-mouse/rat CD126 (IL-6Rα chain, clone D7715A7, #115815, Biolegend), MG-132 Ready Made solution 0.5 μM (#M7449, Sigma Aldrich, 10 mM stock solution in DMSO) IL-12 10 ng/ml (#210-12-10UG, Peprotech, 100 ng/ml), L-1-methyltryptophan 1mM (#44739-1G, Sigma Aldrich, 4 mM stock in complete IMDM).
Antibodies and flow cytometry
For extracellular staining, cells were harvested by centrifugation at 1400 r.p.m for 5 minutes, followed by surface staining for 30 minutes at 4°C in 1x PBS + 0.5% BSA and 2 mM EDTA (MACS buffer) in presence of Fc blocking reagent (clone 2.4G, kindly provided by Louis Boon). Subsequently, surface antigens were stained in MACS buffer for 30 minutes at 4°C. Samples were fixed by incubation in 1x PBS + 1% (v/v) paraformaldehyde (#15714, Electron Microscopy Science).
For cytokines intracellular staining, cells were stimulated using 50 ng/mL PMA (phorbol 12-myristate 13-acetate) (#P1585, Sigma-Aldrich), 800 ng/mL ionomycin (#I9657, Sigma-Aldrich), brefeldin 3 μg/ml (#00-4506-51, Thermo Fisher) in complete IMDM for 4 hours. Subsequently, surface antigens were stained in MACS buffer for 30 minutes at 4°C. Samples were fixed by incubation in 1x PBS + 2% (v/v) paraformaldehyde (#15714, Electron Microscopy Science) for 15 minutes at room temperature, followed by washes in MACS buffer, and permeabilized in MACS buffer + 0.5% (w/v) Saponin (#S7900, Sigma-Aldrich) for 5 minutes at room temperature. Cells were washed and cytokines were stained in MACS buffer + 0.05% Saponin 1 hour at 4 °C. Cells were washed twice in MACS buffer + 0.5% (w/v) Saponin and resuspended in MACS buffer + 1% (v/v) PFA. Cells were analyzed with FACS Fortessa and data was analyzed with FlowJo software (Tree Star).
IDO1 intracellular staining
For IDO1 intracellular staining, cells were harvested by centrifugation at 1400 r.p.m for 5 minutes and washed in FACS buffer. Surface antigens were stained in FACS buffer for 30 minutes at 4°C. Cells were washed once in FACS buffer, followed by a wash in 1x cold PBS. Cells were fixed using 1x PBS + 4% (v/v) PFA for 20 minutes at room temperature. Next, cells were incubated with permeabilization buffer (Saponin 0.1% w/vol, 0.01% BSA in 1X PSB) for 5 minutes at room temperature. Intracellular IDO1 staining was performed in permeabilization buffer containing primary anti-IDO1 antibody (clone 8G-11, #MABS485, Merck, 0.5 μg/sample) or Rat IgG2a,k Isotype Ctrl (Clone RTK2758, #400502, Biolegend 1:400 v/v) for 1 hour at 4°C. After washing with permeabilization buffer, samples were incubated with PE anti-mouse secondary antibody (clone Poly4053, #405307, Biolegend, 1:400 v/v) in permeabilization buffer for 30 minutes at 4°C, and then washed twice with permeabilization buffer. Subsequently, FACS buffer + 1% (v/v) PFA was added to the samples and acquired with FACS Fortessa. Data was analyzed with FlowJo software (Tree Star).
Chromatin immunoprecipitation
At least 4×106 cells were used per Chromatin immunoprecipitation (ChIP). ChIP was carried out as described (Grajales-Reyes et al., 2015). CD24+ and CD172a+ cDCs from FLT3L-treated BM cultures of wild type mice were sorted on day 9. For AhR and RelB ChIP, sorted CD172a+ cDCs were stimulated with LPS 0.25 μg/ml O/N and l-kynurenine 50 μM for 3 hours before crosslinking. For IRF8 ChIP, CD24+ cDCs were incubated in fresh media for 36 hours and CCR7+ CD24+ cDC1 were sorted. Cells were crosslinked for 8 minutes at room temperature by adding of one-tenth of the volume of 11% formaldehyde solution, quenched with 1.25M glycine for 5 minutes at room temperature and washed twice with 1x cold PBS. Following the final wash, supernatants were discarded, and cells were flashed in liquid nitrogen for storage at -80°C. Thawed cells were lysed and sonicated as previously described (Grajales-Reyes et al., 2015). Chromatin was sonicated at Amp 60% for 10 cycles of 20s on and 50s off cycles using SoniPrep 150 (MSE Centrifuge). Sonicated lysates were incubated overnight at 4°C with Dynabeads Protein G (#1000-4D, Thermo Fisher), prepared by blocking 12,5 μl of magnetic beads with 0.5% BSA (w/v) in 1x PBS and bound to 2.5 μg of appropriate antibody. Beads containing protein-DNA complexes were washed with RIPA buffer 4 times. DNA fragments were eluted, and crosslinking was reversed by incubation at 65°C for 6 hours in TE buffer with 1% SDS and 1 mg/ml Proteinase K (#P6556, Sigma Aldrich). DNA was purified with QIAamp DNA Micro Kit (#56304, Qiagen). qPCR was performed using iTaq Universal SYBR Green Supermix (#1725124, BioRad). Anti-IgG immunoprecipitation and 5% input were used as controls. Antibodies used in this study are sheep anti-AhR antibody (#AF6697, R&D System), mouse anti-RelB (clone A-9, #sc-166416, Santa Cruz Botecnology), mouse anti-IRF8 (#sc-6058x, Santa Cruz Biotecnology), sheep IgG isotype control (#5-001-A, R&D System), mouse IgG1K isotype control (clone MG1-45, #401402, Biolegend). PCR primers were developed by using Primer3 (Koressaar and Remm, 2007; Untergasser et al., 2012). Data were analyzed fold enrichment method and normalized by adjusted input: Ct(input)-log2(input fraction-1).
Co-immunoprecipitation
Co-Immunoprecipitation experiments were performed according to the manufacturer’s protocol (ThermoFisher). In brief, treated 5×106 CD172+ cDC2 were stimulated with LPS (250 ng/ml, in complete IMDM) for 12 hours and l-kynurenine (50 μM, in in complete IMDM) for 2 hours. Cells were lysed on ice in Lysis buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% Non-idet P -40 (NP-40), 0.5% sodium deoxycholate, 0.1% SDS) supplemented with 1x complete Protease Inhibitor Cocktail (#11697498001, Roche). Lysates were centrifuged for 15 minutes at 4°C and an aliquot of the supernatant (15 μl) was collected as input control, while the remaining supernatant was incubated overnight at 4°C with Dynabeads Protein G, prepared by blocking 12,5 μl of magnetic beads with 0.5% BSA (w/v) in 1x PBS and bound to 2.5 μg of sheep anti-AhR antibody (#AF6697, R&D System) or sheep IgG isotype control (#5-001-A, R&D System). Immunocomplex was washed three times with washing buffer (25 mM citric acid, 50 mM Dibasic Sodium Posphate dodecahydrate pH 5) and was eluted with Elution buffer (0.1 M Sodium Citrate dihydrate pH 2-3) and Sample buffer 4x. Proteins were run on SDS-PAGE.
Nuclear extract
MEF cells expressing WT AhR or Q377A AhR were stimulated with l-kynurenine (50 μM, in HBSS) for 2 hours. Nuclear and cytoplasmatic fractions were prepared as previously reported (Manni et al., 2020). Cells were washed twice in cold 1x PBS and lysed on ice with Extraction Buffer N (15 mM Tris HCl, pH 7.5, 15 mM NaCl, 60 mM KCl, 5 mM MgCl2, 25 mM sucrose, 0.6% NP-40, 1 mM DTT, 2 mM sodium orthovanadate), followed by centrifugation at 3500 r.p.m for 10 minutes at 4°C. Cytosolic fraction was collected and pellet (nuclear fraction) was washed twice in Extraction Buffer N. Lysates were run on SDS page.
Proximity ligation assay (PLA)
MEF cells expressing WT or Q377A AhR or cDC2 stimulated as indicated in Figure 6D, were fixed 10 minutes with 4% PFA, permeabilized, blocked in BSA 5% in 1 X PBS. Duolink® (#DUO92008, Sigma-Aldrich) was performed, according to the manufacturer's protocol. In brief, primary antibodies, sheep anti-AhR (#AF6697, R&D System) and mouse anti-RelB (clone D-4, #sc-48366) were conjugated with either PLUS (#DUO92009, Sigma-Aldrich) or MINUS (#DUO92010, Sigma-Aldrich) oligonucleotide, to create PLA probes. Samples were incubated overnight at 4°C and, subsequently, ligase solution was added for 30 minutes. The signal was amplified with amplification polymerase solution at 37°C for 100 minutes. Then, a counterstain was carried out using a directly labelled FITC anti-alpha tubulin antibody (Anti-α-Tubulin-FITC antibody, #DM 1A Sigma-Aldrich). Nuclei were counterstained with 4’, 6’-diamidino-2phenylindole (DAPI) (#DUO82040, Sigma-Aldrich). Images were captured using a Zeiss Axio Observer Z1 inverted microscope, equipped with Apotome filter and Axiocam MRm camera detection system Zeiss (Castelli et al., 2020).
Western blotting
Samples were collected, washed in ice-cold 1 X PBS and subsequently resuspended in Lysis buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors, Aprotin (#A1153, Sigma-Aldrich, 2 ug/ml), Leupeptin (#L9783, Sigma-Aldrich, 10 ug/ml), Pepstatin A (#P5318, Sigma-Aldrich, 1 ug/ml), PMSF (#P7625, Sigma-Aldrich, 1 mM), Sodium Fluoride (#215309 Sigma-Aldrich, 10 mM), Sodium Orthovanadate (#450243, Sigma-Aldrich, 1 mM) and EDTA 5 mM. Cells were lysed by incubation for 30 minutes on ice followed by vortexing each 5 minutes, then centrifugated at 12000 r.p.m for 30 minutes at 4°C. Protein lysates were quantified using Bradford protein assay (#500-0006, BioRad). 13 μg of protein lysates were boiled at 95°C for 5 minutes. SDS-PAGE was performed using 10% acrylamide gel. Western blotting was performed by transferring proteins onto a nitrocellulose membrane (#1704159, BioRad). Membranes were blocked in 5% non-fat dry milk (#170-6404, BioRad) in 0.1% Tween 20 TBS (TBST) 1 hour at room temperature. Blocked membranes were probed with primary antibodies overnight at 4 °C. After washing, membranes were subsequently incubated with secondary antibodies for 1 hour at room temperature in 5% non-fat dry milk in TBST. Primary antibodies used in this study are mouse anti-IDO1 (clone 8G-11, #MABS485, Merck, 1 μg/ml in 5% non-fat dry milk TBST), sheep anti-AhR (#AF6697, R&D system, 1:400 in 5% non-fat dry milk 3% BSA TBST), mouse anti- AhR (clone RPT1, #MA1-514, Thermo Fisher Scientific ), rabbit anti-RelB (clone C1E4, #4922, Cell Signaling, 1:1000 in 2% BSA TBST), anti-Socs3 (#2923S, Cell signaling, 1:1000 in 5% non-fat dry milk TBST), mouse anti-β-actin (clone 3A-1, #A3853, Sigma-Aldrich, 1:2000 in 5% non-fat dry milk TBST), mouse anti-β-tubulin (clone AA2, Cat#: T8328, Sigma-Aldrich, 1:2000 in 5% non-fat dry milk TBST) and anti-Lamin B (#PA519468, Thermo Fisher Scientific). Anti-IDO2 and anti-TDO2 were raised in our lab and used at 1 μg/ml in 5% non-fat dry milk TBST. Secondary horseradish peroxidase-linked antibodies were purchased from Thermo Fisher Scientific Laboratories and used as follow: anti-sheep (#31480, 1:5000 v/v), anti-mouse (#31430, 1:10000 v/v), anti-rabbit (#31460, 1:10000 v/v). Membranes were developed using Clarity Western ECL Blotting Substrates (#170-5061, BioRad) and Clarity Max Western ECL Substrate (#170-5062, BioRad). β-tubulin, lamin B and β-actin were used as loading control. Protein quantification was performed by densitometry analysis using ImageJ software and calculated as ratio to loading control (Table S1).
l-Kynurenine uptake assay
l-Kynurenine uptake was performed as previously described (Sinclair et al., 2018). At least 1×106 CD172+ cDC2 were stimulated overnight with LPS (250 ng/ml) in complete IMDM. Cells were harvested, washed in 1x PBS and resuspended in 200 μl of pre-warmed HBSS in FACS tubes and kept in water bath at 37°C. l-kynurenine (800 μM stock, in HBSS), BCH (40 mM stock, in HBSS), and HBSS were pre-warmed at 37°C. BCH (100 μl) were added followed by 100 μl of l-kynurenine or vehicle to appropriate samples. The samples were incubated 4 minutes in water bath at 37°C and uptake was stopped by adding 125 μl PBS + 4% (v/v) PFA for 30 minutes at room temperature in the dark. After fixation, cells were washed twice in 1x PBS + 0.5% (w/v) BSA. Samples were analyzed with FACS Fortessa by using 405 nm laser abs 450/50 BP filter. Data was analyzed with FlowJo software (Tree Star).
Antigen presentation assay
In vitro antigen-presentation assays were performed as described previously (Kretzer et al., 2016). cDC1 from WT or Ido1−/− mice were sorted from day 9 FLT3-L cultures as described above and cultured in complete IMDM for 36 hours. CCR7+ cDC1 were sorted. 1×104 cDC1 were incubated with various doses of soluble OVA (#A5503, Sigma Aldrich) and 2.5×104 CFSE-labeled OT-I CD8+ cells for 3 days. After culture, frequency of CD8+TCR-Vα2+CD44+ cells undergone on CSFE solution was determined by flow cytometry. 2.5×104 WT or Ahr−/− cDC2 were co-cultured with sorted 5×104 OT-II cells labeled with CFSE in complete media at the indicated concentrations of soluble OVA. Cells were cultured at 37°C for 3 days and analyzed by flow cytometry. OT-II proliferation was determined as the percent of CD4+TCR-Vα2+CD44+ cells which had undergone at least one CFSE dilution.
Skin test assay
A skin test assay was used for measurements of major histocompatibility complex class I–restricted delayed-type hypersensitivity (DTH) responses to the HY peptide (WMHHNMDLI, BioFab) in C57BL/6 female recipient mice, as described (Pallotta et al., 2014). For in vivo immunization, 3×105 peptide-loaded cDC2 were combined with a minority fraction (5%) of peptide-loaded of various cDC1 or cDC2 and HY peptide 2 hours at 37°C, followed by intravenous injection into WT female recipient mice. Two weeks later, a DTH response was measured to intra-footpad challenge with the eliciting HY peptide, and results were expressed as the increase in footpad weight of peptide-injected footpads over that of vehicle-injected (internal control) counterparts. The minority cell fraction was constituted by WT and Ido1−/− CCR7− and CCR7+ cDC1, or WT, Ahr−/−or Ido1−/− cDC2, left untreated or treated overnight with specific reagents as reported.
Cytokines analysis
IL-6 (#88-7064-88, Thermo Fisher Scientific), IFN-γ (#88-7314-88, Thermo Fisher Scientific) and TGF-β (#DY1679-05, R&D system) were measured by specific ELISA kits according to the manufacturer’s recommendation. Briefly, specific anti cytokines antibodies were coated in Nunc-Immuno 96 MicroWell solid plates (#422404, Thermo Fisher Scientific). Standard curve and samples were incubated 2 hours in coated plates. Subsequently, a detection antibody (biotinylated antibody) was added to bind the immobilized antigen captured during the first incubation. A streptavidin-HPR antibody was added. Luminescence was read ad 450 nm (subtracting the value of 570 nm as non-specific plate absorbance) on Tecan microplate reader Spark. The detection limits of the assays were 4 pg/ml for IL-6, 15 pg/ml for IFN-γ and 31.3 pg/ml for TGF-β. Multiplex ELISA was performed by using a 23-plex immunoassay (Bio-Plex Pro Mouse Cytokine Grp I Panel 23-plex, #M60009RDPD, BioRad) and a MAGPIX system (Luminex Corporation). All cytokines were normalized to the cell counts.
l-kynurenine determination
IDO1 activity was measured in vitro as the ability to convert tryptophan into l-kynurenine, thus monitoring product concentration in culture supernatants by HPLC (high-performance liquid chromatography). Briefly, supernatants were collected and kept frozen at -20 °C until analysis. l-kynurenine concentrations were measured by HPLC. The detection limit of the assay was 0.05 μM. Plasma from EAE mice were subjected to acidic extraction method and analyzed by HPLC–mass spectrometry for tryptophan metabolites determination. In brief, frozen plasma were extracted by the addition of an ice-cold methanol–H2O 4:1 mixture. Plasma extracts were centrifuged for 15 min at 4000g at 4 °C and the supernatant was removed and used in further preparation for liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis. For the analysis of tryptophan pathway intermediates, in negative ionization mode, the sample was prepared by mixing an aliquot of plasma extract (50 μl) with 250 μl of the ice-cold internal standard solution (in 100% acetonitrile). For the analysis of end products of kynurenine pathway (that is, picolinic, quinolinic and nicotinic acid), in positive ionization mode, 300 μl of the plasma extract was evaporated to dryness (using a CentriVap vacuum concentrator, LabConco), and reconstituted with 75 μl 0.2% formic acid in H2O. Samples were centrifuged and supernatant was injected for LC–MS/MS analysis. LC–MS/MS analyses were performed using a 6495 triple quadrupole mass spectrometer (QqQ) interfaced with a 1290 UHPLC system (Waters) and operated in the dynamic Multiple Reaction Monitoring (dMRM) mode. Quinolinic, picolinic and nicotinic acid were measured in positive electrospray ionization (ESI + MS) mode, using an Acquity HSS T3 column (2.1 × 100 mm, 1.8 μm, Waters). An 11-min gradient was applied starting at 0% B (0.2% formic acid in methanol, 0–2 min) and increasing to 50% (2–4 min), and further to 90% (4–5 min), before returning to starting conditions (5–7 min) and re-equilibrating for 4 min (7–11 min). Mobile phase A was 0.2% formic acid in H2O, the flow rate for this method was 400 μl/min and the sample injection volume was 5 μl. ESI source conditions were set as follows: dry gas temperature 250 °C, nebulizer 35 psi and flow 15 l/min, sheath gas temperature 250 °C and flow 8 l/min, nozzle voltage 1,000 V, and capillary voltage +3,000 V. The cycle time in dMRM mode was 500 ms. Standard calibration curves ranging from 0–50 μM were used for quantification for all organic acids.
RNA extraction, cDNA synthesis, and real-time PCR
Cells were lysed in RLT buffer (#79216, Qiagen) and RNA was isolated using RNeasy Mini Kit (#74104, Qiagen). cDNA was transcribed using Quantitect Reverse transcription kit (#205313, Qiagen) according to manufacturer’s protocol. Gene expression was measured using iTaq Supermix (#1725124, Bio-rad). Data were calculated as the ratio to β-actin expression by relative quantification method (ΔΔct; means ± SD of triplicate determination) and data are presented as normalized transcript expression in the samples relative to normalized transcript expression in control cultures (in which fold change = 1).
Transwell DCs co-culture
cDC1 and cDC2 from WT, Ahr−/− or Ido1−/− mice were sorted from day 10 FLT3-L culture as described above. cDC2 were cultured in 24 well plates (0.5×106) and cDC1 (0.5×106) were seed in trans-well system membranes (#353469, Falcon), 0.4 μM pore size, inserted in 24 well plate in complete IMDM and treated as previously indicated for 36 hours. IDO1 expression was analyzed by intracellular staining as previously described. Cells were gated on CD11C+ MHCIIhiCD24–CD172+ CCR7+ or CD11C+MHCIIhiCD24–CD172+CCR7– cells. Human cDC1 and cDC2 were sorted from PBMCs as describe above. cDC2 were cultured in 24 well plates (0.5×106) and cDC1 (0.5×106) were seed in trans-well system membranes (#353469, Falcon), 0.4 μM pore size, inserted in 24 well plate in complete RPMI and treated as previously indicated for 36 hours. IDO1 expression was analyzed by Western blotting.
Microarray and scRNA-seq analysis
For microarrays, RNA was isolated by NucleoSpin RNA XS (Macherey-Nagel) and amplified with WT Pico System (Affymetrix) and hybridized to Gene Chip Mouse Gene 1.0 ST microarrays (Affymetrix) for 18 hours at 45°C in a Gene Chip Hybridization Oven 640. The data was analyzed with the 21Affymetrix Gene Chip Command Console. Microarray expression data were processed using Command Console (Affymetrix, Inc) and the raw (.CEL), files generated were analyzed using Expression Console software with Affymetrix default RMA Gene analysis settings (Affymetrix, Inc). Probe summarization (Robust Multichip Analysis, RMA), quality control analysis, and probe annotation were performed according to recommended guidelines (Expression Console Software, Affymetrix, Inc.). Data were normalized by robust multi-array average summarization and underwent quartile normalization with ArrayStar software (DNASTAR). scRNA-seq analysis was performed as previously described (Schafflick and Xu, 2020).
Immunofluorescence
At least 50.000 CCR7+ cDC1 from WT and Ido1−/− sorted as previously reported were seeded overnight on microscope slides treated with Poly-L-lysin solution 0.1% (#P8920-100ML, Sigma Aldrich) and treated with LPS or vehicle for 48 hours. The samples were fixed using 1x PBS + 2% (v/v) PFA for 20 minutes at room temperature, permeabilized with 1x PBS + 0.1% Triton X-100 for 10 minutes at room temperature and blocked using 1x PBS + 2% BSA (blocking solution) for 60 minutes at room temperature. After three washes with 1x PBS, the samples were incubated with anti l-kynurenine antibody (clone 3D4-F2, #IS0003, Immusmol, 1:50 v/v) in blocking solution overnight at 4°C. After washing, the slides were incubated with anti-mouse PE secondary Abs (clone Poly4053, # 405307, Biolegend, 1:400 v/v) diluted in PBS + 2% BSA for 1 hour at room temperature. Next, slides were washed and stained with fluorescein isothiocyanate (FITC)-labelled phalloidin (1:250) for actin labeling and counterstained with 4’, 6’-diamidino-2phenylindole DAPI (#P36931, Thermo Fisher Scientific) for nuclei. Images were obtained using Zeiss Axio Observer Z1 inverted microscope. Representative images are shown.
Bone marrow chimera
Bone marrow chimera was performed as described (Briseño et al., 2017, Briseño et al., 2017). Briefly, BM cells from CD45.2 WT or Relb−/− were isolated as described in cell isolation and culture section. Cells were counted and 0.5-1×107 total bone marrow cells were transferred by retro-orbital injection into CD45.1 WT recipient mice 24 hours after whole-body irradiation at 950 rads. Mice were used 6–8 week after bone marrow reconstitution. For mix bone marrow chimeras B6-Ly5.1/Cr mice were γ-irradiated with 950 rad and allowed to recover one day before i.v. injection of 50/50 mixes of Il6−/−/Irf4f/f or Il6−/−/Irf4cKO (Irf4f/fCD11cCre+), Ido1−/−/Irf4f/f or Ido1−/−/Irf4cKO and Ahr−/−/Irf4f/f or Ahr−/−/Irf4cKO bone marrow. Mice rested for eight weeks post bone marrow injection before use in EAE experiments.
Induction of EAE
EAE was induced with 50 μg of myelin oligodendrocyte glycoprotein fragment MEVGWYRSPFSRVVHLYRNGK (MOG35–55 peptide; #crb1000205n Cambridge Research Biochemicals) mixed with incomplete Freund’s Adjuvant (#263910, BD) containing 4 mg/ml Mycobacterium tuberculosis TB H37 Ra (#231141, BD), at a ratio of 1:1 (v/v). Mice received two subcutaneous injections of 100 μl each of the MOG/CFA mix. Mice received a single intraperitoneal injection of pertussis toxin (#180, List Biological Laboratories) at a concentration of 1 ng/μL in 200 μL of PBS. A second injection of pertussis toxin at the same concentration was inoculated two days after the EAE induction. Mice were orally treated with 2.5 mg/mouse of l-kynurenine (#K8625-1G, Sigma Aldrich) dissolved in 1x PBS on alternating days starting at day 2 post-EAE induction. For cDC1 WT and Ido1−/− transfer, EAE was induced in Irf8 Δ32−/− mice. CD24+ cDC1 from FLT3L-treated BM cultures of wild type and Ido1−/− mice were sorted on day 9 and incubated in fresh media for 36 hours. CCR7+ CD24+ cDC1 were sorted and pulsed with 25 μg/ml of MOG 2 hours at 37°C. MOG loaded cDC1 WT and Ido1−/− were washed twice with PBS 1X and intravenously injected at doses of 1 x 106 cells on day 4 and 7 post immunization. Mice were monitored and scored daily thereafter. EAE clinical scores were defined as follows: 0 – no signs, 1 – fully limp tail, 2 – hindlimb weakness, 3 – hindlimb paralysis, 4 – forelimb paralysis, 5 – moribund, as described previously (Rothhammer et al., 2016). Mice were randomly assigned to treatment groups.
Histopathology
Foot from mice treated with HY peptide were removed after two weeks, fixed for 24 h in ethyl alcohol (60%), acetic acid (10%), and chloroform (30%), and embedded in paraffin. Footpad sections were cut at 4 μm and stained with H&E to reveal intra-footpad inflammatory infiltrates. Morphological analysis of paraffin-embedded spinal cord sections (4 μm) from mice treated as in Figure 7 were stained with H&E at day 25 after MOG immunization. Slides were analyzed by light microscopy. The sections were quantified using manual cell counting (Table S2).
Statistical analysis
All statistical tests, comparisons, and sample sizes are included in the Figure Legends. Error bars indicate standard deviation. Statistical analyses were performed using one-way or two-way analysis of variance (ANOVA) with Bonferroni or Tukey’s multiple comparison test unless stated. For the skin test assay paired Student’s t test was used (using at least five mice per group). All data are shown as mean ± SD. In all cases, ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = not statistically significant, p > 0.05. In all cases, the stated ‘‘n’’ value indicates a biological replicate. All other in vitro and in vivo experiments were replicated at least once, and all attempts at replication were successful. All statistical analyses were performed using Prism 9 (GraphPad Software).
Acknowledgments
Supporting fundings: Italian Foundation for Cancer Research (AIRC 19903), Telethon (GGP17094), Prin 2017BZEREZ, and Italian Multiple Sclerosis Foundation (FISM 2019/R-single/012) to F.F; US National Institutes of Health (R01AI150297) to K.M.M.
Author contributions
Conceptualization, M.G. and F.F.; investigation and methodology, M.G., G. Scalisi, C.G.B., G. Manni, P.B., V.D., T.L.M., S.K., D.J.T., T.L.M., G. Mencarelli, D.R., E.P., M. Castelli, M.A.D.F., G. Servillo, M. Calvitti, T.Z., P.P., and K.M.M.; resources, D.F., C.O., N.G., C.C., R.P., L.B., A.B., M. Colonna, and K.M.M.; formal analysis, C.A.X. and G.M.z.H.; supervision and writing—review & editing, K.M.M. and F.F.
Declaration of interests
The authors declare no competing interests.
Published: June 14, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.05.013.
Contributor Information
Kenneth M. Murphy, Email: kmurphy@wustl.edu.
Francesca Fallarino, Email: francesca.fallarino@unipg.it.
Supplemental information
References
- Ardouin L., Luche H., Chelbi R., Carpentier S., Shawket A., Montanana Sanchis F., Santa Maria C., Grenot P., Alexandre Y., Gregoire C., et al. Broad and largely concordant molecular changes characterize tolerogenic and immunogenic dendritic cell maturation in thymus and periphery. Immunity. 2016;45:305–318. doi: 10.1016/j.immuni.2016.07.019. [DOI] [PubMed] [Google Scholar]
- Askew D., Harding C.V. Antigen processing and CD24 expression determine antigen presentation by splenic CD4+ and CD8+ dendritic cells. Immunology. 2008;123:447–455. doi: 10.1111/j.1365-2567.2007.02711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacsi S.G., Hankinson O. Functional characterization of DNA-binding domains of the subunits of the heterodimeric aryl hydrocarbon receptor complex imputing novel and canonical basic helix-loop-helix protein-DNA interactions. J. Biol. Chem. 1996;271:8843–8850. doi: 10.1074/jbc.271.15.8843. [DOI] [PubMed] [Google Scholar]
- Bajaña S., Turner S., Paul J., Ainsua-Enrich E., Kovats S. IRF4 and IRF8 act in CD11c+ cells to regulate terminal differentiation of lung tissue dendritic cells. J. Immunol. 2016;196:1666–1677. doi: 10.4049/jimmunol.1501870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessede A., Gargaro M., Pallotta M.T., Matino D., Servillo G., Brunacci C., Bicciato S., Mazza E.M., Macchiarulo A., Vacca C., et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishnupuri K.S., Alvarado D.M., Khouri A.N., Shabsovich M., Chen B., Dieckgraefe B.K., Ciorba M.A. IDO1 and kynurenine pathway metabolites activate PI3K-Akt signaling in the neoplastic colon epithelium to promote cancer cell proliferation and inhibit apoptosis. Cancer Res. 2019;79:1138–1150. doi: 10.1158/0008-5472.CAN-18-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius A.L., Cella M., Maldonado J., Takai T., Colonna M. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood. 2006;107:2474–2476. doi: 10.1182/blood-2005-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bošnjak B., Do K.T.H., Förster R., Hammerschmidt S.I. Imaging dendritic cell functions. Immunol. Rev. 2022;306:137–163. doi: 10.1111/imr.13050. [DOI] [PubMed] [Google Scholar]
- Böttcher C., Fernández-Zapata C., Schlickeiser S., Kunkel D., Schulz A.R., Mei H.E., Weidinger C., Gieß R.M., Asseyer S., Siegmund B., et al. Multi-parameter immune profiling of peripheral blood mononuclear cells by multiplexed single-cell mass cytometry in patients with early multiple sclerosis. Sci. Rep. 2019;9:19471. doi: 10.1038/s41598-019-55852-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G., Zheng S., Valieris R., Tojal da Silva I., Satija R., Nussenzweig M.C. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med. 2016;213:2861–2870. doi: 10.1084/jem.20161135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño C.G., Gargaro M., Durai V., Davidson J.T., Theisen D.J., Anderson D.A., 3rd, Novack D.V., Murphy T.L., Murphy K.M. Deficiency of transcription factor RelB perturbs myeloid and DC development by hematopoietic-extrinsic mechanisms. Proc. Natl. Acad. Sci. USA. 2017;114:3957–3962. doi: 10.1073/pnas.1619863114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelli M., Piobbico D., Chiacchiaretta M., Brunacci C., Pieroni S., Bartoli D., Gargaro M., Fallarino F., Puccetti P., Soddu S., et al. HOPS/TMUB1 retains p53 in the cytoplasm and sustains p53-dependent mitochondrial apoptosis. EMBO Rep. 2020;21:e48073. doi: 10.15252/embr.201948073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan J.M., Lehar S.M., Bevan M.J. CD8(+) but not CD8(-) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 2000;192:1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Ma T., Yan Z., Zettel K.R., Scott M.J., Liao H., Frank A., Morelli A.E., Sodhi C.P., Hackam D.J., Billar T.R. Toll-like receptor 4 signaling on dendritic cells suppresses polymorphonuclear leukocyte CXCR2 expression and trafficking via interleukin 10 During intra-abdominal sepsis. J. Infect. Dis. 2016;213:1280–1288. doi: 10.1093/infdis/jiv562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio P., Duarte J.H., Ahlfors H., Owens N.D., Li Y., Villanova F., Tosi I., Hirota K., Nestle F.O., Mrowietz U., et al. Activation of the aryl hydrocarbon receptor dampens the severity of inflammatory skin conditions. Immunity. 2014;40:989–1001. doi: 10.1016/j.immuni.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte J.H., Di Meglio P., Hirota K., Ahlfors H., Stockinger B. Differential influences of the aryl hydrocarbon receptor on Th17 mediated responses in vitro and in vivo. PLoS One. 2013;8:e79819. doi: 10.1371/journal.pone.0079819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai V., Bagadia P., Granja J.M., Satpathy A.T., Kulkarni D.H., Davidson J.T., Wu R., Patel S.J., Iwata A., Liu T.T., et al. Cryptic activation of an Irf8 enhancer governs cDC1 fate specification. Nat. Immunol. 2019;20:1161–1173. doi: 10.1038/s41590-019-0450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durai V., Murphy K.M. Functions of murine dendritic cells. Immunity. 2016;45:719–736. doi: 10.1016/j.immuni.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallarino F., Grohmann U., You S., McGrath B.C., Cavener D.R., Vacca C., Orabona C., Bianchi R., Belladonna M.L., Volpi C., et al. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J. Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- Ferris S.T., Durai V., Wu R., Theisen D.J., Ward J.P., Bern M.D., Davidson J.T.t., Bagadia P., Liu T., Briseño C.G., et al. cDC1 prime and are licensed by CD4+ T cells to induce anti-tumour immunity. Nature. 2020;584:624–629. doi: 10.1038/s41586-020-2611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei K., Fredrikson S., Fontana A., Link H. Interleukin-6 is elevated in plasma in multiple sclerosis. J. Neuroimmunol. 1991;31:147–153. doi: 10.1016/0165-5728(91)90020-8. [DOI] [PubMed] [Google Scholar]
- Gaetani L., Boscaro F., Pieraccini G., Calabresi P., Romani L., Di Filippo M., Zelante T. Host and Microbial Tryptophan Metabolic Profiling in Multiple Sclerosis. Front. Immunol. 2020;11:157. doi: 10.3389/fimmu.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W.X., Sun Y.Q., Shi J., Li C.L., Fang S.B., Wang D., Deng X.Q., Wen W., Fu Q.L. Effects of mesenchymal stem cells from human induced pluripotent stem cells on differentiation, maturation, and function of dendritic cells. Stem Cell Res. Ther. 2017;8:48. doi: 10.1186/s13287-017-0499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes P., Soares-da-Silva P. L-dopa transport properties in an immortalised cell line of rat capillary cerebral endothelial cells, RBE 4. Brain Res. 1999;829:143–150. doi: 10.1016/s0006-8993(99)01387-6. [DOI] [PubMed] [Google Scholar]
- Grajales-Reyes G.E., Iwata A., Albring J., Wu X., Tussiwand R., Kc W., Kretzer N.M., Briseño C.G., Durai V., Bagadia P., et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat. Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann U., Fallarino F., Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Vázquez C., Quintana F.J. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity. 2018;48:19–33. doi: 10.1016/j.immuni.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot E., Chevallier A., Barouki R., Coumoul X. The AhR twist: ligand-dependent AhR signaling and pharmaco-toxicological implications. Drug Discov. Today. 2013;18:479–486. doi: 10.1016/j.drudis.2012.11.014. [DOI] [PubMed] [Google Scholar]
- Hacker C., Kirsch R.D., Ju X.S., Hieronymus T., Gust T.C., Kuhl C., Jorgas T., Kurz S.M., Rose-John S., Yokota Y., Zemke M. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- Heink S., Yogev N., Garbers C., Herwerth M., Aly L., Gasperi C., Husterer V., Croxford A.L., Möller-Hackbarth K., Bartsch H.S., et al. Trans-presentation of IL-6 by dendritic cells is required for the priming of pathogenic TH17 cells. Nat. Immunol. 2017;18:74–85. doi: 10.1038/ni.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Bagadia P., Anderson D.A., 3rd, Liu T.T., Huang X., Theisen D.J., O'Connor K.W., Ohara R.A., Iwata A., Murphy T.L., Murphy K.M. High amount of transcription factor IRF8 engages AP1-IRF composite elements in enhancers to direct Type 1 conventional dendritic cell identity. Immunity. 2020;53:759–774.e9. doi: 10.1016/j.immuni.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Naka T., Nakahama T., Chinen I., Masuda K., Nohara K., Fujii-Kuriyama Y., Kishimoto T. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J. Exp. Med. 2009;206:2027–2035. doi: 10.1084/jem.20090560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Baumann H., Freer G., Freudenberg M., Lamers M., Kishimoto T., Zinkernagel R., Bluethmann H., Köhler G. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kowal J., Tkach M. Dendritic cell extracellular vesicles. Int. Rev. Cell Mol. Biol. 2019;349:213–249. doi: 10.1016/bs.ircmb.2019.08.005. [DOI] [PubMed] [Google Scholar]
- Kretzer N.M., Theisen D.J., Tussiwand R., Briseño C.G., Grajales-Reyes G.E., Wu X., Durai V., Albring J., Bagadia P., Murphy T.L., Murphy K.M. RAB43 facilitates cross-presentation of cell-associated antigens by CD8α+ dendritic cells. J. Exp. Med. 2016;213:2871–2883. doi: 10.1084/jem.20160597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbergh M.F.S., Stoorvogel W. Antigen presentation by extracellular vesicles from professional antigen-presenting cells. Annu. Rev. Immunol. 2018;36:435–459. doi: 10.1146/annurev-immunol-041015-055700. [DOI] [PubMed] [Google Scholar]
- Manni G., Mondanelli G., Scalisi G., Pallotta M.T., Nardi D., Padiglioni E., Romani R., Talesa V.N., Puccetti P., Fallarino F., Gargaro M. Pharmacologic induction of endotoxin tolerance in dendritic cells by L-kynurenine. Front. Immunol. 2020;11:292. doi: 10.3389/fimmu.2020.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor A.L., Munn D.H. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezrich J.D., Fechner J.H., Zhang X., Johnson B.P., Burlingham W.J., Bradfield C.A. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minarrieta L., Ghorbani P., Sparwasser T., Berod L. Metabolites: deciphering the molecular language between DCs and their environment. Semin. Immunopathol. 2017;39:177–198. doi: 10.1007/s00281-016-0609-6. [DOI] [PubMed] [Google Scholar]
- Morita S., Kojima T., Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Murphy T.L., Grajales-Reyes G.E., Wu X., Tussiwand R., Briseño C.G., Iwata A., Kretzer N.M., Durai V., Murphy K.M. Transcriptional control of dendritic cell development. Annu. Rev. Immunol. 2016;34:93–119. doi: 10.1146/annurev-immunol-032713-120204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O'Keeffe M., Bahlo M., Papenfuss A., et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Nguyen N.T., Kimura A., Nakahama T., Chinen I., Masuda K., Nohara K., Fujii-Kuriyama Y., Kishimoto T. Aryl hydrocarbon receptor negatively regulates dendritic cell immunogenicity via a kynurenine-dependent mechanism. Proc. Natl. Acad. Sci. USA. 2010;107:19961–19966. doi: 10.1073/pnas.1014465107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuti R., Gargaro M., Matino D., Dolciami D., Grohmann U., Puccetti P., Fallarino F., Macchiarulo A. Ligand binding and functional selectivity of L-tryptophan metabolites at the mouse aryl hydrocarbon receptor (mAhR) J. Chem. Inf. Model. 2014;54:3373–3383. doi: 10.1021/ci5005459. [DOI] [PubMed] [Google Scholar]
- Opitz C.A., Litzenburger U.M., Sahm F., Ott M., Tritschler I., Trump S., Schumacher T., Jestaedt L., Schrenk D., Weller M., et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- Orabona C., Pallotta M.T., Volpi C., Fallarino F., Vacca C., Bianchi R., Belladonna M.L., Fioretti M.C., Grohmann U., Puccetti P. SOCS3 drives proteasomal degradation of indoleamine 2,3-dioxygenase (IDO) and antagonizes IDO-dependent tolerogenesis. Proc. Natl. Acad. Sci. USA. 2008;105:20828–20833. doi: 10.1073/pnas.0810278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta M.T., Orabona C., Bianchi R., Vacca C., Fallarino F., Belladonna M.L., Volpi C., Mondanelli G., Gargaro M., Allegrucci M., et al. Forced IDO1 expression in dendritic cells restores immunoregulatory signalling in autoimmune diabetes. J. Cell. Mol. Med. 2014;18:2082–2091. doi: 10.1111/jcmm.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallotta M.T., Orabona C., Volpi C., Vacca C., Belladonna M.L., Bianchi R., Servillo G., Brunacci C., Calvitti M., Bicciato S., et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 2011;12:870–878. doi: 10.1038/ni.2077. [DOI] [PubMed] [Google Scholar]
- Pasqual G., Chudnovskiy A., Tas J.M.J., Agudelo M., Schweitzer L.D., Cui A., Hacohen N., Victora G.D. Monitoring T cell-dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature. 2018;553:496–500. doi: 10.1038/nature25442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterka M., Voss J.O., Werr J., Reuter E., Franck S., Leuenberger T., Herz J., Radbruch H., Bopp T., Siffrin V., Zipp F. Dendritic cells tip the balance towards induction of regulatory T cells upon priming in experimental autoimmune encephalomyelitis. J. Autoimmun. 2017;76:108–114. doi: 10.1016/j.jaut.2016.09.008. [DOI] [PubMed] [Google Scholar]
- Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M., et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccetti P., Grohmann U. IDO and regulatory T cells: a role for reverse signalling and non-canonical NF-kappaB activation. Nat. Rev. Immunol. 2007;7:817–823. doi: 10.1038/nri2163. [DOI] [PubMed] [Google Scholar]
- Pulendran B. The varieties of immunological experience: of pathogens, stress, and dendritic cells. Annu. Rev. Immunol. 2015;33:563–606. doi: 10.1146/annurev-immunol-020711-075049. [DOI] [PubMed] [Google Scholar]
- Pulendran B., Banchereau J., Burkeholder S., Kraus E., Guinet E., Chalouni C., Caron D., Maliszewski C., Davoust J., Fay J., Palucka K. Flt3-ligand and granulocyte colony-stimulating factor mobilize distinct human dendritic cell subsets in vivo. J. Immunol. 2000;165:566–572. doi: 10.4049/jimmunol.165.1.566. [DOI] [PubMed] [Google Scholar]
- Robertson H., Li J., Kim H.J., Rhodes J.W., Harman A.N., Patrick E., Rogers N.M. Transcriptomic analysis identifies A tolerogenic dendritic cell signature. Front. Immunol. 2021;12:733231. doi: 10.3389/fimmu.2021.733231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer V., Mascanfroni I.D., Bunse L., Takenaka M.C., Kenison J.E., Mayo L., Chao C.C., Patel B., Yan R., Blain M., et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafflick D., Xu C.A., Hartlehnert M., Cole M., Schulte-Mecklenbeck A., Lautwein T., Wolbert J., Heming M., Meuth S.G., Kuhlmann T., et al. Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun. 2020;11:247. doi: 10.1038/s41467-019-14118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W., See P., Shin A., Wasan P.S., et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönheit J., Kuhl C., Gebhardt M.L., Klett F.F., Riemke P., Scheller M., Huang G., Naumann R., Leutz A., Stocking C., et al. PU.1 level-directed chromatin structure remodeling at the Irf8 gene drives dendritic cell commitment. Cell Rep. 2013;3:1617–1628. doi: 10.1016/j.celrep.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Sharma M.D., Pacholczyk R., Shi H., Berrong Z.J., Zakharia Y., Greco A., Chang C.S., Eathiraj S., Kennedy E., Cash T., et al. Inhibition of the BTK-IDO-mTOR axis promotes differentiation of monocyte-lineage dendritic cells and enhances anti-tumor T cell immunity. Immunity. 2021;54:2354–2371.e8. doi: 10.1016/j.immuni.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair L.V., Neyens D., Ramsay G., Taylor P.M., Cantrell D.A. Single cell analysis of kynurenine and System L amino acid transport in T cells. Nat. Commun. 2018;9:1981. doi: 10.1038/s41467-018-04366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittig S.P., van Beek J.J.P., Flórez-Grau G., Weiden J., Buschow S.I., van der Net M.C., van Slooten R., Verbeek M.M., Geurtz P.B.H., Textor J., et al. Human type 1 and type 2 conventional dendritic cells express indoleamine 2,3-dioxygenase 1 with functional effects on T cell priming. Eur. J. Immunol. 2021;51:1494–1504. doi: 10.1002/eji.202048580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H., Couwenberg F., Bakker A.Q., Weijer K., Uittenbogaart C.H. Id2 and Id3 inhibit development of CD34(+) stem cells into predendritic cell (pre-DC)2 but not into pre-DC1. Evidence for a lymphoid origin of pre-DC2. J. Exp. Med. 2000;192:1775–1784. doi: 10.1084/jem.192.12.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M., Hawiger D., Nussenzweig M.C. Tolerogenic dendritic cells. Annu. Rev. Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- Steinman R.M., Hemmi H. Dendritic cells: translating innate to adaptive immunity. Curr. Top. Microbiol. Immunol. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- Sundaram G., Lim C.K., Brew B.J., Guillemin G.J. Kynurenine pathway modulation reverses the experimental autoimmune encephalomyelitis mouse disease progression. J. Neuroinflammation. 2020;17:176. doi: 10.1186/s12974-020-01844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Honma K., Matsuyama T., Suzuki K., Toriyama K., Akitoyo I., Yamamoto K., Suematsu T., Nakamura M., Yui K., Kumatori A. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc. Natl. Acad. Sci. USA. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M.C., Gabriely G., Rothhammer V., Mascanfroni I.D., Wheeler M.A., Chao C.C., Gutiérrez-Vázquez C., Kenison J., Tjon E.C., Barroso A., et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci. 2019;22:729–740. doi: 10.1038/s41593-019-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Tailor P., Yamaoka K., Kong H.J., Tsujimura H., O'Shea J.J., Singh H., Ozato K. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 2005;174(Md):2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- Terness P., Bauer T.M., Röse L., Dufter C., Watzlik A., Simon H., Opelz G. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. J. Exp. Med. 2002;196:447–457. doi: 10.1084/jem.20020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen D.J., Davidson J.T.t., Briseño C.G., Gargaro M., Lauron E.J., Wang Q., Desai P., Durai V., Bagadia P., Brickner J.R., et al. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science. 2018;362:694–699. doi: 10.1126/science.aat5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen D.J., Ferris S.T., Briseño C.G., Kretzer N., Iwata A., Murphy K.M., Murphy T.L. Batf3-dependent genes control tumor rejection induced by dendritic cells independently of cross-presentation. Cancer Immunol. Res. 2019;7:29–39. doi: 10.1158/2326-6066.CIR-18-0138. [DOI] [PubMed] [Google Scholar]
- Torralba D., Baixauli F., Villarroya-Beltri C., Fernández-Delgado I., Latorre-Pellicer A., Acín-Pérez R., Martín-Cófreces N.B., Jaso-Tamame Á.L., Iborra S., Jorge I., et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 2018;9:2658. doi: 10.1038/s41467-018-05077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R., Lee W.L., Murphy T.L., Mashayekhi M., Kc W., Albring J.C., Satpathy A.T., Rotondo J.A., Edelson B.T., Kretzer N.M., et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehori J., Fukase K., Akazawa T., Uematsu S., Akira S., Funami K., Shingai M., Matsumoto M., Azuma I., Toyoshima K., et al. Dendritic cell maturation induced by muramyl dipeptide (MDP) derivatives: monoacylated MDP confers TLR2/TLR4 activation. J. Immunol. 2005;174:7096–7103. doi: 10.4049/jimmunol.174.11.7096. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C.F., Wu D., Goth S.R., Baek J., Lollies A., Domhardt R., Grindel A., Pessah I.N. Aryl hydrocarbon receptor signaling regulates NF-κB RelB activation during dendritic-cell differentiation. Immunol. Cell Biol. 2013;91:568–575. doi: 10.1038/icb.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bubnoff D., Scheler M., Wilms H., Fimmers R., Bieber T. Identification of IDO-positive and IDO-negative human dendritic cells after activation by various proinflammatory stimuli. J. Immunol. 2011;186:6701–6709. doi: 10.4049/jimmunol.1003151. [DOI] [PubMed] [Google Scholar]
- Wohn C., Le Guen V., Voluzan O., Fiore F., Henri S., Malissen B. Absence of MHC class II on cDC1 dendritic cells triggers fatal autoimmunity to a cross-presented self-antigen. Sci. Immunol. 2020;5:eaba1896. doi: 10.1126/sciimmunol.aba1896. [DOI] [PubMed] [Google Scholar]
- Yan Y., Zhang G.X., Gran B., Fallarino F., Yu S., Li H., Cullimore M.L., Rostami A., Xu H. IDO upregulates regulatory T cells via tryptophan catabolite and suppresses encephalitogenic T cell responses in experimental autoimmune encephalomyelitis. J. Immunol. 2010;185:5953–5961. doi: 10.4049/jimmunol.1001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev N., Frommer F., Lukas D., Kautz-Neu K., Karram K., Ielo D., von Stebut E., Probst H.C., van den Broek M., Riethmacher D., et al. Dendritic cells ameliorate autoimmunity in the CNS by controlling the homeostasis of PD-1 receptor(+) regulatory T cells. Immunity. 2012;37:264–275. doi: 10.1016/j.immuni.2012.05.025. [DOI] [PubMed] [Google Scholar]
- Yoneyama H., Matsuno K., Toda E., Nishiwaki T., Matsuo N., Nakano A., Narumi S., Lu B., Gerard C., Ishikawa S., Matsushima K. Plasmacytoid DCs help lymph node DCs to induce anti-HSV CTLs. J. Exp. Med. 2005;202:425–435. doi: 10.1084/jem.20041961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D'Angelo C., Massi-Benedetti C., Fallarino F., et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the microarrays reported in this paper are available on the GEO database with the following accession number: GSE203449 and GSE203450.
Following datasets were downloaded and reanalyzed: microarrays for mature cDC1 (GSE71171) (Ardouin et al., 2016), ChIP-seq datasets for IRF4, IRF8, BATF3, H3K4me1, H3K27ac (GSE66899) (Grajales-Reyes et al., 2015), scRNA-seq datasets for MS and control patients (https://github.com/chenlingantelope/MSscRNAseq2019.git.) (Schafflick and Xu, 2020).