Abstract
The phylogenetic diversity was determined for a microbial community obtained from an in situ growth chamber placed on a deep-sea hydrothermal vent on the Mid-Atlantic Ridge (23°22′ N, 44°57′ W). The chamber was deployed for 5 days, and the temperature within the chamber gradually decreased from 70 to 20°C. Upon retrieval of the chamber, the DNA was extracted and the small-subunit rRNA genes (16S rDNA) were amplified by PCR using primers specific for the Archaea or Bacteria domain and cloned. Unique rDNA sequences were identified by restriction fragment length polymorphisms, and 38 different archaeal and bacterial phylotypes were identified from the 85 clones screened. The majority of the archaeal sequences were affiliated with the Thermococcales (71%) and Archaeoglobales (22%) orders. A sequence belonging to the Thermoplasmales confirms that thermoacidophiles may have escaped enrichment culturing attempts of deep-sea hydrothermal vent samples. Additional sequences that represented deeply rooted lineages in the low-temperature eurarchaeal (marine group II) and crenarchaeal clades were obtained. The majority of the bacterial sequences obtained were restricted to the Aquificales (18%), the ɛ subclass of the Proteobacteria (ɛ-Proteobacteria) (40%), and the genus Desulfurobacterium (25%). Most of the clones (28%) were confined to a monophyletic clade within the ɛ-Proteobacteria with no known close relatives. The prevalence of clones related to thermophilic microbes that use hydrogen as an electron donor and sulfur compounds (S0, SO4, thiosulfate) indicates the importance of hydrogen oxidation and sulfur metabolism at deep-sea hydrothermal vents. The presence of sequences that are related to sequences from hyperthermophiles, moderate thermophiles, and mesophiles suggests that the diversity obtained from this analysis may reflect the microbial succession that occurred in response to the shift in temperature and possible associated changes in the chemistry of the hydrothermal fluid.
Despite the diverse geochemical and temperature gradients that are prevalent at deep-sea hydrothermal vents, relatively little is known about the diversity and ecology of the free-living microbial communities that occupy these fluctuating high-temperature niches. The majority of microbial studies at deep-sea vents have relied on enrichment culturing techniques for growing hyperthermophiles (17, 19) and mesophiles (14, 37). Some biogeochemical studies have elucidated the role of microbial populations such as the sulfur-oxidizing bacteria (45, 53), methane-oxidizing mesophiles (10), and endosymbionts (8, 9) in this unusual ecosystem.
Recently, molecular phylogenetic approaches studying the small-subunit rRNA gene (16S rDNA) have been used to examine the diversity of different hydrothermal communities. As has been reported for other environments (for example, see references 2 and 28), this approach revealed a plethora of novel diversity, previously unknown to deep-sea hydrothermal vents (24, 35, 36, 50). Novel bacterial and archaeal sequences were reported in mesophilic microbial mats associated with Loihi Seamount in Hawaii (34–36), and new lineages have been reported for chimney and hydrothermal fluid samples obtained from hydrothermal vents from seamounts in the western Pacific Ocean and from the Okinawa Basin in Japan (50). Furthermore, several studies have used fluorescent in situ hybridization with 16S rRNA-specific probes to study the distribution of microbial types in hydrothermal sulfide structures (24, 25) and the colonization of epibionts on the vent invertebrate Alvinella pompejana (7).
In this study, we report the microbial diversity associated with a 5-day deployment of an in situ growth chamber (vent cap) on a hydrothermal vent in the Snake Pit hydrothermal field on the Mid-Atlantic Ridge. Surfaces within the chamber provided sites for attachment and growth for microorganisms, and the fluid flow through the chamber was such that the colonizing organisms were continually exposed to hydrothermal fluid. Using a molecular phylogenetic approach, we surveyed the diversity of the community obtained from this deployment. From this single sample, we observed a greater bacterial and archaeal phylogenetic diversity than has been previously reported using classical enrichment culturing techniques.
MATERIALS AND METHODS
Deployment of the in situ growth chamber (vent cap).
An in situ growth chamber or vent cap (Fig. 1) was deployed for 5 days on top of the vent orifice of Les Ruches at Snake Pit on the Mid-Atlantic Ridge (23°22′ N, 44°57′ W) by the DSV Nautile during the 1995 Microsmoke cruise (vent cap deployment 2.1 [VC2.1]; 16 to 21 November 1995). The measured temperature at the vent orifice was 112°C. Snake Pit sits atop a large volcanic ridge that is up to 600 m above the seafloor and runs slightly oblique to the main strike of the Mid-Atlantic Ridge for 40 km (51). The in situ chamber consists of a lower cone-shaped base and an upper cylindrical chamber. Surfaces (1 cm2) such as ceramic, steel, silicon, copper, and glass were attached to a titanium mesh within the chamber. These surfaces provided sites for attachment and colonization of microorganisms. Once the in situ growth chamber was placed on the vent orifice, the chamber was opened using the hydraulic arm of the submersible and by pushing the slide across the base of the cylinder. The fluid flows through the center into the cylindrical chamber and exits at the base of the cylinder through the uneven holes. The observers in the submersible noted the fluid flow through the chamber. A temperature probe and datalogger (HOBO; Deep Sea Power & Light) recorded the temperature inside the growth chamber every 3 s. After 5 days, the slide in the growth chamber was closed by the hydraulic arm of the submersible, minimizing contamination from surrounding seawater as the chamber was brought back to the surface.
FIG. 1.
Diagrammatic representation of the in situ growth chamber or vent cap. The temperature datalogger (A) and the slide mechanism (B) that opens or closes the chamber (C) are shown.
Immediately after the growth chamber was returned to the ship, the surfaces and approximately 250 ml of liquid were aseptically removed and combined. Samples for molecular analysis were stored in 80% ethanol at −80°C until they were further processed.
DNA extraction, PCR, and cloning.
DNA was extracted from 1 ml of the sample by the method of Ausubel et al. (1). The bacterial 16S rDNA was amplified using the universal primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) and the domain Bacteria-specific primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′). Archaeal 16S rDNA was amplified with the universal primer 1492R and the domain Archaea-specific primer 21F (5′-TCCGGTTGATCCYGCCGG-3′ where Y = C or T). The primers had 5′-ATGATGATGATG tails that were required for the cloning vector (see below). The PCR conditions were as described previously (44).
The PCR products were purified using a Gene-Clean kit (Bio 101) according to the manufacturer's instructions. Separate archaeal and bacterial clone libraries were prepared by cloning PCR products into the pAmp1 vector (Gibco BRL). Forty-eight bacterial clones and 45 archaeal clones were screened for inserts by PCR amplification with M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) primers using the same conditions as described above.
Restriction fragment length polymorphism (RFLP) analysis.
PCR-amplified bacterial and archaeal inserts were digested using MspI and HinPI (1 U each) according to the manufacturer's instructions (New England Biolabs, Beverly, Mass.). The DNA fragments were separated by gel electrophoresis on a 3.5% NuSieve GTG (FMC Bioproducts) agarose gel run in TBE (Tris-borate-EDTA) buffer at 4°C. The different banding patterns were noted, and the frequency of similar patterns was scored.
16S rDNA sequencing and analysis.
Unique bacterial and archaeal clones were sequenced by cycle sequencing using fluorescent dideoxy terminators. The sequences were determined on an automated sequencer (model 373; Applied Biosystems Inc.). Both strands were completely sequenced utilizing the following primers: 1492R, 907R (5′-CCGTCAATTCCTTTRAGTTT-3′, where R = A or G), 519R (5′-GWATTACCGCGGCKGCTG-3′, where W = A or T and K = G or T), 515F (5′-GTGCCAGCMGCCGCGGTAA-3′, where M = A or G), 906F (5′-GAAACTTAAAKGAATTG-3′), and 27F for Bacteria and 21F for Archaea.
Sequences were assembled with the AutoAssembler Program (Applied Biosystems, Inc.), and secondary structures were used to confirm the fidelity of the assembled sequences. The results of an initial comparison of the sequences with the GenBank nonredundant database using BLAST (available through the National Center for Biotechnology Information) provided a guide for determining which 16S rRNA sequences to use in the sequence alignments. Sequence alignments were performed using the Genetic Data Environment multiple sequence editor obtained from the Ribosomal Database Project (RDP) (32). Conserved sequence regions and the established secondary structure of 16S rRNA were taken into account to ensure that only homologous nucleotides were compared between sequences. The clones were checked for chimeric sequences using CHIMERA_CHECK from RDP (32) and by manual secondary structure comparisons.
Approximately 1,500 nucleotides were obtained from the assembled sequences, and on average about 1,100 nucleotides were included in the phylogenetic analysis. Similarity matrices were constructed by pairwise analysis using the correction computed by Jukes and Cantor (29). Preliminary evolutionary distance phylogenies and similarity matrices were performed on the aligned sequences (13). Phylogenetic trees were constructed by maximum-likelihood methods using fastDNAml (38) contained within the phylogeny inference package (PHYLIP version 3.3) (18). The bootstrap data for Archaea and Bacteria represent 100 samplings.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the sequences from VC2.1 are AF068782 to AF068824 and AF209779.
RESULTS AND DISCUSSION
Sample characteristics.
After the 5-day deployment at Snake Pit, the chamber was filled with a thick, creamy-white biomass. The surfaces were covered with visible filamentous organisms. Microcolony formation was also visible on the ceramic surfaces. During the deployment, the temperature profile within the chamber showed a gradual drop in temperature from a maximum temperature of 70°C to less than 20°C by the fifth day. The chamber and associated microbial growth may have caused a change in the fluid flow, which in turn may have influenced the temperature within the chamber. Preliminary characterization of the community was done on the ship using fluorescent in situ hybridization (G. Geesey and A.-L. Reysenbach, unpublished results). Fluorescein- and rhodamine-labeled oligonucleotide probes specific for the Archaea, Bacteria, and the Aquificales order (25) were used. Bacteria dominated the community, with members of the Aquificales representing about 10% of the community. Cell morphologies included cocci, rods, and filaments of differing diameters and lengths.
The sample we obtained from this deployment does not necessarily represent a typical community at deep-sea hydrothermal vents. This approach, like enrichment culturing, is limited as it selects for organisms capable of attaching to the surfaces inside the chamber and able to thrive in the fluctuating geochemical environment. Additionally, the sample probably represents the microbial diversity associated with the successional changes that occurred in the community over the 5-day period as the temperature in the chamber decreased. It is possible that the known thermophilic members established early in the deployment and were ultimately replaced by the mesophilic members of the community. In both cases, chemolithotrophs and heterotrophs should be present, the former using the geochemical energy associated with the mixed hydrothermal fluid and the latter utilizing the abundance of organic carbon available at hydrothermal vent environments as a result of the high productivity of micro- and macroorganisms.
Based on 16S rRNA phylogenetic analysis of the growth chamber sample, we obtained more than 38 different phylotypes, spanning both the Archaea (11 phylotypes) and Bacteria (27 phylotypes) and most closely related to known thermophilic and mesophilic heterotrophs and chemolithotrophs. Several novel lineages were detected that have no known close relatives. Additionally, it is likely that the diversity present in the chamber was greater than presented here, as screening for novel phylotypes using RFLP analysis may overlook some of the diversity present and we limited our screen to only 85 clones. Rarefaction curves (52) generated for the VC2.1 clones indicated that the diversity of the bacterial members of the community was greater than presented, whereas the archaeal diversity appeared well represented.
Archaeal diversity.
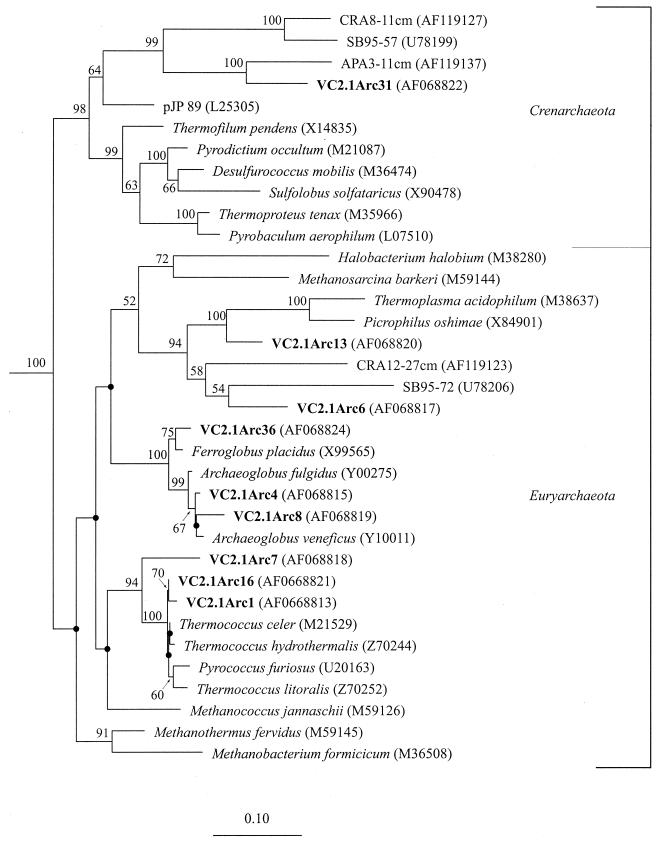
The archaeal sequences were distributed across the Euryarchaeota and Crenarchaeota. The Euryarchaeota were represented by three orders, namely, the Archaeoglobales, Thermococcales, and Thermoplasmales (Fig. 2). The majority of the archaeal sequences (71%) belonged to the Thermococcales (Table 1). Only 5 of the 29 Thermococcales phylotypes were sequenced fully and submitted to GenBank. The Thermococcales sequences identified are very closely related to known Thermococcus spp. with similarities between 94 and 99.4%. The short branch lengths could not be resolved by bootstrap analysis. One phylotype, VC2.1Arc7, clusters with the Thermococcales but is a deeper branch than any of the known Thermococcales isolates.
FIG. 2.
Phylogenetic relationships of archaeal 16S rRNA sequences as determined by maximum-likelihood analysis. Aquifex pyrophilus was used as the outgroup. The numbers at the nodes are the bootstrap values (as percentages). The bootstrap values were less than 50% for the branch points marked with small black circles and no numerical value. Sequences from Snake Pit are marked in bold type, and the remaining sequences were obtained from the RDP (32). The scale bar represents the expected number of changes per nucleotide position.
TABLE 1.
Summary of the archaeal 16S rRNA sequences identified from the growth chamber deployment at Snake Pit, Mid-Atlantic Ridge
| Group | Type sequence(s)a | Other representative(s)b | % of clonesc | Closest relative | % Sequence similarity |
|---|---|---|---|---|---|
| Thermococcales | 71 | ||||
| VC2.1Arc1, -16 | VC2.1Arc5, -35 | Thermococcus spp. | 96.1–97.5 | ||
| VC2.1Arc7 | Thermococcus litoralis | 88.9 | |||
| Archaeoglobales | 22 | ||||
| VC2.1Arc4, -8 | VC2.1Arc2 | Archaeoglobus veneficus | 96.2–98.6 | ||
| VC2.1Arc36 | Archaeoglobus veneficus | 95.1 | |||
| Thermoplasmales | VC2.1Arc13 | 2 | pSSMCA108 | 97.1 | |
| Marine benthic group D (Euryarchaeota) | VC2.1Arc6 | 2 | pMC2A33 | 78.3 | |
| Marine benthic group B (Crenarchaeota) | VC2.1Arc31 | 2 | pMC2A36 | 86.3 |
The type 16S rRNA sequence was used in the maximum-likelihood and bootstrap analyses.
Additional closely related full-length sequences that were submitted to GenBank.
Percentage of clones that are closely related based on RFLP analysis and sequence analysis. This is not a percentage estimate of the relative abundance of these phylotypes in the sample.
The occurrence of Thermococcales at deep-sea hydrothermal vents is widespread, and members of the genus Thermococcus are some of the most numerous newly described hyperthermophiles from deep-sea vents (6, 20, 27, 33). It is therefore not surprising that the Thermococcales were also the most numerous in our archaeal clone library. However, their natural abundance in the vent ecosystem is not known, and in situ oligonucleotide probes will help elucidate this. Clearly, the potential significance of these hyperthermophilic sulfur-reducing heterotrophs in the overall productivity at deep-sea vents needs to be determined.
Twenty-two percent of the archaeal clones grouped among known members of the Archaeoglobales. One of these phylotypes (VC2.1Arc36) was most closely related to Ferroglobus, the only iron-oxidizing member (23) of the otherwise predominantly sulfate-reducing facultatively chemolithotrophic Archaeoglobales (4, 26). Our sequences probably belong to sulfate-reducing and iron-oxidizing Archaea that were present in this community. Few sulfate-reducing Archaea have been isolated from deep-sea vents, and Ferroglobus has been isolated only from shallow marine vents at Vulcano, Italy. Similar sequences were obtained by Takai and Horikoshi (50), which may imply the possible importance and cosmopolitan distribution of this order in deep-sea vent ecosystems.
The Thermoplasmales-related phylotypes obtained in this study and reported by Takai and Horikoshi (50) from Okinawa trough sediments and a chimney sample from Myojin Knoll seamount in Japan adds to our biological inventory of deep-sea diversity. The known genera of the Thermoplasmales, namely, Picrophilus (46), Thermoplasma (47), and Ferroplasma (15), are all acidophiles and restricted to terrestrial solfataras, coal refuse piles, or acid mine drainage areas. A long-standing concern expressed by microbiologists is the apparent absence of thermoacidophiles in deep-sea hydrothermal vent environments. No thermoacidophiles have been isolated from deep-sea hydrothermal vents, although end-member hydrothermal fluid pH is usually below pH 4.5 (55). It is possible that thermoacidophiles cannot tolerate large fluctuations in pH (as probably occurs in the turbulent mixing zones at deep-sea vents). Thermoacidophiles such as Sulfurococcus mirabilis, are unable to maintain a neutral internal pH at 4°C if the surroundings are at a low pH (21). Additionally, it has been suggested that Sulfolobus is unable to grow in saline conditions (49). However, these observations have been limited by results from enrichment culturing, and as has been shown in many other environments, culture-independent approaches provide glimpses into an as yet uncultivated microbial diversity. Based on these Thermoplasmales-related sequences, thermoacidophiles probably do occupy certain niches in the deep-sea hydrothermal ecosystem. How significant these populations are (and whether they occupy a thermophilic or mesophilic niche) has yet to be determined.
Within the Euryarchaeota, several groups of Archaea have been identified with no known representative members as laboratory cultures (12). VC2.1Arc6 clusters with one such lineage, the marine group II and the newly identified marine benthic group D (54) (Table 1, Fig. 2). The deep-sea hydrothermal vent sequences from Myojin Knoll (50) and microbial mat sample sequences from Loihi Seamount (36) are also part of this clade. These sequences and our sequence form a clade with sequences that have exclusively been associated with marine benthic sediments (54).
A second sequence that is affiliated with the marine benthic, nonhydrothermal archaeal sequences is VC2.1Arc31. This phylotype is a crenarchaeotal sequence and is most closely related to sequences obtained from sediment cores collected from the Atlantic abyssal plain (54) and the phylotype pMC2A36 (50) obtained from deep-sea hydrothermal sulfides. The comparison between the signature sequences for marine benthic group B and VC2.1Arc31 are given in Table 2. With one exception, the deviations from the defining signature sequences between Euryarchaeota and Crenarchaeota (56) are the same for marine benthic group B and VC2.1Arc31. At positions 27 and 556, VC2.1Arc31 has a U:G pair which is not seen in marine benthic group B, Euryarchaeota, or Crenarchaeota. The deeply rooted placement of our sequence within this group provides additional support for the high-temperature ancestry of the low-temperature Archaea (11, 54).
TABLE 2.
| Position(s)b | Intradomain nucleotide signature feature fora:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Eury | Cren | MBG B | pMC2A36 | VC2.1Arc31 | MBG D | pMC2A33 | VC2.1Arc6 | |
| 27:556 | G:C | C:G | Cren | Cren | U:G | Eury | Eury | Eury |
| 28:555 | G:Y | C:G | Cren | Cren | Cren | Eury | Eury | Eury |
| 30:553 | Y:R | G:C | Cren | Cren | Cren | Eury | Eury | Eury |
| 34:550 | U:G | C:G | ND:G | Eury | Eury | Eury | Eury | Eury |
| 289:311 | C:G | G:C | A:U (APA3-11) | A:U | A:U | Eury | Eury | Eury |
| Cren (CRA8-27) | ||||||||
| 501:544 | R:Y | C:G | Eury | Eury | Eury | Eury | U:A | Eury |
| 503:542 | C:G | G:C | Cren | Cren | Cren | Eury | Eury | Eury |
| 504:541 | Y:R | G:Y | A:U | A:U | A:U | Eury | Eury | Eury |
| 513:538 | C:G | U:A | Cren | Cren | Cren | G:U | G:U | Eury |
| 518 | C | U | Eury | Eury | Eury | Eury | Eury | Eury |
| 658:747 | Y:R | G:C | Cren | Cren | Cren | Eury | Eury | Eury |
| 692 | U | C | Eury | Eury | Eury | Eury | Eury | Eury |
| 965 | Y | G | Eury | Eury | Eury | Eury | Eury | Eury |
| 1074:1083 | A:C | G:U | Cren | Cren | Cren | Eury | Eury | Eury |
| 1244:1293 | Y:R | R:Y | Cren | Cren | Cren | Eury (U:A) | Eury | Eury |
| 1252 | U | C | Eury | Eury | Eury | Eury | Eury | A |
| 1335 | C | G | Cren | Cren | U | Eury | Eury | Cren |
| 1408 | A | G | Cren | Cren | Cren | Eury | Eury | Eury |
Crenarchaeota (Cren) and Euryarchaeota (Eury) signature sequences where R = purine and Y = pyrimidine. MBG, marine benthic group.
Numbering follows that for the E. coli 16S rRNA sequence.
Bacterial diversity.
The bacterial sequences were primarily affiliated with one of three groups: the ɛ subclass of the Proteobacteria (ɛ-Proteobacteria), the Aquificales, and the recently described genus Desulfurobacterium (30) (Fig. 3). Single clones containing sequences closely related to the Cytophagales (VC2.1Bac22) and the β subclass of the Proteobacteria (VC2.1Bac29) were obtained. Several new lineages were identified (VC2.1Bac16, -35, and -47) with less than 85% similarity between the new sequence and previously identified sequences (Table 3).
FIG. 3.
Phylogenetic relationships of bacterial 16S rRNA sequences as determined by maximum-likelihood analysis. Methanococcus jannaschii was used as the outgroup. The numbers at the nodes are the bootstrap values (as percentages). The bootstrap values were less than 50% for the branch points marked with small black circles and no numerical value. Sequences from the in situ growth chamber deployed at Snake Pit are marked in bold type, and the remaining sequences were obtained from the RDP (32). The scale bar represents the expected number of changes per nucleotide position.
TABLE 3.
Summary of the bacterial sequences identified from the growth chamber deployment at Snake Pit, Mid-Atlantic Ridge
| Group | Type sequence(s)a | Other representative(s)b | % of clonesc | Closest relative | % Sequence similarity |
|---|---|---|---|---|---|
| Aquificales | VC2.1Bac27 | VC2.1Bac10, -11, -28, -33, -39 | 18 | pBB | 92.3–92.6 |
| Desulfurobacterium | VC2.1Bac3, -24, -48 | VC2.1Bac2, -5, -13, -23, -34 | 25 | Desulfurobacterium thermolithotrophum | 93.4–96.0 |
| ɛ-Proteobacteria | VC2.1Bac32 | 3 | PVB_12 | 88.4 | |
| VC2.1Bac1, -31 | VC2.1Bac4 | 9 | SB17 | 90.7–92.6 | |
| VC2.1Bac43, -7, -17, -30 | VC2.1Bac8, -9, -12, -19, -20 | 28 | Unresolved | ||
| ɛ-Proteobacteria | VC2.1Bac29 | 3 | Leptothrix discophora | 96.4 | |
| Flexibacter-Cytophaga-Bacteroides | VC2.1Bac22 | 3 | SB5 | 89.7 | |
| Unresolved | VC2.1Bac16 | 3 | BD2-14 | 79.4 | |
| VC2.1Bac47 | 3 | Thermodesulforhabdus norvegicus | 82.8 | ||
| VC2.1Bac35 | 3 | Nitrococcus mobilis | 83.8 |
The type 16S rRNA sequence was used in the maximum-likelihood and bootstrap analyses.
Additional closely related full-length sequences that were submitted to GenBank.
Percentage of clones that are closely related based on RFLP analysis and sequence analysis. This is not a percentage estimate of the relative abundance of these phylotypes in the sample.
The ɛ-Proteobacteria phylotypes were the most abundant bacterial sequences obtained (Table 3) and can be separated into seven groups based on their relatedness to previously identified lineages. The majority (28%) of these novel phylotypes (VC2.1Bac43, -7, -30, -17, -8, -9, -12, -19, and -20) cluster together as a single clade within the ɛ-Proteobacteria. The remaining proteobacterial sequences are most closely related to sequences identified from other environmental clone libraries. VC2.1Bac32 is most closely related to PVB_12, a phylotype from Loihi Seamount (88.4% similarity). The sequences from VC2.1Bac1, -4, and -31 are most similar to 16S rRNA sequences from a sulfate-reducing and benzene-mineralizing consortium (SB17) (39) and an epibiont from the deep-sea hydrothermal worm Alvinella pompejana (22).
The significance of the ɛ-Proteobacteria at deep-sea hydrothermal vents has only recently been realized (7, 35, 40). Many appear to be epibionts of the deep-sea hydrothermal invertebrates, although their association with their invertebrate host remains speculative. In our study, we identified a clade of sequences that dominated the proteobacterial clones, yet this clade has no known representative in culture yet. Preliminary results from other geographically distinct deep-sea hydrothermal vents indicate that this group may be prevalent in low-temperature mats rich in iron and sulfur precipitates (K. Longnecker and A.-L. Reysenbach, unpublished results). It is possible that these organisms may be important mesophilic or moderately thermophilic microorganisms involved in iron or sulfur cycling at deep-sea hydrothermal vents. Like the ɛ-Proteobacteria division-level lineages recently identified from a hot spring at Yellowstone National Park (28), the sequences from Snake Pit further expand the diversity of ɛ-Proteobacteria identified from high-temperature environments.
From evolutionary distance matrix analyses, VC2.1Bac47 was most closely related to the γ-Proteobacteria, however, maximum-likelihood and bootstrap analyses place this sequence as an unresolved lineage spanning the ɛ- and γ-Proteobacteria. Likewise, bootstrap analysis could not place VC2.1Bac35 confidently within the γ-Proteobacteria, although a similarity matrix placed this sequence most closely to Nitrococcus. This phylotype has an unusual and long (73-bp) insert at position 94 (Escherichia coli numbering).
One β-proteobacterial sequence (VC2.1Bac29) was most closely related to the sheathed iron oxidizer Leptothrix, suggesting that relatives of these sheathed low-temperature iron oxidizers may be present at deep-sea vents. Recent isolations of low-temperature proteobacterial iron oxidizers (16) provide further evidence of the importance of this metabolic group in the iron cycle at deep-sea hydrothermal vents.
Second in abundance to the ɛ-proteobacterial phylotypes were the phylotypes most closely related to the new thermophilic and sulfur-reducing chemolithotrophic bacterial genus and lineage, Desulfurobacterium (30). This organism was first isolated in culture during the same research cruise of 1995. Additionally, isolates were obtained from the growth chamber sample (C. Jeanthon, unpublished results). Fluorescent in situ hybridization analyses of the growth chamber sample revealed that up to 26% of the cells belonged to this novel lineage (E. Corre et al., unpublished results). This is very similar to the percentage of clones obtained for this group (31%) (Table 3). Furthermore, the dominant clones that were implicated as thermophiles are all closely associated with a sulfur-reducing metabolism (both Desulfurobacterium-like and Thermococcus-like) and may imply that this metabolic type plays a significant role in the productivity of deep-sea hydrothermal ecosystems. However, it should be noted that Takai and Horikoshi (50) did not detect a single Thermococcales-related sequence in all the samples they screened for archaeal diversity.
Thirteen percent of the phylotypes grouped with the Aquificales and specifically with the newly identified phylotypes previously reported only from terrestrial thermal springs, namely, OPB13, pBB, and NAK-14 (28, 43, 57). The Aquificales-like sequences identified from Snake Pit were most closely related to pBB (92.3 to 92.6% similarity), a sequence identified from a hot spring in Yellowstone National Park (42). All members of the cultivated Aquificales are microaerophilic hydrogen oxidizers. It is surprising that until recently, only one microaerophile, Pyrolobus fumarii (3), had been reported from deep-sea hydrothermal vents, since mixing of reduced hydrothermal fluid with oxygenated seawater creates microniches with low oxygen concentrations. Based on the sequence obtained for VC2.1Bac27, we have isolated a closely related microaerophile from deep-sea vent samples from the East Pacific Rise at 9° N 104° W (41). Not only do culture-independent assessments help direct enrichment culturing approaches but this new isolate also indicates that microaerophily may be an important process at deep-sea hydrothermal vents. A single phylotype, VC2.1Bac16, that branched deeper within the bacterial 16S rRNA phylogeny than has been previously observed was obtained (Fig. 4). BLAST analysis did not provide any evidence that this sequence was closely related to any other sequence. However, inclusion of three additional sequences (courtesy of Erwan Corre; Corre et al., unpublished), one obtained from another growth chamber deployment (cl15bon, Corre et al., unpublished), one from deep-sea sediments (BD2-14) (31), and one from an oral cavity (X112) (B. J. Paster and F. E. Dewhirst; sequences submitted to GenBank), indicated that VC2.1Bac16 groups with these other sequences that were obtained from very different environments. Using the CHIMERA_CHECK program from the RDP confirmed that these sequences were unlikely to be chimeras. The deeply branching position remains with high bootstrap confidence using maximum-parsimony analysis and neighbor-joining methods (data not shown). Examination of the signature sequences identified by Winker and Woese (56) also show that like Aquifex pyrophilus, VC2.1Bac16 deviates from other Bacteria at positions 340 and 349 by having a G:C pair in this position (Table 4). Additionally, this 16S rRNA sequence has archaeal and not bacterial signatures in positions 923 and 1393, 930 and 1387, 931 and 1386, 933 and 1384, and 962 and 973. The entire clade has a unique nucleotide (A) at position 966, where in Archaea, it is a U, and in the Bacteria, a G occupies the position.
FIG. 4.
Phylogenetic relationships of the deeply branching bacterial 16S rRNA sequences as determined by maximum-likelihood analysis. Methanococcus jannaschii was used as the outgroup. The numbers at the nodes are the bootstrap values (as percentages). Sequences from Snake Pit are marked in bold type, and the remaining sequences were obtained from the RDP (32). Cl15bon represents an unpublished sequence obtained from a second deployment of the in situ growth chamber at Snake Pit (courtesy of E. Corre). The scale bar represents the expected number of changes per nucleotide position.
TABLE 4.
| Position(s)a | Interdomain signature featureb for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Archaea | A. pyrophilus | pBB | cl15bon | X112 | BD2-14 | VC2.1Bac16 | |
| 8 | A | U | A | A | ND | ND | A | A |
| 9:25 | G:C | C:G | G:C | G:C | ND | ND | G:C | G:C |
| 10:24 | R(A):U | Y:Rc | A:U | A:U | ND | ND | A:U | A:U |
| 33:551 | A:U | Y(C):R(G) | A:U | A:U | A:U | A:U | A:U | A:U |
| 52:359 | Y:R | G:C | C:G | C:G | C:G | C:G | C:G | C:G |
| 53:358 | A:U | C:G | A:U | A:U | A:U | A:U | A:U | A:U |
| 113:314 | G:C | C:G | G:C | G:C | G:C | G:C | G:C | G:C |
| 307 | Y/A | G | C | C | U | C | C | U |
| 338 | A | G | A | A | A | A | A | A |
| 339:350 | C:G | G:Y(C) | C:G | C:G | C:G | C:G | C:G | C:G |
| 340:349 | U:A | C:G | C:G | C:G | U:A | U:A | U:A | C:G |
| 361 | R(G) | C | G | G | G | G | G | G |
| 365 | U | A | U | U | U | U | U | U |
| 367 | U | C | G | A | U | U | U | U |
| 377:386 | R(G):Y(C) | Y(C):G | G:C | G:C | G:C | G:C | G:C | G:C |
| 393 | A | G | C | A | A | A | A | A |
| 514:537 | Y(C):R(G) | G:C | C:G | C:G | C:G | C:G | C:G | C:G |
| 549 | C | U | C | C | C | C | C | C |
| 558 | G | Y | A | A | G | G | G | G |
| 585:756 | R(G):Y(C) | C:G | C:G | G:C | C:G | U:A | C:G | C:G |
| 675 | A | U | A | A | A | A | A | A |
| 684:706 | U:A | G(Y):C | G:C | U:A | U:A | U:A | U:A | U:A |
| 716 | A | C | A | A | A | A | A | A |
| 867 | R(G) | Y(C) | G | G | G | G | G | G |
| 923:1393 | A:U | G:C | G:C | A:U | G:C | G:C | G:C | G:C |
| 930:1387 | Y(C):R(G) | A:U | C:G | C:G | A:U | A:U | A:U | A:U |
| 931:1386 | C:G | G:C | C:G | C:G | A:U | A:U | A:U | G:C |
| 933:1384 | G:C | A:U | G:C | G:C | A:U | A:U | A:U | A:U |
| 962:973 | C:G | G:C | C:G | C:G | G:C | G:C | G:C | G:C |
| 966 | G | U | G | G | A | A | A | A |
| 974 | A/C | G | A | A | A | A | A | A |
| 1098 | Y(C) | G | C | C | G | G | A | G |
| 1109 | C | A | C | C | C | C | C | C |
| 1110 | A | G | A | A | A | A | A | A |
| 1194 | U | R(G) | U | U | U | U | U | U |
| 1211 | U | G | U | U | U | U | U | U |
| 1212 | U | A | U | U | U | U | U | U |
| 1381 | U | C | U | U | C | C | C | C |
Numbering follows that for the E. coli 16S rRNA sequence.
The letters in parentheses indicate the dominant form. ND, not determined.
This does not include A:C pairs.
Conclusions.
As the temperature dropped in the vent cap chamber, there was probably a shift in the community structure from hyperthermophiles to moderate thermophiles and then mesophiles. Although physiology cannot be assumed from phylogeny, in many cases the physiology of an organism can be cautiously inferred from the phylogeny. For example, the Aquificales and Thermococcales are represented only by thermophilic genera, the former are generally chemolithotrophic microaerophiles and the latter are all sulfur-reducing heterotrophs. The thermophiles in this study include archaeal members from the Thermococcales, Archaeoglobales, and Thermoplasmales and bacterial members from the Aquificales and the genus Desulfurobacterium. The thermophilic populations were probably replaced by mesophilic heterotrophs and chemolithotrophs, which would probably include the other archaeal and proteobacterial phylotypes obtained in this study.
The limited research on macrofaunal succession at deep-sea hydrothermal vents has been very insightful (48), providing clear community shifts as the vent ecosystem matures. However, whether similar temporal and spatial changes in the microbial communities occur and whether these changes influence macrofaunal succession are entirely unknown. The growth chamber is a good tool to examine temporal changes in the microbial communities at deep-sea vents, as it can be deployed on the same vent for different time periods. Even in this one deployment, the change in the temperature record suggested that changes in the community structure had to occur for the microbial community to respond to the drop in temperature. The temperature shift would cause a shift in the types of geochemical sources available for chemolithotrophic metabolism. The large diversity of organisms obtained from this study may reflect this successional change. In conclusion, the novel diversity obtained from this study will help direct enrichment culturing attempts to better understand the physiological ecology of deep-sea hydrothermal thermophiles and mesophiles.
ACKNOWLEDGMENTS
Special thanks to Daniel Prieur for leadership and guidance as chief scientist during the Microsmoke cruise and to the captain and crew of the R/V Nadir and the DSV Nautile operations group for their technical expertise. The vent cap experiment could not have been accomplished without the innovative design of the growth chamber, and we thank David Lane and Norman Pace for letting us modify and use their instrument. We thank Erwan Corre for sharing his insights into the diversity of the deeply branching bacterial phylotypes and providing sequence information for the manuscript. We thank the members of A.-L. Reysenbach's lab for critically reviewing the manuscript. This research was supported in part by a National Science Foundation-LExEn grant (OCE 9729784) to A.L.R.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 2.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blöchl E, Rachel R, Burggraf S, Hafelbradl D, Jannasch H W, Stetter K O. Pyrolobus fumarii gen. and sp. nov., represents a novel group of archaea extending the upper temperature limit for life to 113°C. Extremophiles. 1997;1:14–21. doi: 10.1007/s007920050010. [DOI] [PubMed] [Google Scholar]
- 4.Burggraf S, Jannasch H W, Nicolaus B, Stetter K O. Archaeoglobus profundus sp. nov., represents a new species within the sulfate-reducing archaebacteria. Syst Appl Microbiol. 1990;13:24–28. [Google Scholar]
- 5.Burggraf S, Olsen G J, Stetter K O, Woese C R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- 6.Canganella F, Jones W J, Gambacorta A, Antranikian G. Thermococcus guaymasensis sp. nov. and Thermococcus aggregans sp. nov., two novel thermophilic Archaea isolated from the Guaymas Basin hydrothermal vent site. Int J Syst Bacteriol. 1998;48:1181–1185. doi: 10.1099/00207713-48-4-1181. [DOI] [PubMed] [Google Scholar]
- 7.Cary S C, Cottrell M T, Stein J L, Camacho F, Desbruyères D. Molecular identification and localization of filamentous symbiotic bacteria associated with the hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol. 1997;63:1124–1130. doi: 10.1128/aem.63.3.1124-1130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanaugh C M. Symbioses of chemoautotrophic bacteria and marine invertebrates from hydrothermal vents and reducing sediments. Bull Biol Soc Wash. 1985;6:373–388. [Google Scholar]
- 9.Childress J J, Lee R W, Sanders N K, Felbeck H, Oros D R, Toulmond A, Desbruyères D, Kennicutt II M C, Brooks J. Inorganic carbon uptake in hydrothermal vent tubeworms facilitated by high environmental pCO2. Nature. 1993;362:147–149. [Google Scholar]
- 10.De Angelis M A, Lilley M D, Olson E J, Baross J A. Methane oxidation in deep-sea hydrothermal plumes of the Endeavour segment of the Juan de Fuca Ridge. Deep-Sea Res Part I. 1993;40:1169–1186. [Google Scholar]
- 11.DeLong E. Archaeal means and extremes. Science. 1998;280:542–543. doi: 10.1126/science.280.5363.542. [DOI] [PubMed] [Google Scholar]
- 12.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeSoete G. A least squares algorithm for fitting additive trees to proximity data. Psychometrika. 1983;48:621–626. [Google Scholar]
- 14.Durand P, Reysenbach A L, Prieur D, Pace N. Isolation and characterization of Thiobacillus hydrothermalis sp. nov., a mesophilic obligately chemolithotrophic bacterium isolated from a deep-sea hydrothermal vent in Fiji Basin. Arch Microbiol. 1993;159:39–44. [Google Scholar]
- 15.Edwards K J, Bond P L, Gihring T M, Banfield J F. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science. 2000;287:1796–1799. doi: 10.1126/science.287.5459.1796. [DOI] [PubMed] [Google Scholar]
- 16.Emerson D, Moyer C. Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol. 1997;63:4784–4792. doi: 10.1128/aem.63.12.4784-4792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erauso G, Reysenbach A L, Godfroy A, Meunier J R, Crump B, Partensky F, Baross J A, Marteinsson V, Barbier G, Pace N R, Prieur D. Pyrococcus abyssi sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Arch Microbiol. 1993;160:338–349. [Google Scholar]
- 18.Felsenstein J. PHYLIP—phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 19.Godfroy A, Lesongeur F, Raguenes G, Querellou J, Antoine E, Meunier J R, Guezennec J, Barbier G. Thermococcus hydrothermalis sp. nov, a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1997;47:622–626. doi: 10.1099/00207713-47-3-622. [DOI] [PubMed] [Google Scholar]
- 20.Godfroy A, Lesongeur F, Raguenes G, Querellou J, Antoine E, Meunier J R, Guezennec J, Barbier G. Thermococcus hydrothermalis sp. nov, a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1997;47:622–626. doi: 10.1099/00207713-47-3-622. [DOI] [PubMed] [Google Scholar]
- 21.Golovacheva R S, Valekho-Roman K M, Troitskii A V. Sulfurococcus mirabilis gen. nov., sp. nov., a new thermophilic archaebacterium with the ability to oxidize sulfur. Mikrobiologiya. 1985;56:100–107. [Google Scholar]
- 22.Haddad A, Camacho F, Durand P, Cary S C. Phylogenetic characterization of the epibiotic bacteria associated with the hydrothermal vent polychaete Alvinella pompejana. Appl Environ Microbiol. 1995;61:1679–1687. doi: 10.1128/aem.61.5.1679-1687.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hafenbradl D, Keller M, Dirmeier R, Rachel R, Rossnagel P, Burggraf S, Huber H, Stetter K O. Ferroglobus placidus gen. nov., sp. nov., a novel hyperthermophilic archaeum that oxidizes Fe2+ at neutral pH under anoxic conditions. Arch Microbiol. 1996;166:308–314. doi: 10.1007/s002030050388. [DOI] [PubMed] [Google Scholar]
- 24.Harmsen H J M, Prieur D, Jeanthon C. Distribution of microorganisms in deep-sea hydrothermal vent chimneys investigated by whole-cell hybridization and enrichment culture of thermophilic subpopulations. Appl Environ Microbiol. 1997;63:2876–2883. doi: 10.1128/aem.63.7.2876-2883.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harmsen H J M, Prieur D, Jeanthon C. Group-specific 16S rRNA-targeted oligonucleotide probes to identify thermophilic bacteria in marine hydrothermal vents. Appl Environ Microbiol. 1997;63:4061–4068. doi: 10.1128/aem.63.10.4061-4068.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber H, Jannasch H, Rachel R, Fuchs T, Stetter K O. Archaeoglobus veneficus sp. nov., a novel facultative chemolithoautotrophic hyperthermophilic sulfite reducer, isolated from abyssal black smokers. Syst Appl Microbiol. 1997;20:374–380. [Google Scholar]
- 27.Huber R, Stohr J, Hohenhaus S, Rachel R, Burggraf S, Jannasch H W, Stetter K O. Thermococcus chitonophagus sp. nov., a novel, chitin-degrading, hyperthermophilic archaeum from a deep-sea hydrothermal vent environment. Arch Microbiol. 1995;164:255–264. [Google Scholar]
- 28.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 30.L'Haridon S, Cilia V, Messner P, Raguénès G, Gambacorta A, Sleytr U B, Prieur D, Jeanthon C. Desulfurobacterium thermolithotrophum gen. nov., sp. nov., a novel autotrophic, sulfur-reducing bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1998;48:701–711. doi: 10.1099/00207713-48-3-701. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Kato C, Horikoshi K. Bacterial diversity in deep-sea sediments from different depths. Biodivers Conserv. 1999;8:659–677. [Google Scholar]
- 32.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeck R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marteinsson V T, Birrien J L, Reysenbach A L, Vernet M, Marie D, Gambacorta A, Messner P, Sleytr U B, Prieur D. Thermococcus barophilus sp. nov., a new barophilic and hyperthermophilic archaeon isolated under high hydrostatic pressure from a deep-sea hydrothermal vent. Int J Syst Bacteriol. 1999;49:351–359. doi: 10.1099/00207713-49-2-351. [DOI] [PubMed] [Google Scholar]
- 34.Moyer C L, Dobbs F C, Karl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rRNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1994;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moyer C L, Tiedje J M, Dobbs F C, Karl D M. Diversity of deep-sea hydrothermal vent Archaea from Loihi Seamount, Hawaii. Deep-Sea Res Part II. 1998;45:303–317. [Google Scholar]
- 37.Nelson D C, Wirsen C O, Jannasch H W. Characterization of large autotrophic Beggiatoa abundant at hydrothermal vents of the Guaymas Basin. Appl Environ Microbiol. 1989;55:2909–2917. doi: 10.1128/aem.55.11.2909-2917.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 39.Phelps C, Kerkof L, Young L. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microb Ecol. 1998;27:269–279. [Google Scholar]
- 40.Polz M F, Cavanaugh C M. Dominance of one bacterial phylotype at a Mid-Atlantic Ridge hydrothermal vent site. Proc Natl Acad Sci USA. 1995;92:7232–7236. doi: 10.1073/pnas.92.16.7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reysenbach A-L, Banta A B, Boone D R, Cary S C, Luther G W., III Microbial essentials at hydrothermal vents. Nature. 2000;404:835. doi: 10.1038/35009029. [DOI] [PubMed] [Google Scholar]
- 42.Reysenbach A-L, Ehringer M, Hershberger K. Microbial diversity at 83°C in Calcite Springs, Yellowstone National Park: another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles. 2000;4:61–67. doi: 10.1007/s007920050008. [DOI] [PubMed] [Google Scholar]
- 43.Reysenbach A-L, Seitzinger S, Kirshtein J, McLaughlin E. Molecular constraints on a high-temperature evolution of early life. Biol Bull. 1999;196:367–372. doi: 10.2307/1542972. [DOI] [PubMed] [Google Scholar]
- 44.Reysenbach A-L, Wickham G, Pace N. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruby E G, Jannasch H W, Deuser W G. Fractionation of stable carbon isotopes during chemoautotrophic growth of sulfur-oxidizing bacteria. Appl Environ Microbiol. 1987;53:1940–1943. doi: 10.1128/aem.53.8.1940-1943.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schleper C, Pühler G, Klenk H-P, Zillig W. Picrophilus oshimae and Picrophilus torridus fam. nov., gen. nov., sp. nov., two species of hyperacidophilic, thermophilic, heterotrophic, aerobic archaea. Int J Syst Bacteriol. 1996;46:814–816. [Google Scholar]
- 47.Segerer A, Stetter K. The genus Thermoplasma. In: Balows A, Trüper H, Dworkin M, Harder W, Shleifer K-H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 712–718. [Google Scholar]
- 48.Shank T M, Fornari D J, Von Damm K L, Lilley M D, Haymon R M, Lutz R A. Temporal and spatial patterns of biological community development at nascent deep-sea hydrothermal vents (9° 50′N East Pacific Rise) Deep-Sea Res Part II. 1998;45:465–515. [Google Scholar]
- 49.Stetter K O. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 1996;18:149–258. [Google Scholar]
- 50.Takai K, Horikoshi K. Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson G, Humphris S, Schroeder B, Sulanowska M, Rona P. Active vents and massive sulfides at 26° N (TAG) and 23′ N (Snake Pit) on the Mid-Atlantic Ridge. Can Mineral. 1988;26:697–711. [Google Scholar]
- 52.Tipper J C. Rarefaction and rarefiction—the use and abuse of a method in paleoecology. Paleobiology. 1979;5:423–434. [Google Scholar]
- 53.Tuttle J H. The role of sulfur-oxidizing bacteria at deep-sea hydrothermal vents. Bull Biol Soc Wash. 1985;6:335–344. [Google Scholar]
- 54.Vetriani C, Jannasch H W, MacGregor B J, Stahl D A, Reysenbach A-L. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Von Damm K L. Controls on the chemistry and temporal variability of seafloor hydrothermal fluids. In: Humphris S, Zierenberg R, Mullineaux L, Thomson R, editors. Seafloor hydrothermal systems: physical, chemical, biological, and geological interactions. Washington, D.C.: American Geophysical Union; 1995. pp. 222–247. [Google Scholar]
- 56.Winker S, Woese C R. A definition of the domains Archaea, Bacteria, and Eucarya in terms of small subunit ribosomal RNA characteristics. Syst Appl Microbiol. 1991;14:305–310. doi: 10.1016/S0723-2020(11)80303-6. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto H, Hiraishi A, Kato K, Chiura H X, Maki Y, Shimizu A. Phylogenetic evidence for the existence of novel thermophilic bacteria in hot spring sulfur-turf microbial mats in Japan. Appl Environ Microbiol. 1998;64:1680–1687. doi: 10.1128/aem.64.5.1680-1687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]