Abstract
Although their etiology varies, tumors share a common trait: the control of an oncogenic transcriptional program that is regulated by interaction of the malignant cells with the stromal and immune cells in the tumor microenvironment (TME). The TME shows a high phenotypic and functional heterogeneity that may be modulated by the interaction with commensal microbes (the microbiota) both systemically and locally. Unlike host cells, the microbiota adapts after environmental perturbations, impacting host-microbes’ interactions. In the liver, the bidirectional relationship within the gut and its associated microbiota creates an interdependent environment. Therefore, the gut microbiota and its metabolites modulate liver gene expression directly and indirectly, causing an imbalance in the gut-liver axis which may result in disease including carcinogenesis.
Keywords: microbiota, tumor microenvironment, dysbiosis, liver cancer, gut-liver axis
Microbiota in the gut-liver-pancreas axis contributes to homeostasis and carcinogenesis
Humans are metaorganisms and our health and ability to survive depend on the presence of commensal microorganisms (microbiota) [1,2], which are resident on epithelial interfaces and particularly abundant in the lower gastro-intestinal tract. A balanced ecology in the gut microbiota contributes to food processing and absorption, regulates host metabolism, and protects against infections, either by preventing expansion of pathogens and pathobionts or by modulating host immunity and maintaining the integrity of the intestinal epithelium. Physically, the liver and the pancreas communicate with the gut via the biliary and pancreatic ducts, while the portal vein transport gut-derived products to the liver. Therefore, the crosstalk between the gut microbiota with liver and pancreas (gut-liver-pancreas axis) may integrate signals as an interconnected system. The complex yet highly coordinated interplay between the host and its microbiota represents a natural ecosystem [3]. Symbiotic interactions such as mutualism, commensalism, predation, parasitism, and competition underlay the crosstalk between microbes, microbial-host, and host-host cells. Thus, cells in the gut-liver axis are in a homeostatic equilibrium, which is altered by environmental perturbations resulting in modulation of transcriptional responses [4] both locally and systemically (Figures 1,2), and contribute to health and diseases, such as cancer [3–12].
Figure 1. Microbes-host interactions regulates homeostasis and disease.
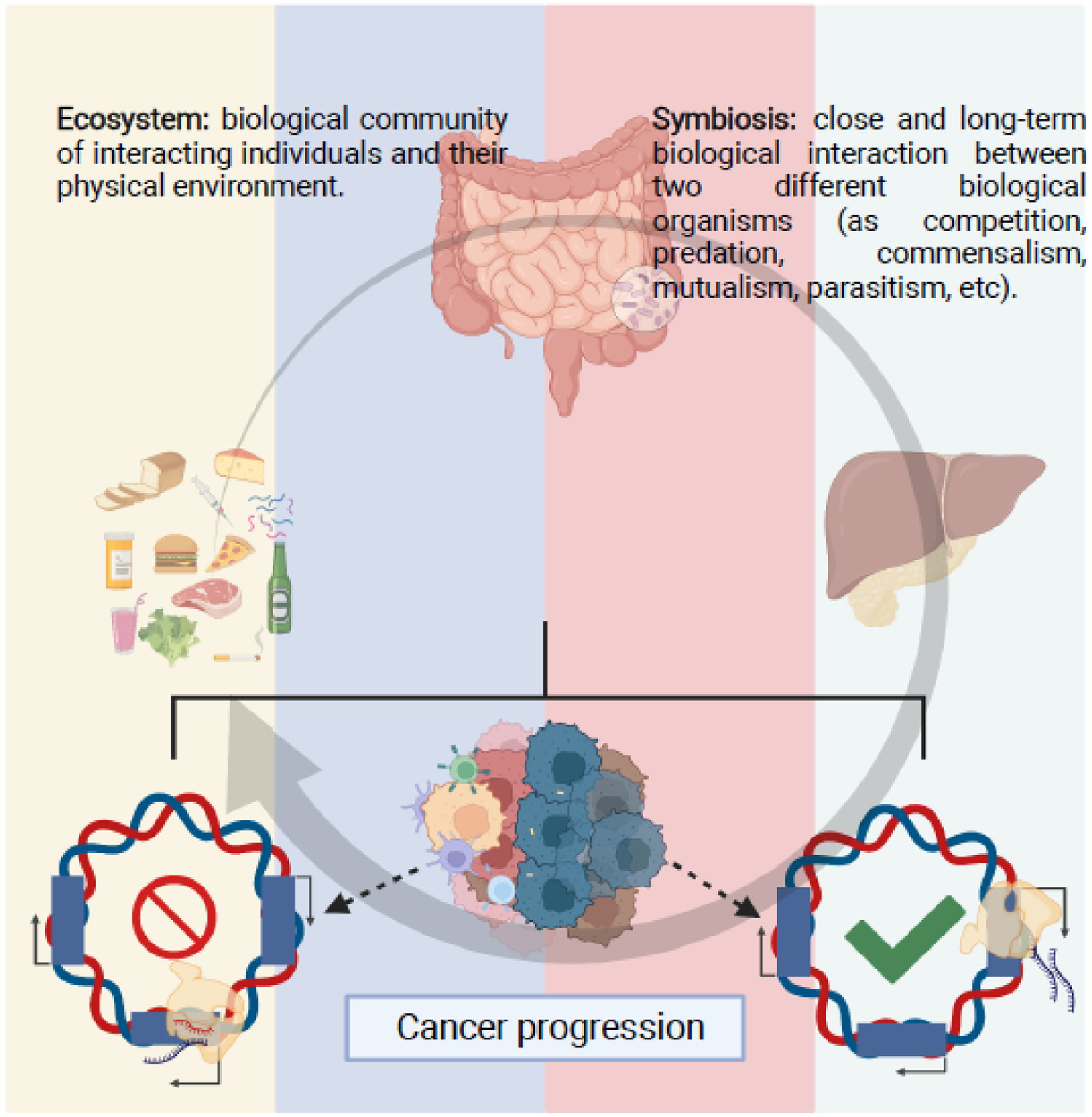
The human body is a metaorganism, in which the large host cells cohabit in a symbiotic relationship with a larger number of microorganisms. The number and type of microbes in different tissues and body system vary greatly, creating tissue specific symbiotic interactions. Microbes are relatively stable, but their relative abundance might rapidly change due to diverse environmental factors, as diet, use of antibiotics, etc. Microbiological communities within the gut and liver-pancreas communicate via symbiotic interactions among themselves and the large host cells and are in a state-of-equilibrium that reminds an ecosystem and benefits both components’ survival and health. Therefore, the microbiota composition in the gut also affects liver and pancreas homeostasis. Alterations of liver physiology, such as non-alcoholic fatty liver disease or viral infections, can modify the microbiota creating conditions predisposing to liver cancer. Bacteria are also present in the tumor microenvironment, mostly growing intracellularly within the tumor cells or infiltrating hematopoietic cells, and, in concert with signals derived from the gut microbiota, may lead to activation of specific genetic programs that may promote or suppress cancer progression, in part by modulating the immune response to the tumor, and defining cancer progression.
Figure 2. Bidirectional relationship within the gut-liver axis regulates homeostasis.
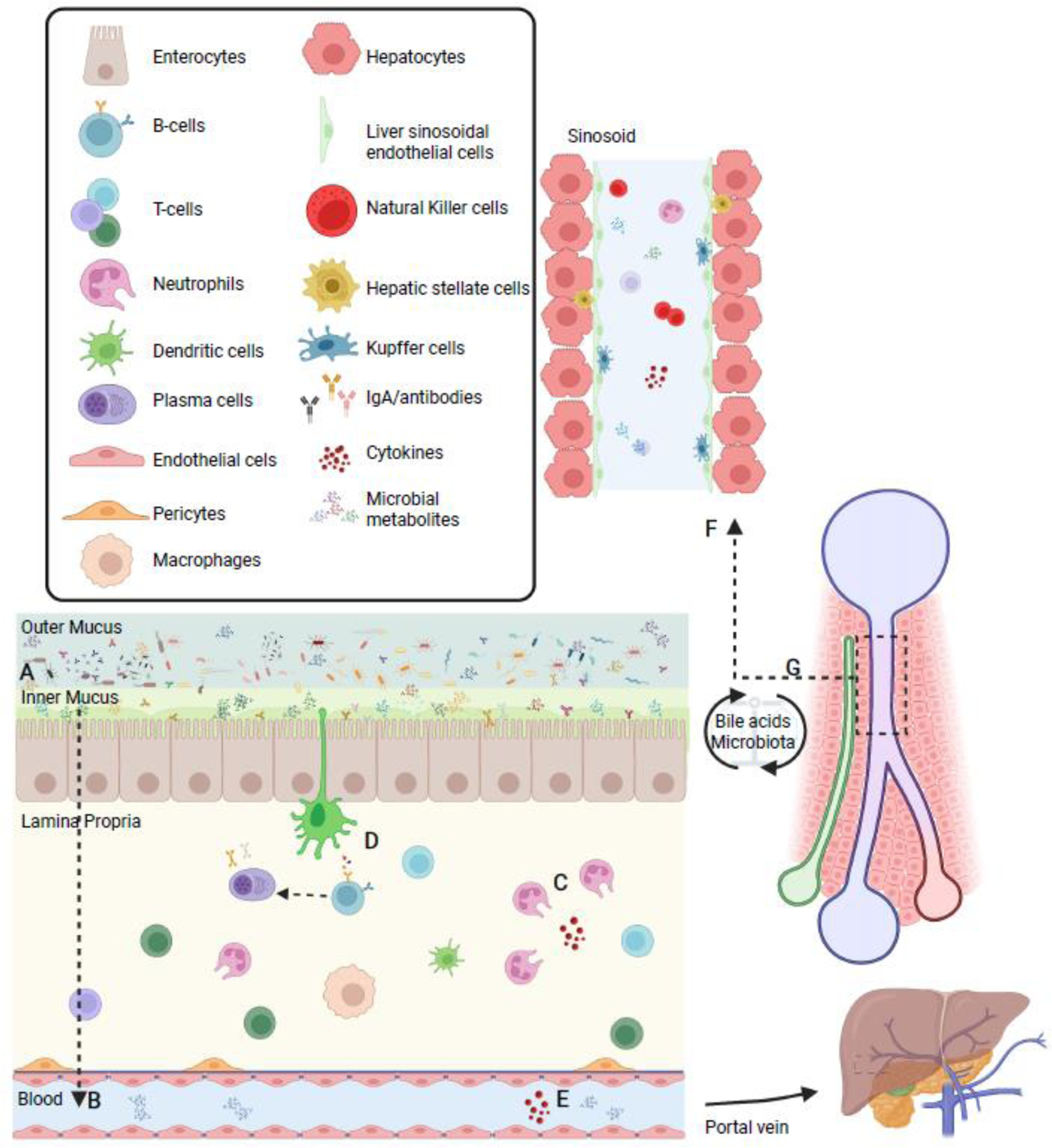
(A) The mucus physically separates the microbiota from the epithelial lining while antimicrobial peptides make the inner mucus almost sterile. (B) As a consequence, interactions between the microbiota and the host are mostly indirect and mediated by metabolic products that may cross the mucus and the epithelial barrier and reach the immune and stromal cells in the lamina propria or, through the lymphatic and blood vasculature, reach the liver and the systemic circulation. (C) Immune cells patrol the epithelium and reinforce the epithelial and mucus barriers through the production of growth factors and cytokines. These products exert a selective pressure for microorganisms. (D) Dendritic cells sense the environment and induce T- and B-cells response leading to production of IgA antibodies, which translocate to the lumen and contribute to mucosal immune protection by regulation of microbial composition and diversity. Influx of metabolic products through the (E) gut vascular barrier reaches the liver via the portal vein. (F) In the liver sinusoids, immune cells scan for foreign material. (G) In turn, the liver communicates with the small intestine by releasing bile acids and other bioactive mediators into the biliary tract; these metabolites can be biotransformed by the abundant microbiota in the terminal ileum and large intestine and in part recirculate to the liver through the portal vein thus possibly affecting both local and systemic functions.
Although studies of the microbiota role in carcinogenesis have originally focused on tumors of the epithelial barrier interfaces such as stomach and colon cancer, pancreatic ductal adenocarcinoma (PDAC) provided much of the evidence of the role of oral and gut microbiota, as well as cancer-associated microbiota on carcinogenesis and identified some of the mechanisms by which bacteria modulate the tumor microenvironment (TME) [13–16] (Box 1). As an interconnected system, the microbe-host crosstalk in the gut-liver axis is expected to be an important factor in hepatocarcinogenesis as it is in PDAC (Box 2). Furthermore, the physiological connection between liver and pancreas urges the question as if what was recently modelled in PDAC can be applied in HCC. Herein, we review the microbiota role on maintaining the gut-liver axis homeostasis and focus on how environmental perturbations may trigger gene responses associated with hepatocarcinogenesis directly (by inducing changes in relative abundance/diversity of microbes) or indirectly (by the action of microbial metabolites). Despite connected responses, this classification is used to discriminate between the correlation between alterations in the microbiome composition and diversity that was already characterized in literature, to description of specific mechanisms associated with the action of microbial metabolites.
Box 1. Pancreatic cancer studies addressed the link between gut dysbiosis, intratumoral bacteria and cancer.
Human pancreatic cancer precursor lesions were shown to be infiltrated by IL-17 producing Th17 cells, accelerating cancer initiation and progression [105]. This is interesting due to the link between IL-17, immune cells, diet and the microbiota. Indeed, several studies have shown an association of PDAC with the composition of the oral microbiome, the increased abundance of oral pathogens such as Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans or the presence of antibodies against oral microbes [106–108]. It was described lower alpha-diversity in pancreatic carcinoma, with increase of lipopolysaccharides-producing bacteria and decrease of butyrate-producing bacteria [13]. Based on the profiles of oral and gut microbiota communities associated with PDAC, non-invasive diagnostic models that distinguish PDAC patients from healthy individuals have been proposed [13,107]. Also, PDAC studies have established that the pancreas, an organ previously considered to be sterile, contains tissue-associated bacteria and fungi that are different and more abundant in pancreatic cancer compared with normal pancreatic tissue [14,16,109]. The bacteria in PDAC are mostly present intracellularly in both immune and cancer cells [110]. The composition of intratumor bacteria is different in each tumor type analyzed and can be used to predict tumor vs normal tissue and tumor type [111]. Although bacteria have been observed in approximately 2/3 of PDAC and in higher number than in most other tumor types, their absolute number in the tumors is only about 1/40 to 1/400 compared with human cells [14,110]. Tumor-associated bacteria in PDAC, compared with other tumors, are rich in gamma-proteobacteria, particularly enterobacterales, and unlike in stomach and colon cancer, are not enriched in fusobacteria [110]. The PDAC -associated microbiota is similar in composition with the duodenal microbiota, a fact that suggests retrograde migration of bacteria via the biliary-pancreatic duct.
Tumor associated microbes have been shown to participate in pancreatic carcinogenesis or resistance to therapy via different mechanisms. For example, bacteria expressing the long isoform of cytidine deaminase such as Gammaproteobacteria metabolize gemcitabine into its inactive form, contributing to drug resistance in PDAC [14]. Intratumoral bacteria may reprogram the TME by inducing myeloid-derived suppressor cells and suppressing M1 macrophage differentiation and CD4+ and CD8+ T cells activation; bacterial ablation by antibiotics treatment reprograms the pancreatic tumor immune microenvironment preventing carcinogenesis and enabling response to immune checkpoint inhibition therapy [16]. However, Riquelme et al. [15] showed that the intratumor bacteria can also be beneficial and that long-term survivors with PDAC exhibit higher microbial alpha-diversity and distinct signature (Pseudoxanthomonas, Streptomyces and Saccharopolyspora) in the tumor associated microbiota, which may induce potent immune cells infiltration and antitumor immunity. The role of microbial modulation in PDAC was recently reviewed [112,113]. These studies support a direct link between extra- and intra-tumor microbiota and susceptibility to development and clinical progression in patients with PDAC.
Box 2. Bacteria are complicit to carcinogenesis via microbes-host and host-host interactions.
Unlike viruses and parasites, only one bacterial species, Helicobacter pylori, has been officially identified as a definite human carcinogen for stomach cancer [114]. However, emerging evidence suggests that bacteria, either residing on the epithelial barrier interfaces or present within the tumor, are complicit to carcinogenesis and tumor progression in local or distant tissues [12,115,116].
Bacteria create a selective pressure in the TME to facilitate tumorigenesis, in part by priming for generation of reactive oxygen species (ROS), affecting the response to changes in pH, competing for limited nutrients, increasing DNA damage and mutagenesis, regulating oncogenes pathways, affecting the metabolism of chemotherapy drugs or modulating immunity [12,14,24,117–122]. Evidence for a role of bacteria in genomic mutation is provided by the observation that Escherichia coli strains carrying the colibactin-producing polyketide-nonribosomal peptide synthase operon (pks) induce a distinct mutational signature in colorectal cancer [123,124]. Microbiota effect on oncogene-induced tumor progression is supported by the observation that mutated p53 is pro-carcinogenic only in distal colon because of the presence of microbially produced gallic acid that prevents mutated p53 to act as a tumor suppressor by disrupting the WNT pathway [125]. Conversely, tumors apply a competitive pressure to local tissue cells that could affect bacteria within the TME, in surrounding tissues and in the gut [116,126].
A competitive dynamics in the TME between host-host neighboring cells, underlying the ecosystem model, has been described in both colon [127–129] and liver [130] carcinogenesis and could provide a substrate for bacteria to modulate the TME. Malignant stem cells secrete factors that promote differentiation of neighboring stem cells into clones harboring cancer-promoting mutations [127–129]. Alterations in the microbiota could represent another trigger combined with other multiple signals involved in the host cell crosstalk, affecting not only the cancer precursor target cells, but all cells by tuning the local tissue environment.
Additionally, tumor-associated bacteria are mostly intracellular in cancer and immune cells, possibly influencing cancer cell signaling and being presented as MHC-restricted peptides on the surface of antigen-presenting cells, thus stimulating the host immunity [103,110,111,115]. Although great progress has been made in improving the technology of identification and analysis of rare bacteria in low biomass normal and tumor tissue sample, these results still need to be interpreted with caution [131,132]. Nevertheless, they support the novel concept that the TME selects the residing microbiota and in turn is influenced by the microbiota [116].
Cascade of interconnected responses associate gut microbiota with liver
The composition of microbiota is established during early infancy and remains relatively stable through adult life. However, the relative abundance of bacterial species may rapidly change due to style of life, diet, disease, infections, and use of antibiotics. Unlike host cells that have a stable genome, the microbiota adapts to environmental changes and modulates host responses by expansion/contraction of microbial species, by occupying different anatomical niches and by mutation and exchange of genetic material. Therefore, the crosstalk between microbiota and host is expected to have a key influence on health and disease. Indeed, while the microbiota plays an important role in the host’s innate and adaptive immune system development since birth [17], the immune system also shapes host-microbe interactions [18]. This process relies on compartmentalization of the gut mucosal surface, microbiota sensing and signal transmission and immune cell priming, to create specific responses and maintain homeostasis [19,20]. Combined, the epithelial barrier, its microbial communities and the local immune system allow not only the tolerance of commensal bacteria in the environment, but also enables a response of the immune system against opportunistic bacteria or microbial products [1,18,20–24].
When these defense mechanisms fail, as due to increased intestinal permeability (leaky gut) or dysbiosis (change on the composition of the microbiota that is associated or causatively linked to disease), there is an influx of bacterial metabolites or previously compartmentalized and potentially harmful microorganisms that may pass the gut vascular barrier (GVB) and extend to the liver via the portal circulation [25]. In the liver, environmental perturbations trigger hepatic immune responses, which are dependent on resident immune cells [25–28] and circulating antigens and endotoxins from the gut microbiota [29] and shaped by immune surveillance linked to the gut-associated lymphoid tissue (GALT) [20], confirming the key role of the microbiota as an interconnected system in liver homeostasis. Small intestinal bacterial overgrowth (SIBO) is also associated with leaky gut and bacterial influx to the liver [30].
Independent of the origin, the increase in the translocation of bacterial products or microbes stimulate a pro-inflammatory response associated with chronic liver diseases [25,30,31]. Conversely, the liver, via the biliary tract, releases in the intestine bile acids and other bioactive mediators, which may undergo biotransformation by the gut microbiota and then be absorbed and released into the systemic circulation [31,32]. This bidirectional relationship explains why disturbances in the intestinal barriers alter the microenvironment in the liver and vice versa (Figure 1), with alteration of the physiological tissue homeostasis that may lead to disease, such as cancer [33,34]. Also, by linking dietary patterns with the microbiome influence on immune and metabolic status [35–37] and cancer therapy responses [38,39], this relationship reveals the link between the microbiota and the liver as a cascade of interconnected responses triggered by local environmental perturbations [30,31]. Collectively, the gut-liver axis ecosystem can trigger both local and distant responses and outline the direct and indirect interactions between the gut microbiota and the liver [20] (Figure 2).
Different triggers correlate with altered microbial relative abundance: consequences within the gut-liver-axis
Healthy intestinal barriers are permeable to some microbial metabolites, but not to most intact microbes [40,41]. However, the influence of diet and other environmental factors rapidly enrich or deplete specific nutrients and bacteria. This effect has consequences for microbial metabolite production and transformation of bile acids, which have the potential to shape the local microenvironment and correlate with the development of chronic liver diseases, including cancer [20,25,30,31,33]. For example, alcohol was shown to induce leaky gut. Together with SIBO, it allows endotoxins to enter the circulation, contributing to liver diseases [31,42,43]. SIBO is one manifestation of gut microbial dysbiosis that is characterized by decreased alpha-diversity and, in a proportion of cases, increased beta-diversity, which might lead to systemic inflammation [44,45]. Indeed, increased bacterial translocation also leads to fibrogenesis and contributes to cirrhosis progression (precursors for HCC) by increasing the exposure of liver cells to microbes and bacterial metabolites or by creating premetastatic niches in the liver that alter the environment to favor the recruitment and proliferation of metastatic cells [25,30,31,46]. Finally, in certain chronic liver diseases, translocation of viable bacteria may lead to immune paralysis in the GALT [30], suggesting a mechanism where changes in the balance of microbes directly influences the local environment. Below we review how gut bacteria may affect liver carcinogenesis directly by spreading to the liver, influencing the environment, or altering the crosstalk within other cells.
Dietary patterns influence the microbiome
Diet alters the gut microbiome [37,47], with consequences for the immune and metabolism landscapes, cancer risk, and response to cancer therapies [38,39,48]. Therefore, the origin and type of ingested food have a key role in modulating the gut microbiome, with consequences for host-microbe interactions [47].
Three predominant variants (enterotypes) have been characterized in the human gut (Bacteroides, Prevotella and Ruminococcus) [49], with different ratios observed in industrialized vs non-industrialized human populations regarding the intake of protein and animal fat (Bacteroides) versus carbohydrates or plant-based food (Prevotella) [50,51]. Industrialization is associated with decreased consumption of naturally rich fiber foods [52], which are important components of a human diet. Dietary fibers are categorized as insoluble (resistant to fermentation) or soluble (can be metabolized by the gut microbiota), such as the prebiotic fiber inulin, which is fermented by gut microbiota into short-chain fatty acids (SCFA) acetate, butyrate, and propionate in the colon. Inulin is a heterogeneous blend of fructose polymers. Fructose intake is converted into acetate and triggers hepatic de novo lipogenesis via microbiota-derived acetate, which reaches the liver via the portal vein [53], confirming the gut influence on the liver environment via diet. In the gut, the effects of inulin on microbiome composition in adult humans were associated with increased Bifidobacterium, Anaerostipes, Faecalibacterium and Lactobacillus spp, and a decrease in the relative abundance of Bacteroides spp in one study [54]. Another study showed an increased abundance of Prevotella, Treponema, and Succinivibrio spp [47,55]. Inversely, a high-fiber diet increased microbiome-encoded glycan-degrading carbohydrate active enzymes without affecting community diversity. In contrast, high-fermented food diet increased microbiome diversity and decreased inflammation [35]. Mechanistically, a fiber-deprived diet was shown to enrich for colonic mucus-degrading bacteria, enhancing Citrobacter rodentium mucus layer transversion and associated colitis [56], suggesting multiple mechanisms by which dietary patterns might influence the gut-liver axis.
Healthy plant-based foods influence gut microbial diversity and composition, including enrichment in butyrate producers, as Roseburia hominis, Agathobaculum butyriciproducens, Faecalibacterium prausnitzii, and Anaerostipes hadrus [57]. Butyrate is four-carbon SCFA produced through microbial fermentation of dietary fibers in the intestinal lumen. Butyrate contributes to mucosal homeostasis and integrity of the gut lining, thereby providing most of the energetic requirement of enterocytes and exerting an anti-inflammatory effect by binding to several G-protein coupled receptors and acting as a histone deacetylase inhibitor [58]. In mouse models, increased dietary fiber protected against colorectal tumorigenesis in a microbiota- and butyrate- dependent manner [59]. Collectively, these studies highlight the profound impact diet has on the microbiome communities and its consequences for the host, depending on the food source that is ingested.
Diet as environmental perturbations leading to liver diseases
It was previously shown that a Mediterranean diet, associated with an enrichment of Bacteroidetes and Firmicutes, and a decrease in Proteobacteria and Bacillota phyla [60,61], can reduce liver fat and is recommended for prevention of nonalcoholic fatty liver disease (NAFLD) [62]. The prolonged consumption of fermentable fiber enriched foods (as soluble fiber inulin or inulin-enriched high-fat diet) in mice prone to dysbiosis leads to inflammation, cholestasis, and HCC [52]. An increase in total bacterial load, reduced diversity, and a specific increase in Proteobacteria and fiber-fermenting bacteria such as Clostridia spp were observed. Interventions to deplete these butyrate-producing bacteria was successful in preventing inulin-enriched HCC, suggesting that modulation of the diet-associated microbiome is a potential avenue to prevent liver cancer.
Finally, high cholesterol/high fat diet sequentially led to stage progression to steatosis, steatohepatitis, fibrosis and eventually NAFLD-HCC in mice, due to gut microbiota dysbiosis. Distinct microbiota composition was enriched along each stage, as Mucispirillum, Desulfovibrio, Anaerotruncus and Desulfovibrionaceae increased sequentially while Bifidobacterium and Bacteroides were depleted towards HCC [39]. Other examples of diet influencing microbes and cancer were recently reviewed [47,61]. Altogether, dietary patterns influence the gut microbiome with potential consequences for the liver by enriching for specific bacteria associated with health or disease.
Microbial diversity associated with liver chronic diseases and hepatocarcinogenesis
Many microbiota products and metabolites are well-known risk factors for liver cancer development. Considering that the environment shapes and selects specific microbiota, it is interesting to speculate which competitive advantages certain species may gain, and how enrichment of specific microbiota could influence progression towards liver cancer. Because it is premature to conclude whether similar or different microbiota-composition may be involved in HCC and Cholangiocarcinoma (CCA) development, we discuss both types of liver cancer in the same section.
It was recently reported that 16S rRNA gene sequencing on HCC of viral and non-viral etiology identified distinct microbial compositions between tumor and non-tumor regions in the liver with species of the Bacteroidetes, Firmicutes and Proteobacteria phyla dominating the tumor-associated microbiota [63]. Ruminococcus gnavus was identified as a signature taxon for HCC patients infected with hepatitis viruses. Recently, the tumor microbiota of HCC patients with cirrhosis showed higher abundance of Stenotrophomonas maltophilia, which associated with senescence-associated secretory phenotype (SASP) in hepatic stellate cells (HSC), confirming the association between dysbiosis and modulation of liver cells [64]. Indeed, it was recently suggested that patients with NAFLD-cirrhosis with or without HCC have distinct gut microbiota composition [65]. Patients with HCC have an increased abundance of Bacteroides and Ruminococcaceae and decreased abundance of Bifidobacterium that correlated with increased levels of fecal calprotectin and evidence of systemic inflammation [65]. Likewise, the gut microbiota in HCC patients showed increasing in genera producing-LPS and decreases in butyrate-producing genera in early HCC as compared to as those with cirrhosis [66]. These data suggest that in HCC patients with NAFLD and cirrhosis, gut microbiota composition and systemic inflammation are correlated and may promote hepatocarcinogenesis.
Conversely, HCC patients progressing from hepatitis B virus (HBV) infection showed increased abundance of anti-inflammatory bacteria (such as Prevotella, Lactobacillus, Bifidobacterium and Faecalibacterium) and reduced pro-inflammatory bacteria (as Escherichia-Shigella and Enterococcus), a gut microbiota composition that is likely associated with HBV infection modulating host immuno-biological pathways [67]. Collectively, these studies highlight the microbial diversity observed in HCC, which represents a correlation between known risk factors and HCC development. As an interconnected system where diet and other environmental factors influence the microbiome, it will be necessary to characterize the local microbiota composition and diversity in multiple sites of evaluation (tumor and non-tumor) and tissues, associated with single-cell analysis and potentially correlated with environmental factors. Also, functional assays highlighting the potential causality of specific bacterial species enrichment are a key step to move past correlations.
Recent studies profiled the microbiota associated within the bile duct. Some bacterial families such as Dietziaceae, Pseudomonadaceae and Oxalobacteraceae [68] dominate the bile duct associated microbiota, showing that unique microbial communities are resident in this anatomic localization. Extrahepatic CCA patients have an abundance of Enterococcus, Streptococcus, Bacteroides, Klebsiella, and Pyramidobacter genera [69]. Also, bile samples of patients with CCA were enriched for Enterobacter, Pseudomonas, and Stenotrophomonas species as compared to patients with choledocholithiasis [70]. Finally, four bacterial genera (Lactobacillus, Actinomyces, Peptostreptococcus, and Alloscardovia) were increased in the gut microbiota of patients with intrahepatic CCA compared to those with HCC, cirrhosis, and healthy individuals [71]. Recently, different gut microbiota signatures have been proposed to distinguish CCA and cholelithiasis. Species of genera Bacteroides, Muribaculaceae_unclassified, Muribaculum, and Alistipes were enriched in CCA while distinct microbial species were enriched in the cholelithiasis group, suggesting changes in microbial association during evolution from benign to malignant hepatobiliary diseases [72].
Collectively, these processes may account for a link between gut bacterial translocation directly leading to the establishment of an inflamed liver environment favorable to liver cancer development and progression. However, it is hard to assume causality uniquely based on correlation with the microbial relative abundance, which is rapidly altered due to multiple environmental factors. Therefore, a comprehensive trans kingdom network analysis comparing HCC and CCA and linking the local gut and tissue microbiota compositions, diversity, and influence of environmental factors on metabolism, immunity, and transcriptional alterations will be crucial for dissecting the causal role of the microbiota in modulating liver carcinogenesis and its mechanisms of action.
Molecular mechanisms of microbiota indirectly inducing liver cancer
To understand the potential implications of specific microbiota within the liver tumor it is necessary to associate the potential mechanisms and the crosstalk between microbes and other cells. In this sense, a recent study characterized the temporal evolution of HCC development in Mdr2-deficent mice that lack the ability to secrete phospholipid into the bile from the liver, thus undergoing cholestasis and HCC development. In these mice, which represent a useful model of inflammation-induced HCC, intestinal dysbiosis induces gut barrier dysfunction that precedes LPS-mediated transcriptional alterations in the liver and consequently HCC development [73]. Furthermore, a transition in the intrahepatic inflammatory gene profile from proinflammatory phenotype in the early phases of liver injury towards an immunosuppressed phenotype in HCC was demonstrated. This change was associated with a redirection of energy source utilization via a switch in microbiome functions from carbohydrate towards amino acid metabolism. This study highlights the indirect influence of alterations in the microbiota on the liver environment (likely through crosstalk within other cells in the environment or microbial metabolites), and its association with liver cancer development and progression.
Alterations in microbial communities were shown to contribute to nonalcoholic fatty liver disease (NAFLD) and its progression to non-alcoholic steatohepatitis (NASH), by affecting hepatic carbohydrate and lipid metabolism, which modulated inflammation [26,43,74]. Modulation of metabolism and inflammation may similarly affect hepatic oncogenesis. Indeed, the gut microbiome is influenced by diet and other environmental factors, and microbial competition by nutrients is a key step towards modulating metabolism and immune responses [35,36,57]. For example, production of SCFA by microbiota-mediated fermentation of dietary fibers has been associated with cholestatic HCC [52]. It was also suggested that translocation of bacterial products may stimulate inflammation and release ROS within the GALT, impacting on the mechanical and secretory barrier and local microbiota [30,31]. These studies highlight the need to continue to conduct systematic and global studies characterizing the diversity and abundance of microbial species in the gut-liver axis as an ecosystem, but also to start dissecting the mechanisms behind those phenotypes.
Bacterial metabolites modulate key liver cells in the environment
Recently, it was suggested that maternal intake of butyrate and glutamine during pregnancy influences the fecal microbial population and metabolites in newborn mice, which are associated with fecal signatures of Bacteroidetes and Clostridia. Also, these newborn mice are resistant to hepatic immune activation, resulting in inflammation and injury of the bile ducts [75]. Mechanistically, the influence by which bacterial metabolites trigger gene responses in host cells may depend on transcriptional alterations within the environment. Indeed, distinct transcriptional signatures in acute liver failure animal models were characterized, and it was suggested that gut microbiota and TLR-signaling activate a MYC-dependent transcriptional program in HSC, Kupffer cells, and liver sinusoidal endothelial cells (LSECs), leading to infiltration of Ly6C-positive inflammatory monocytes and liver failure [76]. Circulating levels of LPS through TLR4 activation also induce HSCs to secrete growth factors, to regulate liver chronic inflammatory status, and suppress apoptosis, processes that are linked to HCC promotion [77]. Activation of the transcriptional regulator YAP1 downstream of LPS-TLR4 interaction in the portal vein area regulates stemness of hepatocytes [78]. Because site localization of hepatocyte turnover is important in the activation of molecular pathways underlaying homeostasis and regeneration [79], it is possible to speculate that LPS could also modulate these mechanisms to influence HCC by regulating local microenvironment reprogramming. In CCA, increased gut permeability induces microbial LPS translocation into the liver, that through a TLR4-dependent mechanism, induces expression of CXCL1 in hepatocytes. This expression in turn leads to the accumulation of CCR2+ polymorphonuclear myeloid-derived suppressor cells (MDSC). [80]
Recently, HCC patients with cirrhosis were shown to have a higher intratumoral abundance of Stenotrophomonas maltophilia, which promoted hepatic fibrosis by inducing expression of SASP factors in senescent HSC and proinflammatory factors via activation of TLR4/NF-κB/NLRP3 pathway, with consequent aggravation of liver fibrosis and progression to HCC [64]. Indeed, HSC proliferation is a key event in the development of liver fibrosis. Finally, bile acids induced HSC proliferation via the activation of the epidermal growth factor receptor [81], highlighting that the gut microbiota modulates the gene expression program of liver cells to promote HCC and CCA.
Diet metabolites influence the liver environment
Diet rapidly alters the human gut microbiome [37]. Indeed, the diet-derived microbial metabolites, p-cresol sulfate, 4-ethylphenyl sulfate, and 4-methylcathechol, have been shown to influence HCC subtypes [82]. The metabolic pathways encoded by the human gut microbiome constantly interact with host gene products through numerous bioactive molecules. For example, nutrient overload increases IL-17A, which in turn induced neutrophil infiltration in white adipose tissue and NASH-induced HCC [83]. Il-17A is a tumor promoting cytokine that regulates alcohol-induced hepatic steatosis, inflammation, fibrosis, and progression to HCC through regulation of inflammatory responses in Kupffer cells and marrow-derived monocytes, and cholesterol synthesis in steatotic hepatocytes [84]. Recently, digoxin, a retinoid orphan receptor gamma t (RORγt) antagonist, reduced IL-17A levels and stabilized body weight [85], suggesting its critical role in metabolic disorders. Also, TNF and IL-17A have been implicated in the development of liver inflammation and fibrogenesis induced by NLRP3 inflammasome activation in myeloid derived cells [86]. Collectively, these studies suggest a mechanistic link between diet metabolites, cytokines, and liver cancer diseases.
Bile acids as messengers for microbiota-gut-liver interactions
Primary bile acids are synthesized within hepatocytes and released into the duodenum and in large part reabsorbed in the small intestine. A small percentage of primary bile acids escape into the colon, where gut commensal bacteria transform into secondary bile acids with multiple important functions for metabolism and host innate immune responses. Indeed, both dietary and microbial bile acids metabolites modulate RORγt positive regulatory T (Treg) cells, contributing to maintain host immunological homeostasis and to improve gut inflammation [87]. Also, bile acid metabolites can control host immune responses by modulating the balance of Th17 and Treg cells.
Alternatively, bile acids can play an active role in high-fat diets [88]. Dietary cholesterol induced gut bacterial metabolite alteration, including increased taurocholic acid and decreased 3-indolepropionic acid, driving NAFLD-HCC in mice. Hence, cholesterol inhibitory therapy and gut microbiota manipulation may be effective strategies for NAFLD-HCC prevention [89]. Finally, two litocholic acid (LCA) derivatives (3-oxoLCA and isoalloLCA) directly affect CD4+Tcells. 3-oxoLCA suppresses Th17 cell differentiation by directly binding to the transcription factor RORγt, while isoalloLCA enhances Treg cell differentiation, confirming that gut microbiota might control host immune responses [90]. For intrahepatic CCA, increases in the glycoursodeoxycholic acid and tauroursodeoxycholic acid plasma-stool ratios and a positive correlation between plasma taurocholic acid and IL-4 was observed, suggesting an interrelationship between gut microbiota, metabolites, cytokines, and bile acids [71]. A recent study of integrative omics revealed a subtype of CCA with elevated bile acid metabolites, dysregulated cholesterol metabolism, and a unique inflammatory response associated with an increased BMI, which suggests a model of obesity-induced gut microbiome dysbiosis to promote hepatocarcinogenesis [91].
Mechanistically, the bile acids/carcinogenesis axis involves bile acids receptors such as farnesoid X receptor (FXR) and G protein-coupled bile acid receptor 1 that could represent important therapeutic targets in cancer [92]. Indeed, bile acids such as DCA was shown to block FXR functions and its ability to suppress intestinal cancer stem cell proliferation, influencing the gut-liver axis homeostasis [32]. Also, activation of the bile acid sensor FXR or the G-protein coupled receptor TGR5 inhibited inflammatory signaling via suppression of the NF-κB dependent signaling pathways and NLRP3-dependendent inflammasome activities [93,94]. Therefore, diet-liver-bile acid-microbiota crosstalk plays an important role in gastrointestinal inflammation and colorectal and liver carcinogenesis, which could be targeted to prevent cancer initiation or progression [92].
Gut microbiome was found to use bile acids as messengers to control a chemokine-dependent accumulation of hepatic natural killer T cells (NKT), influencing anti-tumor immunity in the liver [95]. This process was mediated by alterations in commensal gut bacteria, balance of primary and secondary bile acids, and CXCL16 expression in LSECs, one of the first cells exposed to the gut-derived metabolites in the liver. Conversely, NKT in cooperation with CD8 T cells through an interaction with hepatocytes have been described to promote NASH and NASH-associated HCC [96]. Additionally, the interplay between NK cells and HSCs was recently shown to be a master switch of cancer dormancy and metastasis [97]. A stromal response hampered NK cell- and interferon-γ- mediated maintenance of tumor cell dormancy and induced liver metastases through a process where tissue injury and activated HSCs secrete CXCL12, which retains and renders NK cells quiescent through CXCR4, suppressing immune surveillance and contributing to metastatic outgrowth [97].
Finally, certain bacteria associated with obesity have the capacity to increase the secondary bile acid deoxycholic acid (DCA) [98]. An increase in DCA causes DNA damage in HSCs inducing SASPs leading to the production of inflammatory and tumor-promoting factors as well as COX2-induced immunosuppressive PGE2 in the liver, thus facilitating HCC development. Obesity is also associated with microbiota alterations that result in increased accumulation of the TLR2 agonist lipoteichoic acid (LTA), a major cell wall component in gram-positive bacteria. DCA increases TLR2 expression on HSCs and cooperate with LTA in inducing SASP factors and COX2 that through PGE2 induces immune suppression and facilitates obesity-associated HCC after chemical carcinogen exposure [99]. Other examples of microbes influencing liver diseases and cancer were recently reviewed [25]. The relationship between bile acids, the microbiota and HCC were reviewed elsewhere [100,101]. Altogether, these studies support a model that alterations in the state-of-balance has consequences for connected cells in the environment, confirming the microbiome and its metabolites are key influencers of gene response in the gut and liver, with implications for liver carcinogenesis.
Concluding Remarks
Herein we reviewed the complex interplay of the microbiota influencing HCC, emphasizing the working hypothesis that alterations in microbiota diversity or in microbial-metabolites in the gut-liver axis trigger gene responses in local cells, with consequences for the TME. However, this concept is easier to explain than to test. Until now, most studies have focused on the association of bacteria diversity and their metabolites with the phenotype of target cancer cells, without considering the fine tuning of the environment. When analyzed, alterations in other cells in the environment were often assumed to be due to indirect effects, an unjustified oversimplification. In an ecosystem, individuals interact with other individuals and within the environment, influencing and limiting each other. In the TME, specific microbes influence and are influenced by the other cells (microbes or host cells). Therefore, the ideal scenario would be to move from a target-cell perspective towards a microenvironment/ecosystem-perspective, analyzing direct and indirect alterations mediated by the microbiota within local cells in the environment. Single cell-based and imaging technologies and multi-OMICS studies may help dissecting these complex networks.
Looking forward, it is interesting to speculate whether the anatomical position of the liver in proximity and communication with intestinal mucosa associated microbiota as well as the direct and indirect consequences of the host-microbe crosstalk could be responsible for the high heterogeneity observed in HCC [102] (Outstanding question Box). Furthermore, the known biological and genetic heterogeneity in HCC is usually not taken into consideration for therapeutic decisions. However, a promising clinical trial in advanced HCC (NCT03785210) is evaluating if the immunomodulatory effect induced by Tadalafil, a phosphodiesterase 5 inhibitor (PDE5), combined with oral vancomycin (an antibiotic that has been shown to alter gut commensal bacteria with changes in bile acid metabolism leading to a liver-selective anti-tumor effects [95]) could enhance the therapeutic efficacy of Nivolumab, an anti-PD-1 immune checkpoint inhibitor. In the lab, studies employing single cell and spatial RNA sequencing associated with serum and tissue metabolites characterization in patients should help elucidate these points.
Outstanding question box.
Should we focus our research efforts on comparing higher numbers of patients or evaluating the surrounding cells/tissue?
Is the anatomical position of the liver and direct crosstalk with microbes and microbial-metabolites the major cause of liver cancer heterogeneity?
Can we apply what we learned in other cancers such as pancreatic carcinoma to liver cancer?
Also, it was recently suggested that peptides derived from intracellular bacteria in melanoma can be presented by tumor-associated antigen-presenting cells and elicit an immune response [103]. Due to the key role of the immune response in the liver, it would be interesting to evaluate whether bacterial peptide presentation to immune cells could be harnessed for clinical treatment of HCC.
Because the crosstalk between cancer stem cells and immune cells plays an important role in cancer progression [104], an unanswered question that remains is whether cancer stem cells are easily triggered by microbes/metabolites alterations. Due to physical and physiological connections, it will be important to evaluate the influence of the microbiota on cancer stem cells differentiation and consequences for the gut-liver axis ecosystem, as an integrated system.
The correlation between microbiota and PDAC for patients’ outcome have been more thoroughly investigated than in HCC and have clearly started to identify specific mechanisms, immune or not, by which the microbiota can affect the TME locally or systemically, as well as tumor progression and response to therapy (Box 1). Therefore, some of the strategies used in PDAC studies could be applied to HCC studies (Outstanding question Box). Finally, to better understand the mechanisms underlying the crosstalk between the microbiota and host cells and to obtain clinically useful information, it will be important to focus on multi-organs studies of the gut-liver-pancreas ecosystem as well as on the tissue heterogeneity between patients and between tumor and adjacent normal tissues.
Figure 3. Microbial-metabolites and diversity as triggers for liver cancer progression.
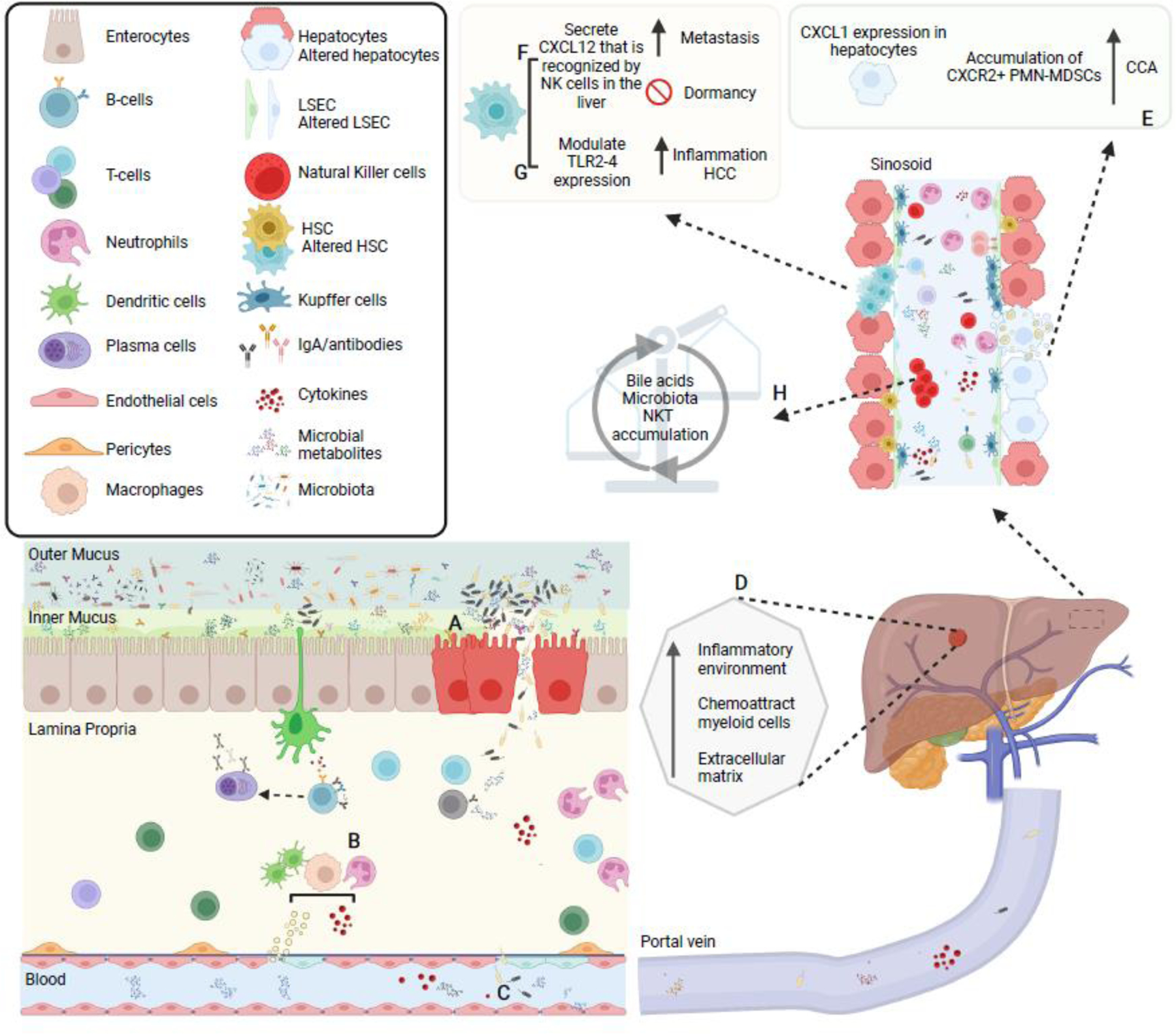
(A) Some bacteria may penetrate the mucus barrier or participate to its degradation, in some cases leading to breaching or disruption of the epithelial barrier, thus allowing (B) direct contact between microbes and microbial-metabolites with immune cells induce pro-inflammatory cytokines production and systemic dissemination. Disruption of the mucus/epithelial barriers may also facilitate (C) translocation of bacteria to the liver (D) where creates a favorable niche for cancer cell seeding. Hence, influx of previously compartmentalized bacteria and microbial products influence local liver cells’ gene expression. For example: (E) hepatocytes may express CXCR1 and induce an accumulation of CRCX2+ polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) creating an immunosuppressive environment to promote cholangiocarcinoma (CCA); (F) activated hepatic stellate cells that exert multiple functions for HCC and cancer metastasis may disrupt the function of NK cells in the liver through CXCL12-CXCR4 interactions, altering NK-cell mediated immunity and promoting breast metastasis into the liver; and (G) other liver immune cells may be activated via LPS-TLR4 or DCA-TLR2 modulation and induce inflammation favoring HCC generation. On the other hand, (H) liver produced bile acids after been modified by gut microbiota may activate a chemokine-dependent accumulation of hepatic NKT cells in the liver, controlling tumor growth.
Highlights.
Humans are metaorganisms composed of large host cells and an equivalent number of commensal microorganisms (the microbiota) residing on all epithelial barrier surfaces but particularly abundant in the lower intestinal tract.
Microbiota composition and microbial-metabolites influence tissue cells in the gut-liver axis.
Alteration of liver physiology, such as non-alcoholic fatty liver disease and viral infections, modify the microbiota and may create predisposing conditions for hepatocarcinogenesis.
Gut microbiota as well as bacteria residing within the tumor itself influence the tumor microenvironment by modulating the gene transcriptional programs in cancer, stromal and inflammatory/immune cells and either promoting or suppressing tumor progression.
Acknowledgements
This work was supported in part by the Intramural Research Program of the National Cancer Institute and by funds from the Canada Research Chair in Transcriptional Genomics (grant #950-231582 to SB). MADS was supported by the CCR/NCI FLEX synergy award “Liver cancer-associated microbiome and its role in tumor progression and therapy” to GT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Schluter J et al. (2020) The gut microbiota is associated with immune cell dynamics in humans. Nature 588, 303–307. 10.1038/s41586-020-2971-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch TC and McFall-Ngai MJ (2011) Metaorganisms as the new frontier. Zoology (Jena) 114, 185–190. 10.1016/j.zool.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foster KR et al. (2017) The evolution of the host microbiome as an ecosystem on a leash. Nature 548, 43–51. 10.1038/nature23292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silveira MAD and Bilodeau S (2018) Defining the Transcriptional Ecosystem. Mol Cell 72, 920–924. 10.1016/j.molcel.2018.11.022 [DOI] [PubMed] [Google Scholar]
- 5.Goldszmid RS and Trinchieri G (2012) The price of immunity. Nat Immunol 13, 932–938. 10.1038/ni.2422 [DOI] [PubMed] [Google Scholar]
- 6.Whisner CM and Athena Aktipis C (2019) The Role of the Microbiome in Cancer Initiation and Progression: How Microbes and Cancer Cells Utilize Excess Energy and Promote One Another’s Growth. Curr Nutr Rep 8, 42–51. 10.1007/s13668-019-0257-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayres JS (2016) Cooperative Microbial Tolerance Behaviors in Host-Microbiota Mutualism. Cell 165, 1323–1331. 10.1016/j.cell.2016.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prager BC et al. (2019) Cancer Stem Cells: The Architects of the Tumor Ecosystem. Cell Stem Cell 24, 41–53. 10.1016/j.stem.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Outschoorn UE et al. (2019) Editorial: Cancer Ecosystems. Front Oncol 9, 718. 10.3389/fonc.2019.00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner J et al. (2019) A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 177, 1330–1345 e1318. 10.1016/j.cell.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kern L et al. (2021) Commensal inter-bacterial interactions shaping the microbiota. Curr Opin Microbiol 63, 158–171. 10.1016/j.mib.2021.07.011 [DOI] [PubMed] [Google Scholar]
- 12.Cullin N et al. (2021) Microbiome and cancer. Cancer Cell. 10.1016/j.ccell.2021.08.006 [DOI] [PubMed] [Google Scholar]
- 13.Ren Z et al. (2017) Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget 8, 95176–95191. 10.18632/oncotarget.18820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geller LT et al. (2017) Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. 10.1126/science.aah5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riquelme E et al. (2019) Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 178, 795–806 e712. 10.1016/j.cell.2019.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pushalkar S et al. (2018) The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 8, 403–416. 10.1158/2159-8290.CD-17-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mishra A et al. (2021) Microbial exposure during early human development primes fetal immune cells. Cell. 10.1016/j.cell.2021.04.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng D et al. (2020) Interaction between microbiota and immunity in health and disease. Cell Res 30, 492–506. 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belkaid Y and Hand TW (2014) Role of the microbiota in immunity and inflammation. Cell 157, 121–141. 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X et al. (2020) The Gut-liver Axis in Immune Remodeling: New insight into Liver Diseases. Int J Biol Sci 16, 2357–2366. 10.7150/ijbs.46405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rooks MG and Garrett WS (2016) Gut microbiota, metabolites and host immunity. Nat Rev Immunol 16, 341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou CB et al. (2021) Gut Microbiota in Cancer Immune Response and Immunotherapy. Trends Cancer 7, 647–660. 10.1016/j.trecan.2021.01.010 [DOI] [PubMed] [Google Scholar]
- 23.Pabst O and Slack E (2020) IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol 13, 12–21. 10.1038/s41385-019-0227-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janney A et al. (2020) Host-microbiota maladaptation in colorectal cancer. Nature 585, 509–517. 10.1038/s41586-020-2729-3 [DOI] [PubMed] [Google Scholar]
- 25.Schwabe RF and Greten TF (2020) Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J Hepatol 72, 230–238. 10.1016/j.jhep.2019.08.016 [DOI] [PubMed] [Google Scholar]
- 26.Betrapally NS et al. (2017) Gut microbiome and liver disease. Transl Res 179, 49–59. 10.1016/j.trsl.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubes P and Jenne C (2018) Immune Responses in the Liver. Annu Rev Immunol 36, 247–277. 10.1146/annurev-immunol-051116-052415 [DOI] [PubMed] [Google Scholar]
- 28.Li X et al. (2021) The immunological and metabolic landscape in primary and metastatic liver cancer. Nat Rev Cancer 21, 541–557. 10.1038/s41568-021-00383-9 [DOI] [PubMed] [Google Scholar]
- 29.Heymann F and Tacke F (2016) Immunology in the liver--from homeostasis to disease. Nat Rev Gastroenterol Hepatol 13, 88–110. 10.1038/nrgastro.2015.200 [DOI] [PubMed] [Google Scholar]
- 30.Wiest R et al. (2014) Pathological bacterial translocation in liver cirrhosis. J Hepatol 60, 197–209. 10.1016/j.jhep.2013.07.044 [DOI] [PubMed] [Google Scholar]
- 31.Tripathi A et al. (2018) The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol 15, 397–411. 10.1038/s41575-018-0011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu T et al. (2019) FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 176, 1098–1112 e1018. 10.1016/j.cell.2019.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Albillos A et al. (2020) The gut-liver axis in liver disease: Pathophysiological basis for therapy. J Hepatol 72, 558–577. 10.1016/j.jhep.2019.10.003 [DOI] [PubMed] [Google Scholar]
- 34.Schroeder BO and Backhed F (2016) Signals from the gut microbiota to distant organs in physiology and disease. Nat Med 22, 1079–1089. 10.1038/nm.4185 [DOI] [PubMed] [Google Scholar]
- 35.Wastyk HC et al. (2021) Gut-microbiota-targeted diets modulate human immune status. Cell 184, 4137–4153 e4114. 10.1016/j.cell.2021.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolte LA et al. (2021) Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 70, 1287–1298. 10.1136/gutjnl-2020-322670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David LA et al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han K et al. (2021) Generation of systemic antitumour immunity via the in situ modulation of the gut microbiome by an orally administered inulin gel. Nat Biomed Eng 5, 1377–1388. 10.1038/s41551-021-00749-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer CN et al. (2021) Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 374, 1632–1640. 10.1126/science.aaz7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chelakkot C et al. (2018) Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med 50, 1–9. 10.1038/s12276-018-0126-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rescigno M (2011) The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol 32, 256–264. 10.1016/j.it.2011.04.003 [DOI] [PubMed] [Google Scholar]
- 42.Scarpellini E et al. (2016) Gut Microbiota and Alcoholic Liver Disease. Rev Recent Clin Trials 11, 213–219. 10.2174/1574887111666160810100538 [DOI] [PubMed] [Google Scholar]
- 43.Bajaj JS (2019) Alcohol, liver disease and the gut microbiota. Nat Rev Gastroenterol Hepatol 16, 235–246. 10.1038/s41575-018-0099-1 [DOI] [PubMed] [Google Scholar]
- 44.Saffouri GB et al. (2019) Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun 10, 2012. 10.1038/s41467-019-09964-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah A et al. (2017) Systematic Review and Meta-Analysis: Prevalence of Small Intestinal Bacterial Overgrowth in Chronic Liver Disease. Semin Liver Dis 37, 388–400. 10.1055/s-0037-1608832 [DOI] [PubMed] [Google Scholar]
- 46.Bertocchi A et al. (2021) Gut vascular barrier impairment leads to intestinal bacteria dissemination and colorectal cancer metastasis to liver. Cancer Cell 39, 708–724 e711. 10.1016/j.ccell.2021.03.004 [DOI] [PubMed] [Google Scholar]
- 47.Makki K et al. (2018) The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 23, 705–715. 10.1016/j.chom.2018.05.012 [DOI] [PubMed] [Google Scholar]
- 48.Steck SE and Murphy EA (2020) Dietary patterns and cancer risk. Nat Rev Cancer 20, 125–138. 10.1038/s41568-019-0227-4 [DOI] [PubMed] [Google Scholar]
- 49.Arumugam M et al. (2011) Enterotypes of the human gut microbiome. Nature 473, 174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu GD et al. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Filippo C et al. (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107, 14691–14696. 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh V et al. (2018) Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 175, 679–694 e622. 10.1016/j.cell.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao S et al. (2020) Dietary fructose feeds hepatic lipogenesis via microbiota-derived acetate. Nature 579, 586–591. 10.1038/s41586-020-2101-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Bastard Q et al. (2020) The effects of inulin on gut microbial composition: a systematic review of evidence from human studies. Eur J Clin Microbiol Infect Dis 39, 403–413. 10.1007/s10096-019-03721-w [DOI] [PubMed] [Google Scholar]
- 55.Kovatcheva-Datchary P et al. (2015) Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab 22, 971–982. 10.1016/j.cmet.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 56.Desai MS et al. (2016) A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 167, 1339–1353 e1321. 10.1016/j.cell.2016.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asnicar F et al. (2021) Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med 27, 321–332. 10.1038/s41591-020-01183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang PV et al. (2014) The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A 111, 2247–2252. 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Donohoe DR et al. (2014) A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov 4, 1387–1397. 10.1158/2159-8290.CD-14-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bifulco M (2015) Mediterranean diet: the missing link between gut microbiota and inflammatory diseases. Eur J Clin Nutr 69, 1078. 10.1038/ejcn.2015.81 [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Montero C et al. (2021) Nutritional Components in Western Diet Versus Mediterranean Diet at the Gut Microbiota-Immune System Interplay. Implications for Health and Disease. Nutrients 13. 10.3390/nu13020699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romero-Gomez M et al. (2017) Treatment of NAFLD with diet, physical activity and exercise. J Hepatol 67, 829–846. 10.1016/j.jhep.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 63.Komiyama S et al. (2021) Profiling of tumour-associated microbiota in human hepatocellular carcinoma. Sci Rep 11, 10589. 10.1038/s41598-021-89963-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu B et al. (2022) Hepatic stellate cell activation and senescence induced by intrahepatic microbiota disturbances drive progression of liver cirrhosis toward hepatocellular carcinoma. J Immunother Cancer 10. 10.1136/jitc-2021-003069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ponziani FR et al. (2019) Hepatocellular Carcinoma Is Associated With Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 69, 107–120. 10.1002/hep.30036 [DOI] [PubMed] [Google Scholar]
- 66.Ren Z et al. (2019) Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 68, 1014–1023. 10.1136/gutjnl-2017-315084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Q et al. (2019) Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog 11, 1. 10.1186/s13099-018-0281-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chng KR et al. (2016) Tissue Microbiome Profiling Identifies an Enrichment of Specific Enteric Bacteria in Opisthorchis viverrini Associated Cholangiocarcinoma. EBioMedicine 8, 195–202. 10.1016/j.ebiom.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saab M et al. (2021) Characterization of biliary microbiota dysbiosis in extrahepatic cholangiocarcinoma. PLoS One 16, e0247798. 10.1371/journal.pone.0247798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dangtakot R et al. (2021) Profiling of Bile Microbiome Identifies District Microbial Population between Choledocholithiasis and Cholangiocarcinoma Patients. Asian Pac J Cancer Prev 22, 233–240. 10.31557/APJCP.2021.22.1.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jia X et al. (2020) Characterization of Gut Microbiota, Bile Acid Metabolism, and Cytokines in Intrahepatic Cholangiocarcinoma. Hepatology 71, 893–906. 10.1002/hep.30852 [DOI] [PubMed] [Google Scholar]
- 72.Zhang T et al. (2021) A Predictive Model Based on the Gut Microbiota Improves the Diagnostic Effect in Patients With Cholangiocarcinoma. Front Cell Infect Microbiol 11, 751795. 10.3389/fcimb.2021.751795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Behary J et al. (2021) Defining the temporal evolution of gut dysbiosis and inflammatory responses leading to hepatocellular carcinoma in Mdr2 −/− mouse model. BMC Microbiol 21, 113. 10.1186/s12866-021-02171-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kolodziejczyk AA et al. (2019) The role of the microbiome in NAFLD and NASH. EMBO Mol Med 11. 10.15252/emmm.201809302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jee JJ et al. (2022) Maternal regulation of biliary disease in neonates via gut microbial metabolites. Nat Commun 13, 18. 10.1038/s41467-021-27689-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kolodziejczyk AA et al. (2020) Acute liver failure is regulated by MYC- and microbiome-dependent programs. Nat Med 26, 1899–1911. 10.1038/s41591-020-1102-2 [DOI] [PubMed] [Google Scholar]
- 77.Dapito DH et al. (2012) Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21, 504–516. 10.1016/j.ccr.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shao C et al. (2021) The stemness of hepatocytes is maintained by high levels of lipopolysaccharide via YAP1 activation. Stem Cell Res Ther 12, 342. 10.1186/s13287-021-02421-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei Y et al. (2021) Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science 371. 10.1126/science.abb1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Q et al. (2021) Gut Microbiome Directs Hepatocytes to Recruit MDSCs and Promote Cholangiocarcinoma. Cancer Discov 11, 1248–1267. 10.1158/2159-8290.CD-20-0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svegliati-Baroni G et al. (2005) Bile acids induce hepatic stellate cell proliferation via activation of the epidermal growth factor receptor. Gastroenterology 128, 1042–1055. 10.1053/j.gastro.2005.01.007 [DOI] [PubMed] [Google Scholar]
- 82.Pomyen Y et al. (2021) Tumor metabolism and associated serum metabolites define prognostic subtypes of Asian hepatocellular carcinoma. Sci Rep 11, 12097. 10.1038/s41598-021-91560-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gomes AL et al. (2016) Metabolic Inflammation-Associated IL-17A Causes Non-alcoholic Steatohepatitis and Hepatocellular Carcinoma. Cancer Cell 30, 161–175. 10.1016/j.ccell.2016.05.020 [DOI] [PubMed] [Google Scholar]
- 84.Ma HY et al. (2020) IL-17 signaling in steatotic hepatocytes and macrophages promotes hepatocellular carcinoma in alcohol-related liver disease. J Hepatol 72, 946–959. 10.1016/j.jhep.2019.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teijeiro A et al. (2021) Inhibition of the IL-17A axis in adipocytes suppresses diet-induced obesity and metabolic disorders in mice. Nat Metab 3, 496–512. 10.1038/s42255-021-00371-1 [DOI] [PubMed] [Google Scholar]
- 86.Wree A et al. (2018) NLRP3 inflammasome driven liver injury and fibrosis: Roles of IL-17 and TNF in mice. Hepatology 67, 736–749. 10.1002/hep.29523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song X et al. (2020) Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature 577, 410–415. 10.1038/s41586-019-1865-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trefflich I et al. (2019) Associations between Dietary Patterns and Bile Acids-Results from a Cross-Sectional Study in Vegans and Omnivores. Nutrients 12. 10.3390/nu12010047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X et al. (2021) Dietary cholesterol drives fatty liver-associated liver cancer by modulating gut microbiota and metabolites. Gut 70, 761–774. 10.1136/gutjnl-2019-319664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hang S et al. (2019) Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148. 10.1038/s41586-019-1785-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chaisaingmongkol J et al. (2017) Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell 32, 57–70 e53. 10.1016/j.ccell.2017.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jia W et al. (2018) Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 15, 111–128. 10.1038/nrgastro.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vavassori P et al. (2009) The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol 183, 6251–6261. 10.4049/jimmunol.0803978 [DOI] [PubMed] [Google Scholar]
- 94.Pols TW et al. (2011) TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab 14, 747–757. 10.1016/j.cmet.2011.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma C et al. (2018) Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360. 10.1126/science.aan5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolf MJ et al. (2014) Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell 26, 549–564. 10.1016/j.ccell.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 97.Correia AL et al. (2021) Hepatic stellate cells suppress NK cell-sustained breast cancer dormancy. Nature. 10.1038/s41586-021-03614-z [DOI] [PubMed] [Google Scholar]
- 98.Yoshimoto S et al. (2013) Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101. 10.1038/nature12347 [DOI] [PubMed] [Google Scholar]
- 99.Loo TM et al. (2017) Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE2-Mediated Suppression of Antitumor Immunity. Cancer Discov 7, 522–538. 10.1158/2159-8290.CD-16-0932 [DOI] [PubMed] [Google Scholar]
- 100.Yu LX and Schwabe RF (2017) The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol 14, 527–539. 10.1038/nrgastro.2017.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jia B and Jeon CO (2019) Promotion and induction of liver cancer by gut microbiome-mediated modulation of bile acids. PLoS Pathog 15, e1007954. 10.1371/journal.ppat.1007954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng H et al. (2018) Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 68, 127–140. 10.1002/hep.29778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kalaora S et al. (2021) Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592, 138–143. 10.1038/s41586-021-03368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bayik D and Lathia JD (2021) Cancer stem cell-immune cell crosstalk in tumour progression. Nat Rev Cancer. 10.1038/s41568-021-00366-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.McAllister F et al. (2014) Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 25, 621–637. 10.1016/j.ccr.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fan X et al. (2018) Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 67, 120–127. 10.1136/gutjnl-2016-312580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Farrell JJ et al. (2012) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588. 10.1136/gutjnl-2011-300784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Michaud DS et al. (2013) Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 62, 1764–1770. 10.1136/gutjnl-2012-303006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aykut B et al. (2019) The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267. 10.1038/s41586-019-1608-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nejman D et al. (2020) The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Poore GD et al. (2020) Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 579, 567–574. 10.1038/s41586-020-2095-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beatty GL et al. (2021) The biological underpinnings of therapeutic resistance in pancreatic cancer. Genes Dev. 10.1101/gad.348523.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chandra V and McAllister F (2021) Therapeutic potential of microbial modulation in pancreatic cancer. Gut. 10.1136/gutjnl-2019-319807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.(1994) Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum 61, 1–241 [PMC free article] [PubMed] [Google Scholar]
- 115.Sepich-Poore GD et al. (2021) The microbiome and human cancer. Science 371. 10.1126/science.abc4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xavier JB et al. (2020) The Cancer Microbiome: Distinguishing Direct and Indirect Effects Requires a Systemic View. Trends Cancer 6, 192–204. 10.1016/j.trecan.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yardeni T et al. (2019) Host mitochondria influence gut microbiome diversity: A role for ROS. Sci Signal 12. 10.1126/scisignal.aaw3159 [DOI] [PubMed] [Google Scholar]
- 118.Ilhan ZE et al. (2017) pH-Mediated Microbial and Metabolic Interactions in Fecal Enrichment Cultures. mSphere 2. 10.1128/mSphere.00047-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Garrett WS (2015) Cancer and the microbiota. Science 348, 80–86. 10.1126/science.aaa4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dzutsev A et al. (2017) Microbes and Cancer. Annu Rev Immunol 35, 199–228. 10.1146/annurev-immunol-051116-052133 [DOI] [PubMed] [Google Scholar]
- 121.Wong-Rolle A et al. (2021) Unexpected guests in the tumor microenvironment: microbiome in cancer. Protein Cell 12, 426–435. 10.1007/s13238-020-00813-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tilg H et al. (2020) The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol 20, 40–54. 10.1038/s41577-019-0198-4 [DOI] [PubMed] [Google Scholar]
- 123.Putze J et al. (2009) Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun 77, 4696–4703. 10.1128/IAI.00522-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pleguezuelos-Manzano C et al. (2020) Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 580, 269–273. 10.1038/s41586-020-2080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kadosh E et al. (2020) The gut microbiome switches mutant p53 from tumoursuppressive to oncogenic. Nature 586, 133–138. 10.1038/s41586-020-2541-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Marshall EA et al. (2021) Functional role of the cancer microbiome in the solid tumour niche. Current Research in Immunology 2, 1–6. 10.1016/j.crimmu.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yum MK et al. (2021) Tracing oncogene-driven remodelling of the intestinal stem cell niche. Nature 594, 442–447. 10.1038/s41586-021-03605-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Neerven SM et al. (2021) Apc-mutant cells act as supercompetitors in intestinal tumour initiation. Nature 594, 436–441. 10.1038/s41586-021-03558-4 [DOI] [PubMed] [Google Scholar]
- 129.Flanagan DJ et al. (2021) NOTUM from Apc-mutant cells biases clonal competition to initiate cancer. Nature 594, 430–435. 10.1038/s41586-021-03525-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Seehawer M et al. (2018) Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 562, 69–75. 10.1038/s41586-018-0519-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dohlman AB et al. (2021) The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe 29, 281–298 e285. 10.1016/j.chom.2020.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dzutsev A and Trinchieri G (2020) Microbial DNA signature in plasma enables cancer diagnosis. Nat Rev Clin Oncol 17, 453–454. 10.1038/s41571-020-0391-1 [DOI] [PubMed] [Google Scholar]


