Abstract
The phylogenetic relationship of Ephemeridae (Insect: Ephemeroptera) remains hotly debated using mitochondrial (mt) genomes. All previously reported mt genomes of Ephemeridae belong to the genus Ephemera. This study provides the first complete mt genome sequence from the genus Hexagenia with an analysis of the mitogenome of Hexagenia rigida Mc Dunnough, 1924 (Ephemeroptera: Ephemeridae) and providing new information to discuss the phylogenetic relationships within Ephemeroptera. The complete mt genome of H. rigida was a circular molecule of 16,159 bp in length, containing 37 genes (2 rRNA genes, 13 protein-coding genes, 22 tRNA genes), which showed the typical mt gene arrangement of insects. The AT content of the whole genome was 70.0% and the length of the control region was 1091 bp. All protein-coding genes used ATN as the start codon, and most PCGs used TAA/TAG as the stop codons excluding COI, COII, ND5 and Cyt b that used T as the stop codon. BI and ML phylogenetic trees constructed from 27 species of 13 families showed that Ephemeridae is a sister clade to the clade Polymitarcyidae.
Keywords: Ephemeridae, mitochondrial genome, phylogeny
Ephemeroptera has a worldwide distribution, occurring on all continents except Antarctica (Ratnasingham & Hebert 2007). As a relatively primitive group of Pterygota, the phylogenetic relationship of Ephemeroptera has always been a research hotspot (Hebert et al. 2003; Ogden and Whiting 2005; Sun et al. 2006; O’Donnell and Jockusch 2008; Ogden et al. 2009; Webb et al. 2012; Saito et al. 2016; Cai et al. 2018; Gao et al. 2018; Ye et al. 2018; Xu et al. 2019; Guan et al. 2021; Xu et al. 2021; Yu et al. 2021). Considerable effort has been devoted to constructing the phylogenetic relationships among Ephemeroptera families based on morphology (Mccafferty 1991; Mccafferty and Edmunds 1979), molecular evidence (Ogden and Whiting 2005), and combined data (Ogden et al. 2009; Xu et al. 2020). However, there are relatively few studies of the phylogenetic relationships within the Ephemeridae. To date, no mitochondrial (mt) genome of genus Hexagenia has been reported. In this study, we sequenced the first mt genome from this genus (Hexagenia rigida Mc Dunnough, 1924) and discuss its phylogenetic relationship within Ephemeridae.
The female imago of H. rigida (Sample Number: WTH201707) was collected by JY Zhang using sweeping net at Carleton University, Ottawa (45°38′ N 75°69′ W), Canada on 15 July 2017. Insects used in this study are not regulated. The sample was identified and stored at −40 °C freezer in the Animal Specimen Museum, College of Life Sciences and Chemistry, Zhejiang Normal University, China. Total genomic DNA was extracted from individual tissues of the sample using an Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China) and stored in the Zhang laboratory (http://mypage.zjnu.edu.cn/ZJY3/zh_CN/index.htm, Zhang JY, zhang3599533@163.com). Universal primers were used to amplify some partial fragments as described in Zhang et al. (2008). Subsequently, the remaining gaps were sequenced by utilizing specific primers according to previously obtained sequences. Manual proofreading and splicing of all nucleotide fragments were conducted using SeqMan in the DNASTAR Package (Burland 2000). Annotation of all mitochondrial genes were identified by the online website MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. 2013). The mt genome was deposited in GenBank with accession number OL678102.
The complete mt genome of H. rigida was 16,159 bp in length, which is similar to all known mt genomes of Ephemeridae. It encoded of 37 genes including 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs), and one control region (CR). The whole mt genome and the control region of H. rigida had a high AT content of 70.0% and 68.7%, respectively. The total length of the PCGs was 11,199 bp and all genes showed a negative AT-skew. Nine of the PCGs (ND2, COI, COII, ATP8, ATP6, COIII, ND3, ND6, and Cyt b) were located on the heavy strand (H-strand), whereas the others (ND5, ND4, ND4L, and ND1) were located on the light strand (L-strand). The start codons of the PCGs in H. rigida were ATG (in COII, COIII, ND5, ND4, ND4L, Cyt b, and ND1), ATT (in ATP8, ND3, and ND6), and ATA (in ND2 and ATP6). The typical stop codons (TAA and TAG) were used in nine PCGs. However, an incomplete stop codon T occurred in four genes (COI, COII, ND5, and Cyt b). It is quite common in insect mt genomes to use an incomplete stop codon. These truncated stop codons are presumed to be completed by post-transcriptional polyadenylation (Ojala et al. 1981). The summed lengths of the 22 tRNAs, two rRNAs and the CR were 1456 bp, 2131 bp, and 1090 bp, respectively. The 16S RNA and 12S RNA genes were located between tRNA-Leu and tRNA-Val, and between tRNA-Val and the CR, respectively. In the whole mt genome, we found nine overlapping areas each ranging from 1 to 8 bp and the gene arrangement was identical to the ancestral insect gene pattern.
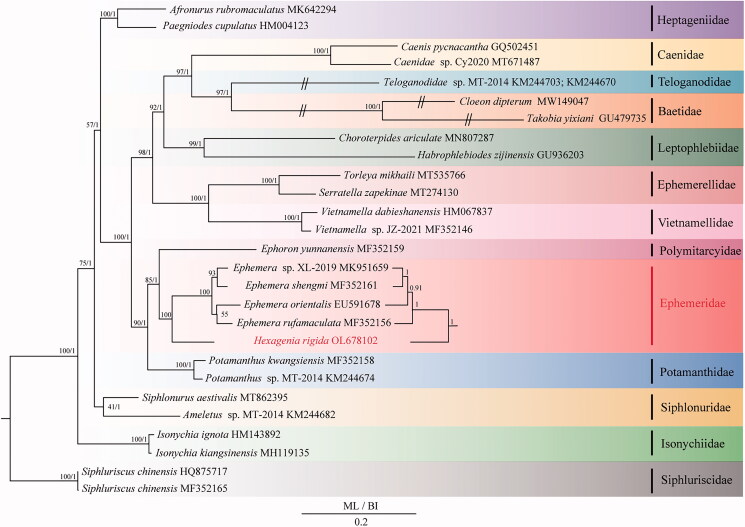
The phylogenetic relationship was constructed by Bayesian inference (BI) using MrBayes 3.1.0 (Huelsenbeck and Ronquist 2001) and maximum-likelihood (ML) using RAxML 8.2.0 (Stamatakis 2014) based on the 13 PCGs. Twenty-seven mt genomes within Ephemeroptera were downloaded from GenBank (Li et al. 2014; Tang et al. 2014; Ye et al. 2018; Cao et al. 2020; Li et al. 2020; Macher et al. 2020; Xu et al. 2020; Yu et al. 2021; Tong et al. 2022) and were used to investigate the phylogenetic relationships. In addition, Siphluriscus chinensis (HQ875717 and MF352165), the most primitive family of the Ephemeroptera, was used as the outgroup. Each alignment was performed using Gblock 0.91b (Castresana 2000) with default settings. The phylogenetic relationship based on BI and ML analyses (Figure 1) indicated that almost all families were monophyletic, including Ephemeridae. According to the results of phylogenetic topologies, Isonychiidae was the basal clade to Ephemeroptera excluding the outgroup Siphluriscidae. After that, Ameletidae and Siphlonuridae were found to be a sister group. Heptageniidae as a sister clade to the remaining Ephemeroptera (Heptageniidae + (((Leptophlebiidae + (Caenidae + (Teloganodidae + Baetidae))) + (Ephemerellidae + Vietnamellidae)) + (Potamanthidae + (Polymitarcyidae + Ephemeridae))). Analysis of phylogenetic revealed that Ephemeridae was shown to be a sister clade to the clade of Polymitarcyidae and H. rigida was a sister clade to genus Ephmera.
Figure 1.
The phylogenetic relationships of BI and ML trees using 28 species of Ephemeroptera, including H. rigida (OL678102) and based on the nucleotide dataset of the 13 mt PCGs. Siphluriscus chinensis (HQ875717 and MF352165) was used as the outgroup. The numbers above branches specify posterior probabilities as determined from BI (left) and bootstrap percentages from ML (right). The GenBank accession numbers of all species are shown in the figure. The long-branch attractions of Baetidae and Teloganodidae have been cut for esthetics.
Funding Statement
This work was supported by the Natural Science Foundation of China [31370042]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Tong Y and Wu L made substantial contributions to the conception or design of the work and drafting the paper; Lin YJ and Ayivi SPG made substantial contributions to analys and interpretation the data; Storey KB, Zhang JY and Yu DN were involved in revising the paper critically for intellectual content; and all authors read the final approval of the version to be published, and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy; and that all authors agree to be accountable for all aspects of the work.
Disclosure statement
The authors declare no conflict of interest.
Data availability statement
The mitochondrial genome data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/nuccore/OL678102] under the accession no. OL678102. The mt genome was obtained by the Sanger method, so no associated “BioProject,” “SRA,” and “Bio-Sample” numbers should be shown.
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Burland TG. 2000. DNASTAR’s Lasergene sequence analysis software. In Bioinformatics Methods and Protocols. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- Cai YY, Gao YJ, Zhang LP, Yu DN, Storey KB, Zhang JY.. 2018. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) and the phylogeny of Ephemeroptera in Pterygota. Mitochondrial DNA Part B. 3(2):577–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SS, Xu XD, Jia YY, Guan JY, Storey KB, Yu DN, Zhang JY.. 2020. The complete mitochondrial genome of Choroterpides apiculata (Ephemeroptera: Leptophlebiidae) and its phylogenetic relationships. Mitochondrial DNA Part B. 5(2):1159–1160. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Gao XY, Zhang SS, Zhang LP, Yu DN, Zhang JY, Cheng HY.. 2018. The complete mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) and its phylogeny. Mitochondrial DNA Part B. 3(1):303–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JY, Zhang ZY, Cao YR, Xu XD, Storey KB, Yu DN, Zhang JY.. 2021. The complete mitochondrial genome of Choroterpes (Euthralus) yixingensis (Ephemeroptera: Leptophlebiidae) and its mitochondrial protein-coding gene expression under imidacloprid stress. Gene. 800:145833–145844. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, Dewaard JR.. 2003. Biological identifications through DNA barcodes. Proc R Soc Lond B. 270(1512):313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755. [DOI] [PubMed] [Google Scholar]
- Li D, Qin JC, Zhou CF.. 2014. The phylogeny of Ephemeroptera in Pterygota revealed by the mitochondrial genome of Siphluriscus chinensis (Hexapoda: Insecta). Gene. 545(1):132–140. [DOI] [PubMed] [Google Scholar]
- Li R, Zhang W, Ma ZX, Zhou CF.. 2020. Novel gene rearrangement pattern in the mitochondrial genomes of Torleya mikhaili and Cincticostella fusca (Ephemeroptera: Ephemerellidae). Int J Biol Macromol. 165(Pt B):3106–3114. [DOI] [PubMed] [Google Scholar]
- Macher JN, Drakou K, Papatheodoulou A, Hoorn B, Vasquez M.. 2020. The mitochondrial genomes of 11 aquatic macroinvertebrate species from Cyprus. Metabarcoding Metagenomics. 4(5):91–96. [Google Scholar]
- McCafferty WP. 1991. Toward a phylogenetic classification of the Ephemeroptera (Insecta): a commentary on systematics. Ann Entomol Soc Am. 84(4):343–360. [Google Scholar]
- McCafferty WP, Edmunds GF.. 1979. The higher classification of the Ephemeroptera and its evolutionary basis. Ann Entomol Soc Am. 72(1):5–12. [Google Scholar]
- O’Donnell BC, Jockusch EL.. 2008. Phylogenetic relationships of leptophlebiid mayflies as inferred by histone H3 and 28S ribosomal DNA. Syst Entomol. 33(4):651–667. [Google Scholar]
- Ogden TH, Gattolliat JL, Sartori M, Taniczek AH, Soldán T, Whiting MF.. 2009. Towards a new paradigm in mayfly phylogeny (Ephemeroptera): combined analysis of morphological and molecular data. Syst Entomol. 34(4):616–634. [Google Scholar]
- Ogden TH, Whiting MF.. 2005. Phylogeny of Ephemeroptera (mayflies) based on molecular evidence. Mol Phylogenet Evol. 37(3):625–643. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G.. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474. [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN.. 2007. Bold: the barcode of life data system (http://www.barcodinglife.org). Mol Ecol Notes. 7(3):355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Jo J, Sekiné K, Bae YJ, Tojo K.. 2016. Phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 46(4):246–259. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Sabo A, Meyer MD, Randolph RP, Jacobus LM, McCafferty WP, Ferris VR.. 2006. Tests of current hypotheses of mayfly (Ephemeroptera) phylogeny using molecular (18S rDNA) data. Ann Entomol Soc Am. 99(2):241–252.2.0.CO;2] [Google Scholar]
- Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes – a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42(22):e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Wu L, Ayivi SPG, Storey KB, Ma Y, Yu DN, Zhang JY.. 2022. Cryptic species exist in Vietnamella sinensis Hsu, 1936 (Insecta: Ephemeroptera) from studies of complete mitochondrial genomes. Insects. 13(5):412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JM, Jacobus LM, Funk DH, Zhou X, Kondratieff B, Geraci CJ, DeWalt RE, Baird DJ, Richard B, Phillips I, et al. ,. 2012. A DNA barcode library for North American Ephemeroptera: progress and prospects. PLOS One. 7(5):e38063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XD, Guan JY, Zhang ZY, Cao YR, Cai YY, Storey KB, Yu DN, Zhang JY.. 2021. Insight into the phylogenetic relationships among three subfamilies within Heptageniidae (Insecta: Ephemeroptera) along with low-temperature selection pressure analyses using mitogenomes. Insects. 12(7):656–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XD, Jia YY, Cao SS, Zhang ZY, Storey KB, Yu DN, Zhang JY.. 2020. Six complete mitochondrial genomes of mayflies from three genera of Ephemerellidae (Insecta: Ephemeroptera) with inversion and translocation of trnI rearrangement and their phylogenetic relationships. PeerJ. 8:e9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XD, Jia YY, Dai XY, Ma JL, Storey KB, Zhang JY, Yu DN.. 2019. The mitochondrial genome of Caenis sp. (Ephemeroptera: Caenidae) from Fujian and the phylogeny of Caenidae within Ephemeroptera. Mitochondrial DNA Part B. 5(1):192–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye QM, Zhang SS, Cai YY, Storey KB, Yu DN, Zhang JY.. 2018. The complete mitochondrial genome of Isonychia kiangsinensis (Ephemeroptera: Isonychiidae). Mitochondrial DNA Part B. 3(2):541–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DN, Yu PP, Zhang LP, Storey KB, Gao XY, Zhang JY.. 2021. Increasing 28 mitogenomes of Ephemeroptera, Odonata and Plecoptera support the Chiastomyaria hypothesis with three different outgroup combinations. PeerJ. 9(1):e11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Zhou CF, Gai YH, Song DX, Zhou KY.. 2008. The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera. Gene. 424(1–2):18–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mitochondrial genome data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/nuccore/OL678102] under the accession no. OL678102. The mt genome was obtained by the Sanger method, so no associated “BioProject,” “SRA,” and “Bio-Sample” numbers should be shown.