Abstract
Respiratory syncytial virus (RSV) is a leading cause of lower respiratory infections in infants and the immunocompromised, yet no efficient therapeutic exists. We have identified the AVG class of allosteric inhibitors of RSV RNA synthesis. Here, we demonstrate through biolayer interferometry and in vitro RNA-dependent RNA polymerase (RdRP) assays that AVG compounds bind to the viral polymerase, stalling the polymerase in initiation conformation. Resistance profiling revealed a unique escape pattern, suggesting a discrete docking pose. Affinity mapping using photoreactive AVG analogs identified the interface of polymerase core, capping, and connector domains as a molecular target site. A first-generation lead showed nanomolar potency against RSV in human airway epithelium organoids but lacked in vivo efficacy. Docking pose-informed synthetic optimization generated orally efficacious AVG-388, which showed potent efficacy in the RSV mouse model when administered therapeutically. This study maps a druggable target in the RSV RdRP and establishes clinical potential of the AVG chemotype against RSV disease.
An orally efficacious antiviral targeting a dynamic interface in respiratory syncytial virus polymerase is identified.
INTRODUCTION
Respiratory syncytial virus (RSV) caused an estimated 33.1 million cases worldwide in 2015 that required 3.2 million hospitalizations and resulted in 59,800 deaths (1). To address this health threat, a number of vaccine and drug candidates were tested clinically. However, inducing lasting vaccine protection turned out to be challenging (2) and the entry inhibitor presatovir that has completed several phase 2b trials has disappointed in subsequent trials (3, 4). Rapid RSV escape from all advanced entry inhibitor candidate classes through mutations mediating pan-resistance (5) further questions clinical potential of RSV entry inhibition. Biopharmaceuticals for immunoprophylaxis have shown promise (6) and the monoclonal anti-RSV antibody palivizumab is approved for human use, but the high cost of antibody therapy has prohibited broad application. Accordingly, palivizumab is predominantly reserved for high-risk patients such as the immunocompromised and infants born prematurely or with congenital heart or lung disease (7).
To overcome the limitations of RSV entry inhibitors (8), drug development efforts targeting the viral RNA-dependent RNA polymerase (RdRP) complex have intensified, motivated by the prospect of a broader window of opportunity through interference with both viral genome replication and transcription. Composed of the viral large (L) and phospho- (P) proteins, all enzymatic centers of the RdRP complex that are necessary for RNA synthesis, polyadenylation, capping, and cap-methylation of viral transcripts are located in the L protein (9). The template for replicase and transcriptase is a nonsegmented, single-stranded 15-kilobase RNA of negative polarity that is encapsidated by the viral nucleoprotein (N) (10). Accordingly, RdRP bioactivity depends on multiple intra- and intermolecular protein-protein interfaces to enable interactions between P and L (11), the P-L complex and the N encapsidating genomic viral RNA (12), P-L and the viral anti-termination cofactor M2-1 (13), and P-L and host cell cofactors (14–16).
In the past decade, structural insight was gained in the spatial organization of the RSV L protein (17, 18), which was in all cases complexed with a fragment of P. Although some experimental RSV polymerase inhibitors are considered to interact with the N (19) or M2-1 proteins (20), most developmental candidates are thought to target the L protein directly (21, 22). The two RSV L structural models offer a clear view of the core RdRP and capping domains. However, these L structures are thought to represent the conformational state of active transcription, based on the position of a “priming” loop away from the polymerase catalytic center. Conformational insight into L in initiation configuration has remained elusive, since flexibility of connector and methyltransferase (MTase) domains located downstream of the capping domain prevented structural characterization. These limitations have impaired the mechanistic understanding of allosteric polymerase inhibitors that are associated with resistance mutations located in the C-terminal regions of L (Fig. 1A), such as AstraZeneca’s inhibitor AZ-27 (23).
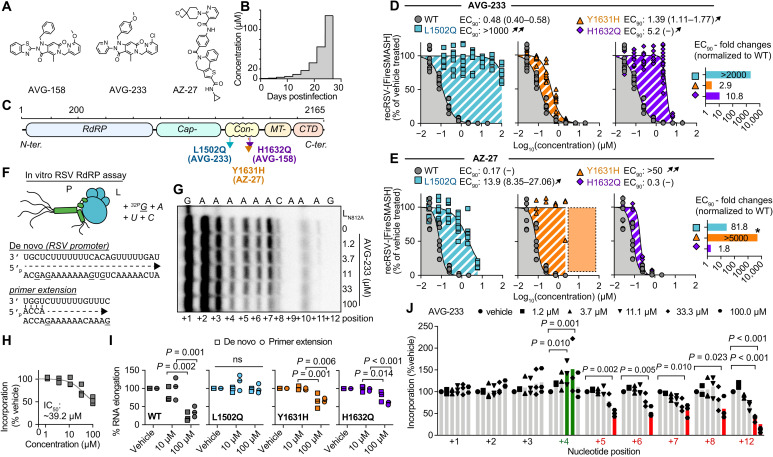
Fig. 1. Resistance and mechanistic profiling of AVG-233.
(A) Chemical structure of AVG-158, AVG-233, and AZ-27. (B) Schematic of the escalating-dose viral adaptation of recRSV-mKate with AVG-158 or AVG-233. (C) Schematics of RSV L with candidate resistance sites for AVG-233 and AVG-158 and a reported resistance site to polymerase inhibitor AZ-27. (D and E) Dose-response inhibition of AVG-233 (D) and AZ-27 (E) against recRSV-mKate harboring L1502Q, Y1631H, or H1632Q substitution. Insets show EC90 fold changes relative to L wild type (WT). Highest AZ-27 concentration tested, 6 μM; dotted area and star based on reported values (23). (F to J) In vitro RSV RdRP assays using synthetic primer/template pairs (G to J) or promoter sequence (I). Representative autoradiogram (G) with densitometric analysis (H). (I) Relative in vitro RNA elongation in the presence of indicated doses of AVG-233 compared to vehicle treated, in the presence of L1502Q, Y1631H, or H1632Q substitution. (J) Densitometric quantitation of in vitro RNA elongation at each incorporated position in the presence of indicated concentration of AVG-233. In all panels, symbols represent independent biological repeats (n = 3), bars represent means, and lines represent four-parameter variable slope regression modeling. Two-way ANOVA with Dunnett’s post hoc tests (I and J).
We have recently identified a unique chemotype that potently blocks RSV RdRP bioactivity (24). A first-generation lead compound of this class, AVG-233, has nanomolar activity against a panel of clinical RSV isolates and has a promising selectivity index [SI = 50% cytotoxic concentration (CC50)/half-maximal effective concentration (EC50)] > 1660. In preparation of formal development, this study has subjected the AVG chemotype to target site identification, characterization of the molecular mechanism of action, and efficacy profiling in primary human airway epithelium (HAE) organoids and in vivo, resulting in the identification of an orally efficacious developmental lead.
RESULTS
We have demonstrated that compounds of the AVG series potently inhibit RSV RNA synthesis (24). To identify the molecular target of this class, we resistance-profiled the chemotype through serial passaging of a recombinant RSV expressing a red fluorescent protein (recRSV-mKate) in three independent lines each in the presence of increasing concentrations of either AVG-233 or an earlier developmental intermediate of the series, AVG-158 (Fig. 1A). Virus populations were considered adapted when inhibitor concentrations exceeding the original EC50 value by 100-fold were tolerated (Fig. 1B).
Unique resistance profile of the AVG series chemotype
Sanger sequencing revealed a unique substitution in the L open reading frame for each lineage that affected either residue 1502 (LL1502Q) or 1632 (LH1632Q) (Fig. 1C). Resistance site LH1632Q is located in immediate proximity to a known LY1631H hotspot that is reportedly involved in escape from several chemically unrelated allosteric RSV polymerase inhibitor classes such as the benzothienoazepines YM-53503 (25), AZ-27 (23), and PC786 (26) and the AstraZeneca inhibitor cpd1 (27). We therefore reengineered LL1502Q, LH1632Q, and LY1631H independently in RSV L expression plasmids and recRSV, and determined susceptibility to both AVG-233 and AZ-27 in minigenome and/or virus dose-response inhibition assays (fig. S1).
In the minigenome assays, the LL1502Q and LH1632Q mutations caused a ≥100-fold increase in EC50, whereas LY1631H had only a minor (approximately fivefold increase) effect (fig. S1). Rebuilding of the mutations in recRSV did not adversely affect viral growth rates and maximal progeny titers reached in multistep growth curves (fig. S2). Consistent with the minireplicon data, LL1502Q caused a >2000-fold increase in EC90 against AVG-233, whereas LY1631H barely conferred resistance to the compound (Fig. 1D). However, the LH1632Q mutation had only a moderated effect in the context of recRSV infection, increasing in EC90 values only by approximately 10-fold. Resistance profiles of AZ-27 drew a mirror image of those of AVG-233; the LY1631H substitution resulted in robust resistance (>2000-fold increase in EC90) consistent with previous reports (23), whereas LH1632Q did not mediate viral escape and LL1502Q had only a moderate effect (Fig. 1E).
To better understand the molecular basis for L inhibition and resistance, we purified RSV P-L polymerase complexes harboring the different substitutions (fig. S3) and subjected the preparations to in vitro RdRP assays using synthetic RNA templates and a 32P-GTP (guanosine triphosphate) tracer (Fig. 1F). Consistent with previous observations (24), AVG-233 dose-dependently impaired RNA elongation of synthetic primer/template RNA pairs (Fig. 1G), exhibiting a ~39 μM median inhibitory concentration (IC50; Fig. 1H). RNA incorporation in vitro was reduced to 36% (±14.6) in the presence of 100 μM AVG-233, but the L1502Q escape mutation restored activity to 98.7% (±12.9), indicating robust resistance (Fig. 1I and fig. S4). By comparison, Y1631H and H1632Q mediated only moderate escape from AVG-233, represented by restoring polymerase activity to 69% (±5.1) and 60% (±4.0) of untreated, respectively. Similar resistance profiles were observed when using a single-stranded RSV promoter as a template to initiate de novo RNA synthesis at the promoter. RNA incorporation after initiation was reduced to 21.7% (±11.8), 96.6% (±4.0), 66.0 (±20.8), and 79.7% (±14.5) when AVG-233 was added to complexes containing L, LL1502Q, LY1631H, or LH1632Q, respectively (Fig. 1I and fig. S5). The arrest of polymerization occurred predominantly after incorporation of four nucleotides and thus after an initial delay (Fig. 1J). However, we noted some variation in the extent of the delay depending on primer/template sequence (fig. S6).
To test for competition of AVG-233 with endogenous RSV polymerase substrates, we added increasing amounts of exogenous nucleosides to the culture medium of RSV-infected cells grown in the presence of a sterilizing concentration of AVG-233 (20 μM). In contrast to the known competitive inhibitor 4′-FlU (22), AVG-233 inhibition of RSV replication was unaffected by exogenous nucleosides, indicating that AVG class compounds block the viral RdRP by a noncompetitive mechanism (fig. S7). These results define a noncompetitive blockage of RNA synthesis after the initiation step as the primary mechanism of L inhibition by AVG-233. Whereas mechanistically similar to AZ-27, the nonoverlapping resistance profiles indicate distinct docking poses.
Positive AVG-233 target identification
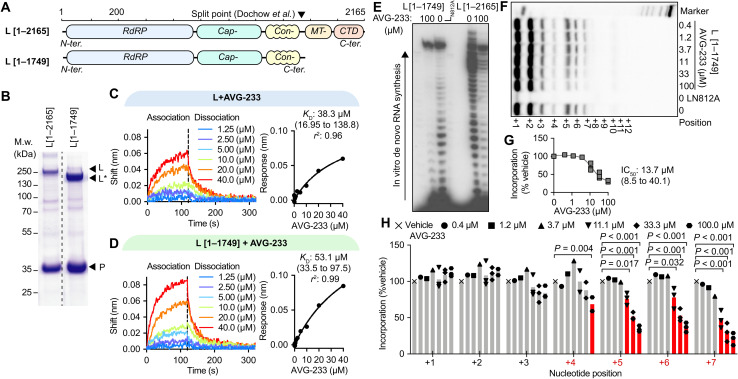
A biologically active RSV polymerase can reconstitute from two independently expressed fragments that are split between the connecting and MTase domains (28) (Fig. 2A), indicating that each of these individual fragments retains folding competence. Using baculovirus/insect cell protein expression, we prepared full-length L and the larger N-terminal fragment of the split L (residues L1–1749) that harbors all of the RdRP domain and all AVG-233 resistance sites (Fig. 2B). Direct binding of AVG-233 to these L preparations was tested through biolayer interferometry (BLI). Both standard L and the L1–1749 fragment bound AVG-233 with similar affinities [dissociation constants (KD’s) were 38.3 μM (16.9 to 138.8) and 53.1 μM (33.5 to 97.5), respectively] (Fig. 2, C and D), providing positive confirmation of L as the molecular target of the compound and demonstrating that the inhibitor binding site is fully present in the L1–1749 fragment.
Fig. 2. Label-free positive target identification of AVG-233.
(A) Schematic of two RSV L targets examined, representing the full-length polypeptide (top) and a folding-competent (28) truncated L1–1749 polypeptide lacking the MTase and C-terminal (CTD) domains. (B) Purification of polymerase complexes specified in (A) after expression in insect cells. Coomassie blue stain after SDS-PAGE fractionation. (C and D) Dose-dependent BLI-based association (120 s) and dissociation (200 s) curves (left) of AVG-233 with full-length RSV L (C) and L1–1749 (D), with nonlinear fit with one-site specific binding (right). (E) In vitro RdRP assay using a synthetic promoter RNA template for de novo RNA synthesis; representative autoradiogram. (F to H) Primer/template-based in vitro RNA elongation assay, assessing bioactivity and AVG-233 inhibition of L1–1749, with representative autoradiogram (F), densitometric quantitation of elongation products (G), and relative quantitation of each RNA product (H). Symbols represent independent biological repeats (n = 3), and bars represent means. Two-way ANOVA with Dunnett’s post hoc tests.
Since this fragment contains the catalytic site for phosphodiester bond formation (17, 18, 29), we explored its ability to synthesize RNA in vitro, using the biochemical RdRP assay setup. Consistent with proper folding into a native conformation, the L1–1749 subunit was RNA synthesis competent in the biochemical assay (Fig. 2E), although with a noticeable drop in processivity after the ninth incorporated nucleotide compared to full-length L. This L fragment was equally susceptible to inhibition by AVG-233 in de novo RNA synthesis (Fig. 2E), confirming productive inhibitor binding.
When testing primer/template-based RNA elongation by the L1–1749 fragment, we observed dose-dependent inhibition by AVG-233 with an IC50 of 13.7 μM (Fig. 2, F and G), which did not substantially differ from the value (IC50 of 39.2 μM) calculated for full-length L (Fig. 1, G and H). AVG-233 suppression of RNA synthesis by the L1–1749 fragment was again delayed, since a significant inhibitory effect was first detectable after incorporation of four nucleotides (Fig. 2H).
Photoaffinity-based AVG-233 target site mapping
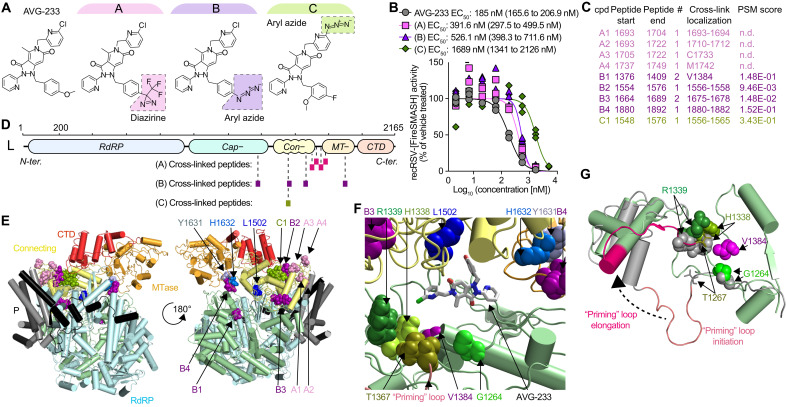
To map the molecular target site of AVG-233, we developed three chemical analogs of the compound capable of carrying diazirine or aryl azide moieties that become covalently reactive when photoactivated through exposure to high-energy ultraviolet light (Fig. 3A). The design of these analogs was guided by the three-dimensional (3D) quantitative structure-activity relationship (3D-QSAR) model that we have developed for the AVG-233 chemotype (24). Consistent with QSAR predictions, all three analogs retained bioactivity in cell-based assays without photoactivation with only minor (approximately 2- to 10-fold) potency penalties (Fig. 3B). Mass spectrometry (MS) analysis after photocrosslinking of these analogs to purified RSV polymerase and trypsin digestion of the covalent complexes identified for each compound a narrow set of peptides (one to four) with covalently bound ligand, consistent with high target site specificity of the cross-links (Fig. 3C). Specifically, analog A labeled a cluster of four peptides that was located in L region 1693–1749, which is thought to be part of a flexible linker between the connecting and MTase domains (Fig. 3D) (30). Analog B also highlighted four peptides, in this case spanning a larger area composed of the L capping (1376–1409), connecting (1554–1576 and 1675–1678), and MTase (1880–1892) domains. Analog C identified a single peptide located in the L connecting domain (1548–1576).
Fig. 3. AVG-233 target site mapping through photoaffinity labeling.
(A) Chemical structures of the AVG-233 analogs synthesized. Photoactivatable groups are highlighted in colored boxes. (B) Dose-response curves of the AVG-233 photoreactive analogs and standard AVG-233 against recRSV-fireSMASh. Symbols represent independent repeats (n = 3). Four-parameter variable slope regression modeling. (C) RSV L-P peptides in close proximity of bound AVG-233, identified through photoaffinity labeling and LC-MS/MS analysis. Specific residue(s) engaged (cross-link localization) and confidence [peptide-spectrum matches (PSM) score] are shown. (D) Schematic representation of the three sets of peptides identified through each photoactivatable AVG-233 analog. (E) Cartoon representation of a structural model of the RSV P-L complex in the putative pre-initiation state, based on RSV P-L reconstruction (PDB 6PZK) with residues (1461–2165) modeled after VSV P-L (PDB 6U1X). Color coding as in Fig. 1. Photocrosslinking target peptides from (C) and resistance mutations from Fig. 1 are highlighted. (F) Molecular docking of AVG-233 into proximity of photocrosslinking targets and resistance sites L1502 and H1632. Residues involved in RNA synthesis (H1338 and R1339) and the putative priming loop are highlighted. (G) Predicted priming loop positions in postulated polymerase initiation and elongation conformations. Labeled are residues identified through photocrosslinking and the adjacent, predicted priming loop pivot residue G1264.
When projected on the structural model of the RSV L core polymerase domain (17, 18), only peptide 1376–1409 could be directly observed due to poor structural resolution of the L C-terminal domain. We therefore generated a homology model of the C-terminal residues of RSV L based on the coordinates reported for the related vesicular stomatitis virus (VSV) and rabies L proteins (31–33). This model posited the AVG-233 resistance mutations and nearly all proximity residues identified through photoaffinity labeling at an interface formed by the L capping, connecting, and MTase domains (Fig. 3E). Data-guided in silico docking of AVG-233 into this site suggested that the compound may broadly restrict structural flexibility of this interface or specifically interfere with conformational rearrangement of the L priming-capping loop (Fig. 3, F and G) that facilitates polymerase switch from RNA synthesis initiation to elongation mode (33) through large spatial movement.
Efficacy of AVG-233 in air-liquid interface cultures of primary epithelium
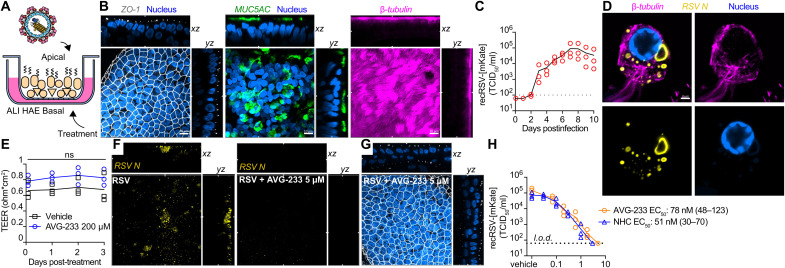
Well-differentiated HAE organoids grown at air-liquid interface (Fig. 4A) represent a premier model to test potency of antivirals directed, for instance, against RSV (34) or influenza viruses (35) in disease-relevant primary human tissues. These cultures develop tight junctions and feature, among others, into mucus-producing goblet and ciliated cells (Fig. 4B and figs. S8 to 12). Following apical infection with recRSV-mKate, we noted sustained, robust virus replication resulting in peak shedding of approximately 1 × 105 infectious particles into the apical space per day over a 10-day experimental window (Fig. 4C). Consistent with previous reports (36), virus replicated predominantly in ciliated cells (fig. S13), forming characteristic (37) cytoplasmic inclusions (Fig. 4D). Our previous studies have also demonstrated that AVG-233 is well tolerated without any signs of cytotoxicity at concentrations of 100 μM. Continued multiday exposure of the airway epithelium cultures to 200 μM compound had no negative effect on tissue organization and tight junction integrity, based on confocal microscopy after ZO-I immunostaining (fig. S14) and stable trans-epithelial electrical resistance (TEER) between the basolateral and apical chambers (Fig. 4E). Basolateral AVG-233 at 5 μM strongly reduced RSV protein expression (Fig. 4F) without detectable negative effect on epithelium integrity (Fig. 4G). Dose escalation resulted in gradual inhibition of apical RSV shedding, reaching sterilizing conditions at approximately 3 μM (Fig. 4H). Regression modeling revealed an EC50 of 0.07 μM, which closely resembled that of N4-hydroxycytidine (NHC) (EC50, 0.05 μM), the parent molecule of the recently authorized broad-spectrum inhibitor molnupiravir (38, 39). These performance parameters correspond to a robust SI > 2850 of AVG-233 in the human airway organoids.
Fig. 4. Efficacy of AVG-233 in well-differentiated HAE (3D-HAE) cells grown at air-liquid interface.
(A) Schematics of 3D-HAE. (B) Confocal imaging of 3D-HAE. Tight junctions immunodetected with anti–ZO-I (white). Mucus-producing goblet cells immunodetected with anti-Muc5AC (green). Ciliated cells immunodetected with anti–β-tubulin (pink). Nuclei stained with Hoechst 35443 (blue). Scale bars, 10 μm. (C) Multistep growth curve of recRSV-mKate in 3D-HAEs. Viral titers were assayed from the apical chambers. Symbols show biological repeats (n = 3); curve connects means ± SD. (D) Immunolabeling of recRSV-fireSMASh–induced inclusion bodies with specific anti-RSV N antibody. Scale bar, 2 μm. (E) Transepithelial electrical resistance of 3D-HAEs exposed basolaterally to 200 μM AVG-233 or vehicle (DMSO) for up to 3 days. Symbols represent means ± SD (n = 3). Two-way ANOVA with Sidak post hoc test. (F) Immunostaining of RSV N in RSV-infected cells with or without AVG-233 5 μM at 3 days postinfection (d.p.i.). Scale as in (B). (G) Immunostaining of tight junctions in RSV-infected cells with AVG-233 5 μM at 3 d.p.i. Scale as in (B). (H) AVG-233 and NHC virus yield reduction against recRSV-mKate in 3D-HAEs. Progeny virus titers were determined 6 days after infection; symbols show biological repeats (n = 3); curve connects mean values. EC50 calculation through four-parameter variable slope regression modeling.
Development of orally efficacious developmental candidates of the AVG series
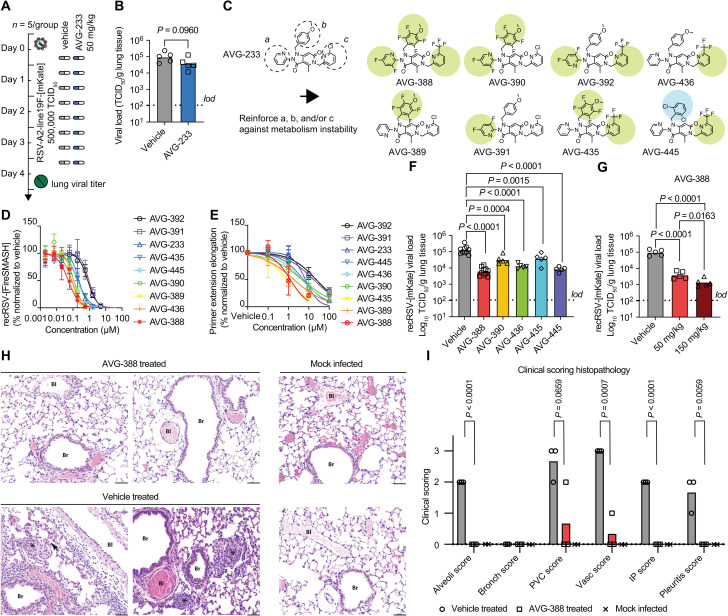
When examining in vivo efficacy of AVG-233 in the mouse model of RSV infection, we noted that neither prophylactic nor therapeutic administration of the compound at a twice-daily dose of up to 200 mg/kg significantly reduced lung virus load (Fig. 5, A and B). We previously reported that AVG series compounds are rapidly metabolized in mice lung microsomes (24). To minimize potential metabolic liabilities, we designed a series of eight analogs with fluorine atoms introduced to block potential sites of oxidative hydroxylation on the (a) and (b) rings, and/or substitution of the chemically labile ortho-chloropyridine (c) ring (40–42) with a more inert ortho-trifluoromethyl pyridine (Fig. 5C). This series retained potent antiviral activity in cell culture (Fig. 5D and table S1) and primary HAE cells (fig. S15 and table S1). Mechanism of action was unchanged from that of AVG-233 (Fig. 5E and fig. S16), and even the substantially modified analog, AVG-388, shared resistance profile (fig. S17 and table S2) and very low cytotoxicity (fig. S18) with AVG-233.
Fig. 5. Identification of orally efficacious developmental analogs of AVG-233.
(A and B) Effect of AVG-233 given orally twice daily at 12 hours postinfection (A) to RSV-infected Balb/c mice on lung titers at 4.5 d.p.i. (B). (C) Chemical structures of AVG-233 analogs with fluoro- or ortho-chloropyridine groups. (D and E) Comparison of AVG fluorine and ortho-chloropyridine analogs on antiviral potency in cell culture against recRSV-mKate (D) and in in vitro primer extension RdRP assays (E). (F and G) Effect of AVG-233 analogs given orally twice daily at 12 hours postinfection and 50 mg/kg (F) or 50 and150 mg/kg (G; AVG-388 only) to recRSV-mKate–infected Balb/cJ mice on lung titers at 4.5 d.p.i. (H and I) Lung histopathology. Representative photomicrographs of lung sections after H&E staining of two animals per treatment group (H) and histopathology scoring (I; n = 3 each for AVG-388– and vehicle-treated groups, n = 2 for mock-infected group). Bl, blood vessel; Br, bronchiole; arrow, interstitial pneumonia; asterisks, alveolitis. Scale bars, 50 μm. The histopathology scoring scale applied is defined in Materials and Methods. Symbols in (B), (F), (G), and (I) show individual biological repeats (independent experiments or individual animals); columns represent sample means. Unpaired t test (B) or one-way ANOVA (F, G, and I) with Dunnett’s (F and G) or Tukey’s (I) multiple comparisons post hoc test; P values are shown.
To assess the effect of these substitutions on in vivo efficacy, we tested the six most potent analogs in the RSV mouse model. Compounds were administered orally at 50 mg/kg body weight in a twice-daily regimen, starting 12 hours after infection (Fig. 5F). Replacing the ortho-chloropyridine in ring (c) of AVG-233 with a trifluoromethyl group (AVG-436) caused a statistically significant reduction in lung viral load of 0.89 (± 0.14) log10 TCID50 (median tissue culture infectious dose)/ml (Fig. 5F and table S3). A slightly lower antiviral effect resulted from fluorine substitutions on both the a and b moieties (AVG-390). Combining these strategies, fluorines on the (a), (b), and (c) rings (AVG-388) yielded strong antiviral efficacy with a lung virus load reduction of 1.3 (± 0.25) log10 TCID50/ml compared to vehicle-treated animals. We did not observe as much benefit from combining fluorines on b and c (AVG-435), but replacing the anisole moiety of AVG-233 with a 1,3 dichlorobenzene (AVG-445) resulted in a lung titer reduction of 1.1 (± 0.16) log10 TCID50/ml, nearly as potent as that seen with AVG-388.
Antiviral efficacy of AVG-388 was dose dependent, resulting in a strong reduction of 1.9 (± 0.23) log10 TCID50/ml when dosed orally at 150 mg/kg twice daily (Fig. 5G and table S3) without any signs of adverse effects (fig. S19). Histopathology analysis of lung tissue extracted 4 days after infection from vehicle-treated animals revealed prominent interstitial pneumonia with multifocal pleuritis and moderate perivascular lymphohistiocytic cuffing (Fig. 5, H and I, and fig. S20). Treatment with AVG-388 at 50 mg/kg initiated 12 hours after infection prevented disease progression to viral pneumonia. Only one animal of the AVG-388 group showed mild vasculitis and perivascular cuffing. These results demonstrating oral efficacy of AVG-388 against RSV confirm successful synthetic optimization of the AVG-233 scaffold and identification of a developmental candidate for the treatment of RSV disease.
DISCUSSION
Our initial characterization of the AVG-233 class revealed that the inhibitor does not block phosphodiester bond formation per se but disturbs initiation of viral RNA synthesis at the promoter (24). This inhibition pattern may reflect pharmacological interference with a predicted conformational rearrangement of the polymerase complex during initiation (43, 44). Three lines of experimental evidence support this view: the AVG class resistance profile, the mechanism of action (MOA) characterization in biochemical RdRP assays, and the photoaffinity labeling–based mapping of the target site.
The primary resistance hotspot of the AVG class, L residue 1502, is positioned at the interface between the large RdRP domains mediating RNA synthesis and the MTase domain required for capping of nascent viral mRNAs. A secondary escape residue, L 1632, likewise locates to this junction, as does residue 1631, which is the primary resistance site for the experimental RSV inhibitors YM-53503 (25), AZ-27 (23), PC786 (26), and cpd1 (27). Cross-resistance among chemically distinct antivirals with comparable MOA is not uncommon. However, distinct resistance profiles of mechanistically related chemotypes predicted to engage the same target domain such as AVG-233 and AZ-27 are rare but open interesting future possibilities for combination therapies. Divergent resistance profiles from mechanistically similar RSV L inhibitors indicate a unique docking pose of the AVG class.
Biochemical RdRP assays using different types of synthetic RNA templates demonstrated that compounds of the AVG class block de novo initiation of RNA synthesis and extension of a paired primer in a synthetic primer/template after the first few nucleotides. This initial delay does not represent an artifact of the RdRP assay, since we found previously that ERDRP-0519, a small-molecule inhibitor of measles virus polymerase that we have developed, completely blocks all phosphodiesterbond formation in the equivalent measles virus RdRP assay (45). Both AVG-233 inhibitory activities were sensitive to the L residue 1502 resistance mutation, indicating that suppressed de novo initiation and impaired RNA elongation are a consequence of a uniform AVG-233 docking pose to the L target. The apparent difference in AVG-233 EC50 values between cell-based and in vitro RdRP assays likely reflects a high representation of bioinactive L complexes in the P-L preparations, which is typical for purified mononegavirus polymerase complexes (46). However, other mechanisms are also conceivable. While absent in cellula, this unproductive material may absorb compound in the biochemical assay without appreciable inhibitory effect. Although less likely, the AVG-233 could alternatively affect transcription, since photocrosslinking proposed a docking pose in which the compound could also interfere with L activities such as cap binding and/or cap methylation. We furthermore cannot fully exclude that AVG-233 may be metabolically modified by cellular enzymes, increasing its target affinity.
Beyond resistance profiling, no physical target site has been mapped for any of the experimental RSV RdRP inhibitors. We closed this knowledge gap for the AVG class, identifying L residues in direct proximity to the docked ligand that spanned an interface between the L capping, connecting, and MTase domains. Confidence in specificity and physiological relevance of the photoaffinity labels comes from three sources: three chemically distinct analogs were generated that differentially interrogate the target site; each of these analogs maintained potent anti-RSV activity; and residues covalently identified by these analogs lined a continuous physical site in the native polymerase complex.
Structural reconstructions of RSV (17, 18), closely related human metapneumovirus (47), and more distantly related paramyxovirus (48) L-P complexes have highlighted a dynamic organization of the C-terminal L domains relative to the polymerase core composed of RdRP and capping domains. On the basis of the mechanistic inhibition profile in the in vitro RdRP assays, the resistance data, and the photoaffinity maps, we considered two mechanistic alternatives as possible molecular basis for AVG-233 inhibition of L: structural lockdown of the MTase domain or positional fixation of the putative priming-capping loop.
Specifically, AVG-233 docking may trap L capping, connecting, and MTase domains in a fixed position relative to each other, resulting in a polymerase complex that is permanently locked in initiation conformation. Preventing relative repositioning of these three domains should impair proper mRNA synthesis, since the MTase must swing away from the product exit channel after cap methylation to allow nascent mRNA elongation. However, our studies revealed that AVG-233 is also active against truncated L1–1749 RdRP complexes lacking the MTase domain, arguing against this model of MTase blockage of the exit channel.
We therefore favor the alternative explanation that AVG-233 prevents reorganization of the L priming-capping loop (49) after incorporation of the first few nucleotides. In RSV L structural models, the loop is retracted downstream of a pivotal residue G1264, which clears a path for the newly synthesized RNA strand to exit the polymerase complex in post-initiation configuration (17, 18). Residue G1264 is located in immediate spatial proximity to V1384, which photoaffinity mapping identified as a direct anchor point for AVG inhibitors, thus positing the docked ligand at the hinge region of the priming-capping loop. Although an actual role of the loop in priming of pneumovirus polymerases has not yet been formally proven (47), we note that a purified mutant RSV RdRP complex bearing a G1264A substitution was unable to elongate RNA beyond the addition of two to three nucleotides (50), thus mimicking the inhibition phenotype of AVG-233.
Despite predicted oral bioavailability in mice (24), in vivo efficacy of AVG-233 was disappointing. Our targeted synthetic program identified a trifluoromethyl substitution in the c ring as instrumental for establishing robust oral efficacy. Since antiviral activity of AVG-233 in cell-based anti-RSV and the in vitro RdRP assays closely resembled that of the efficacious analogs, we hypothesize that the chloro-substituent in ring (c) of AVG-233 presented a metabolic liability in vivo that was overcome with the trifluoromethyl replacement.
Substantiated by a robust SI profile, potent antiviral performance in human tissue organoids, and favorable pharmacokinetic properties, the orally efficacious AVG class leads have strong developmental potential. In addition to its immediate impact as a clinical candidate, the AVG chemotype has identified the interface between the RSV L capping, connecting, and MTase domains as a major druggable site that is likely mechanistically conserved in all mononegavirus polymerase proteins. Considering the available resistance information, we propose that all allosteric RSV RdRP inhibitors interfering with polymerase initiation at the promoter that have been developed to date physically engage this interface. Our results lay the foundation for formal development of the AVG class and the structure-guided identification of companion drugs with overlapping target sites but distinct resistance profiles.
MATERIALS AND METHODS
Experimental design
In this study, we explored the preclinical efficacy of a series of allosteric inhibitors of RSV RdRP using both the mice model and the disease-relevant differentiated primary cells from human bronchial/tracheal epithelium, and we determined the mechanism of action of this class in vitro. The efficacy models were chosen because they jointly provide the closest model available to RSV replication in human lungs and constitute the premier system to evaluate efficacy of drug candidates. We determined the effect of treatment on viral replication at different oral doses in a prophylactic or therapeutic setting. Efficacy was considered as a statistically significant reduction in viral titers in mice lungs and in apical shedding from differentiated human epithelium. Efficacy and cytotoxicity in cell culture were determined in a dose-dependent manner using four-parameter variable slope regression modeling and calculation of 50% maximal efficacious concentrations. The end points were predefined for each experiment. Animals were randomly assigned to each group. Numbers of independent biological repeats for all experiments are specified in the figure legends. No blinding was performed. Complete raw and analyzed numerical data are openly provided in data file S1.
Cell lines, plasmids, and viruses
HEp-2 cells [American Type Culture Collection (ATCC), CCL-23], human embryonic kidney (HEK)–293T (ATCC, CRL-3216), and baby hamster kidney cells (BHK-21; ATCC, CCL-10) stably expressing either T7 polymerase (BSR-T7/5) were grown at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 7.5% heat-inactivated fetal bovine serum. Insect cells from Spodoptera frugiperda (SF9; ATCC, CRL-1711) were propagated in suspension using Sf-900 II serum-free medium (Thermo Fisher Scientific) at 28°C without CO2. Experiments with primary HAE cells involved tissues from the following donors: Primary human bronchial tracheal epithelial cells from a 30-year-old female (“F1” donor, LifeLine Cell Technology, catalog no. LM-0050, lot no. 3123, passage 2) and primary human normal bronchial/tracheal epithelial cells (“M4” donor, Lonza Bioscience, catalog no. CC-2540S, lot no. 0000646466, passage 2) from a 38-year-old male were cultured in BronchiaLife complete cell culture medium (LifeLine Cell Technology, catalog no. LL-0023). Differentiated HAE from F1 donor (3D-HAE) was obtained as described previously (35). Briefly, 33,000 low-passage (<4) cells were seeded in polyester, tissue culture–treated, 0.33-cm2 growth area, 0.4-μm membrane pore size Transwell (Corning, catalog no. 3470). Basal medium was replaced with Pneumacult-ALI (Stemcell Technologies, catalog no. 05001) when confluency was reached, and apical medium was removed. TEER was monitored to validate differentiation with EVOM volt/ohm meter and STX2 electrode (World Precision Instruments). All cell lines used in this study were authenticated and checked for mycoplasma and microbial contamination. Note that the International Cell Line Authentication Committee lists HEp-2 cells as a commonly misidentified cell line; however, their unique ability to efficiently propagate RSV justifies their use to generate and titrate viral stocks. Recombinant RSV strain A2 with line19F and mKatushka or fireSMASh reporter gene, recRSV-mKate or recRSV-fireSMASh, respectively, was rescued and amplified as described previously (51, 52). L1502Q, H1632Q, and Y1631H substitutions were introduced following a protocol described previously (5).
Dose-response antiviral assays
Compound stocks were prepared in dimethyl sulfoxide (DMSO) and, upon dilution in cell culture medium, reached in all wells a final DMSO concentration of 0.1%. For luciferase-based dose-response assays, HEp-2 or primary HAE cells were seeded a day before to reach 50% confluence in 96-well white plates. Threefold serial dilutions of compounds were prepared in triplicate using an automated Nimbus liquid handler (Hamilton) and transferred to the cells. Immediately after addition of compound, cells were infected with recRSV-fireSMASh. Each plate contained four wells each of positive and negative control (medium containing 100 μM cycloheximide or vehicle, respectively). Luciferase activities were determined at 48 hours after transfection using One-GLO buffer (Promega, catalog no. E6130) and a Synergy H1 (BioTek) plate reader. Normalized luciferase activities were analyzed with the following formula: % inhibition = (Signalsample – Signalmin)/(Signalmax – Signalmin) × 100, and dose-response curves were further analyzed by normalized nonlinear regression with variable slope to determine EC50 and 95% confidence intervals (CIs) with Prism 9.0.1 for MacOS (GraphPad).
Minireplicon assays
A set of helper plasmids expressing codon-optimized RSV P, L, N, and M2-1 proteins (A2 strain) under the control of cytomegalovirus promoter and a plasmid expressing the RSV minigenome cassette containing the firefly luciferase reporter, expressed under control of RNA polymerase I promoter, were cotransfected with GeneJuice reagent (MilliporeSigma) following the manufacturer’s instructions in 50% confluent HEK-293T cells or BSR-T7/5 cells as described previously (5). To assay RdRP complex inhibition in dose-response experiments, cells were transfected in 96-well white plates. At 4 hours after transfection, compounds were added to the cells and analyzed as described above.
Cytotoxicity assays
HEp-2 or primary HAE cells were seeded at 50% confluence in 96-well plates and incubated with threefold serial dilution of compound with positive and negative controls as described above. After 48-hour incubation at 37°C, cells were incubated with PrestoBlue (Thermo Fisher Scientific, catalog no. A-13262) for 1 hour at 37°C and fluorescence was measured with an H1 synergy plate reader (BioTek). CC50 and 95% CIs after normalized nonlinear regression and variable slope were determined using Prism 9.0.1 for MacOS (GraphPad).
Growth curves
HEp-2 cells seeded in 12-well plates were infected with recRSV-fireSMASh in the absence or presence of L1502Q, H1632Q, or Y1631H substitution in L, at a multiplicity of infection of 0.1 in three independent replicates. Cells were scrapped and harvested every 12 hours for 4 days following infection. After virus release through freeze-thaw and clarification, viral titers were determined through TCID50 titration with firefly luciferase bioluminescence as the readout, using One-GLO buffer (Promega, catalog no. E6130) and a Synergy H1 (BioTek) plate reader.
Confocal microscopy
3D-HAEs were fixed for 30 min at room temperature with a solution of 4% paraformaldehyde diluted in phosphate-buffered saline (PBS). After permeabilization and blocking with PBS + 3% bovine serum albumin (BSA) + 0.1% Triton X-100 (45 min), cells were incubated at room temperature for 1 hour with primary antibodies diluted in PBS + 0.3% BSA+ 0.05% Tween 20 (wash buffer). After three 5-min washes, cells were incubated with a secondary antibody in wash buffer for 45 min and incubated with Hoechst 34580 (BD Biosciences, catalog no. 565877) (1:1000) in wash buffer for 5 min. Membranes were mounted on glass slides with ProLong Diamond Antifade Reagent (Thermo Fisher Scientific, catalog no. P36970) and imaged with a Zeiss Axio Observer Z.1 and Zeiss LSM 800 + AiryScan module. Image analyses were performed with Zeiss Zen 3.1 Blue software (Windows 10). Representative pictures were taken with a 63× Plan Apochromat (numerical aperture, 1.40, oil) objective. Digital pictures are pseudocolored for optimal presentation. Each sample was reconstructed by stacking 0.22 μm thick optical slices for a total depth of 35 μm, unless stated otherwise in the figure legend.
Apical shed viral titer determination
3D-HAEs were washed apically with PBS without calcium and magnesium. Cells were infected apically with recRSV-mKate (500,000 TCID50) for 2 hours at 37°C. Compound was added in the basal medium (vehicle, 0.1% final DMSO). Shed virus was harvested by incubating the apical side with 200 μl of PBS without calcium and magnesium at 37°C for 30 min. Aliquots were stored at −80°C until titration by standard TCID50 using fluorescence to detect infected cells.
Antibodies
Antibodies used for viral titration (TCID50) were RSV: goat anti-RSV polyclonal antibody (1:1000 dilution) (MilliporeSigma, catalog no. AB1128), followed by donkey anti-goat antibody conjugated with horseradish peroxidase (1:1000 dilution) (Jackson ImmunoResearch, catalog no. 705-035-147). Infected cells were detected using Trueblue peroxidase substrate according to the manufacturer’s instructions (Fisher Scientific, catalog no. 5067428).
Primary antibodies used for confocal microscopy: Adherens junctions were visualized with E-cadherin mouse antibody (1:100 dilution) (BD Biosciences; 610181). Goblet cells were visualized with mouse anti-MUC5AC (1:200 dilution) (Thermo Fisher Scientific, catalog no. MA5-12175). Ciliated cells were visualized with rabbit anti–β IV tubulin recombinant antibody conjugated with Alexa Fluor 647 [EPR16775] (1:100 dilution) (Abcam, catalog no. ab204034). Tight junctions were visualized with mouse anti–ZO-1 (1:50 dilution) (BD Biosciences, catalog no. 610966). RSV-infected cells were visualized either as a whole with goat anti-RSV polyclonal antibody (1:1000 dilution) (MilliporeSigma, catalog no. AB1128) or with a focus on RSV-induced cytoplasmic inclusion bodies using mouse anti-RSV nucleoprotein, clone 130-12H (1:100 dilution) (MilliporeSigma, catalog no. MAB858-3). The following antibodies were used as secondary antibodies as appropriate: rabbit anti-mouse IgG (H+L) cross-adsorbed secondary antibody, Alexa Fluor 488 (1:500 dilution) (Thermo Fisher Scientific, catalog no. A-11059), or donkey anti-goat Alexa Fluor 568 (1:500 dilution) (Thermo Fisher Scientific, catalog no. A-11057).
Protein purification
RSV L + P complexes were prepared as previously described (53, 54). Briefly, codon-optimized sequences of RSV L and a 6×HIS-tagged P were coexpressed in SF9 cells in serum-free medium SF900-II (Thermo Fisher Scientific) from a recombinant baculovirus vector generated with the pFastBac dual system. Cells were harvested at 78 hours after infection and lysed in 50 mM NaH2PO4 (pH 8.0), 150 mM NaCl, 20 mM imidazole, and 0.5% Igepal (MilliporeSigma). After purification through immobilized metal affinity chromatography with Ni-NTA Superflow resin (Qiagen), cells were dialyzed into storage buffer: 20 mM tris-HCl (pH 7.4), 150 mM NaCl, 10% glycerol, and 1 mM dithiothreitol.
In vitro polymerase assay
De novo RNA synthesis using the purified L + P complex was performed as previously described (54). Briefly, 100 to 200 ng of RSV L in complex with P were incubated with 2 μM RNA template corresponding to the 25 nucleotides of the RSV trailer complement sequence (3′ UGCUCUUUUUUUCACAGUUUUUGAU) (Horizon Discovery), 8 mM MgCl2, 1 mM dithiothreitol, 1 mM adenosine triphosphate (ATP), 1 mM uridine triphosphate (UTP), 1 mM cytidine triphosphate (CTP), 50 μM GTP, 10 μCi of α-32P–labeled GTP (Perkin-Elmer), 20 mM tris-HCl (pH 7.4), 15 mM NaCl, and 10% glycerol. Reaction was equilibrated for 10 min at 30°C before addition of L + P complexes and then incubated for 3 hours at 30°C. RNAs were precipitated for 16 hours at −20°C with 2.5 volumes of ice-cold ethanol, 0.1 volume of 3 M sodium acetate, and 625 ng of glycogen (Thermo Fisher Scientific). Pellets were washed with ice-cold 75% ethanol, dried, and resuspended in 50% deionized formamide. After a 3-min denaturation at 95°C, RNAs were separated on 7 M urea/20% polyacrylamide tris borate–EDTA gels and visualized by autoradiography using either CL-XPosure Film (Thermo Fisher Scientific) or a storage phosphor screen BAS IP MS 2040 E (GE Healthcare Life Sciences) and imaged with Typhoon FLA 7000 (GE Healthcare Life Sciences). Densitometry analysis was performed using Fiji 2.0 (55).
3′ Extension assays were performed on the basis of slight modifications of established assays (46, 56, 57). Briefly, 1 μM RNA template (3′ UGGUCUUUUUUGUUUC) and 200 μM 5′ phosphorylated RNA primer (5′ pACCA) (Horizon Discovery) were incubated with 8 mM MgCl2, 1 mM dithiothreitol, 10 μM ATP, 10 μM UTP, 10 μM CTP, 10 μM GTP, 10 μCi of α-32P–labeled GTP (Perkin-Elmer), 20 mM tris-HCl (pH 7.4), 15 mM NaCl, and 10% glycerol and, after 10 min at 30°C, with 100 ng of RSV L in complex with P in a final volume of 5 μl. After 60-min incubation at 30°C, reaction was stopped with 5 μl of deionized formamide with 25 mM EDTA. After denaturation at 95°C, RNAs were separated on 7 M urea/20% polyacrylamide tris borate–EDTA gels and visualized by autoradiography using a storage phosphor screen BAS IP MS 2040 E (GE Healthcare Life Sciences) and imaged with Typhoon FLA 7000 (GE Healthcare Life Sciences). Densitometry analysis was performed using Fiji 2.0 (55).
Biolayer interferometry
Purified RSV L-P complexes were buffer-exchanged for PBS (pH 7.4; room temperature) on PD-10 desalting columns (GE Healthcare), mono-biotinylated with the EZ-Link Sulfo-NHS-SS-Biotin reagent (Thermo Fisher Scientific), and loaded on Super-Streptavidin sensors (Molecular Devices) for 2 hours at 30°C to reach a shift of 1 nm. Uncoupled streptavidin was quenched for 15 min with a solution of 2 mM biocytin. In parallel, a solution of thyroglobulin (1 mg/ml; GE Healthcare) was biotinylated and loaded to 1-nm shift on control sensors. Kinetic experiments were performed at 30°C with 1000 rpm shaking in 96-well plates using the Octet Red 96 system (Fortebio). Biosensors loaded with L-P and thyroglobulin were successively equilibrated for 100 s in assay buffer (PBS, 0.01% BSA, 0.005% Tween 20, and 1% DMSO) (baseline), incubated in a dilution of compound (twofold from 40 nM to 40 μM) for 120 s (association), and then incubated in assay buffer for 200 s (dissociation). Real-time binding kinetics were analyzed and calculated using the Octet Red software package. Raw signal was processed using the double-reference method, by subtracting both the thyroglobulin signal (unspecific signal) and the signal in the absence of compound (drift), after baseline alignment and interstep correction at the dissociation. Kinetic modeling was done by analyzing association and dissociation signals using global fitting with a 1:1 model.
Photolabeling of the AVG-233 binding site
Two micrograms of RSV L-P complexes in PBS was mixed with 100 μM of compound a, b, or c for 5 min on ice and then photocrosslinked for 10 min (compound a) or 45 min (compounds b and c) at 365 nm. Samples with compounds b and c were further treated with the auto-crosslink mode of Stratalinker 1800 (Stratagene). Samples were fractionated on Bolt 4-12% Bis-Tris Plus Gels (Thermo Fisher Scientific) and MES buffer and analyzed by MS.
LC-MS/MS analyses and data processing
Liquid chromatography–MS/MS (LC-MS/MS) analysis was performed by the Proteomics and Metabolomics Facility at the Wistar Institute using a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) coupled with a Nano-ACQUITY UPLC system (Waters). Following procedures originally described in (45), gel bands were excised, digested in-gel with trypsin, and injected onto a UPLC Symmetry trap column (180 μm inside diameter × 2 cm packed with 5-μm C18 resin; Waters). Tryptic peptides were separated by reversed-phase high-performance LC on a BEH C18 nanocapillary analytical column (75 μm inside diameter × 25 cm, 1.7-μm particle size; Waters) using a gradient formed by solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). A 30-min blank gradient was run between sample injections to minimize carryover. Eluted peptides were analyzed by the mass spectrometer set to repetitively scan mass/charge ratio (m/z) from 300 to 2000 in positive ion mode. The full-MS scan was collected at 70,000 resolution followed by data-dependent MS/MS scans at 17,500 resolution on the 20 most abundant ions exceeding a minimum threshold of 20,000. Peptide match was set as preferred, exclude isotopes option, and charge-state screening was enabled to reject unassigned charged ions. Peptide sequences were identified using pFind 3.1.5 (58). MS/MS spectra were searched against a custom database containing Sf9, baculovirus, and hRSV protein sequences. Search parameters include full tryptic specificity with up to three missed cleavages, peptide mass tolerance of 10 parts per million (ppm), fragment ion mass tolerance of 15 ppm, static carboxamidomethylation of Cys, and variable oxidation of Met. In addition, mass addition of 537.117924 (compound a), 470.125788 (compound b), or 484.165901 (compound c) was also considered for all amino acid residues. Consensus identification lists were generated with false discovery rates of 1% at protein and peptide levels.
Molecular modeling and docking
RSV L-P available structure encompasses residue 1–1460 of RSV, likely in a post-initiation conformation [Protein Data Bank (PDB): 6PZK] (18). Modeling of full-length RSV L in putative pre-initiation conformation was performed using I-Tasser server and the RSV L strain A2 (GenBank: AAC14905.1) using as a template related vesicular stomatitis virus L (PDB: 5a22), L + P (PDB: 6U1X), rabies virus L + P (PDB: 6UEB), and human metapneumovirus L C-terminal domain (PDB: 4UCY).
In vivo efficacy testing
Female Balb/cJ mice (The Jackson Laboratory, catalog no. 000651) 6 to 8 weeks of age were housed in an Animal Biosafety Level 2 (ABSL-2) facility (4- to 5-day rest). Mice were divided randomly into groups of five and infected intranasally with 500,000 TCID50 (25 μl per nare) of recRSV-mKate in PBS. Mice were anesthetized with ketamine/xylazine. Treatments were administrated via oral gavage in a 200-μl suspension of 1% methylcellulose in water. Temperature and body weight were determined on a daily and twice-daily basis, respectively. Efficacy studies were terminated at peak viral replication at 4.5 days after infection, mice were euthanized, and lungs were harvested and weighted. Lung viral titers were determined after tissue homogenization with a bead beater with 300 μl of PBS (three bursts of 30 s at 4°C, separated by 30-s rest at 4°C). Homogenates were clarified (5 min at 4°C and 20,000g), aliquoted, and stored at −80°C until titration. Viral titers were determined by TCID50 titration, adjusted to weight (grams) of lung tissue.
Histopathology analysis
For histopathology, mice were subjected to cervical dislocation 4.5 days after infection and lungs were perfused with 10% neutral buffered formalin (NBF) before extraction. Lungs were stored in 10% NBF for 24 hours, followed by incubation in 70% EtOH for 2 days and embedding into wax blocks, using a 21-hour alcohol-xylene-wax embedding sequence. Blocks were sectioned at ~4 μm thickness, and sections were mounted onto microscopy slides and stained with hematoxylin and eosin (H&E). Slides were examined by a board-certified veterinary pathologist, who was blinded to the treatment groups. Lesions were scored according to the following scale. Alveoli, bronchiolar, and pleuritis scores were as follows: 1 = focal, 2 = multifocal, and 3 = multifocal to coalescing; perivascular cuffing: 1 = one layer of leukocytes cuffing vessel, 2 = two to five layers, and 3 = six to nine layers; vasculitis score: 1 = leukocytes infiltrating vessel wall, 2 = leukocytes and smooth muscle cell separation, and 3 = fibrinoid necrosis; interstitial pneumonia: 1 = alveolar septa infiltrated by one leukocyte thickness, 2 = two leukocytes thick, and 3 = three leukocytes thick.
Chemical synthesis
All materials were obtained from commercial suppliers and used without purification, unless otherwise noted. Dry organic solvents, packaged under nitrogen in septum-sealed bottles, were purchased from EMD Millipore and Sigma-Aldrich Co. Reactions were monitored using EMD silica gel 60 F254 TLC plates or using an Agilent 1200 series LCMS system with a diode array detector and an Agilent 6120 quadrupole MS detector. Compound purification was accomplished by liquid chromatography on a Teledyne Isco CombiFlash RF+ flash chromatography system. Nuclear magnetic resonance (NMR) spectra were recorded on an Agilent NMR spectrometer (400 MHz) at room temperature. Chemical shifts are reported in ppm relative to residual solvent signal. The residual shifts were taken as internal references and reported in ppm.
General reaction strategy
An overview of the general synthesis strategy of the AVG chemotype is shown in fig. S21. Dimethyl-1,3-acetonedicarboxylate (6.6 ml, 45.81 mmol) was added to a suspension of 2-hydrazinopyridine (compound 1; fig. S21) (5.0 g, 45.81 mmol) in anhydrous toluene (50 ml) to synthesize compound 2 (fig. S21). The reaction mixture was refluxed for 12 hours. After completion, the reaction mixture was concentrated under reduced pressure. The brown solid product (9.60 g, 90% yield) obtained was washed with diethyl ether and dried in vacuum oven at 50°C and used as such for further reactions.
1H NMR 400 MHz, DMSO-d6, δ 12.45 (s, 1H), 8.43 (d, J = 8 Hz, 1H), 8.02 (pseudo t, J = 8 Hz, 1H), 7.74 (d, J = 8 Hz, 1H), 7.35 (pseudo t, J = 8 Hz, 1H), 5.58 (s, 1H), 3.64 (s, 3H), 3.61 (s, 2H). 13C NMR 100 MHz, DMSO-d6, δ 170.67, 155.88, 153.23, 147.70, 146.90, 140.83, 121.25, 112.97, 88.68, 52.33, 34.99. MS [electrospray ionization and atmospheric pressure ionization (ES-API)] [M + 1]+:234.0.
For synthesis of compound 3 (fig. S21), triethyl orthoacetate (11.8 ml, 64.32 mmol) and acetic acid (0.25 ml, 4.30 mmol) were added to a solution of compound 2 (5.0 g, 21.43 mmol) in anhydrous acetonitrile (50 ml). The reaction mixture was stirred at 70°C for 12 hours. After completion, the reaction mixture was concentrated under reduced pressure and washed with diethyl ether to obtain compound 3 as brownish yellow solid. 1H NMR 400 MHz, DMSO-d6, δ 14.95 (broad s, 1H), 8.43 (d, J = 8 Hz, 1H), 8.26 to 8.16 (m, 2H), 7.42 to 7.40 (m, 1H), 3.78 (s, 2H), 3.60 (s, 3H), 2.31 (s, 3H); 13C NMR 100 MHz, DMSO-d6, δ 190.38, 169.41, 163.57, 150.71, 148.11, 143.64, 141.81, 119.75, 112.63, 102.78, 51.70, 34.78, 28.11. MS (ES-API) [M + 1]+: 276.0.
For synthesis of compound 4 (fig. S21), (6-chloropyridin-2-yl)methanamine dihydrochloride (0.64 g, 3 mmol) and triethylamine (1.25 ml, 9 mmol) were added to a solution of compound 3 (0.8 g, 3 mmol) in anhydrous acetonitrile (20 ml), and the reaction mixture was stirred at room temperature. After 2 hours, 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) (0.9 ml, 6 mmol) was added and stirred at room temperature for another 5 hours. After completion, the reaction mixture was concentrated under reduced pressure and the crude product was purified by flash column chromatography using dichloromethane and methanol as eluent. Product was obtained as yellow solid (0.40 g, 36% yield). 1H NMR 400 MHz, DMSO-d6, δ 11.48 (s, 1H), 8.48 to 8.46 (m, 1H), 8.37 (d, J = 8 Hz, 1H), 7.95 to 7.91 (m, 1H), 7.84 (pseudo t, J = 8 Hz, 1H), 7.42 (d, J = 8 Hz, 1H), 7.28 to 7.22 (m, 2H), 5.68 (s, 1H), 5.37 (s, 2H), 2.83 (s, 3H); 13C NMR 100 MHz, DMSO-d6, 190.38, 169.41, 163.57, 150.71, 148.11, 143.64, 141.87, 119.77, 112.62, 109.58, 102.79, 51.65, 28.11. MS (ES-API) [M + 1]+: 368.0.
For synthesis of analogs A and B (fig. S21), N,N-diisopropylethylamine (0.10 ml, 0.60 mmol) and respective substituted benzyl halide (0.60 mmol) were added to a solution of compound 4 (0.18 g, 0.5 mmol) in anhydrous N,N-dimethylformamide (5 ml), and the reaction mixture was stirred at 50°C for 6 hours. After completion, the reaction mixture was concentrated under reduced pressure and the crude product was purified by flash column chromatography using dichloromethane and methanol as eluent. Desired products were obtained in 25 to 35% isolated yield.
Analog A: 1H NMR 400 MHz, CDCl3, δ 8.54 (d, J = 8 Hz, 1H) 7.85 to 7.78 (m, 2H), 7.61 (pseudo t, J = 8 Hz, 1H), 7.26 to 7.16 (m, 4H), 7.06 to 6.99 (m, 3H), 6.06 (s, 1H), 5.40 (s, 2H), 4.98 (s, 2H), 2.89 (s, 3H); 19F NMR 376 MHz, CDCl3, δ −65.08 (s); 13C NMR 100 MHz, CDCl3 (includes multiplets from 13C-19F couplings), δ 162.91, 162.16, 156.01, 155.20, 154.40, 151.01, 148.65, 148.31, 148.21, 139.62, 138.25, 135.33, 129.31, 129.25, 129.16, 129.09, 126.62, 126.53, 126.45, 123.53, 123.38, 123.27, 121.08, 120.93, 120.64, 120.54, 120.49, 117.17, 101.66, 90.88, 90.71, 77.09, 53.82, 48.14, 15.33. MS (ES-API) [M + 1]+: 565.9.
Analog B: 1H NMR 400 MHz, CDCl3, δ 8.54 (d, J = 8 Hz, 1H), 7.85 (m, 2H), 7.61 (pseudo t, J = 8 Hz, 1H), 7.26 to 7.16 (m, 3H), 7.06 to 6.99 (m, 4H), 6.06 (s, 1H), 5.40 (s, 2H), 4.98 (s, 2H), 2.89 (s, 3H); 13C NMR 100 MHz, CDCl3, δ 162.95, 162.16, 156.09, 155.31, 154.22, 151.01, 148.68, 148.25, 140.28, 139.64, 138.24, 130.44, 130.31, 130.12, 123.38, 120.89, 120.64, 120.49, 119.15, 117.19, 101.90, 91.02, 53.91, 48.17, 15.31. MS (ES-API) [M + 1]+: 498.9.
An overview of the preparation of analog C is shown in fig. S22. Sodium azide (0.32 g, 5.0 mmol) and ammonium chloride (0.26 g, 5 mmol) were added to a solution of compound 5 (0.25 g, 0.5 mmol; fig. S22, prepared as shown in fig. S21 for the synthesis of analogs A and B) in anhydrous dimethylformamide (DMF) (3 ml) and stirred at 110°C for 48 hours. After completion, the reaction mixture was concentrated under reduced pressure. The crude reaction mixture was dissolved in dichloromethane (50 ml) and extracted with water (50 ml) and brine (50 ml). The organic layer was dried over anhydrous sodium sulfate and concentrated under reduced pressure. The crude product was purified by flash column chromatography using dichloromethane and methanol as eluent. Product was isolated as pale yellow solid (0.093 g, 36% yield). 1H NMR 400 MHz, CDCl3, δ 8.56 (ddd, J = 5.2 Hz, 2 Hz, 0.8 Hz, 1H), 8.01 (dd, J = 8 Hz, 0.8 Hz, 1H), 7.84 to 7.80 (m, 1H), 7.72 to 7.70 (m, 1H), 7.61 (dd, J = 12 Hz, 8 Hz, 1H), 7.22 (ddd, J = 5.2 Hz, 2 Hz, 0.8 Hz, 1H), 6.88 (dd, J = 8.4 Hz, 6.4 Hz, 1H), 6.76 to 6.74 (m, 1H), 6.50 to 6.41 (m, 2H), 6.18 (s, 1H), 5.95 (s, 2H), 5.04 (s, 2H), 3.50 (s, 3H), 2.84 (s, 3H); 19F NMR 376 MHz, CDCl3, δ −109.51 to −109.57 (m); 13C NMR 100 MHz, CDCl3 (includes multiplets from 13C-19F couplings) δ 164.90, 162.67, 162.44, 162.10, 158.86, 155.12, 152.77, 148.94, 148.76, 148.17, 137.97, 134.09, 131.88, 120.96, 117.73, 117.40, 114.78, 113.43, 106.79, 106.58, 102.59, 99.03, 98.78, 90.84, 55.31, 48.44, 42.44, 14.86; MS (ES-API) [M + 1]+: 513.0.
Statistical analysis
GraphPad Prism software (v8.3.0 Mac OSX) was used for all statistical analyses. Multiple comparisons were analyzed with one-way or two-way analysis of variance (ANOVA) with Dunnett’s or Sidak’s post hoc tests, as specified in figure legends. Fifty percent and 90% maximal effective concentrations were calculated using four-parameter nonlinear regression modeling. Individual biological replicates (n = 3) are represented as symbols or means with SDs. Significance threshold was set to 0.05. P values are represented as stars (*P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001).
Ethics statement
All animal work was performed in compliance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health as well as the Animal Welfare Act Code of Federal Regulations. Experiments involving mice were approved by the Georgia State University Institutional Animal Care and Use Committee under protocols A17019 and A20012.
Acknowledgments
We thank R. Fearns for the pFastbac Dual RSV L + P plasmid and an aliquot of AZ-27, B. Ludecke for advice with in vitro RdRP assays, H.-Y. Tang and the Wistar Institute Proteomics and Metabolomics Facility for assistance with proteomics analysis, and K. K. Conzelmann for the BSR-T7/5 cell line.
Funding: This work was supported, in part, by NIH grants AI071002 (R.K.P.), AI153400 (R.K.P.), and HD079327 (R.K.P.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Author contributions: Conceptualization: R.K.P. Methodology: R.K.P. Investigation: J.S., C.M.L., M.T., J.-J.Y., M.G., and V.U. Analysis: J.S., C.M.L., M.T., K.S., and R.K.P. Visualization: J.S., C.M.L., and M.T. Funding acquisition: R.K.P. Project administration: R.K.P. Supervision: M.G.N., J.P., J.V., and R.K.P. Writing—original draft: J.S. and R.K.P. Writing—review and editing: R.K.P.
Competing interests: R.K.P. and J.V. are inventors on patent 11084796 and patent application 20210403444 related to this work filed by Georgia State University Research Foundation Inc. (no. 11084796, filed on 10 May 2017, published on 10 August 2021). R.K.P. and J.V. are furthermore inventors on a patent application related to this work filed by Georgia State University Research Foundation Inc. (no. 20210403444, filed on 8 July 2021, published on 30 December 2021). The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The AVG series compounds can be provided by Georgia State University pending scientific review and a completed material transfer agreement. Requests for the AVG series compounds should be submitted to R.K.P.
Supplementary Materials
This PDF file includes:
Figs. S1 to S22
Tables S1 to S3
References
Other Supplementary Material for this manuscript includes the following:
Data file S1
REFERENCES AND NOTES
- 1.Shi T., McAllister D. A., O’Brien K. L., Simoes E. A. F., Madhi S. A., Gessner B. D., Polack F. P., Balsells E., Acacio S., Aguayo C., Alassani I., Ali A., Antonio M., Awasthi S., Awori J. O., Azziz-Baumgartner E., Baggett H. C., Baillie V. L., Balmaseda A., Barahona A., Basnet S., Bassat Q., Basualdo W., Bigogo G., Bont L., Breiman R. F., Brooks W. A., Broor S., Bruce N., Bruden D., Buchy P., Campbell S., Carosone-Link P., Chadha M., Chipeta J., Chou M., Clara W., Cohen C., de Cuellar E., Dang D. A., Dash-Yandag B., Deloria-Knoll M., Dherani M., Eap T., Ebruke B. E., Echavarria M., de Freitas Lazaro Emediato C. C., Fasce R. A., Feikin D. R., Feng L., Gentile A., Gordon A., Goswami D., Goyet S., Groome M., Halasa N., Hirve S., Homaira N., Howie S. R. C., Jara J., Jroundi I., Kartasasmita C. B., Khuri-Bulos N., Kotloff K. L., Krishnan A., Libster R., Lopez O., Lucero M. G., Lucion F., Lupisan S. P., Marcone D. N., McCracken J. P., Mejia M., Moisi J. C., Montgomery J. M., Moore D. P., Moraleda C., Moyes J., Munywoki P., Mutyara K., Nicol M. P., Nokes D. J., Nymadawa P., da Costa Oliveira M. T., Oshitani H., Pandey N., Paranhos-Baccala G., Phillips L. N., Picot V. S., Rahman M., Rakoto-Andrianarivelo M., Rasmussen Z. A., Rath B. A., Robinson A., Romero C., Russomando G., Salimi V., Sawatwong P., Scheltema N., Schweiger B., Scott J. A. G., Seidenberg P., Shen K., Singleton R., Sotomayor V., Strand T. A., Sutanto A., Sylla M., Tapia M. D., Thamthitiwat S., Thomas E. D., Tokarz R., Turner C., Venter M., Waicharoen S., Wang J., Watthanaworawit W., Yoshida L. M., Yu H., Zar H. J., Campbell H., Nair H.; RSV Global Epidemiology Network , Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet 390, 946–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mazur N. I., Higgins D., Nunes M. C., Melero J. A., Langedijk A. C., Horsley N., Buchholz U. J., Openshaw P. J., McLellan J. S., Englund J. A., Mejias A., Karron R. A., Simoes E. A., Knezevic I., Ramilo O., Piedra P. A., Chu H. Y., Falsey A. R., Nair H., Kragten-Tabatabaie L., Greenough A., Baraldi E., Papadopoulos N. G., Vekemans J., Polack F. P., Powell M., Satav A., Walsh E. E., Stein R. T., Graham B. S., Bont L. J.; Respiratory Syncytial Virus Network (ReSViNET) Foundation , The respiratory syncytial virus vaccine landscape: Lessons from the graveyard and promising candidates. Lancet Infect. Dis. 18, e295–e311 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb J., Torres F., Haddad T., Dhillon G., Dilling D., Knoop C., Rampolla R., Walia R., Ahya V., Kessler R., A phase 2b randomized controlled trial of presatovir, an oral RSV fusion inhibitor, for the treatment of respiratory syncytial virus (RSV) in lung transplant (LT) recipients. J. Heart Lung Transplant. 37, S155 (2018). [DOI] [PubMed] [Google Scholar]
- 4.D. Hanfelt-Goade, N. Maimon, A. Nimer, F. Riviere, E. Catherinot, M. Ison, S. Jeong, E. Walsh, A. Gafter-Gvili, S. Nama, A phase 2b, randomized, double-blind, placebo-controlled trial of Presatovir (GS-5806), a novel Oral RSV fusion inhibitor, for the treatment of respiratory syncytial virus (RSV) in hospitalized adults, in C17. New Insights in Acute Pulmonary Infections (American Thoracic Society, 2018), p. A4457. [Google Scholar]
- 5.Yan D., Lee S., Thakkar V. D., Luo M., Moore M. L., Plemper R. K., Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc. Natl. Acad. Sci. U.S.A. 111, E3441–E3449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-RSV Study Group. Pediatrics 102, 531–537 (1998). [PubMed] [Google Scholar]
- 7.Feltes T. F., Cabalka A. K., Meissner H. C., Piazza F. M., Carlin D. A., Top F. H. Jr., Connor E. M., Sondheimer H. M.; Cardiac Synagis Study Group , Palivizumab prophylaxis reduces hospitalization due to respiratory syncytial virus in young children with hemodynamically significant congenital heart disease. J. Pediatr. 143, 532–540 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Aggarwal M., Plemper R. K., Structural insight into paramyxovirus and pneumovirus entry inhibition. Viruses 12, 342 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fearns R., Plemper R. K., Polymerases of paramyxoviruses and pneumoviruses. Virus Res. 234, 87–102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grosfeld H., Hill M. G., Collins P. L., RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J. Virol. 69, 5677–5686 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sourimant J., Rameix-Welti M. A., Gaillard A. L., Chevret D., Galloux M., Gault E., Eleouet J. F., Fine mapping and characterization of the L-polymerase-binding domain of the respiratory syncytial virus phosphoprotein. J. Virol. 89, 4421–4433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galloux M., Gabiane G., Sourimant J., Richard C. A., England P., Moudjou M., Aumont-Nicaise M., Fix J., Rameix-Welti M. A., Eleouet J. F., Identification and characterization of the binding site of the respiratory syncytial virus phosphoprotein to RNA-free nucleoprotein. J. Virol. 89, 3484–3496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fearns R., Collins P. L., Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73, 5852–5864 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burke E., Mahoney N. M., Almo S. C., Barik S., Profilin is required for optimal actin-dependent transcription of respiratory syncytial virus genome RNA. J. Virol. 74, 669–675 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliveira A. P., Simabuco F. M., Tamura R. E., Guerrero M. C., Ribeiro P. G., Libermann T. A., Zerbini L. F., Ventura A. M., Human respiratory syncytial virus N, P and M protein interactions in HEK-293T cells. Virus Res. 177, 108–112 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Richard C. A., Rincheval V., Lassoued S., Fix J., Cardone C., Esneau C., Nekhai S., Galloux M., Rameix-Welti M. A., Sizun C., Eleouet J. F., RSV hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription. PLOS Pathog. 14, e1006920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao D., Gao Y., Roesler C., Rice S., D’Cunha P., Zhuang L., Slack J., Domke M., Antonova A., Romanelli S., Keating S., Forero G., Juneja P., Liang B., Cryo-EM structure of the respiratory syncytial virus RNA polymerase. Nat. Commun. 11, 368 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilman M. S. A., Liu C., Fung A., Behera I., Jordan P., Rigaux P., Ysebaert N., Tcherniuk S., Sourimant J., Eleouet J. F., Sutto-Ortiz P., Decroly E., Roymans D., Jin Z., McLellan J. S., Structure of the respiratory syncytial virus polymerase complex. Cell 179, 193–204.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman J., Abbott E., Alber D. G., Baxter R. C., Bithell S. K., Henderson E. A., Carter M. C., Chambers P., Chubb A., Cockerill G. S., Collins P. L., Dowdell V. C., Keegan S. J., Kelsey R. D., Lockyer M. J., Luongo C., Najarro P., Pickles R. J., Simmonds M., Taylor D., Tyms S., Wilson L. J., Powell K. L., RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrob. Agents Chemother. 51, 3346–3353 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailly B., Richard C. A., Sharma G., Wang L., Johansen L., Cao J., Pendharkar V., Sharma D. C., Galloux M., Wang Y., Cui R., Zou G., Guillon P., von Itzstein M., Eleouet J. F., Altmeyer R., Targeting human respiratory syncytial virus transcription anti-termination factor M2-1 to inhibit in vivo viral replication. Sci. Rep. 6, 25806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cockerill G. S., Good J. A. D., Mathews N., State of the art in respiratory syncytial virus drug discovery and development. J. Med. Chem. 62, 3206–3227 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Sourimant J., Lieber C. M., Aggarwal M., Cox R. M., Wolf J. D., Yoon J. J., Toots M., Ye C., Sticher Z., Kolykhalov A. A., Martinez-Sobrido L., Bluemling G. R., Natchus M. G., Painter G. R., Plemper R. K., 4’-Fluorouridine is an oral antiviral that blocks respiratory syncytial virus and SARS-CoV-2 replication. Science 375, 161–167 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiong-Yip C. L., Aschenbrenner L., Johnson K. D., McLaughlin R. E., Fan J., Challa S., Xiong H., Yu Q., Characterization of a respiratory syncytial virus L protein inhibitor. Antimicrob. Agents Chemother. 58, 3867–3873 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox R. M., Toots M., Yoon J. J., Sourimant J., Ludeke B., Fearns R., Bourque E., Patti J., Lee E., Vernachio J., Plemper R. K., Development of an allosteric inhibitor class blocking RNA elongation by the respiratory syncytial virus polymerase complex. J. Biol. Chem. 293, 16761–16777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudo K., Miyazaki Y., Kojima N., Kobayashi M., Suzuki H., Shintani M., Shimizu Y., YM-53403, a unique anti-respiratory syncytial virus agent with a novel mechanism of action. Antiviral Res. 65, 125–131 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Coates M., Brookes D., Kim Y. I., Allen H., Fordyce E. A. F., Meals E. A., Colley T., Ciana C. L., Parra G. F., Sherbukhin V., Stockwell J. A., Thomas J. C., Hunt S. F., Anderson-Dring L., Onions S. T., Cass L., Murray P. J., Ito K., Strong P., DeVincenzo J. P., Rapeport G., Preclinical characterization of PC786, an inhaled small-molecule respiratory syncytial virus L protein polymerase inhibitor. Antimicrob. Agents Chemother. 61, e00737-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laganas V. A., Dunn E. F., McLaughlin R. E., Tiong-Yip C. L., Yuzhakov O., Isabella V. M., Hill P., Yu Q., Characterization of novel respiratory syncytial virus inhibitors identified by high throughput screen. Antiviral Res. 115, 71–74 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Dochow M., Krumm S. A., Crowe J. E. Jr., Moore M. L., Plemper R. K., Independent structural domains in paramyxovirus polymerase protein. J. Biol. Chem. 287, 6878–6891 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poch O., Sauvaget I., Delarue M., Tordo N., Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8, 3867–3874 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fix J., Galloux M., Blondot M. L., Eleouet J. F., The insertion of fluorescent proteins in a variable region of respiratory syncytial virus L polymerase results in fluorescent and functional enzymes but with reduced activities. Open Virol. J. 5, 103–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horwitz J. A., Jenni S., Harrison S. C., Whelan S. P. J., Structure of a rabies virus polymerase complex from electron cryo-microscopy. Proc. Natl. Acad. Sci. U.S.A. 117, 2099–2107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenni S., Bloyet L. M., Diaz-Avalos R., Liang B., Whelan S. P. J., Grigorieff N., Harrison S. C., Structure of the vesicular stomatitis virus L protein in complex with its phosphoprotein cofactor. Cell Rep. 30, 53–60.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang B., Li Z., Jenni S., Rahmeh A. A., Morin B. M., Grant T., Grigorieff N., Harrison S. C., Whelan S. P. J., Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162, 314–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pickles R. J., Human airway epithelial cell cultures for modeling respiratory syncytial virus infection. Curr. Top. Microbiol. Immunol. 372, 371–387 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toots M., Yoon J.-J., Cox R. M., Hart M., Sticher Z. M., Makhsous N., Plesker R., Barrena A. H., Reddy P. G., Mitchell D. G., Shean R. C., Bluemling G. R., Kolykhalov A. A., Greninger A. L., Natchus M. G., Painter G. R., Plemper R. K., Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 11, eaax5866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L., Peeples M. E., Boucher R. C., Collins P. L., Pickles R. J., Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76, 5654–5666 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rincheval V., Lelek M., Gault E., Bouillier C., Sitterlin D., Blouquit-Laye S., Galloux M., Zimmer C., Eleouet J. F., Rameix-Welti M. A., Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus. Nat. Commun. 8, 563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoon J. J., Toots M., Lee S., Lee M. E., Ludeke B., Luczo J. M., Ganti K., Cox R. M., Sticher Z. M., Edpuganti V., Mitchell D. G., Lockwood M. A., Kolykhalov A. A., Greninger A. L., Moore M. L., Painter G. R., Lowen A. C., Tompkins S. M., Fearns R., Natchus M. G., Plemper R. K., Orally efficacious broad-spectrum ribonucleoside analog inhibitor of influenza and respiratory syncytial viruses. Antimicrob. Agents Chemother. 62, e00766-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.U.S. Food and Drug Administration, DA News Release Coronavirus (COVID-19) Update: FDA Authorizes Additional Oral Antiviral for Treatment of COVID-19 in Certain Adults (2021); https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-oral-antiviral-treatment-covid-19-certain.
- 40.Madhura D. B., Liu J., Meibohm B., Lee R. E., Phase II metabolic pathways of spectinamide antitubercular agents: A comparative study of the reactivity of 4-substituted pyridines to glutathione conjugation. Medchemcomm 7, 114–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Litchfield J., Sharma R., Atkinson K., Filipski K. J., Wright S. W., Pfefferkorn J. A., Tan B., Kosa R. E., Stevens B., Tu M., Kalgutkar A. S., Intrinsic electrophilicity of the 4-methylsulfonyl-2-pyridone scaffold in glucokinase activators: Role of glutathione-S-transferases and in vivo quantitation of a glutathione conjugate in rats. Bioorg. Med. Chem. Lett. 20, 6262–6267 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Inoue K., Ohe T., Mori K., Sagara T., Ishii Y., Chiba M., Aromatic substitution reaction of 2-chloropyridines catalyzed by microsomal glutathione S-transferase 1. Drug Metab. Dispos. 37, 1797–1800 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Duvall J. R., VerPlank L., Ludeke B., McLeod S. M., Lee M. D. IV, Vishwanathan K., Mulrooney C. A., Le Quement S., Yu Q., Palmer M. A., Fleming P., Fearns R., Foley M. A., Scherer C. A., Novel diversity-oriented synthesis-derived respiratory syncytial virus inhibitors identified via a high throughput replicon-based screen. Antiviral Res. 131, 19–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noton S. L., Nagendra K., Dunn E. F., Mawhorter M. E., Yu Q., Fearns R., Respiratory syncytial virus inhibitor AZ-27 differentially inhibits different polymerase activities at the promoter. J. Virol. 89, 7786–7798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox R. M., Sourimant J., Govindarajan M., Natchus M. G., Plemper R. K., Therapeutic targeting of measles virus polymerase with ERDRP-0519 suppresses all RNA synthesis activity. PLOS Pathog. 17, e1009371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jordan P. C., Liu C., Raynaud P., Lo M. K., Spiropoulou C. F., Symons J. A., Beigelman L., Deval J., Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLOS Pathog. 14, e1006889 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pan J., Qian X., Lattmann S., El Sahili A., Yeo T. H., Jia H., Cressey T., Ludeke B., Noton S., Kalocsay M., Fearns R., Lescar J., Structure of the human metapneumovirus polymerase phosphoprotein complex. Nature 577, 275–279 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdella R., Aggarwal M., Okura T., Lamb R. A., He Y., Structure of a paramyxovirus polymerase complex reveals a unique methyltransferase-CTD conformation. Proc. Natl. Acad. Sci. U.S.A. 117, 4931–4941 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogino M., Gupta N., Green T. J., Ogino T., A dual-functional priming-capping loop of rhabdoviral RNA polymerases directs terminal de novo initiation and capping intermediate formation. Nucleic Acids Res. 47, 299–309 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Braun M. R., Deflube L. R., Noton S. L., Mawhorter M. E., Tremaglio C. Z., Fearns R., RNA elongation by respiratory syncytial virus polymerase is calibrated by conserved region V. PLOS Pathog. 13, e1006803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hotard A. L., Shaikh F. Y., Lee S., Yan D., Teng M. N., Plemper R. K., Crowe J. E. Jr., Moore M. L., A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology 434, 129–136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan D., Weisshaar M., Lamb K., Chung H. K., Lin M. Z., Plemper R. K., Replication-competent influenza virus and respiratory syncytial virus luciferase reporter strains engineered for co-infections identify antiviral compounds in combination screens. Biochemistry 54, 5589–5604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ludeke B., Fearns R., The respiratory syncytial virus polymerase can perform RNA synthesis with modified primers and nucleotide analogs. Virology 540, 66–74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noton S. L., Deflube L. R., Tremaglio C. Z., Fearns R., The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLOS Pathog. 8, e1002980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deval J., Hong J., Wang G., Taylor J., Smith L. K., Fung A., Stevens S. K., Liu H., Jin Z., Dyatkina N., Prhavc M., Stoycheva A. D., Serebryany V., Liu J., Smith D. B., Tam Y., Zhang Q., Moore M. L., Fearns R., Chanda S. M., Blatt L. M., Symons J. A., Beigelman L., Molecular basis for the selective inhibition of respiratory syncytial virus RNA polymerase by 2′-Fluoro-4′-chloromethyl-cytidine triphosphate. PLOS Pathog. 11, e1004995 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tchesnokov E. P., Raeisimakiani P., Ngure M., Marchant D., Gotte M., Recombinant RNA-dependent RNA polymerase complex of ebola virus. Sci. Rep. 8, 3970 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chi H., Liu C., Yang H., Zeng W. F., Wu L., Zhou W. J., Wang R. M., Niu X. N., Ding Y. H., Zhang Y., Wang Z. W., Chen Z. L., Sun R. X., Liu T., Tan G. M., Dong M. Q., Xu P., Zhang P. H., He S. M., Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 36, 1059–1061 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Jablonski S. A., Morrow C. D., Mutation of the aspartic acid residues of the GDD sequence motif of poliovirus RNA-dependent RNA polymerase results in enzymes with altered metal ion requirements for activity. J. Virol. 69, 1532–1539 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S22
Tables S1 to S3
References
Data file S1