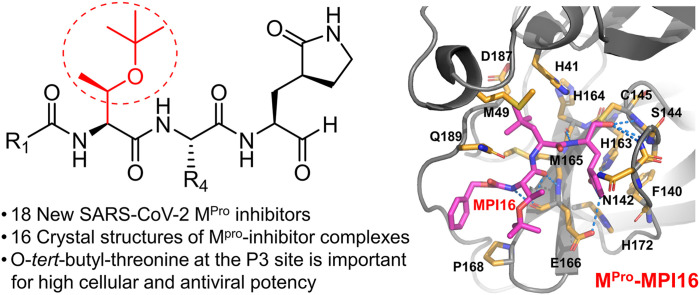
Abstract
As an essential enzyme of SARS-CoV-2, the COVID-19 pathogen, main protease (MPro) is a viable target to develop antivirals for the treatment of COVID-19. By varying chemical compositions at both P2 and P3 positions and the N-terminal protection group, we synthesized 18 tripeptidyl MPro inhibitors that contained also an aldehyde warhead and β-(S-2-oxopyrrolidin-3-yl)-alaninal at the P1 position. Systematic characterizations of these inhibitors were conducted, including their in vitro enzymatic inhibition potency, X-ray crystal structures of their complexes with MPro, their inhibition of MPro transiently expressed in 293T cells, and cellular toxicity and SARS-CoV-2 antiviral potency of selected inhibitors. These inhibitors have a large variation of determined in vitro enzymatic inhibition IC50 values that range from 4.8 to 650 nM. The determined in vitro enzymatic inhibition IC50 values reveal that relatively small side chains at both P2 and P3 positions are favorable for achieving high in vitro MPro inhibition potency, the P3 position is tolerable toward unnatural amino acids with two alkyl substituents on the α-carbon, and the inhibition potency is sensitive toward the N-terminal protection group. X-ray crystal structures of MPro bound with 16 inhibitors were determined. In all structures, the MPro active site cysteine interacts covalently with the aldehyde warhead of the bound inhibitor to form a hemithioacetal that takes an S configuration. For all inhibitors, election density around the N-terminal protection group is weak indicating possible flexible binding of this group to MPro. In MPro, large structural variations were observed on residues N142 and Q189. Unlike their high in vitro enzymatic inhibition potency, most inhibitors showed low potency to inhibit MPro that was transiently expressed in 293T cells. Inhibitors that showed high potency to inhibit MPro transiently expressed in 293T cells all contain O-tert-butyl-threonine at the P3 position. These inhibitors also exhibited relatively low cytotoxicity and high antiviral potency. Overall, our current and previous studies indicate that O-tert-butyl-threonine at the P3 site is a key component to achieve high cellular and antiviral potency for tripeptidyl aldehyde inhibitors of MPro.
Keywords: COVID-19, SARS-CoV-2, Main protease, Covalent inhibitors, Tripeptides
Graphical abstract
1. Introduction
COVID-19 is the current prevailing pandemic that has devastated much of the world. As the COVID-19 pathogen, SARS-CoV-2 uses its membrane Spike protein to recognize the human receptor ACE2 for infection [1,2]. Current COVID-19 vaccines all target this process for the neutralization of the virus. However, the continuous emergence of new viral strains that evade vaccines demands other antivirals to be developed as well. The genome of SARS-CoV-2 encodes a large open reading frame ORF1ab that is translated to two large polypeptides pp1a and pp1b in the human cell host [3,4]. The processing of pp1a and pp1b to 16 nonstructural proteins (nsps) that are functionally critical to the viral replication relies on proteolytic activities of two internal nsp fragments, nsp3 and nsp5 [5]. Nsp5 is also called 3C-like protease and more recently main protease (MPro). Since MPro hydrolyzes 13 out of the total of 16 nsps, it has been considered as a viable target for the development of antivirals. In the past year, a number of papers have been published on the development of peptidyl aldehydes that contain β-(S-2-oxopyrrolidin-3-yl)-alaninal (opal) at the P1 position as potent MPro inhibitors [[6], [7], [8], [9], [10], [11], [12], [13], [14]]. Other inhibitors were developed as well including nirmatrelvir that has been approved by the U.S. Food and Drug Administration for its combined use with ritonavir as a therapy, branded as paxlovid, for COVID-19 and S-217622 that is undergoing clinical trials for COVID-19 in Japan [[15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30]]. However, systematic structure-activity relationship (SAR) studies of opal-based MPro inhibitors are needed. In the current study, we explore variations at the P2 and P3 positions and the N-terminal protection group in opal-based tripeptidyl MPro inhibitors. Our study indicates that O-tert-butyl-threonine at the P3 position in an opal-based MPro inhibitor is critical for achieving high potency inhibiting MPro expressed in human host cells and antiviral potency against SARS-CoV-2.
2. Results
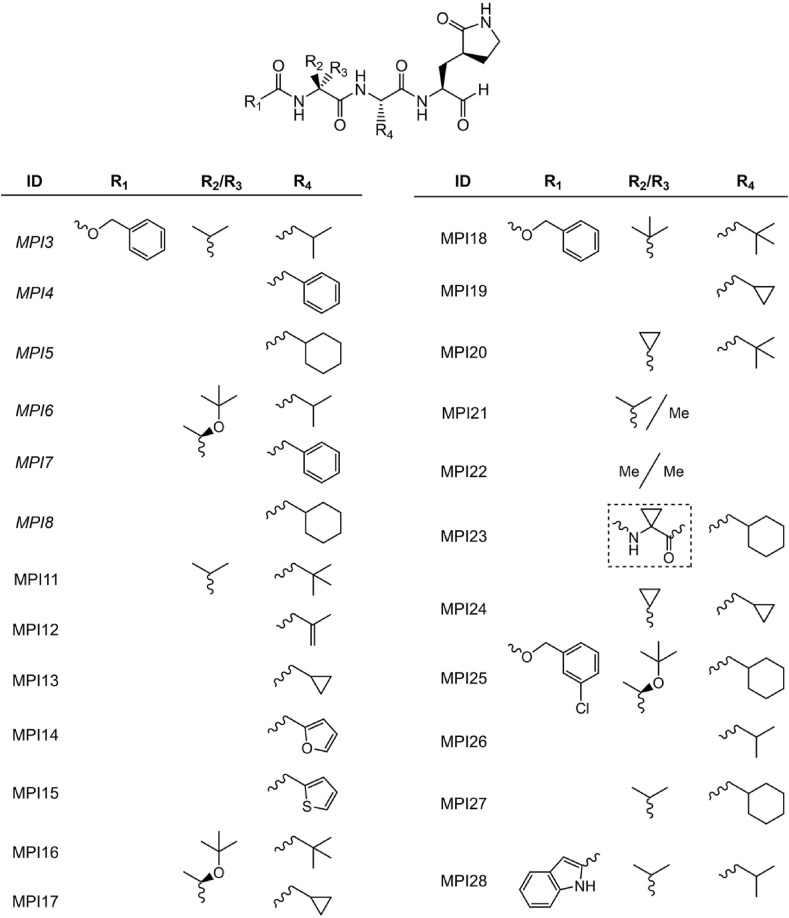
2.1. The design and synthesis of MPI11-28
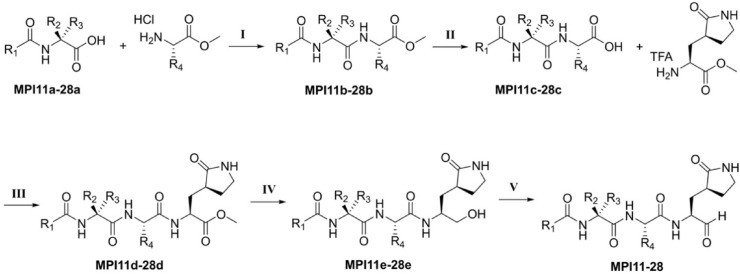
In previous studies, we developed MPI1-10 and characterized both their MPro inhibition potency and their antiviral potency [13,14]. As a tripeptidyl aldehyde, MPI8 shows the highest antiviral potency with an EC50 value of 30 nM to neutralize SARS-CoV-2 (USA-WA1/2020) in Vero E6 cells [31]. MPI3, another tripeptidyl aldehyde, doesn't have high antiviral potency. However, it has the highest in vitro MPro inhibition potency with an IC50 value of 8.5 nM [13]. To explore how substituents at different positions in a tripeptidyl opal-based MPro inhibitor (Fig. 1 ) influence its potency, we decided to carry out a systematic structure-activity relationship (SAR) study. We maintained opal at the P1 position due to its established preferential binding to the MPro P1 binding pocket and its ability to form a covalent adduct with C145, the MPro catalytic cysteine. MPro tolerates leucine and phenylalanine at the P2 position in a substrate. Therefore, we chose to vary this site in our inhibitor design among β-alkyl alanines with sizes between or around leucine and phenylalanine. Chosen alkyl substituents include isopropyl, phenyl, cyclohexyl, t-butyl, isopropenyl, cyclopropyl, 2-furyl, and 2-thienyl. MPro doesn't have a binding pocket for the P3 position in a substrate. However, native MPro substrates have valine or a similar size residue at this position. Based on known opal inhibitors and substrates of MPro, we varied this site among amino acids including valine, O-tert-butyl-threonine, l-cyclopropylglycine, L-tert-butyl-glycine, l-α-methyl-valine, dimethylglycine, and 1-aminocyclopropane-1-carboxylate. The N-terminal protection group was chosen among benzyloxycarbonyl (CBZ), m-chloro CBZ, and indole-2-carboxylate due to their demonstrated contributions to antiviral potency [6,10]. A total of 18 new MPro inhibitors, designated as MPI11-28 shown in Fig. 1, were designed. We synthesized all inhibitors according to the synthetic route shown in Scheme 1 . In this synthesis, a P2 amino acid ester was conjugated with a N-protected P3 amino acid and the afforded product was then hydrolyzed to free its C-terminal carboxylate for a reaction with β-(S-2-oxopyrrolidin-3-yl)-alanine ester. An obtained tripeptidyl ester was reduced to afford a C-terminal alcohol that was then oxidized via Dess-Martin oxidation in a mild condition to make a final product.
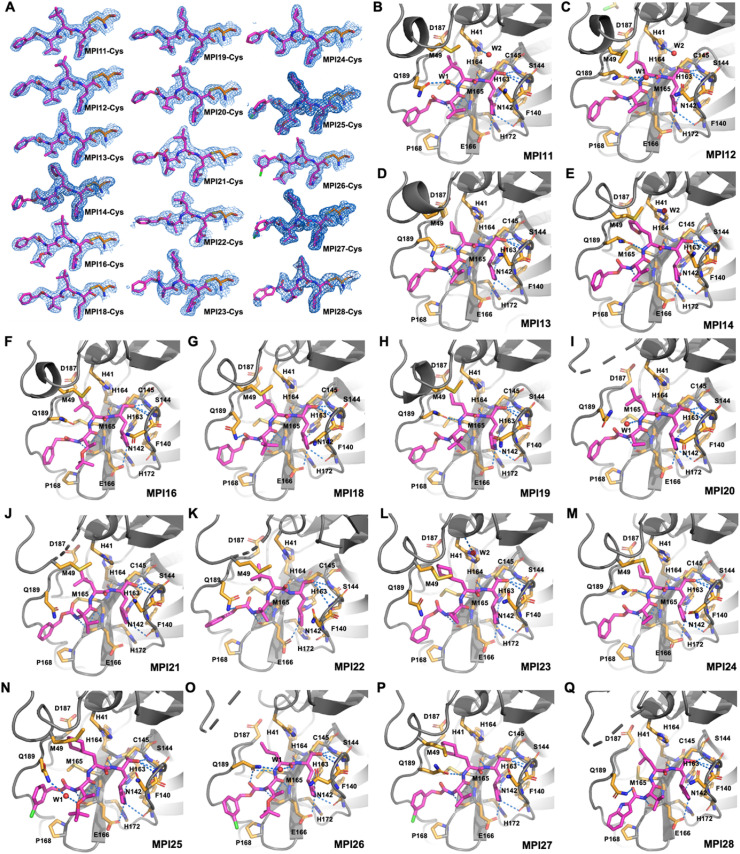
Fig. 1.
Structures of tripeptidyl MPro inhibitors MPI11–28. Inhibitors that have just a single component at R2/R3 shown contain a hydrogen at the R3 position. MPI3-8 were previously developed and are shown for comparison.
Scheme 1.
Reagents and conditions: (I) HATU, DIPEA, DMF; (II) LiOH•H2O, THF/H2O; (III) HATU, DIPEA, DMP; (IV) LiBH4, THF; (V) DMP, DCM.
2.2. Kinetic characterizations of MPI11-28 on their enzymatic inhibition of MPro
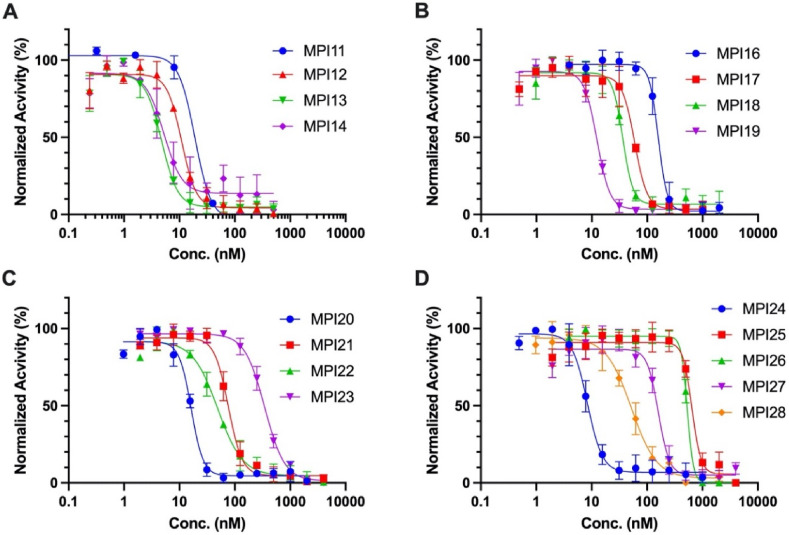
We followed a previously established protocol that uses Sub3 (Dabcyl-KTSAVLQSGFRKME-Edans), a fluorogenic peptide substrate of MPro to determine IC50 values for MPI11-28 [28]. In this assay, we preincubated MPro with an inhibitor for 30 min before Sub3 was added and the fluorescent product formation (Ex: 336 nm/Em: 455 nm) was recorded in a fluorescence plate reader. 30 min incubation time with MPro is a standard procedure that has been used by multiple labs in the determination of IC50 values for MPro inhibitors [11,27,28,32]. Since MPI11-28 are reversible covalent inhibitors, their incubation times with MPro are not expected to significantly influence their determined IC50 values. Tests involving MPI11 in three different incubation times, 15, 30, and 60 min, led to very similar determined IC50 values (Supplementary Fig. 1). Except MPI15 that is insoluble in DMSO and therefore was not characterized, all other inhibitors displayed well traceable inhibition curves as shown in Fig. 2 . We fit all data to a four-parameter variable slope inhibition equation in GraphPad 9.0 to obtain IC50 values for all inhibitors. As shown in Table 1 , MPI11-28 display a large variation of IC50 values that ranges from 4.8 to 650 nM. MPI11-14 all contain valine at the P3 position. They are among the most in vitro potent inhibitors for MPro. MPI13-14 have determined IC50 values around 5 nM. Since 10 nM MPro is the lowest enzyme concentration we can use to do the inhibition analysis, 5 nM is technically the lowest IC50 value we can detect. Therefore, MPro inhibition potency for MPI13-14 is likely higher than the numbers shown in Table 1. In comparison to MPI13-14, MPI11-12 have slightly higher IC50 values. In previous work, we developed another three MPro inhibitors MPI3-5 that also contain a valine at the P3 site. All three have low IC50 values. All 7 inhibitors that have valine at the P3 position and a N-terminal CBZ protection group are among the most potent MPro inhibitors in vitro. A P3 valine apparently favors the enzyme inhibition kinetics. A comparison of all seven inhibitors also reveals that a residue at the P2 position with a size close to leucine leads to better MPro inhibition. β−Cyclopropyl alanine and β-(furan-2yl) alanine that have a more rigid side chain than leucine favor the enzyme inhibition kinetics more. MPI6-8 that have a P3 O-tert-butyl-threonine were developed previously and showed high antiviral potency. We synthesized two more MPro inhibitors with a P3 O-tert-butyl-threonine, MPI16-17. Both have an IC50 value similar to MPI6-8. Varying the P2 position does not seem to significantly change in vitro MPro inhibition potency among this group of inhibitors. To explore further on whether variations at the P3 position lead to different inhibition potency, we developed MPI18-24. Variants include two dialkyl glycines and 1-aminocyclopropane-1-carboxylate that are not standard l-amino acids. Except MPI23 that contains a P3 1-aminocyclopropane-1-carboxylate, all other inhibitors have an IC50 value below 100 nM. The more rigid P3 position in MPI23 likely contributes to its less favorable MPro inhibition kinetics. MPI24 is structurally similar to MPI13. Its enzyme inhibition potency is similar to MPI13 as well. Three inhibitors MPI25-27 that have a N-terminal m-chloro CBZ group were developed and characterized as well. In comparison to their regular CBZ-containing counterpart inhibitors MPI8, MPI6, and MPI5, they display more than 5-fold higher IC50 values. Apparently adding an m-chloro substituent to the terminal CBZ group doesn't improve in vitro enzyme inhibition potency. MPI28 has an N-terminal indole-2-carboxylate. In comparison to its CBZ counterpart inhibitor MPI3, MPI28 has a 6-folder higher IC50 value. In comparison to indole-2-carboxylate, it is evident that CBZ serves as a better N-terminal group for improved in vitro enzyme inhibition potency.
Fig. 2.
Inhibition curves of MPI11-28 on MPro. Triplicate experiments were performed for each compound. For all experiments, 20 or 10 nM MPro was incubated with an inhibitor for 30 min before 10 μM Sub3 was added. The MPro-catalyzed Sub3 hydrolysis rate was determined by measuring linear increase of product fluorescence (Ex: 336 nm/Em: 455 nm) for 5 min.
Table 1.
Determined enzymatic IC50 and cellular EC50 values of MPro inhibitors.
| ID | Enzymatic IC50 (nM) | Cellular EC50 (μM) | Antiviral EC50 (μM) | CC50 (μM) | PDB Entry | ID | Enzymatic IC50 (nM) | Cellular EC50 (μM) | Antiviral EC50 (μM) | CC50 (μM) | PDB Entry |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MPI3 | 8.5b | > 2c | 7JQ0 | MPI18 | 36 | 0.86 | >5e/0.71f/> 5g | >200 | 7RVR | ||
| MPI4 | 15b | > 2c | 7JQ1 | MPI19 | 13 | 1.5 | 7RVS | ||||
| MPI5 | 33b | 0.66c | 7JQ2 | MPI20 | 16 | >2 | 7RVT | ||||
| MPI6 | 60b | 0.12c | 7JQ3 | MPI21 | 77 | >2 | 7RVU | ||||
| MPI7 | 47b | 0.19c | 7JQ4 | MPI22 | 49 | >10 | 7RVV | ||||
| MPI8 | 105b | 0.03c | 0.030e | 109.2 | 7JQ5 | MPI23 | 350 | >10 | 7RVW | ||
| MPI11 | 19 | >2 | 7RVM | MPI24 | 8.4 | >2 | 7RVX | ||||
| MPI12 | 11 | >2 | 7RVN | MPI25 | 650 | 0.14 | 2.2e/1.6f/0.87g | 130.0 | 7RVY | ||
| MPI13 | 4.8d | >2 | 7RVO | MPI26 | 530 | 0.23 | 1.9e/0.65f/1.6g | 182.9 | 7RVZ | ||
| MPI14 | 5.2d | >2 | 7RVP | MPI27 | 160 | 0.63 | >5e/>5f/3.2g | 114.0 | 7RW0 | ||
| MPI15 | n.d.a | n.d.a | – | MPI28 | 52 | >10 | 7RW1 | ||||
| MPI16 | 150 | 0.056 | 1.2e/1.2f/0.58g | >200 | 7RVQ | nirmatrelvir [38] | 66 | 3.4 | 1.30 | ||
| MPI17 | 60 | 0.097 | 1.2e/1.8f/1.1g | >200 | – | GC376 [31] | 30 | >2 | 3.37 |
Its IC50 and EC50 values were not determined (n.d.) due to insolubility.
Data were taken from Ref. [13].
Data were taken from Refs. [31].
Reaching the detection limit.
Antiviral EC50 value for the USA-WA1/2020 strain.
Antiviral EC50 value for the Beta strain.
Antiviral EC50 value for the Delta strain.
2.3. X-ray crystallography analysis of MPro bound with 16 different inhibitors
To characterize our developed inhibitors on their interactions with MPro, we crystallized MPro in its apo form, soaked apo-MPro crystals with different inhibitors and then determined structures of MPro bound with different inhibitors using X-ray crystallography. Among 17 inhibitors that we used to soak crystals, 16 were observable in the active site of MPro. All structures were determined to high resolutions (Table S1). For all inhibitors, their calculated 2Fo-Fc electron density maps in the MPro active site clearly show a covalent interaction with MPro C145 to generate a hemithioacetal (Fig. 3 A). In all formed hemithioacetals, the hemithioacetal alcohol takes an S configuration. In most calculated structures, electron density around inhibitors have defined shapes for modeling the P1, P2 and P3 residues. For MPI12, the isopropenyl group has very weak electron density around its C1 and C3 atoms, indicating that the isopropenyl group is probably freely rotating around its C2 atom in the active site of MPro. For MPI26 and MPI28 that have a P2 leucine, electron density around the side chain isopropyl group is also weak, indicating its flexible conformations at the MPro active site. Although the N-terminal group of inhibitors including MPI11, MPI12, MPI18, and MPI19 were successfully modeled, most calculated structures have relatively weak electron density around the inhibitor N-terminal group, indicating weak binding of the N-terminal group at the active site.
Fig. 3.
Crystal structures of MPro bound with 16 MPIs. (A) Contoured 2Fo-Fc maps at the 1σ level around 16 MPIs and C145 in the active site of MPro. (B–Q) Interactions between 16 MPIs and the MPro active site residues.
In MPro-MPI11 as shown in Fig. 3B, MPI11 forms extensive hydrogen bonds with MPro. The hemithioacetal hydroxyl group is within the hydrogen bond distance to three backbone α-amines in residues 143–145 that form an oxyanion hole to facilitate the enzyme-catalyzed hydrolysis of a substrate. Due to the preferential binding of a negative charged hydroxyl group at this position, it is likely that the hemithioacetal hydroxy group is deprotonated to strongly interact with the three backbone α-amines, explaining the exclusive S configuration of the formed hemithioacetal. The opal side-chain lactam forms three hydrogen bonds with residues E166, N142, and F140 and its backbone α-amine forms a hydrogen bond with H164. The side chain of the P2 β-tert-butyl-alanine (4-methyl-leucine) is involved in van der Waals interactions with MPro residues including M49, H41, M165, D187, and Q189. It is interesting to note that the M49-containing loop has weak electron density around it, indicating flexible conformations of this loop when MPI11 binds to the MPro active site. The MPI11 P2 α-amine interacts with the side chain of Q189 indirectly through two water-bridged hydrogen bonds. The MPro-MPI11 structure shows that the tert-butyl group is within a close van der Waals distance to the Q189-containing loop of MPro. To accommodate a chemical moiety bigger than β-tert-butyl-alanine, MPro will need to rearrange the Q189-containing loop to enlarge the P2 binding pocket. The P3 valine of MPI11 forms two hydrogen bonds with the backbone α-amine and α-carbonyl groups of E166. Its side chain forms van der Waals interactions with the side chain of E166. The N-terminal CBZ of MPI11 interacts hydrophobically with the backbone of the Q189-containing loop and the side chain of P168. However, its weak electron density indicates loose binding. In MPro-MPI11, the side chain of N142 flips away from the opal side chain of MPI11. The MPro-MPI12 structure around the active site is shown in Fig. 3C. MPI12 is involved in hydrogen bonding interactions with MPro mostly similar to MPI11. It forms an S-configuration thiohemiacetal covalent adduct with MPro. Unlike in the MPro-MPI11 structure, Q189 in MPro-MPI12 forms a hydrogen bond directly with the MPI12 P2 α-amine and interacts indirectly with the P2 α-carbonyl oxygen through a water-bridged hydrogen bond network. In MPro-MPI12, the P2 β-isopropenyl-alanine side chain doesn't have a fixed conformation. Additional modification to this side chain to introduce tighter binding to MPro is likely. In MPro-MPI12, N142 adopts a conformation that partially caps the P1 binding pocket in MPro. As in MPro-MPI11, the M49-containing loop has weak electron density around it. The MPro-MPI13 structure around the active site is shown in Fig. 3D. Interactions between MPI13 and MPro are mostly similar to that between MPI12 and MPro. N142 adopts a conformation like the one in MPro-MPI11. Unlike MPI12, the P2 cyclopropyl side chain of MPI13 has defined, strong electron density around it and interacts with the van der Waals distance with side chains from H41, D187 and Q189. This observation explains high in vitro enzymatic inhibition potency for inhibitors including MPI13, MPI19 and MPI24. MPI14 shown in Fig. 3E interacts with MPro mostly similar to MPI11. MPI14 has a P2 β-(2-furanyl)-alanine residue that has defined, strong electron density around the furan side chain. The furan group is in parallel with the H41 imidazole side chain so that the two aromatic groups are likely engaged in a π-π stacking interaction. This π-π stacking interaction likely contributes to the high in vitro enzymatic inhibitor potency of MPI14. In MPro-MPI14, Q189 has weak electron density around its side chain. It is not involved in hydrogen bonding interactions with MPI14 directly or indirectly. In both MPro-MPI13 and MPro-MPI14, the M49-containing loop has weak electron density.
MPI16 differs from MPI11 in its P3 position that is O-tert-butyl-threonine. As shown in Fig. 3F, MPI16 maintains most hydrogen bonding interactions that show in the MPro-MPI11 complex structure. The MPro Q189 side chain amide is 3.5 Å to the P2 α-amine of MPI16. This is around the borderline for a hydrogen bond interaction. In MPro-MPI16, the N142 side chain adopts a conformation that caps the MPI16 P1 opal side chain. Compared to MPI11, MPI16 has an additional P3 O-tert-butyl group. Its location can be clearly refined that shows van der Waals interactions with P168 and E166. MPI18 has a chemical structure very similar to MPI11 except at the P3 position that has an additional methyl group on the β-carbon. Its interactions with MPro as shown in Fig. 3G are also similar to that involving MPI11. Visible differences are at MPro residues N142 and Q189. The N142 side chain adopts a conformation to cap on the MPI18 opal side chain and the Q189 side chain amide forms a hydrogen bond with the N-terminal carbamate carbonyl oxygen atom. MPI19 is structurally similar to MPI13 except an additional methyl group at the P2 β-carbon position. As shown in Fig. 3H, MPI19 interacts with MPro similar to MPI13 except that the N142 side chain in MPro-MPI19 adopts a conformation to cap the MPro P1 binding pocket for the MPI19 P1 opal residue. Except a cyclopropyl group at the P3 position, MPI20 is structurally similar to MPI11. As shown in Fig. 3I, MPI20 interacts with MPro similar to MPI11. A water molecule bridges the interaction between the Q189 side chain amide and the MPI20 P2 α-amine. The N142 side chain caps the MPro P1 binding pocket. In MPro-MPI20, electron density around residues 45–50 is too weak to model these residues, indicating that this loop region adopts a highly flexible conformation. The same region shows weak electron density in MPro-MPI16, MPro-MPI18 and MPro-MPI19.
MPI21 has α-methyl-valine at its P3 position. Compared to MPI11, the additional P3 α-methyl group pushed the N-terminal carbamate of MPI21 to adopt an orientation that rotates about 90° from the one shown in MPro-MPI11 (Fig. 3J). Although the Q189 side chain amide is within the hydrogen bond distance to the MPI21 N-terminal carbamate carbonyl oxygen atom, the geometry is not right. Compared to other inhibitors that have a regular l-amino acid at their P3 position, this different orientation at the N-terminal carbamate in MPI21 apparently leaves space for the Q189 side chain to move deeper toward the MPro active site. In MPro-MPI21, the N142 side chain adopts a conformation similar to the one shown in MPro-MPI11. MPI22 is structurally similar to MPI21 but contains dimethylglycine at its P3 position. The MPro-MPI22 structure at the MPro active site is very similar to MPro-MPI21. The pro-S methyl group at the MPI22 P3 residue pushes the N-terminal carbamate to adopt a conformation that positions its carbamate carbonyl oxygen away from forming a hydrogen bond with the Q189 side chain amide (Fig. 3K). As same as in MPro-MPI21, this new orientation at the N-terminal carbamate leaves space for the Q189 side chain to move deeper toward the MPro active site. MPI23 has a cyclopropyl backbone at its P3 position. Similar to MPI21 and MPI22, this cyclopropyl group pushes the N-terminal carbamate to adopt a conformation that positions the carbamate carbonyl oxygen atom away from forming a hydrogen bond with the Q189 side chain amide through the rotating extend of the carbamate is not as much as in MPI21 and MPI22 (Figure 3L). The P2 position of MPI23 is β-cyclohexyl-alanine. The cyclohexyl group has defined, strong density that indicates a chair conformation. The cyclohexyl group aligns parallelly with respect to the H41 side chain, making two C–H groups from the cyclohexyl group directly interact with the π orbital of the H41 side chain. Compared to MPI11, the P2 residue of MPI23 is larger and pushes the Q189-containing loop to move slightly away from the active site. MPI24 has a structure similar to MPI13. It has a cyclopropyl group compared to an isopropyl group in MPI13 at the P3 β-carbon. The interactions between MPI24 and MPro are also similar to that between MPI13 and MPro (Figure 3M). In MPro-MPI24, The N142 side chain caps the MPro P1 binding pocket and the Q189 side chain amide forms a hydrogen bond with the MPI24 P2 α-amine. In MPro-MPI21, MPro-MPI22, MPro-MPI23, and MPro-MPI24, the M49-containing loop has weak electron density that leads to structures of certain residues in MPro-MPI21 and MPro-MPI22 not able to be refined.
MPI25 is a MPI8 derivative. Its interactions with MPro involve most hydrogen bonds that exist in the MPro-MPI11 structure (Figure 3N). In MPro-MPI25, the N142 side chain points away from the position that can cap the MPro P1 binding pocket and the Q189 side chain amide interacts with the MPI25 P3 side chain oxygen through two water-bridged hydrogen bonds. Since the electron density around the N-terminal group is weak, the chloride position cannot be unambiguously assigned but it is with high confidence that it interacts with the P168 side chain. In MPro-MPI25, The M49-containing loop also has relatively weak electron density so that the M49 side chain conformation cannot be unambiguously assigned either. The O-tert-butyl group in the P3 residue has strong electron density that was used to unambiguously refine its structure that interacts with the P168 and E166 side chains. MPI26 is structurally different from MPI25 only at the P2 residue. As shown in Figure 3O, the binding model of MPI26 at the MPro active site is very similar to MPI25. Visible differences are at MPro residues N142 and Q189. In MPro-MPI26, the N142 side chain adopts a conformation that caps the MPro P1 binding pocket and the Q189 side chain amide forms hydrogen bonds separately with the MPI26 P2 α-amine and the N-terminal carbamate carbonyl oxygen. The electron density around residues 45–50 is also too weak to be used to refine their structures. The electron density around the P2 leucine β-isopropyl group allows only one methyl group can be clearly refined, indicating that the P2 leucine β-isopropyl group may adopt different conformations. MPI27 is structurally different from MPI25 only at its P3 position that is valine. As shown in Figure 3P, MPI27 interacts with MPro similar to MPI25. In MPro-MPI27, the P3 valine side chain packs on top of the E166 side chain to form van der Waals interactions. The Q189 side chain amide forms a hydrogen bond with the MPI27 P2 α-amine. Unlike all other inhibitors, MPI28 has a N-terminal indole-2-carbonyl protection group. Its interactions with MPro shown in Figure 3Q are similar to that in MPro-MPI11. The Q189 side chain amide forms a hydrogen bond with the N-terminal amide oxygen. As in most other MPro-inhibitor structures, the electron density at the N-terminal group is too weak to refine its accurate structure. Like in MPro-MPI26, the electron density at the P2 leucine β-isopropyl group allows the refinement of only one methyl group, indicating flexible binding of this group at the MPro active site. In MPro-MPI28, the electron density around MPro residues 46–50 is too weak to be used to refine their structures. Although some of MPro-inhibitor complexes have refined structures for residues 45–50, all of them have relatively weaker electron density around these residues than other parts of MPro, indicating that the M49-containing loop is flexible after an inhibitor binds to the MPro active site.
2.4. Characterizations of cellular MPro inhibition potency of MPI11-28
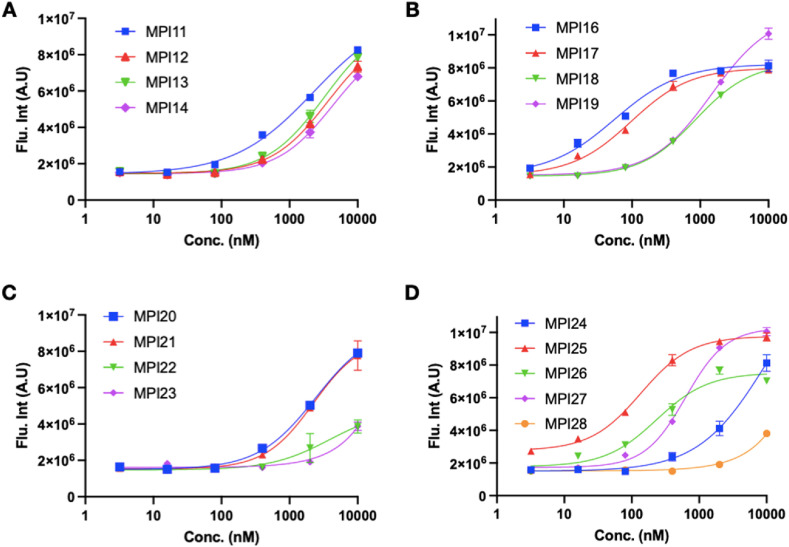
MPro is acutely toxic to a human cell host. Using this unique characteristic, we developed a cell-based analysis to characterize cellular MPro inhibition potency for compounds [31]. A similar assay was also used by others to analyze cellular potency of MPro inhibitors [33,34]. In this assay, a compound with cellular potency inhibits cellular toxicity from the expression of an MPro-eGFP fusion protein, leading to enhanced overall expression of MPro-eGFP that can be characterized by flow cytometry. This assay is more advantageous over an antiviral assay in analyzing the cellular MPro inhibition ability of a compound since it may inhibit host proteases such as TMPRSS2, furin, and cathepsin L that are critical for SARS-CoV-2 infection to provide false positive cellular potency results for MPro inhibition [[35], [36], [37]]. We used this method to characterize a number of investigational MPro inhibitors. Our study showed some inhibitors that have demonstrated antiviral potency do not inhibit MPro directly in a human cell host, indicating different mechanisms for these compounds to achieve antiviral potency [31]. Other advantages of using this assay include quickly priming potent inhibitors for their antiviral assays by bypassing cellular permeability and stability tests and avoiding time-consuming and more complicated antiviral tests that involve the use of a biosafety level 3 facility for compounds with low cellular potency. We adopted this assay to characterize MPI11-28. All inhibitors were tested up to 10 μM in their inhibition of MPro-eGFP that was transiently expressed in human 293T cells. Overall cellular eGFP fluorescence was plotted against the inhibition concentration to obtain their EC50 values. Results are presented in Fig. 4 and Table 1. As shown in Fig. 4A, MPI11-14 exhibited minimal cellular potency to inhibit MPro in 293T cells, which is in significant contrast to their very high in vitro enzyme inhibition potency. Since their inhibition curves do not reach a plateau, their EC50 values are estimated as > 2 μM. MPI16 and MPI17 are the two most potent inhibitors among MPI11-28 on cellular potency with determined EC50 values as 56 nM and 97 nM, respectively (Fig. 4B). In comparison to MPI16 and MPI17, MPI18 and MPI19 have weaker cellular potency with determined EC50 values as 860 nM and 1500 nM, respectively. MPI20-23 all have weak cellular potency (Fig. 4C). MPI22 and MPI23 showed very low inhibition of MPro-eGFP in 293T cells. At 10 μM for these two compounds, MPro-eGFP expression was lower than half of the plateau level of MPro-eGFP that was observed for MPI16-19. For these four inhibitors, their EC50 values are estimated as > 2 μM for MPI20 and MPI21 and > 10 μM for MPI22 and MPI23. Among MPI24-28 (Fig. 4D), MPI25-27 exhibited high cellular potency with EC50 values as 140 nM, 230 nM, and 630 nM respectively. On contrary to their high in vitro enzyme inhibition potency, MPI24 and MPI28 have very weak cellular potency with estimated EC50 values as > 2 μM and >10 μM, respectively. Two reported MPro inhibitors nirmatrelvir [29] and GC376 [7] were tested for in cellulo MPro inhibition potency. Nirmatrelvir has a determined in cellulo EC50 value as 3.4 μM [38], and GC376 shows a in cellulo EC50 value of >2 μM (Table 1) [31]. Their cellular potency are weaker compared to MPI16, MPI17, MPI25, and MPI26.
Fig. 4.
Cellular potency of MPI11-28 in their inhibition of MPro to drive host 293T cell survival and overall MPro-eGFP expression.
2.5. Characterizations of antiviral potency of six selected inhibitors on three SARS-CoV-2 variants
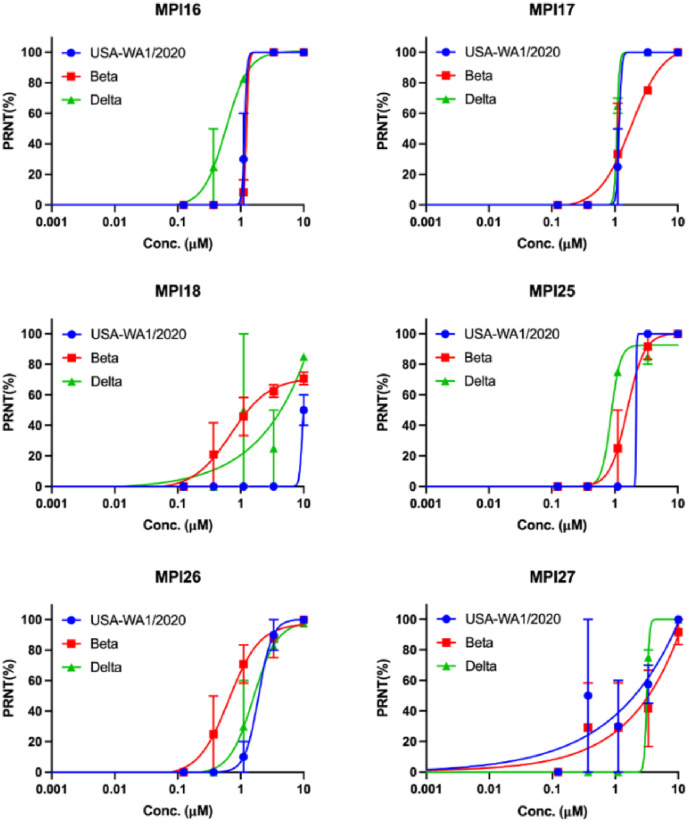
Four most cellularly potent inhibitors MPI16, MPI17, MPI25, and MPI26 that contain a P3 O-tert-butyl-threonine and two mildly cellularly potent inhibitors MPI18 and MPI27 that don't contain a P3 O-tert-butyl-threonine were analyzed on their antiviral potency against three SARS-CoV-2 variants including USA-WA1/2020, Beta and Delta (Fig. 5 ). We conducted plaque reduction neutralization tests in Vero E6 cells for all inhibitors. We infected Vero E6 cells by virus in the presence of an inhibitor at various concentrations for three days and then quantified viral plaque reduction. As presented in the attached figure and table, all four inhibitors that contain a P3 O-tert-butyl-threonine showed high antiviral potency with determined EC50 values around 1 μM and MPI18 and MPI27 that don't have a P3 O-tert-butyl-threonine showed only moderate antiviral potency. It should be noted that MPI5 containing a P3 valine showed mild cellular MPro inhibition potency but potent antiviral activity [13,31]. Its antiviral activity may not only come from the inhibition of MPro but also other mechanisms.
Fig. 5.
Plaque reduction neutralization tests (PRNTs) of MPI16-18 and MPI25-27 on their inhibition of three SARS-CoV-2 strains USA-WA1/2020, Beta and Delta in Vero E6 cells. Two repeats were conducted for each concentration.
2.6. Cytotoxicity of MPI8, MPI16, MPI17, MPI18, MPI25, MPI26 and MPI27
We characterized cytotoxicity for these seven compounds in 293T cells using the MTT assay [39]. Cytotoxicity curves of these inhibitors are shown in Supplementary Fig. S2. Their determined CC50 values are 109.2, >200, >200, >200, 130, 182.9 and 114.0 μM for MPI8, MPI16, MPI17, MPI18, MPI25, MPI26 and MPI27, respectively (Table 1). In term of selectivity index (CC50/antiviral EC50), MPI8, MPI16, MPI17, and MPI18 are around or above 200.
3. Discussion
There are 11 unique proteolytic sites in pp1a and pp1b that are hydrolyzed by MPro. Glutamine is a strictly required residue at the P1 site for all 11 sites. Due to this strict requirement and medicinal chemistry information learnt from the same function enzyme in SARS-CoV, most peptidyl inhibitors that have been developed for MPro have maintained a β-(S-2-oxopyrrolidin-3-yl)-alanine analog at the P1 site for improved potency. In our design, we keep opal at this site for all inhibitors. Our crystallography results show a well-shaped opal side chain in all MPro-inhibitor complexes that fits neatly in the P1 binding pocket of the enzyme. Since opal is optimized for this site, there is small chemical space to manipulate its side chain for improved binding. In all determined structures, the side chain of N142 adopts different conformations that do not necessarily interact with the opal oxopyrrolidine ring. It may be possible to modify the opal oxopyrrolidine ring to introduce interactions such as hydrogen bond(s) with the N142 side chain for improved binding. Among all 11 MPro-targeted proteolytic sites, 9 have a P2 leucine, 1 has a P2 phenylalanine, and 1 has a P2 valine. The enzyme has an apparent preference for a P2 leucine in its substrates. A comparison of in vitro enzyme inhibition potency for all MPIs that we have developed also reveals that MPro prefers a P2 residue with a similar size as leucine in a peptidyl inhibitor for favorable binding. Converting the isopropyl group at the β-carbon of the P2 residue to cyclopropyl and 2-furanyl improves the enzyme inhibition kinetics. Although changing the P2 leucine in an inhibitor to a larger residue such as phenylalanine and β-cyclohexylalanine diminishes in vitro MPro inhibition potency, the level is not significant. It is interesting to see that a β-isoprenyl group at the P2 residue has no fixed conformation but a cyclopropyl group at the same position has a defined conformation in crystal structures. Since inhibitors with a β-cyclopropyl group at the P2 residue have typically high in vitro enzyme inhibition potency, it is possible that adding an additional group such as methyl to the β-carbon of a P2 β-cyclopropyl-alanine will lead to better inhibitors. In the crystal structure of MPro-MPI14, the P2 side chain of MPI14 forms a π-π stacking interaction. This π-π stacking interaction likely contributes to the high in vitro enzymatic inhibition potency of MPI14. Although MPI15 is not soluble, it is expected to have high in vitro enzymatic inhibition potency as well due to a similar -π stacking interaction that can be formed between MPI15 and MPro. Other five-membered rings that contain one or more heteroatoms at the P2 β-carbon may also lead to high in vitro enzymatic inhibition potency since a similar π-π stacking interaction can be formed between them and MPro. In most of our determined MPro-inhibitor complexes, we noticed that the M49-containing loop of MPro has weak electron density, indicating that this loop adopts a flexible structure. Since this loop caps the MPro P2 binding pocket, this flexibility will potentially allow peptide inhibitors with a P2 residue larger than all moieties that have been tested in our inhibitors at this site to be tolerated by MPro. This probability needs to be explored.
MPro shows almost no preference toward the P3 residue in its targeted proteolytic sites in pp1a and pp1b. We explored a variety of chemical variants at this site including dialkyl glycines and 1-aminocyclopropane-1-carboxylate that have no α-proton. Although MPro displays a clear preference for a P3 valine, other substituents at this site are well tolerated. For most inhibitors, their P3 residues can be well refined. Given the diverse structures of tolerable chemical compositions at this site, d-amino acids might be introduced at this site for the development of novel MPro inhibitors as well. This will need to be explored further. One interesting observation is that α,α-disubstituted glycines at the P3 position push their adjacent N-terminal carbamate carbonyl group to adopt an orientation rotating about 90° away from a position that has been observed in MPro-inhibitor structures into the MPro active site. It will be interesting to see whether this observation can be applied to optimize existing MPro inhibitors including nirmatrelvir to achieve better antiviral potency. All our crystal structures show weak electron density around the N-terminal group indicating its weak binding to the enzyme. Adding an m-chloride to the N-terminal CBZ group also leads to significant decrease in in vitro enzyme inhibition potency. So far, our studies indicate that both CBZ and indole-2-carboxyl are not optimal chemical groups for interactions with MPro. Some smaller groups need to be explored at this site. One example will be trifluoroacetamide as seen in nirmatrelvir.
Although most inhibitors we have developed in this study have high in vitro enzyme inhibition potency and two inhibitors have IC50 values reaching the characterization limit, just a few inhibitors show cellular potency to inhibit MPro in 293T cells. Inhibitors that have a P3 O-tert-butyl-threonine all show strong cellular potency. All other P3 substituents lead to relatively weaker cellular potency, although MPI5, MPI18, and MPI27 showed mild cellular potency. Many factors may contribute to this phenomenon. Since all MPIs are peptidyl inhibitors, a small or native P3 residue might be prone to proteolytic degradation by host proteases leading to low cellular potency. This is partially supported by characterizable cellular potency for MPI18-19 that have a P3 β-tert-butyl-alanine. It is also likely that a P3 O-tert-butyl-threonine introduces more favorable cellular permeability than other P3 substituents into an MPro inhibitor. This aspect needs to be further investigated. MPI21-23 that have a P3 dialkylglycine are expected to be more resistant to proteolytic digestion by human proteases than other MPIs. Their low cellular potency might be due to low cellular permeability. Based on cellular potency of MPIs, we can also derive that an N-terminal CBZ group works better than the other two groups that have been tested at this site and a P2 β-cyclohexylalanine favors high cellular potency as well.
In terms of antiviral potency, results for six tested compounds agree well with their cellular potency, supporting the notion that the inhibition of MPro in infected cells is enough to achieve the prevention of viral replication. It should be noted that 293T cells were used in the cellular assay and Vero E6 cells were used in the antiviral assay. Different cell lines may have different enzymes which may contribute to different metabolism of inhibitors, leading to the discrepancy between cellular MPro inhibition potency and antiviral potency. Six tested inhibitors also exhibited relatively low cytotoxicity. With this level of selectivity, potential achievement of their use in containing SARS-CoV-2 in COVID-19 patients is possible. However, all inhibitors have an aldehyde functionality that is potentially metabolically unstable. Exploration of other warheads such as nitrile that was used in nirmatrelvir might potentially solve both metabolic and cytotoxicity concerns.
Author contributions
H. Ji, S. Xu and W. R. Liu conceived the project and directed the study; Y. Ma, Y. R. Alugubelli, X. R. Ma, and K. Khatua synthesized and characterized chemically all inhibitors; K. S. Yang, E. C. Vatansever and G. Yu characterized in vitro IC50 values of all synthesized inhibitors; Z. Z. Geng and C.-C. Cho characterized cellular EC50 values of all synthesized inhibitors; K. S. Yang, L. R. Blankenship, B. Sankaran, and P. Li determined all protein crystal structures; N. Shaabani and R. Allen characterized antiviral EC50 values for selected inhibitors; J. Xiao characterized CC50 for selected inhibitors; H. Ji, S. Xu and W. R. Liu wrote the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by Welch Foundation (grant A-1715), National Institutes of Health (Grants R21AI164088 and R21EB032983), TAMU COS Strategic Transformative Research Program, and Texas A&M X Grants. The ALS-ENABLE beam-lines are supported in part by the National Institutes of Health, National Institute of General Medical Sciences, grant P30 GM124169-01 and the Howard Hughes Medical Institute. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract No. DE-AC02-05CH11231.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2022.114570.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 2.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921 e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11(1):4417. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai W., Jochmans D., Xie H., Yang H., Li J., Su H., Chang D., Wang J., Peng J., Zhu L., Nian Y., Hilgenfeld R., Jiang H., Chen K., Zhang L., Xu Y., Neyts J., Liu H. Design, synthesis, and biological evaluation of peptidomimetic aldehydes as broad-spectrum inhibitors against enterovirus and SARS-CoV-2. J. Med. Chem. 2021 doi: 10.1021/acs.jmedchem.0c02258. [DOI] [PubMed] [Google Scholar]
- 9.Qiao J., Li Y.S., Zeng R., Liu F.L., Luo R.H., Huang C., Wang Y.F., Zhang J., Quan B., Shen C., Mao X., Liu X., Sun W., Yang W., Ni X., Wang K., Xu L., Duan Z.L., Zou Q.C., Zhang H.L., Qu W., Long Y.H., Li M.H., Yang R.C., Liu X., You J., Zhou Y., Yao R., Li W.P., Liu J.M., Chen P., Liu Y., Lin G.F., Yang X., Zou J., Li L., Hu Y., Lu G.W., Li W.M., Wei Y.Q., Zheng Y.T., Lei J., Yang S. SARS-CoV-2 M(pro) inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371(6536):1374–1378. doi: 10.1126/science.abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman R.L., Kania R.S., Brothers M.A., Davies J.F., Ferre R.A., Gajiwala K.S., He M., Hogan R.J., Kozminski K., Li L.Y., Lockner J.W., Lou J., Marra M.T., Mitchell L.J., Jr., Murray B.W., Nieman J.A., Noell S., Planken S.P., Rowe T., Ryan K., Smith G.J., 3rd, Solowiej J.E., Steppan C.M., Taggart B. Discovery of Ketone-based covalent inhibitors of coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020;63(21):12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M(pro) and cathepsin L. Sci. Adv. 2020;6(50) doi: 10.1126/sciadv.abe0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang K.S., Ma X.R., Ma Y., Alugubelli Y.R., Scott D.A., Vatansever E.C., Drelich A.K., Sankaran B., Geng Z.Z., Blankenship L.R., Ward H.E., Sheng Y.J., Hsu J.C., Kratch K.C., Zhao B., Hayatshahi H.S., Liu J., Li P., Fierke C.A., Tseng C.K., Xu S., Liu W.R. A quick route to multiple highly potent SARS-CoV-2 main protease inhibitors. ChemMedChem. 2021;16(6):942–948. doi: 10.1002/cmdc.202000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X.R., Alugubelli Y.R., Ma Y., Vatansever E.C., Scott D.A., Qiao Y., Yu G., Xu S., Liu W.R. MPI8 is potent against SARS-CoV-2 by inhibiting dually and selectively the SARS-CoV-2 main protease and the host cathepsin L. ChemMedChem. 2022;17(1) doi: 10.1002/cmdc.202100456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douangamath A., Fearon D., Gehrtz P., Krojer T., Lukacik P., Owen C.D., Resnick E., Strain-Damerell C., Aimon A., Abranyi-Balogh P., Brandao-Neto J., Carbery A., Davison G., Dias A., Downes T.D., Dunnett L., Fairhead M., Firth J.D., Jones S.P., Keeley A., Keseru G.M., Klein H.F., Martin M.P., Noble M.E.M., O'Brien P., Powell A., Reddi R.N., Skyner R., Snee M., Waring M.J., Wild C., London N., von Delft F., Walsh M.A. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11(1):5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghahremanpour M.M., Tirado-Rives J., Deshmukh M., Ippolito J.A., Zhang C.H., Cabeza de Vaca I., Liosi M.E., Anderson K.S., Jorgensen W.L. Identification of 14 known drugs as inhibitors of the main protease of SARS-CoV-2. ACS Med. Chem. Lett. 2020;11(12):2526–2533. doi: 10.1021/acsmedchemlett.0c00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 18.Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X., Duan Y., Yu J., Yang X., Yang X., Yang K., Liu X., Guddat L.W., Xiao G., Zhang L., Yang H., Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020;27(6):529–532. doi: 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 19.Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol Transl Sci. 2020;3(6):1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathnayake A.D., Zheng J., Kim Y., Perera K.D., Mackin S., Meyerholz D.K., Kashipathy M.M., Battaile K.P., Lovell S., Perlman S., Groutas W.C., Chang K.O. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice. Sci. Transl. Med. 2020;12(557) doi: 10.1126/scitranslmed.abc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu W., Xu M., Chen C.Z., Guo H., Shen M., Hu X., Shinn P., Klumpp-Thomas C., Michael S.G., Zheng W. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol Transl Sci. 2020;3(5):1008–1016. doi: 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunther S., Reinke P.Y.A., Fernandez-Garcia Y., Lieske J., Lane T.J., Ginn H.M., Koua F.H.M., Ehrt C., Ewert W., Oberthuer D., Yefanov O., Meier S., Lorenzen K., Krichel B., Kopicki J.D., Gelisio L., Brehm W., Dunkel I., Seychell B., Gieseler H., Norton-Baker B., Escudero-Perez B., Domaracky M., Saouane S., Tolstikova A., White T.A., Hanle A., Groessler M., Fleckenstein H., Trost F., Galchenkova M., Gevorkov Y., Li C., Awel S., Peck A., Barthelmess M., Schlunzen F., Lourdu Xavier P., Werner N., Andaleeb H., Ullah N., Falke S., Srinivasan V., Franca B.A., Schwinzer M., Brognaro H., Rogers C., Melo D., Zaitseva-Doyle J.J., Knoska J., Pena-Murillo G.E., Mashhour A.R., Hennicke V., Fischer P., Hakanpaa J., Meyer J., Gribbon P., Ellinger B., Kuzikov M., Wolf M., Beccari A.R., Bourenkov G., von Stetten D., Pompidor G., Bento I., Panneerselvam S., Karpics I., Schneider T.R., Garcia-Alai M.M., Niebling S., Gunther C., Schmidt C., Schubert R., Han H., Boger J., Monteiro D.C.F., Zhang L., Sun X., Pletzer-Zelgert J., Wollenhaupt J., Feiler C.G., Weiss M.S., Schulz E.C., Mehrabi P., Karnicar K., Usenik A., Loboda J., Tidow H., Chari A., Hilgenfeld R., Uetrecht C., Cox R., Zaliani A., Beck T., Rarey M., Gunther S., Turk D., Hinrichs W., Chapman H.N., Pearson A.R., Betzel C., Meents A. X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science. 2021;372(6542):642–646. doi: 10.1126/science.abf7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hattori S.I., Higashi-Kuwata N., Hayashi H., Allu S.R., Raghavaiah J., Bulut H., Das D., Anson B.J., Lendy E.K., Takamatsu Y., Takamune N., Kishimoto N., Murayama K., Hasegawa K., Li M., Davis D.A., Kodama E.N., Yarchoan R., Wlodawer A., Misumi S., Mesecar A.D., Ghosh A.K., Mitsuya H. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021;12(1):668. doi: 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo C.J., Chao T.L., Kao H.C., Tsai Y.M., Liu Y.K., Wang L.H., Hsieh M.C., Chang S.Y., Liang P.H. Kinetic characterization and inhibitor screening for the proteases leading to identification of drugs against SARS-CoV-2. Antimicrob. Agents Chemother. 2021;65(4):e02577–20. doi: 10.1128/AAC.02577-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockbaum G.J., Reyes A.C., Lee J.M., Tilvawala R., Nalivaika E.A., Ali A., Kurt Yilmaz N., Thompson P.R., Schiffer C.A. Crystal structure of SARS-CoV-2 main protease in complex with the non-covalent inhibitor ML188. Viruses. 2021;13(2) doi: 10.3390/v13020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaidman D., Gehrtz P., Filep M., Fearon D., Gabizon R., Douangamath A., Prilusky J., Duberstein S., Cohen G., Owen C.D., Resnick E., Strain-Damerell C., Lukacik P., Covid-Moonshot C., Barr H., Walsh M.A., von Delft F., London N. An automatic pipeline for the design of irreversible derivatives identifies a potent SARS-CoV-2 M(pro) inhibitor. Cell Chem Biol. 2021;28(12):1795–1806. doi: 10.1016/j.chembiol.2021.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C.H., Stone E.A., Deshmukh M., Ippolito J.A., Ghahremanpour M.M., Tirado-Rives J., Spasov K.A., Zhang S., Takeo Y., Kudalkar S.N., Liang Z., Isaacs F., Lindenbach B., Miller S.J., Anderson K.S., Jorgensen W.L. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free Energy perturbation calculations. ACS Cent. Sci. 2021;7(3):467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vatansever E.C., Yang K.S., Drelich A.K., Kratch K.C., Cho C.C., Kempaiah K.R., Hsu J.C., Mellott D.M., Xu S., Tseng C.K., Liu W.R. Bepridil is potent against SARS-CoV-2 in vitro. Proc. Natl. Acad. Sci. U. S. A. 2021;118(10) doi: 10.1073/pnas.2012201118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 30.Yang K.S., Leeuwon S.Z., Xu S., Liu W.R. Evolutionary and structural insights about potential SARS-CoV-2 evasion of nirmatrelvir. J. Med. Chem. 2022 doi: 10.1021/acs.jmedchem.2c00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao W., Cho C.-C.D., Geng Z.Z., Shaabani N., Ma X.R., Vatansever E.C., Alugubelli Y.R., Ma Y., Chaki S.P., Ellenburg W.H., Yang K.S., Qiao Y., Allen R., Neuman B.W., Ji H., Xu S., Liu W.R. Evaluation of SARS-CoV-2 main protease inhibitors using a novel cell-based assay. ACS Cent. Sci. 2022;8(2):192–204. doi: 10.1021/acscentsci.1c00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma C., Xia Z., Sacco M.D., Hu Y., Townsend J.A., Meng X., Choza J., Tan H., Jang J., Gongora M.V., Zhang X., Zhang F., Xiang Y., Marty M.T., Chen Y., Wang J. Discovery of di- and trihaloacetamides as covalent SARS-CoV-2 main protease inhibitors with high target specificity. J. Am. Chem. Soc. 2021;143(49):20697–20709. doi: 10.1021/jacs.1c08060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Resnick S.J., Iketani S., Hong S.J., Zask A., Liu H., Kim S., Melore S., Lin F.Y., Nair M.S., Huang Y., Lee S., Tay N.E.S., Rovis T., Yang H.W., Xing L., Stockwell B.R., Ho D.D., Chavez A. Inhibitors of coronavirus 3CL proteases protect cells from protease-mediated cytotoxicity. J. Virol. 2021;95(14) doi: 10.1128/JVI.02374-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iketani S., Forouhar F., Liu H., Hong S.J., Lin F.Y., Nair M.S., Zask A., Huang Y., Xing L., Stockwell B.R., Chavez A., Ho D.D. Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors. Nat. Commun. 2021;12:2016. doi: 10.1038/s41467-021-22362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng Y.W., Chao T.L., Li C.L., Chiu M.F., Kao H.C., Wang S.H., Pang Y.H., Lin C.H., Tsai Y.M., Lee W.H., Tao M.H., Ho T.C., Wu P.Y., Jang L.T., Chen P.J., Chang S.Y., Yeh S.H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2) doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao M.M., Yang W.L., Yang F.Y., Zhang L., Huang W.J., Hou W., Fan C.F., Jin R.H., Feng Y.M., Wang Y.C., Yang J.K. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Tar. 2021;6(1):134. doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alugubelli Y.R., Geng Z.Z., Yang K.S., Shaabani N., Khatua K., Ma X.R., Vatansever E.C., Cho C.-C., Ma Y., Blankenship L., Yu G., Sankaran B., Li P., Allen R., Ji H., Xu S., Liu W.R. The N-terminal carbamate is key to high cellular and antiviral potency for boceprevir-based SARS-CoV-2 main protease inhibitors. bioRxiv. 2021 [Google Scholar]
- 39.Kumar P., Nagarajan A., Uchil P.D. Analysis of cell viability by the MTT assay. Cold Spring Harb. Protoc. 2018;2018(6):469–471. doi: 10.1101/pdb.prot095505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.