Abstract
Inhibitory myeloid cells and their cytokines play critical roles in limiting chimeric antigen receptor T (CART) cell therapy by contributing to the development of toxicities and resistance following infusion. We have previously shown that neutralization of granulocyte-macrophage colony-stimulating factor (GM-CSF) prevents these toxicities and enhances CART cell functions by inhibiting myeloid cell activation. In this report, we study the direct impact of GM-CSF disruption during the production of CD19-directed CART cells on their effector functions, independent of GM-CSF modulation of myeloid cells. In this study, we show that antigen-specific activation of GM-CSFKO CART19 cells consistently displayed reduced early activation, enhanced proliferation, and improved anti-tumor activity in a xenograft model for relapsed B cell malignancies. Activated CART19 cells significantly upregulate GM-CSF receptors. However, the interaction between GM-CSF and its upregulated receptors on CART cells was not the predominant mechanism of this activation phenotype. GM-CSFKO CART19 cell had reduced BH3 interacting-domain death agonist (Bid), suggesting an interaction between GM-CSF and intrinsic apoptosis pathways. In conclusion, our study demonstrates that CRISPR/Cas9-mediated GM-CSF knockout in CART cells directly ameliorates CART cell early activation and enhances anti-tumor activity in preclinical models.
INTRODUCTION
Despite the remarkable activity of CD19-directed chimeric antigen receptor T cell (CART19) therapy in treating B-cell hematologic malignancies,1, 2 limitations include 1) the development of potential life-threatening complications such as neurotoxicity (NT) and cytokine release syndrome (CRS)3, 4 and 2) lack of durable response.5–8 Emerging literature suggests that inhibitory myeloid cells and their cytokines play an important role in inducing CART cell toxicities and contribute to CART inhibition.9–12 Specifically, and of relevance to the work presented in this manuscript, granulocyte-macrophage colony-stimulating factor (GM-CSF) is implicated in the development of NT and CRS after CART19 therapy based on correlative studies from pivotal clinical trials.13, 14
GM-CSF is produced by macrophages, T cells, NK cells, endothelial cells, and fibroblasts and plays several roles in the hematopoietic and immune system.15 GM-CSF plays a role in stimulating stem cells to differentiate into monocytes, granulocytes, and neutrophils.15 GM-CSF also activates monocytes and differentiates them into macrophages and is a component of the immune response to infections.15 Following allogeneic transplantation, GM-CSF has also been demonstrated to drive graft versus host pathology by licensing donor-derived myeloid cells to produce inflammatory mediators such as interleukin 1β.16 GM-CSF was also shown to recruit dendritic cells and promote graft versus host disease, amplifying the activation of alloreactive T cells.16
In an analysis from the pivotal ZUMA-1 clinical trial, which led to FDA approval of the CART19 product, axicabtagene ciloleucel (Axi-cel), in adults with relapsed or refractory large B-cell lymphoma, GM-CSF was the most significantly associated cytokine with the development of CRS and NT.13 Our group identified that neutralization of GM-CSF results in prevention of CRS and NT while enhancing CART19 anti-tumor activity in preclinical models.17 Specifically, our work indicated that GM-CSF neutralization with lenzilumab reduces monocyte activation and decreases inhibitory myeloid cytokines, which in turn ameliorates the development of CRS, prevents neuroinflammation, and improves CART cell anti-tumor activity.17, 18 This preclinical work led to the initiation of the ZUMA-19 study, a phase Ib/II multi-center study of lenzilumab and Axi-cel as sequenced therapy in adults with relapsed or refractory large B-cell lymphoma (NCT04314843).19
While GM-CSF is secreted primarily by myeloid cells and contributes to their activation, it is also produced by T cells.15 We therefore hypothesized that inhibition of GM-CSF during CART cell manufacturing20 may result in reduced GM-CSF levels and decreased monocyte activation. Indeed, we have shown that CRISPR/Cas9 gene editing of GM-CSF in CART cells during the ex vivo expansion process generates GM-CSFKO CART19 cells which produce significantly less GM-CSF while maintaining their effector functions. This work was recently corroborated by a separate, independent preclinical study using TALENs to knock out GM-CSF in CAR-T cells.21 In fact, our results indicate that GM-CSFKO CART19 cells exhibit superior anti-tumor activity in xenograft models in which myeloid cells are lacking. This pointed to a direct impact of GM-CSF on T cells, independent of its role as a mediator of monocyte activation and monocyte-induced T cell inhibition.
To this end, GM-CSF-producing T cells (ThGM) have been identified as a novel subset of T cells with unique phenotypic and functional properties.22 Specifically, ThGM cells were found to induce activation of other T cell subsets, thus amplifying T cell responses.22 ThGM were also demonstrated to be more susceptible to apoptosis and activation-induced cell death (AICD).22
We therefore hypothesized that GM-CSFKO CART cells result in modulation of CART cell activation and enhanced anti-tumor activity independent of the effects of GM-CSF depletion on myeloid cell activation. In this work, we test this hypothesis and study the direct impact of GM-CSF knockout on CART cell function.
MATERIALS/SUBJECTS AND METHODS
Cell lines.
The following cells lines were purchased from ATCC: acute lymphoblastic leukemia cell line Nalm6 (Manassas, VA, USA) and mantle cell lymphoma cell line Jeko-1 (Manassas, VA, USA). Both cell lines (Jeko-1 and Nalm6) were transduced with a firefly luciferase ZsGreen (Addgene, Cambridge, MA, USA) and then sorted to obtain >99% positive population as previously described.17 Cell lines were cultured in R10 or R20 (RPMI 1640, Gibco, Gaithersburg, MD, US), 10% or 20% Fetal Bovine Serum (FBS, Millipore Sigma, Ontario, Canada), respectively, and 1% Penicillin-Streptomycin-Glutamine (Gibco, Gaithersburg, MD, US). Cell lines were kept in culture up to 20 passages, and fresh aliquots were thawed every 7–8 weeks. Cell lines were authenticated by the manufacturer and routinely checked for phenotype by flow cytometry. Cell lines were tested monthly for mycoplasma. The use of recombinant DNA in the laboratory was approved by the Mayo Clinic Institutional Biosafety Committee (IBC).
CART Cells.
Generation of constructs, lentiviral production, titration, GM-CSFKO CART19 cells, and T cell functional experiments were previously described.17, 18 The CSF2 gRNA sequence is GACCTGCCTACAGACCCGCC and the non-targeting (Ctrl) gRNA sequence is GCACTTTGTTTGGCCTACTG.
Sequencing.
RNA isolation and analysis were previously described.17 DNA was isolated using PureLink Genomic DNA Mini Kit (Invitrogen, Carlsbad, CA, USA), prepared with Agilent SureSelectXT (Santa Clara, CA, USA), and sequenced on Illumina HiSeq 4000 (Illumina, San Diego, CA, USA) by the Medical Genome Facility Genome Analysis Core (Mayo Clinic, Rochester, MN, USA). Burrows-Wheeler Aligner23 and Genome Analysis Toolkit24 were used to align reads to GRCh28 and call variants. SAS 9.4 (SAS Institute Inc., Cary, NC, USA) was used to find differences and filter by genomic prevalence (allele frequency ≤1%).25 CRISPR/Cas9 target online predictor (CCTop) off-target predictions were cross-referenced.26 Single nucleotide variants (SNVs) or insertions/deletions (indels) were compared between knockouts and controls. In order to determine disruption efficiency of CRISPR/Cas9 GM-CSF knockout using targeted sequencing through PCR (Polymerase Chain Reaction) and TIDE (Tracking of Indels by Decomposition) analysis, the latter using the available software at https://tide.nki.nl/ as previously described.27 Statistical testing using Wilcoxon signed-rank test performed using GraphPad Prism version 8.1.1 for Windows (GraphPad Software, La Jolla, CA, USA, www.graphpad.com).28
Flow Cytometric Analysis.
Extracellular staining, acquisition, and gating were previously described.17 The following anti-human antibodies were used: anti-CD116 (GM-CSFRα) (clone 4H1) FITC (305906, BioLegend, San Diego, CA, USA), anti-CD131 (GM-CSFRβ) (clone 1C1) PE (306104, BioLegend, San Diego, CA, USA), GM-CSF (clone BVD2–21C11) BV421 (562930, BD Biosciences, San Diego, CA, USA), CD262 (clone DJR2–4) PE (307405, BioLegend, San Diego, CA, USA), CD3 (clone OKT3) BV421 (317344, BioLegend, San Diego, CA, USA), CD3 (clone SK7) APC-H7 (560176, BD Pharmingen, San Jose, CA, USA), CD4 (clone OKT4) FITC (11–0048-42, eBioscience, San Diego, CA, USA), CD8 (clone SK1) PerCP (344707, BioLegend, San Diego, CA, USA), CD3 (clone OKT3) BV650 (317324, BioLegend, San Diego, CA, USA), CD197/CCR7 (clone G043H7) PE (353203, BioLegend, San Diego, CA, USA), CD45RA (clone HI100) APC (304111, BioLegend, San Diego, CA, USA), (CD45 (clone HI30) BV421 (304032, BioLegend, San Diego, CA, USA), CD20 (clone L27) PE (302306, BioLegend, San Diego, CA, USA), CD25 (clone M-A251) PE-Cy7 (557741, BD Biosciences, San Jose, CA, USA), CD69 (clone FN50) BV785 (310932, BioLegend, San Diego, CA, USA), HLA-DR (clone L243) APC-Fire/750 (307657, BioLegend, San Diego, CA, USA). Absolute quantification was obtained using volumetric measurement or CountBright absolute counting beads (C36950, Invitrogen, Carlsbad, CA, USA). Surface expression of CAR was detected by staining with a goat anti-mouse F(ab’)2 antibody (Invitrogen, Carlsbad, CA, USA). Cytometric data were acquired using a CytoFLEX Flow Cytometer (Beckman Coulter, Chaska, MN, USA). Gating was performed using Kaluza version 2.1 (Beckman Coulter, Chaska, MN, USA).
Memory T cell phenotype assay.
Extracellular staining and acquisition were previously described.17 The following antibodies were used: CD197/CCR7 (clone G043H7) PE (353203, BioLegend, San Diego, CA, USA), CD45RA (clone HI100) APC (304111, BioLegend, San Diego, CA, USA), and LIVE/DEAD™ Fixable Aqua Dead Cell Stain Kit (L34966, Invitrogen, Carlsbad, CA, USA). The following gating strategy was used to assess different T cell subsets: Naïve Phenotype (CCR7+CD45RA+), Central Memory /TCM (CCR7+CD45RA−), Effector Memory Cells re-expressing CD45RA/TEMRA (CCR7−CD45RA+) and Effector Memory/TEM (CCR7−CD45RA−).
Apoptosis assays.
GM-CSFWT CART19 or GM-CSFKO CART19 cells were stimulated with PMA/Ionomycin (Millipore Sigma, Ontario, Canada), CD19+ cell line Nalm6, or Cell Therapy Systems Dynabeads CD3/CD28 (Life Technologies, Oslo, Norway) at different time points (0hr, 1hr, 2hr, 4hr, 6hr, 1 day, 2 days, 3 days, 4 days, or 5 days). The following amounts were used for stimulation: 50 ng/mL of PMA, 1 ug/mL of ionomycin, 3:1 ratio of beads:cells, and 1:1 ratio of Nalm6:CART19. Then, cells were spun and washed with flow buffer, followed by incubation in the dark with the following reagents: CD3 (clone SK7) APC-Cy7 (344818, BioLegend, San Diego, CA, USA), Annexin V PE (556421, BD Biosciences, San Jose, CA, USA), 7-AAD (559925, BD Biosciences, San Jose, CA), and 1X annexin binding buffer (1:10 dilution of 10X ABB) (556454, BD Biosciences, San Jose, CA, USA). Then, the expression of Annexin V and 7-AAD on CD3 cells was measured via flow cytometry. For assays including recombinant exogenous hrGM-CSF (78190, Stemcell Technologies, Vancouver, Canada), the following doses were added: 100 and 1000 ng/mL. For exogenous FasL protein (FAL-H5241, ACRO Biosystems, Newark, DE, USA), the following dose was used: 50ng/mL.
Degranulation Assay.
CART19 cells were stimulated with PMA/ionomycin (Millipore Sigma, Ontario, Canada) or CD19+ cell line Nalm6 for 4 hours. The following amounts were used for stimulation 50 ng/mL of PMA, 1 ug/mL of ionomycin, and 1:5 ratio CART19:Nalm6. This assay was performed as previously described,17 and flow cytometric staining for GM-CSF, CD3, and CD69 were assessed by flow cytometry using the antibodies described in the flow cytometry section.
Bid-knockout CART19 with siRNA.
CART19 cells were washed three times in Opti-MEM (Gibco, Gaithersburg, MD, USA) and stimulated with CD3/CD28 beads (Life Technologies, Oslo, Norway) at a 1:3 ratio of beads:cells. 1 million cells were plated per well in a total volume of 1 mL in a 48-well flat bottom plate (Corning, Corning, NY, USA). The siRNA conditions Bid (Invitrogen, Carlsbad, CA, USA), control (Invitrogen, Carlsbad, CA, USA), or no siRNA (Opti-MEM, Gibco, Gaithersburg, MD, USA) were incubated for 5 min with an equal amount of 6% lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) diluted in Opti-MEM. The diluted siRNA conditions were added to the stimulated CART cells at a final dose of 10 pmol/well and incubated for 24 hours. The next day, CART cells were de-beaded and then incubated with irradiated Nalm6 cells (CD19+ antigen) at a 1:1 ratio for 2 hours. Cells were harvested for western blot analysis, and the apoptosis assay was performed.
Western blot.
For immunoblot assays, irradiated CD19+ cell line Nalm6 was co-cultured at a 1:1 ratio with CART19 or GM-CSFKO CART19 cells at different time points (0hr, 1hr, 2hr, 4hr, 6hr). Cell pellets were washed with PBS and lysed in 100μL of RIPA buffer (Thermo Fisher, Waltham, MA, USA), and protein concentration was measured by BCA protein assay (Thermo Fisher, Waltham, MA, USA). SDS-PAGE gels were used to resolve 30μg cell lysates, and proteins were transferred to Nitrocellulose membranes via wet transfer. Nitrocellulose membranes were blocked with 5% BSA in 1X TBS-T wash buffer (10x Tris Buffer Saline [BioRad, Hercules, CA, USA] + Tween® 20 detergent [Millipore Sigma, Ontario, Canada]) for 1 hr at room temperature. Membranes were incubated overnight at 4°C with the following antibodies: BID Antibody (human specific) (2002S, Cell Signaling, Danvers, MA, USA) (dilution 1:1000), Fas (4C3) Mouse mAb (8023S, Cell Signaling, Danvers, MA, USA), and β-Actin (D6A8) Rabbit mAb (8457T, Cell Signaling, Danvers, MA, USA). Membranes were washed with 1X TBS-T and incubated with HRP-conjugated secondary antibodies at a dilution of 1:1000 for 1 hr at room temperature. Blots were revealed using the SuperSignal West Pico Plus Chemiluminescent substrate (Thermo Fisher, Waltham, MA, USA).
Animal Models.
Six-to-eight-week-old non-obese diabetic/severe combined immunodeficient mice bearing a targeted mutation in the interleukin (IL)-2 receptor gamma chain gene (NSG) female mice were purchased from Jackson Laboratories (Jackson Laboratories, Bar Harbor, ME, USA) and then maintained at the Mayo Clinic animal facility. Five mice were used per group to balance statistical power and ethical considerations; these experimental designs were approved by Institutional Animal Care and Use Committee (A00001767). Mice were maintained in an animal barrier space that is approved by the Institutional Biosafety Committee for BSL2+ level experiments (IBC #HIP00000252). Initial blindness was achieved by assigning numbers to mice and samples, although this is ineffective once the dramatic difference in results is observed. Mice were intravenously injected with 1.0×106 luciferase+ JeKo-1 cells. Fourteen days after injection, mice were imaged with a bioluminescent imager using an IVIS® Lumina S5 Imaging System (PerkinElmer, Hopkinton, MA, USA) to confirm engraftment. Imaging was performed 10 minutes after the intraperitoneal injection of 10 μL/g D-luciferin (15 mg/mL, Gold Biotechnology, St. Louis, MO, USA). Mice were then randomized using the rank-order method based on their bioluminescence imaging to receive either untransduced T cells, GM-CSFWT CART19, or GM-CSFKO CART19. Serial bleeding was performed and CD3 was quantified using flow cytometry. Briefly, mouse peripheral blood was lysed using BD FACS Lyse buffer (BD Biosciences, San Jose, CA, USA) and stained with anti-human CD3 APC-Cy7 (344818, BioLegend, San Diego, CA, USA) and anti-mouse CD45 (clone 30-F11) PE (103106, BioLegend, San Diego, CA, USA). Absolute quantification was performed using CountBright absolute counting beads (Invitrogen, Carlsbad, CA, USA). Mice were euthanized for necropsy when moribund, and spleens were harvested. Splenic cells were rinsed with RPMI 1640 (Invitrogen, Carlsbad, CA, USA), broken up with a 20 mL syringe plunger (Coviden, Mansfield, MA, USA), and strained with a 70 uM filter (Falcon, Corning, NY, USA). Then, they were frozen into 3 cryovials with 1 mL of freezing medium containing 10% DMSO (Millipore Sigma, St. Louis, MO, USA) and 90% FBS (Corning, Corning, NY, USA). To analyze subsets, the vials were thawed into RPMI and analyzed using flow cytometry. See memory T cell phenotype assay.
Statistical Analyses.
All statistics were performed using GraphPad Prism version 9.02 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com) or DeSeq2.29 Statistical tests are described in the figure legends. Briefly, one-way ANOVA was used to test the hypotheses for CAR expression, activation, apoptosis, cytokines, phenotype, and proliferation. Two-way ANOVA was used to test the hypotheses for apoptosis and GM-CSFR expression across different timepoints, in vivo tumor burden, and log-rank test was used to test the hypotheses for in vivo survival. Data was not excluded from statistical analysis.
RESULTS
GM-CSF directly contributes to CART cell activation and apoptosis
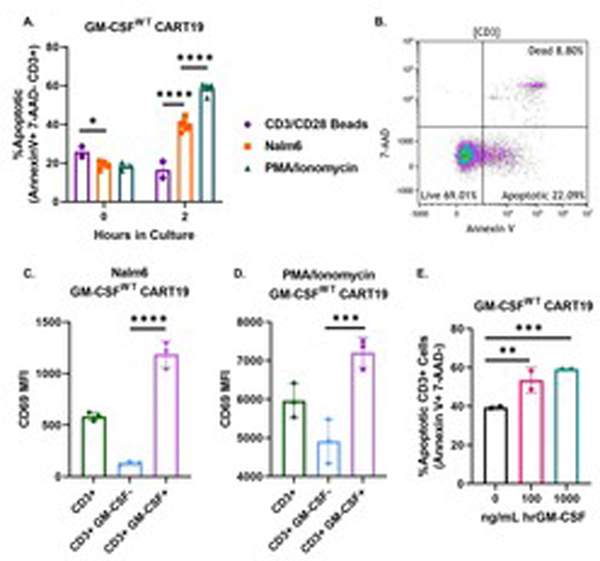
To study CART cell activation, we first assessed CART cell apoptosis as measured by the flow cytometric expression of Annexin V at early activation timepoints following either antigen-specific stimulation through the CAR (with CD19+ Nalm6 cells), non-specific stimulation through the TCR (with CD3/CD28 beads), or non-specific stimulation through Ca2+ influx (with PMA/ionomycin). There were significantly more apoptotic CART19 cells (Annexin V+ 7-AAD−) when CART cells were stimulated via the CAR or PMA/ionomycin compared to non-specific stimulation through the TCR (Fig. 1A–B). Significantly, GM-CSF+ CART19 cells exhibited higher levels of activation through the expression of significantly higher levels of CD69 compared to the GM-CSF− cells with either stimulation method (Fig. 1C–D). Based on these results, we decided to assess whether GM-CSF directly affects apoptosis on CART cells. To do this, we co-cultured CART19 with CD19+ irradiated Nalm6 for 4 hours to activate them through their CAR and then added increasing doses of exogenous GM-CSF (Fig. 1E). In this experiment, we observed that GM-CSF significantly increased CART cell apoptosis in a dose-dependent manner. This suggests that GM-CSF contributes to CART cell activation and apoptosis.
Fig. 1. GM-CSF directly contributes to CART cell activation and apoptosis. (A) GM-CSFWT CART19 cells are more apoptotic when stimulated through the CAR (CD19+ Nalm6) than TCR (CD3/CD28 beads).
CART19 cells were co-cultured with CD19+ cell line Nalm6 (CAR stimulation), CD38/CD28 beads (TCR stimulation), and PMA/ionomycin (Ca2+ influx stimulation). Flow cytometric staining for Annexin V, 7-AAD, and CD3 was performed at 0hr and 2hr (two-way ANOVA; ** p-value <0.01, *** p< 0.001, **** p<0.0001; 4 biological replicates, 2 technical replicates; error bars, SEM). (B) Representative flow plot of showing the expression of GM-CSFWT CART19 showing apoptotic cells. (C-D) GM-CSF+ CART19 cells express higher levels of the activation marker CD69. CART19 cells were co-cultured either in the presence of the CD19+ cell line Nalm6 (C) or PMA/Ionomycin (D) for a 4hr degranulation assay. Flow cytometric analysis of CD3, CD69, and intracellular GM-CSF was performed (one-way ANOVA; *** p-value < 0.001, **** p< 0.0001, 1 biological replicate, 3 technical replicates; error bars, SEM). (E) Exogenous hrGM-CSF increases CART19 cell apoptosis in a dose-dependent manner. CART19 cells were co-cultured with CD19+ cell line Nalm6 and increasing doses of human recombinant GM-CSF (0, 100, and 1000 ng/mL). Flow cytometric staining for Annexin V, 7-AAD, and CD3 was performed after 4 hours (one-way ANOVA; ** p <0.01, *** p <0.001; 1 biological replicate, 2 technical replicates; error bars, SEM).
Knockout of GM-CSF in CART19 cells enhances their antigen-specific proliferation
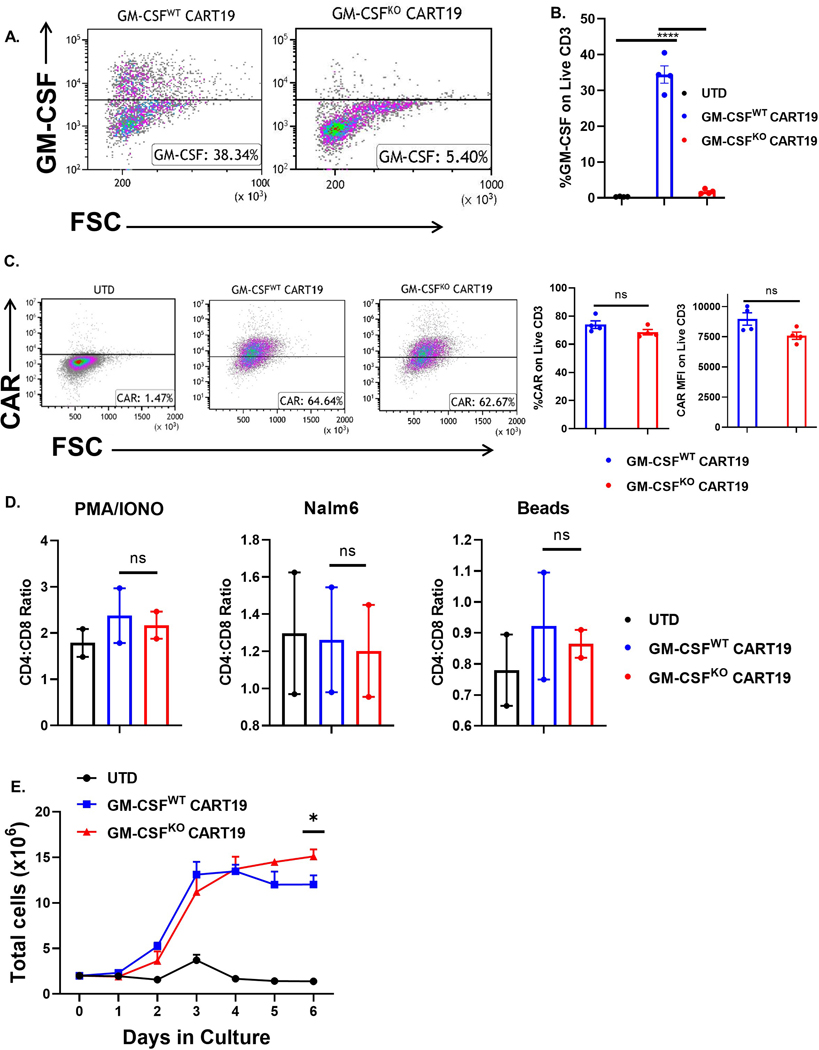
To investigate how GM-CSF affects CART cells, we generated GM-CSFKO CART cells using CRISPR/Cas9 to disrupt GM-CSF (CSF2) in CART19 cells, resulting in CART cells that produced little to no GM-CSF upon antigen-specific activation through their CAR (Fig. 2A–B, see method section, CART cells, supplementary Fig. S1). GM-CSF disruption in CART cells did not affect the transduction efficiency or CAR expression (Fig. 2C), change the composition of CART cell product (CD4:CD8 ratio) at rest or upon activation (Fig. 2D), or alter CART cell antigen-specific killing (supplementary Fig. S2). However, GM-CSFKO CART19 cells exhibited superior antigen-specific proliferation compared to GM-CSFWT CART19 when stimulated through the CAR19 via a co-culture with irradiated CD19+ Nalm6 cells (Fig. 2E). Importantly, while antigen-specific proliferation of GM-CSFKO CART19 and GM-CSFWT CART19 was initially similar, proliferation significantly increased in GM-CSFKO CART19 after 5 days following the initial stimulation.
Fig. 2. GM-CSF knockout via CRISPR/Cas9 does not impair CART19 production and effector functions. (A-B) CRISPR/Cas9 depletion of GM-CSF in CART19 cells produced little to no GM-CSF upon CAR19 stimulation.
Representative flow plot (A) or bar graph (B) showing the levels of GM-CSF detected on live CD3+ cells by intracellular flow cytometric staining upon stimulation with CD19+ cell line Nalm6 for 4 hours (one-way ANOVA, **** p<0.0001; 4 biological replicates, 2 technical replicates; error bars, SEM). (C) CAR19 expression is not impaired by depletion of GM-CSF via CRISPR/Cas9. Representative flow plot showing no differences in CAR19 expression between GM-CSFWT vs GM-CSFKO CART19 cells after CAR19 stimulation via flow cytometric staining (one-way ANOVA, ns = not significant; 4 biological replicates; error bars, SEM). (D) GM-CSF disruption does not affect the composition of CART19 (CD4:CD8 ratio) at rest or upon activation. UTD, GM-CSFWT or GM-CSFKO CART19 cells were co-cultured with either CD19+ cell line Nalm6 (CAR stimulation), CD3/CD28 beads (TCR stimulation) or PMA/ionomycin (Ca2+ influx stimulation) for 5 days, followed by flow cytometric staining of CD4 and CD8 staining (one-way ANOVA, ns = not significant; 2 biological replicates, 2 technical replicates; error bars, SEM). (E) GM-CSFKO CART19 show enhanced delayed proliferation. GM-CSFKO and GM-CSFWT CART19 cells were co-cultured with irradiated CD19+ cell line Nalm6, and cell counts were obtained daily for 6 days (one-way ANOVA, * p < 0.05; 2 biological replicates, 1 technical replicate; error bars, SEM).
GM-CSF editing of CART19 cells is precise and specific
Having shown that GM-CSFKO CART19 cells exhibit enhanced antigen-specific proliferation, we aimed to rule out an off-target effect of the CSF2-directed gRNA. We performed whole exome sequencing (WES) of GM-CSFWT CART19 and GM-CSFKO CART19 cells. Using CRISPR/Cas9 to disrupt CSF2 resulted in a knockout efficiency of 60–70% (supplementary Fig. S3A). WES of the modified cells showed no significant difference in single nucleotide variants (SNV) or indels between GM-CSFKO and control (GM-CSFWT) CART19 cells (supplementary Fig. S3B). WES was only significant for alterations in the intended gene target (supplementary Fig. S3C-D). The high precision and specificity of targeting GM-CSF exon 3 indicated that improved CART function is unlikely due to an off-target effect of the guide RNA, suggesting a direct effect of GM-CSF inhibition on CART cells.
GM-CSFKO CART19 cells exhibit a distinct transcriptomic and immunophenotypic profile
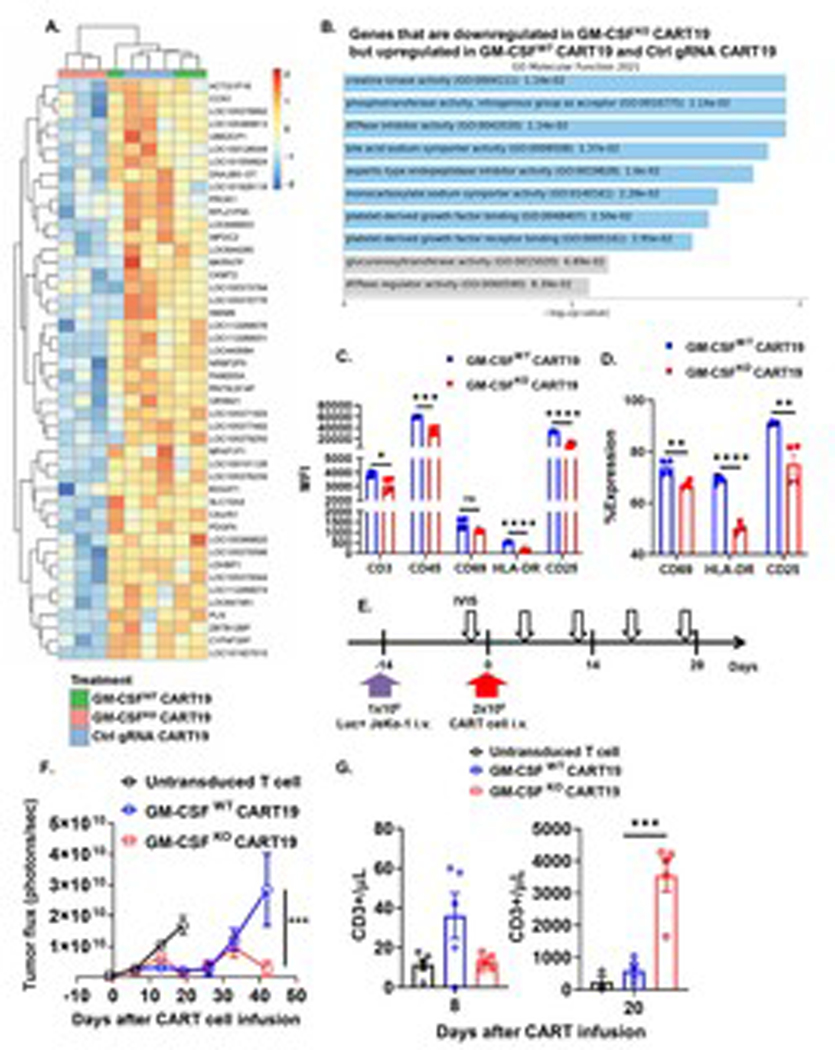
Since our WES data suggest that the improved efficacy of GM-CSFKO CART19 cells is not due to an off-target effect caused by CRISPR/Cas9 genome editing, we decided to examine the different activation pathways in resting GM-CSFKO and GM-CSFWT CART19 cells compared to Ctrl gRNA CART19 at the end of the CART cell manufacturing process (see methods). Transcriptome interrogation revealed 46 genes that were significantly downregulated in resting GM-CSFKO CART19 but upregulated in both resting GM-CSFWT CART19 and Ctrl gRNA CART19 (Fig. 3A). Gene set enrichment analysis demonstrated that the creatine kinase pathway was most significantly altered after GM-CSF disruption in CART19 cells (Fig. 3B), which is associated with the regulation of T cell activation.30 Also of interest is the alteration of CCN1, which regulates Fas-mediated apoptosis.31 These results suggest that GM-CSFKO CART cells have altered susceptibility to activation and potentially apoptosis at their baseline.
Fig. 3. GM-CSF disruption on CART19 modulates early activation and anti-tumor activity.
(A) Comparison of gene expression between GM-CSFKO CART19 cells and control. Differential expression by heatmap on RNA isolated from GM-CSFWT CART19 cells, GM-CSFKO CART19, and non-targeting gRNA control CART19 on day 8 of CART expansion (p-value < 0.01, 3 biological replicates). (B) Activation pathways are enriched in GM-CSFKO CART19. Gene set enrichment analysis of genes that are significantly downregulated in GM-CSFKO CART19 (p-value < 0.01) but upregulated in GM-CSFWT CART19 and control gRNA CART19 using Enrichr (p-value < 0.05). (C-D) GM-CSFKO CART19 cells showed altered expression levels of T cell activation markers. GM-CSFKO or GM-CSFWT CART19 cells were co-cultured with CD19+ Nalm6 at an E:T ratio of 1:5 for 24hrs and flow cytometric staining was performed in order to measure CD3 (A), CD45 (B), CD69 (C- D), HLA-DR (E-F) and CD25 (G-H) (one-way ANOVA; ** p < 0.01, *** p < 0.001, **** p < 0.0001; 2 biological replicates, 2 technical replicates; error bars, SEM). (E-H) GM-CSF disruption reduces early CART cell activation and shows prolonged expansion in an in vivo JeKo-1 relapse xenograft model. Experimental schema showing NSG mice engrafted with the CD19+ luciferase+ cell line JeKo-1 (1 × 106 cells intravenous [i.v.] and randomized to treatment with UTD T cells, GM-CSFWT CART19 cells, and GM-CSFKO CART19 cells (1 × 106 cells i.v.) (E). Tumor burden by bioluminescent imaging over 20 days after CART cell therapy (two-way ANOVA; *** p<0.001; n = 5 mice per group; error bars, SEM) (F). (G) GM-CSFKO CART19 cells exhibit enhanced delayed proliferation in vivo. Peripheral blood analysis of UTD, GM-CSFWT CART19 and GM-CSFKO CART19 cells in which CD3+ cells were quantified (one-way ANOVA, *** p < 0.001; n = 5 mice per group; error bars, SEM).
To validate the transcriptome findings from our RNA-seq, we interrogated the surface expression of various surface activation proteins on GM-CSFKO CART19. Lower activation levels in the edited CART19 were first indicated by a significant difference in CD3 and CD45 expression (Fig. 3C). Twenty-four hours following antigen-specific stimulation, GM-CSFKO CART19 also expressed lower levels of CD69, HLA-DR, and CD25 compared to GM-CSFWT CART19 (Fig. 3D), indicating reduced initial levels of T cell activation. Collectively, our data indicate that GM-CSF disruption in CART cells reduces their initial activation and enhances their antigen-specific proliferation.
GM-CSF disruption in CART19 cells dampens early activation and improves anti-tumor activity
Having demonstrated that GM-CSFKO CART19 cells have altered activation levels based on their transcriptomic and immunophenotypical profiles, we next aimed to study their expansion kinetics and functional profile in vivo. We used a xenograft model for relapsed lymphoma to study the impact of GM-CSF knockout of CART19 in vivo (Fig. 3E). Here, NSG mice were engrafted with the CD19+ mantle cell lymphoma cell line JeKo-1 and then randomized to receive control T cells, GM-CSFKO CART19, or GM-CSFWT CART19. GM-CSFKO CART19 cells exhibited significantly enhanced proliferation (~4-fold difference) after 20 days of treatment and significantly improved anti-tumor activity (Fig. 3F–G). Importantly, GM-CSFKO CART19 cells were also able to prevent late lymphoma relapse compared to GM-CSFWT CART19 during days 30–50 post-treatment. To investigate whether this effect is due to enhanced persistence or a shift into a memory phenotype, we measured the expression of CCR7 and CD45RA on CART19 cells using flow cytometry. There was no difference in CCR7−CD45RA− effector memory T cells (TEM), CCR7+CD45RA− central memory T cells (TCM), CCR7−CD45RA+ effector memory T cells re-expressing CD45RA (TEMRA), or CCR7+CD45RA+ Naïve T cell phenotype in vitro after CART cell manufacturing or in vivo by examining T cell subsets in spleens harvested from our in vivo experiment (see supplementary Fig. S4A–B). This suggests that the enhanced CART efficacy is not related to a shift in memory phenotype.
GM-CSFKO CART19 are intrinsically more resistant to apoptosis
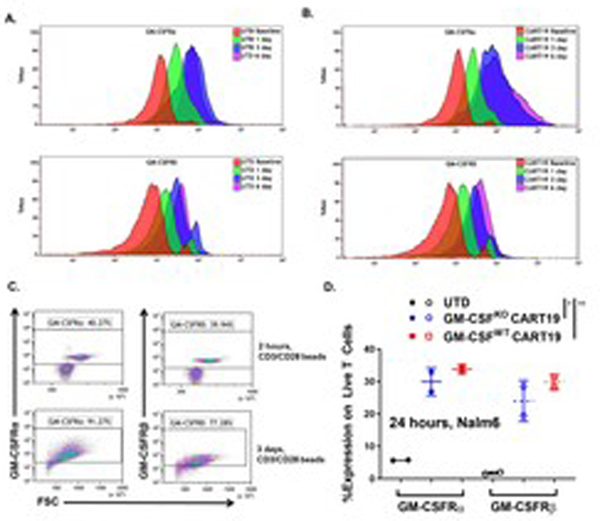
We then investigated the mechanisms by which GM-CSF alters CART cell activation levels. First, we studied whether CART cells express GM-CSF receptors. While resting CART19 cells do not express any GM-CSF receptors (GM-CSFR), our experiments indicate that activated CART cells significantly upregulate both GM-CSFR α and β subunits. This finding was significant when T cells or CART19 cells were non-specifically activated through their T cell receptors (Fig. 4A–B). Additionally, both GM-CSFKO and GM-CSFWT CART19 cells upregulated GM-CSFRα and GM-CSFRβ when activated through the CAR with irradiated CD19+ Nalm6 cells (Fig. 4C–D).
Fig. 4. Activated T cells and CART19 cells express high levels of GM-CSF receptors.
(A-B) GM-CSF receptors (ɑ and β subunits) are activated upon of TCR activation on UTD cells or CART19 cells. Untransduced (UTD) T cells (A) or CART19 cells (B) were stimulated over a 6-day expansion period with CD3/CD28 beads. Flow cytometric analysis was performed on GM-CSF2R α and β subunit expression on days 0, 1, 3 and 6 (representative figures). (C) Representative flow plot of the GM-CSF2R α and β expression on stimulated T cells at 2 hours and 3 days. (D) GM-CSFWT CART19 and GM-CSFKO CART19 express GM-CSF2R α and β when activated through their CAR. UTD, GM-CSFKO and GM-CSFWT CART19 cells were co-cultured with CD19+ cell line Nalm6 and assessed with flow cytometry at 24 hours (two-way ANOVA; ** p-value <0.01, *** p< 0.001, **** p<0.0001; 3 biological replicates, 2 technical replicates; error bars, SEM).
Collectively, our data indicate that 1) GM-CSF-producing T cells are significantly more activated than GM-CSF-negative CART cells, 2) GM-CSFKO CART cells have a distinct transcriptional, immunophenotypical, and functional profile associated with altered levels of activation, and 3) activated CART cells express high levels of GM-CSF receptors. Thus, we aimed to study whether these effects are predominantly through the interaction of GM-CSF with its upregulated receptors. To test this interaction, we generated CART19 cells in the presence of the GM-CSF-neutralizing antibody lenzilumab (see methods, and supplementary Fig. S5). Following generation of CART19 cells in the presence of lenzilumab, we measured their apoptosis after antigen-specific stimulation. There was no difference in apoptotic cells (Annexin+ 7AAD−) between 1) GM-CSFWT CART19 cells generated in the presence of lenzilumab, 2) GM-CSFWT CART19, or 3) GM-CSFWT CART19 co-cultured with lenzilumab either at early timepoints of activation (Fig. 5A) or at extended timepoints (Fig 5B). Furthermore, while a co-culture of exogenous GM-CSF with GM-CSFWT CART19 was associated with increased apoptosis (see Fig. 1E), there was no change in apoptosis of GM-CSFKO CART19 cells when co-cultured with exogenous GM-CSF. This suggested to us that the interaction between GM-CSF and its upregulated receptors is not the predominant mechanism for GM-CSF modulation of CART cell activation (Fig. 5C).
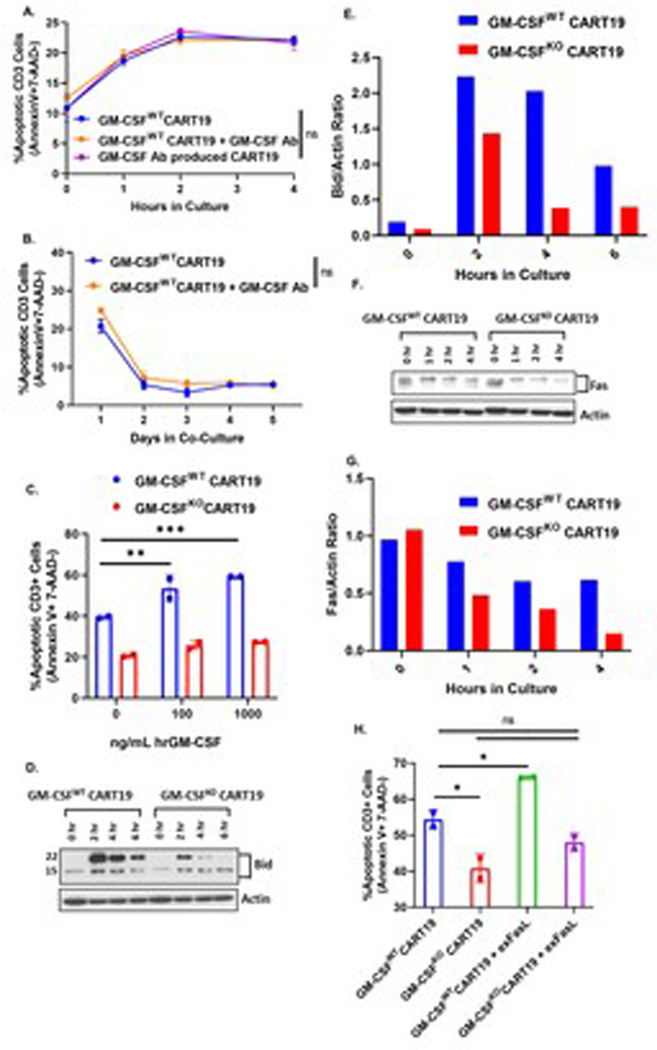
Fig. 5. GM-CSFKO CART19 are intrinsically more resistant to apoptosis. (A-B) Apoptosis is not impaired in CART19 cells that were produced in the presence of anti-GM-CSF antibody.
CART19 or CART19 cells generated in the presence of anti-GM-CSF antibody were co-cultured in the presence of CD19+ cell line Nalm6. CART19 was co-cultured with and without GM-CSF Ab. Flow cytometric staining for Annexin V, 7-AAD, and CD3 was performed at early timepoints (0hr, 1hr, 2hr, and 4hr; A) or later timepoints (1, 2, 3, 4, and 5 days; B) (two-way ANOVA; ns = not significant; 3 biological replicates, 3 technical replicates; error bars, SEM). (C) Exogenous hrGM-CSF increases GM-CSFWT CART19 cell apoptosis but not GM-CSFKO CART19 apoptosis. CART19 cells were co-cultured with the CD19+ cell line Nalm6 and increasing doses of human recombinant GM-CSF (0, 100, and 1000 ng/mL). Flow cytometric staining for Annexin V, 7-AAD, and CD3 was performed after 4 hours (one-way ANOVA; ** p <0.01, *** p <0.001; 1 biological replicate, 2 technical replicates; error bars, SEM). (D-G) GM-CSFKO CART19 cells exhibit reduced Bid and Fas expression. GM-CSFWT CART19 and GM-CSFKO CART19 cells were co-cultured with CD19+ Nalm6. Western blot for Bid was performed at 0hr, 2hr, 4hr, and 6hr (D-E) and western blot for Fas was performed at 0hr, 1hr, 2hr, and 4hr (F-G) (representative figure of 3 biological replicates, 1 technical replicate per biological replicate). (H) Addition of exogenous human Fas ligand (FasL) results in increased apoptosis on GM-CSFWT CART19 cells but not on GM-CSFKO CART19 cells. GM-CSFWT CART19 and GM-CSFKO CART19 cells were co-cultured with CD19+ Nalm6 in the presence or absence of 50 ng/mL of exogenous FasL for 2 hours. Apoptosis was measured via flow cytometric staining of Annexin V, 7-AAD, and CD3 (one-way ANOVA; ns = not significant, * p<0.05; 1 biological replicate, 2 technical replicates; error bars, SEM).
Since our transcriptomic data showed that downregulation of the Fas-regulating gene CCN1 was unique to GM-CSFKO CART19, we wanted to study regulators of activation and apoptosis in GM-CSFKO CART19 cells. Following GM-CSF disruption in CART19 cells, there was a consistent reduction in the pro-apoptotic protein BH3 interacting-domain death agonist (Bid) (Fig. 5D–E). Knockout of Bid in GM-CSFWT CART19 cells resulted in a similar phenotype with reduced apoptosis (supplementary Fig. S6). GM-CSFKO CART19 cells exhibited a consistent reduction of the death receptor Fas (Fig. 5F–G). To study the impact of Fas:FasL interactions on CART19 apoptosis, we added exogenous FasL to both GM-CSFWT and GM-CSFKO CART19. FasL significantly increased the apoptosis of GM-CSFWT CART19 but not GM-CSFKO CART19 (Fig. 5H).
DISCUSSION
In this report, we established the efficacy of GM-CSFKO CART19 cells as a potential independent therapeutic option in B cell malignancies with enhanced anti-tumor activity and reduced GM-CSF levels. We describe a novel mechanism by which GM-CSF affects CART cell function through increased susceptibility to early activation. We identified that activated CART cells upregulate GM-CSF receptors, and that GM-CSF disruption in CART cells ameliorates their early activation and enhances their in vivo proliferation and anti-tumor activity.
Many efforts are ongoing to enhance CART cell in vivo expansion and anti-tumor activity by using strategies to non-specifically stimulate CART cell proliferation or re-stimulate CART cells after infusion, utilizing synthetic biology or combination therapy to edit their exhaustion pathways or prevent their apoptosis.32, 33 However, this often improves CART efficacy at the cost of the CART safety profile: most of these approaches are associated with increased production of inflammatory cytokines and higher risk for the development of CART-associated toxicities such as CRS and NT (efficacy/toxicity linkage). In contrast, our data indicate that GM-CSF knockout in CART cells results in enhanced CART cell proliferation and anti-tumor activity, while being associated with a marked reduction in GM-CSF levels.
Preclinical studies and correlative science from CART19 clinical trials have shown that inhibitory myeloid cells and their cytokines (such as IL-1, IL-6, and GM-CSF) are major players in both inducing CART cell toxicities and limiting anti-tumor effects.9 Depleting IL-6 with monoclonal antibodies is the mainstay for management of CART cell toxicities in the clinic. While we and others have demonstrated that GM-CSF induces monocyte activation and results in both propagation of CRS and inhibition of CART cells,9, 15, 17 a direct impact of GM-CSF on CART cell function independent of myeloid cells has not been reported.
The finding of altered CART activation following GM-CSF disruption in CART cells has significant therapeutic implications. It has become increasingly evident that CART cells are susceptible to apoptosis and AICD,34, 35 and that CART cell apoptosis limits their anti-tumor activity.35 CART cells upregulate Fas, FasL, TRAIL, and TRAIL-R and are prone to Fas- and TRAIL-mediated death when a threshold of cell activation is reached.34, 36, 37 The interaction between Fas-FasL within the CART cells and tumor microenvironment limits both CART persistence and anti-tumor efficacy,32 and genetic engineering of the CAR to include a Fas dominant negative receptor has been shown to enhance anti-tumor activity and persistence in solid tumor models.32 In a different approach, modification of CART cell activation has been accomplished through modulation of CART cell antigen binding by using a lower affinity single chain variable fragment in the CAR design.33 However, these strategies – aimed at ameliorating CART cell apoptosis and nonspecifically enhancing their proliferation – are associated with increased inflammatory cytokine secretion and CART cell-associated toxicities.
Our study builds on two independent observations: correlative science of clinical trials which indicates that GM-CSF is significantly associated with CART cell toxicities, and our prior report which has demonstrated that GM-CSF neutralization results in prevention of CART cell toxicities in preclinical xenograft models while enhancing CART efficacy.17 In the near term, a major limitation to novel strategies aimed at improving CART cell efficacy is maintaining a balance between enhancing efficacy and minimizing toxicity. In contrast, the use of GM-CSFKO CART cells has the potential to break the efficacy/toxicity linkage associated with CART19 therapy in the clinic and can be applied to other CAR constructs.
Our experiments strongly indicate that GM-CSFKO CART19 cells have a distinct transcriptomic and immunophenotypical profile characterized by altered activation upon antigen-specific stimulation. Corroborating this phenotype, GM-CSFKO CART19 cells exhibited an enhanced proliferation and anti-tumor effect in our xenograft models in vivo. These findings are consistent with the knowledge that GM-CSF-producing T cells are exquisitely susceptible to AICD22 and our initial findings that GM-CSF-producing CART19 cells are more activated compared with GM-CSF-negative CART19. While exogenous GM-CSF increased CART19 apoptosis, our study demonstrates that the interaction between GM-CSF and its upregulated receptors in not the predominant mechanism for altered activation of GM-CSFKO CART19. It is possible that different GM-CSF levels result in different activation profile, and we plan to investigate further in a follow-up study. Our investigation also demonstrates that GM-CSFKO CART19 cells are more resistant to Fas-mediated apoptosis. Experiments to elucidate the exact mechanisms of this interaction are ongoing will be reported in a subsequent manuscript.
With respect to limitations, CRISPR/Cas9 editing of CART cells has been reported to induce chromosomal breakage and genomic instability following gene disruption.38 Cas9-directed double strand breaks leads to the initiation of two common repair mechanisms: homology-directed repair and non-homologous end joining (NHEJ). In future applications, if needed, efforts to minimize chromosomal breakage can be employed. Such efforts include using novel Cas proteins such as Cas12a or suppressing NHEJ using gene depletion or microinjection of ssRNAs. Unlike Cas9, Cas12a generates staggered DNA cuts instead of blunt ends, leading to more predictable repair. Our work has indicated that Cas12a results in superior gene knockout to Cas9 in CART cells.39 NHEJ introduces random insertions and deletions and has been shown to be critical in inducing chromosomal translocations in human cells;40 leveraging alternative DNA repair pathways may lead to reduced chromosomal breakage.
As reports continue to demonstrate clinical safety and feasibility of CRISPR-modified CART cells in the clinic,41 CRISPR/Cas9 GM-CSFKO CART19 could represents a potential independent therapeutic strategy in B cell malignancies. These data presented in this report suggest a significant advantage for using GM-CSF knockout CART cells, which results in a direct effect on CART cell efficacy in addition to a reduction of GM-CSF levels, monocyte activation, and CART19-related toxicities. In fact, three patients were recently treated with GM-CSFKO CART cells in a pilot clinical trial. Patients achieved a complete response of the disease without developing CRS or NT.42
In conclusion, our study reveals that GM-CSF disruption in CART19 cells reduces their early activation and thus enhances their proliferation and anti-tumor effects. These findings uncover a new mechanism of resistance to CART cell therapy and importantly illuminate a new avenue to favorably alter CART cell activation through the genetic disruption of GM-CSF.
Competing interests:
SSK is an inventor on patents in the field of CAR immunotherapy that are licensed to Novartis (through an agreement between Mayo Clinic, University of Pennsylvania, and Novartis). MJC, RS, RMS and SSK are inventors on patents in the field of CAR immunotherapy that are licensed to Humanigen (through Mayo Clinic). MH and SSK are inventors on patents in the field of CAR immunotherapy that are licensed to Mettaforge (through Mayo Clinic). SSK receives research funding from Kite, Gilead, Juno, BMS, Novartis, Humanigen, MorphoSys, Tolero, Sunesis/Viracta, Leahlabs, and Lentigen. NEK receives research funding from Acerta Pharma, BMS, Pharmacyclics, MEI Pharma, and Sunesis. NEK has participated in Advisory Board meetings of Cytomx Therapy, Janssen, Juno Therapeutics, Astra Zeneca, and Oncotracker; and on DSMC for Agios and Cytomx Therapeutics. SAP receives research funding from Pharmacyclics, MorphoSys, Janssen, AstraZeneca, TG Therapeutics, BMS, AbbVie, and Ascentage Pharma. SAP has participated in Advisory Board meetings of Pharmacyclics, AstraZeneca, Genentech, Gilead, GlaxoSmithKline, Verastem Oncology, and AbbVie (he was not personally compensated for his participation). CD, DC and OA are employees of Humanigen.
Data and materials availability:
Sequencing data are available at BioProject PRJNA623000.
Supplementary Material
Acknowledgments
Funding: Exact Foundation (SSK), Humanigen (SSK), Mayo Clinic Center for Clinical and Translational Science grant UL1TR002377 (SSK), Mayo Clinic Center for Individualized Medicine (SSK), National Comprehensive Cancer Network (SSK), National Institutes of Health grants K12CA090628 (SSK) and R37CA266344-01 (SSK), Department of Defense grant CA201127 (SSK), and Predolin Foundation (RS and SSK).
References
- 1.June CH, Sadelain M. Chimeric Antigen Receptor Therapy. The New England journal of medicine 2018. Jul 5; 379(1): 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anagnostou T, Riaz IB, Hashmi SK, Murad MH, Kenderian SS. Anti-CD19 chimeric antigen receptor T-cell therapy in acute lymphocytic leukaemia: a systematic review and meta-analysis. Lancet Haematol 2020. Nov; 7(11): e816–e826. [DOI] [PubMed] [Google Scholar]
- 3.Khadka RH, Sakemura R, Kenderian SS, Johnson AJ. Management of cytokine release syndrome: an update on emerging antigen-specific T cell engaging immunotherapies. Immunotherapy 2019. Jul; 11(10): 851–857. [DOI] [PubMed] [Google Scholar]
- 4.Teachey DT, Lacey SF, Shaw PA, Melenhorst JJ, Maude SL, Frey N, et al. Identification of Predictive Biomarkers for Cytokine Release Syndrome after Chimeric Antigen Receptor T-cell Therapy for Acute Lymphoblastic Leukemia. Cancer Discov 2016. Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locke FL, Ghobadi A, Jacobson CA, Miklos DB, Lekakis LJ, Oluwole OO, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. The Lancet Oncology 2019. Jan; 20(1): 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster SJ, Svoboda J, Chong EA, Nasta SD, Mato AR, Anak O, et al. Chimeric Antigen Receptor T Cells in Refractory B-Cell Lymphomas. N Engl J Med 2017. Dec 28; 377(26): 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakemura R, Cox MJ, Hefazi M, Siegler EL, Kenderian SS. Resistance to CART cell therapy: lessons learned from the treatment of hematological malignancies. Leuk Lymphoma 2021. Mar 8: 1–18. [DOI] [PubMed] [Google Scholar]
- 8.Cox LA, Chan J, Rao P, Hamid Z, Glenn JP, Jadhav A, et al. Integrated omics analysis reveals sirtuin signaling is central to hepatic response to a high fructose diet. BMC Genomics 2021. Dec 3; 22(1): 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sterner RM, Kenderian SS. Myeloid cell and cytokine interactions with chimeric antigen receptor-T-cell therapy: implication for future therapies. Curr Opin Hematol 2020. Jan; 27(1): 41–48. [DOI] [PubMed] [Google Scholar]
- 10.Stroncek DF, Ren J, Lee DW, Tran M, Frodigh SE, Sabatino M, et al. Myeloid cells in peripheral blood mononuclear cell concentrates inhibit the expansion of chimeric antigen receptor T cells. Cytotherapy 2016. Jul; 18(7): 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruella M, Klichinsky M, Kenderian SS, Shestova O, Ziober A, Kraft DO, et al. Overcoming the Immunosuppressive Tumor Microenvironment of Hodgkin Lymphoma Using Chimeric Antigen Receptor T Cells. Cancer Discov 2017. Oct; 7(10): 1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain MD, Zhao H, Wang X, Atkins R, Menges M, Reid K, et al. Tumor interferon signaling and suppressive myeloid cells associate with CAR T cell failure in large B cell lymphoma. Blood 2021. Jan 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene Ciloleucel CAR T-Cell Therapy in Refractory Large B-Cell Lymphoma. N Engl J Med 2017. Dec 28; 377(26): 2531–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locke FL, Ghobadi A, Jacobson CA, Miklos DB, Lekakis LJ, Oluwole OO, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol 2019. Jan; 20(1): 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Liu CH, Roberts AI, Das J, Xu G, Ren G, et al. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don’t know. Cell Res 2006. Feb; 16(2): 126–133. [DOI] [PubMed] [Google Scholar]
- 16.Tugues S, Amorim A, Spath S, Martin-Blondel G, Schreiner B, De Feo D, et al. Graft-versus-host disease, but not graft-versus-leukemia immunity, is mediated by GM-CSF-licensed myeloid cells. Sci Transl Med 2018. Nov 28; 10(469). [DOI] [PubMed] [Google Scholar]
- 17.Sterner RM, Sakemura R, Cox MJ, Yang N, Khadka RH, Forsman CL, et al. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood 2019. Feb 14; 133(7): 697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sterner RM, Cox MJ, Sakemura R, Kenderian SS. Using CRISPR/Cas9 to Knock Out GM-CSF in CAR-T Cells. Journal of visualized experiments : JoVE 2019. Jul 22; (149). [DOI] [PubMed] [Google Scholar]
- 19. https://clinicaltrials.gov/ct2/show/NCT04314843.
- 20.Manriquez-Roman C, Siegler EL, Kenderian SS. CRISPR Takes the Front Seat in CART-Cell Development. BioDrugs 2021. Mar; 35(2): 113–124. [DOI] [PubMed] [Google Scholar]
- 21.Sachdeva M, Duchateau P, Depil S, Poirot L, Valton J. Granulocyte-macrophage colony-stimulating factor inactivation in CAR T-cells prevents monocyte-dependent release of key cytokine release syndrome mediators. The Journal of biological chemistry 2019. Apr 5; 294(14): 5430–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Roberts AI, Liu C, Ren G, Xu G, Zhang L, et al. A novel subset of helper T cells promotes immune responses by secreting GM-CSF. Cell Death Differ 2013. Dec; 20(12): 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010. Mar 1; 26(5): 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010. Sep; 20(9): 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature 2015. Oct 1; 526(7571): 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS One 2015; 10(4): e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research 2014; 42(22): e168–e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer V, Boroviak K, Thomas M, Doe B, Riva L, Ryder E, et al. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet 2018. Jul; 14(7): e1007503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15(12): 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Li H, Wang X, Gao X, Liu X. Regulation of T cell development and activation by creatine kinase B. PLoS One 2009; 4(4): e5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juric V, Chen CC, Lau LF. Fas-mediated apoptosis is regulated by the extracellular matrix protein CCN1 (CYR61) in vitro and in vivo. Mol Cell Biol 2009. Jun; 29(12): 3266–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto TN, Lee PH, Vodnala SK, Gurusamy D, Kishton RJ, Yu Z, et al. T cells genetically engineered to overcome death signaling enhance adoptive cancer immunotherapy. The Journal of clinical investigation 2019. Feb 25; 129(4): 1551–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelozzi IM, Castaneda EG, Pohle RVC, Rodriguez FC, Sufi J, Costa PP, et al. Enhanced functionality of low-affinity CD19 CAR T-cells is associated with activation priming and a polyfunctional cytokine phenotype. bioRxiv 2020: 2020.2009.2022.291831. [Google Scholar]
- 34.Tschumi BO, Dumauthioz N, Marti B, Zhang L, Schneider P, Mach JP, et al. CART cells are prone to Fas- and DR5-mediated cell death. J Immunother Cancer 2018. Jul 13; 6(1): 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benmebarek MR, Karches CH, Cadilha BL, Lesch S, Endres S, Kobold S. Killing Mechanisms of Chimeric Antigen Receptor (CAR) T Cells. International journal of molecular sciences 2019. Mar 14; 20(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes-Silva D, Mukherjee M, Srinivasan M, Krenciute G, Dakhova O, Zheng Y, et al. Tonic 4–1BB Costimulation in Chimeric Antigen Receptors Impedes T Cell Survival and Is Vector-Dependent. Cell Rep 2017. Oct 3; 21(1): 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkele A, Johnson AJ, Rolczynski LS, Chang CA, Hoglund V, Kelly-Spratt KS, et al. Functional Tuning of CARs Reveals Signaling Threshold above Which CD8+ CTL Antitumor Potency Is Attenuated due to Cell Fas-FasL-Dependent AICD. Cancer immunology research 2015. Apr; 3(4): 368–379. [DOI] [PubMed] [Google Scholar]
- 38.Manriquez-Roman C, Siegler EL, Kenderian SS. CRISPR Takes the Front Seat in CART-Cell Development. BioDrugs 2021. Feb 27. [DOI] [PubMed] [Google Scholar]
- 39.Siegler EL, Simone BW, Sakemura R, Tapper EE, Horvei P, Cox MJ, et al. Efficient Gene Editing of CART Cells with CRISPR-Cas12a for Enhanced Antitumor Efficacy. Blood 2020; 136(Supplement 1): 6–7.32614958 [Google Scholar]
- 40.Chromosomal translocations are mediated by canonical NHEJ in human cells. Cancer Discov 2014. Nov; 4(11): OF12. [DOI] [PubMed] [Google Scholar]
- 41.Stadtmauer EA, Cohen AD, Weber K, Lacey SF, Gonzalez VE, Melenhorst JJ, et al. First-in-Human Assessment of Feasibility and Safety of Multiplexed Genetic Engineering of Autologous T Cells Expressing NY-ESO −1 TCR and CRISPR/Cas9 Gene Edited to Eliminate Endogenous TCR and PD-1 (NYCE T cells) in Advanced Multiple Myeloma (MM) and Sarcoma. Blood 2019. Nov 13; 134(Supplement_1): 49. [Google Scholar]
- 42.Yi Y, Chai X, Zheng L, Zhang Y, Shen J, Hu B, et al. CRISPR-edited CART with GM-CSF knockout and auto secretion of IL6 and IL1 blockers in patients with hematologic malignancy. Cell Discov 2021. Apr 27; 7(1): 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at BioProject PRJNA623000.