Abstract
Chaetosphaeriaceae is a genera-rich and highly diverse group of fungi with a worldwide distribution in terrestrial and aquatic habitats. Eight fresh collections of Chaetosphaeriaceae were obtained during investigations of hyaline-spored hyphomycetes in China and Thailand. Based on morphological characteristics and phylogenetic analysis of a combined LSU and ITS sequence dataset, Chaetosphaeria obovoidea, Codinaea aseptata, Codinaeella hyalina, Dictyochaeta guizhouensis and Paragaeumannomyces guttulatus were introduced as new species, Codinaea terminalis was reported as new host record, and Codinaea dwaya and Phialosporostilbe scutiformis were introduced as new collections. Phylogenetic analysis in this study revealed that Chaetosphaeria was polyphyletic. Detailed descriptions and illustrations of new taxa and identified species are provided, as well as an updated phylogenetic tree to confirm the placements of these eight new collections.
Keywords: 8 taxa, asexual morph, Chaetosphaeriales, hyaline spore, taxonomy
1. Introduction
Reblova et al. [1] validly established the Chaetosphaeriaceae to accommodate type species Chaetosphaeria Tul. and C. Tul. and their relatives, including Ascocodinaea, Melanochaeta, Melanopsammella, Porosphaerella, Porosphaerellopsis and Striatosphaeria. Subsequently, more new taxa were added to this family [2,3,4,5,6,7,8,9,10]. Reviews of Chaetosphaeriaceae were provided by Lin et al. [9] and Hyde et al. [11]. Forty-three genera were accepted in Chaetosphaeriaceae by Hyde et al. [11]. In recent studies, Zheng et al. [12] included a new genus Phialolunulospora in the family. Réblová et al. [13,14] introduced six new genera namely Arcuatospora, Achrochaeta, Ericiosphaeria, Flectospora, Phialoturbella and Tubulicolla. The lineages of “Catenularia”-like taxa were recognized as a genus Fuscocatenula by Réblová et al. [15]. Five lineages, Codinaeella, Nimesporella, Stilbochaeta, Tainosphaeriella and Xyladelphia, were proposed for “Codinaea”-like fungi by Réblová et al. [16]. Meanwhile, the key to asexual genera of Chaetosphaeriaceae was provided by Maharachchikumbura et al. [17].
Chaetosphaeria (Chaetosphaeriaceae, Chaetosphaeriales), was established by Tulasne and Tulasne [18] with the type species C. innumera Berk. and Broome ex Tul. and C. Tul. It has simple and homogeneous sexual morphs, and complex and diverse asexual morphs [3,4,5,9,10,11,19,20]. Its members have been shown to link with several asexual genera in Chaetosphaeriaceae, including Chloridium, Codinaea, Dictyochaeta and Menispora [9,21,22]. Following the “One Fungus One Name” (1F1N) principle, some Chaetosphaeria species were synonymized under respective anamorphic genera, the latter being older and prioritized, i.e., Cacumisporium, Catenularia, Chloridium, Codinaea, Dictyochaeta, Exserohilum, Gonytrichum and Menispor [4,13,14,15,23,24].
Codinaea was established by Maire [25] and typified by C. aristata Maire. Gamundi et al. (1977) synonymized Codinaea under Dictyochaeta Speg. based on a misidentification of D. fuegiana Speg., the type species of Dictyochaeta. Based on the morphological reexamination of D. fuegiana, Réblová [21] argued that Codinaea and Dictyochaeta were separate genera in Chaetosphaeriaceae. She proposed that Codinaea species have conidia with setulae, while members of Dictyochaeta do not. This proposal was accepted in later studies [11,14,26,27,28,29].
Codinaeella is established by Réblová et al. [16] for “Codinaea”-like fungi based on molecular data and morphological characters of setae and conidiophores, which is typified by Cod. minuta (Tubaki) Réblová and Hern.-Restr. Codinaeella species have a saprobic life mode on various plants and soil [16,30,31]. Réblová et al. [16] accepted eight species in Codinaeella based on molecular data and morphological evidence, and provided a synopsis of accepted species in Codinaeella [16,30,32].
Dictyochaeta was introduced by Spegazzini [32] and typified by D. fuegian. The genus has been considered polyphyletic and phylogenetically unresolved due to its long-term broad delimitation [3,8,9,29,33]. Later, Réblová et al. [14] indicated that Dictyochaeta sensu stricto including D. callimorpha, D. detriticola, D. fuegiana (type species), D. montana, D. querna and D. stratosa formed a monophyletic clade in Chaetosphaeriaceae based on combined ITS and LSU sequence analysis. Furthermore, Réblová et al. [14] reevaluated a generic concept of Dictyochaeta based on multi-loci analyses coupled with morphological comparison and culture characteristics. The keys to the accepted Dictyochaeta species were provided by Kuthubutheen and Nawawi [33] and Réblová et al. [14].
Paragaeumannomyces was typified by P. sphaerocellularis Matsush [34]. Paragaeumannomyces in Chaetosphaeriaceae was confirmed as a monophyletic clade based on the phylogenetic analyses of combined ITS and LSU sequence data [29]. The key to the 16 accepted Paragaeumannomyces species was provided by Réblová et al. [29]. Furthermore, Réblová et al. [13] introduced the new combination Paragaeumannomyces hispidus (Réblová and Seifert) Réblová and Hern.-Restr. (basionym: Chaetosphaeria hispida Réblová and Seifert) based on characteristics of a three-layered ascomatal wall. It is difficult to distinguish Paragaeumannomyces from the similar genus Ericiosphaeria morphologically, as they share semblable morphology in asci, ascospores and setae. However, the two genera can be distinguished based on a number of layers and texture of the ascomatal wall as well as molecular data [6,13,29].
The hyphomycetous genus Phialosporostilbe was verified by Sierra and Portales [35] to accommodate P. turbinate Mercado and J. Mena. The placement of Phialosporostilbe in Chaetosphaeriaceae was confirmed by Yang et al. [36], based on morphological and phylogenetic analyses of the combined ITS and LSU sequence data. The genus is characterized by synnematous conidiophores, monophialidic conidiogenous cells and hyaline, turbinate or cordiform conidia with subapical or apical appendages [36,37,38]. Phialosporostilbe can be distinguished from the similar genus Nawawia by its synnematous conidiophores [35,36,37]. Index Fungorum (May 2022) listed seven epithets under Phialosporostilbe, of which only one species has molecular data available.
In this study, eight chaetosphaeriaceous hyphomycetes were obtained from freshwater and terrestrial habitats in China and Thailand, and eventually identified five novel species (Chaetosphaeria obovoidea, Codinaea aseptata, Codinaeella hyalina, Dictyochaeta guizhouensis and Paragaeumannomyces guttulatus), a new host record Codinaea terminalis and two new collections of Codinaea dwaya and Phialosporostilbe scutiformis. These new isolated data are provided with details (Table 1). Morphological evidence and phylogenetic analysis based on a combined LSU and ITS sequence dataset confirmed the identity of these new collections, providing further evidence on the phylogenetic placement of these taxa in Chaetosphaeriaceae.
Table 1.
Taxa, isolate information and GenBank accession numbers for new collections determined for this study.
| Taxon | Specimen | Status | Country | Host | Habitat | GenBank Accessions | |
|---|---|---|---|---|---|---|---|
| ITS | LSU | ||||||
| Chaetosphaeria obovoidea | HKAS 123765 | H | China | Decaying wood | F | ON502901 | ON502894 |
| Codinaea aseptata | HKAS 123758 | H | China | Decaying wood | F | ON502897 | ON502890 |
| Codinaea dwaya | GZAAS 22-0071 | China | Decaying wood | F | ON502896 | ON502889 | |
| Codinaea terminalis | HKAS 123756 | China | Dead bamboo culms | F | ON502902 | ON502895 | |
| Codinaeella hyalina | HKAS 123757 | H | China | Decaying wood | F | ON502898 | ON502891 |
| Dictyochaeta guizhouensis | HKAS 123753 | H | China | Decaying wood | T | ON502899 | ON502892 |
| Paragaeumannomyces guttulatus | HKAS 123755 | H | China | Dead bamboo culms | F | ON502900 | ON502893 |
| Phialosporostilbe scutiformis | MFLU 22-0077 | Thailand | Decaying wood | F | ON678180 | ON678145 | |
Note: status: H denotes holotype; habitat: F denotes freshwater habitat and T denotes terrestrial habitat.
2. Materials and Methods
2.1. Collection, Isolation and Conservation
Samples of submerged decaying twigs and dead bamboo culms were collected from Chiang Rai Province, Thailand, and Guizhou Province, China. They were packed into plastic bags for transportation to the laboratory, and associated metadata (i.e., date, locality and host) were noted. Fungal colonies on the host surface were examined and observed using a stereomicroscope (Leica EZ4 Microsystems (Schweiz) AG, Singapore). Photomicrographs of micro-morphological characteristics were documented using a Nikon DS-Ri2 digital camera fitted to a Nikon ECLIPSE Ni compound microscope (Nikon, Japan). Measurements were made using the Tarosoft (R) Image Frame Work, and these images used for figures were processed and combined with Adobe Illustrator CS6 (Adobe Systems, San Jose, CA, USA).
Single-spore isolations were made on water agar (WA) and potato dextrose agar (PDA), and germinating conidia were transferred to fresh malt extract agar (MEA) and PDA following the method of Chomnunti et al. [39]. Dried materials were deposited in the herbaria of Mae Fah Luang University (Herb. MFLU), Chiang Rai, Thailand, Herbarium of Cryptogams, Kunming Institute of Botany, Academia Sinica (HKAS), Kunming, China, and Herbarium of Guizhou Academy of Agricultural Sciences (GZAAS), Guiyang, China. Cultures were deposited at Mae Fah Luang University Culture Collection (MFLUCC), Chiang Rai, Thailand, and Guizhou Culture Collection, China (GZCC). Faces of Fungi and Index Fungorum numbers were registered according to the guidelines in Jayasiri et al. [40] and Index Fungorum (2022).
2.2. DNA Extraction, PCR Amplification and Sequencing
Pure cultures were grown on MEA/PDA media at 25 °C for one month. Fresh fungal mycelia were scraped off from the surface of the cultures and transferred to 1.5 mL microcentrifuge tubes. Meanwhile, fungal genomic DNA was extracted with the Biospin Fungus Genomic DNA Extraction Kit (Biospin Fungus Genomic DNA Extraction Kit, BioFlux®, Shanghai, China) following the manufacturer’s instructions. LR0R and LR5 (Vilgalys and Hester 1990) and ITS5 and ITS4 (White et al. 1990) primers were used to amplify the large subunit of the ribosomal DNA (LSU) and the internal transcribed spacer (ITS) gene regions. The amplification reactions were performed in a 50 μL reaction volume, which contained 2 μL of DNA template, 2 μL of each forward and reverse primer (10 μM), 25 μL of 2× Taq PCR Master Mix with blue dye (Sangon Biotech, Shanghai, China) and 19 μL of distilled–deionized water. The following thermo-cycling parameters were used for the LSU and ITS region: initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 45 s, annealing at 56 °C for 50 s, elongation at 72 °C for 1 min and a final extension period for 10 min at 72 °C. The quality of the PCR products was checked on a 1% agarose gel electrophoresis stained with ethidium bromide. Purification and sequencing of PCR products were performed at Sangon Biotech (Shanghai, China) using the same primers.
2.3. Alignments and Phylogenetic Analysis
Original sequences were verified using BioEdit v. 7.1.3.0 [41], and were assembled using SeqMan v. 7.0.0 (DNASTAR, Madison, WI, USA). Consensus sequences were submitted to the NCBI GenBank (Table 1). The new sequences were subjected to BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 1 May 2022) for preliminary determination of the possible species identification range in the GenBank database. Sequences of the ITS and LSU gene were analyzed to assess relationships among species of Chaetosphaeria, Codinaea, Codinaeella, Dictyochaeta, Paragaeumannomyces, Phialosporostilbe and relevant fungi in Chaetosphaeriaceae [6,9,10,12,14,16,20,29,36,42]. Accession numbers for sequences were retrieved from GenBank database and related publication from previous studies (Table 2 and Table S1). The alignments for sequences of each locus were performed with the online multiple alignment program MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/, accessed on 3 May 2022) and then manually verified in BioEdit 7.1.3.0 [43]. The maximum likelihood (ML) and Bayesian inference (BI) analyses inferred phylogenetic relationships, based on the concatenated sequence matrix of a combined LSU and ITS dataset.
Table 2.
Taxa used in this study and their GenBank accession numbers.
| Taxon | Strain | GenBank Accessions | ||
|---|---|---|---|---|
| Status | ITS | LSU | ||
| Achrochaeta talbotii | ICMP 15161 | MT454480 | MT454495 | |
| Adautomilanezia caesalpiniae | CC-LAMIC 102/12 | T | KX821777 | KU170671 |
| Anacacumisporium appendiculatum | HMAS 245593 | T | KP347129 | KT001553 |
| Arcuatospora nouae zelandiae | CBS 109474 | T | MW984569 | MW984552 |
| Arcuatospora seorsa | CBS 147510 | T | MW984572 | MW984555 |
| Brunneodinemasporium brasiliense | CBS 112007 | T | JQ889272 | JQ889288 |
| Brunneodinemasporium jonesii | GZCC 16-0050 | T | KY026058 | KY026055 |
| Calvolachnella guaviyunis | CBS 134695 | T | KJ834524 | KJ834525 |
| Catenularia minor | PRM 900544 | T | MW987827 | MW987822 |
| Catenularia cubensis | SMH 3258 | MW987826 | N/A | |
| Chaetosphaeria acutata | CBS 101312 | AF178553 | AF178553 | |
| Chaetosphaeria biapiculata | SMH 3074 | N/A | AF466065 | |
| Chaetosphaeria caesariata | SMH 2794 | N/A | AF466060 | |
| Chaetosphaeria chalaroides | SMH 2018 | N/A | AY017372 | |
| Chaetosphaeria chlorotunicata | SMH 1565 | T | N/A | AF466064 |
| Chaetosphaeria conirostris | SMH 2183 | N/A | AF466066 | |
| Chaetosphaeria curvispora | CBS 125555 | MH863562 | MH875040 | |
| Chaetosphaeria cylindrospora | SMH 3568 | T | N/A | AY017373 |
| Chaetosphaeria decastyla | SMH 2629 | N/A | AF466068 | |
| Chaetosphaeria dilabens | CBS 712 88 | AF178557 | AF178557 | |
| Chaetosphaeria fennica | CBS 101641 | AF178562 | AF178562 | |
| Chaetosphaeria fusiformis | CBS 101429 | AF178554 | AF178554 | |
| Chaetosphaeria hebetiseta | CBS 102340 | T | AF178549 | AF178549 |
| Chaetosphaeria innumera | M R 1175 | AF178551 | AF178551 | |
| Chaetosphaeria jonesii | MFLUCC 15-1015 | T | NR_154841 | KY212761 |
| Chaetosphaeria lateriphiala | SMH 2629 | N/A | AF466070 | |
| Chaetosphaeria lentomita | MR 1265 | AF178548 | AF178548 | |
| Chaetosphaeria lignomollis | SMH 3015 | T | EU037896 | AF466073 |
| Chaetosphaeria luquillensis | SMH 2973 | N/A | AF466074 | |
| Chaetosphaeria mangrovei | MCD 069 | T | MG813821 | MG813820 |
| Chaetosphaeria metallicans | PDD 92539 | T | NR_119668 | NG_058757 |
| Chaetosphaeria minuta | SMH 3396 | N/A | AF466075 | |
| Chaetosphaeria myriocarpa | CBS 264.76 | AF178552 | AF178552 | |
| Chaetosphaeria preussii | CBS 262.76 | AF178561 | AF178561 | |
| Chaetosphaeria pygmaea | MR 1365 | AF178545 | AF178545 | |
| Chaetosphaeria rivularia | CBS 127686 | T | KR347356 | KR347357 |
| Chaetosphaeria sp. JEH 2019 | JAUCC 2961 | MN619651 | MN607223 | |
| Chaetosphaeria spinosa | SMH 2754 | N/A | AF466079 | |
| Chaetosphaeria sylvatica | SMH 2893 | N/A | AF279419 | |
| Chaetosphaeria tropicalis | SMH 1267 | N/A | AF279418 | |
| Chalarodes obpyramidata | PDD 119364 | MW987828 | MW987823 | |
| Chloridium caesium | CBS 102339 | AF178564 | AF178564 | |
| Chloridium gonytrichii | CBS 195.60 | MH857954 | MH869503 | |
| Chloridium virescens | CBS 152.53 | MH857142 | MH868678 | |
| Codinaea acaciae | OTU5 | KY965397 | N/A | |
| Codinaea amazonensis | MUCL 41171 | OL654076 | OL654133 | |
| Codinaea assamica | CBS 242.66 | MH858788 | MH870426 | |
| Codinaea assamica | CBS 139907 | T | OL654077 | OL654134 |
| Codinaea dwaya | CBS 261.77 | T | OL654078 | OL654135 |
| Codinaea ellipsoidea | MFLUCC 18-1574 | T | MK828628 | MK835828 |
| Codinaea fertilis | CBS 242.66 | OL654079 | OL654136 | |
| Codinaea gonytrichodes | CBS 593.93 | AF178556 | AF178556 | |
| Codinaea lignicola | DLUCC 0899 | T | MK828630 | MK835830 |
| Codinaea pandanicola | KUMCC 16-0153 | T | MH388338 | MH376710 |
| Codinaea paniculata | CBS 145098 | T | MT118230 | MT118201 |
| Codinaea phasma | CBS 147516 | T | OL654081 | OL654138 |
| Codinaea siamensis | CBS 194.96 | OL654082 | OL654139 | |
| Codinaea siamensis | MFLUCC 15-0614 | T | KX609955 | KX609952 |
| Codinaea terminalis | GZCC19-0525 | MW133883 | N/A | |
| Codinaea terminalis | GZCC 18-0085 | T | MN104613 | MN104624 |
| Codinaeella filamentosa | CBS 147265 | OL654083 | OL654140 | |
| Codinaeella lambertiae | CBS 143419 | T | OL654084 | OL654141 |
| Codinaeella lutea | CBS 146618 | T | OL654085 | OL654142 |
| Codinaeella mimusopis | CBS 143435 | T | MH107888 | MH107935 |
| Codinaeella minuta | CBS 298.61 | T | OL654090 | OL654147 |
| Codinaeella minuta | CBS 966.69 | AF178559 | AF178559 | |
| Codinaeella minuta | 417E | MZ078594 | N/A | |
| Codinaeella parvilobata | CBS 144536 | T | OL654100 | OL654157 |
| Codinaeella pini | CBS 138866 | T | KP004465 | KP004493 |
| Codinaeella yunnanensis | MFLUCC 17-0468 | NR_168795 | NG_068630 | |
| Conicomyces pseudotransvaalensis | HHUF 29956 | T | LC001710 | LC001708 |
| Cryptophiale udagawae | GZCC 18-0047 | MN104608 | MN104619 | |
| Dendrophoma cytisporoides | CBS 144107 | MT118234 | MT118205 | |
| Dictyochaeta callimorpha | ICMP 15130 | MT454483 | MT454498 | |
| Dictyochaeta detriticola | ICMP 14948 | T | MT454486 | MT454501 |
| Dictyochaeta fuegiana | ICMP 15153 | T | MT454487 | EF063574 |
| Dictyochaeta montana | CBS 145342 | T | MT454488 | MT454502 |
| Dictyochaeta querna | CBS 146103 | MT454490 | MT454504 | |
| Dictyochaeta sp | CBS 138684 | MT454493 | MT454507 | |
| Dictyochaeta stratosa | CBS 138739 | NR 172308 | MT454505 | |
| Dinemasporium americanum | CBS 127127 | T | JQ889274 | JQ889290 |
| Dinemasporium pseudoindicum | CBS 127402 | T | JQ889277 | JQ889293 |
| Ellisembia aurea | CBS 144403 | T | MH836375 | MH836376 |
| Ellisembia folliculata | CBS 147152 | OL654105 | OL654162 | |
| Eucalyptostroma eucalypti | CBS 142074 | T | KY173408 | KY173500 |
| Flectospora laminata | CBS 112964 | MW984576 | MW984558 | |
| Flectospora sp. | ICMP 23840 | MW984577 | MW984559 | |
| Infundibulomyces cupulatus | BCC 11929 | T | EF113976 | EF113979 |
| Infundibulomyces oblongisporus | BCC 13400 | T | EF113977 | EF113980 |
| Kionochaeta castaneae | GZCC 18-0025 | T | MN104610 | MN104621 |
| Kionochaeta microspora | GZCC 18-0036 | T | MN104607 | MN104618 |
| Leptosporella arengae | MFLUCC 15-0330 | T | MG272255 | MG272246 |
| Leptosporella bambusae | MFLUCC 12-0846 | T | KU940134 | KU863122 |
| Menispora ciliata | CBS 122131 | T | EU488736 | OL654165 |
| Menispora tortuosa | CBS 117552 | OL654110 | OL654168 | |
| Menisporopsis dushanensis | GZCC 18-0084 | T | MN104615 | MN104626 |
| Menisporopsis theobromae | MFLUCC 15 0055 | KX609957 | KX609954 | |
| Multiguttulispora dimorpha | CBS 140002 | MW984582 | MW984564 | |
| Multiguttulispora triseptata | IMI 353690 | MW984584 | MW984566 | |
| Nawawia filiformis | MFLUCC 17-2394 | MH758196 | MH758209 | |
| Neopseudolachnella acutispora | MAFF 244358 | T | AB934065 | AB934041 |
| Neopseudolachnella magnispora | MAFF 244359 | T | AB934066 | AB934042 |
| Nimesporella capillacea | IMI 358908 | T | OL654114 | OL654171 |
| Paliphora intermedia | CBS 896 97 | MH862682 | EF204501 | |
| Paragaeumannomyces abietinus | CBS 145351 | T | MT118235 | MT118206 |
| Paragaeumannomyces albidus | PDD 92537 | T | EU037890 | EU037898 |
| Paragaeumannomyces bombycinus | PDD 92538 | T | EU037892 | N/A |
| Paragaeumannomyces elegans | PDD 118740 | T | MT876580 | N/A |
| Paragaeumannomyces garethjonesii | MFLUCC 15-1012 | T | KY212751 | KY212759 |
| Paragaeumannomyces granulatus | ICMP 15133 | T | MT876575 | MT876577 |
| Paragaeumannomyces lapazianus | SMH 2182 | AY906945 | MT118207 | |
| Paragaeumannomyces longisporus | ANM 1269 | MT118239 | MT118210 | |
| Paragaeumannomyces panamensis | SMH 3596 | T | AY906948 | MT118218 |
| Paragaeumannomyces raciborskii | SMH 3119 | AY906953 | AY436402 | |
| Paragaeumannomyces rubicundus | SMH 3221 | T | MT118242 | MT118224 |
| Paragaeumannomyces sabinianus | ILLS00121384 | T | MT118243 | MT118225 |
| Paragaeumannomyces smokiensis | ILLS00121398 | T | MT118240 | MT118228 |
| Phialosporostilbe scutiformis | MFLUCC 17-0227 | T | MH758194 | MH758207 |
| Phialosporostilbe scutiformis | MFLUCC 18-1288 | MH758199 | MH758212 | |
| Phialoturbella calva | ICMP 23826 | T | MW984585 | MW984567 |
| Phialoturbella lunata | MFLUCC 18-0642 | T | NR 168796 | MK835824 |
| Polynema podocarpi | CBS 144415 | T | MH327797 | MH327833 |
| Pseudodinemasporium fabiforme | CBS 140010 | T | KR611889 | KR611906 |
| Pseudolachnea fraxini | CBS 113701 | T | JQ889287 | JQ889301 |
| Pseudolachnea hispidula | MAFF 244365 | AB934072 | AB934048 | |
| Pseudolachnella asymmetrica | MAFF 244366 | AB934073 | AB934049 | |
| Pseudolachnella scolecospora | MAFF 244379 | AB934086 | AB934062 | |
| Pyrigemmula aurantiaca | CBS 126743 | T | HM241692 | HM241692 |
| Rattania setulifer | GUFCC 15501 | GU191794 | HM171322 | |
| Sporoschisma longicatenatum | MFLUCC 16-0180 | T | KX505871 | KX358077 |
| Sporoschisma mirabile | FMR 11247 | HF677174 | HF677183 | |
| Stilbochaeta aquatica | MFLUCC 15-0983 | T | NR 158452 | MH476569 |
| Stilbochaeta brevisetula | ICMP 22549 | OL654118 | OL654175 | |
| Stilbochaeta cangshanensis | MFLUCC 17-2214 | T | MK828632 | MK835832 |
| Stilbochaeta malaysiana | IMI 312436 | T | OL654121 | OL654178 |
| Striatosphaeria castanea | CBS 145352 | T | MT118244 | MT118229 |
| Striatosphaeria codinaeophora | SMH 1524 | MT118245 | AF466088 | |
| Tainosphaeria jonesii | GZCC 16-0065 | KY026060 | KY026057 | |
| Tainosphaeria siamensis | MFLUCC 15-0607 | T | KX609956 | KX609953 |
| Tainosphaeriella aquatica | MFLUCC 17-2370 | NR_173239 | NG_079564 | |
| Tainosphaeriella thailandensis | MFLUCC 18-1282 | T | MZ161198 | MZ161196 |
| Thozetella nivea | N/A | EU825201 | EU825200 | |
| Thozetella tocklaiensis | CBS 378.58 | T | MH857817 | MH869349 |
| Xyladelphia longiseta | SMH 1725 | OL654131 | AF279416 | |
| Zanclospora iberica | CBS 130426 | T | KY853480 | KY853544 |
Note: status: T denotes ex-type strains; “N/A” indicates no data are available in GenBank.
The website tool “ALTER” (http://www.singgroup. org/ALTER/, accessed on 3 May 2022) was used to convert the aligned fasta file for RAxML analysis [44]. Subsequently, ML analysis was performed using RAxML-HPC v.8 tool via the CIPRES Science Gateway V3.3 (https://www.phylo.org/portal2/home.action, accessed on 3 May 2022) with rapid bootstrap analysis [45,46]. Eventually, 1000 non-parametric bootstrap iterations were run with the GTRGAMMA model, and the final tree was selected amongst suboptimal trees from each run by comparing likelihood scores under the GTR-Gamma substitution model.
The aligned fasta file was converted to the nexus file format for BI analysis using AliView. BI analysis was performed using MrBayes 3.2.7a via CIPRES [45]. The best-fit model of DNA evolution was estimated using MrModeltest v. 2.2 [47]. The Bayesian Markov chain Monte Carlo (BMCMC) sampling method in MrBayes v.3.2.7a determined the posterior probabilities (PPs) [48]. Four simultaneous Markov chains were run for 1 million generations, with trees sampled every 100 generations, resulting in 10,000 trees. The first 2000 trees representing the burn-in phase of the analyses were discarded, and the remaining 8000 trees were used for calculating posterior probabilities (PPs) in the majority-rule consensus tree [49].
Phylogenetic trees were printed with FigTree v. 1.4.0 [50], and the layout was established using Adobe Illustrator CS6 (Adobe Systems, San Jose, CA, USA).
3. Results
3.1. Phylogenetic Analysis
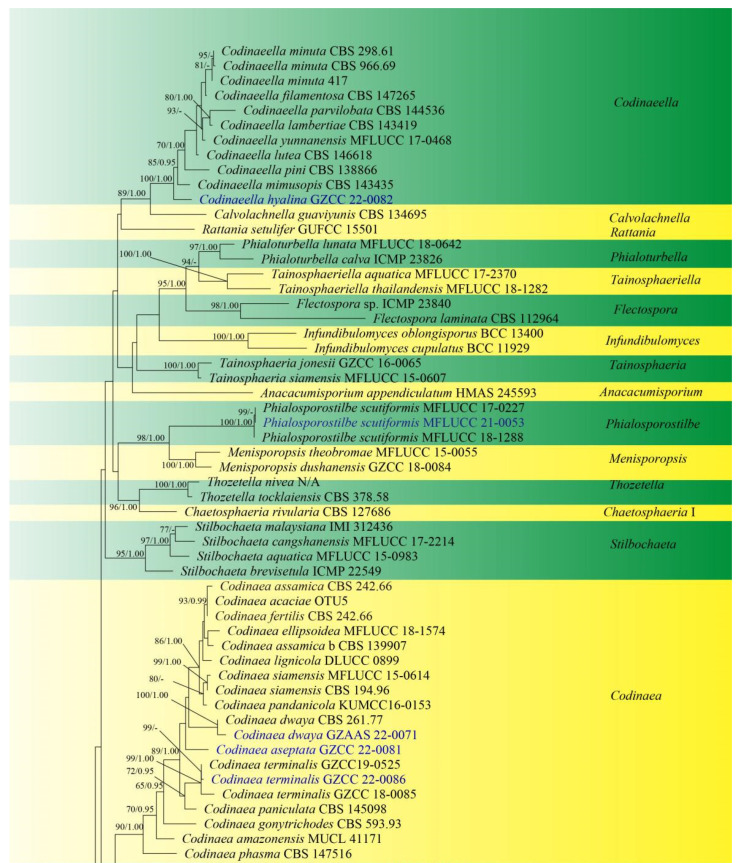
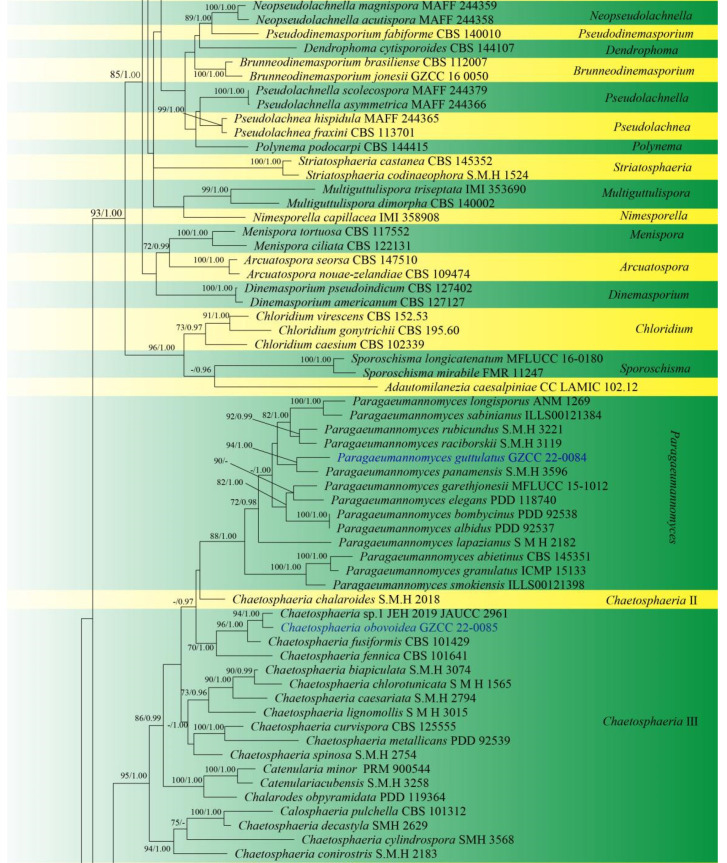
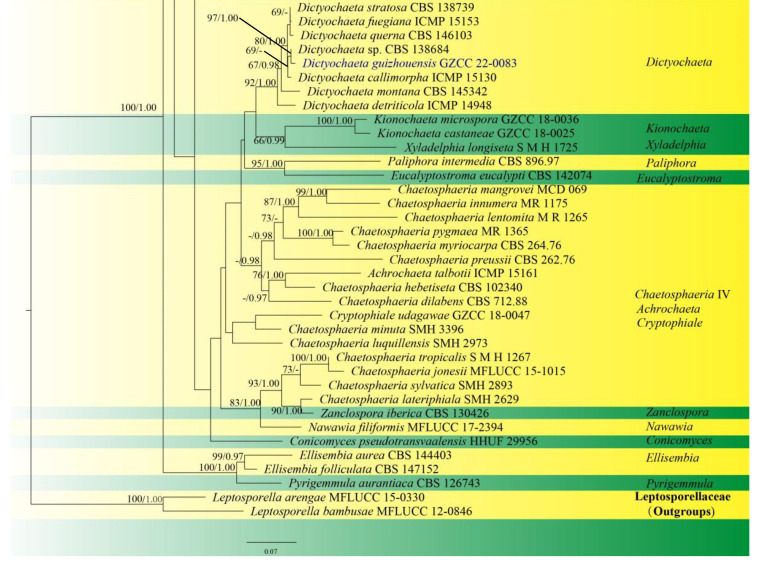
Sequences of LSU and ITS were used to determine the phylogenetic position of the new taxa and collections. The concatenated sequence matrix comprises LSU (903 bp) and ITS (545 bp) sequence data for 151 ingroup taxa of Chaetosphaeriaceae and 2 outgroup taxa, namely Leptosporella arengae (MFLUCC 15-0330) and L. bambusae (MFLUCC 12-0846). The sequence matrix comprises 1448 characters after alignment, including the gap. The matrix had 859 distinct alignment patterns, with 14.02% undetermined characters or gaps. The estimated base frequencies were: A = 0.228939, C = 0.268473, G = 0.306331 and T = 0.196257; substitution rates AC = 1.246064, AG = 1.891246, AT = 1.625669, CG = 0.550487, CT = 6.118919, GT = 1.000000 and Tree-Length = 11.191081; distribution shape parameter α = 0.304587. Bayesian posterior probabilities (PPs) from MCMC were evaluated with a final average standard deviation of split frequencies of 0.009954. Maximum likelihood and Bayesian analyses were conducted, resulting in generally congruent topologies, and the ML analysis result with a final likelihood value of −27,871.044543 is presented in Figure 1.
Figure 1.
The phylogenetic tree generated from ML analysis that is based on a concatenated LSU-ITS dataset for the Chaetosphaeriaceae family. Bootstrap support values for ML equal to or greater than 65% and Bayesian posterior probabilities (PPs) equal to or greater than 0.95 were indicated above or below the nodes as ML/PP. Leptosporella arengae (MFLUCC 15-0330) and L. bambusae (MFLUCC 12-0846) were selected as the outgroup taxa. The newly obtained sequences are indicated in blue.
This phylogenetic study confirmed that the family Chaetosphaeriaceae was a robust clade (100% ML/1.00 PP). There are eight new collections, five of which are in Chaetosphaeria, Codinaeella, Dictyochaeta, Paragaeumannomyces and Phialosporostilbe genera, respectively, while the other three belong to the genus Codinaea, under Chaetosphaeriaceae (Figure 1).
Chaetosphaeria was resolved as a polyphyletic genus in Chaetosphaeriaceae (Figure 1), consistent with many previous studies [6,9,10,13,14,15,29,51,52]. The new species, Chaetosphaeria obovoidea, and three other Chaetosphaeria species are clustered in the subclade of Chaetosphaeria III.
The phylogenetic tree shows that our three new Codinaea collections of Codinaea aseptata sp. nov., and two known species, Co. dwaya and Co. terminalis, clustered with other 16 Codinaea taxa in a monophyletic clade with highly support (90% ML/1.00 PP, Figure 1).
Eight “Codinaea”-like taxa comprised a new species of Codinaeella hyalina and eight previously identified species, representing the Codinaeella clade, which was phylogenetically well-supported by the generic placement (100% ML/1.00 PP, Figure 1). It is consistent with the studies of Réblová et al. [16].
One of the eight fresh collections was identified as a new species in Dictyochaeta species, namely D. guizhouensis. It clustered together with the other six known species and one unidentified Dictyochaeta sp. (CBS 138684) with good support (97% ML/1.00 PP, Figure 1). All species of Dictyochaeta formed into a single monophyletic clade and fit well with the narrow genetic concept of conidia without setulae [14]. Furthermore, the phylogenetic placement of the Dictyochaeta clade and the phylogenetic relationship of taxa within Dictyochaeta presented similar results to those obtained by Réblová et al. [14].
Phylogenetic placements of the Paragaeumannomyces and Phialosporostilbe genera were stable, and those genera were resolved as monophyletic lineages [29,36].
3.2. Taxonomy
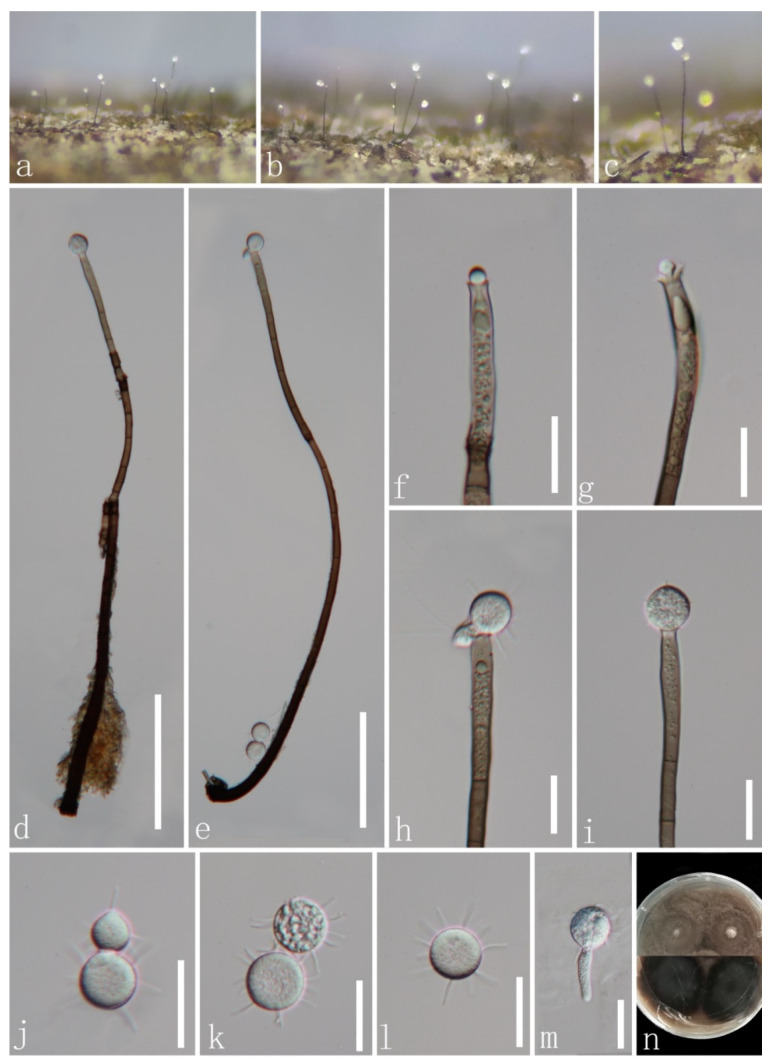
Chaetosphaeria obovoidea J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 2).
Figure 2.
Chaetosphaeria obovoidea (HKAS 123765, holotype). (a,b) Colonies on woody substrate; (c–f) conidiophores with pale gold or red appendant attached; (g–j) conidiogenous cells; (j–l) conidia; (l) germinating conidium; (m,n) culture on PDA from above and below. Scale bars: (c–f) 25 μm; (g–i) 20 μm; (j–l) 10 μm.
Index Fungorum number: IF559696; Facesoffungi number: FoF 11036.
Etymology: referring to obovoid conidia.
Holotype: HKAS 123765.
Saprobic on decaying wood submerged in freshwater habitats. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, scattered, pale brown, with white and gold masses on the apex of conidiophores. Mycelium is composed of partly immersed, partly superficial, hyaline-to-pale brown, septate, branched hyphae. Conidiophores 93–234(–291) μm long, 3.5–5.3 μm wide at the base, macronematous, mononematous, erect, straight or slightly flexuous, occasionally branched, with intercalary conidiogenous loci, cylindrical, solitary, brown at the base, pale brown or subhyaline towards the apex, septate, smooth-walled, with pale gold or red appendants attached. Conidiogenous cells 13–51 × 3–5.5 μm ( = 33.8 × 4.6 μm, n = 20), mono- to polyphialidic, terminal, integrated, cylindrical, with funnel-shaped collarettes, brown or pale brown to subhyaline or hyaline towards the apex. Conidia 10–14.8 × 5–7.2 μm ( = 12.7 × 6 μm, n = 20), amerospores, aseptate, obovoid, pyriform to broadly clavate, rough-walled, aggregated in large and slimy mass, hyaline, round at the apex, tapering at the base and often with a small prominence.
Culture characteristics: Conidia germinating on PDA within 15 h and hyaline germ tube produced from the base of conidia. Colonies growing on PDA at 25 °C reach 17 mm in three weeks, circular, unbonate, entire, with filamentous, dense, aerial mycelium on the surface, white at the center, pale grey at the edge from above; yellowish to greyish brown to pale brown in reverse from the center to the margin of the colony, and do not produce pigmentation in culture.
Material examined: CHINA, Hainan province, Diaoluo Mountain National Nature Reserve, on decaying wood submerged in a stream, 20 August 2021, W.G. Lin, DL2 (HKAS 123765, holotype; GZAAS 22-0076, isotype), ex-type living cultures, GZCC 22-0085, GenBank accession numbers: (LSU) ON502894, (ITS) ON502901.
Notes: In a BLASTn search in GenBank, the closest match to the ITS sequence of our new isolate was Chaetosphaeria sp. (strain JEH-2019) with 97% (MN619651) similarity across 88% of the query sequence. The closest match to the LSU of the new isolate was Chaetosphaeria fusiformis (strain CBS 101429) with 99.21% (AF178554) similarity across 94% of the query sequence. The phylogenetic analysis confirmed that our new isolate of C. obovoidea formed a separate clade within the Chaetosphaeria genus and is a sister clade to Chaetosphaeria sp. (strain JEH-2019) with good support (94% ML/1.00 PP). Thus, we introduced this new isolate as a novel species in the Chaetosphaeria. Additionally, our new species formed a unique asexual morph in the natural substrate, and differs from all existing species of Chaetosphaeria in having macronematous conidiophores with pale gold or red appendants attached, hyaline, aseptate and obovoid conidia with a prominence at the base. This adds complexity and diversity to the asexual morphs of the Chaetosphaeria genus.
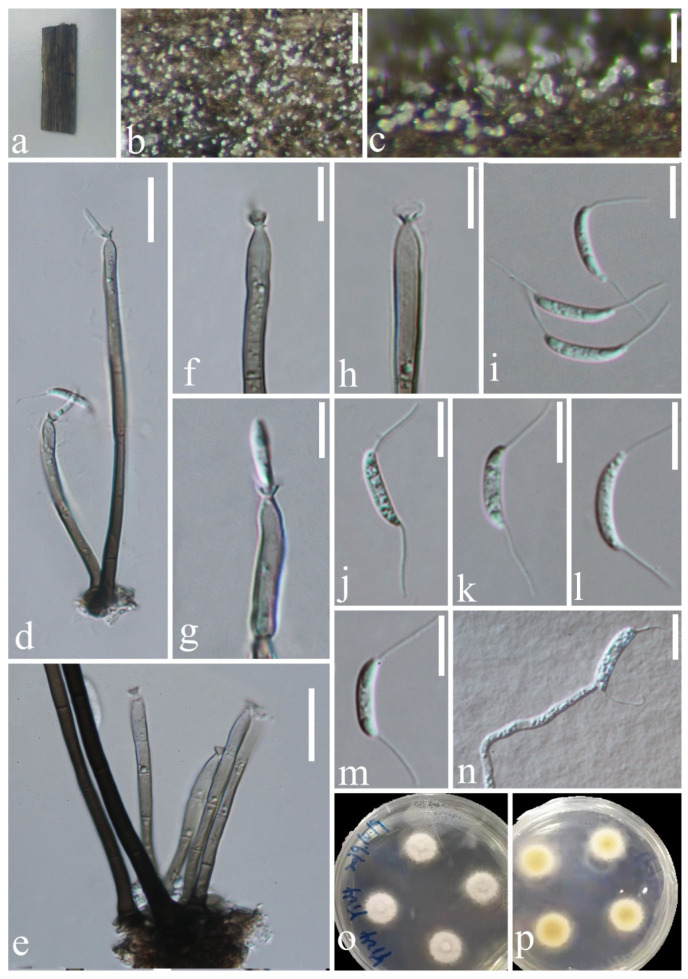
Codinaea aseptata J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 3).
Figure 3.
Codinaea aseptata (HKAS 123758, holotype). (a) Host material; (b,c) colonies on woody substrate; (d,e) setae and conidiophores; (f–h) conidiophores and conidiogenous cells; (i) apex of seta;(j–m) conidia; (n) germinated conidium; (o) culture on PDA from above and below. Scale bars: (d,e) 100 μm; (f,k–n) 10 μm; (g–j) 20 μm.
Index Fungorum number: IF559697; Facesoffungi number: FoF 11038.
Etymology: referring to the aseptate conidia.
Holotype: HKAS 123758.
Saprobic on decaying wood submerged in freshwater habitats. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on the natural substrate effuse, hairy, brown. Mycelium mostly immersed, composed of branched, septate, smooth, brown hyphae. Setae 339–417 μm long, 5–6 μm wide at the base ( = 376.8 × 5.5 μm, n = 20), erect, dark brown at the base, subhyaline towards the apex, septate, unbranched, smooth, rarely fertilizable. Conidiophores 41.6–105.1 μm long, 2–3.74 μm wide at the base ( = 66 × 3 μm, n = 15), macronematous, mononematous, in groups from the mycelial knots associated with the bases of setae, shorter than setae, erect, straight or slightly flexuous, short, cylindrical, unbranched, smooth-walled, septate, pale brown at the base, subhyaline at the apex. Conidiogenous cells 5.7–20 μm ( = 12.2 μm, n = 20) long, monophialidic, integrated, determinate, terminal, cylindrical to cylindrical–lageniform, pale brown at the base, becoming subhyaline to hyaline towards the apex, with flared collarette. Conidia 13.4–16.2 × 2.1–3 μm ( = 14.2 × 2.6 μm, n = 20), acrogenous, solitary, aggregated in slimy droplets, aseptate, cylindrical or long fusiform, curved, with hair-like and 7–10 m-long appendages at both ends, hyaline, smooth-walled, with small guttules.
Culture characteristics: Conidia germinating on PDA within 15 h and germ tubes produced from the base and the upper part. Colonies growing on PDA, reaching 19 mm in diam. in 10 days at 25 °C, circular, flat, entirely to slightly filamentous, section to fan shape at the surface, taupe, becoming white towards the edge from above; yellowish-brown mycelium in the middle and pale-yellow mycelium in the outer ring in reverse, and does not produce pigmentation in culture.
Material examined: CHINA, Hainan Province, Wuzhishan City, Wuzhishan Tropical Rainforest Scenic Area, on decaying wood submerged in a freshwater stream, 15 August 2021, Zheng Luo, WZ48 (HKAS 123758, holotype; GZAAS 22-0072 isotype); ex-type living cultures, GZCC 22-0081. GenBank accession numbers: (LSU) ON502890, (ITS) ON502897.
Notes: Four morphotypes (C1–C4) of Codinaea were provided by Réblová et al. [16]. The morphological characteristics of our new isolate match well with the generic concept of Codinaea and fit well with the morphotype C1. Codinaea aseptata is most similar to Co. terminalis due to having setae in fascicles with conidiophore, phialidic and terminal conidiogenous cells and aseptate, falcate conidia with setulae at both ends [9]. However, our new isolate differs from Co. terminalis in its obviously longer and rarely fertilizable setae. BLAST results of ITS and LSU sequence data are Codinaea acacia OTU5 (96.09% similarity) and Co. paniculate MFLU 34876 (99.43 similarity), respectively. The phylogenetic tree (Figure 1) showed that our new isolate of Cod. aseptata formed an individual lineage in the Codinaea clade, but without statistical support shown in phylogenetic tree (44% ML/-). This may be due to the consideration that many close phylogenetic relatives of our new collection have not yet been discovered. Hence, Codinaea aseptata was introduced as a new species based on its distinct morphology and phylogenetic evidence.
Codinaea dwaya (Subram. and J. Bhat) Réblová and Hern.-Restr., Journal of Fungi 7 (12, no. 1097): 31 (2021) (Figure 4).
Figure 4.
Codinaea dwaya (GZAAS 22-0071). (a–c) Colonies on woody substrate; (d,e) conidiophores and conidia; (f–i) conidiogenous cells; (j–l) conidia; (m) germinating conidium; (n) culture on PDA. Scale bars: (d,e) 100 μm; (f–m) 20 μm.
Index Fungorum number: IF 842192; Facesoffungi number: FoF 11037.
Holotype: MUBL 2351.
Saprobic on decaying wood submerged in freshwater habitats. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, scattered, brown, with glistening masses of conidia on the apex of conidiophores. Mycelium 1.3–2.4 μm wide, mostly immersed, composed of pale brown, septate hyphae. Conidiophores (360–)440–620 × 7.6–14 μm ( = 527.7 × 9.9 μm, n = 20), macronematous, mononematous, erect, straight or slightly flexuous, occasionally branched, cylindrical, solitary, septate, smooth-walled, dark brown, becoming paler brown towards the apex. Conidiogenous cells (17–)35–62.5 × 5.6–8 μm ( = 50.2 × 6.6 μm, n = 20), monophialidic, terminal, integrated, cylindrical, guttulata, with an inconspicuous apical collarette. Conidia in succession by percurrent proliferation from a single fertile locus, spherical, 13.4–17 μm diam. ( = 15.5 μm), amerospores, aseptate, acrogenous, in groups, accumulating in a slimy mass at the tip of the phialide, rough-walled, hyaline, with 7–13 hair-like and 6.4–9.9 μm-long appendages at both ends.
Culture characteristics: Conidia germinating on PDA within 16 h and germ tube produced from conidia. Colonies growing on PDA, reaching 25 mm in diam. in 10 days at 25 °C, circular. Mycelium slightly raised, taupe dark brown with a white protuberance in the center, entirely to slightly filamentous, becoming taupe to dark brown towards the edge in reverse and not producing pigmentation in culture.
Material examined: CHINA, Hainan province, Diaoluo Mountain National Nature Reserve, on decaying wood submerged in a freshwater stream, 20 August 2021, Wei-Guo Lin, DL1-1 (GZAAS 22-0071), living cultures, GZCC 22-0080, GenBank accession numbers: (LSU) ON502889, (ITS) ON502896.
Notes: Following BLASTn searches of NCBI GenBank, the closest matches of the LSU and ITS sequences of our new isolate is Codinaea dwaya (strain CBS 261.77; LSU, OL654135, 99.88% shared identity; ITS, OL654078, 97.79% shared identity). Our new collection fits well with the description of species of Co. dwaya in mononematous conidiophores, integrated, terminal, cylindrical, phialidic conidiogenous cells and hyaline, spherical, aseptate conidia with setulae [16,53,54,55]. Phylogenetically, our isolate grouped together with Co. dwaya (CBS 261.77) with high support value (100% ML/1.00 PP, Figure 1). Thus, we identified this isolate as a new collection of Co. dwaya from a freshwater habitat.
Codinaea terminalis (C.G. Lin and K.D. Hyde) Réblová and Hern.-Restr., Journal of Fungi 7(12, no. 1097): 44 (2021). (Figure 5).
Figure 5.
Codinaea terminalis (HKAS 123756). (a) Host material; (b,c) colonies on host surface; (d,e) conidiophores with conidiogenous cell; (f–h) conidiogenous cells; (i–m) conidia; (n) germinating condium; (o,p) culture on PDA from above and below. Scale bars: (b) 200 μm; (c) 100 μm; (d,e) 20 μm; (f–o) 10 μm.
Index Fungorum number: IF 56706; Facesoffungi number: FoF 06287.
Holotype: MFLU 19-0214.
Saprobic on dead bamboo culms in freshwater habitats. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, gregarious, white, shining, globose aggregated in a large mass. Mycelium mostly immersed, composed of septate, smooth, branched and pale brown hyphae. Setae 99–277 µm long, 3.1–4.7 µm wide at the base ( = 153 × 3.8 µm, n = 25), fertile, straight or slightly flexuous, dark brown at the base, paler towards the apex, septate, smooth-walled, unbranched, cylindrical. Conidiophores 34–62 × 3–4 µm ( = 48.3 × 3.4 µm, n = 20), macronematous, mononematous, erect, in groups from the mycelial knots from the bases of setae, cylindrical, slightly swollen at the base, straight or slightly flexuous, brown at the base becoming pale brown or subhyaline towards the apex, 2–4-septate, unbranched, smooth, guttules. Conidiogenous cells 14.5–36 µm long ( = 24 µm, n = 20), monophialidic, terminal, integrated, cylindrical, brown at the base, pale brown to subhyaline towards the apex, narrowing below the flared and conspicuous collarette. Conidia 13.5–16 × 2.5–3.3 µm ( = 14.5 × 2.9 µm, n = 35), aggregated in a glistening mass of conidia at the tip of conidiophores, amerospores, acrogenous, fusiform, mostly curved, guttules, rough-walled, hyaline, with hair-like and 9.6–11.5 µm-long appendages at both ends.
Culture characteristics: Conidia germinating on PDA within 15 h and germ tubes produced from conidia. Colonies growing on PDA, reaching 19 mm in diam. in 10 days at 25 °C, circular, flat with a protuberance in the center, white, entire margin with thin mycelia; pale yellow to white from center to edge in reverse and not producing pigmentation in culture.
Material examined: CHINA, Guizhou province, Zunyi city, Chishui county, Hushi town, Chishui Alsophila Natural Reserve (28°29′43″ N, 106°0′24″ E), on dead bamboo culms from a stream, 22 September 2019, Jing-Yi Zhang, Y124 (HKAS 123756 = GZAAS 22-0070); living culture, GZCC 22-0086. GenBank accession numbers: (LSU) ON502895, (ITS) ON502902.
Notes: Codinaea terminalis was introduced by Lin et al. [9], isolated from decaying leaves in China. The morphology of our isolate shares similar characters with the ex-type strain Codinaea terminalis (GZCC 18–0085). However, there are some differences in the size of the microstructure such as obviously longer setulae (9.6–11.5 µm vs. 4–9.5 μm). Our new isolate clustered among two Dictyochaeta terminalis strains (GZCC 18–0085 and GZCC19-0525) with strong statistical support (99% ML/1.00 PP, Figure 1). Thus, we identified this isolate as a new host record of Co. terminalis on dead bamboo culms in China.
Codinaeella hyalina J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 6).
Figure 6.
Codinaeella hyalina (HKAS 123757, holotype). (a,b) Colonies on woody substrate; (c,d) conidiophores and conidiogenous cells; (e–g) conidiogenous cells (arrows indicate collarettes); (h–l) conidia; (m) germinating conidium; (n,o) culture on PDA from above and below. Scale bars: (c–e) 20 μm; (f–m) 10 μm.
Index Fungorum number: IF559701; Facesoffungi number: FoF 11039.
Etymology: referring to its hyaline conidia.
Holotype: HKAS 123757.
Saprobic on decaying wood submerged in freshwater habitats. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, mostly single, or arise in groups of 2–3 from knots of hyphal cells, dark brown, with glistening mass of conidia at the apex of conidiophores. Mycelium mostly immersed, composed of pale brown, septate hyphae. Conidiophores 93–143 × 3.4–5.1 μm ( = 121 × 4.4 μm), macronematous, mononematous, single or in small groups, erect, straight or slightly flexuous, smooth, dark brown at the base, becoming paler to subhyaline towards the apex, 2–7-septate, smooth, guttulate. Conidiogenous cells 23.6–68 × 3.2–5.5 μm ( = 50.3 × 4.3 μm), mono- or polyphialidic, with discrete, lateral phialides, integrated, terminal, with lateral openings formed by successive sympodial elongation, cylindrical to cylindrical–lageniform, with funnel-shaped collarettes, brown at the base and becoming subhyaline to hyaline towards the apex, smooth-walled. Conidia 15.9–21.6 × 5.8–7 μm ( = 18.6 × 6.3 μm, n = 20), amerospores, aseptate, often unilateral ventricose, reniform, aggregated in large and slimy mass, rough-walled, hyaline, with hair-like and 4–7 μm-long appendages at both ends.
Culture characteristics: Conidia germinating on PDA within 12 h. and germ tubes produced from conidia. Colonies growing on PDA, slow growth, reaching 20 mm in diam. in 20 days at 25 °C, circular, flat, with dense, white mycelium on the surface with undulate margin, from below olivaceous brown at the center, yellow at the edge and does not produce pigmentation in culture.
Material examined: CHINA, Guizhou province, Zunyi city, Xishui county, Taolin town, Tianlong, on submerged decaying wood in a freshwater stream, 13 February 2021, Jian Ma, TL24 (HKAS 123757, holotype; GZAAS22-0073 isotype), ex-type living culture, GZCC 22-0082. GenBank accession numbers: (LSU) ON502891, (ITS) ON502898.
Notes: Following BLASTn searches, the closest matches of our new isolate Codinaeella hyalina of ITS and LSU sequence data were Codinaeella minuta 417E (MZ078594, 96.57% similarity) and Cod. minuta ATCC 20960 (OL654146, 99.08% similarity), respectively. In the phylogeny (Figure 1), our new isolate formed a separate clade, and is basal to other species of Codinaeella with a high bootstrap support value (100% ML/1.00 PP). Morphologically, our new isolate shares similar characters to Cod. mimusopis in having macronematous, mononematous conidiophores, conidiogenous cell with lateral phialides and hyaline, aseptate conidia with filiform appendages at both ends [16,30]. However, our new species Cod. hyalina differs from Cod. mimusopis by having wider conidiogenous cells (3.2–5.5 μm vs. 3(–3.5) μm) and wider reniform conidia (15.9–21.6 × 5.8–7 μm vs. 16–18(–20) × 2.5–3(–3.5) μm). Additionally, Cod. hyalina differs from existing species of Codinaeella in having obviously wider conidia. Therefore, we introduce Cod. hyalina as a new species based on its distinct morphology and phylogenetic evidence.
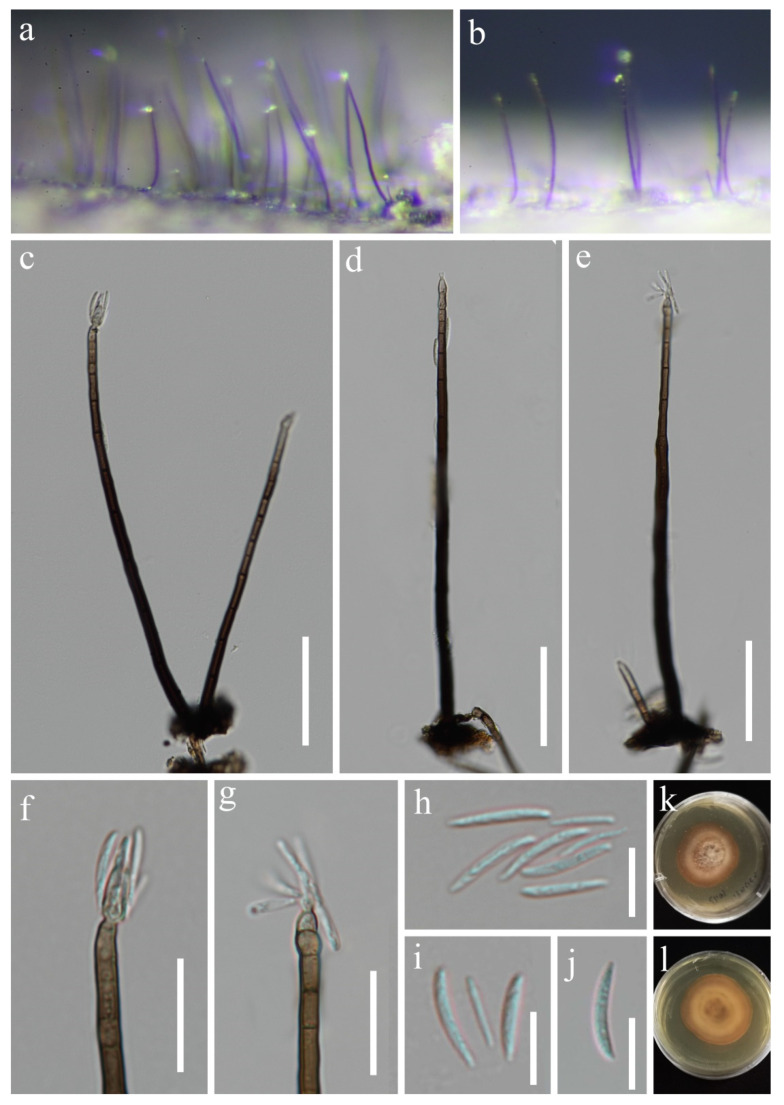
Dictyochaeta guizhouensis J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 7).
Figure 7.
Dictyochaeta guizhouensis (HKAS 123753, holotype). (a,b) Colonies on dead wood; (c–e) conidiophores; (f,g) conidiogenous cells; (h–j) conidia; (k,l) culture on PDA from above and below. Scale bars: (c–e) 50 μm; (f,g) 20 μm; (h–j) 10 μm.
Index Fungorum number: IF559699; Facesoffungi number: FoF 11040.
Etymology: referring to the province “Guizhou”, where this species was collected.
Holotype: HKAS 123753.
Saprobic on dead wood in land. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, gregarious, hairy, brown. Conidiophores 133–240 × 4–6.8 µm ( = 170.7 × 4.8 µm, n = 10), macronematous, mononematous, erect, straight, unbranched, multiseptate, smooth, dark brown at the base, becoming pale brown towards the apex. Conidiogenous cells 8.6–20 × 2.9–4.8 µm ( = 14.8 × 3.8 µm, n = 10), polyphialidic, terminal, integrated, with flared collarette, cylindrical, pale brown to subhyaline or hyaline towards the apex. Conidia 13–16 × 1.5–2.2 µm ( = 15 × 1.8 µm, n = 20), aggregated in a glistening and small mass of conidia at the tip of conidiophores, amerospores, acrogenous, asymmetrical, falcate, straight or slightly curved, often rounded at the apex, tapering toward the basal end, rough-walled, hyaline.
Cultural characteristics: Conidia germinating on PDA within 15 h and germ tubes produced from conidia. Colonies grew on PDA medium at 25 °C, circular, with an entire margin, umbonate, with dense, white mycelium in the middle and yellowish-brown mycelium in the outer ring on the surface; in contrast, they are cream yellow in the middle, while yellowish-brown in the center and the outer ring.
Material examined: CHINA, Guizhou province, Xishui county, Xishui Reserve, 7 September 2020, Yong-Zhong Lu, XSBHq(011) (HKAS 123753, holotype; GZAAS 22-0074, isotype), ex-type living culture, GZCC 22-0083. GenBank accession numbers: (LSU) ON502892, (ITS) ON502899.
Notes: In a BLASTn search in GenBank, the closest match to the LSU and ITS sequences of our new isolate (GZCC 22-0083) exhibited 100% similarity across 100% of the query sequence and 98.49% similarity across 90% of the query sequence to Dictyochaeta sp. (strain CBS 138684), respectively. The phylogenetic tree depicted that our new collection is closely related to unidentified taxon Dictyochaeta sp. (CBS 138684) with 97% ML/1.00 PP support (Figure 1). Dictyochaeta sp. (CBS 138684) was described by Réblová et al. [14] with its sexual and asexual morphs. However, there was insufficient evidence of morphological characteristics due to a sample ageing; therefore, Dictyochaeta sp. (CBS 138684) was treated as an unidentified species [14]. Our new isolate formed an asexual morph in the natural substrate and is characterized by a lack of setae, microconidia and polyphialidic conidiohenous cells with conspicuous collarette, which can be differentiated from Dictyochaeta sp. (CBS 138684). Given the remarkable differences in morphology between our new collection (HKAS 123753) and Dictyochaeta sp. (CBS 138684), we maintain Dictyochaeta sp. as an unidentified species and introduce our new collection as a novel species in Dictyochaeta.
Paragaeumannomyces guttulatus J.Y. Zhang and Y.Z. Lu, sp. nov. (Figure 8).
Figure 8.
Paragaeumannomyces guttulatus (HKAS 123755, holotype). (a) Colonies (arrow) on host surface; (b) seta; (c–e) conidiogenous cell with conidia; (f–k) conidia; (l) germinating conidium; (m,n) colonies on PDA from above and below. Scale bars: (a) 100 μm; (b) 20 μm; (c–l) 10 μm.
Index Fungorum number: IF559700; Facesoffungi number: FoF 11041.
Etymology: referring to its conidia with big guttules.
Holotype: HKAS 123755.
Saprobic on dead bamboo culms in freshwater habitats. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, gregarious. Mycelium composed of partly immersed, partly superficial, septate, pale brown hyphae. Setae scattered over entire colonies, aseptate, dark brown, stiff, pointed, 65–110.5 μm long ( = 85 μm, n = 13). Conidiophores often reduced to conidiogenous cells, semi-macronematous to micronematous, mononematous, flexuous, unbranched, pale to moderately brown, thin-walled. Conidiogenous cells 6.4–10.3 × 5.4–8 μm ( = 8 × 6.5 µm, n = 20), monophialidic, flask-shaped, cup-shaped collarette, hyaline to light brown, guttulate. Conidia 9.4–11.5 μm diam. ( = 10.5 μm, n = 30), enteroblastic, acrogenous, aseptate, hyaline, globose to subglobose or ellipsoid, guttulate, with 2–6 hair-like and 8.8–13.5 µm-long appendages.
Culture characteristics: conidia germinating on PDA within 15 h and the hyaline germ tubes germinates from a point of the conidia. Colonies on MEA at 26 °C, slow growth, reach 5 mm in diam. in 30 days, circular, flat, with entire margin, taupe brown from above, while dark brown in reverse, and do not produce pigmentation in culture.
Material examined: CHINA, Guizhou province, Zunyi city, Chishui county, Hushi town, Chishui Alsophila Natural Reserve (28°29′43″ N, 106°0′24″ E), on dead bamboo culms from a freshwater stream, 22 September 2019, Jing-Yi Zhang, Y132 (HKAS 123755, holotype; GZAAS 22-0076, isotype); ex-type living culture, GZCC 22-0084. GenBank accession numbers: (LSU) ON502893, (ITS) ON502900.
Notes: Following BLASTn searches, the closest match to our new collection Paragaeumannomyces guttulatus is Chaetosphaeria panamensis (LSU, MT118218, 98.46% shared identity; ITS, KY212752, 92.68%). The phylogenetic results confirm that our new isolate P. guttulatus formed a separate clade within Paragaeumannomyces and shared a sister relationship with P. panamensis (S.M.H. 3596) with 94% ML/1.00 PP support (Figure 1). Chaetosphaeria panamensis was introduced by Huhndorf and Fernández [4] with sexual and asexual morphs. Later, Chaetosphaeria panamensis was transferred to the Paragaeumannomyces (P. panamensis) by Réblová et al. [29] based on generic concept and phylogenetic analysis. Morphologically, our new species P. guttulatus formed an asexual morph in the natural substrate and is similar to P. panamensis in having a “Craspedodidymum”-like asexual morph. However, Paragaeumannomyces guttulatus differs from P. panamensis in having bigger conidiogenous cells and smaller and rounder conidia with longer appendages [4,6]. Thus, we introduce P. guttulatus as a new species based on morphological and phylogenetic analyses.
Phialosporostilbe scutiformis N.G. Liu, J. Yang and K.D. Hyde (2019). (Figure 9).
Figure 9.
Phialosporostilbe scutiformis (MFLU 22-0077). (a,b) Colonies on woody substrate; (c) conidiophores; (d,e) conidiogenous cells with conidia; (f,g) conidia; (h) germinated conidium; (i,j) culture on PDA from above and below. Scale bars: (a,b) 200 μm; (c) 100 μm; (d,e) 20 μm; (f–h) 10 μm.
Index Fungorum number: IF555325; Facesoffungi number: FoF 04869.
Holotype: MFLU 18-1502.
Saprobic on decaying wood submerged in freshwater habitats. Sexual morph: undetermined. Asexual morph: hyphomycetous. Colonies on natural substrate superficial, effuse, scattered or gregarious, white, with glistening mass of conidia surround the conidiophores. Mycelium 2.1–3.2 μm ( = 2.7 μm) wide, composed of partly immersed, partly superficial, septate, brown hyphae. Setae 247–358 μm ( = 309 μm), rarely fertilizable, straight or slightly flexuous, dark brown at the base, paler towards the apex, mutiseptate, smooth-walled, unbranched, cylindrical; the lower part is surrounded and compacted by conidiophores. Conidiophores macronematous, synnematous, 11–13.7 μm ( = 12.5 μm) wide at the synnemata, always spread laterally along the upper half of the synnemata, erect, flexuous, unbranched, septate, smooth-walled, dark brown at the base, becoming pale brown and slightly tapering towards the apex, cylindrical. Conidiogenous cells 26–54 × 3.4–5.1 µm ( = 36 × 4.4 μm), monophialidic, terminal, integrated, clavate, thick-walled, rough, brown to subhyaline towards the apex, with an inconspicuous apical collarette. Conidia 6–7.9 μm ( = 7.1 μm, n = 20) long at each above edge, 9–14.4 μm ( = 10.8 μm, n = 20) long at the side, amerospores, acrogenous, round–tetrahedral, triangular from above, rough-walled, hyaline, with (6.5–)9.4–14 µm ( = 10.8 μm, n = 20)-long appendages at each corner.
Cultural characteristics: Conidia germinating on PDA within 15 h and germ tubes produced from the corner of conidia. Colonies on PDA medium, circular, reaching 13 mm in diam. in 10 days at 25 °C, entirely to slightly filamentous, with dense, yellowish-brown mycelium in the middle and thin, slightly yellow mycelium in the edge; yellowish in reverse and not producing pigmentation in culture.
Material examined: THAILAND, Chiang Rai province, on submerged decaying wood in a freshwater stream, 6 March 2021, Jing-Yi Zhang, Y269-2 (MFLU 22-0077); living culture, MFLUCC 22-0053.
GenBank accession numbers: (LSU) ON678145, (ITS) ON678180.
Notes: Phialosporostilbe scutiformis was introduced by Yang et al. [36] based on two specimens collected from freshwater habitats in China and Thailand. A BLASTn search of the ITS and LSU sequence data indicated that our new isolate is closely related to P. scutiformis (MFLUCC 17-0227) with 100% similarity. The morphology of our isolate and the paratype specimen of P. scutiformis (HKAS 102205) are indistinguishable, except that the big guttule in conidia was not observed in our collection. This may be due to the different observation periods. Phylogenetic analysis confirmed that our isolate MFLUCC 22-0053 grouped with Phialosporostilbe scutiformis strains (MFLUCC 17-0227 and MFLUCC 18-1288) with strong bootstrap support (99% ML/1.00 PP, Figure 1). Therefore, we identified this isolate as a new collection of P. scutiformis based on identical morphology and phylogenetic analysis.
4. Discussion
Most Chaetosphaeriaceous species are saprobic on wood or decaying plants from terrestrial and freshwater habitats, sometimes occurring on soil, while some are fungicolous taxa [1,2,6,11,12,14,20,29,56,57,58]. Chaetosphaeria mangrovei was only one species reported from the marine habitat [20]. In this study, five new species among eight newly collections, namely Chaetosphaeria obovoidea, Codinaea aseptata, Codinaeella hyalina, Dictyochaeta guizhouensis and Paragaeumannomyces guttulatus, were documented from China. This increased the number of Chaetosphaeriaceous species and exhibited high diversity, and undiscovered in China. Additionally, three previously known species including a new host record and two new collections were described and identified based on morphological comparison and phylogenetic analysis of a combined LSU and ITS sequence dataset.
The tree topologies resulting from phylogenetic reconstruction revealed Chaetosphaeriaceae are a well-resolved family (Figure 1). However, phylogenetic relationships of some genera in Chaetosphaeriaceae are problematic and uncertain [6,9,13,14,15,16,36]. The type genus Chaetosphaeria (Chaetosphaeriaceae, Chaetosphaeriales) is speciose and phylogenetically unresolved [1,3,4,5,6,9,10,22,29]. Its sexual morph is characterized by black, papillate ascomata, persistent paraphyses, clavate to cylindrical and unitunicate asci with a shallow, refractive, J-apical ring and hyaline, allantoid or ellipsoid, fusiform to filiform curved, septate ascospores with guttules [6,11,17,19,21,57,59]. The asexual morph is described as hyphomycetes with diversity, and is considered to be able to distinguish Chaetosphaeria species [3,10,11,20,23,51,59]. However, the high variability of asexual morphology complicates the phylogenetic placement of Chaetosphaeria [9,10,20,51]. Therefore, further studies on a complicated teleomorph–anamorph relationship are warranted, alongside molecular support to confirm the phylogenetic status of Chaetosphaeria.
Acknowledgments
Jing-Yi Zhang would like to thank Shaun Pennycook (Manaaki Whenua Landcare Research, New Zealand) for advising on the fungal names, and Ning-Guo Liu, Chuan-Gen Lin for the guidance. Jing-Yi Zhang also would like thank the Mae Fah Luang University for granting me the tuition scholarship for my PhD studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8060643/s1. Table S1: taxa used in this study and their GenBank accession numbers.
Author Contributions
Conceptualization, J.-Y.Z.; Formal analysis, J.-Y.Z.; Investigation, J.-Y.Z. and J.M.; Methodology, J.-Y.Z.; Software, J.-Y.Z. and J.M.; Supervision, S.B. and Y.-Z.L.; Writing—original draft, J.-Y.Z.; Writing—review and editing, Y.-P.X., S.B., J.-C.K. and Y.-Z.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study were submitted to GenBank.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was funded by the Science and Technology Foundation of Guizhou Province ([2020]1Y058), the National Natural Science Foundation of China (NSFC 31900020), the China Postdoctoral Science Foundation Project (2020M683657XB) and the Guizhou Province high-level talent innovation and entrepreneurship merit funding project (No. 202104).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reblova M., Barr M.E., Samuels G.J. Chaetosphaeriaceae, a new family for Chaetosphaeria and its relatives. Sydowia. 1999;51:49–70. [Google Scholar]
- 2.Hughes S.J., Kendrick W.B. New Zealand Fungi: 12. Menispora, Codinaea, Menisporopsis. N. Z. J. Bot. 1968;6:323–375. doi: 10.1080/0028825X.1968.10428818. [DOI] [Google Scholar]
- 3.Réblová M., Gams W. Life-history of Ascomycetes: Two new species of Chaetosphaeria with Chloridium and Chloridium-Dictyochaeta anamorphs. Mycoscience. 2000;41:129–138. doi: 10.1007/BF02464321. [DOI] [Google Scholar]
- 4.Huhndorf S.M., Fernández F.A. Teleomorph-anamorph connections: Chaetosphaeria raciborskii and related species, and their Craspedodidymum-like anamorphs. Fungal Divers. 2005;19:23–49. [Google Scholar]
- 5.Fernández F.A., Miller A.N., Huhndorf S.M., Lutzoni F.M., Zoller S. Systematics of the genus Chaetosphaeria and its allied genera: Morphological and phylogenetic diversity in north temperate and neotropical taxa. Mycologia. 2006;98:121–130. doi: 10.1080/15572536.2006.11832718. [DOI] [PubMed] [Google Scholar]
- 6.Perera R., Maharachchikumbura S., Bhat J., Al-Sadi A., Liu J., Hyde K., Liu Z.Y. New species of Thozetella and Chaetosphaeria and new records of Chaetosphaeria and Tainosphaeria from Thailand. Mycosphere. 2016;7:1301–1321. doi: 10.5943/mycosphere/7/9/5. [DOI] [Google Scholar]
- 7.Yang J., Liu J.-K., Hyde K.D., Bhat D.J., Jones E.G., Liu Z.Y. New species of Sporoschisma (Chaetosphaeriaceae) from aquatic habitats in Thailand. Phytotaxa. 2016;289:147–157. doi: 10.11646/phytotaxa.289.2.4. [DOI] [Google Scholar]
- 8.Wei M.-J., Zhang H., Dong W., Boonmee S., Zhang D. Introducing Dictyochaeta aquatica sp. nov. and two new species of Chloridium (Chaetosphaeriaceae, Sordariomycetes) from aquatic habitats. Phytotaxa. 2018;362:187–199. doi: 10.11646/phytotaxa.362.2.5. [DOI] [Google Scholar]
- 9.Lin C.G., McKenzie E.H.C., Liu J.K., Jones E.B.G., Hyde K.D. Hyaline-spored chaetosphaeriaceous hyphomycetes from Thailand and China, with a review of the family Chaetosphaeriaceae. Mycosphere. 2019;10:655–700. doi: 10.5943/mycosphere/10/1/14. [DOI] [Google Scholar]
- 10.Luo Z.L., Hyde K.D., Liu J.K.J., Maharachchikumbura S.S.N., Jeewon R., Bao D.F., Bhat D.J., Lin C.G., Li W.L., Yang J. Freshwater Sordariomycetes. Fungal Divers. 2019;99:451–660. doi: 10.1007/s13225-019-00438-1. [DOI] [Google Scholar]
- 11.Hyde K.D., Norphanphoun C., Maharachchikumbura S.S.N., Bhat D.J., Jones E.B.G., Bundhun D., Chen Y.J., Bao D.F., Boonmee S., Calabon M.S. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 12.Zheng H., Wan Y., Li J., Castañeda-Ruiz R.F., Yu Z. Phialolunulospora vermispora (Chaetosphaeriaceae, Sordariomycetes), a novel asexual genus and species from freshwater in southern China. MycoKeys. 2020;76:17–30. doi: 10.3897/mycokeys.76.57410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Réblová M., Nekvindová J., Hernández-Restrepo M. Reflections on Menisporopsis, Multiguttulispora and Tainosphaeria Using Molecular and Morphological Data. J. Fungi. 2021;7:438. doi: 10.3390/jof7060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Réblová M., Nekvindová J., Kolařík M., Hernández-Restrepo M. Delimitation and phylogeny of Dictyochaeta, and introduction of Achrochaeta and Tubulicolla, genera nova. Mycologia. 2021;113:390–433. doi: 10.1080/00275514.2020.1822095. [DOI] [PubMed] [Google Scholar]
- 15.Réblová M., Nekvindová J., Miller A.N. Phylogeny and taxonomy of Catenularia and similar fungi with catenate conidia. MycoKeys. 2021;81:1–44. doi: 10.3897/mycokeys.81.67785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Réblová M., Kolařík M., Nekvindová J., Réblová K., Sklenář F., Miller A.N., Hernández-Restrepo M. Phylogenetic Reassessment, Taxonomy, and Biogeography of Codinaea and Similar Fungi. J. Fungi. 2021;7:1097. doi: 10.3390/jof7121097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maharachchikumbura S., Hyde K., Jones E., McKenzie E., Bhat J., Hawksworth D., Dayarathne M., Huang S., Norphanphoun C., Senanayake I., et al. Families of Sordariomycetes. Fungal Divers. 2016;79:1–317. doi: 10.1007/s13225-016-0369-6. [DOI] [Google Scholar]
- 18.Tulasne E.L.R., Tulasne C. Selecta Fungorum Carpologia, Ea Documenta et Icones Potissimum Exhibens quae Varia Fructuum et Seminum GENERA in Eodem Fungo Simul aut Vicissim ADESSE Demonstrent. Saraswati Press; West Bengal, India: 1863. pp. 1–319. Xylariei, Valsei, Sphaeriei. Imperiali Typograph. [Google Scholar]
- 19.Ariyawansa H.A., Hyde K.D., Jayasiri S.C., Buyck B., Chethana K.W.T., Dai D.Q., Dai Y.C., Daranagama D.A., Jayawardena R.S., Lücking R. Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274. doi: 10.1007/s13225-015-0346-5. [DOI] [Google Scholar]
- 20.Hyde K., Chaiwan N., Norphanphoun C., Boonmee S., Camporesi E., Chethana K., Dayarathne M., De Silva N., Dissanayake A., Ekanayaka A. Mycosphere notes 169–224. Mycosphere. 2018;9:271–430. doi: 10.5943/mycosphere/9/2/8. [DOI] [Google Scholar]
- 21.Réblová M. Four new species of Chaetosphaeria from New Zealand and redescription of Dictyochaeta fuegiana. Stud. Mycol. 2004;50:171–186. [Google Scholar]
- 22.Seifert K.A., Gams W. The genera of Hyphomycetes–2011 update. Pers. Mol. Phylogeny Evol. Fungi. 2011;27:119–129. doi: 10.3767/003158511X617435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Réblová M., Gams W. Teleomorph-anamorph connections in Ascomycetes. 1. Cylindrotrichum and Cacumisporium anamorphs of Chaetosphaeria. Czech Mycol. 1999;51:1–40. [Google Scholar]
- 24.Huhndorf S.M., Miller A.N., Fernández F.A. Molecular systematics of the Sordariales: The order and the family Lasiosphaeriaceae redefined. Mycologia. 2004;96:368–387. doi: 10.1080/15572536.2005.11832982. [DOI] [PubMed] [Google Scholar]
- 25.Maire R. Fungi Catalaunici: Series altera. Contribution à l’étude de la flore mycologique de la Catalogne. Publ. L’inst. Bot. Barc. 1937;3:1–128. [Google Scholar]
- 26.Li D.W., Kendrick B., Chen J. Two new hyphomycetes: Codinaea sinensis sp. nov. and Parapleurotheciopsis quercicola sp. nov., and two new records from Quercus phillyraeoides leaf litter. Mycol. Prog. 2012;11:899–905. doi: 10.1007/s11557-011-0805-7. [DOI] [Google Scholar]
- 27.Oliveira M.S., Malosso E., Castañeda-Ruiz R.F. A new species and a new combination in Codinaea from Brazil. Mycotaxon. 2015;130:1045–1049. doi: 10.5248/130.1045. [DOI] [Google Scholar]
- 28.Xia J.W., Ma Y.R., Gao J.M., Li Z., Zhang X.G. Codinaea jianfenglingensis sp. nov. and new records from southern China. Mycotaxon. 2015;130:835–841. doi: 10.5248/130.835. [DOI] [Google Scholar]
- 29.Réblová M., Nekvindová J., Fournier J., Miller A.N. Delimitation, new species and teleomorph-anamorph relationships in Codinaea, Dendrophoma, Paragaeumannomyces and Striatosphaeria (Chaetosphaeriaceae) MycoKeys. 2020;74:17–74. doi: 10.3897/mycokeys.74.57824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crous P., Schumacher R.K., Wingfield M.J., Akulov A., Denman S., Roux J., Braun U., Burgess T., Carnegie A., Váczy K. New and interesting fungi. 1. FUSE. 2018;1:169–215. doi: 10.3114/fuse.2018.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lunghini D., Rambelli A., Onofri S. New Codinaea species from tropical forest litter. Mycotaxon. 1982;14:116–124. [Google Scholar]
- 32.Spegazzini C. Algunos hongos de Tierra del Fuego. Physis. 1923;7:9–23. [Google Scholar]
- 33.Kuthubutheen A., Nawawi A. Key to Dictyochaeta and Codinaea species. Mycol. Res. 1991;95:1224–1229. doi: 10.1016/S0953-7562(09)80015-4. [DOI] [Google Scholar]
- 34.Matsushima T. Matsushima mycological memoirs 10. Matsushima Mycol. Mem. 2003;10:1–214. [Google Scholar]
- 35.Sierra Á.M., Portales J.M. Nuevo género de hifomicete fialídico de Cuba. Revista Jard. Bot. Nac. 1985;6:57–60. [Google Scholar]
- 36.Yang J., Liu N.G., Liu J.K., Hyde K.D., Jones E.G., Liu Z.Y. Phylogenetic placement of Cryptophiale, Cryptophialoidea, Nawawia, Neonawawia gen. nov. and Phialosporostilbe. Mycosphere. 2019;9:1132–1150. doi: 10.5943/mycosphere/9/6/5. [DOI] [Google Scholar]
- 37.Bhat D., Kendrick B. Twenty-five new conidial fungi from the Western Ghats and the Andaman Islands (India) Mycotaxon. 1993;49:19–90. [Google Scholar]
- 38.Wu Y.M., Zhang T.Y. New species of Phialosporostilbe and Pleurothecium from soil. Mycotaxon. 2009;110:1–4. doi: 10.5248/110.1. [DOI] [Google Scholar]
- 39.Chomnunti P., Hongsanan S., Aguirre-Hudson B., Tian Q., Peršoh D., Dhami M.K., Alias A.S., Xu J., Liu X.z., Stadler M., et al. The sooty moulds. Fungal Divers. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- 40.Jayasiri S.C., Hyde K.D., Ariyawansa H.A., Bhat J., Buyck B., Cai L., Dai Y.C., Abd-Elsalam K.A., Ertz D., Hidayat I., et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015;74:3–18. doi: 10.1007/s13225-015-0351-8. [DOI] [Google Scholar]
- 41.Hall T.A. BioEdit 5.0.9: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 42.Zheng H., Li J., Guo J.S., Qiao M., Yu Z.F. Anacraspedodidymum submersum sp. nov.(Chaetosphaeriaceae, Chaetosphaeriales), a new species of freshwater hyphomycetes from southwest China. Int. J. Syst. Evol. Micr. 2021;71:004650. doi: 10.1099/ijsem.0.004650. [DOI] [PubMed] [Google Scholar]
- 43.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glez-Peña D., Gómez-Blanco D., Reboiro-Jato M., Fdez-Riverola F. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:14–18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 46.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nylander J.A.A. MrModeltest v2. Program Distributed by the Author. Evolutionary Biology Centre. Uppsala University; Uppsala, Sweden: 2004. [Google Scholar]
- 48.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Boil. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larget B., Simon D.L. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Mol. Boil. Evol. 1999;16:750–759. doi: 10.1093/oxfordjournals.molbev.a026160. [DOI] [Google Scholar]
- 50.Rambaut A., Drummond A. FigTree: Tree Figure Drawing Tool, Version 1.2.2. Institute of Evolutionary Biology, The University of Edinburgh; Edinburgh, UK: 2008. [Google Scholar]
- 51.Liu J.-K., Yang J., Maharachchikumbura S.S., McKenzie E.H., Jones E.G., Hyde K.D., Liu Z.-Y. Novel chaetosphaeriaceous hyphomycetes from aquatic habitats. Mycol. Prog. 2016;15:1157–1167. doi: 10.1007/s11557-016-1237-1. [DOI] [Google Scholar]
- 52.Réblová M., Kolařík M., Nekvindová J., Miller A.N., Hernández-Restrepo M. Phylogeny, global biogeography and pleomorphism of Zanclospora. Microorganisms. 2021;9:706. doi: 10.3390/microorganisms9040706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian C., Bhat D.J. Bahusutrabeeja, a new genus of the hyphomycetes. Can. J. Bot. 1977;55:2202–2206. doi: 10.1139/b77-249. [DOI] [Google Scholar]
- 54.Goh T.K., Hyde K.D., Hodgkiss I.J. Bahusutrabeeja on submerged wood in Hong Kong streams. Mycologia. 2001;93:389–397. [Google Scholar]
- 55.Wu W.P., McKenzie E.H. Obeliospora minima sp. nov. and four other hyphomycetes with conidia bearing appendages. Fungal Divers. 2003;12:223–234. [Google Scholar]
- 56.Jeewon R., Yeung S.Y.Q., Hyde K.D. A novel phylogenetic group within Thozetella (Chaetosphaeriaceae): A new taxon based on morphology and DNA sequence analyses. Can. J. Microbiol. 2009;55:680–687. doi: 10.1139/W08-148. [DOI] [PubMed] [Google Scholar]
- 57.Jones E.B.G., Suetrong S., Cheng W.H., Rungjindamai N., Sakayaroj J., Boonyuen N., Somrothipol S., Abdel-Wahab M.A., Pang K.L. An additional fungal lineage in the Hypocreomycetidae (Falcocladium species) and the taxonomic re-evaluation of Chaetosphaeria chaetosa and Swampomyces species, based on morphology, ecology and phylogeny. Cryptogam. Mycol. 2014;35:119–138. doi: 10.7872/crym.v35.iss2.2014.119. [DOI] [Google Scholar]
- 58.Li C.X., Yu X.D., Dong W., Ming H.D., Saranyaphat B., Zhang H. Freshwater hyphomycetes in Sordariomycetes: Two new species of Tainosphaeria (Chaetosphaeriaceae, Chaetosphaeriales) from Thailand. Phytotaxa. 2021;509:56–68. doi: 10.11646/phytotaxa.509.1.2. [DOI] [Google Scholar]
- 59.Réblová M., Winka K. Phylogeny of Chaetosphaeria and its anamorphs based on morphological and molecular data. Mycologia. 2000;92:939–954. doi: 10.1080/00275514.2000.12061238. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences generated in this study were submitted to GenBank.