Abstract
The gut microbiota is known to play an important role in maintaining gut health through a symbiotic relationship with the host. Altered gut microbiota is a common feature of several diseases of the gastrointestinal tract; however, the causal relationship between microbiota and disease pathogenesis is poorly understood. Necrotising enterocolitis (NEC) and inflammatory bowel disease (IBD) are both severe inflammatory diseases affecting the gastrointestinal tract. Although they affect very different patient populations, with NEC primarily being a disease of prematurity and IBD predominantly affecting adults although children can be affected, they both demonstrate common features of gut microbial dysbiosis and a dysregulated host immune response. By comparing and contrasting the changes in gut microbiota, host immune response and function, we aim to highlight common features in diseases that may seem clinically unrelated. Key areas of interest are the role of pattern recognition receptors in altered recognition and responses to the gut microbiota by the host immune system and the associated dysfunctional gut epithelial barrier. The challenge of identifying causal relationships between microbiota and disease is ever-present; however, considering a disease-agnostic approach may help to identify mechanistic pathways shared across several clinical diseases.
Keywords: NECROTIZING ENTEROCOLITIS, INFLAMMATORY BOWEL DISEASE, IBD, INTESTINAL BACTERIA
Introduction
Approximately 100 trillion (1×1014) microbes colonise the human gastrointestinal (GI) tract, composed of bacteria, fungi, yeasts, viruses, protozoa and archaea.1 2 Over 1000 distinct commensal bacterial species from four dominant phyla: Firmicutes, Bacteroidetes, Actinobacteria and Proteobacteria 3 are found along the entire length of the GI tract, with increasingly dense and varied populations in the colon.4 These complex microbial communities have developed a symbiotic relationship with the human host that is the result of over 500 million years of coevolution.5
The genes and genetic products of the microbiota are collectively referred to as the gut microbiome and they are essential for normal gut function and homoeostasis. With the use of culture-independent sequencing techniques (16S rRNA and shotgun metagenomics) the composition and function of the human gut microbiome is being uncovered. International projects, such as The Human Microbiome Project, have been undertaken to characterise the commensal microbiota in distinct anatomical sites of healthy individuals.3 Although these are still mostly focused on European and North American populations,6 this has provided better understanding of the complex and varied roles the gut microbiota plays in both health and disease.
This review will consider how the gut microbiota develops from early life onwards, its key functions in maintaining gut health and the role of altered gut microbiota in disease. Although altered gut microbiota is a characteristic feature of several diseases, whether there is a causal link remains poorly understood.
Necrotising enterocolitis (NEC) and inflammatory bowel disease (IBD) are both severe inflammatory diseases of the GI tract. They affect very different patient populations, with NEC being a disease of prematurity and IBD primarily affecting those in the second and third decades of life. Although host-specific factors differ in these patient groups, both conditions are associated with distinct changes to the composition of the gut microbiome, and it is postulated that these might drive aberrant function of the gut immune system resulting in destructive inflammation. We describe the role of the gut microbiota in the pathogenesis of these conditions and draw parallels and differences between them. By highlighting features of the immature gut immune system characteristic of those with NEC and those of the genetically susceptible host in IBD we will consider the potential causal role for host–microbe interactions in promoting gut inflammation.
Development of the gut microbiota
Age greatly influences the composition and diversity of the gut microbiota, with key stages throughout life driving substantial shifts in bacterial abundances.7 8 Of these changes, early life factors have a profound impact, with mode of delivery, feeding type and then transitioning to a solid based diet driving alterations in microbiota composition.9 As such, the assembly and trajectory of the gut microbiota is not purely stochastic, but instead follows a patterned development.10 11
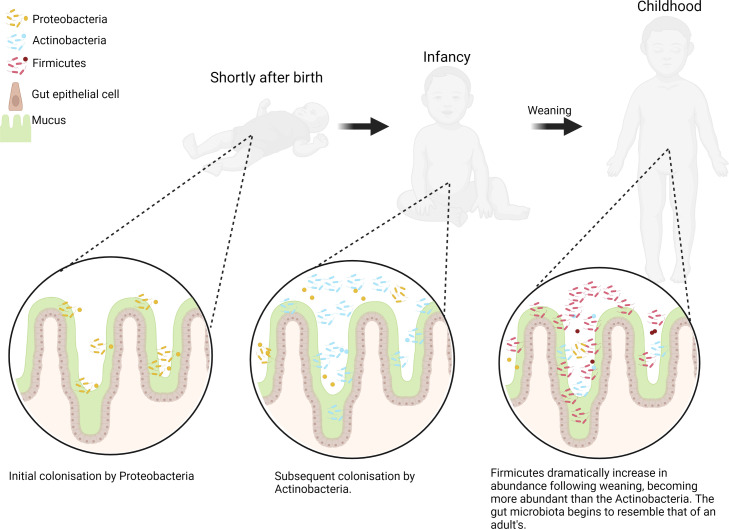
The sequential changes in the initial neonatal gut microbiota composition are well defined and follow a predictable, non-random sequence for healthy, full-term neonates (figure 1).12 13 Initially, the gut is colonised by facultative anaerobes belonging to the Proteobacteria phylum, predominantly the Enterobacteriaceae family of bacteria, which are believed to deoxygenate the gut.9 14 Once an anaerobic environment is formed in the gut, obligate anaerobes begin to colonise, including Bifidobacterium which belong to the Actinobacteria phylum.13–15
Figure 1.
The early development of the gut microbiota. Key changes to the gut microbiota composition are summarised on a phylum level with changes shown at three early life stages: shortly after birth, infancy and postweaning (figure created in BioRender).
Transitioning to a solid based diet from milk is an important milestone in the microbiota development of the neonate and denotes a shift towards a microbiota which resembles that of an adult’s.7 12 16 17 A cross-sectional study of the gut microbiota from new-borns to centenarians shows a dramatic change at 2 years of age whereby the Firmicutes dramatically increase in abundance with the Actinobacteria declining, likely corresponding to the period following weaning.7 Comparisons of the gut microbiota before and after weaning in two distinct geographical cohorts also shows some clear and dramatic changes.16
Much research has been conducted to understand the factors contributing to interindividual variability seen in gut microbiota compositions of adults. In particular, the effect of diet and geography have been studied extensively.18–22 Diets are broadly divided into Western defined diets, comprising relatively lower levels of fibre and higher consumption of refined carbohydrates and fat, and high fibre or Eastern diets.21 The diversity of the gut microbiota community in Western countries is significantly lower than that of non-Western ones with diet thought to be an important determinant of diversity.23
There is a complex symbiotic relationship between the resident microbiota and the host, with a diverse gut microbiota being necessary for normal gut function and health.
Role of the microbiota in normal gut function and protection against pathogens
The organisms comprising the gut microbiota have a symbiotic relationship with the human host and perform several essential functions including regulating host metabolism, synthesis of essential vitamins and protection against pathogens.24 25 In return, these organisms can occupy a nutrient rich environment. The gut microbiota plays an essential role in host digestive function and metabolism, by enabling the extraction of nutrients from substrates that are otherwise indigestible by the host.26 This is primarily achieved through fermentation and anaerobic degradation to maximise the caloric uptake from these ingested nutrients. Metabolites produced through this breakdown of insoluble carbohydrates have a variety of functions in gut homoeostasis. Short chain fatty acids (SCFAs) are the most abundant of these metabolites and include acetate, propionate and butyrate.27 These SCFAs are either used locally as a fuel source by the colonic intestinal epithelial cells (IECs) or absorbed into the portal circulation.28
There are several commensal butyrate-producing species in the colon, with Firmicutes (eg, Faecalibacterium prausnitzii) and Bacteroidetes being the predominant groups.29 Butyrate provides an essential source of energy to the colonic mucosal epithelium, stimulating growth and proliferation of IECs and having an anti-inflammatory effect (table 1).30 31 This anti-inflammatory effect is achieved via several mechanisms including enhancement of intestinal barrier function by increasing mucin production and maintaining the integrity of epithelial tight junctions. Butyrate also reduces expression of inflammatory mediators by IECs via inhibition of the proinflammatory NF-kB pathway.32 33
Table 1.
Role of butyrate in intestinal homoeostasis
| Role in intestinal homoeostasis | Mechanisms | Outcome |
| Anti-inflammatory function | Inhibits histone deacetylases | Pro-inflammatory mediators (TNF-alpha, IL-2, IL-6, IL-8) |
| Inhibits NF-kB pathway |
 Anti-inflammatory mediators (IL-10) Anti-inflammatory mediators (IL-10) |
|
| Energy source | 90% of butyrate taken up by colonocytes for β-oxidation | Butyrate is the primary energy source for colonocytes |
| Protective against colon cancer | Inhibits histone deacetylases (Dachas) |

Apoptosis of colon cancer cells |
| Induces cell cycle arrest |
 Proliferation of colon cancer cells Proliferation of colon cancer cells |
|
| Enhance epithelial barrier function and defence against pathogens | Increased expression of MUC2 gene |
 Mucin production Mucin production |
| Regulates tight junctional proteins |
 Antimicrobial peptide production Antimicrobial peptide production |
|
 Intestinal epithelial permeability Intestinal epithelial permeability |
||
| Anti-diarrhetic | Stimulates Na+ and Cl− coupled transport systems Inhibits secretion of Cl− |
 Sodium, chloride, potassium and water absorption in colon Sodium, chloride, potassium and water absorption in colon |
A summary of the effects of butyrate on gut homoeostasis. The different roles of butyrate in gut homoeostasis are outlined with a description of the mechanisms and outcomes for each role. Blue arrow direction indicates an increase or decrease.
Gut microbial dysbiosis and gut inflammation: a causal relationship?
Dysbiosis can be defined as a compositional and functional alteration in the microbiota in individuals with disease compared with healthy subjects.34 This is a feature of several intestinal and extra-intestinal disorders including IBD and NEC, colorectal cancer, diabetes, multiple sclerosis, obesity and food allergies.25 35–37
NEC is an acute and devastating inflammatory disease of the intestine which leads to gut tissue necrosis primarily affecting preterm infants.38 39 Risk factors include low gestational age and birth weight, formula milk feeding and administration of antibiotics, with varying degrees of risk.40–43
The involvement of gut bacteria in NEC pathogenesis is apparent as the disease is associated with altered gut microbiota in several animal models and human cases of NEC (table 2).39 44–50 Several studies have analysed the relative abundance of bacterial taxa preceding NEC, using a 16S rRNA sequencing approach. A meta-analysis of 14 eligible studies, either cohort or case–control, has found that the phylum Proteobacteria appear to have an increased relative abundance in NEC infants compared with healthy non-NEC control infants.51 Whether this increase in Proteobacteria abundance preceding NEC onset is causative is unknown. It may be in response to other factors which are drivers of NEC such as hypoxia in the gut environment.
Table 2.
A comparison of the major microbial changes seen in the gut microbiotas for IBD and NEC
| IBD | NEC |
| Increased abundance of Proteobacteria | Increased abundance of Proteobacteria |
| Decreased abundance of Firmicutes | Decreased abundance of Firmicutes |
| Increased abundance of Enterobacteriaceae | Decreased abundance of Bacteroidetes |
| Decreased abundance of Faecalibacterium prausnitzii | Increased abundance of Klebsiella |
A comparison of the major microbial changes seen in the gut microbiotas for IBD and NEC. The differences in abundance of some organisms is outlined for IBD and NEC for comparison. Abundances are relative abundances and may be changes associated with each disease before, during or after disease onset.
IBD, inflammatory bowel disease; NEC, necrotising enterocolitis.
IBD, largely comprised of Crohn’s disease (CD) and ulcerative colitis (UC), is another condition associated with inflammation of the GI tract. IBD is associated with specific alterations in the gut microbiota (table 2). Changes frequently reproduced across several studies are that of reduced richness and diversity of both the faecal and mucus-resident microbiome in IBD.52–54 Specifically, reduction in relative abundance of Faecalibacterium prausnitzii and an increase in members of the Proteobacteria phylum such as Escherichia coli are observed in people with IBD compared with healthy individuals.55 56 These differences are seen among members of the same family, including twins, who are discordant for IBD implying that alterations in the gut microbiota are primarily related to disease state.57
Reduced bacterial diversity and shifts in abundance of specific bacterial taxa have also been associated with several chronic inflammatory pathologies, including other immune-mediated conditions such as type 1 diabetes.58 59 The commonality in these findings questions the specificity of dysbiosis in defining disease states. In particular, whether the observed alterations in the microbiota are a cause or consequence of inflammation is yet to be clearly defined.
Role of pattern recognition receptors in promoting gut inflammation
While clear cause and correlation has not been established, there are several mechanisms by which an altered gut microbiota can influence the onset of gut inflammation (figure 2). One important way is by altering the response to recognition of commensal microbes by the host immune system. Genome-wide association studies have identified a variety of target genes implicated in susceptibility to IBD.60 These susceptibility loci and mutations are associated with defects in microbial sensing and clearance.
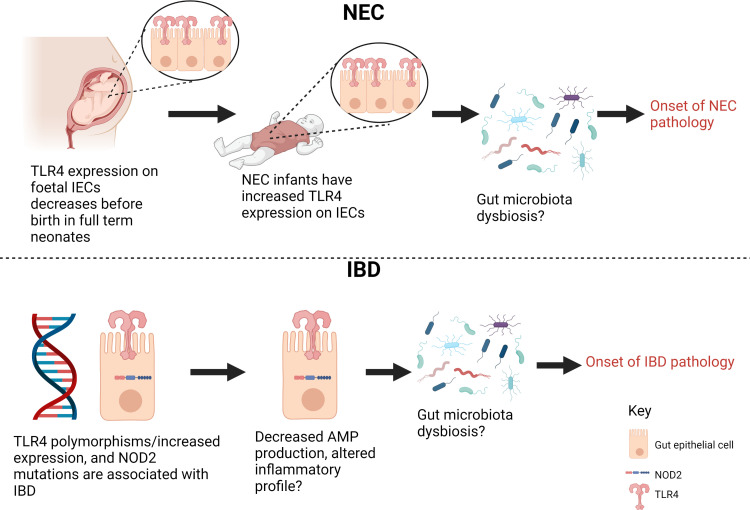
Figure 2.
How pattern recognition receptors expressed on intestinal epithelial cells (IECs) may be involved in necrotising enterocolitis (NEC) and inflammatory bowel disease (IBD) pathology. Alterations to the function and expression of TLR4 on intestinal epithelial cells may beimportant in contributing to the gut microbiota dysbiosis seen during both inflammatorybowel disease and necrotising enterocolitis and subsequently disease pathology. NOD2receptor mutations have also been implicated in IBD pathology via altering the productionof AMPs which may alter the gut microbiota composition leading to dysbiosis and gutinflammation. These are putative mechanisms as indicated by ?. TLR4- Toll-like receptor 4, IECs-Intestinal epithelial cells, AMPs-Antimicrobial peptides, NOD2-Nucleotide-bindingoligomerization domain-containing protein 2. (Figure created in BioRender).
Pattern recognition receptors (PRRs) are integral to the interaction between the host and gut microbiota. They are expressed on IECs and many immune cells and recognise pathogen-associated molecular patterns found in microbes. PRRs are divided into four main families: Toll-like receptors (TLR), nucleotide-binding oligomerisation domain-like receptors (NLR), C-type lectin receptors and RIG-1 like receptors. Altered function and expression of TLRs and NLRs have been specifically implicated in the pathogenesis of IBD and NEC, two conditions linked with microbial dysbiosis.
The role of PRRs, specifically TLR4 have been extensively studied in the pathogenesis of NEC (figure 2).61 TLR4, present on the surface of IECs, recognises and binds to bacterial LPS, which is a component of the outer surface of gram-negative organisms. The binding of LPS to TLR4 initiates the NF-kB signalling pathway leading to the production of inflammatory cytokines.62 Observations that there is increased TLR4 expression on IECs in the foetal gut compared with the full-term neonate have led researchers to consider the role of TLR4 in the aetiology of NEC.63 The reasons for this temporal alteration in TLR4 expression remains unclear, however, TLR4 downregulation may be a mechanism to promote tolerance of gram negative organisms when colonised on birth.63–65
Diet and gut hypoxia are also important in altered gut TLR4 expression. Rat and mouse models of NEC have demonstrated that intestinal TLR4 expression is significantly increased for formula fed hypoxia-stressed litters relative to those which are breastfed and are not hypoxia stressed.46 66 Human infants who develop clinical NEC also show increased intestinal TLR4 expression compared with those who do not.66 This relative increase in TLR4 expression may be related to the immature status of the preterm infant gut and immune system.
Although the aetiology of NEC is poorly defined, it is hypothesised that the immature gut and immune system interact with an altered gut microbiota. This leads to excessive and damaging gut inflammation. Preceding the onset of NEC, there is an increased abundance of gram-negative organisms such as Proteobacteria. These organisms contain LPS in their outer walls, leading to excessive gut inflammation via the LPS-TLR4 interaction.51 67 68
TLR4 polymorphisms have also been shown to be associated with susceptibility to both CD and UC.69 Furthermore, there is increased expression of TLR4 by IECs in both adult and paediatric populations with IBD (figure 2).70 Several studies have demonstrated an increased abundance of Enterobacteriaceae, specifically adherent-invasive E. coli found in tissue biopsies of people with CD.71 72 These organisms contain LPS in their outer walls, suggesting a role for LPS-TLR4 interaction driven inflammation similar to that previously described in NEC.
Another PRR linked to gut inflammation is NOD2. NOD2/CARD15 (nucleotide-binding oligomerisation domain 2) is one of a family of intracellular PRRs which recognise components from the degradation of microbial cell walls such as muramyl dipeptides. Mutations of NOD2, leading to decreased responsiveness to its ligands is associated with for example reduced production of antibacterial peptides, and has been consistently linked with susceptibility to CD and even increased risk of colorectal cancer (figure 3).73–77 NOD2 deficient mice demonstrate a specific mucosa-associated microbial dysbiosis with a higher proportion of Bacteroidetes, lower representation of Firmicutes in the colon and an increase in the relative abundance of pathogenic gram-negative bacterial species.78 In chemically induced mouse models of colitis, this NOD2-mediated dysbiosis can increase risk of inflammation with this risk being transmissible to wild-type mice colonised by faecal transfer with the same dysbiotic microbiota.79 Such changes in the microbiota are mirrored in human studies of CD,52 80 with a decrease in overall species richness, alpha-diversity and depletion of commensal species of the phyla Firmicutes and increased abundance of E. coli.56 Alterations in NOD2 signalling have also been implicated in driving inflammatory responses by promoting the production of inflammatory cytokines thus propagating inflammation. To our knowledge, the role of NOD2 has not been assessed in NEC but alterations in the function of several PRR may contribute to both conditions (figure 3). Furthermore, it is notable that there is cooperation between PRR signalling pathways including NOD2, TLR3, 4 and 9 to enhance inflammatory responses.81–83
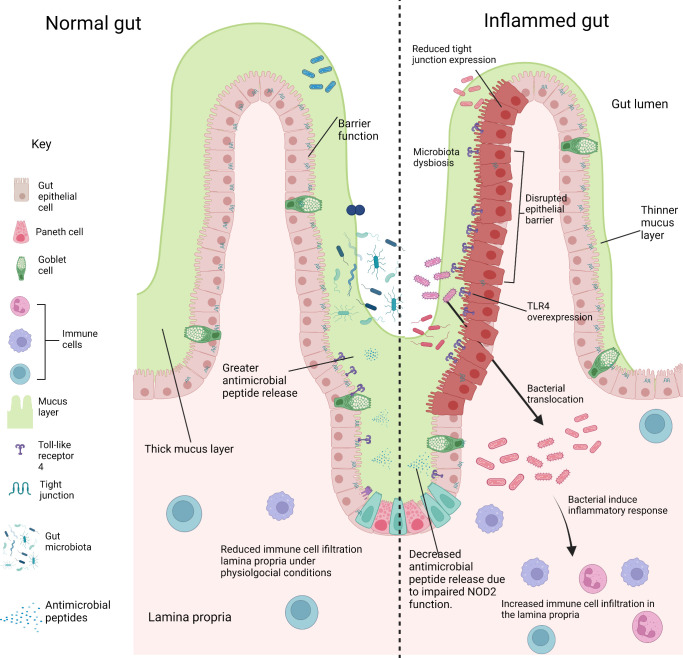
Figure 3.
Comparing the gut environment in physiological and inflammatory conditions. Key features of a healthy normal gut are shown (left), with a thick mucus layer and intactgut epithelium preventing the entry of gut microbiota to the underlying lamina propria. Antimicrobial peptides are secreted by paneth cells into the mucus layer and gut lumen, which regulates the growth of commensals and pathogens. Disruption of these factors leads to gut inflammation (right). The mucus layer is much thinner allowing the association ofbacteria to the epithelium. Gut epithelium barrier function is impaired by the reducedexpression of tight junctions, allowing the translocation of bacteria to the lamina propria. This drives inflammation and the increased infiltration of immune cells compared tophysiological conditions. Reduced expression of antimicrobial peptides occurs due to theimpaired function of Nucleotide-binding oligomerization domain-containing protein 2(NOD2). Reduced antimicrobial peptide secretion leads to bacterial overgrowth in the gutlumen. (Figure created in BioRender).
Altered gut epithelial barrier function in disease
Another mechanism by which the microbiome might influence disease onset is by impairment of gut barrier function and resultant increase in gut barrier permeability preceding the onset of inflammation. The intestinal barrier is formed by a layer of tightly connected IECs and has several mechanisms to reduce the risk of infection by pathogenic organisms. This includes regulation of barrier permeability by tight-junctional proteins, the formation of a mucus layer, to prevent attachment of pathogens and secretion of antimicrobial peptides (figure 3).
TLR4 overexpression in the premature gut and activation by the altered microbiota in NEC negatively impacts the homoeostasis of gut epithelial cells by increasing apoptosis and reducing epithelial cell proliferation, disrupting the ability to repair the intestinal epithelium.66 84 85 A further consequence of TLR4 overexpression is the enhanced recruitment of CD4 +T cells to the gut lamina propria, as seen in mutant mice which selectively overexpress TLR4 on IECs (figure 2).86 This enhanced CD4 +T cell infiltration is also seen in the NEC mouse model for wild-type mice and in the human intestine during NEC, indicating that lymphocyte infiltration in the gut lamina propria may play an important role in NEC pathology.86 In CD, NOD2 mutations have also been linked with altered gut turnover and gut permeability such a tight junctional complex alterations.87 88
Intestinal epithelial hyperpermeability has been a well-documented feature in CD and UC, with mucosal biopsies of patients demonstrating bacterial internalisation in IECs compared with negligible mucosal bacterial load in biopsies of healthy control patients.89 Mouse models with loss of endogenous E-cadherin, which maintains the tight junctions between IECs, developed histological features of CD within 3 months.90 Similarly, in rat and human studies of NEC, there is reduced expression of both E-cadherin gene and protein with associated increase in epithelial permeability. Rho-associated protein kinase (ROCK) is involved with regulation of cadherin function, with activation of ROCK leading to disruption of apical tight junctions.91 In rat models of NEC, increased ROCK1 activation is a feature of early NEC and ROCK1 inhibition being protective in this experimental model.92
The role of the altered microbiota in initiating epithelial barrier dysfunction is still poorly understood. However, the resultant interaction between the microbiota and host clearly plays a role in driving inflammation in both diseases.
Conclusions and future directions
In this review, we have highlighted the critical role of the gut microbiota in supporting host intestinal health and have outlined two inflammatory disorders of the GIT in which the gut microbiota is involved; IBD and NEC. An altered gut microbiota is associated with these diseases and a dysregulation of the immune system is also apparent. However, whether there is a causal relationship between altered gut microbiota and the host immune response is currently unclear, although the presence of a dysfunctional epithelial barrier is an important feature in both diseases.
We have outlined possible mechanisms which begin to explain how a dysbiotic gut microbiota in conjunction with the host immune system may give rise to pathological gut inflammation. In both NEC and IBD, alterations to PRRs and their expression by IECs may be interconnected with the gut microbiota and impaired barrier function which further drives inflammation and inappropriate activation of the immune response.
Targeting these mechanistic pathways may lead to the development of novel therapies for these diseases. Small molecule TLR4 inhibitors have shown promising results, reducing severity of inflammation and disease in animal models of NEC and IBD.93 94 Probiotics, defined as ‘live microorganisms which when administered in adequate amounts confer a health benefit on the host’,95 have shown promise in reducing rates of severe NEC and mortality in preterm infants.96 97 However, evidence of their benefits remains limited, particularly for extremely low birth weight (<1000 g) infants, who are at the greatest risk of developing NEC, neither is there sufficient evidence to determine the optimal probiotic regimen.98 99 In IBD, there is limited evidence of benefit for patients with a lack of well designed and adequately powered studies.
IBD and NEC are both complex and multifactorial gut inflammatory disorders and possible disease mechanisms are only beginning to be understood. Much work has been done to characterise the gut microbiota for both diseases preceding and during disease states which has uncovered specific changes in the gut microbiota. However, whether such changes are driving the onset of such diseases is unclear and could be correlative rather than causative. A better mechanistic understanding of disease will likely arise by moving away from observational study of the gut microbiota and towards studying specific functions of member organisms, thought to be involved in disease.
Footnotes
Twitter: @MissLHancock, @sheencr
Contributors: DS, DE and SC conceptualised and wrote the original draft manuscript. KZC, JM, AB, LH and SC reviewed and edited the original draft manuscript. SC provided oversight and overall supervision for this project.
Funding: DS and DE are both PhD students at the University of Manchester with funding from the Topol Fellowship and Biotechnology and Biological Sciences Research Council (BBSRC), respectively.
Competing interests: None declared.
Provenance and peer review: Commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Scarpellini E, Ianiro G, Attili F, et al. The human gut microbiota and virome: potential therapeutic implications. Dig Liver Dis 2015;47:1007–12. 10.1016/j.dld.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laforest-Lapointe I, Arrieta M-C. Microbial eukaryotes: a missing link in gut microbiome studies. mSystems 2018;3. 10.1128/mSystems.00201-17. [Epub ahead of print: 13 Mar 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007;449:804–10. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bäckhed F, Ley RE, Sonnenburg JL, et al. Host-Bacterial mutualism in the human intestine. Science 2005;307:1915–20. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- 5. Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science 2008;320:1647–51. 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abdill RJ, Adamowicz EM, Blekhman R. Public human microbiome data are dominated by highly developed countries. PLoS Biol 2022;20:e3001536. 10.1371/journal.pbio.3001536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Odamaki T, Kato K, Sugahara H, et al. Age-Related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 2016;16:90. 10.1186/s12866-016-0708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu C, Zhu H, Qiu P. Aging progression of human gut microbiota. BMC Microbiol 2019;19:236. 10.1186/s12866-019-1616-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moore RE, Townsend SD. Temporal development of the infant gut microbiome. Open Biol 2019;9:190128. 10.1098/rsob.190128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coyte KZ, Rao C, Rakoff-Nahoum S, et al. Ecological rules for the assembly of microbiome communities. PLoS Biol 2021;19:e3001116. 10.1371/journal.pbio.3001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rao C, Coyte KZ, Bainter W, et al. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 2021;591:633–8. 10.1038/s41586-021-03241-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015;17:690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 13. Tanaka M, Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int 2017;66:515–22. 10.1016/j.alit.2017.07.010 [DOI] [PubMed] [Google Scholar]
- 14. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J 2017;474:1823–36. 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gritz EC, Bhandari V. The human neonatal gut microbiome: a brief review. Front Pediatr 2015;3:17. 10.3389/fped.2015.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Homann C-M, Rossel CAJ, Dizzell S, et al. Infants' first solid foods: impact on gut microbiota development in two intercontinental cohorts. Nutrients 2021;13:2639. 10.3390/nu13082639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart CJ, Ajami NJ, O'Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018;562:583–8. 10.1038/s41586-018-0617-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clemente JC, Pehrsson EC, Blaser MJ, et al. The microbiome of uncontacted Amerindians. Sci Adv 2015;1:e1500183. 10.1126/sciadv.1500183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaulke CA, Sharpton TJ. The influence of ethnicity and geography on human gut microbiome composition. Nat Med 2018;24:1495–6. 10.1038/s41591-018-0210-8 [DOI] [PubMed] [Google Scholar]
- 20. Partula V, Mondot S, Torres MJ, et al. Associations between usual diet and gut microbiota composition: results from the milieu Intérieur cross-sectional study. Am J Clin Nutr 2019;109:1472–83. 10.1093/ajcn/nqz029 [DOI] [PubMed] [Google Scholar]
- 21. Senghor B, Sokhna C, Ruimy R, et al. Gut microbiota diversity according to dietary habits and geographical provenance. Hum Microb J 2018;7-8:1–9. 10.1016/j.humic.2018.01.001 [DOI] [Google Scholar]
- 22. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Graf D, Di Cagno R, Fåk F, et al. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis 2015;26:26164. 10.3402/mehd.v26.26164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LeBlanc JG, Milani C, de Giori GS, et al. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol 2013;24:160–8. 10.1016/j.copbio.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 25. Kamada N, Seo S-U, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013;13:321–35. 10.1038/nri3430 [DOI] [PubMed] [Google Scholar]
- 26. Sekirov I, Russell SL, Antunes LCM, et al. Gut microbiota in health and disease. Physiol Rev 2010;90:859–904. 10.1152/physrev.00045.2009 [DOI] [PubMed] [Google Scholar]
- 27. Topping DL, Clifton PM. Short-Chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev 2001;81:1031–64. 10.1152/physrev.2001.81.3.1031 [DOI] [PubMed] [Google Scholar]
- 28. Ríos-Covián D, Ruas-Madiedo P, Margolles A, et al. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 2016;7:185. 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 2014;5:e00889. 10.1128/mBio.00889-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gonçalves P, Araújo JR, Di Santo JP. A cross-talk between Microbiota-Derived short-chain fatty acids and the host mucosal immune system regulates intestinal homeostasis and inflammatory bowel disease. Inflamm Bowel Dis 2018;24:558–72. 10.1093/ibd/izx029 [DOI] [PubMed] [Google Scholar]
- 31. Cavaglieri CR, Nishiyama A, Fernandes LC, et al. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci 2003;73:1683–90. 10.1016/S0024-3205(03)00490-9 [DOI] [PubMed] [Google Scholar]
- 32. Suzuki T, Yoshida S, Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr 2008;100:297–305. 10.1017/S0007114508888733 [DOI] [PubMed] [Google Scholar]
- 33. Wong JMW, de Souza R, Kendall CWC, et al. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 2006;40:235–43. 10.1097/00004836-200603000-00015 [DOI] [PubMed] [Google Scholar]
- 34. Levy M, Kolodziejczyk AA, Thaiss CA, et al. Dysbiosis and the immune system. Nat Rev Immunol 2017;17:219–32. 10.1038/nri.2017.7 [DOI] [PubMed] [Google Scholar]
- 35. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15:382–92. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hill DA, Siracusa MC, Abt MC, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med 2012;18:538–46. 10.1038/nm.2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer 2013;13:800–12. 10.1038/nrc3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bellodas Sanchez J, Kadrofske M. Necrotizing enterocolitis. Neurogastroenterol Motil 2019;31:e13569. 10.1111/nmo.13569 [DOI] [PubMed] [Google Scholar]
- 39. Neu J, Pammi M. Necrotizing enterocolitis: the intestinal microbiome, metabolome and inflammatory mediators. Semin Fetal Neonatal Med 2018;23:400–5. 10.1016/j.siny.2018.08.001 [DOI] [PubMed] [Google Scholar]
- 40. Berkhout DJC, Klaassen P, Niemarkt HJ, et al. Risk factors for necrotizing enterocolitis: a prospective multicenter case-control study. Neonatology 2018;114:277–84. 10.1159/000489677 [DOI] [PubMed] [Google Scholar]
- 41. Raba AA, O'Sullivan A, Semberova J, et al. Are antibiotics a risk factor for the development of necrotizing enterocolitis-case-control retrospective study. Eur J Pediatr 2019;178:923–8. 10.1007/s00431-019-03373-0 [DOI] [PubMed] [Google Scholar]
- 42. Rose AT, Patel RM. A critical analysis of risk factors for necrotizing enterocolitis. Semin Fetal Neonatal Med 2018;23:374–9. 10.1016/j.siny.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Samuels N, van de Graaf RA, de Jonge RCJ, et al. Risk factors for necrotizing enterocolitis in neonates: a systematic review of prognostic studies. BMC Pediatr 2017;17:105. 10.1186/s12887-017-0847-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cilieborg MS, Boye M, Mølbak L, et al. Preterm birth and necrotizing enterocolitis alter gut colonization in pigs. Pediatr Res 2011;69:10–16. 10.1203/PDR.0b013e3181ff2a89 [DOI] [PubMed] [Google Scholar]
- 45. Grishin A, Papillon S, Bell B, et al. The role of the intestinal microbiota in the pathogenesis of necrotizing enterocolitis. Semin Pediatr Surg 2013;22:69–75. 10.1053/j.sempedsurg.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jilling T, Simon D, Lu J, et al. The roles of bacteria and TLR4 in rat and murine models of necrotizing enterocolitis. J Immunol 2006;177:3273–82. 10.4049/jimmunol.177.5.3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Millar MR, Linton CJ, Cade A, et al. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J Clin Microbiol 1996;34:2506–10. 10.1128/jcm.34.10.2506-2510.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Niemarkt HJ, De Meij TG, van Ganzewinkel C-J, et al. Necrotizing enterocolitis, gut microbiota, and brain development: role of the brain-gut axis. Neonatology 2019;115:423–31. 10.1159/000497420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ullrich T, Tang Y-W, Correa H, et al. Absence of gastrointestinal pathogens in ileum tissue resected for necrotizing enterocolitis. Pediatr Infect Dis J 2012;31:413–4. 10.1097/INF.0b013e318242534a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peter CS, Feuerhahn M, Bohnhorst B, et al. Necrotising enterocolitis: is there a relationship to specific pathogens? Eur J Pediatr 1999;158:67–70. 10.1007/s004310051012 [DOI] [PubMed] [Google Scholar]
- 51. Pammi M, Cope J, Tarr PI, et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: a systematic review and meta-analysis. Microbiome 2017;5:31. 10.1186/s40168-017-0248-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006;55:205–11. 10.1136/gut.2005.073817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Glymenaki M, Singh G, Brass A, et al. Compositional changes in the gut mucus microbiota precede the onset of Colitis-Induced inflammation. Inflamm Bowel Dis 2017;23:912–22. 10.1097/MIB.0000000000001118 [DOI] [PubMed] [Google Scholar]
- 54. Alipour M, Zaidi D, Valcheva R, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis 2016;10:462–71. 10.1093/ecco-jcc/jjv223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 2008;105:16731–6. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010;139:1844–54. 10.1053/j.gastro.2010.08.049 [DOI] [PubMed] [Google Scholar]
- 58. Kostic AD, Gevers D, Siljander H, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015;17:260–73. 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buttó LF, Haller D. Dysbiosis in intestinal inflammation: cause or consequence. Int J Med Microbiol 2016;306:302–9. 10.1016/j.ijmm.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 60. Jostins L, Ripke S, Weersma RK, et al. Host-Microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hackam DJ, Sodhi CP. Toll-Like receptor-mediated intestinal inflammatory imbalance in the pathogenesis of necrotizing enterocolitis. Cell Mol Gastroenterol Hepatol 2018;6:229–38. 10.1016/j.jcmgh.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 2010;11:373–84. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- 63. Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol 2012;12:9–23. 10.1038/nri3112 [DOI] [PubMed] [Google Scholar]
- 64. Lotz M, Gütle D, Walther S, et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med 2006;203:973–84. 10.1084/jem.20050625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meng D, Zhu W, Shi H. Toll-Like receptor-4 in human and mouse colonic epithelium is developmentally regulated: a possible role in necrotizing enterocolitis. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leaphart CL, Cavallo J, Gribar SC, et al. A critical role for TLR4 in the pathogenesis of necrotizing enterocolitis by modulating intestinal injury and repair. J Immunol 2007;179:4808–20. 10.4049/jimmunol.179.7.4808 [DOI] [PubMed] [Google Scholar]
- 67. Olm MR, Bhattacharya N, Crits-Christoph A. Necrotizing enterocolitis is preceded by increased gut bacterial replication, <em>Klebsiella</em>, and fimbriae-encoding bacteria that may stimulate TLR4 receptors. bioRxiv 2019;558676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Claud EC, Walker WA. Hypothesis: inappropriate colonization of the premature intestine can cause neonatal necrotizing enterocolitis. Faseb J 2001;15:1398–403. 10.1096/fj.00-0833hyp [DOI] [PubMed] [Google Scholar]
- 69. Cheng Y, Zhu Y, Huang X, et al. Association between TLR2 and TLR4 gene polymorphisms and the susceptibility to inflammatory bowel disease: a meta-analysis. PLoS One 2015;10:e0126803. 10.1371/journal.pone.0126803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Szebeni B, Veres G, Dezsõfi A, et al. Increased expression of Toll-like receptor (TLR) 2 and TLR4 in the colonic mucosa of children with inflammatory bowel disease. Clin Exp Immunol 2008;151:34–41. 10.1111/j.1365-2249.2007.03531.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen L, Wang W, Zhou R, et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine 2014;93:e51. 10.1097/MD.0000000000000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Darfeuille-Michaud A, Boudeau J, Bulois P, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004;127:412–21. 10.1053/j.gastro.2004.04.061 [DOI] [PubMed] [Google Scholar]
- 73. Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 2001;411:599–603. 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- 74. Ogura Y, Bonen DK, Inohara N, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001;411:603–6. 10.1038/35079114 [DOI] [PubMed] [Google Scholar]
- 75. Kurzawski G, Suchy J, Kładny J, et al. The NOD2 3020insC mutation and the risk of colorectal cancer. Cancer Res 2004;64:1604–6. 10.1158/0008-5472.can-03-3791 [DOI] [PubMed] [Google Scholar]
- 76. Wehkamp J, Schmid M, Fellermann K, et al. Defensin deficiency, intestinal microbes, and the clinical phenotypes of Crohn’s disease. J Leukoc Biol 2005;77:460–5. 10.1189/jlb.0904543 [DOI] [PubMed] [Google Scholar]
- 77. Elphick D, Liddell S, Mahida YR. Impaired luminal processing of human defensin-5 in Crohn's disease: persistence in a complex with chymotrypsinogen and trypsin. Am J Pathol 2008;172:702–13. 10.2353/ajpath.2008.070755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Al Nabhani Z, Lepage P, Mauny P, et al. Nod2 deficiency leads to a specific and transmissible mucosa-associated microbial dysbiosis which is independent of the mucosal barrier defect. J Crohns Colitis 2016;10:1428–36. 10.1093/ecco-jcc/jjw095 [DOI] [PubMed] [Google Scholar]
- 79. Couturier-Maillard A, Secher T, Rehman A, et al. Nod2-Mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest 2013;123:700–11. 10.1172/JCI62236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Scanlan PD, Shanahan F, O'Mahony C, et al. Culture-Independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J Clin Microbiol 2006;44:3980–8. 10.1128/JCM.00312-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leung C-H, Lam W, Ma D-L, et al. Butyrate mediates nucleotide-binding and oligomerisation domain (NOD) 2-dependent mucosal immune responses against peptidoglycan. Eur J Immunol 2009;39:3529–37. 10.1002/eji.200939454 [DOI] [PubMed] [Google Scholar]
- 82. Tada H, Aiba S, Shibata K-I, et al. Synergistic effect of Nod1 and NOD2 agonists with Toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect Immun 2005;73:7967–76. 10.1128/IAI.73.12.7967-7976.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. van Heel DA, Ghosh S, Hunt KA, et al. Synergy between TLR9 and NOD2 innate immune responses is lost in genetic Crohn's disease. Gut 2005;54:1553–7. 10.1136/gut.2005.065888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mara MA, Good M, Weitkamp J-H. Innate and adaptive immunity in necrotizing enterocolitis. Semin Fetal Neonatal Med 2018;23:394–9. 10.1016/j.siny.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sodhi CP, Neal MD, Siggers R, et al. Intestinal epithelial Toll-like receptor 4 regulates goblet cell development and is required for necrotizing enterocolitis in mice. Gastroenterology 2012;143:708–18. 10.1053/j.gastro.2012.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Egan CE, Sodhi CP, Good M, et al. Toll-Like receptor 4-mediated lymphocyte influx induces neonatal necrotizing enterocolitis. J Clin Invest 2016;126:495–508. 10.1172/JCI83356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Amendola A, Butera A, Sanchez M, et al. Nod2 deficiency is associated with an increased mucosal immunoregulatory response to commensal microorganisms. Mucosal Immunol 2014;7:391–404. 10.1038/mi.2013.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. D'Incà R, Annese V, di Leo V, et al. Increased intestinal permeability and NOD2 variants in familial and sporadic Crohn's disease. Aliment Pharmacol Ther 2006;23:1455–61. 10.1111/j.1365-2036.2006.02916.x [DOI] [PubMed] [Google Scholar]
- 89. Kleessen B, Kroesen AJ, Buhr HJ, Blaut M, et al. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol 2002;37:1034–41. 10.1080/003655202320378220 [DOI] [PubMed] [Google Scholar]
- 90. Hermiston ML, Gordon JI. Inflammatory bowel disease and adenomas in mice expressing a dominant negative N-cadherin. Science 1995;270:1203–7. 10.1126/science.270.5239.1203 [DOI] [PubMed] [Google Scholar]
- 91. Grothaus JS, Ares G, Yuan C, et al. Rho kinase inhibition maintains intestinal and vascular barrier function by upregulation of occludin in experimental necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 2018;315:G514–28. 10.1152/ajpgi.00357.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Buonpane C, Yuan C, Wood D, et al. Rock1 inhibitor stabilizes E-cadherin and improves barrier function in experimental necrotizing enterocolitis. Am J Physiol Gastrointest Liver Physiol 2020;318:G781–92. 10.1152/ajpgi.00195.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Neal MD, Jia H, Eyer B, et al. Discovery and validation of a new class of small molecule Toll-like receptor 4 (TLR4) inhibitors. PLoS One 2013;8:e65779. 10.1371/journal.pone.0065779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tam JSY, Coller JK, Hughes PA, et al. Toll-Like receptor 4 (TLR4) antagonists as potential therapeutics for intestinal inflammation. Indian J Gastroenterol 2021;40:5–21. 10.1007/s12664-020-01114-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hill C, Guarner F, Reid G, et al. Expert consensus document. The International scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014;11:506–14. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- 96. AlFaleh K, Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Evid.-Based Child Health 2014;9:584–671. 10.1002/ebch.1976 [DOI] [PubMed] [Google Scholar]
- 97. Athalye-Jape G, Patole S. Probiotics for preterm infants - time to end all controversies. Microb Biotechnol 2019;12:249–53. 10.1111/1751-7915.13357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Pell LG, Loutet MG, Roth DE, et al. Arguments against routine administration of probiotics for NEC prevention. Curr Opin Pediatr 2019;31:195–201. 10.1097/MOP.0000000000000730 [DOI] [PubMed] [Google Scholar]
- 99. van den Akker CHP, van Goudoever JB, Shamir R, et al. Probiotics and preterm infants: a position paper by the European Society for paediatric gastroenterology hepatology and nutrition Committee on nutrition and the European Society for paediatric gastroenterology hepatology and nutrition Working group for probiotics and prebiotics. J Pediatr Gastroenterol Nutr 2020;70:664–80. 10.1097/MPG.0000000000002655 [DOI] [PubMed] [Google Scholar]