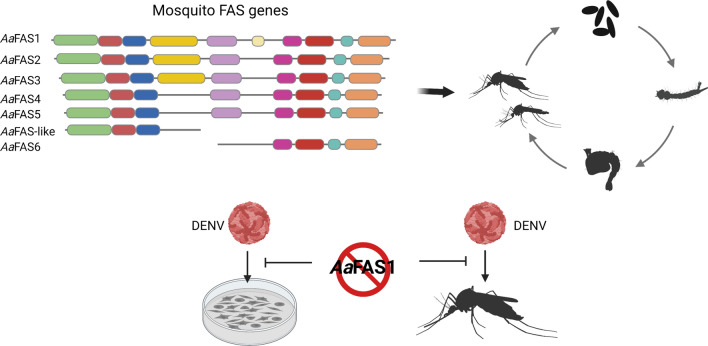
Abstract
Background
Fatty acids are the building blocks of complex lipids essential for living organisms. In mosquitoes, fatty acids are involved in cell membrane production, energy conservation and expenditure, innate immunity, development and reproduction. Fatty acids are synthesized by a multifunctional enzyme complex called fatty acid synthase (FAS). Several paralogues of FAS were found in the Aedes aegypti mosquito. However, the molecular characteristics and expression of some of these paralogues have not been investigated.
Methods
Genome assemblies of Ae. aegypti were analyzed, and orthologues of human FAS was identified. Phylogenetic analysis and in silico molecular characterization were performed to identify the functional domains of the Ae. aegypti FAS (AaFAS). Quantitative analysis and loss-of-function experiments were performed to determine the significance of different AaFAS transcripts in various stages of development, expression following different diets and the impact of AaFAS on dengue virus, serotype 2 (DENV2) infection and transmission.
Results
We identified seven putative FAS genes in the Ae. aegypti genome assembly, based on nucleotide similarity to the FAS proteins (tBLASTn) of humans, other mosquitoes and invertebrates. Bioinformatics and molecular analyses suggested that only five of the AaFAS genes produce mRNA and therefore represent complete gene models. Expression levels of AaFAS varied among developmental stages and between male and female Ae. aegypti. Quantitative analyses revealed that expression of AaFAS1, the putative orthologue of the human FAS, was highest in adult females. Transient knockdown (KD) of AaFAS1 did not induce a complete compensation by other AaFAS genes but limited DENV2 infection of Aag2 cells in culture and the midgut of the mosquito.
Conclusion
AaFAS1 is the predominant AaFAS in adult mosquitoes. It has the highest amino acid similarity to human FAS and contains all enzymatic domains typical of human FAS. AaFAS1 also facilitated DENV2 replication in both cell culture and in mosquito midguts. Our data suggest that AaFAS1 may play a role in transmission of dengue viruses and could represent a target for intervention strategies.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-022-05336-1.
Keywords: Aedes aegypti, Aag2 cells, Fatty acid synthase, FAS, Lipid, Lipid metabolism, Dengue virus, AaegL5 genome assembly
Background
Fatty acid synthase (FAS) is a multifunctional enzyme catalyzing > 40 steps in the de novo fatty acid biosynthesis pathway [1, 2]. It contains seven catalytic and three non-catalytic domains which condense, reduce and dehydrate the three-carbon substrate, malonyl-CoA, into 16–18-carbon fatty acids. These fatty acids are essential building blocks of complex lipids, such as phosphoglycerolipids, glycerolipids and sphingolipids, which are components of cellular membranes and storage lipids, and function as signaling molecules, respectively.
In mosquitoes, fatty acids also play roles in innate immunity, reproduction, development and flight [3–5]. Fatty acids can be acquired or synthesized in both larvae and adult stages. Neonate larvae acquire lipids through the maternal deposition in eggs [6–8] and through consumption of aquatic diets such as diatoms and algae, which are the primary source of polyunsaturated fatty acids [9]. Fatty acids from larval stages can be transferred to the adult stage and some can be deposited in eggs of the first gonotrophic cycle [10]. Adult mosquitoes possess enzymes for de novo synthesis and modification of fatty acids from both sugar (carbohydrate-enriched) and blood (protein-enriched) meals [7, 9, 11]. In the female, fatty acid synthesis is important for metabolism and production of eggs. Transient knockdown (KD) of acetyl-CoA carboxylase (ACC) and fatty acid synthase (FAS), two key enzymes in the de novo fatty acid biosynthesis pathway, led to significantly lower egg production in the first gonotrophic cycle [12]. In addition, eggs produced by ACC-KD mosquitoes lacked eggshells and were nonviable [12].
Apart from its importance to mosquito biology, studies suggest FAS also plays a supportive role for several arboviral infections in both mammalian and mosquito cells [13–16]. Several RNA viruses induce expansion and rearrangement of host cell membranes to support viral genome replication and assembly [17–19]. Studies have shown that FAS facilitates the production of dengue virus serotype 2 (DENV2) infection in both human and mosquito cells, potentially by providing the building blocks for this membrane expansion event [13, 15]. Lastly, studies also reported the elevation of fatty acid abundance in C6/36 (Aedes albopictus) cells and in the Aedes aegypti mosquito midgut during DENV2 infection [15, 20]. These findings suggest that fatty acids are essential for the physiological function of mosquitoes and support DENV2 infection of the mosquito.
Currently, understanding of FAS in mosquitoes and its role in pathogen transmission by the mosquito vector is limited. Here, we describe the molecular and functional characterization of the FAS gene family from Ae. aegypti (AaFAS). We identified seven putative AaFAS genes (AaFAS 1–6 and AaFAS-like) in the AaegL5 genome assembly, characterized the expression of these genes during mosquito development and following consumption of different diets. AaFAS1 has the highest amino acid similarity to human FAS and is the predominant transcript. We investigated the role of AaFAS1 in DENV2 infection in mosquito cells and mosquito vectors using gene KD. We observed a significant reduction of DENV2 replication following AaFAS1-KD in Ae. aegypti cells and a transient reduction of infection in Ae. aegypti midguts at early time points post-infectious blood meal. These results provide insights into the molecular characteristic of AaFASs and their roles during Ae. aegypti development, food source acquisition and arbovirus infection.
Methods
Alignments, conserved motifs and phylogenetic tree
Putative AaFAS sequences from the AaegL5 genome assembly were blasted against the AaegL3 genome assembly retrieved from VectorBase using tBLASTn [21–23]. FAS sequences of Anopheles gambiae, Drosophila melanogaster, Apis mellifera, Homo sapiens, Mus musculus and Saccharomyces cerevisiae were aligned with putative AaFAS sequences using ClustalW [24]. mRNA sequences were retrieved from NCBI and manually curated to confirm the intron/exon boundaries. Conserved FAS motifs were identified by global alignment of vertebrate, invertebrate and yeast proteins using Clustal Omega [25] and Jalview 2.11.1.5 (accession numbers are shown in Additional file 1: Table S1) [26], and conserved amino acids associated with catalytic domains of functional FAS were identified by comparison to sequences reported in published studies [27]. Individual amino acid alignments were also performed between FAS-AaegL5 and FAS-AaegL3 using ClustalW to identify improvements in AaegL5 models.
A Bayesian inference of phylogeny was performed using the amino acid sequence of FAS from Ae. aegypti, An. gambiae, D. melanogaster, A. mellifera, Mus musculus and H. sapiens. Yeast Kexin was used as an outgroup. A sequence alignment with ClustalW was performed prior to tree construction in phylogeny.fr. The substitution model used for the Bayesian inference was Blosum62, and the Markov Chain Monte Carlo parameters included 100,000 generations with sampling every 10 generations, discarding the first 250 trees. The resulting tree was annotated and curated in iTOL [28].
Annotation of protein domains in AaFAS genes
AaFAS amino acid sequences were aligned against the human FAS (NP_004095.4, NCBI) using Clustal Omega to identify the seven catalytic and three noncatalytic domains associated with mammalian FAS. The alignment results were viewed using MView tool [29]. Motifs in the human FAS were identified based on Pfam 31.0 [30], and conserved domains in AaFAS genes were identified by comparative analyses.
Mosquito rearing
Larvae of Ae. aegypti strain Chetumal, originally collected from Yucatan Peninsula in Mexico, were reared on fish food. Adult mosquitoes were reared on 10% sucrose solution and maintained under constant conditions of 28 °C, 80% relative humidity [31].
Blood feeding
Mosquitoes were starved by removal of sucrose solution and water for 24 and 4 h, respectively, prior to blood feeding. Defibrinated sheep blood (Colorado Veterinarian Product) was mixed with 1 mM ATP and placed in an artificial membrane feeder warmed by a 37 °C water jacket. Mosquitoes were allowed to feed for 45–60 min. Only fully engorged mosquitoes were used for the experiment and were reared on 10% sucrose solution and water.
Generating long double-stranded RNA
Long double-stranded RNA (dsRNA) was generated from adult female Ae. aegypti total RNA. Primers were designed to amplify an ~ 500-bp region of the gene of interest (Additional file 1: Table S2). cDNA was generated by reverse transcription (RT) using specific reverse primers and SuperScript III Reverse Transcriptase (Invitrogen). Polymerase chain reaction (PCR) was performed using specific primers containing a 5′ T7 promotor sequence adapted to both forward and reverse primers and Taq polymerase (NEB). PCR products were purified using the GeneJET PCR Purification kit (Thermo Scientific), and in vitro transcription was performed using the MEGAscript T7 kit (Invitrogen) and incubation at 37 °C for 12 h. Following incubation, the product was heated to 75 °C for 5 min and slowly cooled to room temperature for 4 h to dsRNA annealing. Next, dsRNA was treated with DNase (NEB) and purified by phenol–chloroform extraction followed by ethanol precipitation and the purified dsRNA was stored at − 80 °C.
dsRNA knockdown of AaFAS1 in Ae. aegypti
dsRNA was introduced via intrathoracic (IT) injection of adult females at 3–4 days post-eclosion [32]. Mosquitoes were anesthetized at 4 °C on a cold plate. Glass needles were prepared with a vertical pipette puller (P-30, Sutter Instrument Co., Novato, CA), and mosquitoes were IT injected with 3 µg/µl of dsRNA in an injection volume of 69 nl twice (total of ~ 400 ng of dsRNA) using a Nanojet II (Drummond Scientific Company, Broomall, PA). Injected mosquitoes were fed on sucrose solution or blood and reared at 28 °C, 80% relative humidity for 17 days post-injection.
dsRNA knockdown of AaFAS1 gene and DENV2 infection of Aag2 cells
dsRNA KD was performed in RNA interference-competent Ae. aegypti (Aag2) cells. Aag2 cells were cultured in Schneider’s insect medium (Sigma-Aldrich) supplemented with 2 mM L-glutamine, 1% non-essential amino acids and 10% fetal bovine serum (FBS). The cells were seeded in a 48-well plate at 50,000 cells/well for 24 h and subsequently transfected with 260 ng of dsRNA, against AaFAS1, DENV2 (positive KD control) or GFP (negative KD control) genes, mixed with TransIT-2020 Reagent (Mirus) following the manufacturer’s protocol. New medium with 2% FBS was replaced at 6 h post-transfection. Cell viability assays were performed at 2 days post-transfection using resazurin assay.
KD cells were infected with infectious DENV2 expressing a luciferase reporter (DEN-Luc) supplied by C. Rice, Rockefeller University. Cell culture medium was replaced with 300 µl of DEN-Luc supernatant at 48 h post dsRNA transfection, and cells were incubated at 28 °C without CO2. Virus supernatant was removed at 24 h post-infection, and cells were lysed. Luciferase activity was read using the Luciferase Assay System (Promega) as per manufacturer protocol.
Gene expression analyses
Total RNA was extracted from dissected midgut or whole mosquito by TRIzol (Life Tech), and cDNA was produced via reverse transcription using random primers (Life Tech) and SuperScript III Reverse Transcriptase (Invitrogen). Approximately 400 ng of total cDNA was employed for quantitative PCR (qPCR) analyses. Gene-specific primers are listed in Additional file 1: Table S3. β-Actin was used as a reference gene. Relative AaFAS gene expression was assessed by normalization to the levels of the β-actin gene (2−ΔCt). The comparative Ct (2−ΔΔCt) method was used to calculate the relative expression of AaFAS following treatment compared to the control [33].
For RT-PCR assay, total RNA was treated with DNase I, RNase-free (1 U/µl) kit (ThermoFisher) prior to reverse transcription reaction. Purified cDNA was then amplified using Q5® High-Fidelity DNA Polymerase kit (New England BioLabs) with following conditions: 98 °C for 30 s, 35 cycles of 98 °C for 10 s, 68 °C and 72 °C for 2 min and 30 s. Primers are listed in Additional file 1: Table S4.
Virus infection of Ae. aegypti by infectious blood meal
DENV2 serotype 2 strain Jamaica-1409 [34] was cultured in C6/36 cells. Cells were infected with DENV2 at a multiplicity of infection of 0.01 and incubated at room temperature for 1 h. Virus supernatant was removed, and infected cells were cultured in 5 ml total volume of L15 medium supplemented with 3% FBS, 50 μg/ml penicillin–streptomycin and 2 mM L-glutamine. Media were replaced at 7 days post-infection (dpi), and virus supernatant was harvested at 12–14 dpi and immediately used for infectious blood feeding.
To prepare the infectious blood meal, infected cells were scraped from the bottom of the cell culture flask using a cell scraper. A mixture of cells and virus supernatant was added to the defibrinated sheep blood at 1:1 ratio and supplemented with 1 mM ATP. Female mosquitoes (3–7 days post eclosion) were prepared, fed with the infectious blood meal, sorted and reared as mentioned in the blood-feeding section above.
Midgut dissection and plaque titration
Mosquito tissues were collected at multiple days post-exposure to the virus as indicated in the figure legends. Isolated midguts or the mosquito carcass (remainder of the body without midgut) were placed separately into 2-ml safe-lock Eppendorf tubes (Eppendorf) containing 250 µl of mosquito diluent [1 × PBS supplemented with 20% FBS, 50 µg/ml penicillin/streptomycin (Gibco), 50 µg/ml Gentamycin (Gibco) and 2.5 µg/ml Amphotericin B (Gibco)] and a stainless-steel bead [35]. Tissue was homogenized using a Retsch Mixer Mill MM400 at 24 cycles per second for 1 min and centrifuged at 15,000g for 5 min at 4 °C, and supernatant was transferred to a new tube for plaque titration.
Plaque assay was performed on BHK-15 cells. Ten-fold serially diluted viral supernatant was absorbed on the confluent cell layer. After 45 min of absorption, cells were overlaid with 1 × Minimum Essential Media (MEM), 1 × agar supplemented with 2.5% FBS, 25 µg/ml penicillin/streptomycin, 25 µg/ml gentamycin and 1.25 µg/ml amphotericin B, and the cells were incubated at 37 °C with 5% CO2. Cells were stained with 0.033% neutral red (Sigma) in 1 × PBS on day 5 post-infection, and plaques were counted at 24 h post-staining.
Statistical analyses
Statistical analyses comparing gene expression or DENV2 viral load were performed by one-way ANOVA followed by Tukey’s multiple comparison tests. Values of gene expression or virus titer were reported as mean ± SD. Proportions of infection among groups on each day were compared using pairwise χ2 tests with p-values adjusted using a Holm correction for multiple comparisons [36], *p < 0.05, **p < 0.01, ***p < 0.005 and ****p < 0.001.
Results
Molecular characterization of AaFAS genes
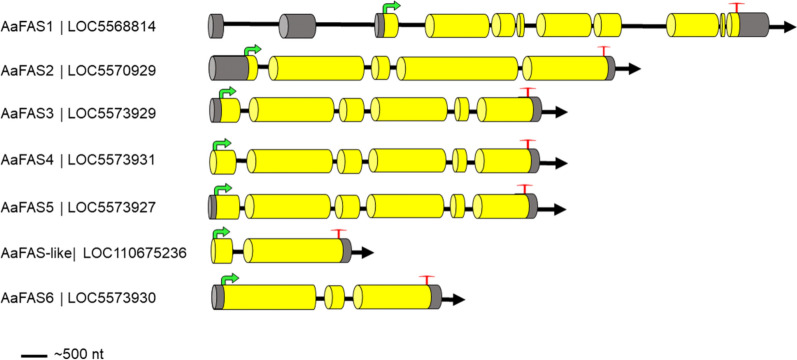
We obtained seven putative AaFAS gene models via manual annotation using the AaegL5 assembly [37]. Previously, five candidate FAS genes (AaFAS1-5) were identified based on the AaegL3 assembly of Nene et al., 2007 [23, 38], and of these, only AaFAS1 and 2 underwent functional studies [12]. The AaegL5 assembly enabled identification of two additional candidate FAS genes (AaFAS6 and AaFAS-like). The corresponding mRNA sequences showing predicted intron/exon structure, and initiation and stop codons are shown in Additional file 2. The AaFAS1 gene model revealed a gene structure comprising 11 exons, while AaFAS2 had 5 exons and AaFAS3-5 had 6 exons (Fig. 1). The incomplete AaFAS-like and AaFAS6 gene models comprised 2 and 3 exons, respectively.
Fig. 1.
Schematic showing the predicted gene structure of the AaFAS gene family. Exons are indicated by yellow cylindrical bars, 5′ and 3′ non-coding exons by dark gray shading, introns by a black line, start codon by green arrow and stop codon by red T
The gene models for AaFAS1-5 appeared to be full length, with an average gene product length of 2360 amino acids (Table 1). AaFAS1-5 possessed features associated with functional FAS, including an initiation methionine, a stop codon and the functional catalytic motifs (DTACSS, EAH and GSVKS) important for ketoacyl synthesis as described by Beedessee et al. [27]. Additionally, AaFAS1-5 containing the YKELRLRGY motif conserved among the FAS genes of vertebrates and invertebrates presents in the polyketide synthase dehydratase domain (Additional file 1: Fig. S1). AaFAS3 lacked six amino acid residues in the 3′ terminus of exon 6. We also identified a total of 127 non-synonymous substitutions in this model compared to its AaegL3 counterpart.
Table 1.
Summary of AaFAS gene family predicted from the Aedes aegypti AaegL5 assembly
| Name | NCBI accession number (AaegL5) | VectorBase accession number (AaegL3) | Max. number of exons | Chro-mo-some | Location | % Ident with human FAS | Length (nucleotides) | Length (amino acids) | No. splice variants/isoforms | Notes on the revised annotation | % identity (AaegL3 and L5) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AaFAS1 | LOC5568814 | AAEL001194 | 11 | 2 | NC_035108.1 (307544012..307601765, complement) | 45.3 | 9732 | 2422 | 1 | 1 SNP in exon_4 | 99.9 |
| AaFAS2 | LOC5570229 | AAEL008160 | 5 | 3 | NC_035109.1 (9993811..10002427, complement) | 36.7 | 8368 | 2385 | 1 | 1 SNP in exon_5 | 100 |
| AaFAS3 | LOC5573929 | AAEL022506 | 6 | 2 | NC_035108.1 (429256663..429264110, complement) | 32.9 | 7144 | 2334 | 1 | 18 SNPs exon_1, 31 SNPs exon_2, 11 SNPs exon_3, 52 SNPs exon_4, 3 SNPs exon_5, and 12 SNPs exon_6. Deletion of six amino acid residues when compared with the AaegL3 orthologue | 94.6 |
| AaFAS4 | LOC5573931 | AAEL002237 | 6 | 2 | NC_035108.1 (429327062..429334421, complement) | 34.6 | 7068 | 2333 | 1 | 4 SNPs exon_2, 10 SNPs exon_4, 6 SNPs exon_6 | 99.1 |
| AaFAS5 | LOC5573927 | AAEL002228 | 6 | 2 | NC_035108.1 (429228612..429236010, complement) | 32.9 | 7097 | 2324 | 1 | 5 SNPs exon_2, 2 SNPs exon_3, 3 SNPs exon_4, 1 SNP exon_6 | 99.5 |
| AaFAS-like | LOC110675236 | – | 2 | 2 | NC_035108.1 (429280401..429282870, complement) | 36.1 | 2409 | 800 | 1 | – | – |
| AaFAS6 | LOC5573930 | – | 3 | 2 | NC_035108.1 (429275401..429279876, complement) | 36.8 | 4353 | 1386 | 1 | – | – |
The AaegL5 annotation is shown in comparison to the AaegL3 gene models reported by Nene et al. [23]
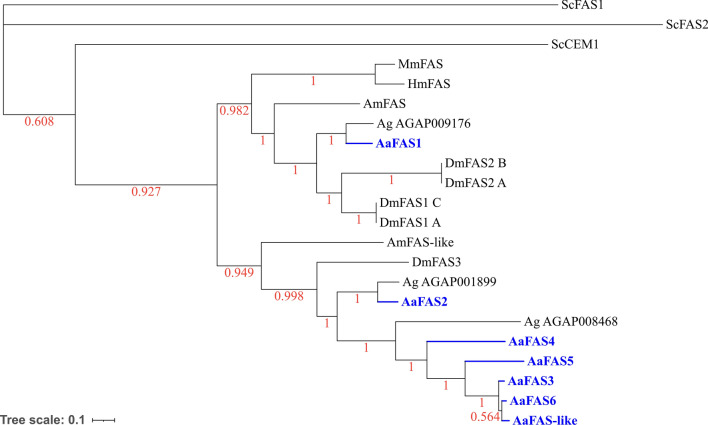
The Bayesian inference supported AaFAS1-5 as paralogues and revealed highest percent amino acid similarity between AaFAS1 and H. sapiens FAS (human FAS) (Fig. 2). Notably, AaFAS1 clustered in a clade comprising the H. sapiens, Mus musculus, A. mellifera FAS, D. melanogaster FAS1 and 2, and an uncharacterized An. gambiae FAS (Fig. 2). Similarly, AaFAS2 clustered in a clade with another uncharacterized An. gambiae FAS. In contrast, AaFAS3, 4, 5, 6 and -like clustered at the most branched portion of the tree, suggesting a recent diversification event. Phylogenetic analyses and amino acid alignment supported AaFAS1-5 as the counterparts of the AaegL3 genome assembly-derived gene models as follows: LOC5568814-AAEL001194; LOC5570229-AAEL008160; LOC5573929-AAEL022506; LOC5573931-AAEL002237 and LOC5573927-AAEL002228 (Fig. 2, Table 1). AaFAS-like and AaFAS6 (LOC110675236 and LOC5573930) were not identified in the AaegL3 assembly, suggesting these models are unique to the AaegL5 assembly.
Fig. 2.
Phylogenetic analysis of AaFAS. Bayesian phylogeny represented with an unrooted tree showing the main relationships between mosquito FAS genes and their counterparts in Ag: Anopheles gambiae, Dm: Drosophila melanogaster, Am: Apis mellifera, Mm: Mus musculus, Hm: Homo sapiens and Sc: Saccharomyces cerevisiae. The branches are supported by posterior probability values. The tree is drawn to scale: amino acid changes per site
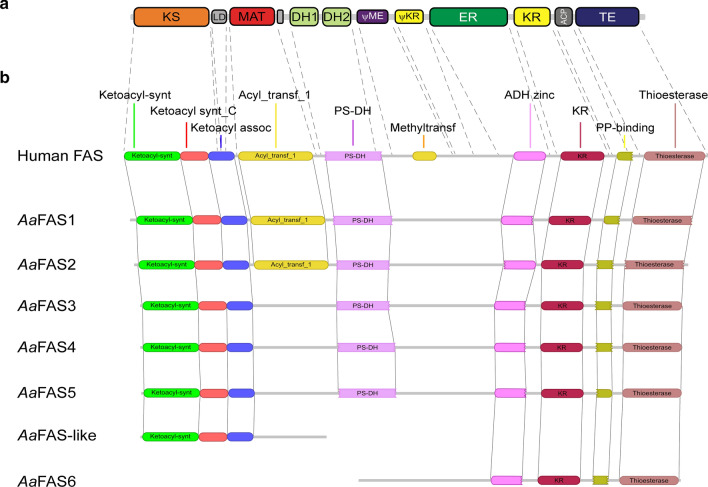
To investigate putative functional domains, we aligned AaFAS sequences to the human FAS using Clustal Omega [25]. Human FAS contains seven catalytic domains and three noncatalytic domains [1]. Collectively, AaFAS genes possessed < 50% amino acid identity to human FAS, and of the seven gene models, AaFAS1 had the highest amino acid identity (45.3%) (Table 1 and Additional file 1: Table S5). Alignment of FAS domains also showed modest sequence identity between human FAS and AaFAS (23.03–63.56%) with greatest similarity for AaFAS1 domains (Additional file 1: Table S5). The linear organization of mammalian FAS domains annotated by Maier et al. is shown in Fig. 3 [1]. Using Pfam 31.0 software, we identified the conservation in linear organization of motifs associated with known functional domains (Fig. 3B). Dotted lines indicate motif boundaries between mammalian FAS domains (Fig. 3A) and AaFAS domains (Fig. 3B). Pfam analysis did not show the presence of functional methyltransferase domains in AaFAS (Fig. 3B), and protein sequence alignment using Clustal Omega showed deletion within pseudo-methyltransferase (ΨME) domains of AaFAS compared to human FAS (16.20–23.03% identity; Additional file 1: Table S5 and Fig. S2).
Fig. 3.
Linear organization of Aedes aegypti FAS genes showing functional domains. (a) Schematic shows linear organization of seven catalytic and three noncatalytic domains of mammalian FAS annotated by Maier et al. [1]. Seven catalytic domains are shown in big squares, and three non-catalytic domains are shown in smaller squares. KS β-ketoacyl synthase; LD linker; MAT malonyl-acetyl transferase; DH dehydratase; ΨME pseudo-methyltransferase; ΨKR pseudo β-ketoacyl synthase; ER enoyl reductase; KR β-ketoacyl synthase; ACP acyl carrier protein and TE thioesterase. b Schematics show conserved domains or motifs of FAS genes and their organization annotated using Pfam 31.0 software. ketoacyl_synt β-ketoacyl synthase; ketoacyl_synt_C β-ketoacyl-acyl carrier protein synthase; ketoacyl_assoc ketoacyl-synthase C-terminal extension; acyl_transf_1 acyl transferase domain; PS-DH polyketide synthase; methyltransf methyltransferase domain; ADH-zinc zinc binding dehydrogenase; KR β-ketoreductase domain; PP-binding phosphopantetheine attachment site; thioesterase thioesterase domain
The gene model of AaFAS-like was 800 amino acids in length and contained all functional catalytic motifs, whereas AaFAS6 was 1386 amino acids in length and lacked catalytic motifs but contained the conserved 3′ motif YKELRLRGY conserved in FAS (Additional file 1: Fig. S1). AaFAS-like contains ketoacyl synthase, ketoacyl synthase_C and ketoacyl-synthase C-terminal extension domains, the first 5′ domains of AaFAS1-5 and human FAS (Fig. 3B), while AaFAS6 contains ADH zinc, β-ketoreductase, PP binding and thioesterase domains, the last four domains located 3′ in AaFAS1-5 and human FAS (Fig. 3B). In the AaegL5 assembly, AaFAS-like and 6 locate on chromosome 2 at positions 429280401-429282870 and 429275401-429279876, respectively. It is possible that AaFAS-like and -6 reflect an error in genome assembly or a gene duplication. However, the molecular data and the inability to detect transcripts associated with either locus (Additional file 1: Fig. S3) suggest that AaFAS-like and 6 represent pseudogenes (Fig. 3B).
FAS expression during Ae. aegypti development
Mosquitoes undergo four developmental stages: egg, larva, pupa and adult. We used RT-qPCR to explore the hypothesis that expression patterns of AaFAS genes vary among these stages. We collected five individual mosquitoes for each of the fourth larval instar, pupa and adult stages.
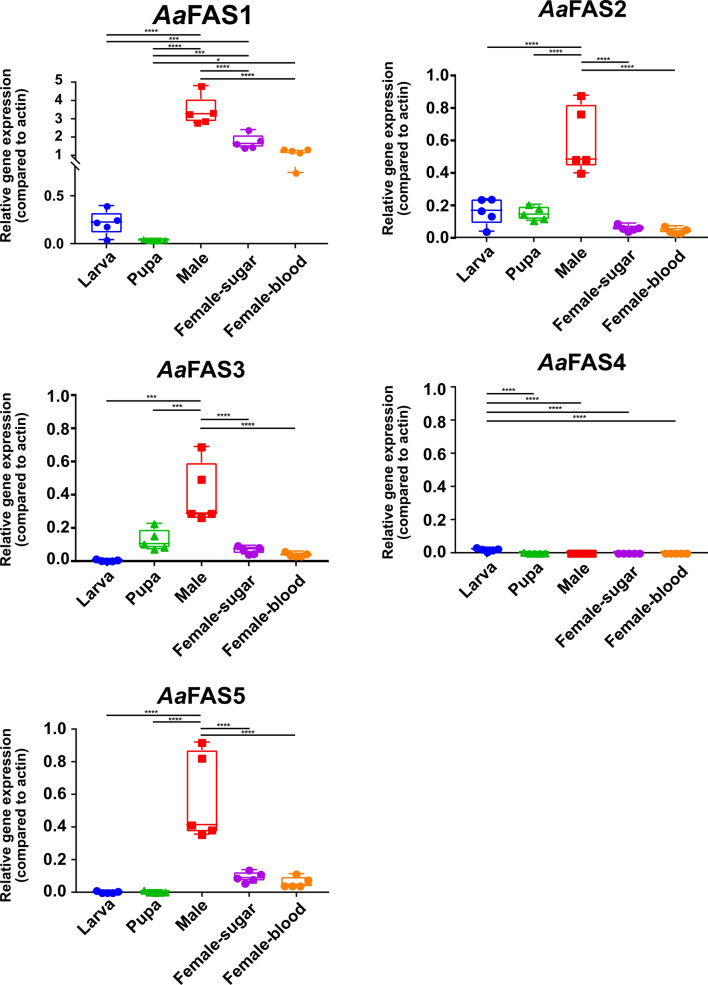
Relative expression analyses revealed negligible AaFAS expression in larval and pupal stages, while we observed the highest expression of all genes except AaFAS4 in adult males (Fig. 4). AaFAS1 was the most predominant FAS expressed in adult mosquitoes. We did not observed differences in expression levels of any AaFAS genes between sugar-fed and 3 days post blood-fed (coinciding with the first gonotrophic cycle) females. The study also revealed negligible AaFAS4 expression in all developmental stages and sexes (Fig. 4).
Fig. 4.
Expression of AaFAS in mosquito developmental stages and sexes. RNA was prepared from five specimens of fourth-instar larvae, pupae, sugar-fed males, sugar-fed females and blood-fed females (3-days pbm) (all adult mosquitoes were collected at the same day time; 8–10 days post eclosion). Samples were subjected to RT-qPCR to assess relative expression of AaFAS1-5 and AaFAS-like. RNA levels between samples were normalized to the β-actin gene using 2−ΔCt method. The boxes show the 25th and 75th percentiles, the whiskers show the minimum and maximum values. The midline indicates the median of the relative gene expression value. The experiment was repeated twice, and representative results from a single experiment are shown
Impact of blood feeding on expression of AaFAS1
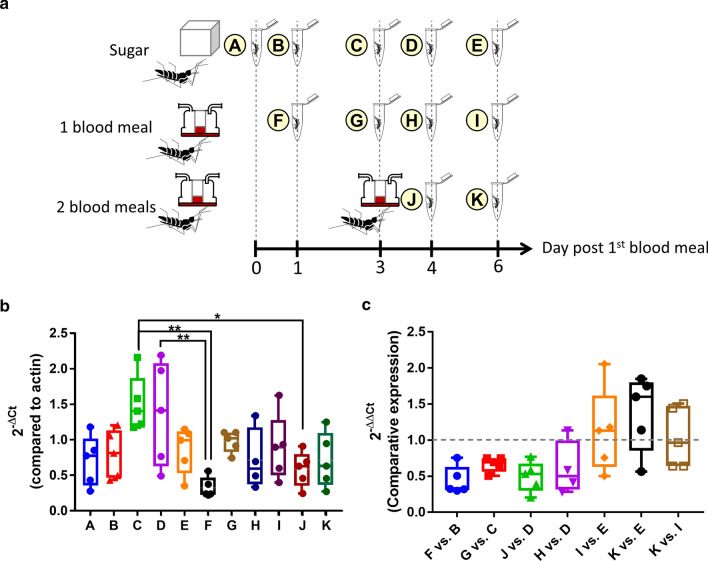
The diet of the female Ae. aegypti typically involves both nectar and blood. The blood meal is rich in proteins and lipids; therefore, this diet may trigger lipolysis, instead of synthesis, to break down lipid molecules. We compared AaFAS1 expression, the predominant AaFAS in adult females, in sugar-fed females versus blood-fed females (feeding once or twice) (Fig. 5). Blood meals were provided only on specific days as shown in Fig. 5A, while mosquitoes from all groups were fed ad lib on 10% sugar diet throughout the experiment. Comparisons of AaFAS1 gene expression from mosquito samples collected on the same day showed no differences among feeding conditions (Fig. 5B). However, when profiled as ratios (Fig. 5C), we observed a slight, but not significant, reduction of AaFAS1 expression in females given a single blood-meal compared to sugar-fed females on days 1, 3 and 4 post-blood meal (pbm) (Fig. 5B: F vs. B, G vs. C and H vs. D). These data suggest that diet may only play a minor role, if any, in the expression of AaFAS1 gene.
Fig. 5.
Expression of AaFAS1 in sugar-fed and blood-fed mosquitoes. a Schematic of the experimental design. Mosquitoes were reared in three different conditions: sugar-fed only, one blood meal, which were fed on day zero, and two blood meals, which were again fed on day 3. Mosquitoes in all conditions were also allowed access to additional sugar and water all the time throughout the experiment. Five mosquitoes from each condition were collected on different days post first blood meal feeding designated in letters A–K. b AaFAS1 expression profile (2−ΔCt; normalized to actin) of each sampling group were shown. c Expression of AaFAS1 was measured by the comparative expression (2−ΔΔCt) method. The samples used for comparisons are shown on the X-axis. Boxes show the 25th and 75th percentiles, whiskers show the minimum and maximum values, and the midline shows the median of the relative gene expression value
Transient knockdown of AaFAS1 gene causing upregulation of other AaFAS genes
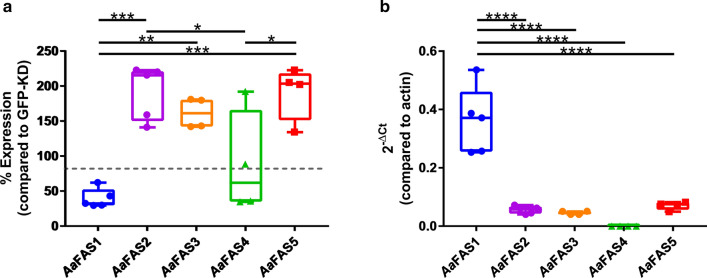
We hypothesized that the redundancy of AaFAS genes may serve as a backup system for the mosquitoes. To test this hypothesis, we employed AaFAS1 loss-of-function studies to investigate the possibility of compensation by other AaFAS genes. Female mosquitoes were IT injected with dsRNA derived from AaFAS1 or GFP (KD control). On day 2 post-dsRNA injection, five mosquitoes were collected for assessment of AaFAS expression (Fig. 6). We observed an approximate 40% reduction in AaFAS1 expression (~ 39.3 ± 13.9%) in AaFAS1-KD mosquitoes compared to the GFP-KD control (Fig. 6A). In AaFAS1-KD mosquitoes, expression levels of AaFAS2, 3 and 5 were 191.7 ± 38.6%, 161.4 ± 21.8% and 191.1 ± 38.9%, respectively, compared to their levels in GFP-KD control, indicating possible compensation for the loss of AaFAS1 transcript. Conversely, the expression of AaFAS4 was 87.71 ± 74.0% compared to AaFAS4 expression in GFP-KD control mosquitoes. To determine whether the upregulation observed in AaFAS2, 3 and 5 could possibly compensate for the loss of AaFAS1 in the AaFAS1-KD mosquitoes, we normalized the level of AaFAS genes to β-actin. We observed modest expression of AaFAS transcripts (5.6 ± 1.44% for AaFAS2, 4.6 ± 0.00% for AaFAS3 and 7.07 ± 0.62% for AaFAS5 compared to β-actin), while these upregulations still did not match the remnant of AaFAS1 expression after the KD effect (36.1 ± 11.6%). These data suggest that other AaFASs may not be able to serve as a backup system for AaFAS1, at least in adult female mosquitoes under transient KD condition.
Fig. 6.
Comparative analyses of AaFAS expression in AaFAS1-KD mosquito. Three-day-old adult female mosquitoes were intrathoracically injected with dsRNA derived from AaFAS1 or GFP mRNA sequence (an irrelevant dsRNA control). Mosquitoes were reared on 10% sugar diet for 2 days post-injection. Five mosquitoes were collected for AaFAS gene expression measurements. a Percent relative expression of AaFAS genes in AaFAS1-KD mosquitoes was compared to GFP-KD control mosquitoes using the comparative Ct (2−ΔΔCt) method. b Gene expression profiles of AaFAS were measured as normalized to β-actin gene expression (2−ΔCt). Boxes show the 25th and 75th percentiles, whiskers show the minimum and maximum values and midline shows median of the relative gene expression value. The experiment was repeated twice, and representative results from a single experiment are shown
Effect of RNAi-induced AaFAS1 knockdown on DENV2 replication in Ae. aegypti cells
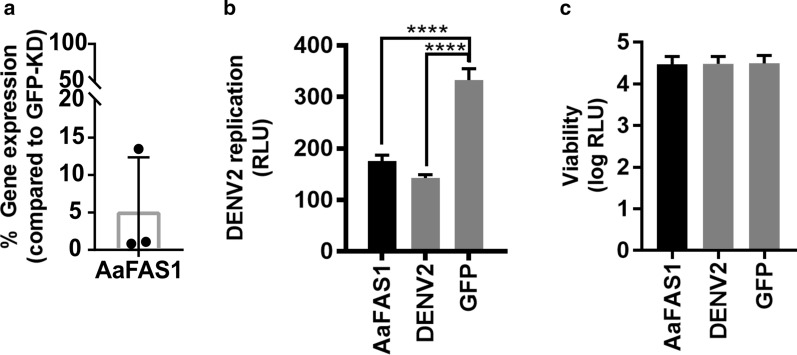
Studies in cell culture have shown that FAS activity is required for flavivirus genome replication [13, 14, 39]. Biochemical inhibition of FAS activity reduced DENV2 replication in both human and mosquito C6/36 cells [13, 15, 16]. The lack of functional RNAi machinery in C6/36 cells hindered the use of transient KD strategy in mosquito cells. However, Ae. aegypti cells, Aag2, have functional RNAi machinery; therefore, we can investigate the role of AaFAS1, the most abundant transcript in female mosquitoes, in DENV2 replication using dsRNA transient KD in these cells [40]. At 48 h post-AaFAS1-KD (time zero of DENV2 infection), the expression level of AaFAS1 in Aag2 cells was 5.13 ± 7.24% compared to AaFAS1 expression in GFP-KD negative KD control cells (Fig. 7A). At 24 h post DENV2 infection (72 h post-KD), we observed significant reduction (P < 0.001) in DENV2 RNA replication in AaFAS1-KD cells compared to the GFP-KD controls, comparable to replication in DENV2-KD (positive KD control) (Fig. 7B), while the KD was not associated with detrimental effects to the cells (Fig. 7C), suggesting that AaFAS1 is required for DENV2 replication in mosquito cells.
Fig. 7.
Assessment of RNAi-induced AaFAS1 knockdown on DENV2 replication in Aag2 cells. a Percent AaFAS1 expression in AaFAS1-KD Aag2 cells compared to the AaFAS1 expression in GFP-KD cells. Expression was measured at 2 days post dsRNA transfection. b RNA replication of luciferase-expressing DENV2 in Aag2 cells treated with dsRNA derived from AaFAS1, DENV2 (positive KD control) or GFP (negative KD control). Cells were transfected with dsRNA for 2 days prior to infection with luciferase-tagged DENV2. At 24 h post infection, cells were lysed and were assayed for luciferase activity (RLU). c Viability of Aag2 cells treated with dsRNA derived from AaFAS1, DENV2 and GFP genes assessed by resazurin assay. All treatment were performed on three biological replicates. The experiment was repeated twice, and representative results from a single experiment are shown
Transient inhibition of AaFAS1 reduced DENV2 infection in the midgut of Ae. aegypti
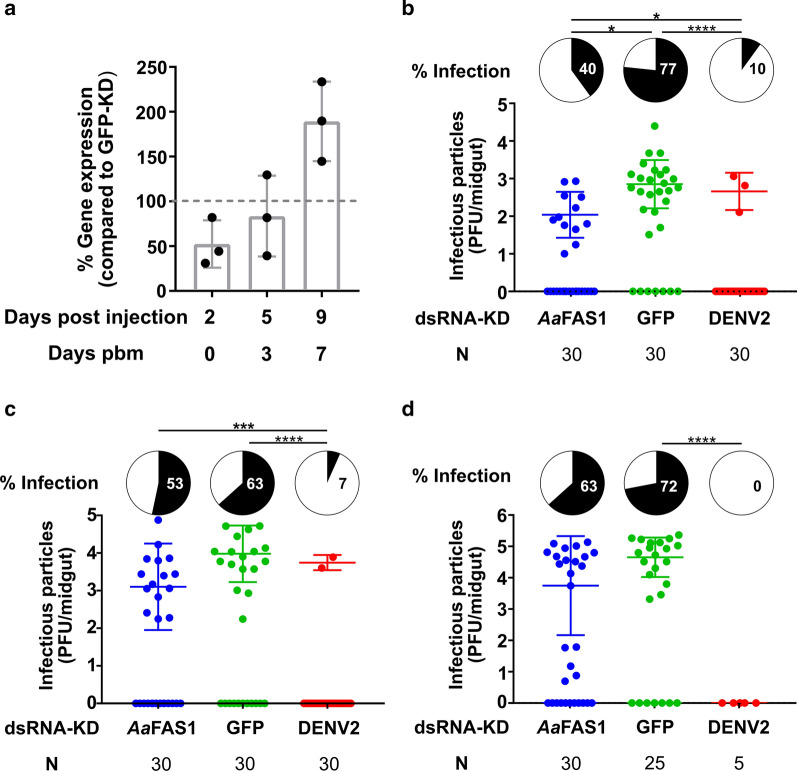
To investigate the role of AaFAS1 in DENV2 replication in vivo, mosquitoes were IT injected with dsRNA derived from AaFAS1 or GFP genes and subsequently exposed to DENV2 infectious blood meal 2 days post-injection (Fig. 8). On days 0, 3 and 7 pbm (corresponding to days 2, 5 and 9 post-dsRNA injection), whole mosquitoes were collected and analyzed for AaFAS1 gene expression (Fig. 8A). On the day of DENV2 infection by blood meal (2 days post-dsRNA injection), the level of AaFAS1 expression was downregulated to 52.36 ± 26.51% relative to GFP-KD group. On day 3 pbm, AaFAS1 expression recovered to 83.52 ± 45.13% and was comparable to the AaFAS1 expression level in the GFP-KD control collected on the same day. On day 7 pbm, AaFAS1 was upregulated to 189.29 ± 44.38%, suggesting a possible over-compensation post KD effect (Fig. 8A).
Fig. 8.
Transient KD of AaFAS1 expression by dsRNA temporarily reduced DENV2 infection in midguts. a Percent AaFAS1 expression in AaFAS1-KD compared to GFP-KD mosquitoes. Mosquitoes were IT injected with ~ 400 ng of dsRNA derived from AaFAS1 or GFP (negative KD control) and fed with a blood meal 2 days post IT injection. On days 2, 5 and 9 post IT injection (days 0, 3 and 7 pbm), 3 pools of 5 mosquitoes from both treatments were collected and analyzed for AaFAS1 expression. b–d Mosquitoes were IT injected with dsRNAs against AaFAS1, GFP and DENV2 (positive KD control) and infected with DENV2 via infectious blood meal at 2 days post injection. Plaque assay was performed on midguts dissected on (b) day 3 and (c) day 7 and (d) carcasses (whole body without midgut) collected on day 14 pbm. Pie charts (black) show percent infected mosquito tissue in each treatment. Pairwise χ2 tests with Holm’s correction for multiple comparisons were used to analyze differences in proportion of infected mosquitoes among groups. Dot plots report virus titer in mosquito tissues. Mean virus titer (infectious particles) was calculated for infected samples only. (i) and (ii) indicate the separation of DENV2 titers in the carcass (day 14) that were produced from the AaFAS1-KD mosquitoes. One-way ANOVA followed by Tukey’s multiple comparison tests were applied to test the differences in virus titer among samples but no significant differences in titers were found
Investigation of DENV2-fed mosquitoes showed that, using pairwise χ2 tests, the percent of infection in the GFP-KD group (negative KD control) was higher compared to the percent infection of DENV2-KD groups (positive KD control) at all time points (Fig. 8B–D and Additional file 1: Table S6), indicating that the negative and positive dsRNA KD controls were effective against DENV2 infection in mosquitoes. Interestingly, we observed significant differences of percent infection between AaFAS1-KD and GFP-KD in midgut samples on day 3 pbm, suggesting that AaFAS1-KD may have a detrimental effect on virus infection in the mosquito midgut (Fig. 8B). However, the inhibitory effect of AaFAS1-KD on DENV2 infection did not persist beyond day 3 pbm since the differences in percent infection between AaFAS1-KD and GFP-KD mosquitoes on days 7 and 14 pbm were not observed (Fig. 8C, D. Additional file 1: Table S6).
Among the mosquitoes that were infected with DENV2 viruses, we did not observe significant differences in viral titers in any treatment at any time point (Fig. 8B–D, Additional file 1: Table S7). However, we observed two distinct populations of viral titers in AaFAS1-KD carcasses on days 14 pbm (Fig. 8D, Additional file 1: Table S7); some with viral titers comparable to GFP-KD control (5.8 × 104 PFU/carcass (i) and some with distinctively lower titers (3 PFU/carcass (ii) (Additional file 1: Table S7). This observation suggests that transient KD of AaFAS1 may have a prolonged effect that impacts dissemination of the virus in mosquitoes.
Discussion
Lipids are essential for a variety of physiological processes in mosquitoes [3, 10, 12, 41, 42]. Mosquitoes not only acquire lipids from maternal (i.e., deposition to eggs) and dietary sources, but they also have the ability to synthesize lipids de novo. In this study, we characterized the expression of the AaFAS gene family, key enzymes in the de novo lipid biosynthesis pathway. Additionally, we investigated the potential role of AaFAS1 in supporting DENV2 replication in the mosquito cell line and the mosquito vector.
In this study, we identified seven putative AaFAS genes (AaFAS1-6 and AaFAS-like) in the AaegL5 assembly based on amino acid similarity to FAS from vertebrates, invertebrates and yeast. Amino acid sequence alignments and domain analyses revealed low amino acid similarity between mosquito and human FAS (< 50%), including the absence of the ΨME domain. This was also observed in other insect FAS, such as D. melanogaster (fruit fly), Bombyx mori (silkworm), A. mellifera (honeybee), Culex pipiens and An. gambiae. While the ΨME domain in human FAS is still present, it lacks the conserve sequence motif for S-adenosyl-methionine (SAM)-dependent methyltransferases, which is highly conserved in bacteria and fungi, resulting in an absence of methyltransferase activity [1]. This may reflect that this domain is unnecessary for metazoan FAS.
Gene duplication is a hallmark of many mosquito gene families and has been proposed as a source of new evolutionary features [23, 38, 43]. Retention of duplicated genes may be indicative of positive/neutral selection and loci associated with a fitness advantage for the mosquito [44]. We performed molecular and preliminary functional characterization of the AaFAS gene family and detected transcripts for five of the seven AaFAS genes. However, we were unable to detect transcripts for either AaFAS6 or AaFAS-like, suggesting that these gene models likely represent pseudogenes or may reflect an issue in the assembly.
Since mosquitoes undergo four distinct developmental stages in their life and these stages possess very distinct habitats and food sources, different AaFAS genes may play roles supporting the unique requirements for FAS in these different life stages. Transcriptional profiles of AaFAS1-5 revealed low expression levels for all AaFAS in larval and pupal stages, suggesting that these genes may not be constitutively active across the mosquito life cycle. We speculated that maternal lipid deposition in eggs during oogenesis (these comprise about 35% of dry egg weight [7]) and larvae diets may serve to support the metabolic needs during these stages [8, 9, 45]. Thus, they may have minimal requirement for de novo fatty acid biosynthesis. Moreover, in male mosquitoes, we observed high levels of expression of all AaFAS, except AaFAS4. Male Ae. aegypti do not blood feed, but solely obtain their diet from plant nectar, honeydew and fruits [46]. Since these diets are high in carbohydrate but low in lipid content, high expression of AaFAS genes in male mosquitoes may reflect a dependency on AaFAS for de novo synthesis of lipids.
While validation of AaFAS genes and their role in de novo fatty acid synthesis is beyond the scope of this study, a previous study by Alabaster et al. [12] showed that transient KD of AaFAS1 and the rate limiting enzyme for fatty acid synthesis, acetyl-coA carboxylase, caused lower numbers of egg deposition and nonviable eggs, respectively. Therefore, this study validated the roles of these enzymes and their importance in mosquito physiology.
In this study, expression analyses also revealed that AaFAS1 is the predominant AaFAS transcript in both male and female mosquitoes. It has the highest amino acid similarity to the human and mouse FAS. Upon AaFAS1-KD in female mosquitoes, we observed a two-fold increase in other AaFAS transcripts, indicating an attempt to compensate for the loss of AaFAS1 expression (Fig. 6A). The expression of these genes may have failed to compensate for the loss of AaFAS1, since the expression levels of these AaFAS transcripts were still lower than the remaining AaFAS1 expression post-KD. Improving the KD efficiency or extending the period of KD of AaFAS1 (such as using CRISPR/Cas9 knockout) may provide further insights into the redundancy of these AaFAS genes.
Since previous studies have demonstrated the importance of FAS activity in flavivirus replication in both human and mosquito cells [13, 15, 16], we wanted to investigate whether AaFAS1 also played an important role in DENV2 infection in the mosquito vector. Indeed, KD of AaFAS1 showed significant inhibition of DENV2 infection in both Ae. aegypti cells in culture and mosquito midguts. However, the inhibitory effect of virus infection in the midgut was only observed on day 3 pbm, as seen by the reduced percent infection in AaFAS1-KD mosquitoes compared to the GFP-KD group. This phenomenon might be caused by the transient KD of AaFAS1 transcripts. Further studies with longer suppression of AaFAS1 expression would be required to demonstrate the prominent impact of AaFAS1 on infection and transmission.
Interestingly, we found an upregulation of AaFAS1 expression (~ 200% increase compared to the AaFAS1 levels in the GFP-KD control) on day 9 post-KD. Further studies are needed to better understand the biological impact of this “rebound” effect as it may have relevance for strategies aimed at suppression of host factors to disrupt pathogen transmission. In this study, we did not observe increased virus infection in these ‘rebound’ mosquitoes above the GFP-KD levels, suggesting that elevated AaFAS1 transcripts may not necessarily equate to increased AaFAS1 activity. Alternately, the ability of AaFAS1 to support DENV2 replication in mosquitoes might have reached its limits.
Additionally, though the expression of AaFAS1 was knocked down only transiently, we observed a prolonged effect on virus transmission. We found a separation of the virus titers into two groups—high (Fig. 8D, i) and low (Fig. 8D, ii)—in the carcass of the AaFAS1-KD mosquitoes. A study by Ye et al. [47], showed that mosquitoes that were IT injected with DENV at 106 PFU expectorated DENV into the saliva at about 102 PFU, while mosquitoes that were IT injected with DENV at 107 PFU expectorated DENV into the saliva at about 104 PFU. These results suggests that viral titer of DENV in saliva may be dependent on the titer in the body (disseminated titers). In our observations, it is possible that mosquitoes with low body titers (group ii) may not be able to transmit the virus efficiently and the KD of AaFAS1 may result in a reduction of transmission potential. This study suggests that biological relevance of low viral titers in carcasses and its impact on transmission dynamics are worthy of further investigation.
Conclusion
Here we present expression analyses of the AaFAS gene family and a focused study of AaFAS1 during DENV2 infection in Ae. aegypti. We annotated seven AaFAS genes from the AaegL5 genome assembly and present evidence to support the function of five genes. Expression data revealed complexities of AaFAS expression between stages and sexes and suggest that AaFAS1 is the predominant transcript in both male and female adult mosquitoes. Sequence homology suggested conservation between mammalian FAS and AaFAS1, and the presence of multiple catalytic domains supports AaFAS1 as a predominant enzyme in the AaFAS family in de novo lipid biosynthesis in Ae. aegypti. In addition, AaFAS1 was found to facilitate DENV2 replication in both cell culture and in Ae. aegypti. In the latter case, it demonstrated the potential to affect vector competency for virus transmission.
Supplementary Information
Additional file 1: Figure S1. Alignment of the conserved YKELRLRGY motif. Figure S2. Amino acid alignment of the pseudo-methyltransferase (ΨME) domain of H. sapiens and Ae. aegypti FAS. Figure S3. RT-PCR assays designed to detect mRNA products of AaFAS-like and AaFAS6. Table S1. List of vertebrate, invertebrate and yeast FAS gene models employed in the present study. Table S2. Primers for generation of dsRNA for knock-down studies. Table S3. Primers for AaFAS expression analyses. Table S4. Primers for RT-PCR assay detecting mRNA products AaFAS-like and AaFAS6. Table S5. Amino acid similarity of FAS domains between H. sapiens and Ae. aegypti. Table S6. Comparisons of percent infection in mosquitoes (P-values). Table S7. Virus titers in mosquito tissues.
Additional file 2: File S1. mRNA and amino acid sequences of 7 AaFAS genes.
Acknowledgements
The authors thank Kenneth E. Olson and Irma Sanchez-Vargas for providing mosquito eggs. We also thank Rebecca Gullberg, Laura St. Clair and Richard J. Kuhn for advice on experimental design, data analysis and critical evaluation of the manuscript and Tach Costello for clerical and administrative support. Graphical Abstract was created using BioRender.
Abbreviations
- ACC
Acetyl-CoA carboxylase
- Ae. aegypti
Aedes aegypti
- AaFAS
Aedes aegypti fatty acid synthase
- bp
Base pair
- cDNA
Complementary deoxyribonucleic acid
- DENV2
Dengue virus serotype 2
- dpi
Days post-infection
- dsRNA
Long double-stranded ribonucleic acid
- FAS
Fatty acid synthase
- FBS
Fetal bovine serum
- GFP
Green fluorescent protein
- IT
Intrathoracic
- KD
Knockdown
- MEM
Minimum essential media
- PBS
Phosphate-buffered saline
- mRNA
Messenger ribonucleic acid
- PCR
Polymerase chain reaction
- qPCR
Quantitative polymerase chain reaction
- pbm
Post-blood meal
- PFU
Plaque forming unit
- ΨME
Pseudo-methyltransferase
- RNA
Ribonucleic acid
- RNAi
Interference ribonucleic acid
- RT
Reverse transcription
- RT-PCR
Reverse transcription polymerase chain reaction
- SNP
Single nucleotide polymorphism
Author contributions
NC, CBS, GR, EL and JMG carried out the experiments. BG performed statistical analyses. NC, CBS, GR, EL, BG, CAH and RP wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by R01AI151166 NIH-NIAID and the Boettcher Foundation Early Career Investigator Awards to RP. GR was funded by the R01AI151166 NIH-NIAID. CBS was supported by Purdue internal monies.
Availability of data and materials
All data and materials were presented in the manuscript and Additional file.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent for publication.
Competing interests
The authors have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nunya Chotiwan, Email: nunya.chotiwan@gmail.com.
Carlos A. Brito-Sierra, Email: cabritos@unal.edu.co
Gabriella Ramirez, Email: Gabriela.Ramirez@colostate.edu.
Elena Lian, Email: elian@rams.colostate.edu.
Jeffrey M. Grabowski, Email: jgrabows25@gmail.com
Babara Graham, Email: barb.graham@colostate.edu.
Catherine A. Hill, Email: hillca@purdue.edu
Rushika Perera, Email: rushika.perera@colostate.edu.
References
- 1.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 2.White SW, Zheng J, Zhang YM, Rock CO. The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 3.Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dadd RH, Kleinjan JE. Essential fatty acid for the mosquito Culex pipiens: arachidonic acid. J Insect Physiol. 1979;25:495–502. doi: 10.1016/s0022-1910(79)80008-6. [DOI] [PubMed] [Google Scholar]
- 5.Stanley DW, Miller JS. Eicosanoid actions in insect cellular immune functions. Entomol Exp Appl. 2006;119:1–13. [Google Scholar]
- 6.Ziegler R. Lipid synthesis by overies and fat body of Aedes aegypti (Diptera: Culicidae) Eur J Entomol. 1997;94:385–391. [Google Scholar]
- 7.Ziegler R, Ibrahim MM. Formation of lipid reserves in fat body and eggs of the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2001;47:623–627. doi: 10.1016/s0022-1910(00)00158-x. [DOI] [PubMed] [Google Scholar]
- 8.Atella GC, Shahabuddin M. Differential partitioning of maternal fatty acid and phospholipid in neonate mosquito larvae. J Exp Biol. 2002;205:3623–3630. doi: 10.1242/jeb.205.23.3623. [DOI] [PubMed] [Google Scholar]
- 9.Sushchik NN, Yurchenko YA, Gladyshev MI, Belevich OE, Kalachova GS, Kolmakova AA. Comparison of fatty acid contents and composition in major lipid classes of larvae and adults of mosquitoes (Diptera: Culicidae) from a steppe region. Insect Sci. 2013;20:585–600. doi: 10.1111/j.1744-7917.2012.01582.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhou GL, Pennington JE, Wells MA. Utilization of pre-existing energy stores of female Aedes aegypti mosquitoes during the first gonotrophic cycle. Insect Biochem Mol Biol. 2004;34:919–925. doi: 10.1016/j.ibmb.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Jenkin HM, McMeans E, Anderson LE, Yang TK. Comparison of phospholipid composition of Aedes aegypti and Aedes albopictus cells obtained from logarithmic and stationary phases of growth. Lipids. 1975;10:686–694. doi: 10.1007/BF02532762. [DOI] [PubMed] [Google Scholar]
- 12.Alabaster A, Isoe J, Zhou G, Lee A, Murphy A, Day WA, et al. Deficiencies in acetyl-CoA carboxylase and fatty acid synthase 1 differentially affect eggshell formation and blood meal digestion in Aedes aegypti. Insect Biochem Mol Biol. 2011;41:946–955. doi: 10.1016/j.ibmb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, et al. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci USA. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martín-Acebes M, Blázquez A, de Oya NJ, Escribano-Romero E, Saiz J. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. Plos ONE. 2011 doi: 10.1371/journal.pone.0024970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, et al. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tongluan N, Ramphan S, Wintachai P, Jaresitthikunchai J, Khongwichit S, Wikan N, et al. Involvement of fatty acid synthase in dengue virus infection. Virol J. 2017;14:28. doi: 10.1186/s12985-017-0685-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsch C, Zeuzem S. RNA-binding activity of hepatitis C virus NS4B: a novel target for small molecule inhibitors. Gastroenterology. 2009;137:2170–2172. doi: 10.1053/j.gastro.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol. 2014;88:4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harak C, Lohmann V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology. 2015;479–480:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chotiwan N, Andre BG, Sanchez-Vargas I, Islam MN, Grabowski JM, Hopf-Jannasch A, et al. Dynamic remodeling of lipids coincides with dengue virus replication in the midgut of Aedes aegypti mosquitoes. PLoS Pathog. 2018;14:e1006853. doi: 10.1371/journal.ppat.1006853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43:D707–13. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 25.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beedessee G, Hisata K, Roy MC, Satoh N, Shoguchi E. Multifunctional polyketide synthase genes identified by genomic survey of the symbiotic dinoflagellate, Symbiodinium minutum. BMC Genom. 2015;16:941. doi: 10.1186/s12864-015-2195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 29.Brown NP, Leroy C, Sander C. MView: a web-compatible database search or multiple alignment viewer. Bioinformatics. 1998;14:380–381. doi: 10.1093/bioinformatics/14.4.380. [DOI] [PubMed] [Google Scholar]
- 30.Mistry J, Bateman A, Finn RD. Predicting active site residue annotations in the Pfam database. BMC Bioinform. 2007;8:298. doi: 10.1186/1471-2105-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett KE, Olson KE, Munoz Mde L, Fernandez-Salas I, Farfan-Ale JA, Higgs S, et al. Variation in vector competence for dengue 2 virus among 24 collections of Aedes aegypti from Mexico and the United States. Am J Trop Med Hyg. 2002;67:85–92. doi: 10.4269/ajtmh.2002.67.85. [DOI] [PubMed] [Google Scholar]
- 32.Mathur G, Sanchez-Vargas I, Alvarez D, Olson KE, Marinotti O, James AA. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2010;19:753–763. doi: 10.1111/j.1365-2583.2010.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 34.Deubel V, Kinney RM, Trent DW. Nucleotide sequence and deduced amino acid sequence of the structural proteins of dengue type 2 virus, Jamaica genotype. Virology. 1986;155:365–377. doi: 10.1016/0042-6822(86)90200-x. [DOI] [PubMed] [Google Scholar]
- 35.Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 37.Lee E, Helt GA, Reese JT, Munoz-Torres MC, Childers CP, Buels RM, et al. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews BJ, McBride CS, DeGennaro M, Despo O, Vosshall LB. The neurotranscriptome of the Aedes aegypti mosquito. BMC Genom. 2016;17:32. doi: 10.1186/s12864-015-2239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang WC, Lin RJ, Liao CL, Lin YL. Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J Virol. 2014;88:6793–6804. doi: 10.1128/JVI.00045-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, et al. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009;5:e1000299. doi: 10.1371/journal.ppat.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Downer RGH, Matthews JR. Patterns of lipid distribution and utilisation in insect. Am Zool. 1976;16:733–745. [Google Scholar]
- 42.Zhou GL, Flowers M, Friedrich K, Horton J, Pennington J, Wells MA. Metabolic fate of [C-14]-labeled meal protein amino acids in Aedes aegypti mosquitoes. J Insect Physiol. 2004;50:337–349. doi: 10.1016/j.jinsphys.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Chau LM, Goodisman MAD. Gene duplication and the evolution of phenotypic diversity in insect societies. Evolution. 2017;71:2871–2884. doi: 10.1111/evo.13356. [DOI] [PubMed] [Google Scholar]
- 44.Helmkampf M, Cash E, Gadau J. Evolution of the insect desaturase gene family with an emphasis on social Hymenoptera. Mol Biol Evol. 2015;32:456–471. doi: 10.1093/molbev/msu315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dadd RH. Essential fatty-acids for the mosquito Culex pipiens. J Nutr. 1980;110:1152–1160. doi: 10.1093/jn/110.6.1152. [DOI] [PubMed] [Google Scholar]
- 46.Wahid I, Sunahara T, Mogi M. Maxillae and mandibles of male mosquitoes and female autogenous mosquitoes (Diptera: Culicidae) J Med Entomol. 2003;40:150–158. doi: 10.1603/0022-2585-40.2.150. [DOI] [PubMed] [Google Scholar]
- 47.Ye YH, Carrasco AM, Frentiu FD, Chenoweth SF, Beebe NW, van den Hurk AF, et al. Wolbachia reduces the transmission potential of dengue-infected Aedes aegypti. PLoS Negl Trop Dis. 2015;9:e0003894. doi: 10.1371/journal.pntd.0003894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Alignment of the conserved YKELRLRGY motif. Figure S2. Amino acid alignment of the pseudo-methyltransferase (ΨME) domain of H. sapiens and Ae. aegypti FAS. Figure S3. RT-PCR assays designed to detect mRNA products of AaFAS-like and AaFAS6. Table S1. List of vertebrate, invertebrate and yeast FAS gene models employed in the present study. Table S2. Primers for generation of dsRNA for knock-down studies. Table S3. Primers for AaFAS expression analyses. Table S4. Primers for RT-PCR assay detecting mRNA products AaFAS-like and AaFAS6. Table S5. Amino acid similarity of FAS domains between H. sapiens and Ae. aegypti. Table S6. Comparisons of percent infection in mosquitoes (P-values). Table S7. Virus titers in mosquito tissues.
Additional file 2: File S1. mRNA and amino acid sequences of 7 AaFAS genes.
Data Availability Statement
All data and materials were presented in the manuscript and Additional file.