Significance
The zygote is a totipotent structure that develops into an embryo with all of the cells needed to produce an entire plant. The BABY BOOM (BBM) transcription factor induces spontaneous asexual embryo development on plant organs when ectopically expressed. Although BBM is at the top of a transcriptional network that promotes asexual embryo development, little is known about its expression and role during zygotic embryogenesis. Here we show in Arabidopsis that BBM regulates the progression of zygotic embryo development and embryo patterning, and division and cellularization of the filial endosperm. In line with its role as a totipotency factor, ectopic BBM expression in the egg cell is also sufficient to induce haploid embryo development in Arabidopsis and dicot crops.
Keywords: Arabidopsis, BABY BOOM, endosperm, parthenogenesis, zygotic embryo
Abstract
The BABY BOOM (BBM) AINTEGUMENTA-LIKE (AIL) AP2/ERF domain transcription factor is a major regulator of plant cell totipotency, as it induces asexual embryo formation when ectopically expressed. Surprisingly, only limited information is available on the role of BBM during zygotic embryogenesis. Here we reexamined BBM expression and function in the model plant Arabidopsis thaliana (Arabidopsis) using reporter analysis and newly developed CRISPR mutants. BBM was expressed in the embryo from the zygote stage and also in the maternal (nucellus) and filial (endosperm) seed tissues. Analysis of CRISPR mutant alleles for BBM (bbm-cr) and the redundantly acting AIL gene PLETHORA2 (PLT2) (plt2-cr) uncovered individual roles for these genes in the timing of embryo progression. We also identified redundant roles for BBM and PLT2 in endosperm proliferation and cellularization and the maintenance of zygotic embryo development. Finally, we show that ectopic BBM expression in the egg cell of Arabidopsis and the dicot crops Brassica napus and Solanum lycopersicon is sufficient to bypass the fertilization requirement for embryo development. Together these results highlight roles for BBM and PLT2 in seed development and demonstrate the utility of BBM genes for engineering asexual embryo development in dicot species.
APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) proteins comprise a large family of plant transcription factors with diverse functions in plant stress response and development (1). The AINTEGUMENTA-LIKE (AIL) clade of AP2 proteins comprises two subclades, one subclade with AINTEGUMENTA (ANT)/AIL1-like proteins and one subclade with BABY BOOM (BBM)-like proteins, including the PLETHORA (PLT) proteins (2). AIL proteins have individual and redundant functions during development, including above- and below-ground meristem initiation, maintenance, and differentiation (3, 4).
BBM is expressed in the embryo and root in Arabidopsis (5), where it regulates development of these structures together with other PLT genes (3). After fertilization, BBM is expressed in the zygotic embryo as early as the four-cell embryo stage (6). BBM is initially expressed throughout the embryo proper and suspensor and then becomes restricted to the embryo root pole around the heart/torpedo stage (5, 6). Mutant phenotypes were not reported in single bbm transfer DNA (T-DNA) insertion mutants, but double bbm plt2 mutant combinations were reported to be embryo-lethal (5), suggesting that BBM and PLT2 have shared roles in embryo development.
Ectopic BBM expression is sufficient to induce asexual embryo development (7, 8), and this trait has been widely exploited to improve plant transformation and clonal propagation (9). BBM expression must therefore be tightly regulated during sexual reproduction to prevent parthenogenesis, the development of the egg cell into an embryo in the absence of fertilization. However, egg cell parthenogenesis is a naturally occurring component of asexual seed reproduction pathways in apomictic plants (10), and BBM-like genes map to the parthenogenesis locus in a number of apomictic grasses, including Pennisetum squalatum and Brachiaria spp. (11–13). In apomictic P. squalatum ASGR-BBML is expressed in the egg cell (14) and expression of ASGR-BBML under its own promoter or an egg-cell-expressed promoter can induce parthenogenesis in sexual grasses and tobacco (11, 15, 16). This trait is not linked to the “apomictic” BBM allele per se but rather to the timing of BBM expression, as heterochronic expression of a (sexual) rice (Oryza sativa) BBM allele in the egg cell is also sufficient to induce parthenogenesis in rice (17). Thus, heterochronic differences in BBM expression can drive both sexual and asexual reproductive pathways.
Here we show that the Arabidopsis BBM gene has a much broader expression pattern and function during seed development than previously reported. BBM is expressed transiently in the chalazal region of the ovule and seed, in the embryo starting from the zygote stage, and in the first few dividing endosperm cells. These expression patterns were reflected in novel BBM functions in endosperm and embryo development that were shared with PLT2. We also show that ectopic BBM expression in the egg cell induces parthenogenesis, a trait that can be enhanced by dmp maternal haploid inducers (18–20). Our findings consolidate previous research showing a role for BBM overexpression in embryo identity induction in somatic tissues with a similar role for BBM during zygotic embryogenesis, and uncover an unexpected role for BBM as a driver of endosperm development in plants.
Results
BBM Is Expressed in the Zygote and the Filial and Maternal Seed Compartments.
In maize and rice, BBM-like genes are expressed in the zygote shortly after fertilization and in the sperm cell before fertilization (21, 22). BBM expression has not been examined in Arabidopsis gametes and the early stages of zygotic embryogenesis, and therefore we made use of available BBM:BBM-GFP (6) and newly developed BBM:BBM-GFP-GUS reporters to examine BBM expression pre- and postfertilization.
First, we determined whether Arabidopsis BBM is expressed in the mature male and female gametophyte. We were unable to detect BBM expression in either the pollen or the egg cell using translational BBM:BBM-GFP-GUS or BBM:BBM-GFP reporter lines (Fig. 1 A and B and SI Appendix, Fig. S1A). However, we did detect weak BBM expression in the chalazal region of a small proportion of ovules starting at stage 12 of flower development (Fig. 1B and SI Appendix, Fig. S1A). These results suggest that BBM is only expressed detectably before fertilization in the sporophytic ovule tissue.
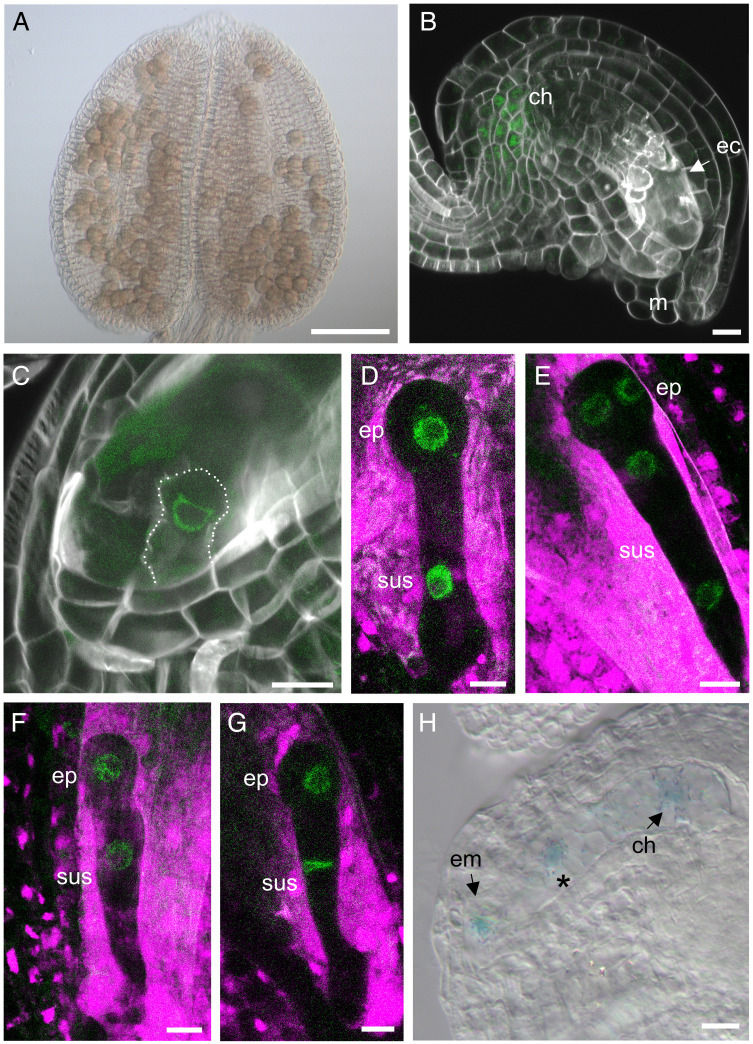
Fig. 1.
Pre- and postfertilization BBM expression. (A) GUS activity (blue) was not observed in BBM:BBM-GFP-GUS anthers at anthesis (floral stage 12). (B) GFP expression (green) in the chalazal region of a BBM:BBM-GFP-GUS ovule at anthesis. GFP was not observed at the position of the egg cell. (C–E) BBM:BBM-GFP expression during seed development. (C) BBM:BBM-GFP expression in the zygote (dashed lines), (D) the one-cell embryo, and (E) the two-cell embryo. (F and G) BBM:BBM-GFP expression during embryo development in reciprocal crosses. (F) GFP expression in the one-cell embryo in a WT × BBM:BBM-GFP cross and (G) in a BBM:BBM-GFP × WT cross. (H) GUS expression in the endosperm nuclei of seeds from a selfed BBM:BBM-GFP-GUS line, 48 hours after pollination (HAP). The blue spot at the micropylar region shows expression in the embryo. (Scale bars: 100 µm in A and 10 µm in B–H.) m, micropylar region; ch, chalazal region; ec, egg cell; em, embryo; ep, embryo proper; sus, suspensor; *, endosperm nuclei.
Next, we examined BBM expression during seed development. Our previous analysis showed BBM:BBM-GFP expression from the four-cell stage of embryo development (6). Reanalysis of BBM:BBM-GFP plants showed that a low level of BBM expression can be detected occasionally in the zygote of selfed seeds (∼2% of ovules) (Fig. 1C), but thereafter BBM expression could be detected consistently in the embryo proper and suspensor of the one- and two-cell-stage embryo (Fig. 1 D and E). Reciprocal crosses between wild-type (WT) and BBM:BBM-GFP lines were used to determine whether BBM shows parent-of-origin expression in the embryo. BBM expression could not be detected in the zygote after crossing but could be observed from the one-cell embryo stage onward, independent of the direction of the cross (Fig. 1 F and G). This result suggests that BBM is expressed from both parental alleles during the first few embryo cell divisions. Our ability to detect BBM expression in the zygote in selfed lines, but not in reciprocal crosses, suggests that BBM expression from a single allele is below the limit of detection.
BBM expression was also observed in the maternal and filial seed compartments. BBM expression was observed in the nucellus up to 48 h after pollination in a small proportion of selfed seeds (SI Appendix, Fig. S1 A and B). In reciprocal crosses between WT and BBM:BBM-GFP-GUS plants, BBM expression in the nucellus was only observed when the BBM:BBM-GFP-GUS line was the female parent (SI Appendix, Fig. S1 C–E), confirming that BBM is expressed in the maternal sporophytic tissues of the seed. We also observed weak BBM expression in endosperm of seeds from self-pollinated BBM:BBM-GFP-GUS lines (Fig. 1H). BBM expression was very low and could only be observed in a small percentage of seeds (8% of seeds, n = 385). When BBM expression was observed it was limited to the primary endosperm nucleus and only rarely observed in the first two endosperm cells (0.3% of seeds). Together these data show that BBM is expressed transiently and at a low level in both the maternal and filial seed tissues.
In summary, our expression analysis showed that BBM is expressed in the embryo much earlier than previously described, starting from the zygote stage, and this expression appears to be biparental, at least during the first few embryo cell divisions. We could not detect BBM expression in the male or female gametes, although we cannot rule out very low levels of BBM expression in these cells. Previously unreported BBM expression patterns in the nucellus and endosperm were also identified. These data suggest early BBM functions in the embryo as well as broader functions during seed development.
Embryo Phenotypes in bbm and plt2 Mutants.
Our results show that BBM is expressed in the zygote, but previous analysis of embryos from segregating PLT2/plt2 bbm or plt2 BBM/bbm T-DNA insertion mutant only reported ∼25% embryo arrest without a detailed description of when embryo arrest took place (5). We therefore performed a more careful analysis using new bbm and plt2 alleles that were generated using CRISPR-Cas9 mutagenesis. We selected three independent mutant alleles for BBM (bbm-cr1, bbm-cr2, and bbm-cr3) and two independent mutant alleles for PLT2 (plt2-cr1 and plt2-cr2), which are referred to generally as bbm-cr and plt2-cr (SI Appendix, Fig. S2). All five mutant alleles lack most or all of the 5′ untranslated region (UTR) and the first exon, including the ATG translation initiation codon. Analysis of BBM and PLT2 expression in their respective single-mutant backgrounds showed that both genes are expressed at WT levels (SI Appendix, Fig. S3). However, given the phenotypes described below, we propose that the bbm-cr and plt-cr mutants are loss-of-function mutants at the protein level (see Discussion).
Mutant embryo phenotypes were not reported for single bbm or plt2 T-DNA insertion lines (5, 6); however, we observed that single bbm-cr and plt2-cr mutant embryos developed faster than embryos from WT plants grown at the same time and under the same conditions (SI Appendix, Table S1). While the majority of WT embryos were at the late heart stage, the majority of plt2-cr and bbm-cr mutants had already reached the torpedo stage. These data suggest that BBM and PLT2 individually control the progression of embryo development in Arabidopsis.
Next, we examined the phenotypes of the progeny of selfed plt2-cr1 BBM/bbm-cr1 and plt2-cr2 BBM/bbm-cr2 lines. Both allele combinations resulted in ∼21 to 28% seeds with embryo defects (Fig. 2 and SI Appendix, Table S2), suggesting that there is no parent-of-origin effect for BBM function during embryo development (Fig. 2). Embryo morphology defects and empty seeds were first observed at 3 days after pollination (DAP), but the proportion of abnormal (presumed mutant) seeds that did not contain a recognizable embryo increased from 6 to 12% at 3 DAP to 78 to 96% at 6 DAP (SI Appendix, Table S2 and Fig. 2 A). These data suggest that bbm-cr plt2-cr embryos are unable to maintain embryo development. We observed a range of morphological defects in segregating progeny of the selfed plt2-cr1 BBM/bbm-cr1 and plt2-cr2 BBM/bbm-cr2 lines. These defects included arrested zygotes, abnormal cell division planes, and abnormally shaped cells in the embryo proper and/or suspensor that became obvious from 3 DAP onward (Fig. 2 B–D and SI Appendix, Fig. S4 and Table S2). For example, at 6 DAP the majority of WT embryos were at the late heart stage (Fig. 2E), while mutant embryos were arrested at the few-celled embryo stage (Fig. 2 B and C) or developed into amorphous multicellular structures (Fig. 2D). We did not observe any abnormal embryos with more than four apical cells. Analysis of multiple 6 DAP replicates showed that the proportion of aborted or abnormal embryos was variable but within the expected segregation ratio for two recessive alleles (SI Appendix, Table S2). This indicates that the embryo abortion phenotype is somewhat stochastic. Similar phenotypes were observed in selfed progenies of bbm-1/BBM plt2-1 T-DNA lines. In these lines, about 25% of the seeds either contained no embryo or embryos that were arrested at the two-celled stage (SI Appendix, Fig. S5). The embryo defects in the bbm-1/BBM plt2-1 T-DNA line were complemented by a BBM:BBM-YFP transgene (SI Appendix, Fig. S5), confirming that BBM and PLT2 mutations are responsible for the observed embryo defects. Together these data indicate that BBM and PLT2 are redundantly required for the progression of Arabidopsis embryo development and embryo patterning.
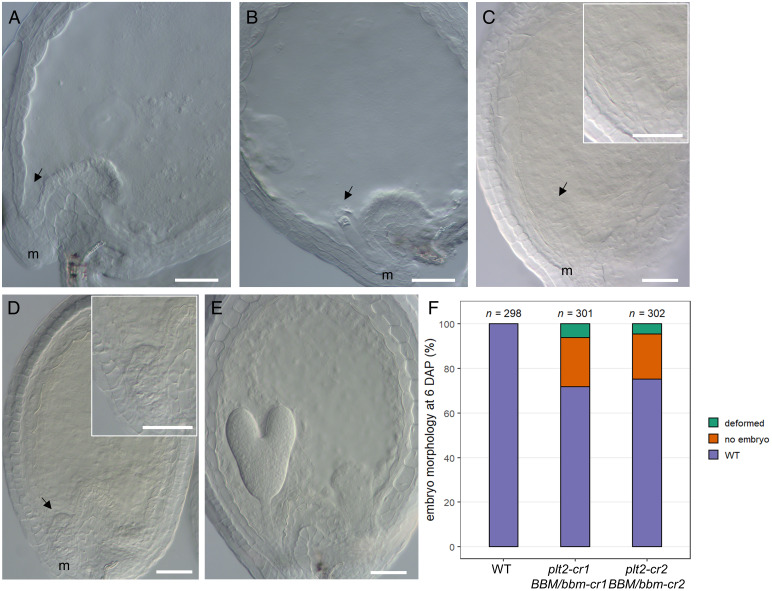
Fig. 2.
Embryo defects in bbm plt2 double mutants. (A–E) DIC microscopy images of seeds with abnormal embryo development in progeny from selfed plt2-cr1 BBM/bbm-cr1 and plt2-cr2 BBM/bbm-cr2 plants around 6 DAP. Seeds (A) lacking an embryo, (B) with an arrested zygote or one-cell embryo, (C) with an arrested and abnormal two-celled, (D) with a multicellular structure, and (E) with a WT-looking late-heart-stage embryo. (F) Proportion of seeds with abnormal embryo development (deformed or no embryo) and WT embryos in siliques from the indicated lines. n = total number of seeds analyzed. Information on the embryo phenotypes is shown in SI Appendix, Table S2. Normal, seeds with a WT embryo; no embryo, seeds lacking an embryo; deformed, multicellular embryos with patterning defects. (Scale bars in A–E: 50 µm.) Arrow, position of (missing) embryo or abnormal ELS; m, micropyle.
BBM and PLT2 Regulate Endosperm Proliferation and Cellularization.
Our results show that BBM is also expressed in the first few endosperm cells, suggesting that BBM has a role in endosperm development. We therefore examined endosperm proliferation and cellularization in bbm-cr and plt2-cr single and double mutants. We did not observe any developmental defects in endosperm proliferation and cellularization in bbm-cr or plt2-cr single-mutant lines (SI Appendix, Fig. S6). By contrast, the progeny of plt2-cr1 BBM/bbm-cr1 and plt2-cr2 BBM/bbm-cr2 seeds showed defects in both processes (Fig. 3). At 1 DAP there were only slight differences in nuclei number between the WT and the segregating double mutants (Fig. 3 A and B), but at 2 DAP the endosperms of the bbm-cr plt2-cr mutant were significantly delayed when compared to the WT (Fig. 3 A and C): In WT plants 100% of 2 DAP seeds already contained more than 32 endosperm nuclei, while for either mutant less than 40% of seeds were at a comparable stage. This indicates that PLT2 and BBM are necessary for timely endosperm proliferation. Next, we evaluated whether plt2 and bbm seeds showed endosperm cellularization defects. We collected seeds of WT, plt2-cr1 BBM/bbm-cr1, and plt2-cr2 BBM/bbm-cr2 at 5, 6, and 7 DAP and evaluated endosperm cellularization using Feulgen staining (Fig. 3 D–I) and chloral hydrate clearings (SI Appendix, Fig. S7 B–J). In WT seeds cellularized endosperm can already be seen at 5 DAP in the micropylar region surrounding the embryo (Fig. 3D). The cellularization then spreads throughout the seed cavity at 6 DAP, and at 7 DAP almost all endosperm is cellular, except for the chalazal region (Fig. 3 E and F). In both plt2 bbm mutants, however, we observed that cellularization was delayed and even at 7 DAP only a small portion of the endosperm had cellularized (Fig. 3 G–I). This phenotype coincided with the lack of an embryo, suggesting that those seeds are mutant for plt2 bbm. Nevertheless, we did occasionally observe seeds lacking an embryo, and thus mutant for plt2 bbm, but with a cellularized endosperm (SI Appendix, Fig. S7A). This indicates that although PLT2 and BBM are necessary for endosperm cellularization, there are likely other redundant factors that contribute to this developmental transition. Overall, our data demonstrate that in addition to their roles in embryo development, PLT2 and BBM are redundantly required for endosperm proliferation and cellularization.
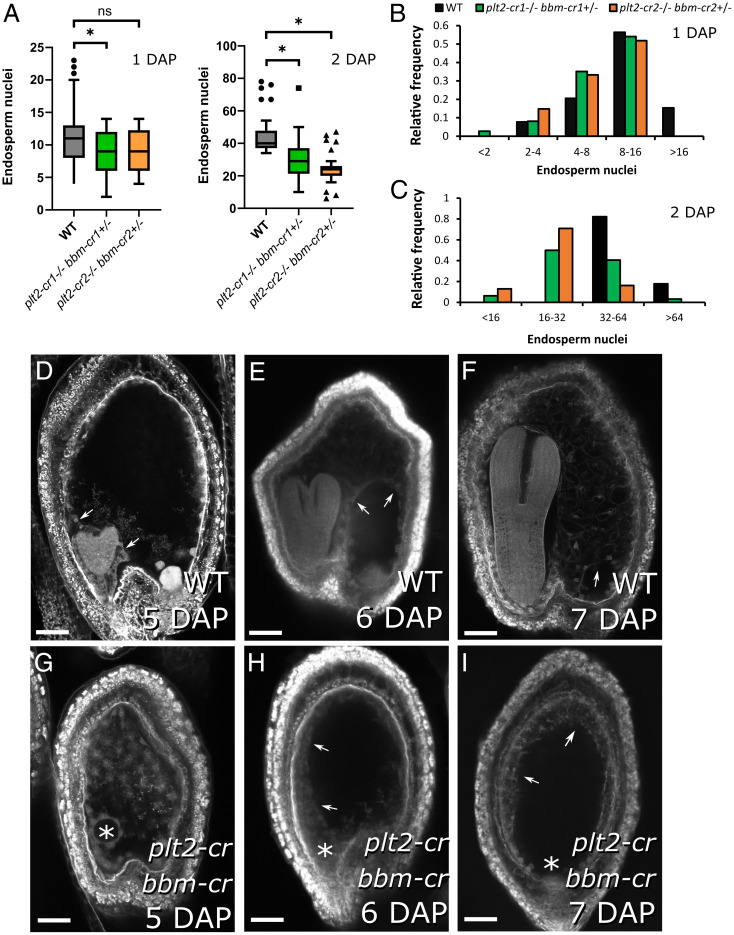
Fig. 3.
Endosperm development is affected in bbm plt2 mutants. (A) Endosperm nuclei counts at 1 and 2 DAP for WT and plt2-cr BBM/bbm-cr double mutants. From 27 to 39 seeds (average of 32) were assayed for each genotype for each time point. Asterisk indicates statistical significance for Mann–Whitney U tests for P < 0.05; ns, not significant. (B and C) Frequency distribution for nuclei number at 1 and 2 DAP. (D–I) Endosperm cellularization status at 5, 6, and 7 DAP in (D–F) WT and (G–I) plt2-cr bbm-cr double mutants. Almost full cellularization of the endosperm cavity is only observable in WT seeds at 6 and 7 DAP (arrows indicate the limits of the cellular endosperm). Note the absence of an embryo in panels G–I (asterisks), indicating that these seeds carry the plt2 bbm genotype. (Scale bars: 50 µm.)
Egg-Cell-Expressed BBM Induces Parthenogenesis.
Expression of BBM genes in the egg cell of sexual and apomictic species is sufficient to bypass the fertilization requirement for embryo development. In monocots, BBM alleles from both apomictic and sexual species induce parthenogenesis (11, 15, 17), while in dicots only the “apomictic” PsASGR-BBML allele has been evaluated (16). We therefore determined whether egg cell expression of a “sexual” BBM allele is also sufficient to induce haploid embryo formation in Arabidopsis. We developed an ectopic BBM expression construct in which the EC1.2en-EC1.1 (referred to here as EC1) promoter (23) fused to an AMV translational enhancer was used to drive egg cell expression of a Brassica napus BBM complementary DNA (cDNA) (BnBBM1). A B. napus BBM cDNA was used instead of the Arabidopsis BBM cDNA to avoid potential gene silencing (24). We then characterized transgenic plants for haploid embryo development in a two-step approach, first in in vitro pistil culture and then in selfed progeny.
We first characterized the primary EC1:AMV:BBM transformants for their ability to produce haploid embryos in vitro. Pistils from WT and EC1:AMV:BBM transformants were excised just before anthesis and cultured in microtiter plates for 7 d. The embryo sac of WT ovules either remained intact (Fig. 4A) or degraded. A variable percentage of both WT and EC1:AMV:BBM ovules also showed autonomous endosperm development after in vitro pistil culture (Fig. 4 B and D and SI Appendix, Table S3). Notably, 2 of the 18 EC1:AMV:BBM lines developed multicellular ectopic structures at the micropylar pole of the embryo sac (Table 1 and Fig. 4 C and D), which were never observed in WT ovules. The EC1:AMV:BBM phenotype was only weakly penetrant (ca. 1.5% of ovules). In most cases, the typical zygotic embryo-like patterning was not observed in the ectopic structures, but we confirmed that the structures express the BBM and LEC1 embryo identity reporters (Fig. 4 E–J).
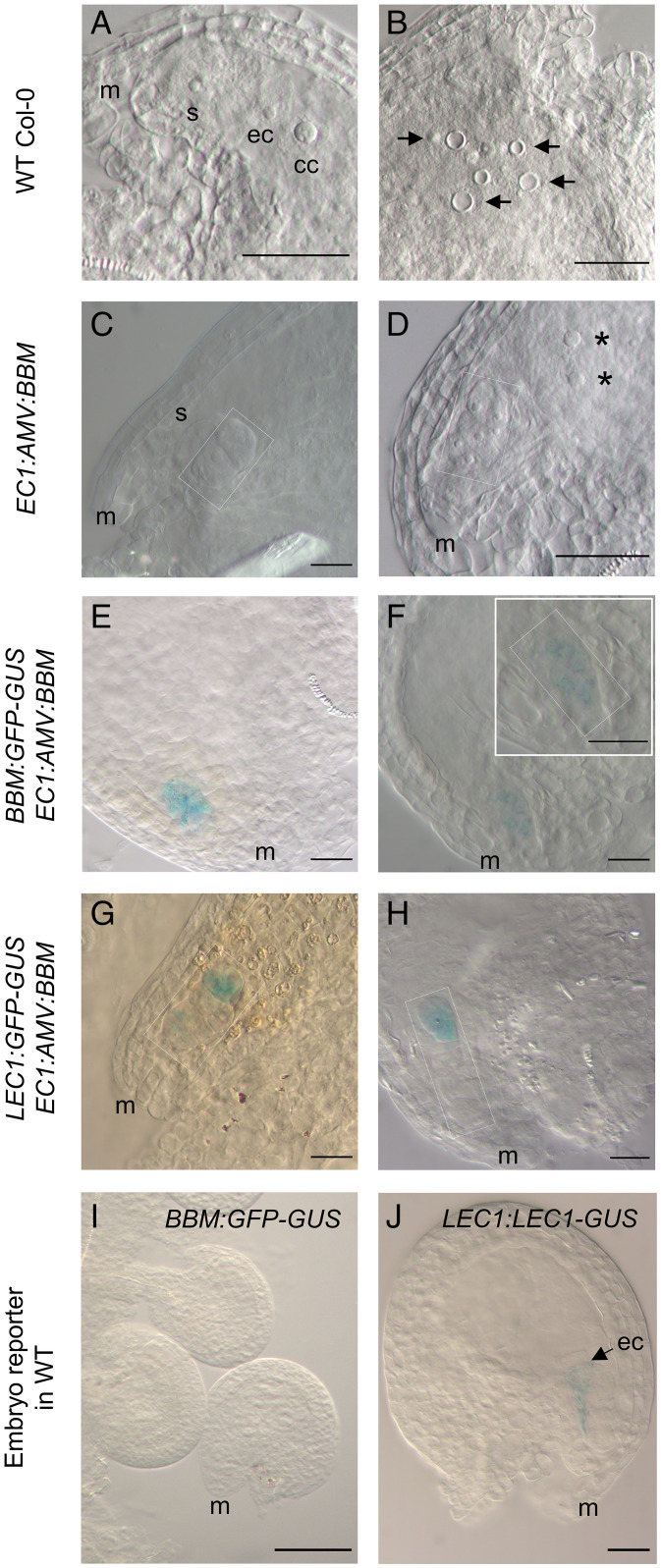
Fig. 4.
Egg-cell-expressed BBM induces parthenogenesis in vitro. (A and B) Ovules from a WT pistil cultured for 7 d. (A) overview of an ovule showing a synergid (s), the egg cell (ec), and the central cell nucleus (cc) and (B) autonomous endosperm development (arrows). (C and D) Ovules from an EC1:AMV:BBM pistil cultured for 7 d. (C) An ovule with an ELS (outlined). (D) Overview of an ovule showing an ELS (outlined) and autonomous endosperm. (E and F) BBM:GFP-GUS expression (blue) in ovules from an EC1:AMV:BBM pistil cultured for 7 d. GUS expression is localized to the micropylar side of the ovule. (F, Inset) A magnification of the micropylar region of the ovule, showing a GUS-stained ELS (outlined). (G and H) LEC1:GFP-GUS expression in ovules from an EC1:AMV:BBM pistil cultured for 10 d. (G) GUS expression is localized to the micropyle side of the ovule in multicellular ELS. (H) GUS expression can been seen in the apical region of an ELS (outlined). (I) BBM:GFP-GUS expression was not detected in WT ovules from 7-d-old pistil cultures. (J) LEC1:LEC1-GUS expression was observed in the egg cell of ovules from 7-d-old WT pistil cultures. ec, egg cell; cc, central cell; m, micropylar region; s, synergid; arrow, central cell nucleus (cc); asterisk, polar nuclei or autonomous endosperm. (Scale bars: 20 µm in A–H and J and 100 µm in I.)
Table 1.
Haploid induction in vitro
| ELS | No. of ovules* | % ELS | |
|---|---|---|---|
| Col-0 | 0 | 321 | 0 |
| EC1:AMV:BnBBM (n = 18 lines) | |||
| #1 | 3 | 192 | 1.6 |
| #2 | 2 | 138 | 1.4 |
| Sum | 5 | 330 | 1.5 ± 0.1 |
| EC1:AMV:BnBBM-GR (n = 22 lines) | |||
| #1 | 1 | 30 | 3.3 |
| #2 | 1 | 29 | 3.4 |
| #7 | 1 | 33 | 3.0 |
| #10 | 1 | 29 | 3.4 |
| #11 | 2 | 34 | 5.8 |
| #12 | 23 | 189 | 12.2 |
| #16 | 2 | 30 | 6.7 |
| #19 | 2 | 39 | 5.1 |
| Sum | 33 | 413 | 5.4 ± 3.1 |
| RPS5A:AMV:BnBBM (n = 9 lines) | |||
| #5 | 1 | 162 | 0.6 |
| #6 | 9 | 81 | 11.1 |
| #7 | 6 | 86 | 7.0 |
| Sum | 16 | 329 | 6.2 ± 5.3 |
*Only ovules that could be cleared were scored. From 19 to 97% of ovules from WT (Col-0) and EC1:AMV:BBM in vitro culture could not be cleared.
Next, we determined whether these ectopic structures are able to form viable haploid embryos by analyzing the phenotype and ploidy of plants derived from selfed EC1:AMV:BBM lines. In the absence of molecular markers, we selected sterile plants (putative haploids) from the selfed progeny and then measured their ploidy. A low proportion (∼0.4%) of haploid seedlings were found among the 2,347 progeny of two homozygous EC1:AMV:BBM lines, while no haploids were identified among the progeny of selfed WT plants (SI Appendix, Fig. S8 and Table S4). Together our in vitro and in planta data suggest that egg-cell-expressed BBM can overcome the requirement of fertilization for embryo initiation but that the haploid embryo induction rate (HIR) is low.
We determined whether additional parameters positively influence BBM-mediated haploid production. One factor that might limit BBM-induced parthenogenesis is selection against lines with relatively high BBM expression, which might have a detrimental effect on embryo sac and/or seed development. To address this point, we used a posttranslationally regulated BBM-GR fusion (EC1:AMV:BBM-GR) to limit BBM activity in time and space (24). Analysis of dexamethasone (DEX)-treated in vitro cultured pistils from independent T1 EC1:AMV:BBM-GR lines showed that there was a clear increase in the number of lines that formed embryo-like structures (ELS), as well as the HIR (Table 1). A second factor that might limit haploid embryo production is the timing of egg-cell promoter expression. If expression is initially activated close to pollination, then the egg cell might be fertilized before parthenogenesis can be induced. The RPS5A promoter is expressed in dividing cells, including the female gametophyte/egg cell (25, 26), and might therefore provide an earlier/longer period of BBM expression than the EC1 promoter. The proportion of RPS5A:AMV:BBM lines and ovules showing haploid embryo development in vitro was higher than when BBM was expressed with the EC1 promoter (Table 1), but no haploid plants were found in the RPS5A:AMV:BBM selfed progenies (SI Appendix, Table S4). In addition to the female gametophyte, RPS5A is also expressed in the seed coat, in embryos from the zygote stage onward and in the endosperm, and thus the broader expression pattern of RPS5A:AMV:BBM in the seed might have a negative effect on seed development.
Next, we evaluated whether delayed pollination also facilitates haploid seed formation by providing the egg cell with a wider window in which to proliferate before fertilization of the central cell. To facilitate crossing, two EC1:AMV:BBM lines that produced haploid embryos were introgressed into the male sterile ms1 mutant (19). The homozygous EC1:AMV:BBM ms1 line was then crossed with pollen from WT Col-0 or from a FAST-Red (OLEO1:OLEO1-RFP) marker line, either immediately after emasculation (day 0) or 2 d later (day 2). FAST-Red is expressed both in the embryo and the endosperm and can be used to distinguish haploid embryos (no FAST-Red expression in embryo, FAST-Red expression in endosperm) from diploid embryos (FAST-Red expression in embryo and endosperm) (19). Putative haploids were screened phenotypically (when WT Col-0 was used as male parent) or for differential FAST-Red expression (when FAST-Red marker line was used as male parent) (SI Appendix, Table S5) and then the ploidy of putative haploids was confirmed by flow cytometry. The haploid induction rate increased significantly when flowers were pollinated on day 2 compared to day 0 (0.1 to 0.4% vs. 1 to 3% HIR) (SI Appendix, Table S5), suggesting that delayed pollination improves BBM-induced haploid embryogenesis.
Enhanced Haploid Embryo Production by Combining BBM and dmp.
Maternal haploid induction using MTL/PLA1/NLD or DMP mutant alleles is an important biotechnology tool for plant breeding (27). In this approach, seeds containing a haploid embryo with only the maternal chromosome complement and triploid endosperm are produced after pollination with mtl/pla1/nld or dmp mutant pollen (19, 28–31). In Arabidopsis, pollination with an atdmp8 atdmp9 double mutant induces maternal haploid embryos (19). We therefore determined whether pollinating EC1:AMV:BBM lines with atdmp8 atdmp9 further enhances haploid embryo induction. Putative haploids from a EC1:AMV:BBM × atdmp8 atdmp9 cross were identified based on differential FAST-Red expression (19) and their ploidy was confirmed by flow cytometry. Approximately 4% of the progeny in ms1 × atdmp8 atdmp9 crosses were haploid compared to 6 to 8% of the progeny from two parthenogenic EC1:AMV:BBM lines (SI Appendix, Table S6). These results show that the combining egg-cell-expressed BBM with the dmp maternal haploid induction system has a synergistic effect on parthenogenesis.
BBM Induces Parthenogenesis in B. napus and Tomato.
To evaluate whether egg-cell-expressed BBM can induce parthenogenesis in dicot crops, we transformed the EC1:AMV:BBM construct to two major dicot crops, B. napus and tomato. Ten independent EC1:AMV:BBM transgenic B. napus lines were crossed with an RPS5A:AMV:GFP marker line to facilitate identification of putative haploid seedlings. RPS5A expression in the root tip was used to distinguish diploid seedlings (green fluorescent protein [GFP]-positive) from haploid seedlings (GFP-negative) in the EC1:AMV:BBM × RPS5A:AMV:GFP cross (SI Appendix, Fig. S9 A and B). We pollinated emasculated EC1:AMV:BBM pistils with RPS5A:AMV:GFP on day 0 after emasculation and selected GFP-negative seedlings as putative haploids. Ploidy analysis of these putative haploids showed that parthenogenic embryos developed in 2 of the 10 lines at a frequency of 0.1% (SI Appendix, Fig. S9C and Table S7). After ploidy analysis, we noticed that many GFP-negative/putative haploid seedlings were true diploids, which might have been due to selection of seedlings with low RPS5A:AMV:GFP expression in the root tip. Given the low frequency of spontaneous haploid production in WT B. napus (0 out of 981 seedlings; SI Appendix, Table S7) (18, 20, 32), our data suggest that egg-cell-expressed BBM also facilitates in vivo haploid embryo development in B. napus.
To evaluate whether an EC1:AMV:BBM × dmp cross can further increase the HIR in B. napus, three independent EC1:AMV:BBM transgenic lines, which showed haploid embryogenesis described above, were crossed with an available bndmp maternal haploid inducer line (20). Putative haploids were identified based on differential FAST-Red expression in the root and cotyledon as described previously (20). Ploidy analysis of these putative haploids suggested that haploid embryos developed at a similar rate in EC1:AMV:BBM × bndmp crosses and WT × bndmp crosses (SI Appendix, Table S8) (20).
We also examined BBM-induced haploid embryo induction in tomato (Solanum lycopersicon) seeds. In tomato, selfed seeds from four independent EC1:AMV:BBM transgenic lines were sown and putative haploids were identified based on their phenotype (smaller size and sterile) (20). Ploidy analysis of these putative haploids showed that haploid embryos developed in one of the four lines at a frequency of 1.4% (SI Appendix, Fig. S10 and Table S9). We and others have shown that spontaneous haploid production in tomato occurs at a very low frequency (from 9 × 10−5 to 4 × 10−4) (20, 33–35), and therefore our data suggest that egg-cell-expressed BBM also induces in vivo haploid embryo development in tomato.
To evaluate whether an EC1:AMV:BBM × dmp cross can further increase the HIR in tomato, a mixture of independent EC1:AMV:BBM T0 transgenic lines were crossed with an available sldmp maternal haploid inducer line (20). The FAST-Red marker was still segregating in the sldmp inducer line, and therefore the putative haploid seedlings were identified based on their phenotypes (20). Ploidy analysis of these putative haploids showed that 2 of the 34 seedlings were true haploids (6%; SI Appendix, Table S9). Then, to analyze haploid embryo induction in the T1 generation, progenies of two T0 EC1:BBM plants (one showed parthenogeneic embryo development [#2] while the other [#1] did not) were pooled and crossed by a sldmp inducer with the homozygous FAST-Red marker. Based on the differential FAST-Red expression in the endosperm and the embryo, we observed an HIR of ∼18% in combination with the sldmp mutant, possibly due to some T1 plants with a homozygous EC1:BBM transgene (SI Appendix, Table S9). The dmp HIR is ∼1.5% in WT tomato crosses (SI Appendix, Table S9) (18), suggesting that egg-cell-expressed BBM in combination with dmp improves haploid seed production above either approach alone.
Discussion
We showed that the Arabidopsis BBM gene is broadly expressed and has novel functions during early seed development. BBM is expressed in the zygote and in the endosperm and transiently in the chalazal region of the ovule (before fertilization) and seed (after fertilization) (Fig. 1). In line with its broader expression pattern, we showed that BBM functions redundantly with PLT2 in promoting embryo progression and viability and endosperm proliferation and cellularization (Fig. 2 and 3). We also described individual functions for BBM and PLT2 in controlling the timing of embryo development (SI Appendix, Table S1). Finally, we showed that ectopic BBM expression in the egg cell of three different dicot plants induces haploid embryo formation (Fig. 4), confirming its role as a key regulator of totipotency in plants.
BBM Is Expressed in Multiple Seed Compartments.
In rice and maize, expression of BBM-like genes can be detected in the zygote as early as 2.5 to 12 h after fertilization (21, 22). BBM-like genes are also expressed in rice sperm cells before fertilization and genetic analysis in rice has shown that sperm expression of BBM-like genes is required for proper embryo development (17). BBM expression has been examined in Arabidopsis using reporter analysis and in situ hybridization, where it was shown to be expressed during embryo and root development (5, 6). BBM transcripts have also been reported in Arabidopsis sperm cells (36), but we were not able to detect BBM expression in mature pollen using BBM reporters (Fig. 1), suggesting that BBM expression levels are below the detection threshold.
We also show very low BBM expression in the zygote of BBM:BBM-GFP progeny and that BBM expression increases dramatically in the one- and two-cell-stage embryo (Fig. 1) These data are in line with transcriptome data showing BBM expression in isolated zygotes and one-cell embryos, but in this transcriptome study BBM is expressed at similar levels at both developmental stages (37). In rice, analysis of mutant combinations of the sperm-expressed BBM-like genes showed that BBM expression in the paternal allele is crucial for early embryogenesis (17). Our results suggest that in Arabidopsis BBM is expressed from both parental alleles in one- and two-celled embryos (Fig. 1), which was confirmed by the segregation ratios of the plt2-cr bbm-cr mutants (Fig. 2). Transcriptome analysis of Arabidopsis zygotes from reciprocal crosses also showed biparental expression of BBM (37). Thus, BBM expression in the embryo might be differently regulated in Arabidopsis and grasses (17, 21).
BBM and PLT2 Function Individually and Synergistically to Regulate Zygotic Embryo Development.
We developed bbm-cr and plt2-cr alleles and showed that the progeny of selfed BBM/bbm-cr plt2-cr plants show ∼25% mutant/embryo lethal phenotypes, as would be expected when the phenotypes are caused by defects in the segregating bbm-cr plt2-cr progeny. We detected normal BBM and PLT2 expression levels in the respective single mutants, suggesting that the defects are due to reduced protein levels. The mutants contain deletions around the 5′ UTR and protein translation start codon suggesting that messenger RNA translation is disturbed in these mutants. In support of this, we showed that the progeny of selfed BBM/bbm plt2 T-DNA mutants, which also show residual BBM expression (5), also had similar embryo arrest and abortion phenotypes and that these phenotypes could be complemented by crossing with a BBM:BBM-YFP line (SI Appendix, Fig. S5). Development of new bbm-cr and plt2-cr lines in which the entire BBM/PLT2 protein coding region is eliminated will be useful to determine if the available CRISPR and T-DNA insertion lines still retain partial BBM/PLT2 activity that allows embryo development to proceed to the few-celled stage.
Single bbm-cr and plt2-cr mutants showed accelerated embryo development (SI Appendix, Table S1), suggesting that BBM and PLT2 function independently to control the progression of embryo development. Both nutrient status and hormones have been shown to influence the progression of embryo development. Embryo growth rates accelerate after endosperm cellularization due to relocation of nutrients from the endosperm to the embryo (38–40), but we did not observe any cellularization defects in the bbm-cr or plt2-cr single mutants (SI Appendix, Fig. S6), while the double bbm-cr plt2-cr mutants showed delayed cellularization (Fig. 3). In Arabidopsis, mutation of the Calcium-Dependent Protein Kinase-Related Kinase 5 gene (AtCRK5) delays embryo development from the globular stage onward and decreases growth of the embryo axis (41). These effects were attributed to reduced gibberellin biosynthesis and changes in auxin distribution due to decreased expression of PIN auxin efflux carriers, but we did not observe any reported gibberellin- or auxin-related embryo defects (42, 43) in the single bbm-cr and plt2-cr mutants. Taken together, our data suggest that BBM and PLT2 control the timing of the progression of embryo development by a different mechanism than previously described.
Our results showing BBM expression starting at the zygote stage (Fig. 1), as well as embryo abortion and few-celled embryo arrest in bbm-cr plt2-cr mutants (Figs. 2 and 3 and SI Appendix, Fig. S4), suggest that BBM is required for embryo initiation and maintenance in Arabidopsis. We were not able to precisely define the stage at which presumed bbm-cr plt2-cr embryos abort, but the presence of seeds without visible embryos at 3 DAP (SI Appendix, Table S2) suggests that embryo abortion occurs starting from the zygote stage. The presence of abnormally patterned embryos with only a few apical and/or basal cells suggests that any embryos that do not initially abort only undergo a few cell divisions before their development arrests.
These results consolidate a large body of data in Arabidopsis and other plants showing spontaneous embryo induction after ectopic BBM expression (Fig. 4 C and D and Table 1) (11, 15–17, 24, 44). BBM is part of a large transcriptional network that regulates embryo identity and development (45). BBM directly regulates expression of the LAFL genes (LEAFY COTYLEDON1 (LEC1), ABI3, FUSCA3, and LEC2). LAFL genes are central regulators of embryo development, as they regulate the maintenance of embryo identity, embryo storage product accumulation, and desiccation tolerance (45–47) and can also induce somatic embryo formation or confer embryo identity when ectopically expressed (48–51). Thus, loss of BBM (and PLT2) expression might simultaneously deregulate expression of many of these key transcription factors and their transcriptional networks, leading to embryo abortion and arrest. Defects in endosperm proliferation and cellularization might also have a negative effect on embryo progression and morphology, as discussed below.
BBM and PLT2 Regulate Endosperm Proliferation and Cellularization.
Analysis of bbm-cr and plt2-cr CRISPR mutants uncovered novel functions for these genes, among which a role in endosperm development. Double bbm-cr plt2-cr mutants showed reduced endosperm proliferation and cellularization (Fig. 3 and SI Appendix, Fig. S7). In Arabidopsis, as in many angiosperms, endosperm development begins with a syncytial (free-nuclear) phase that is followed by cellularization (52, 53). Endosperm cellularization is a critical developmental transition, without which the embryo does not survive (40). In the absence of endosperm cellularization, the embryo arrests around the heart stage (40). However, the lack of endosperm cellularization observed in plt2 bbm double mutants is unlikely to be the cause of the embryo defects in these mutants, as embryos lacking PLT2 and BBM show developmental defects much earlier than the late globular stage. A WT endosperm only begins cellularization at around 5 DAP, at which time point the plt2 bbm mutant embryos already show significant defects, including embryo abortion and arrest. Similarly, it is unlikely that the delayed proliferation of the endosperm observed in plt bbm mutant seeds is linked to the embryo defects, as Arabidopsis endosperm develops autonomously from the embryo; Arabidopsis dmp8 dmp9 mutants that lack an embryo show normal initiation and progression of endosperm development (54). Together, these observations suggest that the phenotypes of plt2 bbm mutant embryos and endosperms are independent of each other.
This bbm plt2 endosperm defect is strikingly similar to that observed in FERTILIZATION INDEPENDENT SEED POLYCOMB REPRESSIVE COMPLEX 2 (FIS-PRC2) mutants. The FIS-PRC2 complex is responsible for the deposition of H3K27me3 marks in the female gametophyte and in the endosperm, and mutations in genes coding for its subunits leads to the failure of endosperm cellularization (40, 55). Our previous chromatin immunoprecipitation sequencing data (6, 56) showed that BBM directly targets the promoter of one of the FIS-PRC2 subunits, FERTILIZATION INDEPENDENT ENDOSPERM (FIE), a WD-40 protein that is homologous to Drosophila enhancer of zeste [E(z)] and that is expressed in both sporophytic and gametophytic tissues (57, 58). BBM binds the FIE gene close to the protein translation start site and the bound region contains three BBM binding motifs (SI Appendix, Fig. S11). Thus, it is possible that BBM and PLT2 redundantly regulate FIE expression during early endosperm development and that reduced FIE expression is (partly) responsible for the observed endosperm defects.
Engineering BBM-Induced Parthenogenesis in Dicots.
Ectopic expression of BBM alleles from sexual and apomictic species induces parthenogenesis in grasses (11, 15, 17). However, BBM-induced parthenogenesis has not been reported in Arabidopsis (15). Here we show that ectopic BBM expression from egg-cell promoters induces haploid embryos development in Arabidopsis (Fig. 4). The parthenogenesis phenotype might have been overlooked in Arabidopsis due to the low HIR in seeds or not induced due to the choice of BBM expression construct. We have shown that BBM-induced somatic embryogenesis requires relatively high levels of BBM protein (24), and thus previous attempts to induce parthenogenesis might have been limited by the promoter choice. We also used the B. napus BBM1 cDNA for ectopic expression, which is less sensitive to silencing in Arabidopsis than the Arabidopsis BBM cDNA (24).
Although the expression levels obtained with the EC1 and RPS5A promoters used in this study were sufficient to induce parthenogenesis, the ectopic expression constructs did not induce highly penetrant embryo induction. On the other hand, ASGR-BBML genes, which are thought to be the causal locus underlying parthenogenesis in apomictic pearl millet, also induce low-penetrance haploid embryo development in sexual grasses (15). Thus, additional factors might be required for efficient engineering of parthenogenesis in sexual plants. We showed that posttranslational activation of the BBM protein (Table 1) and expression of the BBM gene using the RPS5A promoter both enhanced the in vitro HIR, but no viable haploid embryos were produced in RPS5A:AMV:BBM seeds. Both results suggest that BBM expression needs to be finely regulated in seeds for successful parthenogenesis. The EC1 promoter comprises both the EC1.1 and EC1.2 promoters (23). Although EC1.1 expression appears to be restricted to the egg cell during reproduction, EC1.2 (DD45) expression, like RPS5A expression, is much broader (59, 60). As a result, negative selection against high BBM expression levels outside the egg cell might limit the penetrance of haploid induction.
Successful in vitro culture of Arabidopsis fertilized seeds has been described, although high abortion rates and abnormal embryo development were observed (61, 62). All of the ELS that developed in unfertilized in vitro cultured EC1:AMV:BBM lines only underwent a few cell divisions and all had abnormal morphology (Fig. 4 C and D). Abnormal embryo development is also observed when BBM is ectopically expressed in the egg cell of other emasculated plants (11, 17). Likewise, ectopic egg cell expression of RWP-RK domain-containing 2 (RKD2) (63) and loss-of-function mutants in MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) (64) in Arabidopsis induce few-celled parthenogenetic embryos with abnormal morphology. One explanation for the abnormal morphology of these parthenogenic embryos might be the lack of endosperm development. Although we observed autonomous endosperm development in in vitro-cultured WT and EC1:AMV:BBM ovules, only a few (2–8) syncytial endosperm cells were observed. Endosperm is important for embryo development, not only for providing nutrients but also growth regulators (65). Chemical ablation of endosperm induces embryo patterning defects and embryo arrest (66). Viable haploid embryos were obtained from EC1:AMV:BBM plants after selfing, delayed fertilization, or crosses with the dmp haploid inducer (SI Appendix, Tables S4–S6), suggesting that endosperm is also required for proper haploid embryo development.
Haploid seed formation was enhanced when egg-cell-expressed BBM was combined with a dmp maternal haploid inducer in Arabidopsis and tomato (SI Appendix, Tables S6 and S9). In Arabidopsis atdmp8 atdmp9 double mutants, one sperm cell preferentially fertilizes the central cell while the second sperm cell often attaches to but does not fuse with the egg cell, resulting in poor seed set and seed abortion at different stages (54, 67, 68). It is not known how dmp mutation induces haploid embryo formation. Sperm–egg cell adhesion without fertilization or single (central cell) fertilization might both trigger egg cell parthenogenesis (54, 67, 68). Sperm cell adhesion without fertilization might be sufficient to stimulate egg-cell division by relieving the transcriptional repression on BBM and other embryo identity genes in the egg cell, while ectopic/heterochronic expression of embryo identity factors would further enhance this stimulatory effect. Alternatively, preferential fertilization of the central cell in an EC1:BBM × dmp cross might also mimic delayed pollination, allowing the egg cell to divide and avoid hetero-fertilization by a second pollen grain (27, 69).
Finally, we show that egg-cell-expressed BBM induces haploid embryogenesis in two dicot crops. As in Arabidopsis, both the number of lines with haploid seed as well as the HIR in these lines is low, and thus future efforts should focus on improving the HIR, for example by selection of appropriate promoters, by identification of repressors of egg-cell parthenogenesis, or by identification of cis regulatory regions that repress expression of embryo identity genes in the egg cell.
Materials and Methods
Plant Material and Growth Conditions.
All plant materials are in the Col-0 background except the plt2-1 mutant (SGT4287), which is in the Ler background. The Arabidopsis bbm-1 (SALK_097021) mutant; the BBM:BBM-YFP (5) and BBM:BBM-GFP reporters (6); the LEC1:LEC1-GUS reporter (70); and the atdmp8 atdmp9, atms1, bndmp, and sldmp mutants were described previously (19, 20). All plant materials are listed in SI Appendix, Table S10.
Arabidopsis plants were grown in a growth chamber (70% relative humidity) at 20 °C on rock wool plugs (Grodan) that were watered with Hyponex fertilizer (1 g/L) two times per week. Plants were maintained under light-emitting diode light (150 µmol⋅m−2⋅s−1) on a 16 h light/8 h dark day/night cycle. B. napus (71) and tomato (72) plants were grown as previously described.
Vector Construction.
The BBM:BBM-GFP-GUS translational reporter was made by PCR-amplifying a Gateway-compatible Col-0 genomic DNA fragment comprising 3,117 bp upstream of the protein translation start site to the end of the coding region, excluding the stop codon. The BBM:GFP-GUS transcriptional reporter was made in the same way, but using a 3,117-bp promoter fragment. The LEC1:GFP-GUS transcriptional reporter was made in the same way using a 2,708-bp promoter fragment. These PCR amplicons were cloned into a modified pBGWFS7 Gateway binary vector (73) that contains a FAST-Red cassette (OLEO1:OLEO1-RFP) (74) for transgenic plant selection.
RPS5A:AMV:GFP, used for selection of haploid B. napus seedlings, was made by PCR-amplifying a fragment comprising the RPS5A promoter (26) with a reverse primer that also contained the AMV translational enhancer (75). The PCR fragment was inserted into pDONR207 and then the pGreen-GW-eGFP binary vector by Gateway cloning (76).
The EC1:AMV:BnBBM vector was made PCR-amplifying the following DNA fragments with the indicated restriction sites: ‘AscI-EC1:AMV-BamHI,’ ‘BamHI-BnBBM1 cDNA+stop codon-XbaI,’ and ‘XbaI-nos terminator (NOSt)-PacI.’ An EC1:AMV fragment was PCR-amplified from pHEE2E-TRI (23) as described for RPS5A:AMV. NOSt-PacI was amplified from pGreen0029 (77). After restriction digestion, the fragments were ligated into the pGD121 binary vector (78).
A Golden Gate cloning strategy (79) was used to create the EC1:AMV:BnBBM-GR and RPS5A:AMV:BnBBM constructs. First, the PCR fragments ‘EC1:AMV,’ ‘RPS5A:AMV,’ ‘linker,’ and ‘NOSt’ were amplified and cloned into level 0 vectors. Then, the EC1:AMV-linker-NOSt and RPS5A:AMV-linker-NOSt cassettes were produced by a level 1 reaction. Next, the FAST-Red selection cassette and EC1:AMV-linker-NOSt or RPS5A:AMV-linker-NOSt were assembled in vector pICSL4723 (72). To avoid restriction digestion at the BsaI and/or the BpiI sites in the BnBBM1 cDNA, the cDNA with or without a rat glucocorticoid ligand binding domain (GR) fusion were first cloned from 35S:BBM-GR (80) into the AscI site of pGEMT-easy (Promega) and then into the AscI site of the level 2 linker, both using restriction digestion and ligation.
BBM and PLT2 CRISPR-Cas9 mutagenesis constructs were assembled in vector pICSL4723 using the U6-26 promoter for single-guide RNA expression (sgRNAs), RPS5A promoter-driven Arabidopsis codon-optimized human Cas9 (81), and the FAST-Red selection cassette (74). The sgRNAs were cloned into the CRISPR-pink acceptor plasmids (provided by Mark Youles, The Sainsbury Laboratory, Norwich, UK and Synbio Technologies). For Arabidopsis BBM and PLT2 mutagenesis, four sgRNAs were designed to target the 5′ UTR and first exon of each gene. The bbm-cr and plt2-cr CRISPR mutant alleles are shown in SI Appendix, Fig. S2. plt2-cr bbm-cr double mutant lines were obtained by crossing.
Constructs were transformed to Agrobacterium tumefaciens strain C58C1 pMP90 and then to Arabidopsis, B. napus, and tomato. Arabidopsis Col-0 was transformed using the floral dip method (82), except for the EC1:AMV:BBM construct, which was also transformed using the root transformation method (83). B. napus DH12075 was transformed as described in ref. 84. Tomato Micro-Tom was transformed as described in ref. 72.
All cloning and genotyping primers are listed in SI Appendix, Table S11.
Reporter Analysis.
A translational BBM:BBM-GFP reporter line was analyzed for GFP expression during embryo development (6). Confocal analysis of GFP reporter expression was performed as previously described (85). Twelve new BBM:BBM-GFP-GUS reporter lines were analyzed for β-glucuronidase (GUS) expression in the root and embryo, from which three lines were selected for expression analysis. GUS activity assays on ovules and seeds were performed as previously described (86) using 0.5 mM potassium ferri- and ferrocyanide and up to 48-h incubation. After staining, the pistils were blotted on filter paper and then transferred to a microscope slide with HCG clearing solution (water:chloral hydrate:glycerol, 25:55.7:8.3; wt/wt) where the ovules/seeds were separated from the other flower/fruit tissues. The ovules/seeds were cleared for at least 1 h before analysis with differential interference contrast (DIC) microscopy using a Nikon Optiphot microscope. Images were taken with a Zeiss Axiocam 105 camera and processed with the ZEN lite software. To analyze expression in reciprocal crosses with WT, stage-12 flowers were emasculated, hand-pollinated, and then collected for GFP expression or GUS staining at the indicated time points.
BBM:GFP-GUS and LEC1:GFP-GUS reporter lines were used to evaluate embryo identity in ELS. A BBM:GFP-GUS reporter line was crossed and the LEC1:GFP-GUS plasmid transformed to an EC1:AMV:BBM line that induced ELS in vitro. The LEC1:LEC1-GUS reporter (70) is shown as a control for LEC1 expression. The pistils were cultured in vitro, as described below. GUS staining were performed as described above, except that 1.0 mM potassium ferri- and ferrocyanide and overnight staining were used. HCG clearing and DIC imaging were performed as described above. Due to native LEC1 expression in the egg cell, only GUS-expressing multicellular structures were scored as ELS.
Mutant Analysis.
Three bbm CRISPR alleles (bbm-cr1, bbm-cr2, and bbm-cr3) and two plt2 CRISPR alleles (plt2-cr1and plt2-cr2) were selected that contain deletions that span part of the 5′ UTR and first exon of each gene. Single homozygous and segregating homozygous double mutants lacking Cas9 were analyzed for embryo and endosperm defects. Flowers were emasculated, hand-pollinated, and analyzed at the indicated DAP. The same procedure was followed for reciprocal crosses between BBM reporters and WT plants. For clearing of ovules and seeds, whole siliques were fixed with ethanol:acetic acid (9:1) and washed for 10 min in 90% ethanol, followed by 10 min in 70% ethanol and clearing overnight in HCG. DIC images of cleared embryos were captured with a Zeiss Axiocam 105 camera and processed with the ZEN lite software.
Endosperm nuclei were counted in chloral hydrate-cleared seeds under DIC optics using an Olympus BX-51 epi-fluorescence microscope. Images were recorded using a DC View III camera with a 0.63× optical adapter. Fiji was used for image analysis (87); 27 to 39 seeds of each genotype were scored for the amount of endosperm produced. The statistical analyses were done using GraphPad Prism. Feulgen staining of seeds was used for analysis of endosperm cellularization, as described previously (88). Three siliques for each genotype at each time point were scored for endosperm cellularization. The seeds were imaged in a multiphoton Leica Stellaris 8 DIVE with excitation at 800 nm and emission between 565 and 610 nm. The images were acquired using the LAX software and treated using Fiji (87).
qRT-PCR was performed on 7 DAP siliques using RNA isolated with a CTAB protocol (https://opsdiagnostics.com/notes/protocols/ctab_protocol_for_plants.htm) and treated with DNase (TURBO DNA-free kit; Invitrogen). cDNA synthesis was performed with the iScript cDNA synthesis kit (Bio-Rad). qRT-PCR was performed as previously described (24) using the primers shown in SI Appendix, Table S11. Relative gene expression was calculated according to the 2−ΔΔCT method (89) using WT samples as the calibrator and a SAND family gene (At2g28390) (90) as the reference.
Haploid Embryo Induction and Analysis.
Four methods were used to evaluate haploid embryo formation in Arabidopsis: in vitro pistil culture, selfing, delayed pollination, and crosses with the atdmp8 atdmp9 maternal haploid inducer. For in vitro pistil culture assays, inflorescence branches were sterilized in 70% ethanol for 30 s and then washed in sterile water. Stage-12 buds were opened with a small scalpel, then the pistils were removed from the flower and placed in 24-well microtiter plates (Greiner) with four to six pistils and 1.5 mL culture medium (62) per well. For EC1:AMV:BBM-GR fusions 10 µM DEX dissolved in ethanol was added to the well and the same volume of ethanol was added to the WT control pistils. The plates were sealed with plastic wrap and placed at 21 °C in the dark for 7 d, after which the ovules were removed from the pistils and cleared with HCG, as described above. To avoid mistaking an abnormal egg apparatus for ELS, only structures at the micropylar region with four or more cells were scored as ELS.
To evaluate haploid induction after selfing, Arabidopsis EC1:AMV:BBM and RPS5A:AMV:BBM seeds were sown on rockwool and grown to the flowering stage. Putative haploids were selected based on their smaller size and sterility (19, 91) and their ploidy level confirmed by flow cytometry (Iribov), as previously described (19). To facilitate the delayed pollination experiments, male sterile lines were generated by crossing two independent EC1:AMV:BBM lines with an ms1 mutant (19). EC1:AMV:BBM ms1 flowers were tagged upon opening and then pollinated 2 d later with either WT Col-0 or a marker line containing the FAST-Red reporter. Correspondingly, putative haploids were identified either based on phenotype or using differential FAST-Red expression in the embryo and endosperm of mature seeds (19). Flow cytometry of leaf samples was used to confirm the ploidy of putative haploid seedlings.
For haploid identification in B. napus, independent EC1:AMV:BBM lines were crossed with pollen from an RPS5A:AMV:GFP reporter line after emasculation (day 0). Seedlings from these crosses were scored for the presence (putative diploid) or absence (putative haploid) of RPS5A:AMV:GFP expression in the root tip. The ploidy of putative haploid seedlings was confirmed by flow cytometry.
For haploid identification in tomato, selfed seeds from independent EC1:AMV:BBM lines were sown and putative haploids were selected based on their smaller size and sterility (20) and their ploidy level was confirmed by flow cytometry.
For crosses with the Arabidopsis dmp maternal haploid inducer, two independent Arabidopsis EC1:AMV:BBM lines were crossed with pollen from an atdmp8 atdmp9 line carrying the FAST-Red marker (19). Putative haploid and diploid seeds were distinguished using differential FAST-Red expression in seeds. For crosses with the bndmp maternal haploid inducer line (20), three independent EC1:AMV:BBM lines were crossed with pollen from a bndmp line carrying the FAST-Red marker. Putative haploid and diploid seeds were distinguished using differential FAST-Red expression in the root tips and cotyledons of germinating embryos (20). For crosses with the sldmp maternal haploid inducer line (20), four independent EC1:AMV:BBM lines (Micro-Tom background) were pooled and crossed with pollen from a sldmp line (Moneyberg background) carrying a heterozygous FAST-Red marker. As the FAST-Red marker was still segregating, putative haploid and diploid seedlings were distinguished phenotypically as described previously (20). The ploidy of all putative haploid seedlings was confirmed by flow cytometry.
Supplementary Material
Acknowledgments
This work was supported by grants from the Topconsortium voor Kennis en Innovatie (TKI) Tuinbouw & Uitgangsmaterialen (TU-17009 and KV1605-45) and the China Scholarship Council (201506350003).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201761119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Feng K., et al. , Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40, 750–776 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Kerstens M. H. L., Schranz M. E., Bouwmeester K., Phylogenomic analysis of the APETALA2 transcription factor subfamily across angiosperms reveals both deep conservation and lineage-specific patterns. Plant J. 103, 1516–1524 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horstman A., Willemsen V., Boutilier K., Heidstra R., AINTEGUMENTA-LIKE proteins: Hubs in a plethora of networks. Trends Plant Sci. 19, 146–157 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Scheres B., Krizek B. A., Coordination of growth in root and shoot apices by AIL/PLT transcription factors. Curr. Opin. Plant Biol. 41, 95–101 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Galinha C., et al. , PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Horstman A., et al. , AIL and HDG proteins act antagonistically to control cell proliferation. Development 142, 454–464 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Horstman A., Bemer M., Boutilier K., A transcriptional view on somatic embryogenesis. Regeneration (Oxf.) 4, 201–216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bui L. T., et al. , A fern AINTEGUMENTA gene mirrors BABY BOOM in promoting apogamy in Ceratopteris richardii. Plant J. 90, 122–132 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Gordon-Kamm B., et al. , Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants 8, 38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vijverberg K., Ozias-Akins P., Schranz M. E., Identifying and engineering genes for parthenogenesis in plants. Front. Plant Sci. 10, 128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conner J. A., Mookkan M., Huo H., Chae K., Ozias-Akins P., A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. U.S.A. 112, 11205–11210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Worthington M., et al. , A parthenogenesis gene candidate and evidence for segmental allopolyploidy in apomictic Brachiaria decumbens. Genetics 203, 1117–1132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Worthington M., et al. , Translocation of a parthenogenesis gene candidate to an alternate carrier chromosome in apomictic Brachiaria humidicola. BMC Genomics 20, 41 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ke Y., Podio M., Conner J., Ozias-Akins P., Single-cell transcriptome profiling of buffelgrass (Cenchrus ciliaris) eggs unveils apomictic parthenogenesis signatures. Sci. Rep. 11, 9880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conner J. A., Podio M., Ozias-Akins P., Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod. 30, 41–52 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Conner J., Guo Y., Ozias-Akins P., Haploidy in tobacco induced by PsASGR-BBML transgenes via parthenogenesis. Genes (Basel) 11, 1072 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khanday I., Skinner D., Yang B., Mercier R., Sundaresan V., A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Zhong Y., et al. , In vivo maternal haploid induction in tomato. Plant Biotechnol. J. 20, 250–252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong Y., et al. , A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat. Plants 6, 466–472 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Zhong Y., et al. , A genotype independent DMP-HI system in dicot crops. bioRxiv, 2021.06.21.449224 (2021).
- 21.Anderson S. N., et al. , The zygotic transition is initiated in unicellular plant zygotes with asymmetric activation of parental genomes. Dev. Cell 43, 349–358.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Chen J., et al. , Zygotic genome activation occurs shortly after fertilization in maize. Plant Cell 29, 2106–2125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z.-P., et al. , Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16, 144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horstman A., et al. , The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 175, 848–857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maruyama D., et al. , Independent control by each female gamete prevents the attraction of multiple pollen tubes. Dev. Cell 25, 317–323 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Weijers D., et al. , An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128, 4289–4299 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Jacquier N. M. A., et al. , Puzzling out plant reproduction by haploid induction for innovations in plant breeding. Nat. Plants 6, 610–619 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Kelliher T., et al. , MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542, 105–109 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Liu C., et al. , A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 10, 520–522 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Gilles L. M., et al. , Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 36, 707–717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong Y., et al. , Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 5, 575–580 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Thompson K. F., Frequencies of haploids in spring oil-seed rape (Brassica napus). Heredity 24, 318–319 (1969). [Google Scholar]
- 33.Cook R. R., A haploid marglobe tomato. J. Hered. 27, 433–435 (1936). [Google Scholar]
- 34.Koornneef M., et al. , Chromosomal instability in cell- and tissue cultures of tomato haploids and diploids. Euphytica 43, 179–186 (1989). [Google Scholar]
- 35.Hamza S., et al. , Selection for spontaneous tomato haploids using a conditional lethal marker. Theor. Appl. Genet. 86, 657–664 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Borg M., et al. , Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 22, 621–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao P., et al. , Two-step maternal-to-zygotic transition with two-phase parental genome contributions. Dev. Cell 49, 882–893.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Zuma B., Dana M. B., Wang D., Prolonged expression of a putative invertase inhibitor in micropylar endosperm suppressed embryo growth in arabidopsis. Front. Plant Sci. 9, 61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg R. B., de Paiva G., Yadegari R., Plant embryogenesis: Zygote to seed. Science 266, 605–614 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Hehenberger E., Kradolfer D., Köhler C., Endosperm cellularization defines an important developmental transition for embryo development. Development 139, 2031–2039 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Baba A. I., et al. , CRK5 protein kinase contributes to the progression of embryogenesis of Arabidopsis thaliana. Int. J. Mol. Sci. 20, 6120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y., et al. , Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat. Plants 4, 289–298 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Verma S., Attuluri V. P. S., Robert H. S., An essential function for auxin in embryo development. Cold Spring Harb. Perspect. Biol. 13, a039966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dresselhaus T., Jürgens G., Comparative embryogenesis in angiosperms: Activation and patterning of embryonic cell lineages. Annu. Rev. Plant Biol. 72, annurev-arplant-082520-094112 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Tian R., Paul P., Joshi S., Perry S. E., Genetic activity during early plant embryogenesis. Biochem. J. 477, 3743–3767 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos-Mendoza M., et al. , Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54, 608–620 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Jia H., Suzuki M., McCarty D. R., Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip. Rev. Dev. Biol. 3, 135–145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parcy F., et al. , Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lotan T., et al. , Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P., The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7, 373–385 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Stone S. L., et al. , LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown R. C., Lemmon B. E., Nguyen H., Olsen O. A., Development of endosperm in Arabidopsis thaliana. Sex. Plant Reprod. 12, 32–42 (1999). [Google Scholar]
- 53.Boisnard-Lorig C., et al. , Dynamic analyses of the expression of the HISTONE:YFP fusion protein in arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13, 495–509 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong H., Wang W., Sun M.-X., Endosperm development is an autonomously programmed process independent of embryogenesis. Plant Cell 33, 1151–1160 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Kiyosue T., et al. , Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 96, 4186–4191 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li M., et al. , Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis. Plant Physiol. 188, 1095–1110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohad N., et al. , Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo M., Bilodeau P., Dennis E. S., Peacock W. J., Chaudhury A., Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc. Natl. Acad. Sci. U.S.A. 97, 10637–10642 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steffen J. G., Kang I.-H., Macfarlane J., Drews G. N., Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Susaki D., et al. , Dynamics of the cell fate specifications during female gametophyte development in Arabidopsis. PLoS Biol. 19, e3001123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sauer M., Friml J., In vitro culture of Arabidopsis embryos within their ovules. Plant J. 40, 835–843 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Gooh K., et al. , Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev. Cell 34, 242–251 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Kirioukhova-Johnston O., et al. , A RETINOBLASTOMA-RELATED transcription factor network governs egg cell differentiation and stress response in Arabidopsis. bioRxiv [Preprint] (2019). https://www.biorxiv.org/content/10.1101/772400v1 (Accessed 3 June 2022).
- 64.Guitton A.-E., Berger F., Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15, 750–754 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Lafon-Placette C., Köhler C., Embryo and endosperm, partners in seed development. Curr. Opin. Plant Biol. 17, 64–69 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Weijers D., Van Hamburg J.-P., Van Rijn E., Hooykaas P. J. J., Offringa R., Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 133, 1882–1892 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi T., et al. , The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 145, dev170076 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Cyprys P., Lindemeier M., Sprunck S., Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat. Plants 5, 253–257 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Tian X., et al. , Hetero-fertilization together with failed egg-sperm cell fusion supports single fertilization involved in in vivo haploid induction in maize. J. Exp. Bot. 69, 4689–4701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luo X., Ou Y., Li R., He Y., Maternal transmission of the epigenetic ‘memory of winter cold’ in Arabidopsis. Nat. Plants 6, 1211–1218 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Li H., et al. , The histone deacetylase inhibitor trichostatin a promotes totipotency in the male gametophyte. Plant Cell 26, 195–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang R., et al. , Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 9, 1696 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karimi M., Inzé D., Depicker A., GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Shimada T. L., Shimada T., Hara-Nishimura I., A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61, 519–528 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Datla R. S. S., et al. , Improved high-level constitutive foreign gene expression in plants using an AMV RNA4 untranslated leader sequence. Plant Sci. 94, 139–149 (1993). [Google Scholar]
- 76.Zhong S., Lin Z., Fray R. G., Grierson D., Improved plant transformation vectors for fluorescent protein tagging. Transgenic Res. 17, 985–989 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hellens R. P., Anne Edwards E., Leyland N. R., Bean S., Mullineaux P. M., pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832 (2000). [DOI] [PubMed] [Google Scholar]
- 78.de Folter S., et al. , A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 47, 934–946 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S., A modular cloning system for standardized assembly of multigene constructs. PLoS One 6, e16765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Passarinho P., et al. , BABY BOOM target genes provide diverse entry points into cell proliferation and cell growth pathways. Plant Mol. Biol. 68, 225–237 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Fauser F., Schiml S., Puchta H., Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Clough S. J., Bent A. F., Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 83.Vergunst A. C., Waal E. C., Hooykaas P. J. J., “Root transformation by Agrobacterium tumefaciens” in Arabidopsis Protocols (Humana Press, 1998), pp. 227–244. [DOI] [PubMed] [Google Scholar]
- 84.Moloney M. M., Walker J. M., Sharma K. K., High efficiency transformation ofBrassica napus using Agrobacterium vectors. Plant Cell Rep. 8, 238–242 (1989). [DOI] [PubMed] [Google Scholar]
- 85.Soriano M., et al. , Plasticity in cell division patterns and auxin transport dependency during in vitro embryogenesis in Brassica napus. Plant Cell 26, 2568–2581 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sieburth L. E., Meyerowitz E. M., Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9, 355–365 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Batista R. A., Figueiredo D. D., Santos-González J., Köhler C., Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev. 33, 466–476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 90.Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W.-R., Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ravi M., Chan S. W. L., Haploid plants produced by centromere-mediated genome elimination. Nature 464, 615–618 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.