Abstract
Background:
While strong evidence supports adverse maternal and offspring consequences of air pollution, mechanisms that involve the placenta, a key part of the intrauterine environment, are largely unknown. Previous studies of air pollution and placental gene expression were small candidate gene studies that rarely considered prenatal windows of exposure or the potential role of offspring sex. We examined overall and sex-specific associations of prenatal exposure to fine particulate matter (PM2.5) with genome-wide placental gene expression.
Methods:
Participants with placenta samples, collected at birth, and childhood health outcomes from CANDLE (Memphis, TN) (n = 776) and GAPPS (Seattle, WA) (n = 205) cohorts of the ECHO-PATHWAYS Consortium were included in this study. PM2.5 exposures during trimesters 1, 2, 3, and the first and last months of pregnancy, were estimated using a spatiotemporal model. Cohort-specific linear adjusted models were fit for each exposure window and expression of >11,000 protein coding genes from paired end RNA sequencing data. Models with interaction terms were used to examine PM2.5-offspring sex interactions. False discovery rate (FDR < 0.10) was used to correct for multiple testing.
Results:
Mean PM2.5 estimate was 10.5–10.7 μg/m3 for CANDLE and 6.0–6.3 μg/m3 for GAPPS participants. In CANDLE, expression of 13 (11 upregulated and 2 downregulated), 20 (11 upregulated and 9 downregulated) and 3 (2 upregulated and 1 downregulated) genes was associated with PM2.5 in the first trimester, second trimester, and first month, respectively. While we did not find any statistically significant association, overall, between PM2.5 and gene expression in GAPPS, we found offspring sex and first month PM2.5 interaction for DDHD1 expression (positive association among males and inverse association among females). We did not observe PM2.5 and offspring sex interactions in CANDLE.
Conclusion:
In CANDLE, but not GAPPS, we found that prenatal PM2.5 exposure during the first half of pregnancy is associated with placental gene expression.
Keywords: Air pollution, Fine particulate matter, PM2.5, Pregnancy, Placenta, Gene expression
1. Introduction
Strong evidence supports adverse maternal and offspring consequences of air pollution, a pervasive exposure (Klepac et al., 2018; Loftus et al., 2019; Chun et al., 2020; Ni et al., 2021). Recent epidemiologic evidence suggests that prenatal and early-life ambient air pollution exposure, particularly exposure to particulate matter, is associated with pregnancy complications (e.g., preterm birth), pregnancy outcomes (e.g., low birth weight), as well as childhood morbidity (e.g., decreased cognitive function, autism, higher blood pressure and asthma) (Klepac et al., 2018; Loftus et al., 2019; Chun et al., 2020; Ni et al., 2021; Hazlehurst et al., 2021). Mechanisms that explain how air pollution affects mother and offspring, particularly mechanisms that involve the placenta, are largely unknown. The placenta regulates the intrauterine environment that supports the fetus at a time of its highest adaptability and vulnerability (Gude et al., 2004; Barker, 1995). The effect of fetal adaptive responses (growth and programming) to placenta mediated environmental exposures persists throughout life and contributes to childhood and adulthood chronic diseases (Gude et al., 2004; Gluckman et al., 2008; Kuzawa and Quinn, 2009; Bateson et al., 2004).
Evaluating placental effect biomarkers in relation to air pollution will help identify mechanisms by which air pollution could affect development of pregnancy complications or fetal growth/programming. Prior investigations have reported several biomarkers of placental origin related to maternal air pollution exposure (including PM10, black carbon, and PM2.5) (Luyten et al., 2018; Maghbooli et al., 2018; Soto et al., 2017; Tsamou et al., 2018; de Melo et al., 2015). These biomarkers (e.g. DNA adduct and DNA methylation) include signatures of DNA damage as well as genomic, epigenomic, proteomic, metabolomics and exposomic changes that occur in placenta following exposure to ambient air pollution and are closely related to placental function (Luyten et al., 2018). While investigations of placental gene expression can further our understanding of placental functions that are affected by air pollution and related gene-environment interactions, few epidemiological and basic science studies have been conducted on PM2.5 exposure and placental gene expression (Luyten et al., 2018; Saenen et al., 2015; Whyatt et al., 1995; Kingsley et al., 2017; Kim et al., 2018; Deyssenroth et al., 2021). Findings from these studies indicated that exposure to PM2.5 particles, which are small and can get deep into the lung and bloodstream, is associated with changes in placental expression of brain-derived neurotrophic factor (BDNF) (Saenen et al., 2015), synapsin 1 (SYN1) (Saenen et al., 2015), cytochrome P450 1A1 (CYP1A1) (Whyatt et al., 1995), imprinted genes (Kingsley et al., 2017), and genes related to amino acid transport and cellular respiration (Deyssenroth et al., 2021). Limitations of research in this area include inconsistent findings, small sample size, lack of sample diversity, evaluation of limited candidate genes, and lack of examination of multiple windows of exposure during the pregnancy period. While sexual dimorphism in placental gene expression due to normal sex-dependent structural and functional placental differences (including sex chromosomes and epi-mutations) and interactions between fetal sex and the in utero environment shaped by maternal characteristics or exposure (including diet, stress, and air pollution) (Ilekis et al., 2016; Sood et al., 2006; Rosenfeld, 2015; Ghosh et al., 2007; Gonzalez et al., 2018; Buckberry et al., 2014) has been well documented, only one recent study examined genome-wide placental expression and potential effect modification by offspring sex (Kingsley et al., 2017).
The objective of the current study was to address these limitations and determine overall and sex-specific associations of prenatal PM2.5 exposure with placental gene expression.
2. Methods
2.1. Study setting and study participants
The study was conducted as a part of the ECHO-PATHWAYS Consortium. The study population comprised participants of the Conditions Affecting Neurocognitive Development in Early Childhood (CANDLE) and Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) studies. The CANDLE study (N = 1,503), described previously (Sontag-Padilla et al., 2015), is a prospective pregnancy cohort study from the urban South, with participant enrollment between 2006 and 2011. Briefly, women were considered eligible if they were Shelby County, TN residents (majority of which resided in the Memphis metro area), between 16 and 40 years of age, had singleton pregnancies without complications at enrollment, and planned to deliver at a participating study hospital. All women participants of the CANDLE study provided informed consent upon enrollment and research activities were approved by the University of Tennessee Health Sciences Center IRB.
GAPPS was launched by the Seattle Children’s Hospital in 2007 to focus attention and research on reducing the impact of adverse birth outcomes. In 2012, GAPPS initiated a pregnancy biorepository at four medical centers: University of Washington (Seattle, WA), Swedish Medical Center (Seattle, WA), Yakima Valley Memorial Hospital (Yakima, WA), and Loma Linda University Children’s Hospital (Loma Linda, CA). This biorepository served as the basis for the current study. Women were approached by study staff at prenatal checkups or followed up with via phone/email for participation in the study. Eligible participants were >18 years of age or medically emancipated, and confirmed to be pregnant by self-test or by physician’s medical testing. Participants were ineligible for any of the following reasons: unable to provide informed consent, greater than or equal to 37 weeks of gestation, in active labor at the time of recruitment, or received narcotic administration in the 24 h prior to consent. In 2017, eligible participants (N = 1,271) were re-contacted to participate in the ECHO-PATHWAYS study, and recruitment is still ongoing. Eligibility criteria included delivery in Seattle, WA (Swedish Medical Center) or Yakima, WA (Yakima Valley Memorial Hospital), availability of at least one pregnancy urine sample, initial GAPPS enrollment and completion of questionnaire, and GAPPS child currently 4–7 years of age. Study protocols were approved by the Seattle Children’s Hospital and University of Washington Institutional Review Boards.
For the current study, among participants who provided placenta specimen, we excluded participants with stillbirth, participants without address data, and multiple births. Additional exclusion criteria included infants born after severe pregnancy complications, including confirmed clinical chorioamnionitis, oligohydramnios, placental abruption, placental infarction, placenta previa, and fetal chromosomal abnormalities. After these exclusions, participants with placenta samples and childhood health outcomes from the CANDLE (n = 776) and GAPPS (n = 205) cohorts were included in this study.
2.2. Ambient air pollution (PM2.5)
Ambient air pollution was characterized using PM2.5 (μg/m3) estimated using the spatiotemporal models (predicting point-based estimates on a two-week time scale) developed at the University of Washington (Keller et al., 2015; Kirwa et al., 2021). The models utilized monitoring data from regulatory networks supplemented with PM2.5 measurements from intensive research cohort-specific monitors. The model decomposed the space-time field of concentrations into spatially varying long-term averages, spatially varying seasonal and long-term trends, and spatially correlated but temporally independent residuals. Time trends were estimated from observed time series, and spatial smoothing by universal kriging was used to borrow strength between observations (see reference #22 for detailed description of the model). We evaluated different windows of exposure: Trimesters (Trimester 1, 2, and 3), first month of pregnancy (time surrounding implantation and early placental growth), and the last month before delivery. Trimesters were chosen because they correspond to well described developmental milestones. The first month of pregnancy is a time surrounding implantation and early placental growth. The last month before delivery is a time of accelerated fetal growth and placental function that is proximal to delivery (when samples were collected). Notably, previous studies used these exposure windows allowing comparison of findings across studies (Luyten et al., 2018; Maghbooli et al., 2018; Soto et al., 2017; Tsamou et al., 2018; de Melo et al., 2015).
2.3. Placental sample collection
Placental sample collection in the CANDLE study, described previously (Paquette et al., 2021), was as follows. Within 15 min of delivery, a piece of placental villous tissue in the shape of a rectangular prism approximately 2 cm in length, 0.5 cm width and 0.5 cm depth was dissected from the placental parenchyma and cut into four approximately 0.5 cm cubes. The tissue cubes were placed in a 50 ml tube with 20 ml RNAlater and refrigerated at 4 °C overnight (at least 8 h but no >24 h). Each tissue cube was transferred to an individual 1.8 ml cryovial containing fresh RNAlater. The cryovials were stored at −80 °C until the fetal villous tissue was manually dissected, and cleared of maternal decidua. Following dissection, the samples were put into RNAlater and stored at −80 °C.
In the GAPPS study, within 30 min of delivery, 8 mm full-thickness vertical tissue punches from the placental disc were taken and put into 5 ml tubes containing approximately 3 ml of RNAlater and stored at −20 °C before specimens were shipped to the GAPPS facility. Samples were then stored at −0 °C. Punches were thawed and inspected for identifiable fetal-side membranes and maternal sides. The fetal-side of the placental punch was cut-off from the rest of the punch, and divided into 1–3 pieces with mass ranging from 10 mg to 30 mg. Each sample was placed in 1 ml RNAlater and stored at −20 °C until shipped for further processing.
2.4. Sample processing and RNA sequencing
Approximately 30 mg of placental tissue was used for RNA isolation. Placental tissue that was stored in RNAlater at −80 °C was allowed to gradually warm to room temperature. Tissue was removed from tube and dabbed on a Kimwipe to remove excess RNAlater and then placed in tube with 600 μl Buffer RLT Plus containing mercaptoethanol along with 5 mm ball bearings. Tissue was homogenized using a TissueLyser LT instrument (Qiagen, Germantown, MD). RNA was isolated using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Germantown, MD) according to the manufacturer’s recommended protocol. RNA purity was assessed by measuring OD260/230 and OD260/260 ratios with a NanoDrop 8000 Spectrophotometer (Thermo Fischer Scientific, Waltham MA). RNA integrity was determined with a Bioanalyzer 2100 using RNA 6000 Nanochips (Agilent, Santa Clara, CA). Only RNA samples with RNA Integrity Number (RIN) > 7 were used in the RNA-Seq analysis.
All RNA sequencing was performed at the University of Washington Northwest Genomics Center. Total RNA was poly-A enriched to remove ribosomal RNA, and cDNA libraries were prepared from 1 μg of total RNA using the TruSeq Stranded mRNA kit (Illumina, San Diego, CA) and the Sciclone NGSx Workstation (Perkin Elmer, Waltham, MA). Each library was uniquely barcoded and subsequently amplified using a total of 13 cycles of PCR. Library concentrations were quantified using Qubit Quant-it dsDNA High Sensitivity assay fluorometric quantitation (Life Technologies, Carlsbad, CA). Average fragment size and overall quality were evaluated with the DNA1000 assay on an Agilent 2100 Bioanalyzer. Each library was sequenced to an approximate depth of 30 million reads on an Illumina HiSeq sequencer. RNA sequencing quality control was performed using both the FASTX-toolkit (v0.0.13) and FastQC (v0.11.2) (Brown et al., 20172017).
Transcript abundances were estimated by aligning to the GRCh38 transcriptome (Gencode v33) using the quantification program Kallisto (Bray et al., 2016) and condensed to the gene level using the Bioconductor tximport package, scaling to the average transcript length (Soneson et al., 2015). We performed filtering to remove all genes with an average log counts/million counts (log CPM) < 0 and estimated library normalization factors using the Trimmed Mean of M – values (TMM) function from the Bioconductor edgeR package (Robinson and Oshlack, 2010). We analyzed these data using the limma-voom pipeline (Law et al., 2014) from the Bioconductor limma package, which converts the counts/gene to logCPM using the TMM normalization factors and estimates observation-level weights based on the mean-variance relationship. We then fit conventional weighted linear regression models where the observation-level weights adjust for the mean-variance relationship. We included only protein coding genes (>11,000) in the current analyses.
2.5. Other variables
Extensive data was collected in CANDLE and GAPPS using interviewer-administered questionnaires and medical record abstraction. All models were adjusted for potential confounders and precision variables (e.g. experimental variables) including RNA sequencing batch, site (for GAPPS only; Swedish Medical Center in Seattle, WA or Yakima Valley Memorial Hospital in Yakima, WA), maternal age (continuous), race (Black/Others), offspring sex (male/female), season of delivery, calendar year of birth, gestational age (continuous), mode of delivery (cesarean section/vaginal), labor (presence/absence), smoking history (yes/no for current smoking during pregnancy), maternal urine cotinine (binary variable using 200 ng/mL cut off (Schick et al., 2017), maternal education (high school or less/college and above), and pre-pregnancy body mass index (continuous). Offspring sex was examined as a potential modifier of associations.
2.6. Statistical analyses
Cohort-specific descriptive statistics were obtained to understand the characteristics of the study population. We assessed similarities of characteristics of participants included in this analysis with characteristics of the overall respective cohorts (CANDLE and GAPPS). All analyses were cohort-specific. Exposure to PM2.5 during the different windows of pregnancy was estimated as described above. The outcome was genome-wide placental gene expression (only protein coding genes) from sequencing experiments (described above). Differentially expressed mRNAs were identified using conventional weighted linear models with observation-level weights estimated using the limma-voom method based on the relationship between mean and variance for the logCPM values. Significance of the PM2.5 slope parameter was assessed using empirical Bayes adjusted t-statistics (Soneson et al., 2015). Models were fit for each exposure window and individual gene expression, and included all adjustment variables. Complete sample sizes for regression models ranged from 762 to 763 for CANDLE cohort analyses and 164–168 for GAPPS cohort analyses. We selected genes with significantly different expression based on a false discovery rate (FDR) < 0.10 cutoff (Benjamini and Hochberg, 1995). We fitted both overall models as well as models which contain PM2.5-offspring sex interaction terms. FDR < 0.10 of interaction terms were used to determine statistical significance of multiplicative PM2.5-offspring sex interactions.
3. Results
Average maternal ages of participants were 27.3 years (5.5 years standard deviation) and 30.5 years (5.7 years standard deviation) in the CANDLE and GAPPS cohorts, respectively (Table 1). Mean gestational age at birth was 39.0 weeks for CANDLE participants and 38.3 weeks for GAPPS participants. About 56% of CANDLE participants and 2% of GAPPS participants self-identified as Black. Overall, characteristics of the analytic population were similar to characteristics of participants in the respective cohorts (Supplemental Table 1). The range of mean PM2.5 exposure for CANDLE study participants during the different exposure windows was 10.5–10.7 μg/m3 while it was 6.0–6.3 μg/m3 for GAPPS study participants (Table 2).
Table 1.
Selected characteristics of the study population.
| Population Covariates | CANDLE (n = 776) | GAPPS (n = 205) |
|---|---|---|
| Continuous | Mean (SD) | Mean (SD) |
| Maternal Age, years | 27.3 (5.5)* | 30.5 (5.7) * |
| Gestational Age at Birth, weeks | 39.0 (1.5) | 38.3 (3.0) |
| Pre-Pregnancy Body Mass Index, kg/m2 | 27.7 (7.4)* | 26.3 (9.1) * |
| Categorical | N (%) | N (%) |
| Maternal Race/Ethnicity | ||
| White | 292 (37.6) | 164 (80.0) |
| Black/African American | 438 (56.4) | 4 (2.0) |
| Asian | 6 (0.8) | 8 (3.9) |
| Native Hawaiian Pacific Islander | 0 (0) | 0 (0) |
| American Indian/Alaska Native | 0 (0) | 2 (1.0) |
| Other | 37 (4.8) | 13 (6.3) |
| Multiple Race | 3 (0.4) | 3 (1.5) |
| Missing | 11 (5.4) | |
| Offspring Sex | ||
| Male | 381 (49.1) | 108 (52.7) |
| Female | 395 (50.9) | 97 (47.3) |
| Season of Birth | ||
| Spring | 166 (21.4) | 53 (25.9) |
| Summer | 217 (28.0) | 50 (24.4) |
| Fall | 216 (27.8) | 52 (25.4) |
| Winter | 170 (21.9) | 50 (24.4) |
| Missing | 7 (0.9) | |
| Year of Birth | ||
| 2007 | 54 (7.0) | 0 (0) |
| 2008 | 116 (14.9) | 0 (0) |
| 2009 | 209 (26.9) | 0 (0) |
| 2010 | 229 (29.5) | 0 (0) |
| 2011 | 161 (20.7) | 40 (19.5) |
| 2012 | 0 (0) | 66 (32.2) |
| 2013 | 0 (0) | 83 (40.5) |
| 2014 | 0 (0) | 16 (7.8) |
| Missing | 7 (0.9) | |
| Mode of Delivery | ||
| Normal/Vaginal | 470 (60.6) | 114 (55.6) |
| C-section | 306 (39.4) | 71 (34.6) |
| Missing | 20 (9.8) | |
| Presence/Absence of Labor | ||
| Spontaneous, spontaneous augmented, or induced labor | 627 (80.8) | 112 (54.6) |
| No labor | 148 (19.1) | 16 (7.8) |
| Missing | 1 (0.1) | 77 (37.6) |
| Maternal Smoking | ||
| Yes/Cotinine positive (>200 ng/mL) | 56 (7.2) | 0 (0) |
| No/Cotinine negative (<200 ng/mL) | 720 (92.8) | 208 (100) |
| Maternal Education | ||
| Less than high school | 66 (8.5) | 10 (4.9) |
| High school completion | 346 (44.6) | 57 (27.8) |
| Graduated college or technical school | 260 (33.5) | 85 (41.5) |
| Some graduate work or professional degree | 104 (13.4) | 52 (25.4) |
| Missing | 1 (0.5) |
Mean and standard deviations calculated with missing values removed (10 and 3 subjects for maternal age and 2 and 5 for pre-pregnancy BMI in CANDLE and GAPPS respectively.
Table 2.
Distribution of Particulate Matter (PM) 2.5 Exposure in the Study Population.
| Distribution | |||||||
|---|---|---|---|---|---|---|---|
| Exposure Variable | Mean | SD | Min | 25% | Median | 75% | Max |
| CANDLE | |||||||
| PM2.5 – 1st Trimester (μg/ m3); N = 771 | 10.5 | 1.4 | 7.9 | 9.5 | 10.3 | 11.0 | 16.8 |
| PM2.5 – 2nd Trimester (μg/m3); N = 771 | 10.6 | 1.4 | 7.9 | 9.7 | 10.4 | 11.2 | 16.6 |
| PM2.5 – 3rd Trimester (μg/ m3); N = 771 | 10.7 | 1.7 | 7.5 | 9.6 | 10.6 | 11.5 | 17.2 |
| PM2.5 – 1st Month (μg/m3); N = 772 | 10.5 | 2.1 | 5.8 | 9.4 | 10.3 | 11.3 | 19.0 |
| PM2.5 – Last Month (μg/m3); N = 771 | 10.7 | 2.1 | 5.5 | 9.5 | 10.5 | 11.7 | 19.2 |
| GAPPS | |||||||
| PM2.5 – 1st Trimester (μg/ m3); N = 184 | 6.2 | 3.0 | 1.8 | 4.2 | 5.4 | 7.3 | 21.3 |
| PM2.5 – 2nd Trimester (μg/ m3); N = 186 | 6.0 | 3.0 | 2.1 | 3.8 | 5.1 | 7.2 | 16.8 |
| PM2.5 – 3rd Trimester (μg/m3); N = 183 | 6.0 | 3.1 | 2.3 | 3.9 | 5.2 | 7.1 | 20.3 |
| PM2.5 – 1st Month (μg/m3); N = 185 | 6.3 | 3.6 | 1.8 | 3.9 | 5.5 | 7.4 | 22.8 |
| PM2.5 – Last Month (μg/m3); N = 184 | 6.0 | 3.5 | 2.1 | 3.8 | 5.1 | 6.8 | 23.3 |
In the CANDLE study, expression of 13 (11 upregulated and 2 downregulated), 20 (11 upregulated and 9 downregulated), and three genes (2 upregulated and 1 downregulated) was associated with PM2.5 exposure in the first trimester, second trimester, and first month, respectively (FDR < 0.10) (Table 3). The slope parameter for upregulated genes ranged from 1.078 (for RGPD5) to 1.017 (for CLDND1 and THOC7) while the slope parameter for the downregulated genes ranged from 0.988 (for CAPZB and H3-3A) to 0.965 (for SLC15A3 and MT-CYB). These differentially expressed genes included SRSF11 (serine and arginine rich splicing factor 11, up regulated in first and second trimesters), SLC15A3 (solute carrier family 15 member 3, down regulated in first trimester and first month), ANKRD49 (ankyrin repeat domain 49, up regulated in first trimester), ANKRD10 (ankyrin repeat domain 10, up regulated in second trimester), EIF3D (eukaryotic translation initiation factor 3 subunit D, down regulated in second trimester), GABRE (gamma-aminobutyric acid type A receptor epsilon subunit syntaxin 16, up regulated in second trimester), MT-CYB (mitochondrially encoded cytochrome b, down regulated in second trimester), and MT-ND3 (mitochondrially encoded NADH, down regulated in second trimester). We did not observe PM2.5 exposure and offspring sex interactions on placental gene expression in the CANDLE study.
Table 3.
Top Genes for PM2.5 Exposure in CANDLE.
| GAPPS Comparison* | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| ENSEMBL ID | Gene Symbol* | Description | logFC | Fold Change | P-value | FDR P- value | LogFC | P- value | FDR P-value |
| First Trimester (CANDLE) – N = 763 | |||||||||
| ENSG00000168876 | ANKRD49 | ankyrin repeat domain 49 | 0.038 | 1.027 | < 0.001 | 0.039 | −0.015 | 0.168 | 1.000 |
| ENSG00000015568 | RGPD5 | RANBP2 like and GRIP domain containing 5 | 0.108 | 1.078 | < 0.001 | 0.059 | 0.111 | 0.002 | 1.000 |
| ENSG00000080822 | CLDND1 | claudin domain containing 1 | 0.024 | 1.017 | < 0.001 | 0.059 | 0.017 | 0.034 | 1.000 |
| ENSG00000083828 | ZNF586 | zinc finger protein 586 | 0.039 | 1.027 | < 0.001 | 0.059 | −0.021 | 0.103 | 1.000 |
| ENSG00000101161 | PRPF6 | pre-mRNA processing factor 6 | −0.024 | 0.984 | < 0.001 | 0.059 | −0.013 | 0.090 | 1.000 |
| ENSG00000106404 | CLDN15 | claudin 15 | 0.062 | 1.044 | < 0.001 | 0.059 | −0.005 | 0.845 | 1.000 |
| ENSG00000110446 | SLC15A3 | solute carrier family 15 member 3 | −0.051 | 0.965 | < 0.001 | 0.059 | 0.000 | 0.984 | 1.000 |
| ENSG00000116754 | SRSF11 | serine and arginine rich splicing factor 11 | 0.029 | 1.020 | < 0.001 | 0.059 | 0.009 | 0.351 | 1.000 |
| ENSG00000122481 | RWDD3 | RWD domain containing 3 | 0.044 | 1.031 | < 0.001 | 0.059 | −0.015 | 0.269 | 1.000 |
| ENSG00000134744 | TUT4 | terminal uridylyl transferase 4 | 0.028 | 1.020 | < 0.001 | 0.059 | −0.004 | 0.675 | 1.000 |
| ENSG00000144026 | ZNF514 | zinc finger protein 514 | 0.054 | 1.038 | < 0.001 | 0.059 | −0.010 | 0.617 | 1.000 |
| ENSG00000180098 | TRNAU1AP | tRNA selenocysteine 1 associated protein 1 | 0.032 | 1.022 | < 0.001 | 0.068 | −0.003 | 0.767 | 1.000 |
| ENSG00000221944 | TIGD1 | tigger transposable element derived 1 | 0.043 | 1.030 | < 0.001 | 0.098 | −0.032 | 0.084 | 1.000 |
| Second Trimester (CANDLE) – N = 763 | |||||||||
| ENSG00000116754 | SRSF11 | serine and arginine rich splicing factor 11 | 0.038 | 1.027 | < 0.001 | 0.004 | −0.021 | 0.060 | 0.774 |
| ENSG00000088448 | ANKRD10 | ankyrin repeat domain 10 | 0.084 | 1.060 | < 0.001 | 0.010 | −0.023 | 0.288 | 0.860 |
| ENSG00000100353 | EIF3D | eukaryotic translation initiation factor 3 subunit D | −0.023 | 0.984 | < 0.001 | 0.010 | 0.000 | 0.946 | 0.992 |
| ENSG00000102287 | GABRE | gamma-aminobutyric acid type A receptor epsilon subunit | 0.115 | 1.083 | < 0.001 | 0.010 | −0.088 | 0.081 | 0.775 |
| ENSG00000124222 | STX16 | syntaxin 16 | 0.034 | 1.024 | < 0.001 | 0.010 | −0.006 | 0.597 | 0.928 |
| ENSG00000169217 | CD2BP2 | CD2 cytoplasmic tail binding protein 2 | −0.031 | 0.979 | < 0.001 | 0.010 | −0.005 | 0.571 | 0.923 |
| ENSG00000184047 | DIABLO | diablo IAP-binding mitochondrial protein | −0.027 | 0.981 | < 0.001 | 0.018 | −0.005 | 0.476 | 0.902 |
| ENSG00000163945 | UVSSA | UV stimulated scaffold protein A | 0.066 | 1.047 | < 0.001 | 0.026 | −0.030 | 0.241 | 0.844 |
| ENSG00000122026 | RPL21 | ribosomal protein L21 | −0.032 | 0.978 | < 0.001 | 0.029 | 0.008 | 0.529 | 0.921 |
| ENSG00000066923 | STAG3 | stromal antigen 3 | 0.070 | 1.050 | < 0.001 | 0.068 | −0.023 | 0.322 | 0.868 |
| ENSG00000109046 | WSB1 | WD repeat and SOCS box containing 1 | 0.036 | 1.025 | < 0.001 | 0.070 | −0.022 | 0.166 | 0.832 |
| ENSG00000179010 | MRFAP1 | Morf4 family associated protein 1 | −0.019 | 0.987 | < 0.001 | 0.070 | 0.000 | 0.977 | 0.999 |
| ENSG00000198727 | MT-CYB | mitochondrially encoded cytochrome b | −0.052 | 0.965 | < 0.001 | 0.070 | −0.003 | 0.903 | 0.983 |
| ENSG00000198840 | MT-ND3 | mitochondrially encoded NADH; ubiquinone oxidoreductase core subunit 3 | −0.048 | 0.967 | < 0.001 | 0.070 | −0.029 | 0.205 | 0.833 |
| ENSG00000167766 | ZNF83 | zinc finger protein 83 | 0.058 | 1.041 | < 0.001 | 0.076 | 0.015 | 0.072 | 0.858 |
| ENSG00000077549 | CAPZB | capping actin protein of muscle Z-line subunit beta | −0.018 | 0.988 | < 0.001 | 0.093 | 0.006 | 0.331 | 0.880 |
| ENSG00000163041 | H3–3A | H3.3 histone A | −0.017 | 0.988 | < 0.001 | 0.093 | 0.007 | 0.346 | 0.880 |
| ENSG00000167978 | SRRM2 | serine/arginine repetitive matrix 2 | 0.038 | 1.027 | < 0.001 |
0.094 | −0.006 | 0.826 | 0.970 |
| ENSG00000059145 | UNKL | unk like zinc finger | 0.051 | 1.036 | < 0.001 | 0.096 | −0.030 | 0.152 | 0.830 |
| ENSG00000203880 | PCMTD2 | protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 | 0.039 | 1.027 | < 0.001 | 0.098 | −0.031 | 0.049 | 0.767 |
| First Month (CANDLE) – N = 763 | |||||||||
| ENSG00000024862 | CCDC28A | coiled-coil domain containing 28A | 0.019 | 1.013 | < 0.001 | 0.061 | −0.003 | 0.217 | 0.975 |
| ENSG00000110446 | SLC15A3 | solute carrier family 15 member 3 | −0.038 | 0.974 | < 0.001 | 0.061 | 0.004 | 0.689 | 0.960 |
| ENSG00000163634 | THOC7 | THO complex 7 | 0.025 | 1.017 | <0.001 | 0.061 | 0.014 | 0.103 | 0.870 |
Gene symbols and descriptions from HGNC database
Only gene hits with FDR < 0.10 in the CANDLE cohort are shown in this table.
We did not find any association between PM2.5 exposure during any of the exposure windows we considered and placental gene expression in the GAPPS study. However, we identified a potential offspring sex and first month PM2.5 exposure interaction (FDR < 0.10) for DDHD1 (DDHD domain containing 1), an autosomal (Chr14) gene member of the intracellular phospholipase A1 gene family (Fig. 1). Associations between PM2.5 exposure and DDHD1 expression was positive among male infants while it was inverse among female infants. In the CANDLE cohort, associations for DDHD1 were inverse in both male (Log FC = −0.001) and female (Log FC = −0.008) infants and statistically insignificant (unadjusted p-values 0.943 and 0.500, and, FDR 0.99 and 0.91, respectively).
Fig. 1.
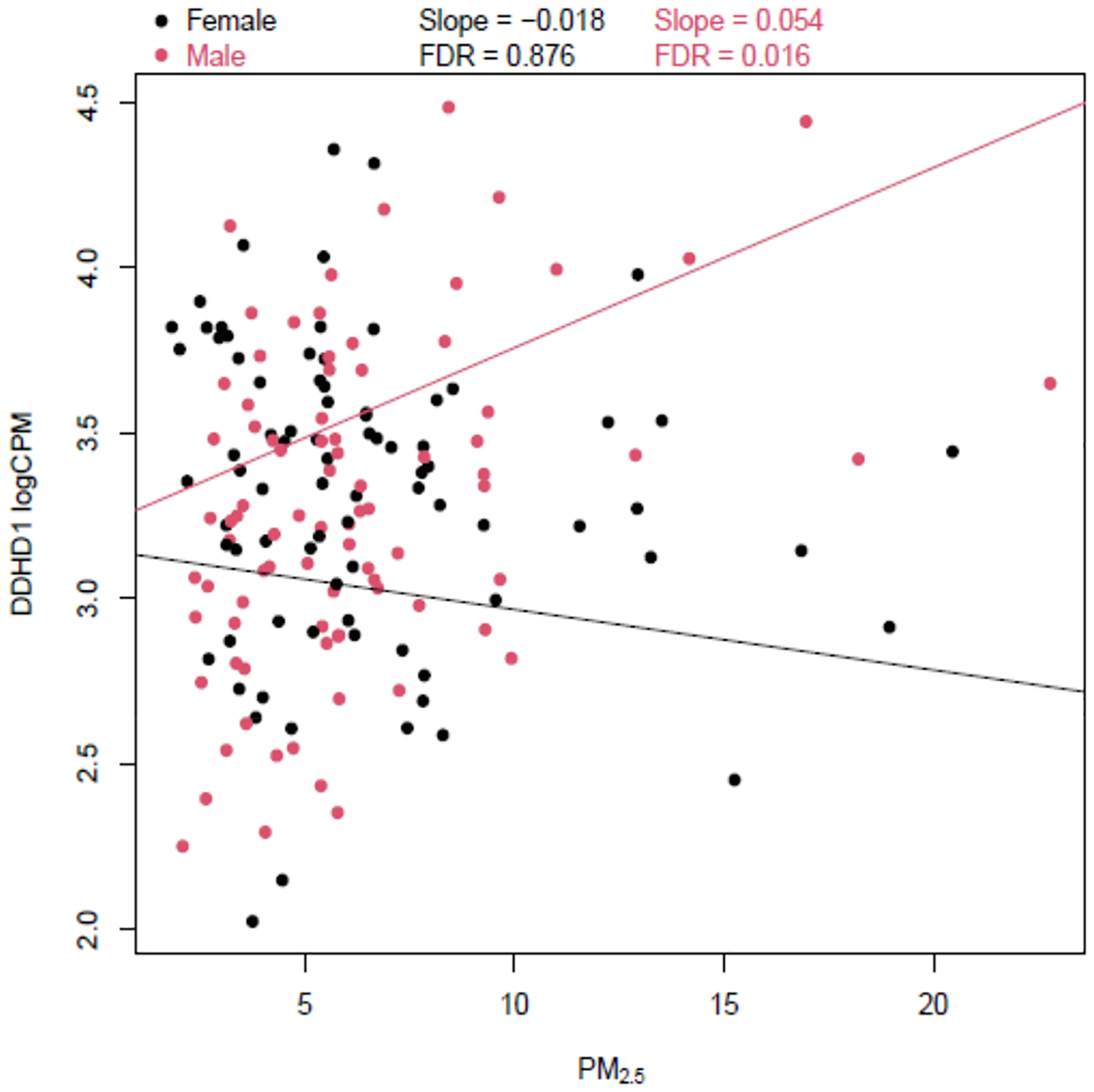
Offspring Sex and First Month PM2.5 Exposure Interaction on DDHD1 Gene Expression in the GAPPS Study.
The top genes for all PM2.5 exposure in the CANDLE and GAPPS studies, rank ordered by p-value are shown in Supplemental Tables 2 and 3. While we did not find differentially expressed genes in the GAPPS study, for a number of the differentially expressed genes in the CANDLE study, similar relationships (though not statistically significant) were observed in the GAPPS study (Table 3). For instance, similar non-statistically significant relationships (trends) were observed in the GAPPS study for first trimester PM2.5 exposure and four (RGPD5, CLDND1, PRPF6, and SRSF11) genes that were differentially expressed in the CANDLE study. In addition, similar, non-statistically significant relationships were observed in the GAPPS study for second trimester PM2.5 exposure and four (CD2BP2, DIABLO, MT-CYB, and MT-ND3) differentially expressed genes in the CANDLE study.
4. Discussion
In this study, conducted among multiple cohorts, we identified differentially expressed genes in placenta at birth that are associated with PM2.5 exposure during the first and second trimesters of pregnancy as well as the first month of pregnancy in the CANDLE cohort. These genes are involved in mRNA splicing (SRSF11), GABAergic signaling (GABRE), transport (SLC15A3), and mitochondrial function (MT-CYB and MT-ND3). Offspring sex and first month PM2.5 exposure interaction on placental expression of DDHD1 was observed in the GAPPS study where associations were positive among males and inverse among females.
Previous studies have examined associations of ambient air pollution, including PM2.5, with placental gene expression. In a study conducted among 70 mother-infant pairs in Krakow, Poland, Whyatt et al. reported that placental CYP1A1 mRNA levels were 3.7-fold higher in placentas from women residing in high compared to low ambient air pollution areas (characterized by PM10) (p = 0.06; Student’s test) (Whyatt et al., 1995). Saenen et al. investigated PM2.5 and brain-derived neurotrophic factor 10 signaling pathway genes involved in neural development, in placenta collected from 90 mother-infant pairs of the ENVIRONAGE birth cohort (Saenen et al., 2015). They found that PM2.5 exposure in the first trimester of pregnancy (particularly during the first month of pregnancy and during the early implantation period) was associated with a 15.9% decrease (p = 0.015) in placental BDNF expression and a 24.3% decrease (p = 0.011) in placental synapsin (SYN1) expression, corresponding to a 5-μg/m3 increase in PM2.5 (Saenen et al., 2015). In the current study, we observed reduction in expression of BDNF in the GAPPS cohort (1.4% decrease per unit PM2.5, p-value = 0.392) but not the CANDLE cohort (1.8% per unit PM2.5, p-value = 0.244), although both associations were not statistically significant. Saenen et al. found a 13.3% reduction in SOS2 expression corresponding to a 5 μg/m3 increase in PM2.5 (p = 0.017) (Saenen et al., 2015). Contrary to this finding, in our CANDLE and GAPPS cohorts, we saw 1.5% and 0.4% statistically insignificant increases in expression of SOS2, per unit PM2.5 (p-values = 0.690 and 0.244 respectively). Kingsley et al. investigated maternal residential air pollution and placental expression of nine candidate birthweight-related and 108 imprinted genes (Kingsley et al., 2017). Among women-infant pairs (N = 410) enrolled in the Rhode Island Child Health Study, they found that PM2.5 was associated with expression of seven birthweight-related (BLCAP, H19, IGF2, MEG3, MEST, PLAGL1, and NNAT) and 41 imprinted (including SLC22A18AS and ANKRD11) genes (Kingsley et al., 2017). In the CANDLE cohort, similar direction of non-significant (p-value > 0.05) associations was observed for IGF2, MEG3, and PLAGL1, increased expression with increased exposure to PM2.5. The direction of non-significant association for NNAT in the CANDLE cohort was different from what was reported before (positive vs. inverse association, respectively). In the GAPPS cohort, directions of non-significant associations were similar to previous reports for IGF2 and NNAT, but not MEST and PLAGL1. In the CANDLE cohort, we identified associations between exposure of PM2.5 in the first trimester and placental expression of ANKRD49 and SLC15A3, genes in the same families as those identified by Kingsley et al. (e.g. SLC22A18AS) which transport a wide range of toxins, common drugs, and nutritional compounds (Walker et al., 20172017). Kingsley et al. also reported associations of PM2.5 with CHD7 that varied by infant sex, with associations in opposite directions for males (positive) and females (inverse) (Kingsley et al., 2017). We did not find similar interactions in the current study (see Supplemental Table 5), although we found offspring sex-PM2.5 interactions on DDHD1 expression in GAPPS. A number of the genes (e.g. CYP1A1, SYN1, BLCAP, H19, and MEG3) that were highlighted by previous studies did not pass our initial gene filtering as their average logCPM was not above 0.
In a recently published study, Deyssenroth et al. investigated birthweight and placental transcriptome-wide network in relation to maternal PM2.5 exposure among participants (N = 149) enrolled in the Rhode Island Child Health Study (Deyssenroth et al., 2021). PM2.5 exposure spanning 12 weeks prior to and 13 weeks into gestation, a growth-restriction exposure window (GREW) was inversely associated with infant birthweight percentiles. The PM2.5 exposure during the GREW period was positively correlated with placental gene expression modules enriched for genes involved in amino acid transport, cellular respiration, cell adhesion, and inversely correlated with modules enriched for genes involved in vasculature development and organ development processes (Deyssenroth et al., 2021). Of these, modules enriched for the amino acid transport and cellular respiration processes (including CCDC53 and EIF5A genes) were additionally correlated with birthweight percentiles (Deyssenroth et al., 2021). They concluded that maternal PM2.5 exposure may alter placental programming of fetal growth with implications for downstream health effects, including birth outcomes, cardiometabolic health, and susceptibility to viral infections (Deyssenroth et al., 2021). Similarities of this recent report with our findings include associations of early (first month), and first and second trimester pregnancy maternal PM2.5 exposure with placental gene expression. More specifically, our study identified differentially expressed genes (CCDC28A and EIF3D) that are in similar families, and have similar functions and are involved in transport, infection response, and mitochondrial processes. These findings highlight the potentially important role of environmental exposure during early and first half of pregnancy on implantation and placental growth, development, and function with implications on the course, complications, and outcomes of pregnancy (Chen et al., 2002).
Available evidence from previous literature supports several of the PM2.5 exposure and placental gene expression associations we found in the current study. Placental expression of SRSF11, a splicing factor, was associated with maternal PM2.5 exposure in our study. Laing et al. have previously reported that PM2.5 exposure can affect mRNA splicing (Laing et al., 2010). More specifically, it activates endoplasmic reticulum (ER) stress sensor IRE1α, but it decreases the activity of IRE1α in splicing the mRNA (Xbp1) encoding the UPR trans-activator X-box binding protein 1 (XBP1) (Laing et al., 2010). XBP1 plays essential roles in normal differentiation and function of specialized cell types and in remodeling cells to adapt to cellular stress (Laing et al., 2010; Zhang and Kaufman, 2008; Ron and Walter, 2007). IRE1α is predominantly activated in placenta and is essential in placental development and function related to vascular endothelial growth factor-A and ER stress (Iwawaki et al., 2009). We also found PM2.5 exposure was related to upregulation of placental expression of GABRE, which is important in GABAergic signaling and addiction to nicotine and morphine. Maternal PM2.5 exposure has been related to expression of genes related to GABAergic signaling and offspring neural development in human and animal studies (Winckelmans et al., 2017; Kulas et al., 2018).
In the GAPPS study, we observed interaction between offspring sex and first month PM2.5 exposure on placental expression of DDHD1. DDHD1 is a gene that codes for a phospholipase enzyme involved in fatty acid metabolism and intimately related to mitochondrial function, bio-energetics, and oxidative stress, similar to the function of the other mitochondrial-function related genes (MT-CYB and MT-ND3) that were associated with PM2.5 exposure in the current study (Tesson et al., 2012; Baba et al., 2014). Interestingly, Fuentes et al. reported that DDHD1 is among genes that is targeted in a sex-specific manner by microRNAs in an acute mouse model of ozone-induced lung inflammation (Fuentes et al., 2018). Given the smaller sample size of the GAPPS study, the PM2.5 exposure, and lack of main effects, further research is needed to confirm this potential sex-specific association of PM2.5 exposure with DDHD1 expression. Since assessing sex-specific associations is important, we have included findings from sex-stratified models for all top hits with main effects (Supplemental Table 4).
Placental gene expression changes in relation to PM2.5 exposure is a relatively understudied area. Maternal PM2.5 exposure can lead to up- or down- regulation of placental gene expression through changes in DNA methylation, DNA damage, oxidative stress, inflammation, or changes in placental perfusion (Ilekis et al., 2016; Janssen et al., 2013; Kingsley et al., 2016; Risom et al., 2005). The current study conducted as part of the NIH-funded ECHO-PATHWAYS Consortium is the largest study, to date, on the topic. Other strengths of the current study include investigations of multiple, well-characterized cohorts, the rigorous air pollution exposure measures (including multiple, temporally-resolved windows of exposure), the state-of-the-art genome-wide transcriptome profiling, adjustment for a large number of potential confounders and precision variables, and evaluation of exposure-offspring sex interactions on placental gene expression.
On the other hand, several limitations deserve mention. The current analyses were conducted among subsets of CANDLE and GAPPS participants. While the purpose of examining the two cohorts separately was in anticipation of replication of findings, inconsistency in findings may have resulted from differences (including demographic differences) in the study populations as well as variation in PM2.5 exposure (such as source, composition, and magnitude) between the cohorts. PM2.5 is a complex mixture of particles and its composition, which varies depending on seasons and regions, may lead to different outcomes, including gene expression (Kim et al., 2018; Bell et al., 2007; Honkova et al., 2018). While the overall size of the study is large, study power may still preclude identification of significant differences in expression of some genes, particularly in the GAPPS study. Placenta gene expression was assessed at delivery - one time point – even though placental gene expression is a dynamic event. We evaluated relationships between placental gene expression at delivery and different windows of exposure during pregnancy (Sitras et al., 2012). Potential differences in placental sample composition and resulting cellular heterogeneity (that includes syncytiotrophoblasts, blood vessels etc.), that is not accounted for in our study, is another limitation of our study. As an uncontrolled confounding, this limitation may result in inaccuracies in the effect estimates of the models (Luyten et al., 2018). We lack single cell data and reference-free approaches for placental transcriptomic analyses to address this concern (Konwar et al., 2019). We used standardized biopsy methods in each cohort to minimize this concern. Finally, generalizability of our findings is limited to populations with similar PM2.5 level and composition exposure. Addressing these limitations are potential areas for future research.
In summary, we found that maternal PM2.5 exposure during first trimester and second trimester was associated with expression of several genes in the placenta in the CANDLE cohort, but not the GAPPS cohort. Our study, along with similar studies, can help characterize distinct biological signatures in placenta that are associated with prenatal exposure to ambient air pollution (Luyten et al., 2018). Better understanding of molecular pathways that are affected by ambient air pollution in placenta, a critical organ of pregnancy, has significant translational potential for development of new preventative and therapeutic measures to improve pregnancy and offspring outcomes (Ilekis et al., 2016).
Supplementary Material
Acknowledgement and Support
This study was conducted by the ECHO PATHWAYS Consortium, funded by the NIH (1UG3OD023271/4UH3OD023271). This manuscript has been reviewed by PATHWAYS for scientific content and consistency of data interpretation with previous PATHWAYS publications. This study was conducted using specimens and data collected and stored on behalf of the Global Alliance to Prevent Prematurity and Stillbirth (GAPPS) Repository and supported by the Urban Child Institute in CANDLE. Air pollution models were constructed with funding support from NIH (P01AG055367, R01ES025888, R56ES026528, and R01ES023500) and the Kresge Foundation Grant No.243365 as well as by STAR research assistance agreements RD831697 (MESA Air), RD83830001 (MESA Air Next Stage), RD83479601 (UW Center for Clean Air Research), and R83374101 (MESA Coarse), awarded by the U. S. Environmental Protection Agency (EPA). This work has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and the EPA does not endorse any products or commercial services mentioned in this publication. Laboratory infrastructure and hardware used for this study was supported by the University of Washington Interdisciplinary Center for Exposures, Diseases, Genomics, and Environment funded by the NIEHS (2P30ES007033). We thank Drs. J. MacIsaac and K. Ramadori from Dr. Michael Kobor’s Laboratory (Center for Molecular Medicine and Therapeutics, University of British Columbia) for performing DNA and RNA extraction. We would also like to thank the participating families and study team members in the GAPPS and CANDLE cohorts.
Footnotes
CRediT authorship contribution statement
Daniel A. Enquobahrie: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. James MacDonald: Methodology, Software, Validation, Formal analysis, Writing – review & editing, Visualization. Michael Hussey: Formal analysis, Visualization, Writing – review & editing. Theo K. Bammler: Methodology, Investigation, Resources, Writing – review & editing. Christine T. Loftus: Conceptualization, Methodology, Data curation, Writing – review & editing. Alison G. Paquette: Conceptualization, Methodology, Writing – review & editing. Nora Byington: Methodology, Investigation, Resources, Writing – review & editing. Carmen J. Marsit: Conceptualization, Methodology, Writing – review & editing. Adam Szpiro: Conceptualization, Methodology, Writing – review & editing. Joel D. Kaufman: Conceptualization, Methodology, Investigation, Resources, Writing – review & editing. Kaja Z. LeWinn: Conceptualization, Writing – review & editing, Funding acquisition. Nicole R. Bush: Conceptualization, Writing – review & editing, Funding acquisition. Frances Tylavsky: Conceptualization, Writing – review & editing, Funding acquisition. Catherine J. Karr: Conceptualization, Methodology, Writing – review & editing, Funding acquisition. Sheela Sathyanarayana: Conceptualization, Methodology, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2022.107310.
References
- Baba T, Kashiwagi Y, Arimitsu N, Kogure T, Edo A, Maruyama T, Nakao K, Nakanishi H, Kinoshita M, Frohman MA, Yamamoto A, Tani K, 2014. Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J. Biol. Chem 289 (16), 11497–11511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJP, 1995. The fetal origins of adult disease. Proc. R. Soc. Lond. B Biol. Sci 262, 37–43. [DOI] [PubMed] [Google Scholar]
- Bateson P, Barker D, Clutton-Brock T, Deb D, D’Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, McNamara J, Metcalfe NB, Monaghan P, Spencer HG, Sultan SE, 2004. Developmental plasticity and human health. Dev. Plast. Hum. Health 430 (6998), 419–421. [DOI] [PubMed] [Google Scholar]
- Bell ML, Dominici F, Ebisu K, Zeger SL, Samet JM, 2007. Spatial and temporal variation in PM2.5 chemical composition in the United States for health effects studies. Environ. Health Perspect 115 (7), 989–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B 57 (1), 289–300. [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L, 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotech, 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brown J, Pirrung M, McCue LA, 2017. FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics, 33, 3137–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckberry S, Bianco-Miotto T, Bent SJ, Dekker GA, Roberts CT, 2014. Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human fetal-maternal interface. Mol. Hum. Reprod 20 (8), 810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Chen JJ, Tzeng C, Li HN, Chang SJ, Cheng YF, Chang CW, Wang RS, Yang PC, Lee YT, 2002. Global analysis of differentially expressed genes in early gestational decidua and chorionic villi using a 9600 human cDNA microarray. Mol. Hum. Reprod 8, 475–484. [DOI] [PubMed] [Google Scholar]
- Chun HeeKyoung, Leung C, Wen SW, McDonald J, Shin HH, 2020. Maternal exposure to air pollution and risk of autism in children: a systematic review and meta-analysis. Environ. Pollut 256, 113307. 10.1016/j.envpol.2019.113307. [DOI] [PubMed] [Google Scholar]
- de Melo JO, Soto SF, Katayama IA, Wenceslau CF, Pires AG, Veras MM, Furukawa LNS, de Castro I, Saldiva PHN, Heimann JC, 2015. Inhalation of fine particulate matter during pregnancy increased IL-4 cytokine levels in the fetal portion of the placenta. Toxicol. Lett 232 (2), 475–480. [DOI] [PubMed] [Google Scholar]
- Deyssenroth MA, Rosa MJ, Eliot MN, Kelsey KT, Kloog I, Schwartz JD, Wellenius GA, Peng S, Hao K.e., Marsit CJ, Chen J, 2021. Placental gene networks at the interface between maternal PM2.5 exposure early in gestation and reduced infant birthweight. Environ. Res 199, 111342. 10.1016/j.envres.2021.111342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes N, Roy A, Mishra V, Cabello N, Silveyra P, 2018. Sex-specific microRNA expression networks in an acute mouse model of ozone-induced lung inflammation. Biol. Sex Differ 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Rankin J, Pless-Mulloli T, Glinianaia S, 2007. Does the effect of air pollution on pregnancy outcomes differ by gender?. A systematic review. Environ. Res 105 (3), 400–408. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Cooper C, Thornburg KL, 2008. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med 359 (1), 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TL, Sun T, Koeppel AF, Lee B, Wang ET, Farber CR, Rich SS, Sundheimer LW, Buttle RA, Chen IYD, Rotter JI, Turner SD, Williams J 3rd, Goodarzi MO, Pisarska MD, 2018. Sex differences in the late first trimester human placenta transcriptome. Biol Sex Differ 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gude NM, Roberts CT, Kalionis B, King RG, 2004. Growth and function of the normal human placenta. Thromb. Res 114 (5–6), 397–407. [DOI] [PubMed] [Google Scholar]
- Hazlehurst MF, Carroll KN, Loftus CT, Szpiro AA, Moore PE, Kaufman JD, Kirwa K, LeWinn KZ, Bush NR, Sathyanarayana S, Tylavsky FA, Barrett ES, Nguyen RHN, Karr CJ, 2021. Maternal exposure to PM2.5 during pregnancy and asthma risk in early childhood: consideration of phases of fetal lung development. Environ Epidemiol 5 (2), e130. 10.1097/EE9.0000000000000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkova K, Rossnerova A, Pavlikova J, Svecova V, Klema J, Topinka J, Milcova A, Libalova H, Choi H, Veleminsky M, Sram RJ, Rossner P, 2018. Gene expression profiling in healthy newborns from diverse localities of the Czech Republic. Environ. Mol. Mutagen 59 (5), 401–415. [DOI] [PubMed] [Google Scholar]
- Ilekis JV, Tsilou E, Fisher S, Abrahams VM, Soares MJ, Cross JC, Zamudio S, Illsley NP, Myatt L, Colvis C, Costantine MM, Haas DM, Sadovsky Y, Weiner C, Rytting E, Bidwell G, 2016. Placental origins of adverse pregnancy outcomes: potential molecular targets: an executive workshop summary of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Am. J. Obstet. Gynecol 215 (1), S1–S46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwawaki T, Akai R, Yamanaka S, Kohno K, 2009. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. PNAS 106 (39), 16657–16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BG, Godderis L, Pieters N, Poels K, Kicinski M, Cuypers A, Fierens F, Penders J, Plusquin M, Gyselaers W, Nawrot TS, 2013. Placental DNA hypomethylation in association with particulate air pollution in early life. Part. Fibre Toxicol 10, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JP, Olives C, Kim S-Y, Sheppard L, Sampson PD, Szpiro AA, Oron AP, Lindström J, Vedal S, Kaufman JD, 2015. A unified spatiotemporal modeling approach for predicting concentrations of multiple air pollutants in the multi-ethnic study of atherosclerosis and air pollution. Environ. Health Perspect 123 (4), 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Cho Y, Song M-K, Lim J-H, Kim JY, Gye MC, Ryu J-C, 2018. Effect of particulate matter 2.5 on gene expression profile and cell signaling in JEG-3 human placenta cells. Environ. Toxicol. 33 (11), 1123–1134. [DOI] [PubMed] [Google Scholar]
- Kingsley SL, Eliot MN, Whitsel EA, Huang Y-T, Kelsey KT, Marsit CJ, Wellenius GA, 2016. Maternal residential proximity to major roadways, birth weight, and placental DNA methylation. Environ. Int 92-93, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley SL, Deyssenroth MA, Kelsey KT, Awad YA, Kloog I, Schwartz JD, Lambertini L, Chen J, Marsit CJ, Wellenius GA, 2017. Maternal residential air pollution and placental imprinted gene expression. Environ. Int 108, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwa K, Szpiro AA, Sheppard L, Sampson PD, Wang M, Keller JP, Young MT, Kim S-Y, Larson TV, Kaufman JD, 2021. Fine-scale air pollution models for epidemiologic research: insights from approaches developed in the multi-ethnic study of atherosclerosis and air pollution (MESA Air). Curr. Environ. Health Rep 8 (2), 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepac P, Locatelli I, Korošec S, Künzli N, Kukec A, 2018. Ambient air pollution and pregnancy outcomes: a comprehensive review and identification of environmental public health challenges. Environ. Res 167, 144–159. [DOI] [PubMed] [Google Scholar]
- Konwar C, Del Gobbo G, Yuan V, Robinson WP, 2019. Considerations when processing and interpreting genomics data of the placenta. Placenta 84, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulas JA, Hettwer JV, Sohrabi M, Melvin JE, Manocha GD, Puig KL, Gorr MW, Tanwar V, McDonald MP, Wold LE, Combs CK, 2018. In utero exposure to fine particulate matter results in an altered neuroimmune phenotype in adult mice. Environ. Pollut 241, 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzawa CW, Quinn EA, 2009. Developmental Origins of Adult Function and Health: Evolutionary Hypotheses. Annu. Rev. Anthropol 38 (1), 131–147. [Google Scholar]
- Laing S, Wang G, Briazova T, Zhang C, Wang A, Zheng Z.e., Gow A, Chen AF, Rajagopalan S, Chen LC, Sun Q, Zhang K, 2010. Airborne particulate matter selectively activates endoplasmic reticulum stress response in the lung and liver tissues. Am. J. Physiol. Cell Physiol 299 (4), C736–C749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK, 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15 (2), R29. 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus CT, Hazlehurst MF, Szpiro AA, Ni Y.u., Tylavsky FA, Bush NR, Sathyanarayana S, Carroll KN, Karr CJ, LeWinn KZ, 2019. Prenatal air pollution and childhood IQ: preliminary evidence of effect modification by folate. Environ. Res 176, 108505. 10.1016/j.envres.2019.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyten LJ, Saenen ND, Janssen BG, Vrijens K, Plusquin M, Roels HA, Debacq-Chainiaux F, Nawrot TS, 2018. Air pollution and the fetal origin of disease: A systematic review of the molecular signatures of air pollution exposure in human placenta. Environ. Res 166, 310–323. [DOI] [PubMed] [Google Scholar]
- Maghbooli Z, Hossein-nezhad A, Adabi E, Asadollah-pour E, Sadeghi M, Mohammad-nabi S, Zakeri Rad L, Malek Hosseini A-A, Radmehr M, Faghihi F, Aghaei A, Omidifar A, Aghababei Y, Behzadi H, Rosenfeld CS, 2018. Air pollution during pregnancy and placental adaptation in the levels of global DNA methylation. PLoS ONE 13 (7), e0199772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Szpiro AA, Young MT, Loftus CT, Bush NR, LeWinn KZ, Sathyanarayana S, Enquobahrie DA, Davis RL, Kratz M, Fitzpatrick AL, Sonney JT, Tylavsky FA, Karr CJ, 2021. Associations of pre- and postnatal air pollution exposures with child blood pressure and modification by maternal nutrition: a prospective study in the CANDLE cohort. Environ. Health Perspect 129, 47004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette AG, MacDonald J, Lapehn S, Bammler T, Kruger L, Day DB, Price ND, Loftus C, Kannan K, Marsit C, Mason WA, Bush NR, LeWinn KZ, Enquobahrie DA, Prasad B, Karr CJ, Sathyanarayana S, 2021. A comprehensive assessment of associations between prenatal phthalate exposure and the placental transcriptomic landscape. Environ. Health Perspect 129 (9), 097003. 10.1289/EHP8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risom L, Møller P, Loft S, 2005. Oxidative stress induced DNA damage by particulate air pollution. Mutat. Res 592 (1–2), 119–137. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A, 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11 (3), R25. 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P, 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol 8 (7), 519–529. [DOI] [PubMed] [Google Scholar]
- Rosenfeld CS, 2015. Sex-specific placental responses in fetal development. Endocrinology 156, 3422–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenen ND, Plusquin M, Bijnens E, Janssen BG, Gyselaers W, Cox B, Fierens F, Molenberghs G, Penders J, Vrijens K, De Boever P, Nawrot TS, 2015. In utero fine particle air pollution and placental expression of genes in the brain-derived neurotrophic factor signaling pathway: An ENVIRONAGE Birth Cohort study. Environ. Health Perspect 123 (8), 834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick SF, Blount BC, Jacob P, Saliba NA, Bernert JT, El Hellani A, Jatlow P, Pappas RS, Wang L, Foulds J, Ghosh A, Hecht SS, Gomez JC, Martin JR, Mesaros C, Srivastava S, St. Helen G, Tarran R, Lorkiewicz PK, Blair IA, Kimmel HL, Doerschuk CM, Benowitz NL, Bhatnagar A, 2017. Biomarkers of exposure to new and emerging tobacco delivery products. Am. J. Physiol. Lung Cell. Mol. Physiol 313 (3), L425–L452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitras V, Fenton C, Paulssen R, Vårtun Å, Acharya G, Oudejans C, 2012. Differences in gene expression between first and third trimester human placenta: a microarray study. PLoS ONE 7 (3), e33294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD, 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4, 1521. 10.12688/f1000research10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag-Padilla L, Burns R, Shih R, et al. The Urban Child Institute CANDLE Study: methodological overview and baseline sample description. RAND Corporation 2015. (http://www.rand.org/pubs/research_reports/RR1336.html.) Accessed May 31, 2021. [Google Scholar]
- Sood R, Zehnder JL, Druzin ML, Brown PO, 2006. Gene expression patterns in human placenta. PNAS 103 (14), 5478–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto S.d.F., Melo J.O.d., Marchesi GD, Lopes KL, Veras MM, Oliveira I.B.d., Souza R.M.d., de Castro I, Furukawa LNS, Saldiva PHN, Heimann JC, Ambrósio CE, 2017. Exposure to fine particulate matter in the air alters placental structure and the renin-angiotensin system. PLoS ONE 12 (8), e0183314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson C, Nawara M, Salih MAM, Rossignol R, Zaki MS, Al Balwi M, Schule R, Mignot C, Obre E, Bouhouche A, Santorelli FM, Durand CM, Oteyza AC, El-Hachimi KH, Al Drees A, Bouslam N, Lamari F, Elmalik SA, Kabiraj MM, Seidahmed MZ, Esteves T, Gaussen M, Monin ML, Gyapay G, Lechner D, Gonzalez M, Depienne C, Mochel F, Lavie J, Schols L, Lacombe D, Yahyaoui M, Al Abdulkareem I, Zuchner S, Yamashita A, Benomar A, Goizet C, Durr A, Gleeson JG, Darios F, Brice A, Stevanin G, 2012. Alteration of fatty-acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet 91, 1051–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsamou M, Vrijens K, Madhloum N, Lefebvre W, Vanpoucke C, Nawrot TS, 2018. Air pollution-induced placental epigenetic alterations in early life: a candidate miRNA approach. Epigenetics 13 (2), 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker N, Filis P, Soffientini U, Bellingham M, O’Shaughnessy PJ, Fowler PA, 2017. Placental transporter localization and expression in the Human: the importance of species, sex, and gestational age differences. Biol. Reprod, 96, 733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyatt RM, Garte SJ, Cosma G, Bell DA, Jedrychowski W, Wahrendorf J, Randall MC, Cooper TB, Ottman R, Tang D, et al. , 1995. CYP1A1 messenger RNA levels in placental tissue as a biomarker of environmental exposure. Cancer Epidemiol. Biomarkers Prev 4, 147–153. [PubMed] [Google Scholar]
- Winckelmans E, Vrijens K, Tsamou M, Janssen BG, Saenen ND, Roels HA, Kleinjans J, Lefebvre W, Vanpoucke C, de Kok TM, Nawrot TS, 2017. Newborn sex-specific transcriptome signatures and gestational exposure to fine particles: findings from the ENVIRONAGE birth cohort. Environ. Health 16, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ, 2008. From endoplasmic-reticulum stress to the inflammatory response. Nature 454 (7203), 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


