Abstract
Extrachromosomal circular DNA (eccDNA) of chromosomal origin is common in eukaryotic cells. Amplification of oncogenes on large eccDNA (ecDNA) can drive biological processes such as tumorigenesis, and identification of eccDNA by sequencing after removal of chromosomal DNA is therefore important for understanding their impact on the expressed phenotype. However, the circular mitochondrial DNA (mtDNA) might challenge the detection of eccDNA because the average somatic cell has hundreds of copies of mtDNA. Here we show that 61.2–99.5% of reads from eccDNA-enriched samples correspond to mtDNA in mouse tissues. We have developed a method to selectively remove mtDNA from total circular DNA by CRISPR/Cas9 guided cleavage of mtDNA with one single-guide RNA (sgRNA) or two sgRNAs followed by exonuclease degradation of the linearized mtDNA. Sequencing revealed that mtDNA reads were 85.9% ± 12.6% removed from eccDNA of 9 investigated mouse tissues. CRISPR/Cas9 cleavage also efficiently removed mtDNA from a human HeLa cell line and colorectal cancer samples. We identified up to 14 times more, and also larger eccDNA in CRISPR/Cas9 treated colorectal cancer samples than in untreated samples. We foresee that the method can be applied to effectively remove mtDNA from any eukaryotic species to obtain higher eccDNA yields.
Keywords: mtDNA removal, CRISPR/Cas9, eccDNA, ecDNA
1. Introduction
Structural variations (SVs) such as DNA amplification and deletions can have striking effects on the phenotypes of eukaryotic cells. This is especially true in cancer cells, where amplification of oncogenes and regulatory elements on eccDNA can drive tumor progression, anti-cancer drug resistance and intra-tumoral genetic heterogeneity [1], [2], [3]. EccDNA describes a group of double and single-stranded covalently closed circular DNA molecules that derive from the nuclear chromosomes but segregate unevenly in mitosis because of the lack of centromeres [4], [5], [6], [7]. EccDNA is also found in healthy cells and has been detected in all the eukaryotes tested so far [8], [9], [10]. While most eccDNA probably never has a phenotypic effect on the hosting cell, some play a role in aging [10], and others allow for adaptation to the selective pressures of yeasts and plants [9], [11]. These findings lead to questions about how eccDNA is formed and maintained as well as how eccDNA affects cell biology and physiology. To address these questions, efficient methods for the purification of eccDNA are needed. One highly precise method to acquire eccDNA is based on the digestion of linear chromosomal DNA from total DNA with an exonuclease, followed by amplification of the enriched circular DNA by rolling circle amplification and subsequent DNA sequencing for genome-wide detection of eccDNA [12], [13], [14], [15], [16], [17], [18].
However, the mitochondrial DNA (mtDNA), which is also a circular structure of DNA, could account for much of the enriched circular DNA as their large amount in humans, plants and even mice germline cells [19], [20], [21]. The average somatic cell has just two copies for most of the nuclear genes or DNA segments but hundreds to thousands of copies of mtDNA. If mtDNA is retained in eccDNA purifications, a significant fraction of sequencing reads is expected to be aligned to the mtDNA genome and necessarily causes waste of sequencing resources. To solve this, the current methods remove mtDNA enzymatically by endonuclease (MssI enzyme in human cell lines) [13]. However, although this procedure removes the majority of mtDNA, it will also affect the large-sized eccDNA, as they are likely to contain the restriction enzyme target size. Other methods rely on ultracentrifugation and cell compartment fractionation to separate nuclear and mitochondrial fractions [22], [23], [24]. The preferred method for mtDNA removal highly depends on the starting material. For example, enzymatic removal of mtDNA is not expected to affect plasma eccDNA much because plasma has small eccDNA with sizes in the range from 100 to 5000 bp [25]. At the same time, this approach will cause degradation of eccDNA in tissues where eccDNA is larger. Thus, a highly specific method, which targets the removal of mtDNA, but keeps other circular DNA intact, is needed.
In the present study, we show how mtDNA can efficiently be removed from total eccDNA by CRISPR/Cas9 in vitro. We tested mouse tissues, the human HeLa cell line and colorectal tumors to develop and demonstrate a method in which mtDNA is removed by Cas9 with one sgRNA or two sgRNAs. Finally, quantitative real-time PCR and next-generation sequencing (NGS) were performed to demonstrate the efficiency of this method. Taken together, this method might be used to remove mtDNA from any kind of tissue and organism and reveals that removal of mtDNA can reduce the cost of sequencing and increase the read coverage of eccDNA deriving from the nuclear genome and help to provide new insight into gene copy-number variation.
2. Material and methods
2.1. Mice tissue, cell culture, and patients samples
Tissues were collected from wild-type C57BL/6NRj male mice immediately after euthanization by cervical dislocation and flash-frozen in liquid nitrogen and stored at −80 °C until further use. In total, 9 different mouse tissues: pancreas, subcutaneous adipose tissue, hippocampus, visceral adipose tissue, cortex, skeletal muscle, skin, whole skin with hair and liver samples (Table S1) were used in this study. Samples were cut and weighted from whole tissues in a −20 °C environment prior to DNA isolation.
The human HeLa cell line was cultured in DMEM medium (ThermoFisher, USA)), supplemented with 10% fetal bovine serum (ThermoFisher, USA) and 1% penicillin–streptomycin (ThermoFisher, USA), in a humidified incubator with 5% CO2 at 37 °C. Cells were grown to 70–90% confluence in 10 cm2 Petri dishes and passaged every 2–3 days to maintain this density.
Fully anonymized tissue samples from 5 patients with colorectal cancer (average age 71.2 years, all males) were used in the study. Two types of tissues: colorectal tumor tissue (adenocarcinoma) and adjacent normal colorectal tissue were collected and confirmed by a pathologist. Samples were cut and stored at −80 °C prior to DNA isolation. The patients had stage I–II colorectal cancer and had the tumor resected at the Department of Gastrointestinal Surgery, Herlev and Gentofte Hospital, Denmark. The patients were included in the Danish REBECCA study.
2.2. DNA isolation and linear DNA removal
Between 9 and 25 mg of mouse tissues (see sample details in S1) and 6 mg of CRC patient's tissue samples were digested overnight at 56 °C with shaking and Proteinase K (QIAGEN, Cat. No. 19131, Germany), and High Molecular Weight DNA (HMW DNA) was isolated the next day using the MagAttract HMW DNA Kit (QIAGEN, Cat. No.67563, Germany). DNA concentration was measured by Qubit HS DNA dsDNA High Sensitivity assay on Qubit 3.0 Fluorometer (Invitrogen). 50 μl total DNA (1–4 μg) were used for linear DNA removal. Linear DNA was removed by ExonucleaseV digestion (NEB, M0345L, USA) for 7 days at 37 °C, adding additional ATP and DNase every day (25 U/day). Afterwards, exonuclease was inactivated at 70 °C for 30 min, and the remaining circular DNA was cleaned using 1.8X AMPure XP magnetic beads (Beckman, Cat. No. A63881). Hereafter referred to as exo-clean DNA. The removal of linear DNA was confirmed by qPCR of the Cox5b gene (Supplementary Fig. 1).
HeLa cells (106) were collected and washed in PBS buffer twice. DNA isolation was performed using the MagAttract HMW DNA Kit (QIAGEN, Cat. No.67563, Germany), following the manufacturer's instructions. 50 μl total DNA (1–4 μg) were used for linear DNA removal. Linear DNA was removed by ExonucleaseV (NEB, M0345L, USA) and incubated at 37 °C for 7 days, adding additional ATP and DNase every day (25 U/day) according to the manufacturer's protocol. Then it was thermally inactivated at 70 °C for 30 min, followed by purification with WAHTS® DNA clean Beads (Vazyme, N411-01, China), and the remaining DNA denoted exo-clean DNA. The removal of linear DNA was confirmed by qPCR of the Cox5b gene (Supplementary Fig. 1).
2.3. Mitochondrial DNA removal with CRISPR/Cas9
For mtDNA removal, we used Cas9 Nuclease, Streptococcus pyogenes (NEB, Cat. No. M0386) in all our experiments, adapting the manufacturer's protocol according to our starting material. All sgRNAs were designed with the online software tool CHOPCHOP and synthesized by the company GenScript (Nanjing, China) or EnGen® sgRNA Synthesis Kit (NEB, E3322, USA). Sequences of sgRNAs are in Table 1. (The G in bold was included to increase the efficiency of sgRNA).
Table 1.
Sequence of sgRNA
| Name | Sequence (5′-3′) |
|---|---|
| mouse-sgRNA1 | GTAGCATGAACGGCTAAACGA |
| mouse-sgRNA2 | GGCCTGATAATAGTGACGCT |
| human-sgRNA1 | GGCTTGGATTAGCGTTTAGA |
| human-sgRNA2 | GCGTAGGGGCCTACAACGTTG |
For experiments where we tested the optimal concentrations of one sgRNA, the assembled reactions contained sgRNA in concentrations 10−7 nM to 102 nM, 10−7 nM to 102 nM of Cas9, 3 μl of 10X NEBuffer 3.1, and nuclease-free water to a final volume of 30 μl. Each reaction was then incubated for 20 min at 25 °C without shaking to allow the sgRNAs to bind the Cas9. Next, 8.5 μl of each assembled reaction were mixed with 15 μl of exo-clean DNA and 1.5 μl of 10X NEBuffer 3.1. This reaction was then incubated for at least 90 min at 37 °C. Afterwards, Cas9 was heat-inactivated for 10 min at 65 °C and allowed to cool off at room temperature. To the same reaction, 0.5 μl of 10X NEBuffer 3.1, 3 μl of 10 mM ATP and 2 μl of ExonucleaseV were added for a final volume of 30.5 μl. ExonucleaseV digestion were incubated overnight at 37 °C and heat-inactivated for 30 min at 70 °C the following day. DNA was purified using AMPure XP magnetic beads (Beckman, Cat. No. A63881).
For all the following experiments with two sgRNAs, the assembled reactions contained 30 nM of each sgRNA, 3 μl of 10X NEBuffer 3.1, 30 nM of Cas9 and nuclease-free water to a final volume of 30 μl. Each reaction was then incubated for 20 min at 25 °C without shaking to allow the sgRNAs to bind the Cas9. Next, 8.5 μl (for CRC tissue samples 17 μl) of the assembled reaction were mixed with 15 μl (for CRC tissue samples 30 μl) of exo-clean DNA and 1.5 μl (for CRC tissue samples 3 μl) of 10X NEBuffer 3.1. This reaction was then treated as described above for reactions with one sgRNA.
For experiments where mtDNA was removed enzymatically, digestions were performed with the restriction enzyme PacI (New England Biolabs, Cat. No.: R0547) and the restriction enzyme MssI (New England Biolabs, Cat. No.: R0560S) following the manufacturer's protocol. The assembled reaction contained 15 μl of exo-clean DNA, 5 μl 10X NEB buffer, 1 μl PacI/ MssI(10U/μl), and nuclease-free water to a final volume of 50 μl. The reaction was then incubated for 1 h at 37 °C and next purified with AMPure XP magnetic beads (Beckman, Cat. No. A63881).
2.4. Quantitative Real-Time PCR (qPCR) and PCR
For mouse qPCR, each reaction was run in a 10 μl reaction system, including 1 μl of sample DNA, 0.25 μl of each primer (5 μM), 5 μl of SYBR Green PCR Master Mix (Cat. No. 4309155, Life Technologies, USA) and 3.5 μl of sterile water. Reactions were run using the following thermal cycling conditions: 95 °C + (95 °C 15 s + 60 °C 15 s + 72 °C 15 s) × 25 cycles + 72 °C. For human cells, each qPCR reaction was run in a 10 µl reaction system, including 1 μl of sample DNA, 0.25 μl of each primer (10 μM), 5 μl of SYBR green (TOYOBO, QPK-201, Japan), 3 μl of RNase-free water under the following thermal cycling conditions: 10 min at 95 °C for pre-incubation, followed by 40 cycles of amplification (30 s at 95 °C, 30 s at 97 °C, 30 s at 72 °C), and completed with 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 min.
For mouse samples, each PCR reaction was run in a 50 µl reaction system, including 1 µl of sample DNA, 2 µl of each primer (10 µM), 5 µl of dNTPs (Cat. No. R0191, ThermoFisher, USA) 5 µl of 10X DreamTaq Buffer and 1 µl of DreamTaq DNA Polymerase (5 U/µl) (Cat. No. EP0705, ThermoFisher, USA). Reactions were run with the following cycling conditions: 15 s at 98 °C, followed by 25 cycles of amplification (15 s at 98 °C, 30 s at 60 °C and 30 s at 72 °C) and 7 min at 72 °C. For human cells, each PCR reaction was run in a 50 µl reaction system, including 1 μl of sample DNA, 1 μl of each primer (10 μM), 25 μl of 2X PCR Master Mix (NEB, #M0541, USA), 22 μl of RNase-free water under the following cycling conditions: 30 s at 98 °C for pre-incubation, followed by 30 cycles of amplification (10 s at 98 °C, 10 s at 56 °C, 20 s at 72 °C), then 5 min at 72 °C for the final extension.
Sequences of qPCR and PCR primers amplifying within the mtDNA are in Table 2:
Table 2.
Sequence of primers
| Name | Sequence |
|---|---|
| Primer-F1 | 5′-CGTGCAAAGGTAGCATAATCAC-3′ |
| Primer-R1 | 5′-TAAGCTCCATAGGGTCTTCTCG-3′ |
| Primer-F2 | 5′-CAACTATACTTTGCCTCGGAGC-3′ |
| Primer-R2 | 5′-TATAGCTTTGAAGAATGCGTGGG-3′ |
| Primer-F3 | 5′-GCCCACTTCCACTATGTCCT-3′ |
| Primer-R3 | 5′-GATTTTGGCGTAGGTTTGGTCT-3′ |
2.5. Rolling-circle amplification
Both exo-clean mouse DNA (with and without mtDNA) and exo-clean human cells DNA were used for Phi29 polymerase amplification by 4BB™ TruePrime® RCA Kit (4basebio, UK), following the manufacturer's protocol. Reactions were incubated at 30 °C for 48 h.
2.6. Sequence library preparation and sequencing
About 500 ng of Phi29 amplified DNA Products were sheared by sonication (Covaris LE220) to generate a median size of 400 bp. Then, 30 ng of fragmented DNA were used as input for library construction using MGIEasy DNA Library Preparation Kit (MGI-BGI, China). The size distribution and quality of each library were examined by the Bioanalyzer 2100 (Agilent). DNA libraries were sequenced (PE150) on the MGISeq-2000 platform (BGI-Qingdao, Qingdao, China). For human cancer samples, samples were multiplexed and sequenced as 2× 150-nucleotide paired-end reads on two lanes (Novaseq 6000, S2 flow-cell) (Rigshospitalet, Denmark).
2.7. Read alignment
Sequencing reads were aligned to the mouse reference genome (mm10 version) and human reference genome (hg38 version) using BWA-MEM v.0.7.17 [26] with the –q option, respectively. Downstream processing analyses of BAM/SAM files were performed using Samtools v1.9 [27]. The percentage of mtDNA reads was calculated according to the following formula:
2.8. Identification of eccDNA in colorectal cancer patient samples
A mapping-based approach was used to identify the chromosomal-derived eccDNAs. We developed a pipeline to process the sequence reads and report a set of identified circles along with several annotations for them. The pipeline included the following steps: 1) sequence data was demultiplexed using the sample barcodes (allowing zero mismatches), 2) the Cutadapt tool was used to trim adapter sequences and low-quality bases (base quality <10), 3) the SeqPrep (https://github.com/jstjohn/SeqPrep) tool was used to merge overlapping read pairs to yield singleton contigs (to help with the detection of small circles). 4) The set of single-ended merged reads and the paired-end unmerged reads were then mapped to the GRCh38 human reference using the BWA-MEM aligner. The read alignments were sorted and indexed using samtools. 5) EccDNAs were then identified by identifying locations with at least 2 pieces of evidence supporting a circle formation. Evidence for an eccDNA circle can be inferred through the presence of chimeric alignments (i.e., the read overlaps a circle boundary and gets chimerically mapped to the two ends), discordant paired-end mapping (reverse-forward orientation of the read pair) or a large soft-clip which mapped to another location indicating a circle boundary. 6) The identified eccDNAs were then assigned three confidence levels: HighQual if the mean coverage in the circle was at least twice the mean coverage in the neighbouring regions and at least 95% of the locations in the putative eccDNA circle were covered; else, MediumQual if the mean coverage did not meet the HighQual criterion but 95% of the locations were still covered, otherwise LowQual. 7) Iterative merging of the eccDNA circles was performed if the circles had a high reciprocal overlap (at least 50%) and the evidence counts merged accordingly. 8) The final list of eccDNAs was annotated with details of the overlapping genes, read support, coverage statistics as well as the identified small variants and reported as a VCF file.
An event was counted as a circle if at least two sequence reads supported the breakpoint of a circle. This also means that recurrent eccDNA such as mtDNA only counted as one event.
2.9. Statistical analysis
All statistical tests were executed by R- 3.6.2. The student's t-test was used to compare two groups.
3. Results
3.1. Workflow of CRISPR/Cas9 in cutting mtDNA
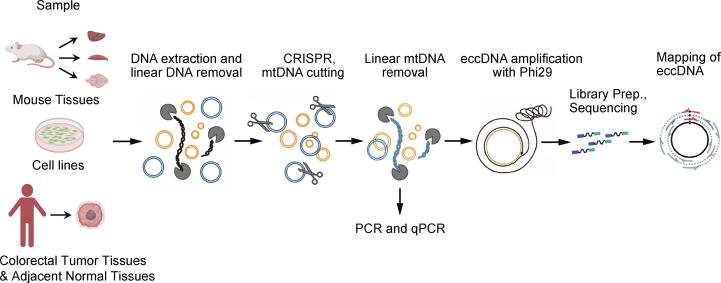
Here we wanted to examine wether mtDNA could be digested specifically by applying CRISPR/Cas9 with sgRNAs targeting mtDNA. As mice and humans are the two most common models for mammals, we tested the system in mice and the human HeLa cell line separately. First, we loaded mtDNA sequences of a mouse and a human on sgRNA design website CHOPCHOP to scan for sgRNA, and selected the top two. BLAST searches against the genomes of a mouse and a human were next used to ensure that the primers had no unspecific binding. Briefly, our overall workflow was as follows, DNA extraction, linear DNA removal, and mtDNA linearization by Cas9 endonuclease followed by exonuclease treatment again to remove the now linear mtDNA. Next, the remaining circular DNA was subjected to PCR and qPCR for mtDNA and linear DNA cutting efficiency test. Finally, eccDNA was amplified by rolling circle amplification to increase the signal, and NGS was performed (Fig. 1).
Fig. 1.
Circle-Seq Workflow with CRISPR/Cas9 mtDNA removal. Total DNA was extracted from mouse tissues, the human HeLa cell line and CRC tissue samples, and then linear DNA (black) was removed by ExonucleaseV. Next, CRISPR/Cas9 was performed to cut mtDNA (blue), followed by linear mtDNA removal, rolling circle amplification, library preparation, next-generation sequencing and mapping of sequence reads for annotation of eccDNA. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Removal of mtDNA in mouse tissues
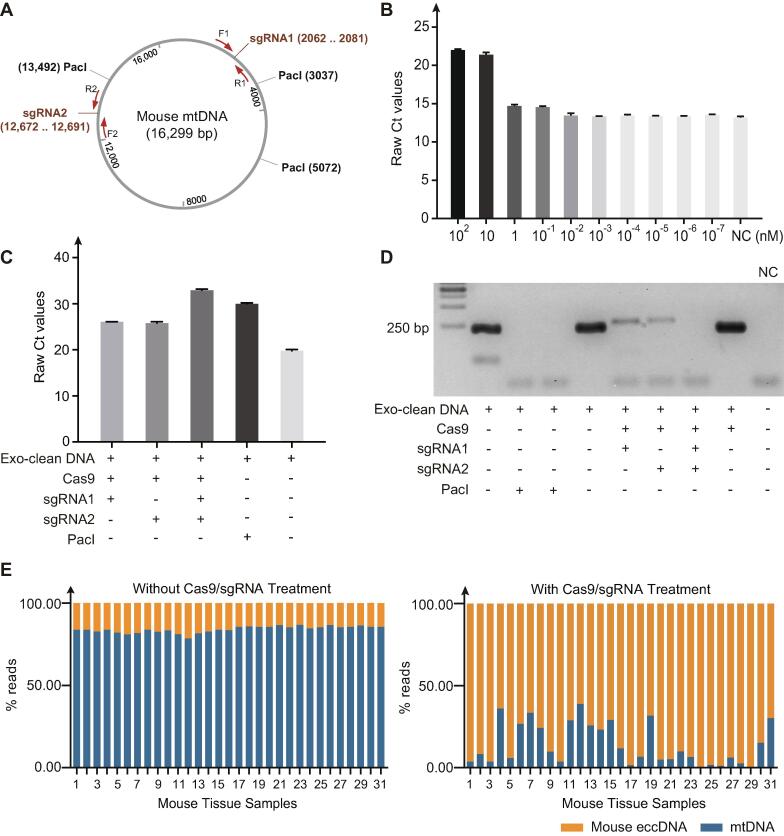
To investigate whether CRISPR/Cas9 could mediate the cleavage of the mouse mtDNA, we designed two sgRNAs to target mouse mtDNA (Fig. 2A, Table 1) and utilized qPCR and PCR to verify the cleavage efficiency. First, to test which concentration of Cas9:sgRNA was most efficient, we incubated exo-clean DNA at different Cas9:sgRNA concentrations, ranging from 102 to 10−7 nM. While there was a similar efficiency for 102 and 10 nM concentrations, there was a drop in efficiency at 1 nM and 10−1 nM and a further reduction at concentrations below 10−2 nM (Fig. 2B). Therefore, we chose to work with 30 nM as indicated in the Cas9 manufacturer's protocol to perform the subsequent experiments. Next, we assessed the effect of multiple sgRNAs on the efficiency by incubating equivalent concentrations of both sgRNAs with Cas9 during assemble reaction. We next used Cas9:sgRNA1/2 complex to incubate with DNA. As shown in Fig. 2C and D, a combination of two sgRNA significantly improved efficiency compared to single sgRNA treatments and compared to restriction enzyme treatment PacI.
Fig. 2.
Removal of mtDNA in mouse tissues. (A) Cleavage site of the two sgRNAs on mouse mtDNA (Table 1). (B) Quantitative real-time PCR (qRT-PCR) confirmation of the cutting efficiency of different Cas9/sgRNA concentrations (primer F1 and R1). Cas9/sgRNA were diluted at the concentration of 102 nM, 10 nM, 1 nM, 10−1 nM, 10−2 nM, 10−3 nM, 10−4 nM, 10−5 nM, 10−6 nM, 10−7 nM, and then subjected to the cleavage reaction of mtDNA. Followed by cleavage efficiency detection with qPCR. All the assays were performed three times. Error bars: mean ± SD of three independent tests. (C) Quantitative real-time PCR (qRT-PCR) confirmation of the CRISPR-Cas9 cleavage efficiency (primer F2 and R2). DNA digested with restriction enzyme PacI was used as a positive control; DNA only treated with cas9 protein was used as a negative control. (D) PCR validation of CRISPR-Cas9 treated DNA (primer F2 and R2). Cas9/sgRNA were diluted at the concentration of 10 nM, and then subjected to the cleavage reaction of mtDNA using sgRNA1, sgRNA2, and both two sgRNAs respectively. Followed by cleavage efficiency detection with PCR. DNA digested only with ExonucleaseV (Exo-clean DNA) was used as a negative control; DNA digested with restriction enzyme PacI was used as a positive control (two technical replicates); H2O was used as non-template control. (E) MtDNA reads mapping ratio with and without CRISPR-Cas9 treatment. DNA was extracted from 9 different mice tissues (Table S1) including 3 pancreas, 2 subcutaneous adipose tissue, 5 hippocampus, 1 visceral adipose tissue, 7 cortex, 5 skeletal muscle, 2 skin, 2 whole skin with hair and 4 liver samples, followed by linear DNA removal, rolling circle amplification, library preparation and next-generation sequencing.
We next tested the efficiency of the mtDNA cleavage on circular DNA purified from a larger set of different mouse somatic tissues, including pancreas, subcutaneous adipose tissue, hippocampus, visceral adipose tissue, cortex, skeletal muscle, skin, whole skin with hair and liver samples, and then performed Circle-Seq to enrich eccDNA [12], [28]. NGS of the enriched and amplified eccDNA revealed that mtDNA presents in the mouse, taking almost 84.03% ± 1.98% (range 61.20–99.46%, Fig. 2E) of sequence reads. After the mtDNA cleavage with Cas9/sgRNA treatment, the reads coverage of mtDNA from mouse eccDNA reads had dropped to an average of 14.10% ± 12.58% (range 0.54–38.80%, Fig. 2E).
3.3. Removal of mtDNA in eccDNA samples from human HeLa cell line
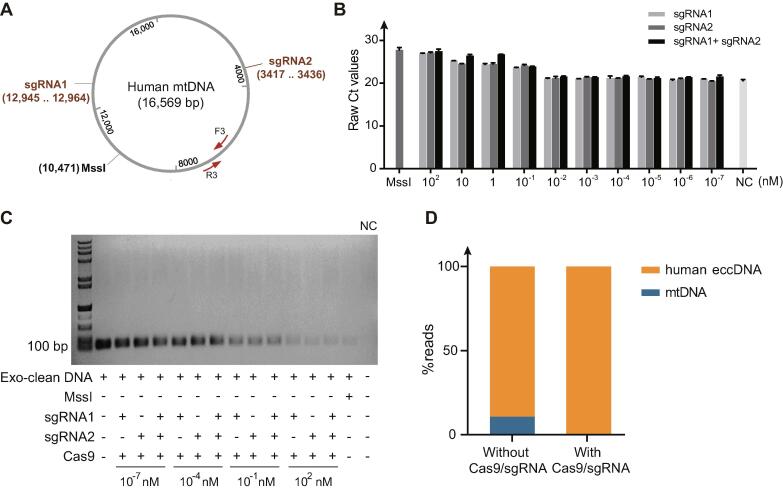
We next tested whether CRISPR/Cas9 guided by sgRNAs also removed mtDNA effectively in eccDNA samples from a human cancer cell line. We first designed two sgRNAs annealing to conserved regions of the human mtDNA (Fig. 3A, Table 1). Total DNA from 106 HeLa cells was next extracted and linear DNA removed by ExonucleaseV prior to CRISPR/Cas9 cleavage of mtDNA. We tested the cleavage efficiency of different concentrations of Cas9/sgRNA by qPCR and found that the maximum mtDNA cleavage rate was at 102 nM sgRNA (Fig. 3B). PCR also supported that 102 nM of sgRNA gave the most efficient cleavage of mtDNA. Moreover, at the concentrations of 102 nM, the Cas9/sgRNA cleavage for removal of mtDNA was just as efficient as digestion with the restriction enzyme MssI (Fig. 3B and C). We also tested the efficiency of CRISPR cleavage in the presence of one sgRNA as well as two sgRNAs and found that the cleavage efficiency of two sgRNAs was much higher than one sgRNA at the concentration of 1 nM and 10 nM, but at the concentration of 102 nM, there was not so much difference (Fig. 3B). Finally, NGS results revealed that for HeLa cells, the percentage of mtDNA reads was 10.74% if mtDNA was not specifically removed, but after CRISPR/Cas9 treatment, the coverage of mtDNA reads had been reduced to 0.01%. (Fig. 3D).
Fig. 3.
Removal of mtDNA in human HeLa cell line. (A) Cleavage site of the two sgRNAs on human mtDNA (Table1). (B) Quantitative real-time PCR (qRT-PCR) confirmation of the CRISPR/Cas9 cleavage efficiency (primer F3 and R3). Cas9/sgRNA were diluted at the concentration of 102 nM, 10 nM, 1 nM, 10−1 nM, 10−2 nM,10−3 nM,10−4 nM, 10−5 nM, 10−6 nM, 10−7 nM, and then subjected to the cleavage reaction of mtDNA. Followed by cleavage efficiency detection with qPCR. All the assays were performed three times. Error bars: mean ± SD of three independent tests. DNA digested with restriction enzyme MssI was used as a positive control. DNA only treated with cas9 protein was used as a negative control. (C) PCR validation of genotypes on CRISPR/Cas9 treated DNA (primer F3 and R3). Cas9/sgRNA were diluted at the concentration of 102 nM, 10−1 nM, 10−4 nM, 10−7 nM, and then subjected to the cleavage reaction of mtDNA using sgRNA1, sgRNA2, and both two sgRNAs, respectively. Followed by cleavage efficiency detection with PCR. The DNA digested only with ExonucleaseV (Exo-clean DNA) was used as a negative control, the DNA digested with restriction enzyme MssI was used as a positive control·H2O was used as non-template control. (D) MtDNA reads mapping ratio with and without CRISPR-Cas9 treatment.
3.4. Effect of removing mtDNA in colorectal cancer patients' samples
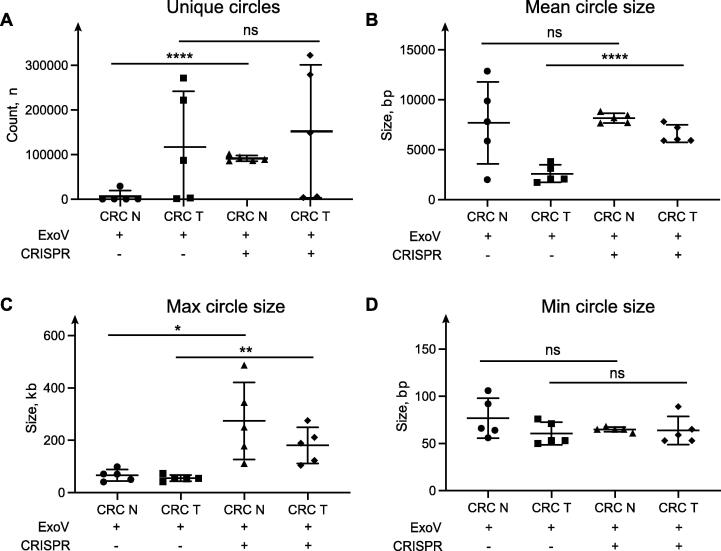
To test the effect of mtDNA removal on human tumor samples, we sequenced 10 tissue samples, 5 from colorectal tumors and 5 from the paired adjacent non-tumorous tissue (normal tissue). Each sample was divided into two parts; one part was treated with CRISPR/Cas9 of 2 sgRNAs, and the other was not. The removal of mtDNA and linear DNA were confirmed by qPCR (Supplementary Fig. 1). Removing mtDNA had a positive effect on the number of unique circles detected in the samples (Fig. 4A), as we found a 14 fold increase (P < 0.0001) of circle count in the normal tissue samples and also more in tumor tissue but not significantly (P = 0.6975) when samples were treated with CRISPR/Cas9. The mean circle size was more consistent among treated samples (Fig. 4B). The effect is also evident regarding detecting big circles (Fig. 4C). We found the average maximal size to be 274,105 bp (SD 19,964) in normal tissue samples when mtDNA was removed, compared to 66,347 bp (SD 131,555) when mtDNA was not removed (207,758 bp difference). In tumor tissue samples, we found the average maximal size to be 180,496 bp (SD 61,807) upon removal of mtDNA compared to 55,334 bp (SD 10,429) without mtDNA removal (125,162 bp difference). CRISPR/Cas9 removal of mtDNA did not influence the assessment of the smallest circles (Fig. 4D). Here, mtDNA circles were only counted as a single eccDNA, and the presence of multiple mtDNA in some samples, therefore, did not influence the mean size of all circles.
Fig. 4.
Profile of circles in human colorectal cancer samples treated with CRISPR/Cas9 and untreated samples. High-quality circles were analyzed. (A) Amount of all the unique circles per sample group. (B) The average size of all the circles per sample group. (C) The biggest circles that were found per sample group. (D) The smallest circles that were found per sample group. CRC T is colorectal cancer samples, CRC N is adjacent normal tissue. ExoV is ExonucleaseV, CRISPR is CRISPR/Cas9. Error bars: mean ± SD of 5 samples. Asterisks indicate significant differences (*P < 0.05; **P < 0.01; ****P < 0.0001; ns: not significant, Student’s t-test).
3.5. A general practice instruction for removal of mtDNA with CRISPR-Cas9
To aid the removal of mtDNA, We have made a general protocol in Box 1, which can be used for both mouse and human eccDNA work.
Protocol for removal of mtDNA with CRISPR-Cas9.
After purifying total DNA with the MagAttract HMW DNA Kit (QIAGEN, Cat. No.67563, Germany), linear DNA is removed from the circular DNA enzymatically, as described before. Cas9 S. pyogenes kit (NEB, Cat. No. M0386) is used in the following experiments.
-
1)
Assemble the following pre-incubation step: 30 nM sgRNA1, 30 nM sgRNA2, 3 µl NEBuffer 3.1 (10×), 30 nM Cas9 Nuclease, add nuclease-free water to a total volume of 30 µl.
-
2)
Incubate the reaction for 20 min at 25 °C without shaking to allow the sgRNAs to bind the Cas9.
-
3)
Assemble the actual cleavage reaction by adding 15 µl exo-clean DNA, 1.5 µl NEBuffer 3.1(10×), 8.5 µl Cas9:sgRNA1/2 complexes to a total volume of 25 µl.
-
4)
Incubate the reaction at 37 °C for 1.5–4 h.
-
5)
Heat-inactivate the reaction 10 min at 65 °C and allow it to cool off at room temperature.
-
6)
Add the exonuclease components to each reaction: 0.5 µl NEBuffer 3.1(10×), 3 µl ATP (10 mM), 2 µl ExonucleaseV (10,000 U/ml) to a final volume of 30.5 µl
-
7)
Incubate the reaction overnight (can be adjusted to 2.5 days if necessary) at 37 °C and heat-inactivate for 30 min at 70 °C the following day.
-
8)
Clean the samples with 1.8X AMPure XP magnetic beads (Beckman, Cat. No. A63881).
4. Discussion
In summary, the present study demonstrates how purification of eccDNA can be optimized by specific removal of mtDNA from eccDNA and how it affects sequencing results. Our results show that mtDNA can efficiently be removed from mouse and human tissues as well as human cell lines using in vitro cleavage by Cas9. The optimal Cas9/sgRNA concentration for in vitro activity and the reaction was firstly determined by qPCR. Here, in both human and mouse samples, two sgRNAs demonstrated better cleavage effectiveness than one sgRNA; therefore, we strongly recommend performing the experiments with two sgRNAs. Given that the cutting efficiency does not differ significantly between concentrations of 10 nM and 100 nM, we suggest using 30 nM as indicated in the Cas9 manufacturer's protocol. We also tested our CRISPR/Cas9 and sgRNA system on CRC patients' samples. Finally, through NGS, we confirmed that this method could considerably improve the sequencing reads coverage, obtained circle amount and circle size of eccDNA, and is as efficient as cleavage of mtDNA with the traditional endonuclease-based Circle-Seq method.
In some studies, endonucleases MssI and PacI were used to assist eccDNA purification from human- and mouse-derived DNA samples, respectively [13], [29]. However, large eccDNA holds a great probability of carrying MssI or PacI restriction sites, causing these circles to be lost during the endonuclease treatment. This situation may be particularly problematic in tumor research where oncogenes amplified on Mb-sized eccDNA (ecDNA) are studied for their role in tumorigenesis [30]. This challenge is solved with the current CRISPR/Cas9 method. Because the method is specific for mtDNA, large circles from other parts of the genome are not expected to be cleaved.
Notably, mtDNAs were not completely removed by our method. This may be attributed to 1) the enzyme kinetics of the Cas9/sgRNAs. Like other catalysts, enzymes provide an alternate pathway from the substrate to the product, but it does not alter the equilibrium between substrates and products, which means it can never achieve an absolute conversion. And 2) single nucleotide polymorphism (SNP) variants within sgRNA binding regions. The challenging biochemical environment in mitochondria can lead to a higher mutation rate than in the nucleus [31] and thereby depletion of the specific sgRNA binding sites. Studies suggested that even within the same mitochondrial haplogroup, differences of roughly 20–80 single-nucleotide polymorphisms (SNPs) are prevalent across mtDNA admixed in random pairings (heteroplasmy) [32], which will result in part of the mtDNA not being recognized by sgRNA.
Previous studies have reported that CRISPR/Cas9 systems have a high potential for off-target activity (especially in vitro experiments) because they have higher promiscuous binding capacities at sites distant from the protospacer-adjacent motif (PAM) region [33], [34], [35]. We have therefore selected the sgRNA based on minimal off-targets, for example, off-targets that typically require more than three mismatches for perfect annealing elsewhere [36]. More advanced genome searches may be required to obtain even better target sites and reduce the number of potential off-target sites associated with various mismatches. In addition, cleavage conditions in vitro, such as sgRNA dosage, may also be optimized to reduce off-target cleavage.
Although here we only tested the effectiveness of this method in mammals, it may also be applied for the study of eccDNA in other species, such as plants. Contrary to the relatively small and homogeneous size of animal mtDNA (typically 14–20 kb), plant mtDNAs are large and diverse in size (200–2,000 kb) [37], [38]. As RCA is biased towards smaller circular DNA, mtDNA may not appear to be a concern in plant eccDNA studies. However, recent research has revealed the possibility that plant mtDNA is made up of a mix of circular subgenomes, linear, and branching molecules [39]. Backert and his colleagues investigated mtDNA from white goosefoot by electron microscopy. They discovered that the majority of big mtDNA molecules are linear or rosette-like, whereas the majority of circular molecules are small [40]. In this case, removal of circular mtDNA should probably be done with several sgRNAs that span across the goosefoot mtDNA.
Also, there might be limitations to the method for analysis of non-model organisms. For example, as mtDNA can break into smaller circles [40], more sgRNA may be needed to degrade mtDNA fully. In addition, the number of mitochondria may be different between tissues and species, which means that further optimization might be required for different situations.
With the advancement of NGS, numerous technologies have been developed for studying eccDNA. For example, whole-genome sequencing data can be used to detect highly amplified eccDNA based on read coverage, discordant read pairs and soft-clipped reads [41]. CIDER-Seq (Circular DNA enrichment sequencing) is another technology for enriching and precisely sequencing circular DNA [42]. ATAC-seq has also been successfully applied for the identification of eccDNA [43]. Finally, the Circle-Seq method developed by our group enables genome-wide detection of eccDNA of all sizes and is not dependent on its copy numbers [12]. The CRISPR/Cas9 based mtDNA removal can be combined with all these methods and will likely lead to more effective use of sequencing resources and greatly improve sequence information from nuclear eccDNA. The method might be applicable for screening of eccDNA in a wide variety of species and beneficial for generating more information regarding biomarkers in different diseases, thereby a promising tool for studies of eccDNA across kingdoms of the tree of life.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The next-generation sequencing of mice tissues and human HeLa cell line were supported by Qingdao-Europe Advanced Institute for Life Sciences. We thank the China National GeneBank for the support in executing the project. We thank all patients for their participation in the study. We also thank Professor Julia Sidenius Johansen, Astrid Zedlitz Johansen and the nurses Tina Bak Sandberg, Maria Kolbech Johansen and Junko Strauss for recruiting patients at Herlev and Gentofte Hospital. “The Bio- and Genome Bank Denmark (Danish Cancer Biobank)” is acknowledged for their assistance with processing and storage of the frozen tissue samples.
Funding
G.A. received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No.801199. B.R and E.Z are also supported by the CIRCULAR VISION project from the European Union's Horizon 2020 program and innovation programme under grant agreement No. 899417. Work in B.R.’s laboratory on eccDNA technology is also funded by the Novo Nordisk Foundation project Atlas of circular DNA (NNF18OC0053139, NNF21OC0072023) and by the Innovation Fund Denmark project CARE-DNA (8088-00049B).
Author's contribution
B.R., G.A. and W.F. conceived the idea and devised the protocol. G.A. and H.P. collected mice tissues and samples. G.A performed the mouse part of the experimental work. W.F. and W.L. performed the human HeLa cell line part of the experimental work. E.Z. performed experimental part and data analysis for colorectal cancer samples. W.F. and W.L. prepared the figures. X.L. analyzed the data. W.F. drafted most of the manuscript. B.R. supervised the study. All authors have contributed to the execution of the experiments and studies. All authors discussed the results and contributed to the final manuscript.
Ethical statement
Mice were housed at the University of Copenhagen, Denmark, according to regulations. The ethical approval was granted from the Danish Veterinary and Food administration (Fødevarestyrelsen) to Jørgen Frank Pind Wojtaszewski (Number 2019-15-0201-01659).
All patients with colorectal cancer gave written informed consent. The study was performed according to the declaration of Helsinki. The REBECCA study protocol was approved by the Ethics Committee of the Capital Region of Denmark (VEK j.nr. H-2-2013-078) and the Danish Data Protection Agency (j. nr. 2007-58-0015, HEH-2014-044, I-suite nr. 02771 and PACTIUS P-2019-614).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.028.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Paulsen T., Kumar P., Koseoglu M.M., Dutta A. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 2018;34:270–278. doi: 10.1016/j.tig.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu S., et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575:699–703. doi: 10.1038/s41586-019-1763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H., et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet. 2020;52:891–897. doi: 10.1038/s41588-020-0678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruitwagen T., Chymkowitch P., Denoth-Lippuner A., Enserink J., Barral Y. Centromeres license the mitotic condensation of yeast chromosome arms. Cell. 2018;175:780–795 e715. doi: 10.1016/j.cell.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey C., Shoura M.J., Mischel P.S., Swanton C. Extrachromosomal DNA-relieving heredity constraints, accelerating tumour evolution. Ann Oncol. 2020;31:884–893. doi: 10.1016/j.annonc.2020.03.303. [DOI] [PubMed] [Google Scholar]
- 6.Hull R.M., Houseley J. The adaptive potential of circular DNA accumulation in ageing cells. Curr Genet. 2020;66:889–894. doi: 10.1007/s00294-020-01069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levan A., Levan G. Have double minutes functioning centromeres? Hereditas. 1978;88:81–92. doi: 10.1111/j.1601-5223.1978.tb01606.x. [DOI] [PubMed] [Google Scholar]
- 8.Gaubatz J.W. Extrachromosomal circular DNAs and genomic sequence plasticity in eukaryotic cells. Mutat Res. 1990;237:271–292. doi: 10.1016/0921-8734(90)90009-g. [DOI] [PubMed] [Google Scholar]
- 9.Koo D.H., et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc Natl Acad Sci U S A. 2018;115:3332–3337. doi: 10.1073/pnas.1719354115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinclair D.A., Guarente L. Extrachromosomal rDNA circles–a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 11.Hull R.M., et al. Transcription-induced formation of extrachromosomal DNA during yeast ageing. PLoS Biol. 2019;17:e3000471. doi: 10.1371/journal.pbio.3000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moller H.D., et al. Genome-wide purification of extrachromosomal circular DNA from eukaryotic cells. J Vis Exp. 2016:e54239. doi: 10.3791/54239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moller H.D., et al. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. 2018;9:1069. doi: 10.1038/s41467-018-03369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moller H.D., Parsons L., Jorgensen T.S., Botstein D., Regenberg B. Extrachromosomal circular DNA is common in yeast. Proc Natl Acad Sci U S A. 2015;112:E3114–E3122. doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz-Lara A., Gent D.H., Martin R.R. Identification of extrachromosomal circular DNA in hop via rolling circle amplification. Cytogenet Genome Res. 2016;148:237–240. doi: 10.1159/000445849. [DOI] [PubMed] [Google Scholar]
- 16.Lanciano S., Zhang P., Llauro C., Mirouze M. Identification of extrachromosomal circular forms of active transposable elements using Mobilome-Seq. Methods Mol Biol. 2021;2250:87–93. doi: 10.1007/978-1-0716-1134-0_7. [DOI] [PubMed] [Google Scholar]
- 17.Mann L., Seibt K.M., Weber B., Heitkam T. ECCsplorer: a pipeline to detect extrachromosomal circular DNA (eccDNA) from next-generation sequencing data. BMC Bioinf. 2022;23:40. doi: 10.1186/s12859-021-04545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su Z., Saha S., Paulsen T., Kumar P., Dutta A. ATAC-Seq-based identification of extrachromosomal circular DNA in mammalian cells and its validation using inverse PCR and FISH. Bio Protoc. 2021;11:e4003. doi: 10.21769/BioProtoc.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pakendorf B., Stoneking M. Mitochondrial DNA and human evolution. Annu Rev Genomics Hum Genet. 2005;6:165–183. doi: 10.1146/annurev.genom.6.080604.162249. [DOI] [PubMed] [Google Scholar]
- 20.Cao L., et al. The mitochondrial bottleneck occurs without reduction of mtDNA content in female mouse germ cells. Nat Genet. 2007;39:386–390. doi: 10.1038/ng1970. [DOI] [PubMed] [Google Scholar]
- 21.Quetier F, Vedel F. Heterogeneous population of mitochondrial DNA molecules in higher plants.
- 22.Defontaine A., Lecocq F.M., Hallet J.N. A rapid miniprep method for the preparation of yeast mitochondrial DNA. Nucleic Acids Res. 1991;19:185. doi: 10.1093/nar/19.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nedeva T., Petrova V., Hristozova T., Kujumdzieva A. A modified procedure for isolation of yeast mitochondrial DNA. Z Naturforsch C J Biosci. 2002;57:960–961. doi: 10.1515/znc-2002-9-1036. [DOI] [PubMed] [Google Scholar]
- 24.Valach M., Tomaska L., Nosek J. Preparation of yeast mitochondrial DNA for direct sequence analysis. Curr Genet. 2008;54:105–109. doi: 10.1007/s00294-008-0200-3. [DOI] [PubMed] [Google Scholar]
- 25.Sin S.T.K., et al. Identification and characterization of extrachromosomal circular DNA in maternal plasma. PNAS. 2020;117:1658–1665. doi: 10.1073/pnas.1914949117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H.W.J.a.G. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. (2013).
- 27.Li H., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moller H.D. Circle-Seq: Isolation and Sequencing of Chromosome-Derived Circular DNA Elements in Cells. Methods Mol Biol. 2020;2119:165–181. doi: 10.1007/978-1-0716-0323-9_15. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., et al. eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature. 2021;599:308–314. doi: 10.1038/s41586-021-04009-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner K.M., et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexeyev M., Shokolenko I., Wilson G., LeDoux S. The maintenance of mitochondrial DNA integrity–critical analysis and update. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Royrvik E.C., Burgstaller J.P., Johnston I.G. mtDNA diversity in human populations highlights the merit of haplotype matching in gene therapies. Mol Hum Reprod. 2016;22:809–817. doi: 10.1093/molehr/gaw062. [DOI] [PubMed] [Google Scholar]
- 33.Cong L., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jinek M., et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moller H.D., et al. CRISPR-C: circularization of genes and chromosome by CRISPR in human cells. Nucleic Acids Res. 2018;46:e131. doi: 10.1093/nar/gky767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duminil J. Mitochondrial genome and plant taxonomy. Methods Mol Biol. 2014;1115:121–140. doi: 10.1007/978-1-62703-767-9_6. [DOI] [PubMed] [Google Scholar]
- 38.Morley S.A., Nielsen B.L. Plant mitochondrial DNA. Front Biosci (Landmark Ed) 2017;22:1023–1032. doi: 10.2741/4531. [DOI] [PubMed] [Google Scholar]
- 39.Chevigny N., Schatz-Daas D., Lotfi F., Gualberto J.M. DNA Repair and the Stability of the Plant Mitochondrial Genome. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21010328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Backert S., Lurz R., Borner T. Electron microscopic investigation of mitochondrial DNA from Chenopodium album (L.) Curr Genet. 1996;29:427–436. doi: 10.1007/BF02221510. [DOI] [PubMed] [Google Scholar]
- 41.Deshpande V., et al. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun. 2019;10:392. doi: 10.1038/s41467-018-08200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehta D., Cornet L., Hirsch-Hoffmann M., Zaidi S.S., Vanderschuren H. Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-Seq. Nat Protoc. 2020;15:1673–1689. doi: 10.1038/s41596-020-0301-0. [DOI] [PubMed] [Google Scholar]
- 43.Kumar P., et al. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv. 2020;6:eaba2489. doi: 10.1126/sciadv.aba2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.