Abstract
Dsb proteins (DsbA, DsbB, DsbC, and DsbD) catalyze formation and isomerization of protein disulfide bonds in the periplasm of Escherichia coli. By using a set of Dsb coexpression plasmids constructed recently, we analyzed the effects of Dsb overexpression on production of horseradish peroxidase (HRP) isozyme C that contains complex disulfide bonds and tends to aggregate when produced in E. coli. When transported to the periplasm, HRP was unstable but was markedly stabilized upon simultaneous overexpression of the set of Dsb proteins (DsbABCD). Whereas total HRP production increased severalfold upon overexpression of at least disulfide-bonded isomerase DsbC, maximum transport of HRP to the periplasm seemed to require overexpression of all DsbABCD proteins, suggesting that excess Dsb proteins exert synergistic effects in assisting folding and transport of HRP. Periplasmic production of HRP also increased when calcium, thought to play an essential role in folding of nascent HRP polypeptide, was added to the medium with or without Dsb overexpression. These results suggest that Dsb proteins and calcium play distinct roles in periplasmic production of HRP, presumably through facilitating correct folding. The present Dsb expression plasmids should be useful in assessing and dissecting periplasmic production of proteins that contain multiple disulfide bonds in E. coli.
The formation of disulfide bonds following polypeptide synthesis contributes to folding and stability of many secretory proteins (3). In Escherichia coli, disulfide bond formation depends on several Dsb proteins found in the periplasm and inner membrane (3, 8, 20, 26, 27). DsbA and DsbC play distinct roles as a disulfide bond-introducing and disulfide bond-isomerizing factor, respectively (26, 30). DsbB and DsbD modulate the DsbA and DsbC activities, respectively (4, 19, 21). Because of this machinery for disulfide bond formation as well as the oxidative environment, the periplasm provides an adequate compartment for expressing proteins with multiple disulfide bonds in E. coli (2, 11, 17, 20). However, periplasmic expression of proteins with multiple disulfide bonds often results in very low yields or inactive products (2, 24, 25, 33). This may result from limited or incorrect formation of disulfide bonds in the target protein (24) because of low activity of disulfide isomerase DsbC (15, 20, 24, see below) and/or complex pattern of disulfide bonds in the target protein (1, 5, 14, 26, 30).
During folding of proteins with multiple cysteine residues in the periplasm, aberrant disulfide bonds may be formed by strongly oxidative protein DsbA (1, 30) and remain uncorrected due to insufficient DsbC activity (15, 20). Thus, proteins with complex patterns of disulfide bonds (14, 26) may form aberrant disulfide bonds and require DsbC-dependent isomerization for correct folding. Misfolded proteins are likely to form insoluble aggregates or products that tend to be degraded in the periplasm (18, 20, 32, 33). The problem of misfolding becomes particularly important for proteins whose tertiary structure formation depends on high disulfide bond-isomerizing activities.
Sone et al. (30) showed that DsbC is indispensable for the formation of correct disulfide bonds in vivo: a certain mutant alkaline phosphatase forms aberrant disulfide bonds in the dsbC null mutant that can be corrected upon overexpression of active DsbC. Thus, overexpression of DsbC is likely to enhance disulfide bond-isomerizing activity, leading to improved expression of correctly folded proteins (25). Indeed, the mutant alkaline phosphatase that failed to be proofread in the DsbC mutant was unstable in vivo but was markedly stabilized upon overexpression of DsbC (30). Certain multiple-disulfide-bonded proteins were also shown to be efficiently expressed in the cytoplasm by overexpressing signal sequenceless DsbC under certain conditions (6). We recently found that overexpressing Dsb proteins can strikingly enhance periplasmic production of human nerve growth factor (NGF) which otherwise aggregates extensively (Y. Kurokawa, H. Yanagi, and T. Yura, submitted for publication). Here, the same Dsb coexpression system was used to analyze another well-characterized target protein, horseradish peroxidase (HRP) isozyme C to further substantiate usefulness of Dsb expression plasmids for periplasmic production of multiple-disulfide-bonded proteins.
HRP is a typical peroxidase, a class of heme proteins acting on hydrogen peroxide and a variety of substrates and plays diverse biological roles (29). It is therefore widely used for experimental, clinical, industrial, or other purposes. HRP has four disulfide bonds formed between nonconsecutive cysteine residues (9, 10) and contains 2 mol of calcium and 1 mol of hemin per mol of protein (29). It has been difficult to obtain active and soluble HRP by using conventional E. coli recombinant expression systems (12, 29). Periplasmic expression of HRP resulted in low yields of active protein (16). We now report that HRP is unstable when expressed in the E. coli periplasm but stabilized appreciably upon overexpression of Dsb proteins. The results revealed significant effects of a set of Dsb proteins in both stabilizing and solubilizing HRP produced and transported to the periplasm in E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strain JM109 was used as the host throughout the experiments. Construction of pAR3-based Dsb coexpression plasmids will be described elsewhere (Kurokawa et al., submitted). Expression of DsbA or DsbC protein was detected by immunoblotting or Coomassie brilliant blue (CBB) staining, respectively. On the other hand, expression of DsbB or DsbD was confirmed by their abilities to complement the defective phenotypes of the respective deletion mutants (Kurokawa et al., submitted). Construction of pTrc99A-derived HRP expression plasmids that produce OmpA-HRP, OmpT-HRP, or MalE-HRP will be described elsewhere (Y. Kurokawa, K. Nishihara, M. Kanemori, H. Yanagi, and T. Yura, unpublished results).
Culture conditions and protein expression.
E. coli JM109 cells carrying an HRP expression plasmid (such as pTrc-OmpA-HRP) and a Dsb expression plasmid were grown to mid-log phase in L broth supplemented wit ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml) at 37°C, and Dsb proteins and HRP were induced by l-arabinose (200 μg/ml) and isopropyl-β-d-thiogalactoside (IPTG) (50 μM), respectively (Kurokawa et al., submitted).
Fractionation and analysis of proteins.
Samples of cells (usually 200 μl) were harvested, fractionated, and analyzed as reported previously (Kurokawa et al., submitted). Briefly, cells were treated with lysozyme in the presence of sucrose and then centrifuged. The supernatant and precipitate were taken as the periplasmic and spheroplast fractions, respectively. Cytoplasmic, membrane, and insoluble protein fractions were obtained by disrupting spheroplasts, followed by fractionation. Whole-cell proteins were prepared separately by directly treating a portion (200 μl) of culture with trichloroacetic acid. Each of the fractions was obtained in sodium dodecyl sulfate (SDS) sample buffer, heated (100°C, 5 min) right away or after trichloroacetic acid precipitation and washing with acetone. Proteins from cells with equal optical densities were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (12.5% polyacrylamide gel) followed by visualization with CBB or by immunoblotting. Antibodies used were prepared against HRP (Nordic Immunological Laboratories), alkaline phosphatase (PhoA; Nordic Immunological Laboratories) or β-lactamase (Bla; 5 Prime-3 Prime, Inc.). Proteins were detected using HRP-conjugated anti-rabbit or anti-mouse antibody and enhanced chemiluminescence kit (Amersham, Inc.), followed by quantification on an Intelligent Quantifier apparatus (BioImage Systems Co.). Native HRP (Toyobo, Osaka, Japan) was used as the standard.
Analysis of protein stability.
The metabolic stability of HRP was determined by the method described previously (23). HRP expression was induced for 1 h, protein synthesis was stopped by adding spectinomycin (final concentration, 500 μg/ml; Sigma-Aldrich), and samples taken at intervals were treated with trichloroacetic acid and analyzed by SDS-PAGE (12.5% polyacrylamide gel) followed by immunoblotting and quantification.
DNA manipulations, media, and buffers.
DNA manipulations and medium and buffer preparation were done essentially as described by Sambrook et al. (28). All chemicals were of analytical grade and supplied by Wako Pure chemical (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
RESULTS
Overexpression of Dsb proteins alleviates growth inhibition caused by HRP production.
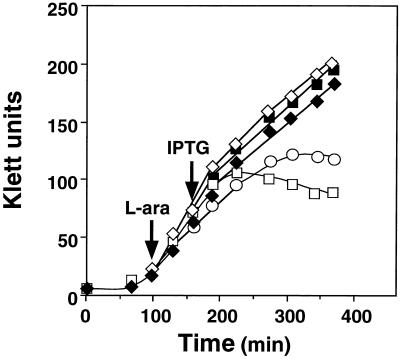
When expression of HRP directed by the trc promoter and several E. coli signal sequences was examined using multicopy expression plasmids, the OmpA signal gave the highest production of HRP compared with other signals (OmpT or MalE) (data not shown). Upon induction of HRP by IPTG, however, growth of cells carrying pTrc-OmpA-HRP was severely inhibited, suggesting that HRP exhibited toxic effects. In contrast, when a full set of Dsb proteins (DsbABCD) was simultaneously overproduced from a pAR3-based plasmid by l-arabinose prior to HRP induction, growth inhibition was mostly reversed (Fig. 1). Similar effects on growth were observed upon overexpression of a pair of Dsb proteins, DsbAC or DsbCD but not DsbAB, suggesting that at least DsbC is required for alleviating growth inhibition (Fig. 1). These results suggested that HRP production inhibits cell growth primarily by overloading DsbC.
FIG. 1.
Effects of HRP production and overexpression of Dsb proteins on cell growth. Derivatives of E. coli strain JM109 harboring both pTrc-OmpA-HRP and pAR3 (vector alone) (○) or pAR3-based plasmids that can express DsbAB (□), DsbAC1 (▪), DsbCD (⋄), or DsbABCD (♦) were grown in L broth to mid-log phase as described in Materials and Methods. Dsb proteins were induced by l-arabinose (L-ara), and after incubation for 1 h, OmpA-HRP was induced by IPTG. Turbidity (Klett units with no. 66 filter) was determined at appropriate intervals and plotted against time.
Dsb coexpression facilitates translocation of HRP into the periplasm.
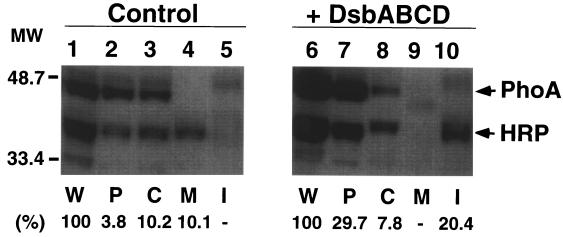
In the absence of Dsb overexpression, periplasmic expression of HRP was low (<5% of whole-cell protein fraction), and appreciable amounts of HRP were found associated with membrane or cytoplasmic fractions (Fig. 2, lanes 3 and 4). When DsbABCD was induced for 1 h before induction of HRP, the amount of HRP produced significantly increased (two- to threefold; lanes 1 and 6). In particular, the fraction of HRP found in the periplasm (P) increased more than 10-fold (about 30% of the whole-cell fraction; lanes 2 and 7), concomitant with a decrease in the membrane fraction and an increase in insoluble aggregates (lanes 4, 5, 9, and 10). A typical periplasmic protein alkaline phosphatase (PhoA) also increased by about twofold (lanes 1 and 6) in the presence of excess Dsb proteins, in part due to stabilization (data not shown); another periplasmic protein β-lactamase (Bla) was not affected (see below). The selective increase in periplasmic PhoA with Dsb coexpression suggests that excess Dsb proteins facilitate transport of at least some secretory proteins into the periplasm. Apparently, only about 25% of HRP produced was recovered by simply adding the HRP in the periplasmic, cytoplasmic, membrane, and insoluble fractions without Dsb coexpression (lanes 1 to 5), whereas about 60% of HRP was recovered with Dsb coexpression (lanes 6 to 10). The low yield of HRP without Dsb coexpression should be contrasted to the >70% recovery of PhoA with or without Dsb coexpression; it seemed likely that proteolysis rather than simple loss during the process of fractionation was involved. These results suggested that HRP produced in the E. coli periplasm is rather unstable but is stabilized upon Dsb overexpression, presumably through facilitating folding of HRP upon translocation in to periplasm, although part of HRP remains insoluble.
FIG. 2.
Effects of DsbABCD coexpression on intracellular localization of OmpA-HRP. A pair of E. coli JM109 strains harboring pTrc-OmpA-HRP and pDbABCD1 (or pAR3 vector) were grown in L broth to mid-log phase, Dsb proteins were induced by l-arabinose (L-ara), and after 30 min, OmpA-HRP was induced by IPTG for 1 h. Cells were collected, and proteins were fractionated as described elsewhere (Kurokawa et al., unpublished). HRP and PhoA (alkaline phosphatase) were analyzed by SDS-PAGE (12.5% polyacrylamide gel) followed by immunoblotting, as described in Materials and Methods. Samples from equivalent culture volumes were applied to the lanes. E. coli JM109 harboring pTrc-OmpA-HRP and pAR3 (control) (lanes 1 to 5) and JM109 harboring pTrc-OmpA-HRP and pDbABCD1 (+ DsbABCD) (lanes 6 to 10) were used. W, P, C, M, and I represent whole-cell, periplasmic, cytoplasmic, membrane, and insoluble protein factions, respectively. The positions of molecular mass standards (in kilodaltons) (protein markers from Bio-Rad) are shown on the left. Values shown below the gels indicate the percent yield of HRP products found in each fraction compared with whole-cell protein.
Dsb overexpression markedly stabilizes HRP.
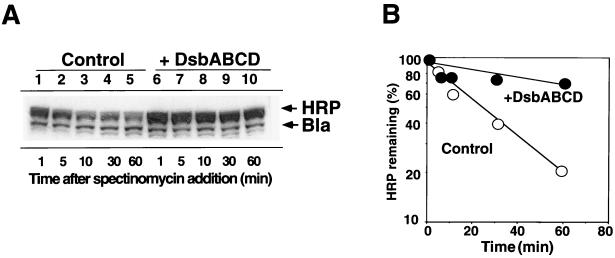
To determine whether overexpression of Dsb proteins stabilizes HRP, stability of HRP produced with or without prior overexpression of Dsb proteins was examined by measuring the amount of HRP remaining after treatment with spectinomycin which stops protein synthesis with a short lag time. Under these conditions, only about 20% of HRP remained after 1 h without Dsb coexpression (half-life of 25 min), whereas about 70% of HRP remained with DsbABCD coexpression (half-life of 120 min) (Fig. 3). In contrast, the stability of Bla and of most proteins remained virtually unchanged during the same period. These results are consistent with the observed increase in the recovery of HRP upon Dsb coexpression as revealed by the above cell fractionation analyses (Fig. 2). Similar extents of HRP stabilization were observed upon coexpression of DsbAC or DsbCD but not DsbAB (data not shown), suggesting that overexpression of DsbC is primarily responsible for stabilization.
FIG. 3.
Effect of DsbABCD coexpression on stability of HRP. A pair of E. coli JM109 strains harboring pTrc-OmpA-HRP and pDbABCD1 (or pAR3 vector) was grown, Dsb proteins and OmpA-HRP were induced as described in the legend to Fig. 2, and spectinomycin was added to stop protein synthesis. Samples were taken at the times indicated (after spectinomycin treatment), and stability of HRP was determined as described in Materials and Methods. (A) Immunoblot analysis of whole-cell proteins. Identical amounts of protein were applied to the lanes. E. coli JM109 harboring pTrc-OmpA-HRP and pAR3 (control) (lanes 1 to 5) and JM109 harboring pTrc-OmpA-HRP and pDbABCD1 (+ DsbABCD) (lanes 6 to 10) were used. (B) The amount of HRP remaining after incubation with spectinomycin as determined in panel A was quantified and plotted against incubation time.
Differential and synergistic effects of Dsb proteins on HRP production.
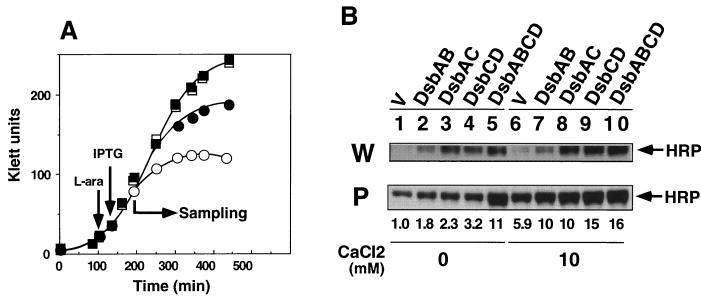
We will show elsewhere that overexpression of DsbCD is effective in enhancing translocation of human NGF into the periplasm (Kurokawa et al., submitted). When the effect of overexpressing various subsets of Dsb proteins on HRP production was examined, coexpression of DsbAB or DsbAC enhanced the yield by twofold over the vector control, whereas that of DsbCD gave threefold enhancement (Fig. 4B, lanes 1 to 4). Under the same conditions, the full set of Dsb proteins DsbABCD enhanced the yield by 11-fold (lane 5). The total amount of HRP produced also increased upon Dsb coexpression (particularly of DsbAC or DsbCD) (lanes 1 to 5). It should be noted that the band intensities for the two fractions (whole-cell and periplasmic fractions) cannot be compared directly (see the legend to Fig. 4). Although overexpression of DsbC seemed to be primarily responsible for the increased yield of HRP due to stabilization, overexpression of all Dsb proteins was apparently required to obtained maximum transport of HRP into the periplasm.
FIG. 4.
Effects of Dsb coexpression and CaCl2 added to the medium on production of HRP. A set of E. coli JM109 strains harboring both pTrc-OmpA-HRP and pAR3-based Dsb expression plasmids (or pAR3 vector) was grown, and expression of Dsb proteins and OmpA-HRP were induced as described in the legend to Fig. 2, except that L broth was supplemented with 10 mM CaCl2 as indicated. Whole-cell (W) and periplasmic (P) protein fractions were prepared and analyzed by SDS-PAGE followed by immunoblotting as described in Materials and Methods. (A) Effects of CaCl2 on growth of JM109 harboring pTrc-OmpA-HRP and pAR3 (control) (○ and ●) or JM109 harboring pTrc-OmpA-HRP and pDbABCD1 (□ and ▪). The strains were grown without CaCl2 (open symbols) or with 10 mM CaCl2 (solid symbols). Turbidity (Klett units with no. 66 filter) was measured and plotted against incubation time. (B) Effects of CaCl2 on HRP production. The whole-cell protein samples presented were diluted fourfold before SDS-PAGE, and the film was exposed for 10 s, instead of the 30-s exposure time for film of periplasmic protein samples. Values shown below the gels indicate the amounts of periplasmic HRP relative to that for the vector control (V) without calcium added (lane 1).
Effect of calcium chloride on HRP production.
Previous in vitro studies showed that active HRP is formed via at least two steps: folding of nascent polypeptide to apoenzyme, which is assisted by calcium, and its conversion to holoenzyme by binding hemin (29, 31). We therefore examined the effect of CaCl2 on folding of HRP in the present study. Addition of 10 mM CaCl2 to L broth partially alleviated the growth inhibition observed upon production of HRP in the absence of Dsb coexpression but was not as effective as that observed with Dsb overexpression (Fig. 4A). CaCl2 without Dsb overexpression (vector control) strikingly enhanced periplasmic production of HRP by five- to sixfold under the conditions used with little effect on total HRP production (Fig. 4B, lanes 1 and 6). Interestingly, it was further enhanced by about twofold upon coexpression on DsbAB or DsbAC and threefold by coexpression of DsbCD, which agreed quantitatively with their effects in the absence of added calcium. Thus, the effects of added calcium and Dsb coexpression appeared to be distinct from each other. On the other hand, coexpression of DsbABCD under the high-calcium condition gave results identical with that of DsbCD, suggesting that excess DsbAB had little additional effect over that of excess DsbCD. The different effects of coexpression of various subsets of Dsb proteins on the total yield of HRP were hardly affected by the added calcium. Calcium chloride (10 mM) was also shown to stabilize HRP significantly under these conditions (data not shown). Other cations such as magnesium had little effect on periplasmic expression of HRP.
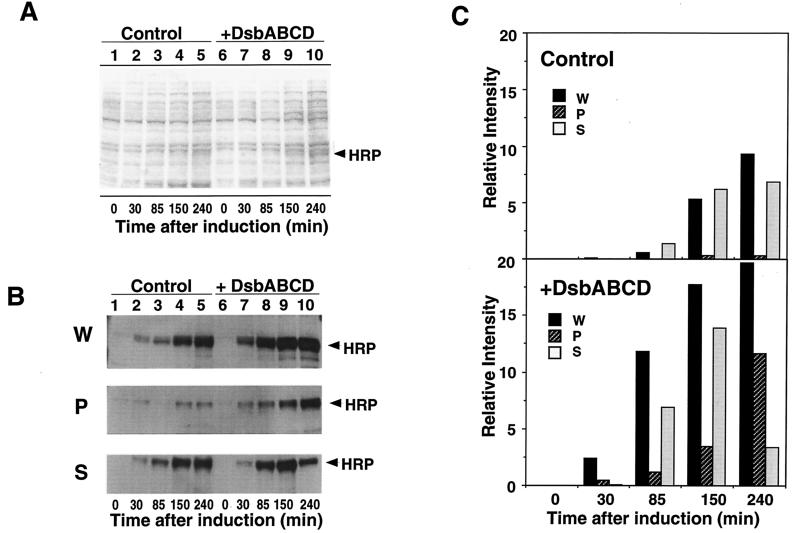
Massive periplasmic production of HRP upon prolonged Dsb coexpression.
To further define the conditions of periplasmic production of HRP in E. coli, we monitored changes in intracellular localization of HRP for 4 h after induction. The total amount of HRP began to increase within 30 min and increased steadily for 3 to 4 h even without Dsb overexpression (Fig. 5B). HRP production in general, and particularly in the periplasm, was markedly accelerated by Dsb overexpression and could be detected even by staining SDS-polyacrylamide gels with CBB by 85 min (Fig. 5A, lane 8). The amount of HRP in the periplasm increased steadily, attaining as much as 60 to 70% of HRP detected in the whole-cell protein fraction after 240 min, concomitant with the decrease in spheroplasts (Fig. 5B and C). In sharp contrast, growth of control cells (without Dsb coexpression) ceased by this time, and the periplasmic expression of HRP was quite low (less than 5%) (Fig. 5B and C).
FIG. 5.
Time course of periplasmic production of HRP. A pair of E. coli JM109 strains harboring pTrc-OmpA-HRP and pDbABCD1 (or pAR3 vector) was grown, and Dsb proteins and OmpA-HRP were induced as described in the legend to Fig. 2. Samples (100 ml) were taken at the times indicated (after HRP induction), fractionated into whole-cell (W), periplasmic (P), and spheroplast (S) fractions, and analyzed as described in Materials and Methods. (A) Analysis of whole-cell proteins by SDS-PAGE followed by CBB staining. (B) Analysis of whole-cell, periplasmic, and spheroplast proteins by immunoblotting. E. coli JM109 harboring pTrc-OmpA-HRP and pAR3 (control) (lanes 1 to 5) and JM109 harboring pTrc-OmpA-HRP and pDbABCD1 (+DsbABCD) (lanes 6 to 10) were used. (C) The relative intensities of HRP bands (B) were quantified and plotted against time after HRP induction.
DISCUSSION
We previously constructed E. coli Dsb expression plasmids that can be used to enhance disulfide bond formation and isomerization capacity and demonstrated its usefulness for improving periplasmic production of soluble NGF which otherwise tends to form aggregates (Kurokawa et al., submitted). We used these expression plasmids to examine production of HRP, which was found to be unstable upon periplasmic expression, and compared the effects of different sets of Dsb proteins. Our results revealed that overexpression of Dsb proteins can markedly increase periplasmic production of HRP, presumably through facilitating protein folding which leads to enhanced translocation into the periplasm as well as to stabilization.
Expression of OmpA-HRP fusion protein severely inhibited cell growth (Fig. 1), most probably due to reduced protein translocation caused by HRP product mainly associated with membranes (Fig. 2). Growth inhibition was largely alleviated either by addition of calcium to the medium (Fig. 4A) or by overexpression of Dsb proteins (Fig. 1), which concomitantly decreased membrane-associated HRP and improved periplasmic production (Fig. 2). Of the subset of Dsb proteins tested, DsbAC or DsbCD (or DsbABCD but not DsbAB) was most effective in preventing growth inhibition (Fig. 1), suggesting that overexpression of DsbC may be primarily responsible for the improved translocation of HRP precursor, presumably through facilitating correct folding in the periplasm and “pulling” the product into the periplasmic compartment. However, Dsb overexpression cannot be a general method of relieving translocation defects, because it was found to exacerbate growth inhibition caused by OmpA-NGF expression, presumably by reducing translocation efficiency of this particular fusion protein (Kurokawa et al., submitted).
Calcium is a normal constituent of HRP and may assist in folding and translocation of HRP, although not as efficiently perhaps because of the limited availability of active Dsb proteins. In the presence of added calcium, overexpression of various sets of Dsb proteins further increased the amount of periplasmic HRP to extents very similar to those observed in the absence of added calcium (Fig. 4B). Under high-calcium conditions, however, overexpression of DsbCD was as effective as that of DsbABCD (Fig. 4B). These results indicate that excess DsbAB is no longer required for maximum effect and that calcium enhances folding of HRP in vivo and in vitro. Whatever the exact mechanisms involved, the growth inhibition observed upon induction of OmpA-HRP appears to be alleviated by facilitating correct folding of HRP and enhancing its translocation into periplasm both by excess Dsb proteins and by the addition of calcium.
Misfolded proteins or proteins with incorrect disulfide bonds may be susceptible to proteolysis and may be degraded by quality control systems of the cells (25). Periplasmic proteolysis may be enhanced by accumulation of misfolded proteins in the periplasm through induction of proteases such as degP (htrA) gene product (7, 20). Overproduction of DsbC most probably facilitates formation of correct disulfide bonds in periplasmic proteins and should provide an effective means of coping with protein misfolding. For example, a mutant alkaline phosphatase that contains aberrant disulfide bonds is highly susceptible to proteolysis in vivo but can be converted to the correctly folded and stable form upon overexpression of DsbC (30). Reduced DsbC was suggested to serve as an effective catalyst of disulfide bond isomerization that requires reduction and rearrangement of disulfide bonds (13). E. coli mutants deficient in genes involved in the maintenance of thiol-disulfide redox potential allow efficient formation of disulfide bonds even in the cytoplasm. In these mutants, oxidative folding of proteins with complex disulfide bonds can be promoted by overexpressing DsbC which lacks signal sequence (6). The active site of DsbC thus expressed in the cytoplasm was reduced, suggesting strongly that DsbC exerted disulfide isomerase activity for correct folding in the cytoplasm (6).
Overexpression of DsbC or eukaryotic protein disulfide isomerase can increase the total yield of heterologous proteins such as bovine pancreatic trypsin inhibitor or insulin-like growth hormone I produced in E. coli (13, 24). These results taken together suggest that overexpression of disulfide isomerase often stabilizes target protein through proofreading disulfide bonds formed. The present results suggest that this mechanism does indeed operate in the case of HRP. The stability of HRP produced was normally low upon periplasmic expression but was markedly enhanced in the presence of excess DsbAC, DsbCD (or DsbABCD) but not DsbAB, suggesting that excess DsbC is primarily responsible for stabilization (Fig. 3). In the present study, maximum yield of periplasmic HRP was obtained by overexpressing the whole set of Dsb proteins (DsbABCD) (Fig. 4A), suggesting that excess disulfide isomerase alone was not sufficient to cope with misfolding of HRP. Thus, excess DsbAB and DsbCD act synergistically in promoting proper folding of HRP. Overexpression of rat protein disulfide isomerase failed to increase the yield of active HRP when expressed as PelB-HRP precursor protein (16) possibly due to insufficient isomerase activity in the oxidative environment of E. coli periplasm (24).
We found that the periplasmic production of HRP steadily increases upon prolonged incubation in the presence of excess Dsb proteins (240 min [Fig. 5]). Although the mechanism of delayed periplasmic accumulation of HRP is unclear at present, the formation of the mature form of HRP in the periplasm may be a slow and rate-limiting process (Fig. 5B and C). In any event, the present findings indicate that excess amounts of DsbC primarily stabilize HRP produced in the periplasm by promoting correct folding through enhanced isomerization of aberrant disulfide bond pairs formed during folding. Of particular importance would be the mechanism by which the oxidative folding of HRP in vivo is catalyzed by DsbC and other Dsb proteins. Special interest is also attached to the mechanism of promoting translocation by excess Dsb proteins, which should differ from that of similar functions of cytoplasmic chaperones. Further analysis of these mechanisms should provide useful information for further understanding and improving periplasmic production of HRP and other multiple-disulfide-bonded proteins.
ACKNOWLEDGMENTS
We thank Koreaki Ito, Yoshinori Akiyama, and Masaaki Kanemori for helpful advice and discussions. We are also grateful to Masako Nakayama, Hideaki Kanazawa, and Seiji Takahara for technical assistance.
REFERENCES
- 1.Alksne L E, Rasmussen B A. Dsb-insensitive expression of CcrA, a metallo-β-lactamase from Bacteroides fragilis, in Escherichia coli after amino acid substitution at two cysteine residues within CcrA. J Bacteriol. 1996;178:4306–4309. doi: 10.1128/jb.178.14.4306-4309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baneyx F. Recombinant protein expression in Escherichia coli. Curr Opin Biotechnol. 1999;10:411–421. doi: 10.1016/s0958-1669(99)00003-8. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell J C. Building bridges: disulphide bond formation in the cell. Mol Microbiol. 1994;14:199–205. doi: 10.1111/j.1365-2958.1994.tb01281.x. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell J C, Lee J O, Jander G, Martin N, Belin D, Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck R, Burtscher H. Expression of human placental alkaline phosphatase in Escherichia coli. Protein Expr Purif. 1994;5:192–197. doi: 10.1006/prep.1994.1030. [DOI] [PubMed] [Google Scholar]
- 6.Bessette P H, Aslund F, Beckwith J, Georgiou G. Efficient folding of protein with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc Natl Acad Sci USA. 1999;96:13703–13708. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betton J M, Boscus D, Missiakas D, Raina S, Hofnung M. Probing the structural role of an alpha beta loop of maltose-binding protein by mutagenesis: heat-shock induction by loop variants of the maltose-binding protein that form periplasmic inclusion bodies. J Mol Biol. 1996;26:140–150. doi: 10.1006/jmbi.1996.0504. [DOI] [PubMed] [Google Scholar]
- 8.Debarbieux L, Beckwith J. Electron avenue: pathways of disulfide bond formation and isomerization. Cell. 1999;99:117–119. doi: 10.1016/s0092-8674(00)81642-6. [DOI] [PubMed] [Google Scholar]
- 9.Fujiyama K, Takemura H, Shibayama S, Kobayashi K, Choi J K, Shinmyo A, Takano M, Yamada Y, Okada H. Structure of the horseradish peroxidase isozyme C genes. Eur J Biochem. 1988;173:681–687. doi: 10.1111/j.1432-1033.1988.tb14052.x. [DOI] [PubMed] [Google Scholar]
- 10.Fujiyama K, Takemura H, Shinmyo A, Okada H, Takano M. Genomic DNA structure of two new horseradish-peroxidase-encoding genes. Gene. 1990;89:163–169. doi: 10.1016/0378-1119(90)90002-9. [DOI] [PubMed] [Google Scholar]
- 11.Georgiou G, Valax P. Expression of correctly folded proteins in Escherichia coli. Curr Opin Biotechnol. 1996;7:190–197. doi: 10.1016/s0958-1669(96)80012-7. [DOI] [PubMed] [Google Scholar]
- 12.Hartmann C, Ortiz de Montellano P R. Baculovirus expression and characterization of catalytically active horseradish peroxidase. Arch Biochem Biophys. 1992;297:61–72. doi: 10.1016/0003-9861(92)90641-9. [DOI] [PubMed] [Google Scholar]
- 13.Joly J C, Leung W S, Swartz J R. Overexpression of Escherichia coli oxidoreductases increases recombinant insulin-like growth factor-I accumulation. Proc Natl Acad Sci USA. 1998;95:2773–2777. doi: 10.1073/pnas.95.6.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joly J C, Swartz J R. In vitro and in vivo redox states of the Escherichia coli periplasmic oxidoreductase DsbA ad DsbC. Biochemistry. 1997;36:10067–10072. doi: 10.1021/bi9707739. [DOI] [PubMed] [Google Scholar]
- 15.Joly J C, Swartz J R. Protein folding activities of Escherichia coli protein disulfide isomerase. Biochemistry. 1994;33:4231–4236. doi: 10.1021/bi00180a017. [DOI] [PubMed] [Google Scholar]
- 16.Lin Z, Thorsen T, Arnold F H. Functional expression of horseradish peroxidase in E. coli by directed evolution. Biotechnol Prog. 1999;15:467–471. doi: 10.1021/bp990037r. [DOI] [PubMed] [Google Scholar]
- 17.Makrides S C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Missiakas D, Betton J M, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–884. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 19.Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missiakas D, Schwager F, Raina S. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. EMBO J. 1995;14:3415–3424. doi: 10.1002/j.1460-2075.1995.tb07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neupert W, Hartl F U, Craig E A, Pfanner N. How do polypeptides cross the mitochondrial membranes? Cell. 1990;63:447–450. doi: 10.1016/0092-8674(90)90437-j. [DOI] [PubMed] [Google Scholar]
- 23.Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl Environ Microbiol. 1998;64:1694–1699. doi: 10.1128/aem.64.5.1694-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ostermeier M, De Sutter K, Georgiou G. Eukaryotic protein disulfide isomerase complements Escherichia coli dsbA mutants and increases in the yield of a heterologous secreted protein with disulfide bonds. J Biol Chem. 1996;271:10616–10622. doi: 10.1074/jbc.271.18.10616. [DOI] [PubMed] [Google Scholar]
- 25.Qiu J, Swartz J R, Georgiou G. Expression of active human tissue-type plasminogen activator in Escherichia coli. Appl Environ Microbiol. 1998;64:4891–4896. doi: 10.1128/aem.64.12.4891-4896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rietsch A, Bessette P, Georgiou G, Beckwith J. Reduction of the periplasmic disulfide bond isomerase, DsbC, occurs by passage of electrons from cytoplasmic thioredoxin. J Bacteriol. 1997;179:6602–6608. doi: 10.1128/jb.179.21.6602-6608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Smith A T, Santama N, Dacey S, Edwards M, Bray R C, Thorneley R N, Burke J F. Expression of a synthetic gene for horseradish peroxidase C in Escherichia coli and folding and activation of the recombinant enzyme with Ca2+ and heme. J Biol Chem. 1990;265:13335–13343. [PubMed] [Google Scholar]
- 30.Sone M, Akiyama Y, Ito K. Differential in vivo roles played by DsbA and DsbC in the formation of protein disulfide bonds. J Biol Chem. 1997;272:10349–10352. doi: 10.1074/jbc.272.16.10349. [DOI] [PubMed] [Google Scholar]
- 31.Tsaprailis G, Chan D W, English A M. Conformational states in denaturants of cytochrome c and horseradish peroxidases examined by fluorescence and circular dichroism. Biochemistry. 1998;37:2004–2016. doi: 10.1021/bi971032a. [DOI] [PubMed] [Google Scholar]
- 32.Walker K W, Gilbert H F. Effect of redox environment on the in vitro and in vivo folding of RTEM-1 beta-lactamase and Escherichia coli alkaline phosphatase. J Biol Chem. 1994;269:28487–28493. [PubMed] [Google Scholar]
- 33.Wulfing C, Pluckthun A. Protein folding in the periplasm of Escherichia coli. Mol Microbiol. 1994;12:685–692. doi: 10.1111/j.1365-2958.1994.tb01056.x. [DOI] [PubMed] [Google Scholar]