Significance
New techniques for probing the RNA structurome in vivo, particularly high-throughput sequencing methods combined with chemical probing, have emerged in recent years. Herein, we introduce “tRNA structure-seq”, which accurately determines the structure of tRNA in vivo using mutational profiling. tRNA structure-seq reveals that the secondary and tertiary structures of certain tRNAs melt under heat stress conditions. It also detects tRNA abundance as well as modification levels and their changes under stress at single-nucleotide resolution. Given that aberrant tRNA folding and modification cause severe human disease, tRNA structure-seq may have medical relevance. Overall, tRNA structure-seq provides comprehensive information regarding RNA structure, modification, and function and is applicable to myriad classes of structured and modified small RNAs.
Keywords: RNA structure, mutational profiling, chemical probing, misfolding, RNA modification
Abstract
RNA structure plays roles in myriad cellular events including transcription, translation, and RNA processing. Genome-wide analyses of RNA secondary structure in vivo by chemical probing have revealed critical structural features of mRNAs and long ncRNAs. Here, we examine the in vivo secondary structure of a small RNA class, tRNAs. Study of tRNA structure is challenging because tRNAs are heavily modified and strongly structured. We introduce “tRNA structure-seq,” a new workflow that accurately determines in vivo secondary structures of tRNA. The workflow combines dimethyl sulfate (DMS) probing, ultra-processive RT, and mutational profiling (MaP), which provides mutations opposite DMS and natural modifications thereby allowing multiple modifications to be identified in a single read. We applied tRNA structure-seq to E. coli under control and stress conditions. A leading folding algorithm predicts E. coli tRNA structures with only ∼80% average accuracy from sequence alone. Strikingly, tRNA structure-seq, by providing experimental restraints, improves structure prediction under in vivo conditions to ∼95% accuracy, with more than 14 tRNAs predicted completely correctly. tRNA structure-seq also quantifies the relative levels of tRNAs and their natural modifications at single nucleotide resolution, as validated by LC-MS/MS. Our application of tRNA structure-seq yields insights into tRNA structure in living cells, revealing that it is not immutable but has dynamics, with partial unfolding of secondary and tertiary tRNA structure under heat stress that is correlated with a loss of tRNA abundance. This method is applicable to other small RNAs, including those with natural modifications and highly structured regions.
RNA structure plays crucial roles in regulation of gene expression via transcription, translation, and RNA processing (1, 2). To do so, RNA molecules fold into specific secondary and tertiary structures. It is thus critically important to accurately determine RNA structure in vivo. To predict RNA structure in vitro and in vivo, multiple approaches have been developed that are semiempirical and use experimental data, for instance (3–7). These approaches rely on two basic strategies: one utilizing structure-specific enzymes, applicable only in vitro, and the other utilizing structure-specific chemicals, applied both in vitro and in vivo. The cleavage/modification sites of these probes are determined by sequencing, which provides information about RNA flexibility and accessibility at single-nucleotide resolution. Dimethyl sulfate (DMS) is one of the most common structure-probing chemicals (8, 9). It methylates the N1 of adenine (m1A), the N3 of cytosine (m3C), and the N7 of guanine (m7G) when the nucleotide is exposed (9). DMS can be used in vivo because it is cell membrane-permeant (7). Reactive sites are determined by library sequencing following reverse transcription (RT), wherein DMS methylation induces either termination or nucleotide mis-incorporation, recorded as either RT stop counts (9) or elevated mutation signals (10), respectively, in next-generation sequencing. These reactivity data are used in structure prediction as experimental restraints that penalize base pairing (11). Using this strategy, we and others have provided insights into in vivo RNA secondary structure (9, 12–14).
Siegfried and coworkers developed SHAPE-MaP, which exploits RT conditions that induce a reverse transcriptase (RT) enzyme to mis-incorporate an incorrect nucleotide opposite a modified nucleotide rather than an RT stop, in so-called “Mutational Profiling” or MaP (10). The mutations in cDNA thus correspond to the SHAPE-reactive sites, with the critical ability to record more than one chemical modification in a single RNA molecule. Rouskin and coworkers extended MaP to DMS (15). Herein, we present a workflow that combines features of MaP and Structure-seq, developed in our labs (4, 9, 13), to facilitate the accurate prediction of highly structured and modified small RNAs, in this case of tRNA (Fig. 1A).
Fig. 1.
tRNA structure-seq: Mutational profiling detects DMS-induced modifications and natural tRNA modifications in yeast tRNAPhe. (A) Workflow for tRNA structure-seq. Detailed procedures and representative results are provided in SI Appendix, Supplementary Text and Fig. S1. (B) DMS reactivity at As and Cs mapped onto the secondary structure. Dashed lines show tertiary contacts (PDB: 1EHZ). Gs with DMS reactivity are circled. (C) DMS reactivities on A, C, G, and U compared between –DMS (teal) and + DMS (red) samples on the yeast tRNAPhe T7 transcript. (D) DMS-reactive guanosines (red) and DMS accessibility (cyan) mapped onto the crystal structure. The Left panel shows the DMS reactivities on G whereas the right panel shows the DMS accessibility to N7G calculated from solvent accessibility with solvent radius set 3Å for DMS. N7 of the guanine base is circled (Inset). Native modified guanosine residues are not depicted. (E) Mutation specificity at DMS-induced modifications from the top 10 DMS-reactive sites across all three types of bases in a T7 transcript. (F) Secondary structure with natural modifications of the native yeast tRNAPhe. Positions with strong mutation rates in –DMS samples are annotated by red arrows. (G) Frequency of the 3 possible misincorporations at natural modification sites, m22G26, yW37, m7G46, and m1A58, obtained from –DMS native yeast tRNAPhe. Each modification is present only once.
Transfer RNA is comprised of four stems, four loops, and the CCA 3′-terminus. It engages in myriad tertiary interactions including parallel-stranded kissing loops between the D- and T-loops (Fig. 1B). Precursor tRNAs undergo tRNA processing and modification to produce the mature tRNAs, which are further aminoacylated for translation (16, 17). The folding of tRNA is enhanced in cellular conditions and it is critical in tRNA maturation and translation (18, 19). Indeed, tRNA variants and defects in tRNA modifications often lead to tRNA misfolding and severe disease (20–22). As such, determining the structure of tRNA genome-wide and in vivo, including during stress, would provide keen insight into tRNA biology; however, doing so requires clearing several technical hurdles. First, tRNA has numerous naturally modified nucleotides, including those that are on the Watson–Crick face such as m1A and 1-methylguanosine (m1G), which results in short cDNA fragments under existing tRNA sequencing methods (23). Second, tRNA is highly structured, which also often terminates RT. Third, tRNA sequences are similar to one another, and thus methods that result in RT stops often yield fragments that are too short (less than 20 nt) to uniquely map to a single reference tRNA gene.
In this study, we developed a new Structure-seq method focused on tRNA, which we call “tRNA structure-seq” (Fig. 1A). We performed two size selection steps during the library preparation, one to obtain full-length tRNA and another to obtain full-length cDNAs, which increase the mapping accuracy and allow for assessment by ShapeMapper of just those single-transcripts that are full-length. After 3′-end-ligation, we used an ultra-processive RT enzyme to perform mutational profiling (MaP) to detect both DMS-induced modifications and natural modifications. tRNA structure-seq predicts tRNA secondary structure with ∼95% accuracy overall, with many tRNAs predicted completely correctly. Our application of tRNA structure-seq to E. coli revealed its tRNA structurome under heat stress and heat shock conditions and uncovered dynamics in tRNA structure and modification that could affect translation and tRNA turnover.
Results
tRNA Structure-seq reports DMS reactivities and natural modifications in yeast tRNAPhe.
To study the in vivo tRNA structurome, we developed tRNA structure-seq, which records DMS reactivity data and natural modifications (Fig. 1A and SI Appendix, Fig. S1). To facilitate misincorporation of nucleotides at DMS- and naturally- modified sites, we used 2 mM Mn2+ in place of Mg2+ in the reaction of Marathon RTase (24). Full-length cDNAs corresponding to the expected length of 3′-end-ligated tRNAs were gel purified (SI Appendix, Fig. S1B). We tested if tRNA structure-seq can detect DMS modification sites using a yeast tRNAPhe T7 transcript, which lacks modified nucleotides (Fig. 1 B–E). Mutation rates, calculated using ShapeMapper2 (25), at A, C, and G in the +DMS sample are significantly higher than the respective –DMS background rates (Fig. 1C). This finding is consistent with the known nucleobase specificity of DMS for the N1 of A, N3 of C, and N7 of G (3), and that modification at all three of these sites, including the Hoogsteen face of G as first reported by Pyle and coworkers for Mararthon RTase with Mn2+ (24), elevates the mutation rate. Likewise, absence of enhanced mutation rate at U’s is consistent with known nucleobase specificity of DMS reaction.
Normalized DMS reactivities (defined in SI Appendix) revealed that six A’s and three C’s in the loops, joining regions, and CCA terminus of the T7 transcript have signals above background (Fig. 1B). We note that while three C’s in stems also had DMS signals above background, one was at the end of the acceptor stem and another near a G•U wobble, providing plausible means for DMS attack. Turning to G’s, these had DMS signals throughout the loops and stems, with the latter likely occurring because N7 of the G resides in the DMS-accessible major groove (Fig. 1D). The DMS-induced modifications caused a 5–9% total rate of mutation at the top 10 DMS-reactive positions, with the actual value depending on whether it was an m1A (9%), m3C(5%), or m7G (5%) DMS modification (Fig. 1E). We note that the dynamic range is broad, with 25 of the 76 bases in tRNA reactive to DMS (Fig. 1 B and D).
Next, we tested if misincorporation levels varied in a native tRNA, which is naturally modified. We performed mutational profiling using –DMS native yeast tRNAPhe (Fig. 1 F–G), which has 12 positions with natural modifications. We observed strong mutation signals at m22G26, Cm32, Gm34, yW37, m7G46, and m1A58 (Fig. 1F). Overall rates of mutation at these naturally modified sites were much higher than at the sites of DMS-induced modification in the tRNAPhe T7 transcript (compare Fig. 1G to Fig. 1E). For example, overall mutation rates at m7G46 and m1A58 in the native tRNAPhe were 22% and 67%, respectively, but only 5% and 9% in the DMS-modified tRNAPhe T7 transcript. Nonetheless, while the rate of mutation varied strongly between natural- and DMS-modifications, the nucleobase specificity of mutation did not vary strongly. For instance, mutation specificity for DMS-induced m1A (top 10) (Fig. 1E) was 6: 2: 1 for changes in the cDNA to T, G, and C, respectively, and 5: 2.4: 1 for the same changes opposite natural m1A58, albeit this natural modification was studied herein on only one tRNA. Likewise, for m7G (top 10), DMS-induced mutation specificity was 4: 1: negligible for changes to A, T, and C, respectively, and 21: negligible: negligible for the same changes opposite natural m7G46. We normalized on the maximum mutational change to estimate the average rate of DMS-induced modification in the in vitro T7 transcript as 15% (=6%A > T/40%A > T * 100%) for m1A (top 10) and 19% (=4%G > A/21%G > A * 100%) for m7G (top 10).
DMS structural restraints significantly improve the accuracy of tRNA structure prediction.
Next, we performed tRNA structure-seq in vitro and in vivo (Fig. 2). We prepared 49 unmodified E. coli tRNA transcripts in vitro by T7 transcription and obtained the in vivo set of E. coli native tRNAs by extraction from cells grown at 37 °C. The experiments were performed with two biological replicates for the tRNA T7 transcripts and three biological replicates for the in vivo tRNAs. Since each replicate showed similar DMS reactivities (SI Appendix, Fig. S2), we combined the replicates and analyzed the combined data, as previously (9, 26).
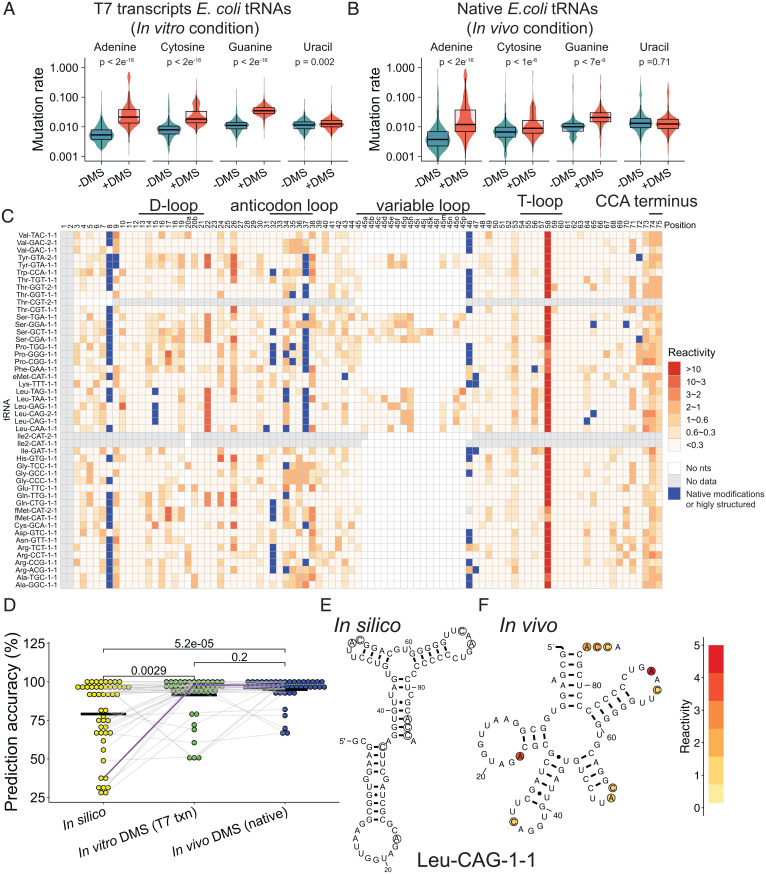
Fig. 2.
DMS reactivities improve tRNA secondary structure prediction accuracy in vitro and in vivo. (A and B) Mutation rates (logarithimic scale) at each of the four nucleotides with –DMS samples (teal) and +DMS samples (red). (A) Mixture of E. coli tRNA T7 transcripts in vitro and (B) E. coli native tRNAs in vivo grown at 37 °C. (C) Normalized reactivity from DMS (heat map) and from natural modifications (or highly structured regions) (blue) from in vivo conditions upon analysis by ShapeMapper 2 (see SI Appendix, Materials and Methods). The 48 tRNA species are provided. “No nts” means no nucleotides exist at this position. “No data” means no reactivity data are available due to low read number. The –DMS background is subtracted. Parallel data for E. coli T7 transcripts in vitro are provided in SI Appendix, Fig. S3. (D) Prediction accuracy for no DMS restraints in silico, T7 transcripts in vitro, and native tRNAs in vivo. Those tRNAs for which DMS data were not available (four and three tRNAs in the in vitro and in the in vivo tRNA datasets, respectively) were excluded from each prediction. Individual values provided in SI Appendix, Table S2. Gray lines show changes in accuracy for the same tRNA. The purple line shows tRNALeu-CAG-1–1. (E and F) Predicted structures of the tRNALeu-CAG-1–1 under (E) in silico and (F) in vivo DMS conditions. Circled nucleotides in (E) are the DMS-reactive nucleotides in (F).
We observed the same trend of A-, C-, and G-specificity of the DMS-induced mutation rate for the large set of E. coli tRNAs both in vitro and in vivo (Fig. 2 A and B), as for yeast tRNAPhe in vitro (Fig. 1C). Given the nucleobase specificity of the DMS reactivity, we analyzed the positional specificity of the reaction in the 48 tRNAs in vitro (SI Appendix, Fig. S3) and in vivo (Fig. 2C). Analysis of the DMS reactivity of these tRNAs revealed a unique pattern of reactivity across all four loops and the CCA terminus both in vitro and in vivo. The anticodon loop spanning positions 32–38, the A at position 58, and the CCA terminus all have high DMS reactivities in vitro and in vivo. Notably, A58 almost always has the highest reactivity of all nucleotides in vitro and in vivo. In the T-loop, A58 forms a reverse Hoogsteen base pair with U54 (see next section) wherein the adenine is flipped 180° (27); this conformation, which can also form in the secondary structure alone (28), exposes the N1 of A58. It is notable that while A58 is not naturally modified in E. coli, it is naturally N1-methylated in yeast tRNAPhe (Fig. 1F); apparently the juxtaposition of this noncanonical base pair predisposes A58 to reaction with either a chemical or a methylating enzyme.
We optimized the ShapeMapper2 separation threshold parameter for Marathon RTase (SI Appendix, Table S1 and Fig. S4) and carried out structure prediction by minimal free energy calculation (Fig. 2D) (29). Reactivities with DMS in vitro and in vivo were used to provide pseudo free energy restraints (see Materials and Methods). Prior to structure prediction, we subtracted out mutations contributed by naturally modified bases and filtered out DMS reactivities at G and U (See SI Appendix, Materials and Methods). Accuracy was judged relative to the genomic tRNA database (gtrnadb.ucsc.edu/index.html), which consists of tRNA secondary structures attained via covariation (30). The mean accuracy of purely in silico prediction was modest at just 79.1% (Fig. 2D, yellow and SI Appendix, Table S2, Bottom) where the canonical cloverleaf structure is equated to 100% accuracy. However, when in vitro or in vivo DMS profiles were incorporated, the mean accuracy of the prediction increased significantly, by 12% (92% prediction accuracy) in vitro and by 16% (95% prediction accuracy) in vivo (Fig. 2D, green and blue and SI Appendix, Table S2, Bottom). The effect on certain RNAs was especially notable. For instance, in silico-predicted tRNALeu-CAG-1–1 scored only 38% structural accuracy, whereas the in vitro and in vivo-predicted tRNALeu-CAG-1–1 improved by 60% to score 98% accuracy, primarily by driving DMS-reactive C35 and three 3′ nucleotides (the discriminator base and CC of the CCA) out of mispairings (Fig. 2D and SI Appendix, Table S2, purple line/highlight; and Fig. 2 E and F). Moreover, when considering the 17 tRNAs with poorest in silico predictions, which is a full 40% of the tRNAs with DMS data, the accuracy increased even more, from 59 to 88% in vitro (an increase of 29%) and from 59 to 93% in vivo (an increase of 34%). Nearly all tRNAs were predicted equally well or much better using DMS restraints than in silico (Fig. 2D, gray lines). Overall, our results indicate that DMS restraints significantly improve the accuracy of tRNA secondary structure prediction, especially for those tRNAs with poor prediction in silico.
tRNA secondary structure partially unfolds under heat stress and heat shock.
Our previous study of RNA structure in rice seedlings showed that heat shock globally unfolds mRNA structure, which promotes degradation and decreased abundance of mRNA (31). The effect of heat on in vivo tRNA structure has not been tested. We carried out tRNA structure-seq under two sets of stress conditions: (i) continuous 47 °C culture referred to as “heat stress” (Fig. 3, “h. stress”) and (ii) 30 °C culture followed by a temperature shift to 47 °C for 20 min referred to as “heat shock” (Fig. 3, “h. shock”); these were compared to continuous growth at 37 °C and 30 °C, respectively.
Fig. 3.
In vivo heat stress in E. coli unfolds the structure of its tRNA. (A) tRNA secondary structure was predicted by minimal free energy calculation using in vivo DMS restraints. Sequencing experiments were performed with three biological repeats (n = 3) and the reads merged. E. coli cultures were grown at 37 °C, h. stress (47 °C), 30 °C, or h. shock (=30 °C → 47 °C for 20 min). Values for individual tRNAs provided in SI Appendix, Table S3. Gray lines show changes in canonical folding of the same tRNA species between different conditions. (The in vivo data points at 37 °C from Fig. 2D are replotted here.) (B) Normalized DMS reactivities in acceptor stem, D-stem, anticodon stem, and T-stem. “Native Folding” (Bottom) are the tRNA species predicted to have canonical structure in the 37 °C and h. stress conditions (i.e., Δaccuracy (A47 °C – A37 °C) = 0% and A37 °C > 95%), whereas “Nonnative Folding” is the tRNA species that were predicted to have a nonnative structure in the h. stress condition (i.e., Δaccuracy (A47 °C – A37 °C) < –7%). (C and D) Normalized DMS reactivities at A58 and C56. (E) Reverse Hoogsteen base pair between U54 and A58 (yellow) and Watson–Crick pair between G19 (blue) and C56 (yellow) in the crystal structure of unmodified E. coli tRNAPhe (PDB: 3L0U). The black circle shows the N1 of A58 is exposed to solvent. The kissing T-loop and D-loop are colored yellow and blue, respectively. (F) Proposed mechanism of partial unfolding of tRNA secondary structure under heat stress involving the acceptor stem and tertiary structure. Red regions have a change in DMS reactivity with temperature.
Structure prediction by the minimal free energy approach supports a heat-induced decrease in native tRNA folding, suggesting tRNA structure is malleable in vivo. This might be expected thermodynamically, but its actual extent has not been assessed for RNAs with such strong native structure as tRNAs. Given that the cloverleaf structure defines a prediction accuracy of 100%, attainment of the cloverleaf structure decreased on average by 7%, from 95 to 88%, in heat stress, and by 4%, from 92 to 88%, upon heat shock (Fig. 3A and SI Appendix, Table S3, Bottom), suggesting that heat partially unfolds tRNA, which leads to alternative structures (predicted tRNA structures in SI Appendix, Fig. S5). Inspection of Fig. 3 indicates severe loss (from 10% to more than 50% loss) of the cloverleaf structure for 9 tRNAs in heat stress and 4 tRNAs in heat shock. We also tested for thermodynamic ensembles of tRNA structure using Rsample, which models multiple RNA secondary structures using thermodynamics combined with structure mapping data (32) (SI Appendix, Fig. S6). The Rsample analysis from all E. coli tRNAs reveals that tRNA has more structural fluctuations under the heat stress condition (SI Appendix, Fig. S6A). For example, at 37 °C tRNAAla-GGC-1–1 folds nearly uniformly into the cloverleaf structure with a folding probability of ∼90% (SI Appendix, Fig. S6B). However, under heat stress, the native fold reduced to ∼59% of the population, with the remainder adopting a rod-like conformation (SI Appendix, Fig. S6C).
Since the heat stress condition shows more significant change in prediction accuracy than the heat shock condition overall with P values of 0.0049 and 0.027, respectively (Fig. 3A, Top), we analyzed the heat stress data in depth. Analysis of DMS reactivities in each stem of those tRNAs predicted to have alternative secondary structures under heat stress (Fig. 3B, columns 2 and 4) revealed that DMS reactivities generally increased in the acceptor stem, while DMS reactivities were largely unchanged in the D-stem, anticodon stem, and T-stem (Fig. 3B). In addition, reactivities at A58 and C56 in the T-loop decreased and increased, respectively (Fig. 3 C and D). The results in the four stems suggest that a major site of unfolding of certain tRNAs at high temperatures is the acceptor stem (Fig. 3B), perhaps because it involves pairing of the most distal nucleotides (Fig. 1C) and perhaps because it is the only stem-loop that has no modified bases (Fig. 1F). Notably, the mean free energy for formation of the acceptor stem in the tRNAs with unfolded acceptor stems is ∼1.0 kcal/mol higher (i.e., weaker) than in the tRNAs with folded acceptor stems consistent with nonnative folders being more sensitive to heat stress.
An inverse dependency of DMS reactivity on temperature occurred at A58 in the T-loop in almost every tRNA (Fig. 3C, P value∼10−12). As mentioned above, A58 is involved in a reverse Hoogsteen pair that leads to hyper-reactivity with DMS (Fig. 2C). Observed loss of A58 DMS reactivity with temperature in the majority of the tRNAs (Fig. 3C, nearly all gray lines) suggests melting of the T-loop in most tRNAs. As mentioned, the D- and T-loops have a kissing tertiary interaction (Figs. 1B and 3E), which depends upon the T-loop conformation (28); thus, loss of the reverse Hoogsteen pair in the T-loop may be associated with loss of the kissing tertiary structure in the tRNA. To test this idea, we examined the DMS reactivity of C56 in the T-loop (Fig. 3D), which participates in the kissing interaction via a Watson–Crick base pair with G19 in the D-loop (Fig. 3E). We found increased reactivities at C56 upon heat stress also in the majority of the tRNAs (Fig. 3D, most gray lines, P value∼10−6), although not in most Watson–Crick structures (Fig. 3B, D-stem, anticodon stem, and T-stem, P values∼0.1–0.4 in heat stress) supporting loss of tertiary structure in heat stress. In sum, we propose that at high temperature the T-loop-D-loop tertiary interaction is broken, at least some of the time, in most tRNAs, and that the acceptor stem melts in a subset of tRNAs (see Fig. 3F for overall model).
tRNA abundance changes upon heat stress.
tRNA structure-seq requires experimental data for + and –DMS. We surmised that we could use the –DMS data for quantification of tRNAs, as we have done for mRNAs (26, 31). Such tRNA abundance information could be helpful for discerning relationships with tRNA unfolding. A classical strategy for tRNA quantification is northern hybridization with a tRNA-specific probe (33), while more recently several quantitative tRNA sequencing methods have been developed, which provide a reference for comparison (34–39) (see Discussion). To test if our tRNA structure-seq data are usable for tRNA quantification, we estimated tRNA expression levels by quantification of the sequence reads mapped to specific tRNA genes using Salmon (40). We observe a clear correlation between our data and the Northern blot data of Dong et al. (SI Appendix, Fig. S7A), which is also consistent with the estimation of tRNA abundance by Wei and coworkers (33, 36) (SI Appendix, Fig. S7B). Thus, tRNA structure-seq appears useful for estimating tRNA abundance.
We then performed differential expression analyses with DE-seq2 (41). The abundance of nine tRNAs differed between 37 °C and heat stress, with five increasing and four decreasing in abundance in the heat stress condition (Fig. 4A). The abundance of the other 36 tRNAs was not significantly changed. In the case of the heat shock condition, we detected only four tRNAs for which abundance changed significantly (Fig. 4B and Discussion). Considering all 44 tRNAs, changes in tRNA abundance have a weak inverse correlation with changes in DMS reactivities (DMSheat stress – DMS37 °C) in the heat-sensitive acceptor stem in tRNA (Fig. 4C). To further assess if these changes in abundance in the heat stress condition were correlated with structural changes in tRNA, we evaluated the predicted secondary structure of the nine tRNAs that differed in expression between 37 °C and heat stress (Fig. 4 D–G and SI Appendix, Fig. S8). At high temperatures, down-regulated tRNAs show nonnative structures and low accuracy (Fig. 4 F and G), while up-regulated tRNAs generally show native, cloverleaf structures (Fig. 4 D and E) with the one exception of Leu-CAG-1–1. Leu-CAG-1–1 is predicted to have alternative structure at high temperature where the As and Cs in the acceptor stem have DMS reactivities, which supports unfolding of the stem. This appears to induce alternative base-pairing in the D-stem and limited base-pairing in the anti-codon stem (SI Appendix, Fig. S8G). Met-CAT-1–1 shows strong reactivity in the 5′-strand of the anticodon stem forming nucleotides (compare SI Appendix, Fig. S8 I–M), while Arg-CCT-1–1 shows reactivity in the 5′-strand of the acceptor stem and D-stem forming nucleotides (compare SI Appendix, Fig. S8 J–N). We previously found that the last base pair in the acceptor stem significantly affects folding activity in tRNA (19). Moreover, the strongly down-regulated tRNAs of Ser-GGA-1–1 and Tyr-GTA-2–1 have reactivity in acceptor stem regions, and both tRNAs have strong DMS activities in the last nucleotide in the acceptor stem (SI Appendix, Fig. S8 O and P), which potentially reduces the tRNA stability with correlated impacts on tRNA abundance. Overall, growth temperature could directly affect tRNA abundance through tRNA unfolding/protein unbinding, favoring ribonuclease access.
Fig. 4.
tRNA structure-seq detects changes in tRNA abundance upon heat stress. Differential expression analysis of tRNA (A) between 37 °C and the h. stress condition and (B) between 30 °C and the h. shock condition are provided, where tRNA structure-seq data (–DMS data, n = 3) were used for the differential expression analysis. (C) Changes in tRNA abundance have a weak inverse correlation with the change in DMS reactivity in the acceptor stem between 37 °C and the h. stress condition. (D–G) Representative predicted secondary structures and statistics of up-regulated (red) or down-regulated (blue) tRNAs are depicted in the heat stress condition depicted with normalized reactivity (see SI Appendix, Fig. S8 for all structures). (D) Arg-CCG-1–1 and (E) Ile-GAT-1–1 are up-regulated at 47 oC, whereas (F) Met-CAT-1–1 and (G) Arg-CCT-1–1 are down-regulated at 47 °C. The arrows show the DMS reactive sites only seen under the h. stress condition.
Finally, we performed differential expression analyses between heat shock and heat stress, with a goal of understanding whether these two treatments affect tRNA structure and expression differently. We found that two tRNAs had an increase in abundance and four had a decrease in heat stress as compared to heat shock (SI Appendix, Fig. S7C). Apparently, tRNA levels can change between these two stress conditions.
tRNA modification levels change upon heat stress.
tRNA is the most heavily modified RNA on a per-base basis (42). In principle, ShapeMapper2 can detect natural modifications in tRNA if they result in mutations in the –DMS cDNA dataset (see SI Appendix, Materials and Methods). Indeed, we were able to detect a significant number of nucleobase modifications, as described earlier (Fig. 1 F and G). When we used just raw in vivo tRNA data (i.e., –DMS), we detected one natural modification in tRNAGly-CCC-1–1, s4U8, but not the other three modifications (SI Appendix, Fig. S9A). In an effort to enhance detection of natural modifications, we tested if subtraction of the in vitro data (–DMS) from the in vivo data (–DMS) for tRNAGly-CCC-1–1 could improve the signal-to-noise (SI Appendix, Fig. S9B). Indeed, mutational signals from natural modifications are not present in the in vitro DMS reactivities as expected (compare Fig. 2C to SI Appendix, Fig. S3). Remarkably, subtraction of the T7 transcript reactivity data led to identification of the other three modifications (SI Appendix, Fig. S9B). It is particularly noteworthy that three of these modifications are not on the Watson–Crick face (D20, m5U54, and Ψ55) yet are still detected using MaP with Marathon.
Next, we analyzed 36 native tRNAs for which we had sufficient in vivo data, i.e., modifications reported in Modomics (42) and at least 5,000 full length reads, to derive modification profiles using the strategy defined above (Fig. 5 A–D and SI Appendix, Fig. S10). As expected, modifications at the Watson–Crick face such as s4U8 and bulky modifications such as 2-methylthio-N6-isopentenyladenosine at position 37 (ms2i6A37) and N6-methyl-N6-threonylcarbamoyladenosine at position 37 (m6t6A37) tend to have a strong mutation signal (Fig. 5 A–D and SI Appendix, Fig. S10, yellow stripe, blue color). In contrast, 2′-O-methyl modifications (e.g., Gm) and dihydrouridines (D), as well as m5U54 and Ψ55 have weak signal (Fig. 5 A–D, yellow stripe, greenish or no colors). In particular, our mutational profiling detected neither of the two dihydrouridines in tRNALeu TAA-1–1 (Fig. 5B) and only one of three dihydrouridines in tRNAThr GGT-1–1 (Fig. 5C). Overall, we detected about 40% of known modified positions in E. coli tRNA (Fig. 5E).
Fig. 5.
Levels of tRNA modification vary under heat stress conditions. (A–D) Normalized reactivity for E. coli tRNA modifications was detected by mutational profiling. “Experiment” shows positions that have high mutation rate (–DMS) whereas “Model” shows positions of the known modification in E. coli. Examples of tRNA mutational profiling for (A) tRNAGly CCC-1–1, (B) tRNALeu TAA-1–1, (C) tRNAThr GGT-1–1, and (D) tRNATyr GTA-1–1 are provided. (E) Mutational profiling is depicted on the secondary structure of tRNA. In the gray box, “known mods” (the denominator) is the number of known natural modifications at particular positions in tRNA according to the Modomics database. The “detected mods” (the numerator) is the number of detected natural modifications by tRNA structure-seq, where detected normalized mutation rates are higher than 2 at the positions. (F) Mutation rates and MS normalized peak areas are correlated. tRNA modification levels estimated by averaged mutational profiling (n = 2) and averaged nucleoside analysis (n = 4). (G–N) Normalized reactivities for overall levels of the known modifications in 36 E. coli tRNAs are plotted. The blue boxes are the standard conditions and the red boxes are the heat conditions. Numerous changes in the individual levels of (G) s4U8, (H) dihydrouridines, (I) Gm18, (J) anticodon modifications at position 32 including Cm32, Um32, and s2C, (K) anticodon modifications at position 34 including cmo5U, Q, ac4C, GluQ, cmnm5Um, mnm5U, cmnm5s2U, mnm5s2U, and Cm, (L) anticodon modifications at position 37 including m2A, ms2i6A, t6A, m6t6A, and m1G, (M) m7G46, and (N) acp3U47 are apparent. All data are found in SI Appendix, Fig. S12.
Historically, levels of tRNA modification have been estimated by nucleoside analysis with LC-MS/MS where tRNA is degraded to nucleosides (43, 44). However, this does not provide information on the site of modification nor on the tRNA identity. We thus tested if the mutation rates in mutational profiling correlate with modification levels based on LC-MS/MS. Four dilution series of native yeast tRNAPhe were prepared, and modification levels were determined by nucleoside analysis with LC-MS/MS or mutational profiling (SI Appendix, Fig. S11). Mutation rate and MS normalized peak area are highly correlated (Fig. 5F and SI Appendix, Fig. S11), indicating that mutational profiling indeed can estimate relative levels of known modifications.
We then applied mutational profiling to test how temperature affects levels of known natural modifications in tRNA. Changes in the levels of modifications in 36 different E. coli tRNAs were estimated from in vivo –DMS data under the four different growth conditions (Fig. 5 G–N and SI Appendix, Fig. S12). We found that levels of s4U and D were statistically constant under heat stress conditions, but that s4U levels increased dramatically under heat shock (Fig. 5 G and H). Modification at position 18 (=2′-O-methylguanine (Gm18)) increased under both heat stress and heat shock conditions (Fig. 5I). Considering the anticodon, levels of modification at positions 34 and 37 were relatively constant with heat (Fig. 5 K and L), but modification at position 32 (=2′-O-methylcytidine (Cm32), 2-thiocytidine (s2C), or 2′-O-methyluridine (Um32),) decreased in the heat stress condition (Fig. 5J). Levels of m7G46 increased under heat stress, while 3-amino-3-carbozypropyluridine at position 47 (acp3U47) decreased quite significantly under the heat stress and increased slightly under heat shock conditions (Fig. 5 M and N). We tracked the extent of modification at specific tRNA sites (SI Appendix, Fig. S12), which were cross-validated by nucleoside analysis (SI Appendix, Figs. S13–S16). The relative changes in modifications calculated from MaP and LC-MS/MS are reasonably correlated (R = 0.63 and P = 3.9e−5) (SI Appendix, Fig. S16E), which supports the quantitative accuracy of mutational profiling.
Discussion
Transfer RNA is one of the most studied RNA molecules due to the importance of the decoding system. Indeed, the cloverleaf secondary structure of tRNA was determined in 1965 and the first crystal structure was solved in 1974 (45, 46). Besides the decoding system, recent studies have revealed that tRNA is related to myriad cellular phenomena such as regulation of the metabolome, gene silencing, and formation of viral particles (47, 48). Despite the extensive long-term studies on tRNA, it has been unclear how tRNA folds in vivo and under various stress conditions. Our techniques help address such outstanding questions. Structure-seq incorporates experimental accessibility data to improve the accuracy of RNA secondary structure prediction and reveals roles of RNA structure in biological processes. Transfer RNA is a small ∼80 nt RNA that plays critical roles in translation including supplying amino acids to the ribosome and decoding codons. It undergoes processing and possesses many posttranscriptional modifications. Loss of tRNA functions causes severe disease in humans including developmental disorders, heart disease, and metabolic diseases (21). It is thus essential to address how tRNA gains and loses its structure and function in the living cell.
Herein, we adapted our Structure-seq method (9, 13, 14) to the tRNA structurome. We find that tRNA structure-seq can predict canonical tRNA structures with 95% average accuracy while revealing that under heat stress conditions many tRNAs undergo a loss in tertiary structure and that the acceptor stem unfolds in a subset of these tRNAs. Using tRNA structure-seq data, we found evidence that tRNA structure also affects tRNA abundance and modification levels.
tRNA structure-seq provides structural information on tRNA under in vivo conditions.
tRNA structure-seq gives insight into the folding of multiple tRNAs under in vivo conditions. This is achieved by DMS probing combined with mutational profiling (MaP) (49). The DMS modifications at single-stranded regions in tRNA can give rise to an RT stop or misincorporation during cDNA synthesis. We used Marathon RTase, a highly processive reverse transcriptase, to promote misincorporations rather than termination in the cDNA at the sites of DMS modification, and conducted size selection of cDNA corresponding to full-length tRNAs. The mutations in cDNA then were quantified by deep sequencing.
We conducted tRNA structurome determination under several conditions: in silico, in vitro, and in vivo under 30 °C, 37 °C, heat stress, and heat shock conditions (Figs. 2D and 3A). Structure prediction, performed by the minimal free energy calculation, revealed that the overall prediction accuracy improved from 79% in silico to 95% in the control in vivo (37 °C) condition. The heat stress conditions, on the other hand, significantly diminished canonical tRNA folding, down to ∼88% on average, and much more for certain tRNAs (Fig. 3A), suggesting that heat stress conditions partially unfold certain tRNA structures. This was supported by enhanced DMS reactivities in acceptor stem and D- and T-loops under heat stress (Figs. 2C and 3).
It is also of interest to consider those tRNAs whose prediction accuracy remains poor under all three conditions: in silico, in vitro, and in vivo. For Gly-GCC-1–1 (accuracy is 67%), the DMS reactive positions look as expected; however, the sequence in the variable loop is incorrectly predicted to base pair with the 5′ sequence in the anticodon stem (SI Appendix, Fig. S5 and Table S3). For Leu-CAG-2–1, accuracy is 100% at 30 °C but drops to 67% at 37 °C and remains near this level under heat stress and shock, suggesting a partial unfolding even at 37 °C (see SI Appendix, Fig. S5 and Table S3). Finally, for Pro-TGG-1–1, as in the case of Gly-GCC-1–1 (above), the sequence in the variable loop can base pair elsewhere, here with the sequence in the T-stem (see SI Appendix, Fig. S5) although it folds closer to the canonical cloverleaf at 30 oC and under stress conditions. Thus, some tRNAs might serve enhanced roles in cold conditions (e.g., Leu-CAG-2-1) while others might do so in heat (e.g., Pro-TGG-1-1).
tRNA structure-seq provides insight into tRNA abundance and modification levels during heat stress and shock.
tRNA structure-seq provides relative levels of tRNAs and their site-specific modifications (Figs. 4 and 5). Our data show that the abundance of both initiator tRNAfMets, which are natively folded under all four conditions (SI Appendix, Table S3), increases upon heat shock (P values∼10−28 and 10−31) (Fig. 4B); strikingly, these were the only tRNAs to increase in abundance under these conditions. This suggests a possible first heat-shock response where more tRNAfMet could enhance the kinetics of the tRNA binding to the P site on the small ribosomal subunit during the formation of the initiation complex. It is also notable that Phe-GAA-1–1 was one of only two tRNAs to decrease in abundance upon heat shock (P value∼10−46), with Ser-GCT-1–1 being the other (P value∼10−19). Notably, Phe-GAA-1–1 is the only tRNA that decodes Phe, suggesting a possible pause in elongation on most mRNAs during the brief heat shock. Indeed, loss of Phe-GAA-1–1 and Ser-GCT-1–1 would halt translation of most proteins near the N terminus, potentially preventing translation and heat-associated protein misfolding (SI Appendix, Fig. S17). Notably, over 99% of all annotated codons in E. coli have one of these three codons, indicating that nearly all protein synthesis would stop. To summarize, during heat shock ribosomes could be strongly recruited to initiate on mRNA, but then stall at Phe-GAA-1–1 and Ser-GCT-1–1 codons, the net effect of which could be protection of the 5′-end of mRNAs and protection against protein misfolding. By contrast, numerous tRNAs changed abundance after the longer heat stress (Fig. 4A) suggesting that many tRNAs may be involved in adaptation to heat stress.
“Modification profiling” has been used to assess modifications in RNA. For example, Kimura and coworkers used RT stop counts and mis-incorporaton rates from the RT reaction to explore tRNA biology wherein their tRNA-seq combined with mass spectrometry enabled identification of a new modification and C-to-Ψ editing in tRNA in V. cholerae (38). Similarly, modification profiling has been used for mapping known RNA modifications such as N6-methyladenosine (m6A), m7G, and 4-acetylcytidine (ac4C) as well as for detecting RNA editing at a transcriptomic level (50–52). Here, we showed that RT in the presence of Mn2+ with subtraction of in vitro data significantly improves the detection sensitivity of natural tRNA modification (SI Appendix, Fig. S9). Our study suggests that mutation rates from MaP are proportional to the modification levels from LC-MS/MS, and therefore the two methods can be combined to identify and quantify modifications (Fig. 5F and SI Appendix, Fig. S16E). At this point, we speculate that mutation rates do not necessarily reflect stoichiometry of modifications in tRNA. For instance, normalized mutation rates of 300 and 20 from different modifications are likely to not mean the stoichiometry of the modifications is 300:20 because mutation rates are likely to be affected by the size of a functional group and its modification site, as well as by the efficiency of ionization in the mass spectrometer (Fig. 5 G–N). Even so, our method allows relative quantification of the same RNA modification as shown in Fig. 5F. Levels of RNA modification previously have been shown to change in response to environmental stress and thus our method may find broad usage. For example, in E. coli levels of the 2′-O-methylated nucleotides in tRNA are up-regulated under antibiotic conditions, while levels of Cm32 and Um32 are down-regulated at 42 °C (53). This down-regulation is consistent with our heat stress data at position 32 for most tRNAs (Fig. 5J), while we see up-regulation upon heat stress/shock for other site-specific modifications such as s4U8/9, Gm18 and m7G46 in certain tRNAs (Fig. 5 and SI Appendix, Fig. S12). Future study will be needed to probe a relationship between individual modifications and tRNA folding.
Comparison of tRNA structure-seq to other genomic RNA mapping methods.
Two genome-wide RNA structure probing techniques, SHAPE-MaP and DMS-MaP, pioneered the use of MaP (8, 10, 49). The Weeks laboratory has applied mutational profiling to ribonucleoproteins (RNPs) by in-cell crosslinking between RNAs and proteins (RNP-MaP), which revealed the RNA-protein interactome of multiple RNPs (54). A recent study from the Rouskin laboratory improved DMS-MaP seq in which they developed the DREEM algorithm that enables detection of alternative RNA structures by identifying mutually exclusive patterns of DMS-induced mutations in sequencing reads and generating multiple clusters of structures in a mathematically rigorous manner (49). These important MaP technologies have led to technical and biological breakthroughs but have not been benchmarked against highly modified RNAs with defined tertiary structures. In contrast, our study focuses on tRNAs, which have crystal structures of the native modified state that enable us to report on prediction accuracy and test defined secondary and tertiary structural elements.
The Narberhaus laboratory recently developed “Lead-seq” that also enables in vivo structure probing of numerous RNAs including tRNAs (55). In their method, chemical cleavage of RNA by Pb2+ causes RT termination, which results in short cDNA fragments. On the one hand, this method improved the prediction accuracy of tRNA secondary structure in vivo, but on the other it lacked signal in a 20 nt stretch at the critical 3′ CCA terminus due to the inability to unambiguously align short cDNA fragments. Furthermore, because it uses a toxic metal ion, Lead-seq may initiate a stress response in the organism, and lead ions are known to bind to tRNA and so could alter their structure (56).
Several other quantitative tRNA sequencing methods have been developed (34–39). Recently, Nedialkova and coworkers developed mim-tRNAseq that quantifies the abundance and modification status of eukaryotic tRNAs by using an optimized TGIRT reaction, considering the positions of known natural modifications, and clustering tRNAs with the same anticodon (39). These methods have each provided important steps forward in in vivo tRNA analysis. In comparison, tRNA structure-seq is unique in that it reports tRNA structure, abundance, and modifications in a single experiment. It uniquely detects nucleoside modifications beyond methylations and does so without fragmenting the tRNA, which aids mapping back to specific tRNA transcripts.
Conclusions.
Overall, our tRNA structure-seq workflow provides multiple fresh insights into how tRNA folds in living cells and how tRNA structure, abundance, and modification levels sense changes in temperature. The malleability of tRNA structure that we report herein could affect any tRNA-related event including tRNA processing, aminoacylation levels, and translation efficiency. To address these events, in the future tRNA structure-seq could be applied to precursor tRNAs, charged tRNAs, organellar tRNA and to ribosome profiling for detailed translation analysis (57). Finally, in the future, the workflow herein could be applied to structured and modified small RNAs beyond tRNA to reveal new facets of RNA biology.
Materials and Methods
Following is a summary of methods, with details in the SI Appendix. In vivo chemical probing was performed with 100 mM DMS at various temperatures for 5 min. Total RNA was extracted by TRIzol. Then, the tRNA fraction was purified by gel separation and extraction. The adapter was ligated to the 3′ end of tRNA, and then RT was performed with Marathon reverse transcriptase in the presence of 2 mM Mn2+. The resultant full-length cDNA was further purified by gel separation and extraction. The purified cDNA was circularized with CircLigase II. Indexing PCR was performed with Q5 high fidelity DNA polymerase for 17 cycles. Libraries were sequenced on an Illumina NextSeq 550. Adapter sequence was trimmed using Cutadapt (58) and resulting sequencing data were analyzed by ShapeMapper2 (25). ShapeMapper outputs were analyzed by custom Python and R scripts. Structure prediction was performed with RNAstructure (29). For nucleoside analysis, tRNA was digested by Benzonase, snake venom phosphodiesterase, and bacterial alkaline phosphatase at 37 °C for 3 h. The reaction mixture was filtered through an Amicon ultracentrifugal filter (Mw 10,000 cut) to remove the enzymes. Nucleoside analysis was performed using Shimadzu Prominence UPLC in line with an AB SCIEX 5600 Triple TOF tandem mass spectrometer.
Supplementary Material
Acknowledgments
We thank the Metabolomics Facility and Genomics Core Facility at Pennsylvania State University for assistance with the LC-MS/MS experiments and next-generation sequencing experiments, respectively. We also thank Dr. Andrew Veenis for help with the DMS accessibility analysis and Professor Paul Babitzke and Dr. Satoshi Kimura for helpful conversations. This work was supported by National Institutes of Health grant R35-GM127064 (to P.C.B.). R.Y. was supported by an Overseas Research Fellowship (201906624) from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201237119/-/DCSupplemental.
Data Availability Statement
Sequencing data, all data for figures, supplementary figures, tables, and supplementary tables are available at Scholar Sphere (https://scholarsphere.psu.edu/resources/fd08962f-fee4-47ad-ae97-0d0562cb29ec).
References
- 1.Cech T. R., Steitz J. A., The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Tapial J., et al. , An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 27, 1759–1768 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehresmann C., et al. , Probing the structure of RNAs in solution. Nucleic Acids Res. 15, 9109–9128 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwok C. K., Tang Y., Assmann S. M., Bevilacqua P. C., The RNA structurome: Transcriptome-wide structure probing with next-generation sequencing. Trends Biochem. Sci. 40, 221–232 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Incarnato D., Oliviero S., The RNA epistructurome: Uncovering RNA function by studying structure and post-transcriptional modifications. Trends Biotechnol. 35, 318–333 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Mustoe A. M., Lama N. N., Irving P. S., Olson S. W., Weeks K. M., RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl. Acad. Sci. U.S.A. 116, 24574–24582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell D. III, Assmann S. M., Bevilacqua P. C., Probing RNA structure in vivo. Curr. Opin. Struct. Biol. 59, 151–158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouskin S., Zubradt M., Washietl S., Kellis M., Weissman J. S., Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding Y., et al. , In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Siegfried N. A., Busan S., Rice G. M., Nelson J. A. E., Weeks K. M., RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deigan K. E., Li T. W., Mathews D. H., Weeks K. M., Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U.S.A. 106, 97–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwok C. K., Ding Y., Tang Y., Assmann S. M., Bevilacqua P. C., Determination of in vivo RNA structure in low-abundance transcripts. Nat. Commun. 4, 2971 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Ritchey L. E., et al. , Structure-seq2: Sensitive and accurate genome-wide profiling of RNA structure in vivo. Nucleic Acids Res. 45, e135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tack D. C., Tang Y., Ritchey L. E., Assmann S. M., Bevilacqua P. C., StructureFold2: Bringing chemical probing data into the computational fold of RNA structural analysis. Methods 143, 12–15 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Zubradt M., et al. , DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan T., Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopper A. K., Nostramo R. T., tRNA processing and subcellular trafficking proteins multitask in pathways for other RNAs. Front. Genet. 10, 96 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagami R., Bingaman J. L., Frankel E. A., Bevilacqua P. C., Cellular conditions of weakly chelated magnesium ions strongly promote RNA stability and catalysis. Nat. Commun. 9, 2149 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leamy K. A., Yamagami R., Yennawar N. H., Bevilacqua P. C., Single-nucleotide control of tRNA folding cooperativity under near-cellular conditions. Proc. Natl. Acad. Sci. U.S.A. 116, 23075–23082 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones C. N., Jones C. I., Graham W. D., Agris P. F., Spremulli L. L., A disease-causing point mutation in human mitochondrial tRNAMet rsults in tRNA misfolding leading to defects in translational initiation and elongation. J. Biol. Chem. 283, 34445–34456 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bednářová A., et al. , Lost in translation: Defects in transfer RNA modifications and neurological disorders. Front. Mol. Neurosci. 10, 135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lant J. T., Berg M. D., Heinemann I. U., Brandl C. J., O’Donoghue P., Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 294, 5294–5308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cozen A. E., et al. , ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo L. T., et al. , Sequencing and structure probing of long RNAs using MarathonRT: A next-generation reverse transcriptase. J. Mol. Biol. 432, 3338–3352 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busan S., Weeks K. M., Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2. RNA 24, 143–148 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tack D. C., Su Z., Yu Y., Bevilacqua P. C., Assmann S. M., Tissue-specific changes in the RNA structurome mediate salinity response in Arabidopsis. RNA 26, 492–511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi H., Moore P. B., The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: A classic structure revisited. RNA 6, 1091–1105 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan C. W., Badong D., Rajan R., Mondragón A., Crystal structures of an unmodified bacterial tRNA reveal intrinsic structural flexibility and plasticity as general properties of unbound tRNAs. RNA 26, 278–289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuter J. S., Mathews D. H., RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11, 129 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan P. P., Lowe T. M., GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su Z., et al. , Genome-wide RNA structurome reprogramming by acute heat shock globally regulates mRNA abundance. Proc. Natl. Acad. Sci. U.S.A. 115, 12170–12175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spasic A., Assmann S. M., Bevilacqua P. C., Mathews D. H., Modeling RNA secondary structure folding ensembles using SHAPE mapping data. Nucleic Acids Res. 46, 314–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong H., Nilsson L., Kurland C. G., Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260, 649–663 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Zheng G., et al. , Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gogakos T., et al. , Characterizing expression and processing of precursor and mature human tRNAs by Hydro-tRNAseq and PAR-CLIP. Cell Rep. 20, 1463–1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y., Silke J. R., Xia X., An improved estimation of tRNA expression to better elucidate the coevolution between tRNA abundance and codon usage in bacteria. Sci. Rep. 9, 3184 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinkard O., McFarland S., Sweet T., Coller J., Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. Nat. Commun. 11, 4104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura S., Dedon P. C., Waldor M. K., Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. Nat. Chem. Biol. 16, 964–972 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behrens A., Rodschinka G., Nedialkova D. D., High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol. Cell 81, 1802–1815.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boccaletto P., et al. , MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basanta-Sanchez M., Temple S., Ansari S. A., D’Amico A., Agris P. F., Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res. 44, e26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamagami R., et al. , Folate-/FAD-dependent tRNA methyltransferase from Thermus thermophilus regulates other modifications in tRNA at low temperatures. Genes Cells 21, 740–754 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Holley R. W., et al. , Structure of a ribonucleic acid. Science 147, 1462–1465 (1965). [DOI] [PubMed] [Google Scholar]
- 46.Robertus J. D., et al. , Structure of yeast phenylalanine tRNA at 3 A resolution. Nature 250, 546–551 (1974). [DOI] [PubMed] [Google Scholar]
- 47.Avcilar-Kucukgoze I., Kashina A., Hijacking tRNAs from translation: Regulatory functions of tRNAs in mammalian cell physiology. Front. Mol. Biosci. 7, 610617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y.-T., Strugatsky D., Liu W., Zhou Z. H., Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150. Nat. Commun. 12, 5513 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomezsko P. J., et al. , Determination of RNA structural diversity and its role in HIV-1 RNA splicing. Nature 582, 438–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linder B., et al. , Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Enroth C., et al. , Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. 47, e126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sas-Chen A., et al. , Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature 583, 638–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galvanin A., et al. , Bacterial tRNA 2′-O-methylation is dynamically regulated under stress conditions and modulates innate immune response. Nucleic Acids Res. 48, 12833–12844 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weidmann C. A., Mustoe A. M., Jariwala P. B., Calabrese J. M., Weeks K. M., Analysis of RNA-protein networks with RNP-MaP defines functional hubs on RNA. Nat. Biotechnol. 39, 347–356 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Twittenhoff C., et al. , Lead-seq: Transcriptome-wide structure probing in vivo using lead(II) ions. Nucleic Acids Res. 48, e71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C., Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry 29, 2515–2523 (1990). [DOI] [PubMed] [Google Scholar]
- 57.Ingolia N. T., Ghaemmaghami S., Newman J. R. S., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data, all data for figures, supplementary figures, tables, and supplementary tables are available at Scholar Sphere (https://scholarsphere.psu.edu/resources/fd08962f-fee4-47ad-ae97-0d0562cb29ec).