Abstract
Nanomedicine design is often a trial-and-error process, and the optimization of formulations and in vivo properties requires tremendous benchwork. To expedite the nanomedicine research progress, data science is steadily gaining importance in the field of nanomedicine. Recently, efforts have explored the potential to predict nanomaterials synthesis and biological behaviors via advanced data analytics. Machine learning algorithms process large datasets to understand and predict various material properties in nanomedicine synthesis, pharmacologic parameters, and efficacy. “Big data” approaches may enable even larger advances, especially if researchers capitalize on data curation methods. However, the concomitant use of data curation processes needed to facilitate the acquisition and standardization of large, heterogeneous data sets, to support advanced data analytics methods such as machine learning has yet to be leveraged. Currently, data curation and data analytics areas of nanotechnology-focused data science, or ‘nanoinformatics’, have been proceeding largely independently. This review highlights the current efforts in both areas and the potential opportunities for coordination to advance the capabilities of data analytics in nanomedicine.
1. Aim of the Review
The medical use of nanomaterials has shown promising advancement in the field of drug delivery. From the first FDA-approved nanodrug Doxil to the clinical use of lipid nanoparticles for mRNA vaccines, numerous nanoparticles can efficiently and safely deliver small molecules, proteins, nucleic acids, or other active pharmaceutical ingredients (APIs).1–3 Encapsulation in nanoparticles improves drug solubility, stability, and potentially facilitates the crossing of biological barriers and the selective targeting to disease sites, when these pharmacologic advantages have not or cannot be engineered into APIs.4 However, designing nanocarriers and optimizing the delivery strategy can be time-intensive, and typically involves a substantial amount of trial and error. Clinical translation of nanomedicines often does not follow pre-clinical development, often due to the complexity and heterogeneity of nanoparticles.5 To address these issues, advanced data analytics methods promise to accelerate and improve the nanomedicine development processes.
While the prediction of nanomaterial behavior in vivo is challenging due to a lack of systematic studies across material types, artificial intelligence (AI) platforms promise to improve and streamline nanomedicine development. Since 2007, when the term ‘nanoinformatics’ was coined,6, 7 there has been an explosion of nanomaterial-related data science efforts, which has generated large datasets with nanomaterial characterizations.7 However, there is a disconnect between the scientists curating the databases and those utilizing AI. As a result, some AI platforms are developed using small-scale, project-specific datasets that lack translational values. In addition, non-standardized reporting metrics make it difficult to compare different material entities, and non-centralized databases are inaccessible to research groups who are not equipped with data mining skills. Herein, the aims of this review are to (1) review the most recent AI studies that capture the epitome of nanomedicine platform development, (2) introduce public databases focused on nanomaterial characterizations, and, most importantly, (3) advocate for a collaboration between the research groups developing AI platforms and nano-informaticians in order to increase the clinical utility of nanomedicine.
2. Introduction to ‘Nanoinformatics’ and Machine Learning
Many science and engineering fields today are becoming more data-driven (ie. data analysis is increasingly powering experimental decision-making). Data science is a multidisciplinary field that combines mathematics and statistics to extrapolate meaningful insights from data.8, 9 As a result of the increasing interest in medical applications of nanotechnology, a new field of data science has emerged. ‘Nanoinformatics’ bridges computer science, information technology, nanotechnology, and medicine to accommodate the increasing investigation for nanomaterial discovery.6 The field encompasses informatics techniques to analyze and process structural and physicochemical properties of nanomaterials and their biological environments with the primary goal of accelerating and facilitating clinical applications.6
Recently, nanomaterial databases have also been under rapid development. This includes standardizing nanomaterial nomenclature, reporting physicochemical properties, and developing quantitative structure-activity relationships (QSAR) to understand efficacy and toxicity mechanisms from in vitro and in vivo results.7, 10 Coupling these analyses with data mining to extract information from literature and patents, data scientists have initiated several nanomedicine libraries to collect, compare and analyze different formulations.11–13 Although computational data mining techniques are increasingly used for data curation, most predictive nanomedicine platforms to date have been developed using manually-curated data, which is low throughput and can pose problems for platform generalizability.14–18 Many platforms have found it difficult to recruit groups to contribute large amounts data, and, as a result, are less impactful than intended. Additionally, as these databases are often structured primarily for manual data analytics, they are not primed for their use as inputs into AI. Therefore, a gap needs to be filled between the nanoinformatics and AI applications in nanotechnology.
AI is a field of computer science in which programs are developed to model cognitive functions of human intelligence, such as learning and problem-solving.19 Machine learning constitutes a subset of AI that learns underlying trends from data to make informed decisions or predictions.19–21 Machine learning platforms typically utilize classic algorithmic approaches, such as clustering, regression, and classification, to make predictions from complex patterns. As input to train machine learning models, descriptive characteristics are curated for each sample, known as a feature vector, and associated with a predicted value.20, 21 However, a constant challenge with developing machine learning platforms is organizing a dataset that will not overfit the algorithm.22, 23 An overfit model is trained to predict the training samples exclusively and will be unsuccessful when challenged with new data.20 Optimal datasets include many samples but few features. Creating large and comprehensive datasets to train any machine learning model accurately remains a challenge for the biological sciences and engineering.24, 25
Because of their ability to deconvolute patterns within datasets, machine learning has become one of the exciting analytic methods to tackle challenges in nanotechnology development. These models have successfully made predictions of physical properties15, 17, 18, 26, 27 and material compositions28–31, in addition to more complex nanomaterial-biological (nano-bio) interactions,14, 32, 33 cellular uptake pathways,33–35 and toxicity profiles.36–38 Robust algorithms require large amounts of comprehensive data for training.20, 21 For this reason, curating nanomedicine properties and experimental results into a centralized database can significantly benefit the future of AI in nanotechnology (Figure 1).
Figure 1.
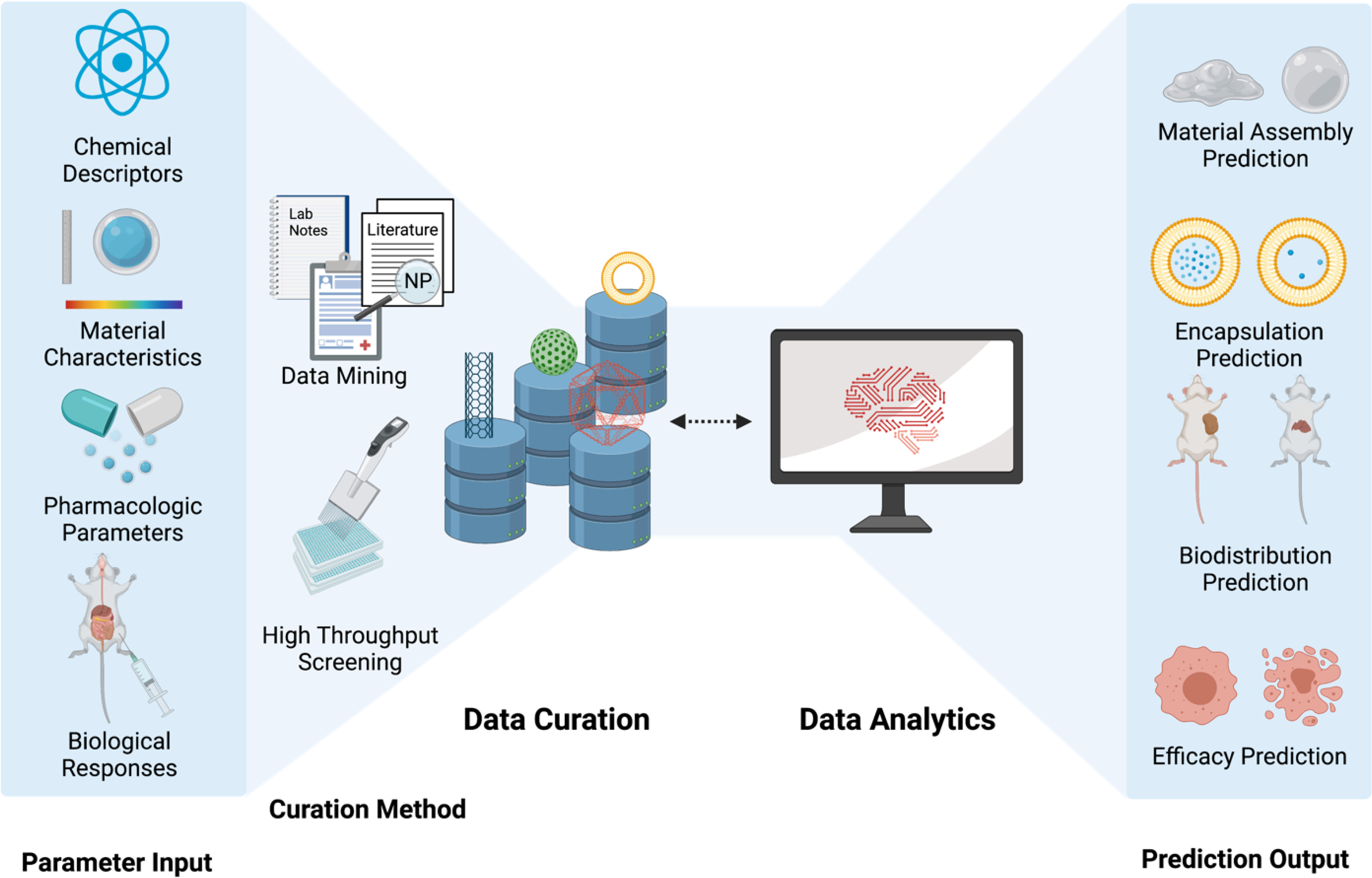
Workflow for nanomedicine data curation and data analytics.
3. Recent advances in nanomedicine using machine learning
Over the past decade, machine learning applications in biomedical science and engineering has gained popularity and changed the landscape of nanomedicine research. According to Web of Science, the number of publications with the keywords “nanomedicine” and “machine learning” or “AI” has increased 10 times within the past 10 years. Nanomedicine research has benefited from various machine learning studies, as many traditional methodologies are unable to delineate the complexity and heterogeneity of nanomedicine entirely. Recent publications have presented many successful ways to seamlessly integrate AI into nanomedicine development in both exploratory and validation stages. Among the compelling stories, AI has been commonly applied to nanomedicine material and synthetic efforts to optimize their design.39, 40 Similarly, AI applications have also been widely used to understand the underlying principles of nanomedicine-based targeted drug delivery and biodistribution.41, 42 While topics on AI guided synthesis,40, 43 strategic delivery,39, 41 and AI-based prediction of particle biodistribution38, 42 have been thoroughly covered in several recent reviews, we want to additionally highlight works in the field of data curation that may benefit nanomedicine development efforts and could facilitate collaborations with other data analytics approaches to improve nanomedicines (Table 1).
Table 1:
Selected studies in nanomedicine development using machine learning, their dataset source, and curation strategy.
| Study | Dataset source | Curation strategy | Training data size | Algorithm | Ref. |
|---|---|---|---|---|---|
| Novel excipient candidates | DrugBank, Drugs@FDA*, | High throughput screening for drug- excipient interactions, Cheminformatics computed by RDkit, and molecular dynamics studies | 2.1 million drug-excipient pairings | Random forest | 29 |
| Nanoparticle protein corona | Literature, UniProt | Data mining for nanoparticle properties and classification, physiochemical descriptions of protein corona. | 56 papers with 178 independent proteins | Random forest | 14 |
| Biological activity prediction | Literature, in-house screening and imaging | Structure-activity relationship, image analysis | 960 SNAs with 17,000 MALDI-MS data points; 1620 samples for immune responses and 301 samples for organ burdens; 1301 micrometastases for image analysis | Random forest, XGBoost; Support Vector Machine | 95, 96, 97 |
Excipients can be sourced under Inactive Ingredient Search for Approved Drug Product
3.1. Using AI to investigate excipients for nanomedicine designs
Most of current nanomedicine research is focused on a handful of material classes. With an increasing demand for nanomaterials to overcome biomedical challenges, discovering novel materials is a major effort in nanomedicine research. While over 50 nanomedicines have been FDA-approved and hundreds more are in clinical trials, only a small fraction of the nanomedicine chemical space has been explored to date.44 Despite a few nanodrugs with novel material entities, such as the nanocrystal Ryanodex or albumin-bound nano-drug Abraxane, the vast majority of FDA-approved nanodrugs are liposomal formulations with similar lipid contents.44–48 Researchers are investigating new material entities to expand the options for nanoformulations in order to overcome multifaceted delivery challenges, such as physical and biological stability, immune surveillance, and reticuloendothelial clearance. Centralized platforms for nanoformulation guidance can potentially accelerate research efforts.
Recent studies have investigated the potential of integrating drug excipients in nanomedicine formulations for increased stability and high drug-loading.49 Previously, we published a self-assembled indocyanine nanoparticle system with high drug-loading carriers for hydrophobic small molecules with robust in vivo activities.50 Through a quantitative structural assembly analysis, we discovered that certain molecular descriptors of small molecules correlate to the formability of the self-assembly system.50 Cheminformatics approaches have also been used to carry hydrophobic compounds in polymeric micelle formulations. Alves et al. showed a workflow for rational design of these polymeric micelle systems through quantitative structure-property relationship modeling.51 The FDA’s Inactive Ingredients Database contains about 3000 exipients from previously approved drug formulation and animal toxicity studies.52 The database provides a thorough set of candidates for nanoformulation design.
Reker et al. recently harnessed machine learning to explore the excipient-drug space and discover novel excipients for self-assembling nanoparticles.29 They generated about 2.1 million drug-excipient combinations, extracting drugs and excipients from the DrugBank and FDA libraries, respectively. Each drug-excipient pair was associated with a feature vector composed of over 4000 features. These features included physicochemical properties of both drug and excipient molecules, as well as parameters of enthalpic, non-covalent interactions between the two molecules, as determined through short molecular dynamics (MD) studies. Machine learning analysis of the large dataset was accomplished using a random forest classifier because of its inherent ability to parse out important features. High-throughput experimental classification using dynamic light scattering (DLS) was used to train and validate the platform. Although the MD-related features highly contributed to accurate co-aggregation predictions and could aid in understanding nanoparticle stability, performing MD for every potential drug-excipient pair is unrealistic due to computational costs. Therefore, the model was re-trained without the MD features and achieved competitive performance, predicting co-aggregate formation with 0.94 accuracy.29 Several predicted co-aggregates from the test set were experimentally validated to verify the platform’s predictive capabilities. Compared to other nanoformulations, the new excipient-drug pairs generally enabled the suspension of higher drug concentrations.
A data analytics-guided formulation design platform could potentially expand the candidates for excipients, which may be needed to limit potential excipient-induced pharmacological side effects via increasing drug loading and enabling rational excipient selection. Although excipients are defined as inactive and nontoxic materials, studies have suggested that they are not all “biologically-inert”. Data analytics showed that excipients in oral medications may cause allergies and intolerances within certain subpopulations, and these excipients are termed adverse reaction-associated inactive ingredients (ARAIIs).53 For instance, cases of polyethylene glycol (PEG) allergy were reported in various medications, and a recent case of postvaccination anaphylaxis caused by PEG allergy necessitated a re-evaluation of these inactive excipients.54 Some excipients carry biological functions that enhance bioavailability of the API. For instance, Pluronic block copolymers modulate activities of drug efflux transporters, such as P-glycoprotein.55 Pottel et al. interestingly found that over 30 known excipients have over 40 biological targets, and combining large-scale computational screening with targeted experimental testing, they demonstrated excipient biological activities both in vitro and in vivo models.28 A major outcome of this work is the need to redefine excipients and explore new biological activities for known excipients.
3.2. Design parameters of nanomaterials in drug delivery
The physicochemical properties of nanomaterials are critical engineering elements that must be tailored for each problem.56 These properties are often tied to nanomaterial colloidal stability, cellular uptake efficiency, tissue targeting, and likely every step of absorption, distribution, metabolism, excretion, and toxicity (ADMET).42, 57–62 A traditional controlled approach to study the impact of the physicochemical properties involves changing one variable of the particles at a time while keeping all others constant, and systematically evaluating the resulting biological efficacy. However, simple linear correlations are not sufficient to fully model the intricate relationship between the physicochemical properties and the fate of nanomedicine in vivo and clinics. Various prototype machine learning algorithms have tackled aspects of nanomaterial design and nanomedicine pharmacokinetics/pharmacodynamics (PK/PD) to streamline optimized nanoformulations.
The most common physicochemical properties of nanomedicines include size, shape, and surface charge, and these properties are frequently used as features in machine learning. Traditional data analysis approaches have found compelling evidence that these physicochemical properties play an important role in biological behaviors or therapeutic efficacy. The size dimension of nanomedicine makes it unique in biological and theranostic applications. Nano-sized materials are broadly defined between 1 to 1000 nanometers, with most nanomedicines less than 100 nm.63 The wide distribution in size enables nanoparticle engineering for various biological potentials and introduces heterogeneity for treatment. For example, to design nanomedicines for cancer therapeutics, a fine balance needs to be achieved between the retention and permeability in tumor tissues: while larger nanoparticle sizes increase the retention time, they decrease exposure to the tumor.64 In addition, the biodistribution of nanomedicine is also impacted by the cutoff sizes of different organs. Nanoparticles with diameters larger than 200 nm accumulated in the spleen, while particles smaller than 50 nm localized in the liver.64–66 Nanoparticles less than 6 nm were filtered by the kidneys,64 and the size from 50 nm to 100 nm nanoparticles have a higher apparent permeability across the blood brain barrier.66 However, many of these studies only considered one type of nanomaterial, and thus the results can drastically change when other physicochemical properties are considered simultaneously.
The shape of the material has been inspired by evolutionary and microbiology but is less commonly studied in a therapeutic aspect.67, 68 Most nanomedicines to date have been classified as spherical nanoparticles, despite a wide range of nanomaterials with lower dimensions such as rods, sheets and other defined shapes, like carbon nanotubes.56 Studies have shown that nanomaterial shape can influence transport motion. Oblate-shaped particles, rather than spheres, are transported in a tumbling manner that enhances the lateral drift of particles towards the blood vessel walls.56, 69, 70 Shape has also been shown to influence clearance via the reticuloendothelial system, which prolongs nanoparticle circulation time and enhances the probability of reaching the target site. Nanoparticle shape can also influence specific cell interactions. The amount of surface area for multivalent interactions can impact the adhesion energy between particles and cells.56 Shape control is difficult in many nanomaterial synthesis methods, but technologies like DNA origami can expand the reservoir of nanomaterial structures. DNA origami involves folding DNA to create 2D and 3D nanoscale structures for drug delivery, biosensing, and biomolecular computing.71, 72 This computer-aided design is highly flexible, tunable, and reproducible as it generates homogenous samples. One study shows that the compactness or aspect ratio of the particle shape can influence the internalization efficiency and kinetics of cellular uptake.73
The surface charge of nanomaterials affects colloidal stability and biological interactions.74 A negative surface charge, or zeta potential, stabilizes particles in a suspension and promotes cellular absorption to the membrane.57 Interestingly, the nanomedicine charge also influences organ targeting. A new technology named selective organ targeting nanoparticles (SORT) demonstrated the potential for organ-specific gene therapy.75 Lipid nanoparticles with increasing concentration of positively charged lipids drive the cargo transport from the liver to the spleen. Eventually, they accumulate in the lung concentration-dependent, whereas adding negatively charged lipids specifically enhanced spleen delivery.75
Surface chemistry is another critical component of nanomaterial design that facilitates interactions within biological systems. Recent analyses have shown that less than 1% of administered nanoparticles reach their target site within murine tumor models.76 Targeting moieties can improve biodistribution, and surface functionalization can mediate biological interactions. Nanoparticle surfaces have been modified with moieties such as antibodies,77, 78 aptamers,79, 80 carbohydrates,81, 82 and polymers83 to modulate physiological stability, cellular uptake mechanisms, and tissue targeting.84 For example, natural product, such as polysaccharides, has been integrated into delivery formulations not only for their stability and synergistic therapy but also for tumor targeting.85, 86 Negatively charged sulfate groups have made certain polysaccharides, such as fucoidan, interesting moieties to assist in the self-assembly of water-insoluble drugs into nanoparticles,85 and its nanomolar binding affinity to P-selectin has been used for targeted therapy in solid tumors.82, 87 Recent work by Alafeef et al investigated the relationship between nanoparticle physicochemical properties and cellular internalization for various types and stages of cancer using machine learning. The results from an artificial neural network model suggest that cellular internalization is more strongly dependent on surface chemistry and cell type than other parameters tested.88
The effect of the protein corona on nanomaterial interactions with biological environments is of increasing interest. Nanoparticle stability, biodistribution, and toxicity can be largely influenced by the proteins and other biomolecules absorbed onto their surface.89–91 Heterogeneity, complex dynamics, and large proteomics profiles all pose a large challenge for protein corona analysis, and it is a suitable application for machine learning-based methods.92 Recent work by Ban et al. harnessed machine learning and meta-analysis to deconvolute the complexities of functional proteins in nanoparticle protein coronas.14 Many factors, including properties of both nanoparticle surface and serum proteins, can influence nano-bio interactions and protein corona formation. Simple linear regressions fail to make accurate predictions. Therefore, the group developed a machine learning platform to investigate the relationship between protein corona formation and targeted cellular uptake, parsing through quantitative and qualitative properties. The first step of the computational analysis was to predict the protein composition and isolate functional components. Since current nanomaterial databases lack comprehensive reports about protein corona, literature mining remains one of the top strategies to obtain sufficient datasets. The authors mined 56 papers for reports of the composition of protein coronas, and a set of nanoparticles with categorized physicochemical properties were generated for machine learning algorithm training. A random forest model was used to accurately identify key proteins in the protein coronas of various nanoparticles to make predictions with a small, heterogeneous dataset.14 Still, overfitting the prediction model was a concern because of the small sample size. In addition to estimating the prediction using a cross-validation approach to combat overfitting, feature importance analysis was used to identify minimal features for model training. In this way, sensitivity was balanced with generalizability. With refined features, the random forest model achieved high R2 values (> 0.7) with low error scores (below 5%). Experimental identification of protein composition verified the machine learning predictions.14
The above study also analyzed the essential factors needed for accurate prediction, indicating that surface modification highly influenced protein interactions.14 Additionally, this analysis emphasized the heterogeneous distribution of factors that influence corona formation in each formulation. Investigating the epitopes of the overall corona, as opposed to specific protein composition, can give insight into binding mechanisms and, therefore, cellular uptake. Functional epitopes of the protein corona can be mistakenly recognized as exogenous matter and cause unwanted immune or inflammatory responses. Protein compositions were tightly correlated (R2 values ≥ 0.8) with uptake efficiencies, pro-inflammatory markers, and immune disturbances.14 This model can predict the physicochemical properties of the protein corona and identify unique fingerprints, thereby guiding nanoparticle design for complex biological environments and specific biological activity.
3.3. Machine learning predictions of nanomedicine biological efficacy
One of the most significant hurdles when predicting nanomedicine biological efficacy is a poor translation from in vitro to in vivo studies. Nanomedicine therapies often do not display therapeutic benefits in vitro because nanoformulations often address immune systems, peripheral tissues, and first-pass metabolism issues. All of these are poorly recapitulated in cell culture. Thus, studies rely on slower in vivo work, and it is often challenging to parse out a single factor from nanoformulation that ultimately contributes to the therapeutic benefits. Machine learning has been incorporated into nano-drug development to expedite the screening process. The research focusing on a particular structure-activity relationship in nanomaterials are currently situation-dependent; most groups chose to incorporate high-quality and specific data points by in-house high-throughput screening. However, recent work also has shown success by mining literature data. Recent work by Yamankurt et al. coupled high-throughput screening with machine learning to assess spherical nucleic acid (SNA) immune activations and identify cancer-vaccine candidates.93 At first glance, the SNA properties did not correspond systematically with biological activity, as measured by TLR9 activation. In a high-throughput fashion, SNAs were rapidly formulated and probed for immune activity using mass spectrometry. Yamankurt and colleagues trained several cross-validated machine learning algorithms, namely linear regression, logistic regression, and non-linear XGBoost, using combinations of SNA properties.93 As the variable to be predicted, each sample was associated with the experimentally measured immune activity. Generally, as the number of features increased, the performance increased, eventually plateauing.
Furthermore, the most predictive features remained the same and distinguishable despite the feature vector size. The team further investigated whether a small, expansive library of SNAs would be sufficient for model training by randomly selecting sample subsets. This work investigated the possibility of limited sample numbers to be used to explore a large design space. Non-linear models achieved higher performance overall (the highest being XGBoost with Q2 = 0.83, where Q2 refers to the R2 of the test set) because of their ability to predict complex trends.93 This machine learning-based analysis captured optimal structure-activity relationships, using minimal SNA synthesis to predict immune activation.
While machine learning algorithms have been increasingly used to model complex nano-bio interactions, comprehensive interpretations and understanding of feature interactions remains a challenge. Yu et al. investigated feature importance and created a feature interaction network in order to overcome some of the interpretation limitations. 94 Through literature searches and manual filtering, the authors built a random forest model (with R2 values > 0.75) to predict immune response and organ burden of a specific nanoparticle treatment. They accomplished a multiway importance analysis for their features to reduce bias. Feature interaction networks are important for modeling the complexity of immune toxicity and give insight into feature relationships, such as synergy or antagonism. The feature interaction network highlighted a clear correlation between nanoparticle properties and immune response or organ burden. For example, the network showed an influence of zeta potential on protein corona formation and nanomaterial uptake, as well as a correlation between the length of nanoparticles and severity of immune response. 94 The improved interpretability of this framework offers specific guidance on nanoparticle design and application.
Another challenge in the nanomedicine field is determining the mechanism of drug release in vivo. When a nanoparticle enters the complex solid tumor microenvironment, it interacts with barriers and cell types differently. To maximize therapeutic benefits in these cases, understanding organ/tissue interactions is critical. Kingston et al. used machine learning image analysis to monitor micrometastasis targeting and predict nanoparticle delivery.95 Tissue imaging with machine learning analysis generated a predictive model for the nanoparticle entry to the micrometastasis. Imaging techniques, including tissue clearing and 3D microscopy, produced complex images from which machine learning methods could extract tremendous amounts of data with single-cell resolution. The machine learning model was trained to differentiate between biological structures such as blood vessels, nuclei, and metastases. This model outperformed other image analysis techniques because it learned important profiles that contribute to structural differentiation from the images. Using these profiles, Kingston and colleagues found a higher degree of nanoparticle accumulation in the micrometastasis than in the primary tumor tissue. Using a Gaussian support vector machine (SVM) algorithm, the platform was expanded to predict nanoparticle delivery in different micrometastasis types. With a Pearson correlation of 0.94, the SVM model successfully predicted the number of cells with nanoparticles, the mean nanoparticle intensity, and the density of nanoparticle-positive cells.95 Micrometastasis imaging and machine learning-enabled accurate nanoparticle delivery predictions based on specific pathophysiology.
3.4. Challenges of machine learning efforts in nanomedicine
While AI efforts have made great strides in informing nanomaterial design, their transformational change has several significant limitations. One of the most ubiquitous challenges those developing predictive platforms face is the need for large, unbiased datasets for model training. AI algorithms need to be fueled by large amounts of data in order to generate robust predictions and distinguish between drug delivery methods.20, 96 Validation, and interpretation of machine learning results are crucial for successful platform development, both of which can be strengthened with comprehensive datasets. Currently, there are two main approaches for consolidating data into valuable datasets for machine learning. Research groups mine literature for published data and create a database for their analysis.14, 15, 97 This technique includes specific keyword searches in literature databases, such as Scopus and Web of Science, and manual inspection to select relevant data. Alternatively, groups have used their experimental data, including previously published data and results from high-throughput experiments.29, 50, 93, 98
With each research study conducting curation efforts independently, many investigations lack generalizability. Standardizing data procurement methods, broadening the net to capture more relevant data sets, and creating an unbiased dataset has proven challenging. For example, nanomaterial size can be reported differently depending on the instrumentation and analysis method. DLS remains the most commonly used method for nanoparticle size measurements. However, the results tend to appear larger than sizes determined by microscopy due to higher sensitivities to particles with larger diameters.99 In addition, analytical methods to extrapolate dimensions from the autocorrelation functions in DLS also vary based on the samples’ volume, number, or intensity. In-house data tend to be specific for one group’s purpose, and the literature is difficult to effectively and comprehensively access. There is a need for broad, standardized databases that are accessible to everyone in order to facilitate robust machine learning analyses. With more attention paid to data curation efforts and increased collaborations between computer, material, and biological scientists, machine learning can be further exploited to strengthen current predictive platforms and uncover new insights from the data produced by many investigators (Figure 2).
Figure 2.
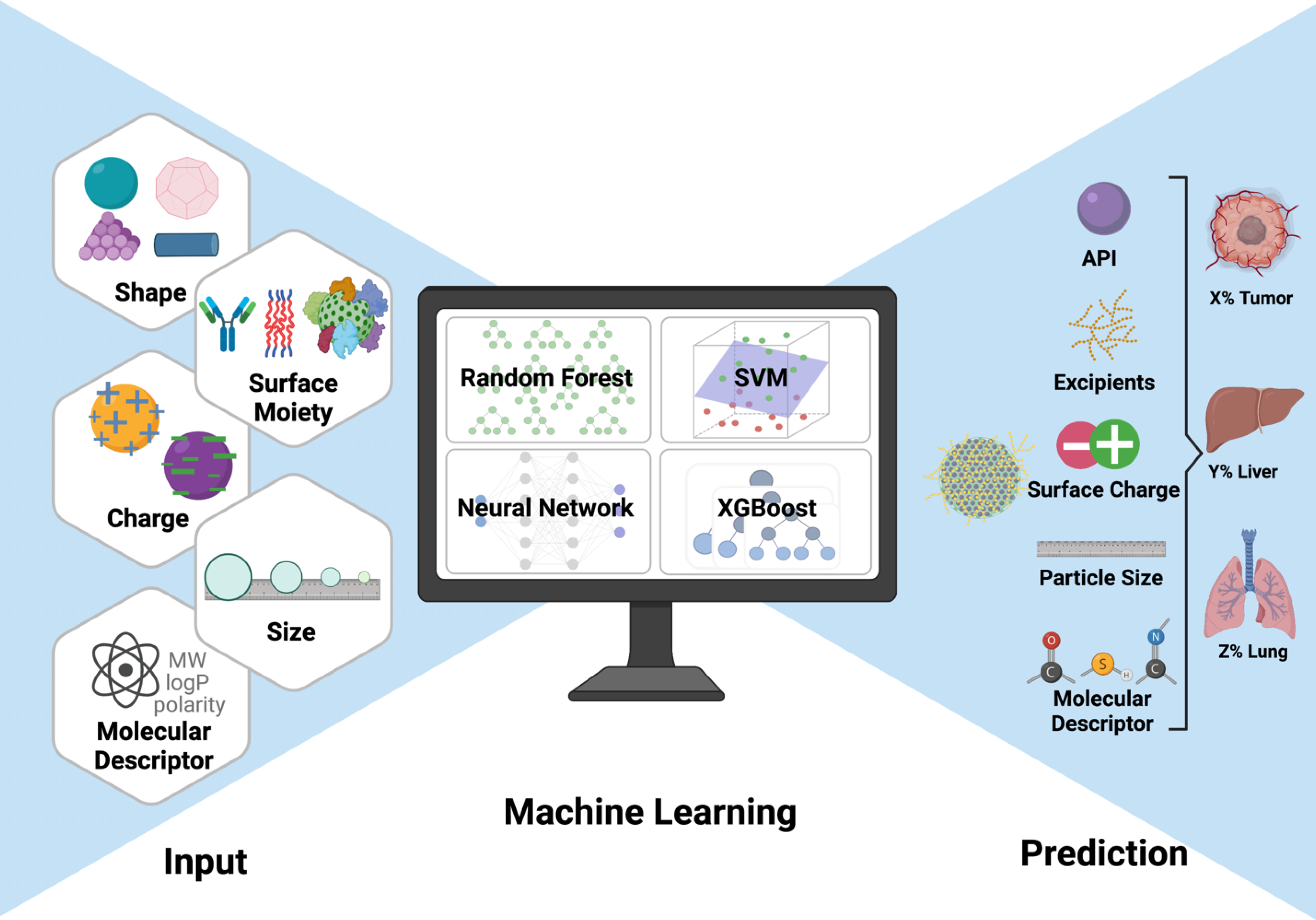
Machine learning platforms are trained using physicochemical and biological features to predict the biodistribution of nanomedicines accurately. The generalization of the platform can promote optimization of the biological efficacy and improvements towards clinical translations of nanomedicines.
4. Data curation for nanomedicine
Recognizing the need for a centralized nanomedicine database to strengthen analyses, nano-informaticians create platforms that accommodate the storage and sharing of heterogeneous nanomedicine data.6, 7, 10 This process falls within a major area of data science - data curation. Historically originated in library science, data curation consolidates data from various sources into one database.100 In addition to categorizing data into a useful presentation, data curators also involves extracting, standardizing, and repurposing the datasets.101 Data curation has been heavily used in other scientific and biomedical fields such as chemistry, polymer sciences, toxicology, and biology.102–107
Data curation for biomedical science research today is critical yet often underestimated. Biomedical data generated from bench to bedside can easily overwhelm the scientists without proper data curation tools. Common databases like The Universal Protein Resource (Uniprot), The Drugbank Database, National Center of Biotechnology Information (NCBI), or The Protein Data Bank (PDB) were all established with tremendous curation efforts and have become inseparable resources for day-to-day biomedical research. Recently, an increasing demand for curating comprehensive, accessible and up-to-date nanomaterial databases is unavoidable for nanomedicine discovery. Access to nanomaterial data is essential for further development and optimization of novel technologies via data-heavy computational methods. Despite many nanomedicine publications and patents, extracting and consolidating relevant information for data analysis has been a challenge. Furthermore, research and industry communities lack the ability to perform efficient data exchange. As a result, research groups curate their own databases based on in-house data or manual literature searches for their specific purpose such as targeting organs, increasing cell uptake, or avoiding toxicity and side effects.76, 108 While individual initiatives for dataset curation have been successful, there is a need to increase collaborations between research laboratories, federal agencies, and private industries in order to encourage data sharing, establish standardization guidelines, and provide sufficient data for analysis platforms.109
4.1. Efforts in nanomedicine data curation
Data curation is necessary to compile and standardize nanomaterial data in order to facilitate the development of robust nanomedicine platforms. This process requires a combined effort between researchers generating the data and curators establishing and maintaining databases.97, 110 To accommodate the increasing amount of nanomaterial information, several nanomedicine/nanomaterial-specific databases have been developed in the past decade. A list of these databases follows (Table 2):
Table 2:
Summary of key databases.
| Database | # Data Points | Data Source | Key Feature | Total citations |
|---|---|---|---|---|
| caNanoLab | >1480 | Data contributed by individual laboratories | Annotated, streamlined property database and design protocol for nanoformulations | 52 |
| Nano | >350,000 | Literature | Comprehensive nanomaterial database | N.A. |
| Public Virtual Nanostructure Simulation | >700 | Data contributed by individual laboratories | Quantitative modeling of nanocomposites. | 21 |
| S2NANO | >33,000 | Data contributed by individual laboratories & Literature | Promoting safety and sustainability in nanomaterials use. | 59 |
| Nanoparticle Information Library | >85 | Data contributed by individual laboratories | Promoting standardization of nanomaterial information | 734 |
| eNanoMapper | >5,500 | Data contributed by individual laboratories & Literature | Virtual platform for accessing tailorable information on nanomaterials | 166 |
| Excipient and small molecule databases | >120 million | Literature | Comprehensive database on small molecules and excipients. | 8 |
caNanoLab
caNanoLab (https://cananolab.nci.nih.gov/caNanoLab/#/) was established by the National Cancer Institute (NCI) in 2007 to generate a comprehensive database of nanotechnologies in the field of cancer care.111 The main goal of caNanoLab is to accumulate data relevant to nanomedicine design, such as pre-clinical materials safety, drug efficacy, nano-bio interactions, and characterization, to assist with the clinical translation of nanomedicine strategies. The caNanoLab database includes complete and comprehensive descriptions of the data in line with data sharing guidelines of the National Institutes of Health (NIH).
The caNanoLab portal enables exploration in three main categories: protocols (135 results), samples (1477 results), and publications (2150 results). The protocol section provides vetted assays for physicochemical, in vitro, and in vivo characterization, as well as sample preparation and synthesis procedures. Furthermore, samples can be filtered by nanomaterial type, functionalization, and function (application). The database contains a broad range of nanomaterial types, including dendrimer, polymer, fullerene, liposome, micelle, metal oxide, nanorod, and carbon nanotube, each associated with characterization parameters.
To ensure that the caNanoLab data is accessible and to promote database development, the NCI Alliance for Nanotechnology in Cancer manages the data curation process with assistance from data coordinators and the NCI data curator.112 NCI-funded nanotechnology grantees are expected to share their data via caNanoLab portal and often name their own data coordinators to do so. Proper annotation methods are detailed on the caNanoLab portal to ensure consistent and comprehensible nanomaterial annotations. The NCI curator verifies NCI grantee data, extracts data from relevant publications, assists with accurate annotations, and organizes the data to maintain a clear and accessible database for users. Coupling manual curation with curation algorithms, caNanoLab has the potential to significantly increase the curation rate while ensuring the quality of the curated data.
Nano
Nano (https://nano.nature.com), founded by Springer Nature to provide solutions for multidisciplinary research. With over 350,000 different nanomaterials, 970,000 nanotechnology-related publications, and 43 million nanotechnology-related patents, Nano consolidates nanotechnology information into one comprehensive database. In addition to manually curated data, Nano includes insights from publications and patents accomplished using AI technology. Furthermore, Nano allows several search tools and advanced filters to explore nanomaterial characterizations and applications. For example, a user can search for a specific result, such as toxicity, of the desired nanomaterial in a disease model of interest. Nano sets the standard for accessible nano-databases by allowing simple navigation using specific queries. However, ‘Nano’ will be retired by June 30th, 2022 as a standalone platform but the data will be incorporated into other existing platforms.
Public Virtual Nanostructure Simulation
The Public Virtual Nanostructure Simulation (PubVINAS) is a nanomaterials database established by the Zhu research group at the Rutgers University-Camden (http://www.pubvinas.com). The PubVINAS database contains curated physicochemical and biological information for over 700 nanocomposites from 11 different materials. The platform also includes an online modeling tool, developed using curated data113 and can generate PDB files for each nanomaterial along with instructive nanostructure annotations. These files can be used to calculate more than 2,000 molecular descriptors to be used in predictive models. As a result, this platform is one of the first databases that promote nanoinformatics and novel nanomaterial design.
S2NANO
Safe & Sustainable Nanotechnology (S2NANO) is a community, established by a research group in the Republic of Korea, with a mission to develop nanotechnology that is safe and sustainable for humans and the environment. The online portal (http://portal.s2nano.org/) share protocols, curated datasets prediction models and general support for safety assessments of nanomaterials. The goal of the S2NANO database is to mitigate concerns regarding nanomaterial hazards and assist with regulatory compliance. Ultimately, S2NANO hopes to facilitate “safety by design” development strategies.
The S2NANO database is comprised of experimental nanomaterial data for over 33,000 nanomaterials, divided into two major components. The “core” database includes physicochemical properties and cytotoxicity data from 46 commercial or synthesized nanomaterials, generated using S2NANO’s measurement and analysis protocols. The “extended” database is composed of curated data from the literature, extracted manually using keyword searches in various literature databases such as Google Scholar, PubMed, and Web of Science. Since multiple research groups contribute to the data curation, the consolidated physicochemical data are verified, and missing values are estimated using the published screening and data gap filling methods.49 Specific materials in the database can be queried and filtered by nanomaterial, regulation, or product information. Each search result contains material descriptors, toxicity results, and physicochemical properties.
The S2NANO interface also provides 14 built-in predictive methods to analyze samples from the database. These platforms utilize algorithms such as random forest, logistic regression, and support vector machine in addition to in silico QSAR to investigate the relationship between physicochemical properties and toxicity results.114 By providing this platform, S2NANO enables users to take advantage of the pre-processing procedures for database standardization in order to properly prepare the data for further analysis.115
Nanoparticle Information Library
The Nanoparticle Information Library (NIL) (http://nanoparticlelibrary.net/index.asp) is a public database created by The National Institute for Occupational Safety and Health (NIOSH), in conjunction with their global partners, to facilitate nanomaterial characterization sharing. The goal of the NIL is to provide a centralized mechanism to catalog nanomaterial information in order to accommodate the increasing and distinct nanomaterial research projects around the world.116 By housing public nanomaterial information, the NIL enables collaborations, provides curated data for research groups, and ensures the safety of the workers handling the nanomaterials.
The database includes information regarding nanomaterial composition, method of production, physicochemical properties, applications, and relevant publications. Nanomaterials can be searched in the library by their structure, element composition, origin, and size range in addition to a regular keyword search. Each result is a publication that used the nanomaterial of interest and includes a report that links relevant figures, the contributing lab, and related publications. NIOSH partners in Oregon State University (OSU) manage the website and characterize nanomaterials to fill the database. Additionally, the NIL team encourages users to submit their own experimental data by contacting the NIOSH representative.
eNanoMapper
The eNanoMapper database (http://www.enanomapper.net/) is framework to search and analyze inter-laboratory nanomaterial data. The platform was developed and administered by a group of 8 partners, with expertise spanning the fields of nanotechnology, information technology, community building, and computational modeling.13, 117–124 With a goal to provide a flexible model that is accessible to any researcher, the eNanoMapper database provides both a convenient interface and relatively simple computational access that can be tailored to a specific need. Comprehensive tutorials and reference pages are included on the website to assist with database navigation. Furthermore, the database is structured in a spreadsheet format to be compatible with data uploads and useful for downloads.
The database is populated with experimental results provided by the contributing partners in addition to data uploaded from research groups.13 Data can be filtered by categories such as nanomaterials, protocols, physicochemical properties, methods, and references. Each search result includes information on the material composition as well as physicochemical properties and toxicity data. Built-in QSAR modeling software is under development to facilitate the use of eNanoMapper database results as inputs. Incorporating flexible data uploading capabilities, data analysis platforms, and security measures in place for data protection, the eNanoMapper database potentially simplifies data curation and nanomaterial analyses.
Excipient and small molecule databases
Recent studies have emphasized the utility of excipients and other small molecules in the design of novel nanoparticles. For this reason, several databases have been established. ZINC (https://zinc.docking.org) is a public database that houses comprehensive information about small molecules. Created by the Irwin and Shoichet Laboratories in the Department of Pharmaceutical Chemistry at the University of California, San Francisco (UCSF),12 ZINC serves as a useful tool for both chemoinformatics and biologists. The platform contains ligand, target, biological activity, and purchasability information of over 120 million purchasable “drug-like” compounds. In addition, the same group founded a smaller excipient-specific database named the Excipients Browser (https://excipients.ucsf.bkslab.org/index). With the goal to enhance drug delivery formulations, the Excipients Browser provides comprehensive information about dosage, administration routes, and biological activities for over 3,000 excipients.
4.2. Data mining: strategies to automate data curation
It has become nearly impossible for individual scientists to thoroughly review the entire scientific literature and gather valuable information, prompting literature mining algorithms. Technological advancements in text and data mining enable researchers to automate specific data curation processes.125–127 These approaches can be used to access literature data to populate nanomaterial databases efficiently. In the biomedical field, automated literature search and text and data mining were used to identify relationships and interactions between diseases and genes128 and proteins and drugs.127 Text and data mining methods rely on natural language processing (NLP), where the computational effort analyzes scientific texts to extract important information.129–131 Several of these tools use unique visual outputs to facilitate understanding of the search result. For example, Coremine (www.coremine.com) and Embase are databases to review >40 million papers on PubMed and summarize valuable information in textual and visual ways such as word clouds and connector plots. In this manner, one can automatically extract useful information from millions of papers by focusing a query with relevant keywords. For example, applying the keywords “Liposomes” and “Drug Delivery Systems” in Coremine yields lists of chemical ingredients, relevant genes, and biological processes (Figure 3). However, in this example, the list does not give the ratio of the lipid combinations or rankings. The biological processes include the most studied techniques in liposomal formulations, such as uptake and membrane fusion. Furthermore, one can use those lists to extract chemical structures further, calculate molecular descriptors, and make predictions.
Figure 3.
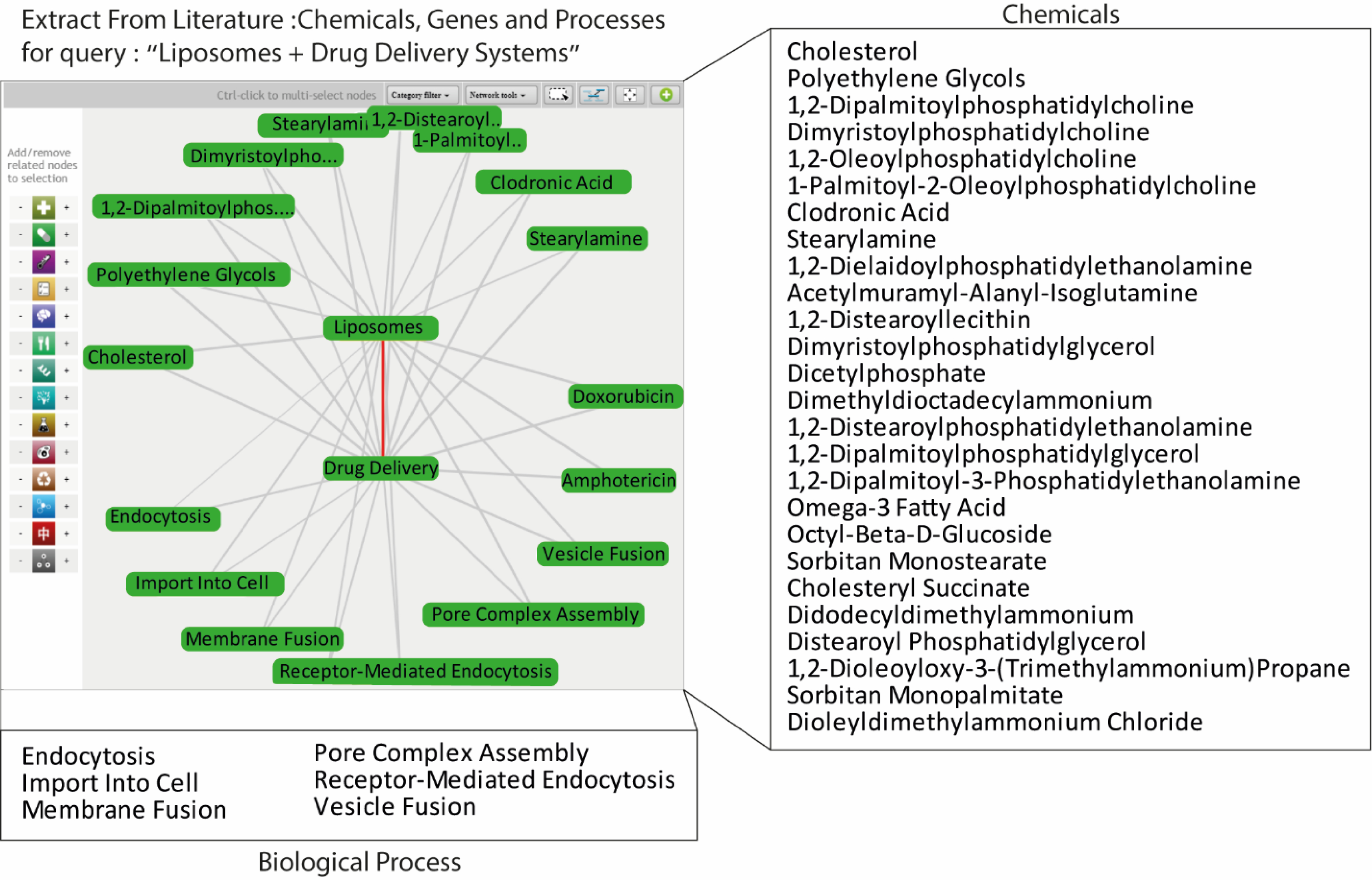
Data mining of chemicals and biological processes related to liposomes using the Coremine database.
Launer-Wachs et al. developed a SPIKE-KBC platform for rapid and personalized knowledge base construction, aimed at both individual researchers and large teams, without prior experience in bio-curation, NLP, or machine learning. In this approach, the basic paradigm is rooted in extractive search (ES), which combines interactive search with NLP-aided knowledge extraction. The extracted knowledge is represented with entities and their relations captured from the literature using the SPIKE engine. The user can then briefly or thoroughly annotate the links for quality control, resulting in a personalized knowledge base. This approach is highly compatible with the main hypotheses in the field of targeted drug delivery, suggesting that a biomaterial A can carry drug B to disease C by actively binding a molecular target D expressed on cell type E with ligand F. In this work, Launer-Wachs et al. used the ES to capture all the known relations between A-F in a relational structure which is shown in Fig 4a. A comprehensive knowledge base for cell-specific drug delivery (CSDD) was constructed using the SPIKE-KBC app, aimed at supporting the broader research community. The extractive search was used to identify pairwise relation candidates from the literature automatically, and then all of the added relations were manually validated by three annotators. This was done to ensure the high precision expected from a public resource. The CSDD consists of 5182 associations between 64 biomaterials, 70 cancers, 20 cell types, 478 drugs, 254 ligands, and 199 targets. It allows researchers to visually navigate through complex hypotheses and assemble new ones (Fig 4b). It also offers the possibility to perform a quantitative meta-analysis of the field, such as the most studied hypotheses of different levels of complexity. For example, the most studied ‘biomaterial-drug-cancer’ trio is ‘Liposomes-Doxorubicin-Breast cancer’, and the most studied ‘cell-type -target cancer trios are ‘Cancer cells-HER2-Breast cancer’ and ‘B cells-CD20-Lymphoma’. This knowledge base can serve as an ever-growing, living review to map the progress of the whole field. The CSDD knowledge base is available at: https://spike-kbc.apps.allenai.org/projects/csdd
Figure 4.
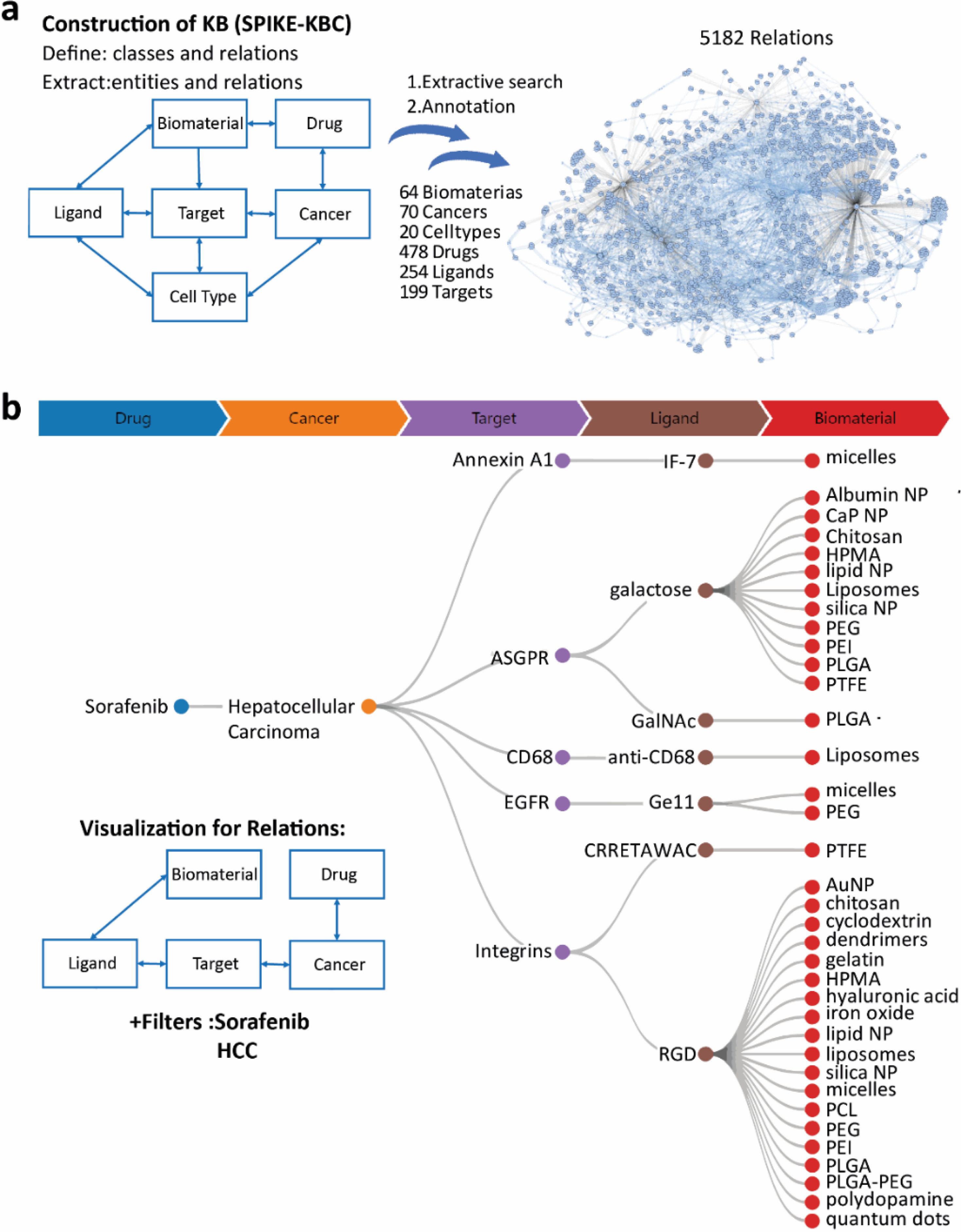
Construction of the cell-specific drug delivery (CSDD) knowledge base. a) Construction scheme and network view of total knowledge base relations between classes. b) Representative visualization and user interface of the tree-view after applying the filter for sorafenib and hepatocellular cancer carcinoma.
4.3. Data curation: challenges and solutions
Multiple nanomaterials databases have been established to provide users with large sets of multiple types of data. However, these platforms have yet to be employed to conduct ‘big data’ analytics studies, which could provide insights for the field as a whole. Applications of these databases are undervalued based on the publication records given the potential of each website (see Table 2). There are several possible reasons that integration of these curated databases into machine learning studies is relatively low. First, the nanomaterial databases are fairly new, and thus many research groups, especially machine learning-focused groups, are still not aware of the existence, size, and potential of these databases. Second, the key features of various databases are different (see Table 2). Depending on the goal of machine learning projects, and the curation strategies of the databases, certain features may not directly translate to usable training sets. The process to extrapolate information from the database can be as costly as traditional data mining methods. Third, unlike measurements like NMR, for small molecules, or the sequence of a nucleic acid, standardized metrics to compare nanomaterials across studies are largely lacking. This has posed a large challenge for standardization efforts. One of the key concerns regarding curation and utilization of these databases is that physicochemical properties of nanomaterials depend on both intrinsic and extrinsic conditions. For instance, nanomedicines formulated in different buffer solutions can greatly affect their surface charge, size, or stability; the site of action for the nanomedicines can change the releasing rate where the discrepancies can occur between acidic environment like tumor microenvironment and neural physiological conditions. Unfortunately, experimental methods and data types are highly dependent on the project, material, and laboratory, and the inability to toggle parameters in the existing databases can be another reason for slow adoption. Fourth, the physicochemical properties of nanomedicines can also be changed by APIs encapsulated. Different APIs used in studies can also contribute to the heterogeneity in the literature data when comparing a specific API with different formulations or a specific formulation with different APIs.
Advances in nanomaterial development are also propelled by inventions of autonomous systems.132 For example, microfluidics technology has brought important control capabilities to nanomedicine synthesis. By precisely controlling the flow rate and temperature, microfluidics drastically decreases batch-to-batch variations during the synthesis and can be easily scaled-up or down based on the needs of the research labs. Microfluidics also potentiates the standardization efforts on reporting some of the nanomaterial synthesis. Parameters like stir rate, time, and temperature now can be accurately quantified in the instrumental settings and make them easily reported and reproduced among labs given a promising quality control for the instruments.133 Other automated technologies can also have great impact on studies of many aspects of nanomedicine research, like high-throughput DLS systems, automated microscopy technology, robotic liquid handling systems, or bioreactors. Experimental automation and digitalization can address data curation challenges and increase the quality of the datasets. However, it is not trivial to streamline all nanomaterial characterizations. Nanomaterials can range from stable entities (e.g., carbon nanotubes) to metastable systems (e.g., drug aggregates), and the latter ones are more prone to changes in environmental factors. Although an in-house screening can generate more homogenous datasets, literature datasets are heterogenous. Subsequent machine learning studies can also be largely affected by the data source. For example, in-house screenings may produce a more predictable model, but they may be challenged by the datasets outside of the research lab.
Input from nanomedicine researchers, database developers, and AI researchers is important for taking advantage of the capabilities of data analytics to improve nanomedicines. Databases should be continually enriched with data from research groups in addition to published data, requiring assistance from individual labs and/or incentives from publishers, funding agencies, or other entities. AI researchers can help structure nanomaterial databases in ways that facilitate data integration into AI platforms. To enable this, data formats should be standardized. For research scientists, reporting thorough synthetic procedures, instrumentation uses, and highlighting the important material features in publications can fundamentally help to improve the data standardizations, quality control, and reproducibility.134 For data scientists, understanding the discrepancies between different instrumentations and focusing on key features that are commonly used in biomedical research is also a starting point for the collective efforts. Organizations and committees around the world have contributed tremendous work to standardize metrics in nanomaterials, and this works should be implemented into research and development in the future.135 Curated databases will be relevant to chemical, structural, biomedical, and computational scientists and will improve our understanding and facilitate the development of nanomedicines.
5. Conclusions and Outlook
In the past decades, the field of nanomedicine has rapidly expanded preclinically and clinically. However, many practical obstacles have delayed the translation of most pre-clinical research into the clinic, including formulation difficulties/scale-up, unsatisfactory PK/PD, and costs.136 We believe these issues can be ameliorated if the field can learn from the massive amounts of data already collected by the field. Recent advances in “big data” analytic approaches, including machine learning, have shown an exciting potential to improve processes in many fields. However, the complexity and heterogeneity of nanomaterials has hindered machine learning efforts to design and clinically translate many potential candidates. On the one hand, comprehensive research done in academic and industry laboratories has been fruitful and resulted in novel nanomedicines entering clinical trials, but, on the other hand, this research has generated enormous amounts of nanomaterial characterization data that has not been fully utilized. As a result, a substantial amount of money is spent funding research to optimize nanoformulations. Currently, AI efforts are primarily developed using data from one or few laboratories. We believe that the integration of data curation and AI platforms will be instrumental for researchers to capitalize on current nanomaterials for the rational and streamlined design of novel nanomedicines.
In this review, we presented the recent capabilities of machine learning algorithms to develop novel nanomedicines and characterize their biological activity and interactions. Machine learning platforms have become popular methods to design novel nanomaterials and predict bioavailability, biocompatibility, cell uptake, treatment efficacy, and toxicity. However, researchers commonly concede that the main limitation with machine learning platform development is the lack of data. Training algorithms to make robust predictions requires enormous amounts of heterogeneous data.
Nanoinformatics researchers have started to focus on nanomedicine data curation. We highlighted several curation initiatives that paved the way for generating comprehensive and accessible databases to facilitate downstream analysis. While these platforms are mostly populated using manually curated data from literature, text, and data mining, algorithms are increasingly used for automatic data curation. These algorithms can extract relevant data from thousands of publications using specific keywords. However, issues remain before these types of databases can be effectively used for most machine learning analyses due to the standardization of data formats, for instance. The field should agree upon some standards such as the best light scattering processing algorithms, such as providing annotation, and considering depositing most data in a central database and integrating text and data mining methodologies. Additionally, funding authorities should continue to improve efforts to encourage scientists to contribute to these databases to promote data curation efforts actively.
The integration of AI platforms will further nanomedicine research and facilitate clinical translation. By creating a community for data sharing and user-friendly platforms, scientists can learn from each other to improve nanomedicines. Moreover, computational design methods can minimize lengthy trial-and-error bench experimentation and reduce animal usage. We believe that a partnership between nano-informaticians and researchers developing intelligent platforms are essential for the success and development of nanomedicines. We hope that this review will encourage such collaborations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barenholz Y, Doxil®--the first FDA-approved nano-drug: lessons learned. J Control Release 2012, 160 (2), 117–34. [DOI] [PubMed] [Google Scholar]
- 2.Hou X; Zaks T; Langer R; Dong Y, Lipid nanoparticles for mRNA delivery. Nat Rev Mater 2021, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anselmo AC; Mitragotri S, Nanoparticles in the clinic: An update post COVID-19 vaccines. Bioeng Transl Med 2021, e10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manzari MT; Shamay Y; Kiguchi H; Rosen N; Scaltriti M; Heller DA, Targeted drug delivery strategies for precision medicines. Nature Reviews Materials 2021, 6 (4), 351–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun D; Zhou S; Gao W, What Went Wrong with Anticancer Nanomedicine Design and How to Make It Right. ACS Nano 2020, 14 (10), 12281–12290. [DOI] [PubMed] [Google Scholar]
- 6.Maojo V; Martin-Sanchez F; Kulikowski C; Rodriguez-Paton A; Fritts M, Nanoinformatics and DNA-Based Computing: Catalyzing Nanomedicine. Pediatric Research 2010, 67 (5), 481–489. [DOI] [PubMed] [Google Scholar]
- 7.Maojo V; Fritts; De La Iglesia D; Cachau; Garcia-Remesal; Mitchell; Kulikowski, Nanoinformatics: a new area of research in nanomedicine. International Journal of Nanomedicine 2012, 3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irizarry RA, The Role of Academia in Data Science Education. 2.1 2020. [DOI] [PMC free article] [PubMed]
- 9.Provost F; Fawcett T, Data Science and its Relationship to Big Data and Data-Driven Decision Making. Big Data 2013, 1 (1), 51–59. [DOI] [PubMed] [Google Scholar]
- 10.Panneerselvam S; Choi S, Nanoinformatics: Emerging Databases and Available Tools. International Journal of Molecular Sciences 2014, 15 (5), 7158–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lijowski M, caNanoLab – A Tool To Benefit Biomedical Nanomaterials Research. Nature Precedings 2010. [Google Scholar]
- 12.Sterling T; Irwin JJ, ZINC 15 – Ligand Discovery for Everyone. Journal of Chemical Information and Modeling 2015, 55 (11), 2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeliazkova N; Chomenidis C; Doganis P; Fadeel B; Grafström R; Hardy B; Hastings J; Hegi M; Jeliazkov V; Kochev N; Kohonen P; Munteanu CR; Sarimveis H; Smeets B; Sopasakis P; Tsiliki G; Vorgrimmler D; Willighagen E, The eNanoMapper database for nanomaterial safety information. Beilstein Journal of Nanotechnology 2015, 6, 1609–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ban Z; Yuan P; Yu F; Peng T; Zhou Q; Hu X, Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proceedings of the National Academy of Sciences 2020, 117 (19), 10492–10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y; Ye Z; Liu X; Wei Z; Qiu F; Li H-F; Zheng Y; Ouyang D, Can machine learning predict drug nanocrystals? Journal of Controlled Release 2020, 322, 274–285. [DOI] [PubMed] [Google Scholar]
- 16.Russo DP; Yan X; Shende S; Huang H; Yan B; Zhu H, Virtual Molecular Projections and Convolutional Neural Networks for the End-to-End Modeling of Nanoparticle Activities and Properties. Analytical Chemistry 2020, 92 (20), 13971–13979. [DOI] [PubMed] [Google Scholar]
- 17.Pellegrino F; Isopescu R; Pellutiè L; Sordello F; Rossi AM; Ortel E; Martra G; Hodoroaba V-D; Maurino V, Machine learning approach for elucidating and predicting the role of synthesis parameters on the shape and size of TiO2 nanoparticles. Scientific Reports 2020, 10 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Youshia J; Ali ME; Lamprecht A, Artificial neural network based particle size prediction of polymeric nanoparticles. European Journal of Pharmaceutics and Biopharmaceutics 2017, 119, 333–342. [DOI] [PubMed] [Google Scholar]
- 19.Panch T; Szolovits P; Atun R, Artificial intelligence, machine learning and health systems. Journal of Global Health 2018, 8 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler KT; Davies DW; Cartwright H; Isayev O; Walsh A, Machine learning for molecular and materials science. Nature 2018, 559 (7715), 547–555. [DOI] [PubMed] [Google Scholar]
- 21.Jordan MI; Mitchell TM, Machine learning: Trends, perspectives, and prospects. Science 2015, 349 (6245), 255. [DOI] [PubMed] [Google Scholar]
- 22.Ghahramani Z, Probabilistic machine learning and artificial intelligence. Nature 2015, 521 (7553), 452–459. [DOI] [PubMed] [Google Scholar]
- 23.Topol EJ, High-performance medicine: the convergence of human and artificial intelligence. Nature Medicine 2019, 25 (1), 44–56. [DOI] [PubMed] [Google Scholar]
- 24.Zhu H, Big Data and Artificial Intelligence Modeling for Drug Discovery. Annual Review of Pharmacology and Toxicology 2020, 60 (1), 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider P; Walters WP; Plowright AT; Sieroka N; Listgarten J; Goodnow RA; Fisher J; Jansen JM; Duca JS; Rush TS; Zentgraf M; Hill JE; Krutoholow E; Kohler M; Blaney J; Funatsu K; Luebkemann C; Schneider G, Rethinking drug design in the artificial intelligence era. Nature Reviews Drug Discovery 2020, 19 (5), 353–364. [DOI] [PubMed] [Google Scholar]
- 26.Curtis C; Mckenna M; Pontes H; Toghani D; Choe A; Nance E, Predicting in situ nanoparticle behavior using multiple particle tracking and artificial neural networks. Nanoscale 2019, 11 (46), 22515–22530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auer A; Strauss MT; Strauss S; Jungmann R, nanoTRON: a Picasso module for MLP-based classification of super-resolution data. Bioinformatics 2020, 36 (11), 3620–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pottel J; Armstrong D; Zou L; Fekete A; Huang X-P; Torosyan H; Bednarczyk D; Whitebread S; Bhhatarai B; Liang G; Jin H; Ghaemi SN; Slocum S; Lukacs KV; Irwin JJ; Berg EL; Giacomini KM; Roth BL; Shoichet BK; Urban L, The activities of drug inactive ingredients on biological targets. Science 2020, 369 (6502), 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reker D; Rybakova Y; Kirtane AR; Cao R; Yang JW; Navamajiti N; Gardner A; Zhang RM; Esfandiary T; L’Heureux J; von Erlach T; Smekalova EM; Leboeuf D; Hess K; Lopes A; Rogner J; Collins J; Tamang SM; Ishida K; Chamberlain P; Yun D; Lytton-Jean A; Soule CK; Cheah JH; Hayward AM; Langer R; Traverso G, Computationally guided high-throughput design of self-assembling drug nanoparticles. Nat Nanotechnol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar R; Le N; Tan Z; Brown ME; Jiang S; Reineke TM, Efficient Polymer-Mediated Delivery of Gene-Editing Ribonucleoprotein Payloads through Combinatorial Design, Parallelized Experimentation, and Machine Learning. ACS Nano 2020, 14 (12), 17626–17639. [DOI] [PubMed] [Google Scholar]
- 31.Yamankurt G; Berns EJ; Xue A; Lee A; Bagheri N; Mrksich M; Mirkin CA, Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nature Biomedical Engineering 2019, 3 (4), 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan X; Zhang J; Russo DP; Zhu H; Yan B, Prediction of Nano–Bio Interactions through Convolutional Neural Network Analysis of Nanostructure Images. ACS Sustainable Chemistry & Engineering 2020, 8 (51), 19096–19104. [Google Scholar]
- 33.Singh AV; Maharjan R-S; Kanase A; Siewert K; Rosenkranz D; Singh R; Laux P; Luch A, Machine-Learning-Based Approach to Decode the Influence of Nanomaterial Properties on Their Interaction with Cells. Acs Appl Mater Inter 2021, 13 (1), 1943–1955. [DOI] [PubMed] [Google Scholar]
- 34.Stillman NR; Kovacevic M; Balaz I; Hauert S, In silico modelling of cancer nanomedicine, across scales and transport barriers. Npj Comput Mater 2020, 6 (1). [Google Scholar]
- 35.Francia V; Montizaan D; Salvati A, Interactions at the cell membrane and pathways of internalization of nano-sized materials for nanomedicine. Beilstein Journal of Nanotechnology 2020, 11, 338–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotzabasaki MI; Sotiropoulos I; Sarimveis H, QSAR modeling of the toxicity classification of superparamagnetic iron oxide nanoparticles (SPIONs) in stem-cell monitoring applications: an integrated study from data curation to model development. RSC Advances 2020, 10 (9), 5385–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rybińska-Fryca A; Mikolajczyk A; Puzyn T, Structure–activity prediction networks (SAPNets): a step beyond Nano-QSAR for effective implementation of the safe-by-design concept. Nanoscale 2020, 12 (40), 20669–20676. [DOI] [PubMed] [Google Scholar]
- 38.Singh AV; Rosenkranz D; Ansari MHD; Singh R; Kanase A; Singh SP; Johnston B; Tentschert J; Laux P; Luch A, Artificial Intelligence and Machine Learning Empower Advanced Biomedical Material Design to Toxicity Prediction. Advanced Intelligent Systems 2020, 2 (12), 2000084. [Google Scholar]
- 39.Poon W; Kingston BR; Ouyang B; Ngo W; Chan WCW, A framework for designing delivery systems. Nat Nanotechnol 2020, 15 (10), 819–829. [DOI] [PubMed] [Google Scholar]
- 40.Pollice R; Dos Passos Gomes G; Aldeghi M; Hickman RJ; Krenn M; Lavigne C; Lindner-D’Addario M; Nigam A; Ser CT; Yao Z; Aspuru-Guzik A, Data-Driven Strategies for Accelerated Materials Design. Acc Chem Res 2021, 54 (4), 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adir O; Poley M; Chen G; Froim S; Krinsky N; Shklover J; Shainsky-Roitman J; Lammers T; Schroeder A, Integrating Artificial Intelligence and Nanotechnology for Precision Cancer Medicine. Adv Mater 2020, 32 (13), e1901989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh AV; Ansari MHD; Rosenkranz D; Maharjan RS; Kriegel FL; Gandhi K; Kanase A; Singh R; Laux P; Luch A, Artificial Intelligence and Machine Learning in Computational Nanotoxicology: Unlocking and Empowering Nanomedicine. Adv Healthc Mater 2020, 9 (17), e1901862. [DOI] [PubMed] [Google Scholar]
- 43.Tao H; Wu T; Aldeghi M; Wu TC; Aspuru-Guzik A; Kumacheva E, Nanoparticle synthesis assisted by machine learning. Nature Reviews Materials 2021, 6 (8), 701–716. [Google Scholar]
- 44.Ventola CL, Progress in Nanomedicine: Approved and Investigational Nanodrugs. P T 2017, 42 (12), 742–755. [PMC free article] [PubMed] [Google Scholar]
- 45.Bhardwaj V; Kaushik A; Khatib ZM; Nair M; McGoron AJ, Recalcitrant Issues and New Frontiers in Nano-Pharmacology. Front Pharmacol 2019, 10, 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang H; Mintri S; Menon AV; Lee HY; Choi HS; Kim J, Pharmacokinetics, pharmacodynamics and toxicology of theranostic nanoparticles. Nanoscale 2015, 7 (45), 18848–18862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miele E; Spinelli GP; Tomao F; Tomao S, Albumin-bound formulation of paclitaxel (Abraxane ABI-007) in the treatment of breast cancer. Int J Nanomedicine 2009, 4, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulbake U; Doppalapudi S; Kommineni N; Khan W, Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 2017, 9 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi J-S; Ha MK; Trinh TX; Yoon TH; Byun H-G, Towards a generalized toxicity prediction model for oxide nanomaterials using integrated data from different sources. Scientific Reports 2018, 8 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shamay Y; Shah J; Işik M; Mizrachi A; Leibold J; Tschaharganeh DF; Roxbury D; Budhathoki-Uprety J; Nawaly K; Sugarman JL; Baut E; Neiman MR; Dacek M; Ganesh KS; Johnson DC; Sridharan R; Chu KL; Rajasekhar VK; Lowe SW; Chodera JD; Heller DA, Quantitative self-assembly prediction yields targeted nanomedicines. Nat Mater 2018, 17 (4), 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alves VM; Hwang D; Muratov E; Sokolsky-Papkov M; Varlamova E; Vinod N; Lim C; Andrade CH; Tropsha A; Kabanov A, Cheminformatics-driven discovery of polymeric micelle formulations for poorly soluble drugs. Sci Adv 2019, 5 (6), eaav9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drugs@FDA: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm.
- 53.Reker D; Blum SM; Steiger C; Anger KE; Sommer JM; Fanikos J; Traverso G, “Inactive” ingredients in oral medications. Sci Transl Med 2019, 11 (483). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuehn BM, Rare PEG Allergy Triggered Postvaccination Anaphylaxis. JAMA 2021, 325 (19), 1931. [DOI] [PubMed] [Google Scholar]
- 55.Kabanov AV; Batrakova EV; Miller DW, Pluronic block copolymers as modulators of drug efflux transporter activity in the blood-brain barrier. Adv Drug Deliv Rev 2003, 55 (1), 151–64. [DOI] [PubMed] [Google Scholar]
- 56.Kinnear C; Moore TL; Rodriguez-Lorenzo L; Rothen-Rutishauser B; Petri-Fink A, Form Follows Function: Nanoparticle Shape and Its Implications for Nanomedicine. Chem Rev 2017, 117 (17), 11476–11521. [DOI] [PubMed] [Google Scholar]
- 57.Ayala V; Herrera AP; Latorre-Esteves M; Torres-Lugo M; Rinaldi C, Effect of surface charge on the colloidal stability and in vitro uptake of carboxymethyl dextran-coated iron oxide nanoparticles. J Nanopart Res 2013, 15 (8), 1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kister T; Monego D; Mulvaney P; Widmer-Cooper A; Kraus T, Colloidal Stability of Apolar Nanoparticles: The Role of Particle Size and Ligand Shell Structure. ACS Nano 2018, 12 (6), 5969–5977. [DOI] [PubMed] [Google Scholar]
- 59.Zhu H; Prince E; Narayanan P; Liu K; Nie Z; Kumacheva E, Colloidal stability of nanoparticles stabilized with mixed ligands in solvents with varying polarity. Chem Commun (Camb) 2020, 56 (58), 8131–8134. [DOI] [PubMed] [Google Scholar]
- 60.Donahue ND; Acar H; Wilhelm S, Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Adv Drug Deliv Rev 2019, 143, 68–96. [DOI] [PubMed] [Google Scholar]
- 61.Yoo J; Park C; Yi G; Lee D; Koo H, Active Targeting Strategies Using Biological Ligands for Nanoparticle Drug Delivery Systems. Cancers (Basel) 2019, 11 (5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Y; Wang J; Jiang K; Chung EJ, Improving kidney targeting: The influence of nanoparticle physicochemical properties on kidney interactions. J Control Release 2021, 334, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albanese A; Tang PS; Chan WC, The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu Rev Biomed Eng 2012, 14, 1–16. [DOI] [PubMed] [Google Scholar]
- 64.Yu W; Liu R; Zhou Y; Gao H, Size-Tunable Strategies for a Tumor Targeted Drug Delivery System. ACS Cent Sci 2020, 6 (2), 100–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poon W; Kingston BR; Ouyang B; Ngo W; Chan WCW, A framework for designing delivery systems. Nature Nanotechnology 2020, 15 (10), 819–829. [DOI] [PubMed] [Google Scholar]
- 66.Brown TD; Habibi N; Wu D; Lahann J; Mitragotri S, Effect of Nanoparticle Composition, Size, Shape, and Stiffness on Penetration Across the Blood-Brain Barrier. ACS Biomater Sci Eng 2020, 6 (9), 4916–4928. [DOI] [PubMed] [Google Scholar]
- 67.Hayashi Y; Miclaus T; Scavenius C; Kwiatkowska K; Sobota A; Engelmann P; Scott-Fordsmand JJ; Enghild JJ; Sutherland DS, Species differences take shape at nanoparticles: protein corona made of the native repertoire assists cellular interaction. Environ Sci Technol 2013, 47 (24), 14367–75. [DOI] [PubMed] [Google Scholar]
- 68.Florez L; Herrmann C; Cramer JM; Hauser CP; Koynov K; Landfester K; Crespy D; Mailänder V, How shape influences uptake: interactions of anisotropic polymer nanoparticles and human mesenchymal stem cells. Small 2012, 8 (14), 2222–30. [DOI] [PubMed] [Google Scholar]
- 69.Blanco E; Shen H; Ferrari M, Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol 2015, 33 (9), 941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toy R; Peiris PM; Ghaghada KB; Karathanasis E, Shaping cancer nanomedicine: the effect of particle shape on the in vivo journey of nanoparticles. Nanomedicine (Lond) 2014, 9 (1), 121–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dey S; Fan C; Gothelf KV; Li J; Lin C; Liu L; Liu N; Nijenhuis MAD; Saccà B; Simmel FC; Yan H; Zhan P, DNA origami. Nature Reviews Methods Primers 2021, 1 (1), 13. [Google Scholar]
- 72.Jiang Q; Liu S; Liu J; Wang ZG; Ding B, Rationally Designed DNA-Origami Nanomaterials for Drug Delivery In Vivo. Adv Mater 2019, 31 (45), e1804785. [DOI] [PubMed] [Google Scholar]
- 73.Wang P; Rahman MA; Zhao Z; Weiss K; Zhang C; Chen Z; Hurwitz SJ; Chen ZG; Shin DM; Ke Y, Visualization of the Cellular Uptake and Trafficking of DNA Origami Nanostructures in Cancer Cells. J Am Chem Soc 2018, 140 (7), 2478–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rasmussen MK; Pedersen JN; Marie R, Size and surface charge characterization of nanoparticles with a salt gradient. Nat Commun 2020, 11 (1), 2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng Q; Wei T; Farbiak L; Johnson LT; Dilliard SA; Siegwart DJ, Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat Nanotechnol 2020, 15 (4), 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WCW, Analysis of nanoparticle delivery to tumours. Nature Reviews Materials 2016, 1 (5), 16014. [Google Scholar]
- 77.Liu S; Chen X; Bao L; Liu T; Yuan P; Yang X; Qiu X; Gooding JJ; Bai Y; Xiao J; Pu F; Jin Y, Treatment of infarcted heart tissue via the capture and local delivery of circulating exosomes through antibody-conjugated magnetic nanoparticles. Nat Biomed Eng 2020, 4 (11), 1063–1075. [DOI] [PubMed] [Google Scholar]
- 78.Juan A; Cimas FJ; Bravo I; Pandiella A; Ocaña A; Alonso-Moreno C, An Overview of Antibody Conjugated Polymeric Nanoparticles for Breast Cancer Therapy. Pharmaceutics 2020, 12 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai C; Gao S; Hu S; Liu X; Li H; Dong J; Huang A; Zhu L; Zhou P; Li S; Shao N, Self-Assembled Multivalent Aptamer Nanoparticles with Potential CAR-like Characteristics Could Activate T Cells and Inhibit Melanoma Growth. Mol Ther Oncolytics 2020, 17, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao Z; Farokhzad OC, Aptamer-functionalized nanoparticles for medical applications: challenges and opportunities. ACS Nano 2012, 6 (5), 3670–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kang B; Opatz T; Landfester K; Wurm FR, Carbohydrate nanocarriers in biomedical applications: functionalization and construction. Chem Soc Rev 2015, 44 (22), 8301–25. [DOI] [PubMed] [Google Scholar]
- 82.Mizrachi A; Shamay Y; Shah J; Brook S; Soong J; Rajasekhar VK; Humm JL; Healey JH; Powell SN; Baselga J; Heller DA; Haimovitz-Friedman A; Scaltriti M, Tumour-specific PI3K inhibition via nanoparticle-targeted delivery in head and neck squamous cell carcinoma. Nat Commun 2017, 8, 14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Begines B; Ortiz T; Pérez-Aranda M; Martínez G; Merinero M; Argüelles-Arias F; Alcudia A, Polymeric Nanoparticles for Drug Delivery: Recent Developments and Future Prospects. Nanomaterials (Basel) 2020, 10 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sanita G; Carrese B; Lamberti A, Nanoparticle Surface Functionalization: How to Improve Biocompatibility and Cellular Internalization. Front Mol Biosci 2020, 7, 587012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tran PHL; Duan W; Tran TTD, Fucoidan-based nanostructures: A focus on its combination with chitosan and the surface functionalization of metallic nanoparticles for drug delivery. International Journal of Pharmaceutics 2020, 575, 118956. [DOI] [PubMed] [Google Scholar]
- 86.Elkordy AA; Haj-Ahmad RR; Awaad AS; Zaki RM, An overview on natural product drug formulations from conventional medicines to nanomedicines: Past, present and future. Journal of Drug Delivery Science and Technology 2021, 63, 102459. [Google Scholar]
- 87.Shamay Y; Elkabets M; Li H; Shah J; Brook S; Wang F; Adler K; Baut E; Scaltriti M; Jena PV; Gardner EE; Poirier JT; Rudin CM; Baselga J; Haimovitz-Friedman A; Heller DA, P-selectin is a nanotherapeutic delivery target in the tumor microenvironment. Sci Transl Med 2016, 8 (345), 345ra87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alafeef M; Srivastava I; Pan D, Machine Learning for Precision Breast Cancer Diagnosis and Prediction of the Nanoparticle Cellular Internalization. ACS Sens 2020, 5 (6), 1689–1698. [DOI] [PubMed] [Google Scholar]
- 89.Pino P. d.; Pelaz B; Zhang Q; Maffre P; Nienhaus GU; Parak WJ, Protein corona formation around nanoparticles – from the past to the future. Materials Horizons 2014, 1 (3), 301–313. [Google Scholar]
- 90.Monopoli MP; Walczyk D; Campbell A; Elia G; Lynch I; Bombelli FB; Dawson KA, Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. J Am Chem Soc 2011, 133 (8), 2525–34. [DOI] [PubMed] [Google Scholar]
- 91.Docter D; Westmeier D; Markiewicz M; Stolte S; Knauer SK; Stauber RH, The nanoparticle biomolecule corona: lessons learned - challenge accepted? Chem Soc Rev 2015, 44 (17), 6094–121. [DOI] [PubMed] [Google Scholar]
- 92.Zanganeh S; Spitler R; Erfanzadeh M; Alkilany AM; Mahmoudi M, Protein corona: Opportunities and challenges. Int J Biochem Cell Biol 2016, 75, 143–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamankurt G; Berns EJ; Xue A; Lee A; Bagheri N; Mrksich M; Mirkin CA, Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nat Biomed Eng 2019, 3 (4), 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yu F; Wei C; Deng P; Peng T; Hu X, Deep exploration of random forest model boosts the interpretability of machine learning studies of complicated immune responses and lung burden of nanoparticles. Sci Adv 2021, 7 (22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kingston BR; Syed AM; Ngai J; Sindhwani S; Chan WCW, Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc Natl Acad Sci U S A 2019, 116 (30), 14937–14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jordan MI; Mitchell TM, Machine learning: Trends, perspectives, and prospects. Science 2015, 349 (6245), 255–60. [DOI] [PubMed] [Google Scholar]
- 97.Ouyang B; Poon W; Zhang YN; Lin ZP; Kingston BR; Tavares AJ; Zhang Y; Chen J; Valic MS; Syed AM; MacMillan P; Couture-Senecal J; Zheng G; Chan WCW, The dose threshold for nanoparticle tumour delivery. Nat Mater 2020, 19 (12), 1362–1371. [DOI] [PubMed] [Google Scholar]
- 98.Russo DP; Yan X; Shende S; Huang H; Yan B; Zhu H, Virtual Molecular Projections and Convolutional Neural Networks for the End-to-End Modeling of Nanoparticle Activities and Properties. Anal Chem 2020, 92 (20), 13971–13979. [DOI] [PubMed] [Google Scholar]
- 99.Stetefeld J; McKenna SA; Patel TR, Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys Rev 2016, 8 (4), 409–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lyngdoh A, 10 - What we leave behind: the future of data curation. In Trends, Discovery, and People in the Digital Age, Baker D; Evans W, Eds. Chandos Publishing: 2013; pp 153–165. [Google Scholar]
- 101.Freitas A; Curry E, Big Data Curation. In New Horizons for a Data-Driven Economy, Springer International Publishing: 2016; pp 87–118. [Google Scholar]
- 102.Brinson LC; Deagen M; Chen W; Mccusker J; Mcguinness DL; Schadler LS; Palmeri M; Ghumman U; Lin A; Hu B, Polymer Nanocomposite Data: Curation, Frameworks, Access, and Potential for Discovery and Design. ACS Macro Letters 2020, 9 (8), 1086–1094. [DOI] [PubMed] [Google Scholar]
- 103.Holinski A; Burke ML; Morgan SL; Mcquilton P; Palagi PM, Biocuration - mapping resources and needs. F1000Research 2020, 9, 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dauga D, Biocuration: A New Challenge for the Tunicate Community. genesis 2015, 53 (1), 132–142. [DOI] [PubMed] [Google Scholar]
- 105.Bento AP; Hersey A; Félix E; Landrum G; Gaulton A; Atkinson F; Bellis LJ; De Veij M; Leach AR, An open source chemical structure curation pipeline using RDKit. Journal of Cheminformatics 2020, 12 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grondin CJ; Davis AP; Wiegers TC; King BL; Wiegers JA; Reif DM; Hoppin JA; Mattingly CJ, Advancing Exposure Science through Chemical Data Curation and Integration in the Comparative Toxicogenomics Database. Environmental Health Perspectives 2016, 124 (10), 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Raccuglia P; Elbert KC; Adler PDF; Falk C; Wenny MB; Mollo A; Zeller M; Friedler SA; Schrier J; Norquist AJ, Machine-learning-assisted materials discovery using failed experiments. Nature 2016, 533 (7601), 73–76. [DOI] [PubMed] [Google Scholar]
- 108.Jones DE; Ghandehari H; Facelli JC, A review of the applications of data mining and machine learning for the prediction of biomedical properties of nanoparticles. Computer Methods and Programs in Biomedicine 2016, 132, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dimitri A; Talamo M, The use of data mining and machine learning in nanomedicine: a survey. Frontiers in Nanoscience and Nanotechnology 2018, 4 (3). [Google Scholar]
- 110.Labouta HI; Asgarian N; Rinker K; Cramb DT, Meta-Analysis of Nanoparticle Cytotoxicity via Data-Mining the Literature. ACS Nano 2019, 13 (2), 1583–1594. [DOI] [PubMed] [Google Scholar]
- 111.Gaheen S; Hinkal GW; Morris SA; Lijowski M; Heiskanen M; Klemm JD, caNanoLab: data sharing to expedite the use of nanotechnology in biomedicine. Computational science & discovery 2013, 6 (1), 014010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morris SA; Gaheen S; Lijowski M; Heiskanen M; Klemm J, Experiences in supporting the structured collection of cancer nanotechnology data using caNanoLab. Beilstein Journal of Nanotechnology 2015, 6, 1580–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yan X; Sedykh A; Wang W; Yan B; Zhu H, Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations. Nature Communications 2020, 11 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ha MK; Trinh TX; Choi JS; Maulina D; Byun HG; Yoon TH, Toxicity Classification of Oxide Nanomaterials: Effects of Data Gap Filling and PChem Score-based Screening Approaches. Scientific Reports 2018, 8 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Furxhi I; Murphy F; Mullins M; Poland CA, Machine learning prediction of nanoparticle in vitro toxicity: A comparative study of classifiers and ensemble-classifiers using the Copeland Index. Toxicology Letters 2019, 312, 157–166. [DOI] [PubMed] [Google Scholar]
- 116.Miller AL; Hoover MD; Mitchell DM; Stapleton BP, The Nanoparticle Information Library (NIL): A Prototype for Linking and Sharing Emerging Data. Journal of Occupational and Environmental Hygiene 2007, 4 (12), D131–D134. [DOI] [PubMed] [Google Scholar]
- 117.Edelweiss Connect. https://www.edelweissconnect.com/.
- 118.National Technical University Of Athens. https://www.ntua.gr/en/.
- 119.in silico toxicology. https://www.in-silico.ch/.
- 120.IDEAconsult Ltd. https://www.ideaconsult.net/.
- 121.Karolinska Institutet. https://ki.se/.
- 122.MISVIK BIOLOGY LTD. https://www.misvik.com/.
- 123.EMBL-EBI. https://www.ebi.ac.uk.
- 124.Maastricht University. https://www.maastrichtuniversity.nl.
- 125.Cohen AM; Adams CE; Davis JM; Yu C; Yu PS; Meng W; Duggan L; Mcdonagh M; Smalheiser NR In Evidence-based medicine, the essential role of systematic reviews, and the need for automated text mining tools, Proceedings of the ACM international conference on Health informatics - IHI ‘10, 2010–01-01; ACM Press: 2010. [Google Scholar]
- 126.Cao C; Liu F; Tan H; Song D; Shu W; Li W; Zhou Y; Bo X; Xie Z, Deep Learning and Its Applications in Biomedicine. Genomics, Proteomics & Bioinformatics 2018, 16 (1), 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zheng S; Dharssi S; Wu M; Li J; Lu Z, Text Mining for Drug Discovery. In Methods in Molecular Biology, Springer; New York: 2019; pp 231–252. [DOI] [PubMed] [Google Scholar]
- 128.Lever J; Zhao EY; Grewal J; Jones MR; Jones SJM, CancerMine: a literature-mined resource for drivers, oncogenes and tumor suppressors in cancer. Nature Methods 2019, 16 (6), 505–507. [DOI] [PubMed] [Google Scholar]
- 129.Lee J; Yoon W; Kim S; Kim D; Kim S; So CH; Kang J, BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ye Z; Tafti AP; He KY; Wang K; He MM, SparkText: Biomedical Text Mining on Big Data Framework. PLOS ONE 2016, 11 (9), e0162721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Beynon R; Leeflang MMG; McDonald S; Eisinga A; Mitchell RL; Whiting P; Glanville JM, Search strategies to identify diagnostic accuracy studies in MEDLINE and EMBASE. Cochrane Database Syst Rev 2013, 2013 (9), MR000022–MR000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Egorov E; Pieters C; Korach-Rechtman H; Shklover J; Schroeder A, Robotics, microfluidics, nanotechnology and AI in the synthesis and evaluation of liposomes and polymeric drug delivery systems. Drug Delivery and Translational Research 2021, 11 (2), 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang H; Zhu YF; Shen YQ, Microfluidics for Cancer Nanomedicine: From Fabrication to Evaluation. Small 2018, 14 (28). [DOI] [PubMed] [Google Scholar]
- 134.Mulvaney P; Parak WJ; Caruso F; Weiss PS, Standardizing Nanomaterials. ACS Nano 2016, 10 (11), 9763–9764. [DOI] [PubMed] [Google Scholar]
- 135.Aublant JM; Clifford CA; Frejafon E; Fujimoto T; Hackley VA; Herrmann J; Hight Walker AR; Kaiser DL; Koltsov DK; Michael DJ; Ono A; Roebben G; Smallwood GJ; Taketoshi N, Response to. ACS Nano 2020, 14 (11), 14255–14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mitragotri S; Burke PA; Langer R, Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat Rev Drug Discov 2014, 13 (9), 655–72. [DOI] [PMC free article] [PubMed] [Google Scholar]


