Abstract
Objective
Necrotising enterocolitis (NEC) is a devastating intestinal disease primarily affecting preterm infants. The underlying mechanisms are poorly understood: mother’s own breast milk (MOM) is protective, possibly relating to human milk oligosaccharide (HMO) and infant gut microbiome interplay. We investigated the interaction between HMO profiles and infant gut microbiome development and its association with NEC.
Design
We performed HMO profiling of MOM in a large cohort of infants with NEC (n=33) with matched controls (n=37). In a subset of 48 infants (14 with NEC), we also performed longitudinal metagenomic sequencing of infant stool (n=644).
Results
Concentration of a single HMO, disialyllacto-N-tetraose (DSLNT), was significantly lower in MOM received by infants with NEC compared with controls. A MOM threshold level of 241 nmol/mL had a sensitivity and specificity of 0.9 for NEC. Metagenomic sequencing before NEC onset showed significantly lower relative abundance of Bifidobacterium longum and higher relative abundance of Enterobacter cloacae in infants with NEC. Longitudinal development of the microbiome was also impacted by low MOM DSLNT associated with reduced transition into preterm gut community types dominated by Bifidobacterium spp and typically observed in older infants. Random forest analysis combining HMO and metagenome data before disease accurately classified 87.5% of infants as healthy or having NEC.
Conclusion
These results demonstrate the importance of HMOs and gut microbiome in preterm infant health and disease. The findings offer potential targets for biomarker development, disease risk stratification and novel avenues for supplements that may prevent life-threatening disease.
INTRODUCTION
Necrotising enterocolitis (NEC) is an inflammation-mediated bowel condition that is a leading cause of death and serious morbidity in preterm infants born before 32 weeks of gestation.1 The mechanisms underlying NEC development are poorly understood, and the lack of specificity of symptoms and tests makes diagnostic certainty difficult. Infants with NEC have enteral feeds stopped and are treated with broad-spectrum antibiotics and may need surgery.2
Receipt of mother’s own breast milk (MOM) is the most protective factor against the development of NEC in preterm infants.3 4 However, infants receiving MOM still develop NEC, suggesting the variable composition of nutrients and other components of breast milk may be important. Human milk oligosaccharides (HMOs) are structurally diverse, complex unconjugated sugars that are not usually present in artificial formula milk.5 HMOs are indigestible to the infant, reaching the lower GI tract intact where they act as growth substrates (ie, prebiotics) for specific bacteria, notably Bifidobacterium spp, thought key to infant health.6–8 HMOs may also protect from enteric organism bloodstream infections due to antimicrobial activity,9 stimulate the immune system,10 enhance gut barrier function11 and act as decoy receptors for pathogens.12 While >150 HMOs have been described, the 19 most abundant represent >95% of the total HMO content.13 HMO profiles are specific to individual mothers and remain relatively stable during lactation.14 Presence of an active FUT2 gene, which is involved in the synthesis of α1–2-fucosylated oligosaccharides, is the main determinant of the HMO profile, termed maternal secretor status.15
Recent work has begun to elucidate the potential contribution of HMOs to preterm infant health. In a neonatal rat model, disialyllacto-N-tetraose (DSLNT), a non-fucosylated but double-sialylated HMO, significantly reduced NEC development and improved NEC-associated mortality rate.16 An association of lower DSLNT concentration in MOM and subsequent higher risk of NEC onset in the infant has since been observed in preterm human studies.17–19 To date, these studies have included very small numbers of infants with NEC (between 4 and 8), with a broad range of NEC phenotypes. Thus, validation in a larger cohort is urgently needed.
Altered gut microbiome development has been associated with NEC in preterm infants. While no specific causative microorganism has consistently been identified, studies have reported a higher relative abundance of Enterobacteriaceae, coupled to lower relative abundance of Bifidobacterium.20–23 Instability of the gut microbiome in infants with NEC has also been reported in longitudinal studies, with more frequent transitions between different preterm gut community types (PGCTs) in NEC.20 These findings were replicated at the site of disease in a study using formalin-fixed paraffin-embedded tissue from infants with NEC matched to non-NEC controls.24 Previous microbiome studies have largely relied on 16S rRNA gene sequencing of the V4 region, which has limited resolution, especially for emerging key organisms of interest for preterm health (ie, Klebsiella and Enterobacter would be classified together as Enterobacteriaceae). Metagenomics may overcome this and recent metagenomic data showed infants who developed NEC had higher relative abundance of Klebsiella and higher replication rates in all bacteria before disease onset.25
In the current study, we performed a combined analysis of maternal HMO profiles and longitudinal development of the infant stool/gut microbiome in a large cohort of preterm infants with NEC and healthy controls matched for gestation, birth weight and day of life (DOL). We then validated our results in an independent cohort using previously published HMO data.17 We hypothesised that differences in maternal HMO profiles and microbiome development may explain why some infants receiving MOM still develop NEC.
METHODS
Cohort
This study included 77 preterm infants (born at <32 weeks of gestation) who were born in or transferred to a single, large tertiary level neonatal intensive care unit in Newcastle upon Tyne, UK, recruited to the ‘Supporting Enhanced Research in Vulnerable Infants Study’ (SERVIS; REC10/H0908/39) with written parental consent covering data and sample collection. Thirty-three infants were diagnosed with definite NEC and 37 non-diseased controls were selected by identifying a healthy infant matched by gestation, birth weight and having a MOM sample available at a corresponding DOL (table 1). Detailed information on feeding and antibiotic use are included in online supplemental tables 1 and 2. Diagnoses were made using an extensive combination of clinical, X-ray and histological findings and blindly agreed by two neonatal clinicians (JEB and NDE). Standard clinical protocols recommended the routine use of supplemental probiotics when more than 30 mL/kg/day of MOM was tolerated for at least 1–2 days: all 33 infants with NEC received MOM and 31 received probiotics. All 37 controls received probiotics. The probiotics administered were either LaBiNIC (Lactobacillus acidophilus, Bifidobacterium infantis and B. bifidum) or Infloran (L. acidophilus and B. bifidum).
Table 1.
Demographics of the analytical cohort with human milk oligosaccharide profile data
| Control | NEC | P value | |
|---|---|---|---|
|
| |||
| Number of patients | 37 | 33 | – |
| Secretors | 25 (68%) | 20 (61%) | 0.544 |
| Male | 14 (38%) | 22 (67%) | 0.016 |
| Vaginal delivery | 25 (68%) | 17 (52%) | 0.171 |
| Gestational age | 25 (24–26) | 25 (24–27) | 0.881 |
| Birth weight | 670 (585–830) | 670 (600–840) | 1.000 |
| Probiotics ever | 37 (100%) | 31 (94%) | 0.855 |
| MOM only | 3 (8%) | 6 (18%) | 0.468 |
| MOM+formula | 11 (30%) | 12 (37%) | |
| MOM+BMF | 10 (27%) | 7 (21%) | |
| MOM+formula+BMF | 13 (35%) | 8 (24%) | |
| DOL breast milk sample | 18 (12–31) | 18 (13–34) | 0.636 |
| DOL disease onset | – | 19 (14–35) | – |
| NEC surgical | – | 16 | – |
Differences between groups were tested applying χ2 test and Wilcoxon post hoc test where applicable.
BMF, breast milk fortifier; DOL, day of life; MOM, mother’s own breast milk; NEC, necrotising enterocolitis.
The median DOL of NEC diagnosis was 19 (IQR 14–35, table 1). A single MOM sample was analysed for each infant, as close to the onset of disease as possible, with control samples matched by DOL (online supplemental figure 1). The median DOL of MOM from NEC cases was 18 (IQR 13–34) and that from controls was also 18 (IQR 12–31). The DOL of the milk sample is the DOL received by the infant and is not necessarily the same day as the mother expressed the milk due to standard practice that often involves milk storage. Metagenomic sequencing of stool samples (n = 644) was performed longitudinally on a subset of 48 infants (including 14 NEC, online supplemental figure 1). These infants were comparable to the full cohort (online supplemental table 3).
Full details of the HMO, metagenome and statistical analysis are described in online supplemental methods.
HMO analysis
The absolute quantification for the 19 most abundant HMOs was determined by high-performance liquid chromatography following derivatisation as per the protocol described by Bode et al.26 Maternal secretor (presence of an active FUT2 gene) status was determined by the presence or near absence of 2′-Fucosyllactose (2′-FL) in the breast milk analysed.
Metagenomes
DNA was extracted from ~0.1 g of stool using the DNeasy PowerSoil Kit (QIAGEN) following the manufacturer’s protocol, and sequencing was performed on the HiSeq X Ten (Illumina) with a read length of 150 bp paired end reads. Processed fastq files were mapped against the MetaPhlan2 marker gene database (mpa_v20_m200).27
Statistical analysis
Statistical analysis of HMO profiles was performed using MetaboAnalyst V.3.0.28 For ordinations, HMO data were normalised by logarithmic transformation and 2000 random permutations were used to test the significance. Multivariate receiver operating characteristic (ROC) curves were generated using linear support vector machine (SVM) classification method coupled with Monte Carlo cross validation.
Correlation between clinical variables and individual HMOs was tested by performing a multivariate adjusted linear model in R V.3.6.3. HMO concentrations were normalised by log transformation prior to analysis, and p values were adjusted by applying the Benjamini and Hochberg correction.29
The cross-sectional cohort of stool samples collected from infants with NEC before diagnosis and matched controls was analysed using MicrobiomeAnalyst.30 31 Permutational multivariate analysis of variance (PERMANOVA) was used to determine the significance of Bray-Curtis principal coordinate analysis. MetagenomeSeq was used to assess differential abundance at the phyla and species level.
Dirichlet Multinomial Mixtures (DMM) clusters samples on the basis of microbial community structure32 and was used to determine the PGCTs from all samples, as performed previously.33 34 Five PGCTs were found to be optimal and were ordered youngest (PGCT-1) to oldest (PGCT-5) based on the average DOL of samples within each PGCT. Analysis was performed at specific time windows, including only a single sample per infant in each time point.
The association of various clinical variables on the HMO and metagenome profiles was tested by applying the function ‘adonis’ of ‘vegan’ V.2.5–6 package35 in R, based on Bray-Curtis dissimilarity and 10 000 permutations. Each test was performed stepwise and p values were adjusted using the Benjamini and Hochberg procedure.29
Random Forest was used for comparing the performance of classification models built using matched cross-sectional datasets.
RESULTS
Association between maternal HMOs and development of NEC in the infant
MOM samples clustered according to maternal secretor status and secretor mothers had a higher total HMO concentration, a higher HMO Shannon diversity and a significantly higher concentration of overall HMO-bound fucose (online supplemental figure 2). Thus, where relevant, we have stratified and adjusted for maternal secretor status in subsequent analyses.
HMO profiles showed significant separation of NEC and control infants (figure 1A; 2000 permutations, p<0.001), and this was consistent when secretor and non-secretor samples were analysed separately (both p<0.001, online supplemental figure 3A,B). Individually, of the 19 HMOs quantified in this study, only DSLNT was significantly different between NEC and controls, with a lower concentration in infants with NEC (adjusted (adj.) p<0.001; figure 1B,C). No significant associations were found in the Shannon diversity of HMOs between NEC and matched controls for the full cohort, or when stratified by maternal secretor status (all p>0.05, online supplemental figure 4).
Figure 1.
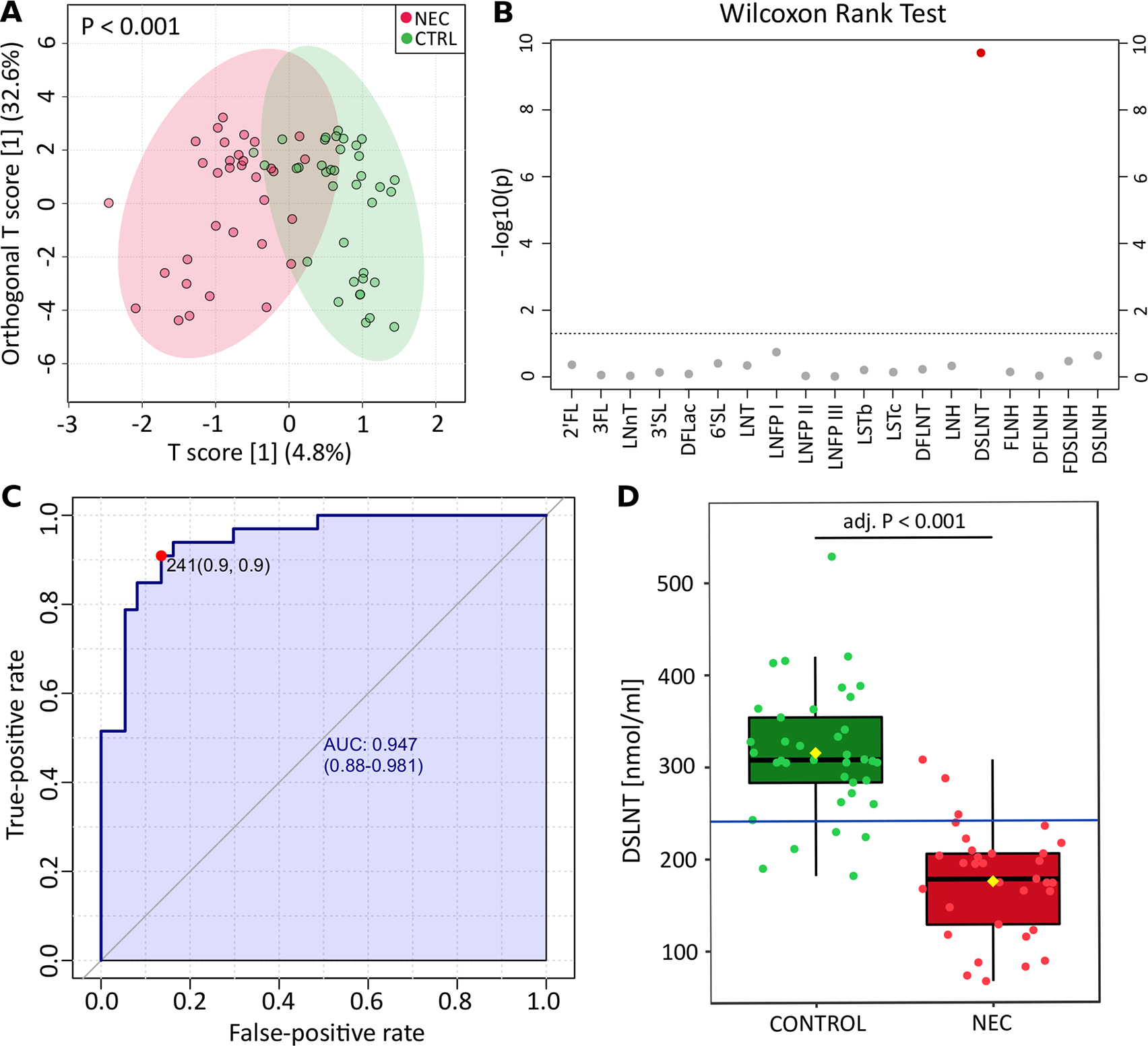
Analysis of HMO profiles and DSLNT concentration in NEC and CTRLs. (A) Orthogonal partial least squares discriminant analysis of maternal HMO profiles fed to infants diagnosed with NEC and CTRLs. The p value was calculated based on 2000 permutations. (B) Visual representation of p values obtained from comparison of individual HMOs between NEC and CTRL groups. Wilcoxon rank-sum test was applied, and p values were adjusted with the false discovery rate algorithm. The line indicates p value=0.05. (C) Univariate receiver operating characteristic curve generated on DSLNT concentration identified 241 nmol/mL as the best threshold for NEC prediction. The performance of the classification is defined by the AUC, specificity (false-positive rate) and sensitivity (true-positive rate). (D), Box plot showing the concentration of DSLNT between NEC and controls. Blue line represents the 241 nmol/mL threshold. 2′-FL, 2′-fucosyllactose; 3′-FL, 3′-Fucosyllactose; 6′SL, 6′-Sialyllactose; adj, adjusted; AUC, area under the curve; CTRL, control; DFLac, difucosyllactose; DFLNH, difucosyllacto-N-hexaose; DFLNT, difucosyllacto-N-tetraose; DSLNH, disialyllacto-n-hexaose; DSLNT, disialyllacto-N-tetraose; FDSLNH, fucodisialyllacto-N-hexaose; FLNH, fucosyllacto-N-hexaose; HMO, human milk oligosaccharide; lacto-N- neotetraose, 3′-SL, 3′-sialyllactose; LNFP, lacto-N-fucopentaose; LNH, lacto-N-hexaose; LNnT; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose; LST, sialyllacto-N-tetraose; NEC, necrotising enterocolitis.
Given that lower DSLNT was associated with NEC independent of secretor status, the utility of this HMO as a biomarker for NEC development was explored. Univariate ROC curve analysis determined that 241 nmol/mL (or 310.93 μg/mL) was the optimal DSLNT concentration in MOM for distinguishing NEC and control infants (figure 1C,D). At this threshold, the area under the curve (AUC) was 0.947, with a sensitivity of 0.9 and a specificity of 0.9, correctly identifying 91% of infants with NEC (below threshold) and 86% of control healthy infants (above threshold).
To test if integration of additional HMOs could improve the classification performance, multivariate ROC curves built on increasing number of HMOs were performed (online supplemental figure 5A). Inclusion of two HMOs (the minimum in multivariate analysis) resulted in the optimal performance, with DSLNT being selected as a discriminatory feature in 100% of permutations (online supplemental figure 5B). 3′-Fucosyllactose (3′-FL) and lacto-N-neotetraose (LNnT) were the second and third most selected features, with a selection frequency of around 30%, being more abundant in cases of NEC. However, the integration of any additional HMOs to DSLNT in the multivariate model resulted in similar performance compared with the univariate model using DSLNT only (AUCs of 0.946 and 0.947, respectively).
To validate the 241 nmol/mL threshold defined in the current study in an independent cohort, we analysed data from Autran et al,17 which contained 8 NEC and 40 matched control infants. Since this study included temporal sampling before disease, we selected the nearest milk sample to NEC onset for each infant and matched the control samples by sample DOL and included only DSLNT concentration. Using a DSLNT threshold of 241 nmol/mL, we found that the MOM sample for 100% (8/8) infants with NEC fell under the threshold, while 60% (24/40) control samples had a DSLNT concentration above 241 nmol/mL (online supplemental figure 5C).
Analysis of HMO profiles stratified by NEC type
We compared medically managed NEC (NEC-M), where infants did not undergo surgery or die from NEC (ie, had less severe disease) with infants with NEC that underwent surgery (NEC-S). NEC-M and NEC-S clustered together and were distinct from matched controls (figure 2A; 2000 permutations, p<0.001). Two HMOs were found to be significantly different, with DSLNT lower in MOM in both NEC-M (adj. p<0.001) and NEC-S (adj. p<0.001) compared with controls (figure 2B). In addition, LNnT in MOM was significantly lower in NEC-S in comparison to both NEC-M (adj. p=0.0016) and matched controls (adj. p=0.0423) (figure 2C).
Figure 2.
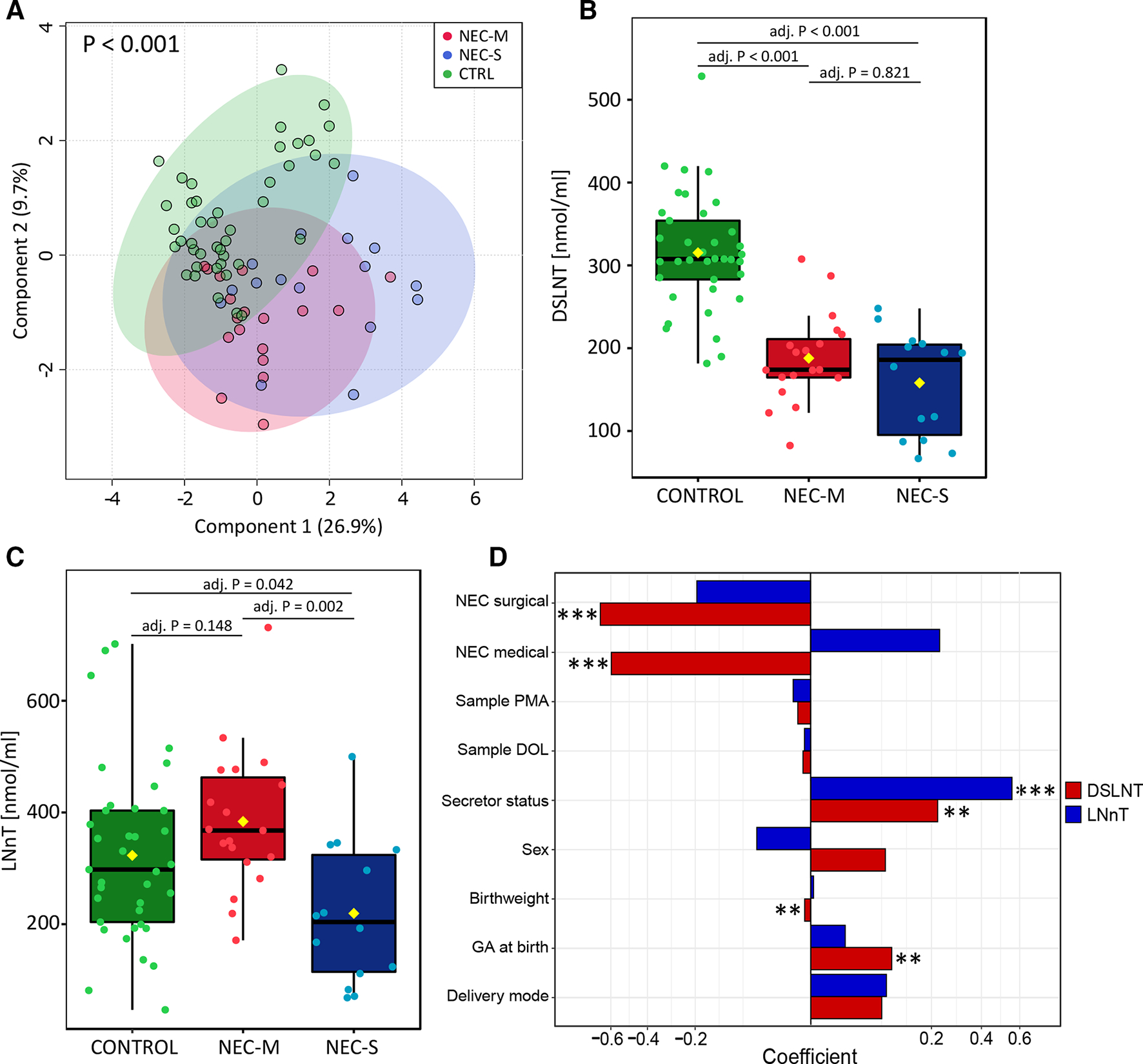
Analysis of HMO profiles with stratification of NEC-M and NEC-S. (A) Partial least squares discriminant analysis of HMO profiles from CTRL, NEC-M and NEC-S infants. NEC-M and NEC-S cluster together and separately from CTRLs (p<0.001). P values were calculated based on 2000 permutations. Box plots of (B) DSLNT and (C) LNnT concentration between CTRL, NEC-M and NEC-S infants. Kruskal-Wallis followed by Dunn’s test using Bonferroni adjustment was applied. (D) Adjusted linear regression model for DSLNT and LNnT including potential clinical confounders. P values were corrected by false discovery rate (FDR). Significant variables are indicated by asterisks: *** denotes FDR p<0.001; ** denotes FDR p>0.01. CTRL, control; DOL, day of life; DSLNT, disialyllacto-N-tetraose; GA, gestational age; HMO, human milk oligosaccharide; LNnT, lacto-N-neotetraose; NEC, necrotising enterocolitis; NEC-M, medically managed NEC; NEC-S, ninfants with NEC that underwent surgery; PMA postmenstrual age.
We subsequently investigated the potential association between DSLNT and LNnT concentrations and clinical variables by applying an adjusted linear model. DSLNT was negatively correlated to both disease types, with coefficients equal to −0.60 for NEC-M (adj. p<0.001) and −0.67 for NEC-S (adj. p<0.001) (figure 2D). However, LNnT was not associated with disease type following adjusted linear modelling (both adj. p>0.05). DSLNT and LNnT were both significantly higher in secretor mothers (adj. p=0.008, adj. p<0.001, respectively). DSLNT in MOM also positively correlated to gestational age (adj. p=0.008) and negatively to birth weight (adj. p=0.008). Neither HMO correlated to sex, delivery mode, postmenstrual age or DOL of the MOM sample (figure 2D and online supplemental figure 6).
Association between infant gut microbiome and development of NEC
We included stool microbiome data on a subset of infants with HMO data, where metagenomic sequencing data were available through an ongoing independent study (the results of which are not yet published). This included 644 stool samples from 34 controls and 14 infants with NEC (online supplemental table 3). To overcome challenges of repeated measures and to compare results with existing published work, we first analysed one stool sample per infant closest to NEC onset (median of 3 days before NEC) and a corresponding control sample matched by DOL (online supplemental figure 1). This cross-sectional analysis showed infants with NEC had significantly lower richness (p=0.027) but comparable Shannon diversity (p=0.443, figure 3A). Bray-Curtis Principal Coordinates Analysis (PCoA) showed no significant difference between the bacterial profiles of NEC and controls (PERMANOVA p=0.182, figure 3B). Analysis at the phylum level showed significantly lower relative abundance of Actinobacteria (adj. p=0.034) and higher relative abundance of Proteobacteria (adj. p=0.034) in infants with NEC (figure 3C). Correspondingly, at the species level, infants with NEC had lower relative abundance of B. longum (adj. p=0.012) and higher relative abundance of Enterobacter cloacae (adj. p=0.012) compared with controls (figure 3D).
Figure 3.
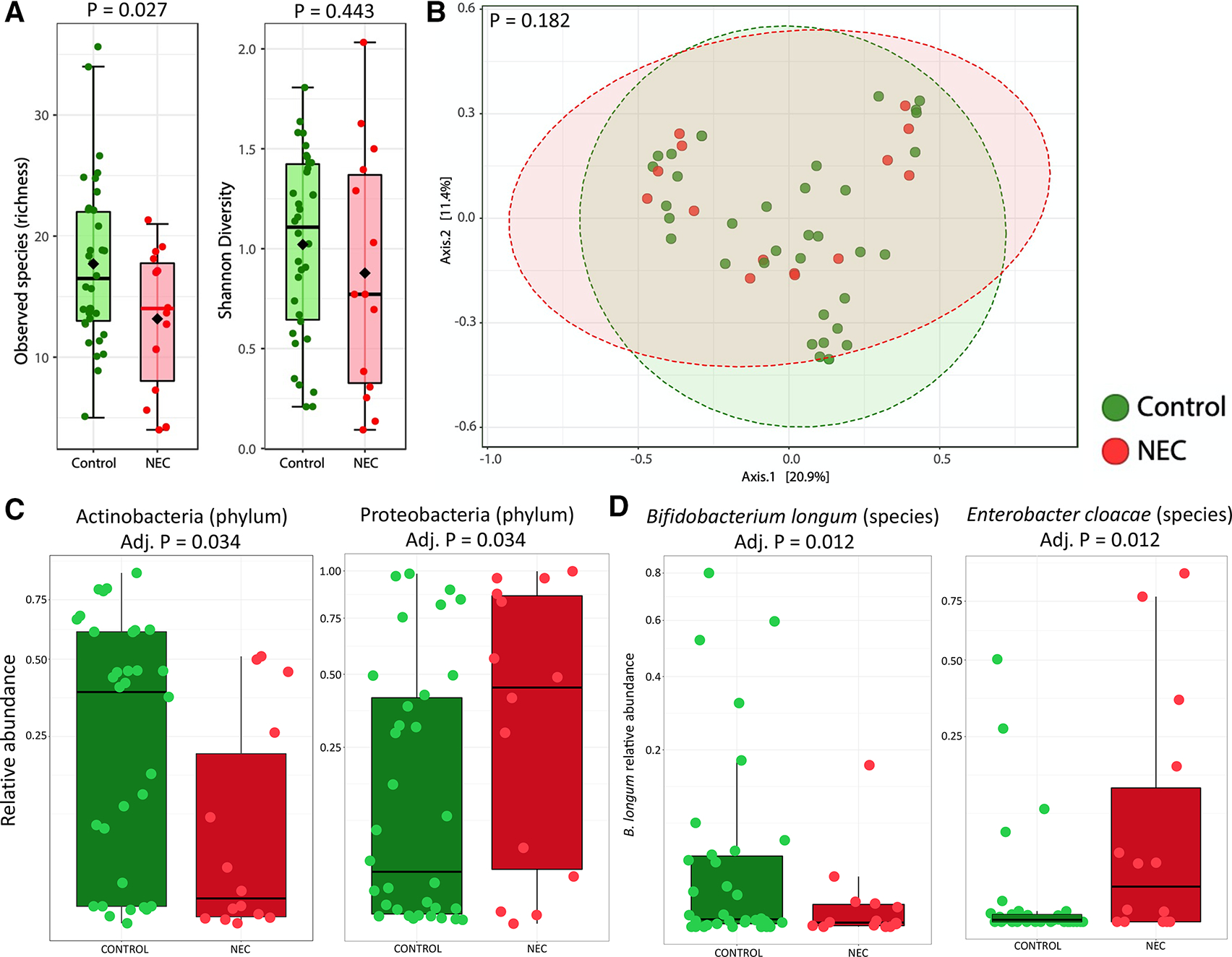
Cross-sectional analysis of preterm stool metagenome profiles between NEC and matched controls. Analysis includes the sample closest NEC onset (median of 3 days prior to NEC) and a corresponding control sample matched by day of life. (A) Alpha diversity based on observed species (richness) and Shannon diversity. (B) Bray-Curtis principal coordinate analysis. (C) Box plots showing the relative abundance of significant phyla. (D) Box plots showing the relative abundance of significant species. NEC, necrotising enterocolitis.
Integrated analysis of HMO and bacterial profiles
DMM clustering was used to determine PGCTs using species-level data, and five PCGTs were deemed optimal (figure 4A). PGCT-1 was characterised by high relative abundance of Staphylococcus spp and Enterococcus faecalis, PGCT-2 had high Escherichia spp; PGCT-3 had high Klebsiella spp; and PGCT-4 and PGCT-5 had high Bifidobacterium spp, with B. breve notably high in PGCT-5. Using the PGCT clusters, we analysed the temporal transition of an infant’s gut microbiome over the first 70 days of life by defining distinct time points and including only one sample per infant at each time point. Based on the distribution of samples across all time points and all clusters, the temporal transition of the microbiome over the first 70 days of life was significantly different in infants in receipt of MOM below the DSLNT threshold of 241 nmol/mL compared with infants above the DSLNT threshold (χ2 test, p<0.001; figure 4B).
Figure 4.
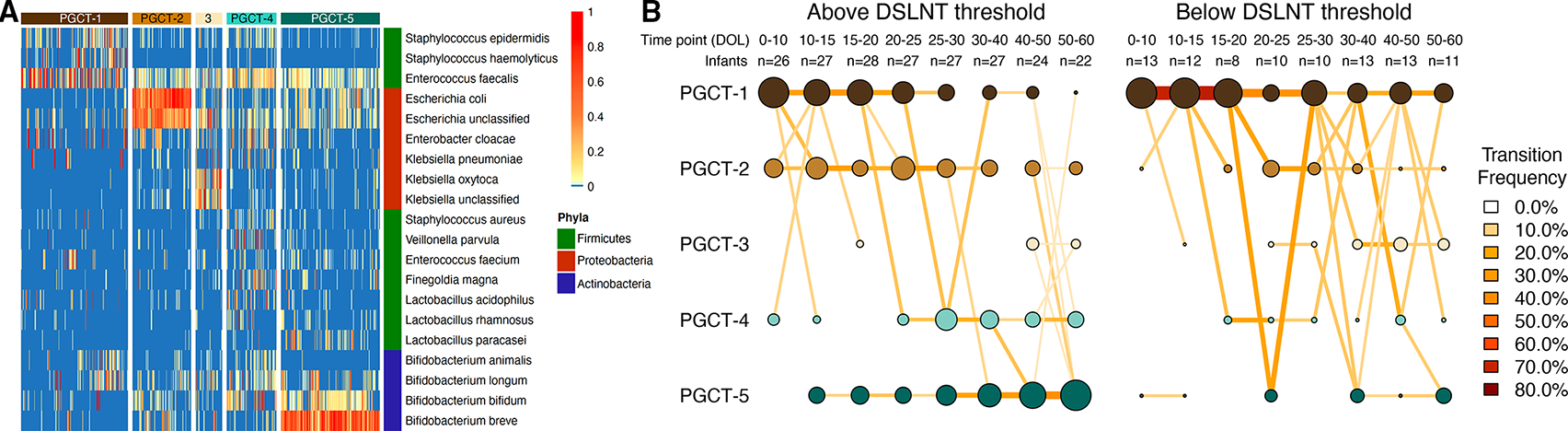
Analysis of PGCTs by infants receiving maternal milk above or below the 241 nmol/mL DSLNT threshold. The entire dataset of 644 samples formed five distinct clusters based on lowest Laplace approximation following Dirichlet multinomial clustering. (A) Heatmap showing the relative abundance of dominant bacterial species within each PGCT cluster. The phyla for each species are also shown. (B) Transition model showing the progression of samples through each PGCT, from day of life 0–60 across eight distinct time points. Plots are separated based on whether the concentration of DSLNT in maternal milk was above or below the 241 nmol/mL threshold. Nodes and edges are sized based on the total counts. Nodes are coloured according to Dirichlet Multinomial Mixtures (DMM) cluster number and edges are coloured by the transition frequency. Transitions with less than 5% frequency are not shown. DSLNT, disialyllacto-N-tetraose; PGCT, preterm gut community type.
The PGCTs were named according to the average age of samples within that cluster, where PGCT-1 contained on average the earliest samples and PGCT-5 on average the latest samples. We compared the number of samples from all time points in only PGCT-1 and PGCT-5 to investigate associations between the MOM DSLNT threshold and gut microbiome development from the typically younger to the typically older PGCTs. Infants receiving MOM with DSLNT level below 241 nmol/mL had significantly more samples remaining within PGCT-1 throughout all time points (78% in PCGT-1 vs 22% in PGCT-5, χ2 test p<0.001), whereas infants receiving MOM with DSLNT above this threshold transitioned from PGCT-1 to PGCT-5 as demonstrated by a similar number of samples in each PGCT across all time points (48% in PCGT-1 vs 52% in PGCT-5, χ2 test p=0.717). In addition to comparing samples from all time points, we next compared samples from the final time point only (ie, DOL 50–60). After correcting for uneven frequency of sampling between groups, at the final time point, infants receiving MOM above the DSLNT threshold were twice as likely to be in PGCT-5 (3/11 samples below vs 12/22 samples above DSLNT threshold; OR 3.20, 95% CI 0.6657 to 15.3819), which was characterised by high relative abundance of Bifidobacterium (figure 4A).
Explained variance and random forest classification of HMO and metagenome data
Using the cross-sectional HMO and cross-sectional metagenome dataset, we sought to determine which clinical factors were most associated with the HMO and the bacterial profiles (figure 5A). Secretor status explained 56% of the variance within HMO profiles (adj. p<0.001), but no other covariate was significantly associated with the HMO profiles. In contrast, the bacterial profiles were significantly associated with both postmenstrual age (R2 0.07, adj. p=0.006) and DOL (R2 0.07, adj. p=0.006), as well as receipt of antibiotics at the time of sampling (R2 0.06, adj. p=0.006) and receipt of probiotics (R2 0.12, adj. p=0.006), but not maternal secretor status (R2 0.02, adj. p=0.58). Together, these findings highlight that HMO and bacterial profiles are influenced by numerous non-overlapping factors related to early life in preterm infants.
Figure 5.
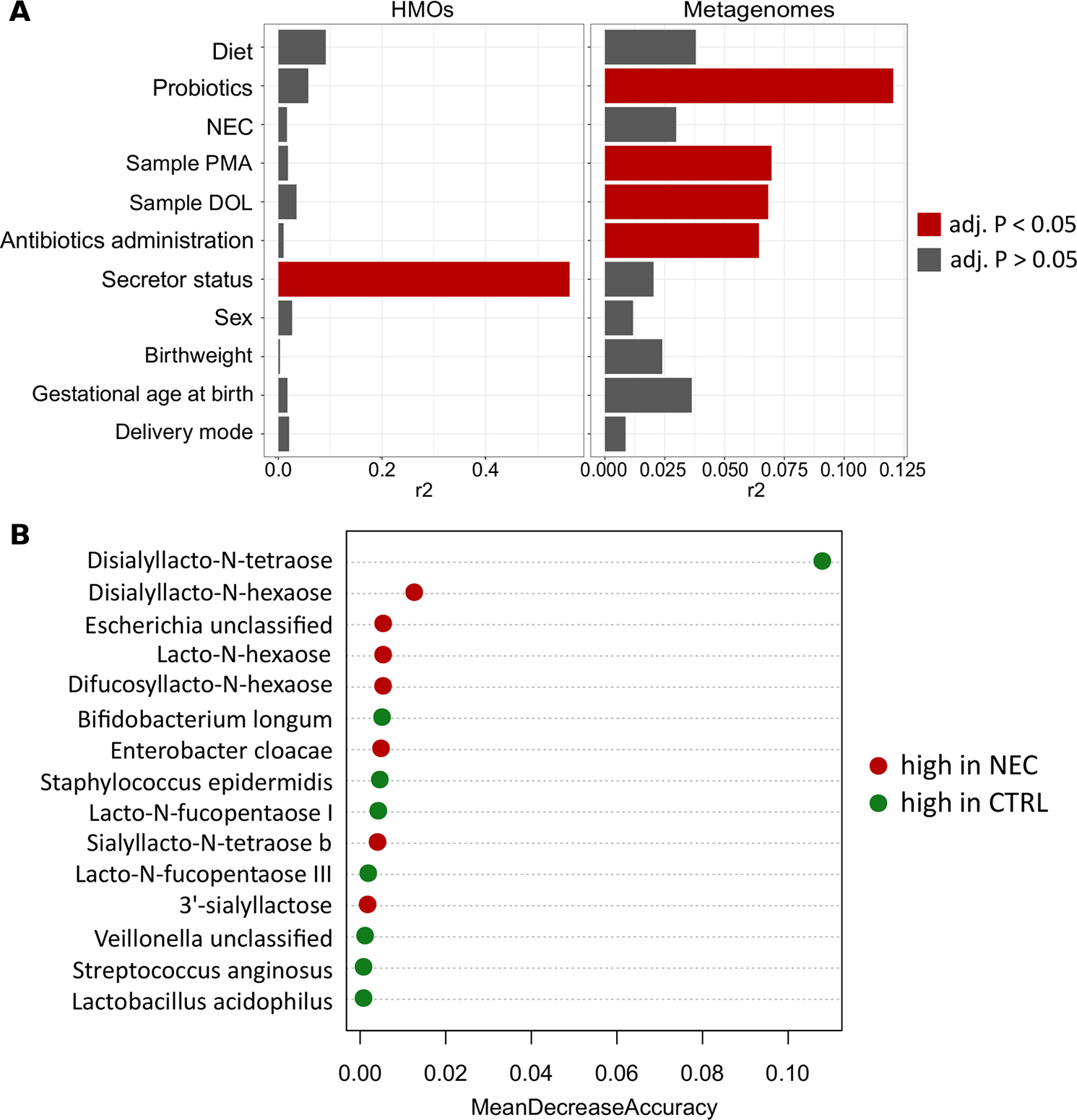
Modelling of cross-sectional HMO and infant stool metagenomic profiles using Adonis and random forest. (A) Horizontal bar plots showing the variance (r2) in maternal HMO and infant stool metagenomic profiles explained by clinical covariates as modelled by univariate Adonis. Variables with a false discovery rate p value of <0.05 are shown in red. (B) Feature importance from combined HMO and metagenome random forest classification model. Mean decrease accuracy value defines the contribution given by a certain feature to classification process. CTRL, control; DOL, day of life; HMO, human milk oligosaccharide; NEC, necrotising enterocolitis; PMA, postmenstrual age.
We compared the performance of random forest classification models built on the cross-sectional subset of HMO profile data, metagenomic sequencing data and the two datasets combined to classify an infant as NEC or healthy, given that all this information is available before onset of disease and could therefore function as a risk stratification system in clinical practice. The HMO profile alone had a classification error of 0.146, with 21% (3/14) NEC and 12% (4/34) control infants misclassified. DSLNT had the greatest contribution to classification with a mean decrease accuracy (MDA) of 0.11. Other HMOs contributing to classification accuracy included lacto-N-hexaose (LNH; MDA=0.012) and difucosyllacto-N-hexaose (DFLNH; MDA=0.011), which were non-significantly higher in infants with NEC. Random forest generated using the metagenomic sequencing data was characterised by a classification error of 0.229, with 43% (6/14) NEC and 15% (5/34) control infants misclassified. E. cloacae was the most important feature guiding the classification (MDA=0.036), with higher relative abundance in infants with NEC, followed by B. bifidum (MDA=0.024) and B. longum (MDA=0.013), which had higher relative abundance in control infants. Combining HMO and metagenome datasets slightly improved the performance compared with using HMOs alone, with 21% (3/14) of infants with NEC and (9%) 3/34 controls misclassified. In this combined model, DSLNT was enriched in controls, and difucosyllacto-N-tetraose (DSLNH) and the relative abundance of Escherichia unclassified were higher in infants with NEC (figure 5B).
DISCUSSION
Receipt of human breast milk and early life gut microbiome development are intrinsically linked and both influence the risk of NEC in preterm infants. Our study represents the largest analysis of HMOs in NEC and the first to integrate HMO and metagenome data. We found DSLNT was present in significantly lower concentrations in MOM fed to infants diagnosed with NEC. Furthermore, lower DSLNT concentrations in MOM were associated with reduced transition into PGCTs typically observed in older infants and lower relative abundance of Bifidobacterium spp.
The HMO results from the current study build on previous findings in humans, showing reduced DSLNT in MOM received by infants developing NEC, independent of maternal secretor status.17–19 This is also supported by rodent studies where total and individual HMOs including 2′-FL and DSLNT have shown a protective effect against NEC development.16 36 37 However, 2′-FL and mixtures of HMOs (one of which included DSLNT) did not show any protection in NEC piglet models.38 39 Importantly from a clinical perspective, in rats, the protection provided by pooled HMOs could be reproduced with DSLNT alone, with specific dependence on its precise structure since closely related sialyllacto-N-tetraose (identical in structure to DSLNT but lacking one sialic acid residue) did not provide protection, suggesting a highly structure-specific mechanism.16 Our findings further extend the evidence for the specificity of DSLNT in the NEC pathway. A threshold level of DSLNT (241 nmol/mL) from a single MOM sample correctly identified 91% of infants with NEC (below threshold) and 86% of control healthy infants (above threshold). Of the three infants who developed NEC despite a DSLNT above the threshold, two had not received MOM in the 3 weeks prior to disease onset, and the remaining infant had a DSLNT concentration of 248 nmol/mL.
Within the validation dataset,17 100% of infants with NEC were correctly classified, but only 60% of controls. Making a robust diagnosis of NEC is difficult, and it is possible that the specific threshold value of DSLNT we identified will have a different predictive value in other populations or where other criteria are used to determine the presence of disease. Our study contains a large number of cases coded clinically as NEC independently validated by blinded review. In addition, our cohort was more homogenous (predominantly white Caucasian) and the concentration of DSLNT was less variable (current study IQR 184–321 nmol/mL vs Autran et al IQR 122–346 nmol/mL) despite using the same analytical platform. Given HMO composition and DSLNT concentrations may be influenced by genetic factors, geographical location, ethnicity40 and seasonality,15 differential thresholds may improve diagnostic performance in other settings. Taken together, this external validation and potential variation in DSLNT concentration by maternal factors underscore the need for large multicentre studies to both refine a universal or stratified threshold for DSLNT concentration in predicting NEC and potentially prospectively identifying milk samples that may benefit from supplementation with synthetically produced DSLNT.
In addition to HMO profiles, our extensive longitudinal stool metagenomic analysis represents one of the largest datasets to date. This extends our previous work,20 33 41 where DMM was used to facilitate analysis of temporal microbiome development and to integrate the HMO DSLNT threshold of 241 nmol/mL with infant gut microbiome profiles. We observed a difference in microbiome development between DSLNT groups, with infants receiving MOM with lower DSLNT tending to have delayed progression into the PGCT typically expected in older infants (ie, PGCT-5). This supports the theory that concentrations of specific HMOs in MOM are associated with differences in gut microbiome development. On the contrary, transition into PGCT-5 was twice as likely in infants receiving MOM with DSLNT above the threshold, which was characterised by high relative abundance of Bifidobacterium spp. Bifidobacterium has previously been linked to health in preterm infants,20 41 42 and our current findings in pre-NEC samples further support the association of reduced Bifidobacterium spp, specifically B. longum, as a risk factor for NEC. In addition, our species-level metagenome data advanced previous associations of Enterobacteriaceae with NEC,21 23 43 showing E. cloacae relative abundance was higher before NEC.
Random forest analysis confirmed the capability of HMO profiles to identify infants who developed NEC and slightly outperformed metagenome profiles by correctly classifying three more NEC cases and one more control. Combining HMO and metagenome data before disease accurately classified 87.5% of infants as healthy or having NEC, with DSLNT and the bacterial species identified as important in the random forest analysis being comparable to the unsupervised analysis in the current study and in previous studies. Further work is needed to determine if DSLNT functions via modulation of the microbiome or by acting directly on the host, such as acting in a structure-specific receptor-mediated way to alter immune functioning and to reduce inflammation leading to necrosis. In the event of the latter, a microbial community with less use of DSLNT could provide an advantage to reducing NEC risk. Taken together, the current findings and recent work highlighting the ability of Bifidobacterium spp to use HMOs is strain specific7 8 underscore the need for further research to better understand the complexity of human milk and other nutritional exposures, including the use of supplements such as prebiotics and probiotics in preterm infants. In addition to therapeutics, the classifiers may provide a basis for the development of biomarkers predicting NEC risk. While additional work is needed, the addition of microbial biomarkers may allow for the most accurate predictions and could inform NEC risk for infants where MOM (and thus HMO information) is not available.
This study involved the largest cohort to date investigating the relationship between HMO composition and NEC development, and includes one of the most extensive longitudinal stool metagenomic analyses of preterm infants. However, there are several limitations and avenues for future work. First, the cross-sectional HMO profiling data precluded assessment of changes within mothers over time and how this may relate to NEC development. The milk sample was selected based on the day of infant feeding, and the actual expression of milk may have occurred several days earlier, which may be important clinically. Current published data suggest that the concentration of HMOs, including DSLNT, is relatively stable over time,14 but validation in longitudinal preterm cohorts is needed. Second, the amount of MOM an infant receives and tolerates each day is variable, and DSLNT exposure is dependent on both concentration and volume. Although this study identifies DSLNT concentration alone may be useful from both a diagnostic and therapeutic perspective, further studies could consider the volume of milk received in addition to concentration. Third, inclusion of metagenome data was opportunistic based on available data, and cost prohibited sequencing all infants in the cohort. As such, the classification accuracy of the model might be impacted by the reduced sample size in comparison to the full cohort, necessitating the need for follow-up analyses in larger cohorts. Despite this, the sample size of 644, including 195 samples from 14 preterm infants who developed NEC, makes this dataset one of the largest published to date. Finally, the gene relative abundance data warrant further investigation, in combination with other experimental approaches, to help inform the HMO use capacity of different strains.
In summary, HMO profiling of MOM coupled to metagenomic sequencing of preterm stool showed that the concentration of a single HMO, DSLNT, was lower in milk received by infants who developed NEC. The lower concentration of DSLNT was associated with altered microbiome development, specifically a reduced progression toward the PGCT typically found in the older infants, which was abundant in Bifidobacterium spp. These results suggest MOM HMO profiling may provide potential targets for biomarker development and disease risk stratification. They may also guide focused donor milk use (eg, prioritise high DSLNT for preterm infants) and novel avenues for supplements that may prevent life-threatening disease.
Supplementary Material
Significance of this study.
What is already known on this subject?
Necrotising enterocolitis (NEC) is one of the leading causes of death in preterm infants.
Maternal human milk oligosaccharides (HMOs), including disialyllacto-N-tetraose (DSLNT), have been associated with protection from NEC development.
Differences in infant gut microbiome development have been linked to infants with NEC and without NEC, but the causative and protective organisms have not been determined.
What are the new findings?
We found for the first time that combined analysis of maternal HMOs and infant gut microbiome can predict NEC.
A specific DSLNT threshold level of 241 nmol/mL had a sensitivity and specificity of 0.9 for NEC and infants receiving milk below this threshold showed abnormal microbiome development.
Infants who developed NEC had significantly lower relative abundance of Bifidobacterium longum and significantly higher relative abundance of Enterobacter cloacae before disease diagnosis.
How might it impact on clinical practice in the foreseeable future?
Our findings demonstrate the importance of maternal HMOs and infant gut microbiome in preterm infants, providing targets for biomarker development, disease risk stratification and novel avenues for supplementing the infant feed.
Acknowledgements
The authors wish to thank the neonatal intensive care staff involved in the sample collection. We are grateful to the families for their willingness to help and support research.
Funding
This work was supported by the MRC Discovery Medicine North Doctoral Training Partnership (to AM and CS), the Newcastle University academic career track scheme (to CS) and Astarte Medical for funding stool sample retrieval and shipment for metagenomic sequencing (to CS). The funders played no part in the study design, analysis, interpretation or reporting.
Footnotes
Competing interests CS declares performing consultancy for Astarte Medical and honoraria from Danone Early Life Nutrition. NDE declares research funding from Prolacta Biosciences US and Danone Early Life Nutrition, and received lecture honoraria from Baxter and Nestle Nutrition Institute, but has no share options or other conflicts. LB is UC San Diego Chair of Collaborative Human Milk Research, endowed by the Family Larsson-Rosenquist Foundation and serves on the foundation’s scientific advisory board. LB is coinventor on patent applications regarding human milk oligosaccharides in prevention of necrotising enterocolitis and other inflammatory disorders. The other authors declare that they have no competing interests.
Patient consent for publication Not required.
Ethics approval Ethics approval was obtained from the County Durham and Tees Valley Research Ethics Committee (REC10/H0908/39) and the parents gave informed consent for stool and data collection.
Provenance and peer review Not commissioned; externally peer reviewed.
Data availability statement Data are available in a public, open access repository. All sequencing data generated and analysed in this study have been deposited in the European Nucleotide Archive under study accession number PRJEB39610. The metagenomic data are publically available and can be accessed online (https://www.ebi.ac.uk/ena/browser/view/PRJEB39610). HMO data are avilable upon request.
Supplemental material This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
REFERENCES
- 1.Masi AC, Stewart CJ. The role of the preterm intestinal microbiome in sepsis and necrotising enterocolitis. Early Hum Dev 2019;138:104854. [DOI] [PubMed] [Google Scholar]
- 2.Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med 2011;364:255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meinzen-Derr J, Poindexter B, Wrage L, et al. Role of human milk in extremely low birth weight infants’ risk of necrotizing enterocolitis or death. J Perinatol 2009;29:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan S, Schanler RJ, Kim JH, et al. An exclusively human milk-based diet is associated with a lower rate of necrotizing enterocolitis than a diet of human milk and bovine milk-based products. J Pediatr 2010;156:562–7. [DOI] [PubMed] [Google Scholar]
- 5.Bode L. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 2012;22:1147–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vatanen T, Franzosa EA, Schwager R, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018;562:589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawson MAE, O’Neill IJ, Kujawska M, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. Isme J 2020;14:635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gotoh A, Katoh T, Sakanaka M, et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci Rep 2018;8:13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ackerman DL, Doster RS, Weitkamp J-H, et al. Human milk oligosaccharides exhibit antimicrobial and antibiofilm properties against group B Streptococcus. ACS Infect Dis 2017;3:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bode L, Kunz C, Muhly-Reinholz M, et al. Inhibition of monocyte, lymphocyte, and neutrophil adhesion to endothelial cells by human milk oligosaccharides. Thromb Haemost 2004;92:1402–10. [DOI] [PubMed] [Google Scholar]
- 11.Wu RY, Li B, Koike Y, et al. Human milk oligosaccharides increase mucin expression in experimental necrotizing enterocolitis. Mol Nutr Food Res 2019;63:e1800658. [DOI] [PubMed] [Google Scholar]
- 12.Gustafsson A, Hultberg A, Sjöström R, et al. Carbohydrate-Dependent inhibition of Helicobacter pylori colonization using porcine milk. Glycobiology 2006;16:1–10. [DOI] [PubMed] [Google Scholar]
- 13.Ramani S, Stewart CJ, Laucirica DR, et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat Commun 2018;9:5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Austin S, De Castro CA, Sprenger N, et al. Human milk oligosaccharides in the milk of mothers delivering term versus preterm infants. Nutrients 2019;11. doi: 10.3390/nu11061282. [Epub ahead of print: 05 Jun 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azad MB, Robertson B, Atakora F, et al. Human milk oligosaccharide concentrations are associated with multiple fixed and modifiable maternal characteristics, environmental factors, and feeding practices. J Nutr 2018;148:1733–42. [DOI] [PubMed] [Google Scholar]
- 16.Jantscher-Krenn E, Zherebtsov M, Nissan C, et al. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 2012;61:1417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Autran CA, Kellman BP, Kim JH, et al. Human milk oligosaccharide composition predicts risk of necrotising enterocolitis in preterm infants. Gut 2018;67:1064–70. [DOI] [PubMed] [Google Scholar]
- 18.Van Niekerk E, Autran CA, Nel DG, et al. Human milk oligosaccharides differ between HIV-infected and HIV-uninfected mothers and are related to necrotizing enterocolitis incidence in their preterm very-low-birth-weight infants. J Nutr 2014;144:1227–33. [DOI] [PubMed] [Google Scholar]
- 19.Hassinger D, Clausen DM, Nitka S, et al. Analysis of Disialyllacto-N-Tetraose (DSLNT) content in milk from mothers of preterm infants. J Hum Lact 2020;890334420904041. [DOI] [PubMed] [Google Scholar]
- 20.Stewart CJ, Embleton ND, Marrs ECL, et al. Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome 2016;4:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sim K, Shaw AG, Randell P, et al. Dysbiosis Anticipating necrotizing enterocolitis in very premature infants. Clin Infect Dis 2015;60:389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward DV, Scholz M, Zolfo M, et al. Metagenomic Sequencing with Strain-Level Resolution Implicates Uropathogenic E. coli in Necrotizing Enterocolitis and Mortality in Preterm Infants. Cell Rep 2016;14:2912–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrow AL, Lagomarcino AJ, Schibler KR, et al. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 2013;1:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart CJ, Fatemizadeh R, Parsons P, et al. Using formalin fixed paraffin embedded tissue to characterize the preterm gut microbiota in necrotising enterocolitis and spontaneous isolated perforation using marginal and diseased tissue. BMC Microbiol 2019;19:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olm MR, Bhattacharya N, Crits-Christoph A, et al. Necrotizing enterocolitis is preceded by increased gut bacterial replication, Klebsiella, and fimbriae-encoding bacteria. Sci Adv 2019;5:eaax5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bode L, Kuhn L, Kim H-Y, et al. Human milk oligosaccharide concentration and risk of postnatal transmission of HIV through breastfeeding. Am J Clin Nutr 2012;96:831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong DT, Franzosa EA, Tickle TL, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 2015;12:902–3. [DOI] [PubMed] [Google Scholar]
- 28.Xia J, Sinelnikov IV, Han B, et al. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res 2015;43:W251–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B 1995;57:289–300. [Google Scholar]
- 30.Chong J, Liu P, Zhou G, et al. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc 2020;15:799–821. [DOI] [PubMed] [Google Scholar]
- 31.Dhariwal A, Chong J, Habib S, et al. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 2017;45:W180–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One 2012;7:e30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart CJ, Embleton ND, Clements E, et al. Cesarean or vaginal birth does not impact the longitudinal development of the gut microbiome in a cohort of exclusively preterm infants. Front Microbiol 2017;8:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018;562:583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jari Oksanen FGB, Friendly M, Kindt R, et al. Vegan: community ecology package. R package version 2.5–6, 2019. Available: https://CRAN.R-project.org/package=vegan [Google Scholar]
- 36.Good M, Sodhi CP, Yamaguchi Y, et al. The human milk oligosaccharide 2′-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br J Nutr 2016;116:1175–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Autran CA, Schoterman MHC, Jantscher-Krenn E, et al. Sialylated galacto-oligosaccharides and 2′-fucosyllactose reduce necrotising enterocolitis in neonatal rats. Br J Nutr 2016;116:294–9. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen SO, Martin L, Østergaard MV, et al. Human milk oligosaccharide effects on intestinal function and inflammation after preterm birth in pigs. J Nutr Biochem 2017;40:141–54. [DOI] [PubMed] [Google Scholar]
- 39.Cilieborg MS, Bering SB, Østergaard MV, et al. Minimal short-term effect of dietary 2’-fucosyllactose on bacterial colonisation, intestinal function and necrotising enterocolitis in preterm pigs. Br J Nutr 2016;116:834–41. [DOI] [PubMed] [Google Scholar]
- 40.McGuire MK, Meehan CL, McGuire MA, et al. What’s normal? oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 2017;105:1086–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart CJ, Embleton ND, Marrs ECL, et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 2017;5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torrazza RM, Ukhanova M, Wang X, et al. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PLoS One 2013;8:e83304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mai V Young CM, Ukhanova M, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One 2011;6:e20647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


