Abstract
Engineered cardiac tissues derived from human induced pluripotent stem cells (iPSCs) are increasingly used for drug discovery, pharmacology and the models of developmental and disease. While there are numerous platforms to engineer cardiac tissues, they often require expensive and non-conventional equipment, and utilize advanced video processing algorithms. As a result, only specialized academic labs have been able to harness this technology. In addition, methodologies and tissue features have been challenging to reproduce between different groups and models. Here, we describe a facile technology (milliPillar) that covers the entire pipeline required for studies of engineered cardiac tissues. We describe methodologies for (i) platform fabrication, (ii) tissue engineering, (iii) electrical stimulation, (iv) automated real-time data acquisition and (v) advanced video analyses. We validate these methodologies and demonstrate the versatility of the platform by showcasing the fabrication of tissues in different hydrogel materials, and using cardiomyocytes derived from different iPSC lines in combination with different types of stromal cells. We also validate the use of electrical stimulation for long term culture (100 days) and provide protocols for automated analysis of force generation and calcium flux using both brightfield and fluorescent imaging. Lastly, we demonstrate the compatibility of the milliPillar platform with electromechanical stimulation to enhance cardiac tissue function. We expect that this resource will provide a valuable and user-friendly tool for the generation and real-time assessment of engineered human cardiac tissues for basic and translational studies.
Keywords: cardiac tissue engineering, organs-on-a-chip, induced pluripotent stem cells, cardiomyocytes, electromechanical stimulation, real-time imaging
INTRODUCTION
Numerous models of human myocardium have been developed in the past few years with the goal to generate minimally functional units that capture the heart patho/physiology in a simplified manner1. Monolayer cultures represent the simplest high-throughput model; however they lack the cell maturity and three-dimensional (3D) tissue environment required for most studies2,3. Microspheres can overcome this limitation while maintaining high throughput, but lack the structural template and mechanical cues, limiting their organization and not allowing direct measurements of contraction force, a critical aspect in disease modeling studies. Although lower in throughput, 3D cardiac tissues represent a promising alternative 4.
Cardiac tissues have been created through the compaction of cell-hydrogel constructs cast around anchoring structures such as wires, posts and pillars to provide mechanical loading that supports tissue maturation5–8. Further maturation has been achieved by electrical stimulation, designed to mimic the electrical pacing of cells in the native heart9,10. With advances in stem cell biology and the use of induced pluripotent stem cells (iPSCs), 3D models of the human heart can be used to study human cardiac biology, gain mechanistic insights into genetic disorders, and inform more effective patient-tailored therapies11.
While methodologies to generate and mature iPSC-derived cardiac tissues have been established by several groups, they are often difficult to set up and require specialized devices8,12,13. The equipment and protocols for electrical stimulation also vary from one study to another. Most importantly, the differences in the injected electrical charge can lead to difficulties in reproducing experiments between labs. With the increasing interest in harnessing these models for cardiac research, there is a clear need for a simple and accessible methodology to generate engineered cardiac tissues that can be adopted by any research laboratory.
Similarly, measurements of tissue functionality are an important point of consideration. Readouts from cardiac tissue models must be quantifiable, standardized, reproducible and relevant to the biological question at hand. With a diversity of existing models, it is imperative to compare tissue functionality across different studies, using absolute values of functional readouts. Quantitative metrics extracted from calcium and brightfield images can provide such values for pharmacological screening and studies of cardiac development and disease. In addition, the quantitative metrics in real-time is of great interest since it allows longitudinal studies, where the same tissue can be repeatedly perturbed and evaluated over time.
To address these needs, we have developed milliPillar; a robust, user-friendly and customizable platform that supports tissue fabrication, electromechanical stimulation, and real-time data acquisition and analysis. milliPillar is designed to be tailored to specific needs of different research groups. We have developed this technology to facilitate adoption of cardiac tissue engineering (especially to those entering the field), allow exchange of experimental resources and data between laboratories, and support further advances in the field. The capabilities of the milliPillar platform and the previously reported cardiac tissue engineering platforms are compared in Table S18,12,14–19.
RESULTS AND DISCUSSION
Platform Overview
The milliPillar platform was designed to accommodate six wells for the cultivation of millimeter-sized tissues composed of iPS-derived cardiomyocytes and supporting stromal cells encapsulated in fibrin or collagen hydrogel (Figure 1, Step I). Cell-laden hydrogels were dispensed into each well and compacted around two flexible pillars that provided mechanical load during tissue contraction. Two electrodes embedded along the platform were connected to a customized electrical stimulator to provide tissues with controlled electrical stimulation (Figure 1, Step II). The bottom of the platform was bonded to a glass slide to allow real-time imaging. Custom video analysis algorithms were developed to measure tissue contractility (by brightfield imaging) and calcium handling (by fluorescent imaging) (Figure 1, Step III). These three steps make milliPillar suitable for a wide variety of research and experimental designs.
Figure 1. The milliPillar pipeline for engineered cardiac tissue fabrication and assessment.
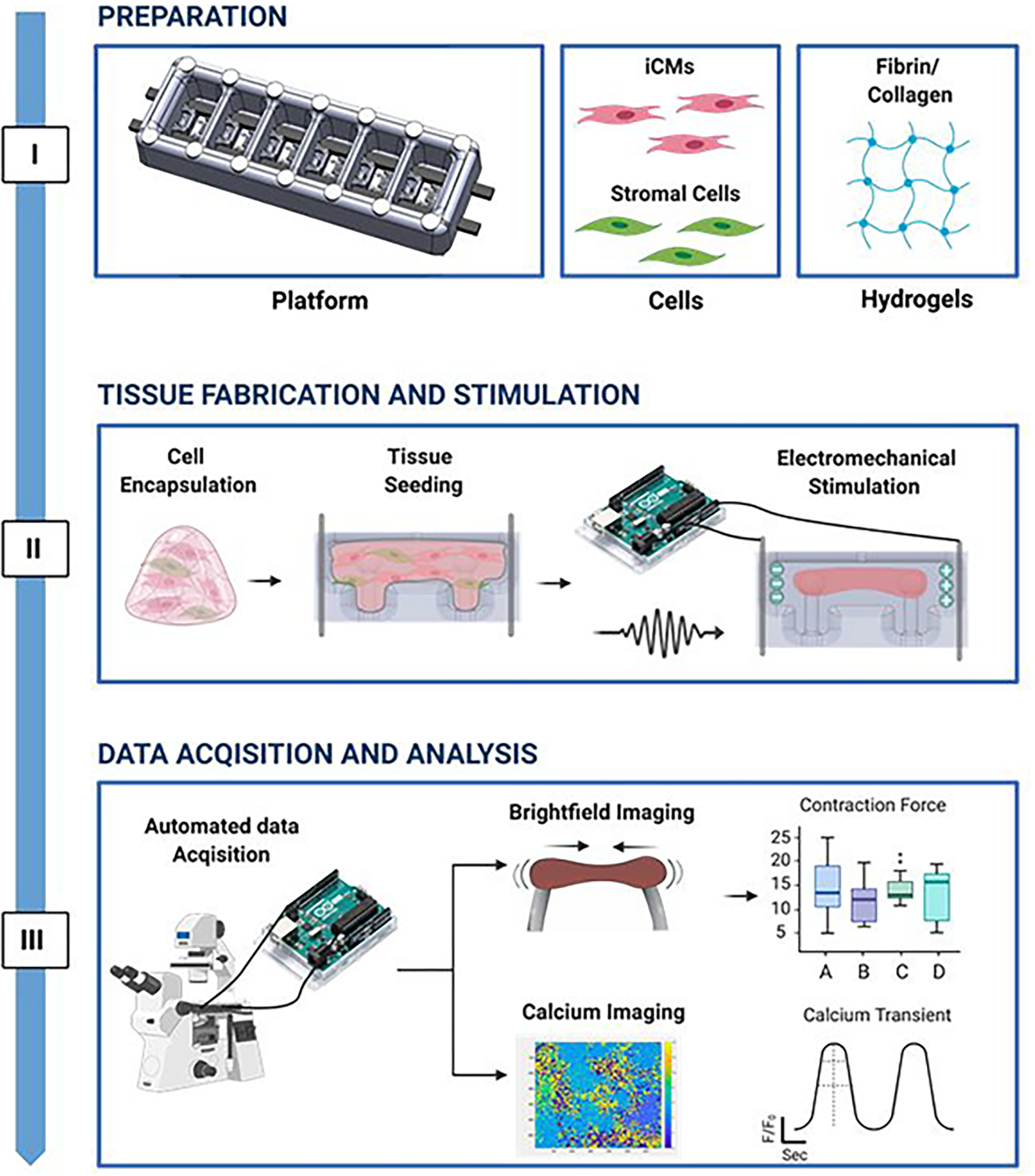
Step I: Preparation of the platform, cells, and hydrogel. Step II: Encapsulation of cardiomyocytes and stromal cells into a hydrogel, seeding tissues, and maturation of cardiac tissues by electromechanical stimulation. Step III: Automated analysis assessing cardiac tissue function using brightfield and calcium imaging.
Fabrication and characterization of milliPillar platform
Platform design.
The milliPillar platform was fabricated by casting polydimethylsiloxane (PDMS) and carbon electrode rods into custom molds and curing in an oven at 65° C (Supplementary Figures S1 and S2), detached from the mold and bonded to a glass slide (Figure 2A–B). PDMS was chosen due to its low cost, ease of manipulation, biocompatibility, and gas exchange properties. However, users must take into consideration that PDMS non-selectively absorbs hydrophobic compounds, including oxygen and many drugs, and therefore could lead to misinterpretation of pharmacological studies and supernatant analyses20. Efforts are in progress to replace PDMS with other biocompatible and inert materials21,22.
Figure 2. milliPillar platform fabrication and mechanical characterization.
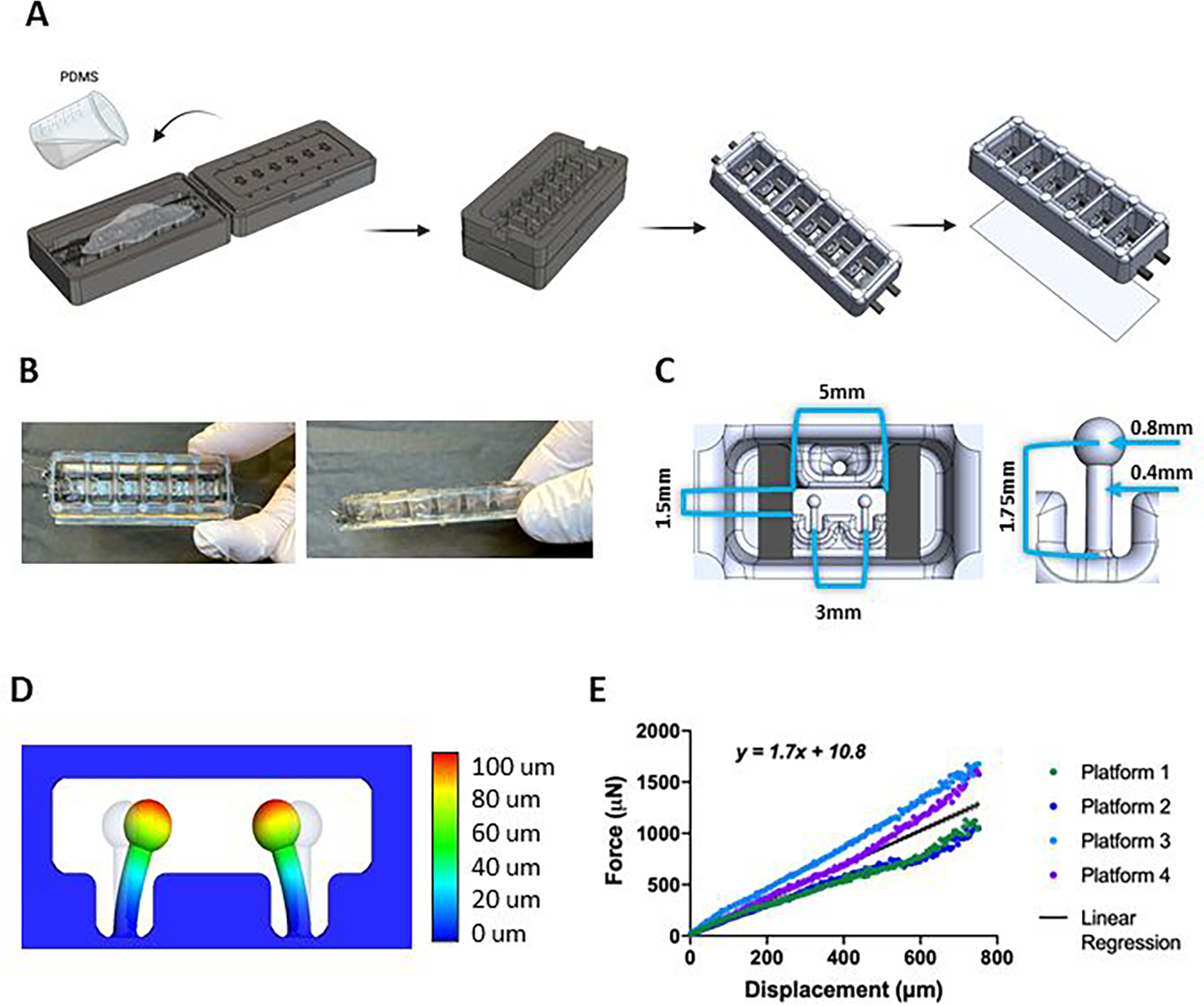
(A) Schematics of the milliPillar platform fabrication. (B) top and side views of the milliPillar platform. (C) Single well and pillar dimensions. (D) Modeling of pillar deflection. (F) Force vs displacement curves of platforms from 4 different fabrication batches.
The platform was designed to accommodate six tissues in separate wells to enable controlled culture conditions for each individual tissue (Figure 2A–B). The mm-size of the tissues was selected to decrease the number of cells10 while still providing sufficient tissue mass for molecular, structural and functional assays and also keeping the platform easy to manipulate under standard laboratory conditions (Figure 2C). Four platforms fit into one standard 4-well plate, enabling 24 tissues to be simultaneously cultured and analyzed (Supplementary Figure S2, bottom panel, step 7). The tissue spacing is compatible with a 96-well configuration to provide compatibility with microplate readers and other standard instrumentation. The dimensions of each culture well (13mm wide × 7mm long × 6 mm high) were designed to accommodate small amounts of culture media (400 μL) in order to maximize the injected electrical charge per unit volume and facilitate studies which may require precious media supplements (e.g. patient serum, exosomes, growth factors and cytokines).
Mechanical characterization of pillars.
Flexible anchors (e.g pillars, posts, wires) provide passive tension for continuous mechanical strain and auxotonic tissue contraction8,12,14–19. We designed the pairs of pillars with a head diameter of 0.8 mm and stem length of 1.75 mm that were placed in a horizontal configuration at the bottom of each well to allow compaction of the tissues around the pillar heads and enable visualization of pillar deflection using an inverted microscope, to accurately calculate tissue force generation (Figure 2C–D). To our knowledge, this is the first use of horizontal pillars, which allow for observation of the tissues movement in two dimensions.
The mechanical properties of the PDMS pillars are sensitive to curing temperature, curing time, ratio of the base material and curing agent and the environmental temperature10. Using a microscale force transducer and linear actuator, we tracked the pillar movement and the force associated with deflection, as well as the pillar deflection at rest (Supplementary Figure S3A). The coefficient derived from linear regression of the force - displacement data allowed for direct calculations of active force and passive tension based on the displacement of the pillars by the cardiac tissues during contraction and rest. Among four independent experiments, the pillars exhibited linear elastic behavior (r2 = 0.834) over the 0–750 μm testing range (Figure 2E). The pillars exhibited no hysteresis, suggesting that both tissue contraction and relaxation can be evaluated with the same coefficient (Supplementary Figure S3B). Importantly, this feature suggests that the same coefficient can be used throughout the study and that longitudinal measurements of the same cardiac tissue can be taken over time without re-calibration.
As anticipated, there was some batch-to-batch variation of material properties (Supplementary Figure S3C). Ideally, a single batch of platforms should be fabricated at the same time for a set of experiments and used soon after fabrication to minimize variability.
Customized electrical stimulation apparatus
Many groups, including our own, have utilized electrical stimulation designed to mimic signals driving cardiac muscle contractions in the heart and have demonstrated improved cell-cell connectivity, alignment and overall tissue function9,12,23–25. In previous studies, we have evaluated and modeled the electric field stimulation used to facilitate cardiac tissue contractions and have shown that carbon is a superior material of choice26. Here, we built upon this model and designed the milliPillar electrodes with carbon rods (6.5mm long × 2mm wide × 1 mm high; 5 mm apart) to create a double bi-layer capacitor. We utilized pure carbon rods as an improvement over electrodes used in the past due to the elimination of resin, to obtain a more porous material with increased surface area and higher conductivity27.
Electrical stimulation of engineered cardiac tissues requires expensive and specialized equipment, not commonly available in most laboratories. To overcome this barrier, we have built upon our previous work10 and developed an inexpensive, user-friendly, and easily customizable electrical stimulation device built from commercially available components and a circuit board (Figures 3A, B). The use of this custom stimulator allows for precise control and measurement of electrical stimulation at any point during tissue studies.
Figure 3. milliPillar custom electrical stimulation apparatus and validation.
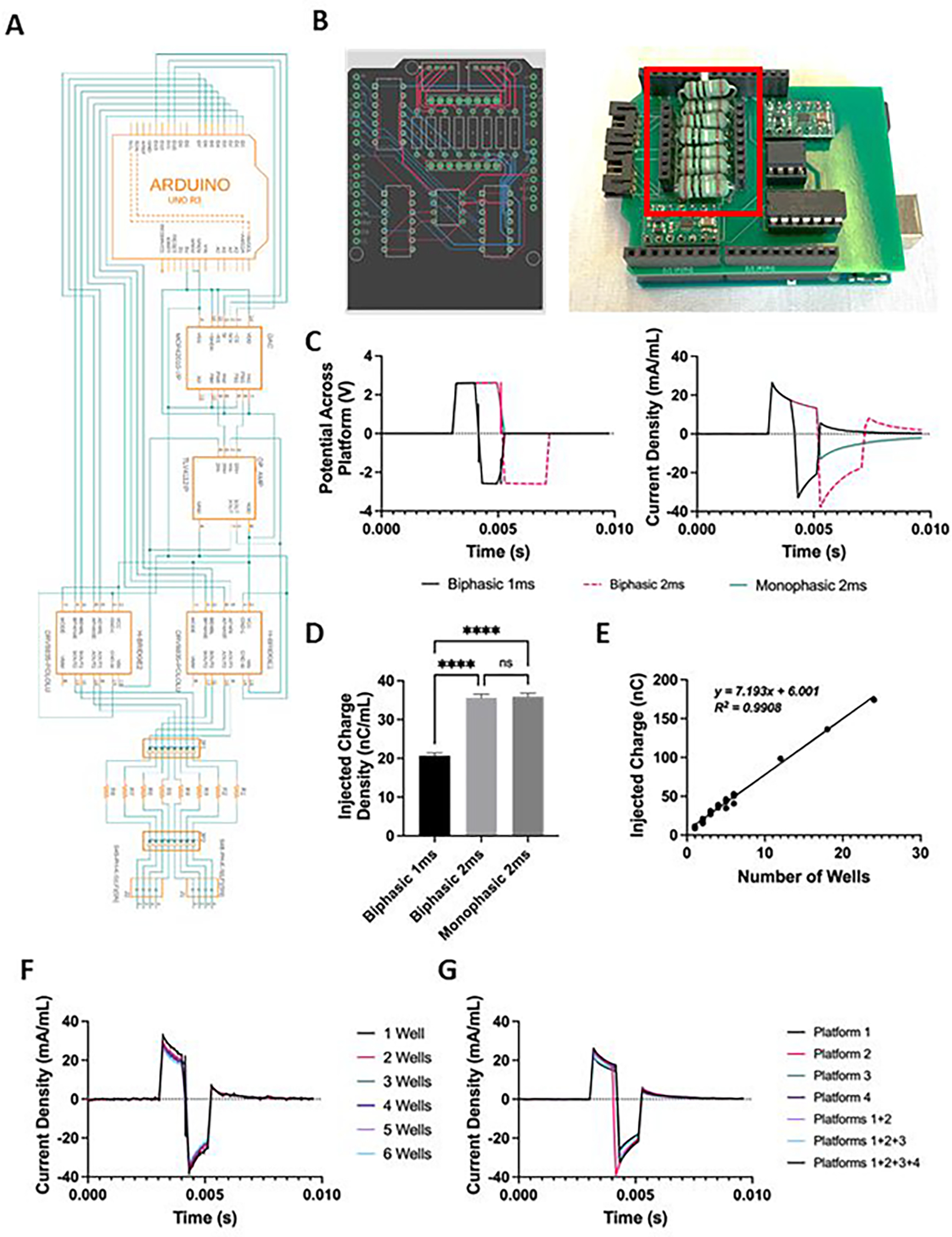
(A) Schematics of the custom electrical stimulator. (B) CAD rendering of the printed circuit board designed for the stimulator (left) and the assembled circuit connected to the Arduino development board (right). The connectors for measuring current across a series of test resistors are highlighted in red. (C) Representative measurements of voltage (left) and current density (right) for a single platform stimulated with 1 ms monophasic, 1 ms biphasic, and 2 ms biphasic waveforms. (D) Quantification of the charge density injected by these three waveforms. (E) Injected charge as a function of the number of wells connected to the stimulator, demonstrating a linear relationship through at least 24 wells. (F-G) Injected current density as a function of time measured through differing numbers of wells (F) and platforms (G), demonstrating consistency across these conditions.
Customizable stimulation regimens.
Customizable stimulation regimens were used (i) to promote the maturation of tissues12,28, and (ii) for functional assessment to reduce variability between tissues with different spontaneous beating frequencies and measure tissue characteristics that require dynamic changes in beating frequencies, such as the force-frequency response and frequency-dependent acceleration of relaxation (FDAR). The built-in electrodes enabled real-time non-destructive functional characterization of the tissues at any point during culture without the need to transfer the tissues to external stimulation devices12.
The electrical stimulator has the capability to generate either biphasic or monophasic waveforms. In addition, we have incorporated a current and voltage measurement system to provide control over the stimulation regimen for the duration of tissue culture (Figure 3B). Routinely, the voltage across and current running through a single tissue were measured during stimulation at 2.5V (generating a 5 V/cm electric field) and 2Hz with varying 2 ms waveforms (Figure 3C). As expected, a monophasic wave maintained 2.5 V across the platform for 2 ms before returning to baseline, while the current density peaked at 30 mA/mL and then decreased exponentially before returning to baseline at 2 ms, in accordance with the theoretical charging of a double bi-layer capacitor. A charge-matched biphasic wave maintained 2.5 V across the platform for 2 ms, followed by −2.5 V for 2 ms, before returning to baseline. The current density followed a similar pattern during monophasic stimulation, but with the current injected in both directions. Charge injection was calculated by integrating the injected current over time. A biphasic wave of 1 ms instead of 2 ms enhanced cardiac tissue function, potentially by reducing cell death mediated by reactive oxygen species (ROS)29. This improved tissue function could be due to the decrease in charge injected by a 1 ms biphasic wave.
The milliPillar stimulator was therefore programmed to electrically stimulate tissues with a 1 ms biphasic wave in long term cultures (Figure 3D). Further studies are warranted to determine the minimum amount of charge necessary to stimulate cardiac tissues for maximal functional enhancement and cardiomyocyte health.
Injected charge is controlled and varies with the number of wells in use.
We have extensively validated the electrical stimulator to ensure that it can provide sufficient charge injection for the milliPillar platform as determined in previous studies27,29. The current across the platform depends on how many wells are in use. Also, there is an additional current running through the system, due to the decreased resistance from additional electrolyte to carbon surface area, at an average of 7.19 ± 0.29 nC per well per pulse (corresponding to 17.9 ± 0.7 nC/mL) according to the linear regression (r2 =0.99) of the injected charge vs the number of wells in use (Figure 3E). In line with these data, the current normalized by the volume of electrolyte was not dependent on the number of wells in use (Figure 3F). Each channel on the stimulator can deliver a controlled charge for 4 platforms (24 wells), maintaining an electric field of 5V/cm (Figure 3G).
Previous studies have shown that the injection of charge into the media containing multiple tissues makes it difficult to control the amount of charge that each tissue is exposed to12,25. As electrical stimulation generates oxidative stress within the cells, and at extremes can create Faradaic currents, it is important to control the injected charge in a manner that preserves the tissue health. The milliPillar platform allows for control over the amount of charge injected into each tissue well, to reproducibly expose each cardiac tissue to the same amount of electrical stimulation, and to detect more nuanced differences between tissues from different experimental groups.
Cardiac tissue assembly using a range of hydrogels, iPS lines and stromal cell sources
Engineered cardiac tissues were fabricated by mixing iPS-derived cardiomyocytes and cardiac fibroblasts into a hydrogel that was then cast into the platform wells. The area in which the cell-laden hydrogel was cast was designed to minimize the tissue size, requiring only 15 μL of hydrogel containing 550,000 cells. During the first 7 days, the cells extensively remodeled the hydrogel and formed compact tissues attached to the pillar heads. The milliPillar platform gives the user the option to use electrical stimulation to promote maturation of cardiac tissues. To evaluate the effect of sustained biphasic electrical stimulation, tissues were stimulated for 21 days with an intensity training regimen (“ramp stimulation”) as previously reported12. Further tissue remodeling and compaction were observed over the time of stimulation (Figure 4A), with a significant decrease in tissue width (0.67 mm ± 0.02 vs 0.52 mm ± 0.01, p <0.0001, Figure 4B).
Figure 4. Generation of cardiac tissues.
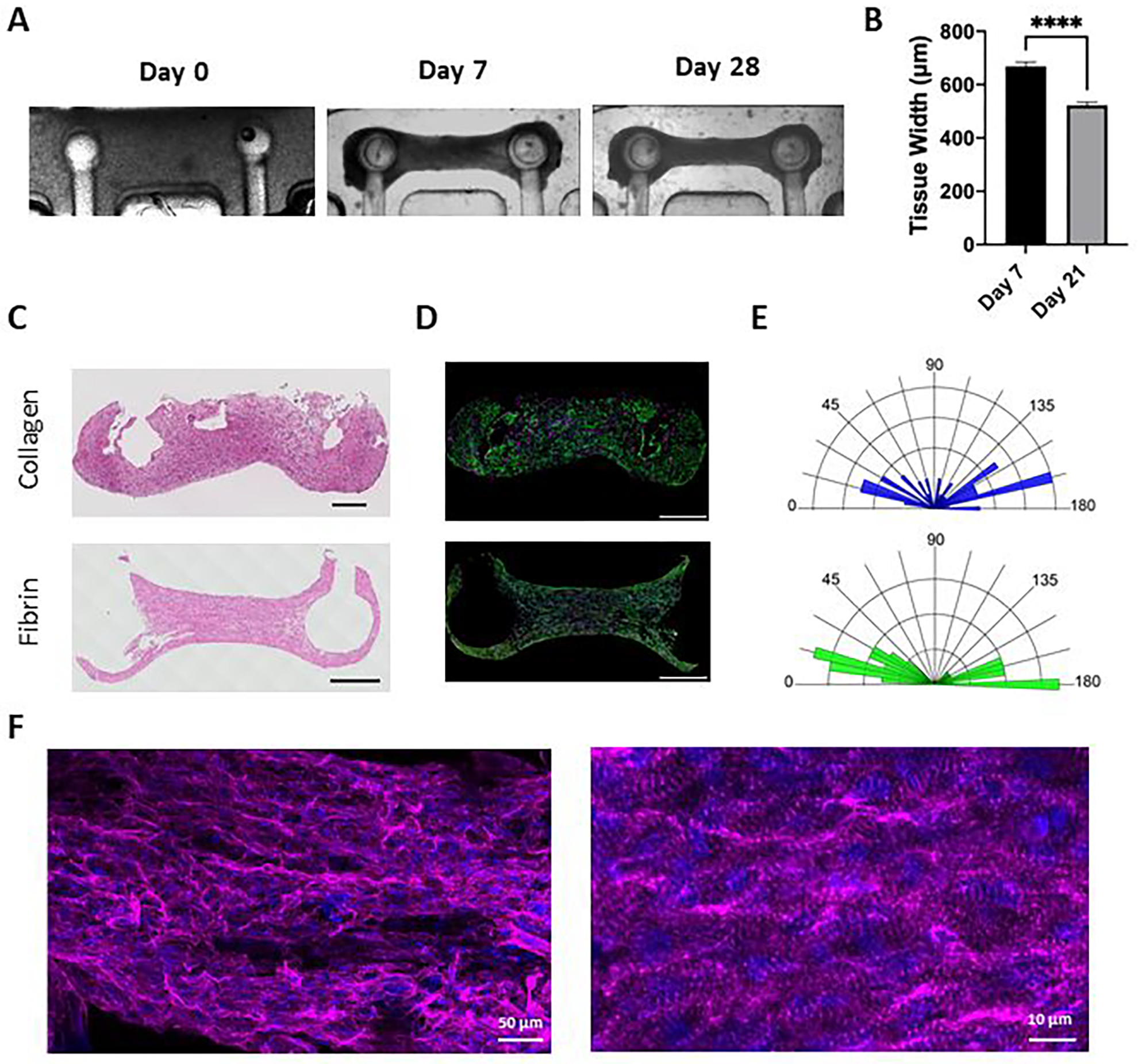
(A) Representative brightfield images of millPillar tissues immediately after seeding into the platform (Day 0), before electrical stimulation (day 7) and at the end of 21 days of ramp electrical stimulation (day 28). (B) Impact of culture period and electrical stimulation on milliPillar tissue width before and after electrical stimulation. Representative images of (C) Hematoxylin and Eosin (H&E) staining and (D) immunostaining for troponin-T (green), vimentin (magenta) and nuclei (blue) staining of milliPillar tissues formed using collagen and fibrin hydrogels and electrically stimulated for 21 days (scale bar, 500 μm). (E) Quantification of iPS-cardiomyocytes alignment in collagen and fibrin tissues. (F) Representative confocal images of fibrin tissues electrically stimulated with ramp stimulation regimen and immunostained for sarcomeric α-actinin (magenta), and nuclei (blue).
It has previously been demonstrated that different hydrogel materials such as collagen, fibrin, and decellularized ECM can support functional cardiac tissue assembly12,30–34. Therefore, we sought to evaluate the versatility of the milliPillar platform by generating cardiac tissues using both collagen and fibrin hydrogels. In both collagen and fibrin, the cells remodeled the hydrogel and formed compact tissues with aligned cardiomyocytes, as observed by histological staining and immunofluorescence imaging (Figure 4C–D). Quantification of cellular alignment demonstrated that the fibrin hydrogel resulted in more cellular alignment than the collagen hydrogels (Figure 4E). High magnification imaging of milliPillar tissues fabricated with fibrin demonstrated the formation of pronounced sarcomeric α-actinin striations and cell alignment indicative of an improved contractile apparatus (Figure 4F).
The choices of iPS-cardiomyocyte lines, types of stromal cells, and ratios of cardiomyocytes to stromal cells differ between protocols. Our goal here was to develop a technology that is customizable and lends the researcher the high flexibility. We thus investigated the ability of the milliPillar platform to support tissue formation using various cell types. Functional tissue assembly was achieved using different iPS-cardiomyocyte lines (BS2, WTC11, and WTC11-GCaMP6f) and different types of stromal cells (cardiac fibroblasts, iPS-cardiac fibroblasts, and dermal fibroblasts) (Supplementary Videos S1, S2 and S3). A previously optimized concentration of 75% iPSC-cardiomyocytes to 25% cardiac fibroblasts was used, however, the ratios of iPSC-cardiomyocytes to other stromal cells may require further optimization.
To demonstrate the potential of the platform for use in long-term studies, we cultured milliPillar tissues for as long as 100 days. Immunofluorescence images revealed pronounced α-actinin striations in these long-term tissue cultures (Supplementary Figure S4A). We also explored the ventricular phenotype of the milliPillar tissues. As expected, MLC2v staining indicated that our culture conditions facilitated a ventricular cardiomyocyte phenotype (Supplementary Figure S4B). While further validation is required, atrial tissues could also be generated by incorporating cells from atrial cardiomyocyte differentiation protocols8,35.
Custom software and hardware for the functional analysis of cardiac tissues
Monitoring cardiac tissue functionality, non-invasively and over extended periods of time, is important to evaluate tissue response to pharmaceutical compounds and environmental signals, and to understand their development. Optical imaging provides an ideal solution since it is non-destructive and could be translated into absolute values of tissue outcomes. Automated data acquisition and streamlined analysis are also important for improving throughput and reducing user error.
To address these needs, we have developed a custom-built stimulator and software to control and synchronize video acquisition, electrical stimulation, and stage positioning using a standard laboratory microscope (Figure 5A). This set up facilitates the assessment of up to 24 tissues simultaneously. The milliPillar platform is complemented by a suite of customized software, based on established methodologies to analyze brightfield videos for measurements of force generation and tissue movement, and calcium imaging for measurements of calcium handling13,25,36. A summary of the metrics obtained using the custom milliPillar software is provided in Supplementary Figure S5.
Figure 5. Custom software and hardware for functional analysis of cardiac tissues.
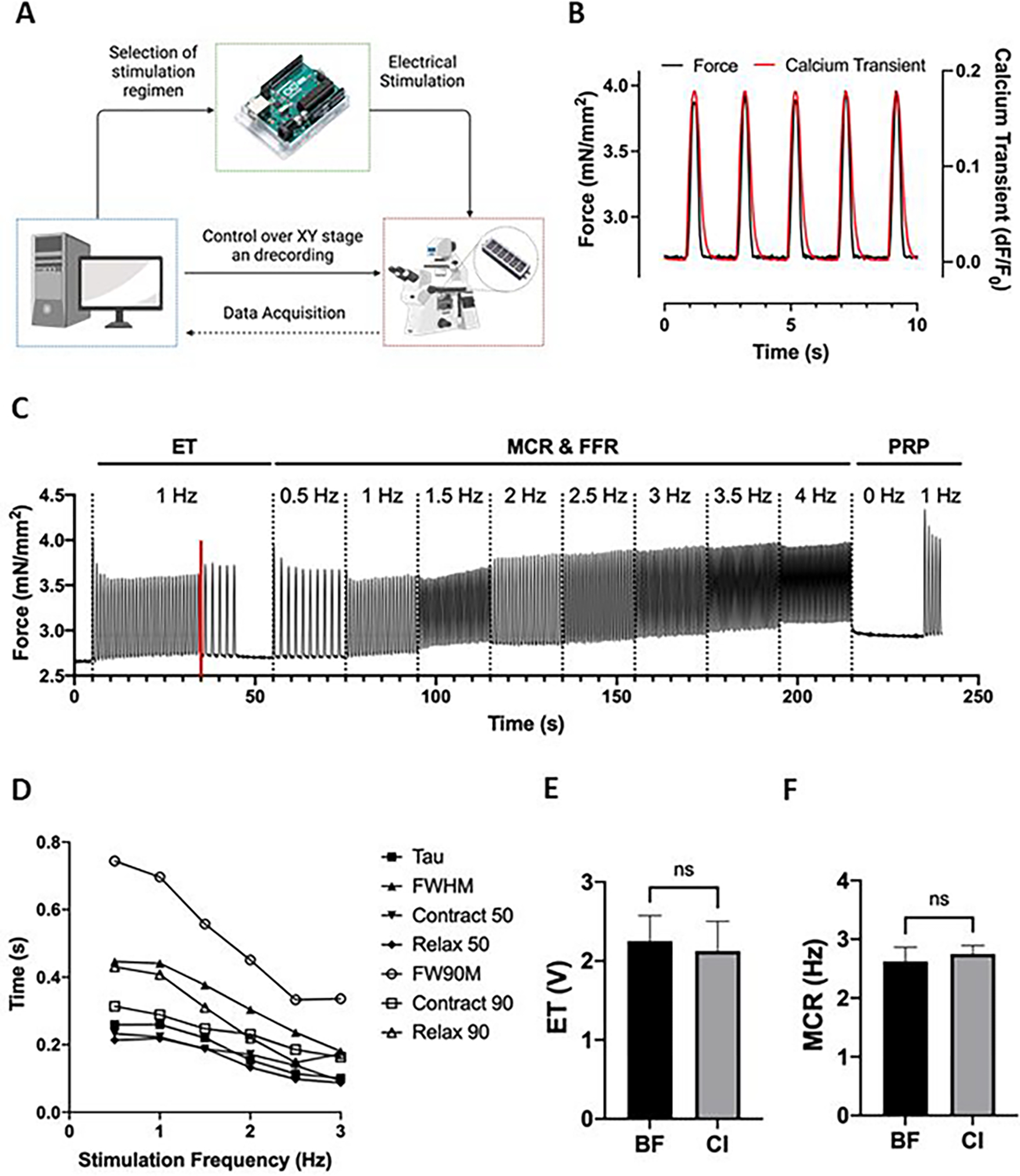
(A) Overview of the system that enables simultaneous and automated tissue stimulation, microscopy video recordings, and microscope stage movement between multiple tissues in the XY axes. (B) Representative traces of calcium signal and force generation of milliPillar tissues stimulated at 0.5 Hz of which absolute force and time measurements are extracted. (C) Representative trace of milliPillar tissues in response to the combined custom stimulation regimen that enables the measurement of ET, MCR, maximum beating frequency, force generation, contraction and relaxation velocity, and the characterization of FFR. (D) Representative measurements of calcium handling parameters (Tau, FWHM, Contract 50, Relax 50, FW90M, Contract 90, and Relax 90) across varying stimulation frequencies. (E) ET and (F) MCR measurements extracted from brightfield (BF) and calcium imaging (CI) videos demonstrate similar values.
Calcium transients directly affect tissue contraction, relaxation and arrhythmogenicity. Therefore, following dynamic changes in calcium handling is key for understanding tissue patho/physiology and responses to different conditions. Here, calcium transients were measured using cardiomyocytes differentiated from the iPS-GCaMP6f cell line that is genetically encoded with a calcium indicator to allow real-time readouts37. However, this cell line is not required, and a calcium dye could be used as well. Measurements of the fluorescent signals of the calcium indicator over time allow for the direct determination of calcium handling parameters.
By directly measuring the deflection of pillars calibrated using known bending coefficients, the milliPillar system enables calculation of absolute values of force generation, active force, and passive tension, rather than the relative approximations generated by some other systems12,38. This method facilitates assessment of contractile force normalized to the cross-section of the tissue area (mN/mm2), which is becoming a requirement for various consortia and regulatory agencies.
Representative calcium and force transients recorded simultaneously at 0.5 Hz are shown in Figure 5B. Studies differ in the types of parameters extracted from calcium transients (e.g., measurement of the contraction side of the calcium transient between 0–90%, 10–90%, or 0–80%)36. We thus provide here the custom code so that the same metrics can be compared across groups.
A stimulation regimen was desined to allow extraction of excitation threshold (ET, minimum voltage required for tissues to capture at 1 Hz while stimulated at 1 Hz) maximum capture rate (MCR, maximum frequency at which the tissues capture when stimulated at 5 V), and post-rest potentiation (PRP, the maximum force generated by tissues upon stimulation after a period of exertion followed by rest) in a single automated recording from which the pillar movement, and thus the force generation, are extracted (Figure 5C). The details of this regimen and representative tissue recordings are provided in Supplementary Figure S6A. A representative video demonstrating accurate tracking of the pillar heads is provided in (Supplementary Video S4. Importantly, the analysis program can detect subtle changes in beating, eliminating the error and bias that may arise from attempting to make such determinations by eye.
Of note, the force generation by tissues within the milliPillar platform is not necessarily the at the maximum level achievable by the tissues, due to the Frank-Starling relationship between cardiomyocyte length and force generation39. The tissues were not stretched to their optimal relaxed length to maximize force generation, since the passive tension and stretch, which correlate to afterload in vivo, cannot be adjusted by the user. This is a trade-off to the ease of longitudinal real-time measurements. At the study endpoint, tissues can be removed from the platform and evaluated from standard force recordings in an organ bath.
Calcium dynamics can also be assessed at different stimulation frequencies (Figure 5D). Notably, calcium transient durations decreased with an increase in stimulation frequency. The versatility of the system to allow stimulation in a wide range of frequencies is important given the frequency dependence of cell phenotypes and drug responses. Such features may only become apparent during stimulation at frequencies that recapitulate bradycardia or tachycardia, emphasizing the importance of recording at dynamic frequencies.
The analysis suite can also be set to calculate the ET and MCR with brightfield or fluorescent imaging. We found that brightfield and fluorescent imaging showed no significant difference in assessing ET and MCR (Figure 5E). With our custom code, the ET and MCR are easier to analyze using calcium transients, due to the high signal to noise and ease of computation. It is important to note that blue light is phototoxic, and overexposure may lead to confounding effects. Since calcium transient features can only be calculated with fluorescent imaging, care must be taken to not overexpose tissues to blue light.
Cardiac tissue heterogeneity can be introduced at any of the steps; thus, baseline imaging is used to quality control the individual tissues to reduce variability. Depending on the question, any metric can be chosen for quality control (e.g. if calcium handling is of more interest the full width half max, FWHM, could be chosen; if force generation is of more interest PRP could be chosen). An example is the use of the full width 90 max (FW90M) for quality control, to identify and exclude the top 10% and bottom 10% of cardiac tissues, and thereby secure normal distribution of tissue functionality (Supplementary Figure S5C).
Electromechanical stimulation for enhanced cardiac tissue function
milliPillar provides the capability to enhance tissue function using a customized electrical stimulator. The stimulator can be used for any stimulation regimen; however, the ramp stimulation has been shown to enhance tissue functionality and maturation12, therefore we used this regimen and tracked the function of the milliPillar tissues over time, analyzing the tissues before and after 21 days of ramp stimulation. As the cells within tissues become electrically coupled, aligned, and beat in unison, they can generate more force perpendicular to the pillar axis.
As expected, milliPillar tissues showed increased force generation post-stimulation, with both 1 Hz force generation (3.22 ± 0.17 mN/mm2 vs 2.07 ± 0.25 mN/mm2, p<0.001, Supplementary Videos S5 and S6) and PRP (4.05 ± 0.14 mN/mm2 vs 2.11 ± 0.25 mN/mm2, p<0.001) (Figure 6A). Additionally, as the electrical stimulation is maturing the cardiomyocytes, they approach higher contraction and relaxation velocities, as observed in vivo. Accordingly, the maximum values of contraction velocity (622 ± 94 μm/s vs 280 ± 4 μm/s, p<0.05) and relaxation velocity (911 ± 162 μm/s vs 263 ± 4 μm/s, p<0.01) also increased with electrical stimulation, demonstrating more mature and physiologically functional tissues (Figure 6B).
Figure 6. Engineered cardiac tissues fabricated and cultured within the milliPillar platform exhibit enhanced tissue function after 21 days of electrical ramp stimulation.
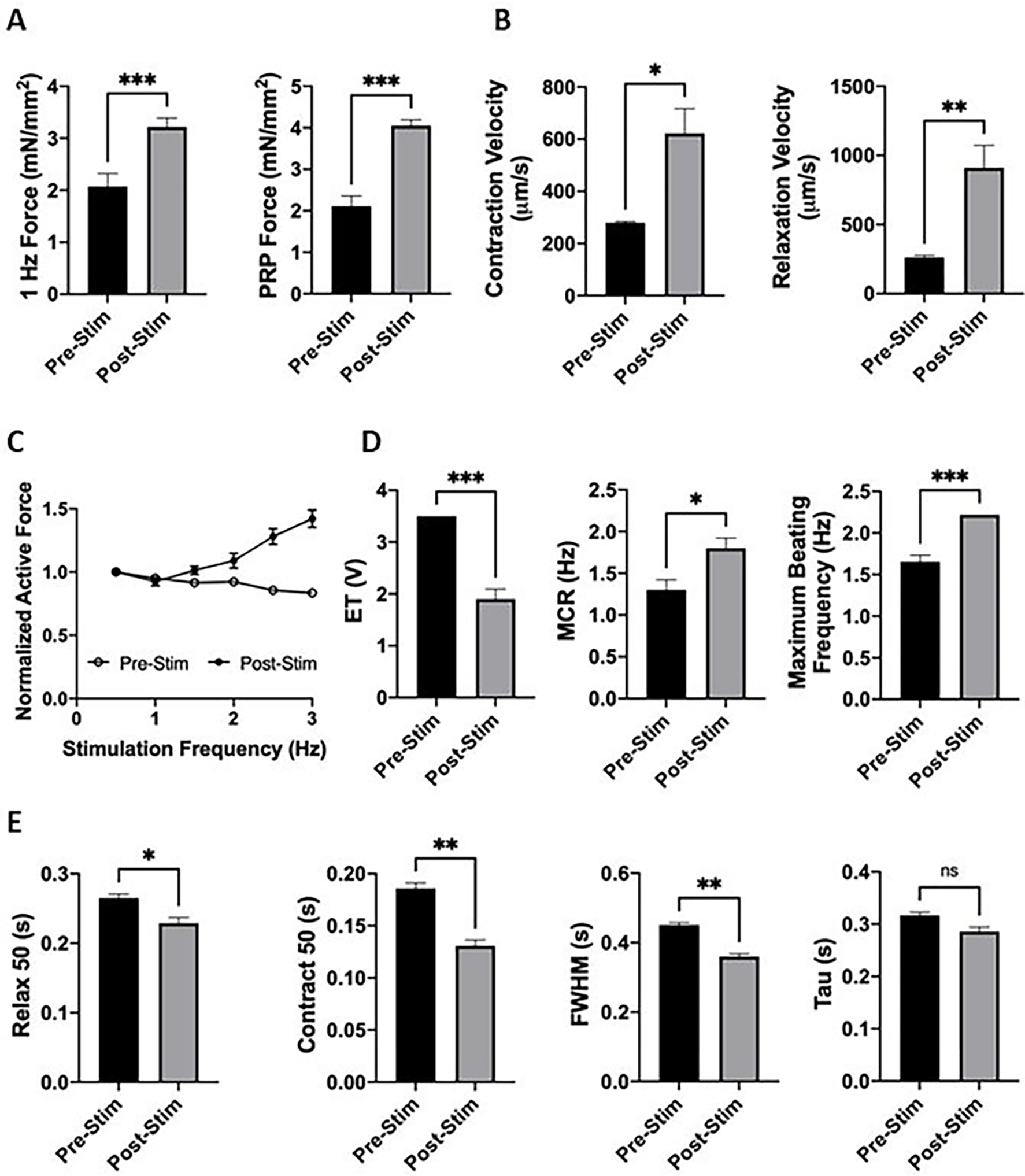
(A) Ramp stimulation led to enhanced force generation measured at 1 Hz stimulation and PRP. (B) Contraction and relaxation velocities increased after electrical stimulation. (C) Positive FFR was observed in tissues after electrical stimulation. (D) Tissue excitability was enhanced after electrical stimulation. (E) Calcium transient durations decreased after electrical stimulation.
Dynamic frequency measurements allowed us to assess the force frequency response of cardiac tissues. As the stimulation frequency increases, the maximum force generation of each contraction also increases, demonstrating a positive FFR, a feature of mature cardiac tissue8,12,40,41. Our data demonstrate that milliPillar tissues before stimulation did not exhibit a positive FFR, and that by applying ramp stimulation we were able achieve a positive FFR, indicating improvement of their functional maturation (Figure 6C).
To evaluate the effect of electrical stimulation on tissue excitability and sensitivity, we measured the ET and MCR. As expected, milliPillar tissues exhibited a markedly lower ET after stimulation (1.9 ± 0.19 V vs 3.5 V, p <0.001), higher MCR (1.8 ± 0.12Hz vs 1.3 ± 0.12 Hz, p<0.05) and higher maximum beating frequency (2.22 Hz vs 1.67 ± 0.08 Hz, p<0.001) (Figure 6D).
Mature ventricular cardiomyocytes exhibit short calcium transients as their sarcoplasmic reticulum is organized and saturated with ryanodine receptors and SERCA2A calcium channels. Mature sarcoplasmic reticulum is able to more rapidly release and uptake calcium during every beat42. In disease or in response to drugs, some or all of the calcium handling metrics (contraction, duration, relaxation) may be altered. Calcium handling is enhanced by electrical stimulation, as shown by decreases in contraction, duration, and relaxation: Contract 50 (0.13 ± 0.005 sec vs 0.186 ± 0.005 sec, p<0.01), Relax 50 (0.229 ± 0.008 sec vs 0.265 ± 0.006 sec, p<0.05), FWHM (0.36 ± 0.008 sec vs 0.45 ± 0.007 sec, p<0.01) (Figure 6E). The decrease in the time constant Tau (0.29 ± 0.009 sec vs 0.32 ± 0.006 sec) was not significant. Longer culture periods under electrical stimulation may lead to a further reduction in Tau.
Taken together, we have demonstrated that electromechanical stimulation in the milliPillar platform led to improved force generation, tissue excitability and calcium handling. We have validated that the mechanical load applied by the milliPillar platform, electrical stimulation by the milliPillar stimulator, and real-time measurements at any point during the protocol allowed maturation of cardiac tissues and assess their function over time.
CONCLUSION
We describe the milliPillar platform, a robust and versatile technology that has been developed and validated to provide a streamlined pipeline for reproduction and utilization of engineered cardiac tissues for in vitro research. We have validated milliPillar’s ability to support functional cardiac tissue assembly using multiple cell lines, cell types and hydrogel materials, in long term cultures (up to 100 days). We characterized the amount of charge injected into each tissue for controlled stimulation leading to advanced maturation and function, and developed the hardware and software necessary for long term electrical stimulation and the functional real-time analysis of tissue function.
By integrating multiple steps required for successful cardiac tissue engineering studies, we believe that milliPillar can help overcome several of the challenges cardiac tissue models currently face. Our goal is to enhance reproducibility and data comparisons across different studies, and to facilitate adoption of cardiac tissue engineering for advanced basic and applied research.
MATERIALS AND METHODS
milliPillar platform design and fabrication
Platforms and molds were designed in SolidWorks (Dassault Systemes, Vélizy-Villacoublay, France). A computer numerical control (CNC) milling machine (Haas OM-2A 2015) was used to make a 3-part mold out of Delran to generate one platform (Supplementary Figure S1 and S2, design files provided in Supplementary File S1). The molds were deburred and subsequently cast with PDMS (SLYGARD 184 Silicone Elastomer Kit, Dow Chemical Company, cat. no. 2065622) three times to clear debris before the initial use. Before casting, the pillar spaces in the molds were cleaned with pressurized air to ensure clearing of the pilar space in the mold. Metal tools should not be used to avoid scratching.
To fabricate platforms, parts #1 and #2 were assembled and carbon rods (Ohio Carbon Blank, Squares & Plates, AR-14, Semi precision, Saw-Cut X=2.63 Y=0.0790 Z=0.0590) were placed into mold part #2. PDMS (10:1 ratio of base to curing agent) was mixed thoroughly, degassed, and cast into part #2. An additional degassing stage was performed for 45 minutes, or until no more bubbles were visible. Next, the top of part #2 was covered with part #3. The assembled molds were then clamped (McMaster, low profile C-clamps, cat. no. 1705A11) with the 1/4” × 4” hex standoff (McMaster, cat. no. 91780A060) in place, topped off with PDMS, and placed into a 65°C oven overnight for curing (Supplementary Figure S2, top panel).
Platforms were removed using a flat tool, by gently separating the sides of the platform from the mold until the platform slid out. PDMS was cut off the ends of the platform and the PDMS film on the rods within the wells was removed with forceps and a scalpel. A 1/32” hole was drilled into the ends of the rods. The platforms were sonicated with 1% Tween-20 in distilled water for 1 hr, rinsed thoroughly with distilled water and allowed to dry in a 65° C oven overnight. The slides were cut using a diamond tipped blade to 25 mm × 60 mm. Platforms were bonded to slides by 5 mBarr Oxygen Plasma treatment (Harrick Plasma, Plasma Cleaner Model PDC-001) for 30 sec. A platinum wire (Superpure Chemical, cat. no. 2805) was threaded through the drilled holes and wrapped around the carbon rods (Supplementary Figure S2, bottom panel).
Pillar Force-Displacement Analysis
The bending of the pillars was simulated with COMSOL Multiphysics (COMSOL, Inc., Stockholm, Sweden) to estimate the displacement of each portion of the pillars throughout a bending cycle. The force required to displace the pillars was determined using a microscale mechanical tester, Microtester MT-LT (CellScale, Ontario, Canada). A 0.4064mm diameter circular tungsten microbeam with platen (1mm × 1mm) was used to displace the pillar head. Before the test, the platform was fixed on the testing stage with clamps. The probe tip with platen was placed adjacent to the pillar head without contact and gradually moved towards the center of the platform at a velocity of 8.5 μm/s. The tip displaced the pillar head and applied the force perpendicular to the original pillar position. All platforms were fabricated according to the protocol mentioned previously. Four to six pillars were tested in each milliPillar platform and four batches of milliPillar platforms were included. The experimental data were fit into a linear equation, generating a force-displacement calibration curve with a coefficient that can be used to calculate active forces and passive tensions based on the position of the pillar head during the experiment.
Electrical stimulation
A custom electrical stimulator was designed to work within the Arduino software and hardware environment. Detailed schematics of the circuit, a full bill of materials, and computer aided design (CAD) files for the printed circuit board are provided in Supplementary File 2. Briefly, the circuit consists of an Arduino Uno Rev3 microcontroller development board (Arduino, cat. no. A000066), a digital potentiometer (Microchip Technology, cat. no. MCP42100-I/P), a power operational amplifier (Texas Instruments, cat. no. TLV4112IP), two dual channel H-bridge motor drivers (Pololu Robotics, cat. no. 2135), and a series of 1 ohm test resistors. The microcontroller sets the stimulation voltage by adjusting the resistance of the digital potentiometer, which is placed between +5V and ground in the circuit. The wiper from the digital potentiometer connects to the power op-amp in a unity gain configuration such that the output of the op-amp maintains the specified voltage but with the capability to supply a much greater current (~300 mA). This output powers the motor drivers and provides the current for stimulation. Each motor driver channel is controlled by two digital outputs from the microcontroller using the driver’s PHASE/ENABLE mode to generate biphasic pulses at ± the specified stimulation voltage supplied to the drivers.
The frequency, duration, and phase offset of these pulses are specified in the Arduino code and can be easily customized. Monophasic stimulation can also be selected instead of biphasic. Each of the four output channels can operate independently at a unique frequency, but all channels share a common output voltage (see readme file). Due to the incorporation of field-effect transistors within the motor drivers, there is a slight voltage drop across the motor drivers that varies in real-time with the output current. We recommend measuring the output voltage after connection to the platform and adjusting if necessary.
Cardiomyocyte differentiation
hiPSCs were obtained through material transfer agreements from B. Conklin, Gladstone Institutes (WTC11 and WTC11-GCaMP6f lines) and from the Stem Cell Core at Columbia University (BS2 line).
Cardiomyocytes were differentiated from all three iPSC lines as previously described43. On Day 10, RPMI-no glucose (Life Technologies, cat. no. 11879020) supplemented with B27 (Thermo Fisher Scientific, cat. no. 17504044) and 213 μg/mL ascorbic acid (Sigma-Aldrich, cat. no. A445), was used to purify the iPSC-CMs population and eliminate potential contaminating mesodermal and endodermal populations. Starvation medium was replaced on day 13 with RPMI-B27 medium supplemented with 213 μg/ml ascorbic acid until day 16.
On day 17 cells were pretreated with rock-inhibitor (y-27632 dihydrochloride, 5 μM) for 4 hours before dissociation. Cells were dissociated by enzyme digestion with collagenase type II (95 U/mL; Worthington, cat. no. LS004176) and pancreatin (0.6 mg/mL; Sigma–Aldrich, cat. no. P7545) in dissociation buffer (Glucose (5.5 mM), CaCl2·2H20 (1.8 mM), KCl (5.36 mM), MgSO4·7H20 (0.81 mM), NaCl (0.1 M), NaHCO3 (0.44 mM), NaH2PO4 (0.9 mM)) on a shaker in a 37°C incubator. After 10 minutes, a 5 mL pipette (or cell scraper) can be used to gently triturate the cells and lift them off the plate. Cells were placed back in the incubator for 10 minutes until dissociated into single cells. With a 5mL pipette, the cardiac suspension was triturated and added to a 50mL conical tube. Cells were gently triturated again to form a homogenous suspension. One volume of RPMI-B27 media was added to the tube and the cells were spun down at 1200 RPM for 5 minutes.
Cell purity of at least 85% is required to ensure reproducibility. Flow cytometry for cTnT+ (BD BioSciences cat. no 565744) was performed prior to cell use for tissue fabrication. Cells can be frozen in freezing media (CryoStor CS10, Stem Cell Technologies, cat. no. 07955) at a concentration of 5–10 million/mL or used right away. If cells were thawed, medium was added dropwise for 60–90 seconds, filled to an appropriate volume, and then spun at room temperature at 100 × g. Cells were kept on ice for tissue making.
Fibroblasts
Primary Human cardiac fibroblasts (NHCF-V; Lonza, cat. no. CC-2904) and dermal fibroblasts (NHDFs; Lonza, cat. no. CC-2509) were cultured according to the manufacturer’s recommendation. iPS-CFs were differentiated according to previously described protocol22.
Engineering and culture of cardiac tissues
Either thawed or fresh iPS-cardiomyocytes (75%) and fibroblasts (25%) were resuspended in RPMI-B27 medium to form a cell mixture. Precise cardiomyocyte purity was determined by flow cytometry for cTnT (BD BioSciences cat. no. 565744) to ensure ratio accuracy.
When fibrin was used to make tissues, the cell mixture was resuspended in fibrinogen by mixing 33 mg/mL human fibrinogen (Sigma-Aldrich, cat. no. F3879) with the cell solution to a final fibrinogen concentration of 5 mg/mL. The volume of each individual tissue was 15 μL and it contained 550,000 cells (370,000 cells/μL). When calculating the amount of fibrinogen to add, the volume of the cell pellet and the volume of the thrombin solution is taken into account. 3 μL of thrombin (2U/mL) were added to each well. Following, 12 μL of fibrinogen-cells solution was dispensed and quickly spread over the entire well with a pipet tip. Tissues were placed in a 37°C incubator for 15–20 minutes to allow gelation in the well. If tissues in more than one platform were formed, fibrin-cell suspensions were kept homogenous by frequent mixing, without introducing bubbles. Pipette tips were replaced after seeding each tissue to prevent residual thrombin crosslinking with the fibrin-cell suspension.
When collagen gel was used, it was prepared according to the manufacturer’s protocol (Advanced Biomatrix, Cat. no. 5279) and mixed with cell mixture for a final concentration of 4 mg/mL Collagen. 15 μL of the cell-gel suspension were added to each well.
400 uL of RPMI-B27 with 213 μg/mL ascorbic acid, 10uM Rock inhibitor and 5 mg/mL 6-aminocaproic acid (Sigma-Aldrich, cat. no. A7824, only necessary for fibrinogen tissues to prevent rapid degradation) was added to the wells. After 1 hour, a 26- gauge needle was used to detach tissues from the walls of the well. 24 hrs later, tissues were detached again and the medium was changed to RPMI-B27 with 213 μg/mL ascorbic acid and 5 mg/mL 6-aminocaproic acid. The medium was changed every other day for 6 days.
On Day 7, 6-aminocaproic acid was removed from the medium and electrical stimulation was started with the stimulator using a 5V/cm biphasic pulse (2ms pulse length, 1 ms per phase) at 2Hz. Tissues were either paced at 2Hz continuously or following the previously reported ramp stimulation regimen12. During the ramp stimulation regimen, the frequency started at 2 Hz and was increased everyday by ⅓ Hz until reaching 6Hz. 6Hz stimulation was maintained for three days, after which stimulation frequency was reduced to 2Hz and maintained until endpoint analysis at day 21. This code is made available in Supplementary File 3. Stimulation voltage and pulse duration were not modified during the stimulation regimen. Medium was changed every other day.
Histology and immunostaining
Whole mount engineered cardiac tissues were fixed and permeabilized in 100% ice cold methanol for 10 min, washed three times in PBSX1, and then blocked for 1 hr at room temperature in PBSX1 with 2% fetal bovine serum (FBS). For tissue sections, tissues were fixed in 4% paraformaldehyde (PFA) for 30 min and washed three times in PBSX1. Whole tissues were placed at the bottom of 15mm square disposable histology base molds and encapsulated in 1 mL molten Histogel (Fisher Scientific, cat. no. 22–110-678). Histogel blocks were cooled according to the manufacturer’s protocol and then fixed with 4% PFA for 30 minutes followed by three washes with PBSX1.
Fixed Histogel blocks were paraffin-embedded and cut into 5 μm -thick sections. Paraffin sections underwent heat mediated antigen retrieval in a pH 6 sodium citrate buffer, permeabilized with 0.01% triton in PBSX1 and then placed into the blocking buffer using 10% FBS. After blocking, the tissues were incubated with primary mouse anti–α-sarcomeric actinin antibody (1:750, Sigma-Aldrich, cat. no. A7811), anti-cardiac troponin T (cTnT, 1:100; Thermo Fisher Scientific, cat. no. MS-295-P1), and vimentin (Abcam, cat. no. ab24525) washed three times, and incubated for 1 hr with secondary antibodies (Millipore Sigma, cat. no. AP194C; Thermo Fischer, cat. no. A-21206; Thermo Fischer, cat. no. A31571). For nuclei detection, the tissues were washed and subsequently incubated with NucBlue (Thermo Fisher, cat. no. R37606). Samples were visualized using a scanning laser confocal microscope (Nikon Eclipse Ti) or a DMi8 microscope (Leica Microsystems)
Calcium imaging
To make tissues with an endogenous marker of cytosolic calcium, we used WTC11-GCaMP6f iPSCs that contain a constitutively expressed GCaMP6f calcium-responsive fluorescent protein inserted into a single allele of the AAVS1 safe harbor locus44. The incorporation of these GCaMP6f cells allows real-time visualization of calcium transients without the need for additional dyes. Tissues were imaged in a live-cell chamber (STX Temp & CO2 Stage Top Incubator, Tokai Hit, Fujinomiya, Japan) using a sCMOS camera (Zyla 4.2, Andor Technology) connected to an inverted fluorescence microscope with a standard GFP filter set (Olympus IX-81). To assess calcium transients in tissues made with non-GCaMP cell lines, cardiac tissues can be incubated with 10 μM fluo-4 AM (Invitrogen, cat. no. F14201) and 0.1% Pluronic F-127 (Sigma-Aldrich, cat. no. P2443) for 45 min at 37 °C.
Tissues were stimulated once with the analysis stimulation regimen to get acclimated for measurement. Tissues were then electrically stimulated, and videos were acquired at 20 frames per second (fps) for 4600 frames (the first 4600 frames described in Supplementary Figure S6A) to measure ET/MCR or 300 frames (stimulated at 1 Hz) to measure calcium flux. 1Hz stimulation calcium parameters may be pulled from the ET/MCR stimulation code, but parameters extracted from different stimulation regiments are not compared (i.e. a tissue stimulated for 4600 frames vs 300 frames will behave differently enough to confound results).
Brightfield imaging
Cardiac tissues were stimulated once with the ET, MCR-FFR custom program to acclimate all tissues for measurement. Videos were then acquired at 20 fps for 4800 frames using a custom program to stimulate cardiac tissues from 0.5 Hz to 4 Hz. The stimulation regimen begins by recording the spontaneous beating activity, followed by 1Hz stimulation at 5 V to measure the ET. The stimulation voltage drops by 0.5 V every 5 seconds so that the analysis program can determine ET as the voltage at which the tissue stops responding to stimulation. For the MCR and FFR, the voltage was fixed at 5V and the stimulation frequency was increased in 0.5 Hz increments every 20 seconds. The program determines the MCR as the frequency at which the tissue stops contracting with every stimulus. To measure PRP, the stimulation was paused for 20 seconds after the MCR/FFR frequency ramp (ending at maximum frequency of 4 Hz) and then resumed at 1 Hz. The PRP is determined as the force generated by the first beat upon the resumption of stimulation.
Calcium Signals
Calcium signals were analyzed from calcium imaging videos recorded at 20 fps. A custom Python script was developed to average the pixel intensities for each frame. This transient was then corrected for fluorescent decay. The SDRR, Tau, FWHM, FW90M, Contract 90, Contract 50, Relax 50, and the Relax 90 were calculated for every transient (Supplementary Figure S5B). An average of every transient over 15 seconds was calculated and exported into a table. When calculating ET and MCR, the custom program provides traces for each stimulation frequency and determines when tissues begin to beat out of sync from stimulation. Manual inspection of each trace is recommended due to the sensitivity of the program to cardiac tissue abnormalities (spiral waves, etc.).
Force Generation
Force generation was analyzed from brightfield videos. A custom Python script was developed to track the motion of the pillar heads and to calculate the force by multiplying the displacement of the pillars with the coefficient determined from the force-displacement calibration curve generated for the pillars. This script uses the correlation tracker class from the dlib C++ library (https://github.com/davisking/dlib)45 to determine the location of the pillar heads in each frame based on initial bounding boxes placed around the pillar heads in the first frame by the user. The script uses the location of the pillar heads to determine the total deflection of the pillar from their equilibrium position without any force applied. The dlib correlation tracker is based on a previously published object-tracking algorithm that uses a cosine correlation filter applied to a histogram of ordered gradients (HOG) feature descriptor for each frame in the recording46.
Statistical Analysis
Statistical analyses were performed using GraphPad prism. Single comparisons of data were assumed to follow a normal distribution and assessed using a one or two-tailed paired Student’s t-test to determine statistical significance. A p-value of <0.05 was considered statistically significant. *P<0.05, **p<0.01, *** p<0.001, ****p<0.0001.
Supplementary Material
ACKNOWLEDGMENT
The authors gratefully acknowledge funding support of NIH (grants EB025765, EB027062, HL076485 to GV-N; T32GM00736 support to TRN) and NSF (grant NSF16478 to GV-N).
DATA AND MATERIAL AVAILABILITY
The most up to date version of our custom software can be found on our GITHUB: https://github.com/GVNLab. CAD files can be found in the supplementary material of this paper and on our Tissue Engineering Resource Center website (TERC) (www.nextgenterc.com/).
REFERENCES
- (1).Ogle BM; Bursac N; Domian I; Huang NF; Menasché P; Murry CE; Pruitt B; Radisic M; Wu JC; Wu SM; Zhang J; Zimmermann W-H; Vunjak-Novakovic G Distilling Complexity to Advance Cardiac Tissue Engineering. Sci. Transl. Med. 2016, 8, 342ps13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Karbassi E; Fenix A; Marchiano S; Muraoka N; Nakamura K; Yang X; Murry CE Cardiomyocyte Maturation: Advances in Knowledge and Implications for Regenerative Medicine. Nat. Rev. Cardiol. 2020, 17, 341–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Giacomelli E; Meraviglia V; Campostrini G; Cochrane A; Cao X; van Helden RWJ; Krotenberg Garcia A; Mircea M; Kostidis S; Davis RP; van Meer BJ; Jost CR; Koster AJ; Mei H; Míguez DG; Mulder AA; Ledesma-Terrón M; Pompilio G; Sala L; Salvatori DCF; Slieker RC; Sommariva E; de Vries AAF; Giera M; Semrau S; Tertoolen LGJ; Orlova VV; Bellin M; Mummery CL Human-IPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-Cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 2020, 2, 862–879.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Campostrini G; Windt LM; van Meer BJ; Bellin M; Mummery CL Cardiac Tissues From Stem Cells. Circ. Res. 2021, 128, 775–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Fink C; Ergün S; Kralisch D; Remmers U; Weil J; Eschenhagen T Chronic Stretch of Engineered Heart Tissue Induces Hypertrophy and Functional Improvement. FASEB J. 2000, 14, 669–679. [DOI] [PubMed] [Google Scholar]
- (6).Boudou T; Legant WR; Mu A; Borochin MA; Thavandiran N; Radisic M; Zandstra PW; Epstein JA; Margulies KB; Chen CS A Microfabricated Platform to Measure and Manipulate the Mechanics of Engineered Cardiac Microtissues. Tissue Eng. Part A 2012, 18, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zimmermann W-H; Melnychenko I; Wasmeier G; Didié M; Naito H; Nixdorff U; Hess A; Budinsky L; Brune K; Michaelis B; Dhein S; Schwoerer A; Ehmke H; Eschenhagen T Engineered Heart Tissue Grafts Improve Systolic and Diastolic Function in Infarcted Rat Hearts. Nat. Med. 2006, 12, 452–458. [DOI] [PubMed] [Google Scholar]
- (8).Zhao Y; Rafatian N; Feric NT; Cox BJ; Aschar-Sobbi R; Wang EY; Aggarwal P; Zhang B; Conant G; Ronaldson-Bouchard K; Pahnke A; Protze S; Lee JH; Davenport Huyer L; Jekic D; Wickeler A; Naguib HE; Keller GM; Vunjak-Novakovic G; Broeckel U; Backx PH; Radisic M A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Radisic M; Park H; Shing H; Consi T; Schoen FJ; Langer R; Freed LE; Vunjak-Novakovic G Functional Assembly of Engineered Myocardium by Electrical Stimulation of Cardiac Myocytes Cultured on Scaffolds. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 18129–18134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ronaldson-Bouchard K; Yeager K; Teles D; Chen T; Ma S; Song L; Morikawa K; Wobma HM; Vasciaveo A; Ruiz EC; Yazawa M; Vunjak-Novakovic G Engineering of Human Cardiac Muscle Electromechanically Matured to an Adult-like Phenotype. Nat. Protoc. 2019, 14, 2781–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tavakol DN; Fleischer S; Vunjak-Novakovic G Harnessing Organs-on-a-Chip to Model Tissue Regeneration. Cell Stem Cell 2021, 28, 993–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ronaldson-Bouchard K; Ma SP; Yeager K; Chen T; Song L; Sirabella D; Morikawa K; Teles D; Yazawa M; Vunjak-Novakovic G Advanced Maturation of Human Cardiac Tissue Grown from Pluripotent Stem Cells. Nature 2018, 556, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Hansen A; Eder A; Bönstrup M; Flato M; Mewe M; Schaaf S; Aksehirlioglu B; Schwoerer AP; Schwörer A; Uebeler J; Eschenhagen T Development of a Drug Screening Platform Based on Engineered Heart Tissue. Circ. Res. 2010, 107, 35–44. [DOI] [PubMed] [Google Scholar]
- (14).Mannhardt I; Breckwoldt K; Letuffe-Brenière D; Schaaf S; Schulz H; Neuber C; Benzin A; Werner T; Eder A; Schulze T; Klampe B; Christ T; Hirt MN; Huebner N; Moretti A; Eschenhagen T; Hansen A Human Engineered Heart Tissue: Analysis of Contractile Force. Stem Cell Rep. 2016, 7, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Jackman CP; Carlson AL; Bursac N Dynamic Culture Yields Engineered Myocardium with Near-Adult Functional Output. Biomaterials 2016, 111, 66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tiburcy M; Hudson JE; Balfanz P; Schlick S; Meyer T; Chang Liao M-L; Levent E; Raad F; Zeidler S; Wingender E; Riegler J; Wang M; Gold JD; Kehat I; Wettwer E; Ravens U; Dierickx P; van Laake LW; Goumans MJ; Khadjeh S; Toischer K; Hasenfuss G; Couture LA; Unger A; Linke WA; Araki T; Neel B; Keller G; Gepstein L; Wu JC; Zimmermann W-H Defined Engineered Human Myocardium With Advanced Maturation for Applications in Heart Failure Modeling and Repair. Circulation 2017, 135, 1832–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Dostanić M; Windt LM; Stein JM; van Meer BJ; Bellin M; Orlova V; Mastrangeli M; Mummery CL; Sarro PM A Miniaturized EHT Platform for Accurate Measurements of Tissue Contractile Properties. J. Microelectromech. Syst. 2020, 29, 881–887. [Google Scholar]
- (18).Thavandiran N; Hale C; Blit P; Sandberg ML; McElvain ME; Gagliardi M; Sun B; Witty A; Graham G; Do VTH; Bakooshli MA; Le H; Ostblom J; McEwen S; Chau E; Prowse A; Fernandes I; Norman A; Gilbert PM; Keller G; Tagari P; Xu H; Radisic M; Zandstra PW Functional Arrays of Human Pluripotent Stem Cell-Derived Cardiac Microtissues. Sci. Rep. 2020, 10, 6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Turnbull IC; Karakikes I; Serrao GW; Backeris P; Lee J-J; Xie C; Senyei G; Gordon RE; Li RA; Akar FG; Hajjar RJ; Hulot J-S; Costa KD Advancing Functional Engineered Cardiac Tissues toward a Preclinical Model of Human Myocardium. FASEB J. 2014, 28, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Radisic M; Loskill P Beyond PDMS and Membranes: New Materials for Organ-on-a-Chip Devices. ACS Biomater. Sci. Eng. 2021, 7, 2861–2863. [DOI] [PubMed] [Google Scholar]
- (21).Borysiak MD; Bielawski KS; Sniadecki NJ; Jenkel CF; Vogt BD; Posner JD Simple Replica Micromolding of Biocompatible Styrenic Elastomers. Lab. Chip 2013, 13, 2773–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Sano E; Mori C; Matsuoka N; Ozaki Y; Yagi K; Wada A; Tashima K; Yamasaki S; Tanabe K; Yano K; Torisawa Y Tetrafluoroethylene-Propylene Elastomer for Fabrication of Microfluidic Organs-on-Chips Resistant to Drug Absorption. Micromachines 2019, 10, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Fleischer S; Shevach M; Feiner R; Dvir T Coiled Fiber Scaffolds Embedded with Gold Nanoparticles Improve the Performance of Engineered Cardiac Tissues. Nanoscale 2014, 6, 9410–9414. [DOI] [PubMed] [Google Scholar]
- (24).Thavandiran N; Dubois N; Mikryukov A; Massé S; Beca B; Simmons CA; Deshpande VS; McGarry JP; Chen CS; Nanthakumar K; Keller GM; Radisic M; Zandstra PW Design and Formulation of Functional Pluripotent Stem Cell-Derived Cardiac Microtissues. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E4698–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Nunes SS; Miklas JW; Liu J; Aschar-Sobbi R; Xiao Y; Zhang B; Jiang J; Massé S; Gagliardi M; Hsieh A; Thavandiran N; Laflamme MA; Nanthakumar K; Gross GJ; Backx PH; Keller G; Radisic M Biowire: A Platform for Maturation of Human Pluripotent Stem Cell–Derived Cardiomyocytes. Nat. Methods 2013, 10, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Tandon N; Cannizzaro C; Chao P-HG; Maidhof R; Marsano A; Au HTH; Radisic M; Vunjak-Novakovic G Electrical Stimulation Systems for Cardiac Tissue Engineering. Nat. Protoc. 2009, 4, 155–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tandon N; Marsano A; Maidhof R; Wan L; Park H; Vunjak-Novakovic G Optimization of Electrical Stimulation Parameters for Cardiac Tissue Engineering. J. Tissue Eng. Regen. Med. 2011, 5, e115–e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Chan Y-C; Ting S; Lee Y-K; Ng K-M; Zhang J; Chen Z; Siu C-W; Oh SKW; Tse H-F Electrical Stimulation Promotes Maturation of Cardiomyocytes Derived from Human Embryonic Stem Cells. J Cardiovasc. Transl. Res. 2013, 6, 989–999. [DOI] [PubMed] [Google Scholar]
- (29).Chiu LLY; Iyer RK; King J-P; Radisic M Biphasic Electrical Field Stimulation Aids in Tissue Engineering of Multicell-Type Cardiac Organoids. Tissue Eng. Part A 2011, 17, 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tiburcy M; Meyer T; Soong PL; Zimmermann W-H Collagen-Based Engineered Heart Muscle. In Cardiac Tissue Engineering: Methods and Protocols; Radisic M, Black LD III, Eds.; Methods in Molecular Biology; Springer: New York, NY, 2014; pp 167–176. [DOI] [PubMed] [Google Scholar]
- (31).Kaiser NJ; Kant RJ; Minor AJ; Coulombe KLK Optimizing Blended Collagen-Fibrin Hydrogels for Cardiac Tissue Engineering with Human IPSC-Derived Cardiomyocytes. ACS Biomater. Sci. Eng. 2019, 5, 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lemoine MD; Mannhardt I; Breckwoldt K; Prondzynski M; Flenner F; Ulmer B; Hirt MN; Neuber C; Horváth A; Kloth B; Reichenspurner H; Willems S; Hansen A; Eschenhagen T; Christ T Human IPSC-Derived Cardiomyocytes Cultured in 3D Engineered Heart Tissue Show Physiological Upstroke Velocity and Sodium Current Density. Sci. Rep. 2017, 7, 5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Edri R; Gal I; Noor N; Harel T; Fleischer S; Adadi N; Green O; Shabat D; Heller L; Shapira A; Gat-Viks I; Peer D; Dvir T Personalized Hydrogels for Engineering Diverse Fully Autologous Tissue Implants. Adv. Mater. 2019, 31, 1803895. [DOI] [PubMed] [Google Scholar]
- (34).Gaetani R; Yin C; Srikumar N; Braden R; Doevendans PA; Sluijter JPG; Christman KL Cardiac-Derived Extracellular Matrix Enhances Cardiogenic Properties of Human Cardiac Progenitor Cells. Cell Transplant. 2016, 25, 1653–1663. [DOI] [PubMed] [Google Scholar]
- (35).Lee JH; Protze SI; Laksman Z; Backx PH; Keller GM Human Pluripotent Stem Cell-Derived Atrial and Ventricular Cardiomyocytes Develop from Distinct Mesoderm Populations. Cell Stem Cell 2017, 21, 179–194.e4. [DOI] [PubMed] [Google Scholar]
- (36).Psaras Y; Margara F; Cicconet M; Sparrow AJ; Repetti GG; Schmid M; Steeples V; Wilcox JAL; Bueno-Orovio A; Redwood CS; Watkins HC; Robinson P; Rodriguez B; Seidman JG; Seidman CE; Toepfer CN CalTrack: High-Throughput Automated Calcium Transient Analysis in Cardiomyocytes. Circ. Res. 2021, 129, 326–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chen T-W; Wardill TJ; Sun Y; Pulver SR; Renninger SL; Baohan A; Schreiter ER; Kerr RA; Orger MB; Jayaraman V; Looger LL; Svoboda K; Kim DS Ultrasensitive Fluorescent Proteins for Imaging Neuronal Activity. Nature 2013, 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Sala L; van Meer BJ; Tertoolen LGJ; Bakkers J; Bellin M; Davis RP; Denning C; Dieben MAE; Eschenhagen T; Giacomelli E; Grandela C; Hansen A; Holman ER; Jongbloed MRM; Kamel SM; Koopman CD; Lachaud Q; Mannhardt I; Mol MPH; Mosqueira D; Orlova VV; Passier R; Ribeiro MC; Saleem U; Smith GL; Burton FL; Mummery CL MUSCLEMOTION. Circ. Res. 2018, 122, e5–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Moss RL; Fitzsimons DP Frank-Starling Relationship. Circ. Res. 2002, 90, 11–13. [PubMed] [Google Scholar]
- (40).Saleem U; Mannhardt I; Braren I; Denning C; Eschenhagen T; Hansen A Force and Calcium Transients Analysis in Human Engineered Heart Tissues Reveals Positive Force-Frequency Relation at Physiological Frequency. Stem Cell Rep. 2020, 14, 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).de Lange WJ; Farrell ET; Kreitzer CR; Jacobs DR; Lang D; Glukhov AV; Ralphe JC Human IPSC-Engineered Cardiac Tissue Platform Faithfully Models Important Cardiac Physiology. Am. J. Physiol. Heart Circ. Physiol. 2021, 320, H1670–H1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Bers DM Calcium Cycling and Signaling in Cardiac Myocytes. Annu. Rev. Physiol. 2008, 70, 23–49. [DOI] [PubMed] [Google Scholar]
- (43).Burridge PW; Matsa E; Shukla P; Lin ZC; Churko JM; Ebert AD; Lan F; Diecke S; Huber B; Mordwinkin NM; Plews JR; Abilez OJ; Cui B; Gold JD; Wu JC Chemically Defined Generation of Human Cardiomyocytes. Nat. Methods 2014, 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Huebsch N; Loskill P; Mandegar MA; Marks NC; Sheehan AS; Ma Z; Mathur A; Nguyen TN; Yoo JC; Judge LM; Spencer CI; Chukka AC; Russell CR; So P-L; Conklin BR; Healy KE Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales. Tissue Eng. Part C Methods 2015, 21, 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).King DE Dlib-Ml: A Machine Learning Toolkit. J. Mach. Learn. Res. 2009, 10, 1755–1758. [Google Scholar]
- (46).Bolme DS; Beveridge JR; Draper BA; Lui YM Visual Object Tracking Using Adaptive Correlation Filters. In 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition; 2010; pp 2544–2550. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


