Graphical abstract
Keywords: Deep neural network, Hypoxia microenvironment, Uveal melanoma, Multi-omics data
Abstract
Uveal melanoma (UM) represents the most common primary intraocular malignancy in adults and is characterized by aggressive behaviors and a lack of targeted therapies. Hypoxia-targeted therapy has become a promising new therapeutic strategy in tumors. Therefore, a better understanding of the tumor hypoxia microenvironment is critical to improve the treatment efficacy of UM. In this study, we conducted an extensive multi-omics analysis to explore the heterogeneity and prognostic significance of the hypoxia microenvironment. We found that UM revealed the most significant degree of intertumoral heterogeneity in hypoxia by quantifying tumor hypoxia compared with other solid tumor types. Then we systematically correlated the hypoxia phenotypes with clinicopathological features and found that hypoxic UM tumors were associated with an increased risk of metastasis, more aggressive phenotypes, and unfavorable clinical outcomes. Integrative multi-omics analyses identified multidimensional molecular alterations related to hypoxia phenotypes, including elevated genome instability, co-occurring of 8q arm gains and loss of chromosome 3, and BAP1 mutations. Furthermore, hypoxic UM tumors could be characterized by increased CD8+ T cell infiltration and decreased naïve B cell and dysregulated metabolic pathways. Finally, we introduced DNN2HM, an interpretable deep neural network model to decode hypoxia phenotypes from multi-omics data. We showed that the DNN2HM improves hypoxia phenotype prediction and robustly predicts tumor aggressiveness and prognosis in different multi-center datasets. In conclusion, our study provides novel insight into UM tumor microenvironment, which may have clinical implications for future rationalized hypoxia-targeted therapy.
1. Introduction
Uveal melanoma (UM), originating from melanocytes in the uvea, is the most common primary intraocular malignancy in adults and represents the second most prevalent subtype of melanoma after cutaneous melanoma [1], [2]. The main risk factors for UM include light eye color, Caucasian ancestry, exposure to ultraviolet light, inherited skin disorders, and specific genetic mutations [3]. The local recurrence rate of UM was low, whereas nearly half of UM patients develop distant metastases after primary tumor treatment [4], [5]. Up to 93% of patients with UM metastases are involved in the liver [6], which has a poor overall survival, and usually dies within one year [7]. However, UM metastases respond poorly to chemotherapy, targeted therapy, and immunotherapy [8]. Currently, no effective treatments are available to prevent the development of UM metastases.
An increased understanding of molecular biology in UM may provide mechanistic, prognostic, and therapeutic insights. More recently, integrative analyses on multi-omics data of UM have identified four molecular subtypes of UM that differ in genomic variation, methylation patterns, gene expression profiles, and clinical outcomes [9]. It shows that different chromosomal abnormalities and gene mutations are closely related to the clinical outcomes of patients. Unlike other melanoma subtypes, UM is a genetically superficial tumor with a remarkably low mutational burden, and the median somatic mutation density is around 1.1 per Mb versus 18 per Mb in cutaneous melanoma [9], [10]. Most UM tumors carry somatic mutations in GNAQ or GNA11, which function by activating the mitogen-activated protein kinase pathway [11]. Loss-of-function mutation or reduced expression of the BAP1 gene is highly correlated with UM metastasis [12]. Loss of one copy of chromosome 3 in UM tumors is associated with an increased risk of metastasis and poor prognosis [13], [14].
In addition, studies have also reported that in UM patients, with the rapid growth of tumor volume, increased intervascular distance, and decreased oxygen content, a hypoxic tumor microenvironment will be generated, resulting in changes in the expression of immune response genes, which further stimulate tumor growth and have a poor prognosis [15]. Tumor hypoxia is one of the metabolic characteristics of the tumor microenvironment, which occurs in about half of solid tumors [16], [17]. Tumor adaptation to this hypoxic environment is associated with genomic instability, tumor propagation, malignant progression, and resistance to therapy [18], [19], [20], [21]. Previous studies have evaluated tumor hypoxia across multiple tumor types and characterized the associated molecular features in the hypoxic tumor microenvironment [22]. Nevertheless, the critical role of the hypoxic tumor microenvironment in UM and its relationship with molecular and clinical characteristics has not been investigated. There is a desperate need to develop promising biomarkers to identify those who may benefit from hypoxia-targeted therapeutic strategies.
In the present study, an extensive multi-omics analysis was conducted to explore the heterogeneity and clinical significance of hypoxia phenotypes in a large cohort of UM tumors across multi-center studies to gain a comprehensive insight into UM tumor microenvironment and refine the classification of UM subtypes for hypoxia-targeted therapy.
2. Material and methods
2.1. Data collection and processing
TCGA multi-omics data and matched clinical data of 80 UM patients were collected from the UCSC Xena data repository (https://xena.ucsc.edu/) [23]. The multi-omics data containing mRNA, lncRNA, miRNA, DNA methylation, single-nucleotide variant (SNV), and copy number variation (CNV) were measured to depict the molecular landscape of UM. The GENCODE human annotation (version 38) was utilized to define lncRNA genes and mRNA genes. The lncRNA/mRNA gene expression profiles were estimated using transcripts per kilobase million. The miRNA expression data was measured with reads per million reads. We integrated the DNA methylation level of CpG sites inside a CpG island using the arithmetic mean method for the methylation profile. Overall survival (OS) and disease-free survival (DFS) were acquired for survival analysis.
Two external independent UM cohorts and corresponding clinical data were retrospectively collected from the publicly available Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), including 63 UM patients from Laurent’s study (accession number GSE22138) [24] and 28 UM patients from van Essen’s study (accession number GSE84976) [25]. Raw microarray data from GSE22138 generated by Affymetrix Human Genome U133 Plus 2.0 Array were processed using the RMA algorithm from the affy R package for background correction and quantile normalization. Raw data from GSE84976 generated by Illumina HumanHT-12 V4.0 expression beadchip were processed using the Lumi R package.
2.2. Hypoxia scoring
The hypoxia status was evaluated using the hypoxia metagene signature derived from Buffa et al., as described previously [26]. The hypoxia score for each tumor was calculated based on the 51 hypoxia metagenes using the single sample Gene Set Enrichment Analysis (ssGSEA) method [27]. Specifically, tumors with a higher gene expression level than the median would be given a plus one score, and conversely, those with a lower value would be given a minus one. This procedure was repeated for each gene in the hypoxia metagene signature. The total scores of all genes were calculated as the hypoxia scores, representing the hypoxic status of tumors (i.e., a higher hypoxia score indicated deficient oxygen in the tumor and vice versa).
2.3. Association between hypoxic tumor microenvironment and clinical phenotypes
Inter-tumoral heterogeneity of hypoxia within each tumor type was assessed using the interquartile range (IQR). High IQR indicated that the tumor was heterogeneous in hypoxia, whereas low IQR was indicative of homogeneity. The difference in hypoxia scores between various clinical groups was evaluated by the Mann-Whitney U test. Tumors were split into two groups (high and low hypoxic tumors) according to the median hypoxia score of all tumors. The Kaplan-Meier method and the log-rank test were used to assess the correlation between the hypoxia groups and survival outcomes in different cohorts.
2.4. Identification of molecular features associated with hypoxic tumor microenvironment
Genome instability was assessed by the percentage of the genome with a copy-number aberration (PGA), which is calculated for each tumor as the proportion of the number of base pairs involved in a copy-number variation for the total length of the human genome. Four molecular distinct copy-number clusters for 80 tumors in the TCGA cohort were obtained as previously described [9]. The nonparametric Kruskal-Wallis test was used to test the difference in hypoxia scores among four copy-number clusters. The significance of the associations between cytogenetic abnormalities, SNV and hypoxia score was evaluated by the Mann-Whitney U test. The associations between PGAs, mRNA, miRNA, lncRNA, methylation and hypoxia scores were assessed using Spearman’s correlation coefficient and subsequent significance testing.
2.5. Functional insights of hypoxia-related genes
Differential gene expression analysis was used to determine hypoxia-related genes. Based on raw count data of mRNA genes, DEseq2 was conducted between the distinct hypoxic subtypes [28]. Top 100 dysregulated genes between hypoxic and normoxic UM tumours, according to adjusted P-value, were retained as hypoxia-related genes. Functional enrichment analysis was performed based on gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases by R package clusterProfiler [29]. We implemented gene set enrichment analysis (GSEA) [30] to determine which type of immune cell highly expressed UM-specific hypoxia genes using a set of pan-cancer metagenes for 28 immune cell subpopulations defined by Charoentong’s study [31].
2.6. Characterization of tumor microenvironment for different hypoxia status
To characterize the tumor metabolic microenvironment, we retrieved 86 metabolic pathways from the KEGG database. These metabolic pathways were grouped into ten subcategories: Amino acid, Lipid, Carbohydrate, Glycan, Cofactors and vitamins, Energy, Xenobiotics, Secondary metabolites, Nucleotide, and Terpenoids and polyketides. We then transferred the gene-level expression data into pathway-level activity profiles. Specifically, we scored the activity of each metabolic pathway from the expression data of the gene set within the pathway using the R package GSVA [32]. A comparison of pathway-level activity profiles between two hypoxic subtypes was performed using R package limma [33], and pathways with P-value less than 0.05 were identified as hypoxia-associated metabolic pathways.
For deciphering the immune microenvironment of UM tumors, we utilized the ESTIMATE method [34] to infer tumor purity (ESTIMATE score), the fraction of stromal and immune cells (stromal score and immune score) in UM tumors. Different distribution of tumor purity, stromal score, and immune score between two hypoxic subtypes were tested using the Mann-Whitney U test. To further evaluate the association between hypoxic subtypes and different immune cell types, we implemented the CIBERSORT tool [35] to quantify the relative levels of 22 distinct immune cell types within a tumor admixture. The difference in cell composition between two hypoxic subtypes was tested using the Mann-Whitney U test for each type of immune cell. Spearman’s correlation coefficient and subsequent significance testing were used to evaluate the association between hypoxia score and cell composition of a specific immune cell.
2.7. Development of an interpretable deep neural network modeling of hypoxia microenvironment (DNN2HM) in UM
To develop a UM-specific hypoxia model, we split the TCGA cohort into the training cohort (50 UM tumors) and the test cohort (30 UM tumors). Each cohort comprises 50% high hypoxic and 50% low hypoxic tumors. We defined UM-specific hypoxia markers as the most relevant features from mRNA, lncRNA, miRNA, and methylation data in the training cohort. For each type of data, the correlation with hypoxia was evaluated for each feature, and the top 100 most relevant features were retained as UM-specific hypoxia markers. Then, we performed a 3-stage stacked autoencoder, a type of artificial neural network, to learn feature structure and reduce the marker numbers to 20 for each type of data. Using deep neural network, single molecular layer-based hypoxia predictors were built by leveraging 20 markers from each molecular layer. Based on 80 UM-specific markers from four data types, we trained a deep neural network (DNN2HM) model to decode the hypoxic microenvironment and distinguish between hypoxic and normoxic tumors.
2.8. Statistical analysis
All statistical analyses were implemented using the R language (version 3.6.3). The Mann-Whitney U test was used to compare two groups and the Kruskal-Wallis test for comparing more than two groups. Spearman’s correlation was used to measure the association between two continuous variables. The Kaplan-Meier method and the log-rank test were used to compare the survival outcome among distinct groups. Univariate Cox regression analysis was conducted to assess the impact of various immune cell compositions on patients’ survival outcomes. The hazard ratio (HR) and 95% confidence interval (CI) were calculated and shown using a forest plot. A value of p < 0.05 is statistically significant.
3. Results
3.1. Hypoxic UM tumor is associated with clinicopathological characteristics and predicted poor prognosis
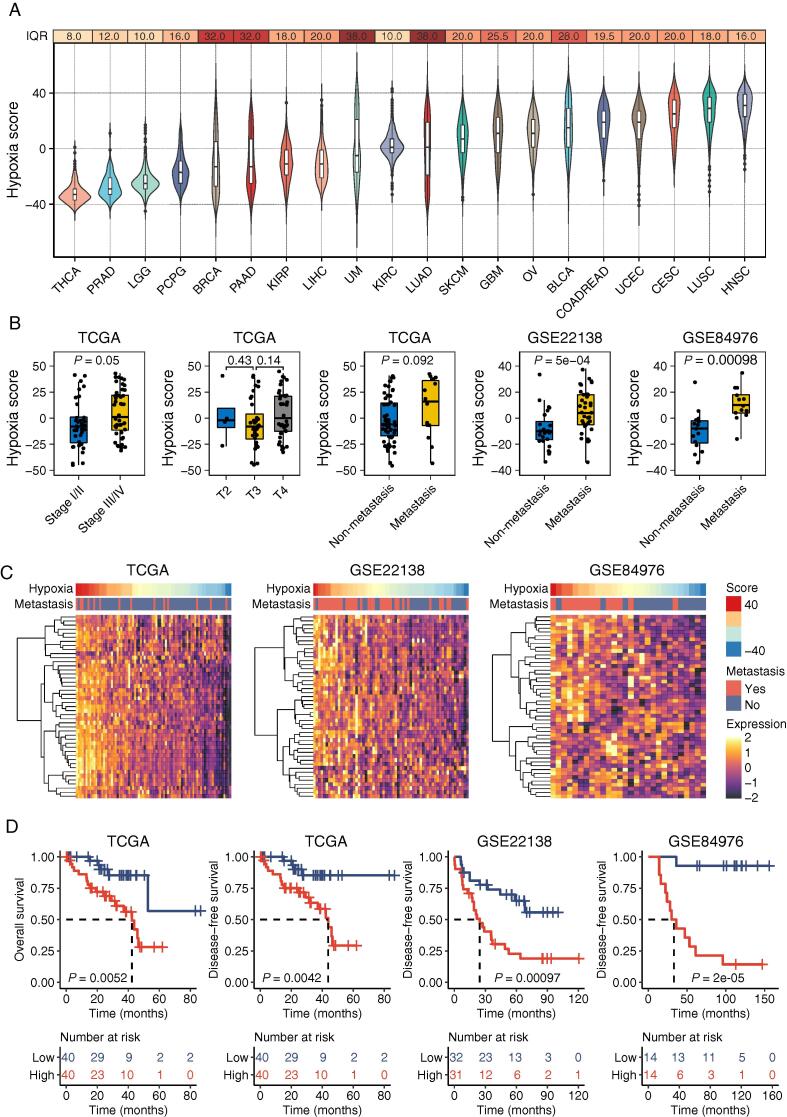
To characterize the hypoxic microenvironment, we calculated the hypoxia scores for 20 types of tumors in the TCGA cohort to represent the degree of hypoxia (Fig. 1A). The degree of intertumoral heterogeneity in hypoxia was evaluated using IQR. Notably, UM tumors were observed to be medium hypoxic (ranked number nine) across all 20 tumor types, nevertheless exhibiting the highest intertumoral variability (IQR = 38), highlighting the highest intertumoral heterogeneity of hypoxic microenvironments among UM tumors compared with other tumor types.
Fig. 1.
Tumor hypoxia patterns in UM. (A) The distribution of hypoxia scores for 20 types of tumors, sorted by median values. The interquartile range (IQR) was calculated to measure the intertumoral variability of hypoxia for each type of tumor. The violin plot depicts the distribution of hypoxia scores using density curves. The boxes show the median ± 1 quartile. Whiskers extend 1.5 times the IQR. (B) Boxplot showing differences of tumor hypoxia pattern between subgroups stratified by clinicopathological characteristics including clinical stages, T stages, and metastasis status (Mann-Whitney U test). (C) Heatmap showing gene expression profiles for Buffa’s hypoxia metagene signature. (D) Kaplan-Meier curves of overall and disease-free survival for high and low hypoxic tumors (Log-rank test). The number of cases and events in a subgroup are shown in the plots.
We next sought to identify clinicopathological features associated with hypoxic patterns. By comparing hypoxic status between patients with different clinicopathological features in different UM cohorts, we observed a strong correlation between tumor hypoxia and the clinical stage of UM tumors. Tumors in early-stage subjects were less hypoxic than tumors in late-stage subjects (P = 0.05, Mann-Whitney U test, Fig. 1B). Specifically, there were no significant differences in hypoxia between the T category but a marginally significant association between tumor hypoxia and metastasis status (P = 0.092, Mann-Whitney U test, Fig. 1B). To further estimate the association between tumor hypoxia and metastasis, we quantified the hypoxia in two independent cohorts (GSE22138 and GSE84976). Strong associations were also observed between tumor hypoxia and metastasis status. Metastatic patients were more likely to have higher hypoxia scores than patients without metastasis (P = 5e–04 in GSE22138 and P = 0.00098 in GSE84976, Mann-Whitney U test, Fig. 1B). As indicating in the expression heatmap of Buffa’s hypoxia metagene signature (Fig. 1C), the expression pattern of hypoxia metagenes differed in the metastatic and non-metastatic groups, and hypoxia metagene is highly expressed in the metastatic group compared with the non-metastatic group (i.e., metastasis group highly expressed hypoxia-related genes). These results suggest that the hypoxia microenvironment within UM tumors was associated with an increased risk of metastasis and more aggressive tumors.
We further examined the correlations of the hypoxia status with the clinical outcome of UM tumors. We divided each cohort into two equal-size subgroups according to the median value of the hypoxia score of all tumors. Kaplan-Meier analysis showed a highly significant difference in overall survival between the high hypoxia (i.e., hypoxic) group and the low hypoxia (i.e., normoxic) group in the TCGA cohort (P = 0.0052, log-rank test, Fig. 1D). Patients with low hypoxia scores are associated with prolonged survival times. Similarly, a significant difference was observed in disease-free survival between the two groups (P = 0.0042 for TCGA cohort, P = 0.00097 for GSE22138 cohort, P = 2e–05 for GSE84976 cohort, log-rank test, Fig. 1D). These results suggest that the hypoxia microenvironment within UM tumors was associated with unfavorable clinical outcomes and thus could be a predictor to help in individualized patient risk stratification.
3.2. The multidimensional molecular features of hypoxic tumor microenvironment in UM
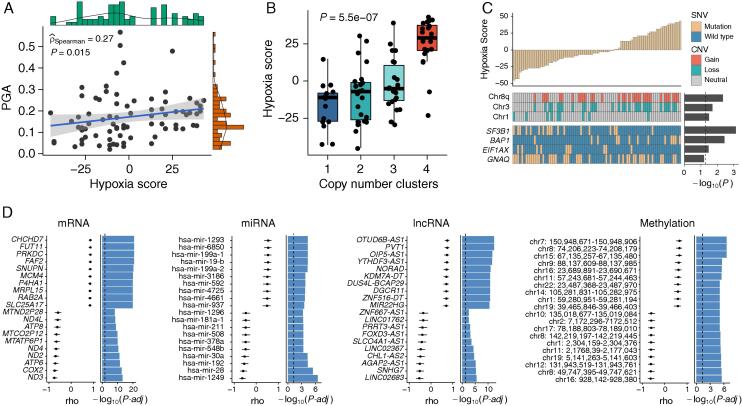
To identify genomic features associated with hypoxia patterns, we first assessed the genome instability using PGA and found that tumor hypoxia was significantly correlated with elevated genome instability (rho = 0.27, P = 0.015, Spearman’s rank correlation test, Fig. 2A). Furthermore, we obtained four distinct molecular subtypes (copy number clusters) from Robertson’s study [9] to assess the correlation between tumor hypoxia and copy number. There were significant differences in hypoxia status among four clusters (P = 5.5e–07, Kruskal-Wallis test, Fig. 2B), and the most hypoxic tumors were enriched in cluster 4, which showed 8q isochromosome (i.e., chromosome 8 with two q arms) in all 20 samples. To identify mutational features associated with tumor hypoxia, we incorporated CNVs and SNVs into the association analysis. We found that gain of 8q arm and loss of one copy of chromosome 3 (monosomy 3, M3) were co-occurring and significantly enriched in hypoxic tumors (P = 0.0045 for 8q arm gain and P = 0.02 for M3, Mann-Whitney U test, Fig. 2C), whereas loss of chromosome 1 was enriched in normoxic tumors.
Fig. 2.
The multidimensional molecular features of tumor hypoxia phenotype. (A) Scatter plot showing the association between tumor hypoxia and genome instability (Spearman’s rho). (B) Boxplot of hypoxia scores among four clusters previously defined (Kruskal-Wallis test). (C) Associations of SNVs and CNVs with tumor hypoxia in UM. (D) Hypoxia-associated multidimensional molecular signatures (Spearman’s rho). Results are shown for the top ten positively and top ten negatively correlated features, according to adjusted P-value. PGA, the percentage of the genome altered by copy-number aberrations; SNV, single-nucleotide variant; CNV, copy number variation.
We included the four most significantly mutated genes with sufficient statistical power to consider the relationships between specific mutational events and tumor hypoxia. Hypoxic UM tumors showed elevated BAP1 somatic point mutations (P = 0.0036, Mann-Whitney U test, Fig. 2C), whereas SF3B1 mutations and EIF1AX mutations were enriched in normoxic UM tumors. These hypoxia-CNV and hypoxia-SNV relationships suggest that tumor hypoxia potentially underpins several aggressive genomic features in UM tumors.
Tumor hypoxia has been demonstrated to impact multi-omics molecular features [36]. To identify molecular features associated with hypoxia at the multi-omics level, we performed an association analysis between four molecular layers (mRNA, miRNA, lncRNA, and methylation) and tumor hypoxia. The top ten positively and negatively correlated features for each molecular layer were retained, as shown in Fig. 2D. Many of them have been shown to be associated with tumor hypoxia in other types of cancers. For instance, mRNA gene FUT11 was recognized in hypoxic conditions in breast cancer [37]. The lncRNA gene PVT1 functioned as competing endogenous RNA for miR-199-5p in non-small cell lung cancer, thus promoting the expression of HIF1A, a master regulator of response to hypoxia conditions [38].
3.3. Functional insights of distinct hypoxic patterns in UM tumors
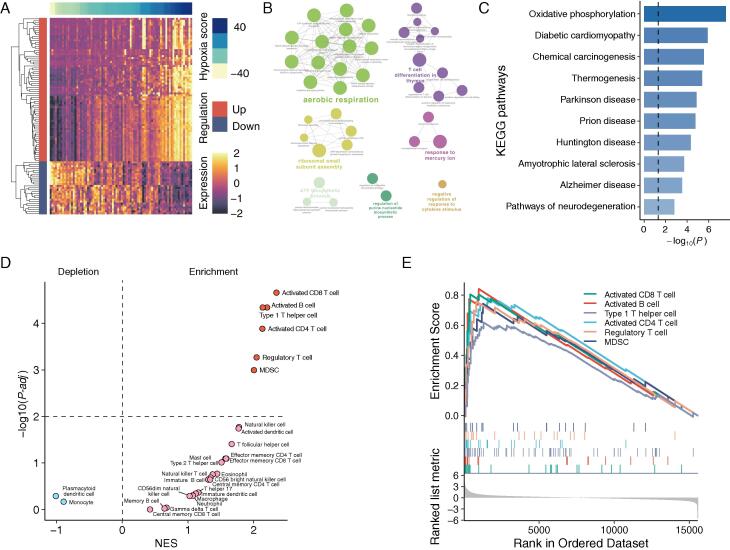
To obtain functional insights into distinct hypoxic patterns in UM tumors, we performed differential gene expression analysis between hypoxic and normoxic tumors and detected 73 up-regulated and 27 down-regulated mRNA genes, referred to as hypoxia-related genes (Fig. 3A; Supplemental Table 1). GO enrichment analysis discovered that hypoxia-related genes are mainly involved in aerobic respiration and T cell differentiation (Fig. 3B). KEGG pathway enrichment analysis found that oxidative phosphorylation, diabetic cardiomyopathy, chemical carcinogenesis, and some neurodegenerative disease-related pathways were the most enriched pathways (Fig. 3C). We also implemented gene set enrichment analysis (GSEA) to determine which type of immune cell highly expressed hypoxia-related genes using pan-cancer metagenes for 28 immune cell subpopulations defined by Charoentong’s study [31]. We observed hypoxia-related genes significantly enriched in activated CD8 T cells, activated B cells, type 1 T helper cells, activated CD4 T cells, regulatory T cells, and myeloid-derived suppressor cells (Fig. 3D-E). These findings indicate that UM-specific hypoxia-related genes may be involved in immune-mediated biological processes besides oxidative phosphorylation.
Fig. 3.
Functional insights of distinct hypoxic patterns in UM tumors. (A) Heatmap representation of differential gene expression between hypoxic and normoxic tumors. (B) Functional enrichment network of GO terms of hypoxia-related genes. (C) Top ten most significant KEGG pathways by enrichment analyses. (D) Volcano plot for enriched (red) and depleted (blue) immune cell subsets in the hypoxic tumors compared with the normoxic tumors. (E) Gene set enrichment analysis plot of activated CD8 T cell, activated B cell, type 1 T helper cell, activated CD4 T cell, regulatory T cell, and myeloid-derived suppressor cell. The vertical-colored short lines in the middle panel of the GSEA plot highlight markers of six enriched immune cell types in the ordered gene list. NES, normalized enrichment score. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Effects of hypoxia on the tumor microenvironment
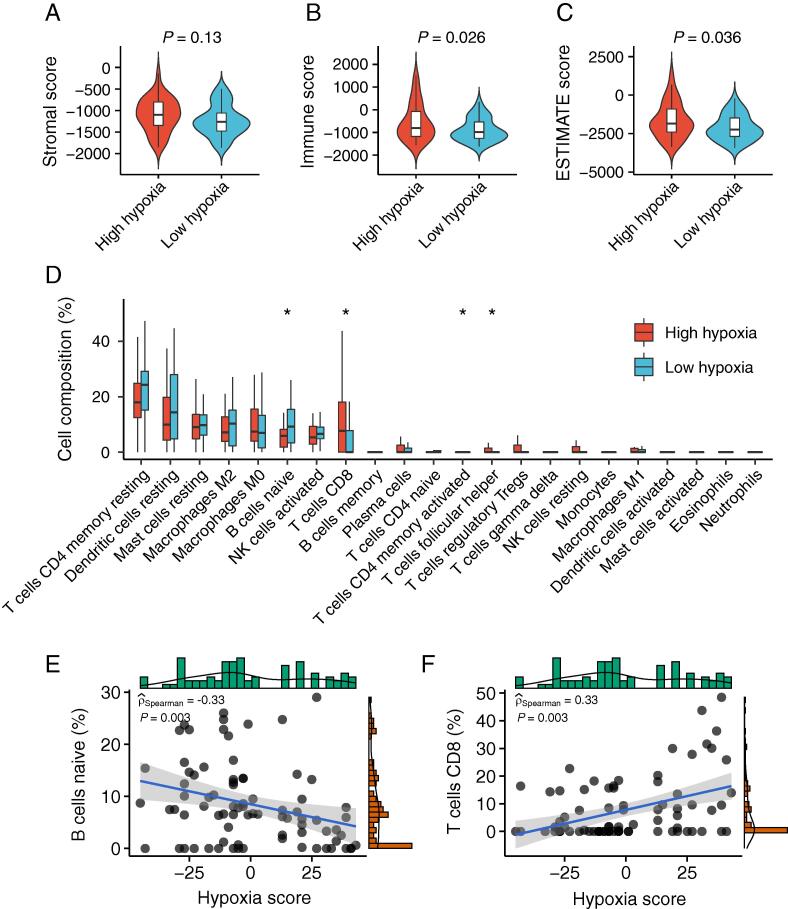
The above results implied relationships between hypoxia and immune-mediated biological processes. We further explored which composition of immune cells had been affected by the hypoxic microenvironment. The ESTIMATE method was performed to infer the ESTIMATE score, stromal score, and immune score as a representation of tumor purity and the fraction of stromal and immune cells. There was no significant difference in stromal scores between hypoxic and normoxic tumors (P = 0.13, Mann-Whitney U test, Fig. 4A), whereas ESTIMATE scores and immune scores were significantly higher in hypoxic tumors than normoxic tumors (P = 0.026 for immune score and P = 0.036 for ESTIMATE score, Mann-Whitney U test, Fig. 4B-C). Specifically, the relative levels of 22 distinct immune cell types were quantified within a tumor admixture. Overall, resting memory CD4 T cell and resting dendritic cell were the most abundant cell types in both hypoxic and normoxic tumors. There were significant differences in the composition of naïve B cell, CD8 T cell, activated memory CD4 T cell, and follicular helper T cell between hypoxic and normoxic tumors (Fig. 4D). We confirmed that the composition of naïve B cells was negatively correlated with hypoxia in the TCGA cohort (P = 0.003, rho = −0.33, Fig. 4E). In contrast, the composition of CD8 T cells was positively correlated with hypoxia in the TCGA cohort (P = 0.003, rho = 0.33, Fig. 4F). These findings demonstrate the effects of hypoxia status on the immune microenvironment and immune cell compositions.
Fig. 4.
TIME characteristics of tumor hypoxia subtypes in UM. (A-C) Boxplot showing differences in the stromal score (A), immune score (B), and ESTIMATE score (C) between high hypoxia and low hypoxia groups (the Mann-Whitney U test). (D) Boxplot showing differences in the relative abundance of 22 distinct immune cell types between high hypoxia and low hypoxia groups (Mann-Whitney U test). (E) Scatter plot showing the correlation between naïve B cell and hypoxia score (Spearman’s rho). (F) Scatter plot showing the correlation between CD8 T cell with hypoxia score (Spearman’s rho).
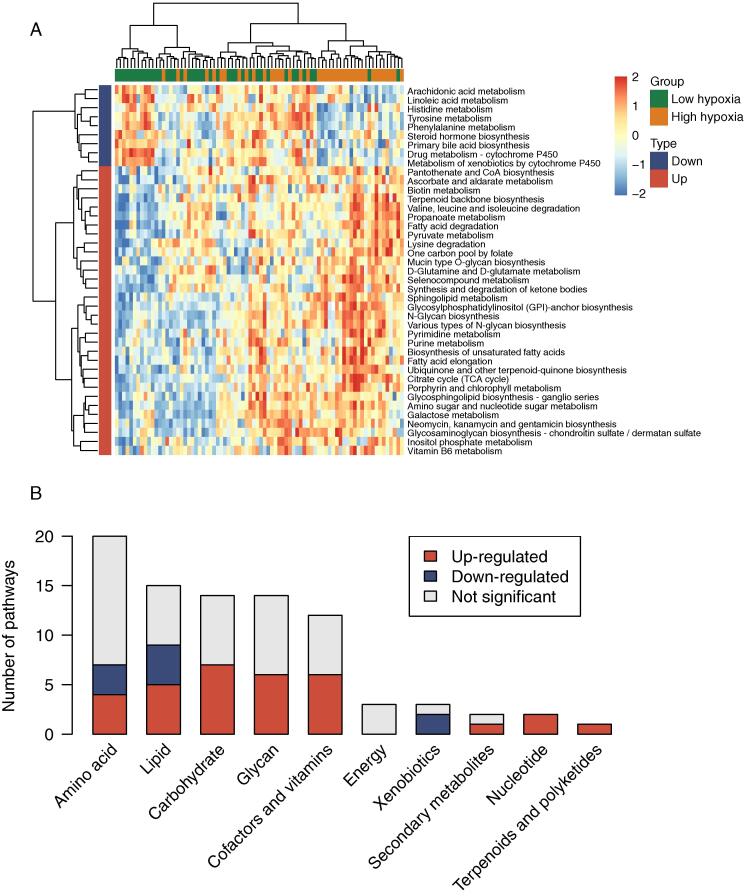
We further investigated the metabolic microenvironment in distinct hypoxia patterns. By comparing 84 pathway-level transcriptional activity profiles between hypoxic tumors and normoxic tumors, we sought to identify which metabolic pathway was dysregulated in hypoxic tumors. GSEA and differential analysis demonstrated that a total of 32 metabolic pathways were significantly up-regulated, whereas nine metabolic pathways were downregulated in hypoxic tumors in reference to normoxic tumors (P < 0.05) (Fig. 5A). These dysregulated pathways encompass most of the metabolic subcategories except energy metabolic pathways (Fig. 5B). In addition, the majority of dysregulated pathways were up-regulated in hypoxic tumors. These results show that hypoxia in aggressive UM tumors might induce metabolic reprogramming to support the increased request of neoplastic cells.
Fig. 5.
Effects of hypoxia on the tumor metabolic microenvironment. (A) Heatmap of pathway-level transcriptional activity profiles for 86 metabolic pathways from the KEGG database. A two-sided P < 0.05 is considered a statistically significant difference using limma. (B) The distribution of dysregulated pathways in ten metabolic subcategories. These dysregulated pathways encompass most of the metabolic subcategories except energy metabolic pathways.
3.5. Effects of hypoxia and immune on patient prognosis
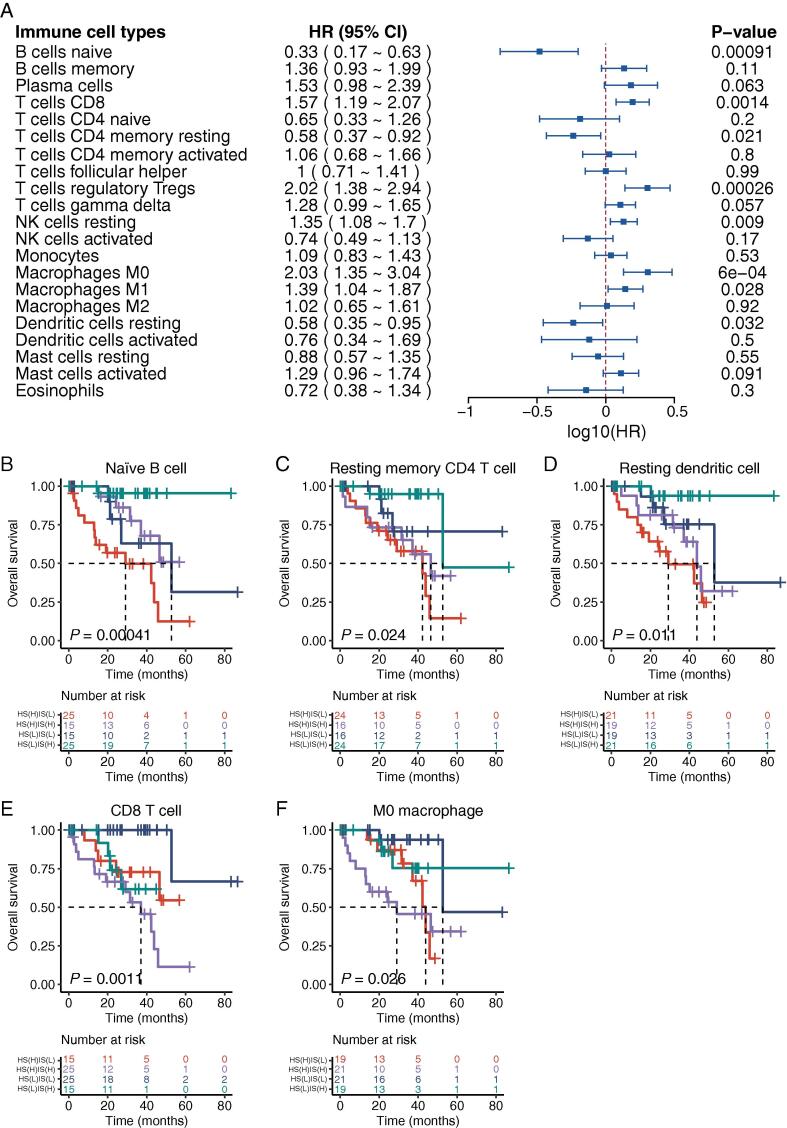
To further depict the immune microenvironment in UM tumors, survival analysis was performed for each type of immune cell to assess the impact of various immune cell compositions on patients’ survival outcomes. As a result, patients with a high fraction of naïve B cell, resting memory CD4 T cell, or resting dendritic cell tend to live longer than patients with a low fraction. Conversely, patients with a high fraction of CD8 T cell, regulatory T cell, resting NK cell, M0 macrophage, and M1 macrophage appear to have a poor survival (Fig. 6A).
Fig. 6.
Effects of hypoxia and immune on patient prognosis. (A) Forest plot depicting the impact of immune cell compositions on patients’ survival outcomes using the univariate Cox regression analysis. HR refers to the hazard ratio for subjects with a high level of immune cell composition compared with other subjects. (B-F) Combining hypoxic patterns and immune cell compositions provides more delicate risk stratification for UM subjects in the TCGA cohort (log-rank test). (B) naïve B cell, (C) resting memory CD4 T cell, (D) resting dendritic cell, (E) CD8 T cell, and (F) M0 macrophage. The number of cases and events in a subgroup are shown in the plots. HR, hazard ratio; CI, confidence interval; HS(L), low hypoxia score; HS(H), high hypoxia score; IS(L), low immune score; IS(H), high immune score.
We next investigated whether a combination of hypoxic patterns and immune compositions could provide a more accurate prediction for survival. All 80 patients from the TCGA cohort were divided into four subcategories by combining the hypoxic patterns (low/high hypoxia score) and immune cell compositions (low/high immune score). Considering the abundance of immune cells, five types of immune cells with relatively high composition in UM tumors were included in further analysis. Overall, significant differences were observed in the Kaplan-Meier survival analysis for all combinations of five types of immune cell composition and hypoxia patterns (Fig. 6B-F). Patients with a high immune cell composition, such as naïve B cell, resting memory CD4 T cell, resting dendritic cell, and low hypoxia score have the longest survival time compared with patients in other combinations. In contrast, patients with low immune cell composition, such as M0 macrophage, CD8 T cell, and low hypoxia score, have the longest survival time compared with patients in other combinations.
3.6. Development of a deep neural network modeling of hypoxic tumor microenvironment in UM
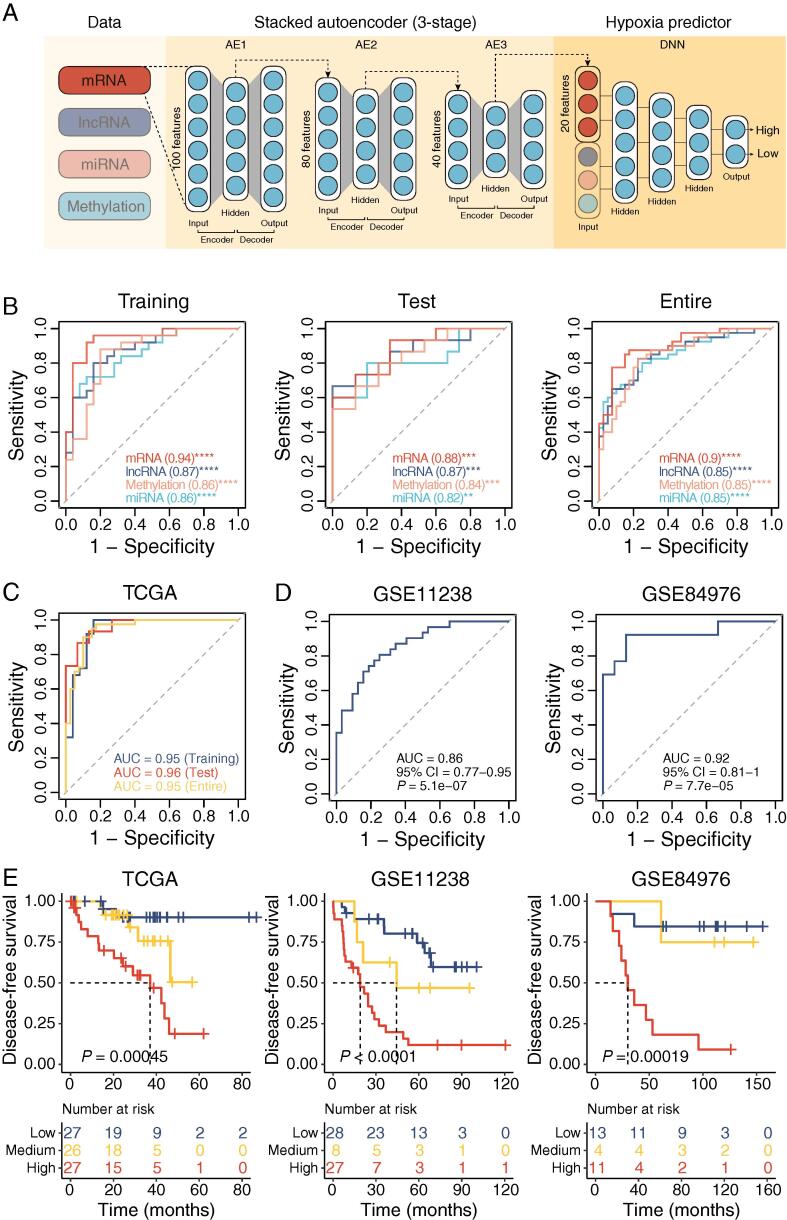
We confirmed hypoxia was a critical microenvironmental factor in aggressive UM tumors, and multi-omics features were associated with distinct hypoxic patterns. We thus integrated multi-omics features using a deep neural network (DNN) framework to develop a hypoxia predictor to model the hypoxic tumor microenvironment (Fig. 7A).
Fig. 7.
Deep neural network performance. (A) Workflow for developing deep neural network (DNN2HM) through integrating multi-omics features. TCGA cohort was split into the training cohort (50 UM tumors) and the test cohort (30 UM tumors). A three-stage stacked autoencoder was performed to learn feature structure from the training cohort and reduce the marker numbers to 20 for each molecular layer. Based on 80 UM-specific markers from four molecular layers, a deep neural network (DNN2HM) was developed to decode the hypoxic microenvironment for UM tumors. (B) Receiver-operating characteristic curves for the single molecular layer-based hypoxia predictors. Receiver-operating characteristic curves for integrated models applied on TCGA cohort (C), GSE11238 cohort, and GSE84976 cohort (D). (E) Kaplan-Meier curves of disease-free survival for high, medium, and low hypoxic tumors predicted by the DNN2HM (Log-rank test). The number of cases and events in a subgroup are shown in the plots. AE, autoencoder; DNN, deep neural network; AUC, area under the curve.
Firstly, single molecular layer-based hypoxia predictors were derived from mRNA, lncRNA, miRNA, and methylation data. As shown in Fig. 7B, single molecular layer-based four hypoxia predictors performed very well in predicting hypoxic patterns in training cohort (area under the curve [AUC] = 0.94 for mRNA model, AUC = 0.87 for lncRNA model, AUC = 0.86 for miRNA model and AUC = 0.86 for methylation model). Similar performance was observed in the test and entire cohorts, indicating that no obvious overfitting was detected in four hypoxia predictors. Two GEO cohorts were included as independent test cohorts to explore the performance of hypoxia predictors further. As only mRNA data available in GEO cohorts, we tested the mRNA-based hypoxia predictor and observed well performance (AUC = 0.86, 95% CI = 0.77–0.95, P = 5.1e-07 in GSE22138 cohort; AUC = 0.92, 95% CI = 0.81–1, P = 7.7e-05 in GSE84976 cohort, Fig. 7D). We developed a comprehensive model (DNN2HM) for predicting hypoxic patterns by combining multi-omics data using a deep learning algorithm. DNN2HM outperformed all single molecular layer-based four hypoxia predictors (AUC = 0.95, 95% CI = 0.88–1, P = 3.4e-08 in training cohort; AUC = 0.96, 95% CI = 0.91–1, P = 8e-06 in test cohort; AUC = 0.95, 95% CI = 0.9–0.99, P = 2.5e-12 in entire cohort, Fig. 7C), highlighting the validity of strategy (i.e., combining multi-omics data). We also performed a survival analysis to investigate whether the DNN2HM could predict survival outcomes. Patients were grouped into three subcategories: low-, medium- and high-risk groups, and significant differences in survival rate among three subcategories were detected in different patient cohorts (P < 0.001 for all three UM cohorts, log-rank test) (Fig. 7E). These results demonstrate that DNN2HM could serve as a promising clinically applicable predictive therapeutic biomarker to improve the clinical benefit of hypoxia-targeted therapy and aid future rationalized precision treatment.
4. Discussion
Tumor hypoxia is an essential feature of the microenvironment in solid tumors [16], [17]. Nevertheless, the hypoxic microenvironment in UM tumors had not been described. In this study, we evaluated the hypoxic patterns of UM tumors and demonstrated the critical role of the hypoxic tumor microenvironment in UM tumors. The relations of tumor hypoxia with metastasis, genomic instability, and specific genomic alterations were determined through association analysis. In addition, altered gene expression and methylation profiles were observed in hypoxic UM tumors. We also found that hypoxia in UM tumors influenced the immune and metabolic microenvironment. Finally, a deep neural network modeling of the hypoxic tumor microenvironment, DNN2HM, was developed for UM patients. Overall, our study provides valuable insights into the heterogeneity of the hypoxic microenvironment in UM tumors.
The development of risk stratification tools concerning cancer survivorship has become one of the priorities for research to inform clinical practice [39]. Many examples exist in risk stratification to assess risk and determine clinical management in UM tumors [40], [41], [42]. For example, a previous study identified four distinct molecular subtypes correlated with differential clinical outcomes [9]. This study quantified the hypoxia score for each UM tumor by leveraging the hypoxia metagene signature derived from Buffa’s study [26]. The degree of intertumoral heterogeneity in hypoxia was evaluated, and UM tumors exhibited the highest intertumoral heterogeneity of hypoxic microenvironments compared with other tumor types. The high degree of hypoxic heterogeneity can pose challenges for personalized medicine [43] and lead to classifying tumor subtypes [44] for accurate risk stratification. We classified UM tumors into hypoxic and normoxic subgroups across three cohorts, and hypoxia-related subgroups showed prognostic associations. We combined hypoxic patterns and immune compositions of UM tumors, and four subcategories were obtained to provide a more delicate classification, suggesting that combining different hypoxia and immune patterns could provide a clinically relevant classifier. Thus, we propose that hypoxia-based risk stratification be considered a screening test to identify those who need further investigation, intervention, or support.
Unlike other melanoma subtypes, UM is a genetically simple tumor with a remarkably low mutational burden, and the median somatic mutation density is around 1.1 per Mb [9]. However, a hostile tumor microenvironment (e.g., a hypoxic tumor environment) may drive the adaptation of a distinctive genomic profile [22]. In UM tumors, tumor hypoxia was correlated with elevated genome instability. Specifically, 8q arm gains and loss of one copy of chromosome 3 (monosomy 3, M3) were co-occurring and significantly enriched in hypoxic tumors, whereas loss of chromosome 1 was enriched in normoxic tumors. For single-nucleotide variants, hypoxic UM tumors showed an elevated rate of BAP1 somatic point mutations, whereas SF3B1 mutations and EIF1AX mutations were enriched in normoxic UM tumors. Some of these chromosome aberrations (e.g., M3) and gene mutations (e.g., Loss-of-function mutations in BAP1 and SF3B1 has been identified in previous studies to correlate strongly with clinical outcome in UM tumors [45], [46], [47].
Nonetheless, the molecular pathways involved in aggressive UM tumors have not been elucidated. Through multi-omics data mining, we identified molecular alterations driven by the hypoxic microenvironment for four different molecular layers (mRNA, miRNA, lncRNA, and methylation). Many of them (e.g., FUT11 and PVT1) have been associated with tumor hypoxia in other types of cancers. Integrating multi-omics features, a hypoxia predictor (DNN2HM) was developed to model the hypoxic tumor microenvironment for UM tumors. These molecular alterations driven by the hypoxic microenvironment provide insights into biological processes responsible for aggressive UM tumors with poor prognoses.
Oxygen is indispensable for cellular metabolism, and hypoxia is common in aggressive tumors that often impact a broad range of hypoxia-associated biological processes, including apoptosis, proliferation, angiogenesis, inflammation, and metabolism [48]. The adaptation to tumor hypoxia is regulated by hypoxia-inducible factors (HIFs), central regulators of many innate and adaptive immunological functions [49]. A typical feature of immune cells is their ability to infiltrate and function in low nutrients and oxygen tissues [50]. In this study, we confirmed the relationships between hypoxia and the composition of various immune cells (e.g., naïve B cell, CD8 T cell, activated memory CD4 T cell, and follicular helper T cell), highlighting the crucial role of HIFs in modulating immune cell functions. In addition, immune cells can be recruited from the oxygen-rich circulatory system into a pathologically hypoxic immune environment (i.e., tumors), thus needing to balance its requirements for energy and molecular biosynthesis. Consequently, HIF signaling changes immune cell function by inducing metabolic reprogramming and unregulated metabolic pathways, such as fatty acid synthesis, the tricarboxylic acid cycle, and amino acid metabolism [51].
UM metastases usually have poor clinical outcomes, and conventional systemic chemotherapy has shown low response rates in UM patients. Severe hypoxia contributes primarily to resistance to anticancer therapies, and blocking HIF activity can improve the response rates of chemotherapy treatment [52]. Therefore, hypoxia-targeted therapy is in desperate need of UM tumors. Our findings suggest that different molecular subtypes of UM tumors had distinct hypoxic patterns, and some of these UM tumors may benefit from hypoxia-targeted therapy. For example, the previously defined molecular subtype, copy number cluster 4, showed the most hypoxic patterns and thus may benefit from hypoxia-targeted treatment. Furthermore, DNN2HM has partitioned UM tumors into three subcategories with varying degrees of hypoxia. Tumors with the highest degree of hypoxia are most likely associated with favorable responses to hypoxia-targeted therapy. However, the sample size of the study was relatively small. Further studies with a larger sample size are necessary to validate our findings.
In conclusion, our study characterized heterogeneous hypoxia phenotypes associated with differential clinical outcomes and therapeutic effects and identified multidimensional hypoxia-associated molecular features. With a more profound understanding of the tumor hypoxia microenvironment, the hypoxia phenotypic characteristics of UM hold the potential to help improve the clinical benefit of hypoxia-targeted therapy, and aid future rationalized precision treatment.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
CRediT authorship contribution statement
Jingting Zhao: Data curation, Formal analysis, Methodology, Writing – original draft, Visualization. Quanyong Yi: Data curation, Formal analysis. Ke Li: Formal analysis, Methodology. Lu Chen: Formal analysis, Methodology. Lijun Dai: Formal analysis, Visualization. Jiayao Feng: Formal analysis. Yan Li: Conceptualization. Meng Zhou: Conceptualization, Funding acquisition, Project administration, Writing – review & editing. Jie Sun: Conceptualization, Funding acquisition, Supervision, Project administration, Writing – review & editing.
Funding
This study was supported by the Scientific Research Foundation for Talents of Wenzhou Medical University (QTJ18029 and QTJ18024). The funders had no roles in study design, data collection and analysis, decision to publish, or manuscript preparation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.034.
Contributor Information
Yan Li, Email: liyan_8809@126.com.
Meng Zhou, Email: zhoumeng@wmu.edu.cn.
Jie Sun, Email: suncarajie@wmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jager M.J., Shields C.L., Cebulla C.M., Abdel-Rahman M.H., Grossniklaus H.E., Stern M.-H., et al. Uveal melanoma. Nat Rev Dis Prim. 2020;6:24. doi: 10.1038/s41572-020-0158-0. [DOI] [PubMed] [Google Scholar]
- 2.Singh A.D., Turell M.E., Topham A.K. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011;118:1881–1885. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Weis E., Shah C.P., Lajous M., Shields J.A., Shields C.L. The association between host susceptibility factors and uveal melanoma: a meta-analysis. Arch Ophthalmol (Chicago, Ill 1960) 2006;124(124):54–60. doi: 10.1001/archopht.124.1.54. [DOI] [PubMed] [Google Scholar]
- 4.Eskelin S., Pyrhönen S., Summanen P., Hahka-Kemppinen M., Kivelä T. Tumor doubling times in metastatic malignant melanoma of the uvea: tumor progression before and after treatment. Ophthalmology. 2000;107:1443–1449. doi: 10.1016/s0161-6420(00)00182-2. [DOI] [PubMed] [Google Scholar]
- 5.Shields C.L., Furuta M., Thangappan A., Nagori S., Mashayekhi A., Lally D.R., et al. Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch Ophthalmol (Chicago, Ill. 1960;2009(127):989–998. doi: 10.1001/archophthalmol.2009.208. [DOI] [PubMed] [Google Scholar]
- 6.Collaborative Ocular Melanoma Study Group. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the Collaborative Ocular Melanoma Study (COMS): COMS report no. 15. Arch Ophthalmol (Chicago, Ill 1960) 2001;119:670–6. 10.1001/archopht.119.5.670. [DOI] [PubMed]
- 7.Diener-West M, Reynolds SM, Agugliaro DJ, Caldwell R, Cumming K, Earle JD, et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch Ophthalmol (Chicago, Ill 1960) 2005;123:1639–43. 10.1001/archopht.123.12.1639. [DOI] [PubMed]
- 8.Khan S., Carvajal R.D. Dual immunological checkpoint blockade for uveal melanoma. J Clin Oncol. 2021;39:554–556. doi: 10.1200/JCO.20.03274. [DOI] [PubMed] [Google Scholar]
- 9.Robertson A.G., Shih J., Yau C., Gibb E.A., Oba J., Mungall K.L., et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32:204–220.e15. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Network C.G.A. Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Raamsdonk C.D., Griewank K.G., Crosby M.B., Garrido M.C., Vemula S., Wiesner T., et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harbour J.W., Onken M.D., Roberson E.D.O., Duan S., Cao L., Worley L.A., et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White V.A., McNeil B.K., Horsman D.E. Acquired homozygosity (isodisomy) of chromosome 3 in uveal melanoma. Cancer Genet Cytogenet. 1998;102:40–45. doi: 10.1016/s0165-4608(97)00290-2. [DOI] [PubMed] [Google Scholar]
- 14.Rodrigues M., Ait Rais K., Salviat F., Algret N., Simaga F., Barnhill R., et al. Association of partial chromosome 3 deletion in uveal melanomas with metastasis-free survival. JAMA Ophthalmol. 2020;138:182–188. doi: 10.1001/jamaophthalmol.2019.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronkhorst I.H.G., Jehs T.M.L., Dijkgraaf E.M., Luyten G.P.M., van der Velden P.A., van der Burg S.H., et al. Effect of hypoxic stress on migration and characteristics of monocytes in uveal melanoma. JAMA Ophthalmol. 2014;132:614–621. doi: 10.1001/jamaophthalmol.2014.43. [DOI] [PubMed] [Google Scholar]
- 16.Weber C.E., Kuo P.C. The tumor microenvironment. Surg Oncol. 2012;21:172–177. doi: 10.1016/j.suronc.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Dhani N., Fyles A., Hedley D., Milosevic M. The clinical significance of hypoxia in human cancers. Semin Nucl Med. 2015;45:110–121. doi: 10.1053/j.semnuclmed.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 18.Bristow R.G., Hill R.P. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 19.Brizel D.M., Dodge R.K., Clough R.W., Dewhirst M.W. Oxygenation of head and neck cancer: changes during radiotherapy and impact on treatment outcome. Radiother Oncol. 1999;53:113–117. doi: 10.1016/s0167-8140(99)00102-4. [DOI] [PubMed] [Google Scholar]
- 20.Rankin E.B., Giaccia A.J. Hypoxic control of metastasis. Science. 2016;352:175–180. doi: 10.1126/science.aaf4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong H., De Marzo A.M., Laughner E., Lim M., Hilton D.A., Zagzag D., et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 22.Bhandari V., Hoey C., Liu L.Y., Lalonde E., Ray J., Livingstone J., et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019;51:308–318. doi: 10.1038/s41588-018-0318-2. [DOI] [PubMed] [Google Scholar]
- 23.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol. 2020;38:675–678. doi: 10.1038/s41587-020-0546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurent C., Valet F., Planque N., Silveri L., Maacha S., Anezo O., et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011;71:666–674. doi: 10.1158/0008-5472.CAN-10-0605. [DOI] [PubMed] [Google Scholar]
- 25.van Essen T.H., van Pelt S.I., Bronkhorst I.H.G., Versluis M., Némati F., Laurent C., et al. Upregulation of HLA expression in primary uveal melanoma by infiltrating leukocytes. PLoS ONE. 2016;11:e0164292. doi: 10.1371/journal.pone.0164292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buffa F.M., Harris A.L., West C.M., Miller C.J. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102:428–435. doi: 10.1038/sj.bjc.6605450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye Y., Hu Q., Chen H., Liang K., Yuan Y., Xiang Y., et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat Metab. 2019;1:431–444. doi: 10.1038/s42255-019-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye I.C., Fertig E.J., DiGiacomo J.W., Considine M., Godet I., Gilkes D.M. Molecular portrait of hypoxia in breast cancer: A prognostic signature and novel HIF-regulated genes. Mol Cancer Res. 2018;16:1889–1901. doi: 10.1158/1541-7786.MCR-18-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang C., Han C., Zhang Y., Liu F. LncRNA PVT1 regulate expression of HIF1α via functioning as ceRNA for miR-199a-5p in non-small cell lung cancer under hypoxia. Mol Med Rep. 2018;17:1105–1110. doi: 10.3892/mmr.2017.7962. [DOI] [PubMed] [Google Scholar]
- 39.Watson E.K., Rose P.W., Neal R.D., Hulbert-Williams N., Donnelly P., Hubbard G., et al. Personalised cancer follow-up: risk stratification, needs assessment or both? Br J Cancer. 2012;106:1–5. doi: 10.1038/bjc.2011.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J., Lu J., Li W. A comprehensive prognostic and immunological analysis of a six-gene signature associated with glycolysis and immune response in uveal melanoma. Front Immunol. 2021;12:738068. doi: 10.3389/fimmu.2021.738068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao Y., Xie J., Chen L., Hu Y., Zhai L., Yuan J., et al. Construction and validation of a novel pyroptosis-related gene signature to predict the prognosis of uveal melanoma. Front Cell Dev Biol. 2021;9:761350. doi: 10.3389/fcell.2021.761350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv X., Ding M., Liu Y. Landscape of infiltrated immune cell characterization in uveal melanoma to improve immune checkpoint blockade therapy. Front Immunol. 2022;13:848455. doi: 10.3389/fimmu.2022.848455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bedard P.L., Hansen A.R., Ratain M.J., Siu L.L. Tumour heterogeneity in the clinic. Nature. 2013;501:355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tellez-Gabriel M., Ory B., Lamoureux F., Heymann M.-F., Heymann D. Tumour heterogeneity: the key advantages of single-cell analysis. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalirai H., Dodson A., Faqir S., Damato B.E., Coupland S.E. Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing. Br J Cancer. 2014;111:1373–1380. doi: 10.1038/bjc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koopmans A.E., Verdijk R.M., Brouwer R.W.W., van den Bosch T.P.P., van den Berg M.M.P., Vaarwater J., et al. Clinical significance of immunohistochemistry for detection of BAP1 mutations in uveal melanoma. Mod Pathol. 2014;27:1321–1330. doi: 10.1038/modpathol.2014.43. [DOI] [PubMed] [Google Scholar]
- 47.Yavuzyigitoglu S., Koopmans A.E., Verdijk R.M., Vaarwater J., Eussen B., van Bodegom A., et al. Uveal melanomas with SF3B1 mutations: A distinct subclass associated with late-onset metastases. Ophthalmology. 2016;123:1118–1128. doi: 10.1016/j.ophtha.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 48.Höckel M., Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 49.Imtiyaz H.Z., Simon M.C. Hypoxia-inducible factors as essential regulators of inflammation. Curr Top Microbiol Immunol. 2010;345:105–120. doi: 10.1007/82_2010_74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krzywinska E, Stockmann C. Hypoxia, metabolism and immune cell function. Biomedicines 2018;6. 10.3390/biomedicines6020056. [DOI] [PMC free article] [PubMed]
- 51.Corcoran S.E., O’Neill L.A.J. HIF1α and metabolic reprogramming in inflammation. J Clin Invest. 2016;126:3699–3707. doi: 10.1172/JCI84431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jing X., Yang F., Shao C., Wei K., Xie M., Shen H., et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18:157. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.